ABSTRACT
Tensins are a family of focal adhesion proteins consisting of four members in mammals (TNS1, TNS2, TNS3 and TNS4). Their multiple domains and activities contribute to the molecular linkage between the extracellular matrix and cytoskeletal networks, as well as mediating signal transduction pathways, leading to a variety of physiological processes, including cell proliferation, attachment, migration and mechanical sensing in a cell. Tensins are required for maintaining normal tissue structures and functions, especially in the kidney and heart, as well as in muscle regeneration, in animals. This Review discusses our current understanding of the domain functions and biological roles of tensins in cells and mice, as well as highlighting their relevance to human diseases.
Keywords: Tensin, CTEN, SH2, PTB, Focal adhesion, Mitral valve prolapse, Cystic kidney, Nephrotic syndrome, Cancer
Summary: This Review discusses current understanding on the domain functions and biological roles of tensins in cells and mice as well as their contributions to disease development.
Introduction
Tensin was first reported in 1991 as an actin-binding focal-adhesion protein containing a Src homology 2 (SH2) domain, a newly identified binding motif specific for phosphotyrosine (pTyr) at the time, suggesting an interesting role of tensin in bridging signal transduction pathways with the cytoskeletal networks (Davis et al., 1991). The first complete cDNA of tensin was isolated from chicken (Lo et al., 1994a), and most of the early studies were performed using recombinant chicken tensin. It took a decade to realize that there are more than one tensin in mammals. The original tensin was then named tensin-1 (TNS1), and three additional members tensin-2 [TNS2; also known as C1-Ten (TENC1)], tensin-3 (TNS3) and C-terminal tensin-like [CTEN; also known as tensin-4 (TNS4)], were subsequently identified, which all have extensive similarity to TNS1 (Chen et al., 2002; Cui et al., 2004; Lo and Lo, 2002). Tensins typically reside at focal adhesions, which connect the extracellular matrix (ECM) to the cytoskeletal networks, mainly through integrin receptors and their associated protein complexes (Critchley, 2000; Geiger et al., 2001; Zamir and Geiger, 2001). Focal adhesions mediate both outside-in and inside-out signaling pathways that regulate cellular events, such as cell attachment, migration, proliferation, apoptosis, gene expression and differentiation (Hynes, 2002; Legate and Fässler, 2009; Winograd-Katz et al., 2014), in response to cues from either the outside environment or within the cell. This Review offers details on our current understanding of tensins at molecular, cellular and animal levels, as well as highlighting their relevance in human diseases.
Domain functions of tensins
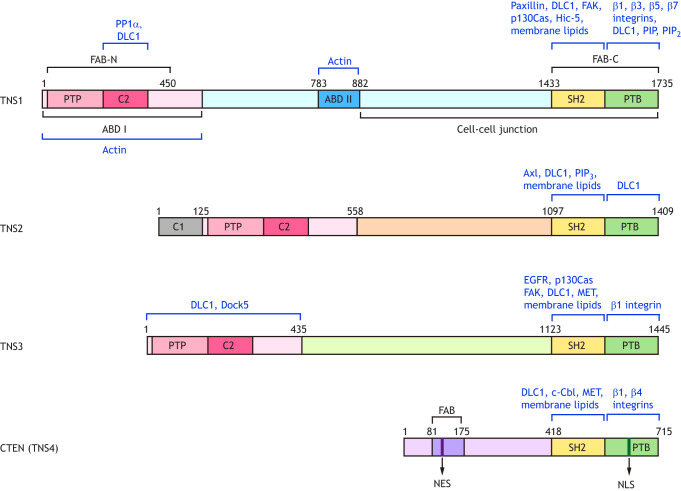
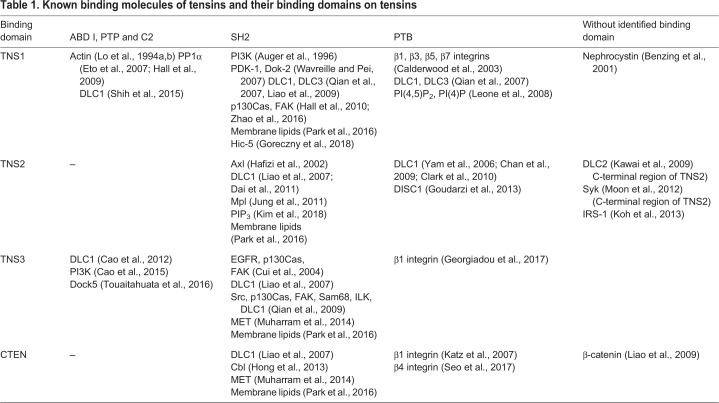
Tensins are large proteins, ranging between 170 kDa and 220 kDa except for CTEN, which is ∼80 kDa (Chen et al., 2002; Cui et al., 2004; Lo et al., 1994a; Lo and Lo, 2002). They are multidomain proteins consisting of homologous protein kinase C conserved region 1 (C1), protein tyrosine phosphatase (PTP), C2, Src homology 2 (SH2) and phosphotyrosine-binding (PTB) domains, in addition to functional domains involved in binding to actin and focal adhesion proteins (Fig. 1) (Lo, 2004). These domains allow tensins to anchor actin cytoskeletons to integrin receptors, and to transduce various types of signaling pathways through their binding partners (Table 1), giving rise to a variety of cellular events.
Fig. 1.
Domain structures of human tensins and their binding partners. The domains of tensins are represented by colored rectangles. The N-terminal regions of TNS1, TNS2 and TNS3 contain the actin-binding domain I (ABD I) that overlaps with the focal-adhesion-binding (FAB-N) site, as well as a PTEN-like protein tyrosine phosphatase (PTP) and C2 domains. The C-terminal regions of all tensins share the Src homology 2 (SH2) and phosphotyrosine-binding (PTB) domains that possess the FAB activity (FAB-C). The ABD II and the sequences required for proper cell–cell junction localization are unique to TNS1. The protein kinase C conserved region 1 (C1) is only present in TNS2. The N-terminal of CTEN (TNS4) contains a unique FAB domain, which includes a nuclear export sequence (NES), whereas a nuclear localization sequence (NLS) is located within the PTB domain. The binding partners of tensins mentioned in the main text are indicated in blue.
Table 1.
Known binding molecules of tensins and their binding domains on tensins
Actin-binding domains
Tensin was initially discovered in a fraction of proteins exhibiting actin-binding activities that co-existed with purified vinculin, another focal adhesion protein (Wilkins and Lin, 1986). By experiments using recombinant full-length TNS1 protein that had been expressed and isolated from a baculoviral expression system and by co-sedimentation, electron microscopy, dynamic light scattering and polymerization assays, TNS1 was shown to directly bind to and crosslink actin filaments, as well as reduce the actin polymerization rate (Lo et al., 1994b). The two actin-binding domains (ABDs) were further defined using bacterially expressed TNS1 fragments. ABD I is located near the N-terminus of TNS1 and binds to the side of actin filaments, whereas ABD II interacts with the barbed end of actin filaments and modulates the actin polymerization rate (Lo et al., 1994b) (Fig. 1). These actin-binding activities were only fully validated in chicken TNS1 (Chuang et al., 1995; Lo et al., 1994b), and the assignment of ABD I in TNS2 and TNS3 was purely based on their high sequence similarities. Nonetheless, the ABD I of TNS3 can interact with Dock5, a guanine nucleotide exchange factor (GEF) for the GTPase Rac, and modulate its activity in osteoclasts (Touaitahuata et al., 2016). The centrally located ABD II of TNS1 is not found in TNS2, TNS3 and CTEN.
Focal-adhesion-binding domains, nuclear localization sequence and nuclear export sequence
As a focal adhesion molecule, TNS1 contains two independent focal-adhesion-binding (FAB) sites. FAB-N is localized within the ABD I region that includes the PTP and C2 domains, near the N-terminus, while FAB-C overlaps with the SH2 and PTB domains at the C-terminus (Chen and Lo, 2003; Hong et al., 2019). Although CTEN only shares the FAB-C site with other tensins, it also contains a second FAB site, but its sequence is different from that in other tensins (Hong et al., 2019). In addition, CTEN contains a nuclear export sequence (NES) within its unique FAB region and a nuclear localization sequence (NLS) within the PTB domain (Hong et al., 2019) (Fig. 1). Intriguingly, this NLS is conserved among tensins and exogenous GFP fused with the SH2-PTB domains of other tensins can be detected at focal adhesions and in the nucleus (Chen and Lo, 2003; Hong et al., 2019 and our unpublished data). However, CTEN is the only member that is currently known to translocate to the nucleus (Hong et al., 2019). It is possible that the other tensins are too large or contain an ‘inhibitory sequence’ so that this NLS is not functional in cells. Alternatively, the NLS sites could only be unmasked and become functional under yet-to-be identified conditions.
C1 domain
C1 domains are found exclusively in all three human TNS2 splice variants (Hong et al., 2016). Because GFP–C1 domain fusion proteins are detected in the nucleus, the C1 domain was thought to guide TNS2 to the nucleus (Hafizi et al., 2010). However, endogenous TNS2 has not been found in the nucleus and all full-length TNS2 isoforms display the same distribution at focal adhesions (Hafizi et al., 2010; Hong et al., 2016). The function of the C1 domain therefore remains unknown.
PTEN-PTP and C2 domains
TNS1, TNS2 and TNS3 harbor a PTEN-like cysteine-based PTP domain and a PTEN homology C2 domain, which is known to bind phospholipids (Zhang and Aravind, 2010) (Fig. 1). Whether the PTP-C2 region of tensin can bind to phospholipids is currently unknown. However, the C2 domain of TNS1 binds to and recruits the serine/threonine protein phosphatase 1α (PP1α) to focal adhesions (Eto et al., 2007). This is unique to TNS1 since its C2 region contains the essential PP1α-binding 299KVXF302 site that is not conserved in other tensins (Hall et al., 2009). Additionally, the C2 region is sufficient for the interaction of TNS1 with the sterile alpha motif (SAM) of deleted in liver cancer 1 (DLC1), a Rho GTPase-activating protein (GAP) and tumor suppressor (Shih et al., 2015). Besides the C2 region, the SH2 and PTB domains of tensins also bind to DLC1 at separate sites (Chen et al., 2012; Dai et al., 2011; Liao et al., 2007; Qian et al., 2007; Shih et al., 2015; Yam et al., 2006), and the interaction between DLC1 and TNS1, TNS2 or TNS3 has been shown to suppress the GAP activity of DLC1 toward RhoA in HEK293 and endothelial cells (Shih et al., 2015). This is in agreement with additional reports showing that TNS1, TNS2, and TNS3 negatively regulate DLC1 GAP activity in fibroblasts, breast cancer cells and lung cancer cells (Clark et al., 2010; Hall et al., 2009; Tripathi et al., 2014). Nevertheless, TNS3 has been reported to enhance the GAP activity of DLC1 in osteoclasts (Touaitahuata et al., 2016), glioblastoma cell lines (Chen et al., 2017) and EGF-treated MCF10A non-malignant mammary cells, which require a specialized medium to grow (Cao et al., 2015, 2012). This might represent a unique function of TNS3 in these cell types or under EGF treatment.
Most cysteine-based PTPs contain the signature motif C-(X)5-R, and the PTP motif in PTEN is 124CKAGKGR. Although TNS3 is the only tensin member that contains both essential cysteine and arginine residues in the signature motif (107CRGGKGR), it has not yet been reported to have PTP activity (Alonso and Pulido, 2016). The PTP motif in TNS1 (113NKGNRGR) lacks the critical cysteine residue is thus considered an inactive PTP (Alonso and Pulido, 2016). Unexpectedly, although the motif in TNS2 (231CKGNKGK) lacks the conserved arginine residue, TNS2 has been demonstrated to have PTP activity that is comparable to that of PTEN in vitro when using pTyr-containing peptides as substrates (Koh et al., 2013). Furthermore, TNS2 can dephosphorylate pTyr-612 residue of insulin receptor substrate-1 (IRS-1), which decreases IRS-1 stability and, in turn, inhibits the activation of Akt and AMP-activated protein kinase (AMPK) pathways (Jeong et al., 2017; Koh et al., 2013), showing the biological function of TNS2 PTP activity.
SH2 and PTB domains
All four tensins contain the closely spaced SH2-PTB tandem domains at their C-termini, which is a unique structural feature of the tensin family (Fig. 1). The SH2 and PTB domains are integral for tensin-mediated pTyr-based signal transduction and anchoring tensins to focal adhesions.
SH2 domains are well-known binding motifs for pTyr, and, as shown more recently, for lipids (Park et al., 2016). SH2 domains interact with lipids through surface cationic patches away from pTyr-binding pockets, allowing SH2 domains to bind to pTyr and lipids independently (Park et al., 2016). The cationic patches may form grooves for specific lipid headgroup recognition or flat surfaces for non-specific plasma membrane (PM) binding. SH2 domains of all tensins bind to PM-mimetic vesicles with Kd values ranging between 180 and 350 nM (Park et al., 2016). The SH2 domain of TNS2 exhibits a high binding affinity for phosphatidylinositol (3,4,5)-triphosphate (PIP3) through a three-lysine cationic patch (K1147, K1155, K1157). Mutations of these lysine residues block the PIP3-binding of TNS2, but not its binding to pTyr nor its PTP activity (Kim et al., 2018). Nonetheless, the phosphorylation levels of IRS-1 Y612 and Akt1 T308/S473 upon insulin stimulation are significantly increased in cells with mutations in these three lysine residues of TNS2, indicating that recognition of PIP3 by the TNS2 SH2 domain is essential for its signaling function (Kim et al., 2018). As expected, the SH2 domains of tensins recruit pTyr-containing proteins, such as epidermal growth factor receptor (EGFR), MET (also known as hepatocyte growth factor receptor), Axl, Src, focal adhesion kinase (FAK; also known as PTK2) and p130Cas (also known as BCAR1), and these interactions transduce signaling cascades that are mediated by protein tyrosine kinases (Cui et al., 2004; Davis et al., 1991; Hafizi et al., 2002; Muharram et al., 2014). Interestingly, unlike the SH2 domains of other proteins that only bind to pTyr sites, the SH2 domains of tensins can also interact with their partners, such as DLC1, when the tyrosine sites are not phosphorylated (Liao et al., 2007). This adds a uniqueness to the SH2 domain of tensins.
The PTB domains of tensins directly interact with the NPXY motifs present in the cytoplasmic tails of integrin β1, β3, β5 and β7 in a pTyr-insensitive fashion (Calderwood et al., 2003; Katz et al., 2007; McCleverty et al., 2007), allowing tensins to bring actin filaments, through their ABDs, to focal adhesion sites. Interestingly, the PTB domain of TNS1 can also bind to lipids, including PI(4)P and PI(4,5)P2, and this binding pocket is distinct from the β-integrin recognition site (Leone et al., 2008), indicating that both SH2 and PTB domains of tensins are able to interact with lipids.
The middle non-conserved region
The middle regions of tensins do not display any sequence similarity to each other and are not expected to share common functions. TNS1 contains ABD II, which interacts with the barbed end of actin filaments (Lo et al., 1994b), and the region spanning amino acids 882–1032 is necessary, but not sufficient, for the localization of human TNS1 to cell–cell junctions (Wu et al., 2019).
The role of tensins in biological processes
Tensins are major components at focal adhesions, which regulate a variety of biological processes in response to external or internal signals. In addition to more anticipated roles of tensins in cell adhesion, migration and proliferation, emerging findings demonstrate the critical functions of tensins in mechanical sensing. Although discussed separately below, these roles of tensins are highly linked to these other cellular events.
Cell adhesion
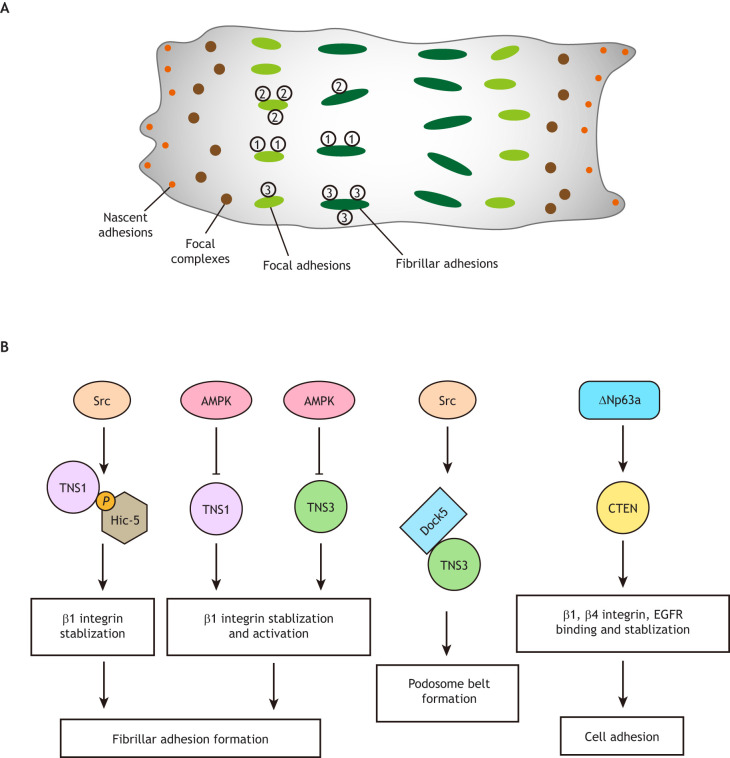
Cell adhesion allows cells to integrate into tissues and provides a platform for bidirectional signal transduction. Dynamic changes in cell adhesion are crucial to morphogenesis and play an essential role in the regulation of fundamental cellular processes, such as cell migration, proliferation and mechanical sensing. Cell–matrix adhesions are mainly provided by integrin-based adhesions, including focal adhesions in most cell types, hemidesmosomes in epithelial cells and podosomes in Rous sarcoma virus-transformed cells, osteoclasts and macrophages (Hynes, 2002). Note that ‘focal adhesion’ is commonly used as an umbrella term to refer to various subtypes of adhesion structures. Fibroblasts bound to the ECM, for example, initiate the formation of nascent adhesions, which develop into dot-like focal complexes, which further mature into larger focal adhesions and then into highly elongated fibrillar adhesions (Parsons et al., 2010) (Fig. 2A). TNS1, TNS2 and TNS3 are found at focal and fibrillar adhesions in fibroblasts, although to different degrees. TNS2 is localized mainly at focal adhesions, while TNS3 is mostly found at fibrillar adhesions, and TNS1 is found at both (Clark et al., 2010).
Fig. 2.
Cell adhesion structures and roles of tensins in cell adhesion. (A) Schematic representation of different subtypes of cell adhesion structures. Adherent cells initially form small nascent adhesions (orange dots), which develop into dot-like focal complexes (brown dots). Focal complexes progressively grow in size and mature into focal adhesions (green ovals), which then transform into elongated fibrillar adhesions (dark green oval). TNS1 (1), TNS2 (2), and TNS3 (3) are found in both focal and fibrillar adhesions. TNS2 is localized mainly in focal adhesions, TNS3 is mostly found in fibrillar adhesions, and TNS1 is distributed in both. (B) Tensins are required for cell adhesion. Src-dependent phosphorylation of Hic-5 interacts with TNS1 to promote β1 integrin stability and fibrillar adhesion maturation. AMP-activated protein kinase (AMPK) negatively regulates TNS1- and TNS3-dependent β1 integrin stabilization and activation, which is critical for fibrillar adhesion formation. TNS3 promotes podosome belt formation through a Src-dependent interaction with Dock5 in osteoclasts. CTEN expression is positively regulated by ΔNp63α, a transcription factor, and promotes cell adhesion through stabilizing β1 integrin, β4 integrin and EGFR.
Tensins regulate cell adhesion (Fig. 2B). It has been shown that expressing a dominant-negative chicken TNS1 fragment impairs fibrillar adhesion formation and fibronectin fibrillogenesis in human fibroblasts (Pankov et al., 2000), although it was later found that the fragment sequence is not conserved in human TNS1 (Clark et al., 2010). Cancer-associated fibroblasts (CAFs) lacking Hic-5 (also known as TGFB1I1), a LIM domain-containing protein, exhibit a disability in forming fibrillar adhesions, which can be rescued by Hic-5 re-expression (Goreczny et al., 2018). This rescue effect requires the mechanosensitive Src-dependent Hic-5 and TNS1 interaction, since the fibrillar adhesion formation in Hic-5 re-expressing CAFs is impaired by TNS1 knockdown or Src inactivation, and is markedly reduced when plated onto soft (polydimethylsiloxane) compared with hard (glass) substrates (Goreczny et al., 2018). Silencing of TNS1 or TNS3 in AMP-activated protein kinase (AMPK)-knockout (knockout for both α1 and α2 subunits) fibroblasts reduces fibrillar adhesion formation and fibronectin fibrillogenesis (Georgiadou et al., 2017). These findings suggest a positive role of TNS1 and TNS3 in promoting fibrillar adhesion formation. However, other reports show that knockdown of TNS1, TNS2, or TNS3, either all together or individually, has no effect on the assembly of fibrillar adhesions in fibroblasts (Clark et al., 2010; Rainero et al., 2015). It is possible that tensins do have a positive role in fibrillar adhesion formation and fibrillogenesis, but their function is compensated by other regulators, such as AMPK, when tensins are downregulated. This may explain the fibrillar adhesion formation defects of TNS1 or TNS3 knockdown that are detected in AMPK-knockout fibroblasts or in CAFs, which are modified by tumor cells to provide them with a favorable microenvironment, but not observed in normal fibroblasts.
Osteoclasts form a specialized cell–matrix adhesion called the sealing zone, which defines the resorption area of the bone. When grown in cell culture, osteoclasts form a unique structure named the podosome belt, instead of sealing zone (Takito et al., 2018). In osteoclasts, TNS3, but not TNS1 or TNS2, binds to and activates Dock5 GEF activity toward Rac and organizes podosomes into the belt. Silencing of TNS3 reduces the formation of the podosome belt and the bone-resorption activity of osteoclasts (Touaitahuata et al., 2016). Interestingly, a similar finding has been reported for p130Cas, a focal adhesion protein that binds to the SH2 domain of TNS3 (Qian et al., 2009), in that p130Cas promotes podosome belt formation through a Src-dependent interaction with Dock5 and activation of Rac activity (Nagai et al., 2013). These findings lead to a mechanistic model whereby, during podosome belt formation, TNS3 recruits pTyr-p130Cas phosphorylated by Src to link the actomyosin network and Dock5 to activate Rac and drive the formation of the podosome belt (Touaitahuata et al., 2016). However, whether TNS3, pTyr-p130Cas and Dock5 do form a complex remains to be investigated.
In the suspension subpopulation of MDA-MB-468 breast cancer cells, loss of cell–matrix adhesion results in a dramatic downregulation of TNS3, whereas TNS1, TNS2 and other main cell matrix adaptor proteins, such as vinculin and the talins, are not affected (Veß et al., 2017). Re-expressing TNS3 in the suspended MDA-MB-468 cells rescues their adhesion, whereas knockdown of TNS3 in the adherent parental MDA-MB-468 cells reduces their attachment (Veß et al., 2017), demonstrating that TNS3 is a positive regulator of cell adhesion.
CTEN expression is directly regulated by ΔNp63α, the predominant isoform of the transcription factor p63 (also known as TP63) in epithelial cells (Yang et al., 2016). Knockdown of ΔNp63α markedly impairs cell adhesion and reduces CTEN level in RWPE-1 non-malignant prostatic epithelial cells, and the reduced cell adhesion is restored by CTEN re-expression (Yang et al., 2016). Silencing of ΔNp63α also decreases MCF-10A cell adhesion and levels of several receptors, including that of β1 integrin, β4 integrin and EGFR (Carroll et al., 2006). Here, re-expression of individual receptor partially rescues the cell adhesion defects caused by ΔNp63α knockdown (Carroll et al., 2006). Interestingly, CTEN is known to both interact with integrins (Katz et al., 2007; Seo et al., 2017) and to inhibit EGFR degradation (Hong et al., 2013), suggesting that CTEN promotes cell adhesion likely by stabilizing β1 integrin, β4 integrin and EGFR.
Migration, invasion and epithelial-to-mesenchymal transition
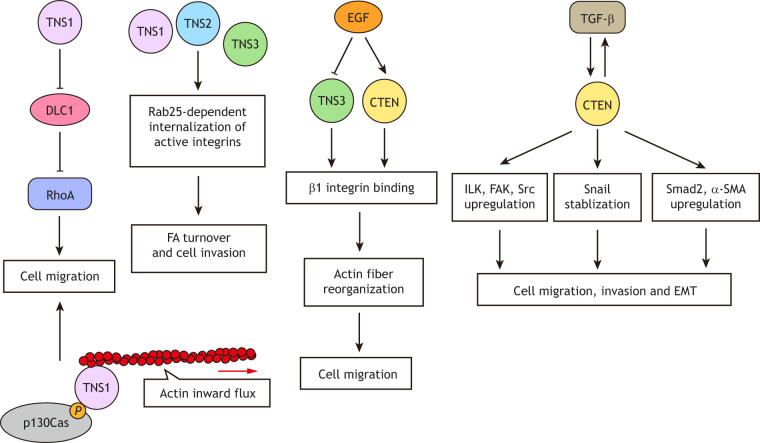
Tensins also play roles in cell migration and invasion (Fig. 3). TNS1-knockout mouse fibroblasts and endothelial cells migrate slower than controls, and overexpression of GFP–TNS1 or GFP–TNS2 promotes HEK293 human embryonic kidney cell migration (Chen et al., 2002; Shih et al., 2015). Both the FAB domains and a functional SH2 of TNS1 are required for promoting migration (Chen and Lo, 2003); this is likely due to their binding to DLC1 and suppressing its GAP activity toward RhoA (Shih et al., 2015), and/or by linking pTyr-p130Cas to the inwardly moving actin cytoskeleton (Zhao et al., 2016). TNS1, TNS2 and TNS3 are critical for Rab25-dependent internalization of active integrins, and this internalization is required for focal adhesion turnover. Thus, knockdown of tensins, either individually or in combination, impairs integrin internalization and results in reduced invasiveness of Rab25-transfected A2780 ovarian cancer cells (Rainero et al., 2015). Depletion of TNS3 in MDA-MB-231 and MDA-MB-468 breast cancer cells suppresses cell invasion and migration (Shinchi et al., 2015; Veß et al., 2017). These results support the idea that TNS1, TNS2 and TNS3 positively regulate cell migration. However, other reports show that TNS1 overexpression reduces MDA-MB-231 cell invasion (Hall et al., 2009), that overexpression of TNS2 or TNS3 inhibits HEK293 cell migration (Hafizi et al., 2005; Martuszewska et al., 2009), and that TNS3 knockdown promotes cell migration in WM793 melanoma, 05MG glioblastoma and MCF10A cells (Cao et al., 2012; Chen et al., 2017; Katz et al., 2007). These findings indicate that TNS1, TNS2 and TNS3 negatively regulate cell migration in these cell lines. Altogether, the roles of these tensins in migration appear to be cell context dependent.
Fig. 3.
Roles of tensins in the regulation of cell migration, invasion and EMT. TNS1 promotes migration by interacting with DLC1, thereby suppressing the GTPase-activating protein (GAP) activity of DLC1 toward RhoA, and/or by linking pTyr-p130Cas to inwardly moving actin cytoskeleton. TNS1, TNS2 and TNS3 are critical for Rab25-dependent internalization of active integrins and this internalization is required for focal adhesion (FA) turnover and cell migration. EGF treatment activates a transcriptional switch that results in CTEN upregulation and TNS3 downregulation. Increased CTEN displaces TNS3 by binding to β1 integrin, but not actin filaments, leading to actin fiber reorganization that favors cell migration. Additionally, CTEN promotes cell migration, invasion and EMT by upregulating transforming growth factor β (TGF-β) and downstream effectors, including ILK, FAK, Src, Snail, Smad2 and α-smooth muscle actin (α-SMA). Moreover, TGF-β also induces CTEN expression, thus forming a positive-feedback loop.
In contrast to the conflicting findings on TNS1, TNS2 and TNS3 in regulating cell migration, CTEN has been consistently reported to promote cell migration and invasion of colon, lung, breast, pancreas, skin, liver and gastric cancer cells (Al-Ghamdi et al., 2011, 2013; Albasri et al., 2011a, 2009; Aratani et al., 2017; Asiri et al., 2019; Bennett et al., 2015; Chan et al., 2015; Katz et al., 2007; Liao et al., 2009; Lo, 2014; Lu et al., 2018; Thorpe et al., 2017). EGF treatment activates a transcriptional switch that results in TNS3 downregulation and CTEN upregulation (Cao et al., 2012; Katz et al., 2007). Upregulated CTEN binds to β1 integrin through its PTB domain, but it lacks the ABD found in other tensins. Therefore, this transcriptional switch allows CTEN to displace TNS3 from actin and disrupt its links to the actin cytoskeleton, leading to actin fiber reorganization and cell migration (Katz et al., 2007). In addition, a functional SH2 domain is essential for promoting cell migration, invasion and colony formation, since a CTEN R474A mutant lacking the SH2-pTyr-binding activity has no effect on migration (Hong et al., 2013). Furthermore, CTEN may promote cell migration, invasion and epithelial-to-mesenchymal transition (EMT) by upregulating integrin-linked kinase (ILK) (Albasri et al., 2011a), FAK (Al-Ghamdi et al., 2013) and Src (Asiri et al., 2019). These three kinases are known focal adhesion proteins that promote migration, invasion and EMT. All these effects are likely attributed to CTEN-mediated upregulation of TGF-β (Lu et al., 2018), a potent inducer of EMT (Katsuno et al., 2013), and Snail (also known as SNAI1) (Thorpe et al., 2017), a transcriptional repressor controlling EMT during embryogenesis and tumor progression (Barrallo-Gimeno and Nieto, 2005). Interestingly, CTEN expression is also enhanced by TGF-β (Lu et al., 2018), thus forming a positive-feedback loop promoting cell migration, invasion and EMT.
Cell proliferation
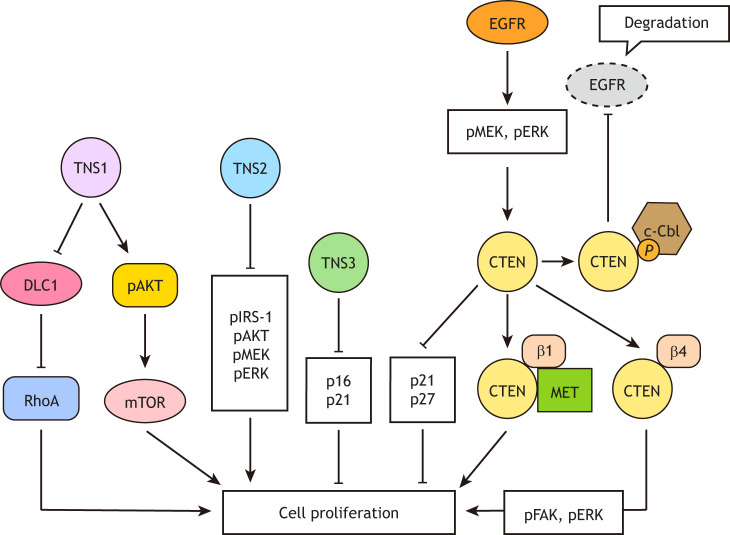
Tensins regulate the proliferation of normal and cancer cells (Fig. 4). Endothelial cells isolated from TNS1-knockout mice proliferate and migrate markedly slower than controls. Concomitantly, the RhoA activity is downregulated in these knockout cells, and this reduction can be restored by further silencing of DLC1, suggesting that TNS1 promotes endothelial cell proliferation and migration through inhibiting DLC1-GAP activity toward RhoA (Shih et al., 2015). TNS1 knockdown also reduces the proliferation of SW620 colon cancer cells (Zhou et al., 2018), and U937 and HL60 acute myeloid leukemia cell lines by suppressing the Akt-mTOR signaling pathway (Sun et al., 2020). TNS2 overexpression reduces HEK293 cell proliferation and survival (Hafizi et al., 2005). Silencing of TNS2 promotes cell proliferation, colony formation and xenograft growth of HeLa cervical cancer cells and A549 lung cancer cells. The levels of phosphorylated IRS-1, Akt family proteins, MEK proteins (MAP2K1 and MAP2K2) and ERK proteins (MAPK3 and MAPK1), and total IRS-1 are significantly increased in these TNS2-knockdown cells (Hong et al., 2016). Similar results have been reported in myotubes (Koh et al., 2013). Taken together, TNS2 negatively regulates cell proliferation likely by suppressing IRS-1, Akt and MEK-ERK signaling pathways.
Fig. 4.
Roles of tensins in the control of cell proliferation. TNS1 promotes cell proliferation through inhibiting the GAP activity of DLC1 toward RhoA and activating the Akt-mTOR signaling pathway. TNS2 negatively regulates cell proliferation by suppressing insulin receptor substrate-1 (IRS-1), Akt family proteins and the Mek-Erk pathway. TNS3 and CTEN prevent the accumulation of cyclin-dependent kinase (CDK) inhibitors (p16, p21 or p27), implicating positive roles of TNS3 and CTEN in cell proliferation. CTEN expression is induced by EGFR activation through the mitogen-activated protein kinase kinase (MEK)-extracellular signal-regulated kinase (ERK) pathway and upregulated CTEN prevents active EGFR from degradation by binding to the E3 ubiquitin ligase c-Cbl and decreasing the ubiquitylation of EGFR. Similarly, CTEN can form a stable complex with MET and β1 integrin (β1) to prevent these receptors from internalization and degradation. Additionally, CTEN can bind to and activate β4 integrin (β4), which triggers FAK and ERK activation, and thus promotes keratinocyte proliferation.
TNS3 knockdown in tonsil-derived mesenchymal stem cells (TMSCs) results in an increase in the cyclin-dependent kinase (CDK) inhibitors p16 and p21 (CDKN2A and CDKN1A), and reduces cell proliferation (Park et al., 2019). Silencing of CTEN also enhances the accumulation of the CDK inhibitors p21 and p27 (CDKN1B) and attenuates RWPE-1 non-malignant prostate epithelial cell proliferation (Wu and Liao, 2018). Additional reports have shown that knockdown of CTEN reduces proliferation in keratinocytes and various cancer cell lines (Hong et al., 2019; Muharram et al., 2014; Seo et al., 2017). These data suggest that TNS3 and CTEN are positive regulators of cell proliferation. Mechanistically, CTEN may promote both cell proliferation and migration by prolonging the duration of signaling cascades induced by receptors, such as EGFR, MET and β1 integrin in cancer cells. CTEN reduces ligand-induced EGFR degradation by binding to the E3 ubiquitin ligase c-Cbl and decreasing the ubiquitylation of EGFR (Hong et al., 2013). CTEN also forms a stable complex with MET and β1 integrin and prevents internalization as well as degradation of these receptors (Muharram et al., 2014). Interestingly, while knockdown of TNS3 or CTEN reduces proliferation, overexpression of TNS3 or CTEN does not further promote cell growth in numerous cell lines (Asiri et al., 2018; Hong et al., 2013, 2019; Martuszewska et al., 2009), except for CTEN in normal keratinocytes and nuclear CTEN in HeLa cells. In human keratinocytes, CTEN binds to and activates β4 integrin, instead of β1 integrin. This interaction triggers FAK and ERK activation and promotes cell proliferation (Seo et al., 2017). Expression of NES-deleted or NLS-tagged CTEN enhances HeLa cell proliferation (Hong et al., 2019). Since CTEN interacts with β-catenin, a cell–cell adhesion protein and transcriptional factor, only in the nucleus (Liao et al., 2009), nuclear CTEN may enhance proliferation by retaining of β-catenin in the nucleus and/or regulating its transcriptional activity.
Mechanical sensing
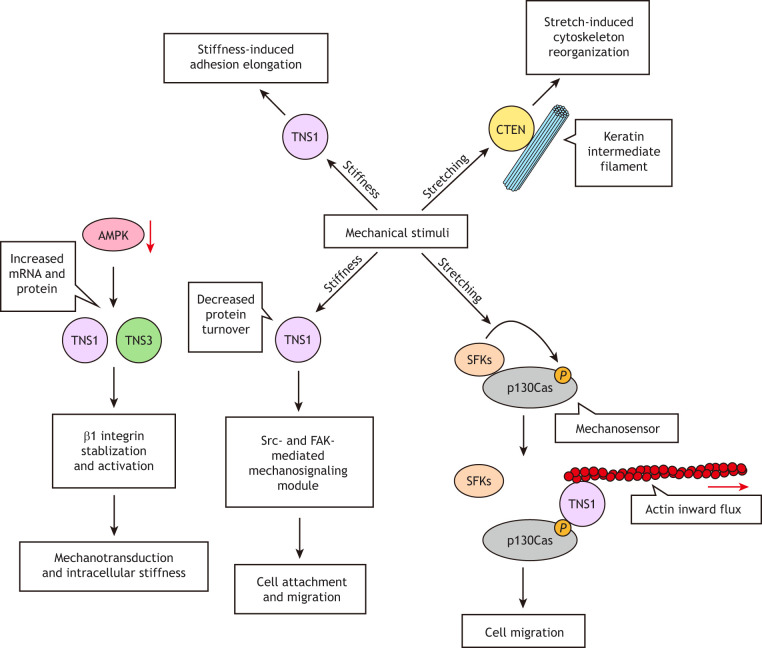
Tensins respond to various mechanical stimuli (Fig. 5). AMPK not only functions as an energy sensor but also inhibits β1 integrin activity by transcriptionally reducing TNS1 and TNS3 levels in fibroblasts (Georgiadou et al., 2017). In AMPK-knockout (α1 and α2 subunits) fibroblasts, upregulated TNS1 and TNS3 bind to and active β-integrins, thus supporting integrin-mediated processes, including cell spreading, ECM assembly, mechanotransduction and intracellular stiffness (Georgiadou et al., 2017). TNS1 silencing markedly shortens the length of fibrillar adhesion in fibroblasts plated on the stiffness-gradient gels, indicating that TNS1 is required for the stiffness-induced adhesion elongation (Barber-Pérez et al., 2020). Additionally, TNS1 senses and responds to the extracellular mechanical stimuli by modifying its protein turnover rate, which is significantly slower with increased substrate stiffness (Stutchbury et al., 2017). This critical sensing response allows for FAK- and Src-mediated tyrosine phosphorylation within focal adhesions and leads to fibroblast spreading and migration (Stutchbury et al., 2017). Consistent with these findings, TNS1 binds to Hic-5 in a Src-dependent and substrate stiffness-sensitive manner, and this TNS1–Hic-5 interaction stabilizes β1 integrins and promotes fibrillar adhesion formation and fibronectin fibrillogenesis in CAFs (Goreczny et al., 2018). Furthermore, TNS1 is a critical effector of p130Cas force sensing. p130Cas becomes tyrosine phosphorylated in response to physical stretching (Sawada et al., 2006); subsequently, TNS1 anchors pTyr-p130Cas to the inwardly moving actin cytoskeleton and mediates the disassociation of p130Cas from focal adhesions, thus promoting fibroblast migration (Zhao et al., 2016). In epithelial cells, both the actin cytoskeleton and keratin intermediate filaments are highly responsive to physical stretching (Cheah et al., 2019). CTEN rapidly accumulates along tension-bearing keratin fibers, but not actin filaments, during stretching. Dissociation of CTEN from tension-free keratin fibers depends on the duration of cell stretch, indicating that physical stretching favors the establishment of stable CTEN–keratin network interactions over time (Cheah et al., 2019). These findings reveal an unexpected role of CTEN in keratin-based mechanotranduction and start shedding light on how the keratin network responds to mechanical stimuli.
Fig.
5. Roles of tensins in mechanical sensing. The metabolic sensor AMPK also has a role in mechanotransduction through regulating tensins. Reduced levels of AMPK induce protein expression of TNS1 and TNS3, which bind to and activate β1 integrin activity, thereby enhancing intracellular stiffness. TNS1 is required for the stiffness-induced elongation of fibrillar adhesions. TNS1 also senses substrate stiffness by altering its protein turnover rate, which in turn modulates FAK- and Src-mediated phosphorylation events linked to cell attachment and migration. Physical stretching induces p130Cas phosphorylation by Src family kinases (SFKs), allowing TNS1 to link pTyr-p130Cas to inwardly moving actin cytoskeletons, thus promoting cell migration. Physical stretching also leads to cytoskeleton network reorganization. CTEN rapidly accumulates along tension-bearing keratin fibers, but not actin filaments in epithelial cells.
Animal models to study tensin function
Genetically modified animal models greatly facilitate the analyses of tensin function in the context of a whole organism. Unlike mammals, which have four tensins, Drosophila melanogaster and Caenorhabditis elegans only possess one tensin each. Intriguingly, worm tensin is more similar to TNS1, TNS2 and TNS3 (Bruns and Lo, 2020), whereas the fly tensin is shorter, similar to CTEN (Lee et al., 2003). Flies lacking tensin display a wing-blister phenotype (Lee et al., 2003; Torgler et al., 2004) and lay abnormally shaped eggs with a decreased hatching rate (Cha et al., 2017). Rescue experiments have demonstrated the requirement of both the N-terminal region and the SH2 domain, but not the PTB domain, for fly tensin to prevent the wing-blister defects (Lee et al., 2003). Nevertheless, the requirement for the PTB domain, the N-terminal region and a functional SH2 domain has been reported in another rescue study (Torgler et al., 2004). Tensin knockout in C. elegans, which has no impact on development and survival, results in slowed defecation and increased pharyngeal pumping rates (Bruns and Lo, 2020). Another C. elegans tensin mutant (ok80), likely expressing a truncated tensin lacking the C-terminus, also shows no physiological abnormality under normal conditions, but displays reduced axon regrowth after injury (Hisamoto et al., 2019).
Mice lacking TNS1 expression develop renal interstitial fibrosis, inflammatory cell infiltration and tubular dilation, which gives rise to cystic kidney disease (Lo et al., 1997). TNS1-knockout mice also show premature skeletal muscle fibers and delayed skeletal muscle regeneration (Ishii and Lo, 2001), as well as enlarged posterior mitral leaflets with abnormal collagen and proteoglycan deposits in the heart (Dina et al., 2015). A deficiency in TNS2 results in the development of glomerular sclerosis, leading to nephrotic syndrome and renal failure; however, this is only the case in specific mouse strains, such as FVB inbred mice, whereas other genetic backgrounds including C57BL/6 and sv129 are apparently normal, indicating that the phenotypes of TNS2-knockout mice are dictated by genetic differences among mouse strains (Cho et al., 2006; Kato et al., 2008; Uchio-Yamada et al., 2013). Furthermore, the SH2 and PTB domains of TNS2, but not its PTP activity, are required for preventing TNS2-knockout renal defects (Marusugi et al., 2016; Sasaki et al., 2020). TNS3-knockout mice die 3 weeks postnatally, showing defects in lung, small intestine and bone development in C57BL/6 and sv129 mixed genetic backgrounds (Chiang et al., 2005). However, when TNS3-knockout mice are backcrossed to either C57BL/6 or sv129 inbred strains, the defects are no longer present (our unpublished observations). Loss of CTEN in mice does not cause obvious defects, likely due to its restrictive expression pattern (our unpublished observation).
These animal studies have demonstrated that individual tensins are not essential for embryonic or tissue development, but are required for maintaining the normal structure and function of the kidney and heart, as well having a role in wound regeneration processes. The results also illustrate the powerful roles of yet-to-be identified genetic factors in dictating phenotypes.
Tensins in human diseases
The involvements of tensins in human diseases are suggested by analyses of animal models, genome-wide association studies (GWAS) and expression patterns in patients. Below, the relevance of tensins in lung function, kidney diseases and cancers are discussed.
Through GWAS, TNS1 has been identified as one of highly susceptible genes for mitral valve prolapse (MVP) (Dina et al., 2015), lung function (Panasevich et al., 2013; Repapi et al., 2010), chronic obstructive lung disease (COPD) (Artigas et al., 2011; Yang et al., 2014) and asthma with hay fever phenotype (Ferreira et al., 2014). The involvement of TNS1 in MVP has been demonstrated in zebrafish and mouse-knockout models (Dina et al., 2015). Aligned with the observation that TNS1-knockout mice develop cystic kidneys, TNS1 is downregulated in patients with autosomal dominant polycystic kidney disease (Dixon et al., 2020).
Mutations in TNS2 along with five other functional associated genes, including DLC1, were reported to be the likely causes of renal malfunction of 17 families with partially treatment-sensitive nephrotic syndrome (Ashraf et al., 2018). An involvement of TSN2 in human nephrotic syndrome is clearly supported by the phenotype of the TNS2-knockout mice (Cho et al., 2006; Uchio-Yamada et al., 2016, 2013). Surprisingly, overexpression of TNS2 by injecting adenoviruses carrying TNS2 into mouse kidneys also leads to nephrotic syndrome (Lee et al., 2017). These mouse studies indicate that both overexpression and lack of TNS2 results in nephrotic syndrome.
The results from the two GWAS suggest that TNS3 is associated with pancreatic cancer (Klein et al., 2018). However, we did not observe the development of pancreatic cancer in our TNS3-knockout mice (our unpublished data). High levels of CTEN are reported to be prognostic markers for patients with melanoma (Sjoestroem et al., 2013), breast cancer (Albasri et al., 2011b), gastric cancer (Aratani et al., 2017; Sakashita et al., 2008; Sawazaki et al., 2017), colorectal cancer (Albasri et al., 2011a), hepatocellular carcinoma (Chen et al., 2014) and lung adenocarcinoma (He et al., 2018; Misono et al., 2019).
Based on the literature, the role of tensins in tumorigenesis appears somewhat controversial. Expression data showing the up- or down-regulation of the different tensins have been reported and are sometimes contradictory. This is likely due to the use of different cohorts, sample numbers, threshold measurements and cancer types, among other reasons. To obtain a clearer picture, we analyzed the expression of each tensin in various types of cancer using datasets and tools at Oncomine (https://www.oncomine.org/) with high stringencies (P-value ≤0.0001, fold change ≥2, gene rank within top 10%) (see Table S1 for a snapshot). TNS1 is downregulated in 12 cancer types and upregulated in five cancer types. TNS2 is only found to be downregulated in 11 cancer types, whereas TNS3 is upregulated in some cancers and downregulated in others. TNS4 is upregulated in colorectal, gastric, lung and pancreatic cancer, but downregulated in kidney cancer and melanoma. Interestingly, overexpression of TNS1 and TNS3 (both ranked in the top 1% of nine and ten datasets, respectively) are found in lymphoma (Table S1). Despite the fact that upregulation of TNS1 was reported to be associated with poor prognosis for colorectal cancer (Burghel et al., 2013; Zhou et al., 2018, 2016), 47 cancer datasets, including four colorectal cancer datasets, at Oncomine show that TNS1 is in the top 1% or 5% of downregulated genes. TNS1 and TNS2 are in the top 1% of downregulated genes in sarcoma, while all TNS members are downregulated in kidney cancer. In lung cancer, TNS1, TNS2 and TNS3 are downregulated, whereas CTEN is upregulated. As an example, we further examined the relevance of this expression patterns for disease prognosis of lung cancer. By using lung adenocarcinoma datasets with nearly 2000 patients collected from KMPlot (https://kmplot.com/analysis/), low levels of TNS1 (P<0.000001), TNS2 (P<0.0005) and TNS3 (P<0.000001) or high level of CTEN (P<0.005) are individually associated with poor prognosis of lung adenocarcinoma. These findings suggest that the expression levels of tensin genes are highly promising prognostic markers for lung adenocarcinoma and warrant the extensive analysis of tensins as biomarkers in other relevant cancers. Based on mutant mouse studies, aberrant expression of a single tensin by itself does not appear to be sufficient to initiate tumor formation, for instance lung cancer, because none of the TNS-knockout mice display a higher rate of tumor formation than the control mice (our unpublished observations). Therefore, tensins are not cancer-driver genes, at least not in mice. Nonetheless, they are likely to play a critical role in accelerating tumor progression and metastasis.
Concluding remarks
Over three decades of studies, we have gained better understanding on overall functions of the tensin family, yet open additional questions to be answered. At the protein level, both the N-terminal and C-terminal regions of tensins possess multiple binding and/or enzymatic activities. How these activities are coordinated within the molecule, and among different tensins, are intriguing questions to be addressed. Animal studies have revealed the critical roles of tensins in the kidney and heart, as well as dramatic impacts of genetic factors on mouse phenotypes. Whether tensins share redundant roles in other tissues or embryogenesis, as well as the identities of genetic factors are interesting questions to be explored. In addition, any disease associations that are implicated by GWAS results remain to be experimentally validated. Finally, the involvement of tensins in various cancers, together with their potential use as diagnosis and prognosis markers, or indeed as therapeutic targets, is another important research avenue and warrant extensive efforts.
Supplementary Material
Acknowledgements
We are grateful to our colleagues and lab members for their comments on the manuscript. This article is dedicated to the memory of Professor Tung-Bin Lo, who fostered and inspired the careers and lives of many.
Footnotes
Funding
Our work in this area is support by grants from the Ministry of Science and Technology, Taiwan (NSC-102-2628-B-002-028-MY3 to Y.-C.L.), the National Institutes of Health, USA (CA102537, CA151366, DK64111 and HL139473 to S.H.L.) and the US Department of Defense (W81XWH-06-1-0074 to S.H.L.).
Contributor Information
Yi-Chun Liao, Email: yliao@ntu.edu.tw.
Su Hao Lo, Email: shlo@ucdavis.edu.
References
- Al-Ghamdi, S., Albasri, A., Cachat, J., Ibrahem, S., Muhammad, B. A., Jackson, D., Nateri, A. S., Kindle, K. B. and Ilyas, M. (2011). Cten is targeted by kras signalling to regulate cell motility in the colon and pancreas. PLoS ONE 6, e20919. 10.1371/journal.pone.0020919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ghamdi, S., Cachat, J., Albasri, A., Ahmed, M., Jackson, D., Zaitoun, A., Guppy, N., Otto, W. R., Alison, M. R., Kindle, K. B.et al. (2013). C-terminal tensin-like gene functions as an oncogene and promotes cell motility in pancreatic cancer. Pancreas 42, 135-140. 10.1097/MPA.0b013e3182557ceb [DOI] [PubMed] [Google Scholar]
- Albasri, A., Seth, R., Jackson, D., Benhasouna, A., Crook, S., Nateri, A. S., Chapman, R. and Ilyas, M. (2009). C-terminal tensin-like (CTEN) is an oncogene which alters cell motility possibly through repression of E-cadherin in colorectal cancer†. J. Pathol. 218, 57-65. 10.1002/path.2508 [DOI] [PubMed] [Google Scholar]
- Albasri, A., Al-Ghamdi, S., Fadhil, W., Aleskandarany, M., Liao, Y.-C., Jackson, D., Lobo, D. N., Lo, S. H., Kumari, R., Durrant, L.et al. (2011a). Cten signals through integrin-linked kinase (ILK) and may promote metastasis in colorectal cancer. Oncogene 30, 2997-3002. 10.1038/onc.2011.26 [DOI] [PubMed] [Google Scholar]
- Albasri, A., Aleskandarany, M., Benhasouna, A., Powe, D. G., Ellis, I. O., Ilyas, M. and Green, A. R. (2011b). CTEN (C-terminal tensin-like), a novel oncogene overexpressed in invasive breast carcinoma of poor prognosis. Breast Cancer Res. Treat. 126, 47-54. 10.1007/s10549-010-0890-3 [DOI] [PubMed] [Google Scholar]
- Alonso, A. and Pulido, R. (2016). The extended human PTPome: a growing tyrosine phosphatase family. FEBS J. 283, 1404-1429. 10.1111/febs.13600 [DOI] [PubMed] [Google Scholar]
- Aratani, K., Komatsu, S., Ichikawa, D., Ohashi, T., Miyamae, M., Okajima, W., Imamura, T., Kiuchi, J., Nishibeppu, K., Kosuga, T.et al. (2017). Overexpression of CTEN relates to tumor malignant potential and poor outcomes of adenocarcinoma of the esophagogastric junction. Oncotarget 8, 84112-84122. 10.18632/oncotarget.21109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artigas, M. S., Wain, L. V., Repapi, E., Obeidat, M., Sayers, I., Burton, P. R., Johnson, T., Zhao, J. H., Albrecht, E., Dominiczak, A. F.et al. (2011). Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am. J. Respir. Crit. Care 184, 786-795. 10.1164/rccm.201102-0192OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf, S., Kudo, H., Rao, J., Kikuchi, A., Widmeier, E., Lawson, J. A., Tan, W., Hermle, T., Warejko, J. K., Shril, S.et al. (2018). Mutations in six nephrosis genes delineate a pathogenic pathway amenable to treatment. Nat. Commun. 9, 1960. 10.1038/s41467-018-04193-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asiri, A., Raposo, T. P., Alfahed, A. and Ilyas, M. (2018). TGFβ1-induced cell motility but not cell proliferation is mediated through Cten in colorectal cancer. Int. J. Exp. Pathol. 99, 323-330. 10.1111/iep.12300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asiri, A., Toss, M. S., Raposo, T. P., Akhlaq, M., Thorpe, H., Alfahed, A., Asiri, A. and Ilyas, M. (2019). Cten promotes Epithelial-Mesenchymal Transition (EMT) in colorectal cancer through stabilisation of Src. Pathol. Int. 69, 381-391. 10.1111/pin.12811 [DOI] [PubMed] [Google Scholar]
- Auger, K. R., Songyang, Z., Lo, S. H., Roberts, T. M. and Chen, L. B. (1996). Platelet-derived growth factor-induced formation of tensin and phosphoinositide 3-kinase complexes. J. Biol. Chem. 271, 23452-23457. 10.1074/jbc.271.38.23452 [DOI] [PubMed] [Google Scholar]
- Barber-Pérez, N., Georgiadou, M., Guzmán, C., Isomursu, A., Hamidi, H. and Ivaska, J. (2020). Mechano-responsiveness of fibrillar adhesions on stiffness-gradient gels. J. Cell Sci. 133, jcs242909. 10.1242/jcs.242909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno, A. and Nieto, M. A. (2005). The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132, 3151-3161. 10.1242/dev.01907 [DOI] [PubMed] [Google Scholar]
- Bennett, D. T., Reece, T. B., Foley, L. S., Sjoberg, A., Meng, X., Fullerton, D. A. and Weyant, M. J. (2015). C-terminal tensin-like protein mediates invasion of human lung cancer cells and is regulated by signal transducer and activator of transcription 3. J. Thorac Cardiovasc. Surg. 149, 369-375. 10.1016/j.jtcvs.2014.08.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzing, T., Gerke, P., Höpker, K., Hildebrandt, F., Kim, E. and Walz, G. (2001). Nephrocystin interacts with Pyk2, p130Cas, and tensin and triggers phosphorylation of Pyk2. Proc. Natl. Acad. Sci. USA 98, 9784-9789. 10.1073/pnas.171269898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns, A. N. and Lo, S. H. (2020). Tensin regulates pharyngeal pumping in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 522, 599-603. 10.1016/j.bbrc.2019.11.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghel, G. J., Lin, W.-Y., Whitehouse, H., Brock, I., Hammond, D., Bury, J., Stephenson, Y., George, R. and Cox, A. (2013). Identification of candidate driver genes in common focal chromosomal aberrations of microsatellite stable colorectal cancer. PLoS ONE 8, e83859. 10.1371/journal.pone.0083859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood, D. A., Fujioka, Y., de Pereda, J. M., García-Alvarez, B., Nakamoto, T., Margolis, B., McGlade, C. J., Liddington, R. C. and Ginsberg, M. H. (2003). Integrin β cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. USA 100, 2272-2277. 10.1073/pnas.262791999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., Voss, C., Zhao, B., Kaneko, T. and Li, S. S.-C. (2012). Differential regulation of the activity of deleted in liver cancer 1 (DLC1) by tensins controls cell migration and transformation. Proc. Natl. Acad. Sci. USA 109, 1455-1460. 10.1073/pnas.1114368109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., Kaneko, T., Li, J. S., Liu, A.-D., Voss, C. and Li, S. S. C. (2015). A phosphorylation switch controls the spatiotemporal activation of Rho GTPases in directional cell migration. Nat. Commun. 6, 7721. 10.1038/ncomms8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, D. K., Carroll, J. S., Leong, C.-O., Cheng, F., Brown, M., Mills, A. A., Brugge, J. S. and Ellisen, L. W. (2006). p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 8, 551-561. 10.1038/ncb1420 [DOI] [PubMed] [Google Scholar]
- Cha, I. J., Lee, J. H., Cho, K. S. and Lee, S. B. (2017). Drosophila tensin plays an essential role in cell migration and planar polarity formation during oogenesis by mediating integrin-dependent extracellular signals to actin organization. Biochem. Biophys. Res. Commun. 484, 702-709. 10.1016/j.bbrc.2017.01.183 [DOI] [PubMed] [Google Scholar]
- Chan, L.-K., Ko, F. C. F., Ng, I. O.-L. and Yam, J. W. P. (2009). Deleted in liver cancer 1 (DLC1) utilizes a novel binding site for tensin2 PTB domain interaction and is required for tumor-suppressive function. PLoS ONE. 4, e5572. 10.1371/journal.pone.0005572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, L.-K., Chiu, Y.-T., Sze, K. M.-F. and Ng, I. O.-L. (2015). Tensin4 is up-regulated by EGF-induced ERK1/2 activity and promotes cell proliferation and migration in hepatocellular carcinoma. Oncotarget 6, 20964-20976. 10.18632/oncotarget.4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah, J. S., Jacobs, K. A., Heinrich, V., Lo, S. H. and Yamada, S. (2019). Force-induced recruitment of cten along keratin network in epithelial cells. Proc. Natl. Acad. Sci. USA 116, 19799-19801. 10.1073/pnas.1911865116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. and Lo, S. H. (2003). Regulation of tensin-promoted cell migration by its focal adhesion binding and Src homology domain 2. Biochem. J. 370, 1039-1045. 10.1042/BJ20021308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Duncan, I. C., Bozorgchami, H. and Lo, S. H. (2002). Tensin1 and a previously undocumented family member, tensin2, positively regulate cell migration. Proc. Natl. Acad. Sci. USA 99, 733-738. 10.1073/pnas.022518699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Liu, C., Ko, F. C. F., Xu, N., Ng, I. O.-L., Yam, J. W. P. and Zhu, G. (2012). Solution structure of the Phosphotyrosine Binding (PTB) domain of human tensin2 protein in complex with deleted in liver cancer 1 (DLC1) peptide reveals a novel peptide binding mode. J. Biol. Chem. 287, 26104-26114. 10.1074/jbc.M112.360206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Zhang, Y., Deng, G., Ma, J., Wu, X., Qu, Y. and Zeng, S. (2014). Correlation between the expression of C-terminal tensin-like protein and the prognosis of hepatocellular carcinoma. J. Cent. South Univ. Med. Sci. 39, 1233-1239. 10.11817/j.issn.1672-7347.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Chen, H.-Y., Lin, L.-T., Wang, M.-L., Laurent, B., Hsu, C.-H., Pan, C.-M., Jiang, W.-R., Chen, P.-Y., Ma, H.-I., Chen, Y.-W.et al. (2017). Musashi-1 enhances glioblastoma cell migration and cytoskeletal dynamics through translational inhibition of tensin3. Sci. Rep. 7, 8710. 10.1038/s41598-017-09504-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, M.-K., Liao, Y.-C., Kuwabara, Y. and Lo, S. H. (2005). Inactivation of tensin3 in mice results in growth retardation and postnatal lethality. Dev. Biol. 279, 368-377. 10.1016/j.ydbio.2004.12.027 [DOI] [PubMed] [Google Scholar]
- Cho, A.-R., Uchio-Yamada, K., Torigai, T., Miyamoto, T., Miyoshi, I., Matsuda, J., Kurosawa, T., Kon, Y., Asano, A., Sasaki, N.et al. (2006). Deficiency of the tensin2 gene in the ICGN mouse: an animal model for congenital nephrotic syndrome. Mamm. Genome 17, 407-416. 10.1007/s00335-005-0167-z [DOI] [PubMed] [Google Scholar]
- Chuang, J. Z., Lin, D. C. and Lin, S. (1995). Molecular cloning, expression, and mapping of the high affinity actin-capping domain of chicken cardiac tensin. J. Cell Biol. 128, 1095-1109. 10.1083/jcb.128.6.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, K., Howe, J. D., Pullar, C. E., Green, J. A., Artym, V. V., Yamada, K. M. and Critchley, D. R. (2010). Tensin 2 modulates cell contractility in 3D collagen gels through the RhoGAP DLC1. J. Cell. Biochem. 109, 808-817. 10.1002/jcb.22460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley, D. R. (2000). Focal adhesions - the cytoskeletal connection. Curr. Opin. Cell Biol. 12, 133-139. 10.1016/s0955-0674(99)00067-8 [DOI] [PubMed] [Google Scholar]
- Cui, Y. M., Liao, Y.-C. and Lo, S. H. (2004). Epidermal growth factor modulates tyrosine phosphorylation of a novel tensin family member, tensin3. Mol. Cancer Res. 2, 225-232. [PubMed] [Google Scholar]
- Dai, K., Liao, S., Zhang, J., Zhang, X. and Tu, X. (2011). Solution structure of tensin2 SH2 domain and its phosphotyrosine-independent interaction with DLC-1. PLoS ONE 6, e21965. 10.1371/journal.pone.0021965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S., Lu, M. L., Lo, S. H., Lin, S., Butler, J. A., Druker, B. J., Roberts, T. M., An, Q. and Chen, L. B. (1991). Presence of an Sh2 domain in the actin-binding protein tensin. Science 252, 712-715. 10.1126/science.1708917 [DOI] [PubMed] [Google Scholar]
- Dina, C., Bouatia-Naji, N., Tucker, N., Delling, F. N., Toomer, K., Durst, R., Perrocheau, M., Fernandez-Friera, L., Solis, J., Le Tourneau, T.et al. (2015). Genetic association analyses highlight biological pathways underlying mitral valve prolapse. Nat. Genet. 47, 1206-1211. 10.1038/ng.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, E. E., Maxim, D. S., Kuhns, V. L. H., Lane-Harris, A. C., Outeda, P., Ewald, A. J., Watnick, T. J., Welling, P. A. and Woodward, O. M. (2020). GDNF drives rapid tubule morphogenesis in a novel 3D in vitro model for ADPKD. J. Cell Sci. 133, jcs249557. 10.1242/jcs.249557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto, M., Kirkbride, J., Elliott, E., Lo, S. H. and Brautigan, D. L. (2007). Association of the tensin N-terminal protein-tyrosine phosphatase domain with the α isoform of protein phosphatase-1 in focal adhesions. J. Biol. Chem. 282, 17806-17815. 10.1074/jbc.M700944200 [DOI] [PubMed] [Google Scholar]
- Ferreira, M. A. R., Matheson, M. C., Tang, C. S., Granell, R., Ang, W., Hui, J., Kiefer, A. K., Duffy, D. L., Baltic, S., Danoy, P.et al. (2014). Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J. Allergy Clin. Immunol. 133, 1564-1571. 10.1016/j.jaci.2013.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger, B., Bershadsky, A., Pankov, R. and Yamada, K. M. (2001). Transmembrane crosstalk between the extracellular matrix and the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2, 793-805. 10.1038/35099066 [DOI] [PubMed] [Google Scholar]
- Georgiadou, M., Lilja, J., Jacquemet, G., Guzmán, C., Rafaeva, M., Alibert, C., Yan, Y., Sahgal, P., Lerche, M., Manneville, J.-B.et al. (2017). AMPK negatively regulates tensin-dependent integrin activity. J. Cell Biol. 216, 1107-1121. 10.1083/jcb.201609066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goreczny, G. J., Forsythe, I. J. and Turner, C. E. (2018). Hic-5 regulates fibrillar adhesion formation to control tumor extracellular matrix remodeling through interaction with tensin1. Oncogene 37, 1699-1713. 10.1038/s41388-017-0074-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi, S., Smith, L. J. M., Schütz, S. and Hafizi, S. (2013). Interaction of DISC1 with the PTB domain of Tensin2. Cell. Mol. Life Sci. 70, 1663-1672. 10.1007/s00018-012-1228-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafizi, S., Alindri, F., Karlsson, R. and Dahlbäck, B. (2002). Interaction of Axl receptor tyrosine kinase with C1-TEN, a novel C1 domain-containing protein with homology to tensin. Biochem. Biophys. Res. Commun. 299, 793-800. 10.1016/S0006-291x(02)02718-3 [DOI] [PubMed] [Google Scholar]
- Hafizi, S., Ibraimi, F. and Dahlbäck, B. (2005). C1-TEN is a negative regulator of the Akt/PKB signal transduction pathway and inhibits cell survival, proliferation, and migration. FASEB J. 19, 971-973. 10.1096/fj.04-2532fje [DOI] [PubMed] [Google Scholar]
- Hafizi, S., Sernstad, E., Swinny, J. D., Gomez, M. F. and Dahlbäck, B. (2010). Individual domains of tensin2 exhibit distinct subcellular localisations and migratory effects. Int. J. Biochem. Cell Biol. 42, 52-61. 10.1016/j.biocel.2009.09.005 [DOI] [PubMed] [Google Scholar]
- Hall, E. H., Daugherty, A. E., Choi, C. K., Horwitz, A. F. and Brautigan, D. L. (2009). Tensin1 requires protein phosphatase-1α in addition to RhoGAP DLC-1 to control cell polarization, migration, and invasion. J. Biol. Chem. 284, 34713-34722. 10.1074/jbc.M109.059592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, E. H., Balsbaugh, J. L., Rose, K. L., Shabanowitz, J., Hunt, D. F. and Brautigan, D. L. (2010). Comprehensive analysis of phosphorylation sites in tensin1 reveals regulation by p38MAPK. Mol. Cell. Proteomics 9, 2853-2863. 10.1074/mcp.M110.003665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, W., Ju, D., Jie, Z., Zhang, A., Xing, X. and Yang, Q. (2018). Aberrant CpG-methylation affects genes expression predicting survival in lung adenocarcinoma. Cancer Med. 7, 5716-5726. 10.1002/cam4.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamoto, N., Shimizu, T., Asai, K., Sakai, Y., Pastuhov, S. I., Hanafusa, H. and Matsumoto, K. (2019). C. elegans tensin promotes axon regeneration by linking the met-like SVH-2 and integrin signaling pathways. J. Neurosci. 39, 5662-5672. 10.1523/Jneurosci.2059-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.-Y., Shih, Y.-P., Li, T., Carraway, K. L., 3rd and Lo, S. H. (2013). CTEN prolongs signaling by EGFR through reducing its ligand-induced degradation. Cancer Res. 73, 5266-5276. 10.1158/0008-5472.Can-12-4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.-Y., Shih, Y.-P., Sun, P., Hsieh, W.-J., Lin, W.-C. and Lo, S. H. (2016). Down-regulation of tensin2 enhances tumorigenicity and is associated with a variety of cancers. Oncotarget 7, 38143-38153. 10.18632/oncotarget.9411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.-Y., Shih, Y.-P., Lo, A. and Lo, S. H. (2019). Identification of subcellular targeting sequences of Cten reveals its role in cell proliferation. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1866, 450-458. 10.1016/j.bbamcr.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes, R. O. (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110, 673-687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Ishii, A. and Lo, S. H. (2001). A role of tensin in skeletal-muscle regeneration. Biochem. J. 356, 737-745. 10.1042/bj3560737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H., Koh, A., Lee, J., Park, D., Lee, J. O., Lee, M. N., Jo, K.-J., Tran, H. N. K., Kim, E., Min, B.-S.et al. (2017). Inhibition of C1-Ten PTPase activity reduces insulin resistance through IRS-1 and AMPK pathways. Sci. Rep. 7, 17777. 10.1038/s41598-017-18081-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, A. S., Kaushansky, A., MacBeath, G. and Kaushansky, K. (2011). Tensin2 is a novel mediator in thrombopoietin (TPO)-induced cellular proliferation by promoting Akt signaling. Cell Cycle. 10, 1838-1844. 10.4161/cc.10.11.15776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, T., Mizuno, S., Taketo, M. M. and Kurosawa, T. M. (2008). The possible involvement of tensin2 in the expression and extension of nephrin by glomerular podocytes in mice. Biomed. Res. 29, 279-287. 10.2220/biomedres.29.279 [DOI] [PubMed] [Google Scholar]
- Katsuno, Y., Lamouille, S. and Derynck, R. (2013). TGF-β signaling and epithelial–mesenchymal transition in cancer progression. Curr. Opin. Oncol. 25, 76-84. 10.1097/CCO.0b013e32835b6371 [DOI] [PubMed] [Google Scholar]
- Katz, M., Amit, I., Citri, A., Shay, T., Carvalho, S., Lavi, S., Milanezi, F., Lyass, L., Amariglio, N., Jacob-Hirsch, J.et al. (2007). A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat. Cell Biol. 9, 961-969. 10.1038/ncb1622 [DOI] [PubMed] [Google Scholar]
- Kawai, K., Seike, J.-I., Lino, T., Kiyota, M., Iwamae, Y., Nishitani, H. and Yagisawa, H. (2009). START-GAP2/DLC2 is localized in focal adhesions via its N-terminal region. Biochem. Biophys. Res. Commun. 380, 736-741. 10.1016/j.bbrc.2009.01.095 [DOI] [PubMed] [Google Scholar]
- Kim, E., Kim, D.-H., Singaram, I., Jeong, H., Koh, A., Lee, J., Cho, W. and Ryu, S. H. (2018). Cellular phosphatase activity of C1-ten/tensin2 is controlled by phosphatidylinositol-3,4,5-triphosphate binding through the C1-ten/tensin2 SH2 domain. Cell. Signal. 51, 130-138. 10.1016/j.cellsig.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, A. P., Wolpin, B. M., Risch, H. A., Stolzenberg-Solomon, R. Z., Mocci, E., Zhang, M., Canzian, F., Childs, E. J., Hoskins, J. W., Jermusyk, A., et al. (2018). Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat. Commun. 9, 556. 10.1038/s41467-018-02942-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, A., Lee, M. N., Yang, Y. R., Jeong, H., Ghim, J., Noh, J., Kim, J., Ryu, D., Park, S., Song, P.et al. (2013). C1-ten is a protein tyrosine phosphatase of insulin receptor substrate 1 (IRS-1), regulating IRS-1 stability and muscle atrophy. Mol. Cell Biol. 33, 1608-1620. 10.1128/Mcb.01447-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. B., Cho, K. S., Kim, E. and Chung, J. (2003). blistery encodes Drosophila tensin protein and interacts with integrin and the JNK signaling pathway during wing development. Development 130, 4001-4010. 10.1242/dev.00595 [DOI] [PubMed] [Google Scholar]
- Lee, J., Koh, A., Jeong, H., Kim, E., Ha, T.-S., Saleem, M. A. and Ryu, S. H. (2017). C1-ten is a PTPase of nephrin, regulating podocyte hypertrophy through mTORC1 activation. Sci. Rep. 7, 12346. 10.1038/s41598-017-12382-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate, K. R. and Fässler, R. (2009). Mechanisms that regulate adaptor binding to beta-integrin cytoplasmic tails. J. Cell Sci. 122, 187-198. 10.1242/jcs.041624 [DOI] [PubMed] [Google Scholar]
- Leone, M., Yu, E. C., Liddington, R. C., Pasquale, E. B. and Pellecchia, M. (2008). The PTB domain of tensin: NMR solution structure and phosphoinositides binding studies. Biopolymers 89, 86-92. 10.1002/bip.20862 [DOI] [PubMed] [Google Scholar]
- Liao, Y.-C., Si, L., deVere White, R. W. and Lo, S. H. (2007). The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J. Cell Biol. 176, 43-49. 10.1083/jcb.200608015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y.-C., Chen, N.-T., Shih, Y.-P., Dong, Y. and Lo, S. H. (2009). Up-regulation of C-terminal tensin-like molecule promotes the tumorigenicity of colon cancer through β-catenin. Cancer Res. 69, 4563-4566. 10.1158/0008-5472.Can-09-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, S. H. (2004). Tensin. Int. J. Biochem. Cell Biol. 36, 31-34. 10.1016/s1357-2725(03)00171-7 [DOI] [PubMed] [Google Scholar]
- Lo, S. H. (2014). C-terminal tensin-like (CTEN): a promising biomarker and target for cancer. Int. J. Biochem. Cell Biol. 51, 150-154. 10.1016/j.biocel.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, S. H. and Lo, T. B. (2002). Cten, a COOH-terminal tensin-like protein with prostate restricted expression, is down-regulated in prostate cancer. Cancer Res. 62, 4217-4221. [PubMed] [Google Scholar]
- Lo, S. H., An, Q., Bao, S. D., Wong, W. K., Liu, Y., Janmey, P. A., Hartwig, J. H. and Chen, L. B. (1994a). Molecular cloning of chick cardiac muscle tensin - full-length cDNA sequence, expression, and characterization. J. Biol. Chem. 269, 22310-22319. 10.1016/S0021-9258(17)31791-X. Go to ISI://WOS:A1994PE09800059. [DOI] [PubMed] [Google Scholar]
- Lo, S. H., Janmey, P. A., Hartwig, J. H. and Chen, L. B. (1994b). Interactions of tensin with actin and identification of its three distinct actin-binding domains. J. Cell Biol. 125, 1067-1075. 10.1083/jcb.125.5.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, S. H., Yu, Q.-C., Degenstein, L., Chen, L. B. and Fuchs, E. (1997). Progressive kidney degeneration in mice lacking tensin. J. Cell Biol. 136, 1349-1361. 10.1083/jcb.136.6.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X., Gao, J., Zhang, Y., Zhao, T., Cai, H. C. and Zhang, T. (2018). CTEN induces epithelial-mesenchymal transition (EMT) and metastasis in non small cell lung cancer cells. PLoS ONE 13, e0198823. 10.1371/journal.pone.0198823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martuszewska, D., Ljungberg, B., Johansson, M., Landberg, G., Oslakovic, C., Dahlbäck, B. and Hafizi, S. (2009). Tensin3 is a negative regulator of cell migration and all four tensin family members are downregulated in human kidney cancer. PLoS ONE 4, e4350. 10.1371/journal.pone.0004350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusugi, K., Nakano, K., Sasaki, H., Kimura, J., Yanobu-Takanashi, R., Okamura, T. and Sasaki, N. (2016). Functional validation of tensin2 SH2-PTB domain by CRISPR/Cas9-mediated genome editing. J. Vet. Med. Sci. 78, 1413-1420. 10.1292/jvms.16-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleverty, C. J., Lin, D. C. and Liddington, R. C. (2007). Structure of the PTB domain of tensin1 and a model for its recruitment to fibrillar adhesions. Protein Sci. 16, 1223-1229. 10.1110/ps.072798707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misono, S., Seki, N., Mizuno, K., Yamada, Y., Uchida, A., Sanada, H., Moriya, S., Kikkawa, N., Kumamoto, T., Suetsugu, T.et al. (2019). Molecular pathogenesis of gene regulation by the miR-150 duplex: miR-150-3p regulates TNS4 in lung adenocarcinoma. Cancers 11, 601. 10.3390/cancers11050601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, K. D., Zhang, X., Zhou, Q. and Geahlen, R. L. (2012). The protein-tyrosine kinase Syk interacts with the C-terminal region of tensin2. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1823, 199-205. 10.1016/j.bbamcr.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muharram, G., Sahgal, P., Korpela, T., De Franceschi, N., Kaukonen, R., Clark, K., Tulasne, D., Carpén, O. and Ivaska, J. (2014). Tensin-4-dependent MET stabilization is essential for survival and proliferation in carcinoma cells. Dev. Cell 29, 421-436. 10.1016/j.devcel.2014.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, Y., Osawa, K., Fukushima, H., Tamura, Y., Aoki, K., Ohya, K., Yasuda, H., Hikiji, H., Takahashi, M., Seta, Y.et al. (2013). p130Cas, Crk-associated substrate, plays important roles in osteoclastic bone resorption. J. Bone Miner. Res. 28, 2449-2462. 10.1002/jbmr.1936 [DOI] [PubMed] [Google Scholar]
- Panasevich, S., Melén, E., Hallberg, J., Bergström, A., Svartengren, M., Pershagen, G. and Nyberg, F. (2013). Investigation of novel genes for lung function in children and their interaction with tobacco smoke exposure: a preliminary report. Acta Paediatr. 102, 498-503. 10.1111/apa.12204 [DOI] [PubMed] [Google Scholar]
- Pankov, R., Cukierman, E., Katz, B.-Z., Matsumoto, K., Lin, D. C., Lin, S., Hahn, C. and Yamada, K. M. (2000). Integrin dynamics and matrix assembly: tensin-dependent translocation of alpha5beta1 integrins promotes early fibronectin fibrillogenesis. J. Cell Biol. 148, 1075-1090. 10.1083/jcb.148.5.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M.-J., Sheng, R., Silkov, A., Jung, D.-J., Wang, Z.-G., Xin, Y., Kim, H., Thiagarajan-Rosenkranz, P., Song, S., Yoon, Y.et al. (2016). SH2 domains serve as lipid-binding modules for pTyr-signaling proteins. Mol. Cell 62, 7-20. 10.1016/j.molcel.2016.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, G. C., Kim, H.-S., Park, H.-Y., Seo, Y., Kim, J. M., Shin, S.-C., Kwon, H.-K., Sung, E.-S., Lee, J.-C. and Lee, B.-J. (2019). Tensin-3 regulates integrin-mediated proliferation and differentiation of tonsil-derived mesenchymal stem cells. Cells-Basel 9, 89. 10.3390/cells9010089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, J. T., Horwitz, A. R. and Schwartz, M. A. (2010). Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633-643. 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, X., Li, G., Asmussen, H. K., Asnaghi, L., Vass, W. C., Braverman, R., Yamada, K. M., Popescu, N. C., Papageorge, A. G. and Lowy, D. R. (2007). Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc. Natl. Acad. Sci. USA 104, 9012-9017. 10.1073/pnas.0703033104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, X., Li, G., Vass, W. C., Papageorge, A., Walker, R. C., Asnaghi, L., Steinbach, P. J., Tosato, G., Hunter, K. and Lowy, D. R. (2009). The tensin-3 protein, including its SH2 domain, is phosphorylated by Src and contributes to tumorigenesis and metastasis. Cancer Cell 16, 246-258. 10.1016/j.ccr.2009.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainero, E., Howe, J. D., Caswell, P. T., Jamieson, N. B., Anderson, K., Critchley, D. R., Machesky, L. and Norman, J. C. (2015). Ligand-occupied integrin internalization links nutrient signaling to invasive migration. Cell Rep. 10, 398-413. 10.1016/j.celrep.2014.12.037 [DOI] [PubMed] [Google Scholar]
- Repapi, E., Sayers, I., Wain, L. V., Burton, P. R., Johnson, T., Obeidat, M., Zhao, J. H., Ramasamy, A., Zhai, G., Vitart, V.et al. (2010). Genome-wide association study identifies five loci associated with lung function. Nat. Genet. 42, 36-44. 10.1038/ng.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita, K., Mimori, K., Tanaka, F., Kamohara, Y., Inoue, H., Sawada, T., Hirakawa, K. and Mori, M. (2008). Prognostic relevance of tensin4 expression in human gastric cancer. Ann. Surg. Oncol. 15, 2606-2613. 10.1245/s10434-008-9989-8 [DOI] [PubMed] [Google Scholar]
- Sasaki, H., Takahashi, Y., Ogawa, T., Hiura, K., Nakano, K., Sugiyama, M., Okamura, T. and Sasaki, N. (2020). Deletion of the tensin2 SH2-PTB domain, but not the loss of its PTPase activity, induces podocyte injury in FVB/N mouse strain. Exp. Anim. 69, 135-143. 10.1538/expanim.19-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada, Y., Tamada, M., Dubin-Thaler, B. J., Cherniavskaya, O., Sakai, R., Tanaka, S. and Sheetz, M. P. (2006). Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127, 1015-1026. 10.1016/j.cell.2006.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawazaki, S., Oshima, T., Sakamaki, K., Aoyama, T., Sato, T., Shiozawa, M., Yoshikawa, T., Rino, Y., Imada, T. and Masuda, M. (2017). Clinical significance of tensin 4 gene expression in patients with gastric cancer. In Vivo 31, 1065-1071. 10.21873/invivo.11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, E. Y., Jin, S.-P., Kim, Y. K., Lee, H., Han, S., Lee, D.-H. and Chung, J. H. (2017). Integrin-β4-TNS4–focal adhesion kinase signaling mediates keratinocyte proliferation in human skin. J. Invest. Dermatol. 137, 763-766. 10.1016/j.jid.2016.10.039 [DOI] [PubMed] [Google Scholar]
- Shih, Y.-P., Sun, P., Wang, A. and Lo, S. H. (2015). Tensin1 positively regulates RhoA activity through its interaction with DLC1. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1853, 3258-3265. 10.1016/j.bbamcr.2015.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinchi, Y., Hieda, M., Nishioka, Y., Matsumoto, A., Yokoyama, Y., Kimura, H., Matsuura, S. and Matsuura, N. (2015). SUV420H2 suppresses breast cancer cell invasion through down regulation of the SH2 domain-containing focal adhesion protein tensin-3. Exp. Cell Res. 334, 90-99. 10.1016/j.yexcr.2015.03.010 [DOI] [PubMed] [Google Scholar]
- Sjoestroem, C., Khosravi, S., Zhang, G., Martinka, M. and Li, G. (2013). C-Terminal tensin-like protein is a novel prognostic marker for primary melanoma patients. PLoS ONE 8, e80492. 10.1371/journal.pone.0080492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutchbury, B., Atherton, P., Tsang, R., Wang, D.-Y. and Ballestrem, C. (2017). Distinct focal adhesion protein modules control different aspects of mechanotransduction. J. Cell Sci. 130, 1612-1624. 10.1242/jcs.195362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Yang, S. and Song, W. (2020). Prazosin inhibits the proliferation and survival of acute myeloid leukaemia cells through down-regulating TNS1. Biomed. Pharmacother. 124, 109731. 10.1016/j.biopha.2019.109731 [DOI] [PubMed] [Google Scholar]
- Takito, J., Inoue, S. and Nakamura, M. (2018). The sealing zone in osteoclasts: a self-organized structure on the bone. Int. J. Mol. Sci. 19, 984. 10.3390/ijms19040984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe, H., Asiri, A., Akhlaq, M. and Ilyas, M. (2017). Cten promotes epithelial-mesenchymal transition through the post-transcriptional stabilization of Snail. Mol. Carcinog. 56, 2601-2609. 10.1002/mc.22704 [DOI] [PubMed] [Google Scholar]
- Torgler, C. N., Narasimha, M., Knox, A. L., Zervas, C. G., Vernon, M. C. and Brown, N. H. (2004). Tensin stabilizes integrin adhesive contacts in Drosophila. Dev. Cell 6, 357-369. 10.1016/S1534-5807(04)00055-3 [DOI] [PubMed] [Google Scholar]
- Touaitahuata, H., Morel, A., Urbach, S., Mateos-Langerak, J., de Rossi, S. and Blangy, A. (2016). Tensin 3 is a new partner of Dock5 that controls osteoclast podosome organization and activity. J. Cell Sci. 129, 3449-3461. 10.1242/jcs.184622 [DOI] [PubMed] [Google Scholar]
- Tripathi, B. K., Qian, X., Mertins, P., Wang, D., Papageorge, A. G., Carr, S. A. and Lowy, D. R. (2014). CDK5 is a major regulator of the tumor suppressor DLC1. J. Cell Biol. 207, 627-642. 10.1083/jcb.201405105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchio-Yamada, K., Sawada, K., Tamura, K., Katayama, S., Monobe, Y., Yamamoto, Y., Ogura, A. and Manabe, N. (2013). Tenc1-deficient mice develop glomerular disease in a strain-specific manner. Nephron Exp. Nephrol. 123, 22-33. 10.1159/000354058 [DOI] [PubMed] [Google Scholar]
- Uchio-Yamada, K., Monobe, Y., Akagi, K.-I., Yamamoto, Y., Ogura, A. and Manabe, N. (2016). Tensin2-deficient mice on FVB/N background develop severe glomerular disease. J. Vet. Med. Sci. 78, 811-818. 10.1292/jvms.15-0442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veß, A., Blache, U., Leitner, L., Kurz, A. R. M., Ehrenpfordt, A., Sixt, M. and Posern, G. (2017). A dual phenotype of MDA-MB-468 cancer cells reveals mutual regulation of tensin3 and adhesion plasticity. J. Cell Sci. 130, 2172-2184. 10.1242/jcs.200899 [DOI] [PubMed] [Google Scholar]
- Wavreille, A.-S. and Pei, D. (2007). A chemical approach to the identification of tensin-binding proteins. ACS Chem. Biol. 2, 109-118. 10.1021/cb600433g [DOI] [PubMed] [Google Scholar]
- Wilkins, J. A. and Lin, S. (1986). A re-examination of the interaction of vinculin with actin. J. Cell Biol. 102, 1085-1092. 10.1083/jcb.102.3.1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winograd-Katz, S. E., Fässler, R., Geiger, B. and Legate, K. R. (2014). The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 15, 273-288. 10.1038/nrm3769 [DOI] [PubMed] [Google Scholar]
- Wu, W.-M. and Liao, Y.-C. (2018). Downregulation of C-terminal tensin-like protein (CTEN) suppresses prostate cell proliferation and contributes to acinar morphogenesis. Int. J. Mol. Sci. 19,, 3190. 10.3390/ijms19103190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z.-Y., Chiu, C.-L., Lo, E., Lee, Y.-R. J., Yamada, S. and Lo, S. H. (2019). Hyperactivity of Mek in TNS1 knockouts leads to potential treatments for cystic kidney diseases. Cell Death Dis. 10, 871. 10.1038/s41419-019-2119-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam, J. W. P., Ko, F. C. F., Chan, C.-Y., Jin, D.-Y. and Ng, I. O.-L. (2006). Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res. 66, 8367-8372. 10.1158/0008-5472.CAN-05-2850 [DOI] [PubMed] [Google Scholar]
- Yang, J., Zhou, H. X., Liang, B. M., Xiao, J., Su, Z. G., Chen, H., Ma, C. L., Li, D. X., Feng, Y. L. and Ou, X. M. (2014). Association of five genetic variants with chronic obstructive pulmonary disease susceptibility and spirometric phenotypes in a Chinese Han population. Respirology 19, 262-268. 10.1111/resp.12212 [DOI] [PubMed] [Google Scholar]
- Yang, K., Wu, W.-M., Chen, Y.-C., Lo, S. H. and Liao, Y.-C. (2016). ΔNp63α transcriptionally regulates the expression of CTEN that is associated with prostate cell adhesion. PLoS ONE 11, e0147542. 10.1371/journal.pone.0147542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir, E. and Geiger, B. (2001). Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 114, 3583-3590. https://www.ncbi.nlm.nih.gov/pubmed/11707510. [DOI] [PubMed] [Google Scholar]
- Zhang, D. and Aravind, L. (2010). Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes. Gene 469, 18-30. 10.1016/j.gene.2010.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z. H., Tan, S. H., Machiyama, H., Kawauchi, K., Araki, K., Hirata, H. and Sawada, Y. (2016). Association between tensin 1 and p130Cas at focal adhesions links actin inward flux to cell migration. Biol. Open 5, 499-506. 10.1242/bio.016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H.-M., Fang, Y.-Y., Weinberger, P. M., Ding, L.-L., Cowell, J. K., Hudson, F. Z., Ren, M., Lee, J. R., Chen, Q.-K., Su, H.et al. (2016). Transgelin increases metastatic potential of colorectal cancer cells in vivo and alters expression of genes involved in cell motility. BMC Cancer 16, 55. 10.1186/s12885-016-2105-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., Zhang, Y., Wu, L., Xie, W., Li, L., Yuan, Y., Chen, Y., Lin, Y. and He, X. (2018). Elevated transgelin/TNS1 expression is a potential biomarker in human colorectal cancer. Oncotarget 9, 1107-1113. 10.18632/oncotarget.23275 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.