ABSTRACT
Chromatin plays an essential role in the nuclear mechanical response and determining nuclear shape, which maintain nuclear compartmentalization and function. However, major genomic functions, such as transcription activity, might also impact cell nuclear shape via blebbing and rupture through their effects on chromatin structure and dynamics. To test this idea, we inhibited transcription with several RNA polymerase II inhibitors in wild-type cells and perturbed cells that presented increased nuclear blebbing. Transcription inhibition suppressed nuclear blebbing for several cell types, nuclear perturbations and transcription inhibitors. Furthermore, transcription inhibition suppressed nuclear bleb formation, bleb stabilization and bleb-based nuclear ruptures. Interestingly, transcription inhibition did not alter the histone H3 lysine 9 (H3K9) modification state, nuclear rigidity, and actin compression and contraction, which typically control nuclear blebbing. Polymer simulations suggested that RNA polymerase II motor activity within chromatin could drive chromatin motions that deform the nuclear periphery. Our data provide evidence that transcription inhibition suppresses nuclear blebbing and rupture, in a manner separate and distinct from chromatin rigidity.
Keywords: Chromatin, Mechanobiology, Nucleus, Nuclear blebbing
Highlighted Article: Inhibitors of transcription suppress both nuclear bleb formation and stabilization without altering nuclear rigidity, actin compression or actin contraction, suggesting new biophysics for nuclear morphology.
INTRODUCTION
Aberrant alterations in transcription and disruptions to nuclear shape are two common cellular hallmarks of human disease. Both are used as prognostic indicators of disease severity in breast, cervical and prostate cancers, among others (Papanicolaou and Traut, 1997; Helfand et al., 2012; Radhakrishnan et al., 2017; Lu et al., 2018). Several studies have suggested that there are interactions between transcription and nuclear shape. In the testosterone-sensitive prostate cancer model cell line LNCaP, testosterone-induced transcription via the androgen receptor results in increased nuclear blebbing (Helfand et al., 2012). Transcription activation via TGFβ1 has also been shown to induce abnormal nuclear shape (Chi et al., 2022). Additionally, active RNA polymerase (pol) II and transcriptionally active genes and chromosomes are enriched in nuclear blebs in cells with lamin B1 knocked down by shRNA and in progeria cells (Shimi et al., 2008; Bercht Pfleghaar et al., 2015). The biochemical and biophysical state of chromatin can control nuclear morphology (Furusawa et al., 2015; Schreiner et al., 2015; Stephens et al., 2018), but it is unclear whether transcription is necessary to induce abnormal nuclear shape in perturbed nuclei. Furthermore, the mechanism of how transcription affects nuclear shape is unknown. Thus, understanding how transcription affects nuclear shape and integrity would provide new insights into these two vital functions that are perturbed in disease.
Chromatin mechanics is essential to the nuclear mechanical response, and it is a key determinant of nuclear shape and stability (Stephens et al., 2019b). Chromatin generates a spring-like elastic response to small mechanical deformations of a few micrometers in size, complementing the robust elastic response to large deformations provided by lamins (Stephens et al., 2017; Hobson et al., 2020; Currey et al., 2022). The rigidity of chromatin is governed by histone modifications (Chalut et al., 2012; Krause et al., 2013; Shimamoto et al., 2017; Stephens et al., 2018, 2019a; Hobson et al., 2020; Nava et al., 2020), histone H1 dynamics (Furusawa et al., 2015; Senigagliesi et al., 2019), chromatin-binding proteins (Schreiner et al., 2015; Wang et al., 2018; Williams et al., 2020 preprint; Strom et al., 2021) and three-dimensional genome organization (Belaghzal et al., 2021). Independently of lamins, chromatin decompaction results in a softer nucleus, which can more easily succumb to external perturbations, such as cytoskeletal forces. Thus, the nucleus can lose its normal shape and form a protrusion called a nuclear bleb (Furusawa et al., 2015; Stephens et al., 2018; Kalinin et al., 2021). Both chromatin and lamin perturbations result in nuclear blebs, which have highly curved surfaces prone to rupture (De Vos et al., 2011; Vargas et al., 2012; Stephens et al., 2018; Xia et al., 2018; Nmezi et al., 2019; Pfeifer et al., 2022). The resulting loss of nuclear compartmentalization via blebbing and rupture causes genomic dysfunction through DNA damage (Denais et al., 2016; Irianto et al., 2016; Raab et al., 2016; Chen et al., 2018; Xia et al., 2018; Stephens et al., 2019a; Shah et al., 2021), global changes to transcription along with transcriptional inhibition within the bleb (De Vos et al., 2011; Helfand et al., 2012), and loss of cell cycle control (Pfeifer et al., 2018). Thus, the physical properties of chromatin, and potentially transcription, can impact nuclear function through nuclear morphology.
Transcription affects both chromatin structure and dynamics, which suggests possible pathways to nuclear blebbing and rupture. Chromosome conformation capture (Hi-C and Micro-C) experiments have indicated that polymerases may aid in the establishment of A/B chromatin compartments typically associated with euchromatin and heterochromatin (Jiang et al., 2020; Zhang et al., 2021), generate characteristic spatial patterns around active genes (Banigan et al., 2023; Zhang et al., 2023) and contribute to enhancer–promoter contacts (Hsieh et al., 2020; Zhang et al., 2023). Furthermore, active transcription is generally associated with an open, decompact chromatin structure, as assessed by assay for transposase-accessible chromatin with sequencing (ATAC-seq) (Buenrostro et al., 2013). However, it has also been suggested that transcription could play a restrictive role on chromatin by locally constraining it via crosslinking (Nagashima et al., 2019) or phase separation (Hnisz et al., 2017). Nonetheless, transcription drives large-scale chromatin dynamics, which manifest as correlated motions of micrometer-sized regions (Zidovska et al., 2013; Shaban et al., 2018, 2020). Simulations suggest that chromatin connectivity combined with the forces generated by polymerase motor activity (∼10 pN per polymerase; Herbert et al., 2008) could generate these dynamics (Liu et al., 2021). The same mechanism could contribute to shape fluctuations of isolated nuclei (Liu et al., 2021), although experiments indicate that transcription inhibition in live cells marginally enhances shape fluctuations (Chu et al., 2017). Thus, transcriptional changes could potentially affect nuclear morphology and compartmentalization through changes to the chromatin-based nuclear mechanical response or large-scale chromatin dynamics.
To investigate the biophysical effects of transcription activity on nuclear blebbing, mechanics and rupture, we used several different transcription inhibitors in wild-type cells and cells with perturbations that increased blebbing. We particularly focused on transcription inhibition by α-amanitin in cells with nuclear blebbing induced by the established mechanisms of the histone deacetylase inhibitor valproic acid (VPA) (Stephens et al., 2018, 2019a; Kalinin et al., 2021). In this way, we assessed the nuclear morphological effects of transcription inhibition under both normal conditions and conditions that typically result in nuclear blebs. Treatment with α-amanitin did not change the population-level prevalence of nuclear blebbing in both mouse embryonic fibroblasts (MEFs) and HT1080 wild-type cells, but it suppressed nuclear blebbing in VPA-treated cells. Micromanipulation force measurements showed that transcription inhibition did not alter nuclear rigidity, suggesting an alternative, dynamic mechanism for nuclear blebbing. Timelapse imaging of cells expressing a GFP-tagged nuclear localization signal (NLS–GFP) to track nuclear blebbing and ruptures revealed that transcription inhibition suppressed nuclear bleb formation and stabilization, even in wild-type cells. This suppression ultimately affected the number of nuclear ruptures that occur per nucleus. We found that active initiating RNA pol II, but not elongating RNA pol II, was enriched in nuclear blebs. These findings could be recapitulated and understood through a polymer simulation model in which chromatin was a crosslinked polymer contained within a polymer shell (the lamina), and transcription activity was represented by motor activity within the chromatin. Thus, we found that transcription inhibition suppresses nuclear blebbing and ruptures without changing the bulk rigidity of chromatin.
RESULTS
α-Amanitin decreases transcriptional activity in both untreated and VPA-treated cells
Transcription is a major nuclear function that affects the biophysical properties of chromatin and the nucleus, and thus could modulate nuclear blebbing and dynamics. To assess this possibility, we inhibited transcription using the RNA pol II inhibitor α-amanitin in wild-type MEFs that were either untreated or treated with the histone deacetylase inhibitor VPA. VPA induces chromatin decompaction via increased euchromatin, which results in a weaker nucleus that blebs and ruptures (Stephens et al., 2018, 2019a).
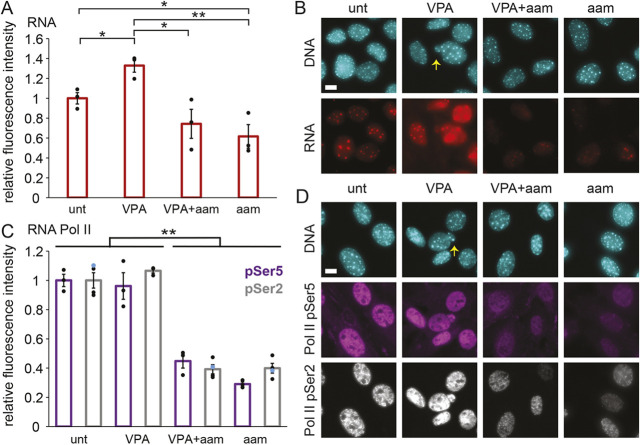
To assay changes in transcription upon α-amanitin treatment, we measured whole nuclear RNA levels using the uridine analog 5-ethynyluridine (EU) and click chemistry (Jao and Salic, 2008). VPA treatment resulted in an increased level of RNA compared to that in untreated (‘unt’) wild-type cells (Fig. 1A,B). Treatment with α-amanitin (‘aam’) drastically reduced nuclear RNA levels in both untreated and VPA-treated wild-type cells. Thus, α-amanitin successfully represses transcription in both untreated and VPA-treated cells.
Fig. 1.
Transcription is decreased upon treatment with the RNA pol II inhibitor α-amanitin in both untreated and VPA-treated MEFs. (A,B) Graph (A) and example images (B) of RNA fluorescence levels in MEFs, labeled via EU Click-iT chemistry (red) and DNA labeled via Hoechst (cyan) for untreated (‘unt’), VPA-treated, VPA- and α-amanitin-treated (‘VPA+aam’), and α-amanitin-treated (‘aam’) cells. (C,D) Graph (C) and example images (D) of immunofluorescence levels of RNA pol II phosphorylated at Ser5 (purple, initiating) and Ser2 (gray, elongating) with DNA labeled via Hoechst (cyan). Yellow arrows denote nuclear blebs in example images. α-Amanitin treatment was for 24 h. Three biological replicates were performed with n>30 cells. Error bars represent standard error. *P<0.05; **P<0.01 (two-tailed unpaired Student's t-test). Scale bars: 10 µm.
Next, we aimed to establish how VPA and α-amanitin affect active RNA pol II levels. We measured the average immunofluorescence intensities of active RNA pol II phosphorylated at Ser5 (initiating; pSer5) and Ser2 (elongating; pSer5) in nuclei. Without α-amanitin, we found no difference in the levels of RNA pol II pSer5 and pSer2 in untreated cells and VPA-treated cells (Fig. 1C,D). As expected, α-amanitin treatment resulted in a drastic 60% or greater decrease of active RNA pol II levels in cells without or with VPA treatment (Fig. 1C,D). The decreased levels of active RNA pol II were also similar between pSer5 and pSer2, suggesting that initiation and elongation are inhibited similarly. Thus, α-amanitin decreases transcription via inhibition of active RNA pol II.
Transcription inhibition suppresses nuclear blebbing
With a defined approach for decreasing transcription, we aimed to determine the effect that transcription inhibition has on nuclear blebbing. Untreated wild-type nuclei normally exhibited a low background level of nuclear blebbing, with 4% of nuclei presenting blebs (Fig. 2A). With VPA treatment, the percentage of nuclei with nuclear blebs increased to 11% (Fig. 2A), consistent with previous reports (Stephens et al., 2018, 2019a). Interestingly, dual treatment with the RNA pol II inhibitor α-amanitin and VPA for 16–24 h resulted in a significant decrease in nuclear blebbing levels from 11% for VPA to 6% for VPA and α-amanitin together, effectively returning nuclear blebbing to wild-type levels (Fig. 2A). The addition of α-amanitin alone did not alter wild-type nuclear blebbing percentages, as they remained at 4%. Thus, transcription inhibition is a significant inhibitor of nuclear blebs in cells with chromatin decompaction induced by VPA.
Fig. 2.
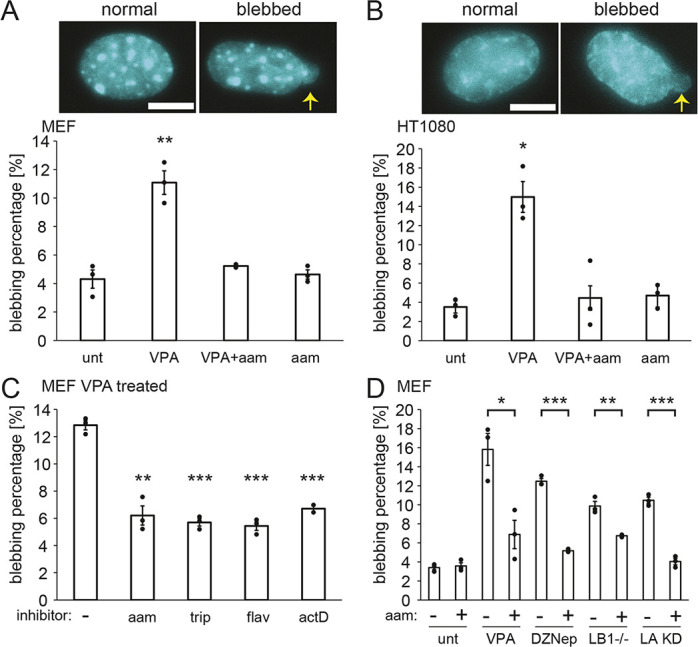
Transcription inhibition suppresses nuclear blebbing across cell types, drugs and perturbations that cause nuclear blebbing. (A,B) Example images and graph of percentages of nuclei that bleb in MEF cells (A) and HT1080 cells (B) for untreated (‘unt’), VPA-treated (VPA), VPA- and α-amanitin-treated (‘VPA+aam’), and α-amanitin-treated (‘aam’) cells. The means of three biological replicates with n=60–200 cells each are shown as dots. Yellow arrows denote nuclear blebs in example images. Scale bars: 10 µm. (C) Graph of percentages of nuclei that blebbed in VPA-treated MEF cells with or without the RNA pol II inhibitors α-amanitin (‘aam’, 24 h), triptolide (‘trip’, 24 h), flavopiridol (‘flav’, 24 h) and actinomycin D (‘actD’, 1.5 h). The means of three technical replicates with n>100 cells each are shown as dots. (D) Graph of percentages of nuclei that blebbed in wild-type cells (‘unt’) and different perturbations without or with the RNA pol II inhibitor α-amanitin (‘aam’). The perturbations were: increased euchromatin (VPA treatment), decreased heterochromatin (DZNep treatment), lamin B1 knockout (LB1−/− cells) and lamin A knockdown (LA KD cells). The means of three biological replicates are shown as dots with n>71 cells each for ‘unt’ and ‘VPA’, and with n>300 cells each for ‘DZNep’, ‘LB1−/−’ and ‘LA KD’. Error bars represent standard error. *P<0.05; **P<0.01; ***P<0.001 (two-tailed unpaired Student's t-test).
Next, we aimed to determine whether the effect of transcription inhibition on nuclear blebbing is a general phenomenon. First, we considered an alternative cell type. Human fibrosarcoma (HT1080) cells also displayed increased levels of nuclear blebbing upon chromatin decompaction by VPA treatment, increasing from 3% to 15% (Fig. 2B), as previously reported (Stephens et al., 2019a). Similar to MEFs, dual treatment of HT1080 cells with α-amanitin and VPA resulted in wild-type levels of nuclear blebbing, indicating that bleb formation resulting from VPA treatment was suppressed.
To determine whether specific transcription inhibition treatments and durations have differing effects on nuclear blebbing, we used different transcription inhibitors with VPA-treated MEFs. We used triptolide (‘trip’) to inhibit transcription initiation, flavopiridol (‘flav’) to inhibit transcription elongation, and the DNA intercalator actinomycin D (‘actD’), which inhibits elongation and acts quickly (Bensaude, 2011). All transcription inhibitor treatments also resulted in significant decreases in nuclear blebbing induced by VPA (P<0.05, Fig. 2C). Moreover, all four transcription inhibitors tested resulted in a similarly low percentage of nuclei exhibiting a bleb (5–6%, P>0.05). Thus, transcription is broadly necessary for increased nuclear blebbing.
To determine the importance of transcription activity to nuclear blebbing more generally, we used alternative methods for nuclear perturbations to induce nuclear blebs. Treatment with the histone methyltransferase inhibitor 3-deazaneplanocin A (DZNep) results in decreased heterochromatin and nuclear stiffness resulting in blebs, similar to VPA treatment (Stephens et al., 2018). DZNep-treated cells presented 12% nuclear blebbing, whereas treatment with DZNep and α-amanitin together suppressed this nuclear blebbing to 5% (Fig. 2D). To compare chromatin-based perturbations to lamin-based perturbations, we studied cells with decreased lamin levels. MEF Lmnb1−/− cells, null for lamin B1, also exhibit a significantly increased level of nuclear blebbing relative to that in wild-type cells (Lammerding et al., 2006; Vargas et al., 2012; Hatch and Hetzer, 2016; Stephens et al., 2019a; Young et al., 2020). Constitutive knockdown (KD) of lamin A (Lmna or LA) in MEFs is also reported to cause nuclear blebbing (Vahabikashi et al., 2022). Both MEF Lmnb1−/− and LA KD cells treated with α-amanitin exhibited decreased levels of nuclear blebbing, dropping significantly from 10% to 6% and 4%, respectively (Fig. 2D). Similar nuclear blebbing suppression occurred for DZNep-treated, Lmnb1−/− and LA KD cells treated with flavopiridol and triptolide (Fig. S1A). Thus, transcription inhibition suppresses nuclear blebbing across different cell types and for both chromatin- and lamin-based perturbations.
Nuclear rigidity is not altered by transcription inhibition
Nuclear blebbing is thought to be driven by disruptions to one or both of the two major nuclear mechanical components, chromatin and lamin A/C (Stephens et al., 2019b; Kalukula et al., 2022). We thus investigated whether transcription inhibition alters nuclear mechanics.
Given that VPA treatment leads to softer nuclei due to increased histone acetylation, we sought to determine whether transcription inhibition suppresses bleb formation by suppressing histone acetylation. To this end, we assayed levels of euchromatin via immunofluorescence for histone H3 lysine 9 acetylation (H3K9ac). With VPA treatment, euchromatin levels increased by ∼50% from untreated wild-type cells (orange bars, Fig. 3A,B), as previously reported (Stephens et al., 2017, 2018). Surprisingly, treatment with α-amanitin and VPA together increased the levels of H3K9ac even more than VPA treatment alone, but α-amanitin treatment alone did not have any effect on H3K9ac levels (orange bars, Fig. 3A,B). Furthermore, addition of α-amanitin did not significantly alter heterochromatin levels as measured by immunofluorescence for H3K9 methylation (H3K9me2,3) (gray bars, Fig. 3B), which are associated with larger nuclear stiffness (Stephens et al., 2017, 2018, 2019a; Strom et al., 2021). These data demonstrate that nuclear bleb suppression in transcriptionally inhibited VPA-treated cells does not result from a decrease in H3K9ac euchromatin or an increase in H3K9me2,3 heterochromatin.
Fig. 3.
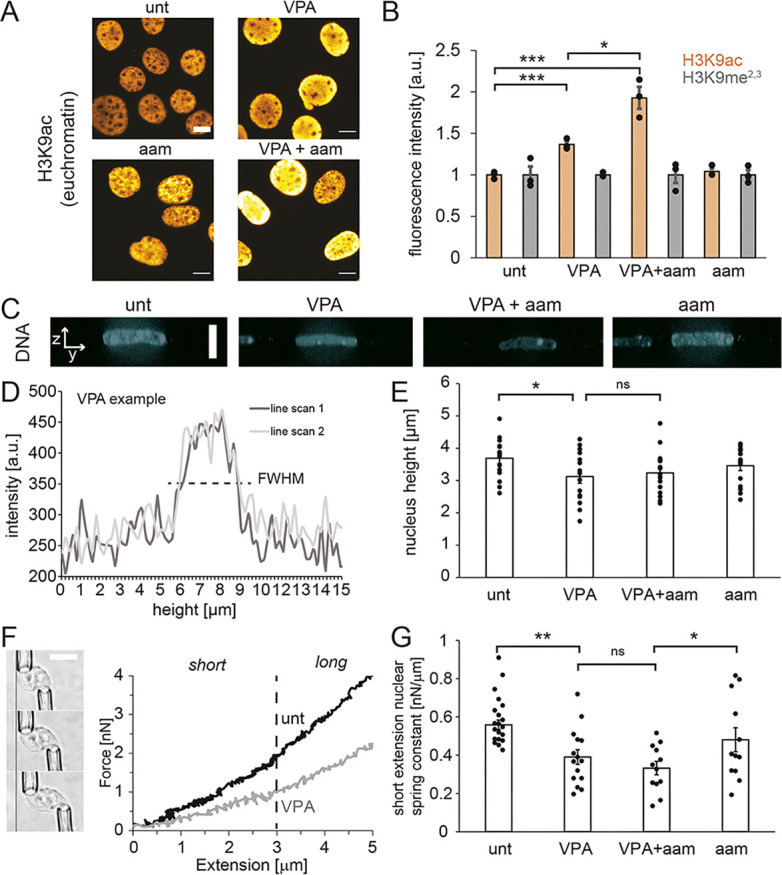
The mechanical properties of the nucleus are not altered by transcription inhibition. (A,B) Example images (A) and graphs (B) of the relative immunofluorescence signal of a euchromatin marker (H3K9ac, orange) and a heterochromatin marker (H3K9me2,3, gray) in untreated (‘unt’), VPA-treated, VPA- and α-amanitin-treated (‘VPA+aam’), and α-amanitin-treated (‘aam’) cells. The means of three biological replicates with n=100–300 cells each are shown as dots. (C) Example images of side and top-down views of Hoechst-labeled nuclei captured using spinning-disk confocal microscopy. There is no yellow dotted line. (D) Example line scans through the side-on image of the nucleus of the VPA-treated cell from panel C, from which the full-width at half maximum (FWHM) values from two different line scans were averaged to determine the height of each nucleus. (E) Individual (dots) and average (bar) measurements of nuclear height for each condition (n=15 nuclei for each condition). (F) Left: example images of a micromanipulation force-extension measurement, with an isolated nucleus from MEF V−/− cells pulled by the ‘pull’ pipette (bottom right) to extend the nucleus and held by the ‘force’ pipette (top left), the deflection of which multiplied by a precalibrated spring constant measures force. Right: example force-extension graph of control (‘unt’) and chromatin-decompacted (‘VPA’) cells over short and long regimes. The dashed line indicates the crossover from the short-extension to the long-extension regime. (G) Graph of the individual (dots) and average (bar) short-extension nuclear spring constants (‘unt’, n=21; ‘VPA’, n=15; ‘VPA+aam’, n=12; ‘aam’, n=12). Long-extension spring constants did not change for all conditions (P>0.05, Fig. S3B). α-Amanitin treatment was for 24 h. Error bars represent standard error. ns, not significant, P>0.05; *P<0.05; **P<0.01 (two-tailed unpaired Student's t-test). a.u., arbitrary units. Scale bars: 10 µm.
We also investigated whether transcription inhibition decreases nuclear blebbing through changes in chromatin-based nuclear rigidity, which could alter the mechanical force balance between the nucleus and cytoskeleton (Stephens et al., 2019b). Nuclear height measurements provide a proxy for this force balance between the nucleus and the actin cytoskeleton compressing the nucleus. Using spinning-disk confocal microscopy of Hoechst 33342 (hereafter Hoechst)-stained nuclei, we measured nuclear height in untreated and VPA-treated cells without or with α-amanitin (Fig. 3C,D). VPA treatment resulted in a statistically significant decrease in the average height of the nucleus (Fig. 3E). This is likely due to nuclear softening from VPA-induced chromatin decompaction (Stephens et al., 2018), allowing further nuclear compression by actin fibers on top of the nucleus (Khatau et al., 2009). Treatment with α-amanitin did not induce additional significant changes in nuclear height in both untreated and VPA-treated cells (Fig. 3E). Actin contraction [as observed by immunofluorescence of phosphorylated MLC2 (pMLC2), encoded by MYL9] was also similar after α-amanitin treatment (Fig. S2). Thus, transcription inhibition appears to have little effect on the overall mechanical force balance between the nucleus and the actin cytoskeleton.
To directly measure whether nuclear rigidity is altered by RNA pol II inhibition, we used micropipette micromanipulation of individual nuclei to measure the nuclear force–extension relation (Stephens et al., 2017; Currey et al., 2022). Micromanipulation force measurements uniquely provide the ability to measure both short extension, chromatin-based nuclear stiffness and lamin-A-based strain stiffening at longer extensions (>3 µm). For ease of isolation, we used MEF vimentin null (V−/−) nuclei, which display nuclear rigidity (Stephens et al., 2017) and nuclear blebbing similar to those of wild-type MEFs (Fig. S3A). VPA-treated V−/− nuclei exhibited a weaker short-extension chromatin-based spring constant than untreated V−/− nuclei (Fig. 3F,G), consistent with previous reports (Stephens et al., 2017, 2018, 2019a). Interestingly, transcription inhibition did not alter chromatin-based nuclear stiffness in V−/− untreated or VPA-treated cells (Fig. 3G). These data agree with the measurements of nuclear height inside live wild-type cells, which also did not change upon transcription inhibition (Fig. 3E). Nuclear strain stiffening in the long-extension lamin-dominated regime also did not significantly change across all conditions (Fig. S3B). However, in all transcription inhibition experiments, we observed wrinkles in the nuclear lamina (Fig. S3C), which could have occurred due to altered tension and buckling in the lamina. Altogether, we found that transcription does not increase chromatin-based nuclear rigidity and does not change overall nuclear stiffness.
Transcription inhibition changes the type and frequency of rupture
Nuclear blebbing often correlates with the loss of nuclear compartmentalization. To determine whether decreased nuclear blebbing upon transcription inhibition also impacts nuclear rupture, we tracked nuclear ruptures in transcription-inhibited cells. Nuclear rupture was observed by the spilling of concentrated NLS–GFP from the nucleus into the cytoplasm (Fig. 4A). By live imaging NLS–GFP cells over 3 h at 2-min intervals, we observed that the percentage of nuclei that ruptured did not change in a statistically significant manner upon transcription inhibition but did exhibit a trend of decreased nuclear ruptures from VPA treatment to VPA co-treated with α-amanitin (19±3% versus 12±2%, indicated as mean±s.e.m., P=0.08, Fig. 4B). Thus, although transcription inhibition in VPA-treated cells decreased nuclear blebbing to wild-type levels, it only had a moderate effect on the percentage of nuclei that underwent nuclear rupture.
Fig. 4.
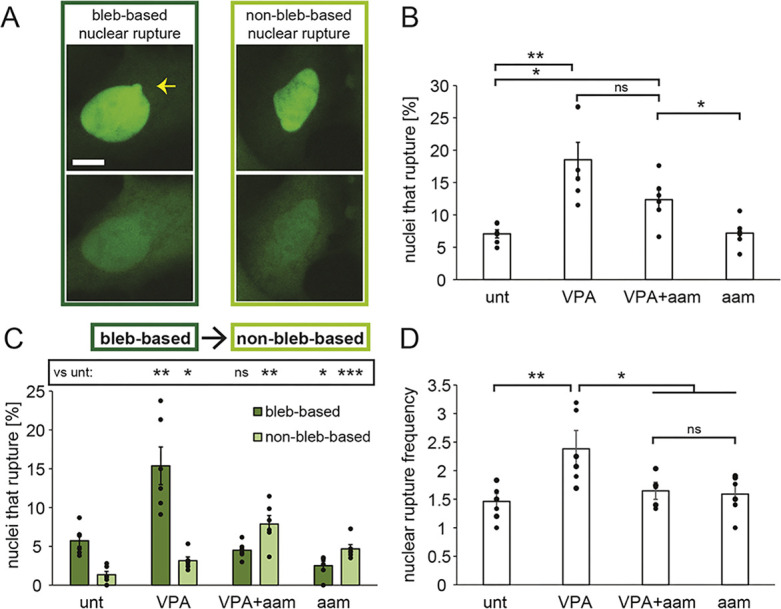
Transcription inhibition alters the type and frequency of ruptures per nucleus. (A) Example images of bleb-based (yellow arrow) and non-bleb-based nuclear ruptures. Scale bar: 10 µm. (B) Graph of the percentages of nuclei that displayed at least one rupture in a 3-h timelapse with 2-min intervals based on NLS–GFP fluorescence in untreated (‘unt’), VPA-treated, VPA- and α-amanitin-treated (‘VPA+aam’), and α-amanitin-treated cells (‘aam’). (C) Graph of the percentages of nuclei that present bleb-based (dark green) or non-bleb-based (light green) nuclear ruptures. (D) Graph of average nuclear rupture frequency which is, for nuclei that rupture, the average number of times a nucleus ruptured during the 3-h timelapse. The averages were calculated from six biological replicates, graphed as dots, and each consisted of n=100–300 cells. α-Amanitin treatment was for 24 h. Error bars represent standard error. ns, not significant, P>0.05; *P<0.05; **P<0.01; ***P<0.001 (two-tailed unpaired Student's t-test).
Nuclear blebs have been reported to be responsible for most nuclear ruptures (Denais et al., 2016; Raab et al., 2016; Patteson et al., 2019; Stephens et al., 2019a), but ruptures can occur in non-blebbed nuclei (Robijns et al., 2016; Chen et al., 2018; Xia et al., 2018; Zhang et al., 2019). It is possible that nuclear rupture in transcription-inhibited cells occurs in non-blebbed nuclei. We therefore counted ruptures as being bleb-based or non-bleb-based for untreated and VPA-treated cells without or with α-amanitin (Fig. 4A,C). In untreated and VPA-treated cells with normal transcription, the majority of nuclear ruptures were bleb-based (>80% of ruptures, dark green, P<0.001, Fig. 4C), in agreement with previous reports (Stephens et al., 2019a). Upon transcription inhibition with α-amanitin, the predominant type of rupture changed to non-bleb-based nuclear rupture (>63% of ruptures, light green, P<0.01, Fig. 4C). A limitation of these measurements is that if a bleb formed, ruptured and was reabsorbed in less than 2 min, it would have appeared as a non-bleb-based rupture. However, based on the long typical lifetime of a nuclear bleb, this error was likely minimal. Thus, even though transcription inhibition suppressed nuclear blebbing, it resulted in a similar percentage of nuclei rupturing, the majority of which were not blebbed.
Nonetheless, of the nuclei that ruptured, we hypothesized that transcription might control how frequently they ruptured. For each nucleus that ruptured, we counted the number of times it ruptured over the 3-h timelapse. Wild-type nuclei that ruptured did so an average of 1.5±0.2 times, whereas VPA-treated nuclei ruptured 2.4±0.4 times over 3 h. Dual treatment with VPA and α-amanitin decreased nuclear rupture frequency to wild-type levels (1.6±0.2 per rupturing nucleus over 3 h), which was also similar to treatment with α-amanitin alone (Fig. 4D). Taken together, the data show that transcription inhibition decreases the number of times an individual nucleus may rupture, possibly by inhibiting bleb formation.
Nuclear bleb formation and stabilization are dependent on transcription activity
Transcription inhibition could inhibit nuclear blebs by preventing either the formation or the stabilization of nuclear blebs. To assay these possibilities, we tracked cells during the timelapse for new bleb formation and whether that bleb was stabilized or reabsorbed. Blebs were counted as stabilized if they remained after NLS–GFP reaccumulated in the nucleus following bleb formation and rupture (Fig. 5A, blue); alternatively, blebs were counted as reabsorbed if they disappeared following nuclear rupture (Fig. 5A, gold). Wild-type cells exhibited formation of nuclear blebs coupled to nuclear ruptures, and the majority of newly blebbed nuclei stabilized to remain blebbed, whereas a minority quickly reabsorbed their bleb and returned to a normal shape (Fig. 5). VPA-treated cells exhibited a similar behavior, but with more nuclear bleb formation than in untreated cells (Fig. 5B). Thus, in cells with normal transcription (untreated and VPA treated), regardless of the level of blebbing, the majority of nuclear blebs that formed were stabilized after formation (Fig. 5C, blue). However, transcription inhibition by α-amanitin drastically decreased nuclear bleb formation (Fig. 5B). When a nuclear bleb did form, it was usually reabsorbed quickly, before NLS–GFP that spilled into the cytoplasm was reaccumulated in the nucleus (Fig. 5C, gold). Furthermore, timelapse imaging in the first 8 h of transcription inhibition treatment showed a larger portion of blebs being reabsorbed back into the nucleus, increasing from 5% in both VPA-treated wild-type and untreated Lmnb1−/− cells to 30% following α-amanitin treatment (Fig. S1B). Thus, both nuclear bleb formation and stabilization decreased drastically upon transcription inhibition in both untreated and VPA-treated wild-type cells, suggesting that transcription activity contributes to nuclear blebbing.
Fig. 5.
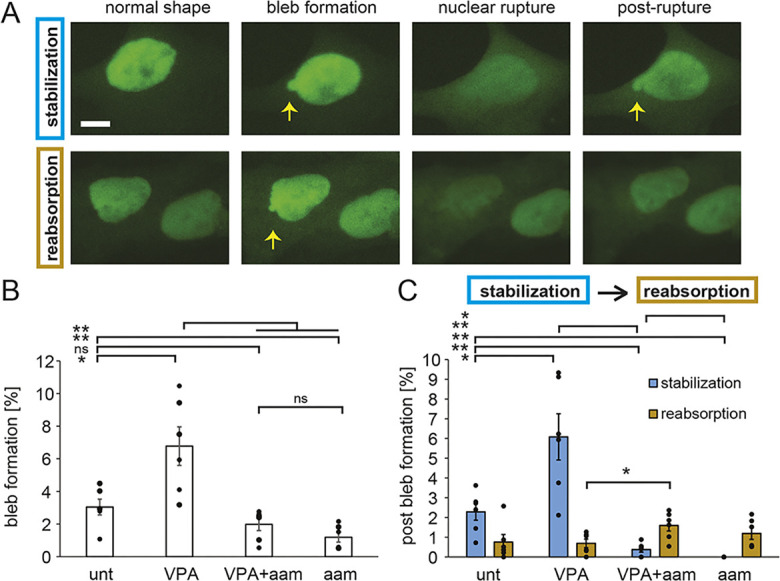
Bleb formation and stabilization are dependent on transcription activity. (A) Example images of stabilization (top, blue) or reabsorption (bottom, gold) of a newly formed bleb tracked via NLS–GFP live-cell imaging. Yellow arrows denote blebs. Scale bar: 10 µm. (B) Graph of the percentages of nuclei that display new nuclear bleb formation in a 3-h timelapse for untreated (‘unt’), VPA-treated, VPA- and α-amanitin-treated (‘VPA+aam’), and α-amanitin-treated (‘aam’) cells. (C) Graph showing the percentages of nuclei that displayed new nuclear bleb formation alongside a nuclear rupture that resulted in either bleb stabilization (blue) or reabsorption (gold) post rupture. The averages were calculated from six biological replicates, graphed as dots, and each consisted of n=100–300 cells. α-Amanitin treatment was for 24 h. Error bars represent standard error. ns, not significant, P>0.05; *P<0.05; **P<0.01 (two-tailed unpaired Student's t-test).
Transcriptional contribution to nuclear bleb composition
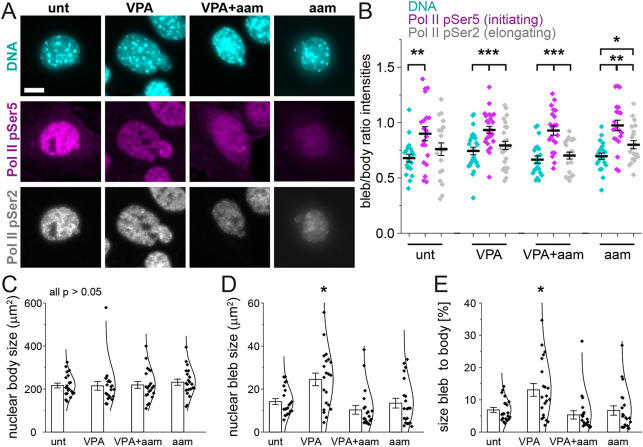
To assess how transcriptional activity aids the formation and stabilization of nuclear blebs, we assayed markers of transcription in the bleb. Previous reports indicate that active and/or phosphorylated RNA pol II is enriched in nuclear blebs (Shimi et al., 2008; Helfand et al., 2012; Bercht Pfleghaar et al., 2015). Using immunofluorescence, we assayed the distribution of active RNA pol II pSer5 (initiation) and pSer2 (elongation) relative to bulk chromatin labeled by Hoechst (Fig. 6A,B). Specifically, we measured the average intensity of each transcription marker and chromatin in the nuclear bleb relative to the nuclear body to compute a ratio. Hoechst intensities revealed that, on average, there was less chromatin in the nuclear bleb relative to the nuclear body across all conditions (∼66%, Fig. 6A,B), in agreement with previous reports (Bercht Pfleghaar et al., 2015; Stephens et al., 2018). Interestingly, RNA pol II pSer5, marking transcription initiation, was significantly enriched in the bleb versus in the nuclear body relative to Hoechst in all conditions (Fig. 6A,B). In contrast, RNA pol II pSer2 marking transcription elongation had decreased intensity in the bleb as compared to in the nuclear body, similar to the corresponding ratio for Hoechst. RNA exhibited an intermediate, increased bleb enrichment compared to DNA, but remained less enriched than in the nuclear body in wild-type cells (∼80%, Table S1). Thus, transcription initiation is enriched in the bleb relative to chromatin and transcription elongation.
Fig. 6.
Transcription initiation is enriched in blebs relative to DNA content and overall supports bleb size. (A) Example images of Hoechst (DNA, cyan), RNA pol II phosphorylated at Ser5 (initiating, pSer5, magenta) and RNA pol II phosphorylated at Ser2 (elongating, pSer2, gray) staining in untreated (‘unt’), VPA-treated, VPA- and α-amanitin-treated (‘VPA+aam’), and α-amanitin-treated (‘aam’) cells. Scale bar: 10 µm. (B) Graph of the single-nucleus bleb intensity/nuclear body intensity ratio. DNA (Hoechst) signal intensity was decreased in blebs, but pol II pSer5 signal intensity in the bleb was enriched relative to DNA signal intensity and had a signal that was similar to that in the main nuclear body. (C–E) Graphs of average nuclear body size (C), nuclear bleb size (D) and average size of the bleb as a percentage of the nuclear body (E) (‘unt’, n=20; ‘VPA’, n=26; ‘VPA+aam’, n=21; ‘aam’, n=21). α-Amanitin treatment was for 24 h. Error bars represent standard error. *P<0.05; **P<0.01; ***P<0.001 (two-tailed unpaired Student's t-test).
To determine whether transcription activity supports nuclear blebs, we assayed nuclear bleb size upon transcription inhibition. The area of the nuclear body and bleb were measured in all conditions. The average size of the nuclear body did not change between untreated and VPA-treated cells without or with α-amanitin (Fig. 6C). However, from untreated (low nuclear blebbing) to VPA-treated (high nuclear blebbing) cells, there was a significant increase in the size of nuclear blebs, suggesting that chromatin decompaction by histone acetylation can grow and stabilize blebs (Fig. 6D). Transcription inhibition by α-amanitin suppressed bleb size in VPA-treated cells, returning the average size to that of untreated wild-type cells (Fig. 6D). Analysis of nuclear bleb size as a percentage of the nuclear body size revealed the same trends, further supporting the notion that changes in bleb size were not due to differences in nuclear body size (Fig. 6E). Taken together, these data suggest that transcription activity stimulates nuclear bleb growth and maintenance of size upon chromatin decompaction via VPA treatment.
Transcriptional motor activity generates nuclear deformations in active polymer simulations
Our data show that inhibition of transcription activity suppresses nuclear bleb formation and stabilization across different cell types and perturbations. Surprisingly, although transcription inhibition suppresses blebbing, it does not alter major cellular components or properties that are widely thought to be responsible for nuclear blebbing (Le Berre et al., 2012; Hatch and Hetzer, 2016; Mistriotis et al., 2019; Stephens et al., 2019b; Kalukula et al., 2022). This list includes nuclear stiffness (Fig. 3G), levels of major euchromatic and heterochromatic H3K9 histone modifications (Fig. 3A,B), nuclear confinement by the actin cytoskeleton (Fig. 3E), and contractile activity within the actin cytoskeleton (Fig. S2), all of which are unaltered by transcription inhibition. We therefore sought to determine whether the observed effects of transcription on nuclear blebbing could be understood through other known biophysical phenomena.
To this end, we adapted a coarse-grained Brownian dynamics polymer simulation model of the nucleus. Earlier realizations of this model have previously been used to study nuclear mechanical response and morphology (Banigan et al., 2017; Stephens et al., 2017; Lionetti et al., 2020; Liu et al., 2021; Strom et al., 2021), and transcription-driven mesoscale chromatin dynamics (Liu et al., 2021) that have been observed experimentally (Zidovska et al., 2013; Shaban et al., 2018, 2020).
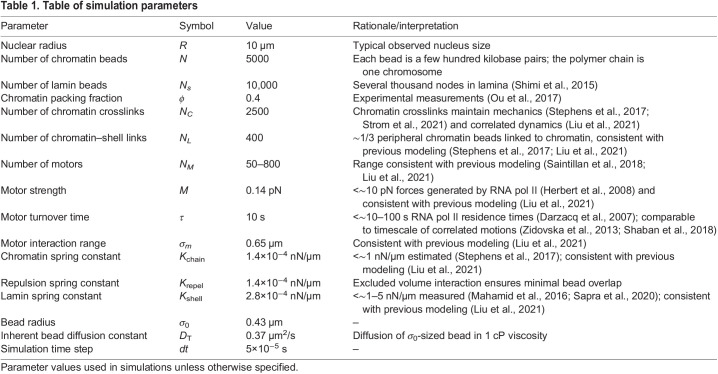
The model is a deformable elastic, polymeric shell representing the nuclear lamina (Fig. 7A, purple) that encapsulates an active polymer chain representing the chromatin fiber (Fig. 7A, blue). The polymer chain is crosslinked by springs to model gel-like chromatin (Strickfaden et al., 2020; Belaghzal et al., 2021; Strom et al., 2021) (Fig. 7A, red). Polymer and shell subunits are also linked by springs (Fig. 7A, green); these linkages lead to a stiffer nucleus in simulations (Strom et al., 2021) and in vivo (Schreiner et al., 2015). We included motor activity to model the effect of transcription, which can drive motion of micrometer-sized chromatin domains in experiments (Zidovska et al., 2013; Shaban et al., 2018, 2020), possibly through the forces that it exerts on chromatin (Liu et al., 2021). In our model, RNA pol II activity is incorporated through extensile motors that repel nearby chromatin, without directly interacting with the lamina (Fig. 7A, gold, inset). This coarse-grained representation of motor activity was intended to qualitatively model the collective activity of many polymerases. We considered motors that generate sub-pN forces, well below the 10 pN forces that may be generated by individual RNA polymerases (Herbert et al., 2008). Further simulation details and parameters are given in Table 1 and in the Materials and Methods.
Fig. 7.
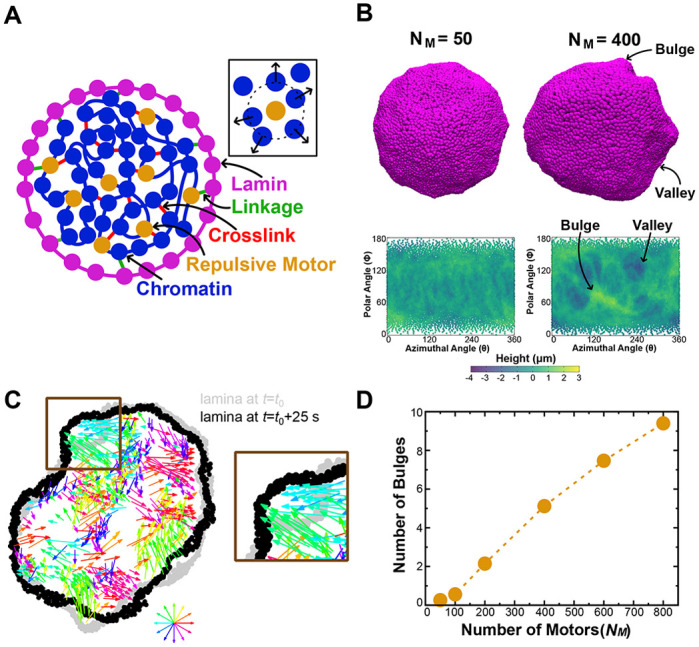
Inhibition of motors decreases nuclear bulge formation. (A) Schematic two-dimensional cross-section of the simulation model. Inset: illustration of a repulsive motor (gold) repelling chromatin subunits within the interaction range. (B) Top: simulation snapshots of simulated nuclei with bulges and valleys for simulations with different numbers (NM) of motors. Bottom: lamina height maps corresponding to the simulation snapshots, showing bulges (green) and valleys (blue). Maps show deviation of the lamina from the mean shell radius at coordinates given by the polar angle θ and azimuthal angle φ. (C) Left: snapshot of a two-dimensional cross-section of a simulation showing the chromatin displacements over 25 s (colored arrows) and changes in nuclear shape. The lamina is shown at the beginning (time t0, gray) and end (time t0+25 s, black) of the 25 s time interval. The color wheel indicates the direction of motion encoded by the corresponding color. The region outlined by the brown box identifies a part of the lamina that bulges outward due to chromatin motion, which is reproduced in the image on the right. (D) Mean number of bulges increases with increasing numbers of motors (NM).
Table 1.
Table of simulation parameters
Because α-amanitin inhibits RNA pol II motor activity, we performed simulations with different numbers of motors (NM) to compare the effects of different levels of activity within chromatin. For most values of NM, simulated nuclei exhibited transient bulges (Fig. 7B, top row), which were generated by correlated coherent motion of the active chromatin (Fig. 7C). We did not observe stable complete blebs in these simulations, likely because this model omits complications such as rupture of lamin-to-lamin bonds and chromatin-to-chromatin linkages. Nonetheless, the bulges suggest that transcriptionally active chromatin may exert forces that generate potential precursors to blebs.
To quantify this observation, we constructed height maps of the lamina (Fig. 7B, see Materials and Methods) and identified bulges as protrusions with a maximum height greater than 1 μm above the mean lamina height. Decreasing NM led to fewer bulges and greater homogeneity in lamina heights (Fig. 7B,D). This observation indicates that chromatin motions generated by motor activity could induce nuclear bulges (Fig. 7C). Inversely, inhibiting transcriptional motors suppresses bulges. These results mirror the experimental findings that inhibiting RNA pol II activity decreases nuclear blebbing (Fig. 2). Overall, the model provides a plausible new mechanism through which transcription inhibition might suppress bleb formation by reducing the prevalence of chromatin-driven bleb precursors generated by correlated chromatin motions.
DISCUSSION
Chromatin is a major mechanical component of the nucleus that aids in the maintenance of both nuclear shape and compartmentalization to protect genomic organization and function. Our experiments show that inhibition of transcription activity suppresses nuclear bleb formation and bleb stabilization after bleb formation (Figs 2 and 5; Fig. S1). However, transcription inhibition does not govern nuclear morphology through changes to bulk nuclear rigidity (Fig. 3C–G) or the actin cytoskeleton (Fig. S2). Rather, transcription inhibition appears to modulate bleb formation and dynamics through an alternative mechanism, which is evident in nuclei that are already less mechanically resilient due to chromatin decompaction or loss of lamin. Transcription initiation appears to be a key feature of nuclear blebs, as it is enriched in blebs (Fig. 6A,B). This decrease in nuclear blebbing also results in fewer nuclear ruptures (Fig. 4), which are an established driver of cellular dysfunction. Building on previous biophysical modeling of the cell nucleus, we simulated the nucleus as a polymeric lamina with an active chromatin polymer interior. These simulations suggest that blebs could arise due to the forces that transcription activity exerts on chromatin, which can manifest as larger chromatin motions and nuclear bulges (Fig. 7). Altogether, these findings demonstrate that transcription contributes to nuclear blebbing and rupture and suggest that nuclear morphology may be altered by chromatin dynamics rather than by chromatin mechanics alone.
Transcription is necessary for bleb formation in perturbed nuclei
Our investigation shows that excess nuclear blebbing in cells with nuclear perturbations does not occur when transcription is inhibited. Perturbations of histone modifications (by VPA and DZNep) or lamin levels (using Lmnb1−/− and LA KD cells) generally increased nuclear blebbing, but these effects were negated by transcription inhibition (by several inhibitors; Fig. 2; Fig. S1). Apparently, the reduction in nuclear rigidity due to either chromatin decompaction or loss of lamins alone was insufficient to increase blebbing dramatically above untreated wild-type levels (Fig. 3) (Stephens et al., 2017, 2018; Vahabikashi et al., 2022). Instead, our data indicate that the effects of transcription on genome spatiotemporal structure and/or dynamics are also required for increased blebbing.
Interestingly, however, transcription inhibition appeared to have no effect on the basal percentage of cells that underwent nuclear blebbing for both MEF and HT1080 cell populations (Fig. 2). These data are consistent with the existence of multiple factors and processes that can drive nuclear shape disruptions. Many reports show that both actin confinement and contraction are also vital to nuclear bleb formation and stabilization (Le Berre et al., 2012; Hatch and Hetzer, 2016; Mistriotis et al., 2019; Pho et al., 2022). However, our data show that neither actin confinement, as measured by nuclear height (Fig. 3E), nor actin contraction, as measured by myosin activity (pMLC2 immunofluorescence; Fig. S2), were altered by transcription inhibition. Therefore, it is likely that these mechanisms act independently, even if they also act synergistically in some scenarios.
The ability of transcription to promote nuclear blebbing is important because of the relationship between blebbing and nuclear rupture, even if blebbing is not strictly necessary for rupture. Observations of blebs generated in transcriptionally active nuclei are important in that they correspond to increases in nuclear rupture (Fig. 4B–D) and, presumably, corresponding nuclear dysfunction, such as DNA damage (Stephens, 2020). The additional observation of non-bleb-based rupture (Fig. 4C) is consistent with previous findings (Chen et al., 2018; Stephens et al., 2019a; Earle et al., 2020; Pho et al., 2022), but indicates that transcription is not essential for nuclear ruptures in the absence of nuclear blebs. Thus, there are likely multiple factors responsible for regulating nuclear integrity and morphology. We have shown that transcription activity is one such factor, and the data suggest that it may act in concert with known mechanisms.
Our findings provide new insights into previous reports that suggested that alterations in transcription could drive aberrant nuclear architecture in perturbed cell types. Two previous studies of cancer cell lines linked activation hormones that increase transcription to nuclear blebbing and shape abnormalities (Helfand et al., 2012; Chi et al., 2022). In prostate cancer cells (LNCaP), stimulation of transcription by testosterone-activated androgen receptor resulted in a threefold increase in nuclear blebbing, correlating with an increase in the Gleason grade of the cancer (Helfand et al., 2012). Similarly, in hepatocarcinoma cells (Huh7), TGFβ1 treatment resulted in the majority of nuclei becoming abnormally shaped (Chi et al., 2022). However, in previous studies, unlike in our investigation, nuclear shape changes arising after transcription induction occurred in concert with altered expression of lamins and reorganization of chromatin and the lamina, which could affect the nuclear mechanical response. Consistently, LNCaP prostate cancer cells have been reported to be softer than wild-type prostate cells (Khan et al., 2018), but required transcription upregulation to induce greater levels of nuclear blebbing. Beyond these factors, TGFβ1 induction also stimulates actin contractility (Inoue-Mochita et al., 2015), which is linked to nuclear deformations (Mistriotis et al., 2019; Pho et al., 2022). In contrast, we observed that histone acetylation (H3K9ac) and methylation (H3K9me2,3), lamin A/C and actin contractility (pMLC2) are largely unaltered by transcriptional changes (Fig. 3A,B; Figs S2 and S3). Thus, our experiments and simulations suggest that even normal transcription activity can stimulate nuclear blebbing and that there is an additional mechanism linking transcription to nuclear morphology, beyond what has been reported previously. We propose that, in parallel to previously observed pathways, transcription-induced changes to chromatin dynamics could lead to the observed nuclear shape disruptions.
Modeling nuclear bleb formation of transcriptionally active nuclei
Our simulations of the nucleus as a polymeric lamina shell enclosing a transcriptionally active, crosslinked chromatin polymer show how transcriptional regulation of genome dynamics could impact nuclear shape. As shown by our experiments, chromatin and transcription activity are key contributors to nuclear blebbing and rupture. Models accounting only for nuclear lamins (Wren et al., 2012; Funkhouser et al., 2013) are unable to explain nuclear shape disruptions linked to chromatin compaction and/or transcription. To that end, our model provides a chromatin-based mechanism for abnormal nuclear morphology (Fig. 7) while also matching other mechanical and dynamical observations of chromatin (Banigan et al., 2017; Stephens et al., 2017; Liu et al., 2021; Strom et al., 2021).
However, our model captures nuclear deformations in the form of transient bulges rather than long-lived blebs, which leaves open the question of precisely how blebs form. The largest bulges are similar to blebs, so the bulges that we observe might be precursors to blebs, which, in turn, might form in a more complicated model with additional features. Notably, linkages, including lamin–lamin bonds, chromatin crosslinks and chromatin–lamina links, are permanent in our model and cannot break. In vivo, these linkages are dynamic and breakable under force (e.g. Cheutin et al., 2003; Festenstein et al., 2003; Kind et al., 2013; Sapra et al., 2020; Vahabikashi et al., 2022), which could contribute to the material failure that leads to nuclear bleb formation. Furthermore, we simulate isolated nuclei, which are not subject to external forces or confinement from the cytoskeleton, which could induce blebbing (Le Berre et al., 2012; Hatch and Hetzer, 2016; Mistriotis et al., 2019; Pho et al., 2022). Inclusion of these features could fully reconcile our model with experiments and allow bulges to develop into blebs.
Bulk nuclear stiffness is not the sole determinant of nuclear blebbing and rupture
Surprisingly, we found that decreased nuclear stiffness alone was not sufficient to cause nuclear blebbing. This was shown by the loss of nuclear blebbing with transcription inhibition in cells with chromatin and/or lamin perturbations (Figs 2 and 3). This observation differs from previous studies, which suggested that the nucleus maintains its shape solely by resisting cytoskeletal and/or other external antagonistic forces (Khatau et al., 2009; Le Berre et al., 2012; Hatch and Hetzer, 2016; Stephens et al., 2018; Earle et al., 2020). Thus, loss of chromatin- and/or lamin-based rigidity could cause the nucleus to succumb to cytoskeletal forces and form a nuclear herniation or bleb. Given that transcription inhibition suppresses the formation and stabilization of nuclear blebs, at least some aspect of nuclear shape deformation appears to be independent of the bulk mechanical strength of the nucleus.
However, force balance between the nucleus and the cytoskeleton appears to govern nuclear rupture to some extent, even in scenarios where transcription is disrupted and blebs are not present (Fig. 4A–C). Regardless of the transcriptional state of the nucleus, nuclei with softer, decompacted chromatin (VPA treated) were more likely to rupture, with or without blebs. Nonetheless, the force balance governing nuclear rupture was apparently affected, but not controlled by the presence of blebs, as nuclei with blebs ruptured more frequently (i.e. multiple times in several hours; Fig. 4D). Therefore, transcriptionally active nuclei that undergo rupture could suffer greater DNA damage (Stephens, 2020) or disruption of the cell cycle (Pfeifer et al., 2018) because they bleb more frequently than less transcriptionally active nuclei. Furthermore, the higher frequency of ruptures agrees with reports that high nuclear curvature, such as that found on the surface of blebs, promotes rupture (Robijns et al., 2016; Stephens et al., 2018; Xia et al., 2018; Nmezi et al., 2019; Pfeifer et al., 2022), perhaps as a result of higher local surface tension in the lamina (Deviri et al., 2017; Xia et al., 2018; Srivastava et al., 2021). Therefore, high nuclear curvature and transcription can promote nuclear rupture, but neither are strictly necessary for ruptures to occur.
Consistently, our experiments showed that most nuclear ruptures in transcription-inhibited cells occur in non-blebbed and relatively normal-shaped nuclei (Fig. 4C). The high curvature of blebs was also found to be dispensable in previous studies observing ruptures without blebbing or substantial nuclear shape fluctuations (Robijns et al., 2016; Chen et al., 2018; Penfield et al., 2018; Earle et al., 2020). This notion suggests that local tensile stress can induce nuclear ruptures without necessarily leading to abnormal nuclear shape or blebbing (Zhang et al., 2019). Therefore, we hypothesize that in VPA-treated cells with transcription inhibition, chromatin is soft such that it does not support the lamina against rupture via cytoskeletal forces, but the loss of transcription prevents the formation and stabilization of blebs.
A plausible mechanism of transcriptional regulation of nuclear blebbing
As transcription promotes blebbing but does not alter whole-nucleus stiffness, we propose that an alternative mechanism might connect these phenomena. Our experiments and modeling suggest that transcription might promote nuclear blebbing by globally altering chromatin dynamics and, consequently, increasing chromatin-driven pushing and pulling on the nuclear lamina.
These effects could arise through the correlated motions of micrometer-sized chromatin domains and/or chromatin density fluctuations that emerge due to transcriptional activity (Shaban et al., 2018, 2020; Liu et al., 2021; Barth et al., 2020). In this scenario, transcription generates relatively large, correlated regions of chromatin motion throughout the nucleus, and some of these may be driven into the lamina, causing local deformation via bulges or blebs. Consistently, previous simulations of isolated nuclei showed that (transcriptional) motor activity in an active, crosslinked polymer can drive correlated polymer (chromatin) dynamics and increase shape fluctuations of the polymeric shell (the lamina) (Liu et al., 2021).
Additionally, polymeric shells, such as the nuclear lamina, subject to tensile or compressive stresses can buckle, thereby undergoing a sudden, dramatic change in shape resembling a first-order phase transition (Paulose and Nelson, 2013; Yong et al., 2013; Banigan et al., 2017). Thus, there may generally be a kinetic barrier to bleb formation, which can be overcome in part by energy generated by transcriptional dynamics throughout the nucleus.
Complementarily to these dynamic mechanisms, chromatin linkages to the lamina might play a role in governing bleb formation. A previous study of lamina-associated chromatin domains showed that transcription of genes tends to detach them from the lamina, whereas transcription inhibition induces lamina attachment of inactivated genes (Brueckner et al., 2020). Based on previous simulations suggesting that chromatin–lamina linkages can help maintain nuclear shape (Banigan et al., 2017; Lionetti et al., 2020), it is possible that linkages induced by transcription inhibition suppress bleb formation. Nonetheless, regulation of linkages might be only a secondary mechanism, as we did not observe any change in nuclear rigidity upon transcription inhibition (Fig. 3), as seen in previous simulations (Strom et al., 2021).
It has also been proposed that transcription affects chromatin–chromatin connections (Nagashima et al., 2019), which, in turn, could alter nuclear rigidity and potentially nuclear morphology. Experiments imaging single nucleosomes in vivo showed that transcription inhibition by α-amanitin or 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) enhanced spatial fluctuations of nucleosomes, suggesting that there were fewer constraints within transcriptionally inactive chromatin. Notably, inhibition by actinomycin D had the opposite effect of suppressing nucleosome fluctuations. We found that transcription inhibition by α-amanitin did not reduce nuclear rigidity (Fig. 3), as we would expect if chromatin crosslinking or bridging by RNA pol II has nucleus-scale effects (Stephens et al., 2017; Strom et al., 2021). These observations suggest that transcriptional effects on crosslinking are not a major contributor to nuclear morphology.
Another clue about the mechanism is the composition of the nuclear bleb. The transcription initiation marker RNA pol II pSer5 is enriched relative to DNA, whereas the transcription elongation marker RNA pol II pSer2 is not (Fig. 6). This clarifies which active RNA pol II is prominently enriched within blebs, which had previously been shown to be enriched in active RNA pol II (Shimi et al., 2008; Helfand et al., 2012; Bercht Pfleghaar et al., 2015). This observation suggests that components associated with transcription initiation are important for bleb stabilization, growth and, possibly, formation. However, the contribution of transcription initiation is presently unclear.
Nonetheless, our results emphasize the importance of chromatin and its constituents in regulating nuclear morphology, even in situations in which other nuclear mechanical components, such as lamins, are unaltered. More generally, regardless of the precise physical mechanism, our study raises the possibility that other chromatin-bound molecular motors, such as condensin, cohesin, topoisomerase and DNA polymerase, might influence nuclear shape through their activities and effects on chromatin dynamics.
MATERIALS AND METHODS
Cell culture
Wild-type, V−/− and Lmnb1−/− MEFs from the Goldman lab (Department of Cell and Molecular Biology, Northwestern University Feinberg School of Medicine, Chicago, IL USA), and human HT1080 cells (ATCC) were grown in Dulbecco's modified Eagle medium (DMEM; Corning) complete with 1% penicillin-streptomycin (Thermo Fisher Scientific) and 10% fetal bovine serum (FBS; HyClone) at 37°C and 5% CO2. For cell culture, glass-bottomed four-well imaging dishes were prepared 48 h before imaging. Cells were passaged every other day into fresh DMEM complete. 100 µl of confluent wild-type and NLS–GFP-expressing [generated stable cell line (Currey et al., 2022)] MEFs were added into the corner of each well and 600 µl of complete DMEM was added carefully in order to flow over the cells and fill the well. Drug treatment was done 22–24 h prior to imaging. VPA (Sigma-Aldrich) was added to a final concentration of 2 mM (20 mM stock solution), α-amanitin (Tocris 4025) at 10 µM (1 mM stock solution), triptolide (Cayman Chemical 11973) at 1 µM and flavopiridol (Cayman Chemical 0580070-2) at 0.5 µM. Actinomycin D (Cayman Chemical 11421) was added at a concentration of 10 µg/ml with an incubation time of no more than 30 min prior to imaging.
Imaging
Images were acquired with Nikon Elements software on a Nikon Instruments Ti2-E microscope with Crest V3 Spinning Disk Confocal, Orca Fusion Gen III camera, Lumencor Aura III light engine, TMC CleanBench air table, a 40× air objective (NA 0.75, working distance 0.66 mm, MRH00401) or a Plan Apochromat Lambda 100× Oil Immersion Objective Lens (NA 1.45, working distance 0.13 mm, field of view 25 mm, MRD71970). Live-cell imaging was possible using a Nikon Perfect Focus System and an Okolab heat, humidity and CO2 stage-top incubator (H301). Images were captured via a 16-bit camera for population images or a 12-bit sensitive camera for timelapse imaging with a 40× air objective (NA 0.75; Nikon MRH00401). For timelapse data, images were taken in 2-min intervals during 3 h with nine fields of view for each condition.
Bleb count
MEF cells were treated with Hoechst 33342 at a dilution of 1:20,000 to 1:40,000 for 15 min before population imaging or imaged of NLS–GFP to analyze nuclear shape. Images were taken with nine fields of view for each condition, more than 100 nuclei were counted, and the percentage of cells showing blebbed nuclei calculated (>100) for each biological replicate (≥3). Nuclei were scored as blebbed if a protrusion 1 µm in diameter or larger was present, as previously outlined in Stephens et al. (2018).
Nuclear rupture analysis
NLS–GFP MEFs were used for nuclear rupture analysis. Image stacks were analyzed using the NIS Elements AR Analysis software (Nikon). For each condition, total nuclei were counted at the first and last frame of the timelapse and averaged. Blebbed nuclei were counted as all nuclei that displayed a nuclear bleb at any time during the timelapse. Nuclear ruptures were determined by a >25% change in the NLS–GFP intensity in the cytoplasm to that in the nucleus using 5×5 µm boxes in each and with the background subtracted. Total nuclei showing a nuclear rupture were counted while differentiating between bleb-based ruptures and non-blebbed ruptures. Bleb-based ruptures were defined as nuclei showing a bleb prior to rupturing, whereas non-blebbed ruptures did not show a bleb on the nucleus. Rupture frequency was calculated by counting and averaging the number of ruptures for each rupturing nucleus. Additionally, blebs formed during timelapse imaging were counted while differentiating between (1) blebs forming, rupturing and thereby stabilizing the bleb, and (2) blebs forming, but disappearing without showing a nuclear rupture, or blebs forming, rupturing but disappearing, and not being stabilized by the rupture. This analysis justifies counting the percentage of blebbed cells only at one or two timepoints as the dynamics of the formation of new blebs are captured. For each condition, three fields of views were analyzed for each replicate. Graphs were made to show the percentage of ruptures as the total number of ruptured nuclei by the total number of nuclei in each field of view.
Nuclear height analysis
Nuclei were imaged using a spinning-disk confocal microscope with a Plan Apochromat Lambda 100× Oil Immersion Objective Lens (NA 1.45, working distance 0.13 mm, field of view 25 mm). 77 images with an axial distance of 0.2 µm were taken as z-slices with a total distance of 15 µm covered. The center of one nucleus was selected at a time and a slice view was created. The fluorescence intensity profile for the z-slice was analyzed using a full-width half-maximum (FWHM) calculation giving the width of the fluorescence intensity peak corresponding to the height of the nucleus. Two measurements of nuclear height were averaged for each nucleus. Ten to 20 nuclei were measured for each condition.
Micromanipulation force measurements
As first described (Stephens et al., 2017) and more recently updated (Currey et al., 2022), vimentin null (V−/−) MEFs were grown in a micromanipulation well (see Currey et al., 2022) to provide low-angle access via micropipettes. V−/− MEF nuclei were isolated from living cells via spray micropipette of the mild detergent Triton X-100 (0.05%) in PBS. The pull micropipette was used to grab the nucleus. The isolated nucleus was then grabbed at the opposite end with a precalibrated force micropipette and suspended in preparation for force-extension measurements. The pull pipette was moved at 50 nm/s to provide a 3 or 6 µm extension to the nucleus. The pull micropipette is tracked to provide the nucleus extension (µm), whereas measurement of the deflection of the force micropipette multiplied by the bending modulus (1.2–2 nN/µm) provides the measure of force (nN). The slope of the force versus extension plot provides the spring constant (nN/µm) for the short chromatin-dominated regime (<3 µm) and long-extension lamin A-dominated strain-stiffening regime (>3 µm). The long-regime spring constant minus the short-regime spring constant provides the measure of lamin A-based strain stiffening.
Immunofluorescence
Cells were grown in eight-well dishes for 48 h prior to fixation. Treatment with drugs was done 24 h prior to fixation, which was done with a solution of 3.3% paraformaldehyde in 0.1% Triton X-100 in PBS for 15 min. Three washing steps were performed with PBS and the last one with PBS containing 0.06% Tween 20 (PBS-T). Primary antibodies were diluted in 10% goat serum in PBS (GPBS) and incubated with the cells for 1 h at 37°C before washing three times with PBS. Secondary antibodies were incubated in GPBS for 1 h at room temperature. The primary antibodies used were: anti-lamin A/C at 1:200 (Cell Signaling Technology, 4C11, mouse monoclonal antibody #4777), anti-H3K9ac at 1:500 (Cell Signaling Technology, C5B11, rabbit monoclonal antibody #9649), anti-H3K9me2,3 at 1:800 (Cell Signaling Technology 5327, mouse monoclonal antibody), anti-pMLC2 at 1:200 (Cell Signaling Technology 3671, rabbit monoclonal antibody), anti-RNA pol II CTD repeat YSPTSPS (phospho S5/pSer5) at 1:1000 (Abcam, 4H8 – ChIP Grade, ab5408, mouse monoclonal antibody) and anti-RNA pol II CTD repeat YSPTSPS (phospho S2/pSer2) at 1:1000 (Abcam, ab5095, rabbit polyclonal antibody). Secondary antibodies were used at a 1:1000 dilution and include Alexa Fluor 488-, 555- or 647-conjugated goat anti-mouse or goat anti-Rabbit IgG (H+L), F(ab′)2 fragment (Cell Signaling Technology, 4408–4414). The nuclei were stained using Hoechst 33342 in PBS at a 1:40,000 dilution for at least 5 min. Cells were kept and imaged in PBS or mounted using Prolong Gold anti-fade mountant (Invitrogen, P36930) and incubated overnight in the dark. Fluorescence intensities were analyzed by measuring single nuclei as regions of interest and subtracting backgrounds as 30×30 pixel areas with no cells. All single nucleus measurements were averaged over multiple fields of view to provide a single average intensity measurement for each experiment, for which there were at least three replicates. For comparison, all intensities were normalized with the mean value for untreated nuclei. For bleb versus body measurements, a region of interest was hand drawn around each and intensities were background subtracted as detailed above.
RNA labeling
RNA labeling was accomplished using Click-iT RNA Alexa Fluor 594 Imaging Kit (Invitrogen, C10330) as previously described (Jao and Salic, 2008). Cells were plated in eight-well plates (Cellvis, C8-1.5H-N), and grown and left untreated or treated with VPA and/or α-amanitin. EU was added at a final concentration of 1 mM to cells and incubated for 1 h. Cells were fixed and permeabilized as described above. After two PBS washes, the 500 µl formulation of the Click-iT reaction cocktail was added and allowed to incubate for 30 min in the dark. The reaction was terminated by removing the solution and washing the cells with the defined Click-iT reaction rinse buffer. The nuclei were stained using Hoechst 33342 in PBS at a 1:40,000 dilution for at least 5 min. The cells were rinsed two more times in PBS. Cells were kept and imaged in PBS or mounted using Prolong Gold anti-fade mountant and incubated overnight in the dark.
Simulation model
For the simulation of the nuclear lamina shell, we first generated 10,000 point particles (or ‘subunits’) in the Fibonacci sequence on a sphere of radius R=10 µm. Given that the nuclear lamina is not a regular lattice (Shimi et al., 2015; Mahamid et al., 2016; Turgay et al., 2017), we then randomized the positions of the particles on the shell with thermal noise. While randomizing subunit positions, we gradually and sequentially increased the size of subunits to σ0 such that no large forces were generated due to overlap. Subunits did not overlap due to a soft repulsive potential given by:
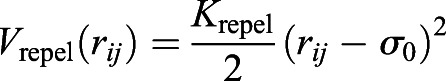 |
for rij≤σ0, where rij is the distance between particles i and j, and Krepel is the spring constant. Once the lamin subunits reached size σ0, we randomized again with the current size to ensure that no remnant of the original lattice survives in the simulated structure. In vivo, the nuclear lamin network has an average coordination number of approximately 4 to 5 (Shimi et al., 2015; Sapra et al., 2020). To model this, we developed an algorithm to connect the lamin subunits such that the average coordination number of the lamin network is around 4.5.
To model the chromatin chain, we first generated a random walk on a face-centered cubic lattice with no overlapping steps. We set a confining spherical boundary condition at radius R on the chain and subsequently decreased the chain size such that it was compacted into the sphere. The Rouse chain subunits are joined by harmonic springs, which also disfavors overlap by a soft repulsive potential:
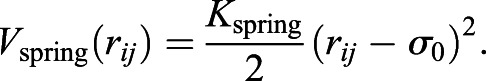 |
We then equilibrated the compacted chain inside the lamina shell. We modeled chromatin–chromatin crosslinks and chromatin-lamina linkages by randomly choosing nearby subunits on the chromatin chain and nuclear lamina and connecting them by harmonic springs. Purely repulsive (‘extensile’) motors were simulated as generating a monopolar (outward) repulsive force on chromatin subunits if they were within the interaction range (σm) of the motor. Chromatin motors did not exert any force on the nuclear lamin subunits.
Simulation methods
The time evolution of the system is governed by overdamped Langevin dynamics:
where ri is position of ith particle; μ = DT/kBT; 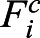 is the conservative force on ith particle, calculated as Fic = Fispring + Firepel;
is the conservative force on ith particle, calculated as Fic = Fispring + Firepel; 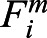 is the non-conservative force due to the repulsive motors that repel chromatin subunits radially outward; DT is the particle diffusion coefficient; and ξ(t) is delta-correlated random noise. Simulation parameters are given in Table 1. We integrated the equation of motion by the Euler–Murayama method.
is the non-conservative force due to the repulsive motors that repel chromatin subunits radially outward; DT is the particle diffusion coefficient; and ξ(t) is delta-correlated random noise. Simulation parameters are given in Table 1. We integrated the equation of motion by the Euler–Murayama method.
In our model, the chromatin chain had 5000 subunits and the nuclear lamina (shell) had 10,000 subunits. There were 400 chromatin–shell linkages and 2500 crosslinkers. Each chromatin or lamin subunit had a diameter of σ=0.43089 µm. The spring constant (both Kspring and Krepel) for the chain was 1.4×10−4 nN/µm and the spring constant for the shell was 2.8×10−4 nN/µm. We evolved the simulation for 107 timesteps, which we took to be 500 s. We ran ten realizations for each NM shown in Fig. 7C, identified and counted bulges for each realization (see below), and averaged over realizations to compute the mean bulge number. Motors stochastically switched off of one subunit and onto another, randomly selected chromatin subunit, with a mean turnover time of 10 s, as in previous simulations (Liu et al., 2021).
Identification of bulges in simulations
To identify bulges and valleys, we first calculated the average shell radius and computed the height of each shell subunit above or below this radius. Subunits with height greater than 1 µm were considered to be part of a bulge.
To identify and count bulges, we projected the shell, using spherical polar coordinates, onto a two-dimensional map of the polar and azimuthal angles of each subunit (θ and φ, respectively). Bulges were first identified by local maxima in subunit heights. To determine the area that a bulge occupied (and thus identify unique bulges), we first centered the map on the local maximum, and then considered heights of subunits within a circle around that point. Subunits with height greater than 1 µm were counted as part of the bulge. If there were subunits within the circle that were part of the bulge, we expanded the circle radius to look for additional subunits in the bulge. After expanding the circle, if no additional nearby subunits had a height greater than 1 µm, we defined the bulge as containing only the subunits that had already been counted. Bulges were defined such that each bulge contained a unique set of subunits (i.e. overlapping bulges were merged to be considered a single bulge).
Supplementary Material
Acknowledgements
We would like to thank Karen Reddy and Bob Goldman labs for MEF lines including Lmnb1−/− and LA KD. We would also like to thank Paul Janmey and Amir Vahabikashi for helpful discussions and Kuang Liu for helpful discussions and advice on implementing the simulation model.
Footnotes
Author contributions
Conceptualization: S.G., A.D.S.; Methodology: I.K.B., M.L.C., S.G., B.V.N., J.M.S., E.J.B., A.D.S.; Formal analysis: I.K.B., M.L.C., S.G., Y.B., B.V.N., M.P.; Investigation: I.K.B., M.L.C., S.G., B.V.N., M.P.; Data curation: I.K.B., M.L.C., S.G., Y.B., B.V.N., M.P.; Writing - original draft: S.G., E.J.B., A.D.S.; Writing - review & editing: A.E.P., J.M.S., S.G., E.J.B., A.D.S.; Supervision: J.M.S., E.J.B., A.D.S., A.E.P.; Project administration: A.D.S.; Funding acquisition: A.E.P., J.M.S., A.D.S.
Funding
I.K.B., M.L.C., Y.B., M.P. and A.D.S. are supported by the National Institutes of Health Pathway to Independence Award (R00GM123195). A.E.P. is supported by the National Institutes of Health (R35GM142963), J.M.S. is supported by the National Science Foundation Division of Materials Research (1832002 and NSF-DMR-2204312), and S.G. is funded by a Syracuse University dissertation fellowship. I.K.B., M.L.C., E.J.B. and A.D.S. are supported by Center for 3D Structure and Physics of the Genome 4DN2 grant (1UM1HG011536) funded by the National Institutes of Health. Deposited in PMC for release after 12 months.
Data availability
All relevant data can be found within the article and its supplementary information.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/lookup/doi/10.1242/jcs.261547.reviewer-comments.pdf
References
- Banigan, E. J., Stephens, A. D. and Marko, J. F. (2017). Mechanics and buckling of biopolymeric shells and cell nuclei. Biophys. J. 113, 1654-1663. 10.1016/j.bpj.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banigan, E. J., Tang, W., van den Berg, A. A., Stocsits, R. R., Wutz, G., Brandão, H. B., Busslinger, G. A., Peters, J.-M. and Mirny, L. A. (2023). Transcription shapes 3D chromatin organization by interacting with loop extrusion. Proc. Natl. Acad. Sci. USA 120, e2210480120. 10.1073/pnas.2210480120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, R., Bystricky, K. and Shaban, H. A. (2020). Coupling chromatin structure and dynamics by live super-resolution imaging. Sci. Adv. 6, eaaz2196. 10.1126/sciadv.aaz2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaghzal, H., Borrman, T., Stephens, A. D., Lafontaine, D. L., Venev, S. V., Weng, Z., Marko, J. F. and Dekker, J. (2021). Liquid chromatin Hi-C characterizes compartment-dependent chromatin interaction dynamics. Nat. Genet. 53, 367-378. 10.1038/s41588-021-00784-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude, O. (2011). Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2, 103-108. 10.4161/trns.2.3.16172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercht Pfleghaar, K., Taimen, P., Butin-Israeli, V., Shimi, T., Langer-Freitag, S., Markaki, Y., Goldman, A. E., Wehnert, M. and Goldman, R. D. (2015). Gene-rich chromosomal regions are preferentially localized in the lamin B deficient nuclear blebs of atypical progeria cells. Nucleus 6, 66-76. 10.1080/19491034.2015.1004256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner, L., Zhao, P. A., van Schaik, T., Leemans, C., Sima, J., Peric-Hupkes, D., Gilbert, D. M. and van Steensel, B. (2020). Local rewiring of genome-nuclear lamina interactions by transcription. EMBO J. 39, e103159. 10.15252/embj.2019103159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. and Greenleaf, W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213-1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalut, K. J., Höpfler, M., Lautenschläger, F., Boyde, L., Chan, C. J., Ekpenyong, A., Martinez-Arias, A. and Guck, J. (2012). Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophys. J. 103, 2060-2070. 10.1016/j.bpj.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N. Y., Kim, P., Weston, T. A., Edillo, L., Tu, Y., Fong, L. G. and Young, S. G. (2018). Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc. Natl. Acad. Sci. USA 115, 10100-10105. 10.1073/pnas.1812622115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin, T., McNairn, A. J., Jenuwein, T., Gilbert, D. M., Singh, P. B. and Misteli, T. (2003). Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299, 721-725. 10.1126/science.1078572 [DOI] [PubMed] [Google Scholar]
- Chi, Y.-H., Wang, W.-P., Hung, M.-C., Liou, G.-G., Wang, J.-Y. and Chao, P.-H. G. (2022). Deformation of the nucleus by TGFβ1 via the remodeling of nuclear envelope and histone isoforms. Epigenet. Chromatin 15, 1. 10.1186/s13072-021-00434-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, F.-Y., Haley, S. C. and Zidovska, A. (2017). On the origin of shape fluctuations of the cell nucleus. Proc. Natl. Acad. Sci. USA 114, 10338-10343. 10.1073/pnas.1702226114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currey, M. L., Kandula, V., Biggs, R., Marko, J. F. and Stephens, A. D. (2022). A versatile micromanipulation apparatus for biophysical assays of the cell nucleus. Cell. Mol. Bioeng. 15, 303-312. 10.1007/s12195-022-00734-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq, X., Shav-Tal, Y., de Turris, V., Brody, Y., Shenoy, S. M., Phair, R. D. and Singer, R. H. (2007). In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14, 796-806. 10.1038/nsmb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos, W. H., Houben, F., Kamps, M., Malhas, A., Verheyen, F., Cox, J., Manders, E. M. M., Verstraeten, V. L. R. M., van Steensel, M. A. M., Marcelis, C. L. M.et al. (2011). Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet. 20, 4175-4186. 10.1093/hmg/ddr344 [DOI] [PubMed] [Google Scholar]
- Denais, C. M., Gilbert, R. M., Isermann, P., McGregor, A. L., te Lindert, M., Weigelin, B., Davidson, P. M., Friedl, P., Wolf, K. and Lammerding, J. (2016). Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353-358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deviri, D., Discher, D. E. and Safran, S. A. (2017). Rupture dynamics and chromatin herniation in deformed nuclei. Biophys. J. 113, 1060-1071. 10.1016/j.bpj.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle, A. J., Kirby, T. J., Fedorchak, G. R., Isermann, P., Patel, J., Iruvanti, S., Moore, S. A., Bonne, G., Wallrath, L. L. and Lammerding, J. (2020). Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat. Mater. 19, 464-473. 10.1038/s41563-019-0563-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festenstein, R., Pagakis, S. N., Hiragami, K., Lyon, D., Verreault, A., Sekkali, B. and Kioussis, D. (2003). Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science 299, 719-721. 10.1126/science.1078694 [DOI] [PubMed] [Google Scholar]
- Funkhouser, C. M., Sknepnek, R., Shimi, T., Goldman, A. E., Goldman, R. D. and Olvera de la Cruz, M. (2013). Mechanical model of blebbing in nuclear lamin meshworks. Proc. Natl. Acad. Sci. USA 110, 3248-3253. 10.1073/pnas.1300215110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa, T., Rochman, M., Taher, L., Dimitriadis, E. K., Nagashima, K., Anderson, S. and Bustin, M. (2015). Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun. 6, 6138. 10.1038/ncomms7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, E. M. and Hetzer, M. W. (2016). Nuclear envelope rupture is induced by actin-based nucleus confinement. J. Cell Biol. 215, 27-36. 10.1083/jcb.201603053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand, B. T., Wang, Y., Pfleghaar, K., Shimi, T., Taimen, P. and Shumaker, D. K. (2012). Chromosomal regions associated with prostate cancer risk localize to lamin B-deficient microdomains and exhibit reduced gene transcription. J. Pathol. 226, 735-745. 10.1002/path.3033 [DOI] [PubMed] [Google Scholar]
- Herbert, K. M., Greenleaf, W. J. and Block, S. M. (2008). Single-molecule studies of RNA polymerase: motoring along. Annu. Rev. Biochem. 77, 149-176. 10.1146/annurev.biochem.77.073106.100741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. and Sharp, P. A. (2017). A phase separation model for transcriptional control. Cell 169, 13-23. 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson, C. M., Kern, M., O'Brien, E. T., III, Stephens, A. D., Falvo, M. R. and Superfine, R. (2020). Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Mol. Biol. Cell 31, 1788-1801. 10.1091/mbc.E20-01-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, T.-H. S., Cattoglio, C., Slobodyanyuk, E., Hansen, A. S., Rando, O. J., Tjian, R. and Darzacq, X. (2020). Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell 78, 539-553.e8. 10.1016/j.molcel.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue-Mochita, M., Inoue, T., Fujimoto, T., Kameda, T., Awai-Kasaoka, N., Ohtsu, N., Kimoto, K. and Tanihara, H. (2015). p38 MAP kinase inhibitor suppresses transforming growth factor-β2-induced type 1 collagen production in trabecular meshwork cells. PLoS ONE 10, e0120774. 10.1371/journal.pone.0120774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto, J., Pfeifer, C. R., Bennett, R. R., Xia, Y., Ivanovska, I. L., Liu, A. J., Greenberg, R. A. and Discher, D. E. (2016). Nuclear constriction segregates mobile nuclear proteins away from chromatin. Mol. Biol. Cell 27, 4011-4020. 10.1091/mbc.E16-06-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao, C. Y. and Salic, A. (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA 105, 15779-15784. 10.1073/pnas.0808480105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., Huang, J., Lun, K., Li, B., Zheng, H., Li, Y., Zhou, R., Duan, W., Wang, C., Feng, Y.et al. (2020). Genome-wide analyses of chromatin interactions after the loss of Pol I, Pol II, and Pol III. Genome Biol. 21, 158. 10.1186/s13059-020-02067-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinin, A. A., Hou, X., Ade, A. S., Fon, G.-V., Meixner, W., Higgins, G. A., Sexton, J. Z., Wan, X., Dinov, I. D., O'Meara, M. J.et al. (2021). Valproic acid-induced changes of 4D nuclear morphology in astrocyte cells. Mol. Biol. Cell 32, 1624-1633. 10.1091/mbc.E20-08-0502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalukula, Y., Stephens, A. D., Lammerding, J. and Gabriele, S. (2022). Mechanics and functional consequences of nuclear deformations. Nat. Rev. Mol. Cell Biol. 23, 583-602. 10.1038/s41580-022-00480-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, Z. S., Santos, J. M. and Hussain, F. (2018). Aggressive prostate cancer cell nuclei have reduced stiffness. Biomicrofluidics 12, 014102. 10.1063/1.5019728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau, S. B., Hale, C. M., Stewart-Hutchinson, P. J., Patel, M. S., Stewart, C. L., Searson, P. C., Hodzic, D. and Wirtz, D. (2009). A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA 106, 19017-19022. 10.1073/pnas.0908686106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind, J., Pagie, L., Ortabozkoyun, H., Boyle, S., de Vries, S. S., Janssen, H., Amendola, M., Nolen, L. D., Bickmore, W. A. and van Steensel, B. (2013). Single-cell dynamics of genome-nuclear lamina interactions. Cell 153, 178-192. 10.1016/j.cell.2013.02.028 [DOI] [PubMed] [Google Scholar]
- Krause, M., te Riet, J. and Wolf, K. (2013). Probing the compressibility of tumor cell nuclei by combined atomic force-confocal microscopy. Phys. Biol. 10, 065002. 10.1088/1478-3975/10/6/065002 [DOI] [PubMed] [Google Scholar]
- Lammerding, J., Fong, L. G., Ji, J. Y., Reue, K., Stewart, C. L., Young, S. G. and Lee, R. T. (2006). Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 281, 25768-25780. 10.1074/jbc.M513511200 [DOI] [PubMed] [Google Scholar]
- Le Berre, M., Aubertin, J. and Piel, M. (2012). Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr. Biol. 4, 1406-1414. 10.1039/c2ib20056b [DOI] [PubMed] [Google Scholar]
- Lionetti, M. C., Bonfanti, S., Fumagalli, M. R., Budrikis, Z., Font-Clos, F., Costantini, G., Chepizhko, O., Zapperi, S. and La Porta, C. A. M. (2020). Chromatin and cytoskeletal tethering determine nuclear morphology in progerin-expressing cells. Biophys. J. 118, 2319-2332. 10.1016/j.bpj.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K., Patteson, A. E., Banigan, E. J. and Schwarz, J. M. (2021). Dynamic nuclear structure emerges from chromatin cross-links and motors. Phys. Rev. Lett. 126, 158101. 10.1103/PhysRevLett.126.158101 [DOI] [PubMed] [Google Scholar]
- Lu, C., Romo-Bucheli, D., Wang, X., Janowczyk, A., Ganesan, S., Gilmore, H., Rimm, D. and Madabhushi, A. (2018). Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab. Invest. 98, 1438-1448. 10.1038/s41374-018-0095-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamid, J., Pfeffer, S., Schaffer, M., Villa, E., Danev, R., Cuellar, L. K., Förster, F., Hyman, A. A., Plitzko, J. M. and Baumeister, W. (2016). Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969-972. 10.1126/science.aad8857 [DOI] [PubMed] [Google Scholar]
- Mistriotis, P., Wisniewski, E. O., Bera, K., Keys, J., Li, Y., Tuntithavornwat, S., Law, R. A., Perez-Gonzalez, N. A., Erdogmus, E., Zhang, Y.et al. (2019). Confinement hinders motility by inducing RhoA-mediated nuclear influx, volume expansion, and blebbing. J. Cell Biol. 218, 4093-4111. 10.1083/jcb.201902057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima, R., Hibino, K., Ashwin, S. S., Babokhov, M., Fujishiro, S., Imai, R., Nozaki, T., Tamura, S., Tani, T., Kimura, H.et al. (2019). Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II. J. Cell Biol. 218, 1511-1530. 10.1083/jcb.201811090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava, M. M., Miroshnikova, Y. A., Biggs, L. C., Whitefield, D. B., Metge, F., Boucas, J., Vihinen, H., Jokitalo, E., Li, X., García Arcos, J. M.et al. (2020). Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell 181, 800-817.e22. 10.1016/j.cell.2020.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nmezi, B., Xu, J., Fu, R., Armiger, T. J., Rodriguez-Bey, G., Powell, J. S., Ma, H., Sullivan, M., Tu, Y., Chen, N. Y.et al. (2019). Concentric organization of A- and B-type lamins predicts their distinct roles in the spatial organization and stability of the nuclear lamina. Proc. Natl. Acad. Sci. USA 116, 4307-4315. 10.1073/pnas.1810070116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, H. D., Phan, S., Deerinck, T. J., Thor, A., Ellisman, M. H. and O'Shea, C. C. (2017). ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, eaag0025. 10.1126/science.aag0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou, G. N. and Traut, H. F. (1997). The diagnostic value of vaginal smears in carcinoma of the uterus. 1941. Arch. Pathol. Lab. Med. 121, 211-224. [PubMed] [Google Scholar]
- Patteson, A. E., Vahabikashi, A., Pogoda, K., Adam, S. A., Mandal, K., Kittisopikul, M., Sivagurunathan, S., Goldman, A., Goldman, R. D. and Janmey, P. A. (2019). Vimentin protects cells against nuclear rupture and DNA damage during migration. J. Cell Biol. 218, 4079-4092. 10.1083/jcb.201902046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulose, J. and Nelson, D. R. (2013). Buckling pathways in spherical shells with soft spots. Soft Mat. 9, 8227. 10.1039/c3sm50719j [DOI] [Google Scholar]
- Penfield, L., Wysolmerski, B., Mauro, M., Farhadifar, R., Martinez, M. A., Biggs, R., Wu, H.-Y., Broberg, C., Needleman, D. and Bahmanyar, S. (2018). Dynein pulling forces counteract lamin-mediated nuclear stability during nuclear envelope repair. Mol. Biol. Cell 29, 852-868. 10.1091/mbc.E17-06-0374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer, C. R., Xia, Y., Zhu, K., Liu, D., Irianto, J., García, V. M. M., Millán, L. M. S., Niese, B., Harding, S., Deviri, D.et al. (2018). Constricted migration increases DNA damage and independently represses cell cycle. Mol. Biol. Cell 29, 1948-1962. 10.1091/mbc.E18-02-0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer, C. R., Tobin, M. P., Cho, S., Vashisth, M., Dooling, L. J., Vazquez, L. L., Ricci-De Lucca, E. G., Simon, K. T. and Discher, D. E. (2022). Gaussian curvature dilutes the nuclear lamina, favoring nuclear rupture, especially at high strain rate. Nucleus 13, 129-143. 10.1080/19491034.2022.2045726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pho, M., Berrada, Y., Gunda, A., Lavallee, A., Chiu, K., Padam, A., Currey, M. L. and Stephens, A. D. (2022). Actin contraction controls nuclear blebbing and rupture independent of actin confinement. bioRxiv 2022.12.01.518663. 10.1101/2022.12.01.518663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab, M., Gentili, M., de Belly, H., Thiam, H.-R., Vargas, P., Jimenez, A. J., Lautenschlaeger, F., Voituriez, R., Lennon-Duménil, A.-M., Manel, N.et al. (2016). ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359-362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- Radhakrishnan, A., Damodaran, K., Soylemezoglu, A. C., Uhler, C. and Shivashankar, G. V. (2017). Machine learning for nuclear mechano-morphometric biomarkers in cancer diagnosis. Sci. Rep. 7, 17946. 10.1038/s41598-017-17858-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robijns, J., Molenberghs, F., Sieprath, T., Corne, T. D. J., Verschuuren, M. and De Vos, W. H. (2016). In silico synchronization reveals regulators of nuclear ruptures in lamin A/C deficient model cells. Sci. Rep. 6, 30325. 10.1038/srep30325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintillan, D., Shelley, M. J. and Zidovska, A. (2018). Extensile motor activity drives coherent motions in a model of interphase chromatin. Proc. Natl. Acad. Sci. USA 115, 11442-11447. 10.1073/pnas.1807073115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra, K. T., Qin, Z., Dubrovsky-Gaupp, A., Aebi, U., Müller, D. J., Buehler, M. J. and Medalia, O. (2020). Nonlinear mechanics of lamin filaments and the meshwork topology build an emergent nuclear lamina. Nat. Commun. 11, 6205. 10.1038/s41467-020-20049-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner, S. M., Koo, P. K., Zhao, Y., Mochrie, S. G. J. and King, M. C. (2015). The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun. 6, 7159. 10.1038/ncomms8159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senigagliesi, B., Penzo, C., Severino, L. U., Maraspini, R., Petrosino, S., Morales-Navarrete, H., Pobega, E., Ambrosetti, E., Parisse, P., Pegoraro, S.et al. (2019). The High Mobility Group A1 (HMGA1) chromatin architectural factor modulates nuclear stiffness in breast cancer cells. Int. J. Mol. Sci. 20, 2733. 10.3390/ijms20112733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban, H. A., Barth, R. and Bystricky, K. (2018). Formation of correlated chromatin domains at nanoscale dynamic resolution during transcription. Nucleic Acids Res. 46, e77. 10.1093/nar/gky269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban, H. A., Barth, R., Recoules, L. and Bystricky, K. (2020). Hi-D: nanoscale mapping of nuclear dynamics in single living cells. Genome Biol. 21, 95. 10.1186/s13059-020-02002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, P., Hobson, C. M., Cheng, S., Colville, M. J., Paszek, M. J., Superfine, R. and Lammerding, J. (2021). Nuclear deformation causes DNA damage by increasing replication stress. Curr. Biol. 31, 753-765.e6. 10.1016/j.cub.2020.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto, Y., Tamura, S., Masumoto, H. and Maeshima, K. (2017). Nucleosome-nucleosome interactions via histone tails and linker DNA regulate nuclear rigidity. Mol. Biol. Cell 28, 1580-1589. 10.1091/mbc.e16-11-0783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi, T., Pfleghaar, K., Kojima, S., Pack, C.-G., Solovei, I., Goldman, A. E., Adam, S. A., Shumaker, D. K., Kinjo, M., Cremer, T.et al. (2008). The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 22, 3409-3421. 10.1101/gad.1735208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi, T., Kittisopikul, M., Tran, J., Goldman, A. E., Adam, S. A., Zheng, Y., Jaqaman, K. and Goldman, R. D. (2015). Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell 26, 4075-4086. 10.1091/mbc.E15-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, N., Nader, G. P. d. F., Williart, A., Rollin, R., Cuvelier, D., Lomakin, A. and Piel, M. (2021). Nuclear fragility, blaming the blebs. Curr. Opin. Cell Biol. 70, 100-108. 10.1016/j.ceb.2021.01.007 [DOI] [PubMed] [Google Scholar]
- Stephens, A. D. (2020). Chromatin rigidity provides mechanical and genome protection. Mutat. Res. 821, 111712. 10.1016/j.mrfmmm.2020.111712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, A. D., Banigan, E. J., Adam, S. A., Goldman, R. D. and Marko, J. F. (2017). Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell 28, 1984-1996. 10.1091/mbc.e16-09-0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, A. D., Liu, P. Z., Banigan, E. J., Almassalha, L. M., Backman, V., Adam, S. A., Goldman, R. D. and Marko, J. F. (2018). Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 29, 220-233. 10.1091/mbc.E17-06-0410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, A. D., Liu, P. Z., Kandula, V., Chen, H., Almassalha, L. M., Herman, C., Backman, V., O'Halloran, T., Adam, S. A., Goldman, R. D.et al. (2019a). Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell 30, 2320-2330. 10.1091/mbc.E19-05-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, A. D., Banigan, E. J. and Marko, J. F. (2019b). Chromatin's physical properties shape the nucleus and its functions. Curr. Opin. Cell Biol. 58, 76-84. 10.1016/j.ceb.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden, H., Tolsma, T. O., Sharma, A., Underhill, D. A., Hansen, J. C. and Hendzel, M. J. (2020). Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell 183, 1772-1784.e13. 10.1016/j.cell.2020.11.027 [DOI] [PubMed] [Google Scholar]
- Strom, A. R., Biggs, R. J., Banigan, E. J., Wang, X., Chiu, K., Herman, C., Collado, J., Yue, F., Politz, J. C. R., Tait, L. J.et al. (2021). HP1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. eLife 10, e63972. 10.7554/eLife.63972.sa2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay, Y., Eibauer, M., Goldman, A. E., Shimi, T., Khayat, M., Ben-Harush, K., Dubrovsky-Gaupp, A., Sapra, K. T., Goldman, R. D. and Medalia, O. (2017). The molecular architecture of lamins in somatic cells. Nature 543, 261-264. 10.1038/nature21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahabikashi, A., Sivagurunathan, S., Nicdao, F. A. S., Han, Y. L., Park, C. Y., Kittisopikul, M., Wong, X., Tran, J. R., Gundersen, G. G., Reddy, K. L.et al. (2022). Nuclear lamin isoforms differentially contribute to LINC complex-dependent nucleocytoskeletal coupling and whole-cell mechanics. Proc. Natl. Acad. Sci. USA 119, e2121816119. 10.1073/pnas.2121816119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, J. D., Hatch, E. M., Anderson, D. J. and Hetzer, M. W. (2012). Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus 3, 88-100. 10.4161/nucl.18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Dreger, M., Madrazo, E., Williams, C. J., Samaniego, R., Hodson, N. W., Monroy, F., Baena, E., Sánchez-Mateos, P., Hurlstone, A.et al. (2018). WDR5 modulates cell motility and morphology and controls nuclear changes induced by a 3D environment. Proc. Natl. Acad. Sci. USA 115, 8581-8586. 10.1073/pnas.1719405115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. F., Surovtsev, I. V., Schreiner, S. M., Nguyen, H., Hu, Y., Mochrie, S. G. J. and King, M. C. (2020). Phase separation enables heterochromatin domains to do mechanical work. bioRxiv 2020.07.02.184127. 10.1101/2020.07.02.184127 [DOI] [Google Scholar]
- Wren, N. S., Zhong, Z., Schwartz, R. S. and Dahl, K. N. (2012). Modeling nuclear blebs in a nucleoskeleton of independent filament networks. Cell. Mol. Bioeng. 5, 73-81. 10.1007/s12195-011-0196-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y., Ivanovska, I. L., Zhu, K., Smith, L., Irianto, J., Pfeifer, C. R., Alvey, C. M., Ji, J., Liu, D., Cho, S.et al. (2018). Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J. Cell Biol. 217, 3796-3808. 10.1083/jcb.201711161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong, E. H., Nelson, D. R. and Mahadevan, L. (2013). Elastic platonic shells. Phys. Rev. Lett. 111, 177801. 10.1103/PhysRevLett.111.177801 [DOI] [PubMed] [Google Scholar]
- Young, A. M., Gunn, A. L. and Hatch, E. M. (2020). BAF facilitates interphase nuclear membrane repair through recruitment of nuclear transmembrane proteins. Mol. Biol. Cell 31, 1551-1560. 10.1091/mbc.E20-01-0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Tamashunas, A. C., Agrawal, A., Torbati, M., Katiyar, A., Dickinson, R. B., Lammerding, J. and Lele, T. P. (2019). Local, transient tensile stress on the nuclear membrane causes membrane rupture. Mol. Biol. Cell 30, 899-906. 10.1091/mbc.E18-09-0604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Übelmesser, N., Josipovic, N., Forte, G., Slotman, J. A., Chiang, M., Gothe, H. J., Gusmao, E. G., Becker, C., Altmüller, J.et al. (2021). RNA polymerase II is required for spatial chromatin reorganization following exit from mitosis. Sci. Adv. 7, eabg8205. 10.1126/sciadv.abg8205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Übelmesser, N., Barbieri, M. and Papantonis, A. (2023). Enhancer-promoter contact formation requires RNAPII and antagonizes loop extrusion. Nat. Genet. 55, 832-840. 10.1038/s41588-023-01364-4 [DOI] [PubMed] [Google Scholar]
- Zidovska, A., Weitz, D. A. and Mitchison, T. J. (2013). Micron-scale coherence in interphase chromatin dynamics. Proc. Natl. Acad. Sci. USA 110, 15555-15560. 10.1073/pnas.1220313110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.