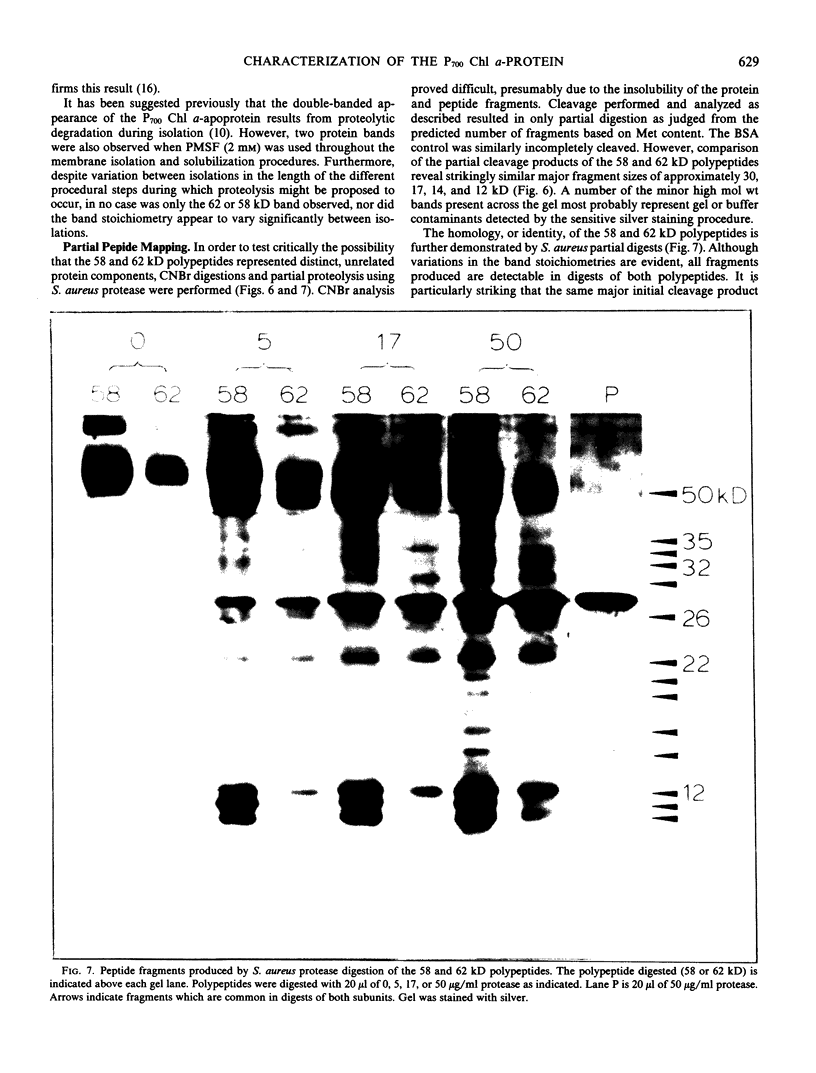
Abstract
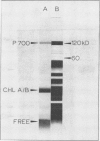
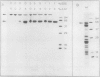
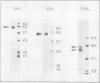
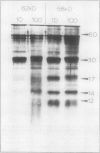
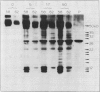
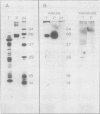
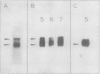
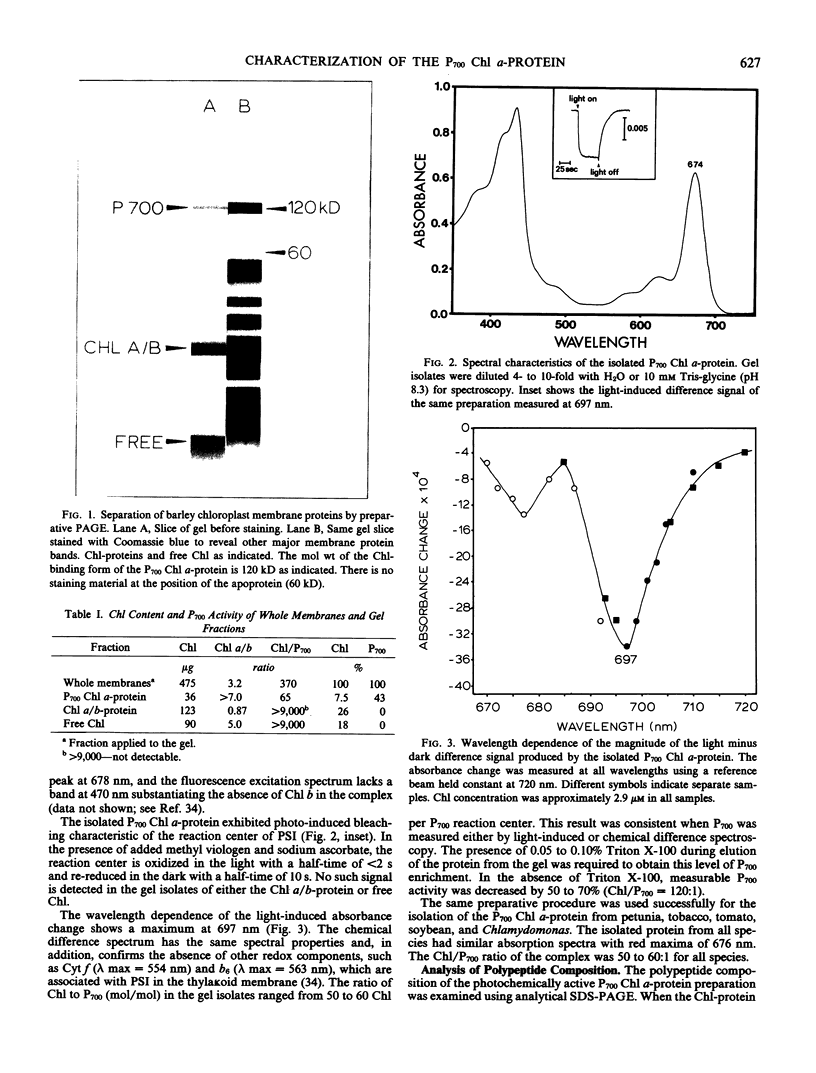
The P700 chlorophyll α-protein was purified by preparative sodium dodecyl sulfate (SDS) gel electrophoresis from SDS-solubilized barley (Hordeum vulgare L., cv Himalaya) chloroplast membranes. After elution from the gel in the presence of 0.05 to 0.1% Triton X-100, the recovered protein had a chlorophyll/P700 ratio of 50 to 60/1 and contained no chlorophyll b or cytochromes. Analysis of the polypeptide composition of the chlorophyll-protein revealed a 58 to 62 kilodalton (kD) polypeptide component but no lower molecular weight polypeptides. The 58 to 62 kD component was further resolved into two distinct polypeptide bands which were subsequently mapped by partial cyanogen bromide digestion and Staphylococcus aureus proteolysis. Based on results from the mapping experiments and other data, we suggest that the two components are conformational variants of a single polypeptide. Measurement of the chlorophyll to protein ratio by quantitative amino acid analysis and consideration of the yield of P700 in the protein isolate suggest that, contrary to previous models (Bengis and Nelson, 1975, 1977), P700in vivo is associated with a minimum of four subunits of approximately 60 kD.
Antibodies raised against the photochemically active chlorophyll-protein complex from barley reacted specifically with the 58 to 62 kD apoprotein. The same preparative electrophoresis procedure was used to isolate photochemically active P700 chlorophyll a-protein from soybean (Glycine max L.), tobacco (Nicotiana tobacum L.), petunia (Petunia × hybrida), tomato (Lycopersicum esculentum), and Chlamydomonas reinhardti. The isolated complex from all species exhibited identical polypeptide compositions and chlorophyll/P700 ratios. Antibodies to the barley protein cross reacted with all species tested demonstrating the highly conserved structure of the apoprotein.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. M., Levine R. P. The relationship between chlorophyll-protein complexes and chloroplast membrane polypeptides. Biochim Biophys Acta. 1974 Jul 25;357(1):118–126. doi: 10.1016/0005-2728(74)90117-0. [DOI] [PubMed] [Google Scholar]
- Anderson J. M. P-700 content and polypeptide profile of chlorophyll-protein complexes of spinach and barley thylakoids. Biochim Biophys Acta. 1980 Jun 10;591(1):113–126. doi: 10.1016/0005-2728(80)90225-x. [DOI] [PubMed] [Google Scholar]
- Arnon D. I. COPPER ENZYMES IN ISOLATED CHLOROPLASTS. POLYPHENOLOXIDASE IN BETA VULGARIS. Plant Physiol. 1949 Jan;24(1):1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengis C., Nelson N. Purification and properties of the photosystem I reaction center from chloroplasts. J Biol Chem. 1975 Apr 25;250(8):2783–2788. [PubMed] [Google Scholar]
- Bengis C., Nelson N. Subunit structure of chloroplast photosystem I reaction center. J Biol Chem. 1977 Jul 10;252(13):4564–4569. [PubMed] [Google Scholar]
- Broglie R., Bellemare G., Bartlett S. G., Chua N. H., Cashmore A. R. Cloned DNA sequences complementary to mRNAs encoding precursors to the small subunit of ribulose-1,5-bisphosphate carboxylase and a chlorophyll a/b binding polypeptide. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7304–7308. doi: 10.1073/pnas.78.12.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. Y. Amino-terminal analysis of polypeptide using dimethylaminoazobenzene isothiocyanate. Anal Biochem. 1980 Mar 1;102(2):384–392. doi: 10.1016/0003-2697(80)90172-4. [DOI] [PubMed] [Google Scholar]
- Chua N. H., Matlin K., Bennoun P. A chlorophyll-protein complex lacking in photosystem I mutants of Chlamydomonas reinhardtii. J Cell Biol. 1975 Nov;67(2PT1):361–377. doi: 10.1083/jcb.67.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Dietrich W. E., Jr, Thornber J. P. The P700-chlorophyll -protein of a blue-green alga. Biochim Biophys Acta. 1971 Sep 7;245(2):482–493. doi: 10.1016/0005-2728(71)90164-2. [DOI] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- Jockusch B. M., Kelley K. H., Meyer R. K., Burger M. M. An efficient method to produce specific anti-actin. Histochemistry. 1978 Apr 4;55(3):177–184. doi: 10.1007/BF00495757. [DOI] [PubMed] [Google Scholar]
- Keegstra K., Cline K. Evidence that Envelope and Thylakoid Membranes from Pea Chloroplasts Lack Glycoproteins. Plant Physiol. 1982 Jul;70(1):232–237. doi: 10.1104/pp.70.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Machold O., Aurich O. Sites of synthesis of chloroplast lamellar proteins in Vicia faba. Biochim Biophys Acta. 1972 Sep 29;281(1):103–112. doi: 10.1016/0005-2787(72)90192-x. [DOI] [PubMed] [Google Scholar]
- Mullet J. E., Burke J. J., Arntzen C. J. Chlorophyll proteins of photosystem I. Plant Physiol. 1980 May;65(5):814–822. doi: 10.1104/pp.65.5.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechushtai R., Nelson N. Purification properties and biogenesis of Chlamydomonas reinhardii photosystem I reaction center. J Biol Chem. 1981 Nov 25;256(22):11624–11628. [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Obata F., Shibata K. Two pigment proteins in spinach chloroplasts. Biochim Biophys Acta. 1966 Feb 7;112(2):223–234. doi: 10.1016/0926-6585(66)90323-2. [DOI] [PubMed] [Google Scholar]
- Remy R., Hoarau J., Leclerc J. C. Electrophoretic and spectrophotometric studies of chlorophyll-protein complexes from tobacco chloroplasts. Isolation of a light harvesting pigment protein complex with a molecular weight of 70,000. Photochem Photobiol. 1977 Aug;26(2):151–158. doi: 10.1111/j.1751-1097.1977.tb07466.x. [DOI] [PubMed] [Google Scholar]
- Shiozawa J. A., Alberte R. S., Thornber J. P. The P700-chlorophyll a-protein. Isolation and some characteristics of the complex in higher plants. Arch Biochem Biophys. 1974 Nov;165(1):388–397. doi: 10.1016/0003-9861(74)90177-5. [DOI] [PubMed] [Google Scholar]
- Thornber J. P. Comparison of a chlorophyll a- protein complex isolated from a blue-green alga with chlorophyll-protein complexes obtained from green bacteria and higher plants. Biochim Biophys Acta. 1969 Feb 25;172(2):230–241. doi: 10.1016/0005-2728(69)90066-8. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]