Abstract
Autism spectrum disorder (ASD) is a complex neurodevelopmental disorder with increasing prevalence. Over 1,000 risk genes have now been implicated in ASD, suggesting diverse etiology. However, the diagnostic criteria for the disorder still comprise two major behavioral domains - deficits in social communication and interaction, and the presence of restricted and repetitive patterns of behavior (RRBs). The RRBs associated with ASD include both stereotyped repetitive movements and other motor manifestations including changes in gait, balance, coordination, and motor skill learning. In recent years, the striatum, the primary input center of the basal ganglia, has been implicated in these ASD-associated motor behaviors, due to the striatum’s role in action selection, motor learning, and habit formation. Numerous mouse models with mutations in ASD risk genes have been developed and shown to have alterations in ASD-relevant behaviors. One commonly used assay, the accelerating rotarod, allows for assessment of both basic motor coordination and motor skill learning. In this corticostriatal-dependent task, mice walk on a rotating rod that gradually increases in speed. In the extended version of this task, mice engage striatal-dependent learning mechanisms to optimize their motor routine and stay on the rod for longer periods. This review summarizes the findings of studies examining rotarod performance across a range of ASD mouse models, and the resulting implications for the involvement of striatal circuits in ASD-related motor behaviors. While performance in this task is not uniform across mouse models, there is a cohort of models that show increased rotarod performance. A growing number of studies suggest that this increased propensity to learn a fixed motor routine may reflect a common enhancement of corticostriatal drive across a subset of mice with mutations in ASD-risk genes.
Keywords: striatum, motor learning, autism spectrum disorder, mouse models, corticostriatal, rotarod, direct pathway, indirect pathway
Introduction
An estimated 1 in 100 children globally have autism spectrum disorder (ASD), and CDC estimates indicate even greater prevalence in America, where roughly 1 in 36 children is diagnosed with ASD (Zeidan et al., 2022; Maenner et al., 2023; Talantseva et al., 2023). As ASD is highly heritable (Sandin et al., 2017), much work has been done in recent years to identify genes that confer risk of developing ASD. Increased accessibility of DNA sequencing has allowed for the identification of hundreds of ASD risk genes, which range widely in the types of proteins for which they code (Satterstrom et al., 2020). Despite this molecular heterogeneity, ASD is still diagnosed through identification of behaviors that fall into two primary domains: deficits in social communication and interaction, and the presence of restricted, repetitive patterns of behavior (RRBs) (APA, 2022).
In individuals with ASD, RRBs can span a range of “lower order” and “higher level” behaviors. “Lower order” motor presentations may include self-stimulation or self-injury like head banging, hand flapping, twirling, lining up or manipulating objects, or repeatedly pressing buttons. “Higher level” repetitive behaviors include rituals, perseverative interests and insistence on sameness in a variety of situations (Caldwell-Harris, 2021). In addition to the repetitive behaviors recognized as core ASD symptoms, other motor presentations can include changes to gross motor skills such as balance, gait and posture, as well as alterations in fine motor skills and motor skill learning (Chukoskie et al., 2013). In studies of balance, individuals with ASD exhibit reduced postural control, in particular when somatosensory or visual challenges are introduced. This could occur when a subject is instructed to close their eyes, stand on one leg, or balance on a swaying platform, for example (Minshew et al., 2004; Travers et al., 2013). Atypical gait, which several studies have reported in individuals with ASD, may occur as a result of difficulties with balance and posture (Chukoskie et al., 2013). While specific changes in gait parameters are heterogenous across studies, a lack of smoothness, irregular trunk movements, and shorter stride length are commonly identified in individuals with ASD (Vernazza-Martin et al., 2005; Weiss et al., 2013). Foundational motor movements such as reaching and grasping have also been shown to be altered in children with ASD (Haswell et al., 2009; David et al., 2012), which may underlie some of the deficits seen in executing gross motor skills like throwing and catching, as well as fine motor skills like buttoning, manipulating small objects, and handwriting (Green et al., 2009; Chukoskie et al., 2013; Battah et al., 2023). Notably, handwriting has been reported to be significantly altered in those with ASD since the earliest descriptions of the disorder (Asperger, 1991). Although the early presence of motor symptoms is highly predictive of later overall ASD symptom severity, this remains an understudied and undertreated symptom domain (Troyb et al., 2016; Zampella et al., 2021). The use of common behavioral assays in tractable animal models of ASD can greatly assist in the identification of circuits that may underlie motor changes in autism.
Increasingly, the basal ganglia, and in particular the striatum, has been implicated in the manifestation of repetitive behaviors in ASD, because of the role of these circuits in motor learning, action selection, and habit formation (Fuccillo, 2016). Indeed, both structural and functional imaging studies identify aberrant striatal morphology and connectivity in individuals with ASD, in some cases strongly correlating with the presentation of repetitive behaviors (Hollander et al., 2005; Estes et al., 2011; Dichter, 2012). Magnetic resonance imaging (MRI) studies in mice support these findings, where a diverse range of genetic ASD mouse models exhibit altered striatal morphology and connectivity (Portmann et al., 2014; Ellegood et al., 2015; Lai et al., 2016; Wang et al., 2016). In this review we will discuss the relationship between striatal function and motor performance in mouse models of ASD, which has been illuminated through the use of a common behavioral assay of motor coordination and learning, the accelerating rotarod.
Mouse models
An increase in the identification of genes implicated in ASD risk paired with the genetic accessibility of animal models has allowed for the development of many genetic mouse models of ASD (Bey and Jiang, 2014). Targeting mutations in these mouse models to risk genes that have been identified in individuals with ASD provides construct validity (where the perturbation used to generate the disease model recapitulates the known etiology of the disease in people) (Nestler and Hyman, 2010). Face validity of these models (where the model displays key clinical manifestations of the disease) is more challenging to achieve given the heterogeneity and variability of ASD presentations in people. That said, a range of assays have been developed with the goal of measuring mouse behaviors analogous to those comprising the symptom domains of ASD (Bey and Jiang, 2014).
For the RRB domain of ASD, mouse behavioral assays primarily fit into the “lower order” and “higher level” domain distinctions detailed above. The former is typically measured with the open-field assay, allowing for detection of changes in general locomotor features such as speed and distance traveled, as well as the presence of motor stereotypies such as repetitive grooming, rearing, circling or jumping (Gandhi and Lee, 2020). Other assays like the marble burying test and the hole board take advantage of natural exploratory mouse behaviors like digging and head poking to detect increased repetition of these spontaneous behaviors (Bey and Jiang, 2014). More complex, “higher level” aspects of RRBs can also be assessed in mice, measuring resistance to change, cognitive inflexibility and perseveration in a range of reversal learning, set-shifting and response extinction tasks (Gandhi and Lee, 2020). The changes to gross motor function and coordination that appear to coincide with the repetitive behavior domain in individuals with ASD can also be assessed in mice using balance beams and commercially available systems for measuring and analyzing gait parameters (e.g., DigiGait, Neurocube) (Simmons et al., 2021). The recent development of deep-learning-based platforms such as DeepLabCut and MoSeq allows for unsupervised, data-driven detection and analysis of mouse behavioral parameters (Mathis et al., 2018; Wiltschko et al., 2020).
One behavioral assay commonly utilized in mouse models, the accelerating rotarod task, can be used both as a measure of gross motor coordination, as well as motor skill learning. Below we will outline the structure and parameters of the rotarod task, the way that learning occurs over the course of trials, and the brain regions and circuits implicated in rotarod performance.
The rotarod task measures motor coordination and learning
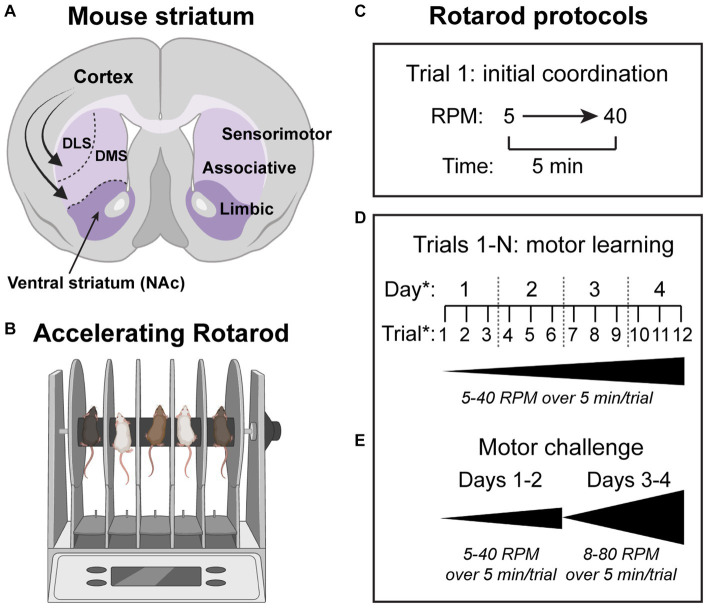
First described in the 1950’s (Dunham and Miya, 1957), the accelerating rotarod task has historically been used as a measure of motor coordination and function in animal models of disease (Hamm et al., 1994; Heng et al., 2008; Lubrich et al., 2022) (Figure 1). However, performance on this test can also be used as a measure of motor skill learning. In the task, mice are trained to walk on a rotating rod as it increases in speed at a constant rate. Protocols utilized in the task vary, but typically the rod increases from 5 to 40 revolutions per minute over the course of 5 min. The latency to fall, or rotate backwards off the rod, is used to determine the terminal velocity in each trial, with increases in this measure indicating better performance. Over several trials, animals exhibit improvement both within a given training day, and over the course of training sessions (Luft and Buitrago, 2005). In this way, initial performance in the task can be isolated as a measure of basic motor coordination, with differences between mouse models at this early stage indicating gross motor deficits or altered baseline motor function. If initial performance is similar, but there are differences in improvement within a given training day and/or across training days, this indicates a difference in motor learning. Many different versions of this extended protocol have been used, ranging from 3 to 5 trials for 1 day up to ten trials a day for 8 days in longer versions of the task (Yin et al., 2009). Most common is to utilize 3–4 trials per day across 3–4 days of testing (Rothwell et al., 2014; Lynch 3rd et al., 2020; Benthall et al., 2021; Le Merrer et al., 2023) (Figure 1).
Figure 1.
Striatal circuits drive motor learning in the accelerating rotarod task. (A) Schematic of a coronal mouse brain section showing the major subdivisions of the striatum (purple). DLS = dorsolateral striatum, DMS = dorsomedial striatum, NAc = nucleus accumbens. Curved arrows depict glutamatergic inputs from the cortex to all striatal subregions. (B) Schematic of the rotarod apparatus used to measure motor coordination and motor learning in rodents. (C–E) Various rotarod protocols have been used. In the simplest version of the task (C), the rod accelerates from 5 to 40 revolutions per minute (RPM) over the course of 5 min. The time to fall off or rotate off the rod is a measure of motor coordination. (D) To measure motor learning, multiple trials are used and the gain in performance from the first to last trial is assessed for each mouse. The number of trials per day and number of testing days can vary. A common version of the task uses three trials per day across four testing days. (E) In some cases, a more challenging version of the task can reveal phenotypes. In this protocol, the rod is accelerated from 8 to 10 RPM up to 80 RPM over 5 min. Schematics in panels A and B were created with bioRender.com.
When given home cage access to a running wheel, animals perform better on the rotarod overall, but the rate of both intra-and intersession improvement remains the same, indicating that increasing performance in the task goes beyond gains in locomotor fitness (Buitrago et al., 2004). Instead, animals develop and optimize a sequence of movements that allows them to stay on the rod at faster speeds, which is exemplified by shifts in gait patterns across training from stepping to running (Buitrago et al., 2004). In some cases, differences in performance between models is only revealed in versions of the task that utilize faster speeds, up to 80 revolutions per minute, which necessitates even greater motor program optimization (Rothwell et al., 2014; DiCarlo et al., 2019; Lynch 3rd et al., 2020; Benthall et al., 2021).
Given the multiphasic nature of the accelerating rotarod task, several brain regions are implicated in task performance, including the cortex (Yang et al., 2009; Fu et al., 2012; Ash et al., 2021b), basal ganglia (Costa et al., 2004; Yin et al., 2009; Durieux et al., 2012), and cerebellum (Sathyamurthy et al., 2020; Simmons et al., 2021). In this review, we highlight the role of the basal ganglia, in particular the striatum, in the motor learning that occurs during rotarod training. Given the involvement of the striatum in a number of other motor learning functions, such as instrumental learning and extinction (Yin et al., 2005, 2006; Santos et al., 2015), active avoidance, response-based procedural learning (Pittenger et al., 2006), and shifting from action-outcome to stimulus–response performance (Hawes et al., 2015), altered rotarod performance, which is easily assessed in mice, likely translates into changes in these more difficult to measure corticostriatal-dependent behaviors. In this way, performance in the accelerating rotarod task is an informative indicator of the function of a frequently altered circuit in mouse models of ASD (Li and Pozzo-Miller, 2020).
Motor learning depends on corticostriatal circuits
The striatum, the main input center of the basal ganglia, is composed of GABAergic striatal projection neurons (SPNs) and local interneurons. SPNs, which make up over 95% of striatal neurons, send their outputs to downstream nuclei via two largely parallel pathways. Dopamine D1-receptor expressing SPNs of the direct pathway (dSPNs) send their primary projections to the substantia nigra pars reticulata and globus pallidus internal segment (SNr/GPi) and broadly facilitate movement when activated in bulk (Kravitz et al., 2010; Gerfen and Surmeier, 2011; Tai et al., 2012). D2-receptor expressing SPNs of the indirect pathway (iSPNs) send their primary projections to the globus pallidus external segment (GPe) and generally inhibit movement or suppress competing actions when activated as a population (Kravitz et al., 2010; Gerfen and Surmeier, 2011; Tai et al., 2012; Calabresi et al., 2014). During behavior, both populations of SPNs are activated in a coordinated way to orchestrate movement and decision-making. SPNs are innervated by a variety of inputs, most notably glutamatergic input from the cortex and thalamus, and dopamine input from the midbrain (Ding et al., 2008; Doig et al., 2010; Gerfen and Surmeier, 2011). Despite overall similar cytoarchitecture, the dorsal and ventral regions of the striatum are thought to be implicated in different functions, with the former controlling motor and cognitive functions, and the latter mediating limbic functions such as appetitive behavior and reward (Voorn et al., 2004) (Figure 1).
Further parsing of striatal regions, based primarily on differences in cortical inputs, implicates the dorsomedial striatum (DMS) as an associative region involved in the initial stages of learning action-outcome pairings and the dorsolateral striatum (DLS) as a sensorimotor region involved in the acquisition of habitual or procedural behaviors (Voorn et al., 2004). In both subregions, SPN ensemble activity and plasticity at striatal synapses is important for a variety of learning tasks, including motor skill learning (Costa et al., 2004; Barnes et al., 2005; Dang et al., 2006; Yin et al., 2009; Kupferschmidt et al., 2019). In the accelerating rotarod task, in vivo electrophysiological recordings showed that neurons in the striatum exhibit task-related activity that is highly correlated with performance (Costa et al., 2004; Barnes et al., 2005). Within the striatum, different subregions exhibit dynamic activity patterns throughout different phases of motor learning. In the DMS, positive modulation of firing rate in task-related SPNs predominantly occurs early in rotarod training, while in DLS, this firing rate modulation occurs after extensive training. Consistent with this, lesions of the DMS impair early learning while lesions of the DLS impair both early and late learning (Yin et al., 2009). Together this work establishes a key role for dorsal striatal circuits in rotarod learning.
While initial work highlighted the importance of the dorsal striatum in motor skill learning, several studies suggest that the ventral striatum may also play a role. In particular, a recent study showed that ablation of iSPNs in the nucleus accumbens (NAc) is sufficient to impair rotarod learning (Le Merrer et al., 2023). In addition, as discussed below, ventral striatal-specific manipulation of some ASD risk genes is sufficient to impact rotarod performance (Rothwell et al., 2014; Platt et al., 2017). This fits within the theory first introduced by Haber and colleagues that the ventral and dorsal striatum interact dynamically over the course of learning (Haber et al., 2000). Just as varying cortical inputs form a gradient across dorsolateral and ventromedial striatum, so too do the inputs to and outputs from dopaminergic substantia nigra. Ventral striatal subregions are proposed to influence behavioral gating in dorsal striatal regions through an ascending “spiral” of information through these striatonigrostriatal connections (Haber et al., 2000; Belin et al., 2007). Dynamic changes in the activity and functional roles of different SPN subtypes across this spiral likely occur during rotarod training.
In terms of the striatal cell types involved in motor learning, studies using ex vivo electrophysiology showed that D2-receptor expressing iSPNs of the DLS undergo significant synaptic potentiation during late training and that administration of a D2R antagonist late in training impairs rotarod performance (Yin et al., 2009). This suggests that plasticity of dorsal striatal indirect pathway activity may be important for rotarod learning. A study using adult neurotoxin-induced ablation of iSPNs throughout the striatum confirmed the importance of iSPNs for rotarod performance, particularly for early learning (Durieux et al., 2012). However, it was also shown that ablation of dSPNs throughout the striatum (Durieux et al., 2012), or selectively in the dorsal striatum (Durieux et al., 2012; Le Merrer et al., 2023), impairs rotarod performance, resulting in severe motor learning deficits. This is consistent with other studies showing that manipulations of dorsal striatal dSPNs can impact rotarod performance (Benthall et al., 2021; Ma et al., 2022). In terms of the ventral striatum, Le Merrer and colleagues showed that ablation of iSPNs (but not dSPNs) in the NAc disrupts rotarod performance (Le Merrer et al., 2023). Furthermore, reducing the excitability of dSPNs in the NAc has also been shown to impair motor learning (Rothwell et al., 2014). Together these studies provide evidence that multiple striatal circuits and subregions are required for motor learning and likely play a coordinated role in motor skill acquisition and maintenance.
The differential roles of striatal sub-regions as well as SPN subtypes during different stages of rotarod learning is likely driven by changes in cortical drive (Yin et al., 2009). Indeed, intact glutamatergic corticostriatal transmission is necessary for rotarod learning. Loss of the presynaptic scaffolding protein RIM1 from corticostriatal neurons, which disrupts excitatory transmission in the dorsal striatum, impairs rotarod learning (Kupferschmidt et al., 2019). In addition, striatal-specific deletion of glutamatergic NMDARs results in a significant deficit in learning in the task (Dang et al., 2006). Taken together, these studies show that changes in the synaptic properties of direct and indirect pathway neurons, throughout dorsal and ventral striatum, shape rotarod performance throughout different stages of the task. Our emerging understanding of the synaptic and circuit mechanisms that underlie rotarod learning make it a useful assay to apply to mouse models of disease.
Altered rotarod performance in mice with mutations in ASD risk genes
Rotarod performance has been assessed across numerous mouse models with mutations in ASD risk genes, making it a useful assay for identifying potential convergent phenotypes. In surveying the literature, we find that many (but not all) ASD mouse models exhibit altered performance in this task, which can include altered initial performance, a global change in performance, or a difference in learning rate across trials (Table 1). One challenge with making general conclusions from this assessment is that multiple different rotarod protocols have been used. While utilizing a rod that increases in speed from 5 to 40 RPM over the course of 5 min per trial is most common, the number of trials implemented per day, and the total number of days of the task vary greatly across studies. In some cases where multiple protocols have been used, mice can show changes in one version of the rotarod task but not another (Rothwell et al., 2014; DiCarlo et al., 2019; Lynch 3rd et al., 2020; Benthall et al., 2021). Therefore, if no phenotype is reported with one rotarod protocol, it’s possible that performance would be altered if the acceleration speed, number of trials, and/or number of testing days were different.
Table 1.
Summary of rotarod performance in mouse models with mutations in ASD risk genes.
| Human gene/CNV | Mouse model | Rotarod phenotype (reference) |
|---|---|---|
| 15q11-13 | patDp/+ (6.3 Mb duplication on chromosome 7) | Similar coordination, enhanced learning (Nakatani et al., 2009) |
| 16p11.2 | Delm (Mills model) | Similar coordination, enhanced learning (Lynch 3rd et al., 2020; Ouellette et al., 2020) Similar coordination, deficit in learning (Yin et al., 2021) |
| Dup/+ (Sult1a1-Spn interval) | Similar coordination, deficit in learning (Arbogast et al., 2016) | |
| 17p11.2 | Dp(11)17/+ | Similar coordination, deficit in learning (Ricard et al., 2010) |
| Df(11)17/+ | Deficit in coordination, similar learning (Ricard et al., 2010) | |
| ARHGAP32 (PX-RICS) | PX-RICS−/− | Deficit in coordination, deficit in learning (Nakamura et al., 2016) |
| ARID1B | Arid1b hKO | Deficit in coordination, deficit in learning (Shibutani et al., 2017) |
| Arid1b+/− | Similar coordination, deficit in learning (Jung et al., 2017) | |
| ARX | Arx(GCG)10 + 7 | Enhanced overall performance (Price et al., 2009) |
| Arxdup24/0 | Increased average latency to fall across 3 trials (Dubos et al., 2018) | |
| ATP1A3 | Atp1a3+/− | Enhanced overall performance (Ikeda et al., 2013) |
| CACNA1G | Cacna1g-Arg1723His-KI+/−, Cacna1g-Arg1723His-KI−/− | Deficit in coordination, deficit in learning (Hashiguchi et al., 2019) |
| CADM1 | Cadm1-KO | Deficit in coordination, deficit in learning (Takayanagi et al., 2010) |
| CDKL5 | Cdkl5−/y | Similar coordination, deficit in learning (Wang et al., 2012; Gao et al., 2020; Adhikari et al., 2022) Deficit in coordination, deficit in learning (Jhang et al., 2017) |
| Cdkl5+/−, Cdkl5−/− | Similar coordination, deficit in learning (Fuchs et al., 2018) | |
| CHD8 | Chd8+/E31T | Enhanced overall performance (Hulbert et al., 2020) |
| Chd8+/− | Similar coordination, enhanced learning (Platt et al., 2017) | |
| CNTNAP2 | Cntnap2−/− | Increased performance on a single trial (Penagarikano et al., 2011) Increased latency to fall from constant speed rotarod (Dawes et al., 2018) |
| CTNNB1 | Bfc/+ | Deficit in coordination, deficit in learning (Tucci et al., 2014) |
| CYFIP1 | Cyfip1+/tm2a(EUCOMM)Wtsi | Similar coordination, deficit in learning (Bachmann et al., 2019) |
| Cyfip+/− | Deficit in coordination, similar learning (Domínguez-Iturza et al., 2019) | |
| DDX3X | Ddx3x+/− | Similar coordination, deficit in learning (Boitnott et al., 2021) |
| DLG4 | Dlg4−/− | Deficit in coordination, deficit in learning (Feyder et al., 2010) |
| DSCAM | Dscamdel17/del17 | Deficit in coordination, deficit in learning (Xu et al., 2011) |
| DYRK1A | mBACtgDyrk1a (186n3) | Similar coordination, deficit in learning (Souchet et al., 2014) |
| EN2 | En2−/− | Similar coordination, deficit in learning (Brielmaier et al., 2012) Deficit in coordination, deficit in learning (Cheh et al., 2006) |
| FOXP2 | Foxp2 R552H/+ | Similar coordination, deficit in learning (Groszer et al., 2008) Deficit in coordination, deficit in learning (French et al., 2012) |
| Foxp2wt/ko | Similar coordination, deficit in learning (Enard et al., 2009) | |
| FMR1 | Fmr1−/− | Similar coordination, enhanced learning (Nolan et al., 2017) Enhanced overall performance (Roy et al., 2011) Similar coordination, deficit in learning (Bhattacharya et al., 2012; Uutela et al., 2012; Li et al., 2023) |
| Fmr1 CGG KI | Similar coordination, deficit in learning (Van Dam et al., 2005) | |
| GABRB3 | Gabrb3−/− | Similar coordination, deficit in learning (DeLorey et al., 1998) |
| p+/m- | Similar coordination, deficit in learning (DeLorey et al., 2011) | |
| p−/m+ | Similar coordination, deficit in learning (DeLorey et al., 2011) | |
| IL1RAPL1 | Il1rapl1 −/Y | Enhanced overall performance (Yasumura et al., 2014) |
| KDM5C | Kdm5c−/y | Decreased performance on a single trial (Scandaglia et al., 2017) |
| KIRREL3 | Kirrel3−/− | Similar coordination, enhanced learning (Hisaoka et al., 2018) |
| LRRC4 | Lrrc4−/− | Similar coordination, deficit in learning (Um et al., 2018) |
| MECP2 | Mecp2-308 | Similar coordination, deficit in learning (De Filippis et al., 2010) |
| Mecp2tm1Tam | Deficit in coordination, deficit in learning (Pelka et al., 2006) | |
| Mecp2tm1.1Jae | Deficit in coordination, deficit in learning (Morello et al., 2018) | |
| Tau-Mecp2 (overexpression) | Similar coordination, deficit in learning (Na et al., 2012) | |
| Mecp2tm1.Bird | Similar coordination, deficit in learning (Pratte et al., 2011) Deficit in coordination, deficit in learning (Kao et al., 2015; Vogel Ciernia et al., 2017) Decreased performance on a single trial (Santos et al., 2007) |
|
| Mecp2T158A | Deficit in coordination, deficit in learning (Goffin et al., 2011) | |
| Mecp2R168X | Decreased average latency to fall across 3 trials (Schaevitz et al., 2013) | |
| Mecp2R294X | Deficit in coordination, deficit in learning (Collins et al., 2022) | |
| Mecp2R306C | Decreased average latency to fall across 3 trials (Lyst et al., 2013) Decreased average latency to fall across 2–4 trials (Ebert et al., 2013) |
|
| Mecp2ΔAT-hook1 | Decreased average latency to fall across 3 trials (Xu et al., 2018) | |
| Mecp2TG (overexpression) | Similar coordination, enhanced learning (Collins et al., 2004, 2022; Sztainberg et al., 2015; Ash et al., 2021a,b) | |
| MYT1L | Myt1l+/− | Similar coordination, deficit in learning (Wohr et al., 2022) |
| NRXN1 | Nrxn1α KO | Similar coordination, enhanced learning (Etherton et al., 2009) |
| Nrxn1α+/ΔExon1, Nrxn1αΔExon1/ΔExon1 | Similar coordination, enhanced learning (Xu et al., 2023) | |
| Nrxn1α+/ΔExon9, Nrxn1αΔExon9/ΔExon9 | Similar coordination, enhanced learning (Xu et al., 2023) | |
| NLGN2 | Nlgn2−/− | Deficit in coordination, similar learning (Blundell et al., 2009) |
| Nlgn2+/− | Similar coordination, enhanced learning (Wohr et al., 2013) | |
| NLGN3 | Nlgn3−/− | Similar coordination, enhanced learning (Rothwell et al., 2014) |
| Nlgn3 R451C KI | Similar coordination, enhanced learning (Chadman et al., 2008; Rothwell et al., 2014; Cao et al., 2022) | |
| Nlgn3mf | Similar coordination, enhanced learning (Yoshida et al., 2021) | |
| NF1 | Nf1+/− | Similar coordination, deficit in learning (van der Vaart et al., 2011) |
| Nf123a−/− | Similar coordination, deficit in learning (Costa et al., 2001) | |
| NRP2 | Nrp2−/− | Similar coordination, deficit in learning (Shiflett et al., 2015) |
| NTNG1 | Ntng2−/− | Enhanced overall performance (Zhang et al., 2016) |
| OTUD7A | Otud7a−/− | Similar coordination, deficit in learning (Yin et al., 2018) |
| PAX5 | Pax5R31Q/− | Deficit in coordination, deficit in learning (Kaiser et al., 2022) |
| PTCHD1 | Ptchd1−/y | Decreased average latency to fall across 3 trials (Ung et al., 2018) |
| PTEN | Ptenm3m4/m3m4 | Deficit in coordination, deficit in learning (Tilot et al., 2014) |
| RAB39B | Rab39b−/− | Decreased average latency to fall across 3 trials (Wang et al., 2023) Similar coordination, deficit in learning (Niu et al., 2020; Zhang et al., 2020) |
| RELN | Reln ΔC-KI | Enhanced overall performance (Sakai et al., 2016) |
| Reln+/rl-Orl | Similar coordination, deficit in learning (Sobue et al., 2018) Deficit in coordination, deficit in learning (Lalonde et al., 2004) |
|
| SCN1A | Scn1a+/− | Similar coordination, deficit in learning (Beretta et al., 2022) |
| Scn1a+/R1407X | Enhanced coordination, similar learning (Ito et al., 2013) | |
| Scn1a+/A1783V | Similar coordination, enhanced learning (Miljanovic et al., 2021) Decreased average latency to fall across 3 trials (Ricobaraza et al., 2019; Fadila et al., 2020) |
|
| SCN2A | Scn2a+/− | Similar coordination, enhanced learning (Lena and Mantegazza, 2019) Deficit in coordination, deficit in learning (Tatsukawa et al., 2019) |
| Scn2a+/K1422E | Similar coordination, enhanced learning (Echevarria-Cooper et al., 2022) | |
| SHANK1 | Shank1−/− | Similar coordination, deficit in learning (Hung et al., 2008; Silverman et al., 2011) |
| SHANK3 | Shank3+/E13 | Similar coordination, enhanced learning (Jaramillo et al., 2017) |
| Shank3+/Δ4-22 | Similar coordination, deficit in learning (Drapeau et al., 2018) | |
| Shank3e4-9/e4-9 | Deficit in coordination, deficit in learning (Wang et al., 2011) | |
| Shank3E13/E13 | Similar coordination, deficit in learning (Jaramillo et al., 2017) | |
| Shank3Δ13-16/Δ13-16 | Similar coordination, deficit in learning (Peixoto et al., 2019) | |
| Shank3fx/fx | Deficit in coordination, deficit in learning (Mei et al., 2016) | |
| Shank3 InsG3680/ InsG3680 | Deficit in coordination, deficit in learning (Speed et al., 2015; Zhou et al., 2016) | |
| Shank3Δ11/Δ11 | Similar coordination, deficit in learning (Vicidomini et al., 2017) | |
| Shank3ΔC/ΔC | Deficit in coordination, deficit in learning (Kouser et al., 2013) | |
| Shank3−/− | Deficit in coordination, deficit in learning (Yang et al., 2012) | |
| SLC6A3 | DATT356M/T356M | Similar coordination, enhanced learning (DiCarlo et al., 2019) |
| SYNGAP1 | Syngap1+/− | Deficit in coordination, deficit in learning (Nakajima et al., 2019) Deficit in coordination, similar learning (Muhia et al., 2010) |
| TOP3B | Top3β−/− | Deficit in coordination, deficit in learning (Rahman et al., 2021) |
| TSC2 | Tsc2+/− | Similar coordination, enhanced learning (Benthall et al., 2021) |
| Tsc2ΔRG | Similar coordination, deficit in learning (Chevere-Torres et al., 2012) | |
| UBE3A | Ube3am−/p+ | Deficit in coordination, deficit in learning (Heck et al., 2008; Leach and Crawley, 2018; Sonzogni et al., 2018; Jiang et al., 1998; Miura et al., 2002; Mulherkar and Jana, 2010; Born et al., 2017; Huang et al., 2013) |
| Ube3am−/p- | Deficit in coordination, deficit in learning (Heck et al., 2008) | |
| Ube3aGenedel | Deficit in coordination, deficit in learning (Syding et al., 2022) | |
| Ube3aOE (overexpression) | Enhanced overall performance (Punt et al., 2022) | |
| Ube3amatT503A (gain of function) | Similar coordination, enhanced learning (Xing et al., 2023) | |
| WDFY3 | Wdfy3+/lacZ | Deficit in coordination, deficit in learning (Le Duc et al., 2019) |
ASD risk genes/CNVs depicted and references for a given model are representative and not exhaustive. Mouse models were chosen using the Simons Foundation Autism Research Initiative (SFARI) Gene mouse models module. Mouse models of ASD risk genes designated as Category 1 (high confidence gene) or Category 2 (strong candidate gene) by SFARI’s gene scoring system were considered. More information about SFARI gene and their gene scoring system can be found at gene.sfari.org. Mouse studies that failed to detect a phenotype are not presented, nor are models that utilized cell-type specific perturbation.
With this caveat noted, we do find a group of models, including mice with loss-of-function mutations in Mecp2, Shank3 and Ube3a, which show consistent deficits in rotarod performance (Table 1). Some of these models exhibit poor performance from the first trial of the task, exemplified by decreased latency to fall from the rod in trial 1 compared to wild-type (WT) controls, owing to baseline deficits in motor coordination (see Table 1 - models with a deficit in coordination). In other models, trial 1 performance resembles that of WT controls, suggesting intact coordination, however, the latency to fall across trials either does not increase, or increases less than WT controls, indicating a deficit in motor learning (see Table 1 - models with a deficit in learning).
In the case of many loss-of-function Mecp2, Shank3 and Ube3a mutations, mice exhibit deficits in both initial coordination and motor learning. The phenotypes observed in these mouse models may reflect the motor deficits that occur in individuals with mutations in these genes (Chukoskie et al., 2013; Troyb et al., 2016; Caldwell-Harris, 2021). Specifically, while motor function can be quite variable across individuals with ASD as a whole, one of the core diagnostic criteria of Rett syndrome, which is caused by loss-of-function mutations in the MECP2 gene, is the deterioration of motor function, often resulting in complete loss of mobility in patients (Chahrour and Zoghbi, 2007). Similarly, patients with Angelman syndrome, a neurodevelopmental disorder caused by mutations in the UBE3A gene, generally exhibit severe motor dysfunction including orthopedic and movement difficulties, walking that is stiff or jerky, and a lack of coordination or development of complex motor skills (Rotaru et al., 2020). A comprehensive clinical assessment of 17 individuals with point mutations in the SHANK3 gene, a gene located within the 22q13.3 chromosomal region implicated in the neurodevelopmental disorder Phelan-McDermid syndrome, identified less severe motor dysfunction than typically seen in the above syndromes; however, nearly all individuals assessed exhibited hypotonia and gait abnormalities (De Rubeis et al., 2018). The identification of motor dysfunction as a common clinical presentation caused by mutations in these genes, alongside the consistently decreased rotarod performance seen in models of these syndromes lends face validity to the rotarod assay.
Notably, while phenotypic analysis of animal models of neuropsychiatric disorders often focuses on identifying deficits, there is a cohort of ASD mouse models that show increased performance on the rotarod task (Table 1). The enhanced performance in these models can either be apparent from initial testing onward or revealed over the course of training. In the remainder of this review, we will focus specifically on these “gain-of-function” cases and discuss how enhanced motor learning may reflect changes in striatal circuit function that could facilitate the development of RRBs.
Enhanced rotarod performance in mice with ASD risk gene mutations
Copy number variations
Many different copy number variations (CNVs) and genomic deletions, duplications or inversions, have been found in individuals with ASD (Takumi and Tamada, 2018). The 16p11.2 variant is one of the most common CNVs associated with ASD (Weiss et al., 2008). Mice with a syntenic 16p11.2 microdeletion (16p11.2 Delm) have been generated and shown to exhibit increased performance on the rotarod, in particular, in a version of the task that utilizes higher speeds (8–80 RPM) (Lynch 3rd et al., 2020). Another 16p11.2 microdeletion mouse model exhibits cellular changes in the striatum including an increased number of iSPNs, increased relative volume of the ventral striatum (in particular the NAc), and excitatory synapse deficits onto SPNs in the NAc. While this mouse model has gross motor alterations such as tremors and gait changes, rotarod performance is unchanged, although the higher speeds utilized in Lynch et al. were not tested (Portmann et al., 2014). Another study found that stride and stance duration in adult 16p11.2 heterozygous mice (16p11.2 Delm) are significantly shorter than in controls, which are features that positively correlate with increased speed (Brunner et al., 2015). These gait changes may contribute to the increased performance on the rotarod task seen in some models of this CNV.
Another CNV implicated in ASD spans the 15q11-13 region, and is most commonly identified as a duplication (Takumi and Tamada, 2018). Mice with a paternal duplication in the 15q11-13 region exhibit increased rotarod performance compared to controls, staying on the rod for significantly longer in every trial after the first, reaching near ceiling performance (Nakatani et al., 2009). Gait assessment in another model of this CNV using a transparent treadmill identified significant changes in the motor program of these mice, which may contribute to their increased performance on the rotarod (Piochon et al., 2014).
Cell adhesion molecules
Several of the rare genetic variants that have been identified as conferring ASD risk impact synaptic cell adhesion molecules, which are proteins involved in the formation and stabilization of synaptic contacts (Betancur et al., 2009). The best characterized synaptic cell adhesion molecules implicated in ASD are those of the neurexin and neuroligin families of proteins. Nrxn1 (neurexin 1a) mutant mice exhibit increased performance on the accelerating rotarod, to the point of near peak performance after ten trials at 4–45 RPM over 5 min (Etherton et al., 2009). This type of enhancement has been observed in another Nrxn1 mutant mouse model as well (Xu et al., 2023). With testing over two additional trials at five times the rate of acceleration (4–45 RPM over 1 min), Nrxn1 knockout (KO) mice continue to perform significantly better than WT mice (Etherton et al., 2009).
Multiple Nlgn3 (neuroligin 3) mutant mouse models also exhibit enhanced performance on the accelerating rotarod, in particular at higher speeds (8–80 RPM) (Chadman et al., 2008; Rothwell et al., 2014; Yoshida et al., 2021; Cao et al., 2022). Video analysis of one such model revealed that Nlgn3 KO mice have reduced variability in their motor performance, streamlining step location, timing, and length significantly more than WT counterparts throughout the task. Variability in these measures negatively correlates with time to fall off the rod, indicating that they represent a valid measure of acquisition of this stereotyped behavior (Rothwell et al., 2014).
Mice lacking another ASD risk gene of the neurexin family, Cntnap2, which encodes a cell adhesion molecule implicated in the stabilization of potassium channels, perform significantly better than WT littermates in a single-trial version of the accelerating rotarod task (Penagarikano et al., 2011), and in a constant speed rotarod task (Dawes et al., 2018). In another study of Cntnap2−/− mice, gait analysis found that KO mice are faster than WT controls. KO mice also exhibit shorter strides, which may contribute to their increased performance (Brunner et al., 2015). A few studies identified alterations in the development or function of inhibitory interneuron populations in the striatum of Cntnap2−/− mice (Penagarikano et al., 2011; Ahmed et al., 2023), a change that may alter SPN excitability and in turn the propensity to form motor routines.
KIRREL3 is an ASD risk gene that codes for a transmembrane protein implicated in synapse formation (Martin et al., 2015). Mice with complete loss of Kirrel3 exhibit enhanced performance on the rotarod, particularly in later trials of the task (Hisaoka et al., 2018). Loss of the ASD risk gene IL1RAPL1 (interleukin 1 receptor accessory protein-like 1), which also encodes a protein that mediates synapse formation, results in enhanced performance on the accelerating rotarod. Il1rapl1−/− mice are able to stay on the rod significantly longer than WT controls for all six trials of the task, demonstrating significantly increased baseline coordination, as well as motor learning (Yasumura et al., 2014).
In the space surrounding synapses, extracellular matrix proteins like reelin aid in the stabilization of cell–cell interactions. Mice with a C-terminal domain mutation in Reln, a gene implicated in a number of neuropsychiatric disorders such as bipolar disorder, schizophrenia, and ASD, exhibit significantly enhanced performance in the accelerating rotarod (Sakai et al., 2016). At the cellular level, another study found that a protocol used to induce synaptic long-term depression (LTD) at corticostriatal synapses in WT mice instead induces long-term potentiation (LTP) in mice with homozygous loss of Reln. This effect is partially explained by a loss of GABAergic tone due to decreased numbers of striatal GABAergic interneurons in Reln mutant mice (Marrone et al., 2006). This enhanced corticostriatal excitability could underlie the increased rotarod performance seen in some Reln mutant models.
mTOR regulators
The mechanistic target of rapamycin (mTOR) serves as a central signaling hub involved in cellular metabolic processes such as protein and lipid synthesis and autophagy (Saxton and Sabatini, 2017). Several genes encoding proteins involved in the mTOR pathway are ASD risk genes, and dysregulation of mTOR signaling may occur in multiple forms of ASD (Winden et al., 2018). TSC2, which codes for an inhibitor of mTOR complex 1 (mTORC1) signaling, is one such ASD risk gene (Curatolo et al., 2015; Davis et al., 2015). Mice with heterozygous loss of Tsc2 have normal initial performance but exhibit increased motor learning on the accelerating rotarod (Benthall et al., 2021). Notably, increased performance is only revealed at higher rotarod speeds (10–80 RPM), as Tsc2+/− mice perform similarly to WT littermates on 5–40 RPM trials (Benthall et al., 2021). This may reflect a ceiling effect, as WT mice can often stay on the rotarod for the entire 5-min trial with speeds up to 40 RPM.
Mice with altered function of Pten, another inhibitor of mTOR signaling, also exhibit changes in rotarod behavior. While global heterozygous loss of Pten does not alter rotarod performance (Clipperton-Allen and Page, 2014), Kwon et al. found that conditional loss of Pten results in increased performance on the accelerating rotarod compared to controls. In this model, Pten loss occurs in a subset of cortical and hippocampal neurons (Kwon et al., 2006). Pten deletion in interneurons is likely not the driver of this enhanced performance, as cell-type specific loss of Pten in parvalbumin (PV) and/or somatostatin (SST) interneurons led to impaired rotarod performance (Shin et al., 2021). Instead, increased local and long range excitatory input onto Pten KO cells in sensory cortex suggests that increased excitatory drive of corticostriatal neurons could underlie increased rotarod performance (Xiong et al., 2012).
Transcriptional and translational regulators
Neural development requires precise coordination of molecular programs and several genes involved in transcriptional and translational control are implicated in ASD (Longo and Klann, 2021). CHD8, which encodes the chromatin remodeling factor chromodomain helicase DNA binding protein 8, has been identified as one of the genes with the strongest association with ASD (Weissberg and Elliott, 2021). Chd8+/− mice perform significantly better than WT counterparts on the accelerating rotarod, regardless of whether mice were trained at 4–40 RPM once a day for 5 days, or three times a day for 2 days (Platt et al., 2017). Enhanced rotarod performance was also observed in a different Chd8 mutant model (Hulbert et al., 2020). In this study, Chd8+/E31T mice performed significantly better than WT controls on all 4 trials of both accelerating (4–40 RPM over 5 min) and steady state (32 RPM) rotarod tasks.
As discussed above, loss of the transcriptional regulator MECP2 results in Rett syndrome, which is characterized by motor deficits in people and in mouse models. However, duplication of the MECP2 locus causes MECP2 duplication syndrome, which is a neurodevelopmental disorder highly comorbid with ASD (Qiu, 2018). In contrast to Mecp2 deficient mice, mice with duplication of Mecp2 exhibit significantly enhanced performance on the rotarod task, a phenotype that has been observed in several different Mecp2 duplication models (Collins et al., 2004; Sztainberg et al., 2015). At the cellular level, following rotarod training, Mecp2 duplication mice have significantly more new dendritic spines, as well as more stabilized spines on layer V pyramidal neurons in the motor cortex (M1), which project to the dorsal striatum (Ash et al., 2021b). These new stabilized spines tend to be located in clusters in Mecp2 duplication mice, a characteristic that has been associated with increased motor skill learning (Yang et al., 2009; Fu et al., 2012). Indeed, Ash et al. found that the formation and stabilization of new spine clusters is significantly correlated with increased performance on the rotarod in both Mecp2 duplication mice and WT controls (Ash et al., 2021b).
To interrogate the molecular mechanisms driving the enhanced rotarod learning in Mecp2 duplication mice, Ash et al. targeted Ras–ERK signaling by intraperitoneally injecting the ERK inhibitor SL327 daily preceding rotarod training. This reversed the enhanced performance of Mecp2 duplication mice, without altering WT performance in the task (Ash et al., 2021a). Together these findings suggest that increased synaptic stability within the corticostriatal sensorimotor loop may underlie enhanced motor learning in the context of Mecp2 duplication. Notably, mice with a loss-of-function mutation in Mecp2 exhibit significantly decreased spine density in pyramidal cells of both motor (Tropea et al., 2009) and somatosensory cortex, as well as altered short-term structural plasticity of spines in the latter region (Landi et al., 2011). These gene dose-dependent changes in synaptic stability within sensorimotor circuitry may contribute to the opposing impact of Mecp2 mutations on rotarod performance.
Mutations in the FMR1 gene result in Fragile X syndrome, a neurodevelopmental disorder with high comorbidity with ASD. FMR1 mutations alter the expression of Fragile X Messenger Ribonucleoprotein (FMRP), an RNA binding protein involved in translational control (Jin and Warren, 2003). In one Fmr1 KO mouse model, accelerating rotarod performance is enhanced compared to WT controls across all three sessions of the task, indicating both enhanced baseline coordination as well as increased learning over trials (Roy et al., 2011). Another Fmr1 model exhibits similar initial coordination as WT controls, but significantly increased learning across the eight trials of the rotarod task (Nolan et al., 2017). Other studies of this model identified changes in striatal endocannabinoid-mediated long-term depression (eCB-LTD) (Maccarrone et al., 2010; Jung et al., 2012), a form of synaptic plasticity altered in other genetic mouse models with enhanced performance in the rotarod task (Martella et al., 2018; Benthall et al., 2021). In the dorsal striatum, eCB-LTD is enhanced at GABAergic synapses in the context of FMRP loss (Maccarrone et al., 2010), whereas eCB-LTD at excitatory synapses in the ventral striatum of Fmr1 KO mice is abolished (Jung et al., 2012). Taken together, this loss of LTD at excitatory synapses and enhanced LTD at inhibitory synapses may culminate in unchecked corticostriatal drive in Fmr1 KO mice, which could underlie the convergent motor phenotype across these models.
Dopamine and rotarod performance
Dopamine is a potent modulator of cortical and striatal synapses (Tritsch and Sabatini, 2012) and functional dopamine signaling is important for motor performance and learning (Packard and Knowlton, 2002). Mice lacking dopaminergic neurons of the substantia nigra pars compacta, leading to 90% reductions in dorsal striatal dopamine, are unable to increase performance on the rotarod over trials, a deficit that is rescued by treatment with the dopamine precursor L-DOPA (Beeler et al., 2010). Several mouse models of ASD exhibit alterations in dopaminergic function (Kosillo and Bateup, 2021). A de novo mutation in SLC6A3, which results in a T356M amino acid substitution in the gene encoding the dopamine transporter (DAT), has been linked to ASD (Neale et al., 2012). In vitro characterization shows that this mutation results in efflux, rather than typical influx, of dopamine when expressed, potentially leading to greater synaptic dopamine (Hamilton et al., 2013). Mice expressing one copy of the Slc6a3 mutation perform similarly to WT controls in early training; however, T356M+/− mice exhibit significantly enhanced performance in later trials of the task. DAT expression levels are normal in these mice, but striatal dopamine (DA) reuptake is impaired and increased extracellular dopamine in the striatum results in increased striatal DA metabolism and reduced striatal DA synthesis (DiCarlo et al., 2019). Appropriate regulation of extracellular dopamine is important for rotarod performance as administration of the dopamine reuptake blocker nomifensine increases performance, while the dopamine agonist apomorphine diminishes performance (Shiotsuki et al., 2010). Nomifensine increases extracellular dopamine (Cragg and Rice, 2004), while apomorphine’s action at presynaptic D2 autoreceptors suppresses dopamine release (Schmitz et al., 2002). The opposing impacts of these drugs on rotarod performance highlight the importance of proper dopamine signaling in motor learning.
Striatal changes drive altered rotarod performance
The majority of studies assessing accelerating rotarod performance have used mouse models with global mutations in ASD risk genes; therefore, the brain region or circuit responsible for the phenotype is difficult to ascertain. As discussed above, striatal circuits have been identified as a key node in rotarod motor learning. To directly test the contribution of striatal neurons to rotarod phenotypes, conditional KO mice have been generated in which the ASD risk gene is manipulated selectively in striatal neurons. These studies have revealed a direct link between striatal function and rotarod performance.
In the case of Nlgn3, conditional deletion in cells of the direct, but not the indirect, pathway of the striatum results in increased rotarod performance (Rothwell et al., 2014). A deficit in inhibition specifically onto dSPNs in these mice, which is expected to enhance excitability of the direct pathway, likely contributes to their increased rotarod performance. Indeed, in WT mice, a manipulation that reduces the activity of indirect pathway cells, which would have the net effect of facilitating direct pathway activation of downstream basal ganglia nuclei, results in increased performance in the task. In addition, rotarod performance is restored to WT levels in Nlgn3 dSPN conditional KO mice via expression of the potassium channel Kir2.1, which decreases dSPN excitability (Rothwell et al., 2014). This study provides compelling evidence that altered balance between the striatal direct and indirect pathways can contribute to altered motor learning in ASD mouse models. Interestingly, conditional deletion of Nlgn3 in the NAc, and not broadly in the dorsal striatum, is sufficient to recapitulate the enhanced rotarod performance (Rothwell et al., 2014). This finding supports a potentially underappreciated role for the ventral striatum in motor learning. A possible explanation for the cell type and anatomical specificity of Nlgn3 deletion is that Nlgn3 is preferentially expressed in dSPNs of the NAc and therefore expected to have a greater effect when disrupted in these cells (Rothwell et al., 2014).
As discussed above, multiple studies of Chd8 mouse models have identified enhanced rotarod performance (Platt et al., 2017; Hulbert et al., 2020). One such study performed gene expression analysis across brain regions in Chd8+/− mice, identifying the NAc as a region with significant gene dysregulation. Following this, Platt et al. injected Chd8-targeting sgRNA into the NAc in a Cas9 knock-in mouse to determine the impact of Chd8 reduction specifically in this region. Similar to the findings in the Nlgn3 study (Rothwell et al., 2014), reduction of Chd8 specifically in the NAc, and not the dorsal striatum, recapitulated the increased rotarod performance seen in constitutive heterozygous mice (Rothwell et al., 2014; Platt et al., 2017). Electrophysiological assessment of these mice found that SPNs of the NAc core have increased frequency and amplitude of spontaneous excitatory synaptic currents, as well as decreased amplitude of miniature inhibitory synaptic currents, suggesting overall increased excitatory drive of SPNs in the region (Platt et al., 2017). A distinction between dSPNs and iSPNs was not made in this study.
While the above studies implicate altered NAc function as a driver of enhanced rotarod performance, a recent study identified increased motor learning in a mouse model with dorsal striatum-selective Tsc1 loss (Benthall et al., 2021). In this study, dSPN-specific deletion of the ASD-risk gene Tsc1 resulted in increased performance on the accelerating rotarod, in particular at higher speeds (10–80 RPM). Mice with loss of Tsc1 in iSPNs did not exhibit changes in rotarod performance, consistent with the findings in Nlgn3 mice (Rothwell et al., 2014; Benthall et al., 2021). The D1-Cre line utilized in this study to target dSPNs is relatively restricted to dSPNs of the dorsal striatum, sparing the majority of NAc cells (Benthall et al., 2021). This, together with the results described above, indicate that altered direct pathway activity in either the dorsal or ventral striatum is sufficient to alter motor learning.
In Tsc1 dSPN KO mice, electrophysiology experiments revealed that Tsc1-KO dSPNs have increased glutamate release probability at cortical inputs, resulting in enhanced corticostriatal drive. This study also found a deficit in eCB-LTD onto Tsc1 KO dSPNs, which may explain the change in presynaptic release probability (Benthall et al., 2021). This prominent form of striatal synaptic depression works through the release of postsynaptic endocannabinoids that act on cortical presynaptic CB1 receptors, ultimately reducing the probability of neurotransmitter release (Kreitzer and Malenka, 2008; Lovinger, 2010). Loss of eCB-LTD onto Tsc1 KO dSPNs likely renders these cells unable to depress excitatory inputs, leading to increased corticostriatal drive over time. Interestingly, a similar deficit in eCB-LTD was identified in the dorsal striatum of a Nlgn3 mutant mouse model that exhibits enhanced performance in the rotarod task (Martella et al., 2018).
Here we have highlighted several examples of mouse models that exhibit enhanced rotarod performance. In a few of these models, striatal-specific manipulation of an ASD-risk gene was sufficient to induce changes in rotarod motor learning. In several studies, synaptic changes were reported that are expected to enhance striatal activation, particularly increase corticostriatal drive and/or excitability of the direct pathway (Rothwell et al., 2014; Platt et al., 2017; Benthall et al., 2021). Given the importance of striatal circuits for not only motor skill leaning but also habit learning etc., it seems plausible that gain-of-function at the neural circuit level facilitates the formation of fixed motor routines or perseverative behaviors. For ASD models where motor deficits predominate, the neural circuitry underlying the presence of repetitive behaviors remains to be established. In these cases, basic motor circuits may be disrupted such that rotarod deficits arise, but gain-of-function in other motor control circuitry likely drives the emergence of RRBs, for example in the case of Shank3 models (Drapeau et al., 2018). Further investigation into the motor phenotypes of ASD mouse models and the synaptic and circuit basis of repetitive behaviors will provide additional insight into this core aspect of ASD.
Other considerations
Genetic background influences rotarod performance
There are several factors beyond a targeted genetic manipulation that contribute to differences in rotarod performance, which should be considered when comparing across ASD mouse models. A study assessing 16 mouse strains from the “Collaborative Cross” (CC), a large panel of inbred mice that captures 90% of the known variation among laboratory mice, found that rotarod performance varies widely across the strains (Mao et al., 2015). Forty-five gene loci associated with rotarod performance were identified using genetic linkage analysis, many of which overlap with human GWAS-nominated genes associated with neuropsychiatric disorders including ASD and ADHD. A similar study assessing a range of behaviors across 10 inbred mouse strains also found wide variability in rotarod performance across strains (Moy et al., 2007). Two of these inbred mouse strains, BTBR T+tf/J (BTBR) and BALB/cByJ (BALB), have been utilized for over a decade as models of ASD, owing to their strong and consistent displays of autism-relevant behaviors, including social behavior deficits and/or repetitive behaviors or stereotypies (Ellegood and Crawley, 2015). While BTBR mice exhibit deficits in the rotarod task, BALB mice perform similarly to C57BL/6J controls (Moy et al., 2007). Since these models are inbred strains that lack known genetic abnormalities and do not recapitulate known genetic causes of human ASD, it can be difficult to link neurodevelopmental changes to ASD-like behavior. However, continued study of these inbred models that demonstrate good face validity for ASD-like manifestations may uncover their potential construct and predictive validity.
Along with genetic background, body weight also significantly impacts rotarod performance, with weight being strongly negatively correlated with performance (Mao et al., 2015). While Mao et al. found that rotarod performance between sexes within a given strain is highly correlated, females tend to have a lower body weight than males, which may alter performance (Mao et al., 2015). Given that significant sex differences exist in both the prevalence and presentation of ASD in humans (APA, 2022), it’s possible that motor coordination and learning could differ across sexes within a given ASD mouse model. Variability in these factors across studies may contribute to some of the heterogeneity in rotarod outcomes reported in the literature (see Table 1).
Environmental risk factors
Here we have focused on rotarod performance in genetic mouse models of ASD; however, there is a growing body of work implicating exposure to certain environmental factors in the manifestation of autism, including factors that illicit an immune response (Meltzer and Van de Water, 2017). In mice, this is often modeled with a maternal immune activation paradigm. Briefly, pregnant dams are directly infected with a pathogen (e.g., influenza virus, Escherichia coli), or injected with a substance that mimics a pathogen to evoke a large immune response. Subsequently, significant immunological, behavioral and neurodevelopmental changes can then be observed in offspring (Careaga et al., 2017). As brain development continues after birth, models of postnatal infections and postnatal immune activation have also been shown to lead to some of these changes (Depino, 2013). While some immune activation mouse models exhibit enhanced performance on the accelerating rotarod (Carlezon Jr et al., 2019), other models have no change in performance (Wei et al., 2012), or deficits in the task (Naviaux et al., 2014).
In utero exposure to certain medications has also been linked to the development of ASD, including the antiepileptic and bipolar medication valproic acid (VPA) (Roullet et al., 2013). The mechanism of VPA that may increase the risk of ASD is unknown, and prenatal exposure to the drug, like other implicated environmental factors, could modify existing genetic risk (Wang et al., 2017). Mouse models of VPA exposure are typically achieved through injection of VPA into a pregnant dam roughly midway through gestation, resulting in offspring that exhibit both neurodevelopmental and behavioral alterations (Roullet et al., 2013). Rotarod phenotypes across studies of VPA models vary, with some identifying enhanced rotarod performance (Hernandez et al., 2023), and others identifying no difference from WT (Gandal et al., 2010), or a deficit in the task (Wang et al., 2018), owing potentially to variability in VPA exposure protocols.
Genetic rat models of ASD
Finally, while this review focuses on mouse models, there is a growing body of literature on genetic rat models of ASD (Berg et al., 2021; Harris et al., 2021; Dey and Chattarji, 2022), including models with mutations in ASD risk genes linked to enhanced rotarod performance discussed above, such as Fmr1 (Till et al., 2015; D’Elia et al., 2022; Schiavi et al., 2023) and Nlgn3 (Thomas et al., 2017; Anstey et al., 2022). However, as genetic accessibility of rat models is a recent development, assessment of rotarod performance in these models remains to be performed. Given the more expansive behavioral repertoire of rats, and the technical benefits that their larger size affords, increased study of genetic rat models of ASD is likely to benefit the understanding of rotarod behavior and motor skill learning in the context of ASD risk gene mutations in the coming years (Ellenbroek and Youn, 2016).
Interactions between the basal ganglia and other motor control circuits
This review highlights the ways that striatal, and in particular corticostriatal, circuits play a role in accelerating rotarod performance. However, cerebellar circuits are also implicated in motor skill learning. Different cerebellar subcircuits are implicated in early versus late stages of motor skill learning, and a number of studies suggest that cerebellar relays exist within or parallel to the cortico-basal ganglia-thalamic loop controlling motor learning (Dayan and Cohen, 2011). In the rotarod specifically, a subpopulation of excitatory cerebellospinal neurons in deep cerebellar nuclei that project to the spinal cord were identified as being necessary for learning, but not the execution, of rotarod behavior (Sathyamurthy et al., 2020). The specific inputs to these neurons are yet unclear, but they may receive direct input from the cortex and/or thalamus. In the context of ASD, the cerebellum is frequently implicated as a region of potential convergent change. Postmortem studies in humans reveal cellular and structural cerebellar abnormalities in individuals with ASD (Schumann and Nordahl, 2011; Bolduc et al., 2012; Ecker et al., 2012). In addition, mouse models that target mutation of ASD risk genes specifically to cerebellar cell types can recapitulate ASD-like phenotypes (Tsai et al., 2012; Hampson and Blatt, 2015). However, we note that in the case of cerebellar-specific ASD risk gene mutations, mouse models most often exhibit deficits in accelerating rotarod performance (Tsai et al., 2012; Reith et al., 2013; Kawamura et al., 2021; Liu et al., 2022), even in cases where constitutive mutation of the gene leads to enhanced performance (Reith et al., 2013; Platt et al., 2017). Thus, while cerebellar circuits may participate in rotarod performance, it seems unlikely that they would contribute to the enhanced rotarod phenotype seen in the mouse models described above. Rather, we posit that synaptic gain-of-function in corticostriatal circuits is more likely to drive increased motor learning in the context of ASD.
Conclusion
As the number and heterogeneity of identified ASD risk genes continues to expand, the utilization of common behavioral assays to identify convergent phenotypes in mouse models of ASD is of great benefit. When it comes to assessing motor symptom domains of ASD in mouse models, the accelerating rotarod task has proved very useful for identifying phenotypes. The task is relatively fast and straightforward to carry out, reveals information about gross motor coordination and motor learning, and has established underlying neural circuitry. Corticostriatal circuits are key regulators of rotarod performance and are increasingly implicated as a point of convergent alteration across a range of mouse models of ASD (Li and Pozzo-Miller, 2020). If these circuits are found to consistently contribute to altered behavior, they represent a potential site for targeted therapeutics, which may be applicable across ASDs of different genetic origin.
Author contributions
KC: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. HB: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NIH grants #R01NS105634, #R01MH130839 and SFARI research grant #514428 to HB and NIH fellowship #F31NS124499 to KC.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Adhikari A., Buchanan F. K. B., Fenton T. A., Cameron D. L., Halmai J., Copping N. A., et al. (2022). Touchscreen cognitive deficits, hyperexcitability and hyperactivity in males and females using two models of Cdkl5 deficiency. Hum. Mol. Genet. 31, 3032–3050. doi: 10.1093/hmg/ddac091, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N. Y., Knowles R., Liu L., Yan Y., Li X., Schumann U., et al. (2023). Developmental deficits of Mge-derived interneurons in the Cntnap2 knockout mouse model of autism spectrum disorder. Front. Cell Dev. Biol. 11:1112062. doi: 10.3389/fcell.2023.1112062, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstey N. J., Kapgal V., Tiwari S., Watson T. C., Toft A. K. H., Dando O. R., et al. (2022). Imbalance of flight-freeze responses and their cellular correlates in the Nlgn3(−/y) rat model of autism. Mol. Autism. 13:34. doi: 10.1186/s13229-022-00511-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- APA (2022). Diagnostic and Statistical Manual of Mental Disorders, APA, Arlington, Va. [Google Scholar]
- Arbogast T., Ouagazzal A. M., Chevalier C., Kopanitsa M., Afinowi N., Migliavacca E., et al. (2016). Reciprocal effects on neurocognitive and metabolic phenotypes in mouse models of 16p11.2 deletion and duplication syndromes. PLoS Genet. 12:e1005709: e1005709. doi: 10.1371/journal.pgen.1005709, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash R. T., Buffington S. A., Park J., Suter B., Costa-Mattioli M., Zoghbi H. Y., et al. (2021a). Inhibition of elevated ras-MAPK signaling normalizes enhanced motor learning and excessive clustered dendritic spine stabilization in the MECP2-duplication syndrome mouse model of autism. eNeuro 8, 1–11. doi: 10.1523/ENEURO.0056-21.2021, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash R. T., Park J., Suter B., Zoghbi H. Y., Smirnakis S. M. (2021b). Excessive formation and stabilization of dendritic spine clusters in the Mecp2-duplication syndrome mouse model of Autism. eNeuro 8, 1–13. doi: 10.1523/ENEURO.0282-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asperger H. (1991). ‘Autistic Psychopathy’ in Childhood. Autism and Asperger Syndrome. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Bachmann S. O., Sledziowska M., Cross E., Kalbassi S., Waldron S., Chen F., et al. (2019). Behavioral training rescues motor deficits in Cyfip1 haploinsufficiency mouse model of autism spectrum disorders. Transl. Psychiatry 9:29. doi: 10.1038/s41398-018-0338-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes T. D., Kubota Y., Hu D., Jin D. Z., Graybiel A. M. (2005). Activity of striatal neurons reflects dynamic encoding and recoding of procedural memories. Nature 437, 1158–1161. doi: 10.1038/nature04053, PMID: [DOI] [PubMed] [Google Scholar]
- Battah H. W., Lotan M., Moran D. S. (2023). The need for a motor assessment tool for children with Autism-An opinion article. Diagnostics (Basel):13: 2095. doi: 10.3390/diagnostics13122095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler J. A., Cao Z. F., Kheirbek M. A., Ding Y., Koranda J., Murakami M., et al. (2010). Dopamine-dependent motor learning: insight into levodopa’s long-duration response. Ann. Neurol. 67, 639–647. doi: 10.1002/ana.21947, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin D., Deroche-Gamonet V., Jaber M. (2007). Cocaine-induced sensitization is associated with altered dynamics of transcriptional responses of the dopamine transporter, tyrosine hydroxylase, and dopamine D2 receptors in C57Bl/6J mice. Psychopharmacology 193, 567–578. doi: 10.1007/s00213-007-0790-3, PMID: [DOI] [PubMed] [Google Scholar]
- Benthall K. N., Cording K. R., Agopyan-Miu A., Wong C. D., Chen E. Y., Bateup H. S. (2021). Loss of Tsc1 from striatal direct pathway neurons impairs endocannabinoid-ltd and enhances motor routine learning. Cell Rep. 36:109511. doi: 10.1016/j.celrep.2021.109511, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta S., Gritti L., Ponzoni L., Scalmani P., Mantegazza M., Sala M., et al. (2022). Rescuing epileptic and behavioral alterations in a Dravet syndrome mouse model by inhibiting eukaryotic elongation factor 2 kinase (eef2K). Mol. Autism. 13:1. doi: 10.1186/s13229-021-00484-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg E. L., Jami S. A., Petkova S. P., Berz A., Fenton T. A., Lerch J. P., et al. (2021). Excessive laughter-like vocalizations, microcephaly, and translational outcomes in the Ube3a deletion rat model of Angelman syndrome. J. Neurosci. 41, 8801–8814. doi: 10.1523/JNEUROSCI.0925-21.2021, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur C., Sakurai T., Buxbaum J. D. (2009). The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 32, 402–412. doi: 10.1016/j.tins.2009.04.003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bey A. L., Jiang Y. H. (2014). Overview of mouse models of autism spectrum disorders. Current Protoc Pharmacol 66, 5.66.1–5.66.26. doi: 10.1002/0471141755.ph0566s66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A., Kaphzan H., Alvarez-Dieppa A. C., Murphy J. P., Pierre P., Klann E. (2012). Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron 76, 325–337. doi: 10.1016/j.neuron.2012.07.022, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J., Tabuchi K., Bolliger M. F., Blaiss C. A., Brose N., Liu X., et al. (2009). Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes Brain Behav. 8, 114–126. doi: 10.1111/j.1601-183X.2008.00455.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitnott A., Garcia-Forn M., Ung D. C., Niblo K., Mendonca D., Park Y., et al. (2021). Developmental and behavioral phenotypes in a mouse model of Ddx3X syndrome. Biol. Psychiatry 90, 742–755. doi: 10.1016/j.biopsych.2021.05.027, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc M. E., Du Plessis A. J., Sullivan N., Guizard N., Zhang X., Robertson R. L., et al. (2012). Regional cerebellar volumes predict functional outcome in children with cerebellar malformations. Cerebellum 11, 531–542. doi: 10.1007/s12311-011-0312-z, PMID: [DOI] [PubMed] [Google Scholar]
- Born H. A., Dao A. T., Levine A. T., Lee W. L., Mehta N. M., Mehra S., et al. (2017). Strain-dependence of the Angelman syndrome phenotypes in Ube3a maternal deficiency mice. Sci. Rep. 7:8451. doi: 10.1038/s41598-017-08825-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brielmaier J., Matteson P. G., Silverman J. L., Senerth J. M., Kelly S., Genestine M., et al. (2012). Autism-relevant social abnormalities and cognitive deficits in engrailed-2 knockout mice. PLoS One 7:e40914. doi: 10.1371/journal.pone.0040914, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D., Kabitzke P., He D., Cox K., Thiede L., Hanania T., et al. (2015). Comprehensive analysis of the 16p11.2 deletion and null Cntnap2 mouse models of Autism Spectrum disorder. PLoS One 10:e0134572. doi: 10.1371/journal.pone.0134572, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitrago M. M., Schulz J. B., Dichgans J., Luft A. R. (2004). Short and long-term motor skill learning in an accelerated rotarod training paradigm. Neurobiol. Learn. Mem. 81, 211–216. doi: 10.1016/j.nlm.2004.01.001, PMID: [DOI] [PubMed] [Google Scholar]
- Calabresi P., Picconi B., Tozzi A., Ghiglieri V., Di Filippo M. (2014). Direct and indirect pathways of basal ganglia: a critical reappraisal. Nat. Neurosci. 17, 1022–1030. doi: 10.1038/nn.3743, PMID: [DOI] [PubMed] [Google Scholar]
- Caldwell-Harris C. L. (2021). An explanation for repetitive motor behaviors in Autism: facilitating inventions via trial-and-error discovery. Front. Psych. 12:657774. doi: 10.3389/fpsyt.2021.657774, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Li J. H., Lin S., Xia Q. Q., Du Y. L., Yang Q., et al. (2022). Nmda receptor hypofunction underlies deficits in parvalbumin interneurons and social behavior in neuroligin 3 R451C knockin mice. Cell Rep. 41:111771. doi: 10.1016/j.celrep.2022.111771, PMID: [DOI] [PubMed] [Google Scholar]
- Careaga M., Murai T., Bauman M. D. (2017). Maternal immune activation and Autism Spectrum disorder: from rodents to nonhuman and human Primates. Biol. Psychiatry 81, 391–401. doi: 10.1016/j.biopsych.2016.10.020, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon W. A., Jr., Kim W., Missig G., Finger B. C., Landino S. M., Alexander A. J., et al. (2019). Maternal and early postnatal immune activation produce sex-specific effects on autism-like behaviors and neuroimmune function in mice. Sci. Rep. 9:16928. doi: 10.1038/s41598-019-53294-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadman K. K., Gong S., Scattoni M. L., Boltuck S. E., Gandhy S. U., Heintz N., et al. (2008). Minimal aberrant behavioral phenotypes of neuroligin-3 R451C knockin mice. Autism Res. 1, 147–158. doi: 10.1002/aur.22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M., Zoghbi H. Y. (2007). The story of Rett syndrome: from clinic to neurobiology. Neuron 56, 422–437. doi: 10.1016/j.neuron.2007.10.001, PMID: [DOI] [PubMed] [Google Scholar]
- Cheh M. A., Millonig J. H., Roselli L. M., Ming X., Jacobsen E., Kamdar S., et al. (2006). En2 knockout mice display neurobehavioral and neurochemical alterations relevant to autism spectrum disorder. Brain Res. 1116, 166–176. doi: 10.1016/j.brainres.2006.07.086, PMID: [DOI] [PubMed] [Google Scholar]
- Chevere-Torres I., Maki J. M., Santini E., Klann E. (2012). Impaired social interactions and motor learning skills in tuberous sclerosis complex model mice expressing a dominant/negative form of tuberin. Neurobiol. Dis. 45, 156–164. doi: 10.1016/j.nbd.2011.07.018, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukoskie L., Townsend J., Westerfield M. (2013). Motor skill in autism spectrum disorders. Int. Rev. Neurobiol. 113, 207–249. doi: 10.1016/B978-0-12-418700-9.00007-1 [DOI] [PubMed] [Google Scholar]
- Clipperton-Allen A. E., Page D. T. (2014). Pten haploinsufficient mice show broad brain overgrowth but selective impairments in autism-relevant behavioral tests. Hum. Mol. Genet. 23, 3490–3505. doi: 10.1093/hmg/ddu057, PMID: [DOI] [PubMed] [Google Scholar]
- Collins A. L., Levenson J. M., Vilaythong A. P., Richman R., Armstrong D. L., Noebels J. L., et al. (2004). Mild overexpression of Mecp2 causes a progressive neurological disorder in mice. Hum. Mol. Genet. 13, 2679–2689. doi: 10.1093/hmg/ddh282, PMID: [DOI] [PubMed] [Google Scholar]
- Collins B. E., Merritt J. K., Erickson K. R., Neul J. L. (2022). Safety and efficacy of genetic Mecp2 supplementation in the R294X mouse model of Rett syndrome. Genes Brain Behav. 21:e12739. doi: 10.1111/gbb.12739, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. M., Cohen D., Nicolelis M. A. L. (2004). Differential corticostriatal plasticity during fast and slow motor skill learning in mice. Curr. Biol. 14, 1124–1134. doi: 10.1016/j.cub.2004.06.053, PMID: [DOI] [PubMed] [Google Scholar]
- Costa R. M., Yang T., Huynh D. P., Pulst S. M., Viskochil D. H., Silva A. J., et al. (2001). Learning deficits, but normal development and tumor predisposition, in mice lacking exon 23a of Nf1. Nat. Genet. 27, 399–405. doi: 10.1038/86898, PMID: [DOI] [PubMed] [Google Scholar]
- Cragg S. J., Rice M. E. (2004). Dancing past the Dat at a Da synapse. Trends Neurosci. 27, 270–277. doi: 10.1016/j.tins.2004.03.011, PMID: [DOI] [PubMed] [Google Scholar]
- Curatolo P., Moavero R., De Vries P. J. (2015). Neurological and neuropsychiatric aspects of tuberous sclerosis complex. Lancet Neurol. 14, 733–745. doi: 10.1016/S1474-4422(15)00069-1, PMID: [DOI] [PubMed] [Google Scholar]
- D’Elia A., Schiavi S., Manduca A., Rava A., Buzzelli V., Ascone F., et al. (2022). Fmr1 deletion in rats induces hyperactivity with no changes in striatal dopamine transporter availability. Sci. Rep. 12:22535. doi: 10.1038/s41598-022-26986-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang M. T., Yokoi F., Yin H. H., Lovinger D. M., Wang Y., Li Y. (2006). Disrupted motor learning and long-term synaptic plasticity in mice lacking Nmdar1 in the striatum. Proc. Natl. Acad. Sci. U. S. A. 103, 15254–15259. doi: 10.1073/pnas.0601758103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- David F. J., Baranek G. T., Wiesen C., Miao A. F., Thorpe D. E. (2012). Coordination of precision grip in 2-6 years-old children with autism spectrum disorders compared to children developing typically and children with developmental disabilities. Front. Integr. Neurosci. 6:122. doi: 10.3389/fnint.2012.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis P. E., Peters J. M., Krueger D. A., Sahin M. (2015). Tuberous sclerosis: a new frontier in targeted treatment of Autism. Neurotherapeutics 12, 572–583. doi: 10.1007/s13311-015-0359-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes J. M., Weir G. A., Middleton S. J., Patel R., Chisholm K. I., Pettingill P., et al. (2018). Immune or genetic-mediated disruption of Caspr2 causes pain hypersensitivity due to enhanced primary afferent excitability. Neuron 97:e10. doi: 10.1016/j.neuron.2018.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan E., Cohen L. G. (2011). Neuroplasticity subserving motor skill learning. Neuron 72, 443–454. doi: 10.1016/j.neuron.2011.10.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippis B., Ricceri L., Laviola G. (2010). Early postnatal behavioral changes in the Mecp2-308 truncation mouse model of Rett syndrome. Genes Brain Behav. 9, 213–223. doi: 10.1111/j.1601-183X.2009.00551.x, PMID: [DOI] [PubMed] [Google Scholar]
- De Rubeis S., Siper P. M., Durkin A., Weissman J., Muratet F., Halpern D., et al. (2018). Delineation of the genetic and clinical spectrum of Phelan-McDermid syndrome caused by Shank3 point mutations. Mol. Autism. 9:31. doi: 10.1186/s13229-018-0205-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorey T. M., Handforth A., Anagnostaras S. G., Homanics G. E., Minassian B. A., Asatourian A., et al. (1998). Mice lacking the beta3 subunit of the Gabaa receptor have the epilepsy phenotype and many of the behavioral characteristics of Angelman syndrome. J. Neurosci. 18, 8505–8514. doi: 10.1523/JNEUROSCI.18-20-08505.1998, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorey T. M., Sahbaie P., Hashemi E., Li W. W., Salehi A., Clark D. J. (2011). Somatosensory and sensorimotor consequences associated with the heterozygous disruption of the autism candidate gene, Gabrb3. Behav. Brain Res. 216, 36–45. doi: 10.1016/j.bbr.2010.06.032, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depino A. M. (2013). Peripheral and central inflammation in autism spectrum disorders. Mol. Cell. Neurosci. 53, 69–76. doi: 10.1016/j.mcn.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Dey R., Chattarji S. (2022). The same stress elicits different effects on anxiety-like behavior in rat models of Fmr1(−/y) and Pten(+/). Behav. Brain Res. 428:113892. doi: 10.1016/j.bbr.2022.113892, PMID: [DOI] [PubMed] [Google Scholar]
- Dicarlo G. E., Aguilar J. I., Matthies H. J., Harrison F. E., Bundschuh K. E., West A., et al. (2019). Autism-linked dopamine transporter mutation alters striatal dopamine neurotransmission and dopamine-dependent behaviors. J. Clin. Invest. 129, 3407–3419. doi: 10.1172/JCI127411, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichter G. S. (2012). Functional magnetic resonance imaging of autism spectrum disorders. Dialogues Clin. Neurosci. 14, 319–351. doi: 10.31887/DCNS.2012.14.3/gdichter, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Peterson J. D., Surmeier D. J. (2008). Corticostriatal and thalamostriatal synapses have distinctive properties. J. Neurosci. 28, 6483–6492. doi: 10.1523/JNEUROSCI.0435-08.2008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig N. M., Moss J., Bolam J. P. (2010). Cortical and thalamic innervation of direct and indirect pathway medium-sized spiny neurons in mouse striatum. J. Neurosci. 30, 14610–14618. doi: 10.1523/JNEUROSCI.1623-10.2010, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Iturza N., Lo A. C., Shah D., Armendáriz M., Vannelli A., Mercaldo V., et al. (2019). The autism-and schizophrenia-associated protein Cyfip1 regulates bilateral brain connectivity and behaviour. Nat. Commun. 10:3454. doi: 10.1038/s41467-019-11203-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau E., Riad M., Kajiwara Y., Buxbaum J. D. (2018). Behavioral phenotyping of an improved mouse model of Phelan-McDermid syndrome with a complete deletion of the Shank3 gene. eNeuro 5, 1–55. doi: 10.1523/ENEURO.0046-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos A., Meziane H., Iacono G., Curie A., Riet F., Martin C., et al. (2018). A new mouse model of Arx dup24 recapitulates the patients’ behavioral and fine motor alterations. Hum. Mol. Genet. 27, 2138–2153. doi: 10.1093/hmg/ddy122, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham N. W., Miya T. S. (1957). A Note on a Simple Apparatus for Detecting Neurological Deficit in Rats and Mice**College of Pharmacy, University of Nebraska, Lincoln 8. J. Am. Pharm. Assoc. Am. Pharm. Assoc. 46, 208–209. doi: 10.1002/jps.3030460322 [DOI] [PubMed] [Google Scholar]
- Durieux P. F., Schiffmann S. N., De Kerchove D’exaerde A. (2012). Differential regulation of motor control and response to dopaminergic drugs by D1R and D2R neurons in distinct dorsal striatum subregions. EMBO J. 31, 640–653. doi: 10.1038/emboj.2011.400, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D. H., Gabel H. W., Robinson N. D., Kastan N. R., Hu L. S., Cohen S., et al. (2013). Activity-dependent phosphorylation of Mecp2 threonine 308 regulates interaction with NcoR. Nature 499, 341–345. doi: 10.1038/nature12348, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echevarria-Cooper D. M., Hawkins N. A., Misra S. N., Huffman A. M., Thaxton T., Thompson C. H., et al. (2022). Cellular and behavioral effects of altered NaV1.2 sodium channel ion permeability in Scn2aK1422E mice. Hum. Mol. Genet. 31, 2964–2988. doi: 10.1093/hmg/ddac087, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C., Suckling J., Deoni S. C., Lombardo M. V., Bullmore E. T., Baron-Cohen S., et al. (2012). Brain anatomy and its relationship to behavior in adults with autism spectrum disorder: a multicenter magnetic resonance imaging study. Arch. Gen. Psychiatry 69, 195–209. doi: 10.1001/archgenpsychiatry.2011.1251, PMID: [DOI] [PubMed] [Google Scholar]
- Ellegood J., Crawley J. N. (2015). Behavioral and neuroanatomical phenotypes in mouse models of Autism. Neurotherapeutics 12, 521–533. doi: 10.1007/s13311-015-0360-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegood J., Nakai N., Nakatani J., Henkelman M., Takumi T., Lerch J. (2015). Neuroanatomical phenotypes are consistent with Autism-like behavioral phenotypes in the 15q11-13 duplication mouse model. Autism Res. 8, 545–555. doi: 10.1002/aur.1469, PMID: [DOI] [PubMed] [Google Scholar]
- Ellenbroek B., Youn J. (2016). Rodent models in neuroscience research: is it a rat race? Dis. Model. Mech. 9, 1079–1087. doi: 10.1242/dmm.026120, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W., Gehre S., Hammerschmidt K., Holter S. M., Blass T., Somel M., et al. (2009). A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cells 137, 961–971. doi: 10.1016/j.cell.2009.03.041, PMID: [DOI] [PubMed] [Google Scholar]
- Estes A., Shaw D. W., Sparks B. F., Friedman S., Giedd J. N., Dawson G., et al. (2011). Basal ganglia morphometry and repetitive behavior in young children with autism spectrum disorder. Autism Res. 4, 212–220. doi: 10.1002/aur.193, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton M. R., Blaiss C. A., Powell C. M., Sudhof T. C. (2009). Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc. Natl. Acad. Sci. U. S. A. 106, 17998–18003. doi: 10.1073/pnas.0910297106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadila S., Quinn S., Turchetti Maia A., Yakubovich D., Ovadia M., Anderson K. L., et al. (2020). Convulsive seizures and some behavioral comorbidities are uncoupled in the Scn1a(A1783V) Dravet syndrome mouse model. Epilepsia 61, 2289–2300. doi: 10.1111/epi.16662, PMID: [DOI] [PubMed] [Google Scholar]
- Feyder M., Karlsson R. M., Mathur P., Lyman M., Bock R., Momenan R., et al. (2010). Association of mouse Dlg4 (Psd-95) gene deletion and human Dlg4 gene variation with phenotypes relevant to autism spectrum disorders and Williams’ syndrome. Am. J. Psychiatry 167, 1508–1517. doi: 10.1176/appi.ajp.2010.10040484, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C. A., Jin X., Campbell T. G., Gerfen E., Groszer M., Fisher S. E., et al. (2012). An aetiological Foxp2 mutation causes aberrant striatal activity and alters plasticity during skill learning. Mol. Psychiatry 17, 1077–1085. doi: 10.1038/mp.2011.105, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M., Yu X., Lu J., Zuo Y. (2012). Repetitive motor learning induces coordinated formation of clustered dendritic spines in vivo. Nature 483, 92–95. doi: 10.1038/nature10844, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuccillo M. V. (2016). Striatal circuits as a common node for Autism pathophysiology. Front. Neurosci. 10:27. doi: 10.3389/fnins.2016.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs C., Gennaccaro L., Trazzi S., Bastianini S., Bettini S., Lo Martire V., et al. (2018). Heterozygous Cdkl5 knockout female mice are a valuable animal model for Cdkl5 disorder. Neural Plast. 2018:9726950. doi: 10.1155/2018/9726950, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal M. J., Edgar J. C., Ehrlichman R. S., Mehta M., Roberts T. P., Siegel S. J. (2010). Validating gamma oscillations and delayed auditory responses as translational biomarkers of autism. Biol. Psychiatry 68, 1100–1106. doi: 10.1016/j.biopsych.2010.09.031, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi T., Lee C. C. (2020). Neural mechanisms underlying repetitive behaviors in rodent models of Autism Spectrum disorders. Front. Cell. Neurosci. 14:592710. doi: 10.3389/fncel.2020.592710, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Irvine E. E., Eleftheriadou I., Naranjo C. J., Hearn-Yeates F., Bosch L., et al. (2020). Gene replacement ameliorates deficits in mouse and human models of cyclin-dependent kinase-like 5 disorder. Brain 143, 811–832. doi: 10.1093/brain/awaa028, PMID: [DOI] [PubMed] [Google Scholar]
- Gerfen C. R., Surmeier D. J. (2011). Modulation of striatal projection systems by dopamine. Annu. Rev. Neurosci. 34, 441–466. doi: 10.1146/annurev-neuro-061010-113641, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin D., Allen M., Zhang L., Amorim M., Wang I. T., Reyes A. R., et al. (2011). Rett syndrome mutation Mecp2 T158A disrupts Dna binding, protein stability and Erp responses. Nat. Neurosci. 15, 274–283. doi: 10.1038/nn.2997, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D., Charman T., Pickles A., Chandler S., Loucas T., Simonoff E., et al. (2009). Impairment in movement skills of children with autistic spectrum disorders. Dev. Med. Child Neurol. 51, 311–316. doi: 10.1111/j.1469-8749.2008.03242.x, PMID: [DOI] [PubMed] [Google Scholar]
- Groszer M., Keays D. A., Deacon R. M., De Bono J. P., Prasad-Mulcare S., Gaub S., et al. (2008). Impaired synaptic plasticity and motor learning in mice with a point mutation implicated in human speech deficits. Curr. Biol. 18, 354–362. doi: 10.1016/j.cub.2008.01.060, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber S. N., Fudge J. L., Mcfarland N. R. (2000). Striatonigrostriatal pathways in primates form an ascending spiral from the shell to the dorsolateral striatum. J. Neurosci. 20, 2369–2382. doi: 10.1523/JNEUROSCI.20-06-02369.2000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton P. J., Campbell N. G., Sharma S., Erreger K., Herborg Hansen F., Saunders C., et al. (2013). De novo mutation in the dopamine transporter gene associates dopamine dysfunction with autism spectrum disorder. Mol. Psychiatry 18, 1315–1323. doi: 10.1038/mp.2013.102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMM R. J., PIKE B. R., O’DELL D. M., LYETH B. G., JENKINS L. W. (1994). The rotarod test: an evaluation of its effectiveness in assessing motor deficits following traumatic brain injury. J. Neurotrauma 11, 187–196. doi: 10.1089/neu.1994.11.187, PMID: [DOI] [PubMed] [Google Scholar]
- Hampson D. R., Blatt G. J. (2015). Autism spectrum disorders and neuropathology of the cerebellum. Front. Neurosci. 9:420. doi: 10.3389/fnins.2015.00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E., Myers H., Saxena K., Mitchell-Heggs R., Kind P., Chattarji S., et al. (2021). Experiential modulation of social dominance in a Syngap1 rat model of Autism Spectrum disorders. Eur. J. Neurosci. 54, 7733–7748. doi: 10.1111/ejn.15500, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi S., Doi H., Kunii M., Nakamura Y., Shimuta M., Suzuki E., et al. (2019). Ataxic phenotype with altered ca(V)3.1 channel property in a mouse model for spinocerebellar ataxia 42. Neurobiol. Dis. 130:104516: 104516. doi: 10.1016/j.nbd.2019.104516, PMID: [DOI] [PubMed] [Google Scholar]
- Haswell C. C., Izawa J., Dowell L. R., Mostofsky S. H., Shadmehr R. (2009). Representation of internal models of action in the autistic brain. Nat. Neurosci. 12, 970–972. doi: 10.1038/nn.2356, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes S. L., Evans R. C., Unruh B. A., Benkert E. E., Gillani F., Dumas T. C., et al. (2015). Multimodal plasticity in dorsal striatum while learning a lateralized navigation task. J. Neurosci. 35, 10535–10549. doi: 10.1523/JNEUROSCI.4415-14.2015, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck D. H., Zhao Y., Roy S., Ledoux M. S., Reiter L. T. (2008). Analysis of cerebellar function in Ube3a-deficient mice reveals novel genotype-specific behaviors. Hum. Mol. Genet. 17, 2181–2189. doi: 10.1093/hmg/ddn117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng M. Y., Detloff P. J., Albin R. L. (2008). Rodent genetic models of Huntington disease. Neurobiol. Dis. 32, 1–9. doi: 10.1016/j.nbd.2008.06.005, PMID: [DOI] [PubMed] [Google Scholar]
- Hernandez A., Delgado-Gonzalez E., Durairaj R. V., Reyes-Haro D., Martinez-Torres A., Espinosa F. (2023). Striatal synaptic changes and behavior in adult mouse upon prenatal exposure to valproic acid. Brain Res. 1815:148461: 148461. doi: 10.1016/j.brainres.2023.148461, PMID: [DOI] [PubMed] [Google Scholar]
- Hisaoka T., Komori T., Kitamura T., Morikawa Y. (2018). Abnormal behaviours relevant to neurodevelopmental disorders in Kirrel3-knockout mice. Sci. Rep. 8:1408. doi: 10.1038/s41598-018-19844-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander E., Anagnostou E., Chaplin W., Esposito K., Haznedar M. M., Licalzi E., et al. (2005). Striatal volume on magnetic resonance imaging and repetitive behaviors in autism. Biol. Psychiatry 58, 226–232. doi: 10.1016/j.biopsych.2005.03.040, PMID: [DOI] [PubMed] [Google Scholar]
- Huang H. S., Burns A. J., Nonneman R. J., Baker L. K., Riddick N. V., Nikolova V. D., et al. (2013). Behavioral deficits in an Angelman syndrome model: effects of genetic background and age. Behav. Brain Res. 243, 79–90. doi: 10.1016/j.bbr.2012.12.052, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert S. W., Wang X., Gbadegesin S. O., Xu Q., Xu X., Jiang Y. H. (2020). A novel Chd8 mutant mouse displays altered ultrasonic vocalizations and enhanced motor coordination. Autism Res. 13, 1685–1697. doi: 10.1002/aur.2353, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung A. Y., Futai K., Sala C., Valtschanoff J. G., Ryu J., Woodworth M. A., et al. (2008). Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J. Neurosci. 28, 1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Satake S., Onaka T., Sugimoto H., Takeda N., Imoto K., et al. (2013). Enhanced inhibitory neurotransmission in the cerebellar cortex of Atp1a3-deficient heterozygous mice. J. Physiol. 591, 3433–3449. doi: 10.1113/jphysiol.2012.247817, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Ogiwara I., Yamada K., Miyamoto H., Hensch T. K., Osawa M., et al. (2013). Mouse with Nav1.1 haploinsufficiency, a model for Dravet syndrome, exhibits lowered sociability and learning impairment. Neurobiol. Dis. 49, 29–40. doi: 10.1016/j.nbd.2012.08.003, PMID: [DOI] [PubMed] [Google Scholar]
- Jaramillo T. C., Speed H. E., Xuan Z., Reimers J. M., Escamilla C. O., Weaver T. P., et al. (2017). Novel Shank3 mutant exhibits behaviors with face validity for autism and altered striatal and hippocampal function. Autism Res. 10, 42–65. doi: 10.1002/aur.1664, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhang C. L., Huang T. N., Hsueh Y. P., Liao W. (2017). Mice lacking cyclin-dependent kinase-like 5 manifest autistic and Adhd-like behaviors. Hum. Mol. Genet. 26, 3922–3934. doi: 10.1093/hmg/ddx279, PMID: [DOI] [PubMed] [Google Scholar]
- Jiang Y. H., Armstrong D., Albrecht U., Atkins C. M., Noebels J. L., Eichele G., et al. (1998). Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 21, 799–811. doi: 10.1016/S0896-6273(00)80596-6, PMID: [DOI] [PubMed] [Google Scholar]
- Jin P., Warren S. T. (2003). New insights into fragile X syndrome: from molecules to neurobehaviors. Trends Biochem. Sci. 28, 152–158. doi: 10.1016/S0968-0004(03)00033-1, PMID: [DOI] [PubMed] [Google Scholar]
- Jung E. M., Moffat J. J., Liu J., Dravid S. M., Gurumurthy C. B., Kim W. Y. (2017). Arid1b haploinsufficiency disrupts cortical interneuron development and mouse behavior. Nat. Neurosci. 20, 1694–1707. doi: 10.1038/s41593-017-0013-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K. M., Sepers M., Henstridge C. M., Lassalle O., Neuhofer D., Martin H., et al. (2012). Uncoupling of the endocannabinoid signalling complex in a mouse model of fragile X syndrome. Nat. Commun. 3:1080. doi: 10.1038/ncomms2045, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser F. M. P., Gruenbacher S., Oyaga M. R., Nio E., Jaritz M., Sun Q., et al. (2022). Biallelic Pax5 mutations cause hypogammaglobulinemia, sensorimotor deficits, and autism spectrum disorder. J. Exp. Med. 219:e20220498. doi: 10.1084/jem.20220498, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao F. C., Su S. H., Carlson G. C., Liao W. (2015). Mecp2-mediated alterations of striatal features accompany psychomotor deficits in a mouse model of Rett syndrome. Brain Struct. Funct. 220, 419–434. doi: 10.1007/s00429-013-0664-x, PMID: [DOI] [PubMed] [Google Scholar]
- Kawamura A., Katayama Y., Kakegawa W., Ino D., Nishiyama M., Yuzaki M., et al. (2021). The autism-associated protein Chd8 is required for cerebellar development and motor function. Cell Rep. 35:108932. doi: 10.1016/j.celrep.2021.108932, PMID: [DOI] [PubMed] [Google Scholar]
- Kosillo P., Bateup H. S. (2021). Dopaminergic dysregulation in syndromic Autism Spectrum disorders: insights from genetic mouse models. Front Neural Circuits 15:700968. doi: 10.3389/fncir.2021.700968, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouser M., Speed H. E., Dewey C. M., Reimers J. M., Widman A. J., Gupta N., et al. (2013). Loss of predominant Shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. J. Neurosci. 33, 18448–18468. doi: 10.1523/JNEUROSCI.3017-13.2013, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz A. V., Freeze B. S., Parker P. R., Kay K., Thwin M. T., Deisseroth K., et al. (2010). Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature 466, 622–626. doi: 10.1038/nature09159, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitzer A. C., Malenka R. C. (2008). Striatal plasticity and basal ganglia circuit function. Neuron 60, 543–554. doi: 10.1016/j.neuron.2008.11.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt D. A., Augustin S. M., Johnson K. A., Lovinger D. M. (2019). Active zone proteins Rim1alphabeta are required for Normal Corticostriatal transmission and action control. J. Neurosci. 39, 1457–1470. doi: 10.1523/JNEUROSCI.1940-18.2018, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C. H., Luikart B. W., Powell C. M., Zhou J., Matheny S. A., Zhang W., et al. (2006). Pten regulates neuronal arborization and social interaction in mice. Neuron 50, 377–388. doi: 10.1016/j.neuron.2006.03.023, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J. K., Lerch J. P., Doering L. C., Foster J. A., Ellegood J. (2016). Regional brain volumes changes in adult male Fmr1-Ko mouse on the Fvb strain. Neuroscience 318, 12–21. doi: 10.1016/j.neuroscience.2016.01.021, PMID: [DOI] [PubMed] [Google Scholar]
- Lalonde R., Hayzoun K., Derer M., Mariani J., Strazielle C. (2004). Neurobehavioral evaluation of Reln-rl-orl mutant mice and correlations with cytochrome oxidase activity. Neurosci. Res. 49, 297–305. doi: 10.1016/j.neures.2004.03.012, PMID: [DOI] [PubMed] [Google Scholar]
- Landi S., Putignano E., Boggio E. M., Giustetto M., Pizzorusso T., Ratto G. M. (2011). The short-time structural plasticity of dendritic spines is altered in a model of Rett syndrome. Sci. Rep. 1:45. doi: 10.1038/srep00045, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Duc D., Giulivi C., Hiatt S. M., Napoli E., Panoutsopoulos A., Harlan De Crescenzo A., et al. (2019). Pathogenic Wdfy3 variants cause neurodevelopmental disorders and opposing effects on brain size. Brain 142, 2617–2630. doi: 10.1093/brain/awz198, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Merrer J., Detraux B., Gandía J., de Groote A., Fonteneau M., de Kerchove d’Exaerde A., et al. (2023). Balance between projecting neuronal populations of the nucleus Accumbens controls social behavior in mice. Biol. Psychiatry. doi: 10.1016/j.biopsych.2023.05.008 [DOI] [PubMed] [Google Scholar]
- Leach P. T., Crawley J. N. (2018). Touchscreen learning deficits in Ube3a, Ts65Dn and Mecp2 mouse models of neurodevelopmental disorders with intellectual disabilities. Genes Brain Behav. 17:e12452. doi: 10.1111/gbb.12452, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena I., Mantegazza M. (2019). Na(V)1.2 haploinsufficiency in Scn2a knock-out mice causes an autistic-like phenotype attenuated with age. Sci. Rep. 9:12886. doi: 10.1038/s41598-019-49392-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Pozzo-Miller L. (2020). Dysfunction of the corticostriatal pathway in autism spectrum disorders. J. Neurosci. Res. 98, 2130–2147. doi: 10.1002/jnr.24560, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. J., Zhang K., Sun T., Guo Y. Y., Yang Q., Liu S. B., et al. (2023). Improvement of learning and memory by elevating brain D-aspartate in a mouse model of fragile X syndrome. Mol. Neurobiol. 60, 6410–6423. doi: 10.1007/s12035-023-03438-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Nanclares C., Simbriger K., Fang K., Lorsung E., Le N., et al. (2022). Autistic-like behavior and cerebellar dysfunction in Bmal1 mutant mice ameliorated by mtorc1 inhibition. Mol. Psychiatry. doi: 10.1038/s41380-022-01499-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo F., Klann E. (2021). Reciprocal control of translation and transcription in autism spectrum disorder. EMBO Rep. 22:e52110. doi: 10.15252/embr.202052110, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovinger D. M. (2010). Neurotransmitter roles in synaptic modulation, plasticity and learning in the dorsal striatum. Neuropharmacology 58, 951–961. doi: 10.1016/j.neuropharm.2010.01.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubrich C., Giesler P., Kipp M. (2022). Motor behavioral deficits in the Cuprizone model: validity of the Rotarod test paradigm. Int. J. Mol. Sci. 23:11342. doi: 10.3390/ijms231911342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luft A. R., Buitrago M. M. (2005). Stages of motor skill learning. Mol. Neurobiol. 32, 205–216. doi: 10.1385/MN:32:3:205 [DOI] [PubMed] [Google Scholar]
- Lynch J. F., 3rd, Ferri S. L., Angelakos C., Schoch H., Nickl-Jockschat T., Gonzalez A., et al. (2020). Comprehensive behavioral phenotyping of a 16p11.2 Del mouse model for neurodevelopmental disorders. Autism Res. 13, 1670–1684. doi: 10.1002/aur.2357, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst M. J., Ekiert R., Ebert D. H., Merusi C., Nowak J., Selfridge J., et al. (2013). Rett syndrome mutations abolish the interaction of Mecp2 with the NcoR/Smrt co-repressor. Nat. Neurosci. 16, 898–902. doi: 10.1038/nn.3434, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Day-Cooney J., Benavides O. J., Muniak M. A., Qin M., Ding J. B., et al. (2022). Locomotion activates Pka through dopamine and adenosine in striatal neurons. Nature 611, 762–768. doi: 10.1038/s41586-022-05407-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarrone M., Rossi S., Bari M., De Chiara V., Rapino C., Musella A., et al. (2010). Abnormal mGlu 5 receptor/endocannabinoid coupling in mice lacking Fmrp and Bc1 Rna. Neuropsychopharmacology 35, 1500–1509. doi: 10.1038/npp.2010.19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner M. J., Warren Z., Williams A. R., Amoakohene E., Bakian A. V., Bilder D. A., et al. (2023). Prevalence and characteristics of Autism Spectrum disorder among children aged 8 years — Autism and developmental disabilities monitoring network, 11 sites, United States, 2020. MMWR Surveill. Summ. 72, 1–14. doi: 10.15585/mmwr.ss7202a1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. H., Langley S. A., Huang Y., Hang M., Bouchard K. E., Celniker S. E., et al. (2015). Identification of genetic factors that modify motor performance and body weight using collaborative Cross mice. Sci. Rep. 5:16247. doi: 10.1038/srep16247, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone M. C., Marinelli S., Biamonte F., Keller F., Sgobio C. A., Ammassari-Teule M., et al. (2006). Altered cortico-striatal synaptic plasticity and related behavioural impairments in reeler mice. Eur. J. Neurosci. 24, 2061–2070. doi: 10.1111/j.1460-9568.2006.05083.x, PMID: [DOI] [PubMed] [Google Scholar]
- Martella G., Meringolo M., Trobiani L., De Jaco A., Pisani A., Bonsi P. (2018). The neurobiological bases of autism spectrum disorders: the R451C-neuroligin 3 mutation hampers the expression of long-term synaptic depression in the dorsal striatum. Eur. J. Neurosci. 47, 701–708. doi: 10.1111/ejn.13705, PMID: [DOI] [PubMed] [Google Scholar]
- Martin E. A., Muralidhar S., Wang Z., Cervantes D. C., Basu R., Taylor M. R., et al. (2015). The intellectual disability gene Kirrel3 regulates target-specific mossy fiber synapse development in the hippocampus. elife 4:e09395: e09395. doi: 10.7554/eLife.09395, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A., Mamidanna P., Cury K. M., Abe T., Murthy V. N., Mathis M. W., et al. (2018). DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289. doi: 10.1038/s41593-018-0209-y, PMID: [DOI] [PubMed] [Google Scholar]
- Mei Y., Monteiro P., Zhou Y., Kim J. A., Gao X., Fu Z., et al. (2016). Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature 530, 481–484. doi: 10.1038/nature16971, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer A., Van De Water J. (2017). The role of the immune system in Autism Spectrum disorder. Neuropsychopharmacology 42, 284–298. doi: 10.1038/npp.2016.158, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miljanovic N., Hauck S. M., Van Dijk R. M., Di Liberto V., Rezaei A., Potschka H. (2021). Proteomic signature of the Dravet syndrome in the genetic Scn1a-A1783V mouse model. Neurobiol. Dis. 157:105423: 105423. doi: 10.1016/j.nbd.2021.105423, PMID: [DOI] [PubMed] [Google Scholar]
- Minshew N. J., Sung K., Jones B. L., Furman J. M. (2004). Underdevelopment of the postural control system in autism. Neurology 63, 2056–2061. doi: 10.1212/01.WNL.0000145771.98657.62, PMID: [DOI] [PubMed] [Google Scholar]
- Miura K., Kishino T., Li E., Webber H., Dikkes P., Holmes G. L., et al. (2002). Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol. Dis. 9, 149–159. doi: 10.1006/nbdi.2001.0463, PMID: [DOI] [PubMed] [Google Scholar]
- Morello N., Schina R., Pilotto F., Phillips M., Melani R., Plicato O., et al. (2018). Loss of Mecp2 causes atypical synaptic and molecular plasticity of Parvalbumin-expressing interneurons reflecting Rett syndrome-like sensorimotor defects. eNeuro 5, 1–19. doi: 10.1523/ENEURO.0086-18.2018, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy S. S., Nadler J. J., Young N. B., Perez A., Holloway L. P., Barbaro R. P., et al. (2007). Mouse behavioral tasks relevant to autism: phenotypes of 10 inbred strains. Behav. Brain Res. 176, 4–20. doi: 10.1016/j.bbr.2006.07.030, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhia M., Yee B. K., Feldon J., Markopoulos F., Knuesel I. (2010). Disruption of hippocampus-regulated behavioural and cognitive processes by heterozygous constitutive deletion of Syngap. Eur. J. Neurosci. 31, 529–543. doi: 10.1111/j.1460-9568.2010.07079.x, PMID: [DOI] [PubMed] [Google Scholar]
- Mulherkar S. A., Jana N. R. (2010). Loss of dopaminergic neurons and resulting behavioural deficits in mouse model of Angelman syndrome. Neurobiol. Dis. 40, 586–592. doi: 10.1016/j.nbd.2010.08.002, PMID: [DOI] [PubMed] [Google Scholar]
- Na E. S., Nelson E. D., Adachi M., Autry A. E., Mahgoub M. A., Kavalali E. T., et al. (2012). A Mouse Model forMeCP2Duplication Syndrome: MeCP2 Overexpression Impairs Learning and Memory and Synaptic Transmission. J. Neurosci. 32, 3109–3117. doi: 10.1523/JNEUROSCI.6000-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima R., Takao K., Hattori S., Shoji H., Komiyama N. H., Grant S. G. N., et al. (2019). Comprehensive behavioral analysis of heterozygous Syngap1 knockout mice. Neuropsychopharmacol Rep 39, 223–237. doi: 10.1002/npr2.12073, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Arima-Yoshida F., Sakaue F., Nasu-Nishimura Y., Takeda Y., Matsuura K., et al. (2016). Px-Rics-deficient mice mimic autism spectrum disorder in Jacobsen syndrome through impaired Gabaa receptor trafficking. Nat. Commun. 7:10861. doi: 10.1038/ncomms10861, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani J., Tamada K., Hatanaka F., Ise S., Ohta H., Inoue K., et al. (2009). Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell(Cambridge,Mass) 137, 1235–1246. doi: 10.1016/j.cell.2009.04.024, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux J. C., Schuchbauer M. A., Li K., Wang L., Risbrough V. B., Powell S. B., et al. (2014). Reversal of autism-like behaviors and metabolism in adult mice with single-dose antipurinergic therapy. Transl. Psychiatry 4:e400. doi: 10.1038/tp.2014.33, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale B. M., Kou Y., Liu L., Ma’ayan A., Samocha K. E., Sabo A., et al. (2012). Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245. doi: 10.1038/nature11011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler E. J., Hyman S. E. (2010). Animal models of neuropsychiatric disorders. Nat. Neurosci. 13, 1161–1169. doi: 10.1038/nn.2647, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu M., Zheng N., Wang Z., Gao Y., Luo X., Chen Z., et al. (2020). Rab39B deficiency impairs learning and memory partially through compromising autophagy. Front. Cell Dev. Biol. 8:598622. doi: 10.3389/fcell.2020.598622, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan S. O., Reynolds C. D., Smith G. D., Holley A. J., Escobar B., Chandler M. A., et al. (2017). Deletion of Fmr1 results in sex-specific changes in behavior. Brain Behav. 7:e00800. doi: 10.1002/brb3.800, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette J., Toussay X., Comin C. H., Costa L. D. F., Ho M., Lacalle-Aurioles M., et al. (2020). Vascular contributions to 16p11.2 deletion autism syndrome modeled in mice. Nat. Neurosci. 23, 1090–1101. doi: 10.1038/s41593-020-0663-1, PMID: [DOI] [PubMed] [Google Scholar]
- Packard M. G., Knowlton B. J. (2002). Learning and memory functions of the basal ganglia. Annu. Rev. Neurosci. 25, 563–593. doi: 10.1146/annurev.neuro.25.112701.142937, PMID: [DOI] [PubMed] [Google Scholar]
- Peixoto R. T., Chantranupong L., Hakim R., Levasseur J., Wang W., Merchant T., et al. (2019). Abnormal striatal development underlies the early onset of behavioral deficits in Shank3B(−/−) mice. Cell Rep. 29:e4. doi: 10.1016/j.celrep.2019.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka G. J., Watson C. M., Radziewic T., Hayward M., Lahooti H., Christodoulou J., et al. (2006). Mecp2 deficiency is associated with learning and cognitive deficits and altered gene activity in the hippocampal region of mice. Brain 129, 887–898. doi: 10.1093/brain/awl022, PMID: [DOI] [PubMed] [Google Scholar]
- Penagarikano O., Abrahams B. S., Herman E. I., Winden K. D., Gdalyahu A., Dong H., et al. (2011). Absence of Cntnap2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell(Cambridge,Mass) 147, 235–246. doi: 10.1016/j.cell.2011.08.040, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piochon C., Kloth A. D., Grasselli G., Titley H. K., Nakayama H., Hashimoto K., et al. (2014). Cerebellar plasticity and motor learning deficits in a copy-number variation mouse model of autism. Nat. Commun. 5:5586. doi: 10.1038/ncomms6586, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger C., Fasano S., Mazzocchi-Jones D., Dunnett S. B., Kandel E. R., Brambilla R. (2006). Impaired bidirectional synaptic plasticity and procedural memory formation in striatum-specific camp response element-binding protein-deficient mice. J. Neurosci. 26, 2808–2813. doi: 10.1523/JNEUROSCI.5406-05.2006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt R. J., Zhou Y., Slaymaker I. M., Shetty A. S., Weisbach N. R., Kim J. A., et al. (2017). Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep. 19, 335–350. doi: 10.1016/j.celrep.2017.03.052, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portmann T., Yang M., Mao R., Panagiotakos G., Ellegood J., Dolen G., et al. (2014). Behavioral abnormalities and circuit defects in the basal ganglia of a mouse model of 16p11.2 deletion syndrome. Cell Rep. 7, 1077–1092. doi: 10.1016/j.celrep.2014.03.036, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratte M., Panayotis N., Ghata A., Villard L., Roux J. C. (2011). Progressive motor and respiratory metabolism deficits in post-weaning Mecp2-null male mice. Behav. Brain Res. 216, 313–320. doi: 10.1016/j.bbr.2010.08.011, PMID: [DOI] [PubMed] [Google Scholar]
- Price M. G., Yoo J. W., Burgess D. L., Deng F., Hrachovy R. A., Frost J. D., Jr., et al. (2009). A triplet repeat expansion genetic mouse model of infantile spasms syndrome, Arx(Gcg)10+7, with interneuronopathy, spasms in infancy, persistent seizures, and adult cognitive and behavioral impairment. J. Neurosci. 29, 8752–8763. doi: 10.1523/JNEUROSCI.0915-09.2009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt A. M., Judson M. C., Sidorov M. S., Williams B. N., Johnson N. S., Belder S., et al. (2022). Molecular and behavioral consequences of Ube3a gene overdosage in mice. JCI Insight 7:e158953. doi: 10.1172/jci.insight.158953, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z. (2018). Deciphering Mecp2-associated disorders: disrupted circuits and the hope for repair. Curr. Opin. Neurobiol. 48, 30–36. doi: 10.1016/j.conb.2017.09.004, PMID: [DOI] [PubMed] [Google Scholar]
- Rahman F. U., Kim Y. R., Kim E. K., Kim H. R., Cho S. M., Lee C. S., et al. (2021). Topoisomerase IIIbeta deficiency induces neuro-behavioral changes and brain connectivity alterations in mice. Int. J. Mol. Sci. 22:12806. doi: 10.3390/ijms222312806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith R. M., Mckenna J., Wu H., Hashmi S. S., Cho S. H., Dash P. K., et al. (2013). Loss of Tsc2 in Purkinje cells is associated with autistic-like behavior in a mouse model of tuberous sclerosis complex. Neurobiol. Dis. 51, 93–103. doi: 10.1016/j.nbd.2012.10.014, PMID: [DOI] [PubMed] [Google Scholar]
- Ricard G., Molina J., Chrast J., Gu W., Gheldof N., Pradervand S., et al. (2010). Phenotypic consequences of copy number variation: insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol. 8:e1000543. doi: 10.1371/journal.pbio.1000543, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricobaraza A., Mora-Jimenez L., Puerta E., Sanchez-Carpintero R., Mingorance A., Artieda J., et al. (2019). Epilepsy and neuropsychiatric comorbidities in mice carrying a recurrent Dravet syndrome Scn1A missense mutation. Sci. Rep. 9:14172. doi: 10.1038/s41598-019-50627-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotaru D. C., Mientjes E. J., Elgersma Y. (2020). Angelman syndrome: from mouse models to therapy. Neuroscience 445, 172–189. doi: 10.1016/j.neuroscience.2020.02.017 [DOI] [PubMed] [Google Scholar]
- Rothwell P. E., Fuccillo M. V., Maxeiner S., Hayton S. J., Gokce O., Lim B. K., et al. (2014). Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cells 158, 198–212. doi: 10.1016/j.cell.2014.04.045, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roullet F. I., Lai J. K., Foster J. A. (2013). In utero exposure to valproic acid and autism--a current review of clinical and animal studies. Neurotoxicol. Teratol. 36, 47–56. doi: 10.1016/j.ntt.2013.01.004 [DOI] [PubMed] [Google Scholar]
- Roy S., Zhao Y., Allensworth M., Farook M. F., Ledoux M. S., Reiter L. T., et al. (2011). Comprehensive motor testing in Fmr1-Ko mice exposes temporal defects in oromotor coordination. Behav. Neurosci. 125, 962–969. doi: 10.1037/a0025920, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K., Shoji H., Kohno T., Miyakawa T., Hattori M. (2016). Mice that lack the C-terminal region of Reelin exhibit behavioral abnormalities related to neuropsychiatric disorders. Sci. Rep. 6:28636. doi: 10.1038/srep28636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin S., Lichtenstein P., Kuja-Halkola R., Hultman C., Larsson H., Reichenberg A. (2017). The heritability of Autism Spectrum disorder. JAMA 318, 1182–1184. doi: 10.1001/jama.2017.12141, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F. J., Oliveira R. F., Jin X., Costa R. M. (2015). Corticostriatal dynamics encode the refinement of specific behavioral variability during skill learning. elife 4:e09423. doi: 10.7554/eLife.09423, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M., Silva-Fernandes A., Oliveira P., Sousa N., Maciel P. (2007). Evidence for abnormal early development in a mouse model of Rett syndrome. Genes Brain Behav. 6, 277–286. doi: 10.1111/j.1601-183X.2006.00258.x, PMID: [DOI] [PubMed] [Google Scholar]
- Sathyamurthy A., Barik A., Dobrott C. I., Matson K. J. E., Stoica S., Pursley R., et al. (2020). Cerebellospinal neurons regulate motor performance and motor learning. Cell Rep. 31:107595. doi: 10.1016/j.celrep.2020.107595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom F. K., Kosmicki J. A., Wang J., Breen M. S., De Rubeis S., An J.-Y., et al. (2020). Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of Autism. Cells 180, 568–584.e23. doi: 10.1016/j.cell.2019.12.036, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R. A., Sabatini D. M. (2017). Mtor signaling in growth, metabolism, and disease. Cells 168, 960–976. doi: 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandaglia M., Lopez-Atalaya J. P., Medrano-Fernandez A., Lopez-Cascales M. T., Del Blanco B., Lipinski M., et al. (2017). Loss of Kdm5c causes spurious transcription and prevents the fine-tuning of activity-regulated enhancers in neurons. Cell Rep. 21, 47–59. doi: 10.1016/j.celrep.2017.09.014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaevitz L. R., Gomez N. B., Zhen D. P., Berger-Sweeney J. E. (2013). Mecp2 R168X male and female mutant mice exhibit Rett-like behavioral deficits. Genes Brain Behav. 12, 732–740. doi: 10.1111/gbb.12070, PMID: [DOI] [PubMed] [Google Scholar]
- Schiavi S., Manduca A., Carbone E., Buzzelli V., Rava A., Feo A., et al. (2023). Anandamide and 2-arachidonoylglycerol differentially modulate autistic-like traits in a genetic model of autism based on Fmr1 deletion in rats. Neuropsychopharmacology 48, 897–907. doi: 10.1038/s41386-022-01454-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz Y., Schmauss C., Sulzer D. (2002). Altered dopamine release and uptake kinetics in mice lacking D2 receptors. J. Neurosci. 22, 8002–8009. doi: 10.1523/JNEUROSCI.22-18-08002.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann C. M., Nordahl C. W. (2011). Bridging the gap between Mri and postmortem research in autism. Brain Res. 1380, 175–186. doi: 10.1016/j.brainres.2010.09.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani M., Horii T., Shoji H., Morita S., Kimura M., Terawaki N., et al. (2017). Arid1b Haploinsufficiency causes abnormal brain gene expression and Autism-related behaviors in mice. Int. J. Mol. Sci. 18:1872. doi: 10.3390/ijms18091872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett M. W., Gavin M., Tran T. S. (2015). Altered hippocampal-dependent memory and motor function in neuropilin 2-deficient mice. Transl. Psychiatry 5:e521. doi: 10.1038/tp.2015.17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Santi A., Huang S. (2021). Conditional Pten knockout in parvalbumin-or somatostatin-positive neurons sufficiently leads to autism-related behavioral phenotypes. Mol. Brain 14:24. doi: 10.1186/s13041-021-00731-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotsuki H., Yoshimi K., Shimo Y., Funayama M., Takamatsu Y., Ikeda K., et al. (2010). A rotarod test for evaluation of motor skill learning. J. Neurosci. Methods 189, 180–185. doi: 10.1016/j.jneumeth.2010.03.026, PMID: [DOI] [PubMed] [Google Scholar]
- Silverman J. L., Turner S. M., Barkan C. L., Tolu S. S., Saxena R., Hung A. Y., et al. (2011). Sociability and motor functions in Shank1 mutant mice. Brain Res. 1380, 120–137. doi: 10.1016/j.brainres.2010.09.026, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D. H., Titley H. K., Hansel C., Mason P. (2021). Behavioral tests for mouse models of Autism: An argument for the inclusion of cerebellum-controlled motor behaviors. Neuroscience 462, 303–319. doi: 10.1016/j.neuroscience.2020.05.010 [DOI] [PubMed] [Google Scholar]
- Sobue A., Kushima I., Nagai T., Shan W., Kohno T., Aleksic B., et al. (2018). Genetic and animal model analyses reveal the pathogenic role of a novel deletion of Reln in schizophrenia. Sci. Rep. 8:13046. doi: 10.1038/s41598-018-31390-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonzogni M., Wallaard I., Santos S. S., Kingma J., Du Mee D., Van Woerden G. M., et al. (2018). A behavioral test battery for mouse models of Angelman syndrome: a powerful tool for testing drugs and novel Ube3a mutants. Mol. Autism. 9:47. doi: 10.1186/s13229-018-0231-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souchet B., Guedj F., Sahun I., Duchon A., Daubigney F., Badel A., et al. (2014). Excitation/inhibition balance and learning are modified by Dyrk1a gene dosage. Neurobiol. Dis. 69, 65–75. doi: 10.1016/j.nbd.2014.04.016 [DOI] [PubMed] [Google Scholar]
- Speed H. E., Kouser M., Xuan Z., Reimers J. M., Ochoa C. F., Gupta N., et al. (2015). Autism-associated insertion mutation (InsG) of Shank3 exon 21 causes impaired synaptic transmission and behavioral deficits. J. Neurosci. 35, 9648–9665. doi: 10.1523/JNEUROSCI.3125-14.2015, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syding L. A., Kubik-Zahorodna A., Nickl P., Novosadova V., Kopkanova J., Kasparek P., et al. (2022). Generation and characterization of a novel Angelman syndrome mouse model with a full deletion of the Ube3a gene. Cells 11:2815. doi: 10.3390/cells11182815, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztainberg Y., Chen H. M., Swann J. W., Hao S., Tang B., Wu Z., et al. (2015). Reversal of phenotypes in Mecp2 duplication mice using genetic rescue or antisense oligonucleotides. Nature 528, 123–126. doi: 10.1038/nature16159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai L. H., Lee A. M., Benavidez N., Bonci A., Wilbrecht L. (2012). Transient stimulation of distinct subpopulations of striatal neurons mimics changes in action value. Nat. Neurosci. 15, 1281–1289. doi: 10.1038/nn.3188, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi Y., Fujita E., Yu Z., Yamagata T., Momoi M. Y., Momoi T., et al. (2010). Impairment of social and emotional behaviors in Cadm1-knockout mice. Biochem. Biophys. Res. Commun. 396, 703–708. doi: 10.1016/j.bbrc.2010.04.165, PMID: [DOI] [PubMed] [Google Scholar]
- Takumi T., Tamada K. (2018). CNV biology in neurodevelopmental disorders. Curr. Opin. Neurobiol. 48, 183–192. doi: 10.1016/j.conb.2017.12.004 [DOI] [PubMed] [Google Scholar]
- Talantseva O. I., Romanova R. S., Shurdova E. M., Dolgorukova T. A., Sologub P. S., Titova O. S., et al. (2023). The global prevalence of autism spectrum disorder: a three-level meta-analysis. Front. Psych. 14:1071181. doi: 10.3389/fpsyt.2023.1071181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsukawa T., Raveau M., Ogiwara I., Hattori S., Miyamoto H., Mazaki E., et al. (2019). Scn2a haploinsufficient mice display a spectrum of phenotypes affecting anxiety, sociability, memory flexibility and ampakine Cx516 rescues their hyperactivity. Mol. Autism. 10:15. doi: 10.1186/s13229-019-0265-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A. M., Schwartz M. D., Saxe M. D., Kilduff T. S. (2017). Sleep/wake physiology and quantitative electroencephalogram analysis of the Neuroligin-3 knockout rat model of Autism Spectrum disorder. Sleep 40, 1–11. doi: 10.1093/sleep/zsx138, PMID: [DOI] [PubMed] [Google Scholar]
- Till S. M., Asiminas A., Jackson A. D., Katsanevaki D., Barnes S. A., Osterweil E. K., et al. (2015). Conserved hippocampal cellular pathophysiology but distinct behavioural deficits in a new rat model of Fxs. Hum. Mol. Genet. 24, 5977–5984. doi: 10.1093/hmg/ddv299, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilot A. K., Gaugler M. K., Yu Q., Romigh T., Yu W., Miller R. H., et al. (2014). Germline disruption of Pten localization causes enhanced sex-dependent social motivation and increased glial production. Hum. Mol. Genet. 23, 3212–3227. doi: 10.1093/hmg/ddu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers B. G., Powell P. S., Klinger L. G., Klinger M. R. (2013). Motor difficulties in autism spectrum disorder: linking symptom severity and postural stability. J. Autism Dev. Disord. 43, 1568–1583. doi: 10.1007/s10803-012-1702-x, PMID: [DOI] [PubMed] [Google Scholar]
- Tritsch N. X., Sabatini B. L. (2012). Dopaminergic modulation of synaptic transmission in cortex and striatum. Neuron 76, 33–50. doi: 10.1016/j.neuron.2012.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropea D., Giacometti E., Wilson N. R., Beard C., Mccurry C., Fu D. D., et al. (2009). Partial reversal of Rett syndrome-like symptoms in Mecp2 mutant mice. Proc. Natl. Acad. Sci. U. S. A. 106, 2029–2034. doi: 10.1073/pnas.0812394106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyb E., Knoch K., Herlihy L., Stevens M. C., Chen C. M., Barton M., et al. (2016). Restricted and repetitive behaviors as predictors of outcome in Autism Spectrum disorders. J. Autism Dev. Disord. 46, 1282–1296. doi: 10.1007/s10803-015-2668-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P. T., Hull C., Chu Y., Greene-Colozzi E., Sadowski A. R., Leech J. M., et al. (2012). Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature 488, 647–651. doi: 10.1038/nature11310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci V., Kleefstra T., Hardy A., Heise I., Maggi S., Willemsen M. H., et al. (2014). Dominant beta-catenin mutations cause intellectual disability with recognizable syndromic features. J. Clin. Invest. 124, 1468–1482. doi: 10.1172/JCI70372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um S. M., Ha S., Lee H., Kim J., Kim K., Shin W., et al. (2018). Ngl-2 deletion leads to autistic-like behaviors responsive to Nmdar modulation. Cell Rep. 23, 3839–3851. doi: 10.1016/j.celrep.2018.05.087, PMID: [DOI] [PubMed] [Google Scholar]
- Ung D. C., Iacono G., Meziane H., Blanchard E., Papon M. A., Selten M., et al. (2018). Ptchd1 deficiency induces excitatory synaptic and cognitive dysfunctions in mouse. Mol. Psychiatry 23, 1356–1367. doi: 10.1038/mp.2017.39, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uutela M., Lindholm J., Louhivuori V., Wei H., Louhivuori L. M., Pertovaara A., et al. (2012). Reduction of Bdnf expression in Fmr1 knockout mice worsens cognitive deficits but improves hyperactivity and sensorimotor deficits. Genes Brain Behav. 11, 513–523. doi: 10.1111/j.1601-183X.2012.00784.x [DOI] [PubMed] [Google Scholar]
- Van Dam D., Errijgers V., Kooy R. F., Willemsen R., Mientjes E., Oostra B. A., et al. (2005). Cognitive decline, neuromotor and behavioural disturbances in a mouse model for fragile-X-associated tremor/ataxia syndrome (Fxtas). Behav. Brain Res. 162, 233–239. doi: 10.1016/j.bbr.2005.03.007, PMID: [DOI] [PubMed] [Google Scholar]
- Van Der Vaart T., Van Woerden G. M., Elgersma Y., De Zeeuw C. I., Schonewille M. (2011). Motor deficits in neurofibromatosis type 1 mice: the role of the cerebellum. Genes Brain Behav. 10, 404–409. doi: 10.1111/j.1601-183X.2011.00685.x, PMID: [DOI] [PubMed] [Google Scholar]
- Vernazza-Martin S., Martin N., Vernazza A., Lepellec-Muller A., Rufo M., Massion J., et al. (2005). Goal directed locomotion and balance control in autistic children. J. Autism Dev. Disord. 35, 91–102. doi: 10.1007/s10803-004-1037-3, PMID: [DOI] [PubMed] [Google Scholar]
- Vicidomini C., Ponzoni L., Lim D., Schmeisser M. J., Reim D., Morello N., et al. (2017). Pharmacological enhancement of mGlu5 receptors rescues behavioral deficits in Shank3 knock-out mice. Mol. Psychiatry 22, 689–702. doi: 10.1038/mp.2016.30, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel Ciernia A., Pride M. C., Durbin-Johnson B., Noronha A., Chang A., Yasui D. H., et al. (2017). Early motor phenotype detection in a female mouse model of Rett syndrome is improved by cross-fostering. Hum. Mol. Genet. 26, 1839–1854. doi: 10.1093/hmg/ddx087, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorn P., Vanderschuren L. J., Groenewegen H. J., Robbins T. W., Pennartz C. M. (2004). Putting a spin on the dorsal-ventral divide of the striatum. Trends Neurosci. 27, 468–474. doi: 10.1016/j.tins.2004.06.006, PMID: [DOI] [PubMed] [Google Scholar]
- Wang I. T., Allen M., Goffin D., Zhu X., Fairless A. H., Brodkin E. S., et al. (2012). Loss of Cdkl5 disrupts kinome profile and event-related potentials leading to autistic-like phenotypes in mice. Proc. Natl. Acad. Sci. U. S. A. 109, 21516–21521. doi: 10.1073/pnas.1216988110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Bey A. L., Katz B. M., Badea A., Kim N., David L. K., et al. (2016). Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat. Commun. 7:11459. doi: 10.1038/ncomms11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Geng H., Liu W., Zhang G. (2017). Prenatal, perinatal, and postnatal factors associated with autism: a meta-analysis. Medicine (Baltimore) 96:e6696. doi: 10.1097/MD.0000000000009492, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Mccoy P. A., Rodriguiz R. M., Pan Y., Je H. S., Roberts A. C., et al. (2011). Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum. Mol. Genet. 20, 3093–3108. doi: 10.1093/hmg/ddr212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Tan J., Guo J., Zheng Y., Han Q., So K. F., et al. (2018). Aberrant development and synaptic transmission of cerebellar cortex in a Vpa induced mouse Autism model. Front. Cell. Neurosci. 12:500. doi: 10.3389/fncel.2018.00500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang D., Jiang Y., Wang Y., Niu M., Wang C., et al. (2023). Loss of Rab39B does not alter Mptp-induced Parkinson’s disease-like phenotypes in mice. Front. Aging Neurosci. 15:1087823. doi: 10.3389/fnagi.2023.1087823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Chadman K. K., Mccloskey D. P., Sheikh A. M., Malik M., Brown W. T., et al. (2012). Brain Il-6 elevation causes neuronal circuitry imbalances and mediates autism-like behaviors. Biochim. Biophys. Acta 1822, 831–842. doi: 10.1016/j.bbadis.2012.01.011 [DOI] [PubMed] [Google Scholar]
- Weiss M. J., Moran M. F., Parker M. E., Foley J. T. (2013). Gait analysis of teenagers and young adults diagnosed with autism and severe verbal communication disorders. Front. Integr. Neurosci. 7:33. doi: 10.3389/fnint.2013.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss L. A., Shen Y., Korn J. M., Arking D. E., Miller D. T., Fossdal R., et al. (2008). Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 358, 667–675. doi: 10.1056/NEJMoa075974, PMID: [DOI] [PubMed] [Google Scholar]
- Weissberg O., Elliott E. (2021). The mechanisms of Chd8 in neurodevelopment and Autism Spectrum disorders. Genes (Basel) 12:1133. doi: 10.3390/genes12081133, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltschko A. B., Tsukahara T., Zeine A., Anyoha R., Gillis W. F., Markowitz J. E., et al. (2020). Revealing the structure of pharmacobehavioral space through motion sequencing. Nat. Neurosci. 23, 1433–1443. doi: 10.1038/s41593-020-00706-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winden K. D., Ebrahimi-Fakhari D., Sahin M. (2018). Abnormal mtor activation in Autism. Annu. Rev. Neurosci. 41, 1–23. doi: 10.1146/annurev-neuro-080317-061747, PMID: [DOI] [PubMed] [Google Scholar]
- Wohr M., Fong W. M., Janas J. A., Mall M., Thome C., Vangipuram M., et al. (2022). Myt1l haploinsufficiency leads to obesity and multifaceted behavioral alterations in mice. Mol. Autism. 13:19. doi: 10.1186/s13229-022-00497-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohr M., Silverman J. L., Scattoni M. L., Turner S. M., Harris M. J., Saxena R., et al. (2013). Developmental delays and reduced pup ultrasonic vocalizations but normal sociability in mice lacking the postsynaptic cell adhesion protein neuroligin2. Behav. Brain Res. 251, 50–64. doi: 10.1016/j.bbr.2012.07.024, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Simon J. M., Ptacek T. S., Yi J. J., Loo L., Mao H., et al. (2023). Autism-linked Ube3A gain-of-function mutation causes interneuron and behavioral phenotypes when inherited maternally or paternally in mice. Cell Rep. 42:112706. doi: 10.1016/j.celrep.2023.112706, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q., Oviedo H. V., Trotman L. C., Zador A. M. (2012). PTEN regulation of local and long-range connections in mouse auditory cortex. J. Neurosci. 32, 1643–1652. doi: 10.1523/JNEUROSCI.4480-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Ho Y., Fasolino M., Medina J., O’brien W. T., Lamonica J. M., et al. (2023). Allelic contribution of Nrxn1α to autism-relevant behavioral phenotypes in mice. PLoS Genet. 19:e1010659. doi: 10.1371/journal.pgen.1010659, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Song P., Huang W., He R., He Y., Zhou X., et al. (2018). Disruption of at-hook 1 domain in Mecp2 protein caused behavioral abnormality in mice. Biochim. Biophys. Acta Mol. basis Dis. 1864, 347–358. doi: 10.1016/j.bbadis.2017.10.022, PMID: [DOI] [PubMed] [Google Scholar]
- Xu Y., Ye H., Shen Y., Xu Q., Zhu L., Liu J., et al. (2011). Dscam mutation leads to hydrocephalus and decreased motor function. Protein Cell 2, 647–655. doi: 10.1007/s13238-011-1072-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Bozdagi O., Scattoni M. L., Wöhr M., Roullet F. I., Katz A. M., et al. (2012). Reduced excitatory neurotransmission and mild Autism-relevant phenotypes in adolescent <em>Shank3</em> null mutant mice. J. Neurosci. 32, 6525–6541. doi: 10.1523/JNEUROSCI.6107-11.2012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Pan F., Gan W. B. (2009). Stably maintained dendritic spines are associated with lifelong memories. Nature 462, 920–924. doi: 10.1038/nature08577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura M., Yoshida T., Yamazaki M., Abe M., Natsume R., Kanno K., et al. (2014). Il1rapl1 knockout mice show spine density decrease, learning deficiency, hyperactivity and reduced anxiety-like behaviours. Sci. Rep. 4:6613. doi: 10.1038/srep06613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Chen W., Chao E. S., Soriano S., Wang L., Wang W., et al. (2018). Otud7a knockout mice recapitulate many neurological features of 15q13.3 microdeletion syndrome. Am. J. Hum. Genet. 102, 296–308. doi: 10.1016/j.ajhg.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Jones N., Yang J., Asraoui N., Mathieu M. E., Cai L., et al. (2021). Delayed motor learning in a 16p11.2 deletion mouse model of autism is rescued by locus coeruleus activation. Nat. Neurosci. 24, 646–657. doi: 10.1038/s41593-021-00815-7, PMID: [DOI] [PubMed] [Google Scholar]
- Yin H. H., Knowlton B. J., Balleine B. W. (2005). Blockade of NMDA receptors in the dorsomedial striatum prevents action-outcome learning in instrumental conditioning. Eur. J. Neurosci. 22, 505–512. doi: 10.1111/j.1460-9568.2005.04219.x [DOI] [PubMed] [Google Scholar]
- Yin H. H., Knowlton B. J., Balleine B. W. (2006). Inactivation of dorsolateral striatum enhances sensitivity to changes in the action-outcome contingency in instrumental conditioning. Behav. Brain Res. 166, 189–196. doi: 10.1016/j.bbr.2005.07.012, PMID: [DOI] [PubMed] [Google Scholar]
- Yin H. H., Mulcare S. P., Hilário M. R. F., Clouse E., Holloway T., Davis M. I., et al. (2009). Dynamic reorganization of striatal circuits during the acquisition and consolidation of a skill. Nat. Neurosci. 12, 333–341. doi: 10.1038/nn.2261, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Yamagata A., Imai A., Kim J., Izumi H., Nakashima S., et al. (2021). Canonical versus non-canonical transsynaptic signaling of neuroligin 3 tunes development of sociality in mice. Nat. Commun. 12:1848. doi: 10.1038/s41467-021-22059-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampella C. J., Wang L. A. L., Haley M., Hutchinson A. G., De Marchena A. (2021). Motor skill differences in Autism Spectrum disorder: a clinically focused review. Curr. Psychiatry Rep. 23:64. doi: 10.1007/s11920-021-01280-6 [DOI] [PubMed] [Google Scholar]
- Zeidan J., Fombonne E., Scorah J., Ibrahim A., Durkin M. S., Saxena S., et al. (2022). Global prevalence of autism: a systematic review update. Autism Res. 15, 778–790. doi: 10.1002/aur.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Goto H., Akiyoshi-Nishimura S., Prosselkov P., Sano C., Matsukawa H., et al. (2016). Diversification of behavior and postsynaptic properties by netrin-G presynaptic adhesion family proteins. Mol. Brain 9:6. doi: 10.1186/s13041-016-0187-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Ma L., Yang M., Shao Q., Xu J., Lu Z., et al. (2020). Cerebral organoid and mouse models reveal a Rab39b-Pi3K-mtor pathway-dependent dysregulation of cortical development leading to macrocephaly/autism phenotypes. Genes Dev. 34, 580–597. doi: 10.1101/gad.332494.119, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Kaiser T., Monteiro P., Zhang X., Van Der Goes M. S., Wang D., et al. (2016). Mice with Shank3 mutations associated with Asd and schizophrenia display both shared and distinct defects. Neuron 89, 147–162. doi: 10.1016/j.neuron.2015.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]