Abstract
Patients with autoimmune polyendocrinopathy syndrome type 1 (APS-1) caused by autosomal recessive AIRE deficiency produce autoantibodies that neutralize type I interferons (IFNs)1,2, conferring a predisposition to life-threatening COVID-19 pneumonia3. Here we report that patients with autosomal recessive NIK or RELB deficiency, or a specific type of autosomal-dominant NF-κB2 deficiency, also have neutralizing autoantibodies against type I IFNs and are at higher risk of getting life-threatening COVID-19 pneumonia. In patients with autosomal-dominant NF-κB2 deficiency, these autoantibodies are found only in individuals who are heterozygous for variants associated with both transcription (p52 activity) loss of function (LOF) due to impaired p100 processing to generate p52, and regulatory (IκBδ activity) gain of function (GOF) due to the accumulation of unprocessed p100, therefore increasing the inhibitory activity of IκBδ (hereafter, p52LOF/IκBδGOF). By contrast, neutralizing autoantibodies against type I IFNs are not found in individuals who are heterozygous for NFKB2 variants causing haploinsufficiency of p100 and p52 (hereafter, p52LOF/IκBδLOF) or gain-of-function of p52 (hereafter, p52GOF/IκBδLOF). In contrast to patients with APS-1, patients with disorders of NIK, RELB or NF-κB2 have very few tissue-specific autoantibodies. However, their thymuses have an abnormal structure, with few AIRE-expressing medullary thymic epithelial cells. Human inborn errors of the alternative NF-κB pathway impair the development of AIRE-expressing medullary thymic epithelial cells, thereby underlying the production of autoantibodies against type I IFNs and predisposition to viral diseases.
Subject terms: Immunogenetics, Infectious diseases
Inborn errors of the alternative NF-κB pathway in humans impair the development of AIRE-expressing medullary thymic epithelial cells, thereby underlying the production of autoantibodies against type I IFNs and predisposition to viral diseases
Main
Autoantibodies neutralizing type I IFNs (AAN-I-IFNs) have been reported in patients treated with type I IFNs, systemic lupus erythematosus (SLE), thymoma or myasthenia gravis4. These autoantibodies were widely thought to be clinically silent, with the notable exception of a 77-year-old woman who had such antibodies and disseminated shingles, reported in 19815,6. Nearly 40 years later, we showed that pre-existing neutralizing AAN-I-IFNs underlie at least 15% of cases of life-threatening COVID-19 pneumonia4,7–11. These autoantibodies were also found to underlie severe adverse reactions to yellow fever live-attenuated viral vaccine12, influenza pneumonia13, MERS pneumonia14 and West Nile virus encephalitis15. AAN-I-IFNs underlie clinical phenocopies of inborn errors of type I IFN immunity, as the same viral diseases have been reported in patients with autosomal-recessive IFNAR1 or IFNAR2 deficiency4,9,11. These autoantibodies block cell-protective antiviral effects of type I IFNs in vitro8,12,13,15 and impair the induction of IFN-stimulated genes (ISGs) in peripheral blood mononuclear cells and nasal mucosae infected with SARS-CoV-2 ex vivo7,16,17. Finally, these autoantibodies are also present in the general population, with the prevalence sharply increasing in individuals over 70 years of age, thereby contributing to the age-related increase in the risk of severe COVID-197,10.
Notably, the production of AAN-I-IFNs can be driven by monogenic inborn errors of immunity (IEIs). These IEIs include autosomal-recessive APS-1, which is caused by germline biallelic deleterious variants of AIRE; immunodysregulation polyendocrinopathy enteropathy X-linked (IPEX) syndrome, caused by deleterious hemizygous variants of FOXP3; and combined immunodeficiency due to biallelic hypomorphic RAG1 or RAG2 variants4. All of these IEIs affect T cell thymic selection, in a T-cell-intrinsic or -extrinsic manner. AIRE deficiency impairs the expression of tissue-specific antigens in medullary thymic epithelial cells (mTECs), enabling autoreactive T cells to escape18,19. FOXP3 deficiency impairs the development of thymic regulatory T (Treg) cells, whereas hypomorphic variants of RAG1 or RAG2, which restrict TCR diversity, also have an effect on thymic architecture and the development of mTECs20–22. The disruption of self-tolerance in the thymus therefore seems to underlie the production of AAN-I-IFNs. Patients with APS-1 display severe multiorgan autoimmunity with a wide range of autoantibodies against tissue-specific antigens18. They also frequently have neutralizing autoantibodies against IL-17A and/or IL-17F that underlie chronic mucocutaneous candidiasis, a disease that is seen in patients with inborn errors of IL-17A/F immunity4. Most, if not all, patients with APS-1 also produce AAN-I-IFNs in early childhood4, and are highly vulnerable to critical COVID-19 pneumonia3 and to severe varicella23.
In mice, the expression of the Aire gene in mTECs is controlled by the alternative (or non-canonical) NF-κB pathway24–26. Once triggered, NIK activates IKKα, leading to the phosphorylation of the full-length NF-κB2 precursor p100 (amino acids 1–900) on serine residues Ser866 and Ser870. This leads to p100 processing to generate the p52 (amino acids 1–405) active form, which preferentially dimerizes with RELB27. This p52–RELB heterodimer migrates to the nucleus, inducing the transcription of target genes involved in lymphoid organ development, germinal centre formation, B cell survival, maturation, homeostasis, mTEC development and osteoclastogenesis27. In resting cells, unprocessed cytoplasmic p100 can form high-molecular-mass complexes by homomultimerization (generating kappaBsomes) through its C-terminal IκB-like domain, thereby inhibiting the DNA-binding activity of almost all NF-κB subunits (referred to as IκBδ function)28. In the mouse thymus, RANK and the alternative NF-κB pathway have a crucial role in mTECs by governing self-tolerance24,26. Deficiencies in mouse Traf6, Ikkα, Map3k14 (encoding NIK) or RelB impair mTEC development and AIRE expression in mTECs29. We tested the hypothesis that human inborn errors of the alternative NF-κB pathway—including autosomal-dominant NF-κB2 disorders, and autosomal-recessive RELB, IKKα and NIK deficiencies—can underlie the production of AAN-I-IFNs, thereby predisposing patients to severe viral diseases, including COVID-19 pneumonia.
Inborn errors of the alternative NF-κB pathway
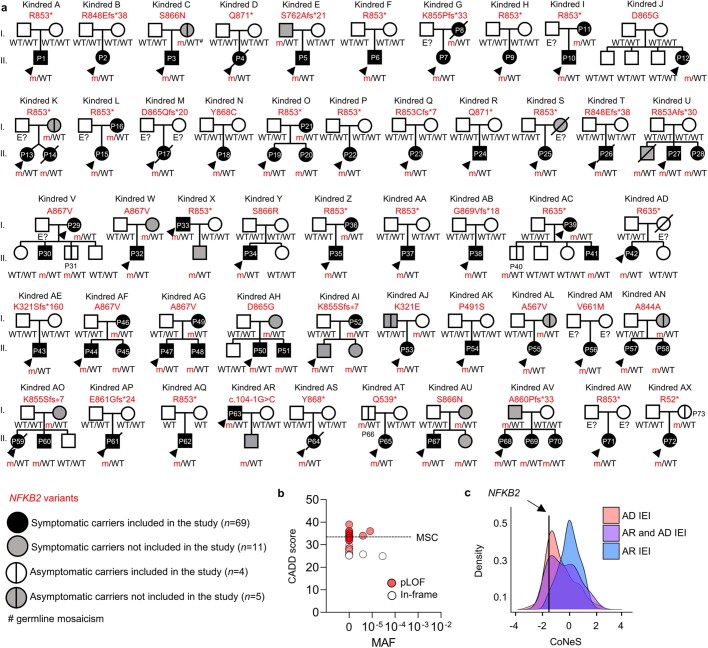
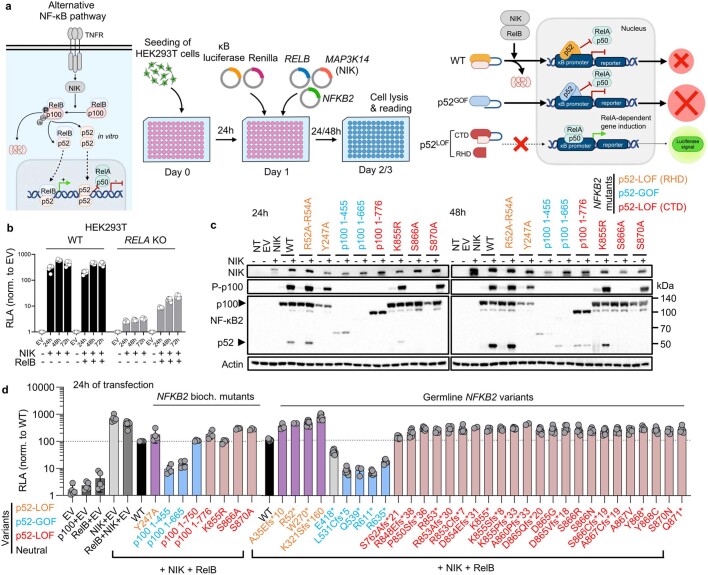
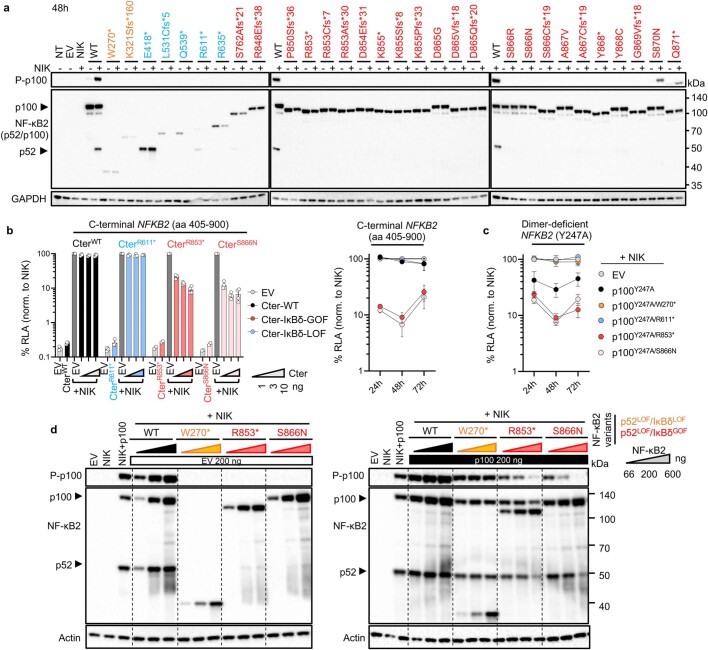
We recruited an international cohort of 73 patients from 50 kindreds heterozygous for 28 different rare (minor allele frequency (MAF) < 0.0001) non-synonymous NFKB2 variants (Extended Data Fig. 1a,b and Supplementary Table 1). Most affected individuals had a predominant phenotype of primary antibody deficiency (PAD) (62 out of 69, 89.9% of these patients). After a comprehensive functional characterization, we identified three types of autosomal-dominant inborn errors of NF-κB2, designated as p52LOF/IκBδLOF in 4 patients heterozygous for NFKB2 variants causing haploinsufficiency of p100 and p52; p52GOF/IκBδLOF in 6 patients heterozygous for NFKB2 variants causing GOF of p52; and p52LOF/IκBδGOF in 57 patients heterozygous for NFKB2 variants that are associated with both transcriptional (p52 activity) LOF due to impaired p100 processing to generate p52, and regulatory activity (IκBδ activity) GOF due to the accumulation of unprocessed p100 (Fig. 1, Supplementary Results 1, Extended Data Figs. 2–5 and Supplementary Figs. 2–6). Six other patients carried a neutral NFKB2 heterozygous variant (hereafter, idiopathic PAD). Among the three inborn errors of NF-κB2, only the p52LOF/IκBδGOF variants severely impaired the alternative NF-κB pathway activation by preventing the nuclear translocation of p52 and RELB (Fig. 1d and Supplementary Results 1). Moreover, the p52LOF/IκBδGOF variants also impaired the formation of p52–RELB heterodimers in patients’ heterozygous fibroblasts, in contrast to fibroblasts that are heterozygous for a p52LOF/IκBδLOF variant (Extended Data Fig. 5c,d). Finally, only patients heterozygous for p52LOF/IκBδGOF variants displayed a unique immunological phenotype associated with B cell lymphopenia and reduced Treg and TFH cell counts (Fig. 2a–c, Supplementary Results 2, Extended Data Fig. 6 and Supplementary Fig. 7). We also enrolled 14 patients with other inborn errors of the alternative NF-κB pathway (autosomal-recessive NIK (n = 2) and autosomal-recessive RELB (n = 8) deficiencies) or upstream receptors (autosomal-recessive BAFF (n = 1) or X-linked recessive CD40L (n = 3) deficiencies) (Extended Data Fig. 2d,e and Supplementary Table 2).
Extended Data Fig. 1. Pedigrees of the 73 patients studied carrying heterozygous NFKB2 variants.
(a) Pedigrees of the patients heterozygous for rare variants of NFKB2. Generations are indicated by Roman numerals (I–II), and each symptomatic carrier included in the study, represented by a black symbol, is indicated as P followed by an Arabic numeral (P1–P73). Grey symbols represent relatives who are symptomatic carriers but for whom no material was available for this study. A vertical bar, within a white or grey symbol, indicates an asymptomatic carrier included or not included (due to a lack of available material), respectively in the study; an arrow indicates the index case; a black diagonal line indicates that the individual is deceased. “E?” indicates individuals of unknown genotype. (b) CADD-MAF (combined annotation-dependent depletion-minor allele frequency) graph of the rare or private NFKB2 variants (n = 28) from the 73 patients recruited. The red and white dots represent pLOF and missense heterozygous NFKB2 variants, respectively. Each score was calculated with CADD version 1.6. The dashed line represents the mutation significance (MSC) cutoff threshold of 33 for NFKB2. (C) CoNeS score of the NFKB2 gene.
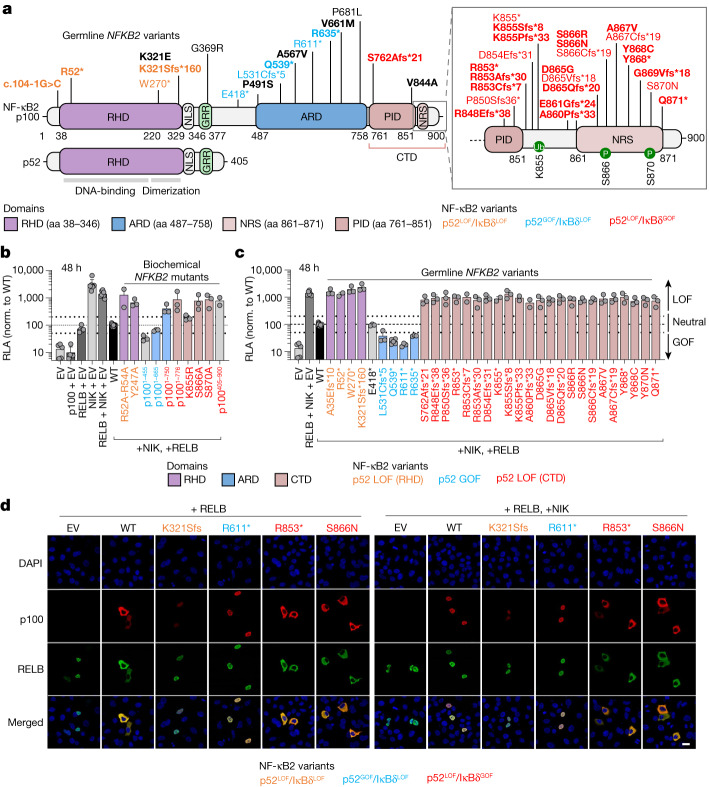
Fig. 1. Functional testing of the NFKB2 alleles by overexpression.
a, Schematic of the NF-κB2 protein (p100 and p52) with the variants, identified in heterozygous patients, that were included in this study (n = 28 variants, shown in bold) or not included here but reported elsewhere (n = 13 variants). The C-terminal domain (CTD) spans amino acids (aa) 760–900. The REL-homology domain (RHD; purple), the ankyrin repeat domain (ARD; blue) and the CTD, including the processing-inhibitory domain (PID) and the NIK-responsive sequence (NRS) (brown), are shown. The NFKB2 variants that are LOF for p52/p52 repression of κB transcriptional activity (p52 activity) and LOF for IκBδ regulatory activity (p52LOF/IκBδLOF) are shown in orange. The variants that are GOF for p52 activity and LOF for IκBδ activity are shown in blue (p52GOF/IκBδLOF). The variants in the CTD that are both LOF for the p52 activity and GOF for the IκBδ regulatory activity (p52LOF/IκBδGOF) are shown in red. Neutral NFKB2 variants are shown in black. b, The relative luciferase activity (RLA) of HEK293T cells transfected with a κB reporter luciferase construct (κB-luc) in the presence or absence of plasmids encoding NIK, RELB and/or p100/NF-κB2 WT or biochemical p100/NF-κB2 mutants reported in previous studies, normalized (norm.) to WT p100/NF-κB2, after 48 h of transfection. Data are mean ± s.d. from three independent experiments. EV, empty vector. c, The RLA of HEK293T cells transfected with a κB-luc vector, in the presence of plasmids encoding NIK, RELB and p100/NF-κB2 WT or the NFKB2 variants included in this study or reported in previous studies, at 48 h after transfection. Data are mean ± s.d. from three independent experiments. d, Subcellular localization of the WT or the NF-κB2 variants used for cotransfection with RELB without (left) or with (right) NIK, as determined by confocal microscopy analysis of HeLa cells. The nuclei were stained with DAPI; p100 and RELB were detected using antibodies recognizing their N-terminal domains. Data shown are representative of two independent experiments. Scale bar, 20 μm.
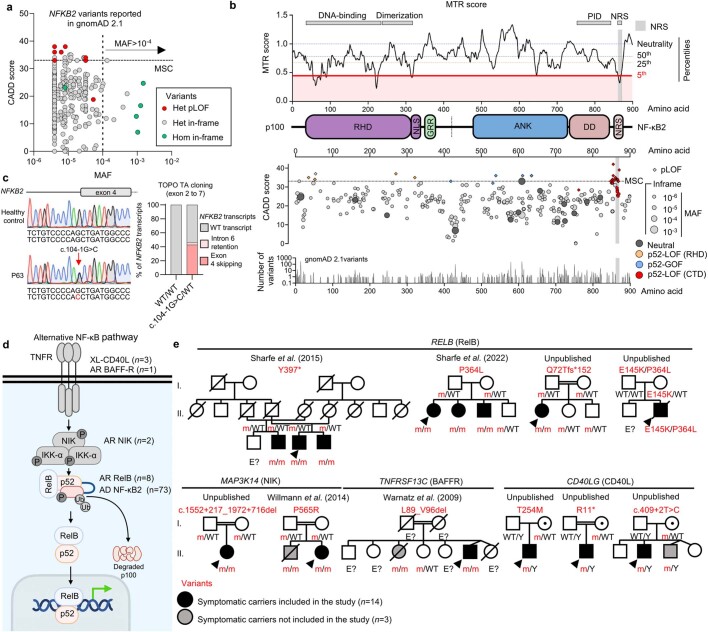
Extended Data Fig. 2. Population genetics and constraint metrics of the NFKB2 gene, and pedigrees of the patients with inborn errors of RelB, NIK, BAFFR and CD40L.
(a) CADD-MAF graph for the NFKB2 variants reported in the gnomAD v2.2.1. The red and grey dots represent monoallelic pLOF and heterozygous in-frame (missense and indel) variants, respectively. The green dots represent homozygous missense variants. The horizontal dashed line represents the mutation significance (MSC) cutoff threshold of 33 for NFKB2. (b) Genomic constrained coding regions across NFKB2, as estimated by the missense tolerance ratio (MTR) score evaluating region-specific intolerance to missense variants. A score <1.0 indicates a lower-than-expected ratio of missense to synonymous variants in the gnomAD v2.0 dataset for the 21 bp window surrounding an amino-acid residue. The horizontal-coloured dashed lines represent the percentiles for the most missense-depleted regions of NFKB2. The NIK-responsive sequence (NRS) is within the 5th percentile for the most missense-depleted regions for NFKB2. The lower graph shows the distribution of the heterozygous NFKB2 variants reported in gnomAD 2.1.1 and from the patients reported in this study, by location within the protein and CADD score. (c) Electropherograms showing the c.104-1 G > C/WT essential splice-site variant carried in the heterozygous state of P63 and a healthy donor (left) and the proportion of transcripts identified by sequencing 100 colonies from TOPO cloning with cDNA from PCR products corresponding to a region spanning exon 2 to 7 in P63 or a healthy donor. (d) Representation of the alternative NF-κB pathway and the patients included. (e) Pedigrees and variants of patients with inborn errors of RelB, NIK, BAFFR and CD40L. A dot within a white symbol indicates an asymptomatic carrier; an arrow indicates the index case; a black diagonal line indicates a deceased individual. “E?” indicates individuals of unknown genotype.
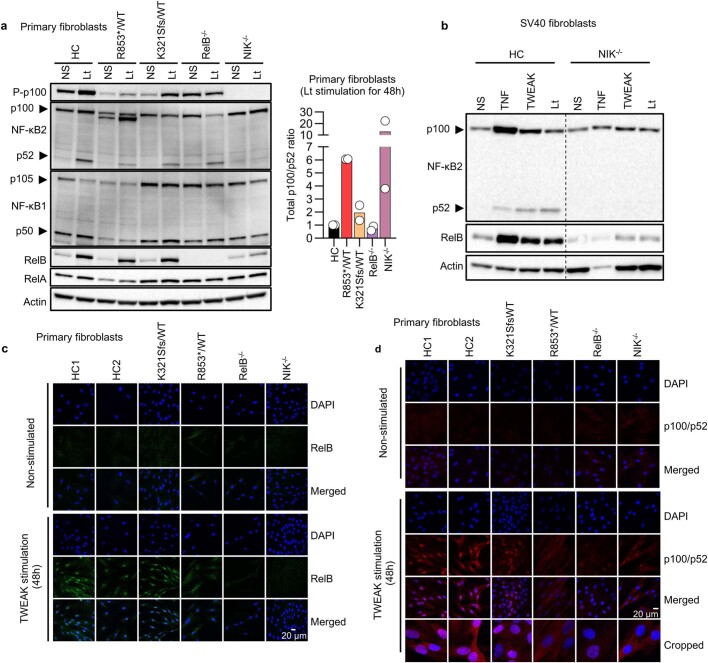
Extended Data Fig. 5. The processing-resistant NFKB2 mutants have enhanced p100-IκBδ activity in heterozygous patients’ cells.
(a) Western blot of P-p100, NF-κB2 (p100/p52), NF-κB1 (p105/p50), RelB, and RelA in primary fibroblasts from one healthy donor (HC), a patient with the p52LOF/IκBδGOF R853*/WT variant, a patient with the p52LOF/IκBδLOF K321Sfs/WT variant, a patient with AR complete (Q72Tfs*152/Q72Tfs*152) RelB (RelB−/−) or (P565R/P565R) NIK (NIK−/−) deficiency, with or without stimulation with LT-α1β2 (Lt) for 48 h (left panel), and a graph depicting total p100/p52 intensity ratio after Lt stimulation (right panel). Bars represent the mean values (± s.d.) from two independent experiments. (b) Western blot showing p100 processing into p52 and RelB induction in total cell extracts of SV40 fibroblasts from a healthy donor (HC) or a patient with AR complete NIK deficiency (NIK−/−) either left non stimulated (NS) or after stimulation for 48 h with TNF, TWEAK, Lt. Data representative of three independent experiments are shown. (c) Confocal microscopy showing the subcellular distribution of RelB in primary fibroblasts from two healthy controls (HC1, HC2), patients with a p52LOF/IκBδLOF K321Sfs/WT or a p52LOF/IκBδGOF R853*/WT NF-κB2/p100 variant, and patients with AR complete RelB (RelB−/−) or NIK (NIK−/−) deficiency, without and with stimulation with TWEAK for 48 h. Data representative of three independent experiments are shown. (d) Confocal microscopy showing the subcellular distribution of p100/p52 in primary fibroblasts from two healthy controls (HC1, HC2), patients with a p52LOF/IκBδLOF K321Sfs*/WT or a p52LOF/IκBδGOF R853*/WT NF-κB2/p100 variant, AR complete RelB deficiency (RelB−/−), or AR complete NIK deficiency (NIK−/−) without and with stimulation with TWEAK for 48 h, with an antibody recognizing the N-terminus of p100. Data representative of three independent experiments are shown. The bottom panels represent magnified images (cropped images).
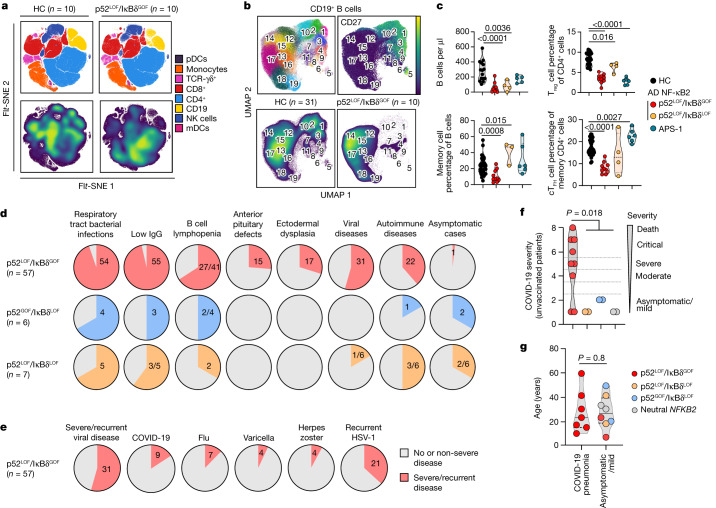
Fig. 2. Distinctive immunological and clinical phenotype of patients with p52LOF/IκBδGOF heterozygous variants.
a, FFT-accelerated interpolation-based (FI) t-distributed stochastic neighbour embedding (t-SNE) analysis of concatenated whole-blood samples from ten patients with p52LOF/IκBδGOF variants, and ten age-matched healthy control individuals (HC), based on cytometry by time of flight (CyTOF) data. t-SNE analysis is not shown for the patients with p52LOF/IκBδLOF variants (n = 4) or APS-1 (n = 6) owing to their lower number. NK, natural killer cells; mDCs and pDCs, myeloid and plasmacytoid dendritic cells, respectively. b, Uniform manifold approximation and projection (UMAP)-based unsupervised clustering analysis of CD19+ B cells from a concatenated group of 10 patients with p52LOF/IκBδGOF variants and 31 age-matched controls (HC), with a heat map showing the mean levels of the surface markers included in the clustering defining 19 distinct metaclusters, CD27 marker expression and the metacluster distribution in healthy control individuals and patients with p52LOF/IκBδGOF variants. c, The number of B cells and the proportions of memory B cells, Treg cells and circulating TFH (cTFH) cells in patients with a p52LOF/IκBδGOF variant (n = 10, red dots, except for the B cell numbers, showing only patients above 6 years of age, n = 9), age-matched controls (n = 27, black dots), patients with a p52LOF/IκBδLOF variant (n = 4, orange dots) and patients with APS-1 (n = 6, green dots). Statistical comparisons were performed using two-tailed Mann–Whitney U-tests. AD, autosomal dominant. d, The proportion and number of patients with p52LOF/IκBδGOF (n = 57), p52GOF/IκBδLOF (n = 6) or p52LOF/IκBδLOF (n = 7, including 4 reported here and 3 previously reported56) NF-κB2 variants with their corresponding clinical manifestations. e, The proportion and number of patients with severe/recurrent (red shape) or no/non-severe (grey shape) viral diseases among the 57 patients with p52LOF/IκBδGOF NF-κB2 variants. f, COVID-19 severity scale for unvaccinated patients with a p52LOF/IκBδGOF (red dots, n = 9), p52LOF/IκBδLOF (orange dots, n = 2), p52GOF/IκBδLOF (blue dots, n = 2) or neutral (grey dots, n = 2) NF-κB2 variant. Statistical comparisons were performed using two-tailed Mann–Whitney U-tests. g, Age at the COVID-19 episode in unvaccinated patients with a p52LOF/IκBδGOF (red dots, n = 9), p52LOF/IκBδLOF (orange dots, n = 2), p52GOF/IκBδLOF (blue dots, n = 2) or neutral (grey dots, n = 2) NF-κB2 variant, as a function of COVID-19 severity. Statistical comparisons were performed using two-tailed Mann–Whitney U-tests.
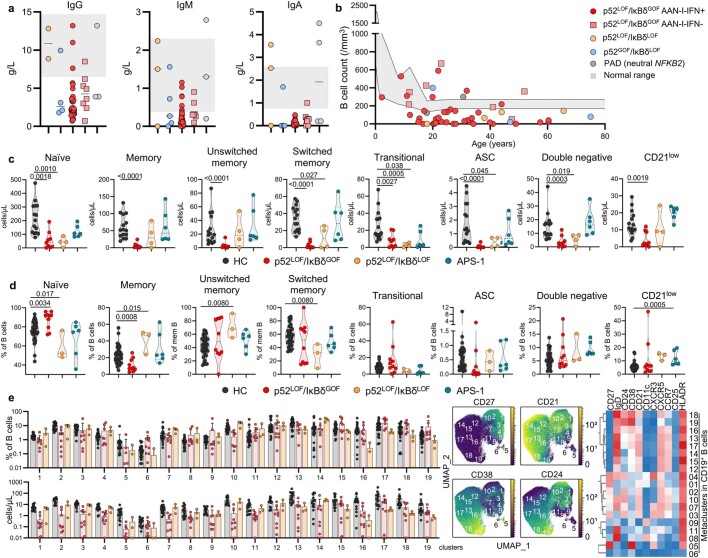
Extended Data Fig. 6. Immunoglobulin level and B cell immunophenotyping of patients with inborn errors of NF-κB2.
(a) Immunoglobulin IgG, IgM, and IgA levels (g/L) in patients with inborn errors of NF-κB2. Normal immunoglobulin distribution corresponds to the grey area. Bars represent the median values. (b) B cell count across ages in patients with p52LOF/IκBδGOF variants with (n = 39, red dots) and without (n = 7, red squares) AAN-I-IFNs, patients with p52LOF/IκBδLOF/WT (n = 3, orange dots), p52GOF/IκBδLOF/WT (n = 4, blue dots), or neutral (PAD, grey dots) NF-κB2 variants. Normal B-cell count for age corresponds to the grey area. (c) Cell numbers among B cell subsets, as determined by CyTOF, in healthy donors (n = 15, black dots), patients with p52LOF/IκBδGOF variants aged ≥ 6 years (n = 9, red dots), patients with p52LOF/IκBδLOF variants (n = 4, orange dots), and APS-1 patients (n = 6, green dots). (d) Proportions of B cell subsets, as determined by CyTOF, in healthy donors (n = 15, black dots), patients with p52LOF/IκBδGOF variants (n = 10, red dots), patients with p52LOF/IκBδLOF variants (n = 3, orange dots), and APS-1 patients (n = 6, green dots). B-cell subset proportions from a patient with a p52LOF/IκBδLOF R52*/WT variant are not shown due to his lack of circulating B cells. (e) Proportions of B cell subsets and absolute counts of B cells identified in the 19 metaclusters in healthy donors (n = 22, black dots), patients with a p52LOF/IκBδGOF variant aged ≥ 6 years (n = 9, red dots), and patients with a p52LOF/IκBδLOF variant (n = 3, orange dots) (left panels); representation of the CD27, CD21, CD38, and CD24 markers on UMAP (middle panels); heatmap showing the mean levels of the surface markers included in the clustering defining 19 distinct metaclusters. The error bars represent the mean values (± s.d.) of each group.
Viral diseases of patients with p52LOF/IκBδGOF
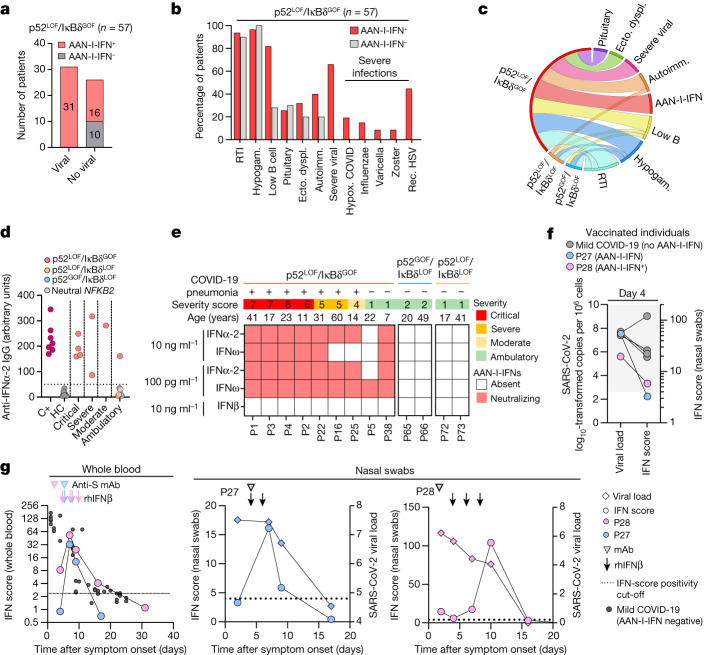
Whereas PAD and autoimmune diseases were reported in patients with any of the three types of autosomal-dominant NF-κB2 deficiency, ectodermal dysplasia and anterior pituitary hormone deficiencies were reported exclusively in patients carrying p52LOF/IκBδGOF variants (Fig. 2d and Supplementary Table 1). Severe or recurrent viral diseases were almost exclusively reported in patients carrying p52LOF/IκBδGOF variants (n = 31 out of 57, 54%) (Fig. 2d,e). This susceptibility could not be explained by immunosuppressive treatments (used in seven patients with p52LOF/IκBδGOF variants) (Supplementary Table 1). The main viral disease reported was recurrent mucocutaneous HSV-1 lesions (n = 21, 37%) (Fig. 2e). Six out of the nine unvaccinated patients and two patients with an unknown vaccination status with p52LOF/IκBδGOF variants developed hypoxaemic COVID-19 pneumonia (NIH scale, 5 to 8, out of 8) after infection with SARS-CoV-2. Three of these patients, aged 17, 23 and 39 years, were admitted to intensive care and two of these individuals (aged 23 and 39 years) died (Fig. 2f). One patient was hospitalized for COVID-19 pneumonia without requiring oxygen supplementation (NIH scale, 4). Eight additional unvaccinated patients developed asymptomatic disease or mild symptoms (NIH scale, 1–2) without pneumonia or hospitalization. These patients carried a p52LOF/IκBδGOF (n = 2, aged 7 and 22 years), p52LOF/IκBδLOF (n = 2, aged 17 and 41 years), p52GOF/IκBδLOF (n = 2, aged 20 and 49 years) or neutral (n = 2, aged 30 and 31 years) NF-κB2 variant (Fig. 2f). COVID-19 severity was not associated with age or treatment (Fig. 2g and Supplementary Table 1). Severe influenza pneumonia was reported in 7 out of the 57 patients with p52LOF/IκBδGOF variants (12%), five of whom required hospitalization and oxygen supplementation, including one patient with acute respiratory distress syndrome (ARDS) and encephalitis (Fig. 2e). Four patients suffered from recurrent (n = 1) or severe (n = 3) varicella (Fig. 2e). All patients with severe varicella were hospitalized, including one with encephalitis and one with severe skin disease requiring acyclovir. The other severe viral diseases observed are indicated in Supplementary Table 1. None of the patients were vaccinated with yellow fever YFV-17D live-attenuated vaccine. All eight of the patients with inborn errors of NF-κB2 who died carried a p52LOF/IκBδGOF variant; six died from suspected or proven viral illnesses, including two from COVID-19. Together, these findings suggest that, in contrast to patients with the other two forms of inborn errors of NF-κB2, patients with p52LOF/IκBδGOF variants present a distinctive syndrome that is strongly associated with the risk of developing PAD and/or a severe viral disease. Conversely, p52/p100 haploinsufficiency (p52LOF/IκBδLOF) and GOF of p52 (p52GOF/IκBδLOF) may underlie humoral deficiency with variable clinical and immunological penetrance, whereas these conditions do not appear to underlie ectodermal, endocrine or viral phenotypes. The milder clinical phenotype associated with these forms may account for the smaller number of patients with such defects identified.
AAN-I-IFNs in patients with p52LOF/IκBδGOF
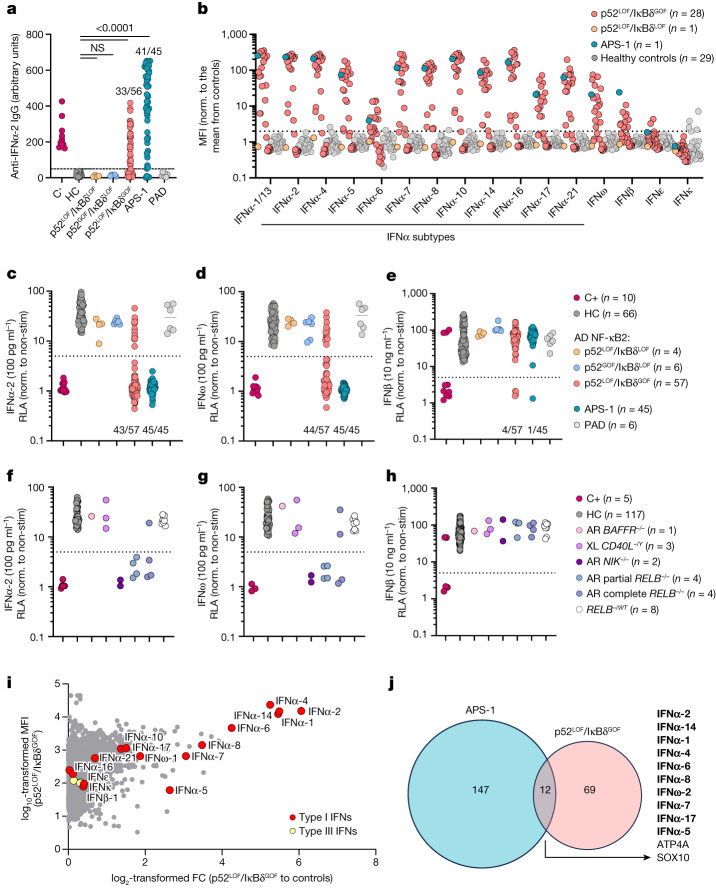
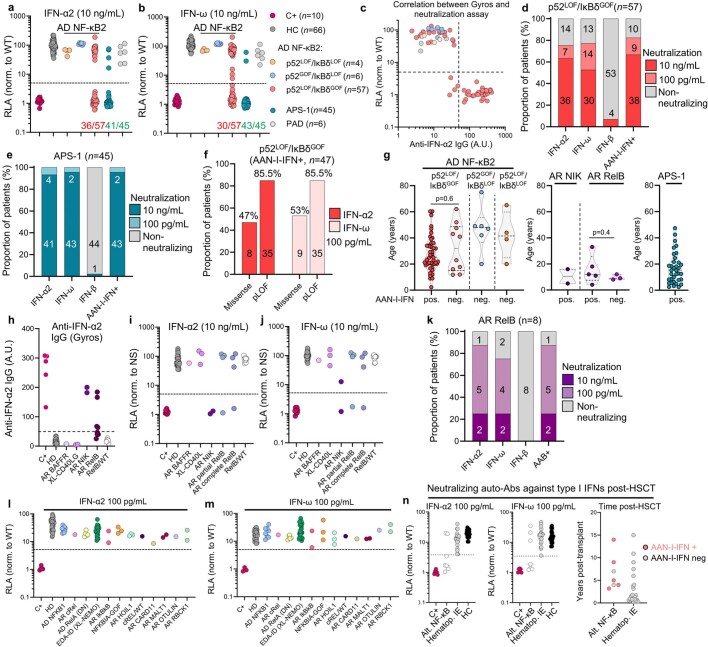
Given the strong susceptibility of patients heterozygous for p52LOF/IκBδGOF to viral diseases, we assessed the presence of AAN-I-IFNs in the plasma of 73 patients heterozygous for a deleterious or neutral variant. We detected high titres (arbitrary units > 50) of anti-IFNα-2 IgG in 33 out of 56 (59%) patients with p52LOF/IκBδGOF variants, 41 out of 45 (91%) patients with APS-1, but none in those carrying p52LOF/IκBδLOF (n = 4) or p52GOF/IκBδLOF (n = 6) alleles, or with idiopathic PAD (n = 6) (Fig. 3a). Moreover, patients with p52LOF/IκBδGOF variants and autoantibodies against IFNα-2 also had detectable autoantibodies against most of the 11 other IFNα subtypes, IFNω and, less frequently, IFNβ, but not against IFNκ or IFNε, as evaluated in a multiplex bead assay (Fig. 3b). We next assessed the neutralization ability of patients’ plasma in the presence of high (10 ng ml−1) or low (100 pg ml−1) concentrations of IFNα-2, IFNω or IFNβ (10 ng ml−1). Overall, 36 out of 57 (65%), 30 out of 57 (53%) and 4 out of 57 (7%) patients with p52LOF/IκBδGOF variants neutralized high concentrations of IFNα-2, IFNω and IFNβ, respectively (Fig. 3e and Extended Data Fig. 7a–c), and 43 out of 57 (75%) and 44 out of 57 (77%) neutralized low concentrations of IFNα-2 or IFNω, respectively (Fig. 3c,d and Extended Data Fig. 7d). For comparison, 41 (91%), 43 (96%) and 1 (2%) out of the 45 patients with APS-1 neutralized IFNα, IFNω and IFNβ, respectively, at a concentration of 10 ng ml−1 (Fig. 3e and Extended Data Fig. 7a, b), and serum from all of these patients neutralized IFNα-2 and IFNω at a concentration of 100 pg ml−1 (Fig. 3c,d and Extended Data Fig. 7e). By contrast, none of the plasma samples from any of the patients with p52LOF/IκBδLOF, p52GOF/IκBδLOF or neutral NFKB2 variants neutralized IFNα-2, IFNω or IFNβ (at 10 ng ml−1 or 100 pg ml−1). The proportion of patients with p52LOF/IκBδGOF variants carrying AAN-I-IFNs was higher among those carrying pLOF variants than among those carrying missense variants but was independent of patient age at testing (P = 0.6) or sex (Extended Data Fig. 7f,g). In ten patients with p52LOF/IκBδGOF variants, no neutralizing autoantibodies against IFNα-2, IFNω or IFNβ could be detected. Seven of them carried the A867V variant (Supplementary Results 3 and Supplementary Fig. 8). In total, plasma samples from 82% (47 out of 57) of the patients with a p52LOF/IκBδGOF variant neutralized IFNα-2 and/or IFNω; the plasma of three of these patients neutralized only IFNα-2, whereas that of four patients neutralized only IFNω, and that of another four patients neutralized IFNα-2, IFNω and IFNβ (Extended Data Fig. 7d and Supplementary Table 4). Overall, we found a strong association between the NFKB2 genotype (p52LOF/IκBδGOF) and the presence of AAN-I-IFNs (Supplementary Fig. 9).
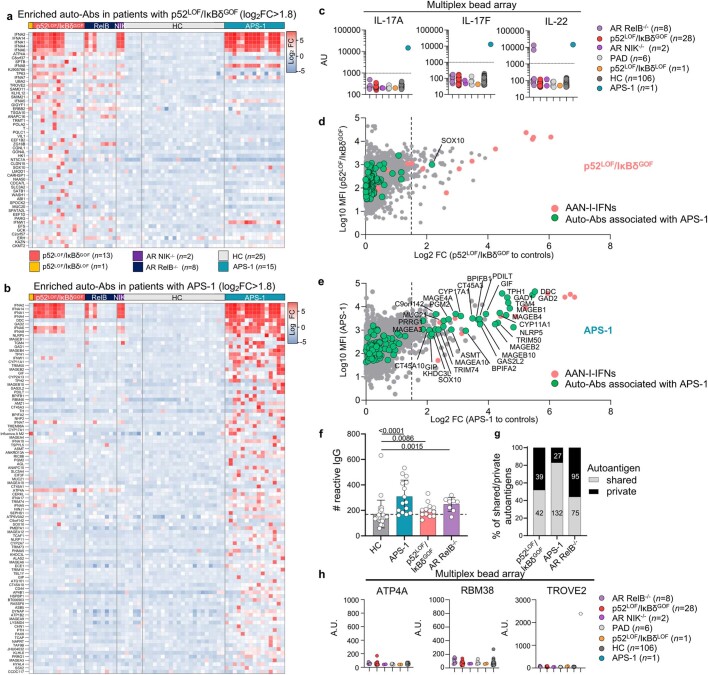
Fig. 3. AAN-I-IFNs detected in patients heterozygous for p52LOF/IκBδGOF variants and patients with inborn errors of RELB or NIK.
a, Detection of IgG autoantibodies against IFNα-2 by Gyros in patients with inborn errors of NF-κB2 with a p52LOF/IκBδLOF (n = 4), p52GOF/IκBδLOF (n = 6) or p52LOF/IκBδGOF (n = 56) variant, patients with APS-1 (n = 45), patients with idiopathic PAD (n = 6), positive control individuals with AAN-I-IFNs (C+, n = 10) or healthy control individuals (HC, n = 25). Data are the mean values from at least three independent experiments. Statistical comparisons were performed using two-tailed Mann–Whitney U-tests. NS, not significant. b, Detection, using a multiplex bead assay, of autoantibodies against the 16 type I IFNs in patients with p52LOF/IκBδGOF (n = 28) or p52LOF/IκBδLOF (n = 1) variants or with APS-1 (n = 1). Values are normalized to the mean fluorescence intensity (MFI) of plasma samples from healthy control individuals (n = 29) for each indicated cytokine. c–e, Luciferase-based neutralization assay to detect autoantibodies neutralizing 100 pg ml−1 IFNα-2 (c), IFNω (d) or 10 ng ml−1 IFNβ (e) in positive-control individuals (n = 10), healthy control individuals (n = 66), patients with a p52GOF/IκBδLOF (n = 6), p52LOF/IκBδLOF (n = 4) or p52LOF/IκBδGOF (n = 57) variant, patients with idiopathic PAD (n = 6) and patients with APS-1 (n = 45). Non-stim., non-stimulated. f–h, Luciferase-based neutralization assay to detect autoantibodies neutralizing 100 pg ml−1 IFNα-2 (f) or IFNω (g) or 10 ng ml−1 IFNβ (h) in patients with autosomal-recessive BAFFR (n = 1), X-linked (XL) CD40L deficiency (n = 3), autosomal-recessive NIK deficiency (n = 2), autosomal-recessive RELB partial or complete deficiency (n = 8) in healthy relatives heterozygous for a null or hypomorphic RELB allele (n = 8), positive control individuals (n = 5) or healthy control individuals (n = 117). All neutralization assay data are presented as the mean of at least two independent experiments. i, Protein microarray showing the distribution of autoantibody reactivity in plasma samples from patients carrying a p52LOF/IκBδGOF variant (n = 13). Data are represented as the fold change (FC) relative to 26 plasma samples from healthy control individuals. Data for HuProt are presented as the mean of at least two technical replicates. j, Representation of the global autoantigen profile of patients with APS-1 and patients with a p52LOF/IκBδGOF variant, with their overlap. Type I IFN autoantigens are highlighted in bold.
Extended Data Fig. 7. AAN-I-IFNs in patients with inborn errors of the alternative NF-κB pathway.
(a-b) Luciferase-based neutralization assay for detecting auto-Abs neutralizing 10 ng/mL IFN-α2 (a) or IFN-ω (b) in patients with the three inborn errors of NF-κB2, APS-1 and PAD patients, positive controls (C+), and healthy controls (HC). (c) Correlation between the detection of auto-Abs against IFN-α2 by Gyros (x-axis) and results for the luciferase-based neutralization assay (y-axis) after stimulation with 10 ng/mL IFN-α2. The dotted line represents the cutoffs for detection (A.U. value > 50) or neutralization (induction <5). (d-e) Proportion of patients with auto-Abs neutralizing type I IFNs at 10 ng/mL or 100 pg/mL among patients with a p52LOF/IκBδGOF variant (d), and APS-1 patients (e). (f) Proportion of patients with AAN-I-IFNs among patients carrying a missense or pLOF p52LOF/IκBδGOF variant. (g) Age distribution of patients with the three inborn errors of NF-κB2, AR RelB or NIK deficiency, or APS-1 according to the presence or absence of AAN-I-IFNs in plasma. (h) Detection of IgG auto-Abs against IFN-α2 by Gyros in patients with inborn errors of NIK, RelB, BAFF and CD40L. (i-j) Luciferase-based neutralization assay for detecting auto-Abs neutralizing 10 ng/mL IFN-α2 (i) or IFN-ω (j) in patients with inborn errors of NIK, RelB, BAFF and CD40L. (k) Proportion of patients with auto-Abs neutralizing type I IFNs at 10 ng/mL or 100 pg/mL in patients with AR RelB deficiency. (l-m) Luciferase-based neutralization assay for detecting auto-Abs neutralizing 100 pg/mL IFN-α2 (l) or IFN-ω (m) in patients with inborn errors of the canonical NF-κB pathway. DN = dominant-negative. (n) Detection of auto-Abs neutralizing 100 pg/mL IFN-α2 or IFN-ω in patients with inborn errors of the alternative NF-κB pathway post-HSCT (n = 7) versus children with inborn errors of T-cell intrinsic immunity [(SCID, n = 3, CID, n = 1), neutrophil-intrinsic immunity (chronic granulomatous disease, CGD, n = 10), cytotoxicity (familial hemophagocytic lymphohistiocytosis, HLH, n = 3), erythrocyte function (β-thalassaemia, n = 3)] who underwent HSCT (Hematop. IE, n = 20) (left panel), with the time interval between HSCT and plasma collection (right panel).
AAN-I-IFNs in NIK or RELB deficiency
We next investigated the presence of AAN-I-IFNs in patients with other inborn errors of the alternative NF-κB pathway. AAN-I-IFNs were detected in the two patients with complete autosomal-recessive NIK deficiency. In one of these patients, the autoantibodies detected neutralized IFNα-2 and IFNω at a concentration of 10 ng ml−1, whereas, in the other, they neutralized IFNα-2 at 10 ng ml−1 and IFNω at 100 pg ml−1 (Fig. 3f–h, Extended Data Fig. 7h–j and Supplementary Fig. 10). AAN-I-IFNs were also detected in patients with autosomal-recessive RELB deficiency (n = 7 out of 8: four patients with partial and three with complete deficiency). These autoantibodies neutralized IFNα-2 and IFNω at 10 ng ml−1 in two patients, and IFNα-2 and/or IFNω at 100 pg ml−1 in five patients (Fig. 3f–h and Extended Data Fig. 7h–k). By contrast, no AAN-I-IFNs were detected in patients with autosomal-recessive BAFFR or X-linked CD40L deficiency, or in the plasma from heterozygous relatives of patients with autosomal-recessive RELB deficiency (n = 8) (Fig. 3f–h and Extended Data Fig. 7h–j). Finally, we tested eight patients with autosomal-dominant NF-κB1 haploinsufficiency, and 32 additional patients with deleterious mutations of 10 different canonical NF-κB pathway-related genes (REL, RELA, IKBKB, IKBKG, NFKBIA, HOIL1, CARD11, MALT1, OTULIN and RBCK1). All of the patients tested negative for AAN-I-IFNs (Extended Data Fig. 7l,m). These autoantibodies were also absent in patients with IEIs associated with defective T follicular helper (TFH) cell function (autosomal-dominant STAT3 deficiency, n = 11), low Treg cell proportions (autosomal-dominant IL6ST deficiency, n = 10; autosomal-recessive ZNF341 deficiency, n = 10), or both low Treg and TFH cell counts (autosomal-recessive CARMIL2 deficiency, n = 16) (Supplementary Fig. 11). Haematopoietic stem cell transplantation (HSCT) cannot cure defects of thymic stromal cells. We therefore hypothesized that AAN-I-IFNs might appear even after transplantation. One of the four patients with autosomal-recessive complete RELB deficiency who had undergone HSCT had neutralizing AAN-I-IFNs before transplantation (at the age of 2 years). Neutralizing AAN-I-IFNs were detected in post-transplant samples from two out of the three other patients with RELB deficiency (Q72Tfs*152 and Y397*, 6 and 2.5 years after HSCT, respectively), whereas no such autoantibodies were detected in a patient with autosomal-recessive c-REL deficiency over a period of 7 years after HSCT, or in children with inborn errors of T-cell-intrinsic or neutrophil-intrinsic immunity or of erythrocyte function (n = 20), up to 15 years after transplantion (Supplementary Table 5 and Extended Data Fig. 7n). These autoantibodies were also detected in the plasma of patients with autosomal-recessive complete NIK deficiency (n = 2 out of 2, 3 and 7 years after HSCT), or with a p52LOF/IκBδGOF variant (n = 1 out of 1, 14 years after HSCT), for whom the available plasma samples were collected exclusively after transplantation (Supplementary Table 5). These results suggest that inborn errors of RELB, NIK and NF-κB2 from the alternative NF-κB pathway underlie the development of AAN-I-IFNs, even after HSCT, whereas defects of the canonical NF-κB pathway do not. Effective functioning of the alternative NF-κB pathway in thymic stromal cells therefore appears to be essential to prevent the generation of AAN-I-IFNs.
Autoantibody profile of patients with p52LOF/IκBδGOF
The presence of autoantibodies against other proteins was assessed in patients with inborn errors of the alternative NF-κB using a panel of around 20,000 human proteins corresponding to a large proportion of the full-length proteome, many of which were in their native conformation (HuProt). Moreover, 15 patients with APS-1 and 25 healthy controls, all sex- and aged-matched with the 13 patients with p52LOF/IκBδGOF variants tested, were included. The IFNα subtypes and IFNω were among the autoantigens with the highest level of enrichment in the 13 patients with p52LOF/IκBδGOF tested relative to control plasma (log2-transformed fold change of >1.5) (Fig. 3i and Extended Data Fig. 8a). This enrichment was specific to the IFNα subtypes and IFNω, but not other type I IFNs (IFNβ, IFNκ or IFNε) or type III IFNs (Fig. 3i). By contrast, autoantibodies against IL-17A, IL-17F and IL-22 (multiplex beads assay) and most of the other autoantigens commonly identified in cohorts of patients with APS-1 (HuProt microarray) were not found in patients with p52LOF/IκBδGOF variants (Extended Data Fig. 8b–e). Patients with p52LOF/IκBδGOF variants had a lower diversity of IgG-binding autoantigens compared with patients with APS-1 (n = 81 and 159 targeted proteins, respectively). Moreover, half (n = 39, 48%) of the enriched reactive autoantigens in patients with p52LOF/IκBδGOF variants were private, whereas a much smaller proportion (n = 27, 17%) of those enriched in patients patients with APS-1 was private (Extended Data Fig. 8f,g). There were only 12 overlapping IgG-reactive autoantigens, 10 of which were IFNω or IFNα subtypes (Fig. 3j). Most of the reactivities other than those to type I IFNs identified in patients with p52LOF/IκBδGOF variants by HuProt were not detected in a multiplex bead assay (Extended Data Fig. 8h), whereas no pituitary, skin or other tissue-specific autoantigens were detected by HuProt in these patients. We confirmed, by classical diagnostic methods, that almost all of the patients (26 out of 30, 87%) with p52LOF/IκBδGOF variants lacked the tissue-specific autoantibodies typically observed in patients with APS-1 (detected in 25 out of 31, 81%) (Supplementary Fig. 12). These data suggest that autoantibodies neutralizing the 12 IFNα subtypes and IFNω are the principal disease-associated autoantibodies detected in patients with inborn errors of the alternative NF-κB pathway.
Extended Data Fig. 8. Narrow autoantibody profiles in patients with p52LOF/IκBδGOF variants.
(a-b) Heat map of the autoantigens with the highest levels of enrichment in patients with a p52LOF/IκBδGOF variant (n = 13, a) or APS-1 patients (n = 15, b), versus patients with AR RelB deficiency (n = 8), AR NIK (n = 2) deficiency, or with APS-1 (n = 15), as determined with protein microarray (HuProt). Results are shown as the mean fluorescence of two technical replicates with a log2 fold-change >1.8 in patients with p52LOF/IκBδGOF variants (a) or APS-1 (b) relative to 25 healthy controls (HC). (c) Detection of IgG auto-Abs against IL-17A, IL-17F, or IL-22 using a multiplex bead array in patients with inborn errors of the alternative NF-κB pathway. Data representative of one independent experiment are shown. (d) Protein microarray distribution of auto-Abs against IFN-α and IFN-ω (red dots) or other autoantigens frequently found targeted in patients with APS-1 (green dots), in patients with a p52LOF/IκBδGOF variant, relative to controls. (e) Protein microarray distribution of auto-Abs against IFN-α and IFN-ω (red dots), or other autoantigens associated with APS-1 (green dots) in APS-1 patients relative to controls. (f) Number of autoreactive IgG in each patient (APS-1, p52LOF/IκBδGOF, RelB−/−) or control, as determined by the sum of autoantigens with a log2 FC > 1.5 relative to the mean value for all healthy controls (HC). The error bars represent the median ± s.d. of the autoreactive IgG in each group. Comparisons done using two-tailed Mann–Whitney test. (g) Proportion of shared (by ≥ 2 patients) and private reactive autoantigens in the group of patients with a p52LOF/IκBδGOF variant, APS-1, or AR RelB deficiency. (h) Detection of auto-Abs against ATP4A, RBM38, or TROVE2 in a multiplex bead array. The white dot indicates the positive control for the detection of anti-TROVE2 auto-Abs. A.U. corresponds to arbitrary units. Data representative of one independent experiment are shown.
AAN-I-IFNs underlie viral susceptibility
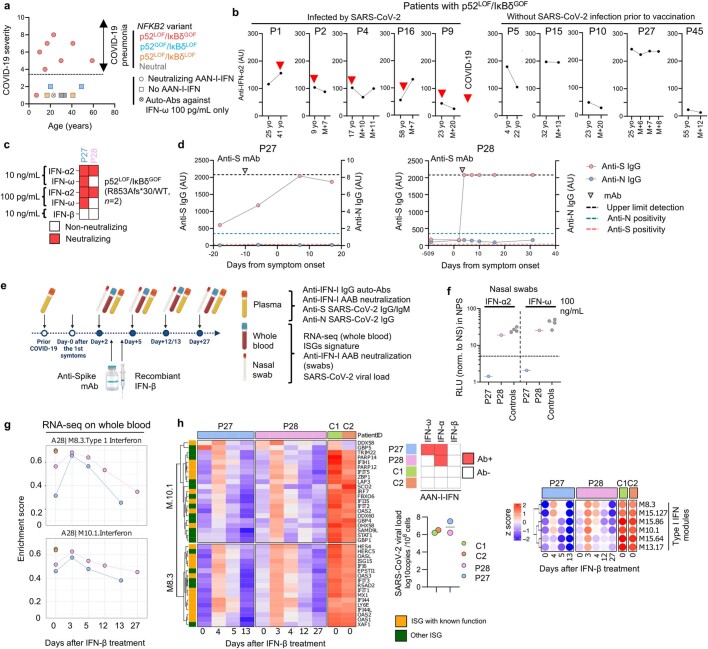
We hypothesized that the susceptibility to viral diseases, including COVID-19, reported in patients with inborn errors of the alternative NF-κB pathway might be at least partly explained by the presence of AAN-I-IFNs. All of the patients (n = 31) with p52LOF/IκBδGOF variants and severe viral infections had AAN-I-IFNs, including all of those with severe forms of COVID-19, influenza, varicella zoster virus or recurrent HSV-1 disease (Fig. 4a). Furthermore, at least one episode of severe or recurrent viral disease was reported in 31 of the 47 (66%) patients with p52LOF/IκBδGOF variants and AAN-I-IFNs, but not in those without such antibodies. With the exception of viral susceptibility and B cell lymphopenia, there were no strong clinical or immunological differences between patients with p52LOF/IκBδGOF variants with and without AAN-I-IFNs (Fig. 4b,c). Two out of the eight patients with autosomal-recessive RELB deficiency developed a severe viral disease (varicella pneumonia, n = 2; and PML, n = 1), and both had autoantibodies neutralizing IFNα and IFNω (Supplementary Table 2). All seven patients with p52LOF/IκBδGOF variants who developed COVID-19 pneumonia during the prevaccination period had neutralizing autoantibodies against both IFNα-2 and IFNω, and experienced critical (n = 4), severe (n = 2) or moderate (n = 1) COVID-19 pneumonia (Fig. 4d,e, Extended Data Fig. 9a and Supplementary Table 6). Plasma samples collected from two of these patients (P1 and P16) before SARS-CoV-2 infection neutralized IFNα-2 and IFNω at a concentration of 10 ng ml−1. These samples were collected up to 16 years before COVID-19, demonstrating that these neutralizing autoantibodies were present before infection and were therefore not triggered by SARS-CoV-2 infection (Extended Data Fig. 9b). These autoantibodies against IFNα and IFNω blocked type I IFN signalling by impairing type I IFN ISG induction in vivo in the blood and upper respiratory tract during COVID-19, which could be rescued by exogenous IFNβ treatment in these patients (Supplementary Results 4, Extended Data Fig. 9c–h and Supplementary Fig. 13). Two other patients were infected without developing pneumonia or requiring hospitalization: one 22-year-old patient with autoantibodies neutralizing only IFNω at the lowest dose of 100 pg ml−1 (P5, S762Afs*21/wild type (WT)) and one 7-year-old patient with autoantibodies neutralizing both IFNα-2 and IFNω at a concentration of 10 ng ml−1 (P38, G869Vfs*18/WT) (Fig. 4d,e). The six infected patients without AAN-I-IFNs received ambulatory care and did not develop pneumonia. They were heterozygous for neutral (A567 and V661M) variants, for the Q539* p52GOF/IκBδLOF variant (n = 2) or for the R52*/WT p52LOF/IκBδLOF variant (n = 2) (Fig. 4d,e and Extended Data Fig. 9a). Furthermore, ten patients with a p52LOF/IκBδGOF variant and pre-existing AAN-I-IFNs encountered SARS-CoV-2 after vaccination (corresponding to the Omicron period, from October 2021 to February 2022) (Supplementary Fig. 14). They received an infusion of anti-SARS-CoV-2 monoclonal antibodies (n = 4, as sotrovimab (n = 3) or tixagevimab/cilgavimab (n = 1)), remdesivir (n = 1) or nirmatrelvir/ritonavir (n = 1) and/or recombinant IFNβ (n = 2) in addition to intravenous immunoglobulin supplementation (n = 10). All of these patients reported asymptomatic to mild (NIH scale, 1–3) COVID-19 without pneumonia (Supplementary Fig. 14 and Supplementary Table 6). P3, who developed critical COVID-19 pneumonia during the first wave of the SARS-CoV-2 pandemic, developed ambulatory disease (NIH score, 2) after vaccination and the therapeutic infusion of sotrovimab. The two patients with p52LOF/IκBδLOF variants (P43 and P63) and three with p52GOF/IκBδLOF variants (P39, P40 and P41) without AAN-I-IFNs had ambulatory disease. Overall, these results indicate that AAN-I-IFNs are clinically important, underlying severe forms of COVID-19 pneumonia and, probably, other severe viral diseases, including influenza pneumonia and severe varicella.
Fig. 4. Susceptibility to COVID-19 pneumonia and other severe viral diseases is strongly associated with the presence of AAN-I-IFNs.
a, The number of patients with a p52LOF/IκBδGOF variant and manifestations of viral diseases as a function of their AAN-I-IFN status. b, Clinical and immunological manifestations in patients with a p52LOF/IκBδGOF variant, as a function of their AAN-I-IFN status. Autoimm., autoimmunity; ecto. dyspl., ectodermal dysplasia; hypogam., hypogammaglobulinaemia; hypox., hypoxaemic; rec., recurrent; RTI, recurrent bacterial respiratory tract infection. c, Chord diagram of the main clinical and immunological manifestations of patients with inborn errors of NF-κB2. d, Anti-IFNα-2 IgG detection by Gyros in positive control individuals (n = 10), healthy control individuals (n = 7), patients with a p52LOF/IκBδGOF (n = 9), p52LOF/IκBδLOF (n = 2), p52GOF/IκBδLOF (n = 2) or neutral (n = 2) NF-κB2 variant and COVID-19, as a function of disease severity. e, Heat map showing the type I IFN neutralization profile of unvaccinated patients during COVID-19, according to disease severity and clinical presentation during infection, including patients with a p52LOF/IκBδGOF (n = 9), p52GOF/IκBδLOF (n = 2) or p52LOF/IκBδLOF (n = 2) variant. The red squares indicate a complete neutralization ability of the plasma for ISRE induction in the luciferase reporter assay system, and the white squares indicate a total absence of neutralizing autoantibody detection in the ISRE–luciferase assay. f, The viral load and IFN score in nasal swabs over the course of SARS-CoV-2 infection in patients with a p52LOF/IκBδGOF variant (n = 2) with AAN-I-IFNs, and in vaccinated individuals with a mild disease and no AAN-I-IFNs (n = 4). g, The IFN score and viral load in whole blood (left) or nasal swabs (right) over the course of SARS-CoV2 infection in patients with a p52LOF/IκBδGOF variant with AAN-I-IFNs (n = 2), or in individuals infected with SARS-CoV-2 presenting only mild disease (n = 36). The vertical arrows indicate the times of recombinant human IFNβ (rhIFNβ) injection and the arrowheads indicate the infusion of monoclonal antibodies (mAbs) against SARS-CoV-2 spike protein.
Extended Data Fig. 9. AAN-I-IFNs prevent ISG induction in blood and the upper respiratory tract during COVID-19, a defect that can be rescued by exogenous IFN-β treatment.
(a) Correlation between age and COVID-19 severity in patients with inborn errors of NF-κB2. The crossed light red square represents a patient with auto-Abs neutralizing only IFN-ω at 100 pg/mL. (b) Changes in the titres of auto-Abs against IFN-α2, as measured by Gyros, with age, in patients with a p52LOF/IκBδGOF variant and COVID-19. Red arrows indicate the onset of COVID-19. (c) Heatmap showing the neutralization profile of P27 et P28 heterozygous for a p52LOF/IκBδGOF variant (R853Afs*30/WT) during COVID-19. (d) Longitudinal follow-up of anti-S and anti-N IgG in P27 and P28 during the course of COVID-19, before and after treatment by the infusion of an anti-S monoclonal Ab (mAb, grey arrow). (e) Overview of the longitudinal investigation of COVID-19 episodes in P27 et P28. (f) Neutralization capacity of the nasal swab from P27 et P28 upon SARS-CoV-2 infection and individuals infected with the omicron variant but without detectable AAN-I-IFNs (controls, n = 4, grey dots). Bars represent the median. (g) Longitudinal IFN module enrichment score during the course of COVID-19 in P27 and P28 and in two age-matched controls infected with SARS-CoV-2. IFN modules M.10.1 and M.8.3 are represented. Values obtained before and after the treatment of P27 and P28 with IFN-β. (h) ISG score induction by IFN module analysis during the course of COVID-19 in P27 and P28, before and after recombinant IFN-β treatment, and in age-matched controls (C1 and C2, n = 2) infected with SARS-CoV-2.
AIRE expression in alternative NF-κB IEIs
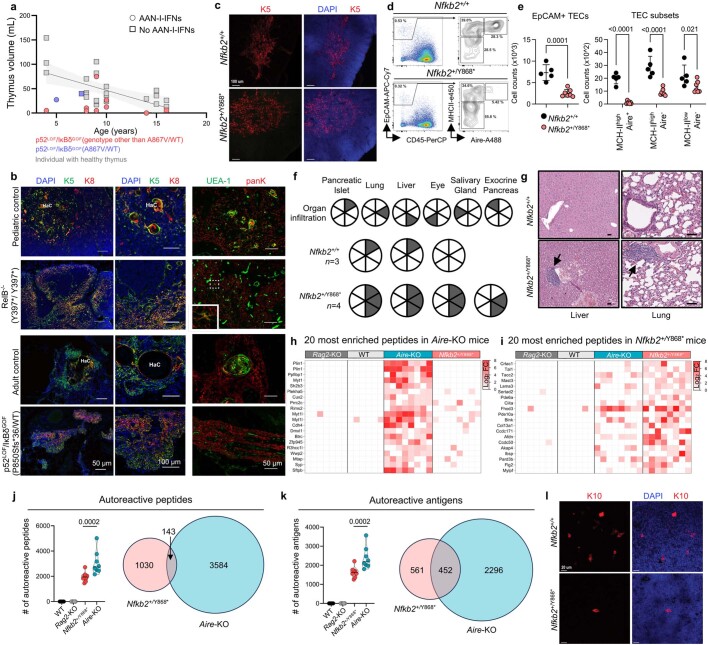
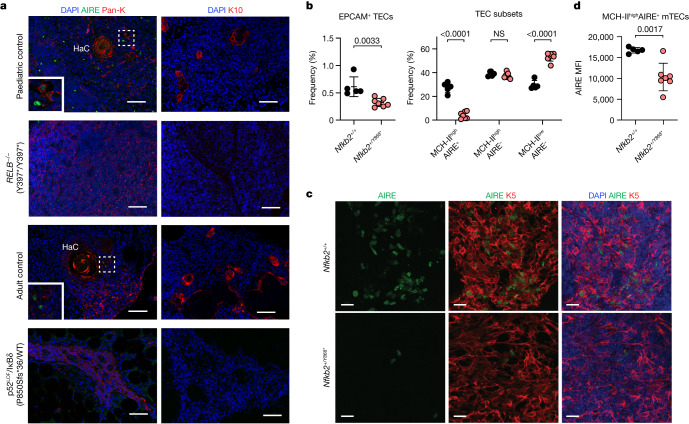
In mice, mTEC development and AIRE expression are dependent on the alternative NF-κB pathway, through NIK and RELB24–26. Consequently, Relb- and Nik-deficient mice, and mice heterozygous for a p52LOF/IκBδGOF variant, display thymic hypoplasia with weak medullary thymic formation, impaired maturation of AIRE-expressing mTECs and tolerance breakdown26,30,31. In human fetal thymuses, NFKB2 and RELB transcripts are highly abundant in AIRE+ mTECs32. However, the impact of deleterious variants affecting the alternative NK-κB pathway on human AIRE expression remains unclear. We hypothesized that patients with inborn errors of NIK, RELB or NF-κB2 develop AAN-I-IFNs due to insufficient AIRE expression in the thymus. An analysis of the thymic volume in patients with p52LOF/IκBδGOF variants (n = 11) aged 4 to 16 years revealed that the total thymic volume was smaller in these patients compared with age-matched controls with conditions unrelated to immunity (Extended Data Fig. 10a). We next analysed thymic biopsy samples from a patient with autosomal-recessive complete RELB deficiency (mutation Y397*/Y397*, biopsy performed at the age of one year, with AAN-I-IFNs) and a deceased patient with a p52LOF/IκBδGOF variant (P850Sfs*36/WT, the sample was collected at the age of 27 years; no plasma sample was available). Immunofluorescence analysis of the thymic tissue sections from these two patients revealed a dysplastic organ with a disorganized corticomedullary architecture and atrophic medulla (Extended Data Fig. 10b). A residual epithelial cell population (pan-keratin-expressing cells) with disorganized keratin 5 (K5)- and keratin 8 (K8)-positive cells was detected in the thymuses of both patients (Extended Data Fig. 10b). mTECs (defined as pan-keratin+UEA-1+ cells) were rare, but not entirely absent, in the thymus of the patient with autosomal-recessive RELB deficiency (Extended Data Fig. 10b). However, no AIRE or keratin 10 (K10)-positive Hassall’s corpuscles were detected (Fig. 5a). These findings suggest that RELB deficiency does not completely block mTEC specification but, rather, prevents differentiation into AIRE-expressing and post-AIRE mTECs. An analysis of the thymus from the adult patient with a p52LOF/IκBδGOF variant showed that, relative to an age-matched control thymus, the thymus tissue from this patient lacked UEA-1+ mTECs, AIRE and Hassall’s corpuscles (Fig. 5a and Extended Data Fig. 10b). These data suggest that the p52LOF/IκBδGOF genotype impaired the maturation of human AIRE-expressing cells. However, thymic involution in this older patient made it difficult to draw definitive conclusions regarding the impact of the mutation on mTEC development earlier in life. Collectively, these data suggest that human p52–RELB heterodimers control the development of mature mTECs and the thymic expression of AIRE, and that inborn errors of the human alternative NF-κB pathway underlie the production of AAN-I-IFNs due to the impaired development of mature AIRE-expressing mTECs.
Extended Data Fig. 10. Thymus volume in p52LOF/IκBδGOF children, and autoimmune pathology and autoreactive IgG profiles in Nfkb2+/Y868* mice.
(a) Estimation of thymus volume in p52LOF/IκBδGOF patients relative to aged-matched controls aged from 3 to 16 years. Black line represents simple linear regression of the control thymus volume with its 95% confidence bands (grey area). Data representative of one independent experiment are shown. (b) Immunofluorescence staining of thymic tissue from age-matched controls and patients with AR complete RelB deficiency or a p52LOF/IκBδGOF variant. HaC, Hassall’s corpuscles. Scale bars, 50 μm (left and right panel) or 100 μm (central panel). (c) Representative confocal microscopy images of the thymic medulla stained for cytokeratin 5 (K5) for the WT controls (Nfkb2+/+, n = 3) or Nfkb2+/Y868* mice (n = 3). Scale bar, 100 μm. Data representative of two independent experiment are shown. (d) Contour plot for Aire expression in mature MHC-II+ mTECs. (e) Absolute number of EpCAM+CD45- thymic epithelial cells (TECs), and TEC subsets in Nfkb2+/+ (n = 5) and Nfkb2+/Y868* (n = 7) mice. Comparisons done using unpaired, parametric, two-tailed Student’s t-test (for EpCAM+ TECs) or two-way non-parametric ANOVA (Sidak’s test) with correction for multiple comparisons (for TEC subsets). Three independent experiments were performed. (f) Summary of lymphocytic cell infiltrates in Nfkb2+/+ (n = 3) and Nfkb2+/Y868* (n = 4) mice. Each circle represents an individual animal, and each slice of the circle represents a tested organ. Lymphocytic infiltrates of the designated organ are indicated by the grey-shaded sections of the circle. One independent experiment was performed. (g) Representative tissue sections stained with hematoxylin and eosin (H&E) showing lymphocytic infiltrates (black arrows) in Nfkb2+/+ or Nfkb2+/Y868* mice. Scale bar, 50 μm. Data representative of one independent experiment are shown. (h-i) Heatmap of the top 20 autoreactivities by degree of enrichment, in Aire-KO (h) or Nfkb2+/Y868* (i) mice relative to WT and Rag2-KO mice. (j-k) Number of autoreactive peptides (j) or antigens (k) displaying enrichment in WT, Rag2-KO, Nfkb2+/Y868* and Aire-KO mice, and representation of the autoreactive peptide and antigen profiles of Nfkb2+/Y868* and Aire-KO mice, with their overlap. Comparisons done using two-tailed Mann–Whitney test. (l) Representative confocal microscopy images of K10 and DAPI staining on Nfkb2+/+ (n = 3) and Nfkb2+/Y868* (n = 3) mouse thymuses. Scale bars, 20 μm. Data representative of two independent experiments are shown.
Fig. 5. Impaired mTEC development and thymic AIRE expression in a patient with autosomal-recessive RELB deficiency, a patient heterozygous for a p52LOF/IκBδGOF NF-κB2 variant and in mice heterozygous for the Y868* NF-κB2 variant.
a, Immunofluorescence staining of thymic tissue from age-matched controls, a patient with autosomal-recessive complete RELB deficiency or heterozygous for a p52LOF/IκBδGOF NF-κB2 variant. AIRE-expressing cells (green) and Hassall’s corpuscles (HaC) are shown on the left. Pan-K, pan-keratin. Staining for K10 (red), defining terminally differentiated corneocyte-like mTECs, is shown on the right. DAPI staining is shown in blue. Scale bars, 50 μm (left) and 100 μm (right). Inset: the controls at a higher magnification. Data shown are representative of one independent experiment. b, The percentage of EPCAM+CD45− thymic epithelial cells (TECs), and the various TEC subsets (defined on the basis of their MHC class II (MHC-II) and AIRE expression) in WT controls (Nfkb2+/+, black dots, n = 5) and mice carrying a heterozygous missense variant homologous to the human Y868* p52LOF/IκBδGOF NF-κB2 variant (Nfkb2+/Y868*, red dots, n = 7). Statistical comparisons were performed using unpaired, parametric, two-tailed Student’s t-tests (EPCAM+ TECs) or two-way nonparametric analysis of variance (ANOVA) (Sidak’s test) with correction for multiple comparisons (TEC subsets). Data are mean ± s.d. Data shown are representative of three independent experiments. c, Representative confocal microscopy images of AIRE (green), K5 (red) and DAPI (blue) of WT (Nfkb2+/+, n = 3, top) and Nfkb2+/Y868* (n = 3, bottom) mouse thymuses. Scale bars, 20 μm. Data shown are representative of two independent experiments. d, Mean fluorescence intensity (MFI) of AIRE expression in mature MHC-IIhighAIRE+ mTECs from WT (n = 5) and Nfkb2+/Y868* (n = 7) mouse thymuses. Statistical comparisons were performed using unpaired, parametric two-tailed Student’s t-tests. Data are mean ± s.d. Data shown are representative of three independent experiments.
Aire expression in Nfkb2+/Y868* mice
We further investigated the role of the p52–RELB heterodimer in mature AIRE+ mTEC development by generating mice carrying a heterozygous variant homologous to the human Y868* p52LOF/IκBδGOF NF-κB2 variant (Nfkb2+/Y868*)31,33. Despite the relatively normal thymic medullary compartmentalization, as shown by K5 immunofluorescence staining (Extended Data Fig. 10c), the cellularity of the thymic epithelium was significantly lower in Nfkb2+/Y868* mice compared with in WT mice, reflecting a dysregulation of mTEC development and homeostasis (Fig. 5b and Extended Data Fig. 10d,e). The proportion of AIRE+ mTECs and absolute counts for this subset were very low, but non-zero, in Nfkb2+/Y868* mice relative to in WT mice, as shown by both flow cytometry and immunofluorescence staining of tissue sections (Fig. 5b,c and Extended Data Fig. 10d,e). The residual AIRE+ mTECs in the Nfkb2+/Y868* mice displayed significantly weaker AIRE expression compared with their WT counterparts (Fig. 5d). Impaired mTEC development in Nfkb2+/Y868* and Nfkb2+/D865G mice causes T cell autoimmunity in multiple organs31. The lymphocytic infiltrates in the Nfkb2+/Y868* mice affected the pancreatic islets, lung and liver, as in NOD Aire-KO mice of the same age34. However, in marked contrast to NOD Aire-KO mice, the salivary glands, exocrine pancreas and retina were spared (Extended Data Fig. 10f,g). Consistently, the autoreactive IgG profiles of Nfkb2+/Y868* mice, like those of their human counterparts, are narrower than that of Aire-KO mice, with only a minimal overlap, as shown by analysis using phage-display immunoprecipitation and sequencing (PhIP–seq) (Extended Data Fig. 10j,k). After their maturation into MHC-IIhighAIRE+ mTECs, these cells can display a downregulation of AIRE expression and give rise to terminally differentiated mTECs (also called post-AIRE mTECs or mimetic cells), with distinctive extrathymic parenchyma-specific features35,36. In mice, AIRE expression and function are required for the development of some terminally differentiated mTECs, as shown by the small proportions of corneocyte-like mTECs in Aire-KO mice35,37,38. We therefore investigated the consequences of impaired p52–RELB heterodimer activation in mouse mTECs on the development of corneocyte-like mTECs by assessing K10 expression. Like their human counterparts, Nfkb2+/Y868* mice had very small numbers of K10-expressing post-AIRE mTECs in the medulla (Extended Data Fig. 10i). These data strongly suggest that the human alternative NF-κB pathway is essential for the development of mature mTECs, as its defects prevent proper AIRE expression and the generation of other AIRE-dependent terminally differentiated mTECs, whereas this pathway appears to be redundant for the development of some other mTEC subsets (MHClowAIRE− cells).
Discussion
We found that human inborn errors of the alternative NF-κB pathway (autosomal-recessive NIK, autosomal-recessive RELB or autosomal-dominant p52LOF/IκBδGOF NF-κB2 disorders) define a new group of IEIs underlying the development of AAN-I-IFNs. The presence of these autoantibodies is consistent with the cellular phenotype found in the patients’ fibroblasts, culminating in defective p52–RELB activity, which may be secondary to the impaired processing of p100 to generate p52 or to a quantitative or qualitative RELB deficiency. By contrast, no AAN-I-IFNs were found in patients who were heterozygous for p100–IκBδ LOF variants, or in patients with inborn errors of the canonical NF-κB pathway. This suggests that a correct NIK-dependent processing of p100 is a key checkpoint for the p52–RELB-dependent activation of the alternative NF-κB pathway that is required to prevent the development of AAN-I-IFNs.
The p52–RELB heterodimers control AIRE expression in mouse mTECs29. Indeed, Map3k14- (encoding NIK), Ikka- or Relb-deficient mice, and Nfkb2+/Y868* mice have strongly reduced thymic AIRE expression26,31,39,40, and the deletion of the enhancer element containing two NF‐κB binding sites upstream from the Aire-coding locus phenocopies Aire deficiency41. We detected no AIRE expression in human thymuses lacking RELB or heterozygous for a p52LOF/IκBδGOF variant. Moreover, the development of terminally differentiated corneocyte-like mTECs was impaired in both humans and mice with defective p52–RELB activity. This finding is consistent with the small size of this population in Aire-KO mice, and the role of AIRE in decreasing chromatin accessibility at NF-κB regulatory elements in mature mTECs, facilitating their terminal differentiation35,37,38,42. Thus, the alternative NF-κB pathway appears to be essential for the development of mature mTECs in humans, and for proper thymic AIRE expression, by ensuring correct p52–RELB activation.
The notable association between the presence of AAN-I-IFNs in patients with human inborn errors of NIK, RELB and NF-κB2 suggests that intact p52–RELB activation is essential to prevent the breakdown of AIRE-dependent central T cell tolerance toward type I IFNs in humans. Causality is supported by several lines of evidence: the development of these AAN-I-IFNs in almost all, if not all, humans with inherited AIRE deficiency, regardless of age or ancestry1,2,23,43–48; the reduced AIRE expression in patients with other germline (hypomorphic RAG1 or RAG2 variants20–22) or somatic (mTEC neoplasia49) conditions underlying the development of these autoantibodies; the impaired development of AIRE-expressing mTECs in both mouse and human disorders of the alternative NF-κB pathway26,31,39,40; the absence of Hassall’s corpuscles in patients with inborn errors of the alternative NF-κB pathway, which mirrors the decreased levels of terminally differentiated corneocytes observed in Nfkb2+/Y868* mice and Aire-KO mice; and the persistence of these AAN-I-IFNs up to 14 years after HSCT with engraftment.
However, it is surprising that AIRE deficiency in patients with inborn errors of the alternative NF-κB pathway leads to such an apparently narrow breakdown of central tolerance, restricted almost exclusively to the 12 IFNα subtypes and IFNω, when pathogenic autoantibodies are considered. This situation contrasts with the immunological and clinical manifestations of patients with APS-1, which only partially overlap those of patients with inborn errors of the NF-κB pathway44,45. The absence of the typical clinical and immunological features of APS-1 other than AAN-I-IFNs in patients with p52LOF/IκBδGOF variants may be attributed to the presence of residual mature mTECs or terminally differentiated mTECs (mimetic cells) that would ensure central tolerance to the other antigens targeted in APS-135,36,50. Conversely, the low blood counts of B and TFH cells in patients with p52LOF/IκBδGOF variants, but not in patients with APS-1, may be a result of the impaired alternative NF-κB pathway in B cells or in non-mTEC stromal cells51. The low Treg cell counts of patients with p52LOF/IκBδGOF variants may result from an impaired alternative NF-κB pathway in T cells or impaired AIRE expression in mTECs31,52,53. The clinical manifestations in patients with p52LOF/IκBδGOF variants also differ from those in patients with the other two forms of autosomal-dominant inborn errors of NF-κB2, probably due to the higher levels of IκBδ activity of the mutant protein (Supplementary Table 7).
Our findings confirm the detrimental consequences of the presence of AAN-I-IFNs for viral susceptibility (COVID-19 pneumonia, influenza pneumonia and herpesvirus diseases)3,7,13,23. Despite their high risk of developing life-threatening COVID-19 pneumonia, unvaccinated patients with inborn errors of the alternative NF-κB pathway displayed a high but incomplete penetrance of hypoxaemic COVID-19 pneumonias, as reported in patients with APS-1 or SLE3,54,55. Additional protective or risk factors may be required in these patients to influence the clinical outcome of COVID-19, such as age or the nature of the AAN-I-IFNs (neutralizing IFNω and/or the 12 IFNα subtypes). Our findings also suggest that a reinforcement of prophylactic or therapeutic interventions can improve the clinical outcome of viral diseases in patients with AAN-I-IFNs, throughout their lives, as these autoantibodies may persist even after HSCT12. Collectively, these results suggest that the human alternative NF-κB pathway controls AIRE expression in mTECs and that human inborn errors of this pathway thereby underlie the development of AAN-I-IFNs and the resulting predisposition to viral infection. They confirm that at least some individuals develop AAN-I-IFNs because of an underlying IEI, suggesting that other genetic aetiologies remain to be discovered in the 0.3% to 2% of individuals under 70 years of age who carry such autoantibodies. The observation that genetic aetiologies of AIRE in cis or in trans that disrupt central T cell tolerance underlie these autoantibodies suggests that as yet undiscovered genetic aetiologies may also affect this process. The genetic study of patients with AAN-I-IFNs may reveal new molecular components in this or other processes. What triggers the rise in autoantibody levels against type I IFNs after the age of 70 years is another related question potentially linked to thymic involution.
Methods
Participants and samples
We enrolled 73 patients with rare variants of NFKB2 though an international collaborative study. Data were collected through an anonymized survey sent to specialists in immunology or paediatrics with reported or unreported patients with these IEIs (Supplementary Table 1). All index cases were genotyped after suspicion of an inborn error of immunity. An analysis of the familial segregation of each NFKB2 variant was performed in all relatives for whom genomic DNA was available. We included all individuals heterozygous for a rare (MAF < 0.0001) NFKB2 non-synonymous variant (detected by Sanger sequencing: n = 19; IEI gene NGS panel: n = 26; whole-exome sequencing (WES): n = 23; or whole-genome sequencing (WGS): n = 5) for whom a plasma/serum sample was also available. Clinical and immunological data were collected with a standardized questionnaire using Microsoft Excel, together with at least one plasma sample. The plasma samples from patient P1 were obtained through the NCT03394053 and NCT03610802 protocols with the approval of the National Institutes of Health institutional review board. We also enrolled 14 patients with other inborn errors of the alternative NF-κB pathway including patients with autosomal-recessive complete NIK deficiency (n = 2 from 2 kindreds57 and unpublished results); autosomal-recessive partial (n = 4 from 2 kindreds) or complete (n = 4 from 2 kindreds) RELB deficiency57–61 (and unpublished results), or the related TNF receptors (TNFR) (autosomal-recessive complete BAFFR deficiency (n = 1)58); or X-linked recessive CD40L deficiency (n = 3 from 3 kindreds (unpublished)). They were detected by Sanger sequencing: n = 7; and WES: n = 7. No plasma from patients with autosomal-recessive IKKα deficiency was available.
Definitions and outcome measures
PAD was defined by the association of hypogammaglobulinaemia and recurrent bacterial respiratory tract infections62. Ectodermal dysplasia was defined by the association of sparse hair, eyebrows, or eyelashes, or nail dysplasia, with or without alopecia areata or totalis.
The severity of COVID-19 was defined according to the NIH ordinal scale, as previously reported8,63. The NIH scale is an eight-point ordinal scale ranging from ambulatory (1, no limitations of activities; 2, limitation in activity), to hospitalized (3, not requiring supplemental oxygen), moderate (4, not requiring supplemental oxygen but requiring ongoing medical care (related to COVID-19 or to other medical conditions)), severe (5, requiring supplemental oxygen) or critical (6, requiring non-invasive ventilation or use of high-flow oxygen devices; 7, receiving invasive mechanical ventilation or extracorporeal membrane oxygenation (ECMO); and 8, death).
Plasmids and mutagenesis
The NFKB2 (encoding p100), RELB and MAP3K14 (encoding NIK) plasmids were obtained from Origen with a C-terminal DDK tag. The κB reporter construct (κB-luc), pGL4.32[luc2P/NF-κB-RE/Hygro] and pRL-SV40 vectors were obtained from a previous study64. Site-directed mutagenesis was performed as previously described64.
Cell culture and transfection
HEK293T cells or HeLa cells (American Type Culture Collection) were maintained in Dulbecco’s modified Eagle medium (DMEM; Gibco) supplemented with 10% FBS (Gibco). Transient transfection was performed using X-tremeGENE 9 DNA Transfection Reagent (Merck) according to the manufacturer’s instructions. The cell lines were regularly tested and were found to be free of mycoplasma contamination.
Functional evaluation of NFKB2 variants
Luciferase reporter assays
The luciferase reporter assay was performed as previously described64. WT HEK293T cells in 96-well plates were transfected with a κB reporter plasmid (100 ng per well), the pRL-SV40 vector (10 ng per well), WT MAP3K15, WT RELB, and a WT or mutant p100 in the presence of X-tremeGENE 9 DNA Transfection Reagent (Merck). After incubation for 24 to 48 h, cells were collected, and luciferase activity was measured with the Dual-Glo Luciferase Assay System (Promega). We considered a deleterious variant to be p52-LOF if its luciferase activity was equivalent to that after cotransfection with EV, RELB and MAP3K14, hypomorphic if this activity was more than half that of the WT allele, and p52 gain-of-function (GOF) if this activity was less than half of that after cotransfection with RELB, MAP3K14 and WT NFKB2.
Western blotting
Whole-cell lysates from HEK293T cells, MDDC, T cell blasts, primary or SV-40-transformed fibroblasts were prepared in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with Complete Protease Inhibitor Cocktail (Roche). Proteins were separated by electrophoresis in 10% PROTEAN TGX Precast Protein Gels (Bio-Rad), and transferred onto Immobilon-P polyvinylidene fluoride membrane (Millipore). All blots were incubated overnight with primary antibodies and developed with the Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). The antibodies used in this study included antibodies against p100/p52 (4882; Cell Signaling Technology, 1:1,000), p105/p50 (N terminus; 3035; Cell Signaling Technology, 1:1,000), p65 (sc-372; Santa Cruz Biotechnology, 1:1,000), RELB (sc-48366; Santa Cruz Biotechnology, 1:800), REL (sc-6955; Santa Cruz Biotechnology, 1:1,000), and the following secondary antibodies: Amersham ECL mouse IgG, HRP-linked whole antibody (from sheep; NA931; GE Healthcare Life Sciences) and Amersham ECL rabbit IgG, HRP-linked whole antibody (from donkey; NA934; GE Healthcare Life Sciences). Uncropped western blots can be found in Supplementary Fig. 1.
Confocal microscopy
HeLa cells were plated on chambered coverslips (80826, iBidi) and were left untransfected or were transiently transfected with a plasmid encoding p100, RELB and/or NIK and/or an empty pCMV6 vector for 48 h. Primary or SV-40 fibroblasts were plated on chamber coverslips and left unstimulated or were stimulated with 100 ng ml−1 Lt or 100 ng ml−1 TWEAK for 48 h. The cells were fixed in 4% formaldehyde in phosphate-buffered saline (PBS), pH 7.4. Cells were incubated overnight at 4 °C with anti-p100/p52 (4882; Cell Signaling Technology, 1:1,000), or RELB (sc-48366; Santa Cruz Biotechnology, 1:800) primary antibodies. The cells were washed three times with 1× PBS and stained by incubation with secondary antibodies for 1 h at room temperature (goat anti-mouse IgG Alexa Fluor 488 (A-11029, 1:250); goat anti-rabbit IgG Alexa Fluor 633 (A-11037, 1:250) before mounting in Prolong-gold and visualization by confocal microscopy (×63 or ×40 oil-immersion lens).
Detection and functional evaluation of anti-cytokine autoantibodies
Gyros
Cytokines, rhIFNα-2 (Miltenyi Biotec, 130-108-984) or rhIFNω (Merck, SRP3061) were first biotinylated with EZ-Link Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific, A39257), according to the manufacturer’s instructions, with a biotin-to-protein molar ratio of 1:12. The detection reagent contained a secondary antibody (Alexa Fluor 647 goat anti-human IgG (Thermo Fisher Scientific, A21445) diluted in Rexip F (Gyros Protein Technologies, P0004825); 1/500 dilution of the 2 mg ml−1 stock to yield a final concentration of 4 µg ml−1). PBS-T 0.01% buffer and Gyros Wash buffer (Gyros Protein Technologies, P0020087) were prepared according to the manufacturer’s instructions. Plasma or serum samples were then diluted 1/100 in PBS-T 0.01% and tested with Bioaffy 1000 CD (Gyros Protein Technologies, P0004253), and Gyrolab X-Pand (Gyros Protein Technologies, P0020520). Cleaning cycles were performed in 20% ethanol.
Luciferase reporter assays
The blocking activity of anti-IFNα-2 and anti-IFNω autoantibodies was determined with a reporter luciferase assay. In brief, HEK293T cells were transfected with a plasmid containing the firefly luciferase gene under the control of the human ISRE promoter in the pGL4.45 backbone, and a plasmid constitutively expressing Renilla luciferase for normalization (pRL-SV40). Cells were transfected in the presence of the X-tremeGene 9 transfection reagent (Sigma-Aldrich, 6365779001) for 24 h. Cells in DMEM (Thermo Fisher Scientific) supplemented with 2% fetal calf serum and 10% healthy donors or patient serum/plasma were either left unstimulated or were stimulated with IFNα-2 (Miltenyi Biotec, 130-108-984) or IFNω (Merck, SRP3061) at 10 ng ml−1 or 100 pg ml−1, or with IFNβ (Miltenyi Biotech, 130-107-888) at 10 ng ml−1 or 1 ng ml−1, or with one of the 13 IFNα subtypes for 16 h at 37 °C. Each sample was tested once for each cytokine and dose in at least two independent experiments. Finally, cells were lysed for 20 min at room temperature and the luciferase levels were measured using the Dual-Luciferase Reporter 1000 assay system (Promega, E1980), according to the manufacturer’s protocol. Luminescence intensity was measured with a VICTOR X Multilabel Plate Reader (PerkinElmer Life Sciences). RLA was calculated by normalizing firefly luciferase activity against Renilla luciferase activity, and then normalizing against non-stimulated conditions. The samples were considered to be neutralizing if the luciferase activity signal, normalized to the non-stimulated conditions, was below 5.
Protein microarray
Protein microarrays (HuProt, CDI laboratories) were incubated in 5 ml blocking buffer, consisting of 2% bovine serum albumin and 0.05% Tween-20 in PBS, for 90 min. The arrays were then incubated overnight in 5 ml blocking buffer per array with serum from a blood donor or patient diluted 1:2,000. Each array was then washed five times, for 5 min each, with 5 ml PBS-T (PBS + 0.05% Tween-20). Alexa Fluor 647 goat anti-human IgG (Thermo Fisher Scientific, A-21445, RRID:AB_2535862) and Dylight 550 goat anti-GST (Columbia Biosciences, D9-1310) were diluted in blocking buffer (1:2,000 and 1:10,000, respectively) and each array was incubated in 5 ml of the resulting mixture for 90 min. Five washes were then conducted as previously described. Incubations and washes were performed on an orbital shaker, with aluminium foil to block out the light during the steps after adding the fluorescent antibodies. Finally, each array was immersed in deionized water three times and centrifuged for approximately 30 s for drying. The arrays were scanned later the same day with an Innoscan 1100AL Fluorescence scanner (Innopsys) using Mapix v.9.1.0 and the resulting images were analysed with the Jan 18-22 Huprot v4.0 Genepix Array List file and either of GenePix Pro v.5.1.0.19 or GenePix Pro 7. Normalization was used to compensate for variation in the signal intensity between experiments. Data from additional healthy donors from separate protein array experiments was included. Signal intensities were extracted from the scanned image with GenePix Pro v.5.1.0.19 and GenePix Pro 7, with the subtraction of the local background. IgG-reactive proteins were identified as proteins with a fluorescence intensity log2[fold change] ≥ 1.5. Autoantigens identified in patients with APS-1 were extracted from previous studies46,47,65. Protein arrays were performed on plasma from 24 patients with inborn errors of the alternative NF-κB pathway, with (n = 15) or without (n = 9) AAN-I-IFNs: p52LOF/IκBδGOF variant (n = 8 and 5), autosomal-recessive RELB (n = 5 and 3) and autosomal-recessive NIK (n = 2 with AAN-I-IFNs) deficiency, and the p52LOF/IκBδLOF variant (n = 1, without AAN-I-IFNs). Moreover, plasma from patients with APS-1 (n = 15) and healthy donors (n = 25) was included, sex- and aged-matched with the 13 patients with the p52LOF/IκBδGOF variant.
Multiplex bead arrays
The method for detecting human IgG in the serum using magnetic beads was described previously66. We used this method with a few modifications, as specified below. The AnteoTech Activation Kit for Multiplex Microspheres (A-LMPAKMM-10) was used in accordance with the manufacturer’s protocol, including the optional blocking, to couple magnetic beads (MagPlex, Luminex) to a panel of 96 analytes including the following commercially available proteins (with 1.5 × 106 beads to 3 μg of the proteins not provided as lysates): IFN-α2, IFN-α1, IFN-α7, IFN-α14, IFN-β1, IFN-ε, IFNω-1, IFN-α5, IL-22, IFN-α6, IFN-α10, IFN-α8, IFN-α16, IFN-α17, IFN-κ, anti-IgG, IFN-α21, IL-17A, TROVE2, RBM38, IFN-α4, IL-17F, and ATP4A. The samples were diluted 1:25 in PBS and then 1:10 in assay buffer (0.05% PBS-T, 3% BSA, 5% milk). Stocks of magnetic beads were sonicated for 1 min before mixing with storage buffer from the activation kit. The diluted samples were centrifuged for 1 min at 3,000 rpm, and 45 μl of each sample was then incubated for 2 h in the dark at room temperature with 5 μl of stock bead solution, with shaking at 650 rpm. The beads were then washed (3 times with PBS-T 0.05%), centrifuged at 2,000 rpm, resuspended in 50 μl 0.2% PFA per well and carefully vortexed. After incubation for 10 min at room temperature and centrifugation at 2,000 rpm, the beads were washed (3 times with PBS-T 0.05%) and incubated with secondary antibodies (Invitrogen, H10104, 2384336) for 30 min at room temperature. Finally, the wash routine described above was repeated, and the beads were dispensed in PBS-T 0.05% before the Luminex FlexMap 3D read out.
Screening for tissue-specific autoantibodies
Plasma samples obtained from patients with APS-1 (n = 31), p52LOF/IκBδGOF (n = 30) or p52LOF/IκBδLOF (n = 4) variants were analysed for the presence of specific autoantibodies in various immunological tests. The anti-tissue autoantibodies on rat tissue test (BioRad/Kallestad, 29020) and the anti-adrenal autoantibodies on primate tissue test (Inova, 508375) were performed using commercially available slides. The detection of anti-intrinsic factor (Thermo Fisher Scientific, Phadia, 14-5668-01), anti-thyroperoxydase (Thermo Fisher Scientific, Phadia, 14-5641-01) and anti-thyroglobulin (Thermo Fisher Scientific, Phadia, 14-5642-02) antibodies was performed using the ELiA technique; the presence of anti-IA2 (Theradiag, 10513417) or anti-21-OH (Theradiag, RL21E/96D) autoantibodies were assessed by ELISA. All of the test procedures were performed once and conducted according to the protocols provided by the kit manufacturers.
Microbiological investigations
The normalized viral load was determined for each sample, by determining the viral load for 1 million cells in the nasopharyngeal swabs by quantitative PCR with reverse transcription using the SARS-CoV-2 R-gene kit (bioMérieux). In brief, nucleic acids were extracted from 0.2 ml nasopharyngeal swab (NPS) with NUCLISENS easyMAG and amplification was performed using the Bio-Rad CFX96 instrument. The viral load was determined with four internally developed quantification standards (QSs) targeting the SARS-CoV-2 N gene: QS1 to QS4, at 1 × 105, 1 × 104, 1 × 103 and 1 × 102 copies per µl, respectively, of a SARS-CoV-2 DNA standard. These QSs were controlled and quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific) and Applied Biosystems QuantStudio 3D Digital PCR. In parallel, NPS were tested using the CELL Control R-GENE kit (amplification of the HPRT1 housekeeping gene; bioMérieux), which contains two quantification standards, QS1 and QS2, at 104 copies per µl (50,000 cells per PCR in our conditions) and 103 copies per µl (5,000 cells per PCR) of DNA standard, respectively, to normalize the viral load according to the amount of sample. Normalized viral load was calculated as log10[copies per 106 cells]. Potential co-infections were investigated using the BioFire Respiratory 2.1 plus Panel (RP2.1plus) detecting 23 respiratory pathogens, including SARS-CoV-2 (bioMérieux).
Blood and nasal IFN score determination
Total RNA was extracted from whole blood into PAXgene tubes using the Maxwell16 LEV simplyRNA Blood kit (Promega) according to the manufacturer’s instructions. Blood IFN score was determined using Nanostring technology as previously described67. For the nasal IFN score, we tested 100 μl nasal pharyngeal swab samples with the IFN prototype, as previously described17. The first prototype of the IFN pouch encompasses four ISGs (IFNα-inducible protein 27, IFI44L, IFN-induced protein with tetratricopeptide repeats 1, radical S-adenosyl methionine domain containing 2) and three housekeeping genes (hypoxanthine phosphoribosyltransferase 1, peptidylprolyl isomerase B and 2,4-dienoyl-CoA reductase 1) for signal normalization. In brief, the pouches were hydrated with the hydration solution. The PAXgene blood or nasal pharyngeal swab samples were mixed with 800 μl of the sample buffer provided with the kit and injected directly into the pouch and run on FilmArray 2.0 and FilmArray Torch instruments (BioFire Diagnostics). Results were delivered within 1 h. Using a research version of the instrument, we determined the real-time quantification cycle values and post-amplification melt peaks for each assay. The normalized expression values for each assay were then calculated with the internal reference genes. Nasal pharyngeal ISG score was calculated using the same method as for PAXgene samples, as previously described67.
CyTOF
The whole-blood mass cytometry panels used were custom produced, and their contents are shown in Supplementary Table 8. Labelled cells were frozen at −80 °C after overnight dead-cell staining, and acquisition was performed on the Helios machine (Fluidigm). All of the samples were processed within 48 h of sampling. The six patients with autosomal-recessive APS-1 included in the analysis were on treatment with JAK inhibitors at the time of blood collection. Data analysis was performed using OMIQ software. The gating strategy for CyTOF immunophenotyping is shown in Supplementary Fig. 15.
Immunostaining of human thymus sections
Thymic biopsy samples were collected from a patient with complete autosomal-recessive RELB deficiency (mutation Y397*/Y397*, P2 from ref. 59) and a deceased patient with a p52LOF/IκBδGOF variant (P850Sfs*36/WT from ref. 68). Tissues were fixed in 4% paraformaldehyde (Thermo Fisher Scientific), washed with PBS and embedded in paraffin. Antigen retrieval was performed on rehydrated tissue by boiling sections in Citra antigen retrieval solution (Biogenex). The sections were blocked by incubation for 30 min at room temperature in CAS-Block (Thermo Fisher Scientific) plus 0.2% Triton X-100 (Sigma-Aldrich), and were then incubated overnight at 4 °C with primary antibodies. The sections were washed with PBS-Tween 0.1% and stained by incubation with a biotinylated secondary antibodies for 1 h at room temperature for AIRE visualization. When necessary, secondary antibody staining was performed at room temperature for 1 h. The sections were washed with PBS-Tween 0.1% and mounted in ProLong Diamond Antifade mounting solution (Thermo Fisher Scientific). Images were acquired on an Apotome microscope (Zeiss). The antibodies used were K8-Alexa647, Rb (EP1628Y), Abcam, ab192468, 1:300; K5 Alexa488, Rb (EP1601Y), Abcam, ab193894, 1:300; AIRE, rat, eBioscience, 14-9534-82, 1:50; pan-keratin, Rb, Abcam, ab9377, 1:200; K10 Alexa647, Rb (EP1607IHCY), Abcam, ab194231, 1:300; UEA-1 biotinylated, Vector Laboratories, B-1065-2, 1:500.
Mice
Nfkb2+/Y868* NOD mice were generated by the Genetics Core Facility at National Jewish Health, Denver Colorado. Both WT littermate control and Nfkb2+/Y868* NOD mice were maintained in specific-pathogen-free facilities at the University of California San Francisco (UCSF) in accordance with the guidelines established by the Institutional Committee on Animal Use and Care (IACUC) and Laboratory Animal Resource Center (LARC). Animal procedures were approved by the IACUC and LARC at UCSF, where mice aged 8–12 weeks, matched for age and sex, were used for tissue collection.
Mouse mTEC isolation and flow cytometry
A previously established mouse thymus tissue-processing and single-cell-isolation protocol was used for flow cytometry analysis69. Single-cell suspensions were incubated with Live/Dead Fixable Blue Dead Cell Stain (Thermo Fisher Scientific) in 1× PBS for 15 min at 4 °C and then washed in PBS. They were blocked by incubation with anti-mouse CD16/CD32 (24G2) antibodies (UCSF Hybridoma Core Facility) for 15 min at 4 °C before cell surface marker staining in FACS buffer for 30 min at 4 °C. Cells were fixed and permeabilized with the FOXP3 staining buffer kit (eBioscience) according to the manufacturer’s protocol for intracellular protein staining. Flow cytometry data were collected on the LSRII Flow Cytometer (BD Biosciences) and analysed using FlowJo v.10.8.1. Antibodies against the following proteins were used: AIRE (5H12, eBioscience, 53593482), CD45 (30-F11, BioLegend, 103130), EPCAM (G8.8, BioLegend, 118218), I-Ak (10-3.6, BioLegend, 109908).
Immunostaining of mouse thymus sections
Mouse thymuses were fixed by incubation in 2% paraformaldehyde (Thermo Fisher Scientific, 28908) in PBS for 2 h at room temperature and were then incubated overnight at 4 °C in 30% (w/v) sucrose (Sigma-Aldrich, S7903-1KG) in PBS. Tissues were embedded in Optimal Cutting Temperature Compound (Tissue-Tek 4583) and stored at −80 °C until sectioning (30–50 μm) on a Cryostat (Leica). Tissue sections on slides were rehydrated in PBS for 5 min before permeabilization in 0.3% Triton X-100 (Sigma-Aldrich), 0.2% BSA (Sigma-Aldrich) and 0.1% sodium azide (Sigma-Aldrich) in PBS with shaking for 45 min at room temperature. The sections were blocked by incubation with BlockAid (Thermo Fisher Scientific, B10710) at room temperature for 1 h. The sections were stained by incubation with primary fluorophore-conjugated antibodies for 1 h at room temperature and washed in 1× PBS. The sections were then stained with DAPI (BioLegend, 422801) for 5 min at room temperature followed by three washes in 1× PBS. Antibodies against the following proteins were used at a dilution of 1:200: AIRE (5H12, eBioscience, 53593482), K5 (EP1601Y, Abcam, 193895), K10 (EP1607IHCY, Abcam, 194231). All of the tissue sections were mounted in ProLong Diamond (Thermo Fisher Scientific) mounting medium. Images were captured on the Leica SP8 (Leica) laser-scanning confocal microscope.
Histology
Organs from age- and sex-matched Nfkb2+/+ and Nfkb2+/Y868* mice (10 to 11 weeks old) were collected and fixed overnight in 10% formalin, and shipped in 70% ethanol to HistoWiz for sectioning, and staining for haematoxylin and eosin. Immune infiltrates of tested organs were confirmed with a blinded observer.
PhIP–seq with a mouse proteome library
The mouse proteome T7 phage-display library, which was described elsewhere70, was used for immunoprecipitation and sequencing to identify autoreactivities. Serum samples from Rag2-KO (n = 5), WT NOD (n = 8), Aire-KO NOD (n = 8) and Nfkb2+/Y868* NOD (n = 8) mice were used in a previously published high-throughput protocol70. We analysed peptide enrichment after PhIP–seq by aligning reads at the protein level with RAPsearch as previously described70. Aligned reads were normalized to 100,000 reads per k-mer (RPK) to account for variable read depth, and log2-transformed fold changes in read counts were calculated for each sample relative to the mean number of read counts in mock immunoprecipitations and Rag2-KO mice. Peptides with z-scores greater than or equal to 3 were considered to be hits, and peptides displaying enrichment in mutant mice were identified as peptides classified as hits in at least three mutant mice and no WT mice. Peptides were labelled with the corresponding protein.
RNA extraction, sequencing and analysis
Total RNA was isolated from whole blood as previously described67. RNA sequencing was performed using the Illumina NovaSeq S2 instruments (2 × 100 bp), at a read depth of 70 million. Single samples were sequenced across two lanes, and the resulting FASTQ files were merged by sample. All FASTQ files passed quality control and the sequences were aligned with the GRCh38 reference genome using STAR (v.2.6.1d). BAM files were converted to a raw count expression matrix using featurecount. Raw count data were normalized using DEseq2 (v.1.40.2). The ensemble IDs targeting multiple genes were collapsed (average), and a final data matrix gene was generated for single-gene set enrichment analysis with the BloodGen3Module gene set71. Statistical analysis was performed on a predefined gene set. Specifically, we used a fixed repertoire of 382 blood transcriptional modules that were thoroughly annotated and characterized functionally as described previously71. In brief, this repertoire of transcriptional modules (BloodGen3) was identified on the basis of co-expression, as measured in a collection of reference blood transcriptome datasets encompassing 16 pathological or physiological states and 985 individual transcriptome profiles. Sets of co-expressed transcripts were derived from a large weighted coclustering network in which edges represented the number of times a pair of genes coclustered in the 16 reference datasets (with a weight of 1 to 16). We calculated an IFN module enrichment score for individual samples by performing single-sample gene set enrichment analysis (ssGSEA) (GSVA package v.1.48.3), with the six IFN-response modules of the BloodGen3Module gene set (1.8.0), aggregate A28 as input. The enrichment scores of individual samples were used for heat-map visualization.
Thymus CT scan
We performed a retrospective assessment of the thymus for those patients for whom a chest CT scan was available. For patients with several scans, we selected the first scan or the scan on which the thymus was largest. Most of the patients’ scans were performed without contrast injection, and the thymic margins were assessed by multiplanar reconstruction. The thymus was measured in three planes: thickness and width in the axial plane through the aortic arch, greatest height in a coronal or sagittal oblique plane. We established a control group matched for age (±1 month) and sex. Three controls were selected per patient. The control group was randomly selected from scans performed at our centre for polytrauma, excluding severe head trauma with coma or neurological disorders and thoracic trauma (so as not to alter mediastinal anatomic reports).
Statistical methods
Data were analysed using GraphPad Prism software v.9.5.0 (GraphPad Software). The statistical significance of quantitative differences between groups was assessed in two-tailed unpaired Mann–Whitney U-tests. The statistical significance of differences between two groups in mouse studies was calculated using unpaired, parametric, two-tailed Student’s t-tests. For comparisons of more than two groups, the statistical significance of differences was calculated using two-way nonparametric ANOVA (Sidak’s test) with correction for multiple comparisons. Only statistically significant comparisons are indicated by their P values. All data are expressed as the mean ± s.d. calculated from at least three independent experiments unless otherwise stated.
Ethics statement
Patients were included in the C18-41 Genetic Predisposition to Severe Infections study approved by the Sud Est II ethics committee (approval no. 2022-A00257-36) in France. All of the enrolled participants provided written informed consent and were collected through protocols conforming to local ethics requirements. Ethics approval was obtained from the Comitato Etico Provinciale (NP 4000—Studio CORONAlab) in Brescia, Italy, the French Ethics Committee Comité de Protection des Personnes, Ile de France II (2010-A00634-35 protocol no. C10-13) and the Rockefeller University Institutional Review Board in New York (protocol no. JCA-0700).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-023-06717-x.
Supplementary information
Supplementary Results 1–4, providing additional information regarding the functional characterization of three types of inborn errors of NF-κB2, immunological and clinical description of the cohort, and Supplementary References.
Uncropped images from the western blots displayed in the indicated figures.
Supplementary Tables 1–8.
Source data
Source Data Fig. 5 and Source Data Extended Data Fig. 10
Acknowledgements
We thank the patients and their families for participating in the study; the members of the Laboratory of Human Genetics of Infectious Diseases for discussions; Y. Nemirovskaya, D. Liu, M. Woollett, L. Lorenzo-Diaz, C. Rivalain, M. Charabieh and T. Kochetkov for administrative and technical assistance; D. Chaussabel for assistance with RNA-seq analysis; the National Facility for Autoimmunity and Serology Profiling at SciLifeLab for technical support. The Laboratory of Human Genetics of Infectious Diseases is supported by the Howard Hughes Medical Institute, the Rockefeller University, the St Giles Foundation, the National Institutes of Health (NIH) (R01AI088364, R01AI127564-06 and R01AI163029), the National Center for Advancing Translational Sciences (NCATS), the NIH Clinical and Translational Science Award (CTSA) programme (UL1 TR001866), the Fisher Center for Alzheimer’s Research Foundation, the Meyer Foundation, the JBP Foundation, the French National Research Agency (ANR) under the ‘Investments for the Future’ programme (ANR-10-IAHU-01), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), the French Foundation for Medical Research (FRM) (EQU201903007798), the ANRS-COV05, ANR GENVIR (ANR-20-CE93-003), ANR AABIFNCOV (ANR-20-CO11-0001), ANR AAILC (ANR-21-LIBA-0002) and ANR AI2D (ANR-22-CE15-0046) projects, the ANR-RHU programme (ANR-21-RHUS-08-COVIFERON), the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 824110 (EASI-genomics), the HORIZON-HLTH-2021-DISEASE-04 programme under grant agreement 01057100 (UNDINE), the Square Foundation, Grandir—Fonds de solidarité pour l’enfance, the Fondation du Souffle, the SCOR Corporate Foundation for Science, the Battersea & Bowery Advisory Group, the French Ministry of Higher Education, Research, and Innovation (MESRI-COVID-19), Institut National de la Santé et de la Recherche Médicale (INSERM) and of Paris Cité University. J.R. was supported by the INSERM PhD programme (poste d’accueil INSERM). J.R., T.L.V., P.B. and Q.R. are supported by the Bettencourt-Schueller Foundation and the MD-PhD programme of Imagine Institute. P.B. was supported by the French Foundation for Medical Research (FRM, EA20170638020) and a ‘Poste CCA-INSERM-Bettencourt’ (with the support of the Fondation Bettencourt-Schueller). Q.P. was supported by the Assistance Publique Hôpitaux de Paris (AP-HP, Année Recherche) and the MD-PhD programme of INSERM (Ecole de l’INSERM Liliane Bettencourt). M.O. was supported by the David Rockefeller Graduate Program, the Funai Foundation for Information Technology (FFIT), the Honjo International Scholarship Foundation (HISF), the New York Hideyo Noguchi Memorial Society (HNMS). Nils Landegren is funded by The Swedish Research Council and the Göran Gustafsson Foundation. B. Keller is funded by Bundesministerium für Bildung und Forschung (GAIN 01GM1910A). Q.P.-H. is supported by the Swedish Research Council, and the Knut and Alice Wallenberg Foundation (KAW). F. Hauck was funded by the Else Kröner-Fresenius Stiftung (EKFS, 2017_A110) and the German Federal Ministry of Education and Research (BMBF, 01GM1910C). L.D.N. and H.C.S. are supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. C.S.M. and S.G.T. are supported by Investigator Grants awarded by the National Health and Medical Research Council of Australia. CIRCA Investigators (A.P-C, A.S, L.D, D.S, M.W, A.K, C.S.M, C.C.G, P.E.G, S.G.T) are funded by grants awarded by the Jeffrey Modell Foundation and the John Brown Cook Foundation. M. Momenilandi was supported by ANRS, Maladies infectieuses émergentes (ECTZ173053). F.R.L. was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM) and by government grants managed by the Agence National de la Recherche as part of the ‘Investment for the Future’ programme (Institut Hospitalo-Universitaire Imagine, grant ANR-10-IAHU-01, Recherche Hospitalo-Universitaire, grant ANR-18-RHUS-0010), the Centre de Référence Déficits Immunitaires Héréditaires (CEREDIH), the Agence National de la Recherche (ANR-14-CE14-0026-01 ‘Lumugene’; ANR-18-CE17-0001 ‘Action’; ANR-22-CE15-0047-02 ‘BREAK-ITP’), the Fondation pour la recherche Médicale (FRM : EQU202103012670). F. Atschekzei was funded by the German Federal Ministry of Education and Research (BMBF, 01GM2206B) and the German Center for Infection Research (DZIF TTU 07.801).
Extended data figures and tables
Extended Data Fig. 3. Functional testing of NFKB2 variants by overexpression.
(a) Schematic representation of the alternative NF-κB pathway and the function of the p52/RelB and p52/p52 heterodimers (left panel); a graphical overview of the luciferase assay for testing the p52 function of the NFKB2 variants (middle panel); and a schematic representation of the functional consequences of the WT, p52GOF and p52LOF variants in the luciferase assay (right panel). (b) Relative luciferase activity (RLA) of WT or RelA-deficient HEK293T cells transfected with a κB reporter construct (κB-luc) in the absence or presence of plasmids encoding NIK and/or RelB for 24, 48 or 72 h. Results are expressed as the RLA normalized against the value for the EV. Bars represent the mean values (± s.d.) from 3 independent experiments performed in technical duplicates. (c) Western blot of HEK293T cells transfected for 24 h (left panel) or 48 h (right panel) in the absence or presence of plasmids encoding NIK and the WT or previously reported biochemical NF- κB2/p100 mutants. Data representative of two independent experiment are shown. (d) Luciferase assay testing the NF-κB2/p100 biochemical mutants (left) or deleterious variants from patients (right), 24 h after transfection. Bars represent the mean values (± s.d.) from more than 3 independent experiments.
Extended Data Fig. 4. Assessments of NIK-dependent p100 processing and p100-IκBδ activity of the NFKB2 variants by overexpression.
(a) Western blot of HEK293T cells transfected for 48 h in the presence or absence of plasmids encoding NIK and WT or mutant NF-κB2, showing phosphorylated Y866-p100 (P-p100) levels, and p100 and p52 expression. Data representative of three independent experiments are shown. (b) Relative luciferase activity (RLA), indicating the p100/NF-κB2-dependent capacity to repress κB transcriptional activity for luciferase in HEK293T cells in the presence or absence of plasmids encoding NIK and various amounts of a plasmid encoding the C-terminal part of p100/NF-κB2 (Cter, aa 405-900) from WT or mutants, 48 h after transfection. The results are expressed as a percentage of the κB-luc RLA after transfection with NIK alone (left panel); the kinetic effect of transfection with NIK either alone or together with a plasmid encoding the various Cter constructs, as assessed by κB-luc transcriptional repression, from 24 to 72 h after transfection is shown in the right panel. Bars represent the mean values (± s.d.) from two independent experiments performed in duplicate. (c) Kinetic effect of transfection with NIK alone or together with a plasmid encoding the dimer-deficient Y247A single (p100Y247A) or double (p100/Y247A/W270*, p100Y247A/R611*, p100Y247A/S866N, or p100Y247A/R853*) mutants, in terms of κB transcriptional repression of luciferase activity from 24 to 72 h after transfection. Results are expressed as a percentage of the κB RLA after transfection with NIK alone. Bars represent the mean values (± s.d.) from two independent experiments performed in duplicate. (d) Western blot of HEK293T cells cotransfected with a plasmid encoding NIK, together with various amounts of a plasmid encoding the WT or mutant NF-κB2 variants, together with a constant amount of empty vector (left panel) or of WT NFKB2 (right panel), showing phosphorylated p100 (P-p100), p100 and p52 expression. Data representative of three independent experiments are shown.
Author contributions
T.L.V., A. Puel, M.S.A., L.D.N. and J.-L. Casanova conceived the research, designed the experiments, interpreted the data and wrote the manuscript. J. Rosain, A. Gervais, M.P.L., S.G., L.B., G.H., Q.P., M.O. and J.B. participated in IFN neutralization assays and/or overexpression assay experiments. A.V.P. and M.S.A. performed human thymus section staining and analysis. X.L. and M.S.A. performed mouse experiments and analysis. A.C., D.E., O.K., M.A.-G. and N.L. performed HuProt microarray and multiplex bead assay experiments and helped with data analysis. F.R. performed viral serological tests. D.R., P.Z. and Y.F. analysed the RNA-seq data. V. B. helped with the CyTOF data analysis. L.B. performed the thymic volume analysis from CT scans. S.T.A. and A. Gervais determined the IFN score for nasal swabs. E.R., J.L.D. and D.Y. performed PhIP–seq with the mouse proteome library. P.G.D., J.-L. Charuel and M. Miyara. performed screening for tissue-specific autoantibodies. A. Pinzon, A.F.G., A.B., A. Servettaz, A.M., S. Sediva, A. Sullivan, A. Guffroy, A.I., A.K., B.K., B.N., B.B., B.P., G.G., C.A., C. Soudée, C. Schuetz., C. Sobacchi, C.M., M.C.P. C.M.R., C.C.R., C.C.G., C.M., C.F., C.H.K., C.A.S., D.S., D.I.B., E.J., E.L., F.H., F.A., F.C., F.R., F.S.-R., F.R.L., G.S., G.H., G.D., G.L.G, H.H., H.K., H.S., H.S.K, H.G., I.D., J.B., J.B.C., J. Raedler, J.B., J.F., J.O., K.B., K.W., L.V., L.B.R., L.B., L.D.N., L.I., L.D., L.R.L.M.R., M.S.G., M. Momenilandi, M.E.M., M.E., M.A.-G., M. Slatter, M. Malphettes, M. Migaud, M.P., M.B., M.O., M.W., M.E., M.D.K., M.H., M. Shahrooei, N.A., N.P., N.M., O.M.D., P.C., P.M., P.F., P.B., P.E.G., M.M., Q.Z., Q.P.H., Q.R., Q.P., R.C., R.D., R.A., R.S.A., S.W., S.D.R., S.K., S.M.H., S.T., S.L., T.N., T.K., T.R., V.L.B, V.L., V.M.H., V.B., Y.Z., W.A.F.G. recruited patients and/or coordinated the clinical study protocol and/or sample collection. L.A. and A.C. performed the statistical analysis. J.-L. Casanova. and A. Puel supervised experiments or analyses. All of the authors discussed and reviewed the manuscript and approved its submission. A. Gervais. and J. Rosain. contributed equally. T.N., M.P.L. and E.R. contributed equally. O.K., PB., C.R and N.L. contributed equally.
Peer review
Peer review information
Nature thanks Shiv Pillai and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
All the data supporting the findings of this study are available within the Article and its Supplementary Information. The gel source data are shown in Supplementary Fig. 1. The RNA-seq data generated in this study have been deposited in the NCBI database under NCBI-SRA project PRJNA989123. All the other data and material supporting the findings of this study are available under a data transfer agreement from the corresponding authors on reasonable request. Source data are provided with this paper.
Competing interests
A. Guichard is an employee of Biomerieux. J.-L. Casanova is listed as an inventor on patent application PCT/US2021/042741, filed on the 22 July 2021 and submitted by The Rockefeller University, that covers the diagnosis of susceptibility to, and treatment of, viral disease and viral vaccines, including COVID-19 and vaccine-associated diseases. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Audrey V. Parent, Xian Liu, Axel Cederholm
These authors jointly supervised this work: Luigi D. Notarangelo, Mark S. Anderson, Jean-Laurent Casanova, Anne Puel
Lists of authors and their affiliations appear online
Contributor Information
Tom Le Voyer, Email: tom.le-voyer@institutimagine.org.
Jean-Laurent Casanova, Email: casanova@rockefeller.edu.
Anne Puel, Email: anne.puel@inserm.fr.
NF-κB Consortium:
Stéphanie Boisson-Dupuis, Eric Oksenhendler, Satoshi Okada, Oana Caluseriu, Mathilde Valeria Ursini, Eric Ballot, Geoffroy Lafarge, Tomas Freiberger, Carlos A. Arango-Franco, and Romain Levy
Extended data
is available for this paper at 10.1038/s41586-023-06717-x.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-023-06717-x.
References
- 1.Levin M. Anti-interferon auto-antibodies in autoimmune polyendocrinopathy syndrome type 1. PLoS Med. 2006;3:e292. doi: 10.1371/journal.pmed.0030292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meager A, et al. Anti-interferon autoantibodies in autoimmune polyendocrinopathy syndrome type 1. PLoS Med. 2006;3:e289. doi: 10.1371/journal.pmed.0030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastard P, et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J. Exp. Med. 2021;218:e20210554. doi: 10.1084/jem.20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puel A, Bastard P, Bustamante J, Casanova J-L. Human autoantibodies underlying infectious diseases. J. Exp. Med. 2022;219:e20211387. doi: 10.1084/jem.20211387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mogensen KE, Daubas P, Gresser I, Sereni D, Varet B. Patient with circulating antibodies to alpha-interferon. Lancet. 1981;318:1227–1228. doi: 10.1016/S0140-6736(81)91460-4. [DOI] [PubMed] [Google Scholar]
- 6.Pozzetto, B., Mogensen, K. E., Tovey, M. G. & Gresser, I. Characteristics of autoantibodies to human interferon in a patient with varicella-zoster disease. J. Infect. Dis.150, 707–713 (1984). [DOI] [PubMed]
- 7.Bastard, P. et al. Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Sci. Immunol. 10.1126/sciimmunol.abl4340 (2021). [DOI] [PMC free article] [PubMed]
- 8.Bastard, P. et al. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science10.1126/science.abd4585 (2020). [DOI] [PMC free article] [PubMed]
- 9.Zhang Q, Bastard P, COVID Human Genetic Effort, Cobat A, Casanova J-L. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature. 2022;603:587–598. doi: 10.1038/s41586-022-04447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manry J, et al. The risk of COVID-19 death is much greater and age dependent with type I IFN autoantibodies. Proc. Natl Acad. Sci. USA. 2022;119:e2200413119. doi: 10.1073/pnas.2200413119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casanova, J. L. & Abel, L. Mechanisms of viral inflammation and disease in humans. Science374, 1080–1086 (2021). [DOI] [PMC free article] [PubMed]
- 12.Bastard P, et al. Auto-antibodies to type I IFNs can underlie adverse reactions to yellow fever live attenuated vaccine. J. Exp. Med. 2021;218:e20202486. doi: 10.1084/jem.20202486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, et al. Autoantibodies against type I IFNs in patients with critical influenza pneumonia. J. Exp. Med. 2022;219:e20220514. doi: 10.1084/jem.20220514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alotaibi F, et al. Type I interferon autoantibodies in hospitalized patients with Middle East respiratory syndrome and association with outcomes and treatment effect of interferon beta‐1b in MIRACLE clinical trial. Influenza Other Respir. Viruses. 2023;17:e13116. doi: 10.1111/irv.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gervais A, et al. Autoantibodies neutralizing type I IFNs underlie West Nile virus encephalitis in ∼40% of patients. J. Exp. Med. 2023;220:e20230661. doi: 10.1084/jem.20230661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Wijst MGP, et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci. Transl. Med. 2021;13:eabh2624. doi: 10.1126/scitranslmed.abh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez J, et al. Early nasal type I IFN immunity against SARS-CoV-2 is compromised in patients with autoantibodies against type I IFNs. J. Exp. Med. 2021;218:e20211211. doi: 10.1084/jem.20211211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng M, Anderson MS. Thymic tolerance as a key brake on autoimmunity. Nat. Immunol. 2018;19:659–664. doi: 10.1038/s41590-018-0128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 20.Cavadini P, et al. AIRE deficiency in thymus of 2 patients with Omenn syndrome. J. Clin. Invest. 2005;115:728–732. doi: 10.1172/JCI200523087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poliani PL, et al. Early defects in human T-cell development severely affect distribution and maturation of thymic stromal cells: possible implications for the pathophysiology of Omenn syndrome. Blood. 2009;114:105–108. doi: 10.1182/blood-2009-03-211029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Ravin SS, et al. Hypomorphic Rag mutations can cause destructive midline granulomatous disease. Blood. 2010;116:1263–1271. doi: 10.1182/blood-2010-02-267583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hetemäki I, et al. Patients with autoimmune polyendocrine syndrome type 1 have an increased susceptibility to severe herpesvirus infections. Clin. Immunol. 2021;231:108851. doi: 10.1016/j.clim.2021.108851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossi SW, et al. RANK signals from CD4+3− inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J. Exp. Med. 2007;204:1267–1272. doi: 10.1084/jem.20062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White AJ, et al. Sequential phases in the development of Aire-expressing medullary thymic epithelial cells involve distinct cellular input. Eur. J. Immunol. 2008;38:942–947. doi: 10.1002/eji.200738052. [DOI] [PubMed] [Google Scholar]
- 26.Akiyama T, et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 2008;29:423–437. doi: 10.1016/j.immuni.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Sun S-C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fusco AJ, et al. The NF-κB subunit RelB controls p100 processing by competing with the kinases NIK and IKK1 for binding to p100. Sci. Signal. 2016;9:ra96. doi: 10.1126/scisignal.aad9413. [DOI] [PubMed] [Google Scholar]
- 29.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat. Rev. Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 30.Burkly L, et al. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- 31.Wirasinha RC, et al. Nfkb2 variants reveal a p100-degradation threshold that defines autoimmune susceptibility. J. Exp. Med. 2021;218:e20200476. doi: 10.1084/jem.20200476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bautista JL, et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat. Commun. 2021;12:1096. doi: 10.1038/s41467-021-21346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tucker E, et al. A novel mutation in the Nfkb2 gene generates an NF-κB2 “super repressor”. J. Immunol. 2007;179:7514–7522. doi: 10.4049/jimmunol.179.11.7514. [DOI] [PubMed] [Google Scholar]
- 34.Jiang W, Anderson MS, Bronson R, Mathis D, Benoist C. Modifier loci condition autoimmunity provoked by Aire deficiency. J. Exp. Med. 2005;202:805–815. doi: 10.1084/jem.20050693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michelson DA, Hase K, Kaisho T, Benoist C, Mathis D. Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells. Cell. 2022;185:2542–2558. doi: 10.1016/j.cell.2022.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michelson DA, Mathis D. Thymic mimetic cells: tolerogenic masqueraders. Trends Immunol. 2022;43:782–791. doi: 10.1016/j.it.2022.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yano M, et al. Aire controls the differentiation program of thymic epithelial cells in the medulla for the establishment of self-tolerance. J. Exp. Med. 2008;205:2827–2838. doi: 10.1084/jem.20080046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bornstein C, et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature. 2018;559:622–626. doi: 10.1038/s41586-018-0346-1. [DOI] [PubMed] [Google Scholar]
- 39.Kinoshita D, et al. Essential role of IκB kinase α in thymic organogenesis required for the establishment of self-tolerance1. J. Immunol. 2006;176:3995–4002. doi: 10.4049/jimmunol.176.7.3995. [DOI] [PubMed] [Google Scholar]
- 40.Kajiura F, et al. NF-κB-inducing kinase establishes self-tolerance in a thymic stroma-dependent manner. J. Immunol. 2004;172:2067–2075. doi: 10.4049/jimmunol.172.4.2067. [DOI] [PubMed] [Google Scholar]
- 41.LaFlam TN, et al. Identification of a novel cis-regulatory element essential for immune tolerance. J. Exp. Med. 2015;212:1993–2002. doi: 10.1084/jem.20151069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koh AS, et al. Rapid chromatin repression by Aire provides precise control of immune tolerance. Nat. Immunol. 2018;19:162–172. doi: 10.1038/s41590-017-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kisand K, et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J. Exp. Med. 2010;207:299–308. doi: 10.1084/jem.20091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puel A, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J. Exp. Med. 2010;207:291–297. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer S, et al. AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell. 2016;166:582–595. doi: 10.1016/j.cell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landegren N, et al. Proteome-wide survey of the autoimmune target repertoire in autoimmune polyendocrine syndrome type 1. Sci. Rep. 2016;6:20104. doi: 10.1038/srep20104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang EY, et al. High-throughput identification of autoantibodies that target the human exoproteome. Cell Rep. Methods. 2022;2:100172. doi: 10.1016/j.crmeth.2022.100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruserud Ø, et al. A longitudinal follow-up of autoimmune polyendocrine syndrome type 1. J. Clin. Endocrinol. Metab. 2016;101:2975–2983. doi: 10.1210/jc.2016-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ströbel P, et al. Deficiency of the autoimmune regulator AIRE in thymomas is insufficient to elicit autoimmune polyendocrinopathy syndrome type 1 (APS-1) J. Pathol. 2007;211:563–571. doi: 10.1002/path.2141. [DOI] [PubMed] [Google Scholar]
- 50.Givony, T. et al. Thymic mimetic cells function beyond self-tolerance. Nature622, 164–172 (2023). [DOI] [PubMed]
- 51.Weih F, Caamaño J. Regulation of secondary lymphoid organ development by the nuclear factor-κB signal transduction pathway. Immunol. Rev. 2003;195:91–105. doi: 10.1034/j.1600-065X.2003.00064.x. [DOI] [PubMed] [Google Scholar]
- 52.Perry JSA, et al. Distinct contributions of aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity. 2014;41:414–426. doi: 10.1016/j.immuni.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malchow S, et al. Aire-dependent thymic development of tumor-associated regulatory T cells. Science. 2013;339:1219–1224. doi: 10.1126/science.1233913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathian, A. et al. Lower disease activity but higher risk of severe COVID-19 and herpes zoster in patients with systemic lupus erythematosus with pre-existing autoantibodies neutralising IFN-α. Ann. Rheum. Dis.10.1136/ard-2022-222549 (2022). [DOI] [PubMed]
- 55.Meisel C, et al. Mild COVID-19 despite autoantibodies against type I IFNs in autoimmune polyendocrine syndrome type 1. J. Clin. Invest. 2021;131:e150867. doi: 10.1172/JCI150867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuehn HS, et al. Novel nonsense gain-of-function NFKB2 mutations associated with a combined immunodeficiency phenotype. Blood. 2017;130:1553–1564. doi: 10.1182/blood-2017-05-782177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willmann KL, et al. Biallelic loss-of-function mutation in NIK causes a primary immunodeficiency with multifaceted aberrant lymphoid immunity. Nat. Commun. 2014;5:5360. doi: 10.1038/ncomms6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warnatz K, et al. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proc. Natl Acad. Sci. USA. 2009;106:13945–13950. doi: 10.1073/pnas.0903543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharfe N, et al. The effects of RelB deficiency on lymphocyte development and function. J. Autoimmun. 2015;65:90–100. doi: 10.1016/j.jaut.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Merico D, Sharfe N, Hu P, Herbrick J-A, Roifman CM. RelB deficiency causes combined immunodeficiency. LymphoSign J. 2015;2:147–155. doi: 10.14785/lpsn-2015-0005. [DOI] [Google Scholar]
- 61.Sharfe, N. et al. NFκB pathway dysregulation due to reduced RelB expression leads to severe autoimmune disorders and declining immunity. J. Autoimmun.10.1016/j.jaut.2022.102946 (2022). [DOI] [PMC free article] [PubMed]
- 62.Tangye, S. G. et al. Human inborn errors of immunity: 2022 update on the classification from the international union of immunological societies expert committee. J. Clin. Immunol.10.1007/s10875-022-01289-3 (2022). [DOI] [PMC free article] [PubMed]
- 63.Zhang, Q. et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science10.1126/science.abd4570 (2020). [DOI] [PMC free article] [PubMed]
- 64.Li J, et al. Biochemically deleterious human NFKB1 variants underlie an autosomal dominant form of common variable immunodeficiency. J. Exp. Med. 2021;218:e20210566. doi: 10.1084/jem.20210566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vazquez SE, et al. Identification of novel, clinically correlated autoantigens in the monogenic autoimmune syndrome APS1 by proteome-wide PhIP-Seq. eLife. 2020;9:e55053. doi: 10.7554/eLife.55053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abolhassani H, et al. X-linked TLR7 deficiency underlies critical COVID-19 pneumonia in a male patient with ataxia-telangiectasia. J. Clin. Immunol. 2022;42:1–9. doi: 10.1007/s10875-021-01151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pescarmona R, et al. Comparison of RT-qPCR and Nanostring in the measurement of blood interferon response for the diagnosis of type I interferonopathies. Cytokine. 2019;113:446–452. doi: 10.1016/j.cyto.2018.10.023. [DOI] [PubMed] [Google Scholar]
- 68.Slade CA, et al. Fatal enteroviral encephalitis in a patient with common variable immunodeficiency harbouring a novel mutation in NFKB2. J. Clin. Immunol. 2019;39:324–335. doi: 10.1007/s10875-019-00602-x. [DOI] [PubMed] [Google Scholar]
- 69.Miller CN, et al. Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature. 2018;559:627–631. doi: 10.1038/s41586-018-0345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rackaityte, E. et al. Validation of a murine proteome-wide phage display library for the identification of autoantibody specificities. Preprint at bioRxiv10.1101/2023.04.07.535899 (2023). [DOI] [PMC free article] [PubMed]
- 71.Altman MC, et al. Development of a fixed module repertoire for the analysis and interpretation of blood transcriptome data. Nat. Commun. 2021;12:4385. doi: 10.1038/s41467-021-24584-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Results 1–4, providing additional information regarding the functional characterization of three types of inborn errors of NF-κB2, immunological and clinical description of the cohort, and Supplementary References.
Uncropped images from the western blots displayed in the indicated figures.
Supplementary Tables 1–8.
Source Data Fig. 5 and Source Data Extended Data Fig. 10
Data Availability Statement
All the data supporting the findings of this study are available within the Article and its Supplementary Information. The gel source data are shown in Supplementary Fig. 1. The RNA-seq data generated in this study have been deposited in the NCBI database under NCBI-SRA project PRJNA989123. All the other data and material supporting the findings of this study are available under a data transfer agreement from the corresponding authors on reasonable request. Source data are provided with this paper.