Abstract
The causes of nigrostriatal cell death in idiopathic Parkinson’s disease are unknown, but exposure to toxic chemicals may play some role. We followed up here on suggestions that bacterial secondary metabolites might be selectively cytotoxic to dopaminergic neurons. Extracts from Streptomyces venezuelae were found to kill human dopaminergic neurons (LUHMES cells). Utilizing this model system as a bioassay, we identified a bacterial metabolite known as aerugine (C10H11NO2S; 2-[4-(hydroxymethyl)-4,5-dihydro-1,3-thiazol-2-yl]phenol) and confirmed this finding by chemical re-synthesis. This 2-hydroxyphenyl-thiazoline compound was previously shown to be a product of a wide-spread biosynthetic cluster also found in the human microbiome and in several pathogens. Aerugine triggered half-maximal dopaminergic neurotoxicity at 3–4 μM. It was less toxic for other neurons (10–20 μM), and non-toxic (at <100 μM) for common human cell lines. Neurotoxicity was completely prevented by several iron chelators, by distinct anti-oxidants and by a caspase inhibitor. In the Caenorhabditis elegans model organism, general survival was not affected by aerugine concentrations up to 100 μM. When transgenic worms, expressing green fluorescent protein only in their dopamine neurons, were exposed to aerugine, specific neurodegeneration was observed. The toxicant also exerted functional dopaminergic toxicity in nematodes as determined by the “basal slowing response” assay. Thus, our research has unveiled a bacterial metabolite with a remarkably selective toxicity toward human dopaminergic neurons in vitro and for the dopaminergic nervous system of Caenorhabditis elegans in vivo. These findings suggest that microbe-derived environmental chemicals should be further investigated for their role in the pathogenesis of Parkinson’s disease.
Keywords: Dopaminergic neurodegeneration, Streptomyces venezuelae, Bacterial toxin, Ferroptosis, Caenorhabditis elegans, Parkinson’s disease
1. Introduction
The increasing number of idiopathic Parkinson’s disease (PD) cases suggests that environmental factors exist that specifically affect dopaminergic (DAergic) neuron viability. Indeed, several industrial chemicals and pesticides have been implicated to contribute to PD pathogenesis (Bellou et al., 2016; Obeso et al., 2017; Tanner, 1992; Terron et al., 2018). Metabolites synthesized by bacteria and fungi are also candidates for initiation and perpetuation of the disease (Dorsey et al., 2018; Fuhrimann et al., 2021; Inamdar et al., 2013).
Although some genes are known to be associated with PD as dominant or recessive factors, at least 90% of all PD cases are of sporadic origin (Obeso et al., 2017; Program, 2021). Several lines of evidence, including twin studies, have shown the critical importance of environmental factors regarding PD onset. Age is clearly the major risk factor for PD, but it is not clear, why part of the aging population develops the disease and a major fraction does not (Costello et al., 2009; De Miranda et al., 2022; Goldman, 2014; Priyadarshi et al., 2001; Yuan et al., 2022).
There is a wealth of epidemiological data connecting pesticide exposure to PD (Kanthasamy et al., 2012). The case is particularly convincing for the pesticide rotenone, and this is well supported by mechanistic studies and animal experiments (Betarbet et al., 2000; De Miranda et al., 2022; Ockleford et al., 2017). Some evidence suggests also a role for dieldrin (Hatcher et al., 2007; Moretto and Colosio, 2011) and high levels of paraquat (McCormack et al., 2005; Ockleford et al., 2017), but the final verdict on these is still pending. Whilst pesticides might be responsible for some of the PD cases in rural areas, overall disease prevalence is unlikely to be explained by exposure to agricultural chemicals (Costello et al., 2009; Firestone et al., 2005; Gorell et al., 1998; Priyadarshi et al., 2001). The same applies to manganese exposure (Austin et al., 2016; Erikson and Aschner, 2019). Another chemical capable of inducing parkinsonism is 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridin (MPTP), but only few cases of drug addicts have been documented (Langston, 2017).
Based on our knowledge on disease etiology, other chemicals may play roles in PD etiology by promoting neuroinflammation or ferroptosis (de Farias et al., 2017; Panicker et al., 2022; Sian-Hulsmann and Riederer, 2021). Neuroinflammatory conditions in the developing brain have been shown to contribute to the loss of dopamine (DA) neurons (Barlow et al., 2007). Similarly, in the adult brain, the activation of glial cells and the presence of certain cytokines have been identified as potent exacerbators of nigrostriatal degeneration in animal models (Schildknecht et al., 2017). However, environmental chemicals triggering such conditions remain elusive. The situation is similar for changes in the homeostasis of free ferric and ferrous ions in cells. Iron can play an important role in DAergic cell death (Devos et al., 2014; Mahoney-Sánchez et al., 2021), but the specific chemical triggers for such changes remain unknown.
Metabolites produced by the microbiome have been considered relevant factors, as they may affect large populations. Indeed, the gut microbiome of PD patients is different from healthy individuals, and the gut microbiome-brain axis has been shown to be altered in PD (De Miranda et al., 2022). Moreover, neurodegenerative changes of the enteric nervous system occur during early stages of PD pathology. They may predate the onset of motor symptoms by years (Braak et al., 2006; Scheperjans et al., 2015; Shannon et al., 2012). In experimental animals, a link between gut microbiota and PD pathology has been established, as gut stool transferred from PD patients promoted motor dysfunction in mice (Sampson et al., 2016).
Some microbial metabolites have been reported to specifically affect DAergic neurons. For instance, the fungal metabolite 1-octen-3-ol was active in a Drosophila melanogaster-based PD model (Inamdar et al., 2013). Moreover, Streptomyces venezuelae was suspected to produce a metabolite, which caused selective DAergic neurodegeneration in Caenorhabditis elegans (Caldwell et al., 2009). The bioactivity of the putative metabolite was characterized extensively, but the exact molecular identity has not been elucidated (Caldwell et al., 2018; Martinez et al., 2015; Ray et al., 2014).
The toxin production of S. venezuelae may be of limited relevance for most PD cases, as these bacteria typically live in soil. However, elucidation of the neurotoxicant structure can help in identifying the genes involved in its synthesis. Biosynthetic clusters for secondary metabolites often use simple primary metabolites (e.g., amino acids) to build complex new molecules (Inahashi et al., 2017; Vior et al., 2018), and the genes for this process can be transmitted horizontally across many bacterial species (Tidjani et al., 2019). Thus, secondary metabolites identified in soil bacteria may also be found in microbes closely associated with human physiology or pathology (Abe et al., 2020; Jiang et al., 2017).
An important condition for the identification of DAergic toxicants are suitable model systems. The use of C. elegans is well established for studies of the molecular pathology of PD (Cooper et al., 2006; Liu et al., 2011; Tucci et al., 2011). The model has also been used successfully in neurotoxicity testing (Braungart et al., 2004; Caito et al., 2013; Vanduyn et al., 2010). A complementary approach makes use of DAergic neurons generated from induced pluripotent stem cells (iPSC), or of LUHMES cells, i.e. conditionally immortalized human DAergic precursor cells that can be triggered to undergo terminal differentiation to post-mitotic neurons (Scholz et al., 2011). The latter cells are highly sensitive to various neurotoxicants, and have been characterized to undergo apoptosis, ferroptosis or selective neurite degeneration, depending on the toxicant used (Delp et al., 2019; Efremova et al., 2015; Gutbier et al., 2018a; Gutbier et al., 2018b).
Being intrigued by a potentially specific DAergic toxicant produced by microbes (Caldwell et al., 2018; Caldwell et al., 2009), we set out to verify this hypothesis and to elucidate the chemical structure. Our initial studies confirmed that S. venezuelae extracts killed human, DAergic neurons, while the related strain S. lividans did not have such an effect. A purification strategy based on a cellular neurotoxicity assay was developed. After identification of the bioactive molecule, it was re-synthesized and tested for selective neurotoxicity to human neurons. A pharmacological rescue screen was undertaken to profile the type of cell death triggered. Finally, we aimed to determine toxicant specificity for DAergic neurons in vivo by using the C. elegans model system.
2. Material and methods
2.1. Materials and chemicals
Dibutyryl cyclic adenosine monophosphate (dBcAMP), fibronectin, fetal calf serum (FCS), Hoechst H-33342, poly-L-ornithine hydrobromide (PLO), resazurin sodium salt, tetracycline, sodium pyruvate, and Triton-X100 were purchased from Sigma Aldrich (Steinheim, Germany). Chemicals and solvents used for extraction, purification and synthesis of aeruginol and derivatives were purchased from Sigma Aldrich (Steinheim, Germany). Nicotinamide-adenine-dinucleotide (NADH) disodium salt was purchase from Carl Roth GmbH + Co. KG (Karlsruhe, Germany). Recombinant human fibroblast growth factor 2 (FGF-2) and recombinant human glial cell derived neurotrophic factor (GDNF) were purchased from R&D Systems (Minneapolis). All cell culture reagents were purchased from Gibco/Fisher scientific (Hampton, New Hampshire, USA) unless otherwise specified. Seahorse materials and reagents were purchased from Agilent (California, United States). Cortex.4U, CNS.4U and their culture media were purchased from Axiogenesis (Köln, Germany). Dihydroaeruginoic acid (DHAA, CAS 143209–04-5) was purchased from Santa Cruz Biotechnology, Inc (Dallas, US). Calcein-AM was purchased from Biomol GmbH (Hamburg, Germany).
2.2. Extraction and purification of Streptomyces metabolites
S. venezuelae (ATCC 10712) and S. lividans (ATCC 19844) were grown in SYZ medium (15 g/l starch, 2 g/l yeast extract, 4 g/l casein peptone, 2 g/l glucose, pH = 6.2) at 30 °C and 160 rpm over 3 days and 350 μl were inoculated on a large (145 × 20 mm) SYZ agar plate (SYZ medium + 15 g/l agar). The plate was incubated for 4 days at 30 °C. The bacteria were scratched off with a spatula from the surface and resuspended in 5 ml SYZ medium. 200 μl of the bacterial suspension were then added onto 40 large (145 × 20 mm) SYZ agar plates and the plates incubated for 6 days at 30 °C. The plates were chopped into pieces and ethyl acetate (EA) was added. The extraction was repeated 3 times with 2 l of EA each and the solvent was evaporated. SepPak fractionation (5 g cartridge, Sep-Pak C18 20 cc, 55–105 μm particle size, Waters) and elution with 0, 30, 60 and 100% methanol (MeOH) yielded an active 60% MeOH fraction. The active fraction was further sub-fractionated by preparative reverse phase HPLC on a C18-AQ column with Reprosil-Pur 120 (250 × 20 mm, 10 μm, Dr. Maisch) using the following gradient: T0min = 50% B, T3min = 50% B, T25min = 95% B, with a flow rate of 15 ml/ min, monitoring absorbance at 195 nm. The solvents were: A = water (+0.1% formic acid (FA)) and B = acetonitrile (+0.1% FA). At 17 min, an active fraction eluted which was further purified via semi-preparative reverse phase HPLC on a C18 column with Reprospher 100 (250 × 10 mm, 5 μm, Dr. Maisch) using a flow rate of 1.5 ml/min, monitoring absorbance at 254 nm. Gradient: T0min = 70% B, T1min = 70% B, T40 min = 95% B. The solvents were: A = water (+0.1% FA) and B = acetonitrile (+0.1% FA). NMR measurements indicated a mixture of two compounds, which were separate again by semi-preparative HPLC using the same column. Gradient: T0min = 30% B, T5min = 30% B, T25min = 80% B, T40 min =95% B. After NMR and MS characterization, aerugine (0.68 mg) and aeruginol (0.46 mg) could be unambiguously confirmed by NMR spectroscopy and mass spectrometry and comparison to literature values.
2.3. LUHMES cell culture
LUHMES cells were cultivated as described previously (Krug et al., 2013; Schildknecht et al., 2013; Scholz et al., 2011). In brief, cells were grown in standard cell culture flasks pre-coated with 50 μg/ml PLO and 1 μg/ml fibronectin in water for at least 3 h at 37 °C. The maintenance culture was kept in proliferation medium consisting of advanced DMEM/F12 with 2 mM L-glutamine, 1x N2-supplement, and 40 ng/ml FGF-2. The cells were incubated at 37 °C with 5% CO2 and passaged trice a week when reaching 75–90% confluence. They were used up to passage 18. For differentiation, the medium was changed to differentiation medium consisting of advanced DMEM/F12 supplemented with 2 mM L-glutamine, 1 mM dBcAMP, 1 μg/ml tetracycline and 2 ng/ml GDNF on day of differentiation 0 (d0).
For toxicity assessment of mature LUHMES neurons, precursor cells were pre-differentiated for 2 days in cell culture flasks pre-coated as described above and differentiation medium. Then, 45′000 cells per well were seeded in differentiation medium into standard 96-well plates pre-coated as described above. They were incubated at 37 °C and 5% CO2 for 4 days before compound treatment. At this time, cells were >99% post-mitotic and had established an extensive neurite network (Scholz et al., 2011) The cell viability was assessed via the Calcein/Hoechst readout, the LDH assay and the resazurin reduction assay.
2.4. Stem cell derived neurons
The human iPSC (Sigma iPSC0028 (EPITHELIAL-1, #IPSC0028) were maintained and differentiated towards immature dorsal root ganglia neurons according to Hoelting et al. (2016) (Hoelting et al., 2016). Briefly, a total of 90,000 cells/cm2 were seeded on Matrigel. The differentiation was initiated with neural differentiation medium (KSR-S; Dulbecco’s modified Eagle’s medium [DMEM/F12] containing 15% knockout serum replacement, 1 × Glutamax, 1 × nonessential amino acids, and 50 μM ß-mercaptoethanol) supplemented with 17.5 ng/ml noggin, 10 μM SB-431642 from day of stem cell differentiation (DoD) DoD0 to DoD5. Starting on DoD2 CHIR99021 (1.5 μM), SU5402 (5 μM) and DAPT (5 μM) were added. From DoD4 onwards, the medium was step-wise replaced by N2-S medium (DMEM/F12 containing 2 mM Glutamax, 0.1 mg/ml apotransferrin, 1.55 mg/ml glucose, 25 mg/ml insulin, 100 mM putrescine, 30 nM selenium, and 20 nM progesterone; 25% N2-S medium on DoD4, 50% N2-S medium on DoD5, 75% N2-S medium on DoD7). On DoD9, the cells were frozen.
Commercial preparation of iPSC-derived neurons (Cortex.4U and CNS.4U) were handled and used as described by the manufacturer (nCardia). In brief, plates were coated with 10 μg/ml PLO, 10 μg/ml laminin and 10 μg/ml fibronectin. Cells were thawed and immediately seeded onto 96-well plates in their respective culture media (Cortex.4U: Cortex.4U™ culture medium with Cortex.4U™ supplement; CNS.4U: Neuro.4U Basal Medium [Ax-M-NBM250] with Neuro-Supplement 2 [Ax-M-DCS-DA]) to be used in neurite outgrowth assays.
2.5. Neurite outgrowth assays
The assays were performed as described previously (Hoelting et al., 2016; Klima et al., 2021; Krug et al., 2013). In brief, 30′000 cells per well were seeded on d2 (LUHMES) / d0 (peripheral neurons, Cortex.4U, CNS.4U) in their respective differentiation media into standard pre-coated 96-well plates. They were incubated for 1 h at 37 °C and 5% CO2 before treatment. 24 h after treatment the Calcein/Hoechst readout was performed.
2.6. Hek293FT, HepG2 and HeLa cell culture
Hek293FT were acquired from ThermoFisher Scientific (cat. no. R70007). HepG2 were acquired from ATCC (HB-8065). HeLa were acquired from ATCC (CCL-2). The cell culture was performed as described previously (Dolde et al., 2021). Briefly, the cell lines were kept in DMEM Glutamax with 10% FCS and 1% penicillin/streptomycin at 37 °C with 5% CO2. They were grown in normal cell culture flasks (Sarstedt) to 75–90% confluence and seeded into standard 96-well plates 24 h prior to compound treatment. The cells were treated by performing a medium change to Advanced DMEM/F12 supplemented with 2 mM L-glutamine containing the indicated compound concentrations.
2.7. Calcein/Hoechst readout
Neurite area (NA) and viability (V) were assessed as described previously (Stiegler et al., 2011). In brief, cells were stained with 1 μM calcein-AM and 1 μg/ml H-33342 for 30 min at 37 °C and 5% CO2. Image acquisition was performed using an Array-Scan VTI HCS Reader (Cellomics, PA, USA) equipped with a Hamamatsu ORCA-ER camera. 10 fields per well were imaged with 2 channels at 20x magnification (2×2 pixel binning). Excitation/emission wavelengths of 365 ± 50/535 ± 45 nm were used for Hoechst detection in channel 1 and 474 ± 40/535 ± 45 nm were used for calcein detection in channel 2.
Nuclei were identified in channel 1 depending on intensity, area, size and shape. Their outlines were expanded by 3.2 μm to define a virtual cell soma area (VCSA), which was bigger than the average cell soma to prevent false positive NAs. All calcein-positive pixels were defined as viable cellular structures (VCSs). The NA was automatically calculated by excluding the VCSAs from the VCSs. Furthermore all nuclei co-localizing with VCSs were defined as alive.
2.8. Additional TMRE/PI readout
Cells were stained either with 50 nM tetramethylrhodamine ethyl ester (TMRE) or 1.5 μM propidium iodide (PI) for at least 30 min at 37 °C and 5% CO2 (Brüll et al., 2020; Krug et al., 2014). Fluorescent imaging was performed (AxioObserver, Zeiss, Germany) with excitation/emission wavelengths of 575 ± 25/640 ± 35 nm.
2.9. LDH assay
The assay was performed as described earlier (Latta et al., 2000). In brief, supernatants were transferred to an un-coated 96-wellplate and cells were lysed in 110 μl of phosphate buffered saline (PBS) with 0.1% Triton-X100 for 1 h at room temperature. Lactate dehydrogenase (LDH) activity was assessed in the supernatant and the cell lysate individually by combining 10 μl sample with 100 μl potassium phosphate buffer pH 7.5 containing 640 μM pyruvate and 240 mM NADH. NADH was quantified by measuring the absorption at 340 nm over 15 min and its decline was proportional to the LDH content. LDH release was calculated as the ratio of LDHsupernatant/LDHsupernatant+lysate.
2.10. Resazurin reduction assay
The assay was performed as described previously (Schildknecht et al., 2009). In brief, cells were stained with 10 μg/ml resazurin and incubated for 30 min at 37 °C and 5% CO2. The fluorescence of resazurin was detected with an excitation wavelength of 530 nm, using a 590 nm long-pass filter to record the emission.
2.11. Seahorse assessment of mitochondrial respiratory parameters
The method was performed as described previously. (Delp et al., 2018) In brief, LUHMES cells were seeded on d2 into pre-coated Agilent Seahorse XFe24 well plates at a density of 100′000 cells/well in differentiation medium. On d3 the cells were analyzed in a Seahorse XF analyzer using the Mito stress test kit in accordance with the manufacturer’s instructions. In brief, 1 h prior to analysis the medium was changed to Seahorse XF base medium supplemented with 18 mM glucose, 2 mM glutamine and 1 mM pyruvate. The oxygen consumption rate (OCR) was measured to determine the basal respiration in untreated cells. After test compound injection the acute response was quantified. Injection of 1 μM oligomycin allowed for the determination of ATP production. 1.5 μM carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP) uncoupled the electron respiratory chain from the ATP synthase and enabled the measurement of the maximal respiration. The addition of 0.5 μM rotenone and 0.5 μM antimycin A completely shut down mitochondrial respiration to quantify non-mitochondrial respiration. Afterwards the cells were stained with 1 μg/ml Hoechst for 30 min at 37 °C. Image acquisition was performed using an Array-Scan VTI HCS Reader (Cellomics, PA, USA) equipped with a Hamamatsu ORCA-ER camera. 25 fields per well were imaged at 20x magnification (2×2 pixel binning). Excitation/emission wavelengths of 365 ± 50/535 ± 45 nm were used for H-33342 detection. Nuclei were identified and quantified depending on intensity, area, size and shape. According to pixel size and well size the total cell count was calculated and used for normalization of the OCR data.
2.12. Bacterial growth assay
Neisseria gonorrhoeae MS11 was cultured in PPM medium (15 g/l Proteose Pepton, 1 g/l soluble starch, 5 g/l NaCl, 4 g/l KH2PO4, 1 g/l K2HPO4, pH 7.5) for liquid culture and on GC plates (BD Difco™ GC Medium Base), both supplemented with 1 % vitamin mix (100 g/l glucose, 10 g/l glutamine, 26 g/l L-cysteine, 100 mg/l carboxylase, 250 mg/l NAD, 500 μl/l Fe(NO3)3, 150 mg/l arginine, 3 mg/l thiamine-HCl, 10 mg/l vitamine B12, 13 mg/l p-amino benzoic acid, 1.1 g/l L-cystine, 1 g/l adenine, 500 mg/l uracil, 30 mg/l guanine). Strains were either cultivated at 37 °C and 220 rpm (liquid medium) or at 37 °C and 5 % CO2 (solid medium).
Enterococcus faecalis was grown in BHI medium (BD Difco Brain Heart Infusion) with 36 g/l supplemented with 20 μg/ml hemin, 20 μg/ml NAD and 15 g/l agar for plates. Enterococcus faecalis strain 438 originates from eye isolate. Neisseria gonorrhoeae strain MS11 was gifted by T. F. Meyer (Max-Planck-Institute for Infection Biology, Berlin, Germany) (Edwards et al., 1984).
Bacteria were grown as described above on solid medium. Prior to substance exposure, bacteria were pre-cultured in liquid medium for at least 2 h. Optical density was determined and a volume equal to an OD550 of 0.2 for Neisseria gonorrhoeae MS11 (equates a final OD550 of 0.04) and an OD600 of 0.2 for Enterococcus faecalis (equates a final OD600 of 0.04) was inoculated into 5 ml of the respective liquid medium. Compound was added at the indicated amounts. Dimethyl sulfoxide (DMSO) level was adjusted to 1 % of final concentration including DMSO control. Samples were incubated for either 10.5 h, until they reached a stationary phase (three measured values in stationary phase) or until they reached an OD of 2.5. Optical density was determined every 0.5 h at OD550 for Neisseria gonorrhoeae MS11 and at OD600 for Enterococcus faecalis. (Szamosvári et al., 2019).
2.13. C. elegans study to determine specific neurodegeneration and behavioral changes
The strain BY200 Pdat-1::gfp (vtIs1), dat-1::GFP(vtls1) was kindly provided by the Blakely laboratory, Vanderbilt University Medical Center and the strain Punc-25:GFP was obtained from the Caenorhabditis Genetics Center (CGC, University of Minnesota, Minneapolis, MN, USA). The worms were maintained in nematode growth medium (NGM) plates that contain: 3 g/l NaCl, 17 g/l agar, 2.5 g/l peptone, 1 mM CaCl2, 5 mg/l cholesterol, 1 mM MgSO4, 25 mM potassium phosphate buffer (pH 6.0) with 1.25 ml nystatin and 50 mg/l streptomycin sulfate) and Escherichia coli (E. coli) OP50 strain was used as food (Brenner, 1974). Synchronization of worms was carried out using extraction solution (0.2% NaOCl, 0.5 M NaOH, in distilled water). The first larval stage (L1) population were obtained by isolating eggs with a 30% sucrose gradient, washed with sterile water, and resuspended in M9 buffer (42 mM Na2HPO4, 22 mM KH2PO4, 8.5 mM NaCl, and 1 mM MgSO4) (Sulston and Hodgkin, 1998).
For the lethality analysis, 40 to 60 BY200 and P unc-25 worms at L4 larval stage were exposed to vehicle or aerugine (8, 40 and 100 μM) in OP50-seeded NGM plates from day 0 until day 8. Worms were scored as dead when they did not respond to a mechanical stimulus. Moreover, worms were transferred to fresh plates for the first three days of treatment, which coincide with the egg laying days to avoid mixing of different generations. All treatments were performed in triplicates and experiments were repeated independently at least three times.
The basal slowing response (BSR) is a behavioral assay that assesses the functionality of the worms’ DAergic system. For this, worms were washed three times with S-basal buffer (100 mM NaCl, 5 mg/L cholesterol, 50 mM potassium phosphate buffer, pH 6.0) to remove residual bacteria, and placed on the center of a 60-mm NGM plates in the absence or presence of an OP50 bacteria ring. After habituation (5 min), the number of body bends of each worm was counted during 20 s to determine its immobility rate. The results were expressed as the difference (Δ) between the number of body bends in the absence (a) and presence (p) of bacteria on fresh plates (Δ = a-p). Given, worms with normal DA content move slower in the presence of bacteria than in the absence of bacteria (Sawin et al., 2000).
On day 2 and day 8, approximately, 20–30 worms per treatment were picked onto slides prepped with a 3% agarose pad, anesthetized with 5 μl of 3 mM levamisole hydrochloride (Sulston et al., 1975) and observed under an Olympus BX41 fluorescence microscope to determine morphology status according to novel Likert-like scale of degeneration as previously described (Benedetto et al., 2010).
2.14. Chemical analysis and identification of aerugine and aeruginol
1H NMR and 13C NMR spectra were either recorded on a Bruker Avance III 400 (400 MHz) or a Bruker Avance III 600 (600 MHz) spectrometer and referenced to the residual proton and carbon signal of the deuterated solvent, respectively.
LC-MS analysis was conducted with a Bruker Amazon SL IonTrap mass spectrometer connected to a 1260 Infinity Agilent Technologies HPLC equipped with a Nucleoshell RP-18 column (50 × 2 mm, 2.7 μm) (Macherey-Nagel, Germany) operating in positive ion mode with a flow rate of 0.5 ml/min. For 30 s the flow was disconnected from the ESI source and flushed into the waste to prevent salts from entering the MS. Mobile phases for the mass spectrometers was A = water (+0.1% FA) and B = acetonitrile (+0.1% FA). The gradient was: T0 min: B = 5%; T5 min: B = 5%; T25 min: B = 95%; T33 min: B = 95%.
Aerugine
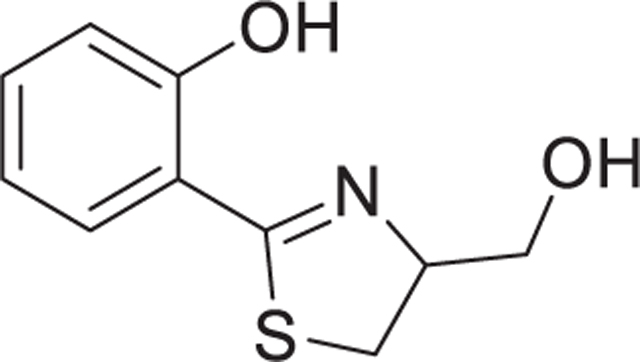
1H NMR (800 MHz, DMSO-d6) δ 12.65 (s, 1H, Ph-OH), 7.41 (t, J = 7.3 Hz, 2H, Ph-H), 6.97 (dd, J = 8.9, 1.2 Hz, 1H, Ph-H), 6.94 (td, J = 7.6, 1.2 Hz, 1H, Ph-H), 5.11 (t, J = 5.6 Hz, 1H, –CH2-OH), 4.83 – 4.78 (m, 1H, –CH(CH2-OH)), 3.63 (m, 2H, –CH2-OH), 3.49 (dd, J = 10.8, 8.9 Hz, 1H, -S-CHH-CH(CH2-OH)), 3.32 – 3.29 (m, 1H, -S-CHH-CH(CH2-OH)).
13C NMR (800 MHz, DMSO-d6) δ 171.05, 158.41, 133.21, 130.32, 119.10, 116.70, 115.82, 77.83, 62.32, 32.58.
HR-ESI-MS: m/z = 210.0580 [M + H]+ (calc. C10H11NO2S = 210.0583, Δppm = 1.4).
1D- and 2D-NMR spectra of aerugine are shown in Fig S2 and spectral data 1.
Aeruginol
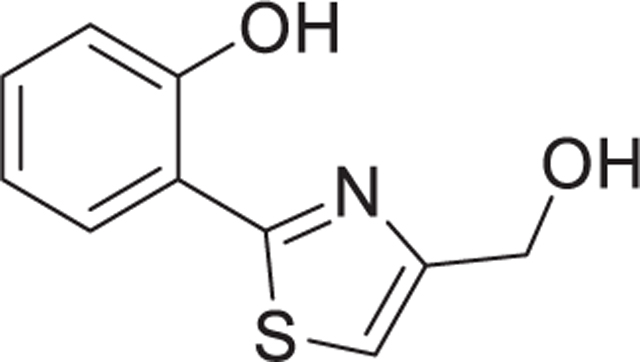
1H NMR (800 MHz, DMSO-d6) δ 11.65 (s, 1H, Ph-OH), 8.01 – 7.95 (m, 1H, Ph-H), 7.44 (s, 1H, -S-CH = C(CH2-OH)), 7.28 (t, J = 7.6 Hz, 1H, Ph-H), 7.01 (d, J = 8.2 Hz, 1H, Ph-H), 6.90 (t, J = 7.4 Hz, 1H, Ph-H), 5.38 (s, 1H, –CH2-OH), 4.62 (s, 2H, –CH2-OH).
13C NMR (800 MHz, DMSO-d6) δ 164.41, 156.55, 155.79, 130.95, 127.24, 119.05, 118.62, 116.84, 114.54, 59.62.
HR-ESI-MS: m/z = 208.0429 [M + H]+ (calc. C10H9NO2S = 208.0427, Δppm = 1.0).
1D- and 2D-NMR spectral data of aeruginol are shown in the supplementary data.
2.15. Synthesis of aerugine
The synthetic steps for aerugine were described by S. Noel et al. (Noel et al., 2011) and A. Zamri et al. (Zamri and Abdallah, 2000) which we used in the following synthetic route:
2.15.1. Synthesis of 2-(2-hydroxyphenyl)-4,5-dihydrothiazole-4-carboxylic acid (1a)
2-Hydroxybenzonitrile (500 mg, 4.2 mmol) was dissolved in MeOH (18.7 ml) and L-cysteine hydrochloride (1.475 g, 8.4 mmol) was added. Then, 0.1 M phosphate buffer was added (pH = 6.4, 18.7 ml). The solution was adjusted to pH =6.4 again with dry K2CO3 and was stirred for 12 h at 60 °C. The white precipitate was filtered off and the flow-through was concentrated. After concentration, the residual solution as diluted with H2O (20 ml). The pH was adjusted to 2 by addition of solid citric acid and the cloudy solution extracted with CH2Cl2 (DCM; 3x). The combined organic layers were dried over MgSO4 and the solvent evaporated to yield the pure compound (738 mg, 79%).
1H NMR (400 MHz, acetone-d6) δ 12.39 (s, 1H, –COOH), 11.56 (s, 1H, Ph-OH), 7.50 – 7.41 (m, 2H, Ph-H), 7.00 – 6.92 (m, 2H, Ph-H), 8.0 Hz, 1H, –CH(COOH)), 5.55 (dd, J = 9.2 Hz, 1H, –CH(COOH)), 3.81 – 3.73 (m, 2H, -S-CH2-CH(COOH)).
13C NMR (400 MHz, acetone-d6) δ 174.49, 171.34, 160.12, 134.43, 131.51, 119.88, 117.87, 116.86, 77.56, 34.14.
2.15.2. Synthesis of methyl 2-(2-hydroxyphenyl)-4,5-dihydrothiazole-4-carboxylate (1b)
Acetylchloride (516 μl, 7.3 mmol) was dissolved in MeOH (4.0 ml) at 0 °C. 2-(2-hydroxyphenyl)-4,5-dihydrothiazole-4-carboxylic acid (1a, 300 mg, 1.3 mmol) was added and the solution was heated to 50 °C for 24 h. The solvent was evaporated and ethyl acetate (10 ml) was added. The organic phase was washed twice with H2O (2 × 10 ml) and brine. The organic layer was dried over MgSO4 and the solvent evaporated to yield the pure compound (263 mg, 82%).
1H NMR (400 MHz, MeOD) δ 7.46 (dd, J = 7.8, 1.6 Hz, 1H, Ph-H), 7.43 – 7.35 (m, 1H, Ph-H), 6.99 – 6.87 (m, 2H, Ph-H), 5.50 – 5.42 (m, 1H, –CH(COOCH3)), 3.82 (s, 3H, –COOCH3), 3.70 – 3.66 (m, 2H, -S-CH2-C(COOCH3)).
13C NMR (400 MHz, MeOD) δ 175.51, 172.25, 160.23, 134.64, 131.81, 120.17, 118.03, 117.23, 78.05, 53.24, 34.42.
2.15.3. Synthesis of aerugine (1)
To 1 ml of tetrahydrofuran was added methyl 2-(2-hydroxyphenyl)-4,5-dihydrothiazole-4-carboxylate (1b, 36.5 mg, 0.16 mmol) and NaBH4 (16.2 mg, 0.43 mmol). The solution was stirred for 15 min until it turned yellow. 45 μl H2O were added dropwise and stirring was continued for 10 min. Then, 635 μl H2O were added and the solution was extracted with diethyl ether (3 × 1 ml). The organic layer was washed with brine and was dried over Na2SO4. The solvent was evaporated and the residue was purified with semipreparative HPLC with a normal phase Si-column with Si Reprosil 100 (250 × 10 mm, 5 μm, Dr. Maisch). The solvents were: A = DCM, B = MeOH. Gradient: isocratic 10% B. At 5 min, aerugine eluted as pure compound (7.8 mg, 23%).
1H NMR (400 MHz, MeOD) δ 7.43 (dd, J = 7.8, 1.7 Hz, 1H, Ph-H), 7.35 (ddd, J = 8.9, 7.2, 1.6 Hz, 1H, Ph-H), 6.93 (dd, J = 8.4, 1.2 Hz, 1H, Ph-H), 6.89 (td, J = 7.5, 1.2 Hz, 1H, Ph-H), 4.89 – 4.77 (m, 1H, –CH (CH2-OH)), 3.83 – 3.77 (m, 2H, –CH2-OH), 3.48 (dd, J = 11.0, 8.7 Hz, 1H, -S-CHH-CH(CH2-OH)), 3.33 (d, J = 3.1 Hz, 1H, -S-CHH-CH(CH2-OH)).
13C NMR (400 MHz, MeOD) δ 173.58, 160.20, 134.06, 131.60, 119.95, 117.81, 117.58, 79.51, 64.23, 33.70.
ESI-MS: m/z = 210.03 (calc. 210.06).
2.16. Synthesis of aeruginol
Aeruginol was synthesized by reduction of IQS according to Fig S3.
2.16.1. Synthesis of IQS
The synthesis was performed according to F. Shang et al. (Shang et al., 2014). In short, 2-bromothiazole-4-carbaldehyde (115 mg, 0.6 mmol), LiCl (53.8 mg, 1.27 mmol) and Pd(PPh3)4 (30 mg, 0.026 mmol) were added to a dry Schlenk tube under N2. Then, 2-hydroxylphenylboronic acid pinacol ester (158 mg, 0.72 mmol), 4 ml dry toluene, 4 ml ethanol and 2 ml 1 M aq. Na2CO3 were added. The mixture was heated to 110 °C and stirred for 23 h. The mixture was concentrated in vacuo and diluted with 20 ml CH2Cl2, washed with 20 ml H2O and 20 ml brine. The organic phase was dried over MgSO4 and the solvent evaporated. Automated flash chromatography with 1:1 hexane:EA yielded a semi-pure product, which was further purified with semipreparative HPLC with a normal phase Si-column with Si Reprosil 100 (250 × 10 mm, 5 μm, Dr. Maisch). The solvents were: A = EA, B = hexane. Gradient: T0min = 30% B, T1min = 30% B, T15min = 0% B. At 4 min, IQS eluted as pure compound (1.35 mg, 1%).
1H NMR (400 MHz, CDCl3): 11.58 (1H, br s, Ph-OH), 10.07 (1H, s, C (CHO)), 8.13 (1H, m, -S-CH = C(CHO)-), 7.63 (1H, m, Ph-H), 7.39 (1H, m, Ph-H), 7.12 (1H, m, Ph-H), 6.96 (1H, m, Ph-H). HR-ESI-MS: m/z = 206.0270 [M + H]+ (calc. 206.0270, Δppm = 0).
2.16.2. Aeruginol
NaBH4 (0.5 mg, 0.02 mmol) was added to a solution of IQS (2.0 mg, 0.01 mmol) in 1 ml THF. The solution was stirred at room temperature for 15 min. 50 μl H2O were added dropwise and stirring was continued for 10 min. Then, 100 μl H2O were added and the solution was dried over Na2SO4. The solvent was evaporated and the reaction yielded a pure compound (1.34 mg, 67%).
1H NMR (400 MHz, DMSO-d6 (400 MHz): 11.42 (1H, br s, Ph-OH), 7.96 (1H, d, J = 7.7,Ph-H), 7.47 (1H, s, -S-504 CH = C(CH2-OH)), 7.31 (1H, t, J = 7.7, Ph-H), 7.02 (1H, d, J = 8.3, Ph-H), 6.94 (1H, t, J = 7.5, Ph-H), 5.38 (1H, br s, –CH2-OH), 4.62 (2H, s, –CH2-OH).
2.17. Data analysis
Unless otherwise stated, all data are expressed as means ± SEM of at least three biological replicates, each of them based on three technical replicates. The methods for statistical analysis are indicated in the respective figure legends.
3. Results and discussion
3.1. Purification of neurotoxic metabolites from Streptomyces venezuelae extracts
Previously, the existence of a microbial metabolite with specific neurotoxic properties had been proposed, but its structural identification remained elusive (Caldwell et al., 2018; Caldwell et al., 2009). To fill this knowledge gap, we sequentially perfomed an activity-guided fractionation of bacterial lysates, using a neurotoxicity test as bioassay (Fig. 1a). As a starting point, we used cultures of S. venezuelae (presumed to be neurotoxic) and cultures of S. lividans (phylogenetically related, yet deemed non-toxic) (Caldwell et al., 2009)) that were extracted with different solvent combinations and subsequently pre-fractionated over reverse-phase C18 Sep-Pak columns. One of the S. venezuelae fractions killed >50% of a DAergic human neuronal cultures (produced from LUHMES cells), while a similarly-prepared extract of S. lividans was non-cytotoxic (Fig. 1b).
Fig. 1.
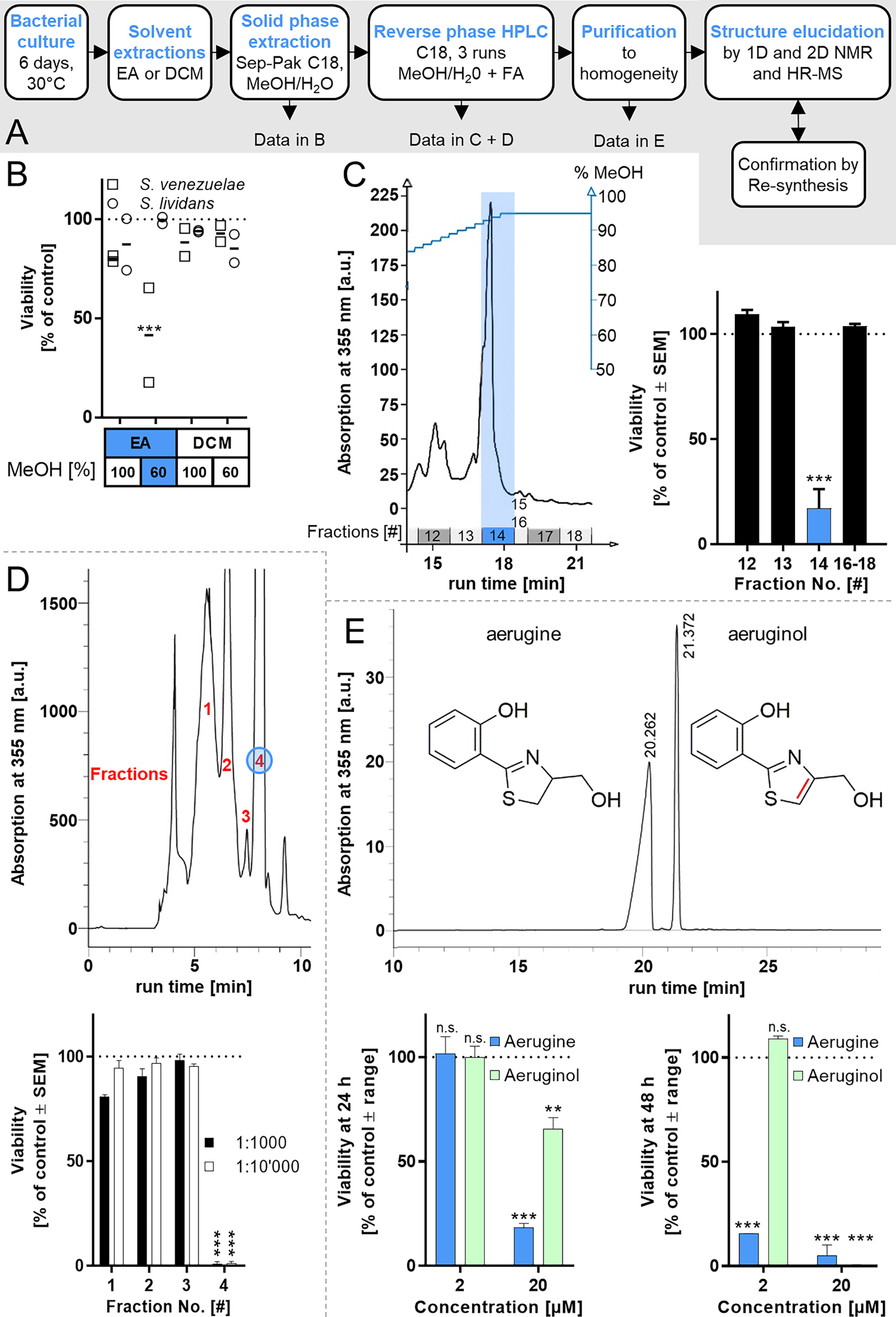
Purification of neurotoxic small molecules from Streptomyces bacterial strains. A Workflow of metabolite identification from bacterial extracts. Subfigures that provide activity data on the respective purification stages are indicated. Information on the purification procedures is given in the subfigure paragraphs below. B-E The neurotoxicity of extracts, fractions and pure compounds was tested on human dopaminergic neurons. After 6 days of differentiation (d6), cultures of LUHMES neurons were exposed to bacterial fractions. The cell viability was assessed by calcein-AM & H-33342 staining, automated fluorescence microscopy and quantification by an image processing algorithm. B Cultures of Streptomyces venezuelae and S. lividans were extracted with ethyl acetate (EA) or dichloromethane (DCM). The crude extracts were fractionated via Sep-Pak C18 and eluted using a stepwise gradient with methanol and water (MeOH = 60% or 100%). Dried and DMSO-reconstituted fractions were tested for effects on neuronal viability. Data from at least two fully independent runs. Viability data was obtained after 24 h of exposure. C The EA/60% MeOH extract of S. venezuelae was further fractionated by reverse phase HPLC (C18 column, MeOH gradient). The collected fractions are indicated in the absorption spectrum (a part of the spectrum is shown, the full spectrum is given in Fig S1; the MeOH gradient started at 50%). Fractions were dried, taken up in DMSO and added to neurons for 48 h (final dilution 1:1000). Viability data are means ± SEM (n = 3). D Fraction 14 of the previous step was further purified by several reverse phase HPLC (C-18) runs in acetonitrile/water eluent. Absorption peaks were collected in fractions as depicted (red numbers in the spectrum). They were tested for their effect on neuronal viability (24 h, at 1:1000 and 1:10000 dilutions). E Two single substances (aerugine and aeruginol) of fraction 4 of the previous purification step were separated and purified to homogeneity. The chemical structures (elucidation details in Fig S2 and spectral data 1) are displayed next to their respective peaks in the elution trail diagram. Note the additional double bond in the thiazolidine ring of aeruginol. The two compounds were assessed in two separate experiments for their effect on neuronal viability after incubation times of 24 h and 48 h. All statistical analyses were performed using a one-way ANOVA with Dunnett’s multiple comparisons test (** = p < 0.005, *** = p < 0.0001).
The bioactive lysate was purified by HPLC runs, until a fraction was obtained that still showed bioactivity (killing neurons) when diluted 10000-fold (Fig. 1c,d; Fig S1). From this fraction, two compounds were each purified to homogeneity by multiple HPLC runs and the structure identified by 1D and 2D NMR spectroscopy in combination with mass spectrometry (Fig S2). Both compounds, aerugine (209.27 g/mol) and aeruginol (207.25 g/mol) were confirmed to be cytotoxic to DAergic neurons in a time- and concentration-dependent manner (Fig. 1e). As aerugine was more potent (low μM range), further work concentrated on this metabolite. Its structure was confirmed by re-synthesis (Fig S3), and an overview of its physicochemical properties was obtained. The compound was predicted to reach intracellular compartments and to cross the blood-brain barrier without need for transporters (Fig S4).
3.2. Biosynthetic pathway of aerugine
S. venezuelae is a soil-living bacterium, and its production of a neurotoxic metabolite may be of little relevance to humans or most other vertebrates. However, genetic elements required for synthesis of secondary metabolites may be wide-spread amongst bacteria. The gut microbiome produces a large variety of metabolites that may influence physiology and pathology of the local and central nervous systems (Baldini et al., 2020; De Miranda et al., 2022; Dodd et al., 2017; Donia and Fischbach, 2015; Nicolas and Nolan, 2022; Sampson et al., 2016; Swann et al., 2020). We used here published literature and databases to identify the likely biosynthetic pathway of aerugine, and to check whether the responsible genetic machinery may also be found in human pathogens or the intestinal flora.
Indeed, the compound identified here, has been described earlier as a byproduct of watasemycin biosynthesis. Moreover, the biosynthetic relevance of the underlying genetic machinery has been proven by detection of aerugine in cultures of S. coelicolor M1152, which were engineered to express the watasemycin gene cluster (Inahashi et al., 2017). Our analysis of the gene sequence of S. venezuelae (ATCC 10712) used in this study confirmed the presence of this non-ribosomal peptide synthetase (NRPS) biosynthetic gene cluster (Fig S5). The synthesis uses the building blocks salicylic acid plus cysteine. Aerugine is likely to be generated from a watasemycin precursor by a non-enzymatic hydrolytic step. A highly homologous biosynthetic machinery is used by Pseudomonas aeruginosa to generate the siderophore pyochelin. Aerugine has been previously described as by-product of pyochelin biosynthesis (Kaplan et al., 2021). In enterobacteriae, the identical precursor is further modified via a mixed NRPS – polyketide synthetase (PKS) involving condensation of a malonyl-CoA and one additional cysteine to the full length product yersiniabactin (Miller et al., 2002). Siderophores of the watasemycin-pyocheline-yersiniabactin family are widespread and generated by biosynthetic machineries that likely enable the production of aerugine as shunt-product. Several producers are found in the human intestine (Pseudomonas sp., E. coli, Yersinia sp., Salmonella sp. etc.) (Fig S6). Within the compound family, aerugine is most-closely related to pulicatins. The latter compounds are methylated in position five of the thiazoline ring. Very little information is available on human-relevant bioactivities. The only suggested target for aerugine is the 5HT2B receptor, from which it displaced radioactive lysergic acid diethylamide at low μM concentrations. However, functional data are not available to our knowledge (Lin et al., 2017).
3.3. Activity confirmation and primary characterization
After having obtained information on aerugine biosynthesis, we compared the bioactivity of purified preparations and pure synthetic product. The cytotoxic potency of extracted compound was similar to that of aerugine prepared by chemical synthesis (Fig. 2a,b), and the EC50 value was 3.1 ± 0.7 μM. Accordingly, all further experiments were performed with synthetic aerugine.
Fig. 2.
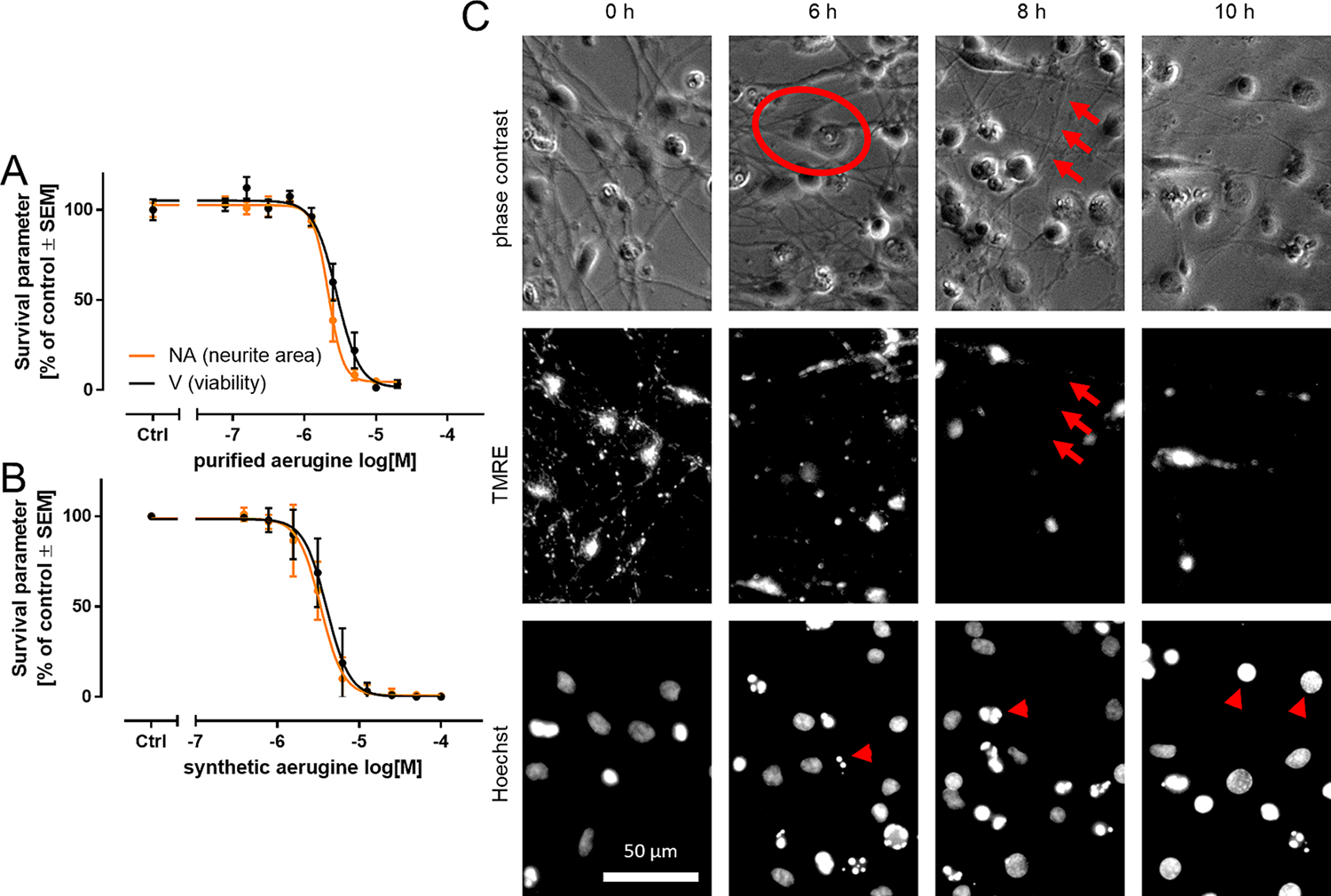
Degeneration of human dopaminergic neurons exposed to aerugine. Cultures of differentiated LUHMES neurons were used on d6 for cytotoxicity testing. Cells were exposed to serial dilutions of (A) aerugine purified from bacterial extracts and (B) synthetic aerugine. The cell viability (V) and neurite area (NA) were assessed 24 h later by calcein-AM & H-33342 staining and high content imaging. Data are presented as means ± SEM of three biological replicates. C LUHMES neurons (d6) were treated with 10 μM aerugine. After the indicated incubation times, they were stained with H-33342 & the mitochondrial membrane potential indicator tetramethylrhodamine ethyl ester (TMRE). Representative images after exposure to aerugine are shown. Cells reacted similarly to purified aerugine. The scale bar represents 50 μm. A flattened cell is circled. Arrows indicate an intact neurite without functional mitochondria. Arrow heads point to features of apoptotic nuclear condensation. Enlarged details are shown in Fig S8.
The standard assay used to evaluate toxic effects on neuronal cultures is based on single cell staining with a live cell dye (calcein). This allows also the quantification of potential effects on neurite structures (Delp et al., 2019; Stiegler et al., 2011). In the 24 h assay setup, aerugine affected neurites to the same extent and at similar concentrations as cell bodies (Fig. 2a,b). Biochemical viability assays that measure the average of a whole culture well (lactate dehydrogenase (LDH)-release and resazurin reduction tests) confirmed the single cell data, although with slightly less sensitivity (Fig S7).
To obtain additional information on the death process triggered by aerugine, we also examined earlier time points after exposure to a fixed concentration of 10 μM. Semi-quantitative information was obtained from phase contrast images, by staining of energized mitochondria with the potential-sensitive dye tetramethylrhodamine ethyl ester (TMRE) and by visualization of the chromatin structure with the DNA-intercalating dye H-33342. After 6 h exposure, clear morphological changes became obvious. While the neurite structure was well-preserved, some of the cell bodies flattened in a way that the nucleus became visible in phase contrast. The normally round/dome-like cells took a flattened “fried egg” structure. In parallel, the DNA condensed and fragmented into small, round and highly-fluorescent spheres, a typical hallmark of apoptosis. Cells with apoptotic nuclei had lost their TMRE stain. At 8 h, more cell bodies looked flattened (“unhealthy”) and the neurites were thinning. Even within structurally intact neurites (no blebbing, sharp contrast) the mitochondrial membrane potential was lost (absence of TMRE staining). At 10 h, the majority of neurites was lost, and most nuclei were condensed (Fig. 2c; Fig S8). This sequence of events is typical of neuronal apoptosis, e.g. under mild excitotoxic conditions, or after exposure to proteasome inhibitors or cytoskeletal poisons (Gutbier et al., 2018b; Leist et al., 1997; Volbracht et al., 2001; Volbracht et al., 1999). The overall kinetics of cell death was rather fast, compared to many other toxicants. This suggests that aerugine directly affects one of the cell biological processes essential for survival. To obtain a more quantitative confirmation of the rapid process, cells were stained with calcein-acetoxymethyl ester and propidium iodide (PI) and observed over time. The staining of nuclei (indication of plasma membrane permeability) by PI was quantified by an imaging algorithm. By 6 h, dead cells increased by 17 ± 6%, by 8 h by 25 ± 5%, and by 10 h, > 95% of the cell nuclei were positive. This confirms the fast cell death process triggered by aerugine (10 μM) in neurons.
We explored here a potential mitochondrial toxicity of aerugine, as mitochondria are one of the most obvious vital structures in neurons, and a large body of evidence points to the role of mitochondrial impairment especially in toxicity to DAergic neuronal death (Delp et al., 2019; Huang et al., 2022; Schildknecht et al., 2017). Initially, we evaluated the effect of aerugine on mitochondrial respiration. The oxygen consumption rate (OCR) in the presence of aerugine was not different from control conditions, even when tested under various metabolic conditions (Fig S9a). Also, when various functional parameters (e.g., basal respiration or ATP production) were calculated, no significant difference was observed (Fig S9b). To confirm these data, we made use of the fact that LUHMES neurons can be made extremely dependent on mitochondrial respiration by exchange of glucose in the medium to galactose. Under such conditions, mitochondrial inhibitors can be detected with up to 1000-fold increased sensitivity (Delp et al., 2019). However, also under this condition, no increased aerugine toxicity was observed (Fig S9c,d). It seems therefore unlikely that aerugine directly affects mitochondrial respiration. More efforts are required in the future to identify the cell biological target process of this neurotoxicant. The clear effects of aerugine on mitochondrial membrane potential during cell death are likely to be caused indirectly.
3.4. Specificity of aerugine neurotoxicity
Although aerugine was identified here based on its activity as neurotoxic (Fig S10a), we could not assure that our findings were neuro-specific. It was also unclear, whether we witnessed an unspecific effect of substituted phenolic compounds or a specific event triggered only by aerugine and very closely-related structures. To address this latter point, we compared aerugine to three structural analogs that were available to us. When the aliphatic OH-group of aerugine was oxidized to an aldehyde (dhIQS) or a carboxylic acid (DHAA), all toxicity was lost (Fig S10b,c). This indicates that there is a specific structural requirement for aerugine to be neurotoxic. The aldehyde of aeruginol (IQS) killed DAergic neurons within 24 h in the 10–25 μM concentration range, i.e. it was about equipotent with aeruginol (Fig S10d). This indicates that some close analogs of aerugine may also be neurotoxic. Further quantitative structure-activity relationship (QSAR) studies are required to better understand the toxicological potential and mechanism of the whole watasemycin-aerugine family.
To investigate the cell type specificity, we first compared LUHMES cells of different maturity. We found that their sensitivity to aerugine increases as they differentiate. The EC50 values for proliferating, immature and mature cells were 13 ± 0.2 μM, 9.3 ± 1.1 μM, and 4.1 ± 0.8 μM (Fig. 3a; Fig S11). Thus, even within the same cell line, the neuronal maturity state increased aerugine sensitivity approximately three-fold. When we examined aerugine toxicity to other commonly used human cell lines (HepG2 – liver-derived, Hek293FT – kidney-derived, HeLa – cervix-derived), no toxicity was observed, even with a prolonged exposure of 72 h (Fig. 3b). To confirm the toxicity to neurons, we obtained various human stem cell-derived cultures. Mixed central nervous system neurons were clearly affected by aerugine (but at higher concentrations), while peripheral nervous system cells showed no effect at concentrations up to 100 μM. Cortical neurons (also representative for the central nervous system, but not containing DAergic neurons) were also sensitive to aerugine, but at about 50x higher concentrations than DA neurons (mature LUHMES) (Fig. 3c). Finally, we also addressed the toxicity to prokaryotes. We used Enterococcus and Neisseria gonorrhoeae as representatives of gram-negative and gram-positive bacteria. They were not affected in viability or growth at 20 μM aerugine, a concentration that would kill all mature DA neurons (Fig. 3d). We conclude from these data that aerugine is not a general, unspecific cytotoxicant. It rather has a considerable specificity for DA neurons.
Fig. 3.
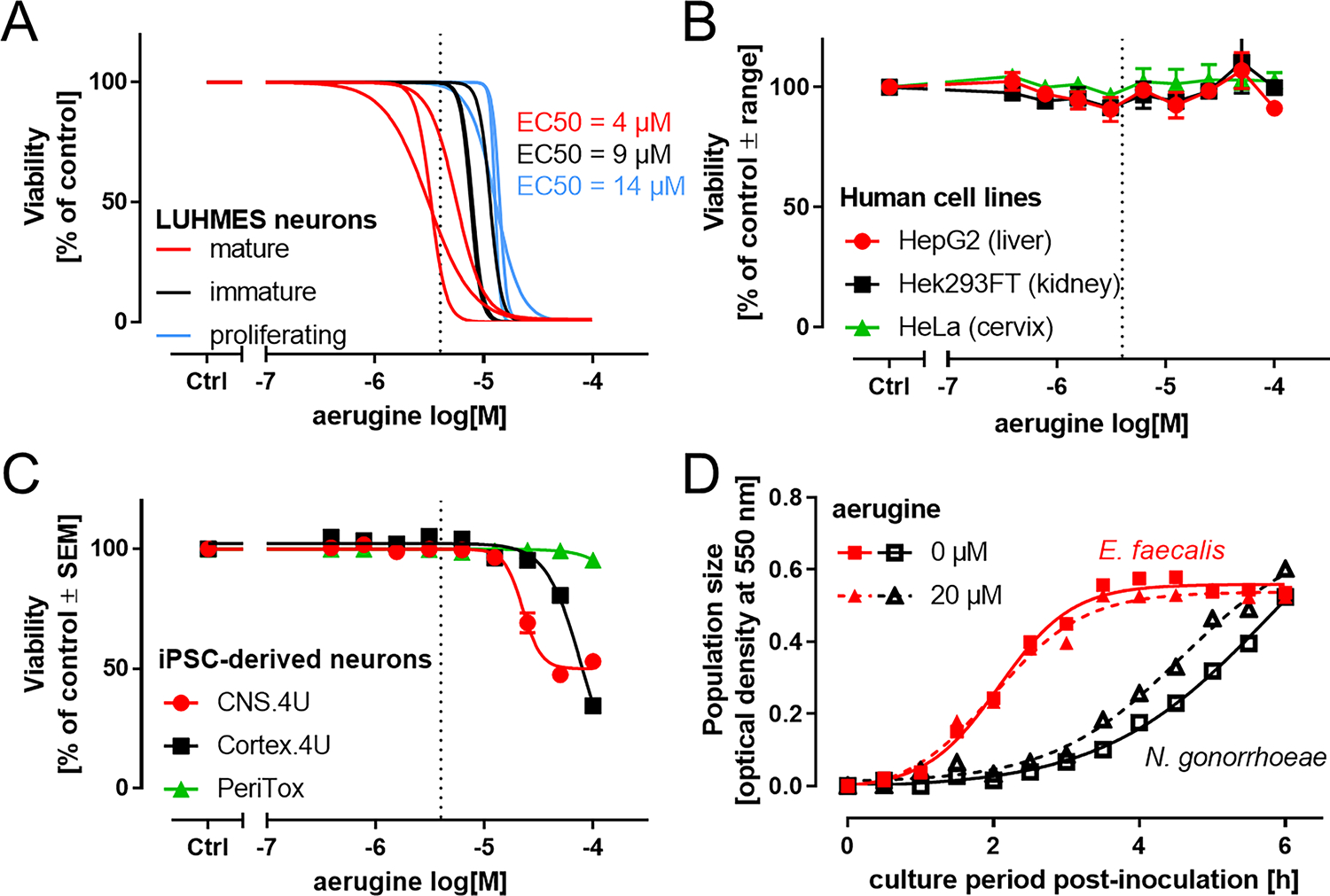
Cell type specificity of aerugine cytotoxicity. A Cultures of LUHMES neurons were used on d0 (proliferating, blue), d2 (immature, black) or d6 (mature, red) of differentiation for cytotoxicity testing. Cells were exposed to nine serial dilutions of synthetic aerugine. The cell viability was assessed 24 h later by calcein-AM & H-33342 staining and high content imaging. Data are presented as sigmoidal curve fits of data points from each different experiment (averaged data points are displayed in Fig S11). The half-maximal cytotoxic potency (EC50) is displayed for the three maturity stages. Dotted lines at 4 μM indicate the average EC50 for mature LUHMES. B Three widely-used human cell lines were cultured as suggested by the supplier/cell bank and used for experiments in 96-well plates. They were treated for 72 h with synthetic aerugine. The overall cell viability was assessed by measuring resazurin reduction in technical triplicates. Data are presented as means ± range of two biological replicates (different experiments). C Three iPSC-derived neuronal cell types were treated for 24 h with synthetic aerugine. The cell viability was assessed 24 h later by calcein-AM & H-33342 staining and high content imaging. Data are presented as means ± SEM of three biological replicates (different experiments). D Lack of cytotoxic or bacteriostatic effects of aerugine (20 μM) on exemplary gram-positive and gram-negative microbes: The growth characteristics of Enterococcus faecalis (gram-positive) and Neisseria gonorrhoeae MS11 (gram-negative) were followed during the log growth phase in their respective culture media. Data are from one biological replicate.
3.5. Pharmacological modulation of aerugine toxicity
A typical feature of specific toxicants is that their effects on cell health/survival can be prevented by inhibitors or modifiers of downstream pathways that are affected. We used here a panel of nine such modulators and studied their effect on the neurotoxicity of aerugine (Fig. 4a). Three iron chelators (used in the low μM range) completely prevented cell death (Fig. 4b). Under these conditions, not just the cell bodies survived, but also the neurite structure was well-preserved (Fig. 4c). Rescue by iron chelators may point to a ferroptotic death mechanism (Lin et al., 2022), but iron may also play a role in other types of cell death, such as apoptosis or lysosomal death (Bogdan et al., 2016; Chen et al., 2019; Terman and Kurz, 2013; Whitnall and Richardson, 2006). Especially, DAergic neuronal death has many associations with iron overload (Devos et al., 2014; Gutbier et al., 2020; Riederer et al., 2021; Whitnall and Richardson, 2006).
Fig. 4.
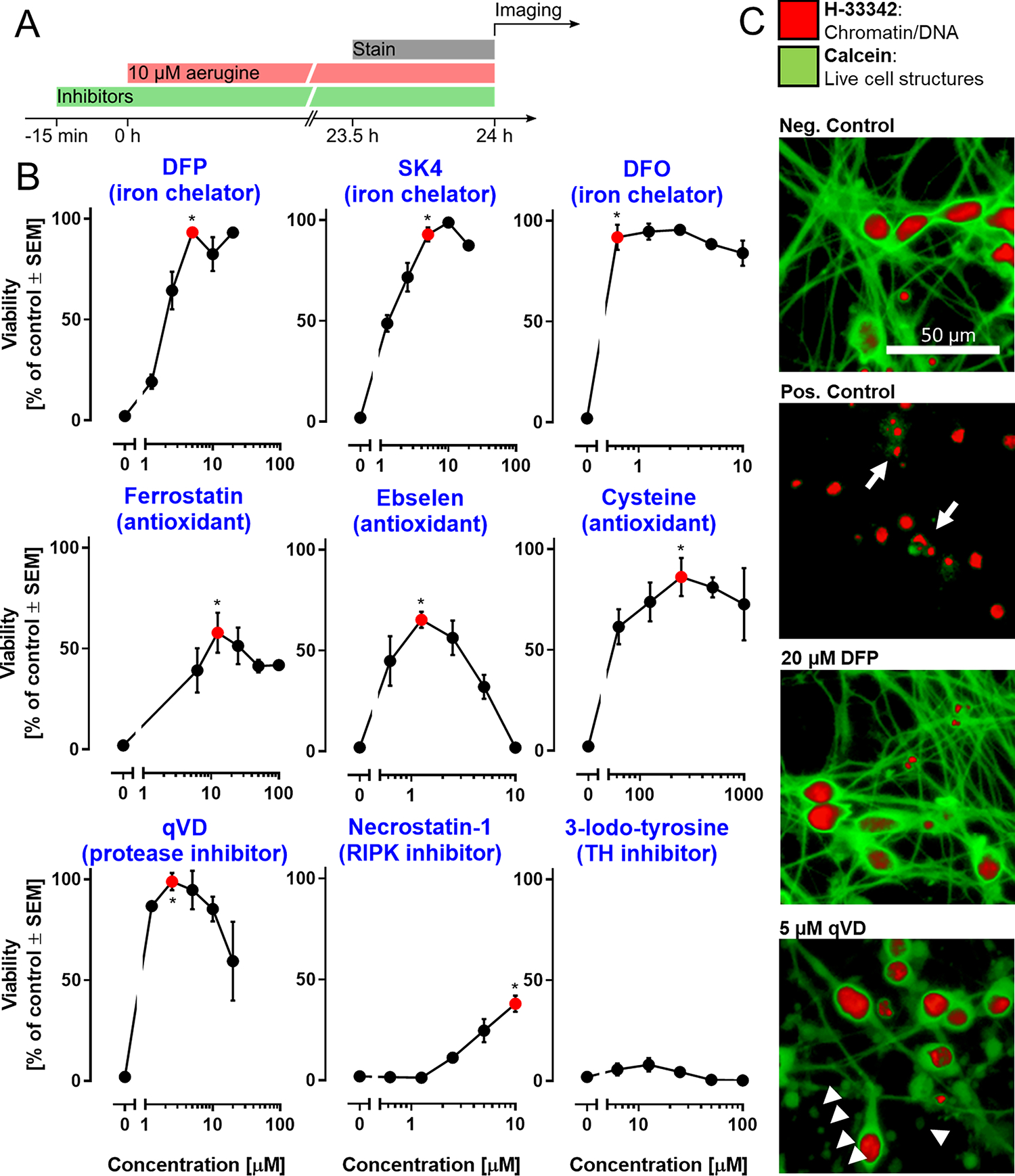
Modulation of aerugine-induced neurodegeneration by mechanistic inhibitors. LUHMES cultures (d6) were used under standard conditions to assess the cytotoxicity of aerugine (10 μM) in the presence of various agents known to interfere with cell death pathways. A Treatment scheme: the respective inhibitors were applied 15 min prior to aerugine treatment (10 μM), and the viability was determined by high-content imaging after 24 h. B Inhibitors were used at different concentrations, as indicated. The main biochemical activity of each agent is indicated in brackets. Data are means ± SEM of at least three biological replicates with 3 technical replicates each. DFP = deferiprone; DFO = deferoxamine; qVD = caspase inhibitor; RIPK = receptor-interacting serine/threonine-protein kinase. For statistical analysis, the lowest concentration of inhibitor that produced the maximal protective effect (red) was compared to the treatment with aerugine alone using a two-tailed t-test (*: p = 0.01). C Exemplary pictures of LUHMES cultures after 24 h treatment: negative control (=DMSO), positive control (=10 μM aerugine), DFP (=20 μM DFP + 10 μM aerugine) and qVD (=5 μM qVD + 10 μM aerugine). Arrows indicate cells with a broken cell membrane (calcein-negative) and condensed nuclei (small H-33342-positive area). Arrow heads indicate disintegrating neurites (chains of calcein-positive blebs). Enlarged details are given in Fig S12.
Iron is a key player in oxidative stress pathways. Consistent with this, three different types of antioxidants (ferrostatin: radical scavenging; ebselen: SOD mimetic; cysteine: glutathione precursor) partially (maximally 60–90%) prevented aerugine neurotoxicity. The pronounced rescue by cysteine may be interpreted as pointing towards ferroptosis (Morris et al., 2018). However, cysteine and related compounds (e.g., N-acetylcysteine) are also anti-apoptotic. For instance, LUHMES cells driven into apoptosis by proteasome inhibitors, can be rescued by cysteine (Gutbier et al., 2018b; Suciu et al., 2023).
A role of apoptosis in aerugine neurotoxicity is suggested by the protective effect of the protease inhibitor qVD. At optimal concentrations (2–4 μM) cell death was completely prevented (rescue of all cell somata). However, damage to the neurites still occurred in the presence of qVD. This set of observation is consistent with a role of apoptotic caspase activation in aerugine toxicity (Fig. 4b,c; Fig S12). In contrast, the necroptosis-inhibitor necrostatin-1 only slightly attenuated cell death. The tyrosine hydroxylase inhibitor 3-iodo-tyrosine had no effect at all. The latter finding suggests that DA autoxidation, a process sometimes associated with DAergic neurotoxicity, plays no major role. While more mechanisms need to be investigated to find the target of aerugine, our pharmacologic intervention data suggest that aerugine triggers defined death-associated pathways, and inhibition of these pathways abolishes neurotoxicity, also in the continued presence of the toxicant.
3.6. In vivo toxicity of aerugine to C. elegans dopaminergic neurons
Our study was triggered by the observation that metabolite extracts of S. venezuelae caused selective DAergic neurotoxicity in the nematode C. elegans (Caldwell et al., 2009). Here, we isolated aerugine as human DAergic neurotoxicant, showed its cell type specificity and the triggering of defined death pathways (inhibited by pharmacological intervention). However, experimental proof is required that aerugine indeed reproduces the effects of bacterial extracts in a viable model, such as C. elegans. Moreover, studies using a complex life organism are an important confirmation of the postulated cell type specificity of aerugine-induced neurotoxicity.
To address this question, we used transgenic worms with green fluorescent protein (GFP) expression targeted to either DA neurons or γ-aminobutyric acid (GABA)-neurons. They were exposed to aerugine at the L4 larval stage for up to one week. In this experimental setup, the number of surviving worms declined over time. However, aerugine neither triggered death during the first 2 days (when there was little natural death), nor did it enhance/accelerate the natural decline over longer time periods (8 days) (Fig. 5a). The GFP-labelling allowed the assessment of DAergic neuron integrity in the worms (4 labelled neurons, including their neurite structure were scored) (Fig. 5b). An assessment after 48 h exposure showed a concentration-dependent degeneration of DA neurons in aerugine-exposed worms, while GABA neurons were not affected, and the worms survived (Fig. 5c,d; Fig S13 for detailed scoring). Up to 50% of the DAergic neurons lost their normal (fully) healthy phenotype, with the majority of effects being detected on the neurite integrity.
Fig. 5.
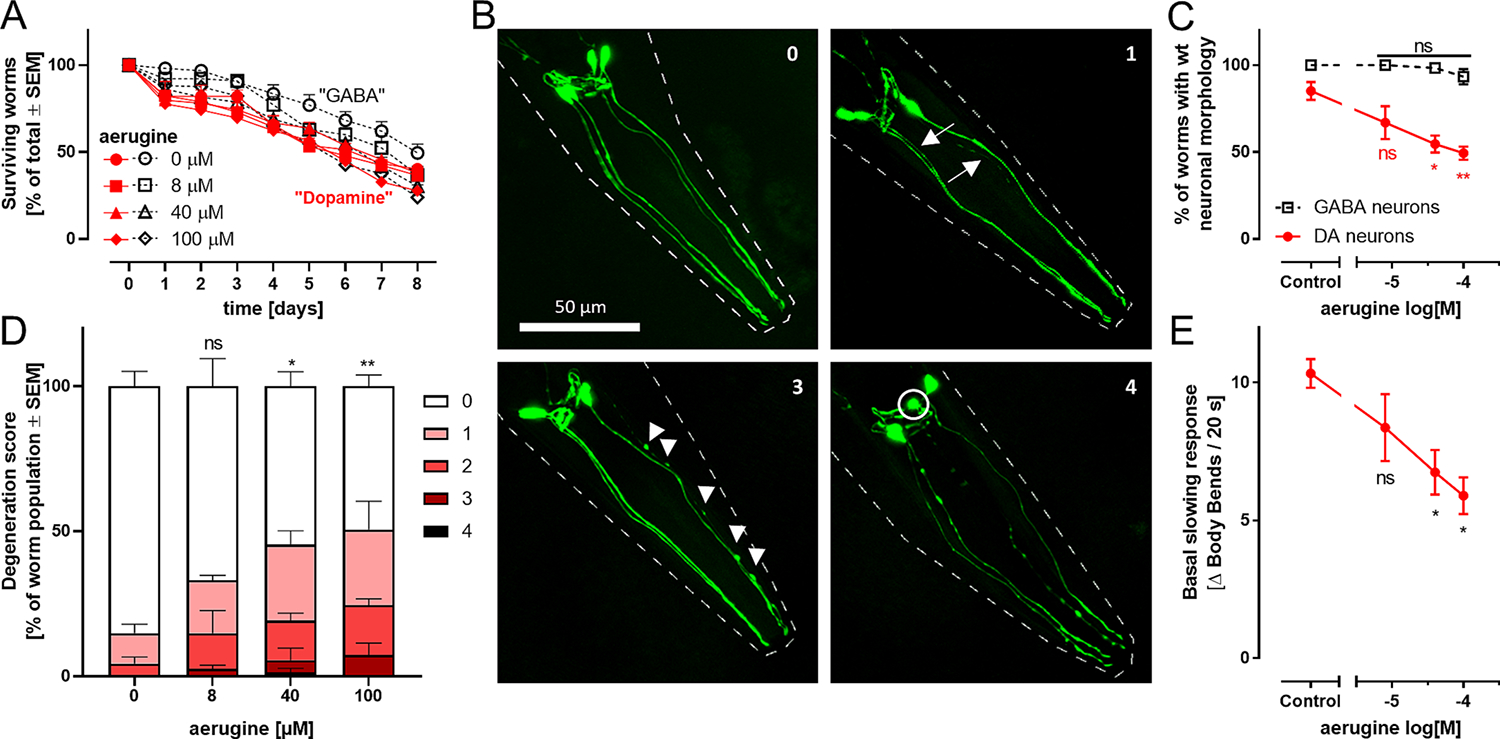
Specific dopaminergic neurodegeneration triggered by aerugine in C. elegans. A The C. elegans worm strains BY200 (containing fluorescently labelled dopamine neurons; label “Dopamine”) and Punc-25 (containing fluorescently labelled γ-aminobutyric acid neurons, label “GABA”) were used at L4 larval stage for an eight day survival test. They were exposed to different aerugine concentrations and counted every 24 h. Data are based on 40–60 worms per group. Differences on a given day were analyzed by ANOVA and found to be non-significant. Two-way ANOVA (time × treatment) indicated a significant trend of decreased survival over time (independent of treatment or genotype). B L4 larval stage worms of both strains were treated for 2 days with aerugine. The GFP-labelled neurons of 20–30 worms per condition were analyzed and scored for specific neurodegeneration. Each worm was assigned a degeneration score (0 for wild type and 1–4 for increasing severity). Representative images illustrating neurodegeneration scores (indicated by numbers in the upper right corner): The pictures show the 4 cephalic dopaminergic neurons in green. The worm outline is indicated by dashed white lines. Arrows highlight thinning dendrites. Arrow heads point out blebs (irregular protrusions of the dendrites). Degenerating cell bodies are circled. Degeneration score 2 looks similar to 3 but has a maximum of 4 blebs. These scores were assessed to quantify the dopaminergic neurodegeneration caused be aerugine in vivo. C Worms with wild type morphology (score 0) were counted. For statistical analysis, a one-way ANOVA with Dunnett’s multiple comparisons test was performed (*: p < 0.05; **: p < 0.01 for difference of aerugine groups vs untreated controls). D The degeneration scores exemplified in B were quantified. The data for DAergic neurons is shown. The data for GABAergic neurons can be found in Fig S13. E The “basal slowing response” was assessed as proxy for the functionality of the worms’ dopaminergic system. It was measured in L4 larval stage BY200 worms treated for 2 days with aerugine. As the slowing response is triggered by food, the body bends per 20 s in the absence and presence of food were counted and the difference (Δ) was calculated as experimental readout. For statistical analysis, a one-way ANOVA with Dunnett’s multiple comparisons test was performed (*: p < 0.05).
To understand the functional consequences of this morphological evaluation, we used a behavioral endpoint strongly dependent on DAergic signaling. We made use of the fact that worms, normally body-bending in their growth medium, move less dynamically, when they are exposed to feed (de Bono and Maricq, 2005; Krum et al., 2021; Sawin et al., 2000). We assessed this “basal slowing response” (BSR) in nematodes exposed to food (bacteria) in the presence or absence of aerugine. A concentration-dependent response was observed, i.e. the BSR was attenuated by the neurotoxicant. This indicates that the function of the DAergic system was disturbed by aerugine.
4. Outlook and conclusions
In summary, our study provides an entirely novel perspective on human health implications of the environmentally-produced bioactive compound aerugine. We first corroborated earlier, well-documented observations (Caldwell et al., 2009), that a microbial-derived agent can trigger pathological processes relevant to idiopathic PD. Next, we proceeded to identify the low molecular weight, stable and neurotoxic chemical postulated in previous studies to be the DAergic toxin (Caldwell et al., 2018; Caldwell et al., 2009). Knowledge on the molecular structure of aerugine allowed us to identify the corresponding biosynthetic gene cluster. An important implication of this work is that not only soil bacteria (S. venezuelae) may be the source of the potent neurotoxicant aerugine, but also that constituents of the normal or of the disease-associated human microbiome (E. coli or P. aeruginosa) may produce this or related compounds.
By using a panel of cell death pathway inhibitors, we identified several compounds that attenuated or even completely blunted the toxicity of aerugine. Both, the data from our study and general knowledge on the iron-chelating properties of aerugine-related compounds (see Fig S6 for structural similarity to pulicatins) are consistent with a cell death mechanism related to iron-enhanced oxidative stress. Oxidative stress plays an important role in many regulated and non-regulated death modes (Dionísio et al., 2021; Orrenius et al., 2007). Long before the predominantly oxidative types of cell death, i.e., ferroptosis and oxeiptosis, have been defined as stand-alone entities (Jiang et al., 2021; Scaturro and Pichlmair, 2019), the generation of reactive oxygen species and the loss of cellular redox-buffers was known as common features of most types of dying cells. This observation covered the full cell death spectrum, ranging from necrosis to apoptosis (Tang et al., 2019).
Iron can take a key role in cellular demise, as it converts hydrogen peroxide, which has a low-reactivity to cellular structures, via the Fenton reaction into the highly reactive and destructive hydroxyl radicals. Low amounts of redox-reactive metals can be detrimental, as they drive the so-called “iron-enhanced Haber-Weiss cycle”. This catalytical role of free ferric and ferrous iron in the production of reactive oxygen species is well-established to occur within live cells (Kehrer, 2000) and it plays an important role in DAergic neurons and in PD (Cerasuolo et al., 2023; Gutbier et al., 2020; Lin et al., 2022). To fully appreciate the potential role of iron in cell damage, it is important to consider various cellular iron pools. The total iron concentration in brain cells is in the low mM range, but most of it is bound to ferritin in a catalytically inactive form (Philpott and Ryu, 2014; Reinert et al., 2019; Wu et al., 2023). Only nM levels are found as free iron ions in the presence of oxygen and at neutral pH. However, cells contain a substantial pool of so-called “labile iron” (Kaplan and Ward, 2013; Lv and Shang, 2018), which describes metal ions complexed to glutathione, glutaredoxin and other cellular components. Such loosely complexed iron can participate in catalytic reactions that generate reactive oxygen species. Moreover, it is well-established that certain chelators even enhance this catalytic activity (Luzzatto et al., 1995; Miller et al., 2016; Szarka et al., 2022). A compound like aerugine may thus cause iron-dependent toxicity by changing availability and complexation of cellular iron, by slightly shifting cellular iron pools or by facilitating the import of additional extracellular iron into the cytosol. Our finding of protection by several antioxidants and iron chelators would be compatible with such a mechanism. To provide further evidence, technologies to quantify distinct iron pools, their redox states and their catalytic activity will be required (Lv and Shang, 2018).
We suggest here that the postulated death mechanism should explain (to some extent) the cell type selectivity. One may question whether iron, which is present in all mammalian cell types, is likely to be a key player in the extremely specific toxicity of aerugine to DAergic neurons. However, a selectivity of iron-mediated cell death is consistent with literature data. For instance, there are large differences in ferroptosis sensitivity of various cell types. While some cells are fully resistant, neurons are a particularly sensitive population (Bouchaoui et al., 2023; Sun et al., 2023). Moreover, DAergic neurons affected by PD are known to have a particularly high iron content (Friedrich et al., 2021). For an overall evaluation, it is important to note that toxicity by oxidative stress is always dependent on the balance between the generation of reactive oxygen species and the buffering by e.g. glutathione and glutathione-dependent enzymatic reactions. Cells differ greatly in their capacity to regenerate glutathione, and neurons are known to be particularly dependent on the uptake of glutathione pre-cursors from extracellular sources. Their own anti-oxidant pool is easily exhausted (Asanuma and Miyazaki, 2021; Ishii et al., 2019; Lotharius et al., 2005). For instance, neurons, including the LUHMES cells used here, recruit a substantial amount of their cysteine from protein turnover. If this is inhibited, glutathione is rapidly depleted (Gutbier et al., 2018b; Suciu et al., 2023). The oxidative stress generated by DA autoxidation may be a further sensitizing factor to explain the selectivity (Sun et al., 2006; Uhl, 2019; Vergo et al., 2007; Xie et al., 2010).
Inhibition of the mitochondrial electron transfer chain (Bose and Beal, 2016; Schapira, 2012) is one of the most-studied pathomechanisms relevant for neurons. On first sight, it may not seem intuitive that DAergic neurons are specifically affected by mitochondrial inhibitors, as most cell types somehow use mitochondria. However, data sets spanning from animal experiments to epidemiological findings support this hypothesis (Schildknecht et al., 2017; Terron et al., 2018), with the most likely explanation being that DAergic neurons depend particularly strongly on mitochondrial function. We considered it necessary to examine this candidate mechanism for aerugine. Using two different experimental approaches, we found that mitochondrial ATP generation is unlikely to be the primary target of the toxicant. Thus, this alternative hypothesis on aerugine toxicity can be excluded.
For many complex human diseases, no laboratory model can cover all aspects. Conversely, it is important for drug discovery to have access to diverse models reflecting different aspects of the pathogenesis. In the field of parkinsonian neurodegeneration, many mitochondrial models have been developed and are in current use. Also, a large panel of genetic models is available. They make use of gene variants found in familial forms of PD (e.g., mutations or altered expression levels of synuclein, parkin, PINK1) (Bose et al., 2022; Chia et al., 2020; Program, 2021) and mimic disturbed proteostasis or impaired mitochondrial function. An aerugine-based model could be an interesting addition that covers complementary aspects. A starting point into this direction could be the exposure of more complex human organoids to the toxin or the probing of species specificity. In parallel, a full toxicological and pharmacological characterization of aerugine and the exploration of related chemical structures are important future research lines. This requires the development of chemical synthesis strategies for related bacterial secondary metabolites and a further characterization of biosynthetic clusters in different microbial communities in humans and the environment.
Supplementary Material
Acknowledgements
We thank our laboratory colleagues, in particular M. Funke, for contributing important infrastructure, fruitful discussions and some data. We gratefully acknowledge funding by the Deutsche Forschungsgemeinschaft (DFG Emmy Noether program, SFB969, TRR353, KoRS-CB). The work was also facilitated by grants from the BMBF (161L0243B, 016LW0146K), and the Land Baden-Württemberg (NAM_ACCEPT). Funding was also received from the European Union’s Horizon 2020 research and innovation program under grant agreements No. 964537 (RISK-HUNT3R), No. 964518 (ToxFree) and No. 101057014 (PARC). M.A. was supported in part by grants from the National Institute of Environmental Health Sciences (NIEHS) R01ES07331 and R01ES10563.
Abbreviations:
- BSR
Basal slowing response
- DA
Dopamine
- dBcAMP
Dibutyryl cyclic adenosine monophosphate
- DCM
Dichloromethane
- DHAA
Dihydroaeruginoic acid, carboxylic acid of aerugine
- dhIQS
Aldehyde of aerugine
- DFO
Deferoxamine
- DFP
Deferiprone
- DMSO
Dimethyl sulfoxide
- DoDx
Day x of stem cell differentiation
- dx
Day x of differentiation
- EA
Ethyl acetate
- EC50
Half-maximal effective concentration
- FA
Formic acid
- FCCP
Carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone
- FCS
Fetal calf serum
- FGF-2
Fibroblast growth factor 2
- GABA
γ-Aminobutyric acids
- GDNF
Recombinant human glial cell derived neurotrophic factor
- GFP
Green fluorescent protein
- Gpx4
Glutathione peroxidase 4
- iPSC
Induced pluripotent stem cells
- IQS
Aldehyde of aeruginol
- L1
First larval stage
- L4
Larval stage 4
- LDH
Lactate dehydrogenase
- MeOH
Methanol
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- NA
Neurite Area
- NADH
Nicotinamide-adenine-dinucleotide
- NGM
Nematode growth medium
- OCR
Oxygen consumption rate
- OD
Optical density
- PBS
Phosphate buffered saline
- PD
Parkinson’s disease
- PI
Propidium iodide
- PLO
Poly-L-ornithine hydrobromide
- QSAR
Quantitative structure-activity relationship
- qVD
modified valyl-aspartate, QVD-OPh
- R
Resazurin reduction
- RIPK
Receptor-interacting serine/threonine-protein kinase
- ROS
Reactive oxygen species
- SEM
Standard error of the mean
- SK4
3-hydroxy-4(1H)-pyridinone
- SOD
Superoxide dismutase
- TH
Tyrosine hydroxylase
- THF
Tetrahydrofuran
- TMRE
Tetramethylrhodamine ethyl ester
- VCSA
Virtual cell some area
- VCS
Viable cellular structures
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Anna-Katharina Ückert: Formal analysis, Investigation, Writing – original draft, Writing – review & editing. Sina Rütschlin: Formal analysis, Investigation, Writing – original draft. Simon Gutbier: Formal analysis, Investigation. Nathalie Christine Wörz: Formal analysis, Investigation. Mahfuzur R. Miah: Formal analysis, Investigation, Writing – original draft. Airton C. Martins: Formal analysis, Investigation, Writing – original draft. Isa Hauer: Investigation. Anna-Katharina Holzer: Formal analysis, Investigation, Writing – original draft. Birthe Meyburg: Investigation. Ann-Kathrin Mix: Formal analysis, Investigation, Writing – original draft. Christof Hauck: Formal analysis, Investigation. Michael Aschner: Formal analysis, Conceptualization, Funding acquisition. Thomas Böttcher: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. Marcel Leist: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2023.108229.
Data availability
Data will be made available on request.
References
- Abe K, Nomura N, Suzuki S, 2020. Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asanuma M, Miyazaki I, 2021. Glutathione and Related Molecules in Parkinsonism. Int. J. Mol. Sci. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RN, Freeman JL, Guilarte TR, 2016. Neurochemistry of lead and manganese. Metallomics 8, 561–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini F, Hertel J, Sandt E, Thinnes CC, Neuberger-Castillo L, Pavelka L, Betsou F, Krüger R, Thiele I, 2020. Parkinson’s disease-associated alterations of the gut microbiome predict disease-relevant changes in metabolic functions. BMC Biol. 18, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow BK, Cory-Slechta DA, Richfield EK, Thiruchelvam M, 2007. The gestational environment and Parkinson’s disease: evidence for neurodevelopmental origins of a neurodegenerative disorder. Reprod. Toxicol. 23, 457–470. [DOI] [PubMed] [Google Scholar]
- Bellou V, Belbasis L, Tzoulaki I, Evangelou E, Ioannidis JP, 2016. Environmental risk factors and Parkinson’s disease: An umbrella review of meta-analyses. Parkinsonism Relat. Disord. 23, 1–9. [DOI] [PubMed] [Google Scholar]
- Benedetto A, Au C, Avila DS, Milatovic D, Aschner M, 2010. Extracellular dopamine potentiates mn-induced oxidative stress, lifespan reduction, and dopaminergic neurodegeneration in a BLI-3-dependent manner in Caenorhabditis elegans. PLoS Genet. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT, 2000. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat. Neurosci. 3, 1301–1306. [DOI] [PubMed] [Google Scholar]
- Bogdan AR, Miyazawa M, Hashimoto K, Tsuji Y, 2016. Regulators of Iron Homeostasis: New Players in Metabolism, Cell Death, and Disease. Trends Biochem. Sci 41, 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose A, Beal MF, 2016. Mitochondrial dysfunction in Parkinson’s disease. J. Neurochem. 139 (Suppl 1), 216–231. [DOI] [PubMed] [Google Scholar]
- Bose A, Petsko GA, Studer L, 2022. Induced pluripotent stem cells: a tool for modeling Parkinson’s disease. Trends Neurosci. 45, 608–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchaoui H, Mahoney-Sanchez L, Garçon G, Berdeaux O, Alleman LY, Devos D, Duce JA, Devedjian JC, 2023. ACSL4 and the lipoxygenases 15/15B are pivotal for ferroptosis induced by iron and PUFA dyshomeostasis in dopaminergic neurons. Free Radic. Biol. Med. 195, 145–157. [DOI] [PubMed] [Google Scholar]
- Braak H, de Vos RA, Bohl J, Del Tredici K, 2006. Gastric alpha-synuclein immunoreactive inclusions in Meissner’s and Auerbach’s plexuses in cases staged for Parkinson’s disease-related brain pathology. Neurosci. Lett. 396, 67–72. [DOI] [PubMed] [Google Scholar]
- Braungart E, Gerlach M, Riederer P, Baumeister R, Hoener MC, 2004. Caenorhabditis elegans MPP+ model of Parkinson’s disease for high-throughput drug screenings. Neurodegener Dis 1, 175–183. [DOI] [PubMed] [Google Scholar]
- Brenner S, 1974. The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüll M, Spreng AS, Gutbier S, Loser D, Krebs A, Reich M, Kraushaar U, Britschgi M, Patsch C, Leist M, 2020. Incorporation of stem cell-derived astrocytes into neuronal organoids to allow neuro-glial interactions in toxicological studies. ALTEX 37, 409–428. [DOI] [PubMed] [Google Scholar]
- Caito SW, Valentine WM, Aschner M, 2013. Dopaminergic neurotoxicity of S-ethyl N, N-dipropylthiocarbamate (EPTC), molinate, and S-methyl-N, N-diethylthiocarbamate (MeDETC) in Caenorhabditis elegans. J. Neurochem. 127, 837–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell KA, Thies JL, Caldwell GA, 2018. No Country for Old Worms: A Systematic Review of the Application of C. elegans to Investigate a Bacterial Source of Environmental Neurotoxicity in Parkinson’s Disease. Metabolites 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell KA, Tucci ML, Armagost J, Hodges TW, Chen J, Memon SB, Blalock JE, DeLeon SM, Findlay RH, Ruan Q, Webber PJ, Standaert DG, Olson JB, Caldwell GA, 2009. Investigating bacterial sources of toxicity as an environmental contributor to dopaminergic neurodegeneration. PLoS One 4, e7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerasuolo M, Di Meo I, Auriemma MC, Trojsi F, Maiorino MI, Cirillo M, Esposito F, Polito R, Colangelo AM, Paolisso G, Papa M, Rizzo MR, 2023. Iron and Ferroptosis More than a Suspect: Beyond the Most Common Mechanisms of Neurodegeneration for New Therapeutic Approaches to Cognitive Decline and Dementia. Int. J. Mol. Sci. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Huang YJ, Cui JT, Song N, Xie J, 2019. Iron Dysregulation in Parkinson’s Disease: Focused on the Autophagy-Lysosome Pathway. ACS Chem. Nerosci. 10, 863–871. [DOI] [PubMed] [Google Scholar]
- Chia SJ, Tan EK, Chao YX, 2020. Historical Perspective: Models of Parkinson’s Disease. Int. J. Mol. Sci. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S, 2006. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science 313, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello S, Cockburn M, Bronstein J, Zhang X, Ritz B, 2009. Parkinson’s disease and residential exposure to maneb and paraquat from agricultural applications in the central valley of California. Am. J. Epidemiol. 169, 919–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono M, Maricq AV, 2005. Neuronal substrates of complex behaviors in C. elegans. Annu. Rev. Neurosci. 28, 451–501. [DOI] [PubMed] [Google Scholar]
- de Farias CC, Maes M, Bonifacio KL, Matsumoto AK, Bortolasci CC, Nogueira AS, Brinholi FF, Morimoto HK, de Melo LB, Moreira EG, Barbosa DS, 2017. Parkinson’s Disease is Accompanied by Intertwined Alterations in Iron Metabolism and Activated Immune-inflammatory and Oxidative Stress Pathways. CNS Neurol. Disord. Drug Targets 16, 484–491. [DOI] [PubMed] [Google Scholar]
- De Miranda BR, Goldman SM, Miller GW, Greenamyre JT, Dorsey ER, 2022. Preventing Parkinson’s Disease: An Environmental Agenda. J. Parkinsons Dis. 12, 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delp J, Gutbier S, Cerff M, Zasada C, Niedenführ S, Zhao L, Smirnova L, Hartung T, Borlinghaus H, Schreiber F, Bergemann J, Gätgens J, Beyss M, Azzouzi S, Waldmann T, Kempa S, Nöh K, Leist M, 2018. Stage-specific metabolic features of differentiating neurons: Implications for toxicant sensitivity. Toxicol. Appl. Pharmacol. 354, 64–80. [DOI] [PubMed] [Google Scholar]
- Delp J, Funke M, Rudolf F, Cediel A, Bennekou SH, van der Stel W, Carta G, Jennings P, Toma C, Gardner I, van de Water B, Forsby A, Leist M, 2019. Development of a neurotoxicity assay that is tuned to detect mitochondrial toxicants. Arch. Toxicol. 93, 1585–1608. [DOI] [PubMed] [Google Scholar]
- Devos D, Moreau C, Devedjian JC, Kluza J, Petrault M, Laloux C, Jonneaux A, Ryckewaert G, Garçon G, Rouaix N, Duhamel A, Jissendi P, Dujardin K, Auger F, Ravasi L, Hopes L, Grolez G, Firdaus W, Sablonnière B, Strubi-Vuillaume I, Zahr N, Destée A, Corvol JC, Pöltl D, Leist M, Rose C, Defebvre L, Marchetti P, Cabantchik ZI, Bordet R, 2014. Targeting chelatable iron as a therapeutic modality in Parkinson’s disease. Antioxid. Redox Signal. 21, 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionísio PA, Amaral JD, Rodrigues CMP, 2021. Oxidative stress and regulated cell death in Parkinson’s disease. Ageing Res. Rev. 67, 101263. [DOI] [PubMed] [Google Scholar]
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, Sonnenburg JL, 2017. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolde X, Karreman C, Wiechers M, Schildknecht S, Leist M, 2021. Profiling of Human Neural Crest Chemoattractant Activity as a Replacement of Fetal Bovine Serum for In Vitro Chemotaxis Assays. Int. J. Mol. Sci. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Fischbach MA, 2015. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science 349, 1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey ER, Sherer T, Okun MS, Bloem BR, 2018. The Emerging Evidence of the Parkinson Pandemic. J. Parkinsons Dis. 8, S3–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, McDade RL, Schoolnik G, Rothbard JB, Gotschlich EC, 1984. Antigenic analysis of gonococcal pili using monoclonal antibodies. J. Exp. Med. 160, 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova L, Schildknecht S, Adam M, Pape R, Gutbier S, Hanf B, Bürkle A, Leist M, 2015. Prevention of the degeneration of human dopaminergic neurons in an astrocyte co-culture system allowing endogenous drug metabolism. Br. J. Pharmacol. 172, 4119–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erikson KM, Aschner M, 2019. Manganese: Its Role in Disease and Health. Met Ions Life Sci. 19. [DOI] [PubMed] [Google Scholar]
- Firestone JA, Smith-Weller T, Franklin G, Swanson P, Longstreth WT Jr., Checkoway H, 2005. Pesticides and risk of Parkinson disease: a population-based case-control study. Arch. Neurol. 62, 91–95. [DOI] [PubMed] [Google Scholar]
- Friedrich I, Reimann K, Jankuhn S, Kirilina E, Stieler J, Sonntag M, Meijer J, Weiskopf N, Reinert T, Arendt T, Morawski M, 2021. Cell specific quantitative iron mapping on brain slices by immuno-μPIXE in healthy elderly and Parkinson’s disease. Acta Neuropathol. Commun. 9, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrimann S, Farnham A, Staudacher P, Atuhaire A, Manfioletti T, Niwagaba CB, Namirembe S, Mugweri J, Winkler MS, Portengen L, Kromhout H, Mora AM, 2021. Exposure to multiple pesticides and neurobehavioral outcomes among smallholder farmers in Uganda. Environ. Int. 152, 106477. [DOI] [PubMed] [Google Scholar]
- Goldman SM, 2014. Environmental toxins and Parkinson’s disease. Annu. Rev. Pharmacol. Toxicol. 54, 141–164. [DOI] [PubMed] [Google Scholar]
- Gorell JM, Johnson CC, Rybicki BA, Peterson EL, Richardson RJ, 1998. The risk of Parkinson’s disease with exposure to pesticides, farming, well water, and rural living. Neurology 50, 1346–1350. [DOI] [PubMed] [Google Scholar]
- Gutbier S, May P, Berthelot S, Krishna A, Trefzer T, Behbehani M, Efremova L, Delp J, Gstraunthaler G, Waldmann T, Leist M, 2018a. Major changes of cell function and toxicant sensitivity in cultured cells undergoing mild, quasi-natural genetic drift. Arch. Toxicol. 92, 3487–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutbier S, Spreng AS, Delp J, Schildknecht S, Karreman C, Suciu I, Brunner T, Groettrup M, Leist M, 2018b. Prevention of neuronal apoptosis by astrocytes through thiol-mediated stress response modulation and accelerated recovery from proteotoxic stress. Cell Death Differ. 25, 2101–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutbier S, Kyriakou S, Schildknecht S, Ückert AK, Brüll M, Lewis F, Dickens D, Pearson L, Elson JL, Michel S, Hubscher-Bruder V, Brandel J, Tetard D, Leist M, Pienaar IS, 2020. Design and evaluation of bi-functional iron chelators for protection of dopaminergic neurons from toxicants. Arch. Toxicol. 94, 3105–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher JM, Richardson JR, Guillot TS, McCormack AL, Di Monte DA, Jones DP, Pennell KD, Miller GW, 2007. Dieldrin exposure induces oxidative damage in the mouse nigrostriatal dopamine system. Exp. Neurol. 204, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelting L, Klima S, Karreman C, Grinberg M, Meisig J, Henry M, Rotshteyn T, Rahnenführer J, Blüthgen N, Sachinidis A, Waldmann T, Leist M, 2016. Stem Cell-Derived Immature Human Dorsal Root Ganglia Neurons to Identify Peripheral Neurotoxicants. Stem Cells Transl. Med. 5, 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Bargues-Carot A, Riaz Z, Wickham H, Zenitsky G, Jin H, Anantharam V, Kanthasamy A, Kanthasamy AG, 2022. Impact of Environmental Risk Factors on Mitochondrial Dysfunction, Neuroinflammation, Protein Misfolding, and Oxidative Stress in the Etiopathogenesis of Parkinson’s Disease. Int. J. Mol. Sci. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inahashi Y, Zhou S, Bibb MJ, Song L, Al-Bassam MM, Bibb MJ, Challis GL, 2017. Watasemycin biosynthesis in Streptomyces venezuelae: thiazoline C-methylation by a type B radical-SAM methylase homologue. Chem. Sci. 8, 2823–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamdar AA, Hossain MM, Bernstein AI, Miller GW, Richardson JR, Bennett JW, 2013. Fungal-derived semiochemical 1-octen-3-ol disrupts dopamine packaging and causes neurodegeneration. PNAS 110, 19561–19566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Warabi E, Mann GE, 2019. Circadian control of BDNF-mediated Nrf2 activation in astrocytes protects dopaminergic neurons from ferroptosis. Free Radic. Biol. Med. 133, 169–178. [DOI] [PubMed] [Google Scholar]
- Jiang X, Ellabaan MMH, Charusanti P, Munck C, Blin K, Tong Y, Weber T, Sommer MOA, Lee SY, 2017. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 8, 15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Stockwell BR, Conrad M, 2021. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22, 266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthasamy A, Jin H, Anantharam V, Sondarva G, Rangasamy V, Rana A, Kanthasamy A, 2012. Emerging neurotoxic mechanisms in environmental factors-induced neurodegeneration. Neurotoxicology 33, 833–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan AR, Musaev DG, Wuest WM, 2021. Pyochelin Biosynthetic Metabolites Bind Iron and Promote Growth in Pseudomonads Demonstrating Siderophore-like Activity. ACS Infect. Dis. 7, 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J, Ward DM, 2013. The essential nature of iron usage and regulation. Curr. Biol. 23, R642–R646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer JP, 2000. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology 149, 43–50. [DOI] [PubMed] [Google Scholar]
- Klima S, Suciu I, Hoelting L, Gutbier S, Waldmann T, Dietrich DR, Leist M, 2021. Examination of microcystin neurotoxicity using central and peripheral human neurons. ALTEX 38, 73–81. [DOI] [PubMed] [Google Scholar]
- Krug AK, Balmer NV, Matt F, Schönenberger F, Merhof D, Leist M, 2013. Evaluation of a human neurite growth assay as specific screen for developmental neurotoxicants. Arch. Toxicol. 87, 2215–2231. [DOI] [PubMed] [Google Scholar]
- Krug AK, Gutbier S, Zhao L, Pöltl D, Kullmann C, Ivanova V, Förster S, Jagtap S, Meiser J, Leparc G, Schildknecht S, Adam M, Hiller K, Farhan H, Brunner T, Hartung T, Sachinidis A, Leist M, 2014. Transcriptional and metabolic adaptation of human neurons to the mitochondrial toxicant MPP(+). Cell Death Dis. 5, e1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krum BN, Martins AC Jr., Queirós L, Ferrer B, Milne GL, Soares FAA, Fachinetto R, Aschner M, 2021. Haloperidol Interactions with the dop-3 Receptor in Caenorhabditis elegans. Mol. Neurobiol. 58, 304–316. [DOI] [PubMed] [Google Scholar]
- Langston JW, 2017. The MPTP Story. J. Parkinsons. Dis. 7, S11–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta M, Künstle G, Leist M, Wendel A, 2000. Metabolic depletion of ATP by fructose inversely controls CD95- and tumor necrosis factor receptor 1-mediated hepatic apoptosis. J. Exp. Med. 191, 1975–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M, Volbracht C, Kühnle S, Fava E, Ferrando-May E, Nicotera P, 1997. Caspase-mediated apoptosis in neuronal excitotoxicity triggered by nitric oxide. Mol. Med. 3, 750–764. [PMC free article] [PubMed] [Google Scholar]
- Lin KJ, Chen SD, Lin KL, Liou CW, Lan MY, Chuang YC, Wang PW, Lee JJ, Wang FS, Lin HY, Lin TK, 2022. Iron Brain Menace: The Involvement of Ferroptosis in Parkinson Disease. Cells 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Smith MD, Concepcion GP, Haygood MG, Olivera BM, Light A, Schmidt EW, 2017. Modulating the Serotonin Receptor Spectrum of Pulicatin Natural Products. J. Nat. Prod. 80, 2360–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hamamichi S, Lee BD, Yang D, Ray A, Caldwell GA, Caldwell KA, Dawson TM, Smith WW, Dawson VL, 2011. Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson’s disease models. Hum. Mol. Genet. 20, 3933–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotharius J, Falsig J, van Beek J, Payne S, Dringen R, Brundin P, Leist M, 2005. Progressive degeneration of human mesencephalic neuron-derived cells triggered by dopamine-dependent oxidative stress is dependent on the mixed-lineage kinase pathway. J. Neurosci. 25, 6329–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzatto E, Cohen H, Stockheim C, Wieghardt K, Meyerstein D, 1995. Reactions of low valent transition metal complexes with hydrogen peroxide. Are they “Fenton-like” or not? 4. The case of Fe(II)L, L = edta; hedta and tcma. Free Radic. Res. 23, 453–463. [DOI] [PubMed] [Google Scholar]
- Lv H, Shang P, 2018. The significance, trafficking and determination of labile iron in cytosol, mitochondria and lysosomes. Metallomics 10, 899–916. [DOI] [PubMed] [Google Scholar]
- Mahoney-Sánchez L, Bouchaoui H, Ayton S, Devos D, Duce JA, Devedjian JC, 2021. Ferroptosis and its potential role in the physiopathology of Parkinson’s Disease. Prog. Neurobiol. 196, 101890. [DOI] [PubMed] [Google Scholar]
- Martinez BA, Kim H, Ray A, Caldwell GA, Caldwell KA, 2015. A bacterial metabolite induces glutathione-tractable proteostatic damage, proteasomal disturbances, and PINK1-dependent autophagy in C. elegans. Cell Death Dis. 6, e1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack AL, Atienza JG, Johnston LC, Andersen JK, Vu S, Di Monte DA, 2005. Role of oxidative stress in paraquat-induced dopaminergic cell degeneration. J. Neurochem. 93, 1030–1037. [DOI] [PubMed] [Google Scholar]
- Miller DA, Luo L, Hillson N, Keating TA, Walsh CT, 2002. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem. Biol. 9, 333–344. [DOI] [PubMed] [Google Scholar]
- Miller CJ, Rose AL, Waite TD, 2016. Importance of Iron Complexation for Fenton-Mediated Hydroxyl Radical Production at Circumneutral pH. Front. Mar. Sci. 3. [Google Scholar]
- Moretto A, Colosio C, 2011. Biochemical and toxicological evidence of neurological effects of pesticides: the example of Parkinson’s disease. Neurotoxicology 32, 383–391. [DOI] [PubMed] [Google Scholar]
- Morris G, Berk M, Carvalho AF, Maes M, Walker AJ, Puri BK, 2018. Why should neuroscientists worry about iron? The emerging role of ferroptosis in the pathophysiology of neuroprogressive diseases. Behav. Brain Res. 341, 154–175. [DOI] [PubMed] [Google Scholar]
- Nicolas S, Nolan YM, 2022. Intermittent fasting to slow down Alzheimer’s disease. Nat Aging. 1. [DOI] [PubMed] [Google Scholar]
- Noel S, Guillon L, Schalk IJ, Mislin GLA, 2011. Synthesis of Fluorescent Probes Based on the Pyochelin Siderophore Scaffold. Org. Lett. 13. [DOI] [PubMed] [Google Scholar]
- Obeso JA, Stamelou M, Goetz CG, Poewe W, Lang AE, Weintraub D, Burn D, Halliday GM, Bezard E, Przedborski S, Lehericy S, Brooks DJ, Rothwell JC, Hallett M, DeLong MR, Marras C, Tanner CM, Ross GW, Langston JW, Klein C, Bonifati V, Jankovic J, Lozano AM, Deuschl G, Bergman H, Tolosa E, Rodriguez-Violante M, Fahn S, Postuma RB, Berg D, Marek K, Standaert DG, Surmeier DJ, Olanow CW, Kordower JH, Calabresi P, Schapira AHV, Stoessl AJ, 2017. Past, present, and future of Parkinson’s disease: A special essay on the 200th Anniversary of the Shaking Palsy. Mov. Disord. 32, 1264–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ockleford C, Adriaanse P, Berny P, Brock T, Duquesne S, Grilli S, Hernandez-Jerez AF, Bennekou SH, Klein M, Kuhl T, Laskowski R, Machera K, Pelkonen O, Pieper S, Smith R, Stemmer M, Sundh I, Teodorovic I, Tiktak A, Topping CJ, Wolterink G, Angeli K, Fritsche E, Leist M, Mantovani A, Menendez P, Price A, Viviani B, Chiusolo A, Ruffo F, Terron A, 2017. Investigation into experimental toxicological properties of plant protection products having a potential link to Parkinson’s disease and childhood leukaemia. EFSA J. 15, e04691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrenius S, Gogvadze V, Zhivotovsky B, 2007. Mitochondrial oxidative stress: implications for cell death. Annu. Rev. Pharmacol. Toxicol. 47, 143–183. [DOI] [PubMed] [Google Scholar]
- Panicker N, Kam TI, Wang H, Neifert S, Chou SC, Kumar M, Brahmachari S, Jhaldiyal A, Hinkle JT, Akkentli F, Mao X, Xu E, Karuppagounder SS, Hsu ET, Kang SU, Pletnikova O, Troncoso J, Dawson VL, Dawson TM, 2022. Neuronal NLRP3 is a parkin substrate that drives neurodegeneration in Parkinson’s disease. Neuron 110, 2422–2437.e2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC, Ryu MS, 2014. Special delivery: distributing iron in the cytosol of mammalian cells. Front. Pharmacol. 5, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshi A, Khuder SA, Schaub EA, Priyadarshi SS, 2001. Environmental risk factors and Parkinson’s disease: a metaanalysis. Environ. Res. 86, 122–127. [DOI] [PubMed] [Google Scholar]
- Program TGPsG, 2021. GP2: The Global Parkinson’s Genetics Program. Mov. Disord. 36, 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Martinez BA, Berkowitz LA, Caldwell GA, Caldwell KA, 2014. Mitochondrial dysfunction, oxidative stress, and neurodegeneration elicited by a bacterial metabolite in a C. elegans Parkinson’s model. Cell Death Dis. 5, e984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert A, Morawski M, Seeger J, Arendt T, Reinert T, 2019. Iron concentrations in neurons and glial cells with estimates on ferritin concentrations. BMC Neurosci. 20, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer P, Monoranu C, Strobel S, Iordache T, Sian-Hülsmann J, 2021. Iron as the concert master in the pathogenic orchestra playing in sporadic Parkinson’s disease. J. Neural Transm. (Vienna) 128, 1577–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK, 2016. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 167, 1469–1480.e1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, Horvitz HR, 2000. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619–631. [DOI] [PubMed] [Google Scholar]
- Scaturro P, Pichlmair A, 2019. Oxeiptosis: a discreet way to respond to radicals. Curr. Opin. Immunol. 56, 37–43. [DOI] [PubMed] [Google Scholar]
- Schapira AH, 2012. Targeting mitochondria for neuroprotection in Parkinson’s disease. Antioxid. Redox Signal. 16, 965–973. [DOI] [PubMed] [Google Scholar]
- Scheperjans F, Aho V, Pereira PA, Koskinen K, Paulin L, Pekkonen E, Haapaniemi E, Kaakkola S, Eerola-Rautio J, Pohja M, Kinnunen E, Murros K, Auvinen P, 2015. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 30, 350–358. [DOI] [PubMed] [Google Scholar]
- Schildknecht S, Pöltl D, Nagel DM, Matt F, Scholz D, Lotharius J, Schmieg N, Salvo-Vargas A, Leist M, 2009. Requirement of a dopaminergic neuronal phenotype for toxicity of low concentrations of 1-methyl-4-phenylpyridinium to human cells. Toxicol. Appl. Pharmacol. 241, 23–35. [DOI] [PubMed] [Google Scholar]
- Schildknecht S, Karreman C, Pöltl D, Efrémova L, Kullmann C, Gutbier S, Krug A, Scholz D, Gerding HR, Leist M, 2013. Generation of genetically-modified human differentiated cells for toxicological tests and the study of neurodegenerative diseases. ALTEX 30, 427–444. [DOI] [PubMed] [Google Scholar]
- Schildknecht S, Di Monte DA, Pape R, Tieu K, Leist M, 2017. Tipping Points and Endogenous Determinants of Nigrostriatal Degeneration by MPTP. Trends Pharmacol. Sci. 38, 541–555. [DOI] [PubMed] [Google Scholar]
- Scholz D, Pöltl D, Genewsky A, Weng M, Waldmann T, Schildknecht S, Leist M, 2011. Rapid, complete and large-scale generation of post-mitotic neurons from the human LUHMES cell line. J. Neurochem. 119, 957–971. [DOI] [PubMed] [Google Scholar]
- Shang F, Muimhneachain EO, Jerry Reen F, Buzid A, O’Gara F, Luong JHT, Glennon JD, McGlacken GP, 2014. One step preparation and electrochemical analysis of IQS, a cell-cell communication signal in the nosocomial pathogen Pseudomonas aeruginosa. Bioorg. Med. Chem. Lett. 24, 4703–4707. [DOI] [PubMed] [Google Scholar]
- Shannon KM, Keshavarzian A, Dodiya HB, Jakate S, Kordower JH, 2012. Is alpha-synuclein in the colon a biomarker for premotor Parkinson’s disease? Evidence from 3 cases. Mov. Disord. 27, 716–719. [DOI] [PubMed] [Google Scholar]
- Sian-Hulsmann J, Riederer P, 2021. The Nigral Coup in Parkinson’s Disease by α-Synuclein and Its Associated Rebels. Cells 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler NV, Krug AK, Matt F, Leist M, 2011. Assessment of chemical-induced impairment of human neurite outgrowth by multiparametric live cell imaging in high-density cultures. Toxicol. Sci. 121, 73–87. [DOI] [PubMed] [Google Scholar]
- Suciu I, Delp J, Gutbier S, Ückert AK, Spreng AS, Eberhard P, Karreman C, Schreiber F, Madjar K, Rahnenführer J, Celardo I, Amelio I, Leist M, 2023. Dynamic Metabolic and Transcriptional Responses of Proteasome-Inhibited Neurons. Antioxidants (Basel) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J, 1998. The Nematode Caenorhabditis elegans. In: Wood WBs (ed.), vol. 17: Cold Spring Harbor Monographs. [Google Scholar]
- Sulston J, Dew M, Brenner S, 1975. Dopaminergic neurons in the nematode Caenorhabditis elegans. J Comp Neurol 163, 215–226. [DOI] [PubMed] [Google Scholar]
- Sun F, Anantharam V, Zhang D, Latchoumycandane C, Kanthasamy A, Kanthasamy AG, 2006. Proteasome inhibitor MG-132 induces dopaminergic degeneration in cell culture and animal models. Neurotoxicology 27, 807–815. [DOI] [PubMed] [Google Scholar]
- Sun J, Lin XM, Lu DH, Wang M, Li K, Li SR, Li ZQ, Zhu CJ, Zhang ZM, Yan CY, Pan MH, Gong HB, Feng JC, Cao YF, Huang F, Sun WY, Kurihara H, Li YF, Duan WJ, Jiao GL, Zhang L, He RR, 2023. Midbrain dopamine oxidation links ubiquitination of glutathione peroxidase 4 to ferroptosis of dopaminergic neurons. J. Clin. Invest. 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann JR, Spitzer SO, Diaz Heijtz R, 2020. Developmental Signatures of Microbiota-Derived Metabolites in the Mouse Brain. Metabolites 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamosvári D, Schuhmacher T, Hauck CR, Böttcher T, 2019. A thiochromenone antibiotic derived from the Pseudomonas quinolone signal selectively targets the Gram-negative pathogen Moraxella catarrhalis. Chem. Sci. 10, 6624–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarka A, Lőrincz T, Hajdinák P, 2022. Friend or Foe: The Relativity of (Anti) oxidative Agents and Pathways. Int. J. Mol. Sci. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Berghe TV, Vandenabeele P, Kroemer G, 2019. The molecular machinery of regulated cell death. Cell Res. 29, 347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner CM, 1992. Occupational and environmental causes of parkinsonism. Occup. Med. 7, 503–513. [PubMed] [Google Scholar]
- Terman A, Kurz T, 2013. Lysosomal iron, iron chelation, and cell death. Antioxid. Redox Signal. 18, 888–898. [DOI] [PubMed] [Google Scholar]
- Terron A, Bal-Price A, Paini A, Monnet-Tschudi F, Bennekou SH, Leist M, Schildknecht S, 2018. An adverse outcome pathway for parkinsonian motor deficits associated with mitochondrial complex I inhibition. Arch. Toxicol. 92, 41–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidjani AR, Lorenzi JN, Toussaint M, van Dijk E, Naquin D, Lespinet O, Bontempsm C, Leblond P, 2019. Massive Gene Flux Drives Genome Diversity between Sympatric Streptomyces Conspecifics. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci ML, Harrington AJ, Caldwell GA, Caldwell KA, 2011. Modeling dopamine neuron degeneration in Caenorhabditis elegans. Methods Mol. Biol. 793, 129–148. [DOI] [PubMed] [Google Scholar]
- Uhl GR, 2019. Dopamine compartmentalization, selective dopaminergic vulnerabilities in Parkinson’s disease and therapeutic opportunities. Ann. Clin. Transl. Neurol. 6, 406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanduyn N, Settivari R, Wong G, Nass R, 2010. SKN-1/Nrf2 inhibits dopamine neuron degeneration in a Caenorhabditis elegans model of methylmercury toxicity. Toxicol. Sci. 118, 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergo S, Johansen JL, Leist M, Lotharius J, 2007. Vesicular monoamine transporter 2 regulates the sensitivity of rat dopaminergic neurons to disturbed cytosolic dopamine levels. Brain Res. 1185, 18–32. [DOI] [PubMed] [Google Scholar]
- Vior NM, Lacret R, Chandra G, Dorai-Raj S, Trick M, Truman AW, 2018. Discovery and Biosynthesis of the Antibiotic Bicyclomycin in Distantly Related Bacterial Classes. Appl. Environ. Microbiol. 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volbracht C, Leist M, Nicotera P, 1999. ATP controls neuronal apoptosis triggered by microtubule breakdown or potassium deprivation. Mol. Med. 5, 477–489. [PMC free article] [PubMed] [Google Scholar]
- Volbracht C, Leist M, Kolb SA, Nicotera P, 2001. Apoptosis in caspase-inhibited neurons. Mol. Med. 7, 36–48. [PMC free article] [PubMed] [Google Scholar]
- Whitnall M, Richardson DR, 2006. Iron: a new target for pharmacological intervention in neurodegenerative diseases. Semin. Pediatr. Neurol. 13, 186–197. [DOI] [PubMed] [Google Scholar]
- Wu Y, Torabi SF, Lake RJ, Hong S, Yu Z, Wu P, Yang Z, Nelson K, Guo W, Pawel GT, Van Stappen J, Shao X, Mirica LM, Lu Y, 2023. Simultaneous Fe(2+)/Fe(3+) imaging shows Fe(3+) over Fe(2+) enrichment in Alzheimer’s disease mouse brain. Sci. Adv. 9, eade7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Li X, Li C, Zhu W, Jankovic J, Le W, 2010. Proteasome inhibition modeling nigral neuron degeneration in Parkinson’s disease. J. Neurochem. 115, 188–199. [DOI] [PubMed] [Google Scholar]
- Yuan X, Tian Y, Liu C, Zhang Z, 2022. Environmental factors in Parkinson’s disease: New insights into the molecular mechanisms. Toxicol. Lett. 356, 1–10. [DOI] [PubMed] [Google Scholar]
- Zamri A, Abdallah MA, 2000. An Improved Stereocontrolled Synthesis of Pyochelin, Siderophore of Pseudomonas aeruginosa and Burkholderia cepacia. Tetrahedron 56, 249–256. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.


