Abstract
The bacterial spirochete Borrelia burgdorferi, the causative agent of Lyme Disease, can disseminate and colonize various tissues and organs, orchestrating severe clinical symptoms including arthritis, carditis, and neuroborreliosis. Previous research has demonstrated that breast cancer tissues could provide an ideal habitat for diverse populations of bacteria, including B. burgdorferi, which is associated with a poor prognosis. Recently, we demonstrated that infection with B. burgdorferi enhances the invasion and migration of triple-negative MDA-MB-231 cells which represent a type of breast tumor with more aggressive cancer traits. In this study, we hypothesized that infection by B. burgdorferi affects the expression of cancer-associated genes to effectuate breast cancer phenotypes. We applied the high-throughput technique of RNA-sequencing on B. burgdorferi-infected MDA-MB-231 breast cancer and normal-like MCF10A cells to determine the most differentially expressed genes (DEG) upon infection. Overall, 142 DEGs were identified between uninfected and infected samples in MDA-MB-231 while 95 DEGs were found in MCF10A cells. A major trend of the upregulation of C-X-C and C-C motif chemokine family members as well as genes and pathways was associated with infection, inflammation, and cancer. These genes could serve as potential biomarkers for pathogen-related tumorigenesis and cancer progression which could lead to new therapeutic opportunities.
Keywords: Lyme disease, breast cancer, DEGs, TNBC, KEGG, REACTOME
Introduction
Breast cancer is the most diagnosed cancer among women worldwide with a 1 in 8 chance of a woman being diagnosed with breast cancer in their lifetime in the United States [1]. It accounts for the leading cause of death due to cancer among women with a 1 in 39 chance of fatality, second only to lung cancer [1]. Among the various sub-types of breast cancer, triple-negative breast cancer (TNBC), which tests negative for estrogen, progesterone, and human epidermal growth factor receptor, presents with a shorter survival and higher mortality rate [2]. TNBC is highly invasive with a recurrence rate of 25% and is not sensitive to endocrine therapy resulting in limited treatment options [2].
Infectious agents stand third among risk factors for cancer development. As per the American Cancer Society, 15–20% of cancers are caused by infectious agents [3]. For example, Mycobacterium tuberculosis has been associated with lung cancer, Helicobacter pylori infection results in increased migration and invasion rates in gastric cancer, and Campylobacter jejuni is correlated with small intestine lymphomas [4].
Recent findings showed that intracellular microbiota promote the survival of circulating breast cancer cells but not primary tumor growth [5]. However, another report shows the colonization of breast cells by the bacterium Fusobacterium nucleatum is associated with accelerated tumor growth [6]. Moreover, a study published in 2018 highlighted bacterial signatures associated with breast cancer, of which one of the spirochetal bacteria, Borrelia burgdorferi, correlated with a poorer prognosis in breast cancer [7]. This data supports our recent publication, where we reported that B. burgdorferi can invade normal and tumorigenic mammary epithelial cells and affect the in vitro invasion and matrix remodeling potential as well as miRNA expression profile of cancer cells [8, 9]. Interestingly, B. burgdorferi was also found to be associated previously with primary cutaneous B cell, non-Hodgkin and Mucosa-Associated lymphoid tissue (MALT) lymphomas [10–12].
B. burgdorferi is the causal organism of Lyme disease, which is a multi-systemic disease with several clinical manifestations like musculoskeletal inflammation and neurological symptoms [10, 11]. The pathogen enters the body via the bite of the Ixodes tick and then disseminates to and colonizes various tissues and organs [10, 11]. Previous research has reported that B. burgdorferi can evade the host immune response using various strategies like altering its membrane proteins, actively suppressing the host immune system, and existing in pleomorphic forms [13–22]. Additionally, B. burgdorferi can avoid detection by the immune system by internalizing into the host cells without affecting cell viability [13]. This ability, in conjunction with cancer cells' indefinite proliferative capacity, could serve as a powerful mechanism to ensure the bacteria's survival within the host organism. Further, our previous study demonstrated that infection of TNBC cells by the spirochete can make the cancer cells potentially more aggressive [8]. Hence, it is imperative to understand the physiological effect of B. burgdorferi on TNBC cells and identify the molecular mechanisms that B. burgdorferi recruits to effectuate these more aggressive cancer traits.
RNA-sequencing is a high-throughput method that allows for the quantitative analysis of gene expression, and it is ideal for measuring differential gene expression in bacteria-infected cells [23]. It was successfully used in the comprehensive mapping of genes with altered expression in response to Borrelia bavariensis infection in human brain microvascular cells [24]. Consistently, many of the differentially expressed genes (DEGs) were found in pathways that are known to be dysregulated in cancer, including breast cancer [23]. Therefore, we applied RNA-sequencing technology followed by gene ontology analysis of normal and triple-negative breast cancer cell lines with the goal to propound the molecular mechanisms that may orchestrate the effect of B. burgdorferi on breast cancer cells.
Materials and methods
Culture techniques and infection with B. burgdorferi
The B31 strain of B. burgdorferi (ATCC 35210, Manassas, VA) was cultured in Barbour-Stoner-Kelly-H (BSK-H) medium (Sigma Aldrich, St Louis, MO), supplemented with 6% rabbit serum (GeminiBio, West Sacramento, CA) in sterile 15 mL glass tubes at 33 °C and 3% carbon dioxide. Low passage numbers (<P6) were used for all experiments.
Normal breast epithelial cells (MCF10A) and triple-negative breast cancer cells (TNBC, MDA-MB-231) were cultured using standard cell culture techniques at 37 °C and 5% carbon dioxide. MDA-MB-231 was cultured in Dulbecco's Modified Eagle Medium (DMEM, Sigma Aldrich, Louis, MO) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin-glutamine (PSG, ThermoFisher Scientific, Waltham, MA), along with 20 μg mL−1 gentamycin. MCF10A was cultivated in Dulbecco's Modified Eagle Medium-F12 (DMEM-F12), (Sigma Aldrich, St Louis, MO) supplemented with 5% Horse Serum (GibcoTM, ThermoFisher, Waltham, MA), 10 μg mL−1 bovine insulin (Sigma Aldrich, St Louis, MO), 100 ng mL−1 cholera toxin (Sigma Aldrich, St Louis, MO), 20 ng mL−1 epidermal growth factor (EGF, ThermoFisher, Waltham, MA), 0.5 mg mL−1 hydrocortisone (Sigma Aldrich, St Louis, MO), and 1% penicillin-streptomycin (PS, GibcoTM, ThermoFisher, Waltham, MA). The cells were serum-deprived for 10 h in serum-free basal growth media supplemented with 1% of the required antibiotic (PSG for MDA-MB-231 and PS for MCF10A). Infection with B. burgdorferi at a multiplicity of infection (MOI) of 60 for 48 h was performed in co-culture media composed of 1/3rd serum- and antibiotic-free growth media and 2/3rd of BSK-H supplemented with 6% rabbit serum. Uninfected cells grown in coculture media were used as a negative control.
Immunofluorescence and confocal microscopy
MCF10A and MDA-MB-231 cells (4 × 104/well) were grown on sterile 4-well chamber slides (ThermoFisher Scientific) for 24 h. The cells were then washed with 1X phosphate buffer saline (PBS, PH 7.4) after 24 h and were infected with B. burgdorferi B31, at a multiplicity of infection (MOI) of 60, resuspended in 1 mL of coculture media. Uninfected cells grown in coculture media were used as a negative control. The uninfected and infected normal and breast cancer epithelial cells were incubated for 48 h at 37°C, 5% CO2. After infection, the co-culture media was removed, and cells were washed twice with 1X PBS pH 7.4. The infected cells were treated with 10 μg mL−1 of Cell tracker CM-Dil dye (ThermoFisher Scientific, C7001) diluted in 1X PBS pH 7.4 to stain the epithelial cells. The cells were incubated at 37°C for 5 min and then transferred to 4°C for 15 min. The dye was then aspirated, and the cells were washed once with 1X PBS (pH 7.4), then fixed using cold methanol for 5 min at −20°C. The methanol was aspirated, and cells were washed once with 1X PBS pH 7.4. The cells were then treated with a 1:200 dilution of a FITC labeled polyclonal rabbit anti-B. burgdorferi antibody (PA-1-73005, Thermo Scientific) to stain B. burgdorferi and incubated at 37°C for 30 min. The cells were then washed 3x with 1X PBS pH 7.4 and a 1:1000 dilution of 4′,6-diamidino-2-phenylindole (DAPI, ThermoFisher Scientific) in 1X PBS pH 7.4 was added to the cells to stain the nucleus of the cells. The cells were incubated for 5 min at room temperature (RT), washed once with 1X PBS pH 7.4, and mounted with VECTASHIELD Antifade Mounting Medium (VECTOR Laboratories, Burlingame, CA). Images were generated at a magnification of 630X using Leica SP8 confocal microscope along with Leica Application Suite X software for image processing (Yale University, West Campus Microscopy Facility, West Haven CT).
RNA extraction, cDNA library prep and RNA-sequencing
Infected cells along with co-culture controls were trypsinized and pelleted by centrifugation at 2,200 rpm for 10 min at room temperature (RT). The medium was aspirated, and the pellet was resuspended in phosphate-buffered saline (PBS, Sigma Aldrich, St Louis, MO) followed by centrifugation at 2,200 rpm for 10 min at RT. RNA was extracted using the RNeasy mini kit (Qiagen, Hilden, Germany) following the manufacturer's instruction and quantitated using the Nanodrop Lite spectrophotometer (ThermoFisher Scientific, Waltham, MA). The total RNA extracted from three independent sets of infected and uninfected samples for each cell line was sent for sequencing at the Yale Center for Genome Analysis (Yale West Campus, Orange, CT).
The integrity of the RNA was determined by running an Agilent Bioanalyzer gel (Agilent, Santa Clara, CA), to measure the ratios of the ribosomal peaks and to assign an RNA integrity/RNA quality number (RIN/RQN) to the samples. Total RNA was subjected to polyA selection using Roche Kapa mRNA Hyper Prep Kit (Roche KAPA Biosystems, Wilmington, MA). Samples with a yield of ≥0.5 ng μL−1 and a size distribution of 150–300 bp were used for sequencing. Samples were sequenced on Illumina NovaSeq600 according to Illumina protocols at the Yale Center for Genome Analysis (Yale West Campus, Orange, CT).
Analysis of differentially expressed genes and gene ontology analysis
The raw sequencing reads were trimmed and filtered to remove low-quality bases using Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic). The trimmed reads were aligned against the human transcriptome using the STAR aligner program (https://github.com/alexdobin/STAR). Read counts for each gene were determined using HTSeq. (https://htseq.readthedocs.io/). Differential expression analysis and visualization was performed using the R package DESeq-2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html).
Two cutoff criteria were used to select differentially expressed genes, a P-value of <0.05 and the log2 FC (fold change) (≥3-fold for MDA-MB-231 and ≥ 2-fold for MCF10A). The differentially expressed genes were run through the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) to determine cellular processes in which the (DEGs) were enriched. Within DAVID, two different databases – Reactome and KEGG, were used for the ontological analysis. For this study, pathways that highlighted ≥10 genes (P-value <0.05) for MDA-MB-231 and those ≥5 genes (P-value <0.05) for MCF10A were further evaluated.
qRT-PCR validation
cDNA was made from 500 ng of RNA extracted from 1 × 106 of MDA-MB-231 and MCF-10A cell lines that were either uninfected or infected or infected with B. burgdorferi, utilizing the Verso cDNA Synthesis Kit (ThermoFisher Scientific, Waltham, MA), using a T100 Thermal Cycler (Bio-Rad, Hercules, CA) following the manufacturer's instruction. cDNA (1000 ng) was then subjected to quantitative real-time PCR using 2x iTaqTM Universal SYBR® Green Supermix and 400 nM of gene-specific primers. Primer sequences and PCR conditions were obtained from PrimerBank – MGH-PGA Harvard University webserver (https://pga.mgh.harvard.edu/primerbank/). The presence of B. burgdorferi was confirmed in infected cells by a previously published 16S rDNA-specific protocol [25]. The quantitation of the obtained data was normalized against GAPDH (housekeeping gene) and the expression fold-change was calculated using the Livak method [26]. All qRT-PCR experiments were performed at least 3 times independently and each at least in four replicates (N = 12). Statistical analysis was performed by the two-tailed Student's T-test of mean comparison using Microsoft Office Excel program (Redmond, WA). Data was normalized to uninfected cells control condition and presented as the mean ± standard error of the mean (SEM).
Ethics statement
Not applicable since only commercially available cell lines were used in the study.
Results
RNA-seq on B. burgdorferi-infected triple-negative breast cancer and normal-like mammary epithelial cells
MDA-MB-231 and MCF10A cells were plated in biological triplicates for 6 h after which they were serum starved for 10 h and then infected with B. burgdorferi at a multiplicity of infection (MOI) of 60 alongside uninfected controls. To verify the successful infection in both cell lines, a B. burgdorferi-specific 16S rRNA qRT-PCR was performed on uninfected and 24, 48, and 72 h B. burgdorferi infected cells from MDA-MB-231 and MCF10A cells. The results showed that as early as 24h, B. burgdorferi 16S rRNA was detected in infected cells of both cell lines but not in uninfected cells. To be able to quantify the data at different time points, the obtained results were normalized using the GAPDH housekeeping gene and compared to 24 h B. burgdorferi infected cells to assess fold change in expression of 16S rRNA for 48h and 72h. After quantification and statistical analysis of the results from three independent experiments, it was concluded that B. burgdorferi efficiently invaded the cells and had active transcription for at least 3 days.
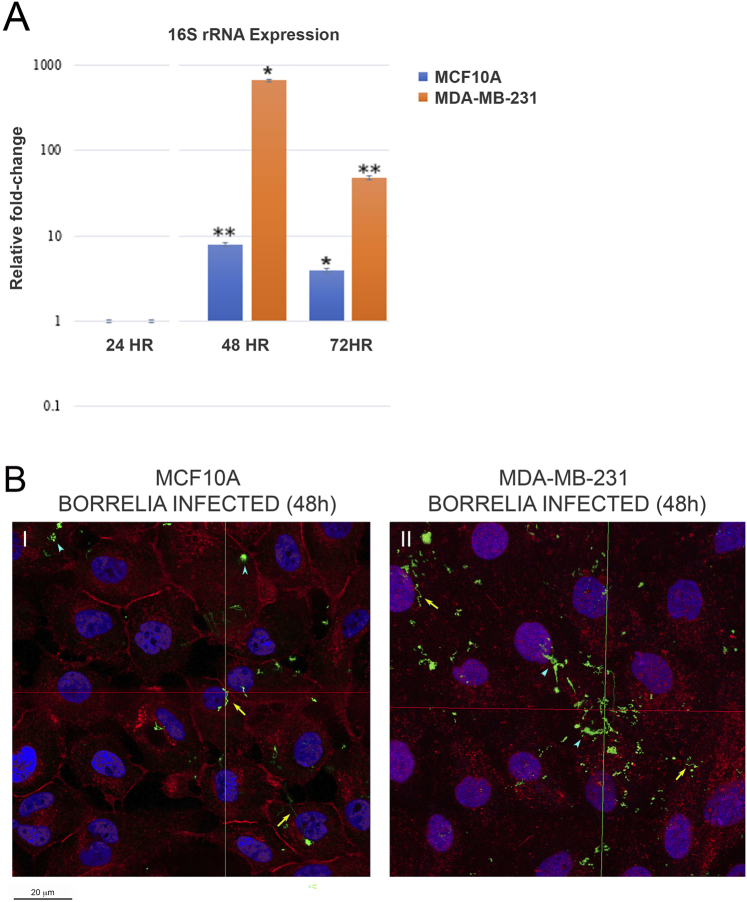
As seen in Fig. 1A, both MCF10A and MDA-MB-231 cells infected with B. burgdorferi showed the highest 16S rRNA level at 48h time point. In MCF10A cells, 16S rRNA expression increase was approx. 8-fold for 48h and 4-fold for 72h post-B. burgdorferi infection, respectively (Fig. 1). Similarly, MDA-MB-231 cells showed the highest increase of ∼650-fold at 48h followed by approximately 50-fold increase at 72h post-infection (Fig. 1A). Based on these experiments, the 48h time point for B. burgdorferi infection was chosen for all subsequent experiments. Figure 1B shows a representative immunohistochemical image of the B. burgdorferi infected MCF10A and MDA-MB-231 at 48h timepoint. Invading spirochetes (yellow arrows), as well as some small B. burgdorferi aggregates (blue arrowheads) visualized by confocal microscopy as described in Methods. In summary, successful infection of MCF10A and MDA-MB-231 cells by B. burgdorferi was evidenced by both qRT-PCR and immunohistochemistry (IHC) methods.
Fig. 1.
Evidence of successful infection of MCF10A and MDA-MB-231 cells by B. burgdorferi using qRT-PCR and immunohistochemistry (IHC) methods. (A) Quantification and statistical analyses of three independent qRT-PCR experiments for the expression levels of 16S rRNA in 48 and 72h B. burgdorferi infected MCF10A and MDA-MB-231 cells. The results were normalized using the GAPDH housekeeping gene and compared to 24h B. burgdorferi-infected cells. The P-value of the data <0.05 (*) and <0.01 (**) is considered significant. (B) Representative confocal microscopy images of (I) MCF-10A and (II) MDA-MB-231 mammalian epithelial cells infected with B. burgdorferi for 48 h h and visualized by IHC and confocal microscopy methods. CM-Dil membrane stain was used to label the cells (red), DAPI was used to stain the nucleus (blue), and polyclonal anti-B. burgdorferi was used to visualize B. burgdorferi (green), as described in the Material and Methods. The representative images were taken at 630X with a scale bar of 20 μm. The yellow arrows represent invading spirochetes, and the blue arrowhead shows some small B. burgdorferi aggregates inside the cells
The RNA extracted from uninfected and B. burgdorferi-infected cells were independently evaluated by the Yale Center for Genome Analysis (YCGA) and found to be of excellent quality as assessed by an RNA quality number or RNA integrity number (RQN/RIN) value of 10 (RQN>7) and an average 28S:18S ratio of 2:1. A principal component analysis (PCA) was conducted using the R package DESeq2. The first principle component (PC1) showed that there was 78% variance between the transcriptomic profiles of control and B. burgdorferi-infected samples for MDA-MB-231 cells and 71% for MCF10A cells suggesting that there are significant differences in gene expression in a large number of genes. The obtained results showed that 142 DEGs between uninfected and infected samples were identified in MDA-MB-231 cells (P < 0.01) and 95 DEGs in MCF10A cells (P < 0.01) (Supplemental Tables S1A and S1B respectively). Within these DEGs, a subset of 40 genes for MDA-MB-231 cells were differentially expressed by over 2-fold, out of which 12 genes were overexpressed by over 3-fold (P < 0.01, Supplemental Table S1A and Table S2). For MCF10A cells, 11 genes were differentially expressed over 2-fold (P < 0.01, Supplemental Table S1B and Table S2).
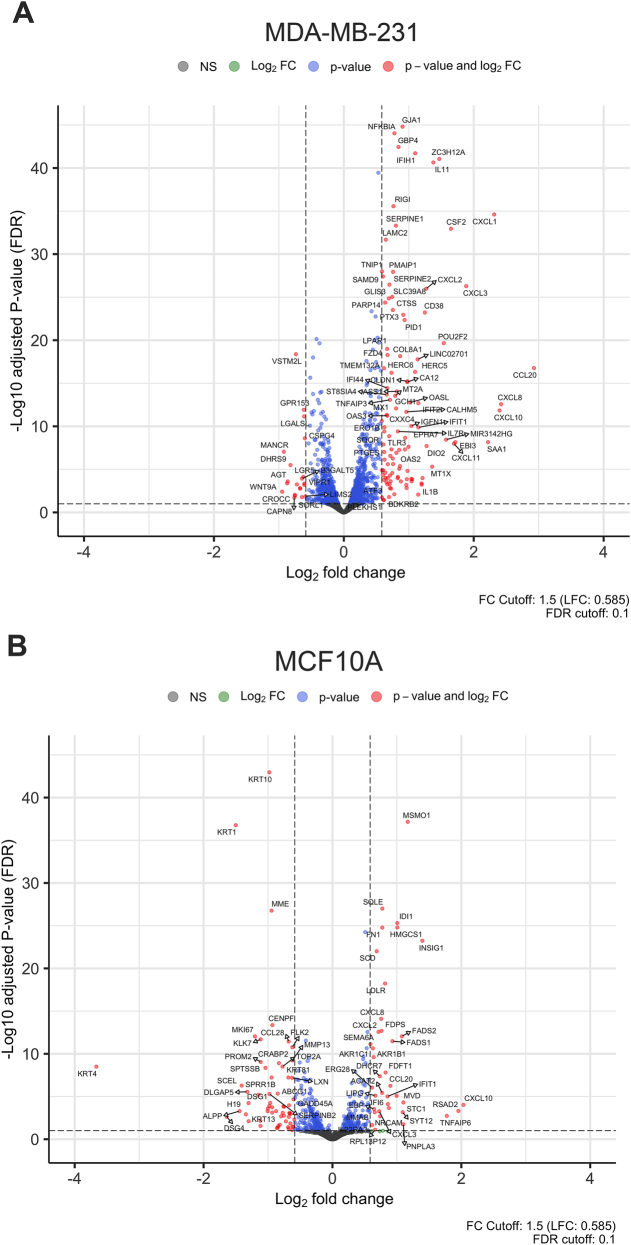
The volcano plot shows the statistically significant primary transcripts identified in the comparisons of uninfected vs. infected MDA-MB-231 and MCF10A cells respectively (Fig. 2, Panels A and B). DEGs with a fold change (FC) ≥ 3 for MDA-MB-231 and FC ≥ 2 for MCF10A cells were further analyzed and validated using qRT-PCR. The general trend for both cell lines was that most DEGs were upregulated (Supplemental Tables S1A and S1B, respectively). As shown in the volcano plots (Fig. 2), with an FC cutoff of 1.5, it was observed that 83% of DEGs were upregulated in MDA-MB-231, whereas 58% of DEGs were upregulated in MCF10A.
Fig. 2.
Dispersion patterns of differentially expressed genes (DEGs): (A) Volcano plot of DEGs in MDA-MB 231 cell line. (B) Volcano plot of DEGs in MCF10A cell line. The x-axis represents the log2 fold change and the y-axis represents the negative log10 of the adjusted P-value. The cutoff criteria were the adjusted P-value ≤0.10 and absolute log2 fold change greater than +1.5 and less than −1.5. Points in red represent genes that exceed both cutoff criteria
qRT-PCR validation for DEGs
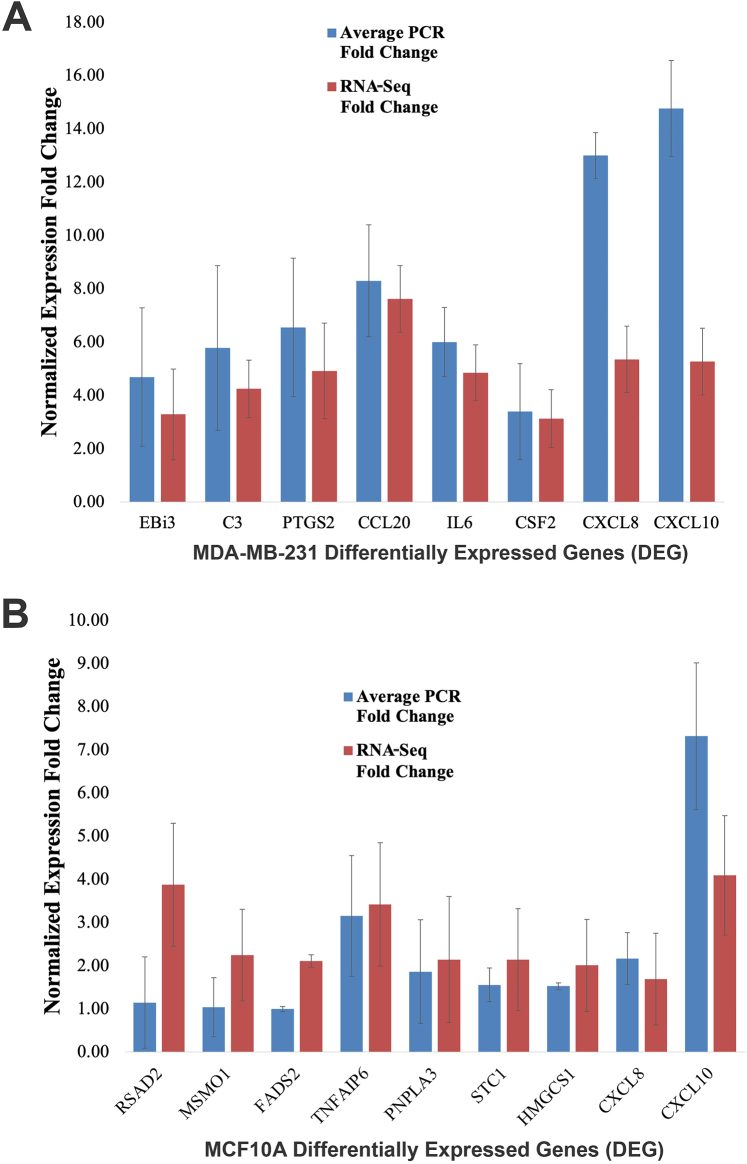
To validate the RNA-seq data, 8 out of the 9 DEGs expressed with FC ≥ 3 in MDA-MB-231 and 9 out of the 11 DEGs that expressed FC ≥ 2 in MCF10A upon B. burgdorferi-infection were analyzed using qRT-PCR analysis. All experiments were conducted in biological and technical triplicates for each gene and compared alongside uninfected controls to determine the expression fold-change of the DEGs. The qRT-PCR analysis demonstrated that all of the DEGs from infected MDA-MB-231 and MCF10A cells had significant upregulation complying with the RNA-seq results (P-values <0.01, Fig. 3A and B). Interestingly CXCL8 and CXCL10 from infected MDA-MB-231 and CXCL10 from infected MCF10A cells were more significantly upregulated in our qRT-PCR than in our RNA-seq experiments. Results obtained in a previous study demonstrated a similar very significant upregulation of CXCL8 and CXCL10 in B. burgdorferi-infected MDA-MB-231 cells using qRT-PCR analysis [23].
Fig. 3.
Confirmation of differentially expressed genes identified by RNAseq analyses in B. burgdorferi infected MDA-MB-231 and MCF10A cell lines by qRT-PCR. 1 × 106 cells of MDA-MB-231 and MCF10A cell lines were infected with B. burgdorferi for 48 h, RNA was extracted, cDNA was synthesized, and subjected to qRT-PCR as discussed in Material and Methods. The expression fold change is normalized to GAPDH, and the delta-delta Ct method is used for quantitative analyses. (A) qRT-PCR and RNA-seq results for eight genes were differentially expressed in MDA-MB-231 while (B) qRT-PCR and RNA-seq results for nine genes were differentially expressed in MCF10A cells upon B. burgdorferi infection
Gene ontology analysis
KEGG and REACTOME pathway enrichment analyses of DEGs in triple-negative breast cancer and normal mammary epithelial cells
To further explore the functional significance of these commonly identified genes, additional analysis was performed using the KEGG and REACTOME pathway enrichment analyses in DAVID which revealed pathways related to inflammation, infection, metabolism, and cancer (Supplementary Table S3A and S3B). This analysis supports the notion that these genes may be involved in critical biological processes implicated in B. burgdorferi-infected TNBC cells.
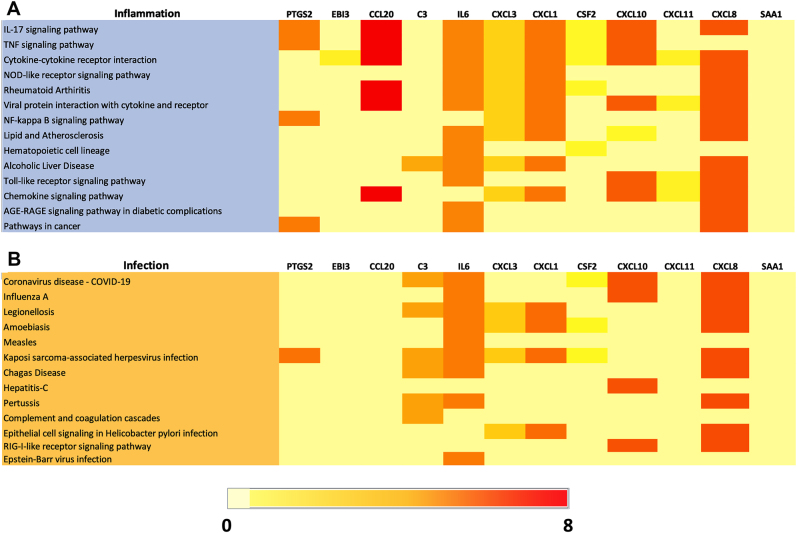
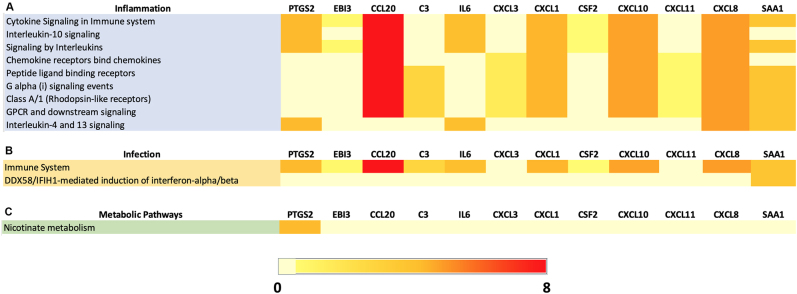
In order to gain insights into the functional implications of DEG genes in MDA-MB-231 cells, pathway analysis was performed using the top 12 genes upregulated upon B. burgdorferi infection (Fig. 4A). The heatmap represents the expression patterns of these genes and their corresponding fold changes in response to B. burgdorferi infection. The color intensity in the heatmap reflects the magnitude of gene expression changes, with red indicating higher expression levels and light yellow indicating lower expression levels. Pathway analysis of these top 12 upregulated genes revealed their involvement in approximately 15 KEGG pathways associated with diverse inflammation processes. Notably, several genes were significantly enriched in signaling pathways including the IL-17, TNF, NOD-like receptor, NF-kB, Toll-like receptor, and chemokine-signaling pathways. From the top 12 genes, the CCL and CXCL-family genes (CCL-20 and CXCL8, 3 & 1) as well as IL6 were the most commonly enriched in all the KEGG inflammatory pathways. Additionally, several of the DEGs were also enriched in pathways implicated in cancer such as PTGS2, IL6 and CXCL8. These findings align with the emerging evidence of B. burgdorferi's influence on gene expression and pathways associated with inflammation and tumorigenesis in MDA-MB-231 cells.
Fig. 4.
Heatmap depicting the expression of top 12 upregulated genes and their fold changes associated with KEGG inflammation (A) and infection (B) pathways from B. burgdorferi-infected MDA-MB-231. Pathways with the Benjamini-Hochberg adjusted P-value ≤0.10 were considered
Figure 3B represents the expression patterns of the same 12 upregulated genes and their corresponding fold changes in response to B. burgdorferi infection associated with diverse infectious processes. Pathway analysis of these top 12 upregulated genes revealed their involvement in approximately 13 KEGG pathways. Notably, one signaling pathway that is the RIG-I like receptor-signaling pathway was significantly enriched. Additionally, pathways involved in pathogenic infection and epithelial cell signaling in H. pylori infection were also observed which shows a potential connection to other pathways implicated in cancers with infectious origins in MDA-MB-231 cells. From the top 12 genes, the CXCL-family genes (CXCL8, 3 and 1) and IL6 were enriched in the majority of the KEGG infection pathways. However, complement 3 (C3), which was involved in a total of six out of the 13 pathways depicts a key role of the complement system pathway in MDA-MB-231 cells infected with B. burgdorferi (Fig. 4B).
To further explore the functional significance of the top 12 upregulated genes in MDA-MB-231 upon B. burgdorferi infection, REACTOME pathway enrichment analysis was conducted which highlighted 9 inflammation, 2 infection, and 1 metabolic pathway. From Fig. 5A, out of the 9 inflammation-related pathways, 6 of which were involved in important signaling processes such as cytokine, interleukin (IL-10, 4 & 13), G alpha (I), and GPCR and downstream signaling pathways. Among the 12 top regulated genes, CXCL8 was involved in all the pathways followed by CXCL10 and 1 and CCL20 being the most common genes expressed in the majority of the inflammatory pathways. Surprisingly SAA1, which was not involved in any of the KEGG pathways, was found to be enriched in 7 out of the 9 inflammation-associated pathways in the REACTOME analysis. Looking further into the infection pathways highlighted in the REACTOME server for MDA-MB-231 (Fig. 5B), only two significant pathways were observed which include the immune system pathway depicting the upregulation of most of the genes except CXCL3 and 11. Also, for the IFIH1-mediated induction of interferon-alpha/beta pathway, only SAA1 was involved indicating that it is an important factor in B. burgdorferi-infected MDA-MB-231 cells. Figure 5C shows the only involvement of PTGS2 in the Nicotinate metabolism pathway which was shown to be associated with inflammatory processes [27]. In summary, REACTOME analyses confirmed that inflammation associated pathways are the most significant pathways induced by B. burgdorferi.
Fig. 5.
Heatmap depicting the expression of top 12 upregulated genes and their fold changes associated with (A) REACTOME inflammation pathways, (B) REACTOME infection pathways and (C) REACTOME metabolic pathways from B. burgdorferi-infected MDA-MB-231. Pathways with the Benjamini-Hochberg adjusted P-value ≤0.10 were considered
The comparative analysis of the effects of B. burgdorferi infection in both normal and cancerous cellular contexts was perpetuated by using MCF10A cells, which serve as a relevant control to discern pathway alterations that are specific to B. burgdorferi-infected TNBC cells, as opposed to general cellular responses to B. burgdorferi.
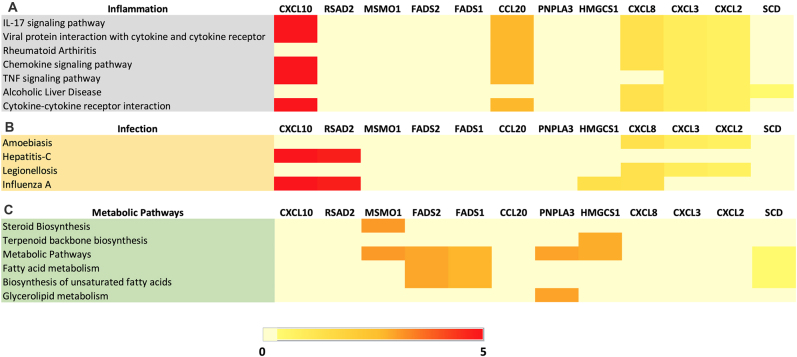
KEGG pathway analysis was performed using the top 12 genes upregulated in MCF10A RNA sequencing data upon B. burgdorferi infection. Pathway analysis of these upregulated genes revealed their involvement in approximately 7 KEGG pathways associated with diverse inflammation processes. The heatmap (Fig. 6A) depicts that IL-17, cytokine-cytokine & TNF signaling pathways which were already indicated in the top 3 pathways from B. burgdorferi-infected MDA-MB-231 cells, were also significantly involved in infected MCF10A cells. From the top 12 genes, the CXCL-family genes (CXCL2, 3, 8 & 10) and CCL20 were the most commonly enriched in the majority of the KEGG inflammatory pathways similar to what was found for MDA-MB-231 cells. Furthermore, Fig. 6B represents the expression patterns of the same 12 upregulated genes and their involvement in approximately 4 KEGG pathways associated with diverse infectious processes. From the top 12 DEG genes for MCF10A, the CXCL-family genes (CXCL2, 3, 8 & 10) and interferon-inducible antiviral protein that belongs to the S-adenosyl-l-methionine superfamily of enzymes, RSAD2 were commonly enriched in the majority of the KEGG infection pathways. Additionally, the KEGG server in DAVID, highlighted fatty acid metabolism and metabolic pathways in general, for MCF10A as shown in Fig. 6C. Interestingly, several DEGs were enriched in pathways related to the biosynthesis of steroids, terpenoid backbone, and unsaturated fatty acids, which could be indicative of a cellular transformation in MCF10A. The most common genes involved in at least 2 of these 5 pathways related to metabolism and biosynthesis are methyl sterol monooxygenase 1 (MSMO1), a member of the fatty acid desaturase gene family-FADS1 & 2, an enzyme involved in lipid metabolism as well as the Patatin-like phospholipase domain-containing protein 3 which is a phospholipase that mediates triacylglycerol hydrolysis in adipocytes.
Fig. 6.
Heatmap depicting the expression of top 12 upregulated genes and their fold changes associated with (A) KEGG inflammation pathways, (B) KEGG infection pathways and (C) KEGG metabolic pathways from B. burgdorferi-infected MCF10A cells. Pathways with the Benjamini-Hochberg adjusted P-value ≤0.10 were considered
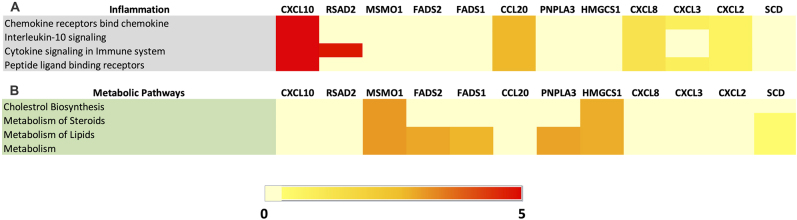
To further explore the functional significance of the top 12 upregulated genes consistently upregulated in MCF10A upon B. burgdorferi infection, REACTOME pathway enrichment analysis was conducted. It surprisingly showed 4 inflammation and 4 metabolic pathways enriched. From Fig. 7A, out of the 4 pathways highlighted for inflammation, one of them is involved in the interleukin-10 signaling pathway. The other important pathways indicate the receptors functioning such as the chemokine receptors and the peptide ligand binding receptors. Among the 12 top-upregulated genes, CXCL2, 8 and 10, and CCL20 were involved in all the pathways, similar to what we have seen in B. burgdorferi-infected MDA-MB-231 cells. CXCL3 was involved in the binding of the chemokine receptors pathway and peptide ligand binding receptors pathway. Also, RSAD2, the interferon-inducible antiviral protein, is involved in cytokine signaling in the immune system. Looking further into the metabolic pathways in the REACTOME server for MCF10A (Fig. 7B), several DEGs were significantly enriched in pathways involved in the metabolism of steroids, lipids, and metabolism in general. Interestingly, several DEGs were involved in pathways related to the biosynthesis of cholesterol which further confirms the chances of cellular transformation in MCF10A. The most common genes include MSMO1, ACAT2 and FDPS in all the metabolic and biosynthesis pathways. In summary, as seen in B. burgdorferi-infected MDA-MB-231 cells, several inflammatory pathways were upregulated in infected MCF10A cells. In addition, multiple metabolic pathways were also significantly influenced by B. burgdorferi infection.
Fig. 7.
Heatmap depicting the expression of top 12 upregulated genes and their fold changes associated with (A) REACTOME inflammation pathways, (B) REACTOME metabolic pathways from B. burgdorferi-infected MCF10A cells. Pathways with the Benjamini-Hochberg adjusted P-value ≤0.10 were considered
Discussion
Our previous studies demonstrated that B. burgdorferi spirochetes are capable of invading both triple-negative breast cancer MDA-MB-231 and the normal mammary epithelial MCF10A cells, increasing migration and invasion of the cancer cells but not the normal mammary epithelial cells [8]. In this study, we found that infection of these cells with B. burgdorferi resulted in the differential expression of 142 genes in MDA-MB-231 cells, out of which 12 genes were overexpressed by over 3-fold. On the contrary, 95 genes were found to be differentially expressed in B. burgdorferi-infected MCF10A cells, of which 11 genes were overexpressed by over 2-fold. Based on the ontological analysis, B. burgdorferi is not only capable of infecting these cells but can also elicit a very significant physiological response from the infected cells ranging from inflammation, infection-response, and altered metabolic pathways.
In this study, a major trend observed in both MDA-MB-231 and MCF10A cells upon B. burgdorferi infection was the upregulation of C-X-C and C-C motif chemokine family members (CXCL1, 2, 3, 8, and 10) and CCL20. Those chemokines have been widely demonstrated to be involved in cancer development [28, 29]. For example, previous studies have linked CXCL1, CXCL8, and CXCL10 to breast cancer development and progression, and they have been associated with tumor growth, metastasis, and angiogenesis [30–32]. In breast cancer cells, CXCL1 expression is lineage-dependent, being highest in triple-negative MDA-MB-231 cells [33]. CXCL1 induces cell proliferation, migration, and epithelial-to-mesenchymal transition (EMT) [33]. It also contributes to metastasis, particularly in bone, lungs, brain, and lymph nodes [34]. CXCL1 is also said to be associated with chemoresistance and radio-resistance, and its increased expression is linked to a worse prognosis in breast cancer patients [30]. In humans, Interleukin-8 (IL-8 or CXCL8) is a granulocytic chemokine with multiple roles within the tumor microenvironment (TME), such as recruiting immunosuppressive cells to the tumor, increasing tumor angiogenesis, and promoting EMT [35]. All these effects of CXCL8 on individual cell types can result in cascading alterations to the TME [36]. It has been reported that CXCL10 also might be related to worse prognosis in TNBC, suggesting that it might be an effective prognostic factor and a potential therapeutic target for TNBC treatment [37].
The most significantly upregulated gene in MDA-MB-231 cell was a C-C motif chemokine family member, CCL20. Recent reports have shown that the CCL20 and its receptor CCR6 are involved in several cancers, including breast cancer, and function by enhancing the migration and proliferation of cancer cells and remodeling the tumor microenvironment [38]. CCL20 chemokine has also been demonstrated to promote self-renewal and maintenance of breast cancer stemness through protein kinase C or alternatively through p38 mitogen-activated protein kinase (MAPK)-mediated activation of p65 nuclear factor kappa B (NF-κB) pathway [38].
Several additional genes identified in this study have a strong association with breast cancer. For example, upregulated EBi3 (subunit of the composite cytokine IL27 and IL35) was shown to correlate with poor outcomes and tumor progression in breast cancer through the increase of IL-27p28 and IL-1235 [39]. Another upregulated gene, complement protein 3 (C3) plays a role in triple-negative breast cancer (TNBC) and infection by exerting diverse effects in both contexts [38, 39]. In the context of infection, C3 is a key component of the innate immune response. When pathogens invade the body, the complement system, including C3, is activated to opsonize and eliminate the invaders [40]. In TNBC, C3 has been implicated in promoting tumor growth, invasion, and metastasis. Its expression is often elevated in TNBC tissues, and studies have shown that C3 contributes to tumor progression by interacting with complement receptors on cancer cells, leading to the activation of downstream signaling pathways that promote proliferation and survival [41–43]. Moreover, C3 can interact with tumor-associated macrophages and immune cells, fostering an immunosuppressive microenvironment that hinders anti-tumor immune responses [43].
Another significant DEG for the B. burgdorferi-infected MDA-MB-231 was the Serum amyloid A (SAA) which is known to be elevated in several malignancies with poor prognosis [44]. Previous studies have reported that SAA is a critical mediator of pro‐inflammatory cytokine production and recruitment of immune cells, and promotes tumor development by facilitating angiogenesis, tumor invasion, and immunosuppression [44, 45].
Triple-negative breast cancer (TNBC) cells have been shown to exhibit dysregulation of various signaling pathways involved in immune responses and pathogen recognition. One of the key pathways implicated in TNBC is the toll-like receptor (TLR) pathway which was identified as an important pathway in B. burgdorferi infection in MDA-MB-231 cells. TLRs play a crucial role in recognizing pathogen-associated molecular patterns (PAMPs) and initiating innate immune responses against invading pathogens, including bacteria like B. burgdorferi [46]. There is mounting evidence associating the TLR pathway and development of breast cancer suggesting that it can serve as a biomarker for breast cancer pathogenesis and progression [47].
Additionally, the nuclear factor-kappa B (NF-κB) pathway was also identified in B. burgdorferi-infected MDA-MB-231 cells. NF-κB, a proinflammatory transcription factor plays a central role in regulating the expression of genes involved in inflammation and cell survival [48]. The activation of (NFκB), is a commonly observed phenomenon in breast cancer and it was suggested that it can serve as a potential therapeutic target [49].
These pathways may collectively contribute to the host's response to B. burgdorferi infection in TNBC cells and potentially affect the tumor microenvironment, influencing cancer progression and immune evasion. Further pathway analysis also identified several additional pathways involved in inflammation and infection. The most common inflammatory pathways in TNBC cells identified by KEGG server were the IL-17 signaling, TNF signaling, and chemokine signaling pathways. The other signaling pathways highlighted by the REACTOME server include cytokine signaling, IL-4, 10, and 13 signaling, G alpha, and GPCR downstream signaling pathways. From the KEGG server, the most common inflammatory pathways in MCF10A cells included the IL-17 signaling, TNF signaling and chemokine signaling pathways. The other signaling pathways depicted by the REACTOME server include IL-10 signaling and cytokine signaling pathways. Several common pathways associated with inflammatory diseases are Rheumatoid Arthritis and Alcoholic Liver disease. From the KEGG server, the most common infectious disease-associated pathways in TNBC cells included COVID-19, Legionellosis, Prion disease, Influenza-A, Pertussis, Measles, Kaposi-sarcoma-associated herpesvirus, and Chagas disease, among others. Additionally, pathways involved in pathogenic E. coli infection and epithelial cell signaling in H. Pylori infection were also observed, showing a potential connection to other infection-related pathways in TNBC cells- MDA-MB-231. The other infection-associated pathways also common to MCF10A include Hepatitis-C and Amoebiasis.
Ontological analysis also revealed the involvement of the DEGs in metabolic pathways in both cell lines. PTGS2 or COX2 which was significantly upregulated in MDA-MB-231 mediates the conversion of arachidonic acid to prostaglandins and has been linked to mutagenesis, angiogenesis, and enhanced cell migration when upregulated [50–52]. Equally important, in MCF10A, several of the DEGs were found to be involved in pathways related to steroid and fatty acid biosynthesis and metabolism including RSAD2, MSMO1, PNPLA3, FADS2, and HMGCS1. Alteration of metabolic pathways is a hallmark of cancer and an early event in tumorigenesis [53], which demands attention as MCF10A cells are non-tumorigenic. The fact that B. burgdorferi can alter these pathways, particularly the ones pertaining to steroid metabolism, can have severe implications, especially in the context of breast cancer as it can be a steroid hormone-driven malignancy in many cases (ER-and PR-positive breast cancer). Alteration of metabolic pathways including de novo synthesis of steroids, HDL remodeling, cholesterol biosynthesis, and metabolism are critical mechanisms in breast cancer development vital for cell growth, migration, and epithelial to mesenchymal transition (EMT), a process vital for cell migration and metastasis [53]. While previous research has yet to demonstrate any phenotypic effects pertaining to MCF10A assuming a cancerous phenotype upon infection with B. burgdorferi, the dysregulation of genes associated with metabolic pathways could serve as an important early diagnostic marker.
Our study relates to a similar transcriptomic analysis of B. bavariensis infected human brain microvascular endothelial cells (hBMECs). This study also revealed several of the genes related to cytokines, chemokines, interferon, and TNF signaling that may cause alteration in the blood-brain barrier (BBB) integrity [24]. While our study focused on investigating the gene expression changes in triple-negative breast cancer (TNBC) cells upon B. burgdorferi infection, the infected endothelial cells of the brain microvasculature represent very similar findings. Some of the most common upregulated genes between hBMECs and MDA-MB-231 include the abovementioned CXCL family genes – CXCL1, CXCL2, CXCL3, CXCL8, and CXCL10, playing significant roles in signal transduction, innate immune response and in the induction of the pattern recognition receptors and downstream signaling cascades. CCL20 was also identified as a common DEG as well as the interleukin family - IL1B and IL6 which were discussed to be responsible for the remodeling of cell cytoskeleton and recruiting leukocytes during the B. burgdorferi invasion. Interestingly C3 and SAA1 were also common markers from the infected endothelial cells and the breast cancer cells in our study suggesting that common pathways are triggered by infection by Borrelia spp in both cell types.
In summary, our whole genome transcriptomic analysis on B. burgdorferi-infected breast cancer and normal mammary epithelial cells revealed that several pathways associated with infection, inflammation, cancer, and metabolism are altered by B. burgdorferi infection of normal and neoplastic cancer cells.
Conclusion
Triple-negative breast cancer is one of the most aggressive types of breast cancer, which has an extremely high treatment burden with no definitive cause, and potentially a wide array of dysregulated cellular and molecular mechanisms. The high prevalence of Lyme disease combined with the ability of B. burgdorferi to infect breast cancer cells prompts the exploration of a potential cause-effect relationship. High-throughput transcriptomic analysis tools such as those used in our study can prove to be extremely effective for preliminary screening to identify molecular markers that the bacteria may be implementing to orchestrate more severe breast cancer phenotypes, particularly in TNBC.
Author Contributions
Conceptualization, V.A.K., A.M. and E.S.; methodology, V.A.K., N.J.P., S.T.; J.Y.S., D.L.K; C.L.M., validation, V.A.K., N.J.P., S.P., D.L.K., J.A.V., J.J., G.O.A., and E.S.; formal analysis, V.A.K., N.J.P., S.P., C.L.M., J.A.V., A.M.; investigation, V.A.K., S.T.; J.Y.S., D.L.K; C.L.M., J.A.V., J.J., resources, E.S.; data curation, V.A.K, N.J.P., S.P., J.A.V., A.M., and E.S.; writing—original draft preparation, V.A.K., and E.S.; writing—review and editing V.A.K., N.J.P., S.T.; J.A.V., J.J., G.O.A., A.M., and E.S.; visualization, V.A.K., N.J.P., S.P., J.A.V., A.M.; supervision, E.S.; project administration, E.S.; funding acquisition, E.S. All authors have read and agreed to the published version of the manuscript. All authors had full access to all data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding sources
This research received no external funding
Data availability statement
The data presented in this study are available at a reasonable request from the corresponding author.
Conflict of interest
The authors have declared that no competing interests exist.
Supplementary materials
Acknowledgment
This work was supported by the Pink Clover Foundation and National Philanthropic Trust. The supporter had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. The authors would like to thank Yale Center for Genome Analysis for conducting the RNA-Sequencing. The authors would also like to thank Dr. Joerg Nikolaus, Ph.D., at the Yale West Campus Microscopy facility for training and supervising the use of the confocal microscope.
References
- 1.Breast cancer Statistics | How common is breast cancer? American Cancer Society. https://www.cancer.org/cancer/breast-cancer/about/how-common-is-breast-cancer.html [Accessed 2 Sep, 2022]. [Google Scholar]
- 2.Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res 2020;22(1):61. 10.1186/s13058-020-01296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Infections that can lead to cancer. American Cancer Society. https://www.cancer.org/cancer/cancer-causes/infectious-agents/infections-that-can-lead-to-cancer.html [Accessed 12 May, 2022]. [Google Scholar]
- 4.Khatun S, Appidi T, Rengan AK. The role played by bacterial infections in the onset and metastasis of cancer. Curr Res Microb Sci 2021;2:100078. 10.1016/j.crmicr.2021.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu A, Yao B, Dong T, Chen Y, Yao J, Li H, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022;185(8):1356–1372. 10.1016/j.cell.2022.02.027. [DOI] [PubMed] [Google Scholar]
- 6.Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun 2020;11(1). 10.1038/s41467-020-16967-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Peck KN, et al. Distinct microbial signatures associated with different breast cancer types. Front Microbiol 2018;9:951. 10.3389/fmicb.2018.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaur G, Sawant JY, Chavan AS, Khatri VK, Liu YH, Zhang M, et al. Effect of invasion of Borrelia burgdorferi in normal and neoplastic mammary epithelial cells. Antibiotics 2021;10(11):1295. 10.3390/antibiotics10111295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debbarma A, Mansolf M, Khatri VA, Valentino JA, Sapi E. Effect of Borrelia burgdorferi on the expression of miRNAs in breast cancer and normal mammary epithelial cells. Microorganisms 2023;11(6):1475. 10.3390/microorganisms11061475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garbe C, Stein H, Dienemann D, Orfanos CE. Borrelia burgdorferi-associated cutaneous B cell lymphoma: clinical and immunohistologic characterization of four cases. J Am Acad Dermatol 1991;24(4):584–590. 10.1016/0190-9622(91)70088-j. [DOI] [PubMed] [Google Scholar]
- 11.Schöllkopf C, Melbye M, Munksgaard L, Smedby KE, Rostgaard K, Glimelius B, et al. Borrelia infection and risk of non-Hodgkin lymphoma. Blood 2008;111(12):5524–5529. 10.1182/blood-2007-08-109611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinden T, Manivel J, and Valen P. Mucosa-Associated Lymphoid Tissue (MALT) lymphoma of the colon in a patient with Borrelia burgdorferi infection (Lyme arthritis). Case Rep Clin Med 2021;(10): 403–411. 10.4236/crcm.2021.1012050. [DOI] [Google Scholar]
- 13.Bobe JR, Jutras BL, Horn EJ, Embers ME, Bailey A, Moritz RL, et al. Recent progress in Lyme disease and remaining challenges. Front Med (Lausanne) 2021;8:666554. 10.3389/fmed.2021.66655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson C, Brissette CA. The Brilliance of Borrelia: mechanisms of host immune evasion by Lyme disease-causing spirochetes. Pathogens 2021;10(3):281. 10.3390/pathogens10030281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brorson Ø, Brorson SH. Transformation of cystic forms of Borrelia burgdorferi to normal, mobile spirochetes. Infection 1997;25(4):240–246. 10.1007/bf01713153. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald AB. Spirochetal cyst forms in neurodegenerative disorders,…hiding in plain sight. Med Hypotheses 2006;67(4):819–832. 10.1016/j.mehy.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 17.Miklossy J, Kasas S, Zurn AD, McCall S, Yu S, McGeer PL. Persisting atypical and cystic forms of Borrelia burgdorferi and local inflammation in Lyme neuroborreliosis. J Neuroinflammation 2008;5:1–18. 10.1186/1742-2094-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sapi E, Bastian SL, Mpoy CM, Scott S, Rattelle A, Pabbati N, et al. Characterization of biofilm formation by Borrelia burgdorferi in vitro. PLoS One 2012;7:1–11. 10.1371/journal.pone.0048277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meriläinen L, Herranen A, Schwarzbach A, Gilbert L. Morphological and biochemical features of Borrelia burgdorferi pleomorphic forms. Microbiology. 2015;161:516–527. 10.1099/mic.0.000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sapi E, Balasubramanian K, Poruri A, Maghsoudlou JS, Socarras KM, Timmaraju AV, et al. Evidence of in vivo existence of Borrelia biofilm in borrelial lymphocytomas. Eur J Microbiol Immunol 2016;6:9–24. 10.1556/1886.2015.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meriläinen L, Brander H, Herranen A, Schwarzbach A, Gilbert L. Pleomorphic forms of Borrelia burgdorferi induce distinct immune responses. Microbes Infect 2016 Jul–Aug;18(7–8):484–495. 10.1016/j.micinf.2016.04.002. Epub 2016 Apr 30. PMID: 27139815. [DOI] [PubMed] [Google Scholar]
- 22.Karvonen K, Nykky J, Marjomäki V, Gilbert L. Distinctive evasion mechanisms to allow persistence of Borrelia burgdorferi in different human cell lines. Front Microbiol 2021;12:711291. 10.3389/fmicb.2021.711291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kukurba KR, Montgomery SB. RNA sequencing and analysis. Cold Spring Harb Protoc 2015;2015(11):951–969. 10.1101/pdb.top084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tkáčová Z, Bhide K, Mochnáčová E, Petroušková P, Hruškovicová J, Kulkarni A, et al. Comprehensive mapping of the cell response to Borrelia bavariensis in the brain microvascular endothelial cells in vitro using RNA-Seq. Front Microbiol 2021;12:760627. 10.3389/fmicb.2021.760627. PubMed PMID: 36612163; PubMed Central PMCID: PMC9818145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sapi E, Kasliwala RS, Ismail H, Torres JP, Oldakowski M, Markland S, et al. The long-term persistence of Borrelia burgdorferi antigens and DNA in the tissues of a patient with Lyme Disease. Antibiotics 2019;8(4):183. 10.3390/antibiotics8040183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25(4):402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Walsh C, Akrap N, Garre E, Magnusson Y, Harrison H, Andersson D, et al. The mevalonate precursor enzyme HMGCS1 is a novel marker and key mediator of cancer stem cell enrichment in luminal and basal models of breast cancer. PLOS One 2020;15(7):e0236187. 10.1371/journal.pone.0236187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu T, Yang W, Sun A, Wei Z, Lin Q. The role of CXC chemokines in cancer progression. Cancers (Basel) 2022 Dec 28;15(1):167. 10.3390/cancers15010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadomoto S, Izumi K, Mizokami A. The CCL20-CCR6 Axis in cancer progression. Int J Mol Sci 2020;21(15):5186. 10.3390/ijms21155186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korbecki J, Bosiacki M, Barczak K, Lagocka R, Brodowska A, Chlubek D, et al. Involvement in tumorigenesis and clinical significance of CXCL1 in reproductive cancers: breast cancer, cervical cancer, endometrial cancer, ovarian cancer and prostate cancer. Int J Mol Sci 2023;24(8):7262. 10.3390/ijms24087262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res 2008;14(21):6735–6741. 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 32.Ejaeidi A, Craft BS, Puneky LV, Lewis RE, Cruse JM. Hormone receptor-independent CXCL10 production is associated with the regulation of cellular factors linked to breast cancer progression and metastasis. Exp Mol Pathol 2015;99(1):163–172. 10.1016/j.yexmp.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Wang N, Liu W, Zheng Y, Wang S, Yang B, Li M, et al. CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-κB/SOX4 signaling. Cell Death Dis 2018;9(9):880. 10.1038/s41419-018-0876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bièche I, Chavey C, Andrieu C, Busson M, Vacher S, Le Corre L, et al. CXC chemokines located in the 4q21 region are up-regulated in breast cancer. Endocr Relat Cancer 2007 Dec;14(4):1039–1052. 10.1677/erc.1.01301. PMID:18045955. [DOI] [PubMed] [Google Scholar]
- 35.Ha H, Debnath B, Neamati N. Role of the CXCL8-CXCR1/2 axis in cancer and inflammatory diseases. Theranostics 2017;7(6):1543–1588. 10.7150/thno.15625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han ZJ, Li YB, Yang LX, Cheng HJ, Liu X, Chen H. Roles of the CXCL8-CXCR1/2 axis in the tumor microenvironment and immunotherapy. Molecules 2021;27(1):137. 10.3390/molecules27010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chuan T, Li T, Yi C. Identification of CXCR4 and CXCL10 as potential predictive biomarkers in triple negative breast cancer (TNBC). Med Sci Monit 2020 Jan 11;26:e918281. 10.12659/MSM.918281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadomoto S, Izumi K, Mizokami A. The CCL20-CCR6 Axis in cancer progression. Int J Mol Sci 2020 Jul 22;21(15):5186. 10.3390/ijms21155186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kourko O, Seaver K, Odoardi N, Basta S, Gee K. IL-27, IL-30, and IL-35: a cytokine triumvirate in cancer. Front Oncol 2019;9:969. Published 2019 Oct 1. 10.3389/fonc.2019.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwak J, Laskowski J, Li H, McSharry M, Sippel T, Bullock B, et al. Complement activation via a C3A receptor pathway alters CD4+ T lymphocytes and mediates lung cancer progression. Cancer Res 2018;78(1):143–156. 10.1158/0008-5472.can-17-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afshar-Kharghan V. The role of the complement system in cancer. J Clin Invest 2017;127(3):780–789. 10.1172/JCI90962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu M, Jia BB, Li MF. Complement C3 and activated fragment C3a are involved in complement activation and anti-bacterial immunity. Front Immunol 2022;13:813173. 10.3389/fimmu.2022.813173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zha H, Wang X, Zhu Y, Chen D, Han X, Yang F, et al. Intracellular activation of complement C3 leads to PD-L1 antibody treatment resistance by modulating tumor-associated macrophages. Cancer Immunol Res 2019;7(2):193–207. 10.1158/2326-6066.CIR-18-0272. PMID: 30514794. [DOI] [PubMed] [Google Scholar]
- 44.Malle E, Sodin-Semrl S, Kovacevic A. Serum amyloid A: an acute-phase protein involved in tumour pathogenesis. Cell Mol Life Sci 2009;66(1):9–26. 10.1007/s00018-008-8321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niu X, Yin L, Yang X, Yang Y, Gu Y, Sun Y, et al. Serum amyloid A 1 induces suppressive neutrophils through the Toll‐like receptor 2–mediated signaling pathway to promote progression of breast cancer. Cancer Sci 2022;113(4):1140–1153. 10.1111/cas.15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cervantes JL, Hawley KL, Benjamin SJ, Weinerman B, Luu SM, Salazar JC. Phagosomal TLR signaling upon Borrelia burgdorferi infection. Front Cell Infect Microbiol 2014;4:55. 10.3389/fcimb.2014.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi S, Xu C, Fang X, Zhang Y, Li H, Wen W. Expression profile of Toll-like receptors in human breast cancer. Mol Med Rep 2019 Nov;20(5):4059–4066. 10.3892/mmr.2019.10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther 2017;2:17023. 10.1038/sigtrans.2017.23. PMID: 29158945; PMCID: PMC5661633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W, Nag SA, Zhang R. Targeting the NFκB signaling pathways for breast cancer prevention and therapy. Curr Med Chem 2015;22(2):264–289. 10.2174/0929867321666141106124315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazhar D, Ang R, Waxman J. COX inhibitors and breast cancer. Br J Cancer 2006;94(3):346–350. 10.1038/sj.bjc.6602942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009;459(7249):1005–1009. 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boland GP, Butt I, Prasad R, Knox WF, Bundred N. COX-2 expression is associated with an aggressive phenotype in ductal carcinoma in situ. Br J Cancer 2004;90(2):423–429. 10.1038/sj.bjc.6601534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pavlova NN, Zhu J, Thompson CB. The hallmarks of cancer metabolism: still emerging. Cell Metab 2022;34(3):355–377. 10.1016/j.cmet.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available at a reasonable request from the corresponding author.