Abstract
Biological aging is a relevant risk factor for chronic diseases, and several indicators for measuring this factor have been proposed, with telomere length (TL) among the most studied. Oxidative stress may regulate telomere shortening, which is implicated in the increased risk. Using a novel estimator for TL, we examined whether adherence to the Mediterranean diet (MedDiet), a highly antioxidant-rich dietary pattern, is associated with longer TL. We determined TL using DNA methylation algorithms (DNAmTL) in 414 subjects at high cardiovascular risk from Spain. Adherence to the MedDiet was assessed by a validated score, and genetic variants in candidate genes and at the genome-wide level were analyzed. We observed several significant associations (p < 0.05) between DNAmTL and candidate genes (TERT, TERF2, RTEL1, and DCAF4), contributing to the validity of DNAmTL as a biomarker in this population. Higher adherence to the MedDiet was associated with lower odds of having a shorter TL in the whole sample (OR = 0.93; 95% CI: 0.85–0.99; p = 0.049 after fully multivariate adjustment). Nevertheless, this association was stronger in women than in men. Likewise, in women, we observed a direct association between adherence to the MedDiet score and DNAmTL as a continuous variable (beta = 0.015; SE: 0.005; p = 0.003), indicating that a one-point increase in adherence was related to an average increase of 0.015 ± 0.005 kb in TL. Upon examination of specific dietary items within the global score, we found that fruits, fish, “sofrito”, and whole grains exhibited the strongest associations in women. The novel score combining these items was significantly associated in the whole population. In the genome-wide association study (GWAS), we identified ten polymorphisms at the suggestive level of significance (p < 1 × 10−5) for DNAmTL (intergenics, in the IQSEC1, NCAPG2, and ABI3BP genes) and detected some gene–MedDiet modulations on DNAmTL. As this is the first study analyzing the DNAmTL estimator, genetics, and modulation by the MedDiet, more studies are needed to confirm these findings.
Keywords: telomere length, DNA methylation, aging, Mediterranean diet, antioxidants, fruits, genetics, GWAS, epigenetics
1. Introduction
Numerous studies have shown an important relationship between diet and chronic diseases [1,2,3,4]. The magnitude of the association varies depending on the disease type analyzed, which is generally higher for cardiovascular diseases, diabetes, metabolic syndrome, and obesity [1,2,3,4,5,6,7,8,9,10]. However, the precise mechanisms by which diet may exert a protective or risk effect against various chronic diseases are not well understood [11]. In general, oxidative stress is crucial in the pathogenesis, progression, and associated complications of the most prevalent chronic diseases [12,13,14,15]. An imbalance between the generation of reactive oxygen species and the antioxidant defense mechanisms typically causes cellular damage and dysfunction. Given that dietary foods can provide a variety of compounds with high antioxidant capacity, this is one of the postulated mechanisms for the protection of certain dietary patterns against these diseases [16,17,18].
In chronic diseases, another common factor in them is age, since the prevalence of these diseases increases with chronological age [19]. Nonetheless, it is widely recognized that a substantial distinction exists between chronological age, which refers to the time that has passed since birth, and biological age, which is a more complex concept developed from pathophysiological assessments that reflect the extent of an individual’s aging process [20,21]. Accelerated aging is the term used to describe the gap between chronological age and biological age [21,22].
The importance of oxidative stress in aging has been demonstrated in several studies [23,24], and there is a growing interest in identifying lifestyle factors (mainly diet) that may potentially mitigate the effects of biological aging in human studies [25,26]. The aging process is a multifaceted phenomenon primarily characterized by impairment in cellular, tissue, and organ functionality, leading to an augmented susceptibility to various diseases. This process encompasses a number of alterations, commonly referred to as “hallmarks of aging” [27]. These hallmarks are being characterized in depth [27,28]. In a recent update, the initial listing expanded from nine to twelve interconnected hallmarks [29]. Among them, we highlight “telomere attrition”, as it can be analyzed more easily in epidemiological studies in humans compared to other hallmarks such as “depletion of stem cells” or “modified intracellular communication” that require greater support of basic laboratory research.
Located at the ends of eukaryotic chromosomes, telomeres are specialized nucleoprotein structures with a protective function [30]. Telomeres consist of repetitive TTAGGG DNA sequences and are associated with a complex of six proteins referred to as Shelterin that includes the telomere repeat factor 1 (TRF1) and 2 (TRF2), protection of telomeres-1 (POT1), TIN2 (TRF1-Interacting Nuclear factor 2), Ras-related protein 1 (RAP1), and the telomere protection protein 1 (TPP1) [30,31]. The DNA component of telomeres undergoes a gradual reduction in length with every cycle of cell division, ultimately leading to the initiation of cellular senescence [30,31,32]. Multiple humans studies have observed that telomere length (TL) decreases with chronological age [33,34]. Similarly, a shorter TL has been associated with a higher risk of cardiovascular disease, mortality and other metabolic diseases [35,36,37,38,39,40]. Therefore, in epidemiological studies, TL has been proposed as a measure of biological aging despite the fact that measuring telomeres can be challenging [30,41]. Although TL may be different depending on the tissue analyzed, it has been shown that examining TL in peripheral leukocytes can be a good overall indicator [42]. The main drawback is the heterogeneity of the classical methodologies used to measure TL in epidemiological studies (i.e., terminal restriction fragment analysis, quantitative polymerase chain reaction, quantitative fluorescence in situ hybridization methods, or single telomere length analysis), which produce varying results [43,44,45]. Using the novel omics methods, Lu et al. developed and validated a new method to measure TL in epidemiological studies [46]. This method is based on the DNA methylation profile of specific cytosine phosphate guanine (CpG) sites derived by machine learning [46]. The DNA methylation estimator of TL (DNAmTL), based on 140 CpGs and validated in leukocytes from several cohorts, outperformed several limitations of the classical techniques in epidemiological studies [46]. Subsequently, this DNAmTL biomarker has been used in other cohorts [47,48,49,50,51], although more studies in diverse populations are needed to better know its external validity.
Moreover, as far as we know, none of the studies published with this biomarker of TL have analyzed the influence of the Mediterranean diet (MedDiet) on it. The MedDiet is recognized as a healthful eating pattern that is high in antioxidants and can have anti-aging benefits [11,52,53,54]. The traditional MedDiet is a dietary pattern that was commonly observed in Greece and Italy throughout the early 1960s [55]. Furthermore, it is important to remember that in Spain, individuals also followed the typical MedDiet [11]. From the initial definition of the MedDiet to the spread of this diet internationally, mainly in the United States, thanks to the work of Dr. Dimitrios Trichopoulos [56] and Dr. Antonia Trichopoulou [57], there has been extensive research and dissemination activity to the general population, characterizing this diet as one of the healthiest in the world. A further detail of this historical evolution of the MedDiet can be found in the review by Sikalidis et al. [55]. The traditional MedDiet is characterized by a preferential use of virgin olive oil, seasonal vegetables, fruits, legumes, nuts and seeds, low red meat consumption and moderate fish consumption [11]. It has been indicated that the overall dietary pattern rather than individual components is important for health and longevity [56]. Thus, the MedDiet can reduce the level of oxidative stress markers and inflammation due to a high abundance of antioxidant compounds such as ω-3 fatty acids and resveratrol, among others [11]. It is important to note that the first meta-analyses on the topic already associated the MedDiet with greater longevity [57] as well as with a lower mortality from all causes and specific causes [58].
However, previous studies examining the association between the MedDiet and telomere length have yielded varying results [59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75]. Despite the conclusion of a meta-analysis that higher adherence to the MedDiet was related to longer TL, the association was heterogeneous, and sex differences were observed [72] with women having more significant associations than men. On the other hand, it is known that genetic factors play a relevant role in TL [76,77,78]; however, the vast majority of previous studies analyzing the impact of the MedDiet on TL did not account for genetic variants [59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75]. Moreover, although multiple GWASs have been taken for TL measured with the classic techniques [76,77,78,79,80,81,82,83,84,85,86], there are barely any GWAS using the DNAmTL variable [51,87]. Thus, little is known about the genetic architecture underlying DNAmTL, and more research is needed. In this study, we first aimed to investigate the association between adherence to the MedDiet and TL in a Spanish Mediterranean population using the DNAmTL estimator, taking into account sex-specific analyses. Our secondary aims were to perform an exploratory GWAS on DNAmTL to identify genetic variants associated with this biomarker and to explore gene–MedDiet modulations on DNAmTL both at the TL-related candidate genes and at the genome-wide level.
2. Materials and Methods
2.1. Participants and Study Design
A total of 414 individuals, comprising both men and women, aged between 55 and 75 years, were analyzed in the present study. These individuals were recruited in the PREDIMED-Plus-Valencia study. The PREDIMED-Plus-Valencia study is one of the field sites for the ongoing multicenter PREDIMED-Plus trial [88]. The participants in this study were selected from primary health care facilities in the Valencia region according to the inclusion and exclusion criteria outlined in the PREDIMED-Plus trial protocol. The Valencia region is situated along the eastern Mediterranean coast of Spain. The subjects selected for participation in the study were adults living in the community, with a body mass index (BMI) ranging from 27 to 40 kg/m2 and exhibiting at least three components of the metabolic syndrome [88]. A total of 465 participants were recruited at the PREDIMED-Plus Valencia site. However, for the purposes of this study, 414 subjects were included. This number represents individuals who consented to participate in the genomics/epigenomics studies and whose DNA samples and methylation workflows met the quality control criteria for the leukocyte DNA methylation analysis, as previously reported [89]. A cross-sectional study using baseline (sociodemographic, dietary, lifestyle, clinical, epigenetic and genetic) data was undertaken. The Human Research Ethics Committee of the University of Valencia provided ethical approval in accordance with the Helsinki Declaration for the study protocols and procedures (approval codes H1373255532771, 15 July 2013; and H1509263926814, 6 November 2017). Written informed consent was obtained from the participants.
2.2. Clinical, Biochemical and Anthropometric Variables
At baseline, demographic information, clinical variables, and medication use were assessed by questionnaire as previously reported [90]. Following a 12 h overnight fast, plasma glucose, total cholesterol, HDL-C, LDL-C, and triglyceride concentrations were measured as previously reported [91]. In addition, we assessed the complete blood count (CBC) in peripheral blood samples. The CBC included the total number of leukocytes as well as the type of white blood cells (neutrophils, eosinophils, basophils, monocytes, and lymphocytes) as previously indicated [92]. Blood pressure was measured by qualified personnel using the study protocol and a validated semiautomatic oscillometer (Omron HEM-705CP, Hoofddorp, The Netherlands) [88]. Anthropometric data (height, weight, and waist circumference) were measured by qualified personnel using calibrated scales, a standardized ribbon, and a wall-mounted stadiometer as previously reported [88]. The BMI was calculated by dividing the individual’s weight in kilograms by their height in meters squared. The condition of obesity was operationally defined as having a BMI equal to or exceeding 30 kg/m2.
2.3. Adherence to the MedDiet and Other Lifestyle Factors
The validated PREDIMED-Plus 17-item score [93], an updated version of the previously described PREDIMED 14-item scale [94], was used to evaluate adherence to the MedDiet. This 17-item questionnaire contained seventeen questions about the MedDiet. The questionnaire was scored as follows: 1 point for each response that captured adherence to the MedDiet, and 0 points for responses that did not. Table S1 displays the full set of questions and responses for MedDiet adherence. In brief, the foods and habits included were as follows: I-1: use only extra virgin olive oil for cooking, salad dressings, and spreads; I-2: vegetable consumption; I-3: fruits; I-4: red meat; I-5: butter, margarine or cream; I-6; sugar-sweetened beverages; I-7: legumes; I-8: fish/shellfish; I-9: pastries, cookies, sweets or cakes; I-10; nuts; I-11: preference for white meat; I-12: “sofrito” (Mediterranean sauce made with tomato, onion, garlic and olive oil); I-13: add preferentially non-caloric artificial sweeteners to beverages instead of sugar; I-14: servings of white bread; I-15: whole grains; I-16: refined cereals; I-17; moderate wine consumption. The greater the score (0 to 17), the greater the adherence to the MedDiet pattern. As previously reported [92], the overall score was categorized into two groups representing low (from 0 to 8 points) and high (from 9 to 17 points) adherence to the MedDiet. Both the overall score and specific food items were analyzed. In addition, a previously described PREDIMED-Plus general questionnaire [88] administered by trained personnel at baseline collected information regarding tobacco use and educational attainment. Those classified as current smokers smoked at least one cigarette, cigar, or a pipe per day. There defined three categories for smoking status: current smokers, ex-smokers, and non-smokers. Lastly, never-smokers and ex-smokers were compared to current smokers. Using the validated REGICOR Short Physical Activity Questionnaire [95], the estimation of total energy expenditure related to leisure-time physical activity involved the summation of the frequency, duration, and intensity of each activity, which was then divided by the number of days in a month (30 days) to provide the value in METmin/day.
2.4. DNA Isolation, DNA Methylation and DNAmTL Calculation
At baseline, DNA was extracted from as described in a prior publication [92]. Double-stranded DNA was quantified using PicoGreen (Invitrogen Corporation, Carlsbad, CA, USA). A subsequent investigation of leukocyte DNA methylation was conducted only on samples containing 500 ng of DNA of good quality. In this study, we employed the Infinium HumanMethylationEPIC BeadChip (850K) array, which was manufactured by Illumina in San Diego, CA, USA. This array allows for the analysis of methylation patterns by examining approximately 850,000 CpG sites. Notably, it covers more than 90% of the probes detected on the 450 K array and includes extra CpG sites [96]. In order to minimize batch effects, the position (sample wells) of the DNA samples on the microarrays were randomized [97]. The arrays underwent additional processing at the Human Genomics Facility located at Erasmus MC in Rotterdam, the Netherlands. The DNA was processed by bisulfite conversion using the Zymo EZ-96 DNA Methylation Kit (Zymo Research, Irvine, CA, USA) and subsequently hybridized to the Illumina EPIC array, following the instructions provided by the manufacturer. The microarrays underwent scanning using an Illumina HiScan machine, resulting in the generation of “*.idat” files. The Human Genomics laboratory conducted quality control processes to evaluate the quality and reliability of the DNA methylation data obtained. These procedures involved utilizing the R packages Minfi (release 3.18), Meffil (release 1.1.1), and ewastools (release 1.7.2) [98,99,100]. In summary, the quality control process aimed to identify samples that had deficiencies or substandard control metrics, specifically in relation to inadequate bisulfite conversion, various forms of suboptimal hybridization, and samples with a low call [92]. A total of 414 samples successfully underwent quality control and were thereafter subjected to analysis for the computation of DNAmTL. Data normalization, including functional normalization and normal-exponential out-of-band (NOOB) correction, was carried out. [92,101]. Beta values were obtained for the corresponding CpG sites [92]. The resulting DNAm beta matrix was uploaded to the online Horvath epigenetic age calculator [102]. The Horvath DNAmTL was used for TL estimation as previously published by Lu et al. [46]. For this estimator, methylation at 140 specific CpG sites are used to derive TL measures in kilobases (kb) according to the initial machine learning model of telomere restricted fragments (TRF)-measured leukocyte TL [46]. Likewise, the age-adjusted estimate of DNAmTL (referred to as DNAmTLadjAge) was derived online by regressing DNAmTL on age, and the resulting raw residual was defined as DNAmTLadjAge [46]. In addition, the same DNAm data were used to calculate the intrinsic epigenetic age acceleration (IEAA) for the Hannum epigenetic clock [103] to estimate the correlation between both epigenetic measures. IEAA is defined as the residual resulting from regressing the DNAm age estimate from Hannum on chronological age and blood cell count estimates [104]. Positive IEAA indicates a higher biological age than the chronological age, whereas negative IEAA values indicate a lower biological age than expected [105]. Furthermore, we assessed the quality of the methylation computations by determining whether the methylation beta value distributions deviated from a gold standard (corSampleVSgoldstandard > 0.80) [102,104]. All 414 samples passed the criterion at >0.90.
2.5. Genetic Analysis
From the isolated DNA of the 414 participants, the University of Valencia conducted high-density genotyping utilizing the Infinium OmniExpress-24 v1.2 BeadChip genotyping array, which captured 713,599 markers (Illumina Inc., San Diego, CA, USA). The genotyping was performed following the manufacturer’s methodology and adhering to previously stated quality criteria [106]. The Genome Studio genotyping module (Illumina, Inc.) was utilized for allele identification and genotype determination. Using conventional analysis pipelines implemented in the Python programming language with the Numpy library modules and PLINK [107,108], cleaning of data was performed. Single nucleotide polymorphisms (SNPs) non-mapped on autosomal chromosomes, SNPs with a minor allele frequency (MAF) of less than 0.01, SNPs that differed from the anticipated Hardy–Weinberg equilibrium with a p-value less than 1.0 × 10−4, and SNPs with a call rate lower than 90% were excluded [106]. After genotyping, we employed two approaches for genetic analysis. In the first, we selected directly measured SNPs in candidate genes previously linked to TL across several GWAS [109]. In the second approach, we explored the association between the genome-wide measured SNPs and DNAmTL in this population. In addition, we conducted post-GWAS analyses utilizing FUMA (Functional Mapping and Annotation) [110] and FORGE2 [111,112] tools.
2.6. Statistical Analyses
First, descriptive statistical analyses were conducted. Proportions were compared using Chi-square testing. The mean values of continuous variables were compared using Student’s t-tests and ANOVA testing. The concentrations of triglycerides were subjected to a logarithmic transformation in order to carry out statistical testing. The association analysis involved several regression models depending on the variable characteristics. We analyzed the raw association between DNAmTL (as continuous) and chronological age at time of blood collection in the whole population and by sex using linear regression. Scatter plots were displayed. Several multivariate regression models were gradually fitted to account for potential confounders, including sociodemographic, clinical, anthropometric and lifestyle variables and DNAmTL as a continuous variable. When considering two categories of TL based on the DNAmTLadjAge, we also employed logistic regression. We placed negative DNAmTLadjAge values in the category of shorter than expected TL based on age, and we placed positive values in the opposite category (longer than expected). As specified in the corresponding analyses, multivariate logistic models were fitted sequentially. The association between SNPs in candidate genes previously associated with TL in multiple GWAS [83] and DNAmTL was analyzed using sex- and age-adjusted linear regression. The genotypes were analyzed using an additive genetic model that considered the minor allele for each SNP. Likewise, the association between DNAmTL and the methylation beta values of each CpG site included in the estimator of Lu et al. [46] was analyzed by multivariate lineal regression models.
Adherence to the MedDiet was examined as continuous as well as categorical when indicated. We also tested the associations with the specific foods of the MedDiet score and derived novel sub-scores based on the results. Statistical analyses were conducted for the entire population and stratified per sex. Additionally, the sex heterogeneity was tested by computing the statistical significance of the interaction term between sex and the variable of adherence to the MedDiet in the corresponding model described in the results.
For the exploratory GWAS to identify genes and SNPs associated with DNAmTL in this population, we fitted general linear models in PLINK [107,108]. Each SNP at the genome-wide level was considered in accordance with an additive pattern, and multivariable adjustment for sex, age and other variables specifically indicated in each model was undertaken. Regression coefficients for the minor allele for each SNP were estimated. For the SNP-based GWAS, we used the conventional threshold of p < 5 × 10−8 for genome-wide statistical significance. Likewise, SNPs with p-values below 1 × 10−5 were also considered suggestive of genome-wide significance. Despite these general considerations, as there are very few GWAS studies that analyzed DNAmTL as the outcome, some tables show SNPs with p-values above this threshold for meta-analysis. A quantile–quantile plot (Q-Q plot) comparing the expected and observed p-values was performed in the R-statistical environment [113], and we used R qqman R library to create Manhattan plots. Despite the fact that all participants were white Caucasians, and no ethnic bias or population stratification was anticipated, we computed the genomic inflation factor (lambda coefficient). Likewise, we adjusted for the top of the principal components derived in the population stratification analysis as previously reported [106]. We used LocusZoom.js to generate locus-specific graphical displays of the position of the selected SNPs in the GWAS to nearby genes and local recombination hotspots [114] as well as to indicate the linkage disequilibrium (LD). In addition to the SNP-based GWAS, we carried out a gene-based GWAS with FUMA [110]. This analysis considers the aggregate effect of multiple SNPs in a single test instead of analyzing each SNP separately [110]. The FUMA platform uses MAGMA (Multi-marker Analysis of GenoMic Annotation) [115] as well as the annotation of the selected genes in biological context [110,116]. In addition, we used FORGEdb, a resource of genomic annotations and an integrated score of functional tools [111,112], to rank the selected loci into biological insight. Finally, we explored some gene–MedDiet interactions in determining DNAmTL. Adherence to the MedDiet was considered as dichotomous as previously reported [92,117]. We chose “a priori” the lead candidate gene SNP for TL, associated with DNAmTL in this population, and examined the significance of the SNP–MedDiet interaction term in a hierarchical multivariable regression model. We also conducted an exploratory genome-wide SNP–MedDiet interaction [117] on DNAmTL. For analyses not involving genome-wide associations, a two-sided p-value 0.05 was considered statistically significant.
3. Results
3.1. Participants Characteristics
Participants were 414 individuals (184 men and 228 women) recruited for the PREDIMED-Plus Valencia study and having high-quality DNA methylation data at the baseline visit. The demographic, clinical, anthropometric, and lifestyle characteristics are presented in Table 1.
Table 1.
Demographic, clinical and lifestyle characteristics of the study population according to sex.
| Total (n = 414) |
Men (n = 186) |
Women (n = 228) |
p | |
|---|---|---|---|---|
| Age at recruitment 1 (years) | 65.1 ± 0.2 | 63.8 ± 0.4 | 66.1 ± 0.3 | <0.001 |
| Age at the baseline visit 2 (years) | 65.7 ± 0.2 | 64.4 ± 0.4 | 66.7 ± 0.3 | <0.001 |
| Weight (Kg) | 84.3 ± 0.7 | 92.3 ± 1.0 | 77.8 ± 0.7 | <0.001 |
| BMI (Kg/m2) | 32.3 ± 0.2 | 32.2 ± 0.3 | 32.4 ± 0.2 | 0.440 |
| Waist circumference (cm) | 105.9 ± 0.5 | 110.9 ± 0.6 | 101.8 ± 0.6 | <0.001 |
| SBP (mm Hg) | 141.9 ± 0.9 | 143.7 ± 1.4 | 140.5 ± 1.2 | 0.076 |
| DBP (mm Hg) | 81.0 ± 0.5 | 82.6 ± 0.7 | 79.6 ± 0.6 | 0.002 |
| Total cholesterol (mg/dL) | 195.7 ± 1.8 | 188.1 ± 2.8 | 202.0 ± 2.3 | <0.001 |
| LDL-C (mg/dL) | 124.3 ± 1.5 | 121.5 ± 2.4 | 126.6 ± 1.9 | 0.096 |
| HDL-C (mg/dL) | 51.6 ± 0.6 | 47.3 ± 0.8 | 55.1 ± 0.7 | <0.001 |
| Triglycerides 3 (mg/dL) | 141.3 ± 2.9 | 139.0 ± 4.0 | 143.1 ± 4.2 | 0.488 |
| Fasting glucose (mg/dL) | 113.4 ± 1.4 | 113.7 ± 2.2 | 113.2 ± 1.8 | 0.875 |
| Physical activity 4 (MET·min/wk) | 1708 ± 78 | 1941 ± 133 | 1519 ± 89 | 0.007 |
| Adherence to MedDiet (17-I) 5 | 8.0 ± 2.8 | 7.9 ± 2.8 | 8.1 ± 2.7 | 0.210 |
| High adherence to MedDiet 6 (≥9) n (%) | 172 (41.5) | 75 (40.3) | 97 (42.5) | 0.648 |
| Educational level | <0.001 | |||
| Primary n (%) | 255 (61.6) | 92 (49.5) | 163 (71.5) | <0.05 |
| Secondary n (%) | 87 (21.0) | 55 (29.6) | 32 (14.0) | <0.05 |
| University n (%) | 72 (17.4) | 39 (21.0) | 33 (14.5) | <0.05 |
| Current smokers n (%) | 48 (11.6) | 30 (16.1) | 18 (7.9) | <0.001 |
| Never smokers n (%) | 188 (45.4) | 32 (17.2) | 156 (68.4) | <0.001 |
| Former smokers n (%) | 178 (43.0) | 124 (66.7) | 54 (23.6) | <0.001 |
| Type 2 diabetes n (%) | 164 (39.6) | 75 (40.3) | 89 (39.0) | 0.790 |
| On metformin n (%) | 128 (30.9) | 60 (32.3) | 68 (29.8) | 0.594 |
| On insulin n (%) | 21 (5.1) | 11 (5.9) | 10 (4.4) | 0.481 |
| Lipid-lowering drugs n (%) | 274 (66.2) | 125 (67.2) | 149 (65.4) | 0.692 |
| Blood pressure medication n (%) | 327 (79.0) | 149 (80.1) | 178 (78.1) | 0.613 |
Values are mean ± SE for continuous variables and number (%) for categorical variables. BMI: body mass index; SBP: systolic blood pressure; DBP: diastolic blood pressure; LDL-C: low-density lipoprotein cholesterol; HDL-C: high-density lipoprotein cholesterol; p: p-value for the comparisons (means or %) between men and women. Student’s t test was used to compare means and Chi-squared tests were used to compare categories. 1 Age in years at recruitment according to the inclusion criteria for age. 2 Age in years at the baseline visit when the blood sample for methylation analysis was drawn. 3 Triglycerides was ln-transformed for statistical testing. 4 Total physical activity was computed. 5 Quantitative 17-item (17-I) questionnaire for adherence to the Mediterranean diet (MedDiet). 6 High adherence to MedDiet ≥ 9 points in the 17-I score.
They were elderly (mean age 65.7 ± 0.2 years at the baseline visit) individuals with metabolic syndrome. We presented the mean age at the recruitment visit in addition to the age at the baseline visit after a run-in period included in the study protocol [88]. During the baseline visit, DNA methylation analysis and all other measurements were performed. The mean BMI was high (32.3 ± 0.2 kg/m2) and did not differ significantly between sexes. There were no sex differences in fasting glucose, medication use, or adherence to the MedDiet. Nonetheless, some variables, including age, total HDL-C, physical activity, educational level, and tobacco use, varied by sex. In the whole population, the prevalence of current, former, and never smokers was 11.6%, 43.0%, and 45.4%, respectively. In addition to the quantitative variable (17 points score) for adherence to the MedDiet, we considered a dichotomous variable indicating high (≥9 points) or low adherence (<9 points) to the MedDiet. No significant differences (p = 0.648) in prevalence were detected in men (40.3% high adherence) versus women (42.5% high adherence).
3.2. Descriptive Measures of DNAmTL in this Population
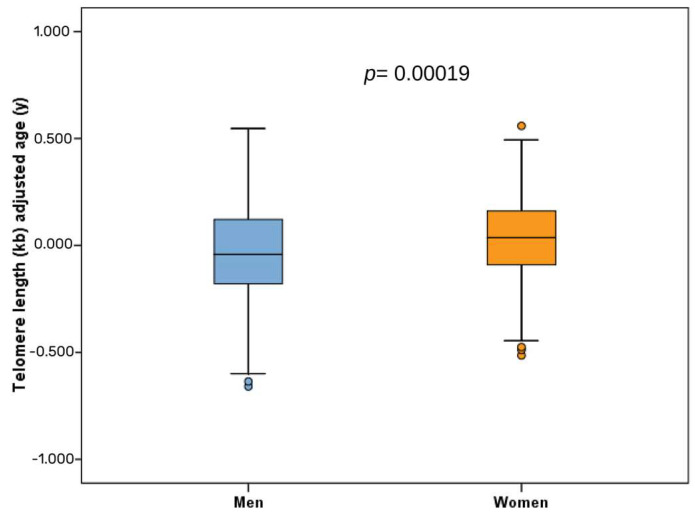
The DNAmTL (raw data) for the whole population was 6.6958 ± 0.0172 kilobases (kb) (Table 2). This estimator possesses the same units (kb) as that of the mean measured TL by the gold standard method of TRF [46]. DNAmTL (raw data) was statistically higher in women than in men (p = 0.025). Moreover, when DNAmTL was adjusted for age (referred to as DNAmTLAdj/Age), this difference by sex was more prominent (p < 0.001) (Table 2). From the calculated DNAmTLAdj/Age, we obtained the number of subjects having shorter TL. A negative value of DNAmTLAdj/Age would indicate DNAmTL that is shorter than expected (53.8% in men versus 41.2% in women; p = 0.013), while a positive value would indicate higher DNAmTL than expected based on age. We also presented in Table 2 another descriptive measure of epigenetic age acceleration (IEAA Hannum) in this population. The mean value of this biomarker was positive in men (indicating higher biological age than the chronological age), whereas a negative mean value for IEAA was found in women, indicating lower biological age than expected (p = 0.011).
Table 2.
Leukocyte telomere length–DNA methylation (DNAmTL), DNAmTL adjusted for age (DNAmTLAdjAge) and intrinsic epigenetic age acceleration (IEAA), based on Hannum’s calculation, per sex.
| Total (n = 414) |
Men (n = 186) |
Women (n = 228) |
p | |
|---|---|---|---|---|
| DNAmTL (Kb) | 6.6958 ± 0.0107 | 6.6693 ± 0.0172 | 6.7174 ± 0.0133 | 0.025 |
| DNAmTLAdjAge 1 (Kb/year) | −0.0018 ± 0.0101 | −0.0431 ± 0.0162 | 0.0321 ± 0.0122 | <0.001 |
| Subjects with shorter TL 2, n (%) | 194 (46.9) | 100 (53.8) | 94 (41.2) | 0.011 |
| IEAA Hannum 3 | −0.0003 ± 0.238 | 0.6540 ± 0.3567 | −0.5364 ± 0.3153 | 0.013 |
Values are mean ± SE for continuous variables and number (%) for categorical variables. DNAmTL: telomere length–DNA methylation; Kb: kilobases; p: p-value for the comparisons (means or %) between men and women. TL: telomere length. 1 DNAmTL adjusted for age. 2 Subjects with shorter telomere length having negative values for the computed DNAmTLAdjAge. 3 Intrinsic epigenetic age acceleration (IEAA) based on Hannum’s calculation.
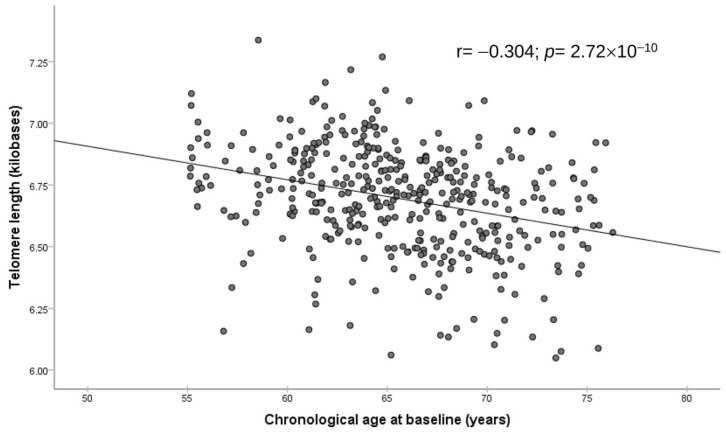
Figure 1 displays the correlation between chronological age at baseline in the whole population (age at the time of blood collection) and DNAmTL (raw values). We observed an inverse association (r = −0.304; p = 2.72 × 10−10). The corresponding regression coefficient indicated that DNAmTL shortened by 0.014 kb per year (95%CI: −0.018 to −0.009; p = 2.72 × 10−10).
Figure 1.
Association between leukocyte telomere length–DNA methylation and chronological age in the whole population (n = 414). Scatter plots with raw values, Pearson correlation coefficient and p-value.
Similar inverse correlations were observed when men and women were analyzed separately (Figure S1, panels A and B). Figure 2 shows the boxplots of DNAmTLAdj/Age in men and women.
Figure 2.
Boxplot of telomere length–DNA methylation adjusted for age (DNAmTLAdjAge) in men and women (n = 414). Raw data of the age-adjusted variable (DNAmTLAdjAge) per sex and the corresponding p-value for the comparison of means between men and women.
Moreover, we detected an expected inverse association between DNAmTL and the other biomarker of age acceleration, the IEAA-Hannum (r = −0.465; p = 1.5 × 10−23 for DNAmTL and r = −0.307; p = 1.9 × 10−10 for DNAmTLAdj/Age), adding validity to this TL estimator.
Next, we analyzed the site-specific association between the 140 CpGs contributing to the estimator [46] and the derived DNAmTL in this population. Table 3 displays the 10 most statistically significant CpG sites associated with the global DNAmTL.
Table 3.
Association between the 10 most significant (top-ranked) methylation sites included in the leukocyte DNAmTL computation algorithm and the estimated DNAmTL in this population.
| CpG Site 1 | Gene Symbol | Chr | BP | p 2 | r 3 |
|---|---|---|---|---|---|
| cg21566642 | NCAPG | 4 | 17813558 | 5.80 × 10−20 | 0.43869 |
| cg14391737 | CCND3 | 6 | 41904398 | 2.25 × 10−16 | 0.39760 |
| cg01940273 | TPI1 | 12 | 6977747 | 1.92 × 10−15 | 0.38594 |
| cg17739917 | DHRS3 | 1 | 12664243 | 4.70 × 10−15 | 0.38093 |
| cg21911711 | DNTT | 10 | 98062687 | 1.81 × 10−12 | −0.34522 |
| cg00475490 | YPEL3 | 16 | 30106682 | 1.02 × 10−10 | −0.31819 |
| cg24859433 | TPST1 | 7 | 65817282 | 4.23 × 10−9 | 0.29048 |
| cg18110140 | CCDC102B | 18 | 66389420 | 4.82 × 10−9 | 0.28946 |
| cg09935388 | SRRM3 | 7 | 75897850 | 7.19 × 10−9 | 0.28630 |
| cg25648203 | C7orf41 | 7 | 30185776 | 9.24 × 10−9 | −0.28430 |
Chr: chromosome; BP: base position in the chromosome (Homo Sapiens GRCh37.p13 genome build). 1 Supplementary Table S2 shows a more complete list of the methylation sites ordered by p-value in this population and also the direct or inverse effect in each site in the original work of Lu et al., 2019 [46]. 2 p-value obtained in this population between the corresponding CpG site and the estimated global DNAmTL (n = 414). Models were adjusted for sex, age, diabetes, body mass index, batch effect and leukocyte cell-types. 3 Partial correlation coefficient in the multivariate adjusted model (described above) for the corresponding methylation site and the global DNAmTL.
As specified, models were multivariable adjusted, and the partial correlation coefficient for each CpG site indicated an inverse or direct correlation between methylation at that site and DNAmTL. The most significant CpG associated with DNAmTL was located on chromosome 4 within the NCAPG (Non-SMC Condensin I Complex Subunit G) gene. Hypermethylation of this site was associated with longer DNAmTL. Likewise, hypermetilation at cg14391737 in the CCND3 (Cyclin D3) gene and at the cg01940273 in the TPI1 (Triosephosphate Isomerase 1) gene were also associated with longer DNAmTL. Table S2 provides additional information on the top-ranked CpGs, including other CpG sites associated with DNAmTL.
3.3. Associations between SNPs in Candidate Genes Related to TL and DNAmTL
Here, we tested the association between SNPs in candidate genes previously associated with telomere length [83] and the estimated DNAmTL in this population. Table 4 shows these associations sorted by statistical significance and including the 20 most significant SNPs.
Table 4.
Association between SNPs in candidate genes for telomere length reported in the literature and the DNAmTL in this population.
| Gene Symbol |
Chr | SNP | BP | MAF | Beta | p 1 |
|---|---|---|---|---|---|---|
| TERT | 5 | rs2075786 | 1266310 | 0.326 | −0.0407 | 0.0078 |
| TERF2 | 16 | rs3785073 | 69401937 | 0.238 | −0.0424 | 0.0086 |
| TERF2 | 16 | rs3743669 | 69398353 | 0.197 | −0.0390 | 0.0179 |
| RTEL1 | 20 | rs6011011 | 62299578 | 0.140 | 0.0551 | 0.0193 |
| TERT | 5 | rs2736100 | 1286516 | 0.472 | −0.0328 | 0.0274 |
| TERF2 | 16 | rs9939870 | 69396585 | 0.315 | −0.0356 | 0.0277 |
| RTEL1 | 20 | rs6011002 | 62297802 | 0.092 | 0.0498 | 0.0375 |
| DCAF4 | 14 | rs17781739 | 73392839 | 0.182 | −0.0309 | 0.0376 |
| RTEL1 | 20 | rs3848668 | 62293272 | 0.055 | 0.0513 | 0.0390 |
| DCAF4 | 14 | rs12885397 | 73408134 | 0.294 | 0.0324 | 0.0545 |
| POT1 | 7 | rs17246404 | 124462661 | 0.222 | −0.0288 | 0.0677 |
| SENP7 | 3 | rs9870022 | 101071429 | 0.215 | −0.0346 | 0.0696 |
| ATM | 11 | rs3218674 | 108115587 | 0.012 | 0.1189 | 0.0696 |
| OBFC1 | 10 | rs3814219 | 105647095 | 0.199 | 0.0282 | 0.0739 |
| PARP1 | 1 | rs8679 | 226548554 | 0.108 | 0.0297 | 0.0780 |
| PARP1 | 1 | rs2271347 | 226549498 | 0.108 | 0.0293 | 0.0804 |
| SENP7 | 3 | rs6772703 | 101232736 | 0.282 | 0.0267 | 0.0831 |
| TERF2 | 16 | rs251796 | 69395434 | 0.196 | 0.0274 | 0.0942 |
| ATM | 11 | rs1800058 | 108160350 | 0.022 | 0.0900 | 0.0951 |
| ZNF208 | 19 | rs12608935 | 22145147 | 0.351 | −0.0236 | 0.0986 |
Chr: chromosome; SNP: single nucleotide polymorphism. BP: base position in the chromosome (Homo Sapiens GRCh37.p13 genome build); MAF: minor allele frequency; Beta: regression coefficient indicates the effect for the minor allele on DNAmTL. Models adjusted for sex and age. 1 p-value obtained in the regression models adjusted for sex and age for each SNP using a genetic additive approach (n = 414).
The corresponding regression coefficients (beta) refer to the least common allele, and models were adjusted for sex and age. The most significant SNP was rs2075786 in the TERT gene (telomerase reverse transcriptase) with p = 0.008. The TL in carriers of the minor allele was 0.0407 kb shorter (per allele) compared to the homozygote for the frequent allele. The second most significant SNP was rs3785073 located in the TERF2 gene (telomeric repeat binding factor 2) with p = 0.0086. For this SNP, the minor allele was also associated with a shorter TL. Statistically significant results have also been obtained for SNPs in the RTEL1 (regulator of telomere elongation helicase 1) and DCAF4 (DDB1 and CUL4 associated factor 4) genes. These significant associations with genes associated with TL contribute to increase the validity of the DNAmTL estimator derived in this population.
In addition, we computed a genetic risk score (GRS) by aggregating the effects of the four candidate genes that were found to be statistically significant. The most significant SNP variants of each gene were chosen for the GRS (TERT-rs2075786, TERF2 rs3785073, RTEL1-rs6011011, and DCAF4-rs17781739) with only one SNP per gene examined. The effect allele linked to a shortened DNAmTL was accounted for when calculating the score. The GRS ranged from 1 to 7. However, the prevalence of the scores 1 and 7 was low. Then, scores 1 and 2 were computed in the same category. Likewise, scores 6 and 7 were considered together. This GRS was strongly associated with the DNAmTL (p < 0.001). Figure S2 shows the association of this score and DNAmTLAdj/Age in a model additionally adjusted for diabetes and BMI.
3.4. Association between Adherence to the MedDiet and Shorter DNAmTL
Following the genetic validation of the DNAmTL estimator, we examined the association between adherence to the MedDiet (as a continuous variable) and DNAmTL, taking into account two distinct categories: subjects whose TL was shorter than expected and subjects whose TL was longer than expected (Table 5). Odds (OR and 95% CI) of having a shortened TL associated with a one-point increase in the MedDiet 17-I score were calculated utilizing multivariable logistic regression. Three multivariable-adjusted models were fitted for the whole population and by sex as indicated. Greater adherence to the MedDiet was associated with a decreased likelihood of having a shorter TL for the entire population (OR = 0.92; 95%CI: 0.86–0.99; p = 0.032 in model 1). In the fully adjusted model (model 3), this association remained statistically significant (p = 0.049).
Table 5.
Association between adherence to the Mediterranean diet (MedDiet) and shorter leukocyte telomere length 1 in the whole population and by sex.
| Total (n = 414) OR (95% CI) |
p | Men (n = 186) OR (95% CI) |
p | Women (n = 228) OR (95% CI) |
p | |
|---|---|---|---|---|---|---|
| Adherence to MedDiet (17-I) 2 | ||||||
| Model 1 3 | 0.92 (0.86–0.99) | 0.032 | 0.96 (0.87–1.07) | 0.444 | 0.89 (0.81–0.98) | 0.028 |
| Model 2 4 | 0.93 (0.86–0.99) | 0.034 | 0.96 (0.87–1.07) | 0.470 | 0.89 (0.81–0.99) | 0.031 |
| Model 3 5 | 0.93 (0.85–0.99) | 0.049 | 0.99 (0.86–1.11) | 0.904 | 0.88 (0.79–0.98) | 0.023 |
OR: odds ratio, indicates the effect for the minor allele on adherence to the Mediterranean diet (MedDiet) and telomere length; CI: confidence interval. Values are OR and 95% CI of having shorter telomere length for a 1 point increase in adherence to MedDiet. 1 Subjects with shorter telomere length are those subjects having negative values for the computed DNAmTLAdjAge (n = 194) (see Table 2). 2 Quantitative 17-item (17-I) questionnaire for adherence to MedDiet. 3 Model 1 adjusted for sex and age. 4 Model 2 adjusted for sex, age, diabetes, and body mass index (BMI). 5 Model 3 adjusted for sex, age, diabetes, BMI, metformin, insulin, lipid-lowering drugs, hypertension medication, systolic blood pressure, education, smoking, and physical activity.
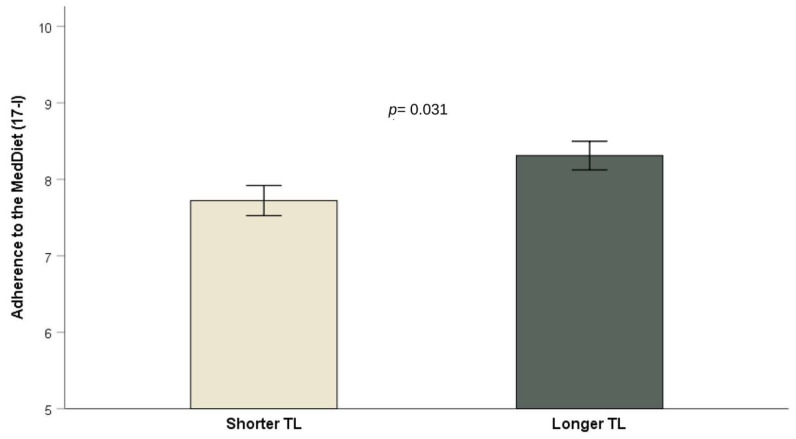
The association, however, only attained statistical significance among women (OR = 0.88; 95%CI: 0.79–0.98; p = 0.023 for model 3) in the stratified analysis by sex. As shown in Figure 3, individuals with shorter TL exhibited lower levels of adherence to the MedDiet score when compared to those with longer TL (p < 0.05).
Figure 3.
Adherence to Mediterranean diet (means ± SE) in subjects with shorter (n = 194) and longer (n = 220) TL. p = 0.031 for difference of means in a model adjusted for sex and age. p = 0.034 for difference of means in a model additionally adjusted for diabetes and BMI. p = 0.044 in the model additionally adjusted for metformin, insulin, lipid-lowering drugs, hypertension medication, systolic blood pressure, education, smoking, and physical activity. Error bars: SE of means.
3.5. Association between Global Adherence to the MedDiet and Consumption of Particular Items with DNAmTL (as a Continuous Variable)
Furthermore, we examined the association between adherence to the MedDiet and DNAmTL as a continuous variable. Table 6 shows the results in the whole population and stratified per sex.
Table 6.
Association between adherence to the Mediterranean diet (MedDiet) and telomere length (DNAmTL) by sex, considering the whole pattern and the score of the 4 MedDiet items (fruit, fish, “sofrito” and whole grains).
| Total (n = 414) |
p | Men (n = 186) |
p | Women (n = 228) |
p | |
|---|---|---|---|---|---|---|
| β (SE) | β (SE) | β (SE) | ||||
| Adherence to MedDiet (17-I) 1 | ||||||
| Model 1 2 | 0.006 (0.004) | 0.095 | −0.003 (0.006) | 0.588 | 0.014 (0.005) | 0.002 |
| Model 2 3 | 0.006 (0.004) | 0.113 | −0.003 (0.006) | 0.648 | 0.014 (0.005) | 0.003 |
| Model 3 4 | 0.005 (0.004) | 0.147 | −0.006 (0.006) | 0.332 | 0.015 (0.005) | 0.003 |
| High adherence to MedDiet (2 categories) 5 | ||||||
| Model 1 2 | 0.042 (0.020) | 0.041 | 0.001 (0.033) | 0.994 | 0.077 (0.025) | 0.003 |
| Model 2 3 | 0.040 (0.020) | 0.048 | 0.003 (0.034) | 0.922 | 0.075 (0.026) | 0.004 |
| Model 3 4 | 0.036 (0.021) | 0.080 | −0.016 (0.034) | 0.637 | 0.078 (0.026) | 0.004 |
| Score of 4 MedDiet items 6 | ||||||
| Model 1 2 | −0.237 (0.055) | <0.001 | −0.204 (0.103) | 0.048 | −0.295 (0.065) | <0.001 |
| Model 2 3 | −0.233 (0.055) | <0.001 | −0.207 (0.104) | 0.047 | −0.290 (0.066) | <0.001 |
| Model 3 4 | −0.217 (0.055) | <0.001 | −0.133 (0.105) | 0.210 | −0.289 (0.067) | <0.001 |
Values are regression coefficients (expressed as kb per unit of adherence variable) and SEs. 1 Quantitative 17-item (17-I) questionnaire for adherence to Mediterranean diet (MedDiet). 2 Model adjusted for sex and age. 3 Model adjusted for sex, age, diabetes, and body mass index (BMI). 4 Model adjusted for sex, age, diabetes, BMI, metformin, insulin, lipid-lowering drugs, hypertension medication, systolic blood pressure (SBP), education, smoking, and physical activity. 5 High adherence to MedDiet ≥ 9 points in the 17-I score compared with low adherence. 6 Score 4: The 4 MedDiet items (fruit, fish, “sofrito” and whole grains) most significantly associated with TL in women (see Supplemental Material). Estimators comparing category 1 (zero adherence) with category 5 (0 to 4 points) are shown.
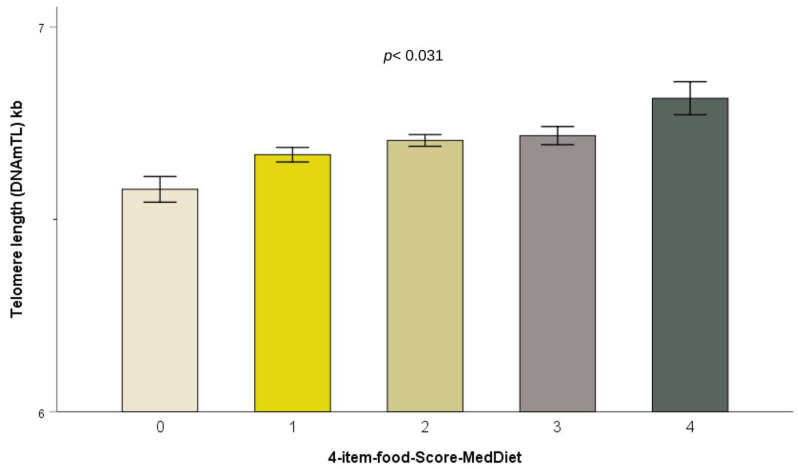
In women, the global score of adherence to the MedDiet (17-I) was found to be significantly associated with longer DNAmTL (p = 0.002 in Model 1 after adjusting for age, and this association persisted in multivariate Model 3 after controlling for age, diabetes, BMI, metformin, insulin, lipid-lowering drugs, hypertension medication, SBP, education, smoking, and physical activity). We found, according to Model 3 in women, that a one-point increase in the MedDiet adherence score resulted in a 0.015 ± 0.0005 kilobases increase in DNAmTL length. In the case of males, no statistically significant association was found. MedDiet was also evaluated as a dichotomous variable. In women, there was a significant association between higher adherence to the MedDiet (score ≥ 9) and longer DNAmTL (0.078 ± 0.026; p = 0.004) compared to lesser adherence. Although the MedDiet pattern is more important as a whole, we examined the particular foods/items of the MedDiet score. Table S3 shows the compliance for each item by sex according to the criteria for adherence to the MedDiet specified in Table S1. Table S4 shows the statistical significance of the associations between each one of the 17 MedDiet items and DNAmTL by sex. In women, we detected statistically significant associations (p < 0.05) with daily fruit intake, fish intake, “sofrito” and whole grain intake. Compliance with the criteria predefined for adherence to the MedDiet for each item was associated with significantly longer DNAmTL. These associations remained statistically significant even after multivariate adjustment in Model 3. We then calculated a new MedDietFood score by adding these four items. The 4-MedDietFood score ranged from 0 to 4 points. Table 6 shows the association between this score and DNAmTL in the whole population and by sex. Beta coefficients for category 1 (zero points) in comparison with category 5 (4 points) were computed. We observed a reduction in DNAmTL in subjects who did not adhere to the four food items as compared to those who adhered completely; results were statistically significant for women and for the whole population in Model 3. In men, Models 1 and 2 were also statistically significant. Therefore, we considered this score for the whole population in further analyses. Figure 4 shows DNAmTL according to the score obtained in the 4-MedDietFood item variable in the whole population.
Figure 4.
Telomere length (DNAmTL) depending on the adherence to the 4-MedDietFood score ranging from 0 to 4. Values are adjusted mean ± SE. p < 0.001 for the lineal trend of the 4-MedDietFood score in Models 1, 2 and 3. Error bars: SE of means.
3.6. Exploratory GWASs for DNAmTL
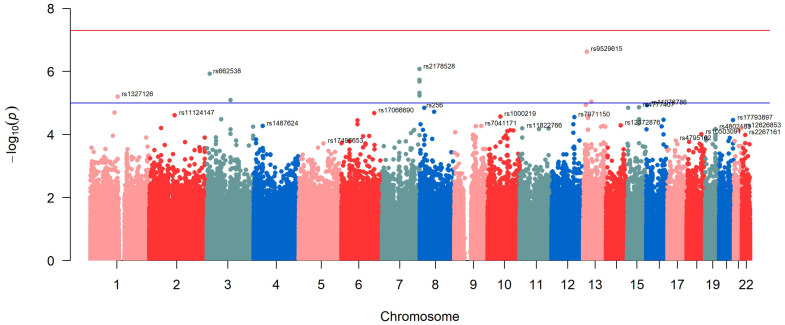
To examine the SNPs associated with the DNAmTL in this population, we initially performed an exploratory SNP-based GWAS. The Manhattan plot corresponding to the model that was adjusted for age and gender is depicted in Figure 5. Figure S3 displays the Q-Q plot for this GWAS as well as the lambda parameter (lambda = 1.004), indicating no statistical inflation or population stratification bias. Ten SNPs surpassed the suggestive level of significance for GWAS (p < 1 × 10−5). Nevertheless, we were unable to identify any SNPs that were significantly associated at the genome-wide level (p < 5 × 10−8).
Figure 5.
Manhattan plot of the GWAS of leukocyte telomere length–DNA methylation using a gene-based approach (adjusted for sex and age). The red line represents the threshold (−log10(5 × 10−8)). The blue line represents the threshold (−log10(1 × 10−5)).
Table 7 shows the details corresponding to the most significantly associated SNPs with DNAmTL in Figure 5, ranked by p-values.
Table 7.
Top-ranked SNPs in the GWAS for DNAmTL in the whole population. Model adjusted for sex and age.
| Chr | SNP | BP | Beta | p 1 | MAF | Gene Symbol |
FORGEdb Score |
|---|---|---|---|---|---|---|---|
| 13 | rs9529615 | 35307066 | 0.07256 | 2.33 × 10−7 | 0.32708 | intergenic | 5 |
| 7 | rs2178528 | 158393488 | −0.07369 | 8.30 × 10−7 | 0.24521 | intergenic | 6 |
| 3 | rs662538 | 13135500 | −0.13220 | 1.18 × 10−6 | 0.02236 | IQSEC1 | 8 |
| 7 | rs11983468 | 158412279 | −0.07504 | 1.86 × 10−6 | 0.19868 | intergenic | 7 |
| 7 | rs11763040 | 158416203 | −0.07571 | 2.06 × 10−6 | 0.20367 | intergenic | 8 |
| 7 | rs1466210 | 158444975 | −0.07824 | 4.78 × 10−6 | 0.19848 | NCAPG2 | 8 |
| 7 | rs7788516 | 158410592 | −0.07127 | 5.85 × 10−6 | 0.20667 | intergenic | 8 |
| 1 | rs1327126 | 115973920 | 0.06324 | 6.27 × 10−6 | 0.39677 | intergenic | 6 |
| 3 | rs7613610 | 100499321 | 0.08068 | 8.09 × 10−6 | 0.27756 | ABI3BP | 7 |
| 13 | rs12427518 | 53519692 | 0.13080 | 9.12 × 10−6 | 0.22903 | intergenic | 10 |
| 13 | rs17504027 | 30587544 | 0.14460 | 1.15 × 10−5 | 0.03015 | LICO00544 | 4 |
| 16 | rs11076786 | 3811596 | 0.10400 | 1.16 × 10−5 | 0.14018 | CREBBP | 8 |
| 15 | rs4777407 | 71826919 | −0.06769 | 1.37 × 10−5 | 0.35144 | THSD4 | 4 |
| 15 | rs4906654 | 26274113 | 0.09824 | 1.42 × 10−5 | 0.25240 | LOC100128714 | 6 |
| 8 | rs256 | 19811967 | 0.08308 | 1.42 × 10−5 | 0.14417 | LPL | 6 |
| 8 | rs7837548 | 61802242 | 0.07462 | 1.89 × 10−5 | 0.27736 | intergenic | 5 |
| 1 | rs11164337 | 102389733 | −0.07336 | 2.01 × 10−5 | 0.46765 | OLFM3 | 4 |
Chr: chromosome; SNP: single nucleotide polymorphism. BP: base position in the chromosome (Homo Sapiens GRCh37.p13 genome build). Beta: regression coefficient indicates the effect for the minor allele on DNAmTL; MAF: minor allele frequency. 1 p adjusted for sex and age for each SNP using a genetic additive approach.
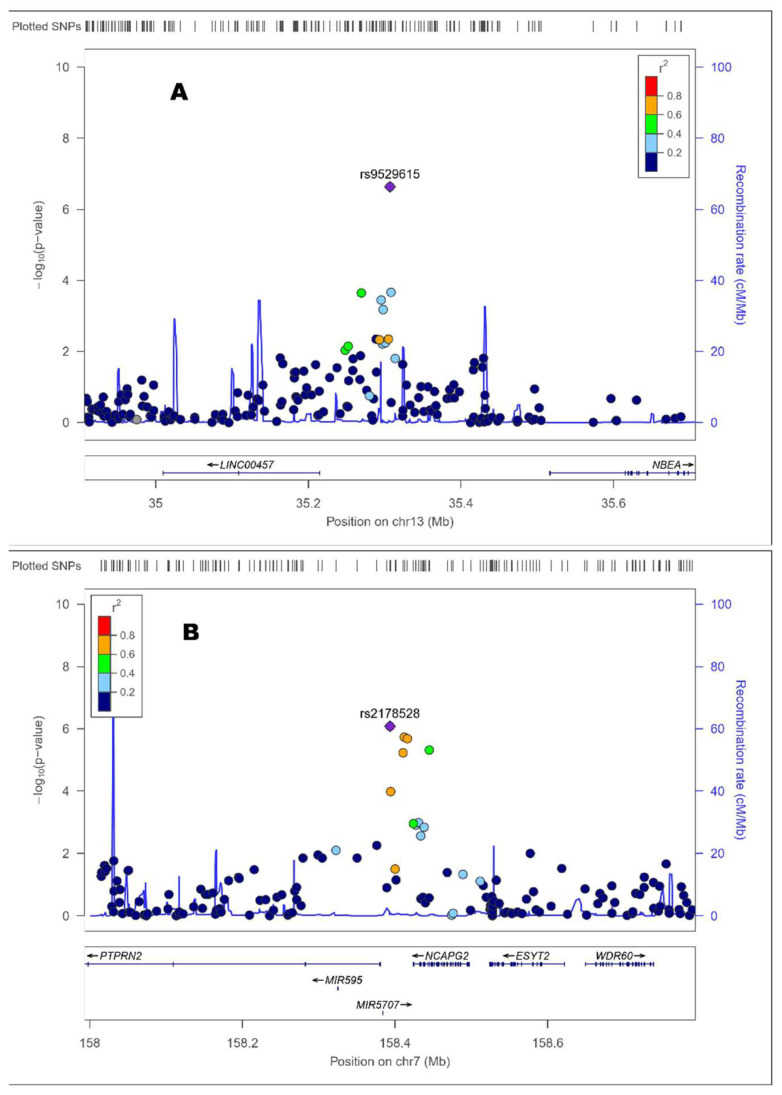
The hit was the rs9529615 SNP located in chromosome 13 at an intergenic position. The minor allele of this SNP was associated with higher DNAmTL (beta: 0.0726; p = 2.3 × 10−7). Figure 6 (panel A) shows the Zoom plot corresponding to the rs9529615 SNP intergenic in chromosome 13. The second most significant SNP was the rs2178528 in chromosome 7. The minor allele for this SNP was associated with shorted DNAmTl (beta: −0.0737; p = 8.3 × 10−7).
Figure 6.
Zoom plots of the selected top-ranked SNPs for the GWAS of DNAmTL in this population (Table 5). The panel shows the following SNPs: (A) rs9529615 (Chr.: 13; p = 2.33 × 10−7); (B) rs2178528 (Chr.: 7; p = 8.30 × 10−7).
The Zoom plot (Figure 6, panel B) shows that this intergenic SNP is located near the PTPRN2 (protein tyrosine phosphatase receptor type N2) gene and near the NCAPG2 (Condensin-2 complex subunit G2) gene. Among the other 10 top-ranked SNPs surpassing the suggestive level for GWAS significance, we detected SNPs in the IQSEC1 (IQ Motif and Sec7 Domain ArfGEF 1), the NCAPG2 and the ABI3BP (ABI Family Member 3 Binding Protein) genes. Figure S4 presents the Zoom plots for the selected intergenic top-ranked SNPs: Panel A (rs11983468 in chromosome 7); Panel B (rs1327126 in chromosome 1); and Panel C (rs12427518 in chromosome 13). Table 7 also shows the FORGEdb Score for each SNP to rank the most significantly associated loci into biological insight. This score ranging from 0 to 10 provides information on associated regulatory elements, transcription factor (TF) binding sites and target genes. The FORGEdb scores are computed based on the presence or absence of five different lines of evidence for regulatory function: (I) DNase I hotspot, marking accessible chromatin (2 points); (II) Histone mark ChIP-seq broadPeak (2 points); (III) TF motif (1 point) and Contextual Analysis of TF Occupancy (CATO) score, marking potential TF binding (1 point); (IV) Activity-by-contact (ABC) interaction, marking gene looping (2 points); and (V) Expression quantitative trait locus (eQTL), marking an association with gene expression (2 points).
Table S5 provides a detailed summary of the global FORGEdb scores computed for the 10 most statistically significant SNPs. In general, these SNPs reached good FORGEdb scores. Among them, we would like to outline the rs12427518 that reached the maximum score of 10, suggesting a large amount of evidence for the functional relevance of this intergenic SNP located between the OLFM4 (Olfactomedin 4) and the PCDH8 (Protocadherin 8) genes.
In addition, to better understand the functionality of these SNPs and related genes, we carried out a functional analysis with the FUMA tools as indicated in the Methods section. Figure S5 shows the gene expression heatmap for the top-ranked selected genes (IQSEC1, NCAPG2 and PTPRN2) using the GTEx V8 (54 tissue-types) dataset and showing the average expression per label (log2 transformed).
Additional adjustment of the model for additional potential confounders did not change significantly the list of the top-ranked SNPs. Table S6 presents the corresponding estimates in the GWAS for DNAmTL adjusted for sex, age, diabetes and BMI.
Next, we carried out a gene-based exploratory GWAS for DNAmTL in this population. Figure S6 displays the gene-based Manhattan plot for the corresponding GWAS. The model was adjusted for sex and age. The 14 most statistically significant genes were annotated. For a gene-based GWAS, the threshold 1 for statistical significance is set at (−log10(2.7 × 10−6)), considering the strict Bonferroni correction. Likewise, a threshold 2 (−log10(1 × 10−4)) at the suggestive level of statistical significance is considered. Several genes in our analysis neared the suggestive level of significance, but none achieved it.
However, we identified some genes, previously detected at the SNP level analysis, such as the ABI3BP or the THSD4 (thrombospondin type 1 domain containing 4). But most importantly, we detected as top-ranked several genes previously characterized as genes being associated with telomeres; these genes include ETHE1 (ETHE1 Persulfide Dioxygenase), BRCA2 (Breast Cancer Type 2 Susceptibility) and RFC2 (Replication Factor C Subunit 2) genes.
3.7. Exploratory Analysis of Gene–MedDiet Interactions in Determining DNAmTL
Finally, we explored the joint contribution of adherence to the MedDiet and the main genetic variants on DNAmTL to identify additive or interactive effects. First, we focused on candidate genes previously reported to be associated with TL (Table 4). In this exploratory analysis, we selected the SNP in candidate genes most statistically significantly associated with DNAmTL (rs2075786 in the TERT gene) and analyzed the interaction with adherence to the MedDiet (as dichotomous variable: low/high), analyzing the interaction term in a hierarchical model additionally adjusted for sex, age, BMI and diabetes. The interaction term did not reach the statistical significance (p = 0.428).
Focusing on the main effects, both the TERT SNP (p = 0.034) and the adherence to the MedDiet (p = 0.047) contributed independently and additively to the DNAmTL. The minor allele was associated with a decreased TL in comparison with the major allele. Thus, homozygous subjects for the minor allele had −0.087 ± 0.035 kb less in the mean DNAmTL than homozygous subjects for the major allele (p = 0.012). This difference can be reduced by a having a high adherence to the MedDiet (≥9 points). This variable was associated with an increase of 0.041 ± 0.020 kb in DNAmTL.
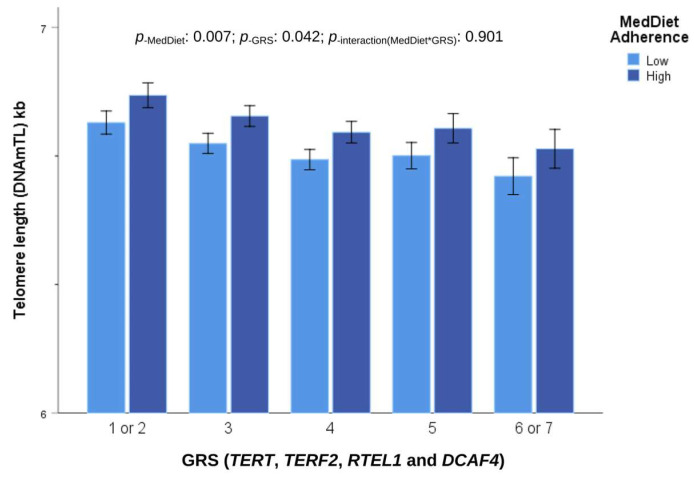
We also analyzed the gene–MedDiet interaction for the combined GRS including SNPs in the genes (TERT, TERF2, RTEL1 and DCAF4; see Figure S2) with adherence to the MedDiet as a dichotomous variable. In a model additionally adjusted for sex, age, BMI and diabetes, no statistical significance was found for the interaction term (p = 0.901), indicating an additive effect. In women, the main effects for adherence to the MedDiet were higher than in men. Figure 7 shows the joint additive effects of the GRS and adherence to the MedDiet on DNAmTL in women.
Figure 7.
Combined effect of adherence to Mediterranean diet (MedDiet), expressed as categories (low and high), and genetics using a genetic risk score(GRS) of TERT, TERF2, RTEL1 and DCAF4, with the effect allele associated with shorter telomere length on DNAmTL in women. Models adjusted for age, diabetes, and BMI. Error bars: SE of means. The symbol “*” means interaction between MedDiet and GRS.
The effect allele of the GRS was associated with reduced DNAmTL (resulting in −0.139 ± 0.055 kb in subjects having six or seven effect alleles in comparison with subjects having one or two effect alleles; p = 0.010). However, this genetic influence can be counteracted by a higher adherence to the MedDiet (associated with an increase in DNAmTL of 0.071 +/− 0.026 kb; p = 0.007).
Likewise, when we examined the gene–MedDiet interaction between the hit SNPs associated with DNDmTL in the GWAS (rs9529615-intergenic; see Table 5) in the same statistical model, we did not find a statistically significant interaction term (p = 0.994).
Furthermore, we explored at the genome-wide level the interaction term between the SNPs and adherence to the MedDiet (two categories) in the whole population. We did not find any gene–MedDiet interaction at the GWAS level of significance but obtained some interactions at the suggestive GWAS level of significance. Figure S7 shows the corresponding Manhattan plot for the statistical significance of the gene–MedDiet interactions terms in determining TL (DNAmTlAdjAge). Among the top-ranked SNPs involved in the gene–MedDiet interaction, there were some intergenic: one in chromosome 1 located in the PATJ (PATJ Crumbs Cell Polarity Complex Component) gene (rs11590158) and another in chromosome 5 located in the ADMTS16 (thrombospondin motifs 16) gene (rs16875241). These gene–diet interactions must be further characterized through additional research employing larger sample sizes.
4. Discussion
To date, one of the most often used biomarkers of aging in epidemiological studies examining risk factors for chronic diseases has been TL [21,30,31,32,33,34,35,36]. However, methodological limitations exist for measuring them in large-scale studies [43,44,45]. In this investigation, we have analyzed the association between adherence to the MedDiet and TL using the new method based on the methylation algorithms published by Lu et al. [46]. To our knowledge, this is the first study analyzing the relationship between adherence to the MedDiet and TL utilizing the DNAmTL estimator. Similarly, this research represents the first investigation of the association between variants at the genome-wide level and DNAmTL in a Mediterranean Spanish population. A few GWAS studies have been published on DNAmTL, such as an investigation including European Americans and African Americans [51], as well as a study focusing on participants recruited in Canada [109]. Considering that linkage disequilibrium and other population characteristics may affect the specific variants most significantly associated in GWASs, it is necessary to conduct studies on multiple populations to assess the level of consistency or heterogeneity in the findings [118]. Similar to the detection of genetic heterogeneity in GWASs, there is also an observed heterogeneity in DNA methylation research based on the ethnic background of the participants [119,120]. Hence, despite the validation and utilization of the DNAmTL estimator in many populations [46,47,48,49,50,51,121,122,123], it is important to obtain specific information related to the Spanish population. This study represents one of the first efforts in this regard. Even while we were unable to measure TL directly in our study, we did use several indicators to test the validity of the estimator DNAmTL in the analyzed subjects. We observed statistically significant correlations between DNAmTL and biological age in the whole population as well as in separate analyses conducted for both men and women. The correlations were relatively high and as expected according to the initial validation study for this estimator [46] using the “gold standard” method of the terminal restriction fragments measured by Southern blotting [44,45,46]. In their study, Lu et al. [46] found that the utilization of DNAmTL for TL assessment showed a stronger association with biological age compared to alternative approaches like qPCR [46]. This can be related to the high coefficient of variation (CV) reported for qPCR and related measurements in previous studies [44,45,123,124,125]. Similarly, in the current study, we observed statistically significant differences in TL according to sex. Specifically, women exhibited longer DNAmTL compared to men, which was expected for this estimator [46]. Another parameter that was assessed during our initial validation process was the IAEE for the Hannum epigenetic clock [103]. This biomarker quantifies the intrinsic epigenetic age acceleration and has shown associations with the incidence of chronic diseases and mortality [99,100,101]. In our population, a statistically significant inverse association was obtained between the estimated DNAmTL and the IAEE biomarker, reinforcing the validity. In addition, we conducted an analysis to examine the relationship between SNPs in candidate genes that have been previously linked to telomere length (TERT, TERF2, RTEL1, DCAF4, POT1, etc.) [109] with DNAmTL. Our findings revealed numerous anticipated statistically significant associations in the expected direction, increasing consistency. In general, despite the limitation of not being able to directly quantify TL in our population, it can be stated that the new biomarker of DNAmTL acts as a reliable estimator of TL in this Mediterranean population, as previously reported by researchers in other populations [46,50,51].
Telomere attrition is a phenomenon that arises from cellular replication and is expedited by diverse environmental conditions, including inflammation and oxidative stress [126,127,128]. Oxidative stress has been recognized as a physiological factor contributing to telomere shortening, which in turn is associated with human aging by several mechanisms [127,128,129,130,131,132,133,134]. Overall, it has been suggested that the repair of oxidative damage in telomeric DNA is comparatively less efficient than in other chromosome regions [126,135]. Additionally, the presence of antioxidants has been found to slow down the loss of telomeres associated with oxidative stress [136,137]. Further investigation in human subjects is necessary to better understand the specific mechanisms involved in vivo [137]. In general, it has been accepted that the process of telomere shortening can be lowered through the adoption of lifestyle choices that are linked to decreased levels of oxidative stress [126,135,138,139]. Focusing on diet, several studies have revealed an association between a healthy food pattern and longer telomeres [65,71,140]. The MedDiet has been reported to stimulate telomerase activity in peripheral blood mononuclear cells, and if combined with moderate exercise, this diet can improve endothelial microvascular and cardiorespiratory functions, which is important for both better health and increased life expectancy [141]. In the present investigation, we have found a statistically significant association between higher adherence to the MedDiet pattern and increased DNAmTL. The observed association showed a greater magnitude in women than men. Although this is the first study to analyze the association between the MedDiet and TL using the methylation algorithm of Lu et al. [46], prior research has already been conducted investigating the relationship between adherence to the MedDiet and TL using other measuring approaches (qPCR, qFISH, or terminal restriction fragment analysis) [54,60,61,63,64,67,70,71,72,73,74,75].
The findings from prior research examining the association between adherence to the MedDiet and LT have displayed notable variability. Multiple factors have played a role in this observation, encompassing the heterogeneity of the populations under examination, differences in the questionnaires employed to assess adherence to the MedDiet, and diverse methodologies employed for measuring telomere length as well as the study design [54,60,61,63,64,67,70,71,72,73,74,75,142]. Multiple studies conducted in populations outside of the Mediterranean region, such as a multiethnic elderly population [60], older Australian men and women [64], Chinese older men and women [69], or a middle-aged population in West Virginia [65], did not find any significant associations between the derived MedDiet pattern and TL. Nevertheless, a meta-analysis [72] that synthesized the findings from multiple studies concluded that a significant association exists between higher adherence to the MedDiet and increased TL, which was primarily observed in women. Our sex-specific analysis confirmed the stronger association between adherence to the MedDiet and TL in women compared to men. The magnitude of this sex difference was more pronounced when analyzing TL as a continuous variable rather than categorizing it into two categories based on shorter and longer TL. More studies are needed to better understand the factors determining these differences. Although in our case, the number of women studied (n = 228) is greater than that of men (n = 186), conferring a greater statistical power in the stratified analyses, it is rather modest, and additional factors must be considered in order to fully account for the observed effects. Previous studies examined men and women jointly [54,60,64,65] or included only women [61], so comparisons are difficult. Furthermore, our study focused on post-menopausal women and older men, thereby limiting our ability to generalize these findings to premenopausal women and young men. Nevertheless, in the National Health and Nutrition Examination Survey (NHANES) carried out in the United States, including data on 4758 men and women aged 20–65 years [142], the statistically significant positive association between the MedDiet score and TL was only observed in women, adding more evidence to the potential extension of the sex-specific differences to younger subjects. However, in a recent study undertaken in the UK Biobank participants [75], a significant direct association between adherence to the MedDiet and TL was observed both in men and women. Hence, there is a need for more targeted investigations that specifically examine the potential disparities between sexes in relation to this association.
Although it is important to take into account that the MedDiet pattern has been noted for its advantageous synergistic effect of the overall dietary pattern, wherein the combined impact of its individual components surpasses their individual contributions [11,60], some studies have specifically investigated the foods that exhibit the strongest association with TL in addition to conducting an overall analysis of the MedDiet effects. In this regard, the results exhibit a greater degree of heterogeneity than the global pattern analysis. Thus, in the Nurses’ Health Study analyzing 4676 women [61], although the authors observed that greater adherence to the MedDiet was associated with longer telomeres, upon further examination of specific food items, they concluded that none of the individual components showed a significant association with TL [61]. In our study, when we examined the specific components of the MedDiet, we observed four remarkable statistically significant associations in women. The relevant food items were fruits, fish consumption, “sofrito” and whole grains. For all of them, compliance with the adherence to the MedDiet for the specific servings was associated with statistically longer TL in women. In men, some of these components tended to be significant or were significant, such as whole grains. Moreover, we created a new score of adherence to these four items of the MedDiet global score, and we obtained statistically significant associations with TL for the whole population and even for men in the statistical model adjusted for age, diabetes and BMI. These findings suggest an increased role of these particular components in the beneficial effects of the MedDiet on TL in both men and women. Previous research examining these particular food items supports our findings [62,71,143,144,145,146,147,148,149,150]. Fruits are known for their substantial content of dietary antioxidant chemicals and fiber, which have been linked to a greater TL in the NHANES study [143]. Fish is a dietary source abundant in omega-3 fatty acids, which have been linked to longer TL in some investigations [147]. Whole grains are known for their high content of dietary fiber, which has been associated with longer telomeres in many studies, including the NHANES study [148] and other research [71,145]. Sofrito, a key component of the MedDiet, is a mix of tomato, onion, garlic, and olive oil, which contains phenolic compounds and carotenoids [149]. It has been demonstrated that sofrito can inhibit oxidative stress [150], contributing to the favorable observed effects.
One strength of our study is the use of a carefully validated score to assess adherence to the MedDiet [93]. Additionally, our study participants reside in a region where the MedDiet is well characterized and there is favorable accessibility to such foods. These characteristics contribute to facilitating good compliance with the global MedDiet score and its individual components among participants who have a preference for it compared to countries outside of the Mediterranean region. While longevity is related to a healthy diet, it is also influenced by a whole range of other factors such as genetics. Thus, another strength of our work compared to the previously published studies on the effects of the MedDiet on TL is that in the present investigation, we have incorporated a comprehensive measurement of genetic factors. None of the previous studies, whether observational or intervention with the MedDiet [54,60,61,63,64,67,70,71,72,73,74,75,142,151], has carried out a multigenic analysis of candidate genes for TL or of GWAS to know the possible influence of the same on the TL. In our study, we have obtained some statistically significant associations between SNPs in candidate genes for TL previously discovered in GWAS [109] in the expected direction (increasing or decreasing TL). We also built a GRS with SNPs in the TERT, TERF2, RTEL1, and DCAF4 candidate genes and determined the cumulative effect on DNAmTL and the MedDiet–GRS interactions. No multiplicative interaction term was observed, but the MedDiet and GRS had additive effects, suggesting a biological modulation [152]. Finally, we conducted an exploratory GWAS to better understand the SNPs and genes associated with DNAmTL. The vast majority of previous GWASs have been carried out using the TL variable measured by various techniques [82,83,84,85,86,89,90,109,153] but without using the DNAmTL indicator. Although by examining SNPs in TL-associated candidate genes, we have obtained significant associations with TERT, TERF1, RTEL1 and DCAF4, the statistical significance of these associations has not been very high. This could be because one limitation of our study is the small sample size compared to the large GWAS meta-analysis. However, another factor that could influence is that the DNAmTL indicator measured additional information to that provided by the classical methods for measuring TL (qPCR, qFish, etc.). This possibility has already been proposed by Lu et al. [46], indicating that DNAmTL and TL may have different patterns of SNP associations. Indeed, in conducting the GWAS in this population, we have found the most significant associations with the suggesting level of association of GWAS (p < 1 × 10−5) with some SNPs other than the TL candidates. The hit has been intergenic on chromosome 13 and has not been previously reported. In addition, we found a very interesting signal for several SNPs on chromosome 7, including the NCAPG2 gene. This gene plays an essential role in chromosome condensation and segregation during mitosis [154]. It has also been involved in DNA damage repair/DNA replication/telomere maintenance in functional studies [155] but has not been identified in previous GWAS. This intriguing gene scored 8 for SNP rs1466210 in the FORGEdb [111,112], suggesting potential functionality. Another top-ranked SNP in the exploratory GWAS was observed in the ABI3BP gene. This gene holds significant interest due to the limited number of prior research that has undertaken GWAS for DNAmTL [51,87]. One of the studies was conducted with 107 individuals, resulting in limited statistical power [87]. The second study was conducted using a sample of 297 individuals of European–American descent and 280 individuals of African–American descent [51]. The ABI3BP gene emerged as one of the highest-ranked genes in the GWAS conducted on African Americans. Hence, we consider our discovery to be an outcome of replication. The ABI3BP gene, encoding mediators of senescence, has previously been implicated in cancer, immunity, and cardiovascular risk [156,157]. However, it has not yet been identified in GWAS for TL. Therefore, additional research with larger sample sizes is necessary to replicate and discover SNPs associated with DNAmTL. Although our study has significant strengths mentioned above, it also has limitations related to the sample size and population characteristics. This investigation has been carried out in an elderly and high-risk cardiovascular population; therefore, the results cannot be extrapolated directly to younger age and more healthy populations. Specific studies of the association between adherence to MedDiet and DNAmTL would need to be carried out in these individuals. Moreover, it is important to conduct further investigations of the sex-specific effects as well as of the gene–MedDiet interactions on TL in diverse populations with enhanced statistical power.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antiox12112004/s1, Table S1: Quantitative 17-item questionnaire for adherence to Mediterranean diet; Figure S1: Association between telomere length–DNA methylation and chronological age in men (A) and in women (B); Table S2: Association between the methylation sites included in the leukocyte DNAmTL computation and the estimated DNAmTL in the population; Figure S2: Telomere length (DNAmTLAdjustAge) depending on the categories of the genetic risk score (GRS) combining the effect alleles (associated with shorter TL) for the SNPs; Table S3: Compliance (%) of adherence to the Mediterranean diet for each item of the 17-I score by sex; Table S4: Association between the particular items (foods/beverages) of the Mediterranean diet score (17-I) with telomere length by sex; Figure S3: Q-Q plot for the genome-wide association study (GWAS) of telomere length–DNA methylation adjusted for sex and age; Figure S4: Zoom plots of the selected top-ranked SNPs for the GWAS of DNAmTL; Table S5: FORGEdb total and detailed scores for the 10 top-ranked SNPs in the GWAS for DNAmTL; Figure S5: Gene expression heatmap based on the dataset GTExV8 (54 tissue types) for average gene expression of genes selected in the GWAS for DNAmTL; Table S6: Coefficients obtained in the SNP-based genome-wide association study (GWAS) for DNAmTL adjusted for additional covariates; Figure S6: Manhattan plot for the gene-based GWAS analysis for DNAmTL. Figure S7: Manhattan plot of the SNP-based GWAS analyzing the interaction between the SNPs and adherence to Mediterranean diet in determining telomere length (DNAmTLAdjustAge).
Author Contributions
Conceptualization, J.V.S., E.M.A. and D.C.; Data curation, E.M.A., R.F.-C. and J.V.S.; Formal analysis, O.C., J.V.S. and D.C.; Investigation, E.M.A., C.O.-A., R.F.-C., E.C.P., R.B., J.F.A., A.P.-F. and O.P.; Methodology, E.M.A., J.V.S., R.F.-C., R.E. and D.C.; Resources, J.V.S., R.E., J.M.O. and D.C.; Software, O.C. and E.M.A.; Supervision, O.P. and J.M.O.; Validation, C.O.-A., R.B., J.I.G., A.P.-F. and O.P.; Writing—original draft, O.C. and D.C.; Writing—review and editing, O.C., E.M.A., J.V.S., C.O.-A., R.F.-C., E.C.P., R.B., J.I.G., R.E., J.F.A., A.P.-F., O.P., J.M.O. and D.C. Writing—review and editing, all the authors. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Human Research Ethics Committee of Valencia University, Valencia (ethical approval code H1373255532771, 15 July 2013; and H1509263926814, 6 November 2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Neither the participants’ consent forms nor ethics approval included permission for open access. However, we follow a controlled data-sharing collaboration model, and data for collaborations will be available upon request pending application and approval. Investigators who are interested in this study can contact the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Funding Statement
This study was partially funded, by the Spanish Ministry of Health (Instituto de Salud Carlos III) and the Ministerio de Economía y Competitividad-Fondo Europeo de Desarrollo Regional (FEDER) (grants CIBER 06/03/035, PI13/00728, PI16/00366, PI19/00781, and OBN22PI02/2023); the Generalitat Valenciana (grants PROMETEO/2021/021 and GV/2021/142); and Grant PID2019-108858RB-I00 funded by AEI 10.13039/501100011033 and by “ERDF A way of making Europe”.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ghorbani Z., Kazemi A., Shoaibinobarian N., Taylor K., Noormohammadi M. Overall, Plant-Based, or Animal-Based Low Carbohydrate Diets and All-Cause and Cause-Specific Mortality: A Systematic Review and Dose-Response Meta-Analysis of Prospective Cohort Studies. Ageing Res. Rev. 2023;90:101997. doi: 10.1016/j.arr.2023.101997. [DOI] [PubMed] [Google Scholar]
- 2.Mente A., Dehghan M., Rangarajan S., O’Donnell M., Hu W., Dagenais G., Wielgosz A., A Lear S., Wei L., Diaz R., et al. Diet, Cardiovascular Disease, and Mortality in 80 Countries. Eur. Heart J. 2023;44:2560–2579. doi: 10.1093/eurheartj/ehad269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Springmann M., Spajic L., Clark M.A., Poore J., Herforth A., Webb P., Rayner M., Scarborough P. The Healthiness and Sustainability of National and Global Food Based Dietary Guidelines: Modelling Study. BMJ. 2020;370:m2322. doi: 10.1136/bmj.m2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang S., Mijatovic J., Li A., Koemel N., Nasir R., Toniutti C., Bell-Anderson K., Skilton M., O’Leary F. Dietary Patterns and Non-Communicable Disease Biomarkers: A Network Meta-Analysis and Nutritional Geometry Approach. Nutrients. 2022;15:76. doi: 10.3390/nu15010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motamedi A., Askari M., Mozaffari H., Homayounfrar R., Nikparast A., Ghazi M.L., Nejad M.M., Alizadeh S. Dietary Inflammatory Index in Relation to Type 2 Diabetes: A Meta-Analysis. Int. J. Clin. Pract. 2022;2022:9953115. doi: 10.1155/2022/9953115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karam G., Agarwal A., Sadeghirad B., Jalink M., Hitchcock C.L., Ge L., Kiflen R., Ahmed W., Zea A.M., Milenkovic J., et al. Comparison of Seven Popular Structured Dietary Programmes and Risk of Mortality and Major Cardiovascular Events in Patients at Increased Cardiovascular Risk: Systematic Review and Network Meta-Analysis. BMJ. 2023;380:e072003. doi: 10.1136/bmj-2022-072003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhandari B., Liu Z., Lin S., Macniven R., Akombi-Inyang B., Hall J., Feng X., Schutte A.E., Xu X. Long-Term Consumption of 10 Food Groups and Cardiovascular Mortality: A Systematic Review and Dose Response Meta-Analysis of Prospective Cohort Studies. Adv. Nutr. 2023;14:55–63. doi: 10.1016/j.advnut.2022.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominguez L.J., Veronese N., Di Bella G., Cusumano C., Parisi A., Tagliaferri F., Ciriminna S., Barbagallo M. Mediterranean Diet in the Management and Prevention of Obesity. Exp. Gerontol. 2023;174:112121. doi: 10.1016/j.exger.2023.112121. [DOI] [PubMed] [Google Scholar]
- 9.Hu H., Zhao Y., Feng Y., Yang X., Li Y., Wu Y., Yuan L., Zhang J., Li T., Huang H., et al. Consumption of Whole Grains and Refined Grains and Associated Risk of Cardiovascular Disease Events and All-Cause Mortality: A Systematic Review and Dose-Response Meta-Analysis of Prospective Cohort Studies. Am. J. Clin. Nutr. 2023;117:149–159. doi: 10.1016/j.ajcnut.2022.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Lee M., Lim M., Kim J. Fruit and Vegetable Consumption and the Metabolic Syndrome: A Systematic Review and Dose-Response Meta-Analysis. Br. J. Nutr. 2019;122:723–733. doi: 10.1017/S000711451900165X. [DOI] [PubMed] [Google Scholar]
- 11.Corella D., Coltell O., Macian F., Ordovás J.M. Advances in Understanding the Molecular Basis of the Mediterranean Diet Effect. Annu. Rev. Food Sci. Technol. 2018;9:227–249. doi: 10.1146/annurev-food-032217-020802. [DOI] [PubMed] [Google Scholar]
- 12.Caturano A., D’Angelo M., Mormone A., Russo V., Mollica M.P., Salvatore T., Galiero R., Rinaldi L., Vetrano E., Marfella R., et al. Oxidative Stress in Type 2 Diabetes: Impacts from Pathogenesis to Lifestyle Modifications. Curr. Issues Mol. Biol. 2023;45:6651–6666. doi: 10.3390/cimb45080420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masschelin P.M., Cox A.R., Chernis N., Hartig S.M. The Impact of Oxidative Stress on Adipose Tissue Energy Balance. Front. Physiol. 2019;10:1638. doi: 10.3389/fphys.2019.01638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreadi A., Bellia A., Di Daniele N., Meloni M., Lauro R., Della-Morte D., Lauro D. The Molecular Link between Oxidative Stress, Insulin Resistance, and Type 2 Diabetes: A Target for New Therapies against Cardiovascular Diseases. Curr. Opin. Pharmacol. 2022;62:85–96. doi: 10.1016/j.coph.2021.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Batty M., Bennett M.R., Yu E. The Role of Oxidative Stress in Atherosclerosis. Cells. 2022;11:3843. doi: 10.3390/cells11233843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cianciosi D., Forbes-Hernández T.Y., Regolo L., Alvarez-Suarez J.M., Navarro-Hortal M.D., Xiao J., Quiles J.L., Battino M., Giampieri F. The Reciprocal Interaction between Polyphenols and Other Dietary Compounds: Impact on Bioavailability, Antioxidant Capacity and Other Physico-Chemical and Nutritional Parameters. Food Chem. 2022;375:131904. doi: 10.1016/j.foodchem.2021.131904. [DOI] [PubMed] [Google Scholar]
- 17.Farhangi M.A., Vajdi M., Fathollahi P. Dietary Total Antioxidant Capacity (TAC), General and Central Obesity Indices and Serum Lipids among Adults: An Updated Systematic Review and Meta-Analysis. Int. J. Vitam. Nutr. Res. 2022;92:406–422. doi: 10.1024/0300-9831/a000675. [DOI] [PubMed] [Google Scholar]
- 18.Mozaffari H., Daneshzad E., Surkan P.J., Azadbakht L. Dietary Total Antioxidant Capacity and Cardiovascular Disease Risk Factors: A Systematic Review of Observational Studies. J. Am. Coll. Nutr. 2018;37:533–545. doi: 10.1080/07315724.2018.1441079. [DOI] [PubMed] [Google Scholar]
- 19.Franceschi C., Garagnani P., Morsiani C., Conte M., Santoro A., Grignolio A., Monti D., Capri M., Salvioli S. The Continuum of Aging and Age-Related Diseases: Common Mechanisms but Different Rates. Front. Med. 2018;5:61. doi: 10.3389/fmed.2018.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diebel L.W.M., Rockwood K. Determination of Biological Age: Geriatric Assessment vs Biological Biomarkers. Curr. Oncol. Rep. 2021;23:104. doi: 10.1007/s11912-021-01097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., Zhang W., Duan Y., Niu Y., Chen Y., Liu X., Dong Z., Zheng Y., Chen X., Feng Z., et al. Progress in Biological Age Research. Front. Public Health. 2023;11:1074274. doi: 10.3389/fpubh.2023.1074274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beltrán-Sánchez H., Palloni A., Huangfu Y., McEniry M.C. Modeling Biological Age and Its Link with the Aging Process. PNAS Nexus. 2022;1:pgac135. doi: 10.1093/pnasnexus/pgac135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liguori I., Russo G., Curcio F., Bulli G., Aran L., Della-Morte D., Gargiulo G., Testa G., Cacciatore F., Bonaduce D., et al. Oxidative Stress, Aging, and Diseases. Clin. Interv. Aging. 2018;13:757–772. doi: 10.2147/CIA.S158513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jomova K., Raptova R., Alomar S.Y., Alwasel S.H., Nepovimova E., Kuca K., Valko M. Reactive Oxygen Species, Toxicity, Oxidative Stress, and Antioxidants: Chronic Diseases and Aging. Arch. Toxicol. 2023;97:2499–2574. doi: 10.1007/s00204-023-03562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J.-K. Antiaging Agents: Safe Interventions to Slow Aging and Healthy Life Span Extension. Nat. Prod. Bioprospect. 2022;12:18. doi: 10.1007/s13659-022-00339-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas A., Belsky D.W., Gu Y. Healthy Lifestyle Behaviors and Biological Aging in the U.S. National Health and Nutrition Examination Surveys 1999–2018. J. Gerontol. A Biol. Sci. Med. Sci. 2023;78:1535–1542. doi: 10.1093/gerona/glad082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The Hallmarks of Aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmauck-Medina T., Molière A., Lautrup S., Zhang J., Chlopicki S., Madsen H.B., Cao S., Soendenbroe C., Mansell E., Vestergaard M.B., et al. New Hallmarks of Ageing: A 2022 Copenhagen Ageing Meeting Summary. Aging. 2022;14:6829–6839. doi: 10.18632/aging.204248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. Hallmarks of Aging: An Expanding Universe. Cell. 2023;186:243–278. doi: 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Blackburn E.H., Epel E.S., Lin J. Human Telomere Biology: A Contributory and Interactive Factor in Aging, Disease Risks, and Protection. Science. 2015;350:1193–1198. doi: 10.1126/science.aab3389. [DOI] [PubMed] [Google Scholar]
- 31.Harley C.B., Futcher A.B., Greider C.W. Telomeres Shorten during Ageing of Human Fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 32.Aguado J., d’Adda di Fagagna F., Wolvetang E. Telomere Transcription in Ageing. Ageing Res. Rev. 2020;62:101115. doi: 10.1016/j.arr.2020.101115. [DOI] [PubMed] [Google Scholar]
- 33.Rizvi S., Raza S.T., Mahdi F. Telomere Length Variations in Aging and Age-Related Diseases. Curr. Aging Sci. 2014;7:161–167. doi: 10.2174/1874609808666150122153151. [DOI] [PubMed] [Google Scholar]
- 34.Ye Q., Apsley A.T., Etzel L., Hastings W.J., Kozlosky J.T., Walker C., Wolf S.E., Shalev I. Telomere Length and Chronological Age across the Human Lifespan: A Systematic Review and Meta-Analysis of 414 Study Samples Including 743,019 Individuals. Ageing Res. Rev. 2023;90:102031. doi: 10.1016/j.arr.2023.102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Meyer T., Nawrot T., Bekaert S., De Buyzere M.L., Rietzschel E.R., Andrés V. Telomere Length as Cardiovascular Aging Biomarker: JACC Review Topic of the Week. J. Am. Coll. Cardiol. 2018;72:805–813. doi: 10.1016/j.jacc.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Martínez P., Blasco M.A. Heart-Breaking Telomeres. Circ. Res. 2018;123:787–802. doi: 10.1161/CIRCRESAHA.118.312202. [DOI] [PubMed] [Google Scholar]
- 37.Allaire P., He J., Mayer J., Moat L., Gerstenberger P., Wilhorn R., Strutz S., Kim D.S.L., Zeng C., Cox N., et al. Genetic and Clinical Determinants of Telomere Length. HGG Adv. 2023;4:100201. doi: 10.1016/j.xhgg.2023.100201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emami M., Agbaedeng T.A., Thomas G., Middeldorp M.E., Thiyagarajah A., Wong C.X., Elliott A.D., Gallagher C., Hendriks J.M.L., Lau D.H., et al. Accelerated Biological Aging Secondary to Cardiometabolic Risk Factors Is a Predictor of Cardiovascular Mortality: A Systematic Review and Meta-Analysis. Can. J. Cardiol. 2022;38:365–375. doi: 10.1016/j.cjca.2021.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Arbeev K.G., Verhulst S., Steenstrup T., Kark J.D., Bagley O., Kooperberg C., Reiner A.P., Hwang S.-J., Levy D., Fitzpatrick A.L., et al. Association of Leukocyte Telomere Length with Mortality Among Adult Participants in 3 Longitudinal Studies. JAMA Netw. Open. 2020;3:e200023. doi: 10.1001/jamanetworkopen.2020.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider C.V., Schneider K.M., Teumer A., Rudolph K.L., Hartmann D., Rader D.J., Strnad P. Association of Telomere Length with Risk of Disease and Mortality. JAMA Intern. Med. 2022;182:291–300. doi: 10.1001/jamainternmed.2021.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fasching C.L. Telomere Length Measurement as a Clinical Biomarker of Aging and Disease. Crit. Rev. Clin. Lab. Sci. 2018;55:443–465. doi: 10.1080/10408363.2018.1504274. [DOI] [PubMed] [Google Scholar]
- 42.Demanelis K., Jasmine F., Chen L.S., Chernoff M., Tong L., Delgado D., Zhang C., Shinkle J., Sabarinathan M., Lin H., et al. Determinants of Telomere Length across Human Tissues. Science. 2020;369:eaaz6876. doi: 10.1126/science.aaz6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin K.-W., Yan J. The Telomere Length Dynamic and Methods of Its Assessment. J. Cell Mol. Med. 2005;9:977–989. doi: 10.1111/j.1582-4934.2005.tb00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai T.-P., Wright W.E., Shay J.W. Comparison of Telomere Length Measurement Methods. Philos. Trans. R. Soc. Lond B Biol. Sci. 2018;373:20160451. doi: 10.1098/rstb.2016.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindrose A.R., McLester-Davis L.W.Y., Tristano R.I., Kataria L., Gadalla S.M., Eisenberg D.T.A., Verhulst S., Drury S. Method Comparison Studies of Telomere Length Measurement Using qPCR Approaches: A Critical Appraisal of the Literature. PLoS ONE. 2021;16:e0245582. doi: 10.1371/journal.pone.0245582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu A.T., Seeboth A., Tsai P.-C., Sun D., Quach A., Reiner A.P., Kooperberg C., Ferrucci L., Hou L., Baccarelli A.A., et al. DNA Methylation-Based Estimator of Telomere Length. Aging. 2019;11:5895–5923. doi: 10.18632/aging.102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearce E.E., Horvath S., Katta S., Dagnall C., Aubert G., Hicks B.D., Spellman S.R., Katki H., Savage S.A., Alsaggaf R., et al. DNA-Methylation-Based Telomere Length Estimator: Comparisons with Measurements from Flow FISH and qPCR. Aging. 2021;13:14675–14686. doi: 10.18632/aging.203126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sehl M.E., Henry J.E., Storniolo A.M., Horvath S., Ganz P.A. The Impact of Reproductive Factors on DNA Methylation-Based Telomere Length in Healthy Breast Tissue. NPJ Breast. Cancer. 2022;8:48. doi: 10.1038/s41523-022-00410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seki Y., Aczel D., Torma F., Jokai M., Boros A., Suzuki K., Higuchi M., Tanisawa K., Boldogh I., Horvath S., et al. No Strong Association among Epigenetic Modifications by DNA Methylation, Telomere Length, and Physical Fitness in Biological Aging. Biogerontology. 2023;24:245–255. doi: 10.1007/s10522-022-10011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hernández Cordero A.I., Yang C.X., Li X., Milne S., Chen V., Hollander Z., Ng R., Criner G.J., Woodruff P.G., Lazarus S.C., et al. Epigenetic Marker of Telomeric Age Is Associated with Exacerbations and Hospitalizations in Chronic Obstructive Pulmonary Disease. Respir. Res. 2021;22:316. doi: 10.1186/s12931-021-01911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jung J., McCartney D.L., Wagner J., Rosoff D.B., Schwandt M., Sun H., Wiers C.E., de Carvalho L.M., Volkow N.D., Walker R.M., et al. Alcohol Use Disorder Is Associated with DNA Methylation-Based Shortening of Telomere Length and Regulated by TESPA1: Implications for Aging. Mol. Psychiatry. 2022;27:3875–3884. doi: 10.1038/s41380-022-01624-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shannon O.M., Ashor A.W., Scialo F., Saretzki G., Martin-Ruiz C., Lara J., Matu J., Griffiths A., Robinson N., Lillà L., et al. Mediterranean Diet and the Hallmarks of Ageing. Eur. J. Clin. Nutr. 2021;75:1176–1192. doi: 10.1038/s41430-020-00841-x. [DOI] [PubMed] [Google Scholar]
- 53.Mazza E., Ferro Y., Pujia R., Mare R., Maurotti S., Montalcini T., Pujia A. Mediterranean Diet In Healthy Aging. J. Nutr. Health Aging. 2021;25:1076–1083. doi: 10.1007/s12603-021-1675-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boccardi V., Esposito A., Rizzo M.R., Marfella R., Barbieri M., Paolisso G. Mediterranean Diet, Telomere Maintenance and Health Status among Elderly. PLoS ONE. 2013;8:e62781. doi: 10.1371/journal.pone.0062781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sikalidis A.K., Kelleher A.H., Kristo A.S. Mediterranean Diet. Encyclopedia. 2021;1:371–387. doi: 10.3390/encyclopedia1020031. [DOI] [Google Scholar]
- 56.Trichopoulos D. In Defense of the Mediterranean Diet. Eur. J. Clin. Nutr. 2002;56:928–929. doi: 10.1038/sj.ejcn.1601521. author reply 930–931. [DOI] [PubMed] [Google Scholar]
- 57.Trichopoulou A. Traditional Mediterranean Diet and Longevity in the Elderly: A Review. Public Health Nutr. 2004;7:943–947. doi: 10.1079/PHN2004558. [DOI] [PubMed] [Google Scholar]
- 58.Sofi F., Cesari F., Abbate R., Gensini G.F., Casini A. Adherence to Mediterranean Diet and Health Status: Meta-Analysis. BMJ. 2008;337:a1344. doi: 10.1136/bmj.a1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marin C., Delgado-Lista J., Ramirez R., Carracedo J., Caballero J., Perez-Martinez P., Gutierrez-Mariscal F.M., Garcia-Rios A., Delgado-Casado N., Cruz-Teno C., et al. Mediterranean Diet Reduces Senescence-Associated Stress in Endothelial Cells. Age. 2012;34:1309–1316. doi: 10.1007/s11357-011-9305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu Y., Honig L.S., Schupf N., Lee J.H., Luchsinger J.A., Stern Y., Scarmeas N. Mediterranean Diet and Leukocyte Telomere Length in a Multi-Ethnic Elderly Population. Age. 2015;37:24. doi: 10.1007/s11357-015-9758-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crous-Bou M., Fung T.T., Prescott J., Julin B., Du M., Sun Q., Rexrode K.M., Hu F.B., De Vivo I. Mediterranean Diet and Telomere Length in Nurses’ Health Study: Population Based Cohort Study. BMJ. 2014;349:g6674. doi: 10.1136/bmj.g6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rafie N., Golpour Hamedani S., Barak F., Safavi S.M., Miraghajani M. Dietary Patterns, Food Groups and Telomere Length: A Systematic Review of Current Studies. Eur. J. Clin. Nutr. 2017;71:151–158. doi: 10.1038/ejcn.2016.149. [DOI] [PubMed] [Google Scholar]
- 63.García-Calzón S., Martínez-González M.A., Razquin C., Arós F., Lapetra J., Martínez J.A., Zalba G., Marti A. Mediterranean Diet and Telomere Length in High Cardiovascular Risk Subjects from the PREDIMED-NAVARRA Study. Clin. Nutr. 2016;35:1399–1405. doi: 10.1016/j.clnu.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 64.Milte C.M., Russell A.P., Ball K., Crawford D., Salmon J., McNaughton S.A. Diet Quality and Telomere Length in Older Australian Men and Women. Eur. J. Nutr. 2018;57:363–372. doi: 10.1007/s00394-016-1326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ventura Marra M., Drazba M.A., Holásková I., Belden W.J. Nutrition Risk Is Associated with Leukocyte Telomere Length in Middle-Aged Men and Women with at Least One Risk Factor for Cardiovascular Disease. Nutrients. 2019;11:508. doi: 10.3390/nu11030508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meinilä J., Perälä M.-M., Kautiainen H., Männistö S., Kanerva N., Shivappa N., Hébert J.R., Iozzo P., Guzzardi M.A., Eriksson J.G. Healthy Diets and Telomere Length and Attrition during a 10-Year Follow-Up. Eur. J. Clin. Nutr. 2019;73:1352–1360. doi: 10.1038/s41430-018-0387-4. [DOI] [PubMed] [Google Scholar]
- 67.Davinelli S., Trichopoulou A., Corbi G., De Vivo I., Scapagnini G. The Potential Nutrigeroprotective Role of Mediterranean Diet and Its Functional Components on Telomere Length Dynamics. Ageing Res. Rev. 2019;49:1–10. doi: 10.1016/j.arr.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 68.Freitas-Simoes T.-M., Cofán M., Blasco M.A., Soberón N., Foronda M., Corella D., Asensio E.M., Serra-Mir M., Roth I., Calvo C., et al. The Red Blood Cell Proportion of Arachidonic Acid Relates to Shorter Leukocyte Telomeres in Mediterranean Elders: A Secondary Analysis of a Randomized Controlled Trial. Clin. Nutr. 2019;38:958–961. doi: 10.1016/j.clnu.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 69.Chan R., Leung J., Tang N., Woo J. Dietary Patterns and Telomere Length in Community-Dwelling Chinese Older Men and Women: A Cross-Sectional Analysis. Eur. J. Nutr. 2020;59:3303–3311. doi: 10.1007/s00394-019-02168-1. [DOI] [PubMed] [Google Scholar]
- 70.Ojeda-Rodríguez A., Zazpe I., Alonso-Pedrero L., Zalba G., Guillen-Grima F., Martinez-Gonzalez M.A., Marti A. Association between Diet Quality Indexes and the Risk of Short Telomeres in an Elderly Population of the SUN Project. Clin. Nutr. 2020;39:2487–2494. doi: 10.1016/j.clnu.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 71.Galiè S., Canudas S., Muralidharan J., García-Gavilán J., Bulló M., Salas-Salvadó J. Impact of Nutrition on Telomere Health: Systematic Review of Observational Cohort Studies and Randomized Clinical Trials. Adv. Nutr. 2020;11:576–601. doi: 10.1093/advances/nmz107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Canudas S., Becerra-Tomás N., Hernández-Alonso P., Galié S., Leung C., Crous-Bou M., De Vivo I., Gao Y., Gu Y., Meinilä J., et al. Mediterranean Diet and Telomere Length: A Systematic Review and Meta-Analysis. Adv. Nutr. 2020;11:1544–1554. doi: 10.1093/advances/nmaa079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fernández de la Puente M., Hernández-Alonso P., Canudas S., Marti A., Fitó M., Razquin C., Salas-Salvadó J. Modulation of Telomere Length by Mediterranean Diet, Caloric Restriction, and Exercise: Results from PREDIMED-Plus Study. Antioxidants. 2021;10:1596. doi: 10.3390/antiox10101596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marti A., Fernández de la Puente M., Canudas S., Zalba G., Razquin C., Valle-Hita C., Fitó M., Martínez-González M.Á., García-Calzón S., Salas-Salvadó J. Effect of a 3-Year Lifestyle Intervention on Telomere Length in Participants from PREDIMED-Plus: A Randomized Trial. Clin. Nutr. 2023;42:1581–1587. doi: 10.1016/j.clnu.2023.06.030. [DOI] [PubMed] [Google Scholar]
- 75.Bountziouka V., Nelson C.P., Wang Q., Musicha C., Codd V., Samani N.J. Dietary Patterns and Practices and Leucocyte Telomere Length: Findings from the UK Biobank. J. Acad. Nutr. Diet. 2023;123:912–922.e26. doi: 10.1016/j.jand.2023.01.008. [DOI] [PubMed] [Google Scholar]
- 76.Andrew T., Aviv A., Falchi M., Surdulescu G.L., Gardner J.P., Lu X., Kimura M., Kato B.S., Valdes A.M., Spector T.D. Mapping Genetic Loci That Determine Leukocyte Telomere Length in a Large Sample of Unselected Female Sibling Pairs. Am. J. Hum. Genet. 2006;78:480–486. doi: 10.1086/500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Delgado D.A., Zhang C., Gleason K., Demanelis K., Chen L.S., Gao J., Roy S., Shinkle J., Sabarinathan M., Argos M., et al. The Contribution of Parent-to-Offspring Transmission of Telomeres to the Heritability of Telomere Length in Humans. Hum. Genet. 2019;138:49–60. doi: 10.1007/s00439-018-1964-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andreu-Sánchez S., Aubert G., Ripoll-Cladellas A., Henkelman S., Zhernakova D.V., Sinha T., Kurilshikov A., Cenit M.C., Jan Bonder M., Franke L., et al. Genetic, Parental and Lifestyle Factors Influence Telomere Length. Commun. Biol. 2022;5:565. doi: 10.1038/s42003-022-03521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lundblad V. Molecular Biology. Telomeres Keep on Rappin’. Science. 2000;288:2141–2142. doi: 10.1126/science.288.5474.2141. [DOI] [PubMed] [Google Scholar]
- 80.Levy D., Neuhausen S.L., Hunt S.C., Kimura M., Hwang S.-J., Chen W., Bis J.C., Fitzpatrick A.L., Smith E., Johnson A.D., et al. Genome-Wide Association Identifies OBFC1 as a Locus Involved in Human Leukocyte Telomere Biology. Proc. Natl. Acad. Sci. USA. 2010;107:9293–9298. doi: 10.1073/pnas.0911494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Codd V., Mangino M., van der Harst P., Braund P.S., Kaiser M., Beveridge A.J., Rafelt S., Moore J., Nelson C., Soranzo N., et al. Common Variants near TERC Are Associated with Mean Telomere Length. Nat. Genet. 2010;42:197–199. doi: 10.1038/ng.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prescott J., Kraft P., Chasman D.I., Savage S.A., Mirabello L., Berndt S.I., Weissfeld J.L., Han J., Hayes R.B., Chanock S.J., et al. Genome-Wide Association Study of Relative Telomere Length. PLoS ONE. 2011;6:e19635. doi: 10.1371/journal.pone.0019635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pooley K.A., Bojesen S.E., Weischer M., Nielsen S.F., Thompson D., Amin Al Olama A., Michailidou K., Tyrer J.P., Benlloch S., Brown J., et al. A Genome-Wide Association Scan (GWAS) for Mean Telomere Length within the COGS Project: Identified Loci Show Little Association with Hormone-Related Cancer Risk. Hum. Mol. Genet. 2013;22:5056–5064. doi: 10.1093/hmg/ddt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Codd V., Nelson C.P., Albrecht E., Mangino M., Deelen J., Buxton J.L., Hottenga J.J., Fischer K., Esko T., Surakka I., et al. Identification of Seven Loci Affecting Mean Telomere Length and Their Association with Disease. Nat. Genet. 2013;45:422–427, 427e1-2. doi: 10.1038/ng.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saxena R., Bjonnes A., Prescott J., Dib P., Natt P., Lane J., Lerner M., Cooper J.A., Ye Y., Li K.W., et al. Genome-Wide Association Study Identifies Variants in Casein Kinase II (CSNK2A2) to Be Associated with Leukocyte Telomere Length in a Punjabi Sikh Diabetic Cohort. Circ. Cardiovasc. Genet. 2014;7:287–295. doi: 10.1161/CIRCGENETICS.113.000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delgado D.A., Zhang C., Chen L.S., Gao J., Roy S., Shinkle J., Sabarinathan M., Argos M., Tong L., Ahmed A., et al. Genome-Wide Association Study of Telomere Length among South Asians Identifies a Second RTEL1 Association Signal. J. Med. Genet. 2018;55:64–71. doi: 10.1136/jmedgenet-2017-104922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qian J., Fischer C., Burhan A., Mak M., Gerretsen P., Kolla N., Al-Chalabi N., Chaudhary Z., Qureshey A., Bani-Fatemi A., et al. GWAS of Biological Aging to Find Longevity Genes in Schizophrenia. Eur. Arch. Psychiatry Clin. Neurosci. 2023 doi: 10.1007/s00406-023-01622-w. [DOI] [PubMed] [Google Scholar]
- 88.Salas-Salvadó J., Díaz-López A., Ruiz-Canela M., Basora J., Fitó M., Corella D., Serra-Majem L., Wärnberg J., Romaguera D., Estruch R., et al. Effect of a Lifestyle Intervention Program with Energy-Restricted Mediterranean Diet and Exercise on Weight Loss and Cardiovascular Risk Factors: One-Year Results of the PREDIMED-Plus Trial. Diabetes Care. 2019;42:777–788. doi: 10.2337/dc18-0836. [DOI] [PubMed] [Google Scholar]
- 89.Codd V., Denniff M., Swinfield C., Warner S.C., Papakonstantinou M., Sheth S., Nanus D.E., Budgeon C.A., Musicha C., Bountziouka V., et al. Measurement and Initial Characterization of Leukocyte Telomere Length in 474,074 Participants in UK Biobank. Nat. Aging. 2022;2:170–179. doi: 10.1038/s43587-021-00166-9. [DOI] [PubMed] [Google Scholar]
- 90.Codd V., Wang Q., Allara E., Musicha C., Kaptoge S., Stoma S., Jiang T., Hamby S.E., Braund P.S., Bountziouka V., et al. Polygenic Basis and Biomedical Consequences of Telomere Length Variation. Nat. Genet. 2021;53:1425–1433. doi: 10.1038/s41588-021-00944-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Coltell O., Asensio E.M., Sorlí J.V., Barragán R., Fernández-Carrión R., Portolés O., Ortega-Azorín C., Martínez-LaCruz R., González J.I., Zanón-Moreno V., et al. Genome-Wide Association Study (GWAS) on Bilirubin Concentrations in Subjects with Metabolic Syndrome: Sex-Specific GWAS Analysis and Gene-Diet Interactions in a Mediterranean Population. Nutrients. 2019;11:90. doi: 10.3390/nu11010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fernández-Carrión R., Sorlí J.V., Asensio E.M., Pascual E.C., Portolés O., Alvarez-Sala A., Francès F., Ramírez-Sabio J.B., Pérez-Fidalgo A., Villamil L.V., et al. DNA-Methylation Signatures of Tobacco Smoking in a High Cardiovascular Risk Population: Modulation by the Mediterranean Diet. Int. J. Environ. Res. Public Health. 2023;20:3635. doi: 10.3390/ijerph20043635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schröder H., Zomeño M.D., Martínez-González M.A., Salas-Salvadó J., Corella D., Vioque J., Romaguera D., Martínez J.A., Tinahones F.J., Miranda J.L., et al. Validity of the Energy-Restricted Mediterranean Diet Adherence Screener. Clin. Nutr. 2021;40:4971–4979. doi: 10.1016/j.clnu.2021.06.030. [DOI] [PubMed] [Google Scholar]
- 94.Schröder H., Fitó M., Estruch R., Martínez-González M.A., Corella D., Salas-Salvadó J., Lamuela-Raventós R., Ros E., Salaverría I., Fiol M., et al. A Short Screener Is Valid for Assessing Mediterranean Diet Adherence among Older Spanish Men and Women. J. Nutr. 2011;141:1140–1145. doi: 10.3945/jn.110.135566. [DOI] [PubMed] [Google Scholar]
- 95.Molina L., Sarmiento M., Peñafiel J., Donaire D., Garcia-Aymerich J., Gomez M., Ble M., Ruiz S., Frances A., Schröder H., et al. Validation of the Regicor Short Physical Activity Questionnaire for the Adult Population. PLoS ONE. 2017;12:e0168148. doi: 10.1371/journal.pone.0168148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pidsley R., Zotenko E., Peters T.J., Lawrence M.G., Risbridger G.P., Molloy P., Van Djik S., Muhlhausler B., Stirzaker C., Clark S.J. Critical Evaluation of the Illumina MethylationEPIC BeadChip Microarray for Whole-Genome DNA Methylation Profiling. Genome Biol. 2016;17:208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jiang Y., Chen J., Chen W. Controlling Batch Effect in Epigenome-Wide Association Study. Methods Mol. Biol. 2022;2432:73–84. doi: 10.1007/978-1-0716-1994-0_6. [DOI] [PubMed] [Google Scholar]
- 98.Murat K., Grüning B., Poterlowicz P.W., Westgate G., Tobin D.J., Poterlowicz K. Ewastools: Infinium Human Methylation BeadChip Pipeline for Population Epigenetics Integrated into Galaxy. Gigascience. 2020;9:giaa049. doi: 10.1093/gigascience/giaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Min J.L., Hemani G., Davey Smith G., Relton C., Suderman M. Meffil: Efficient Normalization and Analysis of Very Large DNA Methylation Datasets. Bioinformatics. 2018;34:3983–3989. doi: 10.1093/bioinformatics/bty476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bhat B., Jones G.T. Data Analysis of DNA Methylation Epigenome-Wide Association Studies (EWAS): A Guide to the Principles of Best Practice. Methods Mol. Biol. 2022;2458:23–45. doi: 10.1007/978-1-0716-2140-0_2. [DOI] [PubMed] [Google Scholar]
- 101.Ross J.P., van Dijk S., Phang M., Skilton M.R., Molloy P.L., Oytam Y. Batch-Effect Detection, Correction and Characterisation in Illumina HumanMethylation450 and MethylationEPIC BeadChip Array Data. Clin. Epigenetics. 2022;14:58. doi: 10.1186/s13148-022-01277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Horvath S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., Sadda S., Klotzle B., Bibikova M., Fan J.-B., Gao Y., et al. Genome-Wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol. Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen B.H., Marioni R.E., Colicino E., Peters M.J., Ward-Caviness C.K., Tsai P.-C., Roetker N.S., Just A.C., Demerath E.W., Guan W., et al. DNA Methylation-Based Measures of Biological Age: Meta-Analysis Predicting Time to Death. Aging. 2016;8:1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smith J.A., Raisky J., Ratliff S.M., Liu J., Kardia S.L.R., Turner S.T., Mosley T.H., Zhao W. Intrinsic and Extrinsic Epigenetic Age Acceleration Are Associated with Hypertensive Target Organ Damage in Older African Americans. BMC Med. Genom. 2019;12:141. doi: 10.1186/s12920-019-0585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fernández-Carrión R., Sorlí J.V., Coltell O., Pascual E.C., Ortega-Azorín C., Barragán R., Giménez-Alba I.M., Alvarez-Sala A., Fitó M., Ordovas J.M., et al. Sweet Taste Preference: Relationships with Other Tastes, Liking for Sugary Foods and Exploratory Genome-Wide Association Analysis in Subjects with Metabolic Syndrome. Biomedicines. 2021;10:79. doi: 10.3390/biomedicines10010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li C., Stoma S., Lotta L.A., Warner S., Albrecht E., Allione A., Arp P.P., Broer L., Buxton J.L., Da Silva Couto Alves A., et al. Genome-Wide Association Analysis in Humans Links Nucleotide Metabolism to Leukocyte Telomere Length. Am. J. Hum. Genet. 2020;106:389–404. doi: 10.1016/j.ajhg.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional Mapping and Annotation of Genetic Associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Breeze C.E., Haugen E., Reynolds A., Teschendorff A., van Dongen J., Lan Q., Rothman N., Bourque G., Dunham I., Beck S., et al. Integrative Analysis of 3604 GWAS Reveals Multiple Novel Cell Type-Specific Regulatory Associations. Genome Biol. 2022;23:13. doi: 10.1186/s13059-021-02560-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Breeze C.E., Haugen E., Gutierrez-Arcelus M., Yao X., Teschendorff A., Beck S., Dunham I., Stamatoyannopoulos J., Franceschini N., Machiela M.J., et al. FORGEdb: A Tool for Identifying Candidate Functional Variants and Uncovering Target Genes and Mechanisms for Complex Diseases. bioRxiv-Genomics. 2023 doi: 10.1101/2022.11.14.516365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Turner S.D. Qqman: An R Package for Visualizing GWAS Results Using Q-Q and Manhattan Plots. JOSS. 2018;3:731. doi: 10.21105/joss.00731. [DOI] [Google Scholar]
- 114.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: Regional Visualization of Genome-Wide Association Scan Results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Watanabe K., Umićević Mirkov M., de Leeuw C.A., van den Heuvel M.P., Posthuma D. Genetic Mapping of Cell Type Specificity for Complex Traits. Nat. Commun. 2019;10:3222. doi: 10.1038/s41467-019-11181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Coltell O., Sorlí J.V., Asensio E.M., Barragán R., González J.I., Giménez-Alba I.M., Zanón-Moreno V., Estruch R., Ramírez-Sabio J.B., Pascual E.C., et al. Genome-Wide Association Study for Serum Omega-3 and Omega-6 Polyunsaturated Fatty Acids: Exploratory Analysis of the Sex-Specific Effects and Dietary Modulation in Mediterranean Subjects with Metabolic Syndrome. Nutrients. 2020;12:310. doi: 10.3390/nu12020310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cruz L.A., Cooke Bailey J.N., Crawford D.C. Importance of Diversity in Precision Medicine: Generalizability of Genetic Associations Across Ancestry Groups Toward Better Identification of Disease Susceptibility Variants. Annu. Rev. Biomed. Data Sci. 2023;6:339–356. doi: 10.1146/annurev-biodatasci-122220-113250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chan M.H.-M., Merrill S.M., Konwar C., Kobor M.S. An Integrative Framework and Recommendations for the Study of DNA Methylation in the Context of Race and Ethnicity. Discov. Soc. Sci. Health. 2023;3:9. doi: 10.1007/s44155-023-00039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kader F., Ghai M. DNA Methylation-Based Variation between Human Populations. Mol. Genet. Genom. 2017;292:5–35. doi: 10.1007/s00438-016-1264-2. [DOI] [PubMed] [Google Scholar]
- 121.Vyas C.M., Sadreyev R.I., Gatchel J.R., Kang J.H., Reynolds C.F., Mischoulon D., Chang G., Hazra A., Manson J.E., Blacker D., et al. Pilot Study of Second-Generation DNA Methylation Epigenetic Markers in Relation to Cognitive and Neuropsychiatric Symptoms in Older Adults. J. Alzheimers Dis. 2023;93:1563–1575. doi: 10.3233/JAD-230093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dobewall H., Keltikangas-Järvinen L., Marttila S., Mishra P.P., Saarinen A., Cloninger C.R., Zwir I., Kähönen M., Hurme M., Raitakari O., et al. The Relationship of Trait-like Compassion with Epigenetic Aging: The Population-Based Prospective Young Finns Study. Front. Psychiatry. 2023;14:1018797. doi: 10.3389/fpsyt.2023.1018797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shi W., Gao X., Cao Y., Chen Y., Cui Q., Deng F., Yang B., Lin E.Z., Fang J., Li T., et al. Personal Airborne Chemical Exposure and Epigenetic Ageing Biomarkers in Healthy Chinese Elderly Individuals: Evidence from Mixture Approaches. Environ. Int. 2022;170:107614. doi: 10.1016/j.envint.2022.107614. [DOI] [PubMed] [Google Scholar]
- 124.Verhulst S., Susser E., Factor-Litvak P.R., Simons M.J.P., Benetos A., Steenstrup T., Kark J.D., Aviv A. Commentary: The Reliability of Telomere Length Measurements. Int. J. Epidemiol. 2015;44:1683–1686. doi: 10.1093/ije/dyv166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Martin-Ruiz C.M., Baird D., Roger L., Boukamp P., Krunic D., Cawthon R., Dokter M.M., van der Harst P., Bekaert S., de Meyer T., et al. Reproducibility of Telomere Length Assessment: An International Collaborative Study. Int. J. Epidemiol. 2015;44:1673–1683. doi: 10.1093/ije/dyu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.von Zglinicki T. Oxidative Stress Shortens Telomeres. Trends Biochem. Sci. 2002;27:339–344. doi: 10.1016/S0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 127.Barnes R.P., Fouquerel E., Opresko P.L. The Impact of Oxidative DNA Damage and Stress on Telomere Homeostasis. Mech. Ageing Dev. 2019;177:37–45. doi: 10.1016/j.mad.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kawanishi S., Oikawa S. Mechanism of Telomere Shortening by Oxidative Stress. Ann. N. Y. Acad. Sci. 2004;1019:278–284. doi: 10.1196/annals.1297.047. [DOI] [PubMed] [Google Scholar]
- 129.von Zglinicki T., Pilger R., Sitte N. Accumulation of Single-Strand Breaks Is the Major Cause of Telomere Shortening in Human Fibroblasts. Free Radic. Biol. Med. 2000;28:64–74. doi: 10.1016/S0891-5849(99)00207-5. [DOI] [PubMed] [Google Scholar]
- 130.von Zglinicki T. Role of Oxidative Stress in Telomere Length Regulation and Replicative Senescence. Ann. N. Y. Acad. Sci. 2000;908:99–110. doi: 10.1111/j.1749-6632.2000.tb06639.x. [DOI] [PubMed] [Google Scholar]
- 131.Zhang W., Peng S.-F., Chen L., Chen H.-M., Cheng X.-E., Tang Y.-H. Association between the Oxidative Balance Score and Telomere Length from the National Health and Nutrition Examination Survey 1999–2002. Oxid. Med. Cell Longev. 2022;2022:1345071. doi: 10.1155/2022/1345071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lin J., Epel E. Stress and Telomere Shortening: Insights from Cellular Mechanisms. Ageing Res. Rev. 2022;73:101507. doi: 10.1016/j.arr.2021.101507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kell L., Simon A.K., Alsaleh G., Cox L.S. The Central Role of DNA Damage in Immunosenescence. Front. Aging. 2023;4:1202152. doi: 10.3389/fragi.2023.1202152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reichert S., Stier A. Does Oxidative Stress Shorten Telomeres in Vivo? A Review. Biol. Lett. 2017;13:20170463. doi: 10.1098/rsbl.2017.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gavia-García G., Rosado-Pérez J., Arista-Ugalde T.L., Aguiñiga-Sánchez I., Santiago-Osorio E., Mendoza-Núñez V.M. Telomere Length and Oxidative Stress and Its Relation with Metabolic Syndrome Components in the Aging. Biology. 2021;10:253. doi: 10.3390/biology10040253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Armstrong E., Boonekamp J. Does Oxidative Stress Shorten Telomeres in Vivo? A Meta-Analysis. Ageing Res. Rev. 2023;85:101854. doi: 10.1016/j.arr.2023.101854. [DOI] [PubMed] [Google Scholar]
- 137.Levy M.A., Tian J., Gandelman M., Cheng H., Tsapekos M., Crego S.R., Maddela R., Sinnott R. A Multivitamin Mixture Protects against Oxidative Stress-Mediated Telomere Shortening. J. Diet. Suppl. 2023:1–18. doi: 10.1080/19390211.2023.2179153. [DOI] [PubMed] [Google Scholar]
- 138.Barragán R., Ortega-Azorín C., Sorlí J.V., Asensio E.M., Coltell O., St-Onge M.-P., Portolés O., Corella D. Effect of Physical Activity, Smoking, and Sleep on Telomere Length: A Systematic Review of Observational and Intervention Studies. J. Clin. Med. 2021;11:76. doi: 10.3390/jcm11010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Buttet M., Bagheri R., Ugbolue U.C., Laporte C., Trousselard M., Benson A., Bouillon-Minois J.-B., Dutheil F. Effect of a Lifestyle Intervention on Telomere Length: A Systematic Review and Meta-Analysis. Mech. Ageing Dev. 2022;206:111694. doi: 10.1016/j.mad.2022.111694. [DOI] [PubMed] [Google Scholar]
- 140.Barcın-Güzeldere H.K., Aksoy M., Demircan T., Yavuz M., Beler M. Association between the Anthropometric Measurements and Dietary Habits on Telomere Shortening in Healthy Older Adults: A-Cross-Sectional Study. Geriatr. Gerontol. Int. 2023;23:565–572. doi: 10.1111/ggi.14620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vidacek N.Š., Nanic L., Ravlic S., Sopta M., Geric M., Gajski G., Garaj-Vrhovac V., Rubelj I. Telomeres, Nutrition, and Longevity: Can We Really Navigate Our Aging? J. Gerontol. A Biol. Sci. Med. Sci. 2017;73:39–47. doi: 10.1093/gerona/glx082. [DOI] [PubMed] [Google Scholar]
- 142.Leung C.W., Fung T.T., McEvoy C.T., Lin J., Epel E.S. Diet Quality Indices and Leukocyte Telomere Length Among Healthy US Adults: Data From the National Health and Nutrition Examination Survey, 1999–2002. Am. J. Epidemiol. 2018;187:2192–2201. doi: 10.1093/aje/kwy124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tucker L.A. Fruit and Vegetable Intake and Telomere Length in a Random Sample of 5448 U.S. Adults. Nutrients. 2021;13:1415. doi: 10.3390/nu13051415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Crous-Bou M., Molinuevo J.-L., Sala-Vila A. Plant-Rich Dietary Patterns, Plant Foods and Nutrients, and Telomere Length. Adv. Nutr. 2019;10:S296–S303. doi: 10.1093/advances/nmz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Valera-Gran D., Prieto-Botella D., Hurtado-Pomares M., Baladia E., Petermann-Rocha F., Sánchez-Pérez A., Navarrete-Muñoz E.-M. The Impact of Foods, Nutrients, or Dietary Patterns on Telomere Length in Childhood and Adolescence: A Systematic Review. Nutrients. 2022;14:3885. doi: 10.3390/nu14193885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ogłuszka M., Lipiński P., Starzyński R.R. Effect of Omega-3 Fatty Acids on Telomeres-Are They the Elixir of Youth? Nutrients. 2022;14:3723. doi: 10.3390/nu14183723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhou M., Zhu L., Cui X., Feng L., Zhao X., He S., Ping F., Li W., Li Y. Influence of Diet on Leukocyte Telomere Length, Markers of Inflammation and Oxidative Stress in Individuals with Varied Glucose Tolerance: A Chinese Population Study. Nutr. J. 2016;15:39. doi: 10.1186/s12937-016-0157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tucker L.A. Dietary Fiber and Telomere Length in 5674 U.S. Adults: An NHANES Study of Biological Aging. Nutrients. 2018;10:400. doi: 10.3390/nu10040400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vallverdú-Queralt A., de Alvarenga J.F.R., Estruch R., Lamuela-Raventos R.M. Bioactive Compounds Present in the Mediterranean Sofrito. Food Chem. 2013;141:3365–3372. doi: 10.1016/j.foodchem.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 150.Storniolo C.E., Sacanella I., Mitjavila M.T., Lamuela-Raventos R.M., Moreno J.J. Bioactive Compounds of Cooked Tomato Sauce Modulate Oxidative Stress and Arachidonic Acid Cascade Induced by Oxidized LDL in Macrophage Cultures. Nutrients. 2019;11:1880. doi: 10.3390/nu11081880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ojeda-Rodriguez A., Alcala-Diaz J.F., Rangel-Zuñiga O.A., Arenas-de Larriva A.P., Gutierrez-Mariscal F.M., Gómez-Luna P., Torres-Peña J.D., Garcia-Rios A., Romero-Cabrera J.L., Malagon M.M., et al. Association between Telomere Length and Intima-Media Thickness of Both Common Carotid Arteries in Patients with Coronary Heart Disease: From the CORDIOPREV Randomized Controlled Trial. Atherosclerosis. 2023;380:117193. doi: 10.1016/j.atherosclerosis.2023.117193. [DOI] [PubMed] [Google Scholar]
- 152.Corella D., Ordovas J.M. Nutrigenomics in Cardiovascular Medicine. Circ. Cardiovasc. Genet. 2009;2:637–651. doi: 10.1161/CIRCGENETICS.109.891366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dorajoo R., Chang X., Gurung R.L., Li Z., Wang L., Wang R., Beckman K.B., Adams-Haduch J., Yiamunaa M., Liu S., et al. Loci for Human Leukocyte Telomere Length in the Singaporean Chinese Population and Trans-Ethnic Genetic Studies. Nat. Commun. 2019;10:2491. doi: 10.1038/s41467-019-10443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ren W., Yang S., Chen X., Guo J., Zhao H., Yang R., Nie Z., Ding L., Zhang L. NCAPG2 Is a Novel Prognostic Biomarker and Promotes Cancer Stem Cell Maintenance in Low-Grade Glioma. Front. Oncol. 2022;12:918606. doi: 10.3389/fonc.2022.918606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gupta R., Somyajit K., Narita T., Maskey E., Stanlie A., Kremer M., Typas D., Lammers M., Mailand N., Nussenzweig A., et al. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell. 2018;173:972–988.e23. doi: 10.1016/j.cell.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lin N., Yao Z., Xu M., Chen J., Lu Y., Yuan L., Zhou S., Zou X., Xu R. Long Noncoding RNA MALAT1 Potentiates Growth and Inhibits Senescence by Antagonizing ABI3BP in Gallbladder Cancer Cells. J. Exp. Clin. Cancer Res. 2019;38:244. doi: 10.1186/s13046-019-1237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hodgkinson C.P., Gomez J.A., Payne A.J., Zhang L., Wang X., Dal-Pra S., Pratt R.E., Dzau V.J. Abi3bp Regulates Cardiac Progenitor Cell Proliferation and Differentiation. Circ. Res. 2014;115:1007–1016. doi: 10.1161/CIRCRESAHA.115.304216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Neither the participants’ consent forms nor ethics approval included permission for open access. However, we follow a controlled data-sharing collaboration model, and data for collaborations will be available upon request pending application and approval. Investigators who are interested in this study can contact the corresponding author.