Abstract
Apiospora, an ascomycetous genus in Apiosporaceae, comprises saprobes, endophytes, and pathogens of humans and plants. They have a cosmopolitan distribution with a wide range of hosts reported from Asia. In the present study, we collected and isolated Apiospora species from Wurfbainia villosa and grasses in Guangdong and Yunnan provinces in China. Multi-locus phylogeny based on the internal transcribed spacer, the large subunit nuclear rDNA, the partial translation elongation factor 1-α, and β-tubulin was performed to clarify the phylogenetic affinities of the Apiospora species. Based on the distinctive morphological characteristics and molecular evidence, Ap. endophytica, Ap. guangdongensis, Ap. wurfbainiae, and Ap. yunnanensis are proposed. Descriptions, illustrations, and notes for the newly discovered species are provided and compared with closely related Apiospora species. An updated phylogeny of Apiospora is presented, along with a discussion on the phylogenetic affinities of ambiguous taxa.
Keywords: Asia, Amphisphaeriales, Apiosporaceae, endophytes, saprobes, taxonomy
1. Introduction
Recent advances in fungal taxonomy and phylogeny have resulted in taxonomic revisions in numerous genera [1,2,3,4], including Apiospora. Apiospora belongs to Apiosporaceae, Amphisphaeriales, Sordariomycetes, and Ascomycota [5]. It was introduced by Saccardo [6], but the typification was not indicated. Subsequently, Clements and Shear [7] designated Apiospora montagnei Sacc. as the type species. However, Crous and Groenewald [8] synonymized Ap. montagnei under Arthrinium arundinis based on the presence of similar characters in their sexual morphs, including multi-locular perithecial stromata and hyaline ascospores surrounded by a thick gelatinous sheath, and also considering that Arthrinium is an older and more commonly referred to name than Apiospora [8,9,10,11]. Crous and Groenewald [8], therefore, treated the sexual genus Apiospora as a synonym of Arthinium on the basis that Arthinium is earlier proposal and in more frequent usage [10,11]. This taxonomic treatment has been followed by several studies [12,13,14]. Subsequently, Pintos and Alvarado [15] re-evaluated the phylogenetic placements of Apiospora and Arthrinium based on multi-locus phylogeny using the internal transcribed spacer (ITS), large subunit nuclear rDNA (LSU), the partial translation elongation factor 1-α (tef1-α), and β-tubulin (tub2) sequence data. The result showed that several Arthrinium species, including the type species Ar. caricicola, form a well-supported but distant clade compared to other Arthrinium species, indicating them into two independent genera. Therefore, the species within this clade were retained in Arthrinium, while other species were transferred to Apiospora [15]. Apiospora is accepted with conidia that are globose to subglobose in the face view and lenticular in the side view with a pale equatorial slit, whereas Arthrinium possesses conidia of various shapes (angular, curved, fusiform, globose, polygonal, and navicular) [15]. The sexual morphs of Apiospora are characterized by immersed, dark brown to black, lenticular, or dome-shaped ascostromata that are erumpent through a longitudinal split, unitunicate, broadly clavate to cylindric-clavate asci, and hyaline ascospores that are 1-septate near the lower end, with or without a sheath [13]. Based on the recent taxonomic treatment and multi-locus phylogenetic analyses, sixty-eight species of Arthrinium were synonymized under Apiospora [14,15,16]. Up to now, 133 epithets are listed under Apiospora in the Index Fungorum [17].
Species of Apiospora are distributed worldwide, mostly from terrestrial and aquatic habitats in Asia [14,17,18]. They are reported as important plant pathogens causing significant damage to economic plants. For example, Apiospora arundinis (previously known as Arthrinium arundinis) is a causal agent of leaf edge spot disease of peach (Prunus persica) in China, with a 20 to 40% disease incidence in two hectares of a severely infected peach orchard [19]. Apiospora arundinis has been commonly reported as the pathogen of Phyllostachys praecox, causing brown culm streak [20]. Apiospora sacchari is reported to cause Barley kernel blight [21], while Ap. phaeospermum is a pathogen causing damping-off disease in wheat [22]. In addition, Apiospora arundinis and A. montagnei have been reported as animal and human pathogens that cause onychomycosis [23,24]. They are also isolated from air and soil, while some are lichen-associated [12,17]. Many Apiospora species are known as saprobes and endophytes on many host plants, including thorny bamboo (Bambusa bambos), bristlegrass (Setaria viridis), loquat (Eriobotrya japonica), windmill palm (Trachycarpus fortunei), and tea (Camellia sinensis) [12,15,16,24,25,26,27,28,29].
In a survey for fungi associated with monocotyledon plants in China, we collected and isolated Apiospora strains from Wurfbainia villosa and grasses in Guangdong and Yunnan provinces. The identifications of Apiospora strains in this study were performed through the combination of ITS, LSU, tef1-α, and tub2 sequence analyses, along with morphological characteristics. A pairwise homoplasy index test was conducted to determine the recombination level within phylogenetically closely related species. The novel Apiospora species were identified, following the guidelines in Jeewon and Hyde [30], Maharachchikumbura et al. [31], and Pem et al. [4].
2. Materials and Methods
2.1. Sample Collection, Observation, and Isolation
Saprobic fungi were collected from dead stems of grasses at the Kunming Institute of Botany, Kunming City, Yunnan Province, China. The samples were placed into zip-lock bags and returned to the laboratory for fungal observation and isolation. The specimens were observed after 2–3 days of inoculation at room temperature using SZ650 (Chongqing Auto Optical Instrument Co., Ltd., Chongqing, China) stereo microscope. Fungal structures (e.g., ascomata, hamathecium, asci, and ascospores) were examined using Nikon Eclipse 80i, connected to the industrial Digital Sight DS-Fi1 (Panasonic, Tokyo, Japan) microscope imaging system. Single spore isolation was performed as described by Senanayake et al. [28]. The germinated spores were grown on potato dextrose agar (PDA: potato 200 g/L, dextrose 15 g/L, agar 15 g/L) and incubated at 25 ± 2 °C for two weeks.
Endophytic fungi were isolated from the healthy leaves of Wurfbainia villosa in Yongning town, Yangjiang City, Guangdong Province, China. The isolation procedures of plant materials were performed as described by Senanayake et al. [28]. Briefly, fresh, healthy leaves were gently rinsed with tap water to eliminate any accumulated particulate matter. The leaves were surface sterilized in 2.5% sodium hypochlorite for 1 min, followed by 75% ethanol for 2 min. The samples were subsequently rinsed three times with sterile water for 3 min each time and air-dried using sterile tissue filter paper. The sterilized leaves were then cut into 0.5 × 0.5 cm pieces using sterile scissors and aseptically transferred onto PDA and incubated at 25 °C [28]. The hyphal tips grown from sterilized leaves after three days of incubation were transferred to fresh PDA for three to four times for purification to obtain a pure culture.
All fungal isolates were preserved on PDA slants and stored at 4 °C and in 15% glycerol. The fungal structures were measured using Tarosoft (R) Image Frame Work program v. 0.9.7. and NIS-Elements BR 5.30.03. The living cultures were deposited in the Zhongkai University of Agriculture and Engineering Culture Collection (ZHKUCC), Guangdong, China. Herbarium specimens were deposited in the Mycological Herbarium of Zhongkai University of Agriculture and Engineering (MHZU), Guangzhou, China. The new species were registered in Faces of Fungi (FoF) (http://www.facesoffungi.org; accessed on 17 October 2023) [32] and Index Fungorum (IF) databases (http://www.indexfungorum.org/names/names.asp; accessed on 17 October 2023). The records of Greater Mekong Subregion fungi will be placed in the GMS database [33].
2.2. DNA Extraction, PCR Amplification, and Sequencing
Fungal mycelia grown on PDA for 5–7 days were collected for Genomic DNA extraction using the MagPure Plant DNA AS Kit, following the manufacturer’s instructions (Guangzhou Magen Biotechnology Co., Ltd., Guangzhou, China). Extracted DNA was stored at −20 °C. The internal transcribed spacer (ITS), large subunit rDNA (LSU), β-tubulin (tub2), and partial translation elongation factor 1–α (tef1-α) were amplified and sequenced using primer ITS1 and ITS4 [34,35], LR5 and LR0R [36], BT2a and BT2b [37], and EF1-728F and EF2 [38,39], respectively.
The 25 µL volume of Polymerase chain reaction (PCR) contains 12.5 µL 2 × Taq Master Mix (buffer, dNTPs, and Taq; Nanjing Vazyme Biotech Co., Ltd., Nanjing, China), 9.5 µL of ddH2O, 1 µL of each primer, and 1 µL of DNA template. The PCR thermal cycle program for ITS and LSU amplification was conducted with an initial denaturation at 95 °C for 3 min, followed by 35 cycles of 94 °C for 30 s; the annealing temperature was 52 °C for 30 s for ITS and LSU; 72 °C for 1 min; and final elongation at 72 °C for 10 min. The annealing temperatures were adjusted to 53.5 °C (30 s) and 55 °C (45 s) for tub2 and tef1-α, respectively. PCR products were purified and sequenced by Tianyi Huiyuan Gene Technology & Services Co. (Guangzhou, China). All sequences generated in this study were submitted to GenBank [40].
2.3. Phylogenetic Analyses
The sequence quality of obtained sequences was assured by checking chromatograms using Bioeidit v. 7.2.3 [41]. Sequences used for phylogenetic analysis were downloaded from GenBank according to the Blastn search of ITS in the GenBank database and following the published literature [16]. A total of 191 sequences were used in the phylogenetic analysis (Table 1). Sporocadus trimorphus strains CFCC 55171 and ROC 113 were used as outgroup taxa. Four loci, ITS, LSU, tef1-α, and tub2, were aligned in MAFFT version v. 7 online program [42] and edited manually where necessary using BioEdit v. 7.2.3 [41]. Alignments were converted to NEXUS format using Alignment Transformation Environment online platform (http://www.sing-group.org/ALTER/; accessed on 17 October 2023).
Table 1.
Details of taxa including their GenBank accession numbers used in the phylogenetic analyses of this study.
| Taxa | Strain Numbers | Substrates | Known Lifestyles | Countries | GenBank Accession Numbers | |||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | tub2 | tef1-α | |||||
| Apiospora acutiapica | KUMCC 20-0210 | Bambusa bambos | Saprobe | China | MT946343 | MT946339 | MT947366 | MT947360 |
| Ap. agari | KUC21333T | Agarum cribrosum | Not mentioned | Republic of Korea | MH498520 | - | MH498478 | MH544663 |
| Ap. agari | KUC21361 | Agarum cribrosum | Not mentioned | Republic of Korea | MH498519 | - | MH498477 | MN868914 |
| Ap. aquatica | S-642 | Submerged wood | Saprobe | China | MK828608 | MK835806 | - | - |
| Ap. arctoscopi | KUC21331T | Egg of Arctoscopus japonicus | Not mentioned | Republic of Korea | MH498529 | - | MH498487 | MN868918 |
| Ap. arctoscopi | KUC21344 | Egg of Arctoscopus japonicus | Not mentioned | Republic of Korea | MH498528 | - | MH498486 | MN868919 |
| Ap. arundinis | CBS 133509 | Aspergillus flavus sclerotium | Saprobe/endophyte | USA | KF144886 | KF144930 | KF144976 | KF145018 |
| Ap. arundinis | CBS 449.92 | Aspergillus flavus sclerotium | Saprobe/endophyte | USA | KF144887 | KF144931 | KF144977 | KF145019 |
| Ap. aurea | CBS 244.83T | - | Saprobe | Japan | AB220251 | KF144935 | KF144981 | KF145023 |
| Ap. balearica | CBS 145129T | Undetermined Poaceae | Saprobe | Spain | MK014869 | MK014836 | MK017975 | MK017946 |
| Ap. bambusicola | MFLUCC 20-0144T | Schizostachyum brachycladum | Saprobe | Thailand | MW173030 | MW173087 | - | MW183262 |
| Ap. biserialis | CGMCC 3.20135T | Bamboo | Saprobe | China | MW481708 | MW478885 | MW522955 | MW522938 |
| Ap. biserialis | GZCC 20-0099 | Bamboo | Saprobe | China | MW481709 | MW478886 | MW522956 | MW522939 |
| Ap. biserialis | GZCC 20-0100 | Bamboo | Saprobe | China | MW481710 | MW478887 | MW522957 | MW522940 |
| Ap. camelliae-sinensis | LC 5007T | Camellia sinensis | Endophyte | China | KY494704 | KY494780 | KY705173 | KY705103 |
| Ap. camelliae-sinensis | LC 8181 | Camellia sinensis | Endophyte | China | KY494761 | KY494837 | KY705229 | KY705157 |
| Ap. chiangraiense | MFLUCC 21-0053T | Dead culms of bamboo | Saprobe | Thailand | MZ542520 | MZ542524 | MZ546409 | - |
| Ap. chromolaenae | MFLUCC 17-1505T | Chromolaena odorata | Saprobe | Thailand | MT214342 | MT214436 | - | MT235802 |
| Ap. cordylines | GUCC 10026 | Cordyline fruticosa | Not mentioned | China | MT040105 | - | MT040147 | MT040126 |
| Ap. cyclobalanopsidis | CGMCC 3.20136T | Cyclobalanopsidis glauca | Saprobe | China | MW481713 | MW478892 | MW522962 | MW522945 |
| Ap. cyclobalanopsidis | GZCC 20-0103 | Cyclobalanopsidis glauca | Saprobe | China | MW481714 | MW478893 | MW522963 | MW522946 |
| Ap. descalsii | CBS 145130T | Ampelodesmos mauritanicus | Saprobe | Spain | MK014870 | MK014837 | MK017976 | MK017947 |
| Ap. dichotomanthi | LC 4950T | Dichotomanthes tristaniicarpa | Saprobe/endophyte | China | KY494697 | KY494773 | KY705167 | KY705096 |
| Ap. dichotomanthi | LC 8175 | Dichotomanthes tristaniicarpa | Saprobe/endophyte | China | KY494755 | KY494831 | KY705223 | KY705151 |
| Ap. dongyingensis | SAUCC 0302T | Leaf of bamboo | Pathogen | China | OP563375 | OP572424 | OP573270 | OP573264 |
| Ap. dongyingensis | SAUCC 0303 | Leaf of bamboo | Pathogen | China | OP563374 | OP572423 | OP573263 | OP573269 |
| Ap. endophytica | ZHKUCC 23-0006T | Wurfbainia villosa | Endophyte | China | OQ587996 | OQ587984 | OQ586062 | OQ586075 |
| Ap. endophytica | ZHKUCC 23-0007 | Wurfbainia villosa | Endophyte | China | OQ587997 | OQ587985 | OQ586063 | OQ586076 |
| Ap. esporlensis | CBS 145136T | Phyllostachys aurea | Saprobe | Spain | MK014878 | MK014845 | MK017983 | MK017954 |
| Ap. euphorbiae | IMI 285638b | Bambusa sp. | Saprobe | Bangladesh | AB220241 | AB220335 | AB220288 | - |
| Ap. fermenti | KUC21289T | Seaweed | Not mentioned | Republic of Korea | MF615226 | - | MF615231 | MH544667 |
| Ap. fermenti | KUC21288 | Seaweed | Not mentioned | Republic of Korea | MF615230 | - | MF615235 | MH544668 |
| Ap. gaoyouensis | CFCC 52301T | Phragmites australis | Saprobe | China | MH197124 | - | MH236789 | MH236793 |
| Ap. gaoyouensis | CFCC 52302 | Phragmites australis | Saprobe | China | MH197125 | - | MH236790 | MH236794 |
| Ap. garethjonesii | KUMCC 16-0202T | Dead culms of bamboo | Saprobe | China | KY356086 | KY356091 | - | - |
| Ap. gelatinosa | KHAS 11962T | Bamboo | Saprobe | China | MW481706 | MW478888 | MW522958 | MW522941 |
| Ap. gelatinosa | GZAAS 20-0107 | Bamboo | Saprobe | China | MW481707 | MW478889 | MW522959 | MW522942 |
| Ap. guangdongensis | ZHKUCC 23-0004T | Wurfbainia villosa | Endophyte | China | OQ587994 | OQ587982 | OQ586060 | OQ586073 |
| Ap. guangdongensis | ZHKUCC 23-0005 | Wurfbainia villosa | Endophyte | China | OQ587995 | OQ587983 | OQ586061 | OQ586074 |
| Ap. guiyangensis | HKAS 102403T | Unidentified grass | Saprobe | China | MW240647 | MW240577 | MW775604 | MW759535 |
| Ap. guizhouensis | LC 5318 | Air in karst cave, bamboo | Airborne/endophyte | China | KY494708 | KY494784 | KY705177 | KY705107 |
| Ap. guizhouensis | LC 5322T | Air in karst cave, bamboo | Airborne/endophyte | China | KY494709 | KY494785 | KY705178 | KY705108 |
| Ap. hainanensis | SAUCC 1681T | Leaf of bamboo | Pathogen | China | OP563373 | OP572422 | OP573268 | OP573262 |
| Ap. hainanensis | SAUCC 1682 | Leaf of bamboo | Pathogen | China | OP563372 | OP572421 | OP573267 | OP573261 |
| Ap. hispanica | IMI 326877T | Beach sand | Saprobe | Spain | AB220242 | AB220336 | AB220289 | - |
| Ap. hydei | CBS 114990T | Culms of Bambusa tuldoides | Saprobe | Hong Kong, China | KF144890 | KF144936 | KF144982 | KF145024 |
| Ap. hydei | KUMCC 16-0204 | Bambusa tuldoides | Saprobe | China | KY356087 | KY356092 | - | - |
| Ap. hyphopodii | MFLUCC 15-0003T | Bambusa tuldoides | Saprobe | China | KR069110 | - | - | - |
| Ap. hyphopodii | KUMCC 16-0201 | Bambusa tuldoides | Saprobe | China | KY356088 | KY356093 | - | - |
| Ap. hysterina | ICPM 6889T | Bamboo | Saprobe | New Zealand | MK014874 | MK014841 | MK017980 | MK017951 |
| Ap. hysterina | CBS 145133 | Bamboo | Saprobe | New Zealand | MK014875 | MK014842 | MK017981 | MK017952 |
| Ap. iberica | CBS 145137T | Arundo donax | Saprobe | Portugal | MK014879 | MK014846 | MK017984 | MK017955 |
| Ap. intestini | CBS 135835T | Gut of a grasshopper | Saprobe | India | KR011352 | MH877577 | KR011350 | KR011351 |
| Ap. intestini | MFLUCC 21-0052 | Gut of a grasshopper | Saprobe | India | MZ542521 | MZ542525 | MZ546410 | MZ546406 |
| Ap. italica | CBS 145138T | Arundo donax | Saprobe | Italy | MK014880 | MK014847 | MK017985 | MK017956 |
| Ap. italica | CBS 145139 | Arundo donax | Saprobe | Italy | MK014881 | MK014848 | MK017986 | - |
| Ap. jatrophae | AMH-9557T | Jatropha podagrica | Saprobe | India | JQ246355 | - | - | - |
| Ap. jatrophae | AMH-9556 | Jatropha podagrica | Saprobe | India | HE981191 | - | - | - |
| Ap. jiangxiensis | LC 4494 | Maesa sp. | Endophyte | China | KY494690 | KY494766 | KY705160 | KY705089 |
| Ap. jiangxiensis | LC 4577T | Maesa sp. | Endophyte | China | KY494693 | KY494769 | KY705163 | KY705092 |
| Ap. kogelbergensis | CBS 113332 | Dead culms of Restionaceae | Saprobe | South Africa | KF144891 | KF144937 | KF144983 | KF145025 |
| Ap. kogelbergensis | CBS 113333T | Dead culms of Restionaceae | Saprobe | South Africa | KF144892 | KF144938 | KF144984 | KF145026 |
| Ap. koreana | KUC21332T | Egg of Arctoscopus japonicus | Not mentioned | Republic of Korea | MH498524 | - | MH498482 | MH544664 |
| Ap. koreana | KUC21348 | Egg of Arctoscopus japonicus | Not mentioned | Republic of Korea | MH498523 | - | MH498481 | MN868927 |
| Ap. lageniformis | KUC21686T | Branch of Phyllostachys pubescens | Not mentioned | Republic of Korea | ON764022 | ON787761 | ON806636 | ON806626 |
| Ap. lageniformis | KUC21687 | Branch of Phyllostachys pubescens | Not mentioned | Republic of Korea | ON764023 | ON787762 | ON806637 | ON806627 |
| Ap. locuta-pollinis | LC 11688 | Brassica campestris | Saprobe | China | MF939596 | - | MF939623 | MF939618 |
| Ap. locuta-pollinis | LC 11683T | Brassica campestris | Saprobe | China | MF939595 | - | MF939622 | MF939616 |
| Ap. longistroma | MFLUCC 11-0479 | Dead culms of bamboo | Saprobe | Thailand | KU940142 | KU863130 | - | - |
| Ap. longistroma | MFLUCC 11-0481T |
Dead culms of bamboo | Saprobe | Thailand | KU940141 | KU863129 | - | - |
| Ap. magnispora | ZHKUCC 22-0001 | Bamboo | Saprobe | China | OM728647 | OM486971 | OM0543544 | OM543543 |
| Ap. malaysiana | CBS 102053T | Macaranga hullettii | Saprobe | Malaysia | KF144896 | KF144942 | KF144988 | KF145030 |
| Ap. marianiae | CBS 148710T | Phleum pratense | Saprobe | Spain | NR_183001 | NG_149092 | - | |
| Ap. marianiae | AP301119 | Phleum pratense | Saprobe | Spain | ON692407 | ON692423 | ON677187 | ON677181 |
| Ap. marii | CBS 497.90T | Beach sands | Saprobe | Spain | AB220252 | KF144947 | KF144993 | KF145035 |
| Ap. marii | DiSSPA_A1 | Beach sands | Saprobe | Spain | MK602320 | - | MK614695 | MK645472 |
| Ap. marina | KUC21328T | Seaweed | Not mentioned | Republic of Korea | MH498538 | - | MH498496 | MH544669 |
| Ap. marina | KUC21353 | Seaweed | Not mentioned | Republic of Korea | MH498537 | - | MH498495 | MN868923 |
| Ap. mediterranea | IMI 326875T | Air | Saprobe | Spain | AB220243 | AB220337 | AB220290 | - |
| Ap. minutispora | 1.70-41 | Mountain soil | Soil | Republic of Korea | LC517882 | - | LC518888 | LC518889 |
| Ap. mori | MFLUCC 20-0181T | Morus australis | Saprobe | Taiwan | MW114313 | MW114393 | - | - |
| Ap. mori | NCYUCC 19-034 | Morus australis | Saprobe | Taiwan | MW114314 | MW114394 | - | - |
| Ap. mukdahanensis | MFLUCC 22-0056T | dead bamboo leave | Saprobe | Thailand | OP377735 | OP377742 | - | OP381089 |
| Ap. multiloculata | MFLUCC 21-0023T | Dead bamboo | Saprobe | Thailand | OL873137 | OL873138 | - | - |
| Ap. mytilomorpha | DAOM 214595T | Andropogon sp. | Saprobe | India | KY494685 | - | - | - |
| Ap. neobambusae | LC 7106T | Leaves of bamboo | Saprobe/endophyte | China | KY494718 | KY494794 | KY705186 | KY806204 |
| Ap. neobambusae | LC 7124 | Leaves of bamboo | Saprobe/endophyte | China | KY494727 | KY494803 | KY705195 | KY806206 |
| Ap. neochinensis | CFCC 53036T | Fargesia qinlingensis | Saprobe | China | MK819291 | - | MK818547 | MK818545 |
| Ap. neochinensis | CFCC 53037 | Fargesia qinlingensis | Saprobe | China | MK819292 | - | MK818548 | MK818546 |
| Ap. neogarethjonesii | KUMCC 18-0192 | Bamboo | Saprobe | China | MK070897 | MK070898 | - | - |
| Ap. neosubglobosa | JHB 006 | Bamboo | Saprobe | China | KY356089 | KY356094 | - | - |
| Ap. neosubglobosa | KUMCC 16-0203T | Bamboo | Saprobe | China | KY356090 | KY356095 | - | - |
| Ap. obovata | LC 4940T | Lithocarpus sp. | Endophyte | China | KY494696 | KY494772 | KY705166 | KY705095 |
| Ap. obovata | LC 8177 | Lithocarpus sp. | Endophyte | China | KY494757 | KY494833 | KY705225 | KY705153 |
| Ap. ovata | CBS 115042T | Arundinaria hindsii | Saprobe | China | KF144903 | KF144950 | KF144995 | KF145037 |
| Ap. paraphaeosperma | MFLUCC 13-0644T | Dead culms of bamboo | Saprobe | Thailand | KX822128 | KX822124 | - | - |
| Ap. phragmitis | CPC 18900T | Phragmites australis | Saprobe | Italy | KF144909 | KF144956 | KF145001 | KF145043 |
| Ap. phyllostachydis | MFLUCC 18-1101T | Phyllostachys heteroclada | Saprobe | China | MK351842 | MH368077 | MK291949 | MK340918 |
| Ap. piptatheri | CBS 145149T | Piptatherum miliaceum | Saprobe | Spain | MK014893 | MK014860 | - | MK017969 |
| Ap. pseudohyphopodii | KUC21680T | Culm of Phyllostachys pubescens | Not mentioned | Republic of Korea | ON764026 | ON787765 | ON806640 | ON806630 |
| Ap. pseudohyphopodii | KUC21684 | Culm of Phyllostachys pubescens | Not mentioned | Republic of Korea | ON764027 | ON787766 | ON806641 | ON806631 |
| Ap. pseudoparenchymatica | LC 7234T | Leaves of bamboo | Endophyte | China | KY494743 | KY494819 | KY705211 | KY705139 |
| Ap. pseudoparenchymatica | LC 8173 | Leaves of bamboo | Endophyte | China | KY494753 | KY494829 | KY705221 | KY705149 |
| Ap. pseudorasikravindrae | KUMCC 20-0208T | Bambusa dolichoclada | Saprobe | China | MT946344 | - | MT947367 | MT947361 |
| Ap. pseudosinensis | CPC 21546T | Leaves of bamboo | Saprobe | Netherlands | KF144910 | KF144957 | - | KF145044 |
| Ap. pseudospegazzinii | CBS 102052T | Macaranga hullettii | Saprobe | Malaysia | KF144911 | KF144958 | KF145002 | KF145045 |
| Ap. pterosperma | CBS 123185 | Lepidosperma gladiatum | Saprobe | Australia | KF144912 | KF144959 | KF145003 | - |
| Ap. pterosperma | CPC 20193T | Lepidosperma gladiatum | Saprobe | Australia | KF144913 | KF144960 | KF145004 | KF145046 |
| Ap. pusillisperma | KUC21321T | Seaweed | Not mentioned | Republic of Korea | MH498533 | - | MH498491 | MN868930 |
| Ap. pusillisperma | KUC21357 | Seaweed | Not mentioned | Republic of Korea | MH498532 | - | MH498490 | MN868931 |
| Ap. qinlingensis | CFCC 52303T | Fargesia qinlingensis | Saprobe | China | MH197120 | - | MH236791 | MH236795 |
| Ap. qinlingensis | CFCC 52304 | Fargesia qinlingensis | Saprobe | China | MH197121 | - | MH236792 | MH236796 |
| Ap. rasikravindrae | LC 8179 | Brassica rapa | Saprobe | China | KY494759 | KY494835 | KY705227 | KY705155 |
| Ap. rasikravindrae | NFCCI 2144T | Soil | Saprobe | Norway | JF326454 | - | - | - |
| Ap. rasikravindrae | MFLUCC 21-0051 | Dead culms of bamboo | Saprobe | Thailand | MZ542523 | MZ542527 | MZ546412 | MZ546408 |
| Ap. rasikravindrae | MFLUCC 21-0054 | Dead culms of Maize | Saprobe | Thailand | MZ542522 | MZ542526 | MZ546411 | MZ546407 |
| Ap. sacchari | CBS 372.67 | Air | Endophyte | - | KF144918 | KF144964 | KF145007 | KF145049 |
| Ap. sacchari | CBS 664.74 | Soil under Calluna vulgaris | Endophyte | Netherlands | KF144919 | KF144965 | KF145008 | KF145050 |
| Ap. saccharicola | CBS 191.73 | Air | Endophyte | Netherlands | KF144920 | KF144966 | KF145009 | KF145051 |
| Ap. saccharicola | CBS 831.71 | - | Endophyte | Netherlands | KF144922 | KF144969 | KF145012 | KF145054 |
| Ap. sargassi | KUC21228T | Sargassum fulvellum | Not mentioned | Republic of Korea | KT207746 | - | KT207644 | MH544677 |
| Ap. sargassi | KUC21232 | Sargassum fulvellum | Not mentioned | Republic of Korea | KT207750 | - | KT207648 | MH544676 |
| Ap. sasae | CBS 146808T | dead culms | Saprobe | Netherlands | MW883402 | MW883797 | MW890120 | MW890104 |
| Ap. septata | CGMCC 3.20134T | bamboo | Saprobe | China | MW481711 | MW478890 | MW522960 | MW522943 |
| Ap. septata | GZCC 20-0109 | bamboo | Saprobe | China | MW481712 | MW478891 | MW522961 | MW522944 |
| Ap. serenensis | IMI 326869T | excipients, atmosphere andhome dust | Saprobe | Spain | AB220250 | AB220344 | AB220297 | - |
| Ap. setariae | MT492005 | Setaria viridis | Saprobe | China | MT492005 | - | MT497467 | MW118457 |
| Ap. setostroma | KUMCC 19-0217T | Dead branches of bamboo | Saprobe | China | MN528012 | MN528011 | - | MN527357 |
| Ap. sichuanensis | HKAS 107008T | dead culm of grass | Saprobe | China | MW240648 | MW240578 | MW775605 | MW759536 |
| Ap. sorghi | URM 93000T | Sorghum bicolor | Endophyte | Brazil | MK371706 | - | MK348526 | - |
| Ap. sp. | ZHKUCC 23-0010 | Wurfbainia villosa | Endophyte | China | OQ588000 | OQ587988 | OQ586066 | OQ586079 |
| Ap. sp. | ZHKUCC 23-0011 | Wurfbainia villosa | Endophyte | China | OQ588001 | OQ587989 | OQ586067 | OQ586080 |
| Ap. sp. | ZHKUCC 23-0012 | Wurfbainia villosa | Endophyte | China | OQ588002 | OQ587990 | OQ586068 | OQ586081 |
| Ap. sp. | ZHKUCC 23-0013 | Wurfbainia villosa | Endophyte | China | OQ588003 | OQ587991 | OQ586069 | OQ586082 |
| Ap. stipae | CBS 146804T | dead culm of Stipa gigantea | Saprobe | Spain | MW883403 | MW883798 | MW890121 | MW890082 |
| Ap. subglobosa | MFLUCC 11-0397T | Dead culms of bamboo | Saprobe | Thailand | KR069112 | KR069113 | - | - |
| Ap. subrosea | LC 7291 | Leaves of bamboo | Endophyte | China | KY494751 | KY494827 | KY705219 | KY705147 |
| Ap. subrosea | LC 7292T | Leaves of bamboo | Endophyte | China | KY494752 | KY494828 | KY705220 | KY705148 |
| Ap. taeanensis | KUC21322T | Seaweed | Not mentioned | Republic of Korea | MH498515 | - | MH498473 | MH544662 |
| Ap. taeanensis | KUC21359 | Seaweed | Not mentioned | Republic of Korea | MH498513 | - | MH498471 | MN868935 |
| Ap. thailandica | MFLUCC 15-0199 | Dead culms of bamboo | Saprobe | Thailand | KU940146 | KU863134 | - | - |
| Ap. thailandica | MFLUCC 15-0202T | Dead culms of bamboo | Saprobe | Thailand | KU940145 | KU863133 | - | - |
| Ap. tropica | MFLUCC 21-0056T | Dead culms of bamboo | Saprobe | Thailand | OK491657 | OK491653 | OK560922 | - |
| Ap. vietnamensis | IMI 99670T | Citrus sinensis | Saprobe | Vietnam | KX986096 | KX986111 | KY019466 | - |
| Ap. wurfbainiae | ZHKUCC 23-0008T | Wurfbainia villosa | Endophyte | China | OQ587998 | OQ587986 | OQ586064 | OQ586077 |
| Ap. wurfbainiae | ZHKUCC 23-0009 | Wurfbainia villosa | Endophyte | China | OQ587999 | OQ587987 | OQ586065 | OQ586078 |
| Ap. xenocordella | CBS 478.86T | Soil from roadway | Soil | Zimbabwe | KF144925 | KF144970 | KF145013 | KF145055 |
| Ap. xenocordella | CBS 595.66 | On dead branches | Saprobe | Misiones | KF144926 | KF144971 | - | - |
| Ap. yunnana | DDQ 00281 | Phyllostachys nigra | Saprobe | China | KU940148 | KU863136 | - | - |
| Ap. yunnana | MFLUCC 15-1002T | Phyllostachys nigra | Saprobe | China | KU940147 | KU863135 | - | - |
| Ap. yunnanensis | ZHKUCC 23-0014T | Grass | Saprobe | China | OQ588004 | OQ587992 | OQ586070 | OQ586083 |
| Ap. yunnanensis | ZHKUCC 23-0015 | Grass | Saprobe | China | OQ588005 | OQ587993 | OQ586071 | OQ586084 |
| Arthrinium austriacum | GZU 345004 | Carex pendula | Saprobe | Austria | MW208928 | - | - | - |
| Ar. austriacum | GZU 345006 | Carex pendula | Saprobe | Austria | MW208929 | MW208860 | - | - |
| Ar. sporophleum | GZU 345102 | Carex firma | Saprobe | Austria | MW208944 | MW208866 | - | - |
| Ar. caricicola | CBS 145127 | Carex ericetorum | Saprobe | China | MK014871 | MK014838 | MK017977 | MK017948 |
| Ar. crenatum | AG19066T | Dead leaves of grass (probably Festuca burgundiana) |
Saprobe | France | MW208931 | MW208861 | - | - |
| Ar. curvatum | AP 25418 | Leaves of Carex sp. | Saprobe | China | MK014872 | MK014839 | MK017978 | MK017949 |
| Ar. japonicum | IFO 30500 | - | Saprobe | Japan | AB220262 | AB220356 | AB220309 | - |
| Ar. japonicum | IFO 31098 | Leaves of Carex despalata | Saprobe | Japan | AB220264 | AB220358 | AB220311 | - |
| Ar. luzulae | AP7619-3T | Luzula sylvatica | Saprobe | Spain | MW208937 | MW208863 | - | - |
| Ar. morthieri | GZU 345043 | Carex pilosa | Saprobe | Austria | MW208938 | MW208864 | - | - |
| Ar. phaeospermum | CBS 114317 | Leaves of Hordeum vulgare | Saprobe | Iran | KF144906 | KF144953 | KF144998 | KF145040 |
| Ar. phaeospermum | CBS 114318 | Leaves of Hordeum vulgare | Saprobe | Iran | KF144907 | KF144954 | KF144999 | KF145041 |
| Ar. puccinioides | CBS 549.86 | Lepidosperma gladiatum | Saprobe | Germany | AB220253 | AB220347 | AB220300 | - |
| Ar. sphaerospermum | CBS 146355 | Probably on Poaceae | Saprobe | Norway | MW208943 | MW208865 | - | - |
| Ar. sporophleum | CBS 145154 | Dead leaves of Juncus sp. | Saprobe | Spain | MK014898 | MK014865 | MK018001 | MK017973 |
| Ar. trachycarpum | CFCC 53039 | Trachycarpus fortune | Pathogen | China | MK301099 | - | MK303395 | MK303397 |
| Ar. urticae | IMI 326344 | - | Saprobe | - | AB220245 | AB220339 | AB220292 | - |
| Nigrospora aurantiaca | CGMCC 3.18130T | Nelumbo sp. | Saprobe | China | KX986064 | KX986098 | KY019465 | KY019295 |
| N. camelliae-sinensis | CGMCC 3.18125T | Camellia sinensis | Endophyte/pathogen | China | KX985986 | KX986103 | KY019460 | KY019293 |
| N. chinensis | CGMCC 3.18127T | Machilus breviflora | Endophyte/pathogen | China | KX986023 | KX986107 | KY019462 | KY019422 |
| N. gorlenkoana | CBS 480.73 | Vitis vinifera | Endophyte/pathogen | Kazakhstan | KX986048 | KX986109 | KY019456 | KY019420 |
| N. guilinensis | CGMCC 3.18124T | Camellia sinensis | Endophyte/pathogen | China | KX985983 | KX986113 | KY019459 | KY019292 |
| N. hainanensis | CGMCC 3.18129T | Musa paradisiaca | Endophyte/pathogen | China | KX986091 | KX986112 | KY019464 | KY019415 |
| N. lacticolonia | CGMCC 3.18123T | Camellia sinensis | Endophyte/pathogen | China | KX985978 | KX986105 | KY019458 | KY019291 |
| N. musae | CBS 319.34 | Musa sp. | Endophyte/pathogen | Australia | MH855545 | KX986110 | KY019455 | KY019419 |
| N. oryzae | LC2693 | Neolitsea sp. | Saprobe | China | KX985944 | KX986101 | KY019471 | KY019299 |
| N. osmanthi | CGMCC 3.18126T | Hedera nepalensis | Endophyte/pathogen | China | KX986010 | KX986106 | KY019461 | KY019421 |
| N. pyriformis | CGMCC 3.18122T | Citrus sinensis | Endophyte/pathogen | China | KX985940 | KX986100 | KY019457 | KY019290 |
| N. rubi | LC2698T | Rubus sp. | Endophyte/pathogen | China | KX985948 | KX986102 | KY019475 | KY019302 |
| N. sphaerica | LC7298 | Nelumbo sp. | Saprobe | China | KX985937 | KX986097 | KY019606 | KY019401 |
| N. vesicularis | CGMCC 3.18128T | Musa paradisiaca | Endophyte | China | KX986088 | KX986099 | KY019463 | KY019294 |
| Sporocadus trimorphus | CFCC 55171T | Rose | Not mentioned | China | OK655798 | OK560389 | OM401677 | OL814555 |
| S. trimorphus | ROC 113 | Rose | Not mentioned | China | OK655799 | OK560390 | OM401678 | OL814556 |
Notes: Newly generated sequences in this study are in blue. “T” indicates ex-type. “-” = information not available. Abbreviations: AMH: Ajrekar Mycological Herbarium, Pune, Maharashtra, India; AP: Alvarado Pintos; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands; CFCC: China Forestry Culture Collection Center, Beijing, China; CGMCC: China General Micro biological Culture Collection; CPC: Culture collection of Pedro Crous, housed at the Westerdijk Fungal Biodiversity Institute; DAOM: Canadian Collection of Fungal Cultures, Ottawa, Canada; GUCC: Guizhou University Culture Collection, Guizhou, China; GZAAS: Guizhou Academy of Agricultural Sciences herbarium, China; GZCC: Guizhou Culture Collection, China; GZU: University of Graz, Austria; HKAS: Herbarium of Cryptogams, Kunming Institute of Botany, Chinese Academy of Sciences, Yunnan, China; ICMP: International Collection of Microorganisms from Plants, New Zealand; IFO: Institute for Fermentation, Osaka, Japan; IMI: Culture collection of CABI Europe UK Centre, Egham, UK; JHB: H.B. Jiang; KUC: the Korea University Fungus Collection, Seoul, Korea; SFC the Seoul National University Fungus Collection; KUMCC: Culture collection of Kunming Institute of Botany, Yunnan, China; LC: Personal culture collection of Lei Cai, housed in the Institute of Microbiology, Chinese Academy of Sciences, China; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; NFCCI: National Fungal Culture Collection of India; SAUCC: Shandong Agricultural University Culture Collection.
Maximum likelihood (ML) and Bayesian inference (BI) analyses were performed in the CIPRES Science Gateway online platform [43] based on the combined ITS, LSU, tef1-α, and tub2 sequence data. The ML analysis was carried out with GTR+G+I evolutionary substitution using RAxML-HPC v.8.2.12 on XSEDE (https://www.phylo.org/; accessed on 17 October 2023) [44], with 1000 rapid bootstrap inferences, followed by a thorough ML search. All free model parameters were estimated by RAxML ML of 25 per site rate categories. The likelihood of the final tree was evaluated and optimized under GAMMA. Bayesian Inference (BI) analysis was conducted using the Markov Chain Monte Carlo (MCMC) method and performed in MrBayes XSEDE (3.2.7a) [45]. Six simultaneous Markov chains were run for 2,000,000 generations, and the trees were sampled for each 100th generation. Phylogenetic trees were visualized in FigTree v. 1.4.0 [46] and formatted using PowerPoint 2010 (Microsoft Corporation, WA, USA).
2.4. Pairwise Homoplasy Index (PHI)
A pairwise homoplasy index (PHI) test [47] was performed using SplitsTree v. 4.15.1 [48] to determine the recombination level within phylogenetically closely related species of the new strains in this study (Apiospora endophytica, A. guangdongensis) with A. arundinis, A. aurea, A. cordylies, and A. hydei. The combined ITS, LSU, tef1-α, and tub2 of these phylogenetically closely related species were applied for PHI test and analyses. The PHI results (Φw) > 0.05 indicated no significant recombination in the dataset. The relationships between our strains with closely related taxa were visualized by constructing splits graphs using Log-Det transformation and split decomposition options using SplitsTree v. 4.15.1.
3. Results
3.1. Phylogeny
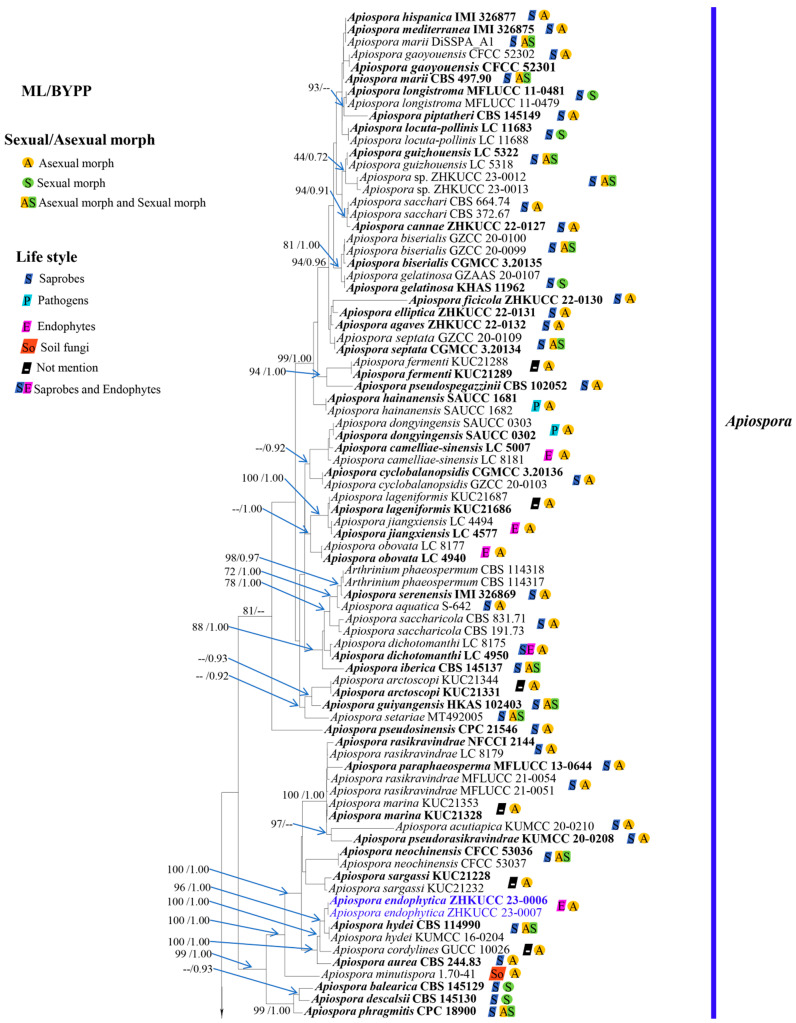
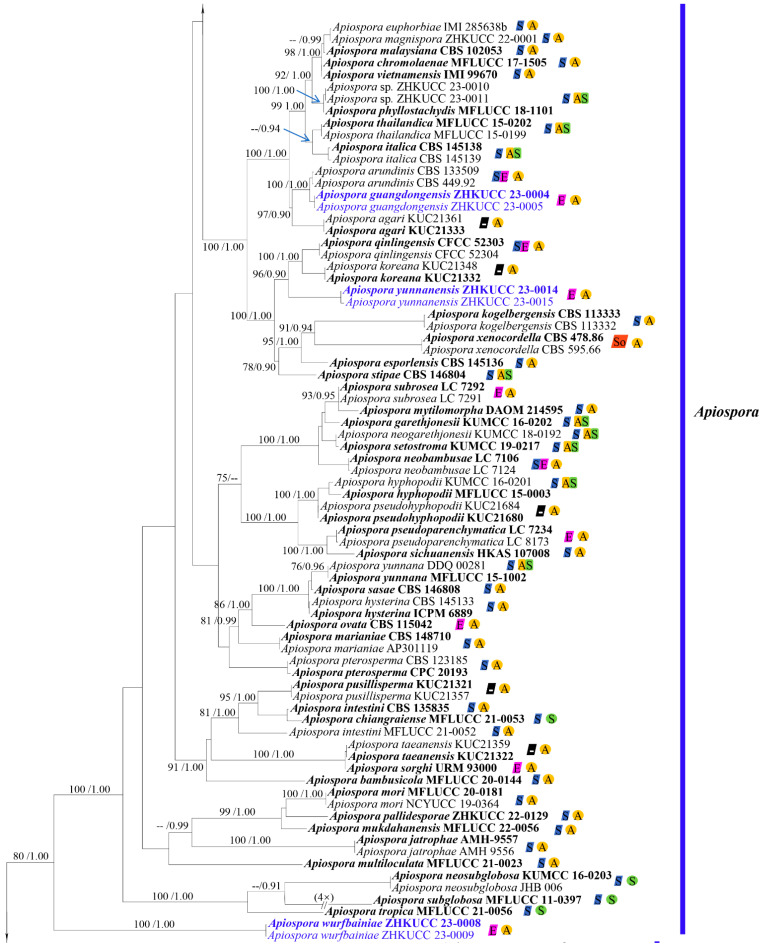
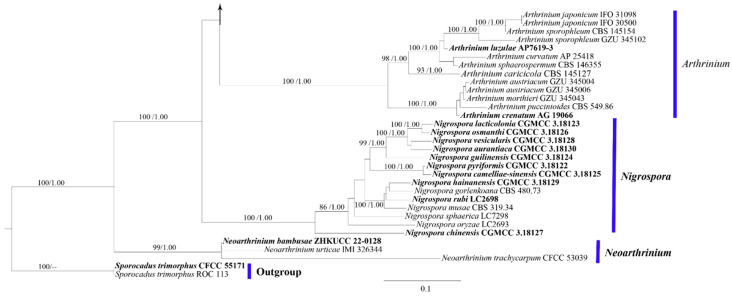
The phylogenetic tree was constructed based on the combined ITS, LSU, tef1-α, and tub2 sequence data of 191 strains (including our new strains), with Sporocadus trimorphus strains CFCC 55171 and ROC 113 as outgroup taxa. There are a total of 2936 characters, including gaps (ITS: 1–772, LSU: 773–1621, tef1-α: 1622–2309, tub2: 2310–2936). The topology of the ML analysis was similar to the BI analysis, and the best-scoring RAxML tree with a final ML optimization likelihood value of -36321.892470 is presented (Figure 1). The matrix had 1805 distinct alignment patterns, with 38.08% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.208057, C = 0.296775, G = 0.242495, T = 0.252673; substitution rates AC = 1.090339, AG = 3.411914, AT = 1.286700, CG = 0.887072, CT = 4.062650, GT = 1.000000; gamma distribution shape parameter α = 0.777262. Phylogenetic analyses showed that our strains belong to Apiospora. The isolates ZHKUCC 23-0010 and ZHKUCC 23-0011 had a close affinity to Apiospora phyllostachydis (MFLUCC 18-1101) with 100% ML bootstrap support and 1.00 BYPP. The isolates ZHKUCC 23-0004 and ZHKUCC 23-0005 formed a sister to A. arundinis (CBS 449.92 and CBS 133509) with 100% ML bootstrap support and 1.00 BYPP. Two isolates of ZHKUCC 23-0014 and ZHKUCC 23-0015 formed a distinct lineage and sister to A. qinlingensis (CFCC 52303 and CFCC 52304) and A. koreana (KUC21332 and KUC21348) with 96% ML bootstrap support and 0.90 posterior probability in BI analysis. The isolates ZHKUCC 23-0012 and ZHKUCC 23-0013 clustered with A. guizhounese (LC 5318 and LC 5322) with low support in ML and BI analyses (44% ML and 0.72 BYPP). The isolates ZHKUCC 23-0006 and ZHKUCC 23-0007 formed a sister to A. hydei (CBS 114990 and KUMCC 16-0204) with 96% ML bootstrap support and 1.00 BYPP. Two isolates, ZHKUCC 23-0008 and ZHKUCC 23-0009, formed a distinct lineage and sister to Apiospora species with 80% ML and 1.00 BYPP (Figure 1).
Figure 1.
Phylogram generated from maximum likelihood analysis (RAxML) of genera in Apiosporaceae based on ITS, LSU, tef1-α, and tub2 sequence data. Maximum likelihood bootstrap values equal or above 75%, and Bayesian posterior probabilities equal or above 0.90 (ML/BYPP) are given at the nodes. A strain number is noted after the species name. The tree is rooted with Sporocadus trimorphus (CFCC 55171) and (ROC 113). Hyphen (-) represents support values below 75% ML and 0.90 BYPP. The ex-type strains are bolded black, and the new isolates are in blue.
3.2. A Pairwise Homoplasy Index
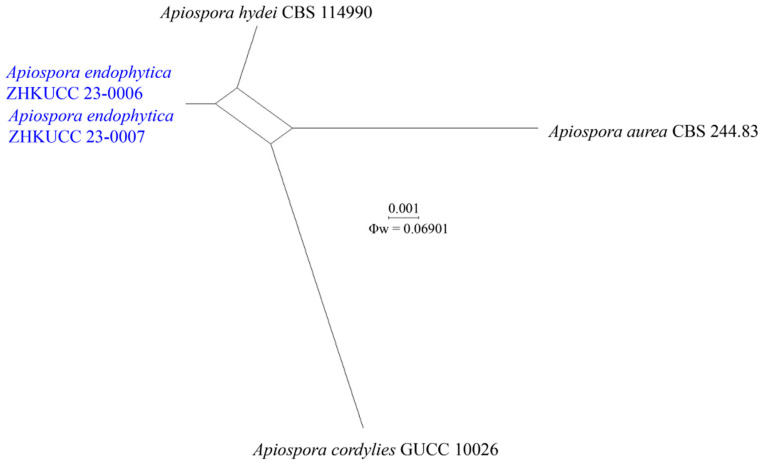
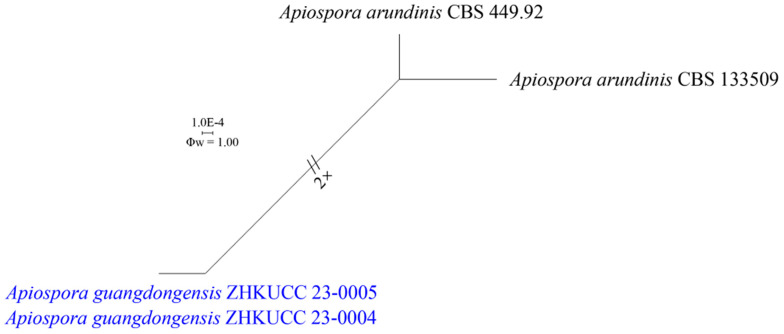
The recombination level within phylogenetically closely related species of generated strains of Apiospora endophytica with A. aurea, A. cordylines, and A. hydei as well as phylogenetically closely related species of A. guangdongensis with A. arundinis were implied in a pairwise homoplasy index (PHI) test using combined ITS, LSU, tef1-α, and tub2 sequence dataset. The PHI result showed that there was no evidence of significant recombination (Φw = 0.06901) among A. endophytica, A. aurea, A. cordylines, and A. hydei with the combined dataset (Figure 2). The A. guangdongensis and A. arundinis has also no significant evidence of recombination (Φw = 1.00) (Figure 3).
Figure 2.
Split graph showing the results of the pairwise homoplasy index (PHI) test of the combined ITS, LSU, tef1-α, and tub2 sequence data between Apiospora endophytica (ZHKU 23-0006, ZHKU 23-0007) with three closely related taxa of A. aurea CBS 244.83, A. hydei CBS 114990, and A. cordylies GUCC 10026 using LogDet transformation and splits decomposition. PHI test result (Φw) = 0.06901 indicates no significant recombination within the dataset (Φw > 0.05). The generated sequences are indicated in blue.
Figure 3.
Split graph showing the results of the pairwise homoplasy index (PHI) test of the combined ITS, LSU, tef1-α, and tub2 sequence data between Apiospora guangdongensis (ZHKUCC 23-0004, ZHKUCC 23-0005) with the closely related taxa of A. arundinis (CBS 449.92, CBS 133509) using LogDet transformation and splits decomposition. PHI test result (Φw) = 1.00 indicates no significant recombination within the dataset (Φw > 0.05). The generated sequences are indicated in blue.
3.3. Taxonomy
Apiospora endophytica C.F. Liao and Doilom, sp. nov. Figure 4.
Figure 4.
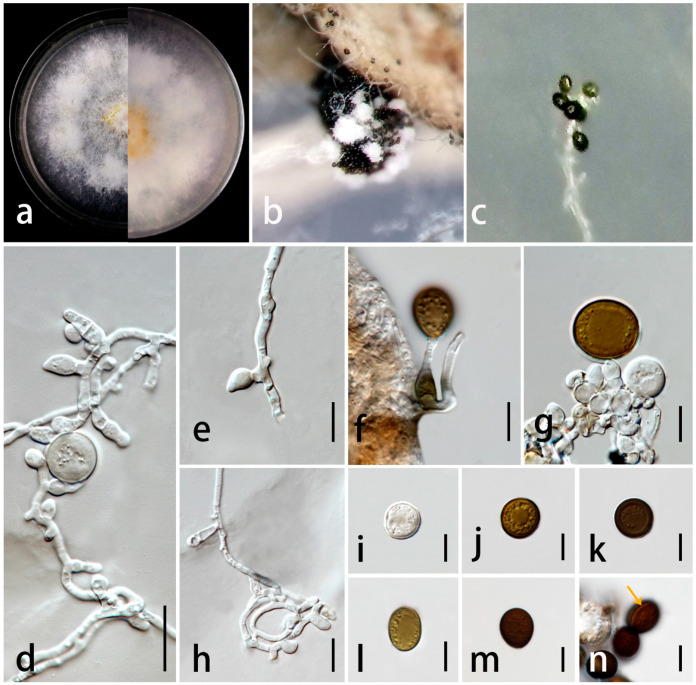
Apiospora endophytica (ZHKU 23-0002, holotype). (a) Upper view and reverse view of culture on PDA. (b,c) Conidia on aerial mycelia on PDA. (d–h) Conidiophores with conidiogenous cells. (i–m) Conidia in the face view. (n) Conidia with germ-slit. Scale bars in (d–n) = 10 μm.
Index Fungorum number: IF900356; Facesoffungi number: FoF14658.
Etymology: The epithet “endophytica” refers to the endophytic lifestyle of the species.
Endophytic in leaves of Wurfbainia villosa. Sexual morph: undetermined. Asexual morph: sporulating on PDA after one month, spore mass visible as black, scattered on white colonies. Hyphae 2–5 μm wide ( = 2.5 μm, n = 30), branched, hyaline to golden brown, septate, smooth-walled. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 4–14 × 2–7 μm ( = 7.5 × 5 μm, n = 35), aggregated in clusters or solitary, hyaline to golden brown, erect, unbranched, cylindrical or clavate, ampulliform or obtriangular, and smooth-walled. Conidia 14–19 × 12–18 μm ( = 17 × 15 μm, n = 30) in the face view, 11–19 × 9–16 μm ( = 15 × 12 μm, n = 20) in the side view, initially hyaline, becoming pale brown to dark brown, globose to subglobose, obovoid to ellipsoidal in the face view, lenticular with a thick equatorial slit in the side view, and smooth-walled. Sterile cells not observed.
Culture characteristics: colonies on PDA reached 2.6 cm in one week at 28 ± 2 °C, fluffy, spreading, with dense, aerial mycelium, composed of small bumps, forming a circle around the center, surface and reverse both golden yellow in the center, and turning white at the edge.
Material examined: China, Guangdong Province, Yangjiang City, Yongning town, 24°40′53″ N 118°41′31″ E, asymptomatic leaves of Wurfbainia villosa (Lour.) Škorničk. and A.D. Poulsen (Zingiberaceae), 1 October 2021, Chunfang Liao, (ZHKU 23-0002, holotype, dried culture); ex-type living culture ZHKUCC 23-0006, ibid., and living culture ZHKUCC 23-0007.
Notes: In the phylogenetic analyses (Figure 1), Ap. endophytica (ZHKUCC 23-0006, ZHKUCC 23-0007) clustered sister to Ap. hydei (CBS 114990 and KUMCC 16-0204) with 96% ML bootstrap support and 1.00 BYPP and formed a distinct lineage separated from Ap. cordylines (GUCC 10026) with 100% ML bootstrap support and 1.00 BYPP) and Ap. aurea (CBS 244.83) by 100% ML bootstrap support and 1.00 BYPP. Morphologically, conidiogenous cells of Ap. endophytica are cylindrical or clavate, ampulliform or obtriangular, while they are subcylindrical to doliiform to lageniform in Ap. hydei. The conidia of Ap. endophytica are dark brown and smooth, while they are brown and roughened in Ap. hydei. In addition, Ap. endophytica has larger conidiogenous cells compared to than those of Ap. hydei (4–14 × 2–7 μm vs. 5–8 × 4–5 μm). Apiospora endophytica differs from Ap. cordylines and Ap. aurea based on the size and shape of conidiogenous cells and conidia (Table 2). The PHI test results indicated no significant recombination between Ap. endophytica and closely related species Ap. aurea (CBS 244.83), Ap. cordylies (GUCC 10026), and Ap. hydei (CBS 114990) (Figure 2). Both morphological and molecular evidence supported Ap. endophytica as a new species.
Table 2.
Synopsis of morphological characteristics of Ap. endophytica and its closely related species.
| Characters | Apiospora Species | |||
|---|---|---|---|---|
| Ap. endophytica | Ap. hydei | Ap. cordylines | Ap. aurea | |
| Host/substrate | Asymptomatic leaf of Wurfbainia villosa | Culms of Bambusa tuldoides | Leaves of Cordyline fruticosa | Air |
| Conidiophores | Reduced to conidiogenous cells | Pale brown, smooth, subcylindrical, transversely septate, branched, 20–40 × 3–5 μm | NA | NA |
| Conidiogenous cells | Aggregated in clusters or solitary, hyaline to golden brown, smoothly, erect, unbranched, cylindrical or clavate, ampulliform or obtriangular, 4–14 × 2–7 μm ( = 7.5 × 5 μm) | Aggregated in clusters, brown, smooth, subcylindrical to doliiform to lageniform, 5–8 × 4–5 μm | Erect, aggregated into clusters, hyaline to pale brown, smooth, doliiform to ampulliform or lageniform, (3–)5–10(–15) × 2.6–5.3 µm ( = 7.0 × 4.5 µm) |
Integrated, polyblastic, denticulate |
| Conidia | Initially hyaline, becoming pale brown to dark brown, globose to subglobose, obovoid to ellipsoidal in the face view, lenticular with a thick equatorial slit in the side view, smooth-walled, 14–19 × 12–18 μm ( = 17 × 15 μm, n = 30) in the face view, 11–19 × 9–16 μm ( = 15 × 12 μm, n = 20) | Brown, roughened, globose in face view, lenticular in the side view, with pale equatorial slit, (15–)17– 19(–22) μm diam. in face view, (10–)11–12(–14) μm diam. in the side view, with a central scar, 1.5–2 μm diam. | Olivaceous to brown, smooth to finely roughened, subglobose to ellipsoidal, 15–19 × 12.5–18.5 µm ( = 17.5 × 15.7 µm) | Solitary, terminal, and sometimes also lateral with a hyaline rim, brown or dark brown, smooth, aseptate, 10–30 × 10–15 μm |
| Reference | This study | [8] | [49] | [50] |
NA: undetermined.
Apiospora guangdongensis C.F. Liao and Doilom, sp. nov. Figure 5.
Figure 5.
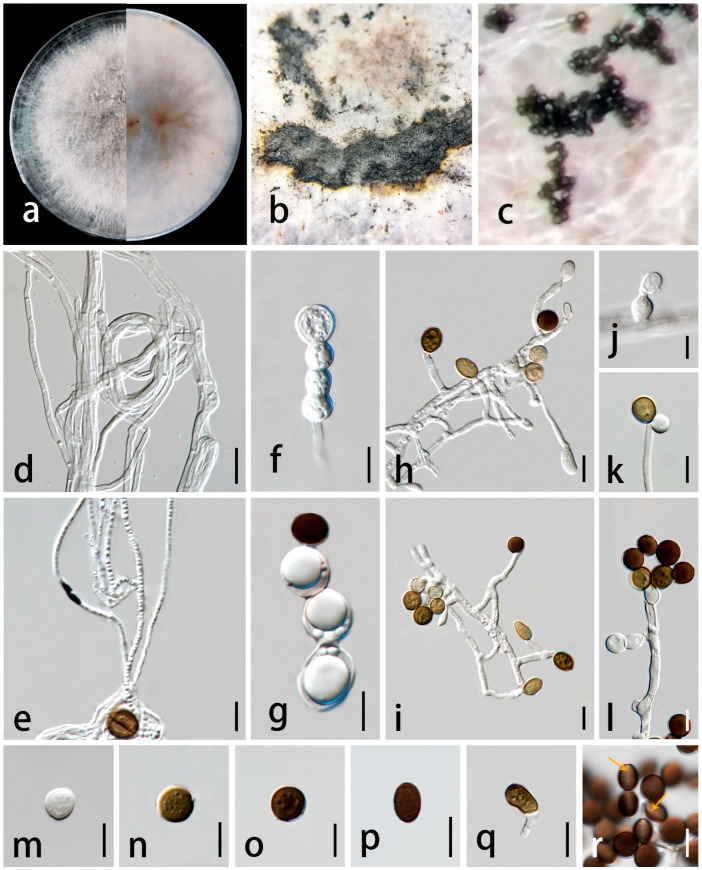
Apiospora guangdongensis (ZHKU 23-0001, holotype). (a) Upper view and reverse view of culture on PDA. (b,c) Conidia on aerial mycelia on PDA. (d,e) Mycelium. (f,g) Chlamydospores. (h–l) Conidiophores with conidiogenous cells. (m–p) Conidia in the face view. (q) Elongated conidia (sterile cells). (r) Conidia with germ-slit (arrows). Scale bars in (d–r) = 10 μm.
Index Fungorum number: IF900357; Facesoffungi number: FoF14659.
Etymology: The epithet “guangdongensis” refers to the locality, Guangdong Province, China where the holotype was collected.
Endophytic in asymptomatic leaves of Wurfbainia villosa. Sexual morph: undetermined. Asexual morph: sporulated on PDA after one month, spore mass visible as black, scattered to aggregated on white colonies. Hyphae 2–3 μm diam. ( = 2.5 μm, n = 30), branched, hyaline, septate, smooth, thin-walled, forming hyphal coils. Conidiophores 45–53 × 2–4 μm ( = 49 × 2.5 μm, n = 30), micronematous, mononematous, erect, solitary, subcylindrical, unbranched, straight or flexuous, hyaline, smooth-walled, sometimes reduced to conidiogenous cells. Conidiogenous cells 4–9 × 2–5 μm ( = 6 × 3.5 μm, n = 30), arising from hyphae, aggregated in clusters or solitary, terminal or lateral, smooth, straight or slightly curved, cylindrical or ampulliform, and sometimes ovate or obpyriform. Conidia 6–9 × 5–9 μm ( = 8 × 7 μm, n = 30) in the face view, 5–8 × 4–6 μm ( = 6.5 × 5 μm, n = 30) in the side view, initially hyaline, becoming pale brown to dark brown, globose to ellipsoidal in face view, lenticular with broad equatorial slit in the side view, aseptate, smooth-walled. Sterile cells 9–16 × 3–8 μm ( = 12 × 5 μm, n = 30), light brown, elongate. Chlamydospores produced in chain, terminal, globose to subglobose, hyaline, smooth-walled.
Culture characteristics: colonies on PDA reaching 6.6 cm in one week at 28 ± 2 °C, floccose, sparse, concentrically spreading, forming aerial mycelia, edge irregular, surface pale brown in center, white at the edge, with punctate or flaky black spores, reverse white to pale brown with some pale brown spot, no pigment.
Material examined: China, Guangdong Province, Yangjiang City, Yongning town, 24°40′53″ N 118°41′31″ E, asymptomatic leaves of Wurfbainia villosa (Lour.) Škorničk. and A.D. Poulsen (Zingiberaceae), 1 October 2021, Chunfang Liao, (ZHKU 23-0001, holotype, dried culture); ex-type cultures ZHKUCC 23-0004, ibid., living culture ZHKUCC 23-0005.
Notes: The phylogenetic analyses showed that Ap. guangdongensis (ZHKUCC 23-0004 and ZHKUCC 23-0005) formed a sister branch to Ap. arundinis with 100% ML bootstrap support and 1.00 BYPP (Figure 1). The morphology of Ap. guangdongensis differs from Ap. arundinis by having shorter conidiogenous cells (4–9 × 2–5 μm vs. 6–12 × 3–4 μm) and larger conidia (6–9 × 5–9 μm vs. (5–)6–7 μm in the face view, 5–8 × 4–6 μm vs. 3–4 μm) [8]. The conidiogenous cells of Ap. guangdongensis are cylindrical or ampulliform, sometimes ovate or obpyriform, while they are ampulliform in Ap. arundinis. The result of the PHI test showed no significant recombination between our isolates and Ap. arundinis (Figure 3). Based on distinct morphological and molecular evidence, we propose Ap. guangdongensis as a new species.
Apiospora wurfbainiae C.F. Liao and Doilom, sp. nov. Figure 6.
Figure 6.
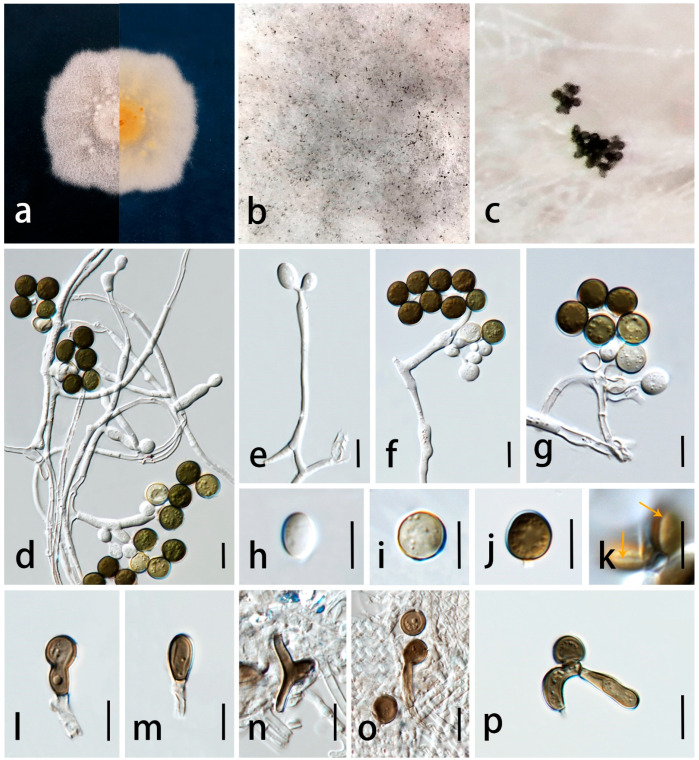
Apiospora wurfbainiae (ZHKU 23-0003, holotype). (a) Upper view and reverse view of culture on PDA. (b,c) Conidia on aerial mycelia on PDA. (d–g) Conidia with conidiogenous cells. (d–j) Conidia. (k) Conidia in the side view with germ-slit (arrows). (l–n) Sterile cells. (o,p) Sterile cell with conidia. Scale bars in (d–p) = 10 μm.
Index Fungorum number: IF900355; Facesoffungi number: FoF14660.
Etymology: The epithet “Wurfbainiae” refers to the host genus Wurfbainia, from which the holotype was collected.
Endophytic in asymptomatic leaves of Wurfbainia villosa. Sexual morph: undetermined. Asexual morph: sporulated on PDA after three months, spore mass visible as black, scattered on colonies. Hyphae 1–3 μm diam. ( = 2 μm, n = 30), branched, hyaline, septate, smooth, forming hyphal coils. Conidiophores reduced to conidiogenous cells, hyaline, smooth, branched. Conidiogenous cells 7–50 × 2–8 μm ( = 22 × 5 μm, n = 60), holoblastic, monoblastic, discrete, hyaline, straight or curved, cylindrical to lageniform, smooth-walled. Conidia 7–9 × 5–9 μm ( = 8 × 7 μm, n = 30) in the face view, 6–9 × 3–6 μm ( = 7 × 4.5 μm, n = 20) in the side view, obovoid, globose to subglobose in face view, lenticular with pale equatorial slit in the side view, initially hyaline, becoming pale brown to dark brown, multi-guttulate, smooth-walled. Sterile cells 8–31 × 2–12 μm ( = 14 × 5 μm, n = 30), light brown, elongated, cylindrical, ovate, triangular-shaped.
Culture characteristics: colonies on PDA reaching 6.8 cm in one week at 28 ± 2 °C, flatted, dense mycelium, edge regular, gray in the center, with some white globular spots from above; pale yellow to gray with some orange spots from below.
Material examined: China, Guangdong Province, Yangjiang City, Yongning town, 24°40′53″ N 118°41′31″ E, asymptomatic leaves of Wurfbainia villosa (Lour.) Škorničk. and A.D. Poulsen (Zingiberaceae), 1 October 2021, Chunfang Liao, (ZHKU 23-0003, holotype, dried culture); ex-type living culture ZHKUCC 23-0008, ibid., living culture ZHKUCC 23-0009.
Notes: Apiospora wurfbainiae shares morphological similarities to Ap. guangdongensis in having globose conidia as well as overlapping conidial size (7–9 × 5–9 μm vs. 6–9 × 5–9 μm in the face view). However, Ap. wurfbainiae has larger conidiogenous cells (7–50 × 2–8 μm vs. 4–9 × 2–5 μm) than Ap. guangdongensis. The sterile cells of Ap. wurfbainiae are elongated, cylindrical, ovate, triangular-shaped while only elongated cells were observed in Ap. guangdongensis.
In the phylogenetic analysis (Figure 1), Ap. wurfbainiae (ZHKUCC 23-0008, ZHKUCC 23-0009) form a distinct subclade which is basal to Apiospora clade with 80% ML and 1.00% BYPP. Further, this subclade is closely related to another subclade consisting of Ap. tropica, Ap. subglobosa, and Ap. neosubglobosa. Morphologically, Ap. tropica, Ap. subglobosa, and Ap. neosubglobosa were described based on their sexual morph but Ap. wurfbainiae was identified solely by its asexual morph, thus their morphological characteristics could not be compared. However, molecular evidence clearly separates Ap. wurfbainiae from other known Apiospora species. Hence, we introduce Ap. wurfbainiae as a novel species.
Apiospora yunnanensis C.F. Liao and Doilom, sp. nov. Figure 7.
Figure 7.
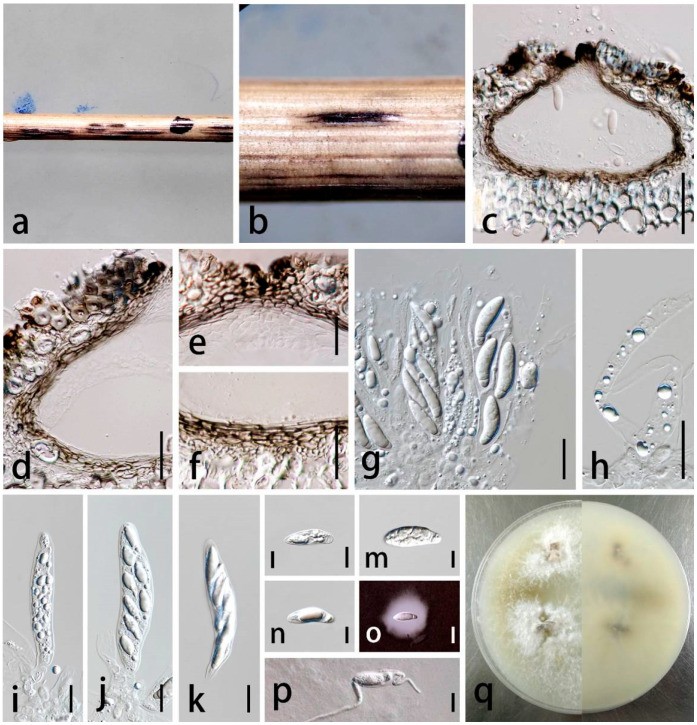
Apiospora yunnanensis (ZHKU 23-0004, holotype). (a,b) Appearance of ascomata on substrate. (c) Vertical section through ascoma. (d) Peridium. (e) Peridium at the top. (f) Peridium at the base. (g) Hamathecium with asci. (h) Hamathecium. (i–k) Asci. (l–n) Ascospores. (o) Ascospore in Indian Ink. (p) Germinated ascospore. (q) Culture characteristics on PDA (left-front, right-reverse). Scale bars in (c–k) = 20 μm, (i–p) = 10 μm.
Index Fungorum number: IF900358; Facesoffungi number: FoF14661.
Etymology: The epithet “yunnanensis” refers to the location, Yunnan Province, China where the holotype was collected.
Saprobic on dead stem of grass. Sexual morph: Ascostromata 750–3600 × 230–420 μm
( = 1590 × 290 μm, n = 20), solitary to gregarious, scattered, immersed to erumpent, with the long axis broken at the top, black, ostiolate. Ascomata 75–155 × 125–245 μm ( = 125 × 200 μm, n = 20), perithecial, immersed, pale brown to black, ampulliform to subglobose with a flattened base in cross-section, 1–2-loculate. Ostiole 35–80 μm wide ( = 54 μm, n = 20), periphysate, central. Peridium 8–26 μm wide ((= 17 μm, n = 50), 2–5-layered, outer layer composed of brown to dark brown, intermixed with host tissue, thick-walled, inner layer composed of hyaline, thin-walled cells of textura angularis. Hamathecium 5–13 μm wide ( = 9 μm, n = 25), composed of hyaline, septate, unbranched paraphyses, embedded in a gelatinous matrix. Asci 70–93 × 15–23 μm ( = 81 × 18 μm, n = 30), 8-spored, unitunicate, broadly cylindrical to clavate, apically rounded, with a pedicel. Ascospores 21–30 × 6–10 μm ( = 23 × 8 μm, n = 50), overlapping 1–2-seriate, clavate to fusiform, 1-septate, composed of a large upper cell and small lower cell, straight to slightly curved near the lower cell, guttulate, hyaline, smooth-walled, and surrounded by a gelatinous sheath. Asexual morph: undetermined.
Culture characteristics: Colonies on PDA reaching 6.0 cm in one week at 28 ± 2 °C, cottony in the center, dense, flat, edge mycelium spars, surface white in center, reverse white to pale brown.
Material examined: China, Yunnan Province, Kunming Institute of botanical garden, 25°02′11″ N 102°42′31″ E, dead stem of grass (Poaceae), 20 July 2019, Chunfang Liao, (ZHKU 23-0004, holotype, dried culture); ex-type living culture ZHKUCC 23-00014, ibid., living culture ZHKUCC 23-00015.
Notes: In the phylogenetic analysis, Ap. yunnanensis (ZHKUCC 23-00014, ZHKUCC 23-00015) formed a distinct branch with Ap. koreana and Ap. qinlingensis with ML = 96%, and BYPP = 0.90% (Figure 1). In comparison between ITS, tef1-α, and tub2 sequence data between our isolate (ZHKUCC 23-00014; ex-type) and Ap. koreana (KUC21332; ex-type), there were differences in 9.44% (51/540 bp), 6.85% (32/467 bp), and 9.31% (38/408 bp), respectively, while the comparison with Ap. qinlingensis (CFCC 52303; ex-type) showed differences in 13.61% (78/573 bp), 21.9% (97/442 bp), and 10.3% (52/505 bp), respectively. The LSU sequence data are currently unavailable for Ap. koreana and Ap. qinlingensis. The morphological characteristics of Ap. yunnanensis cannot be compared with those of its phylogenetically closely related species, as Ap. koreana and Ap. qinlingensis were described based on their asexual morph. While Ap. yunnanensis is currently known only from its sexual morph, attempts to sporulate its conidia on media with pine needles have been unsuccessful.
Morphologically, Ap. yunnanensis is similar to Ap. montagnei in having immersed to erumpent ascostromata, with the long axis broken at the top, broadly cylindrical to clavate asci and clavate to fusiform ascospores. However, Ap. yunnanensis is distinguished from Ap. montagnei by its shorter and wider asci (70–93 × 15–23 μm vs. 72–115 × 14–18 µm) and larger ascospores (20–30 × 6–10 μm vs. 21–25 × 6–8 µm) [15]. The comparison of LSU sequence data from our isolate Ap. yunnanensis (ZHKUCC 23-00014) with the sequences identified as Ap. montagnei ICMP 6967 and AFTOL-ID 951 in NCBI databases revealed differences of 2.24% (18/804 bp) and 2.28% (18/788 bp), respectively. We hereby propose Ap. yunnanensis as a novel species.
4. Discussion
The species diversity of Apiospora has been expanding steadily, especially in China. To date, 40 Apiospora species have been introduced in China, including four novel species in this study [14,16,28,29,51] (Table 1). These four new species, Ap. endophytica, Ap. guangdongensis, Ap. wurfbainiae, and Ap. yunnanensis, are introduced based on morphological characteristics and multi-locus phylogenetic analyses. Based on the host diversity of Apiospora species reported by Monkai et al. [52], it was found that most Apiospora species are associated with Poaceae (63%), including bamboo (31%), non-bamboo (32%), and other plant families (27%). Our study reveals another Apiospora species, Ap. yunnanensis, which was isolated from grass (Poaceae). Furthermore, the additional three species, Ap. endophytica, Ap. guangdongensis, and Ap. wurfbainiae, have been found on W. villosa belonging to the plant family Zingiberaceae. It is likely that W. villosa harbors high Apiospora species diversity. In addition, several Apiospora species have been reported from various monocotyledon plants, including bamboos, Cordyline fruticose, grasses, and Phragmites australis [8,13,49] (this study). It suggested that monocotyledon plants may harbor a high species diversity of Apiospora species.
Our study presents an updated phylogeny for Apiospora species, which is the additional contribution of this study to the previous works. By integrating the recent literature from Pintos et al. [8], Tian et al. [16], and Phukhamsakda et al. [53] with our new collections, we recognize 93 species including four newly discovered species based on multi-locus phylogenetic analyses and morphology. However, the phylogenetic analyses of combined ITS, LSU, tef1-α, and tub2 revealed a close phylogenetic relationship between Ap. hispanica and Ap. mediterranea (Figure 1), which is consistent with the previous studies in Tian et al. [16], Monkai et al. [52], and Phukhamsakda et al. [53]. The comparison of LSU, ITS, and tub2 sequence data showed that Ap. hispanica is identical to Ap. Mediterranea; however, their tef1-α sequence data are currently unavailable in GenBank. Morphologically, Ap. hispanica is similar to Ap. mediterranea by having basauxic, macronematous, and mononematous conidiophores, but it has smaller conidia than Ap. mediterranea (7.5–8.5 × 6.2–7.6 μm vs. 9–9.5 × 7.5–9 μm) [54]. Our phylogenetic result supports the suggestion of Monkai et al. [52] that the morphological reexamination of the type specimens of Ap. hispanica and Ap. mediterranea, including their molecular data from additional genes such as tef1-α, should be investigated to confirm a putative synonymy.
In addition, Ap. marina shares a close phylogenetic affinity with Ap. paraphaeosperma and Ap. rasikravindrae, and these three species clustered sister to Ap. acutiapica and Ap. pseudorasikravindrae with 100% ML and 1.00 BYPP support (Figure 1), which is consistent with the phylogenetic result in Monkai et al. [52]. Morphologically, Ap. marina is similar to Ap. paraphaeosperma and Ap. rasikravindrae by having brown, smooth, globose to elongate conidia, but Ap. marina has smaller conidia than Ap. paraphaeosperma (9.5–)10–12 (−13) × (7.5–)8.0–10 μm vs. 10–19 μm diam.), and Ap. rasikravindrae (9.5–)10–12 (−13) × (7.5–)8.0–10 vs. 10−15 × 6.0−10.5 μm) (Supplementary Table S1). Regarding the aforementioned factors, we suggest that the species boundaries of these ambiguous species should be re-evaluated to confirm the taxonomic status and to facilitate the identification of species grouped in this clade, and that tef1-α and tub2 sequence data from the ex-type of Ap. rasikravindrae (NFCCI 2144) are required. Additionally, there are 41 morphospecies (species without molecular data) listed under Apiospora (Supplementary Table S2). Pintos and Alvarado [15] examined the lectotype for Sphaeria apiospora (=Ap. montagnei, type species of Apiospora) specimens preserved at the PC fungarium, which were collected from Poaceae in lowland Mediterranean habitats. The taxonomic status of the remaining taxa, lacking sequence information and comprehensive morphological descriptions, remains uncertain and requires further investigation.
In this study, we compiled the available information on the sexual/ asexual morph of Apiospora species, including their known lifestyle from the relevant literature (Table 1). According to these data, 12 species have only been reported in their sexual morphs, while 63 species are known solely by their asexual morphs. Additionally, 19 species have been described in both sexual and asexual morphs. The prevalence of Apiospora species is likely to be associated with their asexual morph occurring as saprobic and endophytic lifestyles. On the other hand, the sexual morph is commonly observed from saprobic isolates thus far. Moreover, some Apiospora species have been reported in several lifestyles. For example, Ap. arundinis, Ap. hydei, Ap. thailandica, and Ap. yunnana have been reported in both saprobes and endophytes [8,25,55]. In addition, Ap. arundinis has been known as a saprobe, endophyte and pathogen [56]. The investigation into the potential transition of endophytic or saprobic of Apiospora to alternative lifestyles, such as becoming pathogens, is crucial for understanding their ecological role.
In view of the biological applications, many species of Apiospora produce an interesting bioactive secondary metabolite which could be a promising source of pharmacological and medicinal applications. For instance, a saprobic isolate of Ap. chromolaenae showed antimicrobial activity against Escherichia coli [57]. Apiospora saccharicola and Ap. sacchari isolated from Miscanthus sp. are known to produce industrially important enzymes [58]. Apiospora arundinis and Ap. saccharicola isolated from a brown alga Sargassum sp. produce antimicrobial substances that can inhibit some plant pathogenic fungi [59]. The endophytic Ap. rasikravindrae was isolated from the stem of Coleus amboinicus, which produces a compound with strong antimicrobial and cytotoxic activities [60]. Eijk [61] reported that Ap. sphaerosperma produced a tetrahydroxy anthraquinone pigment and other metabolites, such as ergosterol, succinic acid, and phenolic compounds C18O5. Li et al. [62] conducted whole-genome sequencing of Ap. sphaerosperma and revealed the potential of Ap. sphaerosperma AP-Z13 to synthesize various secondary metabolites based on transcriptomics, proteomics, and metabolomics analyses. However, many novel Apiospora species, including new species in this study, are untapped natural resources and only Ap. sphaerosperma has been the subject of whole-gene sequencing and omics research [62]. The future necessitates further metabolomics analyses to investigate the biological applications of both known and newly discovered Apiospora species, in order to comprehensively explore their biological properties.
Acknowledgments
We would like to express our gratitude to Shaun Pennycook (Landcare Research, New Zealand) for his critical nomenclatural review. We would also like to thank Zhongkai University of Agriculture and Engineering and Mae Fah Luang University for providing research facilities.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof9111087/s1, Supplementary Table S1. Synopsis of morphological characteristics of Ap. marina and its closely related species. Supplementary Table S2. Morphospecies of Apiospora. All data availability was mentioned in the manuscript. The novel taxa were registered in Index Fungorum (http://www.indexfungorum.org/Names/Names.asp, accessed on 26 June 2023) including Index Fungorum numbers IF900357, IF900356, IF900355, IF900358. Final alignment and phylogenetic tree were deposited in TreeBase (https://www.treebase. org/, accessed on 16 October 2023) with submission ID: 30849) and the newly generated sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/submit/, accessed on 26 June 2023) followed as ITS: OQ587994, OQ587995, OQ587996, OQ587997, OQ587998, OQ587999, OQ588000, OQ588001, OQ588002, OQ588003, OQ588004, OQ588005; LSU: OQ587982, OQ587983, OQ587984, OQ587985, OQ587986, OQ587987, OQ587988, OQ587989, OQ587990, OQ587991, OQ587992, OQ587993; tub2: OQ586060, OQ586061, OQ586062, OQ586063, OQ586064, OQ586065, OQ586066, OQ586067, OQ586068, OQ586069, OQ586070, OQ586071; tef1-α: OQ586073, OQ586074, OQ586075, OQ586076, OQ586077, OQ586078, OQ586079, OQ586080, OQ586081, OQ586082, OQ586083, OQ586084.
Author Contributions
Conceptualization, C.L. and M.D.; methodology, C.L.; software, C.L., M.D., I.C.S., K.T. and M.D.; formal analysis, C.L.; investigation, I.C.S.; resources, M.D.; data curation, C.L.; writing—original draft preparation, C.L.; writing—review and editing, C.L., M.D., I.C.S., K.T., W.D., Y.Z. and M.D.; visualization, M.D.; supervision, W.D.; project administration, K.W.T.C.; funding acquisition, M.D. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was supported by the Science and Technology Bureau of Guangzhou City (grant numbers 2023A04J1425 and 2023A04J1426), Guangdong University Key Laboratory for Sustainable Control of Fruit and Vegetable Diseases and Pests (grant number KA21031C502), the High-level Talents in Zhongkai University of Agriculture and Engineering (grant number J2201080102), the Starting Research Fund from Zhongkai University of Agriculture and Engineering, Guangzhou, Guangdong, China (grant number KA22016B746), the National Natural Science Foundation of China (grant number 32200015), and the Innovative team program of the Department of Education of Guangdong Province (grant numbers 2022KCXTD015 and 2022ZDJS020).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Phookamsak R., Liu J.K., McKenzie E.H.C., Manamgoda D.S., Ariyawansa H., Thambugala K.M., Dai D.Q., Camporesi E., Chukeatirote E., Wijayawardene N.N., et al. Revision of Phaeosphaeriaceae. Fungal Divers. 2014;68:159–238. [Google Scholar]
- 2.Hongsanan S., Li Y.M., Liu J.K., Hofmann T., Piepenbring M., Bhat D.J., Boonmee S., Doilom M., Singtripop C., Tian Q., et al. Revision of genera in Asterinales. Fungal Divers. 2014;68:1–68. [Google Scholar]
- 3.Tanaka K., Hirayama K., Yonezawa H., Sato G., Toriyabe A., Kudo H., Hashimoto A., Matsumura M., Harada Y., Kurihara Y., et al. Revision of the Massarineae (Pleosporales, Dothideomycetes) Stud. Mycol. 2015;82:75–136. doi: 10.1016/j.simyco.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pem D., Jeewon R., Chethana K.W.T., Hongsanan S., Doilom M., Suwannarach N., Hyde K.D. Species concepts of Dothideomycetes: Classification, phylogenetic inconsistencies and taxonomic standardization. Fungal Divers. 2021;109:283–319. [Google Scholar]
- 5.Wijayawardene N.N., Hyde K.D., Dai D.Q., Sánchez-García M., Goto B.T., Saxena R.K., Erdoğdu M., Selçuk F., Rajeshkumar K.C., Aptroot A., et al. Outline of Fungi and fungus-like taxa–2021. Mycosphere. 2022;13:53–453. [Google Scholar]
- 6.Saccardo P. Conspectus generum pyrenomycetum italicorum additis speciebus fungorum Venetorum novisvel criticis, systemate carpologico dispositorum. Atti della Societa Veneziana-Trentina-Istrianadi. Scienze Naturali. 1875;4:77–100. [Google Scholar]
- 7.Clements F.E., Shear C.L. The genera of Fungi. H.W. Wilson Company publishing; New York, USA: 1931. pp. 1–496. [Google Scholar]
- 8.Crous P.W., Groenewald J.Z. A phylogenetic reevaluation of Arthrinium. IMA Fungus. 2013;4:133–154. doi: 10.5598/imafungus.2013.04.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pintos Á., Alvarado P., Planas J., Jarling R. Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. hosts. MycoKeys. 2019;49:15–48. doi: 10.3897/mycokeys.49.32115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawksworth D.L., Crous P.W., Redhead S.A., Reynolds D.R., Samson R.A., Seifert K.A., Zhang N. The Amsterdam declaration on fungal nomenclature. IMA Fungus. 2011;2:105–112. doi: 10.5598/imafungus.2011.02.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Réblová M., Miller A.N., Rossman A.Y., Seifert K.A., Crous P.W., Hawksworth D.L., Abdel-Wahab M.A., Cannon P.F., Daranagama D.A., De Beer Z.W., et al. Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales) IMA Fungus. 2016;7:131–153. doi: 10.5598/imafungus.2016.07.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M., Tan X.M., Liu F., Cai L. Eight new Arthrinium species from China. MycoKeys. 2018;34:1–24. doi: 10.3897/mycokeys.34.24221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang N., Liang Y.M., Tian C.M. A novel bambusicolous fungus from China, Arthrinium chinense (Xylariales) Sydowia. 2020;72:77–83. [Google Scholar]
- 14.Feng Y., Liu J.K., Lin C.G., Chen Y.Y., Xiang M.M., Liu Z.Y. Additions to the genus Arthrinium (Apiosporaceae) from bamboos in China. Front. Microbiol. 2021;7:661281. doi: 10.3389/fmicb.2021.661281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pintos Á., Alvarado P. Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Syst. Evol. 2021;7:197–221. doi: 10.3114/fuse.2021.07.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian X.G., Karunarathna S.C., Mapook A., Promputtha I., Xu J., Bao D., Tibpromma S. One new species and two new host records of Apiospora from bamboo and maize in northern Thailand with thirteen new combinations. Life. 2021;11:1071. doi: 10.3390/life11101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Index Fungorum. [(accessed on 17 October 2023)]. Available online: http://www.indexfungorum.org.
- 18.Kwon S.L., Cho M., Lee Y.M., Kim C., Lee S.M., Ahn B.J., Lee H., Kim J.J. Two unrecorded Apiospora species isolated from marine substrates in Korea with eight new combinations (A. piptatheri and A. rasikravindrae) Mycobiology. 2022;50:46–54. doi: 10.1080/12298093.2022.2038857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji Z.L., Zhang S.W., Zhu F., Wan B.X., Liang R.Z. First report of Arthrinium arundinis causing leaf edge spot of peach in China. Plant Dis. 2020;104:3077. doi: 10.1094/PDIS-12-19-2666-PDN. [DOI] [Google Scholar]
- 20.Chen K., Wu X.Q., Huang M.X., Han Y.Y. First report of brown culm streak of Phyllostachys praecox caused by Arthrinium arundinis in Nanjing, China. Plant Dis. 2014;98:1274. doi: 10.1094/PDIS-02-14-0165-PDN. [DOI] [PubMed] [Google Scholar]
- 21.Mavragani D.C., Abdellatif L., McConkey B., Hamel C., Vujanovic V. First report of damping-off of durum wheat caused by Arthrinium sacchari in the semi-arid Saskatchewan fields. Plant Dis. 2007;91:469. doi: 10.1094/PDIS-91-4-0469A. [DOI] [PubMed] [Google Scholar]
- 22.Li B.J., Liu P.Q., Jiang Y., Weng Q.Y., Chen Q.H. First report of culm rot caused by Arthrinium phaeospermum on Phyllostachys viridis in China. Plant Dis. 2016;100:1013. doi: 10.1094/PDIS-08-15-0901-PDN. [DOI] [Google Scholar]
- 23.Dyląg M., Hryncewicz-Gwóźdź A., Jagielski T. Onychomycosis due to Arthrinium arundinis: A case report. Acta Derm. Venereol. 2017;97:860–861. doi: 10.2340/00015555-2673. [DOI] [PubMed] [Google Scholar]
- 24.Ma X., Chomnunti P., Doilom M., Daranagama D.A., Kang J. Multigene phylogeny reveals endophytic Xylariales novelties from Dendrobium species from Southwestern China and Northern Thailand. J. Fungi. 2022;8:248. doi: 10.3390/jof8030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai D.Q., Phookamsak R., Wijayawardene N.N., Li W.J., Bhat D.J., Xu J.C., Taylor J.E., Hyde K.D., Chukeatirote E. Bambusicolous fungi. Fungal Divers. 2017;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- 26.Wang H., Umeokoli B.O., Eze P., Heering C., Janiak C., Müller W.E., Orfali R.S., Hartmann R., Dai H., Lin W., et al. Secondary metabolites of the lichen-associated fungus Apiospora montagnei. Tetrahedron Lett. 2017;58:1702–1705. doi: 10.1016/j.tetlet.2017.03.052. [DOI] [Google Scholar]
- 27.Hyde K.D., Norphanphoun C., Maharachchikumbura S.S.N., Bhat D.J., Jones E.B.G., Bundhun D., Chen Y.J., Bao D.F., Boonmee S., Calabon M.S., et al. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- 28.Senanayake I.C., Rathnayaka A.R., Marasinghe D.S., Calabon M.S., Gentekaki E., Lee H.B., Hurdeal V.G., Pem D., Dissanayake L.S., Wijesinghe S.N., et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere. 2020;11:2678–2754. doi: 10.5943/mycosphere/11/1/20. [DOI] [Google Scholar]
- 29.Jiang N., Tian C.M. The holomorph of Arthrinium setariae sp. nov. (Apiosporaceae, Xylariales) from China. Phytotaxa. 2021;483:149–159. doi: 10.11646/phytotaxa.483.2.7. [DOI] [Google Scholar]
- 30.Jeewon R., Hyde K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere. 2016;7:1669–1677. doi: 10.5943/mycosphere/7/11/4. [DOI] [Google Scholar]
- 31.Maharachchikumbura S.S.N., Chen Y., Ariyawansa H.A., Hyde K.D., Haelewaters D., Perera R.H., Samarakoon M.C., Wanasinghe D.N., Bustamante D.E., Liu J.K., et al. Integrative approaches for species delimitation in Ascomycota. Fungal Divers. 2021;109:155–179. doi: 10.1007/s13225-021-00486-6. [DOI] [Google Scholar]
- 32.Jayasiri S.C., Hyde K.D., Ariyawansa H.A., Bhat D.J., Buyck B., Cai L., Dai Y.C., Abd-Elsalam K.A., Ertz D., Hidayat I., et al. The faces of fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015;74:3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- 33.Chaiwan N., Gomdola D., Wang S., Monkai J., Tibpromma S., Doilom M., Wanasinghe D.N., Mortimer P.E., Lumyong S., Hyde K.D. https://gmsmicrofungi.org : An online database providing updated information of microfungi in the Greater Mekong Subregion. Mycosphere. 2021;12:1513–1526. doi: 10.5943/mycosphere/12/1/19. [DOI] [Google Scholar]
- 34.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Volume 18. Academic Press; Cambridge, MA, USA: 1990. p. 7. [Google Scholar]
- 35.Gardes M., Bruns T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 36.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 40.GenBank. [(accessed on 17 October 2023)]; Available online: http://www.ncbi.nlm.nih.gov/blast/
- 41.Hall T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; Proceedings of the Nucleic Acids Symposium Series; London, UK. 8–12 October 1999; pp. 95–98. [Google Scholar]
- 42.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller M., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the Gateway Computing Environments Workshop; New Orleans, LA, USA. 21 November 2010; pp. 1–8. [Google Scholar]
- 44.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huelsenbeck J.P., Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 46.Rambaut A. FigTree v1.4: Tree Figure Drawing Tool. [(accessed on 17 October 2023)]. Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 47.Philippe H., Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 49.Chen T.Z., Zhang Y., Ming X.B., Zhang Q., Long H., Hyde K.D., Li Y., Wang Y. Morphological and phylogenetic resolution of Arthrinium from medicinal plants in Yunnan, including A. cordylines and A. pseudomarii spp. nov. Mycotaxon. 2021;136:183–199. doi: 10.5248/136.183. [DOI] [Google Scholar]
- 50.Calvo M.A., Guarro J. Arthrinium aureum sp. nov. from Spain. Trans. Br. Mycol. Soc. 1980;75:156–157. doi: 10.1016/S0007-1536(80)80208-7. [DOI] [Google Scholar]
- 51.Yang C.L., Xu X.L., Dong W., Wanasinghe D.N., Liu Y.G., Hyde K.D. Introducing Arthrinium phyllostachium sp. nov. (Apiosporaceae, Xylariales) on Phyllostachys heteroclada from Sichuan province, China. Phytotaxa. 2019;406:91–110. doi: 10.11646/phytotaxa.406.2.2. [DOI] [Google Scholar]
- 52.Monkai J., Phookamsak R., Tennakoon D.S., Bhat D.J., Xu S., Li Q., Xu J., Mortimer P.E., Kumla J., Lumyong S. Insight into the taxonomic resolution of Apiospora: Introducing novel species and records from bamboo in China and Thailand. Diversity. 2022;14:918. doi: 10.3390/d14110918. [DOI] [Google Scholar]
- 53.Phukhamsakda C., Nilsson R.H., Bhunjun C.S., de Farias A.R., Sun Y.R., Wijesinghe S.N., Raza M., Bao D.F., Lu L., Tibpromma S., et al. The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Divers. 2022;114:1–60. doi: 10.1007/s13225-022-00502-3. [DOI] [Google Scholar]
- 54.Larrondo J.V., Calvo M.A. New contributions to the study of the genus Arthrinium. Mycologia. 1992;84:475–478. doi: 10.1080/00275514.1992.12026164. [DOI] [Google Scholar]
- 55.Zeng Y., Ali M.K., He W., Deng L., Yang X., Li X., Pu X., Yang J. Chemical constituents of functional food Amomum villosum to combat human diseases. Curr. Chin. Sci. 2022;2:57–67. doi: 10.2174/2210298101666210923124548. [DOI] [Google Scholar]
- 56.Bon M.C., Goolsby J.A., Mercadier G., Guermache F., Kashefi J., Cristofaro M., Vacek A.T., Kirk A. Detection of a diverse endophyte assemblage within fungal communities associated with the Arundo Leaf Miner, Lasioptera donacis (Diptera: Cecidomyiidae) Diversity. 2023;15:571. doi: 10.3390/d15040571. [DOI] [Google Scholar]
- 57.Mapook A., Hyde K.D., McKenzie E.H.C., Jones E.B.G., Bhat D.J., Jeewon R., Stadler M., Samarakoon M.C., Malaithong M., Tanunchai B., et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed) Fungal Divers. 2020;101:1–75. doi: 10.1007/s13225-020-00444-8. [DOI] [Google Scholar]
- 58.Shrestha P., Ibáñez A.B., Bauer S., Glassman S.I., Szaro T.M., Bruns T.D., Taylor J.W. Fungi isolated from Miscanthus and sugarcane: Biomass conversion, fungal enzymes, and hydrolysis of plant cell wall polymers. Biotechnol. Biofuels. 2015;8:38. doi: 10.1186/s13068-015-0221-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hong J.H., Jang S., Heo Y.M., Min M., Lee H., Lee Y.M., Lee H., Kim J.J. Investigation of marine-derived fungal diversity and their exploitable biological activities. Mar. Drugs. 2015;13:4137–4155. doi: 10.3390/md13074137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Astuti P., Pratoko D.K., Rollando R., Nugroho G.W., Wahyuono S., Hertiani T., Nurrochmad A. Bioactivities of A Major Compound from Arthrinium rasikravindrae an endophytic fungus of Coleus amboinicus. Lour. Fabad J. Pharm. Sci. 2021;46:23–30. [Google Scholar]
- 61.van Eijk G.W. Bostrycin, a tetrahydroanthraquinone pigment and some other metabolites from the fungus Arthrinium phaeospermum. Experientia. 1975;31:783–784. doi: 10.1007/BF01938463. [DOI] [Google Scholar]
- 62.Li S., Tang Y., Fang X., Qiao T., Han S., Zhu T. Whole-genome sequence of Arthrinium phaeospermum, a globally distributed pathogenic fungus. Genomics. 2020;112:919–929. doi: 10.1016/j.ygeno.2019.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.