Summary
Congenital heart disease often arises from perturbations of transcription factors (TFs) that guide cardiac development. ISLET1 (ISL1) is a TF that influences early cardiac cell fate, as well as differentiation of other cell types including motor neuron progenitors (MNPs) and pancreatic islet cells. While lineage specificity of ISL1 function is likely achieved through combinatorial interactions, its essential cardiac interacting partners are unknown. By assaying ISL1 genomic occupancy in human induced pluripotent stem cell-derived cardiac progenitors (CPs) or MNPs and leveraging the deep learning approach BPNet, we identified motifs of other TFs that predicted ISL1 occupancy in each lineage, with NKX2.5 and GATA motifs being most closely associated to ISL1 in CPs. Experimentally, nearly two-thirds of ISL1-bound loci were co-occupied by NKX2.5 and/or GATA4. Removal of NKX2.5 from CPs led to widespread ISL1 redistribution, and overexpression of NKX2.5 in MNPs led to ISL1 occupancy of CP-specific loci. These results reveal how ISL1 guides lineage choices through a combinatorial code that dictates genomic occupancy and transcription.
Keywords: cardiac development, transcription factors, combinatorial code, transcription factor motifs, transcriptional regulation, ISL1, NKX2.5, cell specification, cardiac progenitor
Highlights
-
•
Deep learning (BPNet) identified ISL1-NKX2.5 cardiac-specific complex
-
•
ISL1-NKX2.5 complex drives a cardiac-specific regulatory network
-
•
ISL1-NKX2.5 interaction is necessary for ISL1 cardiac localization
In this article, Deepak Srivastava and colleagues leveraged deep learning (BPNet) to discover that ISL1 forms a cardiac-specific complex with NKX2.5 to drive cardiac gene regulation in human CPs. This interaction is necessary for ISL1’s cardiac-specific localization. These findings reveal how ISL1 achieves a cardiac-specific regulatory role, an essential step for proper understanding of cardiogenesis.
Introduction
Cardiac malformations are typically caused by abnormal specification or morphogenetic events related to specific subsets of developing cardiac cells. Transcription factors (TFs) essential for cardiogenesis have been identified, and mutations in these often underlie human cardiac malformations (Yasuhara and Garg, 2021). However, the precise mechanism by which they guide the gene expression necessary for proper differentiation and morphogenesis remains an active area of investigation.
ISL1 is a TF transiently expressed during emergence and expansion of multipotent progenitor second heart field (SHF) cells, before being downregulated upon further differentiation (Black, 2007). ISL1 activates cardiac transcriptional networks by directly inducing essential cardiac TFs such as MEF2C and GATA factors (Srivastava, 2006). In mice that lack ISL1, the SHF derivative structures are severely reduced because of defects in proliferation, survival, and migration, leading to lethality by embryonic day (E) 10.5 (Cai et al., 2003). In humans, truncating mutations in ISL1 have been associated with SHF defects, including double outlet right ventricle (Wang et al., 2019). A microdeletion of the entire ISL1 allele has been associated with tetralogy of Fallot (Osoegawa et al., 2014), a related outflow tract defect. Thus, ISL1 is critical for establishing and maintaining mammalian SHF cardiac progenitor (CP) populations.
ISL1 is also expressed more broadly, including in neuronal and pancreatic progenitor cells, where it is essential for normal development, suggesting a combinatorial code involving ISL1-associated factors that leads to cell type-specific gene expression and cell fate determination. Within neuronal progenitors, ISL1 interacts with TFs such as the neuronal LHX3 and the ubiquitously expressed LDB1 to regulate genes associated with this fate (Seo et al., 2015). Loss of ISL1 leads to complete absence of motor neurons (MNs) in mice (Pfaff et al., 1996). Similarly, in endodermal progenitors that give rise to the pancreas, ISL1 interacts with the TFs SSBP3 and LDB1 and is required for formation of insulin-producing beta cells (Ediger et al., 2014; Galloway et al., 2015). ISL1 is known to interact with several factors during cardiac differentiation, including LDB1, but this interaction is not cardiac specific (Galloway et al., 2015; Narkis et al., 2012), so is unlikely to be sufficient to guide cardiac-specific gene expression. ISL1 also interacts with the ubiquitously expressed chromatin modifier KDM6B to activate cardiac gene expression (Wang et al., 2016), but the ISL1-associated TFs that lead to cardiac-specific gene expression and subsequent differentiation remain unknown.
Here, we used human induced pluripotent stem cells (hiPSCs) to capture transient ISL1-expressing CP and MN progenitor (MNP) populations (NeuroLINCS Consortium et al., 2021) and investigated how cardiac and neuronal gene expression are differentially regulated by ISL1. We identify NKX2.5 as a critical binding partner for ISL1 to direct cardiac-specific gene expression, and demonstrate how a single TF can have distinct regulatory roles in directing fate decisions based on cellular context.
Results
ISL1 is necessary for instructing cell-fate-specific gene regulation
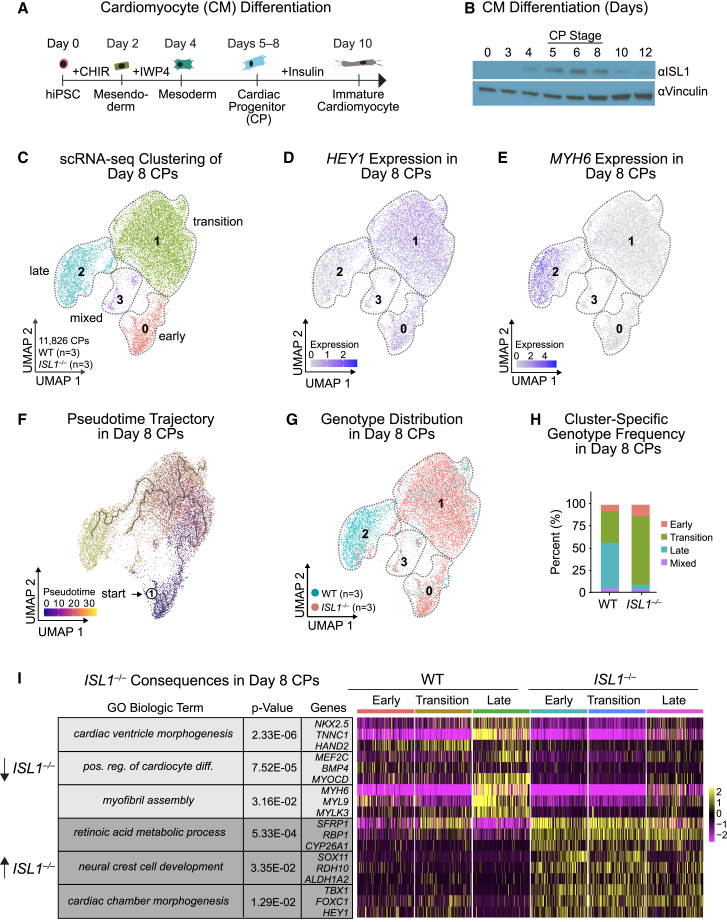
We differentiated hiPSCs into MYH6+:TNNT2+ CPs and subsequently into cardiomyocytes (CMs) using a WNT modulation protocol (Figures 1A and S1A) (Lian et al., 2013). These cells pass through a transient CP-like stage associated with ISL1 enrichment between days 5 and 8 (Figure 1B), followed by beating foci concurrent with ISL1 downregulation, and ultimately a beating monolayer of CMs by day 10 (Video S1).
Figure 1.
ISL1 is required for cardiac cell-fate-specific gene regulation
(A) Schematic of CM differentiation from hiPSCs.
(B) Western blot of ISL1 expression during hiPSC-CM differentiation (days 0–12). Vinculin is the loading control.
(C) Hierarchical UMAP clustering of WT (n = 3 independent experiments) or ISL1−/− (n = 3 independent experiments) day 8 CPs after scRNA-seq.
(D and E) Expression of HEY1 (D) and MYH6 (E) in day 8 CPs, superimposed on the UMAP from (C).
(F) Monocle pseudotime analysis of day 8 CPs. Colors represent arbitrary units of pseudotime.
(G) FeaturePlot display of the distribution of WT and ISL1−/− day 8 CPs across clusters depicted in (C).
(H) Percentages of each genotype in each cluster. Cell numbers for each genotype are normalized to the total number of cells in each cluster.
(I) Heatmap of genes and accompanying biological GO terms significantly dysregulated in scRNA-seq of ISL1−/− as compared with WT day 8 CPs.
We generated an ISL1−/− hiPSC line using two Cas9-gRNAs to excise a portion of the gene starting in exon 2 and ending in exon 5 (Figure S1B). The excision abolished ISL1 RNA and protein expression (Figures S1C and S1D). CMs differentiated from ISL1−/− hiPSCs displayed decreased sarcomeric gene expression at the CP stage (Figure S1A) and a delay in beating induction (Figure S1E and Video S1), similar to previous results (Quaranta et al., 2018).
We performed single-cell RNA sequencing (scRNA-seq) on wildtype (WT) or ISL1−/− progenitor populations at day 8 (Figure 1C). Within the cardiac lineage, we identified four transcriptional signatures (Figures 1C and S1F; Table S1). Expression of more immature markers (FOXC1, HEY1, and SOX4) was higher in clusters 0 and 1, while expression of more differentiated gene markers (e.g., TNNT2, ACTN2, and MYH6) was high in cluster 2 (termed "late" cluster) (Figures 1D, 1E, and S1F). In addition, cluster 0 expressed higher levels of cell replication genes (NUF2, PRC1, and KIF4A) than cluster 1, indicating that cells from cluster 0 are more immature and proliferative; we therefore refer to cluster 0 as the "early" cluster and cluster 1 as the "transition" cluster (Figure S1F). Cluster 3 seems to be a mixture of early and late differentiating cells (termed "mixed") (Figure S1F). Pseudotime analysis confirmed this cell trajectory in silico (Figure 1F).
In the ISL1−/− line, a greater proportion of CPs were in the early and transition clusters compared with the WT line, and fewer cells were in the late cluster (Figures 1G, 1H, and S1G). Within each of the early, transition, and late clusters, we found significant differences between genotypes (Table S1). For example, ISL1−/− CPs were depleted for genes associated with the Gene Ontology (GO) terms of cardiac ventricle morphogenesis (e.g. NKX2.5, TNNC1) and myofibril assembly (e.g. MYH6, MYL9), and enriched for GO terms related to retinoic acid metabolic process (e.g. CYP26A1, ALDH1A2, RBP1) and cardiac chamber morphogenesis, specifically atrial markers (e.g. HEY1, NR2F1) (Figures 1I and S1H; Table S1). This is in agreement with previous findings that ISL1 promotes ventricular cell fate and suppresses atrial identity in vitro (Quaranta et al., 2018).
Complete deletion of ISL1 revealed its early role in cardiogenesis (Figures 1A and 1B), but may have concealed functions in differentiation after cell fate determination. Therefore, we used small interfering RNAs (siRNA) to knockdown (KD) ISL1 specifically during the CP stage, after CP commitment (Figure S1I). Rhodamine-labeled siRNAs targeting ISL1 or a scrambled (Scra) control were introduced on day 5 of CM differentiation, and isolated rhodamine+ cells with significant downregulation of ISL1 were analyzed on day 8 (Figures S1I and S1J). scRNA-seq showed that ISL1 KD at the CP stage did not substantially change cell fate, with ISL1 KD cells clustering with controls (Figure S1K). Next, we generated a high-confidence list of genes potentially regulated by ISL1 in CPs by intersecting the significantly downregulated genes in both the ISL1−/− and ISL1 KD conditions (Table S1). This list included cardiac genes such as MYL9 and TNNT2 (Figures S1L and S1M), confirming our previous ISL1−/− results, suggesting that many dysregulated genes are likely a direct consequence of the absence of ISL1.
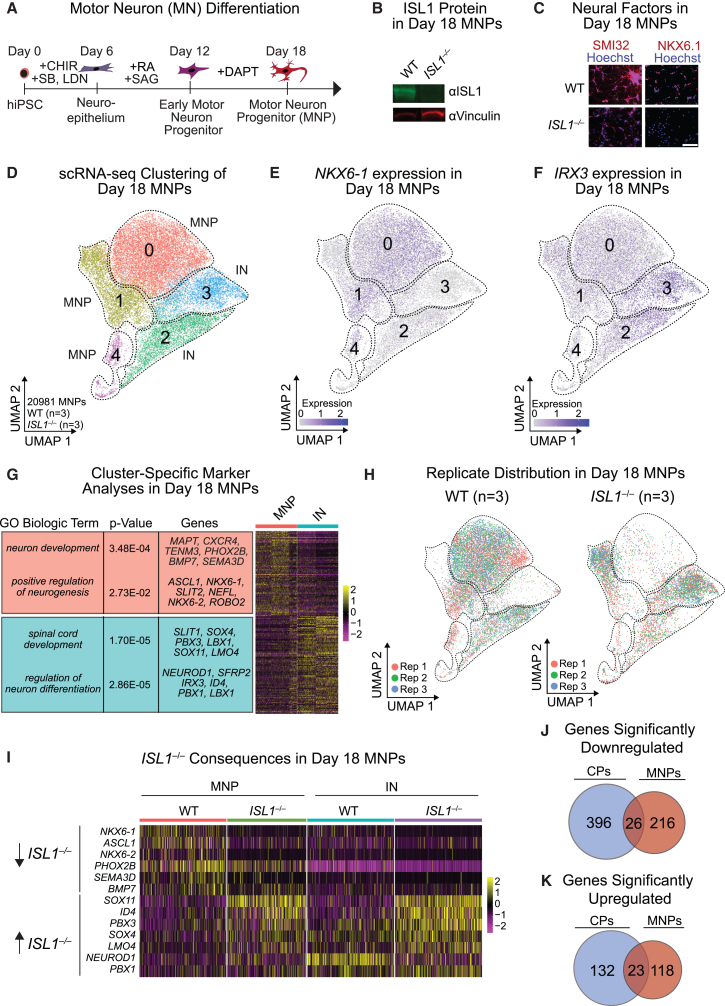
To identify cardiac-specific consequences of ISL1 removal, we differentiated the same ISL1−/− hiPSC line into SMI32- and NKX6.1-expressing MNPs, which form axonal projections by day 18, followed by scRNA-seq (Figures 2A–2C) (NeuroLINCS Consortium et al., 2021). ISL1 deletion led to decreased expression of MN markers NKX6.1 and SMI32, consistent with observations in an ISL1 hypomorphic mouse model (Figure 2C) (Kim et al., 2016).
Figure 2.
scRNA-seq analyses of ISL1 function in MNPs
(A) Schematic of MN differentiation from hiPSCs.
(B) Western blot of ISL1 in WT and ISL1−/− day 18 MNPs. Vinculin served as the loading control.
(C) Immunofluorescence of neural factors SMI32 and NKX6.1 in day 18 MNPs. Scale bar, 100 μM.
(D) Hierarchical UMAP clustering of WT (n = 3 independent experiments) and ISL1−/− (n = 3 independent experiments) scRNA-seq data in day 18 iPSCs differentiated toward MNPs.
(E and F) Expression levels of the MN-related TF NKX6-1 (E) and the IN-related TF IRX3 (F) in day 18 MNPs, superimposed on the UMAP from (D).
(G) Heatmap of genes enriched with accompanying GO terms in each of the MNP or IN clusters depicted in (D).
(H) Replicate (Rep) comparison in WT (n = 3; 14,609 cells) or ISL1−/− (n = 3; 6,372 cells) day 18 MNPs across the clusters depicted in (D).
(I) Heatmap clustering of genes significantly dysregulated in scRNA-seq of ISL1−/−compared with WT day 18 MNPs.
(J) Venn diagram of significantly downregulated genes shared between ISL1−/− day 8 CPs and ISL1−/− day 18 MNPs.
(K) Venn diagram of significantly upregulated genes shared between ISL1−/− day 8 CPs and ISL1−/− day 18 MNPs.
In the MNP lineage, we observed five distinct transcriptional clusters (Figure 2D). All clusters expressed neuron-specific markers, with clusters 0, 1, and 4 also expressing MNP markers such as NKX6.1, NKX6.2, and PHOX2A (termed “MNP” clusters), whereas clusters 2 and 3 had higher expression of interneuron (IN) markers such as IRX3 and PAX2 (termed “IN” clusters) (Figures 2D–2G; Table S2). In the absence of ISL1, cells were more likely to be clustered in the IN clusters rather than MNP clusters (Figure 2H). In addition, we found downregulation of MNP genes, such as NKX6.1 and PHOX2B, and upregulation of IN genes such as PBX1 and LMO4 in ISL1−/− cells (Figure 2I), consistent with observations in knockout ISL1 mouse models (Song et al., 2009). The overlap in differentially regulated genes in CPs and MNPs was small, indicating that ISL1 regulates a unique group of genes and regulatory networks in each cell type (Figures 2J and 2K).
ISL1 genomic localization is lineage specific
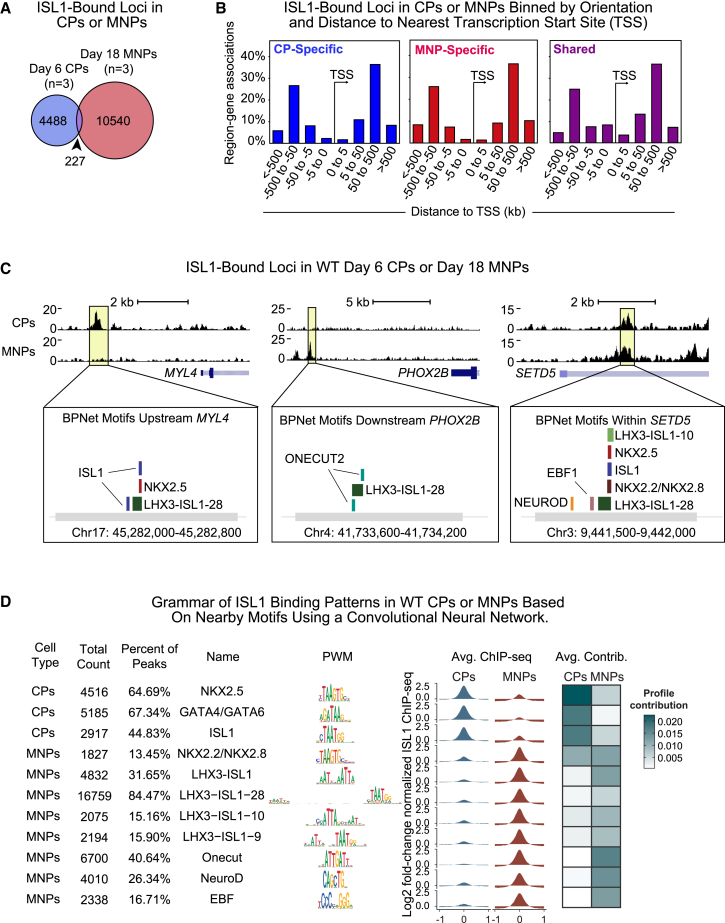
To determine which genes are directly regulated by ISL1 in CPs and MNPs, we used an antibody to endogenous ISL1 to perform chromatin immunoprecipitation followed by sequencing (ChIP-seq) in triplicate (Figure S2A). ISL1 consistently interacted with 4,715 genomic regions in day 6 CPs, and 10,767 regions in day 18 MNPs, with only 227 peaks shared between cell types (Figure 3A). In each cell type, ISL1-bound regions were most frequently distal (>50 kb) to gene transcription start sites (TSSs) (Figure 3B) and characterized as intronic or distal intergenic, within regions with active histone marks and a relative lack of repressive marks, consistent with ISL1 likely binding to distal enhancers (Figures S2B and S2C). Examples of cell-type-specific ISL1 genomic localization are illustrated by CP-specific peaks at the cardiac-related MYL4 locus and MNP-specific peaks at the MN-related PHOX2B locus (Figures 3C and S2D). Some sites were shared between cell types, such as the ISL1 interaction near the SETD5 locus, which encodes a ubiquitous histone methyltransferase (Figure 3C).
Figure 3.
Grammar of distinct ISL1 binding patterns in cardiac or MNPs based on nearby motifs using deep learning
(A) Venn diagram of ISL1 peaks in CPs or MNPs. Overlap is statistically significant (Regione R) (p = 0.001).
(B) Histogram of ISL1-bound peaks based on distance from gene TSSs.
(C) Example ChIP-seq tracks showing ISL1-bound loci with CP-specific, MNP-specific, or shared peaks. Corresponding de novo motifs identified by BPNet that predict cell-type-specific ISL1 binding are shown below. Data shown from a single representative replicate.
(D) De novo motifs identified by BPNet after simultaneous training of the CP-specific or MNP-specific binding profiles. Displayed for each identified motif is: the total count of motifs and percentages, name based on the most likely TF(s) that binds them, the position weight matrices (PWMs), and average ChIP-seq intensity and average contribution to the ISL1 binding predictions as extracted from BPNet.
To interrogate the function of ISL1-occupied loci, we used the Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al., 2010). Significantly enriched GO terms for genes near ISL1-bound CP peaks were associated with cardiac differentiation, such as Cardiac chamber development and Cardiac chamber morphogenesis, whereas significant terms for ISL1 MNP peaks were related to neural differentiation, such as Cranial nerve development and Cell differentiation in spinal cord. GREAT did not detect any significant terms among peaks shared by CPs and MNPs (Figures S2E and S2F).
We expected that ISL1 occupancy might depend on cell-type-specific co-factors. To identify putative co-bound TFs, we visualized TF motifs in the vicinity of the ISL1-bound sites with Hypergeometric Optimization of Motif EnRichment (HOMER) (Li et al., 2009). We combined the top 16 motifs from each cell type (sans ISL1 motif) and compared motif enrichment of this set across CPs and MNPs (Figure S3A). ISL1-bound loci in CPs were enriched in the family of motifs for GATA (including GATA4), TEAD, NKX2 (including NKX2.5), and HAND2. In contrast, ISL1-bound loci in MNPs were enriched in motifs in the family of motifs for NEUROD, CUX, LHX (including NKX6.1), and PHOX2. The enrichment of LHX family motifs in ISL1-bound regions in MNPs is consistent with the known ISL1-LHX3 composite motifs identified in developing MNs (Seo et al., 2015). These data provide evidence for unique enrichment of known TF motifs near ISL1-bound sites in either cell type.
Next, we used the deep learning model BPNet to identify de novo motifs that help to predict ISL1 binding (Avsec et al., 2021). This model can learn complex rules by which combinations of de novo sequence motifs best predict the experimental binding data. BPNet was trained to predict the cell-type-specific binding of ISL1 at all 15,483 regions and optimized by hyperparameter tuning (see experimental procedures, Figure S3B). Interpretation tools were used to systematically extract the sequence motifs from the network (Figure 3D), which matched known motifs recognized by relevant TFs. As expected, the ISL1 motif was among the de novo motifs important for the binding in both cell types and included motifs for the neural TF families ONECUT, NEUROD, and EBF2, all of which have known roles in MN development (Son et al., 2011; Velasco et al., 2017) and an ISL1-containing composite motif that was previously identified to be important in MNPs (Seo et al., 2015). In addition, the NKX2 family motif identified may correspond with NKX2.2 or NKX2.8, both important for neuronal progenitor populations (Jarrar et al., 2015).
Predicted ISL1-associated motifs in CPs belonged to cardiac TF families NKX2 and GATA (Figure 3D), consistent with our HOMER analyses. The GATA motif likely corresponds with binding of the cardiac TFs GATA4 and/or GATA6, while the NKX2 family motif most likely corresponds with the cardiac TF NKX2.5, which is necessary for development of the outflow tract and right ventricle in mice (Lyons et al., 1995; Zhang et al., 2014), similar to ISL1. The average contribution to predicting ISL1 binding was greatest with the NKX2.5 motif. Examples of de novo motifs found in cell-type-specific and shared ISL1-bound regions are illustrated in the ChIP-seq tracks in Figure 3C.
We examined the percentage of ISL1-bound peaks with GATA and/or NKX2.5 motifs and the nature of those sites (Figure S3C) and found cardiac-related GO terms enriched for each category (Figure S3D). This was also the case when the motifs of GATA and NKX2.5 were found in combination or when neither were there. The regions with only the NKX2.5 motif were also enriched in terms related to epithelial-to-mesenchymal transition, which may reflect the role of NKX2.5 in cardiac cell migration. When the NKX2.5 motif was found in combination with the GATA motif, a majority of the terms were exclusively cardiac, demonstrating the importance of combinatorial TFs during cardiac differentiation (Figure S3D).
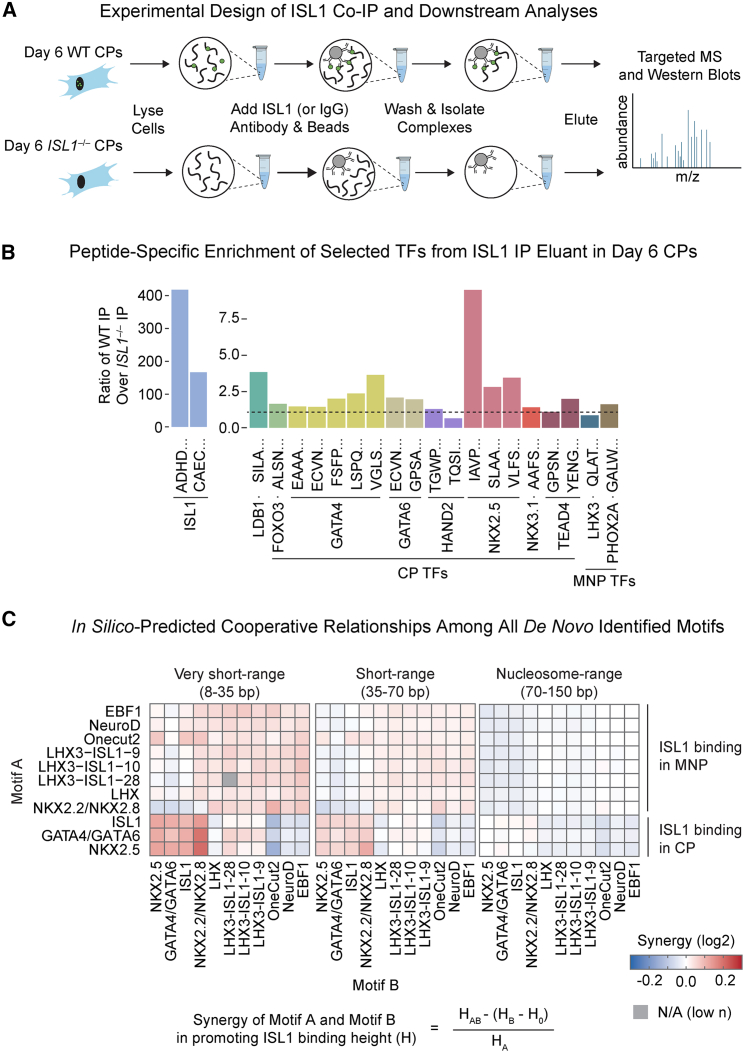
ISL1 forms a DNA-binding complex with NKX2.5
To experimentally identify TFs that cooperate with ISL1 for cardiac fate specification, we used a combination of targeted mass spectrometry (MS) based on selected reaction monitoring (SRM) (Calvo et al., 2011) and co-immunoprecipitation (co-IP) (Figure 4A). We selected TFs for which there were specific known and/or de novo motif enrichment in ISL1 ChIP-seq in WT CPs from our studies (Figures 3D and S3A) and designed targeted SRM assays for unique peptides representing them (Table S3). We immunoprecipitated ISL1 from WT or ISL1−/− CPs as a negative control, followed by SRM, and showed enrichment for ISL1 and LDB1, validating the ISL1 IP. We found no enrichment for PHOX2A or LHX3, indicating the specificity of this method (Figure 4B). Of the cardiac TFs analyzed, NKX2.5 showed the strongest enrichment for several peptides in the ISL1 IP compared with controls, while other TFs including GATA4, GATA6, and TEAD4 showed some enrichment (Figure 4B).
Figure 4.
Proteomics and BPNet identify NKX2.5 and other TFs that guide ISL1 DNA-binding specificity in CPs
(A) Schematic for ISL1 co-IP and subsequent SRM targeted proteomics analyses.
(B) Barplot results from targeted affinity purification followed by MS, depicting enrichment of individual peptides corresponding to cardiac factors, positive controls (ISL1, LDB1), and negative controls (LHX3, PHOX2A) (n = 1 independent experiment).
(C) Cooperative relationship among all de novo identified motifs binned by distance from ISL1 binding predicted by in silico motif mutation.
Since the deep learning model predicted the binding of ISL1 in CPs based on a combination of ISL1, NKX2.5, and GATA4/GATA6 motifs, we asked whether the cooperativity between the de novo-derived motifs was distance dependent by testing how in silico mutated motif patterns would be predicted to disrupt co-binding at specified distances. The strongest synergy was observed when the ISL1 and NKX2.5 motifs were in close proximity of under 35 bp, while no detectable synergy was observed at nucleosome distance (70–150 bp) (Figure 4C). In addition, short-range synergy was observed between the GATA4/GATA6 motif and the ISL1 and NKX2.5 motifs.
To further investigate the cooperativity between ISL1 and NKX2.5 and ISL1 and GATA4, we asked whether these interactions could occur in an alternative cell type in the absence of additional putative cardiac co-factors. In co-IPs from COS-7 cells, exogenously expressed ISL1 interacted strongly with LDB1 (positive control), NKX2.5, and GATA4 (Figure S4A), confirming that these interactions do not require cardiac-specific co-factors. Given the similar cardiac expression pattern and loss-of-function effects between ISL1 and NKX2.5 (Zhang et al., 2014) and the evidence that they genetically interact (Dorn et al., 2015), we focused our investigation on understanding the function of ISL1 and NKX2.5 interaction. To this end, we validated the endogenous ISL1-NKX2.5 interaction in CPs (Figures S4B and S4C).
NKX2.5 co-occupies a majority of ISL1-bound loci in CPs and drives overlapping cardiac gene regulatory networks
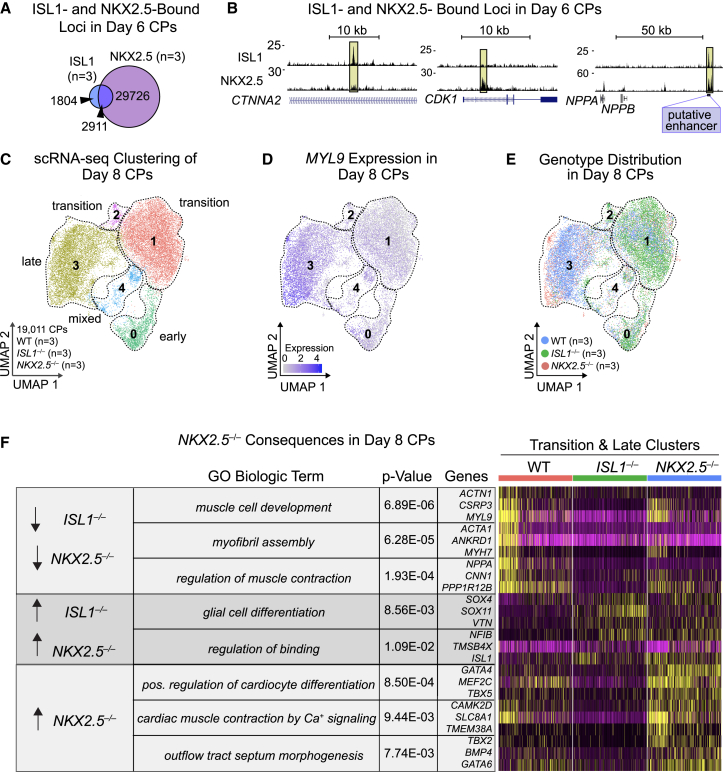
We next tested if NKX2.5 co-localizes to ISL1-bound genomic regions in CPs by ChIP-seq using an antibody to endogenous NKX2.5 in day 6 CPs (Figure S4D). This analysis revealed 32,637 sites. NKX2.5 peaks overlapped with more than 60% of ISL1 peaks in CPs (2,911 loci) (Figures 5A and 5B), but with only 5% of the total MNP ISL1 peaks (483 loci) (Figure S4E). Loci bound only by NKX2.5 tended to be both proximal and distal to TSSs, in contrast with sites only bound by ISL1 or co-bound by both, which were primarily distal sites (Figure S4F). NKX2.5 tended to bind near promoters, yet ISL1-only or co-bound loci tended to be enriched in distal intergenic regions, consistent with a role in regulating enhancers when co-bound (Figure S4G). GREAT analysis showed that peaks co-bound by ISL1 and NKX2.5 were most enriched for cardiac developmental terms such as Cardiac chamber development and Heart morphogenesis, similar to ISL1-only bound loci, but distinct from NKX2.5-only loci (Figure S4H).
Figure 5.
NKX2.5 binds a majority of ISL1 peaks and cooperates with ISL1 to regulate gene expression in CPs
(A) Venn diagram of NKX2.5 and ISL1 ChIP-seq peaks in WT day 6 CPs.
(B) Example ChIP-seq tracks showing loci bound by ISL1, NKX2.5, or both in day 6 CPs. Putative NPPA/NPPB enhancer labeled as related to Figure S5I. Data shown from single representative replicate.
(C) Hierarchical UMAP scRNA-seq clustering of WT, ISL1−/−, and NKX2.5−/− day 8 CPs (n = 3 independent experiments).
(D) Expression of MYL9 in day 8 CPs, superimposed on the UMAP from (C).
(E) FeaturePlot display of the distribution of WT and ISL1−/− day 8 CPs across the clusters depicted in (C).
(F) Heatmap of significantly dysregulated genes with biological GO terms in day 8 ISL1−/− or NKX2.5−/− CPs compared with WT.
To determine the ISL1-binding dependency on NKX2.5, we generated an NKX2.5−/− hiPSC line using CRISPR-Cas9 gene editing (Figures S5A and S5B). As expected, because of the known role of NKX2.5 in repression of ISL1, deletion of NKX2.5 led to upregulation of ISL1 protein (Figure S5C) (Dorn et al., 2015). Given that deletion of NKX2.5 did not grossly disrupt morphological structures in CMs (Figure S5D), we investigated the degree to which NKX2.5 shares a regulatory network with ISL1 in developing CPs by comparing the scRNA-seq profiles of NKX2.5−/− and ISL1−/− CPs (Figure 5C). The day 8 CP cells clustered into the same broad clusters as before, indicated by expression of markers in each cluster and a progression of differentiation validated by pseudotime analysis (Figures 5C, 5D, and S6E; Table S4).
We also observed ISL1 expression throughout all clusters in WT day 8 CPs, yet NKX2.5 expression was enriched in the more differentiated late cluster (Figures S5F and S5G). Therefore, we focused our analyses on the transition and late clusters (Figures 5E, 5F, and S5H; Table S4). Both ISL1 and NKX2.5 were required for proper expression of genes related to Muscle cell development (e.g. CSRP3, MYL9) and Regulation of muscle contraction (e.g. NPPA, CNN1) (Figure 5F). These findings for CPs devoid of NKX2.5−/− are in line with previous bulk RNA-seq findings (Anderson et al., 2018). Without ISL1 or NKX2.5, we observed upregulation of genes related to Glial cell differentiation (e.g. SOX4, SOX11) and Regulation of binding (e.g. NFIB, TMSB4X). The finding of upregulated glial-related markers suggests ISL1 and NKX2.5 might be necessary to repress alternative cell fates; on the other hand, SOX4 and SOX11 are critical for early outflow tract development (Paul et al., 2014) and their upregulation could indicate a critical role for ISL1 and NKX2.5 in suppressing them as CPs proceed through differentiation. Among genes specifically upregulated in the absence of NKX2.5 were markers related to Positive regulation of cardiomyocyte differentiation (e.g. GATA4, TBX5), which could indicate a role for NKX2.5 in blocking differentiation until the correct time in development. In addition, we observed upregulated genes related to Cardiac muscle contraction by Ca+ signaling (e.g. CAMK2D, SLC8A1), in agreement with NKX2.5’s role in suppressing pacemaker cell fate (Dorn et al., 2015).
To test the hypothesis that some cardiac genes are co-regulated by ISL1 and NKX2.5, we evaluated the regulation of NPPA and NPPB, which were significantly downregulated in ISL1−/− and NKX2.5−/− CPs (Figure 5F; Table S4). We assessed activation of a putative NPPA/NPPB regulatory region occupied by both TFs cloned upstream of a luciferase gene (Figure S5I). In COS-7 cells, expression of either ISL1 or NKX2.5 activated the reporter, but the presence of both TFs had an additive effect, indicating that NKX2.5 is likely important for regulating a subset of CP-specific genes when co-bound with ISL1.
NKX2.5 is necessary and sufficient to localize ISL1 to a subset of genomic regions
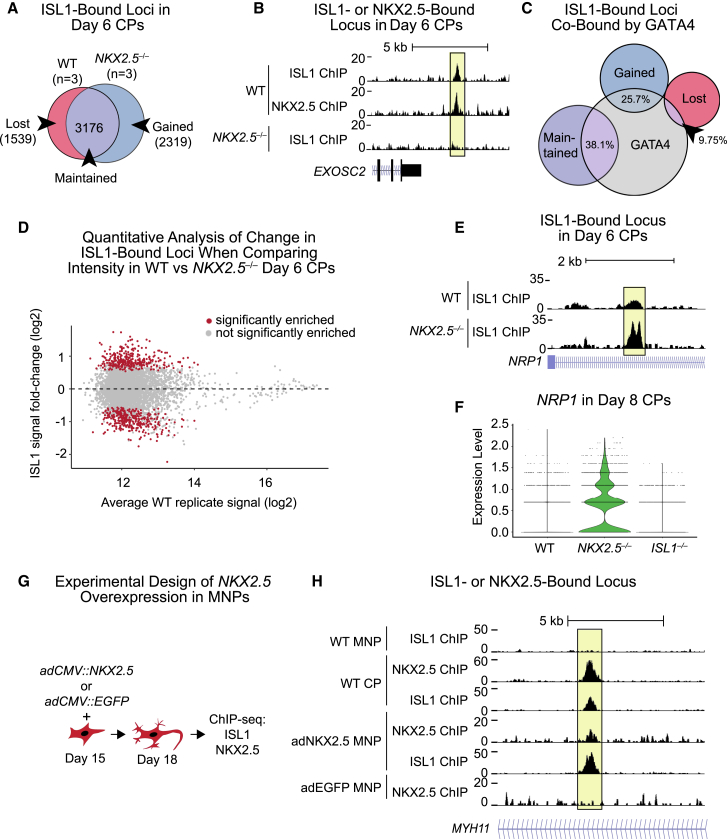
We next tested the contribution of NKX2.5 to the genomic localization of ISL1 by performing ISL1 ChIP-seq in NKX2.5−/− CPs (Figure S6A). In the absence of NKX2.5, we observed 1,539 fewer ISL1-bound peaks (“lost” peaks), while 3,176 peaks remained (“maintained” peaks), and 2,319 additional peaks were detected (“gained” peaks) compared with the WT setting (Figures 6A and 6B). GREAT analysis of the gained peaks showed enrichment of cardiac terms such as Cardiac chamber morphogenesis and Heart morphogenesis, similar to the maintained peaks, suggesting that, in the absence of NKX2.5, ISL1 binds ectopically to alternative cardiac loci (Figure S6B). For example, there is increased ISL1 binding at the TBX5 locus in the absence of NKX2.5, which might drive the aberrant increase of TBX5 expression shown previously (Figures 5F and S6C). In addition to the cardiac terms, Artery development was also enriched, which could indicate ISL1 is also redistributed to other loci regulating genes involved in endothelial or smooth muscle cells, alternative fates of the CPs (Figure S6B). Lost loci were enriched for terms associated with cell polarity, such as Apical constriction and Regulation of Maintenance of Cell Polarity (Figure S6B).
Figure 6.
ISL1 depends on NKX2.5 for DNA localization at a subset of cardiac loci
(A) Venn diagram of ISL1-bound DNA regions that are lost, maintained or gained in NKX2.5−/− day 6 CPs.
(B) Example of an NKX2.5-dependent ISL1-bound peak. Data shown from a single representative replicate.
(C) Venn diagram of ISL1-bound loci from (A) compared with GATA4-bound loci.
(D) Quantitative analysis of change in ISL1 ChIP peak intensity in WT CPs compared with ISL1 binding in NKX2.5−/− day 6 CPs. Regions with statistically significant change using DESeq are in red (p ≤ 0.05); points in gray are not significant. Fold change of ISL1 binding (NKX2.5−/−/NKX2.5+/+) is graphed across ISL1 ChIP signal intensity in WT CPs.
(E) ChIP-seq track of ISL1 detailing increased intensity of ISL1 binding in the absence of NKX2.5 at the NRP1 locus. Data shown from single representative replicate.
(F) Violin plot of NRP1 expression in WT, NKX2.5−/−, and ISL1−/− day 8 CPs (n = 3 independent experiments for each).
(G) Schematic of NKX2.5 experimental overexpression in MNPs.
(H) ChIP-seq tracks of an ISL1 peak gained within the MYH11 locus when NKX2.5 was overexpressed in MNPs. Data shown from a single representative replicate.
We compared ISL1-bound peaks with GATA4-bound loci in day 6 CPs (Figure S6D) (Gonzalez-Teran et al., 2022). Co-bound loci were enriched in terms such as Striated muscle cell differentiation and cardiomyocyte differentiation (Figure S6E). Among the lost, gained, and maintained ISL1-bound loci in NKX2.5−/− CPs for GATA4 binding, a greater portion of the ISL1-bound gained and maintained loci were co-bound with GATA4 compared with the ISL1-bound lost loci (25.7% and 38.1%, compared with 9.75%, respectively) (Figure 6C). Thus, GATA4 may aid in maintaining ISL1 presence at cardiac loci, as well as contribute to redistribution of ISL1 upon loss of NKX2.5.
As a complementary approach, we compared the intensity of the ISL1 peaks identified in WT CPs with those found in NKX2.5−/− CPs (Figure 6D), identifying peaks that had increased intensity, decreased intensity, or no change. Among those with decreased signal, the NKX2.5 motif was proportionally more common than the ISL1 or GATA4/GATA6 motif (Figure S6F), while among the peaks with increased signal, the NKX2.5 motif was proportionally less common than the ISL1 or GATA4/GATA6 motifs. Peaks with increased signal were enriched for GO terms related to the nervous system such as the Facial nerve structural organization and Regulation of axon guidance (Figure S6G). Some genes within these terms are typical of specialized CMs within the sinus node, the electrical pacemaker of the heart. Interestingly, the sinus node myocardium is ISL1 positive yet NKX2.5 negative, and ISL1 is critical for its development (Espinoza-Lewis et al., 2009; Liang et al., 2015). Another locus with increased signal was located within NRP1 (Figure 6E), which is necessary for proper sinus node development (Maden et al., 2012). NRP1 expression was increased in NKX2.5−/− CPs, consistent with increased ISL1 binding (Figure 5F) and supporting the conclusion that NKX2.5 is necessary for proper ISL1 localization to a large number of its binding sites in WT CPs.
Last, we tested if ectopic introduction of NKX2.5 in MNPs could override the effect of ISL1’s native neuronal partners at some loci, redistributing ISL1 to new sites. We overexpressed either EGFP (control) or NKX2.5 in day 15 MNPs and performed ChIP-seq on both ISL1 and NKX2.5 3 days later (Figures 6G, S6H, and S6I). We observed a large overlap in the NKX2.5-bound peaks with ISL1-bound peaks (75%) (Figure S6J). Indeed, despite a mild decrease in ISL1 expression (Figure S6I), ISL1 redistributed to more than 300 new sites typically occupied in CPs but not in MNPs (Figure S6K, p = 0.001), including the MYH11 locus (Figure 6H). Overall, our data indicate that NKX2.5 is both necessary and sufficient for ISL1 genomic localization to a subset of cardiac-enriched loci, although it does not alone alter fate of the MNPs.
Discussion
Here we used hiPSC models, scRNA-seq, ChIP-seq, and the deep learning model BPNet to determine how ISL1 executes cell-type-specific functions in distinct cell types through differentially interacting partners. We found that ISL1 functions in a complex with NKX2.5 to broadly co-occupy cardiac-specific genes, with binding modulated by GATA4 co-occupancy. NKX2.5 is believed to direct ISL1’s localization to hundreds of cardiac-specific loci, and the introduction of NKX2.5 is sufficient to re-localize ISL1 even in neural progenitors. These analyses reveal how tissue-enriched TFs can cooperatively regulate the differentiation of specific cell types.
ISL1 interacts with NKX2.5 to drive cardiac regulatory networks
ISL1 forms transcriptional complexes with TFs LHX3 and LDB1 to facilitate MNP differentiation (Lee et al., 2012) and has been shown to interact with LDB1 in a mouse CP model to facilitate long-range looping (Caputo et al., 2015). However, no cardiac-specific ISL1 interactor had previously been identified.
To analyze ISL1-containing complexes predicted by BPNet analysis, we used SRM and demonstrated that ISL1 forms putative protein complexes with NKX2.5, GATA4, GATA6, and TEAD4. There is previous evidence of complexes that form between several of these factors, such as NKX2.5 and GATA4 (Durocher et al., 1997). In addition, mapping of several cardiac TFs in developing mice showed co-bound loci between GATA4, NKX2.5, and a TEAD protein, suggesting co-regulatory roles for these complexes in the heart (Akerberg et al., 2019). Our data shows that ISL1 is capable of forming proteomic complexes with several of these factors, and we specifically found that the ISL1-NKX2.5 complex co-binds to DNA at hundreds of genomic loci. GATA4 is also often co-localized with ISL1 and NKX2.5 at numerous loci and seems to affect ISL1 localization, particularly in the absence of NKX2.5. Moreover, exogenous expression of NKX2.5 in MNPs was sufficient to re-localize ISL1 to a subset of cardiac loci. Thus, NKX2.5 is necessary for proper ISL1 binding in CPs, although other factors present in cardiac cells likely also contribute.
Genes co-bound by ISL1 and NKX2.5 were largely involved in early cardiac chamber morphogenesis, suggesting the combination dictates early CM gene expression. Comparison of dysregulated genes in NKX2.5−/− CPs and ISL1−/− CPs suggests that early cardiac genes are co-regulated by these factors, and genes uniquely regulated by NKX2.5 are associated with later cardiogenic events. Thus, the cooperativity between ISL1 and NKX2.5 may shift the multipotent progenitor to a CM fate, and downregulation of ISL1 allows progression to a more mature cell type guided by NKX2.5 with its other binding partners.
ISL1 displays lineage-specific regulation of a network of genes
Our data indicate that, in CPs or MNPs, loss of ISL1 results in cells predominantly adopting different cell fates along the same lineage, rather than alternative cell fates. Specifically, CPs lacking ISL1 were more atrial like than ventricular, and MNP-directed differentiation led to more IN-like cells in the absence of ISL1. In addition, there was no significant upregulation of atrial factors upon deletion of ISL1 in MNPs or significant upregulation of IN genes in the ISL1−/− CPs. This suggests there is either redundancy in the repression of such genes, or that an ISL1-independent mechanism represses those transcriptional programs.
In summary, we identified and characterized a cardiac-specific protein complex involving ISL1 that directs CM fate in human CP cells. These results demonstrate how a single TF can have divergent functions in different cell types based on its interaction with cell-type-specific factors, and help to broaden our understanding of how complex cell-specific cues direct TF function.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources, reagents, and materials should be directed to and will be fulfilled by the lead contact, Deepak Srivastava (Deepak.srivastava@gladstone.ucsf.edu).
Materials availability
Requests for materials or resources should be directed to the lead contact, Deepak Srivastava.
Data and code availability
The scRNA-seq have been deposited at GEO:GSE195476. The ChIP-seq datasets have been deposited at GEO:GSE195476. The TSQ datasets have been deposited in the Panorama database at PMID: 29487113. Code for the scRNA-seq data analysis is available at https://github.com/bejmaven/Maven_at_al_2023_SrivastavaLab. Code for the BPNet analyses is available at https://github.com/zeitlingerlab/Maven_ISL1_2022. Additional information required to reanalyze the data reported here is available from the lead contact.
Experimental model and subject details
iPSC culture and differentiation
hiPSCs-CPs and hiPSC-MNPs were generated using previously published methods (Lian et al., 2013; NeuroLINCS Consortium et al., 2021). Full culture conditions and medium formulations can be found in the Supplemental information.
Generation of CRISPR-Cas9 genome-edited hiPSC lines
WTC11 iPSCs were targeted using CRISPR-Cas9 ribonucleic proteins (RNPs), as described (Hultquist et al., 2016; Kim et al., 2014) with minor modifications. Single guide RNAs (sgRNAs) were generated by Dharmacon and Cas9 protein was obtained from QB3 MacroLab (Cas9-nls), and complexed in vitro to form RNPs. Briefly, we electroporated the RNP/sgRNA using the Lonza Nucleofector X-unit 4D-Nucleofector with the Lonza P3 Primary Cell 96-well Nucleofector. Each selected colony was genotyped to confirm homozygosity. To generate the NKX2.5−/− hiPSC line, we used a single sgRNA targeting strategy while the ISL1−/− hiPSC line used two sgRNAs to excise a portion of the ISL1 gene. sgRNAs and oligonucleotides used for genotyping are listed in Table S5.
Adenovirus
Adenoviral human type 5 (dE1/E3) viral particles expressing Ad-NKX2-5 or negative control ad-EF1a-eGFP.
Method details
Immunostaining
Cells were fixed methanol-free diluted paraformaldehyde with water 1:4 (w/v) at room temperature (RT) for 15 min with gentle shaking. Fixed cells were washed twice with Dulbecco’s PBS (DPBS) then exposed to a blocking solution consisting of 5% normal donkey serum and 0.2% Triton X-100 in DPBS for 1 h at RT. Cells were incubated with primary antibody (see the supplemental experimental procedures) at 4°C overnight, washed three times in DPBS, followed by incubation with secondary antibody at RT for 1 h, and washed three times using DPBS containing NucBlue Live ReadyProbes dye. Imaging was performed via a Zeiss S1 Confocal Microscope and image processing done using ImageJ (Schneider et al., 2012).
Whole cell lysate extraction
CP or MNP cell pellets were thawed on ice then resuspended in cold RIPA buffer supplemented with protease and phosphatase inhibitors. Samples were incubated on ice for 10 min, followed by 10 min of gentle rocking at 4°C, then centrifuged at 14,000×g for 20 min at 4°C to isolate whole cell lysate (supernatant).
Western blotting
Western blotting was performed with Bis-Tris Mini Protein Gels following the manufacturer’s protocols and subsequently transferred to nitrocellulose membranes and blocked. Primary antibodies were added and incubated overnight at 4°C. Next, the membrane was washed three times with PBS-T and incubated with appropriate secondary antibodies. Protein detection as performed using the Odyssey Imager or Western Blotting Detection Reagent followed by exposure to film.
RNA extraction, RT-qPCR reaction, and analysis
Cell pellets were lysed in Trizol and RNA extracted with the RNeasy kit and treated with DNase according to manufacturer’s instructions. We converted 1 μg RNA to cDNA with SuperScript III First-Strand Synthesis System. Wes used 1 μL of a 1:50 w/v dilution per qRT-PCR reaction with TaqMan Universal PCR Master Mix. All reactions were run on the 7900HT Fast Real-Time system (Applied Biosystem). TaqMan probes are listed in the Supplemental information. Data shown are averages of at least three biological replicates and three technical replicates, normalized to GAPDH RNA levels.
Single cell RNA-seq
Day 8 CPs or day 18 MNPs were used to generate single cell libraries with the Chromium Single Cell 3′ Library and Gel Bead Kit v3 and Chromium i7 Multiplex Kit. Libraries were sequenced on the Illumina NovaSeq 6000 System or the Illumina NextSeq500 based on the Chromium Single Cell v3 specifications. See the supplemental experimental procedures.
ChIP-seq
Cells were collected from tissue culture plates, cross-linked with paraformaldehyde followed by glycine quenching. For immunoprecipitation, the pellets were thawed and lysed, and the resulting nuclei pellet was sheared. Following incubation with appropriate antibodies, protein complexes were immunoprecipitated with dynabeads and bound proteins eluted. The samples were then processed for sequencing with NEBNext Ultra II DNA Library Kit for Illumina. After sequencing on the Illumina NextSeq500 or the Illumina HiSeq 4000 (UCSF Center for Advanced Technology), samples were analyzed for mapping, filtering, peak calling, and visualization. See the supplemental experimental procedures.
BPNet model training and motif identification
We trained a convolutional neural network using the BPNet code, architecture and loss features consistent with the published approach (Avsec et al., 2021) to explain WT ISL1 ChIP-seq data. See the supplemental experimental procedures.
Analysis of protein complexes
Cells were resuspended in IP buffer, sheared, and then incubated with primary antibodies. Protein complexes were immunoprecipitated using antibody-conjugated beads, followed by washing. Eluates were prepared for western blotting and MS. See the supplemental experimental procedures.
Cloning of overexpression constructs
We used a CMV::3x-FLAG construct (Jäger et al., 2011). Cloning was performed to insert EGFP or the human NKX2.5 downstream of CMV. For CMV::hLDB1-HA, the coding sequence of human LDB1 with a downstream human influenza hemagglutinin (“HA”) tag was assembled into the pcDNA4/TO Vector. For CMV::hISL1-Bio, we cloned in a TEV biotinylation site (“Bio”) and then inserted the human coding sequence of ISL1. Sanger sequencing confirmed all constructs.
Mapped motif pair synergy
To test synergy effects between the TF-MoDISco mapped motifs, we adopted the in silico genomic motif interaction approach described (Avsec et al., 2021). See the supplemental experimental procedures.
Transfection of siRNA
We attached a fluorescent probe to each siRNA with the Label IT siRNA Tracker Reagent Intracellular Localization Kit, CX-Rhodamine, and transfected with Lipofectamine RNAiMAX. To begin, we replated D5 CPs 1:1 into 12-well plates pre-coated with fibronectin (diluted 1:80 with PBS) and added siRNA-lipofectamine complexes into each well. Media were refreshed 24 h later and cells analyzed by flow cytometry 72 h after transfection.
Fluorescence-activated cell sorting
CPs were harvested from plates using accutase, then washed twice with PBS. Cells were strained through a 40-μM mesh (BD Falcon) and sorted on the BD Aria II 5 flow cytometer. Populations with shifted PE Texas Red-A signal, corresponding to Rhodamine, were collected and processed for downstream analysis.
Luciferase assay
The putative NPPA/NPPB regulatory region was synthesized and inserted into the pNL1.1[Nluc] plasmid and co-transfected together with renilla luciferase control pGL4.73[hRluc/SV40], CMV::hISL1-Bio and/or CMV::hNKX2.5-3xFLAG into COS-7 cells using Fugene HD. Luminescence activity was quantified with Dual-Glo Luciferase Assay System. Fold-change shown are averages of at least three biological replicates and four technical replicates, compared with the empty vector controls and then normalized to the renilla luciferase control.
Measuring enrichment of ISL1 peaks
DESeq2 (fitType = local) was used to compare enrichment differences of WT and NKX2.5−/− ISL ChIP-seq signal across peaks from WT ISL1 ChIP-seq in day 6 CPs. Total raw count pileup of WT and NKX2.5−/− ISL1 ChIP-seq in day 6 CPs were collected across every WT ISL1 peak in day 6 CPs across a 1000 bp centered window. Using both an adjusted p value cutoff of p ≤ 0.05 and the reported log2 fold-change enrichment of WT versus NKX2.5−/− ISL1 ChIP-seq signal intensity, peaks were classified as having significantly increased, significantly decreased, or not changed in enrichment upon removal of NKX2.5.
Mapped motif enrichment
Motif enrichment was tested using Bonferroni-corrected chi-square tests across classified ISL1 ChIP-seq peaks (classification described above) measured across WT and NKX.5−/− CPs.
Adenovirus infection of neural cells
Media were changed on day 15 MNPs to stage-appropriate media, and cells were infected drop-wise at an multiplicity of infection (MOI) of 100 viral units per cell. Media was removed and replaced with stage-appropriate media 48 h later. Infection was confirmed by detection of EGFP by fluorescence microscopy 72 h post infection.
Quantification and statistical analysis
Details pertaining to statistical significance and value of n are reported in the methods details, figures and the accompanying figure legends. The level of significance in all figures is represented as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Acknowledgments
We thank Bethany Taylor, Françoise Chanut, and Kathryn Claiborn for editorial assistance; the Gladstone Stem Cell Core, the Gladstone Flow Cytometry Core, the Gladstone Bioinformatic Core, the Gladstone Mass Spectrometry Core, the Gladstone Genomics Core, and the UCSF Center for Advanced Technology (CAT) for their technical expertise and use of equipment; and the members of the Srivastava laboratory and Gladstone community for helpful feedback and discussion.
D.S. is supported by the National Institutes of Health/NHLBI (P01 HL146366, R01 HL057181, R01 HL015100, R01 HL127240), Roddenberry Foundation, L. K. Whittier Foundation, Dario and Irina Sattui, Younger Family Fund, and Additional Ventures. B.E.J.M. was supported by the American Heart Association (19PRE34380715). W.K. was supported by the National Heart, Lung, and Blood Institute, from NIH award number, 5R25HL121037. N.J.K. and R.H. are supported by the National Institutes of Health (P01 HL146366). T.N. is supported by JSPS Overseas Research Fellowship.
Author contributions
B.E.J.M., C.A.G., K.N.I., and D.S. conceived and directed this study with input from R.H., M.C., N.J.K., and J.Z.; CP differentiations: B.E.J.M.; MNP differentiations: B.E.J.M. and W.K.; knockout line generation: B.E.J.M., D.G., and B.G.T.; scRNA-seq library preparation and sequencing: C.A.G. and T.N.; scRNA-seq analyses: B.E.J.M and C.A.G. with input from A.P.; ChIP-seq protocol generation: K.S.K.; ChIP-seq library preparation and sequencing: K.S.K. and B.E.J.M.; ChIP-seq analyses: B.E.J.M. with input from A.P.; BPNet modeling and follow-up: M.W.; desalting and purification of MS samples: M.M.; MS/MS and SRM analyses: B.E.J.M. with input from R.H.; Cloning constructs: B.E.J.M., E.O., and S.R.
Declaration of interests
D.S. is a co-founder and member of the board of directors of Tenaya Therapeutics and has equity in Tenaya Therapeutics. K.N.I. is an employee and shareholder of Tenaya Therapeutics. N.J.K. has received research support from Vir Biotechnology, F. Hoffmann-La Roche, and Rezo Therapeutics. N.J.K. has financially compensated consulting agreements with Maze Therapeutics, Interline Therapeutics, Rezo Therapeutics, and GEn1E Lifesciences, Inc. He is on the Board of Directors of Rezo Therapeutics and is a shareholder in Tenaya Therapeutics, Maze Therapeutics, Rezo Therapeutics, and Interline Therapeutics.
Published: October 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2023.09.014.
Supplemental information
References
- Akerberg B.N., Gu F., VanDusen N.J., Zhang X., Dong R., Li K., Zhang B., Zhou B., Sethi I., Ma Q., et al. A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat. Commun. 2019;10:4907. doi: 10.1038/s41467-019-12812-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.J., Kaplan D.I., Bell K.M., Koutsis K., Haynes J.M., Mills R.J., Phelan D.G., Qian E.L., Leitoguinho A.R., Arasaratnam D., et al. NKX2-5 regulates human cardiomyogenesis via a HEY2 dependent transcriptional network. Nat. Commun. 2018;9:1373. doi: 10.1038/s41467-018-03714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avsec Ž., Weilert M., Shrikumar A., Krueger S., Alexandari A., Dalal K., Fropf R., McAnany C., Gagneur J., Kundaje A., Zeitlinger J. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat. Genet. 2021;53:354–366. doi: 10.1038/s41588-021-00782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.L. Transcriptional pathways in second heart field development. Semin. Cell Dev. Biol. 2007;18:67–76. doi: 10.1016/j.semcdb.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C.-L., Liang X., Shi Y., Chu P.-H., Pfaff S.L., Chen J., Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E., Camafeita E., Fernández-Gutiérrez B., López J.A. Applying selected reaction monitoring to targeted proteomics. Expert Rev. Proteomics. 2011;8:165–173. doi: 10.1586/epr.11.11. [DOI] [PubMed] [Google Scholar]
- Caputo L., Witzel H.R., Kolovos P., Cheedipudi S., Looso M., Mylona A., van IJcken W.F.J., Laugwitz K.-L., Evans S.M., Braun T., et al. The Isl1/Ldb1 Complex Orchestrates Genome-wide Chromatin Organization to Instruct Differentiation of Multipotent Cardiac Progenitors. Cell Stem Cell. 2015;17:287–299. doi: 10.1016/j.stem.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn T., Goedel A., Lam J.T., Haas J., Tian Q., Herrmann F., Bundschu K., Dobreva G., Schiemann M., Dirschinger R., et al. Direct nkx2-5 transcriptional repression of isl1 controls cardiomyocyte subtype identity. Stem Cell. 2015;33:1113–1129. doi: 10.1002/stem.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D., Charron F., Warren R., Schwartz R.J., Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ediger B.N., Du A., Liu J., Hunter C.S., Walp E.R., Schug J., Kaestner K.H., Stein R., Stoffers D.A., May C.L. Islet-1 Is essential for pancreatic β-cell function. Diabetes. 2014;63:4206–4217. doi: 10.2337/db14-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis R.A., Yu L., He F., Liu H., Tang R., Shi J., Sun X., Martin J.F., Wang D., Yang J., Chen Y. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway J.R., Bethea M., Liu Y., Underwood R., Mobley J.A., Hunter C.S. SSBP3 Interacts With Islet-1 and Ldb1 to Impact Pancreatic β-Cell Target Genes. Mol. Endocrinol. 2015;29:1774–1786. doi: 10.1210/me.2015-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Teran B., Pittman M., Felix F., Thomas R., Richmond-Buccola D., Hüttenhain R., Choudhary K., Moroni E., Costa M.W., Huang Y., et al. Transcription factor protein interactomes reveal genetic determinants in heart disease. Cell. 2022;185:794–814.e30. doi: 10.1016/j.cell.2022.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist J.F., Schumann K., Woo J.M., Manganaro L., McGregor M.J., Doudna J., Simon V., Krogan N.J., Marson A. A Cas9 ribonucleoprotein platform for functional genetic studies of HIV-host interactions in primary human T cells. Cell Rep. 2016;17:1438–1452. doi: 10.1016/j.celrep.2016.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Cimermancic P., Gulbahce N., Johnson J.R., McGovern K.E., Clarke S.C., Shales M., Mercenne G., Pache L., Li K., et al. Global landscape of HIV-human protein complexes. Nature. 2011;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrar W., Dias J.M., Ericson J., Arnold H.-H., Holz A. Nkx2.2 and Nkx2.9 are the key regulators to determine cell fate of branchial and visceral motor neurons in caudal hindbrain. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-T., Kim N., Kim H.-K., Lee H., Gruner H.N., Gergics P., Park C., Mastick G.S., Park H.-C., Song M.-R. ISL1-based LIM complexes control Slit2 transcription in developing cranial motor neurons. Sci. Rep. 2016;6 doi: 10.1038/srep36491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim D., Cho S.W., Kim J., Kim J.-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Cuvillier J.M., Lee B., Shen R., Lee J.W., Lee S.-K. Fusion protein Isl1-Lhx3 specifies motor neuron fate by inducing motor neuron genes and concomitantly suppressing the interneuron programs. Proc. Natl. Acad. Sci. USA. 2012;109:3383–3388. doi: 10.1073/pnas.1114515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Zhang Q., Cattaneo P., Zhuang S., Gong X., Spann N.J., Jiang C., Cao X., Zhao X., Zhang X., et al. Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Invest. 2015;125:3256–3268. doi: 10.1172/JCI68257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons I., Parsons L.M., Hartley L., Li R., Andrews J.E., Robb L., Harvey R.P. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Maden C.H., Gomes J., Schwarz Q., Davidson K., Tinker A., Ruhrberg C. NRP1 and NRP2 cooperate to regulate gangliogenesis, axon guidance and target innervation in the sympathetic nervous system. Dev. Biol. 2012;369:277–285. doi: 10.1016/j.ydbio.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkis G., Tzchori I., Cohen T., Holtz A., Wier E., Westphal H. Isl1 and Ldb co-regulators of transcription are essential early determinants of mouse limb development. Dev. Dyn. 2012;241:787–791. doi: 10.1002/dvdy.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NeuroLINCS Consortium. Li J., Lim R.G., Kaye J.A., Dardov V., Coyne A.N., Wu J., Milani P., Cheng A., Thompson T.G., et al. An integrated multi-omic analysis of iPSC-derived motor neurons from C9ORF72 ALS patients. iScience. 2021;24 doi: 10.1016/j.isci.2021.103221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa K., Schultz K., Yun K., Mohammed N., Shaw G.M., Lammer E.J. Haploinsufficiency ofinsulin gene enhancer protein 1(ISL1) is associated with d-transposition of the great arteries. Mol. Genet. Genomic Med. 2014;2:341–351. doi: 10.1002/mgg3.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M.H., Harvey R.P., Wegner M., Sock E. Cardiac outflow tract development relies on the complex function of Sox4 and Sox11 in multiple cell types. Cell. Mol. Life Sci. 2014;71:2931–2945. doi: 10.1007/s00018-013-1523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff S.L., Mendelsohn M., Stewart C.L., Edlund T., Jessell T.M. Requirement for LIM Homeobox Gene Isl1 in Motor Neuron Generation Reveals a Motor Neuron– Dependent Step in Interneuron Differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Quaranta R., Fell J., Rühle F., Rao J., Piccini I., Araúzo-Bravo M.J., Verkerk A.O., Stoll M., Greber B. Revised roles of ISL1 in a hES cell-based model of human heart chamber specification. Elife. 2018;7 doi: 10.7554/elife.31706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S.Y., Lee B., Lee S. Critical roles of the LIM domains of Lhx3 in recruiting coactivators to the motor neuron-specifying Isl1-Lhx3 complex. Mol. Cell Biol. 2015;35:3579–3589. doi: 10.1128/MCB.00335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E.Y., Ichida J.K., Wainger B.J., Toma J.S., Rafuse V.F., Woolf C.J., Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.-R., Sun Y., Bryson A., Gill G.N., Evans S.M., Pfaff S.L. Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development. 2009;136:2923–2932. doi: 10.1242/dev.037986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Velasco S., Ibrahim M.M., Kakumanu A., Garipler G., Aydin B., Al-Sayegh M.A., Hirsekorn A., Abdul-Rahman F., Satija R., Ohler U., et al. A multi-step transcriptional and chromatin state cascade underlies motor neuron programming from embryonic stem cells. Cell Stem Cell. 2017;20:205–217.e8. doi: 10.1016/j.stem.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Y., Guo C., Lu Q., Wang W., Jia Z., Chen P., Ma K., Reinberg D., Zhou C. ISL1 and JMJD3 synergistically control cardiac differentiation of embryonic stem cells. Nucleic Acids Res. 2016;44:6741–6755. doi: 10.1093/nar/gkw301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Song H.-M., Wang F., Zhao C.-M., Huang R.-T., Xue S., Li R.-G., Qiu X.-B., Xu Y.-J., Liu X.-Y., Yang Y.Q. A New ISL1 Loss-of-Function Mutation Predisposes to Congenital Double Outlet Right Ventricle. Int. Heart J. 2019;60:1113–1122. doi: 10.1536/ihj.18-685. [DOI] [PubMed] [Google Scholar]
- Yasuhara J., Garg V. Genetics of congenital heart disease: a narrative review of recent advances and clinical implications. Transl. Pediatr. 2021;10:2366–2386. doi: 10.21037/tp-21-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Nomura-Kitabayashi A., Sultana N., Cai W., Cai X., Moon A.M., Cai C.-L. Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development. Dev. Biol. 2014;390:68–79. doi: 10.1016/j.ydbio.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq have been deposited at GEO:GSE195476. The ChIP-seq datasets have been deposited at GEO:GSE195476. The TSQ datasets have been deposited in the Panorama database at PMID: 29487113. Code for the scRNA-seq data analysis is available at https://github.com/bejmaven/Maven_at_al_2023_SrivastavaLab. Code for the BPNet analyses is available at https://github.com/zeitlingerlab/Maven_ISL1_2022. Additional information required to reanalyze the data reported here is available from the lead contact.