Abstract
Peripheral blood mononuclear cells (PBMCs) are a useful model for biochemical assays, particularly for etiological studies. We describe here a method for measuring DNA repair capacity (DRC) in archival cryogenically preserved PBMCs. To model DRC, we measured γ-H2AX repair kinetics in thawed PBMCs after irradiation with 3 Gy gamma rays. Time-dependent fluorescently labeled γ-H2AX levels were measured at five time points from 1 to 20 h, yielding an estimate of global DRC repair kinetics as well as a measure of unrepaired double strand breaks (DSB) at 20 h. While γ-H2AX levels are traditionally measured by either microscopy or flow-cytometry, we developed a protocol for imaging flow-cytometry (IFC) that combines the detailed information of microscopy with the statistical power of flow methods. The visual imaging component of the IFC allows for monitoring aspects such as cellular health and apoptosis as well as fluorescence localization of the γ-H2AX signal, which ensures the power and significance of this technique. Application of a machine-learning based image classification improved flow cytometry fluorescent measurements by identifying apoptotic cells unable to undergo DNA repair. We present here DRC repair parameters from 18 frozen archival PBMCs and 28 fresh blood samples collected from a demographically diverse cohort of women measured in a high-throughput IFC format. This thaw method and assay can be used alone or in conjunction with other assays to measure etiological phenotypes in cryogenic biobanks of PBMCs.
Graphical Abstract
Introduction
Peripheral blood cells are a valuable source for measuring individual response in biomarker biochemical assays [1–9]. Whole blood is easier to collect, particularly compared to tissue-based biomarkers, from which peripheral blood mononuclear cells (PBMCs) can be cryopreserved for a wide range of uninvasive, non-surgical, future patient clinical studies and large-scale demographic studies of phenotypic differences between case and control populations [10–17]. Short and long-term cryogenic bio-banking of these samples is of particular importance to arrange case-control matching for large demographic cohorts [1, 2, 4–9, 15]. Some challenges of optimizing a thawing protocol for bio-banked cells include cell clumping, cell lysis, removal of the freezing medium, optimization of the content of the thawing medium, and culturing after thaw.
The objective of the present work was to develop a high-throughput DNA repair phenotype assay from archival PBMCs stored for ~20 years to measure DNA repair capacity (DRC). DRC is a measure of a cell’s ability to repair DNA after exposure to genotoxic agents and has been associated with many different cancers in case control and cohort studies (for review see [18]). As cancer initiation is commonly ascribed to mutations in key oncogenes/tumor suppressor genes due to unrepaired or mis-repaired DNA, DRC is of major importance in cancer risk [3–5, 7]. DRC has been measured previously by methods such as the Comet assay [19–22] and end-joining capacity by fluorescent immunoblotting [2]. Conversely, our DRC phenotype assay uses immunodetection of γ-H2AX, a biomarker of global DNA double strand breaks (DSBs) after ionizing radiation [23–27]. H2AX histones are phosphorylated (known as γ-H2AX) and are visualized as foci at the DNA DSB site when fluorescently labeled. Phosphorylated H2AX recruit DNA repair machinery and are dephosphorylated after DNA is successfully repaired [28, 29]. Quantification of fluorescently labeled γ-H2AX foci at serial time points provides an effective measure of the rate of DNA repair and is suitable for high-throughput sample processing optimizations.
In our earlier work, we developed a high-throughput γ-H2AX repair assay for the high-throughput RABiT system [26], that was applied to a demographic study of DRC [30]. We characterized two kinetic repair parameters: an initial fast repair rate (within the first 5 hours after irradiation) and the measurement of residual unrepaired DNA damage after overnight culturing, both of which are considered to comprise a holistic measure of different DNA repair pathways [31, 32]. More recently, we further optimized and extended this assay for fresh whole blood using imaging flow cytometry (IFC) [27]. The advantage of IFC is that it provides both the statistical power of flow-cytometry and the signal specificity conferred by microscopy which can be used for the rapid acquisition and quantification of characteristic γ-H2AX signal. Associated software can also be used to refine image analysis by filtering for cell death and apoptosis as measured by cellular morphological features [33–35]. In order to efficiently characterize and quantify such large sets of sample images, user-guided machine learning sorting algorithms are a valuable tool [36–39].
The primary goal of this study was to establish a high-throughput protocol for measuring DNA repair capacity for thawed cells from biobank archives. To accomplish this, we have completed the following:
Optimized conditions for thawing, irradiation, and cellular recovery after irradiation in a 96-tube, matrix format.
Measured DNA repair kinetics with IFC using the γ-H2AX biomarker for DSBs in thawed PBMC samples and fresh blood samples.
Estimated DRC for these samples using the previously described kinetic repair model of fast repair (1–5 hours after irradiation) and residual damage (20+ hours after irradiation)
Trained a machine learning-based classifier to identify cells damaged from thawing that are unable to undergo repair of DNA damage
Materials and Methods
Sample Information
The Breast Cancer Family Registry (BCFR) is a multi-generational family cohort selected for racial and ethnic diversity and high breast cancer risk due to genetic or environmental factors [2, 40]. Included in this cohort are >12,000 samples from women with no previous breast cancer diagnosis. Previous studies include genotyping of BRCA1/2 mutation status and other cancer risk genes [41, 42]. To begin using this very valuable cohort for cellular phenotype assays such as DRC, we developed this thawing protocol using a subset of BCFR samples from the New York site.
This subset of NYBCFR samples were collected between 1998–2001, with de-identified individual data including: age, body mass index (BMI), race and ethnicity, and smoking status as described previously [2] (and in Supplementary Table 1). After isolating PBMCs from whole blood via Ficoll gradient centrifugation, samples were frozen and preserved according to standard protocols and stored in liquid nitrogen until the thaw assays were performed in 2019. The 19 samples chosen for this optimization study were from a subset of donors studied in a previous DRC assays measuring nucleotide excision repair and DSB repair [1, 2].
Fresh Samples
Fresh peripheral blood samples were collected under Columbia University Irving Medical Center IRB approval IRB-AAAS0059 from 35 healthy volunteers, women between the ages 14–56 years old, with informed consent and a donor response epidemiological survey questionnaire from all participants, including smoking status and alcohol consumption. Blood was collected in BD Vacutainer® sodium heparin coated tubes (Becton Dickinson, Franklin Lakes, NJ) and stored overnight at room temperature in a dark Styrofoam box before proceeding with the γ-H2AX phenotype assay.
Protocol Optimization for Thawing Frozen PBMCs.
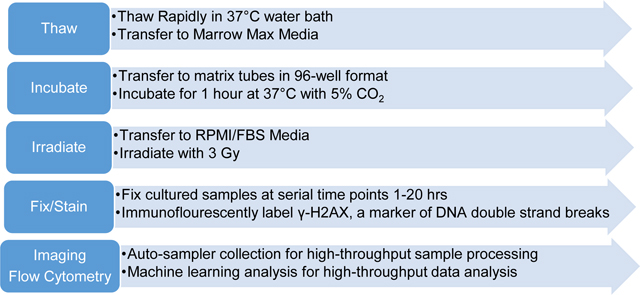
The following media were prepared for thawing the frozen PBMCs: Medium A (RPMI, 30% Fetal Bovine Serum, 1:10,000 dilution Benzonase (Sigma), final concentration 2.5 units/mL) or Medium B (MarrowMax™ (Thermo Fisher Scientific, Waltham, MA) with 1:10,000 dilution of Benzonase (Sigma), 2.5units/mL) and were warmed up to 37°C in a water bath.
Frozen PBMC samples (~1mL in 2mL cryotubes) were thawed in a 37°C water bath for 3 min and transferred into two separate 15mL Falcon tubes with ~ 500 ul volume of cell suspension. Thawing medium A or B was added drop-wise using a transfer pipette every 3–4 sec for the first 2 mL and then filled up to 10mL. Sample were washed a total of 3x by first centrifuging at 300xg for 10 min, removing supernatant, and re-suspending the pellet with 10mL of the corresponding thaw medium. 10 × 1mL cell aliquots were transferred into sterile Matrix™ microtubes (Thermo Scientific™) and incubated at 37°C/5% CO2 for 1 hr. Cells were washed with pre-warmed Medium C (RPMI 1640 with 15% FBS, 1% Pen/Strep). The 10 matrix tubes for each sample are pooled into a fresh, sterile, 15mL Falcon tube. Samples are washed 2x with medium as described above. Samples are re-aliquoted into10×1mL sterile matrix tubes.
γ-H2AX Phenotype Assay
i). Thawed Cell Irradiation:
For each donor 10 × 1 mL aliquots were prepared as follows: two samples were sham irradiated as a non-irradiated control and the remaining 8 were irradiated with 3 Gy γ-rays with a dose rate of 0.72 Gy/min using a Gammacell®40 137Cesium irradiator (Atomic Energy of Canada, Ltd., Chalk River, ON). The 137Cs irradiator was calibrated by nanoDot™ thermoluminescent dosimeter (Landauer, Glenwood IL). Tubes from the irradiated samples were incubated at 37°C/5% CO2 and removed at serial time points of 1, 2, 3, 5, and 20 hrs and the unirradiated controls removed at 2 and 20 hrs. Additional controls were the fluorescent compensations for channel 2 and channel 5, as well as an isotype control.
ii). Fresh Blood Irradiation
For each fresh whole blood (WB) sample, 15 × 0.1mL aliquots of WB were prepared as follows: 6 irradiated and 6 sham-irradiated for removal at serial time points 1, 2, 3, 5, 20, and 24 hours after irradiation, and 2 for compensation and isotype controls as above. Irradiated samples were irradiated with 3 Gy γ-rays as above, at which point all samples were diluted to 1mL with RPMI 1640 media (15% BSA and 1% Pen/Strep) and placed in the incubator at 37°C/5% CO2.
iii). Cell Counts
At the 1 and 20 hr time points, samples were tested for cell counts/viability using the Luna Dual Fluorescence Cell Counter (Logos Bio, Annandale, VA, USA). Briefly, 2μL of Acridine Orange/Propidium Iodine (Logos Bio) were added to 18 μL of cell suspension and counted on a photonslide (Logos Bio), measuring live/dead cells (exposure level 6 for both red and green channels).
iv). Sample Preparation and Labeling of γ-H2AX
At each time point, samples were taken and centrifuged at 300g for 3 min, supernatant removed and the cells re-suspended in 1 mL of ice cold 1% BSA/PBS (from 10x Fisher Scientific, BP399–1). WB samples were incubated with eBioscience™ 1X RBC Lysis Buffer (ThermoFisher Scientific) for 10 min at room temperature. Cells were washed two times with 1% BSA/PBS. After the final wash, cells were re-suspended in 250 μL of ice-cold fix/perm buffer (BD Cytofix/Cytoperm kit #554714) for 20 min on ice, after which 700 μL of Cytofix perm/wash (P/W) buffer was added. Cells were centrifuged and washed with 1 mL of P/W buffer a further two times followed by a final wash with 1% BSA/PBS. Cells collected from all the time points were stored at 4°C until antibody labeling which was typically within the next 24–48 hr.
Samples were stained using Alexa Fluor® 488 mouse anti-H2AX (pS139) (BD #560445) or Alexa Fluor® 488 mouse IgG1κ isotype control (BD #557782) as follows: Cells were centrifuged and washed once with 1mL P/W buffer and then incubated with anti-H2AX (pS139) antibody (final concentration 1:200 dilution) for samples and Ch2 compensation control. An isotype control sample (final concentration 1:200 dilution) and a Ch5 compensation control (P/W buffer only) were also prepared. Samples were incubated for 1 hr at room temperature in a dark drawer. All samples were then washed twice with P/W buffer, followed by 2 washes with 1%BSA in PBS. Samples were stored at 1mL in 1%BSA/PBS at 4°C in matrix tubes until analysis. Cells were centrifuged and 950 μL aspirated. 1 μL of a 1:10 dilution of DRAQ5 (ThermoFisher Cat# 62251) in DPBS was added to each sample. Samples were incubated for >5 min and loaded on the image stream.
v). Sample Acquisition and Analysis by Imaging Flow Cytometry.
Sample acquisition and the initial steps of the γ-H2AX image analysis were built on protocols described previously using imaging flow cytometry using the ImageStream®X MK II (ISXMKII, Amnis, Luminex) [23]. Briefly, for thawed PBMC samples, samples were loaded manually by the user and greater than 500 single cells with Brightfield RMS>50 (“In-focus”) were counted for each time point. For fresh WB samples, 3000 single cells were counted. All samples are collected at 40x magnification, with the 488 nm laser at 200 mW and 750 nm side scatter laser at 1mW, at maximum camera sensitivity, brightfield LED intensity ~35mW. Channel 1 was used to collect brightfield images, channel 2 for AlexaFluor®488 biomarker signal, channel 5 for DRAQ5 nuclear stain and channel 6 for side scatter. For compensations, samples were collected with all channels collected, but with brightfield and the side scatter laser turned off. Compensation coefficients were determined automatically by the IDEAS 6.3 software’s compensation wizard.
Image stream data were analyzed using the IDEAS 6.3.23 software with the machine learning module.
For thawed PBMC samples, first, objects that are out of focus are removed by gating brightfield (channel 1) Gradient RMS >55. Next, single cells were selected using a bivariate plot of area of channel 1 versus aspect ratio channel 1. We then selected an intensity threshold for DRAQ5 fluorescence intensity (channel 5) to exclude cells without an intact nucleus. A “healthy” population of cells was determined using the IDEAS apoptosis wizard, which generated a bivariate plot of contrast in channel 1 versus area of channel 5, masked by a threshold fluorescence value (a measure of nuclear signal that has a minimum fluorescence intensity defined as 50 percent of the maximum signal observed in all cells in the group). We further refined this “healthy” population by setting a morphological cutoff by plotting Area channel 1 versus intensity of side scatter (channel 6), which eliminates granular cells such as monocytes and degraded cells, hence the final assay is of an exclusively lymphocyte population. Finally, a bivariate plot of the machine learning classifiers for identifying “edge staining” and “spotted” cells to identify cells with the foci signal characteristic of true γ-H2AX repair signal.
Whole blood samples were analyzed as above, but with the following differences: a gating step to remove granulocytes was added after the apoptosis wizard, by making a bivariate plot of area channel 1 versus intensity channel 6 (side scatter). Machine learning modestly improved the quality of WB sample kinetic fit parameters, as well, so we used a modified version of the PBMC analysis template with the fresh whole blood samples. Of the 35 blood samples collected, 7 were not included in our results, either because of missing time points or not meeting the quality of fit criteria we set for our model of R2>0.5.
vi). Machine Learning Module
The machine learning classifiers were trained using the IDEAS 6.3 machine learning module, by using user-defined populations of the cells of interest, either edge-stained, or true “spotted” cells, on a large set of cell images. For both classifiers, the user selected cells with clear defined foci for one truth population, but for the “edge stained” classifier, the 2nd truth population were edge stained cells, while in the “spotted” classifier, the 2nd truth population were pan-nuclear stained cells. Classifiers were generated using all image features for all collected channels from all feature categories (size, shape, signal strength, texture, comparison, location).
vii). Quantitative modeling of γ-H2AX fluorescence time courses after irradiation
Median fluorescence intensity (MFI) values of the cells in the 4th quadrant (described in the machine learning classifier section above) were background corrected using 0 Gy controls. For thawed PBMCs, MFI values for hours 1–5 were normalized by subtracting the MFI of a 0 Gy sample at 2 hrs and for the 20-hr time point using its paired 20-hr 0 Gy control. Normalized MFI values at each time point were imported into R 4.1.2 software for analysis[43]. We modeled the time course of normalized MFI after irradiation separately for each donor, using the following equation, where is radiation dose (Gy), is time (hours) after irradiation, Ω is a binary indicator variable for irradiation or not (defined as for and for ), is the signal production term after radiation, is the initial exponential signal decay (repair) rate, and is the residual signal (unrepaired DNA damage) after overnight incubation:
| (1) |
Fitting of the model (Eq. 1) to the median normalized MFI data was performed using non-linear quantile regression (nlrq function in the quantreg R package), which fits median values instead of means. Fitting was done with parallel computing on all available cores, with 50 iterations of the quantile regression with randomized initial parameter values for each donor, and then the model variant with the lowest root-mean-squared error (RMSE) was retained. All kinetic fit parameters (, and ) were restricted to positive values. Samples which generated poor fits (R2<0.5) were not included in statistics summaries. Examination of the poorly fitted samples showed that they tended to exhibit no clear time dependence (i.e. behaved erratically) instead of conforming to biologically-plausible expectations of an initial rise in signal followed by reduction and stabilization at some residual level at longer times.
Results
Cell counts and viability after thawing
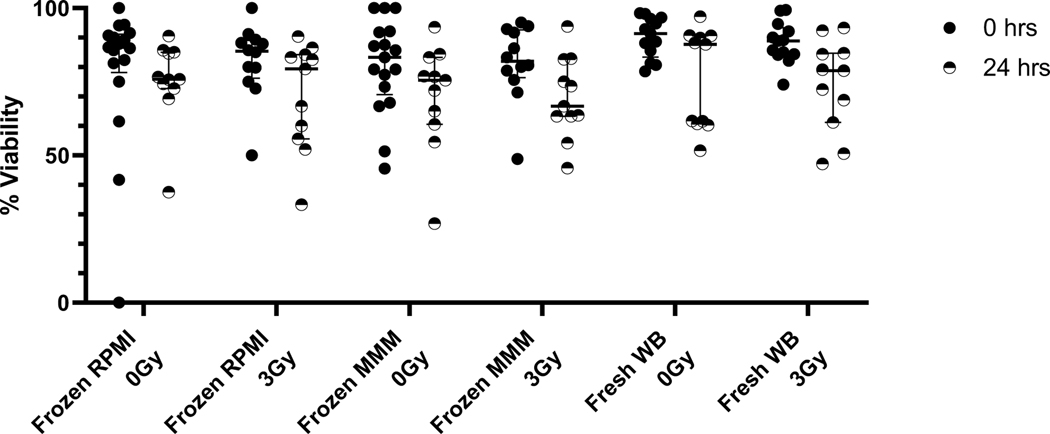
The two thawing methods using either MarrowMax™ Media (MMM) or RPMI/FBS (RPMI) media yielded high viability and a small amount of cell loss in the frozen archival cell samples after irradiation and overnight incubation (Figure 1). Cell viabilities based on Acridine Orange/Propidium Iodine at 1 hr and 20 hrs, after sham or 3 Gy irradiation using the two different media, showed that the media did not have an appreciable effect on cell viability, while viabilities did go down in both groups after 24 hrs of culturing. Median viabilities at 1 hr time points were 87, 85, 83, and 82% for RPMI 0 Gy, RPMI 3 Gy, MMM 0 Gy, MMM 3 Gy, respectively and dropped only slightly at 24 hrs to 76, 79, 76, 67% for the same samples. Freshly collected WB by comparison had median viability of 91% at collection and dropped to 89% after overnight incubation. When we extended the pre-irradiation incubation time after thawing PBMCs in both media from 1 h to 20 h, preliminary results (data not shown) indicated a significant loss in γ-H2AX response after irradiation (0.78x fold change instead of 1.7x fold change for the same donor).
Figure 1.
Cell Viability at 1 hour and 24 hours in thawed PBMCs and freshly collected whole blood measured using acridine orange and propidium iodide on the Luna Dual-Fluorescence Cell Counter tested for both thawing media RPMI1640+30%FBS (“RPMI”) and MarrowMax™ media (“MMM”).
Imaging Flow Cytometry Analysis and Machine Learning
Machine learning selected the appropriate features for the classifiers “Pan-stained” and “Edge-stained” based on the input populations- namely, for “Pan-stained”, most of the features selected to distinguish pan-staining from foci were texture-based for the channel 2 signal, while for “edge-stained”, the signal used a combination of nuclear stain distribution (channel 5) in combination with channel 2 signal localization (e.g., symmetry and lobes) with channel 2 texture-based features. The full composition of each of these features is available upon request.
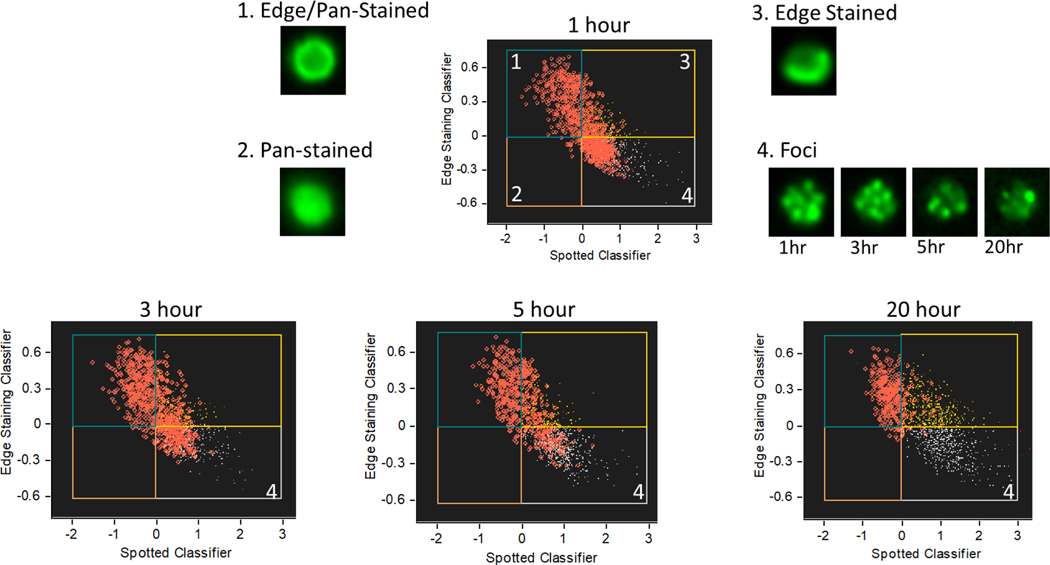
The bivariate plots of the classifiers from a representative sample at 1, 3, 5, and 20-hour time points is shown in Figure 2. The bivariate plot is divided into quadrants representing pan-nuclear and edge-stained cells (Fig. 2.1), pan-nuclear stained cells (Fig. 2.2), edge stained cells (Fig.2.3), and cells that are neither pan-stained nor edge stained (Fig. 2.4), which generally represent cells with true γ-H2AX foci or undamaged cells. Cells with a high threshold of γ-H2AX fluorescence intensity are highlighted as red circles in each time-point’s plot. As DNA damage is repaired, the fraction of high-intensity γ-H2AX foci decreases in the 4th quadrant, but remains high in other quadrants, validating that this is the population of interest in measuring DNA repair. These classifiers were also validated by visual inspection of cell images.
Figure 2.
Development of machine learning classifier. We designed these classifiers to remove false signal from pan-nuclear staining and edge staining. Cells with high γ-H2AX fluorescence intensity are highlighted as red circles. Over time, true γ-H2AX signal (lower right quadrant) goes down. Representative cell images of γ-H2AX fluorescence from each quadrant under 40x magnification are inset.
DNA Repair Capacity Model
Quantitative analysis of DRC is described by the fit parameters , and of our simple time course model (Eq. 1) for normalized median fluorescence intensity after irradiation, while quality of fit is described by RMSE in Figure 4. The average of 18 curve fits in each of the thawing conditions is given in Figure 3, which showed comparable repair behavior (data used for fitting are found in Supplementary Table 2).
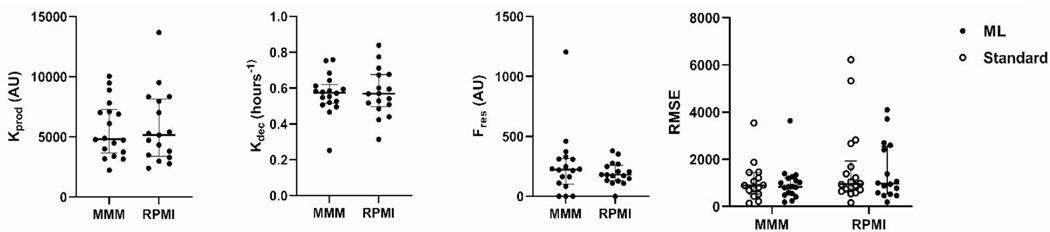
Figure 4.
Kinetic Fit Parameters of 18 archival PBMC samples thawed in either MarrowMax™ Medium (MMM) or RPMI +30%FBS (RPMI), and Root-Mean-Square-Error (RMSE) as a measure of model-comparison for machine learning analysis against the standard analysis. RMSE values were lower by ML analyses under both conditions- In the MarrowMax samples, their mean values were 985 and 1067 for machine learning and standard, respectively and in RPMI samples, their mean values were 1485 and 1683 for machine learning and standard analysis respectively.
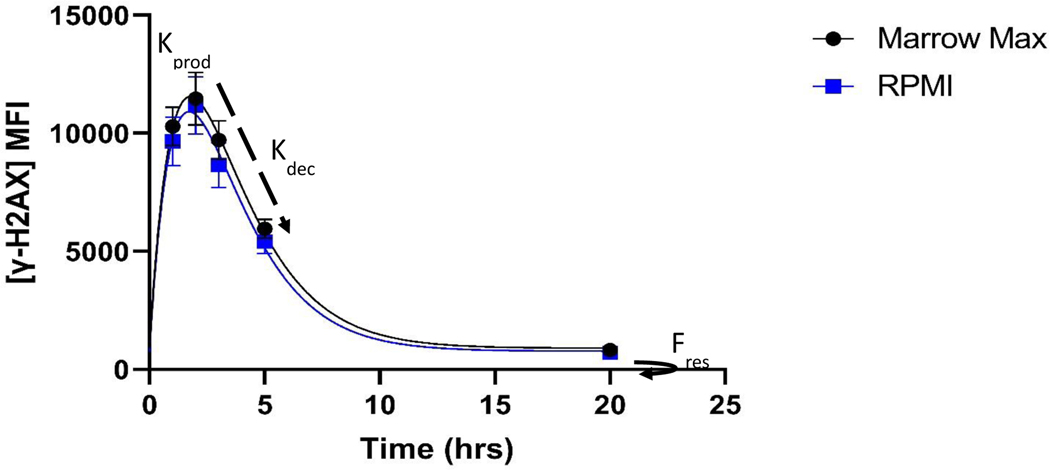
Figure 3.
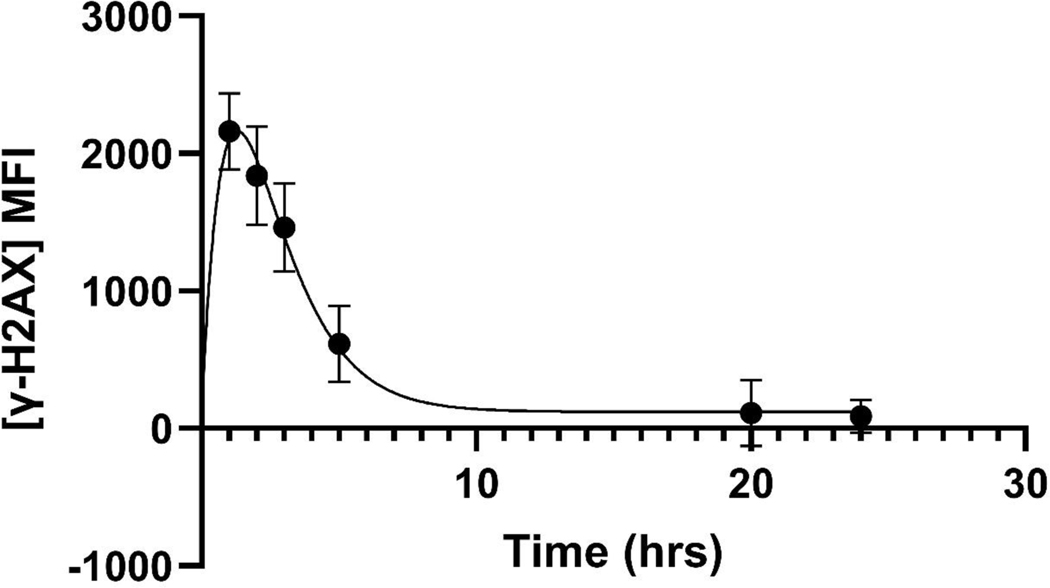
DRC Model fit for two thawing conditions of cryopreserved PBMCs. After irradiation, γ-H2AX fluorescence reaches a peak at 1–2hrs and then rapidly decays up until ~6hours, as described by the fit parameter. This is followed by a slower repair, ultimately leaving a small amount of residual DNA damage, described by the fit parameter. Error bars here are SEM of samples from 18 donors.
Results of kinetic modeling showed similar kinetic fit parameters for both thawing conditions, with median values of (h−1) of 0.57 (95% CI 0.50–0.66) and 0.54 (95% CI 0.46–0.63) and of 290 (95%CI 157–464) and 204 (95%CI 144–378) for MMM and RPMI, respectively. RMSE shows the magnitude of deviations between model predictions and observed data points.
Application of the machine learning-based gating produced a model with lower RMSE in both experimental conditions than the standard analysis model. For future experiments, we prefer MMM because the mean RMSE and deviation in RMSE are lower in that condition than in the RPMI medium. For fresh WB, kinetic modeling was done as above, except that a 0 Gy control was available for each time point. Kinetic measurements from 28 fresh WB samples showed the median value for was 0.64 (h−1) (95% CI 0.44–0.87) and was 14 (95% CI 0–84). Our group previously measured DRC values in fresh WB samples (n = 4 donors), with values for ranging from 0.750–0.892 (h−1) and values of 891–1209 [27]. The average fluorescence values and corresponding model fit for the 28 fresh blood donors is shown in Figure 5.
Figure 5.
Average DNA Repair observed in freshly collected whole blood samples (n= 28), gated to select lymphocytes and refined by machine learning image analysis, and finally fit to the exponential model of
Discussion
We present here a protocol for thawing archival PBMC samples (frozen ~20 years). Building from previously described methods [11, 12, 14, 16, 17], we optimized a cell thawing and recovery protocol and tested DNA repair under these conditions. When working with cryopreserved PBMCs, in contrast to dividing cell lines and stem cells, the experimental assay is limited by the number of terminally, fully differentiated cells stored during sample collection. To our knowledge, this is the first thawing protocol described for archival PBMCs to include a phenotypic functional assay confirming cell activity after thawing, in non-proliferating, naïve, PBMCs. Furthermore, previous studies of viability and cell function after thawing are described for cells stored between several days in the short-term and up to a year, while the cells in the BCFR cohort used in this study are all ~20 years under cryopreservation.
A robust thawing procedure is vital to ensure assay reliability and reproducibility with minimal loss in cell viability. Loss of sample during collection and sample degradation during storage and thawing can interfere with the epidemiological power of a cryopreserved cohort. To solve this problem, some groups have stimulated thawed PBMCs to proliferate using phytohemagglutinin (PHA) and assaying immune cell function using ELISPOT detection of IFNγ [13, 44–46]. By contrast, studies of naïve PBMCs are limited to thawing PBMCs and immediately measuring viability or performing immunophenotyping [12–15]. Additionally, sample quality can vary significantly based on the protocols used and timeline of the collection and freezing of the samples [13]. There are three primary assay types described which have been used to either quantify the success of the thawing process, or identify effects of the freeze-thaw process: cell viability (e.g., trypan blue dye exclusion), immunophenotyping for cell type prevalence, and measuring IFN-γ release as a measure of immune cell function after mitogen stimulation (e.g., PHA). This cohort was entirely from BCRF samples collected at the New York site, with most samples containing greater than one million cells per vial. We obtained kinetic fit parameters for 18 out of 19 of the tested samples, however one sample did not have sufficient cells for all the relevant time points. Samples that contain fewer than 1 million cells are less likely to successfully produce a full repair kinetics curve with appropriate controls, as described here, but may be useful for other types of functional phenotypic studies.
Imaging flow cytometry has a number of advantages that made it integral to developing this phenotypic assay and to validating this thaw procedure. Imaging flow cytometry provides the best of both microscopy and flow-cytometry in the amount of data extracted from each cell, while still measuring enough cells for high statistical significance. This presents a new problem however, in the quantity of data that must be analyzed for each time point, within each dosage, within each donor. Fortunately, analysis of these myriad cell images can be optimized and automated by machine learning methods. The machine learning classifiers we developed reduce fluorescence stemming from noise in the cell recovery/staining processes without decreasing true fluorescence signal. This is quantifiable both in terms of the improved fit-to-model, measured by RMSE, as well as in the observable decrease in “high γ-H2AX fluorescence” cells in some unirradiated cell samples. Another advantage of imaging flow-cytometry is that in fresh blood samples, which contain both granulocytes and lymphocytes, we can avoid the extra steps of immuno-phenotyping by gating cell-types through morphological features alone. This is important, since granulocytes, being more radio-resistant, are not suitable for this DRC assay [47].
We considered several curve-fitting models for the kinetic extrapolations of and , before we ultimately concluded with a quantile regression model, because it is less sensitive to outliers than a least-squares model, which we used in our previous work. While the values measured here from both thawed and the fresh blood samples are comparable to the previously reported fresh blood samples [27], the values are distinctly lower in these fresh blood measurements. MFI values measured for fresh blood here were comparable to those measured previously[27], while MFI values for thawed PBMCs were several fold higher. We believe that higher MFIs in the frozen samples in both irradiated and unirradiated samples were a result of different ratios of anti-H2AX (pS139) to cells/sample, as the whole blood samples have an abundance of granulocytes. In order for to compare between frozen and fresh samples, a bridging step would be necessary to harmonize the maximum fluorescence signals. The lower values observed in the current fresh blood samples compared with the previously measured values is likely a combination of the higher sample number in this set as well as differences in the curve-fitting (least-squares vs quantile regression). While the relative rates of repair are maintained (), the residual signals () after overnight culturing are more sensitive to this change. We expect that a ranking comparison of values to describe DRC from donor to donor would be maintained from one method to the next.
Edge staining and pan-nuclear staining are well-characterized fluorescent staining patterns associated with apoptosis [27, 48–50]. Edge-staining is described as an earlier, pre-apoptotic phenomenon, while pan-nuclear staining is associated with progression of apoptosis, and can actually be used as a marker of apoptosis progression. In the frozen samples, the edge staining effect was more prominent (23% of cells on average) than panstaining (avg 15%) with significant overlap in the pan/edge stained population (avg 12%). In these fresh whole blood samples, the panstaining effect was more prominent (accounting for 22% of cells on average) than the edge staining effect (avg 15%), with significant overlap in the pan/edge stained population (avg 10%). Overall, the amount of cells in these gated categories increased significantly over time, but not with increasing dose, hence the need to remove this type of noise to achieve clear repair signal. Our results suggest similar amounts of apoptosis overall between both fresh WB and cells thawed according to our method. Either way, these phenotypes can be excluded through our machine-learning image recognition classifiers.
Conclusions
We have developed two thaw methods for archival PBMCs, either using the proprietary MarrowMax™ medium or an RPMI based media with high FBS content. Statistical analysis showed that both methods were comparable, but with a possible preference for MarrowMax™. We demonstrated that these thawed cells were able to undergo DNA repair in a manner comparable to freshly drawn WB. To our knowledge, this is the first phenotypic assay of DNA repair functionality in cryopreserved PBMCs and represents a significant step forward for high impact, large-scale, demographic studies using cohorts such as the BCFR. Future work: This thawing protocol can be applied for any metabolic phenotypic assay using multi-color panels including γ-H2AX and a nuclear stain, as the image stream platform can accommodate 4–9 fluorescent channels. This assay technique and validated machine learning classifiers was developed to be scaled up in a large (>400) paired case-control cohort of breast cancer patients. Using this larger cohort, proper model testing could be done to further validate these machine learning classifiers and determine if the differences between apoptotic events (as classified by the edge staining and pan-nuclear staining) are significantly different at different time points or radiation doses. Ultimately, this cohort will be used to test for a correlation between DRC and breast cancer risk.
Supplementary Material
Acknowledgements
The authors would like to thank Ms. Melissa White for recruitment and coordinating blood draws for fresh whole blood donors. The Biomarkers Laboratory that stores BCFR samples is supported by P30ES009089 and P30CA013696. Helen Turner was supported by awards from the National Institute of Allergy and Infectious Diseases [U01 AI148309]. This work was supported by awards from the National Cancer Institute [R01 CA159868, UM1 CA164920, and P30 CA013696] and the National Institute of Environmental Health Sciences [P30 ES009089 and U01 ES029660] and the Breast Cancer Research Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflict of Interest
The authors have no conflict of interest to declare.
Data availability
The data types associated with ImageStream data acquisition are Raw image file (.rif), compensated image file (.cif), and data analysis file (.daf) and require the IDEAS software for analysis. Data from these experiments are available upon request.
References
- 1.Kennedy DO, et al. , DNA repair capacity of lymphoblastoid cell lines from sisters discordant for breast cancer. J Natl Cancer Inst, 2005. 97(2): p. 127–32. [DOI] [PubMed] [Google Scholar]
- 2.Machella N, et al. , Double-strand breaks repair in lymphoblastoid cell lines from sisters discordant for breast cancer from the New York site of the BCFR. Carcinogenesis, 2008. 29(7): p. 1367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slyskova J, et al. , DNA damage and nucleotide excision repair capacity in healthy individuals. Environ Mol Mutagen, 2011. 52(7): p. 511–7. [DOI] [PubMed] [Google Scholar]
- 4.Slyskova J, et al. , Differences in nucleotide excision repair capacity between newly diagnosed colorectal cancer patients and healthy controls. Mutagenesis, 2012. 27(2): p. 225–32. [DOI] [PubMed] [Google Scholar]
- 5.Matta J, et al. , The association of DNA Repair with breast cancer risk in women. A comparative observational study. BMC Cancer, 2012. 12: p. 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong P, et al. , Sensitivity to benzo(a)pyrene diol-epoxide associated with risk of breast cancer in young women and modulation by glutathione S-transferase polymorphisms: a case-control study. Cancer Res, 2001. 61(23): p. 8465–9. [PubMed] [Google Scholar]
- 7.Shi Q, et al. , Reduced DNA repair of benzo[a]pyrene diol epoxide-induced adducts and common XPD polymorphisms in breast cancer patients. Carcinogenesis, 2004. 25(9): p. 1695–700. [DOI] [PubMed] [Google Scholar]
- 8.Bau DT, et al. , DNA double-strand break repair capacity and risk of breast cancer. Carcinogenesis, 2007. 28(8): p. 1726–30. [DOI] [PubMed] [Google Scholar]
- 9.Smith TR, et al. , DNA damage and breast cancer risk. Carcinogenesis, 2003. 24(5): p. 883–9. [DOI] [PubMed] [Google Scholar]
- 10.Ticha O, Moos L, and Bekeredjian-Ding I, Effects of long-term cryopreservation of PBMC on recovery of B cell subpopulations. J Immunol Methods, 2021. 495: p. 113081. [DOI] [PubMed] [Google Scholar]
- 11.Mallone R, et al. , Isolation and preservation of peripheral blood mononuclear cells for analysis of islet antigen-reactive T cell responses: position statement of the T-Cell Workshop Committee of the Immunology of Diabetes Society. Clin Exp Immunol, 2011. 163(1): p. 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran H, et al. , Optimal thawing of cryopreserved peripheral blood mononuclear cells for use in high-throughput human immune monitoring studies. Cells, 2012. 1(3): p. 313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hope CM, et al. , Optimization of Blood Handling and Peripheral Blood Mononuclear Cell Cryopreservation of Low Cell Number Samples. Int J Mol Sci, 2021. 22(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honge BL, et al. , Optimizing recovery of frozen human peripheral blood mononuclear cells for flow cytometry. PLoS One, 2017. 12(11): p. e0187440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holland M, et al. , Separation, banking, and quality control of peripheral blood mononuclear cells from whole blood of melanoma patients. Cell Tissue Bank, 2018. 19(4): p. 783–790. [DOI] [PubMed] [Google Scholar]
- 16.Zhu P, et al. , Detection of tumor-associated cells in cryopreserved peripheral blood mononuclear cell samples for retrospective analysis. J Transl Med, 2016. 14(1): p. 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberg A, et al. , Optimization and limitations of use of cryopreserved peripheral blood mononuclear cells for functional and phenotypic T-cell characterization. Clin Vaccine Immunol, 2009. 16(8): p. 1176–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu HC, et al. , DNA repair phenotype and cancer risk: a systematic review and meta-analysis of 55 case-control studies. Sci Rep, 2022. 12(1): p. 3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Zein RA, et al. , Rapid method for determination of DNA repair capacity in human peripheral blood lymphocytes amongst smokers. BMC Cancer, 2010. 10: p. 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azqueta A, et al. , Comet assay to measure DNA repair: approach and applications. Front Genet, 2014. 5: p. 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins AR and Azqueta A, DNA repair as a biomarker in human biomonitoring studies; further applications of the comet assay. Mutat Res, 2012. 736(1–2): p. 122–9. [DOI] [PubMed] [Google Scholar]
- 22.Au WW, Giri AK, and Ruchirawat M, Challenge assay: A functional biomarker for exposure-induced DNA repair deficiency and for risk of cancer. Int J Hyg Environ Health, 2010. 213(1): p. 32–9. [DOI] [PubMed] [Google Scholar]
- 23.Valdiglesias V, et al. , gammaH2AX as a marker of DNA double strand breaks and genomic instability in human population studies. Mutat Res, 2013. 753(1): p. 24–40. [DOI] [PubMed] [Google Scholar]
- 24.Redon CE, et al. , gamma-H2AX as a biomarker of DNA damage induced by ionizing radiation in human peripheral blood lymphocytes and artificial skin. Adv Space Res, 2009. 43(8): p. 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilch DR, et al. , Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol, 2003. 81(3): p. 123–9. [DOI] [PubMed] [Google Scholar]
- 26.Turner HC, et al. , Adapting the gamma-H2AX assay for automated processing in human lymphocytes. 1. Technological aspects. Radiat Res, 2011. 175(3): p. 282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y, et al. , Development of a high-throughput gamma-H2AX assay based on imaging flow cytometry. Radiat Oncol, 2019. 14(1): p. 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleiner RE, et al. , Chemical proteomics reveals a gammaH2AX-53BP1 interaction in the DNA damage response. Nat Chem Biol, 2015. 11(10): p. 807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stucki M, et al. , MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell, 2005. 123(7): p. 1213–26. [DOI] [PubMed] [Google Scholar]
- 30.Sharma PM, et al. , High throughput measurement of gammaH2AX DSB repair kinetics in a healthy human population. PLoS One, 2015. 10(3): p. e0121083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeggo PA, Geuting V, and Lobrich M, The role of homologous recombination in radiation-induced double-strand break repair. Radiother Oncol, 2011. 101(1): p. 7–12. [DOI] [PubMed] [Google Scholar]
- 32.Horn S, Barnard S, and Rothkamm K, Gamma-H2AX-based dose estimation for whole and partial body radiation exposure. PLoS One, 2011. 6(9): p. e25113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vorobjev IA and Barteneva NS, Quantitative Functional Morphology by Imaging Flow Cytometry. Methods Mol Biol, 2016. 1389: p. 3–11. [DOI] [PubMed] [Google Scholar]
- 34.Vorobjev IA and Barteneva NS, Temporal Heterogeneity in Apoptosis Determined by Imaging Flow Cytometry. Methods Mol Biol, 2016. 1389: p. 221–33. [DOI] [PubMed] [Google Scholar]
- 35.Han Y, et al. , Review: imaging technologies for flow cytometry. Lab Chip, 2016. 16(24): p. 4639–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nassar M, et al. , Label-Free Identification of White Blood Cells Using Machine Learning. Cytometry A, 2019. 95(8): p. 836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg CA, et al. , Exploring dyserythropoiesis in patients with myelodysplastic syndrome by imaging flow cytometry and machine-learning assisted morphometrics. Cytometry B Clin Cytom, 2021. 100(5): p. 554–567. [DOI] [PubMed] [Google Scholar]
- 38.Konieczny M, et al. , Imaging Flow Cytometry to Study Biofilm-Associated Microbial Aggregates. Molecules, 2021. 26(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furrer R, et al. , Remodeling of metabolism and inflammation by exercise ameliorates tumor-associated anemia. Sci Adv, 2021. 7(37): p. eabi4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terry MB, et al. , Cohort Profile: The Breast Cancer Prospective Family Study Cohort (ProF-SC). Int J Epidemiol, 2016. 45(3): p. 683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuhausen SL, et al. , BRCA1 and BRCA2 mutation carriers in the Breast Cancer Family Registry: an open resource for collaborative research. Breast Cancer Res Treat, 2009. 116(2): p. 379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, et al. , Breast cancer risk prediction using a polygenic risk score in the familial setting: a prospective study from the Breast Cancer Family Registry and kConFab. Genet Med, 2017. 19(1): p. 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Development Core Team, R: A language and environment for statistical computing. 2021: Vienna, Austria. [Google Scholar]
- 44.Juhl M, et al. , Cryopreservation of peripheral blood mononuclear cells for use in proliferation assays: First step towards potency assays. J Immunol Methods, 2021. 488: p. 112897. [DOI] [PubMed] [Google Scholar]
- 45.Kreher CR, et al. , CD4+ and CD8+ cells in cryopreserved human PBMC maintain full functionality in cytokine ELISPOT assays. J Immunol Methods, 2003. 278(1–2): p. 79–93. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Mateu E, and Diaz I, Impact of Cryopreservation on Viability, Phenotype, and Functionality of Porcine PBMC. Front Immunol, 2021. 12: p. 765667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heylmann D, et al. , Comparison of DNA repair and radiosensitivity of different blood cell populations. Sci Rep, 2021. 11(1): p. 2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solier S and Pommier Y, The apoptotic ring: a novel entity with phosphorylated histones H2AX and H2B and activated DNA damage response kinases. Cell Cycle, 2009. 8(12): p. 1853–9. [DOI] [PubMed] [Google Scholar]
- 49.Solier S and Pommier Y, The nuclear gamma-H2AX apoptotic ring: implications for cancers and autoimmune diseases. Cell Mol Life Sci, 2014. 71(12): p. 2289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding D, et al. , Induction and inhibition of the pan-nuclear gamma-H2AX response in resting human peripheral blood lymphocytes after X-ray irradiation. Cell Death Discov, 2016. 2: p. 16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data types associated with ImageStream data acquisition are Raw image file (.rif), compensated image file (.cif), and data analysis file (.daf) and require the IDEAS software for analysis. Data from these experiments are available upon request.