Abstract
Brain development requires appropriate regulation of serotonin (5-HT) signaling from distinct tissue sources across embryogenesis. At the maternal-fetal interface, the placenta is thought to be an important contributor of offspring brain 5-HT and is critical to overall fetal health. Yet, how placental 5-HT is acquired, and the mechanisms through which 5-HT influences placental functions, are not well understood. Recently, our group identified a novel epigenetic role for 5-HT, in which 5-HT can be added to histone proteins to regulate transcription, a process called H3 serotonylation. Here, we show that H3 serotonylation undergoes dynamic regulation during placental development, corresponding to gene expression changes that are known to influence key metabolic processes. Using transgenic mice, we demonstrate that placental H3 serotonylation largely depends on 5-HT uptake by the serotonin transporter (SERT/SLC6A4). SERT deletion robustly reduces enrichment of H3 serotonylation across the placental genome, and disrupts neurodevelopmental gene networks in early embryonic brain tissues. Thus, these findings suggest a novel role for H3 serotonylation in coordinating placental transcription at the intersection of maternal physiology and offspring brain development.
Keywords: epigenetics, development, serotonin transporter, H3 serotonylation, placenta
INTRODUCTION
Serotonin (5-hydroxytryptamine, 5-HT) is an essential biogenic monoamine with multipurpose functions, including regulation of fetal brain circuitry that, if disrupted, provides the foundation for behavioral dysfunction later in life1,2. The developing brain requires 5-HT from early embryonic stages, yet an endogenous brain-wide 5-HT source does not emerge until late in gestation3,4, indicating that transport of extraembryonic 5-HT to the conceptus is central to this process. Indeed, previous studies have demonstrated that the placenta, a transient endocrine and metabolic tissue at the maternal-fetal interface, delivers the majority of 5-HT into fetal circulation prior to formation of dorsal raphe nucleus projections throughout the brain5. Placental 5-HT may arise from different pathways, with studies describing conversion from the precursor L-tryptophan via trophoblast expression of the enzyme tryptophan hydroxylase 1 (TPH1)6, transporter-mediated uptake from maternal circulation via the serotonin transporter (SERT/ SLC6A4) on the placental apical membrane7,8, and/or regulation by the organic cation transporter 3 (OCT3/SLC22A3) at the fetoplacental endothelium9–11. Importantly, placental health is critical for fetal health, as indicated by numerous studies showing negative consequences on the fetal brain following placental responses to prenatal/preconception stress, inflammation, and immune activation12–20. Accordingly, 5-HT dysregulation also impacts vasoconstrictive properties of placental blood vessels21,22, as well as proliferation and viability of trophoblast cells23. Thus, neurodevelopment can be influenced by dysregulation of multiple 5-HT-dependent processes in placental tissues, including – but not limited to – monoamine transport. However, the mechanisms through which these 5-HT-dependent functions are regulated, as well as the modes by which placental 5-HT is acquired, are still not well understood.
Recently, a receptor-independent role for select monoamines, including 5-HT and dopamine, termed “monoaminylation,” has been described24–27. Monoaminylation involves the covalent attachment of free monoamine donors to glutamine-containing protein substrates by the enzyme tissue transglutaminase 2 (TGM2)28,29. In particular, monoaminylation using 5-HT as a donor (“serotonylation”) has been demonstrated for proteins in diverse cell types, whereby this serotonyl post-translational modification (PTM) can alter the signaling properties of bound cytosolic substrates30–32. In the nucleus, our group has recently demonstrated that serotonylation occurs on glutamine 5 of histone H3 (H3Q5ser)24. At this site, H3 serotonylation epigenetically regulates transcription either alone or in combination with the neighboring lysine 4 tri-methylation (K4me3) PTM to enhance permissive gene expression through interactions with reader proteins33. The combinatorial H3K4me3Q5ser PTM has been detected in regions throughout the adult brain, where it coordinates relevant gene expression programs upstream of neural differentiation and contributes to sensory processing and stress-induced behavioral plasticity in adult brain, demonstrating diverse roles for this PTM across various functional domains34,35. Moreover, the presence of histone serotonylation in heart, testes and other mouse organs suggest additional actions in peripheral tissues24. In a recent study examining human placental explants, nuclear 5-HT detected in both syncytiotrophoblasts and cytotrophoblast cells was found to be altered by inhibition of both SERT and monoamine oxidase11, suggesting that histone serotonylation may also be dynamically regulated in placental tissues to affect downstream processes, although follow-up studies providing evidence for this phenomenon have not yet been conducted.
Here, we investigated whether histone serotonylation may serve as an epigenetic mechanism for regulating placental gene expression programs capable of ultimately influencing offspring neurodevelopment. We found that expression of H3 serotonylation across both male and female placental development was bidirectionally regulated, with increased PTM enrichment at genomic loci related to important metabolic pathways and decreased patterns reflecting attenuation of cellular proliferation and tissue organization over development. Moreover, we demonstrate that placental 5-HT and H3 serotonylation are reliant on intact 5-HT machinery, where levels of both are reduced in tissues in which the transporters SERT, OCT3, or the enzyme TPH1 were deleted. In these tissues, we further found that SERT deletion most robustly disrupts normal H3 serotonylation patterning across the genome, with decreased enrichment at numerous loci relevant to essential placental processes. Lastly, we observed significant transcriptional abnormalities in neurodevelopmental gene networks downstream of placental changes, which appeared independent of overall 5-HT levels in brain. These findings thus establish histone serotonylation as a previously undescribed epigenetic mechanism that contributes importantly to developmental gene expression programs in placenta; phenomena that, in turn, impact key neurodevelopmental transcriptional networks in the offspring brain.
RESULTS
Roles for histone serotonylation in regulating gene expression programs associated with key placental functions
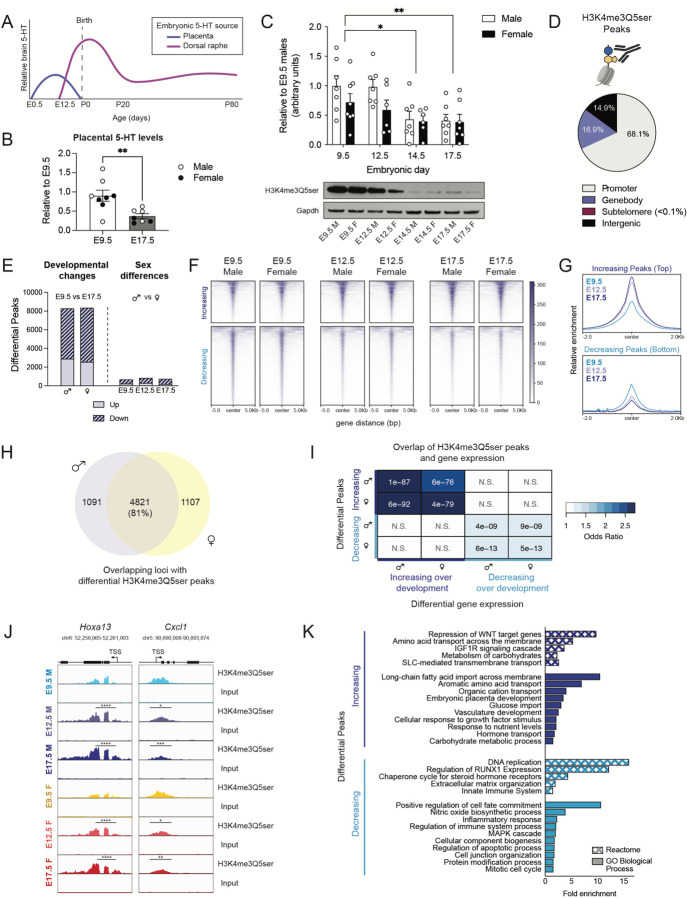
To begin investigating potential roles for 5-HT in placenta that could ultimately impact offspring brain development, we examined developmental 5-HT patterns occurring at E9.5 and E17.5, time points in which brain 5-HT predominantly originates from the placenta vs. dorsal raphe nucleus (DRN, the primary hub of 5-HTergic projection neurons in brain), respectively (Fig. 1A, adapted from Suri et al.36). We found that 5-HT levels in placenta decreased from E9.5 to E17.5 (Fig. 1B), consistent with expected 5-HT contributions from the placenta. Given our recent studies demonstrating covalent binding of 5-HT to nuclear histone proteins, we next used western blotting to assess global levels of the combinatorial serotonyl-PTM in male and female tissues at the same gestational time points. To more precisely detect fluctuations in placental 5-HT-related processes, we examined two additional time points (E12.5 and E14.5) that precede the complete formation of DRN projections throughout the embryonic brain3,4. We found that H3K4me3Q5ser levels decrease in placenta across gestation, with E12.5 appearing to signify the transition point after which time reductions in the mark begin to occur, with no significant effects of sex observed (Fig. 1C, Supplementary Fig. 1). Interestingly, the observed dynamics of histone serotonylation were also found to correspond to the extent of 5-HT supply from placenta to brain (Fig. 1A), suggesting that higher levels of histone serotonylation may regulate crucial placental biology at this mid-gestational window.
Figure 1. H3 serotonylation is associated with developmental gene networks in male and female placenta.
(A) Schematic depicting brain 5-HT levels and tissue of origin, adapted from Suri et al.36 (B) Placental 5-HT levels decrease from E9.5 to E17.5 (unpaired Student’s t-test, t(13) = 3.209, **p = 0.0068), with male and female placental samples clustering together, as noted by circle colors (N=7–8 samples/age). (C) Western blot analysis of H3K4me3Q5ser in male and female placenta tissues at E9.5, E12.5, E14.5 and E17.5 showed a main effect of embryonic age (two-way ANOVA, age F(3,47) = 6.622, p = 0.0008) with no significant effect of sex (F(1,47) = 3.586, p = 0.0644), where histone serotonylation decreased over development (Sidak’s post-hoc test, E9.5 vs E14.5 (*adjusted p = 0.0102); E9.5 vs E17.5 (**adjusted p = 0.0056); E12.5 vs E14.5 (adjusted p = 0.057), E12.5 vs E17.5 (adjusted p = 0.0356), N = 6–8/group). (B, C): Data are normalized to the male E9.5 values and shown as mean ± SEM. (D) Averaged proportion of peaks using annotations from all developmental male and female placentas showed about 68.1% of sites found following H3K4me3Q5ser ChIP-sequencing were located in promoter regions (N = 4 samples/age/sex). (E) There was a ~tenfold greater number of significantly differential peaks comparing E9.5 vs E17.5 in both males and females, compared to sex difference contrasts within embryonic age (p < 0.05, log2(fold change) > 0.1). (F, G) Heatmaps (F) and profiles (G) of differential peaks from E9.5 vs E17.5 comparisons, separated by directionality and centered on genomic regions to show the majority of altered peaks decrease across placental development. (H) Venn diagram depicting the degree of overlap between male and female E9.5 vs E17.5 comparisons using uniquely annotated peaks, indicating developmental changes are largely conserved between sex. (I) Odds ratio analysis of differential H3K4me3Q5ser peaks (from 1E above) and differentially expressed genes (adjusted p < 0.05; N = 4 samples/age/sex) from E9.5 vs E17.5 comparisons show significant association between altered histone serotonylation regulation and gene expression changes. Insert numbers indicate respective p values for each association (N.S., p > 0.05). (J) Representative genome browser tracks of Hoxa13 and Cxcl1 loci for H3K4me3Q5ser (vs respective DNA input) in E9.5, E12.5 and E17.5 male and female placentas (Hoxa13: **** p < 0.0001 relative to E9.5 within sex; Cxcl1: ***p < 0.001, **p < 0.01 relative to E9.5 within sex; *p < 0.05 denotes significant changes in E12.5 vs E17.5 males and E9.5 vs E12.5 females) Each track represents merged signal for 4 samples. (K) Selected Reactome and GO Biological Process pathways for differential peaks displaying significant associations with gene expression between E9.5 vs E17.5 (from 1I above) for male placenta tissues (FDR < 0.05).
As such, we next examined whether H3K4me3Q5ser is enriched at genomic loci relevant to placental functions across development. We performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) in male and female placental tissues at E9.5, E12.5, and E17.5. Following peak calling in all groups, we found that the majority (~68.1%) of H3K4me3Q5ser peaks were annotated to promoter regions, with less than a fifth of peaks each also detected in genebody and distal intergenic regions (~16.9% and ~14.9%, respectively; Fig. 1D), which is consistent with our previous findings in human neurons and rodent brain24,35. To identify differential enrichment sites that may regulate developmental processes, we used Diffbind to compare the earliest and latest gestational time points in our dataset37. In both male and female placental tissues, we identified ~8,000 differentially enriched peaks, with the majority of these peaks for both sexes displaying significantly decreased enrichment from E9.5 to E17.5, corresponding to global western blotting patterns for the mark (Fig. 1E, Supplementary Tables 1–2). As the placenta is largely comprised of cells from the trophoblast lineage, which reflect fetal chromosomal sex38, we also examined potential sex differences in histone serotonylation. Within each developmental stage, we identified several hundred peaks altered between sexes, with E9.5 having the least (Fig. 1E, Supplementary Tables 3–5). Notably, at E12.5 and E17.5, the top 500 peaks showed similar sex differential patterns at the two later gestational ages, but not at E9.5, suggesting that placental sex differences in H3K4me3Q5ser enrichment are established by E12.5 and likely persist until parturition (Supplementary Fig. 2A–C). Annotation of these altered peaks identified sex differential sites throughout the chromosomal complement, with ~5% located on the X and Y chromosomes (Supplementary Fig. 2D–E).
Given the aforementioned patterns, we next evaluated whether developmental changes in placental histone serotonylation were also impacted by sex. Hierarchical clustering of the top 1,000 peaks found to be altered between E9.5 and E17.5 revealed two sets of histone serotonylation changes (up vs. down), with both developmental increases and reductions from E9.5 to E17.5 displaying intermediate enrichment at E12.5, that were similarly expressed in males and females within each time point (Supplementary Fig. 3A–B). Visualization of all 8,274 differential histone serotonylation peaks between E9.5 vs. E17.5 males showed similar enrichment patterns in female placental tissues (Fig. 1F–G, Supplementary Fig. 3C). Comparing the degree of overlap between differential developmental sites following peak annotation, we observed an ~81% overlap of enriched loci between males and females, altogether suggesting that these developmental changes are largely conserved between sexes in placenta (Fig. 1H). We next performed bulk RNA-sequencing to explore the relationship between histone serotonylation changes and gene expression in placenta. In doing so, we identified positive and significant correlations between differential gene expression and changes in serotonylation enrichment across development (Fig. 1I, Supplementary Tables 6–8). We observed greater transcription of gene loci with increasing H3K4me3Q5ser enrichment, as exemplified by the Hoxa13 locus, a transcription factor critical for labyrinth vessel formation crucial for gas and nutrient exchange at the maternal-fetal interface39 (Fig. 1J, Supplementary Fig. 3D). Similarly, decreasing H3K4me3Q5ser enrichment was found to correspond to reduced gene expression, as exemplified by the Cxcl1 locus, a chemokine ligand participant in the unique immune milieu surrounding the allogenic fetal microenvironment40,41 (Fig. 1J, Supplementary Fig. 3E). Altogether, these data indicate that H3K4me3Q5ser likely facilitates permissive transcription in placenta, similar to that of our previous findings in neural cells24. Functional annotation analyses (Reactome, GO Biological Process) of those loci overlapping at sites of H3K4me3Q5ser enrichment and gene expression changes (i.e., from Fig. 1I) further uncovered relevant gene sets to placental biology, including upregulation of vasculature development, nutrient and hormone transport processes over developmental age, and reductions in proliferative, differentiation, and immune processes near gestational term (Fig. 1J, Supplementary Tables 9–10)42.
Placental serotonin levels are mediated by transporter-dependent pathways
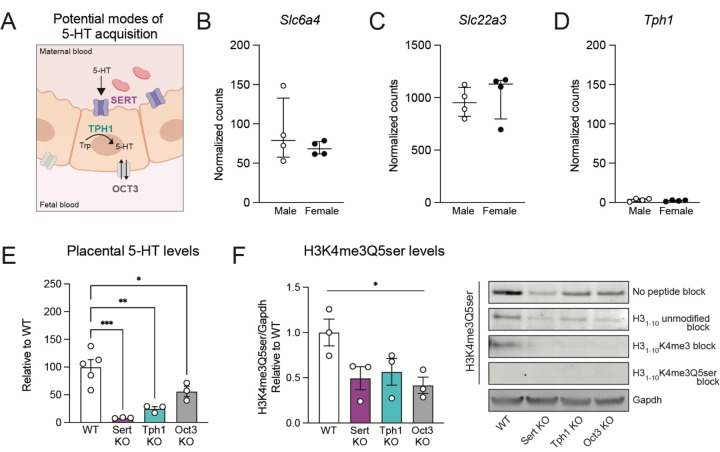
Given suggestive roles of histone serotonylation in regulating the placental transcriptome, we next aimed to understand the source of its intracellular 5-HT donor pool. Prior studies have suggested several potential modes: 1) transporter-dependent mechanisms, via the high-affinity, low-capacity 5-HT uptake transporter encoded by the Slc6a4 gene, SERT and/or the extra-neuronal organic cation transporter OCT3 (encoded by the Slc22a3 gene), which is capable of bidirectional facultative monoamine diffusion43,44; or 2) intrinsic synthesis from tryptophan via trophoblast expression of TPH16 (Fig. 2A). To assess the possibility of active 5-HT acquisition, which may serve as the donor source for the serotonyl-PTM, we chose to evaluate placental tissues at E12.5 given that H3K4me3Q5ser levels are dynamically changing between E9.5 and E17.5 to regulate placental transcriptional processes, and given the formation of a fully differentiated placenta at this stage45. First, to test whether placental 5-HT is transporter-mediated, we took a bioorthogonal metabolic-labelling approach, using propargylated (i.e., alkynylated) serotonin (5-PT) that allows for the immunoprecipitation of 5-PT labelled protein substrates following tissue delivery. Given prior work demonstrating that placental 5-HT depends on SERT function7,11, we hypothesized that 5-PT would similarly be taken up from maternal circulation via SERT. Thus, pregnant mice were injected with 100 nM or 1 μM 5-PT, based upon a reported range of 5-HT levels between basal levels vs. those at sites of thrombosis46, and conceptuses were removed 1 hour post-injection for assessments of 5-PT uptake (Supplementary Fig. 4A). We observed dose-dependent signals of 5-PT-labelled H3 protein in placental extracts (Supplementary Fig. 4B), supporting the hypothesis that histone serotonylation depends on transporter-mediated uptake of 5-HT. Subsequently, we verified placental gene expression of Slc6a4 at E12.5 (Fig. 2B), also observing expression of Slc22a3 (Fig. 2C), but not Tph1 (Fig. 2D), further substantiating our prediction that placental 5-HT is obtained via transporters and is not endogenously synthesized. Notably, while TPH1 is not involved in placental 5-HT generation, global TPH1 knockout (KO) results in an ~80% reduction in circulating 5-HT, which might therefore reduce the availability of 5-HT that could be taken up from circulation47,48.
Figure 2. Placental 5-HT is dependent on SERT-mediated uptake.
(A) Schematic depicting potential modes of placental 5-HT acquisition examined in this study. (B, C) Normalized counts indicating Sert (Slc6a4) and Oct3 (Slc22a3) are expressed in both male and female placental tissues at E12.5, with no differences by sex (unpaired Student’s t-test; Slc6a4: p = 0.3677; Slc22a3: p = 0.5973). (D) The Tph1 gene is not expressed in E12.5 placental tissues. N=4 samples/sex. Data are median ± interquartile range. (E) Assessment of 5-HT levels in E12.5 placental tissues shows significant reductions (one-way ANOVA, F(3,8) = 4.001, p = 0.0004) in Sert KO (Dunnett’s multiple comparisons test; ***adjusted p = 0.0003), Tph1 KO (**adjusted p = 0.0015), and Oct3 KO (*adjusted p = 0.04) tissues. N=3–5/group. (F) Western blot analysis of placental tissues at E12.5, showing reduced H3K4me3Q5ser in Sert KO, Tph1 KO and Oct3 KO tissues (one-way ANOVA, F(3,10) = 15.37, *p = 0.05). Peptide competition assays using H31–10 peptides show selective signal of the serotonyl-PTM epitope is predominantly observed in WT placenta. N = 3/group. Data are mean ± SEM.
To next establish the necessity of transporters for placental 5-HT uptake and histone serotonylation deposition, we utilized transgenic mouse lines with targeted genetic deletions of Slc6a4, Slc22a3 or Tph1. We identified robust 5-HT reductions in placental tissues from all transgenic lines examined, with the greatest loss in 5-HT signal observed in Sert KO tissues (~90%), followed by around 70% reduction of placental 5-HT levels in Tph1 KO, and around 50% reduction in Oct3 KO (Fig. 2E). Thus, we next tested for corresponding reductions in global histone serotonylation levels. Indeed, western blotting revealed overall decreases in H3K4me3Q5ser signal in all three KO lines, which was further confirmed following competition assays with an H31–10 peptide containing the K4me3 PTM (Fig. 2F, Supplementary Fig. 5). In sum, these data demonstrate placental H3 serotonylation’s reliance on 5-HT levels and the integrity of pathways regulating 5-HT entry into this tissue.
SERT deletion downregulates histone serotonylation and disrupts developmental processes in placenta
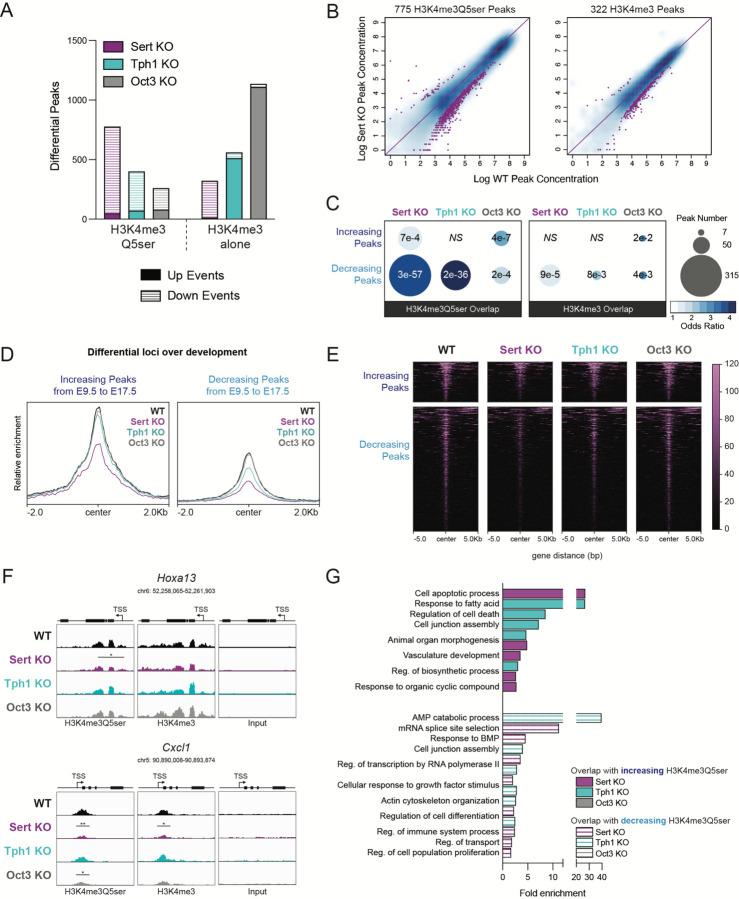
Given histone serotonylation’s dependency on 5-HT transporter function, we next investigated whether knockout of these proteins might alter H3K4me3Q5ser enrichment at key genomic loci known to regulate placental development. Differential peak analysis following ChIP-seq demonstrated that the majority of H3K4me3Q5ser enrichment alterations observed in Sert KO, Tph1 KO, and Oct3 KO placental tissues were decreased compared to age-matched WT controls (Fig. 3A, Supplementary Tables 11–13). To ensure the specificity of H3K4me3Q5ser changes, we additionally performed ChIP-seq for the H3K4me3 mark alone (note that the antibody for H3K4me3 may recognize H3K4me3 both in the presence or absence of H3Q5ser24), which produced a distinct pattern of peak enrichment changes (Fig. 3A, Supplementary Tables 14–16), supporting the notion that histone serotonylation is dependent on tissue 5-HT changes rather than changes in H3K4me3 itself. Consistent with its robust 5-HT reductions, Sert KO similarly had the greatest impact on histone serotonylation peak reductions compared to deletion of OCT3 or TPH1 (Fig. 3B, Supplementary Fig. 6). We next evaluated the extent of overlap between developmentally relevant H3K4me3Q5ser loci that exhibit increased or decreased enrichment over embryonic age (from Fig. 1) with transgenic-mediated reductions in H3K4me3Q5ser or H3K4me3 enrichment. In all KO tissues, H3K4me3Q5ser-enriched loci had significantly greater overlap compared to H3K4me3 alone, with the highest degree of overlap observed for peaks altered by Sert KO (Fig. 3C). Therefore, we next examined those histone serotonylation peaks enriched at genomic loci at the intersection of Sert KO reductions and developmental changes occuring from E9.5 to E17.5. As expected, Sert KO downregulated H3K4me3Q5ser enrichment at these developmentally relevant loci compared to WT, Tph1 KO, and Oct3 KO placental tissues (Fig. 3D–E), as exemplified by the Hoxa13 and Cxcl1 loci (Fig. 3F). Functional annotation analyses of overlapping H3K4me3Q5ser-enriched loci between these multiple datasets demonstrated that SERT and TPH1 (but not OCT3) deletion disrupted important pathways for placental development, including changes in vasculature development, apoptosis, cell differentiation, and immune system processes (Fig. 3G, Supplementary Tables 17–19). In sum, our genomic data indicate that key moderators of the placental 5-HT donor pool lie upstream of histone serotonylation regulation. In particular, we provide evidence that SERT deletion disrupts H3K4me3Q5ser regulation of placental biology that might subsequently impact offspring brain development.
Figure 3. SERT deletion alters placental H3 serotonylation patterning.
(A) Relative to WT, the greatest number of significantly decreased H3K4me3Q5ser peaks was observed in Sert KO placentas, followed by Tph1 KO and Oct3 KO (left; p < 0.05, log2(fold change) > 0.1), where the overall pattern of differential sites diverged from those of H3K4me3 alone (right; N = 3 samples/group). (B) Scatter plots of differential H3K4me3Q5ser (left) and H3K4me3 (right) peaks in Sert KO placentas relative to WT, showing the majority of affected sites are downregulated. (C) Odds ratio analysis examining overlap of significantly reduced H3K4me3Q5ser and H3K4me3 peaks (relative to WT, from 3A) with differential H3K4me3Q5ser sites between E9.5 and E17.5 (from 1E), with bubble size representing number of overlapping loci, indicating SERT deletion has greatest impact on developmentally-regulated sites. Insert numbers denote respective p values for each association (NS, p > 0.05), (D, E) Heatmaps (D) and profiles (E) of differential H3K4me3Q5ser loci between E9.5 and E17.5 that are significantly downregulated in Sert KO placentas, separated by directional changes across development and centered on genomic features. (F) Representative genome browser tracks of Hoxa13 and Cxcl1 loci for H3K4me3Q5ser and H3K4me3 (vs respective DNA input) in WT, Sert KO, Tph1 KO and Oct3 KO placentas (Hoxa13: *p < 0.05 relative to WT; Cxcl1: **p < 0.01, *p < 0.05 relative to WT for each histone modification). Each track represents merged signal for 3 samples. (G) Selected Reactome and GO Biological Process pathways for differential loci (vs WT) overlapping with developmentally regulated H3K4me3Q5ser sites (from 1H). Note: there were no significant pathways enriched for overlapping differential peaks from WT vs Oct3 KO comparisons (FDR < 0.05).
Placental 5-HT and histone serotonylation reductions are associated with changes in neurodevelopmental gene expression programs
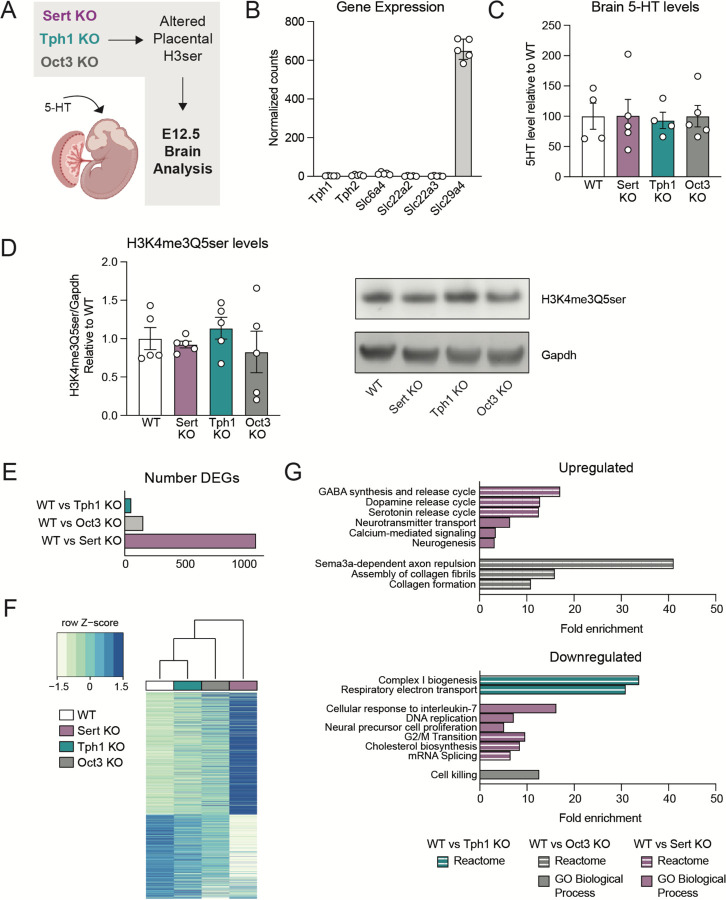
Given that the placenta is the major 5-HT source from early-to-mid gestation, we next sought to understand how brain 5-HT levels might be impacted by these placental changes (Fig. 4A). Importantly, the tissues used were obtained from conventional KO mice; thus we first interrogated whether transgenic-mediated changes alone might impact brain 5-HT. Transcriptomic analysis of embryonic brain tissues showed low levels of Slc6a4, Slc22a3 and Tph1 at E12.5 in WT mice (Fig. 4B), suggesting that SERT and OCT3 are not the major modes of 5-HT entry into the embryonic brain. We also examined gene expression for the neuronal isoform of tryptophan hydroxylase, TPH2 (Tph2), organic cation transporter OCT2 (Slc22a2), and the plasma membrane monoamine transporter PMAT (Slc29a4) to uncover other potential routes through which 5-HT in brain may be incorporated (Fig. 4B). Our data suggest that the E12.5 brain does not express machinery for 5-HT synthesis at this time, indicating that brain 5-HT is likely extrinsically regulated and its uptake may be mediated by the transporter PMAT, as suggested by its high levels of expression. Given that we did not observe significant expression of brain Slc6a4, Slc22a3, or Tph1, which might confound our assessments of placental 5-HT and histone serotonylation effects, we next examined how these placental disruptions might influence brain 5-HT levels. Remarkably, we observed no differences in 5-HT in any KO brain tissues compared to WT (Fig. 4C), similar to other studies6,49. We further examined whether there may be downstream differences in brain H3K4me3Q5ser abundance, but we observed no differences in any KO comparisons vs. WT (Fig. 4D, Supplementary Figure 7). These findings suggest that placental disruptions in 5-HT uptake do not exert direct programming effects in offspring via reductions in 5-HT delivery to the developing brain.
Figure 4. Offspring neurodevelopmental gene expression changes are associated with placental disruptions.
(A) Schematics of study design for investigating E12.5 offspring brain changes. (B) Normalized counts showing gene expression for Tph1, Slc6a4, and Slc22a3 are low compared to that for the transporter PMAT (Slc29a4) in embryonic brain. (C) There is no change in 5-HT levels in E12.5 brains when comparing WT vs KO tissues (one-way ANOVA, F(3,14) = 0.027, p = 0.9938). N=4–5 samples/group. (D) There also are no differences in H3K4me3Q5ser in brain tissues (one-way ANOVA, F(3,16) = 0.5861, p = 0.6328). N=5 samples/group. Data are mean ± SEM. (E) Number of differentially expressed genes from bulk RNA-sequencing comparing WT vs. Sert KO, WT vs. Tph1 KO, WT vs. Oct3 KO brain tissues at E12.5 (adjusted p < 0.05). (F) Hierarchical clustering of all differentially expressed genes relative to WT (adjusted p < 0.05). Expression values are averaged within genotype (N=5–6 samples/group). (G) Selected Reactome and GO Biological Process pathways enriched from differentially expressed genes comparing WT vs KO brain tissues at E12.5 (FDR < 0.05).
Given that SERT and TPH1 deletion both resulted in reduced placental H3K4me3Q5ser enrichment at loci involved in biosynthesis, transport, and vasculature development, we speculated that histone serotonylation might alter other placental functions that could influence the embryonic brain in a 5-HT-independent manner. Thus, to determine the overall impact of such changes on neurodevelopment, we examined embryonic brain tissues using bulk RNA-sequencing at E12.5, a time point that we already established is largely unaffected by transgenic manipulations within the brain itself. We found that the brain transcriptome was robustly altered in Sert KO tissues, with Oct3 KO and Tph1 KO brains also displaying significant regulation (all relative to WT), though to a lesser extent (Fig. 4E–F, Supplementary Tables 20–23). To understand what processes may be impacted in the developing brain, we performed functional annotation analyses (using GO Biological Process and Reactome databases) on differentially expressed genes from all WT vs. KO comparisons. Examining all significantly enriched pathways, we used Revigo to summarize redundant GO terms50, revealing numerous gene sets related to synaptic signaling, monoamine and neurotransmitter regulation, and neuronal proliferation altered in Sert KO brains (Fig. 4G, Supplementary Tables 24–25). There were also significant changes to pathways observed related to collagen formation and apoptosis in Oct3 KO brains, and downregulation of cellular respiration in Tph1 KO brains, which may be indicative of insufficient ‘fuel’ being transported from placenta to the conceptus (Fig. 4G, Supplementary Tables 26–28). In total, these data indicate that even moderate changes to placental 5-HT and histone serotonylation levels appear sufficient to affect important neurodevelopmental processes in the developing fetus.
DISCUSSION
Here, we demonstrated that histone serotonylation likely influences embryonic brain development via epigenetic regulation of the extra-embryonic placental transcriptome. We showed that H3 serotonylation is bidirectionally regulated across embryogenesis, corresponding with gene expression changes and coordination of known placental pathways that are crucial to fetal growth. We further established that SERT is the major mode of 5-HT transport from maternal peripheral circulation to placenta, a process that when disrupted also perturbs normal developmental serotonyl-PTM patterning. Moreover, we found that such disruptions in placental histone serotonylation may have important downstream effects on the embryonic brain transcriptome, supporting placental epigenetics as an exciting mechanism of neurodevelopmental programming that may affect behavioral outcomes and/or disease risk later in life. While the current study illustrates an exciting framework by which the placental 5-HT machinery intersects with chromatin mechanisms to influence offspring outcomes, there are several limitations to the current study that deserve attention. Most notably, given our use of tissues from conventional transgenic KO mice, there may be other tissue contributions involved; however, as maternal stimuli are communicated to the fetus via placental signaling, we propose that the offspring brain outcomes are directly affected by the placental changes observed in this study. Indeed, prior work suggests that increased necrosis in Sert KO and Tph1 KO placentas occurs via 5-HT receptor signaling, which is normally terminated by SERT-mediated uptake23. In the current study, both SERT and TPH1 deletion were found to disrupt H3K4me3Q5ser enrichment at loci involved in cell apoptotic processes, and thus may additionally regulate this phenotype via epigenetic changes. Therefore, further studies selectively targeting histone serotonylation within the placenta will be needed to fully resolve whether such 5-HT-dependent chromatin mechanisms causally contribute to placental dysregulation and/or act in parallel with disrupted receptor signaling.
Furthermore, given the essential role of developmental 5-HT on neuronal patterning, many studies have focused on identifying the mechanism through which placental 5-HT is acquired and transferred to the offspring brain. Debates regarding this source posit that placental 5-HT may derive from a maternal origin via uptake from blood, or endogenous synthesis via metabolism of the precursor L-tryptophan6,51,52. Using genetic targeting of these potential 5-HT sources, our findings support maternal serotonin supply as the major determinant of 5-HT and H3 serotonylation levels in placenta. Indeed, we demonstrated that Tph1 expression is absent in placenta, similar to other studies examining human and rodent tissues11,53,54. For this reason, reductions in placental H3K4me3Q5ser in Tph1 KO tissues may be explained by lowered 5-HT blood levels, due to disrupted 5-HT synthesis in enterochromaffin cells48,55. Therefore, overlapping H3K4me3Q5ser enrichment reductions in Sert KO vs. Tph1 KO tissues likely occur due to a convergence of pathways dependent on 5-HT in maternal blood. In addition to reduced placental uptake via SERT deletion, Sert KO animals have low peripheral 5-HT (due to a deficiency of platelets in taking up 5-HT56) as observed in Tph1 KO, which result in decreased uptake into trophoblast cells, altogether indicating that placental 5-HT is of maternal origin and is not endogenously synthesized within the placenta. Indeed, genetic deletion of SERT eliminates the majority of placental 5-HT at mid-gestation. Residual H3K4me3Q5ser signal in Sert KO tissues, then, likely result from patterning at earlier time points when other modes of 5-HT acquisition may be present (e.g., other transporters and/or transient embryonic synthesis43,44,48,57), or technical artifacts owing to the process of polyclonal antibody generation using H3K4me3Q5ser immunogens. To control for this technical limitation, we additionally performed H3K4me3 ChIP-sequencing and observed that while there were indeed differential sites of overlap between H3K4me3 and H3K4me3Q5ser, differential histone serotonylation could not be accounted for by changes in H3K4me3 alone. Instead, we observed that reduced H3K4me3Q5ser patterns in KO placentas closely corresponded with the extent of 5-HT decreases, suggesting that this PTM depends on donor availability (consistent with our previous biochemical analyses58). It is also worth noting that the overlapping reductions in signal observed between H3K4me3Q5ser and H3K4me3 alone may occur due to previous observations that H3Q5ser inhibits H3K4 demethylase activity, and thus loss of the serotonyl-PTM may additionally destabilize the presence of H3K4me3 at certain loci59.
The developing brain is highly sensitive to placental insults resulting from environmental perturbations and imbalances of specific nutrients, hormones, and other chemical signals38. Using transgenic KO mice, we identified a specific time point in which there was minimal expression of key 5-HT machinery within the brain, allowing us to examine non-cell autonomous effects originating from deletion of SERT, TPH1 or OCT3 in the placenta and/or maternal tissues. Indeed, we detected robust differential gene expression in the E12.5 Sert KO brain, supporting functional responsivity to placental effects. As previously mentioned, we must cautiously interpret these findings given the use of whole-body KO animals. Beginning at E10.5, SERT is detected in embryonic cardiac and liver tissues60, and it is possible that disruptions to these systems may result in excess 5-HT in fetal circulation that also contribute to brain changes. In this way, the effects observed in Tph1 KO brains, though more subtle, provide clearer proof-of-concept evidence that placental 5-HT and histone serotonylation directly impact brain programming, due to restricted non-neuronal Tph1 expression that is not detected until E14.555.
With respect to how precisely placental histone serotonylation changes may mediate brain reprogramming, we did not expect that 5-HT levels would be unaffected in the corresponding KO brains given the robust 5-HT reductions observed in Sert KO and Tph1 KO placentas, though it is notable that other studies have made similar observations6,49. There are several potential explanations: it is possible that the placenta buffers against 5-HT deficiencies, such that the embryo nonetheless attains the necessary amount, or there may be alternate 5-HT sources that compensate for placental insufficiency48. The answer to this question is beyond the scope of the current study, but will be crucial to understanding the complex role of placental 5-HT signaling in developmental brain programming. While we do not detect global histone serotonylation changes within the brain itself, this is likely due to the specific time point examined. For example, SERT expression increases across gestation and is transiently upregulated in the thalamus and hippocampus during early postnatal development, where it is critically necessary for neuronal projection patterning61,62. Moreover, SERT inhibition during early postnatal windows, but not in adulthood, results in behavioral deficits later in life63. Indeed, we postulate that histone serotonylation governs transcriptomic patterns during these select neurodevelopmental windows (as we have described previously in culture systems using neuronal precursor cells and human induced pluripotent stem cell-derived 5-HTergic neurons24), which are the subject of future investigations, but that during early-to-mid embryogenesis, downstream consequences of placental 5-HT disruptions are mediated by non-serotonergic processes in the brain.
Together, our findings establish that placental H3K4me3Q5ser lies at the intersection of maternal 5-HT detection, regulation of tissue transcriptional networks, and offspring brain development, though additional studies will be needed to fully delineate the specific involvement of this histone PTM in modulating tissue-specific functions. Given that the endocrine placenta dynamically regulates H3K4me3Q5ser in response to both SERT disruptions and 5-HT changes in the maternal milieu, outstanding questions regarding the effects of prenatal stress and antidepressant exposures remain. Notably, several studies examining the effects of maternal perturbations observed dysregulation of placental 5-HT15,64–66; therefore, understanding how these triggers may enact negative long-term outcomes on fetal development via placental histone serotonylation changes, how fetal sex impacts these outcomes, and how antidepressant usage may reverse such dysregulated processes, are needed. Moreover, while we show that H3 serotonylation is a dynamic mechanism of developmental regulation within the placenta, a comprehensive catalogue of monoaminylated proteins (including serotonylation of both nuclear and cytoplasmic substrates) and their downstream effects on offspring neurodevelopment may provide further insight into how non-canonical monoamine mechanisms contribute to origins of neurodevelopmental disease risk.
MATERIAL AND METHODS
Animals
Wild-type C57BL6/J mice were purchased from Jackson Laboratories at 8 weeks old, and maintained on a 12-h/12-h light/dark cycle throughout the entirety of the experiment. Mice were provided with ad libitum access to water and food throughout the entirety of the experiment. All animal procedures were done in accordance with NIH guidelines and with approval with the Institutional Animal Care and Use Committee of the Icahn School of Medicine at Mount Sinai. For transgenic tissue studies, wild-type (WT), TPH1-deficient (Tph1-KO)67, SERT-deficient (Sert-KO)68 (Jackson Laboratories, stock #008355) and OCT3-deficient (Oct3-KO)69 (provided by Dr. Ciarimboli), all on C57Bl6/N genetic background, were bred at the MDC animal facility (Berlin, Germany) in individually ventilated cages (Tecniplast, Italy) under specific pathogen-free, standardized conditions in accordance with the German Animal Protection Law. Mice were group-housed at a constant temperature of 21 ± 2°C with a humidity of 65 ± 5%, an artificial 12 hours light/dark cycle, and with free access to water ad libitum. All experimental procedures were performed according to the national and institutional guidelines and have been approved by responsible governmental authorities (Landesamt für Gesundheit und Soziales (LaGeSo), Berlin, Germany).
Timed Breedings
Adult virgin female mice were bred in-house with age-matched males. Copulation plugs were checked every morning within 1 hour after lights on, where confirmation of a plug was designated as E0.5 and signaled the immediate removal of the female to her own cage with a nestlet.
Tissue Collection and Sex Determination
Timed pregnant dams were deeply anesthetized with isoflurane at designated embryonic time points, and conceptuses were isolated from the uterine wall, as previously described65. Placental tissues were hemisected in the transverse plane with removal of decidua cells70, flash frozen on dry ice, and stored at −80°C until further processing. Enriched fetal brain tissues were separated from the head by a single cut above the eye, perpendicular to the anterior-posterior axis. All tissues were flash frozen on dry ice and stored at −80°C until further analyses. Embryonic tails for WT developmental studies were retained for sex determination by Jarid1 genotyping, as previously described71. For KO studies, both male and female tissues were used per genotype after determining there were no sex differences in Slc6a4, Slc22a3, and Tph1 gene expression (Fig. 2B) and due to limited sample n per group.
5-PT Injection and Detection
5-PT was diluted in 1x PBS to 100 nM or 1 µM, representing endogenous levels of 5-HT at basal or inflammatory conditions46. Pregnant mice (E12.5) were injected via tail vein with 5-PT mixtures or vehicle. 1 hour post-injection, conceptuses were removed and placental tissues were collected for further processing. Magnetic streptavidin beads (Thermo Fisher 11205D) were incubated with 10 mM biotin azide (probe condition; Click Chemistry Tools 1265) or 10 mM desthio-biotin (no probe condition; Sigma D1411) on a rotator for 1 hour at 4°C. For copper-click chemistry, placental whole cell lysates containing proteins labelled with the alkyne-functionalized 5-PT were incubated with conjugated beads, 800 µM CuSO4, and 400 µM sodium ascorbate added in that order on a rotator for 1 hour at 4°C in a total volume of 500 µl in 1x PBS. Reactions were stopped by adding EDTA to a final concentration of 20 mM. All samples were washed on a magnetic stand using 0.1M glycine and High Salt Buffer (500mM KCl, 20 mM HEPES, 10 mM MgCl2, 1% NP-40). After the last wash, sample buffer was added to beads and boiled at 95°C for 10 min, followed by gel electrophoresis and incubation with appropriate primary and secondary antibodies.
Serotonin ELISA
Placental or fetal brain tissues were homogenized in cold PBS with 1x protease inhibitor cocktail (Roche). 60 ug of lysate per sample was quantitated using the BCA Protein Assay Kit (Pierce) and mixed 1:1 with assay buffer for measurement. Tissue 5-HT levels were assessed using the Serotonin ELISA Kit according to manufacturer’s instruction (Abcam ab133053).
Western Blotting and Antibodies
Placental or fetal brain tissues were homogenized and sonicated in cold RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% SDS, 1% NP-40) supplemented with 1x protease inhibitor cocktail (Roche). 30 ug of protein per sample was quantitated using the BCA Protein Assay Kit (Pierce) and loaded onto 4–12% NuPage BisTris gels for electrophoresis. Fast transfers were performed using the Trans-Blot Turbo Transfer System (Bio-Rad) for 7 minutes onto nitrocellulose membranes, and blocked in 5% milk or bovine serum albumin (BSA) diluted in 0.1% PBS-T. Membranes were incubated overnight with primary antibodies at 4°C on an orbital shaker. The following day, blots were washed 3x with PBS-T at room temperature, incubated for 1 hour with secondary antibody, and washed again with PBS-T 3x. Bands were detected using either enhanced chemiluminescence (ECL; Millipore) or fluorescence with the ChemiDoc Imaging System (Bio-Rad). Densitometry was used to quantify protein bands via Image J Software and proteins were normalized to total Gapdh. For developmental H3K4me3Q5ser western blots, one sample (run 2x) was removed due to lack of signal, as indicated in Supplementary Figure 1. For peptide competition assays, antibodies were pre-incubated with indicated peptides at 1:3 concentration of peptide to antibody for 1 hour at room temperature. Following pre-incubation, membranes were incubated with the designated antibody/peptide mixture overnight at 4°C on an orbital shaker. The following combinations of antibodies/buffers were used.
| Primary Antibody | Secondary Antibody | Block | Figure |
|---|---|---|---|
| 1:1000 H3K4me3Q5ser (MilliporeSigma ABE2580) | 1:10,000 anti-rabbit (Cytiva NA934V) | 5% milk | 1C, Supp Fig 1 |
| 1:1000 H3K4me3Q5ser (MilliporeSigma ABE2580) | 1:10,000 anti-rabbit (Thermo Fisher A-11010 or A-21235) | 5% BSA | 2F, 4D, Supp Fig 5, 7 |
| 1:10,000 GAPDH (Abcam ab9485) | 1:10,000 anti-rabbit (Cytiva NA934V) | 5% milk | 1C, Supp Fig 1 |
| 1:10,000 GAPDH (Santa Cruz sc-32233) | 1:10,000 anti-Mouse (Thermo Fisher A-21202 or A-11030) | 5% BSA | 2F, 4D, Supp Fig 5, 7 |
| 1:10,000 H3 (Abcam ab1791) | 1:10,000 anti-rabbit (Thermo Fisher A-21235) | 5% BSA | Supp Fig 4 |
Chromatin Immunoprecipitation, ChIP-seq and Analysis
Chromatin from hemisected placental tissues were fixed with 1% formaldehyde rotated for 12 minutes at room temperature and was subsequently quenched using a final concentration of 125mM glycine. Samples were thoroughly homogenized and washed with ice cold PBS. Fixed chromatin was sonicated using a Covaris E220 for 30–60 minutes at 4°C with the following conditions: peak incident power, 140; duty factor, 10%; Cycles/burst, 200; Water level, 0. Equal amounts of chromatin per sample were rotated with select antibodies (2.5 μg antibody/sample of either H3K4me3Q5ser (MilliporeSigma ABE2580) or H3K4me3 (Active Motif 39159)) bound to M-280 Dynabeads at 4°C overnight. The next morning, samples were washed, eluted, and reverse-crosslinked at 65°C. Samples underwent RNA and protein digestion, and DNA was purified using QIAQuick MinElute Spin columns (Qiagen 28140). 1% inputs were removed prior to antibody incubation and purified in parallel with corresponding immunoprecipitates. ChIP-seq libraries were generated using the TruSeq ChIP Library Preparation Kit (Illumina IP-202–1012) according to manufacturer’s protocol and sequenced on an Illumina HiSeq2500 or NovaSeq6000. Raw peaks were aligned to the mm10 mouse genome using the NGS Data Charmer pipeline with default settings (HISAT v.0.1.6b)72. Peak calling was performed using macs2 (v.2.1.1) on individual files with default settings and filtered for peaks with FDR < 0.0573. Differential peak analysis was conducted via pairwise comparisons using the DiffBind package (v3.8.4)37. Differential peaks were filtered first by log2(fold change) > 0.1 and defined by p < 0.05, where log2(fold change) was calculated as log2(E17.5 conc) − log2(E9.5 conc) for developmental comparisons; log2(female conc) − log2(male conc) for sex differences; and log2(KO conc) − log2(WT conc) for transgenic comparisons. These criteria were determined by visual confirmation of differential peaks after inspection of more than 100 sites in the Integrative Genomics Viewer (Broad Institute, v2.11.1). All peaks were annotated to the mm10 genome using the Homer package (v4.10)74. Functional annotation analysis of uniquely annotated loci was conducted using ShinyGO v0.77 with a background of all protein-coding genes in the mm10 genome75, with significant pathways defined by FDR < 0.05 and GO term redundancy reduction using Revigo v1.8.150. Visualization of differential peaks were accomplished using internal functions of the DiffBind package or deepTools v3.5.376.
RNA Isolation, RNA-seq and Analysis
Total mRNA from hemisected placental tissues and embryonic brain tissues were extracted following homogenization in Trizol Reagent (Thermo Fisher) with subsequent clean-up using RNeasy Microcolumns (Qiagen) according to manufacturer’s recommendation. 200ng mRNA per sample was used for RNA-seq library preparation using the TruSeq RNA Library Prep Kit v2 (Illumina RS-122-2001) according to manufacturer’s protocol. Quality control of all libraries were conducted using a Qubit Fluorometer 2.0 (Thermo Fisher) and Bioanalyzer High Sensitivity DNA Analysis (Agilent) prior to sequencing on either an Illumina HiSeq2500 or NovaSeq6000. Raw fastq files containing an average of 20–30 million reads were processed for pseudoalignment and abundance quantification using Kallisto (v.0.46.1) against the EnsemblDB mus musculus (v79)77. To account for unwanted technical variation between batches of animal orders, sample collection, mRNA extraction, and library preparation that are each represented per sample batch, RUVs (v1.32.0) was used with a negative control gene set of total genes identified per sequencing experiment following confirmation that unwanted factors did not correlate with covariates of interest (for all experiments, k=4 was used) as previously described78,79. Next, differential expression analysis was performed using DESeq2 (v1.38.3) and significant genes were defined by adjusted p < 0.0580. Odds ratio overlap analysis was conducted using the GeneOverlap package (v.1.36.0), with significance indicated by p < 0.05. Functional annotation analysis of differentially expressed genes was performed using ShinyGO v0.77 with a background of all protein-coding genes in the mm10 genome, with significant pathways defined by FDR < 0.05 and GO term redundancy reduction using Revigo v1.8.150,75. Importantly, increased Slc6a4 expression was observed in RNA-seq data from Sert KO embryo brains, likely reflecting the aberrant introduction of an internal promoter in the design of this transgenic line and/or increased expression of transcripts that undergo nonsense-mediated decay, as indicated by loss of functional protein (Supplementary Fig. 4). Thus, to ensure nonfunctional increases in Slc6a4 expression did not misleadingly contribute to pathway enrichment data, Slc6a4 was removed from significant differential gene expression lists in WT vs. Sert KO comparisons prior to pathway analysis.
Supplementary Material
Acknowledgements
This work was partially supported by grants from the National Institutes of Health: R01 MH116900 (I.M.), F32 MH126534 (J.C.C.), F31 NS132558 (A.M.C.), as well as funds from the Howard Hughes Medical Institute (I.M.). NA and MB were supported by the EU H2020 MSCA ITN projects “Serotonin and Beyond” (N 953327). All schematics were created with Biorender.com.
Footnotes
Declaration of Interest
The authors declare no competing interests.
Data and Materials Availability
The RNA-seq and ChIP-seq data generated in this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database under accession number GSE246540. We declare that the data supporting findings for this study are available within the article and Supplementary Information. Related data are available from the corresponding author upon reasonable request. No restrictions on data availability apply.
References
- 1.Sodhi M. S. & Elaine S.-B. Serotonin and brain development. Int. Rev. Neurobiol. 59, 111–174 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Brummelte S., Mc Glanaghy E., Bonnin A. & Oberlander T. F. Developmental changes in serotonin signaling: Implications for early brain function, behavior and adaptation. Neuroscience 342, 212–231 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lidov H. G. & Molliver M. E. Immunohistochemical study of the development of serotonergic neurons in the rat CNS. Brain Res Bull 9, 559–604 (1982). [DOI] [PubMed] [Google Scholar]
- 4.Wallace J. A. & Lauder J. M. Development of the serotonergic system in the rat embryo: An immunocytochemical study. Brain Research Bulletin 10, 459–479 (1983). [DOI] [PubMed] [Google Scholar]
- 5.Bonnin A. & Levitt P. Fetal, maternal, and placental sources of serotonin and new implications for developmental programming of the brain. Neuroscience 197, 1–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnin A. et al. A transient placental source of serotonin for the fetal forebrain. Nature 472, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balkovetz D. F., Tiruppathi C., Leibach F. H., Mahesh V. B. & Ganapathy V. Evidence for an Imipramine-sensitive Serotonin Transporter in Human Placental Brush-border Membranes. Journal of Biological Chemistry 264, 2195–2198 (1989). [PubMed] [Google Scholar]
- 8.Prasad P. D. et al. Functional expression of the plasma membrane serotonin transporter but not the vesicular monoamine transporter in human placental trophoblasts and choriocarcinoma cells. Placenta 17, 201–207 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Kekuda R. et al. Cloning and Functional Characterization of a Potential-sensitive, Polyspecific Organic Cation Transporter (OCT3) Most Abundantly Expressed in Placenta *. Journal of Biological Chemistry 273, 15971–15979 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Sata R. et al. Functional analysis of organic cation transporter 3 expressed in human placenta. J Pharmacol Exp Ther 315, 888–895 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Kliman H. J. et al. Pathway of Maternal Serotonin to the Human Embryo and Fetus. Endocrinology 159, 1609–1629 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Hsiao E. Y. & Patterson P. H. Activation of the maternal immune system induces endocrine changes in the placenta via IL-6. Brain Behav Immun 25, 604–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W.-L., Hsiao E. Y., Yan Z., Mazmanian S. K. & Patterson P. H. The placental interleukin-6 signaling controls fetal brain development and behavior. Brain, Behavior, and Immunity 62, 11–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bronson S. L. & Bale T. L. Prenatal stress-induced increases in placental inflammation and offspring hyperactivity are male-specific and ameliorated by maternal antiinflammatory treatment. Endocrinology 155, 2635–2646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goeden N. et al. Maternal Inflammation Disrupts Fetal Neurodevelopment via Increased Placental Output of Serotonin to the Fetal Brain. J Neurosci 36, 6041–6049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan J. C., Nugent B. M. & Bale T. L. Parental Advisory: Maternal and Paternal Stress Can Impact Offspring Neurodevelopment. Biological psychiatry 83, 886–894 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nugent B. M., O’Donnell C. M., Epperson C. N. & Bale T. L. Placental H3K27me3 establishes female resilience to prenatal insults. Nat Commun 9, 2555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergdolt L. & Dunaevsky A. Brain changes in a maternal immune activation model of neurodevelopmental brain disorders. Progress in Neurobiology 175, 1–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cissé Y. M., Chan J. C., Nugent B. M., Banducci C. & Bale T. L. Brain and placental transcriptional responses as a readout of maternal and paternal preconception stress are fetal sex specific. Placenta 100, 164–170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shook L. L., Kislal S. & Edlow A. G. Fetal brain and placental programming in maternal obesity: A review of human and animal model studies. Prenatal Diagnosis 40, 1126–1137 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertrand C. & St-Louis J. Reactivities to serotonin and histamine in umbilical and placental vessels during the third trimester after normotensive pregnancies and pregnancies complicated by preeclampsia. Am J Obstet Gynecol 180, 650–659 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Ugun-Klusek A. et al. Reduced placental vascular reactivity to 5-hydroxytryptamine in preeclampsia and the status of 5HT(2A) receptors. Vascul Pharmacol 55, 157–162 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Hadden C. et al. Serotonin transporter protects the placental cells against apoptosis in caspase 3-independent pathway. J Cell Physiol 232, 3520–3529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farrelly L. A. et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 567, 535–539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lepack A. E. et al. Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 368, 197–201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fulton S. L. et al. Histone H3 dopaminylation in ventral tegmental area underlies heroin-induced transcriptional and behavioral plasticity in male rats. Neuropsychopharmacology 47, 1776–1783 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart A. F., Lepack A. E., Fulton S. L., Safovich P. & Maze I. Histone H3 dopaminylation in nucleus accumbens, but not medial prefrontal cortex, contributes to cocaine-seeking following prolonged abstinence. Mol Cell Neurosci 125, 103824 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mycek M. J., Clarke D. D., Neidle A. & Waelsch H. Amine incorporation into insulin as catalyzed by transglutaminase. Archives of Biochemistry and Biophysics 84, 528–540 (1959). [DOI] [PubMed] [Google Scholar]
- 29.Bader M. Serotonylation: Serotonin Signaling and Epigenetics. Frontiers in Molecular Neuroscience 12, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dale G. L. et al. Stimulated platelets use serotonin to enhance their retention of procoagulant proteins on the cell surface. Nature 415, 175–179 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Walther D. J. et al. Serotonylation of small GTPases is a signal transduction pathway that triggers platelet alpha-granule release. Cell 115, 851–862 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Paulmann N. et al. Intracellular Serotonin Modulates Insulin Secretion from Pancreatic β-Cells by Protein Serotonylation. PLOS Biology 7, e1000229 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan J. C. & Maze I. Nothing Is Yet Set in (Hi)stone: Novel Post-Translational Modifications Regulating Chromatin Function. Trends in Biochemical Sciences 45, 829–844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sardar D. et al. Induction of astrocytic Slc22a3 regulates sensory processing through histone serotonylation. Science 380, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Kachak A. et al. Histone H3 serotonylation dynamics in dorsal raphe nucleus contribute to stress- and antidepressant-mediated gene expression and behavior. 2023.05.04.539464 Preprint at 10.1101/2023.05.04.539464 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suri D., Teixeira C. M., Cagliostro M. K. C., Mahadevia D. & Ansorge M. S. Monoamine-sensitive developmental periods impacting adult emotional and cognitive behaviors. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 40, 88–112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stark R. & Brown G. DiffBind: Differential binding analysis of ChIP-Seq peak data.
- 38.Nugent B. M. & Bale T. L. The omniscient placenta: Metabolic and epigenetic regulation of fetal programming. Frontiers in neuroendocrinology 39, 28–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaut C. A. E., Keene D. R., Sorensen L. K., Li D. Y. & Stadler H. S. HOXA13 Is Essential for Placental Vascular Patterning and Labyrinth Endothelial Specification. PLoS Genetics 4, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun A. E. et al. “Females Are Not Just ‘Protected’ Males”: Sex-Specific Vulnerabilities in Placenta and Brain after Prenatal Immune Disruption. eNeuro 6, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma C. et al. CXCL1 stimulates decidual angiogenesis via the VEGF-A pathway during the first trimester of pregnancy. Mol Cell Biochem 476, 2989–2998 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Woods L., Perez-Garcia V. & Hemberger M. Regulation of Placental Development and Its Impact on Fetal Growth—New Insights From Mouse Models. Frontiers in Endocrinology 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karahoda R. et al. Serotonin homeostasis in the materno-foetal interface at term: Role of transporters (SERT/SLC6A4 and OCT3/SLC22A3) and monoamine oxidase A (MAO-A) in uptake and degradation of serotonin by human and rat term placenta. Acta Physiologica 229, e13478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baković P. et al. Differential Serotonin Uptake Mechanisms at the Human Maternal–Fetal Interface. Int J Mol Sci 22, 7807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenfeld C. S. Placental serotonin signaling, pregnancy outcomes, and regulation of fetal brain development. Biology of Reproduction 102, 532–538 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mössner R. & Lesch K. P. Role of serotonin in the immune system and in neuroimmune interactions. Brain Behav Immun 12, 249–271 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Côté F. et al. Disruption of the nonneuronal tph1 gene demonstrates the importance of peripheral serotonin in cardiac function. Proc Natl Acad Sci U S A 100, 13525–13530 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mordhorst A. et al. Phenylalanine hydroxylase contributes to serotonin synthesis in mice. FASEB J 35, e21648 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Muller C. L. et al. Impact of Maternal Serotonin Transporter Genotype on Placental Serotonin, Fetal Forebrain Serotonin, and Neurodevelopment. Neuropsychopharmacology 42, 427–436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Supek F., Bošnjak M., Škunca N. & Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLOS ONE 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laurent L. et al. Human placenta expresses both peripheral and neuronal isoform of tryptophan hydroxylase. Biochimie 140, 159–165 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Karahoda R. et al. Dynamics of Tryptophan Metabolic Pathways in Human Placenta and Placental-Derived Cells: Effect of Gestation Age and Trophoblast Differentiation. Frontiers in Cell and Developmental Biology 8, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yavarone M. S., Shuey D. L., Sadler T. W. & Lauder J. M. Serotonin uptake in the ectoplacental cone and placenta of the mouse. Placenta 14, 149–161 (1993). [DOI] [PubMed] [Google Scholar]
- 54.Pavličev M. et al. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res 27, 349–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Côté F. et al. Maternal serotonin is crucial for murine embryonic development. Proc Natl Acad Sci U S A 104, 329–334 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mekontso-Dessap A. et al. Deficiency of the 5-hydroxytryptamine transporter gene leads to cardiac fibrosis and valvulopathy in mice. Circulation 113, 81–89 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Basu B. et al. Serotonin in pre-implantation mouse embryos is localized to the mitochondria and can modulate mitochondrial potential. Reproduction 135, 657–669 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Zheng Q. et al. Histone monoaminylation dynamics are regulated by a single enzyme and promote neural rhythmicity. 2022.12.06.519310 Preprint at 10.1101/2022.12.06.519310 (2023). [DOI] [Google Scholar]
- 59.Zhao S. et al. Histone H3Q5 serotonylation stabilizes H3K4 methylation and potentiates its readout. Proc Natl Acad Sci U S A 118, e2016742118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pavone L. M. et al. Fate map of serotonin transporter-expressing cells in developing mouse heart. Genesis 45, 689–695 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Chen X., Petit E. I., Dobrenis K. & Sze J. Y. Spatiotemporal SERT expression in cortical map development. Neurochem Int 98, 129–137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Gregorio R. et al. Sex-biased effects on hippocampal circuit development by perinatal SERT expression in CA3 pyramidal neurons. Development 149, dev200549 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ansorge M. S., Morelli E. & Gingrich J. A. Inhibition of serotonin but not norepinephrine transport during development produces delayed, persistent perturbations of emotional behaviors in mice. J Neurosci 28, 199–207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y. et al. GDM-associated insulin deficiency hinders the dissociation of SERT from ERp44 and down-regulates placental 5-HT uptake. Proc Natl Acad Sci U S A 111, E5697–5705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bronson S. L., Chan J. C. & Bale T. L. Sex-specific neurodevelopmental programming by placental insulin receptors on stress reactivity and sensorimotor gating. Biological psychiatry 82, 127–138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ranzil S. et al. Disrupted placental serotonin synthetic pathway and increased placental serotonin: Potential implications in the pathogenesis of human fetal growth restriction. Placenta 84, 74–83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walther D. J. et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science 299, 76 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Bengel D. et al. Altered brain serotonin homeostasis and locomotor insensitivity to 3, 4-methylenedioxymethamphetamine (‘Ecstasy’) in serotonin transporter-deficient mice. Mol Pharmacol 53, 649–655 (1998). [DOI] [PubMed] [Google Scholar]
- 69.Zwart R., Verhaagh S., Buitelaar M., Popp-Snijders C. & Barlow D. P. Impaired activity of the extraneuronal monoamine transporter system known as uptake-2 in Orct3/Slc22a3-deficient mice. Mol Cell Biol 21, 4188–4196 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pennington K. A., Schlitt J. M. & Schulz L. C. Isolation of Primary Mouse Trophoblast Cells and Trophoblast Invasion Assay. JoVE (Journal of Visualized Experiments) e3202 (2012) doi: 10.3791/3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clapcote S. J. & Roder J. C. Simplex PCR assay for sex determination in mice. Biotechniques 38, 702, 704, 706 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Kim D., Langmead B. & Salzberg S. L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y. et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biology 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heinz S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ge S. X., Jung D. & Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramírez F., Dündar F., Diehl S., Grüning B. A. & Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W187–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bray N. L., Pimentel H., Melsted P. & Pachter L. Near-optimal probabilistic RNA-seq quantification. Nature biotechnology 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Risso D., Ngai J., Speed T. P. & Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 32, 896–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peixoto L. et al. How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets. Nucleic Acids Res 43, 7664–7674 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and ChIP-seq data generated in this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database under accession number GSE246540. We declare that the data supporting findings for this study are available within the article and Supplementary Information. Related data are available from the corresponding author upon reasonable request. No restrictions on data availability apply.