Abstract
Genetically heterogenous cell lines from laboratory mice are promising tools for population-based screening as they offer power for genetic mapping, and potentially, predictive value for in vivo experimentation in genetically matched individuals. To explore this further, we derived a panel of fibroblast lines from a genetic reference population of laboratory mice (the Diversity Outbred, DO). We then used high-content imaging to capture hundreds of cell morphology traits in cells exposed to the oxidative stress-inducing arsenic metabolite monomethylarsonous acid (MMAIII). We employed dose-response modeling to capture latent parameters of response and we then used these parameters to identify several hundred cell morphology quantitative trait loci (cmQTL). Response cmQTL encompass genes with established associations with cellular responses to arsenic exposure, including Abcc4 and Txnrd1, as well as novel gene candidates like Xrcc2. Moreover, baseline trait cmQTL highlight the influence of natural variation on fundamental aspects of nuclear morphology. We show that the natural variants influencing response include both coding and non-coding variation, and that cmQTL haplotypes can be used to predict response in orthogonal cell lines. Our study sheds light on the major molecular initiating events of oxidative stress that are under genetic regulation, including the NRF2-mediated antioxidant response, cellular detoxification pathways, DNA damage repair response, and cell death trajectories.
Keywords: genetics, systems genetics, systems toxicology, high content imaging, cell painting, genetic diversity, genetic mapping, QTL mapping, fibroblasts, arsenic, monomethylarsonous acid, cell morphology, cmQTL, new approach methodologies
Introduction
Cell morphology has served as a useful phenotype for understanding how genetic factors regulate the state of metazoan cells, ranging from yeast to human induced pluripotent stem cells (iPSCs) 1,2. Recent advances in microscopy-based, high-content cellular screening (HCS) have made it cost-effective to analyze cellular phenotypes at scale 3–7. When coupled with machine learning techniques, these technologies enable precise measurements of cellular and sub-cellular morphological traits, which have long been observed in the context of development and disease 8–11.
We and others previously characterized the genetic architecture of ground-state pluripotency and differentiation propensity in genetically diverse mouse embryonic stem cells (mESCs). This work demonstrated that -omics traits like gene expression, chromatin accessibility, and protein levels in genetically diverse cells, especially when combined (multi-omics), provide molecular readouts that can be used to identify the genetic factors regulating cell state 12–15. The correlation of cell morphology traits to these underlying -omics traits offers the potential to quantitatively analyze and delineate how cells respond to genetic and environmental perturbations 16–18. However, multi-omic approaches like these can be expensive, particularly in the context of population-level screens of cell state across many environmental perturbations. Moreover, the utility of cell morphology traits derived from HCS for genetic analysis has not been fully explored, especially in laboratory mouse cells.
In this study, we used cell morphology traits from HCS for genetic analysis of cellular response during acute arsenic exposure. Arsenic is a known carcinogen and a widespread contaminant of groundwater, exposing up to estimated 220 million people worldwide 19. Ingested inorganic arsenic is metabolized through methylation and reducing reactions that generate metabolites including monomethylarsonic acid (MMAV), monomethylarsonous acid (MMAIII), dimethylarsinic acid (DMAV), and dimethylarsinous acid (DMAIII) 20–22. These arsenic metabolites have unique toxicological profiles and urinary ratios that favor the more toxic forms have been linked to disease 23,24. At the cellular level, arsenic exposure induces oxidative stress, DNA damage, and cytotoxicity to varying degrees depending on the metabolites present, the tissue type, and genetic background of the exposed individual. These are the key events that lead to adverse outcomes including cancer or impaired reproduction / development at the population level. Interindividual variation in urinary metabolite ratios from populations exposed to high levels of arsenic have been used in genetic association mapping to identify variants associated with adverse outcomes in sensitive individuals. These studies revealed genes and variants that regulate arsenic metabolism, as well as oxidative stress response and DNA damage repair 25–41. In laboratory mice, the metabolite MMAIII causes DNA damage through oxidative stress and induces tumor development in the kidney 42,43. Given the substantial body of genetic association data for arsenic and our interest in kidney pathophysiology, we sought to evaluate a population-based cellular model and to employ cell morphology traits to access gene by environment interactions for the metabolite MMAIII.
Genetically diverse laboratory mouse resource populations are powerful experimental tools for genetic analysis and they are well established in the study of gene by environment interactions in vivo 44,45. Cell lines from these genetic reference populations offer a new approach methodology wherein genetic screens can be performed ‘in a dish’ to identify haplotypes that confer sensitivity and resilience. Approaches such as these have the potential to reduce the scale of animal studies where informative molecular and/or cellular phenotypes exist. We created a diverse panel of primary fibroblast cell lines from the Diversity Outbred (DO) mouse population 46. DO mice are outbred animals descended from eight inbred mouse strains: A/J (AJ), C57BL/6J (B6), 129S1/SvImJ (129), NOD/ShiLtJ (NOD), NZO/HILtJ (NZO), CAST/EiJ (CAST), PWK/PhJ (PWK), and WSB/EiJ (WSB). These inbred strains represent three sub-species of Mus musculus and thus possess far more genetic variation than traditional mouse crosses, capturing roughly 45 million segregating single nucleotide polymorphisms (SNPs) 46,47.
Using a high content screening (HCS) technique similar to Cell Painting 3, we show that high-dimensional cell morphology phenotypes can be summarized through dose-response modeling to capture latent features that reflect changes in cell state during an acute, arsenic-induced oxidative stress response. We show that these cell state changes vary across genetically diverse cells, revealing both sensitive and resilient individuals to MMAIII-induced cell morphology changes. Using quantitative trait mapping (QTL), we found 854 cell morphology QTL (cmQTL; LOD score > 7.5), which are the genetic loci that regulate the cellular response to arsenical exposure. Additionally, we show that the cmQTL effects are both reproducible and predictive of arsenic sensitivity. At the gene and pathway level, many cmQTL recapitulate genetic associations that have been previously found in human population studies, demonstrating the translational utility of our population-based cellular model. We highlight the roles of Xrcc2 and Txnrd1 alleles that modulate MMAIII-induced cellular death, and we provide new associations for a host of candidate genes that interact with MMAIII.
Results
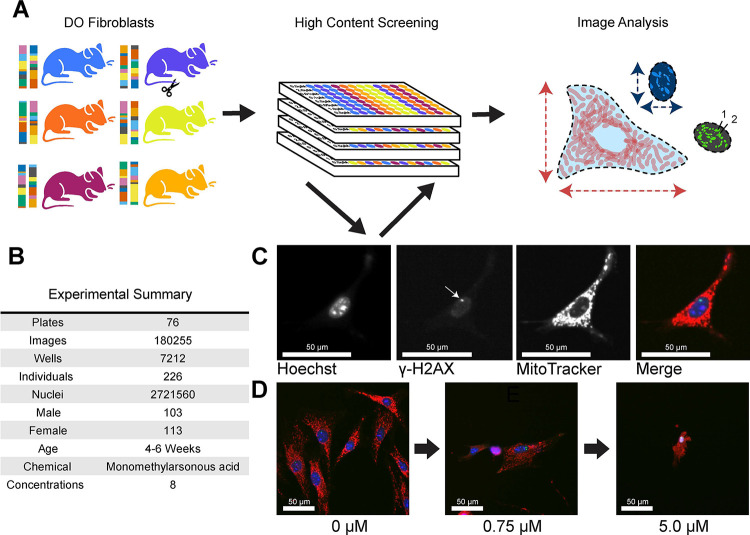
Cell morphology is influenced by genetic variation and environmental factors including chemical exposures 2. Therefore we sought to use morphological traits to quantify the key cellular events that occur during arsenic exposure, and to identify the genetic determinants of cellular sensitivity through a forward genetic screen. We established a population-based cellular model by deriving a panel of tail tip fibroblast lines from the Diversity Outbred (DO) mouse population (n = 600) (Fig. 1A,1B). Tail tip fibroblast cultures can be readily established through minimally invasive techniques, they are adherent, and they can be easily maintained for many passages depending on the age of the donor. Though heterogeneous and tissue specific, fibroblasts are one of the most widespread cell types found in mammals. To observe effects of acute arsenic exposure, we treated 226 of these DO fibroblast lines with eight increasing concentrations of monomethylarsonous acid (MMAIII) across 76 randomized 96-well plates 48. MMAIII is a highly toxic arsenic intermediate that induces oxidative stress associated DNA damage in exposed tissues 49 (Fig. 1A). Based on the genetic architecture of the DO population, we expected this number of individual cell lines would allow us to detect QTL explaining >20% of the phenotypic variance with 90% power 50. To quantify changes in cell morphology associated with oxidative stress and genotoxicity, we used cell stains to label nuclei (Hoechst 33342) and mitochondria (MitoTracker Deep Red), and we used indirect immunolabeling to quantify DNA damage repair (γH2AX) (Fig. 1C). We captured 180,255 images and performed image analysis using Harmony 4.9 to extract 673 image-based, morphological phenotypes from 2,721,560 cells (Fig. 1B).
Figure 1: HCS of MMAIII-exposed DO Fibroblasts.
(A) 600+ primary fibroblasts were derived from Diversity Outbred (DO) mice aged 4–6 weeks. 226 DO fibroblast lines were exposed to 8 concentrations of MMAIII (0 μM, 0.01 μM, 0.1 μM, 0.75 μM, 1.0 μM, 1.25 μM, 2.0 μM, and 5.0 μM). Cell lines were semi-randomly seeded into 96-well plates (4 columns spanning two plates, see Supplementals for more information). Image analysis was performed at the whole well level and summarized across concentrations using dose response modeling.
(B) Table with experimental summary
(C) Example images showing fibroblasts labeled with MitoTracker Deep Red, Hoechst 33342, an anti-gamma γH2AX antibody with a Alexafluor 488 donkey anti-rabbit secondary, and the merged image. Plates were imaged using an Operetta High Content Imager (PerkinElmer) at 20X.
(D) Example merged images showing a fibroblasts’ morphology across three representative doses of MMAIII (0 μM, 0.75 μM, 5.0 μM).
Sources of variation in cell morphology traits
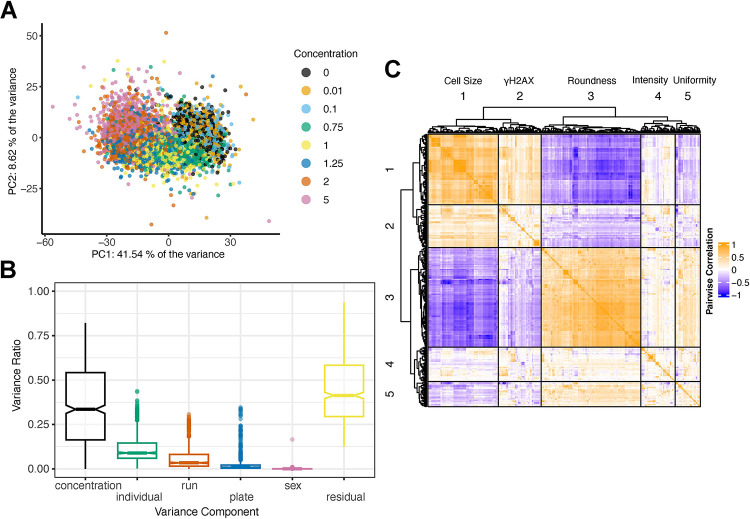
To assess the main drivers of variation in these data, we performed principal components analysis. The first principal component, accounting for 41.5% of the observed variation across all traits was correlated with MMAIII concentration, and there was a clear dose-dependent effect (Fig. 2A). Following Matthew et al. 2, we performed a decomposition of the sources of variation contributing to each trait by fitting a random effects linear model with terms for inter-plate effects (‘plate’), batch effects (12 samples per ‘run’), MMAIII concentration (‘concentration’), DO donor (‘individual’), and the sex of cell donor (sex) (Fig. 2B). Among these factors, arsenic ‘concentration’ explained the most variation, followed by ‘individual’ or donor genetic background. While we randomized DO cell lines by column and MMAIII concentrations by row within a plate, we observed a common HCS finding that inter-plate and inter-run effects also influence variance in measured cellular features (Fig. 2B). Depending on the trait, ‘individual’ explained ~0–40% of the variance with an average of 10%, suggesting that a subset of these traits (those with >20%) would provide sufficient signal for genetic mapping based on the size and architecture of our DO cell population50.
Figure 2: HCS Features are Influenced by MMAIII Concentration and Genetic Background.
(A) Principal Component Analysis (PCA) of the raw image analysis feature dataset colored by the concentration of MMAIII. Among known factors, increasing MMAIII contributed the majority of the variance for both PC1 (41.54 %) and PC2 (8.62 %).
(B) Boxplot showing the aggregated results from variance component analysis (VCA) performed across all cellular features including MMAIII concentrarions (concentration), DO cell lines (individual), each 96-well plate (plate), residual variation, run, and sex.
(D) Heatmap showing the Pearson’s pairwise correlation structure of the all the raw cellular features. Theheatmap and dendrogram were generated using the R package ComplexHeatmap’s Heatmap() function with column_split and row_split each set to 5.
While HCS produces thousands of morphological traits, many of them are highly correlated (Fig. 2C). The correlated groups could be loosely categorized as traits describing ‘cell size’, ‘γH2AX foci’, ‘cell roundness‘, ‘intensity’, and ‘uniformity’ (Fig. 2C). While there are a variety of dimension reduction techniques that take advantage of correlation to summarize high dimensional data, we were most interested in traits exhibiting non-linear, dose-dependent responses.
Dose-response modeling and genetic mapping of cell morphology quantitative trait loci (cmQTL)
Dose-response models are used to define the xenobiotic response profiles of toxicants and drugs. In chemical risk assessment, these models provide benchmark dose estimates, which are the concentrations at which a chemical exposure could pose a health risk 51. To focus on the subset of traits exhibiting dose-dependent responses, we performed dose-response modeling using the drc R package 52 for each cellular trait, individual, and replicate experiment. These models provided quantitative dose-response parameters (DRPs) describing each donor individual’s cellular response including effective concentrations (EC’s), starting/maximum asymptotes, and rates of change (slopes) 53. For example, an individual’s EC50 represents the concentration of MMAIII at which there is a 50% change in a given cellular feature relative to baseline. Following the removal of redundant features and batch effect correction, our dose-response modeling resulted in 5,105 cmDRPs from 568 cellular traits.
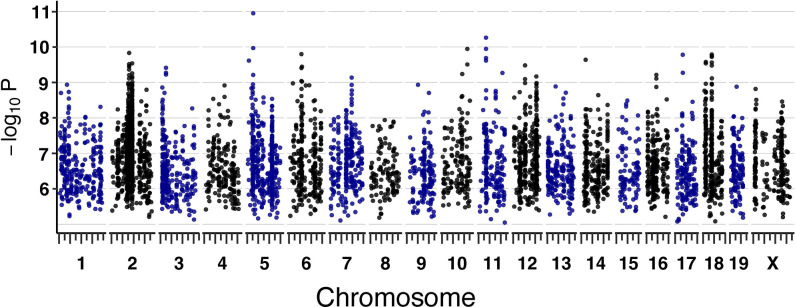
To reveal genetic loci that influence sensitivity to arsenic metabolite MMAIII, we performed quantitative trait loci (QTL) mapping, treating the 5105 cmDRPs as traits (see Methods). To account for the data’s complicated structure and redundancies in the context of multiple testing burden, we calculated a genome-wide false discovery rate (FDR) significance threshold, which resulted in only the maximum peak meeting significance (FDR < 10%) (Fig. 3). Given that this work represents a proof of principle and cmDRPs are potentially noisy as modeled quantities, we also used a lenient significance threshold of LOD score > 7.5, which corresponds to ~80% genome-wide significance threshold in the DO 54. Of the 5105 cmDRPs, 854 possessed suggestive genetic loci associations, with the strongest LOD score being 10.95. We found cmQTL reaching significance on chromosomes 2, 3, 6, 12, 14, 18. Significant response cmQTL included EC’s, slope, and maximum asymptotes, in addition to baseline DRPs, or starting asymptote.
Figure 3: Dose-Response Modeled cmQTL in DO Fibroblasts Exposed to MMAIII.
Summary of cmQTL maximum peaks for 5100 cmDRPs. Each points represents the strength of the genetic association as a LOD score on the y-axis (−log10P) across the mouse genome (x-axis). On the x-axis, long tick marks represent the start of the chromosome and 50 Mbp intervals, while the short tick marks are 25 Mbps.
Candidate cmQTL genes identified using differential gene expression, gene set enrichment, and data integration
To nominate candidate genes and variants within cmQTL, we used several approaches. We generated bulk RNA-Seq data from 16 randomly selected DO fibroblast lines and we used differential expression analysis (DE) to identify expressed genes that showed differential expression in the context of MMAIII exposure (Supp. Table 2). Then, on the resulting set of genes, we used gene set enrichment analysis (GSEA) to identify groups of genes that are functionally related (Supp. Table 3). We interrogated published gene-arsenic interactions through the Comparative Toxicogenomics Database (CTD) 55 and for each DE gene, we quantified the number of interaction annotations in CTD across all curated studies involving MMAIII, MMAV, DMAIII, DMAV, sodium arsenite, sodium arsenate, arsenic, and arsenic trioxide. For any causal variants that exert their effects through gene expression, the contributing haplotypes and direction of their effects will be correlated across eQTL and cmQTL in datasets generated from the same genetic reference population (DO). Therefore, we also correlated the cmQTL allele effects with previous DO eQTL from liver, heart, kidney, striatum, pancreatic islet cells, and mESCs (see Methods). Finally, local SNP association mapping within each cmQTL allowed us to identify the SNPs with the highest LOD scores in each interval.
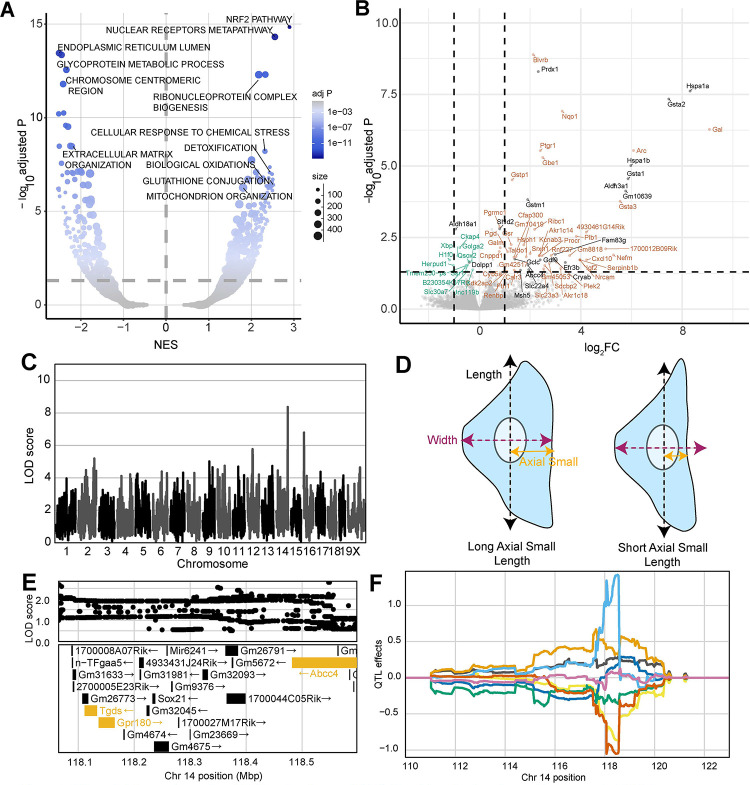
At the pathway level, the most upregulated gene set in dosed samples was ‘NRF2 activation (WP2884)’, which is a well-established response to oxidative stress following arsenical exposure 56–59 (Fig. 4A). NRF2, also known as NFE2L2, is a transcription factor that is shuttled to the nucleus following dissociation from KEAP1 in response to the generation of ROS 60–62. In the nucleus, NFE2L2 binds antioxidant response elements (AREs) upstream of many redox homeostasis and cellular defense genes to drive their transcription in response to stress, including arsenical exposure 56,57,63–66. These data provided multiple lines of evidence supporting Nfe2l2 (Nrf2) as a candidate gene for the cmQTL hotspot that we found on Chr 2 (Fig 3). Our gene expression analysis also revealed five candidate genes for other response cmQTL with LOD scores > 8 (Fig. 4B). Three of the five genes were present within the same CI, including Hspa1b, Hspa1a, and Msh5, with the former two DEGs having over 80 previously defined interactions with arsenicals. Among the other differentially expressed genes we found that 73 (89%) have not previously been associated with MMAIII, though many have been associated with arsenic or other arsenic metabolites.
Figure 4: Differential Expression and cmQTL Together Support MMAIII Glutathione Conjugation and its Export via ABCC4.
(A) Volcano plot showing the normalized effect sizes (NES) and adjusted p-values (−log10 transformed) of the score-based gene set enrichment (GSEA) results from differential expression (DE) analysis across the 0 and 0.75 μM MMAIII exposed DO fibroblasts groups (n = 32, 16 individuals). Expression was filtered based on a median transcript per million ≥ .5 or removed if at least half of the points were below this cutoff. Each point represent a gene set from ‘GO:Component’,‘REACTOME’, ‘KEGG’, ‘WikiPathways’, ‘GO:Tissue’, ‘GO:Molecular Function’ and ‘GO:Biological Process’. The size of each points represents the number of genes within the gene set and the color represents the −log10(adjusted P) (y-axis). Horizontal dashed line indicates the adj. p-value significance threshold (adj. P = 0.05)
(B) Volcano plot showing the log2-fold change (log2FC) and adjusted p-values (−log10adjusted P) for single genes. The horizontal indicates ithe adj. p-value significance threshold (adj. P = 0.05) and the vertical lines represent the ± 1 log2fold change for a point of reference. Points labeled with gene names are significantly differentially expressed (adj. p-value < .05) with effect sizes > 0.75 log2FC or < −0.25 log2FC. Colors represent genes withing cmQTL confidence intervals (black), upregulated (orange) and downregulated (green) DE.
(C) QTL scan for the ‘EC5 Mitosmooth Axial Small length mean per well’ cmQTL with the maximum peak at chromosome 14: 118483436 bp (m38) and a LOD score of 8.36.
(D) Cartoon fibroblast cells depicting the two measurements of cell length (black), width (purple), and axial small width (yellow). Fibroblast on the left has a longer axial small length compared to the fibroblast on the right,
(E) Variant association mapping within the Cl the cmQTL ‘EC5 Mitosmooth Axial Small length mean per well’. Top panel shows the LOD scores of the known, segregating variants in the 8 DO founders (m38). Bottom panel shows the gene models within the respective Cl. Each point represents a variant. Colors indicate whether a gene is expressed > 0.5 TPM (gold) or < 0.5 TPM (black). The arrow indicates the direction of transcription.
(F) Allele effects plot showing the eight DO founders (colors, see Methods) for the ‘EC5 Mitosmooth Axial Small length mean per well’ cmQTL across the surrounding region on chromosome 14 (Mbp).
Natural variation in cellular detoxification pathways partially explains arsenic sensitivity
The other two DEGs within response cmQTL were Cryab and Abcc4, each with ≥ 19 published arsenical interactions (Fig. 4B). SNPs in Abcc4 have been previously associated with sensitivity to arsenic 67. Abcc4 encodes the protein ABCC4/MRP4, which has been shown to export glutathionylated MMAIII from cells 68,69. Glutathione transferases like Gstm1, Gsta1, and Gstp1 were also significantly upregulated in our expression dataset. These genes are members of the glutathione conjugation pathway which is a detoxification pathway that leads to glutathionylation of MMAIII (MMADGIII) (Fig. 4A,4B) 68,70. We found multiple cmQTLs at the Abcc4 locus and they were all for traits related to changes in cell size (i.e., length, compactness) (Fig. 4C). For example, one of these response cmQTL was EC5 of the change in axial small length or the dose at which 5% of the cell population exhibited measurable differences in cell size (defined by the smoothed MitoTracker labeling which captures the cytoplasmic area occupied by mitochondria) (Fig. 4D). Variant association mapping revealed that the highest scoring SNPs in these cmQTLs were within the Abcc4 gene, and the allele effects indicated that changes in cell size (‘shrinkage’) occur at lower doses in individuals with PWK haplotypes compared to those with NZO haplotypes (Fig. 4E,4F). Taken together, these data support a model where sensitivity to arsenic exposure in the DO population is partly regulated by natural variation in the efficiency of MMAIII detoxification.
Xrcc2 haplotypes modulate and predict of cellular responses
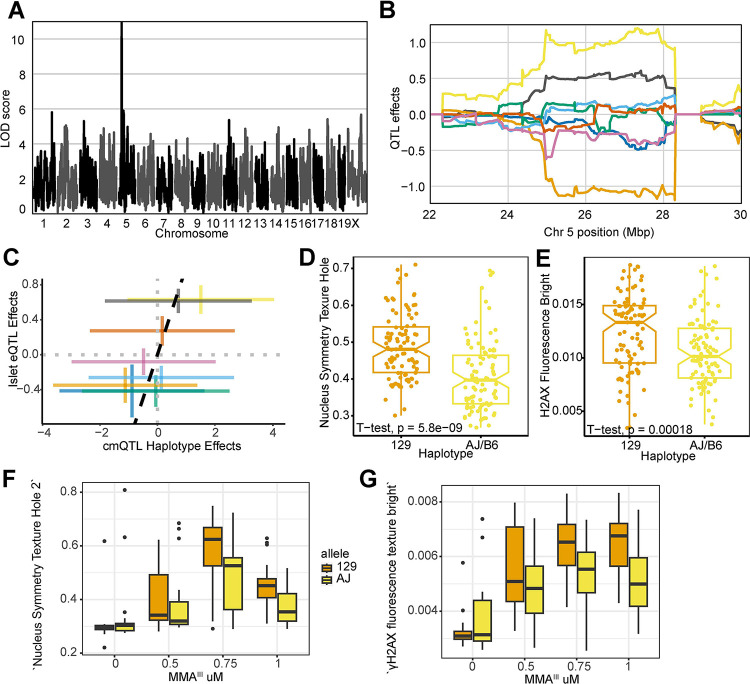
The cmQTL with the highest LOD score was on chromosome 5 at 27,327,254 bp (GRCm38) for the response cmQTL ‘EC90 Nonborder Nucleus Symmetry 02 SER Hole (Hoechst) Mean Per Well’ (Fig. 5A, 5D). Hoechst nuclear fluorescence in cells with the 129 haplotype resembled apoptotic nuclei 71 and were brighter and more uniform than those found in cells with AJ/B6 haplotypes (Fig. 5B, Fig. S1A). The highest associated SNPs for this cmQTL were located in two genes: Actr3b and Xrcc2 (Fig. S1B), however several key points suggest Xrcc2 as the more likely candidate. First, Xrcc2’s paralogs, Xrcc1 72,73 and Xrcc3 74,75 have both been associated with genetic susceptibility to arsenical exposure. Second, knockdowns of Xrcc2 were previously shown to increase both γH2AX intensity and chromosomal abnormalities 76, and Xrcc2 is a member of the Biological Fibroblast Apoptosis (GO:0044346) and DNA Damage Repair pathways (R-MMU-5693532). Lastly, the cmQTL allele effects are highly correlated with an Xrcc2 eQTL in pancreatic islets cells from the same mouse population (Fig. 5C). Taken together, these results suggested that genetic variation at this locus may be mediating DNA damage-induced apoptosis through Xrcc2 expression.
Figure 5: Xrcc2 haplotype modulates chromosomal organization and DNA damage during acute MMAIII exposure.
(A) QTL scan for the ‘EC90 Hoechst Nucleus Symmetry (02) Hole Mean per Well’ cmQTL with the maximum peak at chromosome 5: 27327254 bp(m38) and a LOD score of 10.95.
(B) Allele effects plot showing the eight DO founders (colors, see Methods) for the ‘EC90 Hoechst Nucleus Symmetry (Hoechst) Hole Mean per Well’ cmQTL across the surrounding region on chromosome 5 (Mbp). Colors indicate founder mouse strains: A/J (yellow), C57BL/6J (gray), 129S1/SvImJ (orange), NOD/ShiLtJ (dark blue), NZO/HILtJ (light blue), CAST/EiJ (green), PWK/PhJ (red), and WSB/EiJ (purple)
(C) Pairwise correlation of the haplotype effects of Xrcc2 expression in pancreatic islet cells at chromsome 5:27,327,254 bp (GRCm38) compared to the haplotype effects of ‘EC90 Hoechst Nucleus Symmetry (Hoechst) Hole Mean per Well’. Colors are the same as panel B.
(D) Boxplot showing the significant difference (t-test, p value = 5.8e-9) in ‘Nucleus Symmetry Texture Hole 2’ at 1 μM MMAIII for the top 129 (n = 24; orange) and AJ/B6 (n = 24; yellow) haplotypes in the DO fibroblasts.
(E) Boxplot showing the significant difference (t-test, p value = .00018) in ‘γH2AX fluorescence texture bright’ at 1 μM MMAIII for the top 129 (n = 24) and AJ/B6 (n = 24) haplotypes in the DO fibroblasts.
(F) Boxplot showing the ‘EC90 Hoechst Nucleus Symmetry (02) Hole Mean per Well’ cellular phenotype in a follow-up experiment where DO fibroblasts with 129 (n = 5; orange) and AJ (n = 5; yellow) haplotypes exposed to increasing MMAIII concentrations.
(G) Boxplot showing the ‘γH2AX fluorescence texture bright’ cellular phenotype in a follow-up experiment where DO fibroblasts with 129 (n = 5) and AJ (n = 5) haplotypes exposed to increasing MMAIII concentrations. Colors indicate the DO founder strains (see Methods).
Because of the role in Xrcc2 in DNA damage and apoptosis, we reasoned that γH2AX fluorescence might also be higher in cells with the more sensitive 129 haplotype compared to cells with the more resistant AJ/B6 haplotypes. Indeed, the γH2AX texture ‘bright’ feature was significantly higher in the fibroblasts with the 129 haplotype compared to the AJ/B6 haplotypes (Fig. 5E, Fig. S1A). We sought to assess the reproducibility of these effects, both for the original phenotype and the increase in γH2AX. Taking advantage of our full panel of 600 cell lines, we selected an orthoganal group of lines based on their haplotype at this locus (n = 5 for each allele). Not only were we able to recreate the original nuclear symmetry difference between genetic backgrounds (Fig. 5F), but we also observed the same γH2AX fluorescence effects that were found in the original screen (Fig. 5G). This example shows that genetic variation in Xrcc2 influences sensitivity and that the haplotype effects of cmQTL have predictive value for identifying sensitive individuals.
Non-coding genetic variation influences TXNRD1 cell fate during induced oxidative stress
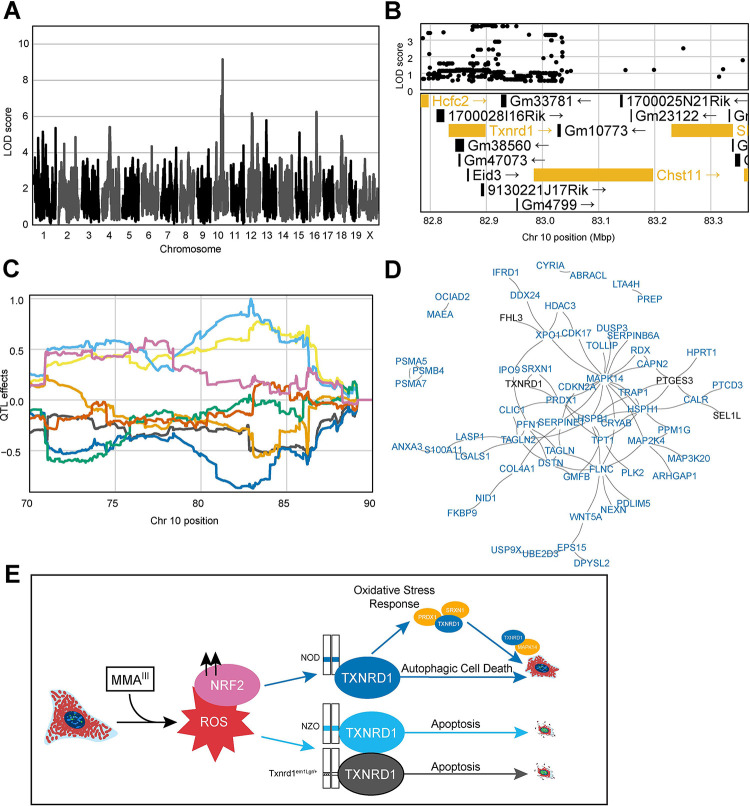
To further investigate how these data could be used for G × E discovery, cmQTL mapping was performed in a subset of cells lacking accumulated DNA damage. Linear classification was performed to separate cells into H2AX positive and negative populations prior to feature extraction. To do this we took advantage of PHENOLogic machine learning algorithms of the Harmony 4.9 software and gated the imaged cells into γH2AX-negative and γH2AX-positive populations prior to feature extraction, dose-response modeling, and mapping. We detected a cmQTL for the rate of MitoTracker area change in γH2AX-negative cells with a LOD score of 9.16 on chromosome 10 (Fig. 6A). This locus was also detected in our original dataset with similar allele effects but with a sub-threshold LOD score (Fig. S2A, S2B, S2C). Upon variant association mapping the highest LOD scoring variants were in the 3’-UTR of the Txnrd1 gene (Fig. 6C), a gene that is highly expressed in fibroblasts and has been previously shown to respond to arsenical exposure via changes in NRF2-mediated expression. Moreover, the reducing capacity of TXNRD1 protein is directly inhibited by MMAIII binding 77,78. As a selenoprotein, the 3’-UTR of Txnrd1 plays a crucial role in recoding a UGA stop codon into a selenocysteine amino acid which is required for function of the TXNRD1 protein as a reducing agent79–81.
Figure 6: Noncoding Variation in Txnrd1 Modulates MMAIII-Induced Cell Death.
(A) QTL scan for the ‘H2AX-negative cells slope Cell Area μm2 mean per well’ cmQTL with the maximum peak at Chromosome 10: 82906780 bp (m38) and a LOD score of 9.16.
(B) Variant association mapping within the Cl the cmQTL ‘H2AX-negative cells slope Cell Area μm2 mean per well’. Top panel shows the LOD scores of the known, segregating variants in the 8 DO founders (GRCm38). Bottom panel shows the gene models within the respective Cl. Each point represents a variant. Colors indicate whether a gene is expressed > 0.5 TPM (gold) or < 0.5 TPM (black). The arrow indicates the direction of transcription.
(C) Allele effects plot showing the eight DO founders (colors, see Methods) for the ‘H2AX-negative cells slope Cell Area μm2 mean per well’ cmQTL across the surrounding region on chromosome 10 (Mbp).
(D) String-db functional enrichment network of the significantly increased protein interactors detected using immunoprecipitation mass spectrometry (IP-MS) in DO fibroblasts with NOD alleles (n = 6) at the maximum locus for the ‘H2AX-negative cells slope Cell Area μm2 mean per well’ cmQTL exposed to 0 and 0.75 μM MMAIII concentrations. Colors indicate whether a protein, or node, was shared with a similar experiment in DO fibroblasts with the NZO allele (n = 5). Black represents shared TXNRD1 interactors, and blue represents unique NOD-TXNRD1 interactors.
(E) Mechanistic summary of allele-specific Txnrd1 responses across the NOD haplotype (blue), NZO haplotype (light blue), and heterozygous SECIS knockout model (Txnrd1em1Lgr/+). Our data suggest DO fibroblasts with the NOD allele have a more robust oxidative stress response upon MMAIII exposure, ultimately succumbing to autophagic cell death represented by increased cell size at medium MMAIII concentrations. In comparison, DO fibroblasts with the NZO allele or the Sec+/− alleles undergo a more apoptotic cell fate as shown by brighter Hoechst 33342 labeling and smaller cells.
To interrogate the plausibility of Txnrd1 as the candidate for these two cmQTL, we performed score-based GSEA using gene expression data from cell lines selected from our collection of 600 lines on the basis of their sensitive (NZO) and resistant (NOD) haplotypes at this locus. We found upregulation of DNA damage and replicative stress gene sets in cells with NZO haplotypes and upregulation of oxidative stress response, p38/MAPK signaling, TGF signaling, RAS signaling, lysosome, and autophagy-related pathways in cells with NOD haplotypes (Supp. Table 4). Among these pathways was nanoparticle triggered autophagic cell death, which can be induced by the treatment of gold, the active component of the TXNRD1 inhibitor auranophin 82. While we didn’t detect a significant difference in Txnrd1 transcript abundance by haplotype, at either concentration (Supp. Table 5), there was a significant difference in protein levels in the unexposed cells (Fig. S2D), and, as expected, TXNRD1 protein levels increased in all arsenic exposed cells. To assess whether TXNRD1 had haplotype specific protein interactions, we performed immunoprecipitation followed by tandem mass spectrometry (IP-MS). Following subtraction of a non-specific binding partner control, we found that compared to healthy, unexposed controls, 0.75 μM MMAIII exposed NOD haplotype cells (n = 6) had a larger number (106) of significant, positive interactors compared to NZO (n=5) TXNRD1 interactors (33). NOD TXNRD1 interacted with proteins involved in oxidative stress (i.e., PRDX1, SRXN1), autophagy/p38 (i.e., MAPK14, TOLLIP), and TP53 related REACTOME pathways, while the NZO TXNRD1 interactors did not show pathway enrichment (Fig. 6D, Supp. Table 6). Considering the gene expression and IP-MS data together, it was evident that in exposed DO fibroblasts, NOD TXNRD1 was involved in autophagy while NZO TXNRD1 was associated with apoptosis. Previous studies of Txnrd1 deficiency have shown disruption of lysosomal-autophagy in favor of apoptotic cell death 83,84, implying that the apoptotic phenotype of cells with NZO haplotypes (NZO-TXNRD1) is akin to that seen with TXNRD1 deficiency. During apoptotic cell death, cell structure and cytoskeleton are quickly degraded, but during autophagy the cytoskeleton is maintained 85–87; providing a basis for our ability to distinguish between these two pathways and to interrogate their genetic regulation using cmQTL. Taken together, these data support a model whereby natural variation in Txnrd1 influences the trajectory of cell death pathways following MMAIII exposure in the DO population (Fig. 6E).
While we did not find coding variants unique to the NZO or NOD Txnrd1 gene, we found that two SNPs private to the NZO haplotype (rs227869362 and rs257393906) in the 3’-UTR were adjacent to the selenocysteine insertion element (SECIS), which is essential for Sec recoding during translation. We also searched publicly available data for structural variants and INDELs in the 3’ UTR but did not find any that were unique to the NZO haplotype 88. To determine the essentiality of this element in vivo, we used CRISPR/cas9 to delete the SECIS in C57BL/6J mice (Txnrd1em1Lgr). While heterozygous mice carrying this deletion were viable and fertile, homozygous mice could not be recovered. Since a full protein knockout of Txndr1 causes recessive embryonic lethality 89, we concluded that deletion of the SECIS element alone is the functional equivalent of a null allele (see Methods). We then isolated tail tip fibroblasts from heterozygous mice and found that the cell area of arsenic exposed Txnrd1em1Lgr/+ fibroblasts more closely resembled fibroblasts with the NZO haplotype than their WT controls (Fig. S2E, S2F). Similarly, nuclear Hoechst 33342 labeling was brighter and more uniform in the Txnrd1em1Lgr/+ nuclei with increasing MMAIII concentration. Taken together, these data highlight the functional importance of non-coding variation in the 3’ UTR of a key selenoprotein in the context of sensitivity to arsenic induced oxidative stress. Detailed molecular and functional studies are needed to determine the impact of single nucleotide variants on sec recoding in Txnrd1. However, there is at least one study demonstrating that naturally occurring and engineered single nucleotide variants in the 3’ UTR of the human selenoprotein, SEP15, influence UGA readthrough and dampen the cellular response to selenium stimulation 90.
Natural genetic variation influences fibroblast morphology
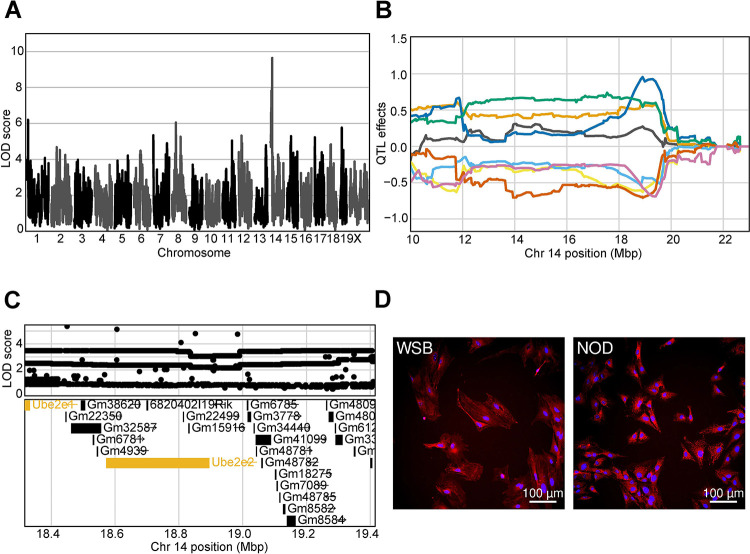
While our primary focus was on population variation in arsenic response, we unexpectedly observed variation in fibroblast morphology in unexposed cells and our genetic analysis revealed multiple loci contributing to this baseline morphological variation (i.e. starting asymptote cmQTL). The highest scoring of these baseline cmQTL (LOD 9.64) was on proximal chromosome 14 (Fig. 7A, Fig. 7b). Several of the top LOD scoring variants were in Ube2e2, which was one of only three protein coding genes expressed in fibroblasts within the confidence interval (Fig. 7C). This cmQTL is for a trait that describes the brightness of Hoechst labeling (i.e., texture feature bright 1 pixel mean per well) which is directly related to the distribution and amount of chromatin in the nucleus (Fig. 7D) 91. The ubiquitin conjugating enzyme E2 (UBE2E2) functions in the nucleus to post-translationally modify proteins that regulate the G1/S phase transition together with Trim28 92, which could explain the difference in Hoechst labeling as mitotic cells accumulate more Hoechst due to their DNA content. This example highlights the role of genetic variation in the regulation of morphology, potentially through variation in basic cellular functions (i.e. cell cycle) providing an exciting avenue for further study.
Figure 7: Genetic variation influences fibroblast morphology at baseline.
(A) QTL scan for the ‘Hoechst 33342 texture bright 1 pixel mean per well’ cmQTL with the maximum peak at chromosome 14: 19401644 bp (GRCm38) and a LOD score of 9.64.
(B) Allele effects plot showing the eight DO founders (colors, see Methods) for the ‘Hoechst 33342 texture bright 1 pixel mean per well’ cmQTL across the surrounding region on chromosome 14 (Mbp).
(C) Variant association mapping within the Cl the cmQTL ‘Hoechst 33342 texture bright 1 pixel mean per well’. Top panel shows the LOD scores of the known, segregating variants in the 8 DO founders (m38). Bottom panel shows the gene models within the respective Cl. Each point represents a variant. Colors indicate whether a gene is expressed > 0.5 TPM (gold) or < 0.5 TPM (black). The arrow indicates the direction of transcription.
(D) Representative images for the two fibroblast lines showing higher Hoechst 33342 texture bright in the sample with the NOD allele at the chromsome 14 locus comapred to the WSB. Nuclei are labeled in blue by Hoechst 33342 labeling and mitochondria are labeled in red by MitoTracker Deep Red. Scale bar indicates 100 μm.
Discussion
Taking advantage of a laboratory mouse genetic reference population, we created a new population-based cellular model for in vitro analysis of gene by environment interactions. Using this model, we performed HCS to quantify morphological cellular features associated with acute MMAIII exposure. We found quantitative variation in these traits across the cell population, and we also found significant variation in the degree to which genetic background could be attributed to this variation (0–40%). We also found significant unexplained residual variation, although the proportion of this contributor to overall variation also varied substantially by trait. Previous studies of cell morphology in genetically diverse cell populations have shown that some traits are prone to high measurement error or experimental variability, especially for features that have high cell to cell variability 1. Since our features are whole well summaries, cell to cell variability is a major contributor to our observed residual variation. We also found that the features with higher residual variation were enriched for γH2AX features and that the mean variance ratio for these features was high compared to the overall mean (0.65 vs. 0.4). This higher residual variance is likely due to the indirect immunolabeling method used for γH2AX detection, which is a multistep staining method that relies on two antibodies and is known to have more experimental variability than direct organelle probes.
We used dose-response modeling to summarize cell morphology changes to increasing MMAIII insult from which we extracted dose-response parameters (DRPs) as latent traits for QTL mapping. However, there are several notable caveats to this approach. First, to induce cell morphology changes that were likely to fit a sigmoidal dose-response curve, we used concentrations of MMAIII that are unlikely to be encountered through environmental or occupational exposures. Other studies have shown that cell morphology was impacted following lower concentration, longer exposures of arsenic 93. Secondly, covariates or Bayesian regression during dose-response modeling could allow for better handling of batch effects in high-content imaging data, however these options were not available in the commonly used drc R package at the time of our study. Lastly, dose-response modeling varies based on the software being used, the model being fit, and as we observed, the genetic background of the samples. Despite these challenges, we identified hundreds of loci where natural genetic variation in the DO founder strains influences the fibroblast responses to MMAIII and baseline fibroblast morphology.
One feature of non-molecular QTL is that while they capture variants with a range of molecular effects (transcriptional or post-transcriptional) they lack a genomic reference point. Thus, a QTL can result from coding variants, noncoding variants, or a combination of both which may influence a cellular trait through a single gene, or multiple, within a QTL region. To refine our cmQTLs and identify candidates, we integrated variant association analysis with orthogonal datasets including gene expression, molecular QTL data from previous DO studies, pathway information, and gene-chemical interaction data from arsenicals through CTD (ctdbase.org). Based on gene-arsenical interactions, we identified 88 genes in our cmQTL that were previously associated susceptibility to arsenic (https://ctdbase.org/). Six genes within our cmQTL including Abcc4, Nfe2l2, Cbs, Gclc, Gstm1, and Xpc contain SNPs affecting the response to As (https://ctdbase.org/). Abcc4 was among the significantly differentially expressed genes fibroblasts which make it an intriguing candidate for the EC5 of the change in axial small length cmQTL. Variants in the 3’ UTR of Abcc4 can regulate its expression through impacting miRNA binding 94. We speculate that unique variants in NZO (rs240728821) and PWK (rs245333533) may be acting in a similar manner. In addition to Abcc4, we found 70 novel gene expression changes based on available MMAIII exposure within CTD.
Like Abcc4, Txnrd1 also has an extensive list of gene-arsenical associations in CTD which provides even greater support for the use of ML during image analysis. The ML-derived cell feature, slope H2AX-negative cell area mm2, was further corroborated by its presence in the original dataset and by CRISPR-deleted SECIS element in the 3’ UTR of Txnrd1 recapitulating the same effect. The essentiality of the SECIS element for sec recoding has been previously demonstrated81. Our breeding data further support the essentiality of this element for fetal development and our genetic data show that in the 3’ UTR of Txnrd1 influences the cell size during acute MMAIII exposure. The gene expression differences between haplotypes at this locus showed more pro-cancer signaling including RAS, TGF, and p38/MAPK signaling in the NOD haplotype compared to the NZO. This coincides with protein interaction data showing increased NOD TXNRD1 affinity for MAPK14 and oxidative stress related proteins compared to NZO, which may explain the resistance to MMAIII-induced morphology changes. Xrcc2’s involvement in the DNA damage pathway may also indicate a cancer-related outcome for the highest cmQTL ‘EC90 Nonborder Nucleus Symmetry 02 SER Hole (Hoechst) Mean Per Well’. This cmQTL region shares conserved synteny with a region significantly associated with susceptibility to arsenic-induced skin lesions in a Bangladeshi population 95.
Fibroblasts are found in many tissues and are involved in disease progression 96. However, the genetic effects in fibroblasts may not recapitulate the same molecular mechanisms of sensitivity and resistance as those found in highly specialized cell types. Primary fibroblast cells are also a limited resource because they will undergo senescence, and they are more difficult to genetically manipulate than pluripotent cells. For these reasons, we have generated induced pluripotent stem cell (iPSCs; n = 284) from this panel for future work. iPSCs also enable differentiation into other cell types, 3-dimensional cell models, organoids, or scaffolded arrays which can be screened across a variety of environmental conditions including other toxicants, drugs, or other culture conditions. It is important to note that while other studies mapping cmQTL were limited by lack of genetic diversity, poor adaptation of some cell types to culture, and the genetic architecture of the population being studied 2,97, we also found that beyond large effect QTL, our study was underpowered despite previous examples showing sample sizes in this range for molecular phenotypes can detect strong QTL 12,54,98. This is the result of experimental and residual sources of variance as described above, as well as the limited extensibility of standard dose-response models to diverse populations. In conclusion, our study demonstrates that dynamic changes in cell morphology ocurring in a population of exposed, genetically diverse cells exhibit predictable dose response relationships. These relationships display interindivual variation and genetic mapping of these relationships unveils the genetic regulation of the molecular initiating events that occur during an acute exposure. Our findings indicate that these loci and their haplotype effects have predictive value for identifying sensitive and resilient individuals in vitro. While further work is needed to explore the applicability of these predictions to in vivo responses, leveraging mouse genetic reference populations presents an exciting opportunity for iterative in vitro screening and precise in vivo testing in matched genetic backgrounds.
Materials and Methods
Fibroblast Derivation
Tail biopsies approximately 2–3 mm were harvested in from adult male and female Diversity Outbred (RRID:IMSR_JAX:009376) mice, aged approximately 4–6 weeks, using a procedure approved by The Jackson Laboratory’s Institutional Animal Care and Use Committee. Samples were initially collected into Advanced RPMI 1640 cell culture media supplemented with 1.0 % Penicillin Streptomycin (P/S), 1.0 % Glutamax-I (Glutamax), 1.0 % MEM Non-Essential Amino Acids (NEAAs), 0.0005% 2-mercaptoethanol (BME). Tail tissue was minced using razor blades and digested with media containing collagenase D at a concentration of 2.5 mg/ml on an orbital shaker at 37°C. The digested samples were further minced using micropipettes ranging from p1000 to p200 and dissociated in RPMI 1640 media containing 1.0 % P/S, 1% Glutamax, 1.0 % non-essential amino acids, .0005% BME, and 10% fetal bovine serum (FBS), hereinafter referred to as fibroblast media, for approximately 3–5 days (passage number 0; P0). All passaging was done using a phosphate buffered saline pH 7.2 (1X; PBS) wash and 0.05% Trypsin-EDTA (Trypsin). Individual Diversity Outbred fibroblast samples were expanded to P5 with reserve samples frozen at approximate densities of 3.5 × 105 cells/ml at passage numbers P2, P3, and P5 in freeze media containing RPMI 1640 with 10% dimethyl sulfoxide (DMSO) and 10% FBS. All DO fibroblast samples were transferred to liquid nitrogen holding tanks for long-term storage after 24 – 48 hours at −80C.
DNA was harvested from spleen tissue for each DO mouse and samples were genotyped using the Giga Mouse Universal Genotyping Array (GigaMUGA; 99). Haplotypes were reconstructed according to the protocol described previously which uses a hidden Markov model to estimate genotype probabilities at each locus for the population 100.
Sample Preparation
Frozen aliquots of P5 fibroblast lines were thawed in fibroblast media and grown for 48 hours in 60 mm tissue culture-treated plates. Viable cell densities were estimated using Trypan Blue (0.4%; Gibco) and a Nexcelom Cellometer Auto T4 Plus Cell Counter. 100 μl of each fibroblast line was seeded into 4 total columns (4 technical replicates) distributed across two CellCarrier Ultra 96-well black, clear bottom, tissue culture treated microplates (PerkinElmer) using the Integra Assist Plus (Integra Biosciences) at a density of ~2500 viable cells/well following randomization across columns. After 24 hours, fibroblast media was replaced by monomethylarsonous acid (MMAIII; Toronto Research Chemicals) containing 100 μL of fibroblast media at concentrations of 0 μM, 0.01 μM, 0.1 μM, 0.75 μM, 1.0 μM, 1.25 μM, 2.0 μM, and 5.0 μM in each row which was randomized across plates.
Following 24-hour exposure, MMAIII media was replaced with MitoTracker Deep Red (200 nM; Invitrogen) containing media and incubated at 37°C for 20 minutes in the 96-well plates. Subsequently, cells were fixed on ice using ice-cold 100 % methanol for 10-minutes. Following 3X PBS washing, cells were bathed in a 1.0 % bovine serum albumin (Fraction V) (BSA), 0.1 % Tween solution overnight at 4°C on a shaker. After ~24 hours, blocking solution was replaced with anti-gamma γH2AX antibody (Abcam, ab11174, 1:2000) in blocking solution and incubated at room temperature for 2 hours on a shaker. Following 3X PBS wash, Alexafluor 488 donkey anti-rabbit secondary antibody (1:2000; Abcam) was added for 1 hour at RT on the shaker. After washing, Hoechst 33342 (1:8000; Abcam), was added to cells and incubated for 10 minutes at RT on the shaker. Plates were subsequently washed, and 100 PBS of media was left in each well for storage at 4°C and imaging.
Automated Image Acquisition
96-well microplates were imaged confocally using an Operetta CLS or Opera Phenix (Fig. S2E,F) equipped with a 20x/1.0 water immersion objective and binning 2. A single z-plane was acquired from 25 contiguous fields per well. Exposure times, focal heights, and excitation power settings for the Operetta CLS screen were: Hoechst 33342 (time: 100 ms, power: 100, height: −5), Alexa 488 (time: 200 ms, power: 100, height: −5), MitoTracker Deep Red (time: 500 ms, power: 100, height: −5). Exposure times, focal heights, and excitation power settings for the Xrcc2 follow-up experiments were: Hoechst 33342 (time: 300 ms, power: 100, height: −6), Alexa 488 (time: 80 ms, power: 100, height: −6), MitoTracker Deep Red (time: 200 ms, power: 100, height: −6). Lastly, exposure times, focal heights, and excitation power settings for the Txnrd1 follow-up experiments were: Hoechst 33342 (time: 100 ms, power: 80, height: −10) and MitoTracker Deep Red (time: 40 ms, power: 50, height: −10).
Image Analysis / Cellular Segmentation
‘Basic’ flatfield corrected images were analyzed and processed using Harmony 4.9 software with PhenoLOGIC (PerkinElmer). Gaussian smoothed images were used for image segmentation, with a focus on 2 main regions of interest (ROIs) including using Hoechst 33342 to define the nucleus, and MitoTracker Deep Red to define the cytoplasm surrounding each nuclear ROI. Fluorescence patterning (i.e. texture) and intensity were measured in the nuclear and cytoplasmic regions using the Hoechst 33342 and MitoTracker Deep Red/MitoTracker Deep Red Gaussian smoothed channels, respectively. Features including nuclear area, Hoechst 33342 intensity, and nucleus edge texture were extracted and represented as mean +/− SD per well.
The second image analysis approach used the PhenoLOGIC machine learning (PerkinElmer) algorithms in the Harmony 4.9 software define sub-populations of cells based on γH2AX/Alexa-488 secondary labeling (γH2AX positive and γH2AX-negative) and MitoTracker Deep Red (stressed and unstressed) prior to feature extraction to generate features including ‘MitoTracker Cell Area in γH2AX negative cells’.
Feature Variance and Relatedness
Principal components analysis was performed on the image analysis features across all concentrations, individuals, and plates using the ‘pca’ function from the R pcaMethods with the option ‘scale = “uv”’. Variance component analysis was performed using the ‘lmer’ function from the R package lme4. The sources of variation included in the model were sex, DO generation (‘generation’), DO donor (‘individual’), 96-well plate (‘plate’), and run (See Equation 1). Variance components were extracted from the model using the function ‘VarCorr’ for each of the random effect (generation, sex, individual, and plate). Residual variance was extracted as the sigma from the model summaries. Ratios of the variance components were determined by dividing each variance component by the sum of all the variance components and the residual variance.
| Equation 1 |
Lastly, the pairwise correlation structure of these data was calculated using the ‘cor’ function in the WGCNA R package with the option ‘use = “pairwise.complete.obs”’. The heatmap was created using the ComplexHeatmap R package with the dendrogram added using the ‘column_split’ and ‘row_split’ options each set to 5. We added terms to the heatmap clusters based on a qualitative examination of the clustered trait names.
Cellular Feature Dose-Response Modeling
We used the drc R package 52 to perform dose-response modeling for each of (insert total number) cellular features. For each of (how many) individuals, we fit 4 technical replicates to the four-parameter log-logistic dose-response model (see Equation 2) using the ‘drm’ function with the ‘fct’ set to ‘LL.4’ and log-normalized cellular features using the ‘bcVal = 0’ option. Model parameters, as shown in Equation 2 52 where represents concentration, including slopes (), upper asymptotes (), lower asymptotes (), and EC50’s () were extracted from the summary of the model fits. Additionally, the ‘ED’ function was used to estimate the EC5, EC10, EC25, EC75, and EC90 for each model fit ‘relative’ to the asymptotes. 4 replicates for each model fit parameter summary were estimated for each DO individual and cellular feature.
| Equation 2 |
LMM / BLUP Estimation
Samples were analyzed on different days, across many 96-well plates, and multiple MMAIII exposures. We summarized the dose-response parameter replicates using Equation 3 accounting for each individual and plate as random effects. We adjusted for potential batch effects across DO progenitors’ concentration response parameters using linear mixed effect models (LMM). We fit the LMM using the ‘lmer’ function from the R package lme4. We modeled each cellular feature as where is the dose-response parameter estimate for a given cellular feature for DO progenitor , modeled with varying intercepts through random effects for mouse/progenitor and 96-well plate is the random error term, assumed to , and is the error variance. Data without the effect of plate were extracted as the best linear unbiased predictors (BLUPs) of the random effect for DO progenitors and used for QTL mapping analysis.
| Equation 3 |
Cellular Feature QTL Mapping
All data were converted to the normal quantiles calculated from the ranked data, i.e., the rank-based inverse normal transformation (rankZ) to force a Gaussian distribution for mapping. QTL mapping was performed using the qtl2 R package. Briefly, a genetic relationship matrix (i.e., kinship matrix) was calculated from the genotype probabilities using the ‘calc_kinship’ function with the ‘leave one chromosome out’ (loco) option for genetic mapping and the “overall” option for heritability estimation. Sex and DO generation were included as covariates following One hot encoding in the LMM for both heritability estimation and QTL mapping.
For QTL mapping, we first tested individual loci spanning the genome for association with each cellular feature (using qtl2’s ‘scan1’ function). We then estimated allele effects at detected QTL as BLUPs (using the ‘scan1blups’ function) to identify the parental haplotypes driving each QTL and their respective directionality. SNP-association mapping was performed using the ‘scan1snps’ function and the known variants across the eight founder strains of the DO. We calculated a genome-wide false discovery rate (FDR = .10) using the permutations (n = 1000) for the ‘EC50 number of nucleì trait as simulated permutations for all 5105 cmDRPs mapped.
Diversity Outbred Fibroblast MMA RNA-seq preparation
32 cell lines, including those with NOD or NZO haplotypes at Chr10:82.89 (GRCm38) were thawed into 60 mm cell culture treated plates and grown to confluency (≥ 0.8 × 106 cells/ml) in DO media. Each cell line was then passaged equally into 2 60 mm cell culture dishes and grown to 75% confluency upon which 1 60 mm dish received 0.75 μM MMAIII containing DO media and 1 60 mm dish received standard DO media. Following 24-hr exposure, both treated and untreated samples were independently collected and snap frozen on dry ice as cell pellets for 15 minutes. Samples were stored at −80°C prior to RNA isolation. RNA was extracted using a NucleoMag RNA Kit (Macherey Nagel) and purified with a KingFisher Flex system (ThermoFisher). Library preparation was enriched for polyA containing mRNA using the KAPA mRNA HyperPrep Kit (Rocher Sequencing and Life Science). Paired end sequencing was performed with a read-length of 150 bp on an Illumina NovaSeq 6000.
Transcriptomic Profiling
Genotypes for each sample were then reconstructed using the genotype by RNA-seq pipeline (GBRS) and aligned to the 8 founder allele-specific genome using GBRS RNA-seq pipeline to quantify read counts for each gene 101 (available through Github at TheJacksonLaboratory/gbrs_nextflow. These expected counts were the input for differential expression between the 0 and 0.75 μM exposures using the R package DEseq2 102. We then used the fgsea R package to perform a score-based gene set enrichment analysis 103. The input for GSEA was the exposure-based log2 fold-change for each gene normalized by its standard error. Gene Ontology (GO), REACTOME, WikiPathways, and Biocarta genesets for mus musculus were obtained via the R package msigdb 104. Additionally, the R package ClusterProfiler was to assess enrichment of the significant differentially expressed gene set based on the outlying alleles for the cmQTL on chromosome 10 (GRCm38) 105.
CTD Database Mining
The Comparative Toxicogenomics database (CTD) was used to identify gene-arsenic interactions previously defined for candidate genes within cmQTL CIs. The gene-arsenic interactions were downloaded for these arsenicals: monomethylarsonic acid (MMAV), monomethylarsonous acid (MMAIII), dimethylarsinic acid (DMAV), dimethylarsinous acid (DMAIII), arsenic trioxide (ATO), sodium arsenite, sodium arsenate, and elemental arsenic (As). NCBI gene ID’s were then merged to Ensembl IDs and their mouse orthologs obtained through Ensembl’s BioMart tool 106. We aggregated the number of ‘Interactions’ for each gene across the arsenicals to get an ‘Interaction Count’ for the genes within cmQTL CIs.
TXNRD1 Relative Abundance
DO fibroblasts were selected based on their genotypes at the Txnrd1 locus representing 6 NOD, 5 NZO, and 4 NOD/NZO haplotypes balanced for both male and female lines. Each line was split into two 60 mm dishes where one 60 mm plate received 0 μM MMAIII containing media (unexposed) while the other contained 0.75 μM MMAIII containing media. After 24 hours, cell pellets split into two vials and snap frozen on dry ice for further processing and liquid chromatography tandem MS (LC-MS/MS) analyses. Protein pellets were resuspended in 150 uL of 50 mM HEPES, pH 7.4, and lysed by passing through a syringe with 28 gauge needle (10 passes), vortexing for 30 seconds, and waterbath sonicating for 5 minutes (30 seconds on, 30 seconds off). Lysates were then clarified via centrifugation at 21,000 × g for 10 minutes at 4°C. Clarified lysates were quantified using a microBCA assay and 20 μg samples were diluted to 50 uL for digestion in 50 mM HEPES, pH 8.2. Samples were then reduced with 10 mM DTT at 37°C for 30 minutes, alkylated with 15 mM IAA at room temperature in the dark for 20 minutes, and trypsin digested overnight at 37°C (trypsin:protein ratio of 1:50). Samples were then cleaned-up using Millipore P10 zip-tips, dried in a vacuum centrifuge, reconstituted in 20 μL of 98% water/2% ACN with 0.1% formic acid, and transferred to mass spec vials. Each sample was analyzed using Thermo Eclipse Tribrid Orbitrap Mass Spectrometer coupled to a nano-flow UltiMate 3000 chromatography system on a Thermo 50 cm EasySpray C18 column as described previously with the exception that the gradient was scaled down to a 90 minute gradient107. TXNRD1 abundance was determined based on the target peptide: IEQIEAGTPGR. Raw peak data was processed using Skyline (version 22.2.1.278) and further analyzed in R. Significance across alleles and concentrations was assessed using permutations (n = 1000) because of the non-normal distributions of the protein levels. All mass spectrometry analysis was performed in the in The Jackson Laboraory (JAX) Mass Spectrometry and Protein Chemistry Service.
Immunoprecipitation Mass Spectrometry (IP-MS)
Immunoprecipitation mass spectrometry (IP-MS) was performed using a rabbit antibody derived against the mouse TXNRD1 protein gifted from Dr. Edward Schmidt to determine TXNRD1 binding partners using the samples and instrumentation described in the ‘TXNRD1 Relative Abundance’ section. M-280 Sheep Anti-Rabbit IgG Dynabeads (Invitrogen, 11203D) were prepared and coupled to the rabbit anti-mouse TXNRD1 antibody according to manufacturer protocol; additional IgG control beads with no TXNRD1 were also prepared as a non-specific binding partner control for the beads. A ratio of 5 ug of antibody to 5 × 107 beads was used. All Dynabeads were then blocked with 5 mg/mL BSA overnight at 4°C during the antibody coupling step. Coupled and control IgG Dynabeads were then bound to 250 μg of protein lysate at room temperature with rotation for one hour. Heterozygous samples were pooled and used as IgG subtractive controls to assess non-specific binding for the beads. All bound bead fractions were clarified with a magnet, then washed three times with Wash Buffer A (10 mM HEPES at pH7.4, 10 mM KCl, 50 mM NaCl, 1 mM MgCl2, NP-40 (0.05% w/v)), followed by two washes with Wash Buffer B (10 mM HEPES at pH7.4, 10 mM KCl, NP-40 (0.05% w/v)). Washed beads were then digested on-bead as described for the relative abundance section above with the exception of 500 ng of trypsin being used. Samples were then purified using a Millipore P10 Zip-tip and prepped for tandem mass spectrometry analysis, both as described above in the relative abundance section. Raw data was analyzed using the Thermo Proteomic Discoverer software as described previously in the JAX Mass Spectrometry and Protein Chemistry Service using standard operating protocols 107.
PPI and Functional Enrichment
We used the string_db R package to assess functional enrichment of proteins binding TXNRD1 to generate protein-protein interaction (PPI) networks for the allele-specific IP-MS results 108. We used a score threshold of ‘400’ to identify functional interactions between TXNRD1 interacting proteins (nodes) across NOD and NZO haplotypes at the chromosome 10 locus which were indicated as edges in the igraph R package visualization. The PPI was colored based on shared (black) and unique (blue) proteins across alleles.
Txnrd1 SECIS deletion
To delete a 200 bp domain containing the SECIS regulatory element of Txnrd1 (MGI:1354175, NCBI Gene: 50493, ENSMUSG00000020250) as well as the flanking regions where 3’ UTR variants are found in NZO haplotypes, we engineered C57BL/6J (The Jackson Laboratory stock #000664, RRID:IMSR_JAX:000664RRID:JAX000664) embryos using CRISPR/Cas9. The SECIS element of murine Txnrd1 is a 75 bp regulatory element ranging from 1967–2042 bp in NM_001042513.1, essential for recoding UGA to specify selenocysteine. Two sets of gRNAs were used (gRNA up 1:GGAGGCTGCAGCATCGCACT, gRNA down 1: GGGTTAATGATACTAGAGAT, gRNA up 2: GAGGCTGCAGCATCGCACTG, gRNA down 2: GGTTAATGATACTAGAGATA) with no repair template. Off-target effects were assessed using the Benchling algorithm (https://benchling.org) and for all guides, potential off target sites were scored <2.0. Two F0 founders (male 5007 and female 5016) carrying the expected 220 bp deletion at chr10:82,896,230–82,896,450 (GRCm38) were identified by PCR. PCR genotyping primers were designed to amplify a 565 bp WT product and a 365 bp deletion product (SECIS_500_FWD 5’ CCTTCCTCTTT CTGCAGATATT 3’, SECIS_500_REV 5’ ACC CAC TTCCACACAGTAAAG 3’). Male founder 5007 was backcrossed to C57BL/6J females and PCR genotyping (primers) was used to identify N1 heterozygous offspring. After two more backcrosses N3 animals were intercrossed to generate N3F1 and N3F2 animals for phenotyping and tail tip fibroblast biopsy. The heterozygous crosses resulted in 320 animals, 211 animals were heterozygous (66%), 109 were wildtype (34%) and 0 were homozygous for the deletion allele. This 2:1 Mendelian ratio (het:WT) was consistent with recessive embryonic lethality of the deletion allele. Targeted oxford nanopore sequencing of the was used to confirm the sequence of the deletion allele and the lack of closely linked off target mutations in the Txnrd1 gene. The resulting strain C57BL/6J-Txnrd1em1Lgr/Lgr was assigned The Jackson Laboratory stock #37668. All experiments using mice were approved by The Jackson Laboratory’s Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgements
We thank Drs. Edward Schmidt and Dr. Justin Prigge, Montana State University for providing the TXNRD1 antibody. We also acknowledge the support of The Jackson Laboratory Mass Spectrometry and Protein Chemistry Service, Protein Sciences, The Jackson Laboratory Genome Technologies Core, and Jackson Laboratory Computational Sciences for their expert assistance. We also thank Dr. Belinda Cornes and Robert Sellers for their computational support during the early stages of this project. Lastly, we thank Dr. Stephen Straub at PerkinElmer for his support including reviewing this manuscript.
Funding
This work was funding by the National Institutes of Health, National Institute of Environmental Health Sciences, R01ES029916 (L.G.R., G.C., and R.K.) and by the NIH Office of Research Infrastructure Programs, Division of Comparative Medicine, P40 OD011102 (L.G.R.). The mass spectrometry-based proteomics was performed at The Jackson Laboratory utilizing a Thermo Eclipse Tribrid Orbitrap mass spectrometer obtained through NIH S10 award (S10 OD026816). Research reported in this publication was partially supported by the National Cancer Institute under award number P30CA034196.
Footnotes
Conflicts of Interest
None to disclose.
Data and Code Availability
All statistical analyses were performed using the R statistical programming language (v4.1.3)109. The data, supplemental tables, and analysis pipelines used to process, analyze, report, and visualize these findings are publicly available (10.6084/m9.figshare.24576181). The raw and processed RNA-seq data are available from Gene Expression Omnibus (GEO) (GSE247877). All images are available from the corresponding authors (C.O.,L.R.) upon reasonable request.
References
- 1.Nogami S., Ohya Y., and Yvert G. (2007). Genetic complexity and quantitative trait loci mapping of yeast morphological traits. PLoS Genet 3, e31. 10.1371/journal.pgen.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthew T., Jatin A., Samira A., Beth A.C., Emily P., Dhara L., Gregory W., Erin W., Aparna N., Tiffany A., et al. (2023). High-dimensional phenotyping to define the genetic basis of cellular morphology. bioRxiv, 2023.2001.2009.522731. 10.1101/2023.01.09.522731. [DOI] [Google Scholar]
- 3.Bray M.-A., Singh S., Han H., Davis C.T., Borgeson B., Hartland C., Kost-Alimova M., Gustafsdottir S.M., Gibson C.C., and Carpenter A.E. (2016). Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nature Protocols 11, 1757–1774. 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor D.L., and Giuliano K.A. (2005). Multiplexed high content screening assays create a systems cell biology approach to drug discovery. Drug Discov Today Technol 2, 149–154. 10.1016/j.ddtec.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 5.Abraham V.C., Taylor D.L., and Haskins J.R. (2004). High content screening applied to large-scale cell biology. Trends Biotechnol 22, 15–22. 10.1016/j.tibtech.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X., Cao X., Perlman Z., and Wong S.T. (2006). A computerized cellular imaging system for high content analysis in Monastrol suppressor screens. J Biomed Inform 39, 115–125. 10.1016/j.jbi.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology 7, R100. 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Underwood J.C., and Crocker J. (1990). Pathology of the Nucleus (Springer). [Google Scholar]
- 9.Zink D., Fischer A.H., and Nickerson J.A. (2004). Nuclear structure in cancer cells. Nature reviews cancer 4, 677–687. [DOI] [PubMed] [Google Scholar]
- 10.Trimmer P.A., Swerdlow R.H., Parks J.K., Keeney P., Bennett J.P. Jr, Miller S.W., Davis R.E., and Parker W.D. Jr (2000). Abnormal mitochondrial morphology in sporadic Parkinson’s and Alzheimer’s disease cybrid cell lines. Experimental neurology 162, 37–50. [DOI] [PubMed] [Google Scholar]
- 11.Herrick J.B. (1910). Peculiar elongated and sickle-shaped red blood corpuscles in a case of severe anemia. Archives of internal medicine 6, 517–521. [Google Scholar]
- 12.Skelly D.A., Czechanski A., Byers C., Aydin S., Spruce C., Olivier C., Choi K., Gatti D.M., Raghupathy N., Keele G.R., et al. (2020). Mapping the Effects of Genetic Variation on Chromatin State and Gene Expression Reveals Loci That Control Ground State Pluripotency. Cell Stem Cell 27, 459–469.e458. 10.1016/j.stem.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byers C., Spruce C., Fortin H.J., Hartig E.I., Czechanski A., Munger S.C., Reinholdt L.G., Skelly D.A., and Baker C.L. (2022). Genetic control of the pluripotency epigenome determines differentiation bias in mouse embryonic stem cells. The EMBO Journal 41, e109445. 10.15252/embj.2021109445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortmann D., Brown S., Czechanski A., Aydin S., Muraro D., Huang Y., Tomaz R.A., Osnato A., Canu G., Wesley B.T., et al. (2020). Naive Pluripotent Stem Cells Exhibit Phenotypic Variability that Is Driven by Genetic Variation. Cell Stem Cell 27, 470–481.e476. 10.1016/j.stem.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aydin S., Pham D.T., Zhang T., Keele G.R., Skelly D.A., Paulo J.A., Pankratz M., Choi T., Gygi S.P., Reinholdt L.G., et al. (2023). Genetic dissection of the pluripotent proteome through multi-omics data integration. Cell Genom 3, 100283. 10.1016/j.xgen.2023.100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haghighi M., Caicedo J.C., Cimini B.A., Carpenter A.E., and Singh S. (2022). High-dimensional gene expression and morphology profiles of cells across 28,000 genetic and chemical perturbations. Nature Methods 19, 1550–1557. 10.1038/s41592-022-01667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tegtmeyer M., Arora J., Asgari S., Cimini B.A., Peirent E., Liyanage D., Way G., Weisbart E., Nathan A., Amariuta T., et al. (2023). High-dimensional phenotyping to define the genetic basis of cellular morphology. bioRxiv, 2023.2001.2009.522731. 10.1101/2023.01.09.522731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohban M.H., Singh S., Wu X., Berthet J.B., Bray M.-A., Shrestha Y., Varelas X., Boehm J.S., and Carpenter A.E. (2017). Systematic morphological profiling of human gene and allele function via Cell Painting. eLife 6, e24060. 10.7554/eLife.24060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podgorski J., and Berg M. (2020). Global threat of arsenic in groundwater. Science 368, 845–850. doi: 10.1126/science.aba1510. [DOI] [PubMed] [Google Scholar]
- 20.Rehman K., and Naranmandura H. (2012). Arsenic metabolism and thioarsenicals. Metallomics 4, 881–892. [DOI] [PubMed] [Google Scholar]
- 21.Challenger F. (1945). Biological methylation. Chemical Reviews 36, 315–361. [Google Scholar]
- 22.Cullen W.R. (2014). Chemical mechanism of arsenic biomethylation. Chemical research in toxicology 27, 457–461. [DOI] [PubMed] [Google Scholar]
- 23.Kuo C.-C., Moon K.A., Wang S.-L., Silbergeld E., and Navas-Acien A. (2017). The Association of Arsenic Metabolism with Cancer, Cardiovascular Disease, and Diabetes: A Systematic Review of the Epidemiological Evidence. Environmental Health Perspectives 125, 087001. doi: 10.1289/EHP577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng C.-H. (2007). Arsenic methylation, urinary arsenic metabolites and human diseases: current perspective. Journal of Environmental Science and Health Part C 25, 1–22. [DOI] [PubMed] [Google Scholar]
- 25.Pierce B.L., Tong L., Argos M., Gao J., Jasmine F., Roy S., Paul-Brutus R., Rahaman R., Rakibuz-Zaman M., Parvez F., et al. (2014). Arsenic metabolism efficiency has a causal role in arsenic toxicity: Mendelian randomization and gene-environment interaction. International Journal of Epidemiology 42, 1862–1872. 10.1093/ije/dyt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamayo L.I., Kumarasinghe Y., Tong L., Balac O., Ahsan H., Gamble M., and Pierce B.L. (2022). Inherited genetic effects on arsenic metabolism: A comparison of effects on arsenic species measured in urine and in blood. Environmental Epidemiology 6, e230. 10.1097/ee9.0000000000000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahsan H., Chen Y., Kibriya M.G., Slavkovich V., Parvez F., Jasmine F., Gamble M.V., and Graziano J.H. (2007). Arsenic Metabolism, Genetic Susceptibility, and Risk of Premalignant Skin Lesions in Bangladesh. Cancer Epidemiology, Biomarkers & Prevention 16, 1270–1278. 10.1158/1055-9965.Epi-06-0676. [DOI] [PubMed] [Google Scholar]
- 28.Rodrigues E.G., Kile M., Hoffman E., Quamruzzaman Q., Rahman M., Mahiuddin G., Hsueh Y., and Christiani D.C. (2012). GSTO and AS3MT genetic polymorphisms and differences in urinary arsenic concentrations among residents in Bangladesh. Biomarkers 17, 240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernández A., and Marcos R. (2008). Genetic variations associated with interindividual sensitivity in the response to arsenic exposure. Pharmacogenomics 9, 1113–1132. 10.2217/14622416.9.8.1113. [DOI] [PubMed] [Google Scholar]
- 30.Faita F., Cori L., Bianchi F., and Andreassi M.G. (2013). Arsenic-Induced Genotoxicity and Genetic Susceptibility to Arsenic-Related Pathologies. International Journal of Environmental Research and Public Health 10, 1527–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen R.J., Argos M., Tong L., Li J., Rakibuz-Zaman M., Islam M.T., Slavkovich V., Ahmed A., Navas-Acien A., Parvez F., et al. (2016). Determinants and Consequences of Arsenic Metabolism Efficiency among 4,794 Individuals: Demographics, Lifestyle, Genetics, and Toxicity. Cancer Epidemiol Biomarkers Prev 25, 381–390. 10.1158/1055-9965.Epi-15-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saint-Jacques N., Parker L., Brown P., and Dummer T.J. (2014). Arsenic in drinking water and urinary tract cancers: a systematic review of 30 years of epidemiological evidence. Environ Health 13, 44. 10.1186/1476-069x-13-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Concha G., Vogler G., Nermell B., and Vahter M. (2002). Intra-individual variation in the metabolism of inorganic arsenic. International archives of occupational and environmental health 75, 576–580. [DOI] [PubMed] [Google Scholar]
- 34.Lovreglio P., D’Errico M.N., Gilberti M.E., Drago I., Basso A., Apostoli P., and Soleo L. (2012). The influence of diet on intra and inter-individual variability of urinary excretion of arsenic species in Italian healthy individuals. Chemosphere 86, 898–905. [DOI] [PubMed] [Google Scholar]
- 35.Gelmann E.R., Gurzau E., Gurzau A., Goessler W., Kunrath J., Yeckel C.W., and McCarty K.M. (2013). A pilot study: The importance of inter-individual differences in inorganic arsenic metabolism for birth weight outcome. Environmental Toxicology and Pharmacology 36, 1266–1275. 10.1016/j.etap.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernández A., Xamena N., Surrallés J., Sekaran C., Tokunaga H., Quinteros D., Creus A., and Marcos R. (2008). Role of the Met287Thr polymorphism in the AS3MT gene on the metabolic arsenic profile. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 637, 80–92. [DOI] [PubMed] [Google Scholar]
- 37.Vahter M. (2000). Genetic polymorphism in the biotransformation of inorganic arsenic and its role in toxicity. Toxicology Letters 112–113, 209–217. 10.1016/S0378-4274(99)00271-4. [DOI] [PubMed] [Google Scholar]
- 38.Steinmaus C., Yuan Y., Kalman D., Rey O.A., Skibola C.F., Dauphine D., Basu A., Porter K.E., Hubbard A., and Bates M.N. (2010). Individual differences in arsenic metabolism and lung cancer in a case-control study in Cordoba, Argentina. Toxicology and applied pharmacology 247, 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierce B.L., Kibriya M.G., Tong L., Jasmine F., Argos M., Roy S., Paul-Brutus R., Rahaman R., Rakibuz-Zaman M., and Parvez F. (2012). Genome-wide association study identifies chromosome 10q24. 32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS genetics 8, e1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karagas M.R., Gossai A., Pierce B., and Ahsan H. (2015). Drinking water arsenic contamination, skin lesions, and malignancies: a systematic review of the global evidence. Current environmental health reports 2, 52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce B.L., Tong L., Argos M., Gao J., Jasmine F., Roy S., Paul-Brutus R., Rahaman R., Rakibuz-Zaman M., and Parvez F. (2013). Arsenic metabolism efficiency has a causal role in arsenic toxicity: Mendelian randomization and gene-environment interaction. International journal of epidemiology 42, 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmad S., Kitchin K.T., and Cullen W.R. (2002). Plasmid DNA damage caused by methylated arsenicals, ascorbic acid and human liver ferritin. Toxicol Lett 133, 47–57. 10.1016/s0378-4274(02)00079-6. [DOI] [PubMed] [Google Scholar]
- 43.Krishnamohan M., and Ng J. (2006). Monomethylarsonous Acid (MMAIII) is Carnogenic in Mice. The Toxicologist, Supplement to Toxicological Sciences 90, 2086. [Google Scholar]
- 44.French J.E., Gatti D.M., Morgan D.L., Kissling G.E., Shockley K.R., Knudsen G.A., Shepard K.G., Price H.C., King D., Witt K.L., et al. (2015). Diversity Outbred Mice Identify Population-Based Exposure Thresholds and Genetic Factors that Influence Benzene-Induced Genotoxicity. Environ Health Perspect 123, 237–245. 10.1289/ehp.1408202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dickson P.E., Ndukum J., Wilcox T., Clark J., Roy B., Zhang L., Li Y., Lin D.T., and Chesler E.J. (2015). Association of novelty-related behaviors and intravenous cocaine self-administration in Diversity Outbred mice. Psychopharmacology (Berl) 232, 1011–1024. 10.1007/s00213-014-3737-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Churchill G.A., Gatti D.M., Munger S.C., and Svenson K.L. (2012). The diversity outbred mouse population. Mammalian genome 23, 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang H., Wang J.R., Didion J.P., Buus R.J., Bell T.A., Welsh C.E., Bonhomme F., Yu A.H.-T., Nachman M.W., and Pialek J. (2011). Subspecific origin and haplotype diversity in the laboratory mouse. Nature genetics 43, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keele G.R. (2023). Which mouse multiparental population is right for your study? The Collaborative Cross inbred strains, their F1 hybrids, or the Diversity Outbred population. G3: Genes, Genomes, Genetics 13, jkad027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tokar E.J., Kojima C., and Waalkes M.P. (2014). Methylarsonous acid causes oxidative DNA damage in cells independent of the ability to biomethylate inorganic arsenic. Arch Toxicol 88, 249–261. 10.1007/s00204-013-1141-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gatti D.M., Svenson K.L., Shabalin A., Wu L.Y., Valdar W., Simecek P., Goodwin N., Cheng R., Pomp D., Palmer A., et al. (2014). Quantitative trait locus mapping methods for diversity outbred mice. G3 (Bethesda) 4, 1623–1633. 10.1534/g3.114.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slob W. (2002). Dose-response modeling of continuous endpoints. Toxicol Sci 66, 298–312. 10.1093/toxsci/66.2.298. [DOI] [PubMed] [Google Scholar]
- 52.Ritz C., Baty F., Streibig J.C., and Gerhard D. (2016). Dose-Response Analysis Using R. PLOS ONE 10, e0146021. 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritz C., Jensen S.M., Gerhard D., & Streibig J.C. (2019). Dose-Response Analysis Using R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keele G.R. (2023). Which mouse multiparental population is right for your study? The Collaborative Cross inbred strains, their F1 hybrids, or the Diversity Outbred population. G3 Genes|Genomes|Genetics 13. 10.1093/g3journal/jkad027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis A.P., Wiegers T.C., Johnson R.J., Sciaky D., Wiegers J., and Mattingly C.J. (2023). Comparative Toxicogenomics Database (CTD): update 2023. Nucleic Acids Res 51, D1257–d1262. 10.1093/nar/gkac833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aono J., Yanagawa T., Itoh K., Li B., Yoshida H., Kumagai Y., Yamamoto M., and Ishii T. (2003). Activation of Nrf2 and accumulation of ubiquitinated A170 by arsenic in osteoblasts. Biochemical and biophysical research communications 305, 271–277. [DOI] [PubMed] [Google Scholar]
- 57.Pi J., Qu W., Reece J.M., Kumagai Y., and Waalkes M.P. (2003). Transcription factor Nrf2 activation by inorganic arsenic in cultured keratinocytes: involvement of hydrogen peroxide. Experimental Cell Research 290, 234–245. 10.1016/S0014-4827(03)00341-0. [DOI] [PubMed] [Google Scholar]
- 58.Lau A., Whitman S.A., Jaramillo M.C., and Zhang D.D. (2013). Arsenic-mediated activation of the Nrf2-Keap1 antioxidant pathway. J Biochem Mol Toxicol 27, 99–105. 10.1002/jbt.21463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janasik B., Reszka E., Stanislawska M., Jablonska E., Kuras R., Wieczorek E., Malachowska B., Fendler W., and Wasowicz W. (2018). Effect of Arsenic Exposure on NRF2-KEAP1 Pathway and Epigenetic Modification. Biol Trace Elem Res 185, 11–19. 10.1007/s12011-017-1219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., and Yamamoto M. (1999). Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes & development 13, 76–86. 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., and Talalay P. (2002). Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proceedings of the National Academy of Sciences 99, 11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang D.D., and Hannink M. (2003). Distinct Cysteine Residues in Keap1 Are Required for Keap1-Dependent Ubiquitination of Nrf2 and for Stabilization of Nrf2 by Chemopreventive Agents and Oxidative Stress. Molecular and Cellular Biology 23, 8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen T., Huang H.C., and Pickett C.B. (2000). Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. The Journal of biological chemistry 275, 15466–15473. 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 64.Banning A., Deubel S., Kluth D., Zhou Z., and Brigelius-Flohé R. (2005). The GI-GPx Gene Is a Target for Nrf2. Molecular and Cellular Biology 25, 4914–4923. doi: 10.1128/MCB.25.12.4914-4923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim Y.C., Masutani H., Yamaguchi Y., Itoh K., Yamamoto M., and Yodoi J. (2001). Hemin-induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. The Journal of biological chemistry 276, 18399–18406. 10.1074/jbc.M100103200. [DOI] [PubMed] [Google Scholar]
- 66.Hayashi A., Suzuki H., Itoh K., Yamamoto M., and Sugiyama Y. (2003). Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein1 in mouse embryo fibroblasts. Biochemical and Biophysical Research Communications 310, 824–829. 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 67.Banerjee M., Marensi V., Conseil G., Le X.C., Cole S.P., and Leslie E.M. (2016). Polymorphic variants of MRP4/ABCC4 differentially modulate the transport of methylated arsenic metabolites and physiological organic anions. Biochem Pharmacol 120, 72–82. 10.1016/j.bcp.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 68.Kala S.V., Kala G., Prater C.I., Sartorelli A.C., and Lieberman M.W. (2004). Formation and Urinary Excretion of Arsenic Triglutathione and Methylarsenic Diglutathione. Chemical Research in Toxicology 17, 243–249. 10.1021/tx0342060. [DOI] [PubMed] [Google Scholar]
- 69.Banerjee M., Carew M.W., Roggenbeck B.A., Whitlock B.D., Naranmandura H., Le X.C., and Leslie E.M. (2014). A novel pathway for arsenic elimination: human multidrug resistance protein 4 (MRP4/ABCC4) mediates cellular export of dimethylarsinic acid (DMAV) and the diglutathione conjugate of monomethylarsonous acid (MMAIII). Molecular pharmacology 86, 168–179. 10.1124/mol.113.091314. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki K.T., Tomita T., Ogra Y., and Ohmichi M. (2001). Glutathione-conjugated Arsenics in the Potential Hepato-enteric Circulation in Rats. Chemical Research in Toxicology 14, 1604–1611. 10.1021/tx0155496. [DOI] [PubMed] [Google Scholar]
- 71.Jia Y., Liu D., Xiao D., Ma X., Han S., Zheng Y., Sun S., Zhang M., Gao H., Cui X., and Wang Y. (2013). Expression of AFP and STAT3 Is Involved in Arsenic Trioxide-Induced Apoptosis and Inhibition of Proliferation in AFP-Producing Gastric Cancer Cells. PLOS ONE 8, e54774. 10.1371/journal.pone.0054774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Breton C.V., Zhou W., Kile M.L., Houseman E.A., Quamruzzaman Q., Rahman M., Mahiuddin G., and Christiani D.C. (2007). Susceptibility to arsenic-induced skin lesions from polymorphisms in base excision repair genes. Carcinogenesis 28, 1520–1525. 10.1093/carcin/bgm063. [DOI] [PubMed] [Google Scholar]
- 73.Chiang C.I., Huang Y.L., Chen W.J., Shiue H.S., Huang C.Y., Pu Y.S., Lin Y.C., and Hsueh Y.M. (2014). XRCC1 Arg194Trp and Arg399Gln polymorphisms and arsenic methylation capacity are associated with urothelial carcinoma. Toxicol Appl Pharmacol 279, 373–379. 10.1016/j.taap.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 74.Andrew A.S., Mason R.A., Kelsey K.T., Schned A.R., Marsit C.J., Nelson H.H., and Karagas M.R. (2009). DNA repair genotype interacts with arsenic exposure to increase bladder cancer risk. Toxicol Lett 187, 10–14. 10.1016/j.toxlet.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kundu M., Ghosh P., Mitra S., Das J.K., Sau T.J., Banerjee S., States J.C., and Giri A.K. (2011). Precancerous and non-cancer disease endpoints of chronic arsenic exposure: the level of chromosomal damage and XRCC3 T241M polymorphism. Mutat Res 706, 7–12. 10.1016/j.mrfmmm.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saxena S., Somyajit K., and Nagaraju G. (2018). XRCC2 Regulates Replication Fork Progression during dNTP Alterations. Cell reports (Cambridge) 25, 3273–3282.e3276. 10.1016/j.celrep.2018.11.085. [DOI] [PubMed] [Google Scholar]
- 77.Meno S.R., Nelson R., Hintze K.J., and Self W.T. (2009). Exposure to monomethylarsonous acid (MMA(III)) leads to altered selenoprotein synthesis in a primary human lung cell model. Toxicol Appl Pharmacol 239, 130–136. 10.1016/j.taap.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ganyc D., Talbot S., Konate F., Jackson S., Schanen B., Cullen W., and Self W.T. (2007). Impact of trivalent arsenicals on selenoprotein synthesis. Environ Health Perspect 115, 346–353. 10.1289/ehp.9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen Q., Chu F.F., and Newburger P.E. (1993). Sequences in the 3’-untranslated region of the human cellular glutathione peroxidase gene are necessary and sufficient for selenocysteine incorporation at the UGA codon. J Biol Chem 268, 11463–11469. [PubMed] [Google Scholar]
- 80.Berry M.J., Banu L., Chen Y.Y., Mandel S.J., Kieffer J.D., Harney J.W., and Larsen P.R. (1991). Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3’ untranslated region. Nature 353, 273–276. 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 81.Berry M.J., Banu L., Harney J.W., and Larsen P.R. (1993). Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. Embo j 12, 3315–3322. 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cox A.G., Brown K.K., Arner E.S.J., and Hampton M.B. (2008). The thioredoxin reductase inhibitor auranofin triggers apoptosis through a Bax/Bak-dependent process that involves peroxiredoxin 3 oxidation. Biochemical pharmacology 76, 1097–1109. 10.1016/j.bcp.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 83.Nagakannan P., Iqbal M.A., Yeung A., Thliveris J.A., Rastegar M., Ghavami S., and Eftekharpour E. (2016). Perturbation of redox balance after thioredoxin reductase deficiency interrupts autophagy-lysosomal degradation pathway and enhances cell death in nutritionally stressed SH-SY5Y cells. Free Radical Biology and Medicine 101, 53–70. 10.1016/j.freeradbiomed.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 84.Lin Y.-X., Gao Y.-J., Wang Y., Qiao Z.-Y., Fan G., Qiao S.-L., Zhang R.-X., Wang L., and Wang H. (2015). pH-Sensitive Polymeric Nanoparticles with Gold(I) Compound Payloads Synergistically Induce Cancer Cell Death through Modulation of Autophagy. Molecular pharmaceutics 12, 2869–2878. 10.1021/acs.molpharmaceut.5b00060. [DOI] [PubMed] [Google Scholar]
- 85.Thorburn A. (2008). Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis 13, 1–9. 10.1007/s10495-007-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Galluzzi L., Maiuri M.C., Vitale I., Zischka H., Castedo M., Zitvogel L., and Kroemer G. (2007). Cell death modalities: classification and pathophysiological implications. Cell Death Differ 14, 1237–1243. 10.1038/sj.cdd.4402148. [DOI] [PubMed] [Google Scholar]
- 87.Lüthi A.U., and Martin S.J. (2007). The CASBAH: a searchable database of caspase substrates. Cell Death Differ 14, 641–650. 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- 88.Ferraj A., Audano P.A., Balachandran P., Czechanski A., Flores J.I., Radecki A.A., Mosur V., Gordon D.S., Walawalkar I.A., Eichler E.E., et al. (2023). Resolution of structural variation in diverse mouse genomes reveals chromatin remodeling due to transposable elements. Cell Genom 3, 100291. 10.1016/j.xgen.2023.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bondareva A.A., Capecchi M.R., Iverson S.V., Li Y., Lopez N.I., Lucas O., Merrill G.F., Prigge J.R., Siders A.M., Wakamiya M., et al. (2007). Effects of thioredoxin reductase-1 deletion on embryogenesis and transcriptome. Free Radic Biol Med 43, 911–923. 10.1016/j.freeradbiomed.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu Y.J., Korotkov K.V., Mehta R., Hatfield D.L., Rotimi C.N., Luke A., Prewitt T.E., Cooper R.S., Stock W., Vokes E.E., et al. (2001). Distribution and functional consequences of nucleotide polymorphisms in the 3’-untranslated region of the human Sep15 gene. Cancer Res 61, 2307–2310. [PubMed] [Google Scholar]
- 91.Gregoire M., Hernandez-Verdun D., and Bouteille M. (1984). Visualization of chromatin distribution in living PTO cells by Hoechst 33342 fluorescent staining. Exp Cell Res 152, 38–46. 10.1016/0014-4827(84)90228-3. [DOI] [PubMed] [Google Scholar]
- 92.Zhang R.-Y., Liu Z.-K., Wei D., Yong Y.-L., Lin P., Li H., Liu M., Zheng N.-S., Liu K., Hu C.-X., et al. (2021). UBE2S interacting with TRIM28 in the nucleus accelerates cell cycle by ubiquitination of p27 to promote hepatocellular carcinoma development. Signal Transduction and Targeted Therapy 6, 64. 10.1038/s41392-020-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Riedmann C., Ma Y., Melikishvili M., Godfrey S.G., Zhang Z., Chen K.C., Rouchka E.C., and Fondufe-Mittendorf Y.N. (2015). Inorganic Arsenic-induced cellular transformation is coupled with genome wide changes in chromatin structure, transcriptome and splicing patterns. BMC Genomics 16, 212. 10.1186/s12864-015-1295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Q., Meng F., Wang L., Mao Y., Zhou H., Hua D., Zhang H., and Wang W. (2017). A polymorphism in ABCC4 is related to efficacy of 5-FU/capecitabine-based chemotherapy in colorectal cancer patients. Scientific reports 7, 7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kibriya M.G., Jasmine F., Parvez F., Argos M., Roy S., Paul-Brutus R., Islam T., Ahmed A., Rakibuz-Zaman M., Shinkle J., et al. (2017). Association between genome-wide copy number variation and arsenic-induced skin lesions: a prospective study. Environmental Health 16, 75. 10.1186/s12940-017-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Plikus M.V., Wang X., Sinha S., Forte E., Thompson S.M., Herzog E.L., Driskell R.R., Rosenthal N., Biernaskie J., and Horsley V. (2021). Fibroblasts: Origins, definitions, and functions in health and disease. Cell 184, 3852–3872. 10.1016/j.cell.2021.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vigilante A., Laddach A., Moens N., Meleckyte R., Leha A., Ghahramani A., Culley O.J., Kathuria A., Hurling C., Vickers A., et al. (2019). Identifying Extrinsic versus Intrinsic Drivers of Variation in Cell Behavior in Human iPSC Lines from Healthy Donors. Cell Rep 26, 2078–2087.e2073. 10.1016/j.celrep.2019.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vincent M., Gerdes Gyuricza I., Keele G.R., Gatti D.M., Keller M.P., Broman K.W., and Churchill G.A. (2022). QTLViewer: an interactive webtool for genetic analysis in the Collaborative Cross and Diversity Outbred mouse populations. G3 Genes|Genomes|Genetics 12. 10.1093/g3journal/jkac146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morgan A.P., Fu C.-P., Kao C.-Y., Welsh C.E., Didion J.P., Yadgary L., Hyacinth L., Ferris M.T., Bell T.A., and Miller D.R. (2016). The mouse universal genotyping array: from substrains to subspecies. G3: Genes, Genomes, Genetics 6, 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Broman K.W., Gatti D.M., Svenson K.L., Sen Ś., and Churchill G.A. (2019). Cleaning Genotype Data from Diversity Outbred Mice. G3 Genes|Genomes|Genetics 9, 1571–1579. 10.1534/g3.119.400165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kwangbom C., Hao H., Daniel M.G., Vivek M.P., Narayanan R., Isabela Gerdes G., Steven C.M., Elissa J.C., and Gary A.C. (2020). Genotype-free individual genome reconstruction of Multiparental Population Models by RNA sequencing data. bioRxiv, 2020.2010.2011.335323. 10.1101/2020.10.11.335323. [DOI] [Google Scholar]
- 102.Love M.I., Huber W., and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alexey A.S. (2016). An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv, 060012. 10.1101/060012. [DOI] [Google Scholar]
- 104.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., and Mesirov J.P. (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carlson M. (2021). org. Mm. eg. db: Genome wide annotation for Mouse. R package version 3.14.0. [Google Scholar]
- 106.Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Austine-Orimoloye O., Azov A.G., Barnes I., Bennett R., et al. (2022). Ensembl 2022. Nucleic Acids Res 50, D988–d995. 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thatcher K., Mattern C.R., Chaparro D., Goveas V., McDermott M.R., Fulton J., Hutcheson J.D., Hoffmann B.R., and Lincoln J. (2023). Temporal Progression of Aortic Valve Pathogenesis in a Mouse Model of Osteogenesis Imperfecta. J Cardiovasc Dev Dis 10. 10.3390/jcdd10080355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452. 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.R Core Team (2021). R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All statistical analyses were performed using the R statistical programming language (v4.1.3)109. The data, supplemental tables, and analysis pipelines used to process, analyze, report, and visualize these findings are publicly available (10.6084/m9.figshare.24576181). The raw and processed RNA-seq data are available from Gene Expression Omnibus (GEO) (GSE247877). All images are available from the corresponding authors (C.O.,L.R.) upon reasonable request.