Abstract
Liver fibrosis is the result of sustained chronic liver injury and inflammation leading to hepatocyte cell death followed by the formation of fibrous scars, which is the hallmark of NASH and alcoholic steatohepatitis and can lead to cirrhosis, HCC, and liver failure. Although progress has been made in understanding the pathogenesis and clinical consequences of hepatic fibrosis, therapeutic strategies for this disease are limited. Preclinical studies suggest that peroxisome proliferator-activated receptor alpha plays an important role in preventing the development of liver fibrosis by activating genes involved in detoxifying lipotoxicity and toxins, transrepressing genes involved in inflammation, and inhibiting activation of hepatic stellate cells. Given the robust preclinical data, several peroxisome proliferator-activated receptor alpha agonists have been tested in clinical trials for liver fibrosis. Here, we provide an update on recent progress in understanding the mechanisms by which peroxisome proliferator-activated receptor alpha prevents fibrosis and discuss the potential of targeting PPARα for the development of antifibrotic treatments.
INTRODUCTION
Hepatic fibrosis is the formation of fibrous scars that arise from sustained wound-healing response to chronic liver injury and inflammation originating from infectious, chemical-induced, cholestatic, and metabolic diseases.[1,2] Early-stage fibrosis is not a disease and causes no harm, but advanced fibrosis leads to cirrhosis, HCC, and liver failure and often requires liver transplantation. The clinical burden of hepatic fibrosis is not only confined to liver-related morbidity and mortality but also affects several extrahepatic organs and regulatory pathways.
Historically, hepatic fibrosis was thought to be a passive and irreversible process resulting from the substitution of hepatic parenchyma with a collagen-rich tissue.[3] However, some studies have demonstrated that even advanced hepatic fibrosis is potentially reversible,[4] and the removal of fibrogenic etiologies is effective in the treatment of hepatic fibrosis resulting from most chronic liver diseases.[5] Since the discovery of HSCs as collagen-producing cells in the liver,[6] key signals responsible for fibrosis have been delineated.[1,2] Although the primary signals that trigger fibrogenesis vary among etiologies, chronic inflammation is a common contributor to myofibroblastic activation that subsequently leads to production and secretion of extracellular matrix components such as collagen and fibronectin to form fibrous scaring.[1,2] It is now clear that activated HSCs are the major, but not the only, source of myofibroblasts.[7] For instance, hepatic myofibroblasts can be formed from portal fibroblasts and mesothelial cells in the presence of the fibrogenic cytokine TGF-β1.[8] Intriguingly, by ectopic expression of specific transcription factors, fibrogenic myofibroblasts can be reprogrammed into hepatocyte-like cells in mice.[9,10] All these pieces of evidence suggest that hepatic fibrosis is a reversible condition.
Steady progress in understanding the pathogenesis of hepatic fibrosis has revealed strategies and therapeutic targets for reversing fibrosis.[5] Preclinical studies suggest that peroxisome proliferator-activated receptor alpha (PPARα, NR1C1) is a potential therapeutic target.[11,12] Notably, the PPARα/γ dual agonist saroglitazar was licensed for the treatment of NASH in India,[13] which is the only approved drug for the treatment of fibrosis. Other PPARα agonists such as fenofibrate and bezafibrate are approved for the treatment of dyslipidemia.[12,14] In this review, we provide an update on recent progress into the mechanisms by which PPARα prevents liver fibrosis and discuss the potential of targeting PPARα for the development of antifibrotic treatments.
PPARα
PPARα along with PPARδ (or PPARβ, NR1C2) and PPARγ (NR1C3) are the 3 PPAR ligand-inducible transcription factors in the nuclear receptor superfamily. The tissue distributions of the 3 PPARs are different with PPARα abundantly expressed in the metabolic active tissues such as the liver, heart, kidneys, and the intestine, while PPARδ is more ubiquitously expressed, and PPARγ is predominantly found in the adipose tissue and immune cells.[15] Functionally, PPARα plays a critical role in hepatic lipid metabolism; PPARδ regulates lipid metabolism, mitochondrial respiration, and thermogenesis; and PPARγ is essential for the formation of adipose tissue and liver.[16]
The 3-dimensional structure of PPARs consists of a DNA-binding domain in the N-terminus and a ligand-binding domain in the C-terminus.[17] Both DNA-binding domain and ligand-binding domain are the most highly conserved regions across the 3 receptor forms.[18] The DNA-binding domain contains 2 zinc finger motifs that can specifically bind peroxisome proliferator response elements (PPREs), a DNA sequence with AGGTCA repeats separated by a single alternative nucleotide (AGGTCA-n-AGGTCA).[19] The ligand-binding domain has an extensive secondary structure comprising 13 α-helices and a small 4-stranded β-sheet.[19] Both natural and synthetic ligands can bind to the ligand-binding domain,[20] and after interaction with specific ligands, PPARs heterodimerize with retinoid X receptor (RXR) and bind to PPREs in the promoter region of target genes to activate transcription of gene batteries involved in normal cellular physiology as well as disease.[18] Notably, in addition to the modulation of gene transcription using PPREs, PPARs were found to repress other transcription factor pathways in a DNA-binding-independent manner.[21] For example, PPARα activation can suppress cytokine IL-1β-induced upregulation of many proinflammatory genes by interfering with NF-κB signaling.[22]
PPARα is the first member of the PPAR family identified in 1990 and is named based on the ability of its agonists, hypolipidemic drugs, or other compounds to induce peroxisome proliferation in rodents.[23] Peroxisomes are membrane-enclosed organelles that contain a complex set of enzymes involved in a variety of metabolic reactions, including the β-oxidation of fatty acids and metabolism of bile acids and cholesterol.[24] Consistently, the target genes of PPARα also play a key role in a plethora of lipid metabolic pathways, including (peroxisomal, microsomal, and mitochondrial) fatty acid oxidation, fatty acid elongation and desaturation, fatty acid binding and transport, triglyceride synthesis and breakdown, lipoprotein metabolism, bile acid metabolism, gluconeogenesis, and various other metabolic pathways.[25,26] As new evidence continues to emerge, the function of PPARα has expanded from being a master regulator of lipid metabolism to multiple aspects, such as modulating liver detoxification processes, suppressing inflammation, and reshaping the immune system.[27]
ROLES AND MECHANISMS FOR PPARα IN PREVENTING LIVER INJURY
Hepatocyte ballooning and multiple forms of hepatocyte death derived from liver injury contribute to the development of liver fibrosis.[28] Through releasing a wide range of signals, injured hepatocytes subsequently trigger inflammation, HSC activation, and fibrogenesis.[28] Since chronic viral infections, alcohol abuse, cholestasis, and metabolic syndrome are the major causative agents leading to liver injury,[1] and current evidence suggests that PPARα is involved in all these injuries through different mechanisms, this section summarizes the potential roles and mechanisms by which PPARα prevents liver injuries in subsections according to different causative agents.
PPARα is involved in virus-induced hepatocellular injury
Chronic HBV/HCV infection represents a major global health problem that can lead to liver fibrosis and cirrhosis.[29,30] The mechanism for this fibrosis is primarily attributed to immune deregulations,[29] but current studies have illustrated that HBV or HCV infection suppresses hepatic PPARα expression and transcriptional activity.[31–33] Mechanisms accounting for this effect include upregulation of microRNA-27 (miR-27) or miR-200c by HCV[33,34] or formation of a complex between HBV X-associated protein 2 (XAP2) and PPARα.[35] In contrast, activation of PPARα and the resultant lipolysis improved HCV-induced liver lesions,[36,37] the PPARα L162V polymorphism tended to favor HBV-induced HCC,[38] and PPARα transrepressive activity on inflammation-repressed HCV entry into hepatocytes.[39] Although these pieces of evidence suggest that PPARα activation may be involved in preventing virus-induced hepatocellular injury, detailed mechanistic studies remain to be explored.
PPARα prevents chemical-induced hepatocellular injury
Hepatocellular injury may stem from a broad range of chemical toxins including ethanol, drugs, and other organic or inorganic toxins. Among them, alcohol consumption is a major risk factor for chronic liver diseases.[40]
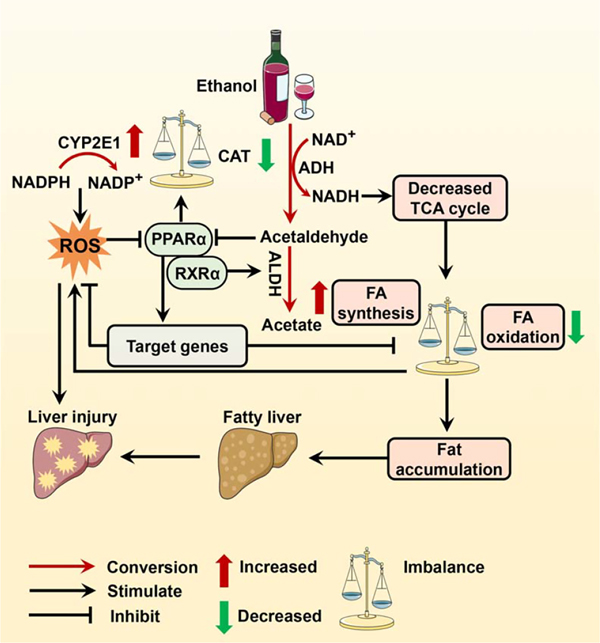
Ethanol can cause hepatocellular injury through a variety of mechanisms. Due to its high energy density and easy availability, ethanol is an important energy source, but it also facilities hepatic fat accumulation through increasing fatty acid synthesis and suppressing fatty acid oxidation, gluconeogenesis, and tricarboxylic acid cycle.[41,42] Under this condition, free fatty acids can promote hepatic lipotoxicity that contributes to the pathogenesis of advanced liver diseases.[43] But a liver injury is not simply caused by hepatic fat accumulation.[44] Growing evidence suggests ethanol as a true hepatic toxin,[45] which leads to the formation of hepatic lesions derived from either ethanol metabolism or ethanol-induced leakage of gut endotoxins.[42,46] In hepatocytes, ethanol is primarily oxidized into acetaldehyde by cytosolic alcohol dehydrogenase, microsomal cytochromes P450 (CYPs), and peroxisomal catalase,[47] and acetaldehyde is rapidly metabolized to acetic acid or acetate by aldehyde dehydrogenase (Figure 1).[47] The metabolite acetaldehyde and CYP2E1-generated reactive oxygen species (ROS) are molecules that can easily react with macromolecules including lipid, protein, RNA, and DNA to promote oxidative stress, lipid peroxidation, hepatocellular damage, and mitochondrial dysfunction.[45,48] Given that acetaldehyde is rapidly detoxified by aldehyde dehydrogenase [47] and the expression of CYP2E1 increases from 4- up to 10-fold after alcohol consumption,[49,50] liver injury may predominantly stem from ROS rather than acetaldehyde. Consistent with this view, ethanol-induced oxidative stress and liver injury in mice are decreased in mice lacking CYP2E1 but restored in CYP2E1-humanized mice,[45] which suggests that CYP2E1-induced oxidative stress is necessary and sufficient to orchestrate liver injury in ethanol-treated mice. In addition, during ethanol metabolism, large amounts of NAD+ are converted to its reduced form NADH,[40] which in turn activates pyruvate dehydrogenase kinase and as a result prevents glucose disposal by inhibiting the pyruvate dehydrogenase complex,[51] which may exacerbate insulin resistance,[52] especially under conditions of lipotoxicity.[43] Taken together, lipotoxicity and oxidative stress may be the root cause of ethanol-induced hepatic lesions attributing to alcoholic liver disease.[53]
FIGURE 1.
Role of PPARα in alcohol metabolism. Ethanol is metabolized by ADH, CYP2E1, and CAT to produce acetaldehyde. Toxic acetaldehyde and CYP2E1-generated ROS can induce hepatocellular injury by inactivating PPARα. PPARα activation enhances detoxification of FFA-derived lipotoxicity by modulating the expression of most key genes involved in FA catabolism and suppresses ROS production by switching alcohol metabolism from the CYP2E1 pathway to the CAT pathway, which restores the balance between oxidants and antioxidants, FA oxidation and synthesis, and thus prevents alcohol-arisen liver injury. Abbreviations: ADH, alcohol dehydrogenase; ALDH, aldehyde dehydrogenase; CAT, catalase; CYP2E1, cytochrome P450 2E1; FA, fatty acid; FFA, free fatty acid; PPARα, peroxisome proliferator-activated receptor alpha; ROS, reactive oxygen species; RXRα, retinoid X receptor.
PPARα activation plays a central role in detoxifying free fatty acid by transcriptionally modulating the expression of most key genes involved in fatty acid catabolism[25,26] and in defense against oxidative stress by increasing the expression of antioxidant enzymes including catalase[54,55] and CuZn-superoxide dismutase (SOD1) (Figure 1).[56] Furthermore, given that Aldh is a target gene of RXRα[57] that is stabilized by PPARα,[58] the indirect upregulation of Aldh by activated PPARα facilitates acetaldehyde removal.[57,59] Therefore, it is proposed that PPARα acts as an initial and central regulator of ethanol-induced hepatocellular injury (Box 1, Figure 1).[53] Consistent with this concept, it was reported that ethanol metabolism might inactivate PPARα, which arises from either acetaldehyde-mediated direct inhibition of the transcriptional and DNA-binding activity of PPARα, [60] or ROS and adenosine-mediated indirect inhibition of PPARα activity by suppressing adiponectin and zinc, respectively,[62–64] or microRNAs such as miR-155-mediated inhibition of PPARα translation,[65] or CYP2E1-induced metabolism of endogenous PPARα ligands.[71] Although it is not yet clear whether inactivation of PPARα exacerbates or triggers ethanol-induced oxidative stress and lipotoxicity,[42,45,72] PPARα activation completely switched alcohol metabolism from the CYP2E1 pathway to the catalase pathway along with suppressed ROS production and accelerated alcohol clearance (Figure 1).[66] Moreover, there is increasing evidence that ethanol diet-fed Ppara-null mice developed more severe liver steatosis and injury,[67,68] whereas upregulation of PPARα by inhibiting its upstream microRNAs abrogated ethanol-induced liver lesions.[65] Activation of PPARα by synthetic or endogenous ligands prevented ethanol-induced liver injury.[68–70] Therefore, PPARα might be a potential target for the detoxification of ethanol hepatotoxicity, thereby preventing the progression of alcoholic liver disease.
Box 1. Key points for PPARα and ethanol-related hepatotoxicity.
Ethanol metabolism inactivates PPARα
Acetaldehyde directly inhibits PPARα transcription and activity.[60,61]
ROS suppresses PPARα activity using adiponectin.[62]
Ethanol upregulates miR-155 prevents PPARα translation.[65]
PPARα activation detoxifies ethanol-related hepatotoxicity
PPARα activation increases the expression of catalase[54,55] and SOD1.[56]
PPARα activation accelerates ethanol clearance via the catalase pathway.[66]
Evidence for PPARα involvement in ethanol-induced hepatotoxicity
In other chemical liver fibrosis models, PPARα activation also serves as a mechanism to prevent hepatocellular toxicity. For example, carbon tetrachloride (CCl4), a widely used chemical in experimental models to induce liver fibrosis,[73] was reported to suppress the expression and activity of PPARα.[74,75] Although CCl4-induced liver injury might be associated with multiple biological processes, pathways, and targets,[76] several PPARα agonists ameliorated hepatocellular injury induced by CCl4 alone or combined with other agents,[70,77] and the activation of PPARα also mediated the protective role of melatonin,[75] 4’-O-methylhonokiol,[78] and apigenin[79] in CCl4-induced liver injury. Similar beneficial effects of PPARα on attenuating liver injury were also reported in fibrosis models induced by other chemicals including acetaminophen,[80,81] alpha-naphthyl isothiocyanate,[82] and thioacetamide.[55] At least 2 potential mechanisms mediate the benefits of PPARα. One mechanism is to increase drug detoxification by upregulating its target drug-metabolizing enzymes including CYP2C8 and CYP3A4.[83,84] Another mechanism may be attributed to the upregulation of antioxidant enzymes,[54–56] thereby preventing oxidative stress that accounts for the hepatotoxicity of all these chemicals.[85–88] Collectively, PPARα activation enhances antioxidant capacity and the detoxification of chemical toxins by CYPs, which may prevent hepatocellular injury and their detrimental effects on hepatic inflammation and fibrosis.
PPARα attenuates lipotoxicity in NAFLD
NAFLD is the most prevalent liver disease worldwide affecting ~1-quarter of the global adult population.[89] Steatosis is the initial stage of this disease, which can progress to NASH, liver fibrosis, liver cirrhosis, and HCC. While detailed mechanisms for progression from steatosis to the more aggressive NASH disorder remain to be fully explored, available data underscore the central role of lipid metabolism dysregulationand lipotoxicity as initiators of hepatocellular lesions and fibrosis.[90,91]
Growing evidence has implicated PPARα activation in preventing fat accumulation and the resultant lipotoxicity and injury (Box 2). Epidemiologic data revealed a negative correlation of hepatic PPARα expression with the severity of steatosis, NASH, and fibrosis,[92] and the single-nucleotide polymorphisms of PPARα (L162V) predispose individuals to the development and progression of NAFLD.[97] Ppara-null mice spontaneously develop hypercholesterolemia and steatosis[94] and have substantially exacerbated fasting-induced elevation of hepatic free fatty acids and liver injury.[95,96] Similarly, hepatocyte-specific PPARα knockout mice fed a standard diet developed NAFLD in old age[98] and promoted high-fat diet-induced liver lesions,[99] suggesting that liver PPARα is crucial for whole-body lipid homeostasis and is a potential therapeutic target for hepatocellular injury and NAFLD.[98]
Box 2. Key points for PPARα involvement in NAFLD.
NAFLD severity is negatively correlated with hepatic PPARα expression.[92]
Fatty acids are endogenous PPARα ligands.[93]
PPARα governs most lipid metabolic processes.[26]
PPARα activation promotes the clearance of lipotoxic fatty acids.[25–27]
PPARα deficiency increases fat accumulation and lipotoxicity.[94–96]
The primary protective role of PPARα against lipotoxicity may arise from its function as a nuclear receptor. It is now clear that a wide range of fatty acids and their derivatives are endogenous PPARα ligands,[93] and many of these ligands activate PPARα within a nanomolar range.[100] This supports the concept that PPARα serves as an intracellular metabolic sensor[101] and fatty acid sensor.[26,27] When certain fatty acids or their metabolic intermediates are present, PPARα will be activated and direct the oxidative degradation of these molecules by controlling gene transcription of enzymes required for lipid catabolism.[26] Currently, PPARα target genes cover almost all lipid metabolic processes,[26] and functional PPREs were identified in the promoters of genes encoding rate-limiting enzymes for fatty acid oxidation, such as CYP4A in microsomal ω-oxidation,[102,103] acyl-CoA oxidase 1 in peroxisomal β-oxidation,[104] and acyl-CoA dehydrogenase medium chain in mitochondrial β-oxidation.[105] Moreover, PPARα transactivates antioxidant enzymes[54–56] and transrepresses inflammation,[21] which further facilitates its function to counteract fibrosis. Consequently, some studies have suggested PPARα as a potential target to develop hypolipidemic and antifibrotic drugs.[106,107]
PPARα suppresses cholestatic liver injury
Cholestasis is a pathologic condition characterized by a decrease in bile flow resulting from impaired bile acid synthesis or obstructed bile acid transport and excretion inside or outside the liver.[108,109] Owing to hydrophobic bile acids being highly toxic when accumulated at high concentrations in hepatocytes,[110] chronic cholestasis contributes to the development of hepatic fibrosis and cirrhosis.[109,111] While cholestatic liver fibrosis may arise from genetic defects, mechanical injury of the bile ducts, and dysregulation of immune responses,[1,112] its molecular mechanisms remain elusive. Growing evidence suggested that the nuclear receptors farnesoid X receptor (FXR, NR1H4) and PPARα coordinately control bile acid homeostasis by regulating the transcription of key proteins involved in bile acid biosynthesis and transport.[113–115] FXR and PPARα should have a central role in the pathogenesis of cholestatic hepatocellular injury and liver fibrosis (Box 3, Figure 2).
Box 3. Key points for PPARα preventing bile acid-induced hepatotoxicity.
PPARα activation reduces bile acid accumulation in hepatocytes
PPARα agonists reduce serum bile acid concentrations.[116,117]
PPARα activation decreases de novo synthesis of bile acid.[118]
Ppara-null mice have increased serum total bile acids.[119]
PPARα activation suppresses bile acid reuptake and reabsorption.[114]
PPARα activation detoxifies bile acids
PPARα activation increases biliary phosphatidylcholine secretion.[120]
PPARα activation enhances bile acid glucuronidation and urinary elimination.[121]
PPARα activation enhances bile acid metabolism and detoxification.[122,123]
PPARα activation prevents bile acid-induced oxidative stress
-
PPARα activation attenuates cholestatic oxidative stress.[124]
Evidence for PPARα involvement in bile acid homeostasis
Bile acids upregulate PPARα gene expression by activating FXR.[113]
Several PPARα target genes contribute to bile acid homeostasis.[120,122,125–129]
Bile acid homeostasis was disrupted in cholic acid treated Ppara-null mice.[130]
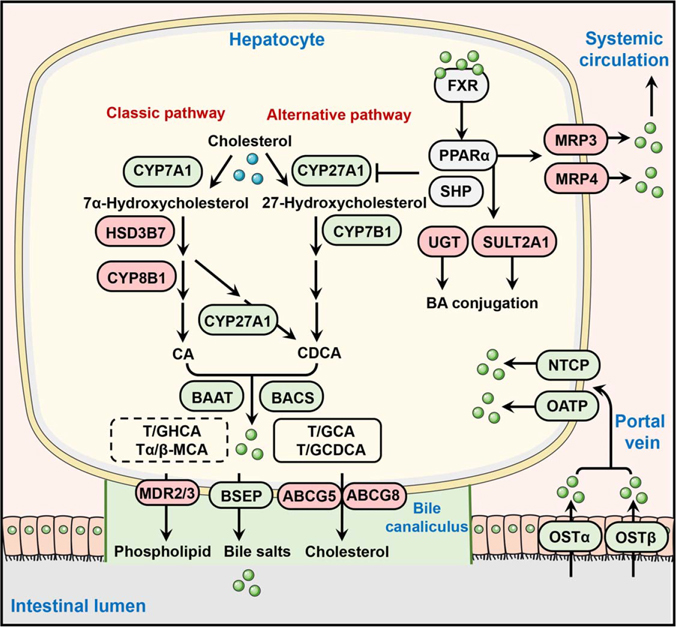
FIGURE 2.
Role of PPARα in the regulation of bile acid synthesis, transport, and metabolism. Bile acids activate the FXR-PPARα axis in hepatocytes to suppress bile acid synthesis primarily by inhibiting CYP7A1 (classic pathway) and CYP27A1 (alternative pathway). The activation of PPARα also promotes bile acid efflux from hepatocytes into the portal vein by induction of MRP3 and MRP4, and into bile canaliculus by induction of MDR2/MDR3 and ABCG5/ABCG8. In addition, PPARα activation represses bile acid reuptake into the liver by downregulating bile acid transporter OATP, NTCP, and OSTβ, as well as enhances the detoxification and metabolism of bile acids through the regulation of some target genes such as UGTs and SULT2A1. PPARα activation upregulates proteins. Filled green circles represent bile acids. Proteins upregulated by PPARα are shaded in red, and those downregulated by PPARα in green. Abbreviations: ABCG, ATP-binding cassette subfamily G; BAAT, bile acid-CoA: amino acid N-acyltransferase; BACS, bile acid-CoA synthetase; BSEP, bile salt export pump; CA, cholic acid; CDCA, chenodeoxycholic acid; CYP7A1, cytochrome P450 7A1; CYP7B1, cytochrome P450 7B1; CYP8B1, cytochrome P450 8B1; CYP27A1, cytochrome P450 27A1; FXR, farnesoid X receptor; HSD3B7, 3 beta-hydroxysteroid dehydrogenase type 7; MDR2/3, multidrug resistance protein 2/3; MRP, multidrug resistance-associated protein; NTCP, sodium/taurocholate cotransporting polypeptide; OATP, organic anion-transporting polypeptide; OSTα/β, organic solute transporter subunit α/β; PPARα, peroxisome proliferator-activated receptor alpha; SULT2A1, sulfotransferase 2A1; UGT, UDP-glucuronosyltransferase.
Through inhibiting the rate-limiting enzymes cholesterol 7α-hydroxylase (CYP7A1) and cytochrome sterol 27-hydroxylase (CYP27A1), the FXR-PPARα axis serves as a negative feedback mechanism to suppress bile acid synthesis (Figure 2).[114,131] Since FXR is a bile acid sensor[132] and PPARα is an FXR target[113] and lipid sensor,[25] the tight regulation of bile acid synthesis enzymes by the FXR-PPARα axis may act as a mechanism to ensure an optimal supply of bile acids in an ever-changing metabolic environment.[133,134] For example, activation of FXR by bile acids (when in excess) suppresses Cyp7a1 transcription through PPARα or other nuclear receptors such as the small heterodimer partner (SHP, NROB2).[115,135] Activation of PPARα by agonists decreased Cyp7a1 mRNA expression and CYP7A1 enzyme activity by about 60%,[118,136] resulting from either PPARα-dependent direct repression of Cyp7a1 transcription as both human and rat Cyp7a1 promoter reporter activities were repressed by activated PPARα,[137,138] or PPARα-dependent decrease in cellular levels of hepatocyte nuclear factor 4α (HNF4α; NR2A1), a positive regulator of Cyp7a1 transcription.[139,140] Fibrates and the PPARα ligand Wy-14,643 also suppressed CYP27A1 mRNA expression and enzyme activity.[118] Notably, Pparα-null mice do not have altered basal CYP7A1 and CYP27A1 levels as compared with wild-type mice,[118,141] but the repressive effect of fibrates on Cyp7a1 and Cyp27a1 transcription was completely abolished,[118,141] which indicates that it is the activation rather than the expression of PPARα that represses Cyp7a1 and Cyp27a1 transcription. In contrast, PPARα agonists increased the expression and activity of CYP8B1 and elevated the ratio of cholic acid to chenodeoxycholic acid.[114,126] All these effects were reversed in Pparα-null mice.[114,126] ChIP-seq data (GSE61817) showed that PPARα had a binding peak at the Cyp8b1 promoter,[142] indicating a direct transcriptional activation of Cyp8b1 by PPARα. This increase in CYP8B1 activity might play a compensatory role in maintaining cholic acid levels,[143] which did not change the effects of PPARα activation on the suppression of overall bile acid synthesis.[143] Consistently, evidence from healthy individuals[116] and primary biliary cirrhosis (PBC) or primary sclerosing cholangitis patients[117] showed that treatment with the PPARα agonist fenofibrate reduced serum bile acid concentrations.
In addition to decreasing bile acid synthesis, activation of PPARα also reduces hepatic bile acid accumulation and toxicity by promoting efflux and suppressing reuptake (Figure 2). After synthesis, bile acids are conjugated and amidated with taurine or glycine to form bile salts, which increase the hydrophilicity of bile acids. Then bile salts, together with phospholipids and cholesterol, are transported from hepatocytes into canaliculi by export pumps, largely by a series of ATP-binding cassette transport proteins (ABCs) including ABCB11/BSEP,[144] ABCB4 (also known as MDR3 in humans or MDR2 in mice),[145] and ABCG5/ABCG8.[146] Some parts of bile salts are secreted from hepatocytes into the portal vein through multidrug resistance-associated proteins (MRPs). In the presence of dietary fat, bile salts are delivered to the duodenum and enter the intestine, where they facilitate fat absorption.[147] After that, 95% of bile acids return to the liver by the process of enterohepatic circulation.[148] Na+-taurocholate cotransporting polypeptide (NTCP/SLC10A1) and organic anion transporting polypeptides (OATPs/SLCOs) are responsible for sinusoidal bile acid reuptake into the hepatocytes.[148] PPARα seems to function as a critical regulator for all these processes. For example, PPARα activation upregulates the expression of genes encoding MRP3, MRP4, ABCB4, and ABCG5/ABCG8,[114,149] while Ppara-null mice have substantially decreased hepatic levels of mRNAs encoding transporters ABCB11, ABCB4, ABCG5, and ABCG8 and consequently resulted in the accumulation of bile acids in the liver during cholic acid challenge.[130] Treatment with PPARα agonist leads to a significant decrease of hepatic NTCP and OATP1 in mice, and these effects are abolished in Ppara-null mice,[114] suggesting PPARα-dependent suppression of bile acid reuptake into the hepatocytes. Moreover, human hydroxysteroid sulfotransferase 2A1 (SULT2A1)[122] and several UDP-glucuronosyltransferases (UGTs)[127–129,150] were identified as encoded by PPARα target genes. Increased levels of UGTs as a result of fibrate treatment were reported to promote bile acid glucuronidation, thereby reducing bile acid toxicity and increasing their urinary elimination.[121] Similarly, upregulation of SULT2A1 promoted bile acid sulfation and protected against bile acid-induced liver damage.[123]
Collectively, PPARα plays a critical role in reducing cholestatic hepatocellular injury. As a target gene of FXR, PPARα contributes to the negative feedback regulation of bile acid synthesis by inhibiting the expression of the rate-limiting enzyme CYP7A1. In addition, activation of PPARα promotes hepatic bile acid efflux and prevents their reuptake into the liver. Some PPARα target genes mediate the detoxification and metabolism of bile acids. Therefore, PPARα activation holds the potential to prevent cholestasis and cholestatic liver fibrosis.
PPARα TRANSREPRESSES INFLAMMATION
Inflammation is a protective response to infection, cellular stress, and injury,[151] and chronic inflammation is a driver of almost all major human diseases.[152] Undoubtedly, sustained exposure to hepatocellular injury and/or toxic agents contributing to the injury may disrupt liver homeostasis and lead to chronic inflammation that has become a common trigger for the development of fibrosis and cirrhosis.[1,28] Since an early finding revealed that Pparα-null mice have an enhanced inflammation response,[153] the crosstalk between PPARα and inflammation has been intensively studied.[27]
The mechanisms by which PPARα interacts with inflammation are complex (Box 4). On one hand, specific inflammatory signals reduce PPARα expression (Figure 3). Consistent with the downregulation of PPARα in patients with NASH[92] or virus hepatitis,[31] proinflammatory cytokines suppress the expression and activity of PPARα. For example, IL-1β was found to decrease PPARα signaling in mice and primary hepatocytes through repression of the Pparα promoter activity through the activation of NF-κB (Figure 3).[154] TNFα could lower Pparα mRNA expression by augmenting the activity of the canonical NF-κB signaling pathway in hepatocytes.[155] In contrast, the pharmacological IKK2 inhibitor AS602868 blocked liver fibrosis and steatohepatitis but activated PPARα and enhanced fatty acid oxidation.[157] These results revealed that NF-κB activation can directly modulate PPARα expression and activity, which orchestrates proinflammatory cytokine-induced interference of PPARα function. This is consistent with the hypothesis that there is a balance between PPARα-mediated anti-inflammatory activity and NF-κB-mediated proinflammatory signaling, and that the activation of NF-κB overwhelms PPARα, which may contribute to increased hepatic inflammation and fibrosis.[166] In addition to NF-κB, cytokines can interfere with PPARα through other mechanisms. IL-6 was reported to decrease PPARα expression and inhibit its transcriptional activity in HepG2 cells through the activation of CCAAT/enhancer-binding protein isoforms (C/EBPs)[156] or signal transduction and activator of transcription isoforms (STATs) (Figure 3).[158] STAT5b downregulates PPARα transcription by up to 80% through inhibition of ligand-independent activation function region-1 (AF-1) transactivation domain.[167] Therefore, under conditions with increased proinflammatory pressure, the functions of PPARα may be suppressed. Given that PPARα is a master regulator of lipid metabolism,[25–27] these findings provide a rationale for explaining that inflammatory responses exacerbate fat accumulation and lipotoxicity.[154]
Box 4. Key points for the interaction of PPARα with inflammation.
Proinflammatory signaling suppresses PPARα expression and activity
PPARα activation suppresses inflammation
PPARα activation promotes the clearance of inflammatory mediators.[25,26,54–57,59]
PPARα activation contributes to anti-inflammatory responses in macrophages.[159]
PPARα activation downregulates inflammatory cytokines.[160]
PPARα activation represses inflammatory transcription factors NF-κB, AP-1, STAT, C/EBPs, and NFAT.[161–165]
Pharmacologically activated PPARα inhibited hepatic inflammatory responses.[21]
Ppara-null enhances inflammatory response.[153]
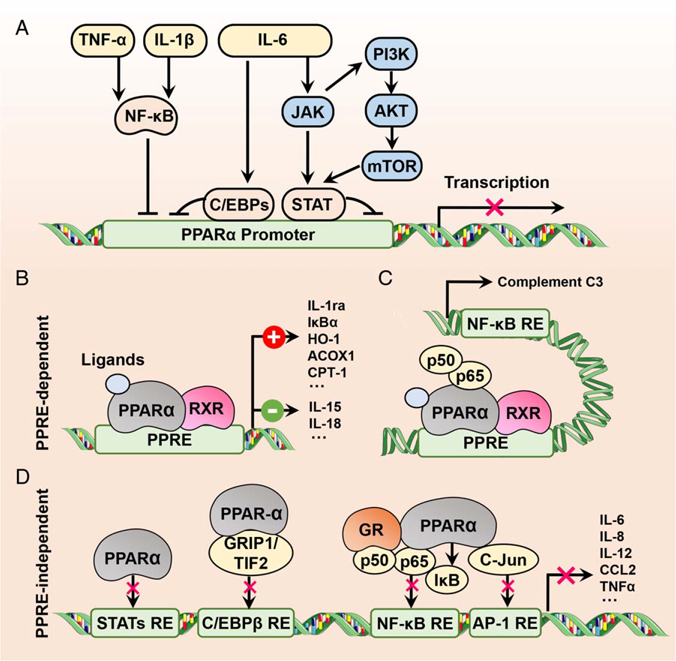
FIGURE 3.
Crosstalk between PPARα and inflammation. (A) Inflammatory cytokine-induced suppression of PPARα activity and/or expression using different proinflammatory transcription factors such as NF-κB, C/EBPs, and STATs. (B) PPARα activation-induced protection against inflammation through downregulating (−) proinflammatory cytokines or upregulating (+) anti-inflammatory factors in a PPRE-dependent manner. (C) Inhibition of NF-κB transcriptional activity by activated PPARα through PPRE-dependent abrogation of p65 binding to an NF-κB response element in the complement C3 promoter. (D) PPRE-independent inhibition of transrepression of proinflammatory cytokines. The transcriptional activity of proinflammatory transcription factors can be directly inhibited by PPARα or indirectly blocked by the interaction of PPARα with proteins. Abbreviations: ACOX1, acyl-CoA oxidase 1; AP-1, activator protein-1; C/EBPs, CCAAT/enhancer-binding protein isoforms; CCL2, C-C motif chemokine ligand 2; CPT-1, carnitine palmitoyltransferase-1; GR, glucocorticoid receptor; GRIP1, glucocorticoid receptor-interacting protein 1; HO-1, heme oxygenase-1; IL-1ra, IL-1 receptor antagonist; IκBα, inhibitor of NF-κB alpha; PPARα, peroxisome proliferator-activated receptor alpha; PPRE, PPAR-response element; RXR, retinoid X receptor; STATs, signal transduction and activator of transcription isoforms; TIF2, transcriptional intermediary factor 2.
On the other hand, PPARα can counteract inflammation from various phases. The inflammatory response has 3 phases: inflammatory inducers (virus, toxins, or tissue injury), inflammatory sensors (macrophages), and inflammatory mediators (cytokines or chemokines).[168] Although there is still insufficient evidence that PPARα can improve viral hepatitis, PPARα activation does contribute to the suppression of inflammatory inducers through different mechanisms (Figure 3). For instance, PPARα agonists facilitate the removal of the harmful metabolite acetaldehyde derived from ethanol via increased aldehyde dehydrogenase activity.[57,59] Activation of PPARα clears lipotoxic fatty acids and their derivatives by transcriptionally promoting the expression of genes involved in fatty acid catabolism.[25,26] Fibrates prevent bile acid-induced hepatotoxicity through modulation of bile acid synthesis, secretion, and reuptake.[14,114] PPARα can induce the expression of catalase[54,55] and SOD1[56] to attenuate ROS production and oxidative stress-induced liver damage.
Moreover, metabolic reprogramming induced by PPARα activation may be involved in the immune response of macrophage.[169] As an inflammatory sensor, macrophage exists in 2 distinct populations: CD68+MARCO+ resident liver macrophages (KC) and recruited CD68+MARCO− monocyte-derived hepatic macrophages, and a scar-associated TREM2+ CD9+ subpopulation of the latter is proinflammatory and profibrogenic and expands in liver fibrosis.[170] Given that the inflammatory macrophage has an enhanced glycolytic metabolism, while the anti-inflammatory macrophage is more dependent on fatty acid oxidation,[169] the increase of fatty acid oxidation by PPARα activation may support anti-inflammatory responses in macrophages.
Finally, the well-known mechanism for the anti-inflammatory role of PPARα is transcriptional repression of proinflammatory mediators (Figure 3). The first clue uncovering a role for PPARα in inflammation came from the finding that PPARα activation acted as a negative feedback mechanism to suppress inflammation.[153] Since then, many studies aimed at revealing the molecular pathways underlying this finding have been performed and have uncovered that more than 39 genes encoding inflammatory mediators are target genes of PPARα.[160] Notably, the majority of these genes are transrepressed by PPARα,[160] except IL-1 receptor antagonist (IL-1ra) that exhibits anti-inflammatory activity and is a direct target gene of PPARα with a functional PPRE present in the promoter,[171] indicating that the anti-inflammatory effects of PPARα may primarily stem from a PPRE-independent mechanism. Indeed, a study with DNA-binding-deficient PPARα mutant (PPARαDISS) mice found that although activated PPARαDISS did not regulate target genes related to fatty acid oxidation, it protected against diet-induced liver damage and blunted inflammatory response and liver fibrosis.[21] Intriguingly, while PPARα expression is lower in human immune cells than in hepatocytes, some anti-inflammatory hormones transcriptionally upregulated PPARα transcript levels in T and B lymphocytes.[172] Macrophage PPARα was found to be a mediator of the anti-inflammatory effects of PPARα agonists,[173] as macrophage-specific Pparα-null mice failed to downregulate the proinflammatory cytokines IL-15 and IL-18.[173] Since proinflammatory mediators are tightly regulated by transcription factors including NF-κB,[174] activator protein-1 (AP-1),[175] STAT,[176] C/EBPs,[177] and nuclear factor of activated T-cells,[178] the interaction of PPARα with these transcription factors may contribute to the mechanism of the PPRE-independent transrepression of inflammation (Figure 3). As expected, growing evidence revealed that PPARα negatively regulates the activity of these transcription factors through different mechanisms.[20] Fibrates were reported to inhibit inflammatory response through PPARα by interfering with NF-κB and AP-1 transactivation capacity involving direct protein-protein interaction with p65 and c-Jun.[164] Palmitoylethanolammide-induced inhibition of NF-κB and subsequent inflammatory effects were proven to be dependent on PPARα, but not PPARγ.[179] It was found that PPARα activation controlled expression of the inhibitory kappa B kinase alpha (IκBα) in human cells and mice[22,] and WY-14,643 enhanced the phosphorylation of IκBα,[180] which was accompanied by a decrease in NF-κB DNA-binding activity.[22] Accumulating evidence indicated that PPARα activation accounted for the decrease in the production of proinflammatory mediators in different cell types through repression of NF-κB.[181,182] In addition, PPARα activators suppressed lipopolysaccharide-stimulated STAT1 phosphorylation and nuclear factor-binding activity, as a result suppressing the release of proinflammatory mediators.[165] The transcriptional activity of STAT5 was negatively regulated by PPARα containing the ligand-independent AF-1 transactivation domain.[161] Fibrates decreased the expression of C/EBPβ.[162,163] In summary, although PPARα has the potential to transactivate anti-inflammatory cytokines, the primary mechanism for the suppression of inflammation by PPARα is transcriptional repression of proinflammatory mediators.
Taken together, there is a balance between PPARα and inflammation (Figure 3). Increased inflammatory mediators may suppress PPARα signaling through different inflammatory transcription factors that potentiate the inflammatory response. This vicious circle is fully manifested under PPARα-deficient conditions. However, many inflammatory mediators are targets of PPARα. Ligand-activated PPARα acts through a transcriptional repression mechanism to decrease the inflammatory response by antagonizing transcription factor-mediated inflammatory mediators, thereby providing a potential rationale for protection against inflammation-associated fibrosis.
PPARα INDIRECTLY SUPPRESSES HSC ACTIVATION
HSCs are nonparenchymal liver pericytes that represent about 10% of all resident liver cells.[183] In normal liver, HSCs exist in a quiescent phenotype and act as the primary depot for storage of vitamin A.[184] Following liver cell damage and immune cell infiltration, HSCs can transdifferentiate into fibrogenic, proliferative, inflammatory, and chemotactic myofibroblasts, which is known as “activation,” and thus functions as a central driver of liver fibrosis.[1] Activation of HSCs is complex and multiple pathways including TGFβ/SMAD, Notch, Wnt/β-catenin, Hedgehog, and Hippo signaling pathways can activate HSCs.[1] A variety of stimuli from metabolic reprogramming, endoplasmic reticulum stress, oxidative stress, or epigenetic modifications can promote HSC activation.[1,185] Extracellular signals from various cells including hepatocytes, liver sinusoidal endothelial cells, macrophage, natural killer cells, and B cells also contribute to HSC activation.[1]
PPARα plays an indirect role in HSC activation (Box 5). Since fatty acids in HSCs primarily serves as an important substrate for the esterification of retinol rather than for energy production,[185] it is generally accepted that HSCs do not express PPARα,[2] a master transcription factor of fatty acid oxidation in hepatocytes.[25,26] In this context, direct activation of PPARα in HSCs should not cause significant effects on HSC function. As expected, evidence for direct activation of HSCs by PPARα is very limited and contradictory. While the inhibition of PPARα and related pathways were reported to orchestrate antioxidant-induced maintenance of quiescent lipocyte phenotype of HSCs,[190,191] suppression of PPARα expression was also found to mediate the promoting role of miR-33a in HSC activation,[192] and inactivation of human HSCs by adiponectin depends on PPARα transcriptional activity.[193] In contrast to PPARα, there was compelling evidence that PPARγ expression and activation consistently contributed to the inactivation of HSCs,[194,195] while PPARγ phosphorylation or loss of PPARγ activates HSCs.[196] These lines of evidence reinforce the view that HSC activation requires loss of lipid droplets, while the recovery of retinoid droplets inhibits HSC activation.[197,198] The latter is facilitated by the activation of lipogenic transcription factors[199,200] and may be potentially antagonized by PPARα. However, lack of convincing evidence for direct HSC activation by PPARα does not imply that PPARα is not involved in the modulation of HSC activation in the liver. Studies from animals and humans consistently suggested that PPARα ligands inhibited myofibroblast transformation and collagen synthesis of HSCs through various pathways,[11,186,201] while these beneficial effects were abolished in Ppara-null mice.[186] Moreover, as mentioned above, PPARα effectively prevents liver damage and inflammation caused by various etiologies, which may indirectly suppress HSC activation.
Box 5. Key points for PPARα in the regulation of HSC activation.
Pharmacologically activated PPARα inhibits HSC activation
Oleoylethanolamide blocks HSC activation.[186]
Loss of PPARα abolishes agonist-induced suppression of HSC activation.[173,186]
PPARα activation suppresses stimuli for HSC activation
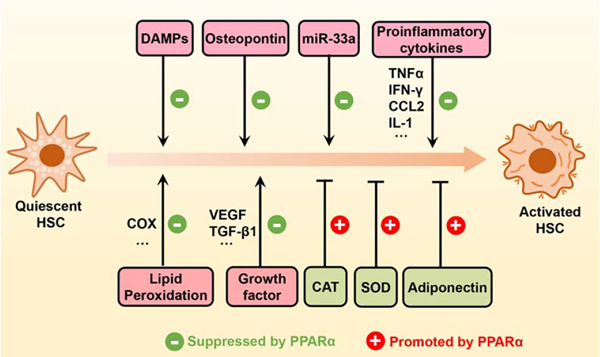
Indeed, current evidence suggested that PPARα modulates HSC activation through regulation of numerous extracellular signals released from different impaired cells (Figure 4). For example, hepatocytes, a major cellular target of hepatic damage and lipid accumulation, are a rich source of extracellular signals for HSC activation. Stressed or dead hepatocytes can activate HSCs through the release of profibrogenic damage-associated molecular patterns[28] or osteopontin,[202] while PPARα is involved in the damage-associated molecular pattern recognition process[187] and fenofibrate reduced serum and tissue levels of osteopontin.[188] Inflammatory cells activate HSCs through the release of proinflammatory cytokines,[203] while hepatic macrophage-specific PPARα disruption prevented the cytokine release[173,] and the transrepressive activity of PPARα was a prerequisite for the suppression of HSC activation.[21] Endothelial cell–mediated growth factor activation, another marker of cellular injury, has the potential to activate HSCs, which is prevented by PPARα agonists.[204] In addition, ROS released from different cells are important mediators to promote HSC activation.[205,206] Genetic disruption of PPARα was found to exacerbate oxidative stress and HSC activation,[68,189] while PPARα ligands ameliorated these effects.[12,55] Collectively, although the direct effects of PPARα on HSC activation remain unclear, various extracellular signals modulated by PPARα have emerged as critical drivers of HSC activation.
FIGURE 4.
PPARα activation indirectly suppresses HSC activation. Upon ligand activation, PPARα can inhibit HSC activation through different mechanisms, such as transcriptional upregulation of catalase, SOD1, and adiponectin, potentiation of the action of miR-33a, and direct or indirect inhibition of various profibrogenic mediators (DAMPs, osteopontin, etc.), proinflammatory cytokines (TNFα, IFN-γ, CCL2, IL-1, etc.), growth factors (VEGF, TGF-β1, etc.), and ROS. Abbreviations: CAT, catalase; CCL2, C-C motif chemokine ligand 2; COX, cyclooxygenase; DAMPs, damage-associated molecular patterns; PPARα, peroxisome proliferator-activated receptor alpha; ROS, reactive oxygen species; SOD1, superoxide dismutase 1.
PPARα MODULATORS IN PRECLINICAL STUDIES
Consistent with the profound role of PPARα in preventing liver injury, inflammation, and HSC activation, preclinical studies have found that activation of PPARα by positive modulators can effectively suppress liver fibrosis, while inhibition of PPARα signaling by negative modulators may deteriorate or trigger fibrosis (Table 1).
TABLE 1.
Main preclinical studies of PPARα-related modulators for liver fibrosis
| Agent | Models | Mechanisms | Effects | Refs. |
|---|---|---|---|---|
| Bezafibrate | Rat | Prevention of HSC activation |




|
207 |
| Feno fibrate | Human | Regulation of vanin-1 |




|
208 |
| Human hepatocytes/Mice | Reduction of cholestasis |




|
(14, 82, 129) | |
| Mice | Regulation of fatty acid metabolism |




|
209 | |
| Rat | Enhancement of catalase activity |




|
55 | |
| Rat | Modulation of inflammatory cytokines |




|
210 | |
| Mice | Reduction of hepatic iron accumulation |




|
211 | |
| Pemafibrate | Mice | — |




|
212 |
| WY14643 | Mice | Enhancement of catalase activity |




|
66, 213 |
| Mice | Inhibition of insulin resistance |




|
214 | |
| Human hepatocytes | Control of catabolism of cytotoxic bile acids |




|
(14, 129) | |
| Mice | Amelioration of hepatic cell death |




|
215 | |
| PPARα/γ agonist | ||||
| Elafibranor | Mice/Rat | PPARa-dependent and -independent mechanisms |




|
216 |
| ZLY16 | Mice | Enhancement of antioxidant enzymes activity |




|
217 |
| MHY2013 | Mice | — |




|
218 |
| Triazo lone derivatives | Mice | — |




|
219 |
| PPARα/γ agonist | ||||
| Chiglitazar | Rat | Improvement of insulin resistance and dyslipidemia |




|
220 |
| Aleglitazar | Rat | Modulation of inflammatory cytokines |




|
221 |
| Saroglitazar | Mice/Rat | Improvement of insulin resistance, modulation of inflammatory cytokines ang adiponectin |




|
222, 223 |
| Pan-PPAR agonist | ||||
| Lanifibranor | Mice | Reduction of proinflammatory macrophages activation |




|
224, 225 |
| MBT1805 | Mice | Reduction of cholestasis |




|
226 |
| Quadruple agonist | ||||
| RLA8 | Mice | Reduction of lipotoxicity and oxidative stress |




|
227 |
| ZLY18 | Mice | Reduction of lipotoxicity and oxidative stress |




|
228 |
| Chemical or oligonucleotide | ||||
| Oleoylethanolamide | Mice | Suppression of SMAD2/3 phosphorylation, α-SMA expression, and myofibroblast transformation |




|
(186) |
| Pirfenidone | Mice | Activation of SIRTl/LKBl/pAMPK |




|
229, 230 |
| Melatonin | Mice | Regulation of circadian clocks |




|
(75) |
| Ascorbic acid | Mice | — |




|
231 |
| Chicory seed extract | Rat/HepG2 cells | — |




|
232 |
| Syringic acid | Mice | Reduction of lipotoxicity |




|
233 |
| Glycyrrhizic acid | Mice | — |




|
234 |
| 2,3,5,4'-tetrahydroxy stilbene-2-O-β-d-glu coside | Mice | Regulation of key regulators of lipid metabolism, inflammation, fibrosis, and oxidative stress |




|
235 |
| 4-O'-methylhonokiol | Mice | Induction of MMPs, AEA, and NAEs and prevention of HSC activation, |




|
(78) |
| Iso-alpha acids | Mice | Reduction of HSC activation and oxidative stress |




|
236 |
| Baicalin | Mice | Regulation of key regulators of lipid metabolism, proinflammatory cytokines, fibrosis markers |




|
237 |
| Betanin | Mice | Downregulation of SREBP-lc |




|
238 |
| Calycosin | Mice | Activation of farnesoid X receptor |




|
239 |
| Naringenin | Mice | Activation of hepatic SIRT1-mediated signaling cascades |




|
240 |
| Conophylline | Mice | Inhibition of hepatic TGF-β level |




|
241 |
| Curcumin | Rat | Regulation of oxidative stress, autophagy and epithelial-mesenchymal transition |




|
242 |
| Quercetin | Rat | Downregulation of the transcription of miR-21 |




|
243 |
| Perfluorooctanoate | Mice | — |




|
244 |
| Alisol B 23-acetate | Mice | Activation of farnesoid X receptor |




|
245 |
| Telmisartan | Mice | Regulation of inflammatory- and fibrosis-related responses |




|
246 |
| Periostin antisense oligonucleotide | Mice | Downregulation of the hepatic periostin expression |




|
247 |
| CircRNA_0046367 | HepG2 cells. | Reduction of miR-34a’s inhibitory effect on PPARα |




|
348 |
| miRNA | ||||
| miR-21 | Mice | Inhibition of PPARα expression |




|
249 |
| miR-27 | Huh7 cells | Repression of PPARα signaling |




|
(33) |
| miR-33a | Hepatic stellate cells LX-2 | Regulation of HSCs trans differentiation and proliferation |




|
(192) |
| miR-155 | Mice | Enhancement of PPRE and PPARα binding and decreased |




|
65 |
| miR-540 | Primary hepatocytes of mice | Inhibition of PPARα expression |




|
250 |



 : positive effect on injury (
: positive effect on injury ( ), steatosis (
), steatosis ( ), inflammation (
), inflammation ( ), or fibrosis (
), or fibrosis ( ).
).



 : neutral or non-available effect on injury (
: neutral or non-available effect on injury ( ), steatosis (
), steatosis ( ), inflammation (
), inflammation ( ), or fibrosis (
), or fibrosis ( ).
).



 : negative effect on injury (
: negative effect on injury ( ), steatosis (
), steatosis ( ), inflammation (
), inflammation ( ), or fibrosis (
), or fibrosis ( ).
).
Abbreviations: PPRE, peroxisome proliferators response elements; PPARα, peroxisome proliferator-activated receptor alpha; Refs., references.
The well-known positive modulators of PPARα are the fibrate class of hyperlipidemic drugs. Current evidence from preclinical studies suggests that PPARα agonists have a comprehensive protective effect on fibrosis (Table 1). For example, the potent experimental PPARα agonist WY14,643 reversed diet-induced or ethanol-induced fibrosis and steatohepatitis in mice[11,70] through enhancing catalase activity,[66,213] inhibiting insulin resistance,[214] ameliorating hepatic cell death,[215] decreasing inflammatory cytokine TNFα,[70] blocking the activation of HSCs and macrophages,[11,70] and suppressing hepatic expression of profibrogenic cytokines osteopontin, TGF-β1, and matrix metalloproteinases.[11,70] Fenofibrate, a clinically used hypolipidemic agent and PPARα agonist, reduced hepatic lipotoxicity,[208] cholestatic liver injury,[14] inflammation,[209] and oxidative stress,[55] and subsequently prevented liver fibrosis derived from different causes.[82,209–211,222] Other fibrates such as bezafibrate[207] also ameliorated fibrosis in different models by reducing the TGF-β-induced myofibroblast differentiation and collagen protein formation. These lines of evidence suggest that PPARα activation can indirectly reverse fibrosis by reducing pro-fibrotic stimuli and activating cells responsible for promoting hepatic fibrosis.
Conventional fibrates are weak PPARα agonists with limited efficacy due to dose-related adverse effects.[251–253] Therefore, several novel agonists were developed that display either more selectivity toward PPARα, or dual PPARα/γ agonist, PPARα/δ agonist, pan-PPARα/γ/δ agonist, and multiple agonist activity toward PPARs and other targets. Among them, pemafibrate, a highly selective PPARα modulator (SPPARMα),[252,254] was proven to decrease fatty liver, inflammation, and fibrogenesis in murine models receiving a high-fat diet,[212] which highlights PPARα as a pivotal pharmacological target for liver fibrosis (Table 1). Elafibranor (GFT505), a dual PPARα/δ agonist, demonstrated liver-protective effects on inflammation and fibrosis in different animal models of NASH and liver fibrosis,[216] and the protective effects were mediated by both PPARα-dependent and PPARα-independent mechanisms,[216] indicating that the effects of PPARα and PPARδ on fibrosis may be synergistic. ZLY16, another novel highly potent PPARα/δ agonist, also decreased liver injury biomarkers, hepatic steatosis, inflammation, ballooning, oxidative stress, and fibrosis, and these effects were more favorable than elafibranor.[217] Moreover, an MHY2013 derivative was also identified as a dual PPARα/δ agonist and displayed antifibrotic effects.[218] A series of novel triazolone derivatives that exhibited more potent and well-balanced PPARα/δ agonistic activity than elafibranor prevented inflammation and liver fibrosis in preclinical models[219] (Table 1). These consistent and comprehensive hepatoprotective effects of PPARα/δ dual agonists indicate that they are promising liver-target drugs for the treatment of fibrosis. Besides, 3 PPARα/γ dual agonists include chiglitazar,[220] aleglitazar,[221] and saroglitazar[222,223] alleviated liver fibrosis in animal models by improving insulin resistance and dyslipidemia, inhibiting the TGF-β/SMAD signaling pathway, or modulating inflammatory cytokines and adiponectin. Given that PPARγ activation can directly inactivate HSCs[194,195] and PPARα indirectly suppresses HSC activation by modulating various extracellular signals, the synergistic protective effects of PPARα and PPARγ on fibrosis are logical. Owing to the potential synergistic effects of different PPAR subtypes, antifibrotic effects of pan-agonists have also been explored. Notably, a pan-PPAR agonist lanifibranor (IVA337) combined the beneficial effects of selective PPAR agonists and displayed an antifibrotic efficacy superior to selective PPARα, PPARδ, or PPARγ agonists.[224,225] MBT1805, a pan-agonist with a balanced PPARα/γ/δ activation effect, relieved α-naphthylisothiocyanate-induced necrosis, vacuolation, and inflammatory infiltration by regulating bile acid synthesis, biotransformation, and transport.[226] Intriguingly, quadruple agonists have also been developed for the treatment of liver fibrosis (Table 1). One example is RLA8, a novel and balanced quadruple agonist for hepatic lipid metabolism and inflammation-related PPARα/γ/δ and G protein-coupled receptor 40 (GPR40). Results from 2 mouse NASH models (induced by a methionine/choline-deficient diet or by a high-fat diet) showed that RLA8 reversed NASH-induced liver damage such as steatosis, inflammation, and fibrosis by reducing lipotoxicity and oxidative stress through activating PPARs and GPR40.[227] Based on this agonist, a newly developed quadruple ZLY18 exhibited far stronger lipid-lowering effects and twice higher metabolic half-life than that of RLA8.[228] In the NASH model, ZLY18 reversed hyperlipidemia to an almost normal level, and significantly decreased hepatocellular ballooning, inflammation, and liver fibrosis even better than RLA8.[228] In addition, ZLY18 is more effective than PPARα agonists in the prevention of CCl4-induced liver fibrosis.[228]
In addition to synthetic PPARα agonists, PPARα seems to mediate the beneficial effects of a series of chemicals or oligonucleotides on ameliorating fibrosis (Table 1). For example, the lipids oleoylethanolamide and palmitoylethanolamide are representative endogenous PPARα ligands. They both significantly reduced inflammatory cytokines, blocked HSC activation, and attenuated liver fibrosis in animal models, and these protective effects were abolished in Pparα-null mice.[186] It was reported that the mechanism of oleoylethanolamide in the inactivation of HSCs is mediated by suppressing SMAD2/3 phosphorylation, α-SMA expression, and myofibroblast transformation.[186] Similar to these ligands, there are a large number of natural or synthetic chemicals that are capable to counteract liver fibrosis and activate or upregulate PPARα, including pirfenidone,[229,230] melatonin,[75] ascorbic acid,[231] chicory seed extract,[232] epigallocatechin gallate,[107] syringic acid,[233] glycyrrhizic acid,[234] 2,3,5,4’-tetrahydroxystilbene-2-O-β-d-glucoside,[235] 4-O’-methylhonokiol,[78] iso-alpha acids,[236] baicalin,[237] betanin,[238] calycosin,[239] naringenin,[240] conophylline,[241] curcumin,[242] quercetin,[243] perfluorooctanoate,[244] alisol B 23-acetate, [245] and telmisartan.[246] Although evidence is still lacking to support whether the antifibrotic effects of these chemicals are PPARα-dependent, these consistent results suggest that PPARα may be a potential common mediator for their protective role in fibrosis. Moreover, some oligonucleotides were reported to prevent fibrosis through upregulating PPARα, such as periostin antisense oligonucleotide[247] and circRNA_0046367.[248]
In contrast, different microRNAs (miRs) that negatively modulate PPARα can trigger hepatic fibrosis (Table 1). A typical example is miR-21 that has been implicated in both hepatic and renal fibrosis.[249,255,256] PPARα is a known target of miR-21,[257] and miR-21 overexpression decreases PPARα levels and signaling, while miR-21 knockout increases PPARα expression and lipid metabolism.[256] Through activating the TGF-β1/SMAD3 pathway[255] or silencing lipid metabolic pathways,[256] miR-21 promoted fibrogenesis by suppressing PPARα. Inhibition of miR-21 expression, on the contrary, restored PPARα expression and reduced liver cell injury, inflammation, and fibrogenesis, while these effects were lost in Ppara-null mice,[249] suggesting the PPARα-dependent profibrogenic role of miR-21. Moreover, caloric restriction[258] and quercetin[243] were found to alleviate fibrosis through suppression of miR-21 and upregulation of PPARα. Therefore, miR-21 may involve in the development of fibrosis by targeting PPARα. Similar to miR-21, other proinflammatory and profibrogenic miRs were also discovered to decrease PPARα, such as miR-27,[33] miR-33a,[192] miR-155,[65] and miR-540.[250]
Collectively, these data reinforce the view that PPARα is a potential therapeutic target for hepatic fibrosis. The discovery of abundant novel agonists, natural chemicals, or oligonucleotides has led to the expectation of effective antifibrotic drugs by targeting PPARα.
PPARα-RELATED AGENTS IN CLINICAL TRIALS
Although liver fibrosis is a serious health problem worldwide and can easily lead to cirrhosis and liver failure, there is no effective strategy available for the treatment except removal of the causative agents or liver transplantation.[5] Substantial progress in preclinical studies targeting PPARα has thrown new light on the development of antifibrotic drugs. Currently, several clinical studies related to PPARα have shown promise and are undergoing further evaluation (Table 2).
TABLE 2.
Main clinical outcomes of PPARα-related drugs for liver fibrosis
| Agent | Clinical Trial # (Phase) | Comments | Effects | Refs. | |||
|---|---|---|---|---|---|---|---|
| PPARa agonist | |||||||
| Bezafibrate | NCT01654731 (III) | In PBC patients (24 months): improved biochemical response, pruritus and liver stiffness. |



|
259 | |||
| NCT02701166 (III) | In PSC and PBC patients (21 days): improved pruritus |



|
260 | ||||
| NCT02937012 (III) | In PBC patients (3–12 months): improved cholestasis |



|
(261, 262) | ||||
| Fenofibrate | NCT02781584 (II) | In NASH patients (2 weeks): improved hypertriglyceridemia |



|
263 | |||
| NCT02354976 (II) | In NAFLD patients (12 weeks): reduced serum triglycerides |



|
264 | ||||
| NCT02891408 (I) | In NASH patients: completed, no results posted. | — | |||||
| NCT00575042 (II) | In PBC patients (48 weeks): biochemical improvement |



|
265 | ||||
| Pemafibrate (K-877) | NCT03350165 (II) | In NAFLD/NASH patients (72 weeks): reduced liver stiffness |



|
266, 267 | |||
| FGF21 mimetics | |||||||
| Pegbelfermin (BMS-986036) | NCT02413372 (II) | In NASH patients (16 weeks): reduced hepatic fat content and liver transaminases, increased serum levels of adiponectin and improved lipid profde. |



|
268 | |||
| NCT03400163 (II) | In NASH patients: completed, no results posted. | — | |||||
| NCT03445208 (I) | In NASH patients: completed, no results posted. | — | |||||
| NCT03486899 (II) NCT03486912 (II) | In NASH patients (24, 48 weeks): improved fibrosis without NASH worsening. |



|
269 | ||||
| NCT03674476 (I) | In NASH patients: completed, no results posted. | — | |||||
| Efruxifermin | NCT03976401 (Ila) | In NASH patients (16 weeks): improved hepatic fat content, NASH, and fibrosis. |



|
270 | |||
| PPARα/γ agonist | |||||||
| Elafibranor | NCT03124108 (11) | In PBC patients (12 weeks): improved serum biochemical response and pruritus. |



|
271 | |||
| NCT04526665(III) | In PBC patients: No results posted. | — | |||||
| NCT01694849 (II) | In NASH patients (52 weeks): resolved NASH (F0-F3) without fibrosis worsening, improved cardiometabolic risk profile. |



|
272 | ||||
| NCT02704403 (III) | In NASH patients (72 weeks): terminated for not achieving NASH resolution without worsening fibrosis |



|
|||||
| PPARα/γ agonist | |||||||
| Saroglitazar | NCT03061721 (II) | In NASH patients (16 weeks): improved ALT, liver fat content, insulin resistance and atherogenic disorders. Side effects: weight gain |



|
273 | |||
| NCT03112681 (II) | In PBC patients (16 weeks): improved serum ALP (primary endpoint) and γGT levels Side effects: increase ALT in some patients |



|
(274, 275) | ||||
| NASH (II) | Very small study demonstrating improved serum lipid and lipoprotein profiles; improvements in hepatocyte ballooning and steatosis; NASH resolution and fibrosis improvement were observed |



|
(13) | ||||
| NCT03863574 (II) | In NASH patients: completed, no results posted. | — | |||||
| Pan-PPAR agonist | |||||||
| Lanifibranor | NCT03008070 (II) | In NASH and F0-F3 fibrosis patients (24 weeks): improved biopsy-confirmed NASH, liver injury, inflammation, and fibrosis. |



|
276, 277 | |||
| NCT03459079 (II) | In NASH patients: No results posted. | — | 225 | ||||
| NCT04849728(III) | In NASH and F0-F3 fibrosis patients: No results posted. | — | |||||


 : positive effect on steatosis (
: positive effect on steatosis ( ), inflammation (
), inflammation ( ), or fibrosis (
), or fibrosis ( ).
).


 : neutral or non-available effect on steatosis (
: neutral or non-available effect on steatosis ( ), inflammation (
), inflammation ( ), or fibrosis (
), or fibrosis ( ).
).
Abbreviations: ALP, alkaline phosphatase; PPRE, peroxisome proliferators response elements; PPARα, peroxisome proliferator-activated receptor alpha; Refs., references.
Given the robust preclinical data, fibrates remain active molecules in the development of antifibrotic drugs through targeting PPARα. As a class of clinically used hypolipidemic drugs since the 1930s, fibrates function to facilitate lipid metabolism and decrease bile acid synthesis. Therefore, it is presumed that fibrates may prevent lipotoxicity and harmful levels of bile acids and may improve liver fibrosis in patients with NAFLD and cholestasis. To develop antifibrotic drugs, 3 fibrates are undergoing assessment in clinical trials (Table 2), including fenofibrate (NCT02891408 in phase 1, NCT02781584, NCT02354976, and NCT00575042 in phase 2), pemafibrate (NCT03350165 in phase 2), and bezafibrate (NCT01654731, NCT02701166, and NCT02937012 in phase 3). In NAFLD patients, fenofibrate (145 mg/d or 200 mg/d) treated for 2 or 12 weeks only effectively mitigated hypertriglyceridemia but not fibrosis,[263,264] while pemafibrate (K-877) treatment significantly improved dyslipidemia[278,279] and reduced magnetic resonance elastography–based liver stiffness,[266] a noninvasive measure of liver fibrosis. Since pemafibrate is one of the novel SPPARMαs that has superior balance of efficacy and safety compared with conventional fibrates,[252] the benefits of pemafibrate on fibrosis indicate that PPARα is a therapeutic target for liver fibrosis, and new SPPARMα may potentially be useful clinically. In patients with PBC and primary sclerosing cholangitis, short-term (21 d) bezafibrate treatment effectively attenuated moderate to severe pruritus.[260] Among patients with PBC who had an inadequate response to ursodeoxycholic acid (UDCA) alone, treatment with bezafibrate in addition to UDCA for 3 months significantly reduced bile acid synthesis and improved serum biliary enzymes, Ig M, cholesterol, and triglyceride concentrations,[261,262] and for 24 months improved biochemical response, pruritus, fatigue, and noninvasive measures of liver fibrosis, including liver stiffness and enhanced liver fibrosis score.[259] Consistently, combination therapy with fenofibrate and UDCA for 48 weeks in PBC patients significantly improved fibrosis as indicated by lower serum alkaline phosphatase (ALP) activity.[265] Collectively, these data from clinical trials suggest that fibrates are potential therapeutic agents for liver fibrosis, especially cholestatic liver fibrosis.
Intriguingly, fibroblast growth factor 21 (FGF21), a protein encoded by the PPARα target gene FGF21, was recently found to be anti-fibrotic and can cause considerable pharmacological benefits on a cluster of metabolic diseases.[280] Several FGF21 mimetics have progressed to early phases of clinical trials (Table 2), such as pegbelfermin (NCT03445208 and NCT03674476 in phase 1, and NCT02413372, NCT03400163, NCT03486899, and NCT03486912 in phase 2) and efruxifermin (NCT03976401 in phase 2). In a phase 2a study in patients with biopsy-confirmed NASH and stage 1–3 fibrosis, treatment with subcutaneously administered pegbelfermin (BMS-986036) for 16 weeks significantly reduced hepatic fat content and liver transaminases, increased serum levels of adiponectin, improved lipid profiles, and attenuated hepatic injury and biomarkers of fibrosis.[268] To expand on the phase 2a results, 2 phase 2b randomized, double-blinded, placebo-controlled studies were designed to assess the efficacy and safety of pegbelfermin in the treatment of patients with NASH and bridging fibrosis or compensated cirrhosis.[269] In another phase 2a study in patients with NASH and fibrosis, administration of efruxifermin through weekly subcutaneous injection for 16 weeks was proven to be safe and efficacious in reducing hepatic fat and markers of liver injury and fibrosis.[270] These data indicate that specific PPARα modulators may improve liver fibrosis through upregulating FGF21.
Based on the hypothesis that agonists with dual or multiple targets will combine the advantages and minimize the side effects caused by selective agonists, the development of modulators with dual or multiple agonist activity toward PPARs and other targets has become a promising strategy for designing effective agents against liver fibrosis. Not surprisingly, given the abundance of preclinical data from mice, several agonists with dual or pan-agonism to PPARs have been tested in phase 2 or phase 3 clinical trials for liver fibrosis (Table 2), including elafibranor (NCT03124108, NCT04526665, NCT01694849, and NCT02704403), saroglitazar (NCT03061721, NCT03112681, and NCT03863574), and lanifibranor (NCT03008070, NCT03459079, and NCT04849728). Although direct evidence for most of these agonists in the regression of liver fibrosis is still being evaluated, current results from clinical trials suggest that they may improve fibrosis through the resolution of causative etiologies or prevention against liver injury.
As a dual PPARα/δ agonist, elafibranor (GFT505) was reported to improve lipid metabolism, insulin resistance, inflammation, and fibrosis in preclinical studies.[281,282] Consistent with these results, in abdominally obese patients, elafibranor significantly reduced fasting plasma triglycerides and enhanced insulin sensitivity.[283,284] Intriguingly, this agonist seems to act in a liver-dependent manner, as it fails to induce PPARα or PPARδ target genes in the skeletal muscle.[283] In the GOLDEN study that is a phase 2 multicenter, double-blinded, randomized controlled trial in 91 adult patients with noncirrhotic NASH, elafibranor (120 mg/d for 1 y) resolved NASH without worsening of fibrosis.[272] Moreover, those patients who achieved resolution of NASH after receiving elafibranor also showed strong reductions in fibrosis, hepatocyte ballooning, lobular steatosis, and the NAFLD activity score (NAS), as well as systemic inflammatory markers such as C-reactive protein, fibrinogen, and haptoglobin.[272] However, in the RESOLVE-IT study which is a phase 3 trial (NCT02704403) in 2000 NASH patients with stage 2–3 fibrosis, elafibranor (120 mg/day for 72 wk) did not show any statistically significant effect on the predefined primary endpoint of NASH resolution without worsening of fibrosis, and therefore, the trial was discontinued. Notably, the reason why elafibranor failed to resolve NASH in this study remains unclear, and it cannot be ruled out that more advanced fibrosis is less “regressible.” In a double-blinded, randomized placebo-controlled phase 2 trial in 45 patients with PBC and incomplete response to UDCA, elafibranor (80 mg/d or 120 mg/d for 12 wk) successfully decreased levels of disease markers, including fibrosis marker serum ALP.[271] Taken together, although the treatment of liver fibrosis with elafibranor has met with a disappointing failure in phase 3 trial in NASH patients, the clinical study of elafibranor in PBC patients has shown promise.
Saroglitazar (ZYH1), a dual PPARα/γ agonist, was shown to improve hepatocyte steatosis, insulin resistance, and inflammation, and prevent the development of fibrosis in animal studies.[77,223,285] In a proof-of-concept study that is a double-blinded phase 2 proof-of-concept trial in 37 patients with PBC and UDCA resistant or intolerant, saroglitazar (2 mg/d or 4 mg/d for 16 wk) led to a rapid and sustained improvement in ALP.[274,275] In a multicenter, prospective, randomized, double-blinded, placebo-controlled proof-of-concept study in 16 adult patients with NASH, treatment with saroglitazar (4 mg and 2 mg for 24 wk) resulted in resolution of steatohepatitis with no worsening of fibrosis[13] and marked improvement in hepatocyte ballooning and steatosis, as well as reduction in serum levels of triglycerides and total cholesterol.[13] In a randomized, controlled, double-blinded phase 2 trial in 106 patients with NAFLD/NASH, saroglitazar at 1, 2, or 4 mg for 16 weeks reduced serum ALT levels by 25.5%, 27.7%, and 45.8%, respectively,[273] and saroglitazar 4 mg also significantly improved liver fat content, fibrosis, insulin resistance, and atherogenic dyslipidemia.[273] Taken together, current evidence from clinical studies suggests that saroglitazar is effective in improvement of liver injury and fibrosis in both patients with PBC or NASH. These findings support further assessment of saroglitazar in phase 3 trials.
Lanifibranor (IVA337) is a moderately potent and well-balanced pan-PPAR agonist,[(106) which] demonstrated excellent antihyperglycemic and hypolipidemic efficacy and anti-fibrotic activity in animal models.[106,286] To assess the therapeutic potential of IVA337, the efficacy and the safety of 2 doses of IVA337 (800 mg, 1200 mg) per day for 24 weeks were evaluated in the NATIVE study which is a phase 2b, double-blinded, randomized, placebo-controlled trial in 247 adult patients with noncirrhotic NASH.[276,277] The results revealed that IVA337 resolved NASH and improved fibrosis according to histological evaluations by biopsy.[277] Based on these findings, a phase 3 study (NCT04849728) is being conducted to evaluate long-term efficacy and safety of lanifibranor in adult NASH patients with stage 2–3 fibrosis.
Collectively, current clinical trials suggest that novel SPPARMα and some agonists with dual or multiple targets including PPARα hold promise for antifibrotic drugs. These data indicate that the development of novel compounds with more selectivity to PPARα or with synergetic agonism to PPARα and other fibrotic molecular targets will be potentially effective strategies for the development of antifibrotic drugs. While the long-term efficacy and safety of these agonists are still under evaluation, these favorable results open a window for the development of effective antifibrotic drugs through targeting PPARα and related molecules.
CHALLENGES AND UNMET NEEDS
Although a great number of PPARα modulators have shown promise in preventing or regressing liver fibrosis in experimental models, there are some challenges and unmet needs that hamper the translation of mouse data into human liver disease treatments (Box 6).
Species differences: in rodents, exposure to PPARα agonists has been associated with peroxisome proliferation, hepatomegaly, and liver cancer, which have not been observed in humans. The reason for this species difference may be attributable to truncated human PPARα splice variants[287] and human PPARα mRNA expression at one-tenth the level of mouse.[288] To address these challenges, a PPARA-humanized mouse strain was created. Current evidence shows that agonist-induced hepatomegaly and hepatocarcinogenesis are abolished in PPARA-humanized mice,[289] suggesting that this mouse strain is a valuable tool for examining species-specific responses to PPARα agonists. However, the application of this strain in fibrosis regression studies is still very limited.
Safety of PPARα agonists: the long-term use of conventional fibrates was reported to increase the risk of cholesterol gallstone and other dose-dependent adverse events in patients.[251–253] Although novel compounds with PPARα agonism display higher efficacy and safety than conventional fibrates, the long-term safety of these compounds remains to be further evaluated.
Complex molecular mechanisms of liver fibrosis: liver fibrosis is increasingly considered as a highly complex disorder with multiple molecular mechanisms driving disease progression. Currently, in addition to PPARα, many other fibrotic molecular targets have been identified,[1] which means that drugs targeting a variety of orthogonal mechanisms may be more effective in resolving liver fibrosis than those targeting a single mechanism. Consistent with this concept, novel compounds with synergistic agonism to PPARα and one or more other molecular targets have been designed, some of which have shown higher antifibrotic efficacy than PPARα-selective agonists in both animal and clinical studies. However, there are still no trials testing the efficacy of drug cocktails targeting several mechanisms in treating liver fibrosis.
Broad functions of PPARα: PPARα regulates extensive physiological functions through modulating the expression of a considerable number of target genes, while not all these targets contribute to the prevention of hepatic fibrosis. The successful suppression of fibrosis by FGF21 mimics in clinical trials[268,270] and selective modulation of PPARα transrepression activity in a preclinical study[21] suggest that it should be a promising strategy for the development of effective antifibrotic drugs through the creation of novel ligands with selective activity for specific PPARα functions. However, such PPARα agonists have not been designed, and the functional selectivity of available agonists remains to be explored. In addition, NASH in humans and mice was associated with the increase of intestinal PPARα and its target gene Fabp1 encoding fatty acid-binding protein (FABP1), while genetic ablation of PPARα or FABP1 specifically in intestine epithelial cells abolished NASH in mice fed a high-fat, high-cholesterol, and high-fructose diet,[290] which suggests that PPARα in different organs plays different roles in the development of fibrosis. Although hepatic PPARα was reported to mediate the major metabolic effects of Wy-14,643,[291] the organ-specific role of most PPARα agonists remains unclear.
Variable composition of myofibroblasts: Since activated HSCs are believed to be the primary source of fibrogenic myofibroblasts in the liver,[7] the cellular mechanisms of liver fibrosis is predominantly HSC centric.[5] Available evidence suggests that the composition of myofibroblasts may vary substantially across etiologies of hepatic fibrosis.[1] However, HSCs do not seem to express PPARα. Although the indirect inhibitory role of PPARα agonists in HSC activation has been intensively studied, the potential role of PPARα in other sources of hepatic myofibroblasts remains to be explored.
Multiple principles of antifibrotic therapy: Several PPARα agonists have shown significant antifibrotic activity in both animal and clinical studies, but the principles underlying different antifibrotic therapies should not be identical. In patients or animal models with mild to moderate fibrosis, the beneficial role of PPARα agonists may stem from attenuating disease progression, whereas in those with advanced fibrosis, their antifibrotic effects may be associated with enhanced matrix degradation. However, while PPARα activation plays protective roles in fibrogenesis through suppression of hepatocellular injury, inflammation, and HSC activation, there is no direct evidence that PPARα agonists can induce matrix degradation.
Etiology-specific effects of PPARα: It is generally believed that the pathogenesis of liver fibrosis varies according to the etiology, which may explain why some PPARα agonists can only improve fibrosis in specific experimental models or patients. In fact, the expression and activity of PPARα are affected by various pathophysiological conditions including disease, stress, hormones, cytokines, and kinases,[27] and these factors may also impact the effects of PPARα modulators on hepatic fibrosis. Hence, it is important to test the efficacy of these modulators in patients with fibrosis derived from different etiologies. However, the etiology-specific effects of different agonists remain to be elucidated.
Insensitive trial endpoints: Recently, the discovery of powerful biomarkers and the successful development of molecular imaging of fibrosis have enabled noninvasive, rapid, and longitudinal readouts of drug efficacy in antifibrotic trials. However, endpoints in current clinical studies evaluating the anti-fibrotic effects of PPARα agonists do not incorporate these noninvasive measures, but instead mainly assess liver histology or serum ALP. Therefore, data on the dynamics of gradual improvement in fibrosis induced by PPARα agonists in patients are still lacking.
Box 6. Key points for challenges and unmet needs.
Species differences between mouse and human PPARα in responses to agonists.
Unclear long-term safety profiles of PPARα agonists.
Lack of synergistic therapeutic strategies against multiple targets in liver fibrosis.
Undefined functional selectivity and organ-specificity of PPARα agonists.
Unknown roles of PPARα in non–HSC-derived myofibroblasts.
Uncertain principles of PPARα agonists in antifibrotic therapy.
Absence of etiology-specific effect of PPARα agonists on liver fibrosis.
Outdated endpoints in PPARα agonist trials.
SUMMARY AND PROSPECTS
Hepatic fibrosis, which arises from a persistent wound-healing response to liver injury, has become a global health problem as it can easily develop into cirrhosis, HCC, and liver failure. However, antifibrotic drug development remains limited. Uncovering the pathophysiological mechanisms of hepatic fibrosis would provide potential therapeutic targets for the development of effective antifibrotic drugs. After 4 decades of steady progress, it is clear now that the pathogenesis of hepatic fibrosis is initiated by liver injury derived from different etiologies, and liver injury triggered inflammation and subsequent HSC activation through various signaling pathways accounting for fibrogenesis.[1] Therefore, a molecular target that could defend against or inhibit all these processes is urgently needed.
PPARα is a ligand-inducible transcriptional modulator of key metabolic processes, as well as inflammation, oxidative stress, and HSC activation. Therefore, PPARα might be a promising therapeutic target for hepatic fibrosis. Dysregulation or suppression of PPARα is a common feature in most causative etiologies of hepatic fibrosis,[31,61,65,92] and PPARα deficiency predisposes animals to hepatic fibrosis in different disease models. In contrast, hepatic PPARα activation seems to act as a modulator of hepatic fibrosis originating from different causative agents by preventing liver injury and subsequent inflammation and HSC activation. As a master regulator of lipid metabolism, PPARα governs the clearance of harmful lipids and the prevention of lipotoxicity. Through transcriptional modulation of bile acid synthesis and drug metabolism, PPARα expression and activity contribute to the detoxification of toxic bile acids and chemicals. The transrepressive activity of PPARα on inflammation is necessary and sufficient to prevent liver fibrosis. Moreover, preclinical studies have revealed a plethora of PPARα modulators, including chemicals or oligonucleotides, that have the potential to impact the development of hepatic fibrosis, which will serve as a resource for the development of antifibrotic drugs.
Collectively, PPARα is a potential therapeutic target for hepatic fibrosis. The development of compounds with selective activity for PPARα or synergetic agonism to PPARα and other targets holds promise to develop effective antifibrotic drugs. These translational approaches are enabling the emergence of precision medicine-based therapies for patients with hepatic fibrosis.
ACKNOWLEDGMENTS
Guolin Li was supported by the National Natural Science Foundation of China (31871198) and the Opening Fund of The National & Local Joint Engineering Laboratory of Animal Peptide Drug Development (Hunan Normal University, National Development and Reform Commission). Fang Wei was supported by the National Natural Science Foundation of China (81903138) and the Natural Science Foundation of Hunan Province (2022JJ30413). Frank J. Gonzalez was supported by the National Cancer Institute Intramural Research Program. The funding sponsors had no role in the writing of the manuscript and in the decision to submit the manuscript for publication.
Abbreviations:
- ABCs
ATP-binding cassette transport proteins
- ACADM
acyl-CoA dehydrogenase medium chain
- ACOX
acyl-CoA oxidase 1
- ADH
alcohol dehydrogenase
- AF-1
activation function region-1
- ALD
alcoholic liver disease
- ALDH
aldehyde dehydrogenase
- ALP
alkaline phosphatase
- AP-1
activator protein-1
- CA
cholic acid
- CAT
catalase
- CCl4
carbon tetrachloride
- CDCA
chenodeoxycholic acid
- C/EBP
CCAAT/enhancer-binding protein isoform
- CYP
cytochromes P450
- DAMP
damage-associated molecular pattern
- DBD
DNA binding domain
- ECM
extracellular matrix
- FABP1
fatty acid binding protein
- FFA
free fatty acid
- GF21
fibroblast growth factor 21
- FXR
farnesoid X receptor
- GPR40
G protein-coupled receptor 40
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HNF4α
hepa tocyte nuclea r fa ctor 4α
- HSC
hepatic stellate cell
- IκBα
inhibitory kappa B kinase alpha
- IL-1β
interleukin-1β
- IL-1ra
interleukin-1 receptor antagonist
- LBD
ligand binding domain
- miR
microRNA
- MRE
magnetic resonance elastography
- MRP
multidrug resistance-associated proteins
- NAD+
nicotinamide adenine dinucleotide
- NADH
reduced form of nicotinamide-adenine dinucleotide
- NAFLD
nonalcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- NFAT
nuclear factor of activated T-cells
- NF-κB
nuclear factor kappa B
- NTCP//SLC10A1
Na+-taurocholate cotransporting polypeptide
- OATPs/SLCOs
organic anion transporting polypeptides
- PBC
primary biliary cirrhosis
- PPARα
peroxisome proliferator-activated receptor alpha
- PPARδ
peroxisome proliferator-activated receptor delta
- PPARγ
peroxisome proliferator-activated receptor gamma
- PPRE
peroxisome proliferators response element
- PSC
primary sclerosing cholangitis
- ROS
reactive oxygen species
- RXR
retinoid X receptor
- SHP
small heterodimer partner
- α-SMA
alpha smooth muscle actin
- SOD1
CuZn-superoxide dismutase
- SPPARMα
selective PPARα modula tor
- STAT
signal transduction and activator of transcription isoform
- SULT2A1
sulfotransferase 2A1
- TGF-β1
transforming growth factor-beta 1
- TNFα
tumor necrosis factor alpha
- XAP2
X-associated protein 2
- UGTs
UDP-glucuronosyltransferases
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts to declare.
REFERENCES
- 1.Kisseleva T, Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2021;18:151–66. [DOI] [PubMed] [Google Scholar]
- 2.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–56. [DOI] [PubMed] [Google Scholar]
- 3.Popper H, Uenfriend S. Hepatic fibrosis. Correlation of biochemical and morphologic investigations. Am J Med. 1970;49:707–21. [DOI] [PubMed] [Google Scholar]
- 4.Soyer MT, Ceballos R, Aldrete JS. Reversibility of severe hepatic damage caused by jejunoileal bypass after re-establishment of normal intestinal continuity. Surgery. 1976;79:601–4. [PubMed] [Google Scholar]
- 5.Friedman SL, Pinzani M. Hepatic fibrosis 2022: unmet needs and a blueprint for the future. Hepatology. 2022;75:473–88. [DOI] [PubMed] [Google Scholar]
- 6.Friedman SL, Roll FJ, Boyles J, Bissell DM. Hepatic lipocytes: the principal collagen-producing cells of normal rat liver. Proc Natl Acad Sci USA. 1985;82:8681–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lua I, Li Y, Zagory JA, Wang KS, French SW, Sevigny J, et al. Characterization of hepatic stellate cells, portal fibroblasts, and mesothelial cells in normal and fibrotic livers. J Hepatol. 2016;64:1137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rezvani M, Espanol-Suner R, Malato Y, Dumont L, Grimm AA, Kienle E, et al. In vivo hepatic reprogramming of myofibroblasts with AAV vectors as a therapeutic strategy for liver fibrosis. Cell Stem Cell. 2016;18:809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song G, Pacher M, Balakrishnan A, Yuan Q, Tsay HC, Yang D, et al. Direct reprogramming of hepatic myofibroblasts into hepatocytes in vivo attenuates liver fibrosis. Cell Stem Cell. 2016;18:797–808. [DOI] [PubMed] [Google Scholar]
- 11.Ip E, Farrell G, Hall P, Robertson G, Leclercq I. Administration of the potent PPARalpha agonist, Wy-14,643, reverses nutritional fibrosis and steatohepatitis in mice. Hepatology. 2004;39:1286–96. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Vilarrupla A, Laviña B, García-Calderó H, Russo L, Rosado E, Roglans N, et al. PPARα activation improves endothelial dysfunction and reduces fibrosis and portal pressure in cirrhotic rats. J Hepatol. 2012;56:1033–9. [DOI] [PubMed] [Google Scholar]
- 13.Siddiqui MS, Idowu MO, Parmar D, Borg BB, Denham D, Loo NM, et al. A phase 2 double blinded, randomized controlled trial of saroglitazar in patients with nonalcoholic steatohepatitis. Clin Gastroenterol Hepatol. 2021;19:2670–. [DOI] [PubMed] [Google Scholar]
- 14.Ghonem NS, Assis DN, Boyer JL. Fibrates and cholestasis. Hepatology. 2015;62:635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbott BD. Review of the expression of peroxisome proliferator-activated receptors alpha (PPAR alpha), beta (PPAR beta), and gamma (PPAR gamma) in rodent and human development. Reprod Toxicol. 2009;27:246–57. [DOI] [PubMed] [Google Scholar]
- 16.Bensinger SJ, Tontonoz P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 2008;454:470–7. [DOI] [PubMed] [Google Scholar]
- 17.Cronet P, Petersen JF, Folmer R, Blomberg N, Sjoblom K, Karlsson U, et al. Structure of the PPARalpha and -gamma ligand binding domain in complex with AZ 242; ligand selectivity and agonist activation in the PPAR family. Structure. 2001;9:699–706. [DOI] [PubMed] [Google Scholar]
- 18.Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409–35. [DOI] [PubMed] [Google Scholar]
- 19.Zoete V, Grosdidier A, Michielin O. Peroxisome proliferator-activated receptor structures: ligand specificity, molecular switch and interactions with regulators. Biochim Biophys Acta. 2007;1771:915–25. [DOI] [PubMed] [Google Scholar]
- 20.Pawlak M, Lefebvre P, Staels B. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol. 2015;62:720–33. [DOI] [PubMed] [Google Scholar]
- 21.Pawlak M, Baugé E, Bourguet W, De Bosscher K, Lalloyer F, Tailleux A, et al. The transrepressive activity of peroxisome proliferator-activated receptor alpha is necessary and sufficient to prevent liver fibrosis in mice. Hepatology. 2014;60:1593–606. [DOI] [PubMed] [Google Scholar]
- 22.Delerive P, Gervois P, Fruchart JC, Staels B. Induction of IkappaBalpha expression as a mechanism contributing to the anti-inflammatory activities of peroxisome proliferator-activated receptor-alpha activators. J Biol Chem. 2000;275:36703–7. [DOI] [PubMed] [Google Scholar]
- 23.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–50. [DOI] [PubMed] [Google Scholar]
- 24.Islinger M, Cardoso MJ, Schrader M. Be different–the diversity of peroxisomes in the animal kingdom. Biochim Biophys Acta. 2010;1803:881–97. [DOI] [PubMed] [Google Scholar]
- 25.Kersten S, Stienstra R. The role and regulation of the peroxisome proliferator activated receptor alpha in human liver. Biochimie. 2017;136:75–84. [DOI] [PubMed] [Google Scholar]
- 26.Kersten S Integrated physiology and systems biology of PPARalpha. Mol Metab. 2014;3:354–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bougarne N, Weyers B, Desmet SJ, Deckers J, Ray DW, Staels B, et al. Molecular actions of PPARalpha in lipid metabolism and inflammation. Endocr Rev. 2018;39:760–802. [DOI] [PubMed] [Google Scholar]
- 28.Schwabe RF, Tabas I, Pajvani UB. Mechanisms of fibrosis development in nonalcoholic steatohepatitis. Gastroenterology. 2020;158:1913–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paquissi FC. Immunity and fibrogenesis: the role of Th17/IL-17 Axis in HBV and HCV-induced chronic hepatitis and progression to cirrhosis. Front Immunol. 2017;8:1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khatun M, Ray RB. Mechanisms underlying hepatitis C virus-associated hepatic fibrosis. Cells. 2019;8:1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dharancy S, Malapel M, Perlemuter G, Roskams T, Cheng Y, Dubuquoy L, et al. Impaired expression of the peroxisome proliferator-activated receptor alpha during hepatitis C virus infection. Gastroenterology. 2005;128:334–42. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Zhu Y, Feng S, Ishida Y, Chiu TP, Saito T, et al. Macrophages activated by hepatitis B virus have distinct metabolic profiles and suppress the virus via IL-1beta to downregulate PPARalpha and FOXO3. Cell Rep. 2022;40:111068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singaravelu R, Chen R, Lyn RK, Jones DM, O’Hara S, Rouleau Y, et al. Hepatitis C virus induced up-regulation of microRNA-27: a novel mechanism for hepatic steatosis. Hepatology. 2014;59:98–108. [DOI] [PubMed] [Google Scholar]
- 34.Ramachandran S, Ilias Basha H, Sarma NJ, Lin Y, Crippin JS, Chapman WC, et al. Hepatitis C virus induced miR200c down modulates FAP-1, a negative regulator of Src signaling and promotes hepatic fibrosis. PLoS One. 2013;8:e70744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sumanasekera WK, Tien ES, Turpey R, Vanden Heuvel JP, Perdew GH. Evidence that peroxisome proliferator-activated receptor alpha is complexed with the 90-kDa heat shock protein and the hepatitis virus B X-associated protein 2. J Biol Chem. 2003;278:4467–73. [DOI] [PubMed] [Google Scholar]
- 36.Read SA, Tay ES, Shahidi M, McLauchlan J, George J, Douglas MW. The mechanism of interferon refractoriness during hepatitis C virus infection and its reversal with a peroxisome proliferator-activated receptor alpha agonist. J Interferon Cytokine Res. 2015;35:488–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rakic B, Sagan SM, Noestheden M, Belanger S, Nan X, Evans CL, et al. Peroxisome proliferator-activated receptor alpha antagonism inhibits hepatitis C virus replication. Chem Biol. 2006;13:23–30. [DOI] [PubMed] [Google Scholar]
- 38.Koytak ES, Mizrak D, Bektas M, Verdi H, Arslan Ergul A, Idilman R, et al. PPAR-alpha L162V polymorphism in human hepatocellular carcinoma. Turk J Gastroenterol. 2008;19:245–9. [PubMed] [Google Scholar]
- 39.Fletcher NF, Sutaria R, Jo J, Barnes A, Blahova M, Meredith LW, et al. Activated macrophages promote hepatitis C virus entry in a tumor necrosis factor-dependent manner. Hepatology. 2014;59:1320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Louvet A, Mathurin P. Alcoholic liver disease: mechanisms of injury and targeted treatment. Nat Rev Gastroenterol Hepatol. 2015;12:231–42. [DOI] [PubMed] [Google Scholar]
- 41.Grunnet N, Kondrup J. The effect of ethanol on the beta-oxidation of fatty acids. Alcohol Clin Exp Res. 1986;10:64S–8S. [DOI] [PubMed] [Google Scholar]
- 42.Ansari RA, Husain K, Rizvi SA. Role of transcription factors in steatohepatitis and hypertension after ethanol: the epicenter of metabolism. Biomolecules. 2016;6:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuschwander-Tetri BA. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology. 2010;52:774–88. [DOI] [PubMed] [Google Scholar]
- 44.Loft A, Alfaro AJ, Schmidt SF, Pedersen FB, Terkelsen MK, Puglia M, et al. Liver-fibrosis-activated transcriptional networks govern hepatocyte reprogramming and intra-hepatic communication. Cell Metab. 2021;33:1685–1700 e1689. [DOI] [PubMed] [Google Scholar]
- 45.Cederbaum AI. Role of CYP2E1 in ethanol-induced oxidant stress, fatty liver and hepatotoxicity. Dig Dis. 2010;28:802–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurman RG, Bradford BU, Iimuro Y, Knecht KT, Arteel GE, Yin M, et al. The role of gut-derived bacterial toxins and free radicals in alcohol-induced liver injury. J Gastroenterol Hepatol. 1998;13(suppl):S39–50. [DOI] [PubMed] [Google Scholar]
- 47.Zakhari S Alcohol metabolism and epigenetics changes. Alcohol Res. 2013;35:6–16. [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, Zhuge J, Wang X, Bai J, Cederbaum AI. Cytochrome P450 2E1 contributes to ethanol-induced fatty liver in mice. Hepatology. 2008;47:1483–94. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi T, Lasker JM, Rosman AS, Lieber CS. Induction of cytochrome P-4502E1 in the human liver by ethanol is caused by a corresponding increase in encoding messenger RNA. Hepatology. 1993;17:236–45. [PubMed] [Google Scholar]
- 50.Tsutsumi M, Lasker JM, Shimizu M, Rosman AS, Lieber CS. The intralobular distribution of ethanol-inducible P450IIE1 in rat and human liver. Hepatology. 1989;10:437–6. [DOI] [PubMed] [Google Scholar]
- 51.Jeoung NH, Harris RA. Role of pyruvate dehydrogenase kinase 4 in regulation of blood glucose levels. Korean Diabetes J. 2010;34:274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeoung NH. Pyruvate dehydrogenase kinases: therapeutic targets for diabetes and cancers. Diabetes Metab J. 2015;39:188–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moriya T, Naito H, Ito Y, Nakajima T. “Hypothesis of seven balances”: molecular mechanisms behind alcoholic liver diseases and association with PPARalpha. J Occup Health. 2009;51:391–403. [DOI] [PubMed] [Google Scholar]
- 54.Shin MH, Lee SR, Kim MK, Shin CY, Lee DH, Chung JH. Activation of peroxisome proliferator-activated receptor alpha improves aged and UV-irradiated skin by catalase induction. PLoS One. 2016;11:e0162628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toyama T, Nakamura H, Harano Y, Yamauchi N, Morita A, Kirishima T, et al. PPARalpha ligands activate antioxidant enzymes and suppress hepatic fibrosis in rats. Biochem Biophys Res Commun. 2004;324:697–704. [DOI] [PubMed] [Google Scholar]
- 56.Inoue I, Goto S, Matsunaga T, Nakajima T, Awata T, Hokari S, et al. The ligands/activators for peroxisome proliferator-activated receptor alpha (PPARalpha) and PPARgamma increase Cu2+,Zn2+-superoxide dismutase and decrease p22phox message expressions in primary endothelial cells. Metabolism. 2001;50:3–11. [DOI] [PubMed] [Google Scholar]
- 57.Gyamfi MA, Kocsis MG, He L, Dai G, Mendy AJ, Wan YJ. The role of retinoid X receptor alpha in regulating alcohol metabolism. J Pharmacol Exp Ther. 2006;319:360–8. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka N, Hora K, Makishima H, Kamijo Y, Kiyosawa K, Gonzalez FJ, et al. In vivo stabilization of nuclear retinoid X receptor alpha in the presence of peroxisome proliferator-activated receptor alpha. FEBS Lett. 2003;543:120–4. [DOI] [PubMed] [Google Scholar]
- 59.Alnouti Y, Klaassen CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci. 2008;101:51–64. [DOI] [PubMed] [Google Scholar]
- 60.Galli A, Pinaire J, Fischer M, Dorris R, Crabb DW. The transcriptional and DNA binding activity of peroxisome proliferator-activated receptor alpha is inhibited by ethanol metabolism. A novel mechanism for the development of ethanol-induced fatty liver. J Biol Chem. 2001;276:68–75. [DOI] [PubMed] [Google Scholar]
- 61.Wan YJ, Morimoto M, Thurman RG, Bojes HK, French SW. Expression of the peroxisome proliferator-activated receptor gene is decreased in experimental alcoholic liver disease. Life Sci. 1995;56:307–17. [DOI] [PubMed] [Google Scholar]
- 62.You M, Considine RV, Leone TC, Kelly DP, Crabb DW. Role of adiponectin in the protective action of dietary saturated fat against alcoholic fatty liver in mice. Hepatology. 2005;42:568–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang X, Zhong W, Liu J, Song Z, McClain CJ, Kang YJ, et al. Zinc supplementation reverses alcohol-induced steatosis in mice through reactivating hepatocyte nuclear factor-4alpha and peroxisome proliferator-activated receptor-alpha. Hepatology. 2009;50:1241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng Z, Borea PA, Varani K, Wilder T, Yee H, Chiriboga L, et al. Adenosine signaling contributes to ethanol-induced fatty liver in mice. J Clin Invest. 2009;119:582–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bala S, Csak T, Saha B, Zatsiorsky J, Kodys K, Catalano D, et al. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol. 2016;64:1378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yue R, Chen GY, Xie G, Hao L, Guo W, Sun X, et al. Activation of PPARalpha-catalase pathway reverses alcoholic liver injury via upregulating NAD synthesis and accelerating alcohol clearance. Free Radic Biol Med. 2021;174:249–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li HH, Tyburski JB, Wang YW, Strawn S, Moon BH, Kallakury BV, et al. Modulation of fatty acid and bile acid metabolism by peroxisome proliferator-activated receptor α protects against alcoholic liver disease. Alcohol Clin Exp Res. 2014;38:1520–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakajima T, Kamijo Y, Tanaka N, Sugiyama E, Tanaka E, Kiyosawa K, et al. Peroxisome proliferator-activated receptor alpha protects against alcohol-induced liver damage. Hepatology. 2004;40:972–80. [DOI] [PubMed] [Google Scholar]
- 69.Zhang D, Tong X, Nelson BB, Jin E, Sit J, Charney N, et al. The hepatic BMAL1/AKT/lipogenesis axis protects against alcoholic liver disease in mice via promoting PPARalpha pathway. Hepatology. 2018;68:883–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nan YM, Kong LB, Ren WG, Wang RQ, Du JH, Li WC, et al. Activation of peroxisome proliferator activated receptor alpha ameliorates ethanol mediated liver fibrosis in mice. Lipids Health Dis. 2013;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Yan T, Wang T, Liu X, Hamada K, Sun D, et al. Crosstalk between CYP2E1 and PPARalpha substrates and agonists modulate adipose browning and obesity. Acta Pharm Sin B. 2022;12:2224–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nanji AA, Dannenberg AJ, Jokelainen K, Bass NM. Alcoholic liver injury in the rat is associated with reduced expression of peroxisome proliferator-alpha (PPARalpha)-regulated genes and is ameliorated by PPARalpha activation. J Pharmacol Exp Ther. 2004;310:417–24. [DOI] [PubMed] [Google Scholar]
- 73.Scholten D, Trebicka J, Liedtke C, Weiskirchen R. The carbon tetrachloride model in mice. Lab Anim. 2015;49:4–11. [DOI] [PubMed] [Google Scholar]
- 74.Xu L, Zheng R, Xie P, Guo Q, Ji H, Li T. Dysregulation of UDP-glucuronosyltransferases in CCl(4) induced liver injury rats. Chem Biol Interact. 2020;325:109115. [DOI] [PubMed] [Google Scholar]
- 75.González-Fernández B, Sánchez DI, Crespo I, San-Miguel B, de Urbina JO, González-Gallego J, et al. Melatonin attenuates dysregulation of the circadian clock pathway in mice with CCl (4)-induced fibrosis and human hepatic stellate cells. Front Pharmacol. 2018;9:556. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Dong S, Chen QL, Song YN, Sun Y, Wei B, Li XY, et al. Mechanisms of CCl4-induced liver fibrosis with combined transcriptomic and proteomic analysis. J Toxicol Sci. 2016;41:561–72. [DOI] [PubMed] [Google Scholar]
- 77.Jain MR, Giri SR, Bhoi B, Trivedi C, Rath A, Rathod R, et al. Dual PPARα/γ agonist saroglitazar improves liver histopathology and biochemistry in experimental NASH models. Liver Int. 2018;38:1084–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Patsenker E, Chicca A, Petrucci V, Moghadamrad S, de Gottardi A, Hampe J, et al. 4-O’-methylhonokiol protects from alcohol/carbon tetrachloride-induced liver injury in mice. J Mol Med (Berl). 2017;95:1077–89. [DOI] [PubMed] [Google Scholar]
- 79.Ji J, Yu Q, Dai W, Wu L, Feng J, Zheng Y, et al. Apigenin alleviates liver fibrosis by inhibiting hepatic stellate cell activation and autophagy via TGF-β1/Smad3 and p38/PPARα pathways. PPAR Res. 2021;2021:6651839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arconzo M, Piccinin E, Moschetta A. Increased risk of acute liver failure by pain killer drugs in NAFLD: focus on nuclear receptors and their coactivators. Dig Liver Dis. 2021;53:26–34. [DOI] [PubMed] [Google Scholar]
- 81.Rivera P, Pastor A, Arrabal S, Decara J, Vargas A, Sánchez-Marín L, et al. Acetaminophen-induced liver injury alters the acyl ethanolamine-based anti-inflammatory signaling system in liver. Front Pharmacol. 2017;8:705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu Z, Li S, Luo J, Luo Y, Dai M, Zheng X, et al. Fenofibrate reverses liver fibrosis in cholestatic mice induced by alpha-naphthylisothiocyanate. Pharmazie. 2021;76:103–8. [DOI] [PubMed] [Google Scholar]
- 83.Makia NL, Goldstein JA. CYP2C8 is a novel target of peroxisome proliferator-activated receptor alpha in human liver. Mol Pharmacol. 2016;89:154–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas M, Burk O, Klumpp B, Kandel BA, Damm G, Weiss TS, et al. Direct transcriptional regulation of human hepatic cytochrome P450 3A4 (CYP3A4) by peroxisome proliferator-activated receptor alpha (PPARalpha). Mol Pharmacol. 2013;83:709–18. [DOI] [PubMed] [Google Scholar]
- 85.Salas-Silva S, Simoni-Nieves A, Razori MV, Lopez-Ramirez J, Barrera-Chimal J, Lazzarini R, et al. HGF induces protective effects in alpha-naphthylisothiocyanate-induced intrahepatic cholestasis by counteracting oxidative stress. Biochem Pharmacol. 2020;174:113812. [DOI] [PubMed] [Google Scholar]
- 86.Shaker ME, Eisa NH, Elgaml A, El-Mesery A, El-Shafey M, El-Dosoky M, et al. Ingestion of mannose ameliorates thioacetamide-induced intrahepatic oxidative stress, inflammation and fibrosis in rats. Life Sci. 2021;286:120040. [DOI] [PubMed] [Google Scholar]
- 87.Du K, Ramachandran A, Jaeschke H. Oxidative stress during acetaminophen hepatotoxicity: sources, pathophysiological role and therapeutic potential. Redox Biol. 2016;10:148–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Unsal V, Cicek M, Sabancilar I. Toxicity of carbon tetrachloride, free radicals and role of antioxidants. Rev Environ Health. 2021;36:279–95. [DOI] [PubMed] [Google Scholar]
- 89.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11–20. [DOI] [PubMed] [Google Scholar]
- 90.Fuchs CD, Radun R, Dixon ED, Mlitz V, Timelthaler G, Halilbasic E, et al. Hepatocyte-specific deletion of adipose triglyceride lipase (adipose triglyceride lipase/patatin-like phospholipase domain containing 2) ameliorates dietary induced steatohepatitis in mice. Hepatology. 2022;75:125–39. [DOI] [PubMed] [Google Scholar]
- 91.Yamaguchi K, Yang L, McCall S, Huang J, Yu XX, Pandey SK, et al. Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohepatitis. Hepatology. 2007;45:1366–74. [DOI] [PubMed] [Google Scholar]
- 92.Francque S, Verrijken A, Caron S, Prawitt J, Paumelle R, Derudas B, et al. PPARalpha gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J Hepatol. 2015;63:164–73. [DOI] [PubMed] [Google Scholar]
- 93.Grygiel-Gorniak B Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications—a review. Nutr J. 2014;13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Costet P, Legendre C, More J, Edgar A, Galtier P, Pineau T. Peroxisome proliferator-activated receptor alpha-isoform deficiency leads to progressive dyslipidemia with sexually dimorphic obesity and steatosis. J Biol Chem. 1998;273:29577–85. [DOI] [PubMed] [Google Scholar]
- 95.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA. 1999;96:7473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Domenici FA, Brochado MJ, Martinelli Ade L, Zucoloto S, da Cunha SF, Vannucchi H. Peroxisome proliferator-activated receptors alpha and gamma2 polymorphisms in nonalcoholic fatty liver disease: a study in Brazilian patients. Gene. 2013;529:326–1. [DOI] [PubMed] [Google Scholar]
- 98.Montagner A, Polizzi A, Fouche E, Ducheix S, Lippi Y, Lasserre F, et al. Liver PPARalpha is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut. 2016;65:1202–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Régnier M, Polizzi A, Smati S, Lukowicz C, Fougerat A, Lippi Y, et al. Hepatocyte-specific deletion of Pparα promotes NAFLD in the context of obesity. Sci Rep. 2020;10:6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hostetler HA, Petrescu AD, Kier AB, Schroeder F. Peroxisome proliferator-activated receptor alpha interacts with high affinity and is conformationally responsive to endogenous ligands. J Biol Chem. 2005;280:18667–82. [DOI] [PubMed] [Google Scholar]
- 101.Monroy-Ramirez HC, Galicia-Moreno M, Sandoval-Rodriguez A, Meza-Rios A, Santos A, Armendariz-Borunda J. PPARs as metabolic sensors and therapeutic targets in liver diseases. Int J Mol Sci. 2021;22:8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kawashima H, Naganuma T, Kusunose E, Kono T, Yasumoto R, Sugimura K, et al. Human fatty acid omega-hydroxylase, CYP4A11: determination of complete genomic sequence and characterization of purified recombinant protein. Arch Biochem Biophys. 2000;378:333–9. [DOI] [PubMed] [Google Scholar]
- 103.Johnson EF, Hsu MH, Savas U, Griffin KJ. Regulation of P450 4A expression by peroxisome proliferator activated receptors. Toxicology. 2002;181–182:203–6. [DOI] [PubMed] [Google Scholar]
- 104.Tugwood JD, Issemann I, Anderson RG, Bundell KR, McPheat WL, Green S. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5’ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 1992;11:433–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gulick T, Cresci S, Caira T, Moore DD, Kelly DP. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci USA. 1994;91:11012–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boubia B, Poupardin O, Barth M, Binet J, Peralba P, Mounier L, et al. Design, synthesis, and evaluation of a novel series of indole sulfonamide peroxisome proliferator activated receptor (PPAR) α/γ/δ triple activators: discovery of lanifibranor, a new antifibrotic clinical candidate. J Med Chem. 2018;61:2246–65. [DOI] [PubMed] [Google Scholar]
- 107.Qian T, Fujiwara N, Koneru B, Ono A, Kubota N, Jajoriya AK, et al. Molecular signature predictive of long-term liver fibrosis progression to inform antifibrotic drug development. Gastroenterology. 2022;162:1210–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hofmann AF. Bile acids: the good, the bad, and the ugly. News Physiol Sci. 1999;14:24–9. [DOI] [PubMed] [Google Scholar]
- 110.Song P, Zhang Y, Klaassen CD. Dose-response of five bile acids on serum and liver bile acid concentrations and hepatotoxicty in mice. Toxicol Sci. 2011;123:359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hofmann AF, Hagey LR. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell Mol Life Sci. 2008;65:2461–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hirschfield GM, Gershwin ME. The immunobiology and pathophysiology of primary biliary cirrhosis. Annu Rev Pathol. 2013;8:303–. [DOI] [PubMed] [Google Scholar]
- 113.Pineda Torra I, Claudel T, Duval C, Kosykh V, Fruchart JC, Staels B. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol Endocrinol. 2003;17:259–72. [DOI] [PubMed] [Google Scholar]
- 114.Xie C, Takahashi S, Brocker CN, He S, Chen L, Xie G, et al. Hepatocyte peroxisome proliferator-activated receptor alpha regulates bile acid synthesis and transport. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:1396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li T, Chiang JY. Regulation of bile acid and cholesterol metabolism by PPARs. PPAR Res. 2009;2009:501739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Trottier J, Caron P, Straka RJ, Barbier O. Profile of serum bile acids in noncholestatic volunteers: gender-related differences in response to fenofibrate. Clin Pharmacol Ther. 2011;90:279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ghonem NS, Auclair AM, Hemme CL, Gallucci GM, de la Rosa Rodriguez R, Boyer JL, et al. Fenofibrate improves liver function and reduces the toxicity of the bile acid pool in patients with primary biliary cholangitis and primary sclerosing cholangitis who are partial responders to ursodiol. Clin Pharmacol Ther. 2020;108:1213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Post SM, Duez H, Gervois PP, Staels B, Kuipers F, Princen HM. Fibrates suppress bile acid synthesis via peroxisome proliferator-activated receptor-alpha-mediated downregulation of cholesterol 7alpha-hydroxylase and sterol 27-hydroxylase expression. Arterioscler Thromb Vasc Biol. 2001;21:1840–5. [DOI] [PubMed] [Google Scholar]
- 119.Dai M, Hua H, Lin H, Xu G, Hu X, Li F, et al. Targeted metabolomics reveals a protective role for basal PPARα in cholestasis induced by α-naphthylisothiocyanate. J Proteome Res. 2018;17:1500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ghonem NS, Ananthanarayanan M, Soroka CJ, Boyer JL. Peroxisome proliferator-activated receptor alpha activates human multidrug resistance transporter 3/ATP-binding cassette protein subfamily B4 transcription and increases rat biliary phosphatidylcholine secretion. Hepatology. 2014;59:1030–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Perreault M, Bialek A, Trottier J, Verreault M, Caron P, Milkiewicz P, et al. Role of glucuronidation for hepatic detoxification and urinary elimination of toxic bile acids during biliary obstruction. PLoS One. 2013;8:e80994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fang HL, Strom SC, Cai H, Falany CN, Kocarek TA, Runge-Morris M. Regulation of human hepatic hydroxysteroid sulfotransferase gene expression by the peroxisome proliferator-activated receptor alpha transcription factor. Mol Pharmacol. 2005;67:1257–67. [DOI] [PubMed] [Google Scholar]
- 123.Kitada H, Miyata M, Nakamura T, Tozawa A, Honma W, Shimada M, et al. Protective role of hydroxysteroid sulfotransferase in lithocholic acid-induced liver toxicity. J Biol Chem. 2003;278:17838–44. [DOI] [PubMed] [Google Scholar]
- 124.Zhao Q, Yang R, Wang J, Hu DD, Li F. PPARalpha activation protects against cholestatic liver injury. Sci Rep. 2017;7:9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jung D, Fried M, Kullak-Ublick GA. Human apical sodium-dependent bile salt transporter gene (SLC10A2) is regulated by the peroxisome proliferator-activated receptor alpha. J Biol Chem. 2002;277:30559–66. [DOI] [PubMed] [Google Scholar]
- 126.Hunt MC, Yang YZ, Eggertsen G, Carneheim CM, Gafvels M, Einarsson C, et al. The peroxisome proliferator-activated receptor alpha (PPARalpha) regulates bile acid biosynthesis. J Biol Chem. 2000;275:28947–53. [DOI] [PubMed] [Google Scholar]
- 127.Senekeo-Effenberger K, Chen S, Brace-Sinnokrak E, Bonzo JA, Yueh MF, Argikar U, et al. Expression of the human UGT1 locus in transgenic mice by 4-chloro-6-(2,3-xylidino)-2-pyrimidinylthioacetic acid (WY-14643) and implications on drug metabolism through peroxisome proliferator-activated receptor alpha activation. Drug Metab Dispos. 2007;35:419–27. [DOI] [PubMed] [Google Scholar]
- 128.Barbier O, Villeneuve L, Bocher V, Fontaine C, Torra IP, Duhem C, et al. The UDP-glucuronosyltransferase 1A9 enzyme is a peroxisome proliferator-activated receptor alpha and gamma target gene. J Biol Chem. 2003;278:13975–83. [DOI] [PubMed] [Google Scholar]
- 129.Barbier O, Duran-Sandoval D, Pineda-Torra I, Kosykh V, Fruchart JC, Staels B. Peroxisome proliferator-activated receptor alpha induces hepatic expression of the human bile acid glucuronidating UDP-glucuronosyltransferase 2B4 enzyme. J Biol Chem. 2003;278:32852–60. [DOI] [PubMed] [Google Scholar]
- 130.Li F, Patterson AD, Krausz KW, Tanaka N, Gonzalez FJ. Metabolomics reveals an essential role for peroxisome proliferator-activated receptor alpha in bile acid homeostasis. J Lipid Res. 2012;53:1625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chiang JYL, Ferrell JM. Discovery of farnesoid X receptor and its role in bile acid metabolism. Mol Cell Endocrinol. 2022;548:111618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tu H, Okamoto AY, Shan B. FXR, a bile acid receptor and biological sensor. Trends Cardiovasc Med. 2000;10:30–5. [DOI] [PubMed] [Google Scholar]
- 133.Chiang JY. Negative feedback regulation of bile acid metabolism: impact on liver metabolism and diseases. Hepatology. 2015;62:1315–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Perino A, Demagny H, Velazquez-Villegas L, Schoonjans K. Molecular physiology of bile acid signaling in health, disease, and aging. Physiol Rev. 2021;101:683–731. [DOI] [PubMed] [Google Scholar]
- 135.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–74. [DOI] [PubMed] [Google Scholar]
- 136.Gbaguidi GF, Agellon LB. The inhibition of the human cholesterol 7alpha-hydroxylase gene (CYP7A1) promoter by fibrates in cultured cells is mediated via the liver x receptor alpha and peroxisome proliferator-activated receptor alpha heterodimer. Nucleic Acids Res. 2004;32:1113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marrapodi M, Chiang JY. Peroxisome proliferator-activated receptor alpha (PPARalpha) and agonist inhibit cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J Lipid Res. 2000;41:514–20. [PubMed] [Google Scholar]
- 138.Cheema SK, Agellon LB. The murine and human cholesterol 7alpha-hydroxylase gene promoters are differentially responsive to regulation by fatty acids mediated via peroxisome proliferator-activated receptor alpha. J Biol Chem. 2000;275:12530–6. [DOI] [PubMed] [Google Scholar]
- 139.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.De Fabiani E, Mitro N, Anzulovich AC, Pinelli A, Galli G, Crestani M. The negative effects of bile acids and tumor necrosis factor-alpha on the transcription of cholesterol 7alpha-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: a novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J Biol Chem. 2001;276:30708–16. [DOI] [PubMed] [Google Scholar]
- 141.Patel DD, Knight BL, Soutar AK, Gibbons GF, Wade DP. The effect of peroxisome-proliferator-activated receptor-alpha on the activity of the cholesterol 7 alpha-hydroxylase gene. Biochem J. 2000;351(pt 3):747–53. [PMC free article] [PubMed] [Google Scholar]
- 142.Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Leiss O, von Bergmann K, Gnasso A, Augustin J. Effect of gemfibrozil on biliary lipid metabolism in normolipemic subjects. Metabolism. 1985;34:74–82. [DOI] [PubMed] [Google Scholar]
- 144.Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, et al. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem. 1998;273:10046–50. [DOI] [PubMed] [Google Scholar]
- 145.Morita SY, Kobayashi A, Takanezawa Y, Kioka N, Handa T, Arai H, et al. Bile salt-dependent efflux of cellular phospholipids mediated by ATP binding cassette protein B4. Hepatology. 2007;46:188–99. [DOI] [PubMed] [Google Scholar]
- 146.Graf GA, Yu L, Li WP, Gerard R, Tuma PL, Cohen JC, et al. ABCG5 and ABCG8 are obligate heterodimers for protein trafficking and biliary cholesterol excretion. J Biol Chem. 2003;278:48275–82. [DOI] [PubMed] [Google Scholar]
- 147.Boyer JL. Bile formation and secretion. Compr Physiol. 2013;3:1035–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dawson PA. Chapter 41—bile formation and the enterohepatic circulation. In: Said HM, ed. Physiology of the Gastrointestinal Tract, 6th ed. Academic Press; 2018:931–56. [Google Scholar]
- 149.Kok T, Wolters H, Bloks VW, Havinga R, Jansen PL, Staels B, et al. Induction of hepatic ABC transporter expression is part of the PPARalpha-mediated fasting response in the mouse. Gastroenterology. 2003;124:160–71. [DOI] [PubMed] [Google Scholar]
- 150.Trottier J, Verreault M, Grepper S, Monte D, Belanger J, Kaeding J, et al. Human UDP-glucuronosyltransferase (UGT) 1A3 enzyme conjugates chenodeoxycholic acid in the liver. Hepatology. 2006;44:1158–70. [DOI] [PubMed] [Google Scholar]
- 151.Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. 2015;160:816–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nathan C Nonresolving inflammation redux. Immunity. 2022;55:592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Devchand PR, Keller H, Peters JM, Vazquez M, Gonzalez FJ, Wahli W. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. [DOI] [PubMed] [Google Scholar]
- 154.Stienstra R, Saudale F, Duval C, Keshtkar S, Groener JE, van Rooijen N, et al. Kupffer cells promote hepatic steatosis via interleukin-1beta-dependent suppression of peroxisome proliferator-activated receptor alpha activity. Hepatology. 2010;51:511–22. [DOI] [PubMed] [Google Scholar]
- 155.Lim WS, Ng DL, Kor SB, Wong HK, Tengku-Muhammad TS, Choo QC, et al. Tumour necrosis factor alpha down-regulates the expression of peroxisome proliferator activated receptor alpha (PPARalpha) in human hepatocarcinoma HepG2 cells by activation of NF-kappaB pathway. Cytokine. 2013;61:266–74. [DOI] [PubMed] [Google Scholar]
- 156.Chew CH, Chew GS, Najimudin N, Tengku-Muhammad TS. Interleukin-6 inhibits human peroxisome proliferator activated receptor alpha gene expression via CCAAT/enhancer-binding proteins in hepatocytes. Int J Biochem Cell Biol. 2007;39:1975–86. [DOI] [PubMed] [Google Scholar]
- 157.Beraza N, Malato Y, Vander Borght S, Liedtke C, Wasmuth HE, Dreano M, et al. Pharmacological IKK2 inhibition blocks liver steatosis and initiation of non-alcoholic steatohepatitis. Gut. 2008;57:655–3. [DOI] [PubMed] [Google Scholar]
- 158.Chew GS, Myers S, Shu-Chien AC, Muhammad TS. Interleukin-6 inhibition of peroxisome proliferator-activated receptor alpha expression is mediated by JAK2- and PI3K-induced STAT1/3 in HepG2 hepatocyte cells. Mol Cell Biochem. 2014;388:25–37. [DOI] [PubMed] [Google Scholar]
- 159.Rigamonti E, Chinetti-Gbaguidi G, Staels B. Regulation of macrophage functions by PPAR-alpha, PPAR-gamma, and LXRs in mice and men. Arterioscler Thromb Vasc Biol. 2008;28:1050–9. [DOI] [PubMed] [Google Scholar]
- 160.Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010;2010:612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shipley JM, Waxman DJ. Down-regulation of STAT5b transcriptional activity by ligand-activated peroxisome proliferator-activated receptor (PPAR) alpha and PPARgamma. Mol Pharmacol. 2003;64:355–64. [DOI] [PubMed] [Google Scholar]
- 162.Kleemann R, Gervois PP, Verschuren L, Staels B, Princen HM, Kooistra T. Fibrates down-regulate IL-1-stimulated C-reactive protein gene expression in hepatocytes by reducing nuclear p50-NFkappa B-C/EBP-beta complex formation. Blood. 2003;101:545–1. [DOI] [PubMed] [Google Scholar]
- 163.Gervois P, Vu-Dac N, Kleemann R, Kockx M, Dubois G, Laine B, et al. Negative regulation of human fibrinogen gene expression by peroxisome proliferator-activated receptor alpha agonists via inhibition of CCAAT box/enhancer-binding protein beta. J Biol Chem. 2001;276:33471–7. [DOI] [PubMed] [Google Scholar]
- 164.Delerive P, De Bosscher K, Besnard S, Vanden Berghe W, Peters JM, Gonzalez FJ, et al. Peroxisome proliferator-activated receptor alpha negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-kappaB and AP-1. J Biol Chem. 1999;274:32048–54. [DOI] [PubMed] [Google Scholar]
- 165.Lee JH, Joe EH, Jou I. PPAR-alpha activators suppress STAT1 inflammatory signaling in lipopolysaccharide-activated rat glia. Neuroreport. 2005;16:829–33. [DOI] [PubMed] [Google Scholar]
- 166.Romics L Jr, Kodys K, Dolganiuc A, Graham L, Velayudham A, Mandrekar P, et al. Diverse regulation of NF-kappaB and peroxisome proliferator-activated receptors in murine non-alcoholic fatty liver. Hepatology. 2004;40:376–85. [DOI] [PubMed] [Google Scholar]
- 167.Zhou YC, Waxman DJ. STAT5b down-regulates peroxisome proliferator-activated receptor alpha transcription by inhibition of ligand-independent activation function region-1 trans-activation domain. J Biol Chem. 1999;274:29874–82. [DOI] [PubMed] [Google Scholar]
- 168.Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140:771–6. [DOI] [PubMed] [Google Scholar]
- 169.Viola A, Munari F, Sanchez-Rodriguez R, Scolaro T, Castegna A. The metabolic signature of macrophage responses. Front Immunol. 2019;10:1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Stienstra R, Mandard S, Tan NS, Wahli W, Trautwein C, Richardson TA, et al. The interleukin-1 receptor antagonist is a direct target gene of PPARalpha in liver. J Hepatol. 2007;46:869–77. [DOI] [PubMed] [Google Scholar]
- 172.Jones DC, Ding X, Daynes RA. Nuclear receptor peroxisome proliferator-activated receptor alpha (PPARalpha) is expressed in resting murine lymphocytes. The PPARalpha in T and B lymphocytes is both transactivation and transrepression competent. J Biol Chem. 2002;277:6838–45. [DOI] [PubMed] [Google Scholar]
- 173.Brocker CN, Yue J, Kim D, Qu A, Bonzo JA, Gonzalez FJ. Hepatocyte-specific PPARA expression exclusively promotes agonist-induced cell proliferation without influence from nonparenchymal cells. Am J Physiol Gastrointest Liver Physiol. 2017;312:G283–g299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu T, Zhang L, Joo D, Sun SC. NF-kappaB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.De Bosscher K, Vanden Berghe W, Haegeman G. The interplay between the glucocorticoid receptor and nuclear factor-kappaB or activator protein-1: molecular mechanisms for gene repression. Endocr Rev. 2003;24:488–522. [DOI] [PubMed] [Google Scholar]
- 176.Banerjee S, Biehl A, Gadina M, Hasni S, Schwartz DM. JAK-STAT signaling as a target for inflammatory and autoimmune diseases: current and future prospects. Drugs. 2017;77:521–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Poli V The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273:29279–82. [DOI] [PubMed] [Google Scholar]
- 178.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–47. [DOI] [PubMed] [Google Scholar]
- 179.Esposito G, Capoccia E, Turco F, Palumbo I, Lu J, Steardo A, et al. Palmitoylethanolamide improves colon inflammation through an enteric glia/toll like receptor 4-dependent PPAR-alpha activation. Gut. 2014;63:1300–12. [DOI] [PubMed] [Google Scholar]
- 180.Huang D, Zhao Q, Liu H, Guo Y, Xu H. PPAR-alpha agonist WY-14643 inhibits LPS-induced inflammation in synovial fibroblasts via NF-kB pathway. J Mol Neurosci. 2016;59:544–3. [DOI] [PubMed] [Google Scholar]
- 181.Crisafulli C, Bruscoli S, Esposito E, Mazzon E, Di Paola R, Genovese T, et al. PPAR-alpha contributes to the anti-inflammatory activity of 17beta-estradiol. J Pharmacol Exp Ther. 2009;331:796–807. [DOI] [PubMed] [Google Scholar]
- 182.Zhao W, Iskandar S, Kooshki M, Sharpe JG, Payne V, Robbins ME. Knocking out peroxisome proliferator-activated receptor (PPAR) alpha inhibits radiation-induced apoptosis in the mouse kidney through activation of NF-kappaB and increased expression of IAPs. Radiat Res. 2007;167:581–91. [DOI] [PubMed] [Google Scholar]
- 183.Friedman SL, Roll FJ. Isolation and culture of hepatic lipocytes, Kupffer cells, and sinusoidal endothelial cells by density gradient centrifugation with Stractan. Anal Biochem. 1987;161:207–18. [DOI] [PubMed] [Google Scholar]
- 184.Blomhoff R, Wake K. Perisinusoidal stellate cells of the liver: important roles in retinol metabolism and fibrosis. FASEB J. 1991;5:271–7. [DOI] [PubMed] [Google Scholar]
- 185.Trivedi P, Wang S, Friedman SL. The power of plasticity-metabolic regulation of hepatic stellate cells. Cell Metab. 2021;33:242–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen L, Li L, Chen J, Li L, Zheng Z, Ren J, et al. Oleoylethanolamide, an endogenous PPAR-α ligand, attenuates liver fibrosis targeting hepatic stellate cells. Oncotarget. 2015;6:42530–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Grabacka M, Pierzchalska M, Plonka PM, Pierzchalski P. The role of PPAR alpha in the modulation of innate immunity. Int J Mol Sci. 2021;22:10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rowbotham SE, Krishna SM, Moran CS, Golledge J. Fenofibrate and telmisartan in the management of abdominal aortic aneurysm. Curr Drug Targets. 2018;19:1241–6. [DOI] [PubMed] [Google Scholar]
- 189.Okiyama W, Tanaka N, Nakajima T, Tanaka E, Kiyosawa K, Gonzalez FJ, et al. Polyenephosphatidylcholine prevents alcoholic liver disease in PPARalpha-null mice through attenuation of increases in oxidative stress. J Hepatol. 2009;50:1236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang Z, Tao Y, Qiu T, Yao X, Jiang L, Wang N, et al. Taurine protected As(2)O(3)-induced the activation of hepatic stellate cells through inhibiting PPARα-autophagy pathway. Chem Biol Interact. 2019;300:123–30. [DOI] [PubMed] [Google Scholar]
- 191.Lu C, Xu W, Shao J, Zhang F, Chen A, Zheng S. Nrf2 induces lipocyte phenotype via a SOCS3-dependent negative feedback loop on JAK2/STAT3 signaling in hepatic stellate cells. Int Immunopharmacol. 2017;49:203–11. [DOI] [PubMed] [Google Scholar]
- 192.Li ZJ, Ou-Yang PH, Han XP. Profibrotic effect of miR-33a with Akt activation in hepatic stellate cells. Cell Signal. 2014;26:141–8. [DOI] [PubMed] [Google Scholar]
- 193.Tardelli M, Claudel T, Bruschi FV, Moreno-Viedma V, Trauner M. Adiponectin regulates AQP3 via PPARα in human hepatic stellate cells. Biochem Biophys Res Commun. 2017;490:51–4. [DOI] [PubMed] [Google Scholar]
- 194.Liu X, Xu J, Rosenthal S, Zhang LJ, McCubbin R, Meshgin N, et al. Identification of lineage-specific transcription factors that prevent activation of hepatic stellate cells and promote fibrosis resolution. Gastroenterology. 2020;158:1728–44.e1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Galli A, Crabb D, Price D, Ceni E, Salzano R, Surrenti C, et al. Peroxisome proliferator-activated receptor gamma transcriptional regulation is involved in platelet-derived growth factor-induced proliferation of human hepatic stellate cells. Hepatology. 2000;31:101–8. [DOI] [PubMed] [Google Scholar]
- 196.Ceni E, Crabb DW, Foschi M, Mello T, Tarocchi M, Patussi V, et al. Acetaldehyde inhibits PPARgamma via H2O2-mediated c-Abl activation in human hepatic stellate cells. Gastroenterology. 2006;131:1235–52. [DOI] [PubMed] [Google Scholar]
- 197.Thoen LF, Guimaraes EL, Dolle L, Mannaerts I, Najimi M, Sokal E, et al. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353–60. [DOI] [PubMed] [Google Scholar]
- 198.Lee TF, Mak KM, Rackovsky O, Lin YL, Kwong AJ, Loke JC, et al. Downregulation of hepatic stellate cell activation by retinol and palmitate mediated by adipose differentiation-related protein (ADRP). J Cell Physiol. 2010;223:648–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pu S, Zhou H, Liu Y, Liu J, Guo Y, Zhou H. Roles of nuclear receptors in hepatic stellate cells. Expert Rev Gastroenterol Hepatol. 2021;15:879–90. [DOI] [PubMed] [Google Scholar]
- 200.Eng FJ, Friedman SL. Transcriptional regulation in hepatic stellate cells. Semin Liver Dis. 2001;21:385–95. [DOI] [PubMed] [Google Scholar]
- 201.Lakatos HF, Thatcher TH, Kottmann RM, Garcia TM, Phipps RP, Sime PJ. The role of PPARs in lung fibrosis. PPAR Res. 2007;2007:71323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhu C, Kim K, Wang X, Bartolome A, Salomao M, Dongiovanni P, et al. Hepatocyte Notch activation induces liver fibrosis in nonalcoholic steatohepatitis. Sci Transl Med. 2018;10:eaat0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Henderson NC, Rieder F, Wynn TA. Fibrosis: from mechanisms to medicines. Nature. 2020;587:555–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Feng X, Gao X, Wang S, Huang M, Sun Z, Dong H, et al. PPAR-alpha agonist fenofibrate prevented diabetic nephropathy by inhibiting M1 macrophages via improving endothelial cell function in db/db mice. Front Med (Lausanne). 2021;8:652558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dehnad A, Fan W, Jiang JX, Fish SR, Li Y, Das S, et al. AGER1 downregulation associates with fibrosis in nonalcoholic steatohepatitis and type 2 diabetes. J Clin Invest. 2020;130:4320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Casini A, Ceni E, Salzano R, Biondi P, Parola M, Galli A, et al. Neutrophil-derived superoxide anion induces lipid peroxidation and stimulates collagen synthesis in human hepatic stellate cells: role of nitric oxide. Hepatology. 1997;25:361–7. [DOI] [PubMed] [Google Scholar]
- 207.Nakagami H, Shimamura M, Miyake T, Shimosato T, Minobe N, Moritani T, et al. Nifedipine prevents hepatic fibrosis in a non-alcoholic steatohepatitis model induced by an L-methionine- and choline-deficient diet. Mol Med Rep. 2012;5:37–40. [DOI] [PubMed] [Google Scholar]
- 208.van Diepen JA, Jansen PA, Ballak DB, Hijmans A, Hooiveld GJ, Rommelaere S, et al. PPAR-alpha dependent regulation of vanin-1 mediates hepatic lipid metabolism. J Hepatol. 2014;61:366–72. [DOI] [PubMed] [Google Scholar]
- 209.Cong WN, Tao RY, Tian JY, Liu GT, Ye F. The establishment of a novel non-alcoholic steatohepatitis model accompanied with obesity and insulin resistance in mice. Life Sci. 2008;82:983–0. [DOI] [PubMed] [Google Scholar]
- 210.Mohamed DI, Elmelegy AA, El-Aziz LF, Abdel Kawy HS, El-Samad AA, El-Kharashi OA. Fenofibrate A peroxisome proliferator activated receptor-α agonist treatment ameliorates Concanavalin A-induced hepatitis in rats. Eur J Pharmacol. 2013;721:35–42. [DOI] [PubMed] [Google Scholar]
- 211.Mandala A, Chen WJ, Armstrong A, Malhotra MR, Chavalmane S, McCommis KS, et al. PPARα agonist fenofibrate attenuates iron-induced liver injury in mice by modulating the Sirt3 and β-catenin signaling. Am J Physiol Gastrointest Liver Physiol. 2021;321:G262–g269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Smati S, Polizzi A, Fougerat A, Ellero-Simatos S, Blum Y, Lippi Y, et al. Integrative study of diet-induced mouse models of NAFLD identifies PPARα as a sexually dimorphic drug target. Gut. 2022;71:807–21. [DOI] [PubMed] [Google Scholar]
- 213.Chen X, Xu Y, Denning KL, Grigore A, Lu Y. PPARalpha agonist WY-14,643 enhances ethanol metabolism in mice: role of catalase. Free Radic Biol Med. 2021;169:283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chou CJ, Haluzik M, Gregory C, Dietz KR, Vinson C, Gavrilova O, et al. WY14,643, a peroxisome proliferator-activated receptor alpha (PPARalpha ) agonist, improves hepatic and muscle steatosis and reverses insulin resistance in lipoatrophic A-ZIP/F-1 mice. J Biol Chem. 2002;277:24484–9. [DOI] [PubMed] [Google Scholar]
- 215.Kim T, Wahyudi LD, Gonzalez FJ, Kim JH. Nuclear receptor PPARalpha agonist Wy-14,643 ameliorates hepatic cell death in hepatic IKKbeta-deficient mice. Biomol Ther (Seoul). 2017;25:504–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Staels B, Rubenstrunk A, Noel B, Rigou G, Delataille P, Millatt LJ, et al. Hepatoprotective effects of the dual peroxisome proliferator-activated receptor alpha/delta agonist, GFT505, in rodent models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Hepatology. 2013;58:1941–52. [DOI] [PubMed] [Google Scholar]
- 217.Zhou Z, Deng L, Hu L, Ren Q, Cai Z, Wang B, et al. Hepatoprotective effects of ZLY16, a dual peroxisome proliferator-activated receptor α/δ agonist, in rodent model of non-alcoholic steatohepatitis. Eur J Pharmacol. 2020;882:173300. [DOI] [PubMed] [Google Scholar]
- 218.Li Z, Xu Y, Cai Z, Wang X, Ren Q, Zhou Z, et al. Discovery of novel dual PPARα/δ agonists based on benzimidazole scaffold for the treatment of non-alcoholic fatty liver disease. Bioorg Chem. 2020;99:103803. [DOI] [PubMed] [Google Scholar]
- 219.Feng Z, Xiang J, Liu H, Li J, Xu X, Sun G, et al. Design, synthesis, and biological evaluation of triazolone derivatives as potent PPARα/δ dual agonists for the treatment of nonalcoholic steatohepatitis. J Med Chem. 2022;65:2571–92. [DOI] [PubMed] [Google Scholar]
- 220.Li PP, Shan S, Chen YT, Ning ZQ, Sun SJ, Liu Q, et al. The PPARalpha/gamma dual agonist chiglitazar improves insulin resistance and dyslipidemia in MSG obese rats. Br J Pharmacol. 2006;148:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tsai HC, Li TH, Huang CC, Huang SF, Liu RS, Yang YY, et al. Beneficial effects of the peroxisome proliferator-activated receptor α/γ agonist aleglitazar on progressive hepatic and splanchnic abnormalities in cirrhotic rats with portal hypertension. Am J Pathol. 2018;188:1608–24. [DOI] [PubMed] [Google Scholar]
- 222.Akbari R, Behdarvand T, Afarin R, Yaghooti H, Jalali MT, Mohammadtaghvaei N. Saroglitazar improved hepatic steatosis and fibrosis by modulating inflammatory cytokines and adiponectin in an animal model of non-alcoholic steatohepatitis. BMC Pharmacol Toxicol. 2021;22:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kumar DP, Caffrey R, Marioneaux J, Santhekadur PK, Bhat M, Alonso C, Koduru SV, et al. The PPAR α/γ agonist saroglitazar improves insulin resistance and steatohepatitis in a diet induced animal model of nonalcoholic fatty liver disease. Sci Rep. 2020;10:9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lefere S, Puengel T, Hundertmark J, Penners C, Frank AK, Guillot A, et al. Differential effects of selective- and pan-PPAR agonists on experimental steatohepatitis and hepatic macrophages(☆). J Hepatol. 2020;73:757–0. [DOI] [PubMed] [Google Scholar]
- 225.Wettstein G, Luccarini JM, Poekes L, Faye P, Kupkowski F, Adarbes V, et al. The new-generation pan-peroxisome proliferator-activated receptor agonist IVA337 protects the liver from metabolic disorders and fibrosis. Hepatol Commun. 2017;1:524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang C, Peng F, Zhong B, Shi Y, Wang X, Jin X, et al. Metabolomic analysis reveals the therapeutic effects of MBT1805, a novel pan-peroxisome proliferator-activated receptor agonist, on alpha-naphthylisothiocyanate-induced cholestasis in mice. Front Pharmacol. 2021;12:732478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Li MH, Chen W, Wang LL, Sun JL, Zhou L, Shi YC, et al. RLA8-A new and highly effective quadruple PPAR-α/γ/δ and GPR40 agonist to reverse nonalcoholic steatohepatitis and fibrosis. J Pharmacol Exp Ther. 2019;369:67–77. [DOI] [PubMed] [Google Scholar]
- 228.Zhou Z, Ren Q, Jiao S, Cai Z, Geng X, Deng L, et al. Discovery of new and highly effective quadruple FFA1 and PPARα/γ/δ agonists as potential anti-fatty liver agents. Eur J Med Chem. 2022;229:114061. [DOI] [PubMed] [Google Scholar]
- 229.Sandoval-Rodriguez A, Monroy-Ramirez HC, Meza-Rios A, Garcia-Bañuelos J, Vera-Cruz J, Gutiérrez-Cuevas J, et al. Pirfenidone is an agonistic ligand for PPARα and improves NASH by activation of SIRT1/LKB1/pAMPK. Hepatol Commun. 2020;4:434–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Gutiérrez-Cuevas J, Sandoval-Rodríguez A, Monroy-Ramírez HC, Vazquez-Del Mercado M, Santos-García A, Armendáriz-Borunda J. Prolonged-release pirfenidone prevents obesity-induced cardiac steatosis and fibrosis in a mouse NASH model. Cardiovasc Drugs Ther. 2021;35:927–38. [DOI] [PubMed] [Google Scholar]
- 231.Lee H, Ahn J, Shin SS, Yoon M. Ascorbic acid inhibits visceral obesity and nonalcoholic fatty liver disease by activating peroxisome proliferator-activated receptor α in high-fat-diet-fed C57BL/6J mice. Int J Obes (Lond). 2019;43:1620–30. [DOI] [PubMed] [Google Scholar]
- 232.Ziamajidi N, Khaghani S, Hassanzadeh G, Vardasbi S, Ahmadian S, Nowrouzi A, et al. Amelioration by chicory seed extract of diabetes- and oleic acid-induced non-alcoholic fatty liver disease (NAFLD)/non-alcoholic steatohepatitis (NASH) via modulation of PPARα and SREBP-1. Food Chem Toxicol. 2013;58:198–209. [DOI] [PubMed] [Google Scholar]
- 233.Ham JR, Lee HI, Choi RY, Sim MO, Seo KI, Lee MK. Anti-steatotic and anti-inflammatory roles of syringic acid in high-fat diet-induced obese mice. Food Funct. 2016;7:689–97. [DOI] [PubMed] [Google Scholar]
- 234.Wang C, Duan X, Sun X, Liu Z, Sun P, Yang X, et al. Protective effects of glycyrrhizic acid from edible botanical glycyrrhiza glabra against non-alcoholic steatohepatitis in mice. Food Funct. 2016;7:3716–23. [DOI] [PubMed] [Google Scholar]
- 235.Xu J, Peng Y, Zeng Y, Hua YQ, Xu XL. 2, 3, 4’, 5-tetrahydroxystilbene-2–0-β-d glycoside attenuates age- and diet-associated non-alcoholic steatohepatitis and atherosclerosis in LDL receptor knockout mice and its possible mechanisms. Int J Mol Sci. 2019;20:1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mahli A, Koch A, Fresse K, Schiergens T, Thasler WE, Schönberger C, et al. Iso-alpha acids from hops (Humulus lupulus) inhibit hepatic steatosis, inflammation, and fibrosis. Lab Invest. 2018;98:1614–26. [DOI] [PubMed] [Google Scholar]
- 237.Zhang J, Zhang H, Deng X, Zhang N, Liu B, Xin S, et al. Baicalin attenuates non-alcoholic steatohepatitis by suppressing key regulators of lipid metabolism, inflammation and fibrosis in mice. Life Sci. 2018;192:46–54. [DOI] [PubMed] [Google Scholar]
- 238.Yahaghi L, Yaghmaei P, Hayati-Roodbari N, Irani S, Ebrahim-Habibi A. Betanin effect on PPAR-α and SREBP-1c expression in NMRI mice model of steatohepatitis with fibrosis. Physiol Int. 2020;107:67–81. [DOI] [PubMed] [Google Scholar]
- 239.Duan X, Meng Q, Wang C, Liu Z, Liu Q, Sun H, et al. Calycosin attenuates triglyceride accumulation and hepatic fibrosis in murine model of non-alcoholic steatohepatitis via activating farnesoid X receptor. Phytomedicine. 2017;25:83–92. [DOI] [PubMed] [Google Scholar]
- 240.Hua YQ, Zeng Y, Xu J, Xu XL. Naringenin alleviates non-alcoholic steatohepatitis in middle-aged Apoe(−/−)mice: role of SIRT1. Phytomedicine. 2021;81:153412. [DOI] [PubMed] [Google Scholar]
- 241.Nakade Y, Sakamoto K, Yamauchi T, Inoue T, Kobayashi Y, Yamamoto T, et al. Conophylline inhibits non-alcoholic steatohepatitis in mice. PLoS One. 2017;12:e0178436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kong D, Zhang Z, Chen L, Huang W, Zhang F, Wang L, et al. Curcumin blunts epithelial-mesenchymal transition of hepatocytes to alleviate hepatic fibrosis through regulating oxidative stress and autophagy. Redox Biol. 2020;36:101600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Alshammari GM, Al-Qahtani WH, AlFaris NA, Alzahrani NS, Alkhateeb MA, Yahya MA. Quercetin prevents cadmium chloride-induced hepatic steatosis and fibrosis by downregulating the transcription of miR-21. Biofactors. 2021;47:489–505. [DOI] [PubMed] [Google Scholar]
- 244.Li X, Wang Z, Klaunig JE. The effects of perfluorooctanoate on high fat diet induced non-alcoholic fatty liver disease in mice. Toxicology. 2019;416:1–14. [DOI] [PubMed] [Google Scholar]
- 245.Meng Q, Duan XP, Wang CY, Liu ZH, Sun PY, Huo XK, et al. Alisol B 23-acetate protects against non-alcoholic steatohepatitis in mice via farnesoid X receptor activation. Acta Pharmacol Sin. 2017;38:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Park JG, Mok JS, Han YI, Park TS, Kang KW, Choi CS, et al. Connectivity mapping of angiotensin-PPAR interactions involved in the amelioration of non-alcoholic steatohepatitis by Telmisartan. Sci Rep. 2019;9:4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kobayashi T, Kanno K, Nguyen PT, Sugiyama A, Kawahara A, Otani Y, et al. Periostin antisense oligonucleotide prevents hepatic steatosis and fibrosis in a mouse model of non-alcoholic steatohepatitis. J Gastroenterol Hepatol. 2020;35:2140–50. [DOI] [PubMed] [Google Scholar]
- 248.Guo XY, Chen JN, Sun F, Wang YQ, Pan Q, Fan JG. circRNA_0046367 prevents hepatoxicity of lipid peroxidation: an inhibitory role against hepatic steatosis. Oxid Med Cell Longev. 2017;2017:3960197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Loyer X, Paradis V, Hénique C, Vion AC, Colnot N, Guerin CL, et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARalpha expression. Gut. 2016;65:1882–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kumar S, Rani R, Karns R, Gandhi CR. Augmenter of liver regeneration protein deficiency promotes hepatic steatosis by inducing oxidative stress and microRNA-540 expression. Faseb j. 2019;33:3825–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kamijo Y Is hepatic peroxisome proliferator-activated receptor alpha essential for the metabolic effects of fibrates? J Gastroenterol Hepatol. 2018;33:978–. [DOI] [PubMed] [Google Scholar]
- 252.Yamashita S, Masuda D, Matsuzawa Y. Pemafibrate, a new selective PPARalpha modulator: drug concept and its clinical applications for dyslipidemia and metabolic diseases. Curr Atheroscler Rep. 2020;22:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ferri N, Corsini A, Sirtori C, Ruscica M. PPAR-alpha agonists are still on the rise: an update on clinical and experimental findings. Expert Opin Investig Drugs. 2017;26:593–602. [DOI] [PubMed] [Google Scholar]
- 254.Sasaki Y, Asahiyama M, Tanaka T, Yamamoto S, Murakami K, Kamiya W, et al. Pemafibrate, a selective PPARα modulator, prevents non-alcoholic steatohepatitis development without reducing the hepatic triglyceride content. Sci Rep. 2020;10:7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Lyu H, Li X, Wu Q, Hao L . Overexpression of microRNA-21 mediates Ang II-induced renal fibrosis by activating the TGF-β1/Smad3 pathway via suppressing PPARα. J Pharmacol Sci. 2019;141:70–8. [DOI] [PubMed] [Google Scholar]
- 256.Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med. 2012;4:121ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ, et al. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci USA. 2011;108:10355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Liu JR, Cai GY, Ning YC, Wang JC, Lv Y, Guo YN, et al. Caloric restriction alleviates aging-related fibrosis of kidney through downregulation of miR-21 in extracellular vesicles. Aging (Albany NY). 2020;12:18052–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Corpechot C, Chazouilleres O, Rousseau A, Le Gruyer A, Habersetzer F, Mathurin P, et al. A placebo-controlled trial of bezafibrate in primary biliary cholangitis. N Engl J Med. 2018;378:2171–81. [DOI] [PubMed] [Google Scholar]
- 260.de Vries E, Bolier R, Goet J, Pares A, Verbeek J, de Vree M, et al. Fibrates for Itch (FITCH) in fibrosing cholangiopathies: a double-blind, randomized, placebo-controlled trial. Gastroenterology. 2021;160:734–43 e736. [DOI] [PubMed] [Google Scholar]
- 261.Honda A, Ikegami T, Nakamuta M, Miyazaki T, Iwamoto J, Hirayama T, et al. Anticholestatic effects of bezafibrate in patients with primary biliary cirrhosis treated with ursodeoxycholic acid. Hepatology. 2013;57:1931–41. [DOI] [PubMed] [Google Scholar]
- 262.Rudic JS, Poropat G, Krstic MN, Bjelakovic G, Gluud C. Bezafibrate for primary biliary cirrhosis. Cochrane Database Syst Rev. 2012;1:CD009145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lawitz EJ, Bhandari BR, Ruane PJ, Kohli A, Harting E, Ding D, et al. Fenofibrate mitigates hypertriglyceridemia in nonalcoholic steatohepatitis patients treated with cilofexor/firsocostat. Clin Gastroenterol Hepatol. 2022;21:143–52.e3. [DOI] [PubMed] [Google Scholar]
- 264.Oscarsson J, Onnerhag K, Riserus U, Sunden M, Johansson L, Jansson PA, et al. Effects of free omega-3 carboxylic acids and fenofibrate on liver fat content in patients with hypertriglyceridemia and non-alcoholic fatty liver disease: a double-blind, randomized, placebo-controlled study. J Clin Lipidol. 2018;12:1390–403 e1394. [DOI] [PubMed] [Google Scholar]
- 265.Levy C, Peter JA, Nelson DR, Keach J, Petz J, Cabrera R, et al. Pilot study: fenofibrate for patients with primary biliary cirrhosis and an incomplete response to ursodeoxycholic acid. Aliment Pharmacol Ther. 2011;33:235–42. [DOI] [PubMed] [Google Scholar]
- 266.Nakajima A, Eguchi Y, Yoneda M, Imajo K, Tamaki N, Suganami H, et al. Randomised clinical trial: pemafibrate, a novel selective peroxisome proliferator-activated receptor alpha modulator (SPPARMalpha), versus placebo in patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2021;54:1263–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Fruchart JC, Hermans MP, Fruchart-Najib J, Kodama T. Selective peroxisome proliferator-activated receptor alpha modulators (SPPARMalpha) in the metabolic syndrome: is pemafibrate light at the end of the tunnel? Curr Atheroscler Rep. 2021;23:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sanyal A, Charles ED, Neuschwander-Tetri BA, Loomba R, Harrison SA, Abdelmalek MF, et al. Pegbelfermin (BMS-986036), a PEGylated fibroblast growth factor 21 analogue, in patients with non-alcoholic steatohepatitis: a randomised, double-blind, placebo-controlled, phase 2a trial. Lancet. 2019;392:2705–17. [DOI] [PubMed] [Google Scholar]
- 269.Abdelmalek MF, Charles ED, Sanyal AJ, Harrison SA, Neuschwander-Tetri BA, Goodman Z, et al. The FALCON program: two phase 2b randomized, double-blind, placebo-controlled studies to assess the efficacy and safety of pegbelfermin in the treatment of patients with nonalcoholic steatohepatitis and bridging fibrosis or compensated cirrhosis. Contemp Clin Trials. 2021;104:106335. [DOI] [PubMed] [Google Scholar]
- 270.Harrison SA, Ruane PJ, Freilich BL, Neff G, Patil R, Behling CA, et al. Efruxifermin in non-alcoholic steatohepatitis: a randomized, double-blind, placebo-controlled, phase 2a trial. Nat Med. 2021;27:1262–71. [DOI] [PubMed] [Google Scholar]
- 271.Schattenberg JM, Pares A, Kowdley KV, Heneghan MA, Caldwell S, Pratt D, et al. A randomized placebo-controlled trial of elafibranor in patients with primary biliary cholangitis and incomplete response to UDCA. J Hepatol. 2021;74:1344–54. [DOI] [PubMed] [Google Scholar]
- 272.Ratziu V, Harrison SA, Francque S, Bedossa P, Lehert P, Serfaty L, et al. Elafibranor, an agonist of the peroxisome proliferator-activated receptor-alpha and -delta, induces resolution of nonalcoholic steatohepatitis without fibrosis worsening. Gastroenterology. 2016;150:1147–59.e1145. [DOI] [PubMed] [Google Scholar]
- 273.Gawrieh S, Noureddin M, Loo N, Mohseni R, Awasty V, Cusi K, et al. Saroglitazar, a PPAR-α/γ agonist, for treatment of NAFLD: a randomized controlled double-blind phase 2 trial. Hepatology. 2021;74:1809–24. [DOI] [PubMed] [Google Scholar]
- 274.Vuppalanchi R, Caldwell SH, Pyrsopoulos N, deLemos AS, Rossi S, Levy C, et al. Proof-of-concept study to evaluate the safety and efficacy of saroglitazar in patients with primary biliary cholangitis. J Hepatol. 2022;76:75–85. [DOI] [PubMed] [Google Scholar]
- 275.Vuppalanchi R, González-Huezo MS, Payan-Olivas R, Muñoz-Espinosa LE, Shaikh F, Pio Cruz-Lopez JL, et al. A multicenter, open-label, single-arm study to evaluate the efficacy and safety of saroglitazar in patients with primary biliary cholangitis. Clin Transl Gastroenterol. 2021;12:e00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Sven MF, Pierre B, Manal FA, Quentin MA, Elisabetta B, Vlad R, et al. A randomised, double-blind, placebo-controlled, multi-centre, dose-range, proof-of-concept, 24-week treatment study of lanifibranor in adult subjects with non-alcoholic steatohepatitis: design of the NATIVE study. Contemp Clin Trials. 2020;98:106170. [DOI] [PubMed] [Google Scholar]
- 277.Francque SM, Bedossa P, Ratziu V, Anstee QM, Bugianesi E, Sanyal AJ, et al. A randomized, controlled trial of the pan-PPAR agonist lanifibranor in NASH. N Engl J Med. 2021;385:1547–58. [DOI] [PubMed] [Google Scholar]
- 278.Ishibashi S, Yamashita S, Arai H, Araki E, Yokote K, Suganami H, et al. Effects of K-877, a novel selective PPARalpha modulator (SPPARMalpha), in dyslipidaemic patients: a randomized, double blind, active- and placebo-controlled, phase 2 trial. Atherosclerosis. 2016;249:36–43. [DOI] [PubMed] [Google Scholar]
- 279.Ishibashi S, Arai H, Yokote K, Araki E, Suganami H, Yamashita S, et al. Efficacy and safety of pemafibrate (K-877), a selective peroxisome proliferator-activated receptor alpha modulator, in patients with dyslipidemia: results from a 24-week, randomized, double blind, active-controlled, phase 3 trial. J Clin Lipidol. 2018;12:173–84. [DOI] [PubMed] [Google Scholar]
- 280.Geng L, Lam KSL, Xu A. The therapeutic potential of FGF21 in metabolic diseases: from bench to clinic. Nat Rev Endocrinol. 2020;16:654–7. [DOI] [PubMed] [Google Scholar]
- 281.Perakakis N, Stefanakis K, Feigh M, Veidal SS, Mantzoros CS. Elafibranor and liraglutide improve differentially liver health and metabolism in a mouse model of non-alcoholic steatohepatitis. Liver Int. 2021;41:1853–66. [DOI] [PubMed] [Google Scholar]
- 282.Briand F, Heymes C, Bonada L, Angles T, Charpentier J, Branchereau M, et al. A 3-week nonalcoholic steatohepatitis mouse model shows elafibranor benefits on hepatic inflammation and cell death. Clin Transl Sci. 2020;13:529–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Cariou B, Hanf R, Lambert-Porcheron S, Zaïr Y, Sauvinet V, Noël B, et al. Dual peroxisome proliferator-activated receptor α/δ agonist GFT505 improves hepatic and peripheral insulin sensitivity in abdominally obese subjects. Diabetes Care. 2013;36:2923–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Cariou B, Zaïr Y, Staels B, Bruckert E. Effects of the new dual PPAR α/δ agonist GFT505 on lipid and glucose homeostasis in abdominally obese patients with combined dyslipidemia or impaired glucose metabolism. Diabetes Care. 2011;34:2008–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Kaul U, Parmar D, Manjunath K, Shah M, Parmar K, Patil KP, et al. New dual peroxisome proliferator activated receptor agonist-Saroglitazar in diabetic dyslipidemia and non-alcoholic fatty liver disease: integrated analysis of the real world evidence. Cardiovasc Diabetol. 2019;18:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Boyer-Diaz Z, Aristu-Zabalza P, Andres-Rozas M, Robert C, Ortega-Ribera M, Fernandez-Iglesias A, et al. Pan-PPAR agonist lanifibranor improves portal hypertension and hepatic fibrosis in experimental advanced chronic liver disease. J Hepatol. 2021;74:1188–99. [DOI] [PubMed] [Google Scholar]
- 287.Gervois P, Torra IP, Chinetti G, Grotzinger T, Dubois G, Fruchart JC, et al. A truncated human peroxisome proliferator-activated receptor alpha splice variant with dominant negative activity. Mol Endocrinol. 1999;13:1535–49. [DOI] [PubMed] [Google Scholar]
- 288.Palmer CN, Hsu MH, Griffin KJ, Raucy JL, Johnson EF. Peroxisome proliferator activated receptor-alpha expression in human liver. Mol Pharmacol. 1998;53:14–22. [PubMed] [Google Scholar]
- 289.Foreman JE, Koga T, Kosyk O, Kang BH, Zhu X, Cohen SM, et al. Species differences between mouse and human PPARalpha in modulating the hepatocarcinogenic effects of perinatal exposure to a high-affinity human PPARalpha agonist in mice. Toxicol Sci. 2021;183:81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Yan T, Luo Y, Yan N, Hamada K, Zhao N, Xia Y, et al. Intestinal peroxisome proliferator-activated receptor alpha-fatty acid-binding protein 1 axis modulates nonalcoholic steatohepatitis. Hepatology. 2022. (In press; ) [2022 Apr 23.doi:doi: 10.1002/hep32538Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Li G, Brocker CN, Xie C, Yan T, Noguchi A, Krausz KW, et al. Hepatic peroxisome proliferator-activated receptor alpha mediates the major metabolic effects of Wy-14643. J Gastroenterol Hepatol. 2018;33:1138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]