Abstract
Non-polyadenylated RNA includes a large subset of crucial regulators of RNA expression and constitutes a substantial portion of the transcriptome, playing essential roles in gene regulation. For example, enhancer RNAs are long non-coding RNAs that perform enhancer-like functions, are bi-directionally transcribed, and usually lack polyA tails. This paper presents a novel method, selSeq, that selectively removes mRNA and pre-mRNA from samples enabling the selective sequencing of crucial regulatory elements, including non-polyadenylated RNAs such as long non-coding RNA, enhancer RNA, and non-canonical mRNA.
Introduction
Noncoding RNAs (ncRNAs), a large subset of which are non-polyadenylated (polyA), are crucial regulators of RNA expression in a diverse range of life forms [1–3]. In mammals, ncRNAs constitute a substantial portion of the transcriptome [1], playing essential roles in gene regulation via chromatin modification transcriptional regulation, and post-transcriptional processing [4]. Long non-coding RNAs (lncRNAs) are the most prevalent and diverse class of ncRNAs and are typically over 200 nucleotides long and lack protein-coding potential [1]. A large subset of lncRNAs is not polyadenylated and regulate gene expression, with abnormal expression associated with several diseases [5–7] and are predictive of outcomes [8]. Enhancer RNAs (eRNAs) are IncRNAs that perform enhancer-like functions [5, 6], are bi-directionally transcribed, and usually lack polyA tails [9]. Due to their low copy numbers [10, 11], eRNAs are challenging to detect, with most being non-polyA. Nevertheless, measuring eRNAs using total RNA-seq has provided valuable insights into transcriptional regulation [12].
Current methods to explore ncRNAs and intermediate messenger RNA splicing reactions, such as total RNA sequencing or microarrays, have severe limitations. For example, microarrays require knowledge of the transcripts of interest a priori and have a small dynamic range. Total RNA sequencing quantifies all RNA in a sample, which, after ribosomal RNA depletion, is dominated by mRNA transcripts and includes a relatively low number of lncRNAs (a typical human cell has 3x103-5x104 lncRNAs, while there are 3x105-1x106 mRNA transcripts). Thus, quantification of rare lncRNAs and isoforms is challenging. Here, we present a novel method that selectively removes polyadenylated RNA (e.g., mRNA and pre-mRNA; and optionally rRNA) from samples, allowing for the comprehensive characterization of low-abundance lncRNAs, eRNAs, and non-canonical mRNA splicing. Our method, selSeq, offers a reliable and sensitive approach to exploring these crucial regulatory elements.
Materials and methods
The protocol described in this peer-reviewed article is published on protocols.io, DOI 10.17504/protocols.io.j8nlkwpk6l5r/v1, and is included for printing as S1 File with this article.
Expected results
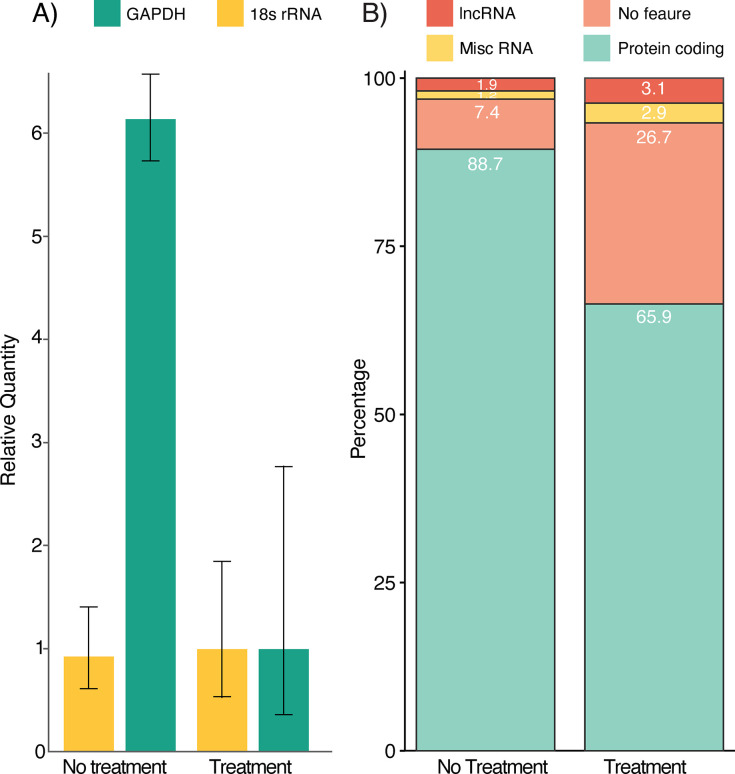
There will be a decrease in the RNA quantity following the completion of the protocol. A reverse transcriptase quantitative PCR can be used to see the reduction of polyA housekeeping genes to non-polyA rRNA (Fig 1A) if the optional rRNA depletion step is not done as part of the protocol. Total RNA sequencing after rRNA depletion shows a large decrease in the proportion of polyA-tailed transcripts as illustrated in Fig 1B, where human liver total RNA (Ambion, USA) was used and aligned to the GENCODE nucleotide sequence of the Human GRCh38.p13 genome assembly version containing all regions, including reference chromosomes, scaffolds, assembly patches, and haplotypes. The proportion of lncRNA, no feature, and miscellaneous RNA have increased substantially, and the protein-coding assignments have decreased, with those remaining likely non-polyadenylated transcripts or degraded polyadenylated transcripts.
Fig 1.
selSeq results showing A) the decrease in polyA tailed GAPDH housekeeping gene and no decrease in non-polyA tailed 18s ribosomal RNA (n = 4; error bars show one standard deviation); and B) total RNA sequencing after rRNA depletion shows an increase in the proportion of long non-coding, unassigned (no feature), and miscellaneous RNA with a corresponding decrease in the protein-coding assignments, the remainder of which are likely non-polyadenylated transcripts.
Supporting information
Sequence data is available on SRA under the accession number PRJNA949687.
(PDF)
Data Availability
Sequence data is available on SRA under the accession number PRJNA949687.
Funding Statement
JZM, 1R01AI153213, National Institute of Allergy and Infectious Diseases (NIAID), https://www.niaid.nih.gov/, The funders did not and will not have a role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol. 2016;937: 3–17. doi: 10.1007/978-3-319-42059-2_1 [DOI] [PubMed] [Google Scholar]
- 2.Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell. 2009;136: 629–641. doi: 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 3.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458: 223–227. doi: 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Reviews Genetics 2009 10:3. 2009;10: 155–159. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 5.Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55. doi: 10.1167/iovs.13-13221 [DOI] [PubMed] [Google Scholar]
- 6.Lekka E, Hall J. Noncoding RNAs in disease. FEBS Letters. 2018. doi: 10.1002/1873-3468.13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J, Wang F, Wu P, Chen Y, Jia Y. Aberrant LncRNA expression in leukemia. Journal of Cancer. 2020. doi: 10.7150/jca.42093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrar JE, Smith JL, Othus M, Huang BJ, Wang Y-C, Ries R, et al. Long Noncoding RNA Expression Independently Predicts Outcome in Pediatric Acute Myeloid Leukemia. J Clin Oncol. 2023; JCO2201114. doi: 10.1200/JCO.22.01114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507. doi: 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbovytska V, Kim SK, Kuybu F, Götze M, Um D, Kang K, et al. Enhancer RNAs stimulate Pol II pause release by harnessing multivalent interactions to NELF. Nat Commun. 2022;13. doi: 10.1038/s41467-022-29934-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465. doi: 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carullo NVN, Phillips RA, Simon RC, Roman Soto SA, Hinds JE, Salisbury AJ, et al. Enhancer RNAs predict enhancer–gene regulatory links and are critical for enhancer function in neuronal systems. Nucleic Acids Res. 2020;48. doi: 10.1093/nar/gkaa671 [DOI] [PMC free article] [PubMed] [Google Scholar]