Summary
Background
Acute rheumatic fever (ARF) is a serious post-infectious sequala of Group A Streptococcus (GAS, Streptococcus pyogenes). In New Zealand (NZ) ARF is a major cause of health inequity. This study describes the genomic analysis of GAS isolates associated with childhood skin and throat infections in Auckland NZ.
Methods
Isolates (n = 469) collected between March 2018 and October 2019 from the throats and skin of children (5–14 years) underwent whole genomic sequencing. Equal representation across three ethnic groups was ensured through sample quotas with isolates obtained from Indigenous Māori (n = 157, 33%), NZ European/Other (n = 149, 32%) and Pacific Peoples children (n = 163, 35%). Using in silico techniques isolates were classified, assessed for diversity, and examined for distribution differences between groups. Comparisons were also made with GAS strains identified globally.
Findings
Genomic analysis revealed a diverse population consisting of 65 distinct sequence clusters. These sequence clusters spanned 49 emm-types, with 11 emm-types comprised of several, distinct sequence clusters. There is evidence of multiple global introductions of different lineages into the population, as well as local clonal expansion. The M1UK lineage comprised 35% of all emm1 isolates.
Interpretation
The GAS population was characterized by a high diversity of strains, resembling patterns observed in low- and middle-income countries. However, strains associated with outbreaks and antimicrobial resistance commonly found in high-income countries were also observed. This unique combination poses challenges for vaccine development, disease management and control.
Funding
The work was supported by the Health Research Council of New Zealand (HRC), award number 16/005.
Keywords: Streptococcus pyogenes, Impetigo, Pharyngitis, Genomics, Children, New Zealand
Research in context.
Evidence before this study
Acute rheumatic fever (ARF) is a serious post-infectious sequala of Group A Streptococcus (GAS, Streptococcus pyogenes) that can lead to permanent heart valve damage and chronic rheumatic heart disease (RHD). In Aotearoa New Zealand, ARF is a major cause of health inequity. Studies specifically focused on GAS skin and throat infections in ARF endemic settings are limited. However, prior studies have shown the distribution and diversity of GAS strains differ by geographical location and socioeconomic factors. Generally emm-type diversity has been shown to be significantly greater in low and middle income countries (LMIC) compared to high-income countries (HIC).
Added value of this study
GAS skin and throat isolates collected from children in Auckland, New Zealand during the 18 months preceding the global COVID-19 pandemic revealed a high degree of genetic diversity in the population at that time. This is illustrated by the identification of 65 distinct sequence clusters within the 469 isolates analysed. The resulting GAS population is a mixture of that seen in LMIC and HIC. For example the most common emm-types were emm1, emm89 and emm12 that have been frequently associated with disease in HICs, yet pattern D isolates that comprise emm-types associated with disease in LMICs accounted for 22% of isolates, and the emm53 pattern D strain was frequently isolated from the skin and throats of Indigenous Māori and Pacific Peoples children.
Implications of all the available evidence
The population structure of GAS in Auckland children, where high strain diversity reminiscent of LMICs is combined with strains associated with outbreaks and antimicrobial resistance in HICs, creates unique challenges for vaccine development as well as disease management and control strategies. The evidence of multiple global introductions, together with local clonal expansion, highlights the need for ongoing surveillance and monitoring of GAS in New Zealand communities.
Introduction
Acute rheumatic fever (ARF) is a serious post-infectious sequala of Group A Streptococcus (GAS, S. pyogenes) that can lead to permanent heart valve damage and chronic rheumatic heart disease (RHD).1 Over 30 million people are estimated to be living with RHD, with the highest burden in low- and middle-income countries (LMIC) and underserved Indigenous communities in some high-income countries (HIC).2 In Aotearoa New Zealand (NZ), ARF is a major cause of health inequity with Indigenous Māori and Pacific Peoples children experiencing some of the highest rates in the world.3 GAS pharyngitis is a well-documented trigger for ARF. However, increasing evidence suggests GAS skin infections also play a role, either directly, or via immune priming whereby repeated GAS exposures contribute to immune dysregulation and ARF.4, 5, 6
Molecular epidemiological studies of GAS have typically relied on single locus emm-typing, which analyzes variability in the 5′ end of the emm gene encoding the M-protein.7 GAS pattern types can be inferred from emm-type (via emm-clustering)8 and have been suggested as a marker of tissue tropism, with pattern A-C associated with throat infections, pattern D with skin and pattern E with either throat or skin.9 The most recent analysis of GAS strains associated with ARF in NZ found a large diversity of emm-types, approximately half of which were classified as pattern D.10 Given the association of pattern D strains with skin infections, these were not considered classical “rheumatogenic” strains. Indeed, the concept of rheumatogenic GAS appears less likely to apply to settings where ARF persists today. In contrast to historical ARF outbreaks that were linked to single strain-types, a broad range of emm-types were associated with ARF in a recent systematic review.11
Since both throat and skin infections may be precursors of ARF, and each of these sites is capable of onward GAS transmission, ARF prevention measures (including future vaccines) should consider both clinical syndromes.12,13 Prior studies have shown the distribution and diversity of GAS strains differs by geographical location and socioeconomic factors. Overall, emm-type diversity is significantly greater in LMICs compared with HICs14,15 and while investigations specifically focused on skin and throat infections in ARF endemic settings are limited, trends are emerging. A higher diversity of emm-types and a larger proportion of pattern D isolates have been associated with both skin and pharyngeal infections in LMIC settings compared with HIC settings.14,16 Moreover, emm-type diversity and the proportion of pattern D isolates was higher in skin and pharyngeal isolates from NZ children living in areas of high social deprivation and elevated ARF risk, compared with areas of low ARF risk.17 This finding suggests the associations between GAS, pattern type and tissue tropism are more complex in ARF endemic areas and further investigation of bacterial factors are needed in these settings.
The high diversity of emm-types in LMIC and ARF endemic settings also presents challenges for vaccine development, particularly candidates based on the M-protein such as the 30-valent vaccine. Due to the high diversity of emm types in LMICs, estimated coverage of this vaccine is considerably lower in LMIC than in HICs for which it was designed.14,15,18 To circumvent coverage limitations, several candidates based on combinations of conserved or less divergent antigens are now in development.19 Comprehensive assessment of antigen carriage and variation for these candidates requires molecular analysis beyond emm-typing, and a fully sequenced Global Atlas of 2000 isolates has been compiled to that end.20 This revealed 15 vaccine components with >99% theoretical coverage across the global population at the time.
Despite the importance of genomics in understanding GAS disease presentation and vaccine development, genomic analysis of GAS from NZ have been limited. Some 300 NZ isolates were included in the Global Atlas,20 and whole-genome sequencing was applied to an outbreak of invasive GAS in an eldercare facility.21 However, the population structure remains largely unexplored, including whether the strains of public health importance such as the M1UK lineage that is associated with upsurges of scarlet fever and invasive GAS disease in the Northern hemisphere and Australia22, 23, 24 is circulating in NZ. Here we describe genomic analysis of GAS isolates from a case-control study that investigated risk-factors for skin and throat infections in NZ school children.25 Sampling isolates from children at high- and low-risk of developing ARF in the year proceeding the global COVID-19 pandemic ensures this data is reflective of usual GAS diversity. This provides a critical baseline from which to explore the impact of NZs COVID-19 elimination strategy, which included strong border entry restrictions and altered the epidemiology of most infectious diseases in NZ including ARF,26,27 on the GAS population.
Methods
Isolate collection and demographics
GAS isolates were obtained from throat or skin swabs collected from Auckland-based children, aged 5–14 years, who presented to primary healthcare with a sore throat or skin sore, between March 2018 to October 2019. Full details of the prospective disease incidence study that investigated the risk-factors associated with GAS skin and throat infections in this cohort has previously been published.25 GAS culture was carried out by Labtests (Auckland, NZ) using standard laboratory methods,17,28 resulting in a collection of 560 GAS isolates, of which 469 GAS isolates were eligible (duplicates from the same participant excluded) and viable for genomic analysis (359 from throat swabs and 110 from skin swabs). Sample quotas ensured equal representation by ethnic group (Māori, Pacific Peoples and NZ European/Other) and isolates were analysed by the prioritized ethnic group of the participant from which they were isolated as described.25 Prioritised ethnic grouping allocates individuals to a single ethnic group based on a prioritised order of Māori, Pacific Peoples, Asian, and NZ European/Other. For example if an individual identifies as being both Māori and NZ European, they will be classified as Māori for the purposes of data analysis. Pacific Peoples in Auckland include many different and varied ethnicities with connections to Pacific Island nations.
Whole-genome sequencing analysis and genome comparison
Genomic DNA was extracted from a single colony using a QIAsymphony™ DSP DNA Mini Kit (Qiagen). Sequence libraries were prepared using NexteraXT and sequenced on the NextSeq500. In silico analysis was conducted using standard pipelines for quality control, genome assembly, phylogenetic analysis, and annotation. Genomes that showed signs of contamination with species other than GAS or was of low quality based on read depth and genome assembly metrics were excluded. Using in silico tools, species-specific typing schemes were assigned and genomes were screened for the presence of virulence and antimicrobial resistance genes. Multi-locus sequence types (MLST) and emm-types were assigned using mlst v2.19.0 (https://github.com/tseemann/mlst) and emmtyper v.0.1.0 (https://github.com/MDU-PHL/emmtyper). The emm patterns were inferred from emm-type. The emm cluster was inferred from emm-type as previously described.8 Virulence and antimicrobial resistance genes were screened via two bioinformatics pipelines, ABRicate (https://bio.tools/ABRicate) and screen_assembly3.20 For both methods a minimum 85% identity and 90% coverage was used to define the presence of key virulence genes and 99% identity, and 99% coverage was used for antimicrobial resistance gene detection.
Genetic relationships were quantified between genomes at both the species and lineage-based level as described previously.13,20 Sequence clusters (SCs) were defined using popPUNK and closely related groups of isolates were defined by a kmer-based (ska) approach using 99% core genome and <20 SNPs. All mapping and variant calling was performed with snippy v4.4.5 (https://github.com/tseemann/snippy) applying a minifrac value of 0.9 and mincov value of 10. Core genome alignments were produced using snippy-core. Maximum likelihood phylogenies were produced using IQ-Tree v1.6.12, using the GTR + F + G4 model with rapid bootstrapping -bb 1000 and -alrt 1000. Phylogenetic trees were visualised with R with ggtree, ggtree-extra packages. Pairwise SNPs were calculated in R using harrietr (v.0.2.3). For lineage and emm-based trees the same parameters as above were applied and trees were corrected for constant sites using snp-sites from the core.full.aln. Baps groups were defined using RHeirBAPS at two levels with a maximum of 50 groups. Context isolates were selected from publicly available WGS sequence data. To maximise the diversity within the comparator dataset isolates were selected that maximised the number of MLST types, emm subtypes and country of origin for a broad and informative view of the NZ isolates in the global context. See the Supplementary Data file for further details.
Epidemiological and statistical analysis
A chi squared goodness of fit test was used to assess whether an emm-type (or emm pattern) was more associated in a particular group. A p-value of 0.05 was considered significant and indicates an emm-type is not proportionally distributed across each group relative to the population proportions. Bonferroni’s correction was applied to adjust for multiple comparisons, with adjusted significances set to p = 0.001 for emm-type and p = 0.017 for emm pattern. Where emm-types are of low prevalence (n < 5) these associations should be treated with caution as are likely influenced by under-sampling. Epidemiological and statistical analysis was performed using RStudio version 1.4.1717.
Estimation of vaccine coverage
Vaccine coverage of the 30-valent StreptAnova vaccine was defined as the proportion of isolates with emm-types that are contained in the vaccine (assuming cross-opsonisation or not) as described.29 Non-vaccine emm-types that had previously reported killing of ≥50% in vitro bactericidal assays performed with strains of the same emm-type were classed as cross-opsonisation (CO) positive.29,30 Non-vaccine emm-types with unknown cross-opsonisation status or <50% killed in the two previously reported cross-opsonisation studies were classed as ‘cross-opsonisation negative’.29,30 There are 33 emm-types defined as cross-opsonisation positive. Vaccine coverage of the TeeVax vaccine was defined as the proportion of isolates covered by the 18 T-antigens included in the recombinant vaccine as well as three T-antigens with high cross-reactivity (T3.1, T18.2, T28.2) for a total of 21 tee-types.31 The tee-types were assigned using a database of bp genes previously reported32 using 99% identity and 99% coverage as cut-offs. TeeVax antigen sequences (T-antigen domains) were screened at the same thresholds. Representative sequences of 12 conserved vaccine antigens contained within leading ‘combination’ vaccine candidates were obtained from UniProt (https://www.uniprot.org) and screened using screen_assembly3.py20 using a 85% identity, 90% coverage threshold. Vaccine coverage for these antigens was defined as the proportion of isolates for which the intact gene was present. Vaccine coverage for peptides was defined as the proportion of isolates for which the peptide was present. The vaccine peptide candidates J8, S2, p∗17 (including the J8.0 and J8.1 alleles) were screened using seqkit locate to find exact matches to the peptide sequences (100% identity).
Ethical approval
Ethics approval for this study was obtained from the NZ Health and Disability Ethics Committee (HDEC) reference 17/NTA/262. Informed consent from a parent or legal guardian and assent (for children) was required for all participants. The ethics, study design, and operation were also reviewed by Māori and Pacific Governance Groups.
Role of the funding source
The funders of this study had no role in the study design, data collection, data analysis, interpretation, or writing of the report.
Results
GAS emm type and disease distributions by ethnic groups
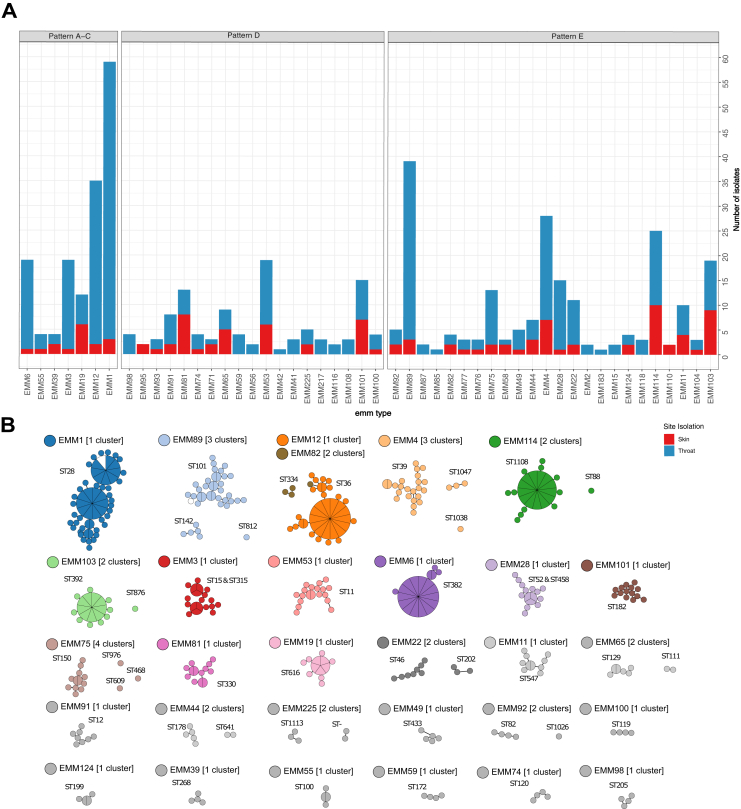
The study included 469 GAS isolates collected from Māori (n = 157), Pacific Peoples (n = 163) and NZ European/Other (n = 149) children aged 5–14 years (Table 1, Supplementary Table S1). Whole genome sequencing identified 49 emm-types and 12 emm clusters (Supplementary Figs. S1 and S2). The five most prevalent emm-types were emm1 (13.0%), emm89 (8.5%), emm12 (7.5%), emm4 (6.0%) and emm114 (5.0%). The Simpson’s reciprocal index (SRI), a measure of diversity, was 20.2 (95% confidence interval [CI], 16.6 to 23.8). This sits between SRI recently described for high- and low-income settings.14 The most common emm-types isolated from Māori and NZ European/Other children groups were somewhat similar. Emm1 (13.3%), emm89 (10.1%) and emm12 (7.6%) were the most prevalent within the Māori group; emm1 (20.0%), emm89 (11.3%) and emm4 (9.3%) were the most prevalent within the NZ European/Other group. In contrast, isolates from the Pacific Peoples group differed with emm103 (6.7%), emm53 (6.7%) and emm12 (6.1%) being the most prevalent (Supplementary Fig. S1). Of the 49 emm-types, 34 were identified in both throat and skin, 13 were exclusively in throat samples, while only two emm-types were identified exclusively in skin samples (Fig. 1a). Seven emm-types had significantly different proportions of isolates in one or more demographic group (p ≤ 0.05) (Supplementary Table S1). Of these comparisons, emm39 and emm77 were exclusively isolated from the Pacific Peoples group, emm53 was more commonly isolated from the Pacific and Māori groups, and emm28 was more commonly isolated from the NZ European/Other group (Supplementary Table S1).
Table 1.
Contingency table for Group A Streptococcus emm patterns by site of isolation and prioritised ethnic group.
| Isolated from | Isolates N (%) |
p-value | ||
|---|---|---|---|---|
| Prioritised ethnicity of children | ||||
| Māori | NZ European/Other | Pacific Peoples | ||
| Throat | ||||
| A–C | 49 (31.2) | 51 (34.2) | 36 (22.1) | 0.381 |
| D | 24 (15.3) | 10 (6.7) | 35 (21.5) | <0.001a |
| E | 52 (33.1) | 59 (39.6) | 43 (26.4) | 0.394 |
| Skin | ||||
| A–C | 6 (3.8) | 2 (1.3) | 8 (4.9) | 0.436 |
| D | 12 (7.6) | 7 (4.7) | 19 (11.7) | 0.537 |
| E | 14 (8.9) | 20 (13.4) | 22 (13.5) | 0.283 |
| Combined throat and skin | ||||
| A–C | 55 (35.0) | 53 (35.6) | 44 (27.0) | 0.305 |
| D | 36 (22.0) | 17 (11.4) | 54 (33.1) | <0.001a |
| E | 66 (42.0) | 79 (53.0) | 65 (39.1) | 0.203 |
| Total isolates | 157 (100) | 149 (100) | 163 (100) | |
Indicates a statistically significant values from the Chi-squared goodness-of-fit test.
Fig. 1.
Overview of the Auckland GAS population. (a) Counts of isolates by emm-type, coloured by site of isolation and faceted by emm pattern type (A–C, D, and E). (b) Networks of minimum weight spanning trees of sequence clusters in the population. Clusters were defined by popPUNK where relative core and accessory distances for each pair were determined based on k-mer comparisons and split-kmer analysis. Nodes are scaled by the number of isolates within a 25 SNP threshold such that larger nodes containing multiple isolates are shown by larger circles with wedges representing multiple isolates within the node. Singleton clusters were excluded, and clusters are coloured by emm type and MLSTs are labelled.
Evaluating emm pattern types showed that Pattern D isolates had a significantly lower prevalence in NZ European/Other group, compared to the Māori and Pacific Peoples groups (Table 1). Pattern D isolates were also found two and 3.5 times more frequently in the throats of Māori and Pacific children in comparison to NZ European/Other children while pattern A-C and E isolates were proportionally distributed across ethnic groups for throat and skin.
The Auckland GAS population comprises a combination of international introductions and geographical restricted lineages
To gain increased resolution of isolate similarity beyond emm-type, WGS and genomic analysis were performed. The 469 isolates formed 65 sequence clusters (SCs), and genomes within each SC shared a lower intragroup distance (0.001% average nucleotide divergence) than they did to any other genome in the dataset (>0.012 average nucleotide divergence). Excluding singletons, 19 emm-types were represented by a single genomic cluster and a dominant MLST type. Strikingly, 11 emm-types were assigned to more than one SC and multiple MLSTs that were too divergent to be considered the same genetic lineage (>4 locus variants) (Fig. 1b). Taken together this shows that the GAS population circulating in Auckland school children is diverse, and that children infected with the same emm-type maybe infected with divergent lineages.
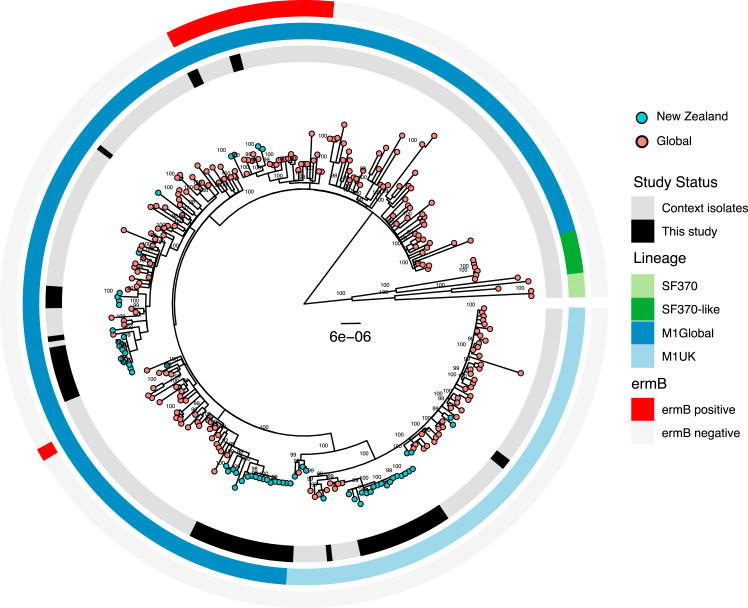
Phylogenetic comparisons of the four most dominant emm-types (emm1, emm89, emm12 and emm4) with global context isolates revealed the GAS population in Auckland children is represented by a mix of global and local lineages. The Auckland emm1 population included multiple M1global lineages (34/59 isolates), M1UK (21/59 isolates) and M1Hong-Kong ermB resistant strains (4/59 isolates). Three large monophyletic clades (two M1global and one M1UK) comprised most of the emm1 isolates in the study population but several smaller clades were also observed across the global context. This indicated there were multiple introductions of emm1 strains into Auckland, as well as local clonal expansion of some of these lineages including M1UK (Fig. 2, Supplementary Fig. S2).
Fig. 2.
Phylogenetic relationship between emm1 GAS strains isolated from this study and global context isolates. Rings a. Black – isolated in this study, Grey – global context isolates. b. Lineage light blue – M1uK, dark blue – MGAS5005, dark green – SF370, light green – SF370-MGAS5005 intermediate.
Similar patterns of multiple introductions and local clonal expansions were also observed in emm4, emm89 and emm12. Briefly, three previously identified clades of emm12 were observed33 and twenty of the emm12 isolates formed a monophyletic clade with other NZ isolates obtained from the GAS Atlas20 suggesting a possible unique emm12 clade in NZ. This unique clade was observed in all ethnic groups. The remaining 15 emm12 isolates had scattered placement throughout the phylogeny intermixed with the global context isolates (Supplementary Fig. S3). The phylogenetic analysis of emm4 and emm89 isolates also supported separation into multiple sequence clusters, with multiple emm89 lineages34 and both chimeric and non-chimeric emm4 isolates identified.35 However, resolution was restricted by the inclusion of those highly divergent isolates (Supplementary Fig. S4a and S5a). Thus separate phylogenies showing only the dominant sequence clusters were examined in more detail (Supplementary Fig. S4b and S5b). Again these phylogenies demonstrate a global representation of emm4 and emm89 are present in Auckland children. These trends were also observed in the less dominant emm-types, as illustrated by multiple ska and popPUNK lineages assigned to isolates of the same emm-type (Supplementary data), suggesting that the mix of highly divergent populations circulating in Auckland is reflective of multiple introductions.
Virulence and antimicrobial resistance
Among the 469 isolates, the most frequently identified resistance determinant was tetM in 74 (16%) of isolates. The remaining resistance determinates including macrolide resistance (ermA, ermB, mefA, msrD) were all found in 1–7 isolates (<1%), indicating that antimicrobial resistance does not appear to be highly prevalent in childhood community streptococcal infections in Auckland (Supplementary Table S4). In contrast, a diverse set of virulence factors were commonly identified and highly prevalent including six Streptodornases (4–100% of isolates), and thirteen superantigens (12–75% of isolates) (Supplementary Table S5). Approximately 30% of all isolates (emm4, emm22, emm89, emm28) lacked the intact hasABC capsule locus—that is they lack intact genes for hasA, hasB or hasC—indicating a high proportion of acapsular strains in circulation.
Estimation of vaccine coverage varies based on formulation and ethnic groups
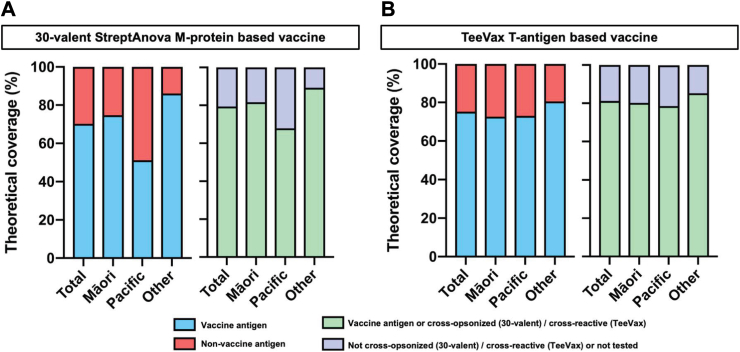
The experimental StreptAnova vaccine is based on 30 unique emm-types of which 21 were observed in this cohort accounting for 70.2% (330/469) of the total isolates (Fig. 3, Supplementary Fig. S1 and Table S6). This increased to 29 emm-types, or 79.4% (373/469) isolates when the potential effect of cross-opsonic antibodies was considered. Across ethnic groups, theoretical vaccine coverage varied significantly and was markedly lower in the Pacific Peoples group (51.2%) than NZ European/Others (86.0%). This deficit was partially corrected if cross-opsonisation of non-vaccine emm-types is assumed, with theoretical coverage increasing significantly in the Pacific Peoples group (67.9%); whereas only modest improvements were observed for Māori (81.6%) and NZ European/Other (89.3%) children (Fig. 3, Supplementary Table S6).
Fig. 3.
Theoretical coverage provided by type-specific vaccines in Auckland GAS isolates. (a) Coverage for the 30-valent StreptAnova vaccine showing the proportion of isolates with emm-types included (blue) or not-included in the vaccine (red), and assuming cross-opsonisation (mint) or not-cross-opsonised or not tested (lilac). (b) Coverage for the TeeVax vaccine showing the proportion of isolates with tee-types included (blue) or not-included in the vaccine (red), and assuming cross-reactivity (mint) or not-cross-reactive or not tested (lilac). Cross-opsonisation and cross-reactivity as reported.29, 30, 31
The experimental TeeVax vaccine is based on 18 tee-types, with a domain of each of the 18 corresponding T-antigens included.31 From a total of 35 tee-types identified in the study population, 18 were theoretically covered by the vaccine accounting for 75.2% (353/469) (Fig. 3, Supplementary Table S7). This increased to 20 tee-types covering 81.2% (381/469) when the potential of reported cross-reactivity was considered. The presence of multiple tee4 subtypes in this study population impacted on TeeVax coverage estimates. Fifty five isolates were classified as tee4, and though a tee4 domain is included in the vaccine, this sequence only matches that found in the tee4.1 subtype and not the tee4.2, 4.3, 4.4, 4.5 and 4.6 subtypes. Cross-reactivity between tee4 subtypes is yet to be tested or reported. In comparison with the 30-valent StreptAnova vaccine coverage (assuming cross-opsonisation or cross-reactivity) TeeVax showed similar coverage for Māori (80.2%), notably higher predicted coverage for the Pacific Peoples group (78.5%) and slightly lower in NZ Europeans/Others (85.2%) (Fig. 3, Supplementary Table S7).
The 12 major antigens from a series of multi-component vaccines in pre-clinical development20 were screened in the study cohort and found at greater than 99% coverage in all three ethnic groups (Table 2). These antigens also exhibited low levels of allelic variation suggesting the potential of these conserved, combination vaccines to provide high coverage across the entire population. Several peptide vaccine formulations are also in varying stages of development including J8 derived from the C-terminal region of the M-protein and Combivax (a bi-valent candidate encompassing a J8 variant peptide named p∗17 and the S2 peptide derived from SpyCEP).37 These components had low to moderate predicted coverage across all three ethnic groups (38–65%). However, for J8 this increased to 82–86% when the reported cross-reactivity between J8 alleles J8.0 and J8.1 was considered.36
Table 2.
Theoretical coverage of combination vaccines in the Auckland GAS isolates.
| Vaccine | Components | Total no. isolates (%) | Māori no. isolates (%) | Pacific Peoples no. isolates (%) | Other no. isolates (%) | Number variants (aa) |
|---|---|---|---|---|---|---|
| GSK-combo | SLO | 468 (99) | 155 (98.7) | 163 (100) | 149 (100) | 36 |
| SpyCEP | 469 (100) | 157 (100) | 163 (100) | 149 (100) | 74 | |
| SpyAD | 469 (100) | 157 (100) | 163 (100) | 149 (100) | 56 | |
| GAC (gacI) | 467 (99.5) | 156 (99.3) | 162 (99.3) | 149 (100) | 15 | |
| 1 antigen present | 469 (100) | 157 (100) | 163 (100) | 149 (100) | – | |
| Spy7 | Spy0651 | 468 (99.7) | 157 (100) | 162 (99.3) | 149 (100) | 61 |
| Spy0762 | 469 (100) | 157 (100) | 163 (100) | 149 (100) | 7 | |
| Spy0942 | 469 (100) | 157 (100) | 163 (100) | 149 (100) | 10 | |
| PulA | 469 (100) | 157 (100) | 163 (100) | 149 (100) | 65 | |
| OppA | 469 (100) | 157 (100) | 163 (100) | 149 (100) | 33 | |
| SpyAD | 469 (100) | 157 (100) | 163 (100) | 149 (100) | 56 | |
| 1 antigen present | 469 (100) | 157 (100) | 163 (100) | 149 (100) | – | |
| Combo5 | TF | 469 (100) | 157 (100) | 163 (100) | 149 (100) | 20 |
| ScpA | 453 (96.5) | 152 (96.8) | 153 (93.8) | 148 (99.3) | 82 | |
| SpyCEP | 469 (100) | 157 (100) | 163 (100) | 149 (100) | 74 | |
| ADI | 469 (100) | 157 (100) | 163 (100) | 149 (100) | 11 | |
| SLO | 468 (99.7) | 155 (98.7) | 163 (100) | 149 (100) | 36 | |
| 1 antigen present | 469 (100) | 157 (100) | 163 (100) | 149 (100) | – | |
| J8 | J8.0 | 189 (40) | 69 (44) | 64 (39) | 56 (38) | 1 |
| J8.1 | 214 (46) | 67 (43) | 71 (44) | 76 (51) | 1 | |
| 1 peptide presenta | 403 (86) | 136 (87) | 135 (83) | 132 (82) | 2 | |
| Combivax | p∗17b | 189 (40) | 69 (44) | 64 (39) | 56 (38) | – |
| S2 | 276 (59) | 102 (65) | 83 (51) | 91 (61) | 2 | |
| 1 peptide present | 327 (70) | 119 (76) | 112 (69) | 96 (64) | – |
Assuming cross-reactivity between J8 and J8.1 as described.36
Allowing 2 mismatches to p∗17 sequence.
Discussion
Whole genome sequencing of GAS skin and throat isolates collected from Auckland children in the 18 months preceding the global pandemic has revealed a high degree of genetic diversity in the population at that time. This is illustrated by 65 distinct sequence clusters within the 469 isolates analysed. These sequence clusters span 49 emm-types, with 11 of these emm-types comprised of several, distinct sequence clusters. The GAS population structure is likely reflective of Auckland demographics, a city that is ethnically diverse with a gradient of social deprivation and a high prevalence of GAS disease.3 The resulting GAS population is a mixture of that seen in both high- and low-income settings globally. For example the most common emm-types were emm1, emm89 and emm12 that have been frequently associated with disease in HIC, yet pattern D isolates that comprise emm-types associated with disease in LMIC14,15,38 accounted for 22% of all isolates, and the emm53 pattern D strain was frequently isolated from the skin and throats of Māori and Pacific children. Furthermore, Simpson’s reciprocal index (SRI), a measure of strain diversity in a population, was 20.2. This sits between the SRI described in a recent study comparing GAS in a HIC (49.3, Sheffield, England) to a LIC (11.1, The Gambia).14
The presence of multiple sequence clusters within certain emm-types, and the diversity of these lineages when compared with global context isolates, suggest that GAS isolated from children living in Auckland is a mix of both global and local lineages. This is clearly illustrated by the emm1 population where M1global, M1UK and M1 Hong-Kong ermB resistant strains were identified alongside specific lineages that have undergone local clonal expansion. In some cases unique local lineages appear to become well established. For example the large, locally expanded emm12 clade observed in this study was previously identified in the NZ isolates included in the GAS Atlas.20 Similar trends were observed across multiple emm-types suggesting that frequent global introductions contribute to the highly divergent GAS population observed. Auckland has a large Pacific community comprised of Pacific Peoples from many different and varied ethnicities, with frequent travel between the island nations in the South Pacific region and Auckland. Prior studies of GAS in Samoa and Fiji have shown diverse emm-types in circulation, with strains belonging to the D4 clustering being the most prevalent.38,39 The D4 cluster is the largest of GAS clusters and comprises 32 emm-types belonging to pattern group D,8 many of which were observed in this study. While the Auckland GAS strain diversity likely results from introductions from various international destinations, the diversity observed in Pacific children where the most common emm-types differed from the other ethnic groups, may also reflect frequent introductions from the South Pacific into the Pacific community within Auckland.
The M1UK lineage has been associated with upsurges of scarlet fever and invasive GAS disease in HIC settings in recent years.22, 23, 24 In this study M1UK comprised 35% of all emm1 isolates suggesting that this lineage was introduced prior to 2018, yet unlike the United Kingdom, did not appear to rapidly expand and become the dominant emm1 lineage leading into the pandemic. In the six years prior to the pandemic (2014 onwards) England experienced increases in scarlet fever notifications,40 and by 2020 M1UK comprised >90% of all emm1 in the United Kingdom.22 In contrast, hospitalisation data indicates scarlet fever did not increase in NZ up to 2018,41 despite the presence of M1UK. It is possible that the diversity of GAS strains and ongoing global introductions selected against rapid M1UK expansion. However, active surveillance of GAS disease other than ARF is lacking in NZ, and it will be important to examine the impacts of the border closures and re-opening throughout 2020–2022 on the prevalence of outbreak strains such as M1UK with further genomic studies.
In line with recent observations in other high ARF prevalence settings,14,38 the proposed tissue tropism of A–C pattern strains dominating throat isolates and pattern D dominating skin isolates was not observed for all groups in this study. Pattern D isolates were more frequently identified in the throats of Māori and Pacific children in comparison to NZ European/Other children, and for Pacific children the proportion of pattern D and pattern A-C isolates from throats was equivalent. This is consistent with prior molecular epidemiology in NZ that found a higher proportion of pattern D isolates in children with elevated ARF risk,17 and similarly suggests that additional microbiological features, beyond pattern type, are associated with tropism. The percentage of pattern D isolates in the ethnic groups was proportional to the predicted coverage of the 30-valent vaccine, which was highest for isolates obtained from NZ European/Other children, intermediate for Māori and lowest for isolates from Pacific children. Predicted coverage of the TeeVax candidate, based on T-antigens was higher than the 30-valent vaccine in Pacific children suggesting that the reduced variation of tee-types compared with emm-types,31 could provide benefit for settings with high GAS strain diversity. However, the molecular epidemiology of tee-types globally is relatively unexplored compared with emm-type. The recent discovery of a new FCT-type (and tee-type) in the Gambia14 points to a need for additional surveillance of tee-types to inform the development of T-antigen based vaccines. There were several previously described, highly conserved, vaccine antigens20 predicted to provide coverage of all strains types identified in this study. As such these antigens, in combination vaccine formulations, would circumvent vaccine coverage challenges in high strain diversity settings such as NZ should they prove efficacious in clinical trials.
The diverse population structure of GAS in Auckland children creates unique challenges for vaccine development, disease management and control. The evidence of multiple global introductions, as well as local clonal expansion, highlights the need for ongoing surveillance and monitoring of GAS in NZ communities. With national and international consortia now focused on accelerating GAS vaccines to prevent serious GAS disease and rheumatic fever,19 surveillance of precursor skin and throat infections using approaches similar to that used in this study will be needed in sentinel sites in the South Pacific region and globally.
Contributors
J.A.L designed the analysis, interpreted the results and co-wrote the original manuscript, J.B project managed the study, interpreted the results and co-wrote the original manuscript, T.J, B.H, T.C and D.L performed investigations, A.A, M.H and D.S.P contributed to Māori and Pacific governance groups and interpreted results, S.T interpreted results, M.G.B and D.A.W initiated and designed the study, obtained funding and interpreted results. N.J.M initiated and designed the study, obtained funding, designed the analysis, interpreted the results and co-wrote the original manuscript. All authors have read and reviewed the manuscript.
Data sharing statement
Sequence reads are available on the NCBI Sequence Read Archive (Bioproject PRJNA985396).
Declaration of interests
The authors have no conflict of interest to declare.
Acknowledgments
We thank the Māori Governance Group, led by Associate Professor Matire Harwood and the Pacific Governance Group, led by Associate Professor Dianne Sika-Paotonu for cultural advice on the study design and operation. We would also like to thank Labtests for collection and storage of the isolates and Professor Thomas Proft and Dr Jacelyn Loh at the University of Auckland for provision of the TeeVax sequences.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lanwpc.2023.100964.
Contributor Information
Jake A. Lacey, Email: jake.lacey@unimelb.edu.au.
Nicole J. Moreland, Email: n.moreland@auckland.ac.nz.
Appendix A. Supplementary data
References
- 1.Carapetis J.R., Beaton A., Cunningham M., et al. Acute rheumatic fever and rheumatic heart disease. Nat Rev Dis Prim. 2016;1:1–24. doi: 10.1038/nrdp.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watkins D.A., Johnson C., Colquhoun S., et al. Global, regional, and national burden of rheumatic heart disease, 1990-2015. N Engl J Med. 2017;377:713–722. doi: 10.1056/NEJMoa1603693. [DOI] [PubMed] [Google Scholar]
- 3.Bennett J., Zhang J., Leung W., et al. Rising ethnic inequalities in acute rheumatic fever and rheumatic heart disease, New Zealand, 2000-2018. Emerg Infect Dis. 2021;27:3–46. doi: 10.3201/eid2701.191791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker M.G., Gurney J., Moreland N.J., et al. Risk factors for acute rheumatic fever: a case-control study. Lancet Reg Health West Pac. 2022;26 doi: 10.1016/j.lanwpc.2022.100508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliver J., Bennett J., Thomas S., et al. Preceding group A streptococcus skin and throat infections are individually associated with acute rheumatic fever: evidence from New Zealand. BMJ Glob Health. 2021;6 doi: 10.1136/bmjgh-2021-007038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorenz N., Ho T.K.C., McGregor R., et al. Serological profiling of group A Streptococcus infections in acute rheumatic fever. Clin Infect Dis. 2021;73 doi: 10.1093/cid/ciab180. [DOI] [PubMed] [Google Scholar]
- 7.Beall B., Facklam R., Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanderson-Smith M., Oliveira D.M., Guglielmini J., et al. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis. 2014;210(8):1325–1338. doi: 10.1093/infdis/jiu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessen D.E., Lizano S. Tissue tropisms in group A streptococcal infections. Future Microbiol. 2010;5:623–638. doi: 10.2217/fmb.10.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson D.A., Smeesters P.R., Steer A.C., et al. M-protein analysis of Streptococcus pyogenes isolates associated with acute rheumatic fever in New Zealand. J Clin Microbiol. 2015;53:3618–3620. doi: 10.1128/JCM.02129-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Crombrugghe G., Baroux N., Botteaux A., et al. The limitations of the rheumatogenic concept for group A Streptococcus: systematic review and genetic analysis. Clin Infect Dis. 2020;70:1453–1460. doi: 10.1093/cid/ciz425. [DOI] [PubMed] [Google Scholar]
- 12.Whitcombe A.L., McGregor R., Bennett J., et al. Increased breadth of Group A Streptococcus antibody responses in children with Acute Rheumatic Fever compared to precursor pharyngitis and skin infections. J Infect Dis. 2022;226(1):167–176. doi: 10.1093/infdis/jiac043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacey J.A., Marcato A.J., Chisholm R.H., et al. Evaluating the role of asymptomatic throat carriage of Streptococcus pyogenes in impetigo transmission in remote Aboriginal communities in Northern Territory, Australia: a retrospective genomic analysis. Lancet Microbe. 2023;4(7):E524–E533. doi: 10.1016/s2666-5247(23)00068-x. [DOI] [PubMed] [Google Scholar]
- 14.Bah S.Y., Keeley A.J., Armitage E.P., et al. Genomic characterization of skin and soft tissue Streptococcus pyogenes isolates from a low-income and a high-income setting. mSphere. 2022;8 doi: 10.1128/msphere.00469-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steer A.C., Law I., Matatolu L., Beall B.W., Carapetis J.R. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 16.Smeesters P.R., Vergison A., Campos D., Aguiar E., Deyi V., Melderenet L. Differences between Belgian and Brazilian group A Streptococcus epidemiologic landscape. PLoS One. 2006;1 doi: 10.1371/journal.pone.0000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williamson D.A., Smeesters P., Steer A.C., et al. Comparative M-protein analysis of Streptococcus pyogenes from pharyngitis and skin infections in New Zealand: implications for vaccine development. BMC Infect Dis. 2016;16:561. doi: 10.1186/s12879-016-1891-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giffard P.M., Tong S.Y.C., Holt D.C., Ralph A.P., Currie B.J. Concerns for efficacy of a 30-valent M-protein-based Streptococcus pyogenes vaccine in regions with high rates of rheumatic heart disease. PLoS Neglected Trop Dis. 2019;13 doi: 10.1371/journal.pntd.0007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walkinshaw D.R., Wright M., Mullin A., Excler J.L., Kim J.H., Steer A.C. The Streptococcus pyogenes vaccine landscape. NPJ Vaccines. 2023;8:16. doi: 10.1038/s41541-023-00609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies M.R., McIntyre L., Mutreja A., et al. Atlas of group A streptococcal vaccine candidates compiled using large-scale comparative genomics. Nat Genet. 2019;51:1035–1043. doi: 10.1038/s41588-019-0417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worthing K.A., Werno A., Pink R., et al. Biphasic outbreak of invasive group A Streptococcus disease in eldercare facility, New Zealand. Emerg Infect Dis. 2020;26:841–848. doi: 10.3201/eid2605.190131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhi X., Li H.K., Li H., et al. Emerging invasive group A Streptococcus M1UK lineage detected by allele-specific PCR, England, 20201. Emerg Infect Dis. 2023;29:1007–1010. doi: 10.3201/eid2905.221887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Putten B.C.L., Vlaminckx B.J.M., de Gier B., Freudenburg-de Graaf W., van Sorge N.M. Group A streptococcal meningitis with the M1UK variant in the Netherlands. JAMA. 2023;329:1791–1792. doi: 10.1001/jama.2023.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies M.R., Keller N., Brouwer S., et al. Detection of Streptococcus pyogenes M1UK in Australia and characterization of the mutation driving enhanced expression of superantigen SpeA. Nat Commun. 2023;14:1051. doi: 10.1038/s41467-023-36717-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett J., Moreland N.J., Zhang J., et al. Risk factors for group A streptococcal pharyngitis and skin infections: a case control study. Lancet Reg Health West Pac. 2022;26:100507. doi: 10.1016/j.lanwpc.2022.100507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Te_Whatu_Ora Reducing rheumatic fever. 2023. https://www.tewhatuora.govt.nz/for-the-health-sector/health-sector-guidance/diseases-and-conditions/rheumatic-fever-guidance/reducing-rheumatic-fever/#rheumatic-fever-report-2022
- 27.Huang Q.S., Wood T., Jelley L., et al. Impact of the COVID-19 nonpharmaceutical interventions on influenza and other respiratory viral infections in New Zealand. Nat Commun. 2021;12:1001. doi: 10.1038/s41467-021-21157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett J., Moreland N.J., Oliver J., et al. Understanding group A streptococcal pharyngitis and skin infections as causes of rheumatic fever: protocol for a prospective disease incidence study. BMC Infect Dis. 2019;19:633. doi: 10.1186/s12879-019-4126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dale J.B., Penfound T.A., Tamboura B., et al. Potential coverage of a multivalent M protein-based group A streptococcal vaccine. Vaccine. 2013;31:1576–1581. doi: 10.1016/j.vaccine.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dale J.B., Penfound T.A., Chiang E.Y., Walton W.J. New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci. Vaccine. 2011;29:8175–8178. doi: 10.1016/j.vaccine.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loh J.M.S., Rivera-Hernandez T., McGregor R., et al. A multivalent T-antigen-based vaccine for Group A Streptococcus. Sci Rep. 2021;11:4353. doi: 10.1038/s41598-021-83673-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steemson J.D., Moreland N.J., Williamson D., Morgan J., Carter P.E., Proft T. Survey of the bp/tee genes from clinical group A streptococcus isolates in New Zealand – implications for vaccine development. J Med Microbiol. 2014;63:1670–1678. doi: 10.1099/jmm.0.080804-0. [DOI] [PubMed] [Google Scholar]
- 33.Davies M.R., Holden M.T., Coupland P., et al. Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat Genet. 2015;47:84–87. doi: 10.1038/ng.3147. [DOI] [PubMed] [Google Scholar]
- 34.Beres S.B., Olsen R.J., Saavedra M.O., et al. Genome sequence analysis of emm89 Streptococcus pyogenes strains causing infections in Scotland, 2010–2016. J Med Microbiol. 2017;66:1765–1773. doi: 10.1099/jmm.0.000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DebRoy S., Li X., Kalia A., et al. Identification of a chimeric emm gene and novel emm pattern in currently circulating strains of emm4 Group A Streptococcus. Microb Genom. 2018;4 doi: 10.1099/mgen.0.000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandey M., Calcutt A., Ozberk V., et al. Antibodies to the conserved region of the M protein and a streptococcal superantigen cooperatively resolve toxic shock-like syndrome in HLA-humanized mice. Sci Adv. 2019;5 doi: 10.1126/sciadv.aax3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandey M., Powell M., Calcutt A., et al. Physicochemical characterisation, immunogenicity and protective efficacy of a lead streptococcal vaccine: progress towards Phase I trial. Sci Rep. 2017;7 doi: 10.1038/s41598-017-14157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell P.T., Tong S.Y.C., Geard N., et al. Longitudinal analysis of group A Streptococcus emm types and emm clusters in a high prevalence setting reveals past infection does not prevent future infection. J Infect Dis. 2019;221(9):1429–1437. doi: 10.1093/infdis/jiz615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taiaroa G., Matalavea B., Tafuna’i M., et al. Scabies and impetigo in Samoa: a school-based clinical and molecular epidemiological study. Lancet Reg Health Western Pac. 2021;6 doi: 10.1016/j.lanwpc.2020.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynskey N.N., Jauneikaite E., Li H.K., et al. Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study. Lancet Infect Dis. 2019;19(11):1209–1218. doi: 10.1016/s1473-3099(19)30446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreland N.J., Webb R.H. Against the trend: a decrease in scarlet fever in New Zealand. Lancet Infect Dis. 2019;19:1285–1286. doi: 10.1016/S1473-3099(19)30617-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.