Abstract
Purpose
Image artefacts continue to pose challenges in clinical molecular imaging, resulting in misdiagnoses, additional radiation doses to patients and financial costs. Mismatch and halo artefacts occur frequently in gallium-68 (68Ga)-labelled compounds whole-body PET/CT imaging. Correcting for these artefacts is not straightforward and requires algorithmic developments, given that conventional techniques have failed to address them adequately. In the current study, we employed differential privacy-preserving federated transfer learning (FTL) to manage clinical data sharing and tackle privacy issues for building centre-specific models that detect and correct artefacts present in PET images.
Methods
Altogether, 1413 patients with 68Ga prostate-specific membrane antigen (PSMA)/DOTA-TATE (TOC) PET/CT scans from 3 countries, including 8 different centres, were enrolled in this study. CT-based attenuation and scatter correction (CT-ASC) was used in all centres for quantitative PET reconstruction. Prior to model training, an experienced nuclear medicine physician reviewed all images to ensure the use of high-quality, artefact-free PET images (421 patients’ images). A deep neural network (modified U2Net) was trained on 80% of the artefact-free PET images to utilize centre-based (CeBa), centralized (CeZe) and the proposed differential privacy FTL frameworks. Quantitative analysis was performed in 20% of the clean data (with no artefacts) in each centre. A panel of two nuclear medicine physicians conducted qualitative assessment of image quality, diagnostic confidence and image artefacts in 128 patients with artefacts (256 images for CT-ASC and FTL-ASC).
Results
The three approaches investigated in this study for 68Ga-PET imaging (CeBa, CeZe and FTL) resulted in a mean absolute error (MAE) of 0.42 ± 0.21 (CI 95%: 0.38 to 0.47), 0.32 ± 0.23 (CI 95%: 0.27 to 0.37) and 0.28 ± 0.15 (CI 95%: 0.25 to 0.31), respectively. Statistical analysis using the Wilcoxon test revealed significant differences between the three approaches, with FTL outperforming CeBa and CeZe (p-value < 0.05) in the clean test set. The qualitative assessment demonstrated that FTL-ASC significantly improved image quality and diagnostic confidence and decreased image artefacts, compared to CT-ASC in 68Ga-PET imaging. In addition, mismatch and halo artefacts were successfully detected and disentangled in the chest, abdomen and pelvic regions in 68Ga-PET imaging.
Conclusion
The proposed approach benefits from using large datasets from multiple centres while preserving patient privacy. Qualitative assessment by nuclear medicine physicians showed that the proposed model correctly addressed two main challenging artefacts in 68Ga-PET imaging. This technique could be integrated in the clinic for 68Ga-PET imaging artefact detection and disentanglement using multicentric heterogeneous datasets.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00259-023-06418-7.
Keywords: PET/CT, Artefacts, Privacy, Deep learning, Federated learning
Introduction
Artefacts in medical imaging are structures that appear in the image but are not present in the patient’s body [1–4]. In whole-body positron emission tomography (PET) imaging, various artefacts can occur for numerous reasons, including the PET image itself and the propagation of artefacts from complementary imaging modalities, such as computed tomography (CT) or magnetic resonance imaging (MRI) in PET/CT and PET/MRI scanners, respectively [1–6]. Attenuation and scatter correction (ASC) during PET image reconstruction are the main steps in which these artefacts can occur [6–8]. In addition, ASC is applied to compensate for photon attenuation and Compton scattering, which is required to achieve quantitative PET imaging [9, 10]. Halo and mismatch artefacts are the most common in PET imaging of gallium-68 (68Ga)-labelled radiopharmaceuticals [1–3, 11, 12]. These artefacts can be easily missed if they are not prominent, but if they are highly effective, they can cause a corrupted image that often requires repeated scans [6, 9, 10]. Nevertheless, in most cases, the repeated scan will not rectify these artefacts, as they are unavoidable in certain cases [1–4, 6, 9, 10].
Mismatch artefacts can occur when there is a discrepancy between PET and anatomical imaging (CT or MRI) and can be caused by voluntary and involuntary organ movement [6, 10, 13–15]. An additional device for mismatch tracking or deformable image registration could partially correct this artefact [13, 14]. Mismatch artefacts result in missing, mislocalization and incorrect quantification of malignant lesions in PET images [6, 10]. This might cause misdiagnosis and change patient management [13, 14, 16]. Different CT acquisition protocols have been proposed to mitigate lung-diaphragm interface mismatch, including end-expiration, mid-expiration and respiratory averaged CT using cinematic CT (4D CT). End-expiration CT acquisition has been suggested to enable better assessment of the lungs on PET images. However, this acquisition reduces anatomical details and might miss small lung nodules in up to 34% of the cases [13, 14]. CT acquisition at mid-expiration is favoured [16]. The 4D CT approach was reported to enhance standardized uptake value (SUV) quantification by more than 50%, compared to mid-expiratory acquisition in malignant lesions [14, 15]. However, it involves higher radiation doses to the patient, and its implementation is not feasible in all centres.
Radiopharmaceutical-related artefacts can occur due to tracer injection, with eventual hot clots or extravasation, which could be interpreted as abnormalities [6]. Foci of increased uptake in the lungs not corresponding to a nodule or any other lung abnormality and axillary lymph nodes with increased uptake ipsilateral to the injection site with extravasation of the radiopharmaceutical are examples of radiopharmaceutical-related artefacts. Halo or photopenic artefacts formed in regions with a high tracer uptake, such as the kidneys, ureters, urinary catheters and bladder, might impede the correct interpretation of PET images. Halo artefacts can be caused by incorrect scatter correction (negative values near high activity regions) during image reconstruction (non-negativity constraint in statistical reconstruction) [11]. This artefact could result in misdiagnosis and often requires repeated scans, since it cannot be recovered using conventional algorithms [17, 18].
In the last few years, various deep-learning (DL)-based algorithms have been developed for quantitative PET image reconstruction [19–21]. Numerous ASC methodologies have been suggested, including scatter and attenuation estimation in the sinogram domain, indirect (attenuation map generation) and direct attenuation correction [6, 10, 22–29]. Furthermore, various DL algorithms have been proposed for artefact correction in single-modality PET, CT or MR imaging [6, 27, 28, 30, 31]. For example, we demonstrated in a previous study the potential of direct ASC framework for artefact correction in 2-deoxy-2-[18F]fluoro-D-glucose (18F-FDG) PET/CT [6, 10, 31]. In addition, DL-based metal artefact reduction in PET/CT in the image and projection domains showed that the DL algorithm outperformed conventional algorithms [32]. Another study successfully used DL for truncation and metallic artefact compensation in PET/MRI [33]. Ultimately, a more recent study investigated MR image artefact disentanglement using unpaired data and a DL algorithm [34].
DL algorithms commonly require a significant sample size for appropriate training [9, 28, 29, 35–40]. Moreover, building a robust DL model requires a large dataset involving a wide range of scanners, image acquisition and reconstruction protocols [9, 28, 29, 35–40]. However, data exchange across hospitals is restricted by ethical and regulatory considerations [9, 28, 29, 35–38]. In addition, all hospitals do not necessarily staffed by AI scientist, having access to computational resources for building centre-based models, and gathering large heterogeneous datasets remains challenging [9, 28, 29, 35–38]. Federated learning (FL) is a technique that allows DL models to be developed on distributed datasets spread across several hospitals or institutes [9, 29, 35–38, 41, 42]. FL aims to train a model that can draw knowledge from various decentralized datasets without moving the data to a single location [9, 29, 35–40]. This is especially helpful if the data are private or sensitive and cannot be shared with a central server or other hospitals [9, 29, 35–38, 43, 44].
There are several approaches to FL, including weighted federated averaging, which involves training a local model on each decentralized dataset and then averaging the model weights across all centres to create a global model [9, 29, 35–43]. In this scenario, the idea is to build one competitive global model with a centralized model [9, 29, 35–43]. Another approach referred to as federated transfer learning (FTL) involves a global model development through decentralized training and then fine-tuning the global model in each centre separately to create a centre-specific personalized model [9, 28, 29, 35–40]. FTL is useful in training DL models considering the high variability and heterogeneity of datasets across different centres, which requires large datasets and centre-specific models [9, 28, 29, 35–38].
As mentioned earlier, image artefacts remain a challenge in clinical molecular imaging, especially mismatch and halo artefacts in 68Ga PET imaging. Correcting these artefacts requires novel algorithmic developments since conventional algorithms have failed to address them adequately. In the current study, we employed differential privacy FTL to manage the data-sharing issue in the clinic for building DL models. In addition, we used FTL approaches to build centre-specific models with prior knowledge of different centres’ data that are specific to each centre. Furthermore, we employed this method for PET image artefact disentanglement on 68Ga-labelled compounds and evaluated its performance qualitatively and quantitatively. Finally, we compared the proposed method with centre-based and centralized algorithms.
Materials and methods
PET image acquisition and preprocessing
The institutional ethics committee of Geneva University Hospital approved this retrospective multi-institution study. This study enrolled a total of 1413 patients with 68Ga-prostate-specific membrane antigen (PSMA)/DOTA-TATE (TOC) PET/CT scans from 3 countries (Switzerland, Iran and Canada), scanned at 8 centres. An experienced nuclear medicine physician reviewed all images to select high-quality and artefact-free PET images for model development (421 clean data out of 1413). Table 1 provides details of the datasets from the different centres. CT-based ASC (CT-ASC) was used for PET image correction. Corrected and non-corrected PET images were converted to SUV units (units changed from Bq/ml to SUV) using injected dose, decay factor and patient’s weights [6, 9, 10, 28, 29]. All PET images were first cropped to the body contour and then zero-padded to the same bounding box (232 × 168 × Z, where Z is the slice number) to preserve both image resolution and body shape. All images were normalized between 0 and 1 using 90% of the histogram by dividing the value of non-ASC and CT-ASC by 2 and 5, respectively [6, 9, 10, 28, 29].
Table 1.
Image acquisition and reconstruction settings in 8 different imaging centres
| Centre | No | Clean | Train | Test | Scanner | Reconstruction | Tracers | Matrix size |
|---|---|---|---|---|---|---|---|---|
| Centre 1 | 70 | 16 | 12 | 4 | Siemens Horizon | PSF + TOF + 3D-OSEM | 68 Ga-PSMA | 180 × 180 |
| Centre 2 | 315 | 71 | 56 | 15 | Siemens Biograph 6 | 3D-OSEM | 68 Ga-PSMA | 168 × 168 |
| Centre 3 | 97 | 49 | 39 | 10 | Siemens mCT | PSF + TOF + 3D-OSEM | 68 Ga-PSMA | 256 × 256 |
| Centre 4 | 97 | 49 | 39 | 10 | Siemens Vision | PSF + TOF + 3D-OSEM | 68 Ga-PSMA | 440 × 440 |
| Centre 5 | 261 | 51 | 40 | 11 | Siemens Biograph 6 | PSF + 3D-OSEM | 68 Ga-PSMA | 168 × 168 |
| Centre 6 | 295 | 57 | 45 | 12 | Siemens mCT | 3D-OSEM | 68 Ga-PSMA | 200 × 200 |
| Centre 7 | 183 | 89 | 71 | 18 | GE Discovery 690 | 3D-OSEM | 68 Ga-DOTA-TATE | 192 × 192 |
| Centre 8 | 95 | 39 | 31 | 8 | GE Discovery IQ | 3D-OSEM | 68 Ga-PSMA | 192 × 192 |
| Total | 1413 | 421 | 333 | 88 | - | - | − | - |
Deep neural network
In the current study, we implemented a modified version of the U2Net [9, 45]. U2Net architecture incorporates residual blocks and deep supervision [9]. In the U2Net architecture, each conventional U-Net block consists of U-Net network, comprising classical U-Net blocks, including convolution, batch normalization, ReLU activation function and up and down sampling with a symmetric encoder and decoder [9, 45]. The input of the deep neural networks in the different scenarios was non-ASC PET, with the target being CT-ASC to generate DL-ASC PET images as output automatically. The network was trained in 2D using the Adam optimizer with a learning rate of 0.001, L2-norm loss and a weight decay of 0.0001 [9, 45]. The network schema is shown in Supplemental Fig. 1. Artefact-free clean data sets were used for train, validation and test set.
Different learning scenarios
Different scenarios were used to develop DL algorithms, including (i) centre-based (CeBa), (ii) centralized (CeZe) and (v) federated transfer learning (FTL).
CeBa
Each hospital independently developed the DL model using its own dataset [9, 28, 29, 35–42]. This training framework has trouble properly adjusting to unobserved new cases [9, 28, 29, 35–42]. Moreover, all hospitals do not necessarily have access to curated and large heterogeneous datasets, computational power and machine learning (ML) developers [9, 28, 29, 35–42].
CeZe
Different hospitals send data to a core hospital to create and develop a global ML model [9, 28, 29, 35–42]. This scenario jeopardizes the privacy of the data and hinders collaboration among different hospitals [9, 28, 29, 35–42].
FTL
The data are decentralized and not shared among hospitals in a FL framework, but the hospitals can work together and exchange DL model parameters to develop a global model [9, 28, 29, 35–42, 46, 47]. Each hospital keeps its own datasets, and the local datasets are used to develop the DL models independently [9, 28, 29, 35–38]. Each hospital sends model updates to a core hospital after training the local model. Then, the central server in the core hospital combines the model updates to create a global DL model. We implemented Gaussian differential privacy FL [9, 28, 29, 35–38, 48, 49] to build the global model using a decentralized data set and then fine-tuned the model for each hospital to develop personalized models for each hospital. More details were provided in the supplementary dataset. All models were implemented in Tensorflow and TensorFlow Federated (TFF) [50] using a server with multiple GPUs (RTX 2080 Ti) [9, 28, 29, 35–38]. The model trained on 80% of the clean data set (20% as the test set) at each hospital, and 10% of the training in each hospital (clean data) was used as a validation set to optimize the hyperparameters of the models.
Evaluation strategies
Two different test sets were used for the evaluation of our proposed method. This includes an artefact-free clean test set (20% of each centre clean dataset) used for quantitative analysis and a second test set consisting of images presenting with artefacts (128 patients) for blind qualitative analysis.
Quantitative analysis
Model performance was evaluated using image-derived PET metrics, including voxel-wise mean absolute error (MAE), mean squared error (MSE), structural similarity index (SSIM) and peak signal-to-noise ratio (PSNR) between ground truth CT-ASC and predicted DL-ASC PET images using artefact-free clean test set [10].
Qualitative assessment of artefacted images
Two nuclear medicine experts blindly performed the qualitative analysis of 258 images from 128 cases. CT-ASC (128 images) and FLT-ASC (128 images) image assessments were performed blindly in terms of image artefacts (1: unacceptable, 2: mild, 3: moderate, 4: minor and 5: none), diagnostic confidence (1: very poor, 2: poor, 3: average, 4: high and 5: excellent) and image quality (1: very poor, 2: poor, 3: average, 4: high and 5: excellent). These analyses were performed separately for different body regions, including the head and neck (including the brain), chest, chest abdomen interval (diaphragm region), abdomen, pelvis and extremities, and whole images (all regions). Moreover, 30 images (15 from FLT-ASC and 15 from CT-ASC) were used to assess the intra-observer variability of each physician. Inter-observer variability was calculated across 256 images.
Statistical analysis
Quantitative analyses were performed on the same clean test sets. Images with artefacts were used only for qualitative analysis. We used the Wilcoxon test to compare image-derived metrics between the different scenarios. The p-value was corrected using the false discovery rate correction method developed by Benjamin Hochberg [51] to provide an adjusted p-value (q-value). To assess consistency, we used the intraclass correlation coefficient (ICC) test based on a two-way mixed effects model for intra-/inter-observer variability assessment. ICC classified as poor reproducibility (ICC < 0.40), fair reproducibility (0.40 < ICC < 0.59), good reproducibility (0.60 < ICC < 0.74), or excellent reproducibility (0.75 < ICC < 1.00) [52]. The McNemar test was used for pairwise comparisons of image quality, artefacts and diagnostic confidence between CT-ASC and FLT-ASC images. The marginal homogeneity test compared these factors’ distributions between CT-ASC and FLT-ASC. We also used generalized linear models (GLMs) to adjust for reader and rate confounders, the centres and scanners and compare the distributions of image quality, artefacts and diagnostic confidence between CT-ASC and FLT-ASC in each region. Statistical analysis was performed using R software version 4.2.0, using a significance level of 0.05.
Results
Quantitative analysis of artefact-free images
Image-based analysis
The three approaches evaluated in this study for 68 Ga-PET imaging (CeBa, CeZe and FTL) resulted in MAE values of 0.42 ± 0.21 (CI 95%: 0.38 to 0.47), 0.32 ± 0.23 (CI 95%: 0.27 to 0.37) and 0.28 ± 0.15 (CI 95%: 0.25 to 0.31), respectively. Regarding SSIM, the FTL approach had a value of 0.80 ± 0.10 (CI 95%: 0.78 to 0.82). According to the Wilcoxon test, there were significant differences between the three approaches, with CeZe achieving 0.75 ± 0.15 (CI 95%: 0.72 to 0.79) and CeBa attaining 0.71 ± 0.15 (CI 95%: 0.68 to 0.74). The results showed that FTL-ASC outperformed CeBa and CeZe (p-value < 0.05). Supplemental Fig. 2 and Supplemental Table 1 compare the different metrics across different approaches in the test set. Supplemental Tables 2 and 3 provide more details for each centre for CeBa and FTL. We continued qualitative analysis of FTL-ASC PET images since FTL resulted in high quantitative accuracy.
Qualitative assessment of artefacted images
Intra and inter-observer variability
The ICC values for image quality, artefacts and diagnostic confidence are shown in Table 2. Both inter- and intra-observer variability showed fair reproducibility in the extremities and good and excellent reproducibility in other regions of the body.
Table 2.
Intra and inter-reader ICC for image quality, diagnostic confidence and image artefacts in all data for both readers
| Intra and Inter-reader analysis | |||
|---|---|---|---|
| Region | Image quality | Diagnostic confidence | Image artefact |
| Intra-Reader Analysis using 30 Images | |||
| Head and neck | 0.85 | 0.88 | |
| Chest | 0.71 | 0.79 | |
| Chest abdomen interval | 0.80 | 0.82 | Motion: 0.86 |
| Abdomen | 0.73 | 0.76 | Halo: 0.88 |
| Pelvis | 0.70 | 0.75 | Halo: 0.82 |
| Extremities | 0.55 | 0.60 | |
| All regions | 0.72 | 0.76 | |
| Inter-Reader Analysis using 256 Images | |||
| Head and neck | 0.66 | 0.66 | |
| Chest | 0.61 | 0.60 | |
| Chest abdomen interval | 0.72 | 0.74 | Motion: 0.73 |
| Abdomen | 0.71 | 0.72 | Halo: 0.82 |
| Pelvis | 0.60 | 0.61 | Halo: 0.74 |
| Extremities | 0.41 | 0.47 | |
| All regions | 0.60 | 0.61 | |
Overall image quality
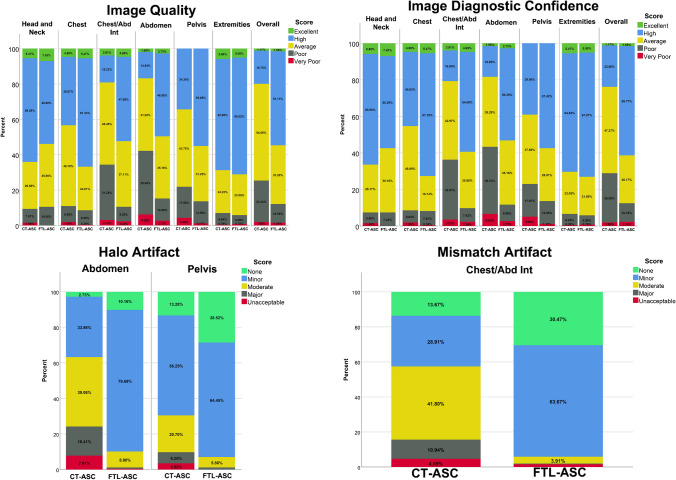
Table 3 (generalized linear model tests) and Supplemental Tables 4–6 (McNemar and marginal homogeneity tests) compare the image quality, artefacts and diagnostic confidence between CT-ASC and FTL-ASC. In addition, the data are visualized in bar plots in Fig. 1. These results demonstrate that image quality improved significantly for the chest/abdomen and abdomen regions. In the pelvic region, image quality improved significantly from 34.38 to 55.08%, while very poor image quality significantly decreased from 4.3 to 1.17%. The overall image quality is significantly improved in all regions of the body (total image). Diagnostic image confidence was significantly improved in the chest, chest/abdomen and abdomen regions.
Table 3.
Comparison of image quality, diagnostic confidence and image artefacts between CT-ASC and FLT-ASC. The reported p-value is based on a generalized linear model after adjustment of ICC for all raters (CT-ASC and FLT-ASC), readers and centres
| Region | Image quality | Diagnostic confidence | Image artefact |
|---|---|---|---|
| Head and neck | 0.566 | 0.711 | - |
| Chest | 0.062 | 0.015 | - |
| Chest abdomen interval | < 0.001 | < 0.001 | Motion: < 0.005 |
| Abdomen | < 0.002 | < 0.001 | Halo: < 0.001 |
| Pelvis | 0.211 | 0.211 | Halo: < 0.05 |
| Extremities | 0.801 | 0.801 | - |
| All regions | 0.001 | 0.001 | - |
Significant value mentioned in the text. P-value less than 0.05
Fig. 1.
Image quality, image diagnostic confidence, halo and mismatch artefact comparison between CT-ASC and FTL-ASC in different regions of the body
Moreover, high and very-poor diagnostic confidence in the pelvic region significantly increased and decreased, respectively, using FTL-ASC. FTL-ASC PET images significantly improved overall diagnostic confidence. The mismatch artefacts in the chest/abdomen region and halo artefacts in the abdomen and pelvis significantly decreased when using FTL-ASC.
Mismatch and halo artefacts
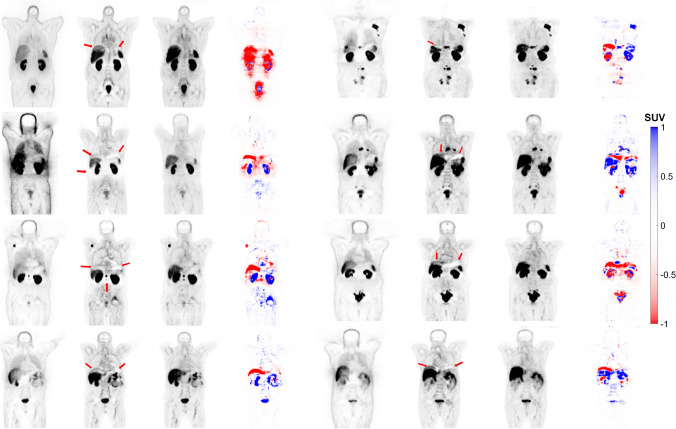
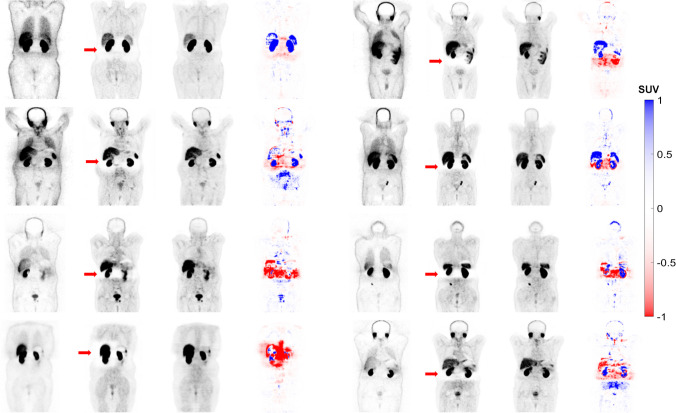
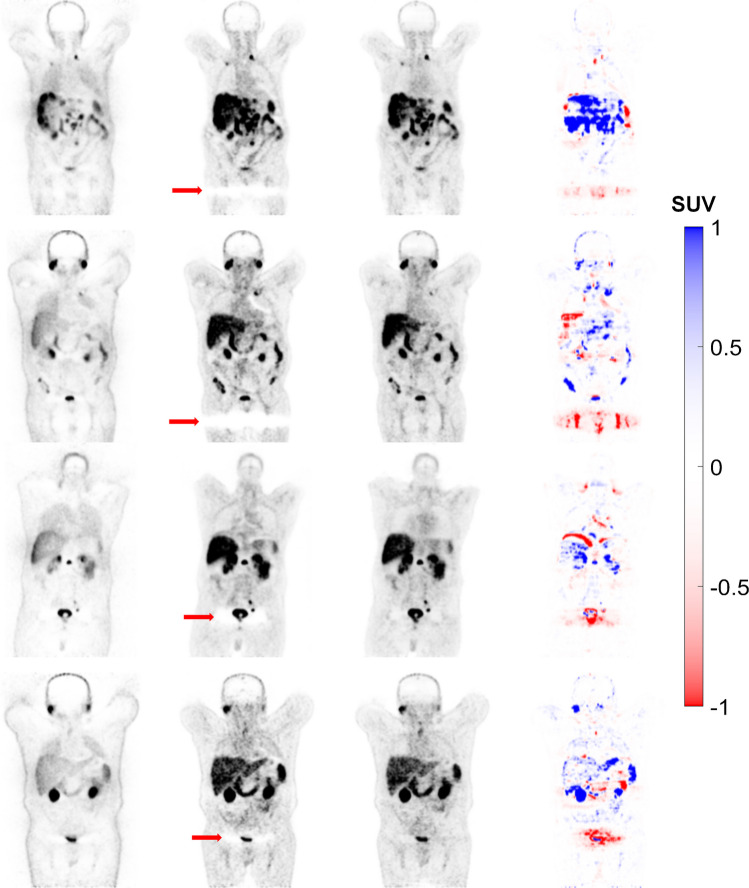
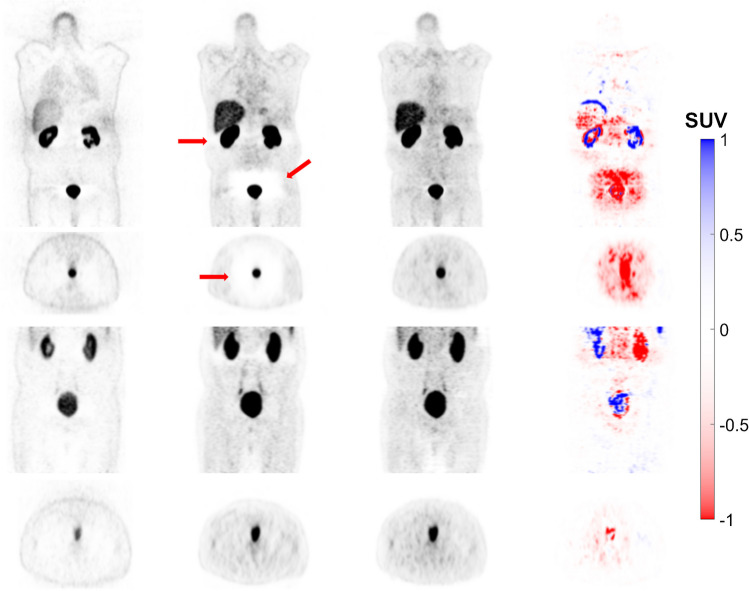
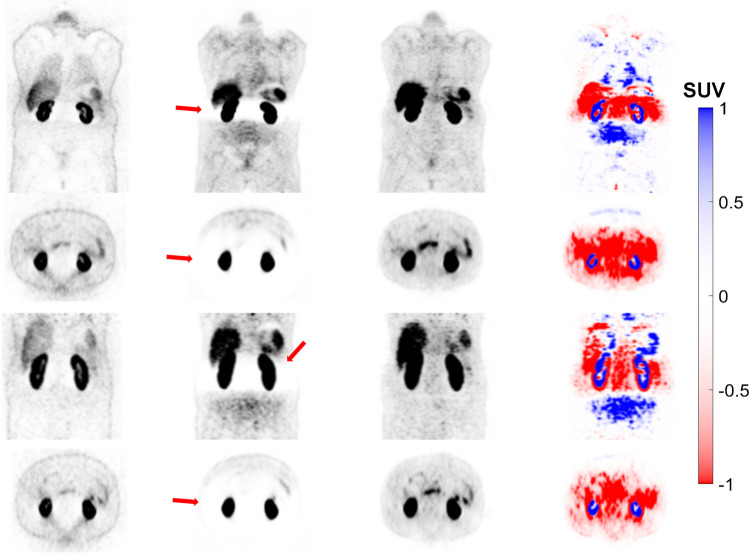
The images generated using the FTL approach successfully corrected the mismatch artefact in the diaphragm region and reduced photopenic artefacts in the lung, liver and spleen region, as shown in Fig. 2. The results of our study demonstrate that FTL-ASC effectively disentangles halo artefacts in the abdominal and pelvic regions (Figs. 3 and 4).
Fig. 2.
Coronal views of 8 representative clinical studies showing from left to right: non-ASC, CT-ASC, FTL-ASC and the difference images of CT-ASC and FTL-ASC. The images generated using the FTL approach successfully corrected the mismatch artefact in the diaphragm region (depicted by the arrows) and reduced photopenic artefacts in the lung, liver and spleen regions in the different cases
Fig. 3.
Coronal views of 8 representative clinical studies showing from left to right: non-ASC, CT-ASC, FTL-ASC and the difference images of CT-ASC and FTL-ASC. FTL-ASC effectively disentangles halo artefacts in the kidney area (depicted by the arrows) in the different cases
Fig. 4.
Coronal views of 4 representative clinical studies showing from left to right: non-ASC, CT-ASC, FTL-ASC and the difference images of CT-ASC and FTL-ASC. FTL-ASC effectively disentangles halo artefacts in the pelvic area (depicted by the arrows) in the different cases
Case study with repeated scans (upon request from the nuclear medicine physician immediately after the initial scan).
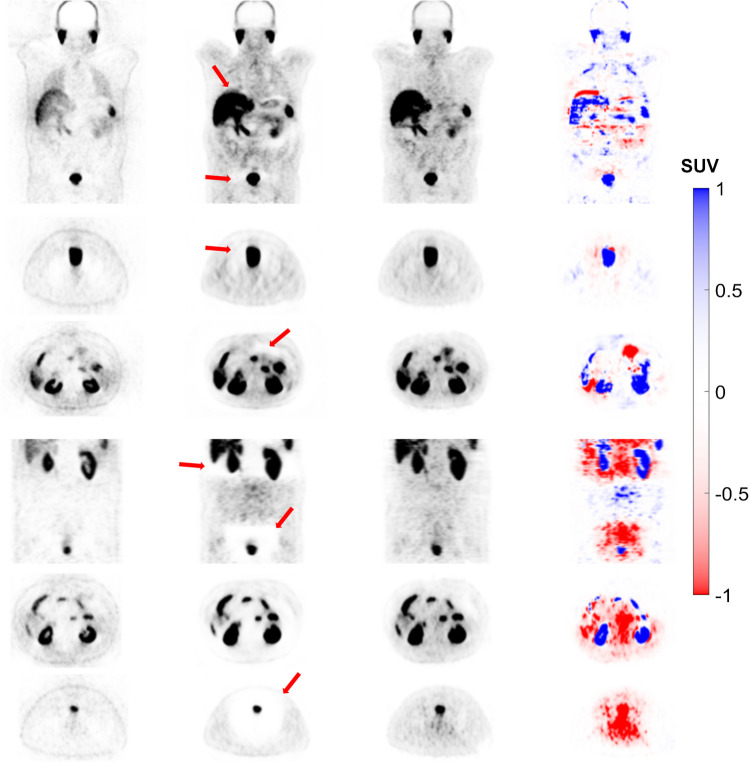
Figure 5 represents a patient with a halo artefact in the pelvic region and the repeated scan performed for this patient in the kidney and pelvic region. FTL-ASC shows high-quality artefact-free images in both scans. This artefact was removed in the repeated CT-ASC scan, and FTL-ASC PET images reported almost similar image quality, diagnostic confidence and pattern as the first scan.
Fig. 5.
Coronal and axial views showing from left to right: non-ASC, CT-ASC, FTL-ASC and the difference images of CT-ASC and FTL-ASC. The top panel shows the initial scan, whereas the bottom panel depicts the repeated scan in the artefacted region (depicted by the arrows). In the repeated CT-ASC scan, this artefact was removed from the images. FTL-ASC produced high-quality artefact-free images in both scans
Figure 6 shows a case with a halo artefact in the kidneys. The repeated scan was performed for this patient in this region owing to low image quality and diagnostic confidence. Unfortunately, the repeated scan could not remove this artefact. The halo artefact is still visible in the same region. However, the FTL-ASC model successfully removed this artefact in both scans.
Fig. 6.
Coronal and axial views showing from left to right: non-ASC, CT-ASC, FTL-ASC and the difference images of CT-ASC and FTL-ASC. The top panel shows the initial scan, whereas the bottom panel depicts the repeated scan in the artefacted region (depicted by the arrows). The repeated scan could not remove this artefact, where the halo artefact remained visible in the same region. However, FTL-ASC successfully removed this artefact in both scans
Figure 7 represents a patient with moderate halo artefacts in the abdomen and pelvic area, with the repeated scan performed for this patient. However, the repeated scan exaggerated the halo artefact resulting in low-quality and low diagnostic confidence images. The FTL-ASC PET image recovered high quality and high confidence for both scans.
Fig. 7.
Coronal and axial views showing from left to right: non-ASC, CT-ASC, FTL-ASC and the difference images of CT-ASC and FTL-ASC. The top panel shows the initial scan, whereas the bottom panel depicts the repeated scan in the artefacted region (depicted by the arrows). The FTL-ASC PET image recovered high quality and high diagnostic confidence for both scans. The halo artefact was exaggerated and produced low-quality low diagnostic confidence images in the repeated scan
Discussion
A single universal model may not be effective due to differences in tracer injected activity, equipment, image acquisition and reconstruction strategies across different hospitals [9, 10, 28, 29, 35–42, 46, 47]. Therefore, it is necessary to create personalized models using large, heterogeneous datasets to overcome this issue [9, 28, 29, 35–38]. In the current study, we utilized differential privacy-preserving FTL to adopt a centralized model for each centre separately, resulting in improved accuracy for ASC in PET images and simultaneously addressing data-sharing privacy issues.
ASCs are the two major corrections based on CT images toward quantitative 68Ga PET imaging [6, 9, 10, 25, 28, 29]. However, mismatch and halo artefacts might appear on 68Ga PET images during this process, leading to potential changes in the diagnosis and prognosis of patients [11, 13, 14, 16–18]. In addition, these artefacts are difficult to detect and correct in real clinical scenarios [6]. The developed model does not require iterative image reconstruction incorporating ASC. In addition, we addressed the data-sharing issue using differential privacy FTL and showed that our model quantitatively outperformed centralized and centre-based models. We then used FTL-ASC for further qualitative analysis. Through quantitative analysis, we observe the impact of radiotracers and scanners on the performance of the models. We observed that FTL enhanced significantly the quantitative accuracy of the models in both situations, outperforming the CeBa approach. Different sources, such as scanner and radiotracer, significantly affect FTL’s effectiveness. Notably, FTL performs more efficiently when different scanners use the same radiotracer, rather than when diverse radiotracers are used on the same scanner. Qualitative analysis revealed the performance of our proposed model for effective mismatch and halo artefact detection and correction in the chest, abdomen and pelvic regions without knowledge of the ground truth in 68Ga PET images.
Different DL-based ASC methods have been proposed for PET, including indirect methods generating attenuation maps from MRI and non-ASC images or using maximum likelihood estimation of activity and attenuation (MLAA) and then using these attenuation maps for ASC during the reconstruction process [21, 27]. For example, Liu et al. [22] used a GAN network to generate pseudo-CT from PET-nonAC in brain PET images, whereas Dong et al. [23] used the same approach in whole-body imaging. In addition, Hwang et al. [24] improved the performance of MLAA by dealing with its main limitations (crosstalk artefacts, slow convergence speed and noisy attenuation maps) through DL for 18F-FDG brain PET and tested this methodology in whole-body PET imaging [25]. The direct DL-based ASC framework directly generates ASC PET images from non-ASC images [9, 10, 21]. Our group first implemented this approach in brain PET imaging [53]. Furthermore, our group [10] and Dong et al. [26] independently assessed the performance of direct ASC in whole-body 18F-FDG PET imaging.
A low injected tracer activity and the high positron range of 68Ga-labelled radiopharmaceuticals produce low-quality images, compared to 18F-labelled compounds, adding challenges to direct ASC [11, 13, 14, 16–18]. At first glance, direct ASC utilizing DL appears as an excessive use of artificial intelligence in PET. However, it not only generates quantitatively and qualitatively corrected PET images but also has some potential, which we emphasized in the current study [10]. The main potential of this approach is mismatch and halo artefact detection and correction of 68 Ga PET images without using anatomical images. In indirect techniques, reconstruction is mandatory to produce ASC PET images, potentially capable of addressing mismatch artefacts using a clean dataset [54, 55]. However, these approaches do not address halo artefacts, as this artefact appears during the reconstruction process and depends mostly on PET images (not CT images). Mismatch and halo artefacts were evaluated in the current study, where they were successfully detected and corrected in different regions of the body. We presented cases in which artefacts appeared in the original 68 Ga PET images, and FTL-ASC successfully removed these artefacts. Moreover, blind analysis performed by physicians showed significant improvement in image quality, confidence in diagnostic accuracy and removal of artefacts. In addition, we showed different scenarios in which repeated scans could be performed to remove these artefacts and observed that DL-based algorithms properly recovered important clinically relevant features. Meanwhile, there were a number of cases in which repeated scans could not remove and even exaggerated these artefacts. In this situation, our proposed DL-based algorithms could disentangle these artefacts without knowledge of the ground truth.
Most previous studies were performed on single-centre datasets, limiting the DL model generalizability [10, 22–26]. A multicentre study could potentially address this issue, where FL is definitely a good option. More recently, we developed a decentralized, federated DL model for ASC of 18F-FDG PET images using 300 clinical studies from 6 different centres [9]. We evaluated two different FL algorithms, parallel and sequential, without a privacy-preserving mechanism, such as the differential privacy implemented in the current study. We reported that both models outperformed the centre-based model and achieved performance levels comparable to the centralized model [9]. The differential privacy-preserving mechanism implemented in the current study can protect the model against various attacks, as FL by itself does not offer that protection [9]. One of the main advantages of FTL is the ability to create personalized DL models using data from various centres without compromising privacy. Furthermore, by enabling DL models to gain knowledge from a wider variety of data, FTL has the potential to enhance the performance of these models.
The use of FL poses additional challenges that need to be considered [39–42, 46, 47]. Decentralized datasets may be heterogeneous due to variations in data acquisition and reconstruction protocols across the different centres [9]. As a result, combining model updates and building a comprehensive global model might be challenging. In CeBa and CeZe training, the data set is small and highly heterogeneous, respectively [9, 36, 38]. In addition, a global FL model would achieve the accuracy of CeZe in the ideal scenario [9]. The current study used FTL algorithms to address the limitations of CeBa, CeZe and FL model development by building a global FL model in the first steps from the heterogeneous multicentric dataset and subsequently developing a centre-specific model using transfer learning [9]. FTL can be computationally demanding because it necessitates coordinating the training process across various centres, aggregating the model updates and providing a centre-specific model in each centre separately [36].
One of the main limitations of the current study is that model development took place on a single server equipped with multiple GPUs [9, 36, 38]. Future research should implement a more realistic computational model addressing the major challenges, such as the communication burden between different centres or the computation power of each centre [9, 36, 38]. Furthermore, various attacks should be designed to assess the performance of the differential privacy-preserving mechanism in protecting the privacy of data in different scenarios [9, 36, 38]. Additionally, since this method was developed based on direct ASC of 68Ga images, it is not suitable for other radiotracers, such as 18F-FDG, as it will fail to generate correct images due to domain and concept shifts [6, 9, 10, 29]. Further studies should be conducted to develop and evaluate the proposed artefact correction techniques for other tracers and other PET image artefacts [6, 9, 10, 29].
Conclusion
We employed a differential privacy-preserving FTL framework for artefact detection and disentanglement in PET imaging of 68Ga-labelled compounds. The proposed approach benefits from using large datasets from multiple centres while preserving patient privacy. Simultaneously, it uses the transfer learning concept, providing site-specific models that outperform centralized and centre-based models. In addition, the qualitative analysis demonstrated that the proposed model correctly addresses two main challenging artefacts in 68Ga-PET imaging.
Supplementary Information
Below is the link to the electronic supplementary material.
Funding
Open access funding provided by University of Geneva The Swiss National Science Foundation supported this work under grant SNSF 320030_176052, the Private Foundation of Geneva University Hospitals under Grant RC-06–01 and the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant RGPIN-2019–06467.
Data availability
Trained models and code are available on GitHub.
Declarations
Research involving human participants
All the procedures performed in the studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was waived for all the individual participants included in the study.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Isaac Shiri and Yazdan Salimi contributed equally to this work.
References
- 1.Sureshbabu W, Mawlawi O. PET/CT imaging artifacts. J Nucl Med Technol. 2005;33:156–61; quiz 63–4. [PubMed]
- 2.Mawlawi O, Pan T, Macapinlac HA. PET/CT imaging techniques, considerations, and artifacts. J Thorac Imaging. 2006;21:99–110. doi: 10.1097/00005382-200605000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Blodgett TM, Mehta AS, Mehta AS, Laymon CM, Carney J, Townsend DW. PET/CT artifacts. Clin Imaging. 2011;35:49–63. doi: 10.1016/j.clinimag.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook GJ, Wegner EA, Fogelman I. Pitfalls and artifacts in 18FDG PET and PET/CT oncologic imaging. Semin Nucl Med. 2004;34:122–133. doi: 10.1053/j.semnuclmed.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Simpson DL, Bui-Mansfield LT, Bank KP. FDG PET/CT: artifacts and pitfalls. Contemp Diagn Radiol. 2017;40:1–7. doi: 10.1097/01.CDR.0000513008.49307.b7. [DOI] [Google Scholar]
- 6.Shiri I, Sanaat A, Salimi Y, Akhavanallaf A, Arabi H, Rahmim A, et al. PET-QA-NET: towards routine PET image artifact detection and correction using deep convolutional neural networks. IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC): IEEE; 2021. p. 1–3.
- 7.Zaidi H, Koral KF. Scatter modelling and compensation in emission tomography. Eur J Nucl Med Mol Imaging. 2004;31:761–782. doi: 10.1007/s00259-004-1495-z. [DOI] [PubMed] [Google Scholar]
- 8.Kinahan PE, Hasegawa BH, Beyer T. X-ray-based attenuation correction for positron emission tomography/computed tomography scanners. Semin Nucl Med. 2003;33:166–179. doi: 10.1053/snuc.2003.127307. [DOI] [PubMed] [Google Scholar]
- 9.Shiri I, Vafaei Sadr A, Akhavan A, Salimi Y, Sanaat A, Amini M, et al. Decentralized collaborative multi-institutional PET attenuation and scatter correction using federated deep learning. Eur J Nucl Med Mol Imaging. 2023;50:1034–1050. doi: 10.1007/s00259-022-06053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiri I, Arabi H, Geramifar P, Hajianfar G, Ghafarian P, Rahmim A, et al. Deep-JASC: joint attenuation and scatter correction in whole-body (18)F-FDG PET using a deep residual network. Eur J Nucl Med Mol Imaging. 2020;47:2533–2548. doi: 10.1007/s00259-020-04852-5. [DOI] [PubMed] [Google Scholar]
- 11.Heußer T, Mann P, Rank CM, Schäfer M, Dimitrakopoulou-Strauss A, Schlemmer HP, et al. Investigation of the halo-artifact in 68Ga-PSMA-11-PET/MRI. PLoS One. 2017;12:e0183329. doi: 10.1371/journal.pone.0183329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magota K, Numata N, Shinyama D, Katahata J, Munakata Y, Maniawski PJ, et al. Halo artifacts of indwelling urinary catheter by inaccurate scatter correction in 18F-FDG PET/CT imaging: incidence, mechanism, and solutions. EJNMMI Phys. 2020;7:66. doi: 10.1186/s40658-020-00333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen-Auerbach M, Yeom K, Park J, Phelps M, Czernin J. Standard PET/CT of the chest during shallow breathing is inadequate for comprehensive staging of lung cancer. J Nucl Med. 2006;47:298–301. [PubMed] [Google Scholar]
- 14.Truong MT, Viswanathan C, Carter BW, Mawlawi O, Marom EM. PET/CT in the thorax: pitfalls. Radiol Clin North Am. 2014;52:17–25. doi: 10.1016/j.rcl.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Pan T, Mawlawi O, Nehmeh SA, Erdi YE, Luo D, Liu HH, et al. Attenuation correction of PET images with respiration-averaged CT images in PET/CT. J Nucl Med. 2005;46:1481–1487. [PubMed] [Google Scholar]
- 16.Carter BW, Betancourt SL, Viswanathan C, Mawlawi O, Marom EM, Truong MT. Potential pitfalls in interpretation of positron emission tomography/computed tomography findings in the thorax. Semin Roentgenol. 2015;50:210–216. doi: 10.1053/j.ro.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Lindemann ME, Guberina N, Wetter A, Fendler WP, Jakoby B, Quick HH. Improving (68)Ga-PSMA PET/MRI of the prostate with unrenormalized absolute scatter correction. J Nucl Med. 2019;60:1642–1648. doi: 10.2967/jnumed.118.224139. [DOI] [PubMed] [Google Scholar]
- 18.Nayak B, Dogra PN, Naswa N, Kumar R. Diuretic 18F-FDG PET/CT imaging for detection and locoregional staging of urinary bladder cancer: prospective evaluation of a novel technique. Eur J Nucl Med Mol Imaging. 2013;40:386–393. doi: 10.1007/s00259-012-2294-6. [DOI] [PubMed] [Google Scholar]
- 19.Zaidi H, El Naqa I. Quantitative molecular positron emission tomography imaging using advanced deep learning techniques. Annu Rev Biomed Eng. 2021;23:249–276. doi: 10.1146/annurev-bioeng-082420-020343. [DOI] [PubMed] [Google Scholar]
- 20.Lee JS. A review of deep learning-based approaches for attenuation correction in positron emission tomography. IEEE Trans Rad Plasma Med Sci. 2021;5:160–184. doi: 10.1109/TRPMS.2020.3009269. [DOI] [Google Scholar]
- 21.Guo R, Xue S, Hu J, Sari H, Mingels C, Zeimpekis K, et al. Using domain knowledge for robust and generalizable deep learning-based CT-free PET attenuation and scatter correction. Nat Commun. 2022;13:5882. doi: 10.1038/s41467-022-33562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB. A deep learning approach for (18)F-FDG PET attenuation correction. EJNMMI Phys. 2018;5:24. doi: 10.1186/s40658-018-0225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong X, Wang T, Lei Y, Higgins K, Liu T, Curran WJ, et al. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Phys Med Biol. 2019;64:215016. doi: 10.1088/1361-6560/ab4eb7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang D, Kim KY, Kang SK, Seo S, Paeng JC, Lee DS, et al. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med. 2018;59:1624–1629. doi: 10.2967/jnumed.117.202317. [DOI] [PubMed] [Google Scholar]
- 25.Hwang D, Kang SK, Kim KY, Seo S, Paeng JC, Lee DS, et al. Generation of PET attenuation map for whole-body time-of-flight (18)F-FDG PET/MRI using a deep neural network trained with simultaneously reconstructed activity and attenuation maps. J Nucl Med. 2019;60:1183–1189. doi: 10.2967/jnumed.118.219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong X, Lei Y, Wang T, Higgins K, Liu T, Curran WJ, et al. Deep learning-based attenuation correction in the absence of structural information for whole-body positron emission tomography imaging. Phys Med Biol. 2020;65:055011. doi: 10.1088/1361-6560/ab652c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMillan AB, Bradshaw TJ. Artificial Intelligence-based data corrections for attenuation and scatter in position emission tomography and single-photon emission computed tomography. PET Clin. 2021;16:543–552. doi: 10.1016/j.cpet.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiri I, Salimi Y, Maghsudi M, Hajianfar G, Jafari E, Samimi R, et al. Deep adaptive transfer learning for site-specific PET attenuation and scatter correction from multi-national/institutional datasets. IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC): IEEE; 2022. p. 1–3.
- 29.Shiri I, Sadr AV, Sanaat A, Ferdowsi S, Arabi H, Zaidi H. Federated learning-based deep learning model for PET attenuation and scatter correction: a multi-center study. IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC): IEEE; 2021. p. 1–3.
- 30.Yang J, Sohn JH, Behr SC, Gullberg GT, Seo Y. CT-less direct correction of attenuation and scatter in the image space using deep learning for whole-body FDG PET: potential benefits and pitfalls. Radiol Artif Intell. 2021;3:e200137. doi: 10.1148/ryai.2020200137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiri I, Salimi Y, Sanaat A, Saberi A, Amini M, Akhavanallaf A, et al. Fully automated PET image artifacts detection and correction using deep neural networks. J Nucl Med 2022. Supplemental 2, p. 3218.
- 32.Arabi H, Zaidi H. Deep learning-based metal artefact reduction in PET/CT imaging. Eur Radiol. 2021;31:6384–6396. doi: 10.1007/s00330-021-07709-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arabi H, Zaidi H. Truncation compensation and metallic dental implant artefact reduction in PET/MRI attenuation correction using deep learning-based object completion. Phys Med Biol. 2020;65:195002. doi: 10.1088/1361-6560/abb02c. [DOI] [PubMed] [Google Scholar]
- 34.Liu S, Thung K-H, Qu L, Lin W, Shen D, Yap P-T. Learning MRI artefact removal with unpaired data. Nat Mach Intell. 2021;3:60–67. doi: 10.1038/s42256-020-00270-2. [DOI] [Google Scholar]
- 35.Shiri I, Amini M, Salimi Y, Sanaat A, Saberi A, Razeghi B, et al. Multi-institutional PET/CT image segmentation using a decentralized federated deep transformer learning algorithm. J Nucl Med 2022. Suppl 2, p. 3348.
- 36.Shiri I, Razeghi B, Vafaei Sadr A, Amini M, Salimi Y, Ferdowsi S, et al. Multi-institutional PET/CT image segmentation using federated deep transformer learning. Comput Methods Programs Biomed. 2023;240:107706. doi: 10.1016/j.cmpb.2023.107706. [DOI] [PubMed] [Google Scholar]
- 37.Shiri I, Showkatian E, Mohammadi R, Razeghi B, Bagheri S, Hajianfar G, et al. Collaborative multi-institutional prostate lesion segmentation from MR images using deep federated learning framework. IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC): IEEE; 2022. p. 1–3.
- 38.Shiri I, Vafaei Sadr A, Amini M, Salimi Y, Sanaat A, Akhavanallaf A, et al. Decentralized distributed multi-institutional PET image segmentation using a federated deep learning framework. Clin Nucl Med. 2022;47:606–617. doi: 10.1097/rlu.0000000000004194. [DOI] [PubMed] [Google Scholar]
- 39.Shyu C-R, Putra KT, Chen H-C, Tsai Y-Y, Hossain KT, Jiang W, et al. A systematic review of federated learning in the healthcare area: from the perspective of data properties and applications. Appl Sci. 2021;11:11191. doi: 10.3390/app112311191. [DOI] [Google Scholar]
- 40.Rahman KJ, Ahmed F, Akhter N, Hasan M, Amin R, Aziz KE, et al. Challenges, applications and design aspects of federated learning: a survey. IEEE Access. 2021;9:124682–124700. doi: 10.1109/ACCESS.2021.3111118. [DOI] [Google Scholar]
- 41.Rieke N, Hancox J, Li W, Milletarì F, Roth HR, Albarqouni S, et al. The future of digital health with federated learning. NPJ Digit Med. 2020;3:119. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaissis GA, Makowski MR, Rückert D, Braren RF. Secure, privacy-preserving and federated machine learning in medical imaging. Nat Machi Intell. 2020;2:305–311. doi: 10.1038/s42256-020-0186-1. [DOI] [Google Scholar]
- 43.Kirienko M, Sollini M, Ninatti G, Loiacono D, Giacomello E, Gozzi N, et al. Distributed learning: a reliable privacy-preserving strategy to change multicenter collaborations using AI. Eur J Nucl Med Mol Imaging. 2021;48:3791–3804. doi: 10.1007/s00259-021-05339-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bercea CI, Wiestler B, Rueckert D, Albarqouni S. Federated disentangled representation learning for unsupervised brain anomaly detection. Nat Mach Intell. 2022;4:685–695. doi: 10.1038/s42256-022-00515-2. [DOI] [Google Scholar]
- 45.Qin X, Zhang Z, Huang C, Dehghan M, Zaiane OR, Jagersand M. U2-Net: going deeper with nested U-structure for salient object detection. Pattern Recognit. 2020;106:107404. doi: 10.1016/j.patcog.2020.107404. [DOI] [Google Scholar]
- 46.Rahman SA, Tout H, Talhi C, Mourad A. Internet of things intrusion detection: centralized, on-device, or federated learning? IEEE Network. 2020;34:310–317. doi: 10.1109/MNET.011.2000286. [DOI] [Google Scholar]
- 47.AbdulRahman S, Tout H, Ould-Slimane H, Mourad A, Talhi C, Guizani M. A survey on federated learning: the journey from centralized to distributed on-site learning and beyond. IEEE Internet Things J. 2020;8:5476–5497. doi: 10.1109/JIOT.2020.3030072. [DOI] [Google Scholar]
- 48.Abadi M, Chu A, Goodfellow I, McMahan HB, Mironov I, Talwar K, et al. Deep learning with differential privacy. Proceedings of the 2016 ACM SIGSAC conference on computer and communications security; 2016. p. 308–18.
- 49.McMahan HB, Ramage D, Talwar K, Zhang L. Learning differentially private recurrent language models. arXiv preprint arXiv:171006963. 2017.
- 50.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. Tensorflow: a system for large-scale machine learning. Osdi: Savannah, GA, USA; 2016. p. 265–83.
- 51.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- 52.Cicchetti DV. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol Assess. 1994;6:284. doi: 10.1037/1040-3590.6.4.284. [DOI] [Google Scholar]
- 53.Shiri I, Ghafarian P, Geramifar P, Leung KH, Ghelichoghli M, Oveisi M, et al. Direct attenuation correction of brain PET images using only emission data via a deep convolutional encoder-decoder (Deep-DAC) Eur Radiol. 2019;29:6867–6879. doi: 10.1007/s00330-019-06229-1. [DOI] [PubMed] [Google Scholar]
- 54.Zhou B, Tsai YJ, Chen X, Duncan JS, Liu C. MDPET: a unified motion correction and denoising adversarial network for low-dose gated PET. IEEE Trans Med Imaging. 2021;40:3154–3164. doi: 10.1109/TMI.2021.3076191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu C, Pierce LA, 2nd, Alessio AM, Kinahan PE. The impact of respiratory motion on tumor quantification and delineation in static PET/CT imaging. Phys Med Biol. 2009;54:7345–7362. doi: 10.1088/0031-9155/54/24/007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Trained models and code are available on GitHub.