Abstract
A cDNA encoding hepcidin was isolated from a library of cDNA from spleen of red sea bream (Chrysophrys major) by expressed sequence tag analysis. The expression of the hepcidin mRNA in various tissues was examined. Challenge of red sea bream with Escherichia coli DH5α elevated hepcidin mRNA levels in spleen, gill, liver, and intestine.
Antimicrobial peptides constitute important components of the innate immune system in many species, including plants, invertebrates, and vertebrates (3, 5, 18). They play an important role in protecting these organisms against microbial invasion. Antimicrobial peptides are widespread in various organisms, and a large number of these molecules have been isolated from invertebrates and vertebrates as well as from plants (4, 10, 11). Few studies have been performed on antimicrobial peptides from teleosts. Pardaxins, misgurin, pleurocidin, and moronecidin, which were found in Moses sole fish (19), in loach (20), in winter flounder (9), and in striped bass (17), are the main antimicrobial peptides so far isolated from teleosts. Cysteine-rich antimicrobial peptides are an important part of the antimicrobial peptide family and have been identified in the hemolymph of crustaceans and the fat bodies of insects. Recently, novel cysteine-rich antimicrobial peptides with low molecular weights have been isolated from human urine (21) and blood (15). These antimicrobial peptides were designated hepcidin because they are expressed predominantly in human liver (21). In fish, few reports on the cloning and expression of hepcidin genes are available (12, 24). In this paper, we report the cloning and expression in various tissues of the hepcidin gene from red sea bream (Chrysophrys major).
A cDNA library was constructed from red sea bream spleen as described previously (7). Sequences of expressed sequence tags (300 to 500 bp) were compared with those in GenBank for identifying hepcidin with the BLAST, version 2.0, program (1, 2). The alignment of the deduced amino acid sequence of hepcidin was performed with ClustalX (26). A phylogenetic tree was constructed by the neighbor-joining method (23) and analyzed with Mega 2 (16). The genomic DNA was isolated from red sea bream liver with a DNA isolation kit (Sangon, Shanghai, People's Republic of China). Intron 1 was amplified with the primer pair RSBhepcN1 (TCAGTGTTGCAGTTGCAGTG) and RSBhepcC1 (TCTCTTCATCTGCAGCAACTG). Intron 2 was amplified with the primer pair RSBhepcN2 (CAATGAGCAATGGCAGCCCA) and RSBhepcC2 (TGCAGCAGGAATCCTCAACG). PCR was performed as described previously (8). 5′ flanking sequences were determined with the LA-PCR kit (Takara, Dalian, People's Republic of China). Primers C1 and C2 were supplied with the kit. The first PCR was performed with primer pair C1 and RSBhepcS1 (5′-CACCTCTGACATCTCTTCATCTGCAGCAACT-3′), and the second PCR was performed with primer pair C2 and RSBhepcS2 (5′-CAACTGCAACACTGAATGTCTTCATCTTAGGA-3′). Challenge of red sea bream with Escherichia coli DH5α was carried out as described previously (8). Twenty-four hours after the injection of bacteria, fish were sacrificed and tissues were removed and kept at −80°C for RNA isolation. Total RNA was isolated with Trizol reagent (GibcoBRL) from the tissues of red sea bream weighing 500 g as described previously (8). The reverse transcription of mRNA and PCR was carried out as described previously (6). Primer pair RSB-hepcN1 (5′-CAATGAGCAATGGCAGCCCA-3′) and RSB-hepcC1 (5′-TGCAGCAGGAATCCTCAACG-3′) was used. Primer pair RSB-18SN1 (5′-GGCAGCGTCCGGGAAACCAAAGTC-3′) and RSB-18SC1 (5′-CCACCCACAGAATCGAGAAAGAGC-3′) was used for 18S rRNA expression as a control.
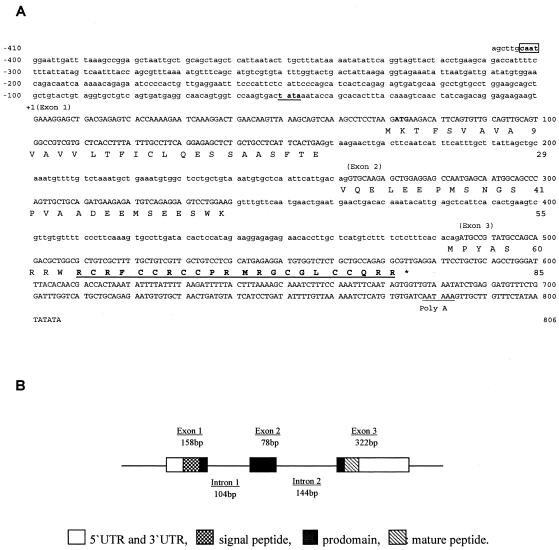
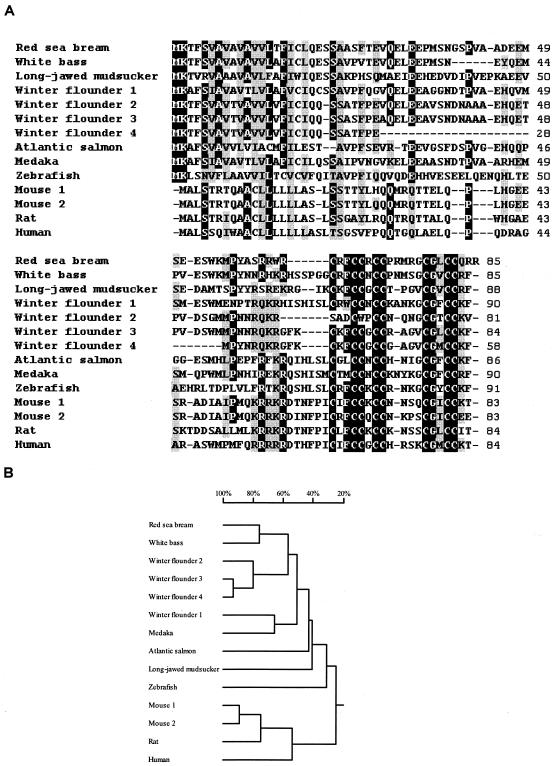
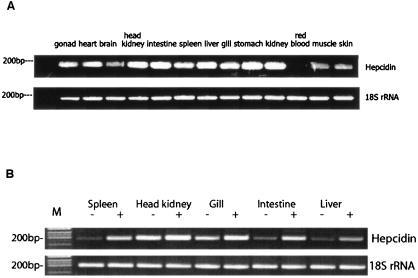
Seventeen cDNA clones among the 2,010 sequenced expressed sequence tags from the library were found to match hepcidin cDNA sequences from other vertebrates and were designated rsbHEPC1. The full-length cDNA is 596 bases long and contains an open reading frame of 255 bases encoding a peptide of 85 amino acids (aa) consisting of a signal peptide of 24 aa, a prodomain of 39 aa, and a mature peptide of 22 aa. The genomic sequence for red sea bream hepcidin and the upstream region was obtained by PCR (Fig. 1A). In the 1,216-bp genomic sequence, three exons and two introns were identified (Fig. 1B). The first exon covers the signal peptide coding sequence and part of the prodomain coding sequence. The mature peptide was encoded by exon 3. The 5′ UTR and 3′ UTR are found in exons 1 and 3, respectively. Analysis of the 5′ flanking region demonstrated the presence of TATA and CAAT boxes at −48 and −401, respectively. Alignment and phylogenetic analysis of the amino acid sequences of the hepcidin from red sea bream and other animals are shown in Fig. 2. The amino acid sequence of red sea bream hepcidin had 65.9, 52.8, 49.4, 48.3, 47.2, 42.2, 39.0, 37.3, 27.9, 25.2. 25.2, 24.1, and 22.8% identity with those of white bass, winter flounder 3, winter flounder 2, medakas, winter flounder 1, long-jawed mudsuckers, winter flounder 4, Atlantic salmon, zebra fish, mouse 1, mouse 2, rats, and humans, respectively. Hepcidin transcripts were highly abundant in pronephros, kidney, intestine, liver, gill, and stomach, abundant in gonad, heart, and spleen, less abundant in brain, muscle, and skin, and undetectable in red blood cells (Fig. 3A). The mRNA levels increased significantly in spleen, gill, liver, and intestine at 24 h after challenge (Fig. 3B).
FIG. 1.
(A) Genomic sequences of red sea bream hepcidin. Uppercase letters, exons; lowercase letters, introns; box, putative transcription factor binding sites. A TATA box and poly(A) signal are underlined. The mature peptide sequence is boldface and underlined. (B) Structure of the red sea bream hepcidin gene.
FIG. 2.
Alignment of the red sea bream hepcidin amino acid sequence with other vertebrate sequences (A) and phylogenetic analysis (B). Numbers to the right of the alignment refer to positions found in the sequences. Identical or similar amino acid residues are black or shaded. Dashes, gaps used to maximize the alignment. Sources of sequences used for comparison, with references for GenBank accession numbers or the number itself in parentheses, are as follows: white bass (24), winter flounder 3 (12), winter flounder 2 (12), medaka (AU178966), winter flounder 1 (12), long-jawed mudsuckers (13), winter flounder 4 (12), Atlantic salmon (12), zebra fish (25), mouse 1 (14), mouse 2 (14), rats (22), and humans (22). Cluster analysis (B) is based on the deduced amino acid sequence of hepcidin from red sea bream and other vertebrates. The scale over the tree refers to the percentage of divergence.
FIG. 3.
Expression of the hepcidin gene in various tissues of normal red sea bream (A) and red sea bream challenged with E. coli DH5α (B). Thirty-five cycles of amplification were used for panel A, and 25 cycles were used for panel B. −, fish injected with phosphate-buffered saline; +, fish injected with E. coli DH5α. 18S rRNA was used as the control.
Similar to those of mammals, the red sea bream hepcidin gene consists of three exons and two introns. The cDNA structure indicates that red sea bream hepcidin is translated as an 85-aa prepropeptide that is cleaved at the amino terminal to a mature peptide of 22 aa residues. Similar to hepcidin from other fish and mammals, red sea bream hepcidin contains eight cysteine residues, which is a characteristic feature of most hepcidins (21, 24). In humans, hepcidin mRNA was predominantly expressed in liver (13, 17). In teleosts, Shike et al. (24) also demonstrated that hepcidin mRNA was predominantly expressed in liver of white sea bass. However, the present study demonstrated that hepcidin mRNA was highly expressed in multiple tissues of red sea bream, which is markedly different from the expression pattern of hepcidin in humans and white bass (21, 24). However, in winter flounder, hepcidin transcripts (type III) were detected in esophagus and cardiac stomach as well as in liver (12). The high similarity in structure to hepcidins from other fish species (12, 24) and non-liver-specific expression of red sea bream hepcidins implied the presence of polymorphism in hepcidin molecules in teleosts. Similar molecular polymorphism of hepcidin cDNA was reported in winter flounder and Atlantic salmon (12). Challenging red sea bream with E. coli DH5α significantly up-regulated the hepcidin expression in spleen, liver, gill, and intestine. Similarly, Pigeon et al. (22) indicated that treatment of mice with lipopolysaccharide significantly elevated the level of hepcidin mRNA in liver. A similar up-regulating effect following bacterial infection was also observed in Atlantic salmon (12) and in striped sea bass (24). These results implied that hepcidin plays an important role in the immune response of red sea bream to infection. The availability of the red sea bream hepcidin gene lays the foundation for generating recombinant hepcidins to elaborate the structure and function of red sea bream hepcidin.
Nucleotide sequence accession numbers.
Clones designated rsbHEPC1 have been assigned GenBank accession number AY452732, and the genomic sequence encoding red sea bream hepcidin and the upstream region was assigned GenBank accession number AY452733.
Acknowledgments
This work was supported by grants from the National Nature Science Foundation of China (40376047), the State 863 High-Technology R&D Project of China (2002AA626010), and the National Major Basic Research Program (973) of China (2004CB117403).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and E. W. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreu, D., and L. Rivas. 1998. Animal antimicrobial peptides: an overview. Biopolymers 47:415-433. [DOI] [PubMed] [Google Scholar]
- 4.Broekaert, W. F., F. R. Terras, B. P. Cammue, and R. W. Osborn. 1995. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol. 108:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cammue, B. P., M. F. De Bolle, H. M. Schoofs, F. R. Terras, K. Thevissen, R. W. Osborn, S. B. Rees, and W. F. Broekaert. 1994. Gene-encoded antimicrobial peptides from plants. Ciba Found. Symp. 186:91-106. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S. L., Y. Hong, S. Scherer, and M. Schartl. 2001. Lack of ultraviolet-light inducibility of the medakafish (Oryzias latipes) tumor suppressor gene p53. Gene 264:197-203. [DOI] [PubMed] [Google Scholar]
- 7.Chen, S. L., M. Y. Xu, S. N. Hu, and L. Li. 2004. Analysis of immune-related genes expressed in red sea bream (Chrysophrys major) spleen. Aquaculture 240:115-130. [Google Scholar]
- 8.Chen, S. L., M. Y. Xu, X. S. Ji, and G. C. Yu. 2004. Cloning and characterization of natural resistance associated macrophage protein (Nramp) cDNA from red sea bream (Chrysophrys major). Fish Shellfish Immunol. 17:305-313. [DOI] [PubMed] [Google Scholar]
- 9.Cole, A. M., R. O. Darouiche, D. Legarda, N. Connell, and G. Diamond. 2000. Characterization of a fish antimicrobial peptide: gene expression, subcellular localization, and spectrum of activity. Antimicrob. Agents Chemother. 44:2039-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Destoumieux, D., P. Bulet, D. Loew, A. V. Dorsselaer, J. Rodriguez, and E. Bachere. 1997. Penaeidins, a new family of antimicrobial peptides isolated from the shrimp Penaeus vannamei. J. Biol. Chem. 272:28398-28406. [DOI] [PubMed] [Google Scholar]
- 11.Destoumieux, D., P. Bulet, J. M. Strub, A. V. Dorsselaer, and E. Bachere. 1999. Recombinant expression and range of activity of penaeidins, antimicrobial peptides from penaeid shrimp. Eur. J. Biochem. 266:335-346. [DOI] [PubMed] [Google Scholar]
- 12.Douglas, S. E., J. W. Gallant, R. S. Liebscher, A. Dacanay, and S. C. M. Tsoi. 2003. Identification and expression analysis of hepcidin-like antimicrobial peptide in bony fish. Dev. Comp. Immunol. 27:589-601. [DOI] [PubMed] [Google Scholar]
- 13.Gracey, A. Y., J. T. Troll, and G. N. Somero. 2001. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc. Natl. Acad. Sci. USA 98:1993-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilyin, G., B. Courselaud, M.-B. Troadec, C. Pigeon, M. Alizadeh, P. Leroyer, P. Brissot, and O. Loreal. 2003. Comparative analysis of mouse hepcidin 1 and 2 genes: evidence for different patterns of expression and co-inducibility during iron overload. FEBS Lett. 542:22-26. [DOI] [PubMed] [Google Scholar]
- 15.Krause, A., S. Neitz, H. J. Magert, A. Schulz, W. C. Forssmann, P. Schulz-Knappe, and K. Adermann. 2000. Leap-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 480:147-150. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 17.Lauth, X., H. Shike, J. C. Burns, M. E. Westerman, V. E. Ostland, J. M. Carlberg, J. C. Van Olst, V. Nizet, S. W. Taylor, C. Shimizu, and P. Bulet. 2002. Discovery and characterization of two isoforms of moronecidin, a novel antimicrobial peptide from hybrid striped bass. J. Biol. Chem. 277:5030-5039. [DOI] [PubMed] [Google Scholar]
- 18.Lehrer, R. I., and T. Ganz. 2002. Defensins of vertebrate animals. Curr. Opin. Immunol. 14:96-102. [DOI] [PubMed] [Google Scholar]
- 19.Oren, Z., and Y. Shai. 1996. A class of highly potent antibacterial peptides derived from pardaxin, a pore-forming peptide isolated from Moses sole fish Pardachirus marmoratus. Eur. J. Biochem. 237:303-310. [DOI] [PubMed] [Google Scholar]
- 20.Park, C. B., J. H. Lee, I. Y. Park, M. S. Kim, and S. C. Kim. 1997. A novel antimicrobial peptide from the loach, Misgurnus anguillicaudatus. FEBS Lett. 411:173-178. [DOI] [PubMed] [Google Scholar]
- 21.Park, C. H., E. V. Valore, A. J. Waring, and T. Ganz. 2001. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 276:7806-7810. [DOI] [PubMed] [Google Scholar]
- 22.Pigeon, C., G. Liyin, B. Courselaud, P. Leroyer, B. Turlin, P. Brissot, and O. Lereal. 2001. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J. Biol. Chem. 276:7811-7819. [DOI] [PubMed] [Google Scholar]
- 23.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 24.Shike, H., X. Lauth, M. E. Westerman, V. E. Ostland, J. M. Carlberg, J. C. Van Olst, C. Shimizu, P. Bulet, and J. C. Burns. 2002. Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Eur. J. Biochem. 269:2232-2237. [DOI] [PubMed] [Google Scholar]
- 25.Shike, H., C. Shimizu, X. Lauth, and J. C. Burns. 2004. Organization and expression analysis of the zebrafish hepcidin gene, an antimicrobial peptide gene conserved among vertebrates. Dev. Comp. Immunol. 28:747-754. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]