Abstract
White adipose tissue (WAT) is important for metabolic homeostasis. We established the differential proteomic signatures of WAT in glucose-tolerant lean and obese individuals and patients with type 2 diabetes (T2D) and the response to 8 weeks of high-intensity interval training (HIIT). Using a high-throughput and reproducible mass spectrometry–based proteomics pipeline, we identified 3773 proteins and found that most regulated proteins displayed progression in markers of dysfunctional WAT from lean to obese to T2D individuals and were highly associated with clinical measures such as insulin sensitivity and HbA1c. We propose that these distinct markers could serve as potential clinical biomarkers. HIIT induced only minor changes in the WAT proteome. This included an increase in WAT ferritin levels independent of obesity and T2D, and WAT ferritin levels were strongly correlated with individual insulin sensitivity. Together, we report a proteomic signature of WAT related to obesity and T2D and highlight an unrecognized role of human WAT iron metabolism in exercise training adaptations.
Proteomics reveals changes in human white adipose tissue induced by type 2 diabetes and high intensity-interval training.
INTRODUCTION
The prevalence of obesity and type 2 diabetes (T2D) is increasing worldwide, with obesity and inactivity being a major driver of insulin resistance and, consequently, the development of T2D. Physical activity play an essential role in the management of both obesity and T2D (1, 2), and exercise training improves whole-body insulin sensitivity, body composition, and cardiorespiratory fitness (V̇O2 max) in both glucose-tolerant lean and obese individuals and patients with T2D (1–6). In humans, this is accompanied by improved lipid droplet distribution and increased content of mitochondria and enzymes involved in insulin signaling to glucose transport and glycogen synthesis in skeletal muscle (3, 7–10). However, whether beneficial adaptations to exercise training occur in human adipose tissue remains largely unknown.
White adipose tissue (WAT) is the major fat-storing organ and serves as the source of numerous secreted adipokines, including leptin and adiponectin, that regulate metabolic processes throughout the body. Obesity-induced insulin resistance is associated with adipose tissue dysfunction, which is characterized by adipocyte hypertrophy, enhanced accumulation of extracellular matrix (ECM) components, fibrosis, and inflammation with increased macrophage infiltration, leading to increased secretion of pro-inflammatory cytokines and reduced secretion of adiponectin (11–16). Furthermore, adipose tissue dysfunction is associated with an impaired mitochondrial oxidative metabolism and increased lipolysis (13, 15–17). These alterations lead to lipid overflow and ectopic lipid deposition in metabolic tissues such as muscle and liver, which, together with low-grade systemic inflammation, are believed to contribute to insulin resistance, and increased risk of T2D (16, 18). However, in humans, the presence of adipose tissue dysfunction in insulin-resistant individuals has mainly been documented by transcriptional profiling (13–15, 17, 19, 20), whereas only few studies, using proteomics analysis of subcutaneous WAT (scWAT), has confirmed a reduced abundance of mitochondrial proteins in insulin-resistant individuals (21, 22). A deeper proteomic characterization of the changes in scWAT associated with obesity and T2D is, therefore, needed to improve our understanding of adipose tissue dysfunction in humans.
Exercise training has been reported to cause adaptations in WAT that may contribute to changes in whole-body metabolism. These changes include an improved transcriptional profile of genes related to mitochondrial function and inflammation, and changes in targeted markers of lipid and glucose metabolism, as demonstrated in rodents, healthy humans, and men with dysglycemia and/or obesity (23–31). However, these studies were mainly performed in rodents or used a hypothesis-based approach focusing on a subset of genes or proteins in the scWAT of humans (20, 29–31). Hence, further unbiased proteomic studies are needed to identify novel markers of exercise training–induced changes in human WAT.
Mass spectrometry (MS)–based proteomics has emerged as a powerful tool for unbiased, system-wide quantification of global and targeted protein analyses (32). Although several studies have revealed important insights into the scWAT tissue proteome, the quantitative readouts at the protein level have been insufficient to capture biological or pathological protein dynamics due to the nature of the technologies used (21, 33–36). With the latest developments in liquid chromatography (LC)–MS instrumentation, sample preparation workflows, and computational workflows, proteomics is emerging as a powerful technology in biomedical and clinical research (32, 37–41). We reasoned that the substantial developments in MS-based proteomics technologies could reveal novel information into the WAT proteome in obesity and T2D and provide an insight into how exercise training influences the adipose tissue proteome.
Here, we applied a high-throughput and high-sensitivity proteomics pipeline to map and compare the WAT proteome of subcutaneous abdominal adipose tissue biopsies obtained from glucose-tolerant lean and obese men and men with T2D at baseline and after 8 weeks of high-intensity interval training (HIIT) combining rowing and cycling exercise.
RESULTS
Metabolic health in obesity and T2D and effects of HIIT
This work represents a prespecified secondary analysis of a nonrandomized intervention study. The clinical and metabolic characteristics were reported recently (6). In brief, fasting serum insulin, plasma glucose and triglycerides levels, and HbA1c were elevated in men with T2D (table S1). Body weight, body mass index (BMI), waist, total fat, and lean body mass were lower in the lean group compared with the other groups. Men with T2D and glucose-tolerant obese and lean men had the same self-reported physical activity level, while V̇O2 max was lower in the diabetic group compared with the other groups but also in the obese versus the lean group. Insulin sensitivity measured as the insulin-stimulated glucose infusion rate (GIR) was ~40% lower in men with T2D compared with both obese and lean men (all, P < 0.01).
As reported (6), 8 weeks of HIIT protocol increased V̇O2 max by 8 to 15% (all, P < 0.01) and slightly reduced body weight and BMI (all, P < 0.05) in all groups (table S1). This was accompanied by a greater loss of total fat mass (1.6 to 2.3 kg; all, P < 0.05) than increase in lean body mass (0.6 to 1.5 kg; all, P < 0.05) in all groups. The HIIT protocol markedly increased insulin sensitivity (~30 to 40%) in all groups. The posttraining insulin sensitivity in men with T2D reached the pretraining levels of insulin sensitivity in the obese and lean men. There were no differences in any of the above HIIT-induced responses between the groups. In men with T2D, the HIIT protocol caused a clinically relevant decrease in HbA1c by ~4 mM.
Comprehensive high-throughput proteome analysis of human WAT
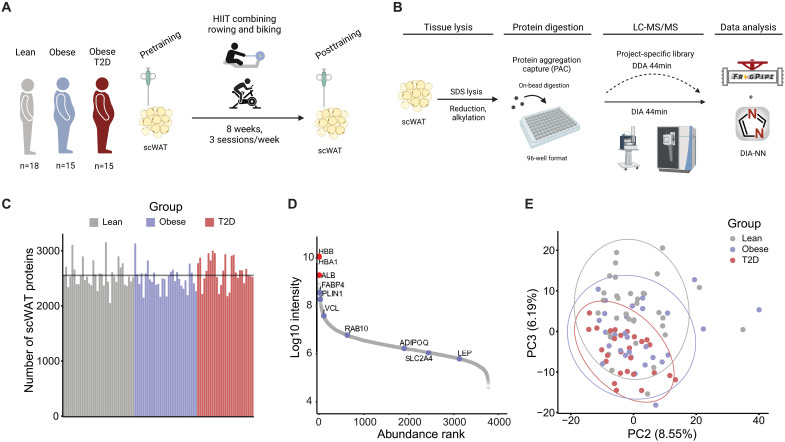
Abdominal scWAT biopsies were collected from lean, obese, and T2D men before and after 8 weeks of HIIT (Fig. 1A). To map the human WAT proteome, we lysed a total of 92 samples in SDS lysis buffer and performed the protein aggregation capture (PAC) method in a 96-well plate format (Fig. 1B) (38). The resulting tryptic peptides were analyzed on an Evosep LC instrument coupled to an Exploris mass spectrometer. Samples were measured by data-independent acquisition (DIA) and matched to a project-specific library (6154 proteins), which led to quantification of 3773 proteins in total (in average 2554 proteins per sample). The abundance range span ~ five orders of magnitude and the quantification included the low abundant adipose-derived hormone leptin (Fig. 1, C and D). Our state-of-the-art workflow enabled an unprecedented proteome depth in highly challenging tissue biopsies of human scWAT using only 44 min analysis time per sample (30 samples/day). The MS runs demonstrated high quantitative reproducibility between runs with a median Pearson correlation coefficient of 0.94 (fig. S1A).
Fig. 1. High-throughput proteome analysis of human scWAT.
(A) Graphic illustration of the study design. (B) Proteomics workflow from sampling and lysis of scWAT to analysis. (C) Number of scWAT proteins quantified per sample in each of the three groups. (D) Dynamic range plot of ranked protein abundance (log10 intensity) showing relevant adipocyte-specific markers (blue dots) and blood contaminants (red dots). (E) Principal component (PC) analysis plot of component 2 (x axis) and component 3 (y axis). Dots are colored by group.
Adipose tissue is a heterogeneous tissue consisting of a variety of different cell types, including adipocytes, immune cells, fibroblasts, and smooth muscle cells (42). When ranking the quantified proteins based on their abundance, as expected, blood contamination markers [hemoglobin subunit beta (HBB), hemoglobin subunit alpha-1 (HBA1), and albumin (ALB)] constituted the most abundant proteins identified, followed by the adipocyte-specific markers [fatty acid binding protein, adipocyte (FABP4), and perilipin-1 (PLIN1)] (Fig. 1D). Proteins known to regulate insulin-stimulated glucose uptake [ras-related protein Rab-10 (RAB10) and glucose transporter type 4 (SLC2A4/GLUT4)] and adipokines involved in whole-body metabolism (adiponectin and leptin) were also identified. For further analysis, we applied stringent filtering and included only proteins (n = 2106), which were quantified in 75% of all samples. Next, we explored the WAT proteome using a principal components analysis (PCA). WAT heterogeneity on PC1 was explained independently of the group (lean, obese, and T2D) (fig. S1C); however, there was a gradual group separation on PC2 (x axis) and PC3 (y axis) (Fig. 1E). Coloring the PCA by individual fat mass (kg) suggests that the PC2 and PC3 separation could be driven partly by adiposity (fig. S1D).
Obesity and T2D related differences in the adipose tissue proteome
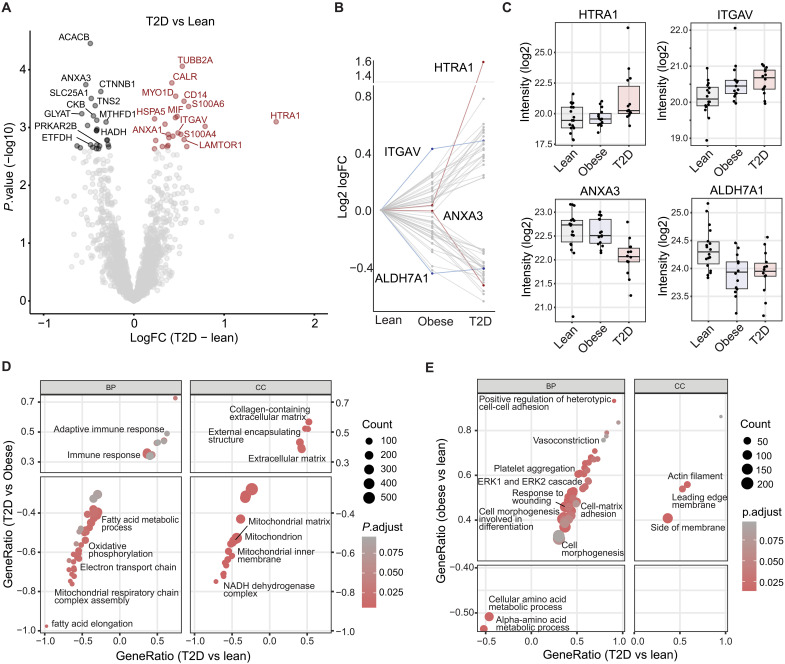
Previous studies have characterized the proteome of isolated human adipocytes and found massive depot-specific differences (43, 44). However, the context of adipocytes in their native environment and the interplay with other cell types is crucial to understand the state of adipose tissue in health and disease. Here, we compared differences at baseline between groups and found 246, 130, and 89 proteins to differ (unadjusted P < 0.05) in the T2D-lean, T2D-obese, and obese-lean comparisons, respectively (data file S1C). Of these, 48 proteins (25 up-regulated and 23 down-regulated) differed significantly between patients with T2D and lean healthy individuals (Fig. 2A and data file S1D) after correction for multiple testing [false discovery rate (FDR) < 0.10)].
Fig. 2. Baseline proteome differences in scWAT from lean, obese, and T2D individuals.
(A) Volcano plot showing baseline (pre) comparison of the WAT proteome between T2D and lean individuals (FDR < 10%). Most regulated proteins are highlighted. (B) Profile plot of significant T2D-lean regulated proteins. Dots connected by a line represent the profile of protein. Proteins with a distinct profile are highlighted in red and blue. (C) Boxplot of highlighted proteins with distinct profile curves between groups. (D and E) GO biological processes (GOBP) and cellular components (GOCC) enrichment analysis. Gene ratios of overlapping terms between T2D-obese (B), obese-lean (C) (y axis), and T2D-lean (x axis) are shown. Dots are colored by adjusted P value and size is based on the number of proteins in the enriched category.
The majority of the down-regulated proteins in T2D are involved in mitochondrial processes, including fatty acid metabolism [acetyl–coenzyme A (CoA) carboxylase 2 (ACACB), glycerol-3-phosphate acyltransferase 1 (GPAM), hydroxyacyl-coenzyme A dehydrogenase (HADH), cAMP-dependent protein kinase type II-beta regulatory subunit (PRKAR2B), and very-long-chain enoyl-CoA reductase (TECR)], tricarboxylic acid (TCA) cycle and electron transport [cytochrome b5 (CYB5A), electron transfer flavoprotein-ubiquinone oxidoreductase (ETFDH), and 2-oxoglutarate dehydrogenase complex component E1 (OGDH)], mitochondrial transport [tricarboxylate transport protein, mitochondrial (SLC25A1)], adenosine triphosphate (ATP) resynthesis/energy shuttling [creatine kinase B-type (CKB)], apoptosis [apoptosis-inducing factor 1, mitochondrial (AIFM1)], and other processes [aldehyde dehydrogenase 7A1 (ALDH7A1), atypical kinase COQ8A, mitochondrial (COQ8A), glycine N-acyltransferase (GLYAT), and C-1-tetrahydrofolate synthase (MTHFD1)]. Regulators of insulin signaling [tensin-2 (TNS2)] and canonical WNT signaling [catenin beta-1 (CTNNB1) and catenin delta-1 (CTNND1)] were also down-regulated in T2D. Several of the up-regulated proteins in T2D are involved in ECM [serine protease (HTRA1), integrin alpha-V (ITGAV), protein S100-A6 (S100A6), and protein S100-A4 (S100A4)], immune response [monocyte differentiation antigen CD14 (CD14), macrophage migration inhibitory factor (MIF), and vitronectin (VTN)], and endoplasmic reticulum (ER) stress [ER chaperone BiP (HSPA5) and calreticulin (CALR)] but also included the lysosomal adaptor protein, regulator complex protein (LAMTOR1), which was recently demonstrated to be essential for the polarization of M2 macrophages (45). Overall, these findings are in line with reported transcriptomic signatures of adipose tissue dysfunction in obesity and T2D in humans (13–15, 17, 19).
There were no statistically significant differences observed in the T2D-obese or obese-lean comparisons when correcting for multiple testing (FDR < 0.1). However, when applying a less stringent P value threshold (unadjusted P < 0.01), several of the T2D-obese–regulated proteins were changing in the same direction as in the T2D-lean comparison (fig. S2, A and B, and data file S1C). Leptin, the major marker of adiposity, was identified and quantified in 63 of 91 samples and showed an expected elevation in the T2D group (T2D versus lean, unadjusted P = 0.002) and a high association with total fat mass (r = 0.59 and P = 1.8 × 10−4) (fig. S2, C and D).
We further investigated the patterns of the 48 proteins that were significantly different between lean men and men with T2D at baseline (data file S1D). The most significantly regulated proteins displayed a gradual increase or decrease in abundance from lean to obese to T2D (Fig. 2B), suggesting that obesity, even in glucose-tolerant individuals, causes changes in the scWAT proteome that are further altered in the same direction during the development of T2D. Notably, some proteins were equally abundant in the lean and obese groups but showed marked changes in abundance in the T2D group (profiles marked in red, Fig. 2B). The most prominent example was HTRA1, which showed a threefold increase (1.6 log2 fold change) in scWAT from patients with T2D (Fig. 2C). We also found that annexin A3 (ANXA3) showed a similar abundance in the lean and obese groups but was markedly decreased in the T2D group (Fig. 2C). A few proteins showed similar abundance in the obese and T2D group, including ITGAV and ALDH7A1, which are an integrin receptor and a semialdehyde dehydrogenase, respectively (profiles marked in blue, Fig. 2B). The differences in these WAT proteins could be mainly obesity driven. To validate our proteomics findings, we determined the protein abundance of HTRA1 and ITGAV by Western blotting in the scWAT samples. The results were consistent with the proteomics data showing between-group differences for both HTRA1 (P = 0.036) and ITGAV (P = 0.003) as well as strong correlations (HTRA1, τ = 0.48 and P = 2.6 × 10−11; ITGAV, τ = 0.25 and P = 5.5 × 10−4) between the proteomics and Western blotting measurements (fig. S2, E to I).
As known from microarray-based transcriptomic studies of tissues in insulin-resistant individuals, most, if not all, differences at the single gene level disappears after correction for multiple testing (46, 47). To gain further insight into cellular and biological processes altered in scWAT in relation to T2D and obesity, respectively, we therefore performed gene set enrichment analysis (GSEA) of gene ontology (GO)–cellular components (CC) and GO–biological processes (BP) using the whole proteome as background. When restricting GO terms to those regulated (FDR < 0.1) in both the T2D-lean and T2D-obese comparisons, we observed that terms related to several mitochondrial processes, including electron transport, oxidative phosphorylation, TCA cycle, and fatty acid metabolism were strongly down-regulated, whereas ECM and immune response related terms were strongly up-regulated in scWAT of patients with T2D (Fig. 2D and data file S1F). This suggests that these changes are highly related to T2D. When comparing the overlap of the regulated GO terms in the Obese-lean and T2D-lean comparisons, we found several terms related to cell adhesion, morphogenesis, and differentiation as well as blood coagulation and wound healing to be up-regulated and terms related to amino acid metabolism to be down-regulated in obese individuals with and without T2D (Fig. 2E and data file S1G).
Correlation of clinical parameters with scWAT proteins
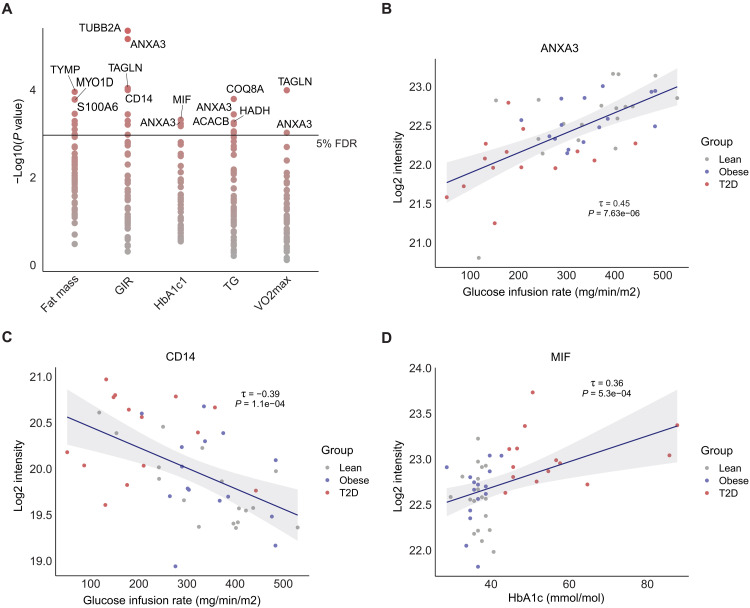
The majority of scWAT proteins altered in the T2D state showed a gradual change in abundance from lean to obese to T2D, suggesting that they may play a role in the progression from healthy to dysfunctional WAT. To reveal molecular individuality linked to the clinical phenotype, we correlated the 48 significantly regulated proteins (T2D-lean) with five clinical variables; total fat mass, insulin sensitivity (GIR), glycemic control (HbA1c), plasma triglycerides, and V̇O2 max and found several significant associations (Fig. 3A and data file S1E). Overall, 18 of the regulated unique proteins correlated with one or more of these five clinical variables. In particular, the protein abundance of tubulin beta-2A (TUBB2A) was strongly negatively associated (τ = −0.46; P < 0.001) with insulin sensitivity (GIR), whereas ANXA3 was strongly positively associated (τ = 0.45 and P < 0.001) with GIR (Fig. 3B). ANXA3 also correlated positively with V̇O2 max and negatively with HbA1c and plasma triglyceride levels (data file S1E). The macrophage infiltration marker, CD14, showed a negative correlation with GIR (τ = −0.39; P = 1.1 × 10−4), consistent with previous research showing that CD14-positive macrophages drive inflammation-induced insulin resistance (Fig. 3C) (48). The protein with the strongest correlation to HbA1c was MIF, mainly driven by the T2D group (τ = 0.36 and P = 5.3 × 10−4) (Fig. 3D). MIF is a secreted protein involved in adipose tissue inflammation and is suggested to stimulate the release of pro-inflammatory cytokines with whole-body systemic effects (49).
Fig. 3. Correlation of clinical parameters with scWAT proteins.
(A) Correlation analysis of significantly regulated proteins at baseline with five clinical parameters. −Log10 (P value) on y axis. Most significant proteins are highlighted. (B and C) Kendall’s rank correlation of ANXA3 and CD14 protein abundance (log2 intensity, y axis) and GIR (mg/min/m2). (D) Kendall’s rank correlation of MIF and HbA1c (mmol/mol). Kendall’s τ coefficient and P value are shown.
Impact of HIIT on scWAT in lean, obese, and T2D individuals
Exercise training improves whole-body insulin sensitivity in both glucose-tolerant lean and obese individuals as well as in patients with T2D (1–6), but the contribution of scWAT to this and other healthy metabolic adaptations is less clear. In the current study, we investigated the effect of 8 weeks of HIIT on abdominal scWAT in healthy lean and obese individuals and in obese patients with T2D. This was assessed by performing MS-based proteomics on scWAT biopsies taken before and after the HIIT intervention, thereby using a paired design.
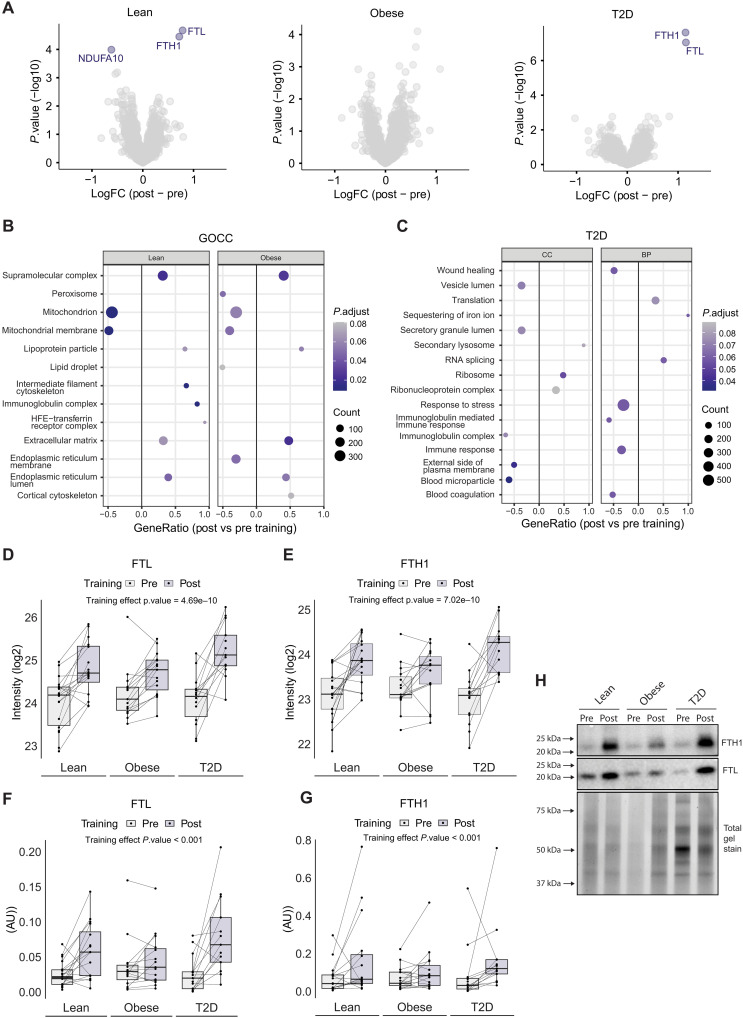
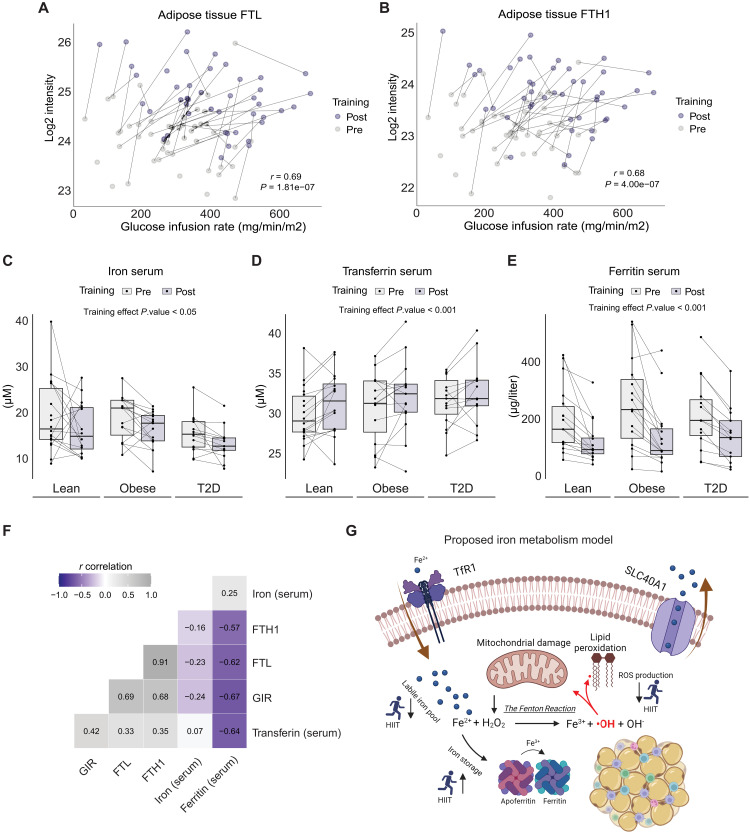
To our surprise, only three proteins, ferritin light chain (FTL), ferritin heavy chain-1 (FTH1), and reduced form of nicotinamide adenine dinucleotide (NADH) dehydrogenase(ubiquinone) 1 alpha subcomplex subunit 10 (NDUFA10) were significantly changed (FDR < 0.10) in response to exercise training in at least one of the groups (Fig. 4A and table S2C). The low impact of exercise training on the human WAT proteome could be attributed to baseline differences within the three groups and the heterogenous nature of exercise training responses.
Fig. 4. Effects of exercise training on the human scWAT.
(A) Volcano plot of group-wise log2 fold changes post- versus pretraining. Significant proteins are highlighted (FDR < 10%). (B) Selected representative GOCC terms significantly changing with exercise training in the lean and obese group. (C) Selected representative GOCC and GOBP terms significantly changing with exercise training in the T2D group. (D and E) Boxplot of FTL and FTH1 protein abundance (log2 intensity) in human scWAT pre/post exercise training. (F and G) Boxplot of FTL and FTH1 protein abundance (arbitrary unit) obtained from Western blot analysis in human scWAT pre/post exercise training. (H) Representative immunoblots from Western blot analysis.
The lean and T2D groups showed a significant increase in scWAT FTL and FTH1 levels (obese: FTL, unadjusted P = 0.007; and FTH1, unadjusted P = 0.064). There was also a significant main effect (FDR < 0.10) of exercise training in the pooled cohort of participants for both FTL and FTH1 (FTL, unadjusted P = 4.69 × 10−10; and FTH1, unadjusted P = 7.02 × 10−10), representing an average increase by 75 and 66%, respectively (Fig. 4, D and E). To validate this observation, we performed a Western blot analysis and verified that both ferritin subunits were exercise-training–responsive proteins in human scWAT. Here, we identified a significant increase in both FTL and FTH1 abundances (both main effect, P < 0.001) after 8 weeks of HIIT (Fig. 4, F to H). A post hoc analysis revealed group-specific differences in the HIIT-mediated changes in FTL abundance as this was only significant in the lean (P = 0.015) and T2D (P < 0.001) groups, confirming the proteomic findings. The proteomics levels of FTL and FTH1 showed strong correlations with the abundances measured by Western blotting (FTL, r = 0.83 and P = 5.01 × 10−12) (fig. S3A). Circulating ferritin can be taken up into cells via the transferrin receptor protein 1 (TfR1); however, WAT TfR1 abundance did not differ between groups or change in response to HIIT when measured using Western blotting (HIIT main effect, P = 0.661) (fig. S3, B and C).
Next, we performed a GSEA of the changes in GO-CC and GO-BP in response to the exercise training protocol. We found considerable similarities between the HIIT-induced changes in scWAT from the lean and obese groups (Fig. 4B and data file S1H). Unexpectedly, several GO terms related to mitochondrial components were down-regulated in the lean and obese groups, whereas ECM and lipoprotein particle components were up-regulated. A lipid droplet GO-CC term was also down-regulated in scWAT from the obese group. This might indicate smaller lipid droplets, which is consistent with the loss of total fat mass observed after HIIT (6). Compared to the lean and obese groups, the T2D group showed a different profile in the HIIT-regulated GO terms. Thus, in the T2D group, we found that terms involved in immune responses, complement activation, stress responses, coagulation, and wound healing, as well as secretory-related terms were down-regulated in response to training (Fig. 4C and data file S1I). This HIIT-induced response could be explained as an alleviation of the elevated levels of GO terms related to immune response and complement activation observed at baseline in the T2D group (Fig. 2D). Terms related to protein synthesis, including ribosome, RNA splicing, and translation were all up-regulated, suggesting a remodeling of the WAT proteomic composition in patients with T2D in response to training. All three groups showed a HIIT-induced increase in small lysosome–related GO-CC terms containing the FTH1 and FTL subunits.
Adaptations to HIIT in scWAT associates with both local and systemic iron metabolism
Intrigued by the HIIT-induced up-regulation of FTL and FTH1 in scWAT, we performed a repeated measures correlation on the abundance of each ferritin subunit in WAT with individual measures of insulin sensitivity (GIR) before and after the 8 weeks of HIIT. We found a highly significant association between the increase in WAT ferritin levels and improvements in insulin sensitivity (FTL, P = 1.81 × 10−7; and FTH1, P = 4 × 10−7) (Fig. 5, A and B). We next measured if there was a systemic change in iron homeostasis by measuring iron content markers in the serum of the study participants. We found an overall effect of HIIT in lowering serum iron levels (main effect, P = 0.013) (Fig. 5C). While serum transferrin, the primary iron-transporting protein in the blood, showed a slight increase (main effect, P < 0.001), there was a remarkable decrease in serum ferritin (87.52 μg/liter; main effect, P < 0.001) in response to HIIT (Fig. 5, D and E). Thus, the HIIT-induced increase in ferritin subunit levels in scWAT is not caused by blood contamination. As circulating ferritin levels have been linked to the risk of obesity and T2D, we performed a repeated measures correlation of serum ferritin with GIR, and FTL and FTH abundance in WAT determined by proteomics. We found serum ferritin to be negatively correlated with insulin sensitivity (r = −0.67 and P = 6.3 × 10−7) (Fig. 5F), which aligns with previous studies (50, 51). In addition, WAT FTL levels negatively correlated with serum ferritin (r = −0.62 and P = 8.17 × 10−6), suggesting a coordinated regulation between whole-body and adipose tissue–specific iron storage capacity. Together, we propose a model where the iron storage capacity of WAT increases in response to HIIT, thereby contributing to a lowered systemic free iron content and a reduction in the intracellular labile free iron pool and cellular oxidative stress (Fig. 5G). This may lead to reduced reactive oxygen species (ROS) production through the Fenton reaction, ultimately reducing mitochondrial damage and lipid peroxidation.
Fig. 5. Exercise training remodels iron homeostasis.
(A and B) Repeated-measures correlation of human scWAT FTL and FTH1 protein expression (log2 intensity) and GIR (mg/min/m2). (C to E) Serum measurements of iron (μM), transferrin (μM), and ferritin (μg/liter). (F) Repeated-measures correlation matrix of parameters related to iron homeostasis. (G) Graphical illustration of a proposed model of iron homeostasis in human scWAT. HIIT elevates intracellular ferritin levels, promoting iron storage and diminishing the free iron labile pool. This reduces Fe2+ oxidation in the Fenton reaction, subsequently leading to fewer radical oxygen species. Such a mechanism could protect the cell from mitochondrial damage and lipid peroxidation.
DISCUSSION
Human WAT has a profound impact on whole-body metabolism and is attracting attention as a therapeutic target in obesity and T2D. Recent evidence, particularly in rodents, suggests that exercise training–induced adaptations in WAT may contribute to the whole-body effects of exercise training (23–26, 28–31). Here, we investigated changes in the human WAT proteomes related to obesity and T2D as well as induced by 8 weeks of HIIT in glucose-tolerant lean and obese individuals and patients with T2D. Using a state-of-the-art high-throughput proteomics technology, we accurately quantified >3500 proteins across all samples. To our knowledge, this is the most comprehensive scWAT proteome from an unprecedentedly large number of lean and obese individuals and patients with T2D. Our analysis revealed multiple obesity- and T2D-related changes in the scWAT proteomes but unexpectedly, few changes in response to HIIT. A majority of the scWAT proteins, and related cellular and biological processes affected by obesity and T2D, showed a gradual change in abundance from the lean to obese to T2D group, suggesting that the scWAT proteome in obese individuals is already in transition to the dysfunctional scWAT proteome observed in patients with T2D. Although HIIT markedly improved whole-body insulin sensitivity, V̇O2 max, and body composition in all groups, it only had minor effects on the scWAT proteome. However, HIIT significantly altered proteins involved in iron homeostasis, providing additional insights into the field.
Transcriptomic studies have reported obesity and T2D related changes in human scWAT (13–15, 17, 19); however, to what extent these alterations are present at the protein level is largely unknown. While previous studies have provided insights into the human scWAT proteome, they generally reported a much lower number of proteins identified and quantified in smaller cohorts of individuals (28, 33–36). The much more comprehensive coverage and quantification of the human scWAT proteome in our study was achieved by an improved sample preparation using the PAC method (38), highly reproducible chromatography (39), and a sensitive mass spectrometer (40). We observed that the obese scWAT proteome appears to exist in a transition state toward the dysfunctional scWAT proteome observed in T2D. Our data indicate that the down-regulation of several mitochondrial processes and up-regulation of ECM and immune response-related pathways is mainly driven by T2D, whereas the up-regulation of proteins involved in cell adhesion, morphogenesis, and growth (e.g., up-regulation of ERK cascade) is driven primarily by obesity. The results from our proteomics study of human scWAT extend the findings in previous transcriptomic studies (13, 15, 17, 19, 21, 52, 53) by showing that increased markers of immune responses, complement activation, and ECM components as well as reduced markers of mitochondrial capacity in scWAT, also at the proteomic level, are crucial factors involved in dysfunctional scWAT in T2D.
Some proteins displayed a discrete rather than continuous expression profile between lean, obese, and patients with T2D. For instance, the abundance of the protein HTRA1, a secreted protease that suppresses adipogenesis and lipid droplet formation in human mesenchymal stem cells (54), was threefold elevated in patients with T2D compared to both lean and obese individuals. An earlier report suggests that HTRA1 is primarily located in blood vessels in WAT and is highly expressed in insulin-resistant individuals (54). Since HTRA1 is a known adipogenesis suppressor, it may act as a late-phase negative regulator to prevent adipocyte differentiation in patients with T2D. Conversely, compared to the lean group, the protein levels of the integrin receptor, ITGAV, were high in both the obese and T2D groups, and they were similar between the obese and T2D group. Since ITGAV is known to modify adipose cell proliferation and differentiation (55), ITGAV is likely involved in the alterations in the adipose cell morphology and function in WAT from obese men and men with T2D. Last, we found ALDH7A1, a protein that protects cells against oxidative stress (56), being down-regulated in obesity and T2D, indicating lower protection against oxidative damage under these pathophysiological conditions.
Associating individual proteomic signatures with clinical phenotypes can be valuable in disease diagnosis and surveillance (57). To this end, we correlated five commonly used clinical parameters with the abundance of scWAT proteins regulated in patients with T2D compared with lean individuals. We found the strongest inverse association between insulin sensitivity and the abundance of TUBB2A. The protein TUBB2A has the highest expression in adipose tissue and has previously been found to be increased in scWAT of insulin-resistant individuals (36, 58). Several of the proteins showing significant associations with the clinical phenotypes are known as secreted proteins, including CD14, MIF, and ANXA3. CD14 is likely to be secreted from infiltrating/resident immune cells, but increased expression of adipocyte CD14 mRNA expression has also been reported in obese individuals (48). MIF is a secreted protein involved in adipose tissue inflammation and is suggested to stimulate the release of pro-inflammatory cytokines with whole-body systemic effects (49). In addition, MIF has been proposed as an effective antidiabetic therapeutic target, as MIF antagonists show great promise to reduce pro-inflammatory cytokines and lower blood glucose levels in mice (59). Last, the secreted protein ANXA3 showed a strong positive correlation with insulin sensitivity. ANXA3 is a negative regulator of adipocyte differentiation and knockdown of ANXA3 in 3T3-L1 adipocytes increased lipid droplet accumulation (60). The role of ANXA3 in the adipose metabolism is unclear, despite its high expression in murine adipose tissue (60). We speculate that the panel of regulated secreted scWAT proteins, which displays a strong correlation with clinical phenotypes, could be used as potential noninvasive plasma biomarkers of whole-body insulin sensitivity or glycemic control in T2D.
Despite a marked beneficial effect of the HIIT protocol on whole-body insulin sensitivity, V̇O2 max, and body composition in glucose-tolerant lean and obese individuals and patients with T2D, only a few significant changes in scWAT protein abundance were observed after HIIT. However, when using GSEA, several GO-CC terms showed similar HIIT-induced responses in the lean and obese individuals. Notably, several groups of proteins representing different components of mitochondria were down-regulated in response to HIIT in these glucose-tolerant groups. Our results extend previous reports of decreased (61) or unchanged mitochondrial respiratory capacity (20, 62) and lack of changes in mitochondrial content and abundance of targeted proteins involved in mitochondrial biogenesis and oxidative phosphorylation in scWAT of lean and obese individuals following exercise training interventions of various durations (6 to 12 weeks), types, and intensities (20, 61–63). Furthermore, in one of these studies (20), exercise training did not improve several other markers of adipose tissue function, such as adipocyte morphology, measures of lipolysis, and markers of inflammation, browning, or adipokines. Our unbiased, large-scale proteomic approach strongly supports these observations suggesting that, e.g., the increase in insulin sensitivity in response to exercise training is not mediated by improvements in known markers of adipose tissue dysfunction, at least not in nondiabetic individuals. However, as speculated in previous studies (20, 61–63), longer intervention periods with intense exercise training and larger reductions in fat mass may be necessary to achieve improvements in these markers of adipose tissue dysfunction.
While GO terms related to mitochondrial processes were unaltered by HIIT in the T2D group, exercise training in this group caused potential beneficial changes in the scWAT proteome. Thus, GO terms related to secretion, immune responses, complement activation, stress responses, coagulation, and wound healing were decreased. This is particularly interesting, as some of these terms were elevated at baseline in T2D or obesity in our study, in line with previous transcriptomic evidence of enhanced inflammatory pathways, including immune responses and complement and coagulation cascades in WAT in obesity and T2D (15, 19, 63, 64). In addition, GO terms such as the ribosome, RNA splicing, and translation were all up-regulated by HIIT in scWAT in the T2D group. These results suggest that exercise training can remodel and improve at least some components of the exaggerated dysfunctional scWAT state observed in individuals with T2D (13).
Iron is essential for several fundamental metabolic functions, including the transportation of oxygen. The risk of developing T2D has been found to be positively associated with iron overload and/or the body iron stores most often measured as serum ferritin (65–70). Serum ferritin is furthermore inversely associated with plasma adiponectin in a cohort of individuals with or at an increased risk of T2D and cardiovascular diseases (71, 72). In the present study, we found no differences in the baseline levels of serum iron, ferritin, transferrin, or WAT ferritin subunits between the groups. This is in line with a study of the expression of iron metabolism-related genes, including FTL, FTH, and TFR, in WAT of lean, overweight, and heavily obese individuals (73). In that study, no basal differences were seen between groups in WAT, whereas the expression of FTL in visceral adipose tissue was increased in the heavily obese group, suggesting depot-specific differences in iron metabolism (73). One study recently explored the effects of exercise training on tissue iron homeostasis in obese men and women and found that HIIT decreased protein ferritin levels in serum, skeletal muscle, and liver (74). In line with existing evidence, we found that HIIT decreased serum ferritin and iron, but unexpectedly, HIIT increased the protein levels of the ferritin subunits in scWAT. The increase in WAT ferritin levels was observed across all groups and this increase in WAT ferritin subunit levels was strongly associated with the improvement in whole-body insulin sensitivity. Notably, a strong negative correlation was observed between serum ferritin and insulin sensitivity in response to HIIT, whereas a positive correlation between WAT ferritin protein levels and insulin sensitivity was seen in response to HIIT. This has not been previously reported. These findings appear to contradict other studies, which have found a negative relationship between WAT ferritin transcript levels and insulin sensitivity (73, 75). However, caution should be exercised when interpreting these baseline differences related to insulin sensitivity and the insulin-sensitizing effect of exercise training. First, in one of the studies, WAT ferritin was measured in visceral adipose tissue biopsies (73), and second, both studies measured mRNA levels rather than the protein abundance. Free intracellular iron increases the translation of ferritin in a posttranscriptional manner (76, 77), making it crucial to determine protein levels, as these may differ from transcript levels, in particular in disease states.
The descriptive nature of the present study only allows us to speculate about the mechanism causing the HIIT-mediated changes in scWAT ferritin abundance. On the basis of the observations from the current study, we propose a model where the iron storage capacity of scWAT increases in response to HIIT. In cultured 3T3-L1 adipocytes, insulin induces a rapid translocation of transferrin receptors to the cell surface following insulin administration (78, 79). The translocation of transferrin receptors and binding of transferrin to the cell surface was associated with an increased uptake of iron (78, 79). Although subsequent studies have found that the trafficking of the transferrin receptor and GLUT4 occurs via different mechanisms in adipocytes (80, 81), improved whole-body insulin sensitivity following HIIT may be associated with increased iron uptake in human scWAT. As the cytosolic ferritin abundance is positively regulated by the intracellular iron levels (82), we therefore suggest that the identified increase in WAT ferritin serves as a protective measure to store iron in an inert form (82, 83), thus preventing it from facilitating the formation of toxic free-radical species that may damage the tissue. Ferritin appears to be important for adipose tissue function as deletion of the Fth gene in mice severely impairs adipose tissue metabolism and disturbs redox balance (84). The protein abundance of FTL and FTH1 has furthermore been found to increase during adipocyte differentiation, which contradicts the notion of WAT ferritin being harmful (85). Moreover, FTL expression in macrophages protects against lipopolysaccharide-induced inflammation and oxidative stress (86), suggesting that both ferritin subunits contribute to iron metabolism. Last, overexpression of ferritin in 3T3-L1 adipocytes shields the cells from iron-induced lipid peroxidation and damage (85). Therefore, it is likely that the HIIT-induced increase in WAT ferritin subunit levels protects adipocytes from oxidative stress and damage. Further studies are warranted to establish a role for the increased scWAT ferritin levels in response to exercise training in humans.
In conclusion, our findings provide additional insights into the scWAT proteome changes associated with obesity and T2D and in response to HIIT combining rowing and cycling. However, further research with larger cohort sizes is needed to validate and build upon these results. Last, the high-throughput proteomics workflow presented here is well suited for larger-scale studies.
MATERIALS AND METHODS
Human participants
The clinical and metabolic results from this nonrandomized intervention study were reported recently (6). In brief, the study included middle-aged, obese men with T2D (n = 15) carefully matched to glucose-tolerant obese (n = 15) and lean (n = 18) men (table S1), all with a low to moderate level of self-reported physical activity, with no difference between groups. The sample size was estimated to detect a difference in insulin sensitivity at baseline between patients with T2D and lean men and an increase in insulin sensitivity in response to exercise training (6). In patients with T2D, use of glucose-lowering agents was restricted to the use of metformin, DPP4 inhibitors, and/or sulphonylureas in addition to blood pressure and lipid-lowering medication (6). The nondiabetic individuals had normal glucose tolerance as evaluated by a 2-hour plasma glucose level below 7.8 mM after an oral glucose tolerance test, an HbA1c within the normal range and no family history of diabetes, and they were not taking any medication. One lean man and three obese men had fasting plasma glucose values corresponding to impaired fasting glucose. All participants had normal results on blood screening tests and ECG. Four participants did not complete the HIIT protocol. Informed consent was obtained from all individuals before participation.
Ethics approval
The study was approved by the Regional Scientific Ethical Committees for Southern Denmark (project ID: S-20170142) and performed in accordance with the Helsinki Declaration II. The study was registered at ClinicalTrials.gov (NCT03500016).
Study design
All participants were examined on 2 days before (days 1 and 2) and after (days 3 and 4) a HIIT protocol combining rowing and cycling as previously reported (6). On days 1 and 3, body composition was assessed by dual-energy x-ray absorptiometry, and V̇O2 max was determined by an incremental exercise test. On days 2 and 4, biochemical characteristics, insulin sensitivity, and substrate metabolism were determined after an overnight fast using a hyperinsulinemic-euglycemic clamp with tracer glucose (2-hour basal period and 3-hour insulin infusion, 40 mU m−2 min−1) combined with indirect calorimetry (6). Day 4 took place 48 hours after the V̇O2 max test. The participants were instructed to refrain from alcohol and caffeine 24 hours before the test days and from physical activity 48 hours before test. Furthermore, participants were informed not to change their dietary habits during the intervention period. In patients with T2D, all medication was withdrawn 1 week before the clamp studies but was otherwise continued during the intervention period. In the present study, we report the prespecified secondary outcome of global changes in protein abundance using proteomics on adipose tissue biopsies. For further details regarding biological samples, antibodies, chemicals, peptides, recombinant proteins, deposited data, software, and algorithms, please refer to our key resources table (table S2).
High-intensity interval training
The training protocol consisted of 8 weeks of supervised HIIT combining rowing and cycling, with three sessions per week. Most training sessions (>95%) took place in the afternoon. In brief, each HIIT session started with a 10-min warm-up period followed by training blocks of 5 × 1 min high-intensity intervals (86 to 88% of max heart rate) on either rowing or cycle ergometers interspersed by 1-min active or resting recovery. Between the training blocks, the participants had a 4-min break in which they shifted from cycling to rowing or vice versa. The number of training blocks was gradually increased from two to five during the intervention period as a block was added every second week. Halfway through the HIIT protocol, the workload was adjusted according to a V̇O2 max test to maintain the relative workload and intensity throughout the training period. The attendance rate to the HIIT sessions was above 95% in all three groups (6).
Sample collection
Abdominal scWAT biopsies were obtained on day 2 and day 4 following the 2-hour basal period of the clamp. The biopsies were obtained using a modified Bergström needle with suction under local anaesthesia (lidocaine) and then blotted free of any visible blood and connective tissue and frozen in liquid nitrogen.
Sample preparation for proteomics analysis
Snap-frozen adipose tissue biopsies were powdered and homogenized in SDS buffer (100 mM tris-HCl, pH 8.5, 1% SDS) with an Ultra Turbax blender (IKA). Homogenates were then boiled at 95°C for 10 min in a Thermomixer (Eppendorf) at 1000 RPM and sonicated using a tip sonicator for 30 s, 1-s on/off at amplitude 50%. Then, 25 U of benzonase were added to each sample and homogenate were incubated in a thermomixer at 37°C for 30 min. Homogenates were centrifuged at 16,000g for 10 min and supernatant lysate was transferred to a new tube and sample protein concentration was determined using DC assay (Thermo Fisher Scientific). Forty micrograms of protein was reduced and alkylated by addition of dithiothreitol and chloroacetamide to a final concentration of 10 and 40 mM, respectively, followed by a 45-min incubation at room temperature without agitation. Proteins were digested following a 96-well adapted PAC protocol (38). In brief, a 1:4 protein to bead ratio was added to the sample lysate and protein aggregation was induced by dispensing acetonitrile (ACN) to a final concentration of 70%, followed by a 10-min incubation without agitation. Then, the sample plates were placed in a magnetic stand (Dynamag Thermo) and beads with protein aggregates were first washed twice with 100% ACN and then twice with 70% EtOH. All washes were performed without removing the plates from the magnetic stand. After discarding the last wash, beads were resuspended in 50 μl of digestion buffer (50 mM tris-HCl, pH 8.5) and 1:500 enzyme to protein ratio of LysC (Wako) was added to the protein mixture and incubated in a Thermomixer for 1 hour at 40°C. Afterward, 1:100 enzyme to protein ratio of trypsin was added to the mixture and proteins were digested overnight at 37°C. The next day, digestion was quenched by addition of 1% trifluoroacetic acid (TFA) in isopropanol. Peptides were cleaned for salts and remaining lipids using styrenedivinylbenzene–reverse phase sulfonate and eluted in 1.25% NH4OH and 80% ACN. Peptides were dried completely and resuspended in 0.1% TFA and 5% ACN and loaded on Evotips.
Proteomics analysis
A total of 200 ng of peptides were loaded on a disposable Evotip C18 trap column (Evosep Biosystem) according to the manufacturer’s instruction. Evotips were wetted with 2-propanol, activated with 0.1% formic acid in ACN and equilibrated with 0.1% formic acid. Peptides were added to the tips and centrifuged at 1000g for 2 min. Evotips were then washed with 0.1% formic acid and stored by adding 200 μl 0.1% formic acid until LC-MS measurements. A project-specific library was generated from a peptide pool of all samples. In brief, 20 μg of peptides were fractionated using high-pH reverse-phase chromatography on a Kinetex 2.6 um EVO C18 100 Å, 150 × 0.3 mm column (Phenomenex) using an EASY-nLC 1200 System (Thermo Fisher Scientific) operating at 1.5 μl/min. A 62-min step gradient from 3 to 60% solvent B [10 mM triethylammonium bicarbonate (TEAB) in 80% ACN] and solvent A (10 mM TEAB in water) was used for separation. A total of 98 fractions were collected and concatenated into 24 fractions. Two hundred nanograms of peptide from each fraction was loaded onto Evotips as described earlier.
Peptides were analyzed by partially eluting from Evotips with <35% ACN and analyzed with Evosep One LC system (Evosep Biosystems) coupled online to an Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific). Eluted peptides were separated on a 15-cm-long PepSep column [150 μm inner diameter packed with 1.5 μm of Reprosil-Pur C18 beads (Dr Maisch)] using standard preset gradient method (44 min, 30 samples per day) and electrosprayed with stainless emitter (30 μm inner diameter) at 2.3 kV. For the single-shot adipose tissue proteome, the data were acquired in DIA mode. Each acquisition cycle consisted of a survey scan at a resolution of 120,000 [normalized automatic gain control (AGC) of 300%; scan range, 350 to 1400 m/z (mass/charge ratio)] followed by 49 DIA cycles with dynamic isolation windows at a resolution of 15,000 where precursor ions were fragmented with high-energy collisional dissociation (HCD) of 27%. The data were acquired in profile mode using positive polarity.
To generate the library, each peptide fractions were measured in data-dependent acquisition (DDA) mode with automatic switching between MS and MS/MS using a top 15 method. MS spectra were acquired in the Orbitrap analyzer with a mass range of 350 to 1400 m/z and 60,000 resolution (normalized AGC target 300%). HCD peptide fragments acquired at 30 normalized collision energy were analyzed at 15000 resolution (normalized AGC target 200%). The data were acquired in profile mode using positive polarity.
Data analysis
For library generation, raw spectra from library DDA and experimental DIA files were processed in FragPipe v19.0 with the DIA_SpecLib_Quant workflow. This was performed with default settings, which include a minimum peptide length of seven amino acids and a maximum of two missed cleavages. M-oxidation and NQ-deamination were set as variable modifications. Raw spectra were searched against the Human Uniprot reviewed FASTA (August 2021 release, 20435 entries). Raw files were then searched with DIA-NN v1.8.2 (beta) workflow in the FragPipe software (37, 41) against the project-specific library with default settings, which include peptide length set to 7 to 30 amino acids and a maximum of two missed cleavages. Carbamidomethylation, N-terminal excision, and M-oxidation were set as variable modifications. Quantification was based on robust LC (high precision) and match between runs was enabled. Precursor FDR was set to 1%.
Differential proteome analysis
The protein group file was imported into R v.4.2.1. Protein intensities were log2 transformed and filtered to be quantified in at least 75% of samples across the entire data. Data normality was checked by plotting histograms and Q-Q plot (see fig. S1B). Before data analysis, one sample was excluded as outlier due to low protein identifications (<1000 IDs). To check data clustering and potential batch effects, a scaled PCA was performed on complete observations (1063 proteins) by the prcomp (stats package) function. This revealed one batch effect related to LC-MS instrumentation. For the PCA (Fig. 1E), this batch effect was removed by the removeBatchEffect (limma package). For analysis of differentially expressed proteins, the batch effect was added as a covariate to the linear mixed model. To test for differentially expressed proteins, the lmFit (Limma) function was applied. The average intersubject correlation was estimated by the duplicateCorrelation function (Limma) and added to the linear model. Empirical Bayes smoothing of standard errors and moderation of t and f statistics were computed by the eBayes function. Contrasts were computed and coefficients were extracted. This allowed us to look for main effects of groups at baseline, main effect of HIIT, and interaction of group and HIIT as well as individual comparisons within groups. P values were adjusted with the Benjamini-Hochberg (BH) method and significance level set to 10% FDR.
Gene set enrichment analysis
GSEA was performed with the clusterProfiler package. All logFC intensities were ranked in a descending order and searched for GO terms (org.Hs.eg.db). Our whole adipose tissue proteome was used as background. The number of permutations was set to 10,000 and P values were corrected by the BH method using an FDR of 10%.
Western blotting
The proteins were separated by SDS-PAGE on appropriate Mini-PROTEAN YGX stain-free gels (Bio-Rad Laboratories, Herlev, DK) and transferred to a polyvinylidene difluoride membrane (Immobilon Transfer Membranes; Millipore, Bagsværd, Denmark) by semidry blotting. The same amount of total lysate protein (7 μg) was loaded for each sample. The Western blots were conducted in a balanced design with samples from all experimental conditions present on all gels and an internal control were included on each gel to allow correction for variation between gels. A standard curve is furthermore included to ensure that quantification of each probed protein is within a linear range. In the Western blot analysis, the relative abundance of a protein is normalized to the stain-free signal from the entire lane. The membrane was blocked in a tris-buffered saline with Tween 20 with milk (3%) or bovine serum albumin (3%) solution and afterward probed with primary antibodies. The protein bands were probed with enhanced chemiluminescence (Merck Millipore, Billerica, MA) and subsequently visualized using the ChemiDoc XRS+ system (Bio-Rad Laboratories, Herlev, DK) and quantified using Image Lab version 6.1 (Bio-Rad Laboratories, Herlev, DK). The abundance of any protein of interest are normalized to the total gel stain of the corresponding lane, which is obtained using the BioRad stain free imaging system (Bio-Rad Laboratories, Herlev, DK).
Serum iron measures
Serum ferritin, transferrin, and iron were determined by routine clinical laboratory methods at Odense University Hospital using a Cobas 8000 (Roche, Basal, Switzerland).
Statistical analysis
The Western blot data and serum measures of iron, transferrin, and ferritin were analyzed using a two-way analysis of variance (ANOVA) with repeated measures and a Student-Newman-Keul post hoc test. Significance was accepted at P < 0.05.
For baseline correlation (presamples), clinical parameters were log2 transformed and only complete pairwise observations were correlated with the cor.test function. Log2-transformed clinical parameters were tested for normality by the Shapiro-Wilk test, and either Kendall’s rank or Pearson correlation test was applied. To calculate the repeated-measures correlation (paired pre-post samples), we applied the rmcorr function as described in (87). Clinical parameters were log2 transformed before the correlation analysis. To assess intersample reproducibility, we performed a Pearson correlation analysis by the cor.test function. Sample correlation coefficients were plotted group-wise in a boxplot (fig. S1A).
Acknowledgments
We thank L. Hansen, C. B. Olsen, and A. S. Petersen at the Steno Diabetes Center Odense, Odense University Hospital for the skilled technical assistance. The Odense Patient data Explorative Network (OPEN) are acknowledged for statistical support and access to data storage in REDCap. MS analyses were performed by the Proteomics Research Infrastructure (PRI) at the University of Copenhagen (UCPH), supported by the Novo Nordisk Foundation (NNF) (grant agreement number NNF19SA0059305).
Funding: This work is supported by an unconditional donation from the Novo Nordisk Foundation (NNF) to NNF Center for Basic Metabolic Research (grant number NNF18CC0034900). This work was partially supported by an EFSD/Novo Nordisk Foundation Future Leaders Awards Program to A.S.D. (NNF19SA058976). The clinical and metabolic parts of this work was supported by grants from the Region of Southern Denmark, Odense University Hospital, the Novo Nordisk Foundation (grant number NNF15OC0015986), University of Southern Denmark, Region of Southern Denmark (19/37137), Christenson-Cesons Family Fund, and from The Sawmill Owner Jeppe Juhl and wife Ovita Juhl Memorial Foundation.
Author contributions: Conceptualization: A.S.D. and K.H.; methodology: J.K.L., R.K., N.S., and R.M.-J.; Investigation: J.K.L., R.K., and G.G.J.; writing–original draft: J.K.L. and R.K.; writing–review and editing: J.K.L., R.K., N.S., R.M.J., G.G.J., M.H.P., M.E.d.A., N.Ø., A.S.D., and K.H.; funding acquisition: A.S.D. and K.H.; resources: ASD and KH; Supervision: ASD and KH.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The R code and data to reproduce all analyses and figures for the published version are uploaded to 10.5281/zenodo.8333816 and up to date code can be found on https://github.com/fpm-cbmr/HIIT_adipose_project.
Supplementary Materials
This PDF file includes:
Figs. S1 to S3
Tables S1 and S2
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Kanaley J. A., Colberg S. R., Corcoran M. H., Malin S. K., Rodriguez N. R., Crespo C. J., Kirwan J. P., Zierath J. R., Exercise/physical activity in individuals with type 2 diabetes: A consensus statement from the American College of Sports Medicine. Med. Sci. Sports Exerc. 54, 353–368 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Hagan C., De Vito G., Boreham C. A. G., Exercise prescription in the treatment of type 2 diabetes mellitus. Sports Med. 43, 39–49 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Vind B. F., Pehmøller C., Treebak J. T., Birk J. B., Hey-Mogensen M., Beck-Nielsen H., Zierath J. R., Wojtaszewski J. F. P., Højlund K., Impaired insulin-induced site-specific phosphorylation of TBC1 domain family, member 4 (TBC1D4) in skeletal muscle of type 2 diabetes patients is restored by endurance exercise-training. Diabetologia 54, 157–167 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Hawley J. A., Hargreaves M., Joyner M. J., Zierath J. R., Integrative biology of exercise. Cell 159, 738–749 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Mogensen M., Vind B. F., Hojlund K., Beck-Nielsen H., Sahlin K., Maximal lipid oxidation in patients with type 2 diabetes is normal and shows an adequate increase in response to aerobic training. Diabetes Obes. Metab. 11, 874–883 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Petersen M. H., de Almeida M. E., Wentorf E. K., Jensen K., Ørtenblad N., Højlund K., High-intensity interval training combining rowing and cycling efficiently improves insulin sensitivity, body composition and VO2max in men with obesity and type 2 diabetes. Front. Endocrinol. 13, 1032235 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phielix E., Meex R., Moonen-Kornips E., Hesselink M. K. C., Schrauwen P., Exercise training increases mitochondrial content and ex vivo mitochondrial function similarly in patients with type 2 diabetes and in control individuals. Diabetologia 53, 1714–1721 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hey-Mogensen M., Hojlund K., Vind B. F., Wang L., Dela F., Beck-Nielsen H., Fernstrom M., Sahlin K., Effect of physical training on mitochondrial respiration and reactive oxygen species release in skeletal muscle in patients with obesity and type 2 diabetes. Diabetologia 53, 1976–1985 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Frøsig C., Rose A. J., Treebak J. T., Kiens B., Richter E. A., Wojtaszewski J. F. P., Effects of endurance exercise training on insulin signaling in human skeletal muscle: Interactions at the level of phosphatidylinositol 3-kinase, Akt, and AS160. Diabetes 56, 2093–2102 (2007). [DOI] [PubMed] [Google Scholar]
- 10.de Almeida M. E., Nielsen J., Petersen M. H., Wentorf E. K., Pedersen N. B., Jensen K., Højlund K., Ørtenblad N., Altered intramuscular network of lipid droplets and mitochondria in type 2 diabetes. Am. J. Physiol. Cell Physiol. 324, C39–C57 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Zatterale F., Longo M., Naderi J., Raciti G. A., Desiderio A., Miele C., Beguinot F., Chronic adipose tissue inflammation linking obesity to insulin resistance and type 2 diabetes. Front. Physiol. 10, 1607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppack S. W., Pro-inflammatory cytokines and adipose tissue. Proc. Nutr. Soc. 60, 349–356 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Sabaratnam R., Skov V., Paulsen S. K., Juhl S., Kruse R., Hansen T., Halkier C., Kristensen J. M., Vind B. F., Richelsen B., Knudsen S., Dahlgaard J., Beck-Nielsen H., Kruse T. A., Højlund K., A signature of exaggerated adipose tissue dysfunction in type 2 diabetes is linked to low plasma adiponectin and increased transcriptional activation of proteasomal degradation in muscle. Cell 11, 2005 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henegar C., Tordjman J., Achard V., Lacasa D., Cremer I., Guerre-Millo M., Poitou C., Basdevant A., Stich V., Viguerie N., Langin D., Bedossa P., Zucker J.-D., Clemant K., Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome Biol. 9, R14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soronen J., Laurila P. P., Naukkarinen J., Surakka I., Ripatti S., Jauhiainen M., Olkkonen V. M., Yki-Järvinen H., Adipose tissue gene expression analysis reveals changes in inflammatory, mitochondrial respiratory and lipid metabolic pathways in obese insulin-resistant subjects. BMC Med. Genomics 5, 9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusminski C. M., Bickel P. E., Scherer P. E., Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat. Rev. Drug Discov. 15, 639–660 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Heinonen S., Buzkova J., Muniandy M., Kaksonen R., Ollikainen M., Ismail K., Hakkarainen A., Lundbom J., Lundbom N., Vuolteenaho K., Moilanen E., Kaprio J., Rissanen A., Suomalainen A., Pietiläinen K. H., Impaired mitochondrial biogenesis in adipose tissue in acquired obesity. Diabetes 64, 3135–3145 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Stern J. H., Rutkowski J. M., Scherer P. E., Adiponectin, leptin, and fatty acids in the maintenance of metabolic homeostasis through adipose tissue crosstalk. Cell Metab. 23, 770–784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Kolk B. W., Saari S., Lovric A., Arif M., Alvarez M., Ko A., Miao Z., Sahebekhtiari N., Muniandy M., Heinonen S., Oghabian A., Jokinen R., Jukarainen S., Hakkarainen A., Lundbom J., Kuula J., Groop P.-H., Tukiainen T., Lundbom N., Rissanen A., Kaprio J., Williams E. G., Zamboni N., Mardinoglu A., Pajukanta P., Pietiläinen K. H., Molecular pathways behind acquired obesity: Adipose tissue and skeletal muscle multiomics in monozygotic twin pairs discordant for BMI. Cell Rep. Med. 2, 100226 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stinkens R., Brouwers B., Jocken J. W., Blaak E. E., Teunissen-Beekman K. F., Hesselink M. K., van Baak M. A., Schrauwen P., Goossens G. H., Exercise training-induced effects on the abdominal subcutaneous adipose tissue phenotype in humans with obesity. J. Appl. Physiol. 125, 1585–1593 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Xie X., Yi Z., Sinha S., Madan M., Bowen B. P., Langlais P., Ma D., Mandarino L., Meyer C., Proteomics analyses of subcutaneous adipocytes reveal novel abnormalities in human insulin resistance. Obesity 24, 1506–1514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fazakerley D. J., Chaudhuri R., Yang P., Maghzal G. J., Thomas K. C., Krycer J. R., Humphrey S. J., Parker B. L., Fisher-Wellman K. H., Meoli C. C., Hoffman N. J., Diskin C., Burchfield J. G., Cowley M. J., Kaplan W., Modrusan Z., Kolumam G., Yang J. Y., Chen D. L., Samocha-Bonet D., Greenfield J. R., Hoehn K. L., Stocker R., James D. E., Mitochondrial CoQ deficiency is a common driver of mitochondrial oxidants and insulin resistance. eLife 7, e32111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stallknecht B., Vinten J., Ploug T., Galbo H., Increased activities of mitochondrial enzymes in white adipose tissue in trained rats. Am. J. Physiol. 261, E410–E414 (1991). [DOI] [PubMed] [Google Scholar]
- 24.Lehnig A. C., Dewal R. S., Baer L. A., Kitching K. M., Munoz V. R., Arts P. J., Sindeldecker D. A., May F. J., Lauritzen H. P. M. M., Goodyear L. J., Stanford K. I., Exercise training induces depot-specific adaptations to white and brown adipose tissue. iScience 11, 425–439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutherland L. N., Bomhof M. R., Capozzi L. C., Basaraba S. A. U., Wright D. C., Exercise and adrenaline increase PGC-1α mRNA expression in rat adipose tissue. J. Physiol. 587 (Pt 7), 1607–1617 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S., Norheim F., Langleite T. M., Gulseth H. L., Birkeland K. I., Drevon C. A., Effects of long-term exercise on plasma adipokine levels and inflammation-related gene expression in subcutaneous adipose tissue in sedentary dysglycaemic, overweight men and sedentary normoglycaemic men of healthy weight. Diabetologia 62, 1048–1064 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Takahashi H., Alves C. R. R., Stanford K. I., Middelbeek R. J. W., Nigro P., Ryan R. E., Xue R., Sakaguchi M., Lynes M. D., So K., Mul J. D., Lee M.-Y., Balan E., Pan H., Dreyfuss J. M., Hirshman M. F., Azhar M., Hannukainen J. C., Nuutila P., Kalliokoski K. K., Nielsen S., Pedersen B. K., Kahn C. R., Tseng Y.-H., Goodyear L. J., TGF-β2 is an exercise-induced adipokine that regulates glucose and fatty acid metabolism. Nat. Metab. 1, 291–303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leggate M., Carter W. G., Evans M. J. C., Vennard R. A., Sribala-Sundaram S., Nimmo M. A., Determination of inflammatory and prominent proteomic changes in plasma and adipose tissue after high-intensity intermittent training in overweight and obese males. J. Appl. Physiol. 112, 1353–1360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandrup C. M., Roland C. B., Egelund J., Nyberg M., Enevoldsen L. H., Kjaer A., Clemmensen A., Christensen A. N., Suetta C., Frikke-Schmidt R., Utoft B. B., Kristensen J. M., Wojtaszewski J. F. P., Hellsten Y., Stallknecht B., Effects of high-intensity exercise training on adipose tissue mass, glucose uptake and protein content in pre- and post-menopausal women. Front. Sports Act. Living 2, 60 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riis S., Christensen B., Nellemann B., Møller A. B., Husted A. S., Pedersen S. B., Schwartz T. W., Jørgensen J. O. L., Jessen N., Molecular adaptations in human subcutaneous adipose tissue after ten weeks of endurance exercise training in healthy males. J. Appl. Physiol. 126, 569–577 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Ahn C., Ryan B. J., Schleh M. W., Varshney P., Ludzki A. C., Gillen J. B., Van Pelt D. W., Pitchford L. M., Howton S. M., Rode T., Hummel S. L., Burant C. F., Little J. P., Horowitz J. F., Exercise training remodels subcutaneous adipose tissue in adults with obesity even without weight loss. J. Physiol. 600, 2127–2146 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aebersold R., Mann M., Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Insenser M., Montes-Nieto R., Vilarrasa N., Lecube A., Simó R., Vendrell J., Escobar-Morreale H. F., A nontargeted proteomic approach to the study of visceral and subcutaneous adipose tissue in human obesity. Mol. Cell. Endocrinol. 363, 10–19 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Boden G., Duan X., Homko C., Molina E. J., Song W., Perez O., Cheung P., Merali S., Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals. Diabetes 57, 2438–2444 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Pérez R., Ortega-Delgado F. J., García-Santos E., López J. A., Camafeita E., Ricart W., Fernández-Real J. M., Peral B., Differential proteomics of omental and subcutaneous adipose tissue reflects their unalike biochemical and metabolic properties. J. Proteome Res. 8, 1682–1693 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Xie X., Yi Z., Bowen B., Wolf C., Flynn C. R., Sinha S., Mandarino L. J., Meyer C., Characterization of the human adipocyte proteome and reproducibility of protein abundance by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. J. Proteome Res. 9, 4521–4534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong A. T., Leprevost F. V., Avtonomov D. M., Mellacheruvu D., Nesvizhskii A. I., MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batth T. S., Tollenaere M. X., Rüther P., Gonzalez-Franquesa A., Prabhakar B. S., Bekker-Jensen S., Deshmukh A. S., Olsen J. V., Protein aggregation capture on microparticles enables multipurpose proteomics sample preparation. Mol. Cell. Proteomics 18, 1027–1035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bache N., Geyer P. E., Bekker-Jensen D. B., Hoerning O., Falkenby L., Treit P. V., Doll S., Paron I., Müller J. B., Meier F., Olsen J. V., Vorm O., Mann M., A novel LC system embeds analytes in pre-formed gradients for rapid, ultra-robust proteomics. Mol. Cell. Proteomics 17, 2284–2296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bekker-Jensen D. B., Martínez-Val A., Steigerwald S., Rüther P., Fort K. L., Arrey T. N., Harder A., Makarov A., Olsen J. V., A compact quadrupole-orbitrap mass spectrometer with FAIMS Interface improves proteome coverage in short LC gradients. Mol. Cell. Proteomics 19, 716–729 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demichev V., Messner C. B., Vernardis S. I., Lilley K. S., Ralser M., DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corvera S., Cellular heterogeneity in adipose tissues. Annu. Rev. Physiol. 83, 257–278 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raajendiran A., Krisp C., De Souza D. P., Ooi G., Burton P. R., Taylor R. A., Molloy M. P., Watt M. J., Proteome analysis of human adipocytes identifies depot-specific heterogeneity at metabolic control points. Am. J. Physiol. Endocrinol. Metab. 320, E1068–E1084 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Hruska P., Kucera J., Pekar M., Holéczy P., Mazur M., Buzga M., Kuruczova D., Lenart P., Kucerova J. F., Potesil D., Zdrahal Z., Bienertova-Vasku J., Proteomic signatures of human visceral and subcutaneous adipocytes. J. Clin. Endocrinol. Metab. 107, 755–775 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura T., Nada S., Takegahara N., Okuno T., Nojima S., Kang S., Ito D., Morimoto K., Hosokawa T., Hayama Y., Mitsui Y., Sakurai N., Sarashina-Kida H., Nishide M., Maeda Y., Takamatsu H., Okuzaki D., Yamada M., Okada M., Kumanogoh A., Polarization of M2 macrophages requires Lamtor1 that integrates cytokine and amino-acid signals. Nat. Commun. 7, 13130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mootha V. K., Lindgren C. M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C., PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Patti M. E., Butte A. J., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R., Landaker E. J., Goldfine A. B., Mun E., DeFronzo R., Finlayson J., Kahn C. R., Mandarino L. J., Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U.S.A. 100, 8466–8471 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernández-Real J. M., del Pulgar S. P., Luche E., Moreno-Navarrete J. M., Waget A., Serino M., Sorianello E., Sánchez-Pla A., Pontaque F. C., Vendrell J., Chacón M. R., Ricart W., Burcelin R., Zorzano A., CD14 modulates inflammation-driven insulin resistance. Diabetes 60, 2179–2186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verschuren L., Kooistra T., Bernhagen J., Voshol P. J., Ouwens D. M., van Erk M., de Vries-van der Weij J., Leng L., van Bockel J. H., van Dijk K. W., Fingerle-Rowson G., Bucala R., Kleemann R., MIF deficiency reduces chronic inflammation in white adipose tissue and impairs the development of insulin resistance, glucose intolerance, and associated atherosclerotic disease. Circ. Res. 105, 99–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shim Y. S., Kang M. J., Oh Y. J., Baek J. W., Yang S., Hwang I. T., Association of serum ferritin with insulin resistance, abdominal obesity, and metabolic syndrome in Korean adolescent and adults: The Korean National Health and Nutrition Examination Survey, 2008 to 2011. Medicine 96, e6179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wrede C. E., Buettner R., Bollheimer L. C., Schölmerich J., Palitzsch K. D., Hellerbrand C., Association between serum ferritin and the insulin resistance syndrome in a representative population. Eur. J. Endocrinol. 154, 333–340 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Nilsson E., Jansson P. A., Perfilyev A., Volkov P., Pedersen M., Svensson M. K., Poulsen P., Ribel-Madsen R., Pedersen N. L., Almgren P., Fadista J., Rönn T., Pedersen B. K., Scheele C., Vaag A., Ling C., Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 63, 2962–2976 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Dahlman I., Forsgren M., Sjogren A., Nordstrom E. A., Kaaman M., Naslund E., Attersand A., Arner P., Downregulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesity and possibly involving tumor necrosis factor-α. Diabetes 55, 1792–1799 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Tiaden A. N., Bahrenberg G., Mirsaidi A., Glanz S., Blüher M., Richards P. J., Novel function of serine protease HTRA1 in inhibiting adipogenic differentiation of human mesenchymal stem cells via MAP kinase-mediated MMP upregulation. Stem Cells 34, 1601–1614 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Morandi E. M., Verstappen R., Zwierzina M. E., Geley S., Pierer G., Ploner C., ITGAV and ITGA5 diversely regulate proliferation and adipogenic differentiation of human adipose derived stem cells. Sci. Rep. 6, 28889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brocker C., Cantore M., Failli P., Vasiliou V., Aldehyde dehydrogenase 7A1 (ALDH7A1) attenuates reactive aldehyde and oxidative stress induced cytotoxicity. Chem. Biol. Interact. 191, 269–277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niu L., Thiele M., Geyer P. E., Rasmussen D. N., Webel H. E., Santos A., Gupta R., Meier F., Strauss M., Kjaergaard M., Lindvig K., Jacobsen S., Rasmussen S., Hansen T., Krag A., Mann M., Noninvasive proteomic biomarkers for alcohol-related liver disease. Nat. Med. 28, 1277–1287 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pujar M. K., Vastrad B., Vastrad C., Integrative analyses of genes associated with subcutaneous insulin resistance. Biomolecules 9, 37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanchez-Zamora Y., Terrazas L. I., Vilches-Flores A., Leal E., Juárez I., Whitacre C., Kithcart A., Pruitt J., Sielecki T., Satoskar A. R., Rodriguez-Sosa M., Macrophage migration inhibitory factor is a therapeutic target in treatment of non-insulin-dependent diabetes mellitus. FASEB J. 24, 2583–2590 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Watanabe T., Ito Y., Sato A., Hosono T., Niimi S., Ariga T., Seki T., Annexin A3 as a negative regulator of adipocyte differentiation. J. Biochem. 152, 355–363 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Dohlmann T. L., Hindsø M., Dela F., Helge J. W., Larsen S., High-intensity interval training changes mitochondrial respiratory capacity differently in adipose tissue and skeletal muscle. Physiol. Rep. 6, e13857 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoffmann C., Schneeweiss P., Randrianarisoa E., Schnauder G., Kappler L., Machann J., Schick F., Fritsche A., Heni M., Birkenfeld A., Niess A. M., Häring H.-U., Weigert C., Moller A., Response of mitochondrial respiration in adipose tissue and muscle to 8 weeks of endurance exercise in obese subjects. J. Clin. Endocrinol. Metab. 105, dgaa571 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Larsen S., Danielsen J. H., Søndergård S. D., Søgaard D., Vigelsoe A., Dybboe R., Skaaby S., Dela F., Helge J. W., The effect of high-intensity training on mitochondrial fat oxidation in skeletal muscle and subcutaneous adipose tissue. Scand. J. Med. Sci. Sports 25, e59–e69 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Elbein S. C., Kern P. A., Rasouli N., Yao-Borengasser A., Sharma N. K., Das S. K., Global gene expression profiles of subcutaneous adipose and muscle from glucose-tolerant, insulin-sensitive, and insulin-resistant individuals matched for BMI. Diabetes 60, 1019–1029 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simcox J. A., McClain D. A., Iron and diabetes risk. Cell Metab. 17, 329–341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bao W., Rong Y., Rong S., Liu L., Dietary iron intake, body iron stores, and the risk of type 2 diabetes: A systematic review and meta-analysis. BMC Med. 10, 119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ford E. S., Cogswell M. E., Diabetes and serum ferritin concentration among U.S. adults. Diabetes Care 22, 1978–1983 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Canturk Z., Cetinarslan B., Tarkun I., Canturk N. Z., Serum ferritin levels in poorly- and well-controlled diabetes mellitus. Endocr. Res. 29, 299–306 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Dymock I. W., Cassar J., Pyke D. A., Oakley W. G., Williams R., Observations on the pathogenesis, complications and treatment of diabetes in 115 cases of haemochromatosis. Am. J. Med. 52, 203–210 (1972). [DOI] [PubMed] [Google Scholar]
- 70.Hilton C., Sabaratnam R., Drakesmith H., Karpe F., Iron, glucose and fat metabolism and obesity: An intertwined relationship. Int. J. Obes. (Lond) 47, 554–563 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wlazlo N., van Greevenbroek M. M., Ferreira I., Jansen E. H. J. M., Feskens E. J. M., van der Kallen C. J. H., Schalkwijk C. G., Bravenboer B., Stehouwer C. D. A., Iron metabolism is associated with adipocyte insulin resistance and plasma adiponectin: The Cohort on Diabetes and Atherosclerosis Maastricht (CODAM) study. Diabetes Care 36, 309–315 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao Z., Li S., Liu G., Yan F., Ma X., Huang Z., Tian H., Body iron stores and heme-iron intake in relation to risk of type 2 diabetes: A systematic review and meta-analysis. PLOS ONE 7, e41641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreno-Navarrete J. M., Novelle M. G., Catalán V., Ortega F., Moreno M., Gomez-Ambrosi J., Xifra G., Serrano M., Guerra E., Ricart W., Frühbeck G., Diéguez C., Fernández-Real J. M., Insulin resistance modulates iron-related proteins in adipose tissue. Diabetes Care 37, 1092–1100 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Ryan B. J., Foug K. L., Gioscia-Ryan R. A., Varshney P., Ludzki A. C., Ahn C., Schleh M. W., Gillen J. B., Chenevert T. L., Horowitz J. F., Exercise training decreases whole-body and tissue iron storage in adults with obesity. Exp. Physiol. 106, 820–827 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McClain D. A., Sharma N. K., Jain S., Harrison A., Salaye L. N., Comeau M. E., Langefeld C. D., Lorenzo F. R., Das S. K., Adipose tissue transferrin and insulin resistance. J. Clin. Endocrinol. Metab. 103, 4197–4208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Munro H. N., Iron regulation of ferritin gene expression. J. Cell. Biochem. 44, 107–115 (1990). [DOI] [PubMed] [Google Scholar]
- 77.Kuriyama-Matsumura K., Sato H., Yamaguchi M., Bannai S., Regulation of ferritin synthesis and iron regulatory protein 1 by oxygen in mouse peritoneal macrophages. Biochem. Biophys. Res. Commun. 249, 241–246 (1998). [DOI] [PubMed] [Google Scholar]
- 78.Tanner L. I., Lienhard G. E., Insulin elicits a redistribution of transferrin receptors in 3T3-L1 adipocytes through an increase in the rate constant for receptor externalization. J. Biol. Chem. 262, 8975–8980 (1987). [PubMed] [Google Scholar]
- 79.Davis R. J., Corvera S., Czech M. P., Insulin stimulates cellular iron uptake and causes the redistribution of intracellular transferrin receptors to the plasma membrane. J. Biol. Chem. 261, 8708–8711 (1986). [PubMed] [Google Scholar]
- 80.Habtemichael E. N., Brewer P. D., Romenskaia I., Mastick C. C., Kinetic evidence that Glut4 follows different endocytic pathways than the receptors for transferrin and α2-macroglobulin. J. Biol. Chem. 286, 10115–10125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wei M. L., Bonzelius F., Scully R. M., Kelly R. B., Herman G. A., GLUT4 and transferrin receptor are differentially sorted along the endocytic pathway in CHO cells. J. Cell Biol. 140, 565–575 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Theil E. C., Coordinating responses to iron and oxygen stress with DNA and mRNA promoters: The ferritin story. Biometals 20, 513–521 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Harrison P. M., Arosio P., The ferritins: Molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta 1275, 161–203 (1996). [DOI] [PubMed] [Google Scholar]
- 84.Blankenhaus B., Braza F., Martins R., Bastos-Amador P., González-García I., Carlos A. R., Mahu I., Faisca P., Nunes J. M., Ventura P., Hoerr V., Weis S., Guerra J., Cardoso S., Domingos A., López M., Soares M. P., Ferritin regulates organismal energy balance and thermogenesis. Mol. Metab. 24, 64–79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Festa M., Ricciardelli G., Mele G., Pietropaolo C., Ruffo A., Colonna A., Overexpression of H ferritin and up-regulation of iron regulatory protein genes during differentiation of 3T3-L1 pre-adipocytes. J. Biol. Chem. 275, 36708–36712 (2000). [DOI] [PubMed] [Google Scholar]
- 86.Fan Y., Zhang J., Cai L., Wang S., Liu C., Zhang Y., You L., Fu Y., Shi Z., Yin Z., Luo L., Chang Y., Duan X., The effect of anti-inflammatory properties of ferritin light chain on lipopolysaccharide-induced inflammatory response in murine macrophages. Biochim. Biophys. Acta 1843, 2775–2783 (2014). [DOI] [PubMed] [Google Scholar]
- 87.Bakdash J. Z., Marusich L. R., Repeated measures correlation. Front. Psychol. 8, 456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3
Tables S1 and S2
Legend for data S1
Data S1