Abstract
The global spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) coronavirus disease (COVID-19) is ongoing. Therefore, effective prevention of virus infection is required. Pyrroloquinoline quinone (PQQ), a natural compound found in various foods and human breast milk, plays a role in various physiological processes and is associated with health benefits. In this study, we aimed to determine the effects of PQQ on preventing coronavirus infections using a proxy Feline Infectious Peritonitis Virus (FIPV; belongs to the coronavirus family). In plaque reduction assays, we showed that pre- and post-PQQ-treated viruses were less infectious. IC50 was 87.9 and 5.1 μM for pre- and post-PQQ-treated viral infections, respectively. These results suggest that PQQ decreased the virion stability and viral replication. RT-qPCR confirmed these results. TEM findings showed that PQQ damaged viral capsids and aggregated viral particles, leading to inhibited virus attachment and entry into the host cells. PQQ was optimized by the addition of ascorbic acid and glutamic acid, which increased the number of redox cycles of PQQ and increased reactive oxygen species production by 14 times. In vitro, PQQ inhibited 3 CLpro/Mpro enzymes (an enzyme critical for viral replication) activity of SARS-CoV-2. Our results demonstrate the antiviral effect of PQQ on coronavirus, mainly by disrupting virion stability and loss of infectivity (occurring outside the host cell), due to increased redox activity. Furthermore, PQQ may hinder viral replication (inside the host cell) by 3 CLpro/Mpro enzyme inhibition. In summary, this study demonstrates the antiviral effect of PQQ and its potential application in coronavirus diseases.
Introduction
Despite mass vaccination and the use of antiviral drugs, the global outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) disease (COVID-19) persists. Therefore, there is an urgent need for effective prevention of coronavirus infections. Recently, the investigation of food ingredients as potential inhibitors of coronavirus infections has gained significant attention.
In recent years, scientific research has focused on the identification of potential antiviral food ingredients. Natural bioactive compounds, found in various foods, mitigate viral infections by bolstering the immune system1,2 or interfering with the viral life cycle.2,3 The results indicate that bioactive compounds may inhibit coronavirus infections. Pyrroloquinoline quinone (PQQ) is a natural compound found in various plant-based foods such as kiwifruit, parsley, green peppers, and fermented soybeans, also human breastmilk.4 PQQ has several health benefits, is recognized as a novel food ingredient, and is commercialized as a food supplement.5
PQQ, a redox cofactor, exhibits powerful antioxidant and anti-inflammatory properties.6−9 In addition, PQQ plays a role in various physiological processes9−12 and has the potential to be a protective agent against inflammation caused by new COVID-19 vaccinations.13 Several studies have evaluated PQQ’s impact on overall health and cellular function; however, limited research has examined its effect against coronavirus infections remains unknown. Therefore, we investigated the potential role of PQQ in preventing coronavirus infections using the Feline Infectious Peritonitis Virus (FIPV; a member of the coronavirus family). In addition, we examined the molecular mechanism of PQQ in preventing viral entry into the host cell.
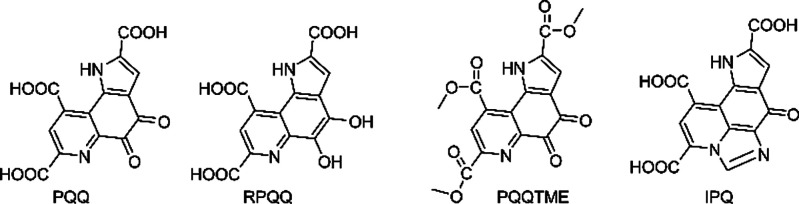
To better understand the association between the PQQ chemical structure and its underlying mechanisms of action, we produced and evaluated several PQQ derivative compounds, including reduced form PQQ (RPQQ), imidazopyrroloquinoline (IPQ), and PQQ trimethylester (PQQ-TME), as shown in Figure 1. PQQ is reduced to RPQQ by the addition of two hydrogen molecules, acquired from ascorbic acid (VC).14 IPQ is a derivative of the reaction between PQQ and amino acids such as glycine and tryptophane.15−17 PQQ-TME is an esterification compound of PQQ where three carbonyl groups are added with methyl ester.18,19 Herein, we utilized PQQ redox modulation to increase the PQQ antiviral activity.
Figure 1.
Chemical structures of PQQ and its derivative compounds. The IUPAC names are as follows: PQQ: 4,5-dioxo-4,5-dihydro-1H-pyrrolo[2,3-f] quinoline-2,7,9-tricarboxylic acid; RPQQ: 4,5-dihydroxy-1H-pyrrolo[2,3-f] quinoline-2,7,9-tricarboxylic acid; PQQ-TME: trimethyl 4,5-dioxo-4,5-dihydro-1H-pyrrolo[2,3-f] quinoline-2,7,9-tricarboxylate; IPQ: 7-oxo-7,10-dihydroimidazo[4,5,1-ij] pyrrolo[2,3-f] quinoline-1,3,9-tricarboxylic acid.
Experimental Section
Cell Culture and Virus Propagation
CRFK cells purchased from JCRB Cell Bank (Osaka, Japan) were maintained in EMEM with l-glutamine and phenol red (Wako; Osaka, Japan) containing 10% fetal bovine serum (Nichirei Bioscience; Tokyo, Japan) and 1% penicillin–streptomycin solution (MP Biomedicals; Ohio, USA) at 37 °C and 5% CO2. FIPV, WSU 79-1146 strain was purchased from ATCC and was propagated in CRFK cells. The experiment was performed in a Biosafety level-2 lab environment. To prepare virus stock, CRFK cells 8 × 106 cell/bottle were cultured with EMEM media in a 175 cm2 cell cultivation flask (vent cap) overnight. After changing the culture media, FIPV were inoculated into the cells by adding 1 mL of stock virus into the cells; the inoculated cells were incubated for 3 days. The medium was collected from the flask. After centrifugation at 3000 rpm for 5 min, the supernatant was filtered using a 0.20 μm disc filter. Collected virus stocks were stored at −80 °C for use in subsequent experiments. The viruses were quantified using the plaque assay method with carboxymethylcellulose (CMC) liquid overlay as described previously20 and in Supporting Information Figure S1. The evaluation of PQQ cytotoxicity in CRKF cell culture is described in the Supporting Information.
Plaque Reduction Assay
Confluent monolayer CRKF cells were precultured in a 24-well plate (2 × 105 cells/well) at 37 °C for 24 h. In the first experiment (virion stability), PQQ (50–400 μM) and FIPV (100 PFU/well) were added into a culture medium and incubated for 1 h at 37 °C. This was considered as the infection solution and was used to inoculate CRFK cells preseeded (2 × 105 cells/well) onto a 24-well plate. The plate was incubated at 37 °C for 1 h. Cells treated with virus only served as the virus control (positive control), and cells not treated with viruses and PQQ served as the cell control (negative control). Subsequently, the infection solution was removed, the wells were washed twice with culture media, and 1 mL of overlay medium (culture medium supplemented with 1% CMC and EMEM) was added into each well. The plate was incubated for 2 days at 37 °C and 5% CO2. In the second experiment (viral replication), FIPV (100 PFU/well) was exposed to PQQ (50–400 μM)-containing culture media and incubated at 37 °C for 1 h. This culture was used as the infection solution. After 1 h, 1 mL of overlay medium was added directly to each well and incubated for 2 days at 37 °C and 5% CO2. PQQ final concentrations were 0.2–25 μM. To quantify the plaque-forming units (PFU), the overlay medium was discarded and the cells were fixed with 4% formaldehyde for 1 h and stained with 0.5% crystal violet for 15 min. Viral plaques were observed under the microscope at 2× magnification, and the plaque numbers were counted to calculate the inhibition rate of virus infection.
Infection Inhibition Assay (RT-qPCR)
FIPV stock culture was diluted with EMEM culture media with the absence (control) or presence of PQQ at varying concentrations. The virus solutions were preincubated at 37 °C for 1 h before each infection step. CRFK cells were seeded (1 × 104 cells/well) onto a 96-well plate and incubated at 37 °C for 24 h. Next, the wells were washed with culture media. For full treatment, PQQ was added to the cell culture 1 h before and after virus infection. For the virion stability test, PQQ was added to the virus solution and preincubated. After infection for 1 h, the cells were washed with culture media. For the post-attachment and post-entry test, the infected cells were washed with culture media and PQQ was added 0 and 4 h post-infection, respectively. The cells were cultured for 24 h, and viral RNAs were quantified using RT-qPCR. First, the cells were lysed using the CellAmp direct RNA prep kit (TakaraBio), according to the manufacturer’s instructions. The cell lysate was stored at −80 °C for up to 1 week until the RT-qPCR experiment. RT-qPCR was performed using the cell lysate as the template with the One Step PrimeScript RT-PCR kit (TakaraBio), according to the manufacturer’s instructions, in a Thermal Cycle Dice Real Time System machine. The thermal cycle conditions were Pattern 1 (Reverse-transcription reaction; hold): 42 °C – 5 min, 95 °C – 10 s; Pattern 2 (PCR; 45 cycles): 95 °C – 5 s, 60 °C – 30 s. The primers used in the reaction targeted the FIPV RNA genome. Primers targeting the GAPDH gene were used for normalization. The sequences of the primers used in this study are listed in Supporting Information Table S1. The Ct values of viral RNA and the reference gene were used to calculate the cell fold change using the 2(−ΔΔCt) method.21
Viral Aggregation Test
The virus stock solution was diluted with the EMEM culture media into an appropriate concentration. The PQQ stock solution was prepared at a 1 mM concentration in PBS buffer. PQQ solution and other tested substances were added into micro test tubes containing 100 μL of virus solution, and PBS buffer was added to obtain a final volume of 1 mL. The control virus was prepared under the same conditions without any tested substance. The mixtures were prepared in triplicates. After the test solution was mixed by gently tapping, the test tubes were incubated at 37 °C for 1 h or the indicated period. Half of each test solution was set aside for the virus capsid integrity test. Virus particles were filtered from the remaining solution using a 1 mL syringe and membrane disc filter (Advantec; Toyo Rashi Kaishi Ltd., Japan) with a pore size of 0.45 or 0.20 μm to remove aggregated viruses. To quantify the viruses in the flow-through solutions, virus RNA genomes were extracted from 10 μL of each sample in duplicates mixed with 10 μL of processing buffer of the CellAmp direct RNA prep kit (TakaraBio) to which RNase inhibitor (TOYOBO) was added. The extraction mixtures were incubated at room temperature (20–25 °C) for 5 min before incubation at 75 °C for 5 min. Extraction mixtures (2 μL) were used as the RNA samples for the One-step RT-PCR experiment. The relative ratio of viral RNA was measured by using the following calculation:
Viral Capsid Integrity Test
To examine whether viruses are still intact after PQQ treatment, virus capsid integrity RT-qPCR was performed as previously described.22 In this test, 500 μM PtCl4 was added to 100 μL of sample solution. The solution was incubated at 4 °C for 30 min in the dark. The virus sample incubated at 95 °C for 15 min was used as the heat-inactivated control to test the reliability of this method. Subsequently, the treated samples underwent RNA extraction using a Viral miniSpin column (Qiagen), according to the manufacturer’s protocol. RT-qPCR was performed in triplicates, and the relative ratio of viral RNA was measured as described in the viral aggregation test section.
Hydrogen Peroxide Assay
The redox reaction between the viral particle and PQQ was examined by evaluating the amount of hydrogen peroxide (H2O2) produced. The concentration of H2O2 in each sample was measured using the Cell Meter Intracellular fluorimetric hydrogen peroxide assay kit (AAT Bioquest) and calculated from a standard curve.
Fluorescence Microscopy Observation and Imaging
The viruses were stained with SYBR Gold (Invitrogen), and viral aggregation was observed with fluorescence microscopy. The virus was treated with PQQ or optimized with PQQ at 37 °C for 1 h. The solutions were then filtered using the Anodisc filter (pore size of 0.02 μm, Whatman, Cytiva). The filter paper was transferred to a glass slide. To prevent drying, samples were covered with glycerol (50% in PBS buffer). The fluorescence images of viral particles were observed and captured using a FLoid Cell Imaging Station under a GFP filter (Thermofisher Scientific).
Transmission Electron Microscopy Observation and Imaging
To observe viral particles with TEM, virus samples were prepared as described in the Supporting Information. To induce hydrophilicity in the carbon surface and enhance the adsorption of virus samples, the 200-mesh copper grid with carbon-coated Formvar films was treated with a glow discharge device (MSP-10, Vacuum Device Inc.) for 60 s. The purified virus sample suspension (5 μL) was deposited onto the grid for 5 min, washed with distilled water, and negatively stained with 2% uranyl acetate for 30 s. After drying, the samples were observed under TEM equipment (H-7650; Hitachi, Ltd., Tokyo, Japan) at an accelerating voltage of 80 kV.
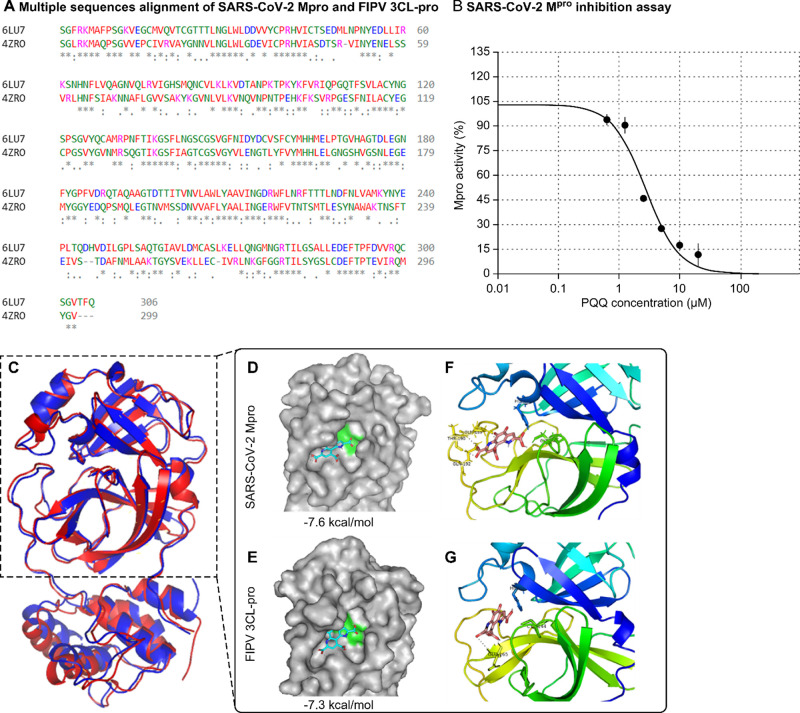
Comparison of Sequence Homology and Protein Structure of SARS-Cov-2 Mpro and FIPV 3CLpro
To compare sequence homology, amino acid sequences of SARS-CoV-2 Main protease (Mpro, also known as 3CL protease) and FIPV 3CLpro were retrieved from the Protein Data Bank [http://www.pdb.org, PDB ID: 6LU7 and 4ZRO respectively]. Sequence alignment was performed using Clustal Omega1 [https://www.ebi.ac.uk/Tools/msa/clustalo/]. The 3D protein structures were aligned and analyzed using PyMOL Molecular Graphic Systems, version 2.4.1 software (Schrodinger).
In Vitro SARS-CoV-2 Mpro Inhibition Assay
The inhibition effect was evaluated by the FRET assay using the SARS-CoV-2 MBP-tagged 3CL Protease Assay Kit (BPS Bioscience; San Diego, USA), in a 96-well black plate, according to the manufacturer’s instructions. Each well of the enzyme reaction mixture contained approximately 100 μg of MBP-tagged protease and 50 μM 3CLpro substrate (DABCYL-KTSAVLQSGFRKME-EDANS) in assay buffer. Each inhibitor compound was added at a fixed concentration and the enzyme mixture was incubated for 30 min at 30 °C before the addition of the substrate. The fluorescence intensity was measured at 30 min and 20 h after substrate addition using a microplate reader (Multimode Plate Reader ARVO X3, PerkinElmer, USA) with a 355 nm/460 nm emission filter. The relative protease activity of each compound was calculated by normalizing to uninterrupted protease activity (without inhibitor). To obtain half maximal inhibitory concentration of the protease activity (IC50), the effect of various concentrations of specific compounds was tested. The data were analyzed by nonlinear regression using the “Quest Graph EC50 Calculator”. AAT Bioquest, Inc., [https://www.aatbio.com/tools/ec50-calculator].
In Silico Molecular Docking Simulation
For visual screening of molecular docking simulation, we operated Autodock Vina (The Scripps Research Institute) employed in the UCSF Chimera 1.14 program (University of California, San Francisco). The three-dimensional structures of SARS-Cov-2 Mpro (PDB ID: 6LU7) and FIPV 3CLpro (PDB ID: 4ZRO) were retrieved from the Protein Data Bank. Co-crystal ligands, peptide-like inhibitors, and water molecules were removed. The protein structure was minimized using the following settings: steepest descent steps = 200; steepest descent step size (Å) = 0.02; conjugate gradient steps = 20; conjugate gradient step size (Å) = 0.02; and update interval = 5 for docking simulation. Hydrogens and AM1-BCC calculation method charges were added during dock prep. For ligands, small molecule compounds used for docking simulation were either retrieved from the PubChem Web site [https://pubchem.ncbi.nlm.nih.gov/] or generated using ChemDraw Professional Chem3D software. Hydrogens were added to the ligands, and the charges were determined. After the docking calculation was completed, the results were visualized and hydrogen bonds between protein and ligands were identified using PyMOL version 2.4.1 software. An in silico study was performed using HP Compaq Elite 8300 PC with Intel Core i7 CPU and Windows 10.
Statistical Analysis
Data analysis was performed using two-tailed Student’s t test or using one-way ANOVA with Dunnett’s post hoc test comparison method. All analyses were performed using commercial SigmaPlot 15.0 software (https://systatsoftware.com/sigmaplot/).
Results
PQQ Reduced Viral Infection of FIPV in CRFK Cells
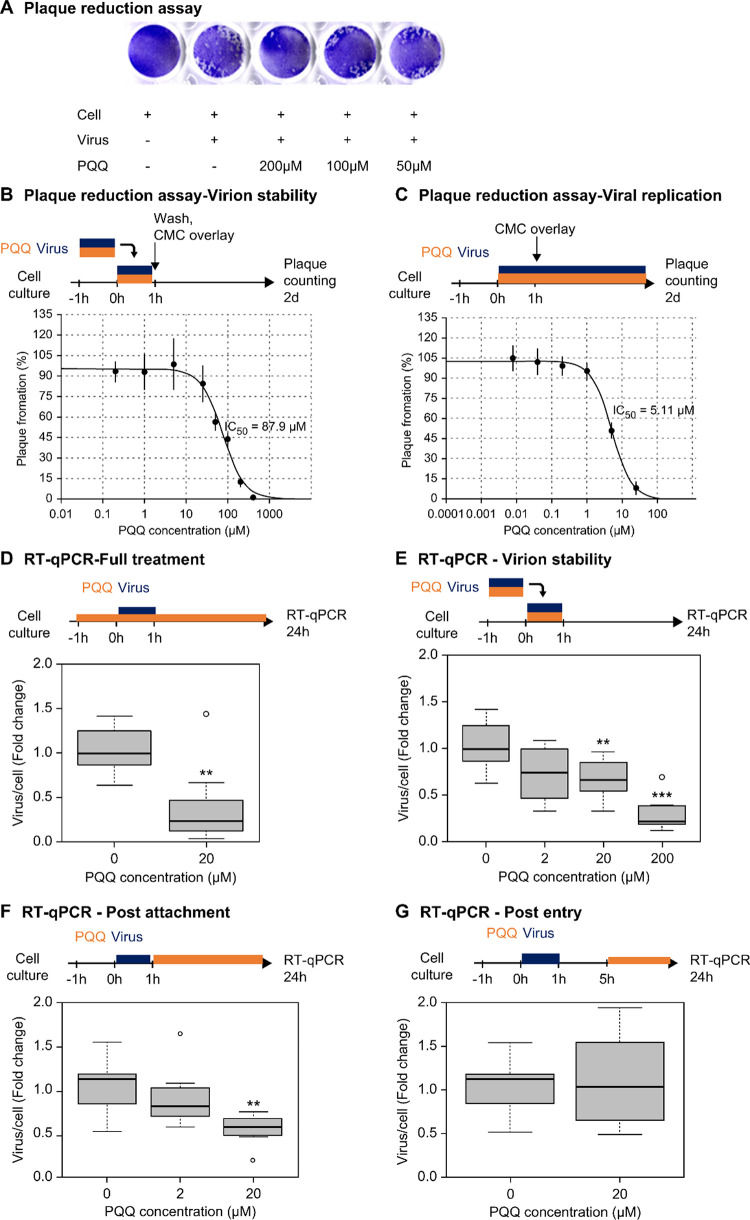
FIPV (WSU 79-1146 strain) causes a severe immune-mediated disease called FIP in domestic and wild cats. We evaluated the inhibitory effect of PQQ on the FIPV infection in CRFK cells. The PQQ 50% cytotoxic concentration (CC50) was 44.8 μM. PQQ concentrations of ≤25 μM did not exhibit significant cytotoxic effects, with less than 10% cell death (Supporting Information Figure S2). Next, inhibition of viral infection was evaluated using the plaque reduction assay, in which the virus is quantified by identifying the number of PFU (Figure 2a). In the first experiment, the viruses were preincubated with PQQ (0.2–400 μM) for 1 h, this infection solution was then inoculated into the cells for 1 h, and subsequently removed from the cell culture. In the second experiment, PQQ (0.04–25 μM) was added during virus infection and was present throughout the incubation. A 50% reduction in plaque formations occurred at PQQ concentrations of 87.9 μM (Figure 2b) and 5.11 μM (Figure 2c) in the first and second experiments, respectively.
Figure 2.
PQQ reduced the viral infection of FIPV in CRFK cells. Representative images of the well plate show the formation of plaque-forming cells 2 days after viral infection (a). Graph depicting plaque formation (%) with different concentrations of PQQ-treated virus infection (b). Graph of plaque formation (%) with different concentrations of PQQ-treated cells (c). PQQ reduced viral RNA copy number in full treatment (d), virion stability (e), virus postattachment (f), and viral postentry (g) treatments of PQQ. The experiments were performed in biological triplicates (n = 3). Asterisks denote a statistically significant difference (**p < 0.01; ***p < 0.001) when compared with the virus control as determined using one-way ANOVA followed by Dunnett’s post hoc test.
We further investigated the effects of PQQ on viral infection by quantifying the viral RNA genome using RT-qPCR. Twenty-four hour PQQ treatment at a concentration of 20 μM significantly reduced the viral copy number to 0.38-fold (Figure 2d). One-hour PQQ treatment at concentrations of 20 and 200 μM significantly reduced the viral copy number to 0.68-fold and 0.30-fold, respectively (Figure 2e). Moreover, 1 h PQQ treatment effectively attenuated 24 h viral replication at a concentration of 20 μM by 0.55-fold (Figure 2f). However, the same concentration of PQQ was found to be ineffective when used to treat infected cells 4 h postinfection (Figure 2g). These results indicate that PQQ is effective as a viral inhibitory treatment during early-stage infection. We hypothesized that these effects were directly related to virion instability, preventing host cell viral entry and replication.
PQQ Aggregated and Disrupted FIPV
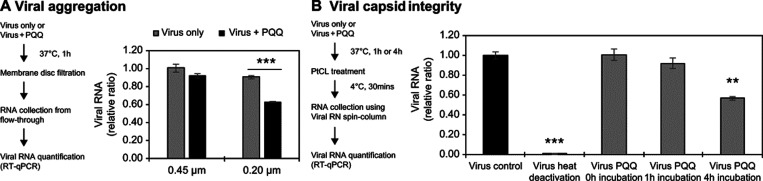
We measured the size of nanoparticle distribution and detected that the mean size of the FIPV viral particles was 86 nm (Supporting Information Table S2 and Figure S3). The PQQ treatment (200 μM) of FIPV (2 × 104 PFU/mL) increased the mean viral particle size to 149 nm, indicating that the viruses were aggregated. To verify the results, we performed membrane filtration using 0.45 and 0.2 μm membrane disks and quantified the virus number in the flow-through solution by RT-qPCR (Figure 3a). In the presence of PQQ, the number of viruses decreased to 0.92-fold and 0.63-fold after membrane filtration at 0.45 and 0.2 μm, respectively. These results show that the aggregated viruses were removed by filtration, verifying that PQQ caused viral aggregation.
Figure 3.
Schematic outline of the experimental procedures and experimental results. The result of viral RNA measurement to examine PQQ effects on viral aggregation (a) and viral capsid integrity (b). Aggregated viral particles larger than 0.45 or 0.2 μm were removed by membrane filtration. The remaining viruses were quantified by RT-qPCR. Viral capsid integrity was evaluated using PtCl4 treatment. Broken capsid allowed PtCl4 binding to viral RNA, which inhibited the amplification of the polymerase reaction during viral RNA measurement. Thus, only RNA from intact viral capsid was measured in RT-qPCR. Incubation with 200 μM PQQ destroyed the viral capsid in a time-dependent manner. The experiments were performed in biological triplicates (n = 3). For the viral aggregation test, asterisks denote a statistically significant difference (***p < 0.001) between virus only and virus + PQQ conditions as determined using two-tailed Student’s t test. For the viral capsid integrity test, asterisks denote a statistically significant difference (**p < 0.01; ***p < 0.001) when compared with the virus control as determined using one-way ANOVA followed by Dunnett’s post hoc test.
To examine the effect of PQQ on virion stability, we performed platinum-based capsid integrity qPCR, a method used to discriminate noninfectious coronavirus on surfaces23 and water samples.22 Following 1 and 4 h of PQQ treatment, the virus viability reduced to 92 and 57%, respectively (Figure 3b). This suggests that PQQ damages the virus particles. Additionally, it shows that viral inactivation occurs in a time-dependent manner.
PQQ Effect was Strategized with VC and Glu Addition
We evaluated the efficacy of PQQ derivative compounds on FIPV treatment (Figure 1). We measured the effects of each compound on viral aggregation using membrane filtration and RT-qPCR methods. PQQ-TME and RPQQ FIPV aggregation effects were greater than that of PQQ (Table 1). In contrast, the aggregation effects of IPQ were lower than that of PQQ. Reducing PQQ with ascorbic acid (VC) increased the aggregation, and the presence of acidic amino acids, such as glutamic acid (Glu), further optimized the viral aggregation effect (Table 1). We confirmed that the optimized PQQ (addition with 5 times weight concentration of VC and Glu) inhibited CRFK virus infection in a plaque assay experiment. One-hour PQQ at 100 μM concentration and optimized PQQ treatment resulted in 57 and 100% reduction in virus infection, respectively. At a concentration of 10 μM, PQQ and optimized PQQ reduced viral infection by 19 and 52%, respectively (Table 2).
Table 1. Percentage of Aggregated Virus and Damaged Capsids Caused by Treatment with PQQ, Its Derivatives, and Its Optimized Conditiona.
| PQQ (200 μM) | RPQQ (200 μM) | IPQ (200 μM) | PQQ-TME(200 μM) | PQQ (200 μM) + VC | Optimized PQQ (200 μM) | |
|---|---|---|---|---|---|---|
| aggregated virus (%) | 10 | 31 | 7 | 65 | 26 | 91 |
| damaged capsid (%) | 14 | 24 | 18 | 25 | 26 | 63 |
PQQ was optimized by the addition of VC and Glu (five times weight amount).
Table 2. Comparison between the Optimized PQQ Mixture and PQQ Alone in Terms of Viral Inhibition (%) and Produced H2O2 Concentration.
| PQQ (100 μM) | Optimized PQQ (100 μM) | PQQ (10 μM) | PQQ (10 μM) + VC | Optimized PQQ (10 μM) | |
|---|---|---|---|---|---|
| viral inhibition (%) (virion stability) | 57 | 100 | 19 | 47 | 52 |
| H2O2 conc. (μM) | 13.8 ± 0.01 | 197.2 ± 6.1 | 13.8 ± 0.02 | 38.0 ± 5.3 | 60.1 ± 5.1 |
| H2O2 conc. fold increased | 1 | 14 | 1 | 3 | 4 |
The changes in the chemical structure of optimized PQQ were examined by using NMR spectroscopy (Supporting Information Table S3). Our data showed that the active species after the addition of VC with or without Glu to PQQ was the reduced form of PQQ (RPQQ). This result suggests that the redox reaction of PQQ, which produces reactive oxygen species (ROS), such as H2O2, can cause viral damage and lead to viral aggregation. We measured the amount of H2O2 produced by PQQ (10 and 100 μM), PQQ with VC, and PQQ with VC and Glu in PBS buffer. The addition of VC and Glu to 10 μM PQQ and 100 μM increased H2O2 concentration by 4-fold and 14-fold, respectively (Table 2).
Virus Observation under Fluorescence and a Transmission Electron Microscope
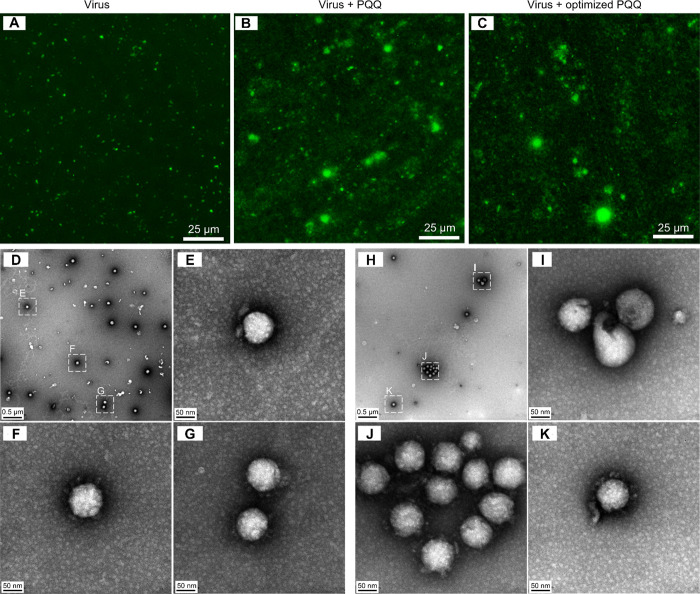
We stained the viruses with SYBR gold to observe viral states using a fluorescence microscope. The viruses were preincubated with PQQ (0–500 μM) for 1 h before 30 min cell infection. Unattached viruses were removed by washing twice. The viruses attached to the host cell were observed (Supporting Information Figure S4), and the fluorescence intensity was measured to quantify the viruses (Supporting Information Figure S5). We found that the virus attachment to the cells was reduced in a PQQ concentration-dependent manner. Furthermore, the stained viruses were observed directly on the Anodisc filter with 0.02 μm pore size (Figure 4a–c). Based on the fluorescence intensity and size, viral aggregation was observed in the presence of PQQ (Figure 4b) and the optimized PQQ (Figure 4c), but not in the control sample (Figure 4a).
Figure 4.
Fluorescence microscopy and TEM images. Fluorescence images of the control virus sample (a), virus treated with PQQ (b) and virus treated with optimized PQQ (c) under a fluorescence microscope with a GFP filter. The viruses were prestained with SYBR Gold before incubated at 37 °C for 1 h with or without PQQ or optimized PQQ. Then, the samples were filtered and observed directly on the Anodisc filter (pore size of 0.02 μm). TEM images of control FIPV viral particles (d–g) and optimized PQQ-treated viral particles (h–k). Viral aggregation was not observed in the control sample (d) but was observed in the optimized PQQ-treated samples (h, j). Broken viral capsids were also observed after PQQ treatment (i).
From TEM observations, viral aggregation was not present in the control virus (Figure 4d–g, Supporting Information, Figure S6). However, in the optimized PQQ-treated sample, viral capsids were destroyed (Figure 4h–k, Supporting Information Figure S7), and more than 10 aggregated viral particles were observed (Figure 4j, Supporting Information Figure S7b). Similar broken viral capsids observation was also obtained using a scanning transmission electron microscope (STEM) (Supporting Information Figure S8). These results verified our hypothesis that PQQ reduced virion stability, preventing host cell infection.
PQQ Inhibited Viral 3CLpro Activity
The 3CLpro, also known as Mpro, is an enzyme that plays a crucial role in the replication of several coronaviruses, including SARS-CoV-2 and FIPV. This enzyme is essential for viral replication because it cleaves the virus-produced polyproteins into functional mature proteins that are required for viral replication and assembly.24,25 The SARS-CoV-2 3CLpro amino acid sequence was highly similar to that of FIPV (44% sequence identity) and had similar properties (97% match) (Figure 5a). FIPV infection of CRFK cells demonstrated that PQQ inhibits viral replication. We used a commercial FRET-based SARS-CoV-2 Mpro inhibitor assay kit to examine the inhibitory effect of PQQ on the enzyme. PQQ decreased Mpro activity in a dose-dependent manner (Supporting Information Table S7), with a half-maximal inhibitory concentration (IC50) of 2.8 μM (Figure 5b).
Figure 5.
PQQ inhibits viral 3 CLpro/Mpro activity. Amino acid sequence alignment of SARS-CoV-2 Mpro (PDB 6LU7) and FIPV 3CLpro (PDB 4ZRO) generated by Cluster Omega Tool (a). An asterisk indicates the position of a single, fully conserved residue. A colon indicates conservation and strongly similar properties between the two groups. A period indicates the conservation of weakly similar properties between groups. PQQ decreased Mpro activity in a dose-dependent manner with a half-maximal inhibitory concentration (IC50) of 2.8 μM in vitro assay (b). Protein structure alignment between SARS-CoV-2 Mpro (red) and FIPV 3CLpro (blue) was generated by PyMOL software (c). Molecular docking of PQQ with SARS-CoV-2 Mpro and PQQ with FIPV 3CLpro predicted by the Autodock Vina program. The interaction of PQQ (blue stick) at the Mpro substrate-binding pocket comprised His41-Cys145 dyad (green region) (d) and the 3CLpro substrate-binding pocket comprised His41-Cys144 dyad (green region) (e). Stereoview of the docked conformation of the PQQ-Mpro complex (f) and PQQ-3CLpro complex (g), showing the possibility of hydrogen bonding with amino acid residues. Images were generated by PyMOL software.
Next, we performed a ligand-protein molecular docking simulation using AutoDock Vina software. PQQ was the ligand and SARS-CoV-2 Mpro and FIPV 3CLpro proteins. Aligned 3D structures of both proteins were generated; these results showed that the protein structures were well conserved (Figure 5c). Docking score results indicate that PQQ has good binding affinity with both proteins at −7.6 kcal/mol (Figure 5d) and −7.3 kcal/mol (Figure 5e). Furthermore, the simulation predicted that PQQ binds to the active site pocket of SARS-CoV-2 Mpro, composed of the His41-Cys145 dyad, and forms polar bonds with Gln189, Thr190, and Gln192 (Figure 5f). PQQ was also predicted to bind in the active site pocket of FIPV 3CL-pro, composed of His41-Cys144 dyad, and form polar bonds with Glu165 (Figure 5g).
Discussion
The safety of PQQ was evaluated by the European Food Safety Authority. A no observed adverse effect level of 100 mg/kg bw per day was determined for PQQ from a 13-week test in rats26 and a 90-day study in humans.5 PQQ is recommended to be consumed by healthy adults as a function food at a basal intake of 20 mg per day, corresponding to 0.29 mg/kg per day for a 70 kg adult.5 Recently, the use of PQQ has been suggested as a new treatment option to manage inflammation caused by COVID-19 vaccination13 and COVID-19 disease27 due to PQQ's anti-inflammatory properties. In nanocurcumin-based formulation, PQQ was found to effectively alleviate the syndrome of COVID-19 inflammation by protecting cardiopulmonary function and mitochondrial homeostasis in response to hypobaric hypoxia.27
However, to the best of our knowledge, no existing studies have examined the antiviral effects of PQQ on coronaviruses, specifically SARS-CoV-2, to prevent the virus infection. We examined the effect of PQQ using FIPV as a proxy to study the effect of PQQ on the SARS-CoV-2 infection. We found that PQQ protects the host cells against viral infection. The protective effect was observed during early PQQ treatment, that is, PQQ was the most effective when used as a treatment immediately to postinfection. Cells treated with PQQ 5 h postinfection were not protected, that is, viral infection persisted. These results suggest that PQQ had a strong antiviral effect during the viral pre-entry stage.
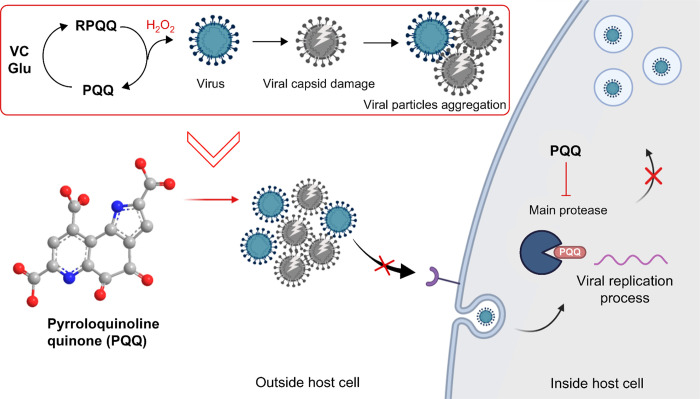
The proposed mechanism of PQQ-induced viral inhibition is summarized in Scheme 1. In this study, we focused on the viral pre-entry protection effect of PQQ. Our microscopy observations reveal that PQQ inhibits viral attachment to host cells. This could be caused by, as observed in this study, PQQ damage and aggregation of viral particles. In addition, in the presence of PQQ, the viral particle size increases. These results are in agreement with the observed reduction in the viral RNA copy number upon PQQ-treated virus membrane filtration. Coronaviruses enter mammalian cells through several endocytosis pathways, including clathrin-mediated endocytosis (CME).28 The particle size affects the entry mechanism as CME can uptake up to approximately 200–300 nm.29,30 Therefore, larger aggregated virus particles would be excluded from this entry process. Furthermore, viral aggregation is reported to limit the diffusion of viral particles that hinder the early stages of effective viral infection.31
Scheme 1. Schematic Illustration of the Proposed Mechanism of PQQ-Induced Inhibition of Coronavirus Infection.
Outside the host cell, PQQ’s redox cycle produces ROS, such as hydrogen peroxide which causes viral capsid damage and viral particle aggregation. Large and aggregated viral particles cannot enter the host cell by endocytosis. Inside the host cell, PQQ inhibits the activity of Mpro, also known as the 3CLpro, an essential enzyme found in coronaviruses. It functions to cleave the large polyprotein, produced by the virus, into smaller functional proteins necessary for viral replication. By inhibiting Mpro activity, PQQ hinders the viral replication process in the host cells. The image was created using Biorender.com.
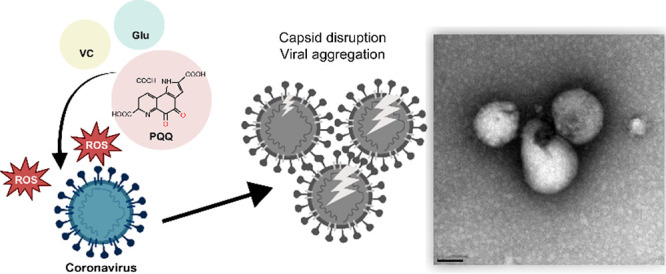
PQQ is a naturally occurring redox cofactor.10 PQQ is relatively stable; thus, the number of potential redox cycles is high. Compared to other antioxidant food compounds, PQQ allows 20,000 catalytic cycles, which is more than 250 times compared to the quercetin, catechin, and epicatechin catalytic cycles.32 We found that the byproduct of the PQQ redox reaction, ROS, damages viral components such as envelope proteins, lipids, and RNA. The efficiency of PQQ derivatives indicates that the quinone structure is important for the PQQ redox reaction. Furthermore, the three carboxylic acid structures did not affect the inhibitory effect. When PQQ is added with the reducing agent VC, the quinone undergoes reduction, thereby adding two electrons (2e−) and two protons (H+) to the quinone molecule. This process results in the formation of hydroxyl groups (−OH), attached to the cyclic structure, and the reduced form of PQQ (RPQQ). RPQQ is easily oxidized and converted back to its quinone form, producing H2O2. Owing to this process, PQQ conditions for redox activation were optimized with the addition of reducing agents (VC and Glu). Protein denaturation is known to promote aggregation, and aggregation is thought to proceed as the viruses are destroyed.
The crystal structure of PQQ and RPQQ has been reported.33 Focusing on hydrogen bonds as an intermolecular interaction, RPQQ has 9 hydrogen bonds, leading to more interactions compared to PQQ with 5 hydrogen bonds. We inferred that the RPQQ forms interaction with amino acids and viral proteins through hydrogen bonds to aggregate them. Based on our result, the addition of amino acid into the PQQ and VC mixture would further optimize viral aggregation condition. However, the mechanism underlying amino acids in viral aggregation remains unknown and requires further study.
PQQ treatment after viral attachment to cells reduced the viral RNA copy number, suggesting that PQQ may interfere with viral replication. We investigated the PQQ viral inhibitory effect on the viral 3CLpro enzyme activity since this enzyme is crucial for viral replication and is used as a target for developing drugs.3 Myriad plant compounds including polyphenols inhibit coronavirus 3CLpro3,.34 We showed that PQQ inhibited 3CLpro activity in vitro and predicted that PQQ could bind into the pocket of the enzyme active site by ligand-protein docking simulation in silico. We hypothesized that PQQ potentially binds to both SARS-CoV-2 Mpro and FIPV 3CLpro, and consequently inhibits viral replication. This result suggests that PQQ could prevent coronavirus replication by inhibiting 3CLpro/Mpro activity in addition to preventing host entry. Further study is required to confirm the inhibitory mechanism.
This study highlights the potential of PQQ as a promising antiviral agent. Although other studies have demonstrated the role of PQQ in modulating the host immune response,6,35−37 we offer promising insights into the potential role of PQQ in combating viral infections, including coronavirus infections. Our data showed that PQQ treatment increased the efficacy of viral infection suppression by polyclonal antibodies targeting the FIPV spike protein (Supporting Information Table S6). The ability of PQQ to enhance antibody action requires further investigation.
PQQ concentrations were detected at 16.4–53.8 nM in human blood serum after a 100 mg PQQ supplement was administered.38 Conversely, in this study, the effective antiviral PQQ concentrations (5–500 μM) were an order of magnitude higher than PQQ concentrations in blood serum, making the amount of PQQ oral intake insufficient. We believe that external usage of the PQQ solution is more suitable for this purpose. The oral cavity is a major reservoir for virus transmission, and mouth rinse can help to reduce viral load.39 Furthermore, anti-SAR-CoV-2 nasal spray has been reported to prevent infection.40 PQQ can be applied for oral and nasal applications. However, this study focused on antiviral effects using in vitro and in silico experiments. Further research including animal studies on viral disease prevention is required to examine the potential of PQQ applications.
Conclusions
Overall, this study demonstrated that the PQQ redox reaction could destroy the coronavirus viral capsid, leading to an increased virus particle aggregation. This would increase the particle size and prevent endocytosis-associated viral entry into the host cell. The PQQ abilities were optimized by the addition of VC and Glu, which increased the number of redox cycles. This study may pave the way for the development of innovative approaches to combat COVID-19 and other coronavirus-related diseases. Indeed, more research is needed to fully understand PQQ mechanisms of action and their potential therapeutic applications.
Acknowledgments
We thank Dr. Manabu Hayatsu from the Division of Microscopic Anatomy, Graduate School of Medical and Dental Sciences, Niigata University, Japan, for providing us with technical assistance for TEM imaging.
Glossary
Abbreviations
- CRFK
Crandell-Rees Feline Kidney
- FIPV
Feline infectious peritonitis virus
- PQQ
pyrroloquinoline quinone
- TME
trimethylester
- RT-qPCR
real-time quantitative polymerase chain reaction
- VC
vitamin C or Ascorbic acid
- Glu
glutamic acid
- ROS
reactive oxygen species
- PFU
plaque-forming unit
- TEM
transmission electron microscope
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c06040.
Materials, cytotoxicity assay, sequences of primer pairs used in RT-qPCR, viral particle size distribution measurement, NMR experiment, fluorescence microscopy experiment, sample preparation for TEM, STEM and SDS-PAGE experiments, additional TEM images, STEM observation and imaging, SDS-PAGE experiment, SARS-CoV-2 Mpro inhibition assay, and FIPV spike1 polyclonal antibody immune test (PDF)
Author Contributions
N.S.M.I. and K.I. participated in planning and performed most of the experiments. T.N. contributed to plaque assay and fluorescence microscopy work. The original manuscript was written by N.S.M.I. through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors are employees of Mitsubishi Gas Chemistry Co. Inc. and declare that the company fully funded this research.
The authors declare the following competing financial interest(s): The authors are employees of Niigata Research Laboratory, Mitsubishi Gas Chemical Co., Inc and declare that the company fully funded this research.
Supplementary Material
References
- Basak S.; Gokhale J. Immunity Boosting Nutraceuticals: Current Trends and Challenges. J. Food Biochem 2022, 46 (3), e13902 10.1111/jfbc.13902. [DOI] [PubMed] [Google Scholar]
- Galanakis C. M.; Aldawoud T. M. S.; Rizou M.; Rowan N. J.; Ibrahim S. A. Food Ingredients and Active Compounds against the Coronavirus Disease (COVID-19) Pandemic: A Comprehensive Review. Foods 2020, 9 (11), 1701. 10.3390/foods9111701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal A.; Jha A. K.; Hazra B. Plant Products as Inhibitors of Coronavirus 3CL Protease. Front Pharmacol 2021, 12, 583387 10.3389/fphar.2021.583387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa T.; Sato K.; Seno H.; Ishii A.; Suzuki O. Levels of Pyrroloquinoline Quinone in Various Foods. Biochem. J. 1995, 307 (2), 331–333. 10.1042/bj3070331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck D.; Bresson J.; Burlingame B.; Dean T.; Fairweather-Tait S.; Heinonen M.; Hirsch-Ernst K. I.; Mangelsdorf I.; McArdle H. J.; Naska A.; Neuhäuser-Berthold M.; Nowicka G.; Pentieva K.; Sanz Y.; Siani A.; Sjödin A.; Stern M.; Tomé D.; Vinceti M.; Willatts P.; Engel K.; Marchelli R.; Pöting A.; Poulsen M.; Schlatter J. R.; de Sesmaisons A.; Van Loveren H. Safety of Pyrroloquinoline Quinone Disodium Salt as a Novel Food Pursuant to Regulation (EC) No 258/97. EFSA J. 2017, 15 (11), e05058 10.2903/j.efsa.2017.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Sun C.; Tao R.; Xu X.; Xu L.; Cheng H.; Wang Y.; Zhang D. Pyrroloquinoline Quinone Decelerates Rheumatoid Arthritis Progression by Inhibiting Inflammatory Responses and Joint Destruction via Modulating NF-ΚB and MAPK Pathways. Inflammation 2016, 39 (1), 248–256. 10.1007/s10753-015-0245-7. [DOI] [PubMed] [Google Scholar]
- Miyauchi K.; Urakami T.; Abeta H.; Shi H.; Noguchi N.; Niki E. Action of Pyrroloquinolinequinol As an Antioxidant Against Lipid Peroxidation in Solution. Antioxid Redox Signal 1999, 1 (4), 547–554. 10.1089/ars.1999.1.4-547. [DOI] [PubMed] [Google Scholar]
- Itoh Y.; Hine K.; Miura H.; Uetake T.; Nakano M.; Takemura N.; Sakatani K. Effect of the Antioxidant Supplement Pyrroloquinoline Quinone Disodium Salt (BioPQQTM) on Cognitive Functions. Adv. Exp. Med. Biol. 2016, 876, 319–325. 10.1007/978-1-4939-3023-4_40. [DOI] [PubMed] [Google Scholar]
- Jonscher K. R.; Chowanadisai W.; Rucker R. B. Pyrroloquinoline-Quinone Is More Than an Antioxidant: A Vitamin-like Accessory Factor Important in Health and Disease Prevention. Biomolecules 2021, 11 (10), 1441. 10.3390/biom11101441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagawa M.; Nakano M.; Ikemoto K. Recent Progress in Studies on the Health Benefits of Pyrroloquinoline Quinone. Biosci Biotechnol Biochem 2016, 80 (1), 13–22. 10.1080/09168451.2015.1062715. [DOI] [PubMed] [Google Scholar]
- Mohamad Ishak N. S.; Ikemoto K.; Kikuchi M.; Ogawa M.; Akutagawa K.; Akagawa M. Pyrroloquinoline Quinone Attenuates Fat Accumulation in Obese Mice Fed with a High-Fat Diet, Daphnia Magna Supplied with a High Amount of Food, and 3T3-L1 Adipocytes. ACS Food Science and Technology 2021, 1 (10), 1979–1989. 10.1021/acsfoodscitech.1c00301. [DOI] [Google Scholar]
- Mohamad Ishak N. S.; Ikemoto K. Pyrroloquinoline-Quinone to Reduce Fat Accumulation and Ameliorate Obesity Progression. Front Mol. Biosci 2023, 10, 1200025 10.3389/fmolb.2023.1200025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boretti A. PQQ Supplementation and SARS-CoV-2 Spike Protein-Induced Heart Inflammation. Nat. Prod. Commun. 2022, 17 (3), 1934578–2210809. 10.1177/1934578X221080929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai K.; Ouchi A.; Nagaoka S.; Nakano M.; Ikemoto K. Pyrroloquinoline Quinone (PQQ) Is Reduced to Pyrroloquinoline Quinol (PQQH2) by Vitamin C, and PQQH2 Produced Is Recycled to PQQ by Air Oxidation in Buffer Solution at PH 7.4. Biosci Biotechnol Biochem 2016, 80 (1), 178–187. 10.1080/09168451.2015.1072462. [DOI] [PubMed] [Google Scholar]
- Urakami T.; Sugamura K.; Niki E. Characterization of Imidazopyrroloquinoline Compounds Synthesized from Coenzyme PQQ and Various Amino Acids. Biofactors 1995, 5 (2), 75–81. [PubMed] [Google Scholar]
- Itoh S.; Mure M.; Suzuki A.; Murao H.; Ohshiro Y. Reaction of Coenzyme PQQ with Amino Acids. Oxidative Decarboxylation, Oxidative Dealdolation (C α – C β Fission) and Oxazolopyrroloquinoline (OPQ) Formation. J. Chem. Soc., Perkin Trans. 2 1992, 8, 1245–1251. 10.1039/P29920001245. [DOI] [Google Scholar]
- Ishida T.; Kawamoto E.; In Y.; Amano T.; Kanayama J.; Doi M.; Iwashita T.; Nomoto K. Formation of Imidazolopyrroloquinoline as Main PQQ Adduct with Amino Acid in Vitro: X-Ray Structural Evidence. J. Am. Chem. Soc. 1995, 117 (11), 3278–3279. 10.1021/ja00116a035. [DOI] [Google Scholar]
- Tsukakoshi K.; Yoshida W.; Kobayashi M.; Kobayashi N.; Kim J.; Kaku T.; Iguchi T.; Nagasawa K.; Asano R.; Ikebukuro K.; Sode K. Esterification of PQQ Enhances Blood-Brain Barrier Permeability and Inhibitory Activity against Amyloidogenic Protein Fibril Formation. ACS Chem. Neurosci. 2018, 9 (12), 2898–2903. 10.1021/acschemneuro.8b00355. [DOI] [PubMed] [Google Scholar]
- Duine J. A.; Frank Jzn J.; Verwiel P. E. J. Structure and Activity of the Prosthetic Group of Methanol Dehydrogenase. Eur. J. Biochem. 1980, 108 (1), 187–192. 10.1111/j.1432-1033.1980.tb04711.x. [DOI] [PubMed] [Google Scholar]
- Mendoza E. J.; Manguiat K.; Wood H.; Drebot M. Two Detailed Plaque Assay Protocols for the Quantification of Infectious SARS-CoV-2. Curr. Protoc. Microbiol. 2020, 57 (1), ecpmc105 10.1002/cpmc.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J.; Schmittgen T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25 (4), 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Cuevas-Ferrando E.; Randazzo W.; Pérez-Cataluña A.; Falcó I.; Navarro D.; Martin-Latil S.; Díaz-Reolid A.; Girón-Guzmán I.; Allende A.; Sánchez G. Platinum Chloride-Based Viability RT-QPCR for SARS-CoV-2 Detection in Complex Samples. Sci. Rep 2021, 11 (1), 18120. 10.1038/s41598-021-97700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ferrando E.; Girón-Guzmán I.; Falcó I.; Pérez-Cataluña A.; Díaz-Reolid A.; Aznar R.; Randazzo W.; Sánchez G. Discrimination of Non-Infectious SARS-CoV-2 Particles from Fomites by Viability RT-QPCR. Environ. Res. 2022, 203, 111831 10.1016/j.envres.2021.111831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q.; Xiong Y.; Zhu G.; Zhang Y.; Zhang Y.; Huang P.; Ge G. The SARS-CoV-2 Main Protease (M pro): Structure, Function, and Emerging Therapies for COVID-19. MedComm (Beijing) 2022, 3 (3), e151 10.1002/mco2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.; Gao Y.; Liu H.; Li H.; Chen W.; Ma J. Advances in Research on 3C-like Protease (3CL) Inhibitors against SARS-CoV-2 since 2020. RSC Med. Chem. 2023, 14 (1), 9–21. 10.1039/D2MD00344A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M.; Takahashi H.; Koura S.; Chung C.; Tafazoli S.; Roberts A. Acute and Subchronic Toxicity Studies of Pyrroloquinoline Quinone (PQQ) Disodium Salt (BioPQQTM) in Rats. Regul. Toxicol. Pharmacol. 2014, 70 (1), 107–121. 10.1016/j.yrtph.2014.06.024. [DOI] [PubMed] [Google Scholar]
- Kushwaha A. D.; Mishra K. P.; Singh M.; Ganju L.; Saraswat D. Nanocurcumin Formulation: A Possible Therapeutic Agent for Post COVID Inflammatory Syndrome. Immunopharmacol Immunotoxicol 2022, 44 (2), 141–146. 10.1080/08923973.2022.2037631. [DOI] [PubMed] [Google Scholar]
- Glebov O. O. Understanding SARS-CoV-2 Endocytosis for COVID-19 Drug Repurposing. FEBS J. 2020, 287 (17), 3664–3671. 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhorn I. S.; Kalicharan R.; Hoekstra D. Lipoplex-Mediated Transfection of Mammalian Cells Occurs through the Cholesterol-Dependent Clathrin-Mediated Pathway of Endocytosis. J. Biol. Chem. 2002, 277 (20), 18021–18028. 10.1074/jbc.M111257200. [DOI] [PubMed] [Google Scholar]
- Canton I.; Battaglia G. Endocytosis at the Nanoscale. Chem. Soc. Rev. 2012, 41 (7), 2718. 10.1039/c2cs15309b. [DOI] [PubMed] [Google Scholar]
- Pradhan S.; Varsani A.; Leff C.; Swanson C. J.; Hariadi R. F. Viral Aggregation: The Knowns and Unknowns. Viruses 2022, 14 (2), 438. 10.3390/v14020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker R.; Chowanadisai W.; Nakano M. Potential Physiological Importance of Pyrroloquinoline Quinone. Altern. Med. Rev. 2009, 14 (3), 268–277. [PubMed] [Google Scholar]
- Ikemoto K.; Mori S.; Mukai K. Synthesis and Crystal Structure of Pyrroloquinoline Quinol (PQQH 2) and Pyrroloquinoline Quinone (PQQ). Acta Crystallogr. B Struct Sci. Cryst. Eng. Mater. 2017, 73 (3), 489–497. 10.1107/S2052520617002281. [DOI] [PubMed] [Google Scholar]
- Bahun M.; Jukić M.; Oblak D.; Kranjc L.; Bajc G.; Butala M.; Bozovičar K.; Bratkovič T.; Podlipnik Č.; Poklar Ulrih N. Inhibition of the SARS-CoV-2 3CLpro Main Protease by Plant Polyphenols. Food Chem. 2022, 373, 131594 10.1016/j.foodchem.2021.131594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X. Feng; Zhai B. Zhong; Hu W. Li; Lou M. Han; Chen Y. Hao; Liu Y. Feng; Chen J. Guo; Mei S.; You Z. Qiang; Liu Z.; Zhang L. Jing; Zhang Y. Hui; Wang Y. Pyrroloquinoline Quinone Ameliorates Diabetic Cardiomyopathy by Inhibiting the Pyroptosis Signaling Pathway in C57BL/6 Mice and AC16 Cells. Eur. J. Nutr. 2022, 61 (4), 1823–1836. 10.1007/s00394-021-02768-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devasani K.; Kaul R.; Majumdar A. Supplementation of Pyrroloquinoline Quinone with Atorvastatin Augments Mitochondrial Biogenesis and Attenuates Low Grade Inflammation in Obese Rats. Eur. J. Pharmacol. 2020, 881, 173273 10.1016/j.ejphar.2020.173273. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Han N.; Zhao K.; Li Y.; Chi Y.; Wang B. Protective Effects of Pyrroloquinoline Quinone against Oxidative Stress-Induced Cellular Senescence and Inflammation in Human Renal Tubular Epithelial Cells via Keap1/Nrf2 Signaling Pathway. Int. Immunopharmacol 2019, 72, 445–453. 10.1016/j.intimp.2019.04.040. [DOI] [PubMed] [Google Scholar]
- Fukuda M.; El-Maghrabey M. H.; Kishikawa N.; Ikemoto K.; Kuroda N. Ultrasensitive Determination of Pyrroloquinoline Quinone in Human Plasma by HPLC with Chemiluminescence Detection Using the Redox Cycle of Quinone. J. Pharm. Biomed Anal 2017, 145, 814–820. 10.1016/j.jpba.2017.08.008. [DOI] [PubMed] [Google Scholar]
- Carrouel F.; Gonçalves L. S.; Conte M. P.; Campus G.; Fisher J.; Fraticelli L.; Gadea-Deschamps E.; Ottolenghi L.; Bourgeois D. Antiviral Activity of Reagents in Mouth Rinses against SARS-CoV-2. J. Dent Res. 2021, 100 (2), 124–132. 10.1177/0022034520967933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrć K.; Milewska A.; Duran E. B.; Botwina P.; Dabrowska A.; Jedrysik M.; Benedyk M.; Lopes R.; Arenas-Pinto A.; Badr M.; Mellor R.; Kalber T. L.; Fernandez-Reyes D.; Schätzlein A. G.; Uchegbu I. F. SARS-CoV-2 Inhibition Using a Mucoadhesive, Amphiphilic Chitosan That May Serve as an Anti-Viral Nasal Spray. Sci. Rep 2021, 11 (1), 20012. 10.1038/s41598-021-99404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.