ABSTRACT
Artificial intelligence (AI) is a science that involves creating machines that can imitate human intelligence and learn. AI is ubiquitous in our daily lives, from search engines like Google to home assistants like Alexa and, more recently, OpenAI with its chatbot. AI can improve clinical care and research, but its use requires a solid understanding of its fundamentals, the promises and perils of algorithmic fairness, the barriers and solutions to its clinical implementation, and the pathways to developing an AI-competent workforce. The potential of AI in the field of nephrology is vast, particularly in the areas of diagnosis, treatment and prediction. One of the most significant advantages of AI is the ability to improve diagnostic accuracy. Machine learning algorithms can be trained to recognize patterns in patient data, including lab results, imaging and medical history, in order to identify early signs of kidney disease and thereby allow timely diagnoses and prompt initiation of treatment plans that can improve outcomes for patients. In short, AI holds the promise of advancing personalized medicine to new levels. While AI has tremendous potential, there are also significant challenges to its implementation, including data access and quality, data privacy and security, bias, trustworthiness, computing power, AI integration and legal issues. The European Commission's proposed regulatory framework for AI technology will play a significant role in ensuring the safe and ethical implementation of these technologies in the healthcare industry. Training nephrologists in the fundamentals of AI is imperative because traditionally, decision-making pertaining to the diagnosis, prognosis and treatment of renal patients has relied on ingrained practices, whereas AI serves as a powerful tool for swiftly and confidently synthesizing this information.
Keywords: artificial intelligence, kidney, machine learning, natural language processing, nephrology
INTRODUCTION
Artificial intelligence (AI) is described as the science and engineering of creating intelligent machines that can mimic human intelligence and learn. Many of us might think that AI is something complex that we do not want to get involved with because we are doctors, we know our speciality, and we do not need any “artificial” help. But fortunately, or unfortunately, this is not an option. We live wrapped in AI: Google, Amazon, Tesla, Alexa, Roomba, Siri, DeepL, facial recognition on our mobile phone, etc., as well as, more recently, OpenAI with its chatbot, ChatGPT, and its customized image designer, DALL·E 2 (Fig. 1). ChatGPT is a large language model (LLM) based on the GPT (Generative Pre-training Transformer) architecture, trained on a dataset of internet text to generate human-like text, while DALL·E 2 is a new AI system that can create realistic images and art from a description in natural language. These astonishing AI tools can help us all. And we say “help” because in most areas of medicine, AI will not replace the professional; rather, whoever understands and can manage it is likely to be a better professional and to make better informed decisions. In this regard, there is much controversy in the USA about the nomenclature, with the American Medical Association using the term “augmented intelligence” [1]. In fact, AI and digital health will augment (not replace) our ability to improve care and maintain health. This means using AI algorithms to augment human intelligence rather than to replace it.
Figure 1:
Examples on how OpenAI is used. (A) Questions and answers from ChatGPT. (B) Proposal to DALL∙E 2: oil paintings of a patient leaving hospital with a new transplanted kidney in his hand.
AI has the potential to improve clinical care and research in nephrology, but its use requires a solid understanding of the fundamentals of AI, the promises and perils of algorithmic fairness, the barriers and solutions to its clinical implementation, and the pathways to developing an AI-competent workforce. To adopt a new technology, clinicians and scientists need to be familiar with its fundamentals, fluent in its vocabulary and nomenclature, and inspired by its potential to improve outcomes for patients with or at risk of kidney disease. Due to the integration of AI into the field of medicine, nephrologists will soon engage with this technology on a daily basis, making it essential for the nephrology community to be well-informed and educated about its implications. Nephrologists around the world need to understand the core concepts of AI and its subtypes, and how the models are created, so that they can critically evaluate them and actively participate in minimizing the current challenges. Given nephrologists’ medical duties and legal responsibilities for the care they provide, they may be understandably reluctant to take action on a patient based on unexplained decisions made by black-box algorithms. The success of any AI-based study will therefore require strong interdisciplinary collaboration between medical experts and computer scientists, always avoiding the black-box feeling which refers to the opaqueness that often perplexes AI users when attempting to elucidate the algorithm's operations. Black-box is directly related with the lack of interpretability of the AI system. Interpretability, also often referred to as explainability, refers to the study of how to understand the decisions of machine learning (ML) systems, and how to design systems whose decisions are easily understood, or interpretable. Interpretable AI systems yield less risk as it can be better identified whether the model makes sense or not, given the observation of parts of the AI in which the reasoning might be flawed (for instance due to confusing variables).
For all these reasons, education on AI should be one of the objectives of nephrology societies (including the European Renal Association). A focus on the basics of AI education for nephrologists will enhance our specialty and improve patient care.
In this review, we aim to provide a very basic knowledge of what AI and more specifically ML is, as well as to outline its challenges and applications in renal medicine.
MACHINE LEARNING
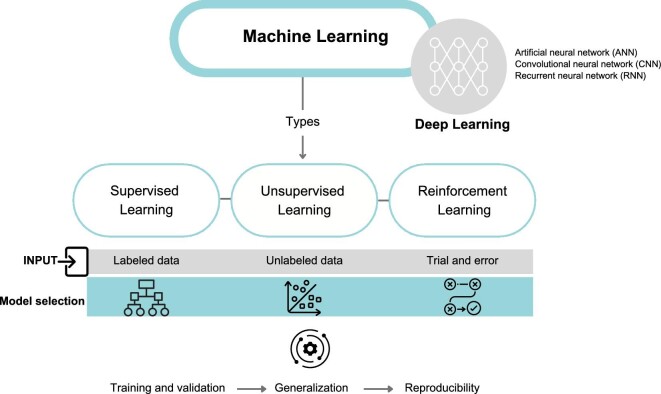
ML is a subset of AI that involves algorithms and statistical models that enable computer systems to improve their performance on a specific task based on the analysis of data (Fig. 2). ML algorithms are designed to learn from training data, identify patterns, and make predictions or decisions without being explicitly programmed to do so. The application to healthcare has been established for decades for classical methods like logistic regressions, but the use of newer methods like neural networks is still incipient, and although the number of publications is quite large, the number of models deployed on the healthcare market are still increasing at a slow pace [2].
Figure 2:
ML, step by step: ML is a type of AI that enables computer systems to learn from training data without explicit programming. DL is a more specific subset of ML that uses algorithms with multiple layers, simulating the complexity of the human brain. In healthcare, popular DL algorithms include artificial neural networks (ANNs), convolutional neural networks (CNNs) and recurrent neural networks (RNNs). There are three main types of ML algorithms: supervised learning, unsupervised learning and reinforcement learning. Supervised learning uses labeled data to train classification models, while unsupervised learning identifies clusters in unstructured data. Reinforcement learning relies on trial and error to learn from feedback. Once the model has been selected and data has been inputted, it must be trained with the data to produce accurate results. The model should then be validated to ensure it can generalize to new data and be reproducible for reliable clinical use.
There are three main types of ML algorithm: supervised learning, unsupervised learning and reinforcement learning.
Supervised learning algorithms learn from labeled data, where the input data are already categorized or labeled with known outcomes. These are commonly used in healthcare applications for tasks such as predicting disease diagnosis or patient outcomes.
Unsupervised learning algorithms, on the other hand, are used to identify patterns in unlabeled data, meaning that the input data are not labeled with known outcomes. These algorithms are used to find structure in data, e.g. identification of clusters of patients with similar disease symptoms or characteristics.
Reinforcement learning algorithms are used to learn through trial and error and involve conveying feedback to the algorithm after this has generated its outputs. In such a scheme, an algorithm could receive feedback from the patient's response to treatment generated by an ML algorithm and adjust the treatment plan accordingly to achieve the best possible outcome.
These algorithms are to be trained on sets of medical data, such as electronic health records or medical images. The quality and quantity of the training data are critical factors for the performance, robustness and general trustworthiness of the devised algorithm. Such algorithms, once trained, can provide a prediction or output such as a diagnosis, a risk score for a particular condition, a natural language response to a patient statement, molecular subtyping or survival data projected from patient trajectories.
One of the most important aspects of training a ML algorithm for clinical applications is the quality and quantity of the training data. The algorithm's ability to make accurate predictions and generalizations depends critically on the quality and representativeness of the data used to train it. If the training data are biased, incomplete or not representative of the population for which this algorithm will be deployed, i.e. if the training data design is poor, the algorithm may not perform well in real-world settings and may even be liable to biased assessments. These defects might not be evident, as they might be the result of a biased workflow, for instance if a segment of the population is less likely to receive a treatment due to systematic or unintentional exclusion of that segment of population (e.g. of racist or economic selection nature).
The concept of generalization refers to how well the algorithm performs on new data on which it has not been trained. If the algorithm is overfitting to the training data, it will not generalize well. Overfitting occurs when the algorithm learns the noise, bias, confounding effects or random variation in the training data instead of the underlying clinical patterns. To prevent overfitting, the algorithm is generally validated on a separate dataset, called the validation dataset. The validation dataset is used to evaluate the performance of the algorithm and ensure that it is not overfitting to the training data, thus ensuring the correct generalization of the algorithm.
Other factors can affect the quality and overall generalization of an ML algorithm, including choice of model, the hyperparameters of the model and the evaluation metrics used to assess the performance of the algorithm. The choice of model depends on the type of data and the task for which the algorithm is being developed. For example, if the algorithm is being developed to analyze endoscopy medical images, a convolutional neural network may be a good choice of model [3]. If the algorithm is being developed to predict outcomes, a logistic regression model may be more appropriate.
Hyperparameters are settings that are used to fine-tune the model and can affect its performance. Examples of hyperparameters include the learning rate, batch size and number of hidden layers for a neural network. The optimal values for hyperparameters can be determined through a process called hyperparameter tuning, where the model is trained and evaluated with different hyperparameters. All ML and AI models have some sort of hyperparameter design [4].
Additional concepts relevant to ML development for medical use are those of reproducibility. ML in health must be reproducible to ensure reliable clinical use. This can be achieved with the publication of open datasets and the provision of code accessibility, which is still an open issue in the case of health-related ML models [5].
Deep learning (DL) is a subset of ML that involves algorithms inspired by the structure and function of the human brain. These algorithms, called artificial neural networks, consist of multiple layers of interconnected nodes that can learn to recognize patterns and make decisions based on input data. DL algorithms are particularly effective in handling unstructured data, such as images, video and natural language, and have been used in applications such as image and speech recognition, natural language processing (NLP), autonomous driving and healthcare [6].
NATURAL LANGUAGE PROCESSING
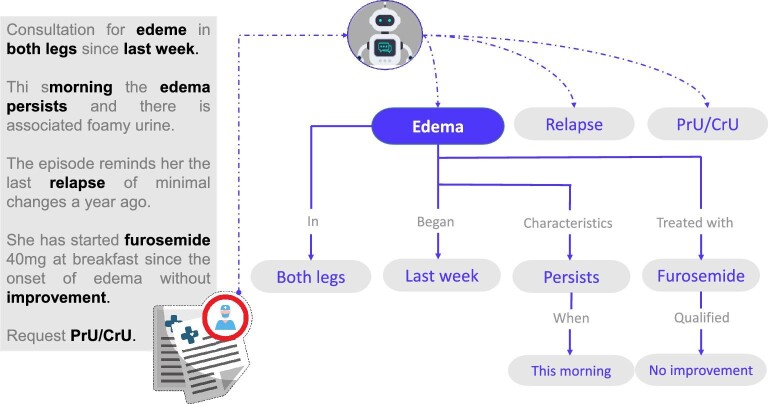
NLP is a field of AI that deals with the interaction between computers and human language. Specifically, NLP aims to enable computers to understand and generate natural language, as well as to perform various tasks such as text generation, text classification, machine translation and information extraction (Fig. 3).
Figure 3:
Use of NLP to extract data from medical records. NLP, a branch of AI, can convert the human language used in medical records, which may include typing errors, into a computer-understandable language in the form of algorithms.
NLP has undergone a significant shift in recent years, moving from a mainly rule-based approach to one that relies heavily on ML. This shift has been instrumental in enabling NLP to achieve better results. Rule-based approaches have significant limitations, such as time-consuming and labor-intensive development, requiring domain experts to spend significant time and effort crafting rules and patterns. Additionally, rule-based systems can struggle to handle the variability and complexity of natural language and do not generalize well to newer domains.
ML techniques have enabled NLP systems to overcome many of these limitations. ML-based systems are trained on datasets, allowing the ML algorithms to learn patterns and associations. ML-based systems adapt to new data and contexts more easily than rule-based systems and can achieve higher levels of accuracy and performance [7]. These ML algorithms, able to learn about language, are referred to as language models. Although language models, statistical representations of language, have been used since 1948 [8] in applications like spell checking, specifically, LLMs [9] have played a key role in improving the quality and effectiveness of embeddings, the numerical representation of a words or documents that captures its context and meaning, and therefore the results in NLP tasks. LLMs are a type of neural network that have been trained on vast amounts of text data, allowing them to capture a wide range of linguistic patterns and relationships. LLMs might be able to capture the subtle differences in meaning between terms like “glomerular” and “proteinuria” that are closely related, as well as between terms like “heart attack” and “lung cancer” that are not.
NLP use cases vary from improving patient care to increasing operational efficiency and reducing errors. In the context of clinical care, NLP is being used to automatically extract relevant information from medical records; for example, diagnoses, medications and laboratory results can be extracted from electronic health records (EHRs) [10] in order to build diverse systems such as clinical decision support [11], patient monitoring, chatbots and virtual assistants [12], and drug safety monitoring [13].
These use cases are built on top of different NLP tasks [14], specific problems that involve processing and understanding human language to be solved. These tasks include, for example, named entity recognition, i.e. the identification and classification of words in a text, such as medical conditions, medications and anatomical locations. Topic modeling, text classification and question answering provide ways to organize and categorize text data, while language translation can facilitate patient engagement and satisfaction by providing medical information in the patient's preferred language. Each task addresses different aspects of text analysis, allowing for a variety of applications in a clinical setting.
USE OF AI FOR KIDNEY DISEASES
Diagnosis
Diagnosis is a critical component of medical practice, as it forms the foundation for subsequent efforts in staging, treatment and prophylaxis. Over the years, clinicians have relied on a variety of tools, including analytical, imaging and histological studies, in addition to semiology, to reach an accurate diagnosis. Furthermore, clinical practice should be based on the best available scientific evidence for each patient.
In today's world, where AI is ubiquitous, it would be a missed opportunity not to utilize its potential in medicine. AI can help clinicians make more accurate and timely diagnoses by analyzing vast amounts of patient data and identifying patterns that may not be visible to the human eye. This technology can also assist in the interpretation of complex imaging studies and histological findings, improving the accuracy of diagnoses.
By incorporating AI into clinical practice, clinicians can make more informed decisions, leading to better patient outcomes. However, it is crucial to ensure that the use of AI in diagnosis is done ethically and transparently, and with appropriate safeguards in place to protect patient privacy and autonomy.
Overall, the use of AI in diagnosis has enormous potential to improve medical practice, and it is important for clinicians to embrace this technology and incorporate it into their daily practice.
Different areas of diagnosis in nephrology have begun to be influenced by the use of AI (Table 1). A random forest algorithm has been developed that enables the early diagnosis of chronic kidney disease (CKD) [15]. Using ML techniques, researchers have identified metabolic signatures associated with pediatric CKD by linking sphingomyelin-ceramide and plasmalogen dysmetabolism with focal segmental glomerulosclerosis [16]. Researchers have also successfully mimicked the ability of nephropathologists to extract diagnostic, prognostic and therapeutic information from native or transplanted kidney biopsies using image recognition [17].
Table 1:
Examples of AI tools in nephrology.
| Field | Study | Scenario | Purpose | Algorithm | Performance | Limitations |
|---|---|---|---|---|---|---|
| AKI | Postop-MAKE [24] | Prediction: patients with normal renal function undergoing cardiac surgery with cardiopulmonary bypass |
Prediction model based on nine preoperative variables (clinical, laboratory, imaging) that predict the risk of developing AKI after surgery | Nanogram. Logistic regression performed with variables selected using LASSO | High discriminatory power: AUC of 0.740 (95% CI 0.726–0.753) in the validation group |
Single-center retrospective study; treatment protocols for these patients could vary from center to center |
| STARZ [26] | Prediction: neonates admitted to NICU Multicenter, prospective |
Prediction model to assess the risk of AKI development in neonates (≤28 days) admitted to NICU on fluids for at least 48 h. Based on 10 variables | Multivariable logistic regression with stepwise backward elimination method | High discriminatory power: the model had an AUC of 0.974 (95% CI 0.958–0.990) |
No cystatin C access to evaluate kidney function in neonates Multicenter but in one single country |
|
| Tomašev et al. (2019) [39] | Prediction: multisite retrospective dataset of 703 782 adult patients (US Department of Veterans Affairs) |
Prediction model developed to calculate the probability of AKI occurring at any stage of severity within the next 48 h. Variables extracted from EHR | Recurrent neural network Deep learning |
Prediction of 55.8% of inpatient episodes of AKI and 90.2% of AKI requiring dialysis with a lead time of up to 48 h. Ratio of two false alerts for every true alert | 90% men Urine output not included Retrospective; no external validation |
|
| Zhang et al. (2019) [35] | Prediction/treatment: AKI patients (n = 6682) with urine output <0.5 mL/kg/h during first 6 h after ICU admission and fluid intake >5 L (US-based critical care database) |
Prediction model used to differentiate between volume-responsive and volume-unresponsive AKI | XGBoost algorithm | AUC 0.860 (95% CI 0.842–0.878) | No external validation Only measurement of short-term outcomes |
|
| CKD | Chan et al. (2021) [40] | Prediction: patients (n = 1146) with prevalent DKD (G3a–G3b with all grades of albuminuria (A1–A3) and G1 and G2 with A2–A3 level albuminuria) |
Prediction risk of ESKD in patients with CKD and type 2 diabetes mellitus Combination of novel biomarkers and data extracted from EHR (KidneyIntelX score). Comparison with traditional models |
ML Random forest model |
AUC for composite kidney endpoint 0.77 compared with an AUC of 0.61 (95% CI 0.60–0.63) for the clinical model | Missing data for urine results |
| Hermsen et al. (2019) [41] | Diagnosis: use of kidney transplant biopsies and nephrectomy samples |
Multiclass segmentation of digitized kidney tissue sections Facilitation of the histological study of samples using digital analysis and comparison of this approach with the performance of expert pathologists |
Deep learning: CNN | The best segmented class was glomeruli. The mean intraclass correlation coefficient for glomerular counting performed by pathologists versus the network was 0.94 | This work focused only on the segmentation of glomeruli and refers to a small sample size | |
| Glomerular disease | IgAN-tool (Asia) [22] | Prediction: patients with IgAN from multiple centers in China (n = 2047) Multicenter, retrospective |
ESKD prediction for IgAN patients Model based on 10 clinical, laboratory, and histological variables |
XGBoost algorithm | High discriminatory power: C-statistic of 0.84 (95% CI 0.80–0.88) for the validation cohort | Study only performed in Asian patients |
| IgAN-tool (EU) [42] | Prediction: IgAN patients (n = 948). Retrospective. Follow-up 89 months |
ESKD prediction for IgAN patients Model based on seven clinical and histological variables including MEST-C score |
Cox regression for variable selection DL: ANN |
AUC 0.82 at 5 years AUC 0.89 at 10 years |
Developed and tested in retrospective cohorts Therapeutic interventions not included |
|
| Inherited kidney disease | Jefferies et al. (2021) [43] | Diagnosis: de-identified health records from a cohort of patients with confirmed Fabry disease (n = 4978) Patients from 50 US states |
Diagnosis of Fabry disease using EHR. ICD-10 codes used AI tool (OM1 Patient FinderTM) (OM1 Inc., Boston, MA) |
ML (model not specified) | AUROC 0.82 Testing in males only: AUROC 0.83 Testing in females only: AUROC 0.82 |
Missings in health records Gender imbalance Not validated in external cohorts outside the USA |
| Potretzke et al. (2023) [18] | Diagnosis/prediction: Patients with ADPKD undergoing MR imaging between November 2019 and January 2021 (N = 170) 1 center: Mayo Clinic |
Evaluate performance in clinical practice of an AI algorithm for MR-derived total kidney volume in ADPKD | DL: CNN | AI algorithm TKV output mean volume difference –3.3%. Agreement for disease class between AI-based and manually edited segmentation high (five cases differed) | Prospective study in different centers to evaluate whether the algorithm is generalizable remains to be elucidated | |
| Dialysis | Barbieri et al. (2016) [34] | Treatment: hemodialysis patients (n = 752) in three different NephroCare centers (Fresenius Medical Care network) across the EU |
Anemia control model to recommend suitable erythropoietic-stimulating agent doses based on patient profiles | DL: ANN | Hb SD decreased (0.97 ± 0.41 g/dL to 0.8 ± 0.29 g/dL) Hb within target 84.1% vs 64.5% |
Not a randomized or blinded controlled trial Short follow-up period to assess outcomes No external validation |
| Zhang et al. (2022) [44] | Diagnosis: digital images of AV accesses before cannulation (1425 AV access images) Cohort of hemodialysis patients from 20 dialysis clinics across six US states |
Classification of vascular access aneurysm as “non-advanced” or “advanced” | DL: CNN | AUROC 0.96 | Real world testing in a demographically diverse population remains to be published Clinical parameters not included |
|
| Lee et al. (2021) [45] | Prediction: analysis of 261 647 hemodialysis sessions (N = 9292) One center (Seoul National University Hospital) |
DL model to predict the risk of intradialytic hypotension using a timestamp-bearing dataset | DL: RNN | AUC 0.94 for prediction of intradialytic hypotension 1 (defined as nadir systolic BP <90 mmHg) | Retrospective cohort. One center Other factors not included (cardiac monitoring, dialysis vintage and medical records) |
|
| Kidney transplant | IBox [23] | Prediction: kidney transplant recipients (n = 7557) from 10 medical centers across Europe and USA |
Prediction of allograft failure Eight functional, histological and immunological prognostic factors combined into a risk score |
Cox regression with boot-strapping for validation | C index 0.18 (95% CI 0.79–0.83) Validation cohorts: Europe: C index 0.81 (95% CI 0.78–0.84) US: C index 0.80 (95% CI 0.76–0.84) |
Emerging predictors post-transplant missing. Adherence not taken into account. Validation in daily clinical practice remains to be analyzed |
ADPKD, autosomal dominant polycystic kidney disease; ANN, artificial neural network; AUC, area under the curve; AUROC, area under the receiver operating characteristic curve; AV access, arteriovenous access; BP, blood pressure; CI: confidence interval; CNN, convolutional neural network; DKD, diabetic kidney disease; ESKD, end-stage kidney disease; Hb, hemoglobin; ICD, International Classification of Diseases; ICU, intensive care unit; IgAN, immunoglobulin A nephropathy; LASSO, least absolute shrinkage and selection operator; MR, magnetic resonance; NICU, neonatal intensive care unit; RNN, recurrent neural network; SD, standard deviation; TKV, total kidney volume; XGBoost, extreme gradient boosting.
An AI-based algorithm has been developed to estimate the total renal volume of patients with autosomal dominant polycystic kidney disease from magnetic resonance imaging scans. This algorithm could aid in disease monitoring and prognostic evaluation [18]. ML algorithms have been developed to identify pathogen-specific fingerprints in peritoneal dialysis patients with bacterial infections. These algorithms can help clinicians make informed treatment decisions and improve patient outcomes [19]. Although tools have been developed to aid in the diagnosis of glomerular diseases, such as ML-based algorithms, the lack of validation in diverse populations has hindered their translation into clinical practice [20].
A recent study demonstrated the potential of using deep phenotyping on EHRs to facilitate genetic diagnosis through clinical exomes [21]. This approach could be particularly useful in nephrology, where the diagnosis of hereditary diseases is still an area that requires further exploration.
Prediction of outcomes
Prediction scores or scales, as we know, have been widely used in the medical field to help clinicians establish criteria for managing patients long before ML became this large in healthcare. In nephrology, various prognostic scores are used to estimate the risk of developing end-stage CKD, with some scores being developed for specific kidney diseases. In order to perform the scoring, traditional statistical methods have been used up to the present. However, the integration of clinical expertise with ML experts opens up new possibilities to provide the best predictions to the patient. When making use of ML tools, like XGBoost, in conjunction with traditional statistical techniques, it becomes more feasible to generate expansive predictive capabilities, even when working with limited data.
Prediction scores typically incorporate both classic risk factors and additional variables such as histology or imaging tests. Some examples of use of ML techniques in the development of these scores (Table 1) include the IgAN-tool [22] and IBox [23]. Scores have also been developed to predict the risk of acute kidney injury (AKI) in different patient populations, such as the postop-MAKE score [24], which estimates the risk of AKI in patients with normal renal function undergoing cardiac surgery, and AKI risk scores in patients with heart failure [25]. Even neonatal patients can benefit from AKI risk assessment using the STARZ score [26].
A common challenge in developing risk scores for kidney diseases is the small size of the patient populations studied, which limits the scope of validation. Moreover, many studies are conducted within a single country, which can further limit validation across ethnicities. To address this issue, it is necessary to conduct and promote multicenter studies with the goal of validating algorithms that can accurately predict outcomes in the future. By facilitating and encouraging collaborative research the algorithms will be validated and tested for effectiveness and become reliable.
Treatment aid
Throughout history, physicians have recognized the uniqueness of each patient and have strived to provide tailored and personalized treatments to deliver the best possible care. This approach emphasizes the importance of interindividual variability and is commonly referred to as precision medicine. The goal of precision medicine is to develop treatment strategies that are specifically tailored to the individual patient, taking into account their unique characteristics, including genetics, lifestyle and environmental factors. By embracing precision medicine, physicians can provide more effective and targeted treatments, leading to better patient outcomes [27].
The large-scale applicability of this concept has been greatly aided by the development of large-scale biological databases, the use of genomics, proteomics and metabolomics, and computational tools that enable massive data analysis. The combination of all these disciplines can guide us in adjusting medical treatment to specific pathological processes and ultimately optimize patient outcomes [27, 28]. Fields such as oncology have been pioneering in the application of ML algorithms to predict response to immunotherapy and to develop targeted therapies based on molecular disease processes that improve outcomes for different types of tumor [29]. Nephrology should be no less forward looking, but there is a long way to go. Still today, targeted therapies for specific etiologies are lacking as improvement of classifications and more specific biomarkers to categorize kidney diseases are needed.
How might we benefit? New therapeutic strategies based on AI models may help us in guiding drug prescription, decreasing variability, achieving a higher percentage of patients on target and avoiding error proneness. ML models may also contribute to clinical trials by identifying high-risk patients who may benefit from new therapies under study as well as those who will not, thus avoiding unnecessary treatments. Some of these objectives may be achieved by using the “digital twins” approach. Health digital twins are defined as virtual representations (“digital twin”) of patients (“physical twin”) that are generated from multimodal patient data, population data and real-time updates on patient and environmental variables [30].
Several ML therapeutic models have been developed in recent years, although most of them are not yet validated for application in daily clinical practice (Table 1). Dialysis is a particularly attractive field for AI application because of its large volume of documented data in EHR [31]. ML models were able to predict dialysis adequacy in chronic hemodialysis patients [32] and could lead to future personalized prescriptions. Different models have been developed to optimize anemia management [33]. Barbieri et al. [34]. developed an artificial neural network which guided the prescription of iron and erythropoietic-stimulating agent doses, resulting in a decrease in hemoglobin variability and an increase in the percentage of hemoglobin values on target. In AKI patients, Zhang et al. [35] developed an XGBoost model that identified patients who would and would not respond to volume-driving treatment strategies.
The IBox system, an accurate and validated algorithm for the prediction of graft failure [23], has laid the groundwork for the use of AI in kidney transplantation. Regarding immunosuppression therapy, achieving a perfect balance between graft survival and chronic immunosuppression-associated complications (i.e. opportunistic infections, malignancies and others) is still a challenge. Different algorithms based on tacrolimus pharmacokinetics and pharmacodynamics [36, 37] have been developed in order to decrease toxicity, but the perfect therapeutic model that brings together genetics, pharmacokinetics and clinical parameters is yet to come.
CHALLENGES OF AI
It is important to carefully consider the risks and challenges associated with AI technology and to put systems in place to mitigate the risks. Some of the challenges are the following.
Data access and quality
Data sets for ML should come from a trusted source of relevant data that is clean, accessible, well managed and secured. Data scarcity is often a shortcoming in nephrology, as many diseases are rare and kidney diseases are generally far less common than other medical conditions.
Data privacy and security
Data protection is also a key concern when using AI in nephrology. Patient data is sensitive and must be protected in accordance with relevant laws and regulations, such as the General Data Protection Regulation (GDPR) in the European Union (EU). Patient data must be anonymized or pseudonymized prior to analysis or storage somewhere other than where it was collected. It is important to ensure that patient data is handled securely and that patients are informed about how their data will be used. Any AI tool or project should have a data management environment for sensitive data.
Bias
The potential for bias in AI algorithms is a further risk that should be considered. If an AI algorithm is trained on biased data, it may perpetuate or even amplify existing biases in the healthcare system. For example, if an AI algorithm is trained on data from predominantly white patients with focal glomerulosclerosis, it may not accurately identify or classify the disease in patients from other racial or ethnic groups. It is important to ensure that AI algorithms are trained on diverse and representative data sets in order to minimize the risk of bias.
Trustworthiness
It is human nature to trust only things that are easy to understand, and doctors are a perfect paradigm of this. One of the critical challenges in implementing AI is the unknown nature of how learning models and a set of inputs can predict the output and provide guidance in a medical intervention. Explainability in AI is needed to provide transparency in the decisions made by AI, as well as in the algorithms that lead to those decisions. This is essential in order to avoid the black-box feel common to many AI tools. Multiple techniques can be used to try avoid this feeling, though. Methodologies like feature importance rank the variables used by an AI model based on their impact on prediction results, revealing how each input contributes to the decision-making process. Although this can shed some light, recently, OpenAI has admitted its lack of full understanding of how ChatGPT works, and the lack of tools to explore newer models’ decision-making process. This kind of lack of understanding has prompted governments to put in place measures to control and limit the expansion of unregulated AI. One tool to improve trustworthiness in the kidney arena is education of the nephrological community in this field.
Computing power
ML and DL are the stepping stones of AI, but they require an ever-increasing number of cores and GPUs to operate efficiently, and these are not readily available everywhere. Achieving the computing power to process the massive amounts of data needed to build AI systems is the biggest challenge the industry has ever faced. And it goes without saying that this computing power comes with a significant environmental footprint. This is a real hurdle for many research projects using AI and has raised concerns in the AI community, leading to calls for more transparency, optimization of training cycles and increased focus on “green AI,” which aims for novel results without increasing computational costs, and ideally reducing it [38].
AI integration
EHRs are relatively new to many healthcare providers. While the EU is supporting efforts to harmonize them and the USA has already introduced some shared patient information, the reality is that interoperability is scarce and embedding AI tools in EHRs is not always feasible in many hospitals.
AI specialists
The integration, deployment and implementation of AI require a specialist, such as a data scientist or data engineer, with a certain level of skills and expertise. One of the main challenges in implementing AI in hospitals or in a research environment is that these experts are expensive and currently quite rare; they are often more willing to join a large company with high salaries than to work in a public environment such as most hospitals or research centers in Europe.
Legal issues
Another risk to consider is the potential for errors in AI algorithms. While AI algorithms can process large amounts of data quickly and accurately, they are not perfect and can make mistakes. It is important to have systems in place to identify and correct errors in AI algorithms in order to ensure the safety and effectiveness of the technology. As mentioned above, for the time being, AI tools will help nephrologists make decisions, but will not replace expert professionals. AI algorithms may also violate laws or regulations, exposing the organization to legal challenges.
EUROPEAN UNION FRAMEWORK
In 2021, the European Commission proposed a regulatory framework that is expected to enter into force in 2023 as a transitional period and in 2024 as a full deployment. It classifies AI technology into four levels of risk: (i) unacceptable risk as a clear threat to safety, (ii) high risk, including transport, health, administrative and law enforcement, among other sectors, (iii) limited risk, such as chatbots, where transparency is key, and (iv) minimal risk, such as AI video games.
Most healthcare AI systems will be placed in the high-risk category and will need to undergo a regulatory process including conformity assessment and compliance specifics. High-risk AI systems considered medical devices will have to be compliant with the Medical Regulation (MDR) and will be registered in an EU database and CE marking will be required.
The European Health Data Space (EHDS) is an initiative officially launched in 2018 by the EU to promote secure and seamless access to health data across the EU. The aim of the EHDS is to create a comprehensive and interoperable platform for sharing health data while ensuring data privacy and protection. The EHDS facilitates the collection and exchange of health data such as electronic health records, research data and patient-generated data among healthcare professionals, researchers and policymakers. It ensures the safety and confidentiality of personal data by complying with the GDPR, which is an extensive data protection regulation applicable to all EU member states that establishes guidelines for the collection, handling and retention of personal data.
Another important component of the EU framework is the Ethical Guidelines for Trustworthy AI, which were formulated by the High-Level Expert Group on AI of the European Commission. The guidelines outline a set of principles and requirements for AI that align with fundamental human rights, ethical values and social well-being. They emphasize the need for AI systems to be transparent, accountable and explainable while respecting data privacy and protection. Furthermore, the guidelines highlight the significance of fairness, non-discrimination and sustainability in the development and deployment of AI, serving as a reference for developers, deployers and users of AI systems across multiple sectors, including healthcare.
The abovementioned guidelines provide a comprehensive framework for AI governance in nephrology and healthcare. While AI algorithms offer significant potential, they are tools for humans, created by humans, and the responsibility to ensure the ethical and responsible use of these technologies remains paramount.
CONCLUSIONS
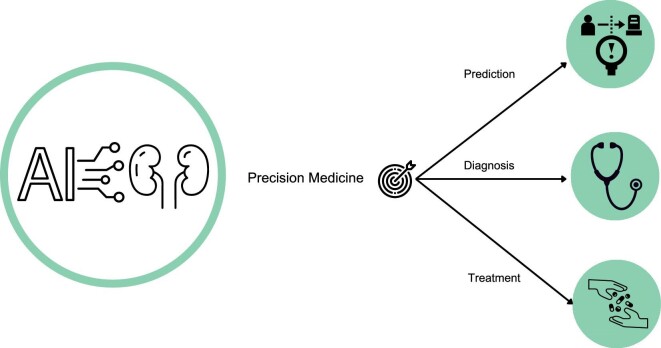
AI has the potential to revolutionize healthcare in several ways, including personalized medicine, early disease detection and improved drug discovery techniques (Fig. 4). Economically, AI can be used to predict patient outcomes, readmission rates and length of hospital stays based on patient data. While AI can significantly improve patient care, it cannot replace the patient–doctor relationship, which is a critical aspect of healthcare.
Figure 4:
The future of nephrology: advancing precision medicine with AI. The field of nephrology aspires to achieve precision medicine by taking into account interindividual variability in prevention, diagnosis and treatment strategy. The use of AI is expected to greatly enhance achievement of this goal in the coming years. AI has the potential to improve patient outcomes and revolutionize clinical management in nephrology. As such, it is a promising paradigm shift that will enable healthcare professionals to provide more personalized and effective patient care.
In nephrology, the use of AI has the potential to bring about significant benefits, but it is crucial to consider the risks and challenges associated with the technology and to develop systems to mitigate these risks. Nephrologists will soon be interacting with AI in their daily practice, making it essential for the nephrology community to be educated about this technology. Understanding the core concepts of AI and how models are created is a prerequisite for effective use of the technology.
While AI will not replace nephrologists, those who can use it effectively will likely become better professionals for their patients. However, it is important to recognize that the integration of AI into clinical practice will require a shift in the traditional roles of healthcare professionals, and that there will be a need for ongoing training and education to ensure that AI is used effectively and ethically.
Contributor Information
Leonor Fayos De Arizón, Nephrology Department, Fundació Puigvert; Institut d'Investigacions Biomèdiques Sant Pau (IIB-Sant Pau); Departament de Medicina, Universitat Autònoma de Barcelona, Barcelona, Spain.
Elizabeth R Viera, Nephrology Department, Fundació Puigvert; Institut d'Investigacions Biomèdiques Sant Pau (IIB-Sant Pau); Departament de Medicina, Universitat Autònoma de Barcelona, Barcelona, Spain.
Melissa Pilco, Nephrology Department, Fundació Puigvert; Institut d'Investigacions Biomèdiques Sant Pau (IIB-Sant Pau); Departament de Medicina, Universitat Autònoma de Barcelona, Barcelona, Spain.
Alexandre Perera, Center for Biomedical Engineering Research (CREB), Universitat Politècnica de Barcelona (UPC), Barcelona, Spain; Networking Biomedical Research Centre in the subject area of Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN), Madrid, Spain; Institut de Recerca Sant Joan de Déu, Esplugues de Llobregat, Barcelona, Spain.
Gabriel De Maeztu, IOMED, Barcelona, Spain.
Anna Nicolau, Neuroelectrics, Barcelona, Spain.
Monica Furlano, Nephrology Department, Fundació Puigvert; Institut d'Investigacions Biomèdiques Sant Pau (IIB-Sant Pau); Departament de Medicina, Universitat Autònoma de Barcelona, Barcelona, Spain.
Roser Torra, Nephrology Department, Fundació Puigvert; Institut d'Investigacions Biomèdiques Sant Pau (IIB-Sant Pau); Departament de Medicina, Universitat Autònoma de Barcelona, Barcelona, Spain.
FUNDING
This work was supported by Instituto de Salud Carlos III, RICORS2040, funded by EU-Next Generation, Mechanism of recuperation and resilience (MRR) and PI22/00361; Fundació la Marató de TV3 (Grant agreement 202036-30); FEDER funds (Grant agreement SPACKDc PMP21/00 109); Spanish Ministry of Economy and Competitiveness (www.mineco.gob.es) PID2021-122952OB-I00, DPI2017-89827-R and Networking Biomedical Research Centre in the subject area of Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN), initiatives of Instituto de Investigación Carlos III (ISCIII).
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY STATEMENT
No new data were generated or analysed in support of this research.
REFERENCES
- 1. Crigger E, Reinbold K, Hanson C. et al. Trustworthy augmented intelligence in health care. J Med Syst 2022;46:12. 10.1007/s10916-021-01790-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wiens J, Saria S, Sendak M. et al. Do no harm: a roadmap for responsible machine learning for health care. Nat Med 2019;25:1337–40. 10.1038/S41591-019-0548-6 [DOI] [PubMed] [Google Scholar]
- 3. Ali S. Where do we stand in AI for endoscopic image analysis? Deciphering gaps and future directions. NPJ Digit Med 2022;5:184. 10.1038/S41746-022-00733-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shankar K, Zhang Y, Liu Y. et al. Hyperparameter tuning deep learning for diabetic retinopathy fundus image classification. IEEE Access 2020;8:118164–73. 10.1109/ACCESS.2020.3005152 [DOI] [Google Scholar]
- 5. McDermott MBA, Wang S, Marinsek N. et al. Reproducibility in machine learning for health research: still a ways to go. Sci Transl Med 2021;13:eabb1655. 10.1126/SCITRANSLMED.ABB1655 [DOI] [PubMed] [Google Scholar]
- 6. Esteva A, Robicquet A, Ramsundar B. et al. A guide to deep learning in healthcare. Nat Med 2019;25:24–9. 10.1038/s41591-018-0316-z [DOI] [PubMed] [Google Scholar]
- 7. Li Q, Yu PS, Peng H. et al. A survey on text classification: from shallow to deep learning. ACM Trans Intell Syst Technol 2020;37:39. 10.1145/1122445.1122456 [DOI] [Google Scholar]
- 8. Shannon CE. A mathematical theory of communication. ACM SIGMOBILE Mob Comput Commun Rev 2001;5:3–55. 10.1145/584091.584093 [DOI] [Google Scholar]
- 9. Vaswani A, Brain G, Shazeer N. et al. Attention is all you need. Adv Neural Inf Process Syst 2017;30. [Google Scholar]
- 10. Hossain E, Rana R, Higgins N. et al. Natural language processing in Electronic Health Records in relation to healthcare decision-making: a systematic review. Comput Biol Med 2023;155:106649. 10.1016/J.COMPBIOMED.2023.106649 [DOI] [PubMed] [Google Scholar]
- 11. Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform 2009;42:760–72. 10.1016/J.JBI.2009.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu L, Sanders L, Li K. et al. Chatbot for health care and oncology applications using artificial intelligence and machine learning: systematic review. JMIR Cancer 2021;7:e27850. 10.2196/27850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo Y, Thompson WK, Herr TM. et al. Natural language processing for EHR-based pharmacovigilance: a structured review. Drug Saf 2017;40:1075–89. 10.1007/S40264-017-0558-6 [DOI] [PubMed] [Google Scholar]
- 14. Collobert R, Weston J, Com J. et al. Natural language processing (almost) from scratch. J Mach Learn Res 2011;12:2493–537. 10.5555/1953048.2078186 [DOI] [Google Scholar]
- 15. Senan EM, Al-Adhaileh MH, Alsaade FW. et al. Diagnosis of chronic kidney disease using effective classification algorithms and recursive feature elimination techniques. J Healthc Eng 2021;2021:1004767. 10.1155/2021/1004767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee AM, Hu J, Xu Y. et al. Using machine learning to identify metabolomic signatures of pediatric chronic kidney disease etiology. J Am Soc Nephrol 2022;33:375–86. 10.1681/ASN.2021040538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Becker JU, Mayerich D, Padmanabhan M. et al. Artificial intelligence and machine learning in nephropathology. Kidney Int 2020;98:65–75. 10.1016/J.KINT.2020.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Potretzke TA, Korfiatis P, Blezek DJ. et al. Clinical implementation of an artificial intelligence algorithm for magnetic resonance-derived measurement of total kidney volume. Mayo Clin Proc 2023;98:689–700. 10.1016/J.MAYOCP.2022.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang J, Friberg IM, Kift-Morgan A. et al. Machine-learning algorithms define pathogen-specific local immune fingerprints in peritoneal dialysis patients with bacterial infections. Kidney Int 2017;92:179–91. 10.1016/J.KINT.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schena FP, Magistroni R, Narducci F. et al. Artificial intelligence in glomerular diseases. Pediatr Nephrol 2022;37:2533–45. 10.1007/S00467-021-05419-8 [DOI] [PubMed] [Google Scholar]
- 21. Son JH, Xie G, Yuan C. et al. Deep phenotyping on electronic health records facilitates genetic diagnosis by clinical exomes. Am J Hum Genet 2018;103:58–73. 10.1016/J.AJHG.2018.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen T, Li X, Li Y. et al. Prediction and risk stratification of kidney outcomes in IgA nephropathy. Am J Kidney Dis 2019;74:300–9. 10.1053/J.AJKD.2019.02.016 [DOI] [PubMed] [Google Scholar]
- 23. Loupy A, Aubert O, Orandi BJ. et al. Prediction system for risk of allograft loss in patients receiving kidney transplants: international derivation and validation study. BMJ 2019;366:l4923. 10.1136/BMJ.L4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu P, Mo Z, Chen Y. et al. Derivation and validation of a model to predict acute kidney injury following cardiac surgery in patients with normal renal function. Ren Fail 2021;43:1205–13. 10.1080/0886022X.2021.1960563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou LZ, Yang XB, Guan Y. et al. Development and validation of a risk score for prediction of acute kidney injury in patients with acute decompensated heart failure: a prospective cohort study in China. J Am Heart Assoc 2016;5:e004035. 10.1161/JAHA.116.004035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wazir S, Sethi SK, Agarwal G. et al. Neonatal acute kidney injury risk stratification score: STARZ study. Pediatr Res 2022;91:1141–8. 10.1038/S41390-021-01573-9 [DOI] [PubMed] [Google Scholar]
- 27. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793–5. 10.1056/NEJMP1500523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med 2019;380:1347–58. 10.1056/NEJMRA1814259 [DOI] [PubMed] [Google Scholar]
- 29. Precision medicine in nephrology. Nat Rev Nephrol 2020;16:615. 10.1038/S41581-020-00360-9 [DOI] [PubMed] [Google Scholar]
- 30. Venkatesh KP, Raza MM, Kvedar JC. Health digital twins as tools for precision medicine: considerations for computation, implementation, and regulation. NPJ Digit Med 2022;5:150. 10.1038/S41746-022-00694-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kotanko P, Zhang H, Wang Y. Artificial intelligence and machine learning in dialysis: ready for prime time? Clin J Am Soc Nephrol 2023;. 18:803–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim HW, Heo SJ, Kim JY. et al. Dialysis adequacy predictions using a machine learning method. Sci Rep 2021;11:15417. 10.1038/S41598-021-94964-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brier ME, Gaweda AE, Aronoff GR. Personalized anemia management and precision medicine in ESA and iron pharmacology in end-stage kidney disease. Semin Nephrol 2018;38:410–7. 10.1016/J.SEMNEPHROL.2018.05.010 [DOI] [PubMed] [Google Scholar]
- 34. Barbieri C, Molina M, Ponce P. et al. An international observational study suggests that artificial intelligence for clinical decision support optimizes anemia management in hemodialysis patients. Kidney Int 2016;90:422–9. 10.1016/J.KINT.2016.03.036 [DOI] [PubMed] [Google Scholar]
- 35. Zhang Z, Ho KM, Hong Y. Machine learning for the prediction of volume responsiveness in patients with oliguric acute kidney injury in critical care. Crit Care 2019;23:7–8. 10.1186/S13054-019-2411-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sridharan K, Shah S. Developing supervised machine learning algorithms to evaluate the therapeutic effect and laboratory-related adverse events of cyclosporine and tacrolimus in renal transplants. Int J Clin Pharm 2023;45:659–68. 10.1007/S11096-023-01545-5 [DOI] [PubMed] [Google Scholar]
- 37. Thishya K, Vattam KK, Naushad SM. et al. Artificial neural network model for predicting the bioavailability of tacrolimus in patients with renal transplantation. PLoS One 2018;13:e0191921. 10.1371/JOURNAL.PONE.0191921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dhar P. The carbon impact of artificial intelligence. Nat Mach Intell 2020;2:423–5. 10.1038/S42256-020-0219-9 [DOI] [Google Scholar]
- 39. Tomašev N, Glorot X, Rae JW. et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 2019;572:116–9. 10.1038/S41586-019-1390-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan L, Nadkarni GN, Fleming F. et al. Derivation and validation of a machine learning risk score using biomarker and electronic patient data to predict progression of diabetic kidney disease. Diabetologia 2021;64:1504–15. 10.1007/S00125-021-05444-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hermsen M, Bel T, Boer MD. et al. Deep learning-based histopathologic assessment of kidney tissue. J Am Soc Nephrol 2019;30:1968–79. 10.1681/ASN.2019020144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schena FP, Anelli VW, Trotta J. et al. Development and testing of an artificial intelligence tool for predicting end-stage kidney disease in patients with immunoglobulin A nephropathy. Kidney Int 2021;99:1179–88. 10.1016/J.KINT.2020.07.046 [DOI] [PubMed] [Google Scholar]
- 43. Jefferies JL, Spencer AK, Lau HA. et al. A new approach to identifying patients with elevated risk for Fabry disease using a machine learning algorithm. Orphanet J Rare Dis 2021;16:518. 10.1186/S13023-021-02150-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang H, Preddie D, Krackov W. et al. Deep learning to classify arteriovenous access aneurysms in hemodialysis patients. Clin Kidney J 2022;15:829. 10.1093/CKJ/SFAB278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee H, Yun D, Yoo J. et al. Deep learning model for real-time prediction of intradialytic hypotension. Clin J Am Soc Nephrol 2021;16:396–406. 10.2215/CJN.09280620 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.