Abstract
Recombinant adeno-associated viruses (rAAVs) are promising gene delivery vectors in the emerging field of in vivo gene therapies. To ensure their consistent quality during manufacturing and process development, multiple analytical techniques have been proposed for the characterization and quantification of rAAV capsids. Despite their indisputable capabilities for performing this task, current analytical methods are rather time-consuming, material intensive, complicated, and costly, restricting their suitability for process development in which time and sample throughput are severe constraints. To eliminate this bottleneck, we introduce here an affinity-based high-performance liquid chromatography method that allows the determination of the capsid titer and the full/empty ratio of rAAVs within less than 5 min. By packing the commercially available AAVX affinity resin into small analytical columns, the rAAV fraction of diverse serotypes can be isolated from process-related impurities and analyzed by UV and fluorescence detection. As demonstrated by both method qualification data and side-by-side comparison with AAV enzyme-linked immunosorbent assay results for rAAV8 samples as well as by experiments using additional rAAV2, rAAV8, and rAAV9 constructs, our approach showed good performance, indicating its potential as a fast, simple and efficient tool for supporting the development of rAAV gene therapies.
Keywords: AAV, high-throughput, gene therapy, adeno-associated, affinity chromatography, fluorescence, process development, capsid titer
Graphical abstract

Introduction
With approval from the European Medicines Agency, the first recombinant adeno-associated virus (rAAV)–based gene therapy product, Glybera, was introduced to the market in 2012, offering treatment for lipoprotein lipase deficiency.1 It was followed by Luxturna in 2017, a medication against an inherited form of retinal dystrophy, being the first rAAV gene therapy product approved by the US Food and Drug Administration. Due to its lack of pathogenicity, the ability to transfer genes into nondividing and proliferating cells,2,3 and the advantage of specific gene delivery to distinct tissues using different serotypes,4,5,6 rAAVs represent a promising gene delivery system in the emerging field of gene therapies.7,8 Wild-type AAV (wtAAV) is a member of the Parvoviridae family. For replication, wtAAV is dependent on adenovirus. In recombinant production of rAAVs, a helper plasmid containing the replication genes of adenovirus is advantageously used. This minimizes the risk of the coproduction of competent rAAV in a target product. Approximately 60 viral proteins of three structural types (VP1, VP2, and VP3) assemble in a ratio of 1:1:10 to form an ∼26-nm nonenveloped capsid with T = 1 icosahedral symmetry.9,10 The capsid incorporates a 4.7-kb single-stranded DNA (ssDNA) that codes for proteins for its replication as well as encapsidation, and is flanked by inverted terminal repeats (ITRs).11 For therapeutic use, the genetic information between the ITRs is replaced by the gene of interest.
The incorporation of the genome during the replication cycle of both wtAAVs and rAAVs is inefficient and results in a high number of empty capsids, which can be problematic.12 They show potential negative effects on gene transduction efficiency and can trigger anticapsid T cell immunity by an increased total capsid load.13,14 However, it was shown that nonfilled capsids can be used as a decoy for preexisting humoral immunity, depending on the patient’s anti-AAV antibody level.15,16 Considering their important role, the amount of filled capsids (%Full) is generally regarded as a critical quality attribute for rAAV products.17,18
Besides controlling the full/empty (F/E) ratio, further challenges arising from the biotechnological production of rAAVs include low product titers, complexity of the product, and a lack of scalability of existing methods. Consequently, there is a high demand for suitable analytical tools to evaluate both product quality and efficiency of upstream- and downstream-processing steps. Several methods have been described in this context. Analytical ultracentrifugation (AUC) is widely regarded as the gold standard for precise capsid characterization, with the ability to discriminate between empty, partially filled, and full capsids.19,20 On the downside, it requires long processing times, trained personnel, and is limited to low throughput. More recently, mass photometry was introduced as a potential alternative to differentiate empty, partially, and normally filled capsids of different serotypes, making it a promising tool for the extended characterization of rAAVs.21 The missing ability to perform high-throughput measurements continues to present a bottleneck in process development, however.
The combination of real-time quantitative/(droplet) digital PCR (qPCR or (d)dPCR) and AAV ELISA is a common orthogonal approach to derive the F/E ratio. For this, AAV ELISA determines the capsid titer, representing the amount of all intact viral capsids, based on a calibration curve using commercially available ELISA kits. Due to the narrow calibration range, usually using concentrations between 1 × 108 and 1× 109 capsids/mL (cp/mL),22 several dilution steps are required for highly concentrated samples with a potential impact on the reproducibility of the method. Together with q/(d)dPCR,23 which is used to quantify the genetic payload, this approach is time-consuming and shows rather low accuracy, with reported coefficients of variation up to 36%.17
High-performance liquid chromatography (HPLC)-based methods are increasingly implemented as appealing alternatives due to their proven suitability in the pharmaceutical industry. Among these methods, size exclusion chromatography coupled to multiangle light scattering detection (SEC-MALS) has been widely implemented for titer and F/E ratio estimation,24 with the drawback of a rather time-intensive data evaluation procedure and the need for relatively pure samples and high titers. Alternatively, anion exchange chromatography (AEX) permits the chromatographic separation of empty and full capsids based on the charge differences of the respective capsids. This is commonly combined with UV or fluorescence detection,25 the latter allowing the measurement of low-concentration samples and reducing the sample consumption per measurement. Although the applicability of AEX has been demonstrated for different serotypes, sufficient separation of F/E capsids remains a challenging task and requires relatively long gradients, amounting to total run times of more than 18 min.26,27 Given the restraints of both SEC-MALS and AEX, these methods are of limited availability for high-throughput testing.
To address these drawbacks, an affinity-based HPLC approach is introduced in this study using the POROS CaptureSelect AAVX affinity resin, which reportedly shows high affinity to various rAAV serotypes.28,29 Using self-packed HPLC columns allows both the elution of the rAAV fraction as a single peak and the subsequent column reequilibration within less than 5 min. Combined with intrinsic fluorescence and UV detection (260/280 nm), the method was applied to determine both the capsid titer and the F/E ratio of different serotypes without the need for physical separation of empty and full capsids. By comparing the obtained results with reference ELISA measurements for a sample set containing rAAV8 at different concentrations and %Full as well as by testing other rAAV constructs, the suitability of the method was demonstrated, thus offering a simple and fast method for high-throughput analysis.
Results
Self-packed AAVX affinity column for rAAV analysis
Among different approaches, AAVX affinity chromatography holds great promise as an efficient and versatile method for the initial purification step in downstream processing of a broad range of rAAV serotypes.29 AAVX applications are currently restricted to the (semi-)preparative scale, because commercially available columns are limited to maximum operating pressures of 30–40 bar and feature minimal column volumes (CVs) of at least 200 μL. To be applied for analytical high-throughput purposes, we envisioned the use of short analytical columns that are capable of eluting the rAAV fraction as a single peak and subsequently reequilibrating the column within a few minutes.
To test the feasibility of this approach, a stainless-steel column (2 × 20 mm) was packed with the AAVX resin by using a standalone HPLC pump. With a total column volume of 60 μL, the self-packed column allowed binding of at least 6 × 1011 capsids according to the manufacturer’s specifications.30 Due to the specified pressure limit of the AAVX resin (100 bar) and the relatively low backpressure of the packed column, the flow rate of the method was set to a maximum of 1.5 mL/min, which in turn allowed run times of less than 5 min. Initial experiments using the self-packed column and nonoptimized eluents (eluent A: 25 mM Tris, 150 mM NaCl, 2 mM MgCl2, pH 7.4; eluent B: 100 mM citric acid, 2 mM MgCl2, pH 2.5) indicated elution of the rAAV fraction as a sharp symmetrical peak (data not shown), thus generally confirming the feasibility of our proposed approach.
Optimization of HPLC method
During the initial experiments using the self-packed AAVX column, the following technical challenges emerged, potentially affecting the reliability of the method:
-
(1)
undesired coelution of product-/process-related impurities (e.g., host cell proteins [HCPs], DNA) with the rAAV fraction due to insufficient washing conditions;
-
(2)
incomplete elution of the rAAVs from the column resulting in carryover between runs and thus compromising the ability to reliably quantify rAAVs;
-
(3)
insufficient equilibration time of the column after elution affecting rAAV binding of the next sample; and
-
(4)
interference signals caused by switching from eluent A to eluent B that coincided with the elution peak of the rAAV fraction, thereby compromising data evaluation and increasing the limit of quantification (LOQ).
To address these issues, we optimized the method by adjusting the eluents used, the flow rates, and the resulting run time.
The binding behavior between the target molecule and the affinity ligand used can be generally modulated by altering the eluent composition (e.g., pH, conductivity) during the chromatographic run.31 The binding of rAAVs onto the column is achieved at neutral pH conditions (pH in the range of 6–8) in combination with relatively low salt concentrations, and their elution takes place at low pH and/or high salt molarities.30 Investigation of different eluent compositions revealed that a pH of 2.5 (using glycine and citric acid as buffer system) in combination with 250 mM MgCl2 resulted in the complete elution of the rAAV8 fractions as a single sharp peak (Figure 1A: 1.8 min), thereby eliminating any issues with sample carryover between runs.
Figure 1.
Chromatograms for stock E (7.2 × 1012 cp/mL, empty capsids, blue) and stock F (8.4 × 1012 cp/mL, 46.9%Full, brown)
Fluorescence emission at 330 nm with an excitation at 280 nm (A), including a close-up on peak apices (A1). (B) UV absorbance at 260 nm and (C) at 280 nm. The UV traces show a significant baseline drift during elution (1.65–2.65 min) due to the absorbance of buffer excipients.
Coelution of non-rAAV-related molecules can arise from the interactions of these components with the stationary phase (i.e., support matrix and ligand), the HPLC/column hardware, or the bound rAAVs. To exclude the occurrence of any nonspecific interactions between potential process-related impurities and the column, which could impair the validity of the results, the supernatant of a HEK293 cell culture (lacking rAAV capsids) was injected as a negative control. Because this sample, containing a wide range of different endogenous proteins, yielded no hints for any meaningful interactions with the column in the absence of rAAV particles, 40 CVs of eluent A (corresponding to 1.5 min) proved sufficient to reach baseline level in the fluorescence detector (FLD) trace. For rAAV8 samples spiked with the cell culture supernatant of a mock HEK293 fermentation, a slight increase in the fluorescence signal in the elution peak was observed, which had a moderate (<20%) impact on method accuracy. However, the resulting variability is still in the range of comparable methods. It may be even further reduced by optimizing the washing conditions after column loading, because this effect can presumably be ascribed to promiscuous binding between the loaded rAAVs and the endogenous proteins in the sample.
Furthermore, initial experiments showed a considerable loss in sample binding capacity during consecutive measurements. This led to the assumption that some remnants of the elution buffer affect subsequent measurements. To overcome this issue, 37.5 CV eluent A (1.4 min) was observed to be sufficient for column reequilibration, thus allowing robust binding of rAAVs.
Changing from eluent A to eluent B during elution caused a system-induced interference peak due to pressure fluctuations following the valve switch. This peak coincided with the actual rAAV signal peak and affected peak integration, especially in the UV trace because UV detection was already impaired by low rAAV concentrations and a baseline drift due to the solvents used (see also UV 260 nm and 280 nm traces in Figures 1B and 1C). By increasing the flow rate from 0 to 1.25 mL/min within 9 s after switching to 100% eluent B, it was possible to separate the system-induced signal (Figures 1B and 1C: 1.65 min) from the rAAV signal peak (1.8 min).
Titer determination by using intrinsic fluorescence
In general, protein fluorescence is achieved by exciting tryptophan residues at a wavelength close to 280 nm. With roughly 2% of the overall amino acids in their primary sequence, tryptophan is remarkably abundant in AAVs compared to other proteins. This makes detection via intrinsic fluorescence a particularly attractive approach, especially in view of the relatively low concentrations of rAAV samples. On the downside, tryptophan fluorescence is notoriously sensitive to its environment, because pH, temperature and polarity of the solvent potentially lead to a change in the maximum emission wavelength and intensity.32
Previous experiments using excitation-emission matrix fluorescence spectroscopy (data not shown) on rAAV2 and rAAV8 samples revealed a maximum emission at ∼330 nm. This finding indicates that some tryptophan residues are buried within the protein and shielded from short-distance interactions with the aqueous solvent.33 The assumption is supported by the fact that neither the change of solvents nor the presence of DNA payload showed an impact on the emission wavelength.
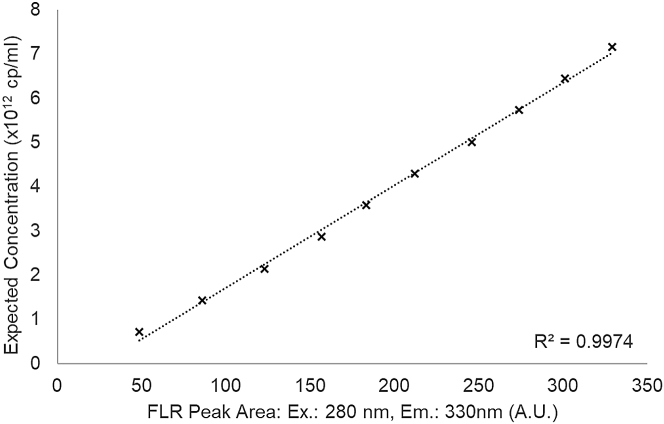
To set the operating range of the affinity HPLC method, the sample injection volume and fluorescence (FLR) sensor photomultiplier tube (PMT) settings were adjusted using a sample of stock E (titer concentration derived by SEC-MALS), such that approximately 90% detector saturation was achieved. The resulting settings (PMT at 9 and an injection volume of 16 μL) were subsequently used for method qualification, which included the measurement of 10 injection amount levels of stock E, spanning from 7 × 1011 to 7 × 1012 cp/mL, both by preparing a manual dilution series and by varying the injection volume in steps of 1.6 μL (Figure S1). For calibration, the expected capsid concentration was plotted against the FLR signal peak area (FLRArea) (Figure 1A) and the coefficient of determination was calculated by linear fitting (Figure 2; coefficient of determination [R2] = 0.997). In both cases, similar results were obtained, rendering them equally suitable for calibration. Moreover, we found that the chosen FLR PMT setting can be applied for measurements within a range of one order of magnitude, with an LOQ of 1.6 × 1010 capsids on column (1 × 1012 cp/mL), representing a typical rAAV titer range after downstream processing. Additional triplicate measurements using different samples of rAAV8 at different injection levels showed high agreement of the associated fluorescence peak area with a relative standard deviation of 1.2%.
Figure 2.
Calibration plot for empty rAAV8 (stock E)
The expected capsid concentration derived by SEC-MALS is plotted against the fluorescence response peak area. Ten samples spanning capsid titers from 7 × 1011 to 7 × 1012 cp/mL were injected and analyzed by affinity HPLC. Slope (A = 2.3 × 1010) and intercept (B = −6.1 × 1011) of the calibration plot were derived from a linear fit.
Using this calibration curve, the estimated capsid titer (titerest.) of a sample was calculated with Equation 1:
| (Equation 1) |
Although we used samples with relatively high rAAV concentrations to confirm the feasibility of the method, we additionally determined the absolute LOQ experimentally. This was achieved by setting the FLR detector PMT to 16 and applying a dilution curve ranging from 3.5 × 109 to 7 × 1010 cp/mL, with an injection volume of 20 μL (Figure S2). By increasing the injection volume (e.g., to 200 μL), the obtained LOQ of 6.1 × 109 cp/mL can be theoretically extended to capsid titers as low as 6.1 × 108 cp/mL. Consequently, the presented method can be applied over a wide rAAV titer range, underlining its suitability for the analysis of in-process controls.
Although the results from this approach were in accordance with reference data for determining the capsid titer in empty rAAV samples, a systematic underestimation of the capsid titer was found for samples containing full virus particles. This effect was investigated in the following section to establish a relationship between the obtained fluorescence signal and the F/E ratio.
Signal correction for genetic payload containing capsids
The estimated capsid titers for both stock E and stock F (containing 46.9 %Full according to SEC-MALS) based on the calibration curve (Figure 2) were compared with the data obtained by SEC-MALS, ELISA, and a bicinchoninic acid (BCA) assay (Table 1).
Table 1.
Comparison of results by different analytical methods
| Method | Sample |
||||||
|---|---|---|---|---|---|---|---|
| Stock E | Stock F | V 01 | V 02 | V 03 | V 04 | V 05 | |
| SEC-MALS | |||||||
| %Full | 0.0 | 46.9 | 46.9a | 0.0a | 13.1a | 36.5a | 25.3a |
| Expected titer (cp/mL × 1012) | 7.2 | 8.4 | 6.1a | 1.4a | 6.3a | 4.8a | 3.3a |
| Affinity HPLC | |||||||
| %Full | 0 | 47 | 46 | NA | 14 | 41 | 24 |
| Estimated titer (cp/mL × 1012) | 7.0 | 5.6 | 4.3 | 1.4 | 6.1 | 3.8 | 2.7 |
| Corrected titer (cp/mL × 1012)b | 7.1 | 8.3 | 6.4 | 1.4 | 6.9 | 5.3 | 3.3 |
| ELISA | |||||||
| Expected titer ( cp/mL × 1012) | 10.0 | 10.0 | 7.3a | 2.0a | 8.4a | 6.0a | 4.2a |
| Obtained titer (cp/mL × 1012) | 9.8 | 9.6 | 6.1 | 2.0 | 7.2 | 6.6 | 4.5 |
| BCA | |||||||
| Protein concentration (μg/mL) | 90.1 | 99.3 | 72.1 | 20.6 | 78.8 | 62.3 | 45.8 |
| Estimated titer (cp/mL × 1012)c | 14.7 | 16.2 | 11.7 | 3.5 | 12.8 | 10.1 | 7.5 |
NA, not applicable.
%Full and expected titers are calculated based on their mixing ratios and the respective analytical results for stocks E and F.
Corrected titers were calculated according to Equation 4.
Protein concentrations in μg/mL were converted into capsid titers by assuming a molecular mass of the rAAV8 of 3.7 MDa.
Whereas SEC-MALS, ELISA, and BCA led to similar or even slightly higher relative capsid titers for stock F compared to stock E (100%–117%), the calculated titerest. using the affinity HPLC method differed significantly (79%). This loss in the expected fluorescence signal was found to linearly correlate with the amount of filled capsids (%Full).
Comparing the chromatograms of stock E and stock F (Figure 1), the elution profiles were highly similar, meaning that both empty and filled rAAVs bound to and eluted from the column in the same manner. Furthermore, no additional peaks occurred during sample loading, column washing, and equilibration, which would indicate the loss of rAAVs. Because the affinity chromatography method has the advantage to elute the rAAVs under steady conditions, the decrease of FLR intensity in filled rAAV capsids can likely be ascribed to the effect of the present DNA on the fluorescence signal. It was therefore decided to determine a correction factor (CF) accounting for the differences in the fluorescence response depending on the F/E ratio. This was achieved by comparing the normalized fluorescence response of stock E and stock F to their respective SEC-MALS references (see supplemental information). Using a Stern-Volmer approach, the fluorescence loss of capsids containing DNA payload was then estimated according to Equation 2:
| (Equation 2) |
By knowing the %Full of a sample and using titerest., it was possible to calculate the corrected capsid titer. This approach was later confirmed using different verification samples during method benchmarking (Table 1).
The use of such a CF was justified by the fluorescence quenching properties of DNA due to molecular interactions with tryptophan residues as well as its propensity to absorb exciting and emitted light, the latter referred to as the inner filter effect (IFE).34,35,36 Thus, although DNA itself contributes only negligibly to intrinsic fluorescence compared to protein,32 there is still the possibility of impairing the measured tryptophan fluorescence. In this regard, the contribution of DNA to fluorescence quenching and/or to an IFE potentially depends on factors such as rAAV serotype, properties of the payload, and the sample matrix and should be confirmed by appropriate control experiments. It is also necessary to control the experimental settings potentially affecting the fluorescence response during the measurement, including differences in buffer composition, pH, or temperature. Finally, the implications of changes regarding the structure and stability of rAAVs during their binding onto stationary phases must be evaluated to exclude a meaningful impact on the fluorescence signal. Thus, we recommend that applying a CF as introduced here should always be supported by sufficient experimental proof to avoid overfitting of the generated data.
UV 260/280 ratio to estimate %Full
Because the %Full of a sample is the basis for using the introduced CF, the UV 260–280 nm ratio () of the elution peak was subsequently leveraged for its approximation.12 In general, DNA and RNA exhibit their maximal absorption at a wavelength of 260 nm, whereas proteins have their maximum at 280 nm. Thus, by acquiring the absorbance values at both wavelengths, the relative amount of DNA/RNA can be determined, which is translatable into the F/E ratio of rAAV particles. However, this method is compromised by its low specificity, because it is incapable of discriminating signals derived from the rAAV capsids and process-related impurities, such as DNA/RNA, HCPs, or other components originating from cell culture harvest and lysis. For this reason, affinity chromatography coupled to UV detection appears as a powerful combination because it enables highly specific binding of the viral vectors while removing potential contaminants.
To test the suitability of measurements for F/E ratio determination in combination with affinity chromatography, we used the full and empty stocks for calibration. After subtracting UV signals of a blank run, was calculated based on the absorbance maximum peak height at each wavelength:
| (Equation 3) |
The obtained were plotted against the %Full derived from reference measurements by SEC-MALS (stock E: 0%, stock F: 46.9%). A linear fit spanning these two points was then used to estimate the %Full of a verification set containing five samples. These samples (V 01–V 05) were prepared with the E and F stocks by mixing them in different ratios and diluting them to different total capsid titers (Table 2).
Table 2.
Preparation of verification samples and their reference SEC-MALS and ELISA values
| Sample | SEC-MALS |
ELISA Titer (cp/mL × 1012) |
Sample composition |
|
|---|---|---|---|---|
| %Full | Titer (cp/mL × 1012) | Stock F; Stock E; Eluent A | ||
| Stock E | 0.0 | 7.2 | 10.0 | NA |
| Stock F | 46.9 | 8.4 | 10.0 | NA |
| V 01 | 46.9a | 6.1a | 7.3a | 0.73 F; 0.27 Eluent A |
| V 02 | 0.0a | 1.4a | 2.0a | 0.20 E; 0.80 Eluent A |
| V 03 | 13.1a | 6.3a | 8.4a | 0.21 F; 0.63 E; 0.16 Eluent A |
| V 04 | 36.5a | 4.8a | 6.0a | 0.45 F; 0.15 E; 0.39 Eluent A |
| V 05 | 25.3a | 3.3a | 4.2a | 0.21 F; 0.21 E; 0.58 Eluent A |
%Full and expected titers are calculated based on their mixing ratios and the respective analytical results for stocks E and F.
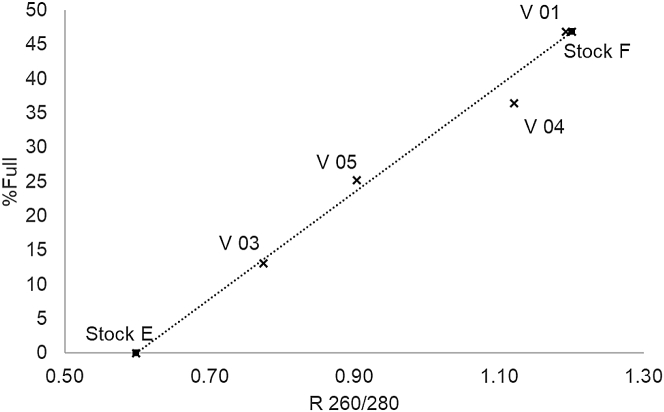
The obtained ratios showed good agreement with the calculated theoretical values (Figure 3). For sample V 02, containing the lowest capsid titer tested (2.2 × 1010 capsids on column), the obtained UV signals were too low to derive . Further experiments using the herein-described setup indicated an LOQ for UV detection at approximately 4.5 × 1010 capsids on column (data not shown).
Figure 3.
Plot of theoretical %Full against the obtained UV 260/280 ratio for verification samples
Linear fit of the two stock solutions was used for method calibration.
Overall, the herein-derived results are consistent with values for measurements reported in the literature.12,24,37 The calculated LOQ may be improved further by using a more sensitive detector, using a higher flow cell volume, or increasing the injection volume. Due to the size and the amount of rAAV particles, light scattering has an influence on the accuracy of UV measurements, and its effect can be minimized by applying scatter correction.37 It is furthermore assumed that varies with the size and composition of the vector DNA and the rAAV serotype of interest. Hence, it is suggested that the effect of different payloads be examined on a case-by-case basis and a calibration curve be prepared for each combination of rAAV serotype and target payload.
By integrating UV acquisition for %Full estimation and using these data for correcting the titer by FLD, we achieved good agreement to SEC-MALS reference measurements. These results were in the following benchmarked against an in-house rAAV capsid ELISA, a well-established technique to achieve titer determination with reasonable sample throughput and costs.
Determination of capsid titers by affinity HPLC and rAAV ELISA
To compare the performance of rAAV ELISA and our affinity HPLC approach, five verification samples (V 01–V 05), composed by mixing stocks E and F at different ratios and capsid concentrations, were analyzed by both methods (Table 1).
As described above, a combination of UV (for obtaining R260/280) and FLR detection was used to estimate the F/E ratio, which was then substituted in Equation 2 to determine the correction factor on the FLR signal. For calibration, stock E (containing only empty capsids) was measured at 10 injections levels and the expected capsid titer (derived from SEC-MALS reference measurements) plotted against the obtained FLR signal (Figure 2). The capsid titer was accordingly calculated (by combining Equations 1 and 2) as follows:
| (Equation 4) |
Capsid determination by AAV ELISA was performed with the commercial Progen AAV8 Titration ELISA using the supplied Kit Control AAV8 (standard) for assay calibration. Due to the different calibration standards deployed for both methods, and consequently, the expected differences in the absolute values, each obtained dataset was normalized with respect to the results for stock E (set to 100%) to allow a relative comparison.
By plotting the expected titer values against the obtained titer for both AAV ELISA (Figure 4A) and AAVX affinity HPLC (Figure 4B) the R2 was calculated. In addition, the root-mean-square error (RMSE) was determined (denoting the square root of the mean square error).
Figure 4.
Comparison of expected and observed capsid titers for ELISA (A) and Affinity HPLC (B)
Titers are displayed in relative values, where the expected titer of stock E is set to 1.0. Assay performance was tested for 5 verification samples, V 01–V 05, and evaluated regarding the R2 and the RMSE.
In regard to both R2 (0.9862 versus 0.9489) and RMSE (4.3% versus 7.1%), the AAVX affinity HPLC method performed slightly better than the AAV ELISA, confirming the suitability of our approach to determine the capsid titer of rAAV samples in a fast and reliable manner. In particular, by preventing any manual sample handling steps and by considerably reducing sample turnaround times, the herein-developed method proved to be a viable alternative for the existing analytical toolbox to support process development. Notably, the columns that we used demonstrated good lifetime, showing no indications of deteriorating performance after at least 350 injections, without the need for any regeneration steps in between. In addition to the analysis of rAAV8, the broader applicability of our method was demonstrated by testing various rAAV constructs, including rAAV2 mGreenLantern (mGL), rAAV8 with a different, not-named (NN), payload, and rAAV9 mGL (Figure S3; Table S1).
The differences between SEC-MALS and the ELISA reference data point at a general problem existing with different methods for the quantification and characterization of gene therapy products: Each method draws conclusions based on different (bio-) chemical or physical properties, leading in some cases to significant discrepancies between the results. Without the availability of appropriate, well-defined reference samples, a quantitative comparison between different analytical techniques remains a major challenge.
Discussion
We introduce here a novel approach for determining the capsid titer and F/E ratio of rAAV samples. By using affinity HPLC, the method enables fast and simple testing of samples across the entire manufacturing process, thereby addressing a major bottleneck for efficient process development of rAAV-based gene therapy products. In contrast to AEX, in which empty and full rAAVs are chromatographically separated, the F/E ratio is here derived from the resulting A260/A280 signals of the rAAV elution peak.
By packing the commercially available POROS CaptureSelect AAVX affinity resin into small analytical columns, the method offers high selectivity toward rAAVs of different serotypes while minimizing the duration of the individual chromatography steps. Besides the resulting total run time of less than 5 min, the absence of any manual sample handling/pretreatment step and the straightforward data evaluation procedure conduce to the short turnaround time of the method, allowing the analysis of over 100 samples per day. Method repeatability was demonstrated by injecting rAAV samples at different concentration levels and varying F/E ratios, leading to highly consistent and robust results between multiple runs.
Although good accuracy and precision for capsid titer determination were achieved in empty capsids using FLD, samples containing full capsids tended to be systematically underestimated. Further experiments revealed a negative correlation between %Full and the obtained capsid content, so that the stock F sample tested in this study (46.9% filled capsids) resulted in a reduction of the fluorescence signal by 21%. Extrapolating this relationship to 100%Full, would lead to a theoretical loss of 40%–50%. It is important to note that this effect originates from the intrinsic properties of FLR detection and is also expected for other chromatography modes such as SEC and ion exchange chromatography. In this regard, the characteristics of our method, leading to the elution of the rAAV fraction under well-controlled conditions, largely exclude a meaningful impact of changing eluent compositions on the FLR response and therefore facilitate data evaluation.
Consequently, a CF was introduced accounting for the %Full dependence of the FLR signal. This CF relies on the F/E ratio of the sample, which was deduced from the A280/A260 ratio obtained by UV detection. Whereas the use of the CF demonstrably improved the reliability of the results for samples containing full capsids, the UV detection squandered the advantages of the more sensitive FLD. It is important to note that this CF must be determined for each rAAV serotype–ssDNA payload combination (Table S1). Moreover, its application is associated with a number of risks due to the dependence of the fluorescence response on factors such as pH, temperature, and structural integrity, and therefore should be verified by sufficient experimental proof.
Overall, the method introduced herein offers a complementary approach for determining the capsid titer based on a calibration curve using reference material with a well-defined composition. Therefore, the choice of the most suitable analytical method for absolute quantification (e.g., SEC-MALS, ELISA and q/(d)dPCR, AUC) has direct implications on the reliability of the method.
Materials and methods
Chemicals and reagents
Tris(hydroxmethyl)-aminoethane (Tris), sodium chloride (NaCl), magnesium chloride hexahydrate (MgCl2× 6H2O), sorbitol, hydrochloric acid (HCl), and citric acid were obtained from Merck (Darmstadt, Germany). Glycine hydrochloride and 10% Pluronic F-68 were purchased from Thermo Fisher Scientific (Waltham, MA).
Mobile phases
Mobile phase A was used for sample dilution, column load, and wash. The buffer contained 25 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% sorbitol, and 0.005% Pluronic F-68 at an adjusted pH of 7.4 using 5 M HCl.
Mobile phase B was used for column elution. It contained 100 mM glycine-HCl, 150 mM citric acid, and 250 mM MgCl2. The pH was adjusted to 2.25 using 5 M NaOH.
All prepared solutions were filtered using 0.22 μm polyethersulfone membrane filters (Millipore Express PLUS, Merck).
Sample preparation
rAAV8 was produced in-house at larger scale and purified using a combination of AAVX affinity chromatography followed by AEX chromatography for further polishing.27,28,38
Concentration determination and preparation of rAAV-containing samples
SEC-MALS
Stock solutions of rAAV empty (stock E) and enriched full (stock F) capsids were characterized using SEC-MALS for the measurement of total capsid titer (cp/mL) and amount of DNA-containing capsids (%Full).24 Analysis was conducted using a Thermo Fisher HPLC Ultimate3000 coupled to a variable wavelength detector (VWD) detector (Thermo Fisher), a DAWN8 MALS detector, and an Optilab dRI detector (Wyatt Technology, Santa Barbara, CA). Samples were loaded onto an SRT SEC-500 column (4.6 × 300 mm) with an additional SRT SEC-500 guard column (4.6 × 50 mm) (Sepax Technologies, Newark, DE) and isocratically eluted using a K3PO4/KCl buffer (pH 6.2) for 30 min. The data evaluation took place using Astra8 (Wyatt Technology). Data normalization and alignment were achieved using BSA and in-house rAAV8 reference material.
ELISA
For method comparison, the samples were evaluated using Progen AAV8 Titration ELISA (Heidelberg, Germany) following the manufacturer’s manual with slight modification. This assay is based on a sandwich ELISA with a biotinylated AAV8 antibody binding to rAAV8 capsids. A streptavidin peroxidase conjugate then reacts with the biotin, resulting in a color reaction proportional to the rAAV8 titer. All of the samples were diluted in 3 steps (1:50,000, 1:100,000, 1:200,000), and each dilution step was measured as a duplicate. The resulting 6 read-outs for each sample were then averaged to determine the final titer. Signal readout was done at 405 nm subtracting a 490-nm background absorbance using the VersaMax Microplate Reader (Molecular Devices, San Jose, CA).
BCA
A BCA Protein Assay (Thermo Fisher) was conducted following the manufacturer’s enhanced protocol using a thermoshaker MKR 13 (Carl Roth GmbH & Co. KG, Karlsruhe, Germany) to keep samples at 60°C for 30 min. For readout at 562 nm, a NanoDrop One (Thermo Fisher) was used.
AAVX-based affinity chromatography
Instrument configuration
Affinity HPLC was performed using an Agilent Technologies 1290 Infinity II LC system (Santa Clara, CA) containing a high-speed binary pump (G7120A), a tempered multisampler set to 10°C (G7167B), a multicolumn thermostat set to 22°C (G7116B), a VWD UV detector (G7114B), and a 1260 Infinity fluorescence detector (G1321B). Because the FLD is more pressure sensitive, it was placed after the UV detector. Detection was achieved using the UV readout set to 280 and 260 nm as well as the FLD with an excitation at 280 nm and emission read at 330 nm. The FLD PMT was adjusted to 9. A 20-μL sample loop was used for sample injection.
Column packing
A 2 × 20 mm C-130B guard column was obtained from Upchurch Scientific (Oak Harbor, WA) and packed with POROS CaptureSelect AAVX affinity resin (Thermo Fisher). Before packing, the resin was treated according to the manufacturer’s manual and reequilibrated in 0.1 M NaCl buffer.
The resin was injected into a reservoir attached to the column inlet. Using a standalone HPLC pump (Agilent, G1311B), the analytical column was packed using a maximum flow rate of 4 mL/min, and the resin was retained at the column outlet. After packing, the reservoir was detached, the excess resin was removed, and the column inlet was closed.
Affinity HPLC measurements
Before measuring the samples, the system was set up as previously described and cleaned with eluent B for 5 min. The column was then equilibrated with eluent A for 30 min at 1.5 mL/min until the temperature and pressure were stable.
If not stated otherwise, for each measurement, 16 μL sample were injected onto the column. To reduce the nonspecific binding of protein, a wash step was implemented using eluent A for 1.6 min at 1.5 mL/min (40 CV). Eluent B was used for sample elution. The gradient used is summarized in Table 3.
Table 3.
Timetable for HPLC method, including the calculated CVs
| Step | Solvent | Total runtime (min) | Flow rate (mL/min) | Volume |
|---|---|---|---|---|
| Injection | Sample | 0 | 16 μL | |
| Wash | Eluent A | 0–1.6 | 1.5 | 40 CV |
| Elution | Eluent B | 1.6–1.75 | 0–1.2 | 1.5 CV |
| 1.75–2.5 | 1.2 | 15 CV | ||
| Equilibration | Eluent A | 2.5–4.0 | 1.5 | 37.5 CV |
Heckel and colleagues present a fast HPLC-based affinity method to determine capsid titer and F/E ratio of rAAV vectors in gene therapy products to support analytics and process development.
A comprehensive description of the capsid titer and CF calculation is provided in the supplemental information.
Data and code availability
Roche Diagnostics GmbH is unable to provide materials, additional datasets, or protocols.
Acknowledgments
We are grateful for valuable discussions and constant support from various members of the laboratories at Roche Diagnostics GmbH in Penzberg, Germany. Special thanks go to Dr. Florian Semmelmann, Helen Seelmann, and Luisa Hilgenfeld (ELISA); Carsten Elger (SEC-MALS); Dr. Thomas Bissinger (sample support); Katrin Heinrich (illustrations), Jasmin Hütten (Graphical Abstract created with biorender.com); and Dr. Aurelia Raducanu and Thomas Vagn Hogg (proofreading).
Author contributions
Conceptualization, J.H., T.G., and A.M.; data curation, J.H.; formal analysis, J.H.; investigation, J.H., T.G., and C.E.; methodology, J.H. and T.G.; project administration, M.L., C.W., and R.R.; resources, M.H.; supervision, M.L. and J. Hubbuch; manuscript draft, J.H. and T.G.; manuscript review and editing, all authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2023.101148.
Supplemental information
References
- 1.Ylä-Herttuala S. Endgame: Glybera Finally Recommended for Approval as the First Gene Therapy Drug in the European Union. Mol. Ther. 2012;20:1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Podsakoff G., Wong K.K., Chatterjee S. Efficient gene transfer into nondividing cells by adeno-associated virus-based vectors. J. Virol. 1994;68:5656–5666. doi: 10.1128/jvi.68.9.5656-5666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flotte T.R., Afione S.A., Zeitlin P.L. Adeno-associated virus vector gene expression occurs in nondividing cells in the absence of vector DNA integration. Am. J. Resp. Cell Mol. 1994;11:517–521. doi: 10.1165/ajrcmb.11.5.7946381. [DOI] [PubMed] [Google Scholar]
- 4.Michelfelder S., Trepel M. Adeno-Associated Viral Vectors and Their Redirection to Cell-Type Specific Receptors. Adv. Genet. 2009;67:29–60. doi: 10.1016/s0065-2660(09)67002-4. [DOI] [PubMed] [Google Scholar]
- 5.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV Serotypes 1–9 Mediated Gene Expression and Tropism in Mice After Systemic Injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 6.Kochergin-Nikitsky K., Belova L., Lavrov A., Smirnikhina S. Tissue and cell-type-specific transduction using rAAV vectors in lung diseases. J. Mol. Med. 2021;99:1057–1071. doi: 10.1007/s00109-021-02086-y. [DOI] [PubMed] [Google Scholar]
- 7.Flotte T.R. Gene Therapy Progress and Prospects: Recombinant adeno-associated virus (rAAV) vectors. Gene Ther. 2004;11:805–810. doi: 10.1038/sj.gt.3302233. [DOI] [PubMed] [Google Scholar]
- 8.Valdmanis P.N., Kay M.A. Future of rAAV Gene Therapy: Platform for RNAi, Gene Editing, and Beyond. Hum. Gene Ther. 2017;28:361–372. doi: 10.1089/hum.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson F.B., Ozer H.L., Hoggan M.D. Structural Proteins of Adenovirus-Associated Virus Type 3. J. Virol. 1971;8:860–863. doi: 10.1128/jvi.8.6.860-863.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Q., Bu W., Bhatia S., Hare J., Somasundaram T., Azzi A., Chapman M.S. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava A., Lusby E.W., Berns K.I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sommer J.M., Smith P.H., Parthasarathy S., Isaacs J., Vijay S., Kieran J., Powell S.K., McClelland A., Wright J.F. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol. Ther. 2003;7:122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 13.Gao K., Li M., Zhong L., Su Q., Li J., Li S., He R., Zhang Y., Hendricks G., Wang J., Gao G. Empty virions in AAV8 vector preparations reduce transduction efficiency and may cause total viral particle dose-limiting side effects. Mol. Ther. Methods Clin. Dev. 2014;1:20139. doi: 10.1038/mtm.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pien G.C., Basner-Tschakarjan E., Hui D.J., Mentlik A.N., Finn J.D., Hasbrouck N.C., Zhou S., Murphy S.L., Maus M.V., Mingozzi F., et al. Capsid antigen presentation flags human hepatocytes for destruction after transduction by adeno-associated viral vectors. J. Clin. Invest. 2009;119:1688–1695. doi: 10.1172/jci36891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mingozzi F., Anguela X.M., Pavani G., Chen Y., Davidson R.J., Hui D.J., Yazicioglu M., Elkouby L., Hinderer C.J., Faella A., et al. Overcoming Preexisting Humoral Immunity to AAV Using Capsid Decoys. Sci. Transl. Med. 2013;5:194ra92. doi: 10.1126/scitranslmed.3005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flotte T.R. Empty Adeno-Associated Virus Capsids: Contaminant or Natural Decoy? Hum. Gene Ther. 2017;28:147–148. doi: 10.1089/hum.2017.29039.trf. [DOI] [PubMed] [Google Scholar]
- 17.Gimpel A.L., Katsikis G., Sha S., Maloney A.J., Hong M.S., Nguyen T.N.T., Wolfrum J., Springs S.L., Sinskey A.J., Manalis S.R., et al. Analytical methods for process and product characterization of recombinant adeno-associated virus-based gene therapies. Mol. Ther. Methods Clin. Dev. 2021;20:740–754. doi: 10.1016/j.omtm.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green E.A., Lee K.H. Analytical methods to characterize recombinant adeno-associated virus vectors and the benefit of standardization and reference materials. Curr. Opin. Biotech. 2021;71:65–76. doi: 10.1016/j.copbio.2021.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burnham B., Nass S., Kong E., Mattingly M., Woodcock D., Song A., Wadsworth S., Cheng S.H., Scaria A., O’Riordan C.R. Analytical Ultracentrifugation as an Approach to Characterize Recombinant Adeno-Associated Viral Vectors. Hum. Gene Ther. Method. 2015;26:228–242. doi: 10.1089/hgtb.2015.048. [DOI] [PubMed] [Google Scholar]
- 20.Maruno T., Ishii K., Torisu T., Uchiyama S. Size Distribution Analysis of the Adeno-Associated Virus Vector by the c(s) Analysis of Band Sedimentation Analytical Ultracentrifugation with Multiwavelength Detection. J. Pharm. Sci. 2023;112:937–946. doi: 10.1016/j.xphs.2022.10.023. [DOI] [PubMed] [Google Scholar]
- 21.Ebberink E.H.T.M., Ruisinger A., Nuebel M., Thomann M., Heck A.J.R. Assessing production variability in empty and filled adeno-associated viruses by single molecule mass analyses. Mol. Ther. Methods Clin. Dev. 2022;27:491–501. doi: 10.1016/j.omtm.2022.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimm D., Kern A., Pawlita M., Ferrari F., Samulski R., Kleinschmidt J. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 1999;6:1322–1330. doi: 10.1038/sj.gt.3300946. [DOI] [PubMed] [Google Scholar]
- 23.Dobnik D., Kogovšek P., Jakomin T., Košir N., Tušek Žnidarič M., Leskovec M., Kaminsky S.M., Mostrom J., Lee H., Ravnikar M. Accurate Quantification and Characterization of Adeno-Associated Viral Vectors. Front. Microbiol. 2019;10:1570. doi: 10.3389/fmicb.2019.01570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIntosh N.L., Berguig G.Y., Karim O.A., Cortesio C.L., De Angelis R., Khan A.A., Gold D., Maga J.A., Bhat V.S. Comprehensive characterization and quantification of adeno associated vectors by size exclusion chromatography and multi angle light scattering. Sci. Rep. 2021;11:3012. doi: 10.1038/s41598-021-82599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khatwani S.L., Pavlova A., Pirot Z. Anion-exchange HPLC assay for separation and quantification of empty and full capsids in multiple adeno-associated virus serotypes. Mol. Ther. Methods Clin. Dev. 2021;21:548–558. doi: 10.1016/j.omtm.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C., Mulagapati S.H.R., Chen Z., Du J., Zhao X., Xi G., Chen L., Linke T., Gao C., Schmelzer A.E., Liu D. Developing an Anion Exchange Chromatography Assay for Determining Empty and Full Capsid Contents in AAV6.2. Mol. Ther. Methods Clin. Dev. 2019;15:257–263. doi: 10.1016/j.omtm.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshi P.R.H., Bernier A., Moço P.D., Schrag J., Chahal P.S., Kamen A. Development of a scalable and robust AEX method for enriched rAAV preparations in genome-containing VCs of serotypes 5, 6, 8, and 9. Mol. Ther. Methods Clin. Dev. 2021;21:341–356. doi: 10.1016/j.omtm.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toueille M., Dejoint L., Attebi E., Cartigny J., Rasle C., Potier S., Rundwasser S., Guianvarc’h L., Lebec C., Hebben M. Development of purification steps for several AAV serotypes using POROS™ CaptureSelect™ AAVX affinity chromatography. Cell Gene Ther. Insights. 2018;4:637–645. doi: 10.18609/cgti.2018.061. [DOI] [Google Scholar]
- 29.Florea M., Nicolaou F., Pacouret S., Zinn E.M., Sanmiguel J., Andres-Mateos E., Unzu C., Wagers A.J., Vandenberghe L.H. High-efficiency purification of divergent AAV serotypes using AAVX affinity chromatography. Mol. Ther. Methods Clin. Dev. 2023;28:146–159. doi: 10.1016/j.omtm.2022.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ScientificTM, T.PorosTM CaptureSelectTM AAVX Affinity.
- 31.Urh M., Simpson D., Zhao K. Affinity Chromatography General Methods. Methods Enzymol. 2009;463:417–438. doi: 10.1016/s0076-6879(09)63026-3. [DOI] [PubMed] [Google Scholar]
- 32.Lakowicz J.R. Springer; 2006. Principles of Fluorescence Spectroscopy; pp. 529–575. [DOI] [Google Scholar]
- 33.Vivian J.T., Callis P.R. Mechanisms of Tryptophan Fluorescence Shifts in Proteins. Biophys. J. 2001;80:2093–2109. doi: 10.1016/s0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arcaya G., Pantoja M.E., Pieber M., Romero C., Tohá J.C. Molecular Interaction of ʟ-Tryptophan with Bases, Ribonucleosides and DNA. Z. Naturforsch. B. 1971;26:1026–1030. doi: 10.1515/znb-1971-1015. [DOI] [PubMed] [Google Scholar]
- 35.Roy S. Fluorescence Quenching Methods to Study Protein–Nucleic Acid Interactions. Methods Enzymol. 2004;379:175–187. doi: 10.1016/s0076-6879(04)79010-2. [DOI] [PubMed] [Google Scholar]
- 36.Brun F., Toulmé J.J., Hélène C. Interactions of aromatic residues of proteins with nucleic acids. Fluorescence studies of the binding of oligopeptides containing tryptophan and tyrosine residues to polynucleotides. Biochemistry-us. 1975;14:558–563. doi: 10.1021/bi00674a015. [DOI] [PubMed] [Google Scholar]
- 37.Werle A.K., Powers T.W., Zobel J.F., Wappelhorst C.N., Jarrold M.F., Lyktey N.A., Sloan C.D.K., Wolf A.J., Adams-Hall S., Baldus P., Runnels H.A. Comparison of analytical techniques to quantitate the capsid content of adeno-associated viral vectors. Mol. Ther. Methods Clin. Dev. 2021;23:254–262. doi: 10.1016/j.omtm.2021.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nass S.A., Mattingly M.A., Woodcock D.A., Burnham B.L., Ardinger J.A., Osmond S.E., Frederick A.M., Scaria A., Cheng S.H., O’Riordan C.R. Universal Method for the Purification of Recombinant AAV Vectors of Differing Serotypes. Mol. Ther. Methods Clin. Dev. 2018;9:33–46. doi: 10.1016/j.omtm.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Roche Diagnostics GmbH is unable to provide materials, additional datasets, or protocols.