Abstract
Predominantly nodal is the most common clinical presentation of peripheral T- (and NK-) cell lymphomas (PTCL), which comprise three main groups of diseases: (i) systemic anaplastic large cell lymphomas (ALCL), whether positive or negative for anaplastic lymphoma kinase (ALK); (ii) follicular helper T-cell lymphomas (TFHL); and (iii) PTCL, not otherwise specified (NOS). Recent advances in the genomic and molecular characterization of PTCL, with enhanced understanding of pathobiology, have translated into significant updates in the latest 2022 classifications of lymphomas. ALK-negative ALCL is now recognized to be genetically heterogeneous, with identification of DUSP22 rearrangements in approximately 20-30% of cases, correlated with distinctive pathological and biological features. The notion of cell-of-origin as an important determinant of the classification of nodal PTCL is best exemplified by TFHL, considered as one disease or a group of related entities, sharing oncogenic pathways with frequent recurrent epigenetic mutations as well as a relationship to clonal hematopoiesis. Data are emerging to support that a similar cell-of-origin concept might be relevant to characterize meaningful subgroups within PTCL, NOS, based on cytotoxic and/or Th1 versus Th2 signatures. The small group of primary nodal Epstein-Barr virus-positive lymphomas of T- or NK-cell derivation, formerly considered PTCL, NOS, is now classified separately, due to distinctive features, and notably an aggressive course. This review summarizes current knowledge of the pathology and biology of nodal-based PTCL entities, with an emphasis on recent findings and underlying oncogenic mechanisms.
Introduction
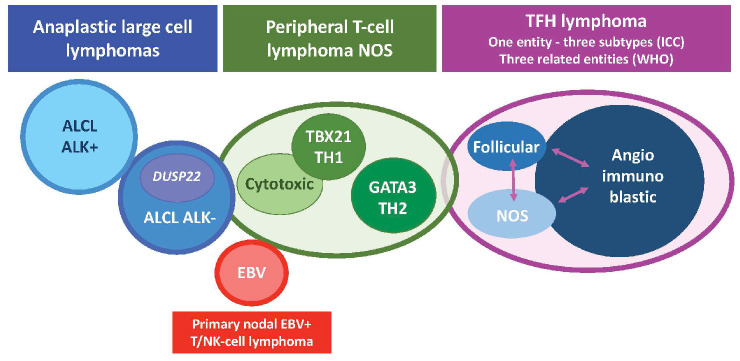
Peripheral T-cell lymphomas (PTCL) collectively refer to neoplasms of mature NK or T cells. They constitute approximately 10% of all lymphomas in the Western world and up to 20% in Asia. PTCL exhibit significant clinical and biological diversity, leading to the recognition of over 30 distinct entities in the latest classification proposals.1,2 These entities can be categorized into three groups based on their typical clinical presentation: involvement of lymph nodes, skin and other extranodal organs, or dissemination/leukemic manifestations. This clinical grouping reflects, to some extent, the distinct cell of origin for each entity, characterized by specific functional, homing, or trafficking properties, as well as varying oncogenic mechanisms. The nodal-based PTCL entities represent the large majority of PTCL cases. They include anaplastic large cell lymphomas (ALCL), categorized as positive or negative for anaplastic lymphoma kinase (ALK+ and ALK–, respectively), lymphomas originating from T-follicular helper (TFH) cells, primary nodal Epstein-Barr virus (EBV)-associated PTCL, and PTCL, not otherwise specified (PTCL, NOS) (Figure 1, Tables 1 and 2).
The two proposed classifications emanated in 2022, the International Consensus Classification (ICC) of mature lymphoid neoplasms1 and the 5th edition of the World Health Organization classification of lymphoid neoplasms (WHO-HAEM5),2 represent updates of the former 2017 revised 4th edition of the World Health Organization classification (WHO-HAEM4R),3 and both maintain the principle of a multiparametric definition of lymphoma entities adopted since 1994. Morphology, immunophenotype, genetic and clinical features as well as the putative normal cellular counterpart are all taken into account for the classification. Research advances elucidating the genomic landscape and molecular characterization of many PTCL, in addition to enhanced understanding of the pathobiology and pathogenesis with information derived from both clinical and experimental models, have translated into significant updates in both classifications. The main adjustments and changes introduced in both proposals reflect similar conceptual shifts. Here, we present a review of our current knowledge of the pathology and biology of nodal-based PTCL, the understanding of oncogenic mechanisms, and discuss future directions.
Figure 1.
Representation of nodal T- and NK-cell lymphoma entities according to the 2022 classifications. NOS: not otherwise specified; TFH: T follicular helper cell; ICC: International Consensus Classification; WHO: World Health Organization; ALCL: anaplastic large cell lymphoma; ALK: anaplastic lymphoma kinase; EBV: Epstein-Barr virus.
Anaplastic large cell lymphomas
ALCL encompass four entities having in common large pleomorphic tumor cells with strong expression of CD30, but being distinct in their pathogenesis, clinical presentation and outcome. The two systemic entities, ALK+ and ALK– ALCL, most often present as nodal-based disease and are considered nodal PTCL, but may show the involvement of a variety of extranodal sites. The other two ALCL entities are extranodal and site-specific. Although not a nodal PTCL entity, breast implant-associated (BIA)-ALCL is described here to highlight both overlapping and distinguishing features compared with ALK– ALCL, and the fact that it may disseminate to lymph nodes.4 Primary cutaneous ALCL (ALK–), classified with other primary cutaneous CD30+ T-cell lymphoproliferative disorders, is not included in this review.1,2
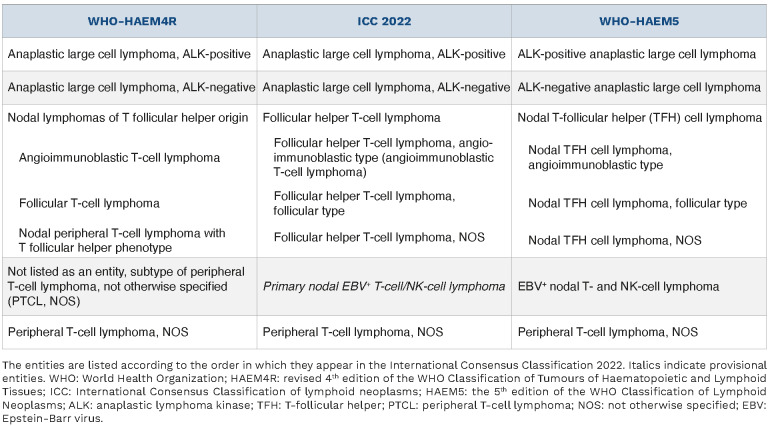
The classification and diagnostic criteria of ALCL entities is essentially identical in the two 2022 classifications (Table 1). Provisional in WHO-HAEM4R, BIA-ALCL is now a definitive entity. Among ALK– ALCL, DUSP22-rearranged (DUSP22-R) cases constitute a distinct genetic subtype according to the ICC 2022, but not in WHO-HAEM5, as discussed below.1,2
Anaplastic large cell lymphomas, ALK-positive
In ALK+ ALCL, there is aberrant expression of the ALK protein secondary to rearrangements involving the ALK gene.3 ALK+ ALCL accounts for 7-9% of (non-cutaneous) PTCL worldwide,5,6 most commonly affects young male patients (median age, 30-35 years), but may occur at any age including in children.7,8 ALK+ ALCL usually presents at an advanced stage, and frequently involves lymph nodes and/or various extranodal sites, including bone, skin and lung.7 ,8 Rare cases, mostly of the small cell variant, present as leukemia.9 ALK+ ALCL has the most favorable prognosis among systemic PTCL, with a long-term overall survival reaching 70-90% in adults and up to 95% in children. This is in part related to the younger age at diagnosis.6-8
ALK+ ALCL comprises several morphological variants,10 all containing “hallmark cells”, with a large, often kidney-shaped nucleus, abundant cytoplasm and a prominent Golgi zone, although in variable proportions.3 In the classic form (common pattern) (Figure 2A-G), hallmark cells are numerous and form cohesive sheets. Nodal infiltration typically shows an intrasinusoidal growth pattern. The small cell and lymphohistiocytic patterns, which are characterized by smaller hallmark cells with a preferential perivascular distribution and a dominant infiltrate of reactive histiocytes, respectively,3 are often encountered together, and have been associated with an adverse prognosis in the pediatric population, but not in adults.11 A rare pattern mimicking nodular sclerosis classic Hodgkin lymphoma (Hodgkin-like pattern) may be diagnostically challenging.12
CD30 expression is strong and diffuse in the classic and Hodgkin-like variants, but may be more focal in the other patterns.3,10 Chimeric ALK protein is by definition expressed, but its subcellular location depends on the translocation partner. The most common t(2;5)(p23;q35) translocation (80% of the cases) results in a NPM1::ALK fusion that shuttles between the nucleus and the cytoplasm, due to the preserved N-terminal portion of NPM1, and is detected by ALK immunohistochemistry in both compartments.13 With alternative partners, ALK staining may be purely cytoplasmic (e.g., ATIC::ALK), membranous (MSN::ALK), or both (TPM3::ALK).14,15 Epithelial membrane antigen (EMA) is typically positive and EBV is always negative. The neoplastic cells often show loss of T-cell receptor (TCR) molecules (“null” immunophenotype) and of several T-cell antigens (typically CD3, CD5 and CD7 commonly negative), while CD2, CD4 and CD43 are more frequently preserved. CD8 is usually negative. Cytotoxic markers (granzyme B, perforin, T-cell intracellular antigen 1 [TIA-1]) are generally expressed.3
Table 1.
The ALK fusion proteins have constitutive tyrosine kinase activity which activates multiple signaling cascades involved in oncogenesis, including the JAK-STAT3 pathway.15,16 Accordingly, nuclear phospho-STAT3 (pSTAT3) is detected by immunohistochemistry in nearly all cases.16,17 In relation to the crucial driver role of ALK fusions, ALK inhibitors have demonstrated high response rates in pediatric and young adult patients with relapsed/refractory ALK+ ALCL.8 Nevertheless, as for ALK-rearranged lung carcinomas, mechanisms of resistance may develop, including acquired ALK mutations.18
Mutational profiling of ALK+ ALCL has revealed NOTCH1 mutations in 20% of the cases. NOTCH1 expression is also induced via STAT3 activation mediated by the ALK fusion protein. In line with these observations, inhibition of the NOTCH1 pathway by γ-secretase inhibitors is effective in vitro and a promising treatment target to be further explored.19 Other recurrently mutated genes in ALK+ ALCL include LRP1B (19%) and TP53 (11%), as well as epigenetic modifier genes such as EP300, KMT2C and KMT2D. TP53 mutations have been associated with an inferior outcome.20
Anaplastic large cell lymphomas, ALK-negative
ALK– ALCL account for 6-15% of PTCL5,6,21 and occur later in life than ALK+ ALCL (median age, 54 years), with a slight male predominance.21 Most patients present with advanced stage disease, and extranodal sites of involvement (50%) include lung, liver and bone.7,22 The 5-year overall survival is, as a whole, worse (∼50% with CHOP-like chemotherapy) for ALK– ALCL patients than for ALK+ patients.7,21
The diagnosis of ALK– ALCL requires typical morphology, with hallmark cells identical to the classic form of ALK+ ALCL, uniformly strong CD30 expression, negativity for ALK expression or ALK rearrangement, and absence of EBV. Tumor cells frequently show loss of T-cell antigens, EMA expression and a cytotoxic phenotype, although not as consistently as observed in ALK+ ALCL.3 Approximately half of all ALK– ALCL express nuclear pSTAT3, as a consequence of translocations or mutations that lead to constitutive JAK-STAT3 activation, including JAK1 and/or STAT3 mutations in 20-30% of the samples.20,22-24 TP53 mutations are observed in 23% of the cases, and an inferior outcome has been reported for ALK– ALCL with TP53 or STAT3 mutations.20
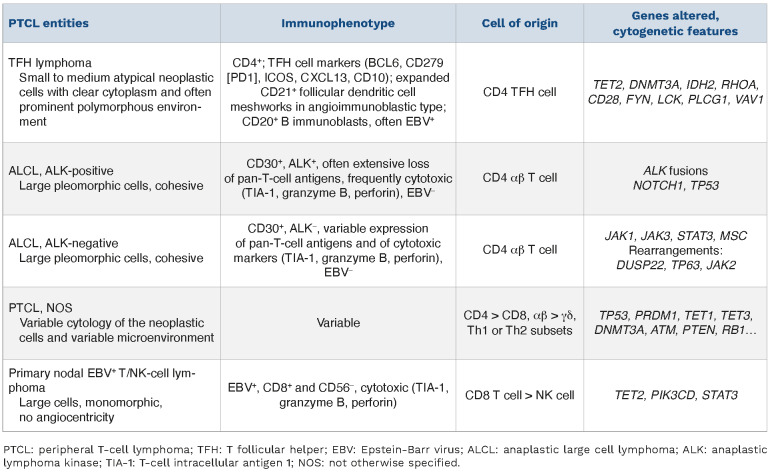
Table 2.
Usual features of the main nodal peripheral T-cell lymphoma entities.
Beyond these shared features, the last decade has brought new data that highlight the genetic heterogeneity of ALK– ALCL, with the identification of recurrent structural aberrations, some of which may influence the clinical outcome. The largest genetic subgroup (20-30% of ALK– ALCL) is characterized by the presence of a rearrangement in the DUSP22 gene,22,25,26 which induces the downregulation of dual specificity phosphatase, thus preventing its inhibitory effect on various signaling pathways. DUSP22-R ALK– ALCL have distinctive biological features, including lack of JAK-STAT3 activation, widespread DNA hypomethylation, expression of the cancer-testis antigens and an immunogenic phenotype.24 One-third of cases have a musculin (MSC) hotspot mutation (E116K), which promotes the CD30–IRF4–MYC axis.27 DUSP22-R ALK– ALCL (Figure 2H-N) frequently demonstrate doughnut-shaped nuclei and, compared to DUSP22 non-rearranged (NR) cases, are more frequently CD3+ but less commonly express EMA and cytotoxic molecules.22,25,26 Conversely, strong and uniform nuclear expression of lymphoid enhancer binding factor 1 (LEF1) is highly associated with DUSP22-R.28 Clinically, the DUSP22-R subgroup has been the subject of a number of recent studies with somewhat disparate results (5-year overall survival: 40%-100%; 5-year progression-free survival: 40-57%), but, although study sizes still remain small, collectively it appears to have a more favorable prognosis, intermediate between ALK+ ALCL and DUSP22-NR ALK– ALCL.22,25,26,29 Consequently, DUSP22-R ALCL is now considered a genetic subtype among ALK– ALCL in the ICC 2022 – which recommends fluorescence in situ hybridization testing if available –, but not in the WHO-HAEM5 classification due to uncertainties around prognosis.1,2
The presence of a TP63-R characterizes a smaller genetic subgroup of ALK– ALCL, accounting for 2-8% of cases and associated with an adverse prognosis.25,26,30 A few cases with JAK2-R have been described, featuring Reed-Stern-berg-like cells in an inflammatory background with eosinophilia, reminiscent of classic Hodgkin lymphoma.31 A Hodgkin-like morphology has also been observed in a subset of cases expressing truncated ERBB4 transcripts.32
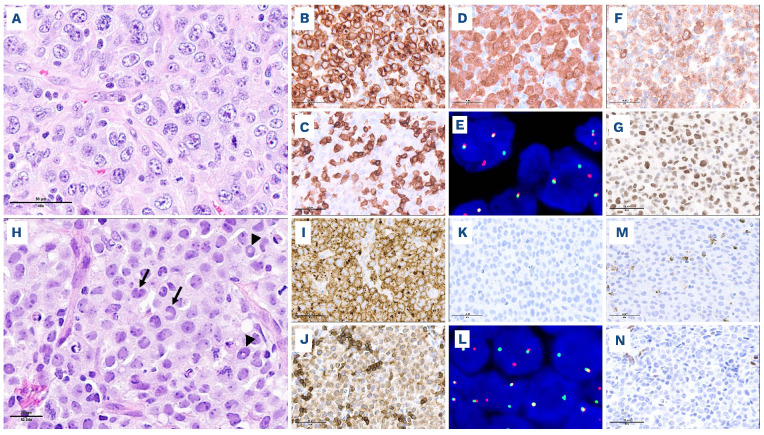
Figure 2.
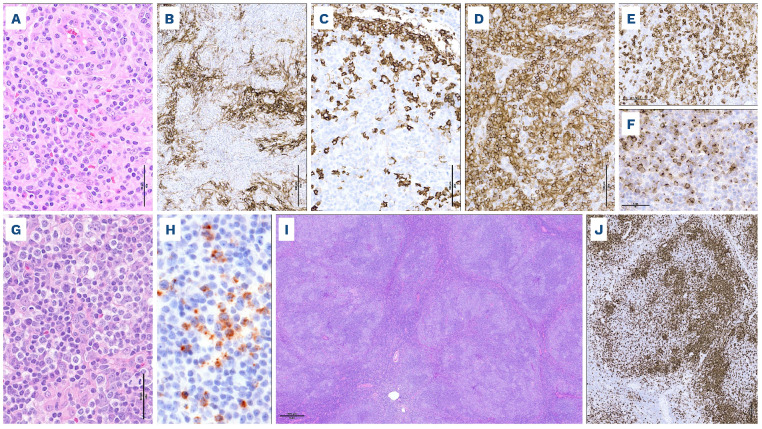
Nodal involvement by systemic anaplastic large cell lymphoma. (A-G) Anaplastic lymphoma kinase (ALK)-positive anaplastic large cell lymphoma (ALCL), classic pattern. This case comprises cohesive sheets of large pleomorphic cells (A, hematoxylin & eosin), shows diffuse strong expression of CD30 (B) and loss of several T-cell antigens including CD3 (C). Nuclear and cytoplasmic expression of ALK protein (D) is indicative of a NPM1::ALK fusion. An ALK gene rearrangement can be confirmed by break-apart fluorescence in situ hybridization (FISH) (E, the rearranged ALK allele is represented by split red and green signals). The cells are positive for cytotoxic markers (F, perforin) and nuclear phospho-STAT3 (G). (H-N) ALK-negative ALCL with DUSP22 rearrangement. This case comprises cohesive sheets of neoplastic cells, including kidney-shaped hallmark cells (H, hematoxylin & eosin, arrows) and cells with doughnut-shaped nuclei (H, arrowheads). The cells are strongly CD30-positive (I), weakly positive for CD3 (J), and ALK-negative (K). Break-apart FISH shows a DUSP22 locus rearrangement (L, the rearranged DUSP22 allele is represented by split red and green signals). The cells are negative for cytotoxic markers (M, T-cell intracellular antigen 1 [TIA-1]) and phospho-STAT3 (N).
Other gene fusions reported in ALK– ALCL involve the tyrosine kinase domain of FRK, ROS1 or TYK2.23,33 Nevertheless, these cases represent very small and heterogeneous subsets of patients, and further data are needed before stratifying DUSP22-NR ALK– ALCL into further genetic subtypes.
Breast implant-associated anaplastic large cell lymphoma
BIA-ALCL is a rare complication of textured breast implants, with a median of 9 years from implantation to diagnosis, and generally an excellent prognosis.34 Recent data suggest that germline BRCA1 or BRCA2 mutations represent a risk factor for developing BIA-ALCL.35
Most often, BIA-ALCL occurs as a peri-prosthetic effusion and is diagnosed on cytological samples.36 In surgical capsulectomies, lymphoma cell deposits are found along the inner surface of the capsule. In the infiltrative forms, tumor cells extend into the capsule and rarely may form a mass.36 Regional lymph node involvement may occur, typically with low tumor burden with sinusoidal and perifollicular growth patterns.37
Although BIA-ALCL resembles systemic ALK– ALCL morphologically and immunophenotypically,36 it develops in a peculiar confined microenvironment, in which bacterial infection, chronic inflammation and hypoxia are believed to be distinctive pathogenic determinants.38 The genetic aberrations acquired in this setting and transformation mechanisms partially overlap with those of systemic ALK– ALCL. The JAK-STAT3 pathway appears to be consistently activated, either by mutations (in 60-90% of cases, most frequently STAT3 and JAK1) or by other mechanisms, as witnessed by the constant pSTAT3 expression.39 In contrast, DUSP22 and TP63 rearrangements have not been found in BIA-ALCL.34,40 Furthermore, loss of 20q13.13, detected by shallow whole-genome sequencing in 66% of cases in one series, is reportedly highly characteristic of BIA-ALCL compared to systemic ALK+ or ALK– ALCL.38 Other recurrent alterations involve epigenetic modifiers, with frequent mutations in KMT2C, CHD2, CREBBP and KMT2D (up to 75% of cases), while TET2 and DNMT3A mutations, typically observed in TFH lymphomas, are uncommonly detected.39 Altogether, the unique clinical association, peculiar pathogenic background and distinctive pathobiological features of BIA-ALCL support its classification as a separate entity.
Follicular helper T-cell lymphoma
T follicular helper (TFH) cells represent a functional subset of CD4+ lymphocytes that are necessary for the formation and maintenance of germinal centers, interacting with germinal center B cells and aiding their differentiation.41 In 2007, immunophenotyping and gene expression profiling analyses identified the TFH cell as the cell of origin of angioimmunoblastic T-cell lymphoma (AITL),42 and this specific cellular derivation has become a cardinal defining feature of the disease. In practice, a TFH phenotype is evaluated by immunohistochemistry by the expression of PD1, ICOS, CXCL13, CD10 and BCL6, which are markers of normal TFH cells, with at least any two positive required for the definition.3 In 2017, the developing concept that TFH cell derivation represents a unifying feature of a larger group of nodal CD4+ T-cell lymphomas led to the creation of an umbrella term “nodal T-cell lymphoma of T follicular helper origin” to encompass AITL, follicular T-cell lymphoma, and nodal PTCL with a TFH phenotype.3
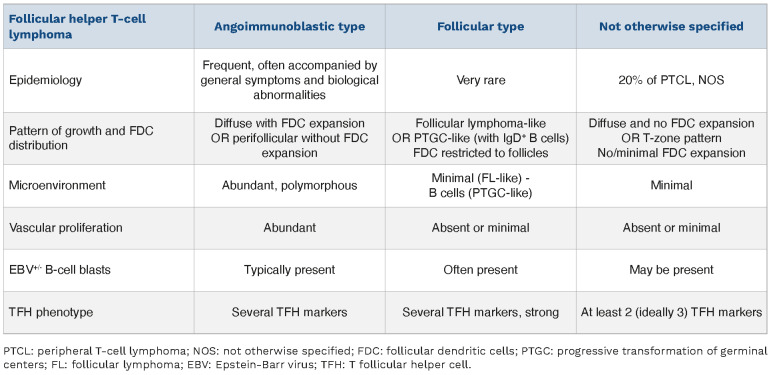
The grouping was supported by the fact that these three entities also share a similar genetic landscape, characterized by recurrent mutations in epigenetic modifier genes, RHOA and other TCR signaling genes.42-48 Identification of a TFH phenotype might also be relevant to treatment decisions.4,49 Therefore the ICC considers one single disease entity, namely follicular helper T-cell lymphoma, with three subtypes, angioimmunoblastic, follicular and NOS (Table 3).50 This entity by definition applies to CD4+ PTCL, implies significant expression of TFH markers, and excludes primary cutaneous CD4+ T-cell lymphoproliferative disorders with a TFH phenotype. The WHO-HAEM5 proposal considers a family of three related entities of nodal T-follicular helper cell lymphomas, angioimmunoblastic-type, follicular-type and NOS types. Collectively, TFH lymphoma(s) outnumber(s) PTCL, NOS in recent epidemiological studies.5,51,52
Follicular helper T-cell lymphoma, angioimmunoblastic type
AITL is the prototypic and most frequent subtype of TFH lymphoma.42 It manifests as a systemic disease in adults, usually in the elderly. Patients present with generalized lymphadenopathy, often with extranodal involvement (e.g., skin, tonsil, liver, spleen,...), systemic symptoms, and various immune abnormalities. The median survival is <3 years, but a subset of patients experience long-term survival.53
Histologically (Figure 3A-F) (for a review, see de Leval et al.54), AITL comprises a polymorphous infiltrate including variable proportions of neoplastic T cells, typically outnumbered by reactive small lymphocytes, histiocytes, im munoblasts, eosinophils and plasma cells. The neoplastic lymphoid cells are usually small to medium-sized, with moderately abundant, clear cytoplasm, but may lack significant atypia. Some cases may contain a larger proportion of neoplastic cells or large cells with abundant, clear cytoplasm. Large B-cell immunoblasts, sometimes resembling Hodgkin and Reed-Sternberg cells, represent a typical component of AITL; they are usually scattered, but sometimes numerous. Some cases rich in histiocytes may resemble lymphoepithelioid (Lennert) lymphoma. Plasma cells may be abundant, and some patients may even present with peripheral blood plasmacytosis. A stromal component is typically present, consisting of a marked proliferation of arborizing high endothelial venules, and abnormal extrafollicular expansion of follicular dendritic cells (FDC) best demonstrated by CD21 or CD23 immunostaining. The lymphoproliferation is usually diffuse (pattern III). In less common instances AITL is associated with hyperplastic or regressive follicles (patterns I and II). Transitions between different patterns have been observed in consecutive biopsies, and a combination of patterns may be seen at one time. Patterns I and II correspond to lower tumor cell burden reflecting partial nodal involvement, but do not correlate with lower stage disease.54-56 Two studies found that a subset of AITL enriched in a B-cell signature was associated with a better outcome,57,58 and interestingly the favorable significance of a B-cell signature was also found in PTCL, NOS in another study.59
Table 3.
Comparison of the three subtypes of follicular helper T-cell lymphoma.
The neoplastic cells of AITL consist of mature CD3+ CD4+ CD8– TCRαβ+ cells. An aberrant T-cell immunophenotype (most commonly reduced or absent surface CD3, CD5 or CD7) is frequently observed, especially by flow cytometry. The neoplastic cells express several TFH markers, including the CXCL13 chemokine; PD1 (CD279), ICOS, CD10 and CD200 membrane receptors; and BCL6 and cMAF transcription factors.60,61 Overall, PD1 and ICOS are more sensitive for identifying the neoplastic TFH cells than CXCL13 or CD10, which are more specific.56 Aberrant co-expression of CD20 and/or partial expression of CD30 by the neoplastic cells is not unusual.62,63 Reactive CD8+ cells are variably abundant and may outnumber the CD4+ neoplastic cells.64 The large B-cell immunoblasts are positive for CD20, PAX5 and CD79a, often CD30, and may also sometimes co-express CD15. They are usually, but not always, infected by EBV (positive for EBV-encoded RNA [EBER] and latent membrane protein 1 [LMP-1]).65 The spectrum of B-cell-derived expansions in AITL also comprises EBV– large B-cell proliferations, and polytypic (or sometimes monotypic) plasma cell expansions. Up to one-third of cases, particularly those with an increased number of B cells, harbor (oligo)clonal rearrangements of the immunoglobulin genes (IG), in addition to monoclonal or oligoclonal T-cell receptor gene (TR) rearrangements. Some patients develop secondary large B-cell lymphomas.66
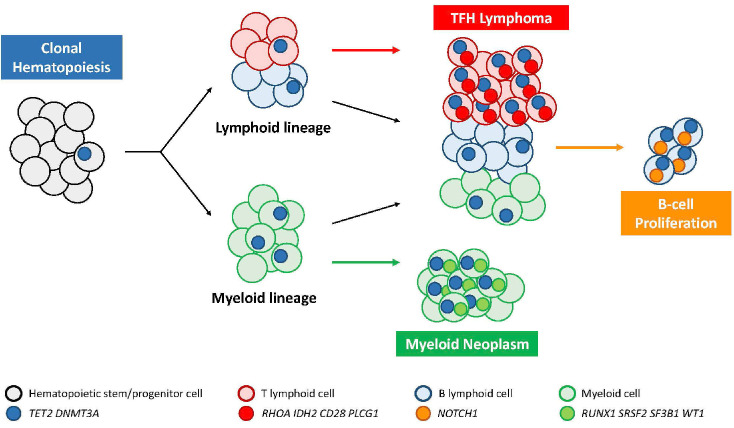
Most cases demonstrate a characteristic mutational landscape that recapitulates a multi-step oncogenic process derived from underlying clonal hematopoiesis (Figure 4).67 The profile of AITL alterations typically consists of epigenetic deregulation (TET2 +/- DNMT3A mutations, occurring at early stages in hematopoietic progenitors, present in about 80% and 30-35% of the cases, respectively),44,46 and second-hit mutations with a more restricted distribution.68 TET2 and DNMT3A mutations are inactivating, TET2 mutations are often multiple (2 or 3), and alterations in both genes tend to co-occur. Their global effect is DNA hypermethylation. DNMT3A mutations were found in one study associated with a shorter progression-free survival,69 and in an experimental mouse model of AITL generated by TET2 inactivation and RHOA G17V, addition of DNMT3A R882H accelerated the development of the disease.70 Second-hit mutations include a hotspot RHOA G17V mutation encoding a dominant negative variant of the protein in up to 70% of cases, and other gain-of-function mutations targeting the TCR pathway (PLCG1, CD28, FYN, PIK3 components, CARD11, etc.).45,47,71 RNA fusions involving CD28 with ICOS or rarely CTLA4, mutually exclusive with CD28 mutations, are detected in a small subset of patients.72 Mouse models have shown that RHOA G17V induces TFH differentiation and autoimmunity, and promotes lymphomagenesis in the presence of TET2 inactivation, indicating the synergistic effect of both mutations.73-75 AITL harboring the RHOA G17V mutation tend to have higher microvessel density, more FDC proliferation and a more pronounced TFH immunophenotype compared to wild-type cases, but no prognostic significance was observed.76,77 One study showed that sensitive RHOA G17V mutation analysis may be valuable for the early diagnosis of TFH lymphoma, as this alteration may be found prior to conclusive histopathological changes.78 IDH2 point mutations at the R172 residue, present in about one-third of cases,79,80 modify IDH2 enzymatic activity resulting in the production of an oncometabolite (2 hydroxyglutarate) ultimately altering DNA and histone methylation.80,81 IDH2-mutated AITL has a characteristic morphology with prominent medium-sized to large clear cells (Figure 3G-H), and tends to show strong CD10 and CXCL13 expression.82 In an AITL mouse model driven by IDH2 and TET2 mutations, the tumors show abundant angiogenesis and plasma cells, and the malignant TFH cells display aberrant transcriptomic and epigenetic programs that impair TCR signaling and alter cross-talk with germinal center B cells, promoting B-cell clonal expansion while decreasing the Fas-FasL interaction and reducing B-cell apoptosis.83 Gains of chromosomes 5 and 21 are frequent, especially in IDH2-mutated cases, and copy number losses in genes regulating the PI3K-AKT-mTOR pathway are enriched in IDH2-wild-type cases.84
Figure 3.
Follicular helper T-cell lymphoma. (A-H) Follicular helper T-cell (TFH) lymphoma, angioimmunoblastic type comprises a polymorphous cellular infiltrate and prominent venules (A, hematoxylin & eosin); immunostains show an irregular proliferation of follicular dendritic cell meshworks (B, CD21), aggregates of small B cells and scattered large blasts positive for CD20 (C), a diffuse infiltrate of CD4+ cells (D), which are positive for TFH markers (E, PD1 and F, CXCL13). In IDH2-mutated cases (G, H), large cells with abundant clear cytoplasm are prominent (G, hematoxylin & eosin), and expression of the IDH2 R172K variant is highlighted by immunohistochemistry (H). (I, J) TFH lymphoma, follicular type is characterized by large nodules resembling progressive transformation of germinal centers at low magnification (I, hematoxylin & eosin), and TFH markers demonstrate aggregates of pale neoplastic cells (J, ICOS).
Detailed genetic analyses of AITL have shown that these lymphomas may contain one or multiple clonal TR gene rearrangements associated with the same TET2 mutation(s), indicating parallel neoplastic evolution from a common TET2-mutant hematopoietic progenitor pool. A biased TRBV usage was also found, suggesting the role of antigenic stimulation in promoting T cells to clonal expansion and malignant transformation.85 Moreover, the study of microdissected T- and B-cell populations has shown that TET2 mutations of AITL clones are frequently present in the B cells as well, indicating that clonal hematopoiesis also generates a large population of mutated mature B cells and, while RHOA and IDH2 mutations are confined to the T-cell compartment, other mutations, notably in NOTCH1, may be identified in the B cells, providing an explanation for the frequently associated B-cell expansions in AITL (Figure 4).86,87 Clonal hematopoiesis appears to be the source of the observed association with myeloid neoplasms in certain patients88 and the increased incidence of myeloid neoplasms after TFH lymphoma-directed therapy, through divergent clonal evolution (Figure 4).4,67
Follicular helper T-cell lymphoma, follicular type
This least common subtype of TFH lymphoma presents either a truly follicular pattern, mimicking follicular lymphoma (FL-like), or more commonly a pattern resembling progressive transformation of germinal centers (PTGC-like). In FL-like cases, nodular aggregates of neoplastic cells are sustained by a meshwork of FDC. In PTGC-like cases (Figure 3I, J), pale aggregates of medium-sized atypical T cells are distributed within expanded mantle zones in large nodules mostly composed of small IgD+ B cells.43,89 TFH lymphoma, follicular type lacks both the extrafollicular expansion of FDC and the proliferation of high endothelial venules characteristic of AITL. The clinical presenting features overlap with those of AITL.89,90 A subset of patients has long-term survival despite sometimes multiple relapses and the prognosis might be slightly better than that of AITL.89,90
The neoplastic cells are CD3+ CD4+ and usually show extensive positivity for most TFH markers (PD1, ICOS, CXCL13, BCL6, CD10, and sometimes CD57).89,91 In one study most cases had at least partial expression of CD30 in the neo-plastic cells.91 A component of large blastic EBV+ or EBV– B cells is often identified, frequently with Reed-Sternberglike morphology and immunophenotype.65,89,91-93
Figure 4.
Oncogenic model of follicular helper T-cell lymphoma in relationship to clonal hematopoiesis. TFH: follicular helper T-cell
The t(5;9)(q33;q22) translocation, resulting in an ITK::SYK fusion, is found in about 20% of the follicular type, and has been reported thus far in only one case of typical AITL.89,94 Based on limited data, the mutational pattern of follicular PTCL appears otherwise to overlap with that of AITL.48
Follicular helper T-cell lymphoma, not otherwise specified
TFH lymphoma, NOS, formerly nodal T-cell lymphoma with a T follicular helper phenotype,3 encompasses cases without specific pathological features, but showing imprints of the TFH signature and/or expression of TFH markers, and/or cases exhibiting some characteristics of AITL (e.g., increased vascularity, presence of EBV+ B-blasts).42,44 According to the WHO-HAEM4R, qualification for a TFH lymphoma required the expression of at least two or ideally three TFH markers (among the 5 recommended for routine testing: PD1, ICOS, CD10, BCL6, CXCL13) by the neoplastic cells, in addition to CD4.3 This criterion is retained in the current proposals (Table 3). Some cases show perifollicular involvement and may mimic marginal zone lymphoma.56 A subset of cases may present as what was previously called the “T-zone variant” of PTCL, NOS, in which there is preserved architecture with residual sometimes hyperplastic B-cell follicles, and interfollicular lymphomatous involvement.95 Since FDC proliferation is generally considered as a typical hallmark of AITL, cases with some FDC expansion are better qualified as tumor-cellrich AITL, but the border between PTCL-TFH and AITL is not well delineated, likely reflecting a biological continuum.48,56 The genetic background of TFH lymphoma, NOS overlaps with that of AITL, including frequent mutations in TET2 and DNMT3A but less frequent RHOA mutations and infrequent IDH2 mutations.44-47,58 According to limited data available, the outcome related to TFH lymphoma, NOS, is similar to that of TFH lymphoma of the angioimmunoblastic type but larger studies are needed.48,96
Primary nodal Epstein-Barr viruspositive T/NK-cell lymphoma
Primary EBV+ nodal T-cell or NK-cell lymphoma was introduced in the WHO-HAEM4R as a variant of PTCL, NOS.3 In the light of novel data confirming its distinctive features from extranodal EBV+ NK/T-cell lymphoma, nasal type (ENKTCL), and supporting specific characteristics, this rare disease is a new entity in the 2022 classifications, with slightly different nomenclatures (Table 1).1,2
Most cases were described in reports from Asia.97-99 Primary nodal EBV+ T/NK-cell lymphoma involves lymph nodes and tends to occur in elderly adults who present with generalized lymphadenopathy, frequent dissemination to the liver or spleen but lack of nasal involvement, sometimes in association with human immunodeficiency virus infection or immunodeficient conditions.56,100 The outcome of patients with primary nodal EBV+ T/NK-cell lymphoma is dismal, being significantly worse than that of patients with ENKTCL or PTCL, NOS.101 Pathological features distinct from ENKTCL include a usually monomorphic large cell morphology, lack of prominent angiocentricity or necrosis, negativity for CD56, positivity for CD8, and more frequent derivation from T cells than from NK cells.100,102 The lymphoma cells are CD3+ CD5–/+ with an activated cytotoxic phenotype, with EBV detected in the majority of tumor cells by in situ hybridization or expression of LMP-1.
Most cases are of T-cell lineage, carry clonally rearranged TR genes associated with 14q11.2 loss, and variably express the TCR.100 Compared with ENKTCL, primary nodal EBV+ T/NK-cell lymphoma is characterized by low genomic instability, upregulation of immune pathways (checkpoint protein PD-L1) that promote immune evasion, and down-regulation of EBV microRNA.101 Few cases have been investigated by high-throughput sequencing; recurrent mutations have been found in TET2, DNMT3A, STAT3, PIK3CD and DDX3X.101,102
Peripheral T-cell lymphoma, not otherwise specified
PTCL, NOS remains defined as a diagnosis of exclusion for those cases of PTCL lacking specific features that qualify for another “specific” PTCL entity. While the definition is unchanged from the previous WHO classifications, subsets of cases formerly included in PTCL, NOS, such as those expressing a TFH phenotype or positive for EBV, are now classified into distinct entities, and therefore the boundaries of PTCL, NOS are narrowing.1-3 Accordingly, while PTCL, NOS has for decades been reported as the most frequent type of PTCL,6 in recent years the reported prevalence of PTCL, NOS is tending to decrease, accounting for 21-27% of PTCL.5,51,52
PTCL, NOS nearly always affects adults.103,104 The presentation is usually nodal, with frequent concurrent extranodal involvement, especially of the skin. Most patients have disseminated disease, constitutional symptoms, intermediate- to high-risk International Prognostic Index score and sometimes blood eosinophilia.103 The overall outcome is 20-30% survival at 5 years.103 A small minority of patients have a preceding lymphoproliferative variant of the hypereosinophilic syndrome105 or chronic lymphocytic leukemia.106
PTCL, NOS is morphologically heterogeneous (Figure 5A-N). Many cases show predominantly medium-sized or large cells with irregular nuclei and prominent nucleoli. Less commonly, others have a predominance of atypical small cells with irregular nuclei.3 Morphological grading is not recommended for clinical purposes, but tumors with a predominance of large cells have been found to have a worse outcome.103 Many cases have an admixture of reactive small lymphocytes, eosinophils, histiocytes, B cells and plasma cells. Any of the microenvironmental components can be dominant, obscure the neoplastic cells, and represent a confounding factor to establishing a correct diagnosis. Cases with a prominent infiltrate of epithelioid histiocytes, referred to as lymphoepithelioid lymphoma (Lennert lymphoma), represented less than 10% of PTCL, NOS in historical series and were associated with an overall better prognosis than other PTCL, NOS.103 In recent years it has turned out that many cases of “Lennert lymphoma” correspond to histiocyte-rich TFH lymphomas.95,107,108 It is unclear at the present time whether the Lennert/lymphohistiocytic lymphomas remaining categorized as PTCL, NOS, which are often derived from CD8+ cells with a non-activated cytotoxic immunophenotype, should be considered as a distinct subgroup of PTCL, NOS.109-111
The neoplastic cells in PTCL, NOS are positive for pan-T-cell antigens (CD3, CD2, CD5, CD7), but one or several of these (most commonly CD5 or CD7) may show reduced or absent expression; they are most commonly CD4+ CD8–, less frequently CD4– CD8+, uncommonly CD4– CD8– or CD4+ CD8+.110 More than 85% of cases express the a|3 TCR, and a minority of cases are either of yö derivation, or TCR-silent.112 Loss of BCL2 expression, observed in 45-60% of the cases, can be a useful marker indicative of T-cell malignancy.113,114 A small proportion of PTCL, NOS (5% or less) express CD20 (or other B-cell markers) in a subset of the neoplastic cells.115 By definition, the neoplastic cells in CD4+ PTCL, NOS must lack a TFH immunophenotype.1,2 CD30 expression is frequent (-30%) and variable.7,62,63 In a study of 141 cases of PTCL, NOS, over 20% of the cases had more than 50% CD30+ tumor cells,62 and staining extent and intensity were higher in cases with large cell morphology. Strong CD30 expression by a majority of the tumor cells is seen occasionally, and raises the need for a differential diagnosis from ALCL.
PTCL, NOS is usually positive for LEF1,63 and negative for BCL6, FOXP3 and TCL1 transcription factors which are, respectively, critical for TFH differentiation and function, related to regulatory T cells, and overexpressed in T-cell prolymphocytic leukemia.116,117 A few cases of FOXP3+ PTCL, NOS have been described in patients negative for HTLV1 infection; these cases were composed of large cells, some had EBV reactivation in bystander cells and the clinical course was aggressive.118
The presence of EBV+ (or EBV–) B cells and plasma cell expansions typical of TFH lymphomas, has been described in PTCL, NOS as well, albeit less frequently; several of these reports, however, antedate the recognition of nodal PTCL of TFH derivation, and therefore their significance is uncertain in the light of the current definition of PTCL, NOS.119,120 Conventional cytogenetics and array-based studies have documented many aberrations and complex patterns of imbalances.121 A whole-genome sequencing study showed that CDKN2A and PTEN deletions are frequent (46% and 26% of the cases, respectively), and may co-occur; this event is specifically associated with PTCL, NOS and never observed in AITL or ALCL.122 CDKN2A deletions, which were associated with shorter survival in that study, are recurrent in the GATA3 molecular subgroup of PTCL, NOS (see below).84
Figure 5.
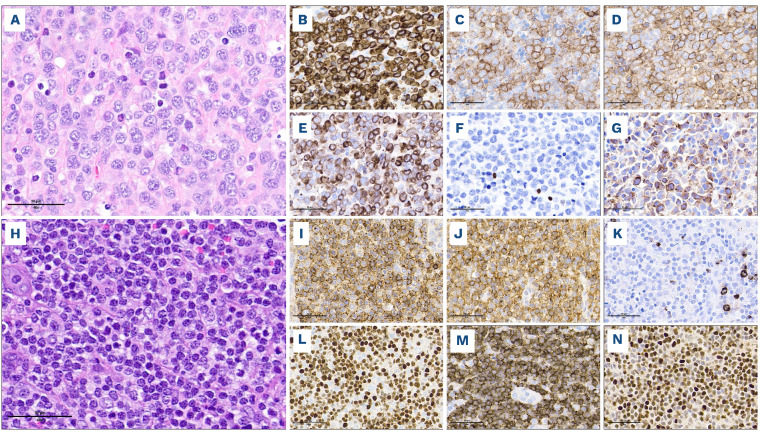
Heterogeneity of peripheral T-cell lymphoma, not otherwise specified. (A-G) Cytotoxic peripheral T-cell lymphoma, not otherwise specified (PTCL, NOS) with large cell morphology and TBX21 (Th1) phenotype. This PTCL, NOS is composed of large pleomorphic cells, with numerous apoptotic bodies (A, hematoxylin & eosin), expresses pan-T-cell markers (B, CD3 and C, CD5), CD4 (D) and cytotoxic markers (E, perforin). Although TBX21 (F) is negative, diffuse expression of CXCR3 (G) subclassifies this lymphoma as PTCL-TBX21. CD30, EBV, GATA3 and CCR4 are also negative (not shown). (H-N) PTCL, NOS with small to medium-sized cells and a GATA3 (Th2) phenotype. This PTCL, NOS comprises diffuse sheets of small to medium-sized cells, with irregular nuclei and occasionally an abundant clear cytoplasm (H, hematoxylin & eosin), is positive for pan-T-cell markers including CD2 (I), and CD4+ (J), and is negative for cytotoxic markers (K, TIA-1). In the absence of TBX21 and CXCR3 expression (not shown), diffuse positivity for GATA3 (L) and CCR4 (M) subclassifies this lymphoma as PTCL-GATA3. P53 protein is diffusely overexpressed (N), reflecting a mutated TP53 gene status (confirmed by sequencing).
Few recurrent translocations and several fusion transcripts have been characterized. Overall, their individual prevalence is low, and they are not specific to PTCL, NOS, as they also occur in other entities, in particular ALK– ALCL and TFH lymphomas.72,123-125 The t(6;14)(p25;q11.2) involving the IRF4 locus, has been reported in clinically aggressive cytotoxic PTCL.126,127 TP63 rearrangements with TBL1XR1 or other partner genes, are associated with an aggressive clinical course and bad outcome, as observed in ALK– ALCL.30 Fusions involving VAV1 (VAV1::MYOF1, VAV1::THAP4, VAV1::S100A7) result in increased activation of VAV1 effector pathways and the oncogenic properties of VAV1::MYOF1 were demonstrated in mice in vivo.128 The FYN::TRAF3IP2 fusion, found in PTCL, NOS and TFH lymphomas, activates the NF-κB pathway.125,129 Fusions involving CD28 (CD28::CTLA4 and CD28::ICOS) occur in PTCL, NOS but are more common in TFH lymphomas and adult T-cell leukemia/lymphoma.72
PTCL, NOS harbors recurrent mutations in epigenetic modifiers, most often TET2 or DNMT3A,130 less commonly SMARCA4 or KMT2D, and in genes related to the TCR signaling pathway, notably activating mutations in PLCG1, CD28 and VAV1.71,102,123,131 Mutations in RHOA and IDH2, recurrent in TFH lymphomas, are essentially absent. Alterations in TP53 (mutations and/or deletions, often biallelic) are detected in 40% of the cases, and portend an adverse prognostic significance.84,132 One study found that alterations in TP53 and/or CDKN2A delineate a group of PTCL, NOS characterized by marked genomic instability, mutations in genes related to immune surveillance and immune evasion (HLA-A, HLA-B, CIITA, CD58, CD274), mutations in transcriptional and post-transcriptional regulators, and a worse out-come.132
As outlined below, research continues into the identification of meaningful subgroups of PTCL, NOS.
Cytotoxic molecule-positive PTCL, NOS. A subset of PTCL, NOS, ranging from 15% to 30-40% of the cases in various series, express one or several cytotoxic granule-associated molecules (TIA-1 and/or granzyme B and/or perforin) indicative of a resting or more commonly activated cytotoxic immunophenotype.109,110,133 Of note, most series of cytotoxic PTCL, NOS have been reported from Asia, and often contained EBV+ cases, which are now classified separately (see above).1,2 In a recently published European cohort of 45 EBV– nodal cytotoxic PTCL,102 the disease affected predominantly males at a median age of 60 years and one-fifth of the patients had a previous history of B-cell lymphoma, solid tumor or underlying immune disorder. Besides a primary nodal presentation, most patients had extranodal disease, and the median survival was only 13 months. Comparison with non-cytotoxic PTCL, NOS in another study demonstrated an inferior overall survival for cytotoxic PTCL, NOS.133 The morphology of these tumors is variable, with predominantly medium-sized to large neoplastic cells and a more or less abundant microenvironment. The neoplastic cells are most commonly CD8+ or CD4– CD8–, less commonly CD4+ or CD4+ CD8+, and in most cases TCR|3F1+ or TCR-silent, rarely TCRYö+.102,110,133 PTCL, NOS with a cytotoxic phenotype express Th1-associated markers, TBX21 or CXCR3, accounting for a subset of PTCL-TBX21 with a more aggressive course (see below); they harbor frequent mutations in epigenetic modifiers, notably in TET2 and DNMT3A, recurrent alterations affecting the TCR and JAK/STAT signaling pathways, including fusions involving VAV1 and CD28, and TP53 mutations in 18% of the cases.102 Cell-of-origin subgroups. Earlier studies suggested that subclasses of PTCL, NOS might be delineated by their immunological profile according to the expression markers associated with Th1 (CXCR3, CCR5, CD134/OX40, CD69, T-bet) or Th2 (CCR4, CXCR4, ST2[L]) differentiation.134,135 These classifiers have not been widely applied due to technical difficulty in assessing the markers, often requiring fresh-frozen tissue, and are lacking validation studies.
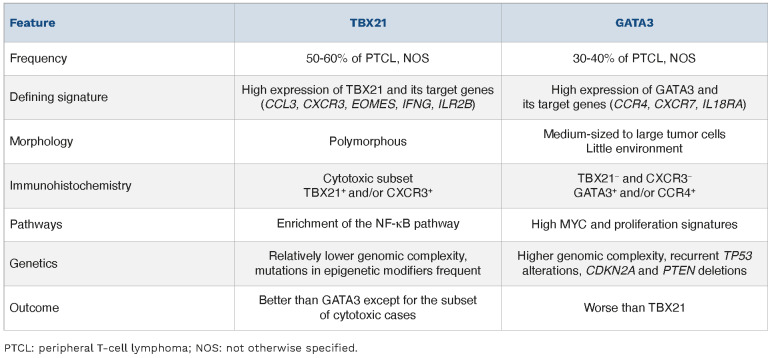
Two molecular subgroups of PTCL, NOS, PTCL-TBX21 and PTCL-GATA3, were identified by transcriptome profiling (Table 4), based on signatures similar to those regulated by the transcription factors TBX21 (T-bet) and GATA3, which are master regulators of Th1 and Th2 differentiation pathways, respectively.136 The TBX21 subgroup shows high expression of TBX21 and its target genes (CCL3, CXCR3, EOMES, IFNG, ILR2B), and enrichment of the NF-κB pathway; conversely the GATA3 subgroup is characterized by high expression of GATA3 and its target genes (CCR4, CXCR7, IL18RA), high MYC and proliferation signatures.136 Histologically, PTCL-TBX21 (Figure 5A-G) tends to be polymorphic with a background of reactive inflammatory cells, including cases of lymphoepithelioid (Lennert) lymphoma, while PTCL-GATA3 (Figure 5H-N) tends to lack a prominent inflammatory microenvironment and shows sheets of medium-sized tumor cells with abundant clear cytoplasm or clusters or sheets of large tumor cells.137 Furthermore, G ATA3 + PTCL, NOS and cases with a cytotoxic phenotype (clustered within the TBX21 subgroup) were found to have a worse outcome than non-cytotoxic TBX21+ tumors.136,138 In addition, subsequent studies showed distinctive genetic features. PTCL-TBX21 has fewer copy number aberrations, and a higher frequency of mutations in epigenetic modifying genes, especially those involved in DNA methylation (TET1, TET3, and DNMT3A). PTCL-GATA3 has greater genomic complexity; frequent losses of TP53, PTEN, RB1, CDKN2A/B, and PRDM1; gains of STAT3 and MYC; and recurrent mutations of TP53 and PRDM1.84 An immunohisto-chemical algorithm has been developed as a surrogate to the gene expression profiling-based classification. The algorithm uses four antibodies to TBX21, CXCR3 (a TBX21 transcriptional target), GATA3, and CCR4 (a GATA3 transcriptional target), which are interpreted sequentially.137 Positivity for TBX21 or CXCR3 in 20% or more of the neoplastic cells defines the TBX21 subgroup (Figure 5A-G). Lymphomas negative or below the threshold for TBX21 markers are classified as GATA3 if 50% or more of the neoplastic cells are GATA3+ or CCR4+ (Figure 5H-N). The GATA3 group identified by immunohistochemistry was similarly associated with an inferior overall survival. Other cases remain unclassified. More recently, a simplified transcriptomic assay based on quantification of 153 selected transcripts dedicated for the molecular diagnosis of the major PTCL entities, including the two subtypes of PTCL, NOS, has been implemented on a digital gene expression profiling platform for routinely processed biopsies.139 The identification of the TBX21 and GATA3 subgroups currently does not have an impact on frontline clinical management of the patients and it remains unclear whether there is differential sensitivity to novel therapies. Thus, it is still considered a research tool and not currently required in standard diagnostic practice.
Table 4.
Comparison of TBX21 and GATA3 subgroups of peripheral T-cell lymphoma, not otherwise specified.
Conclusions and future directions
The current schemes for nodal PTCL classification (Figure 1), in continuity with the previous model, emphasize the role of distinct genetic drivers in the definition of PTCL entities or subtypes, and reinforce the concept of cellular derivation being an important determinant of PTCL biology and of clinical relevance. Advances in the genetic characterization of nodal PTCL have added more or less specific defining features of distinct entities (Table 2). Accordingly, genetic testing is increasingly used for diagnostic purposes, and its role as an aid to clinical decision-making is likely to expand in the near future.140
Among ALCL, the ALK– entity is no longer simply “negative for ALK” but a set of genetically distinct subgroups, which require further characterization to assess their clinical and biological relevance. Gray zones remain around the demarcation between CD30+ PTCL, NOS and ALK– ALCL with, interestingly, some overlap at the genetic level. For example, recurrent JAK2 rearrangements were recently described in PTCL, which had anaplastic features, sometimes ReedSternberg-like cells, frequent CD15 positivity (80%),31 and had been diagnosed in some cases as CD30+ PTCL, NOS and in other cases as ALK– ALCL. Features overlap with cases of PTCL, NOS co-expressing CD30 and CD15 reported earlier.141 While it seems premature to jump to definitive conclusions without collecting additional cases, the question is whether genetics will supersede classic morphological and phenotypic criteria to define ALK– ALCL boundaries. While there are multiple lines of evidence in support of the concept of TFH lymphoma(s), the consideration of one entity versus a group of related disorders differs between the two 2022 classifications, and the distinction between the three types, relying essentially on morphology and immunoarchitecture, may be difficult to apply to a significant fraction of cases showing overlapping features. It is felt that the current definition of the TFH phenotype might be insufficient to capture TFH lymphoma, NOS precisely and distinguish it from PTCL, NOS, and additional criteria, possibly including genetic features, require further research. PTCL, NOS is still viewed as a heterogeneous group of neoplasms that likely do not constitute a single entity, awaiting further identification of meaningful subgroups and substratification. While recent efforts have been made in that direction, and there is growing evidence to substantiate the rationale for molecular or functional subgrouping among PTCL, NOS, there is still a lack of large-scale studies and, at present, these are not recognized as new diagnostic subtypes.
References
- 1.Campo E, Jaffe ES, Cook JR, et al. The International Consensus Classification of mature lymphoid neoplasms: a report from the Clinical Advisory Committee. Blood. 2022;140(11):1229-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alaggio R, Amador C, Anagnostopoulos I, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: lymphoid neoplasms. Leukemia. 2022;36(7):1720-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4th ed. Lyon, France: IARC, 2017. [Google Scholar]
- 4.Ngu HS, Savage KJ. Past, present and future therapeutic approaches in nodal peripheral T-cell lymphomas. Haematologica 2023;108(12):3211-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Leval L, Parrens M, Le Bras F, et al. Angioimmunoblastic T-cell lymphoma is the most common T-cell lymphoma in two distinct French information data sets. Haematologica. 2015;100(9):e361-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vose J, Armitage J, Weisenburger D; International T-cell Lymphoma Project. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26(25):4124-4130. [DOI] [PubMed] [Google Scholar]
- 7.Savage KJ, Harris NL, Vose JM, et al. ALK- anaplastic large-cell lymphoma is clinically and immunophenotypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: report from the International Peripheral T-Cell Lymphoma Project. Blood. 2008;111(12):5496-5504. [DOI] [PubMed] [Google Scholar]
- 8.Rigaud C, Knorr F, Brugieres L, Woessmann W. Diagnosis and management of ALK-positive anaplastic large cell lymphoma in children and adolescents. Best Pract Res Clin Haematol. 2023;36(1):101444. [DOI] [PubMed] [Google Scholar]
- 9.Graetz D, Crews KR, Azzato EM, et al. Leukemic presentation of ALK-positive anaplastic large cell lymphoma with a novel partner, poly(A) binding protein cytoplasmic 1 (PABPC1), responding to single-agent crizotinib. Haematologica. 2019;104(5):e218-e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falini B, Bigerna B, Fizzotti M, et al. ALK expression defines a distinct group of T/null lymphomas ("ALK lymphomas") with a wide morphological spectrum. Am J Pathol. 1998;153(3):875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khanlari M, Li S, Miranda RN, et al. Small cell/lymphohistiocytic morphology is associated with peripheral blood involvement, CD8 positivity and retained T-cell antigens, but not outcome in adults with ALK+ anaplastic large cell lymphoma. Mod Pathol. 2022;35(3):412-418. [DOI] [PubMed] [Google Scholar]
- 12.Vassallo J, Lamant L, Brugieres L, et al. ALK-positive anaplastic large cell lymphoma mimicking nodular sclerosis Hodgkin's lymphoma: report of 10 cases. Am J Surg Pathol. 2006;30(2):223-229. [DOI] [PubMed] [Google Scholar]
- 13.Cordell JL, Pulford KA, Bigerna B, et al. Detection of normal and chimeric nucleophosmin in human cells. Blood. 1999;93(2):632-642. [PubMed] [Google Scholar]
- 14.de Leval L. Approach to nodal-based T-cell lymphomas. Pathology. 2020;52(1):78-99. [DOI] [PubMed] [Google Scholar]
- 15.Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer. 2013;13(10):685-700. [DOI] [PubMed] [Google Scholar]
- 16.Chiarle R, Simmons WJ, Cai H, et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11(6):623-629. [DOI] [PubMed] [Google Scholar]
- 17.Bisig B, de Reynies A, Bonnet C, et al. CD30-positive peripheral T-cell lymphomas share molecular and phenotypic features. Haematologica. 2013;98(8):1250-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hare L, Burke GAA, Turner SD. Resistance to targeted agents used to treat paediatric ALK-positive ALCL. Cancers (Basel). 2021;13(23):6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larose H, Prokoph N, Matthews JD, et al. Whole exome sequencing reveals NOTCH1 mutations in anaplastic large cell lymphoma and points to Notch both as a key pathway and a potential therapeutic target. Haematologica. 2021;106(6):1693-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobello C, Tichy B, Bystry V, et al. STAT3 and TP53 mutations associate with poor prognosis in anaplastic large cell lymphoma. Leukemia. 2021;35(5):1500-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shustov A, Cabrera ME, Civallero M, et al. ALK-negative anaplastic large cell lymphoma: features and outcomes of 235 patients from the International T-Cell Project. Blood Adv. 2021;5(3):640-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sibon D, Bisig B, Bonnet C, et al. ALK-negative anaplastic large cell lymphoma with DUSP22 rearrangement has distinctive disease characteristics with better progression-free survival: a LYSA study. Haematologica. 2023;108(6):1590-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crescenzo R, Abate F, Lasorsa E, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27(4):516-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luchtel RA, Dasari S, Oishi N, et al. Molecular profiling reveals immunogenic cues in anaplastic large cell lymphomas with DUSP22 rearrangements. Blood. 2018;132(13):1386-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrilla Castellar ER, Jaffe ES, Said JW, et al. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood. 2014;124(9):1473-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savage KJ, Slack GW. DUSP22-rearranged ALK-negative anaplastic large cell lymphoma is a pathogenetically distinct disease but can have variable clinical outcome. Haematologica. 2023;108(6):1463-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luchtel RA, Zimmermann MT, Hu G, et al. Recurrent MSC (E116K) mutations in ALK-negative anaplastic large cell lymphoma. Blood. 2019;133(26):2776-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldman AL, Oishi N, Ketterling RP, Ansell SM, Shi M, Dasari S. Immunohistochemical approach to genetic subtyping of anaplastic large cell lymphoma. Am J Surg Pathol. 2022;46(11):1490-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu L, Tang G, Li S, et al. DUSP22 rearrangement is associated with a distinctive immunophenotype but not outcome in patients with systemic ALK-negative anaplastic large cell lymphoma. Haematologica. 2023;108(6):1604-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson JF, Pearce KE, Meyer RG, et al. Fluorescence in-situ hybridisation for TP63 rearrangements in T cell lymphomas: single-site experience of 470 patients and implications for clinical testing. Histopathology. 2020;76(3):481-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzpatrick MJ, Massoth LR, Marcus C, et al. JAK2 rearrangements are a recurrent alteration in CD30+ systemic T-cell lymphomas with anaplastic morphology. Am J Surg Pathol. 2021;45(7):895-904. [DOI] [PubMed] [Google Scholar]
- 32.Scarfo I, Pellegrino E, Mereu E, et al. Identification of a new subclass of ALK-negative ALCL expressing aberrant levels of ERBB4 transcripts. Blood. 2016;127(2):221-232. [DOI] [PubMed] [Google Scholar]
- 33.Hu G, Dasari S, Asmann YW, et al. Targetable fusions of the FRK tyrosine kinase in ALK-negative anaplastic large cell lymphoma. Leukemia. 2018;32(2):565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laurent C, Delas A, Gaulard P, et al. Breast implant-associated anaplastic large cell lymphoma: two distinct clinicopathological variants with different outcomes. Ann Oncol. 2016;27(2):306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghione P, Arcila M, Joseph V, et al. BRCA1/2 mutations impact on the development of breast implant-associated lymphoma (BIA-ALCL) in women with breast cancer reconstructed with textured breast implants. Hematol Oncol. 2023;41(S2):193-194. [Google Scholar]
- 36.Jaffe ES, Ashar BS, Clemens MW, et al. Best pactices guideline for the pathologic diagnosis of breast implant-associated anaplastic large-cell lymphoma. J Clin Oncol. 2020;38(10):1102-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrufino-Schmidt MC, Medeiros LJ, Liu H, et al. Clinicopathologic features and prognostic impact of lymph node involvement in patients with breast implant-associated anaplastic large cell lymphoma. Am J Surg Pathol. 2018;42(3):293-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Leval L. Chromosomes in breast lymphoma. Blood. 2020;136(25):2848-2849. [DOI] [PubMed] [Google Scholar]
- 39.Laurent C, Nicolae A, Laurent C, et al. Gene alterations in epigenetic modifiers and JAK-STAT signaling are frequent in breast implant-associated ALCL. Blood. 2020;135(5):360-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letourneau A, Maerevoet M, Milowich D, et al. Dual JAK1 and STAT3 mutations in a breast implant-associated anaplastic large cell lymphoma. Virchows Arch. 2018;473(4):505-511. [DOI] [PubMed] [Google Scholar]
- 41.Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50(5):1132-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Leval L, Rickman DS, Thielen C, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109(11):4952-4963. [DOI] [PubMed] [Google Scholar]
- 43.de Leval L, Savilo E, Longtine J, Ferry JA, Harris NL. Peripheral T-cell lymphoma with follicular involvement and a CD4+/bcl-6+ phenotype. Am J Surg Pathol. 2001;25(3):395-400. [DOI] [PubMed] [Google Scholar]
- 44.Lemonnier F, Couronne L, Parrens M, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120(7):1466-1469. [DOI] [PubMed] [Google Scholar]
- 45.Palomero T, Couronne L, Khiabanian H, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46(2):166-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakata-Yanagimoto M, Enami T, Yoshida K, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(2):171-175. [DOI] [PubMed] [Google Scholar]
- 47.Vallois D, Dobay MP, Morin RD, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. 2016;128(11):1490-1502. [DOI] [PubMed] [Google Scholar]
- 48.Dobay MP, Lemonnier F, Missiaglia E, et al. Integrative clinicopathological and molecular analyses of angioimmunoblastic T-cell lymphoma and other nodal lymphomas of follicular helper T-cell origin. Haematologica. 2017;102(4):e148-e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bachy E, Camus V, Thieblemont C, et al. Romidepsin plus CHOP versus CHOP in patients with previously untreated peripheral T-cell lymphoma: results of the Ro-CHOP phase III study (conducted by LYSA). J Clin Oncol. 2022;40(3):242-251. [DOI] [PubMed] [Google Scholar]
- 50.Feldman AL, Laurent C, Narbaitz M, et al. Classification and diagnostic evaluation of nodal T- and NK-cell lymphomas. Virchows Arch. 2023;482(1):265-279. [DOI] [PubMed] [Google Scholar]
- 51.Hsi ED, Said J, Macon WR, et al. Diagnostic accuracy of a defined immunophenotypic and molecular genetic approach for peripheral T/NK-cell lymphomas. A North American PTCL study group project. Am J Surg Pathol. 2014;38(6):768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon SE, Song Y, Kim SJ, et al. Comprehensive analysis of peripheral T-cell and natural killer/T-cell lymphoma in Asian patients: a multinational, multicenter, prospective registry study in Asia. Lancet Reg Health West Pac. 2021;10:100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Federico M, Rudiger T, Bellei M, et al. Clinicopathologic characteristics of angioimmunoblastic T-cell lymphoma: analysis of the International Peripheral T-Cell Lymphoma project. J Clin Oncol. 2013;31(2):240-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Leval L, Gisselbrecht C, Gaulard P. Advances in the understanding and management of angioimmunoblastic T-cell lymphoma. Br J Haematol. 2010;148(5):673-689. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez-Justo M, Attygalle AD, Munson P, Roncador G, Marafioti T, Piris MA. Angioimmunoblastic T-cell lymphoma with hyperplastic germinal centres: a neoplasia with origin in the outer zone of the germinal centre? Clinicopathological and immunohistochemical study of 10 cases with follicular T-cell markers. Mod Pathol. 2009;22(6):753-761. [DOI] [PubMed] [Google Scholar]
- 56.Attygalle AD, Cabecadas J, Gaulard P, et al. Peripheral T-cell and NK-cell lymphomas and their mimics; taking a step forward -report on the lymphoma workshop of the XVIth meeting of the European Association for Haematopathology and the Society for Hematopathology. Histopathology. 2014;64(2):171-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iqbal J, Weisenburger DD, Greiner TC, et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood. 2010;115(5):1026-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez M, Alonso-Alonso R, Tomas-Roca L, et al. Peripheral T-cell lymphoma: molecular profiling recognizes subclasses and identifies prognostic markers. Blood Adv. 2021;5(24):5588-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sugio T, Miyawaki K, Kato K, et al. Microenvironmental immune cell signatures dictate clinical outcomes for PTCL-NOS. Blood Adv. 2018;2(17):2242-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bisig B, Thielen C, Herens C, et al. c-Maf expression in angioimmunoblastic T-cell lymphoma reflects follicular helper T-cell derivation rather than oncogenesis. Histopathology. 2012;60(2):371-376. [DOI] [PubMed] [Google Scholar]
- 61.Gaulard P, de Leval L. Follicular helper T cells: implications in neoplastic hematopathology. Semin Diagn Pathol. 2011;28(3):202-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bossard C, Dobay MP, Parrens M, et al. Immunohistochemistry as a valuable tool to assess CD30 expression in peripheral T-cell lymphomas: high correlation with mRNA levels. Blood. 2014;124(19):2983-2986. [DOI] [PubMed] [Google Scholar]
- 63.Onaindia A, Martinez N, Montes-Moreno S, et al. CD30 expression by B and T cells: a frequent finding in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma-not otherwise specified. Am J Surg Pathol. 2016;40(3):378-385. [DOI] [PubMed] [Google Scholar]
- 64.Chen Z, Zhu Q, Deng X, et al. Angioimmunoblastic T-cell lymphoma with predominant CD8+ tumor-infiltrating T-cells is a distinct immune pattern with an immunosuppressive microenvironment. Front Immunol. 2022;13:987227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicolae A, Pittaluga S, Venkataraman G, et al. Peripheral T-cell lymphomas of follicular T-helper cell derivation with Hodgkin/Reed-Sternberg cells of B-cell lineage: both EBV-positive and EBV-negative variants exist. Am J Surg Pathol. 2013;37(6):816-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willenbrock K, Brauninger A, Hansmann ML. Frequent occurrence of B-cell lymphomas in angioimmunoblastic T-cell lymphoma and proliferation of Epstein-Barr virus-infected cells in early cases. Br J Haematol. 2007;138(6):733-739. [DOI] [PubMed] [Google Scholar]
- 67.Lewis NE, Petrova-Drus K, Huet S, et al. Clonal hematopoiesis in angioimmunoblastic T-cell lymphoma with divergent evolution to myeloid neoplasms. Blood Adv. 2020;4(10):2261-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lemonnier F, Gaulard P, de Leval L. New insights in the pathogenesis of T-cell lymphomas. Curr Opin Oncol. 2018;30(5):277-284. [DOI] [PubMed] [Google Scholar]
- 69.Lemonnier F, Safar V, Beldi-Ferchiou A, et al. Integrative analysis of a phase 2 trial combining lenalidomide with CHOP in angioimmunoblastic T-cell lymphoma. Blood Adv. 2021;5(2):539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng J, Wang Z, Pan X, et al. DNMT3A(R882H) accelerates angioimmunoblastic T-cell lymphoma in mice. Oncogene. 2023;42(23):1940-1950. [DOI] [PubMed] [Google Scholar]
- 71.Rohr J, Guo S, Huo J, et al. Recurrent activating mutations of CD28 in peripheral T-cell lymphomas. Leukemia. 2016;30(5):1062-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vallois D, Dupuy A, Lemonnier F, et al. RNA fusions involving CD28 are rare in peripheral T-cell lymphomas and concentrate mainly in those derived from follicular helper T cells. Haematologica. 2018;103(8):e360-e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zang S, Li J, Yang H, et al. Mutations in 5-methylcytosine oxidase TET2 and RhoA cooperatively disrupt T cell homeostasis. J Clin Invest. 2017;127(8):2998-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ng SY, Brown L, Stevenson K, et al. RhoA G17V is sufficient to induce autoimmunity and promotes T-cell lymphomagenesis in mice. Blood. 2018;132(9):935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cortes JR, Ambesi-Impiombato A, Couronne L, et al. RHOA G17V induces T follicular helper cell specification and promotes lymphomagenesis. Cancer Cell. 2018;33(2):259-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ondrejka SL, Grzywacz B, Bodo J, et al. Angioimmunoblastic T-cell lymphomas with the RHOA p.Gly17Val mutation have classic clinical and pathologic features. Am J Surg Pathol. 2016;40(3):335-341. [DOI] [PubMed] [Google Scholar]
- 77.Nagao R, Kikuti Y Y, Carreras J, et al. Clinicopathologic analysis of angioimmunoblastic T-cell lymphoma with or without RHOA G17V mutation using formalin-fixed paraffin-embedded sections. Am J Surg Pathol. 2016;40(8):1041-1050. [DOI] [PubMed] [Google Scholar]
- 78.Dobson R, Du PY, Raso-Barnett L, et al. Early detection of T-cell lymphoma with T follicular helper phenotype by RHOA mutation analysis. Haematologica. 2022;107(2):489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cairns RA, Iqbal J, Lemonnier F, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119(8):1901-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lemonnier F, Cairns RA, Inoue S, et al. The IDH2 R172K mutation associated with angioimmunoblastic T-cell lymphoma produces 2HG in T cells and impacts lymphoid development. Proc Natl Acad Sci U S A. 2016;113(52):15084-15089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang C, McKeithan TW, Gong Q, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126(15):1741-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Steinhilber J, Mederake M, Bonzheim I, et al. The pathological features of angioimmunoblastic T-cell lymphomas with IDH2(R172) mutations. Mod Pathol. 2019;32(8):1123-1134. [DOI] [PubMed] [Google Scholar]
- 83.Leca J, Lemonnier F, Meydan C, et al. IDH2 and TET2 mutations synergize to modulate T follicular helper cell functional interaction with the AITL microenvironment. Cancer Cell. 2023;41(2):323-339. [DOI] [PubMed] [Google Scholar]
- 84.Heavican TB, Bouska A, Yu J, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. 2019;133(15):1664-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yao WQ, Wu F, Zhang W, et al. Angioimmunoblastic T-cell lymphoma contains multiple clonal T-cell populations derived from a common TET2 mutant progenitor cell. J Pathol. 2020;250(3):346-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nguyen TB, Sakata-Yanagimoto M, Asabe Y, et al. Identification of cell-type-specific mutations in nodal T-cell lymphomas. Blood Cancer J. 2017;7(1):e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schwartz FH, Cai Q, Fellmann E, et al. TET2 mutations in B cells of patients affected by angioimmunoblastic T-cell lymphoma. J Pathol. 2017;242(2):129-133. [DOI] [PubMed] [Google Scholar]
- 88.Lemonnier F, Dupuis J, Sujobert P, et al. Treatment with 5-azacytidine induces a sustained response in patients with angioimmunoblastic T-cell lymphoma. Blood. 2018;132(21):2305-2309. [DOI] [PubMed] [Google Scholar]
- 89.Huang Y, Moreau A, Dupuis J, et al. Peripheral T-cell lymphomas with a follicular growth pattern are derived from follicular helper T cells (TFH) and may show overlapping features with angioimmunoblastic T-cell lymphomas. Am J Surg Pathol. 2009;33(5):682-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miyoshi H, Sato K, Niino D, et al. Clinicopathologic analysis of peripheral T-cell lymphoma, follicular variant, and comparison with angioimmunoblastic T-cell lymphoma: Bcl-6 expression might affect progression between these disorders. Am J Clin Pathol. 2012;137(6):879-889. [DOI] [PubMed] [Google Scholar]
- 91.Hartmann S, Goncharova O, Portyanko A, et al. CD30 expression in neoplastic T cells of follicular T cell lymphoma is a helpful diagnostic tool in the differential diagnosis of Hodgkin lymphoma. Mod Pathol. 2019;32(1):37-47. [DOI] [PubMed] [Google Scholar]
- 92.Moroch J, Copie-Bergman C, de Leval L, et al. Follicular peripheral T-cell lymphoma expands the spectrum of classical Hodgkin lymphoma mimics. Am J Surg Pathol. 2012;36(11):1636-1646. [DOI] [PubMed] [Google Scholar]
- 93.Alikhan M, Song JY, Sohani AR, et al. Peripheral T-cell lymphomas of follicular helper T-cell type frequently display an aberrant CD3(-/dim)CD4(+) population by flow cytometry: an important clue to the diagnosis of a Hodgkin lymphoma mimic. Mod Pathol. 2016;29(10):1173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Attygalle AD, Feldman AL, Dogan A. ITK/SYK translocation in angioimmunoblastic T-cell lymphoma. Am J Surg Pathol. 2013;37(9):1456-1457. [DOI] [PubMed] [Google Scholar]
- 95.Agostinelli C, Hartmann S, Klapper W, et al. Peripheral T cell lymphomas with follicular T helper phenotype: a new basket or a distinct entity? Revising Karl Lennert's personal archive. Histopathology. 2011;59(4):679-691. [DOI] [PubMed] [Google Scholar]
- 96.Yoon SE, Cho J, Kim YJ, et al. Comprehensive analysis of clinical, pathological, and genomic characteristics of follicular helper T-cell derived lymphomas. Exp Hematol Oncol. 2021;10(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kato S, Takahashi E, Asano N, et al. Nodal cytotoxic molecule (CM)-positive Epstein-Barr virus (EBV)-associated peripheral T cell lymphoma (PTCL): a clinicopathological study of 26 cases. Histopathology. 2012;61(2):186-199. [DOI] [PubMed] [Google Scholar]
- 98.Asano N, Kato S, Nakamura S. Epstein-Barr virus-associated natural killer/T-cell lymphomas. Best Pract Res Clin Haematol. 2013;26(1):15-21. [DOI] [PubMed] [Google Scholar]
- 99.Jeon YK, Kim JH, Sung JY, Han JH, Ko YH. Hematopathology Study Group of the Korean Society of Pathologists. Epstein-Barr virus-positive nodal T/NK-cell lymphoma: an analysis of 15 cases with distinct clinicopathological features. Hum Pathol. 2015;46(7):981-990. [DOI] [PubMed] [Google Scholar]
- 100.Kato S, Yamashita D, Nakamura S. Nodal EBV+ cytotoxic T-cell lymphoma: a literature review based on the 2017 WHO classification. J Clin Exp Hematop. 2020;60(2):30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wai CMM, Chen S, Phyu T, et al. Immune pathway upregulation and lower genomic instability distinguish EBV-positive nodal T/NK-cell lymphoma from ENKTL and PTCL-NOS. Haematologica. 2022;107(8):1864-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nicolae A, Bouilly J, Lara D, et al. Nodal cytotoxic peripheral T-cell lymphoma occurs frequently in the clinical setting of immunodysregulation and is associated with recurrent epigenetic alterations. Mod Pathol. 2022;35(8):1126-1136. [DOI] [PubMed] [Google Scholar]
- 103.Weisenburger DD, Savage KJ, Harris NL, et al. Peripheral T-cell lymphoma, not otherwise specified: a report of 340 cases from the International Peripheral T-cell Lymphoma Project. Blood. 2011;117(12):3402-3408. [DOI] [PubMed] [Google Scholar]
- 104.Federico M, Bellei M, Marcheselli L, et al. Peripheral T cell lymphoma, not otherwise specified (PTCL-NOS). A new prognostic model developed by the International T cell Project Network. Br J Haematol. 2018;181(6):760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lefevre G, Copin MC, Roumier C, et al. CD3-CD4+ lymphoid variant of hypereosinophilic syndrome: nodal and extranodal histopathological and immunophenotypic features of a peripheral indolent clonal T-cell lymphoproliferative disorder. Haematologica. 2015;100(8):1086-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Trimech M, Letourneau A, Missiaglia E, et al. Angioimmunoblastic T-cell lymphoma and chronic lymphocytic leukemia/small lymphocytic lymphoma: a novel form of composite lymphoma potentially mimicking Richter syndrome. Am J Surg Pathol. 2021;45(6):773-786. [DOI] [PubMed] [Google Scholar]
- 107.Hartmann S, Agostinelli C, Klapper W, et al. Revising the historical collection of epithelioid cell-rich lymphomas of the Kiel Lymph Node Registry: what is Lennert's lymphoma nowadays? Histopathology. 2011;59(6):1173-1182. [DOI] [PubMed] [Google Scholar]
- 108.Kurita D, Miyoshi H, Yoshida N, et al. A clinicopathologic study of Lennert lymphoma and possible prognostic factors: the importance of follicular helper T-cell markers and the association with angioimmunoblastic T-cell lymphoma. Am J Surg Pathol. 2016;40(9):1249-1260. [DOI] [PubMed] [Google Scholar]
- 109.Kagami Y, Suzuki R, Taji H, et al. Nodal cytotoxic lymphoma spectrum: a clinicopathologic study of 66 patients. Am J Surg Pathol. 1999;23(10):1184-1200. [DOI] [PubMed] [Google Scholar]
- 110.Geissinger E, Odenwald T, Lee SS, et al. Nodal peripheral T-cell lymphomas and, in particular, their lymphoepithelioid (Lennert's) variant are often derived from CD8(+) cytotoxic T-cells. Virchows Arch. 2004;445(4):334-343. [DOI] [PubMed] [Google Scholar]
- 111.Gafencu GA, Selicean SE, Petrushev B, et al. Clinicopathological analysis of a case series of peripheral T-cell lymphomas, not otherwise specified, of lymphoepithelioid variant (Lennert's lymphoma). A Central European single-center study. Hum Pathol. 2016;53192-194. [DOI] [PubMed] [Google Scholar]
- 112.Bonzheim I, Geissinger E, Roth S, et al. Anaplastic large cell lymphomas lack the expression of T-cell receptor molecules or molecules of proximal T-cell receptor signaling. Blood. 2004;104(10):3358-3360. [DOI] [PubMed] [Google Scholar]
- 113.O'Malley DP, Chizhevsky V, Grimm KE, Hii A, Weiss LM. Utility of BCL2, PD1, and CD25 immunohistochemical expression in the diagnosis of T-cell lymphomas. Appl Immunohistochem Mol Morphol. 2014;22(2):99-104. [DOI] [PubMed] [Google Scholar]
- 114.Siddiqui F, Perez Silos V, Karube K, et al. B-cell lymphoma-2 downregulation is a useful feature supporting a neoplastic phenotype in mature T-cell lymphomas. Hum Pathol. 2022;125:48-58. [DOI] [PubMed] [Google Scholar]
- 115.Rahemtullah A, Longtine JA, Harris NL, et al. CD20+ T-cell lymphoma: clinicopathologic analysis of 9 cases and a review of the literature. Am J Surg Pathol. 2008;32(11):1593-1607. [DOI] [PubMed] [Google Scholar]
- 116.Bonzheim I, Geissinger E, Tinguely M, et al. Evaluation of FoxP3 expression in peripheral T-cell lymphoma. Am J Clin Pathol. 2008;130(4):613-619. [DOI] [PubMed] [Google Scholar]
- 117.Narducci MG, Pescarmona E, Lazzeri C, et al. Regulation of TCL1 expression in B- and T-cell lymphomas and reactive lymphoid tissues. Cancer Res. 2000;60(8):2095-2100. [PubMed] [Google Scholar]
- 118.Satou A, Asano N, Kato S, et al. FoxP3-positive T cell lymphoma arising in non-HTLV1 carrier: clinicopathological analysis of 11 cases of PTCL-NOS and 2 cases of mycosis fungoides. Histopathology. 2016;68(7):1099-1108. [DOI] [PubMed] [Google Scholar]
- 119.Balague O, Martinez A, Colomo L, et al. Epstein-Barr virus negative clonal plasma cell proliferations and lymphomas in peripheral T-cell lymphomas: a phenomenon with distinctive clinicopathologic features. Am J Surg Pathol. 2007;31(9):1310-1322. [DOI] [PubMed] [Google Scholar]
- 120.Eladl AE, Satou A, Elsayed AA, et al. Clinicopathological study of 30 cases of peripheral T-cell lymphoma with Hodgkin and Reed-Sternberg-like B-cells from Japan. Am J Surg Pathol. 2017;41(4):506-516. [DOI] [PubMed] [Google Scholar]
- 121.de Leval L, Bisig B, Thielen C, Boniver J, Gaulard P. Molecular classification of T-cell lymphomas. Crit Rev Oncol Hematol. 2009;72(2):125-143. [DOI] [PubMed] [Google Scholar]
- 122.Maura F, Dodero A, Carniti C, et al. CDKN2A deletion is a frequent event associated with poor outcome in patients with peripheral T-cell lymphoma not otherwise specified (PTCL-NOS). Haematologica. 2021;106(11):2918-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Abate F, da Silva-Almeida AC, Zairis S, et al. Activating mutations and translocations in the guanine exchange factor VAV1 in peripheral T-cell lymphomas. Proc Natl Acad Sci U S A. 2017;114(4):764-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Drieux F, Ruminy P, Sater V, et al. Detection of gene fusion transcripts in peripheral T-cell lymphoma using a multiplexed targeted sequencing assay. J Mol Diagn. 2021;23(8):929-940. [DOI] [PubMed] [Google Scholar]
- 125.Debackere K, Marcelis L, Demeyer S, et al. Fusion transcripts FYN-TRAF3IP2 and KHDRBS1-LCK hijack T cell receptor signaling in peripheral T-cell lymphoma, not otherwise specified. Nat Commun. 2021;12(1):3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Somja J, Bisig B, Bonnet C, Herens C, Siebert R, de Leval L. Peripheral T-cell lymphoma with t(6;14)(p25;q11.2) translocation presenting with massive splenomegaly. Virchows Arch. 2014;464(6):735-741. [DOI] [PubMed] [Google Scholar]
- 127.Feldman AL, Law M, Remstein ED, et al. Recurrent translocations involving the IRF4 oncogene locus in peripheral T-cell lymphomas. Leukemia. 2009;23(3):574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cortes JR, Filip I, Albero R, et al. Oncogenic Vav1-Myo1f induces therapeutically targetable macrophage-rich tumor microenvironment in peripheral T cell lymphoma. Cell Rep. 2022;39(3):110695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moon CS, Reglero C, Cortes JR, et al. FYN-TRAF3IP2 induces NF-kappaB signaling-driven peripheral T cell lymphoma. Nat Cancer. 2021;2(1):98-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Herek TA, Bouska A, Lone W, et al. DNMT3A mutations define a unique biological and prognostic subgroup associated with cytotoxic T cells in PTCL-NOS. Blood. 2022;140(11):1278-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Manso R, Rodriguez-Pinilla SM, Gonzalez-Rincon J, et al. Recurrent presence of the PLCG1 S345F mutation in nodal peripheral T-cell lymphomas. Haematologica. 2015;100(1):e25-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Watatani Y, Sato Y, Miyoshi H, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33(12):2867-2883. [DOI] [PubMed] [Google Scholar]
- 133.Asano N, Suzuki R, Kagami Y, et al. Clinicopathologic and prognostic significance of cytotoxic molecule expression in nodal peripheral T-cell lymphoma, unspecified. Am J Surg Pathol. 2005;29(10):1284-1293. [DOI] [PubMed] [Google Scholar]
- 134.Jones D, O'Hara C, Kraus MD, et al. Expression pattern of T-cell-associated chemokine receptors and their chemokines correlates with specific subtypes of T-cell non-Hodgkin lymphoma. Blood. 2000;96(2):685-690. [PubMed] [Google Scholar]
- 135.Tsuchiya T, Ohshima K, Karube K, et al. Th1, Th2, and activated T-cell marker and clinical prognosis in peripheral T-cell lymphoma, unspecified: comparison with AILD, ALCL, lymphoblastic lymphoma, and ATLL. Blood. 2004;103(1):236-241. [DOI] [PubMed] [Google Scholar]
- 136.Iqbal J, Wright G, Wang C, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. 2014;123(19):2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Amador C, Greiner TC, Heavican TB, et al. Reproducing the molecular subclassification of peripheral T-cell lymphoma-NOS by immunohistochemistry. Blood. 2019;134(24):2159-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang T, Feldman AL, Wada DA, et al. GATA-3 expression identifies a high-risk subset of PTCL, NOS with distinct molecular and clinical features. Blood. 2014;123(19):3007-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Amador C, Bouska A, Wright G, et al. Gene expression signatures for the accurate diagnosis of peripheral T-cell lymphoma entities in the routine clinical practice. J Clin Oncol. 2022;40(36):4261-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.de Leval L, Alizadeh AA, Bergsagel PL, et al. Genomic profiling for clinical decision making in lymphoid neoplasms. Blood. 2022;140(21):2193-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Barry TS, Jaffe ES, Sorbara L, Raffeld M, Pittaluga S. Peripheral T-cell lymphomas expressing CD30 and CD15. Am J Surg Pathol. 2003;27(12):1513-1522. [DOI] [PubMed] [Google Scholar]