Abstract
In a gene targeting experiment, the generation of a targeting construct often requires complex DNA manipulations. We developed a set of cassettes and plasmids useful for creating targeting vectors to modify the mammalian genome. A positive selection marker cassette (PGK/EM7p-npt), which included dual prokaryotic and eukaryotic promoters to permit consecutive selection for recombination in Escherichia coli and then in mouse embryonic stem cells, was flanked by two FRT-loxP sequences. The PGK/EM7p-npt cassette was placed between the minimum regions of a Tn7 transposable element for insertion into another DNA by means of Tn7 transposase in vitro. We also constructed a plasmid having a loxP-Zeo-loxP cassette between the modified Tn5 outer elements. These cassettes can be integrated randomly into a given genomic DNA through the in vitro transposition reaction, thus producing a collection of genomic segments flanked by loxP sites (floxed) at various positions without the use of restriction enzymes and DNA ligase. We confirmed that this system remarkably reduced the time and labor for the construction of complex gene targeting vectors.
INTRODUCTION
Gene targeting is a powerful method for producing genetically modified animals to study gene function in vivo. This powerful technology allows us to introduce designed mutations into any cloned locus, and to analyze generated mice with the corresponding genetic changes. The resulting phenotypes often provide an insight into the functions of genes. However, a gene targeting experiment is laborious and time-consuming, demanding elaborate techniques for the manipulation of DNA, cells and embryos, and it often takes more than one year from the design of constructs to generation of animals.
Replacement of the native gene with a modified gene fragment in the mammalian genome by homologous recombination is performed to introduce genetic alteration in embryonic stem (ES) cell. One of the limiting steps is the generation of ‘gene targeting vectors’, since the design of a targeting vector must fulfill several requirements. First, efficient homologous recombination in the ES genome requires fairly long (>5 kb, >10 kb, if possible) homologous segments flanking the alteration in the targeting construct (1–4). The use of a positive selection marker is mandatory for selecting candidates having homologous replacement, and it should be placed in a proper region and be designed to be removable afterward. For this purpose, the loxP/Cre (4) and FRT/FLPe (5) systems have been used to remove the selective markers in ES cells or animals. The use of these highly specific recombination systems also led to the second generation gene modification strategy for controlling genetic alterations spatio-temporally in animals (6,7). However, the design and construction of floxed alleles is labor-intensive. The loxP sites must be placed at both ends of the genomic segments to be deleted upon Cre recombinase-mediated recombination, but reasonably far from the ends of the construct for efficient homologous recombination in the ES genome. They must be placed in intron regions to conserve the structure and function of the gene products. In addition, genomic regions containing highly repetitive elements should be excluded from the targeting construct, since these elements hamper efficient and accurate detection of homologous recombination on PCR or Southern hybridization analyses.
The mouse genome project allows us to deduce the structures of loci of interest at the nucleotide sequence level in silico. We searched for single restriction sites that fulfill the requirements described above, and then synthesized loxP segments tagged with the recognition sequence at both ends and integrated them into the targeting vector by using DNA ligase. However, it is not always straightforward to find an appropriate unique recognition site within a given genomic segment. Even when a suitable restriction site exists, it still remains difficult to introduce only a single short loxP segment (∼40 bp) into a large plasmid (>10 kb) by conventional DNA manipulation relying on T4 DNA ligase.
To overcome these limitations, we applied bacterial transposon systems to simplify the construction of complex targeting vectors. Bacterial transposon exhibits minimal sequence preference for their insertions and single insertions within a several hundred kilo base pairs region of a single molecule due to their long-range cis interactions (8). These features can be reconstituted in vitro (9–11). The nature of the in vitro transposition reactions is very powerful for assembling complex targeting constructs, as reported previously (12). Here, we developed a set of plasmids that significantly reduce the time and labor for the construction of gene targeting constructs. We described a typical experiment for modification of the mouse mVam2/Vps41 gene as a model study.
MATERIALS AND METHODS
Antibiotics and bacteria culture
E.coli was cultured in Terrific Broth. Ampicillin sodium salt (Amp), kanamycin sulphate (Kan), chloramphenicol (Chl), tetracycline HCl (Tet) and Zeocin (Zeo) were used at concentrations of 100, 25, 33, 12.5 and 100 μg/ml, respectively. BAC DNA was prepared with a Qiagen large construction kit, and plasmid DNA was prepared with an automated DNA isolation system, PI-50α (KURABO), following the manufacturer's recommendations.
Assembly of pGPS21loxFRTNeo
Plasmid pLox-neo, a PGKp-npt-pA DNA cassette with two loxP sequences in pBluescript II SK+, was a generous gift from Drs Osamu Minowa and Tetsuo Noda. Insertion of a synthetic EM7 promoter (EM7p) for prokaryotes was carried out by site-directed mutagenesis (13) with modifications as follows. The EM7p sequence with homology arms for the PGK promoter (PGKp) region was amplified from an EM7p-containing plasmid, pVgRXR (Invitrogen), by PCR using primer pair PGKEM7S and PGKEM7A (Table 1). The PCR product was annealed to a single-stranded DNA of pLox-neo prepared from an E.coli dut− ung− strain co-infected with M13 KO7 helper phage, and then transformed into a dut+ ung+ strain followed by selection for ampicillin and kanamycin. Plasmids were recovered and then sequenced to verify the loxP-PGK/EM7p-npt-loxP region to obtain pLoxPGKEM7-Neo.
Table 1.
List of oligonucleotides used in this study
| Primer name | Sequence |
|---|---|
| PGKEM7S | 5′-CCTCTTCCTCATCTCCGGGCCTTTCGACCTGTTGACAATTAATCATCGGC-3′ |
| PGKEM7A | 5′-GCAATCCATCTTGTTCAATGGCCGATCCCATGGTGGCCCTCCTATAGTGA-3′ |
| HindFLP-S1 | 5′-TCAAAAGCTTTCTGAAGTTCCTATACTTTC-3′ |
| HindFLP-A1 | 5′-CGGAAAAGCTTTGAAGTTCCTATTCCGAAG-3′ |
| EcoFLP-S1 | 5′-TCAAAAGAATTCTGAAGTTCCTATACTTTC-3′ |
| EcoFLP-A1 | 5′-CGGGAATTCTTTGAAGTTCCTATTCCGAAG-3′ |
| ERVloxSR-5 | 5′-GGGATATCATAACTTCGTATAATGTATGCTATACGAAGTTATGTGTGTCAGTTAGGGTGT-3′ |
| ERVloxSV40-3 | 5′-CCGATATCATAACTTCGTATAGCATACATTATACGAAGTTATCCAGACATGATAAGATAC-3′ |
| loxzeo-Cla | 5′-CCCTAACTGACACACATAACTTCGTAT-3′ |
| loxzeo-Nsi | 5′-ACTCATCAATGCATCTTATCATGTCT-3′ |
| PmeRsr1 | 5′-GTTTAAACGGACCGC-3′ |
| PmeRsr2 | 5′-GGTCCGTTTAAACAGCT-3′ |
| FseSrf1 | 5′-CTAGGATCCGGCCGGCCCGGGC-3′ |
| FseSrf2 | 5′-GATCGCCCGGGCCGGCCGGATC-3′ |
| M13 Fwd23 | 5′-CCCAGTCACGACGTTGTAAAACG-3′ |
| DT-AR | 5′-ACTACTGATTCTAATTGTTTGTGT-3′ |
FRT sequences were inserted into unique HindIII (between the upstream loxP and PGKp) and EcoRI (between the down stream loxP and vector backbone) sites in two steps. First, an FRT segment with HindIII sites at both ends was prepared from yeast 2 μm ori containing plasmid YEp24 by PCR with primers HindFLP-A1 and HindFLP-S1, and then inserted at the HindIII site of pLoxPGKEM7-Neo to yield an intermediate plasmid. Second, an EcoRI-FRT-EcoRI segment was prepared using primer pair EcoFLP-S1 and EcoFLP-A1, and then inserted at the EcoRI site of the intermediate plasmid. The orientations of the two FRT segments were verified by sequencing, and a correctly configured plasmid, pLoxFRTNeo-1 was obtained. pLoxFRTNeo-1 was digested with XhoI and SmaI, polished with T4 DNA polymerase, then subcloned into the SpeI–NotI sites (trimmed with T4 DNA polymerase) of pGPS2.1 (New England Biolabs), introduced into E.coli strain EC100D pir-116 (Epicentre), and then selected on tetracyclin and kanamycin to obtain the final construct, pGPS21loxFRTNeo.
Assembly of pMODloxZeoΔamp3
A loxP-Zeo-loxP DNA segment was amplified with a primer pair ERVloxSR-5 and ERVloxSV40-3 using pVgRXR (Invitrogen) as a template. The PCR product, having a loxP sequence at the both end, a mammalian enhancer/promoter, the EM7 promoter and ZeoR, was digested with EcoRV and then introduced into the ClaI/HindIII sites (flushed with T4 DNA polymerase) of pMOD<MCS> (Epicentre) to yield pMODloxZeo-1. The bla gene of the pMODloxZeo-1 was disrupted by digestion with ScaI and SspI, and trimmed with T4 DNA polymerase, and then a part of mammalian transcriptional element was removed by SphI and PstI digestion to obtain a final plasmid, pMODloxZeoΔamp3.
Assembly of pBSDT-AII
A HincII–XhoI segment of the MC-1 promoter and DT-A region was excised from pMC1DT-A (LifeTec) and inserted into pBluescript II SK− to obtain pBSDT-A. A pair of synthetic oligonucleotides, PmeRsr1 and PmeRsr2 (Table 1), was annealed to obtain a linker, which was then inserted at the SacI–SacII sites of pBSDT-A, and then a second linker (a duplex of FseSrf1 and FseSrf2, see Table 1) was introduced at the BamHI–XbaI sites to generate pBSDT-AII. The structure of multiple cloning site of pBSDT-AII was verified by sequencing.
In vitro transposition reactions
In vitro transposition reaction of pGPS21loxFRTNeo and pMODloxZeoΔamp was performed with TnsABC* (New England Biolabs) and EZ::Tn transposase (Epicentre), respectively, following the manufacturer's instructions. The reaction product was purified by phenol/chloroform extraction and ethanol precipitation, and then introduced into an appropriate E.coli strain by electroporation.
RESULTS AND DISCUSSION
Construction of a transposable KAN/G418 cassette with loxP and FRT sequences
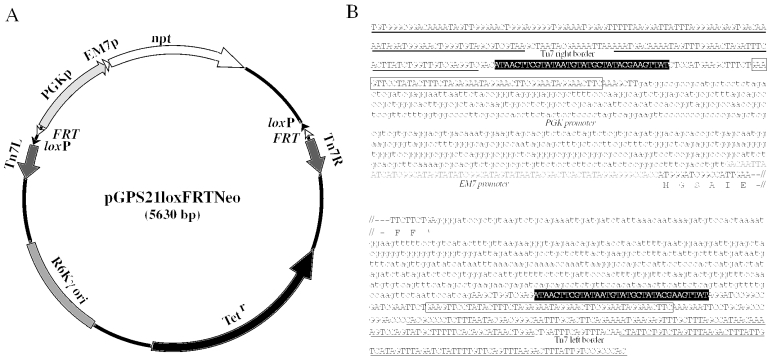
Plasmid pGPS21loxFRTNeo (Figure 1) was designed to introduce a positive selection marker into gene segments by in vitro transposition reaction. This plasmid has several unique features. The Tn5-derived neomycin phosphotransferase gene (npt) was placed downstream of a short fragment of the EM7 promoter (EM7p) and mammalian PGK promoter (PGKp) sequence. This DNA fragment conferred kanamycin resistance (5–50 μg/ml) on E.coli and G418 resistance (100–300 μg/ml) on mouse ES cells, respectively (data not shown), indicating that the PGK/EM7p-npt segment could be used as a positive selection marker in E.coli as well as ES cells.
Figure 1.
Structure of pGPS21loxFRTNeo. pGPS21loxFRTNeo was designed as a donor plasmid for transposase-mediated in vitro integration of a mammalian/bacterial selection marker and FRT, loxP sequences. Characteristic elements of the plasmid are summarized in (A). The plasmid possesses an R6Kγ replication origin, tetracycline resistance (TetR), and transposable elements having FRT-loxP-PGKp/EM7p-npt-FRT-loxP fragments. The fine structure of the transposable elements is shown in (B), except for the middle part of the KAN/G418 resistance gene (npt). The loxP and FRT sequences are shown in closed and open boxes, respectively. The PGK promoter and 3′-termination sequence are indicated by lowercase letters. The EM7 promoter sequence is indicated by gray uppercase letters.
This PGK/EM7p-npt segment is placed between the right and left borders of the Tn7 transposing element, therefore it can be inserted into other DNAs by incubation with a commercially available Tn7 transposase. Upon Flpe-mediated recombination (14), this selectable marker can be removed, leaving one loxP site. The border sequences were placed outside of the two FRT-loxP sequences, therefore transposition introduces a segment of FRT-loxP-PGK/EM7p-npt-FRT-loxP into the target DNA. In vitro transposition events take place almost randomly, and multiple insertions into the same DNA fragment are rare due to the target immunity (8). Plasmid pGPS21loxFRTNeo has R6Kγ replication ori, and common E.coli strains like DH10B and XL1-Blue are not able to maintain this plasmid, but it replicates as a multiple copy plasmid in bacteria expressing the pir-116 protein.
Construction of a transposable cassette with a loxP-Zeo-loxP segment
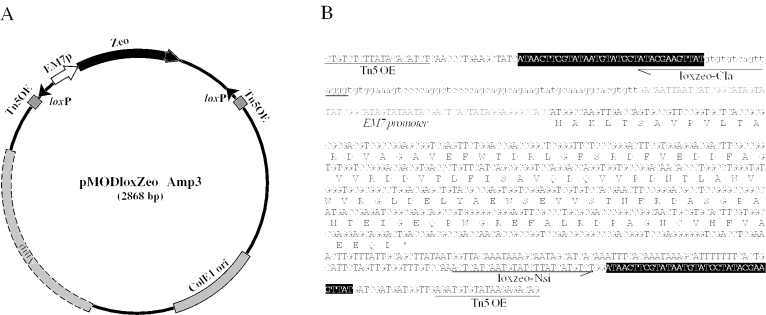
Another plasmid, pMODloxZeoΔamp3 (Figure 2), which has a loxP-Zeo-loxP cassette between modified Tn5 outer elements, was constructed to facilitate the third loxP element insertion. Tn5 and Tn7 require individual machineries for their transposition and both elements can be introduced in the cis configuration. pMODloxZeoΔamp3 has a pUC/Col E1 replication origin, a disrupted ampicillin-resistance gene, and a functional EM7-driven Zeocin-resistance gene, and thus confers ZeoR on common laboratory strains of E.coli.
Figure 2.
Structure of pMODloxZeoΔAmp3. (A) The structure of pMODloxZeoΔAmp3, which serves a transposable element having loxP sequences and Zeocin-resistance marker. This plasmid does not confer ampicillin resistance because its bla gene is truncated, but does confer Zeocin resistance through EM7 promoter-Zeo cassette (B). The nucleotide sequence of the transposable segment is shown. The loxP sequences are indicated by solid boxes. The positions of sequencing primers, lox-Cla and lox-Nsi were indicated by arrows.
pBSDT-AII, a backbone plasmid for gene targeting vectors
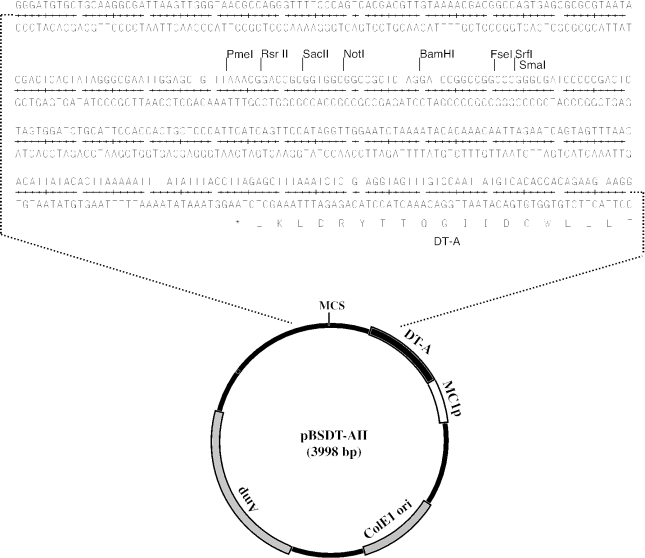
Plasmid pBSDT-AII has several characteristics to serve a backbone function in the final construct: the diphtheria toxin A fragment gene (DT-A) driven by the viral MC-1 promoter functions as a negative selection marker in ES cells (15), and there is a unique cloning site, BamHI, and sites for PmeI, SacII, NotI, FseI and SrfI, which cut infrequently (Figure 3). These provide anchors for restriction mapping of subcloned genomic DNA and, more importantly, they will be unique cutting sites for linearization of the final construct prior to transformation into ES cells. However, the introduction of DT-A and MCS disrupted the coding sequence of the α-fragment of β-galactosidase, and thus blue–white selection on X-gal/IPTG plates is not possible. pBSDTA-II replicates via the ColE1/pUC origin, and thus it is present as a multicopy plasmid in E.coli and confers ampicillin resistance.
Figure 3.
Structure of pBSDT-AII. Plasmid pBSDT-AII functions as the backbone for mouse targeting vectors. It has a unique multiple cloning site (MCS) and a negative selection marker, diphtheria toxin-A fragment (DT-A), to be expressed in mammalian cells through the MC-1 promoter (MC-1p). This plasmid can be propagated in common E.coli strains in the presence of ampicillin (Amp). The nucleotide sequence around the MCS and a part of the DT-A coding region are shown.
Creation of a floxed allele of mouse mVam2/Vps41 gene
We have made several complex targeting constructs by use of the developed plasmids shown above. Here, the strategy for creating an mVam2/Vps41 gene knock-out allele is described as an example (Figure 4). The yeast VAM2/VPS41 is required for normal vacuolar morphogenesis (16–18), and its counterparts are found ubiquitously in mammals (19), higher plants, as well as nematode and insect (20). In Drosophila, a mutation of it causes defective eye pigmentation, and it is suggested to be involved in vesicular trafficking (21). We are interested in the function of the mouse Vam2/Vps41 homologue (mVam2). Because it is predicted to be involved in basic cellular functions like vesicular trafficking, a defect of it may result in lethality in the earliest stages of development. We decided to create a floxed allele that can be inactivated upon excision by Cre recombinase expressed from cell type- and/or stage-specific promoters or genetically modified adenovirus (22).
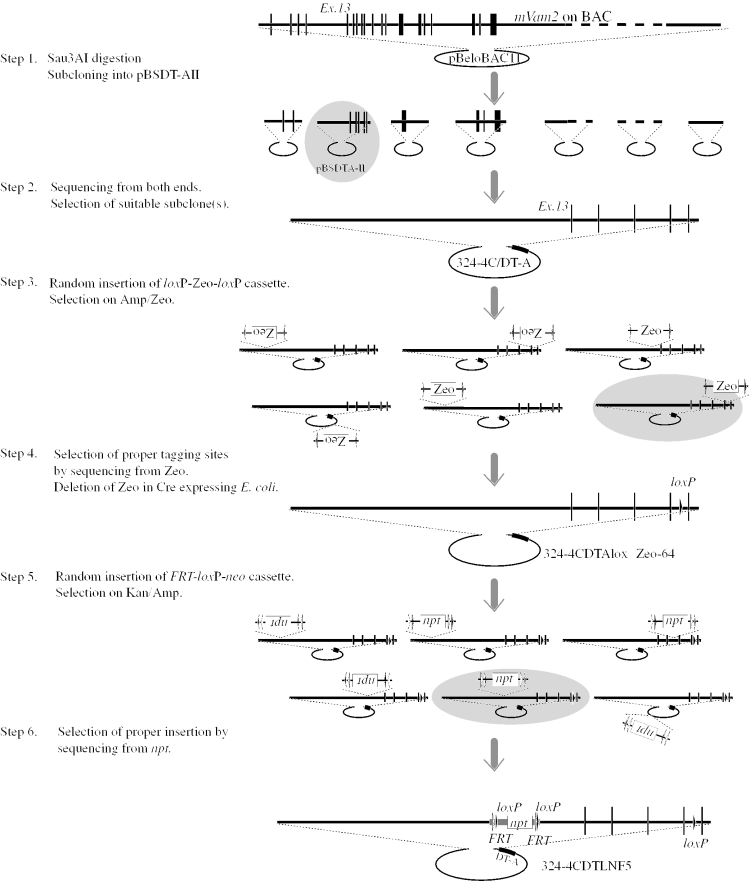
Figure 4.
Diagram of creation of a targeting construct using in vitro transposition reactions. The strategy for a floxed mVam2 allele is drawn schematically. Mouse genomic DNA is indicated by thick lines and exons by vertical lines. The loxP and FRT elements are represented by closed and open triangles. In steps 1, 3 and 5, the selected constructs are indicated by gray ovals.
We identified a BAC clone containing a part of the mVam2 locus from a mouse 129Sv genomic library by PCR. A PCR primer pair directed toward the 3′-noncoding region of the cDNA (GenBank accession no. ABB028843) gave a strong signal of the expected size upon PCR analysis (data not shown) from BAC clone 191m22 in the ES BAC library (Incyte Genomics). The nucleotide sequences of both ends of the BAC insert were determined and compared with a draft of the mouse genome sequence. BAC 191m22 was not enough to cover the entire region of mVam2, but contained the exon 13 to the last exon of the mVam2 locus (Figure 4). The overall scheme of the construction is summarized in Figure 4.
BAC 191m22 was partially digested with Sau3AI, and then the fractionated 10–15 kb fragments were cloned into the BamHI site of pBSDT-AII (Figure 3). Ninety-six colonies were subjected to direct PCR analysis to obtain three individual clones containing exon 14 of mVam2. Through brief restriction mapping and end-sequencing using primer DT-AR or M13 Fwd23 (Table 1), we selected 324-4C/DT-A as a suitable subclone for the construction.
Next, loxP was introduced into 324-4C/DT-A by means of the in vitro transposition reaction. Plasmid 324-4C/DT-A was incubated with pMODloxZeoΔamp3 in the presence of a modified Tn5-derived transposase (EZ::TN transposase; Epicentre). The reaction products were introduced into E.coli XL1-Blue by transformation, and 96 ZeoR, AmpS colonies were isolated. Their plasmids were recovered and subjected to sequencing using primer loxzeo-Cla or loxzeo-Nsi (Table 1 and Figure 2B) to determine the position and orientation of each tagging site. The sequence information also verified the structure and integrity of the subcloned fragment. Then, an appropriate clone with the loxP-Zeo-loxP cassette in a desired position and orientation, 324-4CDTAloxZeo-64, was selected. The plasmid was propagated in E.coli strain BH25.8 expressing Cre recombinase to remove the Zeo sequence and one loxP. Cre-mediated excision was very efficient: we found 100% removal in many experiments (data not shown). The plasmid, 324-4CDTAloxΔZeo-64, was recovered from BH25.8 cells and propagated in DH10B cells, because the quality and quantity of plasmids with BH25.8 cells was not satisfactory, partly due to intermolecular recombination between the loxP sites.
The final step was introduction of the FRT-loxP-PGK/EM7p-npt-FRT-loxP cassette into 324-4CDTAloxΔZeo-64. We performed the second round of the in vitro transposition reaction in this step. Plasmid pGPS21loxFRTNeo was incubated with 324-4CDTAloxΔZeo-64 in the presence of a modified T7 transposase and introduced into E.coli DH10B cells, and selected on ampicillin and kanamycin. We picked up 96 AmpR KanR colonies and then determined the tagging sites by sequencing from both ends of the FRT-loxP-PGK/EM7p-npt-FRT-loxP cassette. The position and orientation of each insertion was determined by comparison with the genome draft and assembled sequence obtained in step 3 (Figure 4).
We chose 324DTLN-F05 as the final targeting vector. It has a 4.2 kb uninterrupted homologous left arm and a short right arm of 1.3 kb. The final construct contains FRT-loxP-PGK/EM7p-npt-FRT-loxP in intron 12, and the third loxP sequence in intron 16. The expression of Flpe recombinase will remove the positive selection marker, npt, leaving two loxP sites in introns 12 and 16. This floxed allele can be inactivated by the expression of Cre recombinase in particular tissues or developmental stages. The plasmid was purified by alkaline-lysis and CsCl ultracentrifugation, linearized with PmeI at the multiple cloning site of the pBSDT-AII backbone and then electroporated into R1 embryonic stem cell (23). We identified 4 ES clones that underwent homologous recombination out of 264 G418R colonies on PCR analysis. These homologous recombinants were introduced into blastocysts of C57Bl/6 mice and we obtained germ line transmitting chimeric mice.
The idea of the transposon-generated gene targeting constructs was reported first by Westphal and Leder (12). We applied the idea to create conditional knock-out constructs that are indispensable for studying the function of genes involved in basic cellular functions including organelle assembly (24) or acidification (25). The described plasmids and transposons are components of the system applicable to any gene of interest, and we could save time and effort in our several ongoing gene targeting projects.
Although most of mouse genome sequence has been determined, there remain ambiguous segments scattering all over the genome. The mVam2 locus, as well as the other genes of our interests, is not assembled into contiguous sequence in the public databases, but several contigs separated by highly repetitive sequences. We avoid repeat sequences in the short homologous arm of the final targeting vector, because they are practically impossible to amplify by PCR thus compromise the screening of ES recombinants. We determined the position and orientation of tagging sites by sequencing the resultant plasmids at steps 3 and 6 (Figure 4) using primers designed to be annealed to the ends of transposons. The 96 clones gave ∼90 reliable sequences of 400–500 bp, in each direction, that cover almost all (10–20 kb) of the cloned segments in both strands, thus we could verify the genomic structure at the nucleotide sequence level.
In the case of the mVam2 gene targeting described here, we first subcloned a genomic segment from BAC into a plasmid, pBSDT-AII, by ‘shot-gun’ strategy involving restriction digestion, size fractionation and ligation. Several groups developed alternative highly efficient strategies for retrieving a large genomic DNA by homologous recombination in E.coli cells having a bacterial or phage-derived recombination system (26,27), and many excellent applications for engineering a large genomic DNA has been developed to date (28–31). We found that the use of such recombineering system eliminates in vitro DNA manipulations. In current projects, we employ bacterial in vivo homologous recombination for step 1 of Figure 4, by using a PCR generated pBSDT-AII fragment flanked by 75 bp arms homologous to the ends of genomic segment to be retrieved. However, we prefer the transposon-mediated approach for insertion of FRT, loxP and neo gene to the recombineering system, because we can obtain the genomic structure of the locus as discussed above. In addition, because many potential candidates having the neo cassette with FRT and loxP at different position can be prepared simultaneously (Figure 4, step 5), there are more choices for assembling different targeting vectors without intensive work. This versatility is an obvious advantage if one construct fails to undergo homologous recombination in ES cells, which is not a rare event in our experience.
While this study was under review, Zhang et al. (32) reported a similar strategy to introduce loxP and selectable markers into targeting constructs by the use of Mu-based transposon. Combination of their and our sets of transposons will be possible because Mu, Tn5 and Tn7 can be integrated into one contiguous DNA segment, adding further layer of gene modification strategy.
Acknowledgments
We wish to thank Drs Osamu Minowa and Tetsuo Noda for providing the loxP-Kan plasmid, and Drs Andreas Nagy, Yujiro Higashi and Hisato Kondoh for ES cell line R1. We also thank Dr Neal G. Copeland for providing the E.coli homologous recombination system. This study was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, and by grants from the Noda Foundation and the Japan Foundation for Applied Enzymology. Funding to pay the Open Access publication charges for this article was provided by MEXT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Deng C., Capecchi M.R. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell. Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasty P., Rivera-Perez J., Bradley A. The length of homology required for gene targeting in embryonic stem cells. Mol. Cell. Biol. 1991;11:5586–5591. doi: 10.1128/mcb.11.11.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu Z., Books J., Kaufman R., Ley T. Long targeting arms do not increase the efficiency of homologous recombination in the beta-globin locus of murine embryonic stem cells. Blood. 2003;102:1531–1533. doi: 10.1182/blood-2003-03-0708. [DOI] [PubMed] [Google Scholar]
- 4.Abuin A., Bradley A. Recycling selectable markers in mouse embryonic stem cells. Mol. Cell. Biol. 1996;16:1851–1856. doi: 10.1128/mcb.16.4.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nature Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 6.Fiering S., Kim C., Epner E., Groudine M. An ‘in-out’ strategy using gene targeting and FLP recombinase for the functional dissection of complex DNA regulatory elements: analysis of the β-globin locus control region. Proc. Natl Acad. Sci. USA. 1993;90:8469–8473. doi: 10.1073/pnas.90.18.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn R., Schwenk F., Aguet M., Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 8.DeBoy R.T., Craig N.L. Tn7 transposition as a probe of cis interactions between widely separated (190 kilobases apart) DNA sites in the Escherichia coli chromosome. J. Bateriol. 1996;178:6184–6191. doi: 10.1128/jb.178.21.6184-6191.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biery M.C., Stewart F.J., Stellwagen A.E., Raleigh E.A., Craig N.L. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 2000;28:1067–1077. doi: 10.1093/nar/28.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goryshin I.Y., Reznikoff W.S. Tn5 in vitro transposition. J. Biol. Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 11.Reznikoff W.S., Goryshin I.Y., Jendrisak J.J. Tn5 as a molecular genetics tool: in vitro transposition and the coupling of in vitro technologies with in vivo transposition. Methods Mol. Biol. 2004;260:83–96. doi: 10.1385/1-59259-755-6:083. [DOI] [PubMed] [Google Scholar]
- 12.Westphal C.H., Leder P. Transposon-generated ‘knock-out’ and ‘knock-in’ gene-targeting constructs for use in mice. Curr. Biol. 1997;7:530–533. doi: 10.1016/s0960-9822(06)00224-7. [DOI] [PubMed] [Google Scholar]
- 13.Kunkel T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchholz F., Angrand P.O., Stewart A.F. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 15.Yagi T., Nada S., Watanabe N., Tamemoto H., Kohmura N., Ikawa Y., Aizawa S. A novel negative selection for homologous recombinants using diphtheria toxin A fragment gene. Anal. Biochem. 1993;214:77–86. doi: 10.1006/abio.1993.1459. [DOI] [PubMed] [Google Scholar]
- 16.Raymond C.K., Howald-Stevenson I., Vater C.A., Stevens T.H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura N., Hirata A., Ohsumi Y., Wada Y. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1997;272:11344–11349. doi: 10.1074/jbc.272.17.11344. [DOI] [PubMed] [Google Scholar]
- 18.Wada Y., Ohsumi Y., Anraku Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J. Biol. Chem. 1992;267:18665–18670. [PubMed] [Google Scholar]
- 19.McVey Ward D., Radisky D., Scullion M.A., Tuttle M.S., Vaughn M., Kaplan J. hVPS41 is expressed in multiple isoforms and can associate with vesicles through a RING-H2 finger motif. Exp. Cell. Res. 2001;267:126–134. doi: 10.1006/excr.2001.5244. [DOI] [PubMed] [Google Scholar]
- 20.Radisky D.C., Snyder W.B., Emr S.D., Kaplan J. Characterization of VPS41, a gene required for vacuolar trafficking and high-affinity iron transport in yeast. Proc. Natl Acad. Sci. USA. 1997;94:5662–5666. doi: 10.1073/pnas.94.11.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warner T.S., Sinclair D.A., Fitzpatrick K.A., Singh M., Devlin R.H., Honda B.M. The light gene of Drosophila melanogaster encodes a homologue of VPS41, a yeast gene involved in cellular-protein trafficking. Genome. 1998;41:236–243. [PubMed] [Google Scholar]
- 22.Wang Y., Krushel L.A., Edelman G.M. Targeted DNA recombination in vivo using an adenovirus carrying the cre recombinase gene. Proc. Natl Acad. Sci. USA. 1996;93:3932–3936. doi: 10.1073/pnas.93.9.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J.C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl Acad. Sci. USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura N., Yamamoto A., Wada Y., Futai M. Syntaxin 7 mediates endocytic trafficking to late endosomes. J. Biol. Chem. 2000;275:6523–6529. doi: 10.1074/jbc.275.9.6523. [DOI] [PubMed] [Google Scholar]
- 25.Sun-Wada G.H., Murata Y., Yamamoto A., Kanazawa H., Wada Y., Futai M. Acidic endomembrane organelles are required for mouse postimplantation development. Dev. Biol. 2000;228:315–325. doi: 10.1006/dbio.2000.9963. [DOI] [PubMed] [Google Scholar]
- 26.Angrand P.O., Daigle N., van der Hoeven F., Scholer H.R., Stewart A.F. Simplified generation of targeting constructs using ET recombination. Nucleic Acids Res. 1999;27:e16. doi: 10.1093/nar/27.17.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee E.C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L., Jenkins N.A., Copeland N.G. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 28.Adams D.J., Biggs P.J., Cox T., Davies R., van der Weyden L., Jonkers J., Smith J., Plumb B., Taylor R., Nishijima I., et al. Mutagenic insertion and chromosome engineering resource (MICER) Nature Genet. 2004;36:867–871. doi: 10.1038/ng1388. [DOI] [PubMed] [Google Scholar]
- 29.Cotta-de-Almeida V., Schonhoff S., Shibata T., Leiter A., Snapper S.B. A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res. 2003;13:2190–2194. doi: 10.1101/gr.1356503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Testa G., Zhang Y., Vintersten K., Benes V., Pijnappel W.W., Chambers I., Smith A.J., Smith A.G., Stewart A.F. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat. Biotechnol. 2003;21:443–447. doi: 10.1038/nbt804. [DOI] [PubMed] [Google Scholar]
- 31.Zhang P., Li M.Z., Elledge S.J. Towards genetic genome projects: genomic library screening and gene-targeting vector construction in a single step. Nature Genet. 2002;30:31–39. doi: 10.1038/ng797. [DOI] [PubMed] [Google Scholar]
- 32.Zhang C., Kitsberg D., Chy H., Zhou Q., Morrison J.R. Transposon-mediated generation of targeting vectors for the production of gene knockouts. Nucleic Acids Res. 2005;33:e24. doi: 10.1093/nar/gni014. [DOI] [PMC free article] [PubMed] [Google Scholar]