Abstract
Pulmonary hypertension (PH) is an extremely malignant pulmonary vascular disease of unknown etiology. ADAR1 is an RNA editing enzyme that converts adenosine in RNA to inosine, thereby affecting RNA expression. However, the role of ADAR1 in PH development remains unclear. In the present study, we investigated the biological role and molecular mechanism of ADAR1 in PH pulmonary vascular remodeling. Overexpression of ADAR1 aggravated PH progression and promoted the proliferation of pulmonary artery smooth muscle cells (PASMCs). Conversely, inhibition of ADAR1 produced opposite effects. High-throughput whole transcriptome sequencing showed that ADAR1 was an important regulator of circRNAs in PH. CircCDK17 level was significantly lowered in the serum of PH patients. The effects of ADAR1 on cell cycle progression and proliferation were mediated by circCDK17. ADAR1 affects the stability of circCDK17 by mediating A-to-I modification at the A5 and A293 sites of circCDK17 to prevent it from m1A modification. We demonstrate for the first time that ADAR1 contributes to the PH development, at least partially, through m1A modification of circCDK17 and the subsequent PASMCs proliferation. Our study provides a novel therapeutic strategy for treatment of PH and the evidence for circCDK17 as a potential novel marker for the diagnosis of this disease.
Key words: Pulmonary hypertension, ADAR1, Circular RNA, Vascular remodeling, Cell proliferation
Graphical abstract
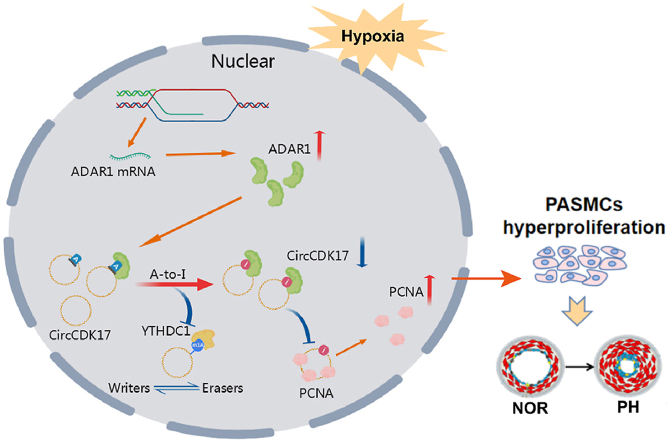
The expression of ADAR1 is elevated under hypoxic condition. ADAR1 binds to circCDK17 and reduces its expression through inhibition of its m1A modification. This further reduces the ubiquitination levels of PCNA and promotes the proliferation of PASMCs, leading to pulmonary vascular remodeling.
1. Introduction
Pulmonary hypertension (PH) is a progressive cardiopulmonary disorder characterized by elevated pulmonary vascular resistance and high pulmonary artery pressure, which eventually results in right ventricular failure and death. The exact cause of PH is still unclear. Hypoxia is the common cause of a variety of common clinical lung diseases, like chronic obstructive pulmonary disease, chronic mountain sickness1, 2, 3, and hypoxic pulmonary hypertension4,5. In particular, more than 140 million people live above 2500 m from sea level and are exposed to chronic hypoxia and some people have symptoms of hypoxic PH, which the same is true for the clinical implications6,7. The basic pathological changes of PH include persistent pulmonary arteriolar contraction, hypoxic pulmonary vascular remodeling (HPVR), and pulmonary vascular fibrosis. Pulmonary artery media thickening is a major pathological process of HPVR mainly caused by excessive proliferation and apoptosis inhibition of pulmonary artery smooth muscle cells (PASMCs)8,9. Therefore, exploring new pathological mechanisms underlying the proliferation of PASMCs and identifying potential therapeutic targets are still an urgent task for PH therapy.
ADAR1, a member of the adenosine deaminase RNA specific (ADAR) family, converts adenosine (A) to inosine (I) in RNA. A-to-I modification is a widespread RNA editing process in mammals10,11. This modification leads to nonsynonymous codon changes in transcription and alternative splicing. The RNA editing enzyme family consists of three major members: ADAR1, ADAR2, and ADAR3. The gene sequences of the three enzymes are highly conserved in vertebrates12,13. ADAR1 is widely expressed and has three subtypes, namely p150, p110, and p80. p150 mainly exists in the cytosol, whereas p110 and p80 are mainly expressed in the nucleus and nucleolus respectively10,14. Studies have shown that both p150 and p110 travel freely between cytoplasm and nucleus15,16. Recent studies have revealed that ADAR1 is involved in the development of various tumors by editing a variety of tumor suppressors and proto-oncogenes. Loss of function of ADAR1 in tumor cells increases tumor sensitivity to immunotherapy17. On the contrary, blockade of ADAR1 was also found to down-regulate smooth muscle contractile protein and attenuate vascular remodeling18. Furthermore, silencing of ADAR1 in hypoxic tumor cells has been shown to promote apoptosis via FGFR219.
In hypoxic situation, an increase of A-to-I RNA editing was found in the 3′UTR of the F11R RNA, to promote F11R expression20. Moreover, ADAR1 also regulated HIF1α signaling by affecting the antisense chain transcripts of HIF1α-AS221. These results indicate that ADAR1 is related to many hypoxia-related diseases of nervous, immune, and cardiovascular systems. However, the role of ADAR1 in hypoxia-induced PH remains unclear.
In eukaryotic cells, circular RNAs (circRNAs) are special non-coding RNAs that produced by reverse splicing of exons of protein-coding genes, sometimes with introns attached to introns22, 23, 24. They exist in mammalian cells and have many important regulatory functions at the post-transcription level25. Emerging evidence indicates that circRNAs are involved in the pathogenesis of many diseases, including PH. As circRNAs are more stable and conserved than linear RNAs, they may serve as better diagnostic markers for PH. Importantly, studies have shown that ADAR1 can regulate circRNAs production by affecting intron splicing26, suggesting that ADAR1 may be involved in regulating dysregulation circRNAs in hypoxic PH, but the mechanism is unclear.
In the current study, we aimed to investigate the role of ADAR1 in the pathogenesis of PH and explore the molecular mechanism of ADAR1 regulating PASMCs hyperproliferation mediated vascular remodeling in PH. Our study provides a novel strategy to treat PH and a new serum biomarker for PH diagnosis.
2. Materials and methods
The authors declare that all supporting data are available within the article and its online-only Supporting Information. Expanded Methods can be found in the online-only Supporting Information.
2.1. Study approval
All human samples were collected with the informed consent of the patients and the procedures were approved by the Ethics Committee for the Use of Human Subjects of the Shenzhen People's Hospital (China) which was in accordance with The Code of Ethics of the Helsinki Declaration of World Medical Association for experiments involving humans. All animal study protocols were reviewed and approved by the Ethics Committee of Shenzhen People's Hospital. This study was in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996).
2.2. Study design
Animals were randomly assigned to the experimental groups. Following the principle of equal opportunity, the random number table method was used to assign each animal. No samples or animals were excluded from analysis. All quantifications (cell viability, cell cycle, proliferation, histology analyses, confocal imaging) were performed in a blind fashion. All figures are representative of at least three experiments unless otherwise noted.
2.3. Clinical sample collection
Lung tissue samples from patients with preoperative hypoxic PH and matched healthy control samples were from the lung transplantation group of Wuxi People's Hospital, Affiliated to Nanjing Medical University, China. Healthy controls collected from donors not suitable for transplantation. Clinical sample information was shown in Supporting Information Table S1.
2.4. Animal experimental protocols
All animals were housed in specific pathogen-free microisolator cages, kept on a 12-h light cycle, and fed autoclaved food and reverse-osmosis water. Mouse hypoxia-PH models were generated by exposing mice (6 weeks old) to 10% oxygen for 4 weeks. For the Sugen-hypoxia (SuHx) model of PH, mice (6 weeks old) were injected subcutaneously with a single dose of the VEGFR inhibitor Su5416 (20 mg/kg, Sigma–Aldrich, USA). Su5416 was suspended in carboxymethylcellulose (CMC) solution (0.5% [w/v] carboxymethylcellulose sodium, 0.9% [w/v] sodium chloride, 0.4% [v/v] polysorbate 80, and 0.9% [v/v] benzyl alcohol indeionized water). Control mice received vehicle injection instead. Animals injected with Su5416 were exposed to chronic hypoxia (10% O2) in the ventilation chamber for 3 weeks and normal oxygen (21% O2) for 2 weeks27.
2.4.1. Serotype 9 adenovirus-associated virus (AAV9) hypoxia PH model
AAV9 overexpression plasmid (rAAV-CMV-ADAR-P2A-EGFP-WPRE-hGH pA) targeting ADAR1 was constructed (GOSV0313962, Genechem, China). 1010‒1011 genome equivalent vectors were prepared in Hank's Balanced Salt Solution (HBSS) with 30 μL28. Six-week-old male C57BL/6 mice (weighing approximately 25 g each) anesthetized with isoflurane were infected with two vectors AAV-ADAR1 and AAV negative control (AAV-NC) via nasal drip, and the infected mice were randomly divided into the normal oxygen group and the hypoxia group (n = 6 mice each), and the hypoxia PH mouse model was created as described above.
2.4.2. 8-Azaadenosine (8Aza) treatment PH model
Six-week-old male C57BL/6 mice (weighing approximately 25 g each) were randomly divided into six groups as follows: normoxic environment plus vehicle (NOR + vehicle) group, normoxic environment plus 8Aza (NOR+8Aza) group, hypoxic environment plus intraperitoneal injection of vehicle (HYP + vehicle) group, hypoxic environment plus intraperitoneal injection of 8Aza (HYP+8Aza) group, hypoxia combined with Su5416 environment plus intraperitoneal injection of vehicle (SuHx + vehicle) and Hypoxia combined with Su5416 environment plus intraperitoneal injection of 8Aza (SuHx+8Aza) (n = 6 mice each). 8Aza (2 mg/kg) (HY-115686, MedChemExpress, USA) was suspended in 10% dimethylsulfoxide, 40% Polyethylene glycol 300, 5% Tween-80, and 45% saline, and injected intraperitoneally every four days for 28 days in hypoxia alone or 35 days SuHx PH models as previously mentioned, respectively29. Control mice received vehicle injections instead.
2.5. Transcriptome sequencing analysis
Total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion) following the manufacturer's protocol. RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The samples with RNA Integrity Number (RIN) ≥ 7 were subjected to the subsequent analysis. The libraries were constructed using TruSeq Stranded Total RNA with Ribo-Zero Gold according to the manufacturer's instructions. Then these libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500 or other platform) and 150 bp/125 bp paired-end reads were generated.
2.6. Cell lines and cell culture
Human pulmonary artery smooth muscle cells (PASMCs) and pulmonary artery endothelial cells (PAECs) were obtained from Meisen Chinese Tissue Culture Collections (Zhejiang, China). Ten milliliters fetal bovine serum (FBS), 5 mL smooth muscle cell growth supplement, and 5 mL penicillin/streptomycin solution in smooth muscle cell medium (1101, ScienCell, USA) was added to PASMCs cultures. Cells were then incubated at 37 °C in a 5% CO2 humidified incubator. PAECs were incubated in endothelial cell medium (1001, ScienCell, USA) containing 25 mL FBS, 5 mL endothelial cell growth factor, and 5 mL penicillin/streptomycin solution at 37 °C in a 5% CO2 humidified incubator. For the hypoxia experiments, cells were placed in the modular incubator chamber (Fisher Scientific, Waltham, MA, USA) with 1% O2.
2.7. Right ventricular systolic pressure (RVSP) and cardiac function measurement
Mice were anesthetized by injection of pentobarbital, and a Millar catheter (SP-836 F) was surgically inserted into the right jugular vein, while changes in RVSP were monitored using Power Lab monitoring equipment. Then, the thoracic cavity of the mice was quickly cut with surgical scissors, the heart and lung tissues were removed, the heart and lung lobes were separated, the right ventricle was slowly separated along the ventricular septum with the tip of surgical scissors, and the degree of right heart hypertrophy was evaluated by weighing.
2.8. Hematoxylin-eosin (HE) staining
Lung tissue sections were deparaffinized, stained in hematoxylin, and differentiated in hydrochloric alcohol. The tissue sections were transferred to eosin solution, dehydrated, permeabilized, and then sealed and dried. The medial wall thickness was calculated with Image J software. The entire vessel area was identified as the total area. The medial wall thickness ratio was calculated as (total area‒lumen area)/total area30.
2.9. Vascular muscularization analysis
The percentage of muscularized pulmonary vessels was determined by dividing the number of partially or fully muscular vessels by the total number counted in the same experimental group. The medial wall thickness of fully muscularized intra-acinar arteries was calculated with ImageJ software. The entire vessel area was identified as the total area. The medial wall thickness ratio was calculated as (total area‒lumen area)/total area30.
2.10. Western blotting analysis
Proteins were extracted using lysis buffer (P0013B, Beyotime, China). Proteins were separated by polyacrylamide gel electrophoresis and electrotransferred to nitrocellulose membranes. The membranes were then incubated with antibodies against proliferating cell nuclear antigen (13,110, PCNA; Cell Signaling Technology; 1:1000 dilution), cyclin A (67,955, Cell Signaling Technology; 1:1000 dilution), cyclin-dependent kinase (67,575-1-lg, CDK1; Proteintech; 1:5000 dilution), β-actin (3700, Cell Signaling Technology; 1:20,000 dilution), ADAR1 (81,284, Cell Signaling Technology; 1:1000 dilution), ALKBH1 (ab126596, Abcam; 1:1000 dilution), YTHDC1 (ab259990, Abcam; 1:1000 dilution) and TRMT6 (16727-1-AP, Proteintech; 1:500 dilution) overnight at 4 °C and followed by incubation with horseradish peroxidase-labeled secondary antibodies at room temperature, and proteins were visualized with enhanced chemiluminescence reagents.
2.11. Quantitative real-time PCR analysis (qPCR)
RNA was extracted by Trizol (Invitrogen, USA) method. Real-time quantitative two-step reverse transcription PCR kit (Applied Biosystems, USA) was used to extract RNA from primary cultured PASMCs, hypoxic mice and PH patients, and the content of target RNA was determined. The cycle number was set in a qPCR instrument (Applied Biosystems, USA), and the data were analyzed by the solubilization curve method. Relevant primer information was shown in Supporting Information Table S2.
2.12. RNA methylation immunoprecipitation (meRIP)
Experiments were performed using GenSeq® MeRIP (CloudSeq Biotech, China) kit according to the manufacturer's protocol. Briefly, RNA was first fragmented into 200 bp fragments, and immunoprecipitated magnetic beads were prepared, resuspended, washed, and incubated with 5 μg anti-1-methyladenosine (m1A) (ab208196, Abcam, USA) or 5 μg anti-6-methyladenosine (m6A) (56,593, Cell Signaling Technology, USA) antibody. Then the fragmented RNA was incubated with magnetic beads at 4 °C, and the magnetic beads were cleaned and centrifuged for RNA extraction and purification.
2.13. RNA pull-down
RNA pull-down assays were performed using the RNA protein pull-down kit (20,164, Thermo Fisher Scientific, USA) according to the manufacturer's protocol. Biotin-labeled probes bound to proteins after binding to magnetic beads. RNA-bound beads were added to cell protein lysates for immunoprecipitation. The magnetic beads were boiled in SDS buffer, and the recovered proteins were then examined using Western blotting and silver staining analysis.
2.14. Silver staining
The silver staining of the Western blotting gel was performed following the protocol of Pierce™ Silver Stain for Mass Spectrometry (24,600, Thermo Fisher Scientific, USA). In brief, the electrophoresis gel was fixed and washed with 30% ethanol for 10 min, followed by washing with water for 10 min. After adding 100 mL of silver dye sensitizing solution (1×), the gel was washed twice with 100 mL of silver solution (1×) and once with water. After adding silver dye stop solution, the gel was then developed color and taken pictures.
2.15. RNA binding protein immunoprecipitation (RIP)
RNA-binding protein immunoprecipitation (RIP) assays were performed using a Magnetic RIP kit (17-700; Millipore, USA) according to the manufacturer's protocol. The cells were collected, and RIP lysate was added to swell and lysate the cells. The magnetic beads were incubated with antibodies, the RIP lysate was centrifuged, and the supernatant was added to RIP immunoprecipitation buffer containing magnetic bead‒antibody complex, incubated overnight at 4 °C, and then washed by centrifugation. The final RNA was purified and subjected to subsequent experiments.
2.16. Dot blotting assay
The sample was dotted onto the nitrocellulose membrane in the center of the grid. The membrane was dried and the nonspecific sites were blocked before incubation with the primary antibody and the secondary antibody. The membrane was then developed under X-rays after incubation with chemiluminescence solution ECL reagent.
2.17. Cell counting kit-8 assay (CCK8)
PASMCs were cultured in 96-well plates, cell transfection was performed when the cell density reached 60%–70%, and the cells were cultured in hypoxia (37 °C, 5% CO2, 3% O2) for 48 h 10 μL of CCK8 (HY-K0301, MedChemExpress, USA) solution was added to each well. The 96-well plates were incubated in a normal cell incubator (37 °C, 5% CO2) for 4 h. The absorbance at 450 nm was measured using a microplate reader.
2.18. Ethynyl-2′-deoxyuridine (EdU) assay
EdU assays were performed using the BeyoClick EdU cell proliferation kit according to the manufacturer's instructions (C0075S, Beyotime, China). In brief, 48 h after transfection of PASMCs in 24-well plates, 2 × EdU working solution was added to the wells, and fluorescence at a wavelength of 488 nm was detected by fluorescence confocal microscopy.
2.19. Cell cycle analysis
Cell cycle progression was detected by Cell Cycle and Apoptosis Analysis Kit (C1052, Beyotime, China). PASMCs treated according to experimental groups were treated with trypsin digestion solution, centrifuged, resuspended in an ethanol solution, and fixed at 4 °C for 24 h. The ethanol solution was removed, resuspended in PBS, and incubated with propidium iodide. The fluorescence values of stained cells and DNA were detected by flow cytometry for statistical analysis.
2.20. RNase R experiment
RNase R treatment Total RNA extracted was divided equally into two aliquots according to the manufacturer's protocol; one aliquot of RNA was incubated without RNase R and one aliquot with 3 U/μg RNase R (RNR07250, Epicenter Biotechnologies, USA) at 37 °C and used for subsequent experiments31.
2.21. Immunohistochemistry
Paraffin sections of mouse lung tissue were deparaffinized to water. Antigen repair, incubation with primary antibody at 4 °C overnight (ADAR1, 81284, Cell Signaling Technology, 1:200 dilution), with biotinylated secondary antibody, and diamine benzidine staining were performed. Fluorescence microscopy observation and statistical processing.
2.22. Cell and tissue immunofluorescence
Transfected PASMCs were cultured for 48 h, washed in 1 × PBS, fixed in 4% paraformaldehyde, lysed with 0.3% Triton, and blocked with 5% bovine serum albumin. Cells were then incubated with primary antibodies at 4 °C overnight (ADAR1, 81284, Cell Signaling Technology, 1:200 dilution; α-SMA, 67735-1-lg, Proteintech, 1:200 dilution), followed by incubation with fluorescent-labeled secondary antibodies. Nuclei were stained with DAPI and observed using a fluorescence microscope. The frozen sections of mouse lung tissues were analyzed in the same manner.
2.23. Fluorescence in situ hybridization (FISH)
Appropriate numbers of PASMCs were cultured in 24-well plates for cell climbing and treated accordingly. The cells were prehybridized after fixation and permeabilization. The corresponding probe storage solution was added to the hybridization solution, and the cells were incubated in a 37 °C incubator overnight. Cells were washed with hybridization wash, stained with DAPI staining solution, and observed using confocal microscopy.
2.24. Quantification and statistical analysis
Representative figures/images reflected the average level of each experiment. Normality of the data (using Shapiro–Wilk test) and the equality of group variance (using Brown–Forsythe test) were performed on all data using GraphPad Prism 8.0 software (GraphPad Software Inc., La Jolla, CA, USA). All data collection and processing in this study are expressed as mean ± standard error (means ± SEM). Single comparisons were made between the two groups using paired or unpaired t-tests. Other data were statistically analyzed using one-way ANOVA or two-way ANOVA with Dunnett's test where appropriate. A result of P < 0.05 was considered statistically significant. All experiments were repeated independently more than three times.
3. Results
3.1. ADAR1 is highly expressed in the lung tissues of PH patients and hypoxic mice
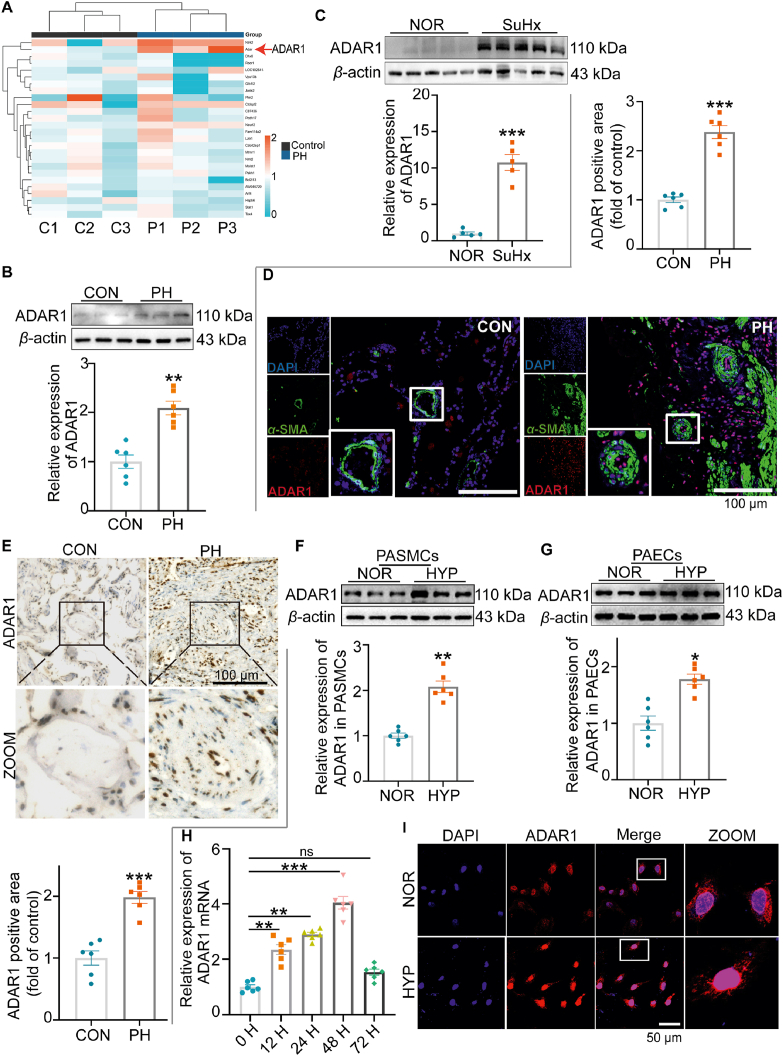
We randomly analyzed PH patient samples from the GEO database (GSE113439) and found that ADAR1 expression was increased in PH patients (Fig. 1A). To confirm this finding, we collected lung tissue samples from PH patients and PH mice induced by hypoxia combined with Su5416 injection (SuHx). Western blotting analysis showed that the expression of ADAR1 was increased in these lung tissues when compared with those in the control groups (Fig. 1B and C).
Figure 1.
ADAR1 was highly expressed in the lung tissues of PH patients and hypoxic mice. (A) The expression of ADAR1 in PH was analyzed with GEO database (GSE113439). (B–C) Western blotting analysis showing that ADAR1 expression was upregulated in the lung tissues of PH patients (B, n = 6) and SuHx-induced PH mice (C, n = 5). (D, E) Immunofluorescence (D) and immunohistochemistry (E) assays showing the expression and distribution of ADAR1 in the lung tissues of PH patients, n = 6. (F, G) Representative Western blots and group data showing the expression of ADAR1 in human PAECs and PASMCs, n = 6. (H) qPCR analysis showing the mRNA expression of ADAR1 in PASMCs, n = 6. (I) Immunofluorescence assay showing the subcellular localization of ADAR1 in PASMCs, n = 6. PASMCs, pulmonary artery smooth muscle cells; PAECs, pulmonary artery endothelial cells; CON (C), control; PH (P), Pulmonary hypertension; NOR, normoxia; HYP, hypoxia. SuHx, Hypoxia combined with Su5416, ns, no significance; H, h; Statistical analysis was performed with one-way ANOVA followed by Dunnett's test or the Student's t-test; All values are presented as mean ± SEM. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Immunofluorescence and immunostaining analysis revealed that ADAR1 was expressed in the pulmonary arteries of PH patients and co-localized with the smooth muscle marker α-SMA, indicating that ADAR1 was expressed in the smooth muscle layer of pulmonary arteries (Fig. 1D and E). To further clarify the cellular distribution of ADAR1, we detected the expression of ADAR1 in PASMCs and pulmonary artery endothelial cells (PAECs), and the results showed that the expression of ADAR1 was increased in both types of vascular cells, and the expression was more significant in PASMCs (Fig. 1F and G). qPCR analysis showed that ADAR1 level was increased in PASMCs upon hypoxia treatment in a time-dependent manner and reached the peak at 48 h (Fig. 1H). Immunofluorescence assay demonstrated that ADAR1 was distributed in both cytoplasm and nucleus in normal situation and hypoxia-induced relocation of more p110 ADAR1 into nucleus (Fig. 1I).
3.2. Overexpression of ADAR1 aggravates hypoxia-induced PH in vivo
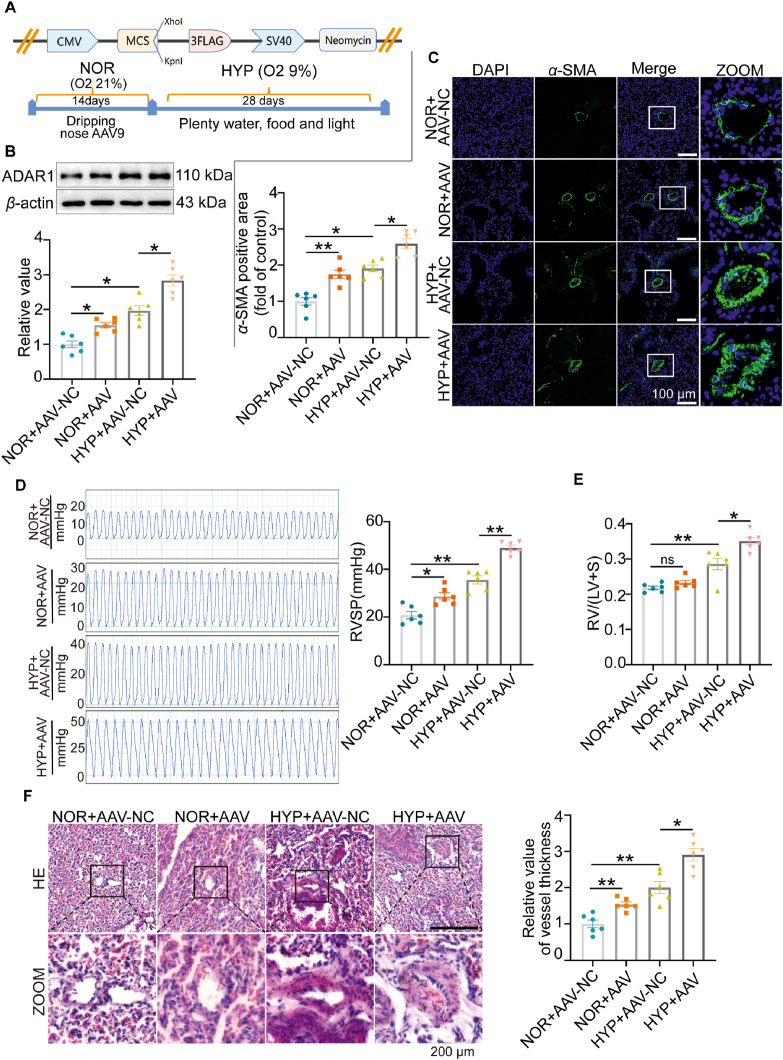
To explore the function of ADAR1 in vivo, we constructed an adeno-associated virus vector 9 (AAV9) to overexpress ADAR1 (OE-ADAR1) in mice exposed to hypoxia (Fig. 2A). Western blotting analysis confirmed that ADAR1 was up-regulated in the mice infected with AAV9 (Fig. 2B). Immunofluorescence assay showed that OE-ADAR1 further exacerbated hypoxia-induced muscularization (Fig. 2C). To clarify whether OE-ADAR1 aggravated hypoxia-induced PH, we evaluated RV/(left ventricle + S) and RV systolic pressure (RVSP). As shown in Fig. 2D and E, OE-ADAR1 increased RVSP under normal conditions and further aggravated both RVSP and RV/(left ventricle + S) in hypoxia situation. This was also confirmed with morphological changes. HE staining showed that OE-ADAR1 aggravated smooth muscle layer thickening in distal or proximal vascular vessel remodeling (Fig. 2F).
Figure 2.
Overexpression of ADAR1 aggravated hypoxia-induced PH in vivo. (A) Schematic illustration showing the construction of mice adeno-associated virus vector 9 (AAV9) plasmid and the treatment protocol. Mice were infected with AAV9 once a day for 14 days in normoxia situation and exposed to hypoxia for 28 days. (B) Representative Western blots and group data showing the expression of ADAR1 upon AAV9 treatment, n = 6. (C) Immunofluorescence assay showing the muscularization of vessels in mice infected with OE-ADAR1-AAV9, n = 6. (D, E) Hemodynamic assay showing that OE-ADAR1 aggravated Right ventricular systolic pressure (RVSP), and right ventricle (RV)/left ventricle (LV)+S weight ratio, n = 6. (F) Hematoxylin-eosin staining (HE) staining assay showing that OE-ADAR1 exacerbated pulmonary artery vascular remodeling, n = 6. NOR, normoxia; HYP, hypoxia, NC, negative control; ns, no significance; Statistical analysis was performed with two-way ANOVA followed by Dunnett's test; All values are presented as mean ± SEM. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.3. Inhibition of ADAR1 reverses hypoxia-induced PH
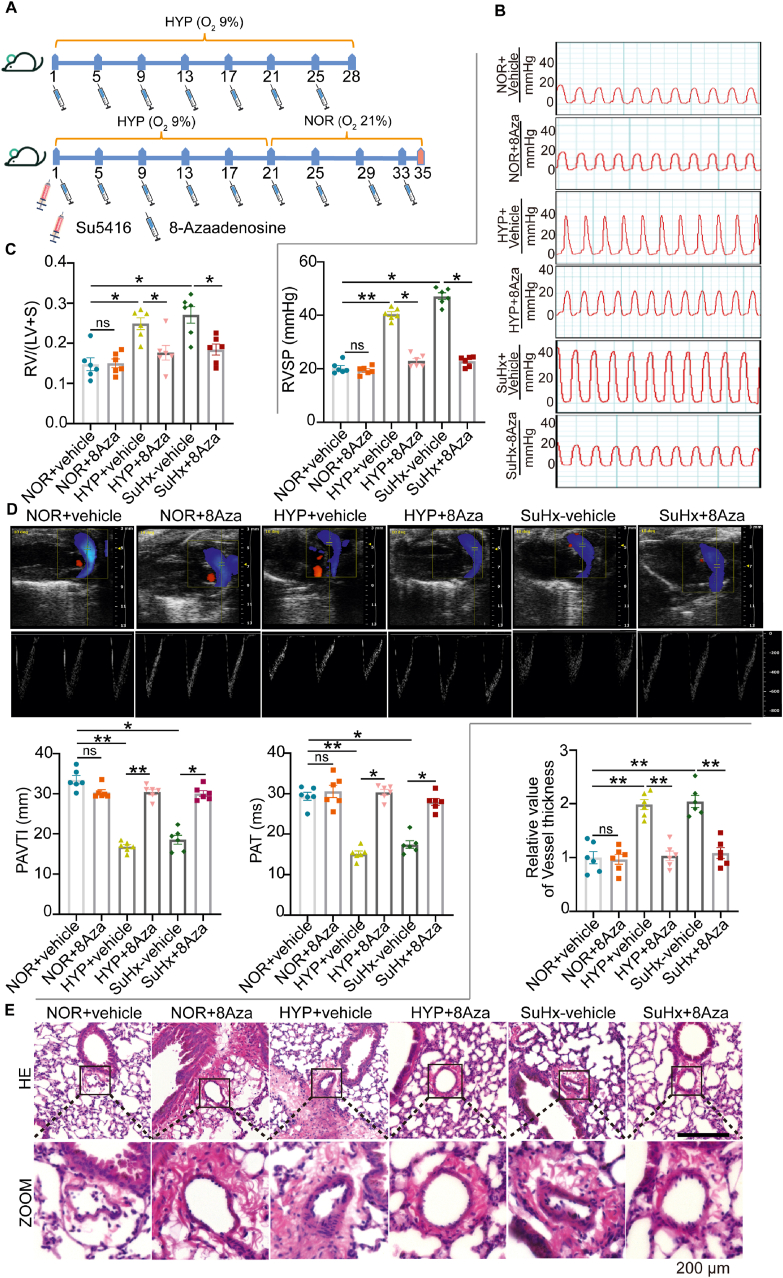
To further clarify the function of ADAR1 in vivo, we established mouse PH models induced by hypoxia alone or by hypoxia combined with Su5416 (SuHx). 8-Azaadenosine (8Aza, 2 mg/kg) was given with intraperitoneal injection every four days (Fig. 3A). As shown in Fig. 3B and C, inhibition of ADAR1 was able to attenuate the increased RV/(left ventricle + S) and RV systolic pressure induced by hypoxia and SuHx. Echocardiography also showed that 8Aza, which alone had no significant effect, significantly improved the impaired pulmonary artery acceleration time (PAT) and pulmonary artery velocity time integral (PAVTI) caused by hypoxia treatment, but there was no significant effect on heart rate (Fig. 3D, Supporting Information Fig. S1). Furthermore, HE staining demonstrated that hypoxia-induced distal or proximal pulmonary vascular remodeling as measured by smooth muscle layer thickening was largely improved by inhibition of ADAR1 (Fig. 3E).
Figure 3.
8Aza reversed hypoxic-induced PH progression. (A) Schematic illustration showing the drug treatment protocol. 8Aza was injected intraperitoneally every four days for 28 days for hypoxia or 35 days for SuHx (hypoxia combined with Su5416) PH models. (B, C) Hemodynamic analysis showing the therapeutic effects of 8Aza on Right ventricular systolic pressure (RVSP), and right ventricle (RV)/left ventricle (LV)+S weight ratio, n = 6. (D) Ultrasound analysis showing that 8Aza significantly attenuated the decreased pulmonary artery acceleration time (PAT) and pulmonary artery time integral (PAVTI) caused by either hypoxia alone or SuHx, n = 6. (E) Hematoxylin-eosin staining (HE) staining showing that 8Aza significantly attenuated the increased median wall thickness of pulmonary vascular vessels caused by either hypoxia alone or SuHx, n = 6. NOR, normoxia; HYP, hypoxia, ns, no significance; Statistical analysis was performed with two-way ANOVA followed by Dunnett's test; All values are presented as mean ± SEM. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.4. ADAR1 regulates the proliferation and cell cycle progression of PASMCs induced by hypoxia
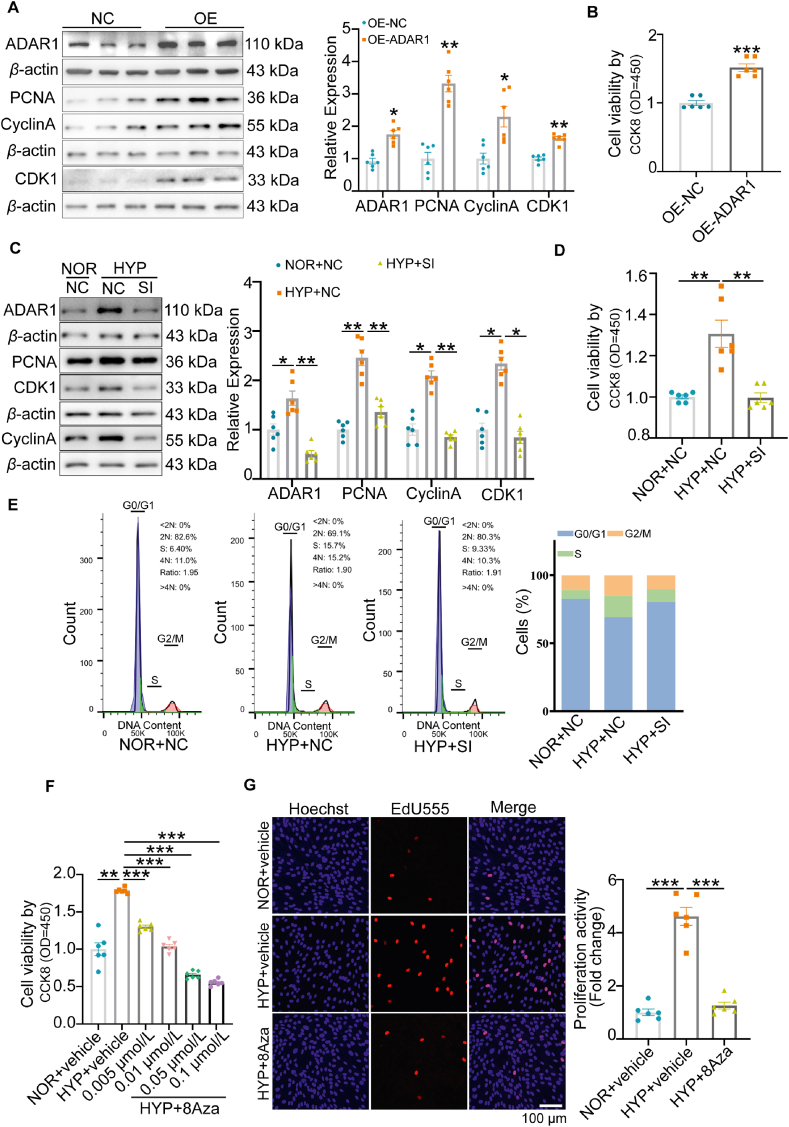
Next, we investigated the effect of ADAR1 on PASMCs proliferation in vitro. Western blotting analysis showed that overexpression of ADAR1 increased the protein levels of proliferating cell nuclear antigen (PCNA), cyclin A, and cyclin-dependent kinase 1 (CDK1) (Fig. 4A), suggesting that ADAR1 promoted PASMCs proliferation. These results were further confirmed with Cell Counting Kit-8 (CCK8) (Fig. 4B) and 5-ethynyl-2-deoxyuridine (EdU) staining (Supporting Information Fig. S2A) assays. On the contrary, silencing of ADAR1 with its siRNA (SI) abolished hypoxia-upregulated expressions of the above proteins (Fig. 4C) and CCK8 (Fig. 4D). In addition, flow cytometry assay showed that knockdown of ADAR1 reversed hypoxia-increased proportion of S-phase and reduced proportion of G0/G1 phase (Fig. 4E). The opposite results were also obtained when ADAR1 was overexpressed in the normoxia situation (Fig. S2B).
Figure 4.
ADAR1 regulated the proliferation and cell cycle progression of PASMCs. (A) and (C) Representative Western blots and group data showing that overexpression of ADAR1 (OE-ADAR1, A) increased, whereas silencing of ADAR1 (C) decreased the protein expression of PCNA, cyclin A and CDK1 in PASMCs, n = 6. (B) and (D) CCK8 analysis showing that OE-ADAR1 increased (B), whereas silencing of ADAR1 (D) decreased cell viability of PASMCs, n = 6. (E) Flow cytometry analysis showing the effect of silencing of ADAR1 on cell cycle of PASMCs, n = 6. (F) CCK8 analysis showing the concentration-dependent effect of 8Aza on hypoxia-stimulated cell viability, n = 6. (G) EdU staining assay showing that 8Aza abolished hypoxia-induced cell proliferation, n = 6. NOR, normoxia; HYP, hypoxia; NC, negative control; SI, siRNA of ADAR1; OE, ADAR1 overexpression plasmid; Statistical analysis was performed with two-way ANOVA followed by Dunnett's test or the Student's t-test; All values are presented as mean ± SEM. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
8Aza, a potent A-to-I editing inhibitor, at 0.005–0.1 μmol/L concentration-dependently decreased hypoxia-upregulated protein expression of PCNA, Cyclin A, and CDK1 measured with Western blotting (Fig. S2C). This resulted in a decrease in cell proliferation, which was further confirmed by CCK8 (Fig. 4F) and EdU (8Aza, 0.005 μmol/L) (Fig. 4G) assays.
3.5. High-throughput transcriptome sequencing reveals the ADAR1-regulated circular RNA-circCDK17
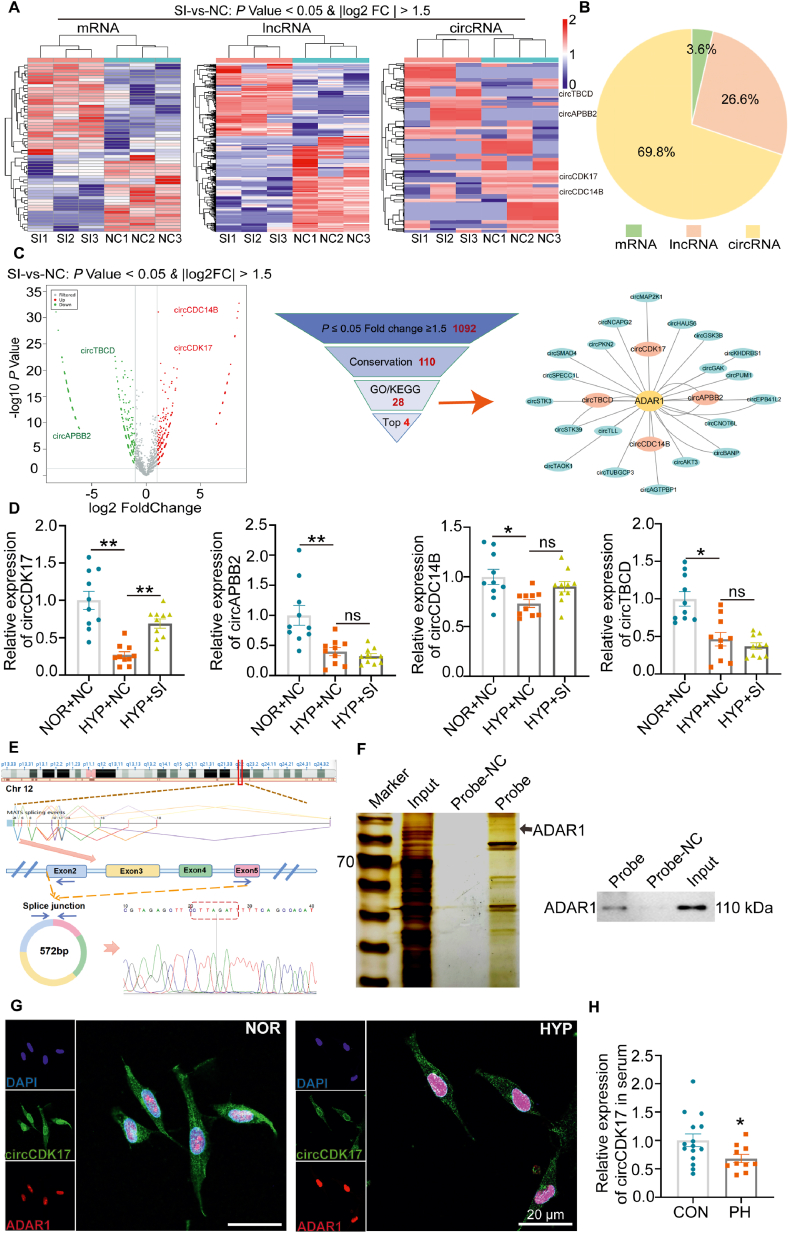
High-throughput whole transcriptome sequencing was used to study the mechanism underlying the effects of ADAR1. It was found that 58 mRNAs, 416 long non-coding RNAs (lncRNAs), and 1092 circRNAs were differentially expressed upon silencing of ADAR1 in PASMCs under hypoxic conditions (Fig. 5A). Among all, circRNAs change accounted for 69.8% (Fig. 5B). These data suggest that ADAR1 may act as an important regulator of circRNAs expression in PH. Functional enrichment analysis (GO) and pathway enrichment analysis (KEGG) (P ≤ 0.05, fold change≥1.5) showed that ADAR1 was involved in several proliferating-related signaling pathways including Hippo signaling pathway, Notch signaling pathway, mTOR signaling pathway, and FoxO signaling pathway. These data suggest that ADAR1 might be involved in PASMCs proliferation in RNA seq data (Figs. S3A and B). Subsequently, we selected circCDK17, circAPBB2, circCDC14B, and circTBCD as ADAR1 regulated candidate genes (Fig. 5C). With specific siRNA targeting the circRNAs cyclization site, we found that ADAR1 mainly regulated the expression of circCDK17 in PASMCs under hypoxia condition (Fig. 5D). In addition, overexpression of ADAR1 also inhibited circCDK17 expression in vitro (Supporting Information Fig. S4A). Subsequently, we further analyzed the correlation between ADAR1 and circCDK17 in the lung tissues of PH patients. The results showed a significant negative correlation between ADAR1 and circCDK17 (Fig. S4B). CircCDK17 is a novel circRNA derived from human chromosome 12 and formed by the cyclization of exons 2-5, and we performed qPCR product sanger sequencing with specific primers designed for the backsplice junction site to verify the accuracy of the junction site. It was found that circCDK17 has a ring structure (Fig. 5E). The expression of convergent and divergent primers designed by circCDK17 in gDNA and cDNA was detected by agarose gel electrophoresis, which further proved the real existence of circCDK17 (Figs. S4C and D). Subsequently, RNase R experiment showed that the addition of RNase R significantly reduced the mRNA expression of β-actin, but had no obvious effect on the expression of circCDK17, which further indicate that circCDK17 was a circular molecule (Fig. S4E).
Figure 5.
ADAR1 regulated circular RNA-circCDK17. (A–B) High-throughput whole transcriptome sequencing showing the RNA changes upon silencing of ADAR1 (A) and the proportion of different types of RNA regulated by ADAR1 (B). (C) Volcanic map of differentially expressed circRNA after silencing of ADAR1, and screening of candidate circRNAs regulated by ADAR1. (D) qPCR analysis showing the expression of circCDK17, circAPBB2, circCDC14B and circTBCB upon silencing of ADAR1, n = 6. (E) Specific primers were designed for circCDK17 cycle-site for qPCR verification, and Sanger sequencing was performed to verify the accuracy of cycle-site. (F) RNA pull-down experiment showing that circCDK17 bound directly to ADAR1, n = 3. (G) Fluorescence in situ hybridization (FISH) showing the colocalization of ADAR1 and circCDK17. (H) qPCR analysis showing the expression of circCDK17 in serum of PH patients (CON = 15, PH = 10). CON, Control; PH, Pulmonary hypertension; NOR, normoxia; HYP, hypoxia; NC, negative control; SI, siRNA of ADAR1; Statistical analysis was performed with two-way ANOVA followed by Dunnett's test or the Student's t-test; All values are presented as mean ± SEM. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
To determine whether ADAR1 can directly bind to circCDK17, we designed a circCDK17-specific biotin probe for RNA pull-down experiment. The expression of ADAR1 was detected in the pull-down products by silver staining and Western blotting analysis (Fig. 5F). In addition, RNA immunoprecipitation (RIP)-qPCR assay with anti-ADAR1 specific antibody detected the expression of circCDK17 in the product (Fig. S4F), and fluorescence in situ hybridization (FISH) assay showed that circCDK17 was distributed in both the nucleus and cytoplasm, and ADAR1 could colocalize with circCDK17 in the nucleus (Fig. 5G). Furthermore, qPCR results showed that the expression of circCDK17 was significantly lowered in serum of PH patients, suggesting that circCDK17 may be used as a valuable marker for the diagnosis of PH (Fig. 5H).
3.6. CircCDK17 inhibits the proliferation and cell cycle progression of PASMCs
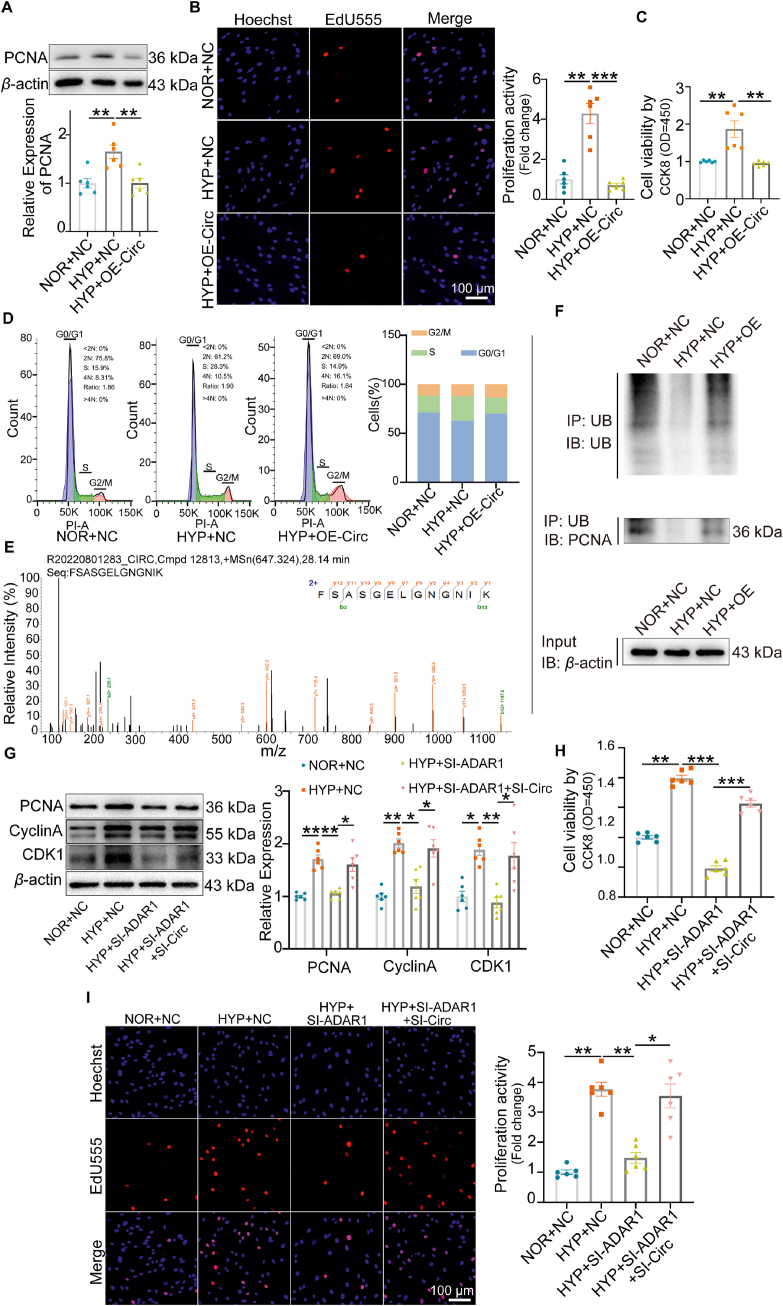
In order to clarify the function of circCDK17, we constructed circCDK17 plasmids and transfected them into PASMCs (Supporting Information Figs. S5A and B). Western blots showed that overexpression of circCDK17 significantly reversed hypoxia-upregulated PCNA expression (Fig. 6A). EdU, CCK8, and flow cytometry results further confirmed that overexpression of circCDK17 reversed cell proliferation and accelerated cell cycle process caused by hypoxia (Fig. 6B‒D). To explore the mechanisms underlying the effects of circCDK17, RNA pull-down product of circCDK17 specific biotin probe was analyzed with mass spectrometry, and the specific peptide of PCNA was detected in the product. PCNA as a downstream effector of circCDK17 was further verified by Western blots (Fig. 6E, Fig. S5C).
Figure 6.
CircCDK17 inhibited the proliferation and cell cycle progression of PASMCs. (A) Western blotting analysis showing that overexpression (OE) of circCDK17 attenuated hypoxia-stimulated PCNA expression, n = 6. (B–C) EdU staining (B) and CCK8 (C) analysis showing that OE-circCDK17 attenuated hypoxia-induced cell proliferation and viability, n = 6. (D) Flow cytometry showing the effect of OE-circCDK17 on cell cycle, n = 6. (E) RNA pull-down combined mass spectrometry showing circCDK17 binds with PCNA specific peptide. (F) Effect of circCDK17 overexpression on PCNA ubiquitination level, n = 3. (G) Western blotting analysis showing that silencing of circCDK17 attenuated the effect of ADAR1 gene knocking-down on protein expression of PCNA, Cyclin A and CDK1, n = 6. (H–I) CCK8 analysis (H) and EdU staining (I) showing that silencing of circCDK17 abolished the effect of ADAR1 deficiency on cell viability and proliferation in the presence of hypoxia, n = 6. NOR, normoxia; HYP, hypoxia; NC, negative control; SI, siRNA; OE, overexpression of circCDK17; Statistical analysis was performed with two-way ANOVA followed by Dunnett's test; All values are presented as mean ± SEM. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
In order to further clarify how circCDK17 regulates PCNA expression, we performed ubiquitination analysis. As shown in Fig. 6F, overexpression of circCDK17 reversed hypoxia-reduced PCNA ubiquitination. These data suggest that circCDK17 regulates the expression of PCNA by influencing its ubiquitination level.
Subsequently, we designed three specific siRNAs for circCDK17 at the cyclization site. The results showed that SI1 had the highest knockdown efficiency, and we also verified whether the SI1 of circCDK17 would affect the expression of its homologous CDK17 mRNA (Figs. S5D and E). We designed the primer sequence of liner CDK17 without the same sequence interval as circCDK17. qPCR results showed that circCDK17 siRNA did not affect the expression of CDK17 mRNA (Fig. S5F). Western blotting, CCK8, and EdU assays showed that silencing of ADAR1 suppressed hypoxia-induced proliferation of PASMCs, whereas circCDK17 siRNA reversed these trends (Fig. 6G‒I). The above results suggest that ADAR1 regulates cell cycle and proliferation through circCDK17.
3.7. ADAR1 regulates m1A modification of circCDK17
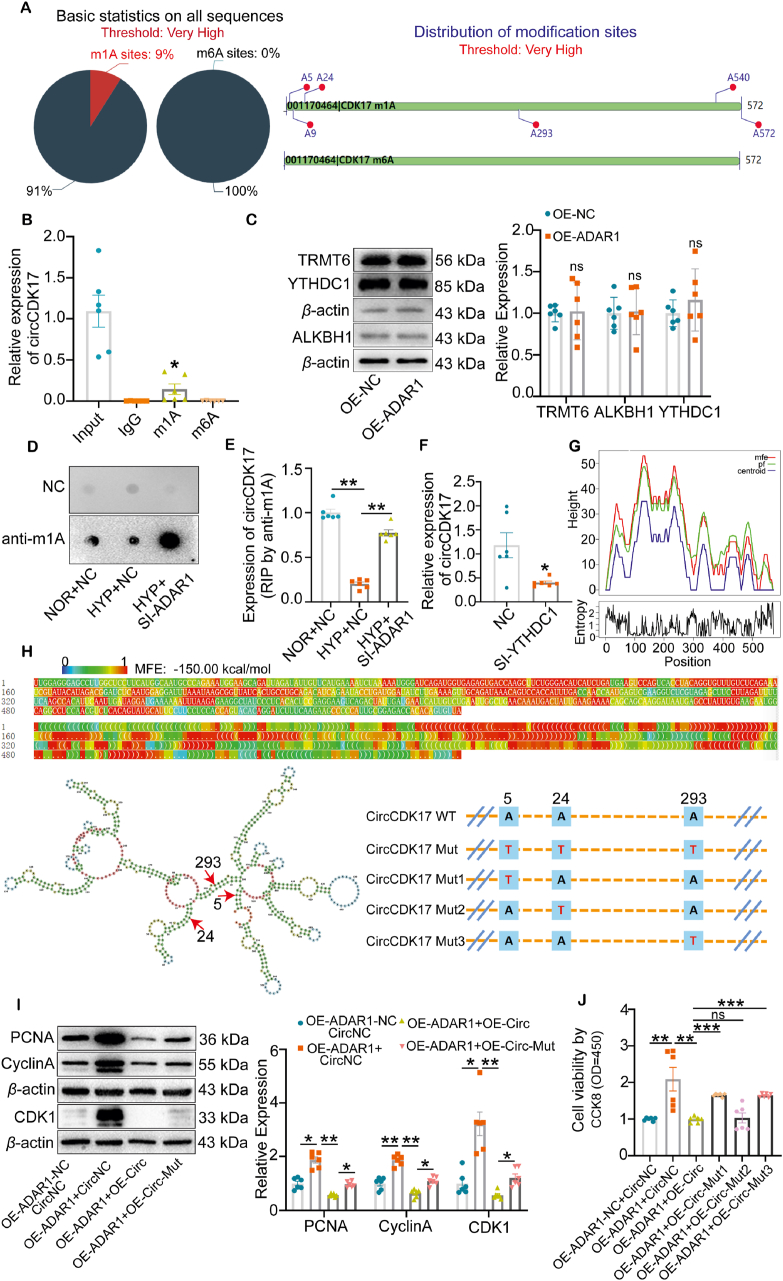
Based on the literatures, we studied whether ADAR1 affects the expression of circCDK17 in PH through N6-methyladenosine (m6A) or m1A modification. Bioinformatics software Deeppromise analysis indicates multiple m1A modification sites but no m6A modification sites in circCDK17 (Fig. 7A, Supporting Information Fig. S6A)32. We then performed methylated RNA immunoprecipitation (meRIP) to immunoprecipitate RNA with m1A and m6A specific antibodies. qPCR analysis showed that the expression of circCDK17 was detected in the precipitation pulled down by m1A specific antibodies, but not in the precipitation of m6A specific antibodies (Fig. 7B). In addition, we found that the m1A modification level of circCDK17 in the lung tissue of PH patients was significantly down-regulated compared with healthy donors (Fig. S6B). These results suggest that circCDK17 was modified by m1A methylation. However, overexpression of ADAR1 did not affect the expression of m1A methylation modification enzymes including TRMT6, ALKBH1, and YTHDC133,34 (Fig. 7C), but markedly increased the level of m1A modification as shown by dot blots (Fig. 7D). meRIP immunoprecipitation showed that ADAR1 silencing reversed hypoxia-induced reduction in m1A antibodies-enriched circCDK17 (Fig. 7E), suggesting that ADAR1 affected the m1A methylation modification level of circCDK17. In addition, we found the presence of YTHDC1 specific peptide in circCDK17 RNA pull-down combined mass spectrometry, which further suggests the presence of m1A modification in circCDK17 (Fig. S6C). qPCR analysis showed that knockdown of Reader-YTHDC1 of m1A reduced the expression of circCDK17, indicating that m1A-modified circCDK17 was more stable (Fig. 7F).
Figure 7.
ADAR1 regulated m1A modification of circCDK17. (A) Online software deeppromise predicts the m1A and m6A modifications of circCDK17 (https://deeppromise.erc.monash.edu/). (B) meRIP-qPCR assay showing the binding of circCDK17 to m1A antibody instead of m6A antibody, n = 6. (C) Western blotting analysis showing protein expression of m1A modifying enzymes in response to ADAR1 overexpression, n = 6. (D) Dot blots showing the modification of m1A upon ADAR1 knockdown. (E) meRIP-qPCR analysis showing the effect of m1A antibody on circCDK17 enrichment in response to the knockdown of ADAR1, n = 6. (F) qPCR analysis showing the expression of circCDK17 upon knockdown of m1A reader YTHDC1, n = 6. (G) Representation of Mountain plot of the MFE (minimum free energy) structure, the partition function (pf), the thermodynamic ensemble of RNA structures, the centroid structure and present the positional entropy for each position (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi). (H) The optimal secondary structure in dot-bracket notation with a minimum free energy of −150.00 kcal/mol is given. RNAfold was combined with the online software catRAPID (http://service.tartaglialab.com/page/catrapid_group) to analyze mutant plasmid sites. (I) Western blotting analysis showing that mutation of all three m1A modification sites attenuated the effects of overexpression of ADAR1 on protein expression of PCNA, Cyclin A, and CDK1, n = 6. (J) CCK8 analysis showing the effect of site mutations of each of the three m1A modification sites on cell viability of PASMCs, n = 6. NOR, normoxia; HYP, hypoxia; NC, negative control; OE, overexpression; Mut, mutant; SI, siRNA; Statistical analysis was performed with two-way ANOVA followed by Dunnett's test or the Student's t-test; All values are presented as mean ± SEM. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
We further investigated whether ADAR1 regulates circCDK17 m1A methylation modification through competitive binding to modification sites. As ADAR1 usually acts on double-stranded RNA, we analyzed the secondary structure of circCDK17 and the interaction site between ADAR1 and circCDK17 using combining RNAfold web server and catRAPID software (Fig. 7G and H; Fig. S6D)35,36. Combined with the three circCDK17 m1A methylation modification sites with high scores in the above Deeppromise, a full m1A methylation sites mutant in circCDK17 (A5T-A24T-A293T) was constructed (Fig. 7H). Western blotting analysis showed that overexpression of circCDK17 reversed the excessive proliferation of PASMCs caused by ADAR1 overexpression, however, this phenomenon was attenuated by the above m1A modification site mutant. These results suggest that ADAR1 mediates the proliferation of PASMCs through m1A modification (Fig. 7I). To identify the potential sites, single-site mutants were constructed (Mut1: A5T; Mut2: A24T; Mut3: A293T). CCK8 analysis showed that both A5T and A293T had significant recovery effects, and the recovery effect seemed to be stronger than A24T (Fig. 7J). These results indicate that ADAR1 regulates circCDK17 m1A methylation and mediates PASMCs proliferation.
4. Discussion
In this study, we found a marked increase of ADAR1 levels in lung tissues of PH patients and animal models as well as hypoxia-treated PASMCs. We demonstrated that ADAR1 upregulation was associated with PASMC proliferation in vitro and in vivo. More importantly, ADAR1 functions by inhibition of circCDK17 m1A methylation. These results suggest that ADAR1 and circCDK17 are involved in hypoxia-induced PH. More interestingly, circCDK17 level in the serum of PH patients was also significantly lower than those of healthy donors. Our data suggest that ADAR1 and circCDK17 may be a key targets and biomarker for treatment and diagnosis of PH.
Abnormal proliferation of PASMCs is the main cause of PH pulmonary vascular remodeling, and the mechanism is extremely complicated. Therefore, it is vital to find the key factors that mediated hypoxia induced PASMCs proliferation. ADAR1-mediated A-to-I modification was significantly increased in lymphoblastoid (LB) cells under hypoxia condition37. In line with this, we also found increased ADAR1 expression in hypoxic PASMCs. Studies have shown that ADAR1 participates in rat primary aortic SMCs phenotypic switching18, and promotes the progression of atherosclerotic cardiovascular disease38, knockdown expression of ADAR1 p110 subtype inhibits glioma cell proliferation39. These studies indicate that ADAR1 may be involved in the hyperproliferation of PASMCs induced by hypoxia. We revealed for the first time in the present study that ADAR1 expression was abnormally higher in PH patients and inhibition of ADAR1 significantly reversed PASMCs proliferation. Thus, ADAR1 is one of hypoxic responders which has a powerful function in PH.
To clarify the biological function of ADAR1 in vivo, we created an ADAR1 overexpression animal model. Interestingly, overexpression of ADAR1 was sufficient to cause pulmonary vascular remodeling and higher RVSP, although not enough to induce right ventricle hypertrophy. However, under hypoxia condition, ADAR1 overexpression further aggravated hypoxia-induced PH and right ventricle hypertrophy. Furthermore, we found that 8Aza, a specific inhibitor of ADAR1, reversed pulmonary vascular remodeling caused by hypoxia and hypoxia combined with Su5416. Our data suggest that ADAR1 is an important mechanism regulating pulmonary artery and right ventricle phenotypic modulation and inhibition of this RNA editing enzyme is an important target to treat PH.
As an important post-transcriptional modification mode, ADAR1-mediated A-to-I RNA editing alters genetic information, thus generating structural and functional diversity of proteins and becoming an important supplement to the central principle. The biological functions of ADAR1 are mainly divided into two types: ADAR1 targets double-stranded RNA (dsRNA)40 and interacts with RNA binding proteins41. Interestingly, recent studies have found that a large number of A-to-I RNA editing sites are located in non-coding regions, suggesting a close relationship between ADAR1 and non-coding RNAs10. For instance, ADAR1 p110, which is highly expressed in liver cancer, inhibited the expression of circRNA42. ADAR1 also antagonized circRNAs expression by melting the stem of the intron26. Compared with other types of RNA, our results suggest that ADAR1 tends to influence circRNAs expression by whole transcriptome sequencing. Thus, the signal axis of ADAR1-circRNA may play a crucial role in the development of PH.
Previous studies have shown that circRNAs are involved in the development of PH43,44. For instance, circRNA CDR1as promotes PASMCs calcification by up-regulating the expression of CAMK2D and CNN3 through sponge adsorption of miR-7-5p45. CircGSAP overexpression also significantly inhibits hypoxia-induced proliferation and promotes apoptosis of pulmonary microvascular endothelial cells46. Circ-calm4 regulates the proliferation and pyroptosis of PASMCs by adsorbing miR-337-3p and miR-124-3p, respectively43,44. However, the upstream mechanism of circRNAs in PH is not clear. In this study, we identified a novel circular RNA-circCDK17 which is regulated by ADAR1 to alleviate PH. We not only find the upstream (ADAR1) but also the downstream of circCDK17. PCNA is closely related to DNA synthesis and plays an important role in the initiation of cell proliferation47. We found in the present study, circCDK17 may combine and interact with PCNA to inhibit its expression and regulate the proliferation of PASMCs. In addition, FISH experiments showed that ADAR1 and circCDK17 had spatial colocalization in the nucleus. Consistent with Western blotting results, ADAR1 was mainly expressed in the p110 subunit of the nucleus in PASMCs. Therefore, ADAR1 functions by influencing circCDK17 in the nucleus of hypoxic PASMCs.
m1A is a new type of methylation modification and also acts on adenine. For this reason, these two processes may compete on adenine with each other. Our results showed that knocking-down ADAR1 increased the m1A modification level of circCDK17. Studies have shown that m1A modification in tRNA regulates translation by improving tRNA stability34, whereas m1A in mRNA and lncRNA regulates RNA processing or protein translation48. As far as we know, there is no relevant report on circRNAs and m1A methylation modification. Whether the m1A methylation modification of circRNAs is involved in the development of PH is still unclear. In this study, we first analyzed the presence of m1A circCDK17 regulated by ADAR1 and verified the presence of m1A modification but not m6A modification in circCDK17 by meRIP experiment. m1A modification is also regulated by corresponding modification enzymes. Importantly, we found that ADAR1 did not affect the expression of m1A modification enzymes, but acted on the m1A site to reduce its modification level. In short, our study reveals for the first time a new mechanism by which ADAR1 functions, which undoubtedly provides new insights into the upstream mechanism of circRNAs.
Interestingly, it has been proved that circRNAs are differentially expressed in plasma between pediatric patients with PH due to congenital heart disease and control subjects49. In the current study, the expression of circCDK17 was found significantly down-regulated in the serum of PH patients. Due to the absence of 3′ and 5′ structures, circRNAs are not sensitive to nucleases and are more stable than traditional linear transcripts. In addition, circRNAs exhibit tissue specificity, temporal specificity, and conservation. Therefore, circRNAs are more suitable to be developed as a biomarker for the diagnosis of PH disease compared to traditional linear RNAs. For the above reasons, we proposed that circCDK17 may serve as a serum biomarker for the diagnosis of PH.
The limitation of this work is that the function of ADAR1 was only studied in hypoxic PH. Given the multiple subtypes of PH, whether ADAR1 and circCDK17 play the same roles in PH in other subtypes remains to be elucidated. In addition, the function of circCDK17 in vivo still needs to be further explored.
In conclusion, we demonstrated for the first time that ADAR1 has a critical regulatory role in PASMC proliferation in vivo and in vitro by the interaction with circCDK17, leading to the change at methylation level of circCDK17 m1A. This finding suggests that ADAR1 may serve as a potential target and circCDK17 may be developed to be a biomarker for diagnosis of PH.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC), China (Grant No 82170064, 82241021 to Xiaowei Nie), Shenzhen Excellent Science and Technology Innovation Talent Development Programme, Shenzhen, China (Grant No. RCJC20210706091946002 to Xiaowei Nie), Shenzhen Science and Technology Program, Shenzhen, China (Grant No. JSGGZD20220822095200001 to Jin-Song Bian), and China Postdoctoral Science Foundation, China (Grant No. 2022M722212 to Junting Zhang).
Author contributions
Junting Zhang, Yiying Li, Xiaowei Nie, and Jin-Song Bian conceptualized the project. Junting Zhang and Yiying Li, designed and performed most of the experiments. Jianchao Zhang, Liu Lu, Yuan Chen, Xusheng Yang, Xueyi Liao, Muhua He, Zihui Jia, and Jun Fan performed or helped in the interpretation and design of some key experiments. Junting Zhang, Yiying Li, Xiaowei Nie, and Jin-Song Bian wrote the original manuscript draft. Yuan Chen and Xusheng Yang collected clinical samples and information. Jianchao Zhang, Muhua He, and Zihui Jia constructed animal models. Junting Zhang, Xiaowei Nie, and Jin-Song Bian acquired funding. Xiaowei Nie and Jin-Song Bian provided resources and supervised the study. All authors provided critical comments on the manuscript.
Conflicts of interest
No conflicts of interest were disclosed.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.07.006.
Contributor Information
Jin-Song Bian, Email: bianjs@sustech.edu.cn.
Xiaowei Nie, Email: niexw2021@mail.sustech.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Hoeper M.M., Al-Hiti H., Benza R.L., Chang S.A., Corris P.A., Gibbs J.S.R., et al. Switching to riociguat versus maintenance therapy with phosphodiesterase-5 inhibitors in patients with pulmonary arterial hypertension (REPLACE): a multicentre, open-label, randomised controlled trial. Lancet Respir Med. 2021;9:573–584. doi: 10.1016/S2213-2600(20)30532-4. [DOI] [PubMed] [Google Scholar]
- 2.Southgate L., Machado R.D., Gräf S., Morrell N.W. Molecular genetic framework underlying pulmonary arterial hypertension. Nat Rev Cardiol. 2020;17:85–95. doi: 10.1038/s41569-019-0242-x. [DOI] [PubMed] [Google Scholar]
- 3.Badesch D.B., McGoon M.D., Barst R.J., Tapson V.F., Rubin L.J., Wigley F.M., et al. Longterm survival among patients with scleroderma-associated pulmonary arterial hypertension treated with intravenous epoprostenol. J Rheumatol. 2009;36:2244–2249. doi: 10.3899/jrheum.081277. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann F., Limper U., Zaha V.G., Reuter H., Zange L., Schulz-Menger J., et al. Evolution of pulmonary hypertension during severe sustained hypoxia. Circulation. 2020;141:1504–1506. doi: 10.1161/CIRCULATIONAHA.119.045192. [DOI] [PubMed] [Google Scholar]
- 5.Ghofrani H.A., Voswinckel R., Reichenberger F., Weissmann N., Schermuly R.T., Seeger W., et al. Hypoxia- and non-hypoxia-related pulmonary hypertension—established and new therapies. Cardiovasc Res. 2006;72:30–40. doi: 10.1016/j.cardiores.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 6.Hassoun P.M. Pulmonary arterial hypertension. N Engl J Med. 2021;385:2361–2376. doi: 10.1056/NEJMra2000348. [DOI] [PubMed] [Google Scholar]
- 7.Penaloza D., Arias-Stella J. The heart and pulmonary circulation at high altitudes: healthy highlanders and chronic mountain sickness. Circulation. 2007;115:1132–1146. doi: 10.1161/CIRCULATIONAHA.106.624544. [DOI] [PubMed] [Google Scholar]
- 8.Song S., Carr S.G., McDermott K.M., Rodriguez M., Babicheva A., Balistrieri A., et al. STIM2 (stromal interaction molecule 2)-mediated increase in resting cytosolic free Ca2 concentration stimulates PASMC proliferation in pulmonary arterial hypertension. Hypertension. 2018;71:518–529. doi: 10.1161/HYPERTENSIONAHA.117.10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montani D., Chaumais M.C., Guignabert C., Günther S., Girerd B., Jaïs X., et al. Targeted therapies in pulmonary arterial hypertension. Pharmacol Ther. 2014;141:172–191. doi: 10.1016/j.pharmthera.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Athanasiadis A., Rich A., Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orlandi C., Barbon A., Barlati S. Activity regulation of adenosine deaminases acting on RNA (ADARs) Mol Neurobiol. 2012;45:61–75. doi: 10.1007/s12035-011-8220-2. [DOI] [PubMed] [Google Scholar]
- 13.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nie Y., Zhao Q., Su Y., Yang J.H. Subcellular distribution of ADAR1 isoforms is synergistically determined by three nuclear discrimination signals and a regulatory motif. J Biol Chem. 2004;279:13249–13255. doi: 10.1074/jbc.M312753200. [DOI] [PubMed] [Google Scholar]
- 15.Fritz J., Strehblow A., Taschner A., Schopoff S., Pasierbek P., Jantsch M.F. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol Cell Biol. 2009;29:1487–1497. doi: 10.1128/MCB.01519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strehblow A., Hallegger M., Jantsch M.F. Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol Biol Cell. 2002;13:3822–3835. doi: 10.1091/mbc.E02-03-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishizuka J.J., Manguso R.T., Cheruiyot C.K., Bi K., Panda A., Iracheta-Vellve A., et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019;565:43–48. doi: 10.1038/s41586-018-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fei J., Cui X.B., Wang J.N., Dong K., Chen S.Y. ADAR1-mediated RNA editing, a novel mechanism controlling phenotypic modulation of vascular smooth muscle cells. Circ Res. 2016;119:463–469. doi: 10.1161/CIRCRESAHA.116.309003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y., Wang Z., Chen X., Wang W., Wang X. ADAR1 silencing-induced HUVEC apoptosis is mediated by FGFR2 under hypoxia stress. Drug Des Dev Ther. 2018;12:4181–4189. doi: 10.2147/DDDT.S181312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Zvi M., Amariglio N., Paret G., Nevo-Caspi Y. F11R expression upon hypoxia is regulated by RNA editing. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma C.P., Liu H., Chang I.Y.F., Wang W.C., Chen Y.T., Wu S.M., et al. ADAR1 promotes robust hypoxia signaling via distinct regulation of multiple HIF-1α-inhibiting factors. EMBO Rep. 2019;20 doi: 10.15252/embr.201847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 23.Chen L., Wang C., Sun H., Wang J., Liang Y., Wang Y., et al. The bioinformatics toolbox for circRNA discovery and analysis. Briefings Bioinf. 2021;22:1706–1728. doi: 10.1093/bib/bbaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 25.Zhou W.Y., Cai Z.R., Liu J., Wang D.S., Ju H.Q., Xu R.H. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:172. doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Abe K., Toba M., Alzoubi A., Ito M., Fagan K.A., Cool C.D., et al. Formation of plexiform lesions in experimental severe pulmonary arterial hypertension. Circulation. 2010;121:2747–2754. doi: 10.1161/CIRCULATIONAHA.109.927681. [DOI] [PubMed] [Google Scholar]
- 28.Halbert C.L., Lam S.L., Miller A.D. High-efficiency promoter-dependent transduction by adeno-associated virus type 6 vectors in mouse lung. Hum Gene Ther. 2007;18:344–354. doi: 10.1089/hum.2006.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Sun Y., Song X., Wang Z., Zhang Y., Zhao Y., et al. Hepatitis B virus evades immune recognition via RNA adenosine deaminase ADAR1-mediated viral RNA editing in hepatocytes. Cell Mol Immunol. 2021;18:1871–1882. doi: 10.1038/s41423-021-00729-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie X., Shen C., Tan J., Wu Z., Wang W., Chen Y., et al. Periostin: a potential therapeutic target for pulmonary hypertension? Circ Res. 2020;127:1138–1152. doi: 10.1161/CIRCRESAHA.120.316943. [DOI] [PubMed] [Google Scholar]
- 31.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z., Zhao P., Li F., Wang Y., Smith A.I., Webb G.I., et al. Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences. Briefings Bioinf. 2020;21:1676–1696. doi: 10.1093/bib/bbz112. [DOI] [PubMed] [Google Scholar]
- 33.Dai X., Wang T., Gonzalez G., Wang Y. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal Chem. 2018;90:6380–6384. doi: 10.1021/acs.analchem.8b01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Zhou J., Li X., Zhang X., Shi J., Wang X., et al. tRNA-m(1)A modification promotes T cell expansion via efficient MYC protein synthesis. Nat Immunol. 2022;23:1433–1444. doi: 10.1038/s41590-022-01301-3. [DOI] [PubMed] [Google Scholar]
- 35.Mathews D.H., Disney M.D., Childs J.L., Schroeder S.J., Zuker M., Turner D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armaos A., Colantoni A., Proietti G., Rupert J., Tartaglia Gian G. catRAPID omics v2.0: going deeper and wider in the prediction of protein–RNA interactions. Nucleic Acids Res. 2021;49:W72–W79. doi: 10.1093/nar/gkab393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nevo-Caspi Y., Amariglio N., Rechavi G., Paret G. A-to-I RNA editing is induced upon hypoxia. Shock. 2011;35:585–589. doi: 10.1097/SHK.0b013e31820fe4b7. [DOI] [PubMed] [Google Scholar]
- 38.Vlachogiannis N.I., Sachse M., Georgiopoulos G., Zormpas E., Bampatsias D., Delialis D., et al. Adenosine-to-inosine Alu RNA editing controls the stability of the pro-inflammatory long noncoding RNA NEAT1 in atherosclerotic cardiovascular disease. J Mol Cell Cardiol. 2021;160:111–120. doi: 10.1016/j.yjmcc.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang B., Hu P., Lin X., Han W., Zhu L., Tan X., et al. PTBP1 induces ADAR1 p110 isoform expression through IRES-like dependent translation control and influences cell proliferation in gliomas. Cell Mol Life Sci. 2015;72:4383–4397. doi: 10.1007/s00018-015-1938-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung H., JJA Calis, Wu X., Sun T., Yu Y., Sarbanes S.L., et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell. 2018;172:811–824.e14. doi: 10.1016/j.cell.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nie Y., Ding L., Kao P.N., Braun R., Yang J.H. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol Cell Biol. 2005;25:6956–6963. doi: 10.1128/MCB.25.16.6956-6963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi L., Yan P., Liang Y., Sun Y., Shen J., Zhou S., et al. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J., Li Y., Qi J., Yu X., Ren H., Zhao X., et al. Circ-calm4 serves as an miR-337-3p sponge to regulate Myo10 (Myosin 10) and promote pulmonary artery smooth muscle proliferation. Hypertension. 2020;75:668–679. doi: 10.1161/HYPERTENSIONAHA.119.13715. [DOI] [PubMed] [Google Scholar]
- 44.Jiang Y., Liu H., Yu H., Zhou Y., Zhang J., Xin W., et al. Circular RNA Calm4 regulates hypoxia-induced pulmonary arterial smooth muscle cells pyroptosis via the Circ-Calm4/miR-124-3p/PDCD6 axis. Arterioscler Thromb Vasc Biol. 2021;41:1675–1693. doi: 10.1161/ATVBAHA.120.315525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma C., Gu R., Wang X., He S., Bai J., Zhang L., et al. circRNA CDR1as promotes pulmonary artery smooth muscle cell calcification by upregulating CAMK2D and CNN3 via sponging miR-7-5p. Mol Ther Nucleic Acids. 2020;22:530–541. doi: 10.1016/j.omtn.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan P., Wu W.H., Gong S.G., Jiang R., Zhao Q.H., Pudasaini B., et al. Impact of circGSAP in peripheral blood mononuclear cells on idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med. 2021;203:1579–1583. doi: 10.1164/rccm.202005-2052LE. [DOI] [PubMed] [Google Scholar]
- 47.González-Magaña A., Blanco F.J. Human PCNA structure, function and interactions. Biomolecules. 2020;10:570. doi: 10.3390/biom10040570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong X., Li X., Yi C. N(1)-Methyladenosine methylome in messenger RNA and non-coding RNA. Curr Opin Chem Biol. 2018;45:179–186. doi: 10.1016/j.cbpa.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., Chen Y., Yao H., Lie Z., Chen G., Tan H., et al. Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm Circ. 2019;9 doi: 10.1177/2045894019888416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.