Abstract
Phytophthora capsici and P. tropicalis are the two species of Phytophthora associated with foot rot disease of black pepper in India. High genetic diversity amongst the Phytophthora species contributes to its wide host range and variability in the virulence pattern. In the present study, genetic diversity of Phytophthora species infecting black pepper was analysed using RAMS (Random Amplified Microsatellites) and REP (Repetitive Extragenic Palindromic)-PCR fingerprinting. Forty-eight isolates, 24 each of P. capsici and P. tropicalis collected from major black pepper growing states, such as Karnataka, Kerala, Tamil Nadu and Goa, were used in the study. The analyses revealed a total of 160 loci of which 150 (93.75%) were polymorphic. UPGMA cluster and PCoA analysis based on combined RAMS and REP-PCR data clearly grouped the P. capsici and P. tropicalis isolates into two clusters which were further divided into four sub-clusters viz., I & II (P. capsici) and III & IV (P. tropicalis). The study clearly indicated that all the isolates were genetically unique and the entire population was heterogeneous. REP-PCR primers showed more polymorphic loci than RAMS primers. Further, sixteen isolates were selected for morphological and infectivity analyses under in vitro conditions. The isolates exhibited varied colony morphology, sporangial shapes and belonged to A1 mating type. Under in vitro conditions, all the sixteen black pepper Phytophthora isolates could infect nutmeg, tomato, chilli, pumpkin, and cucumber and few of the isolates could infect cardamom. None of the isolates could infect coconut, areca nut and vanilla.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-023-03843-1.
Keywords: P. capsici, P. tropicalis, Piper nigrum, RAMS, REP-PCR
Introduction
Black pepper is one of the spice crops with paramount role in the global market and is cultivated across many countries. India being the fifth largest producer accounts for about 66,000 metric tonnes as of 2019 data (FAO 2023). Major producers of black pepper in India are Karnataka, Kerala and Tamil Nadu. In Karnataka, black pepper was cultivated in 211,497 ha with an annual production of 36,000 tonnes followed by Kerala 82,124 ha with 22,000 tonnes and Tamil Nadu 6,576 ha with 1750 tonnes in 2020–21 (Spices Board 2023). According to surveys, the cultivation has been reduced to one-third due to the deleterious disease foot rot (Hema et al. 2007) and it is one amongst the major problems in black pepper production. Being a highly valued commercial crop and its immense trade value in the market, its cultivation is very important and its production constraints are a matter of concern.
Phytophthora is detrimental to its host under epiphytotic conditions and it infects majority of the horticultural crops with economic importance. It is an oomycete soil-borne pathogen which reproduces both sexually and asexually that favours easy propagation and persistence in a population under field conditions. Although sexual reproduction has a major role in genetic recombination, inter-specific hybridization (Donahoo and Lamour 2008), hyphal anastomosis (Stephenson et al. 1974), parasexual recombination (Gu et al. 2000) and migration add to the genetic diversity within the population. The genetic diversity ultimately alters the virulence pattern and rewards the pathogen with better adaptability to adverse environmental variations. The selection pressure and the mutations within the population often lead to the prevalence of fungicide-resistant isolates (Pereira et al. 2020). Unravelling the genetic makeup at the molecular level helps in better understanding of the pathogen diversity and to design a successful management strategy (Burdon 1993).
Genetic diversity studies give an insight on the genetic composition that exists within a population. Variability often directs towards better adaptability of the individuals in the changing environmental conditions. Various approaches reported to analyse the genetic diversity of a population include isozyme analysis (El-Esawi et al. 2017), AFLP (Markert et al. 2010), RAPD (Fu et al 2003), SNP marker (Jara-Arancio et al. 2022), mtDNA analysis (Wang et al. 2009), RAMS and REP-PCR analyses (Truong et al. 2010). The present study employed the benefits of RAMS and REP-PCR as these have proved to be ideal candidates to delineate the genetic diversity due to its precise reproducibility. RAMS (random amplified microsatellites) technique was developed by Zietkiewicz et al. (1994) and it is successfully used to study genetic diversity in fungi (Hantula et al. 1996). RAMS analysis combines the SSR and RAPD techniques to overcome the drawback of each of these techniques and to increase specificity of the analysis.
Short intergenic repeated sequence was initially identified in Escherichia coli (Shirzad-Aski et al. 2016) and Salmonella typhimurium which had highly conserved inverted repeats, such as REP (Repetitive Extragenic Palindromic) and ERIC (Enterobacterial Repetitive Intergenic Consensus) sequence fingerprinting. Since the agarose gel analysis of these PCR products produced unique fingerprints for closely related strains, it can be used to study the genetic diversity between them (de Bruijn 1992). Box elements are the combinations of three subunits, such as boxA, boxB and boxC, which are 59, 45 and 50 nucleotides long, respectively (Bilung et al. 2018; Mishra et al. 2015). Box elements have a role in the regulation of transcription (Borba et al. 2020). Arbitrarily primed PCR fingerprint can be obtained faster, and no prior knowledge on the sequence information is required (Welsh et al. 1991). REP, ERIC, BOX and M13 were used successfully to study the genetic diversity in Phytophthora species (Masanto et al. 2019; Truong et al. 2012).
In India, two species of Phytophthora viz., P. capsici and P. tropicalis are associated with the foot rot disease of black pepper (Bhai et al. 2022; Jeevalatha et al. 2021). Recently, Bhai et al. (2022) studied the diversity amongst the isolates of these two species infecting black pepper through ITS rDNA sequencing, ITS-RFLP, SSCP and MLST analyses. In the present study, we aimed to explore the genetic diversity of P. capsici and P. tropicalis infecting black pepper in India using RAMS and REP-PCR fingerprinting besides identifying potentiality of these isolates to infect other crops.
Materials and methods
Phytophthora cultures and species identification
One hundred and fifty three Phytophthora isolates of black pepper collected from major black pepper growing areas of Kerala, Karnataka, Tamil Nadu and Goa during the period 1997 to 2021 were used in species identification. These cultures were obtained from National Repository of Phytophthora at ICAR—Indian Institute of Spices Research, Kozhikode, Kerala, India. The isolates were cultured in carrot agar medium (Brasier 1967) and maintained at 25 ± 1 °C. Ypt1 gene-based species-specific primers (Jeevalatha et al. 2021) were used in a PCR assay to identify these isolates at species level. The result of species identification was compared with earlier reports of Bhai et al. (2022) who used ITS rDNA sequencing and ITS-RFLP for identification of Phytophthora species infecting black pepper. Forty-eight isolates, 24 isolates each of P. capsici and P. tropicalis listed in Table 1 were selected based on the location (Fig. 1) and year of collection for the genetic diversity analysis.
Table 1.
Details of Phytophthora isolates included in the present study
| S. No. | Name of species | Isolate | Isolation source | Place of collection | State/UT |
|---|---|---|---|---|---|
| 1 | Phytophthora capsici | 98–81 | Black pepper leaf | Kalpakancherry, Malappuram | Kerala |
| 2 | 98–164 | Black pepper leaf | Kargode, Sirsi, Uttara Kannada | Karnataka | |
| 3 | 00–42 | Black pepper | Sirsi, Uttara Kannada | Karnataka | |
| 4 | 01–04 | Black pepper stem | Adivaram, Kozhikode | Kerala | |
| 5 | 02–20 | Rhizosphere soil | Silent Valley, Palakkad | Kerala | |
| 6 | 05–06 | Black pepper spike | Peruvannamuzhi, Kozhikode | Kerala | |
| 7 | 05–14 | Black pepper leaf | Appangala, Kodagu | Karnataka | |
| 8 | 05–19 | Rhizosphere soil | Chettalli, Kodagu | Karnataka | |
| 9 | 06–12 | Black pepper leaf | Irikkur, Kannur | Kerala | |
| 10 | 07–03 | Black pepper leaf | Chelavoor, Kozhikode | Kerala | |
| 11 | 07–06 | Black pepper leaf | Wayanad | Kerala | |
| 12 | 08–01 | Black pepper root | Peruvannamuzhi, Kozhikode | Kerala | |
| 13 | 10–02 | Black pepper leaf | Bommanahally, Bangalore | Karnataka | |
| 14 | 11–13 | Black pepper leaf | Kargunda, Kodagu | Karnataka | |
| 15 | 11–16 | Black pepper leaf | Cherambane, Kodagu | Karnataka | |
| 16 | 13–17 | Rhizosphere soil | Meppadi, Wayanad | Kerala | |
| 17 | 13–21 | Black pepper leaf | Adimali, Idukki | Kerala | |
| 18 | 13–41 | Black pepper leaf | Madikai, Kasaragod | Kerala | |
| 19 | 13–42 | Black pepper leaf | Madikai, Kasaragod | Kerala | |
| 20 | 13–48 | Black pepper root | Goa | Goa | |
| 21 | 18–12 | Black pepper leaf | Idukki | Kerala | |
| 22 | 20–05 | Black pepper leaf | Panniyur, Kannur | Kerala | |
| 23 | 20–06 | Black pepper leaf | Ambalavayal, Wayanad | Kerala | |
| 24 | 21–01 | Black pepper leaf | Panniyur, Kannur | Kerala | |
| 25 | Phytophthora tropicalis | 97–19 | Black pepper stem | Kottakkal, Malappuram | Kerala |
| 26 | 97–55 | Black pepper root | Valparai, Coimbatore | Tamil Nadu | |
| 27 | 98–65 | Black pepper leaf | Valnoor, Kodagu | Karnataka | |
| 28 | 98–93 | Black pepper leaf | Adyanadka, Dakshin Kannad | Karnataka | |
| 29 | 98–175 | Black pepper root | Isloor, Sirsi, Uttara Kannada | Karnataka | |
| 30 | 99–136 | Black pepper leaf | Silver Cloud, Wayanad | Kerala | |
| 31 | 00–38 | Black pepper | Sidapur, Kodagu | Karnataka | |
| 32 | 01–20 | Black pepper | Silent Valley, Palakkad | Kerala | |
| 33 | 03–07 | Black pepper leaf | Adivaram, Kozhikode | Kerala | |
| 34 | 05–17 | Black pepper leaf | Chettalli, Kodagu | Karnataka | |
| 35 | 06–11 | Black pepper berries | Edayannur, Iritti, Kannur | Kerala | |
| 36 | 06–17 | Black pepper leaf | Rasi Estate, Yercaud, Salem | Tamil Nadu | |
| 37 | 08–07 | Black pepper stem | Koorachundu, Kozhikode | Kerala | |
| 38 | 09–01 | Black pepper stem | Peruvannamuzhi, kozhikode | Kerala | |
| 39 | 09–18 | Rhizosphere soil | Pamathatt, Kasaragod | Kerala | |
| 40 | 09–25 | Rhizosphere soil | Kanjirapuzha, Palakkad | Kerala | |
| 41 | 11–20 | Black pepper leaf | Sunnapulikote, Kodagu | Karnataka | |
| 42 | 11–29 | Rhizosphere soil | Wayanad | Kerala | |
| 43 | 11–30 | Rhizosphere soil | Wayanad | Kerala | |
| 44 | 13–23 | Rhizosphere soil | Nedumkandam, Idukki | Kerala | |
| 45 | 13–33 | Rhizosphere soil | Kumali, Idukki | Kerala | |
| 46 | 13–45 | Black pepper leaf | Pilicode, Kasaragod | Kerala | |
| 47 | 13–53 | Black pepper root | Goa | Goa | |
| 48 | 16–06 | Black pepper leaf | Appangala | Karnataka |
Fig. 1.
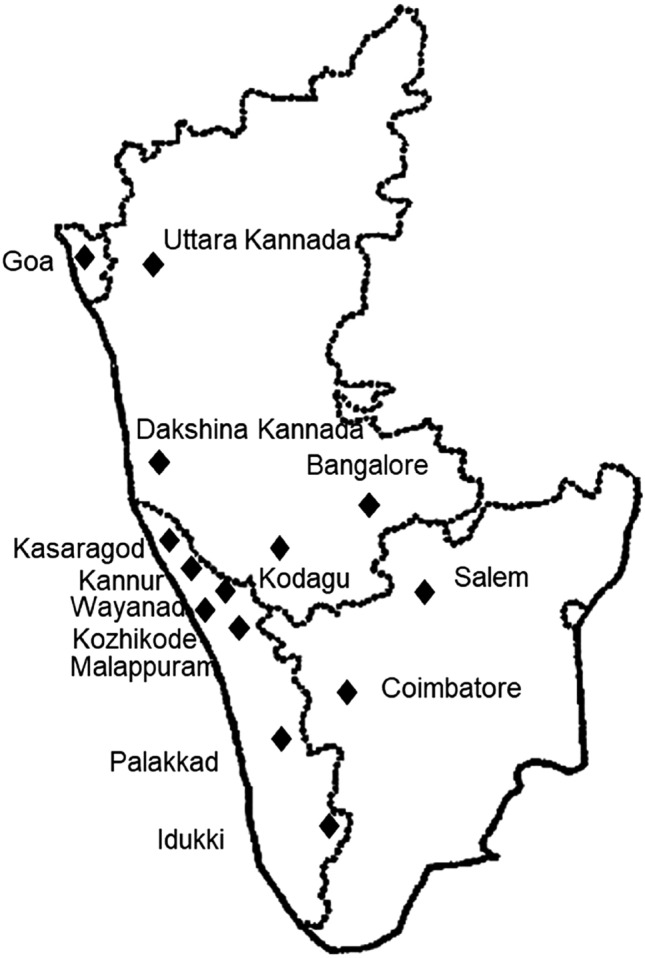
Map of South India showing the location of collection of the isolates across four states: Kerala, Karnataka, Tamil Nadu and Goa
DNA extraction
Agar plugs from 3-day-old cultures were inoculated to Ribeiro’s broth (Erwin and Ribeiro 1996) and actively growing mycelia were harvested on the 4th day, and blot-dried on sterile filter paper. DNA extraction from Phytophthora mycelia was carried out according to Sheji et al. (2009) with slight modifications (Jeevalatha et al. 2021).
RAMS and REP-PCR analyses
The details of primers used for RAMS and REP-PCR analyses are listed in Table 2. PCR amplification was carried out using four RAMS primers in 20 µl reaction volume containing 10 µl of Emerald Amp GT PCR Master Mix (2X Premix; Takara Bio Inc, Shiga, Japan), 0.8 µM of primer and 20 ng of DNA. Thermal cycling was performed in Gradient Thermal Cycler IG-96GEP (iGene Labserve, New Delhi, India) following the thermal cycle conditions given in Table 2 (Hantula et al. 1996). The amplicons were run in 2.5% agarose gel in TAE buffer at 80 V for 3 h and visualised under UV trans-illuminator. PCR amplification using REP-PCR markers was carried out in 25 µl reaction volume containing, 12.5 µl of Emerald Amp GT PCR Master Mix (2X Premix; Takara Bio Inc., Shiga, Japan), 0.4 µM primer and 20 ng DNA template. Thermal cycling was performed using Gradient Thermal Cycler IG-96GEP (iGene Labserve, New Delhi, India) following the temperature profile given in Table 2. The amplicons were run in 1.5% agarose gel in TAE buffer at 80 V for 3 h and visualised under UV trans-illuminator.
Table 2.
Primer sequences and cycle conditions of RAMS and REP-PCR
| Primer | Sequence | Temperature cycle | References |
|---|---|---|---|
| RAMS | |||
| CCA | 5'DD(CCA)5 | 95 °C 10 min 95 °C 30 s 64 °C 45 s 72 °C 2 min 72 °C 7 min- 35 cycles | Hantula et al. (1996) |
| GT | 5'YHY(GT)5G | 95 °C 10 min 95 °C 30 s 53 °C 45 s 72 °C 2 min 72 °C 7 min- 35 cycles | Hantula et al. (1996) |
| ACA | 5'BDB(ACA)5 | 95 °C 10 min 95 °C 30 s 49 °C 45 s 72 °C 2 min 72 °C 7 min- 35 cycles | Hantula et al. (1996) |
| CGA | 5'DHB(CGA)5 | 95 °C 10 min 95 °C 30 s 60 °C 45 s 72 °C 2 min 72 °C 7 min- 35 cycles | Hantula et al. (1996) |
| REP-PCR | |||
| BOX1A | 5′-CTACGGCAAGGCGACGCTGACG-3′ | 95 °C 7 min, 94 °C 1 min, 53 °C 1 min, 65 °C 8 min, 65 °C 16 min- 30 cycles | Versalovic et al. (1991) |
| ERIC1R | 5′-ATGTAAGCTCCTGGGGATTCAC-3′ | ||
| ERIC2 | 5′-AAGTAAGTGACTGGGGTGAGCG-3′ | 95 °C 7 min 94 °C 1 min 52 °C 1 min 65 °C 8 min 65 °C 16 min- 30 cycles | Versalovic et al. (1991) |
| REP1R-I | 5′-IIIICGICGICATCIGGC-3′ | ||
| REP2-I | 5′-ICGICTTATCIGGCCTAC-3′ | 94 °C 7 min 94 °C 1 min 40 °C 1 min 65 °C 8 min 65 °C 16 min- 35 cycles | Versalovic et al. (1993) |
| M13 | 5′-GAGGGTGGCGGTTCT-3′ | 93 °C 3 min 93 °C 1 min 55 °C 1 min 72 °C 1 min 72 °C 10 min- 45 cycles | Zaccardelli et al. (2008) |
Genetic diversity analysis
RAMS and REP-PCR generated bands were scored as either present (1) or absent (0). Strong and reproducible bands were scored and used in the analysis. Genetic diversity analysis was performed for RAMS data alone, REP-PCR data alone as well as combining the scoring patterns of both RAMS and REP-PCR data. Unweighted Pair-Group Method using Arithmetic Averages (UPGMA) clustering was performed using DICE similarity coefficient in NTSYSpc Version 2.10e (Rohlf 2000). Genetic diversity indices were calculated using POPGENE version 1.32 (Yeh et al. 1997). The binary data obtained from RAMS, REP-PCR and combined RAMS & REP-PCR analyses were subjected to principal coordinate analysis (PCoA) using GenAlEx 6.5 software with default parameter settings (Peakall and Smouse 2006, 2012).
Colony morphology and sporangia characters
Sixteen isolates representing different sub-clusters of UPGMA dendrogram constructed based on combined RAMS and REP-PCR data were selected for further study. The pure cultures of the isolates were grown on carrot agar media at 25 ± 1 °C. The colony morphology patterns were photographed on the fifth day after inoculation. The sporangial characters were studied using the 3-day-old cultures. Five mm discs of actively growing cultures were placed in a Petri plate containing sterile distilled water and incubated at 25 ± 1 °C under illumination for 48 h. The sporangial production was checked and photographed using Leica DM 5000 B microscope (Leica Mikrosystems Vertrich GmbH, Germany).
Mating type determination
Determination of mating type of Phytophthora isolates was carried out on 10% clarified V8 juice agar (Li et al. 2017). The isolates were paired with known A1 isolate (ATCC-MYA- 2338) and A2 isolate (ATCC-MYA-4034) of P. capsici obtained from American Type Culture Collection, Manassas, USA (www.atcc.org) and incubated at 25 ± 1 °C in dark for up to 10 days. The plates were observed for the presence of oospores at the junction of the two colonies using Leica DM 5000 B microscope (Leica Mikrosystems Vertrich GmbH, Germany). The pairing between the same isolates served as control. All the pairing experiments were performed in triplicates.
Infectivity analysis
Eight isolates each from P. capsici and P. tropicalis species representing different sub-clusters were selected for infectivity analysis. Infectivity of P. capsici and P. tropicalis isolates was tested on black pepper and also on other crops, such as arecanut, tomato, pumpkin, vanilla, nutmeg, coconut, chilli, cucumber and cardamom, using leaf /fruit inoculation method under in vitro conditions. Either the leaf or the fruit was inoculated with 5 mm disc obtained from 3-day-old actively growing Phytophthora cultures and incubated in moist chamber at 25 ± 1 °C. The experiment was performed in triplicates. The symptom development was monitored for up to 5 days. The length and the width of the lesion were measured, and the area of lesion at 48 h after inoculation was computed (da Silva et al. 2012) using the following formula:
where S is the surface area of lesion, L is the length of lesion and W is the width of lesion. ANOVA was performed with R studio R version 4.2.1 (R Core Team 2022).
Results
Species identification
PCR analysis of 153 Phytophthora isolates using species-specific primers showed that 71 isolates belonged to P. capsici and 82 isolates to P. tropicalis. Of the 92 isolates collected from Kerala across seven districts, 49 isolates were P. capsici and 43 isolates were P. tropicalis. Out of 52 isolates collected from 4 districts of Karnataka, 20 isolates were P. capsici and 32 isolates were P. tropicalis. All the three isolates collected from Tamil Nadu were of P. tropicalis and out of 6 isolates from Goa, 2 were P. capsici and 4 were P. tropicalis. The results of ypt1 gene-based species-specific PCR assay were similar to the results obtained using ITS rDNA sequencing and ITS-RFLP assay to differentiate P. capsici and P. tropicalis isolates of black pepper (Bhai et al. 2022).
RAMS and REP-PCR analyses
RAMS analysis of 48 isolates including P. capsici and P. tropicalis revealed 71 loci, of which Kerala isolates accounted for about 88.73% polymorphism, Karnataka for 83.1%, Tamil Nadu for 11.27% and Goa for 49.3%. The combined data for all states showed 90.14% of polymorphic loci. A total of 89 loci were revealed through REP-PCR analysis and Kerala isolates accounted for 96.63% of polymorphic loci, Karnataka for 78.65%, Tamil Nadu for 8.9% and Goa for 41.57%. Combined results showed that 96.63% of the loci were polymorphic. The combined RAMS and REP-PCR data revealed 160 loci and 93.75% were polymorphic in isolates from all states. Shannon diversity index and Nei’s gene diversity of Kerala population were 0.4414 and 0.2892, respectively and those for Karnataka isolates were 0.4100 and 0.2717, respectively (Table 3). Agarose gel images of RAMS and REP-PCR analyses are given in Supplementary Fig. 1.
Table 3.
Genetic variations and polymorphic features of 24 isolates each of P. capsici and P. tropicalis infecting black pepper
| Marker | No. of isolates | Total no. loci | No. of polymorphic loci (% of total) | Nei gene diversity (h) | Shannon diversity index (I) | Population group* |
|---|---|---|---|---|---|---|
| RAMS | ||||||
| Combined (all states) | 48 | 71 | 64 (90.14) | 0.3204 | 0.4751 | I, II, III, IV |
| Kerala | 30 | 71 | 63 (88.73) | 0.3128 | 0.4654 | I, II, III, IV |
| Karnataka | 14 | 71 | 59 (83.10) | 0.3124 | 0.4605 | I, III, IV |
| Tamil Nadu | 2 | 71 | 8 (11.27) | 0.0467 | 0.0681 | III |
| Goa | 2 | 71 | 35 (49.30) | 0.2042 | 0.2981 | I, IV |
| REP-PCR | ||||||
| Combined (all states) | 48 | 89 | 86 (96.63) | 0.2660 | 0.4158 | I, II, III, IV |
| Kerala | 30 | 89 | 86 (96.63) | 0.2704 | 0.4223 | I, II, III, IV |
| Karnataka | 14 | 89 | 70 (78.65) | 0.2392 | 0.3698 | I, III, IV |
| Tamil Nadu | 2 | 89 | 8 (8.99) | 0.0372 | 0.0544 | III |
| Goa | 2 | 89 | 37 (41.57) | 0.1722 | 0.2514 | I, IV |
| RAMS + REP − PCR | ||||||
| Combined (all states) | 48 | 160 | 150 (93.75) | 0.2901 | 0.4421 | I, II, III, IV |
| Kerala | 30 | 160 | 149 (93.12) | 0.2892 | 0.4414 | I, II, III, IV |
| Karnataka | 14 | 160 | 129 (80.62) | 0.2717 | 0.4100 | I, III, IV |
| Tamil Nadu | 2 | 160 | 16 (10.00) | 0.0414 | 0.0605 | III |
| Goa | 2 | 160 | 72 (45.00) | 0.1864 | 0.2721 | I, IV |
*Population group I and II—P. capsici, III and IV—P. tropicalis
The diversity parameters for each primer were also computed individually (supplementary table1) and it was found that amongst RAMS primers, greater polymorphism was observed for GT (100%) and ACA primer revealed lowest polymorphism (80.95%). Whereas in case of REP- PCR primers, loci amplified by BOX1, M13 and Rep-1R & Rep-2I primers were of 100% polymorphic with 28, 21 and 23 loci, respectively. ERIC1R & ERIC2 primers showed 82.35% polymorphic loci.
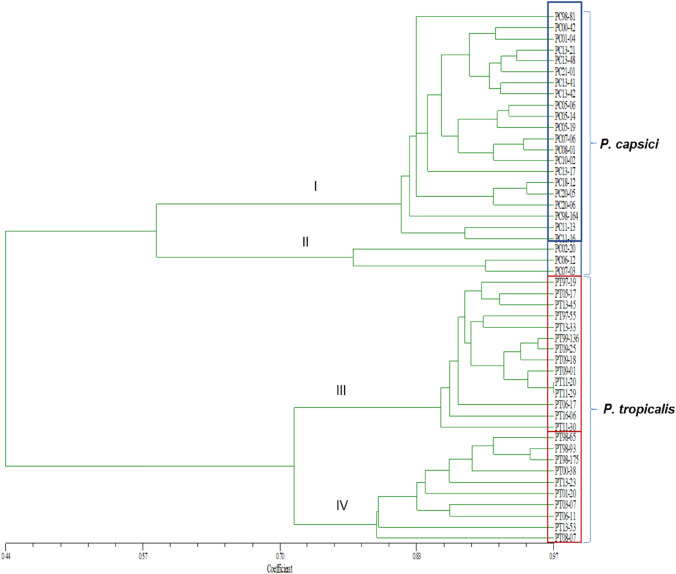
UPGMA cluster analysis based on combined RAMS & REP-PCR data clearly grouped P. capsici and P. tropicalis isolates into two clusters which was further divided into four sub-clusters I, II, III & IV (Fig. 2). P. capsici isolates were grouped in sub-clusters I and II, whereas P. tropicalis spp. were grouped in sub-clusters III and IV. The isolates from each state were distributed randomly in the population group and hence we could not correlate the cluster generated with the geographical origin of the isolates. The isolates which were grouped closer showed up to 97% similarity, which indicated that the isolates were unique and the population was found to be heterogeneous.
Fig. 2.
UPGMA dendrogram of 48 Phytophthora isolates (24 P. capsici and 24 P. tropicalis isolates) based on combined RAMS and REP-PCR data
REP-PCR analysis could alone differentiate P. capsici and P. tropicalis and group them into separate clusters (Supplementary Fig. 2a) as observed in dendrogram based on combined RAMS and REP-PCR data. But, in RAMS dendrogram, slight variation was observed. Sub-cluster II of P. capsici (with three isolates) was found to form major cluster with sub-clusters III and IV of P. tropicalis isolates (Supplementary Fig. 2b). However, four sub-clusters were maintained in RAMS dendrogram also.
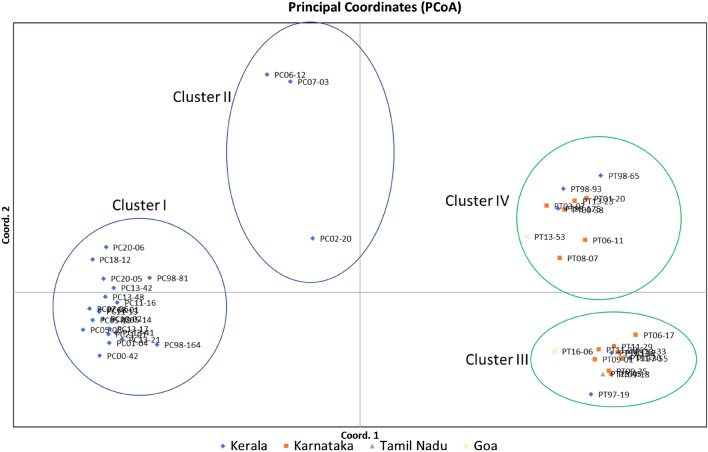
Principal coordinate analysis
PCoA analysis was carried out based on the distance matrix with data standardisation for 71, 89 and 160 loci generated by RAMS, REP-PCR and combined RAMS & REP-PCR, respectively. In case of combined RAMS & REP-PCR, the first three coordinates explained 84.51% cumulative variation. The topology obtained by PCoA analysis was similar to UPGMA dendrogram. The scatter plot divided the 48 isolates into four groups i.e. clusters I & II comprising P. capsici isolates (indicated by blue circle) and clusters III & IV comprising P. tropicalis isolates (indicated by green circle) (Fig. 3). PCoA analysis using RAMS data alone explained 76.40% cumulative variation in first three coordinates. The topology obtained by PCoA analysis was slightly different than UPGMA dendrogram. P. tropicalis isolates were found in two separate clusters as observed in UPGMA dendrogram. However, in case of P. capsici isolates, the isolates were not clustered as observed in UPGMA dendrogram (Supplementary Fig. 3a). PCoA analysis using REP-PCR data alone explained 87.58% cumulative variation in first three coordinates. The topology obtained by PCoA analysis based on REP-PCR data was similar to UPGMA dendrogram (Supplementary Fig. 3b).
Fig. 3.
Principal coordinate analysis (PCoA) plot based on combined RAMS and REP-PCR data for four Phytophthora populations (Kerala, Karnataka, Tamil Nadu and Goa) showing separation of the population into four clusters (P. capsici—Cluster I & II; P. tropicalis—Cluster III & IV)
Colony morphology and sporangia characters
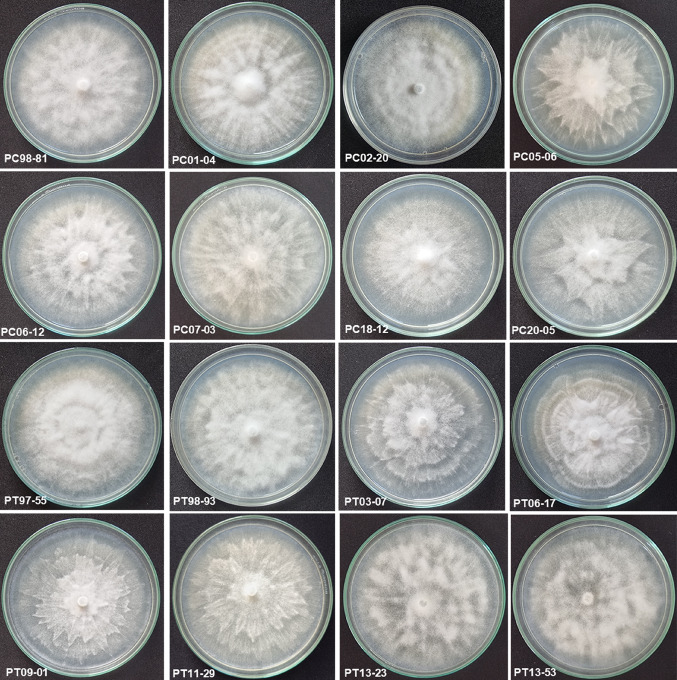
Colony morphology studies of sixteen isolates (8 isolates each of P. capsici and P. tropicalis) showed that the isolates exhibited different growth patterns like modified chrysanthemum (PC98-81, PC05-06, PC07-03 and PT98-93), stellate with concentric rings (PC01-04), cottony with concentric rings (PC02-20), modified stellate (PC06-12, PC18-12, PT03-07, PT09-01 and PT11-29), stellate (PC20-05), modified petaloid (PT06-17) and stellate with cottony mycelium (PT13-23 and PT13-53) (Fig. 4). The sporangia produced by the isolates were papillate and exhibited a variety of shapes like naviculate, globose, elongated, elliptical, pyriform, limoniform, ovoid-obpyriform, ovoid and obpyriform (Fig. 5). There was no correlation between culture morphology, sporangia shape and clustering of isolates.
Fig. 4.
Colony morphology of selected Phytophthora isolates: PC98-81 Modified chrysanthemum; PC01-04 Stellate with concentric rings; PC02-20 Cottony with concentric rings; PC05-06 Modified chrysanthemum; PC06-12 Modified stellate; PC07-03 Modified chrysanthemum; PC18-12 Modified stellate; PC20-05 Stellate; PT97-55 No pattern; PT98-93 Modified chrysanthemum; PT03-07 Modified stellate; PT06-17 Modified petaloid; PT09-01 Modified stellate; PT11-29 Modified stellate; PT13-23 Stellate with cottony mycelia; PT13-53 Stellate with cottony mycelia
Fig. 5.
Microscopic view of sporangia of Phytophthora isolates
Mating type determination
Mating type of Phytophthora isolates was determined using pairing test. Oospores were observed in all the isolates when paired with ATCC-MYA-4034 (A2 mating type) and not with ATCC-MYA-2338 (A1 mating type) which confirmed that all the 48 isolates belonged to A1 mating type.
Infectivity analysis
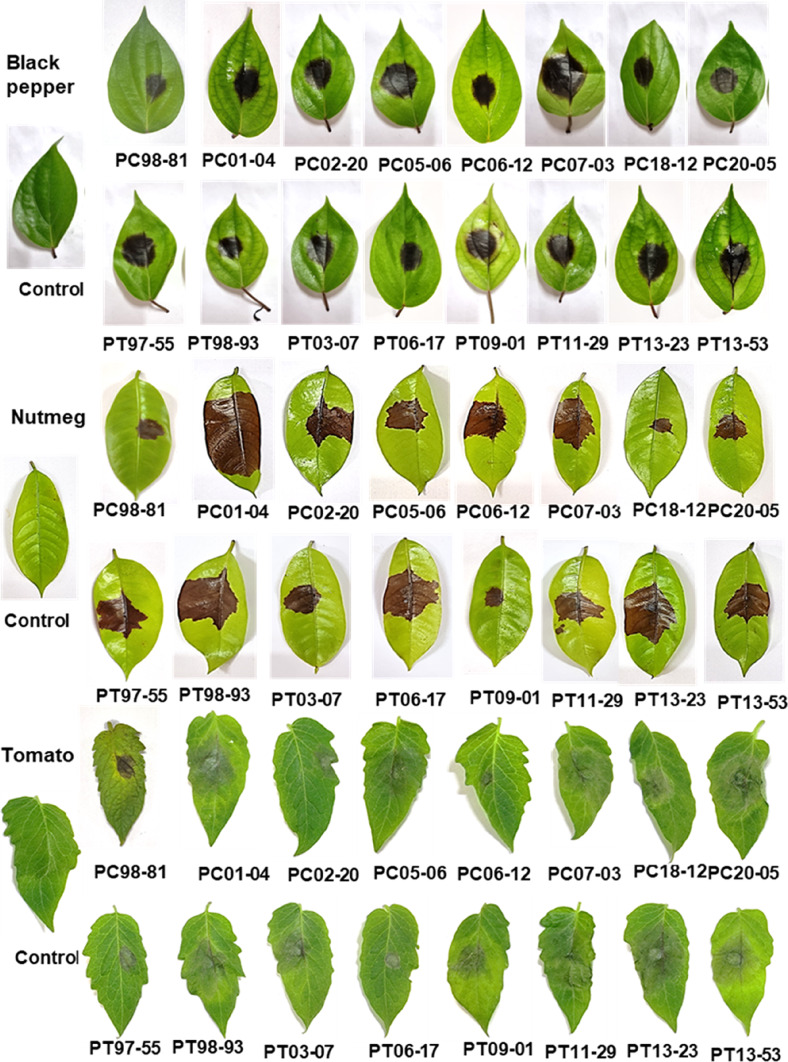
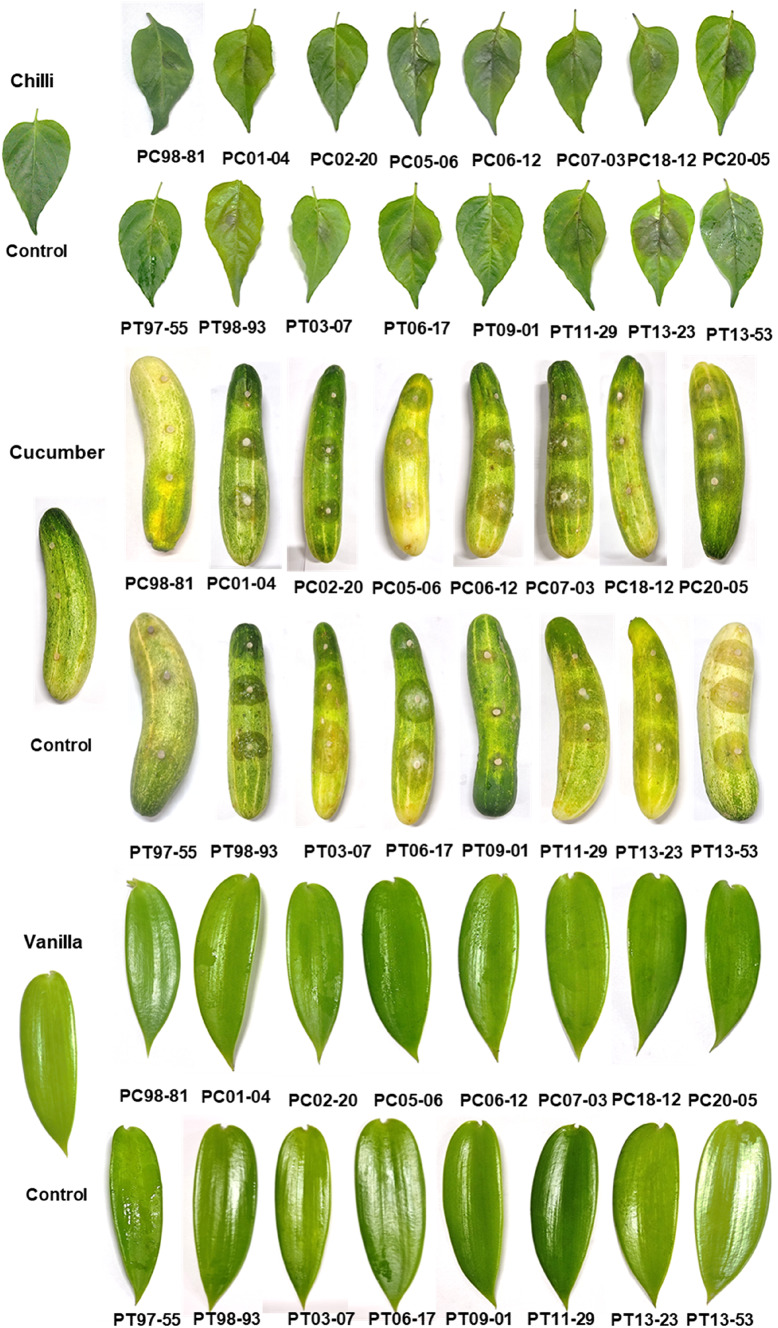
In infectivity analysis, all the 16 Phytophthora isolates from black pepper could infect nutmeg, cucumber, tomato and pumpkin. But only few isolates viz., PC06-12, PC20-05, PT06-17 and PT13-23 could infect cardamom (Fig. 6). Upon inoculation in black pepper, the isolates produced blackish lesions with fimbriate margin. In case of nutmeg, a dark brown-coloured lesion was observed which covered the entire leaf surface at later stage. The symptoms on cucumber appeared as water-soaked lesion, and white cottony mycelial growth was observed at a later stage. A light brownish lesion was observed on tomato leaves and in pumpkin, and water-soaked lesion with sunken spot was observed. Brown-coloured lesion with a yellow margin was observed on cardamom leaves. In black pepper, the isolate 97–55 was highly virulent as it showed larger lesion area (10.54 cm2) followed by 06–12 (8.82 cm2) and 98–81 was least virulent (0.45 cm2). The isolates 13–23, 13–53 and 01–04 displayed a comparable pattern of virulence in nutmeg (10.71 cm2, 10.46 cm2 and 10.33 cm2 respectively), whereas 18–12 was the least virulent (0.77 cm2). The lesion area ranged from 0.41 cm2 to 2.58 cm2 in tomato and there was no significant variation in virulence pattern between the isolates. In chilli, the isolate 13–23 (14.52 cm2) showed high virulence which was on a par with the isolates, 06–17 (14.01 cm2), 07–03 (13.86 cm2) and 03–07 (13.73 cm2). The least virulent isolates were 09–01 (0.79 cm2) and 98–81 (0.64 cm2). In case of pumpkin, isolate 13–53 (9.94 cm2) was highly virulent followed by 06–17 (6.75 cm2), and 97–55 (0.28 cm2) was the least virulent isolate. Not all the isolates could infect cardamom. The isolates, 06–17, 20–05, 13–23 and 05–06 only could infect cardamom with a lesion of 0.21–0.77 cm2 (Table 4).
Fig. 6.
Infectivity analysis of black pepper Phytophthora isolates on various crops (Symptoms photographed at 48 h after inoculation)
Table 4.
ANOVA results showing the lesion area inflicted by Phytophthora isolates on various crops
| Isolates | Black pepper | Nutmeg | Tomato | Chilli | Pumpkin | Cardamom |
|---|---|---|---|---|---|---|
| 98–81 | 0.45e | 1.84gh | 1.21a | 0.64h | 5.75abc | 0.00d |
| 01–04 | 3.61cde | 10.33a | 1.56a | 9.09cd | 2.93bcd | 0.00d |
| 02–20 | 5.21bcd | 4.87cdefg | 0.66a | 4.63efg | 1.78bcd | 0.00d |
| 05–06 | 6.14bcd | 2.90efgh | 1.05a | 5.83ef | 2.76bcd | 0.21c |
| 06–12 | 8.82ab | 6.19bcde | 0.41a | 11.05bc | 2.02bcd | 0.00d |
| 07–03 | 6.07bcd | 9.06ab | 2.58a | 13.86ab | 2.31bcd | 0.00d |
| 18–12 | 4.84bcde | 0.77h | 1.53a | 2.02gh | 1.16cd | 0.00d |
| 20–05 | 4.06cde | 3.59defgh | 2.46a | 5.95ef | 1.89bcd | 0.37b |
| 97–55 | 10.54a | 2.17fgh | 0.66a | 3.77fg | 0.28d | 0.00d |
| 98–93 | 2.75de | 6.50bcd | 2.19a | 7.28de | 1.63bcd | 0.00d |
| 03–07 | 3.37cde | 3.02efgh | 0.90a | 13.73ab | 1.36cd | 0.00d |
| 06–17 | 5.03bcd | 7.63abc | 1.05a | 14.01ab | 6.75ab | 0.77a |
| 09–01 | 6.36abcd | 2.49fgh | 2.13a | 0.7 9h | 2.05bcd | 0.00d |
| 11–29 | 5.29bcd | 5.39cdef | 1.83a | 10.26c | 2.04bcd | 0.00d |
| 13–23 | 4.66bcde | 10.71a | 1.88a | 14.52a | 2.28bcd | 0.22c |
| 13–53 | 7.46abc | 10.46a | 1.72a | 10.44c | 9.94a | 0.00d |
| CV | 44.78 | 32.04 | 126.73 | 20.79 | 89.19 | 91.13 |
| t-value | 2.03 | 2.03 | 2.03 | 2.03 | 2.03 | 2.04 |
| LSD | 3.94 | 2.92 | 3.13 | 2.76 | 4.35 | 0.15 |
No infection in coconut, arecanut and vanilla. Values followed by the same superscript letters in each column are not significantly different from each other as detected by DMRT (p<0.05)
When the isolates were considered, 06–12 and 97–55 were highly virulent in black pepper and chilli but less virulent in the rest of the crops. The isolates, 20–05 and 05–06 were moderately virulent in black pepper and chilli and less virulent in nutmeg and pumpkin. 02–20, 06–17 and 11–29 were moderately virulent in both black pepper and nutmeg, but 02–20 was the least virulent in the rest of the crops, whereas 06–17 and 11–29 were highly virulent in chilli. The isolates, 07–03, 18–12, 09–01, 13–23 and 13–53 were moderately virulent in black pepper, but 07–03, 13–23 and 13–53 were highly virulent in both chilli and nutmeg, whereas 13–53 was highly virulent in pumpkin. Isolates 98–81, 01–04, 98–93 and 03–07 were less virulent in black pepper, but 01–04 was highly virulent in chilli and nutmeg, and 98–93 was moderate. 98–81 was moderately virulent in pumpkin. None of the isolates could infect crops such as coconut, arecanut and vanilla (Fig. 6). The virulence patterns amongst the isolates were also random and no correlation was observed with the UPGMA cluster.
Discussion
Phytophthora foot rot disease has been reported in many countries like Vietnam (Truong et al. 2008), Malaysia (Farhana et al. 2013), Ethiopia (Jibat et al. 2021), where P. capsici, P. tropicalis and P. nicotianae, (Roy et al. 2009) are the causal organisms. In India, the role of two species of Phytophthora has already been elucidated with black pepper foot rot disease. It is also equally important to discern the coexistence of these species within a population. The major black pepper producers in India include Karnataka, Kerala and Tamil Nadu and hence, the present study mainly focussed on the isolates collected from these states. The present study constitutes the first report on genetic diversity of Phytophthora species infecting black pepper in India unravelled through RAMS and REP-PCR analyses.
It was evident from the present study that there existed considerable variations amongst both the species. Size of the sample often influenced the genetic diversity estimation. Based on the availability of the cultures from the national repository, a greater number of isolates were used from Kerala followed by Karnataka i.e. nearly half the quantum. The diversity estimates were greater for Kerala isolates, but when considering the sample size, the Karnataka population was found to be more diverse. The samples collected from Tamil Nadu and Goa were considerably less and hence not sufficient to describe the diversity of these populations.
Through this study, we have identified that the black pepper infecting Phytophthora spp. population in India is highly diverse and the two species of Phytophthora could be clearly delineated using combined RAMS and REP-PCR data. REP-PCR analysis could alone differentiate P. capsici and P. tropicalis and group them into separate clusters as observed in dendrogram based on combined RAMS and REP-PCR data. But, in RAMS dendrogram, slight variation was observed. Similarly, both the markers generated comparable number of polymorphic loci. However, those identified by REP-PCR primers is slightly higher. The topology obtained by PCoA analysis was similar to UPGMA dendrogram, and this indicates the robustness of application of combined RAMS and REP-PCR data or REP-PCR data alone in differentiating the two-sister species infecting black pepper. Several reports are available on the use of REP-PCR fingerprinting in genetic diversity analysis of Phytophthora species. REP-PCR fingerprints detected variations in P. infestans populations and were highly reproducible (Bouws and Finckh 2007). Recently, 81 isolates of P. palmivora in Indonesia and Japan were studied using REP-PCR primers, and the clustering of the isolates was similar to microsatellite markers (Masanto et al. 2019).
All the isolates analysed in this study were unique and the population was found to be heterogeneous. There was no correlation between UPGMA clustering and geographical origin of the isolates. A study from Vietnam on P. capsici infecting black pepper revealed that the P. capsici population in Vietnam is clonal and also no correlation was observed with the geographical origin of the isolates (Truong et al. 2010). The genetic diversity study of P. capsici infecting Capsicum annuum in Bhutan (Rai et al. 2020) could identify polymorphism amongst the isolates and the RAMS clustering corresponded to the geographical origin of the isolates. REP-PCR analysis of P. palmivora from Indonesia and Japan showed similar results i.e. REP-PCR analysis clustered the isolates based on the geographical region rather than host plant or mating type (Masanto et al. 2019). RAMS analysis of P. colocasia from India could identify highly diverse population and the UPGMA cluster did not correspond to geography or phenotype of the isolates (Nath et al. 2016).
All the 48 isolates used in the present study were of A1 mating type. So far, all the Phytophthora isolates infecting black pepper in India were identified as A1 mating type (unpublished data). However, the chance of sexual reproduction amongst the South Indian populations of P. capsici and P. tropicalis cannot be ruled out completely. A2 mating type isolates were reported in P. capsici infecting other crops such as cocoa in southern India (Chowdappa and Chandramohan 1997). In addition, there is also a higher possibility of inter-specific hybridization with A2 mating type of other species such as P. palmivora which is predominant in Kerala and Karnataka (Chowdappa and Chandramohan 1997). The other attributions for the unique genotypes of the isolates in the present study could be mutations and loss of heterozygosity due to mitotic recombination (Goodwin 1997; Truong et al. 2010; Li and Liu 2021).
Colony morphology revealed that the isolates exhibited variety of growth pattern irrespective of species difference, modified chrysanthemum being the most observed pattern. Aragaki and Uchida (2001) reported that P. capsici spp. bear broad sporangia with length-to-breadth ratio less than 1.8 and round sporangial base, whereas the sporangia of P. tropicalis was narrow with breadth ratio greater than 1.8 and tapered sporangial base. As per their study, P. tropicalis spp. produce chlamydospores which is dissimilar to P. capsici, In the present study, elongated sporangia with tapered base and broad sporangia with round base were observed in P. capsici and P. tropicalis, respectively. Production of chlamydospores was observed only in P. tropicalis isolates. All the isolates of P.capsici and P. tropicalis infecting black pepper in India analysed in this study were of A1 mating type.
P. capsici is a volatile pathogen which leaves an egregious effect on its host, and has a broad spectrum of host range including the members of Cucurbitaceae and Solanaceae (Meitz et al. 2010), whereas P. tropicalis is a pathogen of ornamentals (Hong et al. 2006) and fruit trees (Brumat et al. 2022). Amongst the crops selected for the infectivity analysis, P. capsici is reported to infect crops like tomato (Syed-Ab-Rahman et al. 2019), cucumber (Mansfeld et al. 2020), chilli (Majid et al. 2016) and pumpkin (Mohammad and Jose 2018). P. tropicalis could infect seedlings of tomato (Orlikowski et al. 2006) and cucumber (Hong et al. 2008) in vitro. The present study examined the ability of black pepper Phytophthora isolates to infect other crops including plantation, spices and vegetables under in vitro conditions and variability in virulence pattern. Both P. capsici and P. tropicalis were capable of infecting other crops evidenced with the development of symptoms on nutmeg, tomato, cucumber, chilli, pumpkin and cardamom and however, none of the isolates could infect coconut, areca nut and vanilla. It was also observed that there exists a great variability amongst the isolates in exhibiting aggressiveness of the symptom, i.e. the most aggressive isolate on one crop was not aggressive on the other. Neither the most virulent nor the least virulent isolates fall into any specific cluster.
Conclusion
Combined data of RAMS and REP-PCR analyses could clearly differentiate the two species of Phytophthora infecting black pepper and group them into distinct clusters. Amongst the two methods, REP-PCR analysis is better, clearly differentiated P. capsici & P. tropicalis and produced more polymorphic loci than RAMS analysis. All the isolates were genetically unique and the population was diverse irrespective of their place of origin. They showed varied colony morphology, sporangial shapes and belonged to A1 mating type. Infectivity analysis revealed that the black pepper isolates could infect crops, such as tomato, cucumber, chilli, pumpkin, nutmeg and few of the isolates, could infect cardamom under in vitro conditions.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge Director (ICAR-IISR), Head—Division of Crop Protection (ICAR-IISR) for the facilities and support, Mr. K Jayarajan, Chief Technical Officer (ICAR-IISR) for the statistical analysis. University Grants Commission (UGC), Government of India for the fellowship.
Author contributions
FZ—Implementation of the research, analyses and interpretation of results, draft manuscript preparation, AJ—Conceptualization, experimental design, analysis and interpretation of results, manuscript editing, CNB—Analyses and interpretation of results, manuscript editing. All authors reviewed the results and approved the final version of the manuscript.
Data availability
All data generated in this study are available within the paper and also as supplementary information.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the author.
References
- Aragaki M, Uchida J. Morphological distinctions between Phytophthora capsici and P. tropicalis sp. nov. Mycologia. 2001;93:137. doi: 10.1080/00275514.2001.12061285. [DOI] [Google Scholar]
- Bhai RS, Jeevalatha A, Biju CN, Vinitha KB, Cissin J, Rosana OB, Fayad A, Praveena R, Anandaraj M, Eapen SJ. Sympatric occurrence of sibling Phytophthora species associated with foot rot disease of black pepper in India. Braz J Microbiol. 2022;53(2):801–818. doi: 10.1007/s42770-022-00716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilung LM, Pui CF, Su’ut L, Apun K. Evaluation of BOX-PCR and ERIC-PCR as molecular typing tools for pathogenic Leptospira. Dis Markers. 2018;2018(6):1–9. doi: 10.1155/2018/1351634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borba MP, Ballarini AE, Paulo J, Witusk D, Lavin P, Sand SVD. Evaluation of BOX-PCR and REP-PCR as molecular typing tools for Antarctic Streptomyces. Curr Microbiol. 2020;77:3573–3581. doi: 10.1007/s00284-020-02199-6. [DOI] [PubMed] [Google Scholar]
- Bouws H, Finckh MR. Effects of cropping history and origin of seed potatoes on population structure of Phytophthora infestans. Eur J Plant Pathol. 2007;117:313–327. doi: 10.1007/s10658-006-9099-z. [DOI] [Google Scholar]
- Brasier CM (1967) Physiology of reproduction in Phytophthora. Hull, UK: University of Hull, PhD thesis
- Brumat ACL, MayDe Mio LL, Tessmann DJ, Duarte HSS, Auer CG, Dos Santos ÁF. Phytophthora tropicalis: causal agent of persimmon fruit rot in Brazil. J Phytopathol. 2022;170:428–436. doi: 10.1111/jph.13094. [DOI] [Google Scholar]
- Burdon JJ. Genetic variation in pathogen populations and its implications for adaptation to host resistance. In: Jacobs T, Parlevliet JE, editors. Durability of disease resistance. Curr Plant Sci Biotechnol Agric. Dordrecht: Springer; 1993. [Google Scholar]
- Chowdappa P, Chandramohanan R. Occurrence and distribution of mating types of Phytophthora species causing black pod disease of cocoa. Indian Phytopath. 1997;50(2):256–260. [Google Scholar]
- da Silva MR, Martinelli JA, Federizzi LC, Chaves MS, Pacheco MT. Lesion size as a criterion for screening oat genotypes for resistance to leaf spot. Eur J Plant Pathol. 2012;134:315–327. doi: 10.1007/s10658-012-9991-7. [DOI] [Google Scholar]
- de Bruijn FJ. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58(7):2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahoo RS, Lamour KH. Interspecific hybridization and apomixis between Phytophthora capsici and Phytophthora tropicalis. Mycologia. 2008;100(6):911–920. doi: 10.3852/08-028. [DOI] [PubMed] [Google Scholar]
- El-Esawi MA, El-Zaher Mustafa A, Badr S, Sammour R. Isozyme analysis of genetic variability and population structure of Lactuca L. germplasm. Biochem Syst Ecol. 2017;70:73–79. doi: 10.1016/j.bse.2016.11.003. [DOI] [Google Scholar]
- Erwin DC, Ribeiro OK. Phytophthora diseases worldwide. St Paul: American Phytopathological Society Press; 1996. [Google Scholar]
- Farhana MDSN, Bivi MR, Khairulmazmi A, Wong SK, Sariah M. Morphological and molecular characterization of Phytophthora capsici, the causal agent of foot rot disease of black pepper in Sarawak, Malaysia. Int J Agric Biol. 2013;15:1083–1090. [Google Scholar]
- Food and Agriculture Organization of the United Nations (2023) https://www.fao.org/home/en. Accessed 19 April 2023
- Fu C, Qiu Y, Kong H. RAPD analysis for genetic diversity in Changium smyrnioides (Apiaceae), an endangered plant1. Bot Bull Acad Sin (taipei) 2003;44:13–18. [Google Scholar]
- Goodwin SB. The population genetics of Phytophthora. Phytopathology. 1997;87:462–473. doi: 10.1094/PHYTO.1997.87.4.462. [DOI] [PubMed] [Google Scholar]
- Hantula J, Dusabenyagasani M, Hamelin RC. Random amplified microsatellites (RAMS)—a novel method for characterizing genetic variation within fungi. Eur J for Pathol. 1996;26:159–166. doi: 10.1111/j.1439-0329.1996.tb00720.x. [DOI] [Google Scholar]
- Hema M, Kumar R, Singh NP. Volatile price and declining profitability of black pepper in India: disquieting future. Agric Econ Res Rev. 2007;20:61–76. [Google Scholar]
- Hong CX, Richardson PA, Kong P, Jeffers SN, Oak SW. Phytophthora tropicalis isolated from diseased leaves of Pieris japonica and Rhododendron catawbiense and found in irrigation water and soil in Virginia. Plant Dis. 2006;90(4):525. doi: 10.1094/PD-90-0525C. [DOI] [PubMed] [Google Scholar]
- Hong C, Patricia A, Richardson KP. Pathogenicity to ornamental plants of some existing species and new taxa of Phytophthora from irrigation water. Plant Dis. 2008;92(8):1201–1207. doi: 10.1094/PDIS-92-8-1201. [DOI] [PubMed] [Google Scholar]
- Jara-Arancio P, Carvalho CS, Carmona-Ortiz MR, Bustamante RO, Schmidt Villela PM, Andrade SCS, Peña-Gómez FT, González LA, Fleury M. Genetic diversity and population structure of Jubaea chilensis, an endemic and monotype gender from Chile, based on SNP markers. Plants. 2022;11(15):1959. doi: 10.3390/plants11151959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeevalatha A, Biju CN, Bhai RS. Ypt1 gene-based recombinase polymerase amplification assay for Phytophthora capsici and P. tropicalis detection in black pepper. Eur J Plant Pathol. 2021;159:863–875. doi: 10.1007/s10658-021-02211-0. [DOI] [Google Scholar]
- Jibat M, Alo S. Characterization of Phytophthora capsici foot rot disease of black pepper in Ethiopia. J Plant Pathol Microbiol. 2021;12:542. [Google Scholar]
- Li P, Liu D. Genetic diversity amongst asexual and sexual progenies of Phytophthora capsici detected with ISSR markers. Plant Protect Sci. 2021;57:271–278. doi: 10.17221/28/2021-PPS. [DOI] [Google Scholar]
- Li P, Liu D, Guo M, Pan Y, Chen F, Zhang H, Gao Z. A PCR-based assay for distinguishing between A1 and A2 mating types of Phytophthora capsici. J Am Soc Hortic Sci. 2017;142(4):260–264. doi: 10.21273/JASHS04013-17. [DOI] [Google Scholar]
- Majid MU, Awan MF, Fatima K, Tahir MS, Ali Q, Rashid B, Rao AQ, Nasir I, Husnain T. Phytophthora capsici on chilli pepper (Capsicum annuum L.) and its management through genetic and bio-control: a review. ZEMDIRBYSTE. 2016;103:419–430. doi: 10.13080/z-a.2016.103.054. [DOI] [Google Scholar]
- Mansfeld BN, Colle M, Zhang C, Lin YC, Grumet R, Grear JS, Kuhn A. Developmentally regulated activation of defense allows for rapid inhibition of infection in age-related resistance to Phytophthora capsici in cucumber fruit. BMC Genomics. 2020;21:628. doi: 10.1186/s12864-020-07040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markert JA, Champlin DM, Gutjahr-Gobell R, Grear JS, Kuhn A, McGreevy TJ, Roth A, Bagly MJ, Nacci DE. Population genetic diversity and fitness in multiple environments. BMC Evol Biol. 2010;10:205. doi: 10.1186/1471-2148-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masanto HA, Wibowo A, Subandiyah S, Shimizu M, Suga H, Kageyama K. Genetic diversity of Phytophthora palmivora isolates from Indonesia and Japan using rep-PCR and microsatellite markers. J Gen Plant Pathol. 2019;85:367–381. doi: 10.1007/s10327-019-00853-x. [DOI] [Google Scholar]
- Meitz J, Linde C, Thompson LSD, Mcleod A. Phytophthora capsici on vegetable hosts in South Africa: distribution, host range and genetic diversity. Australas Plant Pathol. 2010;39:431–439. doi: 10.1071/AP09075. [DOI] [Google Scholar]
- Mishra RK, Pandey BK, Pathak N, Zeeshan M. BOX-PCR- and ERIC-PCR-based genotyping and phylogenetic correlation among Fusarium oxysporum isolates associated with wilt disease in Psidium guajava L. Biocatal Agric Biotechnol. 2015;4(1):25–32. doi: 10.1016/j.bcab.2014.09.012. [DOI] [Google Scholar]
- Mohammad B, Jose RS (2018) Chemical management of Phytophthora capsicin pumpkin in Illinois. Mod concepts dev agron 1(4)
- Nath VS, Basheer S, Jeeva ML, Veena SS. Genetic and phenotypic characterization of Phytophthora colocasiae in Taro Growing Areas of India. J Plant Pathol Microbiol. 2016;7:383. doi: 10.4172/2157-7471.1000383. [DOI] [Google Scholar]
- Orlikowski LB, Trzewik A, Nowak K, Szkuta G (2006) Phytophthora tropicalis, a new pathogen of ornamental plants in Poland. J Plant Prot Res 46
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira D, McDonald BA, Croll D. The genetic architecture of emerging fungicide resistance in populations of a global wheat pathogen. Genome Biol Evol. 2020;12(12):2231–2244. doi: 10.1093/gbe/evaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai GS, Liew ECY, Guest DI. Survey, identification and genetic diversity of Phytophthora capsici causing wilt of chilli (Capsicum annuum L.) in Bhutan. Eur J Plant Pathol. 2020;158:655–665. doi: 10.1007/s10658-020-02108-4. [DOI] [Google Scholar]
- Rohlf FJ (2000) NTSYS-PC. Numerical Taxonomy and Multivariate Analysis System. Version 2.10e. Department of Ecology and Evolution, State University of New York at Stony Brook, Stony Brook
- Roy SG, Bhattacharyya S, Mukherjee SK, Khatua DC. Molecular identification of Phytophthora spp. affecting some economically important crops in Eastern India through ITS-RFLP and sequencing of the ITS region. J Phytopathol. 2009;157:666–674. doi: 10.1111/j.1439-0434.2008.01537.x. [DOI] [Google Scholar]
- Shej C, Renu SG, Balaji S, Anandaraj M (2009) Ribosomal DNA analysis of three Phytophthora species occurring in India. Indian Phytopathol 62: 155–162. https://epubs.icar.org.in/index.php/IPPJ/article/view/12540
- Shirzad-Aski H, Tabatabaei M. Molecular characterization of Pasteurella multocida isolates obtained from poultry, ruminant, cats and dogs using RAPD and REP-PCR analysis. Mol Biol Res Commun. 2016;5(3):123–132. [PMC free article] [PubMed] [Google Scholar]
- Spices Board (2023) http://indianspices.com/. Accessed 19 April 2023
- Stephenson LW, Erwin DC, Leary JV. Hyphal anastomosis in Phytophthora capsici. Phytopathology. 1974;64:149–150. doi: 10.1094/Phyto-64-149. [DOI] [Google Scholar]
- Syed-Ab-Rahman SF, Xiao Y, Carvalhais LC, Ferguson BJ, Schenk PM. Suppression of Phytophthora capsici infection and promotion of tomato growth by soil bacteria. Rhizosphere. 2019;9:72–75. doi: 10.1016/j.rhisph.2018.11.007. [DOI] [Google Scholar]
- Truong NV, Burgess L, Liew ECY. Prevalence and aetiology of Phytophthora foot rot of black pepper in Vietnam. Australas Plant Pathol. 2008;37:431–442. doi: 10.1071/AP08034. [DOI] [Google Scholar]
- Truong NV, Liew ECY, Burgess LW. Characterisation of Phytophthora capsici isolates from black pepper in Vietnam. Fungal Biol. 2010;114(2–3):160–170. doi: 10.1016/j.funbio.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Truong NV, Burgess LW, Liew ECY. Cross-infectivity and genetic variation of Phytophthora capsici isolates from chilli and black pepper in Vietnam. Australas Plant Pathol. 2012;41:439–447. doi: 10.1007/s13313-012-0136-4. [DOI] [Google Scholar]
- Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J, Kapur V, Mason EO, Shah U, Koeuth T, Lupski JR, Musser JM. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J Infect Dis. 1993;167:850–856. doi: 10.1093/infdis/167.4.850. [DOI] [PubMed] [Google Scholar]
- Wang M, Zhang X, Yang T, Han Z, Yanagimoto T, Gao T. Genetic diversity in the mtDNA control region and population structure in the Sardinella zunasi Bleeker. Afr J Biotechnol. 2009;7:4384–4392. doi: 10.5897/AJB2022.17528. [DOI] [Google Scholar]
- Welsh J, Honeycutt RJ, McClelland M, Sobral BW. Parentage determination in maize hybrids using the arbitrarily primed polymerase chain reaction (AP-PCR) Theor Appl Genet. 1991;82(4):473–476. doi: 10.1007/BF00588601. [DOI] [PubMed] [Google Scholar]
- Yeh F, Boyle T, Yang R, Ye Z, Xiyan JM (1997) POPGENE: Microsoft Window-based International Forestry Research, Edmonton, Canada
- Yu-Huan Gu, Ko WH. Occurrence of parasexual cycle in Phytophthora parasitica following protoplast fusion. Bot Bull Acad Sin (taipei) 2000;41:225–230. [Google Scholar]
- Zaccardelli M, Campanile F, Moretti C, Buonaurio R. Characterization of Italian populations of Xanthomonas campestris pv. campestris using primers based on DNA repetitive sequences. J Plant Pathol. 2008;90:375–381. [Google Scholar]
- Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20(2):176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are available within the paper and also as supplementary information.