Abstract
Microtubules are dynamic cytoskeletal filaments that undergo stochastic switching between phases of polymerization and depolymerization—a behavior known as dynamic instability. Many important cellular processes, including cell motility, chromosome segregation, and intracellular transport, require complex spatiotemporal regulation of microtubule dynamics. This coordinated regulation is achieved through the interactions of numerous microtubule-associated proteins (MAPs) with microtubule ends and lattices. Here, we review the recent advances in our understanding of microtubule regulation, focusing on results arising from biochemical in vitro reconstitution approaches using purified multiprotein ensembles. We discuss how the combinatory effects of MAPs affect both the dynamics of individual microtubule ends, as well as the stability and turnover of the microtubule lattice. In addition, we highlight new results demonstrating the roles of protein condensates in microtubule regulation. Our overall intent is to showcase how lessons learned from reconstitution approaches help unravel the regulatory mechanisms at play in complex cellular environments.
Keywords: microtubule dynamics, microtubule regulation, microtubule-associated proteins, +TIPs, condensates, phase separation, lattice damage, lattice repair
Microtubules are cytoskeletal filaments crucial for a variety of cellular processes, including cell motility, cell division, and intracellular transport. Their hollow cylindrical structures are typically composed of 13 protofilaments assembled from αβ-tubulin heterodimers, with α-tubulin subunits exposed at the minus end and β-tubulin subunits at the plus end. Microtubule ends stochastically switch between phases of growth and shrinkage through a process known as microtubule dynamic instability (1). Microtubule dynamic instability is a nonequilibrium process, powered by the GTPase activity of tubulin. Tubulin heterodimers incorporate into the microtubule polymer with GTP bound to their β-tubulin subunits, which subsequently hydrolyzes, resulting in a polymer lattice composed predominantly of GDP-β-tubulin. However, since GTP hydrolysis occurs with a delay, a growing microtubule maintains a structurally distinct stabilizing ‘cap’ of GTP-tubulin at its end. It is through the loss and regaining of this stabilizing cap that microtubule ends transition from growth to shrinkage (known as microtubule ‘catastrophe’) and from shrinkage to growth (known as microtubule ‘rescue’).
Microtubule dynamic instability is regulated by a complex, interconnected network of microtubule-associated proteins (MAPs). Individual MAPs modulate microtubule growth and shrinkage rates as well as the frequencies of catastrophe and rescue. Due to the number and complexity of interactions among regulators within cells, it is challenging to dissect the specific functions of individual MAPs on microtubules. To circumvent this, bottom-up reductionist approaches are essential—where one starts with a minimal number of components, deciphers the nature and consequence of interactions among those components, and progressively enhances complexity by increasing the number of components in a controlled fashion. The core philosophy underlying the bottom-up reconstitution approach is to reverse-engineer a cell–biological process in a controlled environment outside the cell (2).
The reconstitution of microtubule-based phenomena outside of cells originated from the purification of tubulin and the ability to assemble microtubules in vitro (3, 4, 5), which subsequently led to the discovery of microtubule dynamic instability (1). Shortly thereafter, dynamic instability of reconstituted microtubules was directly visualized at the individual microtubule level using video-enhanced differential interference contrast microscopy (6, 7). Concurrently with the early tubulin purification efforts, a growing catalog of MAPs were being discovered and characterized (3, 8, 9). Although the landmark advancements in the field of reconstitution provided fundamental insights into the microtubule dynamics, there was a caveat—the dynamic parameters observed in vitro were different than those typically observed in cells. The discrepancies between cellular and in vitro observations brought the roles of MAPs into the center stage of microtubule dynamics regulation.
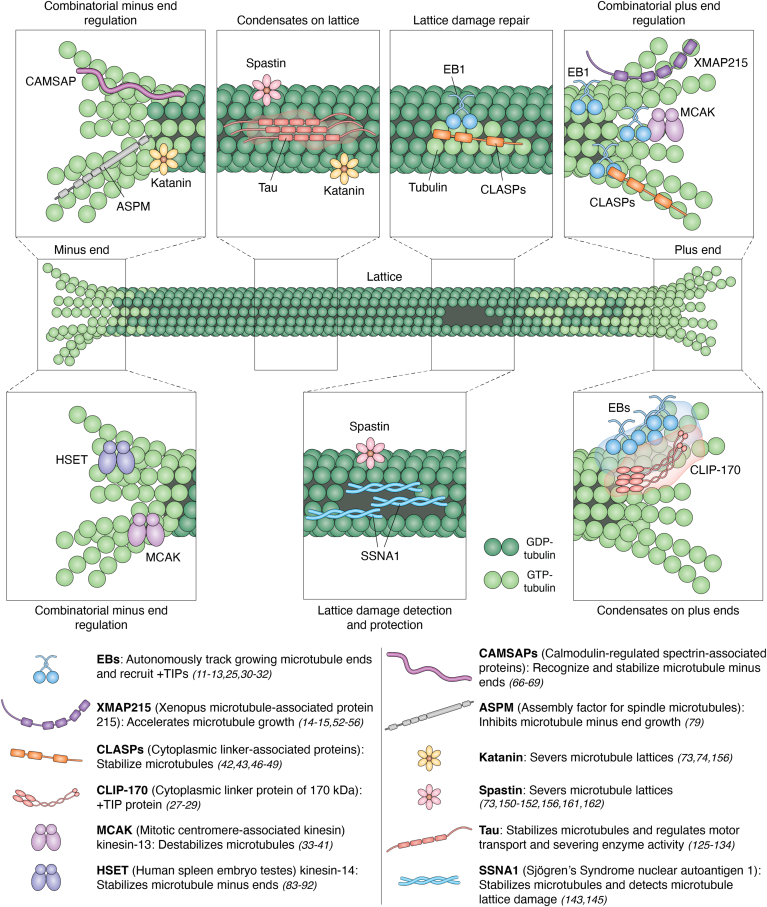
The early reconstitution studies provided a clear pathway for probing the regulation of the microtubule cytoskeleton by MAPs in vitro (10). Since then, enormous progress has been made in understanding how microtubule dynamics are regulated by individual and combinations of MAPs. In this review, we discuss the regulation of dynamic microtubule ends and lattices by key MAPs (Fig. 1), focusing on recent biochemical in vitro reconstitution studies with multiprotein ensembles.
Figure 1.
Combinatorial regulation of microtubule ends and lattices by the collective action of MAPs. MAP, microtubule-associated protein.
Microtubule end dynamics are regulated by ensembles of MAPs
Microtubule dynamics are controlled by an interplay of regulatory proteins acting in concert on dynamic microtubule ends (Fig. 1). Several MAPs autonomously bind microtubule ends. For example, EB-family proteins recognize the GTP-tubulin cap at growing microtubule plus and minus ends (11, 12, 13), and Xenopus microtubule-associated protein 215 (XMAP215)-family proteins use their highly conserved tubulin-binding tumor overexpressed gene domains to recognize tubulin dimers at microtubule plus ends, regardless of their dynamic state (14, 15, 16, 17, 18, 19, 20, 21, 22). Notably, EBs recruit additional plus-end-tracking proteins (+TIPs) to growing microtubule ends that act in conjunction with EBs to modulate microtubule dynamics (23). Indeed, many +TIPs contain a SxIP amino acid sequence motif, which is specifically recognized by the EB homology domain and serves as a microtubule end-localization signal through ‘hitchhiking’ on EBs (24, 25, 26). Other prominent +TIPs, such as cytoplasmic linker protein of 170 kDa (CLIP-170), bind to EBs via a direct interaction between their CAP-Gly domains and the C-terminal EEY motif of EBs (27, 28, 29). Thus, the localization of EBs along the GTP-cap, their ability to interact with many other MAPs, and the multivalency of these interactions all facilitate the formation of higher-order regulatory complexes at growing microtubule ends.
In recent years, a number of in vitro reconstitution studies investigated the combinatorial effects of +TIPs on microtubule dynamics, paying particular attention to proteins that interact with EBs. Although EB proteins directly modulate microtubule dynamics in vitro, strongly promoting microtubule catastrophe and increasing microtubule growth rates (30, 31, 32), these effects are strikingly altered when EBs are combined with other +TIPs. For example, kinesin-13 mitotic centromere-associated kinesin (MCAK), a potent microtubule depolymerase important for spindle organization in dividing cells, contains a SxIP motif that facilitates its interaction with EBs (25, 33, 34, 35, 36, 37, 38, 39, 40). The EB-dependent targeting of MCAK to growing microtubule ends enhances MCAK’s microtubule destabilizing activity in vitro (41). The direct interaction of cytoplasmic linker-associated protein (CLASP) with EBs via CLASPs’ SxIP motifs provides another example of amplified protein activity due to EB-dependent microtubule end targeting (42, 43, 44). CLASPs are a highly conserved family of tumor overexpressed gene-domain proteins that regulate microtubule dynamics in cell division, cell migration, and neuronal development (15, 18, 20, 45, 46). On their own, CLASPs stabilize microtubules by suppressing microtubule catastrophe and promoting microtubule rescue—these effects are dramatically enhanced when CLASP is targeted to the microtubule plus ends by EB (42, 43, 47, 48, 49, 50, 51). Interestingly, in the presence of EB, CLASP protects microtubules against the destabilizing activity of MCAK (42). Such synergistic and antagonistic effects of +TIPs underscore the nuanced balance of EB-mediated microtubule end regulation.
Reconstitutions with multi-MAP ensembles pave the way for recapitulating cellular-like microtubule dynamics in vitro. In cells, microtubules grow significantly faster and transition more frequently between the phases of growth and shrinkage when compared to microtubules polymerized with similar concentrations of purified tubulin in vitro. A landmark study demonstrated that several physiological characteristics of microtubule behavior can be recapitulated in vitro with a three-component module composed of purified tubulin, XMAP215, and XKCM1 (the Xenopus homolog of kinesin-13 MCAK) (10). Here, XMAP215 accelerated microtubule growth, while XKCM1 simultaneously induced frequent catastrophe events. XMAP215’s polymerase activity relies on the preferred stabilization of weakly bound, curved tubulin dimers at growing microtubule ends (52, 53). On its own, XMAP215 increases microtubule growth rate by an order of magnitude (16, 54, 55, 56, 57). Combining XMAP215 with EB1 leads to the further, highly synergistic acceleration of microtubule growth, bringing the rates to the highest levels observed in cells (57). The synergistic effects of XMAP215 and EB1 are allosteric, as XMAP215 and EB1 do not interact directly. Rather, EB-mediated straightening of microtubule protofilaments at the growing microtubule end likely facilitates the release of XMAP215 from an incorporated tubulin dimer, allowing it to more quickly stabilize a subsequent incoming tubulin dimer at the growing end (13, 57). It has also been suggested that EB1 and XMAP215 form a complex with Sentin, another +TIP, which can promote their cooperative effects on microtubule dynamics (58). Although Sentin exhibits XMAP215-dependent plus end tracking activity in vitro, to the best of our knowledge, a direct interaction between Sentin and XMAP215 has not been established. Thus, it is not clear whether the EB1–Sentin–XMAP215 complex formation has a role in microtubule regulation. These findings highlight the need for reconstitution studies with purified proteins, which have the ability to parse apart allosteric (through the microtubule) from direct effects mediated by complex formation. In addition to fast growth and frequent catastrophes, microtubules in cells also undergo periods of pausing and recurrent rescue events (59, 60, 61), behaviors not exhibited by microtubules grown in vitro with tubulin alone nor in the presence of EB1, XMAP215, and XKCM1/MCAK (7, 10, 57). The combination of the D. melanogaster homologs of five MAPs: EB1, XMAP215, MCAK, Sentin, and CLASP (62), resulted in fast growth, rescues, and pausing, thus reconstituting the main characteristics of microtubule plus end dynamics in cells.
The collective regulation of microtubule dynamics by MAPs is not restricted to the microtubule plus ends. In recent years, there has been an increased focus on the interplay of MAPs at microtubule minus ends, which are structurally and biochemically different from the plus ends, and thus exhibit distinct dynamics. Microtubule minus-end regulation is particularly important within acentrosomal microtubule arrays, including those in differentiated epithelial, neuronal, and muscle cells and cortical microtubule arrays in plants (63, 64, 65). Calmodulin-regulated spectrin-associated proteins (CAMSAPs) are a family of proteins that recognize and stabilize growing minus ends and are essential for regulating noncentrosomal microtubule arrays in neuronal and epithelial cells (66, 67, 68, 69, 70, 71, 72). A multicomponent in vitro reconstitution study demonstrated that katanin, a microtubule-severing ATPase (73, 74), interacts with CAMSAPs and restricts the length of CAMSAP-stabilized microtubule stretches, thus antagonizing CAMSAP’s stabilizing activity (69, 75). Conversely, both the mammalian CAMSAP and Patronin, the D. melanogaster homolog of CAMSAP, protect microtubule minus ends from the depolymerase activity of MCAK through steric inhibition (67, 68). Similarly to CAMSAP, SPIRAL2, a plant-specific MAP, autonomously recognizes and stabilizes minus ends and prevents severing by katanin in plants (76, 77). Microtubule minus-end regulation is also important in the spindle poles of dividing cells. Assembly factor for spindle microtubules (ASPM) is a microcephaly associated protein that autonomously recognizes microtubule minus ends in mitotic and meiotic spindles and blocks their growth (78, 79, 80, 81, 82). Interestingly, ASPM forms a complex with katanin, thus bringing katanin’s severing activity in proximity to the minus end and further preventing the minus-end growth (79). Furthermore, kinesin-14 human spleen embryo testes (HSET), a minus-end directed motor involved in spindle formation (83, 84, 85, 86, 87, 88, 89, 90, 91), specifically stabilizes microtubule minus ends by reducing the tubulin off-rate, thereby suppressing minus-end catastrophe (92). HSET’s protective activity was found to be strong enough to resist the destabilizing activity of MCAK at the minus end (92). Notably, ASPM, HSET, MCAK, and katanin are all implicated in microtubule organization at spindle poles—how these four factors with distinct activities interplay to collectively regulate microtubule minus ends has not been investigated.
Another layer of complexity arises when one considers the simultaneous regulation of both microtubule ends. Although microtubules characteristically undergo dynamic instability, treadmilling behavior is also observed in cells where the microtubule minus end shrinks while the plus end simultaneously grows (67, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102). In spindles, microtubule treadmilling may contribute to ‘poleward flux’ where microtubule plus ends grow at kinetochores and the minus ends shrink at the spindle poles (103). Recent in vitro reconstitution experiments backed by in silico predictions showed that robust plus-end-directed microtubule treadmilling emerges as an outcome of dynamics regulation by four MAPs—XMAP215, EB1, CLASP2 and kinesin-13 MCAK—all of which function in spindles and are implicated in poleward flux (104).
Condensation of MAPs adds complexity to microtubule end regulation
Increasing evidence suggests that an additional mechanism underlying MAP-mediated microtubule regulation involves biomolecular condensation (Fig. 1). Condensates are membrane-less compartments that boost reaction kinetics by locally concentrating the chemical components of a reaction inside a condensed confinement (reviewed in (105)). A number of proteins that interact with tubulin have been shown to autonomously form condensates in vitro and co-condense with tubulin to achieve a highly concentrated tubulin phase. Tubulin condensation is implicated in microtubule nucleation from centrosomes (106, 107), within the spindles (108), as well as in branched microtubule nucleation (109, 110).
A series of new studies investigated the condensation of microtubule +TIPs. The propensity for condensation has been particularly addressed for CLIP-170 and EB proteins—core members of the +TIP complex. Overexpression of CLIP-170 in several cell types resulted in the formation of characteristic patches which deform and merge, consistent with biomolecular condensation (111, 112). Purified CLIP-170 was found to readily form condensates in vitro, at concentrations as low as 3 nM (111). CLIP-170 condensates formed independently of microtubules, both in cells and in vitro (111, 112). The condensates were not dissolved by 1,6-hexanediol, a treatment that disrupts hydrophobic interactions, suggesting that the condensation of CLIP-170 does not rely on hydrophobic interactions (111, 113). While the N-terminus region of CLIP-170, containing CAP-Gly domains and a serine-rich region, mediates its interaction with microtubules and is sufficient for robust microtubule plus-end tracking in cells, the ability to form condensates relies on its C-terminal region, containing a zinc-knuckle domain (111, 112). CLIP-170 condensates were also observed to co-condense with tubulin (111), and concentrated CLIP-170/tubulin droplets on the microtubule lattice in cell lysates resulted in new microtubule nucleation (113).
Unlike CLIP-170, condensation of the most prominent +TIP proteins from the EB family required significantly higher protein concentrations (∼micromolar range) and/or use of crowding agents (111, 114, 115). The ability of EB1 to form condensates was found to rely on an intrinsically disordered region (IDR) within its linker region, linking its N-terminal calponin-homology domain essential for microtubule interactions, with its C-terminal EB-homology domain used for dimerization and interactions with other +TIPs (115). Notably, the same unstructured region has been previously implicated in EB1’s tip-tracking ability (116). Along these lines, mutating six conserved lysine/arginine residues to glutamine within the IDR reduced both EB1’s ability to tip-track and form condensates. However, fluorescence recovery after photobleaching analysis of both EB1 and CLIP-170 condensates in these studies demonstrated only a partial turnover of components, with recovery halftimes on the order of tens of seconds, in stark contrast to the previously measured subsecond turnover rates of both CLIP-170 and EB3 at growing microtubule ends in cells and in vitro (30, 41, 117). This finding raises the question of whether and when +TIP comets indeed involve liquid–liquid phase separation in physiological conditions.
When +TIP condensates do form at microtubule tips in cells, what is their functional relevance? Many +TIPs modulate microtubule plus-end dynamics; however, there are a limited number of binding sites at the microtubule end itself. Thus, concentrating selected +TIPs into condensates via multivalent interactions at the microtubule end could dictate the polymerization dynamics. Furthermore, a local increase in tubulin concentration through co-condensation could promote microtubule plus-end growth rate, although tubulin assembly is not considered to be a diffusion-limited process in cellular conditions (118). Notably, even a double depletion of CLIP-170 and EB3 resulted in only a mild (∼20%) reduction in microtubule growth rates in retinal pigment epithelium cells (111). Alternatively, it has been hypothesized that +TIP condensates could have a mechanical role at the microtubule end, forming a stabilizing superstructure to facilitate lateral interactions between microtubule protofilaments, effectively serving as a polymerization chaperone (112). Furthermore, +TIP condensates could play specific mechanical roles in the microtubule–kinetochore attachments (112), an interface where condensation of kinetochore components has been previously reported (119). Interestingly, HeLa cells expressing IDR mutants of EB1 displayed chromosome segregation defects, consistent with the loss of microtubule tip-tracking observed for these mutants, along with their reduced ability to form condensates (115). In fission yeast, the condensation of the EB homolog Mal3 has been implicated in the end accumulation of CLIP-170 homolog TIP1 (114). However, the condensation of Mal3 on microtubules is largely encoded in the IDR2 domain of Mal3, which is specific to yeast and not conserved in mammals. In budding yeast, the condensation of the EB homolog Bim1 and CLIP-170 homolog Bik1 is driven by the assembly of the budding yeast–specific Kar9 network, leading to a formation of a ‘+TIP body’ implicated in establishing microtubule–actin interactions (120). This finding presents another potential role of +TIP condensates in the mechanical coupling of different cytoskeletal systems. Future studies of MAP condensation are likely to uncover new principles underlying higher-order microtubule end regulation.
The interplay of MAPs regulates microtubule lattices
Higher-order complexes of MAPs play regulatory roles along the microtubule lattice in addition to dynamic ends (Fig. 1). Of note, tau, a microtubule-stabilizing protein that aggregates into fibrils in the context of neurodegenerative diseases (121, 122, 123, 124, 125), has been recently found to form patches along the microtubule lattice that merge and slowly turn over, consistent with a liquid condensate phase (126, 127, 128). These patches of tau selectively regulate the movement of motor proteins along the microtubule lattice (127, 128, 129, 130, 131, 132, 133, 134). Tau patches inhibit the processive movements of kinesin-1 and kinesin-3 motor proteins but are more permissive to dynein and kinesin-8 motility (128, 130, 132, 133). The selective modulation of motor movements has also been demonstrated for a number of other microtubule lattice-binding MAPs (132, 133). Additionally, condensates of tau shield the microtubule lattice from damage induced by microtubule severing enzymes and can thus regulate the location and extent of microtubule severing (127, 128).
While severing enzymes are known for their canonical ability to sever microtubule lattices, recent studies highlight that other factors, including motor protein activity and mechanical perturbations, also induce damage and defects within the microtubule lattice (reviewed in (135)). The binding of kinesin-1 motors can expand and change the microtubule lattice structure (136), which in turn has allosteric effects on the binding of other motors and MAPs to the microtubule lattice (137). Furthermore, the walking of motors along the lattice has been reported to induce the removal of tubulin subunits from the lattice (138, 139, 140). Notably, using an optical tweezers assay, it was estimated that ∼30 pN of force is required to extract a tubulin dimer from the lattice (141), considerably larger than typical forces generated by single motors (∼few pN, (142)). Thus, the extraction of tubulin dimers by walking motors is expected to be a relatively rare event. It has been speculated that preexisting lattice defects serve as preferential sites for kinesin-mediated tubulin removal (135). Therefore, the destruction of the microtubule lattice by motors is likely to be context dependent.
Recent studies show that some MAPs directly detect microtubule lattice damage (143, 144). Of note, Sjögren's syndrome nuclear autoantigen 1 (SSNA1) is a MAP implicated in cilium assembly, intraflagellar transport, cell division, and axonal branching (145, 146, 147, 148, 149). SSNA1 specifically localizes to the sites of lattice damage, occurring both spontaneously or induced by spastin, a microtubule-severing enzyme (73, 143, 150, 151, 152). In addition to recognizing the sites of microtubule damage, SSNA1 localization also protects the microtubule lattice against spastin-induced damage. SSNA1 self-associates into higher-order fibrils that bind along the groove between adjacent protofilaments of the microtubule lattice (145, 153). The ability of SSNA1 to form fibrils likely underlies its function on microtubules. Indeed, SSNA1 localizes in stretches on growing microtubule ends where it acts to attenuate microtubule growth and protect against catastrophe (143). Another ciliary protein, centrosome and spindle pole associated protein 1, has recently been shown to recognize microtubule damage and stabilize growing microtubule ends by specifically binding to the microtubule lumen (144). The ability of SSNA1 and centrosome and spindle pole associated protein 1 to both detect microtubule damage and modulate microtubule end dynamics raises the interesting possibility that microtubule ends and damage sites share specific structural features. In support of this, several prominent regulators of microtubule end dynamics, including EB1, CLIP-170, and CLASP2, have been found to localize to microtubule lattice defects (154, 155, 156). The localization of +TIPs to microtubule damage sites may aid with the incorporation of new, GTP-bound tubulin dimers, ultimately resulting in lattice ‘repair’. This activity has been specifically reported for CLASP2 (154, 157). Sites of microtubule lattice repair could enhance microtubule stability, serving as points of subsequent microtubule rescue (158). Indeed, enhanced rescue activity has been reported to occur at lattice defect sites artificially induced by either microtubule-stabilizing drugs or laser irradiation (159, 160). Thus, the regulation of microtubule lattice damage and repair presents a novel mechanism of selective microtubule stabilization.
The balance between microtubule destruction and stabilization is particularly important when considering the physiological function of microtubule-severing enzymes. If stabilized, the microtubule fragments generated through severing may serve as nucleation ‘seeds’ for the growth of new microtubule polymers, resulting in an overall network amplification (73, 156, 161, 162). This mechanism underlies the microtubule array reorientation and amplification that occurs in plants in response to blue light (163, 164, 165). Microtubule network amplification could be further enhanced by recruiting nucleation factors and promoting nucleation from sites of microtubule damage. Overall, the processes of lattice damage, destruction, and repair are likely to have significant functional relevance in the remodeling of microtubule networks in a variety of cellular contexts.
Conclusions
The complex behavior of microtubules in cells arises from the collective actions of a multitude of protein regulators. Without such nuanced regulation, building the intricate and dynamic microtubule-based structures observed in diverse cellular contexts would not be possible. Reconstitution experiments with purified proteins offer a powerful platform to characterize the individual effects of isolated MAPs as well as the collective impact of multi-MAP networks on microtubule dynamics regulation. Studying the activities of MAPs alone and in combination reveals synergistic and antagonistic relationships and emergent behaviors that could not be predicted from the activities of individual MAPs. Further complexity arises through the simultaneous regulation of both microtubule ends by distinct and overlapping networks of MAPs. The microtubule lattice represents yet another point of microtubule regulation. In this light, exciting new research efforts have highlighted the roles of multi-MAP modules in microtubule lattice regulation as well as the roles of biomolecular condensates on microtubule ends and lattices. The multifaceted modes of microtubule regulation emphasize the need for controlled bottom-up approaches to tease apart the relationships occurring between MAPs in the rich environment within cells.
Our understanding of how microtubules are regulated is far from complete. Future studies will likely uncover yet more MAPs and identify the ways in which they integrate into the microtubule regulatory network. Of note, the conformational changes induced by individual MAPs on the microtubule itself can have allosteric effects on microtubule stability and its regulation by other MAPs (57, 137, 166, 167, 168). Furthermore, a variety of tubulin posttranslational modifications as well as the incorporation of different tubulin isotypes within microtubules are now known to define distinct subpopulations of microtubules. These tubulin modifications and variations in isotype composition generate a "code" that is read and modified by MAPs, selectively tuning microtubule regulation and function (169, 170, 171, 172, 173, 174, 175). To date, most reconstitution efforts have focused on the effects of MAPs on individual microtubule dynamics; how their activities interplay to build higher-order microtubule structures is less understood. To this end, increasing the complexity of reconstituted systems will take us one step closer to deciphering the underlying principles governing complex microtubule-based processes in cells.
Conflict of interest
The authors declare no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
E. J. L., M. Z., and S. C. writing–original draft; E. J. L. and M. Z. writing–reviewing and editing.
Funding and additional information
We acknowledge the support from the National Science Foundation grant MCB2018661 and the National Institutes of Health Grant R35GM119552. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Enrique De La Cruz
References
- 1.Mitchison T., Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 2.Liu A.P., Fletcher D.A. Biology under construction: in vitro reconstitution of cellular function. Nat. Rev. Mol. Cell Biol. 2009;10:644–650. doi: 10.1038/nrm2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borisy G.G., Olmsted J.B., Marcum J.M., Allen C. Microtubule assembly in vitro. Fed. Proc. 1974;33:167–174. [PubMed] [Google Scholar]
- 4.Lee J.C., Timasheff S.N. Reconstitution of microtubules from purified calf brain tubulin. Biochemistry. 1975;14:5183–5187. doi: 10.1021/bi00694a025. [DOI] [PubMed] [Google Scholar]
- 5.Weisenberg R.C. Microtubule formation in vitro in solutions containing low calcium concentrations. Science. 1972;177:1104–1105. doi: 10.1126/science.177.4054.1104. [DOI] [PubMed] [Google Scholar]
- 6.Horio T., Hotani H. Visualization of the dynamic instability of individual microtubules by dark-field microscopy. Nature. 1986;321:605–607. doi: 10.1038/321605a0. [DOI] [PubMed] [Google Scholar]
- 7.Walker R.A., O’Brien E.T., Pryer N.K., Soboeiro M.F., Voter W.A., Erickson H.P., et al. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleveland D.W., Hwo S.Y., Kirschner M.W. Purification of tau, a microtubule-associated protein that induces assembly of microtubules from purified tubulin. J. Mol. Biol. 1977;116:207–225. doi: 10.1016/0022-2836(77)90213-3. [DOI] [PubMed] [Google Scholar]
- 9.Sloboda R.D., Rudolph S.A., Rosenbaum J.L., Greengard P. Cyclic AMP-dependent endogenous phosphorylation of a microtubule-associated protein. Proc. Natl. Acad. Sci. U. S. A. 1975;72:177–181. doi: 10.1073/pnas.72.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinoshita K., Arnal I., Desai A., Drechsel D.N., Hyman A.A. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294:1340. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- 11.Maurer S.P., Fourniol F.J., Bohner G., Moores C.A., Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanic M., Stear J.H., Hyman A.A., Howard J. EB1 recognizes the nucleotide state of tubulin in the microtubule lattice. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R., Alushin G.M.M., Brown A., Nogales E. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Bassam J., Kim H., Flor-Parra I., Lal N., Velji H., Chang F. Fission yeast Alp14 is a dose-dependent plus end–tracking microtubule polymerase. Mol. Biol. Cell. 2012;23:2878–2890. doi: 10.1091/mbc.E12-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Bassam J., Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brouhard G.J., Rice L.M. Microtubule dynamics: an interplay of biochemistry and mechanics. Nat. Rev. Mol. Cell Biol. 2018;19:451–463. doi: 10.1038/s41580-018-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farmer V.J., Zanic M. TOG-domain proteins. Curr. Biol. 2021;31:R499–R501. doi: 10.1016/j.cub.2021.01.039. [DOI] [PubMed] [Google Scholar]
- 19.Gard D.L., Becker B.E., Josh Romney S. MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215⧸Dis1 family of microtubule-associated proteins. Int. Rev. Cytol. 2004;239:179–272. doi: 10.1016/S0074-7696(04)39004-2. [DOI] [PubMed] [Google Scholar]
- 20.Slep K.C. The role of TOG domains in microtubule plus end dynamics. Biochem. Soc. Trans. 2009;37:1002–1006. doi: 10.1042/BST0371002. [DOI] [PubMed] [Google Scholar]
- 21.Slep K.C., Vale R.D. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol. Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Breugel M., Drechsel D., Hyman A. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J. Cell Biol. 2003;161:359–369. doi: 10.1083/jcb.200211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akhmanova A., Steinmetz M.O. Microtubule +TIPs at a glance. J. Cell Sci. 2010;123:3415–3419. doi: 10.1242/jcs.062414. [DOI] [PubMed] [Google Scholar]
- 24.Honnappa S., Okhrimenko O., Jaussi R., Jawhari H., Jelesarov I., Winkler F.K., et al. Key interaction modes of dynamic +TIP networks. Mol. Cell. 2006;23:663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 26.Kumar P., Wittmann T. +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 2012;22:418–428. doi: 10.1016/j.tcb.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bieling P., Kandels-Lewis S., Telley I.A., Van Dijk J., Janke C., Surrey T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulinbinding sites. J. Cell Biol. 2008;183:1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixit R., Barnett B., Lazarus J.E., Tokito M., Goldman Y.E., Holzbaur E.L.F. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc. Natl. Acad. Sci. U. S. A. 2009;106:492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galjart N. CLIPs and CLASPs and cellular dynamics. Nat. Rev. Mol. Cell Biol. 2005;6:487–498. doi: 10.1038/nrm1664. [DOI] [PubMed] [Google Scholar]
- 30.Bieling P., Laan L., Schek H., Munteanu E.L., Sandblad L., Dogterom M., et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 31.Komarova Y., Groot C.O.D., Grigoriev I., Gouveia S.M., Munteanu E.L., Schober J.M., et al. Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 2009;184:691–706. doi: 10.1083/jcb.200807179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitre B., Coquelle F.M., Heichette C., Garnier C., Chrétien D., Arnal I. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 2008;10:415–421. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- 33.Cooper J.R., Wagenbach M., Asbury C.L., Wordeman L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat. Struct. Mol. Biol. 2010;17:77–83. doi: 10.1038/nsmb.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desai A., Verma S., Mitchison T.J., Walczak C.E. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 35.Gardner M.K., Zanic M., Gell C., Bormuth V., Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 36.Helenius J., Brouhard G., Kalaidzidis Y., Diez S., Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 37.Hunter A.W., Caplow M., Coy D.L., Hancock W.O., Diez S., Wordeman L., et al. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moores C.A., Milligan R.A. Lucky 13 - microtubule depolymerisation by kinesin-13 motors. J. Cell Sci. 2006;119:3905–3913. doi: 10.1242/jcs.03224. [DOI] [PubMed] [Google Scholar]
- 39.Walczak C.E., Mitchison T.J., Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- 40.Wordeman L. Microtubule-depolymerizing kinesins. Curr. Opin. Cell Biol. 2005;17:82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Montenegro Gouveia S., Leslie K., Kapitein L.C., Buey R.M., Grigoriev I., Wagenbach M., et al. In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr. Biol. 2010;20:1717–1722. doi: 10.1016/j.cub.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 42.Aher A., Kok M., Sharma A., Rai A., Olieric N., Rodriguez-Garcia R., et al. CLASP suppresses microtubule catastrophes through a single TOG domain. Dev. Cell. 2018;46:40–58.e8. doi: 10.1016/j.devcel.2018.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawrence E.J., Arpag G., Norris S.R., Zanic M. Human CLASP2 specifically regulates microtubule catastrophe and rescue. Mol. Biol. Cell. 2018;29:1168–1177. doi: 10.1091/mbc.E18-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akhmanova A., Hoogenraad C.C., Drabek K., Stepanova T., Dortland B., Verkerk T., et al. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 46.Lawrence E.J., Zanic M., Rice L.M. CLASPs at a glance. J. Cell Sci. 2020;133:jcs243097. doi: 10.1242/jcs.243097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Al-Bassam J., Kim H., Brouhard G., van Oijen A., Harrison S.C., Chang F. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence E.J., Chatterjee S., Zanic M. CLASPs stabilize the pre-catastrophe intermediate state between microtubule growth and shrinkage. J. Cell Biol. 2023;222 doi: 10.1083/jcb.202107027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence E.J., Zanic M. Rescuing microtubules from the brink of catastrophe: CLASPs lead the way. Curr. Opin. Cell Biol. 2019;56:94–101. doi: 10.1016/j.ceb.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Majumdar S., Kim T., Chen Z., Munyoki S., Tso S.C., Brautigam C.A., et al. An isolated CLASP TOG domain suppresses microtubule catastrophe and promotes rescue. Mol. Biol. Cell. 2018;29:1359–1375. doi: 10.1091/mbc.E17-12-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slep K.C. A cytoskeletal symphony: owed to TOG. Dev. Cell. 2018;46:5–7. doi: 10.1016/j.devcel.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 52.Ayaz P., Ye X., Huddleston P., Brautigam C.A., Rice L.M. A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ayaz P., Munyoki S., Geyer E.A., Piedra F.-A., Vu E.S., Bromberg R., et al. A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. Elife. 2014;3 doi: 10.7554/eLife.03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farmer V., Arpağ G., Hall S.L., Zanic M. XMAP215 promotes microtubule catastrophe by disrupting the growing microtubule end. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202012144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gard D.L., Kirschner M.W. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasquez R.J., Gard D.L., Cassimeris L. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol. 1994;127:985–993. doi: 10.1083/jcb.127.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zanic M., Widlund P.O., Hyman A.A., Howard J. Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat. Cell Biol. 2013;15:688–693. doi: 10.1038/ncb2744. [DOI] [PubMed] [Google Scholar]
- 58.Li W., Moriwaki T., Tani T., Watanabe T., Kaibuchi K., Goshima G. Reconstitution of dynamic microtubules with Drosophila XMAP215, EB1, and Sentin. J. Cell Biol. 2012;199:849–862. doi: 10.1083/jcb.201206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brittle A.L., Ohkura H. Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 2005;24:1387–1396. doi: 10.1038/sj.emboj.7600629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sousa A., Reis R., Sampaio P., Sunkel C.E. The Drosophila CLASP homologue, Mast/Orbit regulates the dynamic behaviour of interphase microtubules by promoting the pause state. Cell Motil. Cytoskeleton. 2007;64:605–620. doi: 10.1002/cm.20208. [DOI] [PubMed] [Google Scholar]
- 61.Trogden K.P., Rogers S.L. TOG proteins are spatially regulated by Rac-GSK3beta to control interphase microtubule dynamics. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moriwaki T., Goshima G. Five factors can reconstitute all three phases of microtubule polymerization dynamics. J. Cell Biol. 2016;215:357–368. doi: 10.1083/jcb.201604118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Akhmanova A., Hoogenraad C.C. Microtubule minus-end-targeting proteins. Curr. Biol. 2015;25:PR162–R171. doi: 10.1016/j.cub.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 64.Akhmanova A., Steinmetz M.O. Microtubule minus-end regulation at a glance. J. Cell Sci. 2019;132:jcs227850. doi: 10.1242/jcs.227850. [DOI] [PubMed] [Google Scholar]
- 65.Martin M., Akhmanova A. Coming into focus: mechanisms of microtubule minus-end organization. Trends Cell Biol. 2018;28:574–588. doi: 10.1016/j.tcb.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 66.Atherton J., Jiang K., Stangier M.M., Luo Y., Hua S., Houben K., et al. A structural model for microtubule minus-end recognition and protection by CAMSAP proteins. Nat. Struct. Mol. Biol. 2017;24:931–943. doi: 10.1038/nsmb.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodwin S.S., Vale R.D. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143:263–274. doi: 10.1016/j.cell.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hendershott M.C., Vale R.D. Regulation of microtubule minus-end dynamics by CAMSAPs and Patronin. Proc. Natl. Acad. Sci. U. S. A. 2014;111:5860–5865. doi: 10.1073/pnas.1404133111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang K., Hua S., Mohan R., Grigoriev I., Yau K.W., Liu Q., et al. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell. 2014;28:295–309. doi: 10.1016/j.devcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 70.Tanaka N., Meng W., Nagae S., Takeichi M. Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20029–20034. doi: 10.1073/pnas.1218017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toya M., Kobayashi S., Kawasaki M., Shioi G., Kaneko M., Ishiuchi T., et al. CAMSAP3 orients the apical-to-basal polarity of microtubule arrays in epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 2016;113:332–337. doi: 10.1073/pnas.1520638113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yau K.W., van Beuningen S.F., Cunha-Ferreira I., Cloin B.M., van Battum E.Y., Will L., et al. Microtubule minus-end binding protein CAMSAP2 controls axon specification and dendrite development. Neuron. 2014;82:1058–1073. doi: 10.1016/j.neuron.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 73.McNally F.J., Roll-Mecak A. Microtubule-severing enzymes: from cellular functions to molecular mechanism. J. Cell Biol. 2018;217:4057–4069. doi: 10.1083/jcb.201612104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McNally F.J., Vale R.D. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 75.Jiang K., Faltova L., Hua S., Capitani G., Prota A.E., Landgraf C., et al. Structural basis of formation of the microtubule minus-end-regulating CAMSAP-katanin complex. Structure. 2018;26:375–382.e4. doi: 10.1016/j.str.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 76.Fan Y., Burkart G.M., Dixit R. The arabidopsis SPIRAL2 protein targets and stabilizes microtubule minus ends. Curr. Biol. 2018;28:987–994.e3. doi: 10.1016/j.cub.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wightman R., Chomicki G., Kumar M., Carr P., Turner S.R. SPIRAL2 determines plant microtubule organization by modulating microtubule severing. Curr. Biol. 2013;23:1902–1907. doi: 10.1016/j.cub.2013.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Higgins J., Midgley C., Bergh A.-M., Bell S.M., Askham J.M., Roberts E., et al. Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC Cell Biol. 2010;11:85. doi: 10.1186/1471-2121-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang K., Rezabkova L., Hua S., Liu Q., Capitani G., Altelaar A.F.M., et al. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat. Cell Biol. 2017;19:480–492. doi: 10.1038/ncb3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tungadi E.A., Ito A., Kiyomitsu T., Goshima G. Human microcephaly ASPM protein is a spindle pole-focusing factor that functions redundantly with CDK5RAP2. J. Cell Sci. 2017;130:3676–3684. doi: 10.1242/jcs.203703. [DOI] [PubMed] [Google Scholar]
- 81.van der Voet M., Berends C.W.H., Perreault A., Nguyen-Ngoc T., Gönczy P., Vidal M., et al. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat. Cell Biol. 2009;11:269–277. doi: 10.1038/ncb1834. [DOI] [PubMed] [Google Scholar]
- 82.Wakefield J.G., Bonaccorsi S., Gatti M. The Drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J. Cell Biol. 2001;153:637–648. doi: 10.1083/jcb.153.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McDonald H.B., Stewart R.J., Goldstein L.S. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell. 1990;63:1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- 84.Walker R.A., Salmon E.D., Endow S.A. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature. 1990;347:780–782. doi: 10.1038/347780a0. [DOI] [PubMed] [Google Scholar]
- 85.Norris S.R., Jung S., Singh P., Strothman C.E., Erwin A.L., Ohi M.D., et al. Microtubule minus-end aster organization is driven by processive HSET-tubulin clusters. Nat. Commun. 2018;9:2659. doi: 10.1038/s41467-018-04991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mountain V., Simerly C., Howard L., Ando A., Schatten G., Compton D.A. The kinesin-related protein, Hset, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Braun M., Drummond D.R., Cross R.A., McAinsh A.D. The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat. Cell Biol. 2009;11:724–730. doi: 10.1038/ncb1878. [DOI] [PubMed] [Google Scholar]
- 88.Walczak C.E., Verma S., Mitchison T.J. XCTK2: a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J. Cell Biol. 1997;136:859–870. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fink G., Hajdo L., Skowronek K.J., Reuther C., Kasprzak A.A., Diez S. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat. Cell Biol. 2009;11:717–723. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- 90.Wordeman L. How kinesin motor proteins drive mitotic spindle function: lessons from molecular assays. Semin. Cell Dev. Biol. 2010;21:260–268. doi: 10.1016/j.semcdb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.She Z.-Y., Yang W.-X. Molecular mechanisms of kinesin-14 motors in spindle assembly and chromosome segregation. J. Cell Sci. 2017;130:2097–2110. doi: 10.1242/jcs.200261. [DOI] [PubMed] [Google Scholar]
- 92.Strothman C., Farmer V., Arpağ G., Rodgers N., Podolski M., Norris S., et al. Microtubule minus-end stability is dictated by the tubulin off-rate. J. Cell Biol. 2019;218:2841–2853. doi: 10.1083/jcb.201905019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chomicki G., Wightman R., Turner S.R. A specific class of short treadmilling microtubules enhances cortical microtubule alignment. Mol. Plant. 2016;9:1214–1216. doi: 10.1016/j.molp.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 94.Keating T.J., Peloquin J.G., Rodionov V.I., Momcilovic D., Borisy G.G. Microtubule release from the centrosome. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5078–5083. doi: 10.1073/pnas.94.10.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Margolis R.L., Wilson L. Microtubule treadmilling: what goes around comes around. BioEssays. 1998;20:830–836. doi: 10.1002/(SICI)1521-1878(199810)20:10<830::AID-BIES8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 96.Rodionov V., Nadezhdina E., Borisy G. Centrosomal control of microtubule dynamics. Proc. Natl. Acad. Sci. U. S. A. 1999;96:115–120. doi: 10.1073/pnas.96.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodionov V.I., Borisy G.G. Microtubule treadmilling in vivo. Science. 1997;275:215–218. doi: 10.1126/science.275.5297.215. [DOI] [PubMed] [Google Scholar]
- 98.Shaw S.L., Kamyar R., Ehrhardt D.W. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300:1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- 99.Vorobjev I.A., Rodionov V.I., Maly I.V., Borisy G.G. Contribution of plus and minus end pathways to microtubule turnover. J. Cell Sci. 1999;112:2277–2289. doi: 10.1242/jcs.112.14.2277. [DOI] [PubMed] [Google Scholar]
- 100.Waterman-Storer C.M., Salmon E.D. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J. Cell Biol. 1997;139:417–434. doi: 10.1083/jcb.139.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Waterman-Storer C.M., Salmon E.D. Microtubule dynamics:Treadmilling comes around again. Curr. Biol. 1997;7:R369–R372. doi: 10.1016/s0960-9822(06)00177-1. [DOI] [PubMed] [Google Scholar]
- 102.Wittmann T., Bokoch G.M., Waterman-Storer C.M. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J. Cell Biol. 2003;161:845–851. doi: 10.1083/jcb.200303082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cassimeris L. Cell division: eg’ing on microtubule flux. Curr. Biol. 2004;14:R1000–1002. doi: 10.1016/j.cub.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 104.Arpaǧ G., Lawrence E.J., Farmer V.J., Hall S.L., Zanic M. Collective effects of XMAP215, EB1, CLASP2, and MCAK lead to robust microtubule treadmilling. Proc. Natl. Acad. Sci. U. S. A. 2020;117:12847–12855. doi: 10.1073/pnas.2003191117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wiegand T., Hyman A.A. Drops and fibers - how biomolecular condensates and cytoskeletal filaments influence each other. Emerging Top. Life Sci. 2020;4:247–261. doi: 10.1042/ETLS20190174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baumgart J., Kirchner M., Redemann S., Bond A., Woodruff J., Verbavatz J.-M., et al. Soluble tubulin is significantly enriched at mitotic centrosomes. J. Cell Biol. 2019 doi: 10.1083/jcb.201902069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Woodruff J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., Hyman A.A. The centrosome is a selective condensate that Nucleates microtubules by concentrating tubulin. Cell. 2017;169:1066–1077.e10. doi: 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 108.Jiang H., Wang S., Huang Y., He X., Cui H., Zhu X., et al. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell. 2015;163:108–122. doi: 10.1016/j.cell.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.King M.R., Petry S. Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat. Commun. 2020;11:270. doi: 10.1038/s41467-019-14087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Setru S.U., Gouveia B., Alfaro-Aco R., Shaevitz J.W., Stone H.A., Petry S. A hydrodynamic instability drives protein droplet formation on microtubules to nucleate branches. Nat. Phys. 2021;17:493–498. doi: 10.1038/s41567-020-01141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miesch J., Wimbish R.T., Velluz M.-C., Aumeier C. Phase separation of +TIP-networks regulates microtubule dynamics. bioRxiv. 2022 doi: 10.1101/2021.09.13.459419. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu Y.-F.O., Bryant A.T., Nelson N.T., Madey A.G., Fernandes G.F., Goodson H.V. Overexpression of the microtubule-binding protein CLIP-170 induces a +TIP network superstructure consistent with a biomolecular condensate. PLoS One. 2021;16 doi: 10.1371/journal.pone.0260401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jijumon A.S., Bodakuntla S., Genova M., Bangera M., Sackett V., Besse L., et al. Lysate-based pipeline to characterize microtubule-associated proteins uncovers unique microtubule behaviours. Nat. Cell Biol. 2022;24:253–267. doi: 10.1038/s41556-021-00825-4. [DOI] [PubMed] [Google Scholar]
- 114.Maan R., Reese L., Volkov V.A., King M.R., van der Sluis E.O., Andrea N., et al. Multivalent interactions facilitate motor-dependent protein accumulation at growing microtubule plus-ends. Nat. Cell Biol. 2023;25:68–78. doi: 10.1038/s41556-022-01037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Song X., Yang F., Yang T., Wang Y., Ding M., Li L., et al. Phase separation of EB1 guides microtubule plus-end dynamics. Nat. Cell Biol. 2023;25:79–91. doi: 10.1038/s41556-022-01033-4. [DOI] [PubMed] [Google Scholar]
- 116.Xia P., Liu X., Wu B., Zhang S., Song X., Yao P.Y., et al. Superresolution imaging reveals structural features of EB1 in microtubule plus-end tracking. Mol. Biol. Cell. 2014;25:4166–4173. doi: 10.1091/mbc.E14-06-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dragestein K.A., van Cappellen W.A., van Haren J., Tsibidis G.D., Akhmanova A., Knoch T.A., et al. Dynamic behavior of GFP–CLIP-170 reveals fast protein turnover on microtubule plus ends. J. Cell Biol. 2008;180:729–737. doi: 10.1083/jcb.200707203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Odde D.J. Estimation of the diffusion-limited rate of microtubule assembly. Biophys. J. 1997;73:88–96. doi: 10.1016/S0006-3495(97)78050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trivedi P., Palomba F., Niedzialkowska E., Digman M.A., Gratton E., Stukenberg P.T. The inner centromere is a biomolecular condensate scaffolded by the chromosomal passenger complex. Nat. Cell Biol. 2019;21:1127–1137. doi: 10.1038/s41556-019-0376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meier S.M., Farcas A.-M., Kumar A., Ijavi M., Bill R.T., Stelling J., et al. Multivalency ensures persistence of a +TIP body at specialized microtubule ends. Nat. Cell Biol. 2023;25:56–67. doi: 10.1038/s41556-022-01035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee V.M., Goedert M., Trojanowski J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 122.Gao Y.L., Wang N., Sun F.R., Cao X.P., Zhang W., Yu J.T. Tau in neurodegenerative disease. Ann. Transl. Med. 2018;6:175. doi: 10.21037/atm.2018.04.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Iqbal K., Liu F., Gong C.X. Tau and neurodegenerative disease: the story so far. Nat. Rev. Neurol. 2016;12:15–27. doi: 10.1038/nrneurol.2015.225. [DOI] [PubMed] [Google Scholar]
- 124.Morris M., Maeda S., Vossel K., Mucke L. The many faces of tau. Neuron. 2011;70:410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Drechsel D.N., Hyman A.A., Cobb M.H., Kirschner M.W. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol. Biol. Cell. 1992;3:1141–1154. doi: 10.1091/mbc.3.10.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B.T., et al. Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 2017;20:2304–2312. doi: 10.1016/j.celrep.2017.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Siahaan V., Krattenmacher J., Hyman A.A., Diez S., Hernández-Vega A., Lansky Z., et al. Kinetically distinct phases of tau on microtubules regulate kinesin motors and severing enzymes. Nat. Cell Biol. 2019;21:1086–1092. doi: 10.1038/s41556-019-0374-6. [DOI] [PubMed] [Google Scholar]
- 128.Tan R., Lam A.J., Tan T., Han J., Nowakowski D.W., Vershinin M., et al. Microtubules gate tau condensation to spatially regulate microtubule functions. Nat. Cell Biol. 2019;21:1078–1085. doi: 10.1038/s41556-019-0375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chaudhary A.R., Lu H., Krementsova E.B., Bookwalter C.S., Trybus K.M., Hendricks A.G. MAP7 regulates organelle transport by recruiting kinesin-1 to microtubules. J. Biol. Chem. 2019;294:10160–10171. doi: 10.1074/jbc.RA119.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dixit R., Ross J.L., Goldman Y.E., Holzbaur E.L. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319:1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McVicker D.P., Chrin L.R., Berger C.L. The nucleotide-binding state of microtubules modulates kinesin processivity and the ability of tau to inhibit kinesin-mediated transport. J. Biol. Chem. 2011;286:42873–42880. doi: 10.1074/jbc.M111.292987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Monroy B.Y., Sawyer D.L., Ackermann B.E., Borden M.M., Tan T.C., Ori-Mckenney K.M. Competition between microtubule-associated proteins directs motor transport. Nat. Commun. 2018 doi: 10.1038/s41467-018-03909-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Monroy B.Y., Tan T.C., Oclaman J.M., Han J.S., Simó S., Niwa S., et al. A combinatorial MAP code dictates polarized microtubule transport. Dev. Cell. 2020;53:60–72.e4. doi: 10.1016/j.devcel.2020.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vershinin M., Carter B.C., Razafsky D.S., King S.J., Gross S.P. Multiple-motor based transport and its regulation by Tau. Proc. Natl. Acad. Sci. U. S. A. 2007;104:87–92. doi: 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Thery M., Blanchoin L. Microtubule self-repair. Curr. Opin. Cell Biol. 2020;68:144–154. doi: 10.1016/j.ceb.2020.10.012. [DOI] [PubMed] [Google Scholar]
- 136.Peet D.R., Burroughs N.J., Cross R.A. Kinesin expands and stabilizes the GDP-microtubule lattice. Nat. Nanotechnol. 2018;13:386–391. doi: 10.1038/s41565-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Verhey K.J., Ohi R. Causes, costs and consequences of kinesin motors communicating through the microtubule lattice. J. Cell Sci. 2023;136:jcs260735. doi: 10.1242/jcs.260735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Andreu-Carbó M., Fernandes S., Velluz M.-C., Kruse K., Aumeier C. Motor usage imprints microtubule stability along the shaft. Dev. Cell. 2022;57:5–18.e8. doi: 10.1016/j.devcel.2021.11.019. [DOI] [PubMed] [Google Scholar]
- 139.Budaitis B.G., Badieyan S., Yue Y., Blasius T.L., Reinemann D.N., Lang M.J., et al. A kinesin-1 variant reveals motor-induced microtubule damage in cells. Curr. Biol. 2022;32:2416–2429.e6. doi: 10.1016/j.cub.2022.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Triclin S., Inoue D., Gaillard J., Htet Z.M., DeSantis M.E., Portran D., et al. Self-repair protects microtubules from destruction by molecular motors. Nat. Mater. 2021;20:883–891. doi: 10.1038/s41563-020-00905-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kuo Y.W., Mahamdeh M., Tuna Y., Howard J. The force required to remove tubulin from the microtubule lattice by pulling on its α-tubulin C-terminal tail. Nat. Commun. 2022;13:3651. doi: 10.1038/s41467-022-31069-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Howard J. Sinauer Associates, Oxford University Press; Sunderland, MA: 2001. Mechanics of Motor Proteins and the Cytoskeleton. [Google Scholar]
- 143.Lawrence E.J., Arpag G., Arnaiz C., Zanic M. SSNA1 stabilizes dynamic microtubules and detects microtubule damage. Elife. 2021;10 doi: 10.7554/eLife.67282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van den Berg C.M., Volkov V.A., Schnorrenberg S., Huang Z., Stecker K.E., Grigoriev I., et al. CSPP1 stabilizes growing microtubule ends and damaged lattices from the luminal side. J. Cell Biol. 2023;222 doi: 10.1083/jcb.202208062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Basnet N., Nedozralova H., Crevenna A.H., Bodakuntla S., Schlichthaerle T., Taschner M., et al. Direct induction of microtubule branching by microtubule nucleation factor SSNA1. Nat. Cell Biol. 2018;20:1172–1180. doi: 10.1038/s41556-018-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Goyal U., Renvoise B., Chang J., Blackstone C. Spastin-interacting protein NA14/SSNA1 functions in cytokinesis and axon development. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lai C.K., Gupta N., Wen X., Rangell L., Chih B., Peterson A.S., et al. Functional characterization of putative cilia genes by high-content analysis. Mol. Biol. Cell. 2011;22:1104–1119. doi: 10.1091/mbc.E10-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pfannenschmid F., Wimmer V.C., Rios R.-M., Geimer S., Krockel U., Leiherer A., et al. Chlamydomonas DIP13 and human NA14: a new class of proteins associated with microtubule structures is involved in cell division. J. Cell Sci. 2003;116:1449–1462. doi: 10.1242/jcs.00337. [DOI] [PubMed] [Google Scholar]
- 149.Schoppmeier J., Mages W., Lechtreck K.F. GFP as a tool for the analysis of proteins in the flagellar basal apparatus of Chlamydomonas. Cell Motil. Cytoskeleton. 2005;61:189–200. doi: 10.1002/cm.20074. [DOI] [PubMed] [Google Scholar]
- 150.Evans K.J., Gomes E.R., Reisenweber S.M., Gundersen G.G., Lauring B.P. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol. 2005;168:599–606. doi: 10.1083/jcb.200409058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roll-Mecak A., Vale R.D. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 2005;15:650–655. doi: 10.1016/j.cub.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 152.Roll-Mecak A., Vale R.D. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451:363–367. doi: 10.1038/nature06482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rodriguez-Rodriguez M., Trevino M.A., Laurents D.V., Arranz R., Valpuesta J.M., Rico M., et al. Characterization of the structure and self-recognition of the human centrosomal protein NA14: implications for stability and function. Protein Eng. Des. Sel. 2011;24:883–892. doi: 10.1093/protein/gzr050. [DOI] [PubMed] [Google Scholar]
- 154.Aher A., Rai D., Schaedel L., Gaillard J., John K., Liu Q., et al. CLASP mediates microtubule repair by restricting lattice damage and regulating tubulin incorporation. Curr. Biol. 2020;30:2175–2183.e6. doi: 10.1016/j.cub.2020.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.de Forges H., Pilon A., Cantaloube I., Pallandre A., Haghiri-Gosnet A.M., Perez F., et al. Localized mechanical stress promotes microtubule rescue. Curr. Biol. 2016;26:3399–3406. doi: 10.1016/j.cub.2016.10.048. [DOI] [PubMed] [Google Scholar]
- 156.Vemu A., Szczesna E., Zehr E.A., Spector J.O., Grigorieff N., Deaconescu A.M., et al. Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science. 2018;361 doi: 10.1126/science.aau1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Slep K.C. Cytoskeletal repair: microtubule orthopaedics to the rescue. Curr. Biol. 2020;30:R646–R649. doi: 10.1016/j.cub.2020.06.075. [DOI] [PubMed] [Google Scholar]
- 158.Tropini C., Roth E.A., Zanic M., Gardner M.K., Howard J. Islands containing slowly hydrolyzable GTP analogs promote microtubule rescues. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Aumeier C., Schaedel L., Gaillard J., John K., Blanchoin L., Thery M. Self-repair promotes microtubule rescue. Nat. Cell Biol. 2016;18:1054–1064. doi: 10.1038/ncb3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rai A., Liu T., Glauser S., Katrukha E.A., Estevez-Gallego J., Rodriguez-Garcia R., et al. Taxanes convert regions of perturbed microtubule growth into rescue sites. Nat. Mater. 2020;19:355–365. doi: 10.1038/s41563-019-0546-6. [DOI] [PubMed] [Google Scholar]
- 161.Kuo Y.W., Howard J. Cutting, amplifying, and aligning microtubules with severing enzymes. Trends Cell Biol. 2021;31:50–61. doi: 10.1016/j.tcb.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kuo Y.W., Trottier O., Mahamdeh M., Howard J. Spastin is a dual-function enzyme that severs microtubules and promotes their regrowth to increase the number and mass of microtubules. Proc. Natl. Acad. Sci. U. S. A. 2019;116:5533–5541. doi: 10.1073/pnas.1818824116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lindeboom J.J., Nakamura M., Hibbel A., Shundyak K., Gutierrez R., Ketelaar T., et al. A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science. 2013;342 doi: 10.1126/science.1245533. [DOI] [PubMed] [Google Scholar]
- 164.Lindeboom J.J., Nakamura M., Saltini M., Hibbel A., Walia A., Ketelaar T., et al. CLASP stabilization of plus ends created by severing promotes microtubule creation and reorientation. J. Cell Biol. 2019;218:190–205. doi: 10.1083/jcb.201805047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nakamura M., Lindeboom J.J., Saltini M., Mulder B.M., Ehrhardt D.W. SPR2 protects minus ends to promote severing and reorientation of plant cortical microtubule arrays. J. Cell Biol. 2018;217:915–927. doi: 10.1083/jcb.201708130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Grimaldi A.D., Maki T., Fitton B.P., Roth D., Yampolsky D., Davidson M.W., et al. CLASPs are required for proper microtubule localization of end-binding proteins. Dev. Cell. 2014;30:343–352. doi: 10.1016/j.devcel.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Grimaldi A.D., Zanic M., Kaverina I. Encoding the microtubule structure: allosteric interactions between the microtubule +TIP complex master regulators and TOG-domain proteins. Cell Cycle. 2015;14:1375–1378. doi: 10.1080/15384101.2015.1026521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim T., Rice L.M. Long-range, through-lattice coupling improves predictions of microtubule catastrophe. Mol. Biol. Cell. 2019;30:1451–1462. doi: 10.1091/mbc.E18-10-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chen J., Kholina E., Szyk A., Fedorov V.A., Kovalenko I., Gudimchuk N., et al. Alpha-tubulin tail modifications regulate microtubule stability through selective effector recruitment, not changes in intrinsic polymer dynamics. Dev. Cell. 2021;56:2016–2028.e4. doi: 10.1016/j.devcel.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gadadhar S., Bodakuntla S., Natarajan K., Janke C. The tubulin code at a glance. J. Cell Sci. 2017;130:1347. doi: 10.1242/jcs.199471. [DOI] [PubMed] [Google Scholar]
- 171.Janke C., Chloë Bulinski J. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 172.Janke C., Magiera M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 2020;21:307–326. doi: 10.1038/s41580-020-0214-3. [DOI] [PubMed] [Google Scholar]
- 173.Sébastien M., Prowse E.N.P., Hendricks A.G., Brouhard G.J. Doublecortin regulates neuronal migration by editing the tubulin code. bioRxiv. 2023 doi: 10.1101/2023.06.02.543327. [preprint] [DOI] [Google Scholar]
- 174.Roll-Mecak A. The tubulin code in microtubule dynamics and information encoding. Dev. Cell. 2020;54:7–20. doi: 10.1016/j.devcel.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Verhey K.J., Gaertig J. The tubulin code. Cell Cycle. 2007;6:2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]