Abstract
The mechanism of template selection for genome replication in plus-strand RNA viruses is poorly understood. Using the prototypical tombusvirus, Tomato bushy stunt virus (TBSV), we show that recombinant p33 replicase protein binds specifically to an internal replication element (IRE) located within the p92 RNA-dependent RNA polymerase coding region of the viral genome. Specific binding of p33 to the IRE in vitro depends on the presence of a C · C mismatch within a conserved RNA helix. Interestingly, the absence of the p33:p33/p92 interaction domain in p33 prevented specific but allowed nonspecific RNA binding, suggesting that a multimeric form of this protein is involved in the IRE-specific interaction. Further support for the selectivity of p33 binding in vitro was provided by the inability of the replicase proteins of the closely related Turnip crinkle virus and distantly related Hepatitis C virus to specifically recognize the TBSV IRE. Importantly, there was also a strong correlation between p33:IRE complex formation in vitro and viral replication in vivo, where mutations in the IRE that disrupted selective p33 binding in vitro also abolished TBSV RNA replication both in plant and in Saccharomyces cerevisiae cells. Based on these findings and the other known properties of p33 and the IRE, it is proposed that the p33:IRE interaction provides a mechanism to selectively recruit viral RNAs into cognate viral replicase complexes. Since all genera in Tombusviridae encode comparable replicase proteins, these results may be relevant to other members of this large virus family.
Plus-strand RNA viruses replicate their genomes in infected cells by using a replicase complex comprised of viral and host proteins that assembles in association with cellular membranes (1, 4, 15). In infected cells, viral replicases are able to specifically recognize and selectively replicate their cognate viral RNAs from a heterogeneous pool of cellular RNA molecules. In contrast, in vitro studies have shown that many purified viral replicase complexes are able to utilize heterologous promoter or initiation elements quite efficiently (11, 28, 39). These conflicting findings between in vivo and in vitro data have led to models that attribute selective recognition of viral templates to host proteins (4, 13). Phage Qbeta utilizes this type of mechanism whereby the S1 ribosomal protein and elongation factor Tu in the four-subunit replicase complex mediate viral template recognition (3, 13).
There is also evidence that virally encoded proteins can facilitate selective template recruitment to the viral replicase complex. Examples in this category include the 1a protein of Brome mosaic virus and the 126-kDa protein of Tomato mosaic virus (5, 17, 33). However, in both of these cases it is not known whether the respective viral RNAs are recognized directly by these replicase proteins or require assistance from host proteins (6). In Poliovirus, specific binding of 3CD protein to the 5′-terminal cloverleaf-like structure has been reported, and this interaction likely contributes to template selection into replication (9, 38). However, the host poly(C) binding protein 2 also interacts specifically with the same 5′-terminal RNA structure, and it, too, is proposed to be involved in mediating template selection (9, 38). In general, the contribution of viral and cellular proteins to RNA template recognition by cognate replicases is largely unknown in most viral systems.
Tomato bushy stunt virus (TBSV) is the prototypical member of the genus Tombusvirus in the large family Tombusviridae. Its genome encodes two proteins involved in viral RNA replication, the prereadthrough product p33 and the readthrough product p92 (Fig. 1A and B). Both of these proteins are essential for RNA replication (18, 22), they are part of the viral replicase complex (23), and they accumulate in vivo at a ratio of 20:1, respectively (23, 31). The less plentiful p92 functions as the viral RNA-dependent RNA polymerase (RdRp), while the role of the more abundant prereadthrough p33 is undefined (37). In other tombusviruses, the orthologues of TBSV p33 have been shown to be targeted to mitochondrial or peroxisomal membranes, the presumed sites of tombusvirus RNA replication (29). In TBSV, both p33 and p92 are membrane associated (23, 31). RNA-binding domains have also been identified in these proteins (22, 26), and the ability of p33 to interact with itself and p92 has been demonstrated (27). The cumulative data support an essential role for both TBSV p33 and p92 in viral RNA replication, with p92 comprising the catalytic subunit responsible for RNA synthesis and p33 playing a critical but unknown auxiliary role.
FIG. 1.
Specific binding of the recombinant p33 replicase protein to RII(+)-SL in vitro. (A) Schematic representation of the p33 and p92 replicase proteins of TBSV. The sequence of p33 is identical to the N-terminal overlapping (prereadthrough) domain of p92, which also contains the RdRp motifs in its C-terminal domain. p33C representing the C-terminal domain of p33 is shown at the top. p33C includes two functional domains, namely the RNA-binding (RPR motif) and the p33:p33/p92 interaction domains. (B) Schematic representation of TBSV gRNA and DI-72 RNA, a prototypical TBSV replicon with four noncontiguous regions derived from the 5′ end (RI), the p92 ORF (RII), and from the 3′ end (RIII and RIV) of TBSV gRNA. The position of RII(+)-SL is indicated. (C) Efficient binding of recombinant p33C to DI-72(+) RNA requires RII(+). EMSA was performed with 32P-labeled RNA, either the full-length DI-72 or ΔRII derivative, and a decreasing amount of p33C (twofold dilution series starting from 1 μg). Maltose-binding protein (MBP; 1 μg) was used as a negative control to show the migration of the RNA probe in the absence of binding. (D) Systematic analysis of each region of DI-72(+) RNA for binding to recombinant p33C. EMSA was performed with 32P-labeled RNA (50 ng), and 0, 8, and 1 μg of p33C (left, middle, and right lanes, respectively, for each RNA). The free (unbound) probes are indicated by arrowheads, whereas the bound probe with 1 μg of p33C is marked with an asterisk. Note that RII(+) bound to p33C efficiently and RI(+) bound with low efficiency when 1 μg of p33C was used, whereas RIII(+) and RIV(+) did not bind to p33C under this condition. (E) Inefficient binding of the heterologous satC(+) RNA to p33C. EMSA was performed as described for panel C. Note that fusion of TBSV RII(+) to satC(+) makes binding to p33C efficient, demonstrating that RII(+) is sufficient for interaction with p33C.
In this paper we tested the binding of a recombinant TBSV p33 to four conserved regions of the viral genome known to affect replication (37). We demonstrate that p33 binds selectively in vitro to a conserved RNA motif within the p92 coding region of the viral genome. The specific recognition of this RNA element is dependent on a C · C mismatch positioned within a helix, and this key determinant of p33 binding is also essential for TBSV replication in host cells. We propose that this interaction directs viral template recruitment into replication and suggest that this mechanism may also apply to other members of the large family Tombusviridae.
MATERIALS AND METHODS
Expression and purification of recombinant replicase proteins from Escherichia coli.
Expression plasmids for recombinant TBSV, cucumber necrosis virus (CNV), and turnip crinkle virus (TCV) replicase proteins were constructed previously (26-28). The expression construct for TCV p28C was generated by cloning a PCR product obtained with primers 1418 (GAGGAATTCTTGGTAGGAACGGAAGA) and 1419 (GCAGTCTAGACTAGCGGACAAAAGAGAT) by using the full-length TCV clone as a template at the EcoRI and XbaI sites in pMal-c2x (NEB). Expression and purification of the recombinant TBSV, CNV, and TCV replicase proteins were carried out as described earlier (26, 28). Briefly, individual expression plasmids were transformed into E. coli [Epicurion BL21-CodonPlus (DE3)-RIL; Stratagene]. Protein expression was induced at either 37 or 14°C with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 and 8 h, respectively. The recombinant proteins were purified by using an amylose resin column (NEB), as described earlier (26, 28). All protein purification steps were carried out in a cold room. The purified recombinant proteins were analyzed for their purity by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE). The Bio-Rad protein assay, which is based on the Bradford method, was used to measure the amount of purified recombinant proteins. Peptide RPR25 (NH2-TGRPRRRPYAAKIAQVARAKVGYLK-COOH) was synthesized and purified by high-pressure liquid chromatography by United Biochemical Research Inc. (Seattle, Wash.). The purified recombinant NS5B protein (12) was provided by G. Luo (University of Kentucky).
Preparation of RNA templates.
DNA templates for a T7 RNA polymerase reaction were prepared by PCR by using specific primers and DI-72SXP (32) as the PCR template. For the templates where mutation was introduced with the forward oligonucleotide, the first PCR was diluted 50 times and used as a template for a second PCR containing the forward oligonucleotide with the T7 promoter sequence and the reverse oligonucleotide used in the first PCR. The PCR products obtained were purified and used for T7 RNA polymerase-based transcription in the presence of 32P-labeled UTP and unlabeled ATP, CTP, and GTP as described (24). After phenol-chloroform extraction and isopropanol-ammonium acetate precipitation, the obtained RNA probes were checked by 5% PAGE gels containing 8 M urea.
Electrophoretic mobility shift assay (EMSA).
The affinity-purified recombinant proteins (1 μg as the highest amount and twofold dilution series) were incubated with 1 ng of radioactively labeled probe (see above) in a binding buffer (50 mM Tris-HCl [pH 8.2], 10 mM MgCl2, 10 mM dithiothreitol, 10% glycerol, 100 ng of yeast tRNA [Sigma], and 2 U of RNase inhibitor [Ambion])at 25°C for 15 min (26). After the binding reaction, the samples were analyzed by 4 or 5% nondenaturing PAGE in Tris-acetate-EDTA buffer at 200 V in a cold room (26). The gels were dried, exposed, and analyzed in a phosphorimager and quantified by using ImageQuant version 1.2 (Amersham).
Yeast transformation and growth.
Saccharomyces cerevisiae strain INVSc1 (Invitrogen) carrying three plasmids [i.e., pGAD-His92, pGBK-His33, and pYC-DI-72(+)Rz] (23) was grown in synthetic complete medium without uracil, leucine, and tryptophan and containing 2% galactose for 24 h at 30°C (19). Then the cultures were diluted 10-fold with fresh medium (synthetic complete medium without uracil, leucine, and tryptophan) containing 2% galactose and grown at 23°C to an optical density at 600 nm of 0.6 to 0.7 (approximately 24 h). Yeast cells were then harvested by centrifugation at 2,000 × g for 5 min, and the pellet was washed with 20 mM Tris-HCl, pH 8.0, and centrifuged. The pelleted cells were resuspended in 1 ml of 20 mM Tris-HCl (pH 8.0) buffer, followed by centrifugation at 21,000 × g for 1 min and storage of the pellet at −80°C until further use.
Characterization of replicase and p33:DI-72 RNA complexes from yeast.
Frozen yeast cells were homogenized by grinding in liquid nitrogen, followed by the addition of the extraction buffer (200 mM sorbitol, 50 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, yeast protease inhibitor mix; Sigma) and centrifugation at 100 × g for 1.5 min at 4°C. Preparation of the enriched membrane fraction and the in vitro replicase reactions were done as described (19). Because no template was added to the in vitro reaction, the replicase preparation could use only the endogenous template present within the enriched membrane fraction. The RdRp products were extracted by phenol-chloroform, precipitated with isopropanol-ammonium acetate, and analyzed under denaturing conditions (i.e., 5% PAGE gels containing 8 M urea) (24). To test the template activity of wild-type (wt) or ΔC99 DI-72(+) RNAs (a plus-strand construct with a C99 deletion), we performed RdRp assays using tombusvirus RdRp preparations obtained from plants as described (14).
To copurify p33:DI-72 RNA complex based on metal-affinity purification, we used the purification method developed by Panaviene et al. (23). Briefly, the above enriched membrane fraction was resuspended in the extraction buffer containing 1.2 M NaCl; this step was followed by gentle rotation for 20 min at 4°C and centrifugation at 21,000 × g for 15 min at 4°C. The obtained pellet was resuspended in the solubilization buffer (extraction buffer plus 1% Triton X-100 and 5% SB3-10 [caprylyl sulfobetaine; Sigma] and 0.5 M KCl) by gentle rotation for 1 h at 4°C and then centrifuged at 21,000 × g for 15 min at 4°C. After centrifugation, the supernatant was applied to a column containing ProBond resin (Invitrogen) equilibrated with the solubilization buffer. Then the column was rotated for 1 h at 4°C and then washed with two column volumes of the solubilization buffer; this step was followed by washing with the extraction buffer containing 1% Triton X-100 and 5% SB3-10 and then a second wash with the extraction buffer containing 1% Triton-100, 5% SB3-10, and 2 mM imidazole. The recombinant p33 was recovered from the column in the extraction buffer containing 200 mM imidazole and 0.1% Triton X-100 in two-step elution (each in a half-column volume). The obtained p33 preparation was used for Northern and Western blotting as described below.
Total RNA extraction from yeast cells and RNA blot analysis.
To extract total RNA from yeast, equal volumes of RNA extraction buffer (50 mM sodium acetate [pH 5.3], 10 mM EDTA, 1% SDS) and water-saturated phenol were added to the pelleted cells (20). Samples were vortexed, incubated for 4 min at 65°C and then for 2 min on ice, and centrifuged at 21,000 × g for 5 min at room temperature. Total RNA was precipitated with ethanol, resuspended in Tris-EDTA buffer and formamide (in a 1:1 ratio) and heated for 5 min at 85°C, electrophoresed in 1.5% agarose gels, and transferred to Hybond XL membrane (Amersham) (24). Prehybridization and hybridization were done by using ULTRAhyb solution (Ambion) at 68°C according to the supplier's instructions. The 32P-labeled DI-72(−) complementary probe was used for hybridization.
Western blotting.
Aliquots (10 μl) of the enriched membrane fraction from yeast cells or the purified p33 preparation in SDS-PAGE sample loading buffer (30) were heated for 5 min at 85°C, electrophoresed in SDS-8% PAGE gels, and electrotransferred to a polyvinylidene difluoride membrane (Bio-Rad). Membranes were treated with 5% nonfat dry milk solution in Tris-buffered saline buffer (30) also containing 0.1% Tween 20. Incubation with monoclonal anti-His antibodies (Amersham) was carried out for 1 h at room temperature, followed by three 10-min washes with Tris-buffered saline-Tween buffer and incubation for 1 h at room temperature with secondary alkaline phosphatase-conjugated anti-mouse antibody (Sigma). Western blots were developed by using BCIP (5-bromo-4-chloro-3-indolyphosphate) and nitroblue tetrazolium (Sigma).
Viral RNA analysis from plants.
Nicotiana benthamiana plants, inoculated with full-length TBSV RNA transcripts, were used for total RNA isolation from inoculated and systemically infected leaves as described previously (35). Aliquots of total nucleic acid preparations were separated in 1% agarose gels, and TBSV RNAs were detected by electrophoretic transfer to nylon membrane (Hybond-N; Amersham) followed by Northern blot analysis using a 32P-labeled RNA complementary to the 3′ noncoding region of TBSV as a probe. Each experiment was repeated three times.
RESULTS
Recombinant p33 replicase protein binds specifically to an internal replication element in TBSV RNA.
The TBSV replicase shows high template specificity in vivo since only its cognate RNAs are replicated efficiently. This observed selectivity of TBSV could be related to a specific interaction between the viral RNA and a viral protein and/or a host protein. In terms of viral factors, p33 represented a good candidate for the following reasons: (i) it is expressed early in infections from the viral genome (37); (ii) it accumulates to relatively high levels (18, 23, 31); (iii) it is essential for viral RNA replication (18, 22); and (iv) it contains an RNA-binding domain (26). To test if TBSV p33 could specifically recognize TBSV RNA, we performed gel mobility shift experiments by using a highly purified recombinant p33 replicase protein and a prototypical TBSV DI RNA, DI-72. TBSV DI RNAs are excellent model templates for studies on viral RNA replication because they contain all the cis-acting RNA signals, derived from various regions of the genome, that are necessary for amplification by the TBSV replicase (Fig. 1B) (37).
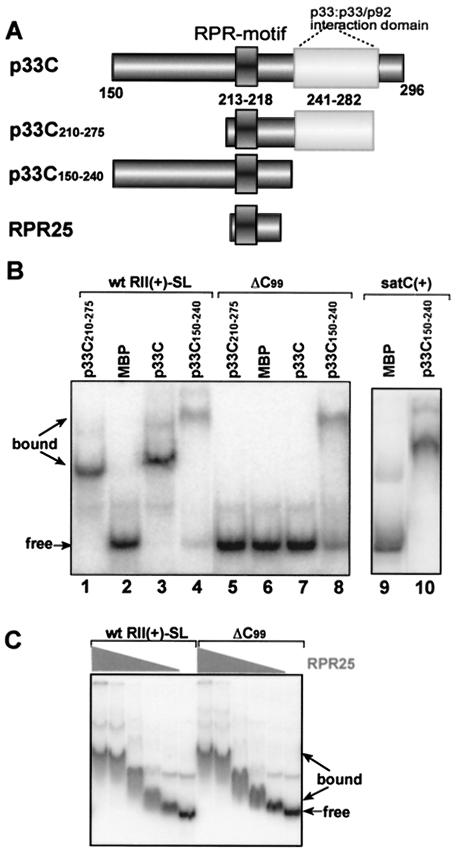
The p33 used in these assays was an N-terminally truncated version of the protein, termed p33C, that in comparison with full-length p33 exhibited improved RNA-binding activity and increased solubility due to the absence a hydrophobic N-terminal segment (Fig. 1A) (26). As shown in Fig. 1C, the recombinant p33C bound efficiently to the full-length DI-72(+) RNA but did not bind to DI-72 lacking region II (DI-72ΔRII). Also, systematic deletion of each of the four conserved regions (RI through RIV) in DI-72(+) RNA revealed that RII was the most important for binding (Fig. 1D). p33C also bound poorly to the heterologous satC(+) of TCV under similar conditions (Fig. 1E). However, fusing RII(+) to satC(+) RNA resulted in efficient binding of the chimeric satC/RII(+) RNA to p33C (Fig. 1E), demonstrating that RII(+) alone is sufficient to confer selective binding of p33 to a heterologous RNA.
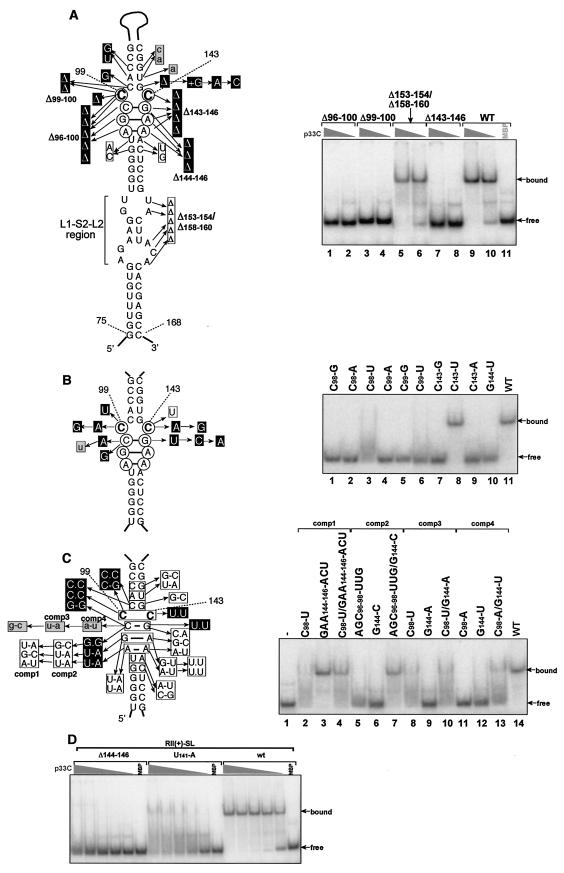
RII(+) corresponds to the readthrough portion of the p92 open reading frame (ORF) and contains an extended stem-loop (SL) structure [RII(+)-SL] (Fig. 1B) that is essential for efficient TBSV DI RNA and genome replication in vivo (accompanying paper [13a]). This RNA element is also known to contain two key functional determinants, a C · C mismatch in loop 3 (L3) and a conformationally flexible L1-S2-L2 element (Fig. 2A and reference 13a). Since these two RNA determinants are important for TBSV RNA replication in vivo (13a), they represented good candidates for p33 binding sites (Fig. 2A). To test this idea, we generated 32P-labeled fragments of RII(+)-SL containing various modifications and used them as ligands in gel mobility shift experiments with p33C. The wt RII(+)-SL bound efficiently to p33C (Fig. 2A, lanes 9 and 10), whereas mutants Δ96-100 (lanes 1 and 2), Δ99-100 (lanes 3 and 4), Δ143-146 (Fig. 2A, lanes 7 and 8,), and Δ144-146 (Fig. 2D) with short deletions within the L3 region did not bind to p33C. In contrast, an RII(+)-SL mutant that targeted the lower L1-S2-L2 region had no effect on p33 binding (Fig. 2A, mutant Δ153-154Δ158-160 in lanes 7 and 8). Therefore, of the two major RNA determinants identified to be important for efficient DI RNA replication in vivo (accompanying paper, reference 13a), only the conserved L3 region appears to be necessary for p33 binding in vitro.
FIG. 2.
A C · C mismatch within the RII(+)-SL is recognized by the recombinant p33. (A) The RII(+)-SL hairpin with the conserved 8-nucleotide-long L3 internal loop (drawn as circled nucleotides) is shown schematically, including the modified or deleted sequences. Nucleotide changes shown in black boxes eliminated in vitro binding to the recombinant p33C, whereas boxed modifications resulted in binding at the wt level. Mutations that affected binding to p33C greatly, but did not eliminate it, are shown in gray boxes with lowercase letters (see U141-A in panel D as an example). Multiple mutations present within the same RNA are boxed together. The nonessential 33-nucleotide-long top part of RII(+)-SL is shown only with a loop. A representative EMSA gel is shown on the right. EMSA was performed with a 32P-labeled RII(+)-SL derivative and a decreasing amount of p33C (from 1 to 0.1 μg of p33C). (B) Effect of single point mutations within the L3 internal loop on binding to the recombinant p33C. Other details are as described for panel A. (C) The effect of compensatory mutations on binding to p33C. To illustrate whether the mutated bases can or cannot maintain base pairing, we show both sides of the SL sequence in the boxes. A representative EMSA gel is shown on the right with selected RNA constructs carrying single or compensatory mutations (also indicated as the series comp1 to comp4). Other details are as described for panel A. (D) A representative EMSA gel showing inefficient (mutant Δ144-146, left, shown in panel A), weak (U141-A, also shown in panel A), and efficient (wt, right) binding of RII(+)-SL derivatives to p33C in vitro. The recombinant protein (the highest amount was 1 μg, shown on the left) was used in a twofold dilution series.
We next conducted further detailed analysis of p33C binding with single, double, and multiple RII(+)-SL mutants in or near the L3 region as shown in Fig. 2. The L3 region is predicted to form an unusual C99 · C143 mismatch, surrounded by strong C-G base pairs from each side and two putative non-Watson-Crick base pairs in the bottom part of the L3 region (13a). All three single mutations at position C99 (Fig. 2B, constructs C99-A, C99-G, and C99-U in lanes 4 to 6) and two of three single mutations at position C143 (Fig. 2B, constructs C143-A and C143-G in lanes 7 and 9) were found to inhibit binding to p33C in vitro. On the contrary, a mutation of C to U at position C143 (Fig. 2B, construct C143-U in lane 8,) did not affect binding to p33C. These data suggest that the C99 · C143 mismatch is essential for binding to p33C and can only be functionally replaced by C99 · U143.
To test if sequences around C99 · C143 could also affect binding, we introduced two types of modifications: single point mutations that not only changed the sequence but also reduced the base pairing potential (Fig. 2A and B) and compensatory mutations that changed the sequence in such a way that base pairs still could be formed flanking the C99 · C143 mismatch (Fig. 2C). These in vitro binding experiments demonstrated that the C100-G142 base pair on top of the C99 · C143 mismatch is essential, because single point mutations, deletion, or addition of a G at these positions interfered with binding (Fig. 2A), whereas replacing it with a G100-C142 base pair did not affect binding (Fig. 2C). Similarly, single mutations in the C98-G144 positions (just below the C99 · C143 mismatch) debilitated binding, except for U98-G144, which was partly permissible (Fig. 2B, lane 3). This mutation likely makes the binding between p33 and RII(+)-SL weak enough that the RNA is released from the p33:RNA complex during the electrophoresis, giving rise to a smeary pattern between the bound and the free forms of the RNA. Compensatory mutations that changed C98-G144 to either U98-A144 (Fig. 2C, lane 10) or A98-U144 (Fig. 2C, lane 13) reduced binding, suggesting that the stability of the base pair is an important factor. Converting the two non-Watson-Crick base pairs in the bottom part of the L3 region to two U-A or G-U base pairs, or to U · U mismatches did not affect binding (Fig. 2C). Also, combined modification of the C98-G144 and the two non-Watson-Crick base pairs to G-C/U-A/U-A or U-A/G-C/A-U base pairs did not have a significant effect on binding (Fig. 2C, lanes 4 and 7). Based on these findings, we conclude that, with the exception of the base pair directly below C99 · C143, the remaining portion of L3 is not essential for binding to p33C.
Specific recognition of RII(+)-SL by p33C requires the p33:p33/p92 interaction domain.
We next wanted to determine which domains within p33C were required for the specific recognition of RII(+)-SL. A 25-amino-acid peptide, named RPR25, that contains only the in vitro defined RNA-binding domain of p33 (termed RPR-motif) (26) recognized both the wt RII(+)-SL and the mutated ΔC99 equally well (Fig. 3C). This is in direct contrast to the selective binding of the wt p33C to the wt RII(+)-SL but not to ΔC99 (Fig. 3B) and suggests that sequences outside of the core RNA-binding domain in p33C contribute to the specificity of RNA recognition. To pursue this idea, we then tested two other truncated forms of p33C that contained the RPR-motif but either included (p33C210-275) or lacked (p33C150-240) the p33:p33/p92 interaction domain (Fig. 3A). Of the two, only p33C210-275 was able to bind to the RNA specifically (Fig. 3B), identifying the p33:p33/p92 interaction domain as a critical element for p33 RNA binding specificity. Consistent with this notion, p33C150-240 lacking the p33:p33/p92 interaction domain was also able to bind efficiently to heterologous satC(+), implying a general loss of selectivity in template recognition (Fig. 3B). Both p33C150-240 and the RPR25 peptide bound efficiently to RNA but did so nonselectively, likely due to the exposure of the positively charged arginine- and proline-rich RNA-binding motif on the surface of these proteins (26).
FIG. 3.
Selective binding of RII(+)-SL by p33 requires the p33:p33/p92 interaction domain. (A) Schematic representation of p33C with the known RNA and protein binding domains. Three derivatives of the recombinant p33C are shown. (B) An EMSA gel shows binding of p33C and its derivatives to either the wt or ΔC99 RII(+)-SL RNA (Fig. 2). Other details are as described in the legend of Fig. 1. (C) Efficient but nonselective binding of the RPR25 peptide to the wt and mutated ΔC99 RII(+)-SL in EMSA. The high-pressure liquid chromatography-purified RPR25 peptide, synthesized chemically, was used in a twofold dilution series starting from 0.05 μg in these assays.
Heterologous viral replicase proteins cannot specifically recognize TBSV RII(+)-SL.
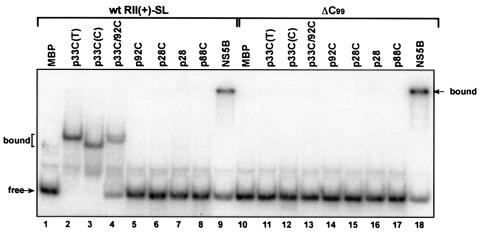
To test whether the C · C mismatch within RII(+)-SL could also be recognized by other viral replicase proteins, we compared the RNA binding activities of seven other purified recombinant viral proteins. These experiments revealed that the p33C protein of the closely related CNV could recognize selectively the wt but not the mutated form of RII(+)-SL (Fig. 4, lanes 3 and 12). This is not surprising, as CNV can efficiently replicate TBSV DI RNA in vivo (20, 36), and its genome also contains a comparably positioned C · C mismatch in its p92 ORF (13a). In contrast, the replicase protein p28 (the homologue of p33) of a related TCV (genus Carmovirus, family Tombusviridae) or its truncated version, p28C, could not recognize RII(+)-SL in vitro (Fig. 4, lanes 7 and 8 and 15 and 16). This is an intriguing observation, because TCV p28 and p28C contain an RPR-motif similar to that of TBSV p33 (26) and TCV replicase preparations can efficiently carry out plus- and minus-strand synthesis of TBSV DI RNAs in vitro (14). This in vitro compatibility is in direct contrast with the inability of TCV to replicate TBSV DI RNAs in vivo (unpublished data). Accordingly, we propose that the defect observed in vivo for this heterologous combination is, at least in part, related to the failure of RII(+)-SL to fit into a putative RNA-binding pocket of TCV p28.
FIG. 4.
Lack of selective recognition of RII(+)-SL by heterologous viral replicase proteins in vitro. The recombinant viral replicase proteins included from left to right: MBP, TBSV p33C [p33C(T)], CNV p33C [p33C(C)], TBSV p92 RdRp with an N-terminal deletion (p33C/92C) (Fig. 1A), TBSV p92 RdRp with deletion of the overlapping domain (p92C) (Fig. 1A), TCV p28 with an N-terminal deletion (p28C), full-length TCV p28, TCV p88 RdRp with deletion of the overlapping domain (p88C) (26), and the HCV NS5B RdRp. All proteins were expressed as fusions to maltose-binding protein (MBP) and purified from E. coli, except NS5B, which did not have a tag and was purified from insect cells. Similar amounts of recombinant proteins were applied in EMSA for binding to either the wt or ΔC99 RII(+)-SL (Fig. 2A).
The C-terminal polymerase domains of TBSV p92 or the related TCV p88 RdRp proteins (termed p92C and p88C, respectively) (Fig. 4, lanes 5, 8, 14, and 17), both of which contain RNA binding regions (26, 28), could not efficiently recognize wt or mutated RII(+)-SL. This suggests that selective template binding is not the function of the RdRp domain. On the contrary, a longer version of the TBSV p92 replicase protein, p33C/92C, that does contain the RPR-motif also present in p33C (and the p33:p33/p92 interaction and RdRp domains) (Fig. 1A) could recognize RII(+)-SL selectively, albeit with reduced efficiency in comparison to p33C (Fig. 4, lanes 4 and 13). Because RNA binding by p92 via its RPR-motif has been shown to be important for replication (18, 22), we propose that binding by the p33/p92 heterodimer to RII(+)-SL in the TBSV RNA might help the corecruitment of all three viral components required for assembly of the viral replicase complex and/or RNA replication.
Another replicase protein, NS5B of hepatitis C virus (HCV), which is distantly related to p92 (16), could recognize RII(+)-SL in vitro, but it could not discriminate between the wt RII(+)-SL or the mutated RII(+)-SL (Fig. 4, lanes 9 and 18). It is known that the NS5B of HCV on its own shows little specificity in RNA binding (12, 34); thus, additional viral or host factors may be required for selective template binding.
Mutations in RII(+)-SL that interfere with p33C binding abolish replication of TBSV RNAs in vivo.
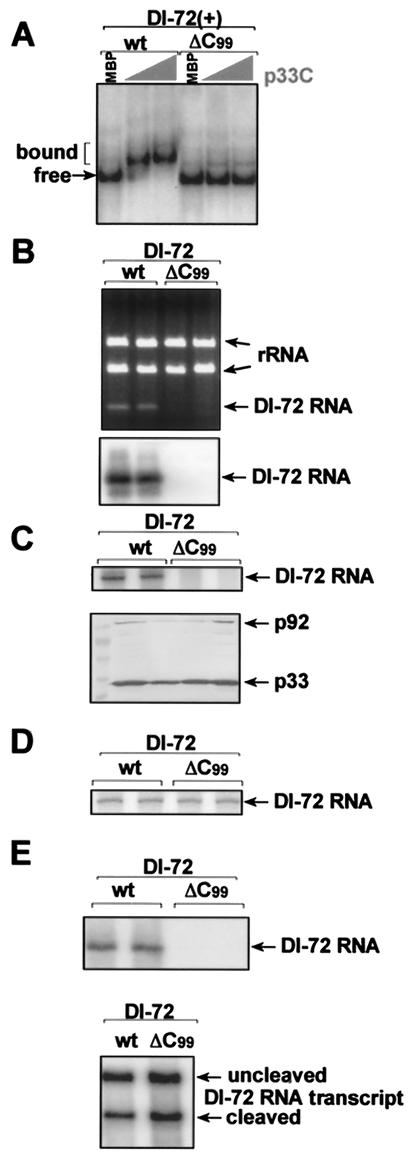
In order to determine whether the RII(+)-SL:p33 interaction observed in vitro is functionally relevant in vivo, we determined the effects of selected RII(+)-SL mutations on RNA template replication in yeast cells and whole plants. TBSV DI RNAs are able to replicate in yeast cells that coexpress p33 and p92 replicase proteins (19). When a full-length DI-72 RNA containing a C99 deletion (Fig. 5B, ΔC99) was tested in yeast, it was unable to replicate. The correlation between lack of p33 binding to DI-72 containing a C99 deletion (Fig. 5A) and the absence of replication in vivo suggests that binding of p33 to RII(+)-SL is critical for DI RNA replication in yeast cells.
FIG. 5.
C99 in RII(+)-SL is essential for replication of DI-72 RNA in yeast. (A) p33C does not bind efficiently to DI-72(+) RNA carrying a ΔC99 mutation in RII(+)-SL in EMSA. Further details are as described in the legend to Fig. 1. (B) Mutant ΔC99 of DI-72 RNA, expressed from the galactose-inducible GAL1 promoter in pYC-DI-72 plasmid, does not replicate at a detectable level in yeast coexpressing CNV p33 and p92 replicase proteins, whereas wt DI-72 RNA was detected in total RNA extract obtained 24 h after induction in an ethidium bromide-stained gel (top) or by Northern blotting (bottom) with probe specific for TBSV RIII(+). (C) The top panel shows a lack of in vitro RNA synthesis by the CNV replicase in the enriched membrane fraction obtained from yeast coexpressing mutant ΔC99 of DI-72RNA and His-tagged CNV p33 and p92, whereas the comparable fraction from wt DI-72-expressing yeast generated DI-72 RNA products. Note that the in vitro replicase products must be derived from copurified RNAs present in the replicase preparations, because no RNA was added to these replicase assays. The Western blot in the bottom panel shows that the CNV p33 and p92 proteins were present in comparable amounts in these replicase preparations. (D) Full-length wt and ΔC99 DI-72(+) RNAs (1 μg of RNA transcripts was added per reaction) are transcribed with comparable efficiency by a template-dependent CNV replicase preparation obtained from CNV-infected plants. (E) Northern blot analysis with 32P-labeled RNA probe was performed to detect copurified DI-72 RNA in p33 preparations obtained from yeast coexpressing His-tagged p33 and p92 and DI-72 RNA (wt or ΔC99) (top panel). Purification was done by using His-tag-based affinity chromatography with the solubilized membrane fraction from yeast. Northern blot analysis with DI-72(+)-specific 32P-labeled RNA probe demonstrates the presence of comparable amounts of ribozyme-cleaved and uncleaved wt or ΔC99 DI-72 RNA transcripts in yeast (bottom panel).
To determine the step in replication that was blocked in the ΔC99 mutant, we used two approaches. First, we tested the template activity of isolated membrane fractions containing tombusvirus replicase complexes (20). Note that in these in vitro replicase assays, we measure replicase activity on endogenous (copurified) RNA templates in the absence of added exogenous templates. We found that preparations containing the endogenous ΔC99 RNA was not transcribed by the tombusvirus replicase (Fig. 5C), whereas those with endogenous wt DI-72 RNA were utilized efficiently. This difference was not due to different amounts of replicase proteins in the two preparations as similar levels of p33 and p92 were detected in the extracts (Fig. 5C). Alternatively, the different results may have been due to the lower levels of ΔC99 in cells (as it did not replicate to a detectable level) and/or the lack of efficient copurification of ΔC99 with the replicase complex.
To determine whether ΔC99 was a competent template for in vitro transcription, an alternative approach with a replicase extract from infected plant tissue was employed. When exogenous wt DI-72 and ΔC99 RNA templates were added to this extract, both RNAs were transcribed into complementary strands with similar efficiencies (Fig. 5D). This indicates that the ΔC99 mutant is indeed a fully competent template for RNA transcription in vitro. For the second approach, we performed p33:RNA copurification experiments from yeast coexpressing His-tagged p33, p92, and DI-72 RNA (wt or ΔC99). After solubilization of enriched membrane fractions, we affinity purified similar amounts of p33 and tested the amount of copurified RNAs in these samples. While we found a detectable amount of wt DI-72 RNA in the affinity-purified p33 sample, the level of copurified ΔC99 RNA was below the detection level by Northern blotting (Fig. 5E, top). Total RNA samples from yeast, however, contained ΔC99 RNA transcripts (Fig. 5E, bottom), excluding the possibility that ΔC99 transcripts were unstable in yeast. Based on these results, we propose that the lack of binding of p33 replicase protein to ΔC99 causes a failure in template recruitment to the replication complex, thereby preventing the replication of the ΔC99 DI-72 RNA.
To test whether the p33:RII(+)-SL interaction is also important for replication of the full-length TBSV genomic RNA (gRNA), we introduced a “silent” C99-to-G substitution in the C99 · C143 mismatch (which corresponds to C1383 · C1427 in gRNA) in RII(+)-SL. This C99-G mutation was shown to interfere with binding of RII(+)-SL to p33C in vitro (Fig. 2B, lane 5), and the same mutation in the viral genome also abolished accumulation of all TBSV RNAs in plants (Fig. 6). Northern blot analysis revealed the lack of accumulation of viral RNAs in both inoculated and upper leaves (Fig. 6). Overall, these experiments demonstrate that recognition of RII(+)-SL by p33 is absolutely critical for tombusvirus RNA replication in both host plants and the yeast model system.
FIG. 6.

C99 in RII(+)-SL is essential for replication of TBSV gRNA in plants. Total RNA extracts from inoculated (I) or systemic (S) leaves of N. benthamiana plants inoculated with either the wt or a mutant TBSV gRNA, which carried a silent C99-G mutation in RII(+)-SL (which represents position C1383 in gRNA) (Fig. 1A), were analyzed by agarose gel electrophoresis (top) or by Northern blotting (bottom). The positions of the gRNA and the two subgenomic RNAs (sgRNAs) are marked with arrows. Others details are as described in the legend of Fig. 5.
DISCUSSION
In this study we show that the TBSV p33 replicase protein can bind specifically to the cognate viral RNA template in vitro [i.e., DI-72(+) RNA] (Fig. 1C). Binding of p33 to DI-72(+) RNA depends on the presence of the conserved RII(+)-SL hairpin, which serves as an internal recognition element. Based on detailed mutagenesis of RII(+)-SL, we consider it unlikely that an extensive RNA sequence motif in RII(+)-SL is recognized by p33. We propose that the major RNA element recognized by p33 is a C99 · C143 mismatch in an internal loop (Fig. 2, L3,) within the RII(+)-SL. Further support for the essential role of the C · C mismatch for specific interaction with p33 comes from in vivo studies that demonstrated the lack of replication of DI-72 RNA in yeast and the TBSV gRNA in plants, which carried a mutation within the C · C mismatch (Fig. 6). It is likely that the ability of p33 replicase protein to specifically recognize a C · C mismatch is conserved in tombusviruses, since all the sequenced members of this genus are predicted to form RII(+)-SL-like structures with the C · C mismatch (not shown). The in vitro binding studies also revealed that (i) nucleotides within the lower portion of L3 do not contribute to the specific recognition of the C · C mismatch by p33 since they can be converted to different base pairs without affecting binding (Fig. 2C), (ii) the formation of the C-G base pairs above and below the C · C mismatch is important for an efficient p33C interaction (Fig. 2B and C), and (iii) other sequences in RII(+)-SL are less important for binding in the in vitro assay. Altogether, our data are in agreement with a role of the specific p33:RII(+)-SL interaction during template selection and recruitment (see model below). In contrast, in vitro replicase studies with added templates showed that a mutation that debilitated the p33:RII(+)-SL interaction did not affect de novo initiation of RNA synthesis (Fig. 5D). Because the above in vitro assay is based on stably assembled replicase complexes, we cannot exclude the possibility that RII(+)-SL is also involved in the initial assembly of the replicase complex. Indeed, both p33 and the viral RNA template are part of the tombusvirus replicase complex purified from yeast cells, and they are also essential for the in vivo assembly of the functional replicase (23). Future study will address the proposed role of RII(+)-SL:p33 interaction in replicase assembly.
How can a C · C mismatch within the extensive RII(+)-SL helix contribute to specific recognition by the RNA-binding p33 replicase protein? We propose that one function of the C99 · C143 mismatch is to open up the central helix in RII(+)-SL to facilitate binding or positioning of the RPR-motif of p33. Accordingly, a mutation of C143 to U, but not A or G mutations, was allowed for in vitro binding, suggesting the requirement of a small pyrimidine at position 143. In contrast, C99 mutation to U, A, or G within the C · C mismatch was not permissible for RII(+)-SL:p33 interaction (Fig. 2), suggesting the existence of base-specific contact between C99 and the RPR-motif of p33. We suggest, therefore, that both sterical (shape and dimension of the internal loop and the helix) and base-specific interactions (position C99) contribute to binding of RII(+)-SL in the cognate RNA to p33. Interestingly, other viral proteins with arginine-rich RNA-binding domains use somewhat similar mechanisms to recognize cognate RNAs. For example, the human immunodeficiency virus Rev protein recognizes purine-purine mismatches within a helix (2), whereas the Tat protein of bovine immunodeficiency virus binds specifically to a hairpin containing two bulged U nucleotides (25). Both proteins bind to the cognate RNAs by recognizing the widened major grooves around the mismatch or bulge and also by specifically interacting with these bases (7).
The minimal requirements for functional RII(+)-SL make one wonder whether some host mRNAs may contain this motif. Although it is currently unknown how common this motif is among host mRNAs, it is important to note that the viral gRNA may have a major advantage for binding to p33 over host mRNAs due to the proximity of the viral RNA and p33 during translation (cis-effect). Also, RII(+)-SL is not the only essential RNA element required for tombusvirus RNA replication. Previous studies demonstrated that a replication silencer element (24) and the minus-strand initiation promoter (gPR) (8, 21) are also required for replication. Therefore, recruitment of some host mRNAs, which are unlikely to contain the replication silencer element and gPR, to the site of tombusvirus replication would not result in their replication at detectable levels.
The in vitro binding experiments exclude the model that the p33 RPR-motif involved in RNA-binding (Fig. 1A) is solely responsible for selective binding to wt RII(+)-SL and discriminating against ΔC99 RII(+)-SL and satC(+). For example, the RPR25 and p33C150-240 peptides carrying the RPR-motif but lacking the p33:p33/p92 interaction domain bound to wt RII(+)-SL, ΔC99 RII(+)-SL, and the unrelated satC(+) RNAs efficiently (Fig. 3). This nonselective RNA binding by the RPR25 and p33C150-240 peptides might be due to surface exposure of an unordered form of the RNA-binding motif in the above peptides (26). This is in contrast with p33-derived proteins carrying both the RNA-binding and the p33:p33/p92 interaction domains, which bound only to wt RII(+)-SL but not to the ΔC99 RII(+)-SL (Fig. 3B) or satC(+) RNA (Fig. 1E). Altogether, the essential role of the p33:p33/p92 interaction domain in selective RNA binding suggests that intermolecular interaction between two or more p33s (and/or possibly p92) proteins results in the formation of an RNA-binding pocket that has high specificity for the C · C mismatch within RII(+)-SL. Heterologous replicase proteins, such as TCV p28, might not be able to bind to RII(+)-SL, because, at least in part, RII(+)-SL might not fit into a putative RNA-binding pocket of TCV p28. This suggestion is in line with the idea that the wt RII(+)-SL:p33 interaction functions at an early step in replication (i.e., template selection) before template replication. This idea is also consistent with other results, which revealed that the in vivo function(s) of RII(+)-SL is required early but is dispensable late in the infection cycle (13a).
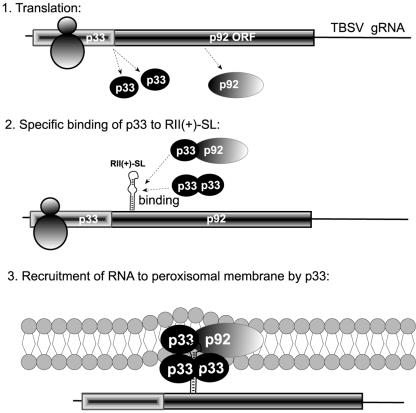
A model for the central role for the RII(+)-SL:p33 interaction in Tombusvirus RNA replication.
Based on the above in vitro and in vivo studies and those in the accompanying article (13a), we propose that the RII(+)-SL, p33, and possibly p92 act together at an early step in replication to facilitate template selection and corecruitment of replication factors. In this model, viral proteins are the central players; however, it cannot be precluded that host proteins also contribute in some manner to template selection. The following multistep model is proposed to describe how this process is perceived to occur (Fig. 7). Following infection, p33 and p92 are translated from the genome, with the former being produced in greater quantities. The RII(+)-SL may be able to form in the genome transiently due to a lower level of ribosome traffic in the readthrough portion of the p92 ORF (13a). The intriguing location of RII(+)-SL within the p92 ORF in TBSV gRNA is also consistent with a cotranslational binding model. When a required threshold concentration of p33 is reached, p33 then interacts productively with RII(+)-SL. The p33 threshold level required may be related to both protein oligomerization requirements and the RNA-protein interaction itself. In this model, recognition of the C · C mismatch is critical for specific binding of p33 to the RNA template and is dependent on protein dimerization or oligomerization (which may also involve p92). The oligomerization requirement suggests that the interface of the protein subunits may form an RNA-binding pocket that, in turn, specifically recognizes the C · C mismatch. The RII(+)-SL:p33 ribonucleoprotein complex formed is then transported to membranes via the membrane targeting signals present in the N terminus of p33 and p92 (29). Recruitment to membranes could also mediate down-regulation of translation (as shown for Brome mosaic virus [10]). In contrast, subgenomic mRNAs transcribed during infection would not be recruited to replication complexes as they lack RII(+)-SL, and they would remain dedicated mRNAs for translation. In this model, the selective binding of p33 (and/or p92) to RII(+)-SL-containing RNAs is proposed to be a primary factor in vivo for the observed specificity of Tombusvirus RNA replication.
FIG. 7.
A model of RNA template-selection and recruitment by the p33 replicase protein in Tombusviruses. First, p33 and a smaller amount of p92 are being synthesized by the ribosome by using the gRNA. Second, p33, likely together with other p33 and/or p92 molecules, binds specifically to RII(+)-SL in the gRNA. Heterologous viral and host RNAs lacking RII(+)-SL will not be selected, resulting in replication of only the cognate RNA. We predict that in order to allow sufficient translation, production of a threshold level of p33 is needed prior to recruitment of the gRNA into replication. Third, stably associated RNA:p33 complex, possibly together with p92 and host factors, is transported to the site of replication (peroxisome-derived membranes [29]). This is predicted to lead to the formation of replication complexes (not shown).
Acknowledgments
We are grateful to G. Luo for providing the purified HCV NS5B protein preparation.
This work was supported by grants from the NSF (MCB0078152) and the University of Kentucky to P.D.N. and from NSERC, PREA, and CRC to K.A.W.
Footnotes
This study is publication no. 04-12-034 of the Kentucky Agricultural Experiment Station.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battiste, J. L., R. Tan, A. D. Frankel, and J. R. Williamson. 1994. Binding of an HIV Rev peptide to Rev responsive element RNA induces formation of purine-purine base pairs. Biochemistry 33:2741-2747. [DOI] [PubMed] [Google Scholar]
- 3.Brown, D., and L. Gold. 1996. RNA replication by Q beta replicase: a working model. Proc. Natl. Acad. Sci. USA 93:11558-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus Protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Draper, D. E. 1999. Themes in RNA-protein recognition. J. Mol. Biol. 293:255-270. [DOI] [PubMed] [Google Scholar]
- 8.Fabian, M. R., H. Na, D. Ray, and K. A. White. 2003. 3′-Terminal RNA secondary structures are important for accumulation of tomato bushy stunt virus DI RNAs. Virology 313:567-580. [DOI] [PubMed] [Google Scholar]
- 9.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 12.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miranda, G., D. Schuppli, I. Barrera, C. Hausherr, J. M. Sogo, and H. Weber. 1997. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J. Mol. Biol. 267:1089-1103. [DOI] [PubMed] [Google Scholar]
- 13a.Monkewich, S., H.-X. Lin, M. R. Fabian, W. Xu, H. Na, D. Ray, O. A. Chernysheva, P. D. Nagy, and K. A. White. 2005. The p92 polymerase coding region contains an internal RNA element required at an early step in tombusvirus genome replication. J. Virol. 79:4848-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy, P. D., and J. Pogany. 2000. Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology 276:279-288. [DOI] [PubMed] [Google Scholar]
- 15.Noueiry, A. O., and P. Ahlquist. 2003. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41:77-98. [DOI] [PubMed] [Google Scholar]
- 16.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287-303. [DOI] [PubMed] [Google Scholar]
- 17.Osman, T. A., and K. W. Buck. 2003. Identification of a region of the tobacco mosaic virus 126- and 183-kilodalton replication proteins which binds specifically to the viral 3′-terminal tRNA-like structure. J. Virol. 77:8669-8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oster, S. K., B. Wu, and K. A. White. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J. Virol. 72:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panavas, T., and P. D. Nagy. 2003. The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis. J. Virol. 77:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panavas, T., and P. D. Nagy. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of tomato bushy stunt virus. Virology 314:315-325. [DOI] [PubMed] [Google Scholar]
- 21.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Analysis of minimal promoter sequences for plus-strand synthesis by the cucumber necrosis virus RNA-dependent RNA polymerase. Virology 296:263-274. [DOI] [PubMed] [Google Scholar]
- 22.Panaviene, Z., J. M. Baker, and P. D. Nagy. 2003. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology 308:191-205. [DOI] [PubMed] [Google Scholar]
- 23.Panaviene, Z., T. Panavas, S. Serva, and P. D. Nagy. 2004. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 78:8254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pogany, J., M. R. Fabian, K. A. White, and P. D. Nagy. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 22:5602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puglisi, J. D., L. Chen, S. Blanchard, and A. D. Frankel. 1995. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science 270:1200-1203. [DOI] [PubMed] [Google Scholar]
- 26.Rajendran, K. S., and P. D. Nagy. 2003. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J. Virol. 77:9244-9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajendran, K. S., and P. D. Nagy. 2004. Interaction between the replicase proteins of Tomato Bushy Stunt virus in vitro and in vivo. Virology 326:250-261. [DOI] [PubMed] [Google Scholar]
- 28.Rajendran, K. S., J. Pogany, and P. D. Nagy. 2002. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 76:1707-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubino, L., and M. Russo. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252:431-437. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Scholthof, K. B., H. B. Scholthof, and A. O. Jackson. 1995. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology 208:365-369. [DOI] [PubMed] [Google Scholar]
- 32.Shapka, N., and P. D. Nagy. 2004. The AU-rich RNA recombination hot spot sequence of Brome mosaic virus is functional in tombusviruses: implications for the mechanism of RNA recombination. J. Virol. 78:2288-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan, M. L., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White, K. A., and T. J. Morris. 1994. Recombination between defective tombusvirus RNAs generates functional hybrid genomes. Proc. Natl. Acad. Sci. USA 91:3642-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White, K. A., and T. J. Morris. 1995. RNA determinants of junction site selection in RNA virus recombinants and defective interfering RNAs. RNA 1:1029-1040. [PMC free article] [PubMed] [Google Scholar]
- 37.White, K. A., and P. D. Nagy. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 78:187-226. [DOI] [PubMed] [Google Scholar]
- 38.Yang, Y., R. Rijnbrand, S. Watowich, and S. M. Lemon. 2004. Genetic evidence for an interaction between a picornaviral cis-acting RNA replication element and 3CD protein. J. Biol. Chem. 279:12659-12667. [DOI] [PubMed] [Google Scholar]
- 39.Yoshinari, S., P. D. Nagy, A. E. Simon, and T. W. Dreher. 2000. CCA initiation boxes without unique promoter elements support in vitro transcription by three viral RNA-dependent RNA polymerases. RNA 6:698-707. [DOI] [PMC free article] [PubMed] [Google Scholar]