Abstract
Human immunodeficiency virus (HIV) infects cells by fusing with cellular membranes. Fusion occurs when the envelope glycoprotein (Env) undergoes conformational changes while binding to cellular receptors. Fusogenic changes involve assembly of two heptad repeats in the ectodomain of the gp41 transmembrane subunit to form a six-helix bundle (6HB), consisting of a trimeric N heptad repeat (N-HR) coiled-coil core with three antiparallel C heptad repeats (C-HRs) that pack in the coiled-coil grooves. Peptides corresponding to the N-and C-HRs (N and C peptides, respectively) interfere with formation of the 6HB in a dominant-negative manner and are emerging as a new class of antiretroviral therapeutics for treating HIV infection. We generated an escape mutant virus with resistance to an N peptide and show that early resistance involved two mutations, one each in the N- and C-HRs. The mutations conferred resistance not only to the selecting N peptide but also to C peptides, as well as other types of N-peptide inhibitors. Moreover, the N-HR mutation altered sensitivity to soluble CD4. Biophysical studies suggest that the 6HB with the resistance mutations is more stable than the wild-type 6HB and the 6HB formed by inhibitor binding to either wild-type or mutant C-HR. These findings provide new insights into potential mechanisms of resistance to HIV peptide fusion inhibitors and dominant-negative inhibitors in general. The results are discussed in the context of current models of Env-mediated membrane fusion.
The envelope glycoprotein (Env) mediates human immunodeficiency virus (HIV) entry by fusing virus to target cells. Env is trimeric on the virion surface, with each monomer consisting of a surface subunit (gp120) and a noncovalently associated transmembrane subunit (gp41). gp120 binding to cellular CD4 and a chemokine receptor triggers fusion-inducing conformational changes in gp41, leading to increased exposure of two heptad repeat motifs (N-HR and C-HR) in the gp41 ectodomain (Fig. 1) and insertion of the fusion peptide into the target membrane (reviewed in references 12 and 15). Subsequently the N- and C-HRs fold in an antiparallel manner to create a six-helix bundle (6HB), composed of a trimeric N-HR coiled-coil core surrounded by three C-HR helices that pack in the grooves of the coiled coil (8, 35, 39). Transition to the thermostable 6HB promotes fusion between viral and cellular membranes (27).
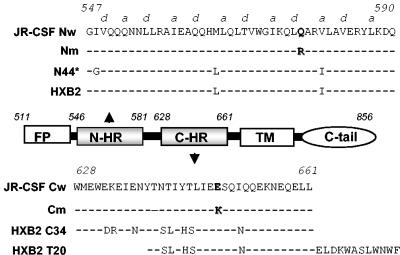
FIG. 1.
Map of gp41 ectodomain and corresponding peptides. The JR-CSF Nw peptide corresponds to wild-type residues in the N-HR in the JR-CSF envelope glycoprotein. a and d indicate positions in the heptad repeat motif. N44* represents the peptide used for selecting the resistant virus. The Nm peptide contains the resistance mutation at position 577. HXB2 indicates residues in the HXB2 clone of HIV. The JR-CSF Cw peptide corresponds to wild-type residues in the C-HR in the JR-CSF envelope glycoprotein. The Cm peptide contains the resistance mutation at position 648. The HXB2 C34 peptide corresponds to C-HR residues in the HXB2 clone of HIV. FP, fusion peptide; TM, transmembrane domain; C-tail, cytoplasmic domain.
Peptides corresponding to the HR of gp41 inhibit HIV infection in vitro and in vivo (reviewed in references 9 and 20). N-HR and C-HR peptides (N and C peptides, respectively) block fusion in a dominant-negative manner by binding to transiently exposed HRs of gp41 during fusion-inducing conformational changes to form a peptide-gp41 6HB (17, 18, 21, 27). The C-peptide inhibitor Enfuvirtide (T20 or DP-178), the first drug in the new class of antiretrovirals called fusion inhibitors, binds the N-HR of gp41 (31, 36). As with other antiretrovirals, however, resistance is a significant clinical problem. In vitro selection studies with T20 (31) or an overlapping C peptide (C34) (1) identified a region in the N-terminal part of the N-HR (residues 33 to 38 in gp41 or residues 544 to 549 in Env, corresponding to the Los Alamos numbering for the reference HXB2 clone) as being important for resistance, with mutations frequently occurring in the highly conserved GIV sequence. Mutations in the same region and extending slightly more C terminal were also generated in patients treated with T20 (32, 38) and were found to occur naturally in treatment-naive subjects whose viruses showed relative resistance to T20 (10, 11). In addition, residues in the C-HR have been shown to increase resistance to T20 in the absence of mutations in the N-HR (19, 29). Increased sensitivity to C peptides has also been correlated with use of the CXCR4 coreceptor in panels of viruses from treatment-naive subjects (11). The mechanisms responsible for this increased sensitivity of X4 viruses is not known, but it has been proposed that gp120-receptor affinities and receptor expression levels may play a role (30).
Combination antiretroviral therapy, preferably involving agents that target different steps of HIV infection, helps to suppress the emergence of resistant viruses. Whereas C-peptide inhibitors target the N-HR (18, 31), N-peptide inhibitors, including the five-helix bundle inhibitor and the chimeric coiled-coil IZN36, appear to bind the C-HR (14, 33). N peptides may additionally bind to the N-HR, forming homologous interactions in a peptide-gp41 coiled coil (3). To further investigate viral determinants for N-peptide resistance and to gain insight into the Env fusion mechanism, we generated an escape mutant virus with resistance to an N-peptide inhibitor. We found that two mutations in gp41, one each in the N- and C-HRs, provided initial steps in conferring resistance to the N peptide. Unexpectedly, these mutations also conferred cross-resistance to C peptides and altered sensitivity to soluble CD4 (sCD4), implying changes in receptor activation of Env. Molecular modeling suggests that the resistance mutations would not directly impair peptide binding to gp41, and biophysical data suggest that the mutations improve endogenous (viral) 6HB stability. These findings provide insights into Env structure-function relationships and activation of fusion and further suggest a potential pathway for resistance to dominant-negative inhibitors involving increased stability of the target protein.
MATERIALS AND METHODS
Cells and plasmids.
293T cells, U87 cells expressing CD4 and CXCR4 (U87-T4-X4) or CD4 and CCR5 (U87-T4-CCR5), the Env-deficient HIV genome reporter plasmid pNL43-Luc-R−E−, and the HXB2 Env expression plasmid pSM-WT were kindly provided by Dan Littman (New York University, New York, N.Y.). The proviral plasmid pLAI(JR-CSF), expressing the LAI genome except with the env gene replaced with JR-CSF env, was engineered by exchanging the KpnI fragments (nucleotides 6431 to 9114) from the LAI genome with nucleotides 6430 to 9107 from the JR-CSF genome. The pJR-CSF-Env expression plasmid was derived from pLAI(JR-CSF) by excising the region from the SphI site (nucleotide 1443) to the PshAI site (nucleotide 4428). An expression vector (pTCLE-G2C) for the N-peptide inhibitor, here called N44*, was a gift from Carl Wild (Panacos Pharmaceuticals, Gaithersburg, Md.,) and Terry Oas (Duke University, Durham, N.C.). PM-1 lymphoid cells expressing CD4, CXCR4, and CCR5 receptors were obtained from Hana Golding (U.S. Food and Drug Administration, Bethesda, Md.).
Peptides and other reagents.
The N-peptide inhibitor N44* (GGVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLKDQ) was expressed in Escherichia coli and purified as previously reported (5). Briefly, the pTCLE-G2C expression plasmid, encoding a TrpLE fusion protein containing residues 540 to 584 from the N-HR of LAI Env, was transformed into BL21(DE3) cells (Novagen), induced with IPTG (isopropyl-β-d-thiogalactopyranoside), and lysed with a French press. Inclusion bodies were pelleted, washed, and dissolved in 70% formic acid prior to cleavage with 50 mg of cyanogens bromide per ml. The dried products were dissolved in 6 M guanidine-HCl and separated by Sephadex G-50 (26- by 70-cm) gel filtration chromatography in 5% acetic acid. Synthetic peptides C34 (WMEWDREINNYTSLIHSLIEESQNQQEKNEQELL), T20 (YTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWF), Cw (WMEWEKEIENYTNTIYTLIEESQIQQEKNEQELL), Cm (WMEWEKEIENYTNTIYTLIEKSQIQQEKNEQELL), Nw (GIVQQQNNLLRAIEAQQHMLQLTVWGIKQLQARVLAVERYLKDQ), and Nm (GIVQQQNNLLRAIEAQQHMLQLTVWGIKQLRARVLAVERYLKDQ) were made by standard 9-fluorenylmethoxy carbonyl chemistry and purified by high-pressure liquid chromatography (CBER Facility for Biotechnology Resources, Bethesda, Md.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analytic high-pressure liquid chromatography indicated that all of the peptides were > 92% pure. All peptides were confirmed to have the expected molecular weight by using matrix-assisted laser desorption ionization-time-of-flight mass spectroscopy and to have inhibitory activity by using infectivity assays. sCD4 was purchased from Progenics Pharmaceuticals (Tarrytown, N.Y.). The five-helix inhibitor was kindly provided by Michael Root (Jefferson University, Philadelphia, Pa.).
Resistant virus.
Viral stocks of HIV type 1 (HIV-1) LAI(JR-CSF) were generated by transfecting pLAI(JR-CSF) into 293T cells by using Fugene 6 (Roche Diagnostics) and collecting filtered culture supernatants after 2 days. Virus was quantified by a reverse transcriptase (RT) assay, as described previously (28), and stored at −80°C. Resistant virus was generated by infecting 106 PM-1 cells with 1.75 × 106 cpm of RT activity from HIV-1 LAI(JR-CSF) virus stock in 4 ml of medium in the presence of 300 nM N44* overnight. The cells were washed twice the next day by centrifugation at 200 × g for 10 min and resuspended at 2 × 105 cells/ml with medium containing the same concentration of peptide. Three days later, supernatants were exchanged with fresh medium containing the peptide, and after the first week, half of the cells and supernatant were removed every 2 days and replaced by an equal volume of peptide-containing medium. Cell supernatants were sampled every 2 days for virus production by RT activity. Supernatants containing the highest level of RT activity were then used for subsequent rounds of infection, using approximately 106 cpm of RT activity and following the same infection protocol as described above except with increasing peptide concentrations as indicated in Fig. 2.
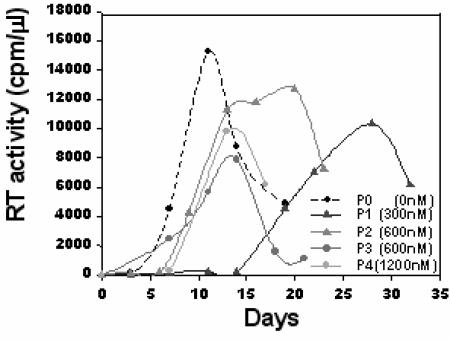
FIG. 2.
Emergence of peptide-resistant virus. P0 shows virus growth in the absence of peptide. P1 shows growth of the culture in the presence of 300 nM N44*. P2 was initiated from supernatant from the P1 culture and shows growth in the presence of 600 nM N44*. P3 was initiated from supernatant from the P2 culture and shows growth in the presence of 600 nM N44*. P4 was initiated from supernatant from the P3 culture and shows growth in the presence of 1,200 nM N44*.
Resistant Env proteins.
Genomic DNA from infected PM1-MC cells was extracted by using the Qiagen DNAeasy kit. The gp41 gene from the proviral genome was amplified by nested PCR with the Elongase enzyme mix (Invitrogen) and the two sets of primers Enf (AAAGAGGAATTCACAGTGGCAATGAGAGTGAAGG)-Enr (CAGGCACTCGAGGTTTTGACCACTTGCCACCCATCTT) and F1 (GAACCATTAGGAGTAGCACCCACCA)-R1 (ATGGAGCAATCACAAGTAGCAATACAG). The PCR product was purified on an agarose gel and digested with the restriction enzymes MfeI and PpuMI. The product was then inserted in the parental Env expression plasmid pJR-CSF-Env at the same sites (nucleotides 2713 and 3458 of the gp41 gene). Each clone was verified by sequencing the entire gp41 gene.
Mutagenesis.
Point mutations in the gp41 genes of JRCSF and HXB2 expression plasmids were created by using the QuickChange site-directed mutagenesis kit (Stratagene). Two sets of complementary primers, TAATACACTCCTTAATTGAAAAGTCGCAAAACCAGCAAG-TTGCTGGTTTTGCGACTTTTCAATTAAGGAGTGTATTA and TCTGGGGCATCAAGCAGCTCCCGCAAGAATCCTGGCTGTGG-CCACAGCCAGGATTCTTGCGCGGAGCTGCTTGATGCCCCAGA, were used to introduce theQ577R and E648K mutations in the C- and N-HRs of the JR-CSF and HXB2 gp41 genes, respectively. Mutations were verified by sequencing the entire env gene. We note that the sequence we found for the wild-type (WT) env gene of JR-CSF differed from that listed in the Los Alamos HIV database. We found two nucleotide changes at positions 7878 and 7879 in the env gene, GC instead of CG, which resulted in an amino acid change from arginine to alanine at residue 22 of gp41. The other sequence discrepancy, G to A at nucleotide 8302 of the env gene, altered the amino acid sequence from glycine to aspartic acid at residue 164 of gp41.
Infectivity inhibition assay.
Pseudovirion stocks with wild-type or mutant Env proteins were generated and assessed for infectivity as previously described (41). Briefly, 3 × 106 293T cells in 10-cm-diameter dishes were cotransfected with 3 μg of the Env expression vector and 2 μg of the Env-deficient viral reporter vector (pNL43-Luc-R−E−). The supernatants were collected at 48 h posttransfection, quantified by p24 enzyme-linked immunosorbent assay (Coulter), and stored at −80°C. One day before infection, 2 × 104 U87-CD4-CCR5 or U87-CD4-X4 cells/well were plated in 96-well dishes in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum, 2 mM glutamine, 1× antibiotics, 1 mM sodium pyruvate, and 1× nonessential amino acids (DMEM+). Cells were infected with pseudovirus supernatants (approximately 200 ng of p24 per ml for HXB2 pseudoviruses and 20 ng of p24 per ml for the JR-CSF pseudoviruses) in the presence of peptide in a total volume of 100 μl supplemented with 8 μg of Polybrene (Sigma) per ml. After 16 h of incubation, the medium containing the peptide was replaced by 100 μl of fresh DMEM+, and cultures were continued for an additional 24 h at 37°C. The cells were then lysed, and infectivity was measured with a luciferase kit (Promega) and luminometry (L-Max; Molecular Devices).
Thermal denaturation studies.
Circular dicrhroism spectra were recorded at 222 nm with a Jasco J 720 instrument equipped with PTC-343 Peltier-type thermostatic cell holder, using a 1-cm-path-length cell with a Teflon stopper (Hellma). Cooling circulating water (20°C) was supplied with a Mulitemp III thermostatic circulator (Amersham Biotech). Stocks of approximately 2.0 mg of N-terminal peptides (in 50 mM sodium formate, pH 3.5) and C-terminal peptides (in 20 mM sodium phosphate, pH 7.0) per ml were mixed (10 μM each) in 20 mM sodium phosphate (pH 7.0) containing 0.2 M NaCl. The salt was required to support solubility of the peptide mixtures. Peptide mixtures (final volume, 0.5 ml) were heated; the temperature was raised 1°C per min with a temperature slope of 20 to 100°C. The step resolution was 1°C, the response time was 1 s, the bandwidth was 1 nm, and the sensitivity 200 m°C. Temperatures at the transition midpoints (melting temperature [Tm]) were estimated from first-derivative plots of the curves.
Statistics.
At least five independent dose-response curves, using multiple batches of each pseudovirus, were generated for each inhibitor. The 50% inhibitory concentrations (IC50s) relative to no inhibitor for each individual pseudovirus were calculated with SoftMax Pro software (Molecular Devices). The geometric mean (GM) IC50 was determined for each pseudovirus and inhibitor. To determine the fold change in resistance relative to WT pseudovirus for each inhibitor, the ratios of GM IC50 for mutants to GM IC50 for the wild type were calculated. The IC50 for each mutant pseudovirus was compared with that for the WT pseudovirus on a logarithmic scale by using Dunnett's multiple-comparison option for one-way analysis of variance platform in JMP 5.1.1 statistical software (SAS Institute). The blocking option with “experiment” as a blocking parameter was used to take into account that values for all three mutants and the WT were measured simultaneously in each individual experiment.
RESULTS
Generating an escape mutant virus.
Parental, wild-type virus was prepared from an infectious molecular clone of a hybrid virus LAI(JR-CSF) consisting of the LAI proviral genome with the env gene replaced by the JR-CSF env gene. This virus uses the CCR5 coreceptor but not CXCR4. A dose-response curve for the selecting N-peptide inhibitor (N44* [Fig. 1]) on PM-1 cells determined that 90% of the RT activity from this virus was inhibited at a concentration of 300 nM, and complete inhibition was observed at concentrations of above 400 nM (not shown). Initial cultures (P1) for selecting escape mutant viruses therefore began at 300 nM N44*. Fifteen days after infection in the presence of 300 nM N44* peptide, RT activity emerged, and it peaked at day 28 (Fig. 2). This supernatant was then used to establish the second passage (P2). Control cultures without peptide (P0) peaked on day 11.
The P2 culture was grown in the presence of 600 nM N44* peptide, and peak RT activity was broad, occurring on days 12 to 20. Supernatant from this culture was used to inoculate the third passage at twice the peptide concentration (1,200 nM). Virus was not detected in this culture after 28 days, so a new third-passage culture (P3) was established from the P2 supernatants at 600 nM to enrich for the resistant virus population. This passage showed peak RT activity on day 14 (Fig. 2) and was used to inoculate the fourth passage (P4), which was grown in the presence of 1,200 nM peptide. RT activity from this culture peaked on day 13. Cellular DNA from this culture was harvested for cloning the env gene from the resistant viruses.
Mapping resistance.
To determine residues responsible for resistance, the env genes from integrated proviruses were sequenced and compared with those from control cultures grown in the absence of selection. Direct sequencing of the entire env gene from cellular proviral DNA in duplicate P4 cultures revealed only two mutations, one each in the N- and C-HRs, which were present in duplicate resistant cultures but not in control cultures. An arginine was substituted for glutamine at position 577 (Q577R, numbering referenced to that for HXB2 strain in the Los Alamos National Laboratory database) in the N-HR, and a lysine was substituted for glutamic acid at position 648 (E648K) in the C-HR (Fig. 1). No consistent mutations were seen in gp120. These results were confirmed by sequencing five Env clones, all of which contained the same two gp41 mutations (not shown). Sequencing of serial passages also showed that the C mutation E648K first emerged at P2 and persisted in subsequent cultures, while the N mutation Q577R appeared at P3 (not shown). It is unclear whether the resistance pathway seen here requires sequential acquisition of these mutations.
The mutations were introduced into the JR-CSF Env expression vector. Env proteins produced from these clones were used to make pseudotypes with an Env-deficient HIV luciferase reporter virus to verify the contributions of each mutation to resistance. WT Env, the reconstructed Env mutants with single mutations in the N-HR (N) and C-HR (C), and the double mutant (NC) Env all expressed high levels of processed Env that were equally well incorporated into pseudovirions (not shown). WT and mutant pseudoviruses, which were normalized for p24, did not show statistically significant differences in single-round infectivity assays, although the pseudovirus containing the N mutation trended towards reduced infectivity (not shown).
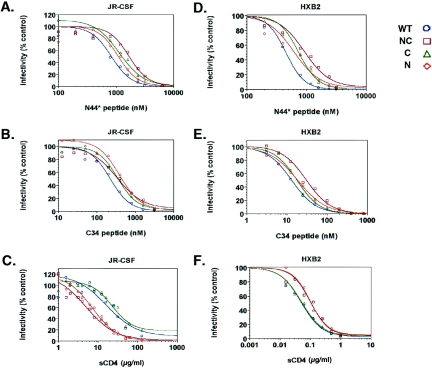
The IC50s for the N44* peptide were determined for each mutant pseudovirus. Pseudoviruses containing the NC and C mutations exhibited approximately 2- and 1.7-fold increases in resistance, respectively, relative to the WT, while the pseudovirus with the N mutation, which did not emerge in culture in the absence of the C mutation, showed a slight increase in resistance that did not reach statistical significance (Fig. 3A and Table 1). The level of resistance conferred by the NC mutations was lower than expected given the near-wild-type kinetics of the replicating resistant virus after several rounds of selection. This finding may relate to the single-round reporter pseudotype assay, which does not amplify resistance in multiple rounds of infection, or to the use of target cells that express high levels of receptors. We also note that the WT Env showed a higher IC50 in the single-round reporter assay (approximately 600 nM) than in the replicating infection assay (approximately 100 nM). We are currently creating infectious molecular clones containing resistant Env proteins to determine resistance levels for replicating virus. Nevertheless, the resistance conferred by the NC and C mutations was statistically significant and likely represents initial steps in resistance that would confer a strong advantage in a replicating culture under selection. Additional selection would probably lead to accumulation of more mutations that increase resistance.
FIG. 3.
Dose-response curves for inhibitors against WT and mutant pseudoviruses containing N, C, or NC resistance mutations. (A) Inhibition of the JR-CSF pseudoviruses with the N44* peptide. (B) Inhibition of the JR-CSF pseudoviruses with the C34 peptide. (C) Inhibition of the JR-CSF pseudoviruses with sCD4. (D) Inhibition of the HXB2 pseudoviruses with the N44* peptide. (E) Inhibition of the HXB2 pseudoviruses with the C34 peptide. (F) Inhibition of the HXB2 pseudoviruses with sCD4. Luciferase activity for each pseudovirus was normalized to its control culture without inhibitor. Results in panels A to C (JR-CSF pseudoviruses) are averages from all experiments. Results in panels D to F (HXB2 pseudoviruses) are representative graphs from at least five independent experiments.
TABLE 1.
Sensitivities of WT and mutant pseudoviruses against a panel of inhibitors
| Pseudovirus | Sensitivitya to:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N44*
|
Five-helix inhibitor
|
C34
|
T20
|
sCD4
|
||||||
| GM (nM) | Ratio (P) | GM (nM) | Ratio (P) | GM (nM) | Ratio (P) | GM (nM) | Ratio (P) | GM (nM) | Ratio (P) | |
| JR-CSF | ||||||||||
| WT | 696 | 1.00 | 131 | 1.00 | 198 | 1.00 | 969 | 1.00 | 23.33 | 1.00 |
| NC | 1,430 | 2.05 (<0.001) | 696 | 5.31 (<0.001) | 324 | 1.63 (0.009) | 1,895 | 1.96 (0.003) | 7.12 | 0.31 (<0.001) |
| N | 897 | 1.29 (0.06) | 276 | 2.11 (<0.001) | 382 | 1.93 (<0.001) | 1,047 | 1.08 (0.9) | 8.34 | 0.36 (<0.001) |
| C | 1,184 | 1.7 (<0.001) | 275 | 2.10 (<0.001) | 311 | 1.57 (0.02) | 1,440 | 1.49 (0.087) | 30.69 | 1.32 (0.3) |
| HXB2 | ||||||||||
| WT | 421 | 1.00 | NDb | 8.95 | 1.00 | ND | 0.062 | 1.00 | ||
| NC | 924 | 2.19 (<0.001) | ND | 25.12 | 2.81 (<0.001) | ND | 0.104 | 1.68 (0.006) | ||
| N | 577 | 1.37 (0.01) | ND | 15.71 | 1.76 (0.02) | ND | 0.101 | 1.63 (0.009) | ||
| C | 668 | 1.58 (<0.001) | ND | 14.46 | 1.62 (0.047) | ND | 0.058 | 0.93 (0.9) | ||
The GM of the 50% inhibitor concentration was calculated for each inhibitor and pseudovirus. The ratio is fold change of the mutant value compared to the WT value, calculated by dividing the GM for each mutant by the GM for the WT. Values shown represent averages from at least five independent experiments. The N44*, C34, and T20 peptides are shown in Figure 1. The five-helix inhibitor is described in Results. Statistically significant changes are in boldface.
ND, not done.
The importance of the combined NC mutations in conferring resistance to N-peptide inhibitors was confirmed by testing other N-peptide inhibitors. One such inhibitor, called the five-helix inhibitor, folds into a 6HB-like structure except that one face of the trimeric N-HR coiled coil is exposed because it lacks a C-HR (33). Significantly, the NC mutations conferred a higher level of resistance to the five-helix inhibitor, exhibiting a 5.3-fold increase in resistance for the double mutant, with each single mutation also conferring significant resistance (Table 1). Similarly, a higher level of resistance (sixfold) was also seen against another stabilized N-HR coiled-coil inhibitor, called IZN36 (14), which contains the N-HR fused to a heterologous coiled-coil trimer (not shown). The reasons for increased resistance against these stabilized, N-coiled-coil trimeric inhibitors compared with the unmodified N44* peptide are unclear, but it could be due to slight differences in the coiled-coil structure imposed by linker or chimeric residues present in the modified constructs but absent in the unmodified N44* peptide. Also, unlike the stabilized N-HR trimers, N44* may be able to bind gp41 in more than one way, because it likely exists as a mixture of monomers, dimers, trimers, and higher-order oligomers (14). Trimeric N44* binds the C-HR, but monomeric (or dimeric) N44* could bind the N-HR by intercalating into the endogenous gp41 coiled coil, as has been previously described for a mutant N peptide (3). Although the occurrence of resistance mutations in both N- and C-HRs might suggest that the N44* peptide targets both N- and C-HRs, our analyses of the resistance residues do not provide clear support for this model, as discussed below.
Determination of sensitivity to other inhibitors.
To further characterize the resistant Env proteins, we assessed sensitivity to other entry inhibitors (Fig. 3B and C). Surprisingly, the N and C mutations also conferred resistance to two overlapping C-peptide fusion inhibitors, C34 and T20 (Fig. 1). For the C34 peptide, each mutation provided an approximately 1.6- to 2-fold increase in resistance (Fig. 3B), but only the double NC mutant provided significant resistance (2-fold) for T20 (Table 1). Cross-resistance between the N- and C-peptide inhibitors was not expected because the peptides bind to different sites of gp41. However, this finding is consistent with the peptide inhibiting the same step. In addition, the N mutation increased sensitivity to inhibition by sCD4. Both the N and NC Env proteins were about 2.5-fold more sensitive to sCD4, while the C Env had no effect (Fig. 3C), indicating that the N mutation dominates the altered phenotype to sCD4.
Transfer of resistance to an X4 virus.
Because the NC mutations appeared to affect global fusogenic properties of Env, we speculated that the resistance mutations identified residues that were generally important in regulating Env-mediated fusion. We tested this hypothesis by introducing the resistance mutations into the HXB2 Env, which uses the CXCR4 coreceptor instead of CCR5, and assessing the transferability of the resistant phenotype. Dose-response curves with the N- and C-peptide fusion inhibitors showed the same pattern of resistance (Fig. 3D and E). Resistance to the N peptide was approximately twofold for the NC pseudovirus (Table 1). Cross-resistance to the C34 peptide was slightly more pronounced for the HXB2 Env, with the NC mutant demonstrating 2.8-fold-increased resistance against the C peptide relative to the WT. As with the JR-CSF Env, the N and NC mutations affected sensitivity to sCD4, with the N mutation again being dominant (Fig. 3F). Curiously, however, the mutations conferred increased resistance to sCD4 instead of increased sensitivity as was seen with the JR-CSF Env. Residue 577, therefore, plays a role in receptor activation in both X4 and R5 strains.
Despite the ability to transfer the resistance phenotype with substitutions in the same two residues, the N mutation, alone or combined with the C mutation, had a severe debilitating effect on function of the HXB2 Env (not shown). Previously, we showed that the Q577R mutation in the HXB2 Env reduced infectivity by >90% (40). We verified this markedly reduced activity when we reconstructed a new HXB2 N Env, along with the HXB2 C and NC Env proteins. All HXB2 pseudoviruses incorporated equal amounts of Env (not shown). Fortunately, the high titer of the HXB2 pseudovirion stocks, even with >90% reduced activity, still allowed us to obtain statistically valid data on sensitivity to inhibitors. Nonetheless, the fitness cost of the N mutation indicates that the HXB2 Env would likely take a different resistance pathway or require accumulation of additional compensatory mutations to restore HXB2 Env activity.
Assessment of the impact of resistance mutations on six-helix interactions.
As the peptides are believed to inhibit gp41 by forming a peptide-gp41 6HB, we modeled the N and C mutations in the 6HB by using coordinates from a high-resolution structure (43) to gain insights into the resistance mechanism. The C mutation 648K involved a “g” residue in the heptad repeat, which faces away from the N and C helices. The N mutation 577R is an “e” residue of the heptad, which lies in the lip of the hydrophobic pocket and does not appear to provide contacts that are critical for docking C-helix residues into the pocket. Modeling thus did not suggest that the resistance mutations would impair peptide binding to gp41. Earlier in vitro mutagenesis studies also showed that position 577 was a notable exception among “e” and “g” residues in the N-HR in tolerating mutations that preserved Env function (41).
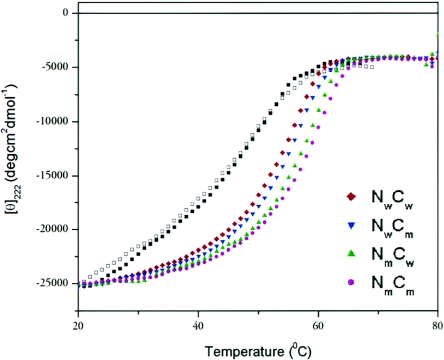
To evaluate how the mutations might affect peptide binding to gp41 and the endogenous (viral) 6HB, we performed thermal denaturation studies using mixtures of synthetic N and C peptides corresponding to wild-type HR (Nw or Cw, respectively), HR containing the N or C resistance mutations (Nm or Cm, respectively), and the selecting N peptide (N44*). All N- and C-peptide mixtures showed circular dichroic spectra with high α-helical content (not shown), consistent with previous reports (24). The wild-type peptide pair (NwCw) was trimeric as determined by sedimentation equilibrium (not shown), and thermal denaturation studies showed that the 6HB exhibited a simple two-state unfolding, with a Tm of ∼55°C (Fig. 4). The melting temperature is considerably lower than other reported measurements for the HIV-1 or simian immunodeficiency virus core gp41 domains (24). This may be due in part to the use of relatively long N peptides that extend beyond contacts with the C peptides. The peptide pairs involving the N44* peptide exhibited complex unfolding, with more than one transition observed. For the N44*Cw and N44*Cm peptide mixtures, these transitions occurred in the ranges of 43 to 49°C and 52 to 57°C, respectively (Fig. 4). The comparatively decreased thermal stabilities of 6HB interactions involving the N44* peptide are likely due to the isoleucine-to-glycine substitution in the second residue of the inhibitor, which may have facilitated the development of resistance.
FIG. 4.
Thermal denaturation studies of equimolar mixtures of the N and C peptides. Circular dichroism measurements were recorded at 222 nm as a function of temperature. Two independent experiments gave identical results. The melting curves for N44*Cw and N44*Cm are indicated by the open and closed squares, respectively; the identities of the other curves are as indicated.
A pattern of increasing thermal stability of the 6HB emerged with the development of the resistance mutations. As shown in Fig. 4, each of the resistance mutations further stabilized the endogenous 6HB. The first mutation appeared in the C-HR. The NwCm peptide pair, which modeled the 6HB of this partially resistant virus, showed a slightly increased Tm relative to wild type (NwCw). The effect of the C mutation on interactions with the N44* peptide is difficult to interpret because of the complex unfolding seen with the N44* peptide mixtures. The second mutation occurred in the N-HR on the background of the C mutation and further increased 6HB stability. The peptide pair modeling this resistant 6HB (NmCm) exhibited the greatest thermal stability, corresponding to a 3.7°C increase in Tm relative to that of the wild-type (NwCw) peptide pair. Notably, the N mutation in gp41 could not affect the 6HB formed by N44* peptide binding to the C-HR (N44*Cw). An additional peptide pair (NmCw) also showed that the N mutation alone increased 6HB thermal stability, but this virus did not emerge in culture. The biological significance of small changes in Tm in peptide models of the 6HB, especially how such changes might relate to the low-level resistance observed here, is unknown. However, these data raise the intriguing possibility that the virus evolved resistance by further stabilizing the endogenous (viral) 6HB, especially in a way that favored the endogenous HR over the inhibitor. The emergence of a stabilized 6HB through combined NC mutations could also contribute to cross-resistance to C peptides.
DISCUSSION
The pathway identified here for early resistance to the N-peptide fusion inhibitor N44* highlights important structure-function relationships in Env that inform treatment strategies and provide insights into Env-mediated fusion. We show that two mutations, one each in the N- and C-HRs, confer a two- to fivefold increase in resistance to several N-peptide fusion inhibitors, as well as cross-resistance to C-peptide fusion inhibitors. The N mutation also altered viral sensitivity to sCD4. Thus, the resistance mutations reveal residues that broadly influence Env activation and fusion, as demonstrated in two Env proteins that use different coreceptors.
What do these findings tell us about mechanisms of resistance and Env-mediated fusion? In theory, several mechanisms of resistance to peptide fusion inhibitors are possible. In one case, resistance could occur directly by mutation of contact residues that impair inhibitor binding, as has been suggested for some T20-resistant viruses (31, 36). Alternatively, resistance could arise indirectly, by reducing inhibitor access or competitiveness to the binding site. Reduced access could reflect (i) steric issues or (ii) decreased time that the binding site is available to the inhibitor due to kinetics of conformational changes and fusion. Reduced inhibitor competitiveness could occur by making contacts in the endogenous 6HB more favorable than those involving the inhibitor. In the case of our escape mutant, our results appear to rule out the direct mechanism. The cross-resistance against the N- and C-peptide inhibitors argues against resistance being due to mutation of contact residues, because the N and C peptides make distinct contacts with gp41. Molecular modeling also does not support a model in which the resistance mutations impair helical interactions. Most convincing, the thermal denaturation studies indicate that the resistance mutations could actually stabilize 6HB interactions rather than destabilize them.
The biophysical experiments introduce thermodynamic considerations into the discussion of resistance. In our peptide models of the 6HB (Fig. 4), the resistance mutations increased the structural stability of the resistant 6HB (NmCm) relative to the wild-type (sensitive) 6HB (NwCw). Interestingly, the mutation in the (noncognate) N-HR benefits the endogenous 6HB (NmCm), but it cannot benefit the 6HB formed by N44* binding to the C-HR (N44*Cm). It thus appears that the virus evolved a way to improve endogenous 6HB stability without improving inhibitor binding. The relevance of this pathway as a mechanism of resistance is supported by a recent study of a patient who developed resistance to T20 (Enfuvirtide) by generating point mutations in the N- and C-HRs (2). Although the mutations reported by Baldwin et al. (2) differ from those described here, it is striking that in vivo resistance to T20 (a C-peptide inhibitor) involved a mutation in the noncognate C-HR that resulted in a 3°C improvement in stability of the resistant 6HB, analogous to our in vitro findings involving an N-peptide fusion inhibitor.
Thermodynamic arguments relating to peptide inhibition suggest a model in which peptide is in direct competition with the endogenous HR at some step of the fusion process. With respect to the increased stability of the resistant 6HB, our data are consistent with an equilibrium model of inhibition by peptide proposed by Caffrey et al. (4). Alternatively, it is thought that the free energy released during formation of 6HB helps overcome the free energy barrier for membrane fusion (27). Stabilization of the endogenous 6HB could simply release more energy to promote fusion pore stabilization and expansion that are needed for complete membrane merge (26), resulting in faster fusion kinetics and less time for the inhibitor to bind. Further experiments are under way to dissect the thermodynamic parameters contributing to enhanced stability of the resistant 6HB and its consequences for resistance.
Our data also support kinetic parameters in the development of resistance. In fact, the biophysical experiments showing that the 6HB involving the N44* peptide inhibitor is less stable than the sensitive (WT) 6HB (NwCw Tm > N44*Cw Tm) indicate that free energy considerations alone do not control inhibition, because N44* remains a potent inhibitor against WT virus. Rather, the data support a kinetic mechanism in which the inhibitor has preferential access to the gp41 target sites during refolding steps in virus entry. Experiments involving sCD4 further suggest kinetic parameters in resistance. In two different strains of HIV, the N mutation altered sensitivity to sCD4, indicating changes in the activation energy for the transition from native to fusion intermediate conformations. For the JR-CSF Env, which is much more resistant to sCD4 than the HXB2 Env, the N mutation increased sensitivity to sCD4, perhaps by destabilizing the native conformation. However, the opposite pattern was observed for the HXB2 Env, which is already easy to activate. This apparent discrepancy is difficult to interpret, but it is tempting to hypothesize that there is an optimal range of Env triggering that reduces the kinetic window for the inhibitor to bind Env. For the JR-CSF Env, easier receptor activation might allow more Env proteins to trigger and organize into fusion pores, speeding up fusion and acquisition of the peptide-resistant 6HB. For the HXB2 Env, reduced hair-trigger activation and consequent premature inactivation of Env might also lead to more efficient fusion kinetics. In this way the N mutation could influence the kinetics of fusion and the time available for the inhibitor to bind gp41. A kinetic model of fusion that is modulated by receptor density and affinity has been previously proposed to explain sensitivity to peptide fusion inhibitors (30).
Indirect models of resistance offer an appealing strategy of escape for dominant-negative inhibitors. Dominant-negative inhibitors mimic interactions in the target protein and would be expected to impose constraints on the development of resistance; mutations that impair inhibitor binding would also likely impair function of the target protein. It is likely that C-HR mutations that reduce binding to N peptides would also destabilize the 6HB comprised of endogenous HR (37). Such destabilizing mutations would also likely reduce infectivity of the mutant virus. A correlation between decreased 6HB stability and reduced infectivity has been reported for other mutations in gp41 (16, 22). Yet T20-resistant viruses can acquire resistance by mutating residues that directly impair peptide binding (2, 31, 36). A stretch of the N-HR including the GIV sequence has been shown to be important for T20 binding to gp41, and this region is frequently mutated in resistant isolates from T20-treated patients (1, 2, 38). A recent clinical study reported that T20-resistant viruses are less fit (23). Residues outside the N-HR, including gp120 sequences, however, have also been shown to modulate sensitivity to T20 (10, 11, 19, 29, 42). Given the mutability of Env, it is likely that several pathways for resistance can emerge, and a pathway may involve several factors that contribute to resistance.
Aside from providing insights into mechanisms of resistance, our escape mutant virus also identified residue 577 as having a role in both activation of Env-mediated fusion and stability of the 6HB. Thus, residue 577 helps to regulate more than one conformation of Env, including the fusogenic 6HB conformation and the native or fusion-intermediate conformations that influence Env activation. This dual effect appears to be analogous to the conformational switch region that has been recently described for the fusion protein (F) of a paramyxovirus (34), in which a region near the C-HR regulates both activation of F-mediated fusion and 6HB stability. Residues in the N-HR region of gp41 have previously been reported to be important for contacts with gp120 (6, 16, 25), further supporting the idea that this residue may be involved in a conformational switch. Notably, residue 577 lies in a hydrophobic pocket region of the N-HR, which has been proposed to be an important region for stabilizing the coiled coil (13) and a good target for developing inhibitors (7). The demonstration that residue 577 tolerates substitutions (40, 41), and indeed mutates under selection as shown here, has implications for drugs targeting this pocket.
In summary, our studies identify new viral determinants that confer cross-resistance to N- and C-type peptide fusion inhibitors and change susceptibility to inhibitors mimicking CD4. The N mutation, which altered sensitivity to sCD4, also shows that this region of N-HR can play a dual role in both receptor activation and 6HB stability. The combined N and C mutations confer more resistance against several inhibitors than the individual mutations (Table 1) and point to an indirect mechanism of resistance, where coupled changes in the N- and C-HRs may regulate several aspects of Env-mediated fusion to provide escape from a dominant-negative inhibitor. The importance of residues 577 and 648 in the development of resistance to peptide fusion inhibitors and the fusion process was not predicted from the 6HB high-resolution structures. Our findings underscore the complexity and plasticity of Env-mediated fusion and some of the challenges in designing fusion inhibitors.
Acknowledgments
We thank Ira Berkower and Marina Zaitseva (U.S. Food and Drug Administration, Bethesda, Md.) for critical reading of the manuscript. We also thank Michael Root (Jefferson University, Philadelphia, Pa.) for the five-helix inhibitor and Vladimir Chizhikov (U.S. Food and Drug Administration) for scientific and sequencing contributions.
This work was supported in part by an Intramural AIDS-Targeted Antiviral Program (IATAP) grant from the National Institutes of Health.
REFERENCES
- 1.Armand-Ugon, M., A. Gutierrez, B. Clotet, and J. A. Este. 2003. HIV-1 resistance to the gp41-dependent fusion inhibitor C-34. Antiviral Res. 59:137-142. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, C. E., R. W. Sanders, Y. Deng, S. Jurriaans, J. M. Lange, M. Lu, and B. Berkhout. 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J. Virol. 78:12428-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bewley, C. A., J. M. Louis, R. Ghirlando, and G. M. Clore. 2002. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled-coil of gp41. J. Biol. Chem. 277:14238-14245. [DOI] [PubMed] [Google Scholar]
- 4.Caffrey, M., J. Kaufman, S. Stahl, P. Wingfield, A. M. Gronenborn, and G. M. Clore. 1999. Monomer-trimer equilibrium of the ectodomain of SIV gp41: insight into the mechanism of peptide inhibition of HIV infection. Protein Sci. 8:1904-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderone, T. L., R. D. Stevens, and T. G. Oas. 1996. High-level misincorporation of lysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli. J. Mol. Biol. 262:407-412. [DOI] [PubMed] [Google Scholar]
- 6.Cao, J., L. Bergeron, E. Helseth, M. Thali, H. Repke, and J. Sodroski. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 67:2747-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 9.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 10.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, Z. Zhang, W. A. O'Brien, L. Ratner, G. M. Shaw, and E. Hunter. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J. Virol. 75:8605-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:F9-F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dwyer, J. J., A. Hasan, K. L. Wilson, J. M. White, T. J. Matthews, and M. K. Delmedico. 2003. The hydrophobic pocket contributes to the structural stability of the N-terminal coiled coil of HIV gp41 but is not required for six-helix bundle formation. Biochemistry 42:4945-4953. [DOI] [PubMed] [Google Scholar]
- 14.Eckert, D. M., and P. S. Kim. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. USA 98:11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 16.Follis, K. E., S. J. Larson, M. Lu, and J. H. Nunberg. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 76:7356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 18.He, Y., R. Vassell, M. Zaitseva, N. Nguyen, Z. Yang, Y. Weng, and C. D. Weiss. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J. Virol. 77:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heil, M. L., J. M. Decker, J. N. Sfakianos, G. M. Shaw, E. Hunter, and C. A. Derdeyn. 2004. Determinants of human immunodeficiency virus type 1 baseline susceptibility to the fusion inhibitors enfuvirtide and T-649 reside outside the peptide interaction site. J. Virol. 78:7582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilby, J. M., and J. J. Eron. 2003. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 348:2228-2238. [DOI] [PubMed] [Google Scholar]
- 21.Koshiba, T., and D. C. Chan. 2003. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J. Biol. Chem. 278:7573-7579. [DOI] [PubMed] [Google Scholar]
- 22.Liu, J., W. Shu, M. B. Fagan, J. H. Nunberg, and M. Lu. 2001. Structural and functional analysis of the HIV gp41 core containing an Ile573 to Thr substitution: implications for membrane fusion. Biochemistry 40:2797-2807. [DOI] [PubMed] [Google Scholar]
- 23.Lu, J., P. Sista, F. Giguel, M. Greenberg, and D. R. Kuritzkes. 2004. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to Enfuvirtide (T-20). J. Virol. 78:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 25.Lu, M., M. O. Stoller, S. Wang, J. Liu, M. B. Fagan, and J. H. Nunberg. 2001. Structural and functional analysis of interhelical interactions in the human immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning mutagenesis. J. Virol. 75:11146-11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markosyan, R. M., F. S. Cohen, and G. B. Melikyan. 2003. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol. Biol. Cell 14:926-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peden, K., and M. A. Martin. 1995. HIV: a practical approach, vol. 1, p. 21-45. IRL Press, Oxford, United Kingdom.
- 29.Poveda, E., B. Rodes, C. Toro, L. Martin-Carbonero, J. Gonzalez-Lahoz, and V. Soriano. 2002. Evolution of the gp41 env region in HIV-infected patients receiving T-20, a fusion inhibitor. AIDS 16:1959-1961. [DOI] [PubMed] [Google Scholar]
- 30.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roman, F., D. Gonzalez, C. Lambert, S. Deroo, A. Fischer, T. Baurith, T. Staub, R. Boulme, V. Arendt, F. Schneider, R. Hemmer, and J. C. Schmit. 2003. Uncommon mutations at residue positions critical for enfuvirtide (T-20) resistance in enfuvirtide-naive patients infected with subtype B and non-B HIV-1 strains. J. Acquir. Immune Defic. Syndr. 33:134-139. [DOI] [PubMed] [Google Scholar]
- 33.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 34.Russell, C. J., K. L. Kantor, T. S. Jardetzky, and R. A. Lamb. 2003. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J. Cell Biol. 163:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trivedi, V. D., S. F. Cheng, C. W. Wu, R. Karthikeyan, C. J. Chen, and D. K. Chang. 2003. The LLSGIV stretch of the N-terminal region of HIV-1 gp41 is critical for binding to a model peptide, T20. Protein Eng. 16:311-317. [DOI] [PubMed] [Google Scholar]
- 37.Wang, S., J. York, W. Shu, M. O. Stoller, J. H. Nunberg, and M. Lu. 2002. Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion. Biochemistry 41:7283-7292. [DOI] [PubMed] [Google Scholar]
- 38.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 40.Weng, Y., and C. D. Weiss. 1998. Mutational analysis of residues in the coiled coil domain of the human immunodeficiency virus type 1 gp41. J. Virol. 72:9676-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weng, Y., Z. Yang, and C. D. Weiss. 2000. Structure-function studies of the self-assembly domain of the human immunodeficiency virus type 1 transmembrane protein. J. Virol. 74:5368-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, L., S. Hue, S. Taylor, D. Ratcliffe, J. A. Workman, S. Jackson, P. A. Cane, and D. Pillay. 2002. Minimal variation in T-20 binding domain of different HIV-1 subtypes from antiretroviral-naive and -experienced patients. AIDS 16:1684-1686. [DOI] [PubMed] [Google Scholar]
- 43.Yang, Z. N., T. C. Mueser, J. Kaufman, S. J. Stahl, P. T. Wingfield, and C. C. Hyde. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 angstrom resolution. J. Struct. Biol. 126:131-144. [DOI] [PubMed] [Google Scholar]