Abstract
Cuproptosis is a novel cell death mechanism caused by copper overload, with FDX1 serving as the key regulator. LncRNAs are known to play a significant role in the aberrant regulation of gene expression in hepatocellular carcinoma (HCC). In this study, we investigated the biological role of the LINC02362/hsa-miR-18a-5p/FDX1 axis in HCC. We first explored the expression pattern, prognostic value, biological functions, drug sensitivity, and immune effect of FDX1. Using bioinformatics techniques, we then predicted several potential target lncRNAs and miRNAs. We identified a lncRNA-miRNA-FDX1 axis based on the ceRNA mechanism. In vitro experiments were conducted to validate the relationship between the lncRNA-miRNA-FDX1 axis and its biological effects in HCC. Finally, we investigated the relationship between the LINC02362/hsa-miR-18a-5p/FDX1 axis and oxaliplatin-induced cuproptosis in HCC. Our findings indicated that FDX1 expression was downregulated in HCC tissues; however, elevated FDX1 expression correlates with improved prognosis and heightened sensitivity to oxaliplatin. We confirmed that LINC02362 binds to and directly regulates the expression of miR-18a-5p, with FDX1 a target of miR-18a-5p. Experimental results suggested that upregulating LINC02362/hsa-miR-18a-5p/FDX1 axis suppressed the proliferation of HCC cells. Furthermore, LINC02362 knockdown led to a reduction in copper concentration and resistance to elesclomol-Cu. We also discovered that augmenting the LINC02362/hsa-miR-18a-5p/FDX1 axis could bolster the sensitivity of HCC to oxaliplatin through cuproptosis. This work presents the LINC02362/hsa-miR-18a-5p/FDX1 axis as a novel pathway that triggers cuproptosis and enhances the sensitivity of HCC to oxaliplatin, presenting a promising therapeutic avenue to combat oxaliplatin resistance in HCC.
Keywords: Hepatocellular carcinoma, cuproptosis, oxaliplatin, lncRNA, FDX1
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related mortality, with an estimated 841,000 cases diagnosed and 782,000 deaths annually [1]. Surgical resection stands as the gold standard for treatment for HCC. However, despite advancements in therapeutic strategies, challenges such as metastasis, recurrence, and drug resistance persist, resulting in a modest overall survival rate.
In recent years, the scientific community has been committed to discovering methods to induce cancer cell death, as it presents a viable approach to cancer treatment. Different types of cell death have been identified based on varying signal cascades and distinct molecular mechanisms, including apoptosis [2], necroptosis [3], pyroptosis [4], and ferroptosis [5]. Tsvetkov et al. introduced cuproptosis, a unique mechanism instigated by copper toxicity, setting it apart from other recognized programmed cell death mechanisms [6]. Their research highlighted that an excess of intracellular copper can lead to the aggregation of lipoylated proteins, loss of iron-sulfur (Fe-S) cluster-containing proteins, and induction of HSP70, cumulatively inducing proteotoxic stress and cytotoxicity. As a key regulator in cuproptosis, FDX1 is an upstream regulator of protein lipoylation. Previous study have shown that FDX1 expression is significantly reduced in multiple cancer types and is correlated with immune infiltration level [7]. While some studies have confirmed the diminished expression of FDX1 in HCC and its correlation with improved survival [7,8], the role of FDX1 in HCC remains elusive and warrants further exploration.
Past research has indicated that copper transporters are fundamental to the biological response to antitumor platinum drugs such as cisplatin, carboplatin, and oxaliplatin [9]. The copper influx transporter CTR1 is pivotal for the cellular uptake of platinum drugs [10-13]. Additionally, proteins like Menkes (ATP7A) and Wilson disease protein (ATP7B) influence the efflux and sequestration of cisplatin, consequently amplifying tumor resistance to platinum drugs [14-18]. The expression levels of the soluble Cu chaperone ATOX1 are also linked to cisplatin sensitivity [19]. Therefore, cuproptosis might play a crucial role in platinum drug-induced cell death in HCC.
LncRNAs, which regulate gene expression across transcriptional, post-transcriptional, and epigenetic levels, are instrumental in various diseases, including cancer [20]. For instance, Liu et al. found that SP1-induced lncRNA DUBR enhances stemness and oxaliplatin resistance in HCC [21], while another study by Liu et al. reported that lncSNHG6 synchronizes cholesterol sensing with mTORC1 activation in HCC [22]. Earlier investigations have also shown that lncRNAs can influence diverse cell death processes in HCC [23-26], reinforcing the idea that lncRNAs are vital modulators of HCC biological phenotypes.
In our study, we first delineated the expression pattern, prognostic implications, copy number, DNA methylation level, functional enrichment analysis, drug sensitivity, and immune effects of FDX1 in HCC. We then employed multiple databases to identify potential lncRNA/miRNA/FDX1 axes and verified their regulatory significance in HCC using in vitro experiments. Additionally, we explored the interplay between the lncRNA/miRNA/FDX1 axis, proliferation, cuproptosis, and oxaliplatin resistance, aiming to uncover potential therapeutic targets and elucidate the relationship between cuproptosis and oxaliplatin resistance in HCC.
Materials and methods
Data collection
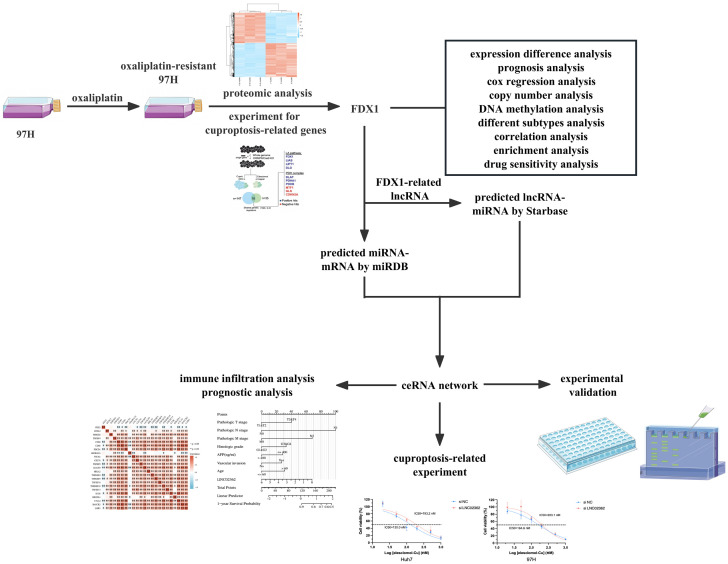
A schematic of the study workflow is illustrated in Figure 1. RNA-seq and miRNA-seq data of 424 samples (comprising 374 tumor and 50 normal samples), along with their clinical data, were sourced from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). Additional RNA-seq data and associated clinical data from HCC samples in the GSE14520 dataset were retrieved from the Gene Expression Omnibus (GEO) database (http://ww.ncbinlm.nih.gov/geo/). Ensembl IDs were processed and converted to gene symbols. The DNA methylation data was downloaded from the UALCAN database (http://ualcan.path.uab.edu). The copy number variation data was downloaded from the cBioPortal database (http://www.cbioportal.org). The gene interaction network was obtained from the String database (https://ngdc.cncb.ac.cn/databasecommons/database/id/62), while lncRNA targets were predicted using LncBase v3 (https://diana.e-ce.uth.gr/lncbasev3/interactions) and miRNA targets from Starbase (https://starbase.sysu.edu.cn/).
Figure 1.
The overall flowchart of the study.
Differential expression analysis
According to the tandem mass tag (TMT) labeling, proteins with FC > 1.2 or FC < 0.833 were considered as differentially expressed proteins (DEPs). RNA-seq data were categorized into low and high FDX1 expression groups from the TCGA HCC cohort. Differential analysis was conducted using the “DESeq2” package [27]. LncRNAs with |logFC| > 1 and an adjusted p-value < 0.05 were deemed as differentially expressed lncRNAs (DElncRNAs).
Function enrichment analysis
To elucidate the underlying mechanism associated with FDX1, gene function enrichment analysis was performed using the Gene Oncology (GO) data (c5.go.v7.5.1.symbols.gmt) and Kyoto Encyclopedia of Genes and Genomes (KEGG) data (c2.cp.kegg.v7.4.symbols.gmt) from the molecular signature dataset. Only results with adjusted p-value < 0.05 were considered statistically significant.
Prognostic analysis
Survival curves between groups were generated by Kaplan-Meier analysis and log-rank test. Univariable analyses and multivariable analyses of risk factors for overall survival (OS), risk factors for progression-free survival (PFS) were performed with Cox regression models. Variables with P < 0.05 from univariable analyses were included in multivariable analyses. Additionally, both univariable and multivariable analyses for OS of hub genes were executed using Cox regression models. Results were visualized as forest plots using the “forestplot” R package. For the construction of the nomogram model, factors related to OS from multivariable Cox regression analysis were utilized. Nomograms were developed using the “rms” and “survival” R packages. A P-value < 0.05 was considered statistically significant for patients.
The role of FDX1 in predicting drug sensitivity
To evaluate the role of FDX1 in estimating the response to HCC treatment, the half-maximal inhibitory concentration (IC50) for oxaliplatin and other standard HCC drugs in the TCGA cohort was calculated using the ‘pRRophetic’ R package [28].
Correlation analysis
Given that the data from RNA-seq, miRNA-seq, and IC50 did not follow a normal distribution, the Spearman correlation was employed for analysis. Results from the Spearman correlation were visualized with the “ggplot2” R package.
Immune infiltrate analysis
Through single-sample gene set enrichment analysis (ssGSEA) with the R package “GSVA” and CIBERSORT databases, we ascertained the infiltration levels of 28 immune cell types and 22 immune cell types individually in TCGA. The stromal score, immune score, and ESTIMATE score from TCGA tumor samples were determined using the R package “ESTIMATE”. Furthermore, the relationship between FDX1 expression and immune checkpoint genes was analyzed using Spearman’s correlation method.
Cell lines and cell culture
The normal liver cell line L02, and human HCC cell lines HepG2, PLC/PRF/5, Hep3B (Cell Bank of the Chinese Academy of Sciences, Shanghai, China), Huh7 (Japanese Cancer Research Resources Bank), MHCC97H and HCCLM3 (Liver Cancer Institute, Fudan University, Shanghai, China) were routinely cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco) containing 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (Invitrogen, USA) at 37°C in humidified air with 5% CO2. Here we used oxaliplatin-resistant MHCC97H (97H-OXA) cell line which was established in our previous study [29]. In brief, MHCC97H cells were exposed to oxaliplatin (Sigma, St. Louis, MO) for three months, with concentrations incrementally increased to 25 μM. 97H-OXA was consistently cultured in 1 μM oxaliplatin to maintain its resistance.
Cell transfection
The LINC02362 small interfering RNA (siRNA) and negative control were purchased from Tsingke Biotechnology Co., Ltd. (Beijing, China). The hsa-miR-18a-5p mimic, negative control, and overexpression plasmid pcDNA-FDX1 were purchased from Genomeditech Co., Ltd. (Shanghai, China). The sequences of these synthesized siRNAs, mimic RNAs, and plasmids are provided in Table S1. Transfections were carried out using Lipofectamine 8000 (Beyotime Biotechnology, China) as per the manufacturer’s instructions.
RNA extraction and quantitative real-time PCR
Total RNA, including miRNA, was extracted from cells using the RNA-Quick Purification Kit (EZBioscience, China). Complementary DNA (cDNA) was synthesized using the 4× EZscript Reverse Transcription Mix II (EZBioscience). For mRNA and lncRNA, single-stranded cDNA was amplified with a 2× SYBR Green qPCR Master Mix (ROX2 plus; EZBioscience). MiRNA analysis was performed using 2× SYBR Green Mix and miDETECT A TrackTM Uni-Reverse Primer (RiboBio Company, Guangzhou, China). The expression of lncRNAs, mRNAs, and miRNAs were normalized to the expression levels of 18S RNA, β-actin, and U6, respectively. Primer sequences are listed in Table S2.
Reagents and antibodies
Tetrathiomolybdate (TTM, E1166) was obtained from Selleckchem (Houston, TX, USA). Z-VAD-FMK (HY-16658B), Necrostatin-1 (Nec-1, HY-15760), Ferrostatin-1 (Fer-1, HY-100579), N-Acetylcysteine (NAC, HY-B0215) and Chloroquine (CQ, HY-17589A) were purchased from MedChemExpress (MCE, China). The following antibodies were used: FDX1 (ab108257, Abcam, UK), GAPDH (AF1186, Beyotime, China).
Western blot analysis
Proteins were extracted from cells with RIPA cell lysis (Beyotime, China) containing 1 mM of phenylmethanesulfonyl fluoride (PMSF, Beyotime). Concentrations were determined using a BCA kit (Beyotime, China). Proteins were run on SDS-PAGE and then blotted into PVDF membranes (Millipore). After blocking with 5% skimmed milk for 1 h, membranes were incubated with primary antibodies at 4°C overnight, followed by incubation with HRP-conjugated secondary antibodies for 1 h at room temperature. Staining was visualized with ECL reagents (Yeasen, China) by the imaging system.
Luciferase reporter assay
Huh7 and MHCC97H cells were seeded into 96-well plates and co-transfected with specific combinations of luciferase expression plasmids: either WT or mutated versions of LINC02362 (pmirGLO-LINC02362-WT or pmirGLO-LINC02362-Mut), and either WT or mutated 3’ untranslated regions (3’UTR) of FDX1 (pmirGLO-FDX1-3’UTR-WT or pmirGLO-FDX1-3’UTR-Mut), in conjunction with either a miR-18a-5p mimic or a negative control. After a 48-hour incubation, both firefly and Renilla luciferase activities were assessed using a dual-luciferase Assay Kit (Yeasen, China) on a GloMax 20/20 Luminometer (Promega, Madison, WI, USA) according to the manufacturer’s protocol.
Measurement of intracellular copper
Cells were seeded in 6-cm plates and incubated overnight; they were then treated with 200 nM elesclomol-Cu (1:1 ratio) for 24 hours. After treatment, cells were collected and resuspended in 120 μL ddH2O. The intracellular copper concentration was then determined using the Copper Assay Kit (E-BC-K775-M, Elabscience, China) according to the manufacturer’s instructions.
ROS assay
Huh7 and MHCC97H cells were seeded in 6-cm plates. After seeding, 200 nM elesclomol-Cu (1:1 ratio) was added and the cells were incubated for 24 hours. Post-incubation, cells were treated with a 10 μM dichlorodihydrofluorescein diacetate probe (Beyotime Biotechnology, China) for 20 minutes at 37°C. Fluorescence intensities, representing reactive oxygen species (ROS) levels, were subsequently captured using a flow cytometer using excitation and emission spectra of 488/525 nm.
Cell viability assay
Approximately 2000 HCC cells per well were seeded into 96-well plates. These cells were then exposed to varying doses of the test reagents for 24 hours. Then, 100 μL of culture medium containing 10% CCK8 solution (Yeasen, China) was added. After a 2-hour incubation at 37°C, the optical density was determined at 450 nm using a spectrophotometer.
Half-maximal inhibitory concentration assay (IC50)
Cells were seeded into 96-well plates at a density of 2000 cells per well. After a 24-hour incubation, elesclomol-Cu (1:1 ratio) or oxaliplatin was introduced at corresponding concentrations for 48 hours. Subsequently, 100 μL of culture medium containing 10% CCK8 reagent (Yeasen, China) was added to each well and the plates were incubated for an additional 2 hours at 37°C. The absorbance was then determined at 450 nm using a Multiskan Spectrum spectrophotometer (Thermo Scientific, USA).
TMT-labelled quantitative proteomics
To discern protein expression differences between the HCC cell line MHCC97H and the oxaliplatin-resistant 97H-OXA, protein extraction and digestion were performed using the filter-aided sample preparation (FASP) method as delineated by Matthias Mann [30]. 100 μg of peptide mixture from each sample was labeled by the TMT reagent according to the manufacturer’s instructions (Thermo Scientific). All samples were analyzed at Shanghai Applied Protein Technology.
Statistical analysis
Data were evaluated using GraphPad Prism 9 and RStudio, along with its related packages. Continuous data, either between two groups or among multiple groups, were compared using the Student’s t-test or one-way ANOVA. For categorical data, comparisons were made using the Chi-square test or Fisher’s exact test. All tests were two-sided, with a p-value < 0.05 indicating statistical significance. Significance levels were denoted as follows: P < 0.05 as *, P < 0.01 as **, and P < 0.001 as ***.
Results
The role of FDX1 in HCC
Recent research has unveiled a novel mechanism wherein copper overload induces cell death, termed “cuproptosis”, with FDX1 being the pivotal regulator of this process [6]. We systematically explored the vital function of FDX1 in HCC through bioinformatics analysis. First, we observed that FDX1 expression is consistently downregulated in HCC tissues from both TCGA and GSE14520 datasets (Figure 2A). Kaplan-Meier survival curves revealed that patients exhibiting elevated FDX1 expression generally experience improved survival outcomes (TCGA-OS: HR=1.306, P=0.0353; GSE14520-OS: HR=1.977, P=0.0008) (Figure 2B). Univariable and multivariable Cox regression analyses were performed on the GSE14520 dataset to assess risk factors for both OS and PFS. As shown in Table S3, expression of FDX1, predicted metastasis risk, main tumor size, multinodular, TNM staging, BCLC staging, and AFP levels correlate with OS in the univariable analysis. The multivariable cox regression analysis confirmed multinodular, TNM staging and BCLC staging as independent predictors. Univariable analysis showed FDX1, predicted metastasis risk, gender, main tumor size, TNM staging and BCLC staging were related to PFS (Table S4). Furthermore, gender, TNM staging, and BCLC staging were singled out as independent predictors in the multivariable analysis. In addition, FDX1 copy number was significantly correlated with mRNA expression level (r=0.34, P < 0.001) (Figure 2C). We also found a decrease in the DNA methylation level of FDX1 in HCC samples (P < 0.001, Figure 2D). The expression of FDX1 was further classified based on several crucial HCC criteria (Figure S1) [31-39]. These results demonstrate that HCC patients with low level expression of FDX1 were generally categorized within the poor prognosis subtype of HCC (including iCluster 1 subclass, subtype A based on NCI proliferation signature, subtype A based on hepatic stem cells signatures, high risk scores based on Seoul National University recurrence signature, cholangiocarcinoma-like, exhibited similarity to non-differentiated RNA clustering phenotypes (Hoshida C2), and high risk scores based on a gene expression signature of 65 genes).
Figure 2.
The role of FDX1 in HCC. A. The mRNA expression level of FDX1 in HCC tissues and normal tissues through the TCGA and GSE14520 database. B. Kaplan-Meier survival curves for HCC patients with high or low FDX1 expression based on TCGA and GSE14520 database. C. The putative copy-number alterations of FDX1 and the relationship between the expression level and the copy number of FDX1 in HCC. D. The correlation of FDX1 expression and DNA methylation level based on TCGA database.
Functional enrichment analyses of FDX1
Using the String database, we constructed an interaction network between FDX1 and its associated genes (Figure S2A). Subsequent GO and KEGG functional enrichment analyses were undertaken to elucidate potential mechanisms linked to FDX1 (Figure S2B, S2C). Interestingly our findings reveal that FDX1, along with its associated genes, predominantly enriches functions associated with cation channels, cation transport, and cation transmembrane transporter related activities.
The role of FDX1 in predicting drug sensitivity
To assess the role of FDX1 in evaluating the response to HCC treatment, some common chemotherapy drugs used during the clinical treatment of HCC were analyzed respective to FDX1 levels using the ‘pRRophetic’ R package [28]. Drugs showing the most significant correlations included Brilanestrant, MIRA-1, PF-4708671, JQ1, Dihydrorotenone, Fulvestrant, SB505124, AZD2014, GSK269962A and RO-3306. Importantly, we identified a marked correlation between FDX1 expression and sensitivity to both cisplatin (r=-0.114, P=0.028) and oxaliplatin (r=-0.138, P=0.008) (Figure S3A-L).
Screening of target lncRNAs and target miRNAs
To identify potential lncRNAs that might regulate FDX1, we conducted a series of bioinformatics analyses. Gene expression profiles from TCGA revealed 3534 differentially expressed lncRNAs (DElncRNAs) in HCC samples compared to normal tissues, 69 FDX1-related lncRNAs, and 965 prognosis-related lncRNAs. From these datasets, we intersected the genes, yielding eight candidates: TMEM220-AS1, AC099508.2, LINC00261, LINC02362, AP001065.3, LINC01093, AC004160.1, and LINC02037 (Figure 3A). Co-expression analysis of these genes demonstrated a strong positive correlation between the eight lncRNAs and FDX1 (TMEM220-AS1, r=0.396, P < 0.001; AC099508.2, r=0.397, P < 0.001; LINC00261, r=0.356, P < 0.001; LINC02362, r=0.336, P < 0.001; AP001065.3, r=0.354, P < 0.001; LINC01093, r=0.345, P < 0.001; AC004160.1, r=0.401, P < 0.001; LINC02037, r=0.384, P < 0.001) (Figure 3B). All eight candidate genes exhibited higher expression in normal tissues (Figure 3C). Kaplan-Meier curves assessed the impact of these lncRNAs on HCC outcomes (TMEM220-AS1, log-rank P < 0.001; AC099508.2, log-rank P < 0.001; LINC00261, log-rank P=0.004; LINC02362, log-rank P=0.011; AP001065.3, log-rank P=0.023; LINC01093, log-rank P=0.044; AC004160.1, log-rank P=0.046; LINC02037, log-rank P=0.050) (Figure 3D). Using lncbase, potential miRNAs that might bind to the eight candidate lncRNAs were predicted. After intersecting potential miRNAs that might bind to FDX1 using Starbase, we identified five candidate miRNAs for HCC (Figure 3E). Figure 3G shows that hsa-miR-140-5p and hsa-miR-576-5p were downregulated in tumor tissues, and only hsa-miR-18a-5p was upregulated in tumor tissues. Kaplan-Meier curves assessed the impact and diagnostic value of these miRNAs on HCC outcomes (hsa-miR-140-5p, log-rank P=0.505; hsa-miR-18a-5p, log-rank P=0.010; hsa-miR-18b-5p, log-rank P=0.106; hsa-miR-23c, log-rank P < 0.001; hsa-miR-576-5p, log-rank P=0.054) (Figure 3H). Based on the mechanism of ceRNA, we identified the LINC02362/hsa-miR-18a-5p/FDX1 axis as highly important in HCC outcomes. Subcellular localization of LINC02362 by the lncLocator platform was predicted to mainly be localized in the cytoplasm (Figure 3F). The Spearman correlation analysis was used to assess the correlation between LINC02362, has-miR-18a-5p, and FDX1 since the expression of these three genes did not conform to normal distribution. Analysis revealed a negative correlation between has-miR-18a-5p and LINC02362 (r=-0.195, P < 0.001) or FDX1 (r=-0.187, P < 0.001), and a positive correlation between LINC02362 and FDX1 (r=0.336, P < 0.001) (Figure 3I).
Figure 3.
Screening of target lncRNAs and target miRNAs. A. The overlapped lncRNAs of between the prognosis-related lncRNAs, FDX1-related lncRNAs and the DElncRNAs in TCGA. B. The co-expression analysis between 8 candidate lncRNAs and FDX1. C. The expression level of 8 candidate lncRNAs between HCC tissues and normal tissues. D. Kaplan-Meier survival curves for HCC patients with high or low lncRNA expression. E. The overlapped miRNAs of between the LncBase predicted miRNAs and the Starbase predicted miRNAs. F. The expression level of 5 miRNAs between HCC tissues and normal tissues. G. Kaplan-Meier survival curves for HCC patients with high or low miRNA expression. H. Analyzed the subcellular localization of the LINC02362 using the lncLocator platform. I. The Spearman correlation analysis between the LINC02362, has-miR-18a-5p, and FDX1. *P < 0.05, **P < 0.01, ***P < 0.001.
Validation of LINC02362/has-miR-18a-5p/FDX1 axis expression in HCC cells
We first evaluated the expression of LINC02362 and FDX1 across six HCC cell lines and normal liver cells. Both genes were frequently downregulated in HCC cell lines (Figure 4A, 4B). After transfecting Huh7 and 97H cells with LINC02362 siRNA, we observed a decrease in both LINC02362 and FDX1 expression in si-LINC02362 cells compared to the si-negative control cells, whereas hsa-miR-18a-5p expression increased (Figure 4C). To further validate the LINC02362/hsa-miR-18a-5p/FDX1 axis, we quantified FDX1 protein expression levels in si-negative control cells, si-LINC02362 cells, mir-negative control cells and hsa-miR-18a-5p mimic cells and found that the protein expression level of FDX1 is significantly downregulated in si-LINC02362 cells compared with si-negative control cells, as well as downregulated in hsa-miR-18a-5p mimic cells compared with mir-negative control cells (Figure 4D). The CCK8 assay further suggested that knockdown of LINC02362 promoted HCC cell proliferation (Figure 4E). Similarly, overexpression of hsa-miR-18a-5p also promoted proliferation in Huh7 and 97H cells (Figure 4F). Finally, dual-luciferase reporter assays in Huh7 cells revealed decreased luciferase activity upon upregulating hsa-miR-18a-5p in cells transfected with either LINC02362 or FDX1 wild-type vectors (Figure 4G).
Figure 4.

Validation of LINC02362/has-miR-18a-5p/FDX1 axis. A, B. The mRNA expression level of LINC02362 and FDX1 in the HCC cell lines and normal live cells by qPCR analysis. C. Compared the mRNA expression level of LINC02362, has-miR-18a-5p and FDX1 between si-negative control cells and si-LINC02362 cells. D. Detected the protein expression levels of FDX1 in HCC cells transfected with LINC02362 siRNA or has-miR-18a-5p mimic by western blot analysis. E. Silencing of LINC02362 expression promoted the growth of HCC cells. F. CCK-8 assays were performed in Huh7 and 97H cells treated with si-NC+miR-NC, si-LINC02362+miR-NC, si-NC+has-miR-18a-5p-mimic, si-LINC02362+has-miR-18a-5p-mimic. G. The luciferase activity of wildtype-LINC02362 or mutant-LINC02362 (wildtype-FDX1 or mutant-FDX1) after co-transfection with miR-18a-5p mimic in Huh7 cells. *P < 0.05, **P < 0.01, ***P < 0.001.
Immune infiltration analysis of LINC02362, has-miR-18a-5p, and FDX1
We explored the relationship between these key genes and immune infiltration levels in the TCGA-LIHC cohort using ssGSEA databases (Figure 5A-D). Intriguingly, LINC02362, hsa-miR-18a-5p, and FDX1 all correlated with NK CD56 bright cells, DC, Th17 cells, TFH, and Tcm. Further, we plotted the relationship between these key genes and stromal score, immune score, and ESTIMATE score using TCGA database (Figure S4). Additionally, we evaluated the correlation between FDX1 mRNA expression and immune checkpoint genes (Figure 5E). The result suggested that FDX1 strongly associates with various immune checkpoint genes, including CD80, CD86, PDCD1, CD276, TNFRSF4, TNFRSF9, TNFRSF18, ICOS, CTLA4, HAVCR2 and LAIR1.
Figure 5.
Immune infiltration analysis of LINC02362, has-miR-18a-5p, and FDX1 in HCC. A-D. The association between LINC02362, has-miR-18a-5p, and FDX1 and different kinds of immune cells in the TCGA-LIHC cohort using ssGSEA databases. E. The heat map for the relationship between the mRNA expression level of FDX1 and immune checkpoint genes in the TCGA-LIHC cohort. *P < 0.05, **P < 0.01, ***P < 0.001.
Prognostic analysis in HCC
We executed univariate and multivariate Cox regression analyses using the TCGA database to evaluate the prognostic potential of LINC02362, hsa-miR-18a-5p, and FDX1 in HCC. Our findings revealed that high expression of LINC02362 might enhance survival rates, whereas high expression of hsa-miR-18a-5p could reduce survival rates in HCC as independent factors (Tables S5, S6, S7). We constructed nomogram models and calibration curves to evaluate the relationship between the LINC02362/has-miR-18a-5p/FDX1 axis and the OS of HCC (Figure 6A-F). Collectively, these results suggest that upregulating LINC02362/has-miR-18a-5p/FDX1 axis may suppress the development of HCC.
Figure 6.
Prognostic analysis of LINC02362, has-miR-18a-5p, and FDX1 in HCC. A, B. Constructed the nomogram models and calibration curves to evaluate the relation between LINC02362 and the OS of HCC. C, D. Constructed the nomogram models and calibration curves to evaluate the relation between has-miR-18a-5p and the OS of HCC. E, F. Constructed the nomogram models and calibration curves to evaluate the relation between FDX1 and the OS of HCC.
Validation of effect of LINC02362/has-miR-18a-5p/FDX1 axis on cuproptosis
To determine if LINC02362 has a role in cuproptosis, which might influence elesclomol-Cu resistance in HCC, we assessed the relative mRNA expression levels of LINC02362, hsa-miR-18a-5p, and FDX1, as well as the relative protein expression of FDX1 in cells treated with elesclomol-Cu for 24 hours. As anticipated, LINC02362 knockdown led to increased hsa-miR-18a-5p levels and decreased FDX1 expression (Figure 7A, 7B). We then determined that LINC02362 knockdown significantly reduced copper concentration in elesclomol-Cu treated HCC cell lines (Figure 7C). Given that prior research indicates oxidative stress is a crucial mechanism of metal-induced toxicity [40], we evaluated ROS levels to understand the oxidative stress induced by elesclomol-Cu. Contrary to expectations, elesclomol-Cu did not significantly increase ROS levels in Huh7 and 97H cells (Figure 7D). Additionally, we measured the IC50 values of elesclomol-Cu (1:1 ratio) in si-NC Huh7 cells, si-LINC02362 Huh7 cells, si-NC 97H cells, and si-LINC02362 97H cells; IC50 values were determined to be 130.0 nM, 193.2 nM, 164.6 nM and 203.1 nM, respectively (Figure 7E). These findings underscore the potential of the LINC02362/hsa-miR-18a-5p/FDX1 axis in promoting cuproptosis in HCC cells.
Figure 7.

Validation of effect of LINC02362/has-miR-18a-5p/FDX1 axis on cuproptosis. A. The mRNA expression level of LINC02362, hsa-miR-18a-5p and FDX1 in si-NC cells, si-NC cells treated with 500 nM elesclomol-Cu (1:1 ratio) for 24 h, si-LINC02362 cells treated with 500 nM elesclomol-Cu (1:1 ratio) for 24 h. B. The protein expression level of FDX1 in si-NC cells, si-NC cells treated with 500 nM elesclomol-Cu (1:1 ratio) for 24 h, si-LINC02362 cells treated with 500 nM elesclomol-Cu (1:1 ratio) for 24 h. C. The intracellular copper levels in si-NC and si-LINC02362 cells treated with 500 nM elesclomol-Cu (1:1 ratio) for 24 h. D. ROS levels in si-NC and si-LINC02362 cells treated with 500 nM elesclomol-Cu (1:1 ratio) for 24 h. E. The IC50 of elesclomol-Cu in si-NC and si-LINC02362 cells were determined using the CCK-8 assay. *P < 0.05, **P < 0.01, ***P < 0.001.
Validation of correlation between LINC02362/has-miR-18a-5p/FDX1 axis and oxaliplatin-resistance
Previous studies have posited a robust relationship between copper transport systems and platinum drugs [9]. In this context, we explored the interplay between the LINC02362/hsa-miR-18a-5p/FDX1 axis and oxaliplatin resistance in HCC. The CCK8 assay results indicated that TTM, a cuproptosis inhibitor, notably mitigated oxaliplatin-induced cell death across Huh7, 97H, and 97H-OXA cells (Figure 8A). qRT-PCR was employed to gauge the mRNA expression levels of cuproptosis-related key genes in 97H and 97H-OXA cells. The results showed differential gene expression, with FDX1 downregulated and MTF1, GLS, and CDKN2A upregulated in 97H-OXA cells (Figure 8B). To elucidate the expression profile of proteins in oxaliplatin-resistant HCC, we performed TMT-labelled quantitative proteomics to identify proteins that were expressed abnormally in parental oxaliplatin-sensitive (97H) and oxaliplatin-resistant (97H-OXA) HCC cells. The results showed that the protein expression level of FDX1 is downregulated in 97H-OXA cells (Figure 8C). Through PPI analysis, we identified proteins significantly associated with FDX1 (Figures S5, S6). KEGG and GO functional enrichment analyses were performed to explore potential FDX1-related mechanisms. Notably, FDX1 and related genes predominantly participate in transmembrane transporter activity, transporter activity, and oxidoreductase activity (Figures S7, S8). To further validate our hypothesis, we studied how oxaliplatin modulates FDX1 expression. Intriguingly, FDX1 protein levels were downregulated in 97H-OXA cells compared to 97H cells (Figure 8D), and FDX1 expression also declined in HCC cells treated with oxaliplatin (Figure 8E). Then we determined the IC50 values of oxaliplatin in si-NC Huh7 cells, si-LINC02362 Huh7 cells, si-NC 97H cells and si-LINC02362 97H cells, which were 8.054 μM, 9.767 μM, 9.832 μM and 12.740 μM, respectively (Figure 8F). We also established FDX1 over-expression (oe) cell lines, confirmed by western blot (Figure S9). The IC50 values of oxaliplatin in Control Huh7 cells, FDX1 oe Huh7 cells, Control 97H cells, and FDX1 oe 97H cells were 10.540 μM, 7.850 μM, 9.929 μM and 7.640 μM, respectively (Figure 8G). TTM (cuproptosis inhibitor) restored cell viability. This was not observed with other cell death inhibitors, including Z-VAD-FMK (apoptosis inhibitor), Necrosulfonamide (necroptosis inhibitor), ferrostatin-1 (ferroptosis inhibitor), N-Acetylcysteine (ROS inhibitor), and CQ (autophagy inhibitor, Figure 8H). Moreover, FDX1 overexpression bolstered resistance to oxaliplatin treatment in HCC cells, as shown by relative cell viability (Figure 8I). Furthermore, the phenomenon raised by FDX1 overexpression was partly recovered by co-treatment with TTM, a cuproptosis inhibitor. Collectively, these findings underscore the pivotal role of the LINC02362/hsa-miR-18a-5p/FDX1 axis in modulating HCC sensitivity to oxaliplatin through cuproptosis.
Figure 8.
Validation of correlation between LINC02362/hsa-miR-18a-5p/FDX1 axis and oxaliplatin-resistance. A. TTM significantly inhibited oxaliplatin-induced cell death in Huh7 cells, 97H cells and 97H-OXA cells. B. The mRNA expression level of the key cuproptosis genes in 97H cells and 97H-OXA cells by qPCR analysis. C. Heat map lists the DEPs related to cuproptosis and volcano plot illustrates all the DEPs involved in 97H cells and 97H-OXA cells using proteomics. D. The FDX1 protein expression level in 97H cells and 97H-OXA cells by western blot analysis. E. The FDX1 protein expression level in cells treated with DMSO or 10 uM oxaliplatin for 24 h by western blot analysis. F. The IC50 of oxaliplatin in si-NC and si-LINC02362 cells were determined using the CCK-8 assay. G. The IC50 of oxaliplatin in Mock and FDX1 oe cells were determined using the CCK-8 assay. H. Mock HCC cells and FDX1-overexpression cells were pretreated overnight with 10 uM DMSO, 20 uM TTM, 30 uM Z-VAD-FMK, 20 uM necrostatin-1, 10 uM ferrostatin-1, 1 mM N-acetylcysteine (NAC), and 10 uM chloroquine, respectively. And then cells were treated with 10 uM oxaliplatin for 48 h with different disposal modes. I. Mock HCC cells, FDX1-overexpression HCC cells, FDX1-overexpression HCC cells incubating with 20 uM TTM were treated with various concentrations of oxaliplatin for 48 h. The relative cell viability was determined by the CCK-8 assays. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
HCC is characterized by a high mortality rate due to its asymptomatic nature, diagnostic challenges, and the absence of effective therapeutic approaches. Despite these challenges, the molecular underpinnings of HCC remain poorly understood, requiring further study to elucidate these mechanisms and drive effective therapeutic breakthroughs. In our study we combined bioinformatics analyses with in vitro experiments to identify a novel therapeutic target for HCC: the LINC02362/FDX1 axis. Our findings indicated that an upregulated LINC02362/FDX1 axis correlates with a more favorable prognosis. Crucially, elevating the LINC02362/FDX1 axis appears to inhibit HCC proliferation and enhance the sensitivity of HCC cells to oxaliplatin through the mechanism of cuproptosis.
Copper, an essential trace element in the human body, is implicated in a variety of tumor-related biological behaviors [41]. Over the years, the relationship between copper and regulated cell death has been a topic of extensive research. Although the potential of copper to induce cell death was proposed as early as the 1980s [42], the exact mechanism wasn’t elucidated until March 2022 by Tsvetkov et al. [6]. This mechanism, termed “cuproptosis”, is believed to interact with components of the mitochondrial tricarboxylic acid (TCA) cycle and involves lipoylation, a conserved post-transcriptional protein modification pathway. Given the liver’s central role in copper metabolism and storage [43] and the elevated copper levels observed in liver cirrhosis, a known risk factor for HCC compared to normal liver tissue [44], we hypothesize that cuproptosis plays a significant role in HCC onset and progression.
Within the cuproptosis mechanism, FDX1 emerges as a pivotal player. It’s postulated to act upstream of the lipoic acid (LA) pathway, influencing protein lipoylation processes. Additionally, FDX1 has been tied closely to elesclomol (ES) sensitivity, as it directly binds with ES-Cu, leading to reduced stability of the Fe-S cluster [45]. FDX1’s vital role in Fe-S cluster biogenesis has been underscored by several studies [46,47]. Notably, FDX1 downregulation in HCC is well-documented, with higher FDX1 levels acting as protective factors [7,8,48]. Today, using bioinformatics analysis to discover novel biomarkers in various cancers has seen success. In light of FDX1’s function in cuproptosis, our study is the first to reveal the potential therapeutic significance of the LINC02362/hsa-miR-18a-5p/FDX1 axis in HCC.
Our methodology began with screening eight candidate lncRNAs from the TCGA database, based on their overlap with DElncRNAs, FDX1-related lncRNAs, and prognosis-related lncRNAs. Subsequent predictions using lncbase and Starbase identified potential miRNAs binding to these lncRNAs and FDX1. Among 5 candidate miRNAs only hsa-miR-18a-5p was found to be upregulated in HCC samples relative to normal samples and was associated with HCC prognosis. These findings align with prior studies highlighting the roles of LINC02362 and hsa-miR-18a-5p in HCC progression [49-51]. Through CCK8 assays, we further corroborated that upregulating the LINC02362/FDX1 axis suppresses HCC cell proliferation. Moreover, we assessed the ability of this axis to induce cuproptosis by evaluating intracellular copper levels and IC50 values.
Oxaliplatin-based regimens are recognized as one of the most effective treatments for advanced HCC [52]. While ferroptosis and apoptosis have been identified as key mechanisms of oxaliplatin-induced cell death [53,54], a notable connection between copper transport systems and platinum drugs, such as cisplatin, carboplatin, and oxaliplatin, has been documented. Recent research by Yang WC et al. suggested that oxaliplatin-resistant colorectal cancer cells exhibit a weakened response to elesclomol-Cu compared to their wild-type counterparts [55]. Based on this, we postulated that cuproptosis is crucial for oxaliplatin-induced cell death in HCC. Our experiments demonstrated that TTM, a copper chelating agent, can partially counteract oxaliplatin-induced cell death by the CCK8 assay. We also observed that LINC02362 knockdown induces oxaliplatin resistance and that oxaliplatin treatment inhibits FDX1 in HCC cells. These findings suggest that oxaliplatin administration could induce cuproptosis. Thus, our research is the first to identify that targeting cuproptosis to improve oxaliplatin treatment efficacy may be a promising strategy to conquer chemoresistance in HCC.
Conclusions
In summary, our research underscores the LINC02362/hsa-miR-18a-5p/FDX1 axis as a novel pathway that suppresses HCC proliferation, driving cuproptosis, and enhances the sensitivity of HCC to oxaliplatin. This pathway holds promise as a new therapeutic target to counteract oxaliplatin resistance in HCC.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 81972889).
Disclosure of conflict of interest
None.
Abbreviations
- HCC
Hepatocellular carcinoma
- Fe-S
iron-sulfur
- TMT
tandem mass tag
- DEPs
differentially expressed proteins
- DElncRNAs
differentially expressed lncRNAs
- GO
Gene Oncology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OS
Overall survival
- IC50
half-maximal inhibitory concentration
- ssGSEA
single-sample gene set enrichment analysis
- TTM
Tetrathiomolybdate
- PFS
progression-free survival
Supporting Information
References
- 1.Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, Bray F. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology. 2020;159:335–349. e315. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinlich R, Oberst A, Beere HM, Green DR. Necroptosis in development, inflammation and disease. Nat Rev Mol Cell Biol. 2017;18:127–136. doi: 10.1038/nrm.2016.149. [DOI] [PubMed] [Google Scholar]
- 4.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B 3rd, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R, Spangler RD, Eaton JK, Frenkel E, Kocak M, Corsello SM, Lutsenko S, Kanarek N, Santagata S, Golub TR. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254–1261. doi: 10.1126/science.abf0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Zeng Y, Guo X, Shen H, Zhang J, Wang K, Ji M, Huang S. Pan-cancer analyses confirmed the cuproptosis-related gene FDX1 as an immunotherapy predictor and prognostic biomarker. Front Genet. 2022;13:923737. doi: 10.3389/fgene.2022.923737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z, Zeng X, Wu Y, Liu Y, Zhang X, Song Z. Cuproptosis-related risk score predicts prognosis and characterizes the tumor microenvironment in hepatocellular carcinoma. Front Immunol. 2022;13:925618. doi: 10.3389/fimmu.2022.925618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnesano F, Natile G. Interference between copper transport systems and platinum drugs. Semin Cancer Biol. 2021;76:173–188. doi: 10.1016/j.semcancer.2021.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Guo Y, Smith K, Petris MJ. Cisplatin stabilizes a multimeric complex of the human Ctr1 copper transporter: requirement for the extracellular methionine-rich clusters. J Biol Chem. 2004;279:46393–46399. doi: 10.1074/jbc.M407777200. [DOI] [PubMed] [Google Scholar]
- 11.Holzer AK, Manorek GH, Howell SB. Contribution of the major copper influx transporter CTR1 to the cellular accumulation of cisplatin, carboplatin, and oxaliplatin. Mol Pharmacol. 2006;70:1390–1394. doi: 10.1124/mol.106.022624. [DOI] [PubMed] [Google Scholar]
- 12.Ishida S, Lee J, Thiele DJ, Herskowitz I. Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc Natl Acad Sci U S A. 2002;99:14298–14302. doi: 10.1073/pnas.162491399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song IS, Savaraj N, Siddik ZH, Liu P, Wei Y, Wu CJ, Kuo MT. Role of human copper transporter Ctr1 in the transport of platinum-based antitumor agents in cisplatin-sensitive and cisplatin-resistant cells. Mol Cancer Ther. 2004;3:1543–1549. [PubMed] [Google Scholar]
- 14.Katano K, Safaei R, Samimi G, Holzer A, Tomioka M, Goodman M, Howell SB. Confocal microscopic analysis of the interaction between cisplatin and the copper transporter ATP7B in human ovarian carcinoma cells. Clin Cancer Res. 2004;10:4578–4588. doi: 10.1158/1078-0432.CCR-03-0689. [DOI] [PubMed] [Google Scholar]
- 15.Mangala LS, Zuzel V, Schmandt R, Leshane ES, Halder JB, Armaiz-Pena GN, Spannuth WA, Tanaka T, Shahzad MM, Lin YG, Nick AM, Danes CG, Lee JW, Jennings NB, Vivas-Mejia PE, Wolf JK, Coleman RL, Siddik ZH, Lopez-Berestein G, Lutsenko S, Sood AK. Therapeutic targeting of ATP7B in ovarian carcinoma. Clin Cancer Res. 2009;15:3770–3780. doi: 10.1158/1078-0432.CCR-08-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Safaei R, Otani S, Larson BJ, Rasmussen ML, Howell SB. Transport of cisplatin by the copper efflux transporter ATP7B. Mol Pharmacol. 2008;73:461–468. doi: 10.1124/mol.107.040980. [DOI] [PubMed] [Google Scholar]
- 17.Samimi G, Katano K, Holzer AK, Safaei R, Howell SB. Modulation of the cellular pharmacology of cisplatin and its analogs by the copper exporters ATP7A and ATP7B. Mol Pharmacol. 2004;66:25–32. doi: 10.1124/mol.66.1.25. [DOI] [PubMed] [Google Scholar]
- 18.Samimi G, Safaei R, Katano K, Holzer AK, Rochdi M, Tomioka M, Goodman M, Howell SB. Increased expression of the copper efflux transporter ATP7A mediates resistance to cisplatin, carboplatin, and oxaliplatin in ovarian cancer cells. Clin Cancer Res. 2004;10:4661–4669. doi: 10.1158/1078-0432.CCR-04-0137. [DOI] [PubMed] [Google Scholar]
- 19.Safaei R, Maktabi MH, Blair BG, Larson CA, Howell SB. Effects of the loss of Atox1 on the cellular pharmacology of cisplatin. J Inorg Biochem. 2009;103:333–341. doi: 10.1016/j.jinorgbio.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 21.Liu S, Bu X, Kan A, Luo L, Xu Y, Chen H, Lin X, Lai Z, Wen D, Huang L, Shi M. SP1-induced lncRNA DUBR promotes stemness and oxaliplatin resistance of hepatocellular carcinoma via E2F1-CIP2A feedback. Cancer Lett. 2022;528:16–30. doi: 10.1016/j.canlet.2021.12.026. [DOI] [PubMed] [Google Scholar]
- 22.Liu F, Tian T, Zhang Z, Xie S, Yang J, Zhu L, Wang W, Shi C, Sang L, Guo K, Yang Z, Qu L, Liu X, Liu J, Yan Q, Ju HQ, Wang W, Piao HL, Shao J, Zhou T, Lin A. Long non-coding RNA SNHG6 couples cholesterol sensing with mTORC1 activation in hepatocellular carcinoma. Nat Metab. 2022;4:1022–1040. doi: 10.1038/s42255-022-00616-7. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, Tong X, Yang W, Xu Q, Huang D, Tu K. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18:28. doi: 10.1186/s12943-019-0957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang L, Huan L, Wang J, Wu Y, Huang S, He X. LncRNA RP11-295G20.2 regulates hepatocellular carcinoma cell growth and autophagy by targeting PTEN to lysosomal degradation. Cell Discov. 2021;7:118. doi: 10.1038/s41421-021-00339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Luo M, Cui X, O’Connell D, Yang Y. Long noncoding RNA NEAT1 promotes ferroptosis by modulating the miR-362-3p/MIOX axis as a ceRNA. Cell Death Differ. 2022;29:1850–1863. doi: 10.1038/s41418-022-00970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, Bao W, Zhang S, Chen B, Zhou X, Zhao J, Shi Z, Zhang T, Chen Z, Wang L, Zheng X, Chen G, Wang Y. LncRNA HEPFAL accelerates ferroptosis in hepatocellular carcinoma by regulating SLC7A11 ubiquitination. Cell Death Dis. 2022;13:734. doi: 10.1038/s41419-022-05173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9:e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin X, Zheng SS, Zhang L, Xie XY, Wang Y, Zhang BH, Wu W, Qiu S, Ren ZG. Identification of long noncoding RNA expression profile in oxaliplatin-resistant hepatocellular carcinoma cells. Gene. 2017;596:53–88. doi: 10.1016/j.gene.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 30.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu; Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341. e1323. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 33.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z, Nevens F, Roskams T, Thorgeirsson SS. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 34.Woo HG, Park ES, Cheon JH, Kim JH, Lee JS, Park BJ, Kim W, Park SC, Chung YJ, Kim BG, Yoon JH, Lee HS, Kim CY, Yi NJ, Suh KS, Lee KU, Chu IS, Roskams T, Thorgeirsson SS, Kim YJ. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14:2056–2064. doi: 10.1158/1078-0432.CCR-07-1473. [DOI] [PubMed] [Google Scholar]
- 35.Woo HG, Lee JH, Yoon JH, Kim CY, Lee HS, Jang JJ, Yi NJ, Suh KS, Lee KU, Park ES, Thorgeirsson SS, Kim YJ. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034–3041. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cairo S, Armengol C, De Reyniès A, Wei Y, Thomas E, Renard CA, Goga A, Balakrishnan A, Semeraro M, Gresh L, Pontoglio M, Strick-Marchand H, Levillayer F, Nouet Y, Rickman D, Gauthier F, Branchereau S, Brugières L, Laithier V, Bouvier R, Boman F, Basso G, Michiels JF, Hofman P, Arbez-Gindre F, Jouan H, Rousselet-Chapeau MC, Berrebi D, Marcellin L, Plenat F, Zachar D, Joubert M, Selves J, Pasquier D, Bioulac-Sage P, Grotzer M, Childs M, Fabre M, Buendia MA. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Sohn BH, Shim JJ, Kim SB, Jang KY, Kim SM, Kim JH, Hwang JE, Jang HJ, Lee HS, Kim SC, Jeong W, Kim SS, Park ES, Heo J, Kim YJ, Kim DG, Leem SH, Kaseb A, Hassan MM, Cha M, Chu IS, Johnson RL, Park YY, Lee JS. Inactivation of hippo pathway is significantly associated with poor prognosis in hepatocellular carcinoma. Clin Cancer Res. 2016;22:1256–1264. doi: 10.1158/1078-0432.CCR-15-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, Watanabe G, Gabriel S, Friedman SL, Kumada H, Llovet JM, Golub TR. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB, Park ES, Lim JY, Heo J, Kim YJ, Kim DG, Kaseb A, Park YN, Wang XW, Thorgeirsson SS, Lee JS. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55:1443–1452. doi: 10.1002/hep.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ercal N, Gurer-Orhan H, Aykin-Burns N. Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem. 2001;1:529–539. doi: 10.2174/1568026013394831. [DOI] [PubMed] [Google Scholar]
- 41.Festa RA, Thiele DJ. Copper: an essential metal in biology. Curr Biol. 2011;21:R877–883. doi: 10.1016/j.cub.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halliwell B, Gutteridge JM. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutsenko S. Dynamic and cell-specific transport networks for intracellular copper ions. J Cell Sci. 2021;134:jcs240523. doi: 10.1242/jcs.240523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poznański J, Sołdacki D, Czarkowska-Pączek B, Bonna A, Kornasiewicz O, Krawczyk M, Bal W, Pączek L. Cirrhotic liver of liver transplant recipients accumulate silver and co-accumulate copper. Int J Mol Sci. 2021;22:1782. doi: 10.3390/ijms22041782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsvetkov P, Detappe A, Cai K, Keys HR, Brune Z, Ying W, Thiru P, Reidy M, Kugener G, Rossen J, Kocak M, Kory N, Tsherniak A, Santagata S, Whitesell L, Ghobrial IM, Markley JL, Lindquist S, Golub TR. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat Chem Biol. 2019;15:681–689. doi: 10.1038/s41589-019-0291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai K, Tonelli M, Frederick RO, Markley JL. Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron-sulfur cluster biosynthesis. Biochemistry. 2017;56:487–499. doi: 10.1021/acs.biochem.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y, Ghosh M, Kovtunovych G, Crooks DR, Rouault TA. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim Biophys Acta. 2012;1823:484–492. doi: 10.1016/j.bbamcr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao C, Yang L, Jin L, Lin W, Zhang F, Huang S, Huang Z. Prognostic and immunological role of cuproptosis-related protein FDX1 in pan-cancer. Front Genet. 2022;13:962028. doi: 10.3389/fgene.2022.962028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z, Ma Y, Pan Y, Zuo S, Zhu H, Yu C, Zhu C, Sun C. Long noncoding RNA RP5-833A20.1 suppresses tumorigenesis in hepatocellular carcinoma through Akt/ERK pathway by targeting miR-18a-5p. Onco Targets Ther. 2019;12:10717–10726. doi: 10.2147/OTT.S219797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li D, Zhou T, Li Y, Xu Y, Cheng X, Chen J, Zheng WV. LINC02362 attenuates hepatocellular carcinoma progression through the miR-516b-5p/SOSC2 axis. Aging (Albany NY) 2022;14:368–388. doi: 10.18632/aging.203813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng Y, Nie P, Xu S. Long noncoding RNA linc00467 plays an oncogenic role in hepatocellular carcinoma by regulating the miR-18a-5p/NEDD9 axis. J Cell Biochem. 2020;121:3135–3144. doi: 10.1002/jcb.29581. [DOI] [PubMed] [Google Scholar]
- 52.Ge S, Huang D. Systemic therapies for hepatocellular carcinoma. Drug Discov Ther. 2015;9:352–362. doi: 10.5582/ddt.2015.01047. [DOI] [PubMed] [Google Scholar]
- 53.Ma L, Xu A, Kang L, Cong R, Fan Z, Zhu X, Huo N, Liu W, Xue C, Ji Q, Li W, Chu Z, Kang X, Wang Y, Sun Z, Han Y, Liu H, Gao X, Han J, You H, Zhao C, Xu X. LSD1-demethylated LINC01134 confers oxaliplatin resistance through SP1-induced p62 transcription in HCC. Hepatology. 2021;74:3213–3234. doi: 10.1002/hep.32079. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Q, Deng T, Zhang H, Zuo D, Zhu Q, Bai M, Liu R, Ning T, Zhang L, Yu Z, Zhang H, Ba Y. Adipocyte-derived exosomal MTTP suppresses ferroptosis and promotes chemoresistance in colorectal cancer. Adv Sci (Weinh) 2022;9:e2203357. doi: 10.1002/advs.202203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang W, Wang Y, Huang Y, Yu J, Wang T, Li C, Yang L, Zhang P, Shi L, Yin Y, Tao K, Li R. 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to promote cuproptosis in colorectal cancer. Biomed Pharmacother. 2023;159:114301. doi: 10.1016/j.biopha.2023.114301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.