Abstract
The c-Jun/AP-1 transcription complex is associated with diverse cellular processes such as differentiation, proliferation, transformation, and apoptosis. These different biological endpoints are likely achieved by the regulation of specific target gene expression. We describe the identification of Ras guanine nucleotide exchange factor 1, Ras-GRF1, by microarray analysis as a c-Jun/AP-1 regulated gene essential for anchorage-independent growth of immortalized rat fibroblasts. Increased Ras-GRF1 expression, in response to inducible c-Jun expression in Rat1a fibroblasts, was confirmed by both real-time PCR and Northern blot analysis. We show that c-Jun/AP-1 can bind and activate the Ras-GRF1 promoter in vivo. A 75-kDa c-Jun/AP-1-inducible protein, p75-Ras-GRF1, was detected, and the inhibition of its expression with antisense oligomers significantly blocked c-Jun-regulated anchorage-independent cell growth. p75-Ras-GRF1 expression occurred with a concomitant increase in activated Ras (GTP bound), and the activation of Ras was significantly inhibited by antisense Ras-GRF1 oligomers. Moreover, p75-Ras-GRF1 could be coprecipitated with a Ras dominant-negative glutathione S-transferase (GST) construct, GST-Ras15A, demonstrating an interaction between p75-Ras-GRF1 and Ras. A downstream target of Ras activation, Elk-1, had increased transcriptional activity in c-Jun-expressing cells, and this activation was inhibited by dominant-negative Ras. In addition, c-Jun overexpression resulted in an increase in phospho-AKT while phosphorylation of ERK1/2 remained largely unaffected. The inhibition of phosphatidylinositol 3-kinase (PI3K)-AKT signal transduction by Ly294002 and wortmannin significantly blocked c-Jun-regulated morphological transformation, while inhibition of basal MEK-ERK activity with PD98059 and U0126 had little effect. We conclude that c-Jun/AP-1 regulates endogenous p75-Ras-GRF1 expression and that c-Jun/AP-1-regulated anchorage-independent cell growth requires activation of Ras-PI3K-AKT signal transduction.
The proto-oncogene c-jun is one of the major components of the transcription factor AP-1 and has a key role in a variety of cellular processes, including growth, differentiation, apoptosis, and survival (2, 4, 11). AP-1 is also implicated in tumorigenic processes such as angiogenesis, deregulated proliferation and apoptosis, invasion, and metastasis (31). AP-1 achieves this functional diversity by binding tetradecanoyl phorbol acetate response elements in the promoters and enhancers of a large number of AP-1-regulated target genes (51). Transformation by oncogenes such as Ras (38) and Raf (18) results in the induction of expression of c-Jun and others in the AP-1 family of proteins (31). c-Jun/AP-1 activity is essential for cellular transformation by the Ras oncoproteins as dominant-negative mutants of c-Jun inhibit Ras-regulated transformation of NIH 3T3 cells (43). Not only is c-Jun required for Ras-induced transformation, its overexpression as a single gene is sufficient to induce transformation of the immortalized rat fibroblast cell line, Rat1a (44). c-Jun overexpression in Rat1a cells induces cell morphology changes (33) which mimic those observed by v-Jun (40) and K-RasV12 (27).
The accepted position of c-Jun/AP-1 during oncogenesis is at the end of signal transduction cascades initiated at the cell membrane by growth factors and cytokines or in the cytoplasm by oncogenes, such as those coding for H-Ras and v-Src (46, 49). The mechanism resulting in increased expression and activation of AP-1 occurs via the activation of the mitogen-activated protein kinase (MAPK) signaling pathways. Erk activation increases Fos expression (25, 29) as well as phosphorylation of Fra1 and Fra2 (25). In this context, the Fos/Fra transcription factors, together with c-Jun, may result in c-jun autoregulation by interacting with the AP-1 binding site in the c-Jun promoter (25, 29). ERK activates the MEF2 transcription factors, which can contribute to c-Jun expression (15, 28). In addition, activation of JNK causes phosphorylation of c-Jun at Ser63 and Ser73 (32). This event is essential for the full activation of c-Jun and its role in cellular transformation (7, 8). Upon activation, c-Jun-containing AP-1 complexes regulate the expression of target genes in both a positive and negative manner and in this way have a role in a diverse set of cellular functions.
To date, few c-Jun/AP-1 target genes have been identified (50, 51). Based on the hypothesis that c-Jun-induced biologic activities are dependent upon transcriptional activation of target genes, we used c-Jun expression under the control of a tet-on vector in Rat1a fibroblasts as a model system to identify such genes. Using the Affymetrix rat oligonucleotide array, RG_U34A, we identified a number of potential c-Jun-regulated candidate genes (34). One of these target genes is the gene coding for Ras-GRF1, a guanine-nucleotide exchange factor (GEF) important in signal transduction via activation of Ras (5, 42, 53). Based on the known function of Ras-GRF1, we evaluated the significance of its regulation by c-Jun/AP-1. Interestingly, we established that c-Jun regulated the expression of p75-Ras-GRF1 that was associated with an increase in GTP-Ras and phosphatidylinositol 3-kinase (PI3K) activity. Both p75-Ras-GRF1 expression and PI3K were essential for c-Jun/AP-1-regulated anchorage-independent growth of rat fibroblasts. Collectively, our data show that c-Jun/AP-1 is a transcriptional regulator of proteins such as p75-Ras-GRF1, which may generate feedback loops for the sustained activation of specific signal transduction pathways required for deregulated cell growth and survival.
MATERIALS AND METHODS
Cell culture and doxycycline induction.
Rat1a-c-Jun4 and Rat1a-c-Myc fibroblasts were grown in Dulbecco's modified Eagle's medium (GIBCO/BRL, Burlington, Ontario, Canada) containing 10% fetal bovine serum and penicillin (100 U/ml)-streptomycin (100 μg/ml) at 37°C in a 5% CO2 incubator. The cells were maintained in selection medium containing 5 μg of blasticidin per ml. For experiments, subconfluent Rat1a-c-Jun4 cells were trypsinized with 0.5% trypsin-0.53 mM EDTA in Hanks balanced salt solution (GIBCO/BRL) and plated in 10% fetal bovine serum (FBS)-containing media in the absence or presence of 2 μg of doxycycline per ml to induce c-jun expression. For nonadherent growth conditions, 106 cells/ml were plated on PolyHeme-coated dishes.
Protein extraction and Western blotting.
Total cell protein was isolated from cells with radioimmunoprecipitation assay (RIPA) lysis buffer (150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 10 mM Tris [pH 7.4], containing 0.1 mM phenylmethylsulfonyl fluoride [PMSF], and 100 μg each of aprotinin and leupeptin per ml). RIPA lysis buffer (with low SDS concentration) was prepared with Tris that was pH adjusted to 7.4 at room temperature. The cells were harvested on ice to prevent protein degradation. The lysed cells were sonicated and centrifuged to remove debris, and protein concentrations were determined with the Bradford assay (Bio-Rad). Samples were stored at −20°C until required. Doxycycline-induced Ras-GRF1 gene expression was monitored by Western blot analysis with the antibody sc224 (Santa Cruz Biotechnology) at dilutions recommended by the supplier. Western blots for tubulin levels (sc9104; Santa Cruz Biotechnology) were done to correct for protein loading. To determine the inhibition of MEK activity by PD98059, and U0126 and PI3K activity with Ly294004 and wortmannin, the following antibodies from Cell Signaling were used: phopsho-Erk1/2 (no. 9101), Erk1/2 (no. 9102), phospho-Akt (no. 9271), and Akt (no. 9272) at dilutions recommended by the manufacturer. Densitometric analysis of Western blots was done with a Bio-Rad scanner and software.
For sense and antisense Ras-GRF1 experiments, 10 μM oligomers (sense, 5′-AACCAAAACTCCCCACATGA-3′; antisense, 5′-TCATGTGGGGAGTTTTGGTT-3′) was added at the time of plating and proteins were harvested 48 h later.
Ras activation assays.
GTP-bound Ras was immunoprecipitated with a Ras activation assay kit (Upstate). All buffers were supplied in the kit, and the manufacturer's protocols were followed. Briefly, cells were lysed in MLB (125 mM HEPES [pH 7.5], 750 mM NaCl, 5% Igepal CA-630, 50 mM MgCl2, 5 mM EDTA, 10% glycerol) and 400 μg of the lysate was precleared with glutathione agarose. The precleared lysate was then incubated with 10 μg of Raf-1 RBD agarose for 1 h at 4°C, and the beads were washed three times with MLB. The samples were then suspended in LDS loading dye containing β-mercaptoethanol and boiled for 5 min. The captured GTP-Ras complexes were loaded on 4 to 12% NuPAGE gels and visualized by immunoblots for Ras (Ras10 antibody; Upstate).
Cell proliferation assays.
To determine anchorage-independent cell growth, 10,000 cells/well in a 96-well plate were grown in 1.5% methylcellulose in 10% FBS-containing media and plated on poly(2-hydroxyethyl methacrylate) (PolyHeme) (Sigma)-treated dishes to prevent adhesion to the dish. Growth in the absence and presence of 2-μg/ml doxycycline, PD98059, U0126, Ly294002, and wortmannin and sense and antisense Ras-GRF oligomers was measured with MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assays (Promega). For sense and antisense Ras-GRF1 experiments, 10 μM oligomers was added to the cell culture medium at the time of plating followed by the addition of 4 μM oligomer daily for an additional 3 days prior the end of the experiment.
To observe colony formation, the cells were stained with 1 mg of p-iodonitrotetrazolium violet per ml for 16 h at 37°C to detect live cells in colonies. The colonies were photographed by bright-field microscopy at a magnification of ×100.
Reporter assays.
Transient transfection assays were performed with FuGene6 reagent (Roche). TRE2-luciferase, Gal-Elk-1, 5× Gal-Luc, and pCMV-RasN17 in FuGene6 reagent were added to cells plated in 60-mm-diameter tissue culture dishes. The pRL-TK plasmid was cotransfected as a control for transfection efficiency. The mouse Ras-GRF1 promoter constructs pGL2-84f, pGL2-84ΔA, and pGL2-84ΔBC were previously described and kindly provided by C. Plass (19). Cells were incubated for 24 h, trypsinized, and plated under nonadherent conditions for 48 h in the absence and presence of 2 μg of doxycycline per ml. Proteins were harvested in passive lysis buffer (Promega), and firefly and Renilla luciferase activity was determined sequentially with the Promega dual luciferase assay kit.
Northern blot analysis.
Total RNA was isolated from cells with Trizol reagent (Life Technologies, Inc.), and 2 μg was used in Northern blot analysis. The Ras-GRF1 probes were generated from IMAGE clones UI-R-Y0-vk-d-08-UI and UI-R-CA1-bje-k-12-0-UI (Research Genetics, Huntsville, Ala.) by PCR with the M13 forward and M13 reverse primers. The clones were sequence confirmed with UI-R-Y0-vk-d-08-UI, corresponding to the C-terminal region of p140-RasGRF1, and UI-R-CA1-bje-k-12-0-UI, which contains part of the N-terminal region. Hybridizations were performed in UltraHyb solution (Ambion) according to the manufacturer's protocol.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as follows. Rat1a-HA-Jun cells expressing hemagglutinin (HA)-tagged c-Jun were seeded at a density of 10 × 106 cells in 150-mm-diameter PolyHeme-coated tissue culture dishes in the presence and absence of 2 μg of doxycycline per ml and incubated for 4 days at 37°C. Protein-DNA complexes were cross-linked with 1% formaldehyde added directly to the culture medium at room temperature for 15 min followed by the addition of 0.125 M glycine, pH 2.5, for 5 min. Cells were pelleted at 200 × g for 5 min at 4°C and washed once with ice-cold phosphate-buffered saline (PBS). The cell pellet was resuspended in 300 μl of lysis buffer (1% SDS, 5 mM EDTA, 50 mM Tris [pH 8], 100 μg of aprotinin per ml, 100 μg of leupeptin per ml, 1 mM PMSF) and incubated on ice for 10 min. The solution was then sonicated three times for 15 s each on maximum power, and cell debris was pelleted by centrifugation for 5 min at 16,000 × g. Twenty microliters of the soluble chromatin was set aside as the input fraction, and the remainder was diluted 1:10 in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris [pH 8], 100 μg of aprotinin per ml, 100 μg of leupeptin per ml, 1 mM PMSF). The diluted soluble chromatin (1 ml) was precleared with 2 μg of sheared herring sperm DNA, 20 μl of preimmune serum, and 45 μl of protein G-agarose beads (50% slurry in 10 mM Tris [pH 8], 1 mM EDTA) for 2 h at 4°C, and the beads were pelleted by centrifugation at 16,000 × g for 1 min. Two micrograms of antibodies to HA (71-5500; Zymed Laboratories, Inc., San Francisco, Calif.) or control rabbit immunoglobulin G (IgG) (sc-2027; Santa Cruz Biotechnology, Santa Cruz, Calif.) was added, and the solution was incubated overnight at 4°C. Following this incubation, 45 μl of protein G-agarose beads and 2 μg of sheared herring sperm DNA were added and this mixture was incubated for an additional 1 h at 4°C. The beads were pelleted by centrifugation and washed sequentially for 10 min each with TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris [pH 8], 150 mM NaCl), TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris [pH 8], 500 mM NaCl), buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris [pH 8]), and TE (10 mM Tris [pH 8], 1 mM EDTA). DNA was eluted from the beads with 100 μl of elution buffer (1% SDS, 0.1 M NaHCO3), and the cross-links were reversed by incubation at 65°C overnight. DNA was purified with the Qiaquick PCR purification kit (QIAGEN, Valencia, Calif.) as per the manufacturer's instructions. One microliter of the ChIP DNA was amplified by real-time PCR with Ras-GRF1 promoter primers F1 (5′-GATCCAGCCAACAGACTAAGA-3′) and R1 (5′-AATGATGCTGCCTTGGGCACA-3′) on an iCycler real-time detection system (Bio-Rad Laboratories, Inc., Hercules, Calif.) with the Quantitect SYBR green PCR kit (QIAGEN, Inc., Valencia, Calif.) as per the manufacturers' instructions. The fold change was calculated by the 2−ΔΔCT method as previously described (35).
RESULTS
Increased Ras-GRF1 expression in c-Jun-expressing cells.
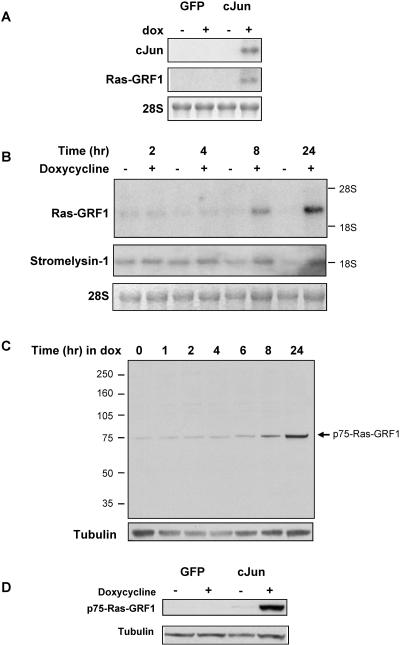
We have previously shown that the conditional expression of c-Jun in Rat1a fibroblasts results in morphology changes and anchorage-independent growth (33). Microarray analysis using the Affymetrix Rat U34A chip to identify c-Jun/AP-1-regulated genes identified significant Ras-GRF1 expression in c-Junexpressing cells relative to that in controls (34). These results were corroborated by Northern blot analysis (Fig. 1A) and quantified by real-time RT-PCR that demonstrated an eightfold increase in Ras-GRF1 expression in doxycycline-treated Rat1a-c-Jun4 cells. Northern blot analysis with a probe containing the C-terminal region of Ras-GRF1 revealed a transcript of approximately 2 to 3 kb was induced in a time-dependent manner after doxycycline-regulated c-Jun expression and reached maximal levels within 24 h (Fig. 1B). Ras-GRF1 mRNA transcripts similar in size have previously been identified in adult and fetal human tissue. These transcripts are shorter than the 5 kb encoding p140-Ras-GRF1 and are proposed to be alternate forms of Ras-GRF1 that retain the 3′ catalytic domain (26). mRNA transcripts were not detected by Northern blot analysis with a probe recognizing the N terminus of Ras-GRF1 (data not shown), suggesting that the Ras-GRF1 transcript induced in c-Jun-expressing Rat1a cells lacks the N-terminal region but retained the C-terminal catalytic domain encoding region. The expression kinetics of c-Jun-regulated Ras-GRF1 was similar to that of the gene coding for stromelysin 1 (Fig. 1B), a previously described c-Jun/AP-1-responsive gene (3), HMGI/Y (30), and other potential AP-1 targets described by us (33, 34). These results suggest a sequence of events, with induced c-Jun protein occurring maximally between 4 and 8 h (33), followed by an increase in Ras-GRF1 mRNA expression starting at approximately 8 h. To determine the levels of Ras-GRF1 protein in c-Jun-expressing cells, Western blot analysis was performed with proteins extracted from Rat1a-c-Jun4 cells grown in the absence and presence of doxycycline for various times. An increase in the expression of a 75-kDa protein, p75-Ras-GRF1, was detected within 8 h of doxycycline treatment (Fig. 1C). The antibody (sc224) used to detect p75-Ras-GRF1 was raised against a peptide mapping near the C terminus of Ras-GRF1 of rat origin. This would suggest that c-Jun/AP-1-induced p75-Ras-GRF1 contains the C-terminal catalytic domain of Ras-GRF1. A corresponding protein was not observed in green fluorescent protein (GFP)-expressing Rat1a-GFP control cells, even after 3 days of doxycline treatment (Fig. 1D). We did not detect other Ras-GRF1 isoforms described previously (13).
FIG. 1.
c-Jun/AP-1 regulates Ras-GRF1 expression in rat fibroblasts. (A) Two micrograms of total RNA from Rat1a-GFP control cells and Rat1a-c-Jun4 cells grown in suspension without (−) and with (+) doxycycline treatment for 3 days was used in Northern blots probing for Ras-GRF1 and c-Jun expression. 28S RNA was used as a loading control. (B) Northern blot analysis of time-dependent induction of Ras-GRF1 and stromelysin expression in response to doxycycline treatment of Rat1a-c-Jun4 cells. (C) Immunoblots showing doxycycline-induced p75-Ras-GRF1 protein expression in Rat1a-c-Jun4 cells. Tubulin was used as a control for loading. (D) Immunoblots of Ras-GRF1 expression in GFP- and c-Jun-expressing cells after 3 days of treatment with doxycycline.
c-Jun/AP-1 binds and activates the Ras-GRF1 promoter.
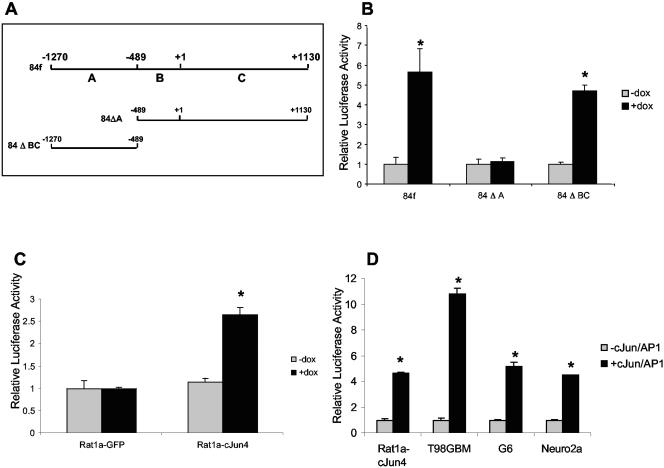
The mouse Ras-GRF1 promoter has previously been identified and contains two putative AP-1 binding sites located at −675 and −424 (Fig. 2A) (19). To determine if c-Jun/AP-1 regulates the Ras-GRF1 promoter, we performed transient transfections into Rat1a-c-Jun4 cells with promoter constructs 84f (−1270/+1130), 84ΔA (−487/+1130), and 84ΔBC(−1270/−487). The results revealed that c-Jun/AP-1 activated the constructs containing the −1270/+1130 and the −1270/−487 promoter regions, while no response was observed with the −487/+1130 construct (Fig. 2B). Activation of the −1270/−487 promoter construct was not observed in Rat1-GFP control cells (Fig. 2C). Comparable results of −1270 to −487 Ras-GRF1 promoter activation in response to the presence of c-Jun/AP-1 were also observed in the human, rat, and mouse glioblastoma cell lines, T98GBM, G6, and Neuro2a, respectively (Fig. 2D). These results imply that c-Jun/AP-1 has a role in the expression of the Ras-GRF1 promoter.
FIG. 2.
Ras-GRF1 promoter activation by c-Jun/AP-1. (A) Mouse Ras-GRF1 promoter constructs used in transient transfection assays. Construct 84f spans from −1270 to +1130 of the promoter region and encompasses both of the putative AP-1 binding sites. Construct 84ΔA includes the −489 to +1130 promoter region, and 84ΔBC contains the −1270 to −489 promoter region. All promoter fragments were cloned into pGL2-Basic. (B) Transient transfection on the Ras-GRF1 promoter constructs into Rat1a-c-Jun4 cells grown with (+dox) and without (−dox) doxycyline. (C) 84ΔBC activity in Rat1a-GFP and Rat1a-c-Jun4 cells grown as described for panel B. (D) 84ΔBC activity in the presence of cotransfected c-Jun in the human, rat, and mouse glioblastoma cell lines, T98GBM, G6, and Neuro2a, respectively, compared to activity in Rat1a-c-Jun4 cells. The results show the mean ± standard deviation of triplicate experiments. *, P < 0.05.
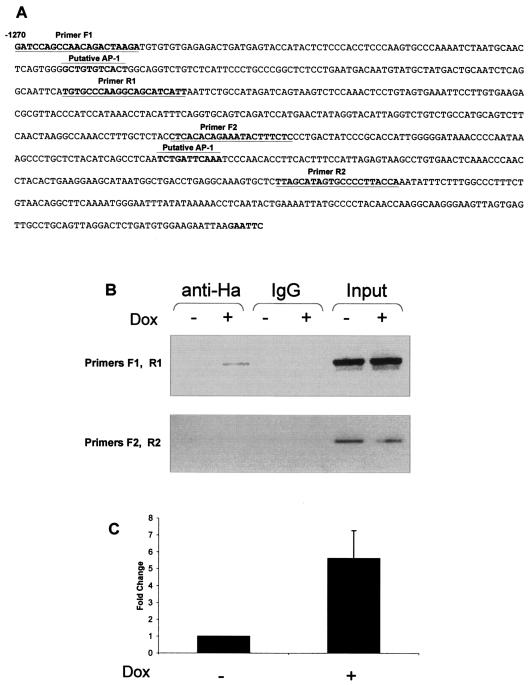
To identify the rat homologous region to the mouse Ras-GRF1 −1270/−487 promoter, we performed a BLAST search with the mouse sequence of the rat genome database (Ensembl Genome Browser). This region showed 80% homology between the rat and mouse sequences, and a search for putative AP-1 binding sites (MatInspector) (41) in the rat sequence revealed two potential sites (Fig. 3A). To verify that c-Jun bound the Ras-GRF1 promoter in Rat1a cells in vivo, we performed ChIP assays with antibodies to HA-tagged Jun with extracts from control and inducible HA-Jun-expressing cells. We used primers flanking the two putative AP-1 binding sites to amplify the immunoprecipitated DNA (Fig. 3A). The results obtained showed that a specific Ras-GRF1 promoter PCR product was obtained with the F1/R1 primer set with anti-HA antibody ChIP DNA (Fig. 3B). This result suggests that c-Jun/AP-1 interacts with the Ras-GRF1 promoter. No PCR product was obtained with the F2/R2 primer set. Quantitative real-time PCR was used to accurately determine the increase in c-Jun/AP-1 binding to the endogenous rat Ras-GRF1 promoter between control and c-Jun-expressing cells. A significant increase in binding of approximately fivefold above the control was identified (Fig. 3C). Taken together, these results provide evidence suggesting that c-Jun/AP-1 binds and activates the Ras-GRF1 promoter in vivo.
FIG. 3.
ChIP assays identify the rat Ras-GRF1 promoter as an AP-1 target gene. (A) The rat Ras-GRF1 promoter equivalent to that of the mouse 84ΔBC region (−1270/−489). Two potential AP-1 binding sites were identified, and primers (F1 and R1 and F2 and R2) flanking these sites were designed for ChIP analysis. (B) ChIP of c-Jun was performed with Rat1a cells with doxycycline-inducible HA-c-Jun. Chromatin from control (−dox) and HA-c-Jun-expressing (+dox) cells was immunoprecipitated with the HA antibody or nonspecific IgG. PCR was performed with primer sets (F1 and R1 and F2 and R2) specific for the rat Ras-GRF1 promoter regions spanning the potential AP-1 binding sites. (C) Real-time PCR analysis using ChIP DNA and primers F1 and R1. The data shown were corrected for nonspecific interactions detected with the IgG control ChIP assay. The results represent the mean ± standard error of five independent ChIP assays. P ≤ 0.05.
p75-Ras-GRF1 expression is necessary for c-Jun-regulated anchorage-independent growth.
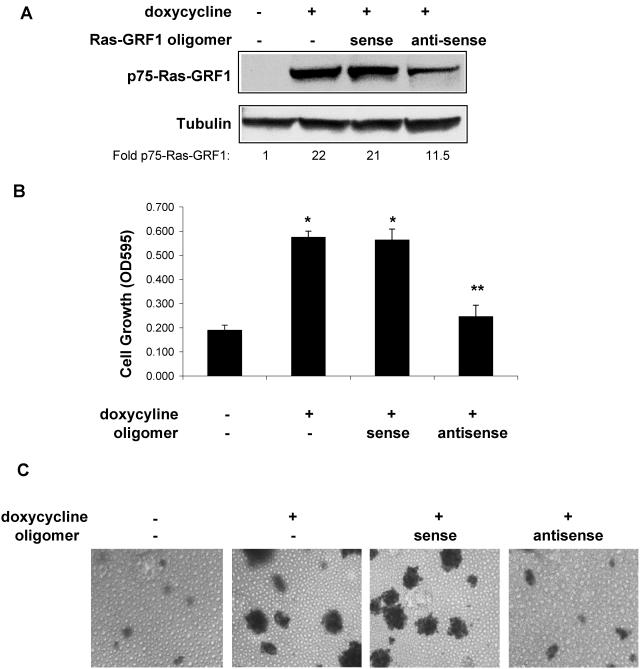
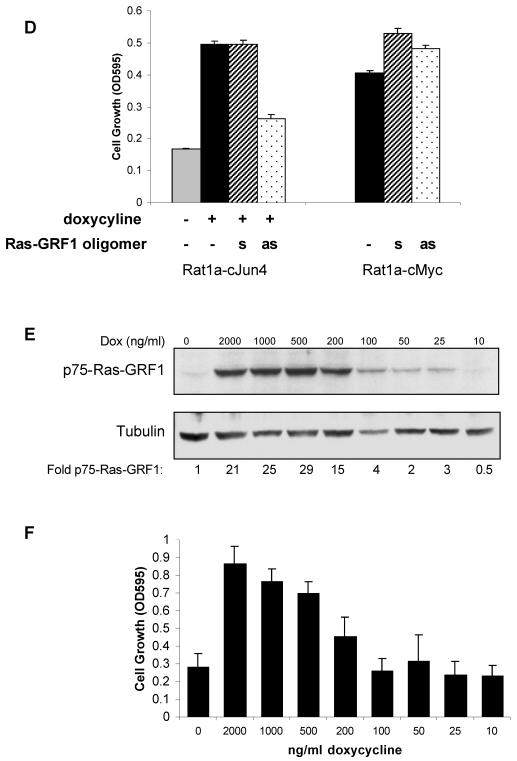
To determine whether p75-Ras-GRF1 was required for anchorage-independent growth of Rat1a-c-Jun4 cells, the cells were treated with 10 μM antisense oligomers to inhibit Ras-GRF1 expression. c-Jun-induced p75-Ras-GRF1 was inhibited approximately 50% by the antisense oligomer, while the sense oligomer had no effect (Fig. 4A). The inhibition of p75-Ras-GRF1 expression was associated with the inhibition of c-Jun-regulated anchorage-independent growth by antisense Ras-GRF1 (Fig. 4B). Cells incubated with the sense oligomer grew similarly to those expressing c-Jun only. Colony formation in the presence of c-Jun was also inhibited by antisense Ras-GRF1 oligomers, while the sense oligomers had little effect (Fig. 4C). To exclude that the Ras-GRF1 antisense oligomers are not general blockers of anchorage-independent growth in transformed Rat1a cells, the effect of these oligomers on the growth of Rat1a cells constitutively expressing the c-Myc oncoprotein (Rat1a-c-Myc) was determined. Antisense oligomers to Ras-GRF1 had no effect on anchorage-independent growth of Rat1a-c-Myc cells (Fig. 4D). This suggested that the antisense oligomer was specifically inhibiting c-Jun-regulated anchorage-independent growth via inhibition of p75-Ras-GRF1 expression. Modulating the amounts of p75-Ras-GRF1 expression by decreasing the concentration of doxycycline (Fig. 4E) resulted in a similar reduction in anchorage-independent cell growth (Fig. 4F). These results imply that p75-Ras-GRF1 expression is necessary for c-Jun-regulated transformation of Rat1a cells.
FIG. 4.
p75-Ras-GRF1 expression is essential for c-Jun/AP-1-regulated anchorage-independent growth. (A) Sense and antisense Ras-GRF1 oligomers (10 μM each) were incubated with nonadherently grown Rat1a-c-Jun4 cells in the presence of 2 μg of doxycycline per ml for 48 h. Proteins were resolved by SDS-polyacrylamide gel electrophoresis and immunoblotted for Ras-GRF1 and tubulin expression. The fold change in p75-Ras-GRF1 compared to the level in the control (without doxycycline) is shown. (B) Anchorage-independent growth of Rat1a-c-Jun4 cells in 1.5% methylcellulose in the presence of doxycycline and 10 μM Ras-GRF1 sense and antisense oligomers for 5 days. Cell growth was measured with MTT assays (Promega) and is representative of the mean ± standard deviation of triplicate samples. *, P ≤ 0.01 compared to growth without doxycycline. Growth in the presence of doxycycline and antisense oligomers was not significantly different from that in the absence of doxycycline (**, P ≥ 0.05). The data shown are representative of three independent experiments. OD595, optical density at 595 nm. (C) Cells grown as described in panel B were stained with p-iodonitrotetrazolium violet for 16 h at 37°C to observe colony formation. (D) Effect of antisense (as) and sense (s) Ras-GRF1 oligomers on anchorage-independent growth of Rat1a-c-Jun4 and Rat1a-c-Myc cells. Rat1a-c-Myc cells contained constitutive c-Myc expression. The cells were grown with sense and antisense oligomers as described for panel B. (E) Western blot analysis for p75-Ras-GRF1 expression in Rat1a-c-Jun4 cells treated with decreasing concentrations of doxycycline. Decreasing the concentration of doxycycline resulted in a decrease in p75-RasGRF1 expression. Tubulin is shown as a loading control. The fold change in p75-RasGRF1 corrected for protein loading compared to the level in the control (without doxycycline) is shown. (F) Anchorage-independent cell growth of Rat1a-c-Jun4 cells grown in the absence and presence of doxycycline at different concentrations for 6 days. Cell growth was measured by MTT assays as described in Materials and Methods. Results are the mean ± standard deviation of growth in 8 wells of a 96-well plate per condition.
Ras-GRF1 is a GEF involved in the cycling of Ras to its active, GTP-bound state. Based on this function, we predicted (i) a Ras-p75-Ras-GRF1 interaction, (ii) the detection of activated Ras, GTP-Ras, and (iii) activation of signal transduction pathways downstream of Ras in c-Jun-overexpressing cells.
p75-Ras-GRF1 interacts with Ras in c-Jun-expressing cells.
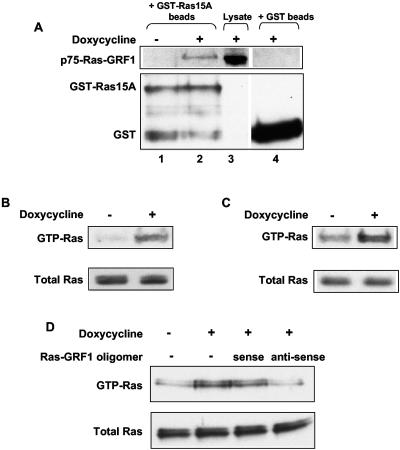
The identity of p75-Ras-GRF1 and its interaction with Ras were confirmed by using GST-Ras15A, a GST-dominant-negative Ras fusion protein that inhibits Ras function by binding GEFs (16, 20) in pull-down assays. GST-Ras15A-agarose beads pulled down p75-Ras-GRF1 in protein lysates from doxycyline-treated Rat1a-c-Jun4 cells, while an equivalent protein band was not detected in the control lysates (−dox) (Fig. 5A, lanes 1 and 2). The interaction between p75-Ras-GRF1 and GST-Ras15A is specific since a corresponding band was not detected in doxycycline-treated extracts incubated with GST-agarose beads alone (Fig. 5A, lane 4). This piece of data shows that p75-Ras-GRF1 is capable of interacting with Ras in c-Jun-expressing cells.
FIG. 5.
p75-Ras-GRF1 is required for the activation of Ras in response to c-Jun/AP-1. (A) p75-Ras-GRF1 and Ras interaction was determined by pull-down experiments with GST-Ras15A-agarose beads. The pulled-down complexes were separated by SDS-PAGE and immunoblotted with antibodies against Ras-GRF1 and GST. Lanes 1 and 2 represent control (without doxycycline) and c-Jun-containing (with doxycycline) extracts incubated with GST-Ras15A-agarose beads, and lane 3 shows p-75-Ras-GRF1 in the total protein lysate. Lane 4 represents c-Jun-containing extracts incubated with GST-agarose beads as a negative control. The top panel shows immunoblots for Ras-GRF1, and the bottom panel shows immunoblots for GST. (B and C) GTP-bound Ras was extracted from control (without doxycycline) and c-Jun-expressing (with doxycycline) Rat1a-c-Jun4 cells grown under adherent (B) and anchorage-independent (C) conditions with Raf-1-RDB agarose beads. The complexes were immunoblotted with a Ras monoclonal antibody. Equal amounts of total Ras were present in control and doxycycline-treated cell lysates. (D) Activated Ras pull down assays using protein extracts from Rat1a-c-Jun4 cells grown in suspension for 48 h with and without doxycycline in the presence of 10 μM sense or antisense Ras-GRF1 oligomers. Immunoblots with Ras antibody are shown. The results are representative of triplicate experiments.
Ras activation requires c-Jun-induced Ras-GRF1 expression.
We next sought to determine if p75-Ras-GRF1 could be connected to activation of Ras in Rat1a-c-Jun4 cells. A significant increase in GTP-bound Ras was detected in protein extracts isolated from c-Jun-expressing cells grown under both adherent (Fig. 5B) and anchorage-independent (Fig. 5C) conditions. Antisense oligomers to Ras-GRF1 inhibited the increase in GTP-Ras, while sense oligomers had little effect (Fig. 5D). Taken together, these results suggest that Ras activation in c-Jun-expressing rat fibroblasts is associated with increased p75-Ras-GRF1 expression. Inhibition of Ras activation by Ras-GRF1 antisense oligomers may explain the inhibition of anchorage-independent growth seen in Fig. 4B.
Elk-1 is activated in the presence of c-Jun overexpression.
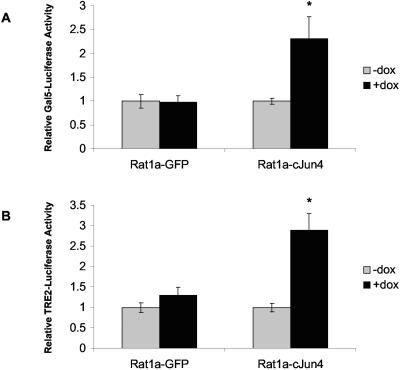
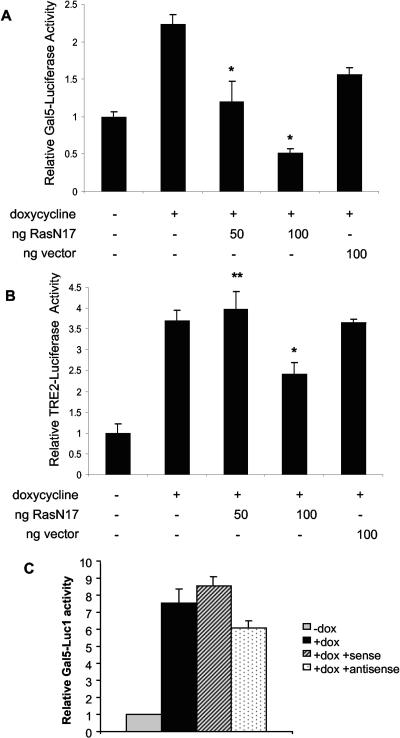
To investigate the downstream effects of Ras activation in c-Jun-expressing cells, the activation of Elk-1 was determined. Rat1a-c-Jun4 and GFP control cells grown with and without doxycycline were transiently cotransfected with a plasmid expressing a Gal4-Elk-1 fusion protein and the 5× Gal-luciferase reporter construct. A significant increase in Elk-1 activity was observed in c-Jun-containing cells and not in GFP-expressing control cells (Fig. 6A). The presence of increased AP-1 in doxycycline-treated Rat1a-c-Jun4 cells was confirmed by the activation of a reporter plasmid containing two AP-1 DNA-binding sites fused to luciferase, TRE2-luciferase (Fig. 6B). Elk-1 activity in c-Jun-expressing cells was significantly inhibited in a dose-dependent manner by cotransfection with dominant-negative RasN17 (Fig. 7A). Similar concentrations of RasN17, however, had less effect on c-Jun/AP-1 activation of the TRE2-luciferase (Fig. 7B). Thus, c-Jun overexpression in Rat1a cells results in Elk-1 activation that requires Ras activity. Cotransfection assays with sense and antisense oligomers to Ras-GRF1 with Elk-1 and the 5× Gal-luciferase reporter revealed a slight inhibition of Elk-1 activation with the antisense oligomer; however, this was not significant (Fig. 7C). Elk-1 activation is one of the far-downstream events of signal transduction pathways; it is hence possible that other factors regulated by c-Jun overexpression may impact its activation.
FIG. 6.
Elk-1 activation is Ras dependent in c-Jun-overexpressing cells. (A) Rat1a-GFP and Rat1a-c-Jun4 cells were transiently cotransfected with plasmids encoding Gal-Elk-1 fusion protein and the reporter construct 5× Gal-Luciferase (Gal5-Luciferase). The cells were grown nonadherently for 48 h in the absence (−) or presence (+) of 2 μg of doxycyline per ml. Renilla luciferase activity was used to control for transfection efficiencies. Luciferase activities were measured with the Promega dual luciferase assay kit. Data are represented as a fold change relative to controls (−dox). *, P ≤ 0.01 relative to controls (−dox). (B) Transient transfection with a construct containing two AP-1 DNA-binding sequences fused to the luciferase reporter gene (TRE2-Luciferase). The results show the mean ± standard deviation of at least three independent experiments, each performed in triplicate. *, P < 0.05.
FIG. 7.
Dominant-negative RasN17 inhibits c-Jun-regulated activation of Elk-1 activity. (A) Rat1a-c-Jun4 cells were transfected with pCMV-RasN17 or pCMV control vector as described in the legend to Fig. 4. Reporter 5× Gal5-Luciferase (Gal5-Luciferase) activation was corrected for transfection efficiency with Renilla luciferase, and the fold change relative to controls (−dox) is shown. *, P < 0.05 compared to with doxycycline. (B) TRE2-luciferase activity in the presence of pCMV-RasN17 and pCMV vector at the concentrations shown. *, P < 0.05, significantly down-regulated compared to with doxycycline; **, P > 0.05, not significantly different from with doxycycline. Results are representative of the mean ± standard deviation of assays performed in triplicate. (C) Elk-1 activation of the 5× Gal5-Luciferase construct in the presence of doxycycline and sense and antisense Ras-GRF1 oligomers. Ras-GRF1 oligomers were added before transfection and included at a concentration of 10 μM for the duration of the assay. Results represent the mean ± standard deviation of assays performed in triplicate.
Ras-PI3K-Akt signaling is activated and required for c-Jun-regulated transformation of Rat1a cells.
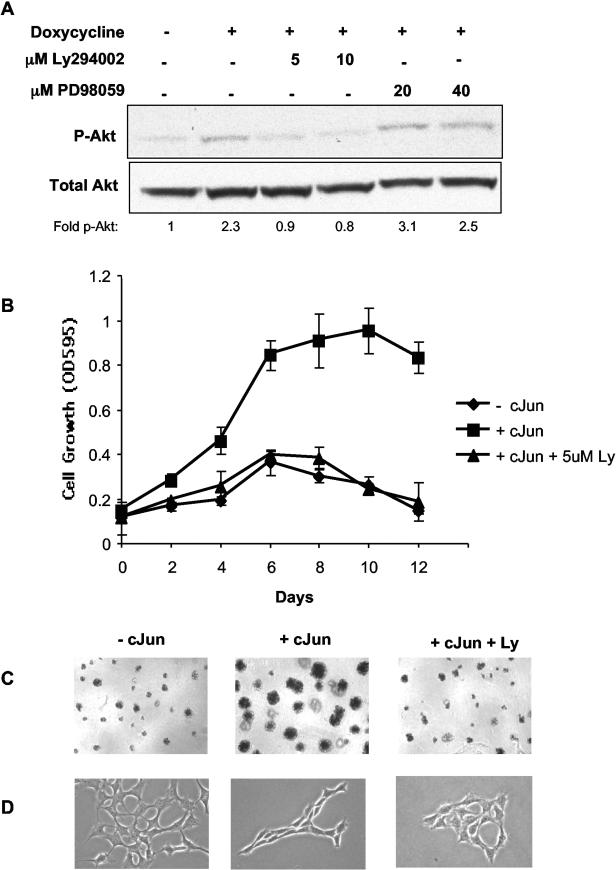
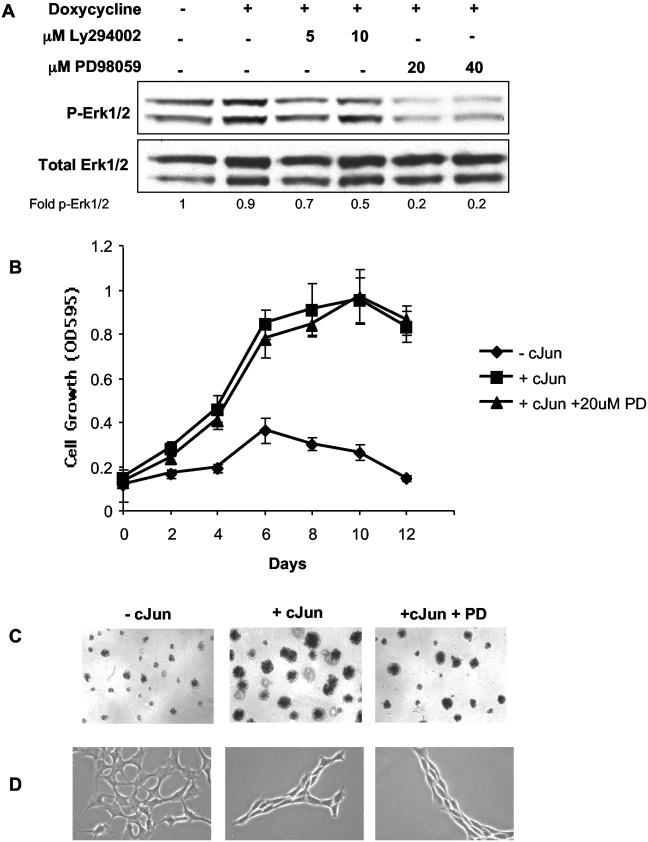
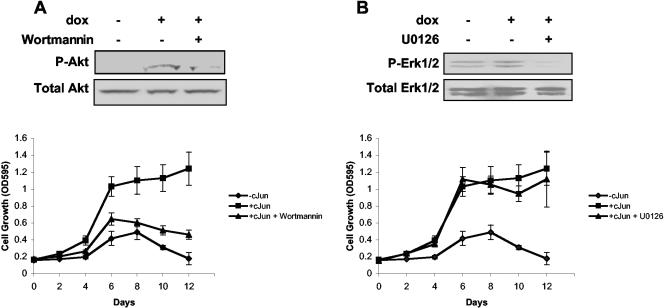
To determine which Ras-mediated signaling pathways are active in c-Jun-expressing cells, we focused on the Ras-MEK-ERK and Ras-PI3K-AKT pathways. Rat1a-c-Jun4 cells (without doxycycline) had a low level of basal phospho-AKT that was increased approximately twofold in cells grown in the presence of doxycycline (i.e., c-Jun overexpressing). This increase could be blocked by the inhibition of PI3K activity with Ly294002 at concentrations of 5 and 10 μM (Fig. 8A). Cells grown in the presence of doxycycline and PD98059 (MEK inhibitor) retained phospho-AKT levels similar to that of cells grown in the presence of doxycycline only. The inhibition of AKT phosphorylation by Ly294002 was associated with inhibition of c-Jun-regulated anchorage-independent growth in Rat1a cells (Fig. 8B). c-Jun-regulated colony formation and morphology changes were similarly inhibited by Ly294002 (Fig. 8C and D). No significant increase in ERK1/2 phosphorylation was observed in c-Jun-expressing cells; however, basal phosphorylation levels could be inhibited similarly by both 20 and 40 μM PD98059 (Fig. 9A). Interestingly, the inhibition of basal ERK phosphorylation by PD98059 had no effect on c-Jun-regulated anchorage-independent growth and colony formation (Fig. 9B and C). PD98059 also had no significant effect on c-Jun-induced cell morphology changes (Fig. 9D). Similar results were obtained with wortmannin, a PI3K inhibitor, and U0126, a MEK inhibitor, at concentrations sufficient to block phosphorylation of AKT and ERK1/2 respectively (Fig. 10A and B). The inhibition of PI3K prevented c-Jun-regulated anchorage-independent growth, while inhibition of MEK had little effect. These results implicate c-Jun in the regulation of AKT phosphorylation and suggest that signaling via the Ras-PI3K-AKT pathway may be activated and required for c-Jun/AP-1-regulated anchorage-independent growth.
FIG. 8.
PI3K activity is required for c-Jun-regulated anchorage-independent cell growth. (A) Total proteins from Rat1a-c-Jun4 cells grown in the absence (−) and presence (+) of 2 μg of doxycycline per ml for 48 h in suspension with Ly294002 and PD98059 at the indicated concentrations were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted with antibodies against p-AKT1 and total AKT1. The fold change in p-AKT1 (p-AKT1 relative to total AKT1) compared to the level in the control (without doxycycline) is shown. (B) Rat1a-c-Jun4 cells were grown in the absence of doxycycline (− cJun), the presence of 2 μg of doxycycline per ml (+ cJun), and the presence of doxycycline and 5 μM Ly294002 (+ cJun + 5uM Ly). Growth in media containing 10% FBS and 1.5% methylcellulose to maintain cells in an anchorage-independent state was assayed with the MTT assay. OD595, optical density at 595 nm. (C) c-Jun-induced colony formation in 1.5% methylcellulose and (D) cell morphology changes are inhibited by 5 μM Ly294002.
FIG. 9.
MEK activation of ERK1/2 is not required for c-Jun-regulated cell growth. (A) Western blots showing p-ERK1/2 and total ERK1/2 protein levels. (B) Rat1a-c-Jun4 cells were grown in the absence (♦) or presence (▪) of 2 μg of doxycycline per ml and the presence (▴) of doxycycline and 20 μM PD98059. Cells were assayed as described in the legend to Fig. 8A. The fold change in p-ERK1/2 (p-ERK1/2 relative to total ERK1/2) compared to that of the control (without doxycycline) is shown. c-Jun-induced colony formation (C) and morphological changes (D) were minimally affected by inhibition of MEK activity with 20 μM PD98059.
FIG. 10.
Effect of wortmannin and U0126 on c-Jun-regulated anchorage-independent growth. (A) Rat1a-c-Jun4 cells were grown in the absence of doxycycline (−cJun), the presence of 2 μg of doxycycline per ml (+cJun), and the presence of doxycycline and 50 nM wortmannin (+cJun + wortmannin). A total of 50 nM wortmannin was sufficient to inhibit phosphorylation of AKT1 and prevent c-Jun-regulated anchorage-independent growth. (B) Cells grown in the absence of doxycycline (−cJun), the presence of 2 μg of doxycycline per ml (+cJun), and the presence of doxycycline and 50 nM U0126 (+cJun + U0126). U0126 at 50 nM prevented phosphorylation of p-ERK1/2 but had little effect on c-Jun-regulated anchorage-independent growth.
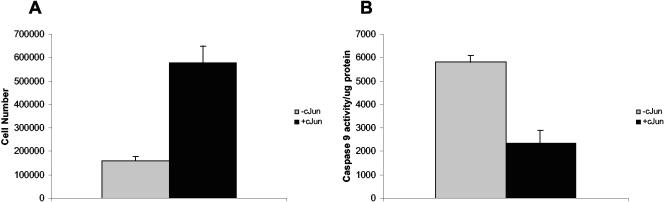
Since the PI3K-AKT pathway is involved in cell survival mechanisms, we addressed whether the effect of c-Jun induction on growth is due to increased proliferation in anchorage-independent conditions, decreased death, or both. Based on our previously published data, we have established that overexpression of c-Jun in Rat1a cells resulted in an increase in proliferation as measured by Trypan blue staining (33) (Fig. 11A). Caspase 9 assays as an indicator of cell death or anoikis revealed that c-Jun-overexpressing cells grown under anchorage-independent growth conditions had decreased caspase 9 activity compared to control cells grown in the absence of doxycycline (Fig. 11B). Taken together, these results imply that overexpression of c-Jun in Rat1a cells grown under anchorage-independent conditions was involved in both an increase in proliferation and suppression of cell death.
FIG. 11.
c-Jun-regulated anchorage-independent growth of Rat1a-c-Jun4 cells is due to increased proliferation and decreased cell death. (A). c-Jun-regulated cell growth was measured by Trypan blue staining. Viable cells were counted after 48 h of growth in suspension without and with 2 μg of doxycycline per ml. Equal numbers of cells were plated at the start of the assay. (B) Caspase 9 assays with protein extracts of Rat1a-c-Jun4 cells grown without and with 2 μg of doxycycline per ml for 48 h. Caspase 9 activity per microgram of protein for duplicate experiments is shown.
DISCUSSION
The conventional position of c-Jun/AP-1 in signal transduction cascades has been closer to the end of MAPK pathways. Signaling via these pathways converges on and activates c-Jun/AP-1 such that downstream events can be initiated (50, 51). Substantial evidence has placed AP-1 downstream of MAPK signaling pathways such as ERK, JNK, and p38 (45, 46) and that activation of c-Jun/AP-1 complexes by phosphorylation is essential to its function as a transcriptional regulator (7). Activation of other members of the AP-1 transcription factor complex such as the Fos family of proteins is thought to be necessary for the autoregulation of c-Jun expression (46). The data presented here demonstrate that c-Jun/AP-1 can regulate the expression of signal transduction molecules such as Ras-GRF1, which are located upstream in the signal transduction cascades. Ras-GRF1 is a GEF that catalyzes the GDP/GTP exchange required for activation of Ras (10, 17, 21). It was initially discovered as a brain-specific 140-kDa protein with its catalytic domain residing in the C-terminal region (37, 47, 52). The functional domains of full-length p140-Ras-GRF1 include a pleckstrin homology domain, a coiled-coil region, a calmodulin-dependent activation domain, the ilimaquinon motif, a DBL homology domain, a protein instability domain, and a CDC25 domain, the catalytic domain (1). In this study, we show that overexpression of c-Jun in rat fibroblasts is associated with an increase in the expression of a 75-kDa form of Ras-GRF1, p75-Ras-GRF1. Without sequence information on the c-Jun-induced p75 protein, we cannot conclusively comment on the Ras-GRF1 domains contained in this protein. Our Northern and Western blot data, however, suggest that p75-Ras-GRF1 may contain the C-terminal region of Ras-GRF1 that contains the CDC25 catalytic domain. This may have arisen due to an alternate splice variant, since Northern blot analysis identified a transcript smaller than that encoding p140-Ras-GRF. Alternative forms of Ras-GRF1, ranging in size from approximately 50 to 140 kDa, have been identified in various tissues and cell lines (14, 22, 26). The biological significance of the smaller forms of Ras-GRF1 is presently unclear. However, overexpression of p75-78, p95, p120, and p140-Ras-GRF1 results in transformation of NIH 3T3 cells (13). In addition, N-terminally truncated versions of Ras-GRF1 that retain the catalytic C-terminal domain appear to be more effective at GDP/GTP exchange of H-Ras than p140-Ras-GRF1 (6).
Based on our results, we propose that c-Jun regulates the expression of p75-Ras-GRF1, which in turn activates Ras and subsequent signal transduction in rat fibroblasts. Ras-mediated signal transduction is increasingly complex and involves its interaction with numerous effector molecules (12). We expected that signal cascades triggered by Ras activation would include the Ras-PI3K-AKT and/or Ras-MEK-ERK pathway. Our findings implicate c-Jun overexpression with an increase in PI3K-AKT1 rather than MEK-ERK activation. This is supported by the observation that the Ras-MEK-ERK pathway appears nonessential for c-Jun-regulated anchorage-independent growth. It is likely that within a given cellular context, such as increased AP-1 activity, the physiological relevance may be that regulation of specific exchange factors results in the activation of specific Ras subpathways. Ras-GRF1 in particular is capable of activating R-Ras, while SOS does not (24, 48). R-Ras in turn activates PI3K rather than MAPK signaling pathways (36). Activation of PI3K by R-Ras is essential for its biological activity in cellular transformation, adhesion, and survival (39). The increase in PI3K activity observed in c-Jun-overexpressing cells is presumably related to survival and transformation under otherwise unfavorable cell growth conditions, such as anchorage independence, which would normally result in cell death, possibly by anoikis. Our finding that MEK-ERK activation is not required for c-Jun-regulated anchorage-independent growth of rat fibroblasts was supported by observations that ERK1/2 phosphorylation is significantly decreased in cells growing under adherent conditions (V. Leaner, unpublished data). These findings are in agreement with those from a recent study showing that transformation of chicken embryo fibroblasts with v-Jun results in activation of Ras and, surprisingly, in deactivation of ERK-MAPK signaling pathways (9). These authors suggest that activation of Ras in v-Jun-expressing cells may be a result of the increased expression of a previously reported v-Jun target gene that codes for heparin-binding epidermal growth factor-like growth factor (HB-EGF) (23). While we have not determined whether HB-EGF is a c-Jun target, our data showing an increase of Ras-GRF1 in response to c-Jun expression and evidence for its interaction with Ras and Ras activation suggest that p75-Ras-GRF1 may be one of the factors contributing to Ras activation in c-Jun-expressing cells. Black et al. (9) also propose that ERK deactivation may be due to inhibition of signaling via Ras-Raf and increased expression of MKP-1 and -2. In support of their results, we have also identified MKP-1 as a c-Jun/AP-1-regulated gene by microarray analysis (34).
To underscore the importance of p75-Ras-GRF1, we found that it is essential for c-Jun-regulated cellular transformation, since inhibition of its expression blocks anchorage-independent growth. We are currently determining whether introduction of p75-Ras-GRF1 as a single gene may cause transformation of Rat1a fibroblasts. This, however, seems unlikely, since the introduction of p140-Ras-GRF1 (kindly provided by L. Feig) into Rat1a cells did not result in anchorage-independent growth (V. Leaner, unpublished observations). This finding was not surprising considering that very few AP-1 target genes have been found to transform cells as single genes (reviewed by Vogt in reference 51). It is thus feasible that p75-Ras-GRF1 expression may be necessary but not sufficient for cellular transformation by c-Jun/AP-1. A combination of c-Jun/AP-1-regulated genes may be required for cellular transformation. These genes likely include c-Jun/AP-1 targets involved in cell cycle, signal transduction, survival, and proliferation. The work presented in this paper introduces an additional mechanism for the role of c-Jun/AP-1 as a regulator of both upstream and downstream signaling events, which may influence cell growth and survival.
Acknowledgments
We thank D. R. Lowy (National Institutes of Health) for helpful discussions and critical review of the manuscript. We also thank Aaron Bell for excellent technical assistance, L. Feig (Tufts University School of Medicine) for the p140-Ras-GRF1 plasmid, E. Prochownik (Children's Hospital of Pittsburgh) for the Rat1a-c-Myc cells, and C. Plass (Ohio State University) for the Ras-GRF1 promoter constructs.
REFERENCES
- 1.Anborgh, P. H., X. Qian, A. G. Papageorge, W. C. Vass, J. E. DeClue, and D. R. Lowy. 1999. Ras-specific exchange factor GRF: oligomerization through its Dbl homology domain and calcium-dependent activation of Raf. Mol. Cell. Biol. 19:4611-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angel, P., E. A. Allegretto, S. T. Okino, K. Hattori, W. J. Boyle, T. Hunter, and M. Karin. 1988. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature 332:166-171. [DOI] [PubMed] [Google Scholar]
- 3.Angel, P., M. Imagawa, R. Chiu, B. Stein, R. J. Imbra, H. J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 4.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 5.Arozarena, I., D. Matallanas, M. T. Berciano, V. Sanz-Moreno, F. Calvo, M. T. Munoz, G. Egea, M. Lafarga, and P. Crespo. 2004. Activation of H-Ras in the endoplasmic reticulum by the RasGRF family guanine nucleotide exchange factors. Mol. Cell. Biol. 24:1516-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baouz, S., E. Jacquet, A. Bernardi, and A. Parmeggiani. 1997. The N-terminal moiety of CDC25(Mm), a GDP/GTP exchange factor of Ras proteins, controls the activity of the catalytic domain. Modulation by calmodulin and calpain. J. Biol. Chem. 272:6671-6676. [DOI] [PubMed] [Google Scholar]
- 7.Behrens, A., W. Jochum, M. Sibilia, and E. F. Wagner. 2000. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene 19:2657-2663. [DOI] [PubMed] [Google Scholar]
- 8.Behrens, A., M. Sibilia, and E. F. Wagner. 1999. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat. Genet. 21:326-329. [DOI] [PubMed] [Google Scholar]
- 9.Black, E. J., M. Walker, W. Clark, A. MacLaren, and D. A. Gillespie. 2002. Cell transformation by v-Jun deactivates ERK MAP kinase signalling. Oncogene 21:6540-6548. [DOI] [PubMed] [Google Scholar]
- 10.Boguski, M. S., and F. McCormick. 1993. Proteins regulating Ras and its relatives. Nature 366:643-654. [DOI] [PubMed] [Google Scholar]
- 11.Bohmann, D., T. J. Bos, A. Admon, T. Nishimura, P. K. Vogt, and R. Tjian. 1987. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science 238:1386-1392. [DOI] [PubMed] [Google Scholar]
- 12.Campbell, S. L., R. Khosravi-Far, K. L. Rossman, G. J. Clark, and C. J. Der. 1998. Increasing complexity of Ras signaling. Oncogene 17:1395-1413. [DOI] [PubMed] [Google Scholar]
- 13.Cen, H., A. G. Papageorge, W. C. Vass, K. E. Zhang, and D. R. Lowy. 1993. Regulated and constitutive activity by CDC25Mm (GRF), a Ras-specific exchange factor. Mol. Cell. Biol. 13:7718-7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cen, H., A. G. Papageorge, R. Zippel, D. R. Lowy, and K. Zhang. 1992. Isolation of multiple mouse cDNAs with coding homology to Saccharomyces cerevisiae CDC25: identification of a region related to Bcr, Vav, Dbl and CDC24. EMBO J. 11:4007-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 16.Chen, S. Y., S. Y. Huff, C. C. Lai, C. J. Der, and S. Powers. 1994. Ras-15A protein shares highly similar dominant-negative biological properties with Ras-17N and forms a stable, guanine-nucleotide resistant complex with CDC25 exchange factor. Oncogene 9:2691-2698. [PubMed] [Google Scholar]
- 17.Cherfils, J., and P. Chardin. 1999. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 24:306-311. [DOI] [PubMed] [Google Scholar]
- 18.Cook, S. J., N. Aziz, and M. McMahon. 1999. The repertoire of Fos and Jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol. Cell. Biol. 19:330-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Puente, A., J. Hall, Y. Z. Wu, G. Leone, J. Peters, B. J. Yoon, P. Soloway, and C. Plass. 2002. Structural characterization of Rasgrf1 and a novel linked imprinted locus. Gene 291:287-297. [DOI] [PubMed] [Google Scholar]
- 20.Feig, L. A. 1999. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1:E25-E27. [DOI] [PubMed] [Google Scholar]
- 21.Feig, L. A., and R. J. Buchsbaum. 2002. Cell signaling: life or death decisions of ras proteins. Curr. Biol. 12:R259-R261. [DOI] [PubMed] [Google Scholar]
- 22.Ferrari, C., R. Zippel, E. Martegani, N. Gnesutta, V. Carrera, and E. Sturani. 1994. Expression of two different products of CDC25Mm, a mammalian Ras activator, during development of mouse brain. Exp. Cell Res. 210:353-357. [DOI] [PubMed] [Google Scholar]
- 23.Fu, S., I. Bottoli, M. Goller, and P. K. Vogt. 1999. Heparin-binding epidermal growth factor-like growth factor, a v-Jun target gene, induces oncogenic transformation. Proc. Natl. Acad. Sci. USA 96:5716-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gotoh, T., Y. Niino, M. Tokuda, O. Hatase, S. Nakamura, M. Matsuda, and S. Hattori. 1997. Activation of R-Ras by Ras-guanine nucleotide-releasing factor. J. Biol. Chem. 272:18602-18607. [DOI] [PubMed] [Google Scholar]
- 25.Gruda, M. C., K. Kovary, R. Metz, and R. Bravo. 1994. Regulation of Fra-1 and Fra-2 phosphorylation differs during the cell cycle of fibroblasts and phosphorylation in vitro by MAP kinase affects DNA binding activity. Oncogene 9:2537-2547. [PubMed] [Google Scholar]
- 26.Guerrero, C., J. M. Rojas, M. Chedid, L. M. Esteban, D. B. Zimonjic, N. C. Popescu, J. Font de Mora, and E. Santos. 1996. Expression of alternative forms of Ras exchange factors GRF and SOS1 in different human tissues and cell lines. Oncogene 12:1097-1107. [PubMed] [Google Scholar]
- 27.Guerrero, S., I. Casanova, L. Farre, A. Mazo, G. Capella, and R. Mangues. 2000. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 60:6750-6756. [PubMed] [Google Scholar]
- 28.Han, T.-H., and R. Prywes. 1995. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol. Cell. Biol. 15:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill, C. S., J. Wynne, and R. Treisman. 1994. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J. 13:5421-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hommura, F., M. Katabami, V. D. Leaner, H. Donninger, T. F. Sumter, L. M. Resar, and M. J. Birrer. 2004. HMG-I/Y is a c-Jun/activator protein-1 target gene and is necessary for c-Jun-induced anchorage-independent growth in Rat1a cells. Mol. Cancer Res. 2:305-314. [PubMed] [Google Scholar]
- 31.Jochum, W., E. Passegue, and E. F. Wagner. 2001. AP-1 in mouse development and tumorigenesis. Oncogene 20:2401-2412. [DOI] [PubMed] [Google Scholar]
- 32.Kallunki, T., T. Deng, M. Hibi, and M. Karin. 1996. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell 87:929-939. [DOI] [PubMed] [Google Scholar]
- 33.Kinoshita, I., V. Leaner, M. Katabami, R. G. Manzano, P. Dent, A. Sabichi, and M. J. Birrer. 2003. Identification of cJun-responsive genes in Rat-1a cells using multiple techniques: increased expression of stathmin is necessary for cJun-mediated anchorage-independent growth. Oncogene 22:2710-2722. [DOI] [PubMed] [Google Scholar]
- 34.Leaner, V. D., I. Kinoshita, and M. J. Birrer. 2003. AP-1 complexes containing cJun and JunB cause cellular transformation of Rat1a fibroblasts and share transcriptional targets. Oncogene 22:5619-5629. [DOI] [PubMed] [Google Scholar]
- 35.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 36.Marte, B. M., P. Rodriguez-Viciana, S. Wennstrom, P. H. Warne, and J. Downward. 1997. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr. Biol. 7:63-70. [DOI] [PubMed] [Google Scholar]
- 37.Martegani, E., M. Vanoni, R. Zippel, P. Coccetti, R. Brambilla, C. Ferrari, E. Sturani, and L. Alberghina. 1992. Cloning by functional complementation of a mouse cDNA encoding a homologue of CDC25, a Saccharomyces cerevisiae RAS activator. EMBO J. 11:2151-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mechta, F., D. Lallemand, C. M. Pfarr, and M. Yaniv. 1997. Transformation by ras modifies AP1 composition and activity. Oncogene 14:837-847. [DOI] [PubMed] [Google Scholar]
- 39.Osada, M., T. Tolkacheva, W. Li, T. O. Chan, P. N. Tsichlis, R. Saez, A. C. Kimmelman, and A. M.-L. Chan. 1999. Differential roles of Akt, Rac, and Ral in R-Ras-mediated cellular transformation, adhesion, and survival. Mol. Cell. Biol. 19:6333-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez, S., E. Vial, H. van Dam, and M. Castellazzi. 2001. Transcription factor ATF3 partially transforms chick embryo fibroblasts by promoting growth factor-independent proliferation. Oncogene 20:1135-1141. [DOI] [PubMed] [Google Scholar]
- 41.Quandt, K., K. Frech, and T. Werner. 1997. Analysis of transcriptional regulatory regions based on detecting transcription factor binding sites and studies of their mutual position. Mol. Biol. (Moscow) 31:749-758. [PubMed] [Google Scholar]
- 42.Quilliam, L. A., J. F. Rebhun, and A. F. Castro. 2002. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog. Nucleic Acid Res. Mol. Biol. 71:391-444. [DOI] [PubMed] [Google Scholar]
- 43.Rapp, U. R., J. Troppmair, T. Beck, and M. J. Birrer. 1994. Transformation by Raf and other oncogenes renders cells differentially sensitive to growth inhibition by a dominant negative c-jun mutant. Oncogene 9:3493-3498. [PubMed] [Google Scholar]
- 44.Schutte, J., J. D. Minna, and M. J. Birrer. 1989. Deregulated expression of human c-jun transforms primary rat embryo cells in cooperation with an activated c-Ha-ras gene and transforms rat-1a cells as a single gene. Proc. Natl. Acad. Sci. USA 86:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 46.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 47.Shou, C., C. L. Farnsworth, B. G. Neel, and L. A. Feig. 1992. Molecular cloning of cDNAs encoding a guanine-nucleotide-releasing factor for Ras p21. Nature 358:351-354. [DOI] [PubMed] [Google Scholar]
- 48.Tian, X., and L. A. Feig. 2001. Basis for signaling specificity difference between Sos and Ras-GRF guanine nucleotide exchange factors. J. Biol. Chem. 276:47248-47256. [DOI] [PubMed] [Google Scholar]
- 49.van Dam, H., and M. Castellazzi. 2001. Distinct roles of Jun:Fos and Jun:ATF dimers in oncogenesis. Oncogene 20:2453-2464. [DOI] [PubMed] [Google Scholar]
- 50.Vogt, P. K. 2002. Fortuitous convergences: the beginnings of JUN. Nat. Rev. Cancer 2:465-469. [DOI] [PubMed] [Google Scholar]
- 51.Vogt, P. K. 2001. Jun, the oncoprotein. Oncogene 20:2365-2377. [DOI] [PubMed] [Google Scholar]
- 52.Wei, W., B. Das, W. Park, and D. Broek. 1994. Cloning and analysis of human cDNAs encoding a 140-kDa brain guanine nucleotide-exchange factor, Cdc25GEF, which regulates the function of Ras. Gene 151:279-284. [DOI] [PubMed] [Google Scholar]
- 53.Yang, H., D. Cooley, J. E. Legakis, Q. Ge, R. Andrade, and R. R. Mattingly. 2003. Phosphorylation of the Ras-GRF1 exchange factor at Ser916/898 reveals activation of Ras signaling in the cerebral cortex. J. Biol. Chem. 278:13278-13285. [DOI] [PubMed] [Google Scholar]