Abstract
Alternative splicing of the locus AβH-J-J generates three functionally distinct proteins: an enzyme, AβH (aspartyl-β-hydroxylase), a structural protein of the sarcoplasmic reticulum membrane (junctin), and an integral membrane calcium binding protein (junctate). Junctin and junctate are two important proteins involved in calcium regulation in eukaryotic cells. To understand the regulation of these two proteins, we identified and functionally characterized one of the two promoter sequences of the AβH-J-J locus. We demonstrate that the P2 promoter of the AβH-J-J locus contains (i) a minimal sequence localized within a region −159 bp from the transcription initiation site, which is sufficient to activate transcription of both mRNAs; (ii) sequences which bind known transcriptional factors such as those belonging to the myocyte enhancer factor 2 (MEF-2), MEF-3, and NF-κB protein families; and (iii) sequences bound by unknown proteins. The functional characterization of the minimal promoter in C2C12 cells and in the rat soleus muscle in vivo model indicates the existence of cis elements having positive and negative effects on transcription. In addition, our data demonstrate that in striated muscle cells the calcium-dependent transcription factor MEF-2 is crucial for the transcription activity directed by the P2 promoter. The transcription directed by the AβH-J-J P2 promoter is induced by high expression of MEF-2, further stimulated by calcineurin and Ca2+/calmodulin-dependent protein kinase I, and inhibited by histone deacetylase 4.
Skeletal muscle contraction involves the transduction of an electrical signal, generated at the neuromuscular junction, into a transient increase of [Ca2+]i within the skeletal muscle fiber via a mechanism which is referred to as excitation-contraction (EC) coupling (35). The sarcoplasmic reticulum (SR) is the intracellular membrane compartment of striated muscle that controls the intracellular calcium concentration and therefore plays an important role in EC coupling (35). The anatomical site of EC coupling is the triad, an intracellular synapse formed by the association of two membrane compartments, the transverse tubules, which are an invagination of the plasmalemma, and the sarcoplasmic reticulum terminal cisternae. The portion of terminal cisternae facing the transverse tubules is called the junctional face membrane. Two major protein constituents of the triad membranes have been identified and shown to play a crucial role in EC coupling (35): the dihydropyridine receptors (DHPRs), localized in T-tubule membrane, and the ryanodine receptors (RyRs), localized in the terminal cisternae of the sarcoplasmic reticulum junctional face membrane. In skeletal muscle the dihydropyridine receptors respond to transverse tubule depolarization by sensing the voltage variation and induce calcium release from the skeletal muscle sarcoplasmic reticulum via direct activation of the RyR. The SR and T tubule membranes are also endowed with a variety of known proteins involved in calcium storage (calsequestrin) (18), signal transduction between the transverse tubule and SR (triadin, junctin, protein kinases, and their regulators) (21, 69), and calcium homeostasis (Ca pump and Na/Ca exchanger) (39). The triad membranes undergo profound structural changes during adaptive responses to disuse and denervation muscle atrophy, suggesting that this membrane compartment could be involved in the mechanism responsible for muscle adaptation and development (44, 54, 58). Muscle disuse seems to induce a complex response entailing a set of transcription factors which are involved in an earlier developmental stage and include high levels of Id and some specific myogenic factors and immediate-early gene products, as well as either down-regulation, e.g., myosin light and heavy chains, or up-regulation, e.g., dihydropyridine receptor, of several muscle-specific genes (12, 22, 1, 44). Moreover, muscle adaptation may entail two distinct pathways, for immediate-early responses and for long-term responses. The genetic hierarchy controlling skeletal muscle myogenesis and differentiation as well as adaptive responses, such as disuse atrophy, compensatory hypertrophy, or denervated degradation, is far from being completely understood at the gene and protein levels (55). Thus, comprehension of the regulation of the expression of the genes involved in EC coupling is crucial not only to understand the mechanism(s) leading to the mechanical properties of muscle fibers under normal and adapting conditions but also to unravel the signaling pathway involved in this adaptation.
The specific regulatory DNA sequences and the relative transcription factors of most of the genes involved in EC coupling, such as Ca2+-ATPase, calsequestrin, ryanodine receptor, triadin, junctin, and dihydropyridyne receptor, are not known. However, little is known about the mechanisms underlying transcriptional control in normal muscles and in muscles during adaptive processes.
We have recently cloned, sequenced, and characterized the AβH-J-J locus, a genomic sequence which generates three functionally distinct proteins which are differentially expressed in various tissues. In addition to the enzyme aspartyl-β-hydroxylase (AβH), this locus encodes junctin a structural protein of SR and the membrane-bound calcium binding protein junctate (60, 61). Junctate is a novel integral calcium binding protein of sarco(endo)plasmic reticulum membrane, which is likely involved in calcium signaling in both excitable and nonexcitable cells (25, 61). Since these proteins are generated by alternative splicing of the primary transcripts of the AβH-J-J locus (60), the transcriptional regulation of the AβH-J-J locus is expected to be quite complex.
To date, little has been known about the promoter sequences regulating the expression of the AβH-J-J locus. In particular, no data are available on the possible promoter activity of regions located upstream of the junctin sequences. To understand how expression of the three main transcripts of the AβH-J-J locus is regulated, we have cloned, sequenced, and analyzed one of the two putative promoter sequences regulating the transcription of junctin, junctate, and aspartyl-β-hydroxylase.
MATERIALS AND METHODS
Cell cultures.
C2C12 mouse muscle myoblast, H9c2 rat heart myoblast, RD human embryonic rhabdomyosarcoma, HepG2 human hepatoblastoma, and HeLa human cervix epithelial carcinoma cell lines were cultured as described previously (27, 28, 49, 57, 67). C2C12-GM cells were cultured in medium supplemented with 10% fetal calf serum (growth medium [GM]), differentiated C2C12-DM cells were obtained by culturing cells in medium supplemented with 2% horse serum plus 10 μg of insulin (Sigma)/ml (differentiation medium [DM]) for 72 h before nuclear extraction, and total RNA purification or chromatin immunoprecipitation (ChIP) assays were performed (52).
RT-PCR.
Total RNA from human adult normal tissue was purchased from BD Biosciences Clontech (Palo Alto, Calif.). Total RNA was harvested from RD, C2C12-GM, or C2C12-DM cell lines by using TRIzol reagent (Invitrogen, Carlsbad, Calif.). cDNA was synthesized from 2 μg of total RNA by the use of ImProm-II (Promega, Madison, Wis.). PCR was performed using a GeneAmp PCR 9600 system (Perkin Elmer) 1/20 of reverse transcription reaction mixture (cDNA), the e2F forward primer (5′-ACC TGC CAG CAG TAC TTT TG-3′), and the e3R reverse primer (5′-TTC CTG AGA GTC CGC CTT TC-3′) or the e2F1 forward primer (5′-TTC ATG GAT TGA AGA AAT CAA AAT G-3′) and the e5R reverse primer (5′-AAT AAA ACT TTG GCA TCA TCC ACA TCA AAA TCT CC-3′) (designed to amplify products of 147 and 255 bp, respectively, and spanning one or more introns to rule out amplification from genomic DNA) (60), 2 U of Taq polymerase, and 33 μM deoxynucleoside triphosphates; the PCR conditions were as follows: 30 or 35 cycles of amplification, respectively, with 15 s of denaturation at 95°C, 30 s of annealing at 62°C, and 20 s of elongation at 72°C. Starting total RNA were normalized by performing reverse transcription-PCR (RT-PCR) of the beta-2 microglobulin housekeeping gene (data not shown).
5′ RACE.
Total RNA (1 μg), purified from human adult skeletal muscle or from RD cells, was subjected to 5′ rapid amplification of cDNA ends (RACE) by the use of a 5′ RACE system (Life Technologies, Gaithersburg, Md.). cDNA was synthesized using the primer e5R, complementary to an exon 5 sequence of the AβH-J-J locus (Fig. 1 and 2) (60), and a poly(G) tail was added by terminal transferase. PCR was performed using this cDNA as the template, 5′ RACE Ambriged anchor primer, and the primer e3R complementary to an exon 3 sequence (60). A dilution of the original PCR was reamplified using 5′ RACE Ambriged universal amplification primer and the nested primer e2R (5′-ATC TTC AGC CAT TTT GAT TTC TTC AAT CC-3′) complementary to an exon 2 sequence. PCR products were purified with Microcon-30 (Millipore, Bedford, Mass.) and subjected to direct sequencing with an ABI PRISM BigDye terminator cycle sequencing ready reaction kit and an ABI PRISM 377 DNA sequencer (PE Applied Biosystems, Foster City, Calif.). Moreover, PCR products were cloned with a pGEM-T Vector System (Promega) and recombinant plasmids were sequenced. The transcription initiation site observed by 5′ RACE was numbered +1, and all the other sites are numbered relative to it.
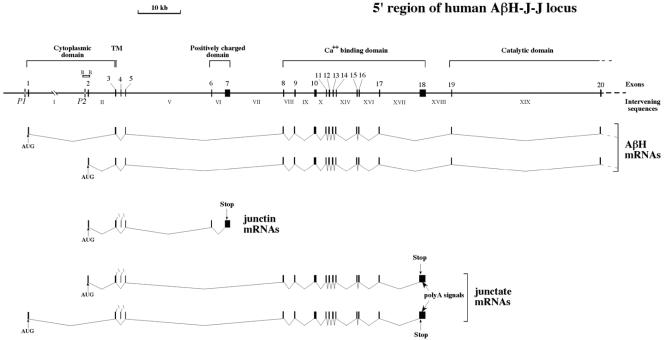
FIG. 1.
Structure of the 5′ end of the human locus for aspartyl-β-hydroxylase, junctin, and junctate. Arabic numbers over black boxes indicate exons. Intervening sequences are indicated in roman numerals. The two putative promoters P1 and P2 are indicated. A schematic representation of aspartyl-β-hydroxylase, junctin, and junctate exon splicing (11, 60) is shown at the bottom of the panel. The cytoplasmic, transmembrane (TM), positively charged, calcium binding, and catalytic domains are indicated. The locations of AUG, stop codons, and poly(A) signals are shown. The BglII (B) plasmid subclone of BAC 1 (60) covering the second exon of the locus is also shown.
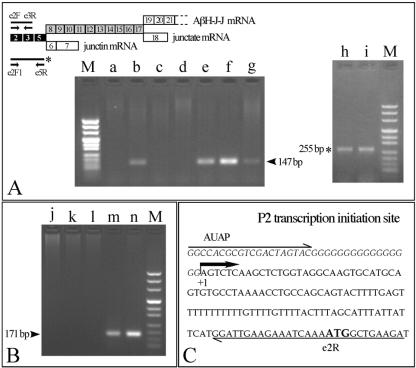
FIG. 2.
(A) In the upper part of the panel, the three transcripts starting from the P2 promoter, the PCR primers, and the 147-bp and the 255-bp (*) PCR products are represented; black boxes indicate exons common to the three transcripts; grey boxes indicate exons common only to aspartyl-β-hydroxylase and junctate. Oligo(dT) RT-PCR was performed with total RNA isolated from human adult normal tissues or cell lines with the e2F/e3R (lanes a to g) or the e2F1/e5R (lanes h and i) primers and analyzed with agarose gel electrophoresis (lower part of panel). Lanes: a, kidney; b, brain; c, adrenal gland; d, liver; e, heart; f, skeletal muscle; g, RD cell line; h, C2C12-GM cells (C2C12 cells cultured in growth medium in the presence of 10% fetal calf serum); i, C2C12-DM cells (C2C12 cells cultured in differentiation medium in the presence of 2% horse serum and 10 μg of insulin/ml); M, pUC Mix Marker 8 (Fermentas). (B) Analysis of 5′ RACE products of AβH-J-J exon 2 starting transcripts. cDNAs were synthesized from RD cells (lane m) or human adult skeletal muscle (lane n) total RNA by using the e5R primer. After addition of a poly(G) tail, PCR was performed with the gene-specific e3R primer. One-fifth of the nested PCRs, after a procedure performed with a gene-specific e2R primer, was analyzed by gel electrophoresis. Lanes j to l, control reactions from RD cell RNA performed in the absence of reverse transcriptase, terminal deoxynucleotidyl transferase, and both, respectively. (C) Nucleotide sequence of RACE products, obtained by direct sequencing or cloning. The P2 transcription initiation site (+1), the primers used for the last PCR, and the translation start site (ATG) are indicated.
Cloning of the AβH-J-J P2 promoter and reporter plasmid construction.
The BglII fragment of the human chromosome 8 BAC 1 clone (60) was cloned into the pUC18 vector and sequenced. The insert encompasses exon 2 of the AβH-J-J locus (Fig. 1) and contains the putative P2 promoter. The sequence corresponding to −686 to +115, relative to the transcription initiation site, was amplified by using a GeneAmp High Fidelity PCR system (Applied Biosystems) and the 5′-ATG TGG TTG TGT AAT CTT TTC CTA GTT C-3′ forward and 5′-CTT CAA TCC ATG AAT AAT AAA TGC TAA AGT-3′ reverse primers, containing a BglII and an NcoI restriction site, respectively. The PCR product was cloned into the BglII/NcoI restriction sites of pGL3-basic firefly luciferase reporter plasmid (Promega). Nine serial deletion constructs were generated from this recombinant plasmid by the use of an Exo III/S1 deletion kit (Fermentas, Vilnius, Lithuania). The sequence of all the constructs was confirmed by DNA sequencing.
Transient transfection and dual luciferase assay.
Cells were seeded at 40 to 60% confluence in 16-mm-diameter wells. Transfection was performed after 24 h by the use of 2 μg of Lipofectamine 2000 (Life Technologies) and 60 ng of pRL-TK vector (Promega), which contains the Renilla luciferase gene as a transfection efficiency control and 1 μg of firefly luciferase reporter plasmid per well (or 0.5 μg each of three constructs in the cotransfections with expression vectors or void vectors). C2C12-DM were seeded at 90% confluence and transfected in medium supplemented with 2% horse serum-10 μg of insulin/ml. Lysates were prepared 24 to 48 h after transfection by adding 100 μl of passive lysis buffer (Dual Luciferase reporter assay system; Promega), and luciferase activity was determined with an analytical luminometer (model TD-20/20; Turner Designs, Sunnyvale, Calif.). Relative luciferase units (RLU) were calculated by determining the ratio of the intensity of the light produced by the firefly luciferase reporter plasmid to that produced by the Renilla luciferase pRL-TK plasmid.
Triplicate luciferase assays of all the samples were performed. Severalfold induction results were determined by calculating the ratio of the firefly luciferase reporter plasmid RLU to that produced by the pGL3-basic vector with basal promoter activity. pcDNAI/A myocyte enhancer factor 2 (MEF-2A), kindly provided by E.N. Olson, has been described by Black et al. (4) and was made by cloning the coding region of human MEF-2A into the polylinker of the pCDNAI/Amp plasmid (Invitrogen, Inc.) (68). A plasmid encoding Myc-tagged histone deacetylase 4 (HDAC4), pHDAC4-Myc, was a kind gift of T. Kouzarides (University of Cambridge, Cambridge, United Kingdom) (43). pHDAC4-Myc Del 119-208 (Δ MEF-2 binding domain [bind. d.]), from which the MEF-2 binding site had been removed, was produced with a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). For the production of FLAG-HDAC4 1-233 (Δ catalytic d.), a fragment encoding amino acids 1 to 233 of human HDAC4 cDNA was inserted into the PCF2H vector (53). In the SRα-calcium-calmodulin-dependent protein kinase I (CaMKI) 1-294 construct, kindly provided by C. D. Kane (Duke University Medical Center, Durham, N.C.), the calmodulin binding domain (Ca2+/CaM independent) was deleted from the cDNA and therefore CaMKI is constitutively active (29). A cDNA fragment encoding the active form of murine calcineurin (that lacks the calmodulin binding and autoinhibitory domains) was inserted into the pBSIISK(+) vector under the control of the myogenin promoter. The Myo 1565-calcineurin expression vector (46) was produced using a cDNA fragment encoding amino acids 1 to 398 of murine calcineurin (active form lacking the calmodulin binding and autoinhibitory domains) inserted into the pBSIISK(+) vector under the control of the myogenin promoter.
Computer-assisted interspecies analyses of sequences corresponding to the P2 promoter of the AβH-J-J locus.
For computer-assisted analyses, we used MacVector sequence analysis software (International Biotechnologies, Inc., New Haven, Ct.) for Macintosh computers. The Mus musculus counterpart of the P2 promoter sequences used in our analyses were from the National Center for Biotechnology Information data bank (accession number AF289199). The rat sequence corresponding to the P2 promoter was obtained after PCR amplification of genomic DNA from a Wistar rat (EMBL nucleotide sequence database accession no. AJ865348). Pustell DNA matrix analyses (16) were performed with the following parameters: window size = 30; strand = both; min % score = 55; jump = 1; hash value = 6.
Gel shift assay.
Nuclear extracts from cell lines or tissues were prepared as described previously (6, 42). Gel shift reactions were carried out as described previously (42) by using, as nonspecific competitors, 1 μg of poly(dI:dC) · poly(dI:dC) and 1 μg of a single-stranded oligonucleotide. Double-stranded oligonucleotides used as probes or cold competitors in gel shift experiments are reported in Table 1. Supershift assays were performed by preincubating nuclear extracts for 20 min with 2 μg of commercially available antibodies specific for MEF-2A (sc-313X), MyoD (sc-304X), and myogenin (sc-12732X) transcription factors (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) before performing standard binding reactions. Human recombinant NF-κB p50 and human purified Sp1 (specific transcription factor 1) transcription factors were purchased from Promega and used in gel shift assays in the presence of 50 ng of poly(dI:dC) · poly(dI:dC) and 50 ng of a single-stranded cold oligonucleotide.
TABLE 1.
Double-stranded synthetic oligonucleotides used for gel-shift assays and for the production of mutated P2 promoter/luciferase constructs
| Oligonucleotide | Source or reference | Sequencesb |
|---|---|---|
| J-kBmer | 5′-AACTGGGAAAAGAATCCCGT-3′ | |
| J-E-box | 5′-TAGGACCAACTGGGAAAAGAATCCCGT-3′ | |
| MyoDmer | 17 | 5′-CCCCCCAACACCTGCTGCCTGA-3′ |
| XDRAbox | 16 | 5′-ACCCTTCCCCTAGCAACAGATGCGTCATCT-3′ |
| NFkBlgmer | sc-2505a | 5′-AGTTGAGGGGACTTTCCCAGGC-3′ |
| Stat1mer | sc-2573a | 5′-CATGTTATGCATATTCCTGTAAGTG-3′ |
| Sp1mer | 7 | 5′-TGAGGCGTGGCCT-3′ |
| J-E-boxM | 5′-GGAGTTAGCTAGGAAGTATCAGGAAAAGAATCCCG-3′ | |
| J-E-boxM1 | 5′-TAGGAAGTACTGGGAAAAGAATCCCGT-3′ | |
| J-EboxM2 | 5′-TAGGACCAACTGGTCGAAGAATCCCGT-3′ | |
| J-EboxM3 | 5′-TAGGACCAACTGATCAAAGAACTACGT-3′ | |
| J-GRmer | 5′-GCACAGTTTGTTCCATTTCA-3′ | |
| GRETIImer | 8 | 5′-AGGATCTGTACAGGATGTTCTAGATCG-3′ |
| MEF-2mer | 17 | 5′-CTCTAAAAATAACTCC-3′ |
| Ets 1mer | sc-2549a | 5′-GGGCTGCTTGAGGAAGTATAAGAAT-3′ |
| J-MEF-3mer | 5′-GTAGTCAGGTCCAATTTCAGT-3′ | |
| J-MEF-3Mmer | 5′-TAGTCAGGTCCAAGCTCAGTTCTAGAGG-3′ | |
| MEF-3mer | 50 | 5′-CCTGGTCAGGTTACAGTGG-3′ |
| MEF-3Mmer | 50 | 5′-CCTGGATATCTATGAGTGG-3′ |
| MEF-3MJmer | 5′-CCTGGTCAGGGCATGAGTGG-3′ | |
| J-MEF-2mer | 5′-TAGCCTCTAAAAATAACTGGTGAG-3′ | |
| J-MEF-2Mmer | 5′-CATTTAGCCTGTAAACATAACTGGTGAG-3′ | |
| J-GRMmer | 5′-GAAAACAAAACTGGCAACGCAGTTTGATACATTTCATTTTTTTTC-3′ |
Santa Cruz Biotechnology, Inc., Santa Cruz, CA.
Nucleotide mutations are underlined.
Site-directed mutagenesis.
Mutagenesis was performed by using a QuikChange site-directed mutagenesis kit (Stratagene). The oligonucleotides J-MEF-2Mmer, J-GRMmer, and J-E-boxM (Table 1), containing mutant binding sites, and their complementary sequences were used to inactivate specific binding in the −159 and −265 pGL3 constructs containing the −159/+115 and −265/+115 promoter sequences numbered with respect to the transcription initiation site. Mutant sequences of the recombinant plasmids were confirmed by DNA sequencing.
In vivo transfection of rat soleus muscles.
Male adult Wistar rats were anesthetized with ketamine, and soleus muscles were injected with 60 μl of 0.9% NaCl, containing 50 μg of firefly luciferase reporter construct, and 5 μg of pRL-TK vector. Transfected skeletal muscle fibers were obtained by electroporation (36, 40, 48). Pulse protocols, i.e., six pulses of 220 V/cm, pulse durations of 20 ms, and interpulse delays of 200 ms, were delivered using an ECM 830 Electro Square Porator (BTX; Genetronics, Inc.). Soleus muscles were removed after 2 days and homogenized in 0.5 ml of passive lysis buffer (Dual Luciferase reporter assay system; Promega), and luciferase activity was determined.
Statistical analysis.
All the data were normally distributed and are presented as means ± standard deviations (SD). Statistical differences between groups were compared using software allowing one-way analyses of variance between groups. Statistical significance was assumed at P < 0.05.
ChIP.
For in vitro chromatin immunoprecipitation (ChIP), a ChIP-IT kit (Active Motif, Carlsbad, Calif.) was used. Two T75 culture flasks of C2C12-DM cells were fixed for 7 min in minimal cell culture medium containing 1% formaldehyde and subsequently rinsed, and the fixation reaction was stopped by adding 10 ml of glycine stop-fix solution. Cells were collected and centrifuged at 4°C for 10 min at 720 × g and resuspended in lysis buffer. After 30 min of lysis and 10 strokes of Dounce homogenization, nuclei collected by 10 min of centrifugation at 2,400 × g were resuspended in shearing buffer. Sonication for ten cycles (pulses of 15 s at 30% amplitude, with a 30-s rest on ice between pulses) was performed with Sonics VibraCell VC 130 to prepare DNA fragments ranging in size from 200 bp to 1,000 bp, followed by centrifugation for 10 min at 13,000 × g. Supernatants were cleared by incubation with salmon sperm DNA-protein G-agarose beads for 2 h at 4°C. Then 10 μl of supernatant was collected and used as the input. Immunoprecipitation was carried out overnight at 4°C with preimmune serum (immunoglobulin G [IgG]) or 3 μg of an antibody raised against MEF-2A (sc-313X; Santa Cruz Biotechnology, Inc.). After centrifugation, washing, and elution, the samples were reverse cross-linked by heating at 65°C overnight and treated with RNase A and proteinase K. DNA was then purified with the DNA minicolumns provided with the kit, and PCR was performed with primers 5′-TGA GTG GGA AGG GCG CAG ATT-3′ and 5′-TAC TGC CAG GAC AAG GGC TCC A-3′, designed to amplify a 151-bp product of the mouse P2 promoter, or primers 5′-GGC ATC AGA TCC CAT CAT AGA CA-3′ and 5′-GGG ACC TAA GTG AGC ATC CTT GA-3′, designed to amplify a 155-bp product of a region about 29 kb upstream of the P2 promoter (negative control), respectively. The PCR conditions were as follows: 30 cycles of amplification with 15 s of denaturation at 95°C, 30 s of annealing at 66°C, and 20 s of elongation at 72°C. The ChIP assay was performed at least three times. Input DNA was diluted 1:100 to be used as the template for a positive-control PCR.
RESULTS
Transcriptional organization of the human AβH-J-J locus and identification of the transcription initiation site proximal to the P2 promoter.
The structural organization of the human AβH-J-J locus is shown in Fig. 1. The scheme presented is based both on results previously reported in detail elsewhere (11, 60) and on newly performed studies employing RT-PCR. The combination of data obtained by PCR amplification and sequencing allowed us to define the splicing events (Fig. 1) as well as the structure of the 5′ region of this locus, which encodes three different proteins, aspartyl-β-hydroxylase, junctin, and junctate (60). The data obtained indicate that the first and second exons are mutually exclusive when mature mRNAs are produced. Moreover, the use of different splice donors has been shown to be involved in the generation of protein diversity by alternative splicing (see lower part of Fig. 1). Our results are in agreement with data from Dinchuk et al. (11) obtained with a homologous mouse system and suggest that at least two promoter sequences (tentatively identified as P1 and P2) are present in the 5′ region of the AβH-J-J locus. In this study, we focused our attention on the P2 promoter, which directs tissue-specific expression of the AβH-J-J locus starting from exon 2. The RT-PCR analysis presented in the upper part of Fig. 2A shows that by using an exon 2-specific forward primer we are able to analyze all the transcripts starting from the P2 promoter. In the case of human tissues and cells we used primers located in the second and third exons, and in the case of murine C2C12 cell line we used primers located in the second and fifth exons, which exhibit a high level of homology in comparisons of human and mouse sequences. The results obtained with kidney, brain, adrenal gland, liver, heart, skeletal muscle, the RD cell line, C2C12-GM cells (C2C12 cells cultured in growth medium), and C2C12-DM cells (C2C12 cells cultured in differentiation medium) show that transcription directed from the P2 promoter is tissue specific. Among the analyzed human tissues, high-level transcription was present only in skeletal muscle, cardiac muscle, and brain (Fig. 2A, lanes f, e, and b). P2 directed transcription was found in RD cells, C2C12-GM, and C2C12-DM cells, with the highest level of expression in differentiated C2C12 cells (Fig. 2A, lane i).
Since the transcription initiation site has not been previously described in detail, we also performed 5′ RACE to precisely map the origin of transcription. This experiment was performed on total RNA isolated from RD cells and human adult skeletal muscle. As shown in Fig. 2B, one band is evident following electrophoresis of 5′ RACE products in both cases (lanes m and n). This band was directly sequenced or cloned and then sequenced, giving identical results in both cases. The sequence of the 5′ RACE products is shown in Fig. 2C and allows mapping the transcription initiation site, which is indicated by a solid arrow. Accordingly, all the nucleotide sequences located upstream from this site were considered potential regulatory regions belonging to the P2 promoter.
Transcriptional activity of the AβH-J-J P2 promoter.
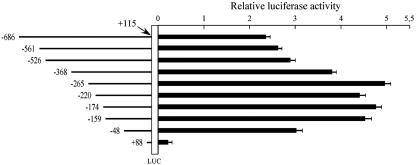
A PCR-generated fragment of the P2 promoter spanning the region from nucleotide −686 to nucleotide +115 with respect to the transcription initiation site was cloned into the firefly luciferase reporter vector pGL3-basic, sequenced, and used to generate progressive deletions for transient transfection experiments. As target DNA, the human genomic BAC 1 clone was used (60). The nine nested deletion constructs shown on the left side of Fig. 3 were generated as described in the Materials and Methods section. Each recombinant plasmid DNA was then transiently transfected into cells of the myogenic C2C12 cell line (27). The vector pGL3-basic, which has no promoter, was used as the control. From the data obtained, we are able to draw the following conclusions. First, the +88 construct does not induce luciferase activity (Fig. 3). Second, luciferase activity was detectable using the −48 construct and increased to the maximum levels with the −159 construct. These data suggest that cis-acting elements activating transcription are present between −159 and +88. Furthermore, the luciferase activity remains at these high levels up to construct −265. Third, luciferase activity was present in the −368 and −526 constructs but at a lower level (P < 0.002 when −368 and −265 constructs are compared and P < 0.0008 when −526 and −368 constructs are compared), suggesting the presence of cis-acting elements inhibiting transcription between −526 and −265. The −526, −561, and −686 constructs exhibited similar activity levels.
FIG. 3.
Promoter activity of serial deletion constructs of the AβH-J-J −686 to +115 nucleotide sequence (the P2 transcription initiation site is referred as +1). C2C12 cells were transfected with sequentially deleted reporter constructs (represented in the left side of the figure). Transient transfection and luciferase assays were performed in triplicate, and the data were normalized to Renilla luciferase activity and reported as a ratio to pGL3-basic RLU (means ± SD). pGL3-basic is a firefly luciferase (LUC) reporter vector with basal promoter activity.
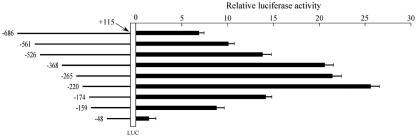
In vivo experiments were performed on male adult Wistar rats. Soleus muscles were injected with the same firefly luciferase reporter constructs, and transiently transfected skeletal muscle fibers were obtained by in vivo administration of electric pulses as described elsewhere (48, 40). After 2 days, soleus muscles were removed and luciferase activities were determined. As shown in Fig. 4, we obtained patterns of reporter activity for soleus muscle very similar to those obtained with C2C12 cells. In particular, constructs −368, −265, and −220 sustained a high level of transcription. Interestingly, lower levels of transcription were obtained using constructs −174 and −159 (P < 0.0001 in comparisons of constructs −220 and −174; P < 0.007 in comparisons of constructs −174 and −159). Furthermore, it should be noted that construct −48 does not support efficient transcription. These data suggest that several cis-acting elements, present between −48 and −220, are involved in the up-regulation of transcription in this experimental system. In addition, as for the experiment employing C2C12 cells (Fig. 3), the luciferase activity sustained by constructs −686, −561, and −526 was significantly lower (P < 0.003 in comparisons to construct −368).
FIG. 4.
Promoter activity of serial deletion constructs of the AβH-J-J −686 to +115 nucleotide sequence in soleus muscle of adult rats. Soleus muscles were injected with 50 μg of serial deletion constructs, and the calf was exposed to electrical pulses. At 48 h after transfection each muscle was homogenized in lysis buffer and assayed for luciferase activity. Transient transfection and luciferase assays were performed in triplicate, and the data were normalized to Renilla luciferase activity and reported as a ratio to pGL3-basic RLU (means ± SD). pGL3-basic is a firefly luciferase (LUC) reporter vector with basal promoter activity.
For the next series of experiments we concentrated on the −265 to +88 sequence, since this exhibited the highest induction of luciferase activity in both systems.
Identification of sequences putatively recognized by nuclear factors within the −265 to +1 P2 region of the AβH-J-J locus.
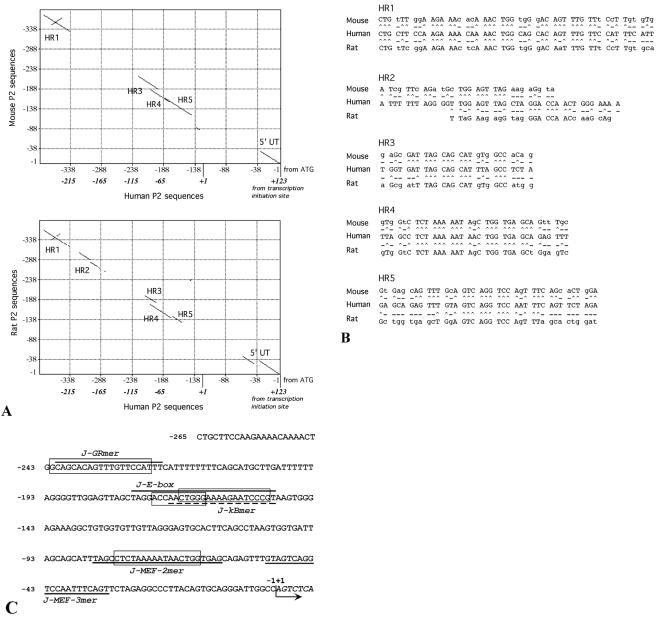
In accordance with the evidence that the region downstream of −265 contains the minimal promoter, the human P2 sequence −265 to +123 was analyzed to find interspecies homologies with mouse and rat counterparts (Fig. 5A). Interspecies homology phylogenetic searching allows identification of evolutionarily conserved regions possibly important for gene regulation, as demonstrated in several research papers (3, 5, 16). This sequence homology study (Fig. 5A) allowed the identification of four regions (HR1, HR3, HR4, and HR5) conserved in mouse, human, and rat sequences and one (HR2) conserved in rat and human sequences. These homologies are analyzed in detail in Fig. 5B. When this analysis is combined with a search for transcription factor binding sites using the TF search program, we identified within these conserved regions sequences homologous to the glucocorticoid receptor element (GRE) (−242 to −224) (8), c-Myb/MyoD (−173 to −164) (47, 50), NF-κB (−161 to −152) (41), and MEF-2 (−80 to −65) (17) consensus boxes. The location of putative transcription factor binding sites, together with the transcription initiation site, is shown in Fig. 5C. The −52 to −32 sequence, underlined in Fig. 5C and relative to HR5, is highly homologous to MEF-3 binding sites present in a large number of skeletal muscle-specific transcriptional enhancers and promoters (24, 50, 56).
FIG. 5.
(A) Dot-matrix comparison of the human AβH-J-J P2 promoter −265 to +123 nucleotide sequence performed with respect to the murine (up; National Center for Biotechnology Information data bank accession number AF289199) and rat (down; EMBL Nucleotide Sequence Database accession number AJ865348) counterpart (−1 to −388 from ATG) by the use of MacVector sequence analysis software. Lines represent homologies between the analyzed sequences; homology regions have been identified as HR1, HR2, HR3, HR4, and HR5. (B) Nucleotide sequences present within the HR regions identified (^, conserved nucleotide; -, nonconserved nucleotide). (C) −265 to +7 P2 promoter sequence. Dark lines and boxes indicate the oligonucleotides used in gel shift assays (Table 1) and the sequences homologous to transcription factors binding sites found within the conserved region depicted in panel A. Homologies were obtained by TFSEARCH version 1.3 software and are as follows: GR-box (glucocorticoid response element), 89%; c-Myb and MyoD boxes, 88 and 80%, respectively, with J-E-box; NF-κB box, 67% with J-kBmer; and MEF-2 box, 90%. The arrow indicates the P2 transcription initiation site (+1).
These boxes, which could be fairly conserved through evolution, were further analyzed by molecular biology approaches.
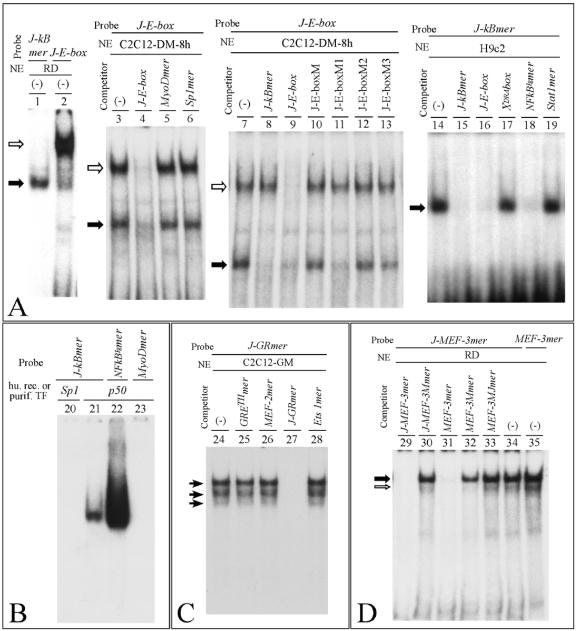
To demonstrate protein-DNA interactions at these sites, band shift assays were performed. Table 1 shows the synthetic and mutant oligonucleotides used for band shift experiments. Figure 6 shows representative examples of the results obtained using the J-E-box and J-kBmer, J-GRmer, and J-MEF-3mer 32P-labeled double-stranded oligonucleotide probes.
FIG. 6.
Binding of nuclear factors to the AβH-J-J promoter 2. (A and B) Band shift assays performed using J-kBmer (−170 to −151), J-E-box (−177 to −151), and NFkBlgmer and MyoDmer (Table 1) probes and 2 to 3 μg of nuclear extracts (NE) from RD, C2C12-DM-8 h (C2C12 cells were induced to differentiate in DM for 8 h and a nuclear extract was prepared), and H9c2 cells or 0.05 μg of human recombinant p50 and purified human Sp1 transcription factors (Promega), as indicated. (-), probe was incubated with nuclear extracts in the absence of competing oligonucleotides. (C and D) Band shift assays performed using J-GRmer (−241 to −222), J-MEF-3mer (−52 to −32), and MEF-3mer (Table 1) probes and 2 to 3 μg of nuclear extracts (NE) from C2C12-GM and RD cell lines. (-), probe was incubated with nuclear extracts in the absence of competing oligonucleotides. In this figure, competing oligonucleotides (Table 1) were added at a 100-fold molar excess. Arrows indicate the specific complexes.
To verify the specificity of the interactions, competitive experiments were performed using (i) unlabeled unrelated oligonucleotides, (ii) homologous sequences recognized by known transcription factors, or (iii) mutations of the binding site under investigation.
The representative data shown in Fig. 6 were obtained from experiments performed after preliminary screening of nuclear extracts from different cell lines (C2C12, RD, and H9c2) to identify nuclear extracts containing the highest levels of the protein(s) able to interact with the oligonucleotides under investigation.
With respect to the J-E-box sequence spanning the region from nucleotide −177 to nucleotide −151, in addition to the canonical E-box (CANNTG) (50), the J-kB sequence is present, exhibiting 67% homology with the asymmetric NF-κB binding site (41). The first conclusion that can be drawn from the results presented in lanes 2 and 3 of Fig. 6 is that at least two complexes are able to bind to the J-E-box probe (white and black arrows). In addition, the J-E-box cold oligonucleotide is able to inhibit both binding activities (Fig. 6, lanes 4 and 9), while the addition of cold J-kBmer (lacking the E-box signal) inhibits only the generation of the fast mobility complex (Fig. 6, lane 8), demonstrating that E-box binding activity generates the lower-mobility retarded band. As far as characterization of binding activity to the E-box site of the J-E-box probe is concerned, we can exclude a direct involvement of the transcription factor MyoD, since cold MyoDmer is unable to inhibit the formation of the low-mobility complex (Fig. 6, lane 5). As expected, the unrelated Sp1mer was also unable to inhibit J-E-box-protein interactions (Fig. 6, lane 6). Accordingly, (i) when 32P-labeled MyoDmer was employed, a different pattern of retarded bands was obtained and (ii) supershift experiments with an antibody against MyoD did not show any alteration of the interaction of nuclear factors with the J-E-box (data not shown). These experiments suggest that nuclear factors other than MyoD bind to the J-E-box element, with the consequent generation of the lower-migrating band. As far as the characterization of the binding activity to the NF-κB-like site of the J-E-box probe is concerned, Fig. 6 (lanes 14 to 19) shows that while competitors containing binding sites for Stat1 and X-box binding proteins are unable to suppress binding of nuclear factors to J-kBmer probe (Fig. 6, lanes 17 and 19), NFkBIgmer suppresses this binding (Fig. 6, lane 18). As expected, J-kBmer and J-E-box are also strongly active as competitors (Fig. 6, lanes 15 and 16).
To characterize the interplay between nuclear factors and the overlapping sites present within the J-E-box, mutant oligonucleotides were used. Figure 6 (lanes 7 to 13) shows that addition of cold mutants altering the conserved E-box sequence (J-kBmer and J-E-boxM1) did not inhibit the formation of the lower-migrating band, when 32P-labeled J-E-box was mixed to nuclear factors from C2C12 cells (Fig. 6, lanes 8 and 11). However, the flanking sequences are also expected to play a role, since the J-E-boxM2 and the J-E-boxM3 mutants do not inhibit the formation of the lower-migrating band (Fig. 6, lanes 12 and 13). As expected, competition was always observed with the wild-type J-E-box (Fig. 6A, lane 9). As far as mutations affecting the NF-κB-like sequence are concerned, those affecting the conserved CTG and AAA stretches of the NF-κB binding sites suppress the binding generating the fast-migrating band. In fact, the oligonucleotides J-E-boxM, J-E-boxM2, and J-E-boxM3 do not bind to NF-κB-like factors, while the J-E-boxM1, carrying an intact NF-κB binding site, interacts with transcription factors generating the fast-migrating complex (Fig. 6, lane 11). These results give strong evidence that proteins belonging to the NF-κB superfamily interact with the NF-κB sites present within the AβH-J-J P2 promoter. This is conclusively shown by the experiment reported in Fig. 6B, showing that the recombinant p50 NF-κB protein binds to the J-kBmer (lane 21). As expected, the binding efficiency is lower than that displayed by the symmetric Igk NF-κB site (lane 22), as already reported in several studies (62). No binding of purified Sp1 transcription factor to the J-kBmer was found (lane 20).
As far as the interactions of J-GRmer with nuclear extracts from C2C12 cells are concerned, the data shown in Fig. 6C demonstrate that the binding is specific, since competition was clearly observed with unlabeled J-GRmer (lane 27) but not with the unrelated oligonucleotides carrying the Ets 1 and MEF-2 binding site (lanes 28 and 26). Surprisingly, the tyrosine-aminotransferase II (TATII)-GRE double-stranded oligonucleotide (8) containing the consensus GRE failed to compete (Fig. 6, lane 25); thus, factors that interact with J-GRmer should be further characterized to establish their relationship, if any, with canonical GRE binding proteins. This result was reproducibly obtained in several experiments, using high amounts of competitor GRETIImer. Control experiments demonstrated high efficiency of binding of 32P-labeled GRETIImer to nuclear extracts. Opportune mutations introduced in J-GRmer (J-GRMmer; see Table 1 for nucleotide sequences) completely abolish the interaction with nuclear factors (data not shown).
Binding to the J-MEF-3mer (Fig. 5C and Table 1) is shown in Fig. 6D, which depicts representative results obtained using the rhabdomyosarcoma RD cell line, expressing MEF-3 binding factors at high levels. The gel shift pathways obtained using 32P-labeled J-MEF-3mer and MEF-3mer (carrying a functional MEF-3 binding site present in the promoter sequence of the slow/cardiac troponin C) (50) are identical (Fig. 6D, lanes 34 and 35). In addition, the binding to 32P-labeled J-MEF-3mer is specifically competed by both cold MEF-3mer and J-MEF-3mer (Fig. 6, lanes 31 and 29). Furthermore, mutations suppressing binding to J-MEF-3mer (lanes 30, 32, and 33) also affect MEF-3mer interaction with nuclear extracts (data not shown). As expected, no inhibition of binding of nuclear factors to MEF-3mer and J-MEF-3mer is obtained with unrelated oligonucleotides carrying SRE or sequences recognized by the Sp1, AP-1, Stat1, and C/EBP transcription factors (data not shown). Taken together, these results support our hypothesis that J-MEF-3mer and MEF-3mer bind to the same nuclear factors.
MEF-2 transcription factors bind to the MEF-2 binding sites of the P2 promoter of the AβH-J-J locus.
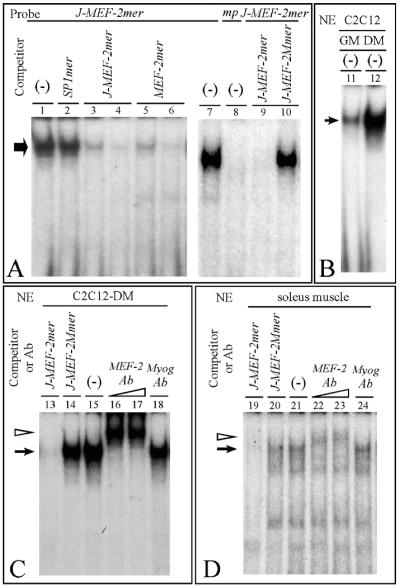
Due to the important role of MEF-2 in muscle-restricted gene expression, the interactions of nuclear proteins with respect to the MEF-2 binding site of the P2 promoter sequences of the AβH-J-J locus were studied in great detail (19). Since J-MEF-2mer contains sequences highly homologous to those recognized by the MEF-2 family factors (including MEF-2A, -B, -C, and -D) (4, 23, 33, 70), competition experiments were first performed using the MEF-2mer oligonucleotide, carrying a canonical MEF-2 binding site (17). Cold MEF-2mer is able to inhibit binding under these experimental conditions (Fig. 7A, lanes 5 and 6). A two-base mutation in the binding site of J-MEF-2mer abolishes formations of gel shift complexes (Fig. 7A, lanes 8 and 10).
FIG. 7.
Identification of MEF-2 family nuclear factors that interact with the AβH-J-J promoter 2. (A and B) Band shift assays were performed using J-MEF-2mer probe (−84 to −61) or mutant probe (mp, J-MEF-2Mmer) (Fig. 7A, lane 8) (Table 1) and 2 μg of nuclear extracts (NE) from RD (A) or C2C12-GM and -DM (B) cells, cultured in growth and differentiation medium, respectively. (-), probes were incubated with nuclear extracts in the absence of competing oligonucleotides. Competing oligonucleotides (Table 1) were added at a 100-fold molar excess with the exception of lanes 3 and 5 (50-fold molar excess). (C and D) Band shift and supershift assays were performed using J-MEF-2mer probe and 2 μg of NE from C2C12-DM or 8 μg of NE from soleus muscle. (-), control samples in the absence of competing oligonucleotides or antibodies; probe was incubated with nuclear extracts in the presence of the indicated competing oligonucleotides (at a 100-fold molar excess) or in the presence of antibodies (Ab) against MEF-2 family members (1 to 2 μg) or against myogenin (2 μg). In this figure, arrows indicate the specific complexes; arrowheads indicate complexes supershifted by antibody.
Figure 7B demonstrates that the MEF-2 binding activity is strongly induced in association with muscle differentiation. In this experiment muscle differentiation was obtained after treatment of C2C12 cells with DM for 3 days.
In addition, supershift experiments were performed using monoclonal antibodies against MEF-2 and nuclear extracts from C2C12-DM cells, demonstrating that addition of the antibody induces a clear supershift (Fig. 7C, lanes 16 and 17). Since this commercial MEF-2A specific antibody cross-reacts with other members of the MEF-2 family, we can conclude that MEF-2 family nuclear factors binds to the J-MEF-2mer probe. An antibody against myogenin, as expected, failed to induce supershift (Fig. 7C, lane 18).
The experiments shown in Fig. 7C were performed in replicate by the use of nuclear extracts from rat soleus muscle. Despite the fact that the binding efficiency in this case is lower (compare panels C and D of Fig. 7), the results are consistent with those obtained using C2C12 cells.
Studies of MEF-2 binding to wild-type and mutated cis elements: effects on transcription.
The finding that MEF-2-related factors bind to the J-MEF-2 box present in the −80 to −65 region of the P2 promoter of the AβH-J-J locus is of great interest for the known tissue-specific up-regulating functions of MEF-2 (37, 9) on the one hand and for the promoter activity of the region between −159 and the transcription initiation site on the other. Since this region contains the MEF-2 binding site (Fig. 5C), it could be considered the “minimal” P2 promoter sequence (Fig. 3 and Fig. 4).
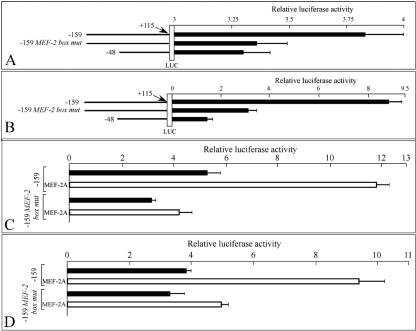
The main goal of the experiment described in Fig. 8 was to confirm the involvement of MEF-2 in the transcriptional regulation of the AβH-J-J locus. We first designed a J-MEF-2mer mutant (J-MEF-2Mmer) unable to bind the MEF-2 family factors. This was demonstrated by the finding that no retarded bands are generated following incubation of nuclear factors to a J-MEF-2Mmer probe (Fig. 7A, lane 8). In agreement, mutated cold J-MEF-2Mmer failed to inhibit MEF-2 proteins and J-MEF-2mer interactions (Fig. 7A, lane 10). Accordingly, we produced a −159 pGL3 deletion construct carrying this MEF-2 binding site mutation (−159 MEF-2 box mut). When the −159 mutant construct was used to transfect C2C12 cells, a decrease in luciferase activity (relative to the wild-type −159 construct) was detected (P < 0.001) (Fig. 8A). To further confirm the role of MEF-2 in the regulation of the promoter we performed in vivo experiments on soleus muscles of male adult Wistar rats. Soleus muscles were injected with the same firefly luciferase reporter constructs shown in Fig. 8A, and luciferase activity was determined. The results are shown in Fig. 8B and conclusively demonstrate that when the −159 mutant construct is used, a highly significant decrease (over 65%) of luciferase activity is obtained with respect to the wild-type construct (P < 0.0008).
FIG. 8.
Transcriptional effect of mutation in the MEF-2 binding site of the AβH-J-J promoter 2. (A) C2C12-GM cells were transfected with wild-type (−159 and −48) and MEF-2 binding site mutant (−159 MEF-2 box mut) AβH-J-J P2 promoter reporter constructs (represented in the left side of the figure). Transient transfection and luciferase assays were performed in triplicate, and the data were normalized to Renilla luciferase activity and are reported as ratios (means ± SD) to the pGL3-basic firefly luciferase (LUC) reporter vector. (B) The same constructs presented in panel A were transiently transfected in rat soleus muscles as described for the experiment whose results are presented in Fig. 4. (C and D) The wild-type and mutant −159 constructs whose results are presented in panels A and B were utilized in the absence (black bars) or in the presence (white bars) of MEF-2A expression vector for C2C12-GM (C) and HeLa (D) cells transfections.
Finally, to unambiguously confirm the role of MEF-2 in the transcription activity directed by the P2 promoter, we used a recombinant plasmid carrying a MEF-2A cDNA under the control of the cytomegalovirus promoter and directing high levels of expression of MEF-2 in transfection experiments (4, 68). When C2C12-DM cells were cotranfected with the −159 construct and the MEF-2A-expressing plasmid, a sharp increase in luciferase activity was observed (Fig. 8C). This result was reproducibly obtained in six independent experiments (P < 0.005). This increase was only barely detectable using the −159 construct carrying the mutated MEF-2 binding site described (lower part of Fig. 8C; −159 MEF-2 box mut). The effects of transfection with the MEF-2A-expressing vector were also detectable using other cell lines, such as HeLa cells (Fig. 8D).
Taken together, these results demonstrate an involvement of the MEF-2 signal and MEF-2 transcription factors in transcriptional regulation of the AβH-J-J locus.
Transcriptional effects of mutations of MEF-2 box, GR box, or E-box.
To obtain information about the importance of some of the identified transcription factor signals of the P2 promoter, we next compared the effects of mutations in the MEF-2 box to those suppressing binding activity of nuclear factors to the GR and E boxes. For this purpose, we designed, synthesized, and tested mutants of GR box and E-box suppressing protein binding to the relative 32P-labeled mers (see nucleotide sequences in Table 1) and produced three −265 constructs carrying the mutated GR box, E-box, and MEF-2 box, respectively. The activity of these constructs was compared to that of the wild-type reporter plasmid after transfection of soleus muscle of adult Wistar rats (Fig. 9).
FIG. 9.
Effect on promoter activity of mutations in the GR box, E-box, or MEF-2 binding site of the AβH-J-J −265 P2 promoter reporter construct. Rat soleus muscles were transiently transfected with wild-type and specific DNA binding site mutants of the AβH-J-J −265 P2 promoter reporter construct as described for the experiments whose results are presented in Fig. 3 and 4.
The results of this experiment clearly indicate that in soleus muscle of adult Wistar rats all mutated constructs exhibited a decreased ability to stimulate luciferase activity, indicating that proteins binding to these boxes play a role in up-regulating transcription (P < 0.0005 for GR box mut; P < 0.003 for E-box mut; P < 0.00002 for MEF-2 mut). When the experiment was conducted with C2C12-GM cells the inhibition of transcription with the MEF-2 mut construct was lower than that found in C2C12-DM cells (data not shown), suggesting that the role of MEF-2-related proteins in regulating the expression of the AβH-J-J locus is mostly evident in differentiating cells.
MEF-2 transcription factors bind chromatin of the P2 promoter.
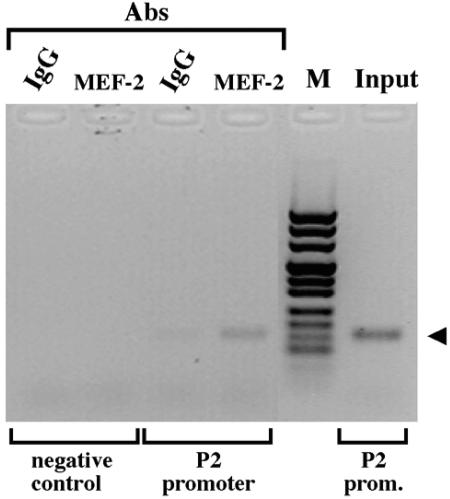
The recently described chromatin immunoprecipitation (ChIP) technology allows formally demonstration in intact cells of the interaction of a given DNA binding protein with promoter region identified by specific PCR primers.
Accordingly, after formaldehyde cross-linking, the chromatin isolated from C2C12-DM cells was sonicated and immunoprecipitated with an anti-MEF-2A antibody. PCR was performed on purified DNA by the use of primers amplifying the AβH-J-J P2 promoter. After 30 PCR cycles, amplification of the AβH-J-J P2 promoter was obtained using only DNA from the MEF-2A-specific immunoprecipitates as the substrate (Fig. 10). No PCR product was obtained using unrelated IgG immunoprecipitation controls. As expected for the ChIP assays, both control IgG and anti-MEF-2 antibody immunoprecipitates contained equal amounts of negative-control sequences that were able to be PCR amplified, but only when the PCR was conducted for a high number of cycles (more than 35; data not shown).
FIG. 10.
Chromatin immunoprecipitation with MEF-2A-specific antibody. C2C12-DM (in differentiation medium for 72 h) cells were formaldehyde cross-linked and processed for ChIP assays with antibodies (Abs) against MEF-2A (Santa Cruz) or IgG (as the control). PCR products, obtained with primers flanking the MEF-2 binding site of the AβH-J-J P2 promoter or with primers amplifying a region 29 kb upstream of the P2 promoter (negative control), were analyzed by gel electrophoresis. Input, control sample before the immunoprecipitation. M, pUC Mix Marker 8 (Fermentas).
Effects of HDACs and CaMK* on MEF-2-directed transcription.
Several studies have demonstrated a strong interplay between MEF-2 and histone deacetylases (HDACs) (33, 37, 43), as well as between MEF-2 and Ca2+/calmodulin-dependent protein kinases (CaMKI and CaMKIV) (51). In this respect, it was recently shown by several laboratories that MEF-2 interacts with class II HDACs and that this interaction leads to repression of the transcriptional activity of MEF-2 (33, 37). CaMKI modulates the interactions between HDACs and MEF-2; in fact, activation of CaMKI leads to HDAC phosphorylation and dissociation of MEF2/HDAC complexes (33).
Accordingly, we determined the effects of plasmids expressing HDAC4 (43) and CaMKI (29) on luciferase activity of the −159 construct.
In the first set of experiments, we used three expression vectors, one for full-length HDAC4 (43) and the other two for mutated HDAC4 lacking either the enzymatic activity domain (Δ catalytic d.) (53) or the MEF-2 binding domain (Δ MEF-2 bind. d.).
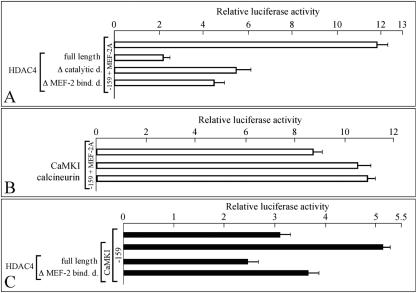
The results shown in Fig. 11A clearly demonstrate that when the C2C12-DM cells are cotransfected with the −159 construct, with MEF-2A- and HDAC4-expressing plasmids a sharp decrease of luciferase activity is observed with respect to transfections carried out with the −159 construct and with MEF-2A. Cotransfections with plasmids encoding mutated HDAC4 (i.e., defective in catalytic activity or in MEF2 binding) show an at least partial restoration of reporter activity. This partial effect is not surprising, since it known that HDAC4 and HDAC5 lacking the catalytic domain and even the MEF-2 binding motifs are still capable of inhibiting MEF-2 dependent transcription (43).
FIG. 11.
Effect of HDAC4, CamKI, and calcineurin on P2 promoter activation by MEF-2A. (A) C2C12-DM cells were transfected with the −159 construct in the presence of MEF-2A expression vector (white bars) and the indicated HDAC4 expression constructs (full-length, wild-type HDAC4; Δ catalytic d., HDAC4 lacking the catalytic domain; Δ MEF-2 bind. d., HDAC4 lacking the MEF-2 binding domain). Transient transfection and luciferase assays were performed in triplicate, and the data were normalized to Renilla luciferase activity and are reported as ratios (means ± SD) to the pGL3-basic firefly luciferase (LUC) reporter vector. (B) The same experiment whose results are presented in panel A was performed with the −159 construct, the MEF-2A expression vector, and the indicated CamKI and calcineurin expression constructs. (C) C2C12-GM cells were transfected with the −159 construct alone or in the presence of CaMKI expression vector and the indicated HDAC4 expression constructs.
To further support the hypothesis that the activity of the P2 promoter depends on MEF-2 activity possibly regulated by HDAC4, the experiment depicted in Fig. 11B was performed. In this experiment, the −159 construct was used in combination with plasmids expressing MEF-2A and CaMKI or calcineurin in C2C12-DM cells. In both cases, an increase in luciferase activity was obtained (Fig. 11B). A sharp increase in luciferase activity was obtained using C2C12-GM cells transfected with the CamKI-expressing vector (Fig. 11C). This increase is not obtained in the presence of expression vectors carrying full-length HDAC4. Vectors carrying HDAC4 lacking MEF-2 binding domains were significantly less effective (Fig. 11C).
DISCUSSION
The aim of the present paper was to investigate one of the two putative promoter sequences regulating the transcription of the AβH-J-J locus. We have been able to (i) define the transcription initiation site of junctin by RACE, (ii) identify a 5′ untranscribed region, possibly involved in the regulation of the expression of the AβH-J-J locus, and (iii) identify transcriptional factors and determine their functional activity in this region in a set of cell lines and also in an in vivo model such as rat soleus muscle.
The major conclusions of the present work are the following: (i) the P2 promoter of the AβH-J-J locus contains sequences recognized by known transcription factors, such as MEF-2, MEF-3, NF-κB, as well as signals for other yet-to-be-characterized proteins; (ii) cis elements were identified with negative and positive effects on transcription; (iii) the minimal promoter is located within −159 nucleotides from the transcription initiation site; and (iv) the nuclear transcription factor MEF-2 is required for transcriptional activation.
The involvement of MEF-2 in the transcriptional activity of the P2 promoter of the AbH-J-J locus is demonstrated by several convergent approaches, including supershift experiments (Fig. 7), in vivo chromatin immunoprecipitation (ChIP) (Fig. 10), and cotransfection of C2C12-DM cells with a plasmid carrying the P2 AβH-J-J minimal promoter and an expression plasmid carrying MEF-2 cDNA under the control of a cytomegalovirus promoter. Moreover, the idea of the involvement of MEF-2 in transcriptional regulation of the AβH-J-J locus is further sustained by experiments employing constructs carrying mutated MEF-2 DNA binding sites. These promoters were found to be less active in sustaining transcription of the luciferase reporter gene both in vitro and in vivo (Fig. 8 and 9). In addition, in striated muscle the promoter containing a mutated MEF-2 binding site is more affected in its transcriptional activity than promoters carrying mutated GR- or E-boxes, suggesting that nuclear proteins binding to the MEF-2 site play a major transcriptional role in this system. This conclusion supports the hypothesis that the muscle-specific MEF-2 DNA binding activity is sufficient to reach maximum levels of transcription (Fig. 8 and 9). However, reproducible differences should be underlined when in vivo (Fig. 4) and in vitro (Fig. 3) systems are compared; first, the severalfold induction of luciferase activity in rat soleus muscles is much higher that that found in C2C12 cells. This is not unexpected, given our RT-PCR data showing that the transcription directed by the P2 promoter is far more sustained in adult muscles than in C2C12 cells. Furthermore, the −48 construct does not sustain transcription in the in vivo system; in contrast, it is active in C2C12 cells. These discrepancies could be due to several factors, such as differences in the differentiation states of the target cells, species, transfection efficiency, and/or transfection strategy.
Our data are in agreement with recent literature showing binding interactions and functional interplay between MEF-2 and histone deacetylase 4 and 5 (33). Accordingly, transfections with constructs expressing HDAC4 decrease transcription directed by the P2 AβH-J-J promoter in the absence or in the presence of transfection with MEF-2-expressing vectors.
These results are consistent with most of the available literature on MEF-2 biological functions. For instance, myogenesis in mouse C2 cells is inhibited by double-stranded phosphorothioate oligodeoxynucleotides containing binding sites recognized by proteins belonging to the MEF-2 superfamily. Exposure of C2 myoblasts to doxorubicin had a profound effect on the expression of regulatory genes critical to the myogenic differentiation program (30); levels of mRNAs for MyoD and myogenin were dramatically reduced. In addition, there was diminished DNA binding activity of the muscle-specific transcription factor MEF-2. These data and other experimental evidence indicate that MEF-2 is involved in the activation of muscle-specific genes, including rat myosin light chain 2 slow, human myoglobin, rat muscle creatine kinase, mouse troponin C slow, and human beta enolase (14, 15, 20, 26, 50). In addition, MEF-2 has been proposed to serve as a nodal point in the mechanisms by which motor nerves control the program of gene expression in skeletal muscle fibers and in the pathway leading to cardiac hypertrophy. High levels of mRNA expression of both junctin and junctate have been observed in cardiac muscle (11, 60). Interestingly, there is experimental evidence demonstrating that the trans-activator factor MEF-2 is involved in the regulation of gene expression in cardiac tissue. MEF-2 would act as a downstream effector of calcium-dependent signaling mediated by calcineurin (32). Recent studies have demonstrated that calcineurin modulates gene transcription not only in cardiac muscle but also in skeletal muscle (65). The calcineurin-dependent signaling has been associated with the adaptive mechanism of cardiac muscle in responses to hypertrophic agonists such as angiotensin II and phenylephrine (45, 13). These two agonists activate calcium release from the cardiac junctional sarcoplasmic reticulum, and the transient increase of calcium levels would in turn activate calcineurin and its downstream effectors NFAT and MEF-2.
In conclusion, the most interesting result emerging from our study is that the calcium-dependent transcriptional factor MEF-2 activates the promoter of two proteins of the junctional sarcoplasmic reticulum membrane in addition to other muscle-specific genes. In agreement with these considerations, transfections with CamKI- and calcineurin-expressing vectors augment transcription directed by the AβH-J-J P2 promoter. These data are well in agreement with observations published elsewhere suggesting that the calcineurin-mediated removal of phosphate groups from MEF-2 augments the potency of the transcriptional activation domain of MEF-2 fused to a heterologous DNA binding domain (65). Whether the observed effect of calcineurin is due to this mechanism of action or to others, including dephosphorylation of NFAT, remains to be determined (37). In respect to CaMKI, it is known that phosphorylation of HDACs by Ca2+/calmodulin-dependent protein kinases leads to dissociation from MEF-2 and increased MEF-2-dependent transcriptional activity.
Note that the relationship between calcium signaling and regulation of the expression of the AβH-J-J locus is not restricted to MEF-2 and other calcium-dependent transcription factors but also involves the biological functions of the proteins encoded by the AβH-J-J locus. In this respect, junctin and junctate are localized in a key membrane compartment involved in the generation of the calcium signal both in skeletal and cardiac muscle.
To our knowledge this is one of the few cases in which a calcium-dependent transcriptional factor influences the expression of gene involved in calcium signaling.
The finding that MEF-2 and MEF-3 binding sites are present in the P2 promoter of the AβH-J-J locus and the data supporting an involvement of MEF-2 in the transcription directed by the P2 promoter are in strong agreement with the muscle-specific expression of junctin and junctate, as reported by several laboratories, including ours. Of great interest is the finding that no homology was found between the P2 promoter sequences and the sequences present in the upstream P1 promoter, as conclusively demonstrated by the results of homology studies performed using MacVector sequence analysis software. In particular, neither MEF-2 nor MEF-3 binding sites are present, while we found Sp1, AP-2, C-EBP, and GATA-1 functional binding sites; this finding supports the hypothesis that this P1 promoter, unlike the P2 promoter, exhibits a strong and broad transcriptional activity (G. Feriotto, F. Zorzato, and R. Gambari, unpublished data).
Therefore, this gene cluster appears to be very appealing for studies of the transcriptional and posttranscriptional regulation of gene expression in eukaryotes, as has been already reported for different eukaryotic gene systems (2, 31), including acetylcholinesterase (38), Pcdh-gamma (63), protocadherin alpha and gamma (59), regulator of G protein signaling GAIP/RGS19 (66), chondrolectin (64), and stromelysin 3 (34).
Our results do not conclusively address the involvement of other factors in the transcription of the AβH-J-J locus, despite the fact that E-box- and GRE-like activities are important for transcription of this locus. In addition, our data do not exclude involvements of protein-protein interactions. This study could be of interest due to strong evidence demonstrating that MEF-2 proteins interact with other transcription factors, including members of the MyoD family of transcriptional activators, to synergistically activate gene expression (10). Similarly, physical interactions between Sp1 and MEF-2 were demonstrated by immunological detection of both proteins in DNA binding complexes formed in vitro by nuclear extracts (20).
In conclusion, the data reported here on the functional characterization of the junctin-junctate promoter in vitro and in vivo suggest the possibility that the calcineurin-dependent MEF-2 response also regulates genes encoding key proteins present in the junctional sarcoplasmic reticulum membrane, a subcellular compartment that makes up the calcium release site of striated muscle cells and that is crucial for delivery of calcium signal to the cell. It is tempting to speculate that the modulation of the expression of junctin may affect the calcium fluxes via the quaternary complex containing the calcium release channel RYR. This in turn may influence activation and inhibition of cellular response to calcium signal, including the coordination of the transcription of genes encoding these proteins. The definition of the transcription factor(s) involved in regulation of genes responsible of calcium signaling will be important for elucidation of the correlation between excitation-contraction and excitation-transcription coupling, two intimately related but functionally distinct mechanisms which are crucial to the functional and structural integrity of striated muscle.
Acknowledgments
This work was supported in part by grants from FIRB and Ministero Università e Ricerca Scientifica e Tecnologica (60 and 40%), the Department of Anesthesia, Kantosspital, Basel, Switzerland, HPRN-CT-2002-00331 from the European Union, the Italian Space Agency, and the Swiss Muscle Foundation. R.G. is supported by grants from the Associazione Italiana per la Ricerca sul Cancro. S.F. is supported by Telethon grant GP0210Y01.
We thank Anemone Gunther and Maria Luisa Piaggio for help in the ChIP procedure.
REFERENCES
- 1.Always, S. E., H. Degens, G. Krishnamurthy, and C. A. Smith. 2002. Potential role for Id myogenic repressors in apoptosis and attenuation of hypertrophy in muscles of aged rats. Am. J. Physiol. Cell Physiol. 283:C66-C76. [DOI] [PubMed] [Google Scholar]
- 2.Ayoubi, T. A., and W. J. Van De Ven. 1996. Regulation of gene expression by alternative promoters. FASEB J. 10:453-460. [PubMed] [Google Scholar]
- 3.Berezikov, E., V. Guryev, R. H. Plasterk, and E. Cuppen. 2004. CONREAL: conserved regulatory elements anchored alignment algorithm for identification of transcription factor binding sites by phylogenetic footprinting. Genome Res. 14:170-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, B. L., J. F. Martin, and E. N. Olson. 1995. The mouse MRF4 promoter is trans-activated directly and indirectly by muscle-specific transcription factors. J. Biol. Chem. 270:2889-2892. [DOI] [PubMed] [Google Scholar]
- 5.Blanchette, M., and M. Tompa. 2002. Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 12:739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blough, E., B. Dineen, and K. Esser. 1999. Extraction of nuclear proteins from striated muscle tissue. BioTechniques 26:202-204. [DOI] [PubMed] [Google Scholar]
- 7.Borgatti, M., I. Lampronti, A. Romanelli, C. Pedone, M. Saviano, N. Bianchi, C. Mischiati, and R. Gambari. 2003. Transcription factor decoy molecules based on a peptide nucleic acid (PNA)-DNA chimera mimicking Sp1 binding sites. J. Biol. Chem. 278:7500-7509. [DOI] [PubMed] [Google Scholar]
- 8.Cairns, W., C. Cairns, I. Pongratz, L. Poellinger, and S. Okret. 1991. Assembly of a glucocorticoid receptor complex prior to DNA binding enhances its specific interaction with a glucocorticoid response element. J. Biol. Chem. 266:11221-11226. [PubMed] [Google Scholar]
- 9.Chen, S. L., S. C. Wang, B. Hosking, and G. E. Muscat. 2001. Subcellular localization of the steroid receptor coactivators (SRCs) and MEF2 in muscle and rhabdomyosarcoma cells. Mol. Endocrinol. 15:783-796. [DOI] [PubMed] [Google Scholar]
- 10.Dechesne, C. A., Q. Wei, J. Eldridge, L. Gannoun-Zaki, P. Millasseau, L. Bougueleret, D. Caterina, and B. M. Paterson. 1994. E-box- and MEF-2-independent muscle-specific expression, positive autoregulation, and cross-activation of the chicken MyoD (CMD1) promoter reveal an indirect regulatory pathway. Mol. Cell. Biol. 14:5474-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinchuk, J. E., N. L. Henderson, T. C. Burn, R. Huber, S. P. Ho, J. Link, K. T. O'Neil, J. R. Focht, M. S. Scully, J. M. Hollis, J. F. Hollis, and P. A. Friedman. 2000. Aspartyl β-hydroxylase (Asph) and an evolutionarily conserved isoform of Asph missing the catalytic domain share exons with junctin. J. Biol. Chem. 275:39543-39554. [DOI] [PubMed] [Google Scholar]
- 12.Eisenberg, B. 1985. Adaptability of ultrastructure in the mammalian muscle. J. Exp. Biol. 115:55-68. [DOI] [PubMed] [Google Scholar]
- 13.Engelhardt, S., P. Boknik, U. Keller, J. Neumann, M. J. Lohse, and L. Hein. 2001. Early impairment of calcium handling and altered expression of junctin in hearts of mice overexpressing the beta1-adrenergic receptor. FASEB J. 15:2718-2720. [DOI] [PubMed] [Google Scholar]
- 14.Esser, K., T. Nelson, V. Lupa-Kimball, and E. Blough. 1999. The CACC box and myocyte enhancer factor-2 sites within the myosin light chain 2 slow promoter cooperate in regulating nerve-specific transcription in skeletal muscle. J. Biol. Chem. 274:12095-12102. [DOI] [PubMed] [Google Scholar]
- 15.Feo, S., V. Antona, G. Barbieri, R. Passantino, L. Cali, and A. Giallongo. 1995. Transcription of the human β enolase gene (ENO-3) is regulated by an intronic muscle-specific enhancer that binds myocyte-specific enhancer factor 2 proteins and ubiquitous G-rich-box binding factors. Mol. Cell. Biol. 15:5991-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feriotto, G., C. Mischiati, N. Bianchi, C. Rutigliano, P. Giacomini, and R. Gambari. 1995. Sequencing of an upstream region of the human HLA-DRA gene containing X' and Y' boxes. Nucleic Acids Res. 23:1671-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friday, B. B., P. O. Mitchell, K. M. Kegley, and G. K. Pavlath. 2003. Calcineurin initiates skeletal muscle differentiation by activating MEF2 and MyoD. Differentiation 71:217-227. [DOI] [PubMed] [Google Scholar]
- 18.Glover, L., S. Quinn, M. Ryan, D. Pette, and K. Ohlendieck. 2002. Supramolecular calsequestrin complex. Eur. J. Biochem. 269:4607-4616. [DOI] [PubMed] [Google Scholar]
- 19.Gossett, L. A., D. J. Kelvin, E. A. Sternberg, and E. N. Olson. 1989. A new myocyte-specific enhancer binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol. Cell. Biol. 9:5022-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grayson, J., R. Bassel-Duby, and R. S. Williams. 1998. Collaborative interactions between MEF-2 and Sp1 in muscle-specific gene regulation. J. Cell Biochem. 70:366-375. [PubMed] [Google Scholar]
- 21.Groh, S., I. Marty, M. Ottolia, G. Prestipino, A. Chapel, M. Villaz, and M. Ronjat. 1999. Functional interaction of the cytoplasmic domain of triadin with the skeletal ryanodine receptor. J. Biol. Chem. 274:12278-12283. [DOI] [PubMed] [Google Scholar]
- 22.Gundersen, K., and J. P. Merlie. 1994. Id-1 as a possible transcriptional mediator of muscle disuse atrophy. Proc. Natl. Acad. Sci. USA 91:3647-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, A., F. Pan, J. C. Stroud, H. D. Youn, J. O. Liu, and L. Chen. 2003. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature 422:730-734. [DOI] [PubMed] [Google Scholar]
- 24.Himeda, C. L., J. A. Ranish, J. C. Angello, P. Maire, R. Aebersold, and S. D. Hauschka. 2004. Quantitative proteomic identification of Six4 as the Trex binding factor in the muscle creatine kinase enhancer. Mol. Cell. Biol. 24:2132-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong, C. S., Y. G. Kwak, J. H. Ji, S. W. Chae, and D. Han Kim. 2001. Molecular cloning and characterization of mouse cardiac junctate isoforms. Biochem. Biophys. Res. Commun. 289:882-887. [DOI] [PubMed] [Google Scholar]
- 26.Horlick, R. A., and P. A. Benfield. 1989. The upstream muscle-specific enhancer of the rat muscle creatine kinase gene is composed of multiple elements. Mol. Cell. Biol. 9:2396-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu, D. K. W., Y. Guo, G. F. Alberts, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, K. A. Peifley, and J. A. Winkles. 1996. Identification of a murine TEF-1-related gene expressed after mitogenic stimulation of quiescent fibroblasts and during myogenic differentiation. J. Biol. Chem. 271:13786-13795. [DOI] [PubMed] [Google Scholar]
- 28.Kageyama, K., Y. Ihara, S. Goto, Y. Urata, G. Toda, K. Yano, and T. Kondo. 2002. Overexpression of calreticulin modulates protein kinase B/Akt signaling to promote apoptosis during cardiac differentiation of cardiomyoblast H9c2 cells. J. Biol. Chem. 277:19255-19264. [DOI] [PubMed] [Google Scholar]
- 29.Kane, C. D., and A. R. Means. 2000. Activation of orphan receptor-mediated transcription by Ca(2+)/calmodulin-dependent protein kinase IV. EMBO J. 19:691-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurabayashi, M., R. Jeyaseelan, and L. Kedes. 1993. Antineoplastic agent doxorubicin inhibits myogenic differentiation of C2 myoblasts. J. Biol. Chem. 268:5524-5529. [PubMed] [Google Scholar]
- 31.Landry, J. R., D. L. Mager, and B. T. Wilhelm. 2003. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 19:640-648. [DOI] [PubMed] [Google Scholar]
- 32.Lim, H. W., and J. D. Molkentin. 1999. Calcineurin and human heart failure. Nat. Med. 5:246-247. [DOI] [PubMed] [Google Scholar]
- 33.Lu, J., T. A. McKinsey, R. L. Nicol, and E. N. Olson. 2000. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 97:4070-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo, D., B. Mari, I. Stoll, and P. Anglard. 2002. Alternative splicing and promoter usage generates an intracellular stromelysin 3 isoform directly translated as an active matrix metalloproteinase. J. Biol. Chem. 277:25527-25536. [DOI] [PubMed] [Google Scholar]
- 35.MacKrill, J. J. 1999. Protein-protein interactions in intracellular Ca2+-release channel function. Biochem. J. 337:345-361. [PMC free article] [PubMed] [Google Scholar]
- 36.Mathiesen, I. 1999. Electropermeabilization of skeletal muscle enhances gene transfer in vivo. Gene Ther. 6:508-514. [DOI] [PubMed] [Google Scholar]
- 37.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27:40-47. [DOI] [PubMed] [Google Scholar]
- 38.Meshorer, E., D. Toiber, D. Zurel, I. Sahly, A. Dori, E. Cagnano, L. Schreiber, D. Grisaru, F. Tronche, and H. Soreq. 2004. Combinatorial complexity of 5′ alternative acetylcholinesterase transcripts and protein products. J. Biol. Chem. 279:29740-29751. [DOI] [PubMed] [Google Scholar]
- 39.Mickelson, J. R., and C. F. Louis. 1996. Malignant hyperthermia: excitation-contraction coupling, Ca2+ release channel, and cell Ca2+ regulation defects. Physiol. Rev. 76:537-592. [DOI] [PubMed] [Google Scholar]
- 40.Mir, L. M., M. F. Bureau, J. Gehl, R. Rangara, D. Rouy, J. M. Caillaud, P. Delaere, D. Branellec, B. Schwartz, and D. Scherman. 1999. High-efficiency gene transfer into skeletal muscle mediated by electric pulses. Proc. Natl. Acad. Sci. USA 96:4262-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mischiati, C., M. Borgatti, N. Bianchi, C. Rutigliano, M. Tomassetti, G. Feriotto, and R. Gambari. 1999. Interaction of the human NF-kappaB p52 transcription factor with DNA-PNA hybrids mimicking the NF-kappaB binding sites of the human immunodeficiency virus type 1 promoter. J. Biol. Chem. 274:33114-33122. [DOI] [PubMed] [Google Scholar]
- 42.Mischiati, C., G. Feriotto, M. Borgatti, P. Giacomini, and R. Gambari. 2000. Characterization of a major histocompatibility complex class II X-box binding protein enhancing Tat-induced transcription directed by the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 74:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell-Felton, H. R., B. Hunter, E. J. Stevenson, and S. C. Kandarian. 2000. Identification of weight-bearing-responsive elements in the skeletal muscle sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA1) gene. J. Biol. Chem. 275:23005-23011. [DOI] [PubMed] [Google Scholar]
- 45.Molkentin, J. D., J. R. Lu, C. L. Antos, B. Markham, J. Richardson, J. Robbins, S. R. Grant, and E. N. Olson. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93:215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Musarò, A., K. J. McCullagh, F. J. Naya, E. N. Olson, and N. Rosenthal. 1999. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 400:581-585. [DOI] [PubMed] [Google Scholar]
- 47.Nagamura-Inoue, T., T. Tamura, and K. Ozato. 2001. Transcription factors that regulate growth and differentiation of myeloid cells. Int. Rev. Immunol. 20:83-105. [DOI] [PubMed] [Google Scholar]
- 48.Nori, A., P. J. Lin, A. Cassetti, A. Villa, K. U. Bayer, and P. Volpe. 2003. Targeting of alpha-kinase-anchoring protein (alpha KAP) to sarcoplasmic reticulum and nuclei of skeletal muscle. Biochem. J. 370:373-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostrovsky, O., E. Bengal, and A. Aronheim. 2002. Induction of terminal differentiation by the c-Jun dimerization protein JDP2 in C2 myoblasts and rhabdomyosarcoma cells. J. Biol. Chem. 277:40043-40054. [DOI] [PubMed] [Google Scholar]
- 50.Parmacek, M. S., H. S. Ip, F. Jung, T. Shen, J. F. Martin, A. J. Vora, E. N. Olson, and J. M. Leiden. 1994. A novel myogenic regulatory circuit controls slow/cardiac troponin C gene transcription in skeletal muscle. Mol. Cell. Biol. 14:1870-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Passier, R., H. Zeng, N. Frey, F. J. Naya, R. L. Nicol, T. A. McKinsey, P. Overbeek, J. A. Richardson, S. R. Grant, and E. N. Olson. 2000. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Investig. 105:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinset, C., D. Montarras, J. Chenevert, A. Minty, P. Barton, C. Laurent, and F. Gros. 1988. Control of myogenesis in the mouse myogenic C2 cell line by medium composition and by insulin: characterization of permissive and inducible C2 myoblasts. Differentiation 38:28-34. [DOI] [PubMed] [Google Scholar]
- 53.Relaix, F., X. J. Wei, X. Wu, and D. A. Sassoon. 1998. Peg3/Pw1 is an imprinted gene involved in the TNF-NFB signal transduction pathway. Nat. Genet. 18:287-291. [DOI] [PubMed] [Google Scholar]
- 54.Salvatori, S., E. Damiani, F. Zorzato, P. Volpe, S. Pierobon, D. Quaglino, Jr., G. Salviati, and A. Margreth. 1988. Denervation-induced proliferative changes of triads in rabbit skeletal muscle. Muscle Nerve 11:1246-1259. [DOI] [PubMed] [Google Scholar]
- 55.Selman, C., and C. Leeuwenburgh. 2003. The role of Id2 and apoptosis during skeletal muscle remodeling. Am. J. Physiol. Regul. Integr. Comp. Physiol. 284:R538-R539. [DOI] [PubMed] [Google Scholar]
- 56.Spitz, F., J. Demignon, A. Porteu, A. Kahn, J. P. Concordet, D. Daegelen, and P. Maire. 1998. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc. Natl. Acad. Sci. USA 95:14220-14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suwanichkul, A., M. L. Cubbage, and D. R. Powell. 1990. The promoter of the human gene for insulin-like growth factor binding protein-1. Basal promoter activity in HEP G2 cells depends upon liver factor B1. J. Biol. Chem. 265:21185-21193. [PubMed] [Google Scholar]
- 58.Takekura, H., N. Kasuga, K. Kitada, and T. Yoshioka. 1996. Morphological changes in the triads and sarcoplasmic reticulum of rat slow and fast muscle fibres following denervation and immobilization. J. Muscle Res. Cell Motil. 17:391-400. [DOI] [PubMed] [Google Scholar]
- 59.Tasic, B., C. E. Nabholz, K. K. Baldwin, Y. Kim, E. H. Rueckert, S. A. Ribich, P. Cramer, Q. Wu, R. Axel, and T. Maniatis. 2002. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell 10:21-33. [DOI] [PubMed] [Google Scholar]
- 60.Treves, S., G. Feriotto, L. Moccagatta, R. Gambari, and F. Zorzato. 2000. Molecular cloning, expression, functional characterization, chromosomal localization, and gene structure of junctate, a novel integral calcium binding protein of sarco(endo)plasmic reticulum membrane. J. Biol. Chem. 275:39555-39568. [DOI] [PubMed] [Google Scholar]
- 61.Treves, S., C. Franzini-Armstrong, L. Moccagatta, C. Arnoult, C. Grasso, A. Schrum, S. Ducreux, M. X. Zhu, K. Mikoshiba, T. Girard, S. Smida-Rezgui, M. Ronjat, and F. Zorzato. 2004. Junctate is a key element in calcium entry induced by activation of InsP3 receptors and/or calcium store depletion. J. Cell Biol. 166:537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, J. K., T. X. Li, Y. F. Bai, and Z. H. Lu. 2003. Evaluating the binding affinities of NF-kappaB p50 homodimer to the wild-type and single-nucleotide mutant Ig-kappaB sites by the unimolecular dsDNA microarray. Anal. Biochem. 316:192-201. [DOI] [PubMed] [Google Scholar]
- 63.Wang, X., H. Su, and A. Bradley. 2002. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 16:1890-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weng, L., D. R. Van Bockstaele, J. Wauters, E. Van Marck, J. Plum, Z. N. Berneman, and J. Merregaert. 2003. A novel alternative spliced chondrolectin isoform lacking the transmembrane domain is expressed during T cell maturation. J. Biol. Chem. 278:19164-19170. [DOI] [PubMed] [Google Scholar]
- 65.Wu, H., F. J. Naya, T. A. McKinsey, B. Mercer, J. M. Shelton, E. R. Chin, A. R. Simard, R. N. Michel, R. Bassel-Duby, E. N. Olson, and R. S. Williams. 2000. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 19:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie, G. X., X. Han, E. Ito, Y. Yanagisawa, K. Maruyama, S. Sugano, Y. Suzuki, Y. Wang, A. Gabriel, S. K. Stevens, J. Mitchell, M. Sharma, and P. P. Palmer. 2003. Gene structure, dual-promoters and mRNA alternative splicing of the human and mouse regulator of G protein signaling GAIP/RGS19. J. Mol. Biol. 325:721-732. [DOI] [PubMed] [Google Scholar]
- 67.Xue, L., J. Wu, W. Zheng, P. Wang, J. Li, Z. Zhang, and T. Tong. 2004. Sp1 is involved in the transcriptional activation of p16(INK4) by p21(Waf1) in HeLa cells. FEBS Lett. 564:199-204. [DOI] [PubMed] [Google Scholar]
- 68.Yu, Y. T., R. E. Breitbart, L. B. Smoot, Y. Lee, V. Mahdavi, and B. Nadal-Ginard. 1992. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 6:1783-1798. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, L., J. Kelley, G. Schmeisser, Y. M. Kobayashi, and L. R. Jones. 1997. Complex formation between junctin, triadin, calsequestrin, and the ryanodine receptor. Proteins of the cardiac junctional sarcoplasmic reticulum membrane. J. Biol. Chem. 272:23389-23397. [DOI] [PubMed] [Google Scholar]
- 70.Zhu, B., and T. Gulick. 2004. Phosphorylation and alternative pre-mRNA splicing converge to regulate myocyte enhancer factor 2C activity. Mol. Cell. Biol. 24:8264-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]