Abstract
The detection of developing antimicrobial resistance (AMR) has become a global issue. The detection of developing antimicrobial resistance has become a global issue. The growing number of AMR bacteria poses a new threat to public health. Therefore, a less laborious and quick confirmatory test becomes important for further investigations into developing AMR in the environment and in clinical settings. This study aims to present a comprehensive analysis and validation of unique and antimicrobial-resistant strains from the WHO priority list of antimicrobial-resistant bacteria and previously reported AMR strains such as Acinetobacter baumannii, Aeromonas spp., Anaeromonas frigoriresistens, Anaeromonas gelatinfytica, Bacillus spp., Campylobacter jejuni subsp. jejuni, Enterococcus faecalis, Escherichia coli, Haemophilus influenzae, Helicobacter pylori, Klebsiella pneumonia subsp. pneumoniae, Pseudomonas aeruginosa, Salmonella enterica subsp. enterica serovar Typhimurium, Thermanaeromonas toyohensis, and Vibrio proteolyticus. Using in-house designed gene-specific primers, 18 different antibiotic resistance genes (algJ, alpB, AQU-1, CEPH-A3, ciaB, CMY-1-MOX-7, CMY-1-MOX-9, CMY-1/MOX, cphA2, cphA5, cphA7, ebpA, ECP_4655, fliC, OXA-51, RfbU, ThiU2, and tolB) from 46 strains were selected and validated. Hence, this study provides insight into the identification of strain-specific, unique antimicrobial resistance genes. Targeted amplification and verification using selected unique marker genes have been reported. Thus, the present detection and validation use a robust method for the entire experiment. Results also highlight the presence of another set of 18 antibiotic-resistant and unique genes (Aqu1, cphA2, cphA3, cphA5, cphA7, cmy1/mox7, cmy1/mox9, asaI, ascV, asoB, oxa-12, acr-2, pepA, uo65, pliI, dr0274, tapY2, and cpeT). Of these sets of genes, 15 were found to be suitable for the detection of pathogenic strains belonging to the genera Aeromonas, Pseudomonas, Helicobacter, Campylobacter, Enterococcus, Klebsiella, Acinetobacter, Salmonella, Haemophilus, and Bacillus. Thus, we have detected and verified sets of unique and antimicrobial resistance genes in bacteria on the WHO Priority List and from published reports on AMR bacteria. This study offers advantages for confirming antimicrobial resistance in all suspected AMR bacteria and monitoring the development of AMR in non-AMR bacteria, in the environment, and in clinical settings.
Keywords: Strain-specific genes, Pathogenic bacterial strains, Antimicrobial resistance, Genome-based detection method, Molecular detection of pathogenic bacteria, Multiplex PCR
1. Introduction
The life forms that inhabit this world range from inconspicuous organisms to large, multicellular, advanced beings. Realizing that the most significant proportion of earth dwellers are microscopic organisms is fascinating. Bacteria are highly cosmopolitan in existence. Even a square area of the soil is colonized by a cocktail of millions of bacteria. However, the diversity and relative abundance of bacterial phyla vary from soil to soil (Gupta et al. 2017). A minor proportion of these bacteria are pathogenic and can result in catastrophic events in humans (Khan et al. 2022). Antimicrobial resistance studies are crucial because they address the growing threat of bacteria and other pathogens becoming resistant to antibiotics (Ventola, 2015). This phenomenon endangers global health, making previously treatable infections deadly (Serwecińska, 2020). Understanding and combating antimicrobial resistance is vital to preserving the effectiveness of our current medical arsenal and ensuring a healthier future (Annunziato, 2019). Antimicrobial resistance genes in bacteria on the WHO priority list are of paramount importance because they pose severe threats to human health. These genes can render antibiotics ineffective, making infections harder to treat, leading to prolonged illness, increased mortality rates, and higher healthcare costs. Urgent research and action are essential to combat this global health crisis (WHO, https://www.who.int/). Therefore, the health and well-being of humans rely on the rapid and early detection of pathogenic organisms. The conventional detection methods of identification of bacteria by isolation and culturing on agar media and confirmation by biochemical and serological testing are cumbersome and time-consuming (Rajapaksha et al. 2019). In addition, the finding that less than 10 % of soil bacteria can be cultured fueled the need for rapid detection techniques. Cutting down the culturing step facilitates the detection of bacteria using PCR amplification (Petti, 2007, Gupta et al., 2017). Diverse forms of PCR, namely, real-time PCR, multiplex PCR, RT-PCR, and droplet digital PCR (ddPCR), are currently used for bacterial detection and quantification. Although techniques such as gene sequencing (Petti, 2007), flow cytometry, optical biosensors, and bioluminescent sensors are available for bacterial detection, PCR remains the most commonly used detection method that utilizes DNA- and RNA-based assays for bacterial identification. Molecular detection methods are rapid and sensitive for bacterial detection, including the identification of emerging pathogens. The sensitivity of the detection assay can be further improved by designing new primers (Rajapaksha et al. 2019). Several gene targets act as important tools in molecular detection assays. The functionally constant, conserved regions of the genes provide universality of the targets annealed by PCR primers. Generally, bacterial identification is performed using the 16S rRNA gene (Petti, 2007). However, the use of strain-specific genes for the identification of bacteria has been reported in recent studies. The detection of bacteria using strain-specific genes proves to be fast, efficient, inexpensive, and reliable. This technique allows the differentiation of bacterial strains that share a significant level of similarity in their morphology and physiology. Here, PCR primers are designed to target the single-copy genes present exclusively in a particular strain. It is worth noting that combining the primer pairs and running multiplexing PCR helps to characterize mixtures of strains simultaneously. This detection assay is highly flexible, as identifying a strain-specific gene helps to determine and distinguish specific bacterial strains (Ferrandis-Vila et al. 2022).
The present study employs the use of strain-specific genes identified in silico to detect and characterize intended bacterial strains from coastal soil samples employing multiplexing PCR. The strain-specific genes of Aeromonas spp., Helicobacter pylori, Campylobacter jejuni, Salmonella enterica, Acinetobacter baumannii, Haemophilus influenzae, Klebsiella pneumonia, and Enterococcus faecalis identified through BV-BRC and BLAST analysis were used to design PCR primers. Multiplexing PCR was used to validate the presence of these bacterial strains.
2. Materials and methods
2.1. Computational analysis for identification of strain-specific genes
Forty-six pathogenic bacterial strains were selected for the study. The unique genes of the respective bacterial strains were obtained through annotation using the Bacterial and Viral Bioinformatics Resource Centre (BV-BRC) webserver (Olson et al. 2022). The NCBI database was screened for previously reported papers and data available on the selected genes. NCBI-BLAST analysis was performed to confirm the identity and specific genes. The uniqueness of the selected genes to a specific bacterial strain was determined from the percentage sequence similarity obtained from BLAST analysis. Fifteen genes were identified to be uniquely expressed individually in 20 bacterial strains and were used in designing primers for laboratory validation (Fig. S1).
2.2. Criteria for selection of the reference genomes and bioprojects
The reference genomes and bioprojects of 46 selected pathogenic bacteria that are highly prevalent in environmental samples were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/). This included Acinetobacter baumannii K09 (NZ_CP043953.1), Aeromonas allosaccharophila FDAARGOS_933 (GCF_016026615.1), Aeromonas allosaccharophila FDAARGOS_933 (NZ_CP065745.1), Aeromonas aquatic AE235 contig7 (NZ_JRGL01000007.1), Aeromonas australiensis CECT 8023 (NZ_CDDH01000062.1), Aeromonas bestiarum GA97-22 Contig0001 (NZ_PPUX01000001.1), Aeromonas bivalvium ZJ19-2 NODE_1 (NZ_NXBQ01000001.1), Aeromonas cavernicola DSM 24474 (NZ_PGGC01000005.1), Aeromonas caviae WP8-S18-ESBL-04 (NZ_AP022254.1), Aeromonas dhakensis 71,431 (NZ_CP084351.1), Aeromonas diversa CDC 2478–85 (NZ_CDCE01000029.1), Aeromonas encheleia NCTC12917 (NZ_LR134376.1), Aeromonas enteropelogenes FDAARGOS_1537 (NZ_CP084358.1), Aeromonas eucrenophila CECT 4224 (NZ_CDDF01000005.1), Aeromonas finlandensis 4287D contig286 (NZ_JRGK01000286.1), Aeromonas fluvialis LMG 24681 (NZ_CDBO01000011.1), Aeromonas hydrophila FDAARGOS_916 (NZ_CP065651.1), Aeromonas jandaei FDAARGOS_986 (NZ_CP066092.1), Aeromonas lacus AE122 Contig147 (NZ_JRGM01000147.1), Aeromonas lusitana MDC 2473 A (NZ_PGCP01000003.1), Aeromonas media TR3_1 (NZ_CP075564.1), Aeromonas molluskorum 848 Cont1 (NZ_AQGQ01000001.1), Aeromonas piscicola LMG 24783 (NZ_CDBL01000052.1), Aeromonas popoffii CIP 105493 (NZ_CDBI01000014.1), Aeromonas rivipollensis G78 G78_contig_29 (NZ_JAAILA010000003.1), Aeromonas rivuli 20-VB00005 (NZ_CP079742.1), Aeromonas salmonicida SRW-OG1(NZ_CP051883.1), Aeromonas sanarellii LMG 24682 (NZ_CDBN01000021.1), Aeromonas schubertii ATCC 43700 Scaffold1 (NZ_LPUO01000001.1), Aeromonas simiae A6 (NZ_CP040449.1), Aeromonas sobria CECT 4245 (NZ_CDBW01000006.1), Aeromonas taiwanensis LMG 24683 (NZ_CDDD01000101.1), Aeromonas tecta CECT 7082 NZ_CDCA01000036.1), Aeromonas veronii FDAARGOS_632 NZ_CP044060.1), Anaeromonas frigoriresistens D2Q NZ_WSFT01000053.1 & NZ_WSFU01000119.1), Campylobacter jejuni subsp. jejuni NCTC 11168 (NC_002163.1), Enterococcus faecalis EnGen0336 (NZ_KB944666.1), Escherichia coli O157 H7 Sakai (NC_002695.2), Haemophilus influenzae 477 (NZ_CP007470.1), Helicobacter pylori MT5135 (NZ_CP071982.1), Klebsiella pneumonia subsp. pneumoniae HS11286 (NC_016845.1), Pseudomonas aeruginosa PAO1 (NC_002516.2), Salmonella enterica subsp. enterica serovar typhimurium LT2 (NC_003197.2), Thermanaeromonas toyohensis ToBE chromosome I (NZ_LT838272.1), and Vibrio proteolyticus NBRC 13287 (NZ_BATJ01000001.1). A total of 46 complete genomes were retrieved in the FASTA file format (Table S1). Bacillus subtilis and Bacillus cereus-group strains were accessed for presence of AMR genes from the BV-BRC web server (https://www.bv-brc.org/) (Olson et al. 2022) for confirmation.
2.3. Annotation of sequence data
The 46 genomes and bioprojects of the selected bacterial strains were annotated on the BV-BRC web server (https://www.bv-brc.org/) (Olson et al. 2022). An individual annotation of each acquired genome was carried out on the BV-BRC Workspace website (Table S1), and some information was retrieved from published literature. Genome-based selective annotation was carried out to identify strain-specific unique genes, specialty genes, domains and motifs, critical pathways, and subsystems.
2.4. Identification of strain-specific unique genes
The termed “specialty” genes contained within the annotated entire reference genomes of each of the different bacterial strains were subjected to a one-by-one examination for the purpose of gene isolation. Through a process known as NCBI-BLAST analysis, the one-of-a-kindness of the gene that was found was determined. The genes that had the lowest percentage of similarity to other gene sequences were chosen. The BV-BRC was consulted to acquire FASTA files containing single-copy gene sequences. The effective publications that were received from the website of the List of Prokaryotic names with Standing in Nomenclature (LPSN) (Parte et al. 2020) were used to initially identify the strain-specific genes of the bacteria that were chosen for the investigation.
2.5. Selective screening for the presence of inter- and intragenus unique genes and detection of point mutations
All 46 inter- and intragenus strains were thoroughly analyzed for the presence of strain-specific unique genes to avoid errors during validation and misinterpretation of results. The presence of similar genes in other strains in the same genus and other strains in another genus were tested and cross-verified. The percentage of similarity among genes was also validated to check for any possible errors. The RIPper - Genome-Wide Repeat-Induced Point (RIP) Mutation Analysis (https://theripper.hawk.rocks/#/home) tool was used to check for point mutations (van Wyk et al., 2019).
2.6. Extraction of DNA from pure cultures
Genomic DNA was extracted from in-house-isolated environmental bacteria from coastal and estuarine soil samples using the phenol:chloroform:isoamyl (25:24:1) (PCA) method (P3803, Merck). This method has been employed on strains of Kocuria, Acinetobacter, Aeromonas, Pseudomonas, Helicobacter, Campylobacter, Enterococcus, Klebsiella, and Salmonella. Quality control strains such as Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Bacillus subtilis were used to obtain DNA samples for the group that served as a control. The extracted DNA was measured with a NanoDrop spectrophotometer (260/280 ratio). Mix 2 mg of a pure culture of bacteria with 100 μl of lysozyme (20 mg/ml) and incubate overnight at 37 °C. After incubation, 15 μl of proteinase K and 40 μl of 10 % SDS were added and brought up to 400 μl with TE buffer. Five microliters of RNase enzyme (100 μg/ml concentration) were added to the tube, which was incubated at 55 °C to 70 °C for four hours with intermittent vortexing. To this mixture, equal volumes (approximately 400 μl) of freshly prepared phenol, chloroform, and isoamyl alcohol (25:24:1) were added and mixed by gentle inversion. The tubes were centrifuged at 10,000 rpm for 30 min. Upon centrifugation, distinct aqueous and organic phases were formed. The aqueous phase (i.e., the upper layer or aqueous layer containing DNA) was carefully transferred to a fresh tube. The volume of 3 M sodium acetate added was 10 % of the total volume of the aqueous phase. Then, 2.5 times ice-cold absolute ethanol was added to the solution and incubated overnight at −20 °C. After incubation, the tubes were centrifuged at 10,000 rpm for 30 min, and the supernatant was discarded. The DNA pellet was washed twice with freshly prepared 70 % ethanol, and the pellet was allowed to air dry before being suspended in 20 μl of TE buffer. The isolated DNA was visualized on a 0.8 % agarose gel (Otal et al., 1991, de Almeida et al., 2013). The DNA concentration in each sample was adjusted to approximately 100 ng/μl for further analysis.
2.7. Design and validation of primers for PCR amplification and confirmation of unique genes using in silico tools
Selected unique genes were used as target genes for the design of primers for polymerase chain reaction (PCR) and subsequent PCR amplification and confirmation of unique genes in a laboratory. A portion of a specific gene was selected, and primers were designed using the Integrated DNA Technologies, Inc., PrimerQuest™ Tool (IDT, 2023a). The expected properties of your oligos before wet laboratory validation for guanine and cytosine (GC) content, melting temperature (Tm), molecular weight, extinction coefficient, µg/OD, nmol/OD, to identify secondary structure potential, to minimize dimerization, and NCBI BLAST™ analysis have been closely examined using the OligoAnalyzer™ Tool (IDT, 2023b) to avoid further errors in experiments during validation. Designed primers were tested for working ability and confirmation of product generation by in silico PCR amplification (Franklin et al. 1996; Rekadwad et al. 2021). The objective of utilizing in silico PCR is to facilitate the acquisition of anticipated PCR outcomes from DNA through the utilization of contemporary bacterial genome sequences (Bikandi et al., 2004, Brown et al., 2005, Rocco et al., 2016).
2.8. Targeted amplification and wet-lab verification of unique genes by polymerase chain reaction (PCR)
PCR amplification and wet-lab verification of the concerned gene were performed by using in-house designed specific forward and reverse primers for genes - AQU-1/cphA2 (aqcp_222-F, aqcp_744-R), CMY-1/MOX (cmy-mox_241-F, cmy-mox_769-R), cephA Family (cephA_195-F, cephA_422-R, cephA_327-R), aqu1 (aqu1_128-F, aqu1_1088-R), alpB (alpB_450_F, alpB_1146_R and alpB_1155_F, alpB_1589_R), ciaB (ciaB_154-F, ciaB_1082-R), rfbU (rfbU_34, rfbU_561), Oxa 51 (oxa51_281-F, oxa51_692-R), ThiU2 (thiU2_157-F, thiU2_356-R), and ebpA (ebpA_242-F, ebpA_892-R), and ECP_4655 (ecp427-F, ecp652-R) (Table 1).
Table 1.
Primers for unique genes found in a selected set of 20 pathogenic and antibiotic-resistant bacteria.
| Gene | Primer F/R | Sequence (5′ to 3′) | Start bp | No of bp | Tm Actual | Total gene bp | ssDNA bp | Product bp | Annealing Temp |
|---|---|---|---|---|---|---|---|---|---|
| AQU-1/cphA2 | aqcp_222-F | GGTCAGCGAGCAGACCCTGTTC | 222 | 22 | 65.8 | 1149 | 927 | – | |
| aqcp_744-R | GCTGGTCTTGATGCCGTAGGCCTC | 744 | 24 | 67.8 | 1149 | 744 | 523 | 63 | |
| CMY-1/MOX | cmy-mox_241-F | GTCAGCGAGCAGACCCTGTTCG | 241 | 22 | 65.8 | 1167 | 1026 | – | |
| cmy-mox_769-R | CCGCCGAGCTGGTCTTGATGCC | 769 | 22 | 67.7 | 1167 | 769 | 529 | 63 | |
| cephA Family | cephA_195-F | GGCGACCTGGACGCCGGATAC | 195 | 21 | 67.6 | 765 | 570 | – | |
| cephA_422-R | TCCGGCAGCCCCTTGCGGGT | 422 | 20 | 67.5 | 765 | 422 | 228 | 64 | |
| cephA_327-R | GGACTTCCAGTAGGCGTTA | 327 | 19 | 56.7 | 765 | 327 | 133 | 53 | |
| aqu1 | aqu1_128-F | AGCACAGGATCCCGGGCATG | 128 | 20 | 63.4 | 1149 | 1021 | – | |
| aqu1_1088-R | CGGTTGGCCAGCATGACGATGC | 1088 | 22 | 65.8 | 1149 | 1088 | 960 | 61 | |
| alpB | alpB_450_F | CCAAGGCAACCTGAGTCTTTAT | 450 | 22 | 58.4 | 1596 | 1146 | – | |
| alpB_1146_R | GAATGTGGGCTTACGCTACTAC | 1146 | 22 | 60.3 | 1596 | 1146 | 696 | 55 | |
| alpB_1155_F | CTTACGCTACTACGGCTTCTTC | 1155 | 22 | 60.3 | 1596 | 434 | – | ||
| alpB_1589_R | CGTAGCCATAGACCCAATACAC | 1589 | 22 | 60.3 | 1596 | 434 | 434 | 57 | |
| ciaB | ciaB_154-F | GCCATACTTAGGCGTTTGATTG | 154 | 22 | 58.4 | 1801 | 1647 | – | |
| ciaB_1082-R | GGAACGACTTGAGCTGAGAATA | 1082 | 22 | 58.4 | 1801 | 1082 | 929 | 57 | |
| rfbU | rfbU_34 | GGTACGGGAATGTGGCAATA | 34 | 20 | 57.3 | 1001 | 977 | – | |
| rfbU_561 | CAACTTGCACCAACAGCTAAA | 561 | 21 | 55.9 | 1001 | 561 | 527 | 53 | |
| Oxa 51 | oxa51_281-F | ATAAGGCAACCACCACAGAAG | 281 | 21 | 57.9 | 801 | 520 | – | |
| oxa51_692-R | GCTGAACAACCCATCCAGTTA | 692 | 21 | 57.9 | 801 | 692 | 411 | 56 | |
| ThiU2 | thiU2_157-F | GCACTTTCCACTTTAGCACTTAC | 157 | 23 | 58.9 | 1301 | 1144 | – | |
| thiU2_356-R | ACCGATACCTTGCCCAATAC | 356 | 20 | 57.3 | 1301 | 356 | 200 | 55 | |
| ebpA | ebpA_242-F | CAGCTCAGCCACCTAAGTTATT | 242 | 22 | 45.5 | 3301 | 2945 | – | |
| ebpA_892-R | ACCGCTATCTGCCAATGTATC | 892 | 21 | 47.6 | 3301 | 892 | 650 | 43 | |
| ECP_4655 | ecp427-F | ATCACCGCAGGATCGTTAATC | 427 | 21 | 57.9 | 901 | 474 | – | |
| ecp652-R | TGGTGCCGGAGAGGTAATA | 652 | 19 | 58.4 | 901 | 652 | 225 | 55.5 |
PCR amplification and wet-lab verification were conducted for the selected genes using specific forward and reverse primers designed in-house. The primer pairs used for each gene are listed in Table 2. The PCR master mix was prepared by combining the eDNA template, forward and reverse primers, dNTPs, Taq DNA polymerase, and PCR buffer. The reaction conditions were set according to the annealing temperature specific to each pair of primers. Positive and negative controls were included in the PCRs using known samples. The reaction components were thoroughly mixed and distributed into PCR tubes. The tubes or plates were then placed into a thermal cycler, and PCR amplification was carried out for 35 cycles. The amplification protocol included an initial denaturation step at 95 °C for 5 min, followed by denaturation at 95 °C for 1 min, annealing at the respective temperature for each set of primers, extension at 72 °C for 45 s, and a final extension at 72 °C for 7 min (Rocco et al. 2016). The annealing temperatures varied for each set of primers used for the amplification of the target genes. The annealing time was set to 45 s at temperatures of 63 °C, 63 °C, 64 °C, 53 °C, 61 °C, 55 °C, 57 °C, 57 °C, 53 °C, 56 °C, 55 °C, 43 °C, and 55.5 °C for the genes AQU-1/cphA2, cmy-1/mox, cephA Family (one forward and two reverse primers, separate PCRs were performed for each reverse primer), aqu1, alpB, ciaB, rfbU, Oxa 51, ThiU2, ebpA, and ECP_4655, respectively. After PCR amplification, the resulting products were analyzed using agarose gel electrophoresis. The DNA bands were visualized under UV light and compared with a DNA ladder (100 bp) to determine the expected sizes for the respective target genes from known quality control strains such as Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Bacillus subtilis. In-house isolated environmental bacteria from coastal soil samples, such Acinetobacter spp. and Kocuria spp., were used for confirmation of in silico results and experimental validation of unique genes (Mullis et al., 1986, Parkhill et al., 2000, Bikandi et al., 2004, Nallapareddy et al., 2006, San Millán et al., 2013; In silico PCR amplification, 2023). Based on PCR specific to the above bacterial taxa and visualization through gel electrophoresis, the cmy-1/mox and aqu1 genes were successfully validated in Pseudomonas aeruginosa, followed by other taxa.
Table 2.
List of scrutinized taxa containing inter- and intragenus strain-specific unique genes.
| Sl. No. | Taxa | NCBI accession number | Previous reports |
|---|---|---|---|
| 1 | Pseudomonas aeruginosa PAO1 | NC_002516.2 | Franklin & Ohman, 1996 |
| 2 | Helicobacter pylori MT5135 | NZ_CP071982.1 | Bai et al., 2002 |
| 3 | Campylobacter jejuni subsp. jejuni NCTC 11168 |
NC_002163.1 |
Parkhill et al., 2000 |
| 4 | Enterococcus faecalis EnGen0336 strain T5 acAro-supercont1.1 | NZ_KB944666.1 | Nallapareddy et al., 2006 |
| 5 | Klebsiella pneumonia subsp. pneumoniae HS11286 |
NC_016845.1 |
No report available |
| 6 | Acinetobacter baumannii K09-14 | NZ_CP043953.1 | Brown et al., 2005 |
| 7 | Salmonella enterica subsp. enterica serovar typhimurium str. LT2 | NC_003197.2 | Xiang et al., 1994 |
| 8 | Haemophilus influenza 477 | NZ_CP007470.1 | No report available |
| 9 | Aeromonas dhakensis 71431 | NZ_CP084351.1 | Wu et al., 2013 |
| 10 | Aeromonas allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | Wang et al., 2021 |
| 11 | Aeromonas enteropelogenes FDAARGOS_1537 | NZ_CP084358.1 | No report available |
| 12 | Aeromonas jandaei FDAARGOS_986 | NZ_CP066092.1 | No report available |
| 13 | Aeromonas veronii FDAARGOS_632 | NZ_CP044060.1 | Ragupathi et al., 2020 |
| 14 | Aeromonas bivalvium ZJ19-2 | NZ_NXBQ01000001.1 | No report available |
| 15 | Aeromonas hydrophila FDAARGOS_916 | NZ_CP065651.1 | Bottoni et al., 2015 |
| 16 | Aeromonas salmonicida SRW-OG1 | NZ_CP051883.1 | No report available |
| 17 | Aeromonas encheleia NCTC12917 | NZ_LR134376.1 | No report available |
| 18 | Aeromonas piscicola LMG 24783 | NZ_CDBL01000052.1 | No report available |
| 19 | Aeromonas caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | No report available |
| 20 | Aeromonas media TR3_1 | NZ_CP075564.1 | Ebmeyer et al., 2019 |
3. Results
3.1. An overview of specialized genes, antibiotic production ability, pathogenicity and reports on pathogenic strains
In the context of bacteria, specialized genes can contribute to their ability to produce antibiotics, enhance pathogenicity, or perform other specialized functions. Almost all analyzed bacterial strains do not have any published reports stating proven antimicrobial resistance or reported unique genes especially in Aeromonas in this study. Those reported have functions in antibiotic inactivation enzyme, beta-lactam resistance gene, Campylobacter invasion antigen B, virulence factor, adherence, biofilm formation, sortase-assembled pili, adhesion, predicted thiazole transporter, outer membrane protein/porin, protein (regulates length and adhesion of type 1 fimbriae, and mediates mannose binding), antiphagocytosis, serum resistance, LPS O-antigen biosynthesis protein, and Tol-Pal system beta propeller repeat protein as per RAST and BV-BRC analysis. Specialized genes refer to specific genes that are unique to certain organisms or have specific functions within 46 selected strains (Table S1), as mentioned above. Strains belonging to the Bacillus subtilis and Bacillus cereus groups were not included due to technical reasons in the main analysis. Bacillus spp. were accessed on the BV-BRC web server for selected sets of genes for the presence of AMR.
Eighteen genes unique to these microorganisms were selected based on criteria adopted for single copy number (Rekadwad et al., 2021): OXA-51, algJ, alpB, AQU-1, cphA2, CEPH-A3, ciaB, CMY-1/MOX, cphA5, cphA7, ebpA, ECP_4655, fliC, MOX-7, MOX-9, CMY-1/MOX, RfbU, ThiU2, and tolB (Table 2). These genes can play a crucial role in the adaptation, survival, and specialized functions of an organism.
3.2. Elucidation of unique genes in a taxon based on single copy number
Based on screening criteria single copy number, selected 20 bacterial strains (out of 46 + strains belonging Bacillus groups) were screened out for the presence of unique taxa-specific genes and selected present single copy number genes highly specific to taxa or groups of similar strains in taxa, such as K09-14, FDAARGOS_933, WP8-S18-ESBL-04, 71431, FDAARGOS_1537, FDAARGOS_916, FDAARGOS_986, TR3_1, LMG 24783, SRW-OG1, FDAARGOS_632, NCTC 11168, EnGen0336 strain T5 acAro-supercont1.1, 477, MT5135, HS11286, PAO1, ZJ19-2, NCTC 12917, and LT2.
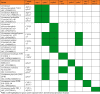
The gene similarity report suggests that the unique genes tolB, alpB, ecp_4655, fliC, oxa-51, rfbU, thiU2, cmy-1-mox-7, aqu-1, cpha2, cepha3, cpha5, cmy-1/mox, and cmy-1-mox-9 were found in selected bacterial strains showing identity with other taxa (Table 3). No point mutations were detected in the selected genes during analysis. This indicates that during pathogenesis, such genes may be acquired by these bacteria either in the environment or through horizontal gene transfer. Further analysis within the genus Aeromonas indicates that cpha5, cmy-1/mox7, and cmy-1/mox7 are unique to the taxon compared to aqu-1, cpha2, cpha3, cpha7, and cmy-1/mox (Table 4).
Table 3.
Presence of 18 unique taxa-specific genes in another genera or domain.
| Gene | Taxon | Gene ID in other taxa, if available | Name and NCBI Accession number of reported taxa |
|---|---|---|---|
| AQU-1, cphA2 | Aeromonas allosaccharophila 71431 (NZ_CP084351.1) | 114286262 (AQU-1) | Camellia sinensis cultivar Shuchazao unplaced genomic scaffold AHAU_CSS_1 Scaffold3615 (NW_021027072.1) |
| AQU-1, cphA2 | Aeromonas dhakensis 71431 (NZ_CP084351.1) | NG_050396.1 (cphA2) | Aeromonas hydrophila AER 19 cphA gene for subclass B2 metallo-beta-lactamase CphA2 (NG_050396.1) |
| AQU-1, cphA2 | Aeromonas hydrophila FDAARGOS_916 (NZ_CP065651.1) | --------- | --------- |
| CEPH-A3 | Aeromonas allosaccharophila FDAARGOS_933 (NZ_CP065745.1) | --------- | --------- |
| CEPH-A3 | Aeromonas enteropelogenes FDAARGOS_1537 (NZ_CP084358.1) | --------- | --------- |
| CEPH-A3 | Aeromonas jandaei FDAARGOS_986 (NZ_CP066092.1) | --------- | --------- |
| CEPH-A3 | Aeromonas veronii FDAARGOS_632 (NZ_CP044060.1) | --------- | --------- |
| CMY-1/MOX | Aeromonas bivalvium ZJ19-2 NODE_1 (NZ_NXBQ01000001.1) | --------- | --------- |
| cphA5 | Aeromonas salmonicida SRW-OG1 (NZ_CP051883.1) | --------- | --------- |
| cphA7, CMY-1/MOX | Aeromonas encheleia NCTC12917 (NZ_LR134376.1) | NG_050400.1 (cphA7) | Aeromonas jandaei ATCC 49568 cphA gene for subclass B2 metallo-beta-lactamase CphA7 (NG_050400.1) |
| cphA7 | Aeromonas piscicola LMG 24783 (NZ_CDBL01000052.1) | NG_050400.1 (cphA7) | Aeromonas jandaei ATCC 49568 cphA gene for subclass B2 metallo-beta-lactamase CphA7 (NG_050400.1) |
| CMY-1-MOX-7 | Aeromonas caviae WP8-S18-ESBL-04 (NZ_AP022254.1) | 18813570 (MOX-7) | Serpula lacrymans var. lacrymans S7.9 unplaced genomic scaffold SERLAscaffold_11 (NW_006763300.1) |
| CMY-1-MOX-9 | Aeromonas media TR3_1 (NZ_CP075564.1) | 18815923 (MOX-9) | Serpula lacrymans var. lacrymans S7.9 unplaced genomic scaffold SERLAscaffold_18 (NW_006763307.1) |
| algJ | Pseudomonas aeruginosa PAO1 (NC_002516.2) | 77219964 | Pseudomonas paraeruginosa strain Cr1 (NZ_CP020560.1) |
| 66491330 | Legionella pneumophila strain C9_S (NZ_CP015941.1) | ||
| 45624672 | Pseudomonas simiae strain PCL1751 (NZ_CP010896.1) | ||
| 78258735 | Butyrivibrio crossotus isolate MGYG-HGUT-01319 (NZ_CABKNR010000014.1) | ||
| 72998054 | Pseudomonas citronellolis strain P3B5 (NZ_CP014158.1) | ||
| 72394459 | Pseudomonas coronafaciens pv. oryzae str. 1_6 (NZ_CP046035.1) | ||
| 61792614 | Pseudomonas avellanae strain CC1416 contig318.1 (NZ_AVEP02000318.1) | ||
| 61708583 | Pseudomonas lactis strain SS101 (NZ_CM001513.1) | ||
| 57261476 | Pseudomonas brassicacearum strain 3Re2-7 (NZ_CP034725.1) | ||
| 45621619 | Pseudomonas simiae strain PCL1751 (NZ_CP010896.1) | ||
| 45522320 | Pseudomonas putida NBRC 14164 (NC_021505.1) | ||
| 1182870 | Pseudomonas syringae pv. tomato str. DC3000 (NC_004578.1) | ||
| 73734174 | Pseudomonas tremae strain PA-1-10F (NZ_CP066270.1) | ||
| 78554965 | Pseudomonas extremaustralis strain DSM 17835 (NZ_LT629689.1) | ||
| 72192903 | Pseudomonas umsongensis strain CY-1 (NZ_CP051487.1) | ||
| 69747833 | Pseudomonas alloputida strain NMI2441_06 (NZ_JAJSPR010000005.1) | ||
| 66647291 | Pseudomonas mandelii strain KGI_MA19 (NZ_CP081178.1) | ||
| 65075219 | Pseudomonas congelans strain DSM 14939 (NZ_FNJH01000002.1) | ||
| 61868467 | Pseudomonas amygdali pv. tabaci str. ATCC 11528 (NZ_CP042804.1) | ||
| 61648697 | Pseudomonas chlororaphis strain qlu-1 (NZ_CP061079.1) | ||
| 58532891 | Pseudomonas asiatica strain RYU5 RYU5_unitig_0 (NZ_BLJF01000001.1) | ||
| 57607844 | Pseudomonas mendocina S5.2 (NZ_CP013124.1) | ||
| 57474020 | Pseudomonas protegens CHA0 (NZ_LS999205.1) | ||
| 55845753 | Pseudomonas tolaasii NCPPB 2192 Ga0070648_11 (NZ_PHHD01000001.1) | ||
| 49870618 | Pseudomonas monteilii strain B5 (NZ_CP022562.1) | ||
| 49614069 | Pseudomonas plecoglossicida strain XSDHY-P (NZ_CP031146.1) | ||
| 77179175 | Pseudomonas guariconensis strain MR119 MR119_8 (NZ_PJCP01000008.1) | ||
| 64093427 | Pseudomonas fulva strain YAB-1 contig13 (NZ_LAWW01000013.1) | ||
| 57399847 | Pseudomonas otitidis strain MrB4 (NZ_AP022642.1) | ||
| 56069949 | Pseudomonas yamanorum strain LBUM636 (NZ_CP012400.2) | ||
| 47554532 | Pseudomonas veronii strain R02 (NZ_CP018420.1) | ||
| 57518971 | Pseudomonas proteolytica strain WS 5126 4_283282_20.4743 (NZ_JAAQXL010000004.1) | ||
| 78504381 | Pseudomonas parafulva NBRC 16636 (NZ_BBIU01000020.1) | ||
| 77277010 | Pseudomonas syringae strain Susan2139 (NZ_CP074578.1) | ||
| 77257756 | Marinobacter salarius strain SMR5 (NZ_CP020931.1) | ||
| 77247859 | Pseudomonas carnis strain NWU Be30 (NZ_JAMKPY010000008.1) | ||
| 77187064 | Pseudomonas capsici strain NCPPB2479 (NZ_JAOXME010000028.1) | ||
| 76211923 | Pseudomonas mediterranea strain DSM 16733 (NZ_LT629790.1) | ||
| 75527915 | Pseudomonas atacamensis strain SM1 (NZ_CP070503.1) | ||
| 75198874 | Pseudomonas kurunegalensis strain T2909-1 1 (NZ_JALKHE010000002.1) | ||
| 75192006 | Pseudomonas siliginis strain OTU6BANIB1 (NZ_CP099598.1) | ||
| 72498365 | Pseudomonas marginalis strain PgKB35 contig2 (NZ_VTFG01000002.1) | ||
| 72478030 | Pseudomonas moraviensis strain LMG 24280 (NZ_LT629788.1) | ||
| 72422112 | Pseudomonas juntendi strain PP_2463 (NZ_CP091088.1) | ||
| 70103785 | Pseudomonas gessardii strain LMG 21604 (NZ_FNKR01000003.1) | ||
| 69858097 | Pseudomonas savastanoi strain MHT1 (NZ_CP076652.1) | ||
| 66761385 | Pseudomonas poae strain LMG 21465 (NZ_LT629706.1) | ||
| 64467080 | Pseudomonas cannabina pv. alisalensis strain MAFF 301419 (NZ_CP067022.1) | ||
| 61932304 | Azotobacter chroococcum strain B3 (NZ_CP011835.1) | ||
| 61881971 | Pseudomonas lundensis strain 2T.2.5.2 (NZ_CP062158.2) | ||
| 61828961 | Pseudomonas synxantha strain R6-28-08 (NZ_CP027756.1) | ||
| 61637163 | Pseudomonas fluorescens strain ATCC 13525 (NZ_LT907842.1) | ||
| 58768986 | Pseudomonas mosselii strain PtA1 (NZ_CP024159.1) | ||
| 58766660 | Pseudomonas mosselii strain PtA1 (NZ_CP024159.1) | ||
| 58694899 | Pseudomonas rhodesiae strain NL2019 (NZ_CP054205.1) | ||
| 57661674 | Pseudomonas gingeri strain A6001 (NZ_JACAOR010000008.1) | ||
| 57633271 | Pseudomonas koreensis strain LMG 21318 (NZ_LT629687.1) | ||
| 57377827 | Pseudomonas azotoformans strain LMG 21611 (NZ_LT629702.1) | ||
| 55644702 | Pseudomonas corrugata strain RM1-1-4 (NZ_CP014262.1) | ||
| 47765936 | Pseudomonas viridiflava strain CFBP 1590 isolate E12-5 (NZ_LT855380.1) | ||
| 45541106 | Pseudomonas cichorii JBC1 (NZ_CP007039.1) | ||
| 42930483 | Pseudomonas alcaligenes strain NEB 585 (NZ_CP014784.1) | ||
| 31709204 | Pseudomonas lurida strain L228 (NZ_CP015639.1) | ||
| 878551 | Pseudomonas aeruginosa PAO1 algX (NC_002516.2) | ||
| alpB | Helicobacter pylori MT5135 (NZ_CP071982.1) | 124639028 | Helicoverpa zea isolate HzStark_Cry1AcR (NC_061469.1) |
| 100125692 | Triticum aestivum cultivar Chinese Spring chromosome 4A (NC_057803.1) | ||
| 542895 | Triticum aestivum cultivar Chinese Spring chromosome 7D (NC_057814.1) | ||
| ciaB | Campylobacter jejuni subsp. jejuni NCTC 11168 (NC_002163.1) | 7411001 | Campylobacter lari RM2100 (NC_012039.1) |
| 78326632 | Arcobacter nitrofigilis DSM 7299 (NC_014166.1) | ||
| 77176464 | Campylobacter ureolyticus strain LMG 6451 (NZ_CP053832.1) | ||
| 66544092 | Campylobacter coli strain FDAARGOS_735 (NZ_CP046317.1) | ||
| 61065105 | Campylobacter fetus strain CFF00A031 (NZ_CP059443.1) | ||
| 61001623 | Campylobacter curvus strain ATCC 35224 (NZ_CP053826.1) | ||
| 60991106 | Campylobacter showae strain ATCC 51146 (NZ_CP012544.1) | ||
| 77266361 | Campylobacter vulpis strain 251/13 (NZ_CP041617.1) | ||
| 56587051 | Campylobacter armoricus strain CCUG 73571 (NZ_CP053825.1) | ||
| 56509597 | Campylobacter hyointestinalis subsp. lawsonii strain CHY5 (NZ_CP053828.1) | ||
| 44004343 | Campylobacter hepaticus strain HV10 (NZ_CP031611.1) | ||
| 39299964 | Aliarcobacter thereius LMG 24486 AA347_contig000001 (NZ_LLKQ01000001.1) | ||
| 905214 | Campylobacter jejuni subsp. jejuni NCTC 11168 (NC_002163.1) | ||
| 66539337 | Helicobacter cinaedi PAGU611 (NC_017761.1) | ||
| 56463744 | Aliarcobacter butzleri ED-1 (NC_017187.1) | ||
| 61153890 | Campylobacter pinnipediorum subsp. pinnipediorum strain RM17261 (NZ_CP012547.1) | ||
| 68759512 | Campylobacter sputorum bv. paraureolyticus LMG 11764 strain LMG 17589 (NZ_CP019684.1) | ||
| 46921585 | Campylobacter lanienae NCTC 13004 (NZ_CP015578.1) | ||
| 74432015 | Campylobacter insulaenigrae NCTC 12927 (NZ_CP007770.1) | ||
| 77230936 | Campylobacter upsaliensis 17-M197059 (NZ_OU701459.1) | ||
| 66287961 | Campylobacter volucris strain LMG 24380 (NZ_CP043428.1) | ||
| 61750346 | Aliarcobacter skirrowii CCUG 10374 (NZ_CP032099.1) | ||
| 57004565 | Helicobacter pullorum strain NCTC13156 (NZ_UGJF01000001.1) | ||
| 56461800 | Aliarcobacter cryaerophilus ATCC 43158 (NZ_CP032823.1) | ||
| 52037112 | Campylobacter helveticus strain ATCC 51209 (NZ_CP020478.1) | ||
| 28663259 | Campylobacter concisus strain ATCC 33237 (NZ_CP012541.1) | ||
| 61924358 | Clostridium innocuum strain ATCC 14501 (NZ_CP048838.1) | ||
| 68118740 | Naegleria fowleri strain ATCC 30894 (NW_025407941.1) | ||
| 54452205 | Macroventuria anomochaeta strain CBS 525.71 (NW_022985375.1) | ||
| ebpA | Enterococcus faecalis EnGen0336 (NZ_KB944666.1) | 1050 | Homo sapiens chromosome 19, GRCh38.p14 (NC_000019.10) |
| 12606 | Mus musculus strain C57BL/6J chromosome 7, GRCm39 (NC_000073.7) | ||
| 110596866 | Homo sapiens chromosome 8, GRCh38.p14 (NC_000008.11) | ||
| 12608 | Mus musculus strain C57BL/6J chromosome 2, GRCm39 (NC_000068.8) | ||
| 19016 | Mus musculus strain C57BL/6J chromosome 6, GRCm39 (NC_000072.7) | ||
| 111832672 | Mus musculus strain C57BL/6J chromosome 5, GRCm39 (NC_000071.7) | ||
| 73389 | Mus musculus strain C57BL/6J chromosome 12, GRCm39 (NC_000078.7) | ||
| 5468 | Homo sapiens chromosome 3, GRCh38.p14 (NC_000003.12) | ||
| 861 | Homo sapiens chromosome 21, GRCh38.p14 (NC_000021.9) | ||
| 5241 | Homo sapiens chromosome 11, GRCh38.p14 (NC_000011.10) | ||
| 56729 | Homo sapiens chromosome 19, GRCh38.p14 (NC_000019.10) | ||
| 4297 | Homo sapiens chromosome 11, GRCh38.p14 (NC_000011.10) | ||
| 1051 | Homo sapiens chromosome 20, GRCh38.p14 (NC_000020.11) | ||
| 13653 | Mus musculus strain C57BL/6J chromosome 18, GRCm39 (NC_000084.7) | ||
| 387173 | Mus musculus strain C57BL/6J chromosome 16, GRCm39 (NC_000082.7) | ||
| 6548 | Homo sapiens chromosome 1, GRCh38.p14 (NC_000001.11) | ||
| 20787 | Mus musculus strain C57BL/6J chromosome 11, GRCm39 (NC_000077.7) | ||
| 6659 | Homo sapiens chromosome 6, GRCh38.p14 (NC_000006.12) | ||
| 5598 | Homo sapiens chromosome 17, GRCh38.p14 (NC_000017.11) | ||
| 723848 | Mus musculus strain C57BL/6J chromosome 4, GRCm39 (NC_000070.7) | ||
| 11091 | Homo sapiens chromosome 9, GRCh38.p14 (NC_000009.12) | ||
| 723814 | Mus musculus strain C57BL/6J chromosome X, GRCm39 (NC_000086.8) | ||
| 57264 | Mus musculus strain C57BL6J chromosome 8, GRCm39 (NC_000074.7) | ||
| 20471 | Mus musculus strain C57BL/6J chromosome 12, GRCm39 (NC_000078.7) | ||
| 28951 | Homo sapiens chromosome 2, GRCh38.p14 (NC_000002.12) | ||
| 13592 | Mus musculus strain C57BL/6J chromosome 14, GRCm39 (NC_000080.7) | ||
| 140815 | Danio rerio strain Tuebingen chromosome 7, GRCz11 (NC_007118.7) | ||
| 18105 | Mus musculus strain C57BL/6J chromosome 13, GRCm39 (NC_000079.7) | ||
| 396728 | Sus scrofa isolate TJ Tabasco breed Duroc chromosome 13 Sscrofa11.1 (NC_010455.5) | ||
| ECP_4655 | Klebsiella pneumoniae subsp. pneumoniae HS11286 (NC_016845.1) | ABG72591.1 | FimH protein precursor [Escherichia coli 536] (ABG72591.1) |
| fliC | Escherichia coli O157 H7 str. Sakai (NC_002695.2) | Many | --------- |
| OXA-51 | Acinetobacter baumannii K09-14 (NZ_CP043953.1) | --------- | --------- |
| RfbU | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (NC_003197.2) | 1238129 | Shigella flexneri 2a str. 301 plasmid pCP301 (NC_004851.1) |
| 1789671 | Escherichia coli O157H7 str. Sakai plasmid pO157 (NC_002128.1) | ||
| ThiU2 | Haemophilus influenzae 477 (NZ_CP007470.1) | --------- | --------- |
| tolB | Vibrio proteolyticus NBRC 13287 (NZ_BATJ01000001.1) | Many | --------- |
Table 4.
Inter- and intrataxa presence of discovered unique genes.
 |
3.3. Unique genes specific to selected strains in the genus Aeromonas
A total of 37 strains among the genus Aeromonas were analyzed for the presence of unique genes and antimicrobial resistance. Of the 37 screened Aeromonas strains, 15 strains possessed various unique and antimicrobial resistance genes belonging to A. allosaccharophila 71431, A. allosaccharophila FDAARGOS_933, A. bivalvium ZJ19-2 NODE_1, A. caviae WP8-S18-ESBL-04, A. dhakensis 71431, A. encheleia NCTC12917, A. enteropelogenes FDAARGOS_1537, A. eucrenophila CECT 4224, A. hydrophila FDAARGOS_916, A. jandaei FDAARGOS_986, A. media TR3_1, A. rivuli 20-VB00005, A. salmonicida SRW-OG1, A. simiae A6, and A. veronii FDAARGOS_632 (Table 5a). Furthermore, 18 genes belong to the genus Aeromonas were disclosed that governs antimicrobial resistance through various mechanisms viz., Acyl-homoserine-lactone synthase (asaI), Type 3 secretion system (ascV), Arsenite oxidase subunit (asoB), Ambler Class beta-lactamase, carbapenem (Ceph-A3), Histidine kinase family (ChpA), CMY beta-lactamase (cmy-1/mox), Cyanophycin synthetase (CphA), OXA β-Lactamases (Oxa-12), 4-amino-6-deoxy-N-Acetyl-D-hexosaminyl-(Lipid carrier) acetyltrasferase (pglD_3), Acetylcholine receptor subunit beta-type acr-2 protein (Acr-2), Aminopeptidase PepA-related protein (PepA), Aminopeptidase Y (Arg, Lys, Leu preference) (UO65), AQU family (Aqu), Inhibitor of invertebrate i-type lysozyme, periplasmic (PliI), Bacteriocin lactacin-F subunit (LafX), Nudix dNTPase - MutT/nudix family protein (DR0274), Transporter 2, ATP binding cassette subfamily B member (TapY2) and T-type phycobiliprotein lyase (CpeT).
Table 5a.
Presence of 18 unique or antimicrobial resistance genes in the Aeromonas.
| Unique/AMR gene | Code | Strains | Taxa | Accession No. | Product size (bp) | Identity (%) in BLAST |
|---|---|---|---|---|---|---|
| Acyl-homoserine-lactone synthase | asaI | 12 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 624 | 0 |
| A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 651 | 1 | |||
| A. caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | 630 | 51 | |||
| A. dhakensis 71,431 | NZ_CP084351.1 | 624 | 24 | |||
| A. encheleia NCTC12917 | NZ_LR134376.1 | 627 | 3 | |||
| A. enteropelogenes FDAARGOS_1537 | NZ_CP084358.1 | 651 | 5 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 624 | 79 | |||
| A. media TR3_1 | NZ_CP075564.1 | 627 | 14 | |||
| A. piscicola LMG 24783 | NZ_CDBL01000052.1 | 624 | 0 | |||
| A. rivuli 20-VB00005 | NZ_CP079742.1 | 657 | 1.9 | |||
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 624 | 37 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 651 | 37 | |||
| Type 3 secretion system | ascV | 5 | A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 2118 | 3 |
| A. diversa CECT 4254 | NZ_CDCE01000029.1 | 2115 | 0 | |||
| A. encheleia NCTC12917 | NZ_LR134376.1 | 2118 | 2 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 2115 | 24 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 2118 | 11 | |||
| Arsenite oxidase subunit | asoB | 15 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 1281 | 0 |
| A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 1320 | 2 | |||
| A. bivalvium ZJ19-2 NODE_1 | NZ_NXBQ01000001.1 | 1278 | 0 | |||
| A. caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | 1281 | 52 | |||
| A. dhakensis 71,431 | NZ_CP084351.1 | 1281 | 27 | |||
| A. encheleia NCTC12917 | NZ_LR134376.1 | 1278 | 3 | |||
| A. enteropelogenes FDAARGOS_1537 | NZ_CP084358.1 | 1281 | 6 | |||
| A. eucrenophila CECT 4224 | NZ_CDDF01000005.1 | 1287 | 0 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 1281 | 71 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 1278 | 13 | |||
| A. media TR3_1 | NZ_CP075564.1 | 1278 | 14 | |||
| A. rivuli 20-VB00005 | NZ_CP079742.1 | 1290 | 1 | |||
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 1281 | 34 | |||
| A. simiae A6 | NZ_CP040449.1 | 1272 | 1 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 1281 | 43 | |||
| Ambler Class beta-lactamase, carbapenem | CEPH-A3 | 4 | A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 765 | 6 |
| A. enteropelogenes FDAARGOS_1537 | NZ_CP084358.1 | 603 | 1 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 765 | 15 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 762 | 47 | |||
| Histidine kinase family | ChpA | 11 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 762 | 0 |
| A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 765 | 6 | |||
| A. caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | 210 | 68.75 | |||
| A. dhakensis 71,431 | NZ_CP084351.1 | 762 | 35 | |||
| A. encheleia NCTC12917 | NZ_LR134376.1 | 663 | 3 | |||
| A. enteropelogenes FDAARGOS_1537 | NZ_CP084358.1 | 603 | 1 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 765 | 64 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 765 | 15 | |||
| A. piscicola LMG 24783 | NZ_CDBL01000052.1 | 765 | 0 | |||
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 762 | 27 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 762 | 47 | |||
| CMY beta-lactamase | CMY-1/MOX | 4 | A. bivalvium ZJ19-2 NODE_1 | NZ_NXBQ01000001.1 | 1170 | 0 |
| A. caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | 1152 | 81 | |||
| A. encheleia NCTC12917 | NZ_LR134376.1 | 1167 | 3 | |||
| A. media TR3_1 | NZ_CP075564.1 | 1152 | 16 | |||
| Cyanophycin synthetase | cphA | 10 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 762 | 0 |
| A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 765 | 6 | |||
| A. dhakensis 71,431 | NZ_CP084351.1 | 762 | 35 | |||
| A. encheleia NCTC12917 | NZ_LR134376.1 | 663 | 3 | |||
| A. enteropelogenes FDAARGOS_1537 | NZ_CP084358.1 | 603 | 1 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 765 | 64 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 765 | 15 | |||
| A. piscicola LMG 24783 | NZ_CDBL01000052.1 | 765 | 0 | |||
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 762 | 27 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 762 | 47 | |||
| OXA β-Lactamases | OXA-12 | 7 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 795 | 0 |
| A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 795 | 4 | |||
| A. dhakensis 71,431 | NZ_CP084351.1 | 795 | 33 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 795 | 78 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 795 | 13 | |||
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 795 | 34 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 795 | 49 | |||
| 4-amino-6-deoxy-N-Acetyl-D-hexosaminyl-(Lipid carrier) acetyltrasferase | pglD_3 | 3 | A. caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | 597 | 40 |
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 609 | 100 | |||
| A. simiae A6 | NZ_CP040449.1 | 645 | 100 | |||
| Acetylcholine receptor subunit beta-type acr-2 protein | Acr-2 | 2 | A. diversa CECT 4254 | NZ_CDCE01000029.1 | 372 | 0 |
| A. encheleia NCTC12917 | NZ_LR134376.1 | 372 | 3.1 | |||
| Aminopeptidase PepA-related protein | PepA | 14 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 1497 | 0 |
| A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 1500 | 2 | |||
| A. bestiarum GA97-22 Contig0001 | NZ_PPUX01000001.1 | 1497 | 1 | |||
| A. caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | 1482 | 52 | |||
| A. dhakensis 71,431 | NZ_CP084351.1 | 1497 | 25 | |||
| A. encheleia NCTC12917 | NZ_LR134376.1 | 1491 | 3 | |||
| A. enteropelogenes FDAARGOS_1537 | NZ_CP084358.1 | 1515 | 5 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 1497 | 70 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 1506 | 12 | |||
| A. media TR3_1 | NZ_CP075564.1 | 1494 | 14 | |||
| A. rivuli 20-VB00005 | NZ_CP079742.1 | 1509 | 1 | |||
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 1497 | 33 | |||
| A. simiae A6 | NZ_CP040449.1 | 771 | 1 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 1500 | 42 | |||
| Aminopeptidase Y (Arg, Lys, Leu preference) | UO65 | 10 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 1068 | 0 |
| A. caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | 1068 | 52 | |||
| A. dhakensis 71,431 | NZ_CP084351.1 | 1068 | 29 | |||
| A. encheleia NCTC12917 | NZ_LR134376.1 | 1068 | 3 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 1068 | 72 | |||
| A. media TR3_1 | NZ_CP075564.1 | 1068 | 14 | |||
| A. rivuli 20-VB00005 | NZ_CP079742.1 | 1077 | 1 | |||
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 1068 | 33 | |||
| A. simiae A6 | NZ_CP040449.1 | 1080 | 1 | |||
| A. tecta CECT 7082 | NZ_CDCA01000036.1 | 1068 | 0 | |||
| AQU family | Aqu | 3 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 1143 | 0 |
| A. dhakensis 71,431 | NZ_CP084351.1 | 1143 | 45 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 1149 | 74 | |||
| Inhibitor of invertebrate i-type lysozyme, periplasmic | PliI | 13 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 438 | 25 |
| A. bivalvium ZJ19-2 NODE_1 | NZ_NXBQ01000001.1 | 438 | 0 | |||
| A. caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | 438 | 52 | |||
| A. dhakensis 71,431 | NZ_CP084351.1 | 438 | 25 | |||
| A. encheleia NCTC12917 | NZ_LR134376.1 | 438 | 3 | |||
| A. enteropelogenes FDAARGOS_1537 | NZ_CP084358.1 | 453 | 5 | |||
| A. eucrenophila CECT 4224 | NZ_CDDF01000005.1 | 438 | 0 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 438 | 72 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 438 | 13 | |||
| A. media TR3_1 | NZ_CP075564.1 | 438 | 13 | |||
| A. rivuli 20-VB00005 | NZ_CP079742.1 | 438 | 3.5 | |||
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 438 | 33 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 438 | 75.9 | |||
| Bacteriocin lactacin-F subunit | LafX | 5 | A. finlandensis 4287D contig286 | NZ_JRGK01000286.1 | 339 | 0 |
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 339 | 57.8 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 339 | 91.6 | |||
| A. lacus AE122 Contig147 | NZ_JRGM01000147.1 | 339 | 0 | |||
| A. rivuli 20-VB00005 | NZ_CP079742.1 | 339 | 100 | |||
| Nudix dNTPase - MutT/nudix family protein | DR0274 | 13 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 558 | 0 |
| A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 564 | 2 | |||
| A. caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | 558 | 52 | |||
| A. dhakensis 71,431 | NZ_CP084351.1 | 558 | 29 | |||
| A. encheleia NCTC12917 | NZ_LR134376.1 | 552 | 3.7 | |||
| A. enteropelogenes FDAARGOS_1537 | NZ_CP084358.1 | 561 | 6 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 558 | 73 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 561 | 12 | |||
| A. media TR3_1 | NZ_CP075564.1 | 558 | 14 | |||
| A. rivuli 20-VB00005 | NZ_CP079742.1 | 579 | 16.6 | |||
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 558 | 33 | |||
| A. tecta CECT 7082 | NZ_CDCA01000036.1 | 558 | 0 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 564 | 43 | |||
| Transporter 2, ATP binding cassette subfamily B member | TapY2 | 4 | A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 282 | 3.3 |
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 276 | 90.9 | |||
| A. piscicola LMG 24783 | NZ_CDBL01000052.1 | 279 | 0 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 282 | 66.6 | |||
| T-type phycobiliprotein lyase | CpeT | 12 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 675 | 0 |
| A. allosaccharophila FDAARGOS_933 | NZ_CP065745.1 | 651 | 2.3 | |||
| A. caviae WP8-S18-ESBL-04 | NZ_AP022254.1 | 666 | 52 | |||
| A. dhakensis 71,431 | NZ_CP084351.1 | 675 | 25 | |||
| A. enteropelogenes FDAARGOS_1537 | NZ_CP084358.1 | 651 | 31.5 | |||
| A. hydrophila FDAARGOS_916 | NZ_CP065651.1 | 675 | 72 | |||
| A. jandaei FDAARGOS_986 | NZ_CP066092.1 | 645 | 11 | |||
| A. media TR3_1 | NZ_CP075564.1 | 660 | 14 | |||
| A. rivuli 20-VB00005 | NZ_CP079742.1 | 684 | 1 | |||
| A. salmonicida SRW-OG1 | NZ_CP051883.1 | 684 | 24 | |||
| A. simiae A6 | NZ_CP040449.1 | 636 | 100 | |||
| A. veronii FDAARGOS_632 | NZ_CP044060.1 | 651 | 53.1 |
We have found that some Aeromonas taxa possesses completely unique and novel genes not showing identity with any other genes in the existing database such as asaI (A. allosaccharophila 71431, and A. piscicola LMG 24783), ascV (A. diversa CECT 4254), asoB (A. allosaccharophila 71431, A. bivalvium ZJ19-2 NODE_1, and A. eucrenophila CECT 4224), ChpA (A. allosaccharophila 71431, and A. piscicola LMG 24783), CMY-1/MOX (A. bivalvium ZJ19-2 NODE_1), cphA (A. allosaccharophila 71431, and A. piscicola LMG 24783), OXA-12 (A. allosaccharophila 71431), Acr-2 (A. diversa CECT 4254), PepA (A. allosaccharophila 71431), UO65 (A. allosaccharophila 71431, and A. tecta CECT 7082) Aqu (A. allosaccharophila 71431), PliI (A. bivalvium ZJ19-2 NODE_1, and A. eucrenophila CECT 4224), DR0274 (A. allosaccharophila 71431, and A. tecta CECT 7082), TapY2 (A. piscicola LMG 24783), and CpeT (A. allosaccharophila 71431) (Table 5b). This suggests that some important strains, such as A. allosaccharophila 71431, A. bivalvium ZJ19-2 NODE_1, A. diversa CECT 4254, A. eucrenophila CECT 4224, A. piscicola LMG 24783, and A. tecta CECT 7082, and other strains, except those that show 95–100 % identity of genes (Table 5a), are potential bacteria to explore for deep analysis to find significant differences among strains belonging to the genus Aeromonas.
Table 5b.
Presence of 15 unique or antimicrobial resistance genes in the Aeromonas.
| Unique/AMR gene | Code | Taxa | Accession No. | Identity (%) in BLAST |
|---|---|---|---|---|
| Acyl-homoserine-lactone synthase | asaI | A. allosaccharophila 71,431 | NZ_CP084351.1 | 0 |
| A. piscicola LMG 24783 | NZ_CDBL01000052.1 | 0 | ||
| Type 3 secretion system | ascV | A. diversa CECT 4254 | NZ_CDCE01000029.1 | 0 |
| Arsenite oxidase subunit | asoB | A. allosaccharophila 71,431 | NZ_CP084351.1 | 0 |
| A. bivalvium ZJ19-2 NODE_1 | NZ_NXBQ01000001.1 | 0 | ||
| A. eucrenophila CECT 4224 | NZ_CDDF01000005.1 | 0 | ||
| Histidine kinase family | ChpA | A. allosaccharophila 71,431 | NZ_CP084351.1 | 0 |
| A. piscicola LMG 24783 | NZ_CDBL01000052.1 | 0 | ||
| CMY beta-lactamase | CMY-1/MOX | A. bivalvium ZJ19-2 NODE_1 | NZ_NXBQ01000001.1 | 0 |
| Cyanophycin synthetase | cphA | A. allosaccharophila 71,431 | NZ_CP084351.1 | 0 |
| A. piscicola LMG 24783 | NZ_CDBL01000052.1 | 0 | ||
| OXA β-Lactamases | OXA-12 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 0 |
| Acetylcholine receptor subunit beta-type acr-2 protein | Acr-2 | A. diversa CECT 4254 | NZ_CDCE01000029.1 | 0 |
| Aminopeptidase PepA-related protein | PepA | A. allosaccharophila 71,431 | NZ_CP084351.1 | 0 |
| Aminopeptidase Y (Arg, Lys, Leu preference) | UO65 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 0 |
| A. tecta CECT 7082 | NZ_CDCA01000036.1 | 0 | ||
| AQU family | Aqu | A. allosaccharophila 71,431 | NZ_CP084351.1 | 0 |
| Inhibitor of invertebrate i-type lysozyme, periplasmic | PliI | A. bivalvium ZJ19-2 NODE_1 | NZ_NXBQ01000001.1 | 0 |
| A. eucrenophila CECT 4224 | NZ_CDDF01000005.1 | 0 | ||
| Nudix dNTPase - MutT/nudix family protein | DR0274 | A. allosaccharophila 71,431 | NZ_CP084351.1 | 0 |
| A. tecta CECT 7082 | NZ_CDCA01000036.1 | 0 | ||
| Transporter 2, ATP binding cassette subfamily B member | TapY2 | A. piscicola LMG 24783 | ||
| T-type phycobiliprotein lyase | CpeT | A. allosaccharophila 71,431 | NZ_CP084351.1 | 0 |
A total of 17 strains belonging to the genera Aeromonas (A. allosaccharophila 71,431 (NZ_CP084351.1), A. allosaccharophila 71,431 (NZ_CP084351.1), A. dhakensis 71,431 (NZ_CP084351.1), A. dhakensis 71,431 (NZ_CP084351.1), A. hydrophila FDAARGOS_916 (NZ_CP065651.1), A. hydrophila FDAARGOS_916 (NZ_CP065651.1), A. allosaccharophila FDAARGOS_933 (NZ_CP065745.1), A. enteropelogenes FDAARGOS_1537 (NZ_CP084358.1), A. jandaei FDAARGOS_986 (NZ_CP066092.1), A. veronii FDAARGOS_632 (NZ_CP044060.1), A. bivalvium ZJ19-2 NODE_1 (NZ_NXBQ01000001.1), A. salmonicida SRW-OG1 (NZ_CP051883.1), A. encheleia NCTC12917 (NZ_LR134376.1), A. encheleia NCTC12917 (NZ_LR134376.1), A. piscicola LMG 24783 (NZ_CDBL01000052.1), A. caviae WP8-S18-ESBL-04 (NZ_AP022254.1), and A. media TR3_1 (NZ_CP075564.1)) were further investigated for disclosed unique genes such as aqu-1, cpha2, aqu-1_d, cpha2, aqu-1_h, cpha2, cepha3_a, cepha3_e, cepha3_j, cepha3_v, cmy-1/mox, cpha5, cpha7_e, cmy-1/mox, cpha7_p, cmy-1-mox-7, and cmy-1-mox-9 to infer either significant differences or similarities among genes (Fig. 1). The heatmap suggests that almost all unique genes in the genus Aeromonas have significant differences rather than similarities among unique genes on a scale of 0 to 1. It has been recorded that aqu1, cepha2, and those showing values less than 0.85 have significant differences among genes.
Fig. 1.
Heatmap of aqu-1, cpha2, aqu-1_d, cpha2, aqu-1_h, cpha2, cepha3_a, cepha3_e, cepha3_j, cepha3_v, cmy-1/mox, cpha5, cpha7_e, cmy-1/mox, cpha7_p, cmy-1-mox-7, and cmy-1-mox-9 in the genus Aeromonas. (Note: 1. Similar genes found in a taxon have been suffixed by the letter of the strain name. For example, the aqu1 gene, if found in multiple taxa, is suffixed with_d in the case of Aeromonas dhakensis. 2. 0 = indicates 100 % difference, while 1 = indicates 100 % similarity).
3.4. Inference from specific validation of novel and unique genes
Antibiotic resistance genes belonging to the genera Aeromonas, Pseudomonas, Helicobacter, Campylobacter, Enterococcus, Klebsiella, Acinetobacter, Salmonella, Bacillus, and Haemophilus have been identified. They found that some unique genes in these strains showed similarity with genes from other taxa with antibiotic production ability or resistance to antibiotics (Table 5b). Aeromonas strains are known pathogens that infect fish, animals and humans. Hence, pathogenic strains belonging to the Aeromonas genera were analyzed in this study for the presence of the abovementioned genes and identified based on single copy 15 unique gene number criteria. These genes play a crucial role in the adaptation, survival, and specialized functions of organisms. Some of these genes were found to be involved in antibiotic resistance, pathogenicity, adherence, biofilm formation, and other functions. This investigation of the genus Aeromonas shows that 15 out of 37 strains were found to possess unique and antimicrobial resistance genes. Additionally, 18 genes related to antimicrobial resistance were identified within the genus. Some strains within the genus Aeromonas showed completely unique and novel genes not found in other strains. Further analysis using a heatmap showed significant differences among the unique genes in the genus Aeromonas.
4. Discussion
The 46 strains chosen were from Aeromonas, Pseudomonas, Helicobacter, Campylobacter, Enterococcus, Klebsiella, Acinetobacter, Salmonella, Haemophilus, and Bacillus genera. They all had 18 single-copy unique genes such as algJ, alpB, AQU-1, CEPH-A3, ciaB, CMY-1-MOX-7, CMY-1-MOX-9, CMY-1/MOX, cphA2, cphA5, cphA7, ebpA, ECP_4655, fliC, OXA-51, RfbU, ThiU2, and tolB (Table 2, Table 3) that were involved in antibiotic resistance, pathogenicity, adherence, and biofilm formation. These suggest that identified genes can play a crucial role in the adaptation, survival, and specialized functions of an organism (Evans and Amyes, 2014, McMillan et al., 2019). According to a few investigations, bacterial strains such as Staphylococcus aureus (Oogai et al. 2011), Helicobacter pylori (Yamaoka, 2010, Wang et al., 2015), Escherichia coli (Bidet et al. 2012), Salmonella spp. (Jennings et al. 2017), Pseudomonas aeruginosa (Olejnickova et al. 2014), and Streptococcus suis (Wu et al., 2013) produce a variety of virulence factors, including toxins, immune-modulating agents, and exoenzymes. Those strains were investigated in this paper, and one of the most interesting findings of the study is the discovery of completely unique and novel genes with significant differences (Xiang et al., 1994, Bai et al., 2002, Wu et al., 2011, Wang et al., 2021) among them and common in all strains were found in one genus, Aeromonas. That means Aeromonas is the hub for all those antimicrobial genes found (Piotrowska and Popowska, 2014, Luo et al., 2022, Dubey et al., 2022) in other genera investigated under this study. This information can be used to better understand the adaptation, survival, and virulence of antimicrobial resistance strains. It can also be used to develop new strategies for preventing and treating antmicrobial resistance and virulence (Beceiro et al. 2013) in Pseudomonas, Helicobacter, Campylobacter, Enterococcus, Klebsiella, Acinetobacter, Salmonella, Haemophilus, Bacillus, and Aeromonas infections. Research from several groups in the last ten years (Martino et al., 2011, Liang et al., 2022, Zhang et al., 2023) backs up the idea that some genes found in other genera were unique to Aeromonas or even to certain strains of Aeromonas. This was the prime reason to explore the genus Aeromonas in the later part of the investigations. The heatmap analysis showed that almost all unique genes in the genus Aeromonas have significant differences rather than similarities. This suggests that the unique genes are highly diverse and may play a role in the diversity of Aeromonas strains. Hence, the findings of this study have a number of potential implications for the prevention, diagnosis, and treatment of antimicrobial-resistant bacteria (Bottoni et al., 2015, Ebmeyer et al., 2019, Ragupathi et al., 2020), not limited to Aeromonas infections. Hence, the identification of unique genes in antimicrobial resistance strains may lead to the development of new diagnostic tools, such as PCR tests, to detect specific unique genes detected in the above genera (Galhano et al. 2021). Furthermore, the identification of unique genes in certain strains could also lead to the development of new therapeutic targets for effective treatments for infections caused by antimicrobial-resistant bacteria in the case of some important diseases such as diabetes, malaria, tuberculosis, AIDS, cancer, etc., (Dadgostar, 2019, Demain and Sanchez, 2009, Qadri et al., 2023). Moreover, the findings of this study may help to understand the process of evolution and acquiring unique genes from taxa. This may help trace the emergence of new strains that may be more virulent or resistant to existing antibiotics.
5. Conclusions
Infections caused by antimicrobial-resistant bacteria such as WHO priority list of antimicrobial-resistant bacteria and previously reported AMR strains such as Acinetobacter baumannii, Aeromonas spp., Anaeromonas frigoriresistens, Anaeromonas gelatinfytica, Bacillus spp., Campylobacter jejuni subsp. jejuni, Enterococcus faecalis, Escherichia coli, Haemophilus influenzae, Helicobacter pylori, Klebsiella pneumonia subsp. pneumoniae, Pseudomonas aeruginosa, Salmonella enterica subsp. enterica serovar Typhimurium, Thermanaeromonas toyohensis, and Vibrio proteolyticus are difficult to detect and treat and have become a global public health threat. This investigation discloses and presents a comprehensive analysis of 46 antimicrobial-resistant strains of 20 pathogenic bacterial taxa. Additionally, two different sets of 18 antibiotic-resistant and unique genes in WHO priority list bacterial strains and in Aeromonas spp., were identified. It was observed that 15 single-copy genes may be suitable for the detection of these pathogenic strains, which belong to 10 different genera, such as Aeromonas, Pseudomonas, Helicobacter, Campylobacter, Enterococcus, Klebsiella, Acinetobacter, Salmonella, Haemophilus, and Bacillus. Identified sets of strain-specific, unique genes that can be used to develop new diagnostic tools to confirm AMR genes in suspected AMR bacteria and track the spread of AMR in non-AMR strains in the environment and clinical settings such as a hospital, laboratories, department, outpatient facility, or primary clinic (medicine, rehabilitation, or wellness), mobile hospitals, and tertiary care hospitals. Thus, this research can be used to develop more effective strategies for surveillance, preventing and combating AMR.
Funding
This research was funded by Yenepoya (Deemed to be University), India (Grant no. YU/SeedGrant/104–2021). The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP2023R134), King Saud University, Riyadh, Saudi Arabia.
CRediT authorship contribution statement
Bhagwan Narayan Rekadwad: Conceptualization, Formal analysis, Project administration, Resources, Supervision, Methodology, Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing, Project administration, Funding acquisition. Nanditha Pramod: Methodology, Data curation, Formal analysis, Visualization, Writing – original draft. Manik Prabhu Narsing Rao: Resources, Data curation, Formal analysis, Writing – review & editing. Abeer Hashem: Resources, Data curation, Formal analysis, Writing – review & editing. Graciela Dolores Avila-Quezada: Resources, Data curation, Formal analysis, Writing – review & editing. Elsayed Fathi Abd_Allah: Project administration, Funding acquisition, Resources, Data curation, Formal analysis, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP2023R134), King Saud University, Riyadh, Saudi Arabia. BNR duly acknowledge funding support received from Yenepoya (Deemed to be University), India (Grant no. YU/SeedGrant/104–2021).
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2023.103869.
Contributor Information
Bhagwan Narayan Rekadwad, Email: rekadwad@yenepoya.edu.in, rekadwad@gmail.com.
Nanditha Pramod, Email: 20368035@pondiuni.ac.in.
Manik Prabhu Narsing Rao, Email: manik.narsing@uautonoma.cl.
Abeer Hashem, Email: habeer@ksu.edu.sa.
Graciela Dolores Avila-Quezada, Email: gdavila@uach.mx.
Elsayed Fathi Abd_Allah, Email: eabdallah@ksu.edu.sa.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Supplementary figure 2.
Workflow of the genome-based robust method for the detection and validation of unique genes in pathogenic and antimicrobial-resistant bacterial strains belonging to various genera. The method utilizes genomic data analysis and polymerase chain reaction (PCR) validation to identify unique genes.
References
- Annunziato G. Strategies to Overcome Antimicrobial Resistance (AMR) making use of non-essential target inhibitors: a review. Int. J. Mol. Sci. 2019;20:5844. doi: 10.3390/ijms20235844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Zhang Y.L., Wang J.D., Lin H.J., Zhang Z.S., Zhou D.Y. Conservative region of the genes encoding four adhesins of Helicobacter pylori: cloning, sequence analysis and biological information analysis. Acad. J. First Med. Coll. p.l.a. 2002;22:869–871. [PubMed] [Google Scholar]
- Beceiro A., Tomás M., Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013;26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidet P., Bonarcorsi S., Bingen E. Virulence factors and pathophysiology of extraintestinal pathogenic Escherichia coli. Arch. Pediatr. 2012;19:S80–S92. doi: 10.1016/S0929-693X(12)71279-4. [DOI] [PubMed] [Google Scholar]
- Bikandi J., San Millán R., Rementeria A., Garaizar J. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR, and endonuclease restriction. Bioinformatics. 2004;20:798–799. doi: 10.1093/bioinformatics/btg491. [DOI] [PubMed] [Google Scholar]
- Bottoni C., Marcoccia F., Compagnoni C., Colapietro M., Sabatini A., Celenza G., Segatore B., Maturo M.G., Amicosante G., Perilli M. Identification of new natural CphA Metallo-β-Lactamases CphA4 and CphA5 in Aeromonas veronii and Aeromonas hydrophila Isolates from municipal sewage in central Italy. Antimicrob. Agents Chemother. 2015;59:4990–4993. doi: 10.1128/AAC.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S., Young H.K., Amyes S.G. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect. 2005;11:15–23. doi: 10.1111/j.1469-0691.2004.01016.x. [DOI] [PubMed] [Google Scholar]
- Dadgostar P. Antimicrobial resistance: implications and costs. Infect. Drug. Resist. 2019;12:3903–3910. doi: 10.2147/IDR.S234610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida I.N., da Silva Carvalho W., Rossetti M.L., Dalla Costa E.R., de Miranda S.S. Evaluation of six different DNA extraction methods for detection of Mycobacterium tuberculosis by means of PCR-IS6110: preliminary study. BMC. Res. Notes. 2013;6:561. doi: 10.1186/1756-0500-6-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain A., Sanchez S. Microbial drug discovery: 80 years of progress. J. Antibiot. 2009;62:5–16. doi: 10.1038/ja.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey S., Ager-Wick E., Peng B., Evensen Ø., Sørum H., Munang'andu H.M. Characterization of virulence and antimicrobial resistance genes of Aeromonas media strain SD/21-15 from marine sediments in comparison with other Aeromonas spp. Front. Microbiol. 2022;13:1022639. doi: 10.3389/fmicb.2022.1022639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeyer S., Kristiansson E., Larsson D.G.J. CMY-1/MOX-family AmpC β-lactamases MOX-1, MOX-2 and MOX-9 were mobilized independently from three Aeromonas species. J. Antimicrob. Chemother. 2019;74:1202–1206. doi: 10.1093/jac/dkz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B.A., Amyes S.G. OXA β-lactamases. Clin. Microbiol. Rev. 2014;27(2):241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandis-Vila M., Tiwari S.K., Mamerow S., Semmler T., Menge C., Berens C. Using unique ORFan genes as strain-specific identifiers for Escherichia coli. BMC Microbiol. 2022;22:135. doi: 10.1186/s12866-022-02508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin M.J., Ohman D.E. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J. Bacteriol. 1996;178:2186–2195. doi: 10.1128/jb.178.8.2186-2195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhano B.S., Ferrari R.G., Panzenhagen P., de Jesus A.C.S., Conte-Junior C.A. Antimicrobial resistance gene detection methods for bacteria in animal-based foods: a brief review of highlights and advantages. Microorganisms. 2021;9:923. doi: 10.3390/microorganisms9050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, A., Gupta, R., Singh, R. L., 2017. Microbes and Environment. In: Principles and Applications of Environmental Biotechnology for a Sustainable Future, in: Singh, R. (Eds.) Applied Environmental Science and Engineering for a Sustainable Future, Springer, Singapore. pp. 43–84.
- IDT, 2023a. PrimerQuest™ Tool, https://sg.idtdna.com/pages/tools/primerquest?, (assessed 15 May 2023).
- IDT, 2023b. OligoAnalyzer™ Tool, https://sg.idtdna.com/pages/tools/oligoanalyzer? (assessed 15 May 2023).
- Jennings E., Thurston T.L.M., Holden D.W. Salmonella SPI-2 Type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host. Microb. 2017;22:217–231. doi: 10.1016/j.chom.2017.07.009. [DOI] [PubMed] [Google Scholar]
- Khan A.K. Pathogenic Bacteria and Medical Treatment. Arch. Clin. Microbiol. 2022;13:200. doi: 10.36648/1989-8436X.22.16.8.200. [DOI] [Google Scholar]
- Liang B., Ji X., Jiang B., Yuan T., Gerile C.L.M., Zhu L., Wang T., Li Y., Liu J., Guo X., Sun Y. Virulence, Antibiotic Resistance, and Phylogenetic Relationships of Aeromonas spp. Carried by Migratory Birds in China. Microorganisms. 2022;11:7. doi: 10.3390/microorganisms11010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Mu K., Zhao Y., Zhang J., Qu Y., Hu D., Jia Y., Dai P., Weng J., Wang D., Yu L. Emergence of blaNDM- 1-Carrying Aeromonas caviae K433 Isolated from patient with community-acquired pneumonia. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.825389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino M.E., Fasolato L., Montemurro F., Rosteghin M., Manfrin A., Patarnello T., Novelli E., Cardazzo B. Determination of microbial diversity of Aeromonas strains on the basis of multilocus sequence typing, phenotype, and presence of putative virulence genes. Appl. Environ. Microbiol. 2011;77:4986–5000. doi: 10.1128/AEM.00708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan E.A., Gupta S.K., Williams L.E., Jové T., Hiott L.M., Woodley T.A., Barrett J.B., Jackson C.R., Wasilenko J.L., Simmons M., Tillman G.E., McClelland M., Frye J.G. Antimicrobial resistance genes, cassettes, and plasmids present in salmonella enterica associated with United States food animals. Front. Microbiol. 2019;10:832. doi: 10.3389/fmicb.2019.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullis K., Faloona F., Scharf S., Saiki R., Horn G., Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1986;51:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- Nallapareddy S.R., Singh K.V., Sillanpää J., Garsin D.A., Höök M., Erlandsen S.L., Murray B.E. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejnickova K., Hola V., Ruzicka F. Catheter-related infections caused by Pseudomonas aeruginosa: virulence factors involved and their relationships. Pathog. Dis. 2014;72:87–94. doi: 10.1111/2049-632X.12188. [DOI] [PubMed] [Google Scholar]
- Olson R.D., Assaf R., Brettin T., Conrad N., Cucinell C., Davis J.J., Dempsey D.M., Dickerman A., Dietrich E.M., Kenyon R.W., Kuscuoglu M., Lefkowitz E.J., Lu J., Machi D., Macken C., Mao C., Niewiadomska A., Nguyen M., Olsen G.J., Overbeek J.C., Parrello B., Parrello V., Porter J.S., Pusch G.D., Shukla M., Singh I., Stewart L., Tan G., Thomas C., VanOeffelen M., Vonstein V., Wallace Z.S., Warren A.S., Wattam A.R., Xia F., Yoo H., Zhang Y., Zmasek C.M., Scheuermann R.H., Stevens R.L. Introducing the bacterial and viral bioinformatics resource centre (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2022;51:D678–D689. doi: 10.1093/nar/gkac1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oogai Y., Matsuo M., Hashimoto M., Kato F., Sugai M., Komatsuzawa H. Expression of virulence factors by Staphylococcus aureus grown in serum. Appl. Environ. Microbiol. 2011;77:8097–8105. doi: 10.1128/AEM.05316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otal I., Martín C., Vincent-Lévy-Frebault V., Thierry D., Gicquel B. Restriction fragment length polymorphism analysis using IS6110 as an epidemiological marker in tuberculosis. J. Clin. Microbiol. 1991;29:1252–1254. doi: 10.1128/jcm.29.6.1252-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Wren B.W., Mungall K., Ketley J.M., Churcher C., Basham D., Chillingworth T., Davies R.M., Feltwell T., Holroyd S., Jagels K., Karlyshev A.V., Moule S., Pallen M.J., Penn C.W., Quail M.A., Rajandream M.A., Rutherford K.M., van Vliet A.H., Whitehead S., Barrell B.G. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Parte A.C., Sardà Carbasse J., Meier-Kolthoff J.P., Reimer L.C., Göker M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020;70:5607–5612. doi: 10.1099/ijsem.0.004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti C.A. Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 2007;44:1108–1114. doi: 10.1086/512818. [DOI] [PubMed] [Google Scholar]
- Piotrowska M., Popowska M. The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann. Microbiol. 2014;64:921–934. [Google Scholar]
- Qadri H., Shah A.H., Alkhanani M., Almilaibary A., Mir M.A. Immunotherapies against human bacterial and fungal infectious diseases: a review. Front. Medicine. 2023;10:1135541. doi: 10.3389/fmed.2023.1135541. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ragupathi N.K., Sethuvel D.P., Anandan S., Murugan D., Asokan K., Neethi Mohan R.G., Vasudevan K., Veeraraghavan B. First hybrid complete genome of Aeromonas veronii reveals chromosome-mediated novel structural variant mcr-3.30 from a human clinical sample. Access Microbiol. 2020;2:acmi000103. doi: 10.1099/acmi.0.000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapaksha P., Elbourne A., Gangadoo S., Brown R., Cozzolino D., Chapman J. A review of methods for the detection of pathogenic microorganisms. Analyst. Royal. Soc. Chem. 2019;144:396–411. doi: 10.1039/c8an01488d. [DOI] [PubMed] [Google Scholar]
- Rekadwad B.N., Li W.J., Rekha P.D. The diversity of unique 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid coding common genes and Universal stress protein in Ectoine TRAP cluster (UspA) in 32 Halomonas species. BMC. Res. Notes. 2021;14:296. doi: 10.1186/s13104-021-05689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco C., Duro I., Di Rosa S., Fagnano M., Fiorentino N., Vetromile A., Adamo P. Composite vs. discrete soil sampling in assessing soil pollution of agricultural sites affected by solid waste disposal. J. Geochem. Expl. 2016;170:30–38. doi: 10.1016/j.gexplo.2016.08.004. [DOI] [Google Scholar]
- San Millán R.M., Martínez-Ballesteros I., Rementeria A., Garaizar J., Bikandi J. Online exercise for the design and simulation of PCR and PCR-RFLP experiments. BMC. Res. Notes. 2013;6:513. doi: 10.1186/1756-0500-6-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwecińska L. Antimicrobials and antibiotic-resistant bacteria: a risk to the environment and to public health. Water. 2020;12:3313. [Google Scholar]
- van Wyk S., Harrison C.H., Wingfield B.D., De Vos L., van der Merwe N.A., Steenkamp E.T. The RIPper, a web-based tool for genome-wide quantification of Repeat-Induced Point (RIP) mutations. PeerJ. 2019;7:e7447. doi: 10.7717/peerj.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola C.L. The antibiotic resistance crisis: part 1: causes and threats. P & t. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hou N., Rasooly R., Gu Y., He X. Prevalence and genetic analysis of chromosomal mcr-3/7 in Aeromonas from U.S. animal-derived samples. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.667406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.Y., Liu X.F., Gao X.Z. Helicobacter pylori virulence factors in development of gastric carcinoma. Future Microbiol. 2015;10:1505–1516. doi: 10.2217/fmb.15.72. [DOI] [PubMed] [Google Scholar]
- Wu C.J., Wang H.C., Chen P.L., Chang M.C., Sunny Sun H., Chou P.H., Ko W.C. AQU-1, a chromosomal class C β-lactamase, among clinical Aeromonas dhakensis isolates: distribution and clinical significance. Int. J. Antimicrob. Agents. 2013;42:456–461. doi: 10.1016/j.ijantimicag.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Wu T., Zhao Z., Zhang L., Ma H., Lu K., Ren W., Liu Z., Chang H., Bei W., Qiu Y., Chen H. Trigger factor of Streptococcus suis is involved in stress tolerance and virulence. Microb. Pathog. 2011;51:69–76. doi: 10.1016/j.micpath.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Xiang S.H., Hobbs M., Reeves P.R. Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J. Bacteriol. 1994;176:4357–4365. doi: 10.1128/jb.176.14.4357-4365.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 2010;7:629–641. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Zhang M., Xu Z., He Y., Zhao X., Cheng H., Chen X., Xu J., Ding Z. The screening of the protective antigens of Aeromonas hydrophila using the reverse vaccinology approach: potential candidates for subunit vaccine development. Vaccines. 2023;11:1266. doi: 10.3390/vaccines11071266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




