Three fundamental issues in postgenomic biology are (i) how the amino acid sequence of a given human protein predicates its structure, function, and regulation; (ii) how a protein is compared to its paralogs, as well as to its orthologs and other homologous proteins in model organisms; and (iii) how related studies contribute to the understanding of human pathology and the development of efficacious diagnostic and therapeutic means. These fascinating issues have inspired us to conduct a comprehensive analysis of information available on class II histone deacetylases (HDACs). In what follows, we will start with a brief description of different classes of HDACs and then compare class II HDACs from yeast and higher organisms in terms of domain organization, function, and regulation. We will also discuss evidence that links class II human HDACs to cardiomyopathy, osteodystrophy, neurodegenerative disorders, and cancer and will propose that, in addition to inhibitors, activators of these HDACs are of potential therapeutic value.
DIFFERENT CLASSES OF HDACs
Lysine acetylation refers to the transfer of an acetyl moiety from acetyl-coenzyme A (CoA) to the ɛ-amino group of a lysine residue. This covalent modification was discovered with histones in the 1960s (30, 137), but its importance only began to be widely considered in the mid-1990s. Acetyllysine is now known to be present in at least 80 other proteins, including ∼40 sequence-specific transcription factors, ∼10 transcriptional coregulators, several viral proteins, α-tubulin, acetyl-CoA synthase, Ku70, and Hsp90 (24, 82, 127, 149). Lysine acetylation is reversible and is controlled by the opposing actions of acetyltransferase and deacetylase in vivo. Since histones were thought to be the major cellular proteins modified by lysine acetylation, most lysine acetyltransferases and deacetylases were initially identified as histone acetyltransferases and HDACs. It is now clear, however, that some of these enzymes also act on other proteins (24, 82, 127, 149).
The first lysine deacetylases, mammalian HDAC1 and yeast Hda1, were identified in 1996 (120, 130). Different proteins were subsequently shown to possess HDAC activity (12, 27, 53, 79, 109, 136). According to phylogenetic analyses as well as sequence homology to yeast Rpd3 (reduced potassium dependency 3), Hda1, and Sir2 (silent information regulator 2), HDACs are grouped into different classes (49, 53, 79, 112, 136). There are 10 known HDACs in Saccharomyces cerevisiae, with Rpd3, Hos1 (Hda1 similar 1), and Hos2 forming class I. Hda1 was considered to be the sole class II member, but a recent phylogenetic analysis suggested that Hos3 is a distant member of this class (49, 120). Sir2 and four Hst (homolog of Sir2, also known as sirtuin) proteins constitute class III. HDACs from other organisms have been similarly grouped. For example, 18 known human HDACs are divided into four classes: I (HDAC1, -2, -3, and -8; homologous to Rpd3), II (HDAC4, -5, -6, -7, -9, and -10; related to Hda1), III (Sirt1, -2, -3, -4, -5, -6, and -7; similar to Sir2) and IV (HDAC11) (12, 49, 53, 109, 136, 150). Class I HDACs display some sequence homology to members of classes II and IV but not to those of class III. In agreement with this, class I, II, and IV HDACs are zinc-dependent enzymes, whereas the deacetylase activity of class III members is NAD+ dependent.
DOMAIN ORGANIZATION OF CLASS II HDACs
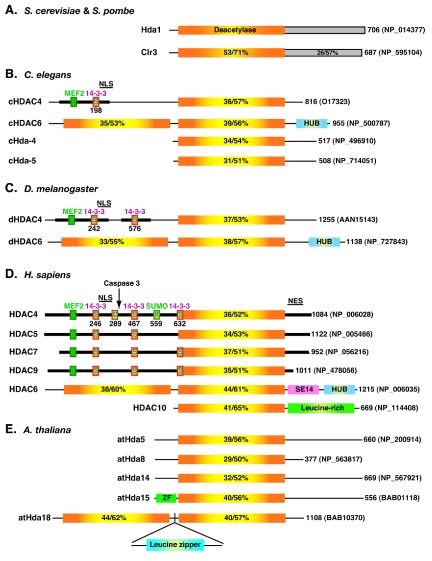
As illustrated in Fig. 1, Hda1-like proteins exist in various eukaryotic organisms. These enzymes possess known and putative deacetylase domains with significant sequence homology to that of Hda1 (identity, 29 to 53%; similarity, 51 to 71%). Hda1 is highly conserved among different yeast species. For example, both the N-terminal deacetylase and the C-terminal domains of Clr3 are highly similar to those of Hda1 (Fig. 1A). In contrast, only the deacetylase domain is conserved in metazoans (Fig. 1A to D). In mammals, there are six Hda1-like proteins: HDAC4 (41, 52, 104, 139), HDAC5 (52, 135), HDAC6 (52, 135), HDAC7 (39, 74), HDAC9 (116, 167), and HDAC10 (38, 56, 75, 132). They are further divided into two subgroups: HDAC4, -5, -7, and -9 form class IIa, whereas the remaining two constitute class IIb (11, 49, 136). Besides their catalytic domains, class IIa mammalian members have sequence similarity in the extended long N-terminal domains and the C-terminal tails. The N-terminal extensions possess conserved motifs important for the function and regulation of class IIa members (Fig. 1D). Unlike mammals, lower organisms, such as Caenorhabditis elegans (Fig. 1B) and Drosophila melanogaster (Fig. 1C), possess only one class IIa member (23, 156). The MEF2 (myocyte enhancer factor 2)-binding site is conserved among the class IIa members from C. elegans and Drosophila (Fig. 1B and C and 2A). C. elegans HDAC4 (cHDAC4) possesses one putative 14-3-3 binding site, whereas D. melanogaster HDAC4 (dHDAC4) contains two (Fig. 1B and C and 2B). Compared to cHDAC4, dHDAC4 displays higher sequence similarity to the nuclear localization signal (NLS) mapped on class IIa human HDACs (Fig. 1D and 2B) (99, 141).
FIG. 1.
Domain organization of Hda1 and related proteins from fission yeast (A), worms (B), flies (C), humans (D), and plants (E). For each protein, only one isoform (usually the full-length isoform) is drawn, with its total number of residues and database accession number shown at the right. The deacetylase domain of Hda1 is depicted with a rectangle containing a yellow center flanked by orange areas. Similar rectangles are used to illustrate the deacetylase domains in Hda1-like proteins, with respective sequence identity/similarity to that of Hda1 listed. The C-terminal domains (shaded rectangles) of Hda1 and Clr3 are homologous. Dark bars, similar N-terminal domains and C-terminal tails of class IIa metazoan HDACs; small green boxes, MEF2-binding motifs; boxes labeled S (for serine), 14-3-3 binding motifs; boxes labeled D (for Asp289) and K (for Lys559), caspase cleavage and sumoylation sites on HDAC4, respectively. NES, nuclear export signal; SE14, SerGlu-containing tetradecapeptide repeats; ZF, zinc finger.
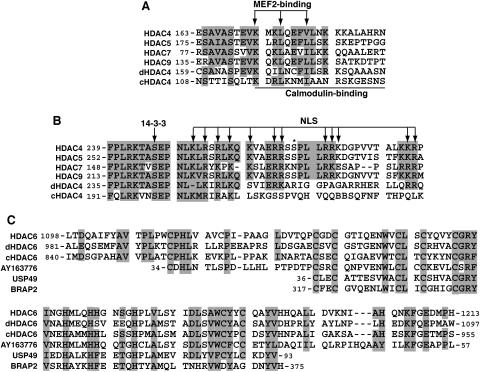
FIG. 2.
(A and B) Sequence alignment of the MEF2-binding motif (A) and the NLS (B) of class IIa HDACs. Residues invariant and highly conserved among at least four sequences are shaded. Arrows, residues known to be essential for the NLS function, 14-3-3 association, or MEF2 binding; solid line, region critical for Ca2+/calmodulin binding; asterisk, potential Dyrk1B phosphorylation site. (C) Sequence similarity between the HUB finger of HDAC6 and motifs in other proteins. Identical and highly conserved residues are shaded. Only the central region of the HUB finger is similar to USP49 and BRAP2. Residues invariant and highly conserved among at least 75% of the sequences are shaded.
Different from class IIa members, HDAC6 possesses two deacetylase domains and a zinc finger motif (Fig. 1) (52, 135). As shown in Fig. 2C, the central part of this motif is similar to regions found in BRAP2 (BRCA1-associated protein 2) and several ubiquitin-specific proteases (USPs), so it has been referred to as DAUP (deacetylase-ubiquitin-specific protease) domain (3), HUB (HDAC6, USP3, and BRCA1-related) finger (11), and Zn-UBP (ubiquitin carboxyl-terminal hydrolase-like zinc finger) (121). Due to its ubiquitin-binding ability, this motif is also known as PAZ (polyubiquitin-associated zinc finger) (64) and BUZ (bound to ubiquitin zinc finger) (78). The domain organization of HDAC6 is conserved in orthologs identified in C. elegans (Fig. 1B) and Drosophila (Fig. 1C) (5). In both organisms, there may be isoforms lacking the HUB finger (5) (NP_500788). There is also a putative plant protein homologous to such isoforms (Fig. 1E, atHda18). This putative homolog also carries a leucine zipper motif within a coiled-coil domain and is an isoform encoded by an uncharacterized sequence (112) (NP_200915). Interestingly, another uncharacterized plant sequence (AY163776) encodes a domain highly homologous to the HUB finger (Fig. 2C). Human HDAC6 possesses a unique SE14-repeat domain (Fig. 1D), which is not intact or present in published sequences of mouse, rat, Drosophila, C. elegans, and Arabidopsis thaliana homologs (10). This domain contains eight consecutive tetradecapeptide repeats and is important for the stable cytoplasmic retention of human HDAC6 (10, 11).
The N-terminal half of HDAC10 (Fig. 1D) is more similar to the first catalytic domain of HDAC6 than to the second (38, 56, 75, 132). The C-terminal half of HDAC10 is leucine rich and shows limited sequence similarity to the second deacetylase domain of HDAC6 (56), but the function remains elusive. The Drosophila genome does not encode an HDAC10 homolog, whereas there are two uncharacterized C. elegans proteins containing C-terminal domains with some similarity to that of HDAC10. In A. thaliana, the atHda5 gene and three uncharacterized sequences encode putative deacetylase domains with high homology to that of HDAC10 (Fig. 1E). The C-terminal part of atHda5 displays limited sequence similarity to that of yeast Hda1 (118). Therefore, in different organisms, unique domains and motifs are linked to Hda1-like domains. Such diverse domain organizations constitute the structural basis for the distinct function and regulation of different class II HDACs (see below).
FUNCTION AND REGULATION OF YEAST CLASS II DEACETYLASES
Deacetylase activity of Hda1 and Hos3.
Hda1 self-associates and interacts with Hda2 and Hda3, two homologous coiled-coil subunits, to form a tetrameric complex (146). Unlike Hda1 alone, this complex is active, so Hda2 and Hda3 are essential for deacetylase activity. Hda1 was reported to mediate the deacetylation of histones H3 and H2B, but not H4 and H2A, in vivo (147). No Hda2 and Hda3 homologs have been found in multicellular organisms, so this complex may be unique to yeast. Like Hda1, Hos3 forms a multiprotein complex in vivo (16). Interestingly, Hos3 itself is active, but this complex is inactive. Therefore, complex formation constitutes an important means for regulating the deacetylase activity of Hda1 and Hos3.
Gene-specific deacetylation by Hda1.
Since chromatin is a polymer, the functional consequence of a deacetylation event is dependent on where it occurs. Hda1 can be recruited to promoters for gene-specific action. DNA-binding transcription factors recruit Tup1 along with Hda1 to deacetylate histones H3 and H2B at the promoters of genes required for mating, oxygen and glucose utilization, DNA damage and osmotic-stress responses, and other inducible cellular programs (124, 147). The deacetylation may synergize with other mechanisms to achieve efficient repression (47, 144, 164).
The DNA-binding transcription factor Sfl1 (suppressor for flocculation 1) recruits Hda1 to the promoter of Flo11 (flocculation 11), a cell surface protein required for adherence, sliding motility, and pseudohyphal development (59). The Flo11 locus is silent in yeast form cells, but under nitrogen starvation, it becomes active for expression and stimulates the formation of filamentous cells. Nitrogen starvation activates protein kinase A and may thus block Hda1 recruitment by Sfl1, suggesting that Hda1 is a signal-responsive transcriptional corepressor. Collaborating with sirtuins, Hda1 regulates yeast cell surface variation, adhesion, and invasiveness (59). A similar scheme may also operate in the opportunistic yeast pathogen Candida albicans, where Hda1 controls colony-type switching (81).
Domain-specific and genome-wide deacetylation by Hda1 and Clr3.
Microarray analyses revealed that Hda1 is not limited to gene-specific deacetylation (138). First, it specifically associates with Hda1-affected subtelomeric (HAST) domains (119). These domains are contiguous chromosomal regions adjacent to telomeric heterochromatin and contain gene clusters required for gluocogenesis and growth under adverse conditions, such as osmotic shock, starvation, and low oxygen, suggesting that Hda1 coordinates the regulation of gene activities within the clusters. Hda1 may be targeted to HAST domains through Tup1, a corepressor that is known to inhibit gene expression at the clusters (84). Second, Hda1 preferentially binds to coding regions that are hypomethylated at Lys4 of histone H3 (9). Such an association may keep the coding regions in hypoacetylated states for transcriptional inhibition. Third, Hda1 carries out low-level deacetylation throughout the genome, and such a global action may be important for decreasing basal transcription (138). Finally, Hda1 is important for controlling mating-type switching in different yeast species (81, 126). Deletion of Clr3 in Schizosaccharomyces pombe alleviates the recombination block at the mat regions (50). In Clr3Δ strains, histone H3 at the mat regions is hyperacetylated at Lys9 and Lys14. Deacetylation of histone H3 at Lys9 may clear the way for methylation and stimulate the association with an HP1-like protein to promote the assembly of silent chromatin (107). Emerging evidence suggests that histone deacetylation mediated by Neurospora crassa Hda1 stimulates DNA methylation, so the deacetylation may synergize with chromatin methylation to stimulate the formation of inactive chromatin. Therefore, through region-specific and global deacetylation, Hda1 promotes transcriptional repression and silencing to regulate the cellular programs required for growth under adverse conditions. This notion also provides a conceptually informative framework for analyzing the function and regulation of Hda1 homologs in higher eukaryotes.
FUNCTION OF CLASS IIA METAZOAN HDACs
Like Hda1, class IIa mammalian HDACs mainly function as transcriptional corepressors (Table 1). In addition to the deacetylase domains (Fig. 1), they possess other repression domains (74, 88, 125, 139, 168). Unlike Hda1, class IIa HDACs do not interact with the Tup1-like proteins Groucho and TLEs (A. H. Wang, S. Stifani, and X.-J. Yang, unpublished observation). Instead, they act through other transcriptional corepressors or bind directly to sequence-specific transcription factors, such as MEF2, NF-AT3c (nuclear factor of activated T cells 3c), and Runx3 (Table 1).
TABLE 1.
Interaction partners of class II HDACs
| Deacetylase | Partner | Function | Reference(s) |
|---|---|---|---|
| Yeast Hda1 | Hda2 | Subunit of the Hda1 complex | 146 |
| Hda3 | Subunit of the Hda1 complex | 146 | |
| TUP1 | Repressor of genes expressed for mating and glucose and oxygen utilization, as well as DNA damage and osmotic stress responses | 147 | |
| Class IIa HDACs | 14-3-3 | Phosphoserine/threonine binding proteins | 54, 76, 100, 140, 161 |
| Calmodulin | Calcium-binding protein | 7, 154 | |
| MEF2 | Regulation of cell differentiation, death, and survival | 76, 93, 104, 135, 139 | |
| SRF | Serum response factor controlling cell growth and differentiation | 29 | |
| Myocardin | Transcriptional coactivator essential for smooth muscle differentiation | 15 | |
| NF-AT3c | Transcription factor required for cell differentiation and immune response | 28 | |
| MRJ | DnaJ-like chaperone protein and corepressor of NF-ATs | 28 | |
| Runx2 and -3 | Runt domain transcription factors important for organogenesis | 72, 133 | |
| TEL (ETV6) | Ets-related transcription factor and leukemic fusion partner of Runx1 | 115 | |
| BCL6 | POZ finger transcription factor important for cell survival and differentiation | 87 | |
| BCoR | BCL6 corepressor important for embryogenesis | 69 | |
| PLZF | POZ finger transcription factor involved in leukemogenesis | 21, 87 | |
| HIF-1α | HIF controlling gene expression during oxygen tension | 77 | |
| CREB2 | Transcription factor regulating gene expression during synaptic growth | 55 | |
| GATA-1 | Transcription factor crucial for erythroid differentiation | 143 | |
| TR2 | Nuclear orphan receptor | 42 | |
| N-CoR | Nuclear receptor corepressor | 66, 74 | |
| SMRT | Nuclear receptor corepressor, highly similar to N-CoR | 66, 74 | |
| ARR19 | Androgen receptor corepressor | 71 | |
| Rea | Repressor of estrogen receptor activity | 85 | |
| RIP140 | Transcriptional corepressor for nuclear receptors | 17 | |
| ANCO-1 | Ankyrin repeat-containing cofactor for p160 nuclear receptor coactivators | 157 | |
| CtBP | E1A C-terminal binding protein, a transcriptional corepressor | 159 | |
| IκBα | Negative regulator of NF-κB | 1 | |
| HP1 | Heterochromatin protein 1, associated with the methyltransferase SUV39H1 | 160 | |
| TIP60 | Histone acetyltransferase | 86 | |
| ICP0 | Immediate-early gene product of the herpes simplex virus | 92 | |
| 53BP1 | p53 binding protein that regulates cell cycling in response to DNA damage | 73 | |
| ETR-A | G protein-coupled endothelin receptor | 86 | |
| HDAC6 | Tubulin | Deacetylating microtubules and increasing cell motility | 57, 68, 97, 108, 111, 163 |
| p150glued/dynactin | Adaptor protein for the molecular motors dyneins and kinesins | 68, 78 | |
| Ubiquitin | Signal for cellular processes, such as protein degradation and endocytosis | 64, 121 | |
| p97/VCP/Cdc48p | AAA ATPase in endoplasmic reticulum-associated proteosomal degradation | 121 | |
| PLAP | Phospholipase A2 activation protein controlling prostaglandin levels and phospholipase activity | 121 | |
| NF-κB (p50/65) | Transcription factor in inflammation and cell growth control | 162 | |
| Runx2 | Transcription factor essential for skeletal development | 145 | |
| LCoR | Transcriptional corepressor for agonist-bound nuclear receptors | 37 | |
| Sumoylated p300 | Transcriptional coregulator with lysine acetyltransferase activity | 45 | |
| PP1 | Phosphatase | 14 | |
| HDAC10 | LCoR | Transcriptional corepressor for ligand-bound nuclear receptors | 37 |
| PP1 | Phosphatase | 14 |
As signal-responsive corepressors of MEF2.
While a single MEF2 protein exists in C. elegans or Drosophila, there are four isoforms in vertebrates, MEF2A, -B, -C, and -D (62, 102). Although they were initially identified as major transcriptional activators for the expression of muscle-specific genes, MEF2s also regulate other cellular programs, including neuronal survival, T-cell apoptosis, and growth factor responsiveness (62, 102). Aberrant MEF2 proteins have been implicated in coronary heart diseases and T-cell lymphoma (142, 155). Since its initial discovery in 1999, the link of class IIa HDACs to MEF2s has been extensively characterized (34, 88, 93, 94, 104, 125, 139, 161). The MEF2-binding site was mapped to a small motif conserved among all four IIa members from mammals (Fig. 1 and 2B) (93, 125, 139). Structural analysis of this motif has revealed that the binding mode resembles that used by the corepressor Cabin1 (60, 61). Importantly, the essential residues are also conserved in HDAC4 proteins from C. elegans and Drosophila (23, 156). For HDAC4 and HDAC5, it has been shown that this motif overlaps with a Ca2+/calmodulin-binding site (Fig. 2B) (7, 154), suggesting that Ca2+/calmodulin binding prevents class IIa HDACs from repressing MEF2-dependent transcription. Ca2+/calmodulin also activates Ca2+/calmodulin-dependent kinases (CaMKs) to promote the nuclear export of these HDACs by phosphorylating them for 14-3-3 binding (see below). Therefore, in a signal-responsive manner, MEF2s recruit class IIa HDACs to repress transcription (Fig. 3). This notion is also consistent with different lines of evidence: (i) exogenous expression of HDAC4, -5, and -9 regulates myoblast differentiation (94, 99, 105, 166); (ii) disruption of the mouse HDAC5 and HDAC9 genes causes cardiac hypertrophy (19, 158); (iii) class IIa HDACs interact with MEF2C and MEF2D to regulate T-cell apoptosis and neuronal survival (22, 32); and (iv) class IIa HDACs promote the sumoylation and perhaps phosphorylation of MEF2C and MEF2D to potentiate transcriptional repression (48). These lines of evidence also show that association with class IIa HDACs converts MEF2s from transcriptional activators into repressors.
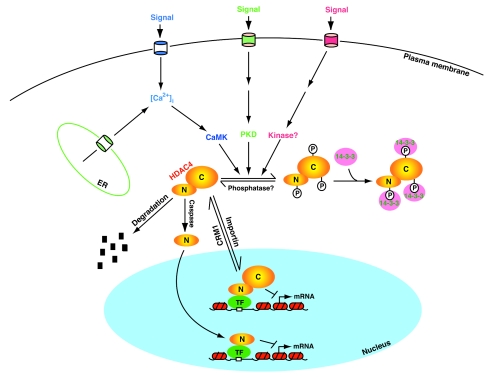
FIG. 3.
Cartoon depicting mechanisms involved in the regulation of HDAC4. It is actively imported and exported; the relative trafficking rate dictates its subcellular distribution. Cell signaling activates CaMK, PKD, and perhaps other kinases, thereby leading to site-specific phosphorylation of HDAC4 and association with 14-3-3 proteins. 14-3-3 binding then shifts the trafficking equilibrium of HDAC4 toward cytoplasmic accumulation. Unknown phosphatases dephosphorylate HDAC4 and dissociate it from 14-3-3 proteins for translocation to the nucleus, where it binds to sequence-specific transcription factors (TF) to repress transcription. The binding can be direct or mediated by a corepressor. Caspase 3 cleaves HDAC4 to generate the N-terminal (N) and C-terminal (C) fragments (Fig. 1D), with the former translocating to the nucleus as a transcriptional corepressor. HDAC4 may also be subject to proteosomal degradation. Dyrk1B and related kinases may phosphorylate HDAC4 and inhibit its nuclear import. Except for a few details, this model can be extended to other class IIa HDACs.
As corepressors of NF-AT3c.
Ca2+/calmodulin also activates the phosphatase calcineurin to reverse inhibitory phosphorylation of NF-ATs. This pathway plays an important role in cardiac hypertrophy and T-cell activation (26). In cardiac muscle cells, NF-AT3c collaborates with transcription factors such as MEF2 and GATA4 to activate transcription. A recent study suggested that HDAC4 interacts with NF-AT3c and MRJ (mammalian relative of DnaJ) to repress transcription (28), suggesting that HDAC4 and other class IIa HDACs serve as corepressors of NF-ATs. It is presently unclear whether this occurs in a signal-dependent manner.
As corepressors of Runt domain transcription factors.
Mammalian Runx transcription factors share DNA-binding domains with Drosophila Runt, a transcription factor important for segmentation (70). Recent studies indicated that HDAC5 binds to and deacetylates Runx3 (72). Related to this, both Runx1 and Runx2 interact with class IIa HDACs (133) (M. Xu and X.-J. Yang, unpublished results). Moreover, dHDAC4 displays segment-specific expression during fly development, and its mutation causes segmental defects (156). Importantly, disruption of the mouse HDAC4 gene causes abnormal skeletal development due to chondrocyte hypertrophy (133). Consistent with this, Runx2 plays an essential role in bone development (36). More interestingly, the human HDAC4 gene is located at chromosome 2q37 (Table 2), a region whose abnormalities have been linked to Albright's hereditary osteodystrophy-like syndrome (20). The other two Runx proteins, Runx1 and Runx3, are important players in development and tumorigenesis (70). Therefore, class IIa HDACs function as corepressors of Runt domain transcription factors. It remains to be investigated whether this is signal dependent.
TABLE 2.
Class II human HDACs and potentially related diseasesa
| HDAC | Chromosome | OMIMb no. | Human disease | Genetic analysis in model organisms |
|---|---|---|---|---|
| HDAC4 | 2q37.2 | 605314 | Osteodystrophy | Segmentational defect in mutant flies; small size, chondrocyte hypertrophy, and exencephaly in null mice |
| HDAC5 | 17q21 | 605315 | Cancer | Cardiac hypertrophy in mouse knockouts |
| HDAC6 | Xp11.22 | 3000272 | Neurodegenerative disorders | Required for formation of aggresomes and immune synapses in cultured cells |
| HDAC7 | 12q13.1 | 606542 | Cardiovascular defects in null mice | |
| HDAC9 | 7p12.1 | 606543 | Peter’s anomaly | Cardiac hypertrophy in mouse knockouts |
| HDAC10 | 22q13.3 | 608544 | Tumor metastasis | Not available |
See text for references.
OMIM, Online Mendelian Inheritance in Man database.
Atypical roles of class IIa HDACs.
In addition to its corepressor role, HDAC7 was recently shown to bind hypoxia-inducible factor 1α (HIF-1α) and activate transcription (77). Moreover, HDAC7 localizes to mitochondria and may regulate the initiation of apoptosis (4). Related to this, HDAC5 interacts with Rea (or Prohibitin), which also associates with mitochondria (85, 128). HDAC4 binds to 53BP1 and may regulate DNA repair (73). An intriguing question to be addressed is whether class IIa HDACs play other noncanonical roles.
REGULATION OF CLASS IIA METAZOAN HDACs
Expression of class IIa HDACs.
Intimately linked to function, HDAC regulation is another important issue (122). The expression of class IIa mammalian HDACs is controlled in a tissue-specific manner. Most of them are highly expressed in skeletal muscle, heart, brain, and thymus (136), which is consistent with their important roles in cell differentiation. Alternative splicing represents an important regulatory mechanism. Multiple isoforms are generated from the HDAC9 gene. One isoform, known as MITR and HDRP, lacks the C-terminal domain and localizes to the nucleus (104, 125, 161, 168), whereas another does not contain the NLS (Fig. 1D and 2B) and is thus constitutively cytoplasmic (115). Alternatively spliced isoforms have also been found with other class IIa members, e.g., dHDAC4 (156).
Deacetylase activity of class IIa HDACs.
As discussed above, Hda2 and Hda3 are required for the deacetylase activity of Hda1 (146). The activity of recombinant HDAC4, expressed in and purified from Sf9 insect cells, is inversely proportional to the expression level (139), so an unidentified factor may be involved in the regulation. Related to this, the deacetylase activity of HDAC4 and HDAC7 was reported to be dependent on associated HDAC3 complexes (39, 40), and sumoylation of HDAC4 was found to be required for its deacetylase activity (80). HDAC4 was shown to coimmunoprecipitate multiple proteins (41), but it is still unclear whether HDAC4 and its homologs interact with other subunits to form stoichiometric multiprotein complexes. If so, identification of such subunits would definitively address the important question of how the deacetylase activity of class IIa HDACs is really controlled in vivo.
Subcellular compartmentalization of class IIa HDACs.
Class IIa mammalian HDACs are present in both the nucleus and the cytoplasm (32, 54, 76, 99, 104, 115, 140). Leptomycin B treatment has firmly established that they are subject to CRM1-dependent nuclear export (41, 54, 76, 99, 104, 140). Consistent with this, these HDACs possess intrinsic nuclear import and export signals for nucleocytoplasmic trafficking (Fig. 1) (99, 101, 141). These signals generate a dynamic trafficking equilibrium, and any alteration of this equilibrium may affect the subcellular distribution (Fig. 3). Among known binding partners, MEF2 and HIF-1α (Table 1) have been shown to promote the nuclear localization of class IIa HDACs (13, 18, 77, 104, 141). By contrast, 14-3-3 proteins stimulate cytoplasmic retention (54, 76, 100, 140). Dependent on site-specific phosphorylation, 14-3-3 proteins bind to these HDACs and exert effects through multiple mechanisms (Fig. 3). First, one major 14-3-3 binding site on class IIa mammalian HDACs overlaps with the NLS (Fig. 1D and 2B), so 14-3-3 binding impedes the access of importin α/β to this NLS (54). This 14-3-3 binding site is conserved in cHDAC4 and dHDAC4 (Fig. 2B) (23, 156), so such a mechanism may also operate in lower metazoans, such as C. elegans and Drosophila. Second, 14-3-3 binding may provide a nuclear export signal in trans to promote the nuclear export of class IIa HDACs. Third, 14-3-3 binding affects the subnuclear localization of class IIa HDACs. For example, upon binding to 14-3-3 proteins, MITR disperses from nuclear dots and yields uniform nuclear redistribution (161). Finally, 14-3-3 association was recently shown to protect HDAC7 from proteosomal degradation (89), which may contribute to its cytoplasmic accumulation.
CaMKs I, II, and IV phosphorylate class IIa mammalian HDACs at the 14-3-3 binding sites (Fig. 1D) and promote nuclear export (22, 29, 76, 90, 99). Compelling evidence suggested that unknown kinases distinct from CaMKs could also do so (158, 166). Indeed, two recent studies demonstrated that protein kinase D1 (PKD1) phosphorylates HDAC5 and HDAC7 at their 14-3-3 binding sites (114, 133a). Since a mutant HDAC4 deficient in 14-3-3 binding is still heavily phosphorylated (11), other kinases may act on distinct sites. Some of these phosphorylation events may promote the nuclear localization of class IIa HDACs. Related to this, oncogenic Ras promotes the nuclear localization of HDAC4 by stimulating its phosphorylation by extracellular signal-regulated kinase 1/2 (169). Subcellular localization of class IIa HDACs is known to be affected by serum starvation (34), membrane depolarization (22, 90), hypoxia (77), and other stimuli (19, 58, 63, 73). Despite their shared regulatory schemes, class IIa members display distinct subcellular localizations (11). As recently reported (77), hypoxia induces the nuclear import of HDAC7, but not HDAC4 or HDAC5. Therefore, distinct kinases may differentially regulate the subcellular localization of class IIa members. Related to this notion, Dyrk1B phosphorylates HDAC5 at a serine residue that is shared by HDAC4 and HDAC9, but not by HDAC7 (Fig. 2B) (31).
Besides phosphorylation, HDAC4, -5, and -9 are subject to ubiquitination and sumoylation (64, 80, 115, 129). These two modifications do not affect the subcellular localization. Caspase 3 cleaves HDAC4 and promotes the nuclear localization of the N-terminal fragment (Fig. 3) (91, 113). This may be specific to HDAC4. Moreover, HDAC7 but not other class IIa members localizes to mitochondria (4), further supporting the notion that class IIa HDACs are also subject to differential regulation. How their sequence divergence (Fig. 1) dictates differential regulation is an important issue deserving more research attention.
FUNCTION AND REGULATION OF CLASS IIB METAZOAN HDACs
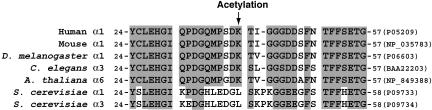
Dependent on its second deacetylase domain, HDAC6 deacetylates α-tubulin (57, 68, 97, 108, 111, 163). The deacetylase domains are highly conserved in HDAC6 proteins from worms and flies, so this mechanism may also operate in other metazoans (Fig. 1). Consistent with this notion, Lys40, the known acetylation site, is found in some α-tubulins from multicellular organisms but not in the only two found in the budding yeast (Fig. 4). Tubulin deacetylation by HDAC6 stimulates cell motility (57, 68). A recent study has established that a subset of acetylated microtubules is necessary for proper organization of the immune synapse, a specialized cell-cell junction formed by antigen-presenting cells and T lymphocytes (123). HDAC6 determines the acetylation kinetics in the immune synapse. In addition to its deacetylase domains, HDAC6 possesses a HUB finger (Fig. 1). For mammalian HDAC6 proteins, it has been shown that this finger binds to ubiquitin (64, 121) and possesses E3 ligase activity (83), suggesting involvement in ubiquitin-dependent pathways. Of relevance, HDAC6 tightly associates with p97, an ATPase known to be involved in endoplasmic reticulum-associated proteosomal degradation (121). HDAC6 directly regulates aggresome formation and cellular management of misfolded proteins (78). Both its deacetylase and ubiquitin-binding activities are required for these processes. Importantly, HDAC6 is present in Lewy bodies associated with neurodegenerative disorders, such as Parkinson's disease (78). Therefore, HDAC6 regulates various processes in the cytoplasm.
FIG. 4.
Sequence comparison of different α-tubulins. The peptides are flanked with the starting and ending positions, and residues invariant among at least five sequences are shaded. The sequence accession numbers are shown in parentheses at the right. Arrow, acetylation site. The acetylatable lysine residue is replaced by leucine in the only two α-tubulins found in budding yeast.
Several lines of evidence suggest that HDAC6 also plays a role in the nucleus. It interacts with several nuclear proteins, including HDAC11 (44), sumoylated p300 (45), transcriptional corepressors such as ETO2 and L-CoR (2, 37), and sequence-specific transcription factors such as NF-kB (162) and Runx2 (145), so the subcellular localization of HDAC6 may be regulated. Indeed, mouse HDAC6 possesses intrinsic nuclear import and export signals for active nucleocytoplasmic trafficking (10, 134). Although early reports showed that HDAC6 is cytoplasmic in Drosophila and mammalian cells (5, 134), a recent study indicated that HDAC6 is nuclear in normal but not transformed epithelial cells (153). How the subcellular localization is regulated is an important issue awaiting further investigation.
HDAC10 is widely expressed (38, 56, 75, 132). It displays weak transcriptional repression potential and localizes to both the nucleus and the cytoplasm. Interestingly, the cytoplasmic localization is resistant to leptomycin B treatment. A recent study showed that the expression level of HDAC10 is reduced in tumors and may serve as a good indicator of poor prognosis in cancer patients (110). HDAC10 is the class II member last identified and least characterized, so further exploration is needed to elucidate its function and regulation.
CLINICAL IMPLICATION
From the above, it is clear that class II human HDACs are molecular targets of clinical relevance (Table 2). These deacetylases play an important role in regulating muscle differentiation, and inactivation of the mouse HDAC5 and HDAC9 genes leads to cardiac hypertrophy (19, 98, 158). Moreover, mice lacking HDAC7 die during midgestation from cardiovascular defects (133). In agreement with these findings, class IIa HDACs interact with MEF2 and NF-AT3c (Table 1), both of which play important roles in regulating cardiac muscle growth. While the human MEF2A gene is mutated in coronary heart diseases (142), the MEF2D gene is rearranged in T-cell lymphoma (155). HDAC5 also targets myocardin, a transcriptional coactivator important for smooth muscle differentiation (15). Abnormal smooth muscle differentiation is linked to atherosclerosis, hypertension, and asthma. In addition, class IIa HDACs interact with Runx transcription factors (Table 1), which play causal roles in tumorigenesis (36, 70). Related to this, HDAC4-null mice display skeletal defects (133). Furthermore, HDAC4 acts downstream from the p53 tumor suppressor (8), HDAC5 induces apoptosis (67), and HDAC9 expression is upregulated in senescent fibroblasts (96).
Like class IIa members, HDAC6 plays an important role in biological processes of clinical relevance. It is required for aggresome formation and is present in Lewy bodies associated with neurodegenerative disorders, such as Parkinson's disease and dementia (78). Moreover, expression levels of class II HDACs decrease as tumors progress, with HDAC6 and HDAC10 being potential prognosis indicators (110, 165). Therefore, available studies indicate that inhibition of class II human HDACs could be detrimental. In light of this, we propose that their activators are of potential therapeutic value.
Since trichostatin A was identified in 1990 as a specific HDAC inhibitor with antitumor activity (151), HDAC inhibitors have emerged as novel agents with potential therapeutic value for the treatment of major human diseases, such as cancer and neurodegenerative disorders (95, 103, 117, 152). One caveat of these agents is toxicity. There are different aspects to this problem: (i) relative sensitivities to normal and abnormal cells, (ii) effects on pathways that are not related to HDACs, and (iii) selectivity of individual HDACs. Most known HDAC inhibitors target class I and II members rather nonselectively, so inhibition of class II HDACs could contribute to toxicity. Thus, inhibitors specific to class I HDACs may be more efficacious. Some progress has been made in developing isoform-specific drugs. Depsipeptide (or FK228) shows some preference to class I over class II HDACs (43), and tubacin specifically targets HDAC6 (57). Equipped with real and virtual screening technologies (6, 51, 131), rational design is driving the exploration of HDAC inhibitors in this promising direction (103).
The drugs theophylline and valproic acid were recently found to target HDACs (25, 46, 116). While the former has been used against asthma for over 70 years, the latter is an established drug for epilepsy. Valproic acid targets members of class I and IIa and was recently tested for cancer treatment (117). In addition, some known HDAC inhibitors are present in diets. For example, two inhibitors of class I and II HDACs, diallyl disulfide and sulforaphane, are present in garlic and broccoli, respectively (35, 106). Related to this, resveratrol, a polyphenol found in red wine and grapes, is an activator of class III deacetylases (65). Therefore, one avenue to find new HDAC inhibitors and activators is to test established drugs and natural products. The presence of HDAC modulators in vegetables and fruits also reinforces the common notion that proper diets are important for disease prevention.
Regulatory pathways of class IIa HDACs (Fig. 3) offer novel points for intervention. For example, it was recently proposed that inhibitors targeting the nuclear export of class IIa HDACs control pathological cardiac gene expression (63). Since some of the regulatory pathways are conserved in worms, flies, and mice, strategies targeting regulation can be developed and tested in these model organisms. The deacetylase domains of class II HDACs are highly conserved (Fig. 1), so model organisms may also be valuable hosts for testing known HDAC modulators and screening for novel ones. Related to this, budding yeast has been successfully used for screening small molecules targeting class III HDACs (6, 51).
CONCLUSION AND PERSPECTIVE
Since Hda1 was identified almost a decade ago (120), research progress on class II HDACs has been amazing. From sequence comparison, it is clear that unique structural domains and motifs are linked to the Hda1-like domains for special function and regulation in diverse organisms (Fig. 1). In metazoans, class IIa HDACs share domain organization, function, and regulation (Fig. 1-2). Like Hda1, they can function as signal-responsive transcriptional corepressors (Table 1). For a majority of the binding partners identified (Table 1), additional experiments will be needed to establish the biological significance. Hda1 also mediates global and domain-wide deacetylation, so an interesting question to be addressed is whether class IIa HDACs play a similar role in multicellular organisms. Deacetylation by Hda1 homologs in S. pombe and Neurospora interplays with chromatin methylation, so an intriguing issue awaiting further investigation is whether class IIa HDACs have an analogous role. They form mysterious dots in the nucleus (33, 76, 104, 140, 148, 161), but the nature and biological significance of these dots still remain obscure. Another characteristic of class IIa HDACs is signal-dependent nucleocytoplasmic trafficking (Fig. 3). As key kinases regulating the trafficking equilibrium, CaMK and PKD are at the centers of cell signaling networks, so it will be important to map the upstream pathways in different biological contexts. Related to this, no responsible phosphatases have been identified yet. The cytoplasmic localization of class IIa HDACs was thought to just sequester them away from their nuclear targets, but a recent study suggested otherwise (4). An important question to be investigated is what roles they play in the cytoplasm. Emerging evidence (Table 2) has challenged the notion that class IIa members are redundant, so it will be necessary to analyze how sequence divergence determines their distinct function and regulation. Unlike class IIa members, HDAC6 is a tubulin deacetylase with an important role in the cytoplasm. Two unaddressed issues are whether it really plays a role in the nucleus and how the deacetylase activity cross talks with ubiquitin-binding ability. Like HDAC6, HDAC10 may have a potentially important role in cancer progression (110, 165), so this deacetylase will certainly receive more research attention in the coming few years.
Functional studies of class II HDACs (Table 2) have provided some explanations on the toxicity observed with HDAC inhibitors that are being clinically evaluated. These studies suggest that inhibitors specific to class I HDACs are more efficacious than known ones and that activators of class II HDACs are of potential therapeutic value. Further analysis of class II HDACs will not only improve our understanding of the aforementioned fundamental question of how the sequence of a protein determines its structure, function, and regulation but will also yield invaluable insight into the molecular basis of related human diseases and illuminate the avenues towards improving the therapeutic value of HDAC inhibitors and activators.
Acknowledgments
This work was supported by funds from the Canadian Cancer Society through the National Cancer Institute of Canada and grants from the Canadian Institutes of Health Research and the Canada Foundation for Innovation (to X.-J.Y.).
REFERENCES
- 1.Aguilera, C., R. Hoya-Arias, G. Haegeman, L. Espinosa, and A. Bigas. 2004. Recruitment of IκBα to the hes1 promoter is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 101:16537-16542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, J. M., J. Nip, D. K. Strom, B. Lutterbach, H. Harada, N. Lenny, J. R. Downing, S. Meyers, and S. W. Hiebert. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21:6470-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amerik, A. Y., S. J. Li, and M. Hochstrasser. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381:981-992. [DOI] [PubMed] [Google Scholar]
- 4.Bakin, R. E., and M. O. Jung. 2004. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. J. Biol. Chem. 279:51218-51225. [DOI] [PubMed] [Google Scholar]
- 5.Barlow, A. L., C. M. van Drunen, C. A. Johnson, S. Tweedie, A. Bird, and B. M. Turner. 2001. dSIR2 and dHDAC6: two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp. Cell Res. 265:90-103. [DOI] [PubMed] [Google Scholar]
- 6.Bedalov, A., T. Gatbonton, W. P. Irvine, D. E. Gottschling, and J. A. Simon. 2001. Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. USA 98:15113-15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, I., C. Bieniossek, C. Schaffitzel, M. Hassler, E. Santelli, and T. J. Richmond. 2003. Direct interaction of Ca2+/calmodulin inhibits histone deacetylase 5 repressor core binding to myocyte enhancer factor 2. J. Biol. Chem. 278:17625-17635. [DOI] [PubMed] [Google Scholar]
- 8.Berns, K., E. M. Hijmans, J. Mullenders, T. R. Brummelkamp, A. Velds, M. Heimerikx, R. M. Kerkhoven, M. Madiredjo, W. Nijkamp, B. Weigelt, R. Agami, W. Ge, G. Cavet, P. S. Linsley, R L. Beijersbergen, and R. Bernards. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428:431-437. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, and S. L. Schreiber. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99:8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertos, N. R., B. Gilquin, M. Chan, T. J. Yen, S. Khochbin, and X. J. Yang. 2004. A tetradecapeptide-repeat domain controls the leptomycin B-resistant cytoplasmic retention of human histone deacetylase 6. J. Biol. Chem. 279:48246-48254. [DOI] [PubMed] [Google Scholar]
- 11.Bertos, N. R., A. H. Wang, and X. J. Yang. 2001. Class II histone deacetylases: structure, function and regulation. Biochem. Cell Biol. 79:243-252. [PubMed] [Google Scholar]
- 12.Blander, G., and L. Guarente. 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73:417-435. [DOI] [PubMed] [Google Scholar]
- 13.Borghi, S., S. Molinari, G. Razzini, F. Parise, R. Battini, and S. Ferrari. 2001. The nuclear localization domain of the MEF2 family of transcription factors shows member-specific features and mediates the nuclear import of histone deacetylase 4. J. Cell Sci. 114:4477-4483. [DOI] [PubMed] [Google Scholar]
- 14.Brush, M. H., A. Guardiola, J. H. Connor, T. P. Yao, and S. Shenolikar. 2004. Deactylase inhibitors disrupt cellular complexes containing protein phosphatases and deacetylases. J. Biol. Chem. 279:7685-7691. [DOI] [PubMed] [Google Scholar]
- 15.Cao, D., Z. Wang, C. L. Zhang, J. Oh, W. Xing, S. Li, J. A. Richardson, D. Z. Wang, and E. N. Olson. 2005. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol. Cell. Biol. 25:364-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carmen, A. A., P. R. Griffin, J. R. Calaycay, S. E. Rundlett, Y. Suka, and M. Grunstein. 1999. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. USA 96:12356-12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castet, A., A. Boulahtouf, G. Versini, S. Bonnet, P. Augereau, F. Vignon, S. Khochbin, S. Jalaguier, and V. Cavailles. 2004. Multiple domains of the receptor-interacting protein 140 contribute to transcription inhibition. Nucleic Acids Res. 32:1957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan, J. K., L. Sun, X. J. Yang, G. Zhu, and Z. Wu. 2003. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 278:23515-23521. [DOI] [PubMed] [Google Scholar]
- 19.Chang, S., T. A. McKinsey, C. L. Zhang, J. A. Richardson, J. A. Hill, and E. N. Olson. 2004. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24:8467-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chassaing, N., P. De Mas, M. Tauber, M. C. Vincent, S. Julia, G. Bourrouillou, P. Calvas, and E. Bieth. 2004. Molecular characterization of a cryptic 2q37 deletion in a patient with Albright hereditary osteodystrophy-like phenotype. Am. J. Med. Genet. 128A:410-413. [DOI] [PubMed] [Google Scholar]
- 21.Chauchereau, A., M. Mathieu, J. de Saintignon, R. Ferreira, L. L. Pritchard, Z. Mishal, A. Dejean, and A. Harel-Bellan. 2004. HDAC4 mediates transcriptional repression by the acute promyelocytic leukaemia-associated protein PLZF. Oncogene 23:8777-8784. [DOI] [PubMed] [Google Scholar]
- 22.Chawla, S., P. Vanhoutte, F. J. Arnold, C. L. Huang, and H. Bading. 2003. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 85:151-159. [DOI] [PubMed] [Google Scholar]
- 23.Choi, K. Y., Y. J. Ji, C. Jee, H. Kim do, and J. Ahnn. 2002. Characterization of CeHDA-7, a class II histone deacetylase interacting with MEF-2 in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 293:1295-1300. [DOI] [PubMed] [Google Scholar]
- 24.Cohen, T., and T. P. Yao. 10 August 2004, posting date. AcK—knowledge reversible acetylation. Sci. STKE 2004:pe42. [Online.] http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;stke.2452004pe42. [DOI] [PubMed] [Google Scholar]
- 25.Cosio, B. G., L. Tsaprouni, K. Ito, E. Jazrawi, I. M. Adcock, and P. J. Barnes. 2004. Theophylline restores histone deacetylase activity and steroid responses in COPD macrophages. J. Exp. Med. 200:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crabtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109(Suppl.):S67-S79. [DOI] [PubMed] [Google Scholar]
- 27.Cress, W. D., and E. Seto. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184:1-16. [DOI] [PubMed] [Google Scholar]
- 28.Dai, Y. S., and J. D. Molkentin. 2003. An MRJ-HDAC4 co-repressor complex suppresses calcineurin-NFAT signaling and cardiomyocyte hypertrophy J. Mol. Cell. Cardiol. J. 35:56. [Google Scholar]
- 29.Davis, F. J., M. Gupta, B. Camoretti-Mercado, R. J. Schwartz, and M. P. Gupta. 2003. Calcium/calmodulin-dependent protein kinase activates serum response factor transcription activity by its dissociation from histone deacetylase, HDAC4. Implications in cardiac muscle gene regulation during hypertrophy. J. Biol. Chem. 278:20047-20058. [DOI] [PubMed] [Google Scholar]
- 30.DeLange, R. J., D. M. Fambrough, E. L. Smith, and J. Bonner. 1969. Calf and pea histone IV. II. The complete amino acid sequence of calf thymus histone IV: presence of epsilon-N-acetyllysine. J. Biol. Chem. 244:319-334. [PubMed] [Google Scholar]
- 31.Deng, X., D. Z. Ewton, S. E. Mercer, and E. Friedman. 2005. Mirk/dyrk1B decreases the nuclear accumulation of class II histone deacetylases during skeletal muscle differentiation. J. Biol. Chem. 80:4894-4905. [DOI] [PubMed] [Google Scholar]
- 32.Dequiedt, F., H. Kasler, W. Fischle, V. Kiermer, M. Weinstein, B. G. Herndier, and E. Verdin. 2003. HDAC7, a thymus-specific class II histone deacetylase, regulates Nur77 transcription and TCR-mediated apoptosis. Immunity 18:687-698. [DOI] [PubMed] [Google Scholar]
- 33.Downes, M., P. Ordentlich, H. Y. Kao, J. G. Alvarez, and R. M. Evans. 2000. Identification of a nuclear domain with deacetylase activity. Proc. Natl. Acad. Sci. USA 97:10330-10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dressel, U., P. J. Bailey, S. C. Wang, M. Downes, R. M. Evans, and G. E. Muscat. 2001. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 276:17007-17013. [DOI] [PubMed] [Google Scholar]
- 35.Druesne, N., A. Pagniez, C. Mayeur, M. Thomas, C. Cherbuy, P. H. Duee, P. Martel, and C. Chaumontet. 2004. Diallyl disulfide (DADS) increases histone acetylation and p21waf1/cip1 expression in human colon tumor cell lines. Carcinogenesis 25:1227-1236. [DOI] [PubMed] [Google Scholar]
- 36.Ducy, P., T. Schinke, and G. Karsenty. 2000. The osteoblast: a sophisticated fibroblast under central surveillance. Science 289:1501-1504. [DOI] [PubMed] [Google Scholar]
- 37.Fernandes, I., Y. Bastien, T. Wai, K. Nygard, R. Lin, O. Cormier, H. S. Lee, F. Eng, N. R. Betos, N. Pelletier, S. Mader, V. K. Han, X. J. Yang, J. H. White. 2003. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol. Cell 11:139-150. [DOI] [PubMed] [Google Scholar]
- 38.Fischer, D. D., R. Cai, U. Bhatia, F. A. Asselbergs, C. Song, R. Terry, N. Trogani, R. Widmer, P. Atadja, and D. Cohen. 2002. Isolation and characterization of a novel class II histone deacetylase, HDAC10. J. Biol. Chem. 277:6656-6666. [DOI] [PubMed] [Google Scholar]
- 39.Fischle, W., F. Dequiedt, M. Fillion, M. J. Hendzel, W. Voelter, and E. Verdin. 2001. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J. Biol. Chem. 276:35826-35835. [DOI] [PubMed] [Google Scholar]
- 40.Fischle, W., F. Dequiedt, M. J. Hendzel, M. G. Guenther, M. A. Lazar, W. Voelter, and E. Verdin. 2002. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9:45-57. [DOI] [PubMed] [Google Scholar]
- 41.Fischle, W., S. Emiliani, M. J. Hendzel, T. Nagase, N. Nomura, W. Voelter, and E. Verdin. 1999. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem. 274:11713-11720. [DOI] [PubMed] [Google Scholar]
- 42.Franco, P. J., M. Farooqui, E. Seto, and L. N. Wei. 2001. The orphan nuclear receptor TR2 interacts directly with both class I and class II histone deacetylases. Mol. Endocrinol. 15:1318-1328. [DOI] [PubMed] [Google Scholar]
- 43.Furumai, R., A. Matsuyama, N. Kobashi, K. H. Lee, M. Nishiyama, H. Nakajima, A. Tanaka, Y. Komatsu, N. Nishino, M. Yoshida, and S. Horinouchi. 2002. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 62:4916-4921. [PubMed] [Google Scholar]
- 44.Gao, L., M. A. Cueto, F. Asselbergs, and P. Atadja. 2002. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 277:25748-25755. [DOI] [PubMed] [Google Scholar]
- 45.Girdwood, D., D. Bumpass, O. A. Vaughan, A. Thain, L. A. Anderson, A. W. Snowden, E. Garcia-Wilson, N. D. Perkins, and R. T. Hay. 2003. p300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11:1043-1054. [DOI] [PubMed] [Google Scholar]
- 46.Gottlicher, M., S. Minucci, P. Zhu, O. H. Kramer, A. Schimpf, S. Giavara, J. P. Sleeman, F. Lo Coco, C. Nervi, P. G. Pelicci, and T. Heinzel. 2001. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 20:6969-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green, S. R., and A. D. Johnson. 2004. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol. Biol. Cell 15:4191-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grégoire, S., and X. J. Yang. 2005. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 25:2273-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregoretti, I. V., Y. M. Lee, and H. V. Goodson. 2004. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338:17-31. [DOI] [PubMed] [Google Scholar]
- 50.Grewal, S. I., M. J. Bonaduce, and A. J. Klar. 1998. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150:563-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grozinger, C. M., E. D. Chao, H. E. Blackwell, D. Moazed, and S. L. Schreiber. 2001. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 276:38837-38843. [DOI] [PubMed] [Google Scholar]
- 52.Grozinger, C. M., C. A. Hassig, and S. L. Schreiber. 1999. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. USA 96:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grozinger, C. M., and S. L. Schreiber. 2002. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 9:3-16. [DOI] [PubMed] [Google Scholar]
- 54.Grozinger, C. M., and S. L. Schreiber. 2000. Regulation of histone deacetylase 4 and 5 transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan, Z., M. Giustetto, S. Lomvardas, J. H. Kim, M. C. Miniaci, J. H. Schwartz, D. Thanos, and E. R. Kandel. 2002. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell 111:483-493. [DOI] [PubMed] [Google Scholar]
- 56.Guardiola, A. R., and T. P. Yao. 2002. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J. Biol. Chem. 277:3350-3356. [DOI] [PubMed] [Google Scholar]
- 57.Haggarty, S. J., K. M. Koeller, J. C. Wong, C. M. Grozinger, and S. L. Schreiber. 2003. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. USA 100:4389-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Halkidou, K., S. Cook, H. Y. Leung, D. E. Neal, and C. N. Robson. 2004. Nuclear accumulation of histone deacetylase 4 (HDAC4) coincides with the loss of androgen sensitivity in hormone refractory cancer of the prostate. Eur. Urol. 45:382-389. [DOI] [PubMed] [Google Scholar]
- 59.Halme, A., S. Bumgarner, C. Styles, and G. R. Fink. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116:405-415. [DOI] [PubMed] [Google Scholar]
- 60.Han, A., J. He, Y. Wu, J. O. Liu, and L. Chen. 2005. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J. Mol. Biol. 345:91-102. [DOI] [PubMed] [Google Scholar]
- 61.Han, A., F. Pan, J. C. Stroud, H. D. Youn, J. O. Liu, and L. Chen. 2003. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature 422:730-734. [DOI] [PubMed] [Google Scholar]
- 62.Han, J., and J. D. Molkentin. 2000. Regulation of MEF2 by p38 MAPK and its implication in cardiomyocyte biology. Trends Cardiovasc. Med. 10:19-22. [DOI] [PubMed] [Google Scholar]
- 63.Harrison, B. C., C. R. Roberts, D. B. Hood, M. Sweeney, J. M. Gould, E. W. Bush, and T. A. McKinsey. 2004. The CRM1 nuclear export receptor controls pathological cardiac gene expression. Mol. Cell. Biol. 24:10636-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hook, S. S., A. Orian, S. M. Cowley, and R. N. Eisenman. 2002. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl. Acad. Sci. USA 99:13425-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howitz, K. T., K. J. Bitterman, H. Y. Cohen, D. W. Lamming, S. Lavu, J. G. Wood, R. E. Zipkin, P. Chung, A. Kisielewski, L. L. Zhang, B. Scherer, and D. A. Sinclair. 2003. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425:191-196. [DOI] [PubMed] [Google Scholar]
- 66.Huang, E. Y., J. Zhang, E. A. Miska, M. G. Guenther, T. Kouzarides, and M. A. Lazar. 2000. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 14:45-54. [PMC free article] [PubMed] [Google Scholar]
- 67.Huang, Y., M. Tan, M. Gosink, K. K. Wang, and Y. Sun. 2002. Histone deacetylase 5 is not a p53 target gene, but its overexpression inhibits tumor cell growth and induces apoptosis. Cancer Res. 62:2913-2922. [PubMed] [Google Scholar]
- 68.Hubbert, C., A. Guardiola, R. Shao, Y. Kawaguchi, A. Ito, A. Nixon, M. Yoshida, X. F. Wang, and T. P. Yao. 2002. HDAC6 is a microtubule-associated deacetylase. Nature 417:455-458. [DOI] [PubMed] [Google Scholar]
- 69.Huynh, K. D., W. Fischle, E. Verdin, and V. J. Bardwell. 2000. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 14:1810-1823. [PMC free article] [PubMed] [Google Scholar]
- 70.Ito, Y. 2004. Oncogenic potential of the RUNX gene family: ‘overview.’ Oncogene 23:4198-4208. [DOI] [PubMed] [Google Scholar]
- 71.Jeong, B. C., C. Y. Hong, S. Chattopadhyay, J. H. Park, E. Y. Gong, H. J. Kim, S. Y. Chun, and K. Lee. 2004. Androgen receptor corepressor-19 kDa (ARR19), a leucine-rich protein that represses the transcriptional activity of androgen receptor through recruitment of histone deacetylase. Mol. Endocrinol. 18:13-25. [DOI] [PubMed] [Google Scholar]
- 72.Jin, Y. H., E. J. Jeon, Q. L. Li, Y. H. Lee, J. K. Choi, W. J. Kim, K. Y. Lee, and S. C. Bae. 2004. Transforming growth factor-beta stimulates p300-dependent RUNX3 acetylation, which inhibits ubiquitination-mediated degradation. J. Biol. Chem. 279:29409-29417. [DOI] [PubMed] [Google Scholar]
- 73.Kao, G. D., W. G. McKenna, M. G. Guenther, R. J. Muschel, M. A. Lazar, and T. J. Yen. 2003. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J. Cell Biol. 160:1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kao, H. Y., M. Downes, P. Ordentlich, and R. M. Evans. 2000. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 14:55-66. [PMC free article] [PubMed] [Google Scholar]
- 75.Kao, H. Y., C. H. Lee, A. Komarov, C. C. Han, and R. M. Evans. 2002. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J. Biol. Chem. 277:187-193. [DOI] [PubMed] [Google Scholar]
- 76.Kao, H. Y., A. Verdel, C. C. Tsai, C. Simon, H. Juguilon, and S. Khochbin. 2001. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276:47496-47507. [DOI] [PubMed] [Google Scholar]
- 77.Kato, H., S. Tamamizu-Kato, and F. Shibasaki. 2004. Histone deacetylase 7 associates with hypoxia-inducible factor 1α and increases transcriptional activity. J. Biol. Chem. 279:41966-41974. [DOI] [PubMed] [Google Scholar]
- 78.Kawaguchi, Y., J. J. Kovacs, A. McLaurin, J. M. Vance, A. Ito, and T. P. Yao. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115:727-738. [DOI] [PubMed] [Google Scholar]
- 79.Khochbin, S., A. Verdel, C. Lemercier, and D. Seigneurin-Berny. 2001. Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 11:162-166. [DOI] [PubMed] [Google Scholar]
- 80.Kirsh, O., J. S. Seeler, A. Pichler, A. Gast, S. Muller, E. Miska, M. Mathieu, A. Harel-Bellan, T. Kouzarides, F. Melchior, and A. Dejean. 2002. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klar, A. J., T. Srikantha, and D. R. Soll. 2001. A histone deacetylation inhibitor and mutant promote colony-type switching of the human pathogen Candida albicans. Genetics 158:919-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kovacs, J. J., C. Hubbert, and T. P. Yao. 2004. The HDAC complex and cytoskeleton. Novartis Found. Symp. 259:170-177. [PubMed] [Google Scholar]
- 84.Kurdistani, S. K., and M. Grunstein. 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4:276-284. [DOI] [PubMed] [Google Scholar]
- 85.Kurtev, V., R. Margueron, K. Kroboth, E. Ogris, V. Cavailles, and C. Seiser. 2004. Transcriptional regulation by the repressor of estrogen receptor activity via recruitment of histone deacetylases. J. Biol. Chem. 279:24834-24843. [DOI] [PubMed] [Google Scholar]
- 86.Lee, H. J., M. Chun, and K. V. Kandror. 2001. Tip60 and HDAC7 interact with the endothelin receptor a and may be involved in downstream signaling. J. Biol. Chem. 276:16597-16600. [DOI] [PubMed] [Google Scholar]
- 87.Lemercier, C., M. P. Brocard, F. Puvion-Dutilleul, H. Y. Kao, O. Albagli, and S. Khochbin. 2002. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J. Biol. Chem. 277:22045-22052. [DOI] [PubMed] [Google Scholar]
- 88.Lemercier, C., A. Verdel, B. Galloo, S. Curtet, M. P. Brocard, and S. Khochbin. 2000. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 275:15594-15599. [DOI] [PubMed] [Google Scholar]
- 89.Li, X., S. Song, Y. Liu, S. H. Ko, and H. Y. Kao. 2004. Phosphorylation of the histone deacetylase 7 modulates its stability and association with 14-3-3 proteins. J. Biol. Chem. 279:34201-34208. [DOI] [PubMed] [Google Scholar]
- 90.Linseman, D. A., C. M. Bartley, S. S. Le, T. A. Laessig, R. J. Bouchard, M. K. Meintzer, M. Li, and K. A. Heidenreich. 2003. Inactivation of the myocyte enhancer factor-2 repressor histone deacetylase-5 by endogenous Ca2+/calmodulin-dependent kinase II promotes depolarization-mediated cerebellar granule neuron survival. J. Biol. Chem. 278:41472-41481. [DOI] [PubMed] [Google Scholar]
- 91.Liu, F., M. Dowling, X. J. Yang, and G. D. Kao. 2004. Caspase-mediated specific cleavage of human histone deacetylase 4. J. Biol. Chem. 279:34537-34546. [DOI] [PubMed] [Google Scholar]
- 92.Lomonte, P., J. Thomas, P. Texier, C. Caron, S. Khochbin, and A. L. Epstein. 2004. Functional interaction between class II histone deacetylases and ICP0 of herpes simplex virus type 1. J. Virol. 78:6744-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lu, J., T. A. McKinsey, R. L. Nicol, and E. N. Olson. 2000. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 97:4070-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu, J., T. A. McKinsey, C. L. Zhang, and E. N. Olson. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6:233-244. [DOI] [PubMed] [Google Scholar]
- 95.Marks, P. A., T. Miller, and V. M. Richon. 2003. Histone deacetylases. Curr. Opin. Pharmacol. 3:344-351. [DOI] [PubMed] [Google Scholar]
- 96.Mason, D. X., T. J. Jackson, and A. W. Lin. 2004. Molecular signature of oncogenic ras-induced senescence. Oncogene 23:9238-9246. [DOI] [PubMed] [Google Scholar]
- 97.Matsuyama, A., T. Shimazu, Y. Sumida, A. Saito, Y. Yoshimatsu, D. Seigneurin-Berny, H. Osada, Y. Komatsu, N. Nishino, S. Khochbin, S. Horinouchi, and M. Yoshida. 2002. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 21:6820-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McKinsey, T. A., and E. N. Olson. 2004. Cardiac histone acetylation—therapeutic opportunities abound. Trends Genet. 20:206-213. [DOI] [PubMed] [Google Scholar]
- 99.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2000. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97:14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21:6312-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27:40-47. [DOI] [PubMed] [Google Scholar]
- 103.Miller, T. A., D. J. Witter, and S. Belvedere. 2003. Histone deacetylase inhibitors. J. Med. Chem. 46:5097-5116. [DOI] [PubMed] [Google Scholar]
- 104.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miska, E. A., E. Langley, D. Wolf, C. Karlsson, J. Pines, and T. Kouzarides. 2001. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 29:3439-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Myzak, M. C., P. A. Karplus, F. L. Chung, and R. H. Dashwood. 2004. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 64:5767-5774. [DOI] [PubMed] [Google Scholar]
- 107.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 108.North, B. J., B. L. Marshall, M. T. Borra, J. M. Denu, and E. Verdin. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11:437-444. [DOI] [PubMed] [Google Scholar]
- 109.North, B. J., and E. Verdin. 2004. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 5:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Osada, H., Y. Tatematsu, H. Saito, Y. Yatabe, T. Mitsudomi, and T. Takahashi. 2004. Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int. J. Cancer 112:26-32. [DOI] [PubMed] [Google Scholar]
- 111.Palazzo, A., B. Ackerman, and G. G. Gundersen. 2002. Tubulin acetylation and cell motility. Nature 421:230. [DOI] [PubMed] [Google Scholar]
- 112.Pandey, R., A. Muller, C. A. Napoli, D. A. Selinger, C. S. Pikaard, E. J. Richards, J. Bender, D. W. Mount, and R. A. Jorgensen. 2002. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30:5036-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Paroni, G., M. Mizzau, C. Henderson, G. Del Sal, C. Schneider, and C. Brancolini. 2004. Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol. Biol. Cell 15:2804-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Parra, M., H. Kasler, T. A. McKinsey, E. N. Olson, and E. Verdin. Protein kinase D1 phosphorylates HDAC7 and induces its nuclear export after TCR activation. J. Biol. Chem., in press. [DOI] [PubMed]
- 115.Petrie, K., F. Guidez, L. Howell, L. Healy, S. Waxman, M. Greaves, and A. Zelent. 2003. The histone deacetylase 9 gene encodes multiple protein isoforms. J. Biol. Chem. 278:16059-16072. [DOI] [PubMed] [Google Scholar]
- 116.Phiel, C. J., F. Zhang, E. Y. Huang, M. G. Guenther, M. A. Lazar, and P. S. Klein. 2001. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 276:36734-36741. [DOI] [PubMed] [Google Scholar]
- 117.Piekarz, R., and S. Bates. 2004. A review of depsipeptide and other histone deacetylase inhibitors in clinical trials. Curr. Pharm. Des. 10:2289-2298. [DOI] [PubMed] [Google Scholar]
- 118.Pipal, A., M. Goralik-Schramel, A. Lusser, C. Lanzanova, B. Sarg, A. Loidl, H. Lindner, V. Rossi, and P. Loidl. 2003. Regulation and processing of maize histone deacetylase Hda1 by limited proteolysis. Plant Cell 15:1904-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang, N. Suka, and M. Grunstein. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437-446. [DOI] [PubMed] [Google Scholar]
- 120.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Seigneurin-Berny, D., A. Verdel, S. Curtet, C. Lemercier, J. Garin, S. Rousseaux, and S. Khochbin. 2001. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 21:8035-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sengupta, N., and E. Seto. 2004. Regulation of histone deacetylase activities. J. Cell Biochem. 93:57-67. [DOI] [PubMed] [Google Scholar]
- 123.Serrador, J. M., J. R. Cabrero, D. Sancho, M. Mittelbrunn, A. Urzainqui, and F. Sanchez-Madrid. 2004. HDAC6 deacetylase activity links the tubulin cytoskeleton with immune synapse organization. Immunity 20:417-428. [DOI] [PubMed] [Google Scholar]
- 124.Smith, R. L., and A. D. Johnson. 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25:325-330. [DOI] [PubMed] [Google Scholar]
- 125.Sparrow, D. B., E. A. Miska, E. Langley, S. Reynaud-Deonauth, S. Kotecha, N. Towers, G. Spohr, T. Kouzarides, and T. J. Mohun. 1999. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 18:5085-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Srikantha, T., L. Tsai, K. Daniels, A. J. Klar, and D. R. Soll. 2001. The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J. Bacteriol. 183:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun, L., L. Liu, X. J. Yang, and Z. Wu. 2004. Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J. Cell Sci. 117:3021-3029. [DOI] [PubMed] [Google Scholar]
- 129.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 130.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 131.Tervo, A. J., S. Kyrylenko, P. Niskanen, A. Salminen, J. Leppanen, T. H. Nyronen, T. Jarvinen, and A. Poso. 2004. An in silico approach to discovering novel inhibitors of human sirtuin type 2. J. Med. Chem. 47:6292-6298. [DOI] [PubMed] [Google Scholar]
- 132.Tong, J. J., J. Liu, N. R. Bertos, and X. J. Yang. 2002. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 30:1114-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vega, R. B., K. Matsuda, J. Oh, A. C. Barbosa, X. Yang, E. Meadows, J. McAnally, C. Pomajzl, J. M. Shelton, J. A. Richardson, G. Karsenty, and E. N. Olson. 2004. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119:555-566. [DOI] [PubMed] [Google Scholar]
- 133a.Vega, R. B., B. C. Harrison, E. Meadows, C. R. Roberts, P. J. Papst, E. N. Olson, and T. A. McKinsey. 2004. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24:8374-8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Verdel, A., S. Curtet, M. P. Brocard, S. Rousseaux, C. Lemercier, M. Yoshida, and S. Khochbin. 2000. Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr. Biol. 10:747-749. [DOI] [PubMed] [Google Scholar]
- 135.Verdel, A., and S. Khochbin. 1999. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J. Biol. Chem. 274:2440-2445. [DOI] [PubMed] [Google Scholar]
- 136.Verdin, E., F. Dequiedt, and H. G. Kasler. 2003. Class II histone deacetylases: versatile regulators. Trends Genet. 19:286-293. [DOI] [PubMed] [Google Scholar]
- 137.Vidali, G., E. Gershey, and V. G. Allfrey. 1968. Chemical studies of histone acetylation. The distribution of ɛ-N-acetyllysine in calf thymus histones. J. Biol. Chem. 243:6361-6366. [PubMed] [Google Scholar]
- 138.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 139.Wang, A. H., N. R. Bertos, M. Vezmar, N. Pelletier, M. Crosato, H. H. Heng, J. Th'ng, J. Han, and X. J. Yang. 1999. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 19:7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang, A. H., M. J. Kruhlak, J. Wu, N. R. Bertos, M. Vezmar, B. I. Posner, D. P. Bazett-Jones, and X. J. Yang. 2000. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20:6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang, A. H., and X. J. Yang. 2001. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 21:5992-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang, L., C. Fan, S. E. Topol, E. J. Topol, and Q. Wang. 2003. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 302:1578-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Watamoto, K., M. Towatari, Y. Ozawa, Y. Miyata, M. Okamoto, A. Abe, T. Naoe, and H. Saito. 2003. Altered interaction of HDAC5 with GATA-1 during MEL cell differentiation. Oncogene 22:9176-9184. [DOI] [PubMed] [Google Scholar]
- 144.Watson, A. D., D. G. Edmondson, J. R. Bone, Y. Mukai, Y. Yu, W. Du, D. J. Stillman, and S. Y. Roth. 2000. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 14:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Westendorf, J. J., S. K. Zaidi, J. E. Cascino, R. Kahler, A. J. van Wijnen, J. B. Lian, M. Yoshida, G. S. Stein, and X. Li. 2002. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21CIP1M/WAF1 promoter. Mol. Cell. Biol. 22:7982-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wu, J., A. A. Carmen, R. Kobayashi, N. Suka, and M. Grunstein. 2001. HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc. Natl. Acad. Sci. USA 98:4391-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu, J., N. Suka, M. Carlson, and M. Grunstein. 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7:117-126. [DOI] [PubMed] [Google Scholar]
- 148.Wu, X., H. Li, E. J. Park, and J. D. Chen. 2001. SMRTe inhibits MEF2C transcriptional activation by targeting HDAC4 and 5 to nuclear domains. J. Biol. Chem. 276:24177-24185. [DOI] [PubMed] [Google Scholar]
- 149.Yang, X. J. 2004. Lysine acetylation and the bromodomain: a new partnership for signaling. BioEssays 26:1076-1087. [DOI] [PubMed] [Google Scholar]
- 150.Yang, X. J., and E. Seto. 2003. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr. Opin. Genet. Dev. 13:143-153. [DOI] [PubMed] [Google Scholar]
- 151.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 152.Yoshida, M., A. Matsuyama, Y. Komatsu, and N. Nishino. 2003. From discovery to the coming generation of histone deacetylase inhibitors. Curr. Med. Chem. 10:2351-2358. [DOI] [PubMed] [Google Scholar]
- 153.Yoshida, N., Y. Omoto, A. Inoue, H. Eguchi, Y. Kobayashi, M. Kurosumi, S. Saji, K. Suemasu, T. Okazaki, K. Nakachi, T. Fujita, and S. Hayashi. 2004. Prediction of prognosis of estrogen receptor-positive breast cancer with combination of selected estrogen-regulated genes. Cancer Sci. 95:496-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Youn, H. D., C. M. Grozinger, and J. O. Liu. 2000. Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J. Biol. Chem. 275:22563-22567. [DOI] [PubMed] [Google Scholar]
- 155.Yuki, Y., I. Imoto, M. Imaizumi, S. Hibi, Y. Kaneko, T. Amagasa, and J. Inazawa. 2004. Identification of a novel fusion gene in a pre-B acute lymphoblastic leukemia with t(1;19)(q23;p13). Cancer Sci. 95:503-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zeremski, M., J. R. Stricker, D. Fischer, S. B. Zusman, and D. Cohen. 2003. Histone deacetylase dHDAC4 is involved in segmentation of the Drosophila embryo and is regulated by gap and pair-rule genes. Genesis 35:31-38. [DOI] [PubMed] [Google Scholar]
- 157.Zhang, A., P. L. Yeung, C. W. Li, S. C. Tsai, G. K. Dinh, X. Wu, H. Li, and J. D. Chen. 2004. Identification of a novel family of ankyrin repeats containing cofactors for p160 nuclear receptor coactivators. J. Biol. Chem. 279:33799-33805. [DOI] [PubMed] [Google Scholar]
- 158.Zhang, C. L., T. A. McKinsey, S. Chang, C. L. Antos, J. A. Hill, and E. N. Olson. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang, C. L., T. A. McKinsey, J. Lu, and E. N. Olson. 2001. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 276:35-39. [DOI] [PubMed] [Google Scholar]
- 160.Zhang, C. L., T. A. McKinsey, and E. N. Olson. 2002. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22:7302-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang, C. L., T. A. McKinsey, and E. N. Olson. 2001. The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis. Proc. Natl. Acad. Sci. USA 98:7354-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang, W., and B. C. Kone. 2002. NF-κB inhibits transcription of the H+-K+-ATPase α2-subunit gene: role of histone deacetylases. Am. J. Physiol. Renal. Physiol. 283:F904-F911. [DOI] [PubMed] [Google Scholar]
- 163.Zhang, Y., N. Li, C. Caron, G. Matthias, D. Hess, S. Khochbin, and P. Matthias. 2003. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 22:1168-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang, Z., and J. C. Reese. 2004. Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279:39240-39250. [DOI] [PubMed] [Google Scholar]
- 165.Zhang, Z., H. Yamashita, T. Toyama, H. Sugiura, Y. Omoto, Y. Ando, K. Mita, M. Hamaguchi, S. Hayashi, and H. Iwase. 2004. HDAC6 expression is correlated with better survival in breast cancer. Clin. Cancer Res. 10:6962-6968. [DOI] [PubMed] [Google Scholar]
- 166.Zhao, X., A. Ito, C. D. Kane, T. S. Liao, T. A. Bolger, S. M. Lemrow, A. R. Means, and T. P. Yao. 2001. The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 276:35042-35048. [DOI] [PubMed] [Google Scholar]
- 167.Zhou, X., P. A. Marks, R. A. Rifkind, and V. M. Richon. 2001. Cloning and characterization of a novel histone deacetylase, HDAC9. Proc. Natl. Acad. Sci. USA 98:10572-10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhou, X., V. M. Richon, R. A. Rifkind, and P. A. Marks. 2000. Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc. Natl. Acad. Sci. USA 97:1056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhou, X., V. M. Richon, A. H. Wang, X. J. Yang, R. A. Rifkind, and P. A. Marks. 2000. Histone deacetylase 4 associates with extracellular signal-regulated kinases 1 and 2, and its cellular localization is regulated by oncogenic Ras. Proc. Natl. Acad. Sci. USA 97:14329-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]