Abstract
DnaK is essential for starvation-induced resistance to heat, oxidation, and reductive division in Escherichia coli. Studies reported here indicate that DnaK is also required for starvation-induced osmotolerance, catalase activity, and the production of the RpoS-controlled Dps (PexB) protein. Because these dnaK mutant phenotypes closely resemble those of rpoS (ς38) mutants, the relationship between DnaK and RpoS was evaluated directly during growth and starvation at 30°C in strains with genetically altered DnaK content. A starvation-specific effect of DnaK on RpoS abundance was observed. During carbon starvation, DnaK deficiency reduced RpoS levels threefold, while DnaK excess increased RpoS levels nearly twofold. Complementation of the dnaK mutation restored starvation-induced RpoS levels to normal. RpoS deficiency had no effect on the cellular concentration of DnaK, revealing an epistatic relationship between DnaK and RpoS. Protein half-life studies conducted at the onset of starvation indicate that DnaK deficiency significantly destabilized RpoS. RpoH (ς32) suppressors of the dnaK mutant with restored levels of RpoS and dnaK rpoS double mutants were used to show that DnaK plays both an independent and an RpoS-dependent role in starvation-induced thermotolerance. The results suggest that DnaK coordinates sigma factor levels in glucose-starved E. coli.
Starvation of Escherichia coli for carbon results in a near-immediate halt in cell division and the establishment of a dormant- or stationary-phase state (3, 35, 36). This state is distinguished from the preceding growth period by a unique physiological resistance to otherwise lethal stresses (13, 25, 52), reflecting the coordinate expression of stationary phase-specific genes. The response to carbon starvation is physiologically unlike that to deprivation of other nutrients, such as nitrogen or phosphorus. Carbon-starved cells rapidly readjust their rate of cell division and cease growth, whereas cells starved for other nutrients, such as nitrogen or phosphorus, respond sluggishly (12, 62). Carbon starvation therefore represents an efficient method for studying the stationary phase and also likely mimics common oligotrophic environmental conditions confronting other bacterial species (43). Regardless of which stimulus precipitates stationary phase, many common features characterize such nongrowing or dormant cells. Consequently, the high-cell-density stationary phase observed in rich undefined media is commonly employed for studies on stationary-phase gene expression and genetics (reviewed in references 17 and 20).
When carbon for growth is limited, protein synthesis largely stops and cell survival and persistence rest on the integrity of protein synthesized at an earlier, prestarvation time. Maintenance of protein structure during growth is determined largely by the activity of protein chaperones, many of which are ATP-dependent enzymes such as DnaK (16, 37). This enzyme promotes structural rearrangements in proteins and can be stimulated by the cochaperones, DnaJ and GrpE. In growing cells, loss of DnaK results in temperature-restricted division and defective expression of heat shock proteins (6, 7). Starvation protein synthesis is also defective in a dnaK mutant (59). Such defects may underlie the observation that DnaK deficiency mitigates starvation-induced physiological readjustments, particularly starvation-induced resistance to heat, oxidation, and reductive division (52). Interestingly, these particular dnaK mutant phenotypes are possessed by rpoS mutants (19, 21, 30, 38, 54). Though a clear role for DnaK in the stationary phase is evident from studies on DnaK deficiency, an excess of this chaperone elicits additional physiological alterations which are again evident only in stationary-phase cells. Overproduction of DnaK is specifically bactericidal in the stationary phase, and this toxicity can be partially ameliorated by cooverproduction of DnaJ (2). The basis of this effect is as yet unclear, though suppressor analysis of stationary-phase DnaK toxicity has led to the recovery of multicopy genes which overcome this effect. One of these has been identified as edd and encodes the first committed enzyme of the Entner-Doudoroff pathway for hexose catabolism (53). Inactivation of this gene by the insertion of a stop codon prevented suppression and suggests a role for this pathway in stationary-phase metabolism.
The RpoS sigma factor controls expression of a diversity of genes, many of which are critical in the stationary phase (20, 33). Specific examples particularly notable because of their role in starvation-induced physiological alterations include katE, encoding catalase HPII (23, 34), and dps (1), also called pexB (31), encoding a small histone-like nonspecific DNA binding protein. However, additional genes involved in osmotolerant growth (22) and low-temperature growth (58) are now also known to be controlled by RpoS. Such observations expand the role of RpoS beyond the stationary phase. Increased expression of stationary-phase RpoS-regulated genes requires an increase in the abundance of RpoS. Such changes reflect alterations in rpoS expression at the level of transcription initiation, translation elongation, and posttranslational events resulting in increased RpoS stability (29, 39, 61). Variation in the allelic state of rssB/sprE indicates that this locus acts as an unlinked negative regulator of RpoS abundance in the exponential phase (45, 51). RpoS abundance during growth also depends in part on the ClpX/P heat shock proteins (56) as well as on the absolute temperature of cultivation (58). Since heat shock of growing cells elicits an increase in the abundance of RpoS, a role for additional regulatory factors of RpoS abundance appears likely (26).
To better understand the significance of the apparent stationary-phase phenotypic overlap between dnaK and rpoS mutants, a dnaK null mutant (49) as well as an otherwise isogenic wild-type and rpoS null mutant strains were analyzed for additional rpoS-like defects. We report here that the dnaK mutant has several of the same phenotypes as rpoS mutants, which result from an apparent defect in RpoS metabolism. Consequently, these findings suggest that DnaK plays only an indirect role in the starvation response. While these studies were under way, a role for DnaK in modulating RpoS levels during heat shock was reported (44). The physiological significance of this observation, however, was not apparent. In addition, it was reported that DnaK deficiency elevated exponential-phase RpoS levels sixfold while decreasing carbon starvation-induced levels only slightly. The physiological significance of these results was not examined. These findings contrast with results obtained with rpoS::lacZ fusions which indicate that RpoS is significantly destabilized during growth and starvation by a mutation in dnaK (44). Failure of an rpoH missense mutant (sidB1) to suppress the dnaK mutant’s effects on RpoS were interpreted as evidence for the exclusion of a role for other heat shock proteins such as ClpP/X from this process. To resolve the interplay between DnaK and RpoS in starvation-related phenotypes, we used additional rpoH mutant alleles and a dnaK rpoS double mutant to study this process. The results solve apparent discrepancies and indicate that dnaK plays two roles in starvation-related phenotypes; one is mediated through RpoS, and the other controls the physiological status of starving cells in an rpoS-independent manner.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cell cultivation.
E. coli strains, plasmids, and phages used in this study are listed in Table 1. Plasmid isolation and subcloning procedures were performed as described previously (2). The pBN15 plasmid- bearing derivative of PBL501, strain PBL504, was constructed by transformation at 30°C. The Dps-producing plasmid pBN49 was constructed by subcloning an 8-kb BamHI-HindIII fragment containing the entire dps gene from phage λ-205 (27) into pACYC184 (9) previously digested with BamHI and HindIII. Phage λ and phage P1 were grown as described previously (4). Strain PBL502 was constructed by generalized transduction with phage P1vir grown on AMS150 (Table 1), which contained a transposon Tn10 insertion in rpoS (katF13::Tn10; 34) to transduce PBL500 to tetracycline resistance. The rpoS deletion strain, PBL503, was constructed by transposon-mediated deletion formation as described previously (5). The extent of the rpoS deletion was mapped by analyzing genes flanking rpoS, including mutS (61.5 min) for the counterclockwise direction and cysC (61.85) for the clockwise direction. The insertion site of the Tn10 in rpoS was identified previously in the 5′ end of the rpoS coding region at base 196 (24). Deletion of the 3′ end of the rpoS gene was indicated by the apparent loss of mutS (10). PBL503, containing the mutS deletion, had a forward mutation rate to streptomycin resistance of 1.14 × 10−7 compared to 3.5 × 10−9 for the otherwise isogenic wild-type parent strain, PBL502. The rpoS deletion did not extend as far as cysC, as PBL503 remained prototrophic. The rpoS deletion is designated ΔmutS-rpoS458.
TABLE 1.
Strains, plasmids, and phages used in this study
| Name | Genotype | Source |
|---|---|---|
| Strains | ||
| AMS150 | katF13::Tn10 | 38 |
| CAG18442 | thr-34::Tn10 | 57 |
| CAG18638 | zhh-21::Tn10 | 57 |
| DH5α | φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA69 | Laboratory stock |
| PBL500 | lacZ::Tn5 lacIQ1 | 52 |
| PBL501 | PBL500; ΔdnaK52::Cmr | 52 |
| PBL502 | PBL500; rpoS-13::Tn10 | P1vir(AMS150) × PBL500 to tetracycline resistance |
| PBL503 | PBL500; ΔmutS-rpoS458 | A tetracycline-sensitive derivative of PBL502 |
| PBL504 | PBL501/pBN15 | This work |
| PBL505 | DH5α/pBN49 | This work |
| PBL603 | PBL500; rpoS13::Tn10 ΔdnaK52::Cmr | P1vir(pBN15/PBL501) × PBL502 to chloramphenicol resistance |
| PBL622 | PBL501; rpoH-1 | This work |
| PBL623 | PBL501; rpoH-2 | This work |
| PBL624 | PBL501; rpoH-3 | This work |
| PBL711 | PBL622/pBN15 | This work |
| PBL713 | PBL622; dnaK+ | This work |
| Plasmids | ||
| pBN15 | Ptac::dnaK+J+ lacI+ bla+ | 52 |
| pBN49 | dps+ | 8-kb BamHI-HindIII fragment from λ-205 subcloned into pACYC184 |
| pACYC184 | Tetr Cmr | Laboratory stock |
| pDS2 | Ptac::rpoH+ bla+ | 18 |
| Phages | ||
| P1vir | Laboratory stock | |
| λ-205 | 27 |
RpoH suppressors of the ΔdnaK52 mutation were isolated as described previously (6). Three independent and spontaneous isolates were mapped by phage P1 transduction and shown to be 50% linked to zhh-21::Tn10. Complementation analysis with the rpoH expression plasmid, pDS2 (18), was used to verify that the mutations were in rpoH. pDS2 restored both the filamentous cell morphology and the reduced viability in all three isolates, characteristic of DnaK deficiency during growth in LB medium (6). To evaluate the starvation-related phenotypes of these rpoH suppressors in otherwise wild-type backgrounds, the ΔdnaK52 mutation in rpoH suppressor 1 was removed by phage P1 cotransduction with the linked insertion, thr-34::Tn10, generating PBL713, and complemented by transformation with plasmid pBN15 (pPtac::dnaK+J+ lacI+ bla+), generating PBL711.
Cell densities of cultures were monitored spectrophotometrically at a wavelength of 600 nm. All cultures were incubated at 30°C in M9 minimal medium (42) containing 0.05% (wt/vol) glucose and shaken at 150 rpm with 1-liter Erlenmeyer flasks in shaking water baths (model G76; New Brunswick). dnaK mutants have been reported to accrue spontaneous mutations during prolonged incubation on a solid medium (6). Care therefore was taken to avoid such events during studies of the unsuppressed dnaK mutant through the use of freshly streaked plates. LB plates were used to determine viable cell counts as described previously (52). Chloramphenicol and tetracycline were added at final concentrations of 25 μg/ml, kanamycin was added at 75 μg/ml, and ampicillin and streptomycin were added at 100 μg/ml. All cultures were maintained in exponential-phase growth by repeated subculturing for a minimum of 10 generations prior to collection of the first exponential-phase sample.
Salt tolerance measurements were performed after 1 day of glucose starvation by adding dry sodium chloride powder, with stirring to a final concentration of 2.5 M. The cultures were then maintained with gentle agitation at 30°C for the duration of the experiment. The number of viable cells per milliliter was determined by plating diluted culture samples onto LB plates in duplicate.
Dps protein purification and antibody production.
A 1-liter culture of strain PBL505, incubated for 5 days at 37°C with shaking in LB medium containing chloramphenicol, was centrifuged for 10 min at 6,000 rpm at 4°C. The remainder of the purification was performed as previously described (1). Briefly, the cell pellet was lysed by passage through a French pressure cell, and the resulting lysate was clarified by ultracentrifugation and fractionated by ammonium sulfate precipitation. The sample was applied to a Sephadex G-200 column, and Dps was recovered in the void volume. The protein was precipitated again with ammonium sulfate and resuspended in a high-salt buffer. The sample was then applied to a Sepharose 6B column, and Dps-containing fractions were recovered and applied to a DNA-cellulose column. Dps was eluted from the column with a linear sodium chloride gradient in which Dps eluted at 200 mM sodium chloride. Purity of the Dps fractions was determined by Coomassie blue R-250 staining.
Anti-Dps polyclonal antibodies were produced by injection of 100 μg of the purified Dps protein into a New Zealand White rabbit, followed by booster shots of equivalent amounts of antigen at two 2-week intervals. Serum was collected after an additional 2 weeks and purified by precipitation with acetone powders as described previously (2) with wild-type exponential-phase cell extracts. Antibodies were purified by affinity chromatography with protein A-Sepharose as described previously (28).
Catalase assays.
The catalase assays were performed by placing 1 ml of cell culture into a prewarmed (30°C), stirred oxygen electrode (Rank Brothers). The cell culture was equilibrated at 30°C for 1 min prior to the addition of hydrogen peroxide to a final concentration of 100 mM. A computer was attached to the oxygen electrode for data collection, and sample readings were made at a rate of three times per second. Oxygen production was measured for a minimum of one min, and the amount produced was calculated as described previously (14). Differential rates of enzyme synthesis were determined as described previously (50, 55). Variation in catalase activity between replicates was less than 4%.
In vivo labeling of proteins and immunoprecipitation.
GroEL heat shock induction ratios were determined as described previously (8). To determine the half-life of RpoS at the onset of starvation, cells were grown in M9 medium containing 0.05% glucose. After reaching the onset of starvation, the culture was labeled with 35S-Translabel (1,000 Ci/mmol; ICN) at 20 μCi of culture per ml for 1 min at 30°C. The sample was chased with 10 mM (each) nonradioactive methionine and cysteine for 20 s at 30°C. Samples consisting of 3.0 × 109 cells were then removed, and protein was precipitated by the addition of ice-cold 50% (wt/vol) trichloroacetic acid (TCA) and kept on ice for 30 min. The precipitated protein was removed by centrifugation at 13,000 × g for 10 min, and the pellet was washed successively with ice-cold solutions of 10% (wt/vol) TCA, 5% (wt/vol) TCA, and 100% acetone. The samples were then dried and resuspended in 0.5 ml of PBS-D (20 mM sodium phosphate, 150 mM sodium chloride [pH 7.4], 1% [vol/vol] Triton X-100, 1% [wt/vol] sodium deoxycholate, and 0.1% [wt/vol] sodium dodecyl sulfate [SDS]). Insoluble protein was removed by centrifugation at 13,000 × g for 10 min. RpoS was immunoprecipitated by the addition of 25 μl of anti-RpoS monoclonal antibody ascitic fluid (1RS1) (47), and the samples were incubated for 2 h at 4°C, with gentle agitation. The antigen-antibody complex was removed by the addition of protein A-Sepharose 4B (Sigma) with 350 μg of protein A per reaction and incubated for 3 h at 4°C with gentle agitation. Samples were then centrifuged at 13,000 × g for 10 min, and the pellet was washed three times in PBS-D. The washed pellets were resuspended in 33 μl in a solution containing 250 mM Tris-HCl (pH 6.8), 2% SDS, 0.75 M 2-mercaptoethanol, 10% glycerol and boiled for 10 min, and 3.3 μl was removed to determine radioactivity. Bromphenol blue was added to the remaining sample to a final concentration of 20 μg/ml prior to analysis by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were detected by autoradiography with Kodak XAR film.
SDS-PAGE and Western blot analysis.
To quantitate total RpoS levels by Western blot analysis, cells from cultures grown at 30°C were concentrated by centrifugation at 13,000 × g, and the resulting cell pellets were resuspended in 25 mM Tris-HCl–40 mM glycine (pH 6.8). Samples were removed to determine protein concentrations by the bicinchoninic acid protein assay (Pierce). The remaining sample was adjusted to 250 mM Tris-HCl (pH 6.8), 2% SDS, 0.75 M 2-mercaptoethanol, 10% glycerol, 20 μg of bromphenol blue per ml, boiled for 10 min, and frozen at −20°C for later use.
Proteins were resolved by SDS-PAGE, with 9% (wt/vol) acrylamide separating gels and 10% (wt/vol) acrylamide stacking gels, as described previously (15), with prestained low-molecular-weight markers (Bio-Rad). Western blots were prepared essentially as described previously (15). Western blots were processed and developed with the ECL reagent system according to the manufacturer’s protocol (Amersham). Western blots were probed with a 1:1,000 dilution of the anti-RpoS antibody 1RS1 (47) and then a 1:1,000 dilution of sheep anti-mouse horseradish peroxidase conjugate (GibcoBRL). As a reference to normalize development of all Western blots for RpoS content, 111 ng of protein of strain PBL504 following 2 h of starvation was included as an internal standard. RpoS signal intensity was linear within the range of detection as determined by regression analysis of PBL504 extracts (r2 = 0.99) analyzed in amounts of 0.111, 0.333, 1.0, 3.0, and 9.0 μg of protein. Chemiluminescence was detected by exposing Kodak XAR film. Exposed films were analyzed by densitometry with a GDS7500 densitometer and gel base/gel blot software (Ultraviolet Products).
Detection of Dps by Western blot analysis was performed as described above with the following two exceptions; proteins were resolved by SDS-PAGE, 14.5% (wt/vol) acrylamide separating gels and 10% (wt/vol) acrylamide stacking gels, and purified Dps was included in all blots as an internal standard for normalization of results. Detection of DnaK by Western blot analysis was performed as described previously (2).
RESULTS
Growth of the dnaK mutant.
Strains containing the ΔdnaK52 mutation can be readily cultured at 30°C. However, even at this reduced temperature, this strain continues to exhibit a reduced growth rate and a filamentous morphology in exponential phase (6, 7). To minimize the possible contribution of growth-related dnaK mutant defects on the starvation response, additional growth conditions which overcame these phenotypes were identified. We have demonstrated previously that the filamentous phenotype is suppressed during growth in a defined medium (52). In addition, more-gentle culture agitation overcame the defect in growth rate. Excessive agitation of the cultures (240 rpm) decreased the specific growth rate (k = 0.28) and final cell densities (1.5 × 108 CFU/ml) of the dnaK mutant cells relative to those of an otherwise isogenic wild-type strain. Consequently, the results presented here utilized the slower agitation rate of 150 rpm. At this rate of agitation, all strains examined had identical specific growth rates (k = 0.693) and cell densities (3.0 × 108 CFU/ml) at the onset of starvation. Stationary phase was elicited by depletion of growth-limiting levels of glucose. Twenty-four hours after the onset of starvation, the variation in cell numbers between the wild type and the mutant strains was less than 15%.
Starvation-induced catalase activity in dnaK and rpoS mutant strains.
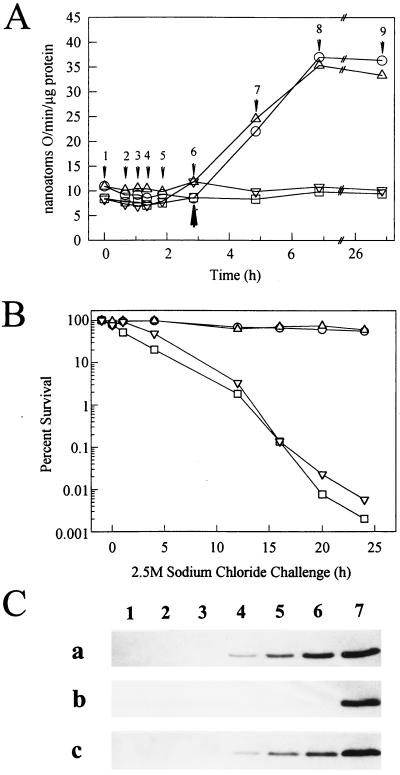
Levels of catalase increase in the stationary phase and result from elevated expression of katE (46). Catalase was therefore examined as an additional indicator of the cellular response to starvation in the dnaK mutant strain. Catalase-specific activity was similar among the four strains during growth (Fig. 1A, arrows 1 through 6). Differential rates of catalase synthesis produced during exponential phase were 9.4 nanoatoms of O/min/μg of protein for the wild-type strain, 8.8 for the dnaK mutant strain, 8.8 for the rpoS mutant strain, and 9.4 for the complemented dnaK mutant strain. In stationary phase, the wild-type and complemented dnaK mutant strains exhibited increased levels of catalase activity reaching 35 nanoatoms of O/min/μg of protein (Fig. 1A). However, the catalase activity of the dnaK and rpoS mutants did not increase in response to starvation and instead remained at prestarvation levels. By 4 h after the onset of starvation, the wild-type strain had 3.5-fold more catalase activity than the rpoS or dnaK mutant strains. The increased catalase activity in these strains remained evident even after 24 h in stationary phase.
FIG. 1.
Catalase activity, salt tolerance, and Dps/PexB levels during growth and starvation in wild-type and dnaK mutant strains. (A) Catalase activity of exponential-phase and glucose-starved cells. The strains are PBL500 (wild type; circles), PBL501 (ΔdnaK52; squares), PBL504 (ΔdnaK52/pPtac::dnaK+J+; triangles), and PBL503 (ΔmutS-rpoS458; inverted triangles). Cell densities were as follows: 0.1 (arrow 1), 0.15 (arrow 2), 0.2 (arrow 3), 0.25 (arrow 4), 0.4 (arrow 5), 0.6 starvation onset (arrow 6), 2 h after onset (arrow 7), 4 h after onset (arrow 8), and 24 h after onset (arrow 9). The onset of starvation is indicated (closed arrow). The values shown are the averages of three replicates. (B) Salt tolerance in glucose-starved cells. Cultures were examined following 24 h of carbon starvation for resistance to killing by exposure to 2.5 M sodium chloride for the times indicated. The strains are indicated as for panel A. (C) Levels of Dps/PexB. The strains used were PBL500 (wild type) (a), PBL501 (ΔdnaK52) (b), and PBL504 (ΔdnaK52/pPtac::dnaK+J+) (c). Western blots were probed with anti-Dps polyclonal antisera. Lanes: 1 and 2, exponential growth (A, arrows 1 and 4); 3, onset of starvation (arrow 6); 4, 2 h after starvation onset (arrow 7); 5, 4 h after onset (arrow 8); 6, 24 h after onset (arrow 9); and 7, purified Dps protein standard.
Starvation responses of dnaK and rpoS mutant strains to sodium chloride exposure.
dnaK mutants are sensitive to sodium chloride exposure in exponential phase (41), while rpoS mutants are sensitive to this stress in the stationary phase (38). However, the response of the dnaK mutant to this challenge in the stationary phase was unknown. To examine this question, the wild type, the otherwise isogenic dnaK mutant, the rpoS mutant, and the complemented dnaK mutant were subjected to 24 h of starvation and then exposed to 2.5 M sodium chloride. The cultures were then incubated further, with continued starvation, and periodically sampled to determine the number of viable cells (Fig. 1B). The wild-type and complemented dnaK mutant strains retained greater than 50% viability over a 24-h period. However, the viability of the dnaK and rpoS mutant strains decreased to less than 0.01% of untreated controls at nearly identical rates of 4.2%/h. These results indicate that DnaK is required for starvation-induced osmotolerance.
Levels of the rpoS-dependent protein, Dps, in starving dnaK mutants.
The results of the sodium chloride tolerance and catalase activity experiments suggested that there may be a more direct link between DnaK and RpoS in starving cells. To explore this possibility further, expression of an RpoS-dependent gene was examined. The dps gene (1, 31) encodes a 19-kDa DNA binding protein whose induction in stationary phase requires rpoS. Cell extracts derived from wild-type, dnaK mutant, and dnaK-complemented cultures in the exponential phase, at the onset of starvation, and at times thereafter were examined for levels of the Dps protein with anti-Dps polyclonal antibodies (Fig. 1C). Dps was readily detected after 2 h following the onset of starvation in the wild-type strain (Fig. 1C, blot a, lane 4) and in the complemented dnaK mutant (Fig. 1C, blot c, lane 4) and increased to maximal levels by 24 h (lane 6). However, Dps was undetectable in the dnaK mutant even 24 h after the onset of starvation (Fig. 1C, blot b, lane 6).
Accumulation of RpoS in a dnaK mutant strain.
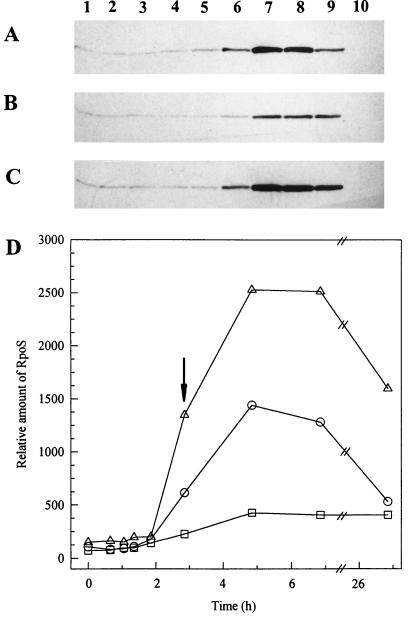
One mechanism to explain the phenotypic overlap between dnaK and rpoS mutants is that DnaK deficiency results in a reduction in RpoS activity or concentration and therefore reduces RpoS-dependent gene expression. This possibility was examined by measuring levels of RpoS in the wild-type strain (Fig. 2A), the dnaK mutant strain (Fig. 2B), and the complemented dnaK mutant strain (Fig. 2C) by Western blot analysis with an RpoS-specific monoclonal antibody. Levels of RpoS during exponential phase in the dnaK mutant strain, when adjusted for total protein, were equal to the otherwise isogenic wild-type strain. In response to starvation, RpoS levels increased 10-fold in the wild-type strain, while in the dnaK mutant strain, rpoS levels increased slightly but overall remained at a lower level. At the onset of starvation in the dnaK mutant strain, RpoS levels were 3.5-fold lower than the levels in the wild-type strain (Fig. 2D). Interestingly, in the complemented dnaK mutant strain, RpoS levels were nearly twofold higher than those in the wild-type strain (Fig. 2D). This last result is consistent with an increase in levels of DnaK which occurs in that strain in stationary phase, resulting from elevated expression from the plasmid-encoded Ptac promoter (2). Western blot analysis indicated that at the time of the maximal increase in RpoS abundance, DnaK levels were fourfold greater than in the wild-type strain (data not shown). Twenty-four hours after the onset of starvation, the level of RpoS in the wild-type and the complemented dnaK mutant strains decreased by 33 and 50%, respectively; however, no change in RpoS levels was observed in the dnaK mutant strain.
FIG. 2.
Accumulation of RpoS in exponential- and stationary-phase wild-type and dnaK mutant cells. Western blots of total cell extracts of PBL500 (wild type) (A), PBL501 (ΔdnaK52) (B), and PBL504 (ΔdnaK52/pPtac::dnaK+J+) (C) probed with anti-RpoS monoclonal antisera. Lanes: 1 to 5, exponential phase (Fig. 1A, arrows 1 through 5); 6, onset of starvation; 7, 2 h after onset; 8, 4 h after onset; 9, 24 h after onset; 10, PBL503 (ΔmutS-rpoS458) 2 h after onset. (D) Densitometric analysis of the bands shown in panels A through C. Symbols: circles, PBL500; squares, PBL501; triangles, PBL504. Five micrograms of protein was loaded per lane. Blots were developed to achieve equivalent band intensity of a constant amount of an RpoS-containing extract (PBL504, lanes not shown). Signal intensity was linear within the range of detection as determined by regression analysis of PBL504 extracts (r2 = 0.99). The onset of glucose starvation is indicated by the arrow.
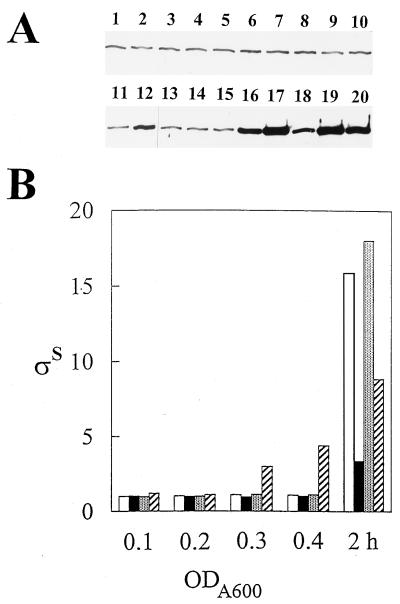
The exponential-phase RpoS levels observed in the dnaK mutant were not elevated relative to those of the otherwise isogenic wild-type strain, in contrast to a previous report (44). In addition, the reduction in starvation-induced RpoS levels reported here was significantly more pronounced (threefold versus 40%) than reported previously. To resolve these differences, the consequence of more-rapid culture agitation on RpoS levels in the dnaK mutant was examined (Fig. 3). Duplicate cultures of the wild type (PBL500) and the dnaK mutant (PBL501) were sampled periodically for RpoS determination, and then one of each of the duplicate cultures was shifted to a more rapid rate of agitation. RpoS levels increased rapidly upon the shift in the dnaK mutant but not in the wild-type strain, indicating that this condition imposes some sort of RpoS-inducing stress. This result is consistent with the reduction in growth exhibited by the dnaK mutant and likely explains why RpoS levels were highly induced in a dnaK mutant strain in a previous study (44).
FIG. 3.
Induction of RpoS levels by increased culture agitation in a dnaK mutant. (A) Western blot of samples. Odd-numbered lanes, PBL500 (wild type); even-numbered lanes, PBL501 (ΔdnaK52). In lanes 1, 2, 5, 6, 9, 10, 13, 14, 17, and 18, cultures were only agitated slowly. In lanes 3, 4, 7, 8, 11, 12, 15, 16, 19, and 20, cultures were agitated slowly and then rapidly. Cell densities at an optical density at 600 nm were 0.1 (lanes 1 through 4), 0.2 (lanes 5 through 8), 0.3 (lanes 9 through 12), 0.4 (lanes 13 through 16), and 2 h after the onset of starvation (lanes 17 through 20). (B) Levels of RpoS determined from the data shown in panel A. Values are normalized to the levels in the wild-type strain at early exponential phase (optical density at 600 nm [ODA600 = 0.1). Open bars, wild type, slow agitation; closed bars, ΔdnaK52, slow agitation; stippled bars, wild type, shifted agitation; hatched bars, ΔdnaK52, shifted agitation.
Role of RpoS in levels of the DnaK protein.
Since RpoS affects the synthesis of many proteins, levels of DnaK were examined in an rpoS mutant to better define the epistatic relationship between these genes. DnaK levels were examined in the wild-type strain and in the rpoS mutant during exponential growth and in response to starvation. Western blot analysis with the anti-DnaK monoclonal antibody 2G5 (28) indicated that the levels of DnaK were not significantly affected by RpoS deficiency either during growth or in response to carbon starvation (data not shown). A starvation-induced, twofold increase in DnaK levels was evident in both strains in comparisons between levels produced during exponential growth and those produced 24 h after the onset of glucose starvation.
RpoS stability in a dnaK mutant strain.
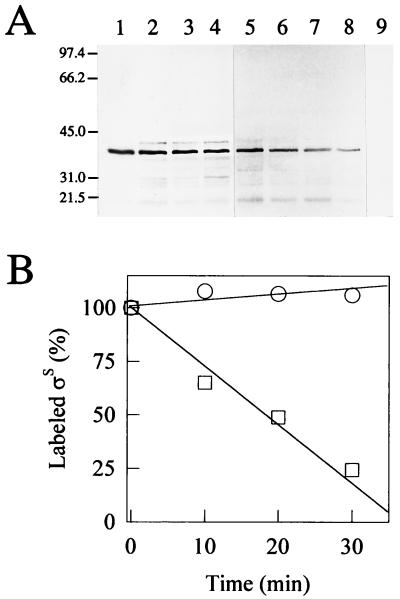
Reduced RpoS accumulation in the starving dnaK mutant strain could reflect defects in either RpoS stability and/or synthesis. To look for DnaK-dependent changes in RpoS protein turnover, the half-life of RpoS was determined. The onset of starvation was selected for analysis because a large difference in RpoS levels was evident in comparisons between the wild type and the dnaK mutant (Fig. 2D). At the onset of starvation, cultures were transiently radiolabeled and RpoS was recovered by immunoprecipitation at specific times thereafter. The immunoprecipitates were analyzed by SDS-PAGE, and the resulting autoradiograms (Fig. 4A) were quantitated (Fig. 4B). In the wild-type strain (Fig. 4A, lanes 1 through 4), RpoS remained fully stable for the duration of the experiment (30 min), while in the dnaK mutant, the half-life of RpoS was approximately 20 min (Fig. 4A, lanes 5 through 8). No protein was detectable when identical procedures were performed on the rpoS deletion mutant PBL503 (Fig. 4A, lane 9). These results indicate that DnaK deficiency reduces starvation-induced accumulation of RpoS by enhancing RpoS turnover.
FIG. 4.
RpoS stability at the onset of starvation in wild-type and dnaK mutant cells. (A) Autoradiograms of RpoS immunoprecipitates. Lanes: 1 through 4, PBL500 (wild type); 5 through 8, PBL501 (ΔdnaK52); 9, PBL503 (ΔmutS-rpoS458). Lanes and the duration of the methionine chase, respectively, were as follows: 1, 5 and 9, 0 min; 2 and 6, 10 min; 3 and 7, 20 min; and 4 and 8, 30 min. (B) Densitometric analysis of the bands shown in panel A. Symbols: circles, PBL500 (wild type); squares, PBL501 (ΔdnaK52). Values given are relative to time 0.
RpoH (ς32) suppressors restore a subset of starvation-induced responses.
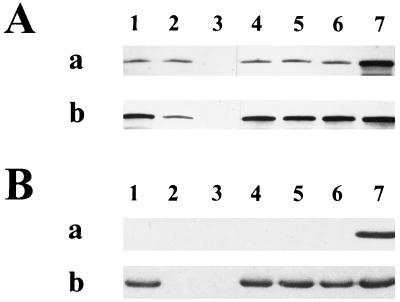
To better understand the relationship between DnaK and RpoS in regulation of the starvation response to carbon deprivation, dnaK mutant suppressors which had restored wild-type levels of RpoS were isolated (Fig. 5A). An internal control consisting of extracts of PBL504 obtained following 2 h of glucose starvation was included in each blot to normalize absolute amounts of RpoS. The rpoH suppressor mutants were recovered as papillae after prolonged incubation on solid medium (6, 8). The use of such strains circumvents RpoS effects on the starvation response, which might otherwise obscure the role of DnaK. Several such mutants were isolated in the ΔdnaK52 PBL501 background and are designated rpoH suppressors—1, 2, or 3. They were mapped by phage P1 cotransduction with linked transposon insertions. To verify their identity further, plasmid pDS2 (18), encoding a Ptac::rpoH gene, was used to complement the suppressor isolates. Such suppressors exhibit a defective heat shock response and can be categorized by the severity of this defect (8). Only those alleles which exhibit no heat shock response at all fail to grow at 43.5°C on solid medium (8). Additional Western blot analysis of these three rpoH suppressor alleles with anti-Dps polyclonal antibodies revealed that levels of Dps were restored to wild-type levels in the dnaK mutant strains containing the rpoH suppressors (Fig. 5B). Starvation-induced levels of catalase production were also restored to wild-type levels in the three suppressor isolates (data not shown). As the restoration of RpoS levels and rpoS-regulated gene expression in the dnaK mutant strain by the rpoH-1 to rpoH-3 suppressors was in apparent contradiction to a previous report (44), the possible allelic difference between these suppressor mutations was examined. The rpoH-1 mutation was selected as a representative and analyzed for heat shock protein induction during heat shock and for growth at elevated temperature (Table 2). The rpoH-1 mutant fails to induce GroEL synthesis following a temperature shift from 30 to 42°C or to form colonies at 43.5°C. It is therefore a stronger suppressor of the dnaK mutation than the allele (sidB1) used previously (8, 44). Taken together, the results presented here and those previously described (44) support the idea that at least some of the effects of the dnaK mutation on starvation-related processes are the result of effects on RpoS mediated by other heat shock proteins. In addition, an rpoS dnaK double mutant starved for 24 h exhibited sensitivity to sodium chloride treatment equivalent to that of mutants containing single mutations in either gene alone, suggesting that the role of dnaK in starvation-induced osmotolerance is mediated entirely through RpoS.
FIG. 5.
Western blot analysis of levels of RpoS and Dps during growth and starvation in wild-type, dnaK, rpoS, and rpoH suppressor strains. (A) Levels of RpoS. Western blot of cell extracts during growth (a) and 24 h after the onset of glucose starvation (b). Blots were probed with anti-RpoS monoclonal antibody. Lanes: 1, PBL500 (wild type); 2, PBL501 (ΔdnaK52); 3, PBL503 (ΔmutS-rpoS458); 4, PBL622 (ΔdnaK52 rpoH-1); 5, PBL623 (ΔdnaK52 rpoH-2); 6, PBL624 (ΔdnaK52 rpoH-3); 7, PBL504 (ΔdnaK52/pPtac::dnaK+ J+) cell extracts from 2 h after the onset of starvation in both panels. (B) Levels of Dps. Western blot of cell extracts during growth (a) and after 24 h of glucose starvation (b). Blots were probed with anti-Dps polyclonal antibodies. Lanes are as indicated for panel A, except that lane 7 contains purified Dps protein standard.
TABLE 2.
Characterization of the rpoH-1 mutant allele
| Strain (genotype) | GroEL inductiona | Growth at 43.5°Cb |
|---|---|---|
| PBL500 (wild type) | 10 | + |
| PBL501 (ΔdnaK52) | 2 | − |
| PBL622 (ΔdnaK52 rpoH-1) | 1 | − |
| PBL713 (rpoH-1) | 1 | − |
The values indicated represent the ratio of GroEL protein produced after 5 min of exposure to 42°C relative to the amount present in cells grown at 30°C prior to the imposition of heat. GroEL levels were determined by densitometry of autoradiograms of 35S-pulse-labeled proteins fractionated by SDS-PAGE.
Growth was determined by the ability of each strain to form single colonies on rich-medium plates. +, growth; −, no growth.
An independent role for DnaK in starvation thermotolerance.
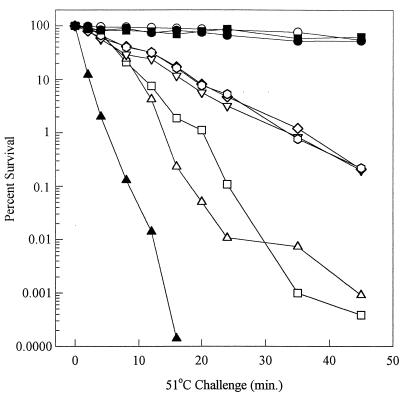
Examination of the thermotolerance of these strains indicates that DnaK and RpoS play important but distinct roles in starvation-induced resistance to heating. Elimination of either dnaK or rpoS in strains which are otherwise isogenic results in severe thermal killing of glucose-starved cells (Fig. 6, open squares and triangles). Wild-type cells, however, exhibited only slight reductions in viable counts for the duration of the 45-min treatment (Fig. 6, open circles). Interestingly, MC4100 (44) is 15-fold more sensitive to heat killing than the wild-type strain (PBL500) used in the present studies (data not shown). All three of the rpoH dnaK mutant suppressor strains exhibited only partial restoration of thermotolerance relative to that of their unsuppressed but otherwise isogenic dnaK mutant parent (Fig. 6, open inverted triangles, hexagons, and diamonds). Thus, these strains remained highly thermosensitive upon starvation for glucose. To test the possibility that continued thermosensitivity results directly from the rpoH mutation, the rpoH mutation in rpoH suppressor 1 was evaluated in a dnaK+ background. Two strain derivatives were constructed, PBL713, in which a wild-type allele of dnaK was introduced by transduction, and PBL711, which contained a plasmid-encoded wild-type dnaK allele. Examination of the starvation-induced thermosensitivity of these strains (Fig. 6, closed circles and closed squares) indicated that the rpoH mutation did not result in thermosensitivity following glucose starvation. Since RpoS levels are restored in the original rpoH suppressor strains, and expression of RpoS-dependent pathways such as Dps and catalase synthesis is normal, continued thermosensitivity of the rpoH dnaK double mutants indicates that DnaK is directly required for starvation thermotolerance. If two distinct pathways for starvation thermotolerance are present, one dependent upon RpoS and the other on DnaK, then a dnaK rpoS double mutant should exhibit greater heat killing than either single-mutant strain alone. Analysis of the thermotolerance of such a strain (Fig. 6, closed triangles) indicated that loss of both genes resulted in significantly greater killing than that observed in the absence of either gene alone. These results indicate that DnaK plays a distinct and RpoS-independent role in starvation thermotolerance.
FIG. 6.
Thermotolerance of glucose-starved wild-type, dnaK, rpoS, and rpoH suppressor strains. Cultures were examined following 24 h of starvation for resistance to killing by exposure to 51°C for the times indicated. The strains were as follows: PBL500 (wild type; open circles), PBL501 (ΔdnaK52; open triangles), PBL503 (ΔmutS-rpoS458; open squares), PBL603 (ΔdnaK52 rpoS13::Tn10; closed triangles), PBL622 (ΔdnaK52 rpoH-1; open diamonds), PBL623 (ΔdnaK52 rpoH-2; open hexagons), PBL624 (ΔdnaK52 rpoH-3; open inverted triangles), PBL713 (dnaK+ transductant of PBL622; closed circles), and PBL711 (PBL622/pPtac::dnaK+J+; closed inverted triangles).
DISCUSSION
The studies reported here focus on the role of DnaK in a stationary phase produced by starvation for carbon. Care was taken to minimize high cell densities in batch cultures through the use of minimal quantities of the limiting nutrient, glucose. Such measures represent an effort to mimic the physiological state confronted by bacteria in natural environments where high cell density conditions are generally thought less likely (43).
The dnaK mutant phenotypes observed in stationary phase result at least in part from a deficiency in the sigma factor RpoS. The reduction in stationary-phase RpoS levels (about threefold) is sufficient to preclude expression of the rpoS-dependent gene, dps (pexB), and therefore possibly other rpoS-dependent genes, such as those involved in mediating resistance to hydrogen peroxide, heat, and sodium chloride excess. This change in RpoS abundance parallels that seen during heat stress administered to growing cells, which is sufficient for changes in RpoS-dependent gene expression (26). A 10-fold increase in RpoS levels over those observed in exponentially growing cells was detected following the onset of starvation in the wild-type strain, PBL500, at 30°C. This is an increase of approximately twofold over those seen previously at 37°C (29, 56) and is consistent with other reports indicating that stationary-phase expression of rpoS is elevated at reduced cultivation temperatures (58). Thus, the pleiotropic spectrum of effects resulting from RpoS deficiency in the dnaK mutant strain offers some explanation for the apparent phenotypic overlap between dnaK and rpoS mutants.
The RpoS-related effects which result from DnaK deficiency are stationary phase specific. RpoS levels appear otherwise normal during exponential-phase growth under the conditions used. Growth conditions were employed to minimize certain dnaK mutant phenotypes. It has been reported that the ΔdnaK52 mutation results in a reduced growth rate (k = 0.28) compared to that of the wild type (k = 0.69) at 30°C (6). We also observed this phenomenon when the ΔdnaK52 mutant was agitated at 200 rpm or greater. In addition, RpoS levels are highly induced by this treatment (44), obscuring subsequent effects resulting from starvation. However, at agitation rates of 150 rpm or less, growth rates were equal to the otherwise isogenic wild-type strain. Growth at 30°C also was employed to avoid the dnaK mutant heat-sensitive growth phenotype (49).
Protein half-life measurements indicated that in the dnaK mutant, RpoS stability was reduced. These results suggest that DnaK somehow stabilizes RpoS in response to carbon starvation. Interestingly, overproduction of DnaK resulting from starvation-mediated derepression of a plasmid-encoded dnaK gene elevated stationary-phase RpoS levels nearly twofold above those in otherwise isogenic wild-type cells. Thus, variation in DnaK abundance results in corresponding changes in the stationary-phase levels of RpoS. Though such changes in RpoS levels were elicited by artificial manipulation of dnaK expression, DnaK levels do increase severalfold in wild-type cells upon carbon starvation as shown here and previously (52). Thus, DnaK may play a crucial role in adjusting stationary-phase RpoS levels in response to carbon starvation.
Overproduction of DnaK is selectively toxic in stationary-phase cells (2). This bactericidal effect was used as the basis of a selection to recover multicopy plasmid suppressors to better understand the toxicity of chaperone excess (53). Identification of these suppressors, such as 6-phosphogluconate dehydratase, may now be explained in part by the observations presented here. Activity of 6-phosphogluconate dehydratase is elevated in the stationary phase (32), while the next enzyme in this pathway, 2-keto-3-deoxy-6-phosphogluconate aldolase, is also increased in stationary phase (48). Perhaps elevated levels of 6-phosphogluconate dehydratase overcome deleterious levels of other enzymes precipitated by DnaK-mediated increases in RpoS by redirecting carbon flux through alternative routes in this pathway.
The results presented here indicate that DnaK controls the levels of the RpoS sigma factor. Since DnaK also controls the levels of the RpoH sigma factor (11), these results indicate that DnaK may coordinate the levels of multiple sigma factors in E. coli. A mechanism for such coordination mediated by a single protein has until now been lacking. However, it would seem critical for the maintenance of balanced gene expression during changing environmental conditions, such as those accompanying starvation.
It is at present unclear how DnaK might affect RpoS stability, though direct and/or indirect mechanisms may be operative. Deficiency of the heat shock protease, ClpX/P, elevates RpoS levels 3.2-fold during exponential-phase growth and 1.7- fold in response to starvation (56). Since starvation increases ClpX/P by 1.5-fold (56), inactivation or blockage of ClpX/P may be required for the starvation-induced increase in RpoS abundance. The epistatic relationship between ClpX/P and RssB further indicates a direct role for ClpX/P in RpoS abundance (51). Thus, the role of DnaK in RpoS stability might be mediated indirectly by altering levels of ClpX/P or RssB. Alternatively, as a direct interaction between DnaK and the heat shock sigma factor RpoH has been reported (40), physical contact between DnaK and RpoS also appears plausible.
RpoH levels are elevated in dnaK mutant strains (60); therefore, compensatory mutations which reduce but do not destroy RpoH activity are selected during prolonged storage on plates (6–8). Such rpoH mutant strains were isolated and studied here to probe the relationship between DnaK and RpoS. These rpoH dnaK double-mutant strains exhibit normal levels of RpoS, suggesting that the effect of the dnaK mutation on RpoS abundance is RpoH linked, perhaps through ClpX/P. Restoration of RpoS levels was accompanied by recovery of catalase and induction of Dps starvation, suggesting that at least these two starvation responses operate in an RpoS-dependent fashion. DnaK apparently indirectly alters their expression by varying levels of RpoS. Starvation-induced thermotolerance was, however, still defective in the rpoH dnaK mutant strains but normal in dnaK+ rpoH derivatives. This result suggests that DnaK plays a distinct and RpoS-independent role in starvation-induced thermotolerance. However, the relative contributions of the two proteins to the response appear quite unequal, suggesting that their actions during the development of the thermotolerant state are mechanistically unrelated.
ACKNOWLEDGMENTS
We thank Carol Gross, Abdul Matin, and Graham Walker for providing strains.
This work was supported by a grant from the Department of Energy to P.B. (DE-FG02-93ER61701).
REFERENCES
- 1.Almiron M, Link A J, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 2.Blum P, Ory J, Bauernfeind J, Krska J. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J Bacteriol. 1992;174:7436–7444. doi: 10.1128/jb.174.22.7436-7444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum, P. H. Molecular genetics of the bacterial stationary phase. In R. Morita (ed.), Bacteria in the oligotrophic environment, with special emphasis on starvation survival, in press. Chapman and Hall, New York, N.Y.
- 4.Blum P H, Jovanovich S B, McCann M P, Schultz J E, Lesley S A, Burgess R R, Matin A. Cloning and in vivo and in vitro regulation of cyclic AMP-dependent carbon starvation genes from Escherichia coli. J Bacteriol. 1990;172:3813–3820. doi: 10.1128/jb.172.7.3813-3820.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochner B R, Huang H-C, Schieven G L, Ames B N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukau B, Walker G C. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate role for heat shock protein in normal metabolism. J Bacteriol. 1989;171:2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukau B, Walker G C. Deletion ΔdnaK52 mutants of Escherichia coli have defects in chromosome segregation and plasmid maintenance at normal growth temperatures. J Bacteriol. 1989;171:6030–6038. doi: 10.1128/jb.171.11.6030-6038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukau B, Walker G C. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 1990;9:4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox E C, Degnen G E, Scheppe M L. Mutator gene studies in Escherichia coli: the mutS gene. Genetics. 1972;72:551–567. doi: 10.1093/genetics/72.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig E A, Gross C A. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- 12.Davis B D, Kuger S M, Tai P C. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliker P R, Frazier W C. Influence of time and temperature of incubation on heat resistance of Escherichia coli. J Bacteriol. 1938;36:83–97. doi: 10.1128/jb.36.1.83-98.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elthon T E, McIntosh L. Identification of the alternative terminal oxidase of higher plant mitochrondria. Proc Natl Acad Sci USA. 1987;84:8399–8403. doi: 10.1073/pnas.84.23.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentry D R, Hernandez V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of the stationary-phase sigma factor ςS is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgopoulos C. The emergence of the chaperone machines. Trends Biochem Genet. 1992;17:295–299. doi: 10.1016/0968-0004(92)90439-g. [DOI] [PubMed] [Google Scholar]
- 17.Goodrich-Blair H, Uria-Nickelsen M, Kolter R. Regulation of gene expression in stationary phase. In: Lin E C C, Simon Lynch A, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman and Hall; 1996. pp. 571–583. [Google Scholar]
- 18.Grossman A D, Straus D B, Walter W A, Gross C A. ς32 synthesis can regulate the synthesis of heat shock proteins in Escherichia coli. Genes Dev. 1987;1:179–184. doi: 10.1101/gad.1.2.179. [DOI] [PubMed] [Google Scholar]
- 19.Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 20.Hengge-Aronis R. Stationary-phase gene regulation. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 21.Hengge-Aronis R, Klein W, Lange R, Rimmele M, Boos W. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J Bacteriol. 1991;173:7918–7924. doi: 10.1128/jb.173.24.7918-7924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengge-Aronis R, Lange R, Henneberg N, Fischer D. Osmotic regulation of rpoS-dependent genes in Escherichia coli. J Bacteriol. 1993;175:259–265. doi: 10.1128/jb.175.1.259-265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanonva A, Miller C, Glinsky G, Eisenstark A. Role of rpoS (katF) in oxyR-independent regulation of hydroperoxidase I in Escherichia coli. Mol Microbiol. 1994;12:571–578. doi: 10.1111/j.1365-2958.1994.tb01043.x. [DOI] [PubMed] [Google Scholar]
- 24.Ivanova A, Renshaw M, Ramareddy V G, Eisenstark A. DNA base sequence variability in katF (putative sigma factor) gene of Escherichia coli. Nucleic Acids Res. 1992;20:5479–5480. doi: 10.1093/nar/20.20.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins D E, Schultz J E, Matin A. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J Bacteriol. 1988;170:3910–3914. doi: 10.1128/jb.170.9.3910-3914.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jishage M, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of ς70 and ς38. J Bacteriol. 1995;177:6832–6835. doi: 10.1128/jb.177.23.6832-6835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 28.Krska J, Elthon T, Blum P. Monoclonal antibody recognition and function of a DnaK (HSP70) epitope found in gram-negative bacteria. J Bacteriol. 1993;175:6433–6440. doi: 10.1128/jb.175.20.6433-6440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange R, Hengge-Aronis R. The cellular concentration of the ςS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 30.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor ςS. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lomovskaya O L, Kidwell J P, Matin A. Characterization of the ς38-dependent expression of a core Escherichia coli starvation gene, pexB. J Bacteriol. 1994;176:3928–3935. doi: 10.1128/jb.176.13.3928-3935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loomis W F, Magasanik B. Nature of the effector of catabolite repression of β-galactosidase in Escherichia coli. J Bacteriol. 1966;92:170–177. doi: 10.1128/jb.92.1.170-177.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowen P C, Hengge-Aronis R. The role of the sigma factor ςS (katF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 34.Lowen P C, Triggs B L. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J Bacteriol. 1984;160:668–675. doi: 10.1128/jb.160.2.668-675.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991;5:3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 36.Matin A, Auger E A, Blum P H, Schultz J E. Genetic basis of starvation survival in nondifferentiating bacteria. Annu Rev Microbiol. 1989;43:293–316. doi: 10.1146/annurev.mi.43.100189.001453. [DOI] [PubMed] [Google Scholar]
- 37.Mayhew M, Hartl F-U. Molecular chaperone proteins. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 922–937. [Google Scholar]
- 38.McCann M P, Kidwell J P, Matin A. The putative ς factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCann M P, Fraley C D, Matin A. The putative ς factor KatF is regulated posttranscriptionally during carbon starvation. J Bacteriol. 1993;175:2143–2149. doi: 10.1128/jb.175.7.2143-2149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarty J S, Rudiger S, Schonfeld H-J, Schneider-Mergener J, Nakahigashi K, Yura T, Bukau B. Regulatory region C of the E. coli heat shock transcription factor, ς32, constitutes a DnaK binding site and is conserved among eubacteria. J Mol Biol. 1996;256:829–837. doi: 10.1006/jmbi.1996.0129. [DOI] [PubMed] [Google Scholar]
- 41.Meury J, Kohiyama M. Role of heat shock protein DnaK in osmotic adaptation of Escherichia coli. J Bacteriol. 1991;173:4404–4410. doi: 10.1128/jb.173.14.4404-4410.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1972. [Google Scholar]
- 43.Morita R. Bioavailability of energy and the starvation state. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 1–23. [Google Scholar]
- 44.Muffler A, Barth M, Marschall C, Hengge-Aronis R. Heat shock regulation of ςS turnover: a role for DnaK and relationship between stress responses mediated by ςS and ς32 in Escherichia coli. J Bacteriol. 1997;179:445–452. doi: 10.1128/jb.179.2.445-452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the ςS subunit of RNA polymerase in Escherichia coli. EMBO J. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- 46.Mulvey M R, Switala J, Borys A, Lowen P C. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990;172:6713–6720. doi: 10.1128/jb.172.12.6713-6720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen L H, Jensen D B, Thompson N E, Gentry D R, Burgess R R. In vitro functional characterization of overproduced Escherichia coli katF/rpoS gene product. Biochemistry. 1993;32:11112–11117. doi: 10.1021/bi00092a021. [DOI] [PubMed] [Google Scholar]
- 48.Nystrom T. Role of guanosine tetraphosphate in gene expression and the survival of glucose or seryl-tRNA starved cells of Escherichia coli. Mol Gen Genet. 1994;245:355–362. doi: 10.1007/BF00290116. [DOI] [PubMed] [Google Scholar]
- 49.Paek K-H, Walker G C. Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol. 1987;169:283–290. doi: 10.1128/jb.169.1.283-290.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmer D T, Blum P H, Artz S W. Effects of hisT mutation of Salmonella typhimurium on translation elongation rate. J Bacteriol. 1983;153:357–363. doi: 10.1128/jb.153.1.357-363.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pratt L A, Silhavy T J. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci USA. 1996;93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rockabrand D, Arthur T, Korinek G, Livers K, Blum P. An essential role for the Escherichia coli DnaK protein in starvation-induced thermotolerance, H2O2 resistance, and reductive division. J Bacteriol. 1995;177:3695–3703. doi: 10.1128/jb.177.13.3695-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rockabrand D, Blum P. Multicopy plasmid suppression of stationary phase chaperone toxicity in Escherichia coli by phosphogluconate dehydratase and the N-terminus of DnaK. Mol Gen Genet. 1995;249:498–506. doi: 10.1007/BF00290575. [DOI] [PubMed] [Google Scholar]
- 54.Sammartano L J, Tuveson R W, Davenport R. Control of sensitivity to inactivation by H2O2 and broad-spectrum near-UV radiation by the Escherichia coli katF locus. J Bacteriol. 1986;168:13–21. doi: 10.1128/jb.168.1.13-21.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schleif R, Hess W, Finkelstein S, Ellis D. Induction kinetics of the l-arabinose operon of Escherichia coli. J Bacteriol. 1973;115:9–14. doi: 10.1128/jb.115.1.9-14.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schweder T, Lee K-H, Lomovskaya O, Matin A. Regulation of Escherichia coli starvation factor (ςS) by ClpXP protease. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sledjeski D D, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 59.Spence J, Cegielska A, Georgopolous C. Role of Escherichia coli heat shock protein DnaK and HtpG (C62.5) in response to nutritional deprivation. J Bacteriol. 1990;172:7157–7166. doi: 10.1128/jb.172.12.7157-7166.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Straus D, Walter W, Gross C A. DnaK, DnaJ and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of ς32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 61.Takayanagi Y, Tanaka K, Takahashi H. Structure of the 5′-upstream region and the regulation of the rpoS gene of Escherichia coli. Mol Gen Genet. 1994;243:525–531. doi: 10.1007/BF00284200. [DOI] [PubMed] [Google Scholar]
- 62.Yamagishi M, Matsushima H, Wada A, Sakagami M, Fujita N, Ishihama A. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]