Abstract
Epigenetic alterations are a primary hallmark of ageing. In mammals, age-related epigenetic changes alter gene expression profiles, disrupt cellular homeostasis and physiological functions and, therefore, promote ageing. It remains unclear whether ageing is also driven by epigenetic mechanisms in invertebrates. Here, we used a pharmacological hypomethylating agent (RG108) to evaluate the effects of DNA methylation (DNAme) on lifespan in an insect—the bumblebee Bombus terrestris. RG108 extended mean lifespan by 43% and induced the differential methylation of genes involved in hallmarks of ageing, including DNA damage repair and chromatin organization. Furthermore, the longevity gene sirt1 was overexpressed following the treatment. Functional experiments demonstrated that SIRT1 protein activity was positively associated with lifespan. Overall, our study indicates that epigenetic mechanisms are conserved regulators of lifespan in both vertebrates and invertebrates and provides new insights into how DNAme is involved in the ageing process in insects.
Keywords: epigenetics, ageing, DNA methylation, insects, sirtuins, bumblebee
1. Introduction
Ageing is a natural process during which organisms experience time-dependent declines in their molecular, cellular and physiological functions [1–4]. Age-related epigenetic changes are one of the primary hallmarks of ageing [2]. Under normal conditions, epigenetic mechanisms mediate gene–environment interactions; thus, they serve an important adaptive function because they enable organisms to phenotypically adjust to environmental conditions [5–7]. However, recent research has shown that the accumulation of epigenetic alterations over time is a particularly important driver of the ageing process because they affect gene expression profiles, consequently leading to disruptions in cellular and physiological homeostasis [8–10]. Accordingly, ageing has been shown to be modulable by manipulating the epigenome. In mammals, restoring early-life epigenetic patterns was shown to promote in vivo rejuvenation by ‘resetting’ transcriptomic profiles and cellular functions [9,10].
The epigenetic mark cytosine DNA methylation (DNAme) is widespread in eukaryotes [11] and has been robustly associated with the ageing process [12,13]. Age-related changes in DNAme have been observed in multiple vertebrate groups, including fishes [14], amphibians [15], birds [16] and mammals [17], suggesting that the association between ageing and this epigenetic mark is evolutionarily conserved. By contrast, to our knowledge, only one study explored the role of DNAme in ageing in invertebrates and reported tentative yet mixed evidence indicating that DNAme might play a role in lifespan regulation in the honeybee Apis mellifera [18]. Therefore, it remains to be unequivocally determined whether DNAme is also involved in ageing in invertebrates and, if so, what are the mechanisms by which this epigenetic mark affects this process.
Epigenetic regulation of lifespan has been consistently shown to be modulated by genes from the highly conserved silent information regulator of transcription (sirtuins, sirt) family, whose members act as master regulators of multiple cellular processes [19,20], including some related to ageing [21], such as DNA damage repair [22], epigenetic regulation of gene expression [23], proteostasis [24] and nutrient sensing [25]. For instance, the protein SIRT1 interacts with DNA-methyltransferase 1 (DNMT1) to regulate DNAme profiles [26]. Experimentally increasing sirt1 expression or SIRT1 activity has been shown to extend lifespan in several model organisms [27], including yeasts [28], nematodes [29], fruit flies [29] and mice [30].
Social Hymenoptera (i.e. ants, social bees and social wasps) are particularly useful models for studying the epigenetic regulation of ageing because the same genetic background can lead to phenotypically distinct groups with contrasting lifespans. The female castes—reproductive queens and non-reproductive workers—provide a dramatic illustration of this point. Queens and workers of social Hymenoptera exhibit the largest intraspecific difference in lifespan ever observed in animals. For instance, in some ant species, queens may live for more than 20 years, while workers die after just a few months [31,32]. Caste fate is determined during larval development and is shaped by environmental and social factors (e.g. food quantity and/or quality, presence of the queen and/or brood) that can cause the same baseline genome to express itself along different developmental paths [33–35]. Furthermore, adult life expectancy can be dynamically influenced by environmental factors, such as diet [36], reproductive status [37], colony size [38], social task [39] or parasite load [40]. Such dramatic plasticity in female lifespan suggests that epigenetic mechanisms could mediate ageing in social Hymenoptera. Indeed, several studies have found an association between individual life expectancy and DNAme patterns in social insects [41–43]. However, research has yet to explore whether this relationship is causal in nature.
Here, we show that pharmacological alterations of DNAme increased worker lifespan in an insect—the bumblebee Bombus terrestris. Experimentally induced longer lifespans were associated with changes in the methylation status of genes involved in several molecular and cellular processes related to ageing, including DNA damage repair, chromatin organization, proteostasis and nutrient sensing, as well as changes in the expression level of the longevity gene sirt1.
2. Methods
(a) . Model organism
The buff-tailed bumblebee (Bombus terrestris) is a particularly well-suited biological model for studying the epigenetic regulation of ageing for several reasons. First, females of this species exhibit lifespan plasticity despite having the same genotype. Thus, lifespan variations must arise from epigenetic differences, at least in part. Second, B. terrestris queens are singly mated and produce workers that are also their closely related daughters (r = 0.75), which greatly reduces the potential for genetic confounding. Third, workers have relatively short lifespans (four to five weeks), facilitating research on survival patterns [44].
In all experiments, we used B. terrestris workers of the same age obtained from 12 different queenright colonies (supplier: Biobest, Westerlo, Belgium). Queenless microcolonies were formed by randomly selecting 5 or 10 workers (depending on the experiment, see below) from the same natal colony to avoid aggressive behaviour. The microcolonies were not given any brood. Previous studies have shown that, in B. terrestris, the presence of five workers is sufficient to consistently reproduce colony social dynamics [45]. The microcolonies were given ad libitum quantities of Salix pollen (Ruchers de Lorraine, Nancy, France) and sugar syrup (Biogluc, Biobest, Westerlo, Belgium). They were maintained in constant darkness (temperature: 26°C, relative humidity: 50–60%). All the assays were performed under red light conditions to minimize disturbances.
(b) . Effect of RG108 treatment on worker lifespan
To investigate the role of DNAme in lifespan regulation, we treated B. terrestris workers with the pharmacological hypomethylating agent RG108 (MedChemExpress, HY-13642). RG108 is a non-nucleoside, specific inhibitor of DNMT1 [46], which has been reported to have a mild, but positive effect on lifespan in honeybees [18] and to show anti-senescent properties in cell cultures [47]. One-week-old workers were exposed to a single dose of RG108. We topically applied 2 µl of RG108 (0.2 mM diluted in DMSO) on the thorax of treatment workers and 2 µl of DMSO (solvent) on the thorax of control workers. We used 14 microcolonies composed of a mix of 10 treated and untread workers each: five treatment workers and five control workers. Treatment and control workers were marked on the wings with different colors. For each microcolony, worker mortality was recorded every 24 h until all the workers had died.
Data normality and heteroscedasticity were tested using the Shapiro–Wilk test and Levene's test, respectively. Mean lifespan and survival curves of workers were compared using the Mann–Whitney U-test and mixed-effects cox model (coxme package in R [48]), respectively. Colony was included as a random factor in mixed-effects model. The statistical analyses were performed using R environment [48].
(c) . Food intake following RG108 treatment
Dietary intake is a robust and highly conserved determinant of lifespan in animals [49,50]. Therefore, we explored whether the hypomethylating agent RG108 affected dietary intake.
Food intake was compared for 50 days between treatment microcolonies (n = 19) and control microcolonies (n = 16). Each microcolony contained five workers and was given sugar syrup (carbohydrate source; Biogluc, Biobest, Westerlo, Belgium) and pollen (protein and lipid source; Salix sp. Rucher de Lorraine, Nancy, France) ad libitum. Food intake was quantified twice a week by weighing the amount of sugar and pollen consumed by the workers. To avoid potential alterations in dietary behaviour due to changes in social structure [51], food consumption was only quantified for colonies containing at least two (out of the five) workers. The amount of food consumed per worker was calculated by dividing the microcolony's total consumption by its number of workers at each intake measurement point.
We used generalized linear models (GLM command in stats package [48]) to evaluate the effect of the RG108 treatment on the intake of sugar syrup and pollen. The latter were normally distributed (model = Gaussian, link = identity). The colony of origin was included as a random factor in the GLM models.
(d) . Genome-wide DNA methylation patterns
We evaluated the genome-wide modifications in DNAme induced by the RG108 treatment by conducting whole-genome bisulfite sequencing (WGBS) of the entire bodies of treatment and control workers from the feeding experiment. To differentiate between the treatment's short- and long-term effects, we compared the methylomes of workers one week and four weeks post treatment. WGBS was performed on the workers' entire bodies; thus, we could not identify tissue-specific changes in DNAme. However, this approach was intentionally adopted to capture either the combined methylation pattern for multiple tissues or tissue-specific methylation patterns that were pronounced enough to produce a signal strong enough to be detected. At each time point, we sampled whole-body DNA from three randomly selected workers in the treatment and control groups for WGBS. The DNA was extracted using a NucleoSpin Tissue Kit (Macherey-Nagel, cat. no. 740952) and was then sent to the Beijing Genomics Institute (BGI) for library preparation and WGBS. Bisulfite conversion and WGBS were performed using an EZ DNA Methylation-Gold Kit (Zymo Research) and a DNBSEQ sequencing platform (DNBSEQ Technology), respectively. WGBS included an unmethylated lambda phage to control for conversion. Sequencing was paired for reads of 100 bp. Read quality was verified using FastQC v. 0.11.9 [52]. Additional trimming of the sequenced reads was performed using TrimGalore v. 0.6.6 [53]. The cleaned reads were aligned with the B. terrestris reference genome (v. 1.2; GCF_910591885.1) using Bismark v. 0.24.0 [54]; only the paired-end alignments with the best unique hits were kept, while duplicates were removed. The recommended default parameters were used for TrimGalore and Bismark. We evaluated alignment quality and read methylation bias using Bismark, Qualimap v. 2.2.1 [55], and MethylDackel v. 0.6.1 (in HTSlib v. 1.16) [56]. Bisulfite converted reads were mapped to the genome with efficiency rates that ranged from 58.7% to 66.7% for workers one week post treatment, and from 50.4% to 67% for workers four weeks post treatment. Final mean coverage for all the alignments was 21.7 ± 36.2 times.
Methylation data were extracted using MethylDackel. Specifically, read methylation bias was estimated using the ‘mbias’ option, then biased positions were removed using the options ‘–methylKit –OT 0,0,0,0 –OB 0,0,0,99’. Differently methylated sites (DMS) were identified using MethylDackel to exclude CpG sites with less than 8 times coverage (-minDepth 8). Differently methylated genes (DMGenes) were identified by retaining low-coverage sites (–minDepth 1) for posterior filtering with methylKit v. 1.24.0 [57].
Differential methylation analyses were performed using R v. 4.2.0 (package = methylKit v 1.24.0 [55]). Workers from treatment and control microcolonies were independently compared for each time point. DMS were identified after only considering sites with a minimum nucleotide coverage of 10× and a maximum nucleotide coverage of 99.9%. To identify DMGenes, gene positions were filtered using the annotated reference genome. Mean methylation levels for the entire gene were compared with a minimum of 10 cytosines to be present (meth_genes$numCsN > = 10) per gene for all sample replicates. The maximum nucleotide coverage was 99.9%. Additionally, we normalized the methylation counts by coverage across replicates (3 per set of conditions). To identify any possible batch effects, we analysed sample clustering and correlation as well as performed a PCA with all the replicates per age category in methylKit. We used the suggested default qvalue of 0.01 as the threshold for significant differences between the treatment and control groups. However, because mean levels of DNAme in B. terrestris are low—typically less than 1%—we established a cut-off in methylation differences that was based on the distribution of our data, excluding extremes. Briefly, we calculated the mean difference between the 1% (hypomethylated) and the 99% (hypermethylated) quartiles of all the methylation data and identified 4% as the minimum cut-off, which is around four times greater than the mean genome methylation level and at least five times greater than the overall mean methylation differences observed between the control and treatment workers (see Results).
Gene ontology (GO) annotations for the B. terrestris genome in the Hymenoptera Genome Database [58,59] (available at hymenoptera.elsiklab.missouri.edu/hgd-go-annotation) were used for the GO term enrichment analyses. Based on the gene IDs (obtained from www.ncbi.nlm.nih.gov/data-hub/gene/taxon/30195/), the GO terms were associated with all the genes. The GO terms for biological processes (BPs) that were enriched for the DMGenes and DMS compared to the entire genome were identified using the R package topGO v. 2.50.0 [60], with the ‘weight01’ algorithm, Fisher's exact test, and alpha level of 0.01.
(e) . Effect of RG108 on sirt1 expression
We compared sirt1 gene expression between treatment and control workers at one week and four weeks post treatment. Expression levels were quantified using reverse transcription quantitative real-time PCR (RT-qPCR). Total RNA was extracted from the whole bodies of six treatment and six control workers at each time point using TRI Reagent (Thermo Fischer Scientific). Relative sirt1 expression was quantified using the ΔΔCt method [61], with the rpl32 gene serving as the standard of reference. The primers were designed using gene sequences from the Hymenoptera Genome Database [58,59] and NCBI primer BLAST software [62].
(f) . Effect of the SIRT1 pharmacological activator and inhibitor on worker lifespan
We sought to confirm the specific influence of SIRT1 on worker lifespan using pharmacological modulators of SIRT1 activity. Starting at one week of age and continuing over their entire lifetime, workers were fed sugar syrup solution that contained either resveratrol—a potent activator of SIRT1 (hereafter called RSV; MedChemExpress, HY-16561; 100 µM in DMSO)—or selisistat—a specific inhibitor of SIRT1 (hereafter called SEL; MedChemExpress, HY-15452; 75 nM in DMSO). Control workers were fed a sugar syrup solution containing DMSO. These feeding regimes were repeated daily over the course of the workers' lifetimes to ensure optimal levels of pharmacological activity. In total, we established 40 microcolonies of five workers each (RSV treatment n = 12, RSV control n = 10, SEL treatment n = 8 and SEL control n = 10). Worker mortality was recorded every 24 h. Statistical analyses were performed as described above for the RG108 treatment.
3. Results
(a) . The hypomethylating agent RG108 extends worker lifespan
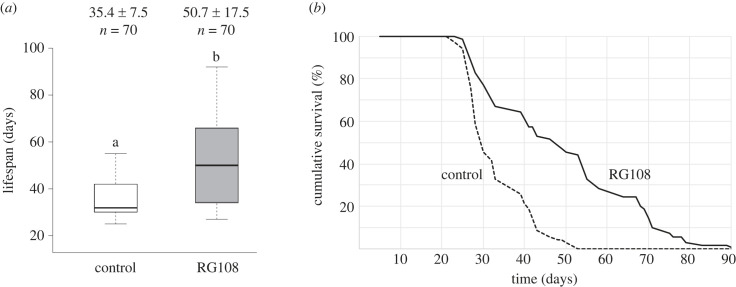
We found that a single topical application of RG108 increased mean worker lifespan by 43% (mean lifespan ± s.d.: control group = 35.4 ± 7.5 days, treatment group = 50.7 ± 17.5 days; Mann–Whitney U-test: U = 1180.5, p < 0.0001; figure 1). Accordingly, the survival curve for the treatment group was significantly shifted to longer lifespans. RG108 also increased maximum lifespan (control group = 55 days, treatment group = 92 days; mixed-effects Cox model: p < 0.0001; figure 1).
Figure 1.
RG108 extends worker lifespan. (a) Boxplots of worker lifespan for the control group (single dose of DMSO solution; white) or the treatment group (single dose of RG108 diluted in DMSO; grey). The mean lifespan ± s.d. and sample size (n) are indicated above each box. The box's midline indicates the median; the box's lower and upper edges are the first and third quartiles, respectively. The whiskers reflect the extreme values. Differences in the letters above the boxes indicate statistically significant differences in mean lifespan (Mann–Whitney U-test: p < 0.0001). (b) Survival curves of workers in the control group (dashed line) and the treatment group (solid line) (mixed-effects Cox model: p < 0.0001).
(b) . Dietary differences do not explain the RG108-mediated increase in lifespan
Despite its effect on lifespan, we did not observed any effect of RG108 on dietary intake: neither sugar nor pollen consumption significantly differed between the two groups (mean total sugar syrup intake per worker ± s.d.: control = 23.36 ± 1.42 g, treatment = 24.34 ± 2.63 g, t1,33= 1.408, p = 0.17 and mean total pollen intake per worker ± s.d. of pollen: control = 1.94 ± 0.15 g, treatment = 1.93 ± 0.29 g, t1,33 = −0.161, p = 0.87; electronic supplementary material, figure S1).
(c) . RG108 treatment induced short- and long-term changes in DNA methylation profiles
Clustering and principal component analyses of WGBS data grouped individuals of the same experimental condition together (i.e. control workers were more similar to each other from a methylomic standpoint than to treatment workers, and vice versa; electronic supplementary material, figure SB and SC).
Genome-wide methylation analyses revealed that treated workers displayed global hypermethylation at both time points, when compared with control workers (electronic supplementary material, figure S2). Furthermore, treatment workers had higher levels of hypermethylation at four weeks than at one week. More specifically, 58% of the differentially methylated single nucleotide polymorphisms (DMS) and 75% of the differentially methylated genes (DMGenes) were hypermethylated at one week, while 62% of DMS and 94% of DMGenes were hypermethylated at four weeks.
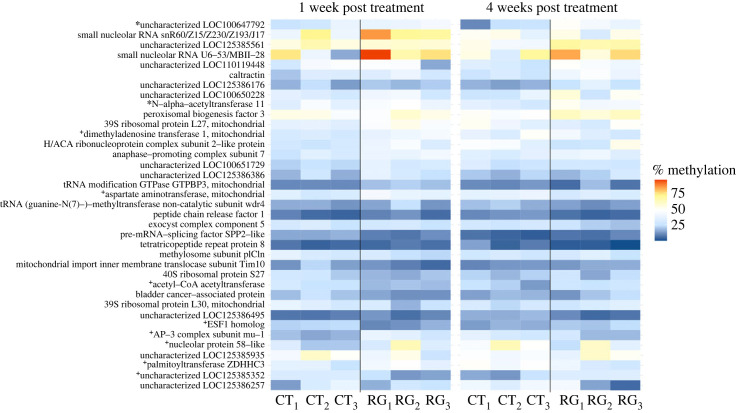
One week post treatment, we observed the presence of 4338 DMS: 1838 were hypomethylated (36.90% average methylation difference between treatment versus control workers), and 2500 were hypermethylated (36.97% average methylation difference between treatment versus control workers) (electronic supplementary material, figure S2). Hypermethylated and hypomethylated cytosines occurred in genes significantly enriched for 31 and 37 GO terms for BPs, respectively. These terms included DNA damage repair, chromatin organization and proteostasis (electronic supplementary material, table S3). When examining entire gene regions, we observed 174 hypermethylated genes (5.69% average methylation difference between treatment versus control workers) and 58 hypomethylated genes (5.83% average methylation difference between treatment versus control workers). The genes that displayed the largest methylation differences are shown in figure 2.
Figure 2.
RG108 induces changes in DNA methylation. Genes with the most pronounced methylation differences between the different experimental conditions (i.e. control workers (CT, DMSO solution) and treatment workers (RG, RG108) at one week or four weeks post treatment) are illustrated. The mean percentage of gene methylation is indicated by the colour scale. *: genes hypermethylated at both time points; +: genes with opposite methylation patterns at the two time points. All the other genes displayed significant patterns of differential methylation at only one of the two time points.
Four weeks post treatment, we observed 6137 DMS: 3781 were hypermethylated (35.28% average methylation difference between treatment versus control workers), and 2356 were hypomethylated (33.60% average methylation difference between treatment versus control workers) (electronic supplementary material, figure S2). The hypermethylated and hypomethylated cytosines were found in genes significantly enriched for 41 and 59 GO terms for BPs, respectively. These terms included DNA damage repair, chromatin organization and proteostasis as well as longevity- and growth-regulating pathways (electronic supplementary material, table S3). The analyses of the DMGenes identified 418 hypermethylated genes (5.77% average methylation difference between treatment versus control workers) and 26 hypomethylated genes (6.71% average methylation difference between treatment versus control workers). The genes that displayed the largest methylation differences are shown in figure 2.
Overall, at both time points, the RG108 treatment induced genome-wide and gene-specific modifications in DNAme that were positively associated with worker lifespan.
(d) . RG108 treatment induces sirt1 overexpression
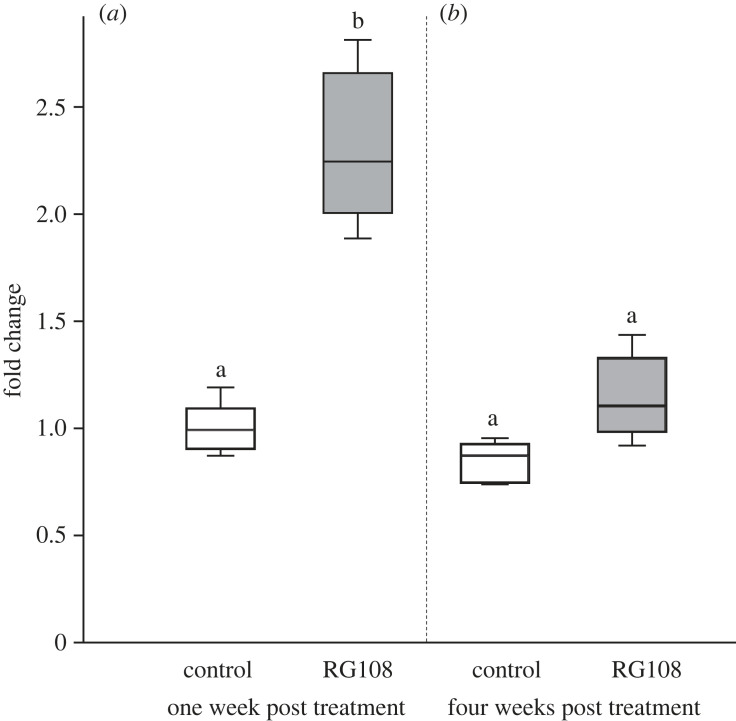
Gene expression of sirt1 was significantly affected by the RG108 treatment in a time-dependent manner (two-way ANOVA: treatment: F1,32 = 84.26, p < 0.0001; time: F1,32 = 57.42, p < 0.0001; interaction: F1,32 = 33.76 p < 0.0001). sirt1 was significantly overexpressed in treatment workers one week after treatment with RG108 (mean fold change ± s.d.: control = 1.00 ± 0.11, treatment = 2.31 ± 0.35, Tukey's post hoc test: p < 0.0001; figure 3). By contrast, there was no difference in sirt1 expression between the two groups at four weeks post treatment (mean fold change ± s.d.: control = 0.85 ± 0.90, treatment = 1.14 ± 0.19, Tukey's post hoc test: p = 0.067; figure 3). This finding indicates that the differential expression of sirt1 induced by the single dose of RG108 did not persist over time, even if the treatment did extend worker lifespan.
Figure 3.
RG108 induces sirt1 overexpression. Boxplots of relative fold changes between control and treatment workers at (a) one-week post treatment and (b) four weeks post treatment (n = 6 for each condition). The box's midline indicates the median; the box's lower and upper edges are the first and third quartiles, respectively. The whiskers reflect the extreme values. Differences in the letters above the boxes indicate statistically significant differences in fold change (two-way ANOVA: ptreatment < 0.0001, ptime < 0.0001, pinteraction < 0.0001; Tukey's post hoc test: p < 0.0001).
(e) . Pharmacological modulators of SIRT1 activity affect worker lifespan
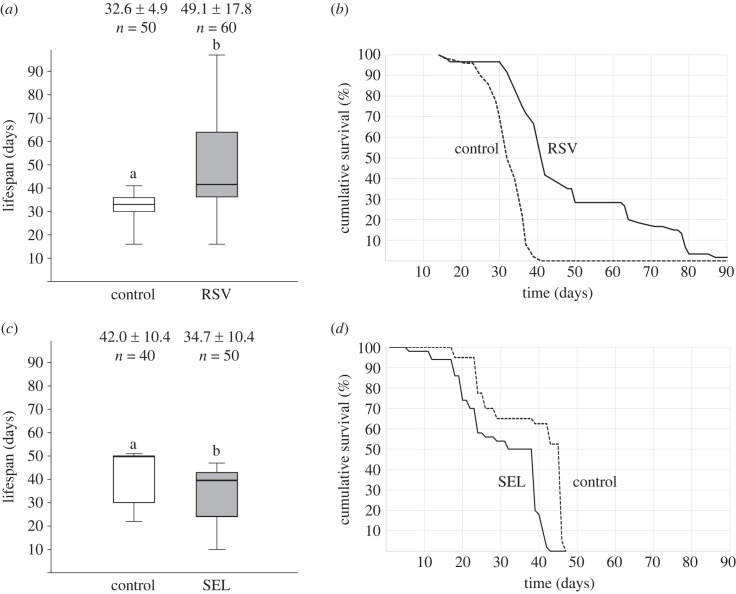
Lifespan of workers that were chronically fed with modulators of SIRT1 protein activity was significantly affected. RSV and SEL had opposite effects on worker lifespan. RSV increased mean worker lifespan by 51% (mean lifespan ± s.d.: control group = 32.6 ± 4.9 days, RSV group = 49.1 ± 17.8 days; Mann–Whitney U-test: U = 406, p < 0.0001; figure 4). Maximum lifespan was also extended (+136% or 97 days total). Conversely, workers fed with SEL had a 17% shorter lifespan than control workers (mean lifespan ± s.d.: control group = 42.0 ± 10.4 days, SEL group = 34.7 ± 10.4 days; Mann–Whitney U-test: U = 474, p < 0.0001; figure 4), and a shorter maximum lifespan (−8% or 47 days total).
Figure 4.
Pharmacological modulation of SIRT1 affects worker lifespan. Boxplots of lifespan for workers fed a daily dietary regime of sugar syrup containing (a) resveratrol diluted in DMSO (RSV) or a control DMSO solution or (c) selisistat diluted in DMSO (SEL) or a control DMSO solution. Mean lifespan ± s.d. and sample size (n) are indicated above each box. The box's midline indicates the median; the box's lower and upper edges are the first and third quartiles, respectively. The whiskers reflect the extreme values. Differences in the letters above the boxes indicate statistically significant differences in mean lifespan (Mann–Whitney U-test: p < 0.0001). Survival curves of workers given the (b) RSV diet (solid line) or control diet (dashed line) or the (d) SEL diet (solid line) or control diet (dashed line) (mixed-effects Cox model: p < 0.0001).
These functional experiments suggest that SIRT1 is involved in lifespan regulation in B. terrestris.
4. Discussion
Using functional experiments and genome-wide DNAme analyses, we found that a single application of RG108, a pharmacological hypomethylating agent, increased the lifespan of one-week-old B. terrestris workers by 43%. Our discovery fits with the results of other studies in mammals, which have shown that various short-term treatments with geroprotectors can have positive, long-lasting effects on longevity [9,63]. This result confirms the previously suggested positive effect of RG108 on longevity in another insect, the honeybee A. mellifera [18]. However, our study goes a step further by analysing the consequences of RG108 on both genome-wide and gene-specific DNAme patterns (see below). Taken together, these findings suggest that DNAme plays a conserved role in lifespan regulation in both vertebrates and invertebrates.
Currently, dietary restriction is the most robust intervention to extend lifespan [49,50]. Food restriction decelerates ageing by preventing changes in DNAme over time [64]. However, our results show that the effects of the RG108 treatment were not mediated via food intake, as previously documented in the honeybee [18]. Instead, our findings suggest that RG108-induced epigenetic changes have effects downstream of food intake that promote longevity.
Consistent with the role of DNAme in lifespan regulation in invertebrates, recent studies have shown that the patterns of this epigenetic mark are influenced by age in the parasitoid wasp Nasonia vitripennis [65] and the water flea Daphnia magna [66]. Our functional analyses revealed that RG108 significantly affected DNAme in the CG context, but not in the non-CG context. This result is unsurprising given that RG108 specifically inhibits DNMT1, which is mostly responsible for CG methylation. On the opposite, non-CG methylation is mostly catalysed by DNMT3 [67], which is not targeted by RG108, thus explaining the similar levels of this type of methylation between treatment and control workers. Therefore, we focused our analysis on CG methylation.
Several complementary explanations for RG108's positive effect on lifespan can be proposed from our WGBS analyses. First, we found that the RG108 treatment induced genome-wide DNA hypermethylation. Given that ageing is known to correlated with global hypomethylation [68], genome hypermethylation could buffer against the progressive loss of DNAme over time. The fact that the hypomethylation agent RG108 increased methylation levels could stem from its influence on genes that directly or indirectly regulate DNAme. For instance, genome hypermethylation might be mediated by crosstalk involving other epigenetic mechanisms, such as histone post-translational modifications [69] or non-coding RNA [70]. Support for this hypothesis comes from our methylomic data, which showed that there were associations with genes encoding proteins involved in histone-related processes in treatment versus control workers at four weeks post treatment and that levels of hypermethylation in treatment workers were higher at four weeks post treatment than at one week post treatment. Alternatively, hypermethylation could result from a rebound effect, where cells respond to RG108 exposure by adjusting the methylation levels of the genome. Interestingly, another hypomethylating agent, 5-Aza-2′-deoxycytidine, has also been found to induce hypermethylation in B. terrestris workers, which was associated with increased colony productivity [71]. By contrast, the same treatment was found to reduce global DNAme level in honeybees [18]. Such differences in the effect of a pharmacological agent were previously reported in Nasonia vitripennis, where the consequences of 5-Aza-dC on DNAme depend on the time of sampling and the tissue [72]. Alternatively, the contrasted effect of RG108 between A. mellifera and B. terrestris might stem from the timing of the treatment (B. terrestris: one-week-old workers; A. mellifera: newly emerged workers). Increased susceptibility to epigenetic modifications at specific time windows was indeed documented in mammals [73], plants [74] and, recently, social Hymenoptera [75,76].
Another, non-mutually exclusive explanation for RG108's impact on worker lifespan is that the agent directly alters the methylation status of the genes involved in lifespan regulation. Consistent with this hypothesis, our enrichment analyses showed that the RG108 treatment affected the methylation status of genes whose GO terms were associated with several key ageing-related processes at both one week and four weeks post treatment, including DNA damage repair, chromatin organization and proteostasis (electronic supplementary material, table S3). (i) Genomic instability is a molecular hallmark of ageing [2], and DNA damage underlies several molecular and cellular mechanisms that promote ageing, such as epigenetic changes and deteriorated proteostasis [77]. Altering the methylation status of genes involved in DNA damage repair could promote longevity by countering ageing-related DNA damage. (ii) Altered chromatin organization and post-translational modifications in histone proteins promote ageing by altering transcriptional profiles [2]. Given that histones modifications act in tandem with DNAme to regulate gene expression [69], RG108 may extend lifespan via a combined effect on different epigenetic marks that serves to ensure the maintenance of epigenomic patterns. (iii) Loss of proteostasis is another hallmark of ageing [2], which refers to alterations in proteomic processes, such as protein synthesis, degradation, post-translational modifications, folding and transport [78]. The enriched GO terms related to proteostasis were mostly trafficking and degradation, which are two key steps in autophagy [79], a cytoprotective process that is strongly linked to longevity [80]. Thus, RG108 could increase lifespan by functionally maintaining autophagy across the lifespan of workers, thus ensuring the prolonged maintenance of cell homeostasis. Finally, at the four weeks mark, the treatment group was characterized by the enrichment in GO terms related to growth- and longevity-regulating pathways, including the nutrient-sensing insulin/insulin-like growth factor signalling (IIS) pathway and the target of rapamycin (TOR) pathway, which are known to influence the trade-off between growth, reproduction, and longevity in many animals [81–84], including social insects [35,85,86]. Deregulation of these nutrient-sensing pathways can drive ageing by altering cell and organismal metabolism [87]. Interestingly, treatment with rapamycin, a TOR inhibitor, in later life stage can increase lifespan in mice [88]. Therefore, the effect of RG108 could be mediated, at least in part, by the methylation of IIS- and TOR-related genes in treatment workers at four weeks post treatment. Altogether, our methylomic analyses indicate that the RG108 treatment induced a combination of global and gene-specific changes in methylation profiles that may act collectively to promote longevity.
The RG108 treatment led to a modest but nonetheless statistically significant difference in sirt1 expression at one week post treatment. This difference was no longer present at the four weeks mark. Given the central role played by sirt1 in cell metabolism, it is not surprising that its expression remains strictly controlled to ensure proper cell function. Mechanistically, it seems likely that the effects of the RG108 treatment arose from an interaction between SIRT1 and DNMT1 proteins. One week after workers experienced the treatment, the hypomethylating effect of RG108 was possibly being counteracted by high induced levels of sirt1 expression. SIRT1 is known to promote the activity of DNMT1, a protein that, in turn, promotes DNA hypermethylation [26]. The resulting hypermethylation would then have become more pronounced four weeks after the treatment, whereas sirt1 expression would have dropped back down to baseline to avoid excessively high methylation levels.
The functional manipulation of SIRT1 activity revealed a positive relationship between SIRT1 activity and lifespan in B. terrestris. While the potent SIRT1 activator RSV increased worker lifespan, the specific SIRT1 inhibitor SEL reduced it. These results do not imply that SIRT1 was solely responsible for the effects of the RG108 treatment on lifespan. Indeed, RSV does not exclusively influence the activity of SIRT1. It also inhibits the growth- and lifespan-regulating pathway TOR [88]. Furthermore, SIRT1 interacts with the IIS [89] and TOR pathways [90], whose components are encoded by genes that we observed to be differentially methylated following RG108 treatment. Thus, it is possible that the two pharmacological treatments that lead to lifespan extension (i.e. RG108 and RSV) do so via the same BPs.
5. Conclusion
To our knowledge, our study is the first to functionally manipulate lifespan in combination with genome-wide DNAme analyses to explore how DNAme is involved in lifespan regulation in an insect. These findings should spark future interest in invertebrates ageing research to determine whether the epigenetic underpinnings of ageing are conserved across animal taxa.
Acknowledgements
We thank Jessica Pearce for proofreading and Cyril Gueydan for thoughtful conversations about the manuscript.
Ethics
This work did not require ethical approval from a human subject or animal welfare committee.
Data accessibility
All data generated or analysed during this study are included in this published article and its electronic supplementary material [91]. WGBS data are accessible in the NCBI SRA database (BioProject PRJNA1036551).
Declaration of AI use
We have not used AI-assisted technologies in creating this article.
Authors' contributions
T.R.: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing—original draft, writing—review and editing; B.M.: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing—original draft, writing—review and editing; N.D.S.A.: formal analysis, methodology, software, validation, visualization, writing—review and editing; S.A.: conceptualization, funding acquisition, project administration, resources, supervision, validation, visualization, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
The authors declare no competing interest.
Funding
This work was supported by the Belgian National Fund for Scientific Research (FRS-FNRS; grant no. J.0004.20F to S.A., postdoctoral fellowship to B.M. and N.D.S.A., and PhD fellowship to T.R.) and the Université Libre de Bruxelles (Fonds d'Encouragement à la Recherche and Actions Blanches to S.A.; Fonds David et Alice Van Buuren et Fondation Jaumotte-Demoulin to T.R.).
References
- 1.Rose MR. 1991. Evolutionary biology of aging. Oxford, UK: Oxford University Press. [Google Scholar]
- 2.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153, 1194-1217. ( 10.1016/j.cell.2013.05.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petralia RS, Mattson MP, Yao PJ. 2014. Aging and longevity in the simplest animals and the quest for immortality. Ageing Res. Rev. 16, 66-82. ( 10.1016/j.arr.2014.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen AA. 2018. Aging across the tree of life: the importance of a comparative perspective for the use of animal models in aging. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 2680-2689. ( 10.1016/j.bbadis.2017.05.028) [DOI] [PubMed] [Google Scholar]
- 5.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. 2009. An operational definition of epigenetics. Genes Dev. 23, 781-783. ( 10.1101/gad.1787609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204-220. ( 10.1038/nrg2719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith ZD, Meissner A. 2013. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204-220. ( 10.1038/nrg3354) [DOI] [PubMed] [Google Scholar]
- 8.Sen P, Shah PP, Nativio R, Berger SL. 2016. Epigenetic mechanisms of longevity and aging. Cell 166, 822-839. ( 10.1016/j.cell.2016.07.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, et al. 2020. Reprogramming to recover youthful epigenetic information and restore vision. Nature 588, 124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang JH, et al. 2023. Loss of epigenetic information as a cause of mammalian aging. Cell 186, 305-326. ( 10.1016/j.cell.2022.12.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916-919. ( 10.1126/science.1186366) [DOI] [PubMed] [Google Scholar]
- 12.Horvath S, Raj K. 2018. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371-384. ( 10.1038/s41576-018-0004-3) [DOI] [PubMed] [Google Scholar]
- 13.Seale K, Horvath S, Teschendorff A, Eynon N, Voisin S. 2022. Making sense of the ageing methylome. Nat. Rev. Genet. 23, 585-605. ( 10.1038/s41576-022-00477-6) [DOI] [PubMed] [Google Scholar]
- 14.Anastasiadi D, Piferrer F. 2020. A clockwork fish: age prediction using DNA methylation-based biomarkers in the European seabass. Mol. Ecol. Resour. 20, 387-397. ( 10.1111/1755-0998.13111) [DOI] [PubMed] [Google Scholar]
- 15.Zoller JA, Parasyraki E, Lu AT, Haghani A, Niehrs C, Horvath S. In press. DNA methylation clocks for clawed frogs reveal evolutionary conservation of epigenetic aging. GeroScience ( 10.1007/s11357-023-00840-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raddatz G, Arsenault RJ, Aylward B, Whelan R, Böhl F, Lyko F. 2021. A chicken DNA methylation clock for the prediction of broiler health. Commun. Biol. 4, 76. ( 10.1038/s42003-020-01608-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu AT, et al. 2023. Universal DNA methylation age across mammalian tissues. Nat. Aging 3, 1-23. ( 10.1038/s43587-023-00462-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardoso-Júnior CA, Guidugli-Lazzarini KR, Hartfelder K. 2018. DNA methylation affects the lifespan of honey bee (Apis mellifera L.) workers–evidence for a regulatory module that involves vitellogenin expression but is independent of juvenile hormone function. Insect Biochem. Mol. Biol 92, 21-29. ( 10.1016/j.ibmb.2017.11.005) [DOI] [PubMed] [Google Scholar]
- 19.Schwer B, Verdin E. 2008. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 7, 104-112. ( 10.1016/j.cmet.2007.11.006) [DOI] [PubMed] [Google Scholar]
- 20.Houtkooper RH, Pirinen E, Auwerx J. 2012. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 13, 225-238. ( 10.1038/nrm3293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longo VD, Kennedy BK. 2006. Sirtuins in aging and age-related disease. Cell 126, 257-268. ( 10.1016/j.cell.2006.07.002) [DOI] [PubMed] [Google Scholar]
- 22.Lagunas-Rangel FA. 2019. Current role of mammalian sirtuins in DNA repair. DNA Repair 80, 85-92. ( 10.1016/j.dnarep.2019.06.009) [DOI] [PubMed] [Google Scholar]
- 23.Watroba M, Dudek I, Skoda M, Stangret A, Rzodkiewicz P, Szukiewicz D. 2017. Sirtuins, epigenetics and longevity. Ageing Res. Rev. 40, 11-19. ( 10.1016/j.arr.2017.08.001) [DOI] [PubMed] [Google Scholar]
- 24.O'Callaghan C, Vassilopoulos A. 2017. Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell 16, 1208-1218. ( 10.1111/acel.12685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guarente L. 2013. Calorie restriction and sirtuins revisited. Genes Dev. 27, 2072-2085. ( 10.1101/gad.227439.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng L, et al. 2011. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol. Cell. Biol. 31, 4720-4734. ( 10.1128/MCB.06147-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonkowski MS, Sinclair DA. 2016. Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 17, 679-690. ( 10.1038/nrm.2016.93) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howitz KT, et al. 2003. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191-196. ( 10.1038/nature01960) [DOI] [PubMed] [Google Scholar]
- 29.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. 2004. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430, 686-689. ( 10.1038/nature02789) [DOI] [PubMed] [Google Scholar]
- 30.Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai SI. 2013. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 18, 416-430. ( 10.1016/j.cmet.2013.07.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller L. 1998. Queen lifespan and colony characteristics in ants and termites. Insectes Soc. 45, 235-246. ( 10.1007/s000400050084) [DOI] [Google Scholar]
- 32.Kramer BH, Schaible R. 2013. Colony size explains the lifespan differences between queens and workers in eusocial Hymenoptera. Biol. J. Linn. Soc. Lond. 109, 710-724. ( 10.1111/bij.12072) [DOI] [Google Scholar]
- 33.Wilson EO. 1971. The insects societies. Cambridge, MA: Harvard University Press. [Google Scholar]
- 34.Evans JD, Wheeler DE. 2001. Gene expression and the evolution of insect polyphenisms. Bioessays 23, 62-68. () [DOI] [PubMed] [Google Scholar]
- 35.Corona M, Libbrecht R, Wheeler DE. 2016. Molecular mechanisms of phenotypic plasticity in social insects. Curr. Opin. Insect. Sci. 13, 55-60. ( 10.1016/j.cois.2015.12.003) [DOI] [PubMed] [Google Scholar]
- 36.Dussutour A, Simpson SJ. 2012. Ant workers die young and colonies collapse when fed a high-protein diet. Proc. R. Soc. B 279, 2402-2408. ( 10.1098/rspb.2012.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Negroni MA, Macit MN, Stoldt M, Feldmeyer B, Foitzik S. 2021. Molecular regulation of lifespan extension in fertile ant workers. Phil. Trans. R. Soc. B 376, 20190736. ( 10.1098/rstb.2019.0736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rueppell O, Kaftanouglu O, Page RE Jr. 2009. Honey bee (Apis mellifera) workers live longer in small than in large colonies. Exp. Gerontol. 44, 447-452. ( 10.1016/j.exger.2009.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tofilski A. 2002. Influence of age polyethism on longevity of workers in social insects. Behav. Ecol. Sociobiol. 51, 234-237. ( 10.1007/s00265-001-0429-z) [DOI] [Google Scholar]
- 40.Beros S, Lenhart A, Scharf I, Negroni MA, Menzel F, Foitzik S. 2021. Extreme lifespan extension in tapeworm-infected ant workers. R. Soc. Open Sci. 8, 202118. ( 10.1098/rsos.202118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucharski R, Maleszka J, Foret S, Maleszka R. 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science 319, 1827-1830. ( 10.1126/science.1153069) [DOI] [PubMed] [Google Scholar]
- 42.Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. 2010. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8, e1000506. ( 10.1371/journal.pbio.1000506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morandin C, Brendel VP, Sundström L, Helanterä H, Mikheyev AS. 2019. Changes in gene DNA methylation and expression networks accompany caste specialization and age-related physiological changes in a social insect. Mol. Ecol. 28, 1975-1993. ( 10.1111/mec.15062) [DOI] [PubMed] [Google Scholar]
- 44.Velthuis HH, Van Doorn A. 2006. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie 37, 421-451. ( 10.1051/apido:2006019) [DOI] [Google Scholar]
- 45.Tasei JN, Aupinel P. 2008. Validation of a method using queenless Bombus terrestris micro-colonies for testing the nutritive value of commercial pollen mixes by comparison with queenright colonies. J. Econ. Entomol. 101, 1737-1742. ( 10.1603/0022-0493-101.6.1737) [DOI] [PubMed] [Google Scholar]
- 46.Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. 2005. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 65, 6305-6311. ( 10.1158/0008-5472.CAN-04-2957) [DOI] [PubMed] [Google Scholar]
- 47.Oh YS, Jeong SG, Cho GW. 2015. Anti-senescence effects of DNA methyltransferase inhibitor RG108 in human bone marrow mesenchymal stromal cells. Biotechnol. Appl. Biochem. 62, 583-590. ( 10.1002/bab.1393) [DOI] [PubMed] [Google Scholar]
- 48.R Core Team. 2022. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/. [Google Scholar]
- 49.Speakman JR, Mitchell SE. 2011. Caloric restriction. Mol. Aspects Med. 32, 159-221. ( 10.1016/j.mam.2011.07.001) [DOI] [PubMed] [Google Scholar]
- 50.Solon-Biet SM, et al. 2014. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 19, 418-430. ( 10.1016/j.cmet.2014.02.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weidenmüller A, Kleineidam C, Tautz J. 2002. Collective control of nest climate parameters in bumblebee colonies. Anim. Behav. 63, 1065-1071. ( 10.1006/anbe.2002.3020) [DOI] [Google Scholar]
- 52.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. See http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 53.Krueger F. 2015. Trim galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. See http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- 54.Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571-1572. ( 10.1093/bioinformatics/btr167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.García-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Götz S, Tarazona S, Dopazo J, Meyer TF, Conesa A. 2012. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28, 2678-2679. ( 10.1093/bioinformatics/bts503) [DOI] [PubMed] [Google Scholar]
- 56.Ryan D. 2017. MethylDackel. GitHub Repository. See https://github.com/dpryan79/MethylDackel.
- 57.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. 2012. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, 1-9. ( 10.1186/gb-2012-13-10-r87) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadd BM, et al. 2015. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 16, 1-32. ( 10.1186/s13059-015-0623-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elsik CG, Tayal A, Diesh CM, Unni DR, Emery ML, Nguyen HN, Hagen DE. 2016. Hymenoptera genome database: integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 44(D1), D793-D800. ( 10.1093/nar/gkv1208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alexa A, Rahnenführer J. 2009. Gene set enrichment analysis with topGO. Bioconductor Improv. 27, 1-26. [Google Scholar]
- 61.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402-408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- 62.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13, 1-11. ( 10.1186/1471-2105-13-134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Juricic P, et al. 2022. Long-lasting geroprotection from brief rapamycin treatment in early adulthood by persistently increased intestinal autophagy. Nat. Aging 2, 824-836. ( 10.1038/s43587-022-00278-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maegawa S, et al. 2017. Caloric restriction delays age-related methylation drift. Nat. Commun. 8, 539. ( 10.1038/s41467-017-00607-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brink K, Thomas C, Jones A, Mallon EB. 2023. An epigenetic clock in an insect model system. bioRxiv, 2023-02. ( 10.1101/2023.02.14.528436) [DOI] [PMC free article] [PubMed]
- 66.Hearn J, Plenderleith F, Little TJ. 2021. DNA methylation differs extensively between strains of the same geographical origin and changes with age in Daphnia magna. Epigenetics Chromatin 14, 1-14. ( 10.1186/s13072-020-00379-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He Y, Ecker JR. 2015. Non-CG methylation in the human genome. Annu. Rev. Genomics Hum. Genet. 16, 55-77. ( 10.1146/annurev-genom-090413-025437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson VL, Jones PA. 1983. DNA methylation decreases in aging but not in immortal cells. Science 220, 1055-1057. ( 10.1126/science.68449) [DOI] [PubMed] [Google Scholar]
- 69.Cedar H, Bergman Y. 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Gen. 10, 295-304. ( 10.1038/nrg2540) [DOI] [PubMed] [Google Scholar]
- 70.Zhao Y, Sun H, Wang H. 2016. Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci. 6, 1-6. ( 10.1186/s13578-016-0109-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pozo MI, Hunt BJ, Van Kemenade G, Guerra-Sanz JM, Wäckers F, Mallon EB, Jacquemyn H. 2021. The effect of DNA methylation on bumblebee colony development. BMC Genom. 22, 1-11. ( 10.1186/s12864-021-07371-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cook N, Parker DJ, Turner F, Tauber E, Pannebakker BA, Shuker DM. 2018. Genome-wide disruption of DNA methylation by 5-aza-2′-deoxycytidine in the parasitoid wasp Nasonia vitripennis. bioRxiv 437202. ( 10.1101/437202) [DOI]
- 73.Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293, 1089-1093. ( 10.1126/science.1063443) [DOI] [PubMed] [Google Scholar]
- 74.Feng S, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA 107, 8689-8694. ( 10.1073/pnas.1002720107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simola DF, et al. 2016. Epigenetic (re) programming of caste-specific behavior in the ant Camponotus floridanus. Science 351, aac6633. ( 10.1126/science.aac6633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glastad KM, Graham RJ, Ju L, Roessler J, Brady CM, Berger SL. 2020. Epigenetic regulator CoREST controls social behavior in ants. Mol. Cell 77, 338-351. ( 10.1016/j.molcel.2019.10.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schumacher B, Pothof J, Vijg J, Hoeijmakers JH. 2021. The central role of DNA damage in the ageing process. Nature 592, 695-703. ( 10.1038/s41586-021-03307-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Labbadia J, Morimoto RI. 2015. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435-464. ( 10.1146/annurev-biochem-060614-033955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rubinsztein DC, Mariño G, Kroemer G. 2011. Autophagy and aging. Cell 146, 682-695. ( 10.1016/j.cell.2011.07.030) [DOI] [PubMed] [Google Scholar]
- 80.Hansen M, Rubinsztein DC, Walker DW. 2018. Autophagy as a promoter of longevity: insights from model organisms. Nat. Rev. Mol. Cell Biol. 19, 579-593. ( 10.1038/s41580-018-0033-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barbieri M, Bonafè M, Franceschi C, Paolisso G. 2003. Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am. J. Physiol. Endocrinol. Metab. 285, E1064-E1071. ( 10.1152/ajpendo.00296.2003) [DOI] [PubMed] [Google Scholar]
- 82.Bluher M, Kahn BB, Kahn CR. 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299, 572-574. ( 10.1126/science.1078223) [DOI] [PubMed] [Google Scholar]
- 83.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Müller F. 2003. Influence of TOR kinase on lifespan in C. elegans. Nature 426, 620. ( 10.1038/426620a) [DOI] [PubMed] [Google Scholar]
- 84.Kaeberlein M, et al. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193-1196. ( 10.1126/science.1115535) [DOI] [PubMed] [Google Scholar]
- 85.Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simões ZL, Maleszka R. 2007. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev. Biol. 7, 1-19. ( 10.1186/1471-213X-7-70) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Korb J, et al. 2021. Comparative transcriptomic analysis of the mechanisms underpinning ageing and fecundity in social insects. Phil. Trans. R. Soc. B 376, 20190728. ( 10.1098/rstb.2019.0728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fontana L, Partridge L, Longo VD. 2010. Extending healthy life span—from yeast to humans. Science 328, 321-326. ( 10.1126/science.1172539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhong LM, et al. 2012. Resveratrol inhibits inflammatory responses via the mammalian target of rapamycin signaling pathway in cultured LPS-stimulated microglial cells. PLoS ONE 7, e32195. ( 10.1371/journal.pone.0032195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang T, Fu M, Pestell R, Sauve AA. 2006. SIRT1 and endocrine signaling. Trends Endocrinol. Metabol. 17, 186-191. ( 10.1016/j.tem.2006.04.002) [DOI] [PubMed] [Google Scholar]
- 90.Ghosh HS, McBurney M, Robbins PD. 2010. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS ONE 5, e9199. ( 10.1371/journal.pone.0009199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Renard T, Martinet B, De Souza Araujo N, Aron S. 2023. DNA methylation extends lifespan in the bumblebee Bombus terrestris. Figshare. ( 10.6084/m9.figshare.c.6949032) [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
Data Availability Statement
All data generated or analysed during this study are included in this published article and its electronic supplementary material [91]. WGBS data are accessible in the NCBI SRA database (BioProject PRJNA1036551).