Abstract
The pandemic caused by the coronavirus SARS-CoV-2 led to a global crisis in the world healthcare system. Despite some progress in the creation of antiviral vaccines and mass vaccination of the population, the number of patients continues to grow because of the spread of new SARS-CoV-2 mutations. There is an urgent need for direct-acting drugs capable of suppressing or stopping the main mechanisms of reproduction of the coronavirus SARS-CoV-2. Several studies have shown that the successful replication of the virus in the cell requires proteolytic cleavage of the protein structures of the virus. Two proteases are crucial in replicating SARS-CoV-2 and other coronaviruses: the main protease (Mpro) and the papain-like protease (PLpro). In this review, we summarize the essential viral proteins of SARS-CoV-2 required for its viral life cycle as targets for chemotherapy of coronavirus infection and provide a critical summary of the development of drugs against COVID-19 from the drug repurposing strategy up to the molecular design of novel covalent and non-covalent agents capable of inhibiting virus replication. We overview the main antiviral strategy and the choice of SARS-CoV-2 Mpro and PLpro proteases as promising targets for pharmacological impact on the coronavirus life cycle.
The review outlines coronavirus SARS-CoV-2 morphology, life cycle, and essential proteins, focusing on a design strategy for dual-acting inhibitors for PLpro and Mpro proteases.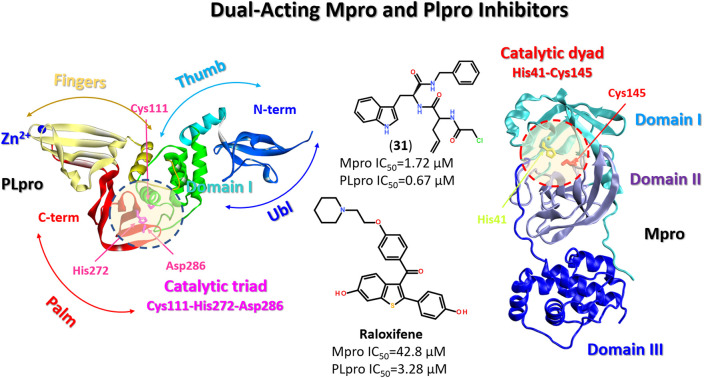
1. Introduction
At the end of 2019, an outbreak of severe acute respiratory syndrome (SARS) led to the global epidemic, which was recognized by the World Health Organization (WHO) as a pandemic on March 11, 2020. SARS and other manifestations of the disease were caused by the SARS-CoV-2 virus, previously not circulating in human intercourse, and received the official name – coronavirus disease 2019 (COVID-19).1
SARS-CoV-2 affected about 770 million people in 2 years, causing respiratory, renal, cardiac, cerebral and intestinal manifestations of the disease, which ranged from mild to fatal consequences.2,3 As of June 20, 2023, 2.5 years after the diagnosis of the disease, about 6.95 million people with a confirmed diagnosis of COVID-19 have died.4
Extensive vaccination campaigns have shown effectiveness against the COVID-19 pandemic in the EU and worldwide. The total number of administered vaccine doses worldwide amounted to more than 13461.3 million doses. As of January 2023, according to the data presented in the EU, the general improvement of the epidemiological situation with COVID-19 continued. Death rates fell to their lowest level in 12 months. Most countries of the world reported a similar picture. The proportion of patients with COVID-19 in hospitals and intensive care units in the EU was 28% of the maximum recorded levels during the pandemic. However, despite improving the epidemiological situation, COVID-19 continues to burden healthcare systems.
Indicators of excessive mortality (deviation of mortality from the expected level) indicate that the consequences of the disease continue to affect the health of the Earth's population. General statistics show vaccinations do not create lasting immunity against the SARS-CoV-2 virus. The disease COVID-19 continues to spread.4,5
On May 5, 2023, WHO stated that COVID-19 is not a Global Public Health Emergency of International Concern (PHEIC). Given that the disease is already well known, it no longer fits the definition of PHEIC, even though it continues. However, this statement does not mean that the pandemic issue itself is over. WHO will develop long-term, ongoing guidance for countries on how to deal with COVID-19 on a continuing basis.4,5
Given the continuous evolution of the virus, regular surveillance is currently being carried out under the auspices of WHO in the world, which consists of detecting and monitoring new variants of SARS-CoV-2. The emergence of variants that pose an increased risk to the global health system has prompted WHO to identify specific variants of the virus as those of interest (VOI) and variants of concern (VOC) to set priorities for decision-making on whether vaccines should be updated or maintained or intensified in response to the increased presence of a particular variant of the SARS-CoV-2 virus (Table 1).5,6
The main known variants of SARS-CoV-2.
| Variant of virus SARS-CoV-2 | The earliest documented case |
|---|---|
| Alfa | Great Britain, September 2020 |
| Beta | North African Republic, May 2020 |
| Gamma | Brazil, November 2020 |
| Delta | India, October 2020 |
| Omicron | Several countries, November 2021 |
Omicron is currently the dominant variant circulating worldwide, accounting for over 98% of viral sequences.7
The main problem with vaccines is the continuous mutation of SARS-CoV-2. Most RNA viruses evolve rapidly. SARS-CoV-2 variants with different phenotypic characteristics, including transmissibility, severity, and immune evasion, continue to emerge rapidly. The Omicron variant discovered at the end of November 2021 marked the beginning of a new phase of the pandemic.8 It is less virulent but more transmissible than earlier variants of SARS-CoV-2.9 Even at the beginning of the discovery, the Omicron variant included three sister lines (BA.1, BA.2 and BA.3).8 Unlike the original SARS-CoV-2 virus, which infected bronchial and lung cells, Omicron BA.1 preferred to replicate in the nasopharynx.10
SARS-CoV-2, like other RNA viruses, shows a significant degree of antigenic evolution, which opens up a sufficient probability of repeated infections. Omicron, for example, has a much higher ability to cause reinfection than any of the previous options. Examining the current trends of Omicron variants, we have publications to expect new waves of infection every additional approximately four months of virus circulation, and going forward SARS-CoV-2 will have a more regular incidence pattern like that of seasonal influenza, leading to an expectation of two to three times the annual burden the flu.10 With the emergence of new variants of SARS-CoV-2 that evade the immune response, vaccines developed against the virus may become less active.8 It means that the issue of the need to develop drugs against COVID-19 continues to be relevant. The importance of the emergence of new therapeutic drugs against coronavirus disease is evident.11
2. General characteristics of the virus SARS-CoV-2
According to the classification of the International Committee on Taxonomy of Viruses (International Committee on Taxonomy of Viruses, ICTV), the SARS-CoV-2 virus belongs to β-coronaviruses.12 The Baltimore classification of viruses is a classification of viruses into groups depending on the type of genomic nucleic acid (DNA, RNA, single-stranded, double-stranded) and the method of its replication.13 In the Baltimore classification system, the SARS-CoV-2 virus belongs to the IV group of viruses containing single-stranded RNA with positive single-stranded RNA viruses (+ssRNA).14,15
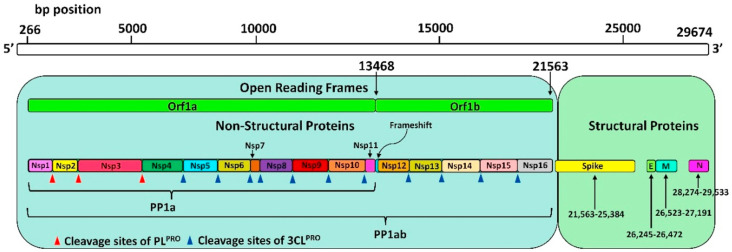
Replication of +ssRNA genomes occurs in the cytoplasm of the host cell in parallel with the assembly of the nucleocapsid, in which the genetic material is packaged.16 The close connection between viral genome replication and nucleocapsid formation means that the genomic RNA and protein components of the virus are highly specific to each other, and the assembly of the +ssRNA virus depends on the interaction between proteins and between proteins and RNA.17,18 Most of the genomic RNA encodes two replicative polyproteins, which are then cleaved during cleavage into 16 nonstructural proteins by viral proteases.14,15 The rest of the SARS-CoV-2 genome encodes four structural proteins: spike protein (S), envelope protein (E), membrane protein (M) and nucleocapsid protein (N).8,16–19
3. Basic strategies of antiviral therapy
In the development of antiviral drugs, two different strategies are used: the influence on the cells of the host organism (indirect methods) or the influence directly on the virus (direct methods).
The indirect methods aim to target proteins of the host cell involved in the virus's life cycle. This approach has a lower probability of developing resistance but significantly higher toxicity.19 Coronaviruses can enter host cells in three ways: receptor-mediated plasma membrane fusion, receptor-mediated endocytosis, or antibody-dependent viral entry. Therefore, receptor proteins on the host cell surface play a crucial role in virus recognizing and attaching host cells for both fusion and endocytosis.20 Preventing binding interactions between SARS-CoV-2 and human angiotensin-converting enzyme II (ACE2) receptors provides example of this strategy21 and a foundation for developing new vaccines against COVID-19.22 Some excellent reviews on this topic has recently appeared.17,19,20
The direct methods affect specifically on the certain components of the virus and its life cycle. These approaches are diverse and characterized by significant specificity and fewer side effects for the body. Viral proteins, such as main protease Mpro and papain-like protease PLpro, as well as other components of the virus at different stages of its life cycle can be the potential targets of the antiviral strategy of the direct approach. General therapeutic approaches to the treatment of viral infections, as a rule, are explicitly directed at viral components. However, such drugs are very sensitive to drug resistance due to rapid changes in the viral genome, especially for RNA viruses. When designing molecules for a pharmaceutical effect on the virus, it is necessary to take the most conservative targets: components of the virus that have fewer mutations and are highly specific for the virus. Reducing the risk of developing resistance in developing drugs focused on more than one viral component.23 This procedure requires particular approaches to the development of antiviral drugs.19
In this review, we will focus on research progress in designing and developing directly acting small-molecule inhibitors against COVID-19, focusing on their chemical structure, structure–activity relationship, and clinical treatments.
4. Life cycle of virus SARS-CoV-2
The life cycle (LC) of the virus includes several main phases:
– Binding.
– Release of the viral genome.
– Genome replication.
– Expression (synthesis) of viral specific structural and non-structural proteins.
– Processing of virion proteins at the maturation stage assembly of the viral capsid.
The first stage of LC is the attachment of the virus to the cell and penetration into the cell. Attachment and penetration can occur by receptor-mediated endocytosis or by direct membrane fusion. With the help of the surface S protein, the SARS-CoV-2 virus attaches to the surface receptor of angiotensin-converting enzyme-2 (ACE2) of the host cell (Fig. 1).19
Fig. 1. SARS-CoV-2 genome encoding 5 structural proteins and 16 non-structural proteins (Nsp) translated into a single polyprotein chain (PP1ab). Red (PLpro) and blue (Mpro) triangles show the cleavage sites of the polyprotein chain by proteases. Reproduced with permission from ref. 24. © 2021 Elsevier B.V. All rights reserved.
The second stage of LC is the release of the viral genome. The viral genome is released in the cell from the viral particle with the help of specific proteases. In this phase, the virus can use the proteases of the infected cell (host cell).
The third stage of LC is the replication of the virus genome. The virus can use the host's machinery to replicate the viral genome in this phase.
The fourth stage of LC is the expression of viral proteins utilizing translation – the process of protein synthesis on the information RNA matrix, which is carried out by the ribosome of the host cell.
A large part of the genomic RNA of the virus encodes two polyproteins – pp1a and pp1ab. The rest of the genome encodes four structural proteins: S-protein, envelope protein (E), membrane protein (M), nucleocapsid (N) protein and others.25
The fifth stage of LC is the stage when the newly formed polyproteins pp1a and pp1ab undergo post-translational modification, so they are proteolyzed (cut) into smaller proteins by two viral proteases, 3C-like protease (3CLpro), also known as main protease (Mpro), and papain-like protease (PLpro). Cleavage products include 16 nonstructural proteins (nsp), including RNA-dependent RNA polymerase (RdRp).25
The sixth stage of LC is the stage of assembly (maturation) of the virus: new viral particles are formed, ready to be released from the host cell.26
5. Molecular architecture of the virus SARS-CoV-2
The development of potential antiviral drugs requires understanding both the life cycle of the SARS-CoV-2 virus and its molecular architecture. State-of-the-art research in cryo-electron tomography (cryoET) has allowed us to visualize intact SARS-CoV-2 virions and gain insight into their structure in their natural state with a resolution close to atomic resolution.12
SARS-CoV-2 virions are approximately spherical or ellipsoidal with an average diameter of 108 ± 8 nm. The outer surface of the virion is covered with surface spike-like proteins (S-protein). The virus's outer membrane contains a membrane protein (M) and an envelope protein (E). The E protein is small, with a mass of 8.5 kDa, forming a membrane channel. The main protein component in the middle of the virion is the nucleocapsid protein N, responsible for binding genomic RNA and packaging it into a ribonucleoprotein (RNP) complex.12,27
The genome of CoVs is 27–32 kb (kilobase pairs) and is the second largest of all RNA virus genomes. Almost two-thirds of genomic RNA has two open reading frames (ORFs): ORF1a and ORF1b at the 5′-end, which are quickly translated into two polyproteins – pp1a and pp1ab, which are converted by two viral proteases into sixteen non-structural proteins (nsp1-16).25 Another third of the genome encodes four structural proteins: spike protein (S), envelope protein (E), membrane protein (M), nucleocapsid protein (N) and other intermediate proteins. ORFs encode these proteins at the 3′-end of genomic RNA.9,12,28
6. Structural and non-structural proteins of SARS-CoV-2
6.1. Spike protein (S protein)
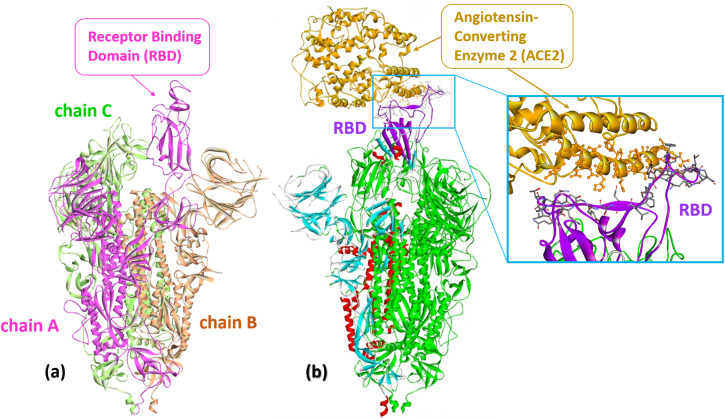
The spike surface protein of SARS-CoV-2 (S protein) is a structural transmembrane glycoprotein located on the surface of the viral particle. It is a trimer with a molecular weight of about 600 kDa (Fig. 2a).29 The S protein recognizes the receptor of angiotensin-converting enzyme 2 (ACE2) on host cells and, through its receptor-binding domain (RBD), ensures the penetration of the virus into the host cell due to the fusion of the membranes of the virus and the host cell (Fig. 2b).12,30,31 Because of its key role in progressing SARS-CoV-2 infection, the S protein is one of the main targets for vaccine and drug developers. The S protein is the antigen of all vaccines currently on the market and elicits a strong immune response in vaccinated and infected individuals.32
Fig. 2. (a) The S protein of SARS-CoV-2 (Omicron S-open-2, PDB: 7WVO): chains A–C with receptor-binding domain (RBD). (b) Cryo-EM structure of SARS-CoV-2 spike protein in complex with angiotensin-converting enzyme 2 (ACE2) shown in gold (PDB: 8HFZ). A close-up representation of the RBD protein and ACE2 interface. The sticks represent the key residues that interact with the ACE2. Numerous atomic connections are made between SARS-CoV-2 RBD (violet) and ACE2 (gold).
The surface of the S protein is highly glycosylated. Such a glycan shell, combined with the flexibility of the spikes of SARS-CoV-2, allows them to scan the host cell's surface and bind to the cellular receptor ACE2, possibly providing protection against neutralizing antibodies.29,32 The S protein is a trimer, in which each monomer consists of two subunits: S1 and S2.
The S1 subunit includes an N-terminal domain (NTD), a receptor-binding domain (RBD), and subdomains associated with the RBD and NTD. In the early stages of infection, the S protein undergoes several conformational changes: the closed initial conformation minimizes the antigenic surface of the virus, and the open conformation at the contact stage with the ACE2 receptor has a much larger available antigenic surface.33 Transitions between open and closed conformations are associated with the opening of the trimer and large movements of the domains, especially the RBD, which are straightened to engage the receptor (Fig. 2a).32,33
The crystal structure of the S protein of SARS-CoV-2 has recently been resolved (PDB ID: 2AJF, 6ZGE, 7BNN).34–36 An example of the S protein of SARS-CoV-2 Omicron (PDB: 7WVO) is shown in Fig. 2a.
The S protein is one of the most immunogenic among other SARS-CoV-2 proteins. The emergence of new Alpha, Beta, Gamma and Delta variants of SARS-CoV-2 was associated with the presence of common S protein mutations in the N-terminal domain (NTD), the receptor-binding domain (RBD) and C-terminal domain (CTD). Mutagenesis studies of SARS-CoV found that two proline substitutions at residues 986 and 987 stabilize the S protein in its pre-fusion form, which elicits a strong immune response.27
The current strategies to inactivate the S protein rely on preventing the viral spike protein from binding to the cellular receptor ACE2 (Fig. 2b).17,22 Considering the allosteric mechanism of RBD-ACE2 binding, it seems that low-molecular weight ligands are not effective in this role.20 Instead, developing efficient RBD-ACE2 blockers is promising alternative.21,37,38 In addition, several neutralizing monoclonal antibodies, such as nCoV617 (PDB: 7E3O),39 B38 and H4 (PDB: 7BZ5)40 were suggested. The S protein stabilized by this double-proline mutation was used to develop mRNA vaccines mRNA-1273 and BNT162b2 by Moderna and Pfizer-BioNTech in December 2020 and January 2021, respectively.41,42 Since 2021, the BNT162b2/Pfizer-BioNTech vaccine has been approved in 85 countries from North and South America and Europe, while mRNA-1273/Moderna has been allowed in 45 countries in Europe and North America.43
A recent study of structural changes in the S protein in the main variations of SARS-CoV-2 revealed several regularities in the evolutionary dynamics of the virus. Spikes of variants acquired amino acid changes linked to increased virulence and immune evasion. In the initial Wuhan variant, the S protein was in a closed form before contact with the ACE2 receptor.12 Such structure may have provided protection against neutralizing antibodies. During the virus evolution, the ability of the spike of SARS-CoV-2 to switch to an open conformation became a competitive advantage because it facilitated binding to the receptor. At the same time, antibodies targeting the open subdomain of the RBD were rare from the point of view of population immunity.
Alpha-, Beta-, and Delta-variants of SARS-CoV-2 are variations with an open S protein structure. The Omicron variant has an already closed or partially closed conformation of the S protein and hides the RBD epitopes that are now recognized by antibodies due to mass vaccination. As a result, the previously circulating Alpha, Beta, and Delta variants are now non-competitive and disappearing at the population level.32
Permanent SARS-CoV-2 mutations are part of the evolutionary development of the virus, and the S protein is its dynamic part that directly responds to the environment and changes. Such responses to changes occurring in the S protein region lead to changes in the immunogenic characteristics of the virus and evasion of the virus from immunity.44
Several drugs that are monoclonal antibodies (mAbs) targeting the SARS-CoV-2 adhesion protein have received emergency use authorization (EUA) from the US Food and Drug Administration (bamlanivimab plus etesemab, cazirivimab plus imdevimab, sotrivab and bebtelovimab). However, they are not currently approved for use in the United States because it is expected that the activity of anti-SARS-CoV-2 mAbs against specific variants and subvariants of the virus may vary dramatically. The COVID-19 Treatment Guideline Panel recommends against the use of monoclonal antibodies against SARS-CoV-2 for the treatment or prevention of COVID-19 because the dominant subvariants of Omicron in the United States are not expected to be susceptible to these products.
The rapid evolution of the viral genome, focused mainly on S-protein mutations, creates problems when using the S-protein as a target for drug discovery. Therefore, the use of highly specific and more mutation-resistant targets is necessary for the development of new drugs against SARS-CoV-2.30
6.2. Nucleocapsid protein (N protein)
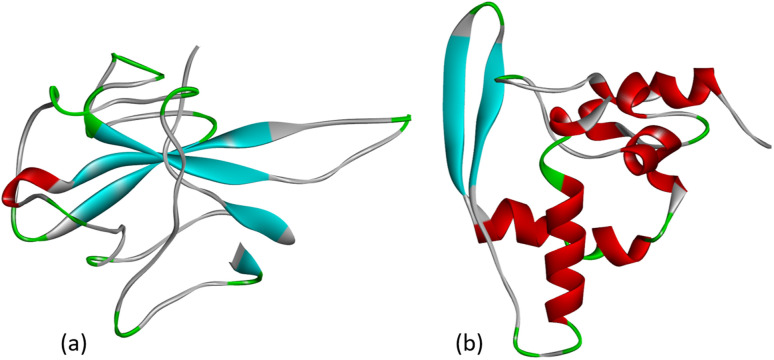
The nucleocapsid protein (N protein) of SARS-CoV-2 plays an important role in the life cycle of the coronavirus. When SARS-CoV-2 enters the host cell, the N protein dissociates from the positive strand (+) of the RNA genome of the virus. After that, the program of replication and expression of the viral genome begins. The N protein is a structurally heterogeneous multidomain RNA-binding protein with a length of 419 amino acids, which has the ability to recognize and bind RNA. The N protein is responsible for the identification and packaging of viral RNA. It binds to the genomic RNA of the virus, forming a ribonucleoprotein complex (RNP). The SARS-CoV-2 N protein consists of three highly disordered regions: the N-arm, the central linker region (LKR), and the C-tail, as well as two structural domains: the N-terminal RNA-binding domain (NTD) and the C-terminal dimerization domain (CTD).27 The crystal structure of the NTD domain and the C-terminal dimerization domain of the SARS-CoV-2 nucleocapsid has currently been characterized as shown in Fig. 3.
Fig. 3. (a) Crystal structure of the N-terminal RNA-binding domain of the nucleocapsid protein (PDB: 6M3M) and (b) the C-terminal dimerization domain of the SARS-CoV-2 nucleocapsid (PDB: 6ZCO).
Because the RNA-binding activity of the N protein is critical for the formation of viral particles, blocking RNA binding is one strategy for therapeutic effects, and N proteins are attractive targets for antiviral drugs.45 Contrary to the spike protein, the structure of which has been affected by mutations, such as Alpha, Beta, Gamma, Delta, Epsilon, and Omicron emerged in a period of the last two years, the N protein sequence alignments revealed ∼98% conserving.45 Since the beginning of the pandemic, some in silico studies have been conducted to identify potential SARS-CoV-2 nucleocapsid inhibitors using molecular docking-based drug repurposing.46,47 At the same time, such research did not lead to the development of drugs of such a focus. One of the reasons for the failure of developing small-molecule therapeutics based on nucleocapsid proteins might be the lack of well-defined binding and catalytic sites.
The N protein plays various important roles in the life cycle of SARS-CoV-2, not only related to the formation of a ribonucleoprotein complex with viral RNA. Further studies of the interaction between SARS-CoV-2 N protein and other viral and host cell proteins under disease conditions are necessary to understand the molecular mechanisms of the effect on the SARS-CoV-2 nucleocapsid.48
6.3. Envelope protein (E protein)
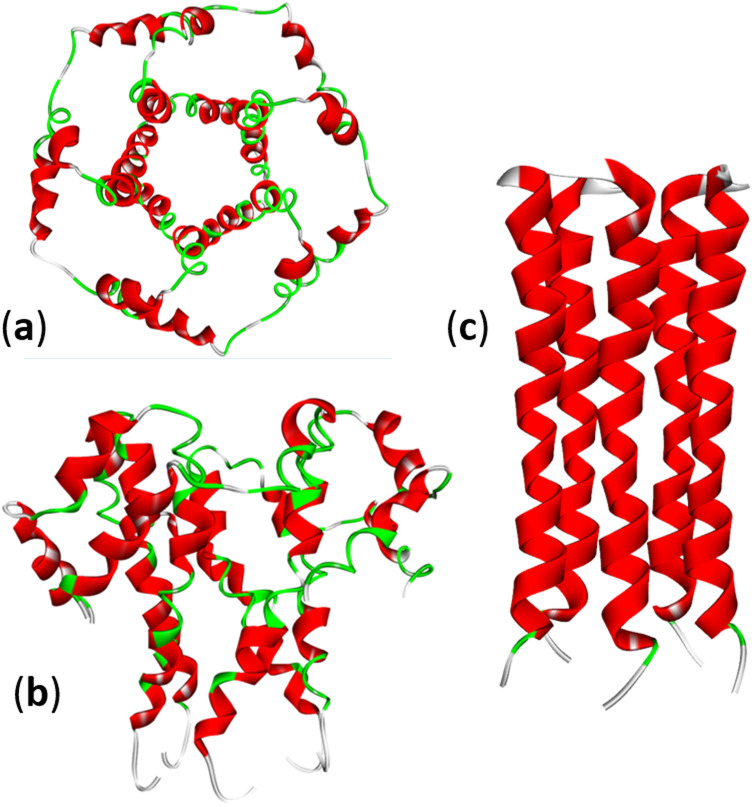
The envelope CoVs protein (E protein) is a small transmembrane protein with 75–109 amino acid residues and a mass of 8–12 kDa, present in small amounts in viral particles. The E protein can homooligomerize to form a cation-selective ion channel (IC) or viroporin (Fig. 4). Many viruses express viroporins; for example, the M2 protein of influenza A. Amantadine is a broad-spectrum viroporin inhibitor previously used to treat influenza A. Amantadine was tested against SARS-CoV-2, but it showed only moderate inhibition of the E protein activity.49
Fig. 4. Structure of the SARS-CoV-2 E-protein pentameric ion channel (PDB: 5X29): (a) top and (b) side views. (c) The pentameric structure of the transmembrane domain of the E-protein of SARS-CoV-2: (PDB: 7K3G).
The use of the E protein as a target for pharmacologic effects on SARS-CoV-2 is not currently being actively considered, but it may be a comprehensive strategy to reduce the emergence of resistance, since the virus is less likely to mutate in the part of the genome that encodes this protein.23
6.4. Membrane protein (M protein)
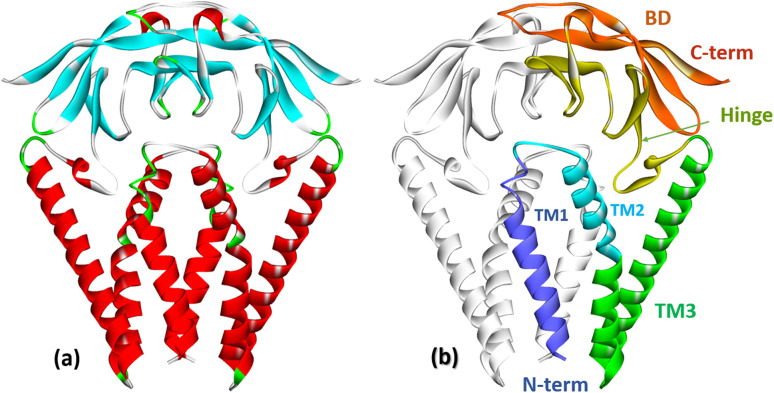
The membrane protein (M protein) is the most abundant protein in the envelope, which directs the process of virion assembly through interaction with other structural proteins. The M protein consists of 221 amino acid residues and has three structural components: the N-terminal part protruding from the membrane; transmembrane domain; the C-terminal part is the final part, which includes two domains (Fig. 5). The M protein forms a mushroom-shaped dimer consisting of two transmembrane three-helical bundles and two intravirion domains (Fig. 5).50
Fig. 5. (a) Crystal structure of the membrane protein (M-protein) of the coronavirus SARS-COV-2 (PDB: 8CTK). (b) Structural components of the M protein: two transmembrane three-helical bundles (TM1, TM2, TM3) and two intravirion Hinge domains and C-terminal β-sheet sandwich domain (BD).50.
The M protein can interact with other structural proteins: the interaction of the M protein with the nucleocapsid (N) protein contributes to the stability of the N protein. The M protein directs the process of viral particle assembly by interacting with other structural proteins.30 It is assumed that the M-protein acts as a framework for the joint assembly of a complex of structural and auxiliary proteins that form both the viral envelope and the viral particle itself.
Taking into account that the M protein is the most abundant protein in the viral envelope, it can be a target for vaccines and therapeutics.51 However, despite the broad abundance, drugs and potential inhibitors for the M protein have not yet been proposed.
6.5. Non-structural proteins
The general genome organization of SARS-CoV-2 includes sixteen non-structural proteins Nsp1–16, which are highly conserved among coronaviruses (Table 2).30 They are encoded by ORF1a/b at the 5′-end and are expressed as a single polyprotein chain Pp1a and Pp1ab, which is further cleaved into individual proteins by internal proteases (Fig. 1). The SARS-CoV-2 genome encodes two different cysteine proteases that are required for the viral proliferation cycle: papain-like protease (PLpro, domain in Nsp3, EC 3.4.22.46) and chymotrypsin-like main protease (3CLpro or Mpro, domain in Nsp5, EC 3.4.22.69). Viral nonstructural proteins are responsible for virulence and survival of the virus. To perform their function, they must be proteolytically separated from the polyprotein chain, thus proteolysis by two viral proteases plays a critical role in the life cycle of SARS-CoV-2.14
Non-structural proteins of SARS-CoV-2 and their functions52.
| Non-structural proteins of SARS-CoV-2 | Functions |
|---|---|
| Nsp1 | Ensures RNA processing and replication (PDB: 6ZOK, 2GDT) |
| Nsp2 | Modulates the host cell survival signalling pathway (PDB: 7EXM) |
| Nsp3 | Separates proteins that are translated (PLpro) (PDB: 6W02, 5Y3E) |
| Nsp4 | Contains a transmembrane domain (TM2) and modifies membranes |
| Nsp5 | The main protease (Mpro), participates in the process of cleavage of polyprotein during replication (PDB: 1WOF, 1Z1J, 2C3C) |
| Nsp6 | Is the transmembrane domain |
| Nsp7 | Has an effect on RNA-dependent RNA polymerase (PDB: 1YSY) |
| Nsp8 | The presence of nsp7, nsp8, and nsp11 affects RNA-dependent RNA polymerase and coronavirus replication/transcription |
| Nsp9 | Functioning as an ssRNA-binding protein |
| Nsp10 | Is crucial for cap-methylation of viral mRNAs (PDB: 2G9T) |
| Nsp11 | Together with nsp7, nsp8 targets RNA-dependent RNA polymerase and coronavirus replication/transcription |
| Nsp12 | Contains RNA-dependent RNA polymerase (RdRp), which is a critical component of coronavirus replication/transcription |
| Nsp13 | The zinc-binding domain of nsp13 is involved in replication and transcription processes (PDB: 6XEZ, 7EIZ) |
| Nsp14 | Proofreading exoribonuclease domain |
| Nsp15 | Has Mn2+-dependent endoribonuclease activity (PDB: 6VWW) |
| Nsp16 | 2′-O-ribosemethyltransferase (PDB: 6WKS) |
Both structural and nonstructural proteins play important roles during different stages of the SARS-CoV-2 life cycle and can be selected as targets for chemotherapy.30 Along with developing small molecule antivirals, significant research progress has been made toward targeting SARS-CoV-2 RNA-dependent RNA polymerase (RdRp), main protease (Mpro or 3CLpro), and papain-like protease (PLpro).53
7. Promising targets for pharmacological action on SARS-CoV-2
7.1. RNA-dependent RNA polymerase (RdRp)
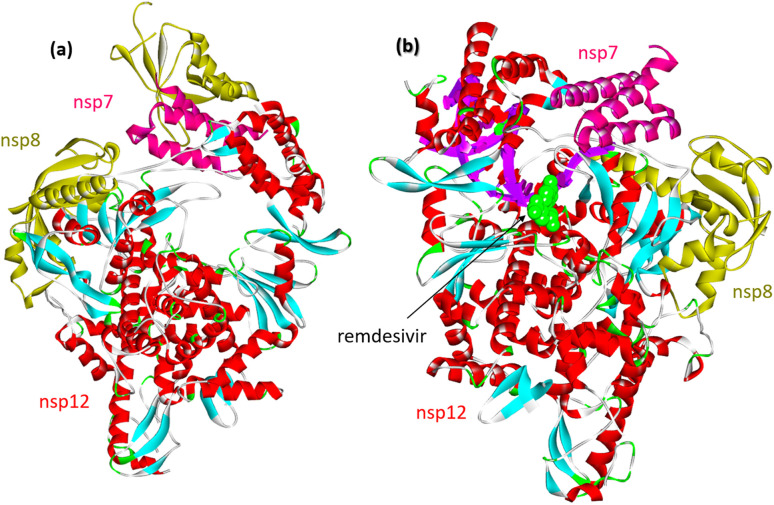
Upon SARS-CoV-2 virus replication, the replication/transcription complex consists of the viral nonstructural proteins nsp7, nsp8, and nsp12.54 The core of the complex is sp12-RNA-dependent RNA polymerase (RdRp). Nsp12 is weakly active by itself. Some studies demonstrated that the presence of nsp7 and nsp8 significantly increased the combination of nsp12 and template RNA. The crystal structure of the nsp12–nsp7–nsp8 complex was recently identified fpr isolated protein and its complex with remdesevir, as shown in Fig. 6.
Fig. 6. (a) Cryo-electron microscope structure of the nsp12–nsp7–nsp8 complex (PDB: 7BV1). (b) The nsp12–nsp7–nsp8 complex associated with the template primer RNA and remdesivir triphosphate (RTP) (PDB: 7BV2).
RNA-dependent RNA polymerase, which catalyzes the synthesis of viral RNA, is a critical component of coronavirus replication/transcription. Therefore, RdRp is believed to be an essential target for developing new antiviral drugs.52,54–58 The first reported inhibitor of SARS-CoV-2 RdRp is Remdesivir, which was identified by a drug repurposing approach (Fig. 7). Remdesivir is a nucleoside analog that inhibits RNA polymerases. Nucleoside analogues usually target viral replication, especially viral DNA or RNA polymerase, and are successfully used in clinical practice in the treatment of viral infections.58,59 In several studies, the RdRp protein was used as a target receptor for the in silico screening for new antiviral drugs.60–66
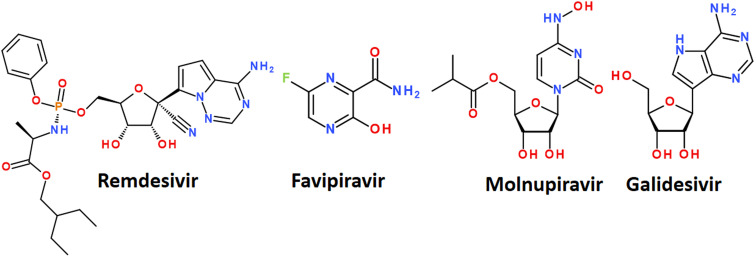
Fig. 7. Some nucleoside analogs available for the treatment of COVID-19.
The mechanism of action of drugs belonging to the nucleoside analogues is based on structural similarity with natural nucleosides/nucleotides.67 However, they have some minor changes in the structure, which are critical for the further fate of the macromolecules in which they are included. Nucleoside analogues are incorporated into the RNA chain synthesized by the virus during replication and disrupt its further assembly.61 It leads to premature chain termination and, hence, the inhibition of virus replication.67 A nucleoside analog is defined as a nitrogenous base attached to a sugar where the nitrogenous base or sugar component is altered in such a way that the molecule is different from that found in nature. Modifications of the nitrogenous bases include halogenation, addition of azido groups, and modifications of the sugar component include ring opening, halogenation, methylation, hydroxylation, or dihydroxylation.68 A number of nucleoside analogues have already passed clinical trials for the treatment of COVID-19 and are on the market. The most successful examples are Remdesivir, Favipiravir, Molnupiravir and Galidesivir (Fig. 7).56,69
Remdesivir has been approved by the USA Food and Drug Administration (FDA) as the first small molecule for an “emergency use permit” for the treatment of COVID-19.70 The full FDA approval was granted in October 2020. Intravenous therapy with this drug is allowed for use in both hospitalized and non-hospitalized patients.70
Remdesivir targets RNA-dependent RNA polymerase, which creates new copies of coronavirus RNA.54,65 Remdesivir mimics the adenine nucleotide, and RNA-dependent RNA polymerase inserts it into the growing RNA molecule instead of adenine.71 This disrupts the normal form of the RNA chain and interrupts its synthesis. In several clinical trials, treatment of COVID-19 with remdesivir resulted in improved health.72 A side-effect is, however, that remdesivir is unstable in the blood and has a limited half-life of less than an hour in humans, requiring a continuous infusion.73
Favipiravir was originally developed against the flu. It belongs to nucleoside analogues and blocks the ability of the virus to copy its genetic material. It is metabolized in cells to favipiravir ribosyltriphosphate (favipiravir RTF) and selectively inhibits RNA-dependent RNA polymerase.
There are evidences that favipiravir may be effective in SARS-CoV-2 infection. At the same time, the WHO, Great Britain, the United States and Canada did not provide recommendations for the use of favipiravir against COVID-19. Some molecular docking studies showed a high affinity of favipiravir to the main Mpro protease of SARS-CoV-2.74,75
Molnupiravir is a nucleoside analogue proposed for oral use in the treatment of early stages of COVID-19. Like favipiravir, it was originally developed as an antiviral drug to treat influenza. The mechanism of action is the accumulation of errors in the viral genome, when the drug is incorporated into the RNA of the virus, causes mutations in the viral RNA and thus stops the replication of the virus. In some countries, the drug has been approved for the treatment of confirmed COVID-19 infections.76
Galidesivir (Galidisvir, BCX4430) is a novel synthetic adenosine analogue, which effects on the inhibition of RNA transcriptional activity was first assessed in a cell-free isolated HCV RdRp assay.77 Galidesivir acts as a nonobligate RNA chain terminator56 and can be administered by both parenteral and oral routes.
A side effect of nucleoside analogues, which limits their clinical treatments, is its ability for disrupting RNA polymerase chains, causing some risks of RNA mutagenesis impairing the human genes.78
Finally, natural and synthetic agents, and the most promising repurposed drugs against SARS-CoV-2 RdRp have recently been reviewed elsewhere.79,80
7.2. Main protease Mpro
Protein kinases are a class of enzyme-kinases that modify specific proteins and are responsible for the activation of various proteins in the cascade of cellular signaling pathways. Due to the significant effect of protein kinases on cell functioning, they are attractive targets for drug development. There are a significant number of available medicinal substances that are inhibitors of protein kinases, and they were primarily investigated as possible therapeutic agents in the treatment of COVID-19.11 Some inhibitors of tyrosine kinase tested by the drug-repurposing algorithm showed activity against COVID-19, such as Baricitinib, Fedratinib, Gilteritinib, Imatinib, Ivermectin, Lonafarnib, Nilotinib, Osimertinib, Ruxolitinib, and Tofacitinib, however, their efficacy was insufficient for direct therapeutic use.81–83
The main attention of the researchers was focused on the main protease of SARS-CoV-2. Since no known human protease recognizes this cleavage site, Mpro is considered an ideal target for therapeutic development.27,84,85 In addition, Mpro active sites are highly conserved among all coronaviruses.52
Nonstructural protein 5 (Nsp5) or main protease Mpro (3C-like protease) is responsible for the cleavage of eleven Mpro-specific sites on the two SARS-CoV-2 polyproteins Pp1a and Pp1ab into 11 nonstructural proteins (nsp4-16) (Fig. 1). MPro consists of three domains: chymotrypsin-like domain I and 3C-protease-like domain II, as well as domain III consisting of 5 α-helices. The MPro binding pocket is located between domains I and II and has a catalytic CysHis dyad (Cys145 and His41). Cysteine sulfur serves as a nucleophile, and the imidazole ring of histidine serves as a general base. MPro targets the recognition sequence Leu–Gln↓(Ser, Ala, Gly)↓ (where ↓ means a cleavage site) (Fig. 8).86
Fig. 8. Mechanism of action of cysteine proteases. Reproduced with permission from ref. 88. © 2020 Elsevier Ltd. All rights reserved.
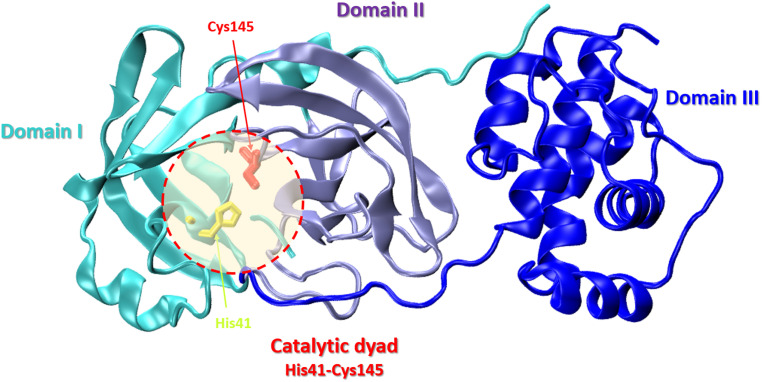
The SARS-CoV-2 Mpro structure consists of three domains: domain I (residues 8–101), domain II (residues 102–184), and domain III (residues 201–303). The third domain is connected to domain II by a long loop region (residues 185–200) (Fig. 9).87
Fig. 9. Crystal structure of SARS-CoV-2 main protease (Mpro) with the catalytic center (His41 and Cys145) (PDB: 6LU7): domain I – cyan, domain II – violet, domain III – blue.
The high-resolution X-ray structures of the Mpro coronavirus protease have been used in many studies to develop antiviral drug candidates targeting SARS-CoV-2.87,89,90
Numerous experimental and theoretical studies have been devoted to design and development of novel noncovalent inhibitors of Mpro enzyme.47,56,84,91–96
Phytochemical extracts, medicinal plants and aromatic herbs, composed of polyphenols, terpinols, flavonols and their glicosides, have revealed a broad spectrum of inhibitory potency and were suggested as potential anti-COVID agents.80,97–103 Several comprehensive reviews and updates on recent development of Mpro inhibitors have been released.104,105 Here we briefly summarize most crucial achievements in this field.
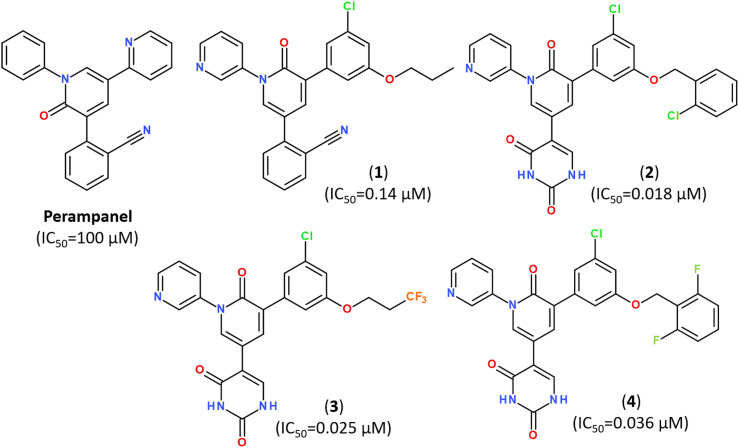
Popular strategies for rational design and discovery of novel inhibitors are often based on re-design, hit-to-lead optimization and hit expansion of available most effective inhibitors.106 Recently, redesigning of know inhibitor perampanel was guided by free energy perturbation MD simulations, kinetic assays and high-resolution X-ray analysis (PDB ID: 7M8M, 7M8N, 7M8O), resulting in a series of noncovalent, nonpeptidic inhibitors with IC50 values in a range of 20–40 nM, as summarized in Fig. 10.107–109
Fig. 10. Structure of Perampanel analogs inhibiting the activity of Mpro.108.
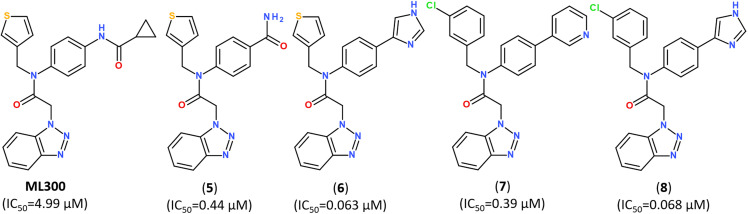
Developing noncovalent inhibitors of a coronavirus 3CLpro in the aftermath of the SARS-CoV-1 outbreak, led to the discovery of inhibitor ML300, which also revealed high activity against SARS-CoV-2.110 Starting from the compound ML300, a structure-based optimization was performed against the SARS-CoV-2 Mpro protease using X-ray analysis of Mpro enzymes in complex with multiple ML300-based inhibitors, including the original probe ML300, resulting in discovery of a series of potent inhibitors with nanomolar IC50 values, as summarized in Fig. 11. The X-ray structures of their complexes with Mpro revealed that the disclosed inhibitors utilize a noncovalent mode of action and noncanonical binding mode (PDB ID: 7MLD, 7MLE, 7MLF).111
Fig. 11. Structure of noncovalent Mpro inhibitors based on ML300 scaffold.111.
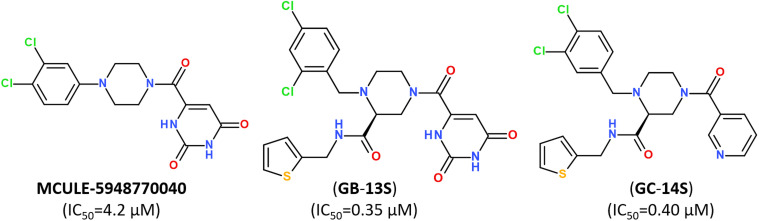
Small-molecule inhibitor MCULE-5948770040 was first identified by scalable high-throughput virtual screening of a targeted compound library of over 6.5 million molecules (Fig. 12).112 A binding mode of newly-discovered inhibitor was validated by using room-temperature X-ray crystallography (PDB: 7LTJ) and biochemical assays estimated its inhibitory activity with a moderate inhibition constant IC50 of 4.2 μM (Fig. 12).
Fig. 12. Structure of non-covalent inhibitor MCULE-5948770040 and its optimized hits.93.
Keeping a 1,2,4-trisubstituted piperazine scaffold, further structure-based rational design was performed and led to the discovery of a series of new potent non-covalent inhibitors, among which GB-13S and GC-14S displayed excellent target selectivity for SARS-CoV-2 Mpro, high antiviral activity, low cytotoxicity (CC50 > 100 μM). Moreover, their X-ray co-crystal structures (PDB ID: 8ACL, 8ACD) proved that these inhibitors occupy multiple sub-pockets by critical non-covalent interactions.93 Recently, novel noncovalent nonpeptidic inhibitor S-217622 (Ensitrelvir) has been discovered via a structure-based drug design (SBDD) strategy, starting from virtual screening and followed by X-ray analysis (PDB ID: 7VTH, 7VU6) and biological evaluation of an in-house compound library (Fig. 13).113
Fig. 13. A noncovalent orally active agent Ensitrelvir (S-217622) and its analogs with strong broad-spectrum anticoronaviral activities.113.
The compound S-217622 exhibited high antiviral activity in vitro against all currently outbreaking SARS-CoV-2 variants, including Delta and Omicron, and showed favorable pharmacokinetic profiles in vivo for once-daily oral dosing, which makes it an universal broad-spectrum antiviral clinical candidate for treating COVID-19.113–115
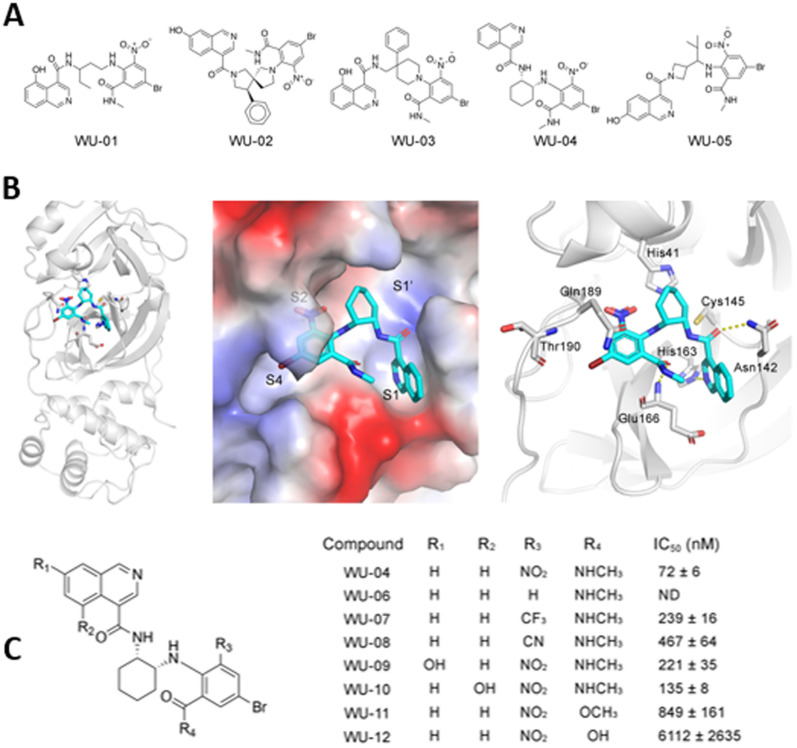
A series of specific, noncovalent inhibitors of Mpro, capable of effectively inhibiting SARS-CoV-2 replications in human cells with EC50 values in the 10 nM range, has been reported.96 WU-04 also inhibits the Mpro of SARS-CoV and MERS-CoV with high potency, similar to that of PF-07321332 (Nirmatrelvir) (Fig. 14).
Fig. 14. (A) Structure of noncovalent inhibitors of SARS-CoV-2 Mpro, containing an isoquinoline ring and a bromophenyl ring. (B) The binding of noncovalent inhibitor WU-04 into the catalytic pocket of SARS-CoV-2 Mpro. (C) Seven hit analogs of WU-04 and their inhibitory activity against SARS-CoV-2 Mpro evaluated using the fluorescent assay. Adapted with permission from ref. 96. Copyright © 2023 The Authors. Published by American Chemical Society.
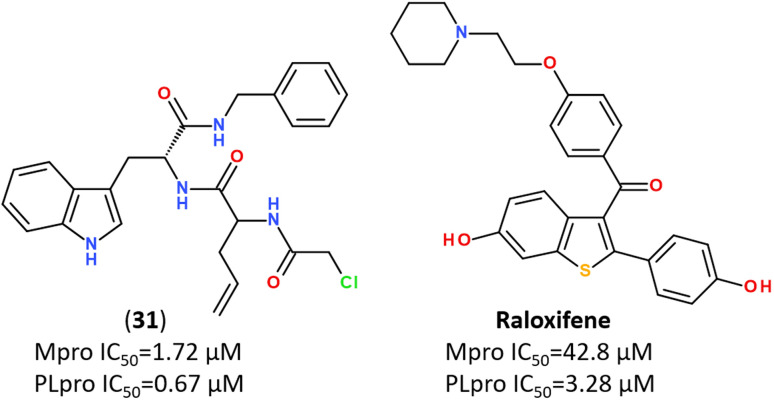
Several promising strategies for developing Mpro inhibitors utilize peptidomimetic scaffolds.83,117–122 A structure-guided design of direct-acting antivirals that exploited the gem-dimethyl effect and inhibited protease Mpro allowed identifying a series of highly potent covalent inhibitors shown in Fig. 15. They inhibited both SARS-CoV-2 and MERS-CoV 3CL proteases in biochemical and cell-based assays. The binding mode of novel inhibitors was confirmed by the X-ray analysis (PDB ID: 8F44, 8E5X).116
Fig. 15. Direct-acting covalent inhibitors of Mpro that exploited the gem-dimethyl effect.116.
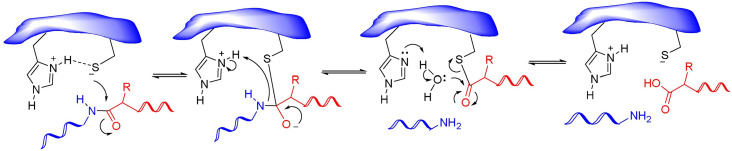
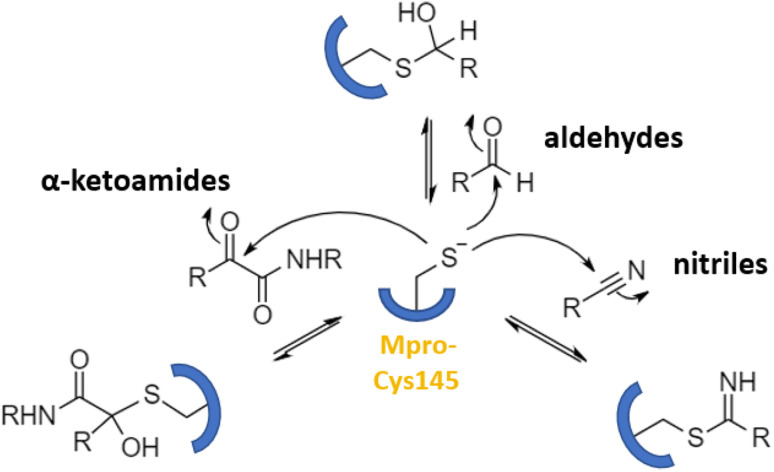
Numerous covalent-acting peptide-like inhibitors of Mpro protease have been discovered and characterized.123–129 Covalent inhibitors possess a reactive functional moiety that forms a temporary covalent bond upon reaction with the nucleophilic cysteine thiolate. Popular reactive groups consist of aldehydes and ketones form hemithioacetals, and nitriles form a reversible thioimidate (Fig. 16).104 Some of covalent Mpro inhibitors are summarized in Fig. 17.
Fig. 16. Different mechanisms of covalent Mpro inhibition, involving α-ketoamides, aldehydes, and nitriles. Reproduced with permission from ref. 104. © 2023 by the authors. Licensee MDPI, Basel, Switzerland.
Fig. 17. Reported peptidomimetic Mpro inhibitors with the high inhibitory activities (μM).120.
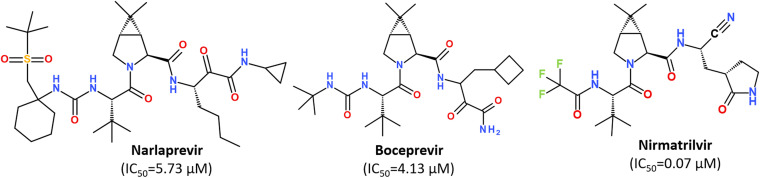
While some FDA-approved HCV drugs, such as Narlaprevir and Boceprevir, have revealed high potency against SARS-CoV-2 too,120,123,130,131 their clinical use was not reported yet (Fig. 18). Currently, Nirmatrelvir (PF-07321332) is only 3CLpro protease inhibitor approved for medical use against SARS-CoV-2 (Fig. 18).132–134 Nirmatrelvir in combination with ritonavir is available as the combination antiviral drug “Paxlovid” for oral use, developed by Pfizer.135,136 Ritonavir is believed to have a synergistic effect on antiviral activity.137,138
Fig. 18. Chemical structure of Narlaprevir, Boceprevir and Nirmatrelvir.120.
Nirmatrelvir is a reversible covalent inhibitor carrying a nitrile warhead that binds to the 3C-like protease of SARS-CoV-2, interrupting the viral replication cycle.90,123,139,140 It has been reported that Nirmatrelvir demonstrated distinct antiviral potency against prevalent variants of SARS-CoV-2, such as Alpha, Beta, Delta, Gamma, Omicron141 and different human coronaviruses.142 Nirmatrelvir has become a promising scaffold for computer-guided design of its better-acting analogs.143 Using a hybrid approach combining machine learning and free energy simulations, six new derivatives were designed by modifying parent nirmatrelvir, so that they bind strongly to SARS-CoV-2 Mpro and might be less toxic to the human body than the original inhibitor.144
7.3. Papain-like protease PLpro
Papain-like protease (PLpro) is a 35 kDa domain of Nsp3, part of a 215 kDa multidomain protein.145 This protease cleaves the peptide bonds between Nsp1 and Nsp2, Nsp2 and Nsp3, and Nsp3 and Nsp4, releasing three proteins: Nsp1, Nsp2, and Nsp3. In addition, PLpro is involved in counteracting the host's immune response during viral infection.146 The SARS-CoV-2 papain-like protease PLpro has long been recognized as a critical enzyme in the virus's life cycle.8
It is highly conserved, found in all coronaviruses, and has low sequence similarity with human enzymes.8 All these features make it a promising target for antiviral drugs.147 Some studies of PLpro led to the identification of many inhibitors specific to SARS-CoV PLpro.53 Moreover, both non-covalent and covalent inhibitors of PLpro have been discovered.56,148–151
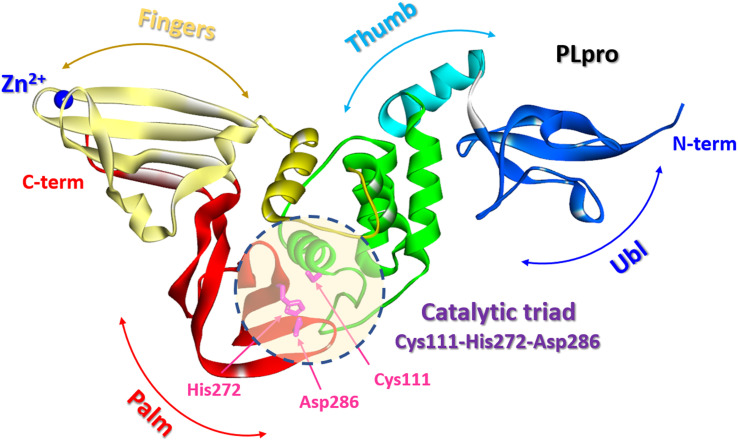
The SARS-CoV-2 PLpro is a protein composed of 315 amino acid residues, with a high content of cysteine residues (3.5%). Several high-resolution structures of free PLpro and its complex with inhibitors have become available (PDB: 7JIR, 7CMD, 6WX4, 6WUU, 6XA9, 6XAA).147,152–154 The PLpro structure contains three domains: a small N-terminal ubiquitin-like (Ubl) domain, a zinc-binding domain, and a catalytic thumb-palm domain, which is sometimes considered as two separate domains (Fig. 19). The zinc-binding domain, which includes four cysteines, coordinates the Zn2+ structural cation in the finger subdomain.53 This domain is believed to maintain the structural integrity of the whole PLpro protein.155
Fig. 19. PLpro structure: terminal ubiquitin-like (Ubl) domain, zinc-binding domain, and catalytic thumb-palm domain. The catalytic active site containing the catalytic triad Cys111-His272-Asp286 (shaded circle) (PDB: 6WX4).
It should be noted that the N-terminal ubiquitin-like (Ubl) domain of PLpro is not specific and has similarities with human cellular DUBs (DUBs are proteases that cleave ubiquitin chains from protein substrates). Ubl inhibitors can affect human DUBs and lead to side effects. For example, Otubain-1 and Otubain-2 are PLpro-like cysteine proteases with the same triad of catalytic Cys/His/Asp, Asn residues.156 This issue might be one of the possible reasons why essential efforts in developing PLpro inhibitors have not yet led to the development of FDA-approved drugs for therapeutic effect on SARS-CoV-2 in humans.
The catalytically active site of cysteine cleavage (also called the active center) is located between the “thumb” – “palm” domains and contains the canonical catalytic triad for cysteine proteases Cys111-His272-Asp286 (ref. 147) (Fig. 19), which recognizes the sequence of Leu-X-Gly-Gly residues found in-between viral proteins nsp1 and nsp2, nsp2 and nsp3, and nsp3 and nsp4 (nsp1/2, nsp2/3, and nsp3/4) and performs their proteolysis – cleaves them after the second Gly.147,157 Of all PLpro cysteines, Cys111 is the most susceptible to oxidation, indicating its unique reactivity toward electrophiles.145 PLpro binds to viral proteins through its active site to fulfill its biological role. The catalytically active site of PLpro is a conservative drug target among SARS-CoV-2 variants. Although some mutations have been identified, all most common mutations are located distal to the binding site.53,158
As mentioned above, from a structural point of view, PLpro seems to be a less attractive target for drug development compared to Mpro because it shares structural similarities with human DUBs. Nevertheless, the growing number of recent studies on PLpro inhibitors indicate that it remains one of the promising targets for the development of small molecule therapeutics against SARS-CoV-2.145,155,159
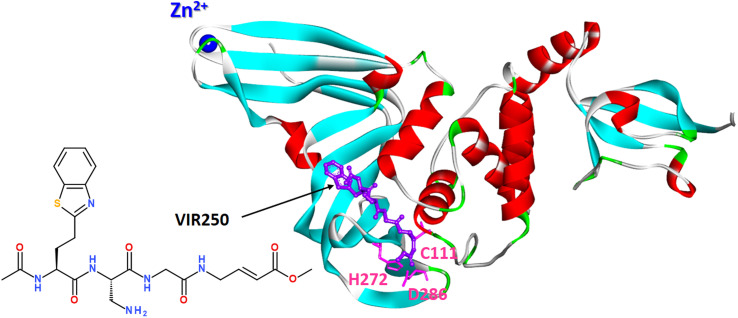
Covalent peptide inhibitors VIR250 and VIR251, consisting of a Gly–Gly mimetic linker and a reactive electrophile, have proved to be active and selectively inhibited the PLpro of SARS-CoV-1 and SARS-CoV-2; however, they exhibited much weaker activity against MERS PLpro and human DUB UCH-L3.152
A molecular basis for the observed inhibitor specificity was provided to resolving X-ray structure of PLpro in complex with VIR250 peptide, as shown in Fig. 20.
Fig. 20. X-ray structure of PLpro in complex with covalent peptide inhibitor VIR250 (PDB: 6WUU).
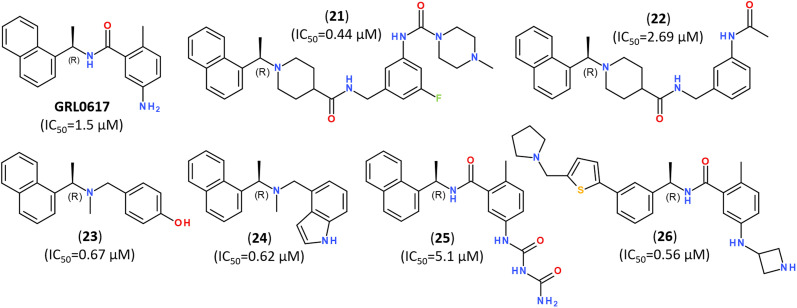
The significant efforts are, however, focused on developing non-covalent inhibitors of PLpro.149 Naphthalene-based non-covalent inhibitor GRL0617 (Fig. 21) was initially developed as an inhibitor of PLpro of SARS-CoV, and it did not show inhibitory activity against other host proteases.160 Earlier trials revealed that the half-maximal inhibitory concentration (IC50) of GRL0617 was 1.5 μM, so that its scaffold became a promising target for developing a series of its analogs.147–149,161,162
Fig. 21. Highly potent inhibitors of PLpro designed on a scaffold of the naphthalene-based inhibitor GRL-0617.
Recent study on hepatitis C virus (HCV) drug repurposing, has demonstrated that four HCV protease inhibitors, such as simeprevir, vaniprevir, paritaprevir, and grazoprevir, inhibited the SARS-CoV-2 PLpro too. So, HCV drugs that acting against PLpro synergized with the viral polymerase inhibitor remdesivir (Fig. 7) to inhibit virus replication, increasing remdesivir's antiviral activity as much as 10-fold.163
It has been shown that some human ubiquitin carboxyl-terminal hydrolase-2 (USP2) inhibitors, such as thiopurine analogs, were capable of inhibiting PLpro proteases. For instance, 6-thioguanine blocks SARS-CoV-2 replication by inhibition of PLpro with an EC50 of approximately 2 mM.164 Using virtual screening combined with in vitro cytotoxicity studies, it was found that 1,3-benzothiazole derivative, known as ZINC9325709 or Z93, revealed the high potency against PLpro.165
In silico screening of a chemical libraries composed of >50k compounds allowed to design and synthesize other structurally diverse lead structures capable of noncovalent broad-spectrum inhibiting of coronavirus PLpro with low in vitro IC50,166–168 as summarized in Fig. 22.
Fig. 22. Structurally diverse inhibitors of PLpro enzyme of SARS-CoV-2.
Beside the synthetic small-molecule inhibitors, some natural compounds, such as flavones, have revealed the inhibitory potency against PLpro enzyme.101,105,169
7.4. Dual and multi-targeting inhibitors of SARS-CoV-2 enzymes
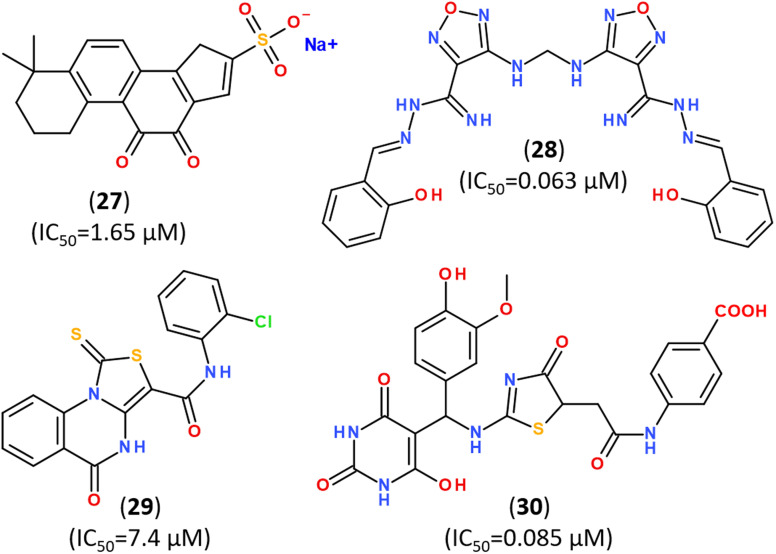
Recent trends in the drug design against COVID-19 are focused on developing multi-target inhibitors for fighting several crucial proteins of SARS-CoV-2 with high potency.167,170–174 The ability to identify inhibitors with dual inhibition against both Mpro and PLpro will increase the chances of success in the development of drug molecules against new variants of SARS-CoV-2.53,175–178 Some studies have already reported compounds identified as dual-acting SARS-CoV-2 protease inhibitors that target both Mpro and PLpro (Fig. 23).179,180
Fig. 23. Dual-targeting inhibitors against Mpro and PLpro enzymes.132,177,180.
In silico screening of 688 naphthoquinoidal derivatives against Mpro protease of SARS-CoV-2 allowed to identify twenty-four derivatives for further biochemical assay evaluation, among which four hits were active against both Mpro and PLpro. These four hits inhibited Mpro with IC50 values between 0.41 μM and 9.0 μM, whereas, three of them inhibited PLpro with IC50 ranging from 1.9 μM to 3.3 μM, respectively.179
Some drug-like furo[2,3-d]pyrimidines and other 74 drug-like compounds were screened as potential active site inhibitors and putative allosteric hotspots modulators of SARS-CoV-2 Mpro and PLpro.181 It has also been reported that some pertinent medicinal plants i.e. Eurycoma harmandiana, Sophora flavescens and Andrographis paniculata contain phyto-compounds, such as terpenoids, flavonoids and their glycosides, act against SARS-COV-2 PLpro and Mpro proteases.80,173,182
A combined approach for in silico screening towards repositioning of FDA-approved drug candidates for anticoronaviral therapy has identified Ouabain, which is an antiarrhythmic drug of plant origin, as a dual inhibitor for both PLpro and Mpro enzymes.183
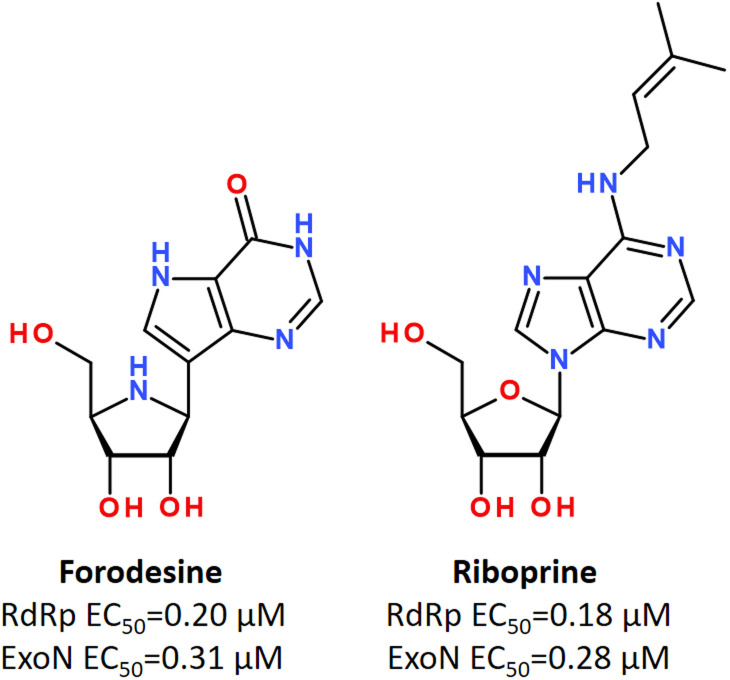
Using in vitro studies and in silico screening, it has been shown that some purine nucleosides inhibiting nucleoside phosphorylase enzymes, such as Forodesine and Riboprine, revealed strong repurposing potential against two broadly conserved SARS-CoV-2 enzymes, such as RNA-dependent RNA polymerase (RdRp) and 3′-to-5′ exoribonuclease (ExoN), irrespective of the SARS-CoV-2 variant type (Fig. 24).184,185
Fig. 24. Forodesine and Riboprine as dual acting inhibitors against RdRp and ExoN enzymes of SARS-CoV-2.184.
Moreover, a newly discovered orally active noncovalent nonpeptidic agent Ensitrelvir (S-217622) (Fig. 13) has also revealed a synergistic triple noncovalent inhibitory activity against the three principal SARS-CoV-2 enzymes, namely: Mpro, RdRp, and ExoN by strong and stabilizing striking the key catalytic pockets of the SARS-CoV-2 RdRp's and ExoN's principal active sites.114
Finally, a recent in silico molecular docking study identified a FDA-approved antiviral agent, Rilpivirine, as a multi-target drug with high inhibitory potency against the Spike Glycoprotein and ACE2 receptors, PLpro and Mpro proteases, as well as RdRp protein, suggesting it may play a critical role in inhibiting the viral replication and assembly of the virus SARS-CoV-2.55
8. Summary and perspectives
The development of antiviral drugs for the treatment of COVID-19 continues to be relevant because of the risk of the appearance of SARS-CoV-2 mutated strains, reducing the effectiveness of existing vaccines. This review provides an overview of recent efforts in researching and developing coronavirus-related therapeutic agents. It includes an overview of coronavirus morphology, life cycle, and key proteins, focusing on antiviral strategies involving small molecule covalent and non-covalent inhibitors. The functional importance of Mpro and PLpro in the viral life cycle and the lack of close homologs in humans identify these proteases as an ideal target for screening of new inhibitors for SARS-CoV-2 replication. Active sites of Mpro are highly conservative for all coronaviruses. The review summarizes the drug-repurposing effort, focused primarily on inhibitors currently known to be effective against key cysteine-like proteases. In particular, Mpro and PLpro proteases of SARS-COV-2 were overviewed as therapeutic targets and their functional significance in the life cycle of the coronavirus was justified. Moreover, this report summarizes critical small molecule compounds discovered during the last three years, suitable for inhibiting several key coronavirus proteins.
The review outlines long-term drug development goals with a particular focus on a design strategy for dual-acting inhibitors that can inhibit both PLpro and Mpro proteases. It is believed that the combined therapy against both Mpro and PLpro inhibitors allows one to prevent the problem of drug resistance due to mutations in the viral genome.
Looking at the current scenario for the discovery of dual-targeting inhibitors, we can conclude that comparing pharmacophore models of Mpro and PLpro proteases may allow the identification of a set of spatial and electronic features, which are crucial for their interaction with biological targets. The combined pharmacophore model should include the pharmacophore features of both Mpro and PLpro models. The use of various selection criteria (stopping rule) makes it possible to determine the points of the best and worst regression model to identify an array of optimal options for the pharmacophore model for dual-acting inhibitors.
Author contributions
Larysa V. Yevsieieva: conceptualization, methodology, writing – original draft. Kateryna O. Lohachova: methodology, writing – original draft, resources. Alexander Kyrychenko: conceptualization, methodology, writing – review & editing. Sergiy M. Kovalenko: conceptualization, methodology, writing – original draft Volodymyr V. Ivanov: conceptualization, methodology, writing – original draft. Oleg N. Kalugin: writing – review & editing, supervision.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors acknowledge Grant No. 42/0062 (2021.01/0062) “Molecular design, synthesis and screening of new potential antiviral pharmaceutical ingredients for the treatment of infectious diseases COVID-19” from the National Research Foundation of Ukraine.
References
- Gil C. Ginex T. Maestro I. Nozal V. Barrado-Gil L. Cuesta-Geijo M. Á. Urquiza J. Ramírez D. Alonso C. Campillo N. E. Martinez A. COVID-19: Drug Targets and Potential Treatments. J. Med. Chem. 2020;63:12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- Meskini M. Rezghi Rami M. Maroofi P. Ghosh S. Siadat S. D. Sheikhpour M. An Overview on the Epidemiology and Immunology of COVID-19. J. Infect. Pub. Health. 2021;14:1284–1298. doi: 10.1016/j.jiph.2021.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavone C. Brusco S. Bertini M. Sportiello L. Rafaniello C. Zoccoli A. Berrino L. Racagni G. Rossi F. Capuano A. Current pharmacological treatments for COVID-19: What's next? Br. J. Pharmacol. 2020;177:4813–4824. doi: 10.1111/bph.15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shargi O. Y. Alzaid S. M. Al-Dosari B. S. Alzubaidi A. S. Adverse events associated with COVID-19 treatment and their possible relationship with patient characteristics: A narrative review. J. Appl. Pharm. Sci. 2023;13:41–49. [Google Scholar]
- Davis H. E. McCorkell L. Vogel J. M. Topol E. J. Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023;21:133–146. doi: 10.1038/s41579-022-00846-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. Hozumi Y. Yin C. Wei G.-W. Decoding SARS-CoV-2 Transmission and Evolution and Ramifications for COVID-19 Diagnosis, Vaccine, and Medicine. J. Chem. Inf. Model. 2020;60:5853–5865. doi: 10.1021/acs.jcim.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. Asghar A. Singh H. N. Faiq M. A. Kumar S. Narayan R. K. Kumar G. Dwivedi P. Sahni C. Jha R. K. Kulandhasamy M. Prasoon P. Sesham K. Kant K. Pandey S. N. SARS-CoV-2 Omicron Variant Genomic Sequences and Their Epidemiological Correlates Regarding the End of the Pandemic: In Silico Analysis. JMIR Bioinform. Biotechnol. 2023;4:e42700. doi: 10.2196/42700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-r. Yin W.-c. Jiang Y. Xu H. E. Structure genomics of SARS-CoV-2 and its Omicron variant: drug design templates for COVID-19. Acta Pharmacol. Sin. 2022;43:3021–3033. doi: 10.1038/s41401-021-00851-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polatoğlu I. Oncu-Oner T. Dalman I. Ozdogan S. COVID-19 in early 2023: Structure, replication mechanism, variants of SARS-CoV-2, diagnostic tests, and vaccine & drug development studies. MedComm. 2023;4:e228. doi: 10.1002/mco2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov P. V. Ghafari M. Beer M. Lythgoe K. Simmonds P. Stilianakis N. I. Katzourakis A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023;21:361–379. doi: 10.1038/s41579-023-00878-2. [DOI] [PubMed] [Google Scholar]
- Zhu W. Shyr Z. Lo D. C. Zheng W. Viral Proteases as Targets for Coronavirus Disease 2019 Drug Development. J. Pharmacol. Exp. Ther. 2021;378:166–172. doi: 10.1124/jpet.121.000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. Rao Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021;19:685–700. doi: 10.1038/s41579-021-00630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. Expression of animal virus genomes. Bacteriol. Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M. Ruggiero A. Squeglia F. Maga G. Berisio R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis. Proofreading and Final Capping. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant A. C. Tian W. Majerciak V. Yang W. Zheng Z.-M. SARS-CoV-2: from its discovery to genome structure, transcription, and replication. Cell Biosci. 2021;11:136. doi: 10.1186/s13578-021-00643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W. Zheng Y. Zeng X. He B. Cheng W. Structural biology of SARS-CoV-2: open the door for novel therapies. Signal Transduction Targeted Ther. 2022;7:26. doi: 10.1038/s41392-022-00884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzato M. Baraldi C. Boscato Sopetto G. Finozzi D. Gentile C. Gentile M. D. Marconi R. Paladino D. Raoss A. Riedmiller I. Ur Rehman H. Santini A. Succetti V. Volpini L. SARS-CoV-2 and the Host Cell: A Tale of Interactions. Front. Virol. 2022;1:815388. [Google Scholar]
- Jackson C. B. Farzan M. Chen B. Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. Xiang R. Huo S. Zhou Y. Jiang S. Wang Q. Yu F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduction Targeted Ther. 2021;6:233. doi: 10.1038/s41392-021-00653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. Shi K. Wang J. Zhang R. Wang Y. Targeting SARS-CoV-2 entry processes: The promising potential and future of host-targeted small-molecule inhibitors. Eur. J. Med. Chem. 2023:115923. doi: 10.1016/j.ejmech.2023.115923. [DOI] [PubMed] [Google Scholar]
- Hanson Q. M. Wilson K. M. Shen M. Itkin Z. Eastman R. T. Shinn P. Hall M. D. Targeting ACE2–RBD Interaction as a Platform for COVID-19 Therapeutics: Development and Drug-Repurposing Screen of an AlphaLISA Proximity Assay. ACS Pharmacol. Transl. Sci. 2020;3:1352–1360. doi: 10.1021/acsptsci.0c00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvarnapathaki S. Chauhan D. Nguyen A. Ramalingam M. Camci-Unal G. Advances in Targeting ACE2 for Developing COVID-19 Therapeutics. Ann. Biomed. Eng. 2022;50:1734–1749. doi: 10.1007/s10439-022-03094-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Mendoza T. The Envelope (E) Protein of SARS-CoV-2 as a Pharmacological Target. Viruses. 2023;15:1000. doi: 10.3390/v15041000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jade D. Ayyamperumal S. Tallapaneni V. Joghee Nanjan C. M. Barge S. Mohan S. Nanjan M. J. Virtual high throughput screening: Potential inhibitors for SARS-CoV-2 PLpro and 3CLpro proteases. Eur. J. Pharmacol. 2021;901:174082. doi: 10.1016/j.ejphar.2021.174082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T. Wendt L. L. Manickam M. Easwaran M. The recent outbreaks of human coronaviruses: A medicinal chemistry perspective. Med. Res. Rev. 2021;41:72–135. doi: 10.1002/med.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. R. Skeate J. G. Kast W. M. Annexin A2 in Virus Infection. Front. Microbiol. 2018;9:2954. doi: 10.3389/fmicb.2018.02954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardenbrook N. J. Zhang P. A structural view of the SARS-CoV-2 virus and its assembly. Curr. Opin. Virol. 2022;52:123–134. doi: 10.1016/j.coviro.2021.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evseeva L. V. Ivanov V. V. Karpina V. R. Kovalenko S. S. Kuznetsov I. E. Langer T. Maes L. J. R. M. Kovalenko S. M. The virtual screening application for searching potential antiviral agents to treat COVID-19 disease. J. Org. Pharm. Chem. 2020;18:3–15. [Google Scholar]
- Herrera N. G. Morano N. C. Celikgil A. Georgiev G. I. Malonis R. J. Lee J. H. Tong K. Vergnolle O. Massimi A. B. Yen L. Y. Noble A. J. Kopylov M. Bonanno J. B. Garrett-Thomson S. C. Hayes D. B. Bortz III R. H. Wirchnianski A. S. Florez C. Laudermilch E. Haslwanter D. Fels J. M. Dieterle M. E. Jangra R. K. Barnhill J. Mengotto A. Kimmel D. Daily J. P. Pirofski L.-a. Chandran K. Brenowitz M. Garforth S. J. Eng E. T. Lai J. R. Almo S. C. Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis. ACS Omega. 2021;6:85–102. doi: 10.1021/acsomega.0c03512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakavandi S. Zare I. VaezJalali M. Dadashi M. Azarian M. Akbari A. Ramezani Farani M. Zalpoor H. Hajikhani B. Structural and non-structural proteins in SARS-CoV-2: potential aspects to COVID-19 treatment or prevention of progression of related diseases. Cell Commun. Signaling. 2023;21:110. doi: 10.1186/s12964-023-01104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J. Ge J. Yu J. Shan S. Zhou H. Fan S. Zhang Q. Shi X. Wang Q. Zhang L. Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Calvaresi V. Wrobel A. G. Toporowska J. Hammerschmid D. Doores K. J. Bradshaw R. T. Parsons R. B. Benton D. J. Roustan C. Reading E. Malim M. H. Gamblin S. J. Politis A. Structural dynamics in the evolution of SARS-CoV-2 spike glycoprotein. Nat. Commun. 2023;14:1421. doi: 10.1038/s41467-023-36745-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Zhang Y. Wu L. Niu S. Song C. Zhang Z. Lu G. Qiao C. Hu Y. Yuen K.-Y. Wang Q. Zhou H. Yan J. Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Li W. Farzan M. Harrison S. C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Benton D. J. Wrobel A. G. Roustan C. Borg A. Xu P. Martin S. R. Rosenthal P. B. Skehel J. J. Gamblin S. J. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2021;118:e2022586118. doi: 10.1073/pnas.2022586118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A. G. Benton D. J. Xu P. Roustan C. Martin S. R. Rosenthal P. B. Skehel J. J. Gamblin S. J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojadzic D. Alcazar O. Chen J. Chuang S.-T. Condor Capcha J. M. Shehadeh L. A. Buchwald P. Small-Molecule Inhibitors of the Coronavirus Spike: ACE2 Protein–Protein Interaction as Blockers of Viral Attachment and Entry for SARS-CoV-2. ACS Infect. Dis. 2021;7:1519–1534. doi: 10.1021/acsinfecdis.1c00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadevi S. Badavath V. N. Thakur A. Yin N. De Jonghe S. Acevedo O. Jochmans D. Leyssen P. Wang K. Neyts J. Yujie T. Blum G. Kobophenol A Inhibits Binding of Host ACE2 Receptor with Spike RBD Domain of SARS-CoV-2, a Lead Compound for Blocking COVID-19. J. Phys. Chem. Lett. 2021;12:1793–1802. doi: 10.1021/acs.jpclett.0c03119. [DOI] [PubMed] [Google Scholar]
- Yang M. Li J. Huang Z. Li H. Wang Y. Wang X. Kang S. Huang X. Wu C. Liu T. Jia Z. Liang J. Yuan X. He S. Chen X. Zhou Z. Chen Q. Liu S. Li J. Zheng H. Liu X. Li K. Yao X. Lang B. Liu L. Liao H.-X. Chen S. Structural Basis of a Human Neutralizing Antibody Specific to the SARS-CoV-2 Spike Protein Receptor-Binding Domain. Microbiol. Spectrum. 2021;9:e01352. doi: 10.1128/Spectrum.01352-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. Wang F. Shen C. Peng W. Li D. Zhao C. Li Z. Li S. Bi Y. Yang Y. Gong Y. Xiao H. Fan Z. Tan S. Wu G. Tan W. Lu X. Fan C. Wang Q. Liu Y. Zhang C. Qi J. Gao G. F. Gao F. Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echaide M. Chocarro de Erauso L. Bocanegra A. Blanco E. Kochan G. Escors D. mRNA Vaccines against SARS-CoV-2: Advantages and Caveats. Int. J. Mol. Sci. 2023;24:5944. doi: 10.3390/ijms24065944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Tenchov R. Smoot J. Liu C. Watkins S. Zhou Q. A Comprehensive Review of the Global Efforts on COVID-19 Vaccine Development. ACS Cent. Sci. 2021;7:512–533. doi: 10.1021/acscentsci.1c00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendler A. Shepherd S. T. C. Au L. Wu M. Harvey R. Wilkinson K. A. Schmitt A. M. Tippu Z. Shum B. Farag S. Rogiers A. Carlyle E. Edmonds K. Del Rosario L. Lingard K. Mangwende M. Holt L. Ahmod H. Korteweg J. Foley T. Barber T. Emslie-Henry A. Caulfield-Lynch N. Byrne F. Deng D. Kjaer S. Song O.-R. Queval C. J. Kavanagh C. Wall E. C. Carr E. J. Caidan S. Gavrielides M. MacRae J. I. Kelly G. Peat K. Kelly D. Murra A. Kelly K. O'Flaherty M. Shea R. L. Gardner G. Murray D. Popat S. Yousaf N. Jhanji S. Tatham K. Cunningham D. Van As N. Young K. Furness A. J. S. Pickering L. Beale R. Swanton C. Gandhi S. Gamblin S. Bauer D. L. V. Kassiotis G. Howell M. Nicholson E. Walker S. Wilkinson R. J. Larkin J. Turajlic S. Functional immune responses against SARS-CoV-2 variants of concern after fourth COVID-19 vaccine dose or infection in patients with blood cancer. Cell. Rep. Med. 2022;3:100781. doi: 10.1016/j.xcrm.2022.100781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scovino A. M. Dahab E. C. Vieira G. F. Freire-de-Lima L. Freire-de-Lima C. G. Morrot A. SARS-CoV-2’s Variants of Concern: A Brief Characterization. Front. Immunol. 2022;13:834098. doi: 10.3389/fimmu.2022.834098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royster A. Ren S. Ma Y. Pintado M. Kahng E. Rowan S. Mir S. Mir M. SARS-CoV-2 Nucleocapsid Protein Is a Potential Therapeutic Target for Anticoronavirus Drug Discovery. Microbiol. Spectrum. 2023;11:e01186. doi: 10.1128/spectrum.01186-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afreen R. Iqbal S. Shah A. R. Afreen H. Vodwal L. Shkir M. In Silico Identification of Potential Inhibitors of the SARS-CoV-2 Nucleocapsid Through Molecular Docking-Based Drug Repurposing. Dr Sulaiman Al Habib Med. J. 2022;4:64–76. [Google Scholar]
- Zhang L. Howland M. Hilgenfeld R. Anderson M. O. Eagon S. Identification of non-covalent SARS-CoV-2 main protease inhibitors by a virtual screen of commercially available drug-like compounds. Bioorg. Med. Chem. Lett. 2021;41:127990. doi: 10.1016/j.bmcl.2021.127990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Chen J. Yu X. Lan H.-Y. Signaling mechanisms of SARS-CoV-2 Nucleocapsid protein in viral infection, cell death and inflammation. Int. J. Biol. Sci. 2022;18:4704–4713. doi: 10.7150/ijbs.72663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala V. S. McKay M. J. Shcherbakov A. A. Dregni A. J. Kolocouris A. Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020;27:1202–1208. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. Nomura N. Muramoto Y. Ekimoto T. Uemura T. Liu K. Yui M. Kono N. Aoki J. Ikeguchi M. Noda T. Iwata S. Ohto U. Shimizu T. Structure of SARS-CoV-2 membrane protein essential for virus assembly. Nat. Commun. 2022;13:4399. doi: 10.1038/s41467-022-32019-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan K. A. Dutta M. Kern D. M. Kotecha A. Voth G. A. Brohawn S. G. Structure of SARS-CoV-2 M protein in lipid nanodiscs. eLife. 2022;11:e81702. doi: 10.7554/eLife.81702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.-Y. Zhao R. Gao L.-J. Gao X.-F. Wang D.-P. Cao J.-M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020;10:587269. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H. Hu Y. Jadhav P. Tan B. Wang J. Progress and Challenges in Targeting the SARS-CoV-2 Papain-like Protease. J. Med. Chem. 2022;65:7561–7580. doi: 10.1021/acs.jmedchem.2c00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W. Mao C. Luan X. Shen D.-D. Shen Q. Su H. Wang X. Zhou F. Zhao W. Gao M. Chang S. Xie Y.-C. Tian G. Jiang H.-W. Tao S.-C. Shen J. Jiang Y. Jiang H. Xu Y. Zhang S. Zhang Y. Xu H. E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameen F. Mamidala E. Davella R. Vallala S. Rilpivirine inhibits SARS-CoV-2 protein targets: A potential multi-target drug. J. Infect. Pub. Health. 2021;14:1454–1460. doi: 10.1016/j.jiph.2021.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannalire R. Cerchia C. Beccari A. R. Di Leva F. S. Summa V. Targeting SARS-CoV-2 Proteases and Polymerase for COVID-19 Treatment: State of the Art and Future Opportunities. J. Med. Chem. 2022;65:2716–2746. doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzadi K. Saba A. Yu M. Liang J. Structure-Based Drug Design of RdRp Inhibitors against SARS-CoV-2. Top. Curr. Chem. 2023;381:22. doi: 10.1007/s41061-023-00432-x. [DOI] [PubMed] [Google Scholar]
- Chien M. Anderson T. K. Jockusch S. Tao C. Li X. Kumar S. Russo J. J. Kirchdoerfer R. N. Ju J. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020;19:4690–4697. doi: 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M. L. Andres E. L. Sims A. C. Graham R. L. Sheahan T. P. Lu X. Smith E. C. Case J. B. Feng J. Y. Jordan R. Ray A. S. Cihlar T. Siegel D. Mackman R. L. Clarke M. O. Baric R. S. Denison M. R. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio. 2018;9:e00221. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hassab M. A. Shoun A. A. Al-Rashood S. T. Al-Warhi T. Eldehna W. M. Identification of a New Potential SARS-COV-2 RNA-Dependent RNA Polymerase Inhibitor via Combining Fragment-Based Drug Design, Docking, Molecular Dynamics, and MM-PBSA Calculations. Front. Chem. 2020;8:584894. doi: 10.3389/fchem.2020.584894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prussia A. J. Chennamadhavuni S. Biostructural Models for the Binding of Nucleoside Analogs to SARS-CoV-2 RNA-Dependent RNA Polymerase. J. Chem. Inf. Model. 2021;61:1402–1411. doi: 10.1021/acs.jcim.0c01277. [DOI] [PubMed] [Google Scholar]
- Byléhn F. Menéndez C. A. Perez-Lemus G. R. Alvarado W. de Pablo J. J. Modeling the Binding Mechanism of Remdesivir, Favilavir, and Ribavirin to SARS-CoV-2 RNA-Dependent RNA Polymerase. ACS Cent. Sci. 2021;7:164–174. doi: 10.1021/acscentsci.0c01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M. Dwivedy A. Mariadasse R. Tiwari S. Kar D. Jeyakanthan J. Biswal B. K. Prediction of Small Molecule Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus-2 RNA-dependent RNA Polymerase. ACS Omega. 2020;5:18356–18366. doi: 10.1021/acsomega.0c02096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazmi M. Motwalli O. In silico virtual screening, characterization, docking and molecular dynamics studies of crucial SARS-CoV-2 proteins. J. Biomol. Struct. Dyn. 2020;39:6761–6771. doi: 10.1080/07391102.2020.1803965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakchaure P. D. Ghosh S. Ganguly B. Revealing the Inhibition Mechanism of RNA-Dependent RNA Polymerase (RdRp) of SARS-CoV-2 by Remdesivir and Nucleotide Analogues: A Molecular Dynamics Simulation Study. J. Phys. Chem. B. 2020;124:10641–10652. doi: 10.1021/acs.jpcb.0c06747. [DOI] [PubMed] [Google Scholar]
- Kumar K. A. Sharma M. Dalal V. Singh V. Tomar S. Kumar P. Multifunctional inhibitors of SARS-CoV-2 by MM/PBSA, essential dynamics, and molecular dynamic investigations. J. Mol. Graphics Modell. 2021;107:107969. doi: 10.1016/j.jmgm.2021.107969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. M. Lamont I. L. Nucleoside Analogues as Antibacterial Agents. Front. Microbiol. 2019;10:952. doi: 10.3389/fmicb.2019.00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordheim L. P. Durantel D. Zoulim F. Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discovery. 2013;12:447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- Mehta N. Mazer-Amirshahi M. Alkindi N. Pourmand A. Pharmacotherapy in COVID-19; A narrative review for emergency providers. Am. J. Emerg. Med. 2020;38:1488–1493. doi: 10.1016/j.ajem.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal V. Mahapure K. S. Bhurwal A. Gupta I. Hassanain S. Makadia J. Madas N. Armaly P. Singh R. Mehra I. O'Horo J. C. Kashyap R. Mortality Benefit of Remdesivir in COVID-19: A Systematic Review and Meta-Analysis. Front. Med. 2021;7:606429. doi: 10.3389/fmed.2020.606429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman R. T. Roth J. S. Brimacombe K. R. Simeonov A. Shen M. Patnaik S. Hall M. D. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi K. Agarwal M. Pahuja I. Bhardwaj B. Rawat M. Bhaskar A. Dwivedi V. P. Combating the challenges of COVID-19 pandemic: Insights into molecular mechanisms, immune responses and therapeutics against SARS-CoV-2. Oxford Open Immunol. 2023;4:iqad001. doi: 10.1093/oxfimm/iqad001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce R. P. Hu V. W. Wang J. The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): an orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations. Med. Chem. Res. 2022;31:1637–1646. doi: 10.1007/s00044-022-02951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P. Rana M. Chowdhury P. DFT and MD simulation investigation of favipiravir as an emerging antiviral option against viral protease (3CLpro) of SARS-CoV-2. J. Mol. Struct. 2021;1246:131253. doi: 10.1016/j.molstruc.2021.131253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshni J. Vaishali R. Ganesh K. S. Dharani N. Alzahrani K. J. Banjer H. J. Alghamdi A. H. Theyab A. Ahmed S. S. S. J. Patil S. Multi-target potential of Indian phytochemicals against SARS-CoV-2: A docking, molecular dynamics and MM-GBSA approach extended to Omicron B.1.1.529. J. Inf. Pub. Health. 2022;15:662–669. doi: 10.1016/j.jiph.2022.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendi P. Razonable R. R. Nelson S. B. Soriano A. Gandhi R. T. First-generation oral antivirals against SARS-CoV-2. Clin. Microbiol. Infect. 2022;28:1230–1235. doi: 10.1016/j.cmi.2022.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T. K. Wells J. Panchal R. G. Stuthman K. S. Garza N. L. Van Tongeren S. A. Dong L. Retterer C. J. Eaton B. P. Pegoraro G. Honnold S. Bantia S. Kotian P. Chen X. Taubenheim B. R. Welch L. S. Minning D. M. Babu Y. S. Sheridan W. P. Bavari S. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T. Zhang L. Current understanding of nucleoside analogs inhibiting the SARS-CoV-2 RNA-dependent RNA polymerase. Comput. Struct. Biotechnol. J. 2023;21:4385–4394. doi: 10.1016/j.csbj.2023.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekheit M. S. Panda S. S. Girgis A. S. Potential RNA-dependent RNA polymerase (RdRp) inhibitors as prospective drug candidates for SARS-CoV-2. Eur. J. Med. Chem. 2023;252:115292. doi: 10.1016/j.ejmech.2023.115292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes A. J. Calado G. P. Fróes Y. N. Araújo S. A. França L. M. Paes A. M. Morais S. V. Rocha C. Q. Vasconcelos C. C. Plant Metabolites as SARS-CoV-2 Inhibitors Candidates: In Silico and In Vitro Studies. Pharmaceuticals. 2022;15:1045. doi: 10.3390/ph15091045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik R. R. Shakya A. K. Aladwan S. M. El-Tanani M. Kinase Inhibitors as Potential Therapeutic Agents in the Treatment of COVID-19. Front. Pharmacol. 2022;13:806568. doi: 10.3389/fphar.2022.806568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S. Yadav S. Banerjee S. Fakayode S. O. Parvathareddy J. Reichard W. Surendranathan S. Mahmud F. Whatcott R. Thammathong J. Meibohm B. Miller D. D. Jonsson C. B. Dubey K. D. Drug Repurposing to Identify Nilotinib as a Potential SARS-CoV-2 Main Protease Inhibitor: Insights from a Computational and In Vitro Study. J. Chem. Inf. Model. 2021;61:5469–5483. doi: 10.1021/acs.jcim.1c00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N. Chen Y.-T. Hsu Y.-C. Hsu N.-Y. Kuo C.-J. Tsai H. P. Kang J.-J. Huang C.-H. Chang S.-Y. Chang Y.-H. Liang P.-H. Yang J.-M. Uncovering Flexible Active Site Conformations of SARS-CoV-2 3CL Proteases through Protease Pharmacophore Clusters and COVID-19 Drug Repurposing. ACS Nano. 2021;15:857–872. doi: 10.1021/acsnano.0c07383. [DOI] [PubMed] [Google Scholar]
- She Z. Yao Y. Wang C. Li Y. Xiong X. Liu Y. Mpro-targeted anti-SARS-CoV-2 inhibitor-based drugs. J. Chem. Res. 2023;47:1–15. doi: 10.1177/17475198231184799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong W. Fischer C. Khan M. B. van Belkum M. J. Lamer T. Willoughby K. D. Lu J. Arutyunova E. Joyce M. A. Saffran H. A. Shields J. A. Young H. S. Nieman J. A. Tyrrell D. L. Lemieux M. J. Vederas J. C. Improved SARS-CoV-2 Mpro inhibitors based on feline antiviral drug GC376: Structural enhancements, increased solubility, and micellar studies. Eur. J. Med. Chem. 2021;222:113584. doi: 10.1016/j.ejmech.2021.113584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkhali R. Koirala P. Rijal S. Mainali A. Baral A. Bhattarai H. K. Structure and Function of Major SARS-CoV-2 and SARS-CoV Proteins. Bioinf. Biol. Insights. 2021;15:1–32. doi: 10.1177/11779322211025876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller D. W. Phillips G. O'Neill H. M. Jedrzejczak R. Stols L. Langan P. Joachimiak A. Coates L. Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:3202. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R. Perera L. Tillekeratne L. M. V. Potential SARS-CoV-2 main protease inhibitors. Drug Discovery Today. 2021;26:804–816. doi: 10.1016/j.drudis.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Lin D. Sun X. Curth U. Drosten C. Sauerhering L. Becker S. Rox K. Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Fang C. Zhang Q. Zhang R. Zhao X. Duan Y. Wang H. Zhu Y. Feng L. Zhao J. Shao M. Yang X. Zhang L. Peng C. Yang K. Ma D. Rao Z. Yang H. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell. 2021:689–693. doi: 10.1007/s13238-021-00883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Y. Kumar S. Zak S. E. Jones K. A. Upadhyay C. Sharma N. Azizi S.-A. Kathayat R. S. Poonam Herbert A. S. Durvasula R. Dickinson B. C. Dye J. M. Rathi B. Kempaiah P. Antiviral evaluation of hydroxyethylamine analogs: Inhibitors of SARS-CoV-2 main protease (3CLpro), a virtual screening and simulation approach. Bioorg. Med. Chem. 2021;47:116393. doi: 10.1016/j.bmc.2021.116393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M. S. Zhang L. Ibrahim M. Zhang K. Sun X. Röske J. Göhl M. Brönstrup M. Cowell J. K. Sauerhering L. Becker S. Vangeel L. Jochmans D. Neyts J. Rox K. Marsh G. P. Maple H. J. Hilgenfeld R. Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease. J. Med. Chem. 2022;65:13328–13342. doi: 10.1021/acs.jmedchem.2c01131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S. Sylvester K. Song L. Claff T. Jing L. Woodson M. Weiße R. H. Cheng Y. Schäkel L. Petry M. Gütschow M. Schiedel A. C. Sträter N. Kang D. Xu S. Toth K. Tavis J. Tollefson A. E. Müller C. E. Liu X. Zhan P. Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity. J. Med. Chem. 2022;65:13343–13364. doi: 10.1021/acs.jmedchem.2c01146. [DOI] [PubMed] [Google Scholar]
- Dampalla C. S. Rathnayake A. D. Galasiti Kankanamalage A. C. Kim Y. Perera K. D. Nguyen H. N. Miller M. J. Madden T. K. Picard H. R. Thurman H. A. Kashipathy M. M. Liu L. Battaile K. P. Lovell S. Chang K.-O. Groutas W. C. Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease. J. Med. Chem. 2022;65:7818–7832. doi: 10.1021/acs.jmedchem.2c00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttens A. Gullberg H. Abdurakhmanov E. Vo D. D. Akaberi D. Talibov V. O. Nekhotiaeva N. Vangeel L. De Jonghe S. Jochmans D. Krambrich J. Tas A. Lundgren B. Gravenfors Y. Craig A. J. Atilaw Y. Sandström A. Moodie L. W. K. Lundkvist Å. van Hemert M. J. Neyts J. Lennerstrand J. Kihlberg J. Sandberg K. Danielson U. H. Carlsson J. Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses. J. Am. Chem. Soc. 2022;144:2905–2920. doi: 10.1021/jacs.1c08402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou N. Shuai L. Zhang L. Xie X. Tang K. Zhu Y. Yu Y. Zhang W. Tan Q. Zhong G. Wen Z. Wang C. He X. Huo H. Gao H. Xu Y. Xue J. Peng C. Zou J. Schindewolf C. Menachery V. Su W. Yuan Y. Shen Z. Zhang R. Yuan S. Yu H. Shi P.-Y. Bu Z. Huang J. Hu Q. Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro. ACS Cent. Sci. 2023;9:217–227. doi: 10.1021/acscentsci.2c01359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey D. Hossain R. Biswas P. Paul P. Islam M. A. Ema T. I. Gain B. K. Hasan M. M. Bibi S. Islam M. T. Rahman M. A. Kim B. Amentoflavone derivatives significantly act towards the main protease (3CLPRO/MPRO) of SARS-CoV-2: in silico admet profiling, molecular docking, molecular dynamics simulation, network pharmacology. Mol. Diversity. 2023;27:857–871. doi: 10.1007/s11030-022-10459-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owis A. I. El-Hawary M. S. El Amir D. Aly O. M. Abdelmohsen U. R. Kamel M. S. Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv. 2020;10:19570–19575. doi: 10.1039/d0ra03582c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badshah S. L. Faisal S. Muhammad A. Poulson B. G. Emwas A. H. Jaremko M. Antiviral activities of flavonoids. Biomed. Pharmacother. 2021;140:111596. doi: 10.1016/j.biopha.2021.111596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahun M. Jukić M. Oblak D. Kranjc L. Bajc G. Butala M. Bozovičar K. Bratkovič T. Podlipnik Č. Poklar Ulrih N. Inhibition of the SARS-CoV-2 3CLpro main protease by plant polyphenols. Food Chem. 2022;373:131594. doi: 10.1016/j.foodchem.2021.131594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzoca M. E. Leuci S. Mignogna M. D. Muzio E. L. Caponio V. C. A. Muzio L. L. Natural compounds may contribute in preventing SARS-CoV-2 infection: a narrative review. Food Sci. Hum. Wellness. 2022;11:1134–1142. doi: 10.1016/j.fshw.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uma Reddy B. Routhu N. K. Kumar A. Multifaceted roles of plant derived small molecule inhibitors on replication cycle of SARS-CoV-2. Microb. Pathog. 2022;168:105512. doi: 10.1016/j.micpath.2022.105512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y. Zhu G.-H. Zhang Y.-N. Hu Q. Wang H.-N. Yu H.-N. Qin X.-Y. Guan X.-Q. Xiang Y.-W. Tang H. Ge G.-B. Flavonoids in Ampelopsis grossedentata as covalent inhibitors of SARS-CoV-2 3CLpro: Inhibition potentials, covalent binding sites and inhibitory mechanisms. Int. J. Biol. Macromol. 2021;187:976–987. doi: 10.1016/j.ijbiomac.2021.07.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer C. Feys J. R. SARS-CoV-2 Mpro Inhibitors: Achieved Diversity, Developing Resistance and Future Strategies. Future Pharmacol. 2023;3:80–107. [Google Scholar]
- Antonopoulou I. Sapountzaki E. Rova U. Christakopoulos P. Inhibition of the main protease of SARS-CoV-2 (Mpro) by repurposing/designing drug-like substances and utilizing nature's toolbox of bioactive compounds. Comput. Struct. Biotechnol. J. 2022;20:1306–1344. doi: 10.1016/j.csbj.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser J. Sedova A. Galanie S. Kneller D. W. Davidson R. B. Maradzike E. Del Galdo S. Labbé A. Hsu D. J. Agarwal R. Bykov D. Tharrington A. Parks J. M. Smith D. M. A. Daidone I. Coates L. Kovalevsky A. Smith J. C. Hit Expansion of a Noncovalent SARS-CoV-2 Main Protease Inhibitor. ACS Pharmacol. Transl. Sci. 2022;5:255–265. doi: 10.1021/acsptsci.2c00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-H. Stone E. A. Deshmukh M. Ippolito J. A. Ghahremanpour M. M. Tirado-Rives J. Spasov K. A. Zhang S. Takeo Y. Kudalkar S. N. Liang Z. Isaacs F. Lindenbach B. Miller S. J. Anderson K. S. Jorgensen W. L. Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations. ACS Cent. Sci. 2021;7:467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh M. G. Ippolito J. A. Zhang C.-H. Stone E. A. Reilly R. A. Miller S. J. Jorgensen W. L. Anderson K. S. Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease. Structure. 2021;29:823–833. doi: 10.1016/j.str.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelly S. Liotta D. Potent SARS-CoV-2 Direct-Acting Antivirals Provide an Important Complement to COVID-19 Vaccines. ACS Cent. Sci. 2021;7:396–399. doi: 10.1021/acscentsci.1c00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlington M. Chun A. Tomar S. Eggler A. Grum-Tokars V. Jacobs J. Daniels J. S. Dawson E. Saldanha A. Chase P. Baez-Santos Y. M. Lindsley C. W. Hodder P. Mesecar A. D. Stauffer S. R. Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: Identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 2013;23:6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. H. Goins C. M. Arya T. Shin W.-J. Maw J. Hooper A. Sonawane D. P. Porter M. R. Bannister B. E. Crouch R. D. Lindsey A. A. Lakatos G. Martinez S. R. Alvarado J. Akers W. S. Wang N. S. Jung J. U. Macdonald J. D. Stauffer S. R. Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CLpro) J. Med. Chem. 2022;65:2880–2904. doi: 10.1021/acs.jmedchem.1c00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde A. Galanie S. Kneller D. W. Ma H. Babuji Y. Blaiszik B. Brace A. Brettin T. Chard K. Chard R. Coates L. Foster I. Hauner D. Kertesz V. Kumar N. Lee H. Li Z. Merzky A. Schmidt J. G. Tan L. Titov M. Trifan A. Turilli M. Van Dam H. Chennubhotla S. C. Jha S. Kovalevsky A. Ramanathan A. Head M. S. Stevens R. High-Throughput Virtual Screening and Validation of a SARS-CoV-2 Main Protease Noncovalent Inhibitor. J. Chem. Inf. Model. 2022;62:116–128. doi: 10.1021/acs.jcim.1c00851. [DOI] [PubMed] [Google Scholar]
- Unoh Y. Uehara S. Nakahara K. Nobori H. Yamatsu Y. Yamamoto S. Maruyama Y. Taoda Y. Kasamatsu K. Suto T. Kouki K. Nakahashi A. Kawashima S. Sanaki T. Toba S. Uemura K. Mizutare T. Ando S. Sasaki M. Orba Y. Sawa H. Sato A. Sato T. Kato T. Tachibana Y. Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19. J. Med. Chem. 2022;65:6499–6512. doi: 10.1021/acs.jmedchem.2c00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltayb W. A. Abdalla M. Rabie A. M. Novel Investigational Anti-SARS-CoV-2 Agent Ensitrelvir “S-217622”: A Very Promising Potential Universal Broad-Spectrum Antiviral at the Therapeutic Frontline of Coronavirus Species. ACS Omega. 2023;8:5234–5246. doi: 10.1021/acsomega.2c03881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawajiri T. Kijima A. Iimuro A. Ohashi E. Yamakawa K. Agura K. Masuda K. Kouki K. Kasamatsu K. Yanagisawa S. Nakashima S. Shibahara S. Toyota T. Higuchi T. Suto T. Oohara T. Maki T. Sahara N. Fukui N. Wakamori H. Ikemoto H. Murakami H. Ando H. Hosoya M. Sato M. Suzuki Y. Nakagawa Y. Unoh Y. Hirano Y. Nagasawa Y. Goda S. Ohara T. Tsuritani T. Development of a Manufacturing Process toward the Convergent Synthesis of the COVID-19 Antiviral Ensitrelvir. ACS Cent. Sci. 2023;9:836–843. doi: 10.1021/acscentsci.2c01203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dampalla C. S. Miller M. J. Kim Y. Zabiegala A. Nguyen H. N. Madden T. K. Thurman H. A. Machen A. J. Cooper A. Liu L. Battaile K. P. Lovell S. Chang K.-O. Groutas W. C. Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV) Eur. J. Med. Chem. 2023;254:115376. doi: 10.1016/j.ejmech.2023.115376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankadara S. Wong Y. X. Liu B. See Y. Y. Tan L. H. Tan Q. W. Wang G. Karuna R. Guo X. Tan S. T. Fong J. Y. Joy J. Chia C. S. B. A head-to-head comparison of the inhibitory activities of 15 peptidomimetic SARS-CoV-2 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2021;48:128263. doi: 10.1016/j.bmcl.2021.128263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W. Zhang B. Jiang X.-M. Su H. Li J. Zhao Y. Xie X. Jin Z. Peng J. Liu F. Li C. Li Y. Bai F. Wang H. Cheng X. Cen X. Hu S. Yang X. Wang J. Liu X. Xiao G. Jiang H. Rao Z. Zhang L.-K. Xu Y. Yang H. Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K. S. Ma X. R. Ma Y. Alugubelli Y. R. Scott D. A. Vatansever E. C. Drelich A. K. Sankaran B. Geng Z. Z. Blankenship L. R. Ward H. E. Sheng Y. J. Hsu J. C. Kratch K. C. Zhao B. Hayatshahi H. S. Liu J. Li P. Fierke C. A. Tseng C.-T. K. Xu S. Liu W. R. A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors. ChemMedChem. 2021;16:942–948. doi: 10.1002/cmdc.202000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C. Sacco M. D. Hurst B. Townsend J. A. Hu Y. Szeto T. Zhang X. Tarbet B. Marty M. T. Chen Y. Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong W. Khan M. B. Fischer C. Arutyunova E. Lamer T. Shields J. Saffran H. A. McKay R. T. van Belkum M. J. Joyce M. A. Young H. S. Tyrrell D. L. Vederas J. C. Lemieux M. J. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewitz L. Dumjahn L. Zhao Y. Owen C. D. Laidlaw S. M. Malla T. R. Nguyen D. Lukacik P. Salah E. Crawshaw A. D. Warren A. J. Trincao J. Strain-Damerell C. Carroll M. W. Walsh M. A. Schofield C. J. Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine. J. Med. Chem. 2023;66:2663–2680. doi: 10.1021/acs.jmedchem.2c01627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller D. W. Li H. Phillips G. Weiss K. L. Zhang Q. Arnould M. A. Jonsson C. B. Surendranathan S. Parvathareddy J. Blakeley M. P. Coates L. Louis J. M. Bonnesen P. V. Kovalevsky A. Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease. Nat. Commun. 2022;13:2268. doi: 10.1038/s41467-022-29915-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi C. Vanet A. Francesconi V. Tagliazucchi L. Tassone G. Venturelli A. Spyrakis F. Mazzorana M. Costi M. P. Tonelli M. Antitarget, Anti-SARS-CoV-2 Leads, Drugs, and the Drug Discovery–Genetics Alliance Perspective. J. Med. Chem. 2023;66:3664–3702. doi: 10.1021/acs.jmedchem.2c01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Monica G. Bono A. Lauria A. Martorana A. Targeting SARS-CoV-2 Main Protease for Treatment of COVID-19: Covalent Inhibitors Structure–Activity Relationship Insights and Evolution Perspectives. J. Med. Chem. 2022;65:12500–12534. doi: 10.1021/acs.jmedchem.2c01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman R. L. Kania R. S. Brothers M. A. Davies J. F. Ferre R. A. Gajiwala K. S. He M. Hogan R. J. Kozminski K. Li L. Y. Lockner J. W. Lou J. Marra M. T. Mitchell Jr L. J. Murray B. W. Nieman J. A. Noell S. Planken S. P. Rowe T. Ryan K. Smith III G. J. Solowiej J. E. Steppan C. M. Taggart B. Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J. Med. Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previti S. Ettari R. Calcaterra E. Di Maro S. Hammerschmidt S. J. Müller C. Ziebuhr J. Schirmeister T. Cosconati S. Zappalà M. Structure-based lead optimization of peptide-based vinyl methyl ketones as SARS-CoV-2 main protease inhibitors. Eur. J. Med. Chem. 2023;247:115021. doi: 10.1016/j.ejmech.2022.115021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Pei R. Li X. Deng W. Xing S. Zhang Y. Zhang C. He S. Sun H. Xiao S. Xiong J. Zhang Y. Chen X. Wang Y. Guo Y. Zhang B. Shang L. The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate. Eur. J. Med. Chem. 2022;238:114458. doi: 10.1016/j.ejmech.2022.114458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B. Joyce R. Tan H. Hu Y. Wang J. SARS-CoV-2 Main Protease Drug Design, Assay Development, and Drug Resistance Studies. Acc. Chem. Res. 2023;56:157–168. doi: 10.1021/acs.accounts.2c00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z. Sacco M. Hu Y. Ma C. Meng X. Zhang F. Szeto T. Xiang Y. Chen Y. Wang J. Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir. ACS Pharmacol. Transl. Sci. 2021;4:1408–1421. doi: 10.1021/acsptsci.1c00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. Ma C. Szeto T. Hurst B. Tarbet B. Wang J. Boceprevir, Calpain Inhibitors II and XII, and GC-376 Have Broad-Spectrum Antiviral Activity against Coronaviruses. ACS Infect. Dis. 2021;7:586–597. doi: 10.1021/acsinfecdis.0c00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. Xiong Y. Zhu G.-H. Zhang Y.-N. Zhang Y.-W. Huang P. Ge G.-B. The SARS-CoV-2 main protease (Mpro): Structure, function, and emerging therapies for COVID-19. MedComm. 2022;3:e151. doi: 10.1002/mco2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. R. Allerton C. M. N. Anderson A. S. Aschenbrenner L. Avery M. Berritt S. Boras B. Cardin R. D. Carlo A. Coffman K. J. Dantonio A. Di L. Eng H. Ferre R. Gajiwala K. S. Gibson S. A. Greasley S. E. Hurst B. L. Kadar E. P. Kalgutkar A. S. Lee J. C. Lee J. Liu W. Mason S. W. Noell S. Novak J. J. Obach R. S. Ogilvie K. Patel N. C. Pettersson M. Rai D. K. Reese M. R. Sammons M. F. Sathish J. G. Singh R. S. P. Steppan C. M. Stewart A. E. Tuttle J. B. Updyke L. Verhoest P. R. Wei L. Yang Q. Zhu Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Hayek-Orduz Y. Vásquez A. F. Villegas-Torres M. F. Caicedo P. A. Achenie L. E. K. González Barrios A. F. Novel covalent and non-covalent complex-based pharmacophore models of SARS-CoV-2 main protease (Mpro) elucidated by microsecond MD simulations. Sci. Rep. 2022;12:14030. doi: 10.1038/s41598-022-17204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb Y. N. Nirmatrelvir Plus Ritonavir: First Approval. Drugs. 2022;82:585–591. doi: 10.1007/s40265-022-01692-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi M. Vakil M. K. Bahmanyar M. Zarenezhad E. Paxlovid: Mechanism of Action, Synthesis, and In Silico Study. BioMed Res. Int. 2022;2022:7341493. doi: 10.1155/2022/7341493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrakis V. Rafailidis P. Trypsianis G. Papazoglou D. Panagopoulos P. The Antiviral Effect of Nirmatrelvir/Ritonavir during COVID-19 Pandemic Real-World Data. Viruses. 2023;15:976. doi: 10.3390/v15040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair H. A. Nirmatrelvir plus ritonavir in COVID-19: a profile of its use. Drugs Ther. Perspect. 2023;39:41–47. doi: 10.1007/s40267-022-00971-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau R. K. Wright S. H. Cherrington N. J. PF-07321332 (Nirmatrelvir) does not interact with human ENT1 or ENT2: Implications for COVID-19 patients. Clin. Transl. Sci. 2022;15:1599–1605. doi: 10.1111/cts.13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duveau D. Y. Thomas C. J. The Remarkable Selectivity of Nirmatrelvir. ACS Pharmacol. Transl. Sci. 2022;5:445–447. doi: 10.1021/acsptsci.2c00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greasley S. E. Noell S. Plotnikova O. Ferre R. Liu W. Bolanos B. Fennell K. Nicki J. Craig T. Zhu Y. Stewart A. E. Steppan C. M. Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants. J. Biol. Chem. 2022;298:101972. doi: 10.1016/j.jbc.2022.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Wang Y. Solanki K. Atre R. Lavrijsen M. Pan Q. Baig M. S. Li P. Nirmatrelvir exerts distinct antiviral potency against different human coronaviruses. Antiviral Res. 2023;211:105555. doi: 10.1016/j.antiviral.2023.105555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohachova K. O. Sviatenko A. S. Kyrychenko A. Ivanov V. V. Langer T. Kovalenko S. M. Kalugin O. N. Computer-aided drug design of novel nirmatrelvir analogs inhibiting main protease of Coronavirus SARS-CoV-2. J. Appl. Pharm. Sci. 2023;13 [Google Scholar]; , accepted for publication
- Tam N. M. Nguyen T. H. Pham M. Q. Hong N. D. Tung N. T. Vu V. V. Quang D. T. Ngo S. T. Upgrading nirmatrelvir to inhibit SARS-CoV-2 Mpro via DeepFrag and free energy calculations. J. Mol. Graphics Modell. 2023;124:108535. doi: 10.1016/j.jmgm.2023.108535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders B. C. Pokhrel S. Labbe A. D. Mathews I. I. Cooper C. J. Davidson R. B. Phillips G. Weiss K. L. Zhang Q. O'Neill H. Kaur M. Schmidt J. G. Reichard W. Surendranathan S. Parvathareddy J. Phillips L. Rainville C. Sterner D. E. Kumaran D. Andi B. Babnigg G. Moriarty N. W. Adams P. D. Joachimiak A. Hurst B. L. Kumar S. Butt T. R. Jonsson C. B. Ferrins L. Wakatsuki S. Galanie S. Head M. S. Parks J. M. Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2. Nat. Commun. 2023;14:1733. doi: 10.1038/s41467-023-37254-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S. Nitsche C. SARS-CoV-2 Papain-Like Protease: Structure, Function and Inhibition. ChemBioChem. 2022;23:e202200327. doi: 10.1002/cbic.202200327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk J. Azizi S.-A. Dvorkin S. Endres M. Jedrzejczak R. Jones K. A. Kang S. Kathayat R. S. Kim Y. Lisnyak V. G. Maki S. L. Nicolaescu V. Taylor C. A. Tesar C. Zhang Y.-A. Zhou Z. Randall G. Michalska K. Snyder S. A. Dickinson B. C. Joachimiak A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021;12:743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H. Liu J. Shen J. Dai J. Xu G. Lu K. Han C. Wang Y. Xu X. Tong Y. Xiang H. Ai Z. Zhuang G. Hu J. Zhang Z. Li Y. Pan L. Tan L. Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2. Cell Chem. Biol. 2021;28:855–865. doi: 10.1016/j.chembiol.2021.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C. Sacco M. D. Xia Z. Lambrinidis G. Townsend J. A. Hu Y. Meng X. Szeto T. Ba M. Zhang X. Gongora M. Zhang F. Marty M. T. Xiang Y. Kolocouris A. Chen Y. Wang J. Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay. ACS Cent. Sci. 2021;7:1245–1260. doi: 10.1021/acscentsci.1c00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim T. M. Ismail M. I. Bauer M. R. Bekhit A. A. Boeckler F. M. Supporting SARS-CoV-2 Papain-Like Protease Drug Discovery: In silico Methods and Benchmarking. Front. Chem. 2020;8:592289. doi: 10.3389/fchem.2020.592289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajbabaie R. Harper M. T. Rahman T. Establishing an Analogue Based In Silico Pipeline in the Pursuit of Novel Inhibitory Scaffolds against the SARS Coronavirus 2 Papain-Like Protease. Molecules. 2021;26:1134. doi: 10.3390/molecules26041134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rut W. Lv Z. Zmudzinski M. Patchett S. Nayak D. Snipas S. J. El Oualid F. Huang T. T. Bekes M. Drag M. Olsen S. K. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti–COVID-19 drug design. Sci. Adv. 2020;6:eabd4596. doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm T. Ebert G. Calleja D. J. Allison C. C. Richardson L. W. Bernardini J. P. Lu B. G. C. Kuchel N. W. Grohmann C. Shibata Y. Gan Z. Y. Cooney J. P. Doerflinger M. Au A. E. Blackmore T. R. van der Heden van Noort G. J. Geurink P. P. Ovaa H. Newman J. Riboldi-Tunnicliffe A. Czabotar P. E. Mitchell J. P. Feltham R. Lechtenberg B. C. Lowes K. N. Dewson G. Pellegrini M. Lessene G. Komander D. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39:e106275. doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. Qin B. Chen P. Zhu K. Hou P. Wojdyla J. A. Wang M. Cui S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B. 2021;11:237–245. doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A.-T. Pandey M. Smith J. R. Ban F. Fernandez M. Cherkasov A. Targeting SARS-CoV-2 papain-like protease in the postvaccine era. Trends Pharmacol. Sci. 2022;43:906–919. doi: 10.1016/j.tips.2022.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar D. Stein M. Binding of SARS-CoV Covalent Non-Covalent Inhibitors to the SARS-CoV-2 Papain-Like Protease and Ovarian Tumor Domain Deubiquitinases. Biomolecules. 2021;11:802. doi: 10.3390/biom11060802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-S. Chang G.-G. Juo C.-G. Lee H.-J. Yeh S.-H. Hsu J. T.-A. Chen X. Papain-Like Protease 2 (PLP2) from Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV): Expression, Purification, Characterization, and Inhibition. Biochem. 2005;44:10349–10359. doi: 10.1021/bi0504761. [DOI] [PubMed] [Google Scholar]
- Hossain M. U. Bhattacharjee A. Emon M. T. H. Chowdhury Z. M. Ahammad I. Mosaib M. G. Moniruzzaman M. Rahman M. H. Islam M. N. Ahmed I. Amin M. R. Rashed A. Das K. C. Keya C. A. Salimullah M. Novel mutations in NSP-1 and PLPro of SARS-CoV-2 NIB-1 genome mount for effective therapeutics. J. Genet. Eng. Biotechnol. 2021;19:52. doi: 10.1186/s43141-021-00152-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitsillou E. Liang J. Ververis K. Hung A. Karagiannis T. C. Interaction of small molecules with the SARS-CoV-2 papain-like protease: In silico studies and in vitro validation of protease activity inhibition using an enzymatic inhibition assay. J. Mol. Graphics Modell. 2021;104:107851. doi: 10.1016/j.jmgm.2021.107851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K. Pegan S. Takayama J. Sleeman K. Coughlin M. Baliji S. Chaudhuri R. Fu W. Prabhakar B. S. Johnson M. E. Baker S. C. Ghosh A. K. Mesecar A. D. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D. Mukherjee R. Grewe D. Bojkova D. Baek K. Bhattacharya A. Schulz L. Widera M. Mehdipour A. R. Tascher G. Geurink P. P. Wilhelm A. van der Heden van Noort G. J. Ovaa H. Müller S. Knobeloch K.-P. Rajalingam K. Schulman B. A. Cinatl J. Hummer G. Ciesek S. Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z. Ratia K. Cooper L. Kong D. Lee H. Kwon Y. Li Y. Alqarni S. Huang F. Dubrovskyi O. Rong L. Thatcher G. R. J. Xiong R. Design of SARS-CoV-2 PLpro Inhibitors for COVID-19 Antiviral Therapy Leveraging Binding Cooperativity. J. Med. Chem. 2022;65:2940–2955. doi: 10.1021/acs.jmedchem.1c01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafna K. White K. Harish B. Rosales R. Ramelot T. A. Acton T. B. Moreno E. Kehrer T. Miorin L. Royer C. A. García-Sastre A. Krug R. M. Montelione G. T. Hepatitis C virus drugs that inhibit SARS-CoV-2 papain-like protease synergize with remdesivir to suppress viral replication in cell culture. Cell Rep. 2021;35:109133. doi: 10.1016/j.celrep.2021.109133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaim C. D. Dwivedi V. Perng Y.-C. Zhao X. Canadeo L. A. Harastani H. H. Darling T. L. Boon A. C. M. Lenschow D. J. Kulkarni V. Huibregtse J. M. 6-Thioguanine blocks SARS-CoV-2 replication by inhibition of PLpro. iScience. 2021;24:103213. doi: 10.1016/j.isci.2021.103213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M. U. Ahmad S. Abdullah I. Froeyen M. Identification of novel human USP2 inhibitor and its putative role in treatment of COVID-19 by inhibiting SARS-CoV-2 papain-like (PLpro) protease. Comput. Biol. Chem. 2020;89:107376. doi: 10.1016/j.compbiolchem.2020.107376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S. Gao X. Tang K. Cai J.-P. Hu M. Luo P. Wen L. Ye Z.-W. Luo C. Tsang J. O.-L. Chan C. C.-Y. Huang Y. Cao J. Liang R. Qin Z. Qin B. Yin F. Chu H. Jin D.-Y. Sun R. Chan J. F.-W. Cui S. Yuen K.-Y. Targeting papain-like protease for broad-spectrum coronavirus inhibition. Protein Cell. 2022;13:940–953. doi: 10.1007/s13238-022-00909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elseginy S. A. Fayed B. Hamdy R. Mahrous N. Mostafa A. Almehdi A. M. Soliman S. S. M. Promising anti-SARS-CoV-2 drugs by effective dual targeting against the viral and host proteases. Bioorg. Med. Chem. Lett. 2021;43:128099. doi: 10.1016/j.bmcl.2021.128099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elseginy S. A. Anwar M. M. In silico analysis of SARS-CoV-2 papain-like protease potential inhibitors. RSC Adv. 2021;11:38616–38631. doi: 10.1039/d1ra07845c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. Ma L. Hu Y. Li X. Yu M. Shang H. Zou Z. Natural biflavones are potent inhibitors against SARS-CoV-2 papain-like protease. Phytochem. 2022;193:112984. doi: 10.1016/j.phytochem.2021.112984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Štekláč M. Zajaček D. Bučinský L. 3CLpro and PLpro affinity, a docking study to fight COVID19 based on 900 compounds from PubChem and literature. Are there new drugs to be found? J. Mol. Struct. 2021;1245:130968. doi: 10.1016/j.molstruc.2021.130968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A. Narwal M. Majowicz S. A. Varricchio C. Toner S. A. Ballatore C. Brancale A. Murakami K. S. Jose J. Identification of SARS-CoV-2 inhibitors targeting Mpro and PLpro using in-cell-protease assay. Commun. Biol. 2022;5:169. doi: 10.1038/s42003-022-03090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S. Chen Y. Lockbaum G. J. Sen S. Chaudhuri S. Reyes A. C. Lee J. M. Kaur A. N. Sultana N. Cameron M. D. Shaffer S. A. Schiffer C. A. Fitzgerald K. A. Thompson P. R. Dual Inhibitors of Main Protease (MPro) and Cathepsin L as Potent Antivirals against SARS-CoV2. J. Am. Chem. Soc. 2022;144:21035–21045. doi: 10.1021/jacs.2c04626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafa Shavira S. H. Fatmaria F. The in-silico potential of Andrographis paniculata phytocompounds as antiviral for the treatment of COVID-19: A systematic review. J. Appl. Pharm. Sci. 2023;13:101–112. [Google Scholar]
- Omage F. B. Madabeni A. Tucci A. R. Nogara P. A. Bortoli M. Rosa A. d. S. Neuza dos Santos Ferreira V. Teixeira Rocha J. B. Miranda M. D. Orian L. Diphenyl Diselenide and SARS-CoV-2: in silico Exploration of the Mechanisms of Inhibition of Main Protease (Mpro) and Papain-like Protease (PLpro) J. Chem. Inf. Model. 2023;63:2226–2239. doi: 10.1021/acs.jcim.3c00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpoot S. Alagumuthu M. Baig M. S. Dual targeting of 3CLpro and PLpro of SARS-CoV-2: A novel structure-based design approach to treat COVID-19. Curr. Res. Struct. Biol. 2021;3:9–18. doi: 10.1016/j.crstbi.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumskiy R. S. Tumskaia A. V. Klochkova I. N. Richardson R. J. SARS-CoV-2 proteases Mpro and PLpro: Design of inhibitors with predicted high potency and low mammalian toxicity using artificial neural networks, ligand-protein docking, molecular dynamics simulations, and ADMET calculations. Comput. Biol. Med. 2023;153:106449. doi: 10.1016/j.compbiomed.2022.106449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl A. C. Godoy A. S. Noske G. D. Nakamura A. M. Gawriljuk V. O. Fernandes R. S. Oliva G. Ekins S. Discovery of PLpro and Mpro Inhibitors for SARS-CoV-2. ACS Omega. 2023;8:22603–22612. doi: 10.1021/acsomega.3c01110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S. A. Banerjee S. Ghosh K. Gayen S. Jha T. Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg. Med. Chem. 2021;29:115860. doi: 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos L. H. Kronenberger T. Almeida R. G. Silva E. B. Rocha R. E. O. Oliveira J. C. Barreto L. V. Skinner D. Fajtová P. Giardini M. A. Woodworth B. Bardine C. Lourenço A. L. Craik C. S. Poso A. Podust L. M. McKerrow J. H. Siqueira-Neto J. L. O'Donoghue A. J. da Silva Júnior E. N. Ferreira R. S. Structure-Based Identification of Naphthoquinones and Derivatives as Novel Inhibitors of Main Protease Mpro and Papain-like Protease PLpro of SARS-CoV-2. J. Chem. Inf. Model. 2022;62:6553–6573. doi: 10.1021/acs.jcim.2c00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sarno V. Lauro G. Musella S. Ciaglia T. Vestuto V. Sala M. Scala M. C. Smaldone G. Di Matteo F. Novi S. Tecce M. F. Moltedo O. Bifulco G. Campiglia P. Gomez-Monterrey I. M. Snoeck R. Andrei G. Ostacolo C. Bertamino A. Identification of a dual acting SARS-CoV-2 proteases inhibitor through in silico design and step-by-step biological characterization. Eur. J. Med. Chem. 2021;226:113863. doi: 10.1016/j.ejmech.2021.113863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi H. Zeynizadeh B. Rimaz M. Green and efficient one-pot three-component synthesis of novel drug-like furo[2,3-d]pyrimidines as potential active site inhibitors and putative allosteric hotspots modulators of both SARS-CoV-2 MPro and PLPro. Bioorg. Chem. 2023;135:106390. doi: 10.1016/j.bioorg.2023.106390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D. Mitra D. Paul M. Chaudhary P. Kamboj A. Thatoi H. Janmeda P. Jain D. Panneerselvam P. Shrivastav R. Pant K. Das Mohapatra P. K. Potential inhibitors of SARS-CoV-2 (COVID 19) proteases PLpro and Mpro/3CLpro: molecular docking and simulation studies of three pertinent medicinal plant natural components. Curr. Res. Pharmacol. Drug Discov. 2021;2:100038. doi: 10.1016/j.crphar.2021.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qayed W. S. Ferreira R. S. Silva J. R. A. In Silico Study towards Repositioning of FDA-Approved Drug Candidates for Anticoronaviral Therapy: Molecular Docking, Molecular Dynamics and Binding Free Energy Calculations. Molecules. 2022;27:5988. doi: 10.3390/molecules27185988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Abdalla M. Forodesine and Riboprine Exhibit Strong Anti-SARS-CoV-2 Repurposing Potential: In Silico and In Vitro Studies. ACS Bio Med Chem Au. 2022;2:565–585. doi: 10.1021/acsbiomedchemau.2c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Abdalla M. Evaluation of a series of nucleoside analogs as effective anticoronaviral-2 drugs against the Omicron-B.1.1.529/BA.2 subvariant: A repurposing research study. Med. Chem. Res. 2023;32:326–341. doi: 10.1007/s00044-022-02970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]