The Fourth International Conference on Bacterial Locomotion and Signal Transduction (BLAST IV) was held 9 to 13 January 1997 in Cuernavaca, Mexico. The meeting attracted 175 scientists in the fields of bacterial motility, environmental sensing, and development. New experimental results and hypotheses in these active and evolving areas of research were aired in a format that encouraged discussion and interaction. Many graduate and postdoctoral students had the opportunity to describe their work in platform talks, and many more presented excellent science in the poster sessions.
This minireview summarizes the information communicated by speakers at the meeting. The wide spectrum of subjects and organisms makes it difficult to introduce the full range of topics adequately. However, since the BLAST conference had its genesis in laboratories studying motility and chemotaxis in Escherichia coli and Salmonella, a brief overview of current knowledge of those processes in these bacteria provides a good starting point.
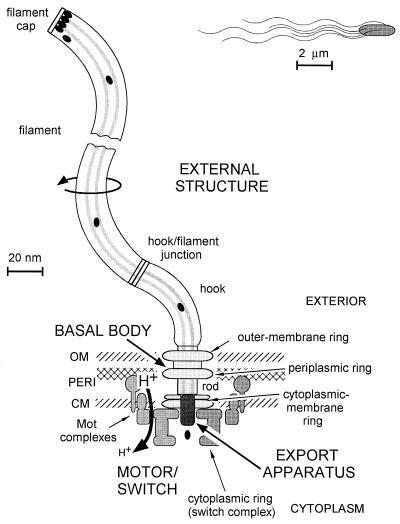
Six to eight flagella arise from random points on the surfaces of E. coli and Salmonella cells (peritrichous flagellation). Cells swim by rotating these flagella (Fig. 1). The flagellar filament is a thin, helical tube of variable length (up to 10 times the length of the cell) that serves as a propeller. The filament joins the flagellar basal body via a hollow, flexible hook, which probably functions as a universal joint. The hook is connected to the rod, a straight tube that forms the driveshaft. Three rings surround the rod: the double MS ring in and just outside the cytoplasmic membrane, the P ring in the peptidoglycan layer, and the L ring embedded in the outer membrane. The flagellar export apparatus secretes the proteins of the rod, hook, and filament in the proper sequence and amounts for them to diffuse through the hollow center of the flagellum to their assembly sites at the distal tip of the growing structure. The export machinery is presumed to be at the cytoplasmic face of the MS ring.
FIG. 1.
Structure of the enteric bacterial flagellum, showing its four major elements: external structure (white); basal body (light gray); motor-switch complex (medium gray); and export apparatus (dark gray). Their various substructures are also shown. Protons flow through the motor down their potential gradient (H+ → h+) to drive motor rotation. External proteins, such as flagellin (black ovals), are selectively passed through the export apparatus, travel down a central channel in the nascent structure, and assemble at its distal end. Most of the flagellum is tiny, so only the filaments are visible at the size scale of the bacterial cell (top right). Abbreviations: OM, outer membrane; PERI, periplasm; CM, cytoplasmic membrane.
A reversible motor powered by the transmembrane proton motive force drives flagellar rotation. The motor-switch complex is mounted on the cytoplasmic face of the MS ring and comprises a bell-shaped structure known as the C ring. The complex contains three proteins (FliG, FliM, and FliN) involved in generation of torque and switching of direction and is believed to rotate along with the MS ring, rod, hook, and filament. The stator of the motor is made up of two proteins that surround the MS ring. The carboxyl-terminal domain of MotB is probably anchored to the cell wall, and the four hydrophobic helices of MotA interact with the amino-terminal membrane-spanning helix of MotB to form a proton-conducting channel through the cytoplasmic membrane. Up to eight independent Mot protein complexes are thought to interact with the motor-switch complex to generate rotation in response to an inward proton current.
A cell typically travels in a three-dimensional random walk. Intervals in which the cells swim in gently curved paths (runs) alternate with briefer periods of chaotic motion (tumbles) that randomly reorient the next run. Cells run when the left-handed flagellar filaments rotate counterclockwise (CCW) and coalesce into a bundle to propel the cell. Cells tumble when the flagella turn clockwise (CW) and disrupt the bundle. Cells extend runs (suppress tumbles) when they head up concentration gradients of attractants or down gradients of repellents.
Spatial gradients are sensed by a temporal mechanism. Cells compare the instantaneous concentration of a compound, measured as percent receptor occupancy, against its concentration a few seconds previously, determined by the level of adaptive methylation of the receptor. The lag between ligand binding and methylation constitutes the “memory” that allows changes in chemoeffector concentrations to be detected.
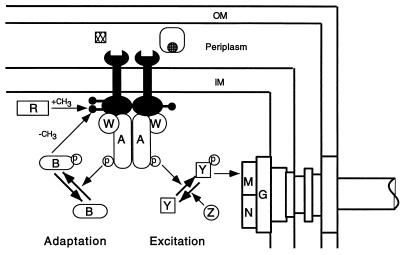
Three classes of proteins are essential for chemotaxis (Fig. 2): transmembrane receptors, cytoplasmic signaling components, and enzymes for adaptive methylation. The classical receptors consist of an amino-terminal transmembrane helix (TM1), a periplasmic ligand interaction domain, a second transmembrane helix (TM2), and a large cytoplasmic signaling and adaptation domain. Crystallographic studies have shown that the periplasmic domain is a four-helix bundle: helix I ascends from TM1, helix II descends toward the membrane, and helix III ascends to join helix IV, which connects to TM2. The receptors are homodimers with or without ligands.
FIG. 2.
Circuit diagram of the chemotactic signaling pathway. The dimeric membrane-spanning chemoreceptors (paired black wrench-like objects) form a ternary complex with two CheA and two CheW polypeptides and stimulate the autokinase activity of CheA. CheA-P can transfer the phosphate to CheY. CheY-P interacts with FliM in the motor-switch complex to induce CW flagellar rotation. The decay of CheY-P is accelerated by CheZ. CheR is a constitutive methyltransferase that methylates certain glutamate residues in the cytoplasmic domains of the receptors. CheB is a methylesterase that is activated by phosphotransfer from CheA-P. CheB-P removes methyl groups from the receptors. In the excitation pathway, some attractant ligands (cross-hatched square) bind directly to the periplasmic domains of the receptors. Others (cross-hatched circle) first bind to substrate-binding proteins, which then interact with the periplasmic domains of the receptors. Attractant binding inhibits stimulation of CheA activity by the receptors. As a consequence, the CheY-P level rapidly falls, and CW flagellar rotation is suppressed. In the adaptation pathway, reduced CheA activity decreases the CheB-P level, although more slowly than the CheY-P level. As methylesterase activity declines, the receptors become more highly methylated. Increased methylation counteracts the attractant-dependent inhibition of CheA activity. As CheA activity rises, the intracellular CheY-P concentration returns to its prestimulus value, and the flagellar motor resumes its prestimulus CW-to-CCW switching ratio. Abbreviations: OM, outer membrane; IM, inner membrane; A, CheA; W, CheW; Y, CheY; Z, CheZ; R, CheR; B, CheB; G, FliG; M, FliM; N, FliN; p, phosphate; CH3, methyl group (shown as lollipop-like objects on the cytoplasmic domains of the receptors). Figure courtesy of Paul Gardina.
Each cytoplasmic domain contains four or five methylatable glutamate residues, and the receptors are therefore also called methyl-accepting chemotaxis proteins (MCPs). Until this meeting, E. coli was known to possess four MCPs (now we know of five), which recognize the indicated attractants: Tsr, serine; Tar, aspartate and maltose; Trg, ribose, galactose, and glucose; and Tap, dipeptides. Salmonella lacks Tap but possesses Tcp, a citrate sensor. Serine, aspartate, and citrate bind directly to the receptors, whereas maltose, ribose, galactose, glucose, and dipeptides bind to their cognate periplasmic binding proteins, which then dock with the appropriate membrane receptors. MCPs also mediate responses to temperature and pH, and they serve as receptors for various repellents.
Communication between the receptors and the flagellar switch involves four proteins: CheA, the histidine protein kinase; CheY, the response regulator; CheW, the receptor-coupling factor; and CheZ, an enhancer of CheY-P dephosphorylation. CheA and CheY constitute a prototype two-component system, although they deviate from the standard paradigm in several ways, most notably in that CheY does not contain a DNA-binding domain nor act as a transcription factor. CheA functions as a dimer, two CheW proteins bind per CheA dimer, and this complex associates with a dimeric receptor. In this ternary complex, CheA autophosphorylation is greatly stimulated, which in turn increases phosphotransfer from CheA-P to CheY. CheY-P binds to FliM in the flagellar motor-switch complex to cause CW rotation. CheZ prevents accumulation of CheY-P by accelerating the decay of its intrinsically unstable aspartyl-phosphate residue. Under steady-state conditions, CheY-P is maintained at a level that generates the random walk.
When an attractant binds to a receptor, it initiates a conformational change that propagates across the membrane and suppresses CheA activity. Levels of CheY-P fall, and cells tumble less often. Thus, cells increase their run lengths as they enter areas of higher attractant concentration. This response does not explain, however, how cells respond to continually increasing attractant concentrations. To accomplish that, adaptation is necessary.
Two enzymes, the methyltransferase CheR and the methylesterase CheB, are necessary for adaptive methylation. CheR is a constitutive enzyme that uses S-adenosylmethionine to methylate glutamate residues in the cytoplasmic domains of the MCPs. CheR binds to some MCPs at their extreme C-terminal tails, an interaction that localizes the methyltransferase to the receptors but is not an integral part of the methylation reactions. CheB is a target for phosphotransfer from CheA, and CheB-P removes methyl groups from the MCPs. In steady state, methyl addition by CheR balances methyl removal by CheB-P to achieve an intermediate level of receptor methylation (0.5 to 1 methyl group per subunit) that maintains run-tumble behavior.
When an attractant binds a receptor and inhibits CheA activity, CheB-P levels fall, although more slowly than CheY-P levels, since CheB-P is not a substrate for CheZ. Increased methylation restores the ability of the receptor to stimulate CheA. Even after basal levels of CheY-P and CheB-P are regained, however, an attractant-bound receptor remains overmethylated, because its properties as a substrate for CheB-P are altered.
This thumbnail sketch should give uninitiated readers a jump start. An extensive background on motility, chemotaxis, and many of the other subjects covered at BLAST IV can be obtained from the relevant chapters in Escherichia coli and Salmonella: Cellular and Molecular Biology (24b). The interested reader should also peruse the literature to find the many papers that have been or will be published with the full accounts of the stories presented here in condensed and digested, but hopefully recognizable, form.
CHEMOSENSING IN NONENTERIC BACTERIA
The chemosensory systems of E. coli and Salmonella are understood in some detail. This meeting featured bacterial subgroups with more-complex sensory systems than those of these enteric bacteria. Such differences may reflect the more-complex environments in which nonenteric organisms live and the greater flexibility of their metabolism.
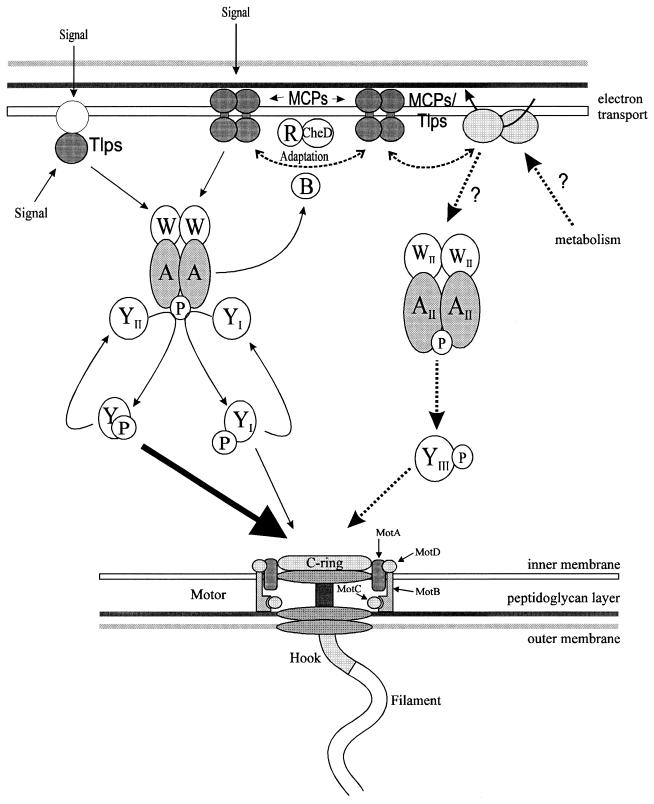
The α subgroup of Proteobacteria is being studied intensively (2), and several common signaling themes emerge (Fig. 3). During chemotaxis, Rhizobium meliloti and Rhodobacter sphaeroides vary their swimming speed. Rüdiger Schmitt (University of Regensburg) presented data suggesting that the two CheY homologs identified in R. meliloti control the speed of flagellar rotation. The R. meliloti flagellum neither stops or switches but changes its rate of rotation. CheY2 mutants swim rapidly and are nonchemotactic. CheY1 mutants swim slowly but, when stimulated, increase speed and exhibit weak chemotactic responses (33). These observations suggest that CheY2 slows motor rotation in response to negative chemotactic stimulation, thereby changing the swimming direction of the cell. The two CheY homologs are phosphorylated by the CheA histidine kinase in vitro. There is no homolog of CheZ, the enzyme that facilitates dephosphorylation of CheY-P in many enteric bacteria. However, in the presence of unphosphorylated CheA and CheY1, CheY1-P is rapidly phosphorylated in vitro. Phosphotransfer studies reveal retro-phosphorylation of CheA by CheY2-P. CheA-P, in turn, generates CheY1-P, which acts as a sink for phosphate and thus functionally assumes the role of a phosphatase. This mechanism of dephosphorylation is new and seems to apply to the other members of the α subgroup of Proteobacteria that contain two CheY homologs and no CheZ.
FIG. 3.
Multiple sensory pathways in α subgroup of Proteobacteria. The diagram presents a composite picture of the current understanding of signaling pathways in these bacteria. In R. centenum, CheA and CheY2 are fused into a single gene product. In all cases, CheY1 and CheY2 may interact to allow signal termination. R. sphaeroides also has a second sensory pathway, shown by the single-headed broken arrows. Abbreviations: R, CheR; B, CheB; W, CheW; A, CheA; P, phosphate; YII, CheY2; YI, CheY1; WII, CheW2; AII, CheA2; YIII, CheY3.
Paul Hamblin (J. P. Armitage’s laboratory, University of Oxford) described an even-more-complex system in the photosynthetic α-subgroup bacterium R. sphaeroides, which has two CheY homologs and cytoplasmic sensory transducers. One operon encodes homologs of the CheA, CheW, and CheR proteins and two CheY homologs. Deletion of these genes hardly affects chemotaxis, but overexpression of the proteins blocks chemotaxis in E. coli. The apparent explanation is that R. sphaeroides contains a second chemotaxis operon with a second cheA gene and a third cheY homolog (17). Strains with this second operon deleted lose phototaxis and chemotaxis, which suggests that this second pathway controls photosensing and chemosensing.
The related bacterium Rhodospirillum centenum produces a single flagellum when free swimming. On surfaces it develops numerous lateral flagella, which allow colonies to spread. Zeyu Jiang (C. Bauer’s laboratory, Indiana University) has identified a chemotaxis operon organized like those of R. meliloti and R. sphaeroides. Mutations in this operon impair both chemotaxis and phototaxis, indicating that no second pathway mediating tactic responses exists. R. centenum produces a free CheY protein and a CheAY fusion protein whose CheY moiety is similar to the CheY domain of the CheAY fusion protein of the gliding bacterium Myxococcus xanthus, a member of the δ subgroup. Thus, CheAY fusion proteins represent an ancient lineage that may play a special role in surface-living bacteria.
The environmentally important bacteria of the genus Pseudomonas metabolize diverse aromatic compounds, including 4-hydroxybenzoate (4-HBA). The catabolic pathways are well understood, but chemosensing is not. Jayna Ditty (C. S. Harwood’s laboratory, University of Iowa) employed transposon mutagenesis to isolate mutants of Pseudomonas putida defective in 4-HBA taxis. The affected gene, pcaK, encodes a protein with 12 predicted membrane-spanning helices. PcaK falls into the major facilitator superfamily (MFS) of transport proteins, like lactose permease (25). Since 4-HBA can enter the cell by diffusion, it is unlikely that the loss of transport per se is responsible for the loss of the chemotactic response. PcaK actively transports 4-HBA when expressed in P. putida or E. coli, although it does not lead to 4-HBA chemotaxis in the latter species. It is not yet clear how PcaK is coupled to the chemotaxis pathway or whether 4-HBA transport is necessary for taxis. However, the large (for the MFS family) cytoplasmic loop between helices 6 and 7 of PcaK is an attractive candidate for interaction with the chemotaxis machinery.
AEROTAXIS
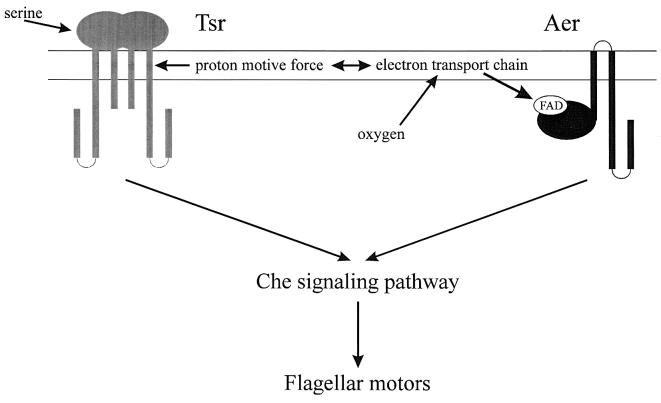
An open reading frame corresponding to a hitherto undetected oxygen sensory transducer, dubbed Aer (4), was described by Sergei Bibikov (J. S. Parkinson’s laboratory, University of Utah). Mutants lacking Aer lose positive aerotaxis but are still repelled by high levels of oxygen. Therefore, a second receptor must mediate the phobic response. Aer has a carboxyl-terminal signaling domain very similar to that of a classical MCP. In contrast, the amino-terminal domain looks like the putative redox-sensing domain of NifL. The hydrophobic central region of Aer may provide a membrane anchor. Aer is expressed at very low levels. When Aer is overexpressed, aerotactic responses are enhanced. Membranes from cells overexpressing Aer exhibit the absorption and fluorescence spectra characteristic of flavin mononucleotide. Thus, Aer is possibly a flavin adenine dinucleotide (FAD)-binding protein that mediates aerotaxis by detecting changes in the redox state of one or more components of the electron transport chain (Fig. 4).
FIG. 4.
Aerotactic signaling. Aer is an FAD-binding protein that is an oxygen sensor for positive aerotaxis in E. coli. Aer may sense the redox state of a component of the electron transport chain and communicate with the chemotaxis signaling pathway through a conserved, MCP-like cytoplasmic domain. Tsr may act as a second sensor for aerotaxis, possibly by measuring the proton motive force.
Mark Johnson (Loma Linda University) pointed out that the Aer protein senses redox potential and internal energy in E. coli, in addition to external oxygen. An aer null strain had diminished but clearly defined responses to oxygen, redox, and energy when chemotaxis was measured in temporal assays. These responses were abolished in an aer tsr double mutant and could be restored by expression of Aer or Tsr from a plasmid in the aer tsr strain (31). A cheB mutant of E. coli, which lacks the receptor methylesterase, is repelled by concentrations of oxygen that attract wild-type cells. The overmethylated Tsr receptor, rather than Aer, mediates these inverted responses, whereas the attractant response to oxygen mediated by Aer dominates in a cheB+ strain. In a cheB mutant, an oxygen-induced increase in proton motive force might cause overmethylated Tsr to generate a CW (tumble) signal unless serine is present, in which case CCW signaling (smooth swimming) would be favored.
METABOLIC CHEMOSENSING
Some nonenteric species, such as R. meliloti and R. sphaeroides, may respond to changes in metabolism and signals generated by receptors. E. coli has been thought to respond only through the MCP or PTS (phosphotransferase) pathway, both of which involve at least some of the Che proteins. Rina Barak (M. Eisenbach’s laboratory, Weizmann Institute of Science), however, reported that E. coli mutants in which all the chemotaxis genes except cheY have been deleted can still respond to aspartate, albeit weakly.
One possible alternate pathway was described by Wolfgang Marwan (D. Oesterhelt’s laboratory, Max Planck-Institut für Biochimie, Martinsreid, Germany). Fumarate can directly alter motor switching in the archaeon Halobacterium salinarium, an effect also seen in flagellated spheroplasts of E. coli (24a). E. coli fumarase mutants were used to show that motor switching is sensitive to intracellular fumarate levels. Some repellents decrease intracellular fumarate by inhibiting fumarase directly, and fumarase mutants do not respond to the repellents indole and benzoate. The only component of the Che pathway required for the fumarate effect was CheY, but a mutant with a nonphosphorylatable CheY still carried out fumarate-dependent switching. There may be two chemotaxis pathways in E. coli: a primitive system linked to metabolism via the citric acid cycle and a second system with dedicated, transmembrane receptors.
FLAGELLAR STRUCTURE, ASSEMBLY, AND FUNCTION
Considerable advances have been made in our understanding of flagellar structure in recent years as electron microscopic and image analysis techniques have become more sophisticated. In the latest development, reported by Dennis Thomas (D. J. DeRosier’s laboratory, Brandeis University) isolated hook-basal body structures in vitreous ice were subjected to the first non-cylindrically-averaged analysis. Individual particles were rotationally aligned by using the hook as reference (hook images had been rotationally aligned previously). That alignment was sustained as successive image sections were analyzed down through the rod and rings. This analysis showed that the rod has a helical symmetry similar to that of the hook and filament and that it has the hollow central channel required for protein export. An analysis of images of the cytoplasmic C ring suggests that it has about 34-fold symmetry.
In enteric bacteria and Caulobacter crescentus, the length of the flagellar hook is closely controlled by a mechanism that involves the FliK protein. Bertha González-Pedrajo (G. Dreyfus’s laboratory, Universidad Nacional Autónoma de México [UNAM]—Mexico City) revealed that a protein homologous to FliK is involved in hook length control in an organism as different as R. sphaeroides, which is photosynthetic, has a single flagellum located in the middle of the cell body, and uses a start-stop motor mechanism (rather than an alternation of CCW and CW flagellar rotation) to carry out chemotaxis.
Most of the mass of a flagellum is extracellular. Therefore, it is not surprising that there are proteins required for export and assembly of these external components. Remarkably, the proteins responsible for export of flagellar components are closely related to proteins responsible for export of virulence factors. One protein needed for flagellar assembly, FliI, is homologous to the catalytic subunit of the F0F1 ATP synthase, which has also been demonstrated recently to be a rotary device. FliI is one of the proteins that has a homolog in virulence systems. The enzymology of FliI in Salmonella was detailed by Fan Fan (R. M. Macnab’s laboratory, Yale University). Despite its similarity to F1, FliI is probably very different mechanistically. FliI has additional sequence at its N terminus that may be important for recognizing exported flagellar proteins, a possibility supported by evidence of interaction between FliI and flagellin (Eugenia Silva-Herzog, G. Dreyfus’s laboratory, UNAM—Mexico City).
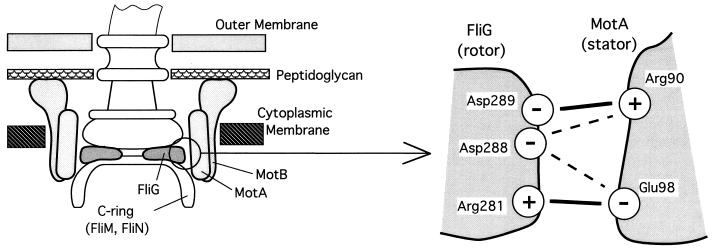
Much interest these days centers around the mechanism of torque generation and switching within the flagellar motor. David Blair (University of Utah) has identified charged residues in the stationary element of the torque generator (the cytoplasmic domain of the membrane protein MotA) and the rotary and switchable element (the peripheral membrane protein, FliG) in the C ring that are important for motility (Fig. 5) (24). Most compellingly, the disruption of motility caused by substitution of a particular negatively charged residue in FliG by a positive residue can be reversed by substitution of a particular positively charged residue in MotA by a negative residue. This result implies that electrostatic interactions between residues in stator and rotor elements may play a crucial role in motor function.
FIG. 5.
Electrostatic interactions between the rotor and stator in the flagellar motor. The diagram on the left shows the flagellar basal body and associated structures as they are thought to be arranged in the E. coli cell envelope. The diagram on the right is a close-up view of the rotor-stator interface that identifies charged residues in the FliG and MotA that have been shown in mutational studies to be important for motor function. Certain combinations of charge-switch (+ to − or vice versa) residue substitutions in FliG and MotA act synergistically in impairing motility, whereas others exhibit mutual suppression in a pattern that indicates that charged residues in FliG interact with charged residues in MotA as shown. Solid lines indicate the primary electrostatic interactions, and broken lines show interactions that are of secondary importance. The precise role of these interactions in torque generation is not known. Figure courtesy of David Blair.
FLAGELLAR GENE REGULATION
The flagellar gene system has been thought to constitute a dedicated genetic unit (regulon), but its complexity appears to have been underestimated. Proteins such as E. coli FlhD, which functions as a positive regulator of the flagellar regulon, are now found to intrude into other aspects of cell function, including cell division. Birgit Prüß (P. Matsumura’s laboratory, University of Illinois—Chicago) explained that the flhD gene, although located in a flagellar gene cluster, encodes a protein that is better described as a global, rather than just flagellar, regulator (28, 29).
Yiping Han (W. Shi’s laboratory, University of California at Los Angeles) described the inverse regulation between motility and virulence in pathogenic bacteria, focusing on the effect of the bvg gene from Bordetella pertussis on flagellar synthesis in E. coli. This pattern of gene expression is consistent with the hypothesis that motility is more valuable outside the host and less so within. Another example of regulatory complexity, presented by Guy LeClerc (B. Ely’s laboratory, University of South Carolina), is a newly discovered set of genes in C. crescentus that are transcribed at the same stage of the cell cycle as flagellin genes yet are under a different control. Their products may play a role in modification of flagellins rather than in their synthesis.
Based on the original studies in E. coli and Salmonella, a dogma has developed. The dogma is that transcription initiation in flagellar operons employs a dedicated sigma factor called ς28 or ςF, which in some cases requires enhancer-like elements as well. It was demonstrated by the group of Nyles Charon (West Virginia University) that this situation does not hold for the pathogenic bacterium Borrelia burgdorferi. This spirochete contains five motility and chemotaxis operons, one of which contains at least 26 flagellar genes (14). All five operons initiate transcription from a ς70 promoter. Karl Klose (J. J. Mekalanos’s laboratory, Harvard University) provided another exception involving ς54 of Vibrio cholerae. This alternate ς factor is required for flagellin synthesis in combination with the response regulator of a two-component signaling system (22). Some aspects of intestinal colonization of the host during pathogenesis also require ς54, and this requirement is independent of the involvement of this ς factor in motility.
SURFACE MOTILITY
The behavior of Proteus mirabilis colonies was captured on a time-lapse video by Jim Shapiro (University of Chicago). Hyperflagellated, elongated swarmer cells differentiate from oligoflagellated, short swimmer cells, migrate outward in groups for a period, and then cease swarming and undergo septation to regenerate swimmers (30). This periodic alternation of swarming and consolidation confers a terraced structure to the colony. Detailed kinetic analysis at different glucose and agar concentrations and mathematical modeling indicate that the clock-like periodicity of P. mirabilis colonies is based on age limits for swarmer cells and collective thresholds for swarmer migration.
Elena Budrene (H. C. Berg’s laboratory, Harvard University) has found conditions under which Proteus swarmers translocate much more rapidly than normal. These swarmers move across the surface in a uniform film within which areas of high cell density develop into dynamic structures, such as rotating spirals or periodic waves. These patterns may reflect the chemotactic responses of cells to spatially nonuniform chemical gradients generated within the swarmer population.
Colonies of Salmonella and E. coli also can swarm if provided with the proper combination of nutrients and agar concentration. A functional chemotaxis system is needed. Swarmer cells elongate and make more flagella, although not as extensively as Proteus.
Rasika Harshey (University of Texas—Austin) described swarming-deficient and swimming- and chemotaxis-competent mutants of Salmonella. Most were affected in lipopolysaccharide or membrane-derived oligosaccharide synthesis, a finding that emphasizes the importance of the cell surface in swarming. The chemotaxis system plays an essential role in swarm cell differentiation, as seen by the abolition of the hyperflagellation response in mutants lacking any one of the Che proteins or lacking both Tar and Tsr. Even mutant cells in which run-tumble behavior is conferred because of counteracting che mutations fail to swarm. Overexpression of the flagellar master operon products FlhC and FlhD restores the ability to hyperflagellate but does not enable Che− cells to swarm. However, cells that express as their sole chemoreceptor a mutant Tsr unable to sense serine do swarm, suggesting that it is the presence of the chemotaxis components, rather than the ability to perform chemotaxis toward nutrients in the external environment, that is necessary for swarming.
Gliding bacteria also move across surfaces but, unlike swarming bacteria, lack flagella. Gliding bacteria are represented in most of the major branches of the eubacteria and are abundant in many environments, but we know remarkably little about the mechanisms of gliding motility. Mark McBride (University of Wisconsin—Milwaukee) presented a genetic analysis of Cytophaga johnsonae (now Flavobacterium johnsoniae) gliding motility. C. johnsonae is a fast glider that can reach speeds of up to 10 μm s−1. A genetic system was developed to study C. johnsonae, and a number of nonmotile mutants were identified. Three genes whose products are required for gliding motility were identified. The gldA gene is homologous to a family of genes that encode ATP-binding cassette (ABC) transporter proteins (1). The gldB gene does not resemble any genes in the database, whereas gldC shares some sequences with genes encoding fungal lectins. The functions of the predicted gene products in cell movement are not yet known.
CELL AGGREGATION AND FRUITING BODY FORMATION IN MYXOCOCCUS XANTHUS
M. xanthus is a slow glider, moving a few micrometers a minute. Cell-cell signaling leads to fruiting body formation in M. xanthus. An A signal is a cell density indicator required to initiate fruiting body formation following starvation. The signal consists of extracellular amino acids and short peptides produced by an extracellular protease. The Asg pathway senses starvation. AsgA is an unusual histidine protein kinase consisting of a CheY-like receiver domain upstream of a kinase domain. AsgB appears to be a DNA-binding protein with a helix-turn-helix motif similar to those of many transcription regulators. The single asgC mutation targets the M. xanthus homolog of ς70. John Davis presented work by Valerie Kessler (L. S. Plamann’s laboratory, Texas A&M University) in which 42 intergenic suppressors of one asgA mutation were identified. Most were bypass mutants with pleomorphic phenotypes.
Heidi Kaplan (University of Texas—Houston) reported a new two-component system composed of the sasS and sasR (formerly hpkA and mrrA, respectively) gene products. This pathway is involved in fruiting body development and appears to respond to a combination of A signal and nutrient deprivation. The sasS null mutants are defective in fruiting body formation and sporulation. The sasS null mutants still form spores, although the spores are defective in long-term survival. The SasR response regulator is an NtrC homolog and is required for transcription from a promoter that belongs to the ς54 family.
The glycerol pathway of sporulation, which bypasses the fruiting body and produces less-stable spores, was outlined by David Zusman (University of California—Berkeley). Glycerol induces β-lactamase and a starvation-independent pathway to spores, suggesting that damage to the cell envelope induces sporulation. This capability provides cells with a protective response to cell wall-threatening (and hence life-threatening) insults from the environment.
Lotte Soggaard-Andersen (University of Odense) studies C factor, which is needed for aggregation during fruiting-body formation. C factor is a cell surface protein that may interact with a C-factor sensor on an adjacent cell, leading to activation of the class II genes required for sporulation, developmental gene expression, and changes in motile behavior. Methylation levels of FrzCD, whose amino-terminal region is an MCP homolog, increase when cells are activated by C factor. This methylation depends on FrzF, the CheR analog in M. xanthus, and a DNA-binding response regulator encoded by a class II gene. C-factor function requires end-to-end cell contact, suggesting that it stimulates FrzCD and decreases reversals, causing cells to line up in chains that march up a gradient of C factor.
What is the aggregation signal in M. xanthus? The frz system clearly plays a role, but the attractant had not been identified. Larry Shimkets (University of Georgia) reported that a purified component from cellular exudates, phosphatidylethanolamine, suppresses cellular reversals and enhances movement up gradients. It also restores aggregation ability to csgA mutants, which lack short-chain alcohol dehydrogenase function and lack the ability to produce the C signal. This is the first reported chemoattractant for the myxobacteria.
Lisa Gorski (Dale Kaiser’s laboratory, Stanford University) discovered that ς54, which works in conjunction with auxiliary transcription factors, is required for expression of one motility gene and five other developmental genes in M. xanthus. This control may provide avenues for regulation by A factor and C factor during the early and middle stages of development, respectively.
In the metabolic gospel according to Phil Youderian (University of Idaho), development seen by a biochemist boils down to turning one set of biopolymers into another. Since starvation induces spore development, M. xanthus faces the problem of conserving intermediary metabolites during this conversion. The A signal consists mostly of ketogenic amino acids catabolized via the tricarboxylic acid cycle. To avoid oxidizing these compounds completely to CO2, M. xanthus, like developing seeds, recycles carbon via the glycosylate pathway. Inhibitors of isocitrate lyase (Icl), the first pathway enzyme, block development, and succinate and glyoxylate, the products of Icl, restore development to a csg mutant. Furthermore, icl disruption mutants form 10,000-fold-fewer spores. Thus, the glyoxylate shunt functions as a carbon-hoarding mechanism that is at the “metabolic heart” of M. xanthus development.
SWIMMING WITHOUT FLAGELLA
As if gliding were not mysterious enough, a decade ago John B. Waterbury (Woods Hole Oceanographic Institute) isolated a cyanobacterium, Synechococcus sp., which swims at 25 μm s−1 without obvious locomotory appendages. To quote loosely Abigail Salyers’ memorable statement on the matter, the ocean is full of these things; flagellated bacteria are the oddballs.
Calcium is required for swimming motility of Synechococcus, and no other divalent cations tested function in its stead. Tom Pitta (H. C. Berg’s laboratory, Rowland Institute) presented data showing that the mammalian calcium channel inhibitors verapamil and nitrendipine inhibit swimming, indicating that voltage-gated calcium channels may be involved in Synechococcus motility (27). The calcium chelators EDTA and EGTA also inhibit swimming, even at low concentrations and in the presence of excess calcium, suggesting that these chelators may also function as calcium ionophores. Thus, a calcium motive force may be required for swimming.
Bianca Brahamsha (Scripps Institute of Oceanography) has identified a large glycoprotein (SWMA) present only in motile strains of Synechococcus (6). It is removed by treatment of the cells with 10 mM EDTA. SWMA is found all over the cell surface. The polypeptide exhibits a classical calcium-binding motif. Synechococcus rotates as it swims, and mutants lacking SWMA do not translocate, although if attached to a surface they still rotate about a point of attachment. Cells lacking SWMA therefore lose thrust but not their motive power. SWMA could be involved directly in translocation or be required for assembly of the motile apparatus, but the facts that it is a calcium-binding protein and that calcium is essential for swimming suggest new avenues for investigating flagella-free swimming.
SIGNAL TRANSDUCTION IN THE ARCHAEA
John Spudich (University of Texas—Houston) presented new results on photosensing in the archaeon Halobacterium salinarium, which contains two transport rhodopsins (bacteriorhodopsin and halorhodopsin) and two sensory rhodopsins (SRI and SRII). Like visual rhodopsin, each of the archaeal rhodopsins has seven membrane-spanning helices, and retinal attaches via a protonated Schiff base to a lysine in helix VII and is buried within the protein. SRI and SRII are tightly bound to transducers HtrI and HtrII, respectively, which are essential for phototaxis. Absorbed light photoisomerizes retinal in the proton pump bacteriorhodopsin, the Schiff base proton is transferred to Asp85 on helix III, and a proton is released to the external environment. The resulting disruption of the salt bridge between the protonated Schiff base and Asp85 opens a channel that allows proton uptake from the cytoplasm. In SRI and SRII, the transducers block the cytoplasmic channel; without their transducers, SRI and SRII become light-driven proton pumps. Asp73 in SRII functions like its homolog of Asp85 in bacteriorhodopsin. A D73N mutant of SRII signals constitutively, implicating disruption of the salt bridge in signaling (34). A unified model for archaeal rhodopsin function proposes that the conformational switch that opens the cytoplasmic channel in bacteriorhopsin also operates in SRI and SRII. HtrI and HtrII convert this change into attractant (inhibition of reversals in swimming direction) and repellent (increase of directional reversals) responses, respectively.
Recently, 13 proteins with homology to the cytoplasmic signaling domain of bacterial MCPs were identified in H. salinarium (36). Alexei Brooun (M. Alam’s laboratory, University of Hawaii) explained that these proteins fall into three subfamilies. Subfamily A has large periplasmic and cytoplasmic domains and two membrane-spanning helices and resembles bacterial chemoreceptors. Subfamily B, which includes HtrI, HtrII, HtrVII, and HtrVIII, has no periplasmic domain but has multiple amino-terminal transmembrane segments. The HtrVIII protein of subfamily B mediates aerotaxis: a strain with the htrVIII gene deleted loses aerotaxis, and a strain overproducing HtrVIII exhibits stronger aerotaxis. The amino-terminal domain of HtrVIII resembles mitochondrial cytochrome c oxidase. (HtrVIII provides an interesting contrast to the FAD-containing oxygen sensor from E. coli.) The cytoplasmic HtrXI protein of subfamily C is involved in chemotaxis to aspartate and glutamate (7). Its methylation levels change as expected upon stimulation with those attractants. It remains to be seen how HtrXI and other cytoplasmic transducers in H. salinarium fit into signaling pathways.
MECHANOSENSING IN PROKARYOTES
Mechanosensing in prokaryotes has been a largely unexplored phenomenon. It was therefore illuminating to hear Paul Moe (C. Kung’s laboratory, University of Wisconsin—Madison) describe experiments with patch-clamped E. coli spheroplasts that discovered two mechanosensitive channels of very large conductance. A 1-nS channel opens at moderate membrane tension, and a 3-nS channel opens only as the membrane approaches a tension that would disrupt the spheroplast. The protein comprising the 3-nS channel, MscL (5), was purified and shown to consist of 136 residues, to contain two membrane-spanning helices, and to form a homohexameric channel. MscL is proposed to function as an emergency “osmotic dumping” device for cells exposed to hypotonic shock. A strain with the mscL gene deleted has no phenotype, presumably because MscL function is redundant with that of the smaller, more easily triggered 1-nS channel. Homologs of MscL are found in a wide range of gram-negative and gram-positive bacteria.
CHEMORECEPTOR STRUCTURE AND FUNCTION
Transmembrane signaling is a central theme in sensory biology. Events triggered by ligands binding at the exterior surface of the cell membrane modulate the activity of the cytosolic domains of the receptors. Determining how ligands bind and how receptors discriminate among them is an important first step in understanding this process.
The Tar chemoreceptor of E. coli recognizes two disparate attractant ligands, aspartate and ligand-associated maltose-binding protein (MBP). Paul Gardina (M. D. Manson’s laboratory, Texas A&M University) used intergenic complementation analysis to show that aspartate and MBP bind across the Tar dimer at one of two rotationally symmetric sites, contacting largely nonoverlapping sets of residues in each subunit. Aspartate binds at the subunit interface, and MBP binds across the dimer apex (13). Aspartate signals primarily through the subunit in which the binding half site makes the larger number of direct contacts with the ligand. This result suggests that a Tar dimer may sense aspartate and maltose simultaneously by responding to aspartate via one subunit and maltose-loaded MBP via the other. Coexpression of different pairs of doubly mutant Tar proteins showed that, depending on the orientation of intact binding sites, aspartate and maltose are sensed independently (signal segregation) or aspartate blocks the maltose response (signal exclusion). Thus, the model may have promise.
The closely related receptors Tar and Tsr provide a good system to study molecular recognition and ligand specificity. Tar does not respond to serine, and Tsr does not respond to aspartate. Tar contacts aspartate via residues in periplasmic helix I and periplasmic helix IV. In or near the ligand-binding site, only three residues differ between Tar and Tsr. Ikuro Kawagishi (Nagoya University) has exchanged these residues, singly and in aggregate. Such manipulations reduced the ability of either receptor to respond to its normal ligand, but in no case was the heterologous ligand recognized. In contrast, replacement of a stretch of 30 residues in helix I and the helix I-helix II loop of Tsr with the equivalent residues of Tar converts Tsr into an aspartate sensor. Thus, residues other than those that participate directly in ligand binding contribute to specificity, probably by determining how helix I is oriented or by differentially positioning critical side chains in helix I.
X-ray crystallographic and nuclear magnetic resonance (NMR) studies of the structures of membrane-spanning chemoreceptors have, for technical reasons, been confined largely to their soluble domains. An exciting new approach developed by Lynmarie Thompson (University of Massachusetts—Amherst) has been the application of solid-state NMR to the Tsr receptor in situ (35). Thus far, ligand-to-protein distances measured with intact, membrane-bound Tsr are consistent with corresponding distances in the crystal structure of the Tar receptor domain, thereby validating the latter and confirming the proposed similarity of the ligand-binding sites of Tar and Tsr. Eventually, this approach should generate entirely new information about the integrated structure and function of the intact receptor.
The structures of the transmembrane domains of the receptors are being probed by electron paramagnetic resonance (EPR) spectroscopy. In work described by Alexander Barnakov (G. L. Hazelbauer’s laboratory, Washington State University), the sulfhydryl groups of cysteine-containing mutants of Trg were used to introduce nitroxide spin-labeled methanethiosulfonate into the purified protein at known locations in TM1. The EPR results indicate that approximately 30 residues of TM1 are in the hydrophobic region of the membrane, that a few residues are close enough (<1.2 nm) in the dimer for their spin labels to interact between TM1 and TM1′, and that TM1 is a dynamic structure. Since only 20 to 24 residues are required to span the hydrophobic layer, TM1 may be tilted or not entirely α-helical, or it may bob up and down in the membrane.
Karen Ottemann (D. E. Koshland’s laboratory, University of California—Berkeley) spoke of attempts to detect ligand-induced movement in the Salmonella Tar receptor. Double cysteine replacements, one in helix I and one in helix IV, were made in the periplasmic domain of Tar. The two Cys substitutions in a pair were about equidistant from the membrane. The proteins were purified and modified with methanethiosulfonate. Although the spin labels were close enough to affect one another and the modified proteins could still mediate normal responses to aspartate in in vitro assays of CheA inhibition and stimulation of methylation, no significant change in the spin label interaction was seen with any of the pairs. The most straightforward conclusion is that the side chains of the selected amino acids on helix I and helix IV move less than 0.1 nm relative to one another upon binding aspartate.
Cysteine mutagenesis was used by Mark Danielson (J. Falke’s laboratory, University of Colorado) to investigate the cytoplasmic domain of Salmonella Tar. Cysteine-scanning mutagenesis encompassed 60 residues, which included the carboxyl-terminal end of the linker region (the α5 helix), a nonhelical gap of 3 residues, and the long α6 helix, which contains the three methylatable glutamates of the K1 peptide. Inhibitory substitutions cluster in the gap and the predicted hydrophobic face of α6. Availability for thiol labeling with fluorescein-5-iodoacetamide disclosed that, in both helices, every seventh residue was often deeply buried, an arrangement that defines a helical packing face. The most inaccessible positions were in the region containing the methylation sites at the carboxyl-terminal end of α6. The methylation sites themselves were located on the charged face of the amphiphilic α6 helix and were all highly accessible. Finally, in vitro experiments assayed the ability of disulfide-cross-linked receptors to stimulate CheA and to block stimulation upon binding aspartate. Nonstimulating receptors could be “locked-off” or structurally disrupted, whereas the C278 and C300 cross-links locked the receptor in an “on” state immune to inhibition by aspartate. Thus, changes at the subunit interface in this region, especially in α6, are critical for control of kinase activity. However, the existence of a number of cross-linked receptors that still allowed essentially normal stimulation and inhibition of CheA indicates that these changes must be subtle.
MODELING CHEMOTAXIS
David Trentham (National Institute of Medical Research, Mill Hill) described experiments done in collaboration with Shahid Khan (Albert Einstein College of Medicine) in which light-induced release of aspartate from a photosensitive “caged” aspartate was used to measure signal amplification in the chemotactic response of E. coli. The response was biphasic, consisting of a rapid increase in CCW bias followed by adaptation. The adaptation time was related to receptor occupancy, and adaptation times were best fitted by assuming the receptor has two binding sites with Kd values of 1.2 and 70 μM. This apparent negative cooperativity affects estimates of gain, which equals the change in bias/change in receptor occupancy. The gain also depended on aspartate levels present in the prestimulus background solution. The value of the gain at zero aspartate background was comparable to that for serine (ca. 5) determined previously (20). It was substantially smaller than the gain calculated earlier (32), using data from tethered cells stimulated with aspartate delivered iontophoretically and assuming that there is a single aspartate-binding site with a Kd value of 5 μM.
The status of computer simulation of bacterial chemotaxis was chronicled by Carl Morton-Firth (D. Bray’s laboratory, University of Cambridge). The original program models all possible reactions in the cell, using the best biochemical data available. It correctly predicts the phenotypes of 36 of 41 strains with different combinations of chemotaxis gene mutations. The five misses and the limitations imposed by the program’s reliance on some simplifying assumptions encouraged the development of a new program. It is a discrete, stochastic simulator that represents all relevant particles in the cell as individual software objects and determines the probability of their interacting to carry out possible reactions. Unlike the earlier program, it is able to model finite numbers of particles and realistic cell volumes, and it displays the stochastic fluctuations in numbers of signaling molecules as a function of time. Each of the two programs has its own strengths and weaknesses, so a combined approach offers the best chance of accurately modeling the behavior of real bacteria.
SUGAR PTS
Milton Saier (University of California—San Diego) emphasized regulation by the PTS. Phosphate moves from phosphoenolpyruvate (PEP) to enzyme I (EnzI) to HPr to EnzIIA to EnzIIB to sugar. EnzIIC forms a sugar-specific pore. EnzIIA, EnzIIB, and EnzIIC can all be in one polypeptide, or they can exist independently. PTS proteins are phosphorylated on histidines, except EnzIIB, which is sometimes phosphorylated on cysteine. Soluble EnzIIAglucose is central to catabolite control in E. coli. Phospho-IIA accumulates in the absence of glucose and stimulates adenylcyclase to boost transcription from promoters that require cyclic AMP (cAMP)-cAMP receptor. When the PTS is actively transporting sugar, phospho-IIA turns over, cAMP levels fall, and catabolite repression ensues. Unphosphorylated IIA inhibits non-PTS transporters like lactose permease to cause inducer exclusion. The PTS of gram-positive bacteria exercises catabolite repression and inducer exclusion very differently. HPr is subject to phosphorylation on a serine residue by a regulatory kinase. Seryl-phosphorylated HPr inhibits several non-PTS permeases and, with the CcpA protein, represses transcription of operons under catabolite control. It also is thought to promote inducer exclusion by more than one mechanism. It strongly inhibits histidyl phosphorylation of HPr and thereby sugar uptake via the PTS. The regulatory kinase, whose activity is regulated by intracellular metabolites, recognizes the tertiary structure of its substrate and belongs to a novel family of protein kinases.
The central role of the PTS in regulation was developed further by Orna Amster-Choder (Hebrew University). The bgl operon of E. coli is regulated by two of its products, BglF and BglG. BglF functions as a membrane-bound EnzII of the PTS, mediating β-glucoside uptake and phosphorylation. It is also a β-glucoside sensor that controls the activity of BglG by reversibly phosphorylating it according to sugar availability. Unphosphorylated BglG dimerizes and acts as a transcription antiterminator. In the absence of β-glucoside, BglF phosphorylates a histidine residue on BglG (9), thus preventing its dimerization and blocking bgl transcription (8). In the presence of β-glucosides, BglF transfers phosphate to the sugar during its uptake and dephosphorylates BglG, thereby allowing transcription of the bgl operon. Remarkably, the same phospho-cysteine (C24) in domain IIB of BglF can transfer phosphate to β-glucoside or to BglG. BglF dimerizes, but dimerization is not induced by β-glucosides and therefore cannot control the sugar-driven signal transduction pathway. However, β-glucosides also trigger formation of a disulfide bond in BglF in a reaction that requires C24.
TWO-COMPONENT RESPONSE REGULATORS
Two-component systems have been a staple of the BLAST meeting. A major development this year was the crystal structure of the NarL response regulator (Fig. 6) (3) presented by Robert Gunsalus (University of California at Los Angeles). The DNA-binding domains of several response regulators, such as UhpA, DegS and others, are homologs of this domain in NarL, which allows their activities to be compared. The regulatory and DNA-binding domains of NarL are joined by a 13-amino-acid tether that is shared by many response regulators that serve as transcription factors. In unphosphorylated NarL, the domains interact to prevent binding to DNA. Phosphorylation is proposed to induce a conformational change that destroys this interaction, freeing the carboxyl-terminal domain to interact with its DNA target.
FIG. 6.
Ribbon diagram of the NarL response regulator. The CheY-like amino-terminal domain (shown in green) has five α helices and a bundle of five β strands. It also possesses a sixth helix, not present in CheY, that forms part of the connection to the carboxyl-terminal domain. The carboxyl-terminal domain (shown in yellow) consists of four α helices; helices 8 and 9 comprise a helix-turn-helix motif responsible for DNA recognition and binding. The aspartate residue that is a target for phosphotransfer from the autokinases NarQ and NarX is shown as a gray and red stick-and-ball figure. Phosphorylation of this residue is proposed to cause a conformational change that unmasks the DNA-binding region, which is blocked by the amino-terminal domain of NarL when the protein is in its unphosphorylated state. Reprinted from Biochemistry (3) with permission of the publisher.
The tether of UhpA is only six amino acids long and thus restricts possible interactions between the domains. Indeed, Bob Kadner (University of Virginia) demonstrated that unphosphorylated UhpA binds DNA perfectly well. Why, then, is phosphorylation required for UhpA activity? The answer seems to be that phosphorylation-induced oligomerization of UhpA on its target promoter allows binding at low-affinity sites to initiate transcription. Mutational evidence suggests that oligomerization depends on sites in helix 1 of the regulatory domain.
Domain interaction in response regulators was first defined in CheB, for which it was shown that the regulatory domain inhibits the methylesterase activity of the carboxyl-terminal domain. The structure of CheB (11) reported by Ann West (University of Oklahoma) (in collaboration with S. P. Djordjevic and A. M. Stock, University of Medicine and Dentistry of New Jersey) reveals that the regulatory domain packs against the catalytic triad of the carboxyl-terminal domain in the unphosphorylated protein. Thus, phosphorylation must induce a large conformational change in the regulatory domain to free the carboxyl-terminal domain for interaction with the methylation sites of the chemoreceptors. The domain interface of CheB differs in orientation by about 90° from that observed for NarL. This observation suggests that different molecular surfaces are utilized for interdomain interactions in different multidomain response regulators.
These three studies of response regulators provide a microcosm of functional diversity. NarL and UhpA differ in the length of the tether that connects the domains. The result is different DNA-binding properties and responses to phosphorylation. The long tether of most response regulators allows many possible combinations of interacting surfaces between the two domains. Thus, a simulation based on the structure of one response regulator may give a completely erroneous picture of a second one, even if it is homologous.
The conformational change induced by phosphorylation of the response regulator is the key to understanding signal transduction in two-component systems. The instability of the aspartyl-phosphate has prevented crystal structure analysis and has complicated interpretation of NMR studies. Christopher Halkides (F. W. Dahlquist’s laboratory, University of Oregon, and P. Matsumura’s laboratory, University of Illinois—Chicago) described a stable charge and size mimic of aspartyl-phosphate designed to circumvent this problem. A D57C mutant of CheY (D57 is the phosphorylated residue) was alkylated with iodoacetate and Triflate to produce a phosphonic acid analog. This derivatized CheY binds FliM and CheZ, and a crystal has been obtained.
In E. coli, the phosphate starvation response is regulated by the PhoR (kinase)-PhoB (response regulator) two-component system. Marion Hulett (University of Illinois—Chicago) described studies demonstrating a Pho regulatory network in Bacillus subtilis that is composed of three two-component systems: PhoP and PhoR, homologs of E. coli PhoB and PhoR; ResD and ResE, regulators of aerobic and anaerobic respiration; and Spo0A, the response regulator for initiation of sporulation. This integrated signal transduction network provides a system for exploring regulatory hierarchies that are imposed on the signals activating the PhoR kinase.
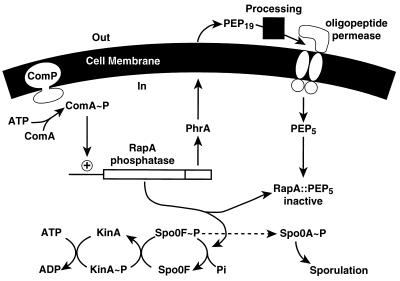
Depending on the system, kinases promote phosphorylation or dephosphorylation of their cognate response regulators. The phosphorelay that controls entry into the sporulation pathway of B. subtilis is also regulated by phosphatases that recognize different signals than the kinases do. Marta Perego (Scripps Research Institute) described the RapA phosphatase that is induced by the competence pathway. RapA dephosphorylates Spo0F-P of the sporulation phosphorelay to preclude sporulation regardless of the activity of kinases phosphorylating Spo0F. RapA activity is regulated by a peptide export-import circuit (26). The rapA transcript includes an open reading frame encoding a 44-amino-acid peptide with an amino-terminal leader sequence. The carboxyl half of this peptide is exported and trimmed by extracellular peptidases down to its last five residues. The activity of these peptidases may be regulated. The oligopeptide permease imports the pentapeptide into the cytoplasm, where it inhibits RapA (Fig. 7). Any alteration of the pentapeptide sequence destroys its function. This export-import circuit may serve as a device to provide temporal control of RapA activity.
FIG. 7.
Control circuit for sporulation initiation in B. subtilis. Initiation of sporulation requires the phosphorylated form of the response regulator Spo0A. Phosphate is delivered to Spo0A via a phosphorelay that involves, in sequence, the KinA sensor kinase, the Spo0F intermediate response regulator, and the Spo0B phosphotransferase (not shown). Transcription of the operon encoding the RapA phosphatase and the PhrA control peptide requires the ComA-P competence regulator. RapA dephosphorylates Spo0F-P, thereby preventing Spo0A-P accumulation and sporulation during expression of competence. After its synthesis, PhrA is exported, proteolytically processed, and reimported into the cell as a pentapeptide by the oligopeptide permease. The PhrA pentapeptide directly and very specifically inhibits RapA activity, allowing sporulation to proceed. PEP19, exported form of PhrA; PEP5, imported pentapeptide. Reprinted from the Proceedings of the National Academy of Sciences of the United States of America (26) with permission of the publisher.
With over 50 different two-component systems operating in E. coli, specificity of kinase-response regulator interactions is crucial to avoid unwanted cross talk. A strain lacking the PhoR and CreC kinases and acetyl phosphate cannot phosphorylate the response regulator PhoB. To examine factors that determine specificity, Barry Wanner (Purdue University) produced VanS, the kinase of the vancomycin resistance system of Enterococcus faecium (15) in such a strain. Transfer of phosphate from VanS to PhoB in vitro occurred at 10−4 of the rate of transfer to the cognate response regulator VanR, and the Pho regulon was expressed only when vanS or phoB was present on a multicopy plasmid. A genetic selection involving integrated plasmids under tight transcriptional control was used to isolate mutants of PhoB that were phosphorylated more efficiently by VanS (16). The mutations obtained affect residues in the proposed recognition helix of PhoB, indicating the utility of this approach for studying kinase-response regulator interactions.
TWO-COMPONENT KINASES
Two-component kinases are as important as response regulators in two-component systems, but much less is known about their structure and function. CheA is the best understood, but its domain structure diverges from those of other kinases, and it has an extra domain involved in receptor binding. Kinases generally function as dimers, and control of dimerization may play a role in regulation of some kinases, especially CheA. Mikhail Levit (J. Stock’s laboratory, Princeton University) showed that all of the determinants for dimerization of CheA are in its catalytic domain, which contains the ATP-binding site (23). Rick Stewart (University of Maryland) suggested that a three-stage mechanism transfers phosphate from CheA to CheY. Kinases fall into two groups in this regard; either they are readily reversible or they also function as phosphatases. CheA falls into the first group.
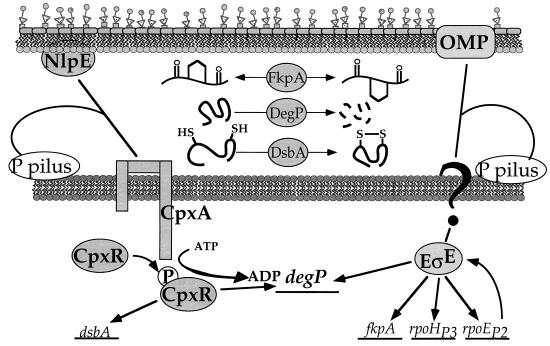
The majority of the kinases of two-component systems are anchored in the cell membrane via variable numbers of transmembrane segments. Perhaps this location reflects their roles as sensors of periplasmic components, although the regulating ligands are in most cases still a mystery. Paul Danese (T. J. Silhavy’s laboratory, Princeton University) reported that the two-component Cpx system, in conjunction with the minor sigma factor ςE, controls production of enzymes involved in protein folding and turnover in the periplasm (Fig. 8) (10). The DegP protease, a peptidyl-prolyl cis to trans isomerase, and an enzyme involved in disulfide-bond formation are all regulated by this system. Cpx represents the first case of a two-component kinase that relays the status of macromolecules on the outside of the membrane to the genetic regulatory mechanism in the cytoplasm. It would be surprising if others did not have similar roles.
FIG. 8.
Model of the Cpx and ςE regulons. The Cpx two-component system is activated by the synthesis of P-pilus subunits and the outer membrane lipoprotein NlpE. The inner membrane sensor CpxA phosphorylates its cognate response regulator, CpxR. CpxR-P stimulates transcription of the degP and dsbA genes. The periplasmic DegP and DsbA proteins are involved in proteolysis and disulfide-bond formation, respectively. An unknown signal transduction pathway (question mark) is postulated to monitor the level of P-pilin and other outer membrane proteins (OMP). This pathway is also proposed to stimulate ςE activity. Transcription from the fkpA, degP, rpoEp2, and rpoHp3 promoters requires ςE. The fkpA gene encodes a periplasmic prolyl isomerase, and rpoE and rpoH encode ςE and ςH, respectively. Reprinted from Genes and Development (10) with permission of the publisher.
A few sensor kinases are soluble cytoplasmic enzymes having nothing to do with membranes. The prototype is NRII (NtrB) of E. coli, which regulates glutamine synthetase expression in response to available nitrogen by regulating the phosphorylation level of the response regulator NRI (NtrC). Phospho-NRI is a transcription factor. NRII is also unusual in that it is regulated solely by modulation of its phosphatase activity. The agent of this regulation is the PII protein, which also controls the activity of the adenyltransferase or adenyl-removing enzyme that modulates the activity of glutamine synthetase. PII activity is regulated by a uridyltransferase or uridyl-removing enzyme and small molecules such as ATP, α-ketoglutarate, and glutamate. Alex Ninfa (University of Michigan) has used genetic and physical-biochemical techniques to probe the different binding interactions of PII with its effectors and its regulatory targets (18, 19). Mutants of PII were obtained that lost the ability to bind to each of the effectors and targets separately, allowing a preliminary map of their binding sites on PII to be constructed.
Two-component systems are not confined to prokaryotes. They have now been identified in Saccharomyces cerevisiae, plants, and the cellular slime mold Dictyostelium discoideum. Stephan Schuster (Max-Planck-Institut für Biochimie) has characterized the DokA protein, which contains a large amino-terminal input domain, a kinase domain, and a carboxyl-terminal response regulator domain. The isolated carboxyl-terminal domain is the first eukaryotic response regulator shown to be phosphorylated by acetyl phosphate (31a). Cells with disruptions of dokA are sensitive to high osmolarity, but DokA is a soluble protein, unlike the osmosensing membrane-bound kinases EnvZ of E. coli and Sln1 of S. cerevisiae. DokA may respond to changes in intracellular solute levels or function in a signaling relay. The target of its regulation may be the cytoskeleton, since dokA amoebae exposed to high osmolarity fail to polymerize myosin or round up into a defensive posture.
TWO-COMPONENT SYSTEMS INVOLVED IN PATHOGENESIS
Salmonella typhi has two additional porins of the OmpC-OmpF family, OmpS1 and OmpS2. Edmundo Calva (UNAM—Cuernavaca) reported that these proteins are normally expressed at low levels, but a deletion of 222 bases upstream of the promoter leads to 400-fold overexpression of OmpS1. Expression of OmpS1 requires the response regulator OmpR, which binds to negative regulatory sites in the deleted region and positive regulatory sites downstream in the promoter downstream of the deletion. Unlike E. coli, S. typhi does not down-regulate OmpC at low osmolarity, although S. typhi OmpF is repressed at high osmolarity. This increased porin repertoire and altered porin regulation may help S. typhi during infection of the host.
Lysis of the cell wall in Staphylococcus aureus is effected by the two-component LytS-LytR system. Ken Bayles (University of Idaho) has shown that this system regulates at least two genes, lrgA and lrgB. The products of the lrgA gene, which encodes a membrane protein of the holin family, and the lrgB gene are thought to be involved in the regulation of murein hydrolase activity. In view of the serious consequences of S. aureus infection and the resistance of this gram-positive organism to virtually all known antibiotics, the Lyt system is an attractive target for pharmacological attack.
Tetracycline resistance in anaerobic, gram-negative Bacteroides spp., another group of potential pathogens, is encoded by a conjugative transposon whose transfer is stimulated by tetracycline via three genes, rteA, rteB, and rteC. RteA and RteB are homologs of the kinases and response regulators, respectively, of two-component systems. Abigail Salyers (University of Illinois—Urbana) has studied the cellular location of these two proteins and found, to everyone’s surprise, that they partition with the outer membrane fraction. These proteins may have a structural role as well.
ADAPTIVE METHYLATION
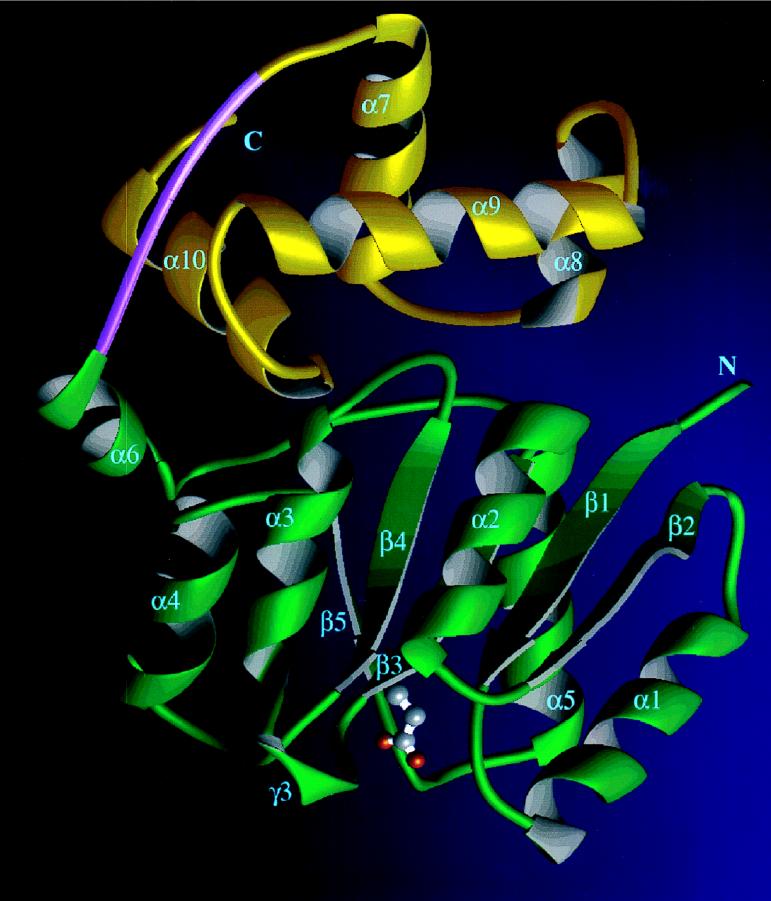
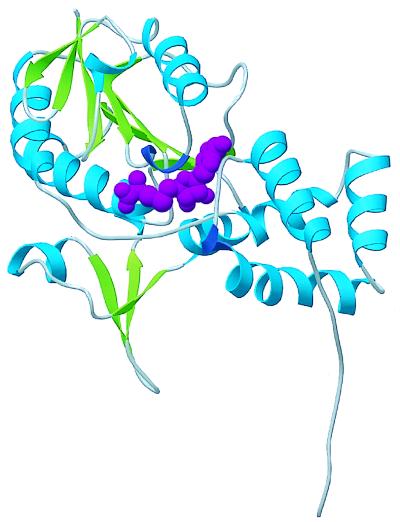
Our understanding of adaptive methylation has reached a new plateau with the structures of the CheB methylesterase, discussed above in “Two-component response regulators” and of the CheR methyltransferase (Fig. 9) (12), solved by Snezana Djordjevic (A. M. Stock’s laboratory, University of Medicine and Dentistry of New Jersey). CheR has a small amino-terminal domain with four α-helices packed in a perpendicular arrangement, and a large α-β domain with a central, seven-stranded β-sheet. The crystal obtained contained the reaction product S-adenosylhomocysteine buried in a nucleotide fold at the domain interface. CheR represents the first high-resolution image of a protein methyltransferase. The structure and biochemical properties of CheR suggest that its reaction mechanism involves neither a metal ion, as in small-molecule methyltransferases, nor a methylcysteinyl intermediate, as in DNA methyltransferases. The surface of CheR predicted to interact with the carboxyl-terminal end of the MCPs is distant from the active site, consistent with the notion that this interaction raises the local CheR concentration at the receptor patch rather than plays a mechanistic role.
FIG. 9.
Ribbon diagram of CheR. The α helices are shown in blue, and the β strands are shown in green. S-adenosylhomocysteine, which was cocrystallized with CheR, appears in magenta. This orientation of the structure places the amino terminus of the protein at the lower right and the carboxyl terminus at the upper left. The α-helical amino-terminal domain and the α-β sandwich of the carboxyl-terminal domain are clearly visible. The carboxyl-terminal domain exhibits homology with other methyltransferases. Inserted between the amino- and carboxyl-terminal domains is a unique, three-stranded β subdomain, which might be involved in interaction with the receptor. Figure courtesy of Snezana Djordjevic.
Chemotaxis in B. subtilis differs from that in E. coli in several important ways. For one thing, B. subtilis has three additional che genes (cheC, cheD, and cheV) but lacks cheZ. For another, in B. subtilis, attractants stimulate CheA kinase, and phospho-CheY promotes CCW flagellar rotation to cause smooth swimming. As John Kirby (G. W. Ordal’s laboratory, University of Illinois—Urbana) made clear, the role of methylation in adaptation is different as well (21). The asparagine receptor McpB demethylates in response to addition and removal of this attractant. Methanol is released with the same kinetics as methyl groups are lost from McpB, so no methyl group transfer is involved, as had been previously postulated. Furthermore, McpB subsequently remethylates in each case, possibly including methylation at new sites. This remethylation following addition of asparagine does not occur in a cheY mutant, a result which implies a direct or indirect interaction between the response regulator CheY and the receptor McpB to control remethylation.
SUMMARY
This review of BLAST IV began with considerations of nonenteric bacteria that behave in strange (to the enterocentric mind) and wonderful ways. To close the circle, it is fitting that it should also end with an organism, like B. subtilis, that does things differently. These examples highlight the fact that we have just begun to unravel the marveously diverse solutions to environmental sensing and adaptive response that have evolved during nearly four billion years of prokaryotic life on earth.
ACKNOWLEDGMENTS
We express our gratitude to all the many people who worked to make BLAST IV possible. We acknowledge in particular Alberto Mendoza, for suggesting that the meeting be held in Cuernavaca; Joe Falke, for serving as chair of the organizing committee; Sandy Parkinson, for heading the program committee; Phil Matsumura, for keeping track of registration and finances and for continuing as CEO of BLAST Inc.; and George Dreyfus, for taking care of arrangements in Cuernavaca. Our special thanks go to Phil Matsumura’s administrative assistant Peggy O’Neill, who saw to it that everything ran as smoothly as possible before, during, and after the meeting.
Funding for BLAST IV was provided by the National Institutes of Health (AI/GM-41086), National Science Foundation (MCB-9603198), Army Research Office (DAAH04-96-1-0465), and Pfizer, Inc.
REFERENCES
- 1.Agarawal S, Hunnicutt D W, McBride M J. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc Natl Acad Sci USA. 1997;94:12139–12144. doi: 10.1073/pnas.94.22.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage J P, Schmitt R. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations on a theme? Microbiology. 1997;143:3671–3682. doi: 10.1099/00221287-143-12-3671. [DOI] [PubMed] [Google Scholar]
- 3.Baikalov I, Schroeder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. The structure of the Escherichia coli response regulator, NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 4.Bibikov S I, Biran R, Rudd K E, Parkinson J S. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blount P, Sukharev S I, Moe P C, Nagle S K, Kung C. Towards an understanding of the structural and functional properties of MscL, a mechanosensitive channel in bacteria. Biol Cell. 1996;87:1–8. [PubMed] [Google Scholar]
- 6.Brahamsha B. An abundant cell-surface polypeptide is required for swimming by the non-flagellated marine cyanobacterium Synechococcus. Proc Natl Acad Sci USA. 1996;93:6504–6509. doi: 10.1073/pnas.93.13.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooun A, Zhang W, Alam M. Primary structure and functional analysis of the soluble transducer protein HtrXI in the archaeon Halobacterium salinarium. J Bacteriol. 1997;179:2963–2968. doi: 10.1128/jb.179.9.2963-2968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Arendts J C, Bader R, Postma P W, Amster-Choder O. BglF, the sensor of the E. coli bgl system, uses the same site to phosphorylate both a sugar and a regulatory protein. EMBO J. 1997;16:4617–4627. doi: 10.1093/emboj/16.15.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Engelberg-Kulka H, Amster-Choder O. The localization of the phosphorylation site of BglG, the response regulator of the E. coli bgl sensory system. J Biol Chem. 1997;272:17263–17268. doi: 10.1074/jbc.272.28.17263. [DOI] [PubMed] [Google Scholar]
- 9a.Creighton, T. E. (ed.). The encyclopedia of molecular biology, in press. R. M. Macnab, Flagella-prokaryotes. John Wiley & Sons, Inc., New York, N.Y.
- 10.Danese P N, Silhavy T J. The sigmaE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 11.Djordjevic, S. P., P. N. Goudreau, Q. Xu, A. M. Stock, and A. H. West. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 12.Djordjevic S P, Stock A M. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure. 1997;5:545–558. doi: 10.1016/s0969-2126(97)00210-4. [DOI] [PubMed] [Google Scholar]
- 13.Gardina P J, Bormans A F, Hawkins M A, Meeker J W, Manson M D. Maltose-binding protein interacts simultaneously and asymmetrically with both subunits of the Tar chemoreceptor. Mol Microbiol. 1997;23:1181–1191. doi: 10.1046/j.1365-2958.1997.3001661.x. [DOI] [PubMed] [Google Scholar]
- 14.Ge Y, Old I G, Saint Girons I, Charon N W. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus ς70 promoter. J Bacteriol. 1997;179:2289–2299. doi: 10.1128/jb.179.7.2289-2299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haldimann A, Fisher S L, Daniels L L, Walsh C T, Wanner B L. Transcriptional regulation of the Enterococcus faecium BM4147 vancomycin resistance gene cluster by the VanS-VanR two-component regulatory system in Escherichia coli. J Bacteriol. 1997;179:5903–5913. doi: 10.1128/jb.179.18.5903-5913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haldimann A, Prahalad M K, Fisher S L, Kim S-K, Walsh C T, Wanner B L. Altered recognition mutants of the response regulator PhoB: a new genetic strategy for studying protein-protein interactions. Proc Natl Acad Sci USA. 1996;93:14361–14366. doi: 10.1073/pnas.93.25.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamblin, P. A., B. A. Maguire, R. N. Grishanin, and J. P. Armitage. Evidence for two chemosensory pathways in Rhodobacter sphaeroides. Mol. Microbiol., in press. [DOI] [PubMed]
- 18.Jiang P, Zucker P, Atkinson M R, Kamberov E S, Tirasophon W, Chandran P, Schefke B R, Ninfa A J. Structure/function analysis of the PII signal transduction protein of Escherichia coli: genetic separation of interactions with protein receptors. J Bacteriol. 1997;179:4342–4353. doi: 10.1128/jb.179.13.4342-4353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang P, Zucker P, Ninfa A J. Probing interactions of the homotrimeric PII signal transduction protein with its receptors by use of PII heterotrimers formed in vitro from wild-type and mutant subunits. J Bacteriol. 1997;179:4354–4360. doi: 10.1128/jb.179.13.4354-4360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan S, Castellano F, Spudich J L, McCray J A, Goody R S, Reid G P, Trentham D R. Excitatory signaling in bacteria probed by caged chemoeffectors. Biophys J. 1993;65:2368–2382. doi: 10.1016/S0006-3495(93)81317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirby J R, Kristich C J, Feinberg S L, Ordal G W. Methanol production during chemotaxis to amino acids in Bacillus subtilis. Mol Microbiol. 1997;24:869–878. doi: 10.1046/j.1365-2958.1997.3941759.x. [DOI] [PubMed] [Google Scholar]
- 22.Klose K E, Mekalanos J J. Differential regulation of multiple flagellins in Vibrio cholerae. J Bacteriol. 1998;180:303–316. doi: 10.1128/jb.180.2.303-316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levit M, Liu Y, Surette M, Stock J. Active site interference and asymmetric activation of the chemotaxis protein histidine kinase CheA. J Biol Chem. 1996;271:32057–32063. doi: 10.1074/jbc.271.50.32057. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd S A, Blair D F. Charged residues in the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol. 1997;266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 24a.Montrone M, Oesterhelt D, Marwan W. Phosphorylation-independent bacterial chemoresponses correlate with changes in the cytoplasmic level of fumarate. J Bacteriol. 1996;178:6882–6887. doi: 10.1128/jb.178.23.6882-6887.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24b.Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. [Google Scholar]
- 25.Nichols N N, Harwood C S. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J Bacteriol. 1997;179:5056–5061. doi: 10.1128/jb.179.16.5056-5061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci USA. 1997;94:8612–8617. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitta T P, Sherwood E E, Kobel A M, Berg H C. Calcium is required for swimming by the nonflagellated cyanobacterium Synechococcus strain WH8113. J Bacteriol. 1997;179:2524–2528. doi: 10.1128/jb.179.8.2524-2528.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prüß B M, Markovic D, Matsumura P. The Escherichia coli flagellar transcriptional activator flhD regulates cell division through induction of the acid response gene cadA. J Bacteriol. 1997;179:3818–3821. doi: 10.1128/jb.179.11.3818-3821.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prüß B M, Matsumura P. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J Bacteriol. 1996;178:668–674. doi: 10.1128/jb.178.3.668-674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rauprich O, Matsushita M, Weijer K, Siegert F, Esipov S, Shapiro J A. Periodic phenomena in Proteus mirabilis swarm colony development. J Bacteriol. 1996;178:6525–6538. doi: 10.1128/jb.178.22.6525-6538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rebbapragada A, Johnson M S, Harding G P, Zuccarelli A J, Fletcher H M, Zhulin I B, Taylor B L. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Schuster S C, Noegel A A, Oehme F, Gerisch G, Simon M I. The hybrid histidine kinase DokA is part of the osmotic response system of Dictyostelium. EMBO J. 1996;15:3880–3889. [PMC free article] [PubMed] [Google Scholar]
- 32.Segall J E, Block S M, Berg H C. Temporal comparisons in bacterial chemotaxis. Proc Natl Acad Sci USA. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sourjik V, Schmitt R. Different roles of CheY1 and CheY2 in the chemotaxis of Rhizobium. Mol Microbiol. 1996;22:427–436. doi: 10.1046/j.1365-2958.1996.1291489.x. [DOI] [PubMed] [Google Scholar]
- 34.Spudich E N, Zhang W, Alam M, Spudich J L. Constitutive signaling of the phototaxis receptor sensory rhodopsin II from disruption of its protonated Schiff base-Asp73 salt bridge. Proc Natl Acad Sci USA. 1997;94:4960–4965. doi: 10.1073/pnas.94.10.4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Balazs Y S, Thompson L K. Solid-state RECOR NMR distance measurements at the ligand site of a bacterial chemotaxis membrane receptor. Biochemistry. 1997;36:1699–1703. doi: 10.1021/bi962578k. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Brooun A, MacCandless J, Banda P, Alam M. Signal transduction in the archaeon Halobacterium salinarium is mediated by a family of thirteen soluble and membrane bound transducers. Proc Natl Acad Sci USA. 1996;93:4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]