Highlights
-
•
Pseudomonas aeruginosa has a relatively large genome and possesses a great genetic versatility which enables it to grow in several different environments, to produce a variety of virulence factors and display antibiotic resistance to the majority of antibiotics.
-
•
P. aeruginosa causes community-acquired as well as hospital borne infections.
-
•
Despite the discovery of several anti-pseudomonas antibiotics, P. aeruginosa causes high morbidity and mortality.
-
•
Antibiotics can be substituted with novel alternatives or used in conjunction with them to treat P. aeruginosa infections.
Keywords: Pseudomonas, Infections, Antibiotics, Resistance, Novel Approaches, Bacteriophages, Immunotherapy, Vaccines
Abstract
Pseudomonas aeruginosa is an aerobic Gram-negative rod-shaped bacterium with a comparatively large genome and an impressive genetic capability allowing it to grow in a variety of environments and tolerate a wide range of physical conditions. This biological flexibility enables the P. aeruginosa to cause a broad range of infections in patients with serious underlying medical conditions, and to be a principal cause of health care associated infection worldwide. The clinical manifestations of P. aeruginosa include mostly health care associated infections and community-acquired infections. P. aeruginosa possesses an array of virulence factors that counteract host defence mechanisms. It can directly damage host tissue while utilizing high levels of intrinsic and acquired antimicrobial resistance mechanisms to counter most classes of antibiotics. P. aeruginosa co-regulates multiple resistance mechanisms by perpetually moving targets poses a significant therapeutic challenge. Thus, there is an urgent need for novel approaches in the development of anti-Pseudomonas agents. Here we review the principal infections caused by P. aeruginosa and we discuss novel therapeutic options to tackle antibiotic resistance and treatment of P. aeruginosa infections that may be further developed for clinical practice.
Graphical abstract
1. Introduction
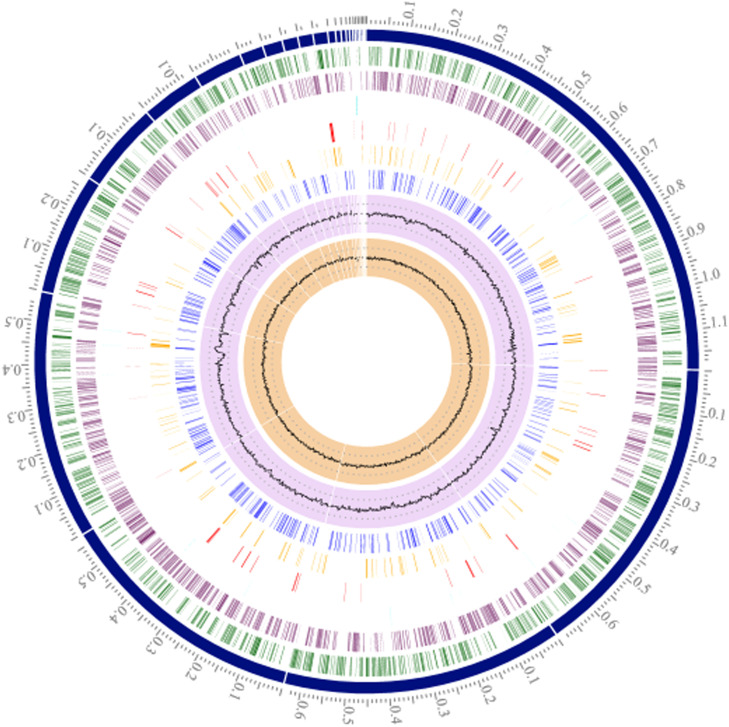
Pseudomonas aeruginosa is a motile, nonfermenting, Gram-negative, rod-shaped [1] and blue-green pigmented bacterium belonging to the family Pseudomonadaceae [2,3]. The bacterium has a relatively large genome (5.5–7 Mbp) (Fig. 1) compared to other bacteria [4], [5], [6] and possesses a great genetic versatility [5,7] which enables it to grow in several different environments, to produce a variety of virulence factors and display antibiotic resistance to the majority of currently available antibiotics [8], [9], [10], [11], [12], [13].
Fig. 1.
Circular view of P. aeruginosa isolate EA31, showing from outer to inner rings: Position on contig in Mb, contigs ordered largest to smallest, forward CDS (green), reverse CDS (purple), noncoding genes (light blue), antimicrobial resistance genes (red), virulence factors (orange), transporters (dark blue), GC content, GC skew. Visualization, annotation and assembly by PATRIC 3.6.8.
The bacterium is commonly isolated from natural resources like soil and surfaces in aqueous environments [14], [15], [16], [17]. P. aeruginosa is also found on the skin of healthy people and has also been isolated from the throat (5%) and stools (3%) of nonhospitalized patients [17]. P. aeruginosa predominantly causes nosocomial infections such as pneumonia [18], infections of the urinary tract (UTIs) [19], wounds [20,21], bones and joints [22], [23], [24] and the bloodstream [[25], [26], [27]]. The bacterium also thrives when the epithelial barrier is damaged [28,29] neutrophil production is depleted [30], mucociliary clearance is altered [29] and in the presence of medical devices [31,32]. P. aeruginosa causes community-acquired infections such as gastrointestinal [33,34], skin and soft tissue infections [22,23] and otitis externa [25,35] and is known to be associated with lower respiratory tract infections in patients with cystic fibrosis [36]. Community-acquired pneumonia is rarely caused by P. aeruginosa; however, Wang et al. [37] reported one case of a healthy individual suffering from pneumonia, thought to be community-acquired, and caused by P. aeruginosa, which lead to septic shock and multiple organ dysfunction syndrome.
Despite the discovery of several anti-Pseudomonas antibiotics [38,39], P. aeruginosa causes high morbidity and mortality [40,41] and remains difficult to treat because of its extensive and emerging antibiotic-resistance mechanisms [41,42]. P. aeruginosa is the second most common cause of ventilator-associated pneumonia in the United States [43], and ranks third among urinary tract infections associated with catheters [44]. It is the fourth leading cause of hospital-associated infections (HAIs) and accounts for 20% of all HAIs in Europe and United States [45] and is responsible for 75% of all deaths of severely burnt patients [46]. Furthermore, P. aeruginosa was found to be the fourth most common cause of mortality in patients with lower respiratory tract infection in India [47].
Alarmingly, the emergence of multidrug-resistant strains of P. aeruginosa is increasing and there are very few treatment options [48], [49], [50]. The World Health Organization (WHO) included carbapenem-resistant P. aeruginosa in the "Critical" category in a list of pathogens published in 2017 which require new antibiotics as a priority. On September 17, 2020, the Infectious Diseases Society of America (IDSA) published the first guidance document on infections caused by P. aeruginosa and categorized as difficult-to-treat resistance (DTR) [51].The resistance to antibiotics among P. aeruginosa strains is a result of the de novo [52,53] emergence of resistance after exposure to antibiotics, patient to patient transfer [[13], [54]] of resistant bacteria and cross-resistance, which can result in multiple-drug-resistant (MDR) P. aeruginosa strains [52,[55], [56], [57]]. A scoping report on antimicrobial resistance in India reported a high prevalence of carbapenem resistance among P. aeruginosa (40%–47%); more than 50% of isolates were also reported resistant to broad spectrum antibiotics such as fluoroquinolones and third-generation cephalosporins [47]. Prakash et al. [9] reported 31.78% MDR P. aeruginosa in studies conducted in hospitals from India. Another study conducted to determine antibiotic resistance in P. aeruginosa isolated from tertiary care hospitals in India reported 47.7% were drug resistant, 50% MDR and 2.3% were extensively drug resistant (XDR) strains with a high level (80%) of carbapenems resistance [58]. Intra et al. [59] identified P. aeruginosa (6.17%) in COVID-19 patients. Xu et al. [60] isolated P. aeruginosa from sputum samples. The isolates were found persistent in respiratory tract of the patient and were resistant to antibiotics.
High clonal diversity is often seen when studying the molecular epidemiology of P. aeruginosa isolates from hospital-acquired infections, CF patients, or the environment. The majority of isolates are connected to distinct genotypes, and a closer check reveals that this is only correct for isolates that are sensitive to antibiotics; isolates that exhibit MDR/XDR characteristics are not included. In fact, there have been numerous MDR/XDR strain epidemic breakout reports and alerts in the hospital setting for decades. The data from these studies and reports has added to the evidence of MDR/XDR global clones, often known as ``high-risk'' clones, spreading in numerous hospitals globally [61].
Colistin-only-sensitive (COS) profiles are frequent in many hospitals across the world, and pan-drug resistance has already been identified [61]. When P. aeruginosa is not susceptible to at least one antibiotic from at least three different antibiotic classes—penicillins, cephalosporins, fluoroquinolones, aminoglycosides, and carbapenems—it is said to as multi-drug resistant. The idea of “difficult-to-treat” resistance was put forth in 2018. DTR is described in this guidance as P. aeruginosa that does not display sensitivity to any of the following drugs: piperacillin-tazobactam, ceftazidime, cefepime, aztreonam, meropenem, imipenem-cilastatin, ciprofloxacin, and levofloxacin.
Multidrug-resistant P. aeruginosa, also known as DTR-P. aeruginosa, typically arises from the interaction of several complex resistance mechanisms, such as reduced expression of outer membrane porins (OprD), hyperexpression of AmpC enzymes, increased activity of efflux pumps, and mutations in penicillin-binding protein targets [51]. P. aeruginosa has been found as a common co-infecting pathogen (23.8%) in COVID-19 patients leading to increase in the severity of the baseline illness [62]. Shafran et al. [63] also reported P. aeruginosa (25%) as a common coinfecting pathogen in COVID-19 patients. Russell et al. [64] identified P. aeruginosa frequently in sputum of COVID-19 patients. A special report on antibiotic resistance in the United States indicate that the rate of hospital-onset MDR P. aeruginosa cases increased by 32% in 2020 as a result of COVID-19 [65].
2. Infectious caused by P. aeruginosa
2.1. Principal infectious caused by P. aeruginosa
Hidron et al. [66] ranked P. aeruginosa on 6th position in the report published in 2006–2007 by the National Healthcare Safety Network (NHSN) at the Centers for Disease Control and Prevention (CDC) as the most common hospital-associated pathogen causing infections. Medical interventions such as mechanical ventilation [67,68], surgery [69], antibiotic therapy [70,71] and chemotherapy [72,73] are the major predisposing factors that may further cause serious P. aeruginosa infections in a hospital environment. The CDC published antimicrobial resistance threats in the USA in 2019 and categorized P. aeruginosa under “Serious threat level”. The CDC also reported 32,600 hospitalized cases and 2,700 deaths leading to $767M attributable healthcare costs in the USA due to P. aeruginosa infections in 2017. Moreover, in a study conducted in a tertiary care hospital in North India in the years from 2012 to 2016, P. aeruginosa was identified as the pathogen in 95% of all infection cases [74]. Infections of P. aeruginosa, causes and complications are summarized in Table 1.
Table 1.
Pseudomonas aeruginosa infections, causes and complications.
| Infections | Causes | Complications |
|---|---|---|
| Bacterial keratitis | Ocular disease, postocular surgery, contamination of the lens or extended lens use [75] | Loss of vision, blindness [76] |
| Ear infection: otitis externa (Swimmers ear) | Contamination of water, prolonged exposure to moisture, insertion of foreign objects [75], ear plugs, ear phones [77]. | Hearing loss [78] |
| Ear infection: chronic suppurative otitis media (CSOM) | Acute otitis media | Lateral sinus thrombosis, meningitis,cerebral abscess, otic hydrocephalus, extradural abscess, encephalitis [79] |
| Skin and soft tissue infections (SSTI) | Contaminated hot tubs or spa pools [80], whirlpools and swimming pools [81,82], leg waxing [81] | Ecthyma gangrenosum, subcutaneous nodules or progressive folliculitis with cellulitis [81] |
| SSTI – Gangrenous cellulitis and necrotizing fasciitis | Trauma or surgery, diabetes, vascular insufficiency [83] | Fulminant skin necrosis [82] |
| Green nail syndrome | Onycholysis, onychotillomania, microtrauma to the nail-fold, chronic paronychia, and associated nail disorders, such as psoriasis [84], diabetes mellitus, immunosuppression [85], frequent exposure to water or moist conditions [86] | Green-black discoloration of the nail bed [86] |
| Burn wound infection | Endogenous flora of the gastrointestinal and or upper respiratory tract [87,88] | Septicaemia [46] |
| Blood stream infections | Nosocomial [89] | Subcutaneous nodules, ecthyma gangrenosum and gangrenous cellulitis [82] |
| Urinary tract infections | Catheterization or surgery [75] | Sepsis in older patients, immunocompromised patients and individuals with diabetes [19] |
| Respiratory tract infections | Cystic fibrosis [90], mechanical ventilation [91], bronchiectasis [92] | Mortality [65,93] |
| Bone and joint infections | Osteomyelitis [94], haematogenous spread or exogenous and endogenous contiguous focus of infection [95] | Persistent infection [96] |
2.2. Burn wound infections
P. aeruginosa causes serious burn infections and sepsis was found as a major cause of deaths in patients [97]. Mortality as high as 75% was reported due to septicaemia caused by Pseudomonas spp. and other bacteria [98]. A study conducted in military hospitals in southern Jordan from June 1990 to May 1998 reported P. aeruginosa as a common cause (42.6%) of invasive burn wound infection [70]. Another study conducted in Al-Kendi hospital from October 2007 to June 2008 reported P. aeruginosa as a common cause (48.9%) of invasive burn wound infection [99]. The estimates by World Health Organization (WHO) reports 265,000 deaths annually due to burn injury, with nearly half of these occurring in the WHO South-East Asia Region [100,101]. Around 7 million people in India suffer from burn injuries every year, resulting in 140,000 deaths [102]. The major factors associated with burn wound infection are thermal destruction of the skin and concomitant depression of the local and systemic host cellular immune response [87,88]. Although burn wound surfaces are sterile immediately following thermal injury, within the first 48 hours microorganisms colonize the wound surface, which is rich in proteins and avascular necrotic tissue [87]. Wounds first become colonized by Gram-positive organisms such as Staphylococcus aureus and Streptococcus pyogenes before infection by P. aeruginosa [98,103,104]. Eventually, other bacteria and yeasts along with P. aeruginosa colonize wounds from patients’ endogenous flora of the gastrointestinal and or upper respiratory tract [87,88]. Recent studies with the burn patients demonstrated that thermal injury results in impaired production of host defence peptides (β-defensins) in tissues surrounding the burn wound, and these peptides play a primary role in defence against P. aeruginosa [105], [106], [107]. The impairment of host immunity and loss of skin integrity allows opportunistic pathogens to enter the body and cause infection. For example, an experiment conducted in animals with partial cutaneous burns resulted in the development of fully grown P. aeruginosa biofilms in 48–72 hours [108]. The biofilm further reduces the effect of treatment of the wound as microorganisms in a biofilm are more resistant to antibiotic treatment [109]. Burn wound infection by P. aeruginosa manifests as a green pigment in subcutaneous fat, which is erythematous and later turns into a black, necrotic, nodular lesion [70]. A retrospective study conducted between April 2007 and January 2010 in the burn unit at Red Cross War Memorial Children's Hospital in Cape Town, South Africa, P. aeruginosa was isolated from 14.5% patients. These isolates were resistant to povidone-iodine (92.5%), piperacillin-tazobactam (36.1%) and tobramycin (3.3%) [110]. A study conducted in India found P. aeruginosa in 54.9% of burn patients with high-level (76.8%) of multidrug resistance [111].
Gonzalez et al. [21] conducted a study to monitor the effect of burn wound exudate on growth and expression of virulence factors of P. aeruginosa. The results of this study indicated that P. aeruginosa can grow in burn wound exudate with a lowered doubling rate compared to its normal control, whereas other common burn wound pathogens failed to grow. An increase in expression of all virulence factors such as quorum sensing, pyocyanin, pyoverdine, elastase, rhamnolipid was also observed when P. aeruginosa was grown in burn wound exudate. This study explained a reason for P. aeruginosa being a predominant pathogen in burn victims.
An intracellular signaling molecules such as 4-hydroxy-2alkylquinolines (HAQs) are involved in iron chelation governs and dictate the infection course, Que et al. [112] for the first time showed that clinically important specimens of P. aeruginosa isolated from active burn wound infection from human patients produce and excrete detectable levels of HAQs.
2.3. Bacterial keratitis
P. aeruginosa causes keratitis in patients with ocular disease, postocular surgery and in individuals who use contact lenses. Most of the contact lens-associated P. aeruginosa infections are due to contamination of the lens or extended lens use—resulting in disruption of the epithelial surface of the cornea that further leads to corneal abrasions [75]. When epithelial barrier function is impaired by using contact lenses for long periods, P. aeruginosa causes an opportunistic infection. P. aeruginosa becomes rapidly internalized by binding to toll-like receptors (TLR5) on the surface of the cornea [113]. Keratitis due to Pseudomonas is characterized by sudden onset, rapid progression of ocular pain, redness, tearing, photophobia, and blurred vision. Clinically, this infection causes corneal epithelial defect and a stromal infiltration that further leads to stromal necrosis and progressive thinning [114]. P. aeruginosa causes potentially blinding condition as a complication of keratitis [115]. P. aeruginosa is responsible for causing bacterial keratitis in 6% to 39% of the cases in the USA [116,117] and 8% to 21% in south India [116]. P. aeruginosa infections are also reported after exposure to ultraviolet rays (260–280 nm) and frequently occur in people who are exposed to sun lamps and individuals who don't use proper shields during welding [118]. Bacterial keratitis may cause loss of vision and, in severe cases, even blindness [76].
2.4. Ear infections
Inflammation or infection of the external auditory canal, referred to as otitis externa or “swimmers ear”, is caused by P. aeruginosa. Otitis externa is associated with contamination of water by P. aeruginosa, prolonged exposure to moisture, and insertion of foreign objects [75]. P. aeruginosa is one of the major organism to cause chronic suppurative otitis media (CSOM), also referred to as chronic active mucosal otitis media, chronic otomastoiditis and chronic tympanomastoiditis. The bacterium causes chronic inflammation of the middle ear and mastoid cavity that further results in recurrent ear discharge [119] or otorrhoea hearing loss through perforation of the tympanic membrane [119,120]. CSOM may cause common ear discharge, hearing loss [121] or, rarely, complications such as fever, otalgia, vertigo, meningitis, facial nerve palsy, and brain abscess. Mittal et al. [122] using human and animal cell based assays demonstrated that otopathogenic P. aeruginosa are able to enter and survive inside macrophages. Molecular mechanism elucidated uptake of bacteria by human and animal cells is dependent on actin polymerization whereas OprF expression plays important role in intracellular survival of P. aeruginosa. Tertiary care centre in Uttarakhand, India, conducted a study to know the prevalence of P. aeruginosa among patients suffering from CSOM. The results of this study indicated P. aeruginosa as a major cause of CSOM (32.1%) with substantial number of MDR strains [119]. More than 700 million cases of CSOM are reported globally per annum, with children below age 5 found to be more vulnerable to P. aeruginosa [120].
2.5. Skin and soft tissue infections (SSTI)
Folliculitis starts with sudden onset of numerous, large, monomorphic, painful papules, and pustules that develop approximately 24 hours after prolonged immersion in contaminated hot tubs or spa pools [80], whirlpools and swimming pools [81,82] or after leg waxing [81]. The lesions often congregate on body parts in contact with contaminated water [82] and usually appear 8–48 hours after exposure [81]. In immunosuppressed patients, folliculitis can further progress to ecthyma gangrenosum. In AIDS patients, P. aeruginosa infection may cause subcutaneous nodules or progressive folliculitis with cellulitis [81]. Another complication found in children is a “hot-foot” syndrome. It is characterized by painful plantar nodules [82].
2.6. SSTI – Gangrenous cellulitis and necrotizing fasciitis
P. aeruginosa infection of the skin and fascial layers is a rare but serious medical condition. It is characterized by rapid and progressive destruction and inflammation that further results in fulminant skin necrosis and death. The spread of necrotizing fasciitis is directly proportional to the thickness of the subcutaneous layer as it moves along the fascial plane. Another complicated infection that P. aeruginosa causes in immunocompromised elderly individuals is necrotising fasciitis: a rare but serious infection of subcutaneous tissue and fascia. A specific variation of necrotising fasciitis is referred to as Fournier's gangrene; P. aeruginosa infection in these patients results in scrotal discomfort and malaise that further lead to perineal pain, swelling, blisters, and necrosis [82].
2.7. Green nail syndrome/chromonychia/Fox-Goldman syndrome
Persistence of pyocyanin in the nail plate and involvement of P. aeruginosa was first described in 1944 by Goldman and Fox and hence named after them [123]. Patients with underlying conditions such as onycholysis, onychotillomania, microtrauma to the nail-fold, chronic paronychia, and associated nail disorders, such as psoriasis [84], diabetes mellitus, immunosuppression [85] and people who are frequently exposed to water or moist conditions suffer from green nail syndrome, as the causative agent, P. aeruginosa, thrives in moist conditions. The condition is characterized by onycholysis and green-black discoloration of the nail bed, it is most often associated with chronic paronychia [86]. Chernosky and Dukes (1963) demonstrated the presence of P. aeruginosa within the nail plate and the green discoloration of the nails is due to pyocyanin produced by P. aeruginosa [82]. Green nail syndrome is commonly restricted to one or two nails with partial or complete involvement of the nail plate [123,85]. The infection is characterized by painless nail plate with erythematous or tender skin around the nail. An infected individual can autologously disseminate the bacterium by scratching or rubbing his or her skin, especially when cutaneous surface is damaged [123]. A retrospective study conducted to investigate fungal coinfection with P. aeruginosa during the period of 2015–2018 reported green nail syndrome commonly affected great toe nail (69.9%) and high prevalence of fungi [124].
2.8. Bacteraemia
Blood stream infections (BSI) caused by P. aeruginosa are often fatal. The bacterium is responsible for 3%–7% of blood stream infection cases with high morbidity and mortality rates (27%–48%) in critically ill patients [125,126]. Wisplinghoff et al. [89] published a nationwide surveillance study on nosocomial BSI in the USA and reported P. aeruginosa as the third most common Gram-negative bacteria causing nosocomial BSI and accounted for 4.3% of all cases. In the ICUs, P. aeruginosa accounted for 4.7% of all cases and was reported as the fifth most common isolate implicated in BSI, and the seventh most common isolate in non-ICU wards, accounting for 3.8% of cases. The great majority of reported crude mortality percentages from large surveillance studies range from 39% to 48%. Systemic P. aeruginosa infections cause subcutaneous nodules, ecthyma gangrenosum and gangrenous cellulitis. Patients with burns and AIDs are more vulnerable to systemic infection by P. aeruginosa [82].
BSI due to P. aeruginosa is an important cause of morbidity and mortality in neutropenic cancer patients. Gudiol et al. [95] retrospectively studied P. aeruginosa BSI cases from January 2006 to May 2018. P. aeruginosa strains isolated from adult neutropenic oncohematological patients, including hematopoietic stem cell transplant recipients, were caused by multi drug resistant (25.4%) and extensively drug resistant bacteria (19.3%). The highest rate of multi drug resistance was observed in Colombia and Argentina. Hickey et al. [126] using Galleria mellonella model demonstrated that the virulence factors such as LecA, RpoN, and proteins involved in cellular metabolism and replication isolated from BSI P. aeruginosa are increased relative to its peripheral counterparts.
2.9. Urinary tract infections (UTIs)
P. aeruginosa is an important uropathogen [19], associated with 7%–12% of all nosocomial urinary tract infections [44,127]. The bacterium usually causes UTIs following catheterization or surgery [75]. UTI is the second most common type of infection in the body [127]. P. aeruginosa was found to be the third most common Gram-negative pathogen causing 7.1% of all nosocomial urinary tract infections in surveillance studies from the Asia-Pacific region in 2009 to 2010 [128]. Bitsori et al. [129] studied P. aeruginosa UTI cases in children with E.coli UTI cases. Results indicated more complications and antibiotic resistance pattern. A retrospective study conducted during September 2012–September 2014 to study mortality rates in hospitalized patients with P. aeruginosa UTIs, reported 17.7% mortalities at 30 days and 33.9% mortalities at 90 days [130]. Shobha et al. [131] collected 107 urine samples from microbiology laboratory during 2015–2016 reported 84.11% P. aeruginosa UTIs. Results of this study indicated majority of UTIs with Pseudomonas species were found in males and observed more in age group more than 60. Studies on nosocomial urinary tract infections occurring in the intensive care unit (ICU) report even higher rates of P. aeruginosa. Studies by national French nosocomial surveillance reported 16% of UTIs were caused by P. aeruginosa and strains isolated in the study also showed higher rates of antimicrobial resistance [132]. A study conducted to demonstrate virulence and antibiotic resistance patterns in P. aeruginosa isolates from UTIs among children in southern Poland reported exoY as most prevalent virulence gene and sensitivity of isolates to beta-lactams, aminoglycosides and colistin; however, large proportion of isolates were resistant to carbapenems and fluoroquinolones [133]. Badamchi et al. [134] conducted a study to detect virulence genes and antimicrobial susceptible pattern in children with UTI in Tehran, reported lasB as most prevalent virulence gene and cefotaxime as a least effective antibiotic. P. aeruginosa is also known to invade epithelial and mast cells [135]. P. aeruginosa urinary infection may cause complications such as sepsis that can be fatal in older patients, immunocompromised patients and individuals with diabetes [19]. Disruption of the epithelial layer during application of a catheter promotes bacterial colonization [75] and the formation of biofilm [127]. Catheter-associated UTI accounts for 20% to 49% of all nosocomial infections; the pathogen enters via extraluminal or by intraluminal route of the catheter [127]. Penaranda et al. [135] demonstrated that P. aeruginosa can survive intracellularly in bladder epithelial cells, sets a stable infection and become tolerant to antibiotics in vivo and in vitro model. Estaji et al. [136] isolated 70 P. aeruginosa strain from patients with UTI hospitalized in different wards in Iran. Findings of this study showed high genetic diversity among the strains isolated from different patients; 35% of these cases were catheter associated UTIs. Isolated strains were also demonstrated resistance to beta-lactam antibiotics and identified with SHV and TEM genes. P. aeruginosa causes 1%–4% UTIs after flexible cystoscopy [137,138]. Sorbets et al. [138] reported an outbreak of UTIs which reached to 10.18% after urinary bladder exploration with contaminated cystoscope.
3. Respiratory tract infections
3.1. Cystic fibrosis
Cystic fibrosis is a common autosomal recessive genetic disease among Caucasians with one in 2,500–4,000 people affected [90,[139], [140], [141], [142]]. The disease is characterized by a mutation in the gene for cystic fibrosis transmembrane conductance regulator (CFTR) protein [90,143]. Dysfunction of the CFTR channel causes hyper sodium absorption [90] and impaired mucociliary clearance [90,140]. In addition to obstructing airways, mucous creates a hypoxic environment that favors the colonization of P. aeruginosa. Other factors that contribute to P. aeruginosa infection are impaired function of antimicrobial peptides, increased availability of bacterial receptors, defective internalization of bacteria by epithelial cells and low levels of defensive agents such as nitric oxide and glutathione [90]. The lung environment in cystic fibrosis patients is different from that of a healthy individual's [141], and factors which affect bacterial colonization are osmotic stress due to the viscous mucus, oxidative and nitrosative stresses due to host responses, sub inhibitory concentrations of antibiotics and the presence of other microorganisms [143]. To overcome these challenges, P. aeruginosa undergoes an evolutionary change to adapt to the cystic fibrosis environment: it adapts to the cystic fibrosis lungs by overproducing the polysaccharide, alginate, and undergoing auxotrophic mutations, loss of motility and elevated mutation rate due to a faulty DNA repair mechanism [143]. Once P. aeruginosa colonizes the lung of a patient with cystic fibrosis, it becomes difficult to eradicate and can be fatal [141,144,145]. During infection, macrophages assemble inflammasome, a cell to cell signaling platform that promotes inflammation. P. aeruginosa from cystic fibrosis patients are unable to induce activation of inflammasomes. This unique mechanism was observed among all the patients and times of infection [146]. LaFayette et al. [147] demonstrated a mechanism by which cystic fibrosis adapted P. aeruginosa lasR mutants induce neutrophil dominant hyper inflammatory response and thereby amplify the inflammation and accelerate disease progression.
3.2. Pneumonia
Pneumonia is caused by P. aeruginosa and is divided into four categories: (1) Hospital-acquired pneumonia, which occurs 48 hours or more after hospitalization; (2) Ventilator-associated pneumonia that develops more than 48 to 72 hours after endotracheal intubation; (3) Health care-associated pneumonia occurs among nonhospitalized patients, who live in a nursing home or long-term care facility, those received intravenous antimicrobial therapy or chemotherapy or wound care, and those who attended a hospital or dialysis clinic in the previous 30 days of the current infection [148], [149], [150] and (4) Community acquired pneumonia. P. aeruginosa is a rare cause of community-acquired pneumonia, but it does occur more frequently in some areas than others. P. aeruginosa is nearly always isolated from elderly patients with concomitant diseases, most notably COPD, rather than from community-dwelling patients. An increase in P. aeruginosa infections in nursing home residents serves as a good illustration of this. There are case reports of CAP in healthy people, but all of them were also smokers [151].
As per the 2004–2006 National REA-RAISIN surveillance data, which reported P. aeruginosa to be the most common cause of pneumonia (25%) among all Gram-negative and -positive pathogens [133]. Weber et al. [91], estimated that more than 90% of the ventilator-associated pneumonia occurred in patients housed in ICUs, whereas 67% of hospital-associated pneumonia occurred in patients not housed in ICUs. P. aeruginosa accounted for 17.5% of all cases in ventilator-associated pneumonia and 9.26% of all cases in hospital-associated pneumonia [91]. Reported percentages of P. aeruginosa implicated in community-acquired pneumonia vary from 0.3% to 11%, whereas it varies from 2.2% to 20% in healthcare associated pneumonia [150,151].
Morello et al. [152] demonstrated the role of LoxA expression using clinical isolates of P. aeruginosa. It was observed that several clinical isolates of P. aeruginosa express LoxA. Gene product when secreted in lungs, it processes a wide range of host polyunsaturated fatty acids that results in production of bioactive lipid mediators. It also inhibits secretion of major chemokines and recruitment of leukocytes. Overall, LoxA expression promotes persistence of bacteria in lung environment and plays important role in pathogenesis. Mutation in mucA gene results in overproduction of alginate polymer, which results in the mucoid phenotype and provides protection to P. aeruginosa. In a retrospective study conducted during 2012–2014, 75 patients with P. aeruginosa pneumonia treated at a tertiary referral hospital in South Korea were studied. The data from this study illustrated that mutation in mucA gene can be considered as an independent predictor of mortality [153].
3.3. Bronchiectasis
Chronic bronchial dilatation is referred to as bronchiectasis. As a result, there is inadequate mucus drainage and increased risk of bacterial infection. Patients with cystic fibrosis, other forms of bronchiectasis, and severe chronic obstructive pulmonary disease are particularly vulnerable to P. aeruginosa since it is an opportunistic bacterium. Despite extensive intravenous antibiotic therapy, P. aeruginosa is seldom eliminated once it develops into a chronic infection in bronchiectasis. Chronic infection is linked to more severe airflow restriction and more extensive lung damage [92]. Davies et al. [92] evaluated the rate of pulmonary function decline in individuals with and without P. aeruginosa infection. The findings imply that P. aeruginosa is a marker of disease severity but does not hasten a decline in pulmonary function. P. aeruginosa is one of the most prevalent bacteria that colonize bronchiectasis in people without cystic fibrosis. P. aeruginosa was estimated to have a persistent colonisation in about 25% of people with non-CF bronchiectasis [154]. In a cross-sectional cohort study with non-CF bronchiectasis in the year 2018, Kwok et al. [154] found that P. aeruginosa was a commonly isolated organism that accounted for about 27% of the entire cohort. In comparison to non-P. aeruginosa colonized patients, these patients had larger sputum volumes, higher FEV1 and FVC, more than three lobes were impacted in 66% of cases, 24% of patients needed hospitalisation, and 18% required long-term macrolide therapy.
3.4. Bone and joint infections
Usually bones and joints are sterile areas, but bacteria can reach such sites by haematogenous spread or exogenous and endogenous contiguous focus of infection [155]. In a study of 454 patients with osteomyelitis, P. aeruginosa was implicated in 4.4% of all cases [94]. Tummala et al. [155] reported P. aeruginosa as the most common pathogen causing osteomyelitis among Gram-negative organisms. Recurrence rate of osteomyelitis caused by P. aeruginosa was reported more than two-fold compared to infection caused by Staphylococcus aureus [94]. P. aeruginosa has been implicated in 10% of all cases of sternoclavicular septic arthritis, for which common risk factors include intravenous drug use, diabetes mellitus, trauma and infected central venous lines [156]. Cerioli et al. [96] retrospectively studied 1,638 implant associated bone joint infection patients over 7 year (2011–2017) period. Ninety patients (5.5%) among them were found infected with P. aeruginosa. During prolonged follow-up, 23 patients experienced treatment failure whereas 7 patients experienced persistent infection and required prolonged antibiotic treatment as long as 3 months. Septic arthritis of wrist joint due to P. aeruginosa is a rare condition in children and characterized by acute onset of fever, swelling and pain [157].
4. Novel approaches to treat Pseudomonas infections
The antimicrobial resistance in P. aeruginosa is increasing steadily; as a result, the treatment of infections caused by P. aeruginosa is extremely challenging. Despite recent advances in the field of anti-bacterials, the number of new antibiotics under clinical development remain limited and their introduction into the market is extremely slow. Thus, there is an urgent medical need for innovative options to be developed for the treatment and management of infections caused by P. aeruginosa. Novel therapeutic options for treating P. aeruginosa infections are summarized in Table 2.
Table 2.
Novel therapies for treating Pseudomonas aeruginosa infections.
| Novel therapeutic agents | Mode of action/target | Reference |
|---|---|---|
| Quorum sensing inhibitors | Signal molecule degradation, preventing accumulation of signal molecules and antagonism of the signals | Chamomile, carrot [158], garlic [159], salicylic acid [160], furanones [161] |
| Immunotherapy | PcrV protein, flagellin, LPS | KB001-A [162], Immunoglobulin Y [163], Panobacumab [164] |
| Iron chelators | Chelation of the environmental iron | Gallium [165] |
| Vaccines | Polysaccharide, PcrV,OprI,Hcp1, FlgE, fructose bisphosphate aldolase, OprH gene product | PcrV-OprI-Hcp1-Trivalent vaccine [166], Polyvalent vaccine [167] |
| Herbal medicine | Exopolysaccharide production, phenazine pyocyanin, rhamnolipids, elastase and alkaline protease | Tanreqing [168], Herba patriniae[169] |
| Phage therapy | Lysis of cell membrane and wall | Pf3R [170], Genetically engineered synthetic phages [171] |
4.1. Quorum sensing inhibition
Bacterial communication can be inhibited using quorum-sensing inhibitors. Production of most of the virulence factors is regulated by quorum sensing and it is a prime therapeutic target [158]. This approach relies on the reduction of virulence rather than killing the pathogen; it may result in a reduction in the evolution of antibiotic resistance and enhance the treatment of multidrug-resistant pathogens [172]. The P. aeruginosa quorum-sensing system is comprised of las and rhl, the gene products of which help bacteria produce virulence factors and form biofilms [160,[173], [174], [175], [176]]. Quorum sensing can be inhibited by different mechanisms such as signal generation, signal molecule degradation, preventing accumulation of signal molecules and antagonism of the signals’ mode of action [158].
Potential candidates having quorum-inhibiting or -quenching activity have been found in chamomile, carrot [158,177], garlic [158,159,177], salicylic acid [158,160,178] and algal furanone [158,161,179]. The effect of garlic formulation on quorum-sensing inhibition was studied in a small pilot, randomized, controlled clinical trial in adults and children with cystic fibrosis and chronic P. aeruginosa infection. The garlic formulation did not cause significant improvement in clinical parameters and there was no reduction in the levels of quorum sensing molecules in plasma and sputum samples [180]. Brominated furanones studied in a reporter-gene assay have been found to be inhibitory against P. aeruginosa quorum-sensing system (las and rhl), resulting in inhibition of production of virulence factor elastase B and biofilm depletion [181]. Plant essential oils, such as clove oil, were found to inhibit swarming motility and quorum sensing [182], [183], [184]. Other synthetic chemicals, such as metabromo-thiolactone and metachloro-thiolactone, inhibited quorum sensing-regulated virulence factor pyocyanin [176]. Alginate oligomer (OligoG CF-5/20) was found to inhibit global regulatory QS signaling and swarming motility in P. aeruginosa [185]. DNAse I treatment in P. aeruginosa biofilms produced in vitro conditions also have been found to reduce biofilm matrix [186].
4.2. Immunotherapy
KB001-A is recombinant anti-Pseudomonas PEGylated monoclonal antibody. KB001-A antibodies inhibit the action of type III secretion system T3SS in P. aeruginosa. KB001-A specifically targets the PcrV protein component of the T3SS tip and blocks its activity [162,187,188]. In 16-week safety and efficacy trial, KB001-A was well-tolerated and found to be safe with no significant unfavorable effects. Overall, treatment caused reduction in sputum inflammatory factor (IL-8) [162].
Anti-Pseudomonas immunoglobulin Y antibodies are produced from chicken egg by immunizing them with P. aeruginosa. These antibodies specifically bind to P. aeruginosa flagellin and decrease their ability to attach to epithelial cells and cause lung infection. A Phase I study was conducted in patients with CF: patients were asked to gargle with IgY antibodies, and the study concluded that none of the patients became chronically colonized and no undesirable side effects were reported [163]. A recent Phase III clinical trial to investigate the anti-Pseudomonas activity of IgY is underway [186]. Another human monoclonal antibody (IgM) targeting the bacterial LPS [189], [190], [191], panobacumab, was reported safe with low recurrence of pneumonia in a phase IIa clinical trial in nosocomial pneumonia [164].
4.3. Iron chelators
P. aeruginosa needs iron for its growth, the formation of biofilm and its survival [192]. It sequesters iron from the environment by secreting the siderophores pyoverdin and pyochelin [193]. The human innate immune system recognizes and blocks biofilm development by secreting lactoferrin. Lactoferrin chelates the iron, causing P. aeruginosa to increase twitching motility rather than forming aggregates and biofilm [194]. Gallium has been found to have antimicrobial activity [165,192,193,195]. Biological systems mistakenly take up gallium as it has an ionic radius similar to Fe3+, but gallium lacks the redox activity of iron and hence it inhibits iron-dependent processes [193,195]. Gallium was also found effective in a mouse lung infection model, with approximately 1,000-fold decrease in the number of bacteria in the lungs [195]. Only one phase I pharmacokinetic study using gallium nitrate with two different doses (100 and 200 mg/m2/day) was conducted in patients with CF, with no adverse effects observed and the study showed promising results for the clinical application of gallium nitrate [195]. A combination of gallium with an antibiotic preparation has been found to enhance the activity of antibiotic: for example, gallium-gentamycin liposomal coencapsulation was found to be more effective than gentamycin alone in eradicating MDR P. aeruginosa growing in planktonic or biofilm community [196,197]. Similarly, the combination of deferasirox and tobramycin was found to significantly prevent the formation of biofilm on CF epithelial cells [197].
4.4. Vaccines
There are no licensed vaccines against P. aeruginosa at present [166,198]. An octavalent, polysaccharide, toxin-conjugate vaccine developed by the Swiss Serum and Vaccine Institute was studied in the European CF community. The study concluded the persistence of antibodies in vaccinated patients with a significantly lower rate of infection. Later, a double-blind, placebo-controlled study conducted to evaluate the safety and efficacy of flagella vaccine in patients with CF, showed a lower risk of P. aeruginosa infection [199]. Another approach used OprF-OprI outer membrane fusion protein as an antigen [198]: human volunteers were vaccinated with systemic, nasal or oral live vaccine followed by a systemic booster and resulted in enhancement of specific IgA antibodies at the pulmonary airway surface; immunization resulted in a rise in serum antibody titers. The study concluded nasal and oral vaccines could be promising candidates for the development of anti-P. aeruginosa immunization [200]. Further phase II studies resulted in significant immunogenic responses against P. aeruginosa [201]. The efficacy of the trivalent vaccine (PcrV-OprI-Hcp1) was studied in mice models and resulted in a significant reduction in acute skin infection and pneumonia due to P. aeruginosa [166]. Wan et al., [202] studied the effect of recombinant vaccine in P. aeruginosa infected mice. This promising vaccine candidate is a flagellar antigen FlgE (reFlgE) isolated from sera of patients recovered from P. aeruginosa infection found to induce a Th2 cell-mediated response. Anti-reFlgE antibodies produced in mice were found to reduce bacterial load and inflammation in mice.
Another innovative new vaccine candidate used Pf phage, a filamentous bacteriophage isolated from chronic diabetic wound has been shown to increase virulence of P. aeruginosa. The vaccine formulation containing peptide from Pf phage coat protein conjugated to the carrier protein CRM197 combined with novel adjuvants and delivery systems shown to induce humoral immunity as well as cell-mediated response against Pf phage peptide. The overall effect provided protection from establishment of P. aeruginosa in mice [203].
An immunoinformatics tool based study predicted an effectiveness of epitope based vaccine against an enzyme fructose bisphosphate aldolase (FBA) produced by P. aeruginosa. This prediction study analyzed possible epitopes for B and T cells. Results indicated 6 MHC-I and four MHC-II promising epitopes. Further in vitro and in vivo studies are required to prove the efficacy of epitope based vaccine [204].
Another novel approach by Cabral et al. [205] studied immunogenicity and protective efficacy of a live vaccine against P. aeruginosa. A vaccine consists of an auxotrophic strain which lacks the key enzyme involved D-glutamate biosynthesis, a key component of bacterial cell wall. The trace amount of glutamate present in in vivo condition doesn't allow bacteria to synthesize cell wall and thereby compromise the growth of the cells without affecting the immunogenic properties of the bacteria. When administered intranasally in mice, a vaccine induced systemic and mucosal antibody production, also stimulated effector memory, central memory, IL-17A-producing CD4+ T cells, and recruited neutrophils and mononuclear phagocytes into the airway mucosa. Intranasal administration also significantly improved survival rate in mice infection model.
Reverse vaccinology approach integrated with bioinformatics tool was used for selection of 52 potential P. aeruginosa antigens. These antigens were conserved in P. aeruginosa genomes from different origin. The combination of selected antigens effectively controlled P. aeruginosa infection in murine pneumonia and acute respiratory infection model [206].
Liu et al. [207] found OprH gene product as a potential vaccine candidate for prevention of lung infection caused by P. aeruginosa.
A novel polyvalent irradiated P. aeruginosa vaccine developed by Ahmed et al. [208] contains inhibited pathogen with functional antigenic expression. Administration of vaccine by intranasal, intramuscular and subcutaneous route followed by challenge test resulted in 95% protective efficacy in murine model.
4.5. Lectin inhibitors
These proteins recognize sugar residues on the cellular surface and permit bacterial cells to cross-link and form aggregates leading to further formation of biofilms [158,209,210]. LecA and LecB have fucose-specific [158,211,212] and galactose-specific binding sites and hence can be blocked by competitive inhibitors [158,212,213]. A randomized trial with a small group of CF patients who received fucose/galactose inhalation treatment resulted in significant reduction of P. aeruginosa colony forming units from sputum and tumor necrosis factor α levels [158].
Production of hypothiocynate is another innate immune defence but, in patients suffering from CF, epithelial cells do not produce thiocyanate [193]. Hypothiocynate is a bactericidal agent produced by oxidative lactoperoxidase-hydrogen peroxide thiocyanate system. This system was found to be defective in patients with CF. The combination of lactoferrin and hypothiocyanate [158,214] was found to be bactericidal and prevent biofilm formation by P. aeruginosa on airway epithelial cells. The combination named ALX-009 (Meveol) was granted orphan drug status in 2009. Studies with combination resulted in a significant decrease in total sputum bacterial density following a single dose. However the number of bacteria was found to increase after treatment was discontinued [215].
4.6. Alternative herbal medicine
Fu et al. [169] constructed a luxCDABE-based reporter system to monitor the expression of 6 key biofilm-associated genes in P. aeruginosa. A library of 36 Chinese herb extracts were screened for their inhibitory properties against the genes involved in biofilm formation and found that the extracts of Herba patriniae displaying significant inhibitory effect on almost all biofilm associated genes and it altered the structure of the mature biofilms. Further experiments also indicated decreased exopolysaccharide production by biofilm forming P. aeruginosa and promoted its swarming motility.
A study conducted to determine antimicrobial activity of five endemic plants (M. macrocarpa, D. loretense, T. impetiginosa, E. camaldulensis and U. tomentosa) commonly used in traditional medicine in the Amazon and sierra regions of Peru exhibited significant in vitro efficacy against P. aeruginosa [216].
A traditional Chinese medicine Tanreqing (TRQ) formula was found to completely inhibit the production of phenazine pyocyanin and moderately inhibit the production of virulence factors such as rhamnolipids, elastase and alkaline protease. A transcriptomic studies indicated that the treatment attenuates the expression of QS-regulated genes in P. aeruginosa [168].
4.7. Phage therapy
A promising approach to treat P. aeruginosa infection is using abundant and self-replicating viruses known as bacteriophages. Bacteriophages are viruses that infect bacteria. Also known simply as phages, bacteriophages are ubiquitous, obligate parasites, which require a bacterial host to replicate. Bacteriophages attach to the host bacterium and hijack the cell's replicative machinery, thus disrupting bacterial metabolism and causing the bacterial host to lyse [217], [218], [219]. Bacteriophages are very species-specific and they form a part of the normal flora of the human body. Bacteriophage therapy does not induce hypersensitivity reactions in patients and acts on targeted bacterial species without affecting the normal bacterial flora. Unlike antibiotics, it is very easy to search for new phages against bacteriophage-resistant bacteria, whereas developing a new antibiotic requires several years. Commercial manufacturing of antibiotics is a complex and costly process, whereas bacteriophages can be produced easily in a cost-effective manner. Bacteriophage treatment can be performed with a very small dose, as phages multiply and increase their number at the site of the infection [220], [221], [222]. Currently, phage therapy is practised in eastern European countries with centres in Warsaw, Poland, and Tbilisi, Georgia. Anti-Pseudomonas cocktails are sold in pharmacies in Georgia and Russia. A few case studies conducted in Belgium and the U.S. reported successful treatment of infections with MDR P. aeruginosa. Very few reports of bacteriophage treatment for P. aeruginosa infection are available in the years from 1990 to 2018. Phages alone, or in combination with antibiotics, were employed to treat infections such as chronic otitis [223], burn wound infection [224,225], UTI [226], sepsis [227], pneumonia, endocarditis, lung infection, bacteraemia [228], and graft infection [229], and resulted in an improvement in conditions of the patients with a concomitant reduction in the number of P. aeruginosa and no recurrent infection [230].
Another approach used nonreplicating phages for delivery of genes encoding proteins toxic to the bacterial pathogen. A genetically engineered filamentous phage (Pf3) of P. aeruginosa was modified by inserting restriction endonuclease gene in place of export protein gene. The variant nonreplicating, nonlytic phage Pf3R was studied in mice infection model and found to reduce endotoxin from target cell thereby increased the survival rate [170].
Pires et al. [171] designed and assembled first genetically engineered synthetic phage. The genome size of phage was reduced by knocking out up to 48% genes encoding hypothetical proteins. The resulting P. aeruginosa phage (vB_PaeP_PE3) was found as efficacious as its wild type. This experiment revealed a novel strategy to clear space from phage genomes in order to introduce genes of interest and potentiate the future treatment of P. aeruginosa infections.
Aghaee et al. [231] proposed combination of phages and antibiotics may increase treatment efficacy and prevent resistance development. An in vitro study involving single phage, mixture of two phages and combination of antibiotic and phages were tested against P. aeruginosa isolated from burn patient. A combination of 2 phages with antibiotic resulted in better efficacy than other formulations.
5. Novel approaches to inhibit Pseudomonas biofilms
A biofilm is a complex matrix of extracellular polymeric substances that includes glycopeptides, lipids and lipopolysaccharides which protect bacteria from extreme conditions. The matrix allows inflow of nutrients, water and signalling molecules [232,233]. Bacteria in a biofilm are found to be 1,000-times less susceptible to antimicrobial therapy than those that are not [109] hence new management strategies are needed for infections caused by P. aeruginosa that result in biofilm development. The exopolysaccharides, Psl, Pel and alginate are major components of biofilms formed by P. aeruginosa [234,235] and play significant roles in adhesion, determination of biofilm architecture, resistance to antibiotics and the host defence mechanism. Polysaccharide Psl helps bacteria in adherence to a new surface, cell migration, and communication with other cells of the biofilm during its early stage of formation. Polysaccharide Psl protects cells against phagocytosis and oxidative stress during infection. Alginate protects biofilm bacteria from opsonophagocytosis, free radicles formed by immune cells and antibiotics [235]. Table 3 summarizes novel approaches to inhibit P. aeruginosa biofilm.
Table 3.
Novel therapies for inhibiting Pseudomonas aeruginosa biofilm.
| Novel inhibitory agents | Mode of action/target | Reference |
|---|---|---|
| Antimicrobial peptides | Kill bacterial cells through both membranolytic and non-membranolytic mechanisms, and by interacting with intracellular targets, such as DNA, RNA, and proteins | LL-37 [236], Peptide 1037 [237], WLBU2 [238], P5 [239], Chensinin-1 [240] |
| Quorum sensing inhibitors | Signal molecule degradation, preventing accumulation of signal molecules and antagonism of the signals, Attenuation of virulence factors | Brominated furanones [171], meta-bromo-thiolactone [241], M64 [242], Itaconimides [243] 3-amino-7-chloro-2-nonylquinazolin-4 (3H)-one (ACNQ) [244], Silver nanoparticle with 4-nitropyridine N-oxide (4NPO) [245] quercetin [246] |
| Iron chelators | Chelation of the iron | desferrioxamine-gallium [186], N,N’-bis (2-hydroxybenzyl) ethylenediamine-N,N’-diacetic acid (HBED) [247] deferoxamine and deferasirox [192] |
| Enzymes | Exopolysaccharide | alginate lyases [248], glucanohydrolases (dextranase and mutanase) [249], glycoside hydrolase (PelAh and PslGH) [250], deoxyribonucleases (e.g., DNase I and Dnase1L2) [251,252] |
| Immunotherapy (Monoclonal antibodies) | Bacterial DNA-binding proteins | Monoclonal antibodies [253] |
| Gaseous agents | Dispersal of biofilm by modification of intracellular c-di-GMP levels | Nitric oxide [254] |
| Photodynamic therapy | Photoinactivation | Tetracationic porphyrin [5,10,15,20-tetrakis (1-methylpyridinium-4-yl)porphyrin tetra-iodide, Tetra-Py+-Me] [255], GD11 [256] |
| Photothermal therapy | Generates localized heat resulting in irreversible damage to bacterial cells | Gold nanoparticles [257] |
| Herbal medicine | expression of biofilm-associated genes (rhlR, rhlA and lasB) | H. patriniae extract [258], Eiekikaryu S, Iribakuga and Hyakujunro [259] |
| Phage therapy | Lysis of cell membrane and wall | IME180 [260], vB_PaeM_SCUT-S1 and vB_PaeM_SCUT-S2 [261], Phage LKA1 O-specific polysaccharide lyase [262], Engineered T7 bacteriophage that encode lactonase enzyme [263] |
6. Conclusions
Infections due to antibiotic-resistant P. aeruginosa are steadily increasing worldwide. Antibiotic resistance will continue to be a challenge with P. aeruginosa because of its high intrinsic resistance and ability to acquire resistance to all classes of antibiotics. The scientific community is making progress in the fields of bioinformatics, microbial genomics, target identification and screening techniques to find new potential therapeutic targets and molecular mechanisms for persistence and antibiotic resistance in P. aeruginosa; however, the number of new antibiotics being developed has fallen sharply.
To combat antibiotic resistance, and to minimize its dissemination, there is an urgent requirement for novel anti-Pseudomonas therapies. Many novel therapeutic agents are under development and are mainly focused on a narrow spectrum, pathogen-specific, anti-virulence, and patient-specific approach. Novel alternative therapies like bacteriophage therapy, iron-chelating agents and immunotherapy have shown promising results in vitro, animal models and in human studies; however, there are still many difficulties and challenges before they can be applied in the clinic. Controlled clinical trials are necessary to prove their safety and efficacy before they are used for routine care. The approach of using combination of antibiotics with alternative therapies will be necessary to overcome the growing problem of resistance.
Acknowledgments
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author Contributions
E.A.: conceptualization, design, supervision, review and editing; N.S.: conceptualization, design, data collection, analysis, writing; P.B.: supervision, review and editing; A.K.: supervision, review and editing; C.S.: supervision, review and editing; L.C.: review and editing.
Acknowledgements
The authors acknowledge the financial support of the Deakin India Research Initiative (DIRI) program initiated between the Reliance Institute of Life Sciences, India and Deakin University, Australia. The authors also gratefully acknowledge Reliance Life Sciences Pvt. Ltd., Mumbai, for financial support for the project.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Available statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics statement
None.
Informed consent
None.
Contributor Information
Arnab Kapat, Email: Arnab.Kapat@ril.com.
Eugene Athan, Email: e.athan@deakin.edu.au.
References
- 1.Wu W., Jin Y., Bai F., et al. In: Molecular Medical Microbiology. 2nd ed. Tang T.W., Sussman M., Liu D., Poxton I., Schwartzman J., editors. Academic Press; Cambridge, MA, USA: 2015. Chapter 41—pseudomonas aeruginosa; pp. 753–767. [DOI] [Google Scholar]
- 2.El-Fouly M.Z., Sharaf A.M., Shahin A.A.M, et al. Biosynthesis of pyocyanin pigment by Pseudomonas aeruginosa. J. Radiat. Res. Appl. Sci. 2015;8(1):36–48. doi: 10.1016/j.jrras.2014.10.007. [DOI] [Google Scholar]
- 3.Brady M.T., Leber A. Principles and Practice of Pediatric Infectious Diseases. Elsevier; 2018. Less Commonly Encountered Nonenteric Gram-Negative bacilli; pp. 855–859. [DOI] [Google Scholar]
- 4.Schmidt K.D., Tümmler B. Römling. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 1996;178(1):85–93. doi: 10.1128/jb.178.1.85-93.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stover C.K., Pham X.Q., Erwin A.L., et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406(6799):959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 6.Lee D.G., Urbach J.M., Wu G., et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome. Biol. 2006;7(10):R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lister P.D., Wolter D.J., Hanson N.D. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009;22(4):582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pathi B., Mishra S.N., Panigrahi K., et al. Prevalence and antibiogram pattern of Pseudomonas aeruginosa in a tertiary care hospital from Odisha, India. Transw. med. J. 2013;1(3):77–80. [Google Scholar]
- 9.Prakash V., Mishra P., Premi H., et al. Increasing incidence of multidrug resistant Pseudomonas aeruginosa in inpatients of a tertiary care hospital. Int. J. Res. Med. Sci. 2014;2(4):1302–1306. doi: 10.5455/2320-6012.ijrms20141111. [DOI] [Google Scholar]
- 10.Ventola C.L. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40(4):277–283. [PMC free article] [PubMed] [Google Scholar]
- 11.Cillóniz C., Gabarrús A., Ferrer M., et al. Community-acquired pneumonia due to multidrug and non-multidrug-resistant Pseudomonas aeruginosa. Chest. 2016;150(2):415–425. doi: 10.1016/j.chest.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen L., Garcia J., Gruenberg K., et al. Multidrug-resistant Pseudomonas infections: hard to treat, but hope on the horizon? Curr. Infect. Dis. Rep. 2018;20(8):23. doi: 10.1007/s11908-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 13.Raman G., Avendano E.E., Chan J., et al. Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob. Resist. Infect. Control. 2018;7:79. doi: 10.1186/s13756-018-0370-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Planet P.J. In: Pseudomonas aeruginosa, in Principles and Practice of Pediatric Infectious Diseases. S Long, C Prober. Fischer M., editor. Elsevier; Toronto, ON, Canada: 2018. pp. 866–870. [Google Scholar]
- 15.Diaz K.E., Remold S.K., Onyiri O., et al. Generalized growth of estuarine, household and clinical isolates of Pseudomonas aeruginosa. Front. Microbiol. 2018;9:305. doi: 10.3389/fmicb.2018.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. Pseudomonas aeruginosa in healthcare settings. Avaliable on: https://www.cdc.gov/hai/organisms/pseudomonas (accessed on 27 August 2018).
- 17.B.H. Iglewski, Pseudomonas. In Medical Microbiology. S Baron (Ed.), University of Texas Medical Branch at Galveston:Galveston, TX, USA,1996. [PubMed]
- 18.Furtado G.H., d'Azevedo P.A., Santos A.F., et al. Intravenous polymyxin B for the treatment of nosocomial pneumonia caused by multidrug-resistant Pseudomonas aeruginosa. Int. J. Antimicrob. Agents. 2017;30(4):315–319. doi: 10.1016/j.ijantimicag.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 19.Shigemura K., Arakawa S., Sakai Y., et al. Complicated urinary tract infection caused by Pseudomonas aeruginosa in a single institution (1999-2003) Int. J. Urol. 2006;13(5):538–542. doi: 10.1111/j.1442-2042.2006.01359.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim M., Christley S., Khodarev N.N., et al. Pseudomonas aeruginosa wound infection involves activation of its iron acquisition system in response to fascial contact. J. Trauma. Acute. Care Surg. 2015;78(4):823–829. doi: 10.1097/TA.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez M.R., Fleuchot B., Lauciello L., et al. Effect of human burn wound exudate on pseudomonas aeruginosa virulence. mSphere. 2016;1(2):e00111–e00115. doi: 10.1128/mSphere.00111-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg R.N., Kennedy D.J., Reilly P.M., et al. Treatment of bone, joint, and soft-tissue infections with oral ciprofloxacin. Antimicrob. Agents Chemother. 1987;31(2):151–155. doi: 10.1128/AAC.31.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laughlin T.J., Armstrong D.G., Caporusso J., et al. Soft tissue and bone infections from puncture wounds in children. West. J. Med. 1997;166(2):126–128. [PMC free article] [PubMed] [Google Scholar]
- 24.Brouqui P., Rousseau M.C., Stein A., et al. Treatment of Pseudomonas aeruginosa-infected orthopedic prostheses with ceftazidime-ciprofloxacin antibiotic combination. Antimicrob. Agents Chemother. 1995;39(11):2423–2425. doi: 10.1128/aac.39.11.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zawacki A., O'Rourke E., Potter-Bynoe G., et al. An outbreak of Pseudomonas aeruginosa pneumonia and bloodstream infection associated with intermittent otitis externa in a healthcare worker. Infect. Control Hosp. Epidemiol. 2004;25(12):1083–1089. doi: 10.1086/502348. [DOI] [PubMed] [Google Scholar]
- 26.Osmon S., Ward S., Fraser V.J., et al. Hospital mortality for patients with bacteremia due to Staphylococcus aureus or Pseudomonas aeruginosa. Chest. 2004;125(2):607–616. doi: 10.1378/chest.125.2.607. [DOI] [PubMed] [Google Scholar]
- 27.Shi Q., Huang C., Xiao T., et al. A retrospective analysis of Pseudomonas aeruginosa bloodstream infections: prevalence, risk factors, and outcome in carbapenem-susceptible and -non-susceptible infections. Antimicrob. Resist. Infect. Control. 2019;8:68. doi: 10.1186/s13756-019-0520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bucior I., Mostov K., Engel J.N. Pseudomonas aeruginosa-mediated damage requires distinct receptors at the apical and basolateral surfaces of the polarized epithelium. Infect. Immun. 2010;78(3):939–953. doi: 10.1128/IAI.01215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Bentzmann S., Plotkowski C., Puchelle E. Receptors in the Pseudomonas aeruginosa adherence to injured and repairing airway epithelium. Am. J. Respir. Crit. Care Med. 1996;154(4):S155–S162. doi: 10.1164/ajrccm/154.4_Pt_2.S155. Pt 2. [DOI] [PubMed] [Google Scholar]
- 30.Chatzinikolaou I., Abi-Said D., Bodey G.P., et al. Recent experience with Pseudomonas aeruginosa bacteremia in patients with cancer: retrospective analysis of 245 episodes. Arch. Intern. Med. 2000;160(4):501–509. doi: 10.1001/archinte.160.4.501. [DOI] [PubMed] [Google Scholar]
- 31.Iversen B.G., Jacobsen T., Eriksen H.M., et al. An outbreak of Pseudomonas aeruginosa infection caused by contaminated mouth swabs. Clin. Infect. Dis. 2007;44(6):794–801. doi: 10.1086/511644. [DOI] [PubMed] [Google Scholar]
- 32.Revdiwala S., Rajdev B.M., Mulla S. Characterization of bacterial etiologic agents of biofilm formation in medical devices in critical care setup. Crit. Care Res. Pract. 2012;2012 doi: 10.1155/2012/945805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills G.Y., Kagan B.M. Effect of oral polymyxin B on Pseudomonas aeruginosa in the gastrointestinal tract. Ann. Intern. Med. 1954;40(1):26–32. doi: 10.7326/0003-4819-40-1-26. [DOI] [PubMed] [Google Scholar]
- 34.Chuang C.H., Wang Y.H., Chang H.J., et al. Shanghai fever: a distinct Pseudomonas aeruginosa enteric disease. Gut. 2014;63(5):736–743. doi: 10.1136/gutjnl-2013-304786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubin J., Yu V.L., Kamerer D.B., et al. Aural irrigation with water: a potential pathogenic mechanism for inducing malignant external otitis? Ann. Otol. Rhinol. Laryngol. 1990;99(2):117–119. doi: 10.1177/000348949009900207. Pt 1. [DOI] [PubMed] [Google Scholar]
- 36.West S.E., Zeng L., Lee B.L., et al. Respiratory infections with Pseudomonas aeruginosa in children with cystic fibrosis: early detection by serology and assessment of risk factors. JAMA. 2002;287(22):2958–2967. doi: 10.1001/jama.287.22.2958. [DOI] [PubMed] [Google Scholar]
- 37.Wang T., Hou Y., Wang R. A case report of community-acquired Pseudomonas aeruginosa pneumonia complicated with MODS in a previously healthy patient and related literature review. BMC Infect. Dis. 2019;19(1):130. doi: 10.1186/s12879-019-3765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neuhauser M.M., Weinstein R.A., Rydman R., et al. Antibiotic resistance among gram-negative bacilli in US intensive care units: implications for fluoroquinolone use. JAMA. 2003;289(7):885–888. doi: 10.1001/jama.289.7.885. [DOI] [PubMed] [Google Scholar]
- 39.National Nosocomial Infections Surveillance System National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control. 2004;32(8):470–485. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 40.Kang C.I., Kim S.H., Kim H.B., et al. Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin. Infect. Dis. 2003;37(6):745–751. doi: 10.1086/377200. [DOI] [PubMed] [Google Scholar]
- 41.Boucher H.W., Talbot G.H., Bradley J.S., et al. Bad bugs, no drugs: no ESKAPE! an update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 42.Centers for Disease Control and Prevention, Multidrug-resistant pseudomonas aeruginosa Avaliableon: www.cdc.gov. (accessed on November 13, 2019).
- 43.Kollef M.H., Chastre J., Fagon J.Y., et al. Global prospective epidemiologic and surveillance study of ventilator-associated pneumonia due to Pseudomonas aeruginosa. Crit. Care Med. 2014;42(10):2178–2187. doi: 10.1097/CCM.0000000000000510. [DOI] [PubMed] [Google Scholar]
- 44.Cole S.J., Records A.R., Orr M.W., et al. Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms. Infect. Immun. 2014;82(5):2048–2058. doi: 10.1128/IAI.01652-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomila A., Carratalà J., Badia J.M., et al. VINCat Colon Surgery Group. Preoperative oral antibiotic prophylaxis reduces Pseudomonas aeruginosa surgical site infections after elective colorectal surgery: a multicenter prospective cohort study. BMC Infect. Dis. 2018;18(1):507. doi: 10.1186/s12879-018-3413-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bang R.L., Sharma P.N., Sanyal S.C., et al. Septicaemia after burn injury: a comparative study. Burns. 2002;28(8):746–751. doi: 10.1016/s0305-4179(02)00183-3. [DOI] [PubMed] [Google Scholar]
- 47.Gandra S., Tseng K.K., Arora A., et al. The mortality burden of multidrug-resistant pathogens in India: a retrospective, observational study. Clin. Infect. Dis. 2019;69(4):563–570. doi: 10.1093/cid/ciy955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obritsch M.D., Fish D.N., MacLaren R., et al. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy. 2005;25(10):1353–1364. doi: 10.1592/phco.2005.25.10.1353. [DOI] [PubMed] [Google Scholar]
- 49.Li B., Webster T.J. Bacteria antibiotic resistance: new challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2018;36(1):22–32. doi: 10.1002/jor.23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Z., Tang J., Khare T., et al. The alarming antimicrobial resistance in ESKAPEE pathogens: can essential oils come to the rescue? Fitoterapia. 2020;140 doi: 10.1016/j.fitote.2019.104433. [DOI] [PubMed] [Google Scholar]
- 51.Tamma P.D., Aitken S.L., Bonomo R.A., et al. Infectious Diseases Society of America 2022 Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa) Clin. Infect. Dis. 2022;75(2):187–212. doi: 10.1093/cid/ciac268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aloush V., Navon-Venezia S., Seigman-Igra Y., et al. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob. Agents Chemother. 2006;50(1):43–48. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng Y., Hodiamont C.J., van Hest R.M., et al. Development of antibiotic resistance during simulated treatment of Pseudomonas aeruginosa in chemostats. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0149310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgan D.J., Rogawski E., Thom K.A., et al. Transfer of multidrug-resistant bacteria to healthcare workers' gloves and gowns after patient contact increases with environmental contamination. Crit. Care Med. 2012;40(4):1045–1051. doi: 10.1097/CCM.0b013e31823bc7c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajkumari N., John N.V., Mathur P., et al. Antimicrobial resistance in Pseudomonas sp. Causing infections in trauma patients: a 6 year experience from a South Asian Country. J. Glob. Infect. Dis. 2014;6(4):182–185. doi: 10.4103/0974-777X.145250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alnour T.M., Ahmed-Abakur E.H. Multidrug resistant Pseudomonas aeruginosa: Medical impact, pathogenicity, resistance mechanisms and epidemiology. JSM Microbiol. 2017;5(3):1046. [Google Scholar]
- 57.Bassetti M., Vena A., Croxatto A., et al. How to manage Pseudomonas aeruginosa infections. Drugs Context. 2018;7 doi: 10.7573/dic.212527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gill J.S., Arora S., Khanna S.P., et al. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant pseudomonas aeruginosa from a tertiary level intensive care unit. J. Glob. Infect. Dis. 2016;8(4):155–159. doi: 10.4103/0974-777X.192962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Intra J., Sarto C., Beck E., et al. Bacterial and fungal colonization of the respiratory tract in COVID-19 patients should not be neglected. Am. J. Infect. Control. 2020;48(9):1130–1131. doi: 10.1016/j.ajic.2020.06.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu C., Liu H., Pan X., et al. Mechanisms for development of ciprofloxacin resistance in a clinical isolate of Pseudomonas aeruginosa. Front Microbiol. 2021;11 doi: 10.3389/fmicb.2020.598291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horcajada J.P., Montero M., Oliver A. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019;32(4):e00031–19. doi: 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qu J., Cai Z., Liu Y., et al. Persistent bacterial coinfection of a COVID-19 patient caused by a genetically adapted Pseudomonas aeruginosa chronic colonizer. Front Cell Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.641920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shafran N., Shafran I., Ben-Zvi H., et al. Secondary bacterial infection in COVID-19 patients is a stronger predictor for death compared to influenza patients. Scientific reports. 2021;11(1):12703. doi: 10.1038/s41598-021-92220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russell C.D., Fairfield C.J., Drake T.M., et al. ISARIC4C investigators. Co-infections, secondary infections, and antimicrobial use in patients hospitalised with COVID-19 during the first pandemic wave from the ISARIC WHO CCP-UK study: a multicentre, prospective cohort study. Lancet Microbe. 2021;2(8):e354–e365. doi: 10.1016/S2666-5247(21)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Centers for Disease Control and Prevention, COVID-19: US Impact on Antimicrobial Resistance, Special Report 2022 (2022). Avaliable on: www.cdc.gov (accessed on 13 November 2019)
- 66.Hidron A.I., Edwards J.R., Patel J., et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention. Infect. Control Hosp. Epidemiol. 2008;29(11):996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 67.Hurley J.C. Incidences of Pseudomonas aeruginosa-associated ventilator-associated pneumonia within studies of respiratory tract applications of polymyxin: testing the stoutenbeek concurrency postulates. Antimicrob. Agents Chemother. 2018;62(8):e00291–18. doi: 10.1128/AAC.00291-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Planquette B., Timsit J.F., Misset B.Y., et al. Pseudomonas aeruginosa ventilator-associated pneumonia. predictive factors of treatment failure. Am. J. Respir. Crit. Care Med. 2013;188(1):69–76. doi: 10.1164/rccm.201210-1897OC. [DOI] [PubMed] [Google Scholar]
- 69.Mermel L.A., McKay M., Dempsey J., et al. Pseudomonas surgical-site infections linked to a healthcare worker with onychomycosis. Infect. Control Hosp. Epidemiol. 2003;24(10):749–752. doi: 10.1086/502125. [DOI] [PubMed] [Google Scholar]
- 70.Al-Akayleh A.T. Invasive burn wound infection. Ann. Burns Fire Disasters. 1999;12(44):204–206. [Google Scholar]
- 71.El Amari E.B., Chamot E., Auckenthaler R., et al. nfluence of previous exposure to antibiotic therapy on the susceptibility pattern of Pseudomonas aeruginosa bacteremic isolates. Clin. Infect. Dis. 2001;33(11):1859–1864. doi: 10.1086/324346. [DOI] [PubMed] [Google Scholar]
- 72.Caselli D., Cesaro S., Ziino O., et al. Multidrug resistant Pseudomonas aeruginosa infection in children undergoing chemotherapy and hematopoietic stem cell transplantation. Haematologica. 2010;95(9):1612–1615. doi: 10.3324/haematol.2009.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morita F., Hirai Y., Suzuki K., et al. The first case of pseudomonas aeruginosa bacteremic pneumonia in a cancer patient receiving pegfilgrastim. Internal medicine. 2017;56(15):2039–2042. doi: 10.2169/internalmedicine.56.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumari M., Khurana S., Bhardwaj N., et al. Pathogen burden & associated antibiogram of Pseudomonas spp. in a tertiary care hospital of India. Indian J. Med. Res. 2019;149(2):295–298. doi: 10.4103/ijmr.IJMR_14_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Streete K., Katouli M. P.aeruginosa: a review of their pathogenesis and prevalence in clinical settings and the environment. Infect. Epidemiol. Med. 2016;2(1):25–32. doi: 10.7508/iem.2016.01.008. [DOI] [Google Scholar]
- 76.Watson S., Cabrera-Aguas M., Khoo P. Common eye infections. Aust. Prescr. 2018;41(3):67–72. doi: 10.18773/austprescr.2018.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McDaniel L.S. Otitis Media and Ear Infections. Bacteria. 2021 [Google Scholar]
- 78.Sander R. Otitis externa: a practical guide to treatment and prevention. Am. Fam. Physician. 2001;63(5):927–942. [PubMed] [Google Scholar]
- 79.Mateus S.P., Ribeiro-Alves M., Salles R.E.B., et al. Mortality and comorbidities in patients with bronchiectasis over a 3-year follow-up. Medicine. 2022;101(52):e32537. doi: 10.1097/MD.0000000000032537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samadi R. Bacterial skin and soft-tissue infections: a systematic approach to diagnosis and treatment. Emerg. Med. Rep. 2004;25(14):161–176. [Google Scholar]
- 81.Zichichi L., Asta G., Noto G. Pseudomonas aeruginosa folliculitis after shower/bath exposure. Int. J. Dermatol. 2000;39(4):270–273. doi: 10.1046/j.1365-4362.2000.00931.x. [DOI] [PubMed] [Google Scholar]
- 82.Wu D.C., Chan W.W., Metelitsa A.I., et al. Pseudomonas skin infection: clinical features, epidemiology, and management. Am. J. Clin. Dermatol. 2011;12(3):157–169. doi: 10.2165/11539770-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 83.Mittal R., Kodiyan J., Gerring R., et al. Role of innate immunity in the pathogenesis of otitis media. Int. J. Infect. Dis. 2014;29:259–267. doi: 10.1016/j.ijid.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chiriac A., Brzezinski P., Foia L., et al. Chloronychia: green nail syndrome caused by Pseudomonas aeruginosa in elderly persons. Clin. Interv. Aging. 2015;10:265–267. doi: 10.2147/CIA.S75525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Müller S., Ebnöther M., Itin P. Green Nail Syndrome (Pseudomonas aeruginosa Nail Infection): two cases successfully treated with topical nadifloxacin, an acne medication. Case Rep. Dermatol. 2014;6(2):180–184. doi: 10.1159/000365863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bae Y., Lee G.M., Sim J.H., et al. Green nail syndrome treated with the application of tobramycin eye drop. Ann. Dermatol. 2014;26(4):514–516. doi: 10.5021/ad.2014.26.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Church D., Elsayed S., Reid O., et al. Burn wound infections. Clin. Microbiol. Rev. 2006;19(2):403–434. doi: 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bielecki P., Glik J., Kawecki M., et al. Towards understanding Pseudomonas aeruginosa burn wound infections by profiling gene expression. Biotechnol. Lett. 2008;30(5):777–790. doi: 10.1007/s10529-007-9620-2. [DOI] [PubMed] [Google Scholar]
- 89.Wisplinghoff H., Bischoff T., Tallent S.M., et al. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004;39(3):309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 90.Davies J.C. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr. Respir. Rev. 2002;3(2):128–134. doi: 10.1016/s1526-0550(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 91.Weber D.J., Rutala W.A., Sickbert-Bennett E.E., et al. Microbiology of ventilator-associated pneumonia compared with that of hospital-acquired pneumonia. Infect. Control. Hosp. Epidemiol. 2007;28(7):825–831. doi: 10.1086/518460. [DOI] [PubMed] [Google Scholar]
- 92.Davies G., Wells A.U., Doffman S., et al. The effect of Pseudomonas aeruginosa on pulmonary function in patients with bronchiectasis. Eur. Respir. J. 2006;28(5):974–979. doi: 10.1183/09031936.06.00074605. [DOI] [PubMed] [Google Scholar]
- 93.Mader J.T., Calhoun J. In: Medical Microbiology. 4th ed. Baron S, editor. University of Texas Medical Branch at Galveston; Galveston (TX): 1996. Bone, joint, and necrotizing soft tissue infections. [PubMed] [Google Scholar]
- 94.Tice A.D., Hoaglund P.A., Shoultz D.A. Risk factors and treatment outcomes in osteomyelitis. J. Antimicrob. Chemother. 2003;51(5):1261–1268. doi: 10.1093/jac/dkg186. [DOI] [PubMed] [Google Scholar]
- 95.Gudiol C., Albasanz-Puig A., Laporte-Amargós J., et al. Clinical Predictive Model of Multidrug Resistance in Neutropenic Cancer Patients with Bloodstream Infection Due to Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2020;64(4) doi: 10.1128/AAC.02494-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cerioli M., Batailler C., Conrad A., et al. Pseudomonas aeruginosa implant-associated bone and joint infections: experience in a regional reference center in France. Front. Med. 2020;(7) doi: 10.3389/fmed.2020.513242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baker C.C., Trunkey D.D., Baker W.J. A simple method of predicting severe sepsis in burn patients. Am. J. Surg. 1980;139(4):513–517. doi: 10.1016/0002-9610(80)90329-3. [DOI] [PubMed] [Google Scholar]
- 98.Bang R.L., Gang R.K., Sanyal S.C., et al. Beta-haemolytic Streptococcus infection in burns. Burns. 1999;25(3):242–246. doi: 10.1016/s0305-4179(98)00167-3. [DOI] [PubMed] [Google Scholar]
- 99.Alwan M.J., Lafta I.J., Hamzah A.M. Bacterial isolation from burn wound infections and studying their antimicrobial susceptibility. Kufa j. vet. Sci. 2011;2(1):121–131. [Google Scholar]
- 100.Argenta A., Demos J. Burn management in the developing world: International Volunteerism. Clin. Plast. Surg. 2017;44(4):875–883. doi: 10.1016/j.cps.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 101.Pujji O.J.S., Nakarmi K.K., Shrestha B., Rai S.M., Jeffery S.L.A. The bacteriological profile of burn wound infections at a tertiary burns center in Nepal. J. Burn Care Res. 2019;40(6):838–845. doi: 10.1093/jbcr/irz096. [DOI] [PubMed] [Google Scholar]
- 102.Bhate-Deosthali P., Lingam L. Gendered pattern of burn injuries in India: a neglected health issue. Reprod. Health Matters. 2016;24(47):96–103. doi: 10.1016/j.rhm.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 103.Lilly H.A., Lowbury E.J., Wilkins M.D., et al. Staphylococcal sepsis in a burns unit. J. Hyg. (Lond) 1979;83(3):429–435. doi: 10.1017/s0022172400026267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Phillips L.G., Heggers J.P., Robson M.C., et al. The effect of endogenous skin bacteria on burn wound infection. Ann. Plast. Surg. 1989;23(1):35–38. doi: 10.1097/00000637-198907000-00007. [DOI] [PubMed] [Google Scholar]
- 105.Dalcin D., Ulanova M. The role of human beta-Defensin-2 in Pseudomonas aeruginosa pulmonary infection in cystic fibrosis patients. Infect. Dis. Ther. 2013;2(2):159–166. doi: 10.1007/s40121-013-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu Y., Li D., Wang Y., et al. Beta-Defensin 2 and 3 promote bacterial clearance of Pseudomonas aeruginosa by inhibiting macrophage autophagy through downregulation of early growth response Gene-1 and c-FOS. Front Immunol. 2018;9:211. doi: 10.3389/fimmu.2018.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu Y., Liu Y., Dong K., et al. Effects of human β-defensin 3 fused with carbohydrate-binding domain on the function of type III secretion system in Pseudomonas aeruginosa PA14. Braz. J. Microbiol. 2020;51(1):29–35. doi: 10.1007/s42770-020-00223-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Harrison-Balestra C., Cazzaniga A.L., Davis S.C., et al. A wound-isolated Pseudomonas aeruginosa grows a biofilm in vitro within 10 hours and is visualized by light microscopy. Dermatol. Surg. 2003;29(6):631–635. doi: 10.1046/j.1524-4725.2003.29146.x. [DOI] [PubMed] [Google Scholar]
- 109.Mah T.F., O'Toole G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 110.Coetzee E., Rode H., Kahn D. Pseudomonas aeruginosa burn wound infection in a dedicated paediatric burns unit. S. Afr. J. Surg. 2013;51(2):50–53. doi: 10.7196/sajs.1134. [DOI] [PubMed] [Google Scholar]
- 111.Bhatt P., Rathi K.R., Hazr S., et al. Prevalence of multidrug resistant Pseudomonas aeruginosa infection in burn patients at a tertiary care centre. Indian J. Burns. 2015;23(1):56. doi: 10.4103/0971-653X.171656. [DOI] [Google Scholar]
- 112.Que Y.A., Hazan R., Ryan C.M., et al. Production of Pseudomonas aeruginosa intercellular small signaling molecules in human burn wounds. J. Pathog. 2011;549302 doi: 10.4061/2011/549302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mahin Samadi P., Gerami P., Elmi A., et al. Pseudomonas aeruginosa keratitis: passive immunotherapy with antibodies raised against divalent flagellin. Iran. J. Basic Med. Sci. 2019;22(1):58–64. doi: 10.22038/ijbms.2018.31499.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weed M.C., Rogers G.M., Kitzmann A.S., et al. Vision loss after contact lens-related Pseudomonas keratitis. Eye Rounds org. 2013 [Google Scholar]
- 115.Chan A., Oo H.H., Stanley P., et al. Recalcitrant Pseudomonas Aeruginosa keratitis with hyphaema. Case Rep. Ophthalmol. 2021;12(1):214–218. doi: 10.1159/000512473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sy A., Srinivasan M., Mascarenhas J., et al. Pseudomonas aeruginosa keratitis: outcomes and response to corticosteroid treatment. Invest. Ophthalmol. Vis. Sci. 2012;53(1):267–272. doi: 10.1167/iovs.11-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang M., Smith W.A., Duncan J.K., et al. Treatment of Pseudomonas keratitis by continuous infusion of topical antibiotics with the Morgan lens. Cornea. 2017;36(5):617–620. doi: 10.1097/ICO.0000000000001128. [DOI] [PubMed] [Google Scholar]
- 118.Yilmaz S., Saklamaz A., Maden A. Pseudomonas keratitis. Ophthalmology. 2006;113(5):883–884. doi: 10.1016/j.ophtha.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 119.Juyal D., Sharma M., Negi V., et al. Pseudomonas aeruginosa and its sensitivity spectrum in chronic suppurative otitis media: a study from Garhwal hills of Uttarakhand State, India. Indian J. Otol. 2017;23:180–184. doi: 10.4103/indianjotol.INDIANJOTOL_31_14. [DOI] [Google Scholar]
- 120.Sahu M.C., Swain S.K., Kar S.K. Genetically diversity of Pseudomonas aeruginosa isolated from chronic suppurative otitis media with respect to their antibiotic sensitivity pattern. Indian J. Otolaryngol. Head Neck Surg. 2019;71(2):1300–1308. doi: 10.1007/s12070-018-1358-8. Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rath S., Das S.R., Padhy R.N. Bayesian analysis of two methods MALDI-TOF-MS system and culture test in otomycosis infection. World J. Otorhinolaryngol. Head Neck Surg. 2019;5(1):6–13. doi: 10.1016/j.wjorl.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mittal R., Lisi C.V., Kumari H., et al. Otopathogenic Pseudomonas aeruginosa enters and survives inside macrophages. Front. Microbiol. 2016;7:1828. doi: 10.3389/fmicb.2016.01828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schwartz R.A., Reynoso-Vasquez N., Kapila R. Chloronychia: the Goldman-Fox Syndrome - implications for patients and healthcare workers. Indian J. Dermatol. 2020;65(1):1–4. doi: 10.4103/ijd.IJD_277_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ohn J., Yu D.A., Park H., et al. Green nail syndrome: analysis of the association with onychomycosis. J. Am. Acad. Dermatol. 2020;83(3):940–942. doi: 10.1016/j.jaad.2020.01.040. [DOI] [PubMed] [Google Scholar]
- 125.Endimiani A., Luzzaro F., Pini B., et al. Pseudomonas aeruginosa bloodstream infections: risk factors and treatment outcome related to expression of the PER-1 extended-spectrum beta-lactamase. BMC Infect. Dis. 2006;6:52. doi: 10.1186/1471-2334-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hickey C., Schaible B., Nguyen S., et al. Increased virulence of bloodstream over peripheral isolates of P. aeruginosa identified through post-transcriptional regulation of virulence factors. Front. Cell Infect. Microbiol. 2018;8:357. doi: 10.3389/fcimb.2018.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mittal R., Aggarwal S., Sharma S., et al. Urinary tract infections caused by Pseudomonas aeruginosa: a mini review. J. Infect. Public Health. 2009;2(3):101–111. doi: 10.1016/j.jiph.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 128.Lu P.L., Liu Y.C., Toh H.S., et al. Epidemiology and antimicrobial susceptibility profiles of Gram-negative bacteria causing urinary tract infections in the Asia-Pacific region: 2009-2010 results from the Study for Monitoring Antimicrobial Resistance Trends (SMART) Int. J. Antimicrob. Agents. 2012;40:S37–S43. doi: 10.1016/S0924-8579(12)70008-0. Suppl. [DOI] [PubMed] [Google Scholar]
- 129.Bitsori M., Maraki S., Koukouraki S., et al. Pseudomonas aeruginosa urinary tract infection in children: risk factors and outcomes. J. Urol. 2012;187(1):260–264. doi: 10.1016/j.juro.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 130.Lamas Ferreiro J.L., Álvarez Otero J., González González L., et al. Pseudomonas aeruginosa urinary tract infections in hospitalized patients: Mortality and prognostic factors. PloS one. 2017;12(5) doi: 10.1371/journal.pone.0178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shobha K.L., Ramachandra L., Rao A.S., et al. Pseudomonas species causing urinary tract infection and its antibiogram at a tertiary care hospital. Asian J. Pharm. Clin. Res. 2017;10(11):50–51. doi: 10.22159/ajpcr.2017.v10i11.20002. [DOI] [Google Scholar]
- 132.Venier A.G., Lavigne T., Jarno P., et al. Nosocomial urinary tract infection in the intensive care unit: when should Pseudomonas aeruginosa be suspected? Experience of the French national surveillance of nosocomial infections in the intensive care unit, Rea-Raisin. Clin. Microbiol. Infect. 2012;18(1):E13–E15. doi: 10.1111/j.1469-0691.2011.03686.x. [DOI] [PubMed] [Google Scholar]
- 133.Pobiega M., Maciag J., Pomorska-Wesolowska M., et al. Urinary tract infections caused by Pseudomonas aeruginosa among children in Southern Poland: Virulence factors and antibiotic resistance. J. Pediatr. Urol. 2016;12(1):36.e1–36.e366. doi: 10.1016/j.jpurol.2015.05.034. [DOI] [PubMed] [Google Scholar]
- 134.Badamchi A., Masoumi H., Javadinia S., et al. Molecular detection of six virulence genes in Pseudomonas aeruginosa isolates detected in children with urinary tract infection. Microb. Pathog. 2017;107:44–47. doi: 10.1016/j.micpath.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 135.Penaranda C., Chumbler N.M., Hung D.T. Dual transcriptional analysis reveals adaptation of host and pathogen to intracellular survival of Pseudomonas aeruginosa associated with urinary tract infection. PLoS Pathog. 2021;17(4) doi: 10.1371/journal.ppat.1009534. Published 2021 Apr 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Estaji M., Tabasi M., Sadeghpour Heravi F., et al. Genotypic identification of Pseudomonas aeruginosa strains isolated from patients with urinary tract infection. Comp. Immunol. Microbiol. Infect. Dis. 2021;65:23–28. doi: 10.1016/j.cimid.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 137.Herr H.W. The risk of urinary tract infection after flexible cystoscopy in patients with bladder tumor who did not receive prophylactic antibiotics. J. Urol. 2015;193(2):548–551. doi: 10.1016/j.juro.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 138.Sorbets E., Evrevin M., Jumas-Bilak E., et al. An outbreak of Pseudomonas aeruginosa urinary tract infections following outpatient flexible cystoscopy. Am. J. Infect. Control. 2019;47(12):1510–1512. doi: 10.1016/j.ajic.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 139.Moffett K.S. P.aeruginosa in patients with cystic fibrosis. Antimicrob. Ther. 2010;1(1) [Google Scholar]
- 140.Mainz J.G., Gerber A., Lorenz M., et al. Pseudomonas aeruginosa acquisition in cystic fibrosis patients in context of otorhinolaryngological surgery or dentist attendance: case series and discussion of preventive concepts. Case Rep. Infect. Dis. 2015 doi: 10.1155/2015/438517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bhagirath A.Y., Li Y., Somayajula D., et al. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm. Med. 2016;16(1):174. doi: 10.1186/s12890-016-0339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Janssens S., Chokoshvilli D., Binst C., et al. Attitudes of cystic fibrosis patients and parents toward carrier screening and related reproductive issues. Eur. J. Hum. Genet. 2016;24(4):506–512. doi: 10.1038/ejhg.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Winstanley C., O'Brien S., Brockhurst M.A. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 2016;24(5):327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Speert D.P., Campbell M.E., Henry D.A., et al. Epidemiology of Pseudomonas aeruginosa in cystic fibrosis in British Columbia, Canada. Am. J. Respir. Crit. Care Med. 2002;166(7):988–993. doi: 10.1164/rccm.2203011. [DOI] [PubMed] [Google Scholar]
- 145.Jiricny N., Molin S., Foster K., et al. Loss of social behaviours in populations of Pseudomonas aeruginosa infecting lungs of patients with cystic fibrosis. PloS one. 2014;9(1):e83124. doi: 10.1371/journal.pone.0083124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huus K.E., Joseph J., Zhang L., et al. Clinical Isolates of Pseudomonas aeruginosa from chronically infected cystic fibrosis patients fail to activate the inflammasome during both stable infection and pulmonary exacerbation. J. Immun. 2016;196(7):3097–3108. doi: 10.4049/jimmunol.1501642. [DOI] [PubMed] [Google Scholar]
- 147.LaFayette S.L., Houle D., Beaudoin T., et al. Cystic fibrosis-adapted Pseudomonas aeruginosa quorum sensing lasR mutants cause hyperinflammatory responses. Sci. Adv. 2015;1(6) doi: 10.1126/sciadv.1500199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.American Thoracic Society; Infectious Diseases Society of America Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 2015;171(4):388–416. doi: 10.1164/rccm.200405-644ST. [DOI] [PubMed] [Google Scholar]
- 149.Williams B.J., Dehnbostel J., Blackwell T.S. Pseudomonas aeruginosa: host defence in lung diseases. Respirology. 2010;15(7):1037–1056. doi: 10.1111/j.1440-1843.2010.01819.x. [DOI] [PubMed] [Google Scholar]
- 150.Chalmers J.D., Taylor J.K., Singanayagam A., et al. Epidemiology, antibiotic therapy, and clinical outcomes in health care-associated pneumonia: a UK cohort study. Clin. Infect. Dis.an off publ. Infect. Dis. Soc. Am. 2011;53(2):107–113. doi: 10.1093/cid/cir274. [DOI] [PubMed] [Google Scholar]
- 151.Jeong B.H., Koh W.J., Yoo H., et al. Performances of prognostic scoring systems in patients with healthcare-associated pneumonia. Clin. Infect. Dis. 2013;56(5):625–632. doi: 10.1093/cid/cis970. [DOI] [PubMed] [Google Scholar]
- 152.Morello E., Pérez-Berezo T., Boisseau C., et al. Pseudomonas aeruginosa Lipoxygenase LoxA contributes to lung infection by altering the host immune lipid signaling. Front. Microbiol. 2019;10:1826. doi: 10.3389/fmicb.2019.01826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jung I.Y., Jeong S.J., Lee K.M., et al. Risk factors for mortality in patients with Pseudomonas aeruginosa pneumonia: clinical impact of mucA gene mutation. Respir. Med. 2018;140:27–31. doi: 10.1016/j.rmed.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 154.Kwok W.C., Ho J.C.M., Tam T.C.C., et al. Risk factors for Pseudomonas aeruginosa colonization in non-cystic fibrosis bronchiectasis and clinical implications. Respir. Res. 2021;22(1):132. doi: 10.1186/s12931-021-01729-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tummala V.S.B., Surapaneni S.B., Pigilam S. Bacteriological study of orthopaedic infections. Int. j. orthop. 2017;3:90–92. [Google Scholar]
- 156.Ross J.J., Shamsuddin H. Sternoclavicular septic arthritis: review of 180 cases. Medicine. 2004;83(3):139–148. doi: 10.1097/01.md.0000126761.83417.29. [DOI] [PubMed] [Google Scholar]
- 157.Jangid M.K., Selim M.A. Community-acquired Pseudomonas aeruginosa wrist joint arthritis in a child-a case report. J. Nepal Paediatr. 2020;40(1):48–51. doi: 10.3126/jnps.v40i1.28854. [DOI] [Google Scholar]
- 158.Hurley M.N., Cámara M., Smyth A.R. Novel approaches to the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. Eur. Respir. J. 2012;40(4):1014–1023. doi: 10.1183/09031936.00042012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fulghesu L., Giallorenzo C., Savoia D. Evaluation of different compounds as quorum sensing inhibitors in Pseudomonas aeruginosa. J. Chemother. 2007;19(4):388–391. doi: 10.1179/joc.2007.19.4.388. [DOI] [PubMed] [Google Scholar]
- 160.Bandara M.B., Zhu H., Sankaridurg P.R., et al. Salicylic acid reduces the production of several potential virulence factors of Pseudomonas aeruginosa associated with microbial keratitis. Invest. Ophthalmol. Vis. Sci. 2006;47(10):4453–4460. doi: 10.1167/iovs.06-0288. [DOI] [PubMed] [Google Scholar]
- 161.Wu H., Song Z., Hentzer M., et al. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J. Antimicrob. Chemother. 2004;53(6):1054–1061. doi: 10.1093/jac/dkh223. [DOI] [PubMed] [Google Scholar]
- 162.Jain R., Beckett V.V., Konstan M.W. KB001-A, a novel anti-inflammatory, found to be safe and well-tolerated in cystic fibrosis patients infected with Pseudomonas aeruginosa. J. Cyst. Fibros. 2018;17(4):484–491. doi: 10.1016/j.jcf.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 163.Kollberg H., Carlander D., Olesen H., et al. Oral administration of specific yolk antibodies (IgY) may prevent Pseudomonas aeruginosa infections in patients with cystic fibrosis: a phase I feasibility study. Pediatr. Pulmonol. 2003;35(6):433–440. doi: 10.1002/ppul.10290. [DOI] [PubMed] [Google Scholar]
- 164.Lu Q., Rouby J.J., Laterre P.F., et al. Pharmacokinetics and safety of panobacumab: specific adjunctive immunotherapy in critical patients with nosocomial Pseudomonas aeruginosa O11 pneumonia. J. Antimicrob. Chemother. 2011;66(5):1110–1116. doi: 10.1093/jac/dkr046. [DOI] [PubMed] [Google Scholar]
- 165.Kaneko Y., Thoendel M., Olakanm O., et al. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 2007;117(4):877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yang F., Gu J., Yang L., et al. Protective efficacy of the trivalent Pseudomonas aeruginosa vaccine candidate PcrV-OprI-Hcp1 in murine pneumonia and burn models. Sci. Rep. 2017;7(1):3957. doi: 10.1038/s41598-017-04029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hassan R., El-Naggar W., Abd El-Aziz A.M., et al. Immunization with outer membrane proteins (OprF and OprI) and flagellin B protects mice from pulmonary infection with mucoid and nonmucoid Pseudomonas aeruginosa. J. Microbiol. Immunol. Infect. 2018;51(3):312–320. doi: 10.1016/j.jmii.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 168.Yang W., Wei Q., Tong Q., et al. Traditional Chinese medicine tanreqing inhibits quorum sensing systems in Pseudomonas aeruginosa. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.517462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fu B., Wu Q., Dang M., et al. Inhibition of Pseudomonas aeruginosa biofilm formation by traditional Chinese medicinal herb Herba patriniae. Biomed. Res. Int. 2017 doi: 10.1155/2017/9584703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hagens S., Habel A., von Ahsen U., et al. Therapy of experimental Pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob. Agents Chemother. 2004;48(10):3817–3822. doi: 10.1128/AAC.48.10.3817-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pires D.P., Monteiro R., Mil-Homens D., et al. Designing P. aeruginosa synthetic phages with reduced genomes. Sci. Rep. 2021;11(1):2164. doi: 10.1038/s41598-021-81580-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pérez-Pérez M., Jorge P., Pérez Rodríguez G., et al. Quorum sensing inhibition in Pseudomonas aeruginosa biofilms: new insights through network mining. Biofouling. 2017;33(2):128–142. doi: 10.1080/08927014.2016.1272104. [DOI] [PubMed] [Google Scholar]
- 173.Müh U., Schuster M., Heim R., et al. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob. Agents Chemother. 2006;50(11):3674–3679. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ishida T., Ikeda T., Takiguchi N., et al. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl. Environ. Microbiol. 2007;73(10):3183–3188. doi: 10.1128/AEM.02233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Adonizio A., Kong K.F., Mathee K. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by South Florida plant extracts. Antimicrob. Agents Chemother. 2008;52(1):198–203. doi: 10.1128/AAC.00612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.O'Loughlin C.T., Miller L.C., Siryaporn A., et al. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA. 2013;110(44):17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Koh C.L., Sam C.K., Yin W.F., et al. Plant-derived natural products as sources of anti-quorum sensing compounds. Sensors. 2013;13(5):6217–6228. doi: 10.3390/s130506217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ahmed S.K.K.S., Rudden M., Smyth T.J., et al. Natural quorum sensing inhibitors effectively downregulate gene expression of Pseudomonas aeruginosa virulence factors. Appl. Microbiol. Biotechnol. 2019;103(8):3521–3535. doi: 10.1007/s00253-019-09618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ghosh R., Das A., Mallik S. Inhibition of quorum sensing in P.aeruginosa: a review. Indian J. Pharm.Sci. 2019;81(5):797–806. doi: 10.36468/pharmaceutical-sciences.573. [DOI] [Google Scholar]
- 180.Smyth A.R., Cifelli P.M., Ortori C.A., et al. Garlic as an inhibitor of Pseudomonas aeruginosa quorum sensing in cystic fibrosis–a pilot randomized controlled trial. Pediatr. Pulmonol. 2010;45(4):356–362. doi: 10.1002/ppul.21193. [DOI] [PubMed] [Google Scholar]
- 181.Shetye G.S., Singh N., Gao X., et al. Structures and biofilm inhibition activities of brominated furanones for Escherichia coli and Pseudomonas aeruginosa. MedChemComm. 2013;4(7):1079–1084. doi: 10.1039/C3MD00059A. [DOI] [Google Scholar]
- 182.Khan M.S., Zahin M., Hasan S., et al. Inhibition of quorum sensing regulated bacterial functions by plant essential oils with special reference to clove oil. Lett. Appl. Microbiol. 2009;49(3):354–360. doi: 10.1111/j.1472-765X.2009.02666.x. [DOI] [PubMed] [Google Scholar]
- 183.Akhand S.S., Pettit R.S., Gardner T.E., et al. New treatments in development for Pseudomonas aeruginosa infections in the lungs of individuals with cystic fibrosis. Orphan Drugs: Res. Revi. 2014;(4):71–81. doi: 10.2147/odrr.s50014. DOI.org/ [DOI] [Google Scholar]
- 184.Rathinam P., Viswanathan P. Anti-virulence potential of eugenol-rich fraction of Syzygium aromaticum against multidrug resistant uropathogens isolated from catheterized patients. Avicenna. J. Phytomed. 2018;8(5):416–431. [PMC free article] [PubMed] [Google Scholar]
- 185.Jack A.A., Khan S., Powell L.C., et al. Oligosaccharide-induced modification of the lasI-lasR and rhlI-rhlR quorum-sensing systems in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018;62(5):e02318–17. doi: 10.1128/AAC.02318-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sharma K K., Pagedar Singh A. Antibiofilm effect of DNase against single and mixed species biofilm. Foods. 2018;7(3):201:42. doi: 10.3390/foods7030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Warrener P., Varkey R., Bonnell J.C., et al. A novel anti-PcrV antibody providing enhanced protection against Pseudomonas aeruginosa in multiple animal infection models. Antimicrob. Agents Chemother. 2014;58(8):4384–4391. doi: 10.1128/AAC.02643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Braddock M. Fighting P.aeruginosa infection in patients with cystic fibrosis: orphan drug designation for a novel PcrV antibody. Expert Opin. Orphan Drugs. 2014:201–204. doi: 10.1517/21678707.2014.882765. [DOI] [Google Scholar]
- 189.Secher T., Fas S., Fauconnier L., et al. The anti-Pseudomonas aeruginosa antibody Panobacumab is efficacious on acute pneumonia in neutropenic mice and has additive effects with meropenem. PloS one. 2013;8(9):e73396. doi: 10.1371/journal.pone.0073396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Que Y.A., Lazar H., Wolff M., et al. Assessment of panobacumab as adjunctive immunotherapy for the treatment of nosocomial Pseudomonas aeruginosa pneumonia. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33(10):1861–1867. doi: 10.1007/s10096-014-2156-1. [DOI] [PubMed] [Google Scholar]
- 191.Smith W.D., Bardin E., Cameron L., et al. Current and future therapies for Pseudomonas aeruginosa infection in patients with cystic fibrosis. FEMS Microbiol. Lett. 2017;364(14) doi: 10.1093/femsle/fnx121. [DOI] [PubMed] [Google Scholar]
- 192.Banin E., Lozinski A., Brady K.M., et al. The potential of desferrioxamine-gallium as an anti-Pseudomonas therapeutic agent. Proc. Natl. Acad. Sci. USA. 2008;105(43):16761–16766. doi: 10.1073/pnas.0808608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Smith D.J., Lamont I.L., Anderson G.J., et al. Targeting iron uptake to control Pseudomonas aeruginosa infections in cystic fibrosis. Eur. Respir. J. 2013;42(6):1723–1736. doi: 10.1183/09031936.00124012. [DOI] [PubMed] [Google Scholar]
- 194.Singh P.K. Iron sequestration by human lactoferrin stimulates P. aeruginosa surface motility and blocks biofilm formation. Biometals. 2004;17(3):267–270. doi: 10.1023/b:biom.0000027703.77456.27. [DOI] [PubMed] [Google Scholar]
- 195.Goss C.H., Kaneko Y., Khuu L., et al. Gallium disrupts bacterial iron metabolism and has therapeutic effects in mice and humans with lung infections. Sci. Transl. Med. 2018;10(460):eaat7520. doi: 10.1126/scitranslmed.aat7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Halwani M., Blomme S., Suntres Z.E., et al. Liposomal bismuth-ethanedithiol formulation enhances antimicrobial activity of tobramycin. Int. J. Pharm. 2008;358(1-2):278–284. doi: 10.1016/j.ijpharm.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 197.Moreau-Marquis S., O'Toole G.A., Stanton B.A. Tobramycin and FDA-approved iron chelators eliminate Pseudomonas aeruginosa biofilms on cystic fibrosis cells. Am. J. Respir. Cell Mol. Biol. 2009;41(3):305–313. doi: 10.1165/rcmb.2008-0299OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tümmler B. Emerging therapies against infections with Pseudomonas aeruginosa. F1000Res. 2019;8:Rev–1371. doi: 10.12688/f1000research.19509.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Döring G., Meisner C., Stern M., et al. A double-blind randomized placebo-controlled phase III study of a Pseudomonas aeruginosa flagella vaccine in cystic fibrosis patients. Proc. Natl. Acad. Sci. USA. 2007;104(26):11020–11025. doi: 10.1073/pnas.0702403104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Baumann U., Mansouri E., von Specht B.U. Recombinant OprF-OprI as a vaccine against Pseudomonas aeruginosa infections. Vaccine. 2004;22(7):840–847. doi: 10.1016/j.vaccine.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 201.Rello J., Valenzuela-Sánchez F., Ruiz-Rodriguez M., et al. Sepsis: a review of advances in management. Adv. Ther. 2017;34(11):2393–2411. doi: 10.1007/s12325-017-0622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wan C., Gao C., Xie Q., et al. Flagella hook protein FlgE is a novel vaccine candidate of Pseudomonas aeruginosa identified by a genomic approach. Vaccine. 2021;39(17):2386–2395. doi: 10.1016/j.vaccine.2021.03.051. [DOI] [PubMed] [Google Scholar]
- 203.Roman-Cruz V.C., Miller S., Buhl C., et al. Adjuvanted bacteriophage vaccine targeting Pseudomonas aeruginosa. J. Immunol. 2020:204. doi: 10.4049/jimmunol.204.Supp.168.10. [DOI] [Google Scholar]
- 204.Elhag M., M.Alaagib R., Ahmed N.M., et al. Design of epitope-based peptide vaccine against pseudomonas aeruginosa fructose bisphosphate aldolase protein using immunoinformatics. J. Immunol. Res. 2020;2020 doi: 10.1155/2020/9475058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cabral M.P., Correia A., Vilanova M., et al. A live auxotrophic vaccine confers mucosal immunity and protection against lethal pneumonia caused by Pseudomonas aeruginosa. PLoS Pathog. 2020;16(2) doi: 10.1371/journal.ppat.1008311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bianconi I., Alcalá-Franco B., Scarselli M., et al. Genome-based approach delivers vaccine candidates against Pseudomonas aeruginosa. Front. Immunol. 2019;9:3021. doi: 10.3389/fimmu.2018.03021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Liu C., Pan X., Xia B., et al. Construction of a protective vaccine against lipopolysaccharide-heterologous pseudomonas aeruginosa strains based on expression profiling of outer membrane proteins during infection. Front. Immunol. 2018;9:1737. doi: 10.3389/fimmu.2018.01737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ahmed M.M., Eljakee J., Mahran T. Development of novel protective polyvalent irradiated pseudomonas. IJPPE. 2021;16:1–10. doi: 10.18052/www.scipress.com/IJPPE.16.1. [DOI] [Google Scholar]
- 209.Sommer R., Wagner S., Rox K., et al. Glycomimetic, orally bioavailable LecB inhibitors block biofilm formation of Pseudomonas aeruginosa. J. Am. Chem. Soc. 2018;140(7):2537–2545. doi: 10.1021/jacs.7b11133. [DOI] [PubMed] [Google Scholar]
- 210.Flockton T.R., Schnorbus L., Araujo A., et al. Inhibition of Pseudomonas aeruginosa biofilm formation with surface modified polymeric nanoparticles. Pathogens. 2019;8(2):55. doi: 10.3390/pathogens8020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Johansson E.M, Crusz S.A., Kolomiets E., et al. Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB. Chem. Biol. 2008;15(12):1249–1257. doi: 10.1016/j.chembiol.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 212.Grishin A.V., Krivozubov M.S., Karyagina A.S., et al. Pseudomonas aeruginosa lectins as targets for novel antibacterials. Acta Naturae. 2015;7(2):29–41. [PMC free article] [PubMed] [Google Scholar]
- 213.Chemani C., Imberty A., de Bentzmann S., et al. Role of LecA and LecB lectins in Pseudomonas aeruginosa-induced lung injury and effect of carbohydrate ligands. Infect. Immun. 2009;77(5):2065–2075. doi: 10.1128/IAI.01204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Savoia D. New perspectives in the management of Pseudomonas aeruginosa infections. Future Microbiol. 2014;9(7):917–928. doi: 10.2217/fmb.14.42. [DOI] [PubMed] [Google Scholar]
- 215.Tunney M.M., Payne J.E., McGrath S.J., et al. Activity of hypothiocyanite and lactoferrin (ALX-009) against respiratory cystic fibrosis pathogens in sputum. The J. Antimicrob. Chemother. 2018;73(12):3391–3397. doi: 10.1093/jac/dky357. [DOI] [PubMed] [Google Scholar]
- 216.Urizer G., AngelAguilar –Luis M., Lama-Odria M.C., et al. Antibacterial activity of five Peruvian medicinal plants against Pseudomonas aeruginosa. Asian Pac. J. Trop. Biomed. 2015;5(11):928–931929. doi: 10.1183/13993003.congress-2018. [DOI] [Google Scholar]
- 217.Sulakvelidze A., Alavidze Z., MorrisJr J.G. Bacteriophage therapy. Antimicrob. Agents. Chemother. 2001;45(3):649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Patel S.R., Verma A.K., Verma V.C., et al. Bacteriophage therapy-looking back in to the future. Méndez-Vilas, A. The battle against microbial pathogens: basic sci, technol adv educ prog.Badajoz: Formatex. 2015:284–294. [Google Scholar]
- 219.Doss J., Culbertson K., Hahn D., et al. A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Viruses. 2017;9(3):50. doi: 10.3390/v9030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Mathur M.D., Vidhani S., Mehndiratta P.L. Bacteriophage therapy: an alternative to conventional antibiotics. J. Assoc. Physicians India. 2003;51:593–596. [PubMed] [Google Scholar]
- 221.Loc-Carrillo C., Abedon S.T. Pros and cons of phage therapy. Bacteriophage. 2011;1(2):111–114. doi: 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Principi N., Silvestri E., Esposito S. Advantages and limitations of bacteriophages for the treatment of bacterial infections. Front. Pharmacol. 2019;10:513. doi: 10.3389/fphar.2019.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wright A., Hawkins C.H., Anggård E.E., et al. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009;34(4):349–357. doi: 10.1111/j.1749-4486.2009.01973.x. [DOI] [PubMed] [Google Scholar]
- 224.Marza J.A., Soothill J..S., Boydell P., et al. Multiplication of therapeutically administered bacteriophages in Pseudomonas aeruginosa infected patients. Burns. 2006;32(5):644–646. doi: 10.1016/j.burns.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 225.Jault P., Leclerc T., Jennes S., et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019;19(1):35–45. doi: 10.1016/S1473-3099(18)30482-1. [DOI] [PubMed] [Google Scholar]
- 226.Khawaldeh A., Morales S., Dillon B., et al. Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J. Med. Microbiol. 2011;60(11):1697–1700. doi: 10.1099/jmm.0.029744-0. [DOI] [PubMed] [Google Scholar]
- 227.Abdul-Hassan H.S., El-Tahan E., Massoud B., et al. Bacteriophage therapy of Pseudomonas burn wound sepsis. Ann. Medit. Burn Club. 1990:262–264. [Google Scholar]
- 228.Aslam S., Courtwright A.M., Koval C., et al. Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am. J. Transplant. 2019;19(9):2631–2639. doi: 10.1111/ajt.15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Chan B.K., Turner P.E., Kim S., et al. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health. 2018;2018(1):60–66. doi: 10.1093/emph/eoy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Haddad L, El, Harb C.P., Gebara M.A., et al. A systematic and critical review of bacteriophage therapy against multidrug-resistant ESKAPE organisms in humans. Clin. Infect. Dis. 2019;69(1):167–178. doi: 10.1093/cid/ciy947. [DOI] [PubMed] [Google Scholar]
- 231.Aghaee B.L., Khan Mirzaei M., Alikhani M.Y., et al. Improving the inhibitory effect of phages against pseudomonas aeruginosa isolated from a burn patient using a combination of phages and antibiotics. Viruses. 2021;13(2):334. doi: 10.3390/v13020334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Alkhulaifi M.M. Using Phage’s to exterminate biofilms. J. Med. Microbiol. Diagnosis. 2017;6(3):259. doi: 10.4172/2161-0703.1000259. [DOI] [Google Scholar]
- 233.Gupta P., Sarkar S., Das B., et al. Biofilm, pathogenesis and prevention- a journey to break the wall: a review. Arch. Microbiol. 2016;198(1):1–15. doi: 10.1007/s00203-015-1148-6. [DOI] [PubMed] [Google Scholar]
- 234.Newman J.W., Floyd R.V., Fothergill J.L. The contribution of Pseudomonas aeruginosa virulence factors and host factors in the establishment of urinary tract infections. FEMS Microbiol. Lett. 2017;364(15) doi: 10.1093/femsle/fnx124. [DOI] [PubMed] [Google Scholar]
- 235.Moradali M.F., Ghods S., Rehm B.H. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Overhage J., Campisano A., Bains M., et al. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008;76(9):4176–4182. doi: 10.1128/IAI.00318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.de la Fuente-Núñez C., Korolik V., Bains M., et al. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012;56(5):2696–2704. doi: 10.1128/AAC.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lin Q., Deslouches B., Montelaro R.C., et al. Prevention of ESKAPE pathogen biofilm formation by antimicrobial peptides WLBU2 and LL37. Int. J. Antimicrob. Agents. 2018;52(5):667–672. doi: 10.1016/j.ijantimicag.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Martinez M., Gonçalves S., Felício M.R., et al. Synergistic and antibiofilm activity of the antimicrobial peptide P5 against carbapenem-resistant Pseudomonas aeruginosa. Biochim. Biophys. Acta Biomembr. 2019;1861(7):1329–1337. doi: 10.1016/j.bbamem.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 240.Yu Z., Kong Y., Luo Z., et al. Anti-bacterial activity of mutant chensinin-1 peptide against multidrug-resistant Pseudomonas aeruginosa and its effects on biofilm-associated gene expression. Exp. Ther. Med. 2019;17(3):2031–2038. doi: 10.3892/etm.2019.7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.O'Loughlin C.T., Miller L.C., Siryaporn A., et al. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. 2013;110(44):17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Maura D., Rahme L.G. Pharmacological inhibition of the Pseudomonas aeruginosa MvfR quorum-sensing system interferes with biofilm formation and potentiates antibiotic-mediated biofilm disruption. Antimicrob. Agents Chemother. 2017;61(12):e01362–17. doi: 10.1128/AAC.01362-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Fong J., Mortensen K.T., Nørskov A., et al. itaconimides as novel quorum sensing inhibitors of Pseudomonas aeruginosa. Front. Cell Infect. Microbiol. 2019;8:443. doi: 10.3389/fcimb.2018.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Singh N., Romero M., Travanut A., et al. Dual bioresponsive antibiotic and quorum sensing inhibitor combination nanoparticles for treatment of Pseudomonas aeruginosa biofilms in vitro and ex vivo. Biomater. Sci. 2019;7(10):4099–4111. doi: 10.1039/c9bm00773c. [DOI] [PubMed] [Google Scholar]
- 245.Liu L., Li J.H., Zi S.F., et al. AgNP combined with quorum sensing inhibitor increased the antibiofilm effect on Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2019;103(15):6195–6204. doi: 10.1007/s00253-019-09905-w. [DOI] [PubMed] [Google Scholar]
- 246.Ouyang J., Sun F., Feng W., et al. Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J. Appl. Microbiol. 2016;120(4):966–974. doi: 10.1111/jam.13073. [DOI] [PubMed] [Google Scholar]
- 247.Mettrick K., Hassan K., Lamont I., et al. The Iron-chelator, N,N'-bis (2-hydroxybenzyl) Ethylenediamine-N,N'-Diacetic acid is an effective colistin adjunct against clinical strains of biofilm-dwelling pseudomonas aeruginosa. Antibiotics 2020;.9(4):144. doi: 10.3390/antibiotics9040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lamppa J.W., Griswold K.E. Alginate lyase exhibits catalysis-independent biofilm dispersion and antibiotic synergy. Antimicrob. Agents Chemother. 2013;57(1):137–145. doi: 10.1128/AAC.01789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Pleszczyńska M., Wiater A., Janczarek M., et al. 1→3)-α-D-Glucan hydrolases in dental biofilm prevention and control: a review. Int. J. Biol. Macromol. 2015;79:761–778. doi: 10.1016/j.ijbiomac.2015.05.052. [DOI] [PubMed] [Google Scholar]
- 250.Baker P., Hill P.J., Snarr B.D., et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2016;2(5) doi: 10.1126/sciadv.1501632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Eckhart L., Fischer H., Barken K.B., et al. DNase1L2 suppresses biofilm formation by Pseudomonas aeruginosa and Staphylococcus aureus. Br. J. Dermatol. 2007;156(6):1342–1345. doi: 10.1111/j.1365-2133.2007.07886.x. [DOI] [PubMed] [Google Scholar]
- 252.Okshevsky M., Regina V.R., Meyer R.L. Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 2015;33:73–80. doi: 10.1016/j.copbio.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 253.Novotny L.A., Jurcisek J.A., Goodman S.D., et al. Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo. EBioMedicine. 2016;10:33–44. doi: 10.1016/j.ebiom.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhu X., Oh H.S., Ng Y.C.B., et al. Nitric oxide-mediated induction of dispersal in pseudomonas aeruginosa biofilms is inhibited by flavohemoglobin production and is enhanced by imidazole. Antimicrob. Agents Chemother. 2018;62(3) doi: 10.1128/AAC.01832-17. e01832-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Beirão S., Fernandes S., Coelho J., et al. Photodynamic inactivation of bacterial and yeast biofilms with a cationic porphyrin. Photochem. Photobiol. 2014;90(6):1387–1396. doi: 10.1111/php.12331. [DOI] [PubMed] [Google Scholar]
- 256.Orlandi V.T., Rybtke M., Caruso E., et al. Antimicrobial and anti-biofilm effect of a novel BODIPY photosensitizer against Pseudomonas aeruginosa PAO1. Biofouling. 2014;30(8):883–891. doi: 10.1080/08927014.2014.940921. [DOI] [PubMed] [Google Scholar]
- 257.Al-Bakri A.G., Mahmoud N.N. Photothermal-induced antibacterial activity of gold nanorods loaded into polymeric hydrogel against pseudomonas aeruginosa biofilm. Molecules. 2019;24(14):2661. doi: 10.3390/molecules24142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Fu B., Wu Q., Dang M., et al. Inhibition of Pseudomonas aeruginosa biofilm formation by traditional Chinese medicinal herb Herba patriniae. Biomed. Res. Int. 2017 doi: 10.1155/2017/9584703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hayashi M., Kaneko H., Yamada T., et al. Chinese herbal medicines and nutraceuticals inhibit Pseudomonas aeruginosa biofilm formation. Access Microbiol. 2021;3(8) doi: 10.1099/acmi.0.000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Mi L., Liu Y., Wang C., et al. Identification of a lytic Pseudomonas aeruginosa phage depolymerase and its anti-biofilm effect and bactericidal contribution to serum. Virus genes. 2019;55(3):394–405. doi: 10.1007/s11262-019-01660-4. [DOI] [PubMed] [Google Scholar]
- 261.Guo Y., Chen P., Lin Z., et al. Characterization of two Pseudomonas aeruginosa viruses vB_PaeM_SCUT-S1 and vB_PaeM_SCUT-S2. Viruses. 2019;11(4):318. doi: 10.3390/v11040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Olszak T., Shneider M.M., Latka A., et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci. Rep. 2017;7(1):16302. doi: 10.1038/s41598-017-16411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Pei R., Lamas-Samanamud G.R. Inhibition of biofilm formation by T7 bacteriophages producing quorum-quenching enzymes. Appl. Environ. Microbiol. 2014;80(17):5340–5348. doi: 10.1128/AEM.01434-14. [DOI] [PMC free article] [PubMed] [Google Scholar]