Abstract
Mycoplasma pneumoniae proteins HMW1-HMW3 collectively are essential for cytadherence, but the function or requirement for each has not been defined. Cytadherence mutant M6 lacks HMW1 because of a frameshift in hmw1 and produces a truncated adherence-associated protein P30 because of a deletion at the 3′ end of p30. Genetic manipulation of this mutant was used to evaluate the role of HMW1 in cytadherence. Mutant M6 was transformed with a recombinant transposon containing a wild-type p30 allele. Transformants synthesized both truncated and full-length P30, from the resident and recombinant alleles, respectively. However, these transformants remained hemadsorption negative, suggesting that HMW1 is required for cytadherence. Wild-type M. pneumoniae cells are generally elongated, tapering to form the attachment organelle at one end of the cell. The cytadhesin protein P1 is normally densely clustered on the mycoplasma surface at this differentiated terminal structure. However, both mutant M6 and M6 transformed with recombinant p30 had a striking ovoid morphology with no tapering at the tip structure, making the attachment organelle indistinguishable. Furthermore, protein P1 was randomly distributed on the mycoplasma surface rather than clustered at a polar location. In contrast, mutant M6 transformed with a recombinant transposon expressing the wild-type hmw1 allele exhibited a near-normal morphology and localized P1 to the attachment organelle. Significantly, M6 transformed with an hmw1 gene truncated slightly at the 3′ end failed to restore proper morphology or P1 localization to the attachment organelle, suggesting a functional importance to the C-terminal domain of HMW1.
The cell wall-less prokaryote Mycoplasma pneumoniae causes tracheobronchitis and “walking pneumonia” in humans. Cytadherence is a critical step in M. pneumoniae colonization of the respiratory mucosa and is mediated largely by the attachment organelle, a polar, tapered extension of the mycoplasma cell that is distinguished by an electron-dense intracytoplasmic core (14, 25). Protein P1 (12) is densely clustered at the attachment organelle, where it binds host cell receptors directly (15, 25). P1 is also found widely scattered on the mycoplasma surface (1), but it is not known whether the adhesin not associated with the tip structure in wild-type mycoplasmas is functional.
The analysis of spontaneously arising noncytadhering mutants (17) has identified other mycoplasma proteins associated with cytadherence (Table 1) (14), but in what is believed to be an accessory role. Protein P30, for example, is likewise localized to the attachment organelle (2). While there is evidence that P30 may function as an adhesin (2), a mutant lacking P30 (II-3 [Table 1]) is able to traffic P1 to the tip structure but remains noncytadherent, raising the possibility that P30 is required in order for P1 to be functional (26). Mutant II-3 also exhibits striking morphological abnormalities, indicating a possible developmental defect (26). Significantly, complementation with a cloned wild-type p30 gene restored cytadherence and a wild-type morphology, underscoring the correlation between P30 and these phenotypic changes (26).
TABLE 1.
Protein profile and hemadsorption phenotype of wild-type and mutant M. pneumoniae strains
| Strain | Result for proteina:
|
HA phenotypea | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P1 | P30 | P30′b | HMW1 | HMW1′c | HMW2 | HMW3 | |||
| Wild type | +++ | +++ | − | +++ | NA | +++ | +++ | +++ | 20 |
| Class I | +++ | +++ | − | +/− | NA | − | +/− | − | 17 |
| II-3 | +++ | − | − | +++ | NA | +++ | +++ | − | 3, 17 |
| II-7 | +++ | − | +++ | +++ | NA | +++ | +++ | − | 3, 17 |
| II-7/P30 | +++ | +++ | +++ | +++ | NA | +++ | +++ | +++ | 25 |
| M6 | +++ | − | +++ | − | NA | +++ | +++ | − | 19 |
| M6/P30 | +++ | +++ | +++ | − | NA | +++ | +++ | − | This study |
| M6/HMW1 | +++ | − | +++ | +++ | NA | +++ | +++ | − | This study |
| M6/HMW1′ | +++ | − | +++ | − | +++ | +++ | +++ | − | This study |
| crl mutantd | +++ | +++ | − | +/− | NA | − | +/− | − | 10, 18 |
+++, protein present; −, protein absent; NA, not applicable; +/−, protein present at significantly reduced level.
P30′, truncated resident P30 resulting from a deletion near the 3′ end of the gene.
HMW1′, truncated recombinant HMW1 resulting from a deletion at the 3′ end of the gene.
crl, cytadherence regulatory locus.
Spontaneous loss of proteins HMW1, HMW2, and HMW3 (class I mutants [Table 1]) (17), on the other hand, is accompanied by an inability to cluster P1 at the attachment organelle (1) and what appears to be slower processing of the leader peptide from the P1 precursor to the mature protein (22). The HMW proteins are components of the Triton X-100-insoluble, mycoplasma cytoskeleton and as such are thought to have a scaffolding or structural role in P1 localization. HMW3 is found at the terminal button of the attachment organelle (29), while HMW1 localizes along the filamentous extensions of the mycoplasma cell (28). Preliminary studies indicate that HMW2 is present near the base of the terminal organelle (6). While their genes have been sequenced, the deduced structural features of HMW1 to HMW3 reveal little about their likely roles as accessory proteins in cytadherence (5, 18, 21).
The focus of this study is HMW1 and its role in P1 trafficking and cytadherence. While antibody accessible on the mycoplasma surface (5, 7, 28), HMW1 is also phosphorylated (4), indicating a likely cytoplasmic domain and, therefore, a transmembrane configuration (7). However, HMW1 is predicted to be largely hydrophilic, with no typical membrane-spanning regions, based upon its deduced amino acid sequence (5), establishing a paradoxical complexity of the membrane topography of HMW1 and shedding no light on probable function. The requirement for HMW1 in cytadherence was recently examined by genetic complementation with recombinant Tn4001 to deliver the cloned hmw1 gene into wild-type and mutant backgrounds (8). Recombinant HMW1 truncated at the C terminus was produced at wild-type levels in a class I mutant but failed to restore cytadherence. For reasons not understood at the time, full-length recombinant HMW1 was never produced at wild-type levels in class I transformants. However, subsequent studies established that the loss of HMW1 in the class I mutant is due to accelerated turnover by what appears to be a proteolytic mechanism targeting the C terminus of the protein (22). The genetic locus responsible for maintaining HMW1 and HMW3 at stable levels in M. pneumoniae was defined by transposon mutagenesis (crl [Table 1]), and transposon insertions were mapped to the gene for HMW2 (10, 18). Because the loss of HMW1 occurs proteolytically in class I and crl mutants, its functional analysis by complementation requires a mutant background in which HMW1 is absent but HMW2 is present.
The cytadherence mutant M6 (Table 1) has a frameshift mutation in the gene for HMW1 and a deletion at the 3′ end of the p30 gene, yielding no HMW1 and a truncated P30 (P30′) (19). In the current study, transposon delivery of recombinant p30 or hmw1 genes into mutant M6 permitted evaluation of HMW1 function more directly. The results clearly indicate a requirement for HMW1 in the development of the attachment organelle and P1 trafficking to this structure and suggest that HMW1 is essential for cytadherence. Furthermore, M6 transformants with recombinant hmw1 having a minor deletion at the 3′ end were indistinguishable from untransformed M6 with respect to morphology and P1 localization, strongly suggesting a functional significance to the C-terminal domain of HMW1.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The M. pneumoniae strains used in this study are summarized in Table 1. All are derived from the virulent, wild-type strain M129-B17 (20). Mycoplasmas were cultured in Hayflick medium (9) at 37°C until mid-log phase and harvested as detailed elsewhere (8). For growth on solid medium to enumerate CFU or isolate individual colonies, mycoplasma suspensions were spread on PPLO agar and incubated at 37°C for 7 to 9 days. Plates were then overlaid with 20% (vol/vol) sheep blood (Carr-Scarborough Microbiological, Inc., Decatur, Ga.) in 1% noble agar (Difco Laboratories; Detroit, Mich.)–saline and incubated for 2 days at 37°C. CFU were determined on the basis of hemolytic plaque number.
Mycoplasma mutants M6 and II-7 were transformed by electroporation (11) with the recombinant Tn4001 derivatives described below. Briefly, mycoplasmas suspended in cold HEPES-sucrose buffer (8 mM HEPES–272 mM sucrose [pH 7.4]) at approximately 107 to 108 CFU in 100 μl were mixed with 3 to 10 μg of recombinant or control pISM2062 (13) and placed on ice for 10 min. After electroporation, the mycoplasmas were plated on PPLO agar containing 18 μg of gentamicin/ml. Individual transformants were carefully picked from plates with sterile Pasteur pipettes and cultured in Hayflick medium containing gentamicin until a color change occurred in the phenol red indicator (generally 7 to 12 days).
Cells of Escherichia coli Sure (Stratagene; La Jolla, Calif.) were grown in Luria broth and prepared as competent cells for transformation (27). Plasmid pISM2062, derived from pMB1 and containing the Staphylococcus aureus transposon Tn4001mod (13), was generously provided by C. Minion (Iowa State University). Tn4001mod contains unique BamHI and SmaI sites 28 bp from the end of the IS256L arm (Fig. 1D). For some constructions, Tn4001mod was altered by creating an EcoRI site in place of the SmaI site by using a short oligonucleotide linker. E. coli cells harboring these plasmids were cultured at 37°C in Luria broth containing ampicillin (50 μg/ml).
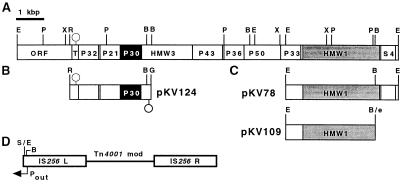
FIG. 1.
Construction of recombinant Tn4001 derivatives containing the cloned wild-type M. pneumoniae gene for P30 or HMW1. (A) Restriction map of the hmw locus. The genes for P30 and HMW1 are designated in black and gray, respectively. The genes encoding HMW3 and ribosomal protein S4 and several predicted open reading frame (ORF) products from this gene cluster are indicated. The circle corresponds to a predicted terminator upstream of p30. B, BamHI; E, EcoRI; P, PstI; R, EcoRV; X, XbaI. (B) pKV124 was constructed as described in detail previously (26). Briefly, an EcoRV-BamHI fragment containing the wild-type p30 gene and an oligonucleotide containing a terminator sequence were cloned into the SmaI site (S) of Tn4001mod (D) (13). G, BglII. (C) Cloning of the intact (pKV78) or truncated (pKV109) hmw1 gene into Tn4001mod. The construction of pKV78 was described in detail previously (8). For the construction of pKV109, the single-stranded end at the BamHI site was made double stranded with the Klenow fragment and deoxynucleoside triphosphates, and an EcoRI linker (e) was added to permit cloning into the EcoRI site of Tn4001mod. This made it possible to orient the insert such that hmw1 was transcribed from an outward-reading promoter in IS256L (8).
Characterization of mycoplasma transformants.
Hemadsorption (HA) correlates well with adherence to respiratory epithelium and was used as an indicator of cytadherence. Transformants were screened microscopically for HA as described elsewhere (8). Mycoplasma proteins were analyzed by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 3% polyacrylamide stacking gel and a 4.5, 12, or 15% polyacrylamide separating gel (8). Samples were heated for 15 min at 68°C prior to electrophoresis. For Western blots (30), proteins were transferred to nitrocellulose membranes (MSI, Westborough, Mass.) and probed with anti-HMW1 serum (8, 28) or monoclonal anti-P30 antibodies (2, 24) as described previously (8). Analysis of M. pneumoniae transformants by Southern blot hybridization was carried out by standard techniques (27). Mycoplasma DNA was extracted with phenol as described previously (8, 21).
SEM and immunoelectron microscopy.
Mycoplasma cell pellets were suspended in fresh Hayflick medium, passed repeatedly through a 25-gauge needle to disperse the cells, and filtered (1.2-μm pore diameter) to remove aggregates. These suspensions were used to inoculate 24-well dishes containing either Formvar-coated, carbon-coated nickel grids (for antibody labeling and transmission electron microscopy [TEM]) or glass coverslips (for scanning electron microscopy [SEM]). Grids and coverslips were precoated with poly-l-Lys (29), to promote attachment of the mutants to these surfaces, and UV sterilized. After 1 to 4 h at 37°C, grids and coverslips were removed and processed for fixation and antibody labeling (28, 29). Briefly, for SEM, coverslips were transferred to fixative (1% paraformaldehyde, 1% glutaraldehyde, 0.1% picric acid, 0.1 M sodium cacodylate [pH 7.4]) for 1 h at 4°C and then were rinsed twice with 25 mM HEPES–0.8% NaCl and once with 0.1 M sodium cacodylate buffer (pH 7.4). Coverslips were then treated sequentially (10 min each) with 30, 50, 70, 85, and 95% ethanol; treated twice with 100% ethanol; and critical point dried. Chromium was evaporated over the samples, which were then examined with a Leo 982 field emission SEM. For TEM, grids were rinsed with 25 mM HEPES–0.8% NaCl (pH 7.4) and fixed in 1% glutaraldehyde–1% paraformaldehyde–0.1% picric acid–0.1 M sodium cacodylate (pH 7.4) for 1 h at 4°C. Grids were then rinsed well with 25 mM HEPES–0.8% NaCl and blocked in 4% calf serum in 0.2 M Tris-HCl (pH 8.2)–0.8% NaCl for at least 30 min at room temperature in a moisture chamber. The primary antibody was mouse monoclonal anti-P1 immunoglobulin (generously provided by S. Geary, University of Connecticut), which was used at a dilution of 1:75 to 1:500 overnight at 4°C. The secondary antibody was 10-nm-diameter colloidal gold-conjugated goat anti-mouse immunoglobulin (Amersham, Arlington Heights, Ill.), which was used at a dilution of 1:10. Grids were rinsed well with 25 mM HEPES–0.8% NaCl, counterstained in 2% aqueous uranyl acetate, and examined with a JEOL 100CX II TEM.
RESULTS AND DISCUSSION
Evidence that HMW1 is required for cytadherence.
Initial attempts at genetic complementation in M. pneumoniae involved transformation of the class I mutant with a recombinant transposon carrying the cloned wild-type hmw1 gene (8). Recombinant HMW1 that was truncated slightly at the C terminus was produced at wild-type levels in this background, but full-length recombinant HMW1 was not. Subsequent studies established that HMW1 is actually synthesized at a normal rate in the class I mutant, but its turnover, by what appears to be a proteolytic mechanism targeting the C terminus of the protein, is accelerated (22). This would account for the inability to restore HMW1 levels in the class I mutant, because recombinant HMW1 would be expected to be turned over by the same mechanism as native HMW1.
The cytadherence mutant M6 produces a truncated P30 due to a deletion in the 3′ end of p30 and lacks HMW1 because of a frameshift in hmw1 (19). The current study utilized mutant M6 in order to explore further the requirement for HMW1 in cytadherence. Transformation of this mutant with recombinant Tn4001 containing the cloned wild-type gene for P30 should restore P30 to wild-type levels (26), yielding a construct lacking only HMW1 and making it possible to assess indirectly the requirement for HMW1 in cytadherence. Figure 1A shows the gene organization of the hmw locus, with the genes for P30 and HMW1 highlighted. Figure 1B indicates the restriction fragment containing the wild-type p30 allele that was cloned into the modified Tn4001 and electroporated into M6 to yield M6/P30 transformants.
Transformants were analyzed by Southern blot hybridization, confirming random insertion of the recombinant transposon and demonstrating the presence of both resident and recombinant p30 alleles (data not shown). Examination of these transformants by Western immunoblotting with P30-specific antibodies established that each synthesized both the recombinant full-length P30 and the resident truncated P30 (Fig. 2A). Previous studies demonstrated that truncation at the C terminus of P30 is sufficient for loss of cytadherence (3). Because it was not known if the truncated P30 was dominant over full-length P30 with respect to cytadherence, we also examined mutant II-7 transformed with recombinant Tn4001 carrying the cloned wild-type gene for P30 (II-7/P30) (26). The II-7/P30 and M6/P30 transformants should differ only in the presence or absence of HMW1, respectively (Table 1). To control for positional effects associated with transposon insertion, several transformants were examined for each construct. As expected, mutants M6 and II-7 and the M6/P30 transformants tested were HA− (data not shown). The inability of M6/P30 transformants to cytadhere might have resulted from interference with the function of full-length recombinant P30 by the truncated resident P30. However, HA is restored in mutant II-7 when transformed with recombinant p30 (26), demonstrating that the wild-type allele is dominant over its truncated counterpart. Therefore, the truncated P30 was probably not responsible for the HA− phenotype of M6/P30 transformants. Taken together, these observations strongly suggest an absolute requirement for HMW1 in cytadherence, although we cannot rule out the presence of another mutation in M6 conferring the HA− phenotype. Attempts to isolate an HMW1+ revertant of M6/P30 in order to address this possibility were unsuccessful, suggesting that reversion of the hmw1 mutation occurs at a lower frequency than reversion in class I mutants (16). Definitive confirmation that HMW1 is required for adherence will require introduction of both the p30 and hmw1 alleles into M6 via recombinant transposon delivery.
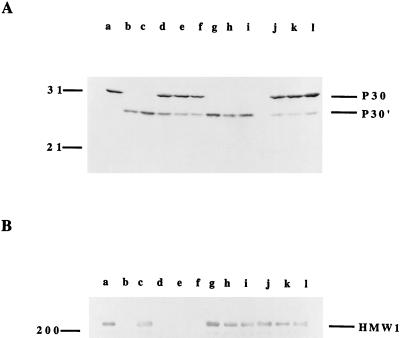
FIG. 2.
Western immunoblot analysis of P30 and HMW1 production in M. pneumoniae transformed with recombinant Tn4001mod. Mycoplasma preparations were analyzed by SDS-PAGE, transferred to nitrocellulose membranes, and probed with antibodies as indicated. (A) Mycoplasma proteins were separated on a 15% polyacrylamide gel, and the corresponding blot was probed with polyclonal anti-P30 antiserum (19). Lanes: a, wild-type M. pneumoniae; b, M6 mutant; c, II-7 mutant; d to f, M6/P30 transformants; g to i, M6/HMW1 transformants; j to l, II-7/P30 transformants. (B) Mycoplasma proteins were separated on a 4.5% polyacrylamide gel, and the corresponding blot was probed with polyclonal anti-HMW1 antiserum (28); the lanes are the same as those in panel A. The positions of protein size standards (in kilodaltons) are shown to the left, while P30, P30′, and HMW1 are indicated to the right.
HMW1 is essential for P1 localization to the attachment organelle.
Efforts to examine HMW1 function by expressing the wild-type hmw1 allele in the class I mutant background were precluded by the accelerated turnover of HMW1 in this mutant due to the loss of HMW2 (8, 22). However, the M6 mutant produces HMW2 and provides a suitable background in which to study phenotypic changes associated with introduction of a wild-type hmw1 allele. Figure 1C shows the restriction fragments containing an intact hmw1 gene or an hmw1 gene slightly truncated at the 3′ end that were cloned into the modified Tn4001 and transformed into mutant M6. The synthesis of HMW1 by M6/HMW1 transformants was established by Western immunoblotting (Fig. 2B) (22). Despite the production of HMW1 at wild-type levels, the M6/HMW1 transformants remained HA−, as expected, because of the lack of full-length P30.
The requirement for HMW1 in the localization of the adhesin P1 to the attachment organelle of M. pneumoniae was examined by immunogold labeling. As expected (1), P1 was generally densely clustered at the terminal organelle in wild-type M. pneumoniae but was also present elsewhere on the mycoplasma surface to a lesser extent (Fig. 3A). In contrast, P1 was randomly distributed on the mycoplasma surface in mutant M6 (Fig. 3C) and M6/P30 transformants (Fig. 3F), which were also morphologically strikingly different from wild-type M. pneumoniae (see below). Some M6/P30 cells exhibited a patchy labeling pattern (Fig. 3F). The significance of this is not clear, but it may reflect a distinction between the clustering of P1 and its localization to the attachment organelle. Full-length P30 is not required for P1 localization (mutant II-7 [Fig. 3D]) (26), suggesting that the inability to cluster P1 at the attachment organelle in mutant M6 was likely due to the absence of HMW1. The distribution of P1 in mutant M6 was similar to that described previously in class I mutants (1). The lack of HMW1 is a common denominator between the class I and M6 mutants, consistent with the conclusion that HMW1 is required for P1 trafficking to the attachment organelle in M. pneumoniae. This conclusion was confirmed directly by transformation of M6 with recombinant hmw1, which restored P1 clustering at the tip structure (Fig. 3E).
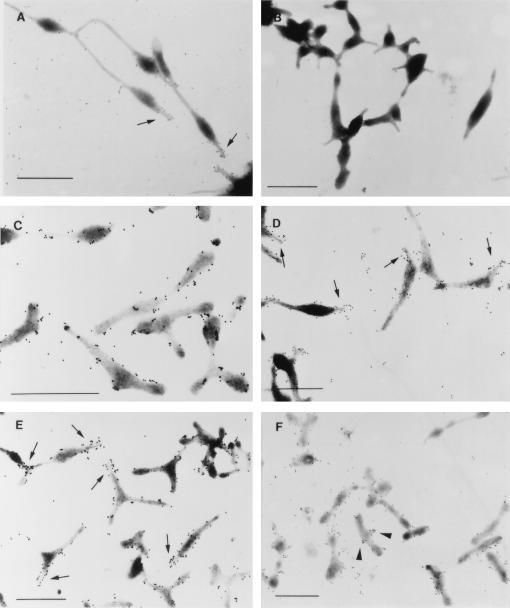
FIG. 3.
Immunogold labelling of wild-type, mutant, and transformant M. pneumoniae strains with anti-P1 antiserum. (A) Wild-type M. pneumoniae. (B) P1− mutant control (17). (C) Mutant M6. (D) Mutant II-7. (E) Mutant M6/HMW1. (F) Mutant M6/P30. Arrows indicate tip structures where high density of labeling was observed. Arrowheads indicate zones of dense antibody labeling on some M6/P30 cells. Bar, 1 μm.
In order to provide a quantitative assessment of P1 distribution, the gold particles labeling approximately 70 to 90 cells each of wild-type, M6 mutant, and M6 transformant M. pneumoniae cells were counted and classified as either polar or not polar. No attempt was made to correct for the substantially greater surface area represented by the not polar category. The counts were averaged for the total number of cells examined. The wild-type and M6/HMW1 cells had more polar than not polar gold particles (8.3 and 7.5 versus 5.6 and 6.0 particles per cell, respectively), despite the much greater surface area represented by the not polar category. In contrast, the M6 mutant had approximately threefold greater numbers of gold particles in the not polar category compared to the polar category (3.67 versus 13.9 particles per cell, respectively). These data were analyzed statistically by one-way analysis of variance and the Student-Newman-Keuls multiple comparisons test. A significant difference (P < 0.001) was noted in polar labeling between M6 and wild-type M. pneumoniae and between M6 and the M6/HMW1 transformant examined. These results reinforce the pattern noted visually, supporting a requirement for HMW1 in the localization of the adhesin P1 to the attachment organelle.
HMW1 is required for development of the attachment organelle.
The striking difference in morphology between wild-type, mutant, and transformant M. pneumoniae cells noted during examination by immunoelectron microscopy (Fig. 3) was evaluated further by SEM. As expected, wild-type cells were characteristically elongated, with a tapered terminal organelle (Fig. 4A). In contrast, mutant M6 (Fig. 4B) and M6/P30 transformants (Fig. 4C) were typically ovoid, with broad, rounded rather than tapered ends and with tip structures not readily identifiable. A near-wild-type morphology was restored when M6 was transformed with the recombinant wild-type hmw1 allele, with transformants exhibiting an elongated shape and tapering at the terminal organelle (Fig. 4D). Thus, a strong correlation was noted between the presence of HMW1 and proper formation of the tapered tip structure.
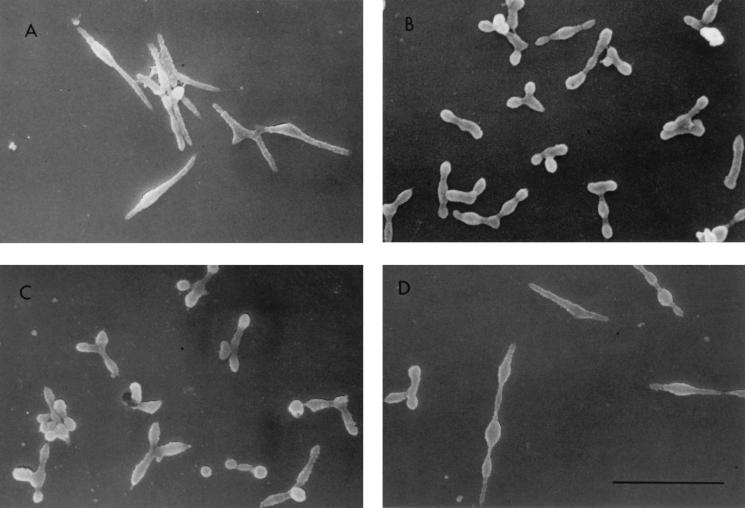
FIG. 4.
SEM analysis of wild-type, mutant, and transformant M. pneumoniae cells. (A) Wild-type M. pneumoniae. (B) Mutant M6. (C) M6/P30. (D) M6/HMW1. The elongated morphology with tapered tip structures was only observed with the wild-type and M6/HMW1 cells. Bar, 2 μm.
HMW1 is a component of the mycoplasma cytoskeleton and localizes largely along the filamentous extensions but not the cell body in M. pneumoniae (28). Based upon this subcellular distribution and its likely transmembrane topography (7), HMW1 may provide a linkage between the mycoplasma membrane and the cytoskeleton that is critical for proper development of the attachment organelle. Furthermore, based upon differences in P1 localization in the M6 mutant and M6/HMW1 transformants, HMW1 could be part of a molecular conduit for directing P1 to the terminal organelle. Significantly, P1 is synthesized as a precursor, the processing of which also appears to be impeded in the class I mutant (22). Preliminary findings suggest that the same is true in M6, but not in M6/HMW1 (23), in which case the loss of HMW1 confers a defect that encompasses both P1 processing and trafficking. Through additional analysis, it should be possible to begin reconstructing the route by which P1 proceeds from synthesis in precursor form to localization at the attachment organelle as a functional adhesin.
The C terminus of HMW1 is required for protein function.
The transformation of mutant M6 with a recombinant transposon expressing the cloned gene for HMW1 restored the ability to localize P1 to the attachment organelle and conferred a near-wild-type morphology. The C terminus of HMW1 is thought to be targeted for proteolytic turnover in class I and crl mutants (22) and might likewise be important in HMW1 function. In order to test this possibility, recombinant Tn4001 encoding a truncated HMW1 lacking 112 amino acid residues at the C terminus (pKV109 [Fig. 1C]) was transformed into mutant M6. Transformants produced truncated HMW1 (HMW1′) at approximately wild-type levels, as determined by Western immunoblotting (Fig. 5A). However, these M6/HMW1′ transformants failed to cluster P1 at the terminal organelle and were morphologically indistinguishable from the M6 mutant (Fig. 5B). Quantitative assessment of P1 distribution in M6/HMW1′ transformants yielded similar values to those noted above for M6 (4.2 polar particles compared to 10.4 nonpolar particles per cell). These observations strongly suggest a functional significance to the C-terminal domain of HMW1 in both P1 trafficking and development of the attachment organelle. This function might involve a direct interaction with P1 as it is translocated to the tip. Alternatively, the C terminus of HMW1 may be required for its targeting to the filamentous extensions of the mycoplasma cell, which might be required for subsequent trafficking of P1. Studies are in progress to distinguish these two possibilities.
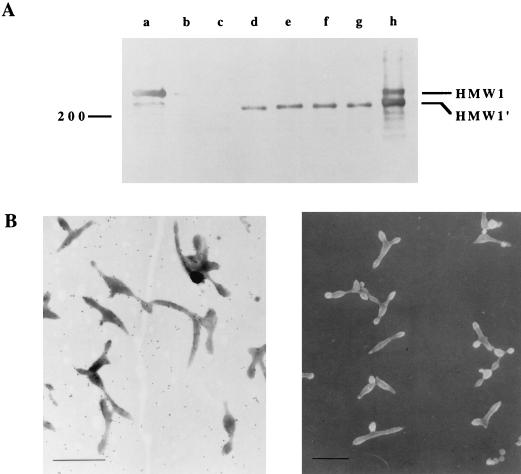
FIG. 5.
Characterization of M6/HMW1′ transformants by Western immunoblot analysis (A) and anti-P1 immunogold labeling and SEM (B). (A) Mycoplasma proteins were separated by SDS-PAGE with a 4.5% polyacrylamide gel and visualized on the corresponding blot by using anti-HMW1 antiserum. Lanes: a, wild-type M. pneumoniae; b, mutant M6; c, mutant M6/P30; d to g, mutant M6/HMW1′ transformants; h, wild-type M. pneumoniae transformed with HMW1′. The 200-kDa size standard is shown to the left, while HMW1 and HMW1′ are indicated to the right. (B, left panel) Immunogold labeling of M6/HMW1′ transformants with anti-P1 antibodies. Note that antibody labeling is scattered and not densely localized at a tip structure. (B, right panel) SEM analysis of M6/HMW1′ transformants. The cell morphology was comparable to that of M6 (Fig. 4B).
In conclusion, the results presented here demonstrate a clear requirement for the cytadherence-associated protein HMW1 in the assembly of the attachment organelle, including the trafficking of the adhesin P1 to this structure. Furthermore, HMW1 truncated at its C terminus lacks this functional capability, focusing additional interest on a region of the protein recently implicated in the regulation of HMW1 levels in the class I mutant (22). Using the techniques employed in the current study, as well as more direct analysis of protein-protein interactions involving HMW1 and the domains thereof, it should be possible to define more clearly how HMW1 effects P1 localization to the terminus and proper development of the attachment organelle.
ACKNOWLEDGMENTS
This work was supported by Public Health Service research grant AI23362 from the National Institute for Allergy and Infectious Diseases to D.C.K.
We thank C. Romero-Arroyo for providing the M6/P30 and II-7/P30 transformants and J. Baseman, R. Herrmann, and S. Geary for sharing antibodies to P30 and P1.
REFERENCES
- 1.Baseman J B, Cole R M, Krause D C, Leith D K. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J Bacteriol. 1982;151:1514–1522. doi: 10.1128/jb.151.3.1514-1522.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baseman J B, Morrison-Plummer J, Drouillard D, Puleo-Scheppke B, Tryon V V, Holt S C. Identification of a 32-kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr J Med Sci. 1987;23:474–479. [PubMed] [Google Scholar]
- 3.Dallo S F, Lazzell A L, Chavoya A, Reddy S P, Baseman J B. Biofunctional domains of the Mycoplasma pneumoniae P30 adhesin. Infect Immun. 1996;64:2595–2601. doi: 10.1128/iai.64.7.2595-2601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dirksen L B, Krebes K A, Krause D C. Phosphorylation of cytadherence-accessory proteins in Mycoplasma pneumoniae. J Bacteriol. 1994;176:7499–7505. doi: 10.1128/jb.176.24.7499-7505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dirksen L B, Proft T, Hilbert H, Plagens H, Herrmann R, Krause D C. Nucleotide sequence analysis and characterization of the hmw gene cluster of Mycoplasma pneumoniae. Gene. 1996;171:19–25. doi: 10.1016/0378-1119(96)00050-9. [DOI] [PubMed] [Google Scholar]
- 6.Fisseha, M., R. Herrmann, K. Birkhead, and D. C. Krause. Unpublished data.
- 7.Hahn, T.-W., and D. C. Krause. Unpublished data.
- 8.Hahn T-W, Krebes K A, Krause D C. Expression in Mycoplasma pneumoniae of the recombinant gene encoding the cytadherence-associated protein HMW1 and identification of HMW4 as a product. Mol Microbiol. 1996;19:1085–1093. doi: 10.1046/j.1365-2958.1996.455985.x. [DOI] [PubMed] [Google Scholar]
- 9.Hayflick, L. 1965. Tissue cultures and mycoplasmas. Tex. Rep. Biol. Med. 23(Suppl. 1):285–303. [PubMed]
- 10.Hedreyda C T, Krause D C. Identification of a possible cytadherence regulatory locus in Mycoplasma pneumoniae. Infect Immun. 1995;63:3479–3483. doi: 10.1128/iai.63.9.3479-3483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedreyda C T, Lee K K, Krause D C. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid. 1993;30:170–175. doi: 10.1006/plas.1993.1047. [DOI] [PubMed] [Google Scholar]
- 12.Hu P-C, Collier A M, Baseman J B. Surface parasitism by Mycoplasma pneumoniae of respiratory epithelium. J Exp Med. 1977;145:1328–1343. doi: 10.1084/jem.145.5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knudtson K L, Minion F C. Construction of Tn4001lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene. 1993;137:217–222. doi: 10.1016/0378-1119(93)90009-r. [DOI] [PubMed] [Google Scholar]
- 14.Krause D C. Mycoplasma pneumoniae cytadherence: unraveling the tie that binds. Mol Microbiol. 1996;20:247–253. doi: 10.1111/j.1365-2958.1996.tb02613.x. [DOI] [PubMed] [Google Scholar]
- 15.Krause D C, Baseman J B. Inhibition of Mycoplasma pneumoniae hemadsorption and adherence to respiratory epithelium by antibodies to a membrane protein. Infect Immun. 1983;39:1180–1186. doi: 10.1128/iai.39.3.1180-1186.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krause D C, Leith D K, Baseman J B. Reacquisition of specific proteins confers virulence in Mycoplasma pneumoniae. Infect Immun. 1983;39:830–836. doi: 10.1128/iai.39.2.830-836.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krause D C, Leith D K, Wilson R M, Baseman J B. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect Immun. 1982;35:809–817. doi: 10.1128/iai.35.3.809-817.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krause D C, Proft T, Hedreyda C T, Hilbert H, Plagens H, Herrmann R. Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J Bacteriol. 1997;179:2668–2677. doi: 10.1128/jb.179.8.2668-2677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Layh-Schmitt G, Hilbert H, Pirkl E. A spontaneous hemadsorption-negative mutant of Mycoplasma pneumoniae exhibits a truncated adhesin-related 30-kilodalton protein and lacks the cytadherence-accessory protein HMW1. J Bacteriol. 1995;177:843–846. doi: 10.1128/jb.177.3.843-846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipman R P, Clyde W A., Jr The interrelationship of virulence, cytadsorption and peroxide formation in Mycoplasma pneumoniae. Proc Soc Exp Biol Med. 1969;131:1163–1167. doi: 10.3181/00379727-131-34061. [DOI] [PubMed] [Google Scholar]
- 21.Ogle K F, Lee K K, Krause D C. Cloning and analysis of the gene encoding the cytadherence phase-variable protein HMW3 of Mycoplasma pneumoniae. Gene. 1991;97:69–75. doi: 10.1016/0378-1119(91)90011-y. [DOI] [PubMed] [Google Scholar]
- 22.Popham P L, Hahn T-W, Krebes K A, Krause D C. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs post-translationally. Proc Natl Acad Sci USA. 1997;94:13979–13984. doi: 10.1073/pnas.94.25.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popham, P. L., and D. C. Krause. Unpublished data.
- 24.Proft T, Herrmann R. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol Microbiol. 1994;13:337–348. doi: 10.1111/j.1365-2958.1994.tb00427.x. [DOI] [PubMed] [Google Scholar]
- 25.Razin S, Jacobs E. Mycoplasma adhesion. J Gen Microbiol. 1992;138:407–422. doi: 10.1099/00221287-138-3-407. [DOI] [PubMed] [Google Scholar]
- 26.Romero-Arroyo, C. E., S. R. James, M. Farmer, M. Willby, and D. C. Krause. Submitted for publication.
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Stevens M K, Krause D C. Localization of the Mycoplasma pneumoniae cytadherence-accessory proteins HMW1 and HMW4 in the cytoskeletonlike triton shell. J Bacteriol. 1991;173:1041–1050. doi: 10.1128/jb.173.3.1041-1050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens M K, Krause D C. Mycoplasma pneumoniae cytadherence phase-variable protein HMW3 is a component of the attachment organelle. J Bacteriol. 1992;174:4265–4274. doi: 10.1128/jb.174.13.4265-4274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]