Abstract
The sequence diversity of 45 Opa outer membrane proteins from Neisseria meningitidis, Neisseria gonorrhoeae, Neisseria sicca, and Neisseria flava indicates that horizontal genetic exchange of opa alleles has been rare between these species. A two-dimensional structural model containing four surface-exposed loops was constructed based on rules derived from porin crystal structure and on conservation of sequence homology within transmembrane β-strands. The minimal continuous epitopes recognized by 23 monoclonal antibodies were mapped to loops 2 and 3. Some of these epitopes are localized on the bacterial cell surface, in support of the model.
The Opa (opacity) proteins are a family of antigenic- and phase-variable outer membrane proteins with a monomer molecular mass of approximately 28 kDa expressed by Neisseria meningitidis, Neisseria gonorrhoeae, and commensal Neisseria species (33, 46). Purified Opa proteins are trimers or tetramers, as determined by gel filtration (2). The expression of certain Opa proteins promotes neisserial adherence to and invasion of epithelial and endothelial cells as well as professional phagocytes (20, 22, 28, 40, 44). Opa proteins can also mediate bacterial aggregation via interactions with lipopolysaccharide (6). Heparan sulfate proteoglycans on epithelial cell surfaces are a target for binding of some Opa proteins, and gonococcal invasion can be blocked by heparin or heparan sulfate (7, 37). The N-terminal domain of some members of the CD66 carcinoembryonic antigen family present on phagocytic cell surfaces is also a target for binding by Opa proteins (8, 16, 41, 42).
Multiple opa loci containing different opa alleles are scattered around the chromosomes of N. gonorrhoeae (11 to 12 opa loci) (4, 10, 20, 33) and N. meningitidis (3 to 4 loci) (3, 18, 25). Earlier comparisons of a limited number of alleles, primarily from two strains of N. gonorrhoeae and one strain of N. meningitidis, have indicated that sequence differences are concentrated in a semivariable region (SV) and two hypervariable regions (HV1 and HV2) within a conserved framework. Sequence differences between opa alleles arise by microevolution: translocations, deletions, point mutations, and import from unrelated neisseriae have been identified in N. meningitidis (17, 18, 25), and translocations have been documented for N. gonorrhoeae (5). The sequence variability of opa alleles is sufficiently large that it has been used for tracing of contacts among patients with gonorrhea (26).
Two slightly different two-dimensional Opa structure models were derived by using protein sequences from two gonococcal strains (4, 36). Both models predicted four surface-exposed loops, the first three of which corresponded to the SV, HV1, and HV2 regions. A few epitopes recognized by murine monoclonal antibodies (MAbs) which are predicted to be exposed on the cell surface have been mapped to the HV1 and HV2 regions (3, 9, 17, 18, 29). Numerous opa sequences from N. meningitidis (17, 25) and the commensal neisseriae Neisseria sicca and Neisseria flava (46) have since been described. We have compared these various sequences to determine whether they can be accommodated by the structural model(s) and whether they provide evidence for horizontal genetic exchange of opa genes between the different species. In addition, we have mapped the minimal binding sites of additional epitopes recognized by bactericidal MAbs in order to localize surface-exposed loops.
MATERIALS AND METHODS
Nomenclature of opa alleles and proteins.
Diverse nomenclatures have been used for opa sequences submitted to GenBank. In addition, the 106 alleles found in release 101 contained a number of incomplete sequences, duplicate sequences with different nomenclatures, and contradictory sequences for supposedly identical alleles. We have assigned arbitrary numbers to each unique allele and SV, HV1, and HV2 region (Table 1) (17, 25), derived from the original allelic or plasmid designations where possible. The complete data set is available upon request from M. Achtman.
TABLE 1.
Unique designations of opa alleles and their variable regions
| opa allelea | Speciesb | Clusterc | GenBank accession no. | Designationd
|
||
|---|---|---|---|---|---|---|
| SV | HV1 | HV2 | ||||
| 41 | Nm C C1938 | Nm-1 | X06445 | 109 | 35 | 37 |
| 46 | Ng JS3 | Ng-3 | X12625 | 113 | 82 | 83 |
| 47 | Ng MS11 | Ng-3 | X60709 | 130 | 77 | 78 |
| 49 | Ng MS11 | Ng-1 | X60711 | 127 | 52 | 53 |
| 65 | Ng VP1 | Ng-2 | Z18940 | 116 | 94 | 95 |
| 66 | Ng VP1 | Ng-1 | Z18941 | 126 | 55 | 56 |
| 67 | Ng VP1 | Ng-1 | Z18942 | 119 | 88 | 89 |
| 68 | Ng VP1 | Ng-1 | Z18936 | 126 | 55 | 57 |
| 69 | Ng F62-SF | Ng-3 | X15780 | 122 | 80 | 81 |
| 70 | Ng MS11 | Ng-1 | X52373 | 117 | 73 | 74 |
| 71 | Ng MS11 | Ng-1 | X52370 | 127 | 58 | 59 |
| 74 | Ng MS11 | Ng-1 | X52368 | 123 | 73 | 74 |
| 75 | Ng MS11 | Ng-2 | X52367 | 124 | 75 | 76 |
| 77 | Ng MS11 | Ng-3 | X52365 | 123 | 84 | 85 |
| 78 | Ng MS11 | Ng-1 | X52371 | 128 | 68 | 69 |
| 79 | Ng MS11 | Ng-1 | X52364 | 120 | 71 | 96 |
| 87 | Ns | Outgroup | U12287 | 132 | 90 | 91 |
| 88 | Nf | Outgroup | U12288 | 131 | 92 | 93 |
| 92 | Nm A | Nm-1 | AF001199 | 101 | 5 | 6 |
| 93 | Nm A | Nm-1 | AF001200 | 102 | 5 | 6 |
| 94 | Nm A | Nm-1 | AF001201 | 100 | 7 | 8 |
| 99 | Nm A | Nm-2 | AF001181 | 100 | 1 | 2 |
| 100 | Nm A | Nm-1 | AF001195 | 100 | 9 | 10 |
| 101 | Nm A | Nm-2 | AF001179 | 109 | 3 | 4 |
| 102 | Nm A | Nm-1 | AF001204 | 103 | 24 | 25 |
| 118 | Nm A | Nm-1 | AF001186 | 103 | 13 | 14 |
| 123 | Nm B 188/87 | Nm-1 | AF016290 | 100 | 38 | 19 |
| 124 | Nm B 188/87 | Nm-1 | AF016288 | 111 | 39 | 40 |
| 126 | Nm B 188/87 | Nm-2 | AF016292 | 106 | 41 | 42 |
| 127 | Nm B 190/87 | Nm-1 | AF016286 | 100 | 43 | 44 |
| 128 | Nm B190/87 | Nm-1 | AF016289 | 106 | 45 | 46 |
| 129 | Nm B190/87 | Nm-1 | AF016285 | 100 | 43 | 48 |
| 130 | Nm B190/87 | Nm-1 | AF016291 | 107 | 49 | 50 |
| 131 | Nm A | Nm-1 | AF001194 | 100 | 9 | 10 |
| 132 | Nm A | Nm-2 | AF001180 | 100 | 1 | 2 |
| 133 | Nm A | Nm-1 | AF001185 | 103 | 13 | 14 |
| 134 | Nm A | Nm-1 | AF001193 | 100 | 15 | 51 |
| 540 | Nm C FAM18 | Nm-1 | X63110 | 103 | 30 | 31 |
| 709 | Ng FA1090 | Ng-2 | X06436 | 118 | 63 | 67 |
| 900 | Nm C FAM18 | Nm-1 | X63111 | 103 | 32 | 97 |
| 1100 | Ng FA1090 | Ng-2 | X06435 | 115 | 63 | 64 |
| 1800 | Nm C FAM18 | Nm-1 | X63109 | 133 | 33 | 98 |
| 5100 | Nm C Z4197 | Nm-1 | U37256 | 109 | 28 | 540 |
| 5200 | Nm C Z4197 | Nm-2 | U37257 | 103 | 22 | 23 |
| 15063 | Ng MS11A | Ng-1 | U13708 | 121 | 61 | 62 |
Unique designation for each allele. Opa proteins implicated in invasion: Opa68, Opa71 (formerly called Opa50) (20), and Opa15063 (43).
Nm, N. meningitidis; Ng, N. gonorrhoeae; Ns, N. sicca; Nf, N. flava. Strain designations are included for N. gonorrhoeae and N. meningitidis serogroup B and C (17) strains. The serogroup B strains 188/87 and 190/87 were isolated in Norway and belong to the ET-5 complex; the serogroup C strains FAM18 and Z4197 are ET-37 complex strains isolated in the USA and Mali, respectively. The serogroup A strains were of subgroups III and IV-1 (25).
Cluster within tree in Fig. 3.
Unique designation based on nucleotide sequence. The amino acid sequences of SV103 and SV106 are identical, whereas the nucleotide sequences differ by 1 bp. These sequences are shown in Fig. 2 as SV-103. The amino acid sequences of HV1-13 and HV1-22 are identical, whereas the nucleotide sequences differ by 1 bp. These sequences are shown in Fig. 2 as HV1-13.
DNA sequences.
Duplicate sequences and sequences representing recombinational or translocation events (4, 17, 25) within the 106 alleles were excluded from analysis to ensure that only unique sequences were compared. Furthermore, only sequences encoding a mature Opa protein were used, thus excluding 15 partial sequences. Contradictory sequences were present for some opa alleles of N. gonorrhoeae MS11. In those cases, the sequences of Bhat et al. (4) were chosen because the PCR amplification and cloning method used by Kupsch et al. (20) has been shown to generate a high frequency of PCR-generated mistakes (25). The final data set consists of 45 sequences (Table 1) and includes 7 sequences from two serogroup B meningococci which have not been published elsewhere. Those sequences were obtained after PCR amplification of chromosomal DNA as described previously (25) and were sequenced by automated dye terminator cycle sequencing (ABI model 377 DNA sequencer) using primers O3510, O80, O82, O83, and O87 (17).
Multiple alignment of protein sequences.
After translation, the amino acid sequences of mature Opa proteins were aligned by using PILEUP (version 9.0; Genetics Computer Group, University of Wisconsin). The alignment was then edited manually, especially in the variable regions, by placing alignment gaps such that they increased the protein sequence similarities.
Sequence analysis.
Alignments stored as an MSF file were analyzed by using a self-written program, PsFind (ftp://novell-del-valle.rz-berlin.mpg.de), which can calculate percent uniformity, defined as the percentage of the most frequent amino acid at each position (excluding gaps introduced by the alignment). PsFind was also used to calculate the properties of the most common amino acid at each position. Phylogenetic trees were calculated in ARB (http://www.mikro.biologie.tu-muenchen.de), using PAM distances and the neighbor-joining method (30). Bootstrap analysis was performed in ARB, using 200 repetitions.
MAbs.
The murine MAbs used (Table 2) include antibodies secreted by five new hybridomas which were generated as previously described (1) after immunization of BALB/c mice with meningococci of serogroup A, subgroup IV-1 (O623) or subgroup III (L614, U506), or of serogroup B, ET-5 complex (192/B8, 210/G9).
TABLE 2.
MAbs which react with Opa proteins
| MAba | Immunoglobulin subclass | Minimal epitope | Specificity |
|---|---|---|---|
| O623 | G2a | NKS | HV1-1 |
| U506 | G1 | NKS | HV1-1 |
| O521b | G3 | NRQD | HV1-1 |
| T116 | NDc | TWKEL | HV1-7 |
| L614 | G1 | KESNYS | HV1-9 |
| P219b | G1 | IY | HV2-2 |
| P322b,d | G3 | TIYNQ | HV2-2 |
| P414b | G2a | TPTIYNQ | HV2-2 |
| W320/15 | G2a | NIP | HV2-2 |
| W124 | ND | NIP | HV2-2 |
| W104 | ND | PTNIPGGT | HV2-2 |
| AB419 | G2b | TTFL | HV2-6 |
| W320/16 | ND | PSGSTT | HV2-8 |
| U205 | G2b | PVPQGPTPK | HV2-10 |
| O516/2b | G2b | KGATQ | HV2-14 |
| P110b,d | G2b | KPTKGAT | HV2-14, HV2-19 |
| P112b,d | G2a | QP | HV2-14 |
| N312 | G1 | QP | HV2-14 |
| P515b,d | G3 | KGAT | HV2-14 |
| P416b,d | G2a | PTKGATQP | HV2-14 |
| P514b,d | G2a | PTKGATQP | HV2-14 |
| 210/G9 | G2a | QPGKL | HV2-14, HV2-19 |
| 192/B8 | G1 | TQPGKLV | HV2-14, HV2-46 |
All have been previously described by Achtman et al. (1) except for D309 (not shown) (2), O521 (18), and 192/B8, 210/G9, L614, O623, and U506 (described for the first time in this report).
Strong immunofluorescence with bacteria expressing Opa proteins recognized by the MAb.
ND, not determined.
Bactericidal at concentration of <8 μg/ml.
Epitope mapping.
Multiple synthetic N-terminally acetylated peptides containing 12 or 10 amino acids were synthesized on pins as previously described (15), using an epitope scanning kit (Cambridge Research Biochemicals) with a dilution aid (Epiguide; Labsystems) and the modifications described elsewhere (23). The pins were screened by enzyme-linked immunosorbent assay (ELISA) for reactivity as previously described (17).
Exposure of epitopes.
Bactericidal activity was tested in microtiter wells as described previously (12), using various concentrations of MAbs in the presence of 20% human complement and the serogroup A, subgroup IV-1 strain C623, which expresses Opa132, Opa136, and Opa137. MAbs were scored as bactericidal when at least 50% of the bacteria were killed by low concentrations compared to control tests lacking antibodies. Immunofluorescence microscopy with live bacteria was performed as described previously (24).
RESULTS
Sequence variation among 45 Opa protein sequences.
Sequence variation has recently been examined among opa alleles from hundreds of N. meningitidis serogroup A, subgroup III strains isolated globally since the 1960s and from representative subgroup IV-1 and IV-2 strains (25). Those sequence variants which represent opa alleles inherited from a common ancestor of these subgroups, or which were subsequently imported by horizontal genetic exchange, were chosen for a comparison of diverse neisserial Opa proteins. The unique opa sequences from two serogroup C strains of the ET-37 complex (17) and from two serogroup B strains of the ET-5 complex, as well as one sequence from strain C1938 of unknown clonal assignment, were included (Table 1). Sequence variants reflecting translocation or recombinational events between opa alleles were excluded from the comparison because they do not represent unique alleles. These 25 sequences were supplemented by 18 unique sequences available in GenBank from N. gonorrhoeae MS11, FA1090, VP1, and JS3 as well as by two sequences from the commensal species N. sicca and N. flava. The resulting list (Table 1) contains those 45 unique opa alleles for which complete sequences encoding the entire mature Opa protein were available. After translation, the amino acid sequences were aligned, with manual addition of gaps within the variable regions to ensure maximal homology, and the amino acid variability at each position was calculated (Fig. 1).
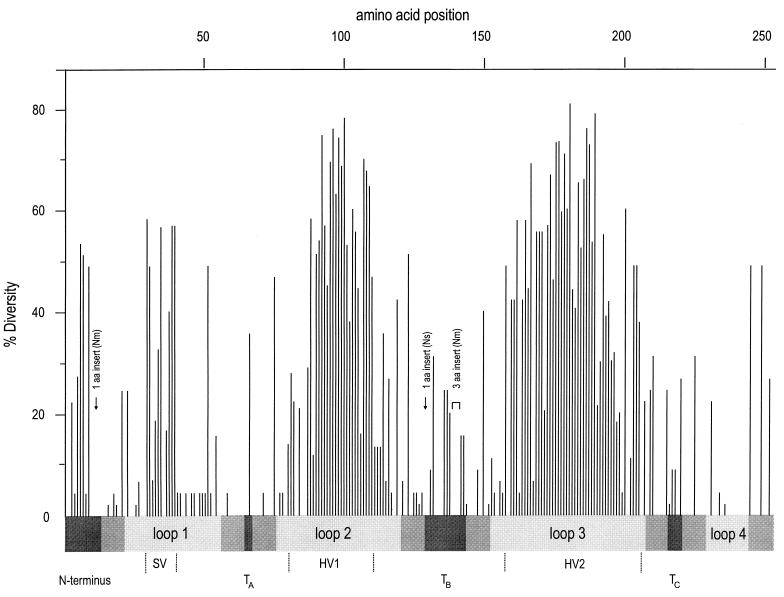
FIG. 1.
Sequence diversity of 45 Opa proteins. The percentage occurrence of the most common amino acid (percent uniformity) was calculated at each position within an alignment of the 45 Opa proteins, using the PsFind program. The percentage diversity consists of percent uniformity subtracted from 100. The regions predicted to correspond to exposed loops on the cell surface are shaded light gray, regions putatively exposed to the periplasm are shaded dark gray, and predicted transmembrane strands are indicated by intermediate shading. The SV, HV1, and HV2 regions were redefined to include the maximal sequence diversity, as indicated aa, amino acid.
The former limits of the variable SV, HV1, and HV2 regions (3, 4, 18, 32) were based on only a few sequences, primarily from N. gonorrhoeae, and need to be redefined as indicated in Fig. 1 and 2 to account for the variability in this data set. Additional sequence variability was concentrated at the N terminus and in the regions assigned to periplasmic turns TB and TC (Fig. 1; see below).
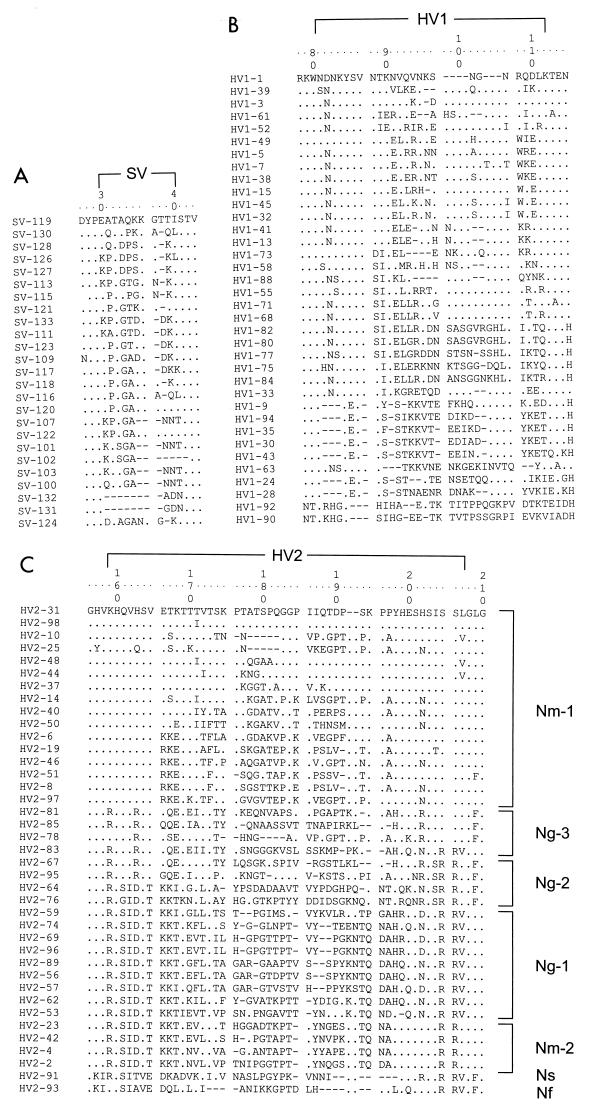
FIG. 2.
Multiple alignments of the SV, HV1, and HV2 regions. Dots indicate identity to the top sequence shown, and gaps introduced to maximize homology are indicated by hyphens. The numbers at the top designate positions in a consensus sequence. (A) SV positions 30 to 40; (B) HV1 positions 81 to 112; (C) HV2 positions 159 to 207. The ordering of the sequences was by maximal visual similarity except for panel C, where the sequences were grouped by phylogenetic protein cluster.
Species-specific differences.
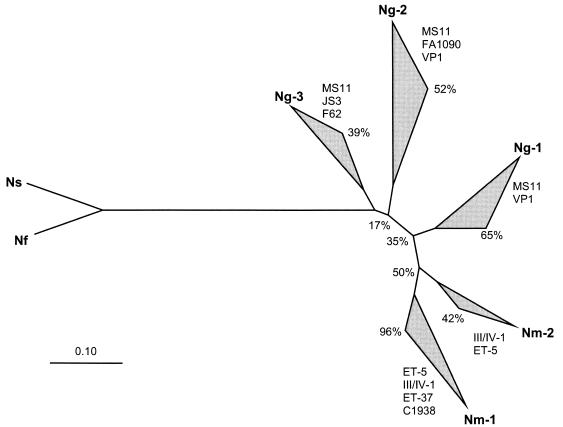
A rooted neighbor-joining (30) phylogenetic tree using N. sicca and N. flava as an outgroup contained two main clusters of sequences from N. meningitidis (Nm-1 and Nm-2) and three clusters of sequences from N. gonorrhoeae (Ng-1, Ng-2, and Ng-3) (Fig. 3). Clusters Nm-1, Ng-1, and Ng-2 were obtained in over 50% of 200 bootstrap repetitions, whereas the other clusters were less robust (Fig. 3). Opa proteins from N. gonorrhoeae MS11 were found in all three Ng clusters, and Opa proteins from N. meningitidis subgroups III/IV-1 or the ET-5 complex were found in both Nm clusters. Similar results were obtained whether the distance algorithm was based on percentage identity or the PAM matrix and when only sequences from the conserved regions were compared (data not shown). The observation that sequences from N. gonorrhoeae and N. meningitidis never clustered together indicates that horizontal genetic transfer of opa alleles between the two pathogenic species is rare. In agreement, the SV, HV1, and HV2 regions from N. meningitidis were also all distinct from those of N. gonorrhoeae (Table 1). The N-terminal portions of all meningococcal Opa proteins contain an additional amino acid (Y) (Fig. 1). Some meningococcal Opa proteins contain a duplication of three amino acids (DKF) at positions 140 to 142.
FIG. 3.
Phylogenetic neighbor-joining tree of 45 Opa proteins, using sequences from commensal neisseriae (Nf, N. flava; Ns, N. sicca) as an outgroup. Clockwise of each cluster are indicated the strains (N. gonorrhoeae, clusters Ng-1 through Ng-3) or subgroups (N. meningitidis, clusters Nm-1 and Nm-2) which they encompassed, as well as confidence levels (percentages) calculated from bootstrap tests with 200 repetitions. The values at the nodes are also bootstrap confidence levels. The genetic distance scale is indicated by a horizontal line at the bottom.
The Opa proteins from N. sicca (opa87) and N. flava (opa88) resembled each other but differed at numerous sites from those of the pathogenic neisseriae, even within otherwise conserved regions (Fig. 4). Pairwise comparisons between Opa proteins from the commensal species and the Opa proteins from the pathogenic neisseriae yielded 50 to 59% identities, whereas Opa proteins from N. meningitidis were 60 to 84% identical to those of N. gonorrhoeae. The SV, HV1, and HV2 regions of the Opa proteins from the commensals were also shorter than those from the pathogenic neisseriae.
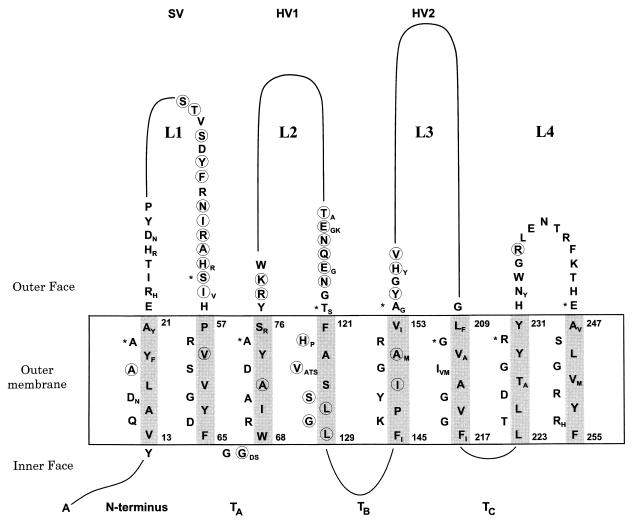
FIG. 4.
Predicted two-dimensional structure of Opa proteins. Variable stretches are indicated by a continuous line, and only details of conserved amino acids are shown. Minor sequence variation is indicated by additional subscript letters next to the most frequent amino acids. Circled amino acids were variable only in the Opa proteins from N. flava or N. sicca. Asterisks indicate the first amino acids predicted to lie outside the outer membrane in the model of Bhat et al. (4). The inner and outer faces of the outer membrane are indicated by horizontal lines. The gray shading indicates the nonpolar side of the eight transmembrane β strands.
Despite the species specificity, certain variable regions were similar between gonococcal and meningococcal proteins, with stretches of identity of up to 13 amino acids. The most similar of these regions were SV-123 (gonococcal opa allele 74 or 77) versus SV-111 (meningococcal Opa124), where 8 of 11 amino acids were identical, HV1-94 (gonococcal Opa65) versus HV1-30 (meningococcal Opa540), where 23 of 25 amino acids were identical, and HV2-78 (gonococcal Opa47) versus HV2-10 (meningococcal Opa100 and Opa131), where 33 of 41 amino acids were identical. These observations are consistent with occasional recombination between the two species. Similarly, after exclusion of the hypervariable regions and two short insertions of one and three amino acids, 145 of 157 amino acids were identical between gonococcal Opa70 and meningococcal Opa900.
Two-dimensional model for the structure of Opa proteins.
Antiparallel amphipathic β strands span the outer membrane within porins (11, 45), and strand prediction has been used to devise two slightly different two-dimensional models of gonococcal Opa proteins (4, 36). Transmembrane strands correspond to the most conserved sequences within outer membrane protein families (19). The C-terminal amino acid of outer membrane proteins is usually a phenylalanine and is preceded by a transmembrane strand (34). Porins characteristically possess short periplasmic turns, containing turn-promoting amino acids and lacking turn-inhibiting amino acids (27), and their transmembrane strands are flanked by aromatic residues (11, 45).
We devised a two-dimensional β-barrel model containing eight transmembrane strands which maximized all of the criteria cited above (Fig. 4). Minor discrepancies to the criteria were the slightly hydrophilic amino acid S125 and turn TB, of 14 amino acids, which is exceptionally long. The model predicts that four hydrophilic loops (L1 to L4) are exposed on the cell surface. The SV region is part of loop L1, HV1 corresponds to L2, and HV2 corresponds to L3. Loop L4 is short and is also strongly conserved. Of the domains predicted to face the periplasm, the N terminus and turns TB and TC show considerable sequence variability. The model (Fig. 4) differs slightly from former models (4, 36) in that the transmembrane strands are of uniform length and the ends of the external loops are translocated by up to three amino acids (marked with asterisks in Fig. 4).
Mapping of cell surface-exposed epitopes.
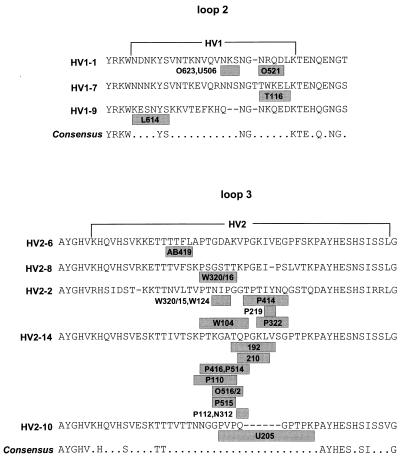
MAbs that react with intact bacteria must recognize epitopes on the cell surface. We have formerly mapped five such epitopes to the regions corresponding to L2 and L3 of Opa proteins from serogroup C meningococci (17) and have now analyzed 26 other MAbs which react specifically with serogroup A meningococci in whole-cell ELISAs and with native Opa proteins purified from those meningococci. Six of these MAbs were bactericidal, and 10 bound to live bacteria, as determined by immunofluorescent light microscopy (Table 2). The specificity of the 26 MAbs correlated with the sequences of individual HV1 or HV2 regions (Table 2). The minimal epitopes recognized by these MAbs were mapped by Pepscan analysis of L2 and L3 of the Opa proteins with which they react. We synthesized 12-mers overlapping by nine amino acids on pins and tested them for reactivity with the 26 MAbs. For 23 MAbs, the results defined epitopes whose limits were further refined by Pepscan with 10-mers overlapping by nine amino acids (data not shown). The locations of the minimal continuous epitopes that these MAbs recognize are summarized in Table 2 and Fig. 5. Only weak reactivity was obtained with two other MAbs (U106 and U214), and a third (D309) reacted with so many peptides that the results were uninterpretable.
FIG. 5.
Minimal continuous epitopes in L2 and L3 recognized by 23 MAbs. The minimal epitopes are shown in gray. The epitopes were defined by Pepscan with 10-mers overlapping by nine amino acids, except that the epitopes recognized by MAbs U205 and L614 were mapped by using 12-mers overlapping by nine amino acids.
Two of the MAbs whose epitopes had been mapped, 192/B8 and 210/G9, reacted with Opa proteins containing HV2-14 from serogroup A meningococci but had been isolated after immunization with serogroup B meningococci expressing Opa128 (HV2-46) and Opa123 (HV2-19), respectively. MAb 192/B8 recognizes TQPGKLV in HV2-14, whereas HV2-46 contains the sequence TVPGKIV (differences are underlined). MAb 210/G9 recognizes the sequence QPGKL in HV2-14, whereas HV2-19 contains the sequence EPGKI. Similarly, P110, raised against serogroup A Opa protein Opa137, also reacted with Opa123 from serogroup B meningococci. The minimal binding site was KPTKGAT in HV2-14 (Opa137), whereas HV2-19 contains KPSKGAT. These results presumably reflect the exchangeability of certain amino acids within minimal epitopes and were not further investigated.
DISCUSSION
The sequences of 45 informative Opa protein sequences from four neisserial species were analyzed in order to test for frequent interspecies recombination and to construct an improved two-dimensional structural model.
The two Opa sequences available from the commensal species, N. sicca and N. flava, differed strongly from sequences from the pathogenic species, N. gonorrhoeae and N. meningitidis (Fig. 3). The commensal sequences possessed shorter SV, HV1, and HV2 regions, corresponding to shorter L1, L2, and L3 (Fig. 2), as well as a large number of different amino acids at otherwise conserved positions (Fig. 4). Additional sequences are necessary to determine whether these differences are typical of the commensal neisseriae.
opa alleles can translocate from one locus to another in N. gonorrhoeae (5) and N. meningitidis (17, 18, 25), leading to formation of mosaic genes, and import of opa alleles from unrelated neisseriae during epidemic spread has also been documented for N. meningitidis (17, 18, 25). Simple cocultivation in the laboratory of different neisserial species suffices to allow DNA transformation of opa alleles (14). Interspecies transfer has been demonstrated for several (non-opa) genes within the neisseriae (citations in reference 21).
In contrast, the results presented here indicate that transfer of opa alleles between neisserial species is rare in nature. In addition to the size differences between Opa proteins from commensal and pathogenic species, all 25 meningococcal Opa proteins contain a unique insertion of one amino acid in the N-terminal region (Fig. 1). Furthermore, Opa proteins from the different species clustered separately within a phylogenetic tree (Fig. 3). These results resemble those found for housekeeping genes (39), for which the lack of genetic overlap between N. gonorrhoeae and N. meningitidis has been interpreted as indicating ecological isolation (31). We note, however, that one example of horizontal genetic exchange between the pathogenic neisseriae has already been described (38) and that the genetic diversity of Opa proteins is likely to be so large that the current sample of 45 sequences is too small to have detected rare genetic exchange.
Two-dimensional structural models based on a limited number of Opa sequences (4, 36) were refined by combining rules based on the properties of amino acids (13, 27) with those derived from porin structures (11, 35, 45) and by minimizing the sequence variability within the nonpolar face of transmembrane β strands. The resulting model (Fig. 4) predicts that the protein traverses the outer membrane eight times, resulting in four hydrophilic loops on the cell surface and terminating at the inner face of the outer membrane. L1, L2, and L3 are highly variable in sequence and correspond to the variable regions called SV, HV1, and HV2. Considerable sequence variability was also found in the N terminus and within turns TB and TC, all predicted to face the periplasmic side of the outer membrane. In light of this degree of variability, the structure of L4 is strikingly constant, suggesting that it might have an important role in protein structure.
The exposure of L2 and L3 to the cell surface was confirmed by mapping continuous epitopes recognized by 23 MAbs to those loops and by showing that some of these MAbs are bactericidal and/or bind to live bacteria (immunofluorescence). No MAbs that bind to L1 and L4 have been described, suggesting that L2 and L3 are immunodominant.
It seems likely that the structures of most Opa proteins will resemble that of the model presented here, especially because the two Opa proteins from N. sicca and N. flava possessed the same structure despite being only 50 to 60% homologous to other Opa proteins. The model also provides the possibility to test the significance of apparent homologies between Opa proteins and proteins from other genera whose genomes are being sequenced.
ACKNOWLEDGMENT
Burkhard Malorny was supported by grant Ac36/6 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Achtman M, Kusecek B, Morelli G, Eickmann K, Wang J, Crowe B, Wall R A, Hassan-King M, Moore P S, Zollinger W. A comparison of the variable antigens expressed by clone IV-1 and subgroup III of Neisseria meningitidis serogroup A. J Infect Dis. 1992;165:53–68. doi: 10.1093/infdis/165.1.53. [DOI] [PubMed] [Google Scholar]
- 2.Achtman M, Neibert M, Crowe B A, Strittmatter W, Kusecek B, Weyse E, Walsh M J, Slawig B, Morelli G, Moll A, Blake M. Purification and characterization of eight class 5 outer membrane protein variants from a clone of Neisseria meningitidis serogroup A. J Exp Med. 1988;168:507–525. doi: 10.1084/jem.168.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aho E L, Dempsey J A F, Hobbs M M, Klapper D G, Cannon J G. Characterization of the opa (class 5) gene family of Neisseria meningitidis. Mol Microbiol. 1991;5:1429–1437. doi: 10.1111/j.1365-2958.1991.tb00789.x. [DOI] [PubMed] [Google Scholar]
- 4.Bhat K S, Gibbs C P, Barrera O, Morrison S G, Jähnig F, Stern A, Kupsch E-M, Meyer T F, Swanson J. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol Microbiol. 1991;5:1889–1901. doi: 10.1111/j.1365-2958.1991.tb00813.x. [DOI] [PubMed] [Google Scholar]
- 5.Bihlmaier A, Römling U, Meyer T F, Tümmler B, Gibbs C P. Physical and genetic map of the Neisseria gonorrhoeae strain MS11-N198 chromosome. Mol Microbiol. 1991;5:2529–2539. doi: 10.1111/j.1365-2958.1991.tb02099.x. [DOI] [PubMed] [Google Scholar]
- 6.Blake M S, Blake C M, Apicella M A, Mandrell R E. Gonococcal opacity: lectin-like interactions between Opa proteins and lipooligosaccharide. Infect Immun. 1995;63:1434–1439. doi: 10.1128/iai.63.4.1434-1439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen T, Belland R J, Wilson J, Swanson J. Adherence of pilus− Opa+ gonococci to epithelial cells in vitro involves heparan sulfate. J Exp Med. 1995;182:511–517. doi: 10.1084/jem.182.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T, Gotschlich E C. CGM1a antigen of neutrophils, a receptor of gonococcal opacity proteins. Proc Natl Acad Sci USA. 1996;93:14851–14856. doi: 10.1073/pnas.93.25.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connell T D, Black W J, Kawula T H, Barritt D S, Dempsey J A, Kverneland K J, Stephenson A, Schepart B S, Murphy G L, Cannon J G. Recombination among protein II genes of Neisseria gonorrhoeae generates new coding sequences and increases structural variability in the protein II family. Mol Microbiol. 1988;2:227–236. doi: 10.1111/j.1365-2958.1988.tb00024.x. [DOI] [PubMed] [Google Scholar]
- 10.Connell T D, Shaffer D, Cannon J G. Characterization of the repertoire of hypervariable regions in the protein II (opa) gene family of Neisseria gonorrhoeae. Mol Microbiol. 1990;4:439–449. doi: 10.1111/j.1365-2958.1990.tb00610.x. [DOI] [PubMed] [Google Scholar]
- 11.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 12.Crowe B A, Wall R A, Kusecek B, Neumann B, Olyhoek T, Abdillahi H, Hassan-King M, Greenwood B M, Poolman J T, Achtman M. Clonal and variable properties of Neisseria meningitidis isolated from cases and carriers during and after an epidemic in the Gambia, West Africa. J Infect Dis. 1989;159:686–700. doi: 10.1093/infdis/159.4.686. [DOI] [PubMed] [Google Scholar]
- 13.Eisenberg D, Weiss M, Terwilliger T C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc Natl Acad Sci USA. 1984;81:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frosch M, Meyer T F. Transformation-mediated exchange of virulence determinants by co-cultivation of pathogenic Neisseriae. FEMS Microbiol Lett. 1992;100:345–350. doi: 10.1111/j.1574-6968.1992.tb14062.x. [DOI] [PubMed] [Google Scholar]
- 15.Geysen H M, Rodda S J, Mason T J, Tribbick G, Schoofs P G. Strategies for epitope analysis using peptide synthesis. J Immunol Methods. 1987;102:259–274. doi: 10.1016/0022-1759(87)90085-8. [DOI] [PubMed] [Google Scholar]
- 16.Gray-Owen S D, Dehio C, Haude A, Grunert F, Meyer T F. CD66 carcinoembryonic antigens mediate interactions between Opa-expressing Neisseria gonorrhoeae and human polymorphonuclear phagocytes. EMBO J. 1997;16:3435–3445. doi: 10.1093/emboj/16.12.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobbs M M, Malorny B, Prasad P, Morelli G, Kusecek B, Heckels J E, Cannon J G, Achtman M. Recombinational reassortment among opa genes from ET-37 complex Neisseria meningitidis isolates of diverse geographic origins. Microbiology. 1998;144:157–168. doi: 10.1099/00221287-144-1-157. [DOI] [PubMed] [Google Scholar]
- 18.Hobbs M M, Seiler A, Achtman M, Cannon J G. Microevolution within a clonal population of pathogenic bacteria: recombination, gene duplication and horizontal genetic exchange in the opa gene family of Neisseria meningitidis. Mol Microbiol. 1994;12:171–180. doi: 10.1111/j.1365-2958.1994.tb01006.x. [DOI] [PubMed] [Google Scholar]
- 19.Jeanteur D, Lakey J, Pattus F. The porin superfamily: diversity and common features. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 363–380. [Google Scholar]
- 20.Kupsch E-M, Knepper B, Kuroki T, Heuer I, Meyer T F. Variable opacity (Opa) outer membrane proteins account for the cell tropisms displayed by Neisseria gonorrhoeae for human leukocytes and epithelial cells. EMBO J. 1993;12:641–650. doi: 10.1002/j.1460-2075.1993.tb05697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiden M C J, Malorny B, Achtman M. A global gene pool in the neisseriae. Mol Microbiol. 1996;21:1297–1298. doi: 10.1046/j.1365-2958.1996.981457.x. [DOI] [PubMed] [Google Scholar]
- 22.Makino S, van Putten J P M, Meyer T M. Phase variation of the opacity outer membrane protein controls invasion by Neisseria gonorrhoeae into human epithelial cells. EMBO J. 1991;10:1307–1315. doi: 10.1002/j.1460-2075.1991.tb07649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGuinness B, Barlow A K, Clarke I N, Farley J E, Anilionis A, Poolman J T, Heckels J E. Deduced amino acid sequences of class I protein (PorA) from three strains of Neisseria meningitidis. Synthetic peptides define the epitopes responsible for serosubtype specificity. J Exp Med. 1990;171:1871–1882. doi: 10.1084/jem.171.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merker P, Tommassen J, Virji M, Sesardic D, Achtman M. Two-dimensional structure of the Opc invasin from Neisseria meningitidis. Mol Microbiol. 1997;23:281–293. doi: 10.1046/j.1365-2958.1997.2051567.x. [DOI] [PubMed] [Google Scholar]
- 25.Morelli G, Malorny B, Müller K, Seiler A, Wang J, del Valle J, Achtman M. Clonal descent and microevolution of Neisseria meningitidis during 30 years of epidemic spread. Mol Microbiol. 1997;25:1047–1064. doi: 10.1046/j.1365-2958.1997.5211882.x. [DOI] [PubMed] [Google Scholar]
- 26.O’Rourke M, Ison C A, Renton A M, Spratt B G. Opa-typing: a high-resolution tool for studying the epidemiology of gonorrhoea. Mol Microbiol. 1995;17:865–875. doi: 10.1111/j.1365-2958.1995.mmi_17050865.x. [DOI] [PubMed] [Google Scholar]
- 27.Paul C, Rosenbusch J P. Folding patterns of porin and bacteriorhodopsin. EMBO J. 1985;4:1593–1597. doi: 10.1002/j.1460-2075.1985.tb03822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rest, R. F., and W. M. Shafer. 1989. Interactions of Neisseria gonorrhoeae with human neutrophils. Clin. Microbiol. Rev. 2(Suppl.):S83–S91. [DOI] [PMC free article] [PubMed]
- 29.Robinson E N, Jr, Clemens C M, McGee Z A, Cannon J G. Immunoelectron microscopic localization of outer membrane proteins II on the surface of Neisseria gonorrhoeae. Infect Immun. 1988;56:1003–1006. doi: 10.1128/iai.56.4.1003-1006.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Spratt B G, Smith N H, Zhou J, O’Rourke M, Feil E. The population genetics of the pathogenic Neisseria. In: Baumberg S, Young J P W, Saunders J R, Wellington E M H, editors. Population genetics of bacteria. Cambridge, England: Cambridge University Press; 1995. pp. 143–160. [Google Scholar]
- 32.Stern A, Brown M, Nickel P, Meyer T F. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell. 1986;47:61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- 33.Stern A, Meyer T F. Common mechanism controlling phase and antigenic variation in pathogenic neisseriae. Mol Microbiol. 1987;1:5–12. doi: 10.1111/j.1365-2958.1987.tb00520.x. [DOI] [PubMed] [Google Scholar]
- 34.Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 35.Tommassen J. Biogenesis and membrane topology of outer membrane proteins in Escherichia coli. In: Op den Kamp J A F, editor. Membrane biogenesis. Berlin, Germany: Springer-Verlag; 1988. pp. 352–373. [Google Scholar]
- 36.van der Ley P. Three copies of a single protein II-encoding sequence in the genome of Neisseria gonorrhoeae JS3: evidence for gene conversion and gene duplication. Mol Microbiol. 1988;2:797–806. doi: 10.1111/j.1365-2958.1988.tb00091.x. [DOI] [PubMed] [Google Scholar]
- 37.van Putten J P M, Paul S M. Binding of syndecan-like cell surface proteoglycan receptors is required for Neisseria gonorrhoeae entry into human mucosal cells. EMBO J. 1995;14:2144–2154. doi: 10.1002/j.1460-2075.1995.tb07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vázquez J A, Berrón S, O’Rourke M, Carpenter G, Feil E, Smith N H, Spratt B G. Interspecies recombination in nature: a meningococcus that has acquired a gonococcal PIB porin. Mol Microbiol. 1995;15:1001–1007. doi: 10.1111/j.1365-2958.1995.tb02275.x. [DOI] [PubMed] [Google Scholar]
- 39.Vázquez J A, De La Fuente L, Berron S, O’Rourke M, Smith N H, Zhou J, Spratt B G. Ecological separation and genetic isolation of Neisseria gonorrhoeae and Neisseria meningitidis. Curr Biol. 1993;9:567–572. doi: 10.1016/0960-9822(93)90001-5. [DOI] [PubMed] [Google Scholar]
- 40.Virji M, Makepeace K, Ferguson D J P, Achtman M, Moxon E R. Meningococcal Opa and Opc proteins: their role in colonization and invasion of human epithelial and endothelial cells. Mol Microbiol. 1993;10:499–510. doi: 10.1111/j.1365-2958.1993.tb00922.x. [DOI] [PubMed] [Google Scholar]
- 41.Virji M, Makepeace K, Ferguson D J P, Watt S M. Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol Microbiol. 1996;22:941–950. doi: 10.1046/j.1365-2958.1996.01551.x. [DOI] [PubMed] [Google Scholar]
- 42.Virji M, Watt S M, Barker S, Makepeace K, Doyonnas R. The N-domain of the human CD66a adhesion molecule is a target for Opa proteins of Neisseria meningitidis and Neisseria gonorrhoeae. Mol Microbiol. 1996;22:929–939. doi: 10.1046/j.1365-2958.1996.01548.x. [DOI] [PubMed] [Google Scholar]
- 43.Waldbeser L S, Ajioka R S, Merz A J, Puaoi D, Lin L, Thomas M, So M. The opaH locus of Neisseria gonorrhoeae MS11A is involved in epithelial cell invasion. Mol Microbiol. 1994;13:919–928. doi: 10.1111/j.1365-2958.1994.tb00483.x. [DOI] [PubMed] [Google Scholar]
- 44.Weel J F L, Hopman C T P, van Putten J P M. In situ expression and localization of Neisseria gonorrhoeae opacity proteins in infected epithelial cells: apparent role of Opa proteins in cellular invasion. J Exp Med. 1991;173:1395–1405. doi: 10.1084/jem.173.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss M S, Abele U, Weckesser J, Welte W, Schiltz E, Schulz G E. Molecular architecture and electrostatic properties of a bacterial porin. Science. 1991;254:1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]
- 46.Wolff K, Stern A. Identification and characterization of specific sequences encoding pathogenicity associated proteins in the genome of commensal Neisseria species. FEMS Microbiol Lett. 1995;125:255–263. doi: 10.1111/j.1574-6968.1995.tb07366.x. [DOI] [PubMed] [Google Scholar]