Graphical abstract
Keywords: IQGAP3, Prostate cancer, Clinical relevance, Prognostic prediction, Overall survival, Immune checkpoint blockade therapy
Highlights
-
•
IQGAP3 network (NW) regulates mitosis in prostate cancer (PC) and other cancers.
-
•
IQGAP3 correlates with PLK1 and TOP2A expression in PC and other cancers.
-
•
SigIQGAP3NW robustly predicts recurrence and poor prognosis in PC and other cancers.
-
•
RELT significantly correlates with immune checkpoint in PC and across human cancers.
-
•
SigQIGAP3NW and RELT predict cancer resistance to immune checkpoint blockage therapy.
Abstract
Introduction
IQGAP3 possesses oncogenic actions; its impact on prostate cancer (PC) remains unclear.
Objective
We will investigate IQGAP3′s association with PC progression, key mechanisms, prognosis, and immune evasion.
Methods
IQGAP3 expression in PC was examined by immunohistochemistry and using multiple datasets. IQGAP3 network was analyzed for pathway alterations and used to construct a multigene signature (SigIQGAP3NW). SigIQGAP3NW was characterized using LNCaP cell-derived castration-resistant PCs (CRPCs), analyzed for prognostic value in 26 human cancer types, and studied for association with immune evasion.
Results
Increases in IQGAP3 expression associated with PC tumorigenesis, tumor grade, metastasis, and p53 mutation. IQGAP3 correlative genes were dominantly involved in mitosis. IQGAP3 correlated with PLK1 and TOP2A expression at Spearman correlation/R = 0.89 (p ≤ 3.069e-169). Both correlations were enriched in advanced PCs and Taxane-treated CRPCs and occurred at high levels (R > 0.8) in multiple cancer types. SigIQGAP3NW effectively predicted cancer recurrence and poor prognosis in independent PC cohorts and across 26 cancer types. SigIQGAP3NW stratified PC recurrence after adjustment for age at diagnosis, grade, stage, and surgical margin. SigIQGAP3NW component genes were upregulated in PC, metastasis, LNCaP cell-produced CRPC, and showed an association with p53 mutation. SigIQGAP3NW correlated with immune cell infiltration, including Treg in PC and other cancers. RELT, a SigIQGAP3NW component gene, was associated with elevations of multiple immune checkpoints and the infiltration of Treg and myeloid-derived suppressor cells in PC and across cancer types. RELT and SigIQGAP3NW predict response to immune checkpoint blockade (ICB) therapy.
Conclusions
In multiple cancers, IQGAP3 robustly correlates with PLK1 and TOP2A expression, and SigIQGAP3NW and/or RELT effectively predict mortality risk and/or resistance to ICB therapy. PLK1 and TOP2A inhibitors should be investigated for treating cancers with elevated IQGAP3 expression. SigIQGAP3NW and/or RELT can be developed for clinical applications in risk stratification and management of ICB therapy.
Introduction
Prostate cancer (PC) is the most frequently diagnosed cancer of men in 112 of 185 countries and remains a major health threat to men [1]. The disease is associated with a heterogeneous prognosis. PCs are categorized by the World Health Organization (WHO) into 5 Grade Groups (GG1-5) based on Gleason score (GS) and prognostic implications: GG1 ≤ GS6, GG2 = GS3 + 4, GG3 = GS4 + 3, GG4 = GS8, and GG5 = GS9 + 10 [2]. Approximately 30% of patients will develop biochemical recurrence (BCR), defined as an increase in serum prostate-specific antigen (PSA) following curative therapies such as radiation and/or surgery [3]. BCR is the significant risk of PC metastasis which is standardly treated with androgen deprivation therapy (ADT). While ADT shows initial effectiveness in treating metastatic PC (mPC), resistance in the form of castration-resistant PC (mCRPC) commonly re-emerges [4], [5] and constitutes a major cause of PC mortality. Mechanisms governing PC recurrence, metastasis, and CRPC progression are complex and involve multiple processes, including cell proliferation, androgen signaling, and immune evasion. In this regard, CRPC can be treated with taxane-based chemotherapy, 2nd generation antiandrogens abiraterone or enzalutamide [5], [6], [7], and/or immunotherapy [8]. The taxane class of drugs disrupts microtubule function, which is required for mitosis.
The IQ motif GTPase-activating scaffold proteins (IQGAPs) are an important platform for cytoskeleton dynamics and cell proliferation in part via their activities in activating small G proteins, PI3K, and ERK [9], [10]. The IQGAP family in humans and mice consists of 3 related proteins: IQGAP1, IQGAP2, and IQGAP3. IQGAP1 and IQGAP2 have been widely researched, including their contributions to cancer biology [9], [10]. IQGAP3 was the latest addition to the IQGAP family in 2007 with its expression enriched in the brain [11]; the current knowledge of IQGAP3 remains lagging compared to our understanding of IQGAP1 and IQGAP2. Nonetheless, accumulative evidence supports IQGAP3′s role in facilitating cell proliferation involving Ras, ERK [12], [13], and cytokinesis [14]. In addition to these cell-based activities, IQGAP3 upregulation and its association with poor prognosis have been reported in multiple cancer types, including hepatocellular carcinoma [15], [16], lung adenocarcinomas [17], colorectal cancer [18], gastric cancer [19], [20], ovarian cancer [21], breast cancer [22], and other cancer types [23]. Among urogenital cancers, IQGAP3 expression is associated with poor prognosis in bladder cancer [23], clear cell renal cell carcinoma (ccRCC) [24], and PC [25]. While a limited number of papers (n = 2) reported that PCs with elevated IQGAP3 mRNA expression were associated with reductions in overall survival (OS) and progress-free survival (PFS) [23], [25], the extent of IQGAP3′s impact, particularly the involvement of its network in PC or tumorigenesis in general, remains largely unexplored.
This study addresses this knowledge gap. We observed the dominant association of IQGAP3 correlative genes with mitosis and chromosome segregation. For instance, the top 8 genes with correlative expression to IQGAP3 expression are NUSAP1 (Spearman R = 0.914, p = 4.75e-196), TPX2 (R = 0.911, p = 4.38e-192), ESPL1 (R = 0.907, p = 1.42e-187), KIF4A (R = 0.906, p = 4.43e-187), KIF20A (R = 0.901, p = 2.37e-181), GTSE1 (R = 0.892, p = 4.91e-173), PLK1 (R = 0.890, p = 3.07e-171), and TOP2A (R = 0.888, p = 5.35e-169). In addition to these correlative genes functioning in chromosome segregation, the impressive correlations observed with PLK1 and TOP2A are particularly appealing owing to their commercially available inhibitors. Furthermore, the high-level correlations between IQGAP3 and PLK1 or TOP2A, and their enrichment in mPC and CRPC as well as in 6 other cancer types imply a therapeutic potential of PLK1 and TOP2A in treating PCs and other cancers with elevated IQGAP3 expression. From the IQGAP3 network (NW), a 13-gene panel (SigIQGAP3NW) was derived. The panel robustly stratifies PC recurrence, associates with alterations in immune cells in PC and other cancer types and predicts response to immune checkpoint blockade (ICB) therapy. We also demonstrated RELT as a novel and effective biomarker of ICB treatment. Collectively, this investigation advances our understanding of PC and tumorigenesis in general.
Materials and methods
Patients
Tissues from PC patients were obtained from Hamilton Health Sciences, Hamilton, Ontario, Canada under approval from the local Research Ethics Board (REB# 11114). Patient details are presented in Table S1.
Data sources
PC datasets used in this research included TCGA, MSKCC, and SU2C organized by CBioPortal [26], [27] and Sawyers’s DNA microarray dataset [28]. TCGA PanCancer Atlas datasets across 32 cancer types were also utilized.
Programs and websites
The tools used in this research included: R2: Genomics Analysis and Visualization Platform (https://r2.amc.nl https://r2platform.com), UALCAN [29], Metascape [30], TISIDB [31], TIDE [32], TIMER [33], CIBERSORT [34], and LinkedOmics [35]. The R glmnet, immunedeconv, tidyverse, rstatix, ggpubr, dplyr, survival, Maxstat, and other packages were used.
Profiling PC-associated immune cells
Immune cells presented in PC and other cancer types were determined from RNA-seq data with CIBERSORT [34], Epic [36], MCP [37], Quantiset [38], xCell, and ssGSEA [39] within the R immunedeconv and SMDIC packages.
Construction of a multigene panel to assess PC biochemical recurrence
The construction was based on our previously published conditions [40], [41]. Briefly, differentially-expressed genes (DEGs, n = 281) concerning IQGAP3 expression were derived from the TCGA PanCancer Atlas PC dataset within cBioPortal [26], [27] (https://www.cbioportal.org/). Training and Testing populations were obtained via randomization at the ratio of 7:3 using R. From these DEGs within the Training dataset, elastic-net logistic regression of Cox model (the glmnet package in R) was used to select a multigene panel to predict BCR. We set the mixing parameter of α at 0.5, performed 6 rounds of selection, and pooled all unique genes from all 6 rounds of selection to form the 13-gene panel SigIQGAP3NW.
Tumor-associated signature score assignment
Coefficient (coef) of SigIQGAP3NW component genes (n = 13) in predicting BCR was produced using multivariate Cox PH regression with the R Survival package. Tumor-associated signature scores were calculated as: Sum (coef1 × Gene1exp + coef2 × Gene2exp + … …+ coefn × Genenexp), where coef1 … coefn are the coefs of individual genes and Gene1exp … … Genenexp are the expression of individual genes.
Immunohistochemistry (IHC)
IHC was performed as we have previously described [42]. Slides were deparaffinized in xylene and cleared in ethanol. Antigens were retrieved via heat treatment in a sodium citrate buffer (pH = 6.0), blocked for non-specific binding sites in PBS containing 1% BSA and 10% normal goat serum (Vector Laboratories) for 1 h. The slides were incubated with an anti-IQGAP3 antibody (ab219354, Abcam; 1:400) overnight at 4 °C, followed by the addition of biotinylated goat anti-rabbit IgG and Vector ABC reagent (Vector Laboratories). Chromogenic reaction (Vector Laboratories) and slide counterstaining with hematoxylin (Sigma Aldrich) were performed. Images were analyzed with ImageScope software (Leica Microsystems Inc.); staining intensity was quantified as H-Scores using the formula [H-Score = (% Positive) × (intensity) + 1] [42]. Secondary antibody alone was used as a negative control.
Ethics statement
All experiments involving animals were performed according to the ethical policies and protocols approved by McMaster University Animal Research Ethics Board (16-06-23).
Generation of CRPC in animal models
LNCaP cells (5x106)-derived subcutaneous (s.c.) xenografts were generated in NOD/SCID mice (The Jackson Laboratory) with tumor volume determined [42]. Tumor growth was monitored by serum PSA levels (PSA kit, Abcam). Surgical castration was performed at a time when tumor size was reaching 100–200 mm3. Serum PSA was determined before and following castration. A rise in serum PSA reflects CRPC growth.
Quantitative real-time PCR
Total RNA was isolated from tumors produced by LNCaP cells in both intact and castrated mice with the Iso-RNA Lysis Reagent (5 PRIME). Reverse transcription was carried out with Superscript III (Thermo Fisher Scientific). Quantitative real-time PCR was run with the ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster, California, USA) using SYBR-green (Thermo Fisher Scientific). Fold changes were calculated using the formula: 2−ΔΔCt. The real-time PCR primers for all 13 SigIQGAP3NW component genes are documented in Table S2.
Statistical analysis
Kaplan-Meier curves and logrank test were conducted by R Survival package and tools provided by cBioPortal. Cox regressions were analyzed using R survival package. ROC (receiver-operating characteristic) profiles were constructed using the PRROC package in R. Multiple t-test with p value adjusted by the Holm method (Holm-Bonferroni method) was carried out using R. Other statistical analyses were performed using specific website programs and by GraphPad Prism 7 and SPSS 26. Data were presented as mean ± SEM/SD. A value of p < 0.05 was considered statistically significant.
Results
IQGAP3 expression at the mRNA but not the protein level correlates with PC pathogenesis and progression
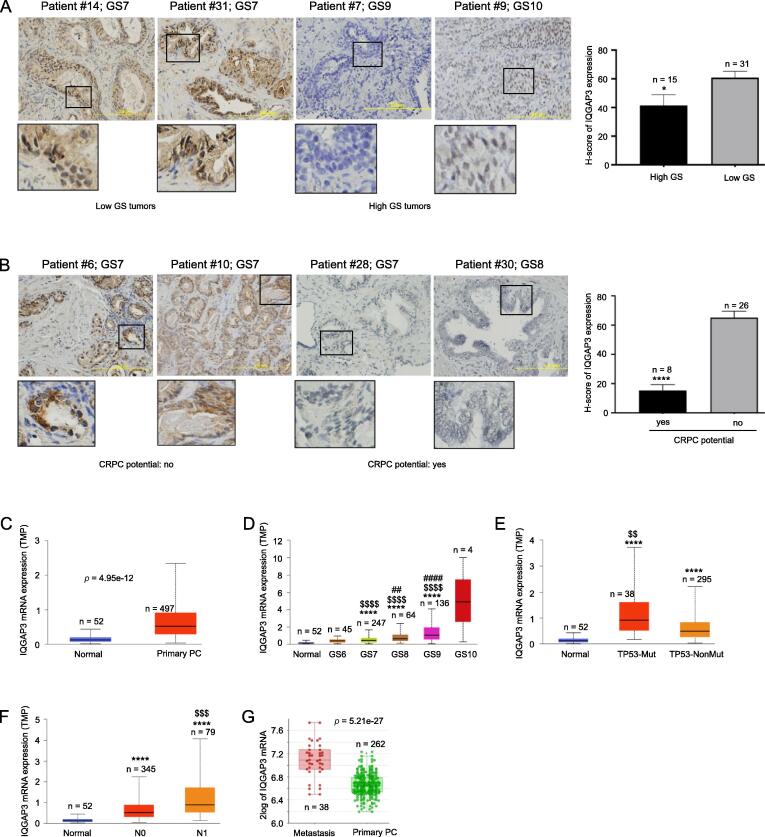
The relevance of IQGAP3 mRNA expression to tumorigenesis has been commonly analyzed across cancer types including PC (23, 25). Nonetheless, little is reported on IQGAP3′s protein expression. We observed a heterogeneous expression of IQGAP3 protein in primary PCs (see IQGAP3 H-scores in Table S1A). Due to the small number of patients, we grouped PC with GS ≤ 8 into the low-grade group and PC with GS9 and 10 into the high-grade group. Typically, GS7 PCs displayed more intensive staining compared to GS9 and GS10 PCs, which is supported by quantification, i.e. a reduction of IQGAP3 protein expression in high-grade PCs (Fig. 1A). Re-assignment of GS8 tumors into the high-grade group did not change the outcome (data not shown). We further analyzed a small set of organ-confined PC with known outcomes of CRPC (CRPC potential). Decreases in IQGAP3 protein expression occurred in primary PCs with positive CRPC potential (Fig. 1B, also see Table S1B for details). A heterogeneous expression pattern for IQGAP3 protein expression in PC was also presented in “The Human Protein Atlas” (https://www.proteinatlas.org/ENSG00000183856-IQGAP3/pathology/prostate + cancer). We acknowledge the small patient population used for this analysis; nonetheless, the data does not support a positive association between IQGAP3 protein expression and PC severity (grade) or progression. These observations were surprising considering the positive associations of IQGAP3 mRNA expression with adverse clinical features of various cancers [10].
Table 1.
Univariate and multivariate Cox analysis of SigIQGAP3NW for PFS in PC.
| Factors |
Univariate Cox Analysis |
Multivariate Cox Analysis |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| Sig 1 | 2.72 | 2.14–3.45 | <2e-16*** | 1.83 | 1.33–2.52 | 0.0002*** |
| Age 2 | 1.02 | 0.99–1.05 | 0.189 | 0.99 | 0.96–1.02 | 0.491 |
| WHO GG2 3 | 3.53 | 0.48–27.64 | 0.219 | 3.33 | 0.43–25.69 | 0.248 |
| WHO GG3 | 5.27 | 0.69–40.57 | 0.110 | 4.01 | 0.51–31.43 | 0.185 |
| WHO GG4 | 9.76 | 1.28–74.61 | 0.028* | 6.44 | 0.82–50.54 | 0.076 |
| WHO GG5 | 21.34 | 2.96–154.5 | 0.002** | 10.6 | 1.4–80.16 | 0.022* |
| Margin 4 | 2.3 | 1.52–3.48 | 8.1e-5*** | 1.26 | 0.8–2.0 | 0.317 |
| Stage 5 | 3.69 | 2.08–6.52 | 7.45e-6*** | 1.59 | 0.83–3.06 | 0.164 |
SigIQGAP3NW score; 2 Age at diagnosis; 3 WHO grade group (GG) 2–5 in comparison to WHO GG1; 4 Compared to no surgical margin status; 5 Stage 3 + 4 in comparison to Stage 1 + 2; *, **, *** for p < 0.05, 0.01, and 0.001 respectively.
Fig. 1.
Association of IQGAP3 with PC tumorigenesis and progression. A., B. Immunohistochemistry staining of IQGAP3 expression in PC. PCs in the B panel were primary PCs with (yes) or without (no) progression to CRPC. Typical images and quantification (mean ± SD/standard deviation) are shown. GS: Gleason score; the marked regions are enlarged 3-fold. * and ****: p < 0.05 and p < 0.0001 respectively by 2-tailed Student’s t test. C-F. Analyses of IQGAP3 mRNA expression (TMP: transcripts per million) in the indicated tissues using the TCGA dataset organized by UALCAN (29). Statistical analyses were performed by UALCAN. ****: p < 0.0001 in comparison to Normal; $$: p < 0.01 compared to PC without TP53 mutation (NonMut) (E), $$$$: p < 0.0001 in comparison to GS6 PCs (D) or PC without lymph node metastasis (N0) (F); ## and ####: p < 0.01 and p < 0.0001 respectively in comparison to GS7 tumors (D). G. The analyses were performed using Sawyers’s DNA microarray dataset (28) within the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nlhttp://r2platform.com).
We subsequently analyzed IQGAP3 mRNA expression following PC pathogenesis. In comparison to normal prostate tissues, primary PC generally expresses substantially elevated IQGAP3 mRNA (Fig. 1C). The magnitude of upregulation correlates with PC severity from GS7 – GS9 (Fig. 1D). The lack of significant increase (p > 0.05) of IQGAP3 mRNA expression in GS10 tumors was likely attributed to the small number of patients. IQGAP3 mRNA expression was further elevated in PCs with TP53 mutation vs those without the mutation (Fig. 1E), lymph node metastasis (Fig. 1F), and distant metastasis (Fig. 1G). Collectively, the above evidence supports the positive associations of IQGAP3 mRNA expression with PC pathogenesis, severity, metastasis, and signaling events.
A potential reason for the difference between the mRNA and protein expression of IQGAP3 during PC pathogenesis could be a potential rapid degradation of the IQGAP3 protein in advanced PC (see Discussion for details). While this possibility needs validation, the above observations nonetheless suggest that the mRNA expression may better reflect IQGAP3′s involvement in PC pathogenesis and prognosis.
Dominant association of IQGAP3 with mitosis and chromosome segregation
Individual factors regulate biological events or processes via network (NW) actions. To identify the major processes or network components of the IQGAP3 network relevant to PC, a set of IQGAP3 correlative genes (genes with their expression correlating with IQGAP3 mRNA expression) were derived from the TCGA PC dataset (n = 497) (Fig. S1) using the LinkedOmics platform [35]. Among these correlative genes, the top enriched GO (gene ontology) biological processes are related to chromosome segregation, DNA replication, spindle organization, and others facilitating DNA metabolism (Fig. 2A); the enrichment is quite robust based on the profiles of core enrichment (Fig. 2B). Similar enrichments were also demonstrated using the KEGG gene sets (Fig. S2A). Consistent with this enrichment pattern, we noticed the top correlative genes, which show robust correlations with IQGAP3, being highly relevant to mitosis and chromosome segregation (Fig. S2B, Table S3A). For instance, the top 8 correlative genes are at Spearman R values of 0.888 to 0.914 and p value ≤ 5.35e-169 (Fig. 2C); NUSAP1 (nucleolar and spindle associated protein 1), TPX2 (TPX2 microtubule nucleation factor), ESPL1 (extra spindle pole bodies like 1, separase), KIF4A (kinesin family member 4A), KIF20A (kinesin family member 20A), GTSE1 (G2 and S-phase expressed 1), PLK1 (polo like kinase 1), and TOP2A (DNA topoisomerase II alpha) all function in chromosome segregation (Table S3B). Both PLK1 and TOP2A promote chromosome segregation in mitosis [43], [44]; TOP2A is required for cancer cell cycle progression [44]. Of importance, inhibition of TOP2 (DNA topoisomerase II) is a classic action of cancer therapy; recently, PLK1 inhibitors have been approved by FDA for cancer therapy [45], [46]. The above evidence thus indicates a potential vulnerability of PCs with elevated IQGAP3 expression to inhibitors targeting either PLK1, TOP2A or both. This concept is further supported by the high-level correlations between IQGAP3 and PLK1 in the TCGA Cell 2015, SU2C, and MSKCC cohorts as well as between IQGAP3 and TOP2A in the TCGA Cell 2015 and SU2C datasets (Fig. S3A). Of relevance, correlations of IQGAP3 with either PLK1 or TOP2A were enriched in high-grade PC and metastatic CRPCs (the SU2C cohort) treated with taxane drugs (Fig. S3A).
Fig. 2.
Association of IQGAP3 correlative genes with mitosis and chromosome segregation in PC. IQGAP3 correlative genes were determined with Spearman correlation using the TCGA dataset (n = 497) organized by LinkedOmics (35). A. Analysis of IQGAP3 correlative genes for the enrichment of GO-BP (gene ontology biological process) gene sets by GSEA. All enrichments were at FDR (false discovery rate) < 0.05. B. Details of GSEA enrichment for the indicated GO-BP gene sets. C. Correlations of IQGAP3 mRNA expression with the indicated gene expression in PC (TCGA dataset, n = 497). D. Genes with expression in PC at Spearman R ≥ 0.6 (Table S3A) were analyzed for enrichment using the Metascape platform (30). The top 10 enriched processes are shown (see Table S3C for detailed enrichment).
Fig. 3.
Top enrichment of mitosis in the IQGAP3 correlative genes across human cancers. IQGAP3 correlative genes were determined in 32 human cancer types using LinkedOmics. Those genes with Spearman R ≥ 0.6 (p < 0.001) in all 32 cancer types were analyzed for pathway enrichment using Metascape. A. Cancer types with Mitotic Cell Cycle being the top (#1) enrichment are shown; the exceptions are THYM and TCGC, in which Mitotic Cell Cycle was enriched as #2 (THYM) and #3 (TCGC) enrichment. The -log10(p) for KICH is indicated. B., C. Cancer types showing the correlations of IQGAP3 mRNA expression with PLK1 and TOP2A mRNA expression. The respective TCGA datasets used for these analyses include - ACC: adrenocortical carcinoma; BLCA: bladder urothelial carcinoma; BRCA: breast invasive carcinoma; CHOL: cholangiocarcinoma; COAD: colon adenocarcinoma; GBM: glioblastoma multiforme; HNSC: head and neck squamous cell carcinoma; KICH: kidney chromophobe; KIPR: kidney renal papillary cell carcinoma; KIRC: kidney renal clear cell carcinoma; LAML: acute myeloid leukemia; LGG: brain lower grade glioma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; MESO: mesothelioma; OV: ovarian serous cystadenocarcinoma; PAAD: pancreatic adenocarcinoma; PCPG: pheochromocytoma and paraganglioma; PRAD: prostate adenocarcinoma; SARC: sarcoma; SKCM: skin cutaneous melanoma; STAD: stomach adenocarcinoma; TGCT: testicular germ cell tumors; THCA: thyroid carcinoma; THYM: thymoma; UCEC: uterine corpus endometrial carcinoma; and UVM: uveal melanoma.
We further analyzed the network components within the top correlative genes, which were defined as Spearman R ≥ 0.6 (n = 140, Table S3A). Among these genes, the mitotic cell cycle process is the top process enriched along with other pathways relevant to mitosis including the PLK1 pathway (Fig. 2D). The correlations of CDK1 (Spearman R = 0.857 and p = 2.24e-144) and its activator CCNA2 (encoding cyclin A2; R = 0.868 and p = 6.42e-153; Table S3A) support the IQGAP3NW-associated enrichment in the mitotic cell cycle process. The protein network connection among these top correlative genes and details of the enrichment are presented in Fig. S3B and Table S3C.
To further investigate the dominant association of IQGAP3NW with the mitotic cell cycle observed in PC, we obtained genes with correlations with IQGAP3 expression at Spearman R ≥ 0.6 in other 32 TCGA cancer types and performed pathway enrichment analysis. Except CESC (cervical squamous cell carcinoma), DLBC (diffused large B-cell lymphoma), ESCA (Esophageal carcinoma), LUSC (lung squamous cell carcinoma), and UCS (uterine carcinoma), the rest of the 27 cancer types revealed “mitotic cell cycle” or “mitotic cell cycle process” as the top enriched process except TGCT (testicular germ cell tumor) and THYM (thymoma) (Fig. 3A). Additionally, numerous cancer types possess high-level correlations (Spearman R ≥ 0.6) of IQGAP3 with PLK1, TOP2A, and both (Fig. 3B, 3C), indicating inhibition of either or both factors as potential therapies for these cancer types with elevated IQGAP3 expression, for instance, ACC (adrenocortical carcinoma) and LGG (low-grade glioma) (Fig. 3B, 3C).
Construction of a multigene panel from IQGAP3-related DEGs
The main participants of the IQGAP3 correlative genes with processes highly relevant to not only PC but across cancer types (Fig. 2, Fig. 3) are consistent with the observed prognostic values of IQGAP3 in numerous cancers [10]. However, to our best knowledge, the prognostic potential of the IQGAP3 network has not been explored in any cancer types. To fulfill this knowledge gap, we first screened IQGAP3-based stratification of BCR risk using a set of cutoff points within the TCGA PanCancer PC dataset (Fig. S4). At the 2.5 SD (standard deviation), the risk stratification was the most effective (Fig. S4). From these two groups, differentially expressed genes (DEGs, n = 281) were obtained, which were defined as q < 0.05 and fold change ≥ |2| (Table S4A). A dataset was retrieved from the TCGA PanCancer PC dataset which contained the expression of these DEGs along with relevant clinical features. We randomized the dataset into a Training (n = 340) and Testing (n = 152) population at the ratio of approximately 7:3 using an R program. With the Training population, covariate selection for predicting PC recurrence (BCR) was performed using Elastic-net within the R glmnet package (see Materials and Methods for details), resulting in SigIQGAP3NW signature consisting of 13 component genes (Table S4B).
We calculated SigIQGAP3NW score for individual tumors as ∑(coefi × Geneiexp)n (coefi: Cox coefficient of genei, Geneiexp: expression of Genei, n = 13). Coefs were generated with the multivariate Cox model. SigIQGAP3NW score effectively stratifies BCR risk in the Training, Testing, and Full cohort (Fig. 4A). SigIQGAP3NW score displays a high level and significant correlation with IQGAP3 expression (Fig. 4B), which supports the construction of SigIQGAP3NW from IQGAP3-related DEGs. Of importance, SigIQGAP3NW effectively stratifies PC recurrence in an independent primary PC MSKCC cohort (Fig. 4C) and survival probabilities in another independent metastatic CRPC (mCRPC) SU2C cohort (Fig. 4D). SigIQGAP3NW scores predict the risk of recurrence in TCGA PanCancer PC dataset (Training, Testing, and Full cohort) and MSKCC cohort as well as the risk of fatality in the SU2C dataset (Fig. 5E). In two independent (MSKCC and SU2C) cohorts, SigIQGAP3NW discriminates PC recurrence (MSKCC) and fatality risk (SU2C) with AUC values as effective or better than its discrimination of PC recurrence in the TCGA dataset (Fig. 5F). Collectively these results validate the prognostic potential of SigIQGAP3NW in PC.
Fig. 4.
SigIQGAP3NW-mediated stratification of PC recurrence and fatality risk. A. Separation of PCs in the Training, Testing, and Full TCGA PanCancer PC dataset with SigIQGAP3NW score. Cutoff points were estimated with Maximally Selected Rank Statistics within the R Maxstat package. Kaplan Meier curves and log-rank test were performed using the R survival package. B. Correlation between IQGAP3 mRNA expression and SigIQGAP3NW score within the TCGA PanCancer PC dataset. C., D. Stratification of PC recurrence in the MSKCC (C) and overall survival probability in mCRPC (SU2C) dataset (D). E. Prediction of PC recurrence (Training, Testing, Full, and MSKCC) and fatality (SU2C) using the respective SigIQGAP3NW scores (continuous data). F. ROC-AUC (receiver operating characteristic-area under the curve) curves for the indicated cohorts.
Fig. 5.
SigIQGAP3NW effectively predicts poor prognosis in a spectrum of human cancers. A. The AUC values of SigIQGAP3NW in discriminating survival probability in all the indicated cancer types except otherwise indicated. The size (n) of TCGA PanCancer datasets used in these analyses is indicated. B-F. SigIQGAP3NW-mediated stratifications of survival probability and recurrence risk for the indicated cancer types are shown.
Consistent with IQGAP3′s biomarker potential in other cancer types (10) and IQGAP3-associated enrichment of mitotic cell cycle process across cancer types (Fig. 3A), SigIQGAP3NW predicts poor OS in 23 cancer types and progression-free survival (PFS) in 3 cancer types all at AUC > 0.5 (Fig. 5A). Even for the 23 cancer types in which SigIQGAP3NW displayed the predictive ability of their fatality risks, SigIQGAP3NW also effectively evaluated their recurrence risk (data not shown). The predictions are at AUC ≥ 0.7 for PFS in THYM as well as for poor OS in ACC, UVM (uveal melanoma), pRCC (papillary renal cell carcinoma), ccRCC, MESO (mesothelioma), UCS (uterine carcinosarcoma), and LAML (acute myeloid leukemia) (Fig. 5A), which once again either matched or outperformed SigIQGAP3NW’s prediction of PC recurrence in the TCGA PanCancer dataset (AUC = 0.7), the dataset in which SigIQGAP3NW was originally constructed in. With optimized cutoff points, SigIQGAP3NW robustly stratifies the fatality risk of ACC, UVM, pRCC, and ccRCC (Fig. 5B-E) as well as the recurrence risk in THYM (Fig. 5F). Collectively, we have validated SigIQGAP3NW’s prognostic biomarker potential not only in independent PC cohorts but also in multiple other cancer types.
Characterization of SigIQGAP3NW
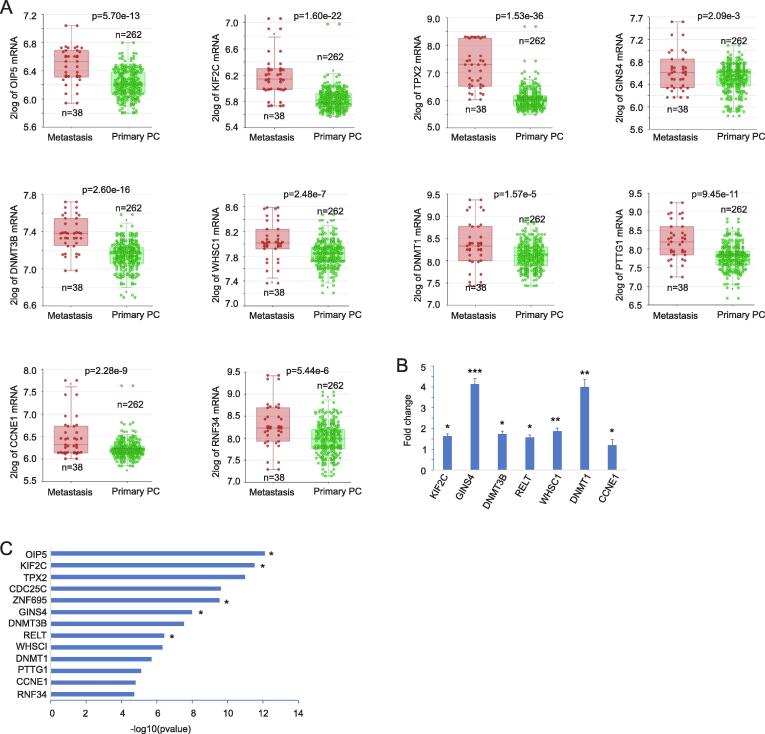
SigIQGAP3NW is a novel multigene prognostic biomarker. Among its 13 component genes, except CDC25C, DNMT3B, PTTG1, CCNE1 and DNMT1, the involvement of other component genes in PC was either limitedly studied or unexplored (OIP5, GINS4, and RNF34) (Table S4B). For OIP5, KIF2C, TPX2, and ZNF695, we provided evidence for their upregulations in PC tissues compared to normal prostate tissues, following advances in PC grades, and in lymph node metastasis compared to non-metastatic PCs (Fig. 6A-D). Increases in OIP5, KIF2C, and TPX2 expression were also associated with p53 mutations in PC (Fig. 6A-C). Significant upregulations of OIP5, KIF2C, TPX2, GINS4, DNMT3B, WHSC1, DNMT1, PTTG1, CCNE1, and RNF34 also occurred in distant metastasis compared to local PCs (Fig. 7A). Furthermore, we demonstrated that LNCaP cell-derived xenografts generated in castrated NOD/SCID mice displayed increases in KIF2C, GINS4, DNMT3B, RELT, WHSC1, DNMT1, and CCNE1 compared to LNCaP cell tumors generated in intact NOD/SCID mice (Fig. 7B), supporting upregulations of these component genes of SigIQGAP3NW in CRPC.
Fig. 6.
Upregulation of SigIQGAP3NW component genes with PC pathogenesis. A-D. Upregulation of OIP5, KIF2C, TPX2, and ZNF695 in PC, PCs with advanced grades, lymph node metastasis, and p53 mutations. Analyses were carried out using the TCGA dataset organized by UALCAN. **: p < 0.01 and ****: p < 0.0001 in comparison to Normal; $, $$, $$$, and $$$$: p < 0.05, 0.01, 0.001, and 0.0001 respectively in comparison to GS6, N0, and None p53 mutation PCs; #, ##, ###, and ####: p < 0.05, 0.01, 0.001, and 0.0001 respectively compared to GS7 PCs. &: p < 0.05 in compared to GS8 tumors.
Fig. 7.
Upregulation of SigIQGAP3NW component genes following PC progression. A. The Sawyers dataset was used to reveal the upregulation for the indicated SigIQGAP3NW component genes in distant PC metastasis. B. LNCaP cell-derived xenografts in either intact mice (n = 6) or castrated mice (n = 6) were produced and used to determine gene expression by real-time PCR for the indicated genes. Gene expression in CRPC xenografts was presented as fold change (mean ± SD) in reference to their expression in tumors produced in intact mice. *, **, and ***: p < 0.05, 0.01, and 0.001 respectively by 2-tailed Student’s t-test. C. All component genes of SigIQGAP3NW predict PC recurrence with respect to their gene expression in univariate Cox analysis. OIP5, KIF2C, ZNF695, GINS4, and RELT remain risk factors of PC recurrence after adjusting for age at diagnosis, WHO GG grade, stage, and surgical margin status.
The common upregulation of the SigIQGAP3NW component genes following PC pathogenesis and progression is in accordance with their functions in multiple pathways relevant to PC (Table S4C). Both KIF2C and TPX2 contribute to mitosis by regulating microtubule functions (see Table S4C for references). OIP5 (Mis18β) is essential in centromere formation following mitosis [47]. CDC25C (cell division cycle 25C) and CCNE1 (cyclin E1) contribute to cell cycle progression (Table S4C). PTTG1 (pituitary tumor transforming gene 1) plays an essential role in sister chromatid separation during mitosis [48]. OIP5, PTTG1, KIF2C, and TPX2 regulate essential aspects of mitosis and are clustered together for their correlations (Fig. S5A); IQGAP3 is within this correlation group (Fig. S5A), supporting IQGAP3′s major role in mitosis in PC. DNMT3B and DNMT1 contribute to the establishment of DNA methylation, a required feature in cell cycle division (Table S4C). WHSC1 is a histone methyltransferase [49]. GINS4 is required to initiate DNA replication via assembly of the GINS complex (Table S4C). RELT promotes activation of the p38 and JNK-MAPK cascade (Table S4C). The function of ZNF695 (zinc finger protein 695) remains unclear. RNF34 is an E3 ubiquitin ligase and inhibits p53 function (Table S4C). The above evidence thus supports the involvement of SigIQGAP3NW in multiple processes relevant to mitosis, DNA metabolism, and others; this property might be the basis for SigIQGAP3NW as a potential prognostic biomarker in PC and other cancer types.
The above concept is supported by the ability of all component genes to predict PC recurrence as individual genes based on their gene expression (Fig. 7C). OIP5, KIF2C, ZNF695, GINS4, and RELT assess PC recurrence risk independently of age at diagnosis, PC grade, tumor stage, and surgical margin (see the legend of Fig. 7C). Consistently, SigIQGAP3NW predicts PC recurrence after adjustment for these clinical features (Table 1). All individual component genes effectively separate PCs into a low-risk and high-risk group of PC recurrence (Fig. S5B). The risk of PC recurrence is at HR (hazard ratio) > 2 for all component genes and HR > 4 for SigIQGAP3NW in the high-risk groups compared to low-risk groups (Fig. S5C).
Properties of immune evasion in PC with high SigIQGAP3NW score
Evading immune response is an essential feature of tumorigenesis and cancer progression. Given the observed prognostic potential of SigIQGAP3NW, its relevance to PC-associated immune alterations is worthy of investigation. For this purpose, we profiled the immune cell populations within PC using the TCGA PanCancer PC RNA-seq data (n = 492) with multiple computational programs, xCell and ssGSEA [39], Epic [36], MCPCounter [37], quanTIseq [38], and CIBERSORT [34]. A set of immune cell changes was detected in multiple programs. Among 24 immune cell populations detected by ssGSEA, SigIQGAP3NW score correlated with the alterations of 16 immune cell types in PC at Spearman R > |0.1| and p < 0.05, including positive correlations with CD8 + T cells, cytotoxic cell, T regulatory (Treg) cells, and Th2 CD4 + T cells (Th2.cells) (Fig. 8A). PCs with high-risk of recurrence stratified by SigIQGAP3NW score cutoff point have enriched content of Th2 cells, Treg cells, and cancer-associated fibroblasts (CAF) (Fig. S6A-C). Treg cells and CAF are known for their contributions to the immunosuppressive microenvironment [50]. In addition, Th2 cells can facilitate immune evasion [51]. The above evidence thus supports a positive correlation of SigIQGAP3NW with PC immune evasion.
Fig. 8.
SigIQGAP3NW correlates with immune cell infiltration. A-F. Immune cell populations in PC, ACC, ccRCC, pRCC, THYM, and UVM were determined from RNA-seq data within the respective TCGA dataset using multiple computational programs. Immune cell contents obtained using ssGSEA were analyzed for correlation (Spearman) with SigIQGAP3NW scores. All correlations were at p < 0.05; only those correlations with Spearman R > |0.1| are indicated. Th2 cells and Treg cells are marked with red, indicating their frequent correlations with SigIQGAP3NW score. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Consistent with SigIQGAP3NW’s robust prediction of poor OS and PFS in ACC, ccRCC, pRCC, UVM, and THYM (Fig. 5), we observed their SigIQGAP3NW scores correlating with immune cell changes (Fig. 8B-F). Interestingly, Th2 cells and Treg cells were positively correlated with SigIQGAP3NW in 4 out of 6 cancer types (PC, ACC, ccRCC, pRCC, THYM, and UVM) (Fig. 8, Fig. S6A-B). SigIQGAP3NW score correlates with Th2 cells at Spearman R = 0.78 (p < 2.2e-16) in ACC (Fig. 8B), suggesting a major role of Th2 cells in ACC immune escape, a feature captured by SigIQGAP3NW. SigIQGAP3NW robustly associates with the content of mesenchymal stem cells (MSCs) in ACC and ccRCC (Fig. S6D, E). MSCs contribute to immune escape in cancer [52]. Collectively, we present a comprehensive set of evidence which suggests a role of SigIQGAP3NW in immune escape in PC and other cancers.
SigIQGAP3NW-mediated prediction of response to ICB (immune checkpoint blockade) therapy
To provide additional support for the positive correlation of SigIQGAP3NW with PC-associated immune tolerance, we observed that RELT, a component gene of SigIQGAP3NW (Table S4B), strongly correlates with the expression of multiple immune checkpoints in PC (Fig. 9A). TGFβ1 promotes immune evasion via facilitating T-cell exclusion [53]. Both ADORA2A (adenosine A2a receptor) and CTLA4 are expressed on tumor-associated T-cell surface and play critical roles in suppressing T-cell response [54]. TIM-3 (HAVCR2) and its ligand LGALS9 (encoding Galactin-9) contribute to the lack of costimulatory signals in dendritic cells (DCs) when DCs present tumor-associated antigens (TAAs) to T cells, leading to T cell anergy [55]. In this regard, RELT mRNA expression significantly correlated with increases in DCs and neutrophils in PC (Fig. 9B). Tumor-associated neutrophil (TAN), macrophages, and myeloid-derived suppressor cells (MDSC) are major cancer-infiltrating myeloid cells, contributing to the immunosuppressive tumor microenvironment (TME) [56]. Additionally, polymorphonuclear MDSCs (PMN-MDSCs), the major group MDSCs, share numerous surface markers (antigens) with neutrophils and are likely differentially related to neutrophils induced by tumors [57], supporting the potential contributions of neutrophils to immune tolerance in PC. MDSCs, Treg cells, and tumor-associated macrophages (TAM) are major immune cells in TME suppressing immune responses [58]. Consistent with this knowledge, RELT expression robustly correlated with Treg and MDSC contents in PC (Fig. 9B). Furthermore, RELT positively correlated with multiple immune checkpoint expression at high levels in multiple cancer types (Fig. S7A) as well as Treg and MDSC infiltration in ccRCC and pRCC (Fig. S7B). Similarly, RELT mRNA expression significantly correlated with TAN and DCs at Spearman R ≥ 0.5 (p ≤ 8.34e-11) in 11 cancer types with a few exceptions (Fig. 9C). The above evidence collectively suggests that RELT enhances immune evasion in PC and multiple cancer types via association with immune checkpoints’ expression, which leads to a functional disarray in T cells, DCs, and neutrophils.
Fig. 9.
Biomarker potential of RELT and SigIQGAP3NW in predicting response to ICB therapy. A. Correlation of RELT mRNA expression with the expression of indicated immune checkpoints in PC. Analysis was performed using the TCGA PC dataset (n = 497) within the TISIDB platform (31). B. Correlations of RELT mRNA expression with neutrophil, dendritic cells (DCs) (graphs were produced by TIMER), Treg cells, and MDSC (graphs were generated by TISIDB). C. Correlations of RELT mRNA expression with neutrophil and dendritic cells in the indicated cancer types (p < 0.0001 for all correlations and p ≤ 8.34e-11 for all correlations at R ≥ 0.5). The analyses were performed using the TIMER platform (33). D., E. The AUC values for RELT, SigIQGAP3NW, and other indicated biomarkers in predicting response to ICB therapy in the indicated cohorts were obtained using TIDE and graphed. F., G. Stratification of responders and non-responders in anti-CTLA4 antibody (lpi: Ipilimumab) naïve melanoma and glioblastoma treated with PD1 blockade. The graphs were produced using TIDE.
In a set of 25 cancer cohorts treated with ICB within the TIDE (tumor immune dysfunction and exclusion) platform [32), RELT predicts resistance to ICB therapy in 14 cohorts at AUC > 0.5 (Fig. S8). Among 4 datasets, the prediction was at AUC > 0.7 (Fig. 9D); RELT assesses resistance to ICB therapy in the “Zhao2019_PD1_Glioblastoma_Post” and “Rupping2021_PD_NSCLC” cohorts more effectively than a set of well-established biomarkers of ICB therapy, including Merck18 [59], INFG (IFNγ) [59], CD8 [60], CD274 (PDL-1) [61], and MSI.score (microsatellite instability) [62] (Fig. 9D). In this context, the perfect discrimination achieved by RELT in predicting Glioblastoma response to PD1 therapy is impressive (Fig. 9D), although further investigations are warranted. These observations are novel and likely significant; nonetheless, its potential as a biomarker of ICB therapy is consistent with RELT’s broad associations with multiple immune checkpoints across cancer types (Fig. S7), which also suggests an association of SigIQGAP3NW with response to ICB therapy.
We subsequently determined this possibility using the TIDE platform [32]. SigIQGAP3NW was found to predict resistance to ICB therapy at AUC ≥ 0.5 in 8 cohorts treated with ICB including HNSC, melanoma, bladder cancer, and gastric cancer (Fig. S8). The prediction achieved by SigIQGAP3NW is better than several published ICB biomarkers, including TMB (tumor mutational burden), T.Clonality, and B.Clonality (Fig. S8). Of importance, SigIQGAP3NW outperformed the most potent ICB biomarkers in predicting resistance of melanoma to PD1 therapy in the “Riadz2017_PD1_Melanoma_lpi.Naive” cohort (Fig. 9E). Compared to RELT, SigIQGAP3NW displays a unique prediction towards HNSC (head and neck squamous cell carcinoma) in the “Uppaluri2020_PD1_HNSC_Post” cohort (comparing Fig. 9E and 9D, Fig. S8), supporting the potential of SigIQGAP3NW as a novel ICB therapy biomarker. SigIQGAP3NW stratifies responders from non-responders in melanoma and glioblastoma (Fig. 9F, G) and SigIQGAP3NW was the only biomarker capable of this stratification in both datasets among a set of published ICB biomarkers (Fig. S9). Collectively, evidence supports SigIQGAP3NW’s biomarker potential in assessing response to ICB therapy.
Discussion
As a late member of the IQGAP family, IQGAP3′s biology has yet to be thoroughly investigated compared to the other two family members. Notably, the oncogenic aspect of IQGAP1 and IQGAP2 but not IQGAP3 has been studied using knockout mice. Nonetheless, the association of IQGAP3 with poor prognosis in multiple cancer types as well as its activities in promoting cancer cell proliferation and xenograft formation have been reported (10). However, the network action underlying IQGAP3′s association with cancer progression remains unclear.
Association of the IQGAP3 network with mitosis and the implications of this association
We report the first comprehensive analysis for IQGAP3′s dominant association with mitosis, chromosome segregation, and DNA metabolism not only in PC but also across many cancer types (Fig. 3A). The associations of IQGAP3 with genes involved in these processes are at impressively high levels with Spearman R approaching to or exceeding 0.9 (Fig. 2C; Fig. 3B, C). To our best knowledge, these observations not only are novel but also possess potential for clinical applications. Namely, with the FDA approved PLK1 inhibitors (https://oncoheroes.com/press-releases-content/2020/10/14/volasertib-a-potential-new-treatment-for-rhabdomyosarcoma-receives-orphan-drug-designation-from-the-us-fda) and the development of less toxic TOP2 inhibitors [63], it will be intriguing to perform clinical trials on PC, ACC, ccRCC, pRCC, and others with elevated IQGAP3 expression with either one or a combination of both inhibitors. In this regard, we have recently reported a correlation between OIP5 and PLK1 (Spearman R = 0.64, p = 3.49e-34) in pRCC, and the PLK1 inhibitor BI2356 potently inhibited pRCC cells with OIP5 overexpression in the development of xenograft tumors [64]. With much higher correlations observed between IQGAP3 and PLK1 in PC and other cancers, as well as the presence of OIP5 in SigIQGAP3NW, it is appealing to examine the therapeutic potential of PLK1 inhibitor with and without inhibition of TOP2A in treating cancers with elevated IQGAP3 expression or high SigIQGAP3NW score.
Predictive potential of the IQGAP3 network towards prognosis and response to ICB therapy
SigIQGAP3NW consists of genes regulating mitosis, chromosome segregation, and DNA metabolism, which recapitulates the major features of IQGAP3 correlative genes in PC. In line with our demonstrated conservation of these features across a spectrum of cancer types, SigIQGAP3NW possesses effective prognostic values towards OS in many cancer types (Fig. 5), despite the panel being initially derived in the context of predicting PFS in PC. SigIQGAP3NW predicts PC recurrence risk in the TCGA PanCancer dataset at AUC of 0.7 (Fig. 4F) and PFS and OS at AUC > 0.7 in both independent PC cohorts (Fig. 4F). A similar trend is observed for additional cancer types including ACC, UVM, KIPR (pRCC), KIRC (ccRCC), and others (Fig. 5A). The high level of robustness observed warrants SigIQGAP3NW’s clinical potential as a prognostic biomarker in multiple cancer types. This concept is supported by not only SigIQGAP3NW-associated prediction of responses to ICB therapy in multiple cohorts and cancer types (Fig. S8), but also its ability to outperform several well-established ICB therapy biomarkers in some cohorts during this process (Fig. 9E). As a component gene of SigIQGAP3NW, RELT displays a novel association with multiple immune checkpoints in PC and other cancer types (Fig. 9A; Fig. S7), providing a rationale for RELT and SigIQGAP3NW as potential ICB biomarkers. As a single gene, RELT’s potency as a biomarker of ICB therapy is novel and intriguing. Additionally, our observations are in accordance with its reported actions in activating the NF-κB pathway [65] and RELT-derived negative impact on the early phase of T-cell responses in animals [66].
Outlook: IQGAP3-related unknowns
The function of another SigIQGAP3NW component gene ZNF695 remains unknown (Table S4C). Given its high correlations (Spearman R > 0.6) with OIP5, KIF2C, CDC25C, and TPX2 (Fig. S5A), it is tempting to suggest its potential roles in mitosis at least in PC. This deserves a future investigation. While IQGAP3 is known to facilitate cell proliferation, its mechanisms in mitosis, chromosome segregation, and DNA metabolism should also be further investigated. This potential is particularly intriguing considering the high level of conservation among IQGAP1, IQGAP2, and IQGAP3 [10]; the cell membrane residence of both IQGAP1 and IQGAP2 in PC [40], [67]; and the largely nuclear localization of IQGAP3 in PC observed in this study (Fig. 1A, B). We noted a disparity between IQGAP3 mRNA and protein expression for its association with PC pathogenesis (Fig. 1); the mechanisms underlying this disparity remain unknown. In addition to the small sample size used to analyze IQGAP3 protein expression, other potential contributing factors include possible reductions in IQGAP3 mRNA translation and rapid IQGAP3 protein degradation in advanced PCs. Among these possibilities, we believe the protein degradation possibility is more likely. This is based on the current knowledge of IQGAP3 in promoting tumorigenesis and the frequent scenarios of functional proteins being dynamically degraded; this is particularly relevant to proteins functioning in cell cycle progression including cyclins and others. Given the major association of IQGAP3 with mitosis observed in this study, it is tempting to suggest that IQGAP3 is under dynamic degradation in advanced PCs. Nonetheless, this possibility requires future investigation.
Outlook: Clinical potential of this research
PC is a highly heterogeneous disease even among human cancers; its heterogeneous nature along with PC’s high prevalence underlines the reactive patient care being complex and high cost [68]. Comprehensive management of PC has been recently recognized as 3PM: predictive, preventive, and personalized medicine [69], [70], [71]. Should our capacity in risk prediction and PC treatment (or personalized medicine) meet the clinical challenges, 3PM will bring substantial benefits to patients and society (at least with the reduction of economic burden). Although numerous PC biomarkers, including serum-based cell-free nucleic acid [72], have been reported, risk stratification for PC needs substantial improvement. Future research on SigIQGAP3NW and its component gene RELT may improve the current ability in predicting PC risk, prognosis for other cancers, and management of ICB therapy. Despite the numerous options available for managing PC patients at different stages, these therapeutic approaches are largely ineffective, particularly for advanced PCs. The association of IQGAP3 with PC progression and particularly its impressively high levels of correlation with PLK1 and TOP2A in advanced PC highlight the potential for future clinical efforts to examine this possibility using PLK1 and TOP2A inhibitors, which are either approved by the FDA or clinically available.
Conclusions
We have demonstrated a comprehensive association of IQGAP3 with PC and other human cancer types via its network actions in mitosis and chromosome segregation. The impressively high correlation levels of IQGAP3 with PLK1 and TOP2A in local PC, mCRPC (the SU2C cohort), ACC, glioma, KIRP, KIRC, and other cancers provide a strong scenario for targeting either PLK1, TOP2A or the combination of both in these cancers with elevated IQGAP3 expression. In this regard, our study may revive the concept of targeting PLK1 in cancer therapy with FDA-approved inhibitors with one additional caveat: consideration for IQGAP3 expression. We constructed a multigene panel SigIQGAP3NW; the panel correlates with IQGAP3 expression, recapitulates the feature of the IQGAP3 network, and robustly predicts the recurrent risk in PC and poor survival in multiple cancer types. SigIQGAP3NW is associated with escaping immune surveillance not only in PC but also in ACC, ccRCC, pRCC, THYM, and UVM. SigIQGAP3NW makes a unique contribution to assessing responses to ICB therapy. As part of the analysis for IQGAP3-derived genes, we also identified RELT as a novel and potent biomarker of ICB therapy. Collectively, our research produced a platform to develop SigIQGAP3NW into a risk prediction tool for PC and multiple cancer types and to launch clinical trials on FDA-approved PLK1 inhibitors and the existing TOP2 cancer therapy in PC.
Data Sharing and Data Accessibility
All data utilized and produced in this study are included in Supplementary materials.
Compliance with Ethical Requirements.
All the animal work protocols performed were approved by McMaster University Animal Research Ethics Board (16-06-23). Collection and analysis of clinical materials were approved by Hamilton Integrated Research Ethics Board (11-3472). TCGA datasets utilized in this research have been approved for research (https://cancergenome.nih.gov/).
CRediT authorship contribution statement
Wenjuan Mei: Conceptualization, Methodology, Formal analysis, Investigation, Writing – review & editing. Ying Dong: Investigation, Formal analysis, Writing – review & editing. Yan Gu: Investigation, Methodology, Formal analysis, Writing – review & editing. Anil Kapoor: Conceptualization, Resources, Investigation, Writing – review & editing. Xiaozeng Lin: Investigation, Writing – review & editing. Yingying Su: Investigation, Writing – review & editing. Sandra Vega Neira: Investigation, Writing – review & editing. Damu Tang: Conceptualization, Methodology, Formal analysis, Resources, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Y.G. is supported by a Postdoctoral Fellowship provided by the Research Institute of St Joe’s Hamilton. Grant support was from CIHR to DT. This research was also supported by funds from Urological Cancer Centre for Research and Innovation (UCCRI), St Joseph’s Hospital, Hamilton, ON L8N 4A6, Canada.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.01.015.
Contributor Information
Wenjuan Mei, Email: wenjuanmei1986@163.com.
Damu Tang, Email: damut@mcmaster.ca.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Gordetsky J., Epstein J. Grading of prostatic adenocarcinoma: current state and prognostic implications. Diagn Pathol. 2016;11:25. doi: 10.1186/s13000-016-0478-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaorsky N.G., Raj G.V., Trabulsi E.J., Lin J., Den R.B. The dilemma of a rising prostate-specific antigen level after local therapy: what are our options? Semin Oncol. 2013;40(3):322–336. doi: 10.1053/j.seminoncol.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Semenas J., Allegrucci C., Boorjian S.A., Mongan N.P., Persson J.L. Overcoming drug resistance and treating advanced prostate cancer. Curr Drug Targets. 2012;13(10):1308–1323. doi: 10.2174/138945012802429615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ojo D., Lin X., Wong N., Gu Y., Tang D. Prostate Cancer Stem-like Cells Contribute to the Development of Castration-Resistant Prostate Cancer. Cancers (Basel) 2015;7(4):2290–2308. doi: 10.3390/cancers7040890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bono J.S., Logothetis C.J., Molina A., Fizazi K., North S., Chu L., et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scher H.I., Fizazi K., Saad F., Taplin M.E., Sternberg C.N., Miller K., et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 8.Chaturvedi S., Garcia J.A. Novel agents in the management of castration resistant prostate cancer. Journal of carcinogenesis. 2014;13:5. doi: 10.4103/1477-3163.128185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedman A.C., Smith J.M., Sacks D.B. The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep. 2015;16(4):427–446. doi: 10.15252/embr.201439834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai Q., Ain Q., Rooney M., Song F., Zipprich A. Role of IQ Motif-Containing GTPase-Activating Proteins in Hepatocellular Carcinoma. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.920652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S., Watanabe T., Noritake J., Fukata M., Yoshimura T., Itoh N., et al. IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth. J Cell Sci. 2007;120(Pt 4):567–577. doi: 10.1242/jcs.03356. [DOI] [PubMed] [Google Scholar]
- 12.Nojima H., Adachi M., Matsui T., Okawa K., Tsukita S., Tsukita S. IQGAP3 regulates cell proliferation through the Ras/ERK signalling cascade. Nat Cell Biol. 2008;10(8):971–978. doi: 10.1038/ncb1757. [DOI] [PubMed] [Google Scholar]
- 13.Fang X., Zhang B., Thisse B., Bloom G.S., Thisse C. IQGAP3 is essential for cell proliferation and motility during zebrafish embryonic development. Cytoskeleton (Hoboken) 2015;72(8):422–433. doi: 10.1002/cm.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adachi M., Kawasaki A., Nojima H., Nishida E., Tsukita S. Involvement of IQGAP family proteins in the regulation of mammalian cell cytokinesis. Genes Cells. 2014;19(11):803–820. doi: 10.1111/gtc.12179. [DOI] [PubMed] [Google Scholar]
- 15.Qian E.N., Han S.Y., Ding S.Z., Lv X. Expression and diagnostic value of CCT3 and IQGAP3 in hepatocellular carcinoma. Cancer Cell Int. 2016;16:55. doi: 10.1186/s12935-016-0332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y., Qin N., Zhou Q., Chen Y., Huang S., Chen B., et al. Role of IQGAP3 in metastasis and epithelial-mesenchymal transition in human hepatocellular carcinoma. J Transl Med. 2017;15(1):176. doi: 10.1186/s12967-017-1275-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu K., Zhang X., Li F., Xiao D., Hou Y., Zhu S., et al. Frequent alterations in cytoskeleton remodelling genes in primary and metastatic lung adenocarcinomas. Nat Commun. 2015;6:10131. doi: 10.1038/ncomms10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z., Li X., Ma J., Li D., Ju H., Liu Y., et al. Integrative Analysis of the IQ Motif-Containing GTPase-Activating Protein Family Indicates That the IQGAP3-PIK3C2B Axis Promotes Invasion in Colon Cancer. OncoTargets and therapy. 2020;13:8299–8311. doi: 10.2147/OTT.S257729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oue N., Yamamoto Y., Oshima T., Asai R., Ishikawa A., Uraoka N., et al. Overexpression of the Transmembrane Protein IQGAP3 Is Associated with Poor Survival of Patients with Gastric Cancer. Pathobiology. 2018;85(3):192–200. doi: 10.1159/000481890. [DOI] [PubMed] [Google Scholar]
- 20.Jinawath N., Shiao M.S., Chanpanitkitchote P., Svasti J., Furukawa Y., Nakamura Y. Enhancement of Migration and Invasion of Gastric Cancer Cells by IQGAP3. Biomolecules. 2020;10(8) doi: 10.3390/biom10081194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dongol S., Zhang Q., Qiu C., Sun C., Zhang Z., Wu H., et al. IQGAP3 promotes cancer proliferation and metastasis in high-grade serous ovarian cancer. Oncol Lett. 2020;20(2):1179–1192. doi: 10.3892/ol.2020.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua X., Long Z.Q., Guo L., Wen W., Huang X., Zhang W.W. IQGAP3 Overexpression Correlates With Poor Prognosis and Radiation Therapy Resistance in Breast Cancer. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.584450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan Y., Jiang X., Tang L., Yang H., Wang J., Zhang D., et al. Comprehensive Analyses of the Immunological and Prognostic Roles of an IQGAP3AR/let-7c-5p/IQGAP3 Axis in Different Types of Human Cancer. Front Mol Biosci. 2022;9 doi: 10.3389/fmolb.2022.763248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng Q., Li C.X., Long D., Lin X. IQGAP3 May Serve as a Promising Biomarker in Clear Cell Renal Cell Carcinoma. Int J Gen Med. 2021;14:3469–3484. doi: 10.2147/IJGM.S316280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng X., Xu H., Yi X., Zhang T., Wei Q., Li H., et al. Tumor-antigens and immune landscapes identification for prostate adenocarcinoma mRNA vaccine. Mol Cancer. 2021;20(1):160. doi: 10.1186/s12943-021-01452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1 doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B., et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ru B., Wong C.N., Tong Y., Zhong J.Y., Zhong S.S.W., Wu W.C., et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 32.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasaikar S.V., Straub P., Wang J., Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife. 2017;6. [DOI] [PMC free article] [PubMed]
- 37.Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finotello F., Mayer C., Plattner C., Laschober G., Rieder D., Hackl H., et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11(1):34. doi: 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu Y., Lin X., Kapoor A., Li T., Major P., Tang D. Effective Prediction of Prostate Cancer Recurrence through the IQGAP1 Network. Cancers (Basel) 2021;13(3) doi: 10.3390/cancers13030430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X., Kapoor A., Gu Y., Chow M.J., Peng J., Major P., et al. Construction of a Novel Multigene Panel Potently Predicting Poor Prognosis in Patients with Clear Cell Renal Cell Carcinoma. Cancers (Basel) 2020;12(11) doi: 10.3390/cancers12113471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan J., Ojo D., Kapoor A., Lin X., Pinthus J.H., Aziz T., et al. Neural Cell Adhesion Protein CNTN1 Promotes the Metastatic Progression of Prostate Cancer. Cancer Res. 2016;76(6):1603–1614. doi: 10.1158/0008-5472.CAN-15-1898. [DOI] [PubMed] [Google Scholar]
- 43.McKinley K.L., Cheeseman I.M. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158(2):397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grue P., Grasser A., Sehested M., Jensen P.B., Uhse A., Straub T., et al. Essential mitotic functions of DNA topoisomerase IIalpha are not adopted by topoisomerase IIbeta in human H69 cells. J Biol Chem. 1998;273(50):33660–33666. doi: 10.1074/jbc.273.50.33660. [DOI] [PubMed] [Google Scholar]
- 45.Gutteridge R.E., Ndiaye M.A., Liu X., Ahmad N. Plk1 Inhibitors in Cancer Therapy: From Laboratory to Clinics. Mol Cancer Ther. 2016;15(7):1427–1435. doi: 10.1158/1535-7163.MCT-15-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raab C.A., Raab M., Becker S., Strebhardt K. Non-mitotic functions of polo-like kinases in cancer cells. Biochim Biophys Acta. 2021;1875(1) doi: 10.1016/j.bbcan.2020.188467. [DOI] [PubMed] [Google Scholar]
- 47.Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., et al. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12(1):17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Perramon M., Jimenez W. Pituitary Tumor-Transforming Gene 1/Delta like Non-Canonical Notch Ligand 1 Signaling in Chronic Liver Diseases. Int J Mol Sci. 2022;23(13) doi: 10.3390/ijms23136897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Want M.Y., Tsuji T., Singh P.K., Thorne J.L., Matsuzaki J., Karasik E., et al. WHSC1/NSD2 regulates immune infiltration in prostate cancer. J Immunother Cancer. 2021;9(2) doi: 10.1136/jitc-2020-001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez H., Hagerling C., Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32(19–20):1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammad H., Debeuf N., Aegerter H., Brown A.S., Lambrecht B.N. Emerging Paradigms in Type 2 Immunity. Annu Rev Immunol. 2022;40:443–467. doi: 10.1146/annurev-immunol-101320-030339. [DOI] [PubMed] [Google Scholar]
- 52.Sanchez N., Miranda A., Funes J.M., Hevia G., Perez R., de Leon J. Oncogenic transformation tunes the cross-talk between mesenchymal stem cells and T lymphocytes. Cell Immunol. 2014;289(1–2):174–184. doi: 10.1016/j.cellimm.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 53.Tauriello D.V.F., Palomo-Ponce S., Stork D., Berenguer-Llergo A., Badia-Ramentol J., Iglesias M., et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–543. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 54.Karoon Kiani F., Izadi S., Ansari Dezfouli E., Ebrahimi F., Mohammadi M., Chalajour H., et al. Simultaneous silencing of the A2aR and PD-1 immune checkpoints by siRNA-loaded nanoparticles enhances the immunotherapeutic potential of dendritic cell vaccine in tumor experimental models. Life Sci. 2022;288 doi: 10.1016/j.lfs.2021.120166. [DOI] [PubMed] [Google Scholar]
- 55.Morad G., Helmink B.A., Sharma P., Wargo J.A. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell. 2021;184(21):5309–5337. doi: 10.1016/j.cell.2021.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geissmann F., Manz M.G., Jung S., Sieweke M.H., Merad M., Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327(5966):656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coffelt S.B., Wellenstein M.D., de Visser K.E. Neutrophils in cancer: neutral no more. Nat Rev Cancer. 2016;16(7):431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- 58.De Sanctis F., Adamo A., Cane S., Ugel S. Targeting tumour-reprogrammed myeloid cells: the new battleground in cancer immunotherapy. Semin Immunopathol. 2022 doi: 10.1007/s00281-022-00965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R., et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen P.L., Roh W., Reuben A., Cooper Z.A., Spencer C.N., Prieto P.A., et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov. 2016;6(8):827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishino M., Ramaiya N.H., Hatabu H., Hodi F.S. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14(11):655–668. doi: 10.1038/nrclinonc.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huyghe N., Benidovskaya E., Stevens P., Van den Eynde M. Biomarkers of Response and Resistance to Immunotherapy in Microsatellite Stable Colorectal Cancer: Toward a New Personalized Medicine. Cancers (Basel) 2022;14(9) doi: 10.3390/cancers14092241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matias-Barrios V.M., Radaeva M., Song Y., Alperstein Z., Lee A.R., Schmitt V., et al. Discovery of New Catalytic Topoisomerase II Inhibitors for Anticancer Therapeutics. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.633142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chow M.J., Gu Y., He L., Lin X., Dong Y., Mei W., et al. Prognostic and Therapeutic Potential of the OIP5 Network in Papillary Renal Cell Carcinoma. Cancers (Basel) 2021;13(17):4483. doi: 10.3390/cancers13174483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao W., Chen Q., Li S., Jia X., Xu L., Wei L. RELT promotes the growth of esophageal squamous cell carcinoma by activating the NF-kappaB pathway. Cell Cycle. 2021;20(13):1231–1241. doi: 10.1080/15384101.2021.1924451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi B.K., Kim S.H., Kim Y.H., Lee D.G., Oh H.S., Han C., et al. RELT negatively regulates the early phase of the T-cell response in mice. Eur J Immunol. 2018;48(10):1739–1749. doi: 10.1002/eji.201847633. [DOI] [PubMed] [Google Scholar]
- 67.Xie Y., Yan J., Cutz J.C., Rybak A.P., He L., Wei F., et al. IQGAP2, A candidate tumour suppressor of prostate tumorigenesis. BBA. 2012;1822(6):875–884. doi: 10.1016/j.bbadis.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 68.Ellinger J., Alajati A., Kubatka P., Giordano F.A., Ritter M., Costigliola V., et al. Prostate cancer treatment costs increase more rapidly than for any other cancer-how to reverse the trend? EPMA J. 2022;13(1):1–7. doi: 10.1007/s13167-022-00276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kucera R., Pecen L., Topolcan O., Dahal A.R., Costigliola V., Giordano F.A., et al. Prostate cancer management: long-term beliefs, epidemic developments in the early twenty-first century and 3PM dimensional solutions. EPMA J. 2020;11(3):399–418. doi: 10.1007/s13167-020-00214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goldstein E., Yeghiazaryan K., Ahmad A., Giordano F.A., Frohlich H., Golubnitschaja O. Optimal multiparametric set-up modelled for best survival outcomes in palliative treatment of liver malignancies: unsupervised machine learning and 3 PM recommendations. EPMA J. 2020;11(3):505–515. doi: 10.1007/s13167-020-00221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mazurakova A., Samec M., Koklesova L., Biringer K., Kudela E., Al-Ishaq R.K., et al. Anti-prostate cancer protection and therapy in the framework of predictive, preventive and personalised medicine - comprehensive effects of phytochemicals in primary, secondary and tertiary care. EPMA J. 2022;13(3):461–486. doi: 10.1007/s13167-022-00288-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crigna A.T., Samec M., Koklesova L., Liskova A., Giordano F.A., Kubatka P., et al. Cell-free nucleic acid patterns in disease prediction and monitoring-hype or hope? EPMA J. 2020;11(4):603–627. doi: 10.1007/s13167-020-00226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.