Abstract
With the rapid growth of the fruit industry worldwide, it is important to assess adulteration to ensure the authenticity and the safety of fruit products. The DNA barcoding approach offers a quick and accurate way of identifying and authenticating species. In this study, we developed reference DNA barcodes (rbcL, ITS2, and trnH-psbA) for 70 cultivated and wild tropical fruit species, representing 43 genera and 26 families. In terms of species recoverability, rbcL has a greater recoverability (100%) than ITS2 (95.7%) and trnH-psbA (88.6%). We evaluated the performance of these barcodes in species discrimination using similarity BLAST, phylogenetic tree, and barcoding gap analyses. The efficiency of rbcL, ITS2, and trnH-psbA in discriminating species was 80%, 100%, and 93.6%, respectively. We employed a multigene-tiered approach for species identification, with the rbcL region used for primary differentiation and ITS2 or trnH-psbA used for secondary differentiation. The two-locus barcodes rbcL + ITS2 and rbcL + trnH-psbA demonstrated robustness, achieving species discrimination rates of 100% and 94.3% respectively. Beyond the conventional species identification method based on plant morphology, the developed reference barcodes will aid the fruit agroindustry and trade, by making fruit-based product authentication possible.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-023-03848-w.
Keywords: DNA barcoding, Chloroplast DNA, Species identification, rbcL, ITS2, trnH-psbA
Introduction
Tropical fruits, a botanically diverse group native to tropical climates (Underhill 2003), historically found their place in small-scale farming or were gathered from the wild for personal consumption. However, recent years have witnessed a significant transformation, turning tropical fruit production into a large and rapidly expanding industry. Mango, mangosteen, guava, pineapple, avocado, and papaya are among the major tropical fruits traded in the international markets (FAO 2022). Advancements in transportation systems and refrigerated storage have played a pivotal role in driving the industry’s growth, enabling efficient global consumption and widespread exportation (Underhill 2003). In 2021, Asia, Africa, Central and South America, the Caribbean, and Oceania emerged as the primary exporters of tropical fruits, collectively contributing to the world export of 8379 thousand tons (FAO 2022). Notably, significant quantities of tropical fruits were annually imported by major players, such as the United States of America, the European Union, and China, with these top importers acquiring more than 5369 thousand tons (FAO 2022). While a substantial portion of tropical fruits is consumed fresh, the establishment of processing industries has added another dimension to the industry (Underhill 2003). To date, the tropical fruit industry is supporting the socioeconomic well-being of approximately 160 million people in the tropics (Zhu et al. 2022).
Malaysia is characterized by its rich flora diversity, known as one of the world’s principal resources for tropical fruit trees (Saw et al. 1991), with a total of 520 tropical fruit species that produce edible fruits or seeds reported from its diverse ecosystems (Milow et al. 2014). In general, tropical fruit species can be categorized as cultivated and wild. Among the 64 widely cultivated tropical fruits in the country, 21 major fruit species are cultivated on a commercial scale, including mango, mangosteen, guava, rambutan, banana, pineapple, durian, and jackfruit (Rozhan 2019). In terms of wild tropical fruits, 76 species that produce wild edible fruits were recorded based on an inventory from the 50-ha research plot of Pasoh Forest Reserve in Malaysia (Saw et al. 1991). The wild mangoes, Mangifera (Anacardiaceae), mangosteen, Garcinia (Clusiaceae), rambutans, Nephelium (Sapindaceae), and the forest breadfruits, Artocarpus (Moraceae) are among the main families that make up more than half the fruit tree flora (Saw et al. 1991). Besides being important contributors to human living and livelihoods, most wild edible fruits also serve as an important food source for animals, as reported in a recent compilation of 160 species of tropical fruit species in Malaysia (Zawiah et al. 2019). Furthermore, tropical fruits are a significant contributor to the country’s agricultural gross domestic product. Annually, the industry generates over US$250 million in export profits and provides employment for more than 260,000 Malaysians (Rozhan 2019).
In the worldwide food market, tropical fruit products are widely available to consumers. Fruit juice is among the most steadily growing industries across developing and developed countries (Dasenaki and Thomaidis 2019). Dietary supplements derived from fruits have also gained substantial popularity, positioned as premium food items renowned for their remarkable health benefits, ranging from obesity treatment and cancer prevention to anti-aging and combating various chronic diseases (Hayamizu et al. 2003; Watanapokasin et al. 2010; Kong et al. 2021). In view of the growing market, ensuring the authenticity and the safety of fruit products become imperative, necessitating robust regulatory systems to detect and prevent adulteration and fraud (Jandrić et al. 2017). Notably, significant strides have been taken in leveraging advanced technologies and analytical methodologies, including chromatography, spectroscopy, and DNA-based techniques, to assess the authenticity of fruit products, signifying a crucial advancement in ensuring the authenticity of these products in the market (Dasenaki and Thomaidis 2019).
Since its inception in 2003, DNA barcoding has been a focal point of extensive research for species identification and authentication. This innovative approach relies on a concise standard genomic region, universally present in target lineages, possessing sufficient sequence variation to effectively discriminate among species (Hebert et al. 2003). The Plant Working Group of the Consortium for the Barcode of Life (CBOL) took a significant step by proposing two coding regions from the plastid genome, rbcL and matK as a “core barcode” for land plants, to be supplemented with additional regions, such as trnH–psbA, atpF–atpH, psbK–psbI, trnL intron, and ITS (CBOL Plant Working Group 2009). Beyond the candidates, other portion of the genome, such as the second internal transcribed spacer (ITS2) from nuclear ribosomal DNA, is highly recommended and has been used in plant barcoding studies (Chen et al. 2010; Duan et al. 2019, Jones et al. 2021). Over the past decades, numerous research teams have piloted large datasets to evaluate the universality, sequence quality, and discriminatory power of the candidate barcode regions (Kress and Erickson 2007; Hollingsworth et al. 2009; China Plant BOL Group et al. 2011). Recognizing differences in their efficiency, a multigene-tiered approach is widely adopted, whereby a first-tier coding region common across the land plants provides resolution at a certain rank (e.g., family or genus), and a more variable (coding or non-coding) region provides resolution up to the species level (Newmaster et al. 2006). This tiered strategy ensures a comprehensive and accurate approach to DNA barcoding, catering to the diverse resolution requirements across different levels of taxonomy.
Despite the importance of tropical fruits in global food trade and the assessment of fruit product authenticity, the availability of DNA barcodes for tropical fruits remains notably limited. Presently, only a handful of DNA barcodes for fruit trees have been documented, primarily within genera, such as Ananas (Hidayat et al. 2012), Annona (Larranaga and Hormaza 2015), Garcinia (Anerao et al. 2021), Selenicereus (Huy et al. 2021), and Mangifera (Suparman et al. 2013; Fitmawati et al. 2016; Kumar et al. 2022). Recognizing this gap, this project was initiated with the purpose of facilitating species identification and fruit product authenticity assessment. We present a simple and accessible approach to barcode the cultivated and wild tropical edible fruits and evaluate the efficiency of species discrimination.
Materials and methods
Sample collection and DNA extraction
All fruit reference samples were obtained from the living collections of the Kepong Botanical Garden (N03.22440 E101.61578), Fruit Arboretum (N03.23043 E101.63397) of the Forest Research Institute Malaysia (FRIM), and the 50-ha forest research plot in Pasoh Forest Reserve (N02.98255, E102.31050). A total of 264 samples were collected, representing 70 fruit tree species, 43 genera, and 26 families. Details of the collected reference samples are listed in Table S1. Leaf samples were collected from two to 14 individuals per species. At least two samples per species were included to increase reliability and enable intraspecific variation detection. Total DNA was extracted according to Murray and Thompson (1980) with a single modification (2 × CTAB) and further purified using a High Pure PCR Template Preparation Kit (Roche Diagnostics GmbH, Penzberg, Germany). For each sample, a properly pressed, labeled, and mounted voucher specimen was made for future reference.
PCR amplification and sequencing of DNA barcoding markers
In this study, we employed refined DNA barcodes, rbcL and trnH-psbA used in tropical plants (Tnah et al. 2019), and supplemented with ITS2, which has proved remarkably effective at discriminating diverse plant groups (Chen et al. 2010). The universal primer pairs of rbcL, ITS2, and trnH-psbA used for amplification and sequencing are listed in Table S2. The three candidate DNA barcodes were amplified separately through a 10 μl PCR reaction, consisting of Q5 High-Fidelity 2X Master Mix (New England Biolabs, Ipswich, MA, USA), 0.5 μM of each primer, and approximately 10 ng of template DNA. The PCR reactions were performed in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA), for an initial denaturing step of 30 s at 98 °C, 35 cycles of 98 °C for 10 s, 50 °C annealing temperature for 30 s, and 72 °C for 30 s, followed by a final extension of 72 °C for 2 min. The PCR products were subsequently purified with ExoSAP-IT Express (Affymetrix, Cleveland, OH, USA). The cycle sequence reactions of the purified products were bi-directionally using the BigDye Terminator Sequencing Kit (Applied Biosystems) based on the standard dideoxy-mediated chain termination method. The sequencing thermal profile consists of 95 °C for 1 min, followed by 50 cycles of 95 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min. The sequenced products were then purified using BigDye XTerminator Purification Kit (Applied Biosystems) prior to electrophoresis on an ABI 3130xl Genetic Analyzer (Applied Biosystems).
Analysis of species recoverability and species discrimination
Nucleotide sequences were edited and assembled using SEQUENCHER version 5.4.6 (Gene Codes Corporation, Ann Arbor, MI, USA). Heterozygous bases found in the chromatograms were manually assessed and assigned IUPAC (International Union of Pure Applied Chemistry) codes. All DNA sequences obtained in this study have been deposited in GenBank (Table S1). Species recoverability (the ability to successfully retrieve a sequence from a species) of rbcL, ITS2, and trnH-psbA was compared among 264 samples of 70 fruit species. In order to ensure the accuracy of sequence identification, the edited barcodes were cross-checked with the nucleotide sequences deposited in the GenBank database. The performance of barcode markers in species discrimination was evaluated using similarity-based (Basic Local Alignment Search Tool, BLAST), nearest genetic distance (Neighbor-joining, NJ), and barcoding gap methods. For each individual data set, the discrimination ability for each barcode region was assessed as a percentage of the total number of species correctly identified divided by the total number of species examined.
In the similarity BLAST analysis, all sequences obtained from the study were formatted as both database and query, and subsequently queried individually against the database. When the query has a 100% match to the conspecific sequences, species discrimination is considered successful. On the contrary, species discrimination is considered a failure when it matches numerous species. As an alternative to sequence similarity-based identification, the NJ tree (Saitou and Nei 1987) was constructed using the program MEGA 11 (Tamura et al. 2021) based on sequence divergence estimated with Kimura 2-parameter (Kimura 1980). The multiple sequence alignments of ITS2 and trnH-psbA were performed using MAFFT (Multiple Alignment using Fast Fourier Transform; Katoh and Standley 2013). Bootstrapping analysis was conducted based on 1000 replicates to evaluate the robustness of the node for each tree. The number of species with conspecific individuals successfully clustered under respective monophyletic groups in the phylogenetic tree was calculated. In the barcoding gap assessment, pairwise Kimura 2-parameter distances were calculated within and between species using MEGA 11. For species with two or more individuals, the barcoding gap was assessed by plotting the maximum intraspecific distance against the minimum interspecific distance. The percentage of species having a barcoding gap was calculated based on those with the minimum interspecific distance greater than the maximum intraspecific distance.
Results and discussion
Reference DNA barcodes of tropical fruits
Despite the desirability of using DNA barcoding broadly in plant identification, the lack of a reliable DNA barcoding reference library is often the main impediment to its application. Therefore, the initial and crucial step involves the development of trustworthy reference barcodes. Securing access to accurately identified specimens from arboretums, botanical gardens, and forest research plots is indispensable for this purpose. In this study, fruit tree species spanning various families—Anacardiaceae, Fabaceae, Sapindaceae, Meliaceae, Moraceae, Myrtaceae, Phyllanthaceae, and others—are integral components of the living collections at FRIM and Pasoh Forest Reserve. Within Malaysia’s tropical fruit diversity, approximately 160 species with major or medium economic status contribute to both self-consumption and market demands (Milow et al. 2014). The ongoing effort aims to expand the DNA barcode reference library to encompass a more extensive array of local fruit species in future.
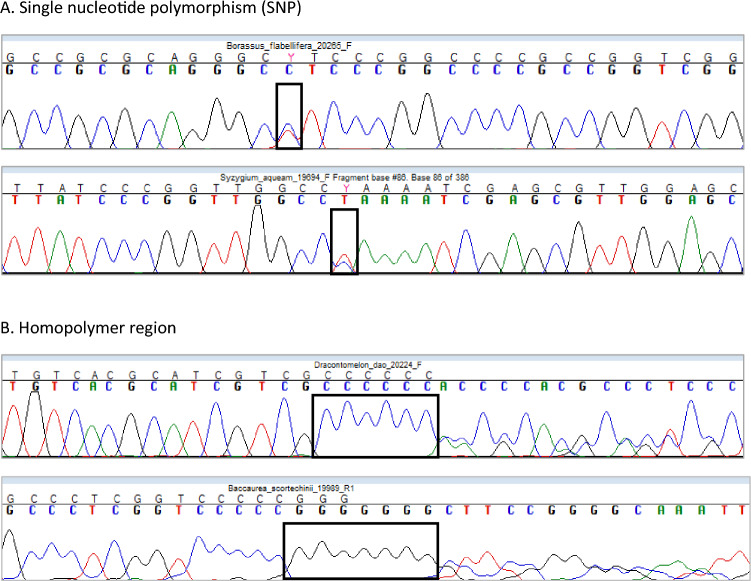
We successfully generated a total of 264 rbcL barcodes (70 species), 253 ITS2 barcodes (67 species), and 223 trnH-psbA barcodes (62 species) (Fig. 1). The PCR products for rbcL had a consistent length of 605 bp across all species (GenBank accession numbers OK052728-OK052798). The length of the ITS2 sequence was 221–495 bp (GenBank accession numbers OK052867-OK052949). For ITS2, high-quality DNA barcodes were obtained; nevertheless, single-nucleotide polymorphisms (SNPs) and homo-polymers were observed in some species (Fig. 2). The intra-genomic variability of ITS2 resulted in a number of SNPs in the region ranging from 1 to 11, which were recoded with IUPAC codes. Partial readable sequences were observed in five species, due to homo-polymers composed of mononucleotide repeats, which caused premature termination of sequencing and resulted in unidirectional reads. The alignment length of trnH-psbA ranged from 215 to 664 bp (GenBank accession numbers OK052799-OK052866). The region was not amplified well in certain genera (e.g., Aglaia, Artocarpus, Borassus, Dracontomelon, Durio, and Lansium) and had homo-polymers that were difficult to sequence accurately.
Fig. 1.
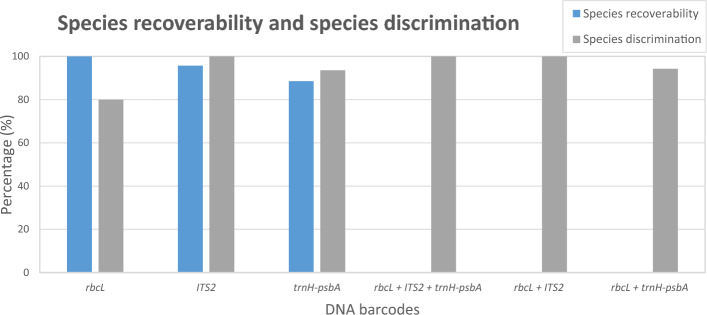
Evaluation of species recoverability and species discrimination is depicted, comparing the efficacy of single barcodes, (rbcL, ITS2, trnH-psbA) and multigene-tiered barcodes, (rbcL + ITS2 + trnH-psbA, rbcL + ITS2 and rbcL + trnH-psbA) through similarity BLAST. Species recoverability is represented by the percentage of successfully sequenced species within each barcode, while species discrimination assesses the capability of the barcodes to achieve identification at the species level
Fig. 2.
The study reveals various types of polymorphisms in ITS2 regions. Chromatograms illustrate examples of (A) single-nucleotide polymorphism (SNP) and (B) homo-polymer regions found in different species, as indicated in the corresponding box
Based on the results, out of the 70 species, rbcL has a higher species recoverability (100%) than ITS2 (95.7%) and trnH-psbA (88.6%) (Fig. 1). All three regions were recovered for 60 species, except for a small number of species in the ITS2 and trnH-psbA. In the cases reported on rbcL (de Vere et al. 2012) and trnH-psbA (Sass et al. 2007), empirical studies revealed that the addition of multiple primer combinations did not improve the overall recoverability. Moreover, as studies have shown that intra-genomic variability of ITS2 is common in plants (Song et al. 2012), further primer combinations might increase the risk of amplifying different copies within this multi-copy marker (Jones et al. 2021).
Evaluation of species discrimination (BLAST, phylogenetic tree, and barcoding gap)
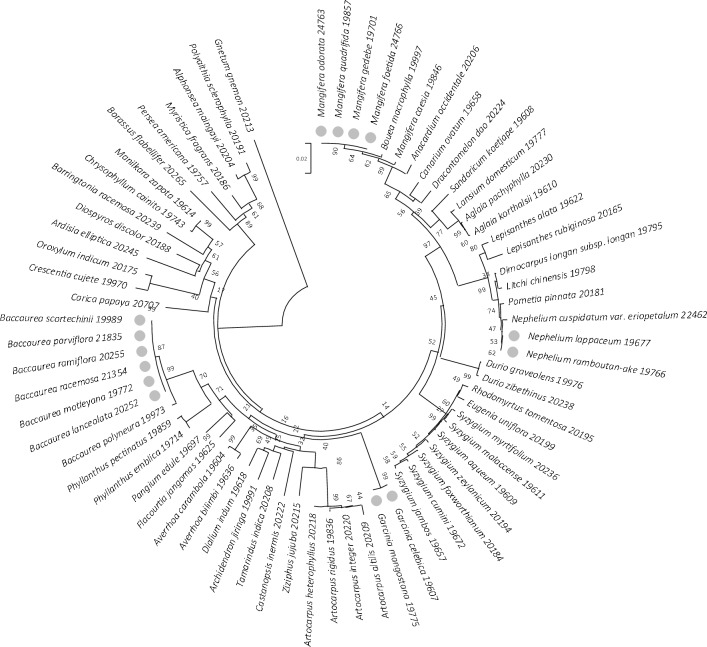
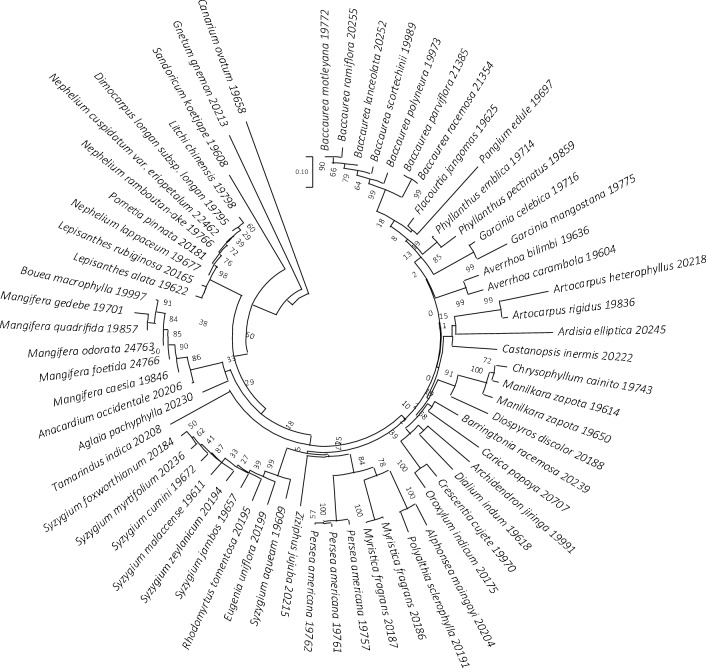
In the present study, the rbcL emerged as the most effective region with good recovery and high sequence quality. Serving as the primary region in our multigene-tiered approach, it establishes a valuable backbone for the barcode dataset. In GenBank, the region provides a good baseline for comparison with other plant species, as it is one of the most characterized plastid coding regions with wide representation from all major groups. The evaluation of similarity BLAST (Fig. 1) and Neighbor-Joining trees (Figs. 3, 4, 5) revealed the discriminatory efficiency of rbcL, ITS2, and trnH-psbA as 80%, 100%, and 93.6%, respectively. Notably, rbcL demonstrates modest resolution at the species level, successfully discriminating 80% of the species, with the exception of 14 species from four genera (Baccaurea, Garcinia, Mangifera, and Nephelium) exhibiting low interspecific variation (Fig. 3). This observation aligns with the slow evolutionary rate of the rbcL region, making it the least divergent plastid gene in flowering plants (Kress et al. 2005). Consistent with other studies, such as those conducted by the US National Herbarium (Kress and Erickson 2007) and the UK flora (Jones et al. 2021), rbcL exhibits a modest discrimination ability, recorded at 76.3% and 64%, respectively.
Fig. 3.
Unrooted Neighbor-joining tree of 70 fruit species based on rbcL sequences, supported by 1000 bootstrap replicates. The circle denotes 14 unresolved species. Each species name is accompanied by the corresponding DNA ID number of the sample
Fig. 4.
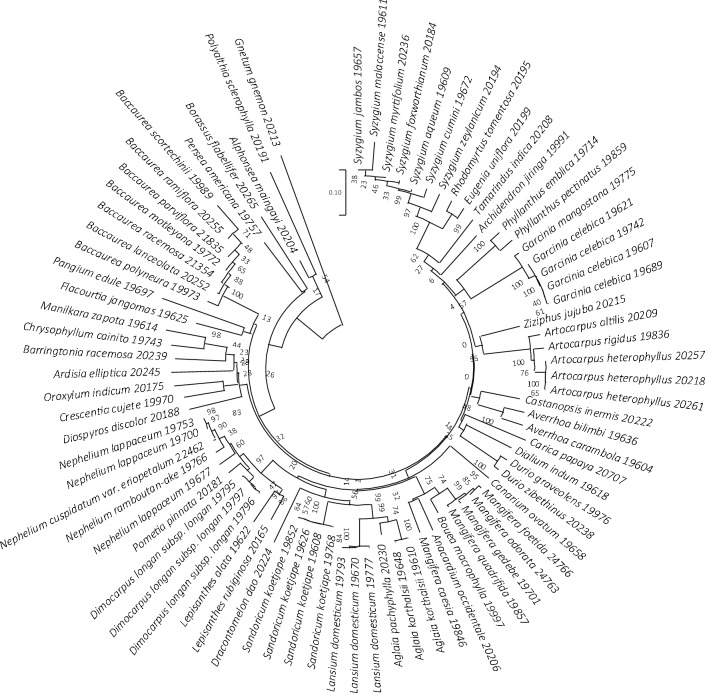
Unrooted Neighbor-joining tree of 67 fruit species based on ITS2 sequences, supported by 1000 bootstrap replicates. Species with more than one individual exhibited the presence of intraspecific variations. Each species name is accompanied by the corresponding DNA ID number of the sample
Fig. 5.
Unrooted Neighbor-joining tree of 62 fruit species based on trnH-psbA sequences, supported by 775 bootstrap replicates. Species with more than one individual exhibited the presence of intraspecific variations. Each species name is accompanied by the corresponding DNA ID number of the sample
In the assessment of the barcoding gap, the correlation between minimum interspecific and maximum intraspecific distances demonstrated high discrimination ability of the three regions (Fig. 6, Table S3). The study unveils that 80%, 97%, and 93.4% of species exhibit barcoding gaps in rbcL, ITS2, and trnH-psbA, respectively (Table S3)—a crucial criterion for establishing a reliable barcode. This alignment with the fundamental DNA barcoding principle, advocates for interspecific divergence to surpass intraspecific variation (Hebert et al. 2003). Calculated mean interspecific distances based on rbcL, ITS2, and trnH-psbA were 0.0143, 0.0958 and 0.1092, respectively. Despite their lower recovery rates, ITS2 and trnH-psbA barcodes exhibited higher species discrimination, outpacing rbcL in the interspecific genetic distance. Remarkably, both ITS2 and trnH-psbA are by far the best barcodes with the highest ability of species discrimination, and have been shown in numerous studies to be effective in the construction of DNA barcoding resources (Kress and Erikson 2007; Chen et al. 2010; Duan et al. 2019).
Fig. 6.
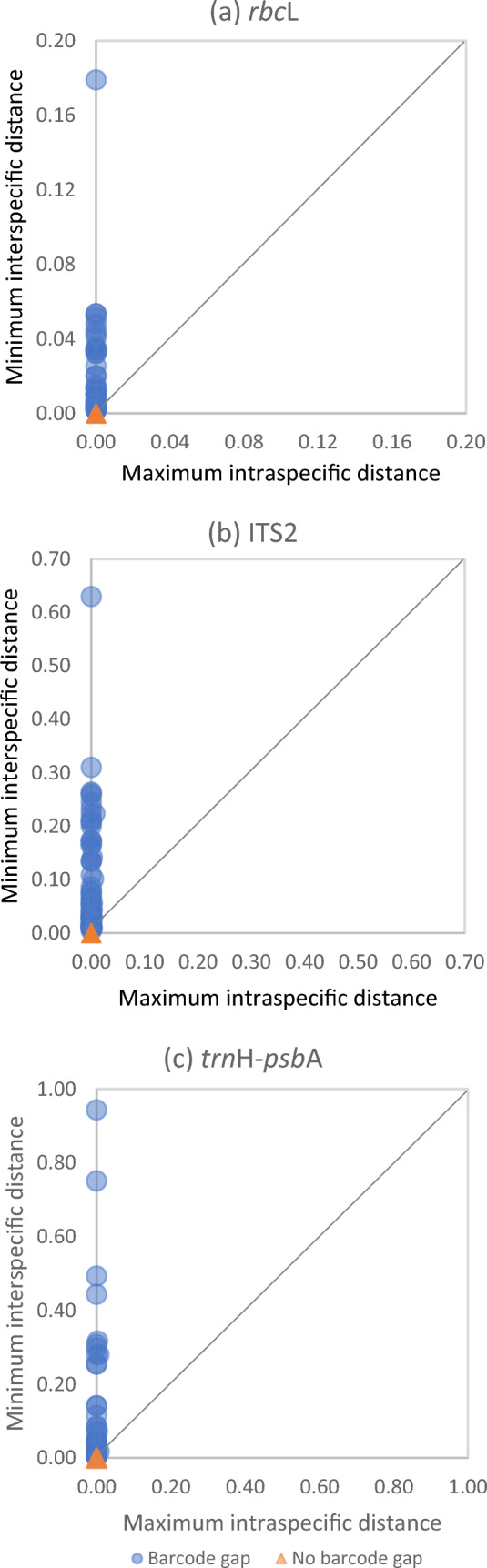
The plot of minimum interspecific vs. maximum intraspecific distances in the barcoding dataset of (a) rbcL, (b) ITS2, and (c) trnH-psbA. Pairwise comparisons are calculated from the multiple alignment of each barcode. The diagonal indicates points of equality, with circles above the 1:1 line signifying the presence of a barcode gap
Finding an effective and robust species-level identification barcode across land plants remains a pressing challenge. Considering the ease of recovery, rbcL is the preferred option but it is hampered by lower species discrimination. If species discrimination greater than 80% is required, then there is a need to adopt a multigene-tiered approach, with the second or third tier being more taxon-specific. In our study, the implementation of three-locus plant barcodes (rbcL + ITS2 + trnH-psbA) yielded marginal or no improvement over two-locus plant barcodes (rbcL + ITS2 or rbcL + trnH-psbA) in terms of discriminated species (Fig. 1). This finding aligns with similar observations by Kress et al. (2010), who achieved 93% species discrimination using two-locus (rbcL + trnH-psbA) and three-locus (rbcL + matK + trnH-psbA) barcodes in a Puerto Rican Forest plot. In terms of cost efficiency, we opted for a two-locus approach rather than a three-locus. Besides, the two-locus plant barcodes, e.g., rbcL + trnH-psbA (Kress and Erickson 2007; Ferri et al. 2015; Tnah et al. 2019), rbcL + trnL-F (de Groot et al. 2011), ITS2 + trnH-psbA (Duan et al. 2019), among others, have been extensively used to establish reference libraries for various important plants.
In the context of two-locus plant barcodes, the universal core locus of the rbcL region serves as a foundation, while ITS2 and trnH-psbA are interchangeably employed as a second tier region. The independent application of the second tier alignment for a limited number of unresolved taxa mitigates difficulties in aligning highly divergent genera. In our study, both ITS2 and trnH-psbA regions exhibited a sufficient number of autapomorphies, elevating their discrimination abilities to 100% and 94.3%, respectively (Fig. 1). Despite the fact that ITS2 had a higher species recovery rate, trnH-psbA was included as a complimentary second tier region. This study revealed instances where certain species were non-amplifiable in ITS2 but were amplified perfectly using trnH-psbA (e.g., Myristica fragrans & Litchi chinensis). Likewise, with the use of trnH-psbA alone, species recoverability in the genera Aglaia, Artocarpus, Borassus, Dracontomelon, Durio, and Lansium was limited. Recognizing the variation in recoverability and the limitation of individual DNA barcodes, the flexibility in choosing a second tier locus offers a substantial advantage. The approach has the advantage of keeping the complementary second tier regions at two for any particular group of taxa, thus maintaining the current efficiency of the system (Newmaster et al. 2006).
Applications of the reference DNA barcodes
While facilitating species identification, reference DNA barcodes play a pivotal role as indispensable tools for taxonomic exploration and phylogenetic analysis. Their utility becomes particularly evident when distinguishing closely related species that share highly similar morphological characteristics. The absence of diagnostic flowers and fruits during the juvenile stage presents challenges in making rapid and unequivocal identification, thereby impeding studies in diversity evaluation, germplasm collection, conservation, and management (Larranaga and Hormaza 2015). In plant genera where diagnostic features are lacking and morphological plasticity complicates the assessment of phylogenetic relationships, as seen in the Mangifera genus (Fitmawati et al. 2016), DNA barcodes provide a crucial solution. Discrepancies between morphological characteristics (Kostermans and Bompard 1993) and molecular studies (rbcL; Suparman et al. 2013, ITS; Fitmawati et al. 2016) highlight the necessity for taxonomic revision. In our current study, the cluster analysis of three Mangifera species based on ITS2 (Fig. 4) supports the previous molecular classification (Fitmawati et al. 2016). The acquisition of molecular data through DNA barcodes is thus imperative for supporting and supplementing morphology-based taxonomic classifications in such instances.
Considering the substantial global consumption of fruit juices and products, the detection and the prevention of adulteration and fraud are important and should be given attention. It is crucial to establish robust control systems to safeguard consumers against impure and deceitfully presented fruit products (Dasenaki and Thomaidis 2019). When a query sample has been processed or is in powder form, taxonomic identification is impossible, let alone verification of product authenticity, unless there are unique biochemical markers for the particular species. DNA barcoding offers a solution to overcome this limitation. The technique has proven effective in identifying fruit residues in various products, including juices (Faria et al. 2013; Wu et al. 2018), pectin (Barcaccia et al. 2015), plant oil (Spaniolas et al. 2008), yogurt (Ortola-Vidal et al. 2007), purees, chocolates, and cookies (Sakai et al. 2010). Moreover, in an increasingly health-conscious population, tropical fruits are seen as an appropriate source of nutrition, added variety, and exotic appeal (Underhill 2003). As such, the DNA barcodes are useful for authentication assessment and plant-based adulterant detection, pertaining to fruit-based nutritional products.
Conclusions
Given the importance of tropical fruits in socioeconomic development, extensive research to catalog fruit genetic resources is needed to strengthen the industry. The establishment of reference DNA barcodes for 70 tropical fruit tree species in this study allows us to use DNA barcoding techniques for species identification and authenticity assessment of fruit products. We evaluated the effectiveness of the multigene tier approach, with the two-locus barcodes, rbcL + ITS2 and rbcL + trnH-psbA demonstrating excellent resolution in species discrimination. In concert with conventional species identification methods rooted in plant morphology, the established reference barcodes hold immense potential for the agroindustry and food trade. Their utilization will not only enhance the accuracy of species identification but also fortify fruit-based product authentication in the industries.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Ahmad Farhan Nuri Rasli, Mohamad Izham Mohd Ariffin, Mohamad Aidil Noordin, Sharifah Talib, Ghazali Jaafar, Yahya Marhani, Ramli Ponyoh, Yasri Baya, Soo Pei Sin, and late Suryani Che Seman for their excellent assistance in the field and laboratory.
Funding
This study was supported by The Government of Malaysia under the 11th Malaysia Plan.
Data availability
All data generated in this study are available within the paper and also as supplementary information.
Declarations
Conflict of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Accession Numbers: GenBank accessions of rbcL (OK052728–OK052798), ITS2 (OK052867–OK052949) and trnH-psbA (OK052799–OK052866).
References
- Anerao J, Jha V, Shaikh N, Shivalkar A, Nityanand A, Sawant D, Rao GR, Mangaonkar K, Deodhar M, Desai N. DNA barcoding of important fruit tree species of agronomic interest in the genus Garcinia L. from the Western Ghats. Genet Resour Crop Evol. 2021;68(8):3161–3177. doi: 10.1007/s10722-021-01177-6. [DOI] [Google Scholar]
- Barcaccia G, Lucchin M, Cassandro M. DNA barcoding as a molecular tool to track down mislabeling and food piracy. Diversity. 2015;8(1):2. doi: 10.3390/d08010002. [DOI] [Google Scholar]
- CBOL Plant Working Group A DNA barcode for land plants. Proc Natl Acad Sci USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Yao H, Han J, Liu C, Song J, Shi L, Zhu Y, Ma X, Gao T, Pang X, Luo K, Li Y, Li X, Jia X, Lin Y, Leon C. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China Plant BOL Group. Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, Chen ZD, Zhou SL, Chen SL, Yang JB. Comparative analysis of a large dataset indicates that internal transcribed spacer ( ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA. 2011;49:19641–19646. doi: 10.1073/pnas.1104551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasenaki ME, Thomaidis NS. Quality and authenticity control of fruit juices - a review. Molecules. 2019;24(6):1014. doi: 10.3390/molecules24061014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot GA, During HJ, Maas JW, Schneider H, Vogel JC, Erkens RH. Use of rbcL and trnL-F as a two-locus DNA barcode for identification of NW-European ferns: an ecological perspective. PLoS ONE. 2011;6(1):e16371. doi: 10.1371/journal.pone.0016371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vere N, Rich TC, Ford CR, Trinder SA, Long C, Moore CW, Satterthwaite D, Davies H, Allainguillaume J, Ronca S, Tatarinova T. DNA barcoding the native flowering plants and conifers of Wales. PLoS ONE. 2012;7(6):e37945. doi: 10.1371/journal.pone.0037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Wang W, Zeng Y, Guo M, Zhou Y. The screening and identification of DNA barcode sequences for Rehmannia. Sci Rep. 2019;9(1):1–12. doi: 10.1038/s41598-019-53752-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2022) Major tropical fruits statistical compendium 2021. Rome
- Faria MA, Magalhães A, Nunes ME, Oliveira MB. High resolution melting of trnL amplicons in fruit juices authentication. Food Control. 2013;33(1):136–141. doi: 10.1016/j.foodcont.2013.02.020. [DOI] [Google Scholar]
- Fay MF, Swensen SM, Chase MW. Taxonomic affinities of Medusagyne oppositifolia (Medusagynaceae) Kew Bull. 1997;52:111–120. doi: 10.2307/4117844. [DOI] [Google Scholar]
- Ferri G, Corradini B, Ferrari F, Santunione AL, Palazzoli F, Alu M. Forensic botany II, DNA barcode for land plants: which markers after the international agreement? Forensic Sci Int: Genet. 2015;15:131–136. doi: 10.1016/j.fsigen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Fitmawati F, Hayati I, Sofiyanti N. Using ITS as a molecular marker for Mangifera species identification in Central Sumatra. Biodiversitas. 2016;17(2):653–656. doi: 10.13057/biodiv/d170238. [DOI] [Google Scholar]
- Hayamizu K, Ishii Y, Kaneko I, Shen M, Okuhara Y, Shigematsu N, Tomi H, Furuse M, Yoshino G, Shimasaki H. Effects of Garcinia cambogia (Hydroxycitric Acid) on visceral fat accumulation: a double-blind, randomized, placebo-controlled trial. Curr Ther Res Clin Exp. 2003;64(8):551–567. doi: 10.1016/j.curtheres.2003.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidayat T, Abdullah FI, Kuppusamy C, Samad AA, Wagiran A. Molecular identification of Malaysian pineapple cultivar based on internal transcribed spacer region. APCBEE Proc. 2012;4:146–151. doi: 10.1016/j.apcbee.2012.11.025. [DOI] [Google Scholar]
- Hollingsworth ML, Andra Clark AL, Forrest LL, Richardson J, Pennington RT, Long DG, Cowan R, Chase MW, Gaudeul M, Hollingsworth PM. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour. 2009;9(2):439–457. doi: 10.1111/j.1755-0998.2008.02439.x. [DOI] [PubMed] [Google Scholar]
- Huy TG, Men TT, Thi NPA, Khang DT. Identification of dragon fruit (Selenicereus) species in Mekong Delta based on DNA barcode sequences. Biodiversitas. 2021;22(10):4216–4222. doi: 10.13057/biodiv/d221012. [DOI] [Google Scholar]
- Jandrić Z, Islam M, Singh DK, Cannavan A. Authentication of Indian citrus fruit/fruit juices by untargeted and targeted metabolomics. Food Control. 2017;72:181–188. doi: 10.1016/j.foodcont.2015.10.044. [DOI] [Google Scholar]
- Jones L, Twyford AD, Ford CR, Rich TC, Davies H, Forrest LL, Hart ML, McHaffie H, Brown MR, Hollingsworth PM, De Vere N (2021) Barcode UK: a complete DNA barcoding resource for the flowering plants and conifers of the United Kingdom. Mol Ecol Resour 21(6):2050–2062 [DOI] [PubMed]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kong YR, Jong YX, Balakrishnan M, Bok ZK, Weng JKK, Tay KC, Goh BH, Ong YS, Chan KG, Lee LH, Khaw KY. Beneficial role of Carica papaya extracts and phytochemicals on oxidative stress and related diseases: a mini review. Biology (basel) 2021;10(4):287. doi: 10.3390/biology10040287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostermans AJGH, Bompard JM. The Mangoes: Their Botany, Nomenclature, Horticulture and Utilization. London: Academic Press; 1993. [Google Scholar]
- Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH–psbA spacer region. PLoS One. 2007;2:e508. doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA. 2005;102(23):8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Kaushik RA, Jain D, Saini VP, Babu SR, Choudhary R, Ercisli S. Genetic diversity among local mango (Mangifera indica L.) germplasm using morphological, biochemical and chloroplast DNA barcodes analyses. Mol Biol Rep. 2022;49:3491–3501. doi: 10.1007/s11033-022-07186-7. [DOI] [PubMed] [Google Scholar]
- Larranaga N, Hormaza JI. DNA barcoding of perennial fruit tree species of agronomic interest in the genus Annona (Annonaceae) Front Plant Sci. 2015;6:589. doi: 10.3389/fpls.2015.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin RA, Wagner WL, Hoch PC, Nepokroeff M, Pires JC, Zimmer EA, Sytsma KJ. Family-level relationships of Onagraceae based on chloroplast rbcL and ndhF data. Am J Bot. 2003;90:107–115. doi: 10.3732/ajb.90.1.107. [DOI] [PubMed] [Google Scholar]
- Milow P, Malek SB, Edo J, Ong HC. Malaysian species of plants with edible fruits or seeds and their valuation. Int J Fruit Sci. 2014;14(1):1–27. doi: 10.1080/15538362.2013.801698. [DOI] [Google Scholar]
- Murray M, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmaster SG, Fazekas AJ, Ragupathy S. DNA barcoding in land plants: evaluation of rbcL in a multigene tiered approach. Botany. 2006;84(3):335–341. [Google Scholar]
- Ortola-Vidal A, Schnerr H, Rojmyr M, Lysholm F, Knight A. Quantitative identification of plant genera in food products using PCR and Pyrosequencing technology. Food Control. 2007;18(8):921–927. doi: 10.1016/j.foodcont.2006.04.013. [DOI] [Google Scholar]
- Rozhan AD (2019) Trends in production, trade, and consumption of tropical fruit in Malaysia. FFTC Agricultural Policy Platform (FFTC-AP). Accessed in 31 May 2021, https://ap.fftc.org.tw/article/1381.
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Ishihata K, Nakano S, Yamada T, Yano T, Uchida K, Nakao Y, Urisu A, Adachi R, Teshima R, Akiyama H. Specific detection of banana residue in processed foods using polymerase chain reaction. J Agric Food Chem. 2010;58(14):8145–8151. doi: 10.1021/jf100675c. [DOI] [PubMed] [Google Scholar]
- Sass C, Little DP, Stevenson DW, Specht CD. DNA barcoding in the Cycadales: testing the potential of proposed barcoding markers for species identification of cycads. PLoS One. 2007;2(11):e1154. doi: 10.1371/journal.pone.0001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saw LG, LaFrankie JV, Kochummen KM, Yap SK. Fruit trees in a Malaysian rain forest. Econ Bot. 1991;45(1):120–136. doi: 10.1007/BF02860057. [DOI] [Google Scholar]
- Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot. 2005;92(1):142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Song J, Shi L, Li D, Sun Y, Niu Y, Chen Z, Luo H, Pang X, Sun Z, Liu C, Lv A. Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA. PLoS One. 2012;7(8):e43971. doi: 10.1371/journal.pone.0043971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaniolas S, Bazakos C, Ntourou T, Bihmidine S, Georgousakis A, Kalaitzis P. Use of lambda DNA as a marker to assess DNA stability in olive oil during storage. Eur Food Res Technol. 2008;227(1):175–179. doi: 10.1007/s00217-007-0707-8. [DOI] [Google Scholar]
- Suparman PA, Hidayat T. Phylogenetic analysis of Mangifera based on rbcL sequences, chloroplast DNA. Sci Papers Ser B Hort. 2013;57:235–240. [Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tnah LH, Lee SL, Tan AL, Lee CT, Ng KK, Ng CH, Nurul-Farhanah Z. DNA barcode database of common herbal plants in the tropics: a resource for herbal product authentication. Food Control. 2019;95:318–326. doi: 10.1016/j.foodcont.2018.08.022. [DOI] [Google Scholar]
- Underhill S (2003) Fruits of tropical climates: commercial and dietary importance. In: Caballero B (ed) Encyclopedia of food sciences and nutrition, 2nd edn. Academic Press: Oxford, UK, pp. 2780–2785
- Watanapokasin R, Jarinthanan F, Jerusalmi A, Suksamrarn S, Nakamura Y, Sukseree S, Uthaisang-Tanethpongtamb W, Ratananukul P, Sano T. Potential of xanthones from tropical fruit mangosteen as anti-cancer agents: caspase-dependent apoptosis induction in vitro and in mice. Appl Biochem Biotechnol. 2010;162(4):1080–1094. doi: 10.1007/s12010-009-8903-6. [DOI] [PubMed] [Google Scholar]
- Wu Y, Li M, Yang Y, Jiang L, Liu M, Wang B, Wang Y. Authentication of small berry fruit in fruit products by DNA barcoding method. J Food Sci. 2018;83(6):1494–1504. doi: 10.1111/1750-3841.14177. [DOI] [PubMed] [Google Scholar]
- Zawiah N, Nik Zanariah NM, Othaman H, Norwati M (2019) Simbiosis flora & fauna. Forest Research Institute Malaysia, Selangor, Malaysia, p 408
- Zhu Z, Johnson J, Zaman QU, Wang H. Challenges and opportunities to improve tropical fruits in Hainan. China Trop Plants. 2022;1(1):1. doi: 10.48130/TP-2022-0013. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are available within the paper and also as supplementary information.