Abstract
Structural and mechanistic studies on human odorant receptors (ORs), key in olfactory signaling, are challenging because of their low surface expression in heterologous cells. The recent structure of OR51E2 bound to propionate provided molecular insight into odorant recognition, but the lack of an inactive OR structure limited understanding of the activation mechanism of ORs upon odorant binding. Here, we determined the cryo-electron microscopy structures of consensus OR52 (OR52cs), a representative of the OR52 family, in the ligand-free (apo) and octanoate-bound states. The apo structure of OR52cs reveals a large opening between transmembrane helices (TMs) 5 and 6. A comparison between the apo and active structures of OR52cs demonstrates the inward and outward movements of the extracellular and intracellular segments of TM6, respectively. These results, combined with molecular dynamics simulations and signaling assays, shed light on the molecular mechanisms of odorant binding and activation of the OR52 family.
Subject terms: Cryoelectron microscopy, Molecular modelling
Limited insight existed on human odorant receptors (ORs). Here, the authors present structures of consensus OR52 in its inactive and active forms, shedding light on the activation mechanism of the OR52 family.
Introduction
Humans perceive numerous odors from the environment. Olfaction, or the sense of smell, is initiated by the stimulation of odorant receptors (ORs) by odorants1,2. Humans have approximately 400 subtypes of ORs expressed on the surface of olfactory sensory neurons in a singular expression manner3–5. They recognize small-molecule odorants and induce the depolarization of olfactory sensory neurons6,7. The pairing of odorants and their cognate ORs has a combinatorial nature, allowing a person to smell far more types of odors than the number of ORs1,6,8.
ORs, which are members of class A G protein-coupled receptors (GPCRs), transduce downstream signals via the olfactory type G protein (Golf), which is highly homologous to Gs9,10. Both Golf and Gs stimulate adenylyl cyclase to produce cyclic AMP (cAMP), which activates the cAMP-dependent pathway11. Despite their classification within the class A GPCR subfamily, ORs do not possess all the conserved motifs that are characteristic of class A GPCRs. Notably, they lack the highly conserved residue W6.48 (superscript numbers refer to the GPCRdb numbering scheme12) and well-established P5.50-I3.40-F6.44 motif, which is critical for agonist-induced activation13,14.
Structural studies of mammalian ORs are challenging because of their limited surface expression in heterologous cells. Overexpression of ORs leads to robust endoplasmic reticulum retention and aggregation15,16. Extensive efforts have been made to overcome this problem, such as co-expression with receptor-transporting protein families, optimization of signal sequences, and large-scale mutagenesis screening17–23. The “consensus strategy”, which was previously applied to other proteins to improve thermostability24,25 has also been used to promote membrane trafficking of ORs, whereby the most frequent amino acids in a specific protein family are introduced to each residue position26. This approach has been successful in improving the surface expression of several human consensus ORs representing OR1, 2, 4, 5, 6, 10, 51, and 52 families while maintaining their odorant recognition abilities23.
Several subtypes of the human OR52 family, including OR52A5, OR52B2, OR52E1, OR52E8, and OR52L1, are known to recognize carboxylic acids27,28, and the consensus OR52 (OR52cs), representing 26 members of the human OR52 family (Supplementary Fig. 1), is also responsive to carboxylic acid odorants23. Although the recently published structure of propionate (PPI)-bound OR51E2 has provided insights into the molecular basis for the binding of carboxylic acid odorants29, the molecular mechanism of OR activation by odorant binding remains unclear as there is no OR structure in an inactive state for comparison with the active structure.
Herein, we aimed to elucidate the mechanisms of odorant binding and activation of the OR52 family using OR52cs as a representative. By comparing the ligand-free (apo) and octanoate (OCA)-bound states of OR52cs, we discovered a unique mechanism involving the large inward movement (7.4 Å) of the extracellular segment of TM6 upon OCA binding, in contrast to the 2-3 Å TM6 movement of non-olfactory class A GPCRs upon agonist binding. Our structural study, together with sequence analysis, mutagenesis studies, and molecular dynamics (MD) simulations, revealed a distinctive activation mechanism of the human OR52 family in response to carboxylic acid odorants with long hydrocarbon tails.
Results
Structure of the OCA–OR52cs–Gs–Nb35 complex
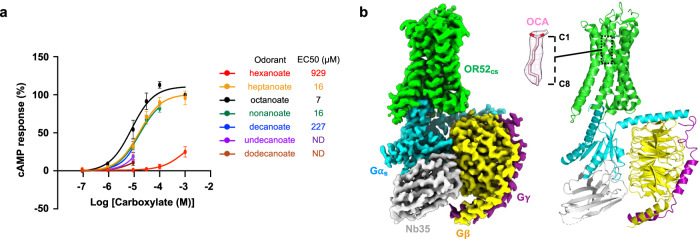
To identify the optimal odorant for OR52cs, we performed a downstream signaling assay on OR52cs. Among the various lengths of carboxylic acid odorants tested (ranging from hexanoate to dodecanoate), OCA, which showed the lowest EC50 value, was selected for our structural study (Fig. 1a). Subsequently, we verified the substitutability of Gαolf with Gαs in its interaction with OR52cs by bioluminescence resonance energy transfer (BRET) assays and structure determination of Gαolf in the GTPγS-bound state at 2.9 Å resolution (Supplementary Fig. 2 and Supplementary Table 1). Structural comparison highlighted the high similarity between Gαolf and Gαs particularly in the C-terminal α helix, which is a major GPCR binding site (Supplementary Fig. 2). Finally, we purified the complex of full-length OR52cs, Gs, and Nb35 (the complex-stabilizing nanobody30) in the presence of OCA, and solved the cryo-EM structure of the OCA–OR52cs–Gs–Nb35 complex at a global resolution of 2.97 Å (Fig. 1b, Supplementary Fig. 3 and Supplementary Table 2). After local refinement of the receptor, a 3.09 Å resolution receptor-focused map was obtained, in which most residues of OR52cs were well resolved and a clear density of OCA was identified within the transmembrane pocket (Fig. 1b).
Fig. 1. Overall structure of the OCA–OR52cs–Gs–Nb35 complex.
a Dose-response curves of OR52cs for carboxylic acid odorants of different lengths; hexanoate (red), heptanoate (orange), octanoate (OCA) (black), nonanoate (green), decanoate (blue), undecanoate (purple), and dodecanoate (brown). Each data point represents the mean ± standard error of the mean (S.E.M.) from n = 3 independent experiments except for OCA (n = 5 independent experiments). b Cryo-EM map (left) and model (right) of the OCA–OR52cs–Gs–Nb35 complex. OR52cs, Gαs, Gβ1, Gγ2, and Nb35 are colored green, cyan, yellow, magenta, and light-gray, respectively. OCA is shown as pink sticks, with the surrounding density map displayed at a threshold of 0.336 in a zoomed view.
Similar to other previously reported GPCR–Gs structures, the C-terminus α5 helix of Gαs is inserted into the cytoplasmic cavity of OR52cs (Supplementary Fig. 4). The OR-conserved VAIC sequences, DRY motif, and several residues in the intracellular loop (ICL) regions (E5912.52, R13334.52, Y13434.53, and S232ICL3) are involved in interactions with Gαs (Supplementary Fig. 4). Multiple sequence alignment showed that most of these residues are highly conserved across human ORs, suggesting the conserved binding interface between OR and Gαs/Gαolf (Supplementary Fig. 4).
Overall structural features of OR52cs
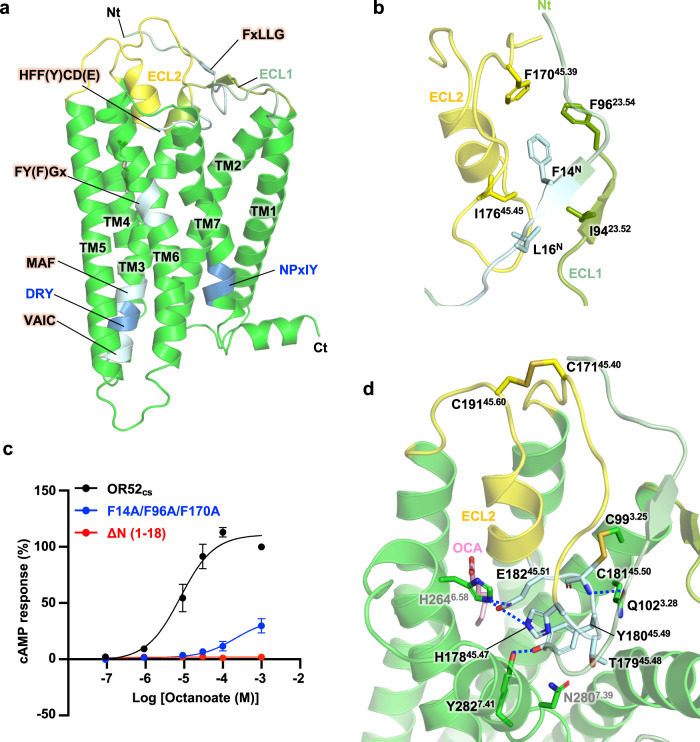
OR-specific conserved motifs, such as the FxLLG motif in the N-tail and the HFF(Y)CD(E) motif in ECL22,31, are essential for maintaining the structural integrity of OR52cs (Fig. 2a). The residue F14N within the FxLLG motif forms an aromatic interaction network with two other phenylalanine residues in ECL1 and ECL2 (F9623.54 and F17045.39) (Fig. 2b). These three Phe residues are highly conserved in human ORs (F/Y) (Supplementary Fig. 5). Substitution of these residues with alanine resulted in a significantly reduced cAMP response upon OCA treatment (Fig. 2c). Another conserved residue, L16N, forms a stable interaction network with I9423.52 and I17645.45, which are also conserved in human ORs (Fig. 2b). Notably, the hydrophobic interaction network mediated by the N-tail appears to be crucial for functional OR52cs expression as demonstrated by the significant decrease in surface expression and, consequently, loss of downstream signaling in the N-terminally truncated OR52cs mutant (Fig. 2c and Supplementary Table. 3). The ECL2 region of OR52cs containing the HTYCE motif is embedded within the transmembrane pocket formed by TMs 3, 4, 6, and 7 (Fig. 2d). This U-shaped conformation of ECL2 is stabilized by the HTYCE motif-mediated interactions: H17845.47–H2646.58, T17945.48–N2807.39, Y18045.49–Y2827.41, and E18245.51–H2646.58, all of which are conserved in the OR52 family (Fig. 2d and Supplementary Fig. 5). The ECL2 conformation is further stabilized by two disulfide bonds (C17145.40– C19145.60 and C18145.50– C993.25) (Fig. 2d and Supplementary Fig. 6). This TM-embedded conformation of ECL2 is likely to impede odorant entry into the central ligand-binding pocket from the extracellular side. Details of odorant binding to OR52cs are discussed below.
Fig. 2. Structural features of OR52cs.
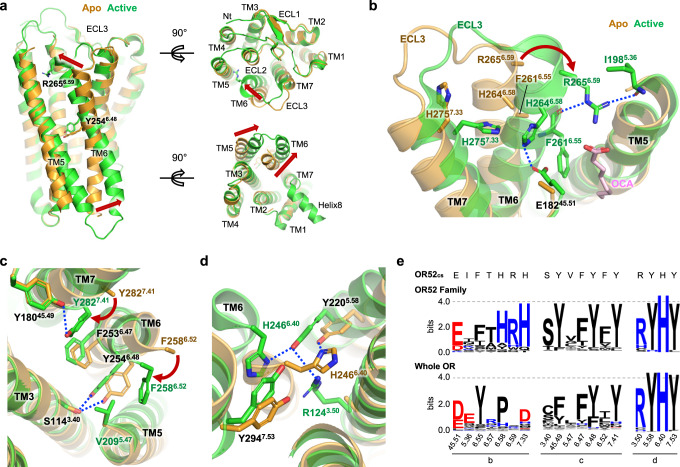
a Overall structure of OR52cs highlighting conserved motifs. The OR-specific conserved motifs are colored in light blue, and the conventional motifs present in class A GPCRs (DRY and NPxIY motifs) are colored in blue. N-tail, ECL1, and ECL2 are colored in pale green, split pea, and yellow, respectively. The TM helices and structural motifs are labeled. b Detailed interactions among the N-tail, ECL1, and ECL2. Residues interacting with F14N and L16N in the conserved FxLLG motif are shown as sticks. c Dose-dependent cAMP response curves of the N-tail deletion mutant (ΔN(1–18)) and the F14A/F96A/F170A mutant. Each data point represents the mean ± standard error of the mean (S.E.M.) from n = 3 independent experiments. d Interactions of ECL2 with TMs 3, 6, and 7. The two disulfide bonds, C993.25-C18145.50 and C17145.40-C19145.60, are shown as sticks. Residues in the conserved HTYCE motif are colored in light blue. Highly conserved residues in ORs are labeled in black, whereas less conserved residues are labeled in gray.
When comparing our structure with the recently published structure of OR51E2, we observed very similar overall structures between them, the consensus OR and the native OR, with a RMSD of 1.5 Å for 289 Cα atoms (Supplementary Fig. 6)29. Furthermore, our structural analysis of OR52cs, in combination with extensive sequence analysis of ORs, suggested that the interaction network observed in this structure is conserved in the human OR52 family (Supplementary Fig. 1), highlighting that the OR52cs structure presented here provides a comprehensive view of the overall architecture of the human OR52 family.
Odorant recognition by OR52cs
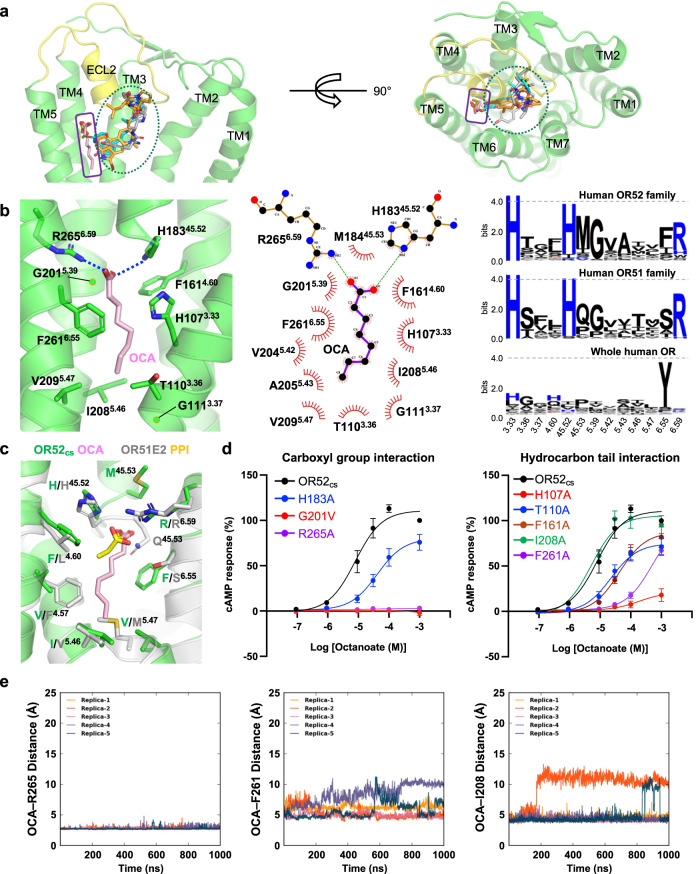
The odorant-binding pocket in OR52cs is formed by TMs 3, 4, 5, and 6 and distantly from TMs 1, 2, and 7, unlike the orthosteric ligand-binding pockets of most class A GPCRs (Fig. 3a). Notably, the TM-embedded ECL2 of OR52cs partially overlaps with this orthosteric ligand-binding site of non-olfactory class A GPCRs (Fig. 3a). In the structure of PPI-bound OR51E2, the odorant binding site is located similarly to that in OR52cs (Fig. 3a), supporting the notion that the architecture of the odorant-binding pocket in OR52cs is representative of the carboxylate-recognizing OR52 family.
Fig. 3. Odorant binding site of OR52cs.
a Comparison of ligand-binding sites in OR52cs, OR51E2, and non-olfactory class A GPCRs. Agonists bound to OR51E2 (PDB: 8F76), β2-adrenergic receptor (PDB:3SN6), dopamine D2 receptor (PDB:6VMS), and μ-opioid receptor (PDB:6DDF) are shown as yellow, cyan, orange, and gray sticks, respectively, and OCA are shown as pink sticks. The orthosteric ligand-binding site and OCA binding site are indicated by blue ellipse and purple square, respectively. The OR52cs structure is shown in cartoon representation, and TM6 and TM7 are removed in the side view on the left panel for clarity. b OCA and its interacting residues within 4.5 Å of OCA are shown as sticks, and the Cα atom of Gly is represented as a green ball. The polar interactions are indicated as dashed blue lines (left). The interaction between OCA and OR52cs was analyzed and visualized using LigPlot+ v.2.2.7 (middle). Sequence conservation of the residues constituting the odorant binding pocket in the human OR52 family, OR51 family, and whole ORs is depicted using WebLogo 3 (right). c Structural alignment of OR52cs with OR51E2. OR51E2 is shown as light-gray, and PPI is shown as a yellow stick. Residues within 4.0 Å of the odorants are shown as sticks. d Dose-dependent cAMP response curves of OR52cs (black) and mutants by OCA treatment. The EC50 values for each curve are summarized in Supplementary Table 3. The mean ± S.E.M. from n = 5 independent experiments (OR52cs) and n = 3 independent experiments (mutants) are shown as symbols and error bars, respectively. e All-atom MD simulations of the OCA–OR52cs–Gs system. The distances between OCA and R2656.59, F2616.55, and I2085.46, respectively, were plotted over the 1 µs simulation time for five replicas.
OCA contains a negatively charged carboxyl group, like PPI, but harbors a longer hydrophobic hydrocarbon moiety (Fig. 3b). The carboxyl group of OCA forms electrostatic interactions with R2656.59 and H18345.52 of OR52cs (Fig. 3c). The importance of R2656.59 for OCA binding was demonstrated by the complete loss of downstream signaling in the R2656.59A mutant (Fig. 3d). In addition, all-atom MD simulations showed that the interaction distance between the carboxyl group of OCA and R2656.59 is maintained within a distance of an average of 2.8 Å in 1 μs simulations (Fig. 3e). Notably, R2656.59 is highly conserved in human OR51/52 families, recognizing carboxylic acids as odorants32–34, but not in other OR families (Fig. 3b), suggesting that this Arg residue is responsible for the specific recognition of carboxylic acid odorants in OR51/52 members. In addition, octanol, which contains a hydroxyl group instead of a carboxyl group at the C1 position, failed to activate OR52cs, highlighting the essential role of the carboxyl group of the odorant in OR52cs activation (Supplementary Fig. 7). The conserved G2015.39 in the OR51/52 families plays a critical role in constituting the odorant binding site. In both our structure and the PPI-OR51E2 structure, the Cα atom of G5.39 is within 4 Å of the carboxyl oxygen atom of the odorants (Fig. 3b), indicating that a side chain at this position can cause steric clashes with the carboxyl group of the odorant. Indeed, the replacement of Gly with Val (G2015.39V) in OR52cs resulted in the loss of OCA signaling (Fig. 3d).
The hydrocarbon moiety of OCA is surrounded by hydrophobic residues. In particular, F2616.55 forms an extensive hydrophobic interaction network with multiple carbon atoms of OCA, and H1073.33 forms van der Waals contacts with the C3 and C5 carbons of OCA (Fig. 3b). The substitution of these residues with Ala significantly reduced downstream signaling (Fig. 3d and Supplementary Table. 3). At the bottom of the ligand binding site, T1103.36, G1113.37, I2085.46, and V2095.47 are closely located within a distance range of 3.5 to 4.5 Å from the C8 atom of OCA, forming van der Waals contacts with each other (Fig. 3b). Unlike the stable contact between the carboxyl group of OCA and R2656.59, the distances between the hydrocarbon tail of OCA and its interacting hydrophobic residues showed some fluctuations in the 1 μs all-atom MD simulations (Fig. 3e). This observation may be attributed to the fact that the ligand-binding pocket is not specifically designed only for OCA, but can also accommodate other odorants such as heptanoate and nonanoate (Fig. 1a).
A comparison of the odorant binding pockets of OR52cs and OR51E2 indicates that while the carboxyl groups of PPI and OCA interact similarly with the conserved R6.59 in each OR, the orientation of the hydrocarbon tails is slightly different in the two structures. In OR51E2, L1584.60 provides contact for the hydrocarbon tail and stabilizes the slightly lateral orientation of PPI. In contrast, the corresponding residue in OR52cs is F1614.60, which restricts the lateral orientation of OCA (Fig. 3c). The downward orientation of OCA tail is stabilized by extensive van der Waals contacts with F2616.55, whereas the corresponding residue S2586.55 of OR51E2 participates in polar interaction with the carboxyl group of PPI (Fig. 3c).
In a previous report, the F1554.57A mutation of OR51E2 showed selectivity for longer-chain fatty acids13. Our structural analysis suggested that the residue at position 5.47 may also affect the selectivity for fatty acid chain lengths. Indeed, F1554.57 and M2065.47 in OR51E2 are replaced with smaller side chains in OR52cs, V1584.57 and V2095.47, respectively (Fig. 3c). Our cAMP assay showed that the V1584.57F mutation of OR52cs greatly reduced the downstream signal for OCA and enhanced selectivity for heptanoate (Supplementary Fig. 8). Double mutation of V1584.57F/V2095.47M of OR52cs showed increased responses to shorter chain fatty acids such as hexanoate, but the preference for OCA was maintained (Supplementary Fig. 8). This result implies that the selectivity for odorant chain length is determined by the combinatorial effect of amino acids constituting the odorant-binding pocket, rather than being dictated by a single residue or two.
Structure of apo state OR52cs
The structure of OCA-bound OR52cs revealed that it would be difficult for an odorant with a long hydrophobic tail to approach the TM pocket from the extracellular side without undergoing large conformational changes as the binding pocket is occluded in the active structure (Supplementary Fig. 9). To understand the mechanism by which a fatty acid odorant with a long hydrocarbon tail reaches the ligand-binding pocket, we aimed to determine the structure of OR52cs in the apo state, without a bound ligand. For the structural study, we utilized a fiducial marker strategy, which has previously been used to determine the apo state structure of FZD5 using cryo-EM35. The four-helical bundle fusion partner of thermostabilized apocytochrome b562 (bRIL) was inserted into ICL3 of OR52cs (OR52cs-bRIL). This chimeric protein, which showed neither basal activity nor OCA-induced activity, was purified in complex with a Fab recognizing bRIL for the cryo-EM study (Supplementary Figs. 10, 11).
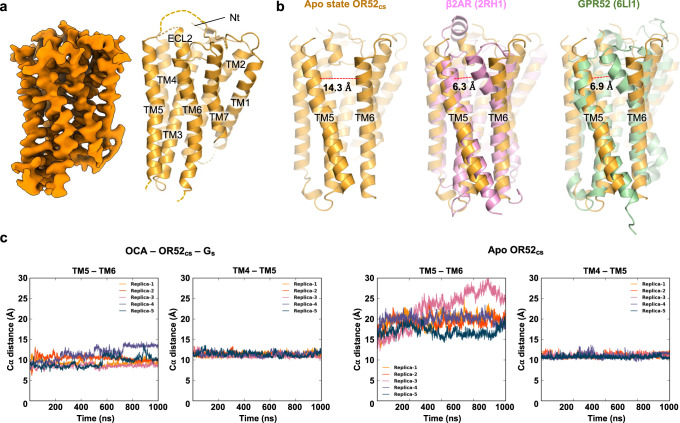
To date, only a few class A GPCR structures have been reported in the apo state, such as structures of rhodopsin and constitutively active GPR5236,37, likely due to the structural flexibility of class A GPCRs in the absence of ligand38–40. With a large dataset, extensive 3D classification, and local refinement of the receptor, we successfully obtained a cryo-EM map at approximately 4 Å for model building of the apo state OR52cs (Fig. 4a, Supplementary Fig. 10 and Supplementary Table 2).
Fig. 4. Structural characteristics of the apo state OR52cs.
a Cryo-EM map and model of the apo state OR52cs are shown. b The structures of carazolol-bound β2AR (PDB: 2RH1, pink)42 and apo state GPR52 (PDB: 6LI1, palegreen)37 are aligned against TM5 of the apo structure of OR52cs (orange). The distance between the extracellular regions of TM5 and TM6 was measured between Cα atoms of residues at positions 5.40 and 6.56 in each structure, and is displayed with a red dashed line. c All-atom MD simulations were performed for OCA–OR52cs–Gs (left) and apo state OR52cs (right). The fluctuations in the distance between TM5(L2025.40) and TM6(L2626.56), and the distance between TM4(L1644.63) and TM5(K1955.33) are plotted during the simulation time. Five replicas were simulated independently for each system. The simulation time is 1 µs for both systems.
The apo structure of OR52cs, which aligned with the AlphaFold2 prediction model41 with a RMSD of 1.1 Å for 242 Cα atoms (Supplementary Fig. 12), reveals an unprecedented feature— a large opening between the extracellular parts of TM5 and TM6 that is not seen in other class A GPCRs in their inactive states42. Indeed, the distance between L2025.40 and L2626.56 reaches 14 Å in the apo state of OR52cs, compared to 6–7 Å in most class A GPCRs (Fig. 4b). All-atom MD simulations of the apo state OR52cs showed considerable fluctuations in the distance between TM5 and TM6, whereas the distance between these helices remained stable in the active state (Fig. 4c). In the apo state, the extracellular segment of TM6 (residues 255-266) makes only a few contacts with TM7 residues, whereas the intracellular segment of TM6 (residues 237-254) forms close contacts with residues on TM5. C2436.37 and H2466.40 form close contacts with Y2205.58, and I2506.44 interacts with I2165.54 (Supplementary Fig. 11). In the middle of TM6, Y2546.48 forms hydrogen bonds with S1143.40 (Supplementary Fig. 11). These interactions stabilize the inactive structure, where the G protein binding pocket is closed (Supplementary Table. 4).
Structural changes in OR52cs upon odorant binding
Comparison of the apo and active structures of OR52cs provided insights into conformational changes upon receptor activation (Fig. 5a). Notably, TM6 underwent substantial conformational changes, with an inward movement at the extracellular end (7.4 Å shift of Cα6.59 atom) and an outward movement at the intracellular end (9.0 Å shift of Cα6.34 atom) upon OCA binding, acting Y2546.48 of the conserved FYxP motif as the pivot (Fig. 5a). In addition, TM6 exhibited a 2–3 Å upward shift. As the TM6 conformation changes, the interaction network that stabilizes the inactive conformation is rearranged.
Fig. 5. Comparison of the apo and active states of OR52cs.
a An overlay of the apo (orange) and active (green) states of OR52cs is presented in the side (left), extracellular (upper right), and intracellular (bottom right) views, with movements of the TMs highlighted by red arrows. Y2546.48 of the FY6.48xP motif that act as a pivot and R2656.59 are shown as sticks. b Residues participating in interactions that stabilize the extracellular segments of TM6 and TM7 are shown as sticks. Polar interactions are represented as dashed blue lines. The rotation of R2656.59 upon activation is indicated by a red arrow. The side chains of F2616.55, H2646.58, and R2656.59 are unresolved in the apo structure. c Interactions between F2586.47 and V2095.47 and between Y2827.41 and Y18045.49, which are observed only in the active state, are shown, with the rotation of F2586.47 and Y2827.41 upon activation, indicated by red arrows. OCA is not shown for clarity. d The interaction network of TM3-TM6-TM7 formed upon receptor activation is shown. e Conservation of key residues stabilizing the active conformation, highlighted in (b–d), is depicted using WebLogo 3.
Notably, R2656.59, a critical residue for OCA binding, exhibits a 7.4 Å inward movement towards the odorant-binding pocket upon OCA binding, and its inward conformation was stabilized by multiple interactions in the active structure. R2656.59, engaged with the carboxyl group of OCA, forms polar interactions with the carbonyl groups of I1985.36 and F2616.55, and van der Waals interactions with I1985.36 (Fig. 5b). The inward conformational shift of the extracellular part of TM6 is further stabilized by the formation of hydrogen bonds of H2646.58 with E18245.51 and H2757.33 (Fig. 5b). F2586.52 underwent an inward movement to form a hydrophobic interaction with V2095.47 (Fig. 5c), although the side chains of both residues were not well-resolved in the apo state. Upon OCA binding, Y2546.48 of the FYxP motif shifted upward by approximately 2.8 Å, while retaining its hydrogen bond with S1143.40 (Fig. 5c). F2536.47 also did not undergo a large conformational change except for a 2.4 Å upward movement, and interacted with Y2827.41, which underwent a large inward movement upon activation (Fig. 5c). In turn, Y2827.41 forms a polar contact with Y18045.49 of the conserved HS(T)YCD(E) motif (Fig. 5c). At the intracellular part of TM6, H2466.40 undergoes a 4.5 Å outward movement with a downward rotamer change, to form a van der Waals contact with Y2947.53 of the NPxIY motif (Fig. 5d). Y2205.58 moved inward by 3.4 Å, to form a hydrogen bond with R1243.50 in the DRY motif (Fig. 5d). The importance of these interactions for OR52cs activation was demonstrated by reduced efficacy or potency of Ala mutations of each interacting residue (Supplementary Fig. 13). Given the high conservation of these interacting residues in the OR52 family, it is plausible that these conformational changes upon activation could occur in native human OR52 members (Fig. 5e and Supplementary Fig. 1).
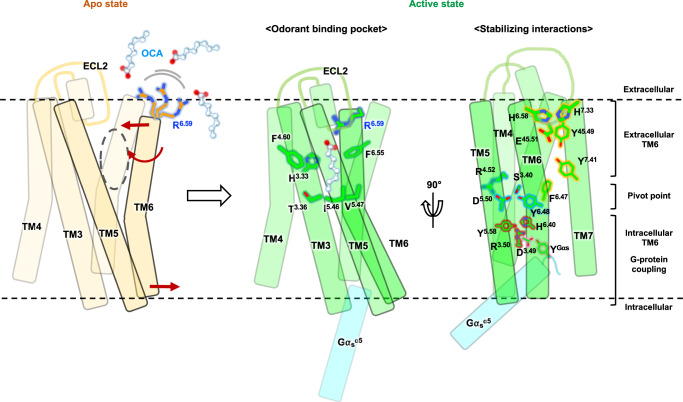
The observation of a large opening between TM5 and TM6 in the apo structure prompted us to hypothesize that an odorant with a long hydrophobic tail could approach the odorant binding pocket through this opening. To gain a deeper understanding of the odorant binding mechanism in OR52cs, we performed extensive all-atom MD simulations. Initially, we attempted to observe OCA entering the odorant binding pocket of OR52cs using the apo structure as a starting model. However, this was not successful, possibly because our simulation timescale was not sufficiently long to capture such events. Therefore, we conducted all-atom MD simulations of the OCA-bound active state OR52cs (with and without Gs) to understand how OCA exits the pocket. In the 20 µs simulation without Gs, we observed that the extracellular part of TM6 moved outward after 1 µs simulation time (Supplementary Fig. 14). As a result, OCA escaped from the pocket through this wide opening between TM5 and TM6, a characteristic structural feature of the apo-state OR52cs. In this state, R2656.59 and F2616.55, which initially pointed toward the odorant binding pocket, reoriented toward the lipid bilayer due to the TM6 rotation. Even when OCA was out of the pocket, R2656.59 still captured the carboxyl group of OCA whereas F2616.55 lost interactions with the hydrocarbon tail of OCA. Interestingly, OCA captured by R2656.59 re-entered the TM pocket around 8 µs simulation time, although it was not properly positioned in the odorant binding pocket because of the lipid entering the TM pocket (Supplementary Fig. 14). Combining these observations, a probable OCA binding mechanism can be inferred: OCA is first captured by R2656.59, and then enters the odorant binding pocket of the apo-state OR52cs. The conformational changes of OR52cs upon OCA binding stabilizes the active structure of OR52cs, which is further potentiated by G protein binding (Fig. 6). Intriguingly, during the same simulation timescale, OCA in OR52cs bound to Gs did not escape the receptor.
Fig. 6. Mechanism of OR52cs activation by OCA binding.
Schematic representation of the activation mechanism of OR52cs upon OCA binding. In the apo state (left, light yellow), there is a wide opening between TM5 and TM6, possibly acting as an entrance for OCA. The highly conserved R6.59 on the extracellular end of TM6 is flexible, represented as multiple conformations. Inward and outward movements of the extracellular and intracellular regions of TM6 and TM6 rotation upon OCA binding are indicated by red arrows. In the active state (green), OCA-mediated interactions in the odorant-binding pocket (middle), and distinct interaction networks that stabilize the active conformation (right) are presented. The active conformation is further stabilized by interactions between OR52cs and G protein (cyan). OCA molecules are shown in the ball-and-stick model. Figure was generated using ChimeraX and Powerpoint.
Discussion
In this study, we present a structural analysis of the OR52 family using a consensus strategy (represented by OR52cs) to overcome the low expression of ORs in heterologous cells. Our research demonstrates the structural characteristics of the odorant-binding pocket of OR52cs, which can accommodate carboxylic acids with carbon lengths of 7–9. Furthermore, we unveiled the odorant binding mechanism by extensive all-atom MD simulations and a comparative study of the apo and active structures. We highlight the importance of conserved residues in the OR52 family, particularly R6.59, in recognizing the carboxyl group of OCA, and hydrophobic residues, F4.60 and F6.55, in interacting with the hydrocarbon tail of OCA.
Based on our OR52cs structure, we inferred the odorant specificity of some OR52 family members, including OR52N2, OR52N5, OR52E4, OR52E5, and OR52L1, via a sequence analysis of the amino acids constituting the odorant binding pocket (Supplementary Table. 5). However, our attempts to verify odorant specificity using cAMP assays with fatty acids ranging from pentanoate to undecanoate were unsuccessful under our experimental conditions, likely due to their limited surface expression (Supplementary Fig. 15). As OR52L1 was previously reported to be responsive to pentanoate28, we conducted computational modeling of pentanoate-bound OR52L1 using our OCA–OR52cs structure as a template. This model showed that most residues involved in odorant binding are well conserved in both structures (Supplementary Fig. 15). Regarding OR52E5, an orphan OR, our computational simulations for various lengths of fatty acids imply that the optimal range could be from octanoate to decanoate, based on the stability of the odorant-binding pocket during 200 ns all-atom MD simulations (Supplementary Fig. 15). Although these computational models should be experimentally verified in the future, we believe that our OR52cs structure can be used for computational modeling and virtual screening to deorphanize orphan OR52 family members43.
The apo structure of OR52cs in this study provides an interesting structural feature of OR52cs in the absence of ligand, which is distinct from previously reported class A GPCR structures in the apo- or inactive states. Notably, the extracellular segment of TM6 is located 14 Å away from TM5, resulting in a wide opening between TM5 and TM6. This unique feature suggests that the hydrophobic tail of OCA in the lipid bilayer may access the transmembrane pocket of OR52cs through this opening. Upon OCA binding, we observe substantial inward and outward movements of the extracellular and intracellular parts of TM6, respectively, with Y2546.48 serving as a pivot point. These conformational changes are stabilized by the interactions between OCA and the transmembrane pocket of OR52cs, as well as the interactions enabled by the repositioning of F2586.52, H2646.58, R2656.59, and Y2827.40. Subsequently, G protein coupling at the cytoplasmic region further stabilizes the active conformation of OCA-bound OR52cs. Importantly, this study provides direct evidence of substantial TM6 conformational changes upon activation, distinguishing it from the established activation mechanism of class A GPCRs. It is noteworthy that the amino acids involved in the activation process are highly conserved in the OR51/52 families but not in all ORs, implying a potential diversity of mechanisms in ORs with varying odorant types.
Extensive all-atom MD simulations suggest that hydrophobic odorants may enter and exit the odorant pocket through an opening between TM 5 and TM6. In the absence of Gs, MD simulation of OCA-bound OR52cs revealed the escape of OCA from the pocket in 2 µs through the opening between TM5 and TM6, and concomitantly, the intrusion of phospholipids into this space, which restricts the inward movement of TM6 (Supplementary Fig. 16). In a longer simulation time even after OCA completely escapes the odorant pocket, we could observe dynamic exchange of phospholipids, which leads to a dynamic TM5-TM6 gap, providing a pathway for OCA to access the binding pocket. When Gs is engaged, the OCA-bound active conformation of OR52cs is maintained in our simulation time (10 µs), preventing phospholipid intrusion. This indicates that both odorant binding and G protein coupling are required for stabilizing OR52cs in its fully active conformation, which is capable of transmitting downstream signals. While our MD simulation data, together with the hydrophobic nature of the OCA tail supports the plausibility of the lateral entry pathway, further studies are needed to understand the odorant entry mechanism. Of note, within the in vivo system, odorant-binding proteins that transport hydrophobic odorants are present within nasal mucus44,45.
Overall, this study provides valuable insights into the activation mechanism and odorant specificity of the OR52 family, and paves the way for further experimental and computational studies to unravel the mechanisms underlying OR function.
Methods
Construct design
Full-length human Gαs (short isoform) with the C3S mutation was cloned into the pFastBac HT B vector. For the Gβ and Gγ subunits, full-length wild-type human Gβ1 and Gγ2 were cloned into the pFastBac Dual expression vector. N-terminal 6xHis-tag followed by a human rhinovirus 3C (HRV 3C) protease site was introduced into the plasmids harboring Gαs and Gβ1 for affinity purification.
For the structural study of apo state OR52cs, the chimeric construct of full-length OR52cs (1–313)23 was designed to insert bRIL between A226 and L227 with a linker sequence derived from adenosine receptor A2a, as previously described35. For purification of active state OR52cs, full-length OR52cs (1–313) was used. Each construct was subcloned into the pFastBac HT B vector with a Lucy signal sequence (MRPQILLLLALLTLGLA)17 and FLAG peptide at the N-terminus. Each construct also included an eGFP with 10xHis-tag, which was fused to the C-terminus of OR52cs with a HRV 3C protease cleavage sequence.
For cAMP assay, CRE luciferase assay, and surface ELISA, the full-length OR52cs construct was cloned into the pcDNA3.1 vector. The Lucy signal sequence and FLAG peptide sequence were added to the N-terminus. Mutant constructs were generated by site-directed mutagenesis. For BRET assay, eYFP was added at the C-terminus of OR52cs with a GGGGS linker. Rluc was added to Gα for resonance partner (after L99 for Gαs, and I100 for Gαolf, respectively, with GGGGS linker back and forth).
Sequence analysis
The amino acid sequences of human ORs were obtained from HORDE46 and pseudogenes were excluded for sequence analysis. The sequences of 388 intact human ORs or 26 members of OR52 family were aligned using MAFFT47 with L-INS-i strategy. MEGAX48 was used for phylogenetic tree generation with maximum-likelyhood method. The sequence conservation was visualized by WebLogo 349.
Purification of OR52cs-bRIL
OR52cs-bRIL was overexpressed in Spodoptera frugiperda (Sf9, Expression Systems, 94-00lF) cells using the Bac-to-Bac baculovirus expression system (Invitrogen). Cells were harvested 60 h after infection and lysed with a buffer containing 20 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 175 µg mL−1 benzamidine, and 10 µM leupeptin using a dounce homogenizer. OR52cs-bRIL was extracted from the membrane fraction with solubilization buffer containing 20 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1% (w/v) lauryl maltose neopentyl glycol (LMNG, Anatrace) and 0.1% (w/v) cholesteryl hemisuccinate (CHS, Sigma-Aldrich), 1 mM PMSF, 175 µg mL−1 benzamidine, and 10 µM leupeptin. After centrifugation, the supernatant was loaded onto a Ni-NTA column (Qiagen) pre-equilibrated with wash buffer consisting of 20 mM Tris-Cl (pH 8.0), 10 mM imidazole, 150 mM NaCl, 0.005% (w/v) LMNG, and 0.0005% (w/v) CHS. After washing the column with wash buffer, OR52cs was eluted with 250 mM imidazole buffer. The eluted sample was loaded onto anti-FLAG affinity (M1) column (Sigma-Aldrich) pre-equilibrated with LMNG buffer (20 mM Tris-Cl (pH 8.0), NaCl 150 mM, 0.005% (w/v) LMNG and 0.0005% (w/v) CHS) supplemented with 2 mM CaCl2. OR52cs-bRIL was eluted with LMNG buffer containing 0.1 mg ml−1 FLAG peptide and 4 mM EDTA, and further purified with size exclusion chromatography (SEC) using a Superdex 200 10/300 column (Cytiva), which was pre-equilibrated with LMNG buffer. The peak fractions were collected and used to form the complex with bRIL-specific Fab.
Purification of bRIL-specific Fab
Genes encoding the bRIL-specific Fab VH and VL domains35 were subcloned into the pFastBac Dual vector and Fab was expressed in BTI-Tn-5B1-4 (High Five, Expression Systems, 94-002 F) cells using the Bac-to-Bac expression system. Cells were harvested 72 h after infection, and the supernatant was collected and loaded onto a Ni-NTA resin pre-equilibrated with wash buffer containing 20 mM HEPES (pH 7.5), 200 mM NaCl, and 10 mM imidazole. After washing the column with wash buffer, Fab was eluted with 250 mM imidazole buffer and subsequently loaded onto a HiLoad 26/200 Superdex 200 column (Cytiva) for further purification.
Purification of OR52cs-bRIL–Fab complex
Purified OR52cs-bRIL and bRIL-specific Fab were mixed at a molar ratio of 1:1.2 and incubated on ice for 30 min. The complex was purified with SEC using a Superdex 200 10/300 column (Cytiva) which was pre-equilibrated with 20 mM Tris-Cl (pH 8.0), 150 mM NaCl, 0.0025% (w/v) LMNG and 0.00025% (w/v) CHS to remove excess Fab. The peak fractions were concentrated to 9 mg ml−1 and used to prepare cryo-EM grids.
Purification of the Nb35
Nanobody-35(Nb35) with a 6xHis-tag at its C-terminus was expressed in Escherichia coli (E. coli) Rosetta (DE3, Novagen) cells and purified as previously described50. Briefly, Nb35 was purified with Ni-NTA column, followed by SEC with Superdex 200 10/300 column (Cytiva). The peak fractions were collected and stored at 4 °C until use.
Purification of OCA–OR52cs–Gs–Nb35 complex
For purification of the OR52cs–Gs–Nb35 complex, OR52cs, human Gαs, human Gβ1, and human Gγ2 were co-expressed in Sf9 insect cells using a Bac-to-Bac expression system. Cells were harvested 60 h after infection and lysed with a buffer containing 20 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1 mM PMSF, benzamidine, and leupeptin. After centrifugation, the pellet was resuspended in a buffer containing 20 mM Tris-Cl (pH 8.0), 150 mM NaCl, 10 µg ml−1 Nb35, 25 mU ml−1 apyrase, 2 mM octanoic acid (OCA, Sigma-Aldrich), 1 mM PMSF, benzamidine, and leupeptin, to form the complex in the membrane, and the membrane was solubilized with 1% (w/v) LMNG (Anatrace) and 0.1% (w/v) CHS at 4 °C for 2 h. After centrifugation at 21,671 × g for 15 min, the supernatant was loaded onto a Ni-NTA column and the column was washed with LMNG buffer containing 10 mM imidazole and 2 mM OCA. The complex was eluted with 200 mM imidazole and loaded onto an M1 column pre-equilibrated with LMNG buffer containing 2 mM CaCl2 and 2 mM OCA. The resin was washed with the same buffer, and the protein complex was eluted with LMNG buffer supplemented with 0.1 mg ml−1 FLAG peptide, 3 mM EDTA, and 2 mM OCA. The protein complex was confirmed by SDS-PAGE gel and concentrated for cryo-EM grid preparation.
Purification of olfactory Gα subunit (Gαolf)
To purify Gαolf, 6xHis-Gαolf was co-expressed with GST-Ric8b in Sf9 cells. Cells were harvested 48 h after infection and lysed with lysis buffer (20 mM Tris-Cl, (pH 8.5), 100 mM NaCl, 1 mM EDTA, and protease inhibitors). After centrifugation (21,671 × g, 20 min, 4 °C), the supernatant was loaded onto a Ni-NTA column. After column washing, Gαolf and Ric8b complexes were eluted with elution buffer (20 mM Tris-Cl (pH 8.5), 150 mM NaCl, 300 mM imidazole, 0.1 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP)). The eluted fractions were collected and further purified using the Hitrap Q anion-exchange column (Cytiva). The peak fractions were pooled and concentrated, and GTPγS and MgCl2 were added to separate Gαolf from Ric8b. After 1 h incubation at room temperature, the reaction mixture was loaded onto Hitrap Q and Gαolf was separately eluted from the column. Purified Gαolf was concentrated to 8 mg ml−1 and used for crystallization.
Crystallization, data collection, and structure determination of Gαolf
GTPγS-bound Gαolf was crystallized by hanging drop vapor diffusion at 22 °C with reservoir solution consisting of 0.2 M MgCl2 and 18% PEG 3350. Crystals were flash-cooled in liquid nitrogen using 20% ethylene glycol as a cryoprotectant.
Diffraction data were collected at 100 K on beamline 5 C at the Pohang Accelerator Laboratory (PAL, Korea) and processed with the XDS package51. The structure of Gαolf was solved by molecular replacement with Phaser52, using the GTPγS-bound Gαs structure (PDB:1AZT) as a search model. Iterative cycles of manual rebuilding with Coot53 and refinement with PHENIX54 were performed to obtain the final model, which was validated with Molprobity55. The final model was deposited in the PDB with PDB code 8HTG.
Cryo-EM grid preparation and data collection
3.5 µl of the purified OR52cs-bRIL–Fab and OCA–OR52cs–Gs–Nb35 complexes (9 mg ml−1 and 5 mg ml−1, respectively) were applied to the glow-discharged 300 mesh holey carbon grid (Quantifoil R1.2/1.3) (SPI) pre-coated with poly-L-lysine (Sigma-Aldrich) and 300 mesh R1.2/1.3 UltraAuFoil grid (SPI), respectively. Each grid was blotted for 3 s with a blot force of 5 at 4 °C, 100% humidity, and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). The grids were initially screened with FEI Glacios (Thermo Fisher Scientific) equipped with a Falcon 4 detector (Thermo Fisher Scientific) at the Center for Macromolecular and Cell Imaging at Seoul National University (SNU CMCI, Korea).
Images of the OR52cs-bRIL–Fab complex were collected using a Titan Krios G4 (Thermo Fisher Scientific) at the Institute of Basic Science (IBS, Korea), equipped with a K3 BioQuantum (Gatan) at a magnification of 105,000X with a calibrated pixel size of 0.848 Å. Movies were collected with a total dose of 68.5 e− A−2 and 57 frames per micrograph with a defocus ranging from −0.7 to −1.9 μm.
Data for the OCA–OR52cs–Gs–Nb35 complex were collected using a Titan Krios G4 at the Institute of Membrane Proteins (IMP, Korea), equipped with a K3 BioQuantum at a magnification of 105,000× with a calibrated pixel size of 0.851 Å. Movies were collected at a total dose of 60.2 e− A−2 and 50 frames per micrograph with a defocus ranging from −0.8 to −2.0 μm.
Cryo-EM data processing and 3D reconstruction
Image stacks were subjected to beam-induced motion correction using the patch motion correction, and the contrast transfer function (CTF) parameters for each non-dose-weighted micrograph were determined using patch CTF estimation. Data processing was mostly performed using cryoSPARC56.
For the OR52cs-bRIL–Fab complex, 21,345 movies were used for data processing using cryoSPARC v4.2.0. Particles were initially selected using a Blob picker. Templates were generated by performing several rounds of two-dimensional (2D) classification and template selection. The final selected particles were used for Topaz training57, and the extracted particles were used to reconstruct the 3D volume. Multiple rounds of Ab-initio reconstruction and heterogeneous refinement were performed to select suitable particles. Non-uniform refinement58 from the selected 440,134 particles resulted in a Fab-focused map with a global resolution of 3.44 Å. A mask covering OR52cs was generated using the Chimera built-in Segger tool and masked 3D classification was performed to isolate the particles that could reconstruct intact TMs of OR52cs. The selected 173,732 particles were beam-tilt corrected by global CTF refinement and reached to 3.66 Å resolution map (volume-1) with intact TMs of OR52cs. Local refinement with an OR52cs-focused mask produced a map with a resolution of 3.74 Å. The additional 3D classification was performed to discard bad particles, and the final 142,861 particles went through local refinement which produced a OR52cs-focused map (volume-2) with a resolution of 3.39 Å. The resolution of the final map was also estimated by RELION v3.1.1 with each half maps59, which results in resolution of 3.6 Å. The output volume-2 was used for the structure determination of OR52cs. Because volume-2 was focused on OR52cs and not sufficient for model building of bRIL, volume-1 was used for structure determination of linker and bRIL. The detailed description of the cryo-EM data collection parameters is provided in Supplementary Table 2.
For the OCA–OR52cs–Gs–Nb35 complex, 4798 movies were used for data processing by cryoSPARC v3.2.2. Particles were selected by 2D classification, and selected particles were used for the Topaz training. 493,826 particles from the Topaz extract were separated by several rounds of 2D classification and heterogeneous refinement, and 74,356 particles were further refined by non-uniform refinement. Because the map has a preferred orientation issue, new templates were created for template picking to excavate the top- and bottom-view particles. Duplicate particles were removed and remaining particles were classified using heterogeneous refinement. The final selected particle sets were further subjected to beam-induced motion correction using a local motion correction tool and refined by non-uniform refinement to a global resolution of 2.97 Å. Masks covering OR52cs or Gs–Nb35 were generated using Chimera built-in Segger tool. Local refinement was performed to refine the OR52cs and Gs–Nb35 parts. Finally, 126,896 particles were used to yield a 3D map with a local resolution of 3.09 Å for OR52cs and 2.84 Å for Gs–Nb35. These maps were combined using the Vop maximum tool built-in Chimera, and the resulting map was used for model building. The detailed description of the cryo-EM data collection parameters is provided in Supplementary Table 2.
Model building and refinement
For the OR52cs-bRIL–Fab complex, the protein sequence of OR52cs was submitted as the input protein sequence of AlphaFold2.ipynb41, and one of the created models was used as the initial model. For the OCA–OR52cs–Gs–Nb35 complex, the same AlphaFold model was used as the initial model. Models of the Gs heterotrimer and Nb35 were derived from the crystal structure of the active β2AR–Gs protein complex (PDB:3SN6). Model building and refinement were performed by iterative cycles of refinement with PHENIX60 and manual rebuilding with Coot53. For the OR52cs-bRIL–Fab complex, the quality of OR52cs model was assessed by Q-score calculating using MapQ software61. To validate the current OCA conformation and pose in our structure, we checked the conformational stability of OCA and the agreement of the OCA structure with the density by running the Rosetta relax application (dual-space relax)62. The criteria for conformational instability of the ligand can be assessed by high RMSD values (more than 2 Å) during the simulations. Relax runs were repeated ten times with and without the electron density information. In both types of simulations, a strong convergence towards the pre-relaxed original conformation (RMSD < 0.8 Å) was observed from the sampled ligand conformations, supporting that our original conformation is stable and agrees well with the electron density. Refinement statistics are presented in Supplementary Table. 2. In the final model of the active structure, the N-terminal seven residues (1–7) and the C-terminal three residues (311-313) of OR52cs, and α-helical region (47–193) as well as the N-terminal nine residues (1–9) and some loop regions (235–249, 281–294, and 308–319) of Gαs were not modeled due to poor map quality. In the case of the apo structure, the N-terminal 11 residues (1-10, 18), ICL1 (53-56), part of ECL2 (169–173, 190–193), helix 8, and the C-tail (295–313) were not modeled.
BRET assay
All signaling assays were performed in Hana3A cells22, which were kindly provided by Dr. Matsunami. Cells were seeded on 6-well clear plate (SPL) and co-transfected with OR52cs-eYFP, Gαs-Rluc or Gαolf-Rluc, Gβ1 and Gγ2 at a 5:1:1:1 ratio. 48 h after infection, cells were collected and resuspended with PBS. 500 μM of OCA was treated to the cells, and cells were transferred to 96-well white microplate (SPL). Coelenterazine H (Nanolight) was added to each well at a final concentration of 10 μM and luminescence was measured using a Tristar 2 LB 942 multimode reader (Berthold). Data were analyzed using GraphPad Prism 9.4.1.
cAMP signaling assay
Hana3A cells were seeded on 96-well white microplates and transfected with OR52cs or the corresponding mutants and cAMP sensor (Promega). The medium was exchanged with CO2-independent medium (Gibco) supplemented with 0.375 mg ml−1 D-luciferin (Nanolight) 48 h after infection, and cells were incubated in the dark at 20 °C for 2 h. Basal luminescence was detected using a Tristar 2 LB 942 multimode reader (Berthold) until it reached a plateau. Subsequently, various concentrations of odorants and forskolin (Sigma-Aldrich) were added to each well, and luminescence was measured again. Data were analyzed and plotted using GraphPad Prism 9.4.1.
CRE assay
Hana3A cells were seeded on 96-well white microplates (SPL) and transfected with plasmids encoding CRE-luciferase, OR52cs or R2656.59A mutant, Renilla luciferase, RTP1S, and Ric8b at a 5:1:1:1:1:1 ratio 24 h after seeding. 48 h after infection, cells were treated with odorants and incubated at 37 °C for 4 h. Luminescence was measured using a Tristar 2 LB 942 multimode reader (Berthold). All values were divided by the Renilla luciferase activity to normalize the effect of cell confluence. All data were analyzed using GraphPad Prism 9.4.1.
ELISA-based surface expression assay
Hana3A cells were seeded on 96-well clear microplates (NEST) and transfected with plasmids encoding OR52cs or its corresponding mutants. All the constructs contained a FLAG tag at the N-terminus. Cells were treated with 4% paraformaldehyde for fixation 48 h after infection and washed with PBS. After 2 h of incubation with blocking solution (5% bovine serum albumin (BSA, Bovogen) in PBS), rabbit anti-FLAG antibody (Cell Signaling Technology, D6W5B, 1:1,000 dilution) was treated at 4 °C overnight. Cells were washed extensively with PBS and treated with goat anti-rabbit horseradish peroxidase (HRP)-conjugated antibody (Enzo Life Sciences, ADI-SAB-300-J, 1:1000 dilution). After 2 h of incubation, the TMB solution (Thermo Fisher Scientific) was added to each well, and when a blue color appeared, 1 M HCl was added to quench the reaction. The absorbance was measured at 450 nm using a FlexStation3 microplate reader (Molecular Devices). For normalization, Janus green solution (0.2% (w/v), TCL) was added to the cells and mixed with 0.1 M HCl. The absorbance was measured at 595 nm. The normalized expression level of the receptor at the cell surface was calculated as the ratio between the absorbance at 450 nm and 595 nm (A450/A595).
All-atom MD simulations of apo and OCA-bound states of OR52cs
Three model systems were prepared for all-atom MD simulation: OCA–OR52cs–Gs (139 × 139 × 171 Å3), OCA–OR52cs (91 × 91 × 113 Å3), and apo OR52cs (90 × 90 × 116 Å3), with the total numbers of atoms (and water molecules) of 308,264 (221,229), 85,555 (17,823), and 87,389 (18,441), respectively (Supplementary Table 6). The receptor was embedded into a model membrane composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and cholesterol (4:1). For the G protein, three lipidations were introduced into the Gαs and Gγ proteins, i.e., N-myristoylation (Gly2 of Gαs), S-palmitoylation (Cys3 of Gαs), and S-geranylgeranylation (C68 of Gγ). We used the CHARMM-GUI PDB Reader & Manipulator, Ligand Reader & Modeler, Membrane Builder, and Input Generator63–68.
The CHARMM36(m) force field69,70 was utilized for lipids and proteins, and CGenFF71 was used for the OCA ligand. For solvation, 0.15 M KCl was included in the TIP3P water model72,73. The van der Waals interactions were switched off smoothly between 10 and 12 Å by a force-based switching function74, and the particle-mesh Eward method75 with a mesh size of ~1 Å was used to calculate the long-range electrostatic interactions. The SHAKE algorithm76 was used for constraining bond lengths including hydrogen atoms, and the temperature and pressure were set to 303.15 K and 1 bar, respectively, with Langevin dynamics with a friction coefficient of 1 ps−1 and Monte Carlo barostat77. Following the CHARMM-GUI six-step equilibration procedure78, NVT simulations (constant particle number, volume, and temperature) with positional and dihedral restraints were performed for equilibration by gradually alleviating the force constant, and then NPT (constant particle number, pressure, and temperature) simulations were conducted for production run without restraints with 4 fs time-step using the hydrogen mass repartitioning method79,80. Periodic boundary condition was applied for all simulations, and we conducted five independent simulations for each system for effective sampling. Simulations were performed at least 1 μs for five replicas using OpenMM simulation package81.
Based on the OpenMM simulations, we utilized Anton2 to extend our investigation of the systems’ dynamics at a longer time scale up to 20 µs and 10 µs for OCA-OR52cs and OCA-OR52cs-Gs, respectively82. In Anton2 simulations, we used a time step of 2 fs and saved frames every 240 ps, where NPT ensemble with Nosé-Hoover method and Multigrator integrator were used for temperature and pressure coupling, respectively83,84. Although we conducted extensive simulations for these systems, the sampling may vary when reproduced.
Docking and MD relaxation of different lengths of carboxylic acids
The OR52E5 receptor structure was predicted using AlphaFold241 supported by the OCA–OR52cs–Gs structure as a template, and refined using Rosetta FastRelax62. To model the odorant–OR52E5 interaction, complex structures for various ligands ranging from hexanoate to dodecanoate were created by using Rosetta GALigandDock85. To estimate the stability of the docked structures, MD relaxations were carried out using pmeMD_cuda from AMBER20 in GPUs86. CHARMM-GUI was used for input generation in the same manner as that described in the previous section. POPE and POPC lipid bilayers were mixed at 1:1 and TIP3P water72,73 at 0.15 M KCl were employed. The Amber force field AMBER19SB, LIPID17, and GAFF2 were applied to the protein, lipids, and ligand. 200 ns production run was performed at 303 K. Changes in the receptor structure were analyzed using the CPPTRAJ utility in AMBER87. As a measure for the structural stability of each receptor–ligand complex, the RMSD of the binding pocket residues (H107, T110, F161, H183, A202, V205, F258, and R262) from the initial docked structure during the simulation trajectory was measured. Structural modeling of pentanoate–OR52L1 was conducted by using the same procedure as for OR52E5.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Dr. Matsunami (Duke University) for kindly providing Hana3A cells. This work was supported by Samsung Science and Technology Foundation under Project Number SSTF-BA1901-09 (to H.-J.C.), and National Science Foundation, USA (MCB-2111728 to W.I.). We thank our research assistant, Bo Ram Seo, for her experimental support. We also thank Dr. Bum Han Ryu (Institute for Basic Science (IBS), Korea) and Dr. Su Jeong Kim (Institute of Membrane Proteins (IMP), Postech, Korea) for supporting data collection at IBS and IMP cryo-EM facilities, respectively, and Global Science experimental Data hub Center (GSDC) and KREONET at Korea Institute of Science and Technology Information (KISTI) and the data analysis hub, Olaf at the IBS Research Solution Center for computing resources. Anton 2 computer time was provided by PSC through Grant R01GM116961 from the National Institutes of Health. The Anton 2 machine at PSC was generously made available by D.E. Shaw Research.
Author contributions
C.C. and J.B. equally contributed to this work. C.C. and J.B. performed protein purification and cryo-EM data collection with help from I.B. C.C. determined the two structures of OR52cs with help from J.B. and J.K. C.C., J.B. and H.-J.C. performed structural analysis of OR52cs. J.B., C.C., K.K. and W.-K.H. performed cell-based signaling assays. S.K. and W.I. performed all-atom MD simulations and their analysis, and S.L., C.S. and H.P. performed ligand docking and its analysis. H.K. determined the crystal structure of Gαolf. H.-J.C. conceived and directed the study. C.C., J.B. and H.-J.C. wrote the manuscript with contributions from all authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The coordinate and structure factor of GTPγS-bound Gαolf were deposited in the Protein Data Bank under accession number of 8HTG. The coordinates of OR52cs in the apo (with and without bRIL) and in complex with OCA, Gs, and Nb35 were deposited in the Protein Data Bank under accession numbers of 8J46, 8WW7 and 8HTI, respectively. The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession numbers of EMD-35971 (globally refined cryo-EM density containing apo state OR52cs, bRIL, and bRIL-Fab), EMD-37336 (OR52cs-focused refined cryo-EM density of apo state OR52cs), EMD-35010, EMDB-35770 (composite (OR52cs-focused and Gs-focused merged) cryo-EM density of OCA–OR52cs–Gs–Nb35 complex), EMD-35772 (OR52cs-focused refined cryo-EM density of OCA–OR52cs–Gs–Nb35 complex), and EMD-35773 (Gs-focused refined cryo-EM density of OCA–OR52cs–Gs–Nb35 complex). The initial and final configurations obtained from 1-µs all-atom MD simulations and extended simulations from Anton2 of all model systems are available at https://github.com/sek24/natcomm2023. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chulwon Choi, Jungnam Bae.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-023-43983-9.
References
- 1.Buck LB. The molecular architecture of odor and pheromone sensing in mammals. Cell. 2000;100:611–618. doi: 10.1016/S0092-8674(00)80698-4. [DOI] [PubMed] [Google Scholar]
- 2.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-X. [DOI] [PubMed] [Google Scholar]
- 3.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/S0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 4.Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–696. doi: 10.1016/S0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez I. Singular expression of olfactory receptor genes. Cell. 2013;155:274–277. doi: 10.1016/j.cell.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 6.DeMaria S, Ngai J. The cell biology of smell. J. Cell Biol. 2010;191:443–452. doi: 10.1083/jcb.201008163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Firestein S. How the olfactory system makes sense of scents. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- 8.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/S0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 9.Simon MI, Strathmann MP, Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 10.Jones DT, Reed RR. Golf: an olfactory neuron specific-G protein involved in odorant signal transduction. Science. 1989;244:790–795. doi: 10.1126/science.2499043. [DOI] [PubMed] [Google Scholar]
- 11.Liu HY, Wenzel-Seifert K, Seifert R. The olfactory G protein G(alphaolf) possesses a lower GDP-affinity and deactivates more rapidly than G(salphashort): consequences for receptor-coupling and adenylyl cyclase activation. J. Neurochem. 2001;78:325–338. doi: 10.1046/j.1471-4159.2001.00422.x. [DOI] [PubMed] [Google Scholar]
- 12.Isberg V, et al. Generic GPCR residue numbers—aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou, Q. et al. Common activation mechanism of class A GPCRs. eLife8, e50279 (2019). [DOI] [PMC free article] [PubMed]
- 14.Hauser AS, et al. GPCR activation mechanisms across classes and macro/microscales. Nat. Struct. Mol. Biol. 2021;28:879–888. doi: 10.1038/s41594-021-00674-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu M, Echeverri F, Moyer BD. Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors. Traffic. 2003;4:416–433. doi: 10.1034/j.1600-0854.2003.00097.x. [DOI] [PubMed] [Google Scholar]
- 16.McClintock TS, et al. Functional expression of olfactory-adrenergic receptor chimeras and intracellular retention of heterologously expressed olfactory receptors. Mol. Brain Res. 1997;48:270–278. doi: 10.1016/S0169-328X(97)00099-5. [DOI] [PubMed] [Google Scholar]
- 17.Shepard BD, Natarajan N, Protzko RJ, Acres OW, Pluznick JL. A cleavable N-terminal signal peptide promotes widespread olfactory receptor surface expression in HEK293T cells. PLoS One. 2013;8:e68758. doi: 10.1371/journal.pone.0068758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Y, et al. Extracellular loop 2 of G protein-coupled olfactory receptors is critical for odorant recognition. J. Biol. Chem. 2022;298:102331. doi: 10.1016/j.jbc.2022.102331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukutani, Y. et al. Hot spot mutagenesis improves the functional expression of unique mammalian odorant receptors. Int. J. Mol. Sci. 23, 277 (2021). [DOI] [PMC free article] [PubMed]
- 20.Sharma, R. et al. Olfactory receptor accessory proteins play crucial roles in receptor function and gene choice. eLife6, e21895 (2017). [DOI] [PMC free article] [PubMed]
- 21.Hague C, et al. Olfactory receptor surface expression is driven by association with the beta2-adrenergic receptor. Proc. Natl. Acad. Sci. USA. 2004;101:13672–13676. doi: 10.1073/pnas.0403854101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 23.Ikegami K, et al. Structural instability and divergence from conserved residues underlie intracellular retention of mammalian odorant receptors. Proc. Natl. Acad. Sci. USA. 2020;117:2957–2967. doi: 10.1073/pnas.1915520117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohl A, et al. Designed to be stable: crystal structure of a consensus ankyrin repeat protein. Proc. Natl. Acad. Sci. USA. 2003;100:1700–1705. doi: 10.1073/pnas.0337680100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozuka K, Nakano S, Asano Y, Ito S. Partial consensus design and enhancement of protein function by secondary-structure-guided consensus mutations. Biochemistry. 2021;60:2309–2319. doi: 10.1021/acs.biochem.1c00309. [DOI] [PubMed] [Google Scholar]
- 26.Lehmann M, Pasamontes L, Lassen SF, Wyss M. The consensus concept for thermostability engineering of proteins. Biochim. Biophys. Acta. 2000;1543:408–415. doi: 10.1016/S0167-4838(00)00238-7. [DOI] [PubMed] [Google Scholar]
- 27.Jimenez RC, et al. The mutational landscape of human olfactory G protein-coupled receptors. BMC Biol. 2021;19:21. doi: 10.1186/s12915-021-00962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatelain, P., Veithen, A. Olfactory receptors involved in the perception of sweat carboxylic acids and the use thereof. Patent PCT/EP2013/061243 (2014).
- 29.Billesbølle CB, et al. Structural basis of odorant recognition by a human odorant receptor. Nature. 2023;615:742–749. doi: 10.1038/s41586-023-05798-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zozulya, S., Echeverri, F., Nguyen, T. The human olfactory receptor repertoire. Genome Biol.2, 1–12 (2001). [DOI] [PMC free article] [PubMed]
- 32.Halperin Kuhns VL, et al. Characterizing novel olfactory receptors expressed in the murine renal cortex. Am. J. Physiol. Renal Physiol. 2019;317:F172–F186. doi: 10.1152/ajprenal.00624.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanz G, Schlegel C, Pernollet JC, Briand L. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem. Senses. 2005;30:69–80. doi: 10.1093/chemse/bji002. [DOI] [PubMed] [Google Scholar]
- 34.Mainland JD, Li YR, Zhou T, Liu WL, Matsunami H. Human olfactory receptor responses to odorants. Sci. Data. 2015;2:150002. doi: 10.1038/sdata.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsutsumi, N. et al. Structure of human Frizzled5 by fiducial-assisted cryo-EM supports a heterodimeric mechanism of canonical Wnt signaling. eLife9, e58464 (2020). [DOI] [PMC free article] [PubMed]
- 36.Kang Y, et al. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature. 2018;558:553–558. doi: 10.1038/s41586-018-0215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin X, et al. Structural basis of ligand recognition and self-activation of orphan GPR52. Nature. 2020;579:152–157. doi: 10.1038/s41586-020-2019-0. [DOI] [PubMed] [Google Scholar]
- 38.Gusach A, et al. Beyond structure: emerging approaches to study GPCR dynamics. Curr. Opin. Struct. Biol. 2020;63:18–25. doi: 10.1016/j.sbi.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Nygaard R, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR Dynamics: Structures in Motion. Chem. Rev. 2017;117:139–155. doi: 10.1021/acs.chemrev.6b00177. [DOI] [PubMed] [Google Scholar]
- 41.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballante F, Kooistra AJ, Kampen S, Graaf CD, Carlsson J. Structure-based virtual screening for ligands of G protein–coupled receptors: what can molecular docking do for you? Pharmacol. Rev. 2021;73:1698–1736. doi: 10.1124/pharmrev.120.000246. [DOI] [PubMed] [Google Scholar]
- 44.Schiefner A, Freier R, Eichinger A, Skerra A. Crystal structure of the human odorant binding protein, OBPIIa. Proteins. 2015;83:1180–1184. doi: 10.1002/prot.24797. [DOI] [PubMed] [Google Scholar]
- 45.Tegoni M, et al. Mammalian odorant binding proteins. Biochim. Biophys. Acta. 2000;1482:229–240. doi: 10.1016/S0167-4838(00)00167-9. [DOI] [PubMed] [Google Scholar]
- 46.Olender T, Nativ N, Lancet D. HORDE: comprehensive resource for olfactory receptor genomics. Methods Mol. Biol. 2013;1003:23–38. doi: 10.1007/978-1-62703-377-0_2. [DOI] [PubMed] [Google Scholar]
- 47.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pardon E, et al. A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch W. xds. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix. refine. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Punjani A, Rubinstein JL, Fleet DJ, Brubaker M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 57.Bepler T, et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods. 2019;16:1153–1160. doi: 10.1038/s41592-019-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Punjani A, Zhang H, Fleet DJ. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods. 2020;17:1214–1221. doi: 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 59.Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife7, e42166 (2018). [DOI] [PMC free article] [PubMed]
- 60.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D: Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pintilie G, et al. Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat. Methods. 2020;17:328–334. doi: 10.1038/s41592-020-0731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim S, et al. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 2017;38:1879–1886. doi: 10.1002/jcc.24829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee J, et al. CHARMM-GUI membrane builder for complex biological membrane simulations with glycolipids and lipoglycans. J. Chem. Theory Comput. 2019;15:775–786. doi: 10.1021/acs.jctc.8b01066. [DOI] [PubMed] [Google Scholar]
- 65.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu EL, et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jo S, et al. CHARMM-GUI PDB manipulator for advanced modeling and simulations of proteins containing nonstandard residues. Adv. Protein Chem. Struct. Biol. 2014;96:235–265. doi: 10.1016/bs.apcsb.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park, S.-J., Kern, N., Brown, T., Lee, J., & Im, W. CHARMM-GUI PDB Manipulator: various PDB structural modifications for biomolecular modeling and simulation. J. Mol. Biol. 435, 67995 (2023). [DOI] [PMC free article] [PubMed]
- 69.Hatcher E, Guvench O, Mackerell AD. CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J. Phys. Chem. B. 2009;113:12466–12476. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang J, et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vanommeslaeghe K, MacKerell AD., Jr. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 73.Durell SR, Brooks BR, Ben-Naim A. Solvent-induced forces between two hydrophilic groups. J. Phys. Chem. 2002;98:2198–2202. doi: 10.1021/j100059a038. [DOI] [Google Scholar]
- 74.Dion M, Rydberg H, Schroder E, Langreth DC, Lundqvist BI. van der Waals density functional for general geometries. Phys. Rev. Lett. 2004;92:246401. doi: 10.1103/PhysRevLett.92.246401. [DOI] [PubMed] [Google Scholar]
- 75.Essmann U, et al. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 76.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 77.Åqvist J, Wennerström P, Nervall M, Bjelic S, Brandsdal BO. Molecular dynamics simulations of water and biomolecules with a Monte Carlo constant pressure algorithm. Chem. Phys. Lett. 2004;384:288–294. doi: 10.1016/j.cplett.2003.12.039. [DOI] [Google Scholar]
- 78.Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao Y, et al. CHARMM-GUI supports hydrogen mass repartitioning and different protonation states of phosphates in lipopolysaccharides. J. Chem. Inf. Model. 2021;61:831–839. doi: 10.1021/acs.jcim.0c01360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hopkins CW, Le Grand S, Walker RC, Roitberg AE. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 81.Eastman P, et al. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comp. Biol. 2017;13:e1005659. doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shaw, D. E. et al. Anton 2: Raising the Bar for Performance and Programmability in a Special-Purpose Molecular Dynamics Supercomputer. In SC ‘14: In Proc. of the International Conference for High Performance Computing, Networking, Storage and Analysis (2014).
- 83.Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/PhysRevA.31.1695. [DOI] [PubMed] [Google Scholar]
- 84.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. doi: 10.1063/1.447334. [DOI] [Google Scholar]
- 85.Park H, Zhou G, Baek M, Baker D, DiMaio F. Force field optimization guided by small molecule crystal lattice data enables consistent sub-angstrom protein-ligand docking. J. Chem. Theory Comput. 2021;17:2000–2010. doi: 10.1021/acs.jctc.0c01184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Case, D. et al. AMBER 2020: University of California. San Francisco, (2020).
- 87.Roe DR, Cheatham TE., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinate and structure factor of GTPγS-bound Gαolf were deposited in the Protein Data Bank under accession number of 8HTG. The coordinates of OR52cs in the apo (with and without bRIL) and in complex with OCA, Gs, and Nb35 were deposited in the Protein Data Bank under accession numbers of 8J46, 8WW7 and 8HTI, respectively. The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession numbers of EMD-35971 (globally refined cryo-EM density containing apo state OR52cs, bRIL, and bRIL-Fab), EMD-37336 (OR52cs-focused refined cryo-EM density of apo state OR52cs), EMD-35010, EMDB-35770 (composite (OR52cs-focused and Gs-focused merged) cryo-EM density of OCA–OR52cs–Gs–Nb35 complex), EMD-35772 (OR52cs-focused refined cryo-EM density of OCA–OR52cs–Gs–Nb35 complex), and EMD-35773 (Gs-focused refined cryo-EM density of OCA–OR52cs–Gs–Nb35 complex). The initial and final configurations obtained from 1-µs all-atom MD simulations and extended simulations from Anton2 of all model systems are available at https://github.com/sek24/natcomm2023. Source data are provided with this paper.