Abstract
Background
Methods to improve stratification of small (≤15 mm) lung nodules are needed. We aimed to develop a radiomics model to assist lung cancer diagnosis.
Methods
Patients were retrospectively identified using health records from January 2007 to December 2018. The external test set was obtained from the national LIBRA study and a prospective Lung Cancer Screening programme. Radiomics features were extracted from multi-region CT segmentations using TexLab2.0. LASSO regression generated the 5-feature small nodule radiomics-predictive-vector (SN-RPV). K-means clustering was used to split patients into risk groups according to SN-RPV. Model performance was compared to 6 thoracic radiologists. SN-RPV and radiologist risk groups were combined to generate “Safety-Net” and “Early Diagnosis” decision-support tools.
Results
In total, 810 patients with 990 nodules were included. The AUC for malignancy prediction was 0.85 (95% CI: 0.82–0.87), 0.78 (95% CI: 0.70–0.85) and 0.78 (95% CI: 0.59–0.92) for the training, test and external test datasets, respectively. The test set accuracy was 73% (95% CI: 65–81%) and resulted in 66.67% improvements in potentially missed [8/12] or delayed [6/9] cancers, compared to the radiologist with performance closest to the mean of six readers.
Conclusions
SN-RPV may provide net-benefit in terms of earlier cancer diagnosis.
Subject terms: Non-small-cell lung cancer, Translational research
Introduction
The optimal management of indeterminate lung nodules is a common clinical problem [1]. While some may represent early lung cancers, the vast majority are benign [2]. Several guidelines have been devised to aid management, which uses size as a key determinant of cancer risk [3–7]. A nodule diameter cut-off of 15 mm has been adopted to dichotomise nodules into “small” or “large” subgroups [8, 9]. For larger nodules, existing guidelines can correctly identify lung cancer in 34–50% of patients referred for further investigation in screening studies [7, 10]. However, predicting malignancy in small nodules is problematic, as approximately 15% of participants will be recalled for early repeat CT in lung cancer screening, and the overwhelming majority will turn out to be benign [11]. At the same time, some patients will experience delayed diagnosis of lung cancer and stage progression due to the growth of nodules initially regarded as benign or indeterminate while undergoing surveillance [12]. There is, therefore, a need to better stratify small lung nodules, where clinical management dilemmas are frequent.
Subsequently, a number of automated machine-learning algorithms have been developed to improve lung nodule classification [13–17], though few studies have solely examined small nodules. Given that the malignancy risk and management may differ for this group and emerging concerns about AI model ‘hidden stratification’, we propose that lung nodule predictive models may be improved by training on specific size groups [9, 18]. Furthermore, few studies explain how automated algorithms will be applied in clinical practice. Therefore, we aimed to develop a radiomics-based nodule classification algorithm using a number of novel approaches: (1) we developed the algorithm using small lung nodules (≤15 mm) only. (2) We compared model performance to expert thoracic radiologists and developed a decision-support tool to be used in combination with radiologist interpretation to aid early cancer detection. (3) We extracted radiomics features from the peri-nodule lung parenchyma based on evidence that tumour immune cells are also found in this region [19]. 4) We validated the algorithm in an external test set, including prospective lung cancer screening CTs.
Materials and methods
Patients
Health Regulatory Authority (HRA) and Research Ethics Committee (REC) approvals were obtained for this retrospective observational study (18/HRA/0434). Informed consent was not required. Patients were identified using (1) institutional databases of lung pathology reports between January 2007 and December 2018 and (2) records of lung multidisciplinary team meetings held between January 2015 and December 2018. All data were link-anonymised.
Inclusion criteria:
5–15 mm solid lung nodules on CT imaging
Biopsy-confirmed ground-truth, OR:
Radiologically confirmed benignity based on shrinkage on serial CT or volumetric stability at 1 year4
Exclusion criteria:
Inability to confirm the location of the lung nodule corresponding to the pathology report
CT slice thickness >3 mm
Calcified, cavitating, subsolid, mediastinal, endotracheal, or endobronchial nodules.
Cancer within the last 5 years or lung metastases.
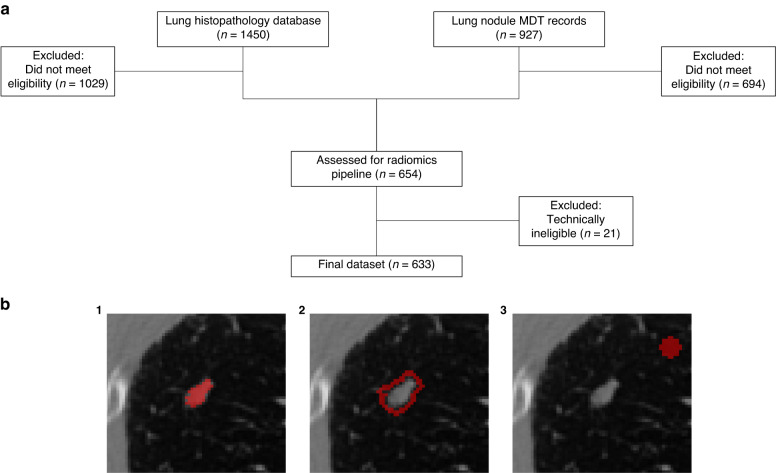
Study recruitment is shown in Fig. 1a. A maximum of five nodules per scan were included, and the final internal dataset contained 736 solid nodules from 633 patients. Internal data were divided randomly into training (609 nodules) and test sets (127 nodules) using the sample.split function in R, grouped by patient ID to avoid data leakage. An external test set of 254 nodules was derived from a lung cancer screening programme (n = 84) and the multi-centre LIBRA study (n = 170) [20, 21]. Clinicodemographic data are shown in Table 1.
Fig. 1. Recruitment diagram and segmentation labels.
a Study recruitment diagram. Databases of lung histopathology (n = 1450) and lung nodule MDT records (n = 927) were used to identify eligible patients. Following exclusion based on eligibility criteria (n = 1723) and technical limitations of CT images (n = 21), the final internal dataset consisted of 633 patients with 736 nodules. b Cropped, axial plane CT images showing binary segmentation masks (red) for nodule regions. The primary lung nodule was segmented (1) and then expanded by 2 mm isotropically to create a spherical annulus structure (2). An 8 × 8 mm spherical background structure (3) was segmented 15 mm away from the primary lesion.
Table 1.
Patient clinicodemographic information.
| Characteristic | Training | Test | External test |
|---|---|---|---|
| Age, mean (SD) | 64.62 (11.90) | 66.43 (9.49) | 68.72 (8.64) |
| Gender, No. (%) | |||
| Male | 285 (46.80) | 50 (39.37) | 111 (43.70) |
| Female | 324 (53.20) | 77 (60.63) | 143 (56.30) |
| Smoking, No. (%) | |||
| Y | 345 (58.87) | 84 (66.14) | 197 (77.56) |
| N | 241 (41.13) | 43 (33.86) | 57 (22.44) |
| Nodule diameter, Mean (SD), mm | 10.98 (3.30) | 11.64 (2.94) | 9.49 (2.49) |
| CT with contrast, No. (%) | |||
| Y | 348 (57.14) | 60 (47.24) | 92 (36.22) |
| N | 261 (42.86) | 67 (52.75) | 162 (63.78) |
| Malignancy, No. (%) | |||
| Y | 305 (50.08) | 72 (56.69) | 63 (24.80) |
| N | 304 (49.92) | 55 (43.31) | 191 (75.20) |
Characteristics are displayed at the nodule level. SD standard deviation. Smoking data were missing for 23 nodules.
Radiological visual nodule interpretation
The internal test set was also evaluated independently by six thoracic radiologists (HR, JB, CR, GR, MS and SD). One reader was a resident with 3 years of thoracic experience, and five were consultant thoracic radiologists with between 5- and 23-years experience (mean 11.2, SD 7.46). Readers provided a visual assessment of malignancy risk using an ordinal scale: 1–2: definitely or probably benign; 3: indeterminate, 4–5: probably or highly likely to be malignant. Inter-reader agreement was calculated using Krippendorf’s-Alpha metric [22].
CT image segmentation and radiomics feature extraction
Nodules were segmented in ITK-Snap by CA (http://www.itksnap.org/) and reviewed by a thoracic radiologist (AD). The following segmentation masks were generated: (1) the lung nodule; (2) a 2 mm isotropic dilation around the nodule, termed the annulus structure [23, 24]; (3) an 8 mm isotropic sphere 15 mm away from the nodule, termed the background structure (Fig. 1b).
Resampling to isotropic voxel parameters is recommended by the Imaging Biomarker Standardisation Initiative to allow comparison across scan cohorts. Images and segmentations were resampled to isotropic voxel dimensions of 1 × 1 × 1, which are commonly used by other groups and models [25]. Cubic spline and nearest neighbour interpolation were used for scan or mask resampling, respectively. Although there is no clear consensus on the superiority of one interpolation method over another, higher-order methods such as cubic spline are generally preferable for scan images to avoid undesirable image smoothing [26].
Features were extracted using TexLab 2.0 (MATLAB 2015b) as described previously [27, 28].
Statistical analysis
Statistical analyses were performed in R Studio (v2021-09-20). All tests were two-sided, with statistical significance defined as p < 0.05. Corrections for multiple comparisons were performed using the Benjamini–Hochberg/false-discovery rate (FDR) method. Due to the exploratory nature of this work, a formal power calculation was not performed. Comparisons of model AUCs were performed using the DeLong test, which is non-parametric and makes no assumptions of underlying data distribution.
Previously described feature-reduction measures were implemented [27] (Supplementary Fig. 1). 1998 radiomic features were scaled using Z-standardisation . Univariable logistic regression was performed to select those with a p value < 0.001 after FDR correction (469 features). A LASSO model was fit using ten-fold cross-validation to select the optimal lambda (lambda.1se), and the weighted sum of the 5 selected features gave the small nodule radiomics predictive vector (SN-RPV). One annulus feature and four lesion-derived features were selected, with no background-derived features selected for the final model (Supplementary Fig. 1c and Supplementary Table 1). K-means clustering was used to group training and test set patients into low, intermediate or high-risk groups according to the SN-RPV (Supplementary Fig. 1d).
Receiver operating characteristic (ROC) curves were constructed to assess SN-RPV and radiologist performance in predicting malignancy as determined by the AUC. 95% confidence intervals were obtained by resampling with 1000 bootstrap iterations. Threshold-based performance metrics were reported using the ROC cutoff, which maximised the training-set Youden index (SN-RPV = −0.1887086). The Youden index is considered a balanced metric which gives equal weight to both sensitivity and specificity. Test-set radiologist predictions were converted to a binary classification of malignant using a threshold of ≥4 (probably malignant).
Integration of radiomics with radiologist interpretation
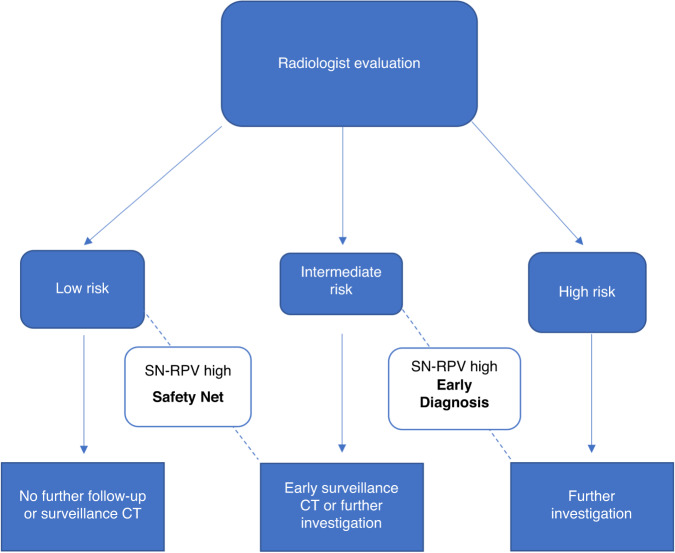
We developed a decision-support algorithm to assess the impact of integrating the radiomics model (SN-RPV) with radiologist interpretations (Fig. 2). We calculated the modelled impact of the decision-support algorithm on rates of potentially missed cancers and delayed cancer diagnoses. Potentially missed cancers were defined as proven malignant nodules categorised as low risk (risk score 1–2) by radiologists. Delayed diagnosis cancers were defined as proven malignant nodules categorised as intermediate risk (risk score 3) by radiologists.
Fig. 2. Decision-support tool scenarios.
In cases where the radiologist categorises a nodule as low-risk, a high-risk SN-RPV triggers the “Safety Net”, prompting earlier surveillance or investigation. In cases where the radiologist classifies a nodule as indeterminate, a high-risk SN-RPV triggers the “Early Diagnosis” pathway, prompting further investigation rather than surveillance. Cases evaluated as high-risk by the radiologist are not affected by the proposed decision support method.
To give the most representative comparator to clinical practice, calculations were performed using risk scores for the radiologist with performance closest to the mean of the six readers.
Results
Patient and nodule characteristics
Patient, nodule and scan characteristics in the training and test sets are shown in Table 1. Of the internal dataset, 49% of patients had benign and 51% had malignant nodules. The overall mean nodule diameter was 11.14 (3.22) mm. The internal data partitions were well matched, though in the test set, the proportion of malignant nodules was higher (57% vs. 50%), and the proportion of contrast-enhanced CT scans was lower (47% vs. 57%). The external test data included lower proportions of malignant nodules (25%) and contrast-enhanced scans (36%).
SN-RPV and radiologist performance
The SN-RPV AUCs for malignancy prediction were 0.85 (95% CI: 0.82–0.88), 0.78 (95% CI: 0.70–0.86) and 0.78 (95% CI: 0.58–0.92) in the training, internal and external test sets respectively (Table 2). In the internal test set, the model performance metrics were as follows: accuracy 73% (95% CI: 65–81), sensitivity 0.86, specificity 0.56, PPV 0.72 and NPV 0.76. In the external test set, the model performance metrics were as follows: accuracy 75% (95% CI: 69–80), sensitivity 0.63, specificity 0.79, PPV 0.50, NPV 0.87.
Table 2.
Performance metrics for the small-nodule radiomics predictive vector, diameter and volume models.
| AUC | Accuracy | Sens | Spec | PPV | NPV | |
|---|---|---|---|---|---|---|
| Training | ||||||
| SN-RPV | 0.85 (0.82–0.88) | 78% (74–81%) | 0.86 | 0.69 | 0.74 | 0.83 |
| Diameter | 0.79 (0.75–0.82) | 74% (70–77%) | 0.75 | 0.73 | 0.71 | 0.76 |
| Volume | 0.78 (0.75–0.82) | 73% (69–77%) | 0.80 | 0.66 | 0.70 | 0.77 |
| Test | ||||||
| SN-RPV | 0.78 (0.70–0.86) | 73% (65–81%) | 0.86 | 0.56 | 0.72 | 0.76 |
| Diameter | 0.69 (0.60–0.78) | 64% (56–72%) | 0.65 | 0.64 | 0.73 | 0.55 |
| Volume | 0.77 (0.68–0.85) | 77% (69–84%) | 0.83 | 0.69 | 0.78 | 0.76 |
| External | ||||||
| SN-RPV | 0.78 (0.58–0.92) | 75% (69–80%) | 0.63 | 0.79 | 0.50 | 0.87 |
| Diameter | 0.68 (0.61–0.76) | 70% (64–75%) | 0.38 | 0.80 | 0.39 | 0.80 |
| Volume | 0.80 (0.73–0.86) | 69% (62–74%) | 0.83 | 0.64 | 0.43 | 0.92 |
95% confidence intervals are displayed in brackets. SN-RPV radiomics predictive vector, AUC area under the curve, PPV positive predictive value, NPV negative predictive value.
The AUC values of the six radiologists ranged from AUC 0.68–0.81 (Table 3). There was moderate inter-reader variability amongst radiologist classifications (Krippendorf Alpha: 0.58, Supplementary Fig. 2c). There was no statistically significant difference between the SN-RPV and the highest-performing radiologist (DeLong’s p = 0.56).
Table 3.
Performance metrics for radiologists in the test set.
| Metric | R1 | R2 | R3 | R4 | R5 | R6 |
|---|---|---|---|---|---|---|
| Accuracy | 65% (56–73%) | 69% (60–76%) | 61% (52–70%) | 74% (65–81%) | 71% (62–79%) | 64% (55–72%) |
| AUC | 0.74 (0.65–0.72) | 0.75 (0.67–0.83) | 0.68 (0.58–0.76) | 0.79 (0.71–0.86) | 0.81 (0.73–0.88) | 0.73 (0.65–0.81) |
| Sensitivity | 0.61 | 0.71 | 0.51 | 0.76 | 0.68 | 0.53 |
| Specificity | 0.69 | 0.65 | 0.75 | 0.71 | 0.75 | 0.78 |
| PPV | 0.72 | 0.73 | 0.73 | 0.77 | 0.78 | 0.76 |
| NPV | 0.58 | 0.63 | 0.54 | 0.70 | 0.64 | 0.56 |
Numerators and denominators are shown in parentheses. 95% CIs are shown in brackets. R Reader, AUC area under the curve, PPV positive predictive value, NPV negative predictive value.
Clinical benchmarking
In the external test set, the SN-RPV performance was statistically significant when compared to transaxial diameter (AUC 0.78 vs. AUC 0.68, DeLong’s p = 0.02) but not 3D volume (AUC 0.78 vs. AUC 0.80, DeLong’s p = 0.24). The relative performance of the SN-RPV and models based on diameter or 3D volume are compared in Table 2. In multivariable analysis, volume (p = 0.03), upper lobe location (0.003) and non-smoking status (0.008) were statistically significant predictors of malignancy (Table 4). The SN-RPV was non-significant (p = 0.06).
Table 4.
Multivariable logistic regression including clinical features.
| Variable | Beta | OR | P value |
|---|---|---|---|
| Age | 0.02 | 1.02 (0.0002–1.38) | 0.45 |
| Diameter | 0.11 | 1.12 (0.88–1.42) | 0.35 |
| Brock | −0.008 | 0.99 (0.92–1.07) | 0.83 |
| Volume | 0.0007 | 1.00 (1.00–1.001) | 0.03* |
| RPV | 0.64 | 1.90 (0.99–3.77) | 0.06 |
| UL | 1.35 | 3.87 (1.62–9.71) | 0.003* |
| Male sex | −0.60 | 0.55 (0.26–1.13) | 0.12 |
| Non-smoker | −1.32 | 0.27 (0.09–0.67) | 0.008* |
| Previous cancer | 0.42 | 1.52 (0.67–3.39) | 0.31 |
| Emphysema | −0.60 | 0.55 (0.23–1.23) | 0.16 |
UL upper lobe. *p < 0.05.
The RPV was moderately correlated with 3D volume (feature name SNS_vol, r = 0.68), as were the individual features Annulus_GLCM_Entrop_LLL (r = 0.66) and Lesion_GLCM_InfCo2_LHL (r = 0.66) in Pearson’s correlation.
We performed a post-hoc analysis to investigate whether the addition of volume to our predictive models would improve performance. A concatenated feature vector including the 5 SN-RPV features plus SNS_vol (3D volume) was used as input to an XGBoost decision-tree classifier. In the test set, the combined classifier improved the AUC by 0.01 to 0.79 (95% CI: 0.71–0.86) but did not improve accuracy (73% [95% CI: 64–80%]).
To assess the impact of IV contrast on performance, we developed and validated separate models using contrast and non-contrast-only scans with a 70% training/test split. For the contrast-only model, the training (n = 285), test (n = 123) and external-test (n = 92) set AUCs were 0.86 (95% CI: 0.81–0.91), 0.76 (95% CI: 0.64–0.87) and 0.69 (95% CI: 0.57–0.80), respectively. For the non-contrast-only model, the training (n = 230), test (n = 98), and external-test (n = 162) set AUCs were 0.84 (95% CI: 0.79–0.88), 0.83 (95% CI: 0.74–0.89) and 0.76 (95% CI: 0.66–0.85).
Performance of decision support tools: integration of SN-RPV and radiologist interpretation
Results of the K-means clustering analysis to group training and test set RPVs into low, intermediate and high-risk groups are shown in Supplementary Fig. 1d. The radiologist decision-support scenarios are shown in Fig. 2. The impact of the ‘Safety Net’ and ‘Early Diagnosis’ tools for the radiologist with the mean score (R2) in the test cohort is shown in Supplementary Table 2.
The radiologist categorised 12/72 (16.67%) lung cancers as benign or probably benign. Eight of the 12 cancers (66.67%) would have undergone closer surveillance CT or earlier investigation using the modelled “Safety Net” decision support algorithm combining radiologist evaluation and SN-RPV (Fig. 2).
Ten out of 55 (18.18%) benign nodules were classified as indeterminate by the radiologist and would have undergone surveillance CT based on nodule management algorithms. The safety net decision support algorithm would have led to surveillance CT for an additional 2/55 (3.6%) benign nodules that were classified as low risk by the radiologist.
Nine lung cancers from 72 malignant nodules (12.5%) were categorised as indeterminate by the radiologist (Supplementary Table 2) and would have ordinarily undergone surveillance CT. Use of the “Early Diagnosis” decision-support algorithm would have identified 6/9 lung cancers (66.67%) as high risk, providing the opportunity for earlier investigation. The radiologist categorised 19/55 (34.5%) benign nodules as probably or highly likely to be malignant, leading to potentially unnecessary further investigation based on the nodule management algorithm. Of the 55 benign nodules, four (7%) were categorised as indeterminate by the radiologist but high risk by the SN-RPV and would have undergone additional potentially unnecessary investigation using the decision support tool.
To explore how the SN-RPV could integrate with volume in clinical practice, we dichotomised test-set nodules into low or high-volume groups using a 300 mm3 threshold to match the BTS guidelines [4]. Of the 36 nodules with a volume < 300 mm3, there were 9 cancers (25%). 8 out of 9 (89%) cancers had an intermediate radiomics risk score and would have been upgraded from CT surveillance (according to BTS guidelines) to further investigation (after integration with SN-RPV risk) group, with the limitation that 10 of the 28 (36%) benign nodules would also be upgraded.
Discussion
Small pulmonary nodules present a challenge for clinicians. The SN-RPV, developed in 736 ≤ 15 mm lung nodules, identified malignant nodules with a test-set AUC of 0.78 (95% CI: 0.59–0.92) and had comparable performance to the panel of radiologists (mean radiologist AUC 0.75 [95% CI: 0.67–0.83]). For cases evaluated as low or intermediate risk by the radiologist, the “Safety Net” and “Early Diagnosis” decision support scenarios would have led to a modelled reduction in missed and delayed cancers of 66.67% [8/12 and 6/9], respectively. The model was validated in an external test set, including prospective screening patients, with an AUC of 0.78 (95% CI: 0.71–0.83).
SN-RPV was derived from the lesion and surrounding lung parenchyma, which may capture important biological processes, such as peri-tumoural stromal reactions and immune-cell infiltration, and this region is a key component of lung cancer radiotherapy planning [19, 29–31]. Moreover, the model was trained on heterogeneous scans, which may improve applicability over those developed using only screening trial data.
Recently several machine-learning studies for nodule classification have been reported, with AUCs ranging from 0.72 to 0.92 [16, 17, 32, 33]. Most existing studies include a range of nodule sizes, but our tool is focused on small lung nodules, which form the bulk of management uncertainty. For example, among nodules categorised with risk scores between 2 and 4 by radiologists, there was a substantial spread of both benign and malignant nodules for all readers (Supplementary Fig. 2c). Although the SN-RPV performed better than transaxial diameter, it was not superior to volume alone. In multivariable analysis, the SN-RPV had a high weight (0.64) but was statistically insignificant (p 0.06), though close to significance. It appears that by dichotomising nodules in this fashion, we have discovered that volume is a strong predictor of malignancy in small nodules that we were not able to surpass using textural analysis. Interestingly, volume was not a stronger predictor than radiomics models in our recent study of ≥15 mm nodules [21]. Two of the SN-RPV features were moderately correlated with volume.
Few prior studies have demonstrated how automated tools might be incorporated into clinical practice, but we have demonstrated potential net benefits from clinical decision integration [34].
The 5 features retained by the LASSO method relate to heterogeneity, as assessed by annulus or tumour-derived grey-level co-occurrence matrices (GLCM). These features provide a spatial representation of homogeneity/heterogeneity of pairs of image voxels with predefined grey-level intensities in orthogonal directions. Within the tumour adjacent annulus, GLCM entropy, the degree of randomness, was a predictive feature. Taken together, the SN-RPV may reflect various biological features known to contribute to intra-tumoural heterogeneity, such as genomic instability, hypoxia, stromal reaction and immune infiltration [19, 35, 36].
There are some other limitations to consider. Firstly, the model requires image pre-processing and manual segmentation steps, which would require automation prior to clinical implementation. Secondly, the model has been evaluated in a retrospective setting only, and additional prospective validation is needed. Thirdly, subsolid nodules and ground glass opacities were excluded and would require separate models for stratification. There is also some debate within the radiomics community regarding the best approach to developing models from contrast and non-contrast enhanced images. While it is recognised that contrast administration may affect radiomics feature reproducibility and model performance, there are limited test-re-test data available to accurately assess the impact of such effects. One study of 104 patients undergoing CT/PET scans found that around 55% of radiomics features had moderate to good (ICC ≥ 0.75) reproducibility between contrast and non-contrast protocols, but this may not be directly applicable to other studies [37]. We argue that a model trained on both contrast and non-contrast images could be more applicable to ‘real world’ data, with an advantage over models trained purely on homogeneous, non-contrast nodule surveillance scans. We have taken this approach in other published models which attained high performance [21]. The utility of training on mixed contrast/non-contrast data is supported by a study of 412 patients by Yang et al. [38]. In this work, models for predicting EGFR mutation status in lung cancer patients performed better in non-contrast or contrast-only test sets when trained on mixed data than if trained purely on one group [38]. To test that the use of mixed contrast models has not unduly affected the SN-RPV performance, we trained and validated separate models on non-contrast and contrast-only cohorts. The discrimination of these models, as assessed by the AUC, was not superior to the SN-RPV.
In summary, we have developed a tool to predict lung cancer in small lung nodules, which has been validated in a large external test set. The SN-RPV is as good as radiologists’ overall assessments and can reduce missed cancers. We found that volume is a very powerful predictor for small nodules. When using the clinically applicable threshold of <300 mm3, the SN-RPV model was able to find intermediate-risk patients in this group and could potentially provide a net benefit in combination with volume. Prospective evaluation of this model, particularly in comparison to existing volume-based assessments, is warranted before clinical use.
Supplementary information
Author contributions
BH: Data collection, analysis and interpretation, paper preparation and editing. CA: Data collection, analysis and interpretation, manuscript preparation and editing. MI: Data analysis and interpretation. KL-R: Data analysis. IP, AN, SK, PLS, PLM, CM, TB, EG, JH, AC, SJ, MM and SP: Data collection. HR, JB, CAR, GR, MS and SD: Radiology reads. EOA, RWL and AD: Study design, project supervision, paper feedback and editing.
Funding
This manuscript represents independent research funded by: (1) the Royal Marsden Partners Cancer Alliance, (2) the Royal Marsden Cancer Charity, (3) the National Institute for Health Research (NIHR) Biomedical Research Centre at the Royal Marsden NHS Foundation Trust and The Institute of Cancer Research, London, (4) the National Institute for Health Research (NIHR) Biomedical Research Centre at Imperial College, London, (5) Cancer Research UK (C309/A31316). (6) The European Regional Development Fund and Higher Education Funding Council for England.
Data availability
The radiomics data generated in this study are deposited into the Mendeley database under the accession code 10.17632/rxn95mp24d.1. The R scripts for model development are provided in notebook format at: https://github.com/dr-benjamin-hunter/Small-nodule-radiomics.
Competing interests
Professor Devaraj reports personal fees from Brainomix, Roche, and Boehringer Ingelheim and has stock options in Brainomix. Dr Lee is funded by the Royal Marsden NIHR BRC, Royal Marsden Cancer Charity and SBRI (including QURE.AI). RL’s institution receives compensation for time spent in a secondment role for the lung health check programme and as a National Specialty Lead for the National Institute of Health and Care Research. He has received research funding from CRUK, Innovate UK (co-funded by GE Healthcare, Roche and Optellum), SBRI, RM Partners Cancer Alliance and NIHR (co-applicant in grants with Optellum). He has received honoraria from CRUK. The remaining authors declare no competing interests.
Ethics Approval and consent to participate
Health Regulatory Authority (HRA) and Research Ethics Committee (REC) approvals were obtained for the presented study (18/HRA/0434). Informed consent was not required. The study was performed in accordance with the Declaration of Helsinki.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Benjamin Hunter, Christos Argyros.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02480-y.
References
- 1.Gould MK, Tang T, Liu ILA, Lee J, Zheng C, Danforth KN, et al. Recent trends in the identification of incidental pulmonary nodules. Am J Respir Crit Care Med. 2015;192:1208–14. doi: 10.1164/rccm.201505-0990OC. [DOI] [PubMed] [Google Scholar]
- 2.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larici AR, Farchione A, Franchi P, Ciliberto M, Cicchetti G, Calandriello L, et al. Lung nodules: size still matters. Eur Respir Rev. 2017;26:170025. doi: 10.1183/16000617.0025-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin DR, Callister MEJ. The British Thoracic Society guidelines on the investigation and management of pulmonary nodules. Thorax. 2015;70:794–8. doi: 10.1136/thoraxjnl-2015-207221. [DOI] [PubMed] [Google Scholar]
- 5.Lam S, Bryant H, Donahoe L, Domingo A, Earle C, Finley C, et al. Management of screen-detected lung nodules: a Canadian partnership against cancer guidance document. Can J Respir Crit Care Sleep Med. 2020;4:236–65. [Google Scholar]
- 6.Gould MK, Donington J, Lynch WR, Mazzone PJ, Midthun DE, Naidich DP, et al. Evaluation of individuals with pulmonary nodules: when is it lung cancer? Diagnosis and management of lung cancer, 3rd ed: American college of chest physicians evidence-based clinical practice guidelines. Chest. 2013;143:e93S. doi: 10.1378/chest.12-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horeweg N, van Rosmalen J, Heuvelmans MA, van der Aalst CM, Vliegenthart R, Scholten ET, et al. Lung cancer probability in patients with CT-detected pulmonary nodules: a prespecified analysis of data from the NELSON trial of low-dose CT screening. Lancet Oncol. 2014;15:1332–41. doi: 10.1016/S1470-2045(14)70389-4. [DOI] [PubMed] [Google Scholar]
- 8.Lung Rads | American College of Radiology. https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/Lung-Rads.
- 9.Zhang R, Tian P, Chen B, Zhou Y, Li W. Predicting lung cancer risk of incidental solid and subsolid pulmonary nodules in different sizes. Cancer Manag Res. 2020;12:8057–66. doi: 10.2147/CMAR.S256719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field JK, Duffy SW, Baldwin DR, Whynes DK, Devaraj A, Brain KE, et al. UK Lung Cancer RCT Pilot Screening Trial: Baseline findings from the screening arm provide evidence for the potential implementation of lung cancer screening. Thorax. 2016;71:161–70. doi: 10.1136/thoraxjnl-2015-207140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crosbie PA, Balata H, Evison M, Atack M, Bayliss-Brideaux V, Colligan D, et al. Implementing lung cancer screening: baseline results from a community-based ‘Lung Health Check’ pilot in deprived areas of Manchester. Thorax. 2019;74:405–9. doi: 10.1136/thoraxjnl-2017-211377. [DOI] [PubMed] [Google Scholar]
- 12.Mascalchi M, Picozzi G, Falchini M, Vella A, Diciotti S, Carrozzi L, et al. Initial LDCT appearance of incident lung cancers in the ITALUNG trial. Eur J Radiol. 2014;83:2080–6. doi: 10.1016/j.ejrad.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Kang G, Liu K, Hou B, Zhang N. 3D multi-view convolutional neural networks for lung nodule classification. PLoS ONE. 2017;12:e0188290. doi: 10.1371/journal.pone.0188290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyu, J & Ling, SH. Using multi-level convolutional neural network for classification of lung nodules on CT images. in Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS vols 2018-July 686-9 (Institute of Electrical and Electronics Engineers Inc., 2018). [DOI] [PubMed]
- 15.Shaffie A, Soliman A, Fraiwan L, Ghazal M, Taher F, Dunlap N, et al. A generalized deep learning-based diagnostic system for early diagnosis of various types of pulmonary nodules. Technol Cancer Res Treat. 2018;17:1533033818798800. doi: 10.1177/1533033818798800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ardila, D, Kiraly, AP, Bharadwaj, S, Choi, B, Reicher, JJ, Peng, L, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med.10.1038/s41591-019-0447-x (2019). [DOI] [PubMed]
- 17.Massion PP, Antic S, Ather S, Arteta C, Brabec J, Chen H, et al. Assessing the accuracy of a deep learning method to risk stratify indeterminate pulmonary nodules. Am J Respir Crit Care Med. 2020;202:241–9. doi: 10.1164/rccm.201903-0505OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seah J, Tang C, Buchlak QD, Milne MR, Holt X, Ahmad H, et al. Do comprehensive deep learning algorithms suffer from hidden stratification? A retrospective study on pneumothorax detection in chest radiography. BMJ Open. 2021;11:e053024. doi: 10.1136/bmjopen-2021-053024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuben A, Zhang J, Chiou SH, Gittelman RM, Li J, Lee WC, et al. Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat Commun. 2020;11:1–13. doi: 10.1038/s41467-019-14273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartlett EC, Kemp SV, Ridge CA, Desai SR, Mirsadraee S, Morjaria JB, et al. Baseline results of the West London lung cancer screening pilot study—impact of mobile scanners and dual risk model utilisation. Lung Cancer. 2020;148:12–19. doi: 10.1016/j.lungcan.2020.07.027. [DOI] [PubMed] [Google Scholar]
- 21.Hunter B, Chen M, Ratnakumar P, Alemu E, Logan A, Linton-Reid K, et al. A radiomics-based decision support tool improves lung cancer diagnosis in combination with the Herder score in large lung nodules. EBioMedicine. 2022;86:104344. doi: 10.1016/j.ebiom.2022.104344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes AF, Krippendorff K. Answering the call for a standard reliability measure for coding data. Commun Methods Meas. 2007;1:77–89. doi: 10.1080/19312450709336664. [DOI] [Google Scholar]
- 23.Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–91. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 24.Beig N, Khorrami M, Alilou M, Prasanna P, Braman N, Orooji M, et al. Perinodular and intranodular radiomic features on lung CT images distinguish adenocarcinomas from granulomas. Radiology. 2019;290:783–92. doi: 10.1148/radiol.2018180910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Compter I, Verduin M, Shi Z, Woodruff HC, Smeenk RJ, Rozema T, et al. Deciphering the glioblastoma phenotype by computed tomography radiomics. Radiother Oncol. 2021;160:132–9. doi: 10.1016/j.radonc.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Hatt M, Krizsan AK, Rahmim A, Bradshaw TJ, Costa PF, Forgacs A, et al. Joint EANM/SNMMI guideline on radiomics in nuclear medicine. Eur J Nucl Med Mol Imaging. 2022;50:352–75. doi: 10.1007/s00259-022-06001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arshad MA, Thornton A, Lu H, Tam H, Wallitt K, Rodgers N, et al. Discovery of pre-therapy 2-deoxy-2- 18 F-fluoro-D-glucose positron emission tomography-based radiomics classifiers of survival outcome in non-small-cell lung cancer patients. Eur J Nucl Med Mol Imaging. 2019;46:455–66. doi: 10.1007/s00259-018-4139-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu H, Arshad M, Thornton A, Avesani G, Cunnea P, Curry E, et al. A mathematical-descriptor of tumor-mesoscopic-structure from computed-tomography images annotates prognostic- and molecular-phenotypes of epithelial ovarian cancer. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-019-08718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bremnes RM, Dønnem T, Al-Saad S, Al-Shibli K, Andersen S, Sirera R, et al. The role of tumor stroma in cancer progression and prognosis: Emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J Thorac Oncol. 2011;6:209–17. doi: 10.1097/JTO.0b013e3181f8a1bd. [DOI] [PubMed] [Google Scholar]
- 30.Whittaker Brown SA, Padilla M, Mhango G, Powell C, Salvatore M, Henschke C, et al. Interstitial lung abnormalities and lung cancer risk in the national lung screening trial. Chest. 2019;156:1195–203. doi: 10.1016/j.chest.2019.06.041. [DOI] [PubMed] [Google Scholar]
- 31.Radiotherapy for lung cancer RCR consensus statements.
- 32.Baldwin DR, Gustafson J, Pickup L, Arteta C, Novotny P, Declerck J, et al. External validation of a convolutional neural network artificial intelligence tool to predict malignancy in pulmonary nodules. Thorax. 2020;75:306–12. doi: 10.1136/thoraxjnl-2019-214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binczyk, F, Prazuch, W, Bozek, P & Polanska, J. Radiomics and artificial intelligence in lung cancer screening. Transl. Lung Cancer Res.10.21037/tlcr-20-708 (2021). [DOI] [PMC free article] [PubMed]
- 34.Lv W, Wang Y, Zhou C, Yuan M, Pang M, Fang X, et al. Development and validation of a clinically applicable deep learning strategy (HONORS) for pulmonary nodule classification at CT: a retrospective multicentre study. Lung Cancer. 2021;155:78–86. doi: 10.1016/j.lungcan.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Ramón y Cajal S, Sesé M, Capdevila C, Aasen T, De Mattos-Arruda L, Diaz-Cano SJ, et al. Clinical implications of intratumor heterogeneity: challenges and opportunities. J Mol Med (Berl) 2020;98:161. doi: 10.1007/s00109-020-01874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanduleanu S, Jochems A, Upadhaya T, Even AJG, Leijenaar RTH, Dankers FJWM, et al. Non-invasive imaging prediction of tumor hypoxia: A novel developed and externally validated CT and FDG-PET-based radiomic signatures. Radiother Oncol. 2020;153:97–105. doi: 10.1016/j.radonc.2020.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Jha AK, Mithun S, Jaiswar V, Sherkhane UB, Purandare NC, Prabhash K, et al. Repeatability and reproducibility study of radiomic features on a phantom and human cohort. Sci Rep. 2021;11:1–12. doi: 10.1038/s41598-021-81526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X, Liu M, Ren Y, Chen H, Yu P, Wang S, et al. Using contrast-enhanced CT and non-contrast-enhanced CT to predict EGFR mutation status in NSCLC patients—a radiomics nomogram analysis. Eur Radiol. 2022;32:2693. doi: 10.1007/s00330-021-08366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The radiomics data generated in this study are deposited into the Mendeley database under the accession code 10.17632/rxn95mp24d.1. The R scripts for model development are provided in notebook format at: https://github.com/dr-benjamin-hunter/Small-nodule-radiomics.