Abstract
Fatty acids (FAs) are major components of membranes and contribute to cellular energetic demands. Besides, FAs are precursors of signaling molecules, including oxylipins and other oxidized fatty acids derived from the activity of lipoxygenases. In addition, non-canonical modified fatty acids, such as nitro-fatty acids (NO2-FAs), are formed in animals and plants. The synthesis NO2-FAs involves a nitration reaction between unsaturated fatty acids and reactive nitrogen species (RNS). This review will focus on recent findings showing that, in plants, NO2-FAs such as nitro-linolenic acid (NO2-Ln) and nitro-oleic acid (NO2-OA) have relevant physiological roles as signaling molecules in biotic and abiotic stress, growth, and development. Moreover, since there is controversy on mechanisms of action of NO2-FAs as signaling molecules, we will provide evidence showing why this aspect needs further evaluation.
Keywords: Nitro-fatty acids, Reactive nitrogen species, Nitric oxide
Main conclusion
Nitro fatty acids (NO2-FA)have relevant physiological roles as signaling molecules in biotic and abiotic stress, growth, and development, but the mechanism of action remains controversial. The two main mechanismsinvolving nitric oxide release and thiol modification are discussed.
Introduction
Nitro-fatty acids (NO2-FAs) are the product of the reaction between nitric oxide (NO)− and nitrite (NO2−)-derived reactive nitrogen species (RNS) and unsaturated fatty acids. The formation of nitro-lipids was initially proposed based on the observation that NO inhibited lipid oxidation propagation reactions (Rubbo et al. 1994). The resulting species remained elusive, but the incorporation of a NO2 group into linoleic acid was reported (O’Donnell et al. 1999). Nitrated linoleic acid was then reported in humans (Baker et al. 2004; Lima et al. 2002). NO2-FAs were later detected in plants (Fazzari et al. 2014) and proposed as signal molecules involved in oxidative stress responses, plant development, and defense responses (Mata-Perez et al. 2016a; Arruebarrena Di Palma et al. 2020; Vollár et al. 2020; Di Fino et al. 2020).
Mechanisms of NO2-FA formation
The signaling events triggered by NO2-FAs have been extensively studied, contrasting with the lower number of reports evaluating the formation and characterization of endogenous species. In animals, reactions in the gastric compartment promoted by dietary or salivary NO2− are the major contributor to systemic levels (Delmastro-Greenwood et al. 2015). A second mechanism includes NO-driven nitration reactions during inflammation (Bonacci et al. 2012; Vitturi et al. 2013; Villacorta et al. 2018). A third mechanism includes the reaction of NO2−-derived species during ischemia/reperfusion events. Independent of the mechanism, the nitration of fatty acids occurs through an initial addition of the nitrogen dioxide radical (•NO2) to a double bond with the formation of a ß-nitroalkyl radical (Trostchansky and Rubbo 2008; Schopfer et al. 2019). Figure 1 summarizes the chemical mechanisms of NO2-FA formation. Thus, the different biological scenarios in which NO2-FA are formed are defined by the mechanism involved in the formation of •NO2. This has important consequences, as the different mechanisms involve reaction milieus that differ on hydrophobicity, the rate and level of different nitrogen oxides formation, oxygen tension, and contributions of other oxidants and antioxidants. These factors will modulate the rates, yields, and products formed during the nitration reactions. The main nitration pathways relevant to biological conditions are depicted in different colours in Fig. 1:
Fig. 1.
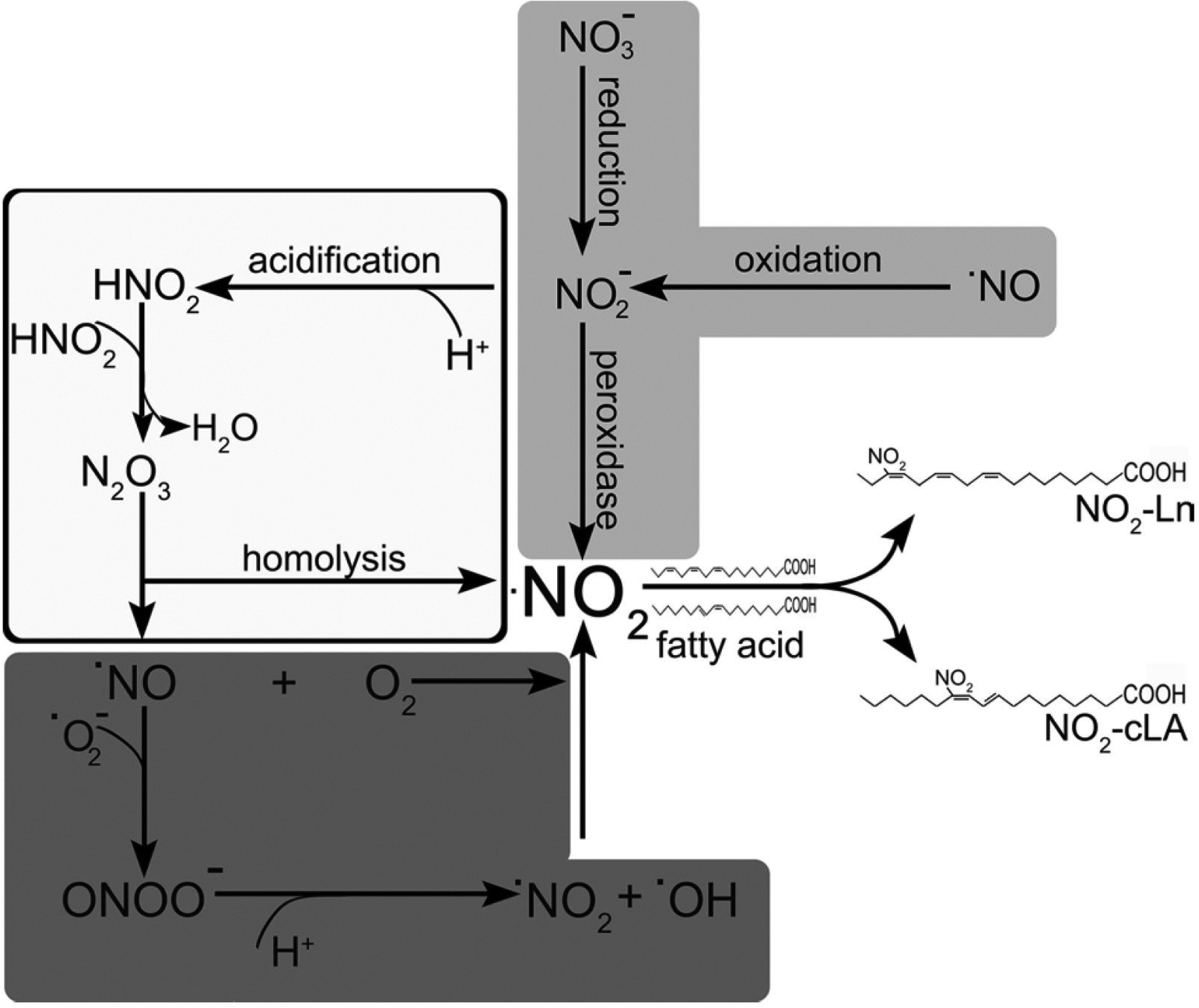
Role of reactive nitrogen species in fatty acid nitration. Nitration of fatty acids occurs via the addition of •NO2 (nitrogen dioxide radical) to a double bond. NO-dependent nitration (dark grey boxes). Under oxidative conditions with concomitant formation superoxide anion (O2•−) and •NO, these molecules react at diffusion-limited reaction rates to yield peroxynitrite (ONOO−). Through protonation and then homolysis, ONOO− produces •NO2. Under conditions with high local NO concentrations and the presence of oxygen, the auto-oxidation of NO leads to the formation of nitrogen dioxide. Heme peroxidase-catalyzed formation (light grey boxes). NO2−, the proximal product of •NO oxidation and nitrate (NO3−) reduction, can be oxidized to •NO2 in a one-electron oxidation reaction catalyzed by heme peroxidases. Acid-catalyzed formation (white boxes). The protonation of NO2− to HNO2 under acidic conditions (pKa 3.4) leads to its dimerization to form dinitrogen tetroxide (H2N2O4), which, upon dehydration, forms N2O3. This species is unstable and undergoes homolysis to form •NO and •NO2
NO-dependent nitration: NO plays a central role in the formation of nitrated fatty acids. Under oxidative conditions with concomitant formation of superoxide anion (O2•−) and •NO, these molecules react at diffusion-limited reaction rates to yield peroxynitrite (ONOO−). Upon protonation to peroxynitrous acid at physiological pH values, ONOO—can undergo homolytic cleavage to form (•NO2 and •OH). Under conditions with high local •NO concentrations and the presence of oxygen, the auto-oxidation of NO leads to the formation of nitrogen dioxide.
Heme peroxidase-catalyzed formation of •NO2: NO2 , the proximal product of •NO oxidation and nitrate (NO3−) reduction, can be oxidized to •NO2 in a one-electron oxidation reaction catalyzed by heme peroxidases (van der Vliet et al. 1997).
Acid-catalyzed formation of •NO2: The protonation of NO2− to HNO2 under acidic conditions (pKa 3.4) leads its dimerization to form dinitrogen tetroxide (H2N2O4), which, upon dehydration, forms N2O3. This species is unstable and undergoes homolysis to form NO and •NO2 (Vitturi et al. 2015).
The nitration of fatty acids proceeds through the addition of the •NO2 radical to a double bond present in fatty acids. This is an extremely fast reaction, but results in an unstable product and the •NO2 is consequently eliminated with the reformation of the double bond (Pryor et al. 1981; Gallon and Pryor 1993). This futile reaction can lead to double bond isomerization, as demonstrated for arachidonic acid (Balazy and López-Fernández 2003). When the addition reaction occurs on a conjugated double bond, the intermediary product is stabilized by resonance, providing on opportunity for other reactions to occur. Through a series of complex reactions, this radical is finally oxidize to a nitroalkene (Fazzari et al. 2014). Consequently, nitration yields are largely increased (> 105 fold) in the presence of conjugated double bonds (Bonacci et al. 2012). In plants, similar scenarios can be foreseen; however, it is unclear whether any of the mechanisms mentioned above play a role or if these reactions take place in specific cellular or subcellular compartments.
NO2-FAs detection
Nitrated fatty acids were initially described in plants as cysteine adducts of nitro-oleic acid (NO2-OA) (50 pmol NO2-OA-Cys/g FW) and as conjugated nitro-linoleic acid (NO2-cLA) (9- and 12-NO2-cLA isomers), as described in olive fruit and extra virgin olive oil (EVOO), respectively (Fazzari et al. 2014). Later, in Arabidopsis thaliana, nitrolinolenic acid (NO2-Ln) was detected in seeds (11 pmol/g FW), 14-old days seedling (3.8 pmol/g FW) and in leaves of 45-day old plants (0.54 pmol/g FW) (Mata-Pérez et al. 2016a). The same group later reported the presence of NO2-Ln in rice (Oryza sativa) and Pea (Pisum sativum). However, some methodological details were missing in the manuscript (e.g., growth plant condition, stage of the plants, techniques for isolation and detection of NO2-FAs), making the interpretation of the results more difficult (Mata-Pérez et al. 2017). Recently, another group reported the presence of NO2-OA in Brassica napus seeds (1.69 nmol/g FW) and 7-day-old seedling plants (0.66 nmol/g FW) (Vollár et al. 2020). The published data makes it clear that the highest content of NO2-FAs as free fatty acid (non-esterified) is found in seeds, likely due to the high fatty acid content found in seeds (Mata-Pérez et al. 2016a; Vollár et al. 2020). In mammals, the levels of esterified NO2-FAs account for over 95% of total circulating levels, with accumulation in the adipose tissue (Fazzari et al. 2017). Thus, these reports provide support to the presence and regulation of NO2-FAs levels in plants.
Despite these findings, the detection, characterization, and quantification of NO2-FAs in plants have proven extremely challenging. Several factors have contributed to this fact and are analyzed below:
Formation of Michael addition products:
NO2-FAs form reversible covalent adducts with nucleophilic amino acids of proteins, particularly cysteine and histidine (Rudolph et al.2009). The cellular cytoplasm is a highly reducing environment and, in animals, the cysteine content in proteins amounts to 10–50 mM and glutathione (GSH) levels are between 4 and 12 mM (Rudolph et al. 2009; Turell et al. 2013). In animal-derived cells, it has been calculated that approximately 99% of NO2-cLA is adducted, leaving only a small proportion of the molecule free for detection (Turell et al. 2017). Moreover, the proportion of free fatty acids in plant cells is low compared to the total fraction of fatty acid esterified to membranes phospholipids and triglycerides. Thus, to account for the presence of NO2-FA in membranes, investigators treated the lipid extracts with a lipase cocktail (Mata-Pérez et al. 2016a, 2018).
Matrix effects:
Plant samples are very rich in pigments of lipidic nature, such as chlorophylls and carotenoids, that can affect extraction yields and further reduce detection of NO2-FAs by dampening ionization when measured by liquid chromatography with tandem mass spectrometry (LC–MS/ MS) (Mata-Pérez et al. 2018). Although specific experiments to address this problem have not been conducted, the high content of these pigments might be partially responsible for the low concentration detected in pigmented tissues compared to non-pigmented tissues.
Currently, the number of identified nitrated lipid species in plants (NO2-OA, NO2-Ln and NO2-cLA) is smaller than those reported in humans (Salvatore et al. 2020). NO2-cLA is the most abundant NO2-FA detected in human urine and plasma, followed closely by NO2-cLn (Salvatore et al. 2020; Bonacci et al. 2012). In vitro assays revealed that conjugated LA (cLA) yielded 105 greater nitration products than LA (Bonacci et al. 2012). The presence of NO2-cLA in olive is intriguing given the scarcity of cLA when compared to other fatty acids (Fazzari et al. 2014). Apart from the cLA isomers, the isomers of a-linolenic acid (Ln) containing conjugated dienes and trienes are the second most prevalent species in humans (Salvatore et al. 2020). Conjugated linolenic acid (cLn) isomers are present at high levels in certain plant species and are the primary natural source of conjugated trienes (Hennessy et al. 2016), providing an interesting and abundant pool of nitration targets. Fatty acids with conjugated triene systems present in plants are mainly represented by (i) eleostearic acid (9-cis,11-trans,13trans-octadecatrienoic) found in a tung plant Aleurites fordii (Takagi and Itabashi 1981) and chinese bitter gourd Momordica charantia (Liu et al. 1997); (ii) Calendic acid (trans8,trans-10,cis-12-octadecatrienoic acid) found throughout the genus Calendula (Qiu et al. 2001) and (iii) Punicic acid (cis-9,trans-11,cis-13-octadecatrienoic acid) found in pomegranate seeds (Punicia granatum) (Takagi and Itabashi 1981) and in Trichosanthes, Mormodica balsamina, Fevillea trilobata oil and Ecballium elaterium (Hennessy et al. 2016). Thus, the presence of cLn in different plant species makes them attractive candidates for searching and identifying new nitro-fatty acids in plant species.
Mechanisms of NO2-FA action
Nitro-lipids are electrophilic molecules that participate in reversible Michael addition with proteins involved in cell signaling, metabolism, and transcription (Schopfer et al. 2019). The main intracellular targets are soft nucleophiles such as cysteine present in proteins and/or GSH, but proteomic approaches also revealed the formation of histidine adducts (Schopfer et al. 2011). In animals and humans, the mechanism of action of NO2-FAs involves nitro-alkylation, a post-translational modification that regulates stability, activity, and localization of the target proteins (Rubbo and Radi 2008; Rudolph and Freeman 2009; Geisler and Rudolph 2012). Nitro-alkylation is a reversible process with the ability to regulate physiological processes by modulating the activity of transcription factors and enzymatic reactions. In plants, the first evidence of nitro-alkylation was reported in olive fruit, where NO2-OA was found adducted to cysteine residues (Fazzari et al. 2014). The significance of nitroalkylation reactions in plants was later established by the targeting of cytosolic Ascorbate peroxidase (APX) in Arabidopsis (Aranda-Caño et al. 2019).
Nitro-alkylation resembles the mode of action of reactive electrophile species (RES) produced by plant cells under different stresses (Farmer and Davoine 2007; Farmer and Mueller 2013). Interestingly, transcriptional and physiological responses are shared between RES and NO2-FA. First, the RES-oxilipins OPDA (12-oxophytodienoic acid) (Muench et al. 2016) and electrophilic isothiocyanates Øverby et al. 2015; Ferber et al. 2020) induced the activation of heat shock factors and transcription of several heat shock (HS) proteins as described for NO2-Ln (Mata-Pérez et al. 2016a). In addition, inhibition of primary root growth and cellular cycle was reported for RES-oxylipins, electrophilic isothiocyanates (Almeras et al. 2003; Mueller et al. 2008; Pauwels et al. 2008; Åsberg et al. 2015; Urbancsok et al. 2017, 2018) and NO2-OA treatments in Arabidopsis (Di Fino et al. 2020). Considering the similarities between these reactive electrophilic species, future studies should focus on evaluating possible common signaling and/or mechanisms of action on plant physiology and divergent activities between these reactive species. More research is necessary to expand our understanding of target proteins and shed light on the signaling pathway in which NO2-FA participate.
Another mechanism of action reported for NO2-FAs relates to their ability to act as NO donors. This mechanism was first described in biochemical systems, cell culture, ex vivo and in vitro, and later translated to plant physiology (Schopfer, et al. 2005; Lima et al. 2003; Gorczynski et al. 2007; Mata-Pérez et al. 2016b, c; Grippo et al. 2020; Sánchez-Calvo et al. 2013). Nonetheless, the mechanism(s) whereby NO2-FAs release NO have not been definitively established and remain controversial. A possible mechanism may include a modified Nef reaction, where, under neutral aqueous conditions, NO2-FAs form a nitronate anion, that upon a sequence of rotonation and deprotonation steps forms a nitroso intermediate characterized by a weak C–N bond that decomposes to release NO (Schopfer, et al. 2005). Using electronparamagnetic resonance (EPR) spectroscopy, NO2-cLA, NO2-LA and NO2-OA have been shown to decompose and generate NO in aqueous solutions. By comparing the EPR signal intensities of spin-adducts, NO2-cLA generated a significantly reater yield of NO than NO2-LA and NO2-OA (Grippo et al. 2020). In vitro, NO2-LA releases less than 0.1% of NO in an aqueous solution. The NO release was inhibited in the presence of non-ionic detergent micelles as hydrophobic environments stabilize NO2-LA (Schopfer et al. 2005). Other studies demonstrated that both NO2-OA isomers yields, under optimal conditions, less than 0.1% of NO, with a 46 times lower release compared to NO donor SNP when using a 50 times higher concentration of NO2-OA (Gorczynski et al. 2007). In an aqueous solution, NO2-Ln releases NO with a peak within the first hour but 6 times lower than the rate reported for the NO donor GSNO (Mata-Pérez et al. 2016b; Ederli et al. 2009). The deprotonation of the bisallylic carbon is the first step in releasing NO; consequently, NO2-Ln and NO2-LA display higher NO release rates than NO2-OA. Nonetheless, the rates and yields observed in biochemical systems remain very low. These results clearly show that nitro-alkenes have an objectively lower ability to release NO than SNP or GSNO donors. More importantly, NO2-FA NO release reactions compete against in vivo alkylation reactions. The high intracellular thiol concentration, the rapid formation of GSH and protein adducts, and the increased stability by incorporation into phospholipids and glycerolipids further reduces their ability to release NO (Schopfer et al. 2019). However, subcellular compartmentalization and/or enzymatic reactions could promote NO release under specific environments and cellular contexts.
NO formation was detected in vivo in Arabidopsis leaves and roots treated with 1 mM NO2-Ln (Sánchez-Calvo et al. 2013), and in primary root treated with 100 μM of NO2-Ln (Mata-Pérez et al. 2016c). Curiously, those NO2-Ln treatments triggered different physiological responses than those triggered by NO donors (further discussed in the next section). A dual response was reported in Brassica seeds treated with NO2-OA: at 50 μM, NO2-OA did not release NO; however, at 100 μM, NO2-OA NO was formed, while 500 μM NO2-OA did not produce NO (Vollár et al. 2020). In contrast, we failed to detect NO release from Arabidopsis roots or tomato culture cells treated with NO2-OA (Arruebarrena Di Palma et al. 2020; Di Fino et al. 2020). In aggregate, while all nitro-alkenes are electrophilic and react with cellular nucleophiles, some of them might release NO under specific conditions. The elucidation of the mechanism of action for NO2-FA under physiological conditions will require considering NO release and Michael addition reactions and incorporate new approaches to dissect the individual contributions of these pathways to the biological response.
Differentiation from NO-dependent activities
In higher plants, NO is involved in a wide range of biological processes, including germination, plant metabolism, senescence, cellular death, stomatal movement, photosynthesis, gravitropism, and primary root growth (Del Castello et al. 2020; Lamattina et al. 2003). Some molecular targets of NO2-FAs are chemically similar to those described for NO signaling, including redox-sensitive cysteine residues. This signaling crossover complicates the analysis of the responses obtained with NO donors and NO2-FAs and the evaluation of the mechanism responsible for the biological actions. NO participates in seed germination regulation by interacting with the ABA signaling during seed dormancy (Liu and Zhang 2009). For instance, pharmacological approaches demonstrated that most known NO donors promote dormancy-releasing and subsequent germination, while NO scavengers favour dormancy (Bethke et al. 2007). It was reported that in Brassica napus, NO2-OA releases NO, which in turn increases germination rate (Vollár et al. 2020). However, this effect was only observed when seeds were treated with 100 μM NO2-OA, with higher or lower concentrations failing to increase NO levels and affect germination. Further work will be necessary to elucidate the mechanisms of action and the signaling pathway involved in this process.
NO upregulates APX activity through S-nitrosation of the conserved cysteine 32 residue (Correa-Aragunde et al. 2013; Begara-Morales et al. 2014; Yang et al. 2015). Conversely, in vitro NO2-Ln nitro-alkylate APX’s histidine residues inhibiting its enzymatic activity (Aranda-Caño et al. 2019). However, although NO2-Ln has been proposed to act through NO release (Mata-Pérez et al. 2016b), S-nitrosation of the APX cysteine residue upon in vitro NO2-Ln treatments has not been reported (Aranda-Caño et al. 2019). The overlapping activities as an NO donor and alkylation agent highlight the complication of establishing a mechanism of action and remind us to be cautious when considering NO2-Ln exclusively as a NO donor.
NO inhibits primary root growth in Arabidopsis thaliana (Fernández-Marcos et al. 2011). Seedlings treated with NO donors show an imbalance in auxin levels (decrease in auxin response reporter DR5:GUS activity) due to NO-dependent alteration of the auxin efflux protein PIN-FORMED1 localization in the cellular membrane of meristematic cells (Fernández-Marcos et al. 2011). Arabidopsis seedlings treated with NO2-OA showed primary root length inhibition without affecting endogenous NO levels and auxin homeostasis (Di Fino et al. 2020). In contrast, NO2-OA reduced root growth by inhibiting the cell cycle in the meristem zone (Di Fino et al. 2020). This provides further evidence that NO and NO2-FAs can induce the same morphological responses through independent signaling pathways.
RNAseq analysis of Arabidopsis cell suspensions treated with NO2-Ln showed that several upregulated genes are related to the heat shock response (Mata-Pérez et al. 2016a). This response was first reported in NO2-FAs-treated endothelial cells (Kansanen et al. 2009). To study whether NO2-Ln modulates gene expression via exclusive NO production, we compared RNAseq data from NO2-Ln (Mata-Pérez et al. 2016a) vs. NO treated Arabidopsis leaves (Hussain et al. 2016) (Fig. 2). Among the most representative shared genes, heat shock protein (HSPs) expression was strongly up-regulated while RNA transcription factors were down-regulated (Fig. 2, a complete set of data is shown in Supplemental Table S1, and a complete list of GO terms for biological processes in Supplemental Table S2). Despite this similarity, it is worth mentioning that only 122 genes are shared between both treatments and that the expression of 194 genes regulated by NO2-Ln was unaffected by the NO treatment under the analyzed conditions. Altogether, the evidence presented above points towards a NO2-FA-specificmechanism of action in different plant physiological systems.
Fig. 2.
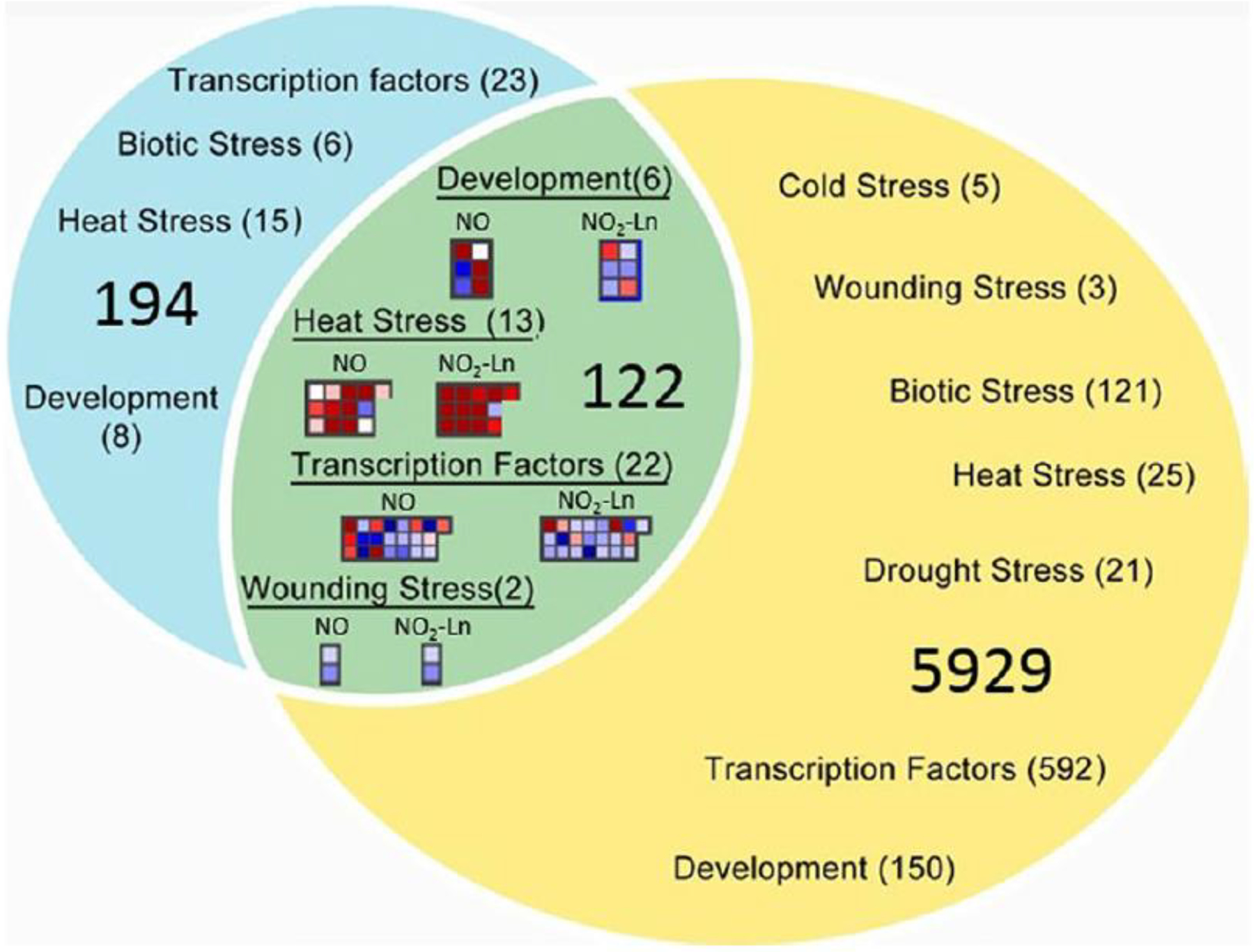
Overlap of genes whose expression is affected by treatments with NO2-Ln and NO. Venn Diagram showing the overlap of differential expressed genes (p-value<0.05 and log2=1 or=− 1) between treatment with NO2-Ln (blue circle) and treatment with NO (yellow circle) was performed with MapMan Software. On the overlapping area are the principal groups of genes shared between the treatments. Those groups of genes were extracted from the BIN analysis performed with MapMan Software. On the unshared area genes related to stress, development and transcription whose expression is only affected in one of the two treatments. A complete list can be found in Supplemental Table 1. RNA seq data was obtained from: Arabidopsis cells treated with from 100 μM of NO2-Ln for 1 h (Mata-Pérez et al. 2016a) and Arabidopsis leaves infiltrated with 1 mM CysNO for 6 h (Hussain et al. 2016)
NO2-OA as regulators of ROS and Ca2+
A recent report showed that exogenous application of NO2-OA induced ROS production on both tomato cell suspensions and tomato and Arabidopsis leaves (Fig. 3, Arruebarrena Di Palma et al. 2020). Pharmacological and reverse genetic experiments showed that NO2-OA-induced ROS were generated specifically by NADPH oxidase RBOH isoform D. The NADPH oxidase RBOHD is responsible for the rapid and robust production of ROS upon pathogen-associated molecular patterns perception (Kadota et al. 2015). In line with the activation of RBOHD, we tested the expression of genes related to pathogen responses observing up-regulation of SlPR1a, SlHSR203J and SlPAL (Arruebarrena Di Palma et al. 2020). Multilayered regulations tightly control RBOHD activity. It is mainly activated via direct binding of Ca2+ to EF-hand motifs, by Ca2+-dependent and independent protein kinases, and by the signal lipid phosphatidic acid (PA) (Kadota et al. 2015; Zhang et al. 2009). No Nitric Oxide was detected under these experimental conditions, suggesting that NO2-FA-derived NO was not a primary contributor to the observed biological responses. Moreover, as RBOHD was reported to be negatively regulated by NO via S-nitrosation of Cys890 (Yun et al. 2011), a NO role is unlikely as we observed enzymatic activation upon treatment with NO2-OA (Arruebarrena Di Palma et al. 2020). In addition, RBOHD and F were required for NO2-OA-dependent induction of stomatal closure (Fig. 3, Arruebarrena Di Palma et al. 2020). However, the mechanism involved in the NADPH oxidase activation by NO2-OA remains unclear, and it could be hypothesized that NO2-FA form adducts directly with RBOHD protein regulating its activity. Nevertheless, we found that calcium and protein kinase activation are required to induce maximal ROS production upon NO2-OA treatments (Arruebarrena Di Palma et al. 2020). Thus, it might be possible that NO2-FA directly regulate protein kinases and/or Ca2+ channels and, therefore, Ca2+-dependent protein kinases. Cytosolic-free calcium elevation is an early signal in the developmental process and response to abiotic and biotic stresses in plants. Membrane-localized Ca2+ permeable ion channels modulate the influx of Ca2+ leading to elevation, in cytosolic Ca2+ concentrations (Kong et al. 2020). Results obtained in our lab using leaf discs that express the cytosolic calcium sensor aequorin showed that [Ca+2]cyt increases within 60–90 min in a dose-dependent manner between 5 and 25 μM NO2-OA treatments (unpublished result). The specific target and the mechanistic regulation of NO2-FA on calcium intracellular concentrations have not been explored, but cys residues in target proteins might be key molecular targets. Recently, the H2O2-induced Ca2+ increase 1 (HPCA1, a leucine-rich-repeat receptor kinase) was shown to act as an extracellular H2O2 sensor (Wu et al. 2020). Interestingly, HPCA1 contains four Cys residues acting as conserved redox-active sites. The molecular mechanism of H2O2 sensing involves the oxidation of these cysteines that induce a conformational change of the receptor. Nitrolipid targeting and regulation of HPCA1 might hold the key to understanding the relationship between Ca2+ and NO2-FA.
Fig. 3.
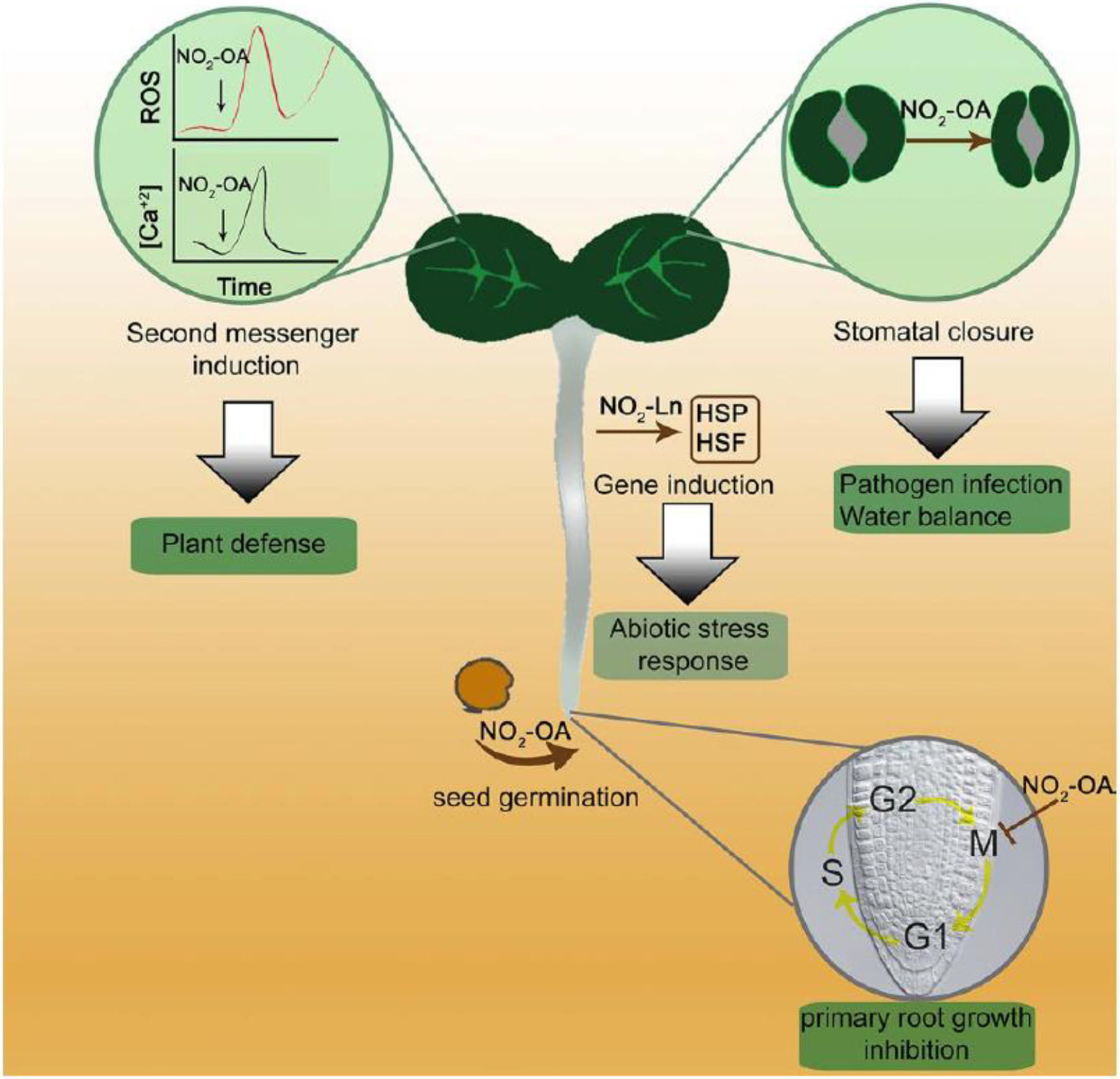
Schematic representation of the physiological responses induced by NO2-FAs in Arabidopsis. Exogenous application of nitrooleic acid (NO2-OA) induces both reactive oxygen species (ROS) and cytosolic calcium levels in Arabidopsis leaf discs (Arruebarrena Di Palma et al. 2020). In addition, NO2-OA induces stomatal closure (Arruebarrena Di Palma et al. 2020), seed germination (Vollár et al. 2020), and cell cycle regulation (Di Fino et al. 2020). Another NO2-FA, nitro-linolenic acid (NO2-Ln) induces a specific set of genes that codify for heat shock proteins (HSP) and heat shock factors (HSF) in Arabidopsis cell cultures (Mata-Peréz et al. 2016a). The green box shows the biological process involved. Arrow indicates induction
Redox regulators as targets of NO2-FAs?
Activation or deactivation of signaling pathways and enzymatic activities mediated by NO2-FA involves the reaction with cysteines of various cellular targets (Schopfer et al. 2019). A well-described signaling pathway regulated by NO2-FAs in mammals is Nfr2/Keap1 (nuclear factor (erythroid-derived 2)-like 2)/kelch-like ECH-associated protein 1). Keap1 functions as a negative regulator of Nrf2 by keeping Nrf2 in the cytoplasmic compartment, promoting ubiquitination and downstream degradation by the proteasome. Under metabolic and inflammatory stress conditions, the formation of NO2-FA leads to the nitro-alkylation of Keap1 and destabilization of Nfr2-Keap1 complex. This inhibits Nrf2 proteasomal degradation and causes nuclear translocation and accumulation of newly synthesized Nrf2 protein, inducing the gene transcription of antioxidant and detoxification enzymes that protect against oxidative stress, inflammation, and drug toxicity (Magesh et al. 2012; Suzuki et al. 2013; Joshi et al. 2012; Kansanen et al. 2011). The central redox regulator NPR1 (non expressor of pathogenesis-related genes 1) is the plant counterpart of the mammalian Nfr2Keap1 pathway, with which they share a common redox sensing mechanism (González-Bosch 2018). Both are activated by modifications (oxidation or alkylations) of their thiol groups that promote their nuclear translocation and activation of defense genes. In both pathways, proteasomal degradation prevents their activation and transcription activity (González-Bosch 2018). Arabidopsis NPR1 has 17 cysteines, with 10 of them being highly conserved among different species (Mou et al. 2003). Upon pathogen infection or accumulation of salicylic acid (SA), changes in cellular redox potential lead to the reduction of cysteines through the activity of thioredoxins (TRX-h3 and TRX-h5), promoting NPR1 translocation to the nucleus (Withers and Dong 2016). Unlike Nrf2, NPR1 does not contain a DNA-binding domain but acts as a transcriptional coactivator by interacting with several transcriptional factors leading to the expression f pathogenesis-related (PR) proteins (Withers and Dong 2016). Indeed, Farmer and Mueller (Farmer and Mueller 2013) proposed NPR1 as a possible target of RES. Considering the similarities between both signaling mechanisms, it will be interesting to focus future research on NPR1 as a possible target of NO2-FA action in plants. Another relevant cellular target reported in animals is glutathione (GSH) (Schopfer et al. 2019). Through enzymatic and non-enzymatic reactions, GSH plays an active role in plant cell redox control systems. Glutathione protects cells against oxidative damage induced by environmental challenges (Hernández et al. 2017). The concentration of GSH in Arabidopsis leaf cells is between 0.05 mM in vacuole to near 15 mM in mitochondria (Gill et al. 2013), making it a likely target of NO2-FA in plants. Exogenous treatment of tomato cell suspension with NO2-OA results in the formation of GS-NO2-OA adducts, with a concomitant reduction in GSH cellular concentration (Arruebarrena Di Palma et al. 2018). Also, it was reported that the release of 15N-labeled NO from exogenously applied 15NO2-Ln (1 mM) induces the generation of GS15NO (Mata-Pérez et al. 2020). The authors indicated that the formation of NO from NO2-Ln probably led to the modulation of the intracellular GSH pool, although the exact mechanism of GSNO formation from NO2-Ln and GSH remains to be elucidated. Moreover, GSH was reported to interplay with different plant hormones like JA, MeJA, SA, and ethylene (Hasanuzzaman et al. 2017). Although GSH has multiple roles in plant physiology, the pathways or mechanisms involved in transducing GSH-induced responses have only been partially addressed (Hasanuzzaman et al. 2017). Given the concentration and reactivity of GSH with NO2-FA, it would be important to establish the role that adduct formation plays in modulating plant signaling and physiology.
Final considerations
NO2-FA has emerged as new molecules regulating physiological processes in plants. Different NO2-FA were identified and detected in a variety of species. Our group has expanded the knowledge about this molecule on plant physiology, studying both their role in development (Di Fino et al. 2020) as well as signaling (Arruebarrena Di Palma et al. 2020) (Fig. 3). Depending on the nitrolipid, the exogenous application may exert a role as an electrophile (NO2-OA), an NO donor (NO2-Ln), or both. The identification of 342 protein adducted to NO2-FA (working as an electrophile) has been reported, but unfortunately, their identities have not been revealed so far (Aranda-Caño et al. 2019). The study of nitrolipid function in non-model plants and the identification of the cellular targets of NO2-FA will shed light on its mechanism of action and the plant physiological responses that they might regulate.
Supplementary Material
Acknowledgements
This work was supported by the UNMdP, ConsejoNacional de Investigaciones Científicas y Técnicas, Agencia Nacionalde Promoción Científica y Tecnológica, ANPCyT; PICT-Raíces 20130800 and PICT 2017-0601 (to AML), PICT 2016-3084 (to AADP) andNIH GM125944 and DK112854 (to FJS).
Declarations
Conflict of interest FJS acknowledges financial interest in Creegh Inc. No other competing financial interests are noted.
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00425-021-03777-z.
Data availability
All data generated or analyzed during this study arealready published. Data sharing does not apply to this article as nodatasets were generated during the current study. The comparison ofthe two different already published RNAseq is, however, presented asa supplemental table and available to the readers.
References
- Akaike T, Nishida M, Fujii S (2013) Regulation of redox signallingby an electrophilic cyclic nucleotide. J Biochem 153:131–138. 10.1093/jb/mvs145 [DOI] [PubMed] [Google Scholar]
- Almeras E, Stolz S, Vollenweider S, Reymond P, Mene-Saffrane L, Farmer EE (2003) Reactive electrophile species activate defensegene expression in Arabidopsis. Plant J 34:205–216. 10.1046/j.1365-313x.2003.01718.x [DOI] [PubMed] [Google Scholar]
- Aranda-Caño L, Sánchez-Calvo B, Begara-Morales JC, Chaki M,Mata-Pérez C, Padilla MN, Valderrama R, Barroso JB (2019) Post-translational modification of proteins mediated by nitro-fattyacids in plants: nitroalkylation. Plants (basel) 8(4):82. 10.3390/plants8040082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruebarrena Di Palma A, Di Fino LM, Salvatore SR, D’Ambrosio JM, Grozeff GEG, García-Mata C, Schopfer F, Laxalt AM (2018) Nitro-oleic acid induced reactive oxygen species formation andplant defense signaling in tomato cell suspensions. BioRxiv 2:e263 [Google Scholar]
- Arruebarrena Di Palma A, Di Fino LM, Salvatore SR, D’Ambrosio JM, García-Mata C, Schopfer FJ, Laxalt AM (2020) Nitrooleic acid triggers ROS production via NADPH oxidase activation in plants: A pharmacological approach. J. Plant Physiol 246247:153128. 10.1016/j.jplph.2020.153128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åsberg SE, Bones AM, Øverby A (2015) Allyl isothiocyanate affectsthe cell cycle of Arabidopsis thaliana. Front Plant Sci 6:364. 10.3389/fpls.2015.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker PR, Schopfer FJ, Sweeney S, Freeman BA (2004) Red cellmembrane and plasma linoleic acid nitration products: synthesis, clinical identification, and quantitation. Proc Natl Acad Sci 101(32):11577–11582. 10.1073/pnas.0402587101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazy M, López-Fernández J (2003) Isomerization and nitro-oxidationof arachidonic acid by NO2. Adv Exp Med Biol 525:173–176. 10.1007/978-1-4419-9194-2_36 [DOI] [PubMed] [Google Scholar]
- Begara-Morales JC, Sánchez-Calvo B, Chaki M, Valderrama R, Mata-Pérez C, López-Jaramillo J, Padilla MN, Carreras A, Corpas FJ, Barroso JB (2014) Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J Exp Bot 65(2):527–538. 10.1093/jxb/ert396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Libourel IG, Aoyama N, Chung YY, Still DW, Jones RL(2007) The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary forseed dormancy. Plant Physiol 143(3):1173–1188. 10.1104/pp.106.093435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacci G, Baker PR, Salvatore SR, Shores D, Khoo NK, Koenitzer JR, Vitturi DA, Woodcock SR, Golin-Bisello F, Cole MP, Watkins S, St Croix C, Batthyany CI, Freeman BA, Schopfer FJ (2012)Conjugated linoleic acid is a preferential substrate for fatty acidnitration. J Biol Chem 287(53):44071–44082. 10.1074/jbc.M112.401356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Aragunde N, Foresi N, Delledonne M, Lamattina L (2013)Auxin induces redox regulation of ascorbate peroxidase 1 activityby S -nitrosylation/denitrosylation balance resulting in changes ofroot growth pattern in Arabidopsis. J Exp Bot 64(11):3339–3349. 10.1093/jxb/ert172 [DOI] [PubMed] [Google Scholar]
- Del Castello F, Nejamkin A, Foresi N, Lamattina L, Correa-Aragunde N (2020) Chimera of globin/nitric oxide synthase: toward improving nitric oxide homeostasis and nitrogen recycling and availability. Front Plant Sci 11:575651. 10.3389/fpls.2020.575651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmastro-Greenwood M, Hughan KS, Vitturi DA, Salvatore SR,Grimes G, Potti G, Shiva S, Schopfer FJ, Gladwin MT, Freeman BA, Gelhaus Wendell S (2015) Nitrite and nitrate-dependent generation of anti-inflammatory fatty acid nitroalkenes. Free RadicalBiol Med 89:333–341. 10.1016/j.freeradbiomed.2015.07.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fino LM, cerrudo I, salvatore sr, schopfer fj, garcía-mata c, laxalt am(2020) exogenous nitro-oleic acid treatment inhibits primary rootgrowth by reducing the mitosis in the meristem in Arabidopsis thaliana. Front Plant Sci 11:1059. 10.3389/fpls.2020.01059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederli L, Reale L, Madeo L, Ferranti F, Gehring C, Fornaciari M,Romano B, Pasqualini S (2009x) NO release by nitric oxidedonors in vitro and in planta. Plant Physiol Biochem 47(1):42–48. 10.1016/j.plaphy.2008.09.008 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Davoine C (2007) Reactive electrophile species. Curr OpinPlant Biol 10(4):380–386. 10.1016/j.pbi.2007.04.019 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Mueller MJ (2013) ROS-mediated lipid peroxidation andRES-activated signaling. Ann Rev Plant Biol 64(1):429–450. 10.1146/annurev-arplant-050312-120132 [DOI] [PubMed] [Google Scholar]
- Fazzari M, Trostchansky A, Schopfer FJ, Salvatore SR, Sánchez-Calvo B, Vitturi D, Valderrama R, Barroso JB, Radi R, Freeman BA, Rubbo H (2014) Olives and olive oil are sources of electrophilicfatty acid nitroalkenes. PLoS ONE 9(1):e84884. 10.1371/journal.pone.0084884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzari M, Khoo NK, Woodcock SR, Jorkasky DK, Li L, Schopfer FJ, Freeman BA (2017) Nitro-fatty acid pharmacokinetics in the adipose tissue compartment. J Lipid Res 58(2):375–385. 10.1194/jlr.M072058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber E, Gerhards J, Sauer M, Krischke M, Dittrich MT, Müller T, Berger S, Agnes F, Mueller MJ (2020) Chemical priming byisothiocyanates protects against intoxication by products of themustard oil bomb. Front Plant Sci 11:887. 10.3389/fpls.2020.00887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marcos M, Sanz L, Lewis DR, Muday GK, Lorenzo O(2011) Nitric oxide causes root apical meristem defects andgrowth inhibition while reducing PIN-FORMED 1 (PIN1)dependent acropetal auxin transport. Proc Natl Acad Sci USA 108(45):18506–18511. 10.1073/pnas.1108644108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallon AA, Pryor WA (1993) The identification of the allylic nitriteand nitro derivatives of methyl linoleate and methyl linolenateby negative chemical ionization mass spectroscopy. Lipids 28(2):125–133. 10.1007/BF02535776 [DOI] [PubMed] [Google Scholar]
- Geisler AC, Rudolph TK (2012) Nitroalkylation a redox sensitive signaling pathway. Biochem Biophys Acta 1820(6):777–784. 10.1016/j.bbagen.2011.06.014 [DOI] [PubMed] [Google Scholar]
- Gill SS, Anjum NA, Hasanuzzaman M, Gill R, Trivedi DK, Ahmad I,Pereira E, Tuteja N (2013) Glutathione and glutathione reductase: a boon in disguise for plant abiotic stress defense operations. PlantPhysiol Biochem 70:204–212. 10.1016/j.plaphy.2013.05.032 [DOI] [PubMed] [Google Scholar]
- González-Bosch C (2018) Priming plant resistance by activation ofredox-sensitive genes. Free Radical Biol Med 122:171–180. 10.1016/j.freeradbiomed.2017.12.028 [DOI] [PubMed] [Google Scholar]
- Gorczynski MJ, Huang J, Lee H, King SB (2007) Evaluation ofnitroalkenes as nitric oxide donors. Bioorg Med Chem Lett 17(7):2013–2017. 10.1016/j.bmcl.2007.01.016 [DOI] [PubMed] [Google Scholar]
- Grippo V, Mojovic M, Pavicevic A, Kabelac M, Hubatka F, Turanek J, Zatloukalova M, Freeman BA, Vacek J (2020) Electrophilic characteristics and aqueous behavior of fatty acid nitroalkenes. Redox Biol 38:101756. 10.1016/j.redox.2020.101756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanuzzaman M, Nahar K, Anee TI, Fujita M (2017) Glutathionein plants: biosynthesis and physiological role in environmentalstress tolerance. Physiol Mol Biol Plants Int J Funct Plant Biol 23(2):249–268. 10.1007/s12298-017-0422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy AA, Ross PR, Fitzgerald GF, Stanton C (2016) Sourcesand bioactive properties of conjugated dietary fatty acids. Lipids 51(4):377–397. 10.1007/s11745-016-4135-z [DOI] [PubMed] [Google Scholar]
- Hernández JA, Barba-Espín G, Diaz-Vivancos P (2017) Glutathione mediated biotic stress tolerance in plants. In: Hossain M, Mostofa M, Diaz-Vivancos P, Burritt D, Fujita M, Tran LS (eds) Glutathione in plant growth, development, and stress tolerance. Springer, Cham [Google Scholar]
- Hussain A, Mun BG, Imran QM, Lee SU, Adamu TA, Shahid M, Kim KM, Yun BW (2016) Nitric Oxide Mediated Transcriptome Profiling Reveals Activation of Multiple Regulatory Pathways in Arabidopsis thaliana. Front Plant Sci 7:975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G, Johnson JA (2012) The Nrf2-ARE pathway: a valuable therapeutic target for the treatment of neurodegenerative diseases. Recent Pat CNS Drug Dis 7(3):218–229. 10.2174/57488912803252023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Zipfel C (2015) Regulation of the NADPHoxidase RBOHD during plant immunity. Plant Cell Physiol 56(8):1472–1480. 10.1093/pcp/pcv063 [DOI] [PubMed] [Google Scholar]
- Kansanen E, Jyrkkänen HK, Volger OL, Leinonen H, Kivelä AM, Häkkinen SK, Woodcock SR, Schopfer FJ, Horrevoets AJ, Ylä-Herttuala S, Freeman BA, Levonen AL (2009) Nrf2-dependent and-independent responses to nitro-fatty acids in human endothelialcells: identification of heat shock response as the major pathwayactivated by nitro-oleic acid. J Biol Chem 284(48):33233–33241. 10.1074/jbc.M109.064873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansanen E, Bonacci G, Schopfer FJ, Kuosmanen SM, Tong KI,Leinonen H, Woodcock SR, Yamamoto M, Carlberg C, YläHerttuala S, Freeman BA, Levonen AL (2011) Electrophilic nitrofatty acids activate NRF2 by a KEAP1 cysteine 151-independentmechanism. J Biol Chem 286(16):14019–14027. 10.1074/jbc.M110.190710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Xu L, Jamieson P (2020) Plant sense: the rise of calcium channels. Trends Plant Sci 25(9):838–841. 10.1016/j.tplants.2020.06.002 [DOI] [PubMed] [Google Scholar]
- Lamattina L, García-Mata C, Graziano M, Pagnussat G (2003) Nitricoxide: the versatility of an extensive signal molecule. Annu RevPlant Biol 54:109–136. 10.1146/annurev.arplant.54.031902.134752 [DOI] [PubMed] [Google Scholar]
- Lima ES, Di Mascio P, Rubbo H, Abdalla DS (2002) Characterizationof linoleic acid nitration in human blood plasma by mass spectrometry. Biochemistry 41(34):10717–10722. 10.1021/bi025504j [DOI] [PubMed] [Google Scholar]
- Lima ES, Di Mascio P, Abdalla DS (2003) Cholesteryl nitrolinoleate,a nitrated lipid present in human blood plasma and lipoproteins. J Lipid Res 44(9):1660–1666. 10.1194/jlr.M200467-JLR200 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang J (2009) Rapid accumulation of NO regulates ABA catabolism and seed dormancy during imbibition in Arabidopsis. PlantSignal Behav 4(9):905–907. 10.4161/psb.4.9.9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Hammond EG, Nikolau BJ (1997) In vivo studies of the biosynthesis of [alpha]-eleostearic acid in the seed of momordicacharantia L. Plant Physiol 113(4):1343–1349. 10.1104/pp.113.4.1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magesh S, Chen Y, Hu L (2012) Small molecule modulators of Keap1Nrf2-ARE pathway as potential preventive and therapeutic agents. Med Res Rev 32(4):687–726. 10.1002/med.21257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Pérez C, Sánchez-Calvo B, Padilla MN, Begara-Morales JC, Luque F, Melguizo M, Jimenez-ruiz J, Fierro-Risco J, Penas-Sanjuan A, Valderrama R, Corpas FJ, Barroso JB (2016a) Nitro-fattyacids in plant signaling: nitro-linolenic acid induces the molecularchaperone network in Arabidopsis. Plant Physiol 170:686–701. 10.1104/pp.15.01671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Pérez C, Sánchez-Calvo B, Begara-Morales JC, Carreras A,Padilla MN, Melguizo M, Valderrama R, Corpas FJ, Barroso JB(2016b) Nitro-linolenic acid is a nitric oxide donor. Nitric OxideBiol Chem 57:57–63. 10.1016/j.niox.2016.05.003 [DOI] [PubMed] [Google Scholar]
- Mata-Pérez C, Sánchez-Calvo B, Begara-Morales JC, Padilla MN, Valderrama R, Corpas FJ, Barroso JB (2016c) Nitric oxide releasefrom nitro-fatty acids in Arabidopsis roots. Plant Signal Behav 11(3):e1154255. 10.1080/15592324.2016.1154255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Pérez C, Sánchez-Calvo B, Padilla MN, Begara-Morales JC, Valderrama R, Corpas FJ, Barroso JB (2017) Nitro-fatty acids in plantsignaling: new key mediators of nitric oxide metabolism. RedoxBiol 11:554–561. 10.1016/j.redox.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Pérez C, Padilla MN, Sánchez-Calvo B, Begara-Morales JC, Valderrama R, Corpas FJ, Barroso JB (2018) Nitro-fatty acid detection in plants by high-pressure liquid chromatography coupled totriple quadrupole mass spectrometry. In: Mengel A, Lindermayr C (eds) Nitric oxide. Methods in molecular biology, vol 1747. Humana Press, New York: [DOI] [PubMed] [Google Scholar]
- Mata-Pérez C, Padilla MN, Sánchez-Calvo B, Begara-Morales JC,Valderrama R, Chaki M, Aranda-Caño L, Moreno-González D, Molina-Díaz A, Barroso JB (2020) Endogenous biosynthesis ofs-nitrosoglutathione from nitro-fatty acids in plants. Front PlantSci 11:962. 10.3389/fpls.2020.00962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquiredresistance regulate NPR1 function through redox changes. Cell 113(7):935–944. 10.1016/s0092-8674(03)00429-x [DOI] [PubMed] [Google Scholar]
- Mueller S, Hilbert B, Dueckershoff K, Roitsch T, Krischke M, Mueller MJ, Berger S (2008) General detoxification and stress responsesare mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20(3):768–785. 10.1105/tpc.107.054809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench M, Hsin CH, Ferber E, Berger S, Mueller MJ (2016) Reactiveelectrophilic oxylipins trigger a heat stress-like response throughHSFA1 transcription factors. J Exp Bot 67(21):6139–6148. 10.1093/jxb/erw376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell VB, Eiserich JP, Chumley PH, Jablonsky MJ, Krishna NR,Kirk M, Barnes S, Darley-Usmar VM, Freeman BA (1999) Nitration of unsaturated fatty acids by nitric oxide-derived reactivenitrogen species peroxynitrite, nitrous acid, nitrogen dioxide, andnitronium ion. Chem Res Toxicol 12:83–92 [DOI] [PubMed] [Google Scholar]
- Øverby A, Bævre MS, Thangstad OP, Bones AM (2015) Disintegrationof microtubules in Arabidopsis thaliana and bladder cancer cellsby isothiocyanates. Front Plant Sci 6:6. 10.3389/fpls.2015.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A (2008) Mapping methyljasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci 105(4):1380–1385. 10.1073/pnas.0711203105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor WA, Lightsey JW (1981) Mechanisms of nitrogen dioxidereactions: initiation of lipid peroxidation and the production ofnitrous Acid. Science 214(4519):435–437. 10.1126/science.214.4519.435 [DOI] [PubMed] [Google Scholar]
- Qiu X, Reed DW, Hong H, MacKenzie SL, Covello PS (2001) Identification and analysis of a gene from Calendula officinalisencoding a fatty acid conjugase. Plant Physiol 125(2):847–855. 10.1104/p.125.2.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbo H, Radi R (2008) Protein and lipid nitration: role in redoxsignaling and injury. Biochim Biophys Acta 1780:1318–1324. 10.1016/j.bbagen.2008.03.007 [DOI] [PubMed] [Google Scholar]
- Rubbo H, Radi R, Trujillo M, Telleri R, Kalyanaraman B, Barnes S, Kirk M, Freeman BA (1994) Nitric oxide regulation of superoxide and peroxynitrite-dependent lipid peroxidation. Formationof novel nitrogen-containing oxidized lipid derivatives. J BiolChem 269:26066–26075 [PubMed] [Google Scholar]
- Rudolph V, Freeman BA (2009) Cardiovascular consequences whennitric oxide and lipid signaling converge. Circ Res 105(6):511–522. 10.1161/CIRCRESAHA.109.202077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph V, Schopfer FJ, Khoo NK, Rudolph TK, Cole MP, Woodcock SR, Bonacci G, Groeger AL, Golin-Bisello F, Chen CS, Baker PR, Freeman BA (2009) Nitro-fatty acid metabolome:saturation, desaturation, beta-oxidation, and protein adduction. J Biol Chem 284(3):1461–1473. 10.1074/jbc.M802298200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore SR, Rowart P, Schopfer FJ (2020) Mass spectrometry-basedstudy defines the human urine nitrolipidome. Free Rad Biol Med 10.1016/j.freeradbiomed.2020.10.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Calvo B, Barroso JB, Corpas FJ (2013) Hypothesis: Nitrofatty acids play a role in plant metabolism. Plant Sci Int J ExpPlant Biol 199–200:1–6. 10.1016/j.plantsci.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Schopfer FJ, Khoo N (2019) Nitro-fatty acid logistics: formation, biodistribution, signaling, and pharmacology. Trends EndocrinolMetab 30(8):505–519. 10.1016/j.tem.2019.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer FJ, Baker PR, Giles G, Chumley P, Batthyany C, Crawford J, Patel RP, Hogg N, Branchaud BP, Lancaster JR, Freeman BA(2005) Fatty acid transduction of nitric oxide signaling. Nitrolinoleic acid is a hydrophobically stabilized nitric oxide donor. JBiol Chem 280(19):19289–19297. 10.1074/jbc.M414689200 [DOI] [PubMed] [Google Scholar]
- Schopfer FJ, Cipollina C, Freeman BA (2011) Formation and signaling actions of electrophilic lipids. Chem Rev 111(10):5997–6021. 10.1021/cr200131e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Motohashi H, Yamamoto M (2013) Toward clinicalapplication of the Keap1-Nrf2 pathway. Trends Pharmacol Sci 34(6):340–346. 10.1016/j.tips.2013.04.005 [DOI] [PubMed] [Google Scholar]
- Takagi T, Itabashi Y (1981) Occurrence of mixtures of geomet-ricalisomers of conjugated octadecatrienoic acids in some seed oils: analysis by open-tubular gas liquid chromatography and high performance liquid chromatography. Lipids 16(7):546–551 [Google Scholar]
- Trostchansky A, Rubbo H (2008) Nitrated fatty acids: mechanisms offormation, chemical characterization, and biological properties. Free Radic Biol Med 44:1887–1896. 10.1016/j.freeradbiomed.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Turell L, Radi R, Alvarez B (2013) The thiol pool in human plasma: thecentral contribution of albumin to redox processes. Free RadicalBiol Med 65:244–253. 10.1016/j.freeradbiomed.2013.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turell L, Vitturi DA, Coitiño EL, Lebrato L, Möller MN, Sagasti C, Salvatore SR, Woodcock SR, Alvarez B, Schopfer FJ (2017) Thechemical basis of thiol addition to nitro-conjugated linoleic acid, a protective cell-signaling lipid. J Biol Chem 292(4):1145–1159. 10.1074/jbc.M116.756288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbancsok J, Bones AM, Kissen R (2017) Glucosinolate-derived isothiocyanates inhibit Arabidopsis growth and the potency dependson their side chain structure. Int J Mol Sci 18(11):2372. 10.3390/ijms18112372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbancsok J, Bones AM, Kissen R (2018) Arabidopsis mutantsimpaired in glutathione biosynthesis exhibit higher sensitivitytowards the glucosinolate hydrolysis product allyl-isothiocyanate. Sci Rep 8(1):1–13. 10.1038/s41598-018-28099-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vliet A, Eiserich JP, Halliwell B, Cross CE (1997) Formationof reactive nitrogen species during peroxidase-catalyzed oxidation of nitrite. A potential additional mechanism of nitric oxide-dependent toxicity. J Biol Chem 272(12):7617–7625. 10.1074/jbc.272.12.7617 [DOI] [PubMed] [Google Scholar]
- Villacorta L, Minarrieta L, Salvatore SR, Khoo NK, Rom O, Gao Z, Berman RC, Jobbagy S, Li L, Woodcock SR, Chen YE, Freeman BA, Ferreira AM, Schopfer FJ, Vitturi D (2018) In situ generation, metabolism and immunomodulatory signaling actions ofnitro-conjugated linoleic acid in a murine model of inflammation. Redox Biol 15:522–531. 10.1016/j.redox.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitturi DA, Chen CS, Woodcock SR, Salvatore SR, Bonacci G, Koenitzer JR, Stewart NA, Wakabayashi N, Kensler TW, Freeman BA, Schopfer FJ (2013) Modulation of nitro-fatty acid signaling: prostaglandin reductase-1 is a nitroalkene reductase. J Biol Chem 288(35):25626–25637. 10.1074/jbc.M113.486282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitturi DA, Minarrieta L, Salvatore SR, Postlethwait EM, Fazzari M, Ferrer-Sueta G, Lancaster JR, Freeman BA, Schopfer FJ (2015) Convergence of biological nitration and nitrosation via symmetrical nitrous anhydride. Nat Chem Biol 11(7):504–510. 10.1038/nchembio.1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollár M, Feigl G, Oláh D, Horváth A, Molnár Á, Kúsz N, Ördög A,Csupor D, Kolbert Z (2020) Nitro-oleic acid in seeds and differently developed seedlings of Brassica napus L. Plants (basel,Switzerland) 9(3):406. 10.3390/plants9030406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers J, Dong X (2016) Posttranslational modifications of NPR1: asingle protein playing multiple roles in plant immunity and physiology. PLoS Pathog 12(8):e1005707. 10.1371/journal.ppat.1005707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Chi Y, Jiang Z et al. (2020) Hydrogen peroxide sensor HPCA1 isan LRR receptor kinase in Arabidopsis. Nature 578(7796):577–581. 10.1038/s41586-020-2032-3 [DOI] [PubMed] [Google Scholar]
- Yang H, Mu J, Chen L, Feng J, Hu J, Li L, Zhou JM, Zuo J (2015) S-nitrosylation positively regulates ascorbate peroxidase activityduring plant stress responses. Plant Physiol 167(4):1604–1615. 10.1104/pp.114.255216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun BW, Feechan A, Yin M, Saidi NB, Le Bihan T, Yu M, Moore JW, Kang JG, Kwon E, Spoel SH, Pallas JA, Loake GJ (2011) S-nitrosylation of NADPH oxidase regulates cell death in plantimmunity. Nature 478:264–268 [DOI] [PubMed] [Google Scholar]
- Zhang H, Fang Q, Zhang Z, Wang Y, Zheng X (2009) The role ofrespiratory burst oxidase homologues in elicitor-induced stomatal closure and hypersensitive response in Nicotiana benthamiana. JExp Bot 60(11):3109–3122. 10.1093/jxb/erp146 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study arealready published. Data sharing does not apply to this article as nodatasets were generated during the current study. The comparison ofthe two different already published RNAseq is, however, presented asa supplemental table and available to the readers.


