Abstract
Cytochrome c oxidase (CcO) is part of the respiratory chain and contributes to the electrochemical membrane gradient in mitochondria as well as in many bacteria, as it uses the energy released in the reduction of oxygen to pump protons across an energy-transducing biological membrane. Here, we use time-resolved serial femtosecond crystallography to study the structural response of the active site upon flash photolysis of carbon monoxide (CO) from the reduced heme a3 of ba3-type CcO. In contrast with the aa3-type enzyme, our data show how CO is stabilized on CuB through interactions with a transiently ordered water molecule. These results offer a structural explanation for the extended lifetime of the CuB-CO complex in ba3-type CcO and, by extension, the extremely high oxygen affinity of the enzyme.
Time-resolved serial crystallography studies display active site dynamics of cytochrome c oxidase.
INTRODUCTION
Cellular respiration is the process in all living cells where high-energy electrons from the breakdown of food are transferred through a chain of redox-active transmembrane protein complexes from low-potential donors to high-potential acceptors. The energy released in this process is used to translocate protons from the negative (N-) side of the membrane to the positive (P-) side, thereby maintaining a proton gradient across an energy-transducing biological membrane. This gradient supplies energy for various processes in the cell including the generation of adenosine triphosphate. The terminal protein complex of the respiratory chain in mitochondria as well as in many bacteria is cytochrome c oxidase (CcO), where the final electron acceptor oxygen is reduced to water. In this reaction, four electrons are transferred from the P-side of the membrane and four protons from the N-side of the membrane to the heme a3-CuB binuclear center that makes up the active site, where one molecule of oxygen is reduced to two molecules of water. At the same time, additional protons are translocated through CcO from the N- to the P-side of the membrane, contributing further to the transmembrane proton gradient (1–3).
The CcO enzyme family is classified into three classes on the basis of cofactor composition and proton conducting pathways—the A-, B, and C-type CcOs. The A-type enzymes include the mitochondrial and bacterial aa3-type CcOs that contain a low-spin heme a that feeds electrons to the binuclear center. Protons for the oxygen chemistry as well as protons to be pumped are transferred through two proton-conducting pathways termed the D- and the K-pathway (4–7). In aa3-type bovine CcO, an additional third proton pathway, the H-pathway, has been suggested to play a role in the transfer of pumped protons (8), although the H-pathway has been shown not to be relevant for proton pumping in other aa3-type CcOs (9–11). For each O2 molecule reduced, the aa3-type CcOs translocate four protons across the membrane, one for each electron that is transferred to the active site. The ba3-type CcO from the thermophilic bacterium Thermus thermophilus is the most studied of the B-type enzymes. Because of the very high oxygen affinity of the enzyme, the host bacterium is able to thrive under low-oxygen conditions (12, 13). It contains a low-spin heme b in place of heme a as the intermediate electron acceptor, and only one proton-conducting pathway has been identified, referred to as the K-pathway analog as it overlaps in space with the K-pathway of the A-type CcOs. It has been suggested that ba3-type CcO pumps protons with a lower stoichiometry compared to the aa3-type CcOs, with ~0.5 protons per electron transferred to oxygen (14, 15), although this observation may be explained by a higher sensitivity of the enzyme to back leakage of protons driven by the proton motive force (16).
Although extremely well studied using a range of spectroscopic, structural, and computational methods [see for example (17–28)], the detailed mechanism by which CcO couples the reduction of oxygen to proton pumping remains elusive. One promising approach to determine the transient structural changes that control proton pumping is time-resolved serial femtosecond crystallography (TR-SFX) (29, 30). Serial femtosecond crystallography (SFX) was developed at x-ray free electron lasers (XFELs), which are x-ray facilities that produce extremely high-intensity x-ray pulses of a few femtoseconds in duration. In an SFX experiment, x-ray diffraction data are collected as a continuous stream of micrometer-sized protein crystals are passed through the x-ray beam. When an x-ray pulse intersects a microcrystal at random orientation, this gives rise to one diffraction image. Many thousands of diffraction images are collected, and those containing diffraction spots are identified, indexed, and merged into a complete dataset (31). SFX allows protein structures to be studied at room temperature and free of x-ray radiation induced damage, something that is of particular importance for metal-containing proteins (32, 33). Moreover, TR-SFX makes it possible to collect x-ray diffraction data at a chosen time delay after reaction initiation and thereby follow the detailed structural rearrangements taking place within a protein during a catalytic reaction in real time (34–38) [for recent reviews, see (39, 40)].
During the last few years, the first results from SFX studies of CcO have been reported. The SFX structure of oxidized ba3-type CcO has been solved (41), displaying a single-atom species in the active site as opposed to the previously suggested peroxide ligand (42). A room temperature SFX structure has also been presented of bovine (aa3-type) CcO with a carbon monoxide (CO) ligand bound in the active site (43). In an attempt to capture an intermediate state of the oxygen reaction cycle, SFX data were collected 8 s after mixing reduced crystals of bovine CcO with an O2 saturated buffer using a mixing jet. This resulted in a structure interpreted as a ferryl intermediate, the so-called PR state of the enzyme, based on spectroscopic data and structural refinement of the CuB and heme a3 Fe ligand distances. This interpretation, however, requires that the reaction was slowed almost a million-fold, from tens of microseconds in solution to several seconds in the crystalline form (44).
Photodissociation of CO bound to heme a3 of CcO is commonly used to probe the dynamics of the active site of the enzyme, because CO rebinding to the binuclear center is thought to mimic O2 binding to the active site. After CO is dislodged from heme a3 by a short laser flash, it has been shown to transiently bind to CuB on a femtosecond timescale. In the case of bovine aa3-type CcO, the CuB─CO bond is broken on a microsecond timescale whereupon CO leaves the active site, and later recombines with heme a3 on a timescale of ~10 ms (45, 46). By contrast, the CO response is drastically different in the case of ba3-type CcO. Spectroscopic evidence suggests that the CO molecule remains bound to CuB for ~30 to 50 ms and then returns to heme a3 without leaving the active site pocket (18, 47). The structural basis for this extended lifetime of the CuB-CO species is not understood.
An XFEL-based time-resolved x-ray diffraction study of structural changes associated with photodissociation of CO from the active site of bovine CcO has been reported. Difference electron density maps were recorded 20 ns and 100 μs after flash photolysis of the CO ligand from the reduced active site by translating larger crystals, cooled to 4°C, between each x-ray pulse (48). The data displayed that the CO molecule was transferred from heme a3 to CuB within 20 ns and further released from the active site by 100 μs, in agreement with previous time-resolved infrared spectroscopy studies in solution (45, 46).
Here, we present TR-SFX results after photodissociation of CO from reduced CO-bound ba3-type CcO. Our data reveal that 2 ms after flash photolysis, the heme a3─CO bond is broken and the CO molecule is instead coordinating CuB. Notably, the CO movement is coordinated with the transient appearance of a water molecule within the active site that likely stabilizes the CuB─CO bond, thereby explaining the longer lifetime of the CuB─CO bound state in the ba3-type enzyme. Our results thus highlight how the enzyme’s structural response upon CO disassociation from heme a3 in ba3-type CcO is distinct from that of the bovine aa3-type enzyme.
RESULTS
Room-temperature reduced CO-bound structure of ba3-type CcO
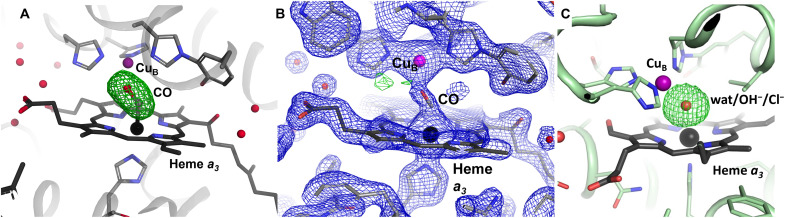
X-ray diffraction data were collected from the reduced CO-bound CcO enzyme without laser illumination (the so-called dark state) to a resolution of 2.0 Å. The data give rise to an elongated electron density in the unbiased Fo-Fc map located in the active site (Fig. 1A), into which a carbon monoxide molecule coordinating with the heme a3 Fe can be accurately modeled (Fig. 1B). As a comparison, the oxidized SFX structure displays a spherical Fo-Fc electron density map feature corresponding to a single-atom species, interpreted as a water molecule or hydroxide ion bound in the active site (Fig. 1C) (41), although a heavier atom such as chloride remains a possibility (49). With carbon monoxide bound to the active site, the Fe-Cu distance is 4.9 Å, which is slightly longer than the distance of 4.8 Å for the oxidized SFX structure of ba3-type CcO. The reduced-carbon monoxide bound state of the enzyme is also confirmed by the optical absorbance spectrum of the prepared microcrystals (fig. S1).
Fig. 1. Active site structure of ba3–type CcO in the reduced CO-bound and oxidized states from SFX data.
(A) The positive Fo-Fc omit map density (green), calculated without the active site ligand, contoured at 4.5 σ for the reduced CO-bound structure. (B) The reduced CO-bound structure with a CO molecule modeled into the active site. The 2Fo-Fc electron density (blue) is contoured at 2 σ and the Fo-Fc electron density (positive in green, negative in red) is contoured at 3.5 σ around the active site ligand. (C) The positive Fo-Fc omit map density (green), calculated without the active site ligand, contoured at 4.5 σ for the oxidized structure (PDB ID 5NDC).
The reduced CO-bound SFX structure can be compared with a previously solved structure of reduced CO-bound ba3-type CcO using synchrotron radiation at cryogenic temperature (50) [Protein Data Bank (PDB) ID 3QJQ, 2.9 Å resolution]. In that case, the Fe-Cu distance is longer at 5.1 Å, and the CO molecule is modeled in a conformation that is less perpendicular with the heme plane (with a Fe-C-O angle of 125.9° compared to 143.2° for the SFX structure; Fig. 2A). However, a Fo-Fc electron density map calculated for the cryo structure without the carbon monoxide in the active site displays a somewhat elongated electron density that stretches rather straight between the Fe and Cu atoms, and there is scope for modeling the CO molecule in alternative conformations. It may be that the binding position of the ligand has been compromised by the x-ray exposure during the experiment, because synchrotron radiation studies are susceptible to radiation-induced damage, whereas with an XFEL the femtosecond x-ray pulses are too short for such chemically induced changes to have time to evolve (32, 33, 35, 51).
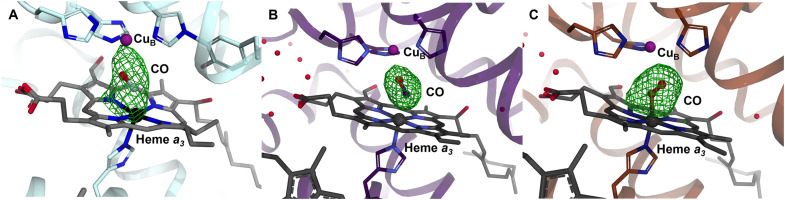
Fig. 2. Active site structure of ba3–type and bovine CcOs in the reduced CO-bound state from synchrotron and XFEL data.
(A) Fo-Fc omit map for the reduced ba3-type CcO-CO structure solved from synchrotron data collected at cryo temperature (PDB ID 3QJQ). (B) Fo-Fc omit map for the reduced bovine CcO-CO structure solved using XFEL data collected at 5°C from larger crystals that were translated between each exposure (PDB ID 5X1F). (C) Fo-Fc omit map for the reduced bovine CcO-CO structure solved using SFX data collected at room temperature (PDB ID 5 W97). The positive Fo-Fc omit map densities (green), calculated without CO modeled in the active sites, are contoured at 4.0 σ.
Two structures of bovine CcO in the reduced CO-bound form have also been solved using XFEL radiation. In one experiment, x-ray diffraction data were collected at Spring-8 Angstrom Compact Free Electron Laser (SACLA) at 5°C on a number of larger crystals that were translated in space after each exposure so as to avoid multiple exposures at the same position of each crystal (48) (PDB ID 5X1F, 2.2 Å resolution). In another study, SFX was used to collect a dataset from microcrystals of reduced CO-bound bovine CcO (43) (PDB ID 5 W97, 2.3 Å resolution). Although the Fe-Cu distance is identical at 5.3 Å in both structures, the CO molecule is modeled in very different conformations (Fig. 2, B and C), with that recovered from lager crystals being more consistent with our observations in the ba3-type enzyme (Fig. 1A). The Fo-Fc electron density map of the active site without the CO molecule modeled is somewhat elongated in the case of the 5X1F structure (Fig. 2B), similar to what we observe (Fig. 1A), but appears more spherical in the 5W97 structure (Fig. 2C). In the case of the 5X1F (XFEL) structure, on-line static visible absorption spectra were measured before and after x-ray diffraction data collection to confirm the state of the enzyme in the crystals (48).
Structural response of the active site upon CO flash photolysis
Time-resolved x-ray diffraction data were recorded 2 ms after a pump laser pulse was used to photodissociate CO from heme a3 in the active site of ba3-type CcO. In these experiments, an lipidic cubic phase (LCP) microjet delivered microcrystals across the XFEL beam as a continuous stream, and for every third XFEL pulse (i.e., the data-collection sequence was dark, dark, laser-on, with an XFEL repetition rate of 30 Hz) microcrystals were illuminated using a 5-ns laser pulse of 532 nm in wavelength containing 200 μJ per pulse and focused into a full-width half-maximum (FWHM) spot of 80 μm. This corresponds nominally to a mean fluence F = 2000 mJ cm−2 across the FWHM, which gives an average value of F ‧ σ/hν = 242 throughout a hypothetical 20-μm-thick microcrystal after adjusting for its optical density, where σ532nm is the absorption cross section of the resting state given by σ = ɛ ‧ 1000 ‧ ln(10)/A, where the extinction coefficient ɛ532nm = 13,000 Mol−1 cm−1, A = 6.02 × 1023 Mol−1 is Avogadro’s number, and hν is Planck’s constant multiplied by the pump laser frequency (52). This ratio provides an upper limit for how many photons are absorbed per protein molecule if one assumes that the optical properties of the sample are ideal, that there are no scattering losses, and that all accessible states have the same cross section as the resting state. When studying ultrafast structural dynamics in biomolecules, it has been argued that single-photon excitation is desirable (52–54), but this goal has yet to be achieved in practice (40). Because the photodissociation of CO from a heme group is a subpicosecond process (55) and the pump laser pulse duration used in the experiment was 5 ns, the photodissociation event will be single photon but excess photons are absorbed by the heme groups and will further heat the protein. From this perspective, F ‧ σ/hν = 242 implies a temperature jump of ~42 K, where ΔT = 10−3 × F ‧ σ ‧ A ‧ [CcO]/C; C is the heat capacity of the LCP grown crystal and is estimated to be ≈ 3.8 J ml−1 (40) and the protein concentration in crystals [CcO] = 0.0029 Mol. However, the experimental data do not suggest that a large temperature jump was induced in the microcrystals. The unit cell parameters of the dark-only, dark1, dark2, and light datasets do not indicate any temperature-induced expansion (fig. S2). Moreover, the statistics for the dark and illuminated datasets are similar (e.g., the indexing rate is 13.4% for dark and 11.1% for laser-on; table S1), and small differences in other crystallographic parameters may arise from the dark dataset being twice as large as the illuminated dataset. Because the XFEL beam was not attenuated, the LCP microjet was vaporized with each and every XFEL pulse (56) and the light-scattering properties of the LCP microjet will be affected by this process, potentially lowering the product F ‧ σ/hν by an unknown extent. Moreover, a model for the cooling of a cylinder when fit to measured temperature changes from protein sample in solution (57) and extrapolated to a 75-μm LCP microjet suggests that the sample has cooled substantially by Δt = 2 ms, and this estimate is also supported by other arguments (58). Our structural results agree very well with previous spectroscopic results that show the transient binding of CO to CuB on a millisecond timescale (18).
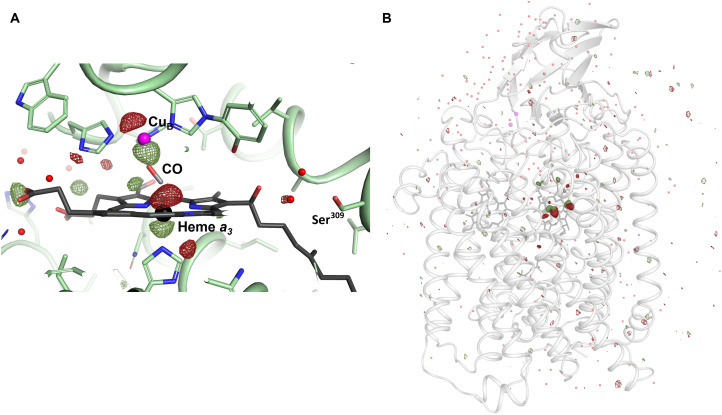
To visualize the major differences between the dark-state (reduced CO-bound, no interleaved light images) structure and the photoactivated-state (laser-on) structure, the Fo(2 ms) − Fo(dark) isomorphous difference Fourier electron density map was calculated (Fig. 3, A and B). The difference electron density map shows strong negative and positive features above and below CuB (Fig. 3A), corresponding to a movement of CuB toward heme a3. This motion of CuB is consistent with what was observed in time-resolved x-ray diffraction studies of bovine CcO upon CO flash photolysis (48). Similar negative and positive features are visible on either side of the heme a3 Fe and indicate an out-of-plane movement of the Fe toward the proximal side of the heme. An out-of-plane bending is associated with the reduced unliganded form of heme a3 (59). Moreover, a weaker positive electron density feature is visible adjacent to the CO ligand, indicating a partial migration of CO away from the heme a3 Fe. Very few additional strong difference electron density features were visible in the Fo(2 ms) − Fo(dark) difference map (Fig. 3B), establishing that the light-induced structural changes were localized around the active site of the enzyme.
Fig. 3. Isomorphous difference Fourier electron density map.
Fo(2 ms) − Fo(dark) isomorphous difference Fourier electron density map displaying differences between the dark (reduced CO-bound) state and the activated (light) state, contoured at 3.5 σ. Positive density is shown in dark green and negative density in dark red. (A) Zoomed-in view of the heme a3–CuB active site. (B) Zoomed-out view of the protein.
The extrapolated structure-factor amplitudes were calculated by extrapolating to 100% occupancy of the photoactivated state (60), which avoids phase bias toward the photoactivated state model by assuming that there is no phase difference between the dark- and photoactivated-state structure factors. To determine the fraction activated state, the extrapolation of light data was iterated from 10 to 95% occupancy in steps of 5% using the method by Genick (60), followed by 20 cycles of rigid body refinement. On the basis of visual inspection of the resulting electron density maps, it was concluded that the laser-on data correspond to approximately 30% photodissociated state and 70% dark state. The resulting Fext-Fc map (Fig. 4A) was used to guide initial placement of the CO molecule in the photoactivated state. Partial-occupancy refinement was used to model the detailed structural response upon light activation, with selected active site cofactors and residues [where the strongest features of the isomorphous Fo(2 ms) − Fo(dark) difference Fourier electron density map were seen] allowed to adopt a second conformation with 30% occupancy, and the dark-state structural model was held fixed with 70% occupancy. During structural refinement, CO-CuB and CO–water 197 distances were restrained to keep the CO molecule in a chemically reasonable position. The approximate position of the CO molecule in the photoactivated state was confirmed by the Fo-Fc map calculated with CO modeled in the dark-state conformation at 70% occupancy but without the photoactivated-state CO molecule present (fig. S3A). From this structural analysis, we conclude that 2 ms after laser irradiation, the CO molecules has moved from a position coordinating with the heme a3 Fe in the dark state to a conformation that is almost coplanar with the heme plane and with the carbon atom of CO coordinating to CuB (Fig. 4B), although the exact conformation of the CO molecule cannot be determined from the data. The refinement statistics are detailed in table S1.
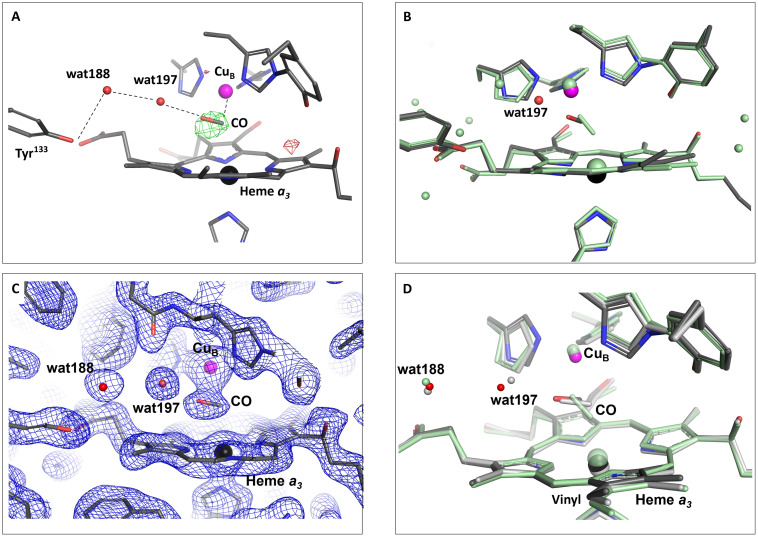
Fig. 4. Comparison of reduced-CO bound state with activated state of ba3-type CcO.
(A) Activated-state structure obtained from partial occupancy refinement with the extrapolated difference electron density map indicating the position of the CO molecule after flash photolysis. The positive (green) and negative (red) Fext-Fc electron density map, calculated using the extrapolated structure factors without water molecules and active site ligand included in the model, are displayed at 4 σ. The link between the CO molecule and Tyr133 is indicated. (B) The dark-state structure (green) overlayed on the 2-ms activated-state structure from partial occupancy refinement (protein, gray; heme a3 iron, black; CuB, purple; new water molecule, red). (C) Activated-state structure refined using the extrapolated structure factors. The 2Fext-Fc electron density map is displayed at 1.5 σ. (D) The dark-state structure (green) and the activated-state structure refined using extrapolated structure factors (light gray) overlayed on the 2-ms activated-state structure from partial occupancy refinement (protein, dark gray; heme a3 iron, black; CuB, purple; water molecules, red).
As a result of this reconfiguration of the active site ligand, both CuB and the heme a3 Fe move in tandem, and the Fe-Cu separation appears to remain at 4.9 Å in the photoactivated state (see further discussion below). This can be compared with structural models of bovine CcO where the Fe-Cu distance is slightly reduced upon photodissociation of CO (43, 48, 61). In an x-ray diffraction study using synchrotron light at cryogenic temperature, lower-resolution structures of reduced ba3-type CcO with CO bound to the heme a3 Fe before illumination, and coordinating with CuB after illumination, indicated a decrease in the Fe-Cu distance from 5.1 to 4.7 Å upon illumination (50), which is not supported by the data presented here. However, infrared spectroscopy measurements of the illuminated crystals showed evidence of x-ray–induced radiation damage that may have compromised the structure of the active site (50).
Moreover, we observe differences in the active site water structure upon laser irradiation. Specifically, in the dark-state reduced CO-bound structure, a water molecule (wat188) that is not present in the oxidized ba3-type CcO structure resides within hydrogen bonding distance of Tyr133. In the photoactivated state structure, the migration of CO from the heme a3 Fe to CuB is accompanied by further ordering of this water together with the ordering of an additional water molecule (wat197, which is present in the dark-state structure at very low, <10%, occupancy) that creates a link between Tyr133 and the CO molecule when bound to CuB (Fig. 4A). Thus, the altered active site water structure acts to stabilize the CuB-CO complex.
As an alternative approach to determine the structural response upon light activation, the extrapolated structure factors were calculated through the program package Xtrapol8 with the Fgenick method (62). Using Xtrapol8, the occupancy was determined to 31% compared to the previously determined occupancy of 30% based on manual inspection of the maps. The structure factors extrapolated to 100% occupancy were then used to refine the structure corresponding to the photoactivated state with REFMAC5 (63), resulting in stable R-factors (table S1) and a good quality electron density map (Fig. 4C). The main features of the activated-state structure agree well independent of whether partial occupancy or extrapolated structure factor refinement was used (Fig. 4D and fig. S4). In particular, the breaking of the heme a3 Fe─CO bond and the translation of the CO from Fe toward CuB is clear in both refined activated-state structures. Moreover, the ordering of wat188 and the appearance of the additional water molecule wat197 that creates a link between the CO molecule and Tyr133 is evident also in the extrapolated map. The refined position of the CO molecule is shifted slightly away from the two water molecules in the structure refined using extrapolated structure factors and does not overlap completely with that from partial occupancy refinement (Fig. 4D and fig. S4), although the exact orientation of the CO molecule is difficult to determine from either of the refined maps. A difference between the partial occupancy refined structural model and the model from extrapolated structure factors is that in the first case, CuB moves closer to the heme group, whereas in the second case it remains closer to the dark-state position. This results in a Fe-Cu distance of 4.9 Å for the partial occupancy refined structure and 5.1 Å for the structure refined using extrapolated structure factors, although this difference may fall within coordinate errors. The strong negative and positive features in the Fo(2 ms) − Fo(dark) isomorphous difference Fourier electron density map above and below CuB (see Fig. 3A) support a movement of CuB toward heme a3 and thus a shorter Fe-Cu distance. The CO carbon atom coordination to CuB is 2.0 Å in the partial occupancy refined structure and 2.4 Å in structure refined using extrapolated structure factors (fig. S4), as a result of the different positions of CuB in the two structures and the slight variation in position of the CO molecule.
Additional structural changes associated with CO binding to heme a3 Fe and migration toward CuB
Overall, the structure of reduced-CO bound CcO is very similar to the oxidized CcO structure previously solved using SFX (41) (PDB ID 5NDC) with a Cα-atom root mean square deviation of 0.14 Å. There are only subtle differences between the two room-temperature SFX structures considering amino acid conformations of the proton translocation pathway, the K-pathway analog. There is, however, an ordering of a water molecule in the reduced-CO bound structure within hydrogen-bonding distance of the heme a3 hydroxyl farnesyl alcohol, Ser309, and the main chain carbonyl oxygen of Val305 near the active site at the end of the proton pathway. This water position has only low occupancy in the oxidized structure and was not previously modeled. The dynamics of the proton pathway and the fact that it is influenced by the state of the active site are further manifested by the observation of a weak negative Fo(2 ms) − Fo(dark) difference Fourier electron density map feature (Fig. 3A) on a neighboring atomic position between the hydroxyl farnesyl alcohol and Ser309.
We observe only minor structural differences upon comparing the reduced CO-bound CcO structure presented here with the reduced structure collected using synchrotron radiation at cryogenic temperature (PDB ID 3EH5, 2.8-Å resolution). These are mainly located around the heme a3 propionates and the associated water cluster where additional water molecules are modeled in the SFX structures, likely because of the higher resolution. Care should be taken not to overinterpret structural differences upon comparing models resulting from x-ray diffraction data collected at room temperature with those from cryogenic temperature. In the photoactivated state structure, we observe that CO migration toward CuB triggers a twisting motion of the A-ring propionate, slightly increasing the interaction between the propionate and Asp372. The twisting of the A-ring propionate is observed also in the structure refined using extrapolated structure factors, although in this case the distance to Asp372 remains the same as in the dark state. This structural region is proposed to harbor the so-called proton-loading site, a protonatable group with alternating access to the N- and P-sides of the membrane during the catalytic cycle that is key for proton pumping (41, 64). In addition, the partial occupancy refinement results suggest that a minor twist of the heme a3 C-ring vinyl group occurs upon light activation. This conformational change is not visible in the structure refined using extrapolated structure factors, however (Fig. 4D). A twisting motion of the vinyl group has previously been suggested to drive conformational changes related to proton pumping in bovine CcO (43, 48, 61). On the basis of the lack of notable features in the Fo(2 ms) − Fo(dark) difference electron density map (Fig. 3B), a potential twist of the heme a3 C-ring vinyl does not appear to trigger further conformational changes in the structural region between heme a3 and heme b.
DISCUSSION
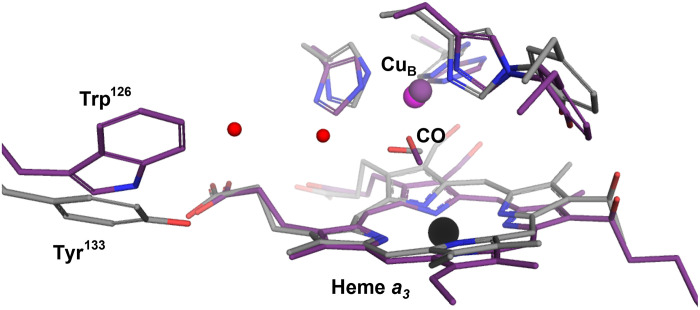
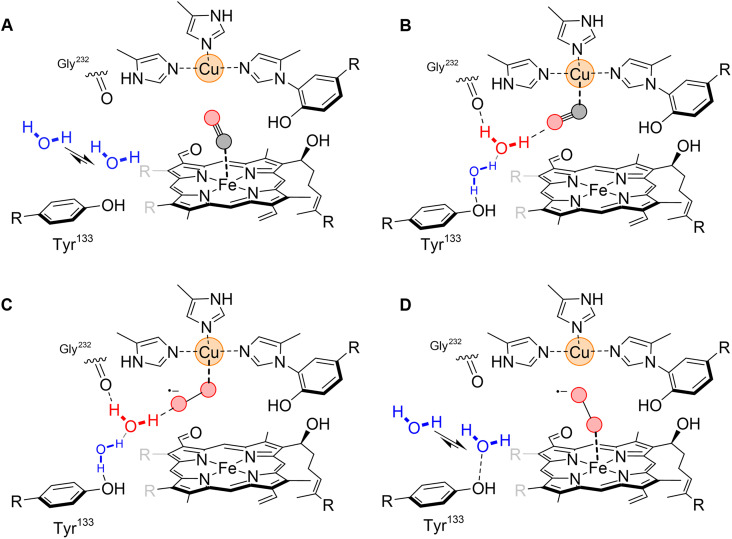
The ba3-type CcO differs from the aa3-type CcOs, including the bovine enzyme, in its affinity for oxygen. This functional difference relates to the physiological requirements of the ba3-type CcO as the expression of this enzyme allows the thermophilic bacteria T. thermophilus to thrive under low-oxygen conditions (12, 13, 65). O2 binding to the active site of ba3-type CcO is 10 times faster than to the aa3-type bovine CcO, with rates of 109 and 108 M−1 s−1, respectively, in the absence of CO blocking the active site (66). This has been attributed to structural differences around the active site, including a tryptophan residue in the aa3-type enzymes (Trp126 in bovine CcO) that restricts access through the oxygen channel to the active site, which is replaced by a tyrosine (Tyr133) in the ba3-type enzyme (Fig. 5). The importance of this difference was nicely confirmed in a time-resolved optical absorption spectroscopy study of a ba3-type CcO variant in which Tyr133 was mutated to a tryptophan residue, resulting in five times slower ligand binding as compared to the wild-type enzyme (67). It is generally believed that CO binding to the binuclear center triggers a similar structural response as binding of the natural ligand oxygen. The TR-SFX data presented here illustrate how the active site water structure of ba3-type CcO responds to the migration of CO from heme a3 to CuB. The CO transfer is associated with the transient ordering of a water molecule that creates a hydrogen-bond network between the CO molecule and Tyr133 (Fig. 6, A and B). Thus, our results show that the tyrosine residue, in addition to allowing fast access of oxygen to the active site, also has a role in stabilizing the CuB coordinated CO molecule via two water molecules, which is manifested as a longer lifetime of the CuB-CO complex in the ba3-type CcO (18, 47). Our structural data thus suggest how the natural substrate oxygen is efficiently trapped in the active site through a stabilized CuB interaction (Fig. 6, C and D) and thereby offers a structural rational for the high O2 affinity of the ba3-type CcO compared to the aa3-type enzymes. On the basis of transient visible absorbance spectroscopy and resonance Raman measurements, CuB has previously been suggested to act as a docking site for oxygen in the ba3-type CcO (68). A water molecule is present at a similar position to wat197 in the structure of oxidized ba3-type CcO solved at cryo-temperature (42), but in that case, it was observed to be hydrogen bonding with the peroxide molecule modeled in the active site, again illustrating how a water molecule in this position may act to stabilize different active site ligands. This water position is not occupied in any of the bovine CcO structures solved to date.
Fig. 5. Structural comparison of ba3-type and bovine CcO.
ba3-type CcO structure (gray) in the activated state with CO coordinating with CuB overlayed on the corresponding bovine CcO structure (PDB ID 5X1B, purple) collected 20 ns after flash photolysis. Water molecules of the ba3-type CcO are shown in red.
Fig. 6. Proposed model for stabilization of CO and O2 by CuB.
(A) Schematic illustration of the CO ligating to the ferrous haem a3. A water molecule (blue) close to Tyr133 is in equilibrium between two different positions. (B) Upon photodissociation of the carbon monoxide, CO moves closer to CuB. This is accompanied by the appearance of an additional water molecule (red) in the active site and further ordering of the first water molecule (blue). The cuprous carbonyl complex is stabilized by a hydrogen-bond interaction with the second water molecule (red), explaining the high CO affinity of CuB in the ba3-type CcO. (C) Transient binding of O2 to CuB. The stabilizing hydrogen bond between the water molecule (red) and oxygen would be even stronger in the case of the cupric superoxide than for CO, effectively trapping oxygen in the active site. (D) O2 bound to heme a3. Porphyrin and carbon hydrogens have been omitted for clarity.
It has been suggested that the H-proton pathway in combination with the proton storage site above the heme a3 propionates and a hydrogen-bond network connecting the two make up the proton pumping machinery in bovine CcO (8). Specifically, the proposed proton pumping mechanism involves a switch between the so-called closed structure, (observed in structures of the oxidized state of the enzyme) and the so-called open structure (observed in structures of the reduced enzyme). In the open structure, the water channel that is part of the H-proton pathway is open and allows protons to be transferred through the pathway. The main conclusion drawn from the time-resolved XFEL study of bovine CcO, where x-ray diffraction data were collected at 20 ns and 100 μm after flash photolysis of the CO ligand from the reduced active site using larger crystals cooled to 5°C, was that CO release from CuB drives the transition from the closed structure to the open structure (48). Similarly, x-ray diffraction structures resulting from cryo-temperature trapping experiments targeting the O, E, P, and F intermediates of bovine CcO are interpreted in terms of fraction open and closed structures (69–71). Conversely, our TR-SFX data collected 2 ms after flash photolysis of CO from ba3-type CcO do not suggest any major conformational changes in the structural region between heme b and heme a3 as no notable features in the Fo(2 ms) − Fo(dark) isomorphous difference Fourier electron density map are observed outside of the active site region (Fig. 3B). Moreover, our activated-state structure refined using extrapolated structure factors does not indicate any noteworthy structural changes in this region.
In the study presented here, we show how the transient CuB-CO state of ba3-type CcO is stabilized by the ordering of a new water molecule in the active site. On the basis of this observation, we offer a structural rational for the longer lifetime of CuB-CO state in ba3-type compared with the aa3-type CcOs. TR-SFX experiments have also been performed where microcrystals of reduced bovine CcO were mixed with O2 saturated buffer 8 s before the crystals intercepted the x-ray beam. From the resulting electron density map, a structure was presented that was suggested to correspond to one of the reaction intermediate states (44). To deduce the detailed proton pumping mechanism of CcO, it is likely that other reaction triggering methods are required that allow diffraction data with higher temporal resolution to be collected that is out of reach using a simple mixing approach. With current developments of TR-SFX allowing a variety of reaction triggering approaches to be explored, it may become possible to advance from trapping studies at cryogenic temperature resulting in a mixture of different states (69–71) as well as mixing experiments with a time resolution of seconds (44) to obtain truly time-resolved structural information describing the enzymatic reduction of molecular oxygen to water by CcO.
METHODS
Protein expression and purification
Production of recombinant ba3-type CcO was performed as previously described (41) with the following modifications. Ni–nitrilotriacetic acid purification was followed by a one-step dialysis in 5 mM Hepes, 0.05% dodecyl-β-d-maltoside (DDM) (pH 8.0) at 4°C overnight. The protein was aliquoted in Eppendorf tubes and spun down at maximum speed using a microcentrifuge (Eppendorf 5424R) for 30 to 40 min to remove precipitated protein. The protein was then loaded onto a prepacked anion exchange column (HiPrep DEAE FF 16/10 GE Healthcare Life Science) equilibrated with 20 mM tris-HCl and 0.05% DDM (pH 7.6). The protein was washed with 120 ml of buffer containing 20 mM tris-HCl and 0.05% DDM (pH 7.6) followed by 40 ml of 20 mM tris-HCl, 0.05% DDM, and 12 mM NaCl (pH 7.6) and subsequently eluted with 20 mM tris-HCl, 0.05% DDM, and 50 mM NaCl (pH 7). Protein purity was estimated by absorbance spectroscopy where the oxidized heme b absorbance peak was compared with the total protein absorbance (Abs414/Abs280). If the ratio did not reach ≥0.70 after the anion-exchange purification step, size exclusion chromatography was performed (Superdex 200 Increase 10/300 GL, GE Healthcare) using the anion exchange elution buffer above. The protein was concentrated to 20 to 25 mg/ml and stored at 4°C until crystallization.
Crystallization
ba3-type CcO protein at a concentration of 20 to 25 mg/ml was reduced with sodium dithionite to a final concentration of 5 mM and reconstituted in LCP. The previously described method for large-scale microcrystallization of LCP in plates (72) was used with the following modifications. Crystallizations were performed in 9-well glass plates (Hampton Research), 24-well TPP tissue culture plates, or 2-ml glass vials where 20-μl strings of LCP were added to 0.5- to 1-ml precipitant solution in each well or vial. The plates were covered with ClearVue plastic sheets (Molecular Dimensions), and the vials were sealed with a septum-containing lid. The crystallization procedure was performed in an anaerobic chamber (Coy Laboratory Products, USA) in an environment of 95 to 97% nitrogen gas and 3 to 5% hydrogen gas. All solutions including protein, reducing agent, and crystallization buffer were purged with nitrogen gas before entry into the anaerobic chamber. Plastic laboratory equipment was incubated in the chamber before use. Crystals of 10 to 20 μm in size in the longest dimension appeared within 2 days in 100 mM 2-morpholinoethanesulfonic acid, 34 to 38% polyethylene glycol, molecular weight 400 (v/v), and 1.4 M NaCl (pH 5.3) after incubation at 19° to 23°C in the anaerobic chamber.
After crystal formation was complete, additional sodium dithionite at a final concentration of 1 mM was added to the precipitant solution to ensure full reduction of the microcrystals. CO was bound to the reduced enzyme by pooling crystal-containing LCP into vials together with the precipitant solution and purging the sealed vials with CO gas for 3 to 4 min. The microcrystals were then incubated for 30 to 60 min under CO atmosphere. Subsequently, the microcrystals were packed in 500-μl Hamilton syringes as described in (72) within the anaerobic chamber, limiting the time of exposure to the N2/H gas to 1 to 2 min to ensure that CO remained bound to the enzyme.
Absorbance spectroscopy
The reduced CO-bound state of ba3-type CcO in microcrystals were prepared as described above. Ultraviolet-visible (UV-vis) spectroscopy of the crystal containing LCP was measured as described previously (41) with the following modifications. Five microliters of LCP microcrystals was added to a gas-tight CaF2 glass cell with a path length of 0.2 mm within the anaerobic chamber. The absorption spectrum of the reduced state of the enzyme displays a characteristic shoulder at 445 nm, which disappears upon binding of CO. The fact that the 445-nm shoulder is absent in the prepared microcrystals confirmed that the enzyme within the crystals was in the reduced CO-bound state (fig. S1). Using this method, it was established that the microcrystals were stable in the reduced CO-bound state for at least 5 days after being packed into gas-tight Hamilton syringes.
Sample preparation, injection, and data collection
Sample preparation and injector loading were performed in an anaerobic chamber at the SACLA. The microcrystals were homogenized and topped-up with CO by mixing the crystals with CO-saturated monoolein in a 56:8 ratio (microcrystals:monoolein). The microcrystals were loaded into a sample cartridge by a syringe adapter and spun down in a microcentrifuge for 10 s to deplete gas bubbles in the LCP. The loaded sample cartridge was assembled with a high-viscosity injector (34, 73) with a 75-μl-sized nozzle. The opening at the nozzle tip was sealed with a plug of vacuum grease, which prevented CO from diffusing out of the LCP-environment through the nozzle tip during mounting of the injector. The vacuum grease plug was extruded from the injector at the start of sample injection. The assembled injector was transferred to the beamline in an airtight heat-sealable plastic bag together with an oxygen indicator tablet, which confirmed that the anaerobic atmosphere in the bag was stable for several hours. The injector was mounted in a helium chamber that is part of the Diverse Application Platform for Hard X-ray diffractioN In SACLA (DAPHNIS) (74).
SFX diffraction data were collected at 7.5 keV with an x-ray repetition rate of 30 Hz and a pulse duration of <10 fs at the BL3 beamline at SACLA. The x-ray pulses were focused to 1.5 μm (height) by 1.3 μm (width) by Kirkpatrick-Baez mirrors, where a pulse energy of 95 μJ was estimated at the sample position. The microcrystals were injected into the x-ray beam using a high-performance liquid chromatography pump. The sample-to-detector distance was 50 mm, and diffraction images were collected using an eight-sensor module charge-coupled device detector (75). The average radiation dose within the exposed area of the crystal was calculated to 7 MGy using the program RADDOSE-XFEL (76). The dark (reduced CO-bound state) dataset was collected at a flow rate of 0.5 μl/min without interleaved light images.
To elucidate time-resolved changes associated with CO dissociation from heme a3 of ba3-type CcO, the sample was illuminated with a 5-ns laser pulse of 532-nm wavelength containing 200 μJ per pulse and focused into a spot of 80 μm (FWHM). This corresponds nominally to a mean fluence F = 2000 mJ cm−2 across the FWHM. Taking an absorbance cross section σ532 = 13,000 M−1 cm−1 gives the dimensionless product F ‧ σ/hν = 260, which is reduced to 242 if adjusted for the optical density of a typical 20-μm crystal ([CcO] = 2.9 mM, optical density ≈ 0.1). The 1/e penetration depth of the crystals by light of 532 nm is 115 μm. A time delay of 2 ms was set between the laser pulse and the x-ray probe. Data were collected at a flow rate of 1.5 μl/min with “dark1,” “dark2,” and “light” identifiers, where the crystals were illuminated every third XFEL pulse, until sufficient number of images were obtained. This flow rate corresponds to a distance of 200 μm between each x-ray pulse.
Data processing
The data were processed using the Cheetah software (77) using a pipeline adapted for SACLA (78), and images containing diffraction spots were selected for indexing and merging by CrystFEL (31). Datasets collected from different injector runs were analyzed for the correct geometry using a detector distance optimization script (79) in the program CrystFEL. Briefly, for each stream file, the SD of the unit-cell parameters was calculated for different detector distances and the most optimal sample-to-detector distance (corresponding to the lowest SD) was selected for each injector run. Last, based on the identifier, the optimized files were separated into dark-only (for resting state CO-bound ba3-type CcO, collected without interleaved light images), dark1, dark2, and light datasets, and corresponding stream files were catenated for further analysis. Of these four subsets, dark and light were considered for structure determination, refinement, and elucidating time-resolved structural changes. For the reduced CO-bound (dark-only) dataset, a total of 201,274 images were collected, 41,059 images were identified as containing diffraction spots, and 26,999 of these were successfully indexed and merged, giving an average crystal hit rate of 20.4% and an indexing rate of 13.4%. For the 2-ms time-delay (laser-on) dataset, ~97,500 images were collected, out of which 13,096 showed diffraction and 10,844 were indexable, hence the hit rate and indexing rate were ~13.4 and ~11.1%, respectively. Reflection intensities from the indexed images were merged and scaled using Partialator without partiality correction. Statistics from data collection and refinement are detailed in table S1.
Structure determination and refinement
The structures (dark and laser-on) were solved by molecular replacement with Phaser using the SFX structure of oxidized ba3-type CcO (PDB ID 5NDC) as a search model. Both models were built in Coot (80) and refined to a resolution of 2.0 Å in REFMAC5 (63), using an x-ray weighting term of 0.05 and anisotropic displacements with three rigid-body motion groups (see paragraph below for details on the laser-on model). The final Rwork/Rfree values were 20.1% / 22.5% for the CO-bound dark structure with 99.9% of the side chains within allowed Ramachandran regions. To elucidate the structural changes in the photoactivated state at 2 ms following pump-laser illumination, an isomorphous Fo(2 ms) − Fo(dark) difference Fourier electron density map was calculated at 2.0-Å resolution using the observed structure factor amplitudes and phases from the CO-bound dark state model (Fig. 3, A and B).
To determine the fraction activated state in the 2-ms data, the extrapolation of light data was iterated from 10 to 95% occupancy in steps of 5% using the method by Genick (60). The extrapolated structure factors Fext are calculated according to: Fext = α(Folight − Fodark) + Fodark, where α is the reciprocal of the occupancy value, and Folight and Fodark are the observed structure factors from the light-activated dataset and the dark-only dataset, respectively. This was followed by 20 cycles of rigid body refinement in REFMAC5 (63). An occupancy of 30% was estimated on the basis of visual inspection of the resulting 2Fext–Fc and the Fext–Fc maps, where an underestimation of the occupancy would result in a map displaying the resting conformation of the CO molecule. The Fext–Fc map displayed an approximate orientation of the CO molecule in the active site of the protein after flash photolysis (Fig. 4A). To model the photoactivated state, partial-occupancy refinement was used using Phenix (81) where the heme a3–CuB active site (excluding the heme farnesyl chain), surrounding residues (Tyr133, His233, Tyr237, His282, His283, and His384) and the CO molecule were selected [on the basis of the major features in the Fo(2 ms) − Fo(dark) difference Fourier map] and allowed to adopt a second conformation with 30% occupancy while the dark-state structure was held fixed. During structural refinement, CO-CuB and CO–wat197 distances in the photoactivated state were restrained at 2.05 and 2.6 Å, respectively, to keep the CO molecule in a chemically reasonable position. The approximate position of the CO molecule and wat197 in the photoactivated state was supported by the Fo(2 ms) − Fc map calculated with CO modeled in the dark-state confirmation at 70% occupancy but without the photoactivated state CO molecule or water molecule present (fig. S3A), or by only omitting the water molecule (fig. S3B). The estimate of a photoactivated state fraction of ~30% was confirmed by comparing B-factors of active site residues in the dark and photoactivated states.
As an alternative approach to determine the structural response upon light activation, the extrapolated structure factors were calculated through the program package Xtrapol8 with the Fgenick method (62). Using Xtrapol8, the occupancy was determined by the distance-analysis method where the occupancy was iterated from 0.26 in 10 evenly spaced steps of 0.01, yielding an optimal value of 0.31, with a small variability from run to run. The structure factors extrapolated to 100% occupancy were then used in 20 cycles of overall B-factor restrained refinement using REFMAC5 (63) as implemented in the Xtrapol8 pipeline, keeping the remaining parameters default. See table S1 for data collection and refinement statistics.
Acknowledgments
XFEL experiments were conducted at BL3 of SACLA with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (proposal nos. 2018B8068 and 2019A8077). Crystal screening experiments were performed at the BioMAX beamline of MAX IV Laboratory (proposal nos. 20180257 and 20180258). We are grateful for assistance by the staff of SACLA and SPring-8 and acknowledge computational support from SACLA HPC system and Mini-K super computer system. We thank T. Nakane for assistance with initial data processing. This work was supported by the X-ray Free-Electron Laser Priority Strategy Program (MEXT).
Funding: G.B. acknowledges funding from the Swedish Research Council (grant nos. 2017-06734 and 2021-05662) and the Carl Trygger Foundation (grant CTS 16:79). This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 789030). R.N. acknowledges funding from the Swedish Research Council (grant no. 2015-00560) and the European commission Marie Curie Training Network X-probe. S.I. acknowledges support from the Platform Project for Supporting Drug Discovery and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED (grant no. JP21am0101070).
Author contributions: G.B. and R.N. conceived the experiment, which was designed with input from C.S., S.G., R.A., E.N., and S.I. Protein production and crystallization was performed by C.S., S.G., R.A., J.J., P.Bå., O.U., D.Z., and E.D. UV-vis characterization was done by C.S., R.A., M.P., and G.B. The oxygen-free environment for crystallization, transportation, and injection was enabled by C.S., R.A., E.N., R.T., E.N., T.Ta., A.Y., T.A., M.Sug., H.T., R.Y., K.O., M.F., T.To., S.I., and G.B. TR-SFX experiments were prepared for and performed at SACLA by C.S., S.G., R.A., J.J., P.Bå., O.U., P.D., D.Z., E.S., E.N., R.T., T.Ta., M.Sug., M.Suz., T.M., H.N., R.B., P.Bö., E.D., G.H., G.O., S.O., K.T., O.N., R.N., and G.B. Data analysis and structural refinement were performed by C.S., S.G., A.V., and G.B. The manuscript was prepared by C.S., S.G., J.J., A.V., R.N., and G.B. with additional input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The atomic coordinates and structure factors files for the resting-state dark, partial occupancy refined activated-state, and extrapolated structure factor refined activated-state datasets are available under accession codes 8 K65, 8AJZ, and 8K6Y, respectively, in the Protein Data Bank (www.pdb.org). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Table S1
REFERENCES AND NOTES
- 1.P. Mitchell, Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148 (1961). [DOI] [PubMed] [Google Scholar]
- 2.M. K. Wikstrom, Proton pump coupled to cytochrome c oxidase in mitochondria. Nature 266, 271–273 (1977). [DOI] [PubMed] [Google Scholar]
- 3.V. R. Kaila, M. I. Verkhovsky, M. Wikström, Proton-coupled electron transfer in cytochrome oxidase. Chem. Rev. 110, 7062–7081 (2010). [DOI] [PubMed] [Google Scholar]
- 4.P. Adelroth, M. S. Ek, D. M. Mitchell, R. B. Gennis, P. Brzezinski, Glutamate 286 in cytochrome aa3 from Rhodobacter sphaeroides is involved in proton uptake during the reaction of the fully-reduced enzyme with dioxygen. Biochemistry 36, 13824–13829 (1997). [DOI] [PubMed] [Google Scholar]
- 5.M. Brändén, H. Sigurdson, A. Namslauer, R. B. Gennis, P. Adelroth, P. Brzezinski, On the role of the K-proton transfer pathway in cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 98, 5013–5018 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.M. Wikström, A. Jasaitis, C. Backgren, A. Puustinen, M. I. Verkhovsky, The role of the D- and K-pathways of proton transfer in the function of the haem-copper oxidases. Biochim. Biophys. Acta 1459, 514–520 (2000). [DOI] [PubMed] [Google Scholar]
- 7.K. Ganesan, R. B. Gennis, Blocking the K-pathway still allows rapid one-electron reduction of the binuclear center during the anaerobic reduction of the aa3-type cytochrome c oxidase from Rhodobacter sphaeroides. Biochim. Biophys. Acta 1797, 619–624 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.S. Yoshikawa, A. Shimada, Reaction mechanism of cytochromecoxidase. Chem. Rev. 115, 1936–1989 (2015). [DOI] [PubMed] [Google Scholar]
- 9.A. Malkamaki, B. Meunier, M. Reidelbach, P. R. Rich, V. Sharma, The H channel is not a proton transfer path in yeast cytochrome c oxidase. Biochim. Biophys. Acta. Bioenerg. 1860, 717–723 (2019). [DOI] [PubMed] [Google Scholar]
- 10.M. L. Bjorck, J. Vilhjalmsdottir, A. M. Hartley, B. Meunier, L. Nasvik Ojemyr, A. Marechal, P. Brzezinski, Proton-transfer pathways in the mitochondrial S. cerevisiae cytochrome c oxidase. Sci. Rep. 9, 20207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. Marechal, J. Y. Xu, N. Genko, A. M. Hartley, F. Haraux, B. Meunier, P. R. Rich, A common coupling mechanism for A-type heme-copper oxidases from bacteria to mitochondria. Proc. Natl. Acad. Sci. U.S.A. 117, 9349–9355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J. A. Keightley, B. H. Zimmermann, M. W. Mather, P. Springer, A. Pastuszyn, D. M. Lawrence, J. A. Fee, Molecular genetic and protein chemical characterization of the cytochrome ba3 from Thermus thermophilus HB8. J. Biol. Chem. 270, 20345–20358 (1995). [DOI] [PubMed] [Google Scholar]
- 13.H. Han, J. Hemp, L. A. Pace, H. Ouyang, K. Ganesan, J. H. Roh, F. Daldal, S. R. Blanke, R. B. Gennis, Adaptation of aerobic respiration to low O2 environments. Proc. Natl. Acad. Sci. U.S.A. 108, 14109–14114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.H. Y. Chang, J. Hemp, Y. Chen, J. A. Fee, R. B. Gennis, The cytochrome ba3 oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proc. Natl. Acad. Sci. U.S.A. 106, 16169–16173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.A. Kannt, T. Soulimane, G. Buse, A. Becker, E. Bamberg, H. Michel, Electrical current generation and proton pumping catalyzed by the ba3-type cytochrome c oxidase from Thermus thermophilus. FEBS Lett. 434, 17–22 (1998). [DOI] [PubMed] [Google Scholar]
- 16.V. Rauhamaki, M. Wikstrom, The causes of reduced proton-pumping efficiency in type B and C respiratory heme-copper oxidases, and in some mutated variants of type A. Biochim. Biophys. Acta 1837, 999–1003 (2014). [DOI] [PubMed] [Google Scholar]
- 17.M. Karpefors, P. Adelroth, Y. Zhen, S. Ferguson-Miller, P. Brzezinski, Proton uptake controls electron transfer in cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 95, 13606–13611 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O. Einarsdottir, W. McDonald, C. Funatogawa, I. Szundi, W. H. Woodruff, R. B. Dyer, The pathway of O2 to the active site in heme–copper oxidases. Biochim. Biophys. Acta 1847, 109–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. A. Yu, T. Egawa, K. Shinzawa-Itoh, S. Yoshikawa, S. R. Yeh, D. L. Rousseau, G. J. Gerfen, Radical formation in cytochrome c oxidase. Biochim. Biophys. Acta 1807, 1295–1304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. Svensson-Ek, J. Abramson, G. Larsson, S. Tornroth, P. Brzezinski, S. Iwata, The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J. Mol. Biol. 321, 329–339 (2002). [DOI] [PubMed] [Google Scholar]
- 21.T. Tsukihara, H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono, S. Yoshikawa, The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144 (1996). [DOI] [PubMed] [Google Scholar]
- 22.T. Soulimane, G. Buse, G. P. Bourenkov, H. D. Bartunik, R. Huber, M. E. Than, Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from thermus thermophilus. EMBO J. 19, 1766–1776 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.J. A. Lyons, D. Aragao, O. Slattery, A. V. Pisliakov, T. Soulimane, M. Caffrey, Structural insights into electron transfer in caa3-type cytochrome oxidase. Nature 487, 514–518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S. Zong, M. Wu, J. Gu, T. Liu, R. Guo, M. Yang, Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 28, 1026–1034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. Iwata, C. Ostermeier, B. Ludwig, H. Michel, Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature 376, 660–669 (1995). [DOI] [PubMed] [Google Scholar]
- 26.F. Kolbe, S. Safarian, Z. Piorek, S. Welsch, H. Muller, H. Michel, Cryo-EM structures of intermediates suggest an alternative catalytic reaction cycle for cytochrome c oxidase. Nat. Commun. 12, 6903 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. R. Blomberg, Mechanism of oxygen reduction in cytochrome c oxidase and the role of the active site tyrosine. Biochemistry 55, 489–500 (2016). [DOI] [PubMed] [Google Scholar]
- 28.P. Saura, D. Riepl, D. M. Frey, M. Wikstrom, V. R. I. Kaila, Electric fields control water-gated proton transfer in cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 119, e2207761119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.H. N. Chapman, P. Fromme, A. Barty, T. A. White, R. A. Kirian, A. Aquila, M. S. Hunter, J. Schulz, D. P. DePonte, U. Weierstall, R. B. Doak, F. R. Maia, A. V. Martin, I. Schlichting, L. Lomb, N. Coppola, R. L. Shoeman, S. W. Epp, R. Hartmann, D. Rolles, A. Rudenko, L. Foucar, N. Kimmel, G. Weidenspointner, P. Holl, M. Liang, M. Barthelmess, C. Caleman, S. Boutet, M. J. Bogan, J. Krzywinski, C. Bostedt, S. Bajt, L. Gumprecht, B. Rudek, B. Erk, C. Schmidt, A. Homke, C. Reich, D. Pietschner, L. Struder, G. Hauser, H. Gorke, J. Ullrich, S. Herrmann, G. Schaller, F. Schopper, H. Soltau, K. U. Kuhnel, M. Messerschmidt, J. D. Bozek, S. P. Hau-Riege, M. Frank, C. Y. Hampton, R. G. Sierra, D. Starodub, G. J. Williams, J. Hajdu, N. Timneanu, M. M. Seibert, J. Andreasson, A. Rocker, O. Jonsson, M. Svenda, S. Stern, K. Nass, R. Andritschke, C. D. Schroter, F. Krasniqi, M. Bott, K. E. Schmidt, X. Wang, I. Grotjohann, J. M. Holton, T. R. Barends, R. Neutze, S. Marchesini, R. Fromme, S. Schorb, D. Rupp, M. Adolph, T. Gorkhover, I. Andersson, H. Hirsemann, G. Potdevin, H. Graafsma, B. Nilsson, J. C. Spence, Femtosecond X-ray protein nanocrystallography. Nature 470, 73–77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.I. Schlichting, Serial femtosecond crystallography: the first five years. IUCrJ 2, 246–255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.T. A. White, R. A. Kirian, A. V. Martin, A. Aquila, K. Nass, A. Barty, H. N. Chapman, CrystFEL: A software suite for snapshot serial crystallography. J. Appl. Cryst. 45, 335–341 (2012). [Google Scholar]
- 32.T. Y. Teng, K. Moffat, Primary radiation damage of protein crystals by an intense synchrotron X-ray beam. J. Synchrotron Radiat. 7, 313–317 (2000). [DOI] [PubMed] [Google Scholar]
- 33.M. Amin, A. Badawi, S. S. Obayya, Radiation damage in XFEL: Case study from the oxygen-evolving complex of Photosystem II. Sci. Rep. 6, 36492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.E. Nango, A. Royant, M. Kubo, T. Nakane, C. Wickstrand, T. Kimura, T. Tanaka, K. Tono, C. Song, R. Tanaka, T. Arima, A. Yamashita, J. Kobayashi, T. Hosaka, E. Mizohata, P. Nogly, M. Sugahara, D. Nam, T. Nomura, T. Shimamura, D. Im, T. Fujiwara, Y. Yamanaka, B. Jeon, T. Nishizawa, K. Oda, M. Fukuda, R. Andersson, P. Bath, R. Dods, J. Davidsson, S. Matsuoka, S. Kawatake, M. Murata, O. Nureki, S. Owada, T. Kameshima, T. Hatsui, Y. Joti, G. Schertler, M. Yabashi, A. N. Bondar, J. Standfuss, R. Neutze, S. Iwata, A three-dimensional movie of structural changes in bacteriorhodopsin. Science 354, 1552–1557 (2016). [DOI] [PubMed] [Google Scholar]
- 35.T. Tosha, T. Nomura, T. Nishida, N. Saeki, K. Okubayashi, R. Yamagiwa, M. Sugahara, T. Nakane, K. Yamashita, K. Hirata, G. Ueno, T. Kimura, T. Hisano, K. Muramoto, H. Sawai, H. Takeda, E. Mizohata, A. Yamashita, Y. Kanematsu, Y. Takano, E. Nango, R. Tanaka, O. Nureki, O. Shoji, Y. Ikemoto, H. Murakami, S. Owada, K. Tono, M. Yabashi, M. Yamamoto, H. Ago, S. Iwata, H. Sugimoto, Y. Shiro, M. Kubo, Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate. Nat. Commun. 8, 1585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.P. Mehrabi, E. C. Schulz, R. Dsouza, H. M. Muller-Werkmeister, F. Tellkamp, R. J. D. Miller, E. F. Pai, Time-resolved crystallography reveals allosteric communication aligned with molecular breathing. Science 365, 1167–1170 (2019). [DOI] [PubMed] [Google Scholar]
- 37.R. Dods, P. Bath, D. Morozov, V. A. Gagner, D. Arnlund, H. L. Luk, J. Kubel, M. Maj, A. Vallejos, C. Wickstrand, R. Bosman, K. R. Beyerlein, G. Nelson, M. Liang, D. Milathianaki, J. Robinson, R. Harimoorthy, P. Berntsen, E. Malmerberg, L. Johansson, R. Andersson, S. Carbajo, E. Claesson, C. E. Conrad, P. Dahl, G. Hammarin, M. S. Hunter, C. Li, S. Lisova, A. Royant, C. Safari, A. Sharma, G. J. Williams, O. Yefanov, S. Westenhoff, J. Davidsson, D. P. DePonte, S. Boutet, A. Barty, G. Katona, G. Groenhof, G. Branden, R. Neutze, Ultrafast structural changes within a photosynthetic reaction centre. Nature 589, 310–314 (2021). [DOI] [PubMed] [Google Scholar]
- 38.P. Skopintsev, D. Ehrenberg, T. Weinert, D. James, R. K. Kar, P. J. M. Johnson, D. Ozerov, A. Furrer, I. Martiel, F. Dworkowski, K. Nass, G. Knopp, C. Cirelli, C. Arrell, D. Gashi, S. Mous, M. Wranik, T. Gruhl, D. Kekilli, S. Brunle, X. Deupi, G. F. X. Schertler, R. M. Benoit, V. Panneels, P. Nogly, I. Schapiro, C. Milne, J. Heberle, J. Standfuss, Femtosecond-to-millisecond structural changes in a light-driven sodium pump. Nature 583, 314–318 (2020). [DOI] [PubMed] [Google Scholar]
- 39.A. R. Pearson, P. Mehrabi, Serial synchrotron crystallography for time-resolved structural biology. Curr. Opin. Struct. Biol. 65, 168–174 (2020). [DOI] [PubMed] [Google Scholar]
- 40.G. Branden, R. Neutze, Advances and challenges in time-resolved macromolecular crystallography. Science 373, (2021). [DOI] [PubMed] [Google Scholar]
- 41.R. Andersson, C. Safari, R. Dods, E. Nango, R. Tanaka, A. Yamashita, T. Nakane, K. Tono, Y. Joti, P. Bath, E. Dunevall, R. Bosman, O. Nureki, S. Iwata, R. Neutze, G. Branden, Serial femtosecond crystallography structure of cytochrome c oxidase at room temperature. Sci. Rep. 7, 4518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.T. Tiefenbrunn, W. Liu, Y. Chen, V. Katritch, C. D. Stout, J. A. Fee, V. Cherezov, High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment. PLOS ONE 6, e22348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.I. Ishigami, N. A. Zatsepin, M. Hikita, C. E. Conrad, G. Nelson, J. D. Coe, S. Basu, T. D. Grant, M. H. Seaberg, R. G. Sierra, M. S. Hunter, P. Fromme, R. Fromme, S. R. Yeh, D. L. Rousseau, Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature. Proc. Natl. Acad. Sci. U.S.A. 114, 8011–8016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.I. Ishigami, A. Lewis-Ballester, A. Echelmeier, G. Brehm, N. A. Zatsepin, T. D. Grant, J. D. Coe, S. Lisova, G. Nelson, S. Zhang, Z. F. Dobson, S. Boutet, R. G. Sierra, A. Batyuk, P. Fromme, R. Fromme, J. C. H. Spence, A. Ros, S. R. Yeh, D. L. Rousseau, Snapshot of an oxygen intermediate in the catalytic reaction of cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 116, 3572–3577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R. B. Dyer, O. Einarsdottir, P. M. Killough, J. J. Lopezgarriga, W. H. Woodruff, Transient binding of photodissociated carbon monoxide to CuB+ of eukaryotic cytochrome oxidase at ambient temperature. Direct evidence from time-resolved infrared spectroscopy. J. Am. Chem. Soc. 111, 7657–7659 (1989). [Google Scholar]
- 46.O. Einarsdottir, R. B. Dyer, D. D. Lemon, P. M. Killough, S. M. Hubig, S. J. Atherton, J. J. Lopez-Garriga, G. Palmer, W. H. Woodruff, Photodissociation and recombination of carbonmonoxy cytochrome oxidase: dynamics from picoseconds to kiloseconds. Biochemistry 32, 12013–12024 (1993). [DOI] [PubMed] [Google Scholar]
- 47.K. Koutsoupakis, S. Stavrakis, E. Pinakoulaki, T. Soulimane, C. Varotsis, Observation of the equilibrium CuB-CO complex and functional implications of the transient heme a3 propionates in cytochrome ba3-CO from Thermus thermophilus. Fourier transform infrared (FTIR) and time-resolved step-scan FTIR studies. J. Biol. Chem. 277, 32860–32866 (2002). [DOI] [PubMed] [Google Scholar]
- 48.A. Shimada, M. Kubo, S. Baba, K. Yamashita, K. Hirata, G. Ueno, T. Nomura, T. Kimura, K. Shinzawa-Itoh, J. Baba, K. Hatano, Y. Eto, A. Miyamoto, H. Murakami, T. Kumasaka, S. Owada, K. Tono, M. Yabashi, Y. Yamaguchi, S. Yanagisawa, M. Sakaguchi, T. Ogura, R. Komiya, J. Yan, E. Yamashita, M. Yamamoto, H. Ago, S. Yoshikawa, T. Tsukihara, A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase. Sci. Adv. 3, e1603042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.L. Powers, M. Lauraeus, K. S. Reddy, B. Chance, M. Wikstrom, Structure of the binuclear heme iron-copper site in the quinol-oxidizing cytochrome aa3 from Bacillus subtilis. Biochim. Biophys. Acta 1183, 504–512 (1994). [DOI] [PubMed] [Google Scholar]
- 50.B. Liu, Y. Zhang, J. T. Sage, S. M. Soltis, T. Doukov, Y. Chen, C. D. Stout, J. A. Fee, Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: a combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B). Biochim. Biophys. Acta 1817, 658–665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.A. Ebrahim, T. Moreno-Chicano, M. V. Appleby, A. K. Chaplin, J. H. Beale, D. A. Sherrell, H. M. E. Duyvesteyn, S. Owada, K. Tono, H. Sugimoto, R. W. Strange, J. A. R. Worrall, D. Axford, R. L. Owen, M. A. Hough, Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins. IUCrJ 6, 543–551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.G. Nass Kovacs, J. P. Colletier, M. L. Grunbein, Y. Yang, T. Stensitzki, A. Batyuk, S. Carbajo, R. B. Doak, D. Ehrenberg, L. Foucar, R. Gasper, A. Gorel, M. Hilpert, M. Kloos, J. E. Koglin, J. Reinstein, C. M. Roome, R. Schlesinger, M. Seaberg, R. L. Shoeman, M. Stricker, S. Boutet, S. Haacke, J. Heberle, K. Heyne, T. Domratcheva, T. R. M. Barends, I. Schlichting, Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin. Nat. Commun. 10, 3177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.R. J. D. Miller, O. Pare-Labrosse, A. Sarracini, J. E. Besaw, Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin in the multiphoton regime and biological relevance. Nat. Commun. 11, 1240 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.T. R. M. Barends, S. Bhattacharyya, A. Gorel, G. Schiro, C. Bacellar, C. Cirelli, J.-P. Colletier, L. Foucar, M. L. Grünbein, E. Hartmann, M. Hilpert, P. J. M. Johnson, M. Kloos, G. Knopp, B. Marekha, K. Nass, G. N. Kovacs, D. Ozerov, M. Stricker, M. Weik, R. B. Doak, R. L. Shoeman, C. J. Milne, M. Huix-Rotllant, M. Cammarata, I. Schlichting, Influence of pump laser fluence on ultrafast structural changes in myoglobin. bioRxiv 2022.2011.2022.517513 [Preprint]. 22 November 2022. 10.1101/2022.11.22.517513. [DOI]
- 55.T. R. Barends, L. Foucar, A. Ardevol, K. Nass, A. Aquila, S. Botha, R. B. Doak, K. Falahati, E. Hartmann, M. Hilpert, M. Heinz, M. C. Hoffmann, J. Kofinger, J. E. Koglin, G. Kovacsova, M. Liang, D. Milathianaki, H. T. Lemke, J. Reinstein, C. M. Roome, R. L. Shoeman, G. J. Williams, I. Burghardt, G. Hummer, S. Boutet, I. Schlichting, Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science 350, 445–450 (2015). [DOI] [PubMed] [Google Scholar]
- 56.B. Stauch, V. Cherezov, Serial femtosecond crystallography of G protein-coupled receptors. Annu. Rev. Biophys. 47, 377–397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.M. Andersson, E. Malmerberg, S. Westenhoff, G. Katona, M. Cammarata, A. B. Wohri, L. C. Johansson, F. Ewald, M. Eklund, M. Wulff, J. Davidsson, R. Neutze, Structural dynamics of light-driven proton pumps. Structure 17, 1265–1275 (2009). [DOI] [PubMed] [Google Scholar]
- 58.A. M. Wolff, E. Nango, I. D. Young, A. S. Brewster, M. Kubo, T. Nomura, M. Sugahara, S. Owada, B. A. Barad, K. Ito, A. Bhowmick, S. Carbajo, T. Hino, J. M. Holton, D. Im, L. J. O’Riordan, T. Tanaka, R. Tanaka, R. G. Sierra, F. Yumoto, K. Tono, S. Iwata, N. K. Sauter, J. S. Fraser, M. C. Thompson, Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography. Nat. Chem. 15, 1549–1558 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.B. Liu, Y. Chen, T. Doukov, S. M. Soltis, C. D. Stout, J. A. Fee, Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme. Biochemistry 48, 820–826 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.U. K. Genick, Structure-factor extrapolation using the scalar approximation: theory, applications and limitations. Acta Crystallogr. D Biol. Crystallogr. 63, 1029–1041 (2007). [DOI] [PubMed] [Google Scholar]
- 61.K. Muramoto, K. Ohta, K. Shinzawa-Itoh, K. Kanda, M. Taniguchi, H. Nabekura, E. Yamashita, T. Tsukihara, S. Yoshikawa, Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate. Proc. Natl. Acad. Sci. U.S.A. 107, 7740–7745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.E. De Zitter, N. Coquelle, P. Oeser, T. R. M. Barends, J.-P. Colletier, Xtrapol8 enables automatic elucidation of low-occupancy intermediate-states in crystallographic studies. Commun. Biol. 5, 640 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.G. N. Murshudov, P. Skubak, A. A. Lebedev, N. S. Pannu, R. A. Steiner, R. A. Nicholls, M. D. Winn, F. Long, A. A. Vagin, REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.C. von Ballmoos, N. Gonska, P. Lachmann, R. B. Gennis, P. Adelroth, P. Brzezinski, Mutation of a single residue in the ba3 oxidase specifically impairs protonation of the pump site. Proc. Natl. Acad. Sci. U.S.A. 112, 3397–3402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.C. Koutsoupakis, T. Soulimane, C. Varotsis, Discrete Ligand Binding and Electron Transfer Properties of ba3-Cytochrome c Oxidase from Thermus thermophilus: Evolutionary Adaption to Low Oxygen and High Temperature Environments. Acc. Chem. Res. 52, 1380–1390 (2019). [DOI] [PubMed] [Google Scholar]
- 66.O. Einarsdottir, C. Funatogawa, T. Soulimane, I. Szundi, Kinetic studies of the reactions of O(2) and NO with reduced Thermus thermophilus ba(3) and bovine aa(3) using photolabile carriers. Biochim. Biophys. Acta 1817, 672–679 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.W. McDonald, C. Funatogawa, Y. Li, I. Szundi, Y. Chen, J. A. Fee, C. D. Stout, O. Einarsdottir, Ligand access to the active site in Thermus thermophilus ba(3) and bovine heart aa(3) cytochrome oxidases. Biochemistry 52, 640–652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.T. Egawa, Y. Chen, J. A. Fee, S. R. Yeh, D. L. Rousseau, The rate-limiting step in O(2) reduction by cytochrome ba(3) from Thermus thermophilus. Biochim. Biophys. Acta 1817, 666–671 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.A. Shimada, Y. Etoh, R. Kitoh-Fujisawa, A. Sasaki, K. Shinzawa-Itoh, T. Hiromoto, E. Yamashita, K. Muramoto, T. Tsukihara, S. Yoshikawa, X-ray structures of catalytic intermediates of cytochrome c oxidase provide insights into its O2 activation and unidirectional proton-pump mechanisms. J. Biol. Chem. 295, 5818–5833 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.A. Shimada, F. Hara, K. Shinzawa-Itoh, N. Kanehisa, E. Yamashita, K. Muramoto, T. Tsukihara, S. Yoshikawa, Critical roles of the CuB site in efficient proton pumping as revealed by crystal structures of mammalian cytochrome c oxidase catalytic intermediates. J. Biol. Chem. 297, 100967 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.I. Ishigami, S. Russi, A. Cohen, S. R. Yeh, D. L. Rousseau, Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase. J. Biol. Chem. 298, 101799 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R. Andersson, C. Safari, P. Bath, R. Bosman, A. Shilova, P. Dahl, S. Ghosh, A. Dunge, R. Kjeldsen-Jensen, J. Nan, R. L. Shoeman, M. Kloos, R. B. Doak, U. Mueller, R. Neutze, G. Branden, Well-based crystallization of lipidic cubic phase microcrystals for serial X-ray crystallography experiments. Acta Crystallogr. D Struct. Biol. 75, 937–946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Y. Shimazu, K. Tono, T. Tanaka, Y. Yamanaka, T. Nakane, C. Mori, K. Terakado Kimura, T. Fujiwara, M. Sugahara, R. Tanaka, R. B. Doak, T. Shimamura, S. Iwata, E. Nango, M. Yabashi, High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure. J. Appl. Cryst. 52, 1280–1288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.K. Tono, E. Nango, M. Sugahara, C. Song, J. Park, T. Tanaka, R. Tanaka, Y. Joti, T. Kameshima, S. Ono, T. Hatsui, E. Mizohata, M. Suzuki, T. Shimamura, Y. Tanaka, S. Iwata, M. Yabashi, Diverse application platform for hard X-ray diffraction in SACLA (DAPHNIS): application to serial protein crystallography using an X-ray free-electron laser. J. Synchrotron Radiat. 22, 532–537 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.T. Kameshima, S. Ono, T. Kudo, K. Ozaki, Y. Kirihara, K. Kobayashi, Y. Inubushi, M. Yabashi, T. Horigome, A. Holland, K. Holland, D. Burt, H. Murao, T. Hatsui, Development of an X-ray pixel detector with multi-port charge-coupled device for X-ray free-electron laser experiments. Rev. Sci. Instrum. 85, 033110 (2014). [DOI] [PubMed] [Google Scholar]
- 76.J. L. Dickerson, P. T. N. McCubbin, E. F. Garman, RADDOSE-XFEL: Femtosecond time-resolved dose estimates for macromolecular X-ray free-electron laser experiments. J. Appl. Cryst. 53, 549–560 (2020). [Google Scholar]
- 77.A. Barty, R. A. Kirian, F. R. Maia, M. Hantke, C. H. Yoon, T. A. White, H. Chapman, Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Cryst. 47, 1118–1131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.T. Nakane, Y. Joti, K. Tono, M. Yabashi, E. Nango, S. Iwata, R. Ishitani, O. Nureki, Data processing pipeline for serial femtosecond crystallography at SACLA. J. Appl. Cryst. 49, 1035–1041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.K. Nass, R. Cheng, L. Vera, A. Mozzanica, S. Redford, D. Ozerov, S. Basu, D. James, G. Knopp, C. Cirelli, I. Martiel, C. Casadei, T. Weinert, P. Nogly, P. Skopintsev, I. Usov, F. Leonarski, T. Geng, M. Rappas, A. S. Dore, R. Cooke, S. Nasrollahi Shirazi, F. Dworkowski, M. Sharpe, N. Olieric, C. Bacellar, R. Bohinc, M. O. Steinmetz, G. Schertler, R. Abela, L. Patthey, B. Schmitt, M. Hennig, J. Standfuss, M. Wang, C. J. Milne, Advances in long-wavelength native phasing at X-ray free-electron lasers. IUCrJ 7, 965–975 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.P. Emsley, B. Lohkamp, W. G. Scott, K. Cowtan, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.P. D. Adams, P. V. Afonine, G. Bunkoczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger, P. H. Zwart, PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Table S1