Abstract
In this study, we cloned the Pseudomonas aeruginosa zwf gene, encoding glucose-6-phosphate dehydrogenase (G6PDH), an enzyme that catalyzes the NAD+- or NADP+-dependent conversion of glucose-6-phosphate to 6-phosphogluconate. The predicted zwf gene product is 490 residues, which could form a tetramer with a molecular mass of ∼220 kDa. G6PDH activity and zwf transcription were maximal in early logarithmic phase when inducing substrates such as glycerol, glucose, or gluconate were abundant. In contrast, both G6PDH activity and zwf transcription plummeted dramatically when bacteria approached stationary phase, when inducing substrate was limiting, or when the organisms were grown in a citrate-, succinate-, or acetate-containing basal salts medium. G6PDH was purified to homogeneity, and its molecular mass was estimated to be ∼220 kDa by size exclusion chromatography. Estimated Km values of purified G6PDH acting on glucose-6-phosphate, NADP+, and NAD+ were 530, 57, and 333 μM, respectively. The specific activities with NAD+ and NADP+ were calculated to be 176 and 69 μmol/min/mg. An isogenic zwf mutant was unable to grow on minimal medium supplemented with mannitol. The mutant also demonstrated increased sensitivity to the redox-active superoxide-generating agent methyl viologen (paraquat). Since one by-product of G6PDH activity is NADPH, the latter data suggest that this cofactor is essential for the activity of enzymes critical in defense against paraquat toxicity.
Pseudomonas aeruginosa is a gram-negative bacillus that is virtually ubiquitous throughout nature. It is also an opportunistic pathogen of humans, most notably those who have cystic fibrosis or whose immune systems have been compromised (e.g., as a result of burns or cancer chemotherapy) (11, 16, 55). The organism has a remarkable capacity to utilize a wide range of carbon sources for growth under a variety of environmental conditions. Although tricarboxylic acid (TCA) cycle intermediates such as succinate are preferentially utilized by P. aeruginosa, the organism readily catabolizes glucose. In contrast to the facultative organism Escherichia coli, P. aeruginosa does not metabolize glucose via the Embden-Meyerhof pathway because it does not possess 6-phosphofructokinase (36). Thus, the catabolism of glucose by P. aeruginosa requires its conversion to glyceraldehyde-3-phosphate and pyruvate via the Enter-Doudoroff enzymes 6-phosphogluconate dehydratase (Edd) and 2-keto-3-deoxy-6-phosphogluconate aldolase (Eda) (36). Depending on the physiological conditions, glucose is converted to 6-phosphogluconate by one of two routes, one of which is oxidative and the other of which is phosphorylative. The direct oxidative route involves oxidation of glucose to gluconate and 2-ketogluconate in the periplasm via membrane-bound glucose and gluconate dehydrogenases (29). Recently both oxidative routes have been shown to be physiologically significant with the isolation of mutants blocked in either gluconate or 2-ketogluconate utilization (67a). Alternatively, the phosphorylative route involves uptake of glucose by an inducible transport system where, once inside the organism, it is phosphorylated by glucokinase and next converted to 6-phosphogluconate by glucose-6-phosphate dehydrogenase (G6PDH; EC 1.1.1.49), the product of the zwf gene (29).
Interestingly, a zwf mutant of E. coli was reported to be more sensitive to the redox-active, superoxide (O2−)-generating agent methyl viologen (paraquat) (19). It was then postulated by Liochev and Fridovich (38) that this sensitivity was attributed to a reduced level of NADPH, a cofactor necessary for the activity of glutathione reductase (3) and alkylhydroperoxide reductase (32), enzymes which combat paraquat-mediated oxidative stress.
In this study, we describe the cloning and characterization of the zwf gene of P. aeruginosa PAO1. We demonstrate that inactivation of the zwf gene does not allow mutant organisms to grow on mannitol as the sole carbon source. In addition, we demonstrate that the zwf gene is under tight control, being highly transcribed in the presence of inducing agents such as glycerol and glucose and weakly transcribed in the presence of the TCA cycle intermediate succinate. In addition, we demonstrate that G6PDH activity is important in resistance to the O2−-generating agent paraquat.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All P. aeruginosa and E. coli strains used in this study are listed in Table 1 and were maintained on Luria (L)-agar plates containing 10 g of tryptone, 5 g of yeast extract, 5 g of NaCl, and 15 g of Bacto Agar per liter. Frozen stocks were stored indefinitely at −80°C in a 1:1 mixture of 25% glycerol and stationary-phase culture grown in L broth.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α-MCR | F−lacZΔM15 recA1 hsdR17 supE44 Δ(lacZYA argF) | Bethesda Research Laboratories |
| SM10 | Kmr, mobilizer strain | 63 |
| P. aeruginosa | ||
| PAO1 | Wild type | 27 |
| PAO9010 | Gmrzwf::Gmr | This study |
| PAO9011 | Gmrzwf::lacZ-Gmr (sense orientation) | This study |
| PAO9012 | Gmrzwf::lacZ-Gmr (antisense orientation) | This study |
| Plasmids | ||
| pBluescript KS−/+ | Extended polylinker pUC derivative | Stratagene |
| pNOT19 | Apr, pUC19 + NotI site | 59 |
| pMOB3 | Apr Cmr KmrsacB oriT mob | 59 |
| pUCGM | Apr Gmr, pUC19 + 850-bp Gmr cassette | 60 |
| pZ1918 | Apr, pUC19/18 with 3.2-kb lacZ cassette | 61 |
| pZ1918G | Apr Gmr, pZ1918 with Gmr cassette from pUCGM immediately downstream of the lacZ gene | H. P. Schweizer |
| pEX30 | Apr, broad-host-range expression vector | |
| pEX100T | Apr, mobilizable oriT sacB vector for construction of mutants | 62 |
| pUCP22 | Apr, broad-host-range cloning vector | 68 |
| pPZ523 | Apr, pUCP22 with 2.5-kb SacII fragment, complements P. aeruginosa PFB98 (zwf-1) | This study |
| pJFM1 | Apr, pBluescript SK− with 2.5-kb SacII fragment containing zwf | This study |
| pJFM2 | Apr, pBluescript SK− with 1.6 kb zwf PCR product | This study |
| pJFM3 | Apr, AflIII-EcoRI fragment of 1.6-kb zwf PCR product in NcoI and EcoRI sites of pEX30 | This study |
| pJFM4 | Apr, 1.7-kb PvuII zwf fragment in SmaI site of pEX100T | This study |
| pJFM5 | Apr Gmr, pJFM4 with 4.0-kb lacZ-Gmr cassette in BamHI site of zwf in sense orientation | This study |
| pJFM6 | Apr Gmr, pJFM4 with 4.0-kb lacZ-Gmr cassette in BamHI site of zwf in antisense orientation | This study |
| pJFM7 | Apr, pUCP22 with 1.0-kb PstI-BamHI fragment of zwf | This study |
| pJFM8 | Apr, pJFM7 with 3.2-kb lacZ in StuI site of zwf | This study |
| pJFM9 | Apr, pNOT19 with ∼1-kb PstI-BamHI fragment of zwf | This study |
| pJFM10 | Apr, pJFM1 with ∼850-bp Gmr cassette in HincII site of zwf | This study |
| pJFM11 | pJFM2 with 5.8-kb NotI oriT sacB fragment from pMOB3 | This study |
Abbreviations used for genetic markers are as described by Holloway et al. (27). mob, mobilization site (ColE1); Tra+, conjugative phenotype; oriT, origin of transfer (RK2); Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance.
Growth conditions.
All bacteria were grown from single-colony isolates in either L broth or a basal salts medium (69) supplemented with 20 to 60 mM selected carbon sources. Liquid cultures were grown at 37°C with shaking at 300 rpm or on a roller wheel at 70 rpm unless otherwise indicated. Culture volumes were 1/10 of the total Erlenmeyer flask volume to ensure proper aeration. All agar media were solidified with 1.5% Bacto Agar.
Cloning and sequence analysis of P. aeruginosa PAO1 zwf.
Steps involved in the cloning of the P. aeruginosa PAO1 zwf gene are described in Results. All DNA sequencing was performed by the dideoxy method on double-stranded DNA (Sequenase 2.0; U.S. Biochemical, Cleveland, Ohio). Sequence obtained by this method was confirmed on both strands by using a PRISM Dye Deoxy terminator cycle sequencing kit and analyzed on an ABI model 373A DNA sequencer. Additional DNA sequencing was provided by the Biotechnology Program of the East Carolina University School of Medicine. Oligonucleotides for DNA sequencing reactions and PCR analysis were synthesized in the DNA Core Facilities, Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati College of Medicine, or were provided by the Biotechnology Program, East Carolina School of Medicine. Sequence analysis was performed with Sequencher 3.0 (Gene Codes Corp., Ann Arbor, Mich.), MacVector 4.1.1 (Eastman Chemical Co., New Haven, Conn.), or Gene Runner (Hastings Software, Inc.). Amino acid alignments were performed by using either the BLASTP program provided by the National Center for Biotechnology Information (1) or the Align Plus 3 global alignment program (Sci-Ed Software, Durham, N.C.). Potential transcription start sites were identified by using the Neural Network Promoter Prediction program (http://www-hgc.lbl.gov/projects/promoter.html [54]).
Manipulation of recombinant DNA.
Plasmid DNA was transformed into either E. coli DH5α-MCR (Gibco-BRL, Gaithersburg, Md.) or E. coli SM10 (63). 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg/ml) was added to agar medium to detect the presence of insert DNA via blue/white selection. Restriction endonucleases, alkaline phosphatase, and T4 DNA ligase were used as specified by the vendor (Gibco-BRL); Plasmid DNA was isolated on a small scale by the alkaline lysis method described by Maniatis et al. (43) or on a large scale by using a plasmid isolation kit (Qiagen). Restriction fragments were recovered from agarose gels by using SeaPlaque low-melting-point agarose (FMC BioProducts, Rockland, Maine).
Construction of the G6PDH overexpression vector, pJFM3.
PCR primers ZWF-START (5′-GTAACAACACATGTCTGATGTCCGCGTTCT-3′) upstream of the P. aeruginosa zwf gene and KS (5′-TCGAGGTCGACGGTATC-3′) of pBluescript KS− were used to PCR amplify the zwf gene from pJFM1, and this fragment was cloned into the EcoRV site of pBluescript KS−. This plasmid, designated pJFM2, was digested with AflIII and EcoRI, and the resulting ∼1.6-kb zwf fragment was ligated into the NcoI/EcoRI-cut expression vector pEX30 (61a), forming pJFM3. Plasmid pJFM3 was then used for overproduction and purification of G6PDH as described below.
Purification of P. aeruginosa G6PDH.
Overproduction of G6PDH in P. aeruginosa PAO1 was accomplished after the following steps. Bacteria harboring the zwf overexpression vector pJFM3 were grown in 4 liters of L broth containing carbenicillin (400 μg/ml) to an optical density at 600 nm (OD600) of 0.3. The synthesis of T7 polymerase was then induced by the addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the bacteria were allowed to grow for an additional 3 h at 37°C. The bacteria were pelleted by centrifugation at 10,000 × g for 15 min, washed in 0.9% saline, and resuspended in 50 mM Tris-HCl (pH 7.8) containing mercaptoethanol (0.02%), lysozyme (0.02%), and the protease inhibitors phenylmethylsulfonyl fluoride (0.5 mM), leupeptin (0.5 μM), and pepstatin (0.5 μM). The suspension was incubated on ice for 1 h and subjected to three freeze (−80°C)-thaw (37°C) cycles to aid in breakage of the cells and further disrupted in a French pressure cell at 12,000 lb/in2 at 4°C. Unbroken cells and cell debris were clarified by ultracentrifugation at 100,000 × g for 1 h at 4°C. The clarified extract was brought to 38% saturation with ammonium sulfate and allowed to incubate at 4°C for 17 h, and the precipitated protein was clarified by centrifugation at 10,000 × g for 20 min. The precipitate was dissolved in 20 mM potassium phosphate buffer (KPi), pH 6.8, and dialyzed against six 1-liter changes of the same buffer at 4°C. This solution was filtered through a 0.22-μm-pore-size filter (Nalgene) and concentrated with an Amicon YM-100 membrane. The retentate, containing G6PDH, was passed over a 2- by 18-cm DE-52 column (Whatman International Ltd., Kent, England) and eluted with a 20 to 400 mM gradient of KPi (pH 6.8). After concentration of the G6PDH-containing fractions as described above, sample was loaded on a 1.5- by 6-cm hydroxyapatite column and eluted with a 20 to 150 mM gradient of KPi. Finally, G6PDH was purified from two smaller contaminating proteins by passage through a 3.5- by 100-cm Bio-Gel 150 gel filtration column equilibrated with 20 mM KPi (pH 6.8) at 4°C while maintaining a flow rate of 0.2 ml/min. Purified G6PDH was then stored on ice at 0°C.
Construction of a P. aeruginosa zwf mutant.
The strategy for insertional mutagenesis of the zwf gene was based on the sucrose counterselection technique (59). To accomplish this, a ∼1-kb PstI-BamHI fragment from pJFM1 was ligated into pNOT19 (59), forming pJFM9. This plasmid was linearized with HincII, a unique site within the zwf gene, and ligated to a SmaI-cut ∼850-bp gentamicin resistance (Gmr) cassette from pUCGM (60), creating pJFM10. This plasmid was linearized with NotI and ligated to the 5.8-kb oriT sacB-containing fragment of pMOB3 (59), creating pJFM11. After biparental mating of E. coli SM10 harboring pJFM11 and P. aeruginosa PAO1, plasmid integration into the genome by homologous recombination was assessed by selection on Pseudomonas isolation agar (PIA)-gentamicin plates. Isolated Gmr colonies were picked and grown in L broth until mid-log phase, and serial dilutions were plated on PIA-gentamicin plates containing 5% sucrose. Candidate zwf mutants were confirmed by Southern blot and G6PDH activity gel analyses.
Paraquat sensitivity assays.
Bacteria were grown for 17 h at 37°C under aerobic conditions in L broth without NaCl containing 400 μg of carbenicillin per ml and 0.2 mM IPTG. Aliquots (2.5 μl) of these suspensions were added to 2.5 ml of the same medium containing increasing concentrations of paraquat and incubated on a roller wheel at 90 rpm for 17 h at 37°C. The final absorbance of the appropriately diluted suspensions was recorded on a Beckman DU 20 (Fullerton, CA) spectrophotometer at 600 nm.
Cell extract preparation, nondenaturing gel electrophoresis, and biochemical assays.
Cell extracts of mid-logarithmic-phase organisms or overnight-grown bacteria were prepared from cultures harvested by centrifugation at 10,000 × g for 10 min at 4°C. Bacteria were washed twice in ice-cold 50 mM sodium phosphate buffer (pH 7.0) and sonicated in an ice water bath for 10 s with a model W-225 sonicator (Heat-Systems, Inc., Farmington, N.Y.) at setting 5. The sonicate was then clarified by centrifugation at 13,000 × g for 10 min at 4°C. Cell extract for native gel electrophoresis was prepared as described above except that 50 mM Tris-HCl (pH 7.8) was used as the diluent. G6PDH activity was monitored by the production of NADPH at 340 nm in 1-ml reaction mixtures containing 1 mM glucose-6-phosphate, 0.4 mM NADP+, and cell extract, using a Beckman DUP spectrophotometer equipped with an NGI (Elk Grove Village, Ill.) Servogor chart recorder unless otherwise indicated. G6PDH activity staining in 7.5% nondenaturing gels was performed with the same reagents as in the spectrophotometric assay described above except for the addition of 0.16 mM phenazine methosulfate and 0.18 mM nitroblue tetrazolium (37). Paraquat:NAD(P)H oxidoreductase activity was assayed by two methods. The first involved following the oxidation of NADH or NADPH spectrophotometrically at 340 nm (39). The second involved activity staining in 10% nondenaturing gels soaked in a mixture of 50 mM Tris-HCl (pH 7.5), 4 mM paraquat, 1 mM nitroblue tetrazolium, and either 0.2 mM NADH or NADPH (40). β-Galactosidase assays were performed on either chloroform-sodium dodecyl sulfate (SDS)-treated bacteria or cell extracts, using o-nitrophenyl-β-d-thiogalactopyranoside (ONPG) as the substrate; the results were expressed as international units (micromoles of ONPG hydrolyzed/minute/milligram of protein), using a millimolar extinction coefficient for ONPG of 3.1 (45). Catalase activity was monitored by following the decomposition of 18 mM H2O2 at 240 nm (4, 6, 25). Superoxide dismutase (SOD) activity was monitored by following the autoxidation of pyrogallol at 320 nm (52), a modification of the original method described by Marklund and Marklund (44). Protein concentrations were estimated by the method of Bradford (5), using bovine serum albumin fraction V (Sigma) as the standard.
Nucleotide sequence accession number.
The sequence shown in Fig. 1 has been assigned GenBank accession no. AF029673.
FIG. 1.
DNA sequence of the zwf gene of P. aeruginosa PAO1. Coding sequences are uppercase, noncoding sequences are lowercase. The zwf coding sequence starts at position 966. The putative ribosome binding (Shine-Dalgarno) sequence is underlined prior to the ATG start codon. A predicted ς70-like promoter is bracketed, with the −35 and −10 regions underlined, and is based on the Neural Network Promoter Prediction program (54). The asterisk indicates the zwf stop codon.
RESULTS
DNA sequence analysis of the P. aeruginosa zwf gene.
The P. aeruginosa PAO1 zwf gene was initially cloned on an 11-kb BamHI fragment that also contained the eda gene, encoding keto-3-deoxy-6-phosphogluconate aldolase (64). The zwf gene was localized to a ∼2.5-kb SacII fragment. The predicted zwf gene product is 490 residues, which could form a tetramer with an molecular mass of ∼220 kDa (Fig. 1). A putative ribosome binding (Shine-Dalgarno) site (GGttGG) was identified 9 bp upstream of the zwf ATG start codon. The estimated size of the translated G6PDH monomer was ∼55.6 kDa, with a pI of 12.6.
Amino acid similarity with other G6PDH proteins.
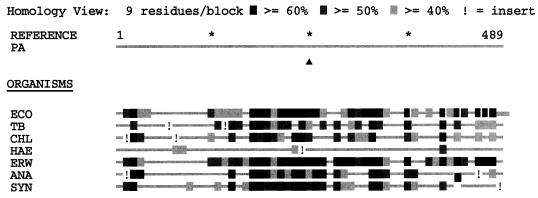
The P. aeruginosa G6PDH was aligned with seven other G6PDH proteins (identified by GenBank accession number and, in brackets, reference) from E. coli (M55005 [57]), Mycobacterium tuberculosis [unpublished]), (Z95844). Chlamydia trachomatis (U83195 [unpublished]), Haemophilus influenzae (U32737 or L42023 [15]), Erwinia chrysanthemi (X74866 [28]), Anabaena sp. strain PCC 7120 (U33282 [48]), and Synechococcus sp. strain PCC 7942 (U33285 or X64768 [58]), using the Align Plus 3 multiple protein alignment program (Fig. 2). The P. aeruginosa G6PDH revealed the greatest identity with G6PDH proteins from E. chrysanthemi (55% identity) and E. coli (54% identity). The weakest homology was demonstrated with the G6PDH of H. influenzae (19% identity). The remaining G6PDH proteins were >40% identical.
FIG. 2.
Alignment of G6PDH proteins from various organisms. G6PDH proteins were retrieved from GenBank and aligned by using the global alignment program Align Plus 3. Abbreviations: PA, P. aeruginosa; ECO, E. coli; TB, M. tuberculosis; CHL, C. trachomatis; HAE, H. influenzae; ERW, E. chrysanthemi; ANA, Anabaena sp.; SYN, Synechococcus sp. (B) Homology blocks of each G6PDH protein.
Construction of an isogenic zwf mutant of P. aeruginosa PAO1.
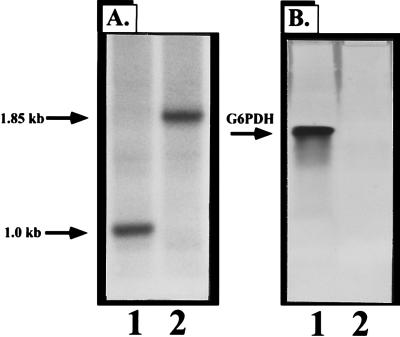
G6PDH-deficient P. aeruginosa PAO1 strains called PFB103 (zwf-2) and PFB98 (zwf-1) were initially constructed by chemical mutagenesis (50, 56). These strains are incapable of utilizing mannitol as the sole carbon source and cannot catabolize glucose via the phosphorylative pathway because of a deficiency in G6PDH activity (50, 56). Here we elected to construct an isogenic zwf mutant of wild-type strain PAO1 containing a selectable antibiotic resistance marker, the details of which are given in Materials and Methods. Insertional inactivation of zwf was first confirmed by Southern analysis on a sucroser Gmr zwf mutant called PAO9010. Genomic DNA from the wild-type strain and PAO9010 was cut with PstI/BamHI, transferred to nitrocellulose, and probed with a ∼1-kb PstI-BamHI zwf fragment. Insertional inactivation of the zwf gene was confirmed by demonstration of a band of the predicted size (1.85 kb [Fig. 3A, lane 2]) relative to wild-type DNA (∼1.0 kb [Fig. 3A, lane 1]). Insertional inactivation of the zwf gene was also confirmed by monitoring G6PDH activity in nondenaturing gels. Figure 3B (lane 2) demonstrates an absence of G6PDH activity band in the mutant organism, while activity is clearly evident in the wild-type strain (lane 1).
FIG. 3.
Construction of a P. aeruginosa isogenic zwf mutant PAO9010. (A) Genomic DNA was digested with PstI and BamHI and transferred to nitrocellulose. Southern blot analysis was performed with a 32P-labeled 1.0-kb PstI/BamHI zwf fragment. Lane 1, PAO1 (wild type); lane 2, PAO9010 (zwf mutant). (B) G6PDH activity gel (7.5%) of cell extracts. Lane 1, PAO1 (wild type); lane 2, PAO9010 (zwf mutant).
Transcription of the zwf gene and G6PDH activity are maximal in logarithmic phase.
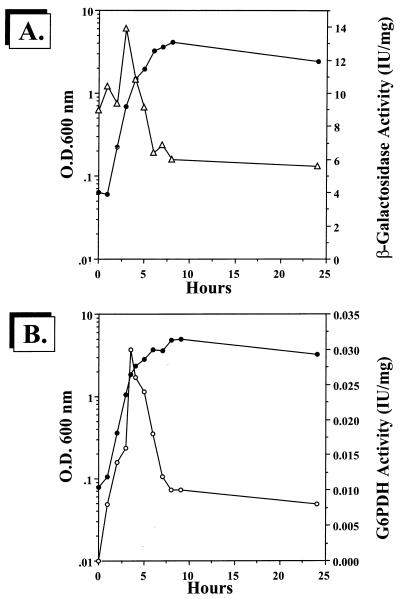
To examine the regulation of zwf in P. aeruginosa, we used both transcriptional and translational approaches. To monitor zwf transcription, a 1.7-kb PvuII zwf-containing fragment from pJFM1 was cloned into the unique SmaI site in pEX100T (62). This plasmid, pJFM4, was cut with BamHI and ligated to a 4.0-kb BamHI-cut promoterless lacZ-Gmr cassette from pZ1918G (61a), resulting in either pJFM5 (sense orientation) or pJFM6 (antisense orientation). These plasmids were conjugated into P. aeruginosa PAO1 via biparental mating and selected on PIA containing gentamicin (300 μg/ml). Genetic confirmation of the zwf::lacZ-Gmr fusion mutant strain PAO9011 was elucidated after sucrose counterselection (as with strain PAO9010) and Southern analysis (data not shown). As shown in Fig. 4A, zwf (sense) transcription rose dramatically in early to mid-logarithmic phase, followed by a marked drop in activity when the organisms entered late log to stationary phase. Antisense transcriptional activity throughout the entire growth phase was negligible (data not shown). G6PDH enzymatic activity paralleled that of the zwf transcriptional analysis in Fig. 4A, with activity rapidly rising in the early stages of growth and falling markedly at the later stages (Fig. 4B).
FIG. 4.
Analysis of zwf transcription (A) and G6PDH activity (B) throughout a normal aerobic growth phase. Wild-type bacteria and strains PAO9011 (zwf::lacZ-Gmr, sense orientation) and PAO9012 (zwf::lacZ-Gmr, antisense orientation) were grown aerobically at 37°C in L broth. At intervals, organisms were harvested and specific activities for β-galactosidase and G6PDH were measured. •, OD600; ▵, β-galactosidase specific activity of strain PAO9011; ○, G6PDH activity.
Effect of carbon source on zwf transcription.
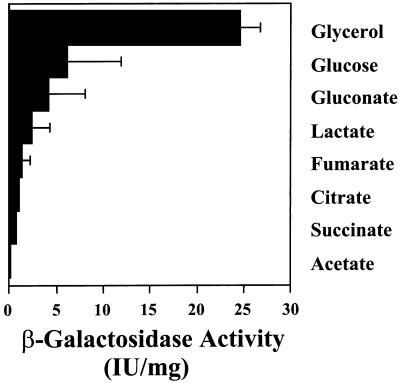
G6PDH activity is induced when organisms are grown in minimal medium supplemented with either glucose, gluconate, or glycerol, highly reduced substrates. In pseudomonads, G6PDH activity is repressed by organic acids via catabolite repression (39; for a review, see reference 10), and this effect has been assumed to be at the level of transcription. To test this assumption, we elected to determine the level of zwf transcription in zwf::lacZ-Gmr fusion strain PAO9011 (constructed above) grown in media containing a variety of carbon sources. As shown in Fig. 5, there is a broad range of transcriptional control of the zwf gene. Growth on glycerol, gluconate, and glucose allowed for the highest levels of zwf transcription. In contrast, growth on organic acids reduced zwf transcription, with succinate and acetate evoking the strongest catabolite repression.
FIG. 5.
Effect of carbon source of zwf transcription. P. aeruginosa PAO9011, with zwf::lacZ integrated into the chromosome (single copy), was grown aerobically at 37°C in minimal medium containing various carbon sources: glycerol (40 mM), glucose (20 mM), gluconate (20 mM), lactate (40 mM), fumarate (30 mM), citrate (20 mM), succinate (30 mM), and acetate (60 mM). Cell suspensions were assayed in early log phase for β-galactosidase activity as previously described (45).
Purification of P. aeruginosa G6PDH and enzyme kinetics.
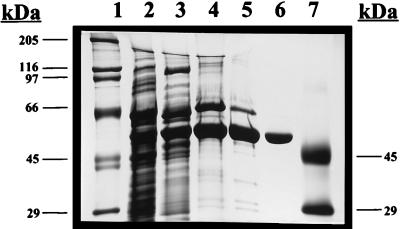
To determine the kinetic parameters of the P. aeruginosa G6PDH, we purified G6PDH from wild-type bacteria overexpressing the enzyme. We used a four-step procedure involving (i) ammonium sulfate precipitation of cell extracts, (ii) DE-52 anion exchange, (iii) hydroxyapatite chromatography, and (iv) Bio-Gel 150 gel filtration. Figure 6 demonstrates the stages of purification using SDS-polyacrylamide gel electrophoresis (PAGE). Clearly, the most significant purification step was DE-52 anion-exchange chromatography (Fig. 6, lane 4). Some proteins which bound to DE-52 did not bind to hydroxyapatite (Fig. 6, lane 5). The smaller contaminating proteins still present after hydroxyapatite chromatography were resolved easily with the Bio-Gel 150 matrix, while G6PDH, because of its large size (estimated at ∼220 kDa), eluted from the column at the void volume (Fig. 6, lane 6). We obtained approximately 500 μg of purified G6PDH, a small portion of which was used for the kinetic analyses described below.
FIG. 6.
Analysis of the purification of P. aeruginosa PAO1 G6PDH by SDS-PAGE (10% gel). Lane 1, high-molecular-weight protein standard; lane 2, crude cell extract (45 μg); lane 3, 50% ammonium sulfate cut (30 μg); lane 4, DE-52 column eluate (10 μg); lane 5, hydroxyapatite column eluate, (7 μg); lane 6, Bio-Gel 150 column eluate (2 μg); lane 7, low-molecular-weight protein standard.
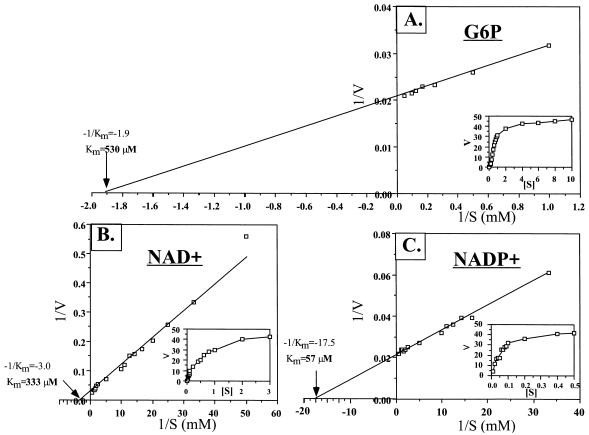
Many G6PDH enzymes have been purified from organisms or tissues from various phyla. Unlike the P. aeruginosa (35) and several other bacterial G6PDH enzymes, however, many G6PDH enzymes cannot use both NAD+ and NADP+ as cofactors for enzyme activity. To determine the specificity of the P. aeruginosa G6PDH toward G6P, NAD+, and NADP+, we performed a kinetic analysis using each compound. Double-reciprocal Lineweaver-Burk plots of enzymatic activity as a function of G6P, NAD+, and NADP+ concentration are shown in Fig. 7. The estimated Km values for G6P (with NADP+ as the cofactor), NAD+, and NADP+ were 530, 333, and 57 μM, respectively. The specific activities for NAD+ and NADP+ were calculated to be 176 and 69 μmol/min/mg.
FIG. 7.
Estimation of Km for glucose-6-phosphate (G6P; A), NAD+ (B), and NADP+ (C) with purified P. aeruginosa G6PDH. Rates were measured in terms of an increase in absorbance at 340 nm at 22°C. Initial rates as a function of either glucose-6-phosphate, NAD+, or NADP+ are presented above on reciprocal coordinates, using 4.2 nM purified P. aeruginosa G6PDH.
An absence of G6PDH confers enhanced sensitivity to paraquat.
The intracellular redox status of all living cells is governed in part by the levels of NAD(P)+ relative to NAD(P)H. NADPH is an essential cofactor for glutathione reductase (3) and alkylhydroperoxide reductase (32), enzymes important in combating oxidative stress (9). Since by-products of G6PDH activity are NADH and NADPH, and reduced levels of these cofactors would limit the efficacy of such enzymes, we predicted that a zwf mutant would be more sensitive to paraquat. However, for paraquat to cause oxidative stress, it must first be reduced, followed by autoxidation of the paraquat monocation radical (PQ*) by oxygen, creating O2. Paraquat reduction within E. coli is catalyzed by three paraquat:NADPH oxidoreductases (39). It would be predicted that the activity of such an enzyme(s) under aerobic conditions would, in part, dictate susceptibility or resistance to paraquat. Because the P. aeruginosa G6PDH enzyme also generates NADH (35), it is possible that paraquat can also be reduced by enzymes utilizing this cofactor that may not utilize NADPH. To monitor the activities of NADH- and NADPH-dependent paraquat oxidoreductases and to ensure that wild-type and zwf mutant levels of these enzymes were similar, cell extracts of both strains were subjected to nondenaturing PAGE followed by activity staining for both enzymes. As shown in Fig. 8, P. aeruginosa possesses both NADPH (Fig. 8A)- and NADH (Fig. 8B)-dependent paraquat oxidoreductases; there were two of the latter and at least five of the former. The addition of PQ (lanes 5 to 8 in Fig. 8A and B) triggered a 1.2-fold increase in oxidoreductase activity, which was measured spectrophotometrically (data not shown). Thus, based on the activity gel, spectrophotometric measurement, and linear scanning densitometry (data not shown), we conclude that wild-type and zwf mutant levels of both paraquat-reducing enzymes were similar. Furthermore, the activities of the important antioxidants SoD and catalase were identical in both wild-type and zwf mutant organisms (data not shown).
FIG. 8.
Analysis of wild-type and zwf mutant cell extracts for paraquat:NADPH and paraquat:NADH oxidoreductase activity. P. aeruginosa PAO1 and PAO9010 containing either pEX30 (vector control) or the zwf expression vector pJFM3 were grown in L broth minus NaCl containing 400 μg of carbenicillin per ml and 0.2 mM IPTG until mid-logarithmic phase (OD600 = 0.6), and the culture was divided into two parts. One set was treated with 100 μM paraquat (PQ), and the other served as a control. These suspensions were incubated aerobically for an additional hour at 37°C prior to preparation of cell extracts. Extracts were separated by nondenaturing PAGE on 7.5% polyacrylamide gels and stained for paraquat:NADPH (A) and paraquat:NADH (B) oxidoreductase activities as previously described (40). Lanes 1 through 4, mid-logarithmic-phase organisms; lanes 5 through 8, mid-logarithmic-phase organisms plus 100 μM paraquat for 1 h. Lanes 1 and 5, PAO1(pEX30); lanes 2 and 6, PAO1(pJFM3); lanes 3 and 7, PAO1 zwf(pEX30); lanes 4 and 8, PAO1 zwf(pJFM3).
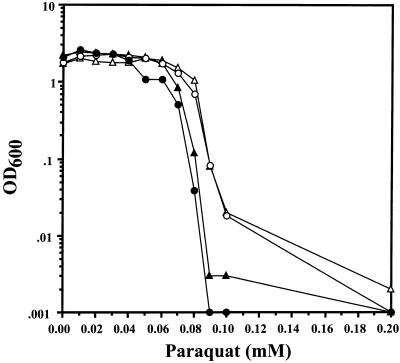
Finally, to test whether a G6PDH deficiency increases the susceptibility of P. aeruginosa to paraquat, wild-type and zwf mutant strains containing the zwf overexpression vector pJFM3 or the control vector pEX30 were grown in L broth in the absence of NaCl and exposed to increasing concentrations of paraquat. Our rationale for not supplementing L broth with NaCl in this experiment is that NaCl has been shown to interfere with paraquat for its receptor on the cell surface and thus restricts its antibiotic efficacy (34). As shown in Fig. 9, wild-type organisms were resistant to 60 μM paraquat whereas the zwf mutant demonstrated increased sensitivity at concentrations of 40 μM and greater. When the zwf gene was provided in trans in both the zwf mutant and wild-type strains, there was enhanced resistance to paraquat which exceeded that of wild-type organisms.
FIG. 9.
Effect of paraquat on growth of wild-type and zwf mutant P. aeruginosa. P. aeruginosa PAO1 and PAO9010 harboring either pEX30 (control vector) or pJFM3 (zwf overexpression plasmid) were grown in L broth (without NaCl) containing 400 μg of carbenicillin per ml and 0.2 mM IPTG for 17 h at 37°C under aerobic conditions. Aliquots (2.5 μl) were added to 2.5 ml of the same media containing increasing concentrations of paraquat and incubated on a roller wheel at 90 rpm for 17 h at 37°C. The final OD600 nm was recorded. The data are representative of four different experiments. ▴, PAO1(pEX30); •, PAO9010(pEX30); ▵, PAO1(pJFM3); ○, PAO9010(pJFM3).
DISCUSSION
P. aeruginosa is capable of utilizing myriad carbon sources for growth. Although it prefers TCA cycle intermediates (e.g., succinate or citrate), it also readily metabolizes glucose. A key enzyme involved in glucose metabolism is G6PDH, which converts glucose-6-phosphate to 6-phosphogluconate. 6-Phosphogluconate is further metabolized by the obligatory Entner-Doudoroff pathway (29). In this study, we have cloned and characterized the zwf gene encoding G6PDH to better understand the physiological role of this enzyme in normal aerobic metabolism.
Regulation of the P. aeruginosa zwf gene was found to be under tight transcriptional control. The transcription of zwf, as well as G6PDH activity, was greatest during mid-logarithmic phase and then rapidly decreased to basal levels. This was not surprising since substrates triggering elevated zwf transcription (e.g., glucose) are likely depleted once organisms begin to enter the stationary phase (41). Furthermore, P. aeruginosa and other Pseudomonas species exhibit a strong catabolite repression control (10, 42), and this pattern of induction/repression is a hallmark of such an event. Catabolite repression control of glucose-6-phosphate activity is normally striking, as evidenced by an approximate 90 to 99% reduction in activity when cells are grown on organic acids (66). We observed a similarly wide range of transcriptional control for the chromosomal zwf::lacZ fusion and now conclude that catabolite repression control of the zwf gene is at the level of transcription. Interestingly, when the zwf::lacZ fusion was introduced on a multicopy plasmid (pUCP22 at ∼30 copies/cell [61b]), β-galactosidase activity was reduced only ∼50% during growth on organic acids compared to glucose, glycerol, or gluconate (data not shown). This is strikingly similar to the HexC phenotype (64), where a cloned promoter for the hex regulon genes edd and gap is proposed to titrate a hex regulon repressor, leading to increased basal activity of all five hex regulon enzymatic activities (Zwf, Edd, Eda, Gap, and Glk) (12, 64, 65). This result indicates the presence of a Hex repressor binding site upstream of the zwf gene, for which there is in vitro evidence (51).
Unlike eukaryotic G6PDH enzymes, which can utilize only NADP+ as a cofactor, many bacterial enzymes, including those from Leuconostoc mesenteroides (13), Acetobacter suboxydans (8), A. hansenii (53), Azotobacter vinelandii (2), P. aeruginosa (this study and reference 35), P. fluorescens (37), P. (Burkholderia) cepacia (7), and P. multivorans (67), utilize both NAD+ and NADP+. Of particular interest was the remarkably low Km of the P. aeruginosa G6PDH for its substrate, glucose-6-phosphate (530 μM). Estimated Km values range from 2.3 to 2.7 mM for glucose-6-phosphate of purified or partially purified G6PDH enzymes or the NAD+/NADP+-cofactored G6PDH from the related organism P. fluorescens (37) to 5 mM in the other related species P. multivorans (67) and Burkholderia (formerly Pseudomonas) cepacia (7). Furthermore, the cofactor specificity for the P. aeruginosa G6PDH was nearly sixfold greater for NADP+ (Km = 57 μM) than for NAD+ (Km = 333 μM) but the specific activity was greater with NAD (176 versus 69 U/mg). One particular advantage for possessing an enzyme with such superb efficiency is that the valuable cellular reducing equivalents NADH and NADPH, by-products of G6PDH activity, are essential cofactors for hundreds of enzymes involved in catabolic or anabolic processes and cellular defense and could be produced in environments where glucose is limiting.
The role of G6PDH in susceptibility to paraquat was also dissected. Resistance to reactive oxygen intermediates in P. aeruginosa is governed in part by antioxidant enzymes, including iron- and manganese-cofactored SOD (21–26) and at least two heme catalases (6, 21). One compound which exacerbates the production of O2− and H2O2 within aerobic bacteria is paraquat (18, 20). In E. coli, transcriptional control of the zwf gene is governed by the soxRS regulon (for a review, see reference 14) and increased upon exposure to paraquat (18, 33). It was postulated that an increase in G6PDH activity is brought about because of the need for NADPH, required as a cofactor for glutathione reductase (3) and alkylhydroperoxide reductase (32) activities. The latter enzymes are important in combating paraquat-mediated oxidative stress (9). The sensitivity of our P. aeruginosa zwf mutant to paraquat is consistent with an unpublished observation in E. coli, where a zwf mutant demonstrated increased sensitivity to both the related redox-active compound menadione and H2O2 (17). Although the enhanced sensitivity of our P. aeruginosa zwf mutant was modest, enhanced resistance could be conferred by overexpression of intracellular G6PDH (Fig. 9). Still, sensitivity to paraquat is likely dependent on the presence or absence of multiple cellular factors, some of which include SOD (24), catalase (6), methionine sulfoxide reductase (46, 47), DNA repair systems (30, 31), and various regulatory proteins (e.g., SoxRS [14]).
Although the P. aeruginosa gor gene encoding glutathione reductase has been cloned (49), no mutants are currently available. Similarly, an ahp (alkylhydroperoxide reductase [32]) homolog has not been identified in this organism. It will be important to construct gor and, if possible, ahp mutants of P. aeruginosa to determine whether inactivation of these genes increases susceptibility to various forms of oxidative stress.
ACKNOWLEDGMENTS
J.-F.M. and P.W.H. contributed equally to completion of this project.
This work was supported in part by grant AI-32085 from the National Institutes of Health (D.J.H.), Cystic Fibrosis grant HASSET96PO (D.J.H.), and Departmental Start-Up Funds from the Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati College of Medicine.
Mary Beth Dail and Ann Covert-Rinaldi provided excellent technical support for the subcloning and sequencing of the zwf gene.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson B M, Anderson C D. Purification and characterization of Azotobacter vinelandii glucose-6-phosphate dehydrogenase: dual coenzyme specificity. J Bacteriol. 1995;321:94–100. doi: 10.1006/abbi.1995.1372. [DOI] [PubMed] [Google Scholar]
- 3.Asnis R E. A glutathione reductase from Escherichia coli. J Biol Chem. 1955;213:77–85. [PubMed] [Google Scholar]
- 4.Beers R F, Jr, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brown S M, Howell M L, Vasil M L, Anderson A, Hassett D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cacciapuoti A F, Lessie T G. Characterization of the fatty acid-sensitive glucose 6-phosphate dehydrogenase from Pseudomonas cepacia. J Bacteriol. 1977;132:555–563. doi: 10.1128/jb.132.2.555-563.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheldelin V H. Metabolic pathways in microorganisms. New York, N.Y: John Wiley & Sons; 1961. [Google Scholar]
- 9.Christman M F, Morgan R, Jacobson F S, Ames B N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 10.Collier D N, Hager P W, Phibbs P V., Jr Catabolite repression control in the Pseudomonads. Res Microbiol. 1996;147:551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- 11.Cross A, Allen J R, Burke J, Ducel G, Harris A, John J, Johnson D, Lew M, MacMillan B, Skalova R, Wenzel R, Tenney J. Nosocomial infections due to Pseudomonas aeruginosa: review of recent trends. Rev Infect Dis. 1983;5:S837–S845. doi: 10.1093/clinids/5.supplement_5.s837. [DOI] [PubMed] [Google Scholar]
- 12.Cuskey S M, Wolff J A, Phibbs P V, Jr, Olsen R H. Cloning of genes specifying carbohydrate catabolism in Pseudomonas aeruginosa and Pseudomonas putida. J Bacteriol. 1985;162:865–871. doi: 10.1128/jb.162.3.865-871.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeMoss R D. Glucose 6-phosphate and 6-phosphogluconic dehydrogenases from Leuconostoc mesenteroides. Methods Enzymol. 1955;1:328–334. [Google Scholar]
- 14.Demple B. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon-a review. Gene. 1996;179:53–57. doi: 10.1016/s0378-1119(96)00329-0. [DOI] [PubMed] [Google Scholar]
- 15.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 16.Govan J R W, Harris G S. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986;3:302–308. [PubMed] [Google Scholar]
- 17.Greenberg J T. Adaptive response to oxidative stress in Escherichia coli. Ph.D. thesis. Cambridge, Mass: Harvard University; 1989. [Google Scholar]
- 18.Greenberg J T, Demple B. A global response induced in Escherichia coli by redox-cycling agents overlaps with that induced by peroxide stress. J Bacteriol. 1989;171:3933–3939. doi: 10.1128/jb.171.7.3933-3939.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg J T, Monach P, Chou J H, Josephy P D, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan H M, Fridovich I. Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch Biochem Biophys. 1979;196:385–395. doi: 10.1016/0003-9861(79)90289-3. [DOI] [PubMed] [Google Scholar]
- 21.Hassett D J, Charniga L, Bean K A, Ohman D E, Cohen M S. Antioxidant defense mechanisms in Pseudomonas aeruginosa: resistance to the redox-active antibiotic pyocyanin and demonstration of a manganese-cofactored superoxide dismutase. Infect Immun. 1992;60:328–336. doi: 10.1128/iai.60.2.328-336.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassett D J, Howell M L, Ochsner U, Johnson Z, Vasil M, Dean G E. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator (Fur) in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J Bacteriol. 1997;179:1452–1459. doi: 10.1128/jb.179.5.1452-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassett D J, Howell M L, Sokol P A, Vasil M, Dean G E. Fumarase C activity is elevated in response to iron deprivation and in mucoid, alginate-producing Pseudomonas aeruginosa: cloning and characterization of fumC and purification of native FumC. J Bacteriol. 1997;179:1442–1451. doi: 10.1128/jb.179.5.1442-1451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassett D J, Schweizer H P, Ohman D E. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassett D J, Sokol P, Howell M L, Ma J-F, Schweizer H P, Ochsner U, Vasil M L. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake and altered aerobic metabolism. J Bacteriol. 1996;178:3996–4003. doi: 10.1128/jb.178.14.3996-4003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassett D J, Woodruff W A, Wozniak D J, Vasil M L, Cohen M S, Ohman D E. Cloning of the sodA and sodB genes encoding manganese and iron superoxide dismutase in Pseudomonas aeruginosa: demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J Bacteriol. 1993;175:7658–7665. doi: 10.1128/jb.175.23.7658-7665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Molecular analysis of the Erwinia chrysanthemi region containing the kdgA and zwf genes. Mol Microbiol. 1994;11:67–75. doi: 10.1111/j.1365-2958.1994.tb00290.x. [DOI] [PubMed] [Google Scholar]
- 29.Hunt J C, Phibbs P V., Jr Regulation of alternative peripheral pathways of glucose catabolism during aerobic and anaerobic growth of Pseudomonas aeruginosa. J Bacteriol. 1983;154:793–802. doi: 10.1128/jb.154.2.793-802.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imlay J A, Linn S. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J Bacteriol. 1986;166:519–527. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imlay J A, Linn S. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bacteriol. 1987;169:2967–2976. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobson F S, Morgan R W, Christman M F, Ames B N. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage: purification and properties. J Biol Chem. 1989;264:1488–1496. [PubMed] [Google Scholar]
- 33.Kao S M, Hassan H M. Biochemical characterization of a paraquat tolerant mutant of Escherichia coli. J Biol Chem. 1985;260:10478–10481. [PubMed] [Google Scholar]
- 34.Kitzler J W, Fridovich I. Effects of salts on the lethality of paraquat. J Bacteriol. 1986;167:346–349. doi: 10.1128/jb.167.1.346-349.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lessie T, Neidhardt F C. Adenosine triphosphate-linked control of Pseudomonas aeruginosa glucose-6-phosphate dehydrogenase. J Bacteriol. 1967;93:1337–1345. doi: 10.1128/jb.93.4.1337-1345.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lessie T G, Phibbs P V., Jr Alternative pathway of carbohydrate utilization in pseudomonads. Annu Rev Microbiol. 1984;38:359–387. doi: 10.1146/annurev.mi.38.100184.002043. [DOI] [PubMed] [Google Scholar]
- 37.Lessmann D, Schmiz K-L, Kurz G. d-Glucose-6-phosphate dehydrogenase (Entner-Doudoroff enzyme) from Pseudomonas fluorescens. Eur J Biochem. 1975;59:545–559. doi: 10.1111/j.1432-1033.1975.tb02481.x. [DOI] [PubMed] [Google Scholar]
- 38.Liochev S I, Fridovich I. Effects of overproduction of superoxide dismutase on the toxicity of paraquat toward Escherichia coli. J Biol Chem. 1991;266:8747–8750. [PubMed] [Google Scholar]
- 39.Liochev S I, Fridovich I. Paraquat diaphorases in Escherichia coli. Free Radical Biol Med. 1994;16:555–559. doi: 10.1016/0891-5849(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 40.Liochev S I, Hausladen A, Beyer W F, Fridovich I. NADPH: ferredoxin oxidoreductase acts as a paraquat diaphorase and is a member of the soxRS regulon. Proc Natl Acad Sci USA. 1994;91:1328–1331. doi: 10.1073/pnas.91.4.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma J-F, Phibbs P V, Jr, Hassett D J. Glucose stimulates alginate production and algD transcription in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1997;148:217–221. doi: 10.1111/j.1574-6968.1997.tb10291.x. [DOI] [PubMed] [Google Scholar]
- 42.MacGregor C H, Wolff J A, Arora S K, Hylemon P B, Phibb P V., Jr . Catabolite repression control in Pseudomonas aeruginosa. In: Galli E, Silver S, Witholt B, editors. Pseudomonas: molecular biology and biotechnology. Washington, D.C: American Society for Microbiology; 1992. pp. 198–206. [Google Scholar]
- 43.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 44.Marklund S, Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur J Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 45.Miller J H. A short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Press; 1992. pp. 72–74. [Google Scholar]
- 46.Moskovitz J, Berlett B S, Poston J M, Stadtman E R. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci USA. 1997;94:9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moskovitz J, Rahman M A, Strassman J, Yancey S O, Kushner S R, Brot N, Weissbach H. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newman J, Karakaya H, Scanlan D J, Mann N H. A comparison of gene organization in the zwf region of the genomes of the cyanobacteria Synechococcus sp. PCC 7942 and Anabaena sp. PCC 7120. FEMS Microbiol Lett. 1995;133:187–193. doi: 10.1111/j.1574-6968.1995.tb07882.x. [DOI] [PubMed] [Google Scholar]
- 49.Perry A C, Ni Bhriain N, Brown N L, Rouch D A. Molecular characterization of the gor gene encoding glutathione reductase from Pseudomonas aeruginosa: determinants of substrate specificity among pyridine nucleotide-disulphide oxidoreductases. Mol Microbiol. 1991;5:63–71. [PubMed] [Google Scholar]
- 50.Phibbs P V, Jr, McGowen S M, Feary T W, Blevins W T. Mannitol and fructose catabolic pathways of Pseudomonas aeruginosa carbohydrate-negative mutants and pleiotropic effects of certain enzyme deficiencies. J Bacteriol. 1978;133:717–728. doi: 10.1128/jb.133.2.717-728.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Proctor W D, Arora S K, Hager P W, Phibbs P V., Jr . Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Integration host factor and the putative repressor HexR bind the hexC locus of Pseudomonas aeruginosa PAO1, abstr. K-95. [Google Scholar]
- 52.Prohaska J R. Changes in tissue growth, concentrations of copper, iron, cytochrome oxidase and superoxide dismutase subsequent to dietary or genetic copper deficiency in mice. J Nutr. 1983;113:2048–2058. doi: 10.1093/jn/113.10.2048. [DOI] [PubMed] [Google Scholar]
- 53.Ragunathan S, Levy H R. Purification and characterization of the NAD-preferring glucose 6-phosphate dehydrogenase from Acetobacter hansenii (Acetobacter xylinum) Arch Biochem Biophys. 1994;310:360–366. doi: 10.1006/abbi.1994.1179. [DOI] [PubMed] [Google Scholar]
- 54.Reese M G, Eeckman F H. Novel neural network algorithms for improved eukaryotic promoter site recognition. Presented at the Seventh International Genome Sequencing and Analysis Conference, Hilton Head, S.C. 1995. [Google Scholar]
- 55.Reynolds H Y, Levine A S, Wood R E, Zierdt C H, Dale D C, Pennington J E. Pseudomonas aeruginosa infections: persisting problems and current research to find new therapies. Ann Intern Med. 1975;82:819–831. doi: 10.7326/0003-4819-82-6-819. [DOI] [PubMed] [Google Scholar]
- 56.Roehl R A, Feary T W, Phibbs P V., Jr Clustering of mutations affecting central pathway enzymes of carbohydrate catabolism in Pseudomonas aeruginosa. J Bacteriol. 1983;156:1123–1129. doi: 10.1128/jb.156.3.1123-1129.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowley D L, Wolf R E J. Molecular characterization of the Escherichia coli K-12 zwf gene encoding glucose-6-phosphate dehydrogenase. J Bacteriol. 1991;173:968–977. doi: 10.1128/jb.173.3.968-977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scanlan D J, Newman J, Sebaihia M, Mann N H, Carr N G. Cloning and sequence analysis of the glucose-6-phosphate dehydrogenase gene from the cyanobacterium Synechococcus PCC 7942. Plant Mol Biol. 1992;19:877–880. doi: 10.1007/BF00027085. [DOI] [PubMed] [Google Scholar]
- 59.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 60.Schweizer H P. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 61.Schweizer H P. Two plasmids, X1918 and Z1918, for easy recovery of the xylE and lacZ reporter genes. Gene. 1993;134:89–91. doi: 10.1016/0378-1119(93)90178-6. [DOI] [PubMed] [Google Scholar]
- 61a.Schweizer, H. P. Unpublished data.
- 61b.Schweizer, H. P. Personal communication.
- 62.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 63.Simon R, Priefer U, Puehler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 64.Temple L, Cuskey S M, Perkins R E, Bass R C, Morales N M, Christie G E, Olsen R H, Phibbs P V., Jr Analysis of cloned structural genes and regulatory genes for carbohydrate utilization in Pseudomonas aeruginosa PAO. J Bacteriol. 1990;172:6396–6402. doi: 10.1128/jb.172.11.6396-6402.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Temple L, Sage A, Christie G E, Phibbs P V., Jr Two genes for carbohydrate catabolism are divergently transcribed from a region of DNA containing the hexC locus in Pseudomonas aeruginosa. J Bacteriol. 1994;176:4700–4709. doi: 10.1128/jb.176.15.4700-4709.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tiwari N P, Campbell J J R. Enzymatic control of the metabolic activity of Pseudomonas aeruginosa grown in glucose or succinate media. Biochim Biophys Acta. 1969;192:395–401. doi: 10.1016/0304-4165(69)90388-2. [DOI] [PubMed] [Google Scholar]
- 67.Vander Wyk J C, Lessie T G. Purification and characterization of the Pseudomonas aeruginosa glucose-6-phosphate dehydrogenase active with nicotinamide adenine dinucleotide. J Bacteriol. 1974;120:1033–1042. doi: 10.1128/jb.120.3.1033-1042.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67a.Wallace, W. H., P. W. Hager, and P. V. Phibbs. Unpublished data.
- 68.West S E H, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 69.Wolff J A, MacGregor C H, Eisenberg R C, Phibbs P V., Jr Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J Bacteriol. 1991;173:4700–4706. doi: 10.1128/jb.173.15.4700-4706.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]