Dear Editor,
Since the start of the COVID-19 pandemic, thousands of neutralizing antibodies (NAbs) targeting different epitopes of the spike protein have been identified and characterized.1–3 However, the emergence of the Omicron variant and its subvariants has raised global concern due to their resistance to most reported monoclonal antibodies and dramatically decreases the immune efficacy of current vaccines.4,5 A newly emerging subvariant XBB.1.16 that is rapidly disseminating on a global scale has been associated with a previously unreported symptom of red or pink eye.6 Compared to XBB.1.5, XBB.1.16 harbors two additional mutations, E180V and K478R, which are hypothesized to confer enhanced transmissibility and antigenic escape.7 The enhanced capacity of XBB.1.16 for transmission and immune evasion poses a greater threat to public health. Therefore, continuous efforts toward the rapid isolation and engineering of antibodies with cross-reactivity and potently neutralizing epitope specificities are essential to aid in the containment of COVID-19 in the face of viral genomic alterations.
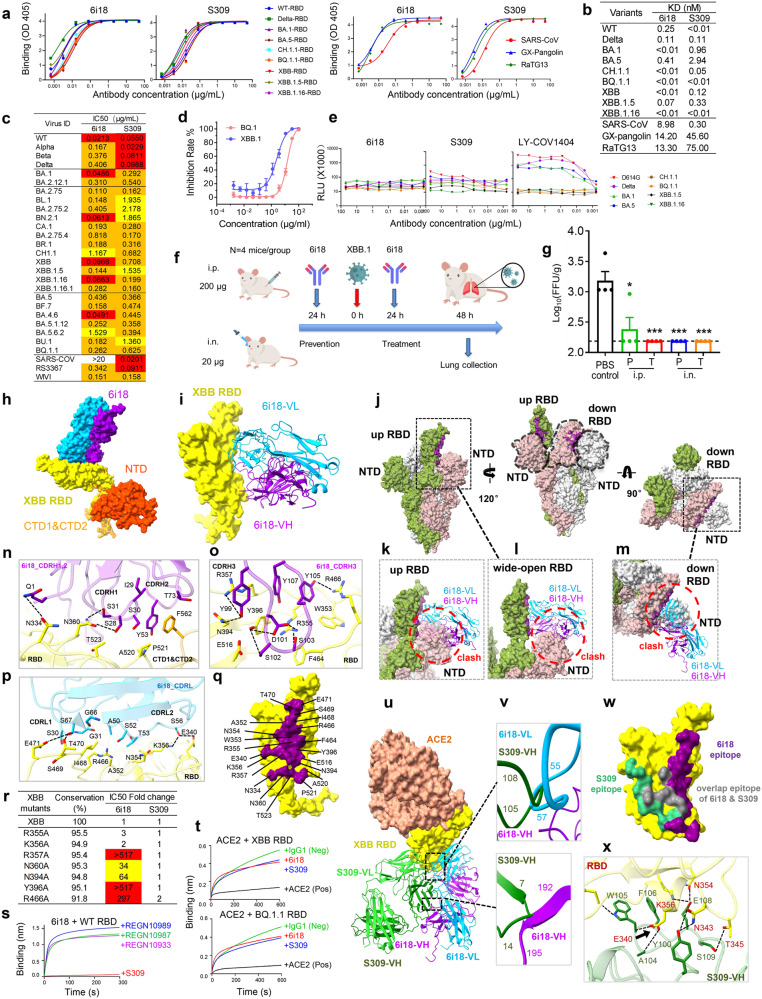
Here, we report the identification of a broadly neutralizing monoclonal antibody (mAb), 6i18 (IGHV4-59, IGKV1-12), derived from a convalescent COVID-19 patient.8 6i18 exhibited strong binding ability to the RBDs of the ancestral SARS-CoV-2 strain and the variants of concern (VOCs), including Delta, BA.1, BA.5, CH.1.1, BQ.1.1, XBB, XBB.1.5, and XBB.1.16 variants (Fig. 1a, left). 6i18 also bound RBDs from other sarbecoviruses like SARS-CoV, GX-pangolin, and RaTG13 (Fig. 1a, right). Moreover, 6i18 exhibits extraordinarily high affinity for various Omicron variants, with KD of 0.01 nM for the RBDs of BA.1, BQ.1.1, XBB, XBB.1.5 and XBB.1.16 (Fig. 1b; Supplementary information, Fig. S1).9 The binding affinity of 6i18 exceeded that of the cross NAb S309 against the Omicron RBDs.
Fig. 1. 6i18 inhibits SARS-CoV-2 variants through a novel mechanism of disrupting S trimer integrity.
a Binding of 6i18 to the RBDs of Omicron variants as assessed by ELISA. S309 was used as a control. b Binding affinity of 6i18 and S309 to the RBDs of Omicron variants as determined by BLI. c Neutralization breadth and potency of 6i18 against a panel of 28 pseudotype SARS-CoV-2 variants. S309 was used as a control. IC50 values between 1–10 μg/mL are colored yellow, 0.1–1 μg/mL are colored orange, and 0.01–0.1 μg/mL are colored red. d Neutralization of the authentic viruses BQ.1 and XBB.1 by 6i18. e ADE of 6i18. LY-CoV1404 and S309 were used as positive and negative controls, respectively. f Schematic overview of the assessment of prophylactic and therapeutic efficacy of 6i18 against XBB.1 infection in mice. g The prophylactic (P) and therapeutic (T) efficacy of 6i18 through intraperitoneal (i.p.) or intranasal (i.n.) routes. Viral titers of XBB.1 in the lung of mice was determined by FFA. h Cryo-EM map of the XBB S in complex with 6i18 with the VL domain colored in blue and VH domain in violet, and the RBD in yellow and NTD in range red. i Structure of XBB S RBD–6i18. The RBD is displayed in yellow surface mode. The heavy chain and light chain of 6i18 are shown as ribbons colored in cornflower blue and violet, respectively. j Apo Omicron S (PDBID: 7WVN) in three different views, showing the 6i18 epitope in purple. Binding of 6i18 (light chain in blue, heavy chain in purple) to an XBB S monomer with up RBD (k), wide-open RBD (PDBID: 7WHK) (l), and down RBD (m). In all cases, 6i18 clashes with the NTD of the adjacent monomer (indicated by red dashed circles). n–p The detailed interactions of 6i18 CDRL, CDRH1,2, CDRH3 with XBB.1 RBD. Residues participating in interactions are represented as sticks. Polar interactions are indicated as dotted lines. q Close-up view of the 6i18 epitope on the RBD. The residues involved in the interaction are labeled. r Neutralization of 6i18 against Omicron XBB RBD single mutants involved in 6i18 binding. Fold change is calculated as the IC50 of the mutant/the IC50 of WT. Mutants that decreased the neutralization of 6i18 with fold change values between 10–100 are highlighted in yellow, and values > 100 are highlighted in red. s Binding of 6i18 to the SARS-CoV-2 RBD in competition with S309 and SARS-CoV-2 specific mAbs REGN10989, REGN10987, and REGN10933 as measured by BLI. t The binding of ACE2 to the RBDs of XBB and BQ.1.1 in competition with 6i18 (red) and S309 (blue). The HIV-1 mAb VRC01 was used as an IgG1 isotype negative control (green), and the addition of ACE2 was used as a positive control (black). u Epitopes of 6i18 and S309 are outside of RBM (PDBID: 7WVP, Omicron S RBD with ACE2). There is steric hindrance between 6i18 and S309 (PDBID: 7XCK). v Close-up view of clashes between 6i18 and S309. w Epitope of 6i18 overlaps with S309 (gray), but 6i18 (purple) binds RBD in a different direction compared to S309 (green). x Detailed interaction between XBB S RBD and S309, inferred from Omicron S RBD with S309 (PDBID: 7XCK).
We measured the neutralizing breadth and potency of 6i18 using a panel of 28 pseudoviruses expressing the spike (S) protein of SARS-CoV-2 variants as well as other sarbecoviruses. 6i18 neutralized all the SARS-CoV-2 variants tested, including BQ.1.1, XBB.1.5, XBB.1.16, and XBB.1.16.1, and exhibited greater potency in neutralizing the BA.2.75 subvariants compared to S309 (Fig. 1c). Remarkably, 6i18 cross-neutralized SARS-CoV and the bat SARS-related coronaviruses (SARSr-CoVs) WIV1 and Rs3367. Moreover, 6i18 effectively neutralized the authentic XBB.1 and BQ.1 variant (Fig. 1d). Therefore, 6i18 represents a promising candidate therapeutic reagent for the most resistant Omicron variants, XBB.1.16 and BQ.1.1.
6i18 effectively neutralized the wild-type SARS-CoV-2, but exhibited diminished neutralizing potency against the Alpha, Beta, and Delta variants, while S309 showed reduced neutralizing activity against all Omicron variants (Fig. 1c). We evaluated the neutralization breadth of 6i18 and S309 against a panel of 47 circulating single mutants. Both 6i18 and S309 potently neutralized most circulating single mutants, but 6i18 displayed decreased neutralization against G446V and D614G mutants (21- and 15-fold; Supplementary information, Table S1 and Fig. S2). The Alpha, Beta, and Delta variants possess the D614G mutation, possibly explaining the reduced neutralizing ability of 6i18 against these variants. S309 exhibited resistance to S371F mutant (>2252 fold) or reduced neutralization against T307E, F374A, and P681R mutants (14–30 fold, Supplementary information, Table S1). All Omicron variants carry the S371F mutation (Supplementary information, Fig. S2), accounting for the 10-fold lower neutralizing potency of S309 against Omicron variants compared to wild-type, Alpha, Beta, and Delta variants (Fig. 1c).
The influence of antibody-dependent enhancement (ADE) induced by different virus variants in FcR-positive cell line (Raji B cells) was analyzed. 6i18 and S309 did not enhance infection with any of the tested VOCs, while the control mAb LY-COV1404 enhanced the infection of D614G, Delta, BA.1, and BA.5 variants, but not the BQ.1.1 or XBB sublineages (Fig. 1e).
The prophylactic and therapeutic efficacy of 6i18 was assessed in BALB/c wild-type mice infected with the authentic XBB.1 variant, which possesses an additional G252V mutation compared to XBB. Six-week-old mice were administered with 6i18 at a dose of 200 μg/each intraperitoneally (i.p.) or 20 μg/each intranasally (i.n.) either 24 h before or 24 h after XBB.1 infection (Fig. 1f). Lungs were collected 48 h post-infection to quantify viral titers using a focus-forming assay (FFA). Following XBB.1 challenge, no or only very mild clinical symptoms were observed, but viral particles were detected in lung samples from the PBS control group (Fig. 1g). 6i18 significantly reduced lung viral titers in both prophylactic (P) and therapeutic (T) groups compared to the control group (P < 0.05). Moreover, no significant difference in viral titers was observed between mice treated through the i.p. and i.n. routes (Fig. 1g).
To investigate the neutralization mechanism of 6i18, we purified the prefusion-stabilized SARS-CoV-2 Omicron XBB S ectodomain trimer complexed with 6i18 (Omicron S-6i18). Negative stain images revealed that the XBB S trimers were disassembled upon 6i18 binding (Supplementary information, Fig. S3a–c). We then determined cryo-EM structure of the complex. Consistent with negative stain results, cryo-EM showed only one particle state: the XBB S monomer complexed with 6i18 (Fig. 1h). The structure of the XBB S monomer (residues 31–700) with 6i18 Fab region was determined at 3.30 Å resolution (Fig. 1h, i; Supplementary information, Table S2 and Fig. S4).
Structural comparison of the XBB S-6i18 complex with apo-Omicron S (7WVN, Fig. 1j) and the wide-open RBD of Omicron S (7WHK, Fig. 1l) reveals steric clashes between 6i18 and the N-terminal domain (NTD) of adjacent monomers upon 6i18 binding to the “up” RBD conformation (Fig. 1k), the ‘wide-open’ RBD conformation (Fig. 1l), and the “down” RBD conformation (Fig. 1m). This indicates that 6i18 induces movement between RBD and NTD of S, resulting in S trimer disassembly.
The binding between 6i18 and the RBD occludes 1083 Å2 of surface area, engaging a total of 21 residues within the RBD domain. The interaction between 6i18 and RBD is largely driven by extensive hydrophilic and hydrophobic interactions between CDRH1, CDRH2 and CDRH3 of heavy chain, and CDRL1, CDRL2 and β4 of light chain of 6i18 and RBD (Fig. 1n–p). Residues N334, E340, A352, W353, N354, R355, K356, R357, N360, N394, Y396, F464, R466, I468, S469, T470, E471, E516, A520, P521, and T523 of the RBD are involved in this interaction (<4 Å), forming 15 hydrogen bonds (H-bonds) (Fig. 1q). Among these, R355 forms salt bridge with D101 of CDRH3, while side chains of K356, R357, N360, N394, Y396 and R466 form H-bonds with S28, S31 of CDRH1, Y99, D101, S103, Y105, Y107 of CDRH3, and A50, S52, S56 of CDRL2. In addition, Y396 forms hydrophobic interactions with Y99 and Y107 of CDRH3 (Fig. 1n–p). The residues involved in the epitope are highly conserved (ranging from 91.6% to 98.7%) among different SARS-CoV-2 variants (Supplementary information, Fig. S5a, b), explaining the broad-spectrum neutralization of 6i18.
To elucidate the key epitope of 6i18, we chose seven residues with medium or long side chains involved in H-bond interactions with 6i18 and generated seven single-point mutants. Mutations at positions N360 and N394 resulted in a substantial decrease in neutralization by 6i18 (10- to 100-fold, Fig. 1r). Mutations at residues R357, Y396, and R466 conferred high-level resistance to neutralization by 6i18 (greater than 100-fold). However, all these mutants remained sensitive to S309 (Fig. 1r). Bioinformatics analysis of sequences deposited in the GISAID database reveals that residues R357, N360, N394, Y396, and R466 represent evolutionarily conserved positions,7 with sequence conservation exceeding 94.5% for R357, N360, N394, and Y396, and 91.8% for R466 across variants ranging from Jan 2020 through present circulating strains (Fig. 1r). Therefore, we conclude that 6i18 engages a highly conserved epitope encompassing residues R357, N360, N394, Y396, and R466, which differs from the epitope bound by S309.
We performed competition binding assays using biolayer interferometry (BLI) to characterize the epitope of 6i18. 6i18 competed with the cross-NAb S309 for binding to the RBD of SARS-CoV-2, whereas no competition was observed between 6i18 and the SARS-CoV-2-specific mAbs REGN10989, REGN10987, or REGN10933 (Fig. 1s).1 The differential competition profiles indicate that 6i18 recognizes a conserved site involved in the cross-neutralizing activity of S309, but does not share the binding specificity of SARS-CoV-2-directed antibodies. Moreover, similar to S309, 6i18 did not compete with ACE2 for binding to the RBDs of XBB and BQ.1.1 variants (Fig. 1t), suggesting that the epitope of 6i18 does not overlap with the ACE2-binding site. Superposition of the 6i18 Fab–XBB S RBD complex structure over the ACE2–Omicron S RBD confirms that 6i18 targets an epitope distinct from the RBM, without clashing with ACE2-binding site (Fig. 1u), which is consistent with the competition assay (Fig. 1t). Compared to the S309–Omicron S RBD complex structure, 6i18 binding causes slight steric hindrance and competes with S309 for binding to XBB (Fig. 1v). Although the 6i18 epitope overlaps with S309, 6i18 binds RBD in a different direction compared to S309, and the 6i18 epitope is also wider than the S309 epitope (Fig. 1w). Additionally, 6i18 forms 15 H-bonds with XBB S RBD (Fig. 1n–p), while S309 forms only 8 H-bonds with XBB RBD (Fig. 1x). This likely explains 6i18’s higher binding affinity for the RBDs of Omicron variants compared to S309 (Fig. 1b).
Chi et al. classified the RBD antibodies into several classes, among which H-RBD class antibodies have been shown to cause destruction of the S trimers.10 Structural comparison shows that 6i18 binds to the RBD surface similarly to H-RBD class antibodies, but with more residues on the RBD face toward the NTD side (E340–N360) and fewer residues on the RBD flank (R454–L492) (Supplementary information, Fig. S5c). This interaction causes greater steric hindrance with the NTD, even when the RBD is in a wide-open conformation. Consequently, 6I18 binding may induce trimer disassembly more easily. This hypothesized mechanism was supported by the neutralization results, which showed that 6i18 exhibits stronger neutralizing activity against the XBB.1.16 variant than the H-RBD class antibodies (Supplementary information, Fig. S5d). The enhanced potency of 6i18 neutralization confirms its unique ability to destabilize the S trimer.
In summary, this research presents an in-depth characterization of a novel broadly neutralizing antibody, 6i18, which recognizes a novel site of vulnerability on Omicron S proteins and blocks infection through a new mechanism disrupting S trimerization. 6i18 highlights potential strategies for engineering variant-resistant countermeasures and enhancing our understanding of S elements susceptible to neutralization in highly evolved SARS-CoV-2 strains. Further investigation into the neutralization mechanism of 6i18 will also provide important insights into the development of broadly NAbs against SARS-CoV-2.
Supplementary information
Acknowledgements
We thank the Center of Cryo-Electron Microscopy, Fudan University, for the support on cryo-EM data collection. This work was supported by the National Natural Science Foundation of China (32370995 and 31771008 to J.H. and 3197014 to Z.C.), the Shanghai Municipal Science and Technology Major Project (ZD2021CY001 to J.H.), the Ministry of Science and Technology of China (2021YFC2302500 to L.S.), the National Youth Talent Support Program (2022 to F.W.), the “Shuguang Program” of Shanghai Education Development Foundation and Shanghai Municipal Education Commission (22SG16 to F.W.), Guangzhou Laboratory (SRPG22-003 to L.S.), the National Science and Technology Major Projects of China (2017ZX10202102 to J.H. and 2018ZX10301403 to F.W.).
Author contributions
J.H., L.S., and F.W. conceived and designed the manuscript. J.H., F.W., Y.M., Y.D.W., J.C., P.R., and L.L. sorted B cells and cloned antibodies, constructed SARS-CoV-2 pseudovirus mutants, performed neutralization assay, ELISA, and BLI. L.S., Q.M., A.H., and Z.C. performed the structural studies. J.Z., Z.Z., and Y.Q.W. performed authentic virus experiments. Y.W. supervised the project. J.H., Y.M., Y.D.W., L.S., A.H., and Z.Z. analyzed the data. J.H., L.S., F.W., Y.D.W., and A.H. wrote the manuscript.
Data availability
The coordinates and maps associated with data reported in this manuscript have been deposited to the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) under accession numbers EMD-36322 and PDB 8JIO, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yunping Ma, Qiyu Mao, Yingdan Wang, Zhaoyong Zhang.
Contributor Information
Jincun Zhao, Email: zhaojincun@gird.cn.
Fan Wu, Email: wufan@fudan.edu.cn.
Lei Sun, Email: llsun@fudan.edu.cn.
Jinghe Huang, Email: Jinghehuang@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-023-00880-6.
References
- 1.Hansen J, et al. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, et al. Cell Discov. 2022;8:104. doi: 10.1038/s41421-022-00463-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westendorf K, et al. Cell Rep. 2022;39:110812. doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Y, et al. Nature. 2023;614:521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, et al. Cell. 2023;186:279–286.e278. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamasoba D, et al. Lancet Infect. Dis. 2023;23:655–656. doi: 10.1016/S1473-3099(23)00278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, et al. Bioinformatics. 2022;38:1735–1737. doi: 10.1093/bioinformatics/btab856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu F, et al. JAMA Intern. Med. 2020;180:1356–1362. doi: 10.1001/jamainternmed.2020.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinto D, et al. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 10.Chi X, et al. Cell Discov. 2023;9:37. doi: 10.1038/s41421-023-00535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and maps associated with data reported in this manuscript have been deposited to the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) under accession numbers EMD-36322 and PDB 8JIO, respectively.