Li et al. demonstrate that activated BLK phosphorylates TOLLIP and promotes its dissociation from IRAK1 to enable efficient inflammatory response. This study uncovers uncharacterized roles of BLK in regulating TLR/IL-1R–mediated inflammatory responses and suggests the possibility of targeting this kinase for alleviation of excessive inflammation.
Abstract
TLR/IL-1R signaling plays a critical role in sensing various harmful foreign pathogens and mounting efficient innate and adaptive immune responses, and it is tightly controlled by intracellular regulators at multiple levels. In particular, TOLLIP forms a constitutive complex with IRAK1 and sequesters it in the cytosol to maintain the kinase in an inactive conformation under unstimulated conditions. However, the underlying mechanisms by which IRAK1 dissociates from TOLLIP to activate TLR/IL-1R signaling remain obscure. Herein, we show that BLK positively regulates TLR/IL-1R–mediated inflammatory response. BLK-deficient mice produce less inflammatory cytokines and are more resistant to death upon IL-1β challenge. Mechanistically, BLK is preassociated with IL1R1 and IL1RAcP in resting cells. IL-1β stimulation induces heterodimerization of IL1R1 and IL1RAcP, which further triggers BLK autophosphorylation at Y309. Activated BLK directly phosphorylates TOLLIP at Y76/86/152 and further promotes TOLLIP dissociation from IRAK1, thereby facilitating TLR/IL-1R–mediated signal transduction. Overall, these findings highlight the importance of BLK as an active regulatory component in TLR/IL-1R signaling.
Introduction
Recognition of pathogen-associated molecular patterns (PAMPs) is mediated by germline-encoded pattern recognition receptors (PRRs) in the extracellular environments and intracellular compartments, leading to subsequent activation of the innate immune system that executes the first line of host defensive responses for eliminating infectious microbes (Akira et al., 2006; Blasius and Beutler, 2010; Hu and Shu, 2020). In particular, active interleukin-1β (IL-1β) is a vital proinflammatory cytokine that serves as a key “gatekeeper” for both local and systemic inflammation and elicits pleiotropic effects upon PAMP challenge (Dinarello, 2011; Sims and Smith, 2010). Upon binding of IL-1β to IL-1 receptor type 1 (IL1R1), IL1R1 accessory protein (IL1RAcP) is recruited to form a high-affinity IL1R1-IL1RAcP heterodimer (Huang et al., 1997; Lin et al., 2018; Wesche et al., 1997b). The activated receptor complex immediately recruits the adaptor protein myeloid differentiation factor 88 (MyD88) via their Toll/IL-1R (TIR) domains (Gurung et al., 2017; Wesche et al., 1997a). Subsequently, MyD88 creates a scaffold for the further recruitment of several serine/threonine kinases, including IL-1R–associated kinase (IRAK) 4, IRAK2, and IRAK1, to form a signal transduction complex termed Myddosome (De Nardo et al., 2018; Ferrao et al., 2014; Zhou et al., 2013). During the assembly of Myddosome, IRAK4 first undergoes autophosphorylation, and activated IRAK4, in turn, phosphorylates IRAK1, enabling further autophosphorylation of IRAK1 within the ProST region to reach a maximally phosphorylated state, which is accepted as a hallmark of IRAK1 activation (Cao et al., 1996; Kollewe et al., 2004; Li et al., 2002). Hyperphosphorylated IRAK1 simultaneously undergoes oligomerization (Kollewe et al., 2004; Neumann et al., 2007; Williams et al., 2005). Subsequently, activated IRAK1 dissociates from Myddosome and associates with the E3 ubiquitin ligase TRAF6. TRAF6 then catalyzes K63-linked autoubiquitination and further recruits the TAK1-TAB1/2/3 complex, resulting in the activation of TAK1 (Conze et al., 2008; Windheim et al., 2008). Ultimately, activated TAK1 phosphorylates inhibitor of nuclear factor κ-B kinases and mitogen-activated protein kinases (MAPKs), leading to the activation of transcription factors NF-κB and AP1 and the induction of numerous proinflammatory cytokines and chemokines (Chen et al., 2012; Lee et al., 2002; Mizukami et al., 2002).
To prevent tissue damage caused by excessive inflammation in infectious processes, IL-1R signaling is tightly controlled by endogenous negative regulators at multiple levels (Lee et al., 2011; Lin et al., 2018; Rao et al., 2005). For example, IRAK-M functions as an inducible negative regulator of IRAK4 to restrain its activation (Kobayashi et al., 2002). In addition, Toll-interacting protein (TOLLIP) was initially identified as an adaptor protein that executes a notably inhibitory role in IL-1R signaling cascade (Burns et al., 2000; Li et al., 2004; Liu et al., 2015; Parmar et al., 2018). Under unstimulated conditions, TOLLIP forms a constitutive complex with IRAK1 and restricts its phosphorylation, the process of which is mediated by the coupling of ubiquitin to endoplasmic reticulum degradation (CUE) domain of TOLLIP and the death domain of IRAK1 (Burns et al., 2000; Ohnuma et al., 2005; Zhang and Ghosh, 2002). Upon IL-1β stimulation, IRAK1 dissociates from TOLLIP to signal downstream effectors for proinflammatory responses (Capelluto, 2012; Kowalski and Li, 2017). Moreover, TOLLIP interacts with the cytoplasmic TIR domains of IL1R1, TLR2, and TLR4 and participates in intracellular trafficking and endolysosomal degradation of IL1R1 (Brissoni et al., 2006; Zhang and Ghosh, 2002). Consistently, Tollip-deficient mice have dysregulated inflammatory cytokine production in response to IL-1β stimulation, suggesting that TOLLIP exerts crucial inhibitory functions in controlling the extent of inflammatory responses (Didierlaurent et al., 2006). Although TOLLIP is classically demonstrated as a pivotal intracellular IRAK1 repressor in the absence of infection, the factors that determine the dissociation of TOLLIP from IRAK1 upon IL-1β stimulation have not been well defined.
Toll-like receptors (TLRs) are an evolutionarily conserved and well-characterized family of PRRs (Martin and Wesche, 2002; O’Neill and Dinarello, 2000; Song and Lee, 2012). Depending on their cellular localization or the corresponding PAMPs they recognize, TLRs can be divided into two subgroups. The first subgroup comprises TLR1, TLR2, TLR4, TLR5, and TLR6, which are localized on the cell surface and largely recognize structurally conserved components of various pathogens, such as lipopolysaccharide (LPS) and glycoproteins. The second subgroup comprises TLR3, TLR7, TLR8, and TLR9, which are expressed within intracellular vesicles and mainly recognize microbial nucleic acids (Barton and Medzhitov, 2003; Imler and Hoffmann, 2001; Li et al., 2020). Each TLR detects distinct PAMPs derived from bacteria, viruses, fungi, parasites, and protozoa. Upon ligand binding, TLRs, in concert with other PRRs, initiate host immune responses against invading pathogens by secreting abundant proinflammatory cytokines and type I interferons (Akira et al., 2001; Kawai and Akira, 2011). In addition, all TLRs, except TLR3, recruit the same adaptor protein MyD88 to initiate MyD88-dependent inflammatory responses, and the specific signal transduction mechanisms are similar to those mediated by IL-1R described above (Blasius and Beutler, 2010; Kumar et al., 2009; Martin and Wesche, 2002; Xiong et al., 2020).
B lymphoid tyrosine kinase (BLK) was initially identified as a B cell–specific member of the Src family of tyrosine kinases (Akerblad and Sigvardsson, 1999). However, recent studies discovered that BLK is expressed in a variety of cell types outside of the B cell lineage, including immune and non-immune cells, such as plasmacytoid dendritic cells (pDCs), progenitor cell populations from the bone marrow and thymus, IL-17–producing γδ T cells, CD4− CD8− αβ T cells, pancreatic β cells, epithelial cancer cells, and so forth (Laird et al., 2010; Malek et al., 1998; Samuelson et al., 2014). Structurally, BLK consists of N-terminal Src homology 2 and 3 domains and a C-terminal protein kinase domain. Recently, genome-wide association studies have shown that BLK risk alleles confer susceptibility to autoimmune disorders through dysregulation of a proinflammatory cytokine network (Han et al., 2009; Jiang et al., 2019; Samuelson et al., 2014; Simpfendorfer et al., 2012). In addition, the transcription factor NF-κB interacts with BLK gene to regulate its transcription during B cell activation (Zwollo et al., 1998). Moreover, BLK-mediated phosphorylation of cGAS at tyrosine 215 facilitates its cytosolic retention to suppress DNA damage and tumorigenesis (Liu et al., 2018). However, it is still largely unknown how abnormal BLK expression eventually leads to autoimmune disease.
Here, we found that BLK positively regulated TLR/IL-1R–mediated signal transduction. BLK directly catalyzed TOLLIP phosphorylation at tyrosine 76/86/152 to promote TOLLIP dissociation from IRAK1 and subsequent signaling cascade. In conclusion, our study reveals an essential role for BLK in TLR/IL-1R signaling and provides insights into the mechanisms that ensure efficient inflammatory response upon microbial infection.
Results
BLK positively regulates TLR/IL-1R–mediated inflammatory signaling
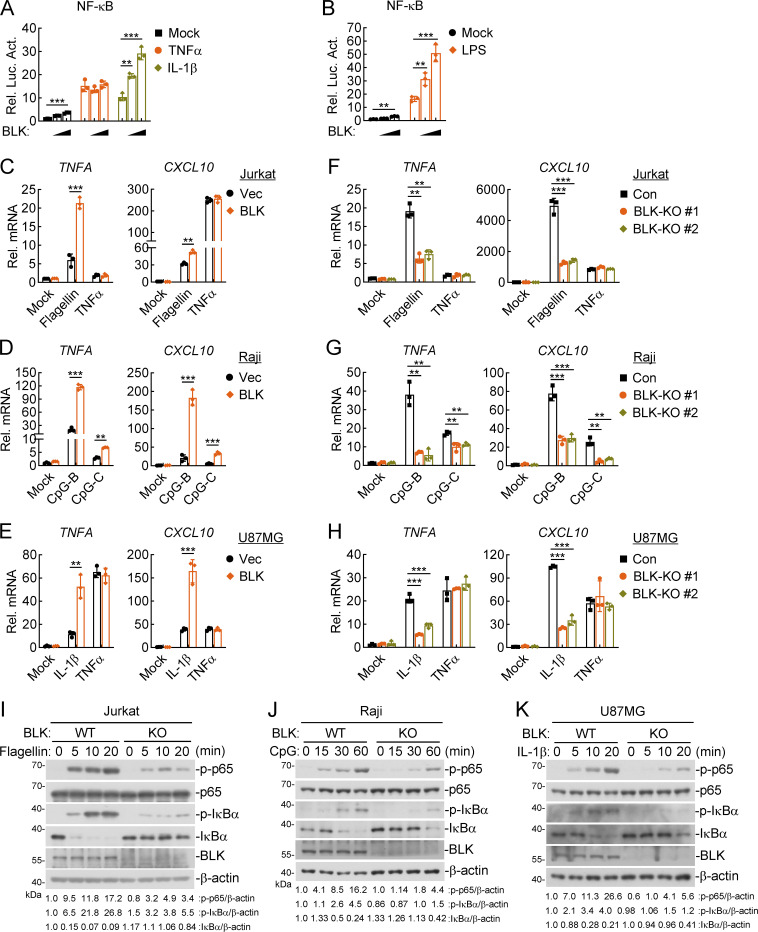
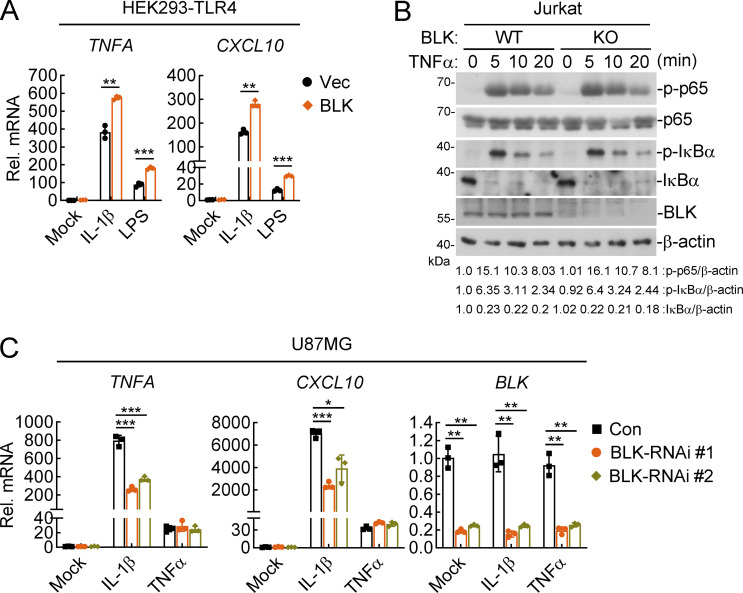
In light of the pivotal roles of tyrosine kinases in innate immune and inflammatory responses (Gurung et al., 2017), we attempted to identify proteins that could regulate NF-κB signaling by screening tyrosine kinase-related human cDNA expression plasmids by reporter assays. These efforts led to the identification of BLK as a candidate regulator. As shown in Fig. 1 A, overexpression of BLK activated NF-κB promoter and potentiated IL-1β– but not TNFα-induced NF-κB activation in a dose-dependent manner. Since the signal transduction downstream of IL-1R and all TLRs except TLR3 is similar, we wondered whether BLK is also involved in TLR-initiated signaling. Stimulation with the TLR4 ligand LPS revealed that overexpression of BLK also enhanced TLR4-mediated NF-κB activation in HEK293 cells stably expressing TLR4 (HEK293-TLR4; Fig. 1 B). Consistently, overexpression of BLK promoted the transcription of TNFA and CXCL10 genes in response to IL-1β, LPS, flagellin (a ligand for TLR5), and CpG-B/C (ligands for TLR9) treatment in different cell types. However, overexpression of BLK showed little effect on TNFα-triggered transcription of TNFA and CXCL10 genes (Fig. 1, C–E; and Fig. S1 A). To investigate whether endogenous BLK regulates TLR/IL-1R–mediated signaling, we constructed two guide RNA (gRNA) plasmids that targeted human BLK gene and downregulated its expression. BLK deficiency markedly inhibited IL-1β–, flagellin-, CpG-B/C–, but not TNFα-induced transcription of TNFA and CXCL10 genes (Fig. 1, F–H), and phosphorylation of p65 and IκBα (Fig. 1, I–K; and Fig. S1 B). In addition, we also designed two siRNAs that downregulated the transcription of BLK gene to different levels. Knockdown of BLK significantly decreased IL-1β– but not TNFα-induced transcription of TNFA and CXCL10 genes (Fig. S1 C). Consequently, these results suggest that BLK positively regulates TLR/IL-1R–mediated inflammatory signaling.
Figure 1.
BLK positively regulates TLR/IL-1R–mediated inflammatory signaling. (A and B) Effects of BLK on TNFα-, IL-1β–, and LPS-induced activation of NF-κB. U87MG (A) or HEK293-TLR4 cells (B) (1 × 105) were cotransfected with NF-κB reporter (0.002 μg), pRL-TK (Renilla luciferase) reporter (0.01 μg), and increased amounts of BLK expression plasmids (0.01, 0.02 μg) for 24 h. Cells were then left untreated or treated with TNFα (20 ng/ml), IL-1β (20 ng/ml), or LPS (100 ng/ml) for 10 h before luciferase assays. (C–E) Effects of BLK on flagellin-, TNFα-, CpG-B/C–, and IL-1β–induced transcription of downstream genes. Jurkat (C), Raji (D), or U87MG (E) cells were transduced with vector (Vec) or BLK expression plasmids by lentivirus-mediated gene transfer to establish stable cell lines. Cells (2 × 105) were left untreated or treated with flagellin (0.1 μg/ml), TNFα (20 ng/ml), CpG-B/C (1 μM), or IL-1β (20 ng/ml) for 3 h before qPCR analysis. (F–H) Effects of BLK deficiency on flagellin-, TNFα-, CpG-B/C–, and IL-1β–induced transcription of downstream genes. Jurkat (F), Raji (G), or U87MG (H) cells were transduced with control (Con) or the indicated gRNA plasmids targeting BLK gene by the CRISPR/Cas9 method to establish stable cell lines. BLK-deficient and control cells (2 × 105) were treated with the indicated stimuli for 3 h before qPCR analysis. (I–K) Effects of BLK deficiency on flagellin-, CpG-B–, and IL-1β–induced phosphorylation of p65 and IκBα. BLK-deficient and control Jurkat (I), Raji (J), or U87MG (K) cells (2 × 105) (BLK-KO #1 plasmids were used) were left untreated or treated with flagellin (0.1 μg/ml), CpG-B (1 μM), or IL-1β (20 ng/ml) for the indicated times before immunoblot analysis. KO, knockout. Graphs show mean ± SD (n = 3 biological replicates in A and B, n = 3 technical replicates in C–H) from one representative experiment. **P < 0.01, ***P < 0.001 (unpaired, two-tailed Student’s t test). Data are representative of three independent experiments with similar results. Source data are available for this figure: SourceData F1.
Figure S1.
BLK positively regulates IL-1β–, LPS-, but not TNFα-induced inflammatory responses. (A) Effects of BLK on IL-1β– and LPS-induced transcription of downstream genes. HEK293-TLR4 cells (2 × 105) were transfected with BLK expression plasmids for 24 h. Cells were then left untreated or treated with IL-1β (20 ng/ml) or LPS (100 ng/ml) for 3 h before qPCR analysis. (B) Effects of BLK deficiency on TNFα-induced phosphorylation of p65 and IκBα. BLK-deficient and control Jurkat cells (2 × 105; BLK-KO #1 plasmids were used) were left untreated or treated with TNFα (20 ng/ml) for the indicated times before immunoblot analysis. KO, knockout. (C) Effects of BLK knockdown on TNFα- and IL-1β–induced transcription of downstream genes. U87MG cells (2 × 105) were transfected with the indicated siRNA (final concentration, 40 nM). 48 h later, cells were left untreated or treated with TNFα (20 ng/ml) or IL-1β (20 ng/ml) for 3 h before qPCR analysis. Graphs show mean ± SD (n = 3 technical replicates in A and C) from one representative experiment. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired, two-tailed Student’s t test). Data are representative of three independent experiments with similar results. Source data are available for this figure: SourceData FS1.
Blk is essential for TLR/IL-1R–mediated inflammatory response in vivo
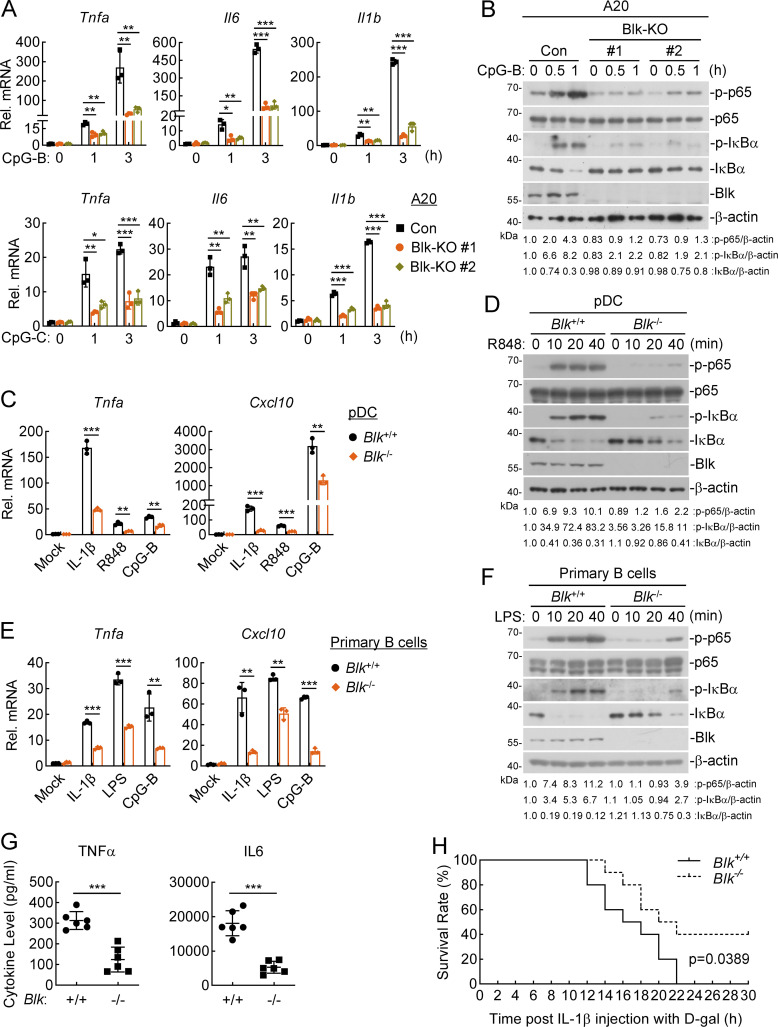
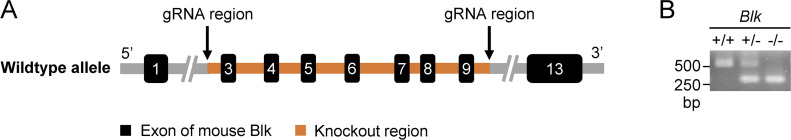
We next explored whether Blk regulates TLR/IL-1R–mediated inflammatory signaling in murine cells. The results in A20 cells, an immortalized murine B lymphoma cell line, showed that Blk deficiency attenuated CpG-B/C–induced transcription of Tnfa, Il6, and Il1b genes and phosphorylation of p65 and IκBα (Fig. 2, A and B). To further investigate the roles of murine Blk in TLR/IL-1R–mediated inflammatory response, we generated Blk-deficient mice by the CRISPR/Cas9 system (Fig. S2 A). The successful deletion of Blk gene in the knockout mice was verified by genotyping analysis (Fig. S2 B).
Figure 2.
Blk deficiency attenuates TLR/IL-1R–mediated inflammatory responses in vivo. (A and B) Effects of Blk deficiency on CpG-B/C–induced transcription of downstream genes and phosphorylation of p65 and IκBα. Murine A20 cells were transduced with control (Con) or the indicated gRNA plasmids targeting Blk gene by the CRISPR/Cas9 method to establish stable cell lines. Blk-deficient and control cells (2 × 105) were left untreated or treated with CpG-B/C (1 μM) for the indicated times before qPCR (A) and immunoblot (B) analyses. KO, knockout. (C and D) Effects of Blk deficiency on IL-1β–, R848-, and CpG-B–induced transcription of downstream genes and phosphorylation of p65 and IκBα. pDCs derived from the spleens of Blk+/+ and Blk−/− mice (1 × 105) were stimulated with murine IL-1β (20 ng/ml), R848 (10 μg/ml), or CpG-B (1 μM) for 3 h before qPCR analysis (C), or stimulated with R848 (10 μg/ml) for the indicated times before immunoblot analysis (D). (E and F) Effects of Blk deficiency on IL-1β–, LPS- and CpG-B–induced transcription of downstream genes and phosphorylation of p65 and IκBα. Primary B cells derived from the spleens of Blk+/+ and Blk−/− mice (5 × 105) were seeded in 12-well plates for 48 h in the presence of anti-lgM/IgG (5 μg/ml) and anti-CD40 antibody (1 μg/ml). Cells were then stimulated with murine IL-1β (20 ng/ml), LPS (100 ng/ml), or CpG-B (1 μM) for 3 h before qPCR analysis (E), or stimulated with LPS (100 ng/ml) for the indicated times before immunoblot analysis (F). (G) Effects of Blk deficiency on IL-1β–induced serum cytokine levels. Sex- and age-matched Blk+/+ and Blk−/− mice (n = 6 for each group) were injected i.p. with murine IL-1β (150 μg/kg) for 2 h before serum cytokines were measured by ELISA. (H) Effects of Blk deficiency on IL-1β–induced inflammatory death. Sex- and age-matched Blk+/+ and Blk−/− mice (n = 10 for each group) were injected i.p. with murine IL-1β (150 μg/kg) plus D-gal (0.5 mg/g) per mouse, and mouse survival was recorded every 1 h. Graphs show mean ± SD (n = 3 technical replicates in A, C, and E, n = 6 biological replicates in G) from one representative experiment. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired, two-tailed Student’s t test). For the mouse survival study in H, Kaplan–Meier survival curves were generated and analyzed by the log-rank test. Data are representative of at least two independent experiments with similar results. Source data are available for this figure: SourceData F2.
Figure S2.
Genotyping of Blk-deficient mice. (A) Blk gene targeting strategy. Exons 3–9 and part of the intron of Blk gene were deleted by the CRISPR/Cas9 method. The arrow marks the sequence position gRNA targeted. (B) Genotyping of Blk−/− mice. PCR analysis of genomic DNA to identify wild-type, heterozygous, and homozygous mice. Source data are available for this figure: SourceData FS2.
In addition to the essential functions in antibody production for eliminating pathogenic infection, B lymphocytes have gained remarkable attention due to their ability to regulate innate immune responses by secreting inflammatory cytokines (Pore et al., 2016). Evidence supports a critical role of TLR signaling via the MyD88 adaptor protein in B lymphocyte activation for therapeutic suppression of autoimmune inflammation, thus identifying potential candidates that regulate the TLR-MyD88 pathway in B lymphocytes is clinically significant (Neves et al., 2010). Moreover, pDCs are specialized sensors of bacterial nucleic acids and conserved structural components, such as CpG DNA and LPS, and major producers of inflammatory cytokines that promote host defense by priming innate immune responses (Kim et al., 2010). Activation of pDCs upon pathogen stimuli has been shown to be dependent on the TLR-MyD88 pathway and was thought to have a critical role in viral elimination (van der Sluis et al., 2022). To date, whether and how TLR-MyD88 signaling is regulated by Blk in murine primary cells remain poorly investigated. We first isolated pDCs and primary B cells from the spleens of Blk−/− mice and their wild-type littermates and then stimulated these cells with IL-1β or TLR ligands. As expected, Blk deficiency impaired IL-1β–, R848-, LPS-, and CpG-B–triggered transcription of Tnfa and Cxcl10 genes (Fig. 2, C and E), and phosphorylation of p65 and IκBα (Fig. 2, D and F). These results suggest that Blk deficiency attenuates TLR/IL-1R–mediated inflammatory responses in murine cells.
To further evaluate the importance of Blk in vivo, age- and sex-matched Blk+/+ and Blk−/− mice were injected intraperitoneally (i.p.) with murine IL-1β. We found that IL-1β–induced levels of serum cytokines in Blk−/− mice, including TNFα and IL6, were obviously lower than those of wild-type mice (Fig. 2 G). Consistently, after injection with murine IL-1β plus D-galactosamine hydrochloride (D-gal), Blk−/− mice experienced a lower percentage of lethality within 30 h compared with their wild-type counterparts (Fig. 2 H). These data suggest that Blk plays an important role in positive regulation of IL-1β–triggered inflammatory response in vivo.
BLK associates with TOLLIP
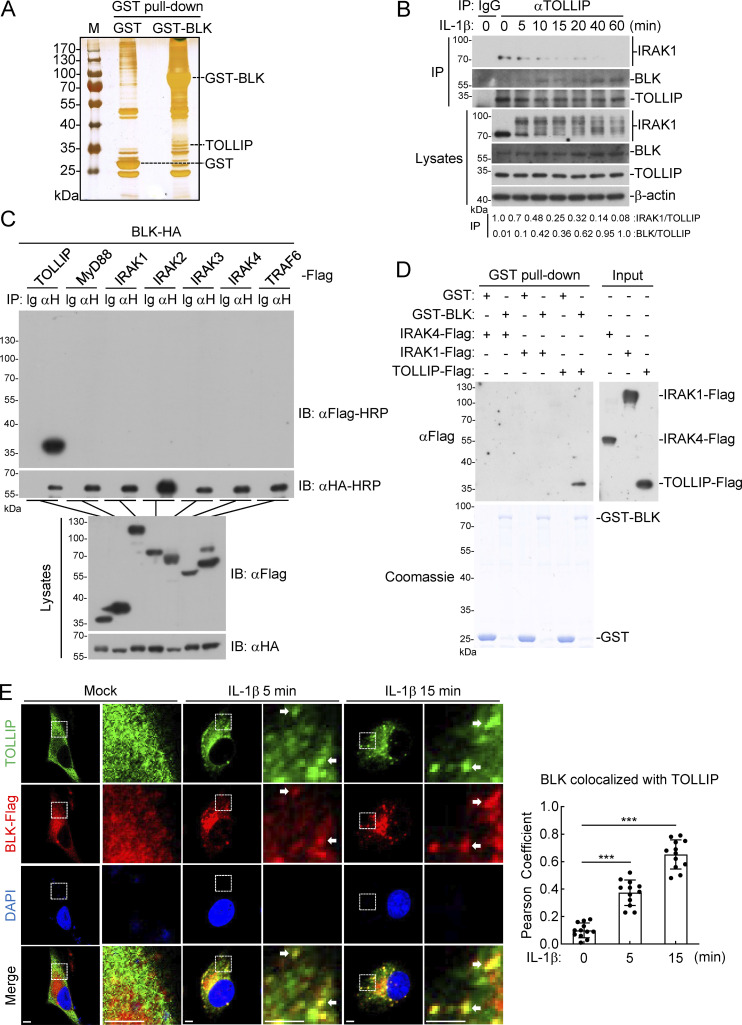
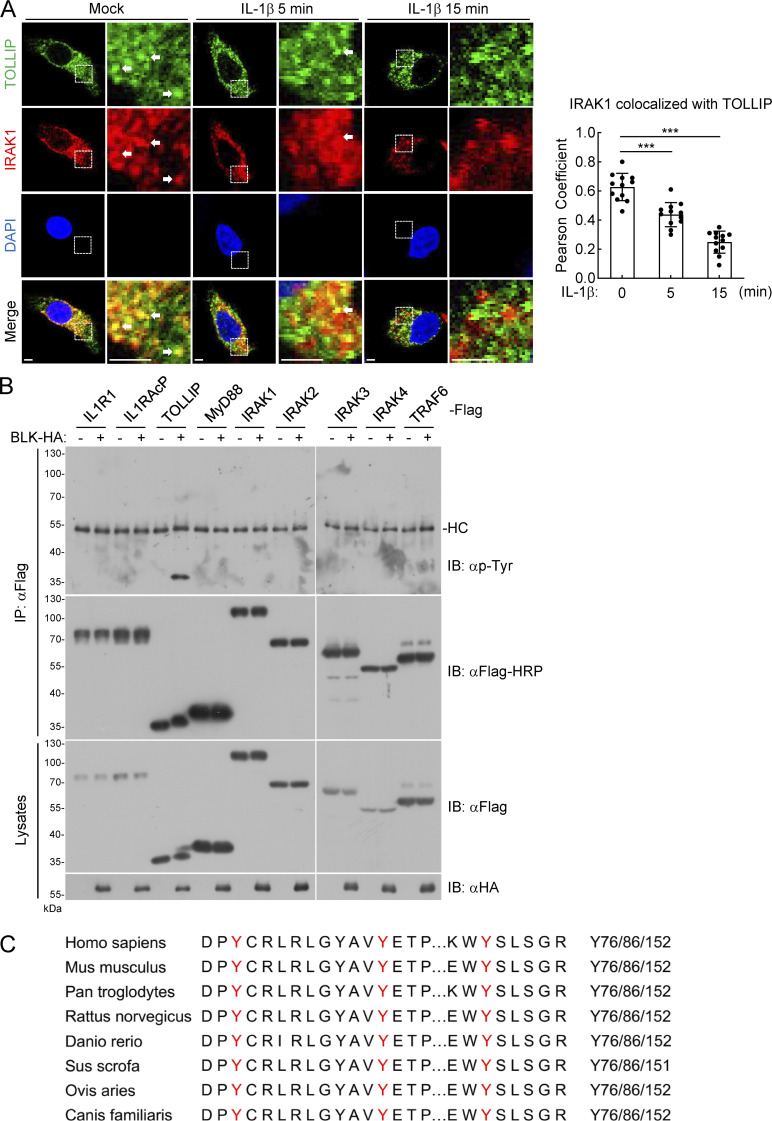
Next, we tried to elucidate the mechanism underlying how BLK regulates TLR/IL-1R signaling. In the hope of uncovering the protein interactors responsible for BLK function, we used affinity purification and mass spectrometry to characterize the BLK interactome. TOLLIP was among the convincing interacting proteins (Fig. 3 A and Table S1). Previous studies have demonstrated that TOLLIP serves as a key intracellular negative regulator of TLR/IL-1R signaling network by interacting with IRAK1 and concomitantly suppressing its kinase activity to prevent excessive proinflammatory response (Burns et al., 2000; Parmar et al., 2018). Consistent with the well-presented studies above, our endogenous coimmunoprecipitation experiments indicated that TOLLIP was constitutively associated with IRAK1 in unstimulated cells, and their association was promptly decreased upon IL-1β stimulation. Nevertheless, the interaction between TOLLIP and BLK gradually increased during IL-1β stimulation (Fig. 3 B). We further examined the relationship in coimmunoprecipitation experiments and confirmed that BLK specifically interacted with TOLLIP but not other components of the Myddosome, including MyD88, IRAK1, IRAK2, IRAK3, IRAK4, and TRAF6 (Fig. 3 C). Moreover, in vitro GST pull-down assays using prokaryotically expressed BLK and purified TOLLIP or the aforementioned IRAKs showed that BLK directly bound to TOLLIP but not IRAKs (Fig. 3 D). Consistently, our confocal microscopy experiments showed that under normal physiological conditions, TOLLIP was dispersed through the cytosol and obviously colocalized with IRAK1 but not visibly colocalized with BLK. Following IL-1β stimulation, TOLLIP gradually colocalized with BLK but had less colocalization with IRAK1 (Fig. 3 E and Fig. S3 A). Statistical analysis of colocalization images also confirmed these results (Fig. 3 E and Fig. S3 A). Taken together, these data suggest that BLK directly interacts with TOLLIP upon IL-1β stimulation.
Figure 3.
BLK associates with TOLLIP. (A) Identification of the BLK interactome by silver staining and mass spectrometry methods. Silver staining showed GST-associated factors (Control) and GST-BLK–associated factors, respectively. TOLLIP was among the convincing interacting factors. (B) Endogenous interaction between TOLLIP and IRAK1 or BLK. U87MG cells (2 × 107) were left untreated or treated with IL-1β (20 ng/ml) for the indicated times. Coimmunoprecipitation (IP) and immunoblot (IB) analysis were performed with the indicated antibodies. (C) BLK interacts with TOLLIP in the mammalian overexpression system. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 24 h. Coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. (D) BLK directly binds to TOLLIP. Prokaryotically expressed and purified GST-BLK protein coupled to glutathione sepharose beads was incubated with purified Flag-tagged IRAK1, IRAK4, or TOLLIP proteins for 3 h at 4°C and then subjected to in vitro GST pull-down assays. (E) BLK colocalizes with TOLLIP upon IL-1β stimulation (left panel). U87MG cells stably expressing BLK (1 × 105) were left untreated or treated with IL-1β (20 ng/ml) for the indicated times, then fixed with 4% PFA and stained with anti-Flag and anti-TOLLIP antibodies before confocal microscopy. The arrow marks the colocalized pixels. Scale bars, 2 μm. Quantitative analysis of colocalization of BLK with TOLLIP (right panel). Statistical analysis was based on colocalization images using ImageJ software. Graphs show mean ± SD (n = 12 cells from three individual images). ***P < 0.001 (unpaired, two-tailed Student’s t test). Data are representative of at least two independent experiments with similar results. Source data are available for this figure: SourceData F3.
Figure S3.
BLK mediates tyrosine phosphorylation of TOLLIP. (A) IRAK1 colocalizes with TOLLIP under resting conditions. U87MG cells (1 × 105) were left untreated or treated with IL-1β (20 ng/ml) for the indicated times, then fixed with 4% PFA, and stained with anti-IRAK1 and anti-TOLLIP antibodies before confocal microscopy (left panel). The arrow marks the colocalized pixels. Scale bars, 2 μm. Statistical analysis of colocalization of IRAK1 with TOLLIP was based on colocalization images using ImageJ software (right panel). Graphs show mean ± SD (n = 12 cells from three individual images). ***P < 0.001 (unpaired, two-tailed Student’s t test). (B) BLK mediates tyrosine phosphorylation of TOLLIP but not other adaptors. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 24 h. Coimmunoprecipitation (IP) and immunoblot (IB) analysis were performed with the indicated antibodies. HC, heavy chain. (C) Sequence alignment of TOLLIP from the indicated species. The sequences correspond to aa74–157 of human TOLLIP. The conserved tyrosine residues are highlighted in red. Data are representative of three independent experiments with similar results. Source data are available for this figure: SourceData FS3.
BLK directly phosphorylates TOLLIP at tyrosine 76/86/152
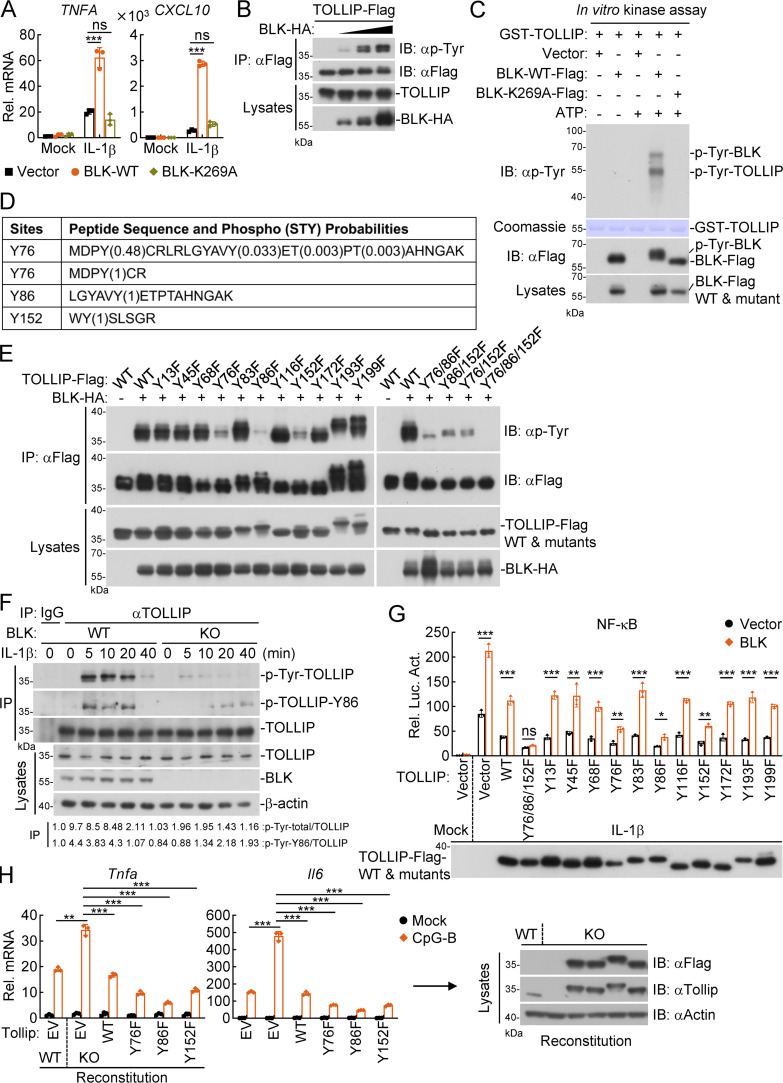
It has been demonstrated that residue K269 of BLK is responsible for ATP binding, and K269A alteration causes an almost complete absence of BLK kinase activity (Oda et al., 1999). We next determined whether BLK kinase activity is required for its regulation of TLR/IL-1R signaling. As shown in Fig. 4 A, overexpression of BLK potently enhanced IL-1β–induced transcription of TNFA and CXCL10 genes, whereas its enzymatically inactive mutant BLK(K269A) showed little effect. In addition, BLK specifically increased tyrosine phosphorylation of TOLLIP but not other adaptors in a dose-dependent manner in the mammalian overexpression system (Fig. 4 B and Fig. S3 B). In vitro kinase assays indicated that wild-type BLK but not BLK(K269A) mediated tyrosine phosphorylation of TOLLIP (Fig. 4 C). These results suggest that BLK catalyzes tyrosine phosphorylation of TOLLIP in a kinase activity-dependent manner.
Figure 4.
Phosphorylation of TOLLIP at Y76/86/152 is essential for its regulatory function in TLR/IL-1R signaling. (A) Effects of BLK and its mutant on IL-1β–induced transcription of downstream genes. U87MG cells (2 × 105) were transfected with BLK or BLK(K269A) plasmids for 24 h. Cells were then left untreated or treated with IL-1β (20 ng/ml) for 3 h before qPCR analysis. (B) BLK mediates tyrosine phosphorylation of TOLLIP in a dose-dependent manner. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 24 h. Coimmunoprecipitation (IP) and immunoblot (IB) analysis were performed with the indicated antibodies. (C) BLK catalyzes tyrosine phosphorylation of TOLLIP in vitro. Purified Flag-tagged BLK and its mutant coupled with Protein G sepharose were subjected to in vitro kinase assays with purified GST-TOLLIP. (D) Identification of potential tyrosine phosphorylation sites of TOLLIP by mass spectrometry. The in vitro phosphorylated TOLLIP in C was subjected to mass spectrometry. The list shows the phosphorylated peptide sequences and phosphorylation (STY) probabilities. (E) BLK mediates tyrosine phosphorylation of TOLLIP at Y76, Y86, and Y152. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 24 h. Coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. (F) Effects of BLK deficiency on IL-1β–triggered TOLLIP Y86 phosphorylation. BLK-deficient and control U87MG cells (2 × 107) were left untreated or treated with IL-1β (20 ng/ml) for the indicated times. Coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. KO, knockout. (G) Effects of BLK and all TOLLIP mutants co-transfection on IL-1β–triggered NF-κB activation. U87MG cells (1 × 105) were cotransfected with NF-κB reporter (0.002 μg), pRL-TK reporter (0.01 μg), BLK plasmids (0.05 μg), either TOLLIP or its mutants (0.05 μg) for 24 h and then treated with IL-1β (20 ng/ml) for 10 h before luciferase assays. The lower blot shows the expression levels of TOLLIP and its mutants as detected by anti-Flag antibody. (H) Effects of reconstitution of Tollip-deficient cells with Tollip or its mutants on CpG-B–induced transcription of downstream genes. A20 cells were transduced with vector or the gRNA plasmids targeting Tollip gene by the CRISPR/Cas9 method to establish the stable cell lines with puromycin (1 μg/ml) selection. Wild-type and Tollip-deficient A20 cells were then transduced with empty vector (EV), Tollip, or its mutants by lentivirus-mediated gene transfer to establish the stable cell lines with blasticidin S (10 μg/ml) selection. Subsequently, the indicated cell lines (2 × 105) were left untreated or treated with CpG-B (1 μM) for 3 h before qPCR analysis. The blots show the expression levels of Tollip and its mutants in the indicated cell lines as detected by anti-Flag or anti-Tollip antibodies, respectively. Graphs show mean ± SD (n = 3 technical replicates in A and H, n = 3 biological replicates in G) from one representative experiment. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant (unpaired, two-tailed Student’s t test). Data are representative of three independent experiments with similar results. Source data are available for this figure: SourceData F4.
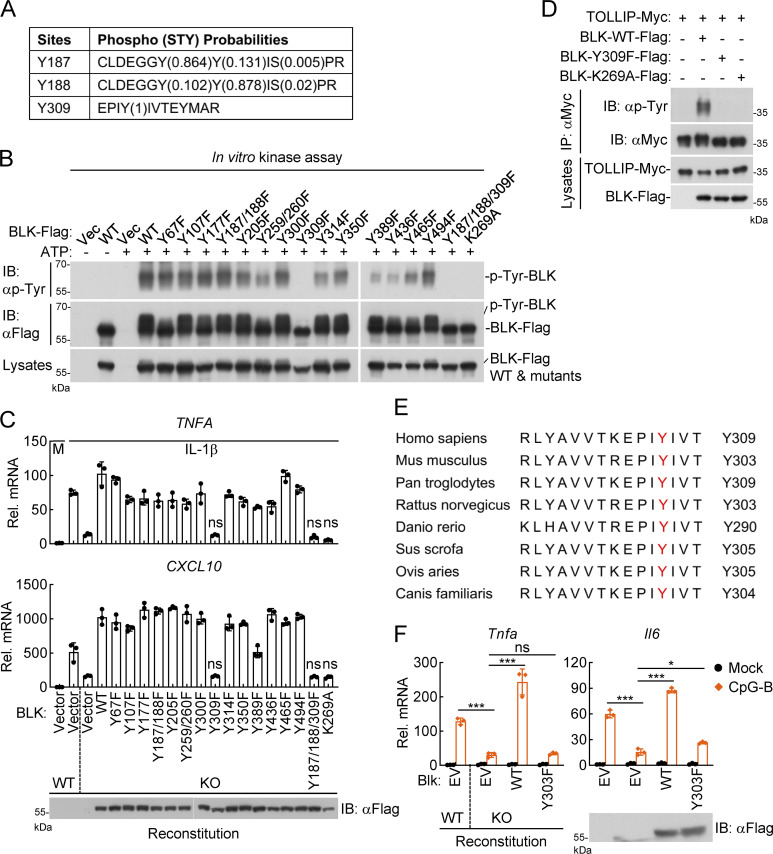
To identify potential residues of TOLLIP that are modified by BLK, the in vitro phosphorylated TOLLIP in Fig. 4 C was subjected to mass spectrometry. The analyzed results indicated that three tyrosine residues of human TOLLIP, Y76, Y86, and Y152, were phosphorylated by BLK (Fig. 4 D and Table S2). Next, we individually mutated all 11 tyrosine residues within TOLLIP to phenylalanine. As shown in Fig. 4 E, mutation of Y76, Y86, or Y152 obviously inhibited BLK-mediated phosphorylation of TOLLIP. Strikingly, the phosphorylation level of TOLLIP(Y76/86/152F) mutant was almost completely abrogated. To verify that BLK indeed targets TOLLIP Y76/86/152 for phosphorylation, we further obtained an antibody produced by immunizing rabbits with a synthetic peptide corresponding to residues surrounding Y86 of human TOLLIP protein, in which the Y86 site is phosphorylated. As shown in Fig. 4 F, BLK deficiency profoundly attenuated IL-1β–triggered tyrosine phosphorylation of endogenous TOLLIP. Notably, TOLLIP Y86 phosphorylation was also impaired. These experiments consistently demonstrate that BLK directly phosphorylates TOLLIP at Y76, Y86, and Y152.
We wondered whether BLK-mediated phosphorylation of TOLLIP is indispensable for its regulatory function in TLR/IL-1R signaling. Reporter assays indicated that TOLLIP and all its mutants inhibited IL-1β–triggered NF-κB activation, and co-transfection of BLK with all TOLLIP mutants except TOLLIP(Y76/86/152F) alleviated their inhibitory effects on NF-κB activation (Fig. 4 G). Additionally, we noticed that among all TOLLIP mutants, TOLLIP(Y76/86/152F) had a much more pronounced inhibitory effect on NF-κB activation (Fig. 4 G). Intriguingly, sequence analysis showed that Y76, Y86, and Y152 of TOLLIP were conserved in various species (Fig. S3 C). Therefore, we next investigated whether Y76, Y86, and Y152 of Tollip have similar roles in murine cells. As shown in Fig. 4 H, Tollip deficiency potentiated CpG-B–induced transcription of Tnfa and Il6 genes in A20 cells. Reconstitution of Tollip-deficient cells with Tollip and its mutants inhibited CpG-B–induced transcription of downstream genes. Importantly, compared with Tollip-reconstituted cells, CpG-B–induced inflammatory responses were much lower in its mutant-reconstituted cells. Altogether, these results suggest that BLK catalyzes tyrosine phosphorylation of TOLLIP at Y76, Y86, and Y152, which is essential for the regulatory function of TOLLIP in TLR/IL-1R signaling.
IL-1β signaling triggers BLK activation
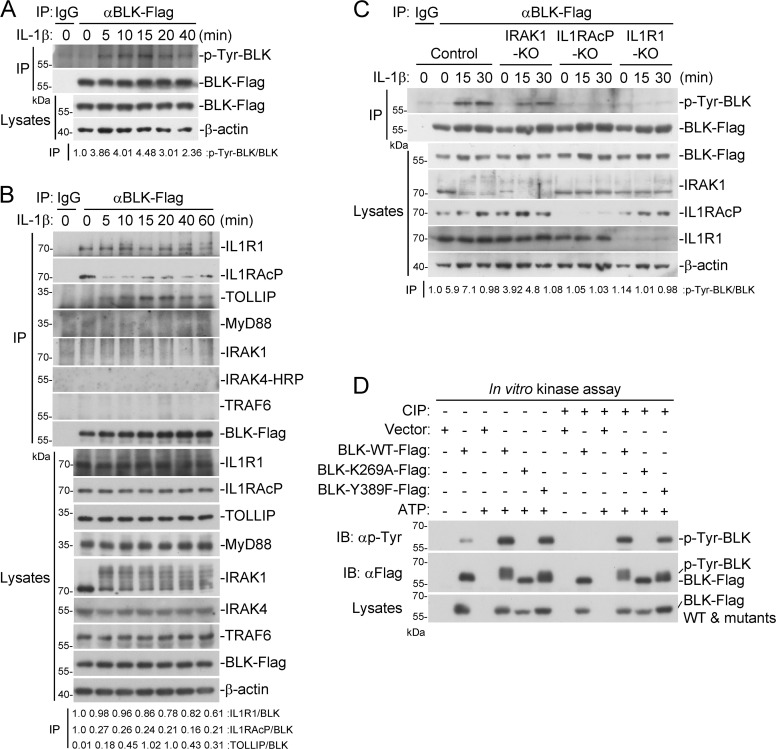
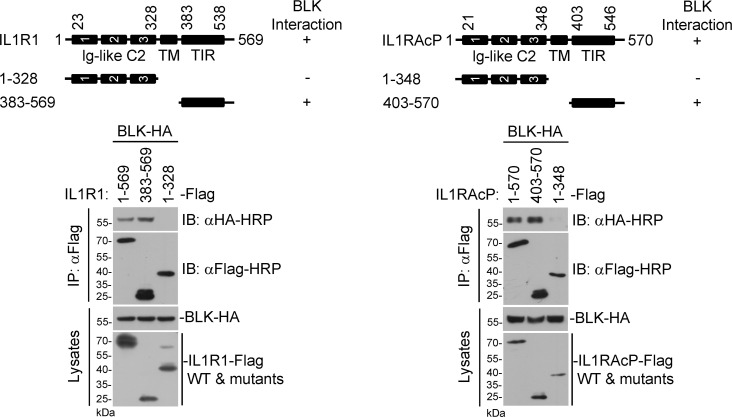
Next, we investigated whether BLK is phosphorylated following IL-1β stimulation. As shown in Fig. 5 A, BLK underwent tyrosine phosphorylation upon IL-1β treatment. We then explored the underlying mechanism by which IL-1β stimulation triggers BLK phosphorylation. As shown in Fig. 5 B, BLK was constitutively associated with IL1R1 and IL1RAcP but not other components of the Myddosome in unstimulated cells. Following IL-1β stimulation, BLK dynamically interacted with TOLLIP. Domain mapping experiments indicated that the cytoplasmic TIR domains of IL1R1 and IL1RAcP were responsible for their interaction with BLK (Fig. S4). Moreover, as shown in Fig. 5 C, individual deficiency of IL1R1 or IL1RAcP abolished IL-1β–induced BLK phosphorylation, whereas deficiency of downstream kinase IRAK1 had no significant effect on BLK phosphorylation. These results indicate that the formation of IL-1β/IL1R1/IL1RAcP heterocomplex triggers BLK phosphorylation.
Figure 5.
Preassociation of BLK with IL1R1 and IL1RAcP is necessary for BLK autophosphorylation. (A) BLK undergoes tyrosine phosphorylation following IL-1β stimulation. U87MG cells stably expressing BLK (2 × 107) were left untreated or treated with IL-1β (20 ng/ml) for the indicated times. Coimmunoprecipitation (IP) and immunoblot (IB) analysis were performed with the indicated antibodies. (B) BLK is constitutively associated with IL1R1 and IL1RAcP under unstimulated conditions. U87MG cells stably expressing BLK (2 × 107) were left untreated or treated with IL-1β (20 ng/ml) for the indicated times. Coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. (C) IL1R1 or IL1RAcP deficiency abolishes IL-1β–induced BLK phosphorylation. U87MG cells were transduced with control or gRNA plasmids targeting IRAK1, IL1R1, or IL1RAcP genes by the CRISPR/Cas9 method to establish stable cell lines. The indicated cell lines (2 × 105) were left untreated or treated with IL-1β (20 ng/ml) for the indicated times before immunoblot analysis. KO, knockout. (D) BLK catalyzes autophosphorylation in a kinase activity-dependent manner. Purified Flag-tagged BLK and its mutants coupled with Protein G sepharose were subjected to in vitro BLK autophosphorylation assays without or with CIP treatment. The detailed procedures are shown in Materials and methods. Data are representative of three independent experiments with similar results. Source data are available for this figure: SourceData F5.
Figure S4.
BLK interacts with the cytoplasmic TIR domains of IL1R1 and IL1RAcP. Domain mapping of the interaction between BLK and IL1R1 or IL1RAcP. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 24 h. Coimmunoprecipitation (IP) and immunoblot (IB) analysis were performed with the indicated antibodies. −: no interaction, +: interaction. TM, transmembrane. Data are representative of three independent experiments with similar results. Source data are available for this figure: SourceData FS4.
We noticed that wild-type BLK but not BLK(K269A) could be phosphorylated in vitro (Fig. 4 C). As shown in Fig. 5 D, in vitro kinase assays further showed that wild-type BLK but not BLK(K269A) underwent tyrosine phosphorylation, and the shifted band with a higher molecular weight represents phosphorylated BLK. Previously, it has been reported that Y389F alteration reduces BLK autophosphorylation, which is crucial for the suppression of chronic myeloid leukemia development (Zhang et al., 2012). Unexpectedly, the BLK(Y389F) mutant still underwent autophosphorylation, which implied that the Y389 site may be dispensable for BLK autophosphorylation (Fig. 5 D). In addition, phosphorylation could be removed by treatment with calf intestinal alkaline phosphatase (CIP). Dephosphorylated BLK and BLK(Y389F) pretreated with CIP could restore their tyrosine phosphorylation in an in vitro kinase reaction (Fig. 5 D). Since BLK was constitutively associated with the cytoplasmic TIR domains of IL1R1 and IL1RAcP (Fig. 5 B and Fig. S4), the formation of IL-1β/IL1R1/IL1RAcP heterocomplex would bring the cytoplasmic TIR domains of IL1R1 and IL1RAcP into close proximity, which could be responsible for BLK autophosphorylation. These results suggest that BLK undergoes autophosphorylation upon IL-1β stimulation.
Autophosphorylation of BLK at tyrosine 309 is crucial for its regulation of TLR/IL-1R–mediated inflammatory pathway
To identify specific residues of BLK autophosphorylation, the in vitro phosphorylated BLK in Fig. 5 D was further analyzed by mass spectrometry. As shown in Fig. 6 A and Table S3, three potential sites of BLK, including Y187, Y188, and Y309, were phosphorylated. Subsequently, we constructed BLK mutants in which all tyrosine residues were individually mutated to phenylalanine. In vitro kinase assays suggested that mutations of these tyrosine residues had varying degrees of effects on BLK autophosphorylation. Somewhat surprisingly, mutation of Y187/188 to phenylalanine had only a slight effect on BLK autophosphorylation. However, the Y309F alteration resulted in a dramatic reduction or almost complete absence of BLK autophosphorylation (Fig. 6 B). Thus, these findings suggest that BLK catalyzes autophosphorylation at Y309. We next explored whether autophosphorylation of BLK at Y309 is required for its ability to modulate TLR/IL-1R signaling. As shown in Fig. 6 C, reconstitution of BLK-deficient cells with all BLK mutants except Y309F restored IL-1β–induced transcription of TNFA and CXCL10 genes. IL-1β stimulation induced the transcription of TNFA and CXCL10 genes to comparable levels in BLK-deficient and BLK(Y309F)-reconstituted cells. Moreover, BLK(Y309F) failed to mediate tyrosine phosphorylation of TOLLIP (Fig. 6 D). These results suggest that BLK autophosphorylation is conducive to tyrosine phosphorylation of TOLLIP for inflammatory cytokine production.
Figure 6.
Autophosphorylation of BLK at Y309 is critical for its regulation of TLR/IL-1R–mediated inflammatory signaling. (A) Identification of potential autophosphorylation sites of BLK by mass spectrometry. The in vitro phosphorylated BLK in Fig. 5 D was subjected to mass spectrometry. The list shows the phosphorylated peptide sequences and phosphorylation (STY) probabilities. (B) BLK catalyzes autophosphorylation mainly at Y309. Purified Flag-tagged BLK and its mutants coupled with Protein G sepharose were subjected to in vitro BLK autophosphorylation assays. (C) Effects of reconstitution of BLK-deficient cells with BLK and its mutants on IL-1β–induced transcription of downstream genes. BLK-deficient and control U87MG cells (2 × 105) were transfected with the same amount of BLK or its mutants for 24 h. Cells were then left untreated or treated with IL-1β (20 ng/ml) for 3 h before qPCR analysis. The lower blot shows the expression levels of BLK and its mutants as detected by anti-Flag antibody. KO, knockout. (D) BLK(Y309F) fails to mediate tyrosine phosphorylation of TOLLIP. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 24 h. Coimmunoprecipitation (IP) and immunoblot (IB) analysis were performed with the indicated antibodies. (E) Sequence alignment of BLK from the indicated species. The sequences correspond to aa 298–312 of human BLK. The conserved tyrosine residues are highlighted in red. (F) Effects of reconstitution of Blk-deficient cells with Blk or its mutant on CpG-B–induced transcription of downstream genes. Blk-deficient A20 cells were transduced with Blk or Blk(Y303F) plasmids by lentivirus-mediated gene transfer to establish the stable cell lines with blasticidin S (10 μg/ml) selection. The indicated cell lines (2 × 105) were then left untreated or treated with CpG-B (1 μM) for 3 h before qPCR analysis. The lower blot shows the expression levels of Blk and Blk(Y303F) in the indicated cell lines as detected by anti-Flag antibody. Graphs show mean ± SD (n = 3 technical replicates in C and F) from one representative experiment. *P < 0.05, ***P < 0.001, ns, not significant (unpaired, two-tailed Student’s t test). Data are representative of three independent experiments with similar results. Source data are available for this figure: SourceData F6.
Sequence alignment found that Y309 of BLK, which corresponds to Y303 in murine Blk, is conserved across multiple species (Fig. 6 E). We speculated that the regulation of TLR/IL-1R signaling by murine Blk may also depend on its Y303 residue. To verify this hypothesis, we reconstituted wild-type Blk or its mutant Blk(Y303F) into Blk-deficient A20 cells via a lentivirus-mediated gene transfer approach. Quantitative PCR (qPCR) experiments indicated that reconstitution with Blk but not Blk(Y303F) restored transcriptional induction of Tnfa and Il6 genes after CpG-B stimulation (Fig. 6 F). Collectively, these results suggest that autophosphorylation at Y309 or Y303 is crucial for human or murine BLK to regulate TLR/IL-1R signaling.
BLK promotes TOLLIP dissociation from IRAK1 by competitively binding to TOLLIP
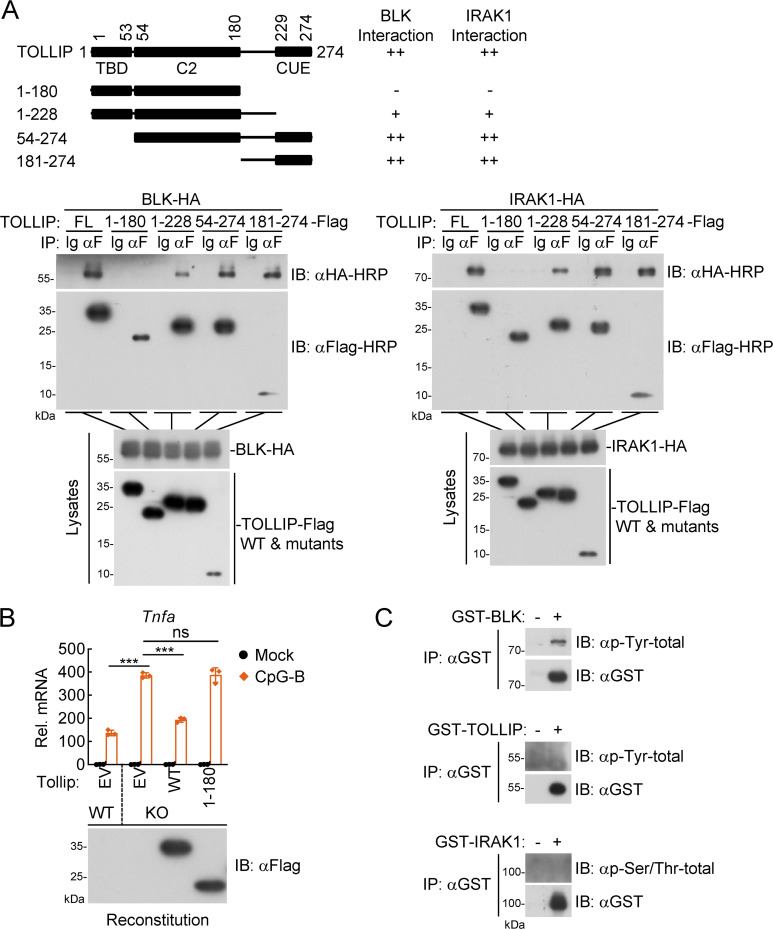
Because BLK autophosphorylation and subsequent tyrosine phosphorylation of TOLLIP were crucial for the activation of TLR/IL-1R signaling, we next attempted to decipher the underlying mechanism by which activated BLK targets TOLLIP to execute its function. As shown in Fig. S5 A, domain mapping experiments demonstrated that the C-terminus (aa 181–274) containing CUE domain of TOLLIP was responsible for its association with BLK and IRAK1, indicating that TOLLIP shares an overlapping region to bind BLK and IRAK1. Moreover, as shown in Fig. S5 B, reconstitution of Tollip-deficient A20 cells with Tollip(1–180) mutant potentiated CpG-B–induced transcription of Tnfa gene. These results suggest that the CUE domain of TOLLIP is crucial for its inhibitory function in TLR/IL-1R signaling.
Figure S5.
The C-terminus (aa181–274) containing CUE domain of TOLLIP is responsible for its association with BLK and IRAK1. (A) Domain mapping of the interaction between TOLLIP and BLK or IRAK1. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 24 h. Coimmunoprecipitation (IP) and immunoblot (IB) analysis were performed with the indicated antibodies. −: no interaction, +: weak interaction, ++: strong interaction. FL, full length. (B) Effects of reconstitution of Tollip-deficient cells with Tollip or Tollip(1–180) on CpG-B–induced transcription of downstream genes. Wild-type and Tollip-deficient A20 cells were transduced with empty vector (EV), Tollip, or Tollip(1–180) by lentivirus-mediated gene transfer to establish the stable cell lines with blasticidin S (10 μg/ml) selection. Subsequently, the indicated cell lines (2 × 105) were left untreated or treated with CpG-B (1 μM) for 3 h before qPCR analysis. The lower blot shows the expression levels of Tollip and Tollip(1–180) in the indicated cell lines as detected by anti-Flag antibody. Graphs show mean ± SD (n = 3 technical replicates) from one representative experiment. ***P < 0.001, ns, not significant (unpaired, two-tailed Student’s t test). KO, knockout. (C) Detection of the phosphorylation status of BLK, TOLLIP, and IRAK1. The purified recombinant proteins BLK, TOLLIP, and IRAK1 were subjected to coimmunoprecipitation and immunoblot analysis. Data are representative of three independent experiments with similar results. Source data are available for this figure: SourceData FS5.
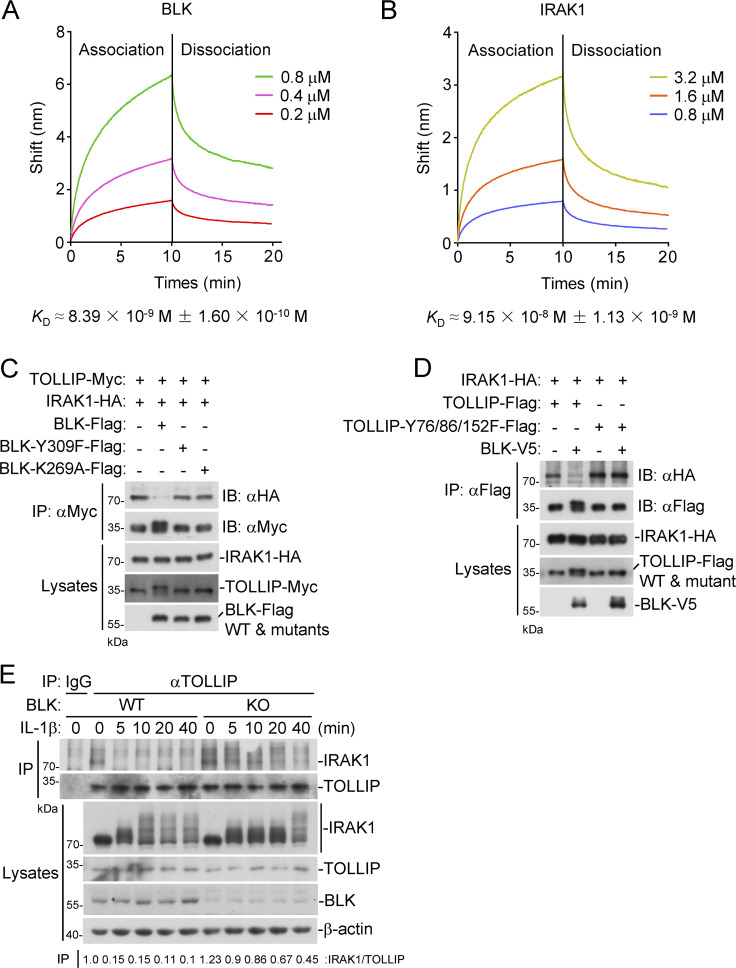
Subsequently, we prepared recombinant BLK, IRAK1, and TOLLIP from bacteria and purified them to homogeneity, and first detected the phosphorylation status of these recombinant proteins. As shown in Fig. S5 C, purified BLK underwent autophosphorylation, whereas neither TOLLIP nor IRAK1 was phosphorylated. We then examined the binding affinities of BLK and IRAK1 to TOLLIP by Octet biolayer interferometry. The results indicated that both BLK and IRAK1 could bind to TOLLIP, but the binding affinity of TOLLIP to BLK (KD ≈ 8.39 × 10−9 M ± 1.60 × 10−10 M) was ∼10-fold higher than that of IRAK1 (KD ≈ 9.15 × 10−8 M ± 1.13 × 10−9 M; Fig. 7, A and B). In addition, coimmunoprecipitation experiments further indicated that wild-type BLK, but not its mutants BLK(Y309F) or BLK(K269A), dramatically inhibited the association of TOLLIP and IRAK1 (Fig. 7 C). Intriguingly, BLK failed to affect the interaction between TOLLIP(Y76/86/152F) and IRAK1 (Fig. 7 D). Moreover, cotransfection of BLK and TOLLIP gave rise to a shifted band with a higher molecular weight of TOLLIP, which represents phosphorylated TOLLIP (Fig. 7, C and D). These data indicated that kinase activity and autophosphorylation of BLK were required for its ability to dissociate TOLLIP from IRAK1. As expected, endogenous coimmunoprecipitation showed that BLK deficiency impaired IL-1β–induced dissociation of IRAK1 from TOLLIP (Fig. 7 E). Taken together, these results suggest that BLK promotes the dissociation of TOLLIP from IRAK1 by competitively binding to TOLLIP.
Figure 7.
BLK promotes TOLLIP dissociation from IRAK1 by competitively binding to TOLLIP. (A and B) TOLLIP-binding affinities of BLK and IRAK1. ForteBio Octet Red system was used to examine the binding affinities of recombinant BLK (A) and IRAK1 (B) to TOLLIP. The vertical and horizontal axes represent the light shift distance (nm) and association/dissociation times, respectively. (C) Effects of BLK and its mutants on the association of TOLLIP and IRAK1. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 24 h. Coimmunoprecipitation (IP) and immunoblot (IB) analysis were performed with the indicated antibodies. (D) Effects of BLK on the association of TOLLIP(Y76/86/152F) and IRAK1. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 24 h. Coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. (E) Effects of BLK deficiency on IL-1β–induced dissociation of IRAK1 from TOLLIP. BLK-deficient and control U87MG cells (2 × 107) (BLK-KO #1 plasmids were used) were left untreated or treated with IL-1β (20 ng/ml) for the indicated times before coimmunoprecipitation and immunoblot analysis. KO, knockout. Data are representative of three independent experiments with similar results. Source data are available for this figure: SourceData F7.
BLK facilitates TLR/IL-1R–mediated signal transduction
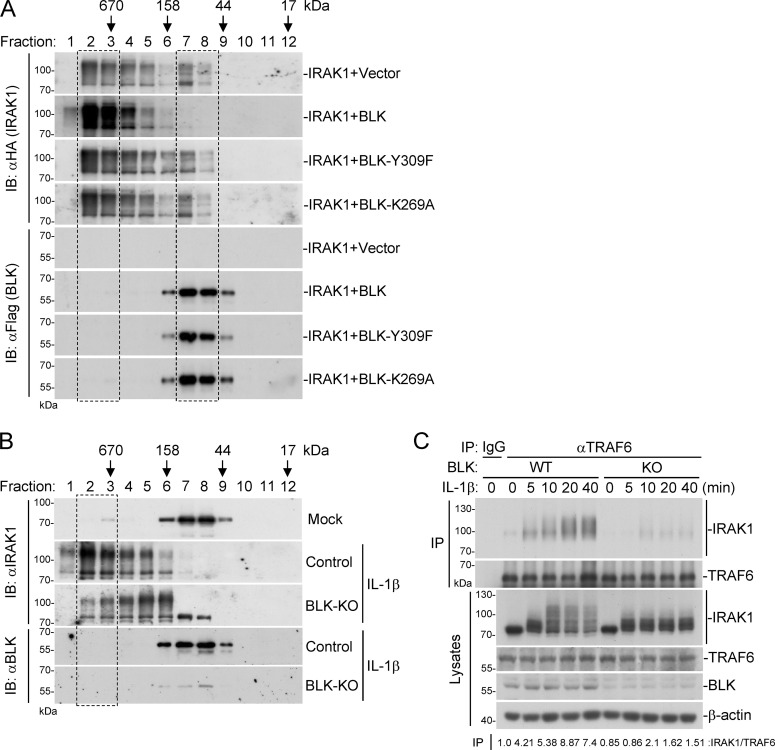
Upon IL-1β stimulation, IRAK1 is instantly recruited to the active receptor complex, enabling IRAK4-mediated phosphorylation of IRAK1 and further autophosphorylation at multiple sites (De Nardo et al., 2018; Kollewe et al., 2004). Simultaneously, IRAK1 undergoes oligomerization and participates in the assembly of Myddosome (Neumann et al., 2007). Hyperphosphorylated IRAK1 subsequently dissociates from IL-1Rs and interacts with the E3 ubiquitin ligase TRAF6, leading to the activation of NF-κB and MAPK pathways (Conze et al., 2008; Kollewe et al., 2004; Williams et al., 2005). We wondered whether BLK targets TOLLIP to further modulate the above process. Therefore, we carried out gel filtration experiments to detect the oligomerization of IRAK1. As shown in Fig. 8 A, in the mammalian overexpression system, IRAK1 mainly existed in the form of oligomers and was eluted in the high-molecular-weight fractions. In addition, a portion of IRAK1 was also eluted in the low-molecular-weight fractions, which represent monomeric IRAK1. Cotransfection of IRAK1 with wild-type BLK but not its mutants BLK(Y309F) or BLK(K269A) promoted distribution of monomeric IRAK1 to the high-molecular-weight fractions (Fig. 8 A, upper panels, dashed box). Intriguingly, the majority of BLK and its mutants were eluted in the low-molecular-weight fractions, which represent monomeric or dimeric BLK, and were almost absent in the high-molecular-weight fractions (Fig. 8 A, lower panels, dashed box), indicating that BLK was not directly associated with oligomerized IRAK1. Conversely, endogenous IRAK1 mostly existed as monomers or dimers in unstimulated cells. Upon IL-1β stimulation, monomeric or dimeric IRAK1 was efficiently converted to oligomers. More importantly, BLK deficiency decreased the ratio of IRAK1 distributed in the high-molecular-weight fractions, suggesting that BLK was required for the oligomerization of IRAK1 (Fig. 8 B, upper panels, dashed box). Furthermore, IL-1β–induced hyperphosphorylation of IRAK1 and subsequent recruitment of IRAK1 to TRAF6 were markedly impaired in BLK-deficient cells in comparison with wild-type cells (Fig. 8 C). Collectively, these results suggest that BLK targets TOLLIP to promote TLR/IL-1R–mediated signal transduction.
Figure 8.
BLK facilitates TLR/IL-1R–mediated signal transduction. (A) Effects of BLK and its mutants on the oligomerization of IRAK1. HEK293 cells (4 × 107) were transfected with IRAK1 and BLK or its mutants for 24 h. Cells were collected and lysed, followed by gel filtration. Fraction size was calibrated with the gel filtration standard (151-1901; Bio-Rad). (B) Effects of BLK deficiency on IL-1β–induced oligomerization of endogenous IRAK1. BLK-deficient and control U87MG cells (1 × 108) were untreated or treated with IL-1β (20 ng/ml) for 10 min before gel filtration. Fraction size was calibrated with the gel filtration standard. (C) Effects of BLK deficiency on IL-1β–induced hyperphosphorylation of IRAK1 and subsequent recruitment of IRAK1 to TRAF6. BLK-deficient and control U87MG cells (2 × 107) (BLK-KO #1 plasmids were used) were left untreated or treated with IL-1β (20 ng/ml) for the indicated times before coimmunoprecipitation (IP) and immunoblot (IB) analysis. Data are representative of three independent experiments with similar results. Source data are available for this figure: SourceData F8.
Discussion
Antimicrobial inflammatory response is primarily initiated via the initial sensing of distinct PAMPs by host PRRs. Among innate immune PRRs, TLR/IL-1Rs have unique capacities to sense the initial infection and initiate potent inflammatory responses through the activation of intracellular signal transduction pathways (Gabay et al., 2010; Kawai and Akira, 2011). In this study, we identified BLK as a pivotal modulator of TLR/IL-1R signaling. Functionally, overexpression of BLK potentiated the transcription of downstream proinflammatory genes in response to IL-1β, LPS, flagellin, and CpG-B/C, but not TNFα treatment in different cell types, whereas BLK deficiency had opposite effects. These results suggest that BLK positively regulates TLR/IL-1R–mediated inflammatory signaling. Although BLK has a limited expression lineage in mice, it is relatively abundant in B lymphocytes and pDCs (Akerblad and Sigvardsson, 1999; Samuelson et al., 2014). Our results indicated that serum cytokines induced by IL-1β, including TNFα and IL6, were markedly decreased in Blk−/− mice in comparison with wild-type mice. Consistently, after injection with IL-1β plus D-gal, Blk−/− mice experienced a lower percentage of lethality compared with their wild-type counterparts. Several lines of evidence collectively point to an important role of BLK in positive regulation of TLR/IL-1R–mediated inflammatory response in vivo.
To prevent aberrant and persistent inflammatory responses that cause tissue damage, mammalian cells have evolved sophisticated regulatory mechanisms to avoid the hyperactivation of TLR/IL-1R signaling. For example, MARCH3 attenuates IL-1β–triggered inflammation by mediating K48-linked polyubiquitination and degradation of IL1R1 (Lin et al., 2018). Unlike other inducible negative regulators, TOLLIP is a constitutive suppressor of IRAK1 that keeps IRAK1 inactive and prevents it from forming higher-order complexes in resting cellular contexts, thereby limiting the magnitude of proinflammatory cytokine production. Upon IL-1β stimulation, TOLLIP disassociates from IRAK1, which triggers IRAK1 phosphorylation at the serine residues in its catalytic pocket (Burns et al., 2000). In line with this observation, our results indicated that TOLLIP robustly interacted and colocalized with IRAK1 in unstimulated cells, whereas their association was rapidly decreased upon IL-1β stimulation. Conversely, IL-1β–induced interaction between TOLLIP and BLK gradually increased. Further experiments suggested that BLK catalyzed tyrosine phosphorylation of TOLLIP at Y76, Y86, and Y152. The antibody specifically detecting TOLLIP Y86 phosphorylation verified that BLK deficiency indeed impaired IL-1β–induced tyrosine phosphorylation of TOLLIP. More importantly, reporter assays indicated that BLK-mediated tyrosine phosphorylation of TOLLIP was indispensable for its regulatory function in TLR/IL-1R signaling. Intriguingly, we found that mutation of tyrosine residues within TOLLIP to phenylalanine affected its molecular weight. Compared with wild-type TOLLIP, mutants including Y116F, Y152F, and Y172F had lower molecular weights, whereas mutants including Y83F, Y86F, Y193F, and Y199F had higher molecular weights (Fig. 4, E and G). This seemingly confusing result may be attributed to the fact that mutation of the above tyrosine residues alters the conformation and charge of TOLLIP, thereby affecting their migration in the gel.
Autophosphorylation of kinases is critical for their stability and physiological function (Dorsey et al., 2009; Kollewe et al., 2004). It is worth emphasizing that the Y309F alteration resulted in a dramatic reduction or almost complete absence of BLK autophosphorylation. Reconstitution of BLK-deficient cells with BLK(Y309F) failed to restore IL-1β–induced inflammatory response. Moreover, BLK(Y309F) failed to mediate tyrosine phosphorylation of TOLLIP, indicating that BLK autophosphorylation occurred prior to BLK-mediated tyrosine phosphorylation of TOLLIP. Although we cannot rule out the possibility that BLK also undergoes autophosphorylation at Y187 or Y188 residues, BLK autophosphorylation is mainly contingent on the Y309 residue, highlighting a prominent role of BLK self-activation in the regulation of TLR/IL-1R signaling.
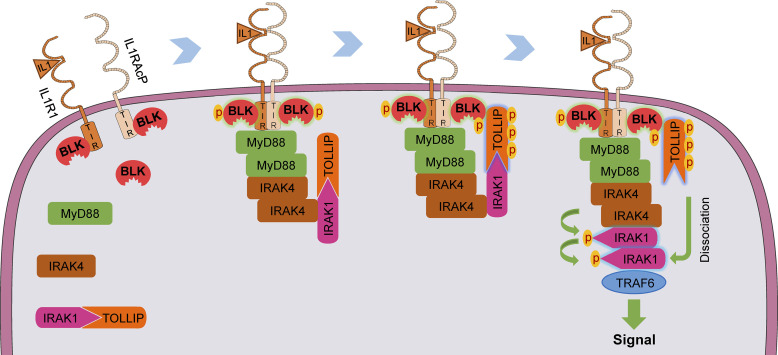
We note that the precise mechanisms of how IL-1β induces TOLLIP dissociation from IRAK1 remain unclear. In this study, Octet biolayer interferometry assays indicated that TOLLIP exhibited a higher affinity for BLK than for IRAK1. Furthermore, wild-type BLK but not its mutants Y309F or K269A dramatically inhibited the association of TOLLIP and IRAK1. Consistently, BLK deficiency impaired IL-1β–induced dissociation of IRAK1 from TOLLIP. These lines of evidence suggest that BLK dissociates TOLLIP from IRAK1 via a competitive mechanism, which relies on the kinase activity of BLK. It is conceivable that BLK is constitutively associated with the cytoplasmic TIR domains of IL-1Rs and TLRs under resting conditions. IL-1β treatment rapidly induces the formation of IL-1β/IL1R1/IL1RAcP heterocomplex, which brings the cytoplasmic TIR domains of IL1R1 and IL1RAcP into close proximity, leading to trans-phosphorylation and activation of BLK. Subsequently, the receptor complex recruits MyD88, IRAK4, IRAK1, and TOLLIP. Activated BLK directly interacts with and phosphorylates TOLLIP at Y76/86/152, which may lead to its conformation change. This event facilitates TOLLIP dissociation from IRAK1 and the exposure of phosphorylation sites within the ProST region of IRAK1, thus enabling hyperphosphorylation and oligomerization of IRAK1 and subsequent downstream inflammatory response (Fig. 9). Certainly, clarifying the detailed mechanisms of BLK autophosphorylation, as well as the association and dissociation between BLK-TOLLIP-IRAK1 ternary complex by more sensitive and direct techniques, such as crystallography or cryo-electron microscopy, warrants further exploration in the future.
Figure 9.
A schematic presentation for the role of BLK in regulating TLR/IL-1R–mediated inflammatory response. Under unstimulated conditions, BLK is constitutively associated with the cytoplasmic TIR domains of IL1R1 and IL1RAcP. Upon binding of IL-1β to IL1R1, IL1RAcP is recruited to form a high-affinity heterodimer, which results in the conformational change of TIR domains and further triggers BLK autophosphorylation. Simultaneously, the receptor complex recruits MyD88, IRAKs, and TOLLIP to form a signal transduction complex termed Myddosome. Activated BLK phosphorylates TOLLIP and further promotes TOLLIP dissociation from IRAK1 by competitively binding to the CUE domain of TOLLIP, thus enabling downstream signaling cascade.
As shown in Fig. 5 B and Fig. S4, our data indicated that BLK was constitutively associated with the cytoplasmic TIR domains of IL1R1 and IL1RAcP, which was essential for BLK activation. The cytoplasmic TIR domains of IL1Rs and TLRs are evolutionarily conserved in their function. These receptors (excluding TLR3) all recruit adaptor MyD88 in a TIR:TIR interaction manner to initiate downstream inflammatory response. Because BLK has a similar regulatory function in IL-1R– and TLR-mediated signal transduction, we speculate that BLK is also associated with TLRs prior to its activation. However, our study lacks direct evidence to suggest that BLK is preassociated with the cytoplasmic TIR domains of TLRs, which is a limitation of this study in clarifying the regulatory mechanism of BLK. Therefore, it is important to perform additional experiments in future studies, including endogenous coimmunoprecipitation and in vitro GST pull-down assays to further validate and characterize the interaction model between BLK and IL-1Rs/TLRs. In light of the pivotal roles of TLR/IL-1R signaling in immune defense and inflammatory response, it will be meaningful to explore the roles of BLK in IL-1β–relevant autoimmune diseases in future studies. In summary, our study provides a mechanistic basis for how tyrosine phosphorylation functions in inflammatory signaling.
Materials and methods
Reagents, antibodies, and cells
Dual-specific luciferase assay kit (E1980; Promega); RNAiso plus (9109; Takara); SYBR Green (172-5274; Bio-Rad); HiScript II Q RT SuperMix (R222-01; Vazyme); CIP (M0290S; NEB); polybrene (TR-1003-G; Millipore); Recombinant human TNFα and IL-1β (210-TA and 201-LB, respectively; R&D Systems); Recombinant murine IL-1β (211-11B-100; Perotech); LPS, flagellin, R848, ODN BW006 (CpG-B), and ODN 2395 (CpG-C; tlrl-b5lps, tlrl-stfla, tlrl-r848, tlrl-bw006, and tlrl-2395, respectively; Invivogen); D-gal (G0500; Sigma-Aldrich); DAPI (D9542; Sigma-Aldrich); Protein G sepharose and glutathione sepharose beads (17-0618-05 and 17-0756-01, respectively; GE Healthcare); Silver Stain for Mass Spectrometry (24600; Thermo Fisher Scientific); Mouse Plasmacytoid Dendritic Cell Isolation Kit (130-107-093; Miltenyi); Mouse CD19 MicroBeads (130-121-301; Miltenyi); ELISA kits for murine TNFα and IL6 (430904 and 431304, respectively; BioLegend); and HRP Conjugation Kit (ab102890; Abcam) were purchased from the indicated manufacturers.
Monoclonal antibodies against Flag (F3165, mouse, western blot [WB] 1:2,000, immunofluorescence [IF] 1:500) and β-actin (A2228, mouse, WB 1:5,000) were purchased from Sigma-Aldrich; MyD88 (AF3109, goat, WB 1:500) was purchased from R&D Systems; BLK (3262, rabbit, WB 1:500), IRAK1 (4359, rabbit, WB 1:1,000), IRAK4 (4363, rabbit, WB 1:1,000), TRAF6 (8028, rabbit, WB 1:1,000), Myc (2,276, mouse, WB 1:2,000), GST (2,624, mouse, WB 1:2,000), BLK (66002, rabbit, WB 1:500), phospho-Tyrosine (P-Tyr-100; 9411, mouse, WB 1:500), phospho-p65 (S536; 3033, rabbit, WB 1:500), and phospho-IκBα (S32/36; 9246, mouse, WB 1:500) were purchased from Cell Signaling Technology; TOLLIP (ab187198, rabbit, WB 1:1,000, IF 1:200) was purchased from Abcam; HA (TA100012, mouse, WB 1:2,000) was purchased from OriGene; p65 (sc-372, rabbit, WB 1:2,000) and IL-1R1 (sc-393998, mouse, WB 1:500) were purchased from Santa Cruz Biotechnology; Pan Phospho-Serine/Threonine (AP1067, mouse, WB 1:500), IL1R1 (A5727, rabbit, WB 1:500), IL1RAcP (A22286, rabbit, WB 1:500), BLK (A7427, rabbit, WB 1:500), and V5 (AE017, mouse, WB 1:2,000) were purchased from ABclonal Technology; IRAK1 (66653-1-Ig, mouse, IF 1:200) was purchased from Proteintech; Mouse anti-IκBα sera (WB 1:2,000) were kindly provided by Professor Hong-Bing Shu from Wuhan University, Wuhan, China (Chen et al., 2012; Li et al., 2011). Purified rabbit polyclonal antibody phospho-TOLLIP (Y86; WB 1:500) was raised specifically against a modified peptide YAVY(P)ETPTAHNG and generated by Atagenix Technology Co., Ltd. The secondary antibody goat anti-mouse (31430, WB 1:10,000) and goat anti-rabbit (31460, WB 1:10,000) IgG conjugated to HRP were purchased from Pierce. The secondary antibody rabbit anti-goat (AS029, WB 1:10,000) IgG conjugated to HRP were purchased from ABclonal Technology. The secondary antibody goat anti-rabbit (R37116, IF 1:2,000) IgG conjugated to Alexa Fluor 488 and goat anti-mouse (R37121, IF 1:2,000) IgG conjugated to Alexa Fluor 594 were purchased from Thermo Fisher Scientific.
HEK293 cells were purchased from ATCC; U87MG, Jurkat, Raji, and A20 cells were kindly provided by Procell Life Science & Technology Co., Ltd. Mycoplasma contamination was checked in all used cells.
Constructs
Overexpression plasmids were constructed into pCMV14 or pRK vectors by standard molecular cloning methods. The point mutation plasmids were constructed by site-directed mutagenesis methods. TOLLIP (aa: 1–180), TOLLIP (aa: 1–228), TOLLIP (aa: 54–274), and TOLLIP (aa: 181–274) are different truncations of TOLLIP. BLK, TOLLIP, or IRAK1 was constructed into pGEX-6P-1 with a GST tag in its N-terminus. BLK was constructed into pRK with a GST tag in its N-terminus. BLK was constructed into pLX304 with a V5 tag in its C-terminus. Human or murine BLK, TOLLIP, or their mutants were constructed into pLOV with a Flag tag in its C-terminus. Human or murine BLK, TOLLIP, MyD88, IRAKs, TRAF6, or their mutants were constructed into pRK with a Flag, HA, or Myc tag in its C-terminus. All gRNAs were constructed into lentiCRISPR v2. NF-κB reporter plasmids were previously described (Li et al., 2020; Xu et al., 2005). All of the above original empty vectors were kindly provided by Professor Hong-Bing Shu from Wuhan University. All of the above eukaryotic and prokaryotic expression plasmids were constructed in our laboratory.
Transfection and reporter assays
HEK293-TLR4 or U87MG cells were seeded on 48-well plates and transfected on the following day by standard calcium phosphate precipitation or Lipofectamine 2000. To normalize the transfection efficiency, 10 ng of pRL-TK (Renilla luciferase) reporter plasmid was added to each transfection. Empty control plasmid was added to ensure that each transfection received the same amount of total plasmid DNA. Luciferase assays were performed using a dual-specific luciferase assay kit. Firefly luciferase activities were normalized on the basis of Renilla luciferase activities.
RNA interference
The siRNA duplexes targeting BLK gene were chemically synthesized by GenePharma. The siRNA duplexes were transfected into cells with PepMute siRNA transfection reagent (SignaGen Laboratories) according to the manufacturer’s instructions. The corresponding RNAi oligonucleotide sequences were as follows:
BLK-RNAi #1: 5′-AAGCGACTGTCATCAAGTA-3′
BLK-RNAi #2: 5′-AGGTGGTTCTTTAGATCAC-3′
Negative control (NC): 5′-TTCTCCGAACGTGTCACGT-3′.
RNA extraction and qPCR
Total RNA was isolated from cells using RNAiso plus reagent. After reverse-transcription with HiScript II Q RT SuperMix, the cDNA was diluted 50-fold and subjected to qPCR analysis to measure mRNA levels of the tested genes. Data shown were the relative abundance of the indicated mRNAs normalized to that of GAPDH. Gene-specific primer sequences were as follows:
GAPDH: 5′-GACAAGCTTCCCGTTCTCAG-3′ (forward) and 5′-GAGTCAACGGATTTGGTCGT-3′ (reverse)
TNFA: 5′-GCCGCATCGCCGTCTCCTAC-3′ (forward) and 5′-CCTCAGCCCCCTCTGGGGTC-3′ (reverse)
CXCL10: 5′-GGTGAGAAGAGATGTCTGAATCC-3′ (forward) and 5′-GTCCATCCTTGGAAGCACTGCA-3′ (reverse)
BLK: 5′-AGGAAAAGCCGATCAAAGAGAAG-3′ (forward) and 5′-CCACCACGAAATGCTTGTCT-3′ (reverse)
Gapdh: 5′-ACGGCCGCATCTTCTTGTGCA-3′ (forward) and 5′-ACGGCCAAATCCGTTCACACC-3′ (reverse)
Tnfa: 5′-GGTGATCGGTCCCCAAAGGGATGA-3′ (forward) and 5′-TGGTTTGCTACGACGTGGGCT-3′ (reverse)
Il6: 5′-TCTGCAAGAGACTTCCATCCAGTTGC-3′ (forward) and 5′-AGCCTCCGACTTGTGAAGTGGT-3′ (reverse)
Cxcl10: 5′-ATCATCCCTGCGAGCCTATCCT-3′ (forward) and 5′-GACCTTTTTTGGCTAAACGCTTTC-3′ (reverse)
Il1b: 5′-CGGACCCCAAAAGATGAAGGGCTG-3′ (forward) and 5′-AGCTGCCACAGCTTCTCCACA-3′ (reverse)
Blk: 5′-CAGTTTGGCGAAGTCTGGATGG-3′ (forward) and 5′-AGACGAACCAGCCTCTCATGCT-3′ (reverse).
Coimmunoprecipitation and immunoblot analysis
Cells were lysed in l ml NP-40 lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40) supplemented with protease and phosphatase inhibitors. The lysate was centrifuged at 12,000 rpm for 15 min at 4°C. For each immunoprecipitation, a 0.4 ml aliquot of lysate supernatant was incubated with 0.5–2 μg of the indicated antibody or control IgG and 25 μl of a 1:1 slurry of Protein G sepharose at 4°C for at least 2 h. The sepharose beads were then washed three times with lysis buffer containing 500 mM NaCl. The precipitates were resuspended in 60 μl of 2 × SDS loading buffer, boiled for 10 min, and further analyzed by standard immunoblot. Briefly, protein samples separated on SDS-PAGE were electrotransferred onto nitrocellulose membrane (Invitrogen) and blocked with 10% skim milk (Sigma-Aldrich) in 1 × TBST (TBS supplemented with 0.1% Tween 20). Subsequently, the blots were incubated with the indicated primary antibodies for 2 h at room temperature. The membranes were washed for 10 min thrice with 1 × TBST and further incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. The membranes were then washed for 10 min thrice with 1 × TBST. Development was conducted with a chemiluminescence-based method (ECL reagent; Thermo Fisher Scientific) using x-ray films (Carestream). ImageJ software was used to perform the quantitative analysis on immunoblots.
Mass spectrometry analysis
HEK293 cells (2 × 107) were transfected with GST-BLK or GST plasmids for 24 h. Cells were then treated with IL-1β (20 ng/ml) for 0.5 h before lysis in NP-40 buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40) supplemented with protease and phosphatase inhibitors. GST-BLK–associated factors were immunoprecipitated from lysates using glutathione sepharose beads. Immunoprecipitated proteins were detected by immunoblot analysis followed by silver staining according to the manufacturer’s instructions. Gel bands were separated and further subjected to mass spectrometry analysis.
In vitro tyrosine phosphorylation assays
Recombinant GST-TOLLIP protein expressed in Escherichia coli Rosetta strain was purified with glutathione sepharose and eluted from beads with elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM reduced glutathione). For in vitro kinase assays, Flag-tagged BLK and its mutants expressed in HEK293 cells were purified with Protein G sepharose and then incubated with purified GST-TOLLIP (50 μg) in an equal volume of 2 × reaction buffer (100 mM Tris-HCl, 20 mM MgCl2, 1 mM Na3VO4, 4 mM DTT, pH 7.2), and 1 mM ATP was added prior to incubation for 1 h at 30°C. The reaction was stopped by adding 1/5 vol of 6 × SDS loading buffer and boiling for 10 min at 95°C before immunoblot analysis.
For BLK autophosphorylation assays, Flag-tagged BLK and its mutants expressed in HEK293 cells were purified with Protein G sepharose and then incubated with 50 μl of 1 × reaction buffer (50 mM Tris-HCl, 10 mM MgCl2, 0.5 mM Na3VO4, 2 mM DTT, pH 7.2), and 1 mM ATP was added prior to incubation for 1 h at 30°C. The reaction was stopped by adding 1/5 vol of 6 × SDS loading buffer and boiling for 10 min at 95°C before immunoblot analysis.
For CIP treatment assays, Flag-tagged BLK and its mutants expressed in HEK293 cells were purified with Protein G sepharose and then incubated with 50 μl of 1 × reaction buffer, and 1 mM ATP was added prior to incubation for 1 h at 30°C. Subsequently, 1 μl CIP was added to the above reaction and incubated at 37°C for 30 min. The sepharose beads were washed three times and then resuspended again in 50 μl of 1 × reaction buffer, and 1 mM ATP was added prior to incubation for 1 h at 30°C. The reaction was stopped by adding 1/5 vol of 6 × SDS loading buffer and boiling for 10 min at 95°C before immunoblot analysis.
Mice
Blk−/− mice with the C57BL/6J background were generated utilizing the CRISPR/Cas9 method. The strategy for construction of the targeting vector is illustrated in Fig. S2 A. Exons 3–9 and part of the intron of Blk gene were targeted by four specific gRNAs, which led to the deletion of exons 3–9 (∼7.8 kb) of Blk coding sequence. Genotyping was performed by PCR with the following primers: P1: 5′-GGCAACTCAGAGAGGAAAGG-3′ (forward), P2: 5′-TTGAAAACTGCAGCCAGGAG-3′ (reverse), and P3: 5′-CTAGAGGACACTGGAGAGCC-3′ (reverse). Amplification of the WT allele produced a 545 bp fragment, and amplification of the disrupted allele produced a 323 bp fragment. All animals were handled in strict accordance with good animal practices according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China. 8–10-wk-old, age- and sex-matched mice were used in all experiments. All mouse studies were approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (approval number LVRIAEC-2023-035).
Isolation of primary B cells and pDCs
Spleens were obtained from 2-mo-old C57BL/6J background mice. A suspension of single splenocytes was obtained by passage of the spleens through a 40-μm mesh size cell strainer after lysis of red blood cells with ACK lysis buffer.
Primary B lymphocytes were purified by positive selection utilizing mouse CD19 MicroBeads according to the manufacturer’s protocol. Purity was phenotypically validated by flow cytometry using an anti-mouse CD19-APC antibody and was >95% in all experiments. Positively selected cells were seeded in 12-well plates in the presence of anti-lgM/IgG (5 μg/ml) and anti-CD40 antibody (1 μg/ml) for 48 h. Then, primary B lymphocytes were stimulated with the corresponding stimuli for the indicated times before qPCR and immunoblot analyses.
Splenic pDCs were purified by negative magnetic selection using the Mouse Plasmacytoid Dendritic Cell Isolation Kit and then stimulated with the corresponding stimuli for the indicated times before qPCR and immunoblot analyses.
ELISA
Age- and sex-matched Blk+/+ and Blk−/− mice were injected i.p. with murine IL-1β (150 μg/kg) for 2 h. The blood was collected and incubated for 1 h at 37°C and then incubated overnight at 4°C. The next day, blood was centrifuged for 10 min at 3,000 rpm/min. The serum was collected from the supernatant. The concentration of serum cytokines was detected with ELISA kits according to the manufacturer’s instructions.
IL-1β injection
Age- and sex-matched Blk+/+ and Blk−/− mice were injected i.p. with murine IL-1β (150 μg/kg) plus D-gal (0.5 mg/g) per mouse. The survival rate of the injected mice was recorded every 1 h.
CRISPR/Cas9 knockout
Double-stranded oligonucleotides corresponding to the target sequences were cloned into the lentiCRISPR v2 vector. Then, this constructed plasmid (10 μg) was cotransfected with two packaging plasmids (LH1 [7.5 μg] and LH2 [5 μg]) into HEK293 cells. The culture medium was replaced with fresh medium without antibiotics 12 h after transfection. After an additional 36 h, the medium containing lentiviral particles was filtered (0.22-μm filter; Millipore) and used to infect Jurkat, Raji, U87MG, or A20 cells in the presence of polybrene (8 μg/ml). The infected cells were selected with puromycin (1 μg/ml) or blasticidin S (10 μg/ml) for at least 6 d before additional experiments were performed. The corresponding gRNA oligonucleotide sequences were as follows:
human BLK gRNA #1: 5′-AAGTAGCAAAAAGCCGGACA-3′
human BLK gRNA #2: 5′-CCGATCAAAGAGAAGGACAA-3′
human IL1R1 gRNA #1: 5′-AATAGTCTTCCCCTAGCACT-3′
human IL1R1 gRNA #2: 5′-ATTACAGATCAATTGTATCT-3′
human IL1RAcP gRNA #1: 5′-ACTGGGGACTAGACACCATG-3′
human IL1RAcP gRNA #2: 5′-GACGTACGTTTCATCTCACC-3′
human IRAK1 gRNA #1: 5′-ACACGGTGTATGCTGTGAAG-3′
human IRAK1 gRNA #2: 5′-AGGAGTACATCAAGACGGGA-3′
mouse Blk gRNA #1: 5′-GGTAAACACCCCGAAGTGGA-3′
mouse Blk gRNA #2: 5′-CCTGGGTGCGGATCTTCACG-3′
mouse Tollip gRNA #1: 5′-GAATTATGGCATGACTCGTA-3′
mouse Tollip gRNA #2: 5′-TAGAACGAGTCCACACCTGG-3′.
Establishment of BLK stable-expressing cell lines
HEK293 cells plated on 100-mm dishes were cotransfected with pMSCV-BLK (10 μg) and two packaging plasmids (Gag-pol [10 μg] and vesicular stomatitis virus-glycoprotein [3 μg]). The detailed procedure is similar to the CRISPR/Cas9 knockout method described above.
Confocal microscopy and image processing and analysis
Cells were stimulated with IL-1β for the indicated times, fixed with 4% paraformaldehyde for 10 min, and permeabilized with 0.1% Triton X-100 in PBS for 10 min at 4°C. First, cells were blocked with 1% BSA in PBS for 1 h and then incubated with the indicated primary antibodies (1:200) for 2 h at room temperature, followed by 3 × 10-min washes in PBS. Subsequently, cells were incubated with secondary antibodies (1:2,000, Alexa Fluor 488 and 594) for 1 h at room temperature and then stained with DAPI (1:2,000) for 2 min, followed by 3 × 10-min washes in PBS. Finally, coverslips were mounted onto slides using ProLong Gold Antifade Mountant and allowed to cure overnight at room temperature before cell imaging. Image acquisition was performed using a confocal microscope (HD25; Nikon) equipped with Plan-Apochromat 60×/1.42 oil objective (NA 0.95 and WD 0.15 mm; Nikon) and microscope camera (Digital Sight 10; Nikon). Digital images were exported using the NIS-Elements Viewer software and processed for contrast and brightness using the Photoshop v3.2 software. Quantitative analysis of colocalization images was performed using the open-source Fiji (ImageJ) software.
Recombinant protein purification and in vitro GST pull-down assays
Recombinant GST-BLK, GST-TOLLIP, and GST-IRAK1 proteins expressed in E. coli Rosetta strain were purified with glutathione sepharose and eluted from beads with elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM reduced glutathione). For pull-down assays, Flag-tagged IRAK1, IRAK4, and TOLLIP proteins expressed in HEK293 cells were purified with Protein G sepharose and eluted from beads with 3×Flag peptide (1 mg/ml). Then, the proteins were added to the purified recombinant GST-BLK protein coupled to glutathione sepharose beads and incubated for 3 h at 4°C. Subsequently, the beads were washed and boiled. The eluates/inputs were fractionated by SDS-PAGE and detected by Coomassie staining and immunoblot.
Gel filtration chromatography
Cotransfection of IRAK1 with BLK or its mutants into HEK293 cells. 24 h after transfection, collected cells (4 × 107 cells per sample) were lysed in lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, supplemented with protease and phosphatase inhibitors) and sonicated for 2 min. Cell lysates were centrifuged at 13,000 rpm, 4°C for 30 min. Subsequently, cell supernatants were filtered (0.45-μm filter; Millipore) and subjected to gel filtration chromatography using Superdex 200 Increase 10/300 GL column and separation buffer (50 mM Tris, pH 7.4, 150 mM NaCl). Fraction collector collected 1 ml per fraction for 12 fractions after balancing with separation buffer of 1/5 column volume. A gel filtration standard (151-1901; Bio-Rad) was also run to calibrate the fractions.
Wild-type and BLK-deficient U87MG cells (1 × 108 cells per sample) were stimulated with IL-1β (20 ng/ml) for 10 min. Subsequently, cells were collected and lysed in lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, supplemented with protease and phosphatase inhibitors) and sonicated for 2 min. Then, cell lysates were centrifuged and subjected to gel filtration chromatography as described above.
ForteBio Octet Red bio-layer interferometry
Binding affinity between TOLLIP and BLK or IRAK1 was measured by the ForteBio Octet Red system (ForteBio, Inc). Streptavidin biosensors were first loaded with biotinylated TOLLIP for 300 s, baselined in equilibration buffer (PBS + 0.02% Tween20 + 0.1% BSA) for 120 s, and then loaded with BLK or IRAK1 for 600 s. Subsequently, the associated BLK or IRAK1 was dissociated in equilibration buffer for 600 s. Association and dissociation of BLK or IRAK1 to TOLLIP were monitored with 200 μl of equilibration buffer by the spectral shift. The affinity constants were calculated with the Octet Red software.
Statistical analysis
Unpaired, two-tailed Student’s t test was used for statistical analysis between two groups. For the mouse survival study, Kaplan–Meier survival curves were generated and analyzed by the log-rank test. Data distribution was assumed to be normal, but this was not formally tested. Statistical differences were evaluated using GraphPad Prism 8 software. P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows the effect of BLK on IL-1β–, LPS-, and TNFα-induced inflammatory responses. Fig. S2 shows the construction strategy of Blk−/− mice. Fig. S3 shows the phosphorylation of TOLLIP mediated by BLK. Fig. S4 shows the interaction domains between BLK and IL1R1 or IL1RAcP. Fig. S5 shows the interaction domains between TOLLIP and BLK or IRAK1. Table S1 shows the interacting proteins of BLK identified by mass spectrometry. Table S2 shows the phosphorylated peptide sequences and phosphorylation sites of TOLLIP identified by mass spectrometry. Table S3 shows the phosphorylated peptide sequences and phosphorylation sites of BLK identified by mass spectrometry.
Supplementary Material
shows the interacting proteins of BLK identified by mass spectrometry.
shows the phosphorylated peptide sequences and phosphorylation sites of TOLLIP identified by mass spectrometry.
shows the phosphorylated peptide sequences and phosphorylation sites of BLK identified by mass spectrometry.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
is the source file for Fig. S5.
Acknowledgments
We thank Ding Gao from the Center for Instrumental Analysis and Metrology at Wuhan Institute of Virology for help with the biolayer interferometry experiments.
This work was supported by the grants from the National Key R&D Program of China (2021YFD1800300, awarded to Z.-X. Zhu, and 2021YFD1801300, awarded to H.-X. Zheng), the National Natural Science Foundation of China (32202781, awarded to W.-W. Li), the Fundamental Research Funds for the Central Universities (awarded to H.-X. Zheng and Z.-X. Zhu), the Key Technologies R&D Program of Gansu Province (21ZD3NA001, awarded to H.-X. Zheng, 20ZD7NA006, awarded to H.-X. Zheng, and 19ZD2NA001, awarded to H.-X. Zheng), the Natural Science Foundation of Gansu Province (22JR5RA034, awarded to W.-W. Li), the Open Competition Program of Top 10 Critical Priorities of Agricultural Science and Technology Innovation for the 14th Five-Year Plan of Guangdong Province (2023SDZG02, awarded to H.-X. Zheng), the Earmarked Fund for CARS-35 (awarded to H.-X. Zheng) and CARS-39-13 (awarded to H.-X. Zheng), the Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-CSLPDCP-2023002, awarded to H.-X. Zheng, and CAAS-ASTIP-2023-LVRI, awarded to H.-X. Zheng), and the Basic Scientific Research Fund of Lanzhou Veterinary Research Institute (1610312021009, awarded to W.-W. Li).
Author contributions: Conceptualization: H.-X. Zheng, Y.-Y. Wang, and W.-W. Li. Formal analysis: W.-W. Li and X.-X. Fan. Funding acquisition: H.-X. Zheng, Y.-Y. Wang, and W.-W. Li. Investigation: W.-W. Li, X.-X. Fan, Z.-S. Xu, Z.-X. Zhu, Z.-Y. Zhu, X.-J. Cao, D.-S. Pei, Y.-Z. Wang, and J.-Y. Zhang. Supervision: H.-X. Zheng and Y.-Y. Wang. Writing–original draft: W.-W. Li and X.-X. Fan. Writing–review & editing: W.-W. Li, X.-X. Fan, Z.-S. Xu, Z.-X. Zhu, Y.-Y. Wang, and H.-X. Zheng.
Data availability
All data, plasmids, and reagents generated in this study are available upon reasonable request.
References
- Akerblad, P., and Sigvardsson M.. 1999. Early B cell factor is an activator of the B lymphoid kinase promoter in early B cell development. J. Immunol. 163:5453–5461. 10.4049/jimmunol.163.10.5453 [DOI] [PubMed] [Google Scholar]
- Akira, S., Takeda K., and Kaisho T.. 2001. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675–680. 10.1038/90609 [DOI] [PubMed] [Google Scholar]
- Akira, S., Uematsu S., and Takeuchi O.. 2006. Pathogen recognition and innate immunity. Cell. 124:783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Barton, G.M., and Medzhitov R.. 2003. Toll-like receptor signaling pathways. Science. 300:1524–1525. 10.1126/science.1085536 [DOI] [PubMed] [Google Scholar]
- Blasius, A.L., and Beutler B.. 2010. Intracellular toll-like receptors. Immunity. 32:305–315. 10.1016/j.immuni.2010.03.012 [DOI] [PubMed] [Google Scholar]
- Brissoni, B., Agostini L., Kropf M., Martinon F., Swoboda V., Lippens S., Everett H., Aebi N., Janssens S., Meylan E., et al. 2006. Intracellular trafficking of interleukin-1 receptor I requires Tollip. Curr. Biol. 16:2265–2270. 10.1016/j.cub.2006.09.062 [DOI] [PubMed] [Google Scholar]
- Burns, K., Clatworthy J., Martin L., Martinon F., Plumpton C., Maschera B., Lewis A., Ray K., Tschopp J., and Volpe F.. 2000. Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nat. Cell Biol. 2:346–351. 10.1038/35014038 [DOI] [PubMed] [Google Scholar]
- Cao, Z., Henzel W.J., and Gao X.. 1996. IRAK: A kinase associated with the interleukin-1 receptor. Science. 271:1128–1131. 10.1126/science.271.5252.1128 [DOI] [PubMed] [Google Scholar]
- Capelluto, D.G.S. 2012. Tollip: A multitasking protein in innate immunity and protein trafficking. Microbes Infect. 14:140–147. 10.1016/j.micinf.2011.08.018 [DOI] [PubMed] [Google Scholar]
- Chen, R., Li M., Zhang Y., Zhou Q., and Shu H.B.. 2012. The E3 ubiquitin ligase MARCH8 negatively regulates IL-1β-induced NF-κB activation by targeting the IL1RAP coreceptor for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA. 109:14128–14133. 10.1073/pnas.1205246109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conze, D.B., Wu C.J., Thomas J.A., Landstrom A., and Ashwell J.D.. 2008. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-kappaB activation. Mol. Cell. Biol. 28:3538–3547. 10.1128/MCB.02098-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nardo, D., Balka K.R., Cardona Gloria Y., Rao V.R., Latz E., and Masters S.L.. 2018. Interleukin-1 receptor-associated kinase 4 (IRAK4) plays a dual role in myddosome formation and Toll-like receptor signaling. J. Biol. Chem. 293:15195–15207. 10.1074/jbc.RA118.003314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didierlaurent, A., Brissoni B., Velin D., Aebi N., Tardivel A., Käslin E., Sirard J.C., Angelov G., Tschopp J., and Burns K.. 2006. Tollip regulates proinflammatory responses to interleukin-1 and lipopolysaccharide. Mol. Cell. Biol. 26:735–742. 10.1128/MCB.26.3.735-742.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello, C.A. 2011. A clinical perspective of IL-1β as the gatekeeper of inflammation. Eur. J. Immunol. 41:1203–1217. 10.1002/eji.201141550 [DOI] [PubMed] [Google Scholar]
- Dorsey, F.C., Rose K.L., Coenen S., Prater S.M., Cavett V., Cleveland J.L., and Caldwell-Busby J.. 2009. Mapping the phosphorylation sites of Ulk1. J. Proteome Res. 8:5253–5263. 10.1021/pr900583m [DOI] [PubMed] [Google Scholar]
- Ferrao, R., Zhou H., Shan Y., Liu Q., Li Q., Shaw D.E., Li X., and Wu H.. 2014. IRAK4 dimerization and trans-autophosphorylation are induced by Myddosome assembly. Mol. Cell. 55:891–903. 10.1016/j.molcel.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay, C., Lamacchia C., and Palmer G.. 2010. IL-1 pathways in inflammation and human diseases. Nat. Rev. Rheumatol. 6:232–241. 10.1038/nrrheum.2010.4 [DOI] [PubMed] [Google Scholar]
- Gurung, P., Fan G., Lukens J.R., Vogel P., Tonks N.K., and Kanneganti T.D.. 2017. Tyrosine Kinase SYK licenses MyD88 adaptor protein to instigate IL-1α-mediated inflammatory disease. Immunity. 46:635–648. 10.1016/j.immuni.2017.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J.W., Zheng H.F., Cui Y., Sun L.D., Ye D.Q., Hu Z., Xu J.H., Cai Z.M., Huang W., Zhao G.P., et al. 2009. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 41:1234–1237. 10.1038/ng.472 [DOI] [PubMed] [Google Scholar]
- Hu, M.M., and Shu H.B.. 2020. Innate immune response to cytoplasmic DNA: Mechanisms and diseases. Annu. Rev. Immunol. 38:79–98. 10.1146/annurev-immunol-070119-115052 [DOI] [PubMed] [Google Scholar]
- Huang, J., Gao X., Li S., and Cao Z.. 1997. Recruitment of IRAK to the interleukin 1 receptor complex requires interleukin 1 receptor accessory protein. Proc. Natl. Acad. Sci. USA. 94:12829–12832. 10.1073/pnas.94.24.12829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imler, J.L., and Hoffmann J.A.. 2001. Toll receptors in innate immunity. Trends Cell Biol. 11:304–311. 10.1016/S0962-8924(01)02004-9 [DOI] [PubMed] [Google Scholar]
- Jiang, S.H., Athanasopoulos V., Ellyard J.I., Chuah A., Cappello J., Cook A., Prabhu S.B., Cardenas J., Gu J., Stanley M., et al. 2019. Functional rare and low frequency variants in BLK and BANK1 contribute to human lupus. Nat. Commun. 10:2201. 10.1038/s41467-019-10242-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, T., and Akira S.. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 34:637–650. 10.1016/j.immuni.2011.05.006 [DOI] [PubMed] [Google Scholar]
- Kim, T., Pazhoor S., Bao M., Zhang Z., Hanabuchi S., Facchinetti V., Bover L., Plumas J., Chaperot L., Qin J., and Liu Y.J.. 2010. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA. 107:15181–15186. 10.1073/pnas.1006539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, K., Hernandez L.D., Galán J.E., Janeway C.A. Jr., Medzhitov R., and Flavell R.A.. 2002. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 110:191–202. 10.1016/S0092-8674(02)00827-9 [DOI] [PubMed] [Google Scholar]
- Kollewe, C., Mackensen A.C., Neumann D., Knop J., Cao P., Li S., Wesche H., and Martin M.U.. 2004. Sequential autophosphorylation steps in the interleukin-1 receptor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. J. Biol. Chem. 279:5227–5236. 10.1074/jbc.M309251200 [DOI] [PubMed] [Google Scholar]
- Kowalski, E.J.A., and Li L.. 2017. Toll-interacting protein in resolving and non-resolving inflammation. Front. Immunol. 8:511. 10.3389/fimmu.2017.00511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, H., Kawai T., and Akira S.. 2009. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 388:621–625. 10.1016/j.bbrc.2009.08.062 [DOI] [PubMed] [Google Scholar]
- Laird, R.M., Laky K., and Hayes S.M.. 2010. Unexpected role for the B cell-specific Src family kinase B lymphoid kinase in the development of IL-17-producing γδ T cells. J. Immunol. 185:6518–6527. 10.4049/jimmunol.1002766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.W., Han S.I., Kim H.H., and Lee Z.H.. 2002. TAK1-dependent activation of AP-1 and c-Jun N-terminal kinase by receptor activator of NF-kappaB. J. Biochem. Mol. Biol. 35:371–376. 10.5483/bmbrep.2002.35.4.371 [DOI] [PubMed] [Google Scholar]
- Lee, Y.S., Park J.S., Kim J.H., Jung S.M., Lee J.Y., Kim S.J., and Park S.H.. 2011. Smad6-specific recruitment of Smurf E3 ligases mediates TGF-β1-induced degradation of MyD88 in TLR4 signalling. Nat. Commun. 2:460. 10.1038/ncomms1469 [DOI] [PubMed] [Google Scholar]
- Li, S., Strelow A., Fontana E.J., and Wesche H.. 2002. IRAK-4: A novel member of the IRAK family with the properties of an IRAK-kinase. Proc. Natl. Acad. Sci. USA. 99:5567–5572. 10.1073/pnas.082100399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., Hu J., and Li L.. 2004. Characterization of Tollip protein upon lipopolysaccharide challenge. Mol. Immunol. 41:85–92. 10.1016/j.molimm.2004.03.009 [DOI] [PubMed] [Google Scholar]
- Li, Q., Yan J., Mao A.P., Li C., Ran Y., Shu H.B., and Wang Y.Y.. 2011. Tripartite motif 8 (TRIM8) modulates TNFα- and IL-1β-triggered NF-κB activation by targeting TAK1 for K63-linked polyubiquitination. Proc. Natl. Acad. Sci. USA. 108:19341–19346. 10.1073/pnas.1110946108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.W., Nie Y., Yang Y., Ran Y., Luo W.W., Xiong M.G., Wang S.Y., Xu Z.S., and Wang Y.Y.. 2020. Ubiquitination of TLR3 by TRIM3 signals its ESCRT-mediated trafficking to the endolysosomes for innate antiviral response. Proc. Natl. Acad. Sci. USA. 117:23707–23716. 10.1073/pnas.2002472117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H., Gao D., Hu M.M., Zhang M., Wu X.X., Feng L., Xu W.H., Yang Q., Zhong X., Wei J., et al. 2018. MARCH3 attenuates IL-1β-triggered inflammation by mediating K48-linked polyubiquitination and degradation of IL-1RI. Proc. Natl. Acad. Sci. USA. 115:12483–12488. 10.1073/pnas.1806217115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Zhang H., Wu X., Ma D., Wu J., Wang L., Jiang Y., Fei Y., Zhu C., Tan R., et al. 2018. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 563:131–136. 10.1038/s41586-018-0629-6 [DOI] [PubMed] [Google Scholar]
- Liu, Y., Zhang Q., Ding Y., Li X., Zhao D., Zhao K., Guo Z., and Cao X.. 2015. Histone lysine methyltransferase Ezh1 promotes TLR-triggered inflammatory cytokine production by suppressing Tollip. J. Immunol. 194:2838–2846. 10.4049/jimmunol.1402087 [DOI] [PubMed] [Google Scholar]
- Malek, S.N., Dordai D.I., Reim J., Dintzis H., and Desiderio S.. 1998. Malignant transformation of early lymphoid progenitors in mice expressing an activated Blk tyrosine kinase. Proc. Natl. Acad. Sci. USA. 95:7351–7356. 10.1073/pnas.95.13.7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M.U., and Wesche H.. 2002. Summary and comparison of the signaling mechanisms of the Toll/interleukin-1 receptor family. Biochim. Biophys. Acta. 1592:265–280. 10.1016/S0167-4889(02)00320-8 [DOI] [PubMed] [Google Scholar]
- Mizukami, J., Takaesu G., Akatsuka H., Sakurai H., Ninomiya-Tsuji J., Matsumoto K., and Sakurai N.. 2002. Receptor activator of NF-kappaB ligand (RANKL) activates TAK1 mitogen-activated protein kinase kinase kinase through a signaling complex containing RANK, TAB2, and TRAF6. Mol. Cell. Biol. 22:992–1000. 10.1128/MCB.22.4.992-1000.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, D., Kollewe C., Resch K., and Martin M.U.. 2007. The death domain of IRAK-1: An oligomerization domain mediating interactions with MyD88, Tollip, IRAK-1, and IRAK-4. Biochem. Biophys. Res. Commun. 354:1089–1094. 10.1016/j.bbrc.2007.01.104 [DOI] [PubMed] [Google Scholar]
- Neves, P., Lampropoulou V., Calderon-Gomez E., Roch T., Stervbo U., Shen P., Kühl A.A., Loddenkemper C., Haury M., Nedospasov S.A., et al. 2010. Signaling via the MyD88 adaptor protein in B cells suppresses protective immunity during Salmonella typhimurium infection. Immunity. 33:777–790. 10.1016/j.immuni.2010.10.016 [DOI] [PubMed] [Google Scholar]
- O’Neill, L.A., and Dinarello C.A.. 2000. The IL-1 receptor/toll-like receptor superfamily: Crucial receptors for inflammation and host defense. Immunol. Today. 21:206–209. 10.1016/S0167-5699(00)01611-X [DOI] [PubMed] [Google Scholar]
- Oda, H., Kumar S., and Howley P.M.. 1999. Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination. Proc. Natl. Acad. Sci. USA. 96:9557–9562. 10.1073/pnas.96.17.9557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma, K., Yamochi T., Uchiyama M., Nishibashi K., Iwata S., Hosono O., Kawasaki H., Tanaka H., Dang N.H., and Morimoto C.. 2005. CD26 mediates dissociation of Tollip and IRAK-1 from caveolin-1 and induces upregulation of CD86 on antigen-presenting cells. Mol. Cell. Biol. 25:7743–7757. 10.1128/MCB.25.17.7743-7757.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar, N., Chandrakar P., Vishwakarma P., Singh K., Mitra K., and Kar S.. 2018. Leishmania donovani exploits Tollip, a multitasking protein, to impair TLR/IL-1R signaling for its survival in the host. J. Immunol. 201:957–970. 10.4049/jimmunol.1800062 [DOI] [PubMed] [Google Scholar]
- Pore, D., Matsui K., Parameswaran N., and Gupta N.. 2016. Cutting edge: Ezrin regulates inflammation by limiting B cell IL-10 production. J. Immunol. 196:558–562. 10.4049/jimmunol.1502098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, N., Nguyen S., Ngo K., and Fung-Leung W.P.. 2005. A novel splice variant of interleukin-1 receptor (IL-1R)-associated kinase 1 plays a negative regulatory role in Toll/IL-1R-induced inflammatory signaling. Mol. Cell. Biol. 25:6521–6532. 10.1128/MCB.25.15.6521-6532.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson, E.M., Laird R.M., Papillion A.M., Tatum A.H., Princiotta M.F., and Hayes S.M.. 2014. Reduced B lymphoid kinase (Blk) expression enhances proinflammatory cytokine production and induces nephrosis in C57BL/6-lpr/lpr mice. PLoS One. 9:e92054. 10.1371/journal.pone.0092054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpfendorfer, K.R., Olsson L.M., Manjarrez Orduño N., Khalili H., Simeone A.M., Katz M.S., Lee A.T., Diamond B., and Gregersen P.K.. 2012. The autoimmunity-associated BLK haplotype exhibits cis-regulatory effects on mRNA and protein expression that are prominently observed in B cells early in development. Hum. Mol. Genet. 21:3918–3925. 10.1093/hmg/dds220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, J.E., and Smith D.E.. 2010. The IL-1 family: Regulators of immunity. Nat. Rev. Immunol. 10:89–102. 10.1038/nri2691 [DOI] [PubMed] [Google Scholar]
- Song, D.H., and Lee J.O.. 2012. Sensing of microbial molecular patterns by Toll-like receptors. Immunol. Rev. 250:216–229. 10.1111/j.1600-065X.2012.01167.x [DOI] [PubMed] [Google Scholar]
- van der Sluis, R.M., Cham L.B., Gris-Oliver A., Gammelgaard K.R., Pedersen J.G., Idorn M., Ahmadov U., Hernandez S.S., Cémalovic E., Godsk S.H., et al. 2022. TLR2 and TLR7 mediate distinct immunopathological and antiviral plasmacytoid dendritic cell responses to SARS-CoV-2 infection. EMBO J. 41:e109622. 10.15252/embj.2021109622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche, H., Henzel W.J., Shillinglaw W., Li S., and Cao Z.. 1997a. MyD88: An adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 7:837–847. 10.1016/S1074-7613(00)80402-1 [DOI] [PubMed] [Google Scholar]
- Wesche, H., Korherr C., Kracht M., Falk W., Resch K., and Martin M.U.. 1997b. The interleukin-1 receptor accessory protein (IL-1RAcP) is essential for IL-1-induced activation of interleukin-1 receptor-associated kinase (IRAK) and stress-activated protein kinases (SAP kinases). J. Biol. Chem. 272:7727–7731. 10.1074/jbc.272.12.7727 [DOI] [PubMed] [Google Scholar]
- Williams, K.L., Lich J.D., Duncan J.A., Reed W., Rallabhandi P., Moore C., Kurtz S., Coffield V.M., Accavitti-Loper M.A., Su L., et al. 2005. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-tumor necrosis factor alpha-and Mycobacterium tuberculosis-induced pro-inflammatory signals. J. Biol. Chem. 280:39914–39924. 10.1074/jbc.M502820200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windheim, M., Stafford M., Peggie M., and Cohen P.. 2008. Interleukin-1 (IL-1) induces the Lys63-linked polyubiquitination of IL-1 receptor-associated kinase 1 to facilitate NEMO binding and the activation of IkappaBalpha kinase. Mol. Cell. Biol. 28:1783–1791. 10.1128/MCB.02380-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, M.G., Xu Z.S., Li Y.H., Wang S.Y., Wang Y.Y., and Ran Y.. 2020. RNF152 positively regulates TLR/IL-1R signaling by enhancing MyD88 oligomerization. EMBO Rep. 21:e48860. 10.15252/embr.201948860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., and Shu H.B.. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 19:727–740. 10.1016/j.molcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Zhang, G., and Ghosh S.. 2002. Negative regulation of toll-like receptor-mediated signaling by Tollip. J. Biol. Chem. 277:7059–7065. 10.1074/jbc.M109537200 [DOI] [PubMed] [Google Scholar]
- Zhang, H., Peng C., Hu Y., Li H., Sheng Z., Chen Y., Sullivan C., Cerny J., Hutchinson L., Higgins A., et al. 2012. The Blk pathway functions as a tumor suppressor in chronic myeloid leukemia stem cells. Nat. Genet. 44:861–871. 10.1038/ng.2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., Yu M., Fukuda K., Im J., Yao P., Cui W., Bulek K., Zepp J., Wan Y., Kim T.W., et al. 2013. IRAK-M mediates Toll-like receptor/IL-1R-induced NFκB activation and cytokine production. EMBO J. 32:583–596. 10.1038/emboj.2013.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwollo, P., Rao S., Wallin J.J., Gackstetter E.R., and Koshland M.E.. 1998. The transcription factor NF-kappaB/p50 interacts with the blk gene during B cell activation. J. Biol. Chem. 273:18647–18655. 10.1074/jbc.273.29.18647 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the interacting proteins of BLK identified by mass spectrometry.
shows the phosphorylated peptide sequences and phosphorylation sites of TOLLIP identified by mass spectrometry.
shows the phosphorylated peptide sequences and phosphorylation sites of BLK identified by mass spectrometry.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
is the source file for Fig. S5.
Data Availability Statement
All data, plasmids, and reagents generated in this study are available upon reasonable request.